Abstract
Objective
We previously reported a statistically significant association of SCARB1 intronic SNP rs10846744 with common carotid intimal-medial artery thickness (CCIMT) in each of the four Multi-Ethnic Study of Atherosclerosis (MESA) racial/ethnic groups (Caucasian, Chinese, African American and Hispanic).
Methods and Results
Using an expanded sample of 7,936 MESA participants, phenotyped for measures of subclinical atherosclerosis (SCA), incident myocardial infarction (MI) and cardiovascular disease (CVD), and genotyped through the SNP Health Association Resource (SHARe) project, we have now examined the genetic association of these phenotypes with 126 genotyped and imputed SCARB1 SNPs. We also performed stratified analyses to examine whether SCARB1 SNP effects differed by sex. Our analysis of the full MESA cohort provides strong evidence for association of rs10846744 with common carotid intimal-medial thickness (P=1.04E-4 in combined analysis of all four MESA racial/ethnic groups). In sex-stratified analysis, we observed statistically significant association of rs10846744 with incident CVD events in males (P=0.01). Examining analytical results from the Myocardial Infarction Genetics Consortium for replication, we observed further support for the association of rs10846744 with MI.
Conclusion
The SCARB1 SNP, rs10846744, exerts a major effect on SCA and incident CVD in humans.
Keywords: genetics, lipids, cardiovascular disease, genomics
The SCARB1 gene encodes scavenger receptor class B type I (SR-BI) protein, characterized as a high-density lipoprotein (HDL) receptor1. In humans, multiple studies have reported association of common SCARB1 single nucleotide polymorphisms (SNPs) with HDL cholesterol levels2–5. Existing studies also indicate the effects of SCARB1 variants on HDL levels may vary by gender3, 6, 7. Specifically, the earlier work by Acton et al.6 showed variability in associations of LDL and HDL to SCARB1 SNPs based on gender.
In mice, SR-BI gene manipulation studies have demonstrated its antiatherogenic properties, with SR-BI/apolipoprotein-E double knockout mice developing complex coronary artery disease, myocardial infarction, and heart failure8. In humans, there have been reports of association of SCARB1 variants with measures of subclinical atherosclerosis (SCA)9, 10, as well as with coronary heart disease11. After identifying association of a SCARB1 SNP (rs838880) with HDL, Teslovich et al.4 further examined whether the same SNP was associated with coronary artery disease, but found no evidence of association. We recently took a different approach, directly examining association of SCARB1 SNPs with measures of SCA (coronary artery calcification [CAC], common carotid intimal-medial artery thickness [CCIMT] and internal carotid intimal-medial thickness [internal IMT]) in the Multi-Ethnic Study of Atherosclerosis (MESA), using a subset of 2,757 participants typed for selected candidate gene SNPs9. We found carriers of the G allele of SNP rs10846744 had significantly lower CCIMT as compared with carriers of the C allele in all MESA racial/ethnic groups (Caucasian P=0.05, Chinese P=0.02, African American P=0.03, and Hispanic P=0.03), with strong evidence for association in pooled analysis combining all racial/ethnic groups (P=0.0002). The significant association of rs10846744 with SCA was not influenced by traditional atherosclerosis risk factors, such as lipids (including HDL-C), hypertension, body mass index, and fasting glucose levels.
To replicate these associations and better characterize the relationship between SCARB1 variants with SCA and incident cardiovascular (CVD) events, we analyzed SCARB1 genotypes available through genome-wide association scan data on 8,224 consenting MESA participants, obtained through the NHLBI SNP Health Association Resource (SHARe) project. We began by examining association of the SCARB1 SNP rs10846744 with measures of SCA in the full MESA cohort, and then went on to assess association of 126 (genotyped and imputed) SCARB1 SNPs in MESA participants with the same measures of SCA. The sample for which we report on genetic associations in the current study includes, but is substantially larger than, the study population reported in our previous manuscript9. Furthermore, we extended our earlier investigation to include clinical outcomes of incident MI and CVD events. We also performed sex-stratified analyses to permit differential SCARB1 SNP effects by sex. Finally, we performed in silico replication using published results from the Myocardial Infarction Genetics (MIGen) Consortium12.
Methods
Study Design
MESA is a longitudinal study of subclinical cardiovascular disease and risk factors that predict progression to clinically overt cardiovascular disease or progression of the subclinical disease. The first clinic visits occurred in 2000–2002 in 6,814 participants recruited from six field centers across the United States, and all participants were free of CVD at the baseline exam. Approximately 38% of the recruited participants are White, 28% African-American, 22% Hispanic, and 12% Asian, predominantly of Chinese descent. MESA has been enhanced by many ancillary studies focused on specific phenotypic and exposure domains 13. One ancillary study (MESA Family Study, MESAFS) recruited African American and Hispanic family members specifically for genetic studies. Another ancillary study (MESA Air) evaluated the effects of air pollution on atherosclerosis risk.
Genotype Data
Participants recruited by the original MESA cohort (6,814), and MESAFS (2,128 from 528 families) and MESA Air (5,479 from MESA, 257 external cohort, and 490 from MESAFS) were genotyped in 2009 using the Affymetrix Human SNP array 6.0. SNPs were filtered for SNP level call rate < 95% and individual level call rate < 95%, and monomorphic SNPs were removed. Examining the distribution of heterozygosity rates across all genotyped SNPs, we observed a generally uniform distribution between 0 – 53%, with less than 0.01% of SNPs having heterozygosity > 53%. Thus, we removed all SNPs with heterozygosity > 53%. The cleaned genotypic data was deposited with MESA phenotypic data into dbGaP as the MESA SHARe project (study accession phs000209, http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000209.v7.p2) for 8,224 consenting individuals (2,685 Caucasian, 777 Chinese, 2,588 non-Hispanic African-American, and 2,174 Hispanic) with 897,981 SNPs passing study specific quality control (QC).
Principal component analysis
Prior to population structure analysis, we first constructed subsets of typed SNPs, thinned for linkage disequilibrium (LD) within each of the four MESA cohorts. To prevent the principal components from being dominated by regions of known long-range LD, we first removed from consideration regions of known long-range linkage disequilibrium among Caucasians14, including HLA, a chromosome 8 inversion, and a region on chromosome 11. We then thinned for local LD within an unrelated subset of the MESA Hispanic cohort using the PLINK15 option “--indep-pairwise” to create a subset of typed SNPs thinned for pairwise R2 no more than 0.2 in a 100- SNP window, moving the windows 25 SNPs at a time. LD thinning was performed separately within each racial/ethnic group, resulting in a subset of 114,035 SNPs for Caucasians; 98,353 SNPs for Chinese; 99,716 SNPs for African Americans; and 61,194 for Hispanics.
Using our LD-thinned subsets constructed separately for each of the four MESA racial/ethnic groups, we performed Principal Component Analysis (PCA) as implemented in the program SMARTPCA16, 17 from the software package EIGENSTRAT to compute principal components (PCs) of ancestry for unrelated subsets of individuals, constructed by removing inferred first degree relatives from the analysis. In computing the PCs, we performed additional LD correction by using results of regression on the previous 5 SNPs as input to the PCA (SMARTPCA option “nsnpldregress”), and performed 5 iterations of outlier removal in which we removed individuals with computed values more than 10 standard deviations from mean along the top 6 PCs of ancestry. We constructed histograms and QQ-plots to assess symmetry and normality of the distribution of loadings for each of the resulting PCs to determine the optimal number of PCs to include in genetic association analysis.
Selection of SNPs for genetic association analysis
To perform genetic association analysis for SNPs in the SCARB1 gene region, we began by selecting SNPs of interest in the region. We combined all SCARB1 SNPs investigated in our previous publication9 with SNPs reported for the SCARB1 gene region in dbSNP to identify a total of 1215 SNPs in the SCARB1 gene region. Of these 1215 SNPs, 49 were genotyped on Affy 6.0 and passed genotype QC. IMPUTE version 2.1.0 was used to perform imputation for the MESA SHARe participants (chromosomes 1–22) using HapMap Phase I and II - CEU+YRI+CHB+JPT as the reference panel (release #22 - NCBI Build 36 (dbSNP b126)) (only the CEU reference panel was used for imputation in Caucasian participants), and we could impute another 108 SCARB1 SNPs in MESA Caucasians, 80 SNPs in Chinese, 80 SNPs in African Americans, and 80 SNPs in Hispanics. Allele frequencies were calculated separately within each racial/ethnic group, and only those SNPs with minor allele frequencies (MAF) > 0.01 were included in genetic association analysis. We further filtered imputed SNPs based on imputation quality > 0.5, using the observed versus expected variance quality metric18, and filtered genotyped SNPs for Hardy-Weinberg Equilibrium (HWE) P-value ≥ 10−5. After applying these filters, we had 90, 99, 111, and 107 SNPs remaining for genetic association analysis in MESA Caucasians, Chinese, African Americans, and Hispanics, respectively, and a total of 126 unique SNPs across all four racial/ethnic groups.
Phenotyping of MESA participants
Measures of SCA examined included the presence or absence of CAC as a binary marker of SCA, and ultrasound measurements of intimal-medial thickness in millimeters (mm) for CCIMT and ICIMT.
Cardiovascular events were adjudicated by a MESA committee which included cardiologists, physician epidemiologists and neurologists. A detailed description of the cardiovascular event adjudication process has already been published19. In MESA, we define CVD events to include incident myocardial infarction, definite angina, probable angina (if followed by coronary artery bypass grafting and percutaneous coronary intervention), resuscitated cardiac arrest, stroke, stroke death, CHD death or other CVD death. For the purposes of this study we examined probable or confirmed CVD events (“CVD-All”), confirmed CVD events (“CVD-Hard”), and incident MI.
Genetic association analysis in MESA
To select individuals to be included in analysis, we began with the full MESA cohort. We stratified by racial/ethnic group, and eliminated those individuals with top principal components of ancestry > 3.5 SD from the mean within any racial/ethnic group. To allow study site to be included as a covariate in genetic association analysis within each racial/ethnic group, we restricted the data set to individuals from study sites with data available for at least 20 individuals of that racial/ethnic group. For each of the phenotypic analyses, we then restricted the data set to individuals with data available for the particular phenotype of interest. For quantitative traits, outliers were defined as individuals with phenotypic values more then 3.5 SD from the mean, with the mean and SD calculated separately for each of the stratified analyses performed. Analyses of CCIMT and ICIMT were performed on the log-scale, which yielded approximately normal phenotypic distributions.
We began with stratified analyses within each racial/ethnic group. For analysis of Caucasian and Chinese, and where there were insufficient families with phenotypes among African American and Hispanics, we first constructed an unrelated subset of individuals by selecting at most one individual from each pedigree, and performed linear regression of quantitative phenotypes or logistic regression of dichotomous phenotypes in R20. For analysis of phenotypes with a substantial familial component among African American and Hispanic cohorts, we performed analysis using an additive model with a linear mixed-effects model for quantitative traits, or generalized estimating equations for dichotomous traits, to account for familial relationships as implemented in the package R/GWAF21.
In all analyses, we began with a basic model (Model 1) including age, sex, study site and PCs of ancestry. (Based on our examination of PCs within each racial/ethnic group, as described above, we used 3 PCs for analysis of Caucasians, 1 PC for Chinese, 1 PC for African Americans, and 3 PCs for Hispanics). To examine sensitivity of our results to other known risk factors for atherosclerosis, we also performed genetic association analysis under a second model (Model 2) including all covariates from Model 1, with additional adjustment for HDL cholesterol (HDL-C), LDL cholesterol (LDL-C), triglycerides, body mass index (BMI), fasting glucose, and hypertension status. To account for multiple comparisons in each of the stratified analyses, we used a strict Bonferroni correction for the number of SNPs remaining in analysis after filter on MAF > 0.01, imputation quality > 0.5, and HWE P-value ≥ 10−5 in each racial/ethnic group.
Following stratified analyses, we performed meta-analysis to combine results across all four racial/ethnic groups. We performed fixed effect meta-analysis to combine estimated effects and standard errors from stratified analyses, as implemented in METAL22. To account for multiple comparisons, we used a strict Bonferroni correction for each phenotype based on the total number of SNPs included in meta-analysis of that trait.
In addition to the main analyses, we further performed sex-stratified analysis. The sex-stratified association analysis followed the basic procedure used for our main analyses, beginning with sex-specific stratified analyses for each racial/ethnic group, and combining the sex-specific results by meta-analysis. As for pooled analysis, we performed sex-stratified analyses using two statistical models where Model 1 included the basic covariates age, study site, and principal components of ancestry, while Model 2 included all covariates from Model 1, with additional adjustment for HDL-C, LDL-C, triglycerides, BMI, fasting glucose, and hypertension status.
To assess genetic heterogeneity seen in stratified analysis of the four MESA racial/ethnic groups, we performed a test of heterogeneity using Cochran’s Q and also examined the inconsistency metric I2 which quantifies the proportion of total variation across studies due to heterogeneity rather than chance23. We also used Cochran’s Q to examine heterogeneity in estimated genetic effects by sex.
Linkage disequilibrium in the SCARB1 gene region
To examine LD in the SCARB1 gene region, we made use of all genotyped SNPs on the Affy 6.0 array available through MESA SHARe (see Selection of SNPs for genetic association analysis above). For each of the four MESA racial/ethnic groups, we produced a graphical display of LD among these genotyped SNPs using the software Haploview24 (Supplementary Figures 1–4), and LD is reported using r2 for each pair of SNPs.
Results
Characteristics of the study sample
Baseline characteristics, atherosclerosis risk factors, and primary phenotypes for our investigation, including measures of SCA (CCIMT, ICIMT, and CAC [present / absent]) and counts of clinical events (cardiovascular disease [CVD] – All, CVD – Hard, and myocardial infarction [MI]), are summarized separately for each racial/ethnic group in Table 1. Sample sizes reflect the inclusion of individuals from the original MESA cohort, combined with those from the ancillary MESA Family and MESA Air studies (described in Methods). Counts of participants with data available are reported separately for each category of phenotypic observations (participant characteristics, lipid levels, SCA, or clinical events). The relatively lower numbers of African American and Hispanic samples with data available for clinical events reflects the fact that these data were recorded for the original MESA cohort only, and are not available for individuals recruited through the MESA Family and MESA Air ancillary studies.
Table 1:
Characteristics of study population.
| Caucasian | Chinese | African American | Hispanic | |
|---|---|---|---|---|
| Participant characteristics* | ||||
| No. subjects | 2526 | 775 | 2529 | 2106 |
| Women | 1320 (52.3) | 394 (50.8) | 1419 (56.1) | 1137 (51.8) |
| Age, years | 63 [54, 71] | 62 [53, 71] | 60 [53, 68] | 60 [52, 68] |
| BMI, kg/m2 | 27.1 [24.2, 30.4] | 23.8 [21.8, 26.0] | 29.5 [26.1, 33.8] | 28.6 [25.9, 32.0] |
| Fasting glucose, mmol/L | 4.8 [4.5, 5.3] | 5.1 [4.8, 5.6] | 5.1 [4.7, 5.7] | 5.2 [4.7, 5.8] |
| Hypertension | 975 (38.6) | 292 (37.8) | 1519(60.1) | 870 (41.3) |
| Lipid levels* | ||||
| No. subjects | 2520 | 772 | 2518 | 2098 |
| HDL-C, mmol/L | 1.3 [1.1, 1.6] | 1.2 [1.0, 1.5] | 1.3 [1.1, 1.6] | 1.2 [1.0, 1.4] |
| LDL-C, mmol/L | 3.0 [2.5, 3.5] | 3.0 [2.5, 3.4] | 3.0 [2.5, 3.5] | 3.1 [2.5, 3.6] |
| Total cholesterol, mmol/L | 5.0 [4.5, 5.6] | 4.9 [4.4, 5.4] | 4.9 [4.3, 5.5] | 5.1 [4.5, 5.7] |
| Triglycerides, mmol/L | 1.3 [0.9, 1,9] | 1.4 [1.0, 2.0] | 1.0 [0.7, 1.4] | 1.5 [1.1, 2.2] |
| Subclinical atherosclerosis | ||||
| No. subjects | 2526 | 773 | 2529 | 2099 |
| Common IMT, mm | 0.84 [0.73, 0.97] | 0.81 [0.71, 0.92] | 0.86 [0.75, 0.99] | 0.81 [0.71, 0.93] |
| Internal IMT, mm | 0.89 [0.71, 1.38] | 0.73 [0.60, 0.94] | 0.91 [0.70, 1.30] | 0.84 [0.68, 1.19] |
| CAC present / absent | 1433 (56.7) | 392 (50.7) | 1085 (42.9) | 962 (45.8) |
| Clinical events | ||||
| No. subjects | 2523 | 773 | 1610 | 1445 |
| Cardiovascular disease - All | 173 (6.9) | 33 (4.3) | 94 (5.8) | 92 (6.4) |
| Cardiovascular disease - Hard | 115 (4.6) | 19 (2.5) | 64 (4.0) | 68 (4.7) |
| Myocardial Infarction | 63 (2.5) | 10 (1.3) | 22 (1.4) | 36 (2.5) |
Data are presented as n (%) for binary measures or median [IQR] for continuous measure.
Summary statistics are reported for the subset of individuals with data available for at least one of the subclinical atherosclerosis measurements or clinical events.
In examining baseline characteristics across MESA racial/ethnic groups, we observed lower BMI among Chinese participants (median 23.8 kg/m2) compared to Caucasian (median 27.1 kg/m2), African American (median 29.5 kg/m2), and Hispanic participants (median 28.6 kg/m2). We further observed a higher rate of hypertension among African Americans participants (60.1%) compared to Caucasian (38.6%), Chinese (37.8%), and Hispanic participants (41.3%).
Rates of clinical events (CVD – All, CVD – Hard and MI) were higher among Caucasians (6.9%, 4.6% and 2.5%, respectively) and Hispanics (6.4%, 4.7%, and 2.5%, respectively) compared to African Americans (5.8%, 4.0%, and 1.4%, respectively) and Chinese (4.3%, 2.5%, and 1.3%, respectively). Within each of the MESA racial/ethnic groups, we also observed rates of clinical events consistently higher in males than females. For example, rates of CVD – Hard in MESA Caucasian, Chinese, African American, and Hispanic male participants were 5.2%, 2.9%, 4.7%, and 6.3%, respectively, compared to 3.9%, 2.0%, 3.3%, and 3.2%, respectively, in females. Rates of MI across the four MESA racial/ethnic groups followed the same trend with respect to sex (3.3%, 1.6%, 2.3%, and 3.7%, respectively, in males versus 1.7%, 1.0%, 0.006%, and 1.3%, respectively, in females).
Genetic association of Common Carotid IMT with SNP rs10846744
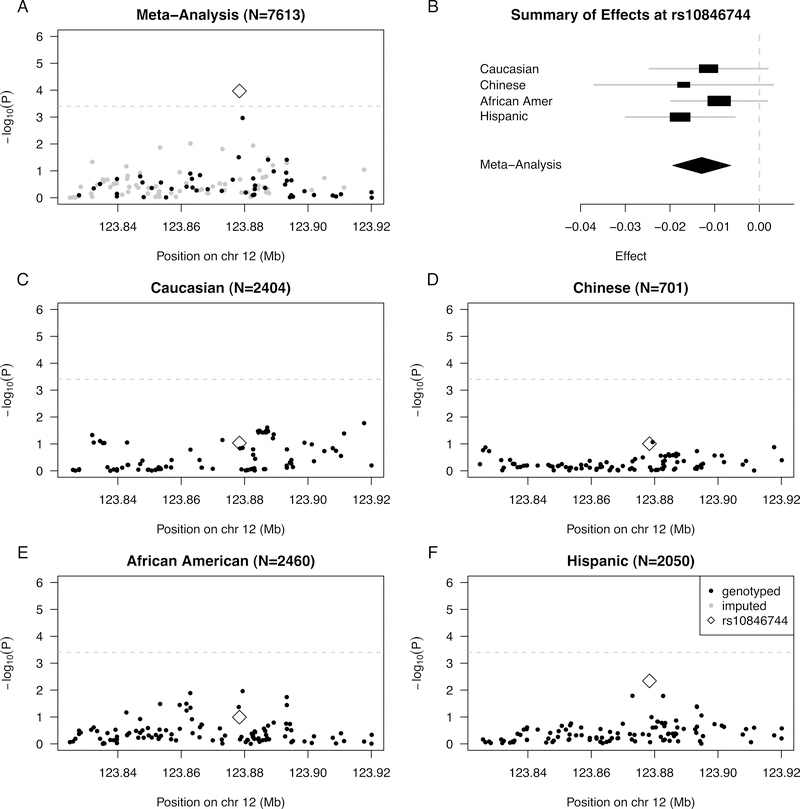
SNP rs10846744 was previously reported as the SCARB1 SNP most statistically significantly associated with CCIMT in our previous analysis of a subset of individuals from MESA9. In the current analysis, rs10846744 was again statistically significantly associated with log CCIMT in meta-analysis (P=1.07E-04), including adjustment for age, sex, study site, and PCs of ancestry within each ethnic group (Model 1), even after strict Bonferroni correction for the number of SNPs under consideration (α*=0.05/126= 0.0004) (Table 2, Figure 1A). The strength of association of rs10846744 with log CCIMT decreased only slightly in meta-analysis (P=5.61E-04) after additional adjustment for known risk factors of atherosclerosis (Model 2). In stratified analysis, this association was strongest in Hispanics (nominal p=0.005).
Table 2:
Linear regression summary statistics for the association of SCARB1 SNP rs10846744 and log CCIMT by MESA racial/ethnic group and in meta-analysis, under multiple models of adjustment.
| Model 1 | Model 2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Group | Allele frequency | N | Beta | SE | P-value | N | Beta | SE | P-value |
| Caucasian | 0.82 | 2405 | −0.011 | 0.007 | 0.092 | 2367 | −0.010 | 0.007 | 0.115 |
| Chinese | 0.43 | 701 | −0.017 | 0.010 | 0.098 | 687 | −0.019 | 0.010 | 0.056 |
| African American | 0.38 | 2457 | −0.009 | 0.005 | 0.102 | 2425 | −0.007 | 0.005 | 0.206 |
| Hispanic | 0.65 | 2050 | −0.018 | 0.006 | 0.005 | 2001 | −0.015 | 0.006 | 0.016 |
| Meta-analysis | −0.013 | 0.003 | 1.07E-04 | −0.011 | 0.003 | 5.61E-04 | |||
Allele frequencies and estimated effects are reported for the effect allele G (versus the reference allele C). Model 1 includes adjustment for age, sex, study site, and principal components of ancestry. Model 2 includes all covariates in Model 1, with the addition of HDL-C, LDL-C, triglycerides, BMI, fasting glucose, and hypertension status. Estimated effect sizes are presented based on an additive genetic model with 1 degree of freedom, for the log-transformed phenotype common IMT (mm).
Figure 1:
Summary of regional association for SNPs in the SCARB1 gene region with common IMT (modeled on a log scale): (A) Strength of association versus SNP position on chromosome 12 based on meta-analysis to combine results from the four MESA racial/ethnic groups; (B) Forest plot of effects (with 95% CIs) reported in stratified analysis of the four MESA racial/ethnic groups; and Strength of association versus SNP position on chromosome 12 based on stratified analysis of (C) Caucasians, (D) Chinese, (E) African Americans, and (F) Hispanic. In plots (A) and (C-F), genotyped SNPs are indicated as solid black dots, imputed SNPs as solid gray dots, the genotyped SNP rs10846744 as an open black diamond, and horizontal dashed gray lines indicate a conservative Bonferroni-threshold for statistical significance based on multiple testing of 126 SNPs.
We examined evidence for heterogeneity in genetic effects of rs10846744 on log CCIMT across the four MESA racial/ethnic groups. The rs10846744 allele G was associated with lower levels of CCIMT in all four racial/ethnic groups (Table 2, Figure 1B), with little evidence for heterogeneity performed in METAL23 under the base model (Model 1, heterogeneity P=0.727, heterogeneity I2=0).
Many of the individuals included in our current MESA sample were also included in the MESA Candidate Gene cohort (the original report of association of rs10846744 with CCIMT9). We therefore examined the association of rs10846744 with CCIMT in the subset of individuals (n=4,994) who were not part of the original MESA Candidate Gene cohort (Supplementary Table I). A stratified analysis was conducted for each racial/ethnic group, as well as meta-analysis combining all racial/ethnic groups, using the same regression models applied for association analysis of the complete MESA cohort (Table 2). For each of the four racial/ethnic groups, the estimated effect for the G allele of rs10846744 was in the same direction as reported in our analysis of the MESA Candidate Gene cohort9. In meta-analysis, the direction of effect was also in agreement with our previous report, and the overall association under Model 1 was nominally significant for the single candidate SNP (P=0.034).
Genetic association analysis of subclinical atherosclerosis traits for SCARB1 SNPs
We conducted analysis of 126 genotyped or imputed SCARB1 SNPs in MESA SHARe for CAC (presence/absence), CCIMT, and ICIMT. Given the strength of association for rs10846744 with CCIMT, we first determined whether other SCARB1 SNPs demonstrated stronger association with CCIMT, and whether the association profile varied by racial/ethnic group (Figure 1). Our results support rs10846744 as providing the strongest evidence for association in meta-analysis of common carotid IMT. No other SCARB1 SNPs reached the Bonferroni threshold for common carotid IMT, either in meta-analysis or in analyses stratified by racial/ethnic group (Supplementary Tables II–VI, available at http://people.virginia.edu/~am3xa/data/). We performed analyses stratified by sex for each of the four racial/ethnic groups, as well as combined by meta-analysis. We did not observe any additional SNPs exhibiting statistically significant association with CCIMT after multiple testing correction.
We further examined association of SCARB1 SNPs with CAC and ICIMT, in analyses stratified by racial/ethnic group (Supplementary Tables 2–5), and by meta-analysis to combine the results of these stratified analyses (Supplementary Table 6). We did not observe any statistically significant results after correction for multiple testing. We also failed to observe a significant association of additional SCARB1 SNPs in sex-stratified analyses.
Genetic association of incident MI and CVD events with rs10846744
As a prospective cohort study designed to investigate the prevalence and progression of subclinical CVD, individuals with previous evidence of CVD were excluded at the time of MESA enrollment beginning in July 200013. As a result, the number of clinical CVD events reported for the MESA Candidate Gene cohort was too small to warrant formal analysis of incident CVD at that time9. In the current analyses, there is a larger number of genotyped individuals with longer follow-up time (increased number of incident CVD events) to permit examination of the association of rs10846744 with incident CVD. In meta-analysis of all individuals, there were no statistically significant associations with any clinical cardiovascular disease events (Table 3). In stratified analysis by sex using the baseline model (Model 1), there were statistically significant associations in males with MI (β=−0.410, SE=0.180, P=0.023) and CVD – Hard (β=−0.354, SE=0.138, P=0.010). These results correspond to odds ratios for MI of 0.664 (95% CI 0.466 – 0.944) and for CVD – Hard of 0.702 (95% CI 0.536 – 0.920) per copy of the effect allele G versus the reference allele C. Results were very similar under the model with additional adjustment for known risk factors of atherosclerosis (Model 2), with slightly stronger statistical significance and estimated effect sizes. There were no nominally significant results for females. We did not observe statistically significant heterogeneity in genetic effects of rs10846744 in males as compared to females for incident MI (Model1, heterogeneity P=0.166) or for CVD – Hard (Model 1, heterogeneity P=0.067).
Table 3:
Summary statistics from meta-analysis to combine all four MESA racial/ethnic groups in association analysis of the SCARB1 SNP rs10846744 and three clinical outcomes of interest (MI, CVD – Hard, and CVD – All) under multiple models of adjustment. Results are presented for pooled analysis of males and females, as well as sex-stratified analysis.
| Model 1 | Model 2 | ||||||
|---|---|---|---|---|---|---|---|
| Trait | Analysis | Beta | SE | P-value | Beta | SE | P-value |
| Myocardial infarction | All | −0.247 | 0.153 | 0.106 | −0.227 | 0.156 | 0.147 |
| Sex-stratified: Male only | −0.408 | 0.180 | 0.024 | −0.420 | 0.185 | 0.023 | |
| Sex-stratified: Female only | 0.087 | 0.319 | 0.785 | 0.107 | 0.350 | 0.759 | |
| CVD - Hard | All | −0.164 | 0.106 | 0.123 | −0.125 | 0.110 | 0.256 |
| Sex-stratified: Male only | −0.358 | 0.138 | 0.010 | −0.381 | 0.145 | 0.009 | |
| Sex-stratified: Female only | 0.052 | 0.174 | 0.764 | 0.110 | 0.187 | 0.557 | |
| CVD - All | All | −0.119 | 0.089 | 0.179 | −0.080 | 0.092 | 0.384 |
| Sex-stratified: Male only | −0.206 | 0.113 | 0.068 | −0.187 | 0.118 | 0.114 | |
| Sex-stratified: Female only | −0.014 | 0.148 | 0.924 | 0.028 | 0.158 | 0.858 | |
Estimated effects are presented for an additive genetic association model on rs10846744, coded as the number of copies of the effect allele G (versus the reference allele C).
Model 1 includes adjustment for age, sex, study site, and principal components of ancestry for pooled analysis of males and females; sex is not included as a covariate in sex-stratified analyses. Model 2 includes all covariates in Model 1, with the addition of HDL-C, LDL-C, triglycerides, BMI, fasting glucose, and hypertension status.
The estimated effect of rs10846744 on the risk of MI is stronger in African American males (β=−0.664, SE=0.442), Chinese males (β=−0.618, SE=0.726) and Caucasian males (β=−0.555, SE=0.266) compared to Hispanic males (β=0.011, SE=0.330) (Table 4). The estimated effect of rs10846744 on CVD – Hard was strongest in African American (β=−0.509, SE=0.284) and Caucasian males (β=−0.422, SE=0.223) compared to Chinese (β=−0.193, SE=0.511) and Hispanic males (β=−0.187, SE=0.250).
Table 4:
Racial/ethnic-specific summary statistics for the association of SCARB1 SNP rs10846744 and two clinical outcomes of interest (MI and CVD – Hard) in stratified analysis of males only, under multiple models of adjustment.
| Model 1 | Model 2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Trait | Group | N | Beta | SE | P-value | N | Beta | SE | P-value |
| Myocardial infarction | Caucasian | 1172 | −0.555 | 0.266 | 0.037 | 1152 | −0.577 | 0.270 | 0.033 |
| Chinese | 350 | −0.618 | 0.726 | 0.394 | 342 | −0.773 | 0.860 | 0.369 | |
| African American | 732 | −0.664 | 0.422 | 0.116 | 723 | −0.618 | 0.427 | 0.148 | |
| Hispanic | 641 | 0.011 | 0.330 | 0.973 | 626 | 0.012 | 0.341 | 0.971 | |
| CVD - Hard | Caucasian | 1172 | −0.422 | 0.223 | 0.058 | 1152 | −0.449 | 0.232 | 0.053 |
| Chinese | 350 | −0.193 | 0.511 | 0.706 | 342 | −0.360 | 0.621 | 0.562 | |
| African American | 732 | −0.509 | 0.284 | 0.073 | 723 | −0.513 | 0.292 | 0.078 | |
| Hispanic | 641 | −0.187 | 0.250 | 0.455 | 626 | −0.189 | 0.264 | 0.474 | |
Estimated effects are presented for an additive genetic association model on rs10846744, coded as the number of copies of the effect allele G (versus the reference allele C).
Model 1 includes adjustment for age, study site, and principal components of ancestry. Model 2 includes all covariates in Model 1, with the addition of HDL-C, LDL-C, triglycerides, BMI, fasting glucose, and hypertension status.
We further investigated the full set of 126 SNPs in the SCARB1 region to determine whether any SNPs showed strong association with incident CVD in meta-analysis across all four MESA racial/ethnic groups (Supplementary Table 6). We did not identify SNPs surpassing the Bonferroni threshold for statistical significance (α*=0.05/126= 0.0004) in meta-analysis of all individuals or in sex-stratified analyses.
Replication through the Myocardial Infarction Genetics Consortium
After observing statistically significant association of rs10846744 with incident MI and CVD in MESA, we sought in silico replication through the Myocardial Infarction Genetics (MIGen) Consortium12. The MIGen consortium recently completed genome-wide association study of 2,967 cases of early-onset MI (in men ≤ 50 years old or women ≤ 60 years old) and 3,075 age- and sex- matched controls free of MI, all of European ancestry, with genetic association results for ~2.5 million directly genotyped and imputed SNPs. From these results, we examined association of the SCARB1 SNP rs10846744 with early-onset MI. We observed the G allele of rs10846744 was associated with reduced risk of premature MI (odds ratio = 0.910; 95% CI 0.833 – 0.993, P=0.043).
Discussion
We previously performed an association study of 43 SCARB1 SNPs with measures of SCA in 2,757 individuals from the MESA cohort9, detecting a significant association of SCARB1 variation with CCIMT. Here, we extend our previous work by: 1) using an expanded set of 7,936 phenotyped individuals from the MESA study with genotypes available for 126 SCARB1 (genotyped and imputed) SNPs through MESA SHARe, and 2) examining an extended set of phenotypes including incident MI and CVD, in addition to the subclinical measures of atherosclerosis examined previously. Using the larger set of individuals now available to us, we confirmed that rs10846744 exhibits the strongest association with CCIMT, validating our report of association for the same SNP in our previous work9. This finding is independent of well-known epidemiologic risk factors for atherosclerosis, including lipids (TC, LDL, TG and HDL-C), BMI, fasting glucose, and hypertension status.
We further report a novel association of rs10846744 with incident MI and CVD in males from the MESA cohort. Subsequent in silico replication of the incident MI association through the MIGen consortium validated the associations observed in MESA, and demonstrated the association of rs10846744 with MI can be seen in studies with mean age of MI ranging from 41 years (in MIGen) to > 60 years in MESA. We note the estimated effect for the SCARB1 rs10846744 SNP seen in MIGen is notably lower than that observed in males from the MESA cohort; however, the MIGen result is based on a combined sample of males and females. Observed differences in estimated effect size for MIGen versus MESA may also reflect differences in the two study samples, with the MIGen focused on early MI, with mean age at the time of MI being 41 years for men and 47 years for women12, while MESA participants were a relatively older cohort, with mean baseline age over 60 years in all racial/ethnic groups (Table 1) and no pre-existing MI or CVD at the time of recruitment.
We now report that SCARB1 SNP rs10846744 is the most significant SNP associated with CCIMT and incident MI and CVD (in males) even after examining an expanded set of 126 SNPs available to us through dense genome-wide genotyping, as well as imputation of additional SNPs based on HapMap Phase I and II data. Our results are consistent with those recently reported by Grallert et al.25, whereby these investigators found a significant association of rs10846744 with prevalent CHD/CAD in the CARDIoGRAM consortium. Our results, and those of Grallert et al.25, offer strong evidence that rs10846744 is either in strong LD with the causal variant (Supplementary Figures 1–4), or is itself a causal variant underlying these important associations.
The rs10846744 SNP is located within intron 1 of the SCARB1 gene (Chromosome 12, position 125312425, dbSNP). This SNP is not located at a traditional alternative splice site. Results from our (A Rodriguez, unpublished) laboratory confirm the absence of alternative mRNA transcripts in carriers of the G allele. A search of publically available microRNA databases also failed to identify this SNP as either a seed or target of microRNA regulation (A Rodriguez, unpublished). A bioinformatics screen using UCSC Genome Bioinformatics website (http://genome.ucsc.edu) revealed that the region contains DNase I hypersensitivity clusters and enhancer-promoter histone markers, suggestive of a region that might exert cis or trans regulatory effects.
A mechanism by which rs10846744 could affect atherosclerosis is by acting as an enhancer within intron 1. As an enhancer it could possibly affect expression of SR-BI or alter expression of a distant gene. No correlation between rs10846744 genotype and levels of SR-BI RNA or protein in macrophages isolated from subjects with hyperalphalipoproteinemia has been identified (A Rodriguez, unpublished), suggesting that this SNP is not directly regulating activity of the SCARB1 promoter. Based on the work by Suchindran et al.26 it might be possible that rs10846744 is an enhancer that exerts a distal effect on expression of Lp-PLA2. These investigators examined genetic determinants that were significantly associated with either Lp-PLA2 activity or mass, and identified rs10846744 as being negatively correlated with Lp-PLA2 activity and mass. As we observed here and previously9 for CCIMT, the effect of rs10846744 on Lp-PLA2 activity and mass is independent of lipids, including HDL-C levels. Thus, while SR-BI exerts an independent effect on HDL-C levels in humans27, the effect of the rs10846744 variant on SCA and incident CVD is likely mediated via non-lipid pathways and through other SCARB1 dependent processes, such as affecting endothelial function28 or inflammatory pathways26.
The risk of premature death due to CAD has been shown in SR-BI null mice on an apoE KO background29. While there are now a number of epidemiological studies showing a significant association of SCARB1 variants with lipid levels30–32, only the recent publication by Grallert et al.25 and our current study have shown an association between rs10846744 variants and clinical CVD. We previously showed a significant association between rs10846744 and SCA in a smaller cohort of MESA participants9. We have now replicated this earlier study, again observing that rs10846744 is significantly associated with lower CCIMT in carriers of the G allele.
In conclusion, SCARB1 exerts an important influence on cardiovascular health in humans. The mechanism by which the intronic rs10846744 SCARB1 SNP affects carotid intimal medial thickness and clinical cardiovascular events is not yet established and warrants further study.
Supplementary Material
Acknowledgments
A. Sources of funding: MESA and the MESA SHARe project are conducted and supported by contracts N01-HC-95159 through N01-HC-95169 and RR-024156 from the National Heart, Lung, and Blood Institute (NHLBI). Funding for MESA SHARe genotyping was provided by NHLBI Contract N02‐HL‐6‐4278. MESA Family is conducted and supported in collaboration with MESA investigators; support is provided by grants and contracts R01HL071051, R01HL071205, R01HL071250, R01HL071251, R01HL071252, R01HL071258, R01HL071259. Dr. Rodriguez is funded by a NIH grant, HL075646. The authors thank the participants of the MESA study, the Coordinating Center, MESA investigators, and study staff for their valuable contributions. A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org.
Footnotes
B. None of the authors report a financial conflict.
Disclosures: None.
References
- 1.Acton S, Rigotti A, Landschulz KT, Xu S, Hobbs HH, Krieger M. Identification of scavenger receptor sr-bi as a high density lipoprotein receptor. Science. 1996;271:518–520 [DOI] [PubMed] [Google Scholar]
- 2.Edmondson AC, Braund PS, Stylianou IM, Khera AV, Nelson CP, Wolfe ML, Derohannessian SL, Keating BJ, Qu L, He J, Tobin MD, Tomaszewski M, Baumert J, Klopp N, Doring A, Thorand B, Li M, Reilly MP, Koenig W, Samani NJ, Rader DJ. Dense genotyping of candidate gene loci identifies variants associated with high-density lipoprotein cholesterol. Circ Cardiovasc Genet. 2011;4:145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts CG, Shen H, Mitchell BD, Damcott CM, Shuldiner AR, Rodriguez A. Variants in scavenger receptor class b type i gene are associated with hdl cholesterol levels in younger women. Hum Hered. 2007;64:107–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee JY, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RY, Wright AF, Witteman JC, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJ, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BW, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O’Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PK, Lucas G, Luben R, Loos RJ, Lokki ML, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, Konig IR, Khaw KT, Kaprio J, Kaplan LM, Johansson A, Jarvelin MR, Janssens AC, Ingelsson E, Igl W, Kees Hovingh G, Hottenga JJ, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Doring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJ, de Faire U, Crawford G, Collins FS, Chen YD, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Adair LS, Taylor HA Jr.,Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrade FM, Fiegenbaum M, Almeida S, Hutz MH. [influence of genetic combinations on hdl-c levels in a southern brazilian population]. Arq Bras Cardiol. 2010;95:430–435 [DOI] [PubMed] [Google Scholar]
- 6.Acton S, Osgood D, Donoghue M, Corella D, Pocovi M, Cenarro A, Mozas P, Keilty J, Squazzo S, Woolf EA, Ordovas JM. Association of polymorphisms at the sr-bi gene locus with plasma lipid levels and body mass index in a white population. Arterioscler Thromb Vasc Biol. 1999;19:1734–1743 [DOI] [PubMed] [Google Scholar]
- 7.Richard E, von Muhlen D, Barrett-Connor E, Alcaraz J, Davis R, McCarthy JJ. Modification of the effects of estrogen therapy on hdl cholesterol levels by polymorphisms of the hdl-c receptor, sr-bi: The rancho bernardo study. Atherosclerosis. 2005;180:255–262 [DOI] [PubMed] [Google Scholar]
- 8.Trigatti BL, Krieger M, Rigotti A. Influence of the hdl receptor sr-bi on lipoprotein metabolism and atherosclerosis. Arterioscler Thromb Vasc Biol. 2003;23:1732–1738 [DOI] [PubMed] [Google Scholar]
- 9.Naj AC, West M, Rich SS, Post W, Kao WH, Wasserman BA, Herrington DM, Rodriguez A. Association of scavenger receptor class b type i polymorphisms with subclinical atherosclerosis: The multi-ethnic study of atherosclerosis. Circ Cardiovasc Genet. 2010;3:47–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox CS, Cupples LA, Chazaro I, Polak JF, Wolf PA, D’Agostino RB, Ordovas JM, O’Donnell CJ. Genomewide linkage analysis for internal carotid artery intimal medial thickness: Evidence for linkage to chromosome 12. Am J Hum Genet. 2004;74:253–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez-Esparragon F, Rodriguez-Perez JC, Hernandez-Trujillo Y, Macias-Reyes A, Medina A, Caballero A, Ferrario CM. Allelic variants of the human scavenger receptor class b type 1 and paraoxonase 1 on coronary heart disease: Genotype-phenotype correlations. Arterioscler Thromb Vasc Biol. 2005;25:854–860 [DOI] [PubMed] [Google Scholar]
- 12.Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, Mannucci PM, Anand S, Engert JC, Samani NJ, Schunkert H, Erdmann J, Reilly MP, Rader DJ, Morgan T, Spertus JA, Stoll M, Girelli D, McKeown PP, Patterson CC, Siscovick DS, O’Donnell CJ, Elosua R, Peltonen L, Salomaa V, Schwartz SM, Melander O, Altshuler D, Merlini PA, Berzuini C, Bernardinelli L, Peyvandi F, Tubaro M, Celli P, Ferrario M, Fetiveau R, Marziliano N, Casari G, Galli M, Ribichini F, Rossi M, Bernardi F, Zonzin P, Piazza A, Yee J, Friedlander Y, Marrugat J, Lucas G, Subirana I, Sala J, Ramos R, Meigs JB, Williams G, Nathan DM, MacRae CA, Havulinna AS, Berglund G, Hirschhorn JN, Asselta R, Duga S, Spreafico M, Daly MJ, Nemesh J, Korn JM, McCarroll SA, Surti A, Guiducci C, Gianniny L, Mirel D, Parkin M, Burtt N, Gabriel SB, Thompson JR, Braund PS, Wright BJ, Balmforth AJ, Ball SG, Hall AS, Linsel-Nitschke P, Lieb W, Ziegler A, Konig I, Hengstenberg C, Fischer M, Stark K, Grosshennig A, Preuss M, Wichmann HE, Schreiber S, Ouwehand W, Deloukas P, Scholz M, Cambien F, Li M, Chen Z, Wilensky R, Matthai W, Qasim A, Hakonarson HH, Devaney J, Burnett MS, Pichard AD, Kent KM, Satler L, Lindsay JM, Waksman R, Knouff CW, Waterworth DM, Walker MC, Mooser V, Epstein SE, Scheffold T, Berger K, Huge A, Martinelli N, Olivieri O, Corrocher R, McKeown P, Erdmann E, Konig IR, Holm H, Thorleifsson G, Thorsteinsdottir U, Stefansson K, Do R, Xie C, Siscovick D. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41:334–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bild DE, Bluemke DA, Burke GL, Detrano R, Diez Roux AV, Folsom AR, Greenland P, Jacob DR Jr., Kronmal R, Liu K, Nelson JC, O’Leary D, Saad MF, Shea S, Szklo M, Tracy RP. Multi-ethnic study of atherosclerosis: Objectives and design. Am J Epidemiol. 2002;156:871–881 [DOI] [PubMed] [Google Scholar]
- 14.Price AL, Weale ME, Patterson N, Myers SR, Need AC, Shianna KV, Ge D, Rotter JI, Torres E, Taylor KD, Goldstein DB, Reich D. Long-range ld can confound genome scans in admixed populations. Am J Hum Genet. 2008;83:132–135; author reply 135–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. Plink: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909 [DOI] [PubMed] [Google Scholar]
- 18.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. Mach: Using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeboah J, Folsom AR, Burke GL, Johnson C, Polak JF, Post W, Lima JA, Crouse JR, Herrington DM. Predictive value of brachial flow-mediated dilation for incident cardiovascular events in a population-based study: The multi-ethnic study of atherosclerosis. Circulation. 2009;120:502–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; 2010 [Google Scholar]
- 21.Chen MH, Yang Q. Gwaf: An r package for genome-wide association analyses with family data. Bioinformatics. 2010;26:580–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willer CJ, Li Y, Abecasis GR. Metal: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of ld and haplotype maps. Bioinformatics. 2005;21:263–265 [DOI] [PubMed] [Google Scholar]
- 25.Grallert H, Dupuis J, Bis JC, Dehghan A, Barbalic M, Baumert J, Lu C, Smith NL, Uitterlinden AG, Roberts R, Khuseyinova N, Schnabel RB, Rice KM, Rivadeneira F, Hoogeveen RC, Fontes JD, Meisinger C, Keaney JF Jr.,Lemaitre R, Aulchenko YS, Vasan RS, Ellis S, Hazen SL, van Duijn CM, Nelson JJ, Marz W, Schunkert H, McPherson RM, Stirnadel-Farrant HA, Psaty BM, Gieger C, Siscovick D, Hofman A, Illig T, Cushman M, Yamamoto JF, Rotter JI, Larson MG, Stewart AF, Boerwinkle E, Witteman JC, Tracy RP, Koenig W, Benjamin EJ, Ballantyne CM. Eight genetic loci associated with variation in lipoprotein-associated phospholipase a2 mass and activity and coronary heart disease: Meta-analysis of genome-wide association studies from five community-based studies. Eur Heart J. 2012;33:238–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suchindran S, Rivedal D, Guyton JR, Milledge T, Gao X, Benjamin A, Rowell J, Ginsburg GS, McCarthy JJ. Genome-wide association study of Lp-PLA(2) activity and mass in the Framingham Heart Study. PLoS Genet 2010;6:e1000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.West M, Greason E, Kolmakova A, Jahangiri A, Asztalos B, Pollin TI, Rodriguez A. Scavenger receptor class B type I protein as an independent predictor of HDL cholesterol levels in subjects with hyperalphalipoproteinemia. J Clin Endocrinol Metab 2009;94:1451–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mineo C, Shaul PW. Role of high-density lipoprotein and scavenger receptor B type I in the promotion of endothelial repair. Trends Cardiovasc Med 2007;17:156–161 [DOI] [PubMed] [Google Scholar]
- 29.Zhang S, Picard MH, Vasile E, Zhu Y, Raffai RL, Weisgraber KH, Krieger M. Diet-induced occlusive coronary atherosclerosis, myocardial infarction, cardiac dysfunction, and premature death in scavenger receptor class B type I-deficient, hypomorphic apolipoprotein ER61 mice. Circulation 2005;111:3457–3464 [DOI] [PubMed] [Google Scholar]
- 30.McCarthy JJ, Lewitzky S, Reeves C, Permutt A, Glaser B, Groop LC, Lehner T, Meyer JM. Polymorphisms of the HDL receptor gene associated with HDL cholesterol levels in diabetic kindred from three populations. Hum Hered. 2003;55:163–170 [DOI] [PubMed] [Google Scholar]
- 31.McCarthy JJ, Lehner T, Reeves C, Moliterno DJ, Newby LK, Rogers WJ, Topol EJ; Genequest investigators. Association of genetic variants in the HDL receptor, SR-BI, with abnormal lipids in women with coronary artery disease. J Med Genet. 2003;40:453–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osgood D, Corella D, Demissie S, Cupples LA, Wilson PW, Meigs JB, Schaefer EJ, Coltell O, Ordovas JM. Genetic variation at the scavenger receptor class B type I gene locus determines plasma lipoprotein concentrations and particle size and interacts with type 2 diabetes: the Framingham Study. J Clin Endocrinol Metab 2003;88:2869–2879 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.