Summary
Fusarium head blight (FHB) is a disease of the floral tissues of wheat and barley for which highly resistant varieties are not available. Thus, there is a need to identify genes/mechanisms that can be targeted for the control of this devastating disease. Fusarium graminearum is the primary causal agent of FHB in North America. In addition, it also causes Fusarium seedling blight. Fusarium graminearum can also cause disease in the model plant Arabidopsis thaliana. The Arabidopsis–F. graminearum pathosystem has facilitated the identification of targets for the control of disease caused by this fungus. Here, we show that resistance against F. graminearum can be enhanced by flg22, a bacterial microbe‐associated molecular pattern (MAMP). flg22‐induced resistance in Arabidopsis requires its cognate pattern recognition receptor (PRR) FLS2, and is accompanied by the up‐regulation of WRKY29. The expression of WRKY29, which is associated with pattern‐triggered immunity (PTI), is also induced in response to F. graminearum infection. Furthermore, WRKY29 is required for basal resistance as well as flg22‐induced resistance to F. graminearum. Moreover, constitutive expression of WRKY29 in Arabidopsis enhances disease resistance. The PTI pathway is also activated in response to F. graminearum infection of wheat. Furthermore, flg22 application and ectopic expression of WRKY29 enhance FHB resistance in wheat. Thus, we conclude that the PTI pathway provides a target for the control of FHB in wheat. We further show that the ectopic expression of WRKY29 in wheat results in shorter stature and early heading time, traits that are important to wheat breeding.
Keywords: Arabidopsis thaliana, flg22 peptide, Fusarium head blight, microbe‐associated molecular pattern, PTI, wheat, WRKY29
Introduction
The ascomycetous fungus Fusarium graminearum (hereafter referred to as Fg) is an important phytopathogen. In wheat (Triticum aestivum) and barley (Hordeum vulgare), Fg is the primary causal agent of Fusarium head blight (FHB) disease which affects floral tissues (Bai and Shaner, 2004; McMullen et al., 1997a; Xu and Nicholson, 2009). In addition, it also causes Fusarium seedling blight. FHB epidemics in the past have resulted in $0.3–3 billion in losses (Bai and Shaner, 2004; Johnson et al., 2003; Wilson et al., 2017). FHB adversely impacts grain yield and quality. Mycotoxins, for example deoxynivalenol (DON), which accumulate in infected grains, further limit grain acceptability for human and animal consumption (Bai and Shaner, 2004; McMullen et al., 1997b; Wilson et al., 2017). Monogenic gene‐for‐gene‐type resistance is not available for FHB. In many cultivated wheat varieties, resistance to FHB is derived from the cultivar Sumai 3 and its derivatives (Bai and Shaner, 2004). Sumai 3‐derived resistance is a quantitative trait that limits fungal spread from the infection site. The non‐availability of highly resistant wheat and barley cultivars, the practical difficulties with the timing of fungicide application during anthesis and the high humidity conditions when disease threat is the highest further constrain efforts to control FHB (McMullen et al., 1997b; Pirgozliev et al., 2003).
The genes and mechanisms that contribute to the basal resistance to Fg offer targets for molecular breeding and genetic engineering of FHB resistance. For example, salicylic acid (SA) signalling, which contributes to basal resistance to FHB in wheat and barley (Diethelm et al., 2014; Hao et al., 2018; Makandar et al., 2006, 2012, 2015), is a target for enhancing FHB resistance. FHB resistance in wheat was enhanced by the constitutive expression of NPR1 (NON‐EXPRESSOR OF PR GENES 1), which is a key regulator of SA signalling, and NPR1‐like genes in wheat (Gao CS et al., 2013; Makandar et al., 2006; Yu et al., 2017). Furthermore, natural variations at two homeologous NPR1‐like genes located on the long arm of chromosomes 2A and 2D were associated with resistance to FHB in winter wheat (Diethelm et al., 2014). FHB resistance was also enhanced in transgenic wheat that accumulated higher levels of SA as a result of the constitutive expression of PAD4, a positive modulator of SA accumulation (Makandar et al., 2015). FHB resistance was also enhanced in barley plants that overexpressed ICS, a gene that encodes an isochorismate synthase, which synthesizes SA (Hao et al., 2018). In contrast, RNA interference (RNAi)‐mediated repression of ICS in barley compromised FHB resistance (Hao et al., 2018).
Pattern‐triggered immunity (PTI) is another process that can be targeted to promote disease resistance. PTI, which involves a complex set of physiological and molecular responses in the plant, including reactive oxygen species (ROS) accumulation and callose deposition, is induced in response to the recognition of conserved microbe‐associated molecular patterns (MAMPs) by pattern recognition receptors (PRRs) located on the plant cell surface (Bigeard et al., 2015; Li et al., 2016). PTI is an important contributor to non‐host resistance in plants (Bigeard et al., 2015). Some well‐studied MAMPs include the bacterial flagellar protein flagellin and elongation factor EF‐Tu, which are perceived by the cognate PRRs FLS2 (FLAGELLIN‐SENSITIVE 2) and EFR (EF‐Tu RECEPTOR), respectively (Bigeard et al., 2015). A 22‐amino‐acid long region of flagellin, epitomized by flg22 from Pseudomonas aeruginosa, is sufficient for the activation of PTI via FLS2 (Gomez‐Gomez and Boller, 2000), whereas an 18‐amino‐acid long epitope of EF‐Tu, represented by elf‐18 from Escherichia coli, is sufficient for PTI activation through EFR (Zipfel et al., 2006). The polysaccharide chitin, which is a major component of fungal cell walls, is another MAMP (Sánchez‐Vallet et al., 2015). In Arabidopsis thaliana, LysM (extracellular lysin motifs)‐containing receptor‐like kinases have been implicated in chitin signalling and resistance against fungal pathogens (Sánchez‐Vallet et al., 2015; Wan et al., 2008). Similarly, in rice (Oryza sativa, Os), chitin fragments are perceived by the LysM domain‐containing OsCEBiP (chitin elicitor binding protein) and OsCERK1 (Kaku et al., 2006; Shimizu et al., 2010). In barley, the LysM domain‐containing HvCERK1 (Chitin Elicitor Receptor Kinase 1) is required for plant response to chitin (Karre et al., 2017). Wheat leaves are also responsive to chitin and flg22, both of which induce the expression of wheat homologues of chitin‐ and flg22‐responsive Arabidopsis genes, including TaPUB23‐like and TaWRKY23‐like (Schoonbeek et al., 2015). In addition, the expression of Arabidopsis EFR is sufficient to confer elf‐18 recognition and to enhance resistance in wheat against the bacterial pathogen Pseudomonas syringae pv. oryzae (Schoonbeek et al., 2015), therefore suggesting the conservation of PTI signalling mechanisms between Arabidopsis and wheat.
There is significant overlap in the genes that are up‐regulated by different MAMPs (Gust et al., 2007; Wan et al., 2008; Zipfel et al., 2006), thus signifying the activation of convergent signalling pathways by these discrete MAMPs, which control the expression of a common set of PTI‐associated genes, although with different dynamics and amplitudes (Li et al., 2016). In Arabidopsis, WRKY29, which encodes a WRKY family transcription factor, is one such gene that is up‐regulated by both flg22 and chitooligosaccharide (Asai et al., 2002; Wan et al., 2008). This convergence of signalling associated with different MAMPs has led to the suggestion that MAMPs are perceived as general danger signals and that plants do not distinguish between different microbes via the defence signalling induced by different MAMPs (Zipfel et al., 2006). Therefore, it is expected that PTI activation should confer cross‐protection against pathogens in different kingdoms. Indeed, ectopic application of the bacterial MAMP, flg22, enhances resistance in Arabidopsis to the fungal pathogen Botrytis cinerea (Ferrari et al., 2007; Galletti et al., 2011). Moreover, the application of chitooligosaccharide promotes resistance in Arabidopsis to the bacterial pathogen Pseudomonas syringae pv. tomato DC3000 (Wan et al., 2008). Cross‐protection also extends to Fg. Chaturvedi et al. (2012) showed that prior treatment with a bacterial pathogen promotes resistance against Fg in Arabidopsis, which has been utilized in several studies as a model plant to characterize the physiological and molecular aspects of plant defence against Fg (Chen et al., 2006, 2009; Cuzick et al., 2008; Makandar et al., 2006, 2010, 2015; Nalam et al., 2015; Savitch et al., 2007; Skadsen and Hohn, 2004; Urban et al., 2002; Van Hemelrijck et al., 2006). Fg can infect leaves and inflorescences of Arabidopsis.
The PTI pathway has been implicated as a major player in the resistance to Fusarium ear rot in the maize inbred line BT‐1 (Wang et al., 2016). Similarly, basal resistance to FHB in barley requires HvCERK1 (Karre et al., 2017), thus suggesting that the PTI pathway is engaged during Fg infection. The aim of this study was to determine whether PTI can be targeted to enhance resistance against Fg. We show that Arabidopsis can be protected against Fg infection by flg22‐mediated induction of PTI via FLS2. This resistance to Fg infection conferred by flg22 requires WRKY29 which, when constitutively expressed in Arabidopsis, confers a high level of resistance to Fg. We further demonstrate that flg22 application and constitutive expression of Arabidopsis WRKY29 confer enhanced resistance to FHB in wheat, which is accompanied by stronger expression of PTI‐associated genes, thus supporting our suggestion that the PTI pathway is a target for enhancing resistance to FHB.
Results
Fg infection induces WRKY29 expression in A. thaliana
The expression of WRKY29 was used as a molecular marker of PTI to test whether Fg infection induces a PTI‐like mechanism in Arabidopsis. Fg was infiltrated into Arabidopsis leaves and WRKY29 expression was monitored by real‐time reverse transcription‐polymerase chain reaction (RT‐PCR). flg22 peptide‐treated leaves provided the positive control for WRKY29 expression. In addition, expression of the Fg‐ and flg22‐responsive PATHOGENESIS‐RELATED 1 (PR1) (Asai et al., 2002; Makandar et al., 2006; Yi et al., 2014) was monitored as a positive control for the two treatments. Expression of PR1, which encodes a cysteine‐rich secretory protein, has been used as an excellent molecular marker for the activation of SA signalling in plants. As shown in Fig. 1A, PR1 and WRKY29 expression were up‐regulated in Fg‐ and flg22‐treated leaves compared with the untreated and mock‐inoculated controls, thus confirming the activation of downstream signalling by these treatments. Fg infection also resulted in the accumulation of hydrogen peroxide (H2O2), another hallmark of PTI (Fig. 1B). Taken together, these results suggest that Arabidopsis responds to Fg infection by stimulating a PTI‐like response.
Figure 1.
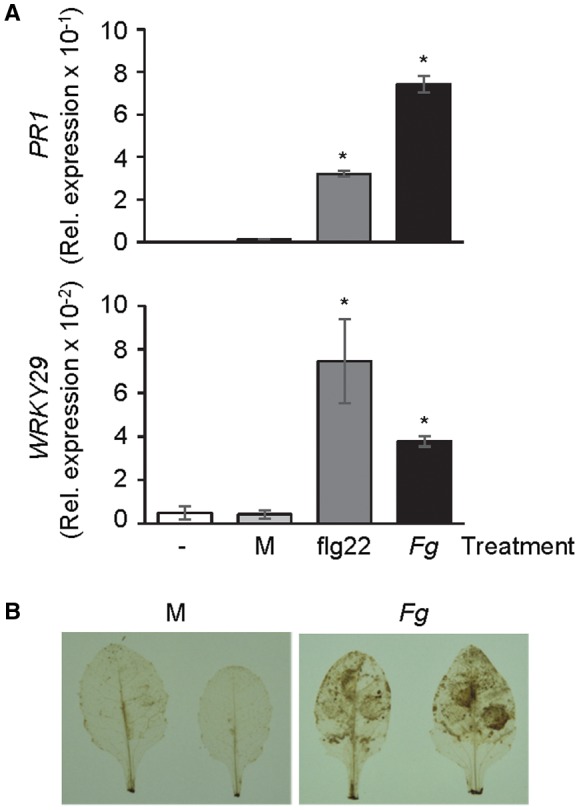
Induction of pattern‐triggered immunity (PTI) markers in flg22‐treated and Fusarium graminearum (Fg)‐inoculated Arabidopsis leaves. (A) PR1 and WRKY29 expression, relative to At1g07940, in flg22‐treated and Fg‐inoculated plants at 24 h post‐treatment. Top: real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of PR1 expression in leaves of wild‐type Arabidopsis accession Columbia plants infiltrated with 50 ng flg22 peptide and in plants inoculated with Fg. Untreated and mock (M)‐inoculated plants provided the controls. Bottom: real‐time RT‐PCR analysis of WRKY29 expression in the above samples. Error bars represent the standard error (SE) (n = 5). Asterisks above the bars indicate values that are significantly different (P < 0.05; t‐test) from the mock‐inoculated plants. (B) 3,3′‐Diaminobenzidine (DAB) staining to monitor H2O2 accumulation in mock (M)‐ and Fg‐inoculated Arabidopsis leaves. Leaves were stained with DAB at 18 h post‐inoculation. Brown deposits indicate H2O2 accumulation.
The flg22 peptide induces resistance against Fg infection in A. thaliana and wheat
To determine whether the PTI pathway can be targeted to enhance resistance against Fg, we tested whether pretreatment of Arabidopsis leaves with the flg22 peptide is capable of augmenting resistance to Fg. Leaves of wild‐type (WT) Arabidopsis accession Columbia plants were infiltrated with flg22 peptide to activate PTI; 24 h later, the same leaves were inoculated with Fg and disease severity was scored at 5 days post‐inoculation (dpi). As shown in Fig. 2A, Fg disease severity was significantly lower in flg22‐treated leaves than in mock‐treated leaves, thus suggesting that an flg22‐activated mechanism can enhance resistance against Fg. We further tested basal resistance to Fg in transgenic Arabidopsis engineered to express a chimeric PR1‐flg22 construct that expresses flg22 fused to the C‐terminus of PR1. As mentioned above, PR1 is a secretory protein that accumulates in the apoplast (Gu and Innes, 2012; Pečenková et al., 2017; Watanabe et al., 2013). Furthermore, the activation of SA signalling promotes the export of PR1 into the apoplast (Wang et al., 2005). Thus, the PR1‐flg22 fusion is expected to deliver flg22 into the apoplast, where it should be perceived by FLS2 to activate PTI. As shown in Fig. 2B, Fg disease severity was lower in leaves of two independently derived PR1‐flg22‐expressing transgenic lines compared with the WT control (Figs 2B and S1, see Supporting Information). Disease severity was also lower in the inflorescence of PR1‐flg22 lines compared with the WT control (Fig. 2B). These results confirm that an flg22‐activated mechanism can confer resistance to Fg infection in Arabidopsis.
Figure 2.
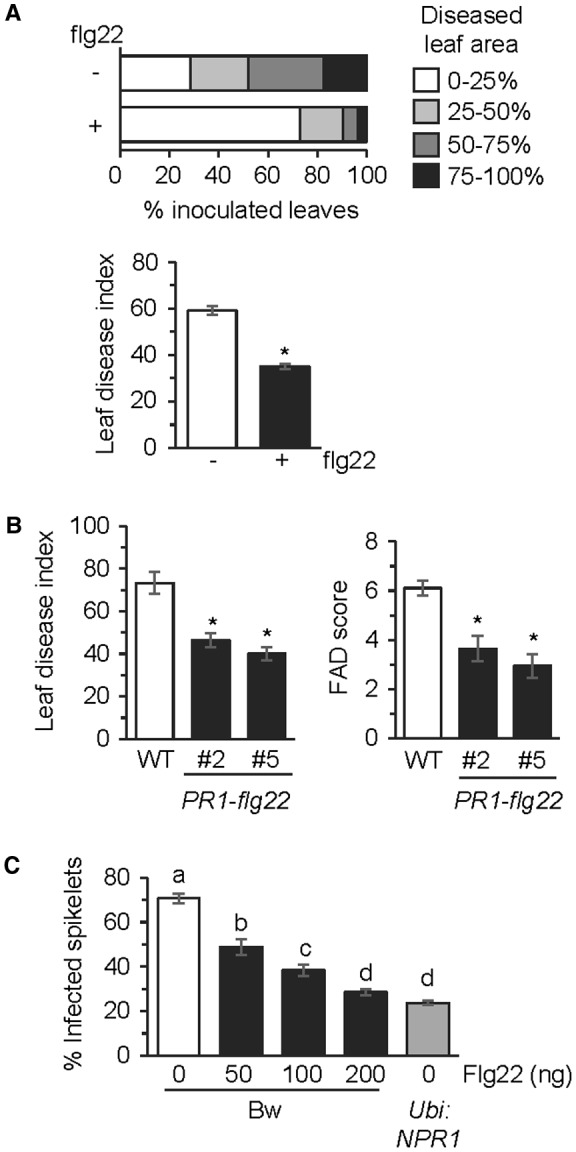
flg22 application enhances resistance to Fusarium graminearum in Arabidopsis and wheat. (A) Top: F. graminearum disease severity in wild‐type (WT) Arabidopsis accession Columbia leaves treated with 50 ng of flg22 (+) and as control with water (–). Fungal inoculation was conducted 24 h after flg22 treatment and disease severity was monitored at 5 days post‐inoculation (dpi) with the fungus (n = 50). Bottom: leaf disease index in the above experiment. All values are means ± standard error (SE) (n = 50). Asterisks above the bars indicate values that are significantly different from the water‐treated control plants (P < 0.05; χ 2 test). (B) Left: leaf disease index in F. graminearum‐inoculated WT accession Columbia plant and two independent PR1‐flg22 transgenic lines that are in the FLS2 background. All values are the means ± SE (n = 50). Asterisks above the bars indicate values that are significantly different from WT (P < 0.05; χ 2 test). Right: Fusarium Arabidopsis Disease (FAD) score in inflorescences of the WT and PR1‐flg22 lines in the FLS2 background. All values are the means ± SE (n = 30). Asterisks above the bars indicate values that are significantly different from WT (P < 0.05; t‐test). (C) Fusarium head blight severity in flg22‐treated wheat cv. Bobwhite (Bw). The spikelets were treated with the indicated amounts of flg22 peptide at 24 h prior to fungal inoculation. The Ubi:NPR1 transgenic plant, which is in the cultivar Bw background, provided the FHB‐resistant control. Disease severity was monitored at 21 dpi. All values are the means ± SE (n = 10). Different letters above the bars indicate values that are significantly different from each other (P < 0.05; Tukey’s test).
We further tested whether flg22 application was capable of promoting FHB resistance in wheat. Varying amounts of the flg22 peptide dissolved in 10 µL of water were applied with a syringe to two central spikelets of each spike of the spring wheat cultivar Bobwhite; 24 h later, these spikelets were inoculated with Fg and FHB disease severity was monitored 21 days later. A Ubi:NPR1 wheat line, which is in the Bobwhite background and constitutively expresses the Arabidopsis NPR1 gene from the maize Ubiquitin promoter to increase FHB resistance (Makandar et al., 2006, 2012), provided the disease‐resistant control for this experiment. As shown in Fig. 2C, pretreatment with flg22 peptide enhanced FHB resistance in the wheat cultivar Bobwhite. The resistance‐promoting effect of flg22 exhibited a dose‐dependent response. At the highest level of 200 ng, the FHB resistance‐promoting effect of flg22 was comparable with that observed in the Ubi:NPR1 line. Taken together, these experiments with Arabidopsis and wheat signify the potential for targeting PTI to enhance resistance against Fg.
FLS2 is required for flg22‐induced resistance to Fg infection in A. thaliana
To confirm that the flg22‐induced resistance to Fg was indeed a result of the activation of PTI, the ability of flg22 to enhance resistance to Fg in the fls2 mutant was studied. WRKY29 expression was monitored as a molecular marker for the activation of PTI. As shown in Fig. 3A, although flg22 treatment, compared with mock treatment, was effective in inducing WRKY29 expression in the WT plant, flg22 was unable to induce WRKY29 expression in the fls2 mutant, thus confirming the requirement of FLS2 for the flg22‐induced expression of WRKY29. Compared with the WT, the Fg resistance‐promoting effect of flg22 was not observed in the fls2 mutant (Fig. 3B). The Fg disease severity in leaves of the flg22‐treated fls2 mutant was comparable with that in the mock‐treated fls2 mutant and significantly higher than that in flg22‐treated WT plants. Experiments with the PR1‐flg22 chimera also confirmed the importance of FLS2 to flg22‐induced resistance to Fg, which was lacking in the fls2 mutant background compared with the FLS2 background (Fig. 3C).
Figure 3.
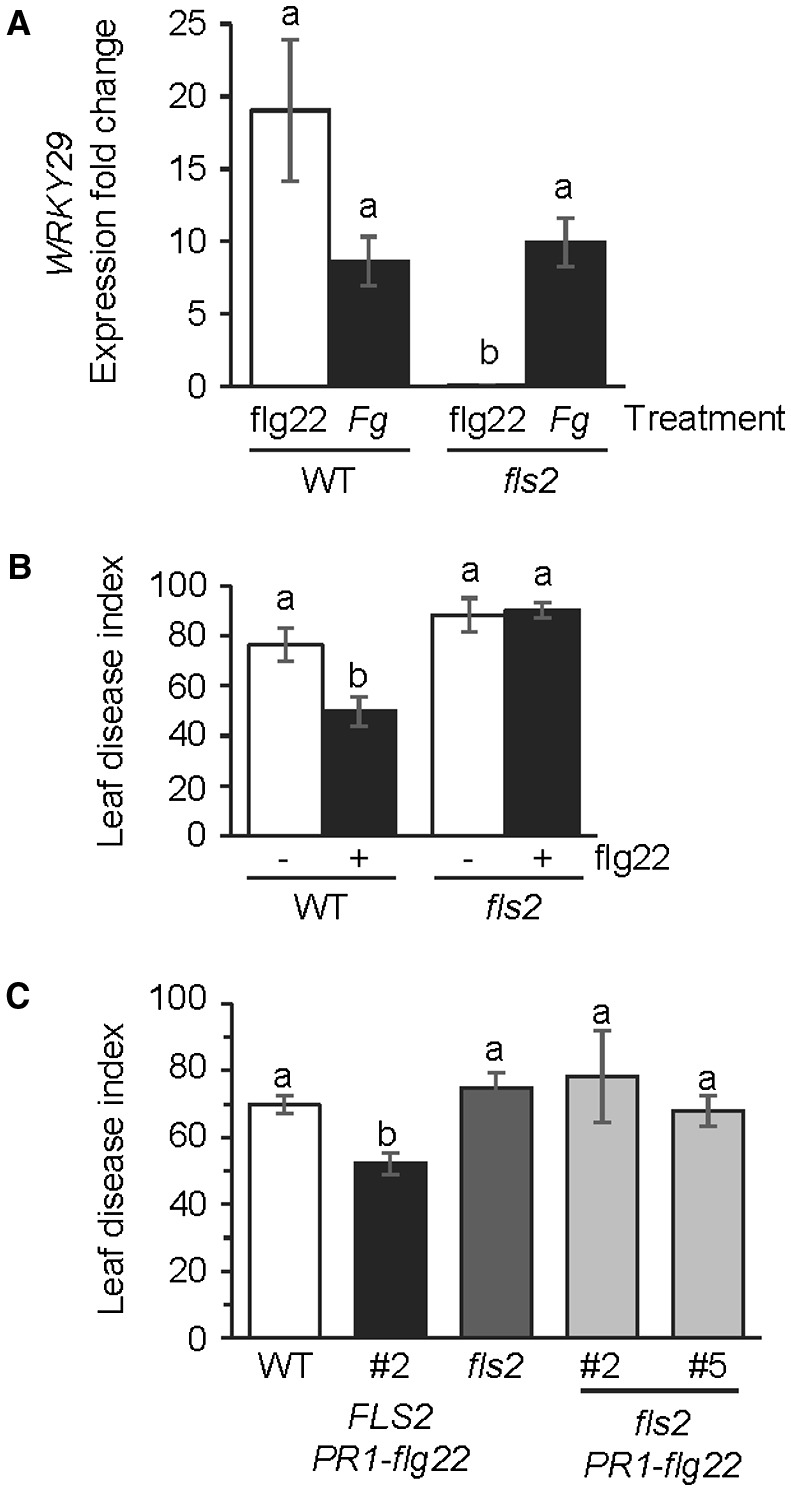
FLS2 function is essential for the flg22‐induced resistance to Fusarium graminearum (Fg) in Arabidopsis. (A) Real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) evaluation of fold induction of WRKY29 expression in flg22‐treated and Fg‐inoculated leaves of wild‐type (WT) and fls2 mutant plants, relative to expression in the corresponding mock‐inoculated leaves. Gene expression was monitored 24 h post‐treatment, with the expression of At1g07940 providing the control. All values are the means ± standard error (SE) (n = 5). Different letters above the bars indicate values that are significantly different from each other (P < 0.05; Tukey’s test). (B) Leaf disease index in WT accession Columbia and fls2 mutant leaves treated with 50 ng of flg22 (+) or as control with water (–). Fungal inoculation was conducted 24 h after flg22 treatment and disease severity was monitored at 5 days post‐inoculation (dpi) with the fungus. All values are the means ± SE (n = 50). Different letters above the bars indicate values that are significantly different from each other (P < 0.05; Tukey’s test). (C) Leaf disease index in Fg‐inoculated WT accession Columbia plant, PR1‐flg22 transgenic line #2 in the FLS2 background, the fls2 mutant and two independent PR1‐flg22 transgenic lines in the fls2 mutant background. Disease severity was monitored at 5 dpi. All values are the means ± SE (n = 50). Different letters above the bars indicate values that are significantly different from each other (P < 0.05; Tukey’s test).
Although the leaf disease index, which reflects the average disease severity across the different disease categories (see Experimental procedures), was not significantly different between the WT and fls2 plants that were not treated with flg22 (Fig. 3B,C), we repeatedly observed significant differences (P < 0.05; χ 2 test) in the distribution of the four disease categories in the fls2 mutant compared with the WT. We therefore suggest that there is a subtle influence of the FLS2 allele on the basal resistance to Fg in Arabidopsis.
flg22‐induced resistance to Fg in A. thaliana requires NPR1 and WRKY29
SA signalling has an important function in Arabidopsis and wheat defence against Fg (Diethelm et al., 2014; Gao CS et al., 2013; Makandar et al., 2006, 2010, 2012, 2015; Yu et al., 2017). In Arabidopsis, SA signalling is also induced in response to flg22 (Tsuda et al., 2008; Yi et al., 2014). Furthermore, SA stimulates FLS2 expression and SA analogues prime the induction of flg22‐triggered responses, including flg22‐triggered up‐regulation of WRKY29 expression (Pick et al., 2012; Yi et al., 2014). In contrast, flg22‐triggered responses, including WRKY29 expression, are attenuated in the SA biosynthesis sid2 mutant (Yi et al., 2014). To determine whether SA signalling is critical for the flg22‐conferred resistance to Fg, we tested the ability of flg22 to promote resistance to Fg in the SA‐insensitive npr1 mutant. 35S:NPR1 plants, which constitutively express NPR1 from the Cauliflower mosaic virus 35S promoter (Cao et al., 1998; Makandar et al., 2006), provided the Fg‐resistant control for this experiment. As shown in Fig. 4A and reported previously (Makandar et al., 2010), Fg disease severity was higher in leaves of the npr1 mutant than in the WT plant. Furthermore, flg22 was unable to enhance resistance to Fg in the npr1 mutant compared with the WT, thus confirming that flg22‐induced PTI cannot bypass the need for SA signalling in defence against Fg. Similarly, WRKY29 function was required for defence against Fg (Fig. 4A). Compared with the WT, Fg disease severity was higher in the wrky29 mutant and comparable with that in the npr1 mutant. Furthermore, unlike in the WT, flg22 was unable to promote resistance in the wrky29 mutant. These results provide further confirmation that the flg22‐conferred resistance to Fg is mediated through genetic components that function downstream of the FLS2/flg22 receptor/ligand pair. The higher level of disease in the wrky29 mutant compared with the WT further indicates that a WRKY29‐dependent defence mechanism(s) is critical for basal resistance to Fg.
Figure 4.
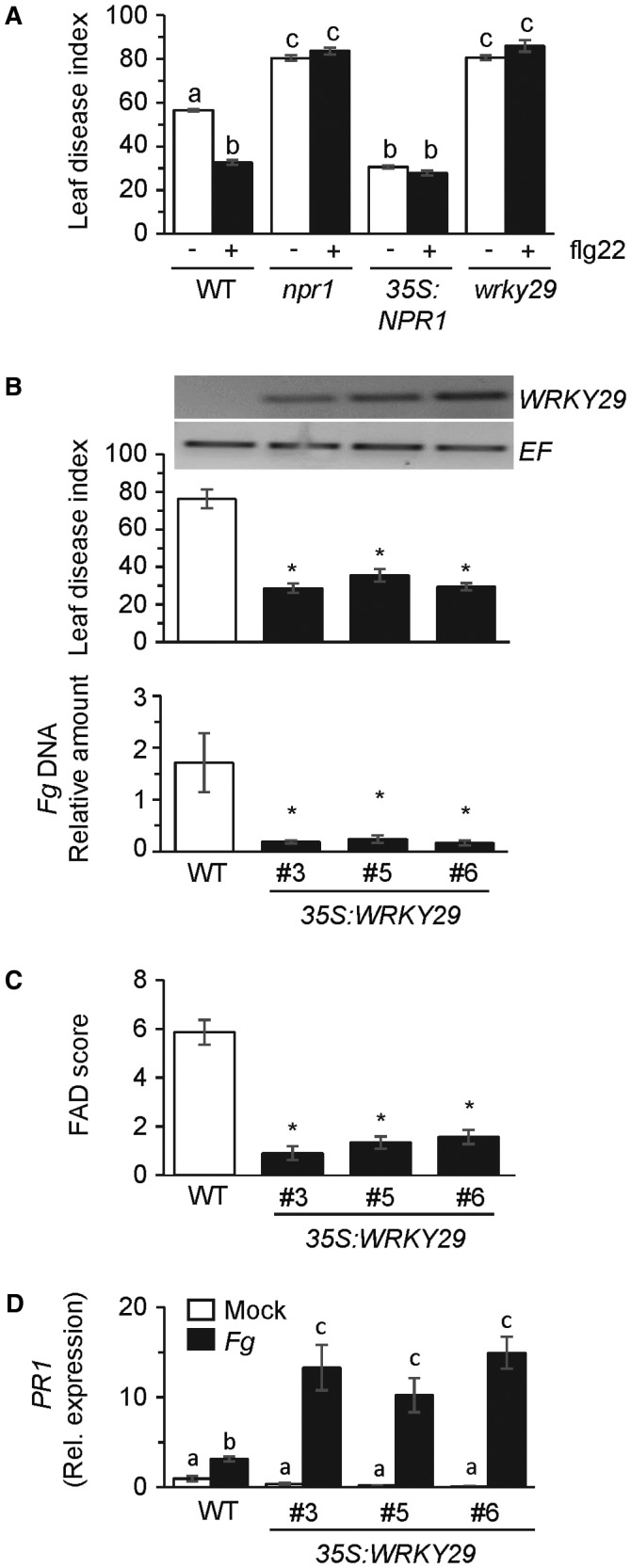
Constitutive expression of WRKY29 in Arabidopsis promotes resistance to Fusarium graminearum (Fg). (A) Leaf disease index in Fg‐inoculated wild‐type (WT) accession Columbia, the npr1 mutant, a 35S:NPR1 transgenic line in which the NPR1 coding sequence is expressed from the 35S promoter, and a wrky29 mutant. Leaves were treated with 50 ng of flg22 peptide, or mock treated, 24 h prior to fungal inoculation. Disease was monitored at 5 days post‐inoculation (dpi). All values are the means ± standard error (SE) (n = 50). Different letters above the bars indicate values that are significantly different from each other (P < 0.05; Tukey’s test). (B) Top: reverse transcription‐polymerase chain reaction (RT‐PCR) demonstration of WRKY29 expression and, as control, At1g07940 (EF) expression in leaves of WT accession Columbia and three independent 35S:WRKY29 lines in which the WRKY29 coding sequence is expressed from the 35S promoter. Middle: leaf disease index in WT accession Columbia and 35S:WRKY29 lines inoculated with Fg. Disease was monitored at 5 dpi. All values are the means ± SE (n = 50). Asterisks above the bars indicate values that are significantly different from the WT (P < 0.05; χ 2 test). Bottom: real‐time PCR analysis (×10−2) of DNA content of Fg nahG gene relative to the Arabidopsis ACT8 gene. All values are means ± SE (n = 4) in leaves of WT Col‐0 and the 35S:WRKY29 plants at 4 dpi with Fg. Asterisks above the bars indicate values that are significantly different from the WT (P < 0.05; t‐test). (C) Fusarium Arabidopsis Disease (FAD) score for the WT accession Columbia and 35S:WRKY29 transgenic lines. All values are the means ± SE (n = 30). Asterisks above the bars indicate values that are significantly different from the WT (P < 0.05; t‐test). (D) Real‐time RT‐PCR evaluation of PR1 expression in mock‐ and Fg‐inoculated leaves of WT accession Columbia and 35S:WRKY29 transgenic lines. Gene expression relative to expression of the control gene At1g07940 was monitored at 24 h post‐treatment. All values are the means ± SE (n = 4). Different letters above the bars indicate values that are significantly different from each other (P < 0.05; Tukey’s test).
Constitutive expression of WRKY29 enhances resistance to Fg in A. thaliana
We further tested the feasibility of engineering the PTI pathway to enhance resistance against Fg by developing plants that constitutively express WRKY29, which encodes a transcription factor that is common to PTI induced by flg22 and chitin (Asai et al., 2002; Wan et al., 2008). The Cauliflower mosaic virus 35S promoter was used to constitutively express WRKY29 in Arabidopsis (Fig. 4B). Compared with the WT plant, Fg disease severity and fungal accumulation, which was monitored by comparing the accumulation of Fg NahG gene DNA, were significantly lower in leaves of all three independently derived 35S:WRKY29 plants (Fig. 4B). Similarly, Fg disease severity was also lower in the inflorescence tissues of 35S:WRKY29 plants compared with the WT (Fig. 4C). These results provide further proof‐of‐concept that the PTI pathway is amenable for engineering resistance to Fg. Compared with the WT, basal expression of the SA‐ and flg22‐responsive PR1 gene was not altered in 35S:WRKY29 plants (Fig. 4D). However, fungal infection resulted in significantly stronger induction of PR1 in 35S:WRKY29 plants than in the WT (Fig. 4D), therefore indicating that constitutive expression of WRKY29 promotes robust activation of defence responses. In contrast, constitutive expression of WRKY29 did not result in stronger accumulation of H2O2 in response to fungal infection (Fig. S2A, see Supporting Information).
Wheat engineered to express WRKY29 exhibits enhanced resistance to FHB and seedling blight
To study the feasibility of targeting WRKY29 expression for the engineering of FHB resistance in wheat, we developed transgenic wheat plants containing a chimeric Ubi:WRKY29 construct, which constitutively expresses the Arabidopsis WRKY29 coding sequence (CDS) from the maize Ubiquitin promoter. Three independently derived Ubi:WRKY29 transgenic lines that stably express WRKY29 were identified (Fig. 5A). All three lines showed significantly higher level of resistance to FHB compared with the control cv. Bobwhite. Disease spread and fungal growth were restricted in the Ubi:WRKY29 plants compared with the non‐transgenic Bobwhite (Fig. 5A,B). In addition, the accumulation of the mycotoxin DON was also significantly lower in the transgenic Ubi:WRKY29 lines than in the control cv. Bobwhite (Fig. 5C). Basal expression of the SA‐responsive TaPR1.2, as well as the PTI marker genes TaWRKY70 and TaPUB‐23‐like (Kage et al., 2017; Schoonbeek et al., 2015), was very low and not altered in the Ubi:WRKY29 plants compared with the non‐transgenic Bobwhite (Fig. 6A), thus suggesting that, as in Arabidopsis, constitutive expression of WRKY29 is not sufficient to constitutively activate PTI. However, in response to Fg infection, WRKY29 expression conferred strong expression of TaPR1.2, TaWRKY70 and TaPUB‐23‐like in the Ubi:WRKY29 relative to non‐transgenic Bobwhite plants (Fig. 6A). In contrast, as in Arabidopsis, constitutive expression of WRKY29 in wheat did not promote stronger H2O2 accumulation in response to fungal infection (Figs 6B and S2B).
Figure 5.
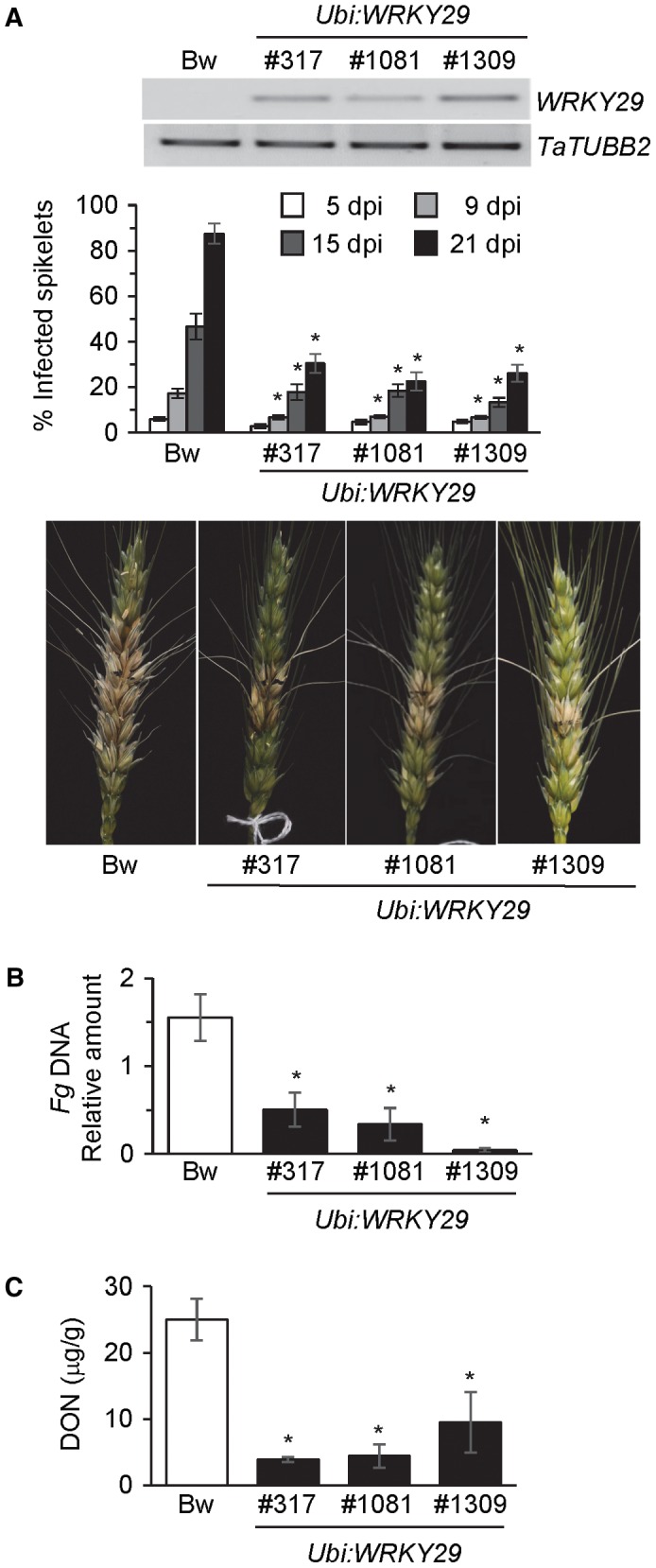
Ectopic expression of WRKY29 in wheat promotes resistance to Fusarium head blight. (A) Top: reverse transcription‐polymerase chain reaction (RT‐PCR) demonstration of WRKY29 expression in leaves of wheat cv. Bobwhite (Bw) and three independent Ubi:WRKY29 transgenic wheat lines in which the Arabidopsis WRKY29 coding sequence is expressed from the maize Ubi promoter. Middle: FHB disease severity in wheat cv. Bw and Ubi:WRKY29 lines in the Bw background. Disease progression was monitored at 5, 9, 15 and 21 days post‐inoculation (dpi) of spikes. All values are the means ± standard error (SE) (n = 12). Asterisks above the bars indicate values that are significantly different from the WT for that particular time point (P < 0.05; t‐test). Bottom: photograph showing disease spread in a representative spike from wheat cv. Bw and the Ubi:WRKY29 lines. Photographs were taken at 21 dpi. (B) Real‐time PCR analysis of the DNA content of the Fusarium graminearum (Fg) NahG gene relative to the wheat TUB2 gene in wheat spikes at 2 dpi with Fg. All values are the means ± SE (n = 3). Asterisks above the bars indicate values that are significantly different from Bw (P < 0.05; t‐test). (C) Deoxynivalenol (DON) content (µg/g seed) in Fg‐inoculated wheat cv. Bw and the Ubi:WRKY29 transgenic wheat lines. All values are the means ± SE (n = 3). Each sample included 3–5 g of seeds collected from two to three spikes derived from separate plants. Asterisks above the bars indicate values that are significantly different from Bw (P < 0.05; t‐test).
Figure 6.
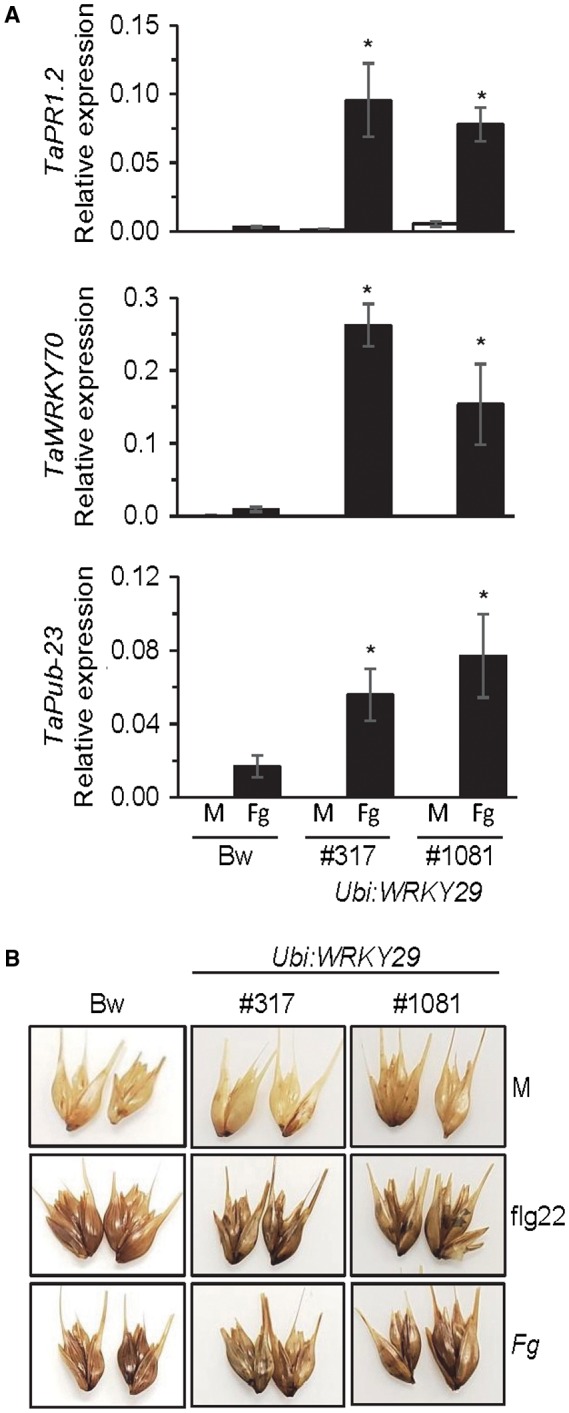
Impact of ectopic WRKY29 expression on wheat defence responses. (A) Real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) evaluation of TaPR1.2, TaWRKY70 and TaPUB23‐like gene expression in mock (M) and Fusarium graminearum (Fg)‐inoculated spikes of the wheat cv. Bobwhite (Bw) and Ubi:WRKY29 transgenic wheat lines #317 and #1081. Gene expression was monitored relative to expression of the control gene TaTUB2. All values are the means ± standard error (SE) (n = 3). Asterisks above the bars indicate values that are significantly different from Bw (P < 0.05; t‐test). (B) 3,3′‐Diaminobenzidine (DAB) staining for H2O2 accumulation in mock (M)‐, flg22‐ and Fg‐treated spikelets of the wheat cv. Bw and two independent Ubi:WRKY29 transgenic lines #317 and #1081. In (A) and (B), spikelets for RNA extraction and DAB staining were harvested at 48 h post‐inoculation (hpi).
The Ubi:WRKY29 plants also demonstrated elevated resistance to Fusarium seedling blight disease (Fig. 7). Taken together, the above results validate our suggestion that the PTI mechanism provides an excellent target for enhancing plant resistance against Fg.
Figure 7.
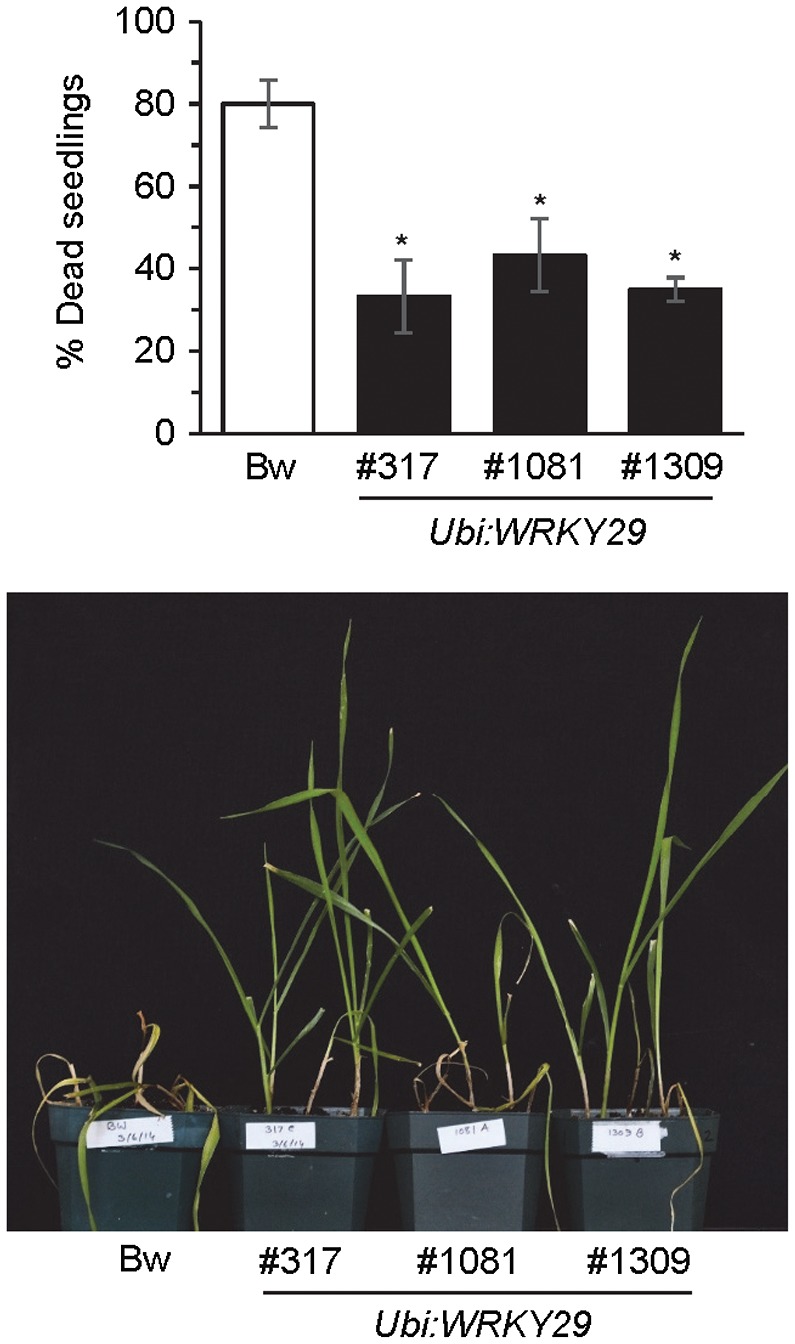
WRKY29 expression in wheat promotes resistance to Fusarium seedling blight. Top: percentage of dead seedlings at 2 weeks post‐inoculation of seedlings of wheat cv. Bobwhite (Bw) and three Ubi:WRKY29 transgenic lines with Fusarium graminearum. All values are the means ± standard error (SE) (n = 20). Asterisks above the bars indicate values that are significantly different from Bw (P < 0.05; t‐test). Bottom: photograph showing phenotype in representative seedlings of F. graminearum‐inoculated wheat cv. Bw and the Ubi:WRKY29 transgenic wheat lines. The photograph was taken at 10 days post‐inoculation (dpi).
Impact of constitutive WRKY29 expression on agronomic and growth parameters of wheat
As shown in Fig. 8A, the Ubi:WRKY29 wheat plants were 25% shorter than the non‐transgenic Bobwhite plants. This was paralleled by an earlier heading time in the Ubi:WRKY29 plants compared with Bobwhite (Fig. 8B). No significant impact on other agronomic parameters, such as the number of spikes produced per plant, the number of seeds produced per spike and seed yield per plant, was observed in Ubi:WRKY29 wheat compared with the control Bobwhite plants (Fig. 8A).
Figure 8.
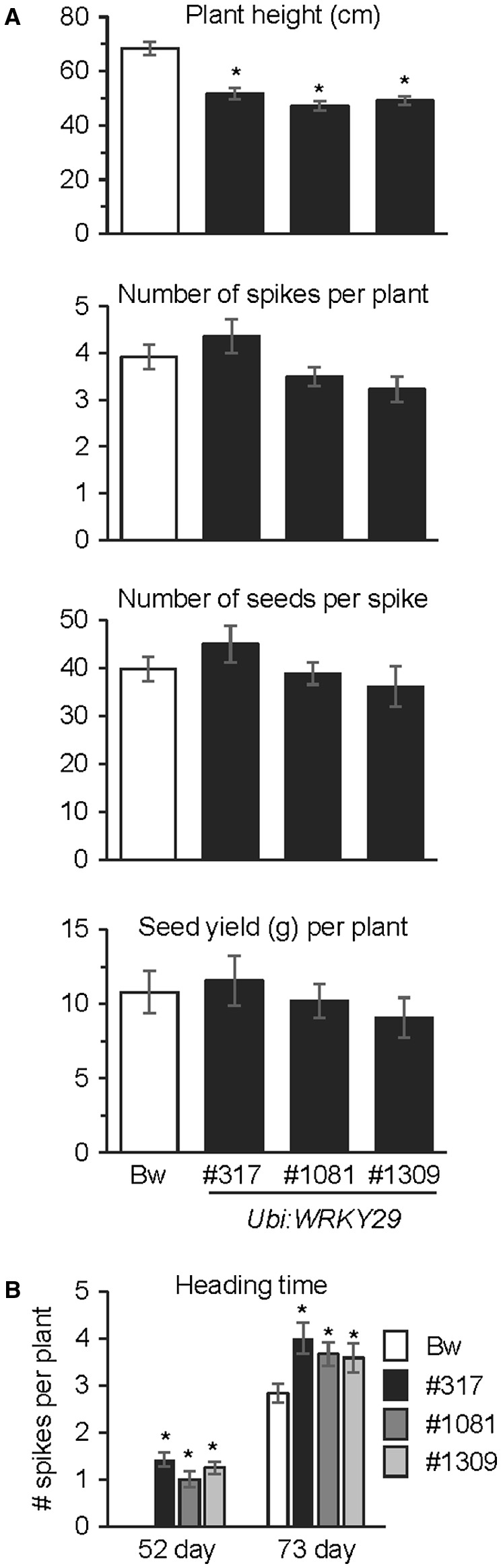
Agronomic and growth parameters in wheat plants expressing WRKY29. (A) Comparison of the average plant height (cm), number of spikes per plant, number of seeds per spike and seed yield (g) per plant for the wheat cv. Bobwhite (Bw) and three independent Ubi:WRKY29 transgenic lines. A minimum of 10 plants per line were analysed to obtain data on plant height, spike numbers and seed yield. The number of seeds per spike was calculated for 10 spikes from each line. (B) Heading time differences between Ubi:WRKY29 transgenic wheat and the non‐transgenic cv. Bw. The number of spikes in each plant was determined at 52 and 73 days after sowing of the seeds. In (A) and (B), all values are the means ± standard error (SE). Asterisks above the bars indicate values that are significantly different from Bw (P < 0.05; t‐test).
Discussion
In the absence of monogenic gene‐for‐gene resistance, current control measures for FHB in wheat and barley involve the use of partially resistant varieties combined with fungicide application and management practices (Bai and Shaner, 2004; Wilson et al., 2017). Limited knowledge of plant mechanisms that can be targeted to enhance FHB resistance has constrained progress on the development of FHB‐resistant wheat and barley varieties. Previously, using an approach that utilized the interaction between Arabidopsis and Fg to identify genes and mechanisms that contribute to defence against Fg, and transgenic validation of the ability of these genes/mechanisms to control Fg infection in Arabidopsis and wheat, we showed that the SA signalling pathway provides a target for enhancing FHB resistance (Makandar et al., 2006, 2010, 2015, 2012). Recent studies have confirmed that genes associated with SA biosynthesis in barley and alleles at NPR1‐like genes in winter wheat are associated with basal resistance to FHB (Diethelm et al., 2014; Hao et al., 2018). Utilizing a similar approach, here we demonstrate that the PTI pathway provides a target for enhancing resistance against Fg. We show that resistance against Fg can be enhanced in wheat and Arabidopsis by the bacterial MAMP flg22. In Arabidopsis, the flg22‐conferred resistance to Fg required the PTI‐associated PRR FLS2. Resistance against Fg was also enhanced in wheat and Arabidopsis by the constitutive expression of WRKY29, which encodes a transcription factor that is associated with PTI in Arabidopsis. Expression of the Arabidopsis WRKY29 CDS also restricted DON accumulation in Ubi:WRKY29 transgenic wheat. Compared with the non‐transgenic Bobwhite, the Ubi:WRKY29 wheat plants responded to Fg infection with stronger expression of TaPR1.2 and the PTI marker genes TaWRKY70 and TaPUB‐23‐like, thus suggesting stronger activation of PTI responses. These results, when considered together with the results of Kage et al. (2017), who showed that the TaWRKY70 gene contributes to basal resistance to FHB, demonstrate that the PTI pathway can be targeted to enhance resistance against Fg.
The fact that Fg infection stimulates WRKY29 expression to comparably high levels in the WT and fls2 mutant suggests that another pathway, presumably involving another PRR and its cognate ligand, stimulates PTI leading to WRKY29 expression in Fg‐inoculated Arabidopsis. The Fg infection‐derived elicitor that induces WRKY29 could potentially be chitin. Previously, a chitooligosaccharide was shown to induce WRKY29 expression in Arabidopsis and to promote resistance against fungal and bacterial pathogens (Wan et al., 2008). Chitosan treatment also promoted resistance to seed‐borne Fg infection in wheat (Bhaskara Reddy et al., 1999). Chitosan promoted the accumulation of lignin precursors and phenolics that have antimicrobial activity and could potentially contribute to resistance (Bhaskara Reddy et al., 1999). Chitin also induced the expression of wheat homologues of PTI‐associated genes (Schoonbeek et al., 2015). More recently, a metabolo‐transcriptomic approach in barley identified HvCERK1, a predicted chitin elicitor receptor kinase encoding gene, to be involved in defence against FHB (Karre et al., 2017). In Arabidopsis, WRKY29 expression is also induced by the Fusarium T‐2 toxin and other type A trichothecenes (Nishiuchi et al., 2006). These toxins, or derived metabolites, could also potentially function as elicitors of PTI in plants infected with Fg.
WRKY genes include a large family of plant‐specific DNA‐binding proteins that contain the conserved WRKYGQK sequence together with a zinc‐finger‐like motif (Eulgem et al., 2000; Pandey and Somssich, 2009). Several of these WRKY proteins are involved in the stress response, including plant defence against pathogens. Although some are positive regulators, others are negative regulators of the stress response (Pandey and Somssich, 2009). In Arabidopsis, WRKY29 is associated with PTI and defence against pathogens (Asai et al., 2002; Wan et al., 2008). Our results indicate an important role for WRKY29 in the control of Fg infection. Not only was WRKY29 expression up‐regulated in response to Fg infection, but, compared with the WT, Fg disease severity was higher in the wrky29 mutant and lower in plants constitutively expressing WRKY29. As Arabidopsis WRKY29 was capable of similarly enhancing disease resistance in wheat, we propose that wheat contains the downstream machinery that is regulated by WRKY29. WRKY homologues are present in wheat and barley, and some of these WRKYs have been shown to confer stress tolerance when constitutively expressed in heterologous systems (Liu et al., 2014; Niu et al., 2012; Wang et al., 2017, 2015). Recently, a wheat WRKY gene, TaWRKY70 (also known as TaWRKY45), was shown to be required for basal resistance to FHB (Kage et al., 2017). TaWRKY70 is associated with QTL‐2DL, which limits FHB severity by the control of fungal spread from the site of initial infection (Kage et al., 2017). TaWRKY70 expression is up‐regulated in FHB‐resistant near‐isogenic lines (NILs) compared with susceptible NILs. TaWRKY70 expression is also up‐regulated in response to infections with Puccinia triticina, which causes leaf rust, and the powdery mildew fungus Blumeria graminis (Bahrini et al., 2011b). Wheat plants overexpressing TaWRKY70 exhibit enhanced resistance to FHB, powdery mildew and leaf rust (Bahrini et al., 2011a, b), thus suggesting that TaWRKY70 is involved in defence against a variety of fungal infections. TaWRKY70 has been suggested to regulate the expression of genes involved in the synthesis of metabolites that are associated with resistance to fungi (Kage et al., 2017). Our results indicate that Arabidopsis WRKY29, when expressed in wheat, promotes the stronger activation of the PTI pathway, leading to the expression of TaWRKY70.
FLS2 is not required for the Fg‐induced up‐regulation of WRKY29 or for the flg22‐induced resistance to Fg in Arabidopsis. Furthermore, the Arabidopsis leaf disease index, which reflects the average of disease severity across the different leaf disease categories, was not significantly different between the Fg‐inoculated WT and fls2 mutant, thus suggesting that FLS2 does not have a major contribution to basal resistance to Fg in Arabidopsis. However, a significant difference (P < 0.05; χ 2 test) was observed between the WT and fls2 mutant for the relative distribution of leaves over the four disease categories, suggesting a subtle impact of FLS2 on basal resistance to Fg. Plant‐associated microbes are known to prime plant defences (Conrath et al., 2015; Pieterse et al., 2014). This priming of FLS2‐dependent defences by random plant‐associated microbes may influence basal resistance to Fg in Arabidopsis.
The results presented here, taken together with the knowledge that TaWRKY70 and the LysM domain‐containing HvCERK1 are required for basal resistance to FHB in wheat and barley, respectively (Kage et al., 2017; Karre et al., 2017), lead us to propose that genes associated with the PTI pathway are good candidates for the development of FHB‐resistant wheat and barley. Alternatively, factors that can induce the PTI pathway could also promote FHB resistance. However, the impact of PTI pathway activation needs to be tested on additional biotypes of Fg to determine whether it is effective against the different chemotypes of Fg, as well as other FHB‐causing Fusarium species. Ubi:WRKY29 wheat also exhibits reduced plant height and faster heading time, without any detrimental effects on yield. Height and heading time are traits that are important to wheat breeding (Hedden, 2003; Wilhelm et al., 2013). Thus, the pathway targeted by WRKY29 has the potential to influence additional beneficial traits for wheat breeding.
Experimental Procedures
Cultivation of Arabidopsis and wheat
A peat‐based soil mix (Fafard #2, Sungro, Agawam, MA) was used to cultivate Arabidopsis and wheat. Arabidopsis was cultivated as described previously (Nalam et al., 2016) in growth chambers programmed for 22 ºC and a 14‐h light (80–100 µE/m2/s) and 10‐h dark regime. The soil was autoclaved for 1 h prior to use. Arabidopsis npr1‐1, wrky29 (CS3024690) and fls2‐101 mutants, and the 35S:NPR1 transgenic lines and the wheat Ubi:NPR1 transgenic line in the cultivar Bobwhite, have been described previously (Cao et al., 1998; Li et al., 2017; Makandar et al., 2006; Pfund et al., 2004). Generation of the Arabidopsis 35S:WRKY29 and 35S:PR1‐flg22 lines and wheat Ubi:WRKY29 lines is described below. Wheat was cultivated in a glasshouse in which natural sunlight was supplemented with halogen lamps to provide a minimum of 14 h exposure to light. The glasshouse was programmed for day/night‐time temperatures of 21 ºC and 18 ºC, respectively.
Pathogen strains, culture conditions and plant infection
Half‐strength potato dextrose medium (Difco Laboratories, Detroit, MI, USA) was used for the growth and maintenance of Fg isolate Z‐3639, and carboxymethylcellulose (CMC) medium was used to promote sporulation, as described previously (Nalam et al., 2016). The fungus was cultivated at 28 °C. Fungal inoculation of Arabidopsis leaves involved infiltration of a suspension of fungal mycelial fragments through the abaxial surface with a needleless syringe (Nalam et al., 2016). Approximately 4‐week‐old Arabidopsis plants were used for leaf assays and 6–7‐week‐old plants for inflorescence assays. Disease severity was scored at 5 dpi, unless stated otherwise. Depending on the extent of chlorosis, leaves were grouped into four categories: Category I, chlorosis covering <25% of leaf area; Category II, chlorosis covering 25%–50% of leaf area; Category III, chlorosis covering 50%–75% of leaf area; Category IV, chlorosis covering 75%–100% of leaf area. A minimum of 50 leaves of each genotype/treatment were analysed for each experiment. The percentage of leaves in each category was used to calculate the leaf disease index, as described previously (Nalam et al., 2016). Inoculations of Arabidopsis inflorescences with Fg macroconidia and the disease rating [expressed as the Fusarium Arabidopsis Disease (FAD) score] were conducted as described previously (Nalam et al., 2016).
Inoculation of wheat spikelets with Fg macroconidia and disease evaluation were performed as described previously (Makandar et al., 2006, 2012). Briefly, at the anthesis stage, two central spikelets were inoculated with 10 µL of a suspension containing 300 fungal macroconidia. High humidity was maintained for 3 days by covering the inoculated spikes with a moistened zip‐lock bag. Over time, the fungal infection spread out to the other spikelets within each spike. Disease spread was monitored at periodic intervals. The final reading was taken at 21 dpi and the disease severity was calculated as the percentage of diseased spikelets.
To study the effect of wheat genotypes on Fusarium seedling blight, seeds of the indicated lines were soaked with an Fg macroconidial suspension for 24 h. In addition, after germination, seedlings were spray inoculated with a macroconidial suspension (100 000 macroconidia/mL) and covered for 3 days. The percentage of surviving seedlings was determined at 14 dpi.
flg22 peptide treatment
A needleless syringe was used to infiltrate 50 ng of flg22 peptide (QRLSTGSRINSAKDDAAGLQIA; Alpha Diagnostic, International Inc., San Antonio, TX; Cat# FLG22‐p‐1) dissolved in 20 µL of water through the abaxial surface of Arabidopsis leaves. Water‐infiltrated leaves provided the controls. After 24 h, the leaves were harvested for RNA isolation or treated with Fg mycelial fragments to monitor the impact of PTI activation on fungal disease. For experiments with wheat, flg22 peptide (50, 100 and 200 ng), dissolved in 10 µL of water, and water as control, were applied with a Hamilton syringe to the central spikelet of each spike.
Mycotoxin analysis
DON content in wheat grains was determined as described previously (Fuentes et al., 2005; Mirocha et al., 1998).
Arabidopsis and wheat transgenics
The 35S:NPR1 Arabidopsis and Ubi:NPR1 wheat plants used in this study have been described previously (Cao et al., 1998; Makandar et al., 2006). Three PCR steps were used to generate the PR1‐flg22 chimera, such that the flg22 peptide fused to the apoplast‐localized PR1 protein could be delivered to the extracellular space. In the first PCR, cDNA prepared from Arabidopsis leaves was used to amplify the PR1 CDS (At2g14610) with the primers PR1‐CDS‐F(BamH1) and flg22(21)‐AtPR1(12)‐R to give a 513‐bp amplicon containing the PR1 CDS, without the stop codon, followed by the coding information for the first seven amino acids of flg22. In a second PCR, the flg22‐F and flg22‐R 66‐mer oligos, which contain the coding information for the flg22 peptide (QRLSTGSRINSAKDDAAGLQIA), were mixed with the PR1(21)‐flg22(12)‐F and flg22‐R(ClaI) primers (Table S1, see Supporting Information) which, on PCR, yielded a 99‐bp product which, at one end, contained the information for the last seven amino acids of PR1 fused to flg22 with a stop codon included at the end of the flg22 CDS. The products of the previous two PCRs were mixed in equal proportions and used in PCR with the primers PR1‐CDS‐F(BamH1) and flg22‐R(ClaI) (Table S1) to yield a 570‐bp product that includes the PR1 CDS fused at its C‐terminal end to the flg22 CDS with a stop codon included. This final product was cloned into the pCR8/GW/TOPO entry vector (Life Technologies, Carlsbad, CA; www.lifetechnologies.com) from which the insert was mobilized into the destination binary vector pMDC32 with the Gateway LR clonase system (Life Technologies; www.lifetechnologies.com). The resultant pMDC32:PR1‐flg22 plasmid contains the PR1‐flg22 chimera between the Cauliflower mosaic virus 35S promoter and Agrobacterium tumefaciens nos (nopaline synthase) gene terminator. This construct was transformed into Arabidopsis accession Columbia and the fls2 mutant by the floral dip method (Zhang et al., 2006). Hygromycin‐resistant transformants containing the 35S:PR1‐flg22 chimera were selected in the presence of hygromycin (25 mg/L). The presence of the insert was confirmed by PCR and expression was tested by RT‐PCR.
The WRKY29 (At4g23550) CDS was amplified from cDNA prepared from flg22‐inoculated leaves with the primers WRKY29‐CDS‐F(BamHI) and WRKY29‐CDS‐R(ClaI) (Table S1). The resultant amplicon was cloned into the pMDC32 vector via the pCR8/GW/TOPO intermediate, as described above for 35S:PR1‐flg22. The resultant plasmid pMDC32‐WRKY29 contains the WRKY29 CDS flanked on its 5′ end by the 35S promoter and at the 3′ end by the nos terminator. This construct was transformed into Arabidopsis accession Columbia by the floral dip method (Zhang et al., 2006). Hygromycin‐resistant transformants were selected as described above for 35S:PR1‐flg22. The presence of the 35S:WRKY29 transgene in Arabidopsis was monitored by PCR conducted with 35S‐F and WRKY29‐R(ClaI) primers (Table S1).
The Ubi:WRKY29 chimeric gene was generated by cloning the WRKY29 CDS amplicon, generated as described above, in the pJS406 backbone (Makandar et al., 2006). The resultant pSS:Ubi:WRKY29 plasmid contains the WRKY29 CDS flanked on the 5′ end by the maize Ubi gene promoter plus intron (Christensen and Quail, 1996) and on the 3′ end by the nos terminator. To generate transgenic wheat, the pSS:Ubi:WRKY29 plasmid and the bar selectable marker containing plasmid pAHC20 (Christensen and Quail, 1996) were co‐bombarded into embryos from the spring wheat cv. Bobwhite, and wheat plants were regenerated as described previously (Anand et al., 2003). Glufosinate (Liberty; Bayer Crop Sciences, Research Triangle, NC, USA) resistance conferred by the bar gene was used as the selectable marker for transgenic wheat. The presence of the Ubi:WRKY29 transgene was monitored by PCR using the WRKY29‐F and WRKY29‐R primers (Table S1). PCR conditions included a 5‐min denaturation at 95 ºC, followed by 50 cycles of 95 ºC for 45 s, 55 ºC for 45 s and 72 ºC for 60 s, with a final extension of 72 ºC for 7 min.
DNA and RNA isolation
DNA for PCR and genotyping was extracted from leaf tissue as described previously (Nalam et al., 2016). RNA was extracted from frozen tissues using an acidic guanidinium thiocyanate–phenol–chloroform mix (Chomczynski and Sacchi, 1987).
RT‐PCR and real‐time PCR
After removal of DNA with RQ1 RNase‐free DNase (Promega, Madison, WI, USA), the purified RNA was used for cDNA synthesis with oligo‐dT 18‐mer primer (New England Biolabs, Ipswich, MA, USA) and GoScript™ reverse transcriptase (Promega). The cDNA was subsequently utilized for RT‐PCR and quantitative real‐time RT‐PCR.
The primer pairs WRKY29‐qRT‐F plus WRKY29‐qRT‐R, and EF‐qRT‐F plus EF‐qRT‐R (Table S1), were used for RT‐PCR to monitor the expression of Arabidopsis WRKY29 and At1g07940, respectively. At1g07940, which encodes an elongation factor related to EF‐1α, was previously identified as a gene that is very suitable for the normalization of gene expression (Czechowski et al., 2005). The PCR conditions for WRKY29 included a 3‐min denaturation at 95 ºC, followed by 25 cycles of 95 ºC for 30 s, 58 ºC for 30 s and 72 ºC for 30 s, with a final extension of 72 ºC for 5 min. The PCR conditions for At1g07940 included a 3‐min denaturation at 95 ºC, followed by 25 cycles of 95 ºC for 30 s, 55 ºC for 30 s and 72 ºC for 30 s, with a final extension of 72 ºC for 5 min. RT‐PCR was used to monitor the expression of PR1‐flg22 from the 35S:PR1‐flg22 construct in Arabidopsis. The primer pairs PR1‐CDS‐F(BamHI) and flg22‐R(ClaI) (Table S1) were used in the PCR. Expression of the Arabidopsis ACT8 gene was monitored as control with the primer pair ACT8‐RT‐F plus ACT8‐RT‐R (Table S1). The PCR conditions included a 3‐min denaturation at 95 ºC, followed by 30 cycles of 95 ºC for 30 s, 55 ºC for 30 s and 72 ºC for 45 s, with a final extension of 72 ºC for 7 min. To monitor WRKY29 expression from the Ubi:WRKY29 construct in wheat, the primer pairs WRKY29‐F plus WRKY29‐R (Table S1) were used for amplification in the RT‐PCRs. Expression of the wheat TaTUBB2 (Tubulin beta‐2) gene was used as control for RT‐PCR. The primer pair TaTUBB2‐F and TaTUBB2‐R (Table S1) was used in the PCR. The PCR conditions for WRKY29 expression derived from the Ubi:WRKY29 chimera in wheat included a 3‐min denaturation at 95 ºC, followed by 40 cycles of 95 ºC for 30 s, 60 ºC for 30 s and 72 ºC for 40 s, with a final extension of 72 ºC for 5 min. The PCR conditions for TUBB2 included a 3‐min denaturation at 95 ºC, followed by 40 cycles of 95 ºC for 30 s, 58 ºC for 30 s and 72 ºC for 30 s, with a final extension of 72 ºC for 5 min.
Quantitative real‐time RT‐PCR was performed with Sybr Green PCR Master Mix (Bio‐Rad Laboratories, Hercules, CA, USA) on an Eco qPCR system (Illumina, San Diego, CA) using the following amplification conditions: 10‐min polymerase activation and denaturation at 95 °C, 40 cycles of 95 °C for 10 s, 58 °C for 30 s and 72 °C for 30 s. This was followed by a product melt to confirm a single PCR product. The level of individual gene expression was normalized to that of At1g07940 by subtracting the cycle threshold value for At1g07940 from the cycle threshold value of the test genes. Fold induction, when calculated, was relative to expression in the mock‐treated plants. The primer pairs WRKY29‐qRT‐F plus WRKY29‐qRT‐R, and EF‐qRT‐F plus EF‐qRT‐R (Table S1), were used for real‐time RT‐PCR to monitor the expression of Arabidopsis WRKY29 and At1g07940, respectively. For qRT‐PCR analysis of wheat TaPR1.2, TaWRKY70 and TaPUB‐23‐like, the primer pairs TaPR1.2‐F plus TaPR1.2‐R, TaWRKY70‐F plus TaWRKY70‐R, and TaPUB‐23‐F plus TaPUB‐23‐R, respectively, were used (Table S1). The expression of these genes was normalized to that of TaTUBB2.
Histological examination for H2O2 accumulation
In situ accumulation of H2O2 was monitored by staining leaves with 3,3′‐diaminobenzidine (DAB; Sigma‐Aldrich, St Louis, MO, USA) using a protocol developed by Thordal‐Christensen et al. (1997) as modified by Gao X et al. (2013). Briefly, Arabidopsis leaves were infiltrated with Fg, whereas wheat leaves were first pierced with a needle and Fg spores were placed on the pierced site. Leaves infiltrated with water and untreated leaves provided the controls. The treated plants were left in the growth room until ready for harvest. On harvest, leaves were immersed in DAB solution (1 mg/mL; pH 3.8). Gentle vacuum was applied for 20 min to infiltrate the DAB solution into the leaves. Leaves immersed in DAB solution were covered in foil and incubated at room temperature. After 8 h, the DAB solution was discarded. For destaining, Arabidopsis leaves were boiled in 95% ethanol for 20 min, whereas wheat leaves were boiled for 30 min. Destained leaves were stored in 70% ethanol. A similar process was used for staining wheat spikes, except that the destaining utilized 70% ethanol and was conducted overnight in a shaker at 70 °C. The destained leaves were observed under a light microscope.
Statistical analysis
Two‐tailed Student’s t‐test was used to determine the significance of variance (P < 0.05) when comparing two treatments or genotypes. The χ 2 test was used to determine whether the differences between disease categories on Arabidopsis leaves were significantly different (P < 0.05) between two genotypes and treatments. Analysis of variance (ANOVA) following the General Linear Model, followed by Tukey’s multiple comparison test, was used to determine the significance of variance (P < 0.05; Minitab v15; www.minitab.com) when comparing multiple genotypes and/or treatments with each other.
Accession numbers
At4g23550 (Arabidopsis WRKY29), At5g46330 (Arabidopsis FLS2), At1g07940 (Arabidopsis GTP‐binding Elongation factor Tu family), At2g14610 (Arabidopsis PR1), At1g49240 (Arabidopsis ACT8), U76745 (wheat TUBB2), AJ007349 (wheat PR1.2), BQ743320 (wheat PUB‐23‐like), AB603890 (wheat WRKY70), FGSG_0811 (Fg NahG).
Conflicts of Interest
The authors declare that there are no conflicts of interest.
Supporting information
Fig. S1 Expression of the PR1‐flg22 chimera in transgenic Arabidopsis. Top: reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of the PR1‐flg22 chimeric transcript and, as control, the Arabidopsis ACT8 gene in the wild‐type (WT) accession Columbia and two independent PR1‐flg22 lines #2 and #5 in the FLS2 background. Bottom: PR1‐flg22 and ACT8 expression in the fls2 mutant and two independent PR1‐flg22 lines #2 and #5 that are in the fls2 mutant background.
Fig. S2 3,3′‐Diaminobenzidine (DAB) staining for H2O2 accumulation in Fusarium graminearum‐inoculated Arabidopsis and wheat leaves. (A) H2O2 accumulation in F. graminearum‐inoculated leaves of wild‐type (WT) accession Columbia and three independent 35S:WRKY29 transgenic Arabidopsis lines. (B) H2O2 accumulation in F. graminearum‐inoculated leaves of wheat cv. Bobwhite (Bw) and two independent Ubi:WRKY29 transgenic lines #317 and #1081. In (A) and (B), leaves were harvested for DAB staining at 2 and 6 h post‐inoculation.
Table S1 Primers used in this study.
Acknowledgements
The authors would like to thank Sarah Oswald, Neeha N. Alam, Nicholas Canoy and Elena Shulaev for assistance with plant genotyping. This work was supported by funding from the US Department of Agriculture Agreement #59‐0790‐8‐060 to J.S. and H.N.T., Agreement #59‐0206‐7‐006 to J.S. and Agreement #59‐0206‐4‐023 to Y.D. as cooperative projects with the US Wheat & Barley Scab Initiative. S.T.A. was supported by graduate and tuition assistantship from the University of North Texas.
References
- Anand, A. , Schmelz, E.A. and Muthukrishnan, S. (2003) Development of a lesion‐mimic phenotype in a transgenic wheat line overexpressing genes for pathogenesis‐related (PR) proteins is dependent on salicylic acid concentration. Mol. Plant–Microbe Interact. 16, 916–925. [DOI] [PubMed] [Google Scholar]
- Asai, T. , Tena, G. , Plotnikova, J. , Willmann, M.R. , Chiu, W.‐L. , Gomez‐Gomez, L. , Boller, T. , Ausubel, F.M. and Sheen, J. (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature, 415, 977–983. [DOI] [PubMed] [Google Scholar]
- Bahrini, I. , Ogawa, T. , Kobayashi, F. , Kawahigashi, H. and Handa, H. (2011a) Overexpression of the pathogen‐inducible wheat TaWRKY45 gene confers disease resistance to multiple fungi in transgenic wheat plants. Breed. Sci. 61, 319–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrini, I. , Sugisawa, M. , Kikuchi, R. , Ogawa, T. , Kawahigashi, H. , Ban, T. and Handa, H. (2011b) Characterization of a wheat transcription factor, TaWRKY45, and its effect on Fusarium head blight resistance in transgenic wheat plants. Breed. Sci. 61, 121–129. [Google Scholar]
- Bai, G.H. and Shaner, G. (2004) Management and resistance in wheat and barley to Fusarium head blight. Annu. Rev. Phytopathol. 42, 135–161. [DOI] [PubMed] [Google Scholar]
- Bhaskara Reddy, M.V. , Arul, J. , Angers, P. and Couture, L. (1999) Chitosan treatment of wheat seeds induces resistance to Fusarium graminearum and improves seed quality. J. Agric. Food Chem. 47, 1208–1216. [DOI] [PubMed] [Google Scholar]
- Bigeard, J. , Colcombet, J. and Hirt, H. (2015) Signaling mechanisms in pattern‐triggered immunity (PTI). Mol. Plant, 8, 521–539. [DOI] [PubMed] [Google Scholar]
- Cao, H. , Li, X. and Dong, X. (1998) Generation of broad spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc. Natl. Acad. Sci. USA, 95, 6531–6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi, R. , Venables, B. , Petros, R.A. , Nalam, V. , Li, M. , Wang, X. , Takemoto, L.J. and Shah, J. (2012) An abietane diterpenoid is a potent activator of systemic acquired resistance. Plant J. 71, 161–172. [DOI] [PubMed] [Google Scholar]
- Chen, X. , Steed, A. , Harden, C. and Nicholson, P. (2006) Characterization of Arabidopsis thaliana–Fusarium graminearum interactions and identification of variation in resistance among ecotypes. Mol. Plant Pathol. 7, 391–403. [DOI] [PubMed] [Google Scholar]
- Chen, X. , Steed, A. , Travella, S. , Keller, B. and Nicholson, P. (2009) Fusarium graminearum exploits ethylene signalling to colonize dicotyledonous and monocotyledonous plants. New Phytol. 182, 975–983. [DOI] [PubMed] [Google Scholar]
- Christensen, A.H. and Quail, P.H. (1996) Ubiquitin promoter‐based vectors for high‐level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 5, 213–218. [DOI] [PubMed] [Google Scholar]
- Chomczynski, P. and Sacchi, N. (1987) Single‐step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem. 162, 156–159. [DOI] [PubMed] [Google Scholar]
- Conrath, U. , Beckers, G.J.M. , Langenbach, C.J.G. and Jaskiewicz, M.R. (2015) Priming for enhanced defense. Annu. Rev. Phytopathol. 53, 97–119. [DOI] [PubMed] [Google Scholar]
- Cuzick, A. , Lee, S. , Gezan, S. and Hammond‐Kosack, K.E. (2008) NPR1 and EDS11 contribute to host resistance against Fusarium culmorum in Arabidopsis buds and flowers. Mol. Plant Pathol. 9, 697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski, T. , Stitt, M. , Altmann, T. , Udvardi, M.K. and Scheible, W.‐R. (2005) Genome‐wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diethelm, M. , Schmolke, M. , Groth, J. , Friedt, W. , Schweizer, G. and Hartl, L. (2014) Association of allelic variation in two NPR1‐like genes with Fusarium head blight resistance in wheat. Mol. Breed. 34, 31–43. [Google Scholar]
- Eulgem, T. , Rushton, P.J. , Robatzek, S. and Somssich, I.E. (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. [DOI] [PubMed] [Google Scholar]
- Ferrari, S. , Galletti, R. , Denoux, C. , De Lorenzo, G. , Ausubel, F.M. and Dewdney, J. (2007) Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is independent of salicylic acid, ethylene, or jasmonate signaling but requires PHYTOALEXIN DEFICIENT3 . Plant Physiol. 144, 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes, R.G. , Mickelson, H.R. , Busch, R.H. , Dill‐Macky, R. , Evans, C.K. , Thompson, W.G. , Wiersma, J.V. , Xie, W. , Dong, Y. and Anderson, J.A. (2005) Resource allocation and cultivar stability in breeding for Fusarium head blight resistance in spring wheat. Crop Sci. 45, 1965–1972. [Google Scholar]
- Gao, C.S. , Kou, X.‐J. , Li, H.P. , Zhang, J.B. , Saad, A.S.I. and Liao, Y.C. (2013) Inverse effects of Arabidopsis NPR1 gene on Fusarium seedling blight and Fusarium head blight in transgenic wheat. Plant Pathol. 62, 383–392. [Google Scholar]
- Gao, X. , Li, F. , Li, M. , Kianinejad, A.S. , Dever, J.K. , Wheeler, T.A. , Li, Z. , He, P. and Shan, L. (2013) Cotton GhBAK1 mediates Verticillium wilt resistance and cell death. J. Integr. Plant Biol. 55, 586–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletti, R. , Ferrari, S. and De Lorenzo, G. (2011) Arabidopsis MPK3 and MPK6 play different roles in basal and oligogalacturonide‐ or flagellin‐induced resistance against Botrytis cinerea . Plant Physiol. 157, 804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez‐Gomez, L. and Boller, T. (2000) FLS2: an LRR receptor‐like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell, 5, 1003–1011. [DOI] [PubMed] [Google Scholar]
- Gu, Y. and Innes, R.W. (2012) The KEEP ON GOING protein of Arabidopsis regulates intracellular protein trafficking and is degraded during fungal infection. Plant Cell, 24, 4717–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust, A.A. , Biswas, R. , Lenz, H.D. , Rauhut, T. , Ranf, S. , Kemmerling, B. , Götz, F. , Glawischnig, E. , Lee, J. , Felix, G. and Nürnberger, T. (2007) Bacteria‐derived peptidoglycans constitute pathogen‐associated molecular patterns triggering innate immunity in Arabidopsis. J. Biol. Chem. 282, 32338–32348. [DOI] [PubMed] [Google Scholar]
- Hao, Q. , Wang, W. , Han, X. , Wu, J. , Lyu, B. , Chen, F. , Caplan, A. , Li, C. , Wu, J. , Wang, W. , Xu, Q. and Fu, D. (2018) Isochorismate‐based salicylic acid biosynthesis confers basal resistance to Fusarium graminearum in barley. Mol. Plant Pathol. 19, 1995–2010. 10.1111/mpp.12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden, P. (2003) The genes of the green revolution. Trends Genet. 19, 5–9. [DOI] [PubMed] [Google Scholar]
- Johnson, D.D. , Flaskerud, G.K. , Taylor, R.D. and Satyanarayana, V. (2003) Quantifying economic impacts of Fusarium head blight in wheat In:Fusarium Head Blight of Wheat and Barley (Leonard, K.J.andBushnell W.R., eds), pp. 461–483. St. Paul, MN: American Pytopathology Society. [Google Scholar]
- Kage, U. , Yogendra, K.N. and Kushalappa, A.C. (2017) TaWRKY70 transcription factor in wheat QTL‐2DL regulates downstream metabolite biosynthetic genes to resist Fusarium graminearum infection spread within spike. Sci. Rep. 7, 42596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaku, H. , Nishizawa, Y. , Ishii‐Minami, N. , Akimoto‐Tomiyama, C. , Dohmae, N. , Takio, K. , Minami, E. and Shibuya, N. (2006) Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA, 103, 11086–11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karre, S. , Kumar, A. , Dhokane, D. and Kushalappa, A.C. (2017) Metabolo‐transcriptome profiling of barley reveals induction of chitin elicitor receptor kinase gene (HvCERK1) conferring resistance against Fusarium graminearum . Plant Mol. Biol. 93, 247–267. [DOI] [PubMed] [Google Scholar]
- Li, B. , Meng, X. , Shan, L. and He, P. (2016) Transcriptional regulation of pattern‐triggered immunity in plants. Cell Host Microbe, 19, 641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Williams, B. and Dickman, M. (2017) Arabidopsis B‐cell lymphoma2 (Bcl‐2)‐associated athanogene 7 (BAG7)‐mediated heat tolerance requires translocation, sumoylation and binding to WRKY29. New Phytol. 214, 695–705. [DOI] [PubMed] [Google Scholar]
- Liu, D. , Leib, K. , Zhao, P. , Kogel, K.‐H. and Langen, G. (2014) Phylogenetic analysis of barley WRKY proteins and characterization of HvWRKY1 and ‐2 as repressors of the pathogen‐inducible gene HvGER4c. Mol. Genet. Genomics, 289, 1331–1345. [DOI] [PubMed] [Google Scholar]
- Makandar, R. , Essig, J.S. , Schapaugh, M.A. , Trick, H.N. and Shah, J. (2006) Genetically engineered resistance to Fusarium head blight in wheat by expression of Arabidopsis NPR1 . Mol. Plant–Microbe Interact. 19, 123–129. [DOI] [PubMed] [Google Scholar]
- Makandar, R. , Nalam, V. , Chaturvedi, R. , Jeannotte, R. , Sparks, A.A. and Shah, J. (2010) Involvement of salicylate and jasmonate signaling pathways in Arabidopsis interaction with Fusarium graminearum . Mol. Plant–Microbe Interact. 23, 861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makandar, R. , Nalam, V.J. , Chowdhury, Z. , Sarowar, S. , Klossner, G. , Lee, H. , Burdan, D. , Trick, H.N. , Gobbato, E. , Parker, J.E. and Shah, J. (2015) The combined action of ENHANCED DISEASE SUSCEPTIBILITY1, PHYTOALEXIN DEFICIENT4 and SENESCENCE‐ASSOCIATED101 promotes salicylic acid‐mediated defenses to limit Fusarium graminearum infection in Arabidopsis thaliana . Mol. Plant–Microbe Interact. 28, 943–953. [DOI] [PubMed] [Google Scholar]
- Makandar, R. , Nalam, V.J. , Lee, H. , Trick, H.N. , Dong, Y. and Shah, J. (2012) Salicylic acid regulates basal resistance to Fusarium head blight in wheat. Mol. Plant–Microbe Interact. 25, 431–439. [DOI] [PubMed] [Google Scholar]
- McMullen, M. , Jones, R. and Gallenberg, D. (1997a) Scab of wheat and barley: a re‐emerging disease of devastating impact. Plant Dis. 81, 1340–1348. [DOI] [PubMed] [Google Scholar]
- McMullen, M.P. , Schatz, B. , Stover, R. and Gregoire, T. (1997b) Studies of fungicide, application timing, and application technologies to reduce Fusarium head blight and deoxynivalenol. Cereal Res. Commun. 25, 779–780. [Google Scholar]
- Mirocha, C.J. , Kolaczkowski, E. , Xie, W. , Yu, H. and Jelen, H. (1998) Analysis of deoxynivalenol and its derivatives (batch and single kernel) using gas chromatography/mass spectrometry. J. Agric. Food Chem. 46, 1414–1418. [Google Scholar]
- Nalam, V.J. , Alam, S. , Keereetaweep, J. , Venables, B. , Burdan, D. , Lee, H. , Trick, H.N. , Sarowar, S. , Makandar, R. and Shah, J. (2015) Facilitation of Fusarium graminearum infection by 9‐lipoxygenases in Arabidopsis and wheat. Mol. Plant–Microbe Interact. 28, 1142–1152. [DOI] [PubMed] [Google Scholar]
- Nalam, V.J. , Sarowar, S. and Shah, J. (2016) Establishment of a Fusarium graminearum infection model in Arabidopsis leaves and floral tissues. Bio‐protocols, 6, e1877 http://www.bio-protocol.org/e1877. [Google Scholar]
- Nishiuchi, T. , Masuda, D. , Nakashita, H. , Ichimura, K. , Shinozaki, K. , Yoshida, S. , Kimura, M. , Yamaguchi, I. and Yamaguchi, K. (2006) Fusarium phytotoxin trichothecenes have an elicitor‐like activity in Arabidopsis thaliana, but its activity differed significantly among their molecular species. Mol. Plant–Microbe Interact. 19, 512–520. [DOI] [PubMed] [Google Scholar]
- Niu, C.‐F. , Wei, W. , Zhou, Q.‐Y. , Tian, A.‐G. , Hao, Y.‐J. , Zhang, W.‐K. , Ma, B. , Lin, Q. , Zhang, Z.‐B. , Zhang, J.‐S. and Chen, S.‐Y. (2012) Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 35, 1156–1170. [DOI] [PubMed] [Google Scholar]
- Pandey, S.P. and Somssich, I.E. (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol. 150, 1648–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pečenková, T. , Pleskot, R. and Žárský, V. (2017) Subcellular localization of Arabidopsis pathogenesis‐related 1 (PR1) protein. Int. J. Mol. Sci. 18, 825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfund, C. , Tans‐Kersten, J. , Dunning, F.M. , Alonso, J.M. , Ecker, J.R. , Allen, C. and Bent, A.F. (2004) Flagellin is not a major defense elicitor in Ralstonia solanacearum cells or extracts applied to Arabidopsis thaliana . Mol. Plant–Microbe Interact. 17, 696–706. [DOI] [PubMed] [Google Scholar]
- Pick, T. , Jaskiewicz, M. , Peterhansel, C. and Conrath, U. (2012) Heat shock factor HsfB1 primes gene transcription and systemic acquired resistance in Arabidopsis. Plant Physiol. 159, 52–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M.J. , Zamioudis, C. , Berendsen, R.L. , Weller, D.M. , Van Wees, S.C.M. and Bakker, P.A.H.M. (2014) Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375. [DOI] [PubMed] [Google Scholar]
- Pirgozliev, S.R. , Edwards, S.G. , Hare, M.C. and Jenkinson, P. (2003) Strategies for the control of Fusarium head blight in cereals. Eur. J. Plant Pathol. 109, 731–742. [Google Scholar]
- Sánchez‐Vallet, A. , Mesters, J.R. and Thomma, B.P.H.J. (2015) The battle for chitin recognition in plant–microbe interactions. FEMS Microbiol. Rev. 39, 171–183. [DOI] [PubMed] [Google Scholar]
- Savitch, L.V. , Subramaniam, R. , Allard, G.C. and Singh, J. (2007) The GLK1 ‘regulon’ encodes disease defense related proteins and confers resistance to Fusarium graminearum in Arabidopsis . Biochem. Biophys. Res. Commun. 359, 234–238. [DOI] [PubMed] [Google Scholar]
- Schoonbeek, H.‐J. , Wang, H.‐H. , Stefanato, F.L. , Craze, M. , Bowden, S. , Wallington, E. , Zipfel, C. and Ridout, C.J. (2015) Arabidopsis EF‐Tu receptor enhances bacterial disease resistance in transgenic wheat. New Phytol. 206, 606–613. [DOI] [PubMed] [Google Scholar]
- Shimizu, T. , Nakano, T. , Takamizawa, D. , Desaki, Y. , Ishii‐Minami, N. , Nishizawa, Y. , Minami, E. , Okada, K. , Yamane, H. , Kaku, H. and Shibuya, N. (2010) Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64, 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skadsen, R.W. and Hohn, T.M. (2004) Use of Fusarium graminearum transformed with gfp to follow infection patterns in barley and Arabidopsis. Physiol. Mol. Plant Pathol. 64, 45–53. [Google Scholar]
- Thordal‐Christensen, H. , Zhang, Z. , Wei, Y. and Collinge, D.B. (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley–powdery mildew interaction. Plant J. 11, 1187–1194. [Google Scholar]
- Tsuda, K. , Sato, M. , Glazebrook, J. , Cohen, J.D. and Katagiri, F. (2008) Interplay between MAMP‐triggered and SA‐mediated defense responses. Plant J. 53, 763–775. [DOI] [PubMed] [Google Scholar]
- Urban, M. , Daniels, S. , Mott, E. and Hammond‐Kosack, K. (2002) Arabidopsis is susceptible to the cereal ear blight fungal pathogens Fusarium graminearum and Fusarium culmorum . Plant J. 32, 961–973. [DOI] [PubMed] [Google Scholar]
- Van Hemelrijck, W. , Wouters, P.F.W. , Brouwer, M. , Windelinckx, A. , Goderis, I.J.W.M. , De Bolle, M.F.C. , Thomma, B.P.H.J. , Cammue, B.P.A. and Delauré, S.L. (2006) The Arabidopsis defense response mutant esa1 as a model to discover novel resistance traits against Fusarium diseases. Plant Sci. 171, 585–595. [Google Scholar]
- Wan, J. , Zhang, X.‐C. , Neece, D. , Ramonell, K.M. , Clough, S. , Kim, S.Y. , Stacey, M.G. and Stacey, G. (2008) A LysM receptor‐like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis . Plant Cell, 20, 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D. , Weaver, N.D. , Kesarwani, M. and Dong, X. (2005) Induction of protein secretory pathway is required for systemic acquired resistance. Science, 308, 1036–1040. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Tao, F. , Tian, W. , Guo, Z. , Chen, X. , Xu, X. , Shang, H. and Hu, X. (2017) The wheat WRKY transcription factors TaWRKY49 and TaWRKY62 confer differential high‐temperature seedling‐plant resistance to Puccinia striiformis f. sp. tritici . PLoS ONE, 12, e0181963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Zeng, J. , Li, Y. , Rong, X. , Sun, J. , Sun, T. , Li, M. , Wang, L. , Feng, Y. , Chai, R. , Chen, M. , Chang, J. , Li, K. , Yang, G. and He, G. (2015) Expression of TaWRKY44, a wheat WRKY gene, in transgenic tobacco confers multiple abiotic stress tolerances. Front. Plant Sci. 6, 615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Zhou, Z. , Gao, J. , Wu, Y. , Xia, Z. , Zhang, H. and Wu, J. (2016) The mechanisms of maize resistance to Fusarium verticillioides by comprehensive analysis of RNA‐seq data. Front. Plant Sci. 7, 1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, S. , Shimada, T.L. , Hiruma, K. and Takano, Y. (2013) Pathogen infection trial increases the secretion of proteins localized in the endoplasmic reticulum body of Arabidopsis. Plant Physiol. 163, 659–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm, E.P. , Boulton, M.I. , Al‐Kaff, N.S. , Balfourier, F. , Bordes, J. , Greenland, A.J. , Powell, W. and Mackay, I.J. (2013) Rht‐1 and Ppf‐D1 associations with height, GA sensitivity, and days to heading in a worldwide bread wheat collection. Theor. Appl. Genet. 126, 2233–2243. [DOI] [PubMed] [Google Scholar]
- Wilson, W.W. , McKee, G. , Nganje, W. , Dahl, B. and Bangsund, D. (2017) Economic impact of USWBSI’s Scab initiative to reduce FHB. Agribusiness and Applied Economics, 774 https://ageconsearch.umn.edu/record/264672. [Google Scholar]
- Xu, X. and Nicholson, P. (2009) Community ecology of fungal pathogens causing wheat head blight. Annu. Rev. Phytopathol. 47, 83–103. [DOI] [PubMed] [Google Scholar]
- Yi, S.Y. , Shirasu, K. , Moon, J.S. , Lee, S.‐G. and Kwon, S.‐Y. (2014) The activated SA and JA signaling pathways have an influence on flg22‐triggered oxidative burst and callose deposition. PLoS One, 9, e88951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G. , Zhang, X. , Yao, J. , Zhou, M.P. and Ma, H. (2017) Resistance against Fusarium head blight in transgenic wheat plants expressing the ScNPR1 gene. J. Phytopathol. 165, 223–231. [Google Scholar]
- Zhang, X. , Henriques, R. , Lin, S.S. , Niu, Q.W. and Chua, N.H. (2006) Agrobacterium‐mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protocols, 1, 641–646. [DOI] [PubMed] [Google Scholar]
- Zipfel, C. , Kunze, G. , Chinchilla, D. , Caniard, A. , Jones, J.D. , Boller, T. and Felix, G. (2006) Perception of the bacterial PAMP EF‐Tu by the receptor EFR restricts Agrobacterium‐mediated transformation. Cell, 125, 749–760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Expression of the PR1‐flg22 chimera in transgenic Arabidopsis. Top: reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of the PR1‐flg22 chimeric transcript and, as control, the Arabidopsis ACT8 gene in the wild‐type (WT) accession Columbia and two independent PR1‐flg22 lines #2 and #5 in the FLS2 background. Bottom: PR1‐flg22 and ACT8 expression in the fls2 mutant and two independent PR1‐flg22 lines #2 and #5 that are in the fls2 mutant background.
Fig. S2 3,3′‐Diaminobenzidine (DAB) staining for H2O2 accumulation in Fusarium graminearum‐inoculated Arabidopsis and wheat leaves. (A) H2O2 accumulation in F. graminearum‐inoculated leaves of wild‐type (WT) accession Columbia and three independent 35S:WRKY29 transgenic Arabidopsis lines. (B) H2O2 accumulation in F. graminearum‐inoculated leaves of wheat cv. Bobwhite (Bw) and two independent Ubi:WRKY29 transgenic lines #317 and #1081. In (A) and (B), leaves were harvested for DAB staining at 2 and 6 h post‐inoculation.
Table S1 Primers used in this study.


