Abstract
Summary
Xylella fastidiosa is a Gram‐negative bacterial plant pathogen with an extremely wide host range. This species has recently been resolved into subspecies that correlate with host specificity. This review focuses on the status of X. fastidiosa pathogenic associations in plant hosts in which the bacterium is either endemic or has been recently introduced. Plant diseases associated with X. fastidiosa have been documented for over a century, and much about what is known in the context of host–pathogen interactions is based on these hosts, such as grape and citrus, in which this pathogen has been well described. Recent attention has focused on newly emerging X. fastidiosa diseases, such as in olives.
Taxonomy
Bacteria; Gammaproteobacteria; family Xanthomonadaceae; genus Xylella; species fastidiosa.
Microbiological properties
Gram‐negative rod (0.25–0.35 × 0.9–3.5 μm), non‐flagellate, motile via Type IV pili‐mediated twitching, fastidious.
Host range
Xylella fastidiosa has a broad host range that includes ornamental, ecological and agricultural plants belonging to over 300 different species in 63 different families. To date, X. fastidiosa has been found to be pathogenic in over 100 plant species. In addition, it can establish non‐symptomatic associations with many plants as a commensal endophyte. Here, we list the four distinct subspecies of X. fastidiosa and some of the agriculturally relevant diseases caused by them: X. fastidiosa ssp. fastidiosa causes Pierce's disease (PD) of grapevine (Vitis vinifera); X. fastidiosa ssp. multiplex causes almond leaf scorch (ALS) and diseases on other nut and shade tree crops; X. fastidiosa ssp. pauca causes citrus variegated chlorosis (CVC) (Citrus spp.), coffee leaf scorch and olive quick decline syndrome (OQDS) (Olea europaea); X. fastidiosa ssp. sandyi causes oleander leaf scorch (OLS) (Nerium oleander). Significant host specificity seemingly exists for some of the subspecies, although this could be a result of technical biases based on the limited number of plants tested, whereas some subspecies are not as stringent in their host range and can infect several plant hosts.
Disease symptoms
Most X. fastidiosa‐related diseases appear as marginal leaf necrosis and scorching of the leaves. In the case of PD, X. fastidiosa can also cause desiccation of berries (termed ‘raisining’), irregular periderm development and abnormal abscission of petioles. In olive trees affected with OQDS, leaves exhibit marginal necrosis and defoliation, and overall tree decline occurs. Plants with ALS and OLS also exhibit the characteristic leaf scorch symptoms. Not all X. fastidiosa‐related diseases exhibit the typical leaf scorch symptoms. These include CVC and Phony Peach disease, amongst others. In the case of CVC, symptoms include foliar wilt and interveinal chlorosis on the upper surfaces of the leaves (similar to zinc deficiency), which correspond to necrotic, gum‐like regions on the undersides of the leaves. Additional symptoms of CVC include defoliation, dieback and hardening of fruits. Plants infected with Phony Peach disease exhibit a denser, more compact canopy (as a result of shortened internodes, darker green leaves and delayed leaf senescence), premature bloom and reduced fruit size. Some occlusions occur in the xylem vessels, but there are no foliar wilting, chlorosis or necrosis symptoms .
Useful websites
http://www.piercesdisease.org/; https://pubmlst.org/xfastidiosa/; http://www.xylella.lncc.br/; https://nature.berkeley.edu/xylella/; https://ec.europa.eu/food/plant/plant_health_biosecurity/legislation/emergency_measures/xylella-fastidiosa_en
Keywords: citrus, grapevine, olive, Pierce's disease, tyloses, xylem
Introduction
In the late 1800s, Newton B. Pierce described a severe grapevine disease occurring in Anaheim, California. This disease was initially called Anaheim disease (Pierce, 1892) and was first attributed to a viral infection because: (i) no causal microorganism could be cultured from the infected vines; and (ii) the disease was graft transmissible. However, electron micrographs indicated bacterial‐like bodies in the xylem of infected plants (Hopkins and Mollenhauer, 1973), and a bacterium was subsequently cultured from infected plants (Davis et al., 1978). Koch's postulates were then completed, ultimately confirming Xylella fastidiosa as the causal agent of what was eponymously named Pierce's disease (PD). In addition to PD, this bacterium has been implicated as the causal agent of many other significant plant diseases (Fig. 1). The severity of the citrus variegated chlorosis (CVC) epidemic in Brazil (Bove and Ayres, 2007) and PD epidemic in southern California (Siebert, 2001) prompted sequencing of the 9a5c CVC strain genome (Simpson et al., 2000), followed by the Temecula 1 PD strain of X. fastidiosa (Van Sluys et al., 2003). Notably, X. fastidiosa was the first plant‐pathogenic bacterium to have its genome sequenced. Subsequently, genomes of other X. fastidiosa isolates have been sequenced (Chen et al., 2010, 2016; Giampetruzzi et al., 2015; Guan et al., 2014), providing a robust database for genome mining and comparative genomics.
Figure 1.

Host symptoms associated with Xylella fastidiosa infection. (A) Leaf scorching. (B) Berry desiccation or ‘raisining’ (asterisk) and green islands (white arrow) associated with Pierce's disease (PD). (C) Limb dieback and (D) foliar symptoms in olive quick decline syndrome (OQDS). (E) Foliar symptoms of almond leaf scorch. (F) Foliar symptoms of oleander leaf scorch (OLS). (G) Foliar symptoms of citrus variegated chlorosis (CVC). Photo credits: (A, B) Philippe Rolshausen, University of California, Riverside, CA, USA; (C, D, G) Rodrigo Kruegner, US Department of Agriculture Agricultural Research Service; (E) David Doll, University of California Cooperative Extension, Merced, CA, USA .
Xylella fastidiosa colonizes two environments: the plant host xylem and the mouthparts of its insect vector. Much of what is known about X. fastidiosa biology has been performed with California isolates of the subspecies fastidiosa, the grapevine host and two prominent insect vectors in California: Homalodisca vitripennis, the glassy‐winged sharpshooter (GWSS), and/or Graphocephala atropunctata, the blue–green sharpshooter (BGSS). Thus, the bulk of the literature base on X. fastidiosa is in the context of the PD pathosystem in California. What has been learned in the grape–GWSS/BGSS pathosystem may be applicable to other plant systems. However, we must be mindful that there may be important differences in the way in which X. fastidiosa interacts with plant hosts and insect vectors found in different environments. Thus, it may be difficult or impossible to extrapolate findings from one pathosystem to another, or one epidemic to another, without taking into consideration the ecological context of each scenario (Almeida and Nunney, 2015). This article focuses primarily on the PD pathosystem in California (Fig. 2).
Figure 2.
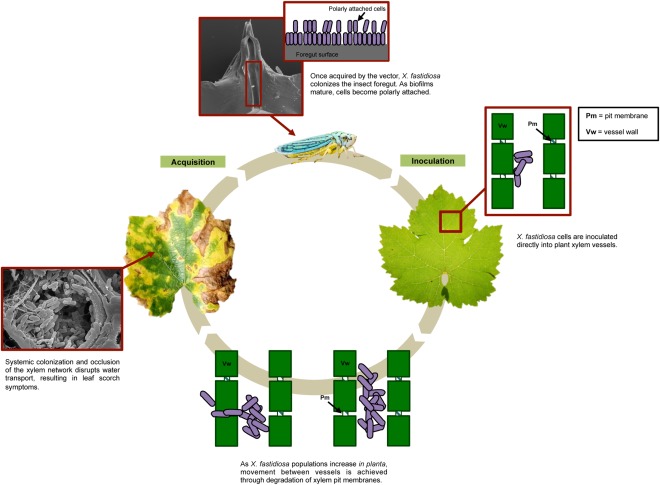
Pierce's disease cycle.
Historically, X. fastidiosa was limited to the Americas; however, recently, it has been identified in Europe, most notably in olives (Olea europaea) in the Apulia region of southern Italy. The spread of X. fastidiosa and its associated diseases is primarily attributed to human activity that introduces infected plant material, or effective insect vectors, to a new region (Almeida and Nunney, 2015). There is also evidence which suggests that X. fastidiosa has a marked capacity to engage in interstrain recombination that can effectively result in new strains with different host ranges from the original parent strains (Nunney et al., 2014).
Host Range
Xylella fastidiosa's broad host range includes both monocots and dicots [European Food Safety Authority (EFSA), 2016)]. Although all X. fastidiosa isolates were initially assigned to the same species, they have been further classified to the subspecies level (Scally et al., 2005; Schuenzel et al., 2005). Three clades of X. fastidiosa have been identified in North America, corresponding to the different subspecies: X. fastidiosa ssp. fastidiosa, which is found in grapevines, almond and alfalfa; X. fastidiosa ssp. multiplex, which can be found in almond, peach, plum and oak; and X. fastidiosa ssp. sandyi, which, thus far, has only been found in oleander. Xylella fastidiosa ssp. pauca is another subspecies which is primarily found in citrus and coffee in South America (Scally et al., 2005; Schuenzel et al., 2005). The strain associated with olive quick decline syndrome (OQDS) in Italy, aptly referred to as the CoDiRo strain (an abbreviation of the Italian name for OQDS), is genetically related to the pauca subspecies (Giampetruzzi et al., 2015; Marcelletti and Scortichini, 2016), and is the current subject of intense research focus on X. fastidiosa in Europe. Furthermore, the bacterium can reside in a wide range of plants as a harmless endophyte and has been detected in hundreds of asymptomatic plant species (Chatterjee et al., 2008a; Hopkins and Purcell, 2002; Raju et al., 1980). Most X. fastidiosa strains do not move systemically in symptomless hosts (Hill and Purcell, 1995a; Purcell and Saunders, 1999), but these plants could still serve as sources of inoculum (Hill and Purcell, 1997). Because of the large number of hosts, the list of plant species described provides prominent examples of plant hosts in which X. fastidiosa has a pathogenic interaction and is not meant to be a comprehensive list.
Pierce's Disease in California: An Epidemiological Case Study
Xylella fastidiosa is spread by xylem‐feeding insect vectors belonging to the sharpshooter subfamily of leafhopper (Cicadellidae) and spittlebug (Cercopidae) families (Hewitt et al., 1946). These vectors can feed on both woody and herbaceous hosts. There is no evidence of vector specificity; presumably, an implicated vector in any region is capable of transmitting any strain of X. fastidiosa, whether it is endemic or invasive (Almeida and Nunney, 2015). Because some sharpshooters fly long distances and feed on many different hosts, they can introduce X. fastidiosa into an array of host plants. Xylella fastidiosa has a persistent and propagative (but non‐circulative) relationship in adult sharpshooter insects, which is the only described vector–microbe relationship of this sort. An adult insect remains infective throughout its life, whereas infective nymphs lose the ability to transmit when they moult and shed the cuticular lining of the foregut (Purcell and Finlay, 1979). These sharpshooters acquire X. fastidiosa by feeding on infected hosts and can immediately transmit X. fastidiosa to a new plant host with no latent period (Fig. 2). A possible explanation for the absence of a latent period is the potential ability of sharpshooters to transiently hold a column of fluid containing X. fastidiosa cells in their stylets without swallowing it, allowing the fluid to be immediately egested during feeding (Backus et al., 2015). Over time, the bacteria colonize the insect foregut (namely the cibarium and precibarium regions) by attaching to the cuticular lining and forming biofilms (Brlansky et al., 1983). Inoculation occurs through a combined egestion/salivation mechanism that dislodges the bacteria from the cuticular lining of the foregut.
Because the bulk of the epidemiological data has been collected in the context of PD in California, this section focuses primarily on the dynamics of PD epidemiology in California as a case study, with the caveat that the epidemiology of X. fastidiosa diseases in other areas may not mirror that of PD in California. Vector and strain behaviour and differences in climate and plant species must be taken into account in other ecological contexts. PD has been in California for over a century, but it spread slowly and did not cause significant damage in most viticultural areas of the state. However, in the late 1990s, PD became an acute concern in California. To understand why the disease incidence increased to epidemic levels, one must consider the behaviour of the insect vectors. Historically, BGSS was the most important vector in California coastal vineyards, whereas the red‐headed (Carneocephala fulgida) and green (Draeculacephala minerva) sharpshooters were most significant in the San Joaquin Valley (Hewitt et al., 1949). These native vectors overwinter in riparian habitats (e.g. stream banks) and, in the spring and summer, move into vineyards where they feed on grapevines. Native sharpshooters exclusively feed on succulent new growth (Purcell, 1975). Once in the plant, the bacteria can move acropetally from the point of inoculation into new growth, as well as basipetally down to the vine cordon and trunk, where they overwinter. Infections that occur in spring or early summer become chronic because the bacteria have adequate time to migrate to these permanent parts of the vine (i.e. the cordon and trunk are not pruned annually). Infections occurring late in the season do not have a chance to become chronic because severe winter pruning removes infected parts of the vine before X. fastidiosa can migrate into the cordon to overwinter.
Because they fly from their preferred habitats only into the edge of vineyards, native sharpshooters probably do not spread PD from one vine to another (Purcell, 1974, 1975). In the northern coastal regions of California, PD persists mainly in ‘PD hot spots’ along parts of the vineyard adjacent to riparian areas. The dominant vector in this region is the native BGSS, which enters vineyards from these riparian areas in the spring and summer, following overwintering, to feed and reproduce. PD incidence is highest in these hot spots, creating what is known as the PD ‘edge effect’ (Hopkins and Purcell, 2002), meaning that infection remains primarily along the edges of the vineyard. The lack of spread within vineyards and limited persistence in vines from year to year were essential features of PD epidemiology, because this limited occurrence and distribution of the disease (Hopkins and Purcell, 2002). Under these conditions, PD is considered to be a monocyclic disease.
The epidemiology of PD changed drastically in the southern regions of California as a result of the introduction of the polyphagous GWSS. It is hypothesized that GWSS was introduced into California from Texas on nursery stock. GWSS and X. fastidiosa are endemic to Texas and the southeastern USA, and are the primary reasons why these regions lack a major Vitis vinifera grape industry. GWSS was first spotted in 1990 in southern California. Soon after, PD reached epidemic proportions, causing millions of dollars of damage to Temecula Valley vineyards (Blua et al., 1999). Furthermore, the presence of GWSS brought about a new X. fastidiosa‐related disease in southern California called oleander leaf scorch (OLS). The disease was first noticed in the Palm Springs–Indio area of Riverside County in 1994 and spread to adjacent areas in the following years (Blua et al., 1999).
This change in PD epidemiology is a result of differences in the feeding, flying and breeding behaviour of GWSS. Unlike native sharpshooters, GWSS can feed on both succulent new growth and woody stems. Woody stems are closer to the cordons and main trunk, and bacteria introduced here require less time to reach the cordon and trunk to overwinter. GWSS can also feed on dormant vines, which can introduce bacteria directly into the permanent part of the vine. Therefore, the feeding behaviour of GWSS increases the incidence of chronic infections. In addition, GWSS is a much stronger flier than the native sharpshooters and can fly faster and further into the vineyard. The final property of GWSS that contributes to the increase in PD is its breeding behaviour. In contrast with native sharpshooters, which preferentially feed and reproduce on riparian plants, GWSS can reproduce on numerous plant species, with special preference for citrus. Citrus groves support enormous populations of these insects and, in southern California, many citrus groves are planted adjacent to vineyards (Perring et al., 2001). This unfortunate situation allows GWSS easy access to vineyards and increases the spread and incidence of PD. With the establishment of GWSS, PD became so severe in southern California that it threatened to eliminate viticulture in some areas. Its potential impact in central and northern grape‐growing regions of California remains unknown. Most of the economically important diseases caused by X. fastidiosa occur mainly in parts of North America that do not experience cold winters (Hopkins and Purcell, 2002). Therefore, if GWSS becomes established in northern California, the incidence and distribution of PD could reach the same levels as experienced in the southern part of the state.
Epidemiology of X. fastidiosa in Europe
Xylella fastidiosa was recently identified in olive trees affected with OQDS in Salento (Lecce district) in the Apulia region in southern Italy. The disease has rapidly spread throughout the region. This represents the first confirmed detection of X. fastidiosa in the European Union (EU) (Loconsole et al., 2014). An understanding of OQDS epidemiology is just emerging, but, thus far, the meadow spittlebug, Philaenus spumarius, has been identified as the primary insect vector (Saponari et al., 2014). Based on multi‐locus sequence tagging analyses, the strain found in Italy is most closely related to subspecies pauca and was probably introduced in contaminated plant material originating from Costa Rica (EFSA Panel on Plant Health, 2015; EFSA, 2016). Xylella fastidiosa is a regulated quarantine pathogen in the EU and, since 2015, coffee plant imports from Costa Rica and Honduras have been forbidden (Almeida, 2016). Following the initiation of a monitoring programme, X. fastidiosa has also been identified in Corsica, Mallorca and the south of France, although there has not been a widespread epidemic in these areas (Almeida, 2016). Three isolates were obtained in France from coffee plants propagated in glasshouses. One of the isolates was characterized as being closely related to both the sandyii and fastidiosa subspecies. The other two isolates were most closely related to subspecies pauca implicated in OQDS in Apulia, but with some divergence (Jacques et al., 2015). In Corsica and the south of France, isolates were found to be different from subspecies pauca (Almeida, 2016). Following regulations set forth by the EU, countries infested with X. fastidiosa have been directed to eradicate infected plant material and suppress insect populations. However, because of the widespread epidemic in Apulia and the geographical distribution of olive plantings, complete eradication is probably not possible in this area (Strona et al., 2017). Moreover, the implementation of these regulations in Italy has proven to be difficult because of a cultural and scientific gridlock that has occurred, caused by the cultural significance of olive trees in Italy (Abbott, 2015; Almeida, 2016; Stokstad, 2015). OQDS threatens to not only cause enormous economic damage to the olive industry, but to also reshape the cultural landscape in Italy's olive‐growing regions. The list of additional vectors and plant hosts in southern Italy is unknown, and epidemiological modelling indicates that southern Italy is becoming a reservoir for X. fastidiosa (Strona et al., 2017). All known xylem‐feeding insects in Europe are considered as potential vectors of the bacterium (EFSA Panel on Plant Health, 2015), further reiterating the risk of X. fastidiosa spread into other countries and plant hosts in Europe.
Economic Impact
Because X. fastidiosa causes diseases in economically valuable crops, the occurrence of these epidemics is typically accompanied by substantial economic consequences. The California table, raisin and wine grape industries were valued at $4.95 billion in 2015 [California Department of Food and Agriculture (CDFA), 2015]. Following the introduction of GWSS into southern California, losses were estimated to be $37.9 million annually (Siebert, 2001). Despite the presence of the PD/GWSS Control Program, established in 2001, PD costs producers over $100 million per year in crop loss and replanting expenses. This amount excludes an additional $50 million per year spent on preventative measures, including vector control (Alston et al., 2015). Alston et al. (2013) evaluated the costs and benefits of PD research in the wine grape industry, and estimated that PD would cost an additional $189 million per year if the PD Control Program (PDCP) ended. Therefore, the benefits of PD research clearly outweigh the costs in terms of preventing another major outbreak.
Similar to the effect of PD on commercial grape production, CVC affects all major commercial sweet orange cultivars (Goncalves et al., 2012). It is considered to be one of the most important diseases affecting the Brazilian citrus industry, which accounts for 30% of sweet orange production and 85% of exports of frozen orange juice concentrate worldwide (Goncalves et al., 2014; Rodrigues et al., 2013). Currently, 40% of citrus plants in Brazil are affected by CVC, and economic losses caused by the disease can reach $120 million annually (Goncalves et al., 2014, 2012).
The recent outbreak of OQDS in Italy presents a formidable threat to European agriculture, with olive farming in southern Italy's Apulia region accounting for nearly 40% of Italy's olive oil production (Strona et al., 2017). EFSA recently stated that the dissemination of X. fastidiosa to other European countries is very likely, as many other olive‐growing regions (e.g. Spain, Portugal, Greece) match the climatic conditions of Apulia (Luvisi et al., 2017). Currently, nearly 10 000 ha of olive trees have been destroyed, which amounts to millions of dollars in damage (Martelli et al., 2016). Thus, the identification and evaluation of risk reduction options are crucial (Pereira, 2015).
Host Colonization
Generally speaking, the ability to form a robust surface‐attached community (biofilm) directly correlates with virulence for many bacterial pathogens (Danhorn and Fuqua, 2007). The formation of a biofilm plays an important role in the colonization of the insect and plant environments for X. fastidiosa. The surface in this case is either the wall of the xylem (Fig. 3A) or the cuticular lining of the insect foregut. Xylella fastidiosa has never been found outside of the plant host or its insect vector. In the insect environment, it is clear that the attachment and formation of biofilm‐like structures within the foregut are tightly associated with the acquisition, retention and, ultimately, transmission of X. fastidiosa by sharpshooters (Killiny and Almeida, 2014). However, the role of biofilm formation in the plant environment is much more nuanced. In heavily infected plants, X. fastidiosa biofilms are abundant (Fig. 3A) (Newman et al., 2003; Roper et al., 2007a); thus, attachment to the xylem wall is a key aspect of establishment in planta. However, to move systemically in the plant, the bacteria must enter an exploratory, motile state. There are several examples of mutants that are defective in surface/cell–cell attachment, but display hypervirulent phenotypes (Guilhabert and Kirkpatrick, 2005). It has been proposed that X. fastidiosa forms biofilms partially to attenuate its own virulence by controlling rampant systemic colonization. Logically, it would benefit the bacterium not to kill its host immediately, and this model also explains how X. fastidiosa can exist as a commensal endophyte in many plant species (Chatterjee et al., 2008a; Guilhabert and Kirkpatrick, 2005; Roper and Lindow, 2014). To be effectively acquired from the plant by the insect, the bacterium must be in the adhesive state. Taken together, this requires that X. fastidiosa fluctuate between an exploratory (non‐adhesive) state and an insect‐acquirable (adhesive) state. In this article, we focus on the latest information regarding the factors governing plant and insect colonization. [For previous reviews on the topic of X. fastidiosa plant and insect colonization, particularly with regard to adhesion, see Chatterjee et al. (2008a) and Roper and Lindow (2014).]
Figure 3.
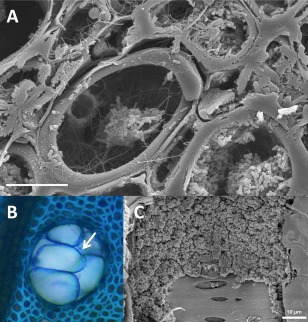
Vascular occlusions and depositions associated with Pierce's disease (PD) in grapevine. (A) Bacterial aggregates in xylem of petioles. (B) Tyloses fully occluding grapevine xylem (white arrow). (C) Amorphous gels coating xylem vessel walls in PD‐infected grapevines. Photo credit: (C) Qiang Sun, University of Wisconsin, Madison, WI, USA.
Plant colonization
Xylella fastidiosa produces prolific amounts of outer membrane vesicles (OMVs) relative to other bacterial species. For X. fastidiosa, OMVs hinder attachment to glass and xylem surfaces, suggesting that they serve to prevent attachment during the exploratory phase of xylem colonization. Blocking attachment is a novel function described for OMVs in bacteria (Ionescu et al., 2014). Afimbrial haemagglutinin adhesins, HxfA and HxfB, facilitate cell–cell attachment (Guilhabert and Kirkpatrick, 2005) and are found in both the outer membrane of X. fastidiosa cells and as part of OMVs that are shed from the cells (Voegel et al., 2010). Loss‐of‐function mutations in both the hxfA and hxfB genes result in hypervirulent phenotypes after inoculation into grapevines, supporting the hypothesis that X. fastidiosa uses adhesion to control the dynamics of systemic colonization to attenuate its own virulence. XatA, an autotransported protein related to surface adhesion and cell–cell aggregation, is also found in OMVs (Matsumoto et al., 2012), but a knockout mutation in xatA resulted in a significant decrease in virulence, rather than the hypervirulent phenotype that resulted from mutations in the haemagglutinin adhesins. It is tempting to speculate that the alteration of OMV composition and OMV production is a mechanism used by X. fastidiosa to adjust its adhesiveness during plant colonization to either facilitate systemic plant movement or enable insect acquisition.
Non‐proteinaceous, cell surface, carbohydrate‐based molecules are also important for the association of X. fastidiosa with plants, specifically lipopolysaccharide (LPS) and exopolysaccharide (EPS). The LPS molecule comprises a conserved lipid A‐core oligosaccharide component and a variable O‐antigen. Truncation of the long‐chain, two‐linked, rhamnose‐rich O‐antigen affects the net surface charge (measured as the zeta potential) of the X. fastidiosa cell and has a significant impact on surface attachment and cell–cell aggregation in both the plant and insect environments (Clifford et al., 2013; Rapicavoli et al., 2015). In plants, it was deduced that truncation of the O‐antigen caused a hyperattaching phenotype, but a defect in cell–cell attachment. The consequences were cells that could attach to a proxy plant surface (glass), but were unable to build upon themselves to form a robust biofilm. This resulted in a thinner and topographically rougher biofilm than the wild‐type X. fastidiosa biofilm. Furthermore, this truncation severely impaired virulence and colonization in grapevine (Clifford et al., 2013).
Xylella fastidiosa produces an EPS, termed fastidian gum. Fastidian gum is predicted to be similar in structure to xanthan gum (da Silva et al., 2001), but the polymer has yet to be fully characterized. Spatiotemporal localization of EPS during the different stages of biofilm formation indicated that there is very little EPS associated with the initial biofilm stages (microcolony formation), but EPS can be seen intercalating the mature biofilm structure. Thus, EPS provides a greater contribution to the maintenance of three‐dimensional biofilm architecture, and stability of the mature biofilm, as opposed to initial surface attachment (Roper et al., 2007a). Findings by Souza et al. (2006) corroborate this, as EPS mutants in the CVC strain of X. fastidiosa were not affected in surface attachment, but showed a reduced capacity to form biofilms. Immunolocalization in PD‐infected grapevines showed that an EPS matrix was associated with X. fastidiosa cell aggregates in the plant xylem vessels, where it also contributed to xylem vessel occlusion (Roper et al., 2007a). Xylella fastidiosa mutants compromised in EPS production showed reduced virulence in grapes (Killiny et al., 2013).
Systemic movement in planta
The xylem in grapevines comprises many interconnected vessels that are finite, but variable in length, and any one vessel rarely spans the entire length of the stem (Thorne et al., 2006b; Tyree and Zimmermann, 2002). These xylem vessels are connected to each other by scalariform bordered pits that separate vessel elements from one another via pit membranes, which are composed of primary plant cell wall material (cellulose microfibrils embedded in a meshwork of hemicellulose and pectin) (Buchanan et al., 2000; Choat et al., 2004). The pores within the pit membrane range from 5 to 20 nm in diameter and allow for the flow of water and small solutes from one vessel to another, but block air embolisms and pathogen movement (Stevenson et al., 2004; Tyree and Zimmermann, 2002). Xylella fastidiosa is too large to passively move through the pores of the pit membranes (Mollenhauer and Hopkins, 1974), and uses enzymatic degradation of the pit membranes as its primary means of achieving systemic colonization. Xylella fastidiosa utilizes an endo‐polygalacturonase (endo‐PG) to digest polymers with a pectin backbone and requires this endo‐PG to systemically colonize grapevines. A deletion mutation in the single copy gene encoding endo‐PG resulted in a complete loss of pathogenicity (Roper et al., 2007b). Interestingly, effective degradation of the pit membranes requires a concerted effort with an endo‐PG and at least one endoglucanase (EGase), EngXCA2, indicating that effective degradation of the plant cell wall meshwork is a cooperative effort between at least two classes of cell wall‐degrading enzyme (CWDE) (Perez‐Donoso et al., 2010). EngXCA2 is a prototypical EGase that degrades both carboxymethylcellulose and xyloglucan substrates (Perez‐Donoso et al., 2010). engXCA1 (PD1856) and egl (PD2061) putatively encode two additional EGases, but these have not been fully characterized. Xylella fastidiosa also possesses three genes that encode S8 subtilisin‐like serine protease autotransporters (PD0218, PD0313 and PD0950) (Tripathi and Sowdhamini, 2008), and their role in systemic movement in planta is still being investigated.
Xylella fastidiosa lacks flagella, but uses Type IV pili as its only currently known means of motility in the xylem. This surface‐based motility allows the bacterium to migrate basipetally against the flow of xylem sap, which is quite remarkable given the high shear rate of fluid in the xylem that can occur during plant transpiration (De La Fuente et al., 2007; Meng et al., 2005). Calcium enhances the twitching motility via PilY1, a Type IV pilus structural protein. It has been proposed that calcium concentrations help to regulate the twitching motility in the xylem as ionic concentrations of calcium fluctuate in the xylem sap (Cruz et al., 2012, 2014). Movement in the acropetal position could be entirely passive if the bacterium uses the transpiration stream to move towards the tip of the plant.
Regulation of host colonization
It is estimated that only 10%–15% of the vessels are heavily colonized by X. fastidiosa (Newman et al., 2003) and that the majority of colonized vessels contain only a few cells. Too many cells in one vessel would block or limit the flow of xylem sap in that particular vessel and, presumably, the nutrients necessary for bacterial survival (Newman et al., 2003). The current model of cell density‐regulated host colonization (Chatterjee et al., 2008a) indicates that, when vessels become heavily occluded, the Rpf high cell density system down‐regulates genes conducive to systemic spread (i.e. motility and CWDEs) via the accumulation of a diffusible signalling factor (DSF) (Newman et al., 2004). This accumulation of DSF activates the RPF signalling system and enhances the production of various adhesins that increase cell ‘stickiness’ (Chatterjee et al., 2008a). This would allow the cells in heavily occluded vessels to enter the insect‐acquirable state to increase the chances of acquisition and, subsequently, transmission to a new plant by an insect vector. OMV production, which is also linked to adhesiveness, is regulated by cyclic di‐guanosine monophosphate (di‐GMP) signalling via the Rpf DSF (Ionescu et al., 2014). Disruption of the Rpf system results in hyper‐vesiculation (Ionescu et al., 2014), making X. fastidiosa less adhesive and, subsequently, hypervirulent in planta (Newman et al., 2004). [Please see Chatterjee et al. (2008b) and Roper and Lindow (2014) for reviews on the gene regulation hierarchy in X. fastidiosa.]
Insect colonization (acquisition and transmission)
Xylella fastidiosa is the only known insect‐transmitted plant pathogen that is persistent within the sharpshooter vector, but non‐circulative (Backus and Morgan, 2011). It is semi‐persistent in nymphs, i.e. the bacterium is lost after each vector moult, but persistent in non‐moulting adult vectors (Backus and Morgan, 2011; Chatterjee et al., 2008a). Xylella fastidiosa is also propagative within vectors, allowing the insects to transmit the pathogen for months after acquisition from an infected plant (Hill and Purcell, 1995b). There is no evidence of trans‐stadial or trans‐ovarial transmission, meaning that the pathogen does not transfer through insect life stages or to offspring, respectively (Janse and Obradovic, 2010). In contrast with X. fastidiosa strain specificity in plant hosts, there is a lack of vector–X. fastidiosa strain specificity (Damsteegt et al., 2006).
The transmission of X. fastidiosa consists of three steps: (i) pathogen acquisition from an infected host plant; (ii) retention of the pathogen within the vector; and (iii) transmission of the pathogen to a susceptible plant host (Killiny and Almeida, 2014). Vectors can acquire X. fastidiosa and immediately transmit it to a new host, indicating that there is no latent period required for transmission (Purcell and Finlay, 1979). On acquisition, X. fastidiosa attaches to and forms biofilms within the foregut of the insect (Killiny and Almeida, 2009, 2014). Cells initially attach laterally, which increases the cell surface area in contact with the insect cuticle. In the later stages of biofilm formation, the cells become polarly attached to the foregut cuticle, presumably to allow for maximal cell surface exposure to the nutrient‐dilute xylem sap (Almeida and Purcell, 2006; Chatterjee et al., 2008a; Killiny and Almeida, 2009). Xylella fastidiosa colonizes two major regions of the foregut: the precibarium and the cibarium. Although the cibarium is probably the reservoir for bacterial inoculum within the foregut, transient colonization of the precibarium is associated with the transmission of X. fastidiosa to plants (Almeida and Purcell, 2006; Backus and Morgan, 2011; Rapicavoli et al., 2015).
The vector foregut is an extremely turbulent environment, as xylem sap flow rates have been estimated to reach nearly 8 cm/s (Retchless et al., 2014). Therefore, attachment to the foregut is not a trivial process, and relies on numerous factors at the vector–pathogen interface. Several surface‐associated bacterial components, such as Type I pili, haemagglutinin adhesins and fimbriae, contribute to the adhesive properties of X. fastidiosa that are necessary for colonization in insect vectors (Chatterjee et al., 2008b; Feil et al., 2007; Killiny and Almeida, 2009; Newman et al., 2004). In addition, cell surface polysaccharides, namely LPS and EPS, also play a role in the acquisition process. The major surface‐associated polysaccharide, LPS, contributes to the early stages of acquisition of X. fastidiosa by BGSS. A mutant strain with a truncated O‐antigen was impaired in attachment to the cuticular lining of the insect foregut, and thus acquisition of the pathogen was disrupted (Rapicavoli et al., 2015). EPS has also been implicated in the insect transmission of X. fastidiosa by BGSS, as mutants impaired in EPS production were poorly transmitted (Killiny et al., 2013). It is not clear whether this inefficiency is a result of defects in the initial acquisition of X. fastidiosa or the retention of cells over time.
Secretion Systems
Unlike many other phytopathogenic bacteria, X. fastidiosa does not possess a Type III secretion system, a system commonly used to secrete protein effectors that enhance virulence and/or suppress plant defences (Dow and Daniels, 2000). However, X. fastidiosa does possess operons containing the genes necessary to produce components of the Type I, II, IV and V secretion systems (Simpson et al., 2000; Van Sluys et al., 2003). The Type I secretion system is a general secretory pathway that has many functions, including multidrug resistance efflux and the secretion of various proteases, hydrolases and toxins (Delepelaire, 2004). The Type I translocator channel is ATP dependent and comprises two inner membrane proteins that attach to TolC, a periplasm/outer membrane protein which is highly conserved amongst many phytopathogenic bacteria (Holland et al., 2005). A mutation in tolC caused a loss of pathogenicity (Reddy et al., 2007). However, bacteria could not be isolated from grapevines inoculated with the mutant strain, suggesting that the tolC mutation is lethal in planta. The mutant strain also displayed a high level of sensitivity to several phytochemicals, detergents and crude plant extracts, highlighting the necessity of TolC for multidrug efflux and the viability of X. fastidiosa in plants.
Xylella fastidiosa possesses a 12‐protein Type II secretion system (T2SS) that is closely related to those found in Xanthomonas campestris pv. campestris and Xanthomonas oryzae pv. oryzae (Jha et al., 2005). The T2SS is made up of three parts: the outer membrane pore, the inner membrane complex and the pseudopilin complex (Sandkvist, 2001). Pseudopilin formation is driven by XpsE, an ATPase that facilitates the stacking of pseudopilin subunits that push the loaded protein out into the extracellular environment (Jha et al., 2005). Proteins secreted by T2SS are often proteases and plant CWDEs that utilize a secretion signal to direct their transport (Sandkvist, 2001). The majority of the proteases and CWDEs in X. fastidiosa are predicted to be secreted via the T2SS. Mutations in xpsE in X. fastidiosa cause a non‐pathogenic phenotype similar to the pglA mutant (M. C. Roper, unpublished data), indicating a strong connection between T2SS and the secretion of critical CWDEs in X. fastidiosa.
The Type V secretion system comprises a family of autotransporter proteins that transport themselves out of the cell without requiring any energy input (van Ulsen et al., 2014). Xylella fastidiosa has six genes predicted to encode members of the AT‐1 autotransporter family, one of which is XatA (Matsumoto et al., 2012). Its C‐terminal helper domain interacts with the bacterial outer membrane and with OMVs, and its passenger domain is released into the extracellular environment. A xatA mutant formed smaller bacterial aggregates and produced less biofilm relative to the wild‐type parent strain, indicating an important role for XatA in X. fastidiosa adhesion. When inoculated into grapevines, the mutant strain did not induce symptoms of PD, or systemically colonize plants, and was compromised in colonization at the point of inoculation (Matsumoto et al., 2012). The Type IV secretion system in X. fastidiosa has not been characterized and currently nothing is known regarding its function.
A list of known X. fastidiosa virulence/pathogenicity factors can be found in Table 1.
Table 1.
List of virulence/pathogenicity factors for Xylella fastidiosa.
| PD number | Gene | Function | Mutant phenotype in virulence/pathogenicity | Reference(s) |
|---|---|---|---|---|
| Adhesins | ||||
| PD0058 | fimF | Fimbrial adhesin: cell–cell aggregation, biofilm formation | Reduced | Feil et al. (2007) |
| PD0062 | fimA | Fimbrial adhesin: cell–cell aggregation, biofilm formation | Reduced | Feil et al. (2007) |
| PD0731 | xadA | Afimbrial adhesin: cell surface attachment, biofilm formation | Reduced | Feil et al. (2007) |
| PD0986 | Putative haemagglutinin‐like protein | Reduced | Zhang et al. (2015) | |
| PD1792 | hxfB | Haemagglutinin: cell–cell aggregation | Reduced | Feil et al. (2007) |
| Enhanced | Guilhabert and Kirkpatrick (2005) | |||
| PD2118 | hxfA | Haemagglutinin: cell–cell aggregation | Enhanced | Guilhabert and Kirkpatrick (2005) |
| Quorum sensing | ||||
| PD0233 | rpfB | DSF synthesis, quorum sensing | Reduced | Almeida et al. (2012) |
| PD0279 | cgsA | Cyclic di‐GMP synthase A, quorum sensing | Reduced | Chatterjee et al. (2010) |
| PD0406 | rpfC | Quorum sensing | Reduced | Chatterjee et al. (2008b) |
| PD0407 | rpfF | Quorum sensing | Enhanced | Chatterjee et al. (2008b) |
| Secretion systems | ||||
| PD0732 | xpsE | Type II secretion system: ATPase | Loss | M. C. Roper (unpublished data) |
| PD1964 | tolC | Type I secretion system | Loss | Reddy et al. (2007) |
| Polysaccharide synthesis | ||||
| PD0814 | wzy | Lipopolysaccharide synthesis | Reduced | Clifford et al. (2013) |
| PD1391 | gumH | Exopolysaccharide synthesis | Loss | Killiny et al. (2013) |
| PD1394 | gumD | Exopolysaccharide synthesis | Loss | Killiny et al. (2013) |
| Hydrolytic enzymes | ||||
| PD0956 | prtA | Serine protease | Enhanced | Gouran et al. (2016) |
| PD1485 | pglA | Polygalacturonase: pit membrane degradation | Loss | Roper et al. (2007b) |
| PD1703 | lesA/lipA | Lipase/esterase | Reduced | Zhang et al. (2015) and |
| Nascimento et al. (2016) | ||||
| PD1826 | chiA | Chitinase | Loss | Labroussaa et al. (2017) |
| PD1851 | engXCA2 | Endoglucanase: pit membrane degradation | Reduced | M. C. Roper (unpublished data) |
| PD1856 | engXCA1 | Endoglucanase: pit membrane degradation | Reduced | M. C. Roper (unpublished data) |
| Membrane proteins | ||||
| PD0528 | xatA | Cell–cell aggregation, biofilm formation | Reduced | Matsumoto et al. (2012) |
| PD0843 | tonB1 | Iron and vitamin B12 transport | Reduced | Cursino et al. (2009) |
| Stress response | ||||
| PD1380 | csp1 | Cold shock protein | Reduced | Burbank and Stenger (2016) |
| Metabolism | ||||
| PD1311 | Acyl‐coenzyme A (acyl‐CoA) synthetase | Loss | Hao et al. (2017) | |
| Toxins | ||||
| PD0928 | zot | Putative Zot‐like toxin | Reduced | Zhang et al. (2015) |
| Regulatory systems | ||||
| PD0848 | pilL | Type IV pilus: regulates twitching | Reduced | Cursino et al. (2011) |
| PD1099 | dinJ/relE | Toxin–anti‐toxin system, regulatory system | Enhanced | Burbank and Stenger (2017) |
| PD1100 | ||||
| PD1284 | algU | Alternate sigma factor: pathogenicity‐related genes, stress response genes | Reduced | Shi et al. (2007) |
| PD1386 | xhpT | Two‐component regulatory system: pathogenicity‐related genes, EPS synthesis | Reduced | Voegel et al. (2013) |
| PD1671 | Two‐component regulatory system: pathogenicity‐related genes | Enhanced | Cursino et al. (2015) | |
| PD1678 | phoQ | Two‐component regulatory system: pathogenicity‐related genes, survival genes | Loss | Pierce and Kirkpatrick (2015) |
| PD1679 | phoP | Two‐component regulatory system: pathogenicity‐related genes, survival genes | Loss | Pierce and Kirkpatrick (2015) |
| PD1984 | gacA | Two‐component regulatory system: pathogenicity‐related genes | Reduced | Shi et al. (2009) |
DSF, diffusible signalling factor; EPS, exopolysaccharide; GMP, guanosine monophosphate.
Plant Host Responses to X. fastidiosa
The obstruction of xylem vessels by gums and tyloses is a common plant response designed to restrict the movement and systemic infection of vascular pathogens (Clerivet et al., 2000; Sun et al., 2008; Tyree and Zimmermann, 2002; Yadeta and Thomma, 2013). Tyloses are outgrowths of xylem‐associated parenchyma cell walls into the neighbouring vessel and are found in abundance in the late stages of X. fastidiosa infection (when disease symptoms are evident) (Fig. 3B). Tyloses result from the weakening of pit membranes by the activity of CWDEs in the vessel lumen and/or the realignment of new cellulose microfibrils that are being added to the plant cell wall as it expands in the presence of ethylene (Esau, 1977). High ethylene levels have been reported in grape tissues infected with X. fastidiosa (Perez‐Donoso et al., 2007; Sun et al., 2007), which can partially explain the extensive induction of tylose formation during PD. Amorphous gels also occur in the xylem of PD‐infected grapevine (Fig. 3C) and it is presumed that these are of plant origin.
Tyloses can efficiently block X. fastidiosa movement in the stem vasculature and prevent systemic infection if the disassembly of pit membranes in the vessel–xylem parenchyma interface occurs more rapidly than the breakdown of the intervessel pit membrane system (Sun et al., 2013). Tyloses form more rapidly in PD‐resistant species of grapevine, such as species of native muscadine and wild grapevines (i.e. V. rotundifolia and V. arizonica, respectively), than in susceptible species (V. vinifera). The formation of these occlusions restricts the movement of X. fastidiosa into vessel segments beyond the inoculation point (Fry and Milholland, 1990a, 1990b; Mollenhauer and Hopkins, 1976; Sun et al., 2013). Conversely, it appears that the enzymatic breakdown of pit membranes facilitated by X. fastidiosa takes place more quickly in the PD‐susceptible genotype, which allows the pathogen to move systemically before tyloses seal the vessels. Although tylose formation could be a successful mechanism for the restriction of bacterial spread, it has the negative side effect of blocking water flux within the affected xylem vessel (Stevenson et al., 2004). Indeed, whether tyloses in X. fastidiosa‐infected grapevines contribute to disease resistance (i.e. prevention of pathogen movement) or susceptibility (i.e. by reduction of water movement) continues to be a matter of debate (Cantu et al., 2016). Numerous observations have confirmed that extensive tylose formation in compatible X. fastidiosa–host interactions has an impact on plant water transport, which, in turn, worsens the symptoms of the disease (Choi et al., 2013; Machado and Tyree, 1994; Perez‐Donoso et al., 2007; Sun et al., 2013). In addition, X. fastidiosa cells and/or their secretions may also contribute to the blockage of vascular tissues (Fig. 3A). It is well established that plants affected by X. fastidiosa infections are more vulnerable to water stress, possibly as a result of the decrease in xylem hydraulic conductivity, a consequence of stomatal closure and blockage of the vessels (de Souza et al., 2009).
It has been proposed that PD symptoms result from the water deficit caused by xylem occlusions, instead of the bacterial infection itself. However, this hypothesis has been challenged in several studies. For example, Thorne et al. (2006a) exposed ‘Chardonnay’ grapevines to severe water stress, inoculation with X. fastidiosa and a combination of both treatments to determine whether symptoms of PD mimicked those of water deficit. The results showed that visual symptoms of PD were qualitatively and quantitatively different from those caused by water stress. Xylella fastidiosa‐inoculated vines showed several PD‐specific symptoms, such as green islands, matchstick petioles and patchy leaf necrosis, whereas vines exposed to water deficit did not develop any of these hallmark symptoms. It is worth noting that vines used in this comparative study were subjected to moderate and severe water stress conditions for a relatively short period of time (91 days); some of the similarities between PD and drought symptoms could be more evident when studying vines under chronic low‐level water deficit. In another study, Choi et al. (2013) evaluated the transcriptional response of ‘Cabernet Sauvignon’ grapevines to X. fastidiosa inoculation, water deficit and a combination of the two stresses. This report indicated that gene expression changes associated with bacterial inoculation were clearly distinct from those related to water stress. Notably, vines experiencing both pathogen infection and water deficit presented more pronounced expression changes of genes involved in X. fastidiosa‐unique responses, which supports the idea of a direct interaction between PD and grapevine water stress.
Plants can respond to pathogen attack by the induction of defence responses, including phytoalexins, pathogenesis‐related (PR) proteins, oxidative reactions and structural reinforcements (i.e. callose and suberin deposition). Phytoalexins are generally low‐molecular‐weight secondary metabolites with antimicrobial properties that accumulate on pathogen perception. Phenolics, a class of secondary metabolites that are derived from the shikimic acid and phenylpropanoid pathways, are considered to be important phytoalexins in many plant species (Nicholson and Hammerschmidt, 1992). Maddox et al. (2010) examined the induction of phenolic compounds in ‘Thompson Seedless’ grapevines inoculated with X. fastidiosa during a period of 6 months. At 2 months, they observed that X. fastidiosa‐inoculated vines showed increased levels of phenolics (e.g. catechins, procyanidins and stilbenoids) in the xylem sap and neighbouring tissues compared with uninoculated controls. In addition, higher levels of lignin and condensed tannin precursors were found in the xylem cell walls of the inoculated vines. Amongst the detected phenolic compounds, catechol, caffeic acid and resveratrol showed stronger anti‐X. fastidiosa activities when evaluated using in vitro agar dilution assays. However, phenolic accumulation in the grapevines was transient because, at 4 months, the levels of these compounds were equal between X. fastidiosa‐inoculated and control vines, and, at 6 months, when the inoculated vines showed severe PD symptoms, their phenolic levels had dropped significantly. The study concluded that phenolic production in grapevine corresponds to an early response to X. fastidiosa and that it may delay or limit systemic infections, but is not sufficient to successfully control the pathogen. These observations may partially explain why particular grapevine cultivars that fail to induce phenolics to high concentrations, such as ‘Chardonnay’ or ‘Crimson Seedless’, display PD symptoms much sooner than other cultivars (Wallis and Chen, 2012). The accumulation of phenolic compounds has also been reported as a response to X. fastidiosa in CVC‐resistant citrus (de Souza et al., 2007a, 2007b). The induction of genes involved in phenylpropanoid and flavonoid biosynthesis occurs as quickly as 1 day after inoculation in ‘Ponkan’ mandarin (Citrus reticulata Blanco), a CVC‐resistant citrus variety (Rodrigues et al., 2013).
PR proteins are rapidly induced in response to biotic stress and have a variety of antimicrobial properties (van Loon et al., 2006), e.g. contact toxicity, defence signalling and the inhibition of pathogen enzymatic activity. Yang et al. (2011) analysed differential protein accumulation after X. fastidiosa inoculation in two siblings [9621–67 (highly PD resistant) and 9621–94 (highly PD susceptible)] from a cross of V. rupestris × V. arizonica. This study detected a strong induction of a thaumatin‐like protein (TLP, PR protein class 5) at 12 weeks post‐inoculation only in the PD‐resistant genotype, suggesting a role of this protein in limiting X. fastidiosa infection. A separate study also identified the presence of TLPs in the xylem sap of a PD‐tolerant Vitis species in response to X. fastidiosa inoculation (Basha et al., 2010). Strong up‐regulation of several PR proteins (e.g. PR protein class 1) was observed at 30 days post‐inoculation with X. fastidiosa in ‘Ponkan’ mandarin when compared against a CVC‐susceptible genotype, indicating that these proteins may be necessary for disease resistance in citrus (Campos et al., 2007).
Control of PD
The most widely used methods of control for PD are: (i) severe pruning and roguing of infected vines; and (ii) control of the insect vector via insecticide applications. These insecticides are critical for the control of PD in California and are based primarily on systemic conventional insecticides (i.e. imidacloprid). Coordinated area‐wide insecticide sprays for H. vitripennis were able to restore grape production to economically viable levels in the Temecula Valley. Growers often apply within‐vineyard applications of the insecticide imidacloprid to complement the area‐wide control applications to control H. vitripennis. It is not clear whether within‐vineyard applications are necessary for the control of PD or if a grower can rely solely on area‐wide management for sufficient PD control. One study indicated that disease incidence was less in vineyards in which within‐vineyard applications were applied, but these benefits may take several growing seasons to become apparent (Daugherty et al. 2015). In addition, modelling of vector behaviour in the context of climate change suggests that warmer temperatures would favour an increase in PD incidence (Daugherty et al., 2017).
A robust grapevine breeding programme has also been established to evaluate germplasm for PD resistance. Although no V. vinifera genotypes resistant to PD have been identified, genetic sources of disease resistance are present in the Vitis germplasm. Wild Vitis populations with varying degrees of PD resistance or tolerance have been identified in the southeastern USA and northern Mexico (Krivanek et al., 2005). Marker‐assisted selection has been successfully used to introduce PD defence genes from wild Vitis genotypes into economically important V. vinifera varieties. One of the genes responsible for the non‐vinifera resistance was mapped on chromosome 14 and named ‘Pierce's disease resistance 1’ (PdR1) (Krivanek et al., 2006). Marker‐assisted selection has been used to introgress this resistance trait into V. vinifera genotypes. The idea is that these PD‐resistant vines can be planted in PD hotspots in northern California.
The availability of genetic transformation approaches for V. vinifera has opened up the possibilities of using biotechnology to improve PD resistance in susceptible genotypes (Aguero et al., 2005; Dandekar et al., 2012; Lindow et al., 2014). Because of the negative perception of genetically modified organisms, various strategies have been suggested to use biotechnology for traditional breeding and viticulture practices. One example is trans‐grafting, which refers to the grafting of non‐transgenic scions to transgenic rootstocks (Song et al., 2015). This approach is based on the hypothesis that rootstocks expressing different PD resistance‐associated transgenes could potentially confer resistance to the non‐transgenic scion. A transgenic grapevine rootstock variety engineered to express X. fastidiosa DSF synthase developed fewer PD symptoms and decreased the systemic movement of the bacterium within the plant in glasshouse trials (Lindow et al., 2014). As discussed above, the Rpf DSF cell–cell signalling system regulates adhesiveness (amongst other X. fastidiosa phenotypes), and it is hypothesized that overexpression of the signalling factor that influences adhesion locks the bacteria in the adhesive state and decreases systemic movement of the pathogen. In addition, a transgenic rootstock variety expressing a pear polygalacturonase‐inhibiting protein (PGIP) (which presumably inhibits the PG used by X. fastidiosa to move systemically in the vine) showed tolerance to PD, but not complete resistance, in glasshouse trials. Transgenic rootstock vines expressing a protein chimera consisting of the lytic antimicrobial peptide, cercoprin B, linked to human neutrophil elastase, which binds to the X. fastidiosa outer membrane protein (MopB), exhibited fewer PD symptoms, but were not completely resistant to PD (Dandekar et al., 2012). Moreover, stacking the resistance genes (or transgenes) mentioned above could also be accomplished by crossing plants of the same rootstock variety. Therefore, if transgenic rootstock strategies for PD defence prove to be effective, it is likely that only a few particular rootstock genotypes would be needed for deployment with numerous V. vinifera grape scion varieties (Cantu et al., 2016).
Future Perspectives
As a result of its capacity to undergo interstrain recombination and its incredibly wide host range, X. fastidiosa remains a formidable threat to many susceptible agricultural crops and, perhaps, in plant species in which a pathogenic relationship has yet to be described. Because there are no effective control measures that target the bacterium itself, this is an open area of research. Current efforts are turning towards the natural chemistries produced by grapevine‐associated microbes as potential control measures for PD‐infected vines (Aldrich et al., 2015). This research is centred on understanding the grapevine's dependence on its natural microbiota and how that affects pathogen invasion and disease outcomes. This requires a forward‐thinking, holistic vision of sustainable crop productivity that can only be arrived at through a systems‐level understanding of the diverse interacting components in any given pathosystem.
In addition, there is very little known about how X. fastidiosa interfaces with the plant host immune system. Current work is focused on LPS as a pathogen‐associated molecular pattern (PAMP) and how grapevines differentially perceive full‐length and truncated versions of the O‐antigen molecule with respect to PAMP‐triggered immunity (J. Rapicavoli et al, unpublished data). There are still many unanswered questions regarding X. fastidiosa biology and its interactions with its plant hosts, and several pertinent ones are summarized in Box 1.
Box 1. Questions to be answered for X. fastidiosa and its hosts.
From the plant perspective
How does the plant immune system interface with X. fastidiosa in the xylem?
What are the specific and/or shared mechanisms underlying the susceptibility of different hosts? How can we translate the knowledge we've gained in grapevines to other X. fastidiosa pathosystems?
Many X. fastidiosa plant hosts are non‐symptomatic. Can we block/remove these susceptibility factors in susceptible hosts? Can we learn from non‐symptomatic interactions to make grape and other susceptible hosts tolerant?
From the pathogen perspective
X. fastidiosa lacks a Type Ill secretion system and its effectors. Do the Type I, II, IV, and V systems secrete effectors that play role(s) in pathogenicity?
How does X. fastidiosa interface with other microbes in the host plant's phytobiome?
Acknowledgements
The authors would like to thank David Doll, Rodrigo Krugner, Philippe Rolshausen and Qiang Sun for providing images of disease symptoms. This work was supported, in part, by the California Department of Food and Agriculture grant agreement numbers 15–0218‐SA and 14–014‐SA and the University of California Agricultural Experiment Station.
In memory of Professor Bruce Kirkpatrick, respected scholar, mentor and friend. You will always be missed.
References
- Abbott, A. (2015). Italian scientists under investigation after olive‐tree deaths. Nature, doi: 10.1038/nature.2015.19078 . [DOI]
- Aguero, C.B. , Uratsu, S.L. , Greve, C. , Powell, A.L.T. , Labavitch, J.M. , Meredith, C.P. and Dandekar, A.M. (2005) Evaluation of tolerance to Pierce's disease and Botrytis in transgenic plants of Vitis vinifera L. expressing the pear PGIP gene. Mol. Plant Pathol. 6, 43–51. [DOI] [PubMed] [Google Scholar]
- Aldrich, T.J. , Rolshausen, P.E. , Roper, M.C. , Reader, J.M. , Steinhaus, M.J. , Rapicavoli, J. , Vosburg, D.A. and Maloney, K.N. (2015) Radicinin from Cochliobolus sp. inhibits Xylella fastidiosa, the causal agent of Pierce's Disease of grapevine. Phytochemistry, 116, 130–137. [DOI] [PubMed] [Google Scholar]
- Almeida, R.P.P. (2016) ECOLOGY. Can Apulia's olive trees be saved? Science, 353, 346–348. [DOI] [PubMed] [Google Scholar]
- Almeida, R.P.P. and Nunney, L. (2015) How do plant diseases caused by Xylella fastidiosa emerge? Plant Dis. 99, 1457–1467. [DOI] [PubMed] [Google Scholar]
- Almeida, R.P.P. and Purcell, A.H. (2006) Patterns of Xylella fastidiosa colonization on the precibarium of sharpshooter vectors relative to transmission to plants. Ann. Entomol. Soc. Am. 99, 884–890. [Google Scholar]
- Almeida, R.P.P. , Killiny, N. , Newman, K.L. , Chatterjee, S. , Ionescu, M. and Lindow, S.E. (2012) Contribution of rpfB to cell‐to‐cell signal synthesis, virulence, and vector transmission of Xylella fastidiosa . Mol. Plant–Microbe Interact. 25, 453–462. [DOI] [PubMed] [Google Scholar]
- Alston, J.M. , Fuller, K.B. , Kaplan, J.D. and Tumber, K.P. (2013) Economic consequences of Pierce's disease and related policy in the California winegrape industry. J. Agr. Resource Econ. 38, 269–297. [Google Scholar]
- Alston, J.M. , Fuller, K.B. , Kaplan, J.D. and Tumber, K.P. (2015) Assessing the returns to R&D on perennial crops: the costs and benefits of Pierce's disease research in the California winegrape industry. Aust. J. Agr. Resource Econ. 59, 95–115. [Google Scholar]
- Backus, E.A. and Morgan, D.J.W. (2011) Spatiotemporal colonization of Xylella fastidiosa in its vector supports the role of egestion in the inoculation mechanism of foregut‐borne plant pathogens. Phytopathology, 101, 912–922. [DOI] [PubMed] [Google Scholar]
- Backus, E.A. , Shugart, H.J. , Rogers, E.E. , Morgan, J.K. and Shatters, R. (2015) Direct evidence of egestion and salivation of Xylella fastidiosa suggests sharpshooters can be “Flying Syringes”. Phytopathology, 105, 608–620. [DOI] [PubMed] [Google Scholar]
- Basha, S.M. , Mazhar, H. and Vasanthaiah, H.K. (2010) Proteomics approach to identify unique xylem sap proteins in Pierce's disease‐tolerant Vitis species. Appl. Biochem. Biotechnol. 160, 932–944. [DOI] [PubMed] [Google Scholar]
- Blua, M. , Phillips, P. and Redak, R. (1999) A new sharpshooter threatens both crops and ornamentals. Calif. Agr. 53, 22–25. [Google Scholar]
- Bove, J.M. and Ayres, A.J. (2007) Etiology of three recent diseases of citrus in Sao Paulo State: sudden death, variegated chlorosis and huanglongbing. IUBMB Life, 59, 346–354. [DOI] [PubMed] [Google Scholar]
- Brlansky, R.H. , Timmer, L.W. , French, W.J. and McCoy, R.E. (1983) Colonization of the sharpshooter vectors, Oncometopia nigricans and Homalodisca coagulata, by xylem‐limited bacteria. Phytopathology, 73, 530–535. [Google Scholar]
- Buchanan, B.B. , Gruissem, W. and Jones, R.L. (2000). Biochemistry & Molecular Biology of Plants. Rockville, MD: American Society of Plant Physiologists. [Google Scholar]
- Burbank, L.P. and Stenger, D.C. (2016) A temperature‐independent cold‐shock protein homolog acts as a virulence factor in Xylella fastidiosa . Mol. Plant–Microbe Interact. 29, 335–344. [DOI] [PubMed] [Google Scholar]
- Burbank, L.P. and Stenger, D.C. (2017) The DinJ/RelE toxin–antitoxin system suppresses bacterial proliferation and virulence of Xylella fastidiosa in grapevine. Phytopathology, 107, 388–394. [DOI] [PubMed] [Google Scholar]
- Campos, M.A. , Rosa, D.D. , Teixeira, J.É.C. , Targon, M.L.P. , Souza, A.A. , Paiva, L.V. , Stach‐Machado, D.R. and Machado, M.A. (2007) PR gene families of citrus: their organ‐specific biotic and abiotic inducible expression profiles based on ESTs approach. Genet. Mol. Biol. 30, 917–930. [Google Scholar]
- Cantu, D. , Roper, M.C. , Powell, A.L. and Labavitch, J.M. (2016) Problematic crops: 1. Grape: to long life and good health: untangling the complexity of grape diseases to develop pathogen‐resistant varieties. In: Plant Pathogen Resistance Biotechnology (David Collinge., Eds), p. 193 Hoboken, New Jersey: John Wiley and Sons, Inc. [Google Scholar]
- California Department of Food and Agriculture (CDFA) (2015) California agricultural production statistics. Available at https://www.cdfa.ca.gov/statistics/.
- Chatterjee, S. , Almeida, R.P.P. and Lindow, S. (2008a) Living in two worlds: the plant and insect lifestyles of Xylella fastidiosa . Ann. Rev. Phytopathol. 46, 243–271. [DOI] [PubMed] [Google Scholar]
- Chatterjee, S. , Wistrom, C. and Lindow, S.E. (2008b) A cell–cell signaling sensor is required for virulence and insect transmission of Xylella fastidiosa . Proc. Natl. Acad. Sci. USA, 105, 2670–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, S. , Killiny, N. , Almeida, R.P.P. and Lindow, S.E. (2010) Role of cyclic di‐GMP in Xylella fastidiosa biofilm formation, plant virulence, and insect transmission. Mol. Plant–Microbe Interact. 23, 1356–1363. [DOI] [PubMed] [Google Scholar]
- Chen, J. , Xie, G. , Han, S. , Chertkov, O. , Sims, D. and Civerolo, E.L. (2010) Whole genome sequences of two Xylella fastidiosa strains (M12 and M23) causing almond leaf scorch disease in California. J. Bacteriol. 192, 4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. , Wu, F. , Zheng, Z. , Deng, X. , Burbank, L.P. and Stenger, D.C. (2016) Draft genome sequence of Xylella fastidiosa subsp. fastidiosa Strain Stag's Leap. Genome Announc. 4, e00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choat, B. , Jansen, S. , Zwieniecki, M.A. , Smets, E. and Holbrook, N.M. (2004) Changes in pit membrane porosity due to deflection and stretching: the role of vestured pits. J. Exp. Bot. 55, 1569–1575. [DOI] [PubMed] [Google Scholar]
- Choi, H.K. , Iandolino, A. , da Silva, F.G. and Cook, D.R. (2013) Water deficit modulates the response of Vitis vinifera to the Pierce's Disease pathogen Xylella fastidiosa . Mol. Plant–Microbe Interact. 26, 643–657. [DOI] [PubMed] [Google Scholar]
- Clerivet, A. , Deon, V. , Alami, I. , Lopez, F. , Geiger, J.P. and Nicole, M. (2000) Tyloses and gels associated with cellulose accumulation in vessels are responses of plane tree seedlings (Platanus × acerifolia) to the vascular fungus Ceratocystis fimbriata f. sp platani. Trees, 15, 25–31. [Google Scholar]
- Clifford, J.C. , Rapicavoli, J.N. and Roper, M.C. (2013) A rhamnose‐rich O‐antigen mediates adhesion, virulence, and host colonization for the xylem‐limited phytopathogen Xylella fastidiosa . Mol. Plant–Microbe Interact. 26, 676–685. [DOI] [PubMed] [Google Scholar]
- Cruz, L.F. , Cobine, P.A. and De La Fuente, L. (2012) Calcium increases Xylella fastidiosa surface attachment, biofilm formation, and twitching motility. Appl. Environ. Microbiol. 78, 1321–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz, L.F. , Parker, J.K. , Cobine, P.A. and De la Fuente, L. (2014) Calcium‐enhanced twitching motility in Xylella fastidiosa is linked to a single PilY1 homolog. Appl. Environ. Microbiol. 80, 7176–7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cursino, L. , Li, Y.X. , Zaini, P.A. , De La Fuente, L. , Hoch, H.C. and Burr, T.J. (2009) Twitching motility and biofilm formation are associated with tonB1 in Xylella fastidiosa . FEMS Microbiol. Lett. 299, 193–199. [DOI] [PubMed] [Google Scholar]
- Cursino, L. , Galvani, C.D. , Athinuwat, D. , Zaini, P.A. , Li, Y.X. , De La Fuente, L. , Hoch, H.C. , Burr, T.J. and Mowery, P. (2011) Identification of an operon, Pil‐Chp, that controls twitching motility and virulence in Xylella fastidiosa . Mol. Plant–Microbe Interact. 24, 1198–1206. [DOI] [PubMed] [Google Scholar]
- Cursino, L. , Athinuwat, D. , Patel, K.R. , Galvani, C.D. , Zaini, P.A. , Li, Y. , De La Fuente, L. , Hoch, H.C. , Burr, T.J. and Mowery, P. (2015) Characterization of the Xylella fastidiosa PD1671 gene encoding degenerate c‐di‐GMP GGDEF/EAL domains, and its role in the development of Pierce's disease. PLoS One, 10, e0121851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damsteegt, V.D. , Brlansky, R.H. , Phillips, P.A. and Roy, A. (2006) Transmission of Xylella fastidiosa, causal agent of citrus variegated chlorosis, by the glassy‐winged sharpshooter, Homalodisca coagulata . Plant Dis. 90, 567–570. [DOI] [PubMed] [Google Scholar]
- Dandekar, A.M. , Gouran, H. , Ibanez, A.M. , Uratsu, S.L. , Aguero, C.B. , McFarland, S. , Borhani, Y. , Feldstein, P.A. , Bruening, G. , Nascimento, R. , Goulart, L.R. , Pardington, P.E. , Chaudhary, A. , Norvell, M. , Civerolo, E. and Gupta, G. (2012) An engineered innate immune defense protects grapevines from Pierce disease. Proc. Natl. Acad. Sci. USA, 109, 3721–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danhorn, T. and Fuqua, C. (2007) Biofilm formation by plant‐associated bacteria. Annu. Rev. Microbiol. 61, 401–422. [DOI] [PubMed] [Google Scholar]
- Daugherty, M.P. , O'Neill, S. , Byrne, F. , and Zeilinger, A. (2015). Is vector control sufficient to limit pathogen spread in vineyards?. Environ. Entomol, 44, 789–797. [DOI] [PubMed] [Google Scholar]
- Daugherty, M. , Zeilinger, A.R. and Almeida, R. (2017) Conflicting effects of climate and vector behavior on the spread of a plant pathogen. Phytobiomes, 1, 46–53. [Google Scholar]
- Davis, M.J. , Purcell, A.H. and Thomson, S.V. (1978) Pierce's disease of grapevines: isolation of the causal bacterium. Science, 199, 75–77. [DOI] [PubMed] [Google Scholar]
- De La Fuente, L. , Burr, T.J. and Hoch, H.C. (2007) Mutations in type I and type IV pilus biosynthetic genes affect twitching motility rates in Xylella fastidiosa . J. Bacteriol. 189, 7507–7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delepelaire, P. (2004) Type I secretion in gram‐negative bacteria. Biochim. Biophys. Acta, 1694, 149–161. [DOI] [PubMed] [Google Scholar]
- Dow, J.M. and Daniels, M.J. (2000) Xylella genomics and bacterial pathogenicity to plants. Yeast, 17, 263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau, K. (1977) Anatomy of Seed Plants, 2nd edn. New York: Wiley. [Google Scholar]
- European Food Safety Authority (EFSA) (2016) Update of a database of host plants of Xylella fastidiosa: 20 November 2015. EFSA J. 14, 4378. [Google Scholar]
- European Food Safety Authority (EFSA) Panel on Plant Health. (2015) Scientific opinion on the risks to plant health posed by Xylella fastidiosa in the EU territory, with the identification and evaluation of risk reduction options. EFSA J. 13, 3989. [Google Scholar]
- Feil, H. , Feil, W.S. and Lindow, S.E. (2007) Contribution of fimbrial and afimbrial adhesins of Xylella fastidiosa to attachment to surfaces and virulence to grape. Phytopathology, 97, 318–324. [DOI] [PubMed] [Google Scholar]
- Fry, S. and Milholland, R. (1990a) Multiplication and translocation of Xylella fastidiosa in petioles and stems of grapevine resistant, tolerant, and susceptible to Pierce's disease. Phytopathology, 80, 61–65. [Google Scholar]
- Fry, S. and Milholland, R. (1990b) Response of resistant, tolerant, and susceptible grapevine tissues to invasion by the Pierce's disease bacterium, Xylella fastidiosa . Phytopathology, 80, 66–69. [Google Scholar]
- Giampetruzzi, A. , Chiumenti, M. , Saponari, M. , Donvito, G. , Italiano, A. , Loconsole, G. , Boscia, D. , Cariddi, C. , Martelli, G.P. and Saldarelli, P. (2015) Draft genome sequence of the Xylella fastidiosa CoDiRO Strain. Genome Announc. 3, e01538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves, F.P. , Stuchi, E.S. , Lourenço, S.A. , Kriss, A.B. , Gottwald, T.R. , and Amorim, L. (2014). The effect of irrigation on development of citrus variegated chlorosis symptoms. Crop Prot, 57, 8–14. [Google Scholar]
- Goncalves, F.P. , Stuchi, E.S. , Lourenco, S.A. , Hau, B. and Amorim, L. (2012) Relationship between sweet orange yield and intensity of citrus variegated chlorosis. Plant Pathol. 61, 641–647. [Google Scholar]
- Gouran, H. , Gillespie, H. , Nascimento, R. , Chakraborty, S. , Zaini, P.A. , Jacobson, A. , Phinney, B.S. , Dolan, D. , Durbin‐Johnson, B.P. , Antonova, E.S. , Lindow, S.E. , Mellema, M.S. , Goulart, L.R. and Dandekar, A.M. (2016) The secreted protease PrtA controls cell growth, biofilm formation and pathogenicity in Xylella fastidiosa . Sci. Rep. 6, 31 098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, W. , Shao, J. , Zhao, T. and Huang, Q. (2014) Genome sequence of a Xylella fastidiosa strain causing mulberry leaf scorch disease in Maryland. Genome Announc. 2, e00916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilhabert, M.R. and Kirkpatrick, B.C. (2005) Identification of Xylella fastidiosa antivirulence genes: hemagglutinin adhesins contribute to X. fastidiosa biofilm maturation and colonization and attenuate virulence. Mol. Plant–Microbe Interact. 18, 856–868. [DOI] [PubMed] [Google Scholar]
- Hao, L. , Johnson, K. , Cursino, L. , Mowery, P. and Burr, T.J. (2017) Characterization of the Xylella fastidiosa PD1311 gene mutant and its suppression of Pierce's disease on grapevines. Mol. Plant Pathol. 18, 684–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt, W.B. , Houston, B.R. , Frazier, N.W. and Freitag, J.H. (1946) Leafhopper transmission of the virus causing Pierce's disease of grape and dwarf of alfalfa. Phytopathology, 36, 117–128. [PubMed] [Google Scholar]
- Hewitt, W.B. , Frazier, N.W. , Freitag, J.H. and Winkler, A.J. (1949) Pierce's disease investigations. Calif. Agr. 19, 207–264. [Google Scholar]
- Hill, B.L and Purcell, A.H. (1995a) Multiplication and movement of Xylella fastidiosa within grapevine and four other plants. Phytopathology, 85, 1368–1372. [Google Scholar]
- Hill, B.L. and Purcell, A.H. (1995b) Acquisition and retention of Xylella fastidiosa by an efficient vector, Graphocephala atropunctata . Phytopathology, 85, 209–212. [Google Scholar]
- Hill, B.L. and Purcell, A.H. (1997) Populations of Xylella fastidiosa in plants required for transmission by an efficient vector. Phytopathology, 87, 1197–1201. [DOI] [PubMed] [Google Scholar]
- Holland, I.B. , Schmitt, L. and Young, J. (2005) Type 1 protein secretion in bacteria, the ABC‐transporter dependent pathway. Mol. Membr. Biol. 22, 29–39. [DOI] [PubMed] [Google Scholar]
- Hopkins, D.L. and Mollenhauer, H.H. (1973) Rickettsia‐like bacterium associated with Pierce's disease of grapes. Science, 179, 298–300. [DOI] [PubMed] [Google Scholar]
- Hopkins, D.L. and Purcell, A.H. (2002) Xylella fastidiosa: cause of Pierce's disease of grapevine and other emergent diseases. Plant Dis. 86, 1056–1066. [DOI] [PubMed] [Google Scholar]
- Ionescu, M. , Zaini, P.A. , Baccari, C. , Tran, S. , da Silva, A.M. and Lindow, S.E. (2014) Xylella fastidiosa outer membrane vesicles modulate plant colonization by blocking attachment to surfaces. Proc. Natl. Acad. Sci. USA, 111, E3910–E3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques, M.‐A. , Denancé, N. , Legendre, B. , Morel, E. , Briand, M. , Mississipi, S. Durand, K., Olivier, V., Portier, P., Poliakoff, F. and Crouzillat, D. (2015) New coffee plant‐infecting Xylella fastidiosa variants derived via homologous recombination. Appl. Environ. Microbiol. 82, 1556–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janse, J.D. and Obradovic, A. (2010) Xylella fastidiosa: its biology, diagnosis, control and risks. J. Plant Pathol. 92, S35–S48. [Google Scholar]
- Jha, G. , Rajeshwari, R. and Sonti, R.V. (2005) Bacterial type two secretion system secreted proteins: double‐edged swords for plant pathogens. Mol. Plant–Microbe Interact. 18, 891–898. [DOI] [PubMed] [Google Scholar]
- Killiny, N. and Almeida, R.P.P. (2009) Xylella fastidiosa afimbrial adhesins mediate cell transmission to plants by leafhopper vectors. Appl. Environ. Microbiol. 75, 521–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killiny, N. and Almeida, R.P.P. (2014) Factors affecting the initial adhesion and retention of the plant pathogen Xylella fastidiosa in the foregut of an insect vector. Appl. Environ. Microbiol. 80, 420–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killiny, N. , Martinez, R.H. , Dumenyo, C.K. , Cooksey, D.A. and Almeida, R.P.P. (2013) The exopolysaccharide of Xylella fastidiosa is essential for biofilm formation, plant virulence, and vector transmission. Mol. Plant–Microbe Interact. 26, 1044–1053. [DOI] [PubMed] [Google Scholar]
- Krivanek, A. , Famula, T. , Tenscher, A. and Walker, M. (2005) Inheritance of resistance to Xylella fastidiosa within a Vitis rupestris × Vitis arizonica hybrid population. Theor. Appl. Genet. 111, 110–119. [DOI] [PubMed] [Google Scholar]
- Krivanek, A.F. , Riaz, S. and Walker, M.A. (2006) Identification and molecular mapping of PdR1, a primary resistance gene to Pierce's disease in Vitis . Theor. Appl. Genet. 112, 1125–1131. [DOI] [PubMed] [Google Scholar]
- Labroussaa, F. , Ionescu, M. , Zeilinger, A.R. , Lindow, S.E. and Almeida, R.P. (2017) A chitinase is required for Xylella fastidiosa colonization of its insect and plant hosts. Microbiology, 163, 502. [DOI] [PubMed] [Google Scholar]
- Lindow, S. , Newman, K. , Chatterjee, S. , Baccari, C. , Iavarone, A.T. , and Ionescu, M. (2014) Production of Xylella fastidiosa diffusible signal factor in transgenic grape causes pathogen confusion and reduction in severity of Pierce's disease. Mol Plant Microbe Interact, 27, 244‐254. [DOI] [PubMed] [Google Scholar]
- Loconsole, G. , Potere, O. , Boscia, D. , Altamura, G. , Djelouah, K. , Elbeaino, T. , Frasheri, D. , Lorusso, D. , Palmisano, F. , Pollastro, P. , Silletti, M.R. , Trisciuzzi, N. , Valentini, F. , Savino, V. and Saponari, M. (2014) Detection of Xylella fastidiosa in olive trees by molecular and serological methods. J. Plant Pathol. 96, 7–14. [Google Scholar]
- van Loon, L.C. , Rep, M. and Pieterse, C.M. (2006) Significance of inducible defense‐related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. [DOI] [PubMed] [Google Scholar]
- Luvisi, A. , Nicolì, F. and Bellis, L.D. (2017) Sustainable management of plant quarantine pests: the case of olive quick decline syndrome. Sustainability, 9, 659. [Google Scholar]
- Machado, J.‐L. and Tyree, M.T. (1994) Patterns of hydraulic architecture and water relations of two tropical canopy trees with contrasting leaf phenologies: Ochroma pyramidale and Pseudobombax septenatum . Tree Physiol. 14, 219–240. [DOI] [PubMed] [Google Scholar]
- Maddox, C.E. , Laur, L.M. and Tian, L. (2010) Antibacterial activity of phenolic compounds against the phytopathogen Xylella fastidiosa . Curr Microbiol. 60, 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcelletti, S. and Scortichini, M. (2016) Xylella fastidiosa CoDiRO strain associated with the olive quick decline syndrome in southern Italy belongs to a clonal complex of the subspecies pauca that evolved in Central America. Microbiology, 162, 2087–2098. [DOI] [PubMed] [Google Scholar]
- Martelli, G.P. , Boscia, D. , Porcelli, F. and Saponari, M. (2016) The olive quick decline syndrome in south‐east Italy: a threatening phytosanitary emergency. Eur. J. Plant Pathol. 144, 235–243. [Google Scholar]
- Matsumoto, A. , Huston, S.L. , Killiny, N. and Igo, M.M. (2012) XatA, an AT‐1 autotransporter important for the virulence of Xylella fastidiosa Temecula1. Microbiologyopen, 1, 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, Y. , Li, Y. , Galvani, C.D. , Hao, G. , Turner, J.N. , Burr, T.J. and Hoch, H.C. (2005) Upstream migration of Xylella fastidiosa via pilus‐driven twitching motility. J Bacteriol. 187, 5560–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollenhauer, H.H. and Hopkins, D.L. (1974) Ultrastructural study of Pierce's Disease bacterium in grape xylem tissue. J. Bacteriol. 119, 612–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollenhauer, H.H. and Hopkins, D.L. (1976) Xylem morphology of Pierce's disease‐infected grapevines with different levels of tolerance. Physiol. Plant Pathol. 9, 95–96. [Google Scholar]
- Nascimento, R. , Gouran, H. , Chakraborty, S. , Gillespie, H.W. , Almeida‐Souza, H.O. , Tu, A. , Rao, B.J. , Feldstein, P.A. , Bruening, G. , Goulart, L.R. and Dandekar, A.M. (2016) The type II secreted lipase/esterase LesA is a key virulence factor required for Xylella fastidiosa pathogenesis in grapevines. Sci. Rep. 6, 18 598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, K.L. , Almeida, R.P. , Purcell, A.H. and Lindow, S.E. (2003) Use of a green fluorescent strain for analysis of Xylella fastidiosa colonization of Vitis vinifera . Appl. Environ. Microbiol. 69, 7319–7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, K.L. , Almeida, R.P.P. , Purcell, A.H. and Lindow, S.E. (2004) Cell–cell signaling controls Xylella fastidiosa interactions with both insects and plants. Proc. Natl. Acad. Sci. USA, 101, 1737–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson, R.L. and Hammerschmidt, R. (1992) Phenolic compounds and their role in disease resistance. Annu. Rev. Phytopathol. 30, 369–389. [Google Scholar]
- Nunney, L. , Hopkins, D.L. , Morano, L.D. , Russell, S.E. and Stouthamer, R. (2014) Intersubspecific recombination in Xylella fastidiosa strains native to the United States: infection of novel hosts associated with an unsuccessful invasion. Appl. Environ. Microbiol. 80, 1159–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, P.S. (2015) Xylella fastidiosa: a new menace for Portuguese agriculture and forestry. Revista de Ciências Agrárias, 38, 149–154. [Google Scholar]
- Perez‐Donoso, A.G. , Greve, L.C. , Walton, J.H. , Shackel, K.A. and Labavitch, J.M. (2007) Xylella fastidiosa infection and ethylene exposure result in xylem and water movement disruption in grapevine shoots. Plant Physiol. 143, 1024–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Donoso, A.G. , Sun, Q. , Roper, M.C. , Greve, L.C. , Kirkpatrick, B. and Labavitch, J.M. (2010) Cell wall‐degrading enzymes enlarge the pore size of intervessel pit membranes in healthy and Xylella fastidiosa infected grapevines. Plant Physiol. 152, 1748–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perring, T. , Farrar, C. and Blua, M. (2001) Proximity to citrus influences Pierce's disease in Temecula Valley vineyards. Calif. Agr. 55, 13–18. [Google Scholar]
- Pierce, B.K. and Kirkpatrick, B.C. (2015) The PhoP/Q two‐component regulatory system is essential for Xylella fastidiosa survival in Vitis vinifera grapevines. Physiol. Mol. Plant Pathol. 89, 55–61. [Google Scholar]
- Pierce, N.B. (1892). The California vine disease: a preliminary report of investigations. US Government Printing Office, Washington D.C.
- Purcell, A. (1974) Spatial patterns of Pierce's disease in the Napa Valley. Am. J. Enol. Vitic. 25, 162–167. [Google Scholar]
- Purcell, A.H. (1975) Role of the blue–green sharpshooter, Hordnia circellata, in the epidemiology of Pierce's disease of grapevines. Environ. Entomol. 4, 745–752. [Google Scholar]
- Purcell, A.H. and Finlay, A. (1979) Evidence for noncirculative transmission of Pierce's disease bacterium by sharpshooter leafhoppers. Phytopathology, 69, 393–395. [Google Scholar]
- Purcell, A.H. and Saunders, S.R. (1999) Fate of Pierce's disease strains of Xylella fastidiosa in common riparian plants in California. Plant Dis. 83, 825–830. [DOI] [PubMed] [Google Scholar]
- Raju, B. , Goheen, A. , Teliz, D. and Nyland, G. (1980) Pierce's disease of grapevines in Mexico. Plant Dis. 64, 280–282. [Google Scholar]
- Rapicavoli, J.N. , Kinsinger, N. , Perring, T.M. , Backus, E.A. , Shugart, H.J. , Walker, S. and Roper, M.C. (2015) O‐Antigen modulates insect vector acquisition of the bacterial plant pathogen Xylella fastidiosa . Appl. Environ. Microbiol. 81, 8145–8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, J.D. , Reddy, S.L. , Hopkins, D.L. and Gabriel, D.W. (2007) TolC is required for pathogenicity of Xylella fastidiosa in Vitis vinifera grapevines. Mol. Plant–Microbe Interact. 20, 403–410. [DOI] [PubMed] [Google Scholar]
- Retchless, A.C. , Labroussaa, F. , Shapiro, L. , Stenger, D.C. , Lindow, S.E. , and Almeida, R.P. (2014). Genomic insights into Xylella fastidiosa interactions with plant and insect hosts In: Genomics of Plant‐Associated Bacteria, pp. 177–202. Berlin: Springer‐Verlag. [Google Scholar]
- Rodrigues, C.M. , de Souza, A.A. , Takita, M.A. , Kishi, L.T. and Machado, M.A. (2013) RNA‐Seq analysis of Citrus reticulata in the early stages of Xylella fastidiosa infection reveals auxin‐related genes as a defense response. BMC Genomics, 14, 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper, M.C. and Lindow, S. (2014). Xylella fastidiosa: insights into the lifestyle of a xylem‐limited bacterium. In: Virulence Mechanisms of Plant Pathogenic Bacteria (de la Fuente, L. and Wang, N., eds), pp. 307–320. St. Paul, MN: American Phytopathological Society Press.
- Roper, M.C. , Greve, L.C. , Labavitch, J.A. and Kirkpatrick, B.C. (2007a) Detection and visualization of an exopolysaccharide produced by Xylella fastidiosa in vitro and in planta . Appl. Environ. Microbiol. 73, 7252–7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper, M.C. , Greve, L.C. , Warren, J.G. , Labavitch, J.M. and Kirkpatrick, B.C. (2007b) Xylella fastidiosa requires polygalacturonase for colonization and pathogenicity in Vitis vinifera grapevines. Mol. Plant–Microbe Interact. 20, 411–419. [DOI] [PubMed] [Google Scholar]
- Sandkvist, M. (2001) Biology of type II secretion. Mol. Microbiol. 40, 271–283. [DOI] [PubMed] [Google Scholar]
- Saponari, M. , Loconsole, G. , Cornara, D. , Yokomi, R.K. , De Stradis, A. , Boscia, D. , Bosco, D. , Martelli, G.P. , Krugner, R. , and Porcelli, F. (2014) Infectivity and transmission of Xylella fastidiosa by Philaenus spumarius (Hemiptera: Aphrophoridae) in Apulia, Italy. J. Econ. Entomol. 107, 1316–1319. [DOI] [PubMed] [Google Scholar]
- Saponari, M. , Boscia, D. , Loconsole, G. , Palmisano, F. , Savino, V. , Potere, O. and Martelli, G.P. (2014) New hosts of Xylella fastidiosa strain CODIRO in Apulia. J. Plant Pathol. 96, 611. [Google Scholar]
- Scally, M. , Schuenzel, E.L. , Stouthamer, R. and Nunney, L. (2005) Multilocus sequence type system for the plant pathogen Xylella fastidiosa and relative contributions of recombination and point mutation to clonal diversity. Appl. Environ. Microbiol. 71, 8491–8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuenzel, E.L. , Scally, M. , Stouthamer, R. and Nunney, L. (2005) A multigene phylogenetic study of clonal diversity and divergence in North American strains of the plant pathogen Xylella fastidiosa . Appl. Environ. Microbiol. 71, 3832–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, X.Y. , Dumenyo, C.K. , Hernandez‐Martinez, R. , Azad, H. and Cooksey, D.A. (2007) Characterization of regulatory pathways in Xylella fastidiosa: genes and phenotypes controlled by algU . Appl. Environ. Microbiol. 73, 6748–6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, X.Y. , Dumenyo, C.K. , Hernandez‐Martinez, R. , Azad, H. and Cooksey, D.A. (2009) Characterization of regulatory pathways in Xylella fastidiosa: genes and phenotypes controlled by gacA . Appl. Environ. Microbiol. 75, 2275–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert, J. (2001). Economic impact of Pierce's disease on the California grape industry. In: California Department of Food and Agriculture Pierce's Disease Research Symposium. 111–116.
- da Silva, F.R. , Vettore, A.L. , Kemper, E.L. , Leite, A. and Arruda, P. (2001) Fastidian gum: the Xylella fastidiosa exopolysaccharide possibly involved in bacterial pathogenicity. FEMS Microbiol. Lett. 203, 165–171. [DOI] [PubMed] [Google Scholar]
- Simpson, A.J.G. , Reinach, F.C. , Arruda, P. , Abreu, F.A. , Acencio, M. , Alvarenga, R. , Alves, L.M.C. , Araya, J.E. , Baia, G.S. , Baptista, C.S. , Barros, M.H. , Bonaccorsi, E.D. , Bordin, S. , Bove, J.M. , Briones, M.R.S. , Bueno, M.R.P. , Camargo, A.A. , Camargo, L.E.A. , Carraro, D.M. , Carrer, H. , Colauto, N.B. , Colombo, C. , Costa, F.F. , Costa, M.C.R. , Costa‐Neto, C.M. , Coutinho, L.L. , Cristofani, M. , Dias‐Neto, E. , Docena, C. , El‐Dorry, H. , Facincani, A.P. , Ferreira, A.J.S. , Ferreira, V.C.A. , Ferro, J.A. , Fraga, J.S. , Franca, S.C. , Franco, M.C. , Frohme, M. , Furlan, L.R. , Garnier, M. , Goldman, G.H. , Goldman, M.H.S. , Gomes, S.L. , Gruber, A. , Ho, P.L. , Hoheisel, J.D. , Junqueira, M.L. , Kemper, E.L. , Kitajima, J.P. , Krieger, J.E. , Kuramae, E.E. , Laigret, F. , Lambais, M.R. , Leite, L.C.C. , Lemos, E.G.M. , Lemos, M.V.F. , Lopes, S.A. , Lopes, C.R. , Machado, J.A. , Machado, M.A. , Madeira, A.M.B.N. , Madeira, H.M.F. , Marino, C.L. , Marques, M.V. , Martins, E.A.L. , Martins, E.M.F. , Matsukuma, A.Y. , Menck, C.F.M. , Miracca, E.C. , Miyaki, C.Y. , Monteiro‐Vitorello, C.B. , Moon, D.H. , Nagai, M.A. , Nascimento, A.L.T.O. , Netto, L.E.S. , Nhani, A. , Nobrega, F.G. , Nunes, L.R. , Oliveira, M.A. , de Oliveira, M.C. , de Oliveira, R.C. , Palmieri, D.A. , Paris, A. , Peixoto, B.R. , Pereira, G.A.G. , Pereira, H.A. , Pesquero, J.B. , Quaggio, R.B. , Roberto, P.G. , Rodrigues, V. , Rosa, A.J.D. , de Rosa, V.E. , de Sa, R.G. , Santelli, R.V. , Sawasaki, H.E. , da Silva, A.C.R. , da Silva, A.M. , da Silva, F.R. , Silva, W.A. , da Silveira, J.F. , Silvestri, M.L.Z. , Siqueira, W.J. , de Souza, A.A. , de Souza, A.P. , Terenzi, M.F. , Truffi, D. , Tsai, S.M. , Tsuhako, M.H. , Vallada, H. , Van Sluys, M.A. , Verjovski‐Almeida, S. , Vettore, A.L. , Zago, M.A. , Zatz, M. , Meidanis, J. , Setubal, J.C. and Org, X.F.C. (2000) The genome sequence of the plant pathogen Xylella fastidiosa . Nature, 406, 151–157. [DOI] [PubMed] [Google Scholar]
- Song, G.Q. , Walworth, A.E. and Loescher, W.H. (2015) Grafting of genetically engineered plants. J. Am. Soc. Hortic. Sci. 140, 203–213. [Google Scholar]
- de Souza, A.A. , Takita, M.A. , Coletta, H.D. , Campos, M.A. , Teixeira, J.E.C. , Targon, M.L.P.N. , Carlos, E.F. , Ravasi, J.F. , Fischer, C.N. and Machado, M.A. (2007a) Comparative analysis of differentially expressed sequence tags of sweet orange and mandarin infected with Xylella fastidiosa . Genet. Mol. Biol. 30, 965–971. [Google Scholar]
- de Souza, A.A. , Takita, M.A. , Coletta, H.D. , Targon, M.L.P.N. , Carlos, E.F. , Locali‐Fabris, E.C. , Amaral, A.M. , Freitas‐Astua, J. , Silva‐Pinhati, A.C.O. , Boscariol‐Camargo, R.L. , Berger, I.J. , Rodrigues, C.M. , Reis, M.S. and Machado, M.A. (2007b) Analysis of expressed sequence tags from Citrus sinensis L. Osbeck infected with Xylella fastidiosa . Genet. Mol. Biol. 30, 957–964. [Google Scholar]
- de Souza, A.A. , Takita, M.A. , Amaral, A. , Coletta‐Filho, H. and Machado, M.A. (2009) Citrus responses to Xylella fastidiosa infection, the causal agent of citrus variegated chlorosis. Tree For. Sci. Biotech. 2, 957–964. [Google Scholar]
- Souza, L.C.A. , Wulff, N.A. , Gaurivaud, P. , Mariano, A.G. , Virgilio, A.C.D. , Azevedo, J.L. and Monteiro, P.B. (2006) Disruption of Xylella fastidiosa CVC gumB and gumF genes affects biofilm formation without a detectable influence on exopolysaccharide production. FEMS Microbiol. Lett. 257, 236–242. [DOI] [PubMed] [Google Scholar]
- Stevenson, J.F. , Matthews, M.A. and Rost, T.L. (2004) Grapevine susceptibility to Pierce's disease I: relevance of hydraulic architecture. Am. J. Enol. Vitic. 55, 228–237. [Google Scholar]
- Stokstad, E. (2015) Food security . Italy's olives under siege. Science, 348, 620. [DOI] [PubMed] [Google Scholar]
- Strona, G. , Carstens, C.J. and Beck, P.S.A. (2017) Network analysis reveals why Xylella fastidiosa will persist in Europe. Sci. Rep. 7, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Q. , Rost, T.L. , Reid, M.S. and Matthews, M.A. (2007) Ethylene and not embolism is required for wound‐induced tylose development in stems of grapevines. Plant Physiol. 145, 1629–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Q. , Rost, T.L. and Matthews, M.A. (2008) Wound‐induced vascular occlusions in Vitis vinifera (Vitaceae): tyloses in summer and gels in winter. Am. J. Bot. 95, 1498–1505. [DOI] [PubMed] [Google Scholar]
- Sun, Q. , Sun, Y.L. , Walker, M.A. and Labavitch, J.M. (2013) Vascular occlusions in grapevines with Pierce's Disease make disease symptom development worse. Plant Physiol. 161, 1529–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne, E.T. , Stevenson, J.F. , Rost, T.L. , Labavitch, J.M. and Matthews, M.A. (2006a) Pierce's disease symptoms: comparison with symptoms of water deficit and the impact of water deficits. Am. J. Enol. Vitic. 57, 1–11. [Google Scholar]
- Thorne, E.T. , Young, B.M. , Young, G.M. , Stevenson, J.F. , Labavitch, J.M. , Matthews, M.A. and Rost, T.L. (2006b) The structure of xylem vessels in grapevine (Vitaceae) and a possible passive mechanism for the systemic spread of bacterial disease. Am. J. Bot. 93, 497–504. [DOI] [PubMed] [Google Scholar]
- Tripathi, L.P. and Sowdhamini, R. (2008) Genome‐wide survey of prokaryotic serine proteases: analysis of distribution and domain architectures of five serine protease families in prokaryotes. BMC Genomics, 9, 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyree, M.T. and Zimmermann, M.H. (2002). Hydraulic architecture of whole plants and plant performance In: Xylem Structure and the Ascent of Sap (T.E. Timell., eds), pp. 175–214. Springer, New York. [Google Scholar]
- van Ulsen, P. , Rahman, S. , Jong, W.S. , Daleke‐Schermerhorn, M.H. and Luirink, J. (2014) Type V secretion: from biogenesis to biotechnology. Biochim. Biophys. Acta, 1843, 1592–1611. [DOI] [PubMed] [Google Scholar]
- Van Sluys, M.A. , de Oliveira, M.C. , Monteiro‐Vitorello, C.B. , Miyaki, C.Y. , Furlan, L.R. , Camargo, L.E.A. , da Silva, A.C.R. , Moon, D.H. , Takita, M.A. , Lemos, E.G.M. , Machado, M.A. , Ferro, M.I.T. , da Silva, F.R. , Goldman, M.H.S. , Goldman, G.H. , Lemos, M.V.F. , El‐Dorry, H. , Tsai, S.M. , Carrer, H. , Carraro, D.M. , de Oliveira, R.C. , Nunes, L.R. , Siqueira, W.J. , Coutinho, L.L. , Kimura, E.T. , Ferro, E.S. , Harakava, R. , Kuramae, E.E. , Marino, C.L. , Giglioti, E. , Abreu, I.L. , Alves, L.M.C. , do Amaral, A.M. , Baia, G.S. , Blanco, S.R. , Brito, M.S. , Cannavan, F.S. , Celestino, A.V. , da Cunha, A.F. , Fenille, R.C. , Ferro, J.A. , Formighieri, E.F. , Kishi, L.T. , Leoni, S.G. , Oliveira, A.R. , Rosa, V.E. , Sassaki, F.T. , Sena, J.A.D. , de Souza, A.A. , Truffi, D. , Tsukumo, F. , Yanai, G.M. , Zaros, L.G. , Civerolo, E.L. , Simpson, A.J.G. , Almeida, N.F. , Setubal, J.C. and Kitajima, J.P. (2003) Comparative analyses of the complete genome sequences of Pierce's disease and citrus variegated chlorosis strains of Xylella fastidiosa . J. Bacteriol. 185, 1018–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voegel, T.M. , Doddapaneni, H. , Cheng, D.W. , Lin, H. , Stenger, D.C. , Kirkpatrick, B.C. and Roper, M.C. (2013) Identification of a response regulator involved in surface attachment, cell–cell aggregation, exopolysaccharide production and virulence in the plant pathogen Xylella fastidiosa . Mol. Plant Pathol. 14, 256–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voegel, T.M. , Warren, J.G. , Matsumoto, A. , Igo, M.M. and Kirkpatrick, B.C. (2010) Localization and characterization of Xylella fastidiosa haemagglutinin adhesins. Microbiology, 156, 2172–2179. [DOI] [PubMed] [Google Scholar]
- Wallis, C.M. and Chen, J. (2012) Grapevine phenolic compounds in xylem sap and tissues are significantly altered during infection by Xylella fastidiosa . Phytopathology, 102, 816–826. [DOI] [PubMed] [Google Scholar]
- Yadeta, K. and Thomma, B.P.H.J. (2013) The xylem as battleground for plant hosts and vascular wilt pathogens. Front. Plant Sci. 4, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L. , Lin, H. , Takahashi, Y. , Chen, F. , Walker, M.A. and Civerolo, E.L. (2011) Proteomic analysis of grapevine stem in response to Xylella fastidiosa inoculation. Physiol. Mol. Plant Pathol. 75, 90–99. [Google Scholar]
- Zhang, S.J. , Chakrabarty, P.K. , Fleites, L.A. , Rayside, P.A. , Hopkins, D.L. and Gabriel, D.W. (2015) Three new Pierce's disease pathogenicity effectors identified using Xylella fastidiosa biocontrol strain EB92–1. PLos One, 10, e0133796. [DOI] [PMC free article] [PubMed] [Google Scholar]


