Abstract
Reports regarding the frequency of SMAD4 loss in human head and neck squamous cell carcinoma (HNSCC) vary significantly. We have shown that SMAD4 deletion contributes to HNSCC initiation and progression. Therefore, accurately detecting genetic SMAD4 loss is critical to determine prognosis and therapeutic interventions in personalized medicine. We developed a SMAD4 fluorescence in situ hybridization (FISH) assay to identify chromosomal SMAD4 loss at the single cell level of primary HNSCC specimens and patient derived xenograft (PDX) tumors derived from HNSCCs. SMAD4 heterozygous loss was detected in 35% of primary HNSCCs and 41.3% of PDX tumors. Additionally, 4.3% of PDX tumors had SMAD4 homozygous loss. These frequencies of SMAD4 loss were similar to those in The Cancer Genome Atlas (TCGA). However, we identified significant heterogeneities of SMAD4 loss (partial or complete) among cells within each tumor. We also found that aneuploidy (monosomy and polysomy) contributed greatly to how to define chromosomal SMAD4 deletion. Furthermore, in cultured PDX tumors, SMAD4 mutant cells outcompeted SMAD4 wildtype cells, resulting in establishing homogenous SMAD4 mutant HNSCC cell lines with partial or complete genomic SMAD4 loss, suggesting a survival advantage of SMAD4 mutant cells. Taken together, our study reveals inter- and intra-tumor heterogeneities of SMAD4 chromosomal loss in HNSCCs. Further, SMAD4 FISH assay provides a platform for future clinical diagnosis of SMAD4 chromosomal loss that potentially serves as a molecular marker for prognosis and therapeutic intervention in cancer patients.
Keywords: Aneuploidy, Chromosomal SMAD4 deletion, genomic instability
1 ∣. INTRODUCTION
Among all cancers worldwide, head and neck cancer is the sixth most common cancer type.1,2 In the head and neck region, squamous cell carcinoma (HNSCC) represents over 90% of all head and neck cancers.3 It is estimated that over 75% of HNSCCs are associated with chronic tobacco and alcohol use worldwide.4 Human papillomavirus (HPV) associated HNSCC accounts for approximately 30-70% of all cases in developed countries.5,6 SMAD4 protein is a tumor suppressor that regulates proliferation, apoptosis, and genomic stability.7-9 SMAD4 genetic loss was initially identified in 30-50% of pancreatic cancers10-12 and colon cancers13-15; SMAD4 loss does not play a role in initiation but promotes metastasis in pancreatic11,16 and colon cancers.17 In contrast, SMAD4 loss is detected in early stage human HNSCC and initiates HNSCC development in mice.8 We have shown that SMAD4 null HNSCCs have extensive genomic instability, hyperproliferation, little apoptosis, and TGFβ- and SMAD3-dependent inflammation.8 Given the significant functional impact of SMAD4 loss in HNSCC development, it is important to accurately determine the true frequency of SMAD4 loss in human HNSCC. Reported rates of SMAD4 loss in HNSCCs vary significantly. SMAD4 point mutations occur in less than 5% of cases18 and SMAD4 loss rates by IHC staining in HNSCC varies from 12% up to 67.8% SMAD4 protein loss19-21; however, antibody specificity remains unclear. In contrast, SMAD4 loss of heterozygosity (LOH) is reported in ~30-50% of HNSCCs.22-24 These variations make it difficult to define if SMAD4 loss affects clinical outcome. For example, reduced SMAD4 expression in HNSCC is linked with poor clinical outcomes in one study,20 but another study did not find such an association.21 Recent TCGA data shows that, similar to what has been observed for other cancer types, large chromosome deletion of 18q is the most common cause of SMAD4 loss in HNSCC.25 Here, we present a technique alternative to sequencing for detecting chromosomal SMAD4 loss at the single cell level in HNSCCs. Our study identified significant intra- and inter-tumor heterogeneity of SMAD4 genetic loss that would be missed by genomewide sequencing of bulk tumors. Our study also provides an approach that can be developed into a Clinical Laboratory Improvement Amendments (CLIA)-certified assay to use for prognosis and personalized medicine.
2 ∣. MATERIALS AND METHODS
2.1 ∣. SMAD4 FISH
A probe set was developed including genomic sequences from SMAD4, mapped at 18q21.2 (chr 18:48,478,290-48,634,496) labeled with SpectrumRed (SR2016-10-19) (Empire Genomics, Buffalo, NY), and centromere 18 (CEN1826) labeled with SpectrumGreen. Formalin-fixed, paraffin-embedded tissue sections were processed as previously reported.27 Fluorescence microscopy with Z-stacking was used to survey the entire tumor section in three dimensions. SMAD4 and CEN18 FISH signals were counted in 50-100 nuclei from at least 4-5 tumor areas, each including 4-5 microscope fields under the 100× objective. Quantification of these signals was averaged per specimen. In cell lines, quantification was performed in interphase and metaphase cells.
2.2 ∣. SMAD4 genomic status
The ratio of SMAD4 signals to CEN18 control signals was used to determine the tumor's SMAD4 FISH status (SMAD4/CEN18) per the following criteria: gain: SMAD4/CEN18 ratio ≥ 1.2; wildtype: SMAD4/CEN18 ratio < 1.2 but ≥ 0.8; hemizygous: SMAD4/CEN18 ratio < 0.8 > 0; homozygous loss: SMAD4/CEN18 ratio = 0. These criteria were applied to primary HNSCCs (n = 20) and PDX tumors (n = 46). The prevalence of SMAD4 FISH genotypes were compared to TCGA human HNSCC provisional sequencing data (n = 496). The TCGA HNSCC dataset was queried for wildtype, gained copy (GAIN), heterozygous loss (HET-LOSS), and homozygous deletion (HOMDEL) of SMAD4 via cBioPortal. Copy numbers of SMAD4 or CEN18 were defined as follows: polysomy: ≥ 2.50; disomy: < 2.50 but ≥ 1.50; monosomy: < 1.50 but >0.50; nullisomy (complete loss): < 0.50. Copy number criteria were applied to cells and tumors. SMAD4 and CEN18 copy numbers were paired (SMAD4.CEN18) for polysomy analysis of cells and tumors.
2.3 ∣. Specimens and cell lines
Primary HNSCC and PDX tumors were from consented patients at the University of Colorado Cancer Center and approval by the Colorado Multiple Institutional Review Board. Normal karyotype lymphoblastic cells (Coriell Institute for Medical Research, Camden, NJ) and HNSCC cell lines were grown in complete DMEM. HNSCC cell lines: CAL27 and FaDu were purchased from ATCC (Manassas, VA). PDX derived cell lines were generated at the University of Colorado as previously reported.28 PDX derived cell lines were grown in RMK Primary Cell Line Media comprised of 45% DMEM (ThermoFisher Scientific, Waltham, MA) #11965-092, 45% DMEM/F12 #11330-032 (ThermoFisher Scientific), 10% FBS, hEGF [10 ng/mL], transferrin [5 μg/mL], insulin [5 μg/mL], and hydrocortisone [400 ng/mL] sterile filtered (0.22 μm). Normal Oral Keratinocytes were grown in Defined Keratinocytes–SFM 10744019 (ThermoFisher Scientific) supplemented with EGF [2 ng/mL].
2.4 ∣. qRT-PCR
At 90% confluence, cells were lifted with 0.4% TrypLE Select (ThermoFisher Scientific, #12563029). Trypsin was neutralized with RMk media, and cells were spun down at 800 rpm for 5 min at 4°C. RNeasy Plus Mini Kit (Qiagen, Hilden, Germany, # 74136) was used to extract RNA from the cells. Total RNA was used to detect SMAD4 expression with TaqMan probes (ThermoFisher Scientific) for SMAD4 (FAM: Hs00232068_m1) and GAPDH (VIC: 4326317E). The Brilliant II QRT-PCR 1-step Master Mix kit (Agilent, Santa Clara, CA, #600809) was used for each 50 ng reaction of RNA on a 96 well plate (BioRad, Hercules, CA, #2239441). Ct values were used to determine relative fold change by 2 −ΔΔCt.
2.5 ∣. Statistical analysis
Statistical analyses were carried out using Graphpad software (PRISM 7). SMAD4 and CEN18 copy number average and standard deviation were determined by quantification of 50-100 nuclei per specimen. Inter-specimen heterogeneity was inferred from the standard deviation. Mean standard error was applied for all error bars. Comparison between subpopulations of SMAD4 and CEN18 for statistical significance was carried out by one-way ANOVA. Comparison between patient derived cell lines for relative SMAD4 mRNA were carried out by student t-test. *: P < 0.05; ****: P < 0.0001.
3 ∣. RESULTS
3.1 ∣. Frequency of genomic loss of SMAD4 in human HNSCC
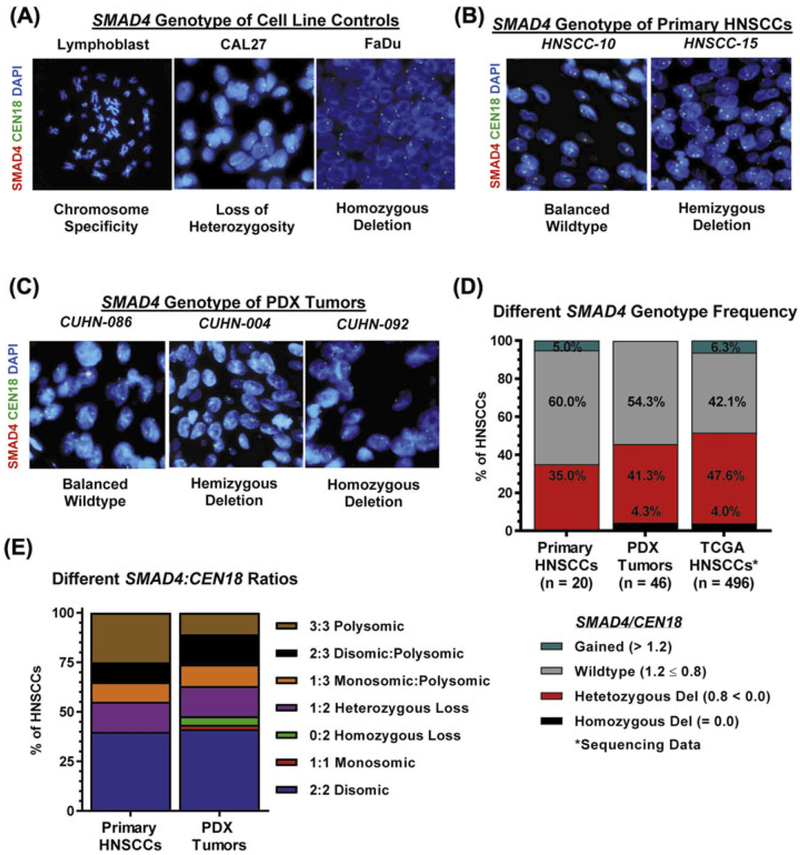
A lymphoblastic cell line with a normal karyotype was used for validation of the SMAD4 probe set (Figure 1A). We also validated the probe set using two HNSCC cell lines with known SMAD4 deletion: CAL27 that lacks one SMAD4 allele and possesses an additional nonsense point mutation in the remaining SMAD4 allele (LOH), and FaDu with homozygous SMAD4 deletion.29 The FISH assay identified one SMAD4 allele per cell in CAL27 and no SMAD4 alleles in FaDu cells (Figure 1A), indicating that the SMAD4 probe set was suitable for detecting genomic SMAD4 deletions of alleles but not point mutations.
FIGURE 1.
Genomic SMAD4 status in HNSCCs. SMAD4 FISH of (A) lymphoblastoid cells with normal karyotype, CAL27 HNSCC with loss of heterozygosity by single point nonsense mutation to show one copy of genomic SMAD4, and FaDu HNSCCs with homozygous deletion of SMAD4. B) Representative images of primary HNSCCs designated SMAD4 balanced-wildtype (HNSCC-10) and hemizygous deletion (HNSCC-15) by SMAD4 FISH. C) Representative images of PDX tumors designated SMAD4 balanced-wildtype (CUHN-086), hemizygous deletion (−004), and homozygous deletion (−092) by SMAD4 FISH. D) SMAD4 FISH: Prevalence of SMAD4 genotypes across primary HNSCC (n = 20) and PDX tumors (n = 46) compared to TCGA HNSCCs (n = 496) sequencing data. SMAD4 loss includes hemizygous/heterozygous loss and homozygous loss. E) Frequency of different SMAD4/CEN18 ratios across primary HNSCCs and PDX tumors. [Color figure can be viewed at wileyonlinelibrary.com]
We performed SMAD4 FISH assay on primary HNSCC specimens (n = 20, Figure 1B, Supplementary Table S1) and PDX tumors (n = 46, Figure 1C, Supplementary Table S2) derived from HNSCC patients. SMAD4 hemizygous deletion was found in 35.0% of primary tumors (Figure 1D) and in 41.3% of PDX tumors. Homozygous SMAD4 loss was found in 4.3% PDX tumors. The frequency of SMAD4 loss was similar to what was found by a TCGA query (Figure 1D) where either heterozygous (47.6%) or homozygous (4.0%) deletion of SMAD4 occurred (ie, 51.6% in combination).30,31 SMAD4 was gained (>2 copies) in one primary HNSCC (5%), a frequency consistent with the SMAD4 gain (6%) found by TCGA (Figure 1D). Together, these analyses show a prevalence of genomic SMAD4 loss in HNSCCs.
We further examined copy numbers of SMAD4 and CEN18 by FISH on all primary HNSCCs (Supplementary Table S3) and PDX tumors (Supplementary Table S4). By considering both SMAD4 and CEN18 copy numbers per tumor to assess for SMAD4 loss, we identified seven different SMAD4/CEN18 patterns (Figure 1E). In total, CEN18 polysomy occurred in 45% of primary HNSCCs and 37% of PDX tumors (Figure 1E). In contrast, SMAD4 polysomy occurred at nearly half of the frequency, that is, 25% of primary HNSCCs and 10.9% of PDX tumors (Figure 1E), suggesting that some of these tumors have lost at least one copy of SMAD4. Indeed, cases with the ratio of SMAD4/CEN18 < 0.8 occurred in 35% of primary HNSCCs and 45.7% of PDX tumors (Figure 1E). One PDX tumor lost one copy of both SMAD4 and CEN18 (monosomic loss, Figure 1E). The unbalanced increase in CEN18 copy number (reflecting increased chromosomal copies) is potentially due to chromosomal instability caused by SMAD4 loss.8 Therefore, individual cells must be assessed for copy number of chromosome 18 to determine the actual copy loss of SMAD4.
3.2 ∣. Inter- and intra-tumor heterogeneity of chromosome 18 and SMAD4 in HNSCC
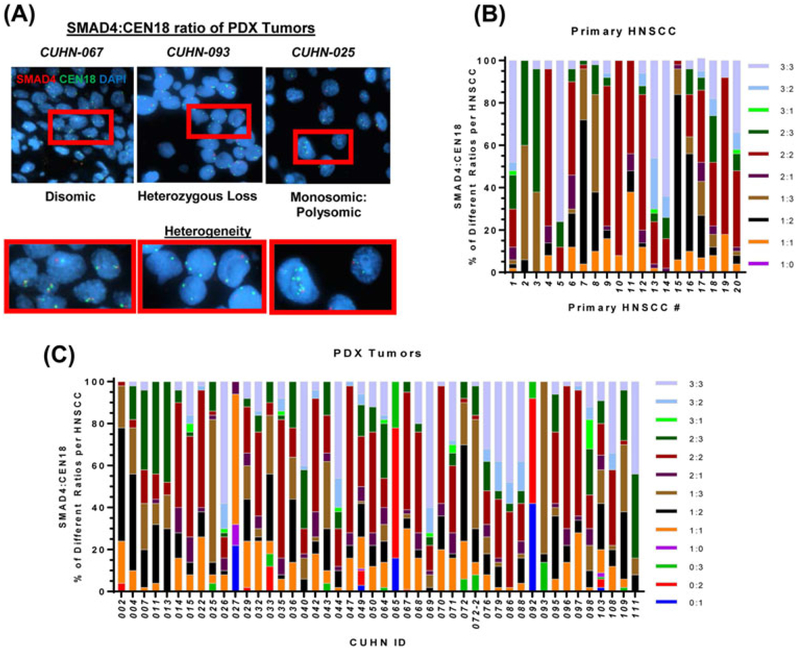
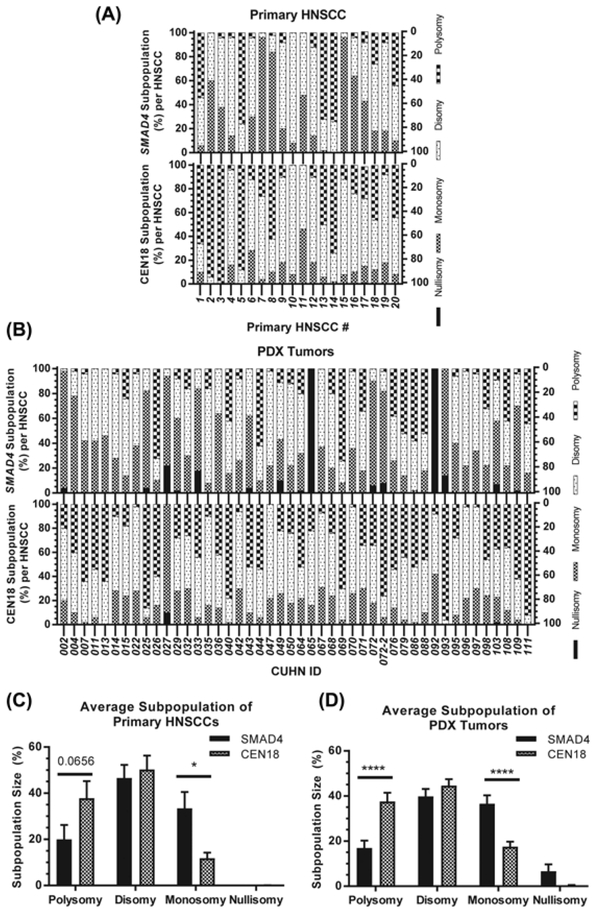
To further examine inter-cellular and inter-tumoral heterogeneity of SMAD4 genotypes, we compared cellular SMAD4/CEN18 ratios within and across tumors (Figure 2A). Among 10 different SMAD4/CEN18 ratios in primary HNSCCs (Figure 2B) and 13 different SMAD4/CEN18 ratios in PDX tumors (Figure 2C), we measured the subpopulation size (%) of each SMAD4/CEN18 cell population within each tumor. It was difficult to determine a dominant subpopulation due to high heterogeneity within and across all primary HNSCCs and PDX tumors (Figures 2B and 2C). Therefore, we compared subpopulation size (%) of SMAD4 or CEN18 per primary HNSCC (Figure 3A) and PDX tumor (Figure 3B). In both primary HNSCCs (Figure 3C) and PDX tumors (Figure 3D), SMAD4 loss subpopulations were significantly more abundant than CEN18 loss subpopulations. In contrast, SMAD4 polysomic subpopulations were smaller than CEN18 polysomic subpopulations in primary HNSCC (Figure 3C) and PDX tumors (Figure 3D). There was no difference between SMAD4 disomic and CEN18 disomic subpopulations (Figures 3C and 3D). Overall, SMAD4 loss correlated with CEN18 polysomy (Figure 3) suggesting that genomic instability associated with SMAD4 loss8 leads to increased accumulation of centromere 18 in primary HNSCCs and PDX tumors.
FIGURE 2.
Intra- and Inter-tumor heterogeneity of genomic SMAD4 and centromere 18. A) Representative SMAD4 FISH images of CUHN-067, −093, and −025 from left to right showing heterogeneity. Red boxes are magnified images. Subpopulations were defined by variations in copy numbers for SMAD4 and CEN18, to identify 10 unique subpopulations across all 20 primary HNSCCs (B) and 13 unique subpopulations across all 46 PDX tumors (C). The SMAD4:CEN18 subpopulations were organized by size (%) within each tumor specimen. [Color figure can be viewed at wileyonlinelibrary.com]
FIGURE 3.
Intra- and Inter-tumor heterogeneity SMAD4 and CEN18 copy numbers. SMAD4 (top) and CEN18 (bottom) subpopulation size (%) per primary (n = 20) HNSCC (A) and PDX (n = 46) tumor (B). Comparison of SMAD4 and CEN18 subpopulation size (%) averaged across all 20 primary HNSCCs (B) and all 46 PDX tumors (D). Standard error was used for error bars and one-way ANOVA was conducted for statistical analysis and P-value was determined. * < 0.05, **** < 0.0001, absent = non-significant difference
3.3 ∣. SMAD4 loss is enriched in cancer cell lines derived from PDX with heterogeneous SMAD4 loss
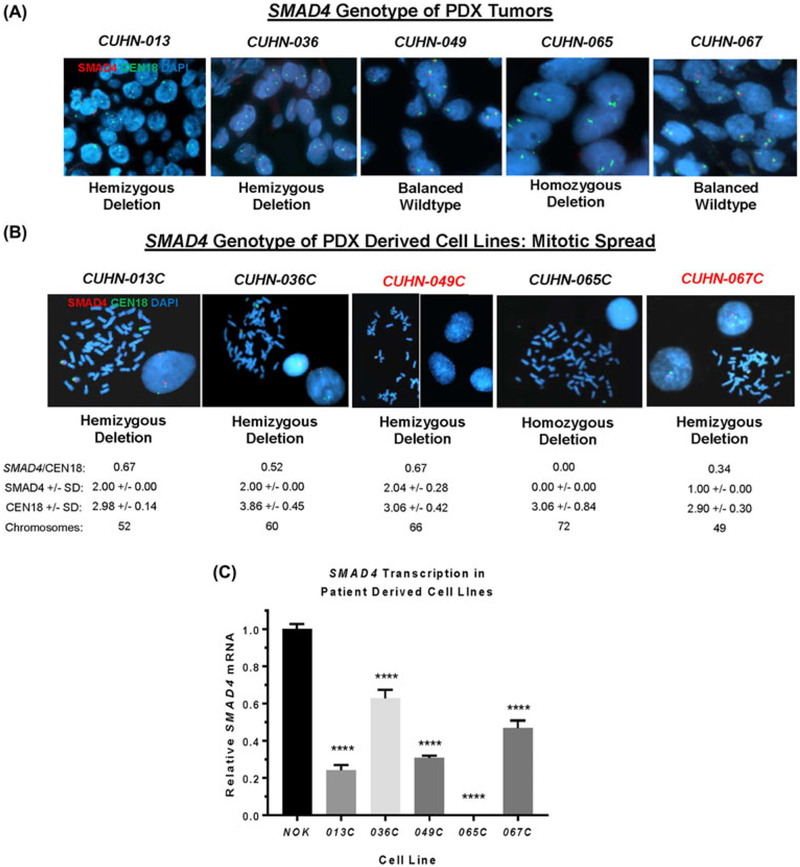
Five cell lines were established from PDX tumors (Figure 4). Three of the parental tumors (CUHN013, −036, and −065) had SMAD4 deletions and two were SMAD4 wild-type (CUHN-049 and −067) (Figure 4A). To determine if the SMAD4 status of established cell lines faithfully replicates the SMAD4 status of parental tumors, we performed SMAD4 FISH on the derived cell lines. While CUHN-013, −036, and −065 cell lines were consistent with parental PDX tumors, CUHN-049 and −067 cell lines drifted from wild-type SMAD4 in parental PDXs to SMAD4 hemizygous deletions (Figure 4B), presumably due to enrichment from a small population of tumor cells with hemizygous SMAD4 loss in CUHN-049 and −067 PDX tumors (Figure 4A). We assessed the mitotic spread of cells to determine ploidy status (Figure 4). All five cell lines had aneuploidy with an average chromosome count of 59.4 per cell line (Figure 4B). These cell lines had an average SMAD4/CEN18 ratio of 0.44 with SMAD4 loss relative to polyploidy CEN18. There was no variance (standard deviation = 0) of SMAD4 in four out of five cell lines, suggesting a homogeneous cell population. One cell line (049C) had a standard deviation of 0.28 for SMAD4 variants, suggesting that cells with heterozygous SMAD4 loss largely outcompeted cells with wild-type SMAD4 from the parental tumor. Loss of SMAD4 expression, partially, or completely, in these lines was confirmed at the mRNA level compared to normal oral keratinocytes (Figure 4C). These data suggest that SMAD4 loss by genomic deletion and reduced expression may enable these cells to permanently establish themselves in vitro.
FIGURE 4.
Loss of genomic SMAD4 is enriched in established HNSCC cell lines. A) Representative images of each parental PDX tumor and SMAD4 genotype by FISH. B) Representative images of PDX derived cell lines in mitotic spread and SMAD4 genotype by FISH. SMAD4/CEN18 ratio, SMAD4 copy number +/− standard deviation, CEN18 copy number ± standard deviation, and chromosome count per cell line. C) qRT-PCR: SMAD4 transcription relative to GAPDH per cell line compared to normal oral keratinocytes. Standard t-test: **** = P-value <0.0001. [Color figure can be viewed at wileyonlinelibrary.com]
4 ∣. DISCUSSION
Loss of SMAD4 is sufficient to initiate HNSCC in a mouse model8; however, reports regarding rates of SMAD4 loss vary significantly.19-21 In this report, we used a FISH assay to determine genomic SMAD4 status in primary and PDX HNSCCs. We found that SMAD4 loss primarily occurred by heterozygous deletion in primary and PDX HNSCCs, similar to TCGA HNSCC sequencing data. Therefore, SMAD4 FISH would provide a reliable but more rapid and cost-effective method for detecting SMAD4 genomic loss at the single cell level compared with genomewide sequencing. Given the significant intratumor heterogeneities of SMAD4 loss in HNSCCs identified from the current study, it would be difficult to determine if SMAD4 heterozygous deletion correlates with HNSCC patient prognosis in either untreated patients or patients with the same treatment regimen(s). However, studies from mouse models have revealed that heterozygous SMAD4 loss is sufficient to increase SCC susceptibility,8,32 and that a small population of heterozygous SMAD4 HNSCC cells were able to outcompete with wild-type SMAD4 cells within the same tumor (Figure 4). Therefore, it is important to monitor HNSCCs with SMAD4 heterozygous loss. Further, the remaining SMAD4 allele in heterozygous SMAD4 HNSCCs can be lost through LOH,22-24 mRNA loss and protein loss.8
Although ploidy in tissue samples could not be assessed by either sequencing or FISH without mitotic spreads, we were able to assess SMAD4 and CEN18 copy numbers in our FISH assay. Copy number assessment revealed inter-tumor heterogeneity as noted by seven different SMAD4/CEN18 copy number variants across all HNSCC specimens examined. We also found that tumors gained genetic copies of centromere 18 more frequently than tumors gained genetic copies of SMAD4, this may be explained by SMAD4 loss-associated genomic instability and subsequent accumulation of centromeres as previously described.8 Genomic instability can also increase tumor heterogeneity.33
Intra-tumor SMAD4/CEN18 heterogeneity was also common with 6.27 ± 1.97 different copy number variants within the same tumor although nuclear truncation may account for some variation in tissue samples. As expected, no tumor was homogeneous across all 66 specimens. It was difficult to compare SMAD4/CEN18 copy number variant populations due to high intra-tumor heterogeneity. By separating the SMAD4 copy number variant populations from CEN18 copy number variant populations per tumor, it is apparent that most tumors had lost one or more copies of SMAD4, but gained copies of CEN18. On average, the polysomy CEN18 cell population was significantly larger than the polysomy SMAD4 cell population. In contrast, the cell population with SMAD4 loss was significantly larger than the cell population with CEN18 loss, this further supported SMAD4-associated genomic instability resulting in accumulation of centromeres.
The use of PDX-derived cell lines allowed us to confirm that CEN18 copy numbers are consistent with chromosomal numbers in chromosomal spreads, hence future molecular diagnosis could use CEN18 as a surrogate marker for ploidy status to more accurately evaluate SMAD4 copy number loss by FISH. Intriguingly, all five cell lines had partial or complete SMAD4 loss suggesting a survival advantage for SMAD4 mutant tumor cells. This may explain why SMAD4 mutant HNSCCs are resistant to therapies primarily targeting tumor growth and survival.34,35 Conversely, SMAD4 mutant tumor cells are shown to be sensitive to certain DNA damaging agents.3,36 Therefore, the SMAD4 FISH analysis described here will provide a simple and cost-effective molecular diagnosis for solid tumors to readily determine genomic SMAD4 deletion status and future clinical studies will determine if partial or complete loss of genomic SMAD4 serves as a biomarker for prognosis and therapeutic interventions.
Supplementary Material
ACKNOWLEDGMENTS
Technical support from the Molecular Pathology Shared Resource of the University of Colorado Cancer Center (NCI P30 CA046934). ALH is supported by Continuing Umbrella of Research Experiences (CURE) Supplement to the National Research Service Award Institutional Training Grant (T32CA174648). XJW is supported by VA merit award (I01 BX003232). XJW and AJ are supported by NIH R01 DE024371. SDK is supported by the Paul Calabresi Career Development Award for Clinical Oncology (K12, CA086913). DR is supported by the Marsico Endowment fund for research. We also thank an anonymous donor to the Head and Neck Research Program for support of this work.
Funding information
VA, Grant number: I01 BX003232; NIH, Grant numbers: K12CA086913, P30 CA046934, R01 DE024371, T32CA174648; National Research Service Award Institutional Training, Grant number: (T32CA174648); Paul Calabresi Career Development Award, Grant number: (K12, CA086913)
Abbreviations:
- CUHN
University of Colorado Head and Neck
- FISH
fluorescence in situ hybridization
- HNSCC(s)
head and neck squamous cell carcinoma(s)
- HPV
human papillomavirus
- LOH
loss of heterozygosity
- PDX
patient derived xenograft
- TCGA
The Cancer Genome Atlas
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article.
REFERENCES
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. [DOI] [PubMed] [Google Scholar]
- 2.Bertsch NS, Bindler RJ, Wilson PL, Kim AP, Ward B. Medication therapy management for patients receiving oral chemotherapy agents at a community oncology center: a pilot study. Hosp Pharm. 2016;51:721–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mendenhall WM, Mancuso AA, Amdur RJ, Stringer SP, Villaret DB, Cassisi NJ. Squamous cell carcinoma metastatic to the neck from an unknown head and neck primary site. Am J Otolaryngol. 2001;22:261–267. [DOI] [PubMed] [Google Scholar]
- 4.Vigneswaran N, Williams MD. Epidemiologic trends in head and neck cancer and aids in diagnosis. Oral Maxillofac Surg Clin North Am. 2014; 26:123–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jansen L, Buttmann-Schweiger N, Listl S, et al. Differences in incidence and survival of oral cavity and pharyngeal cancers between Germany and the United States depend on the HPV-association of the cancer site. Oral Oncol. 2018;76:8–15. [DOI] [PubMed] [Google Scholar]
- 6.Hocking JS, Stein A, Conway EL, et al. Head and neck cancer in Australia between 1982 and 2005 show increasing incidence of potentially HPV-associated oropharyngeal cancers. Br J Cancer. 2011; 104:886–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malkoski SP, Wang XJ. Two sides of the story? Smad4loss in pancreatic cancer versus head-and-neck cancer. FEBS Lett. 2012;586:1984–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bornstein S, White R, Malkoski S, et al. Smad4 loss in mice causes spontaneous head and neck cancer with increased genomic instability and inflammation. J Clin Invest. 2009;119:3408–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reiss M, Santoro V, deJonge RR, Vellucci VF. Transfer of chromosome 18 into human head and neck squamous carcinoma cells: evidence for tumor suppression by Smad4/DPC4. Cell Growth Differ. 1997;8:407–415. [PubMed] [Google Scholar]
- 10.Izeradjene K, Combs C, Best M, et al. Kras(G12D) and Smad4/Dpc4 haploinsufficiency cooperate to induce mucinous cystic neoplasms and invasive adenocarcinoma of the pancreas. Cancer Cell. 2007;11: 229–243. [DOI] [PubMed] [Google Scholar]
- 11.Bardeesy N, Cheng KH, Berger JH, et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20:3130–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aguirre AJ, Nowak JA, Camarda ND, et al. Real-time genomic characterization of advanced pancreatic cancer to enable precision medicine. Cancer Discov. 2018;8:1096–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahn SA, Schutte M, Hoque AT, et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271: 350–353. [DOI] [PubMed] [Google Scholar]
- 14.Takaku K, Oshima M, Miyoshi H, Matsui M, Seldin MF, Taketo MM. Intestinal tumorigenesis in compound mutant mice of both Dpc4 (Smad4) and Ape genes. Cell. 1998;92:645–656. [DOI] [PubMed] [Google Scholar]
- 15.Kitamura T, Kometani K, Hashida H, et al. SMAD4-deficient intestinal tumors recruit CCR1+ myeloid cells that promote invasion. Nat Genet. 2007;39:467–475. [DOI] [PubMed] [Google Scholar]
- 16.lacobuzio-Donahue CA, Fu B, Yachida S, et al. DPC4 gene status of the primary carcinoma correlates with patterns of failure in patients with pancreatic cancer. J Clin Oncol. 2009;27:1806–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyaki M, lijima T, Konishi M, et al. Higher frequency of Smad4 gene mutation in human colorectal cancer with distant metastasis. Oncogene. 1999;18:3098–3103. [DOI] [PubMed] [Google Scholar]
- 18.Agrawal N, Frederick MJ, Pickering CR, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukuchi M, Masuda N, Miyazaki T, et al. Decreased Smad4 expression in the transforming growth factor-beta signaling pathway during progression of esophageal squamous cell carcinoma. Cancer. 2002;95: 737–743. [DOI] [PubMed] [Google Scholar]
- 20.Natsugoe S, Xiangming C, Matsumoto M, et al. Smad4 and transforming growth factor betal expression in patients with squamous cell carcinoma of the esophagus. Clin Cancer Res. 2002;8:1838–1842. [PubMed] [Google Scholar]
- 21.Xie W, Aisner S, Baredes S, Sreepada G, Shah R, Reiss M. Alterations of Smad expression and activation in defining 2 subtypes of human head and neck squamous cell carcinoma. Head Neck. 2013;35:76–85. [DOI] [PubMed] [Google Scholar]
- 22.Takebayashi S, Ogawa T, Jung KY, et al. Identification of new minimally lost regions on 18q in head and neck squamous cell carcinoma. Cancer Res. 2000;60:3397–3403. [PubMed] [Google Scholar]
- 23.Kim SK, Fan Y, Papadimitrakopoulou V, et al. DPC4, a candidate tumor suppressor gene, is altered infrequently in head and neck squamous cell carcinoma. Cancer Res. 1996;56:2519–2521. [PubMed] [Google Scholar]
- 24.Snijders AM, Schmidt BL, Fridlyand J, et al. Rare amplicons implicate frequent deregulation of cell fate specification pathways in oral squamous cell carcinoma. Oncogene. 2005;24:4232–4242. [DOI] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Devilee P, Slagboom P, Cornelisse CJ, Pearson PL. Sequence heterogeneity within the human alphoid repetitive DNA family. Nucleic Acids Res. 1986;14:2059–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varella-Garcia M Stratification of non-smallcell lung cancer patients for therapy with epidermal growth factor receptor inhibitors: the EGFR fluorescence in situ hybridization assay. Diagn Pathol. 2006;1:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keysar SB, Astling DP, Anderson RT, et al. A patient tumor transplant model of squamous cell cancer identifies PI3K inhibitors as candidate therapeutics in defined molecular bins. Mol Oncol. 2013;7:776–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu W, Schonleben F, Li X, Su GH. Disruption of transforming growth factor beta-Smad signaling pathway in head and neck squamous cell carcinoma as evidenced by mutations of SMAD2 and SMAD4. Cancer Lett. 2007;245:163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu X, Brodie SG, Yang X, et al. Haploid loss of the tumor suppressor Smad4/Dpc4 initiates gastric polyposis and cancer in mice. Oncogene. 2000;19:1868–1874. [DOI] [PubMed] [Google Scholar]
- 33.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. [DOI] [PubMed] [Google Scholar]
- 34.Cheng H, Fertig EJ, Ozawa H, et al. Decreased SMAD4 expression is associated with induction of epithelial-to-mesenchymal transition and cetuximab resistance in head and neck squamous cell carcinoma. Cancer Biol Ther. 2015;16:1252–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ozawa H, Ranaweera RS, Izumchenko E, et al. SMAD4 loss is associated with cetuximab resistance and induction of MAPK/JNK activation in head and neck cancer cells. Clin Cancer Res. 2017;23:5162–5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haeger SM, Thompson JJ, Kalra S, et al. Smad4 loss promotes lung cancer formation but increases sensitivity to DNA topoisomerase inhibitors. Oncogene. 2016;35:577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.