Summary
An up‐regulated gene derived from Bamboo mosaic virus (BaMV)‐infected Nicotiana benthamiana plants was cloned and characterized in this study. BaMV is a single‐stranded, positive‐sense RNA virus. This gene product, designated as NbTRXh2, was matched with sequences of thioredoxin h proteins, a group of small proteins with a conserved active‐site motif WCXPC conferring disulfide reductase activity. To examine how NbTRXh2 is involved in the infection cycle of BaMV, we used the virus‐induced gene silencing technique to knock down NbTRXh2 expression in N. benthamiana and inoculated the plants with BaMV. We observed that, compared with control plants, BaMV coat protein accumulation increased in knockdown plants at 5 days post‐inoculation (dpi). Furthermore, BaMV coat protein accumulation did not differ significantly between NbTRXh2‐knockdown and control protoplasts at 24 hpi. The BaMV infection foci in NbTRXh2‐knockdown plants were larger than those in control plants. In addition, BaMV coat protein accumulation decreased when NbTRXh2 was transiently expressed in plants. These results suggest that NbTRXh2 plays a role in restricting BaMV accumulation. Moreover, confocal microscopy results showed that NbTRXh2‐OFP (NbTRXh2 fused with orange fluorescent protein) localized at the plasma membrane, similar to AtTRXh9, a homologue in Arabidopsis. The expression of the mutant that did not target the substrates failed to reduce BaMV accumulation. Co‐immunoprecipitation experiments revealed that the viral movement protein TGBp2 could be the target of NbTRXh2. Overall, the functional role of NbTRXh2 in reducing the disulfide bonds of targeting factors, encoded either by the host or virus (TGBp2), is crucial in restricting BaMV movement.
Keywords: Bamboo mosaic virus, thioredoxin, virus‐induced gene silencing, virus movement
Introduction
Bamboo mosaic virus (BaMV) is a single‐stranded, positive‐sense RNA virus belonging to the Potexvirus genus of α‐Flexiviridae (Lin et al., 1992). The genome contains 6366 nucleotides with a 5′ m7GpppG cap structure (Lin et al., 1994) and approximately 300 adenylates at the 3′ end (Chen et al., 2005). During infection, BaMV can generate two major subgenomic RNAs of 2 and 1 kb in length (Lin et al., 1992). The BaMV genome consists of five open reading frames (ORFs). ORF1 encodes a 155‐kDa replicase comprising a capping enzyme domain (Huang et al., 2004; Li et al., 2001a), a helicase‐like domain with 5′‐triphosphatase activity (Li et al., 2001b) and an RNA‐dependent RNA polymerase (RdRp) domain (Li et al., 1998). ORFs 2–4 constitute a triple gene block (TGB), encoding proteins (TGBp1, TGBp2, and TGBp3) involved in viral movement (Lin et al., 2004, 2006). ORF5 encodes a viral coat protein (CP) associated with symptom expression, virion assembly (DiMaio et al., 2015; Lan et al., 2010) and viral movement (Lee et al., 2011).
Plant viruses generally move from cell to cell through the plasmodesmata (PD) (Kumar et al., 2015; Morozov and Solovyev, 2003; Verchot‐Lubicz, 2005) with the help of viral‐encoded proteins to enlarge the plasmodesmal size exclusion limit (SEL) (Heinlein, 2015; Kumar et al., 2015; Lucas and Lee, 2004). Tobacco mosaic virus (TMV) encodes a 30‐kDa protein that promotes viral movement (Melcher, 2000). This 30‐kDa movement protein (MP) has been demonstrated to bind single‐stranded nucleic acids, located at the PD, and to increase the plasmodesmal SEL (Atkins et al., 1991; Citovsky et al., 1990; Oparka et al., 1997; Tomenius et al., 1987; Wolf et al., 1989). In contrast with TMV, some viruses encode multiple proteins for viral movement, such as the TGB proteins in potexviruses, Pecluvirus, Pomovirus, Foveavirus, Hordeivirus, Carlvirus and Allexivirus (Morozov and Solovyev, 2003; Park et al., 2014; Verchot‐Lubicz, 2005). The multifunctional TGBp1 of potexviruses has been demonstrated to show RNA‐binding and NTPase/helicase activities (Morozov et al., 1999) and to promote viral translation (Atabekov et al., 2000; Rodionova et al., 2003). TGBp1 is a silencing suppressor (Bayne et al., 2005; Chiu et al., 2010) and allows an increase in the plasmodesmal SEL of Nicotiana benthamiana and N. tabacum (Howard et al., 2004; Lough et al., 1998, 2000). TGBp2 and TGBp3 of potexviruses are associated with the endoplasmic reticulum (ER) (Krishnamurthy et al., 2003; Mitra et al., 2003). TGBp2 of BaMV has been observed to bind viral RNA (Hsu et al., 2009). Two conserved cysteine (Cys) residues (C109 and C112), which have been proposed to form a disulfide bond, have been reported to play a key role in BaMV movement (Hsu et al., 2008; Tseng et al., 2009).
Two models for the movement of Potexvirus‐like viruses have been proposed (Verchot‐Lubicz et al., 2010). The first model is based on the association of TGBp3 with the cortical ER, in which virions, viral replicase and TGBp3 are recruited to membrane‐bound bodies (MBBs) (Bamunusinghe et al., 2009; Ju et al., 2005; Krishnamurthy et al., 2003; Wu et al., 2011). TGBp2 and TGBp3 gather at post‐ER vesicles and may be associated with a CP complex (Ju et al., 2005). These vesicles carry infectious materials and are necessary for cell‐to‐cell movement (Hsu et al., 2009; Tseng et al., 2009). Furthermore, TGBp1 and virions, or virion‐like particles, assemble into a ribonucleoprotein (RNP) complex and travel across the PD. TGBp3, derived from the ER and MBBs, may associate with the transfer of viral RNA or the TGBp1–vRNA–CP complex to transport vesicles. In the second model, TGBp2 promotes the budding of new vesicles containing TGBp3 from the ER. TGBp3 mediates the TGBp1–vRNA‐CP complex, which associates with TGBp2 and TGBp3 to move towards the PD; this movement requires β‐1,3‐glucanase to degrade callose at the PD (Verchot‐Lubicz et al., 2010).
Thioredoxins (Trxs) are small proteins of approximately 12 kDa in size. In Arabidopsis thaliana, Trxs are categorized into six major groups: h, f, m, o, x and y (Reichheld et al., 2002). Trxs contain a conserved WCXPC active site to catalyse the dithiol/disulfide exchange reaction (Tarrago et al., 2010). In general, plant Trxs are involved in redox regulation (Meyer et al., 2005). The reduction of Trx h involves two major systems: NADPH‐Trx reductase (NTR), which mediates the reduction of Trx h by NADPH and is termed the NTR/Trx h system (NTS) (Joudrier et al., 2005), and the glutathione/glutaredoxin system (Gelhaye et al., 2003). In addition to reducing disulfide bonds, Trx h moves from cell to cell (Ishiwatari et al., 1998; Meng et al., 2010) and is required for plant immunity (Tada et al., 2008). Nicotiana tabacum Trx h3 (NtTRXh3) overexpression in tobacco can enhance resistance to infection caused by Cucumber mosaic virus and TMV (Sun et al., 2010).
In this study, we identified a potential plant defence gene against BaMV. The protein sequence of this gene contains a Trx‐conserved motif, WCXPC. We designated this protein as NbTRXh2. Moreover, one of the factors involved in BaMV cell‐to‐cell movement is targeted by NbTRXh2, which results in viral movement restriction. The possible target and mechanism of NbTRXh2 are discussed.
Results
Cloning of the Trx gene NbTRXh2 from N. benthamiana
An up‐regulated complementary DNA (cDNA) fragment, ACGT4, was identified post‐BaMV inoculation through cDNA‐amplified fragment length polymorphism. We observed that ACGT4 influenced BaMV accumulation in N. benthamiana plants (Cheng et al., 2010). After ACGT4 had been cloned into a Tobacco rattle virus (TRV)‐based silencing vector to knock down NbTRXh2 expression in plants, followed by BaMV inoculation, we observed that BaMV accumulation increased in knockdown plants compared with control plants (Cheng et al., 2010). The ACGT4 sequence presented a 100% match to an expressed sequence tag (EST) when compared with the N. benthamiana database. To obtain the full‐length gene for further study, we performed the rapid amplification of 5′ cDNA ends (5′ RACE) with a primer based on the known sequence. The full‐length sequence of ACGT4 contains 841 nucleotides (Fig. 1) and a complete ORF that can encode a 139‐amino‐acid polypeptide (Fig. 1). Further analysis of this gene indicated that it showed 47% and 48% identity to Trx h3 in N. tabacum (NtTRXh3; accession number AF435818) and Trx h9 in Arabidopsis (AtTRXh9; accession number NP_001078124), respectively (Fig. 1). The conserved signature peptide of the active site for the Trx h family, WCXPC (Gelhaye et al., 2004; Meyer et al., 2002), was localized in the in silico translated polypeptide (Fig. 1). These observations signify that ACGT4 may be one of the Trx h genes in N. benthamiana. Because the partial sequence of NbTRXh1 (GenBank accession ACV52592) has been reported, we designated this gene as NbTRXh2 (accession number KJ200294). The sequence identity between NbTRXh2 and its homologues in N. tabacum (NtTRXh3) or Arabidopsis (AtTRXh9) is not sufficiently high (all less than 50%), suggesting the presence of another gene in N. benthamiana, which has higher identity to NtTRXh3 and AtTRXh9 in N. tabacum and Arabidopsis, respectively. Accordingly, we compared the sequence of NtTRXh3 with the transcriptome database of N. benthamiana (Nakasugi et al., 2013; Zhao et al., 2013) and retrieved a partial sequence, designated as NbTRXh9, which shared 63% identity with the sequence of NtTRXh3 (Fig. 2A). Furthermore, we performed a phylogenetic analysis of all available Trx h genes from N. benthamiana, N. tabacum and Arabidopsis (Fig. 2B). The analysis revealed that NbTRXh2 identified in this study is unique. Although the sequence of NbTRXh9 is incomplete, we found that the sequence of NbTRXh9 in N. benthamiana is closer to those of NtTRXh3 and AtTRXh9 than to that of NbTRXh2 identified in this study (Fig. 2A).
Figure 1.
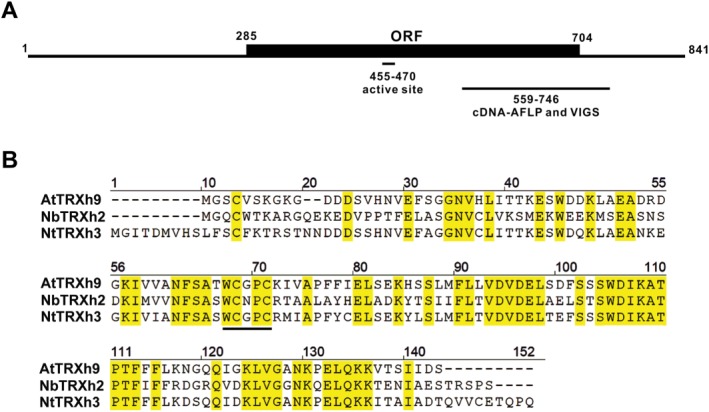
The gene organization of NbTRXh2 from Nicotiana benthamiana and the alignment with its orthologues in other species. (A) Full‐length NbTRXh2 with its open reading frame (ORF; nucleotides 285–704) is indicated. The region of the active site (nucleotides 455–470), the cDNA fragment derived from cDNA‐amplified fragment length polymorphism (AFLP), and the region used for the knockdown experiment (nucleotides 559–746) are indicated. (B) The alignment of the amino acid sequence of NbTRXh2 and its orthologues from other species (NtTRXh3: accession number AF435818 from Nicotiana tabacum; AtTRXh9: accession number NP_001078124 from Arabidopsis thaliana). The conserved active site (WCXPC) of TRX h proteins is indicated with a bold underline. VIGS, virus‐induced gene silencing.
Figure 2.
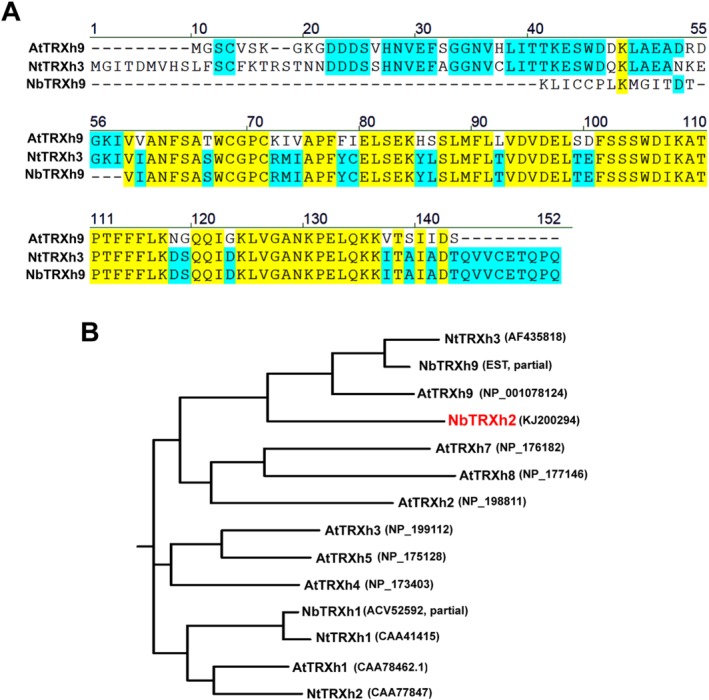
Multiple sequence alignment and the phylogenetic analyses of selected Trx h genes from Nicotiana benthamiana (Nb), N. tabacum (Nt) and Arabidopsis thaliana (At). All the analyses were performed via the Biology WorkBench V3.2 platform (http://workbench.sdsc.edu). (A) The alignment used clustalw with default parameters. (B) The rooted phylogenetic tree was generated with default DRAWGRAM with the selected alignment option.
NbTRXh2 can specifically influence the accumulation of BaMV
To identify the possible role of NbTRXh2 in the viral infection cycle, BaMV was inoculated onto NbTRXh2‐knockdown N. benthamiana plants. No morphological changes were observed in knockdown plants compared with control plants (TRV vector carrying luciferase gene, Fig. S1, see Supporting Information). The accumulation levels of the BaMV CP in the inoculated leaves were examined at 1, 3, 5 and 7 days post‐inoculation (dpi) by performing Western blot analysis. The results demonstrated that, compared with control plants, the reduction in NbTRXh2 expression enhanced BaMV CP accumulation by 1.8‐ and two‐fold at 5 and 7 dpi, respectively, in NbTRXh2‐knockdown plants (Fig. 3A). However, no difference was observed at 1 and 3 dpi between control and NbTRXh2‐knockdown plants. We determined the knockdown efficiency through real‐time reverse transcription‐polymerase chain reaction (RT‐PCR), and observed that NbTRXh2 expression in knockdown plants and protoplasts was approximately 37% of that of their control counterparts (Fig. 3B).
Figure 3.
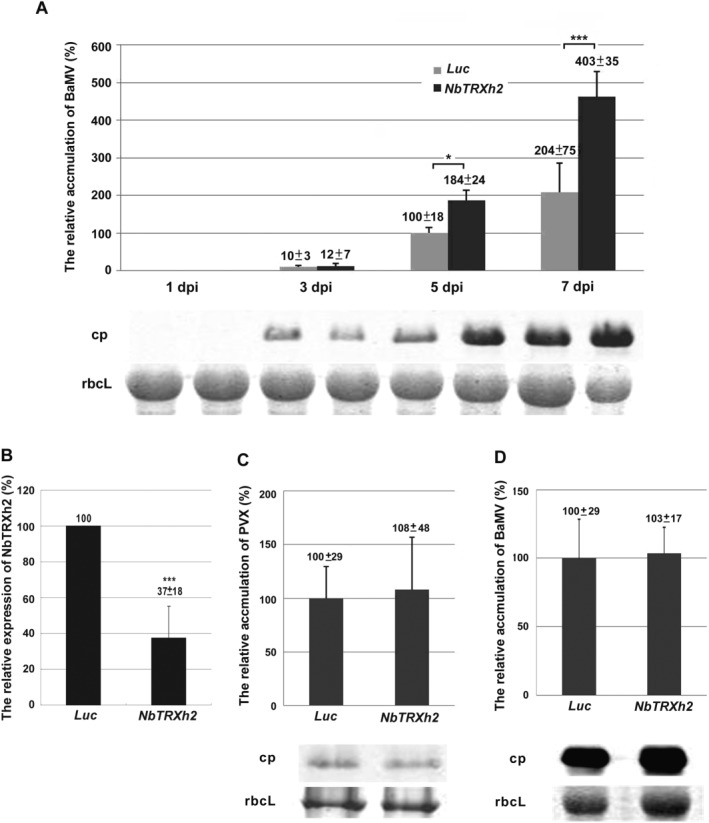
The accumulation of viral coat protein in NbTRXh2‐knockdown plants or protoplasts. Total proteins were extracted from Barley mosaic virus (BaMV)‐inoculated Luc‐ or NbTRXh2‐knockdown plants in (A) and protoplasts in (D), and subjected to Western blot analysis. The accumulation levels of BaMV coat protein in Luc‐knockdown plants at 5 days post‐inoculation (dpi) in (A) and in Luc‐knockdown protoplasts at 48 hpi in (D) were set as 100%. (B) The total RNAs were extracted from the knockdown plants as indicated and used to determine the knockdown efficiency by real‐time reverse transcription‐polymerase chain reaction (RT‐PCR). The numbers shown above the bars are the average and standard error of the relative expression of NbTRXh2 from at least three independent experiments with at least two plants for each experiment. (C) The accumulation levels of Potato virus X (PVX) in Luc‐ and NbTRXh2‐knockdown Nicotiana benthamiana leaves at 7 dpi were also analysed by Western blot analysis. The coat protein accumulation levels of each virus in the Luc‐knockdown control plants were set as 100%. The numbers are the average levels of coat protein with the standard deviation in (A), (C) and (D) obtained from at least three independent experiments with a total of 5–7 plants for each treatment. Luc, Luciferase‐knockdown plants; NbTRXh2, NbTRXh2‐knockdown plants; cp, coat protein; rbcL, RuBisCo large subunit (the loading control for normalization). Asterisks indicate a statistically significant difference of the indicated group analysed by Student's t‐test (*P < 0.05, ***P < 0.001).
To determine whether a reduction in NbTRXh2 expression causes a similar effect in other closely related viruses, we inoculated Potato virus X (PVX) into control and NbTRXh2‐knockdown plants. The results indicated that the PVX CP accumulation level did not differ substantially between the control and NbTRXh2‐knockdown plants (Fig. 3C). We also examined the gene expression profiles of NbTRXh2 after BaMV, PVX and Cucumber mosaic virus (CMV) inoculation. After BaMV inoculation, the expression pattern of NbTRXh2 remained up‐regulated and was higher, even at 7 dpi. By contrast, after PVX and CMV inoculation, the expression pattern was up‐regulated initially at 1 dpi (the expression was approximately three‐fold after CMV inoculation) and then decreased gradually to the control level at 7 dpi (Fig. S2, see Supporting Information). These results indicate that the interference of NbTRXh2 in the accumulation of BaMV is specific.
Reduction in NbTRXh2 expression enhances BaMV movement
To determine whether the enhancement of BaMV accumulation in NbTRXh2‐knockdown plants is caused by an improvement in viral RNA replication or virus movement, we inoculated BaMV virion RNA into NbTRXh2‐knockdown protoplasts to exclude the route of viral cell‐to‐cell movement. We noted no substantial difference in BaMV CP accumulation between the control and NbTRXh2‐knockdown protoplasts at 24 h post‐inoculation (hpi; Fig. 3D). This result and the observation of no significant difference in the inoculated leaves at 3 dpi (Fig. 3A) suggest that the enhancement of BaMV accumulation in NbTRXh2‐knockdown plants is not caused by improved viral RNA replication. It is most probably caused by enhanced cell‐to‐cell movement. To substantiate this hypothesis, we inoculated control and NbTRXh2‐knockdown plants with the BaMV infectious plasmid pCBG which carries the green fluorescent protein (GFP) reporter gene in the viral genome (Lin et al., 2006). GFP signal foci, representing the cell‐to‐cell movement of BaMV, were examined by fluorescence microscopy at 5 dpi. Microscopy results revealed a greater number of foci that were larger in size in NbTRXh2‐knockdown plants than in control plants (Fig. 4). The size difference among infected foci could not be observed at early infection time points, such as from 1.5 to 3 dpi (Fig. S3, see Supporting Information). These results thus validate our hypothesis that NbTRXh2 inhibits the cell‐to‐cell movement, but not the replication, of BaMV in N. benthamiana.
Figure 4.
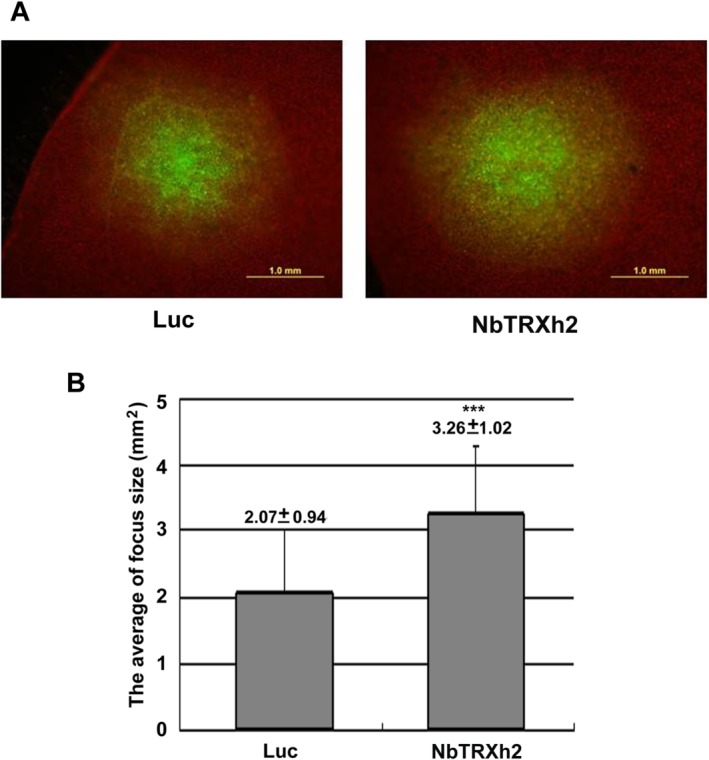
Cell‐to‐cell movement of Barley mosaic virus (BaMV) in Luc‐ and NbTRXh2‐knockdown plants. (A) Areas of green fluorescent protein (GFP) foci on pCBG (infectious BaMV cDNA viral vector which can express GFP)‐inoculated Nicotiana benthamiana leaves at 5 days post‐inoculation, measured by fluorescence microscopy. Bar length, 1.0 mm. (B) Statistical analysis of the results obtained from (A); y‐axis is the GFP focus size (mm2). The numbers shown above the bars are the average and standard deviation of 24 and 35 foci from Luc‐ and NbTRXh2‐knockdown plants, respectively. Asterisks indicate statistically significant differences of the indicated group analysed by Student's t‐test (***P < 0.001).
NbTRXh2 is associated with the membrane
The subcellular localization of the Trx h family members could be cytosolic (Gelhaye et al., 2004) or associated with the membrane (Meng et al., 2010). AtTRXh9 targets the plasma membrane through myristoylation on Gly2 (the second position from the N‐terminus) and possibly through palmitoylation on Cys4 (Meng et al., 2010). The sequence of NbTRXh2 contains a G2/C4 myristoylation/palmitoylation motif, which is similar to the sequence of AtTRXh9 (Fig. 1) (Maurer‐Stroh et al., 2002; Meng et al., 2010; Smotrys and Linder, 2004). Thus, similar to AtTRXh9, NbTRXh2 is probably associated with the membrane. We examined the possibility of myristoylation on G2 using Myristoylator (http://ca.expasy.org/tools/myristoylator), a prediction program. According to the results, NbTRXh2 has high confidence for myristoylation, with the positive and negative score values being 0.972196 and 0.0274927, respectively. To analyse the localization of NbTRXh2, an entire ORF sequence was fused to orange fluorescent protein (OFP) and transiently expressed in N. benthamiana plants. We determined that NbTRXh2‐OFP was localized along the cell edge of epidermal cells (Fig. 5A) and that protoplasts (Fig. 5B) were most likely located on the plasma membrane. To confirm that NbTRXh2‐OFP is membrane associated, we separated cell extracts into membrane and cytoplasmic fractions through centrifugation and then conducted Western blot analysis. The results indicated that NbTRXh2‐OFP was dominant in the membrane fraction, whereas OFP only was mostly distributed in the soluble fraction (Fig. 5C).
Figure 5.
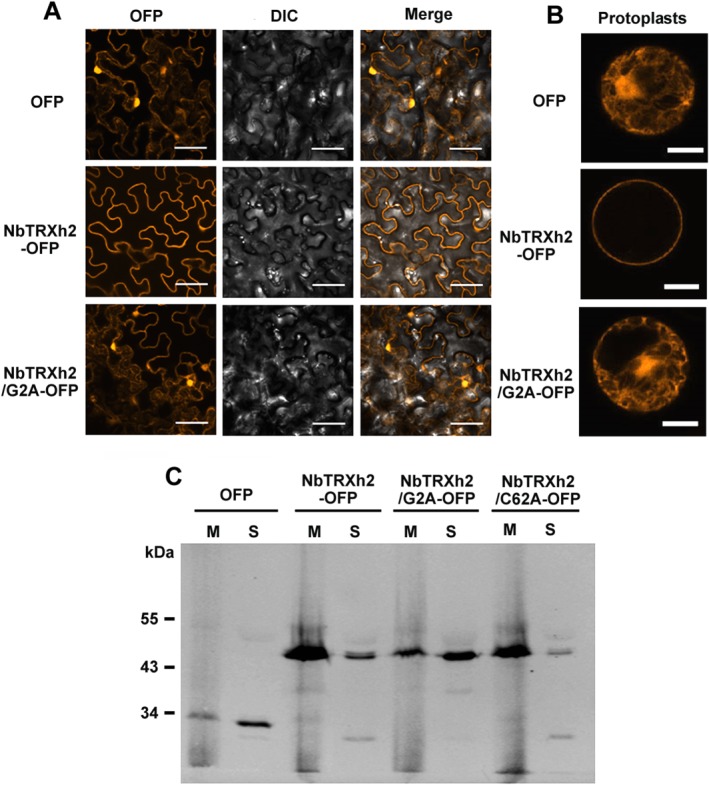
Localization of NbTRXh2‐OFP and its derivatives in Nicotiana benthamiana cells. Orange fluorescent protein (OFP), NbTRXh2‐OFP and NbTRXh2/G2A‐OFP were transiently expressed by agro‐infiltration in N. benthamiana plants (A) and in protoplasts (B) and detected by an Olympus Fluoview FV1000 confocal microscope with 543‐nm laser excitation. DIC, differential interference contrast. (C) OFP, NbTRXh2‐OFP and the two mutants NbTRXh2/G2A‐OFP and NbTRXh2/C62A‐OFP in the soluble (S) or membrane (M) fraction were detected by Western blot analysis. The primary antibody was the rabbit anti‐OFP antibody, and the secondary antibody was fluorescently labelled goat anti‐rabbit immunoglobulin G (IgG) antibody. The molecular weight of the protein bands is indicated to the left of the gel.
Because the NbTRXh2 sequence was predicted to contain no transmembrane domain, the most likely mechanism for the association of NbTRXh2 with the membrane is through the lipid‐anchoring modification. To examine whether NbTRXh2 might be myristoylated at the Gly2 site for a membrane linkage, similar to its Arabidopsis homologue AtTRXh9, we constructed a substitute mutant with Gly2 changed to Ala2 (NbTRXh2/G2A‐OFP) and transiently expressed it in N. benthamiana. The subcellular localization of NbTRXh2/G2A‐OFP was similar to that of OFP only, wherein fluorescent signals were not only demonstrated at the edge of the cells, but also distributed to other organelles (Fig. 5A,B). The fractionation result indicated that, similar to OFP only, NbTRXh2/G2A‐OFP was enriched in the soluble fraction (Fig. 5C). Overall, the results suggest that NbTRXh2 is a membrane‐associated protein (can be the plasma membrane) and uses myristoylation on Gly2 to target the membrane.
Transiently expressed NbTRXh2‐OFP reduces BaMV accumulation
The results of NbTRXh2‐knockdown experiments suggest that NbTRXh2 expression plays a role in restricting BaMV accumulation in N. benthamiana. To further support this hypothesis, we inoculated BaMV virion onto N. benthamiana leaves for 2 days and transiently expressed NbTRXh2‐OFP or OFP through agro‐infiltration onto BaMV‐inoculated leaves for another 3 days. The results revealed that, compared with OFP‐expressing plants, BaMV accumulation decreased by 36% in NbTRXh2‐OFP‐expressing plants at 5 dpi (Fig. 6).
Figure 6.
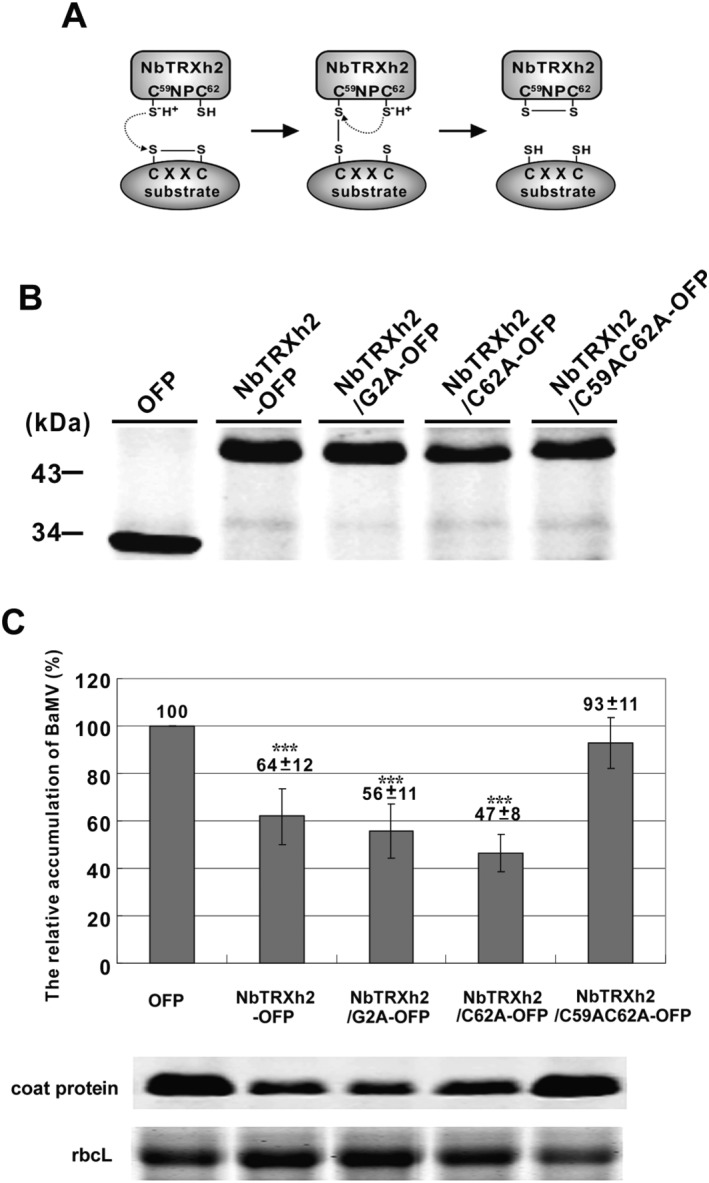
The effects of the expression of NbTRXh2 and its derivatives on Barley mosaic virus (BaMV) infection. (A) A schematic representation of the reaction mechanism of the thioredoxin NbTRXh2 with its proposed substrate BaMV TGBp2. (B) Western blot analysis of the transiently expressed orange fluorescent protein (OFP), NbTRXh2‐OFP, NbTRXh2/G2A‐OFP, NbTRXh2/C62A‐OFP and NbTRXh2/C59AC62A‐OFP by agro‐infiltration on Nicotiana benthamiana leaves as indicated. The total proteins were extracted and separated onto a 12% sodium dodecylsulfate (SDS)‐polyacrylamide gel. The primary antibody was rabbit anti‐OFP antibody and the secondary antibody was the goat anti‐rabbit immunoglobulin G (IgG) antibody. The molecular weight is indicated to the left of the gel. (C) The relative accumulation of BaMV coat protein on the inoculated leaves detected at 5 days post‐inoculation with NbTRX1‐OFP, NbTRXh2/G2A‐OFP, NbTRXh2/C62A‐OFP, NbTRXh2/C59AC62A‐OFP or OFP (as a control) transiently expressed on the virus‐inoculated leaves for the last 2 days. The data determined from Western blots were normalized with the large subunit of RuBisCO (rbcL) as the loading control stained with Coomassie blue. The numbers shown above the bars are the average and standard error of at least three independent experiments. Asterisks indicate statistically significant difference of the indicated group analysed by Student's t‐test (***P < 0.001).
Membrane targeting was not essential for the viral inhibition of NbTRXh2 because the mutant NbTRXh2/G2A‐OFP could reduce BaMV accumulation by approximately 44%. To determine whether two key Cys residues at the catalytic site of NbTRXh2 were involved in BaMV accumulation, we generated two catalytic site (WC59NPC62, Figs 1B and 6A) mutants with a single‐ and double‐amino‐acid substitution, NbTRXh2/C62A‐OFP and NbTRXh2/C59AC62A‐OFP, respectively. The results showed that NbTRXh2/C62A‐OFP expression reduced BaMV accumulation by 53%. By contrast, the double‐substitution mutant NbTRXh2/C59AC62A‐OFP failed to interact with any target and lost its inhibitory activity (Fig. 6C). Because the C‐terminal Cys residue (C62) of the catalytic site was crucial for the dissociation of the mixed disulfide complex (Collet and Messens, 2010), the dissociation could be blocked when Cys mutated into alanine (Ala) (Balmer et al., 2004). Accordingly, NbTRXh2/C62A‐OFP could form a moderately stable complex with the target protein, and NbTRXh2/C59AC62A‐OFP could not interact with its targets. These results indicate that the activity of NbTRXh2 plays a role in inhibiting BaMV accumulation.
NbTRXh2 targets the viral MP TGBp2
Because Trx activity reduces the disulfide bond of target proteins, the possible role of NbTRXh2 is to diminish the disulfide linkage on one or more of the factors required for BaMV movement. The disulfide bond in this protein can be vital to the maintenance of its proper structure and assist BaMV movement. Two conserved Cys residues, Cys‐109 and Cys‐112, in the BaMV MP TGBp2 have been proposed to form a disulfide bond and have been demonstrated to be involved in cell‐to‐cell movement (Tseng et al., 2009). Therefore, the BaMV MP TGBp2 can be a potential target of NbTRXh2. To test this hypothesis, we examined whether TGBp2 interacts with NbTRXh2.
We constructed three histidine (His)‐tagged NbTRXh2 derivatives (His‐NbTRXh2, His‐NbTRXh2/C62A and His‐NbTRXh2/C59AC62A), and overexpressed them in Escherichia coli. BaMV MPs, namely GFP‐TGBp2 and GFP‐TGBp1, were transiently expressed in N. benthamiana plants. Protein extracts containing GFP, GFP‐TGBp1 or GFP‐TGBp2 were passed through anti‐GFP magnetic beads, followed by flow through E. coli extracts containing the expressed His‐NbTRXh2 and its derivatives. The eluents were examined using Western blot analysis (Fig. 7). The results demonstrated that NbTRXh2 and NbTRXh2/C62A strongly interacted with GFP‐TGBp2 (Fig. 7, lanes 7 and 8 on the top panel); nevertheless, the beads could only bind to a small portion of the detergent‐solubilized, membrane‐associated GFP‐TGBp2 expressed in N. benthamiana (Fig. 7, lanes 7–9 of the middle two panels). The less efficient binding of GFP‐TGBp2 with the anti‐GFP beads could be a result of the interference of detergent with solubilized membrane‐associated proteins compared with that of soluble proteins, GFP and GFP‐TGBp1. The double mutant NbTRXh2/C59AC62A could also interact with GFP‐TGBp2, but its interaction efficiency was lower than that of the other two (Fig. 7, lanes 7–9 of the top panel). These results suggest that the interaction between NbTRXh2 and its substrates may not depend solely on the Cys–Cys disulfide bond. The soluble forms of GFP‐TGBp1 and GFP‐only were not expressed in N. benthamiana (0.05% of the total protein loaded for input in Fig. 7); however, most of the soluble GFP‐TGBp1 was bound to anti‐GFP beads (Fig. 7, lanes 4–6; expected to be approximately 7.5‐fold compared with the input if completely bound to beads). Although no substantial interaction of NbTRXh2 or its derivatives with GFP or GFP‐TGBp1 was determined, a weak interaction might be present between NbTRXh2 and TGBp1 (lane 4 on the top panel).
Figure 7.
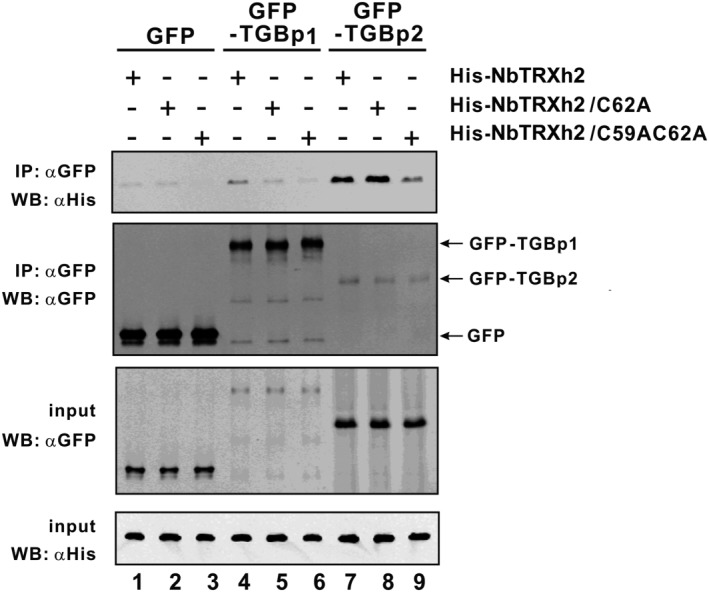
Co‐immunoprecipitation of Barley mosaic virus (BaMV) movement protein with NbTRXh2 and its derivatives. The green fluorescent protein (GFP)‐fused BaMV movement proteins (GFP‐TGBp1 and GFP‐TGBp2) were transiently expressed in Nicotiana benthamiana and subjected to immunoprecipitation by magnetic anti‐GFP beads. The pull‐down GFP or GFP‐fused proteins interacting with NbTRXh2 and its derivatives are indicated. The immunoprecipitated proteins and their expression levels (indicated as input) were analysed by Western blot analysis using anti‐His and anti‐GFP antibodies.
Discussion
In this study, we identified a potential defence gene against BaMV from N. benthamiana, designated as NbTRXh2. The expression of this gene was induced in N. benthamiana after BaMV inoculation. Although the novel targets of NbTRXh2 remain unclear, at least one of the substrates, TGBp2 of BaMV, is involved in the movement of BaMV. The transient expression of NbTRXh2 reduced BaMV accumulation in plants (Fig. 6). We speculate that a disulfide bond in the viral movement‐related protein, either from the virus or host, is the target of NbTRXh2. The reducing form of this protein might fail to efficiently move the viral RNA complex from cell to cell. However, this effect was not observed in PVX, another member of the potexviruses.
To establish a successful infection in its host, a viral plant pathogen must contend with numerous defence mechanisms. Host proteins, which act against viral pathogens, identified thus far mostly restrict the viral replication steps in the infection cycle (Cheng et al., 2009; Huh and Paek, 2013; Ishibashi et al., 2010; Lin et al., 2012; Park et al., 2013). Studies identifying proteins that inhibit viral movement are scarce. Callose (a β‐1,3‐glucan) deposition at the PD has been reported to be a critical factor in viral cell‐to‐cell movement (Balmer et al., 2004). The counter defence for PVX is the use of TGBp2, which interacts with TGBp2‐interacting host proteins and then acts together with β‐1,3‐glucanase. The complex is then translocated to the neck of the PD, resulting in callose deposition (Fridborg et al., 2003). A microtubule‐associated protein, derived from Arabidopsis, AtMPB2C, has been demonstrated to negatively regulate Oilseed rape mosaic virus movement by aligning cortical microtubules (Ruggenthaler et al., 2009). The host protein ribulose‐1,5‐bisphosphate carboxylase/oxygenase small subunit has been demonstrated to interact with the MP of Tomato mosaic virus and to be involved in Tm‐22‐mediated resistance (Lin et al., 2011; Zhao et al., 2013). In addition, chloroplast proteins, ATP‐synthase γ‐subunit and Rubisco activase have been revealed to interact with the replicase of TMV and restrict viral intercellular movement (Bhat et al., 2013).
Once BaMV infects its hosts, viral RNA interacts with the nucleus‐encoded chloroplast phosphoglycerate kinase and is shuttled into the chloroplast for replication (Cheng et al., 2013a). The details of the replication process remain unknown. Nonetheless, subgenomic RNAs encoding TGBp2 and TGBp3 are accumulated in the cytoplasm and presumably translated at the ER (Verchot‐Lubicz et al., 2010). The membrane‐associated TGBp2 and TGBp3 have been revealed to either form the RNP complex and move along the ER network, or associate with membrane vesicles and move along the actin filament towards the PD (Chou et al., 2013). A factor associated with vesicle formation, identified from N. benthamiana, NbRabGAP1, has been shown to be involved in BaMV viral trafficking (Huang et al., 2013). Therefore, membrane vesicle formation might be necessary for the movement of BaMV. The cytoplasmic C‐terminus of TGBp2, containing a disulfide bond (C109–C112), can be crucial for the interaction with TGBp3 and the RNP complex (viral RNA, CP, TGBp1 and replicase) (Chou et al., 2013; Lee et al., 2011; Park et al., 2013; Wu et al., 2011). NbTRXh2, a membrane‐associated protein, is translated in the cytoplasm and targets the membrane, possibly through the myristoylation at Gly2, which is similar to that in AtTRXh9 (Meng et al., 2010). However, membrane targeting is not involved in the interaction of NbTRXh2 with the substrate required for BaMV movement. The possible explanation of this observation is that NbTRXh2 interacts with its substrate immediately after translation before modification. Although the substrate is not clear, the MP TGBp2 of BaMV is most likely to be one of the novel substrates for NbTRXh2 (Fig. 7). Among all the members of potexviruses, two conserved Cys residues are present at the C‐terminus of TGBp2. The substitution of either of these two Cys residues by Ala restricts virus movement in BaMV (Tseng et al., 2009). According to the topology of TGBp2, the C‐terminus is towards the outside of the membrane on the cytosol (Hsu et al., 2008). Therefore, the two conserved Cys residues have been proposed to be oxidized to form a disulfide bond; nevertheless, no evidence has been provided. Notably, the arrangement of the two conserved Cys residues in TGBp2 of BaMV (109CXXC112) is different from that in TGBp2 of PVX (106CXC108). Thus, whether the sequence CXC in PVX cannot form a disulfide bond, or this type of disulfide bond cannot be a target of NbTRXh2, should be investigated in the future.
Experimental Procedures
NbTRXh2 knockdown and virus infection
The technique of knocking down the expression of the NbTRXh2 gene has been reported previously (Liu et al., 2002; Ruiz et al., 1998). In brief, the cDNA fragment ACGT4, derived from cDNA‐AFLP (Fig. 1), was cloned into the pTRV2 vector and electroporated into the Agrobacterium tumefaciens C58C1 for infiltration, as described previously (Cheng et al., 2010). After knockdown, approximately 200 ng of BaMV or 500 ng of PVX virion were mechanically inoculated onto the fourth leaf above the infiltrated leaves. The inoculated leaves were harvested and homogenized with Laemmli buffer at 5 dpi.
RNA extraction and real‐time PCR
Approximately 100 mg of leaf was ground to a powder with liquid nitrogen. The leaf powder was mixed with STE buffer [400 mm Tris‐HCl, pH 8.0, 400 mm NaCl and 40 mm ethylenediaminetetraacetic acid (EDTA)], 1% sodium dodecylsulfate (SDS) and 3.3 mg/mL bentonite and an equal volume of phenol–chloroform. The RNAs were ethanol precipitated with NH4OAc, washed, dried and dissolved in 30 µL of deionized H2O.
The cDNA synthesis reaction was performed following the manufacturer's instructions using ImProm‐II™ Reverse Transcriptase (Promega, Carlsbad, CA, USA) with the primers Oligo dT25. The relative expression levels of NbTRXh2 were determined by real‐time RT‐PCR with the specific primers NbTRXh2 + 1 (5′‐GATGGGACAGTGATGGACTAA‐3′) and NbTRXh2‐215 (5′‐GTTCATGATAGGCTAGTGCT‐3′). The expression of the actin gene was used as an internal control for normalization with the primer set: β‐actin 5′ primer (5′‐GATGAAGATACTCACAGAAAGA‐3′) and β‐actin 3′ primer (5′‐GTGGTTTCATGAATGCCAGCA‐3′). Real‐time PCR was performed in a 20‐μL reaction containing 1000 times dilution of SYBR green I (Cambrex Bio Science Rockland Inc., ME, USA), 0.6 mm primer, 0.2 mm deoxynucleotide triphosphate (dNTP), 10 mm Tris‐HCl, pH 8.8, 1.5 mm MgCl2, 50 mm KCl, 0.1% Triton X‐100, 2 μL of cDNA and 3 units of Taq DNA polymerase (Promega). Reactions were carried out in a RotorGene 3000 (Corbett Research, Sydney, Australia) with data acquisition at 72 °C. All samples were run at least three times.
Western blot analysis
Total proteins were extracted from the inoculated leaves or protoplasts and separated on 12% SDS‐polyacrylamide gels. The proteins smaller than 40 kDa on the gels were transferred onto nitrocellulose membranes (Protran BA 85; Schleicher & Schuell, Dassel, Germany) and probed with the laboratory‐generated primary antiserum (1 : 5000 dilution in use) from rabbits against GFP, BaMV or PVX. The proteins larger than 40 kDa on the gels were stained with Coomassie brilliant blue for the loading control.
Protoplast isolation and inoculation
Protoplasts were isolated from NbTRXh2‐knockdown N. benthamiana 10 days after Agrobacterium infiltration. In brief, approximately 2 g of the knockdown leaves were digested with pectinase (0.6 mg/mL) and cellulose (12 mg/mL) at 25 ºC overnight. The mesophyll protoplasts were isolated from the interface zone between the mannitol‐MES buffer (0.55 m, pH 5.7) and sucrose (0.55 m, pH 5.7). After a few washes, the protoplasts were examined under a fluorescent microscope after staining with fluorescein diacetate. Approximately 4 × 105 cells were inoculated with 1 µg of BaMV viral RNA as described previously (Cheng et al., 2013a; Tsai et al., 1999). The protoplasts were resuspended in culture medium (1 µm CuSO4, 1 µm KI, 10 mm MgSO4, 0.2 mm KPO4, 10 mm KNO3, 10 mm CaCl2, 0.03% cephaloridine, 0.001% gentamycin, in 0.55 m mannitol‐MES) and incubated at 25 ºC under constant light for 24 h.
Cell extract fractionation
For the fractionation assay, 1 g of infiltrated leaves was homogenized with 2 mL of extraction buffer (50 mm Tris‐HCl, pH 7.6, 15 mm MgCl2, 120 mm KCl, 20% glycerol, 0.1% β‐mercaptoethanol and the protease inhibitor) and spun at 4 °C at 2000 rpm (Eppendorf 5415D, Hauppauge, NY, USA) for 10 min (Laliberte et al., 2007). The supernatant was collected and centrifuged at 4 °C at 11 151 g (Beckman TL‐100, Indianapolis, IN, USA) for 35 min to separate the cytosol fraction (supernatant) and the membrane fraction (pellet). The membrane fraction was resuspended in 2 mL of extraction buffer. Portions of both fractions were then mixed with 4 × sample buffer (250 mm Tris‐HCl, pH 6.8, 40% glycerol, 0.02% bromophenol blue and 10% β‐mercaptoethanol) and boiled for 5 min. The proteins were separated on SDS‐polyacrylamide gel and examined by Western blotting.
5′ RACE
The cDNA preparation and 5′ RACE were performed using the SMARTer™ RACE cDNA Amplification Kit (Clontech Laboratories, Inc., Mountain View, CA, USA) with the forward primer, GSP1, supplied from the kit and the two reverse primers, NbTRX5′RACE3 (5′‐GCTTAATATGCATAGAAACACGTAAAAA‐3′) and NbTRX5′RACE4 (5′‐GTTGGAGCAGTAAAACGCAAAATTAAAGTGAC‐3′) designed on the basis of the sequence of N. benthamiana EST (NBREL2_JP_004_E09_19MAY2004_071), which was 100% matched with the ACGT4 cDNA‐AFLP fragment sequence.
Analysis of BaMV cell‐to‐cell movement
The fourth leaf above the infiltrated leaf of NbTRXh2‐ and Luc‐knockdown plants was mechanically inoculated with 10 µg of pCBG in which the infectious BaMV RNA carrying the subgenomic promoter to express GFP protein can be transcribed from the CaMV 35S promoter (Lin et al., 2006). At 5 dpi, the green fluorescence as an infected focus when BaMV moved out of the infected cell was detected by an Olympus IX71 inverted microscope (Tokyo, Japan) using a set of filters: an excitation filter (460‐495 nm) and an emission filter (510 nm), with a chromatin mirror (505 nm). The area of green fluorescent foci was then calculated by ImageJ software (http://rsbweb.nih.gov/ij/).
Transient expression of NbTRXh2‐OFP fusion protein and its derivatives
To construct the vector to express NbTRXh2‐OFP fusion protein, the ORF of NbTRXh2 was amplified by PCR with the primers NbTRXh2/F (5′‐GGATCCATGGCAAGTATGACGGGTGGACAACAGATGGGTTCTAGAATGGGACAGTGCTGGACTAAGGC‐3′) and NbTRXh2Cter(–)KpnI (5′‐GGGTACCACTCGGGGACCTTGTAG‐3′) containing the XbaI and KpnI sites (in italic), respectively. To create the G2A mutant, the primers NbTRXh2G2A (5′‐GTTCTAGAATGGCTCAGTGCTGGACTAAGGC‐3′; XbaI site in italic) and NbTRXh2Cter(–)KpnI were used to amplify the DNA fragment. To generate the active site mutants, the forward primers NbTRXh2C62A (5′‐GGTGTAACCCCGCTAGAACAGCAGCA‐3′) and NbTRXh2C59A/C62A (5′‐GCGTCATGGGCTAACCCCGCTAGAACA‐3′) and the reverse primer NbTRXh2Cter(–)KpnI were used to synthesize the megaprimers. These megaprimers were used for the second PCR with the upstream primer NbTRXh2/F to generate the single‐ and double‐amino‐acid substitution mutants. The DNA fragments were released from the vector by digestion with XbaI and KpnI, and cloned into the OFP‐containing vector pBin‐OFP reconstructed from pmKO2‐S1 (MBL International, Woburn, MA, USA). pBin/NbTRXh2‐OFP and its derivatives were transformed into the Agrobacterium C58C1 strain. Agrobacterium containing pBin/NbTRXh2‐OFP and its derivatives were cultured and infiltrated into BaMV‐infected N. benthamiana leaves at 2 dpi. After 3 days of infiltration, protein expression was examined by Western blot analysis with anti‐OFP antibody (produced by Yao‐Hong Biotechnology Inc., New Taipei City, Taiwan).
Localization of NbTRXh2 and its derivatives
The transient expression of NbTRXh2‐OFP, its derivatives or OFP with HcPro on N. benthamiana leaves is described in the previous section. The image was taken at 3 days post‐infiltration of the leaf tissue or the protoplasts with a confocal laser scanning microscope (FV1000, Olympus) using a 40× objective lens. For the fractionation assay, the soluble and membrane fractions were separated as described previously (Cheng et al., 2013b).
Construction and expression of His‐tagged NbTRXh2 derivatives
To express NbTRXh2 and its derivatives in E. coli, the DNA fragments with wild‐type NbTRXh2 or mutant sequences were amplified with the primer set BamHI‐TRXh1 5′(+) (5′‐GAATTCATGGGTTCTAGAATGGGACA‐3′) and XhoI‐TRXh1 3′(–) (5′‐GCTCGAGAGCTATGCATCCAACGCGTT‐3′), and cloned into pGEM‐T easy vector (Promega). The cloned DNA fragments were verified by sequencing, digested with BamHI and XhoI, and subcloned into the pET‐29a(+) vector (Novagen, Madison, WI, USA).
To produce the recombinant His‐tagged NbTRXh2 protein and its derivatives, E. coli containing the suitable plasmid was grown and induced to express the protein as described previously (Vijayapalani et al., 2012). The cells were harvested and resuspended in dilution buffer (10 mm Tris‐HCl, pH 7.5, 150 mm NaCl) containing the protease inhibitor cocktail (Roche Diagnostics GmbH, Mannheim, Germany), followed by disruption. The soluble recombinant proteins were collected by centrifugation at 17 500 g for 10 min.
Construction of GFP‐fused BaMV MPs
To create GFP‐fused BaMV MPs, the TGBp1 and TGBp2 genes were amplified by PCR using primer sets: TGBp1(+)SacI (5′‐GGAGCTCATGGATAACCGGATAA‐3′) and TGBp1(–)SacI (5′‐GGAGCTCTCAGGTGGTCTGGCCAG‐3′) for TGBp1; and TGBp2(+)SacI (5′‐GGAGCTCATGGACCAGCCTCTTCA‐3′) and TGBp2(–)SacI (5′‐GGAGCTCTTAGCATGGTGGGTGAT‐3′) for TGBp2. The amplified fragments were cloned into pGEM‐T easy vector (Promega), verified by sequencing and subsequently subcloned into SacI‐digested pBI‐N‐mGFP vector, which was modified from pBI‐mGFP (Cheng et al., 2013a) with the addition of a start codon and a deletion stop codon.
Co‐immunoprecipitation
Agro‐infiltrated leaves (approximately 10 g each), collected from transiently expressing GFP, GFP‐TGBp1 or GFP‐TGBp2 N. benthamiana plants, were homogenized in 20 mL of dilution buffer (10 mm Tris‐HCl, pH 7.5, 150 mM NaCl) containing the protease inhibitor cocktail (Roche Diagnostics), and centrifuged at 1000 g for 10 min to remove the debris. The soluble proteins (GFP and GFP‐TGBp1) in the supernatant (S30) were harvested by centrifugation at 30 000 g for another 35 min. The integral membrane protein, GFP‐TGBp2, in the pellet (P30) was subsequently solubilized with 4 mL of 0.1% sarcosyl at 4 ºC for 1 h. The detergent‐solubilized proteins were collected after centrifugation at 30 000 g for 35 min. For the analysis by sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE), the input was 10 μL out of a total of 20 mL (0.05%) extract for GFP and GFP‐TGBp1, and 10 μL out of a total of 4 mL (0.25%) for GFP‐TGBp2 in Fig. 6.
The transiently expressed GFP or GFP‐fused proteins (300 μL) were magnetically immobilized on 10 μL Chromotek‐GFP‐Trap M beads (Allele Biotechnology & Pharmaceuticals, San Diego, CA, USA) at 4 ºC for 1 h according to the manufacturer's instructions. The beads were washed twice with 200 μL of dilution buffer containing 0.5 mm EDTA and mixed with the cell lysates derived from E. coli which expressed His‐NbTRXh2 or its derivatives at 4 ºC for 1 h. After incubation, the complex‐containing beads were washed sequentially with dilution buffer, dilution buffer plus 500 mm NaCl, and dilution buffer plus 0.1% SDS. Finally, the complex‐containing beads were resuspended in 40 μL of 2 × SDS‐sample buffer and 10 μL of each sample were loaded on the gel for analysis.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 The morphology of NbTRXh2‐knockdown and control (Luc‐knockdown) plants. No significant differences were observed between NbTRXh2‐knockdown and control plants. The positive control is the phytoene desaturase‐knockdown plant indicated as PDS. The negative control is the luciferase‐knockdown plant indicated as Luc.
Fig. S2 The expression profiles of NbTRXh2 and viral protein accumulation in Nicotiana benthamiana plants after virus inoculation. Four‐week‐old plants were inoculated with 500 ng of Barley mosaic virus (BaMV), Potato virus X (PVX) or Cucumber mosaic virus (CMV). The total RNAs and proteins were extracted from inoculated leaves at 1, 3, 5 and 7 days post‐inoculation (dpi) and used to determine the expression levels of NbTRXh2 by real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) in (A) and the accumulation levels of the viral coat protein (CP) by Western blotting in (B). The numbers in the table shown under the statistic bars are the average and standard deviation (n = 3) of the relative expression of NbTRXh2. M, mock‐inoculated plants; B, BaMV‐inoculated plants; P, PVX‐inoculated plants; C, CMV‐inoculated plants. The expression level of the mock‐inoculated plant (M1) was set as 100%. Asterisks indicate statistically significant differences of the indicated group analysed by Student's t‐test (*P < 0.05; ***P < 0.001).
Fig. S3 Cell‐to‐cell movement of Barley mosaic virus (BaMV) in Luc‐ and NbTRXh2‐knockdown plants. (A) Areas of green fluorescent protein (GFP) foci on pCBG (500 ng of the infectious BaMV cDNA viral vector which can express GFP)‐inoculated Nicotiana benthamiana leaves (about 4‐week‐old plants) at 1.5 and 3 days post‐inoculation, measured by fluorescence microscopy. Bar length, 0.2 mm. (B) Statistical analysis of the results obtained from (A); y‐axis is the GFP focus size (mm2). The numbers shown above the bars are the average and standard deviation of 10 foci from Luc‐ and NbTRXh2‐knockdown plants. Asterisks indicate statistically significant differences of the indicated group analysed by Student's t‐test (***P < 0.001).
Acknowledgements
We are very grateful to Dr Chang‐Hsien Yang (Graduate Institute of Biotechnology, National Chung Hsing University, Taichung, Taiwan) for providing the pBI‐N‐mGFP vector. The confocal laser scanning microscopy was supported by the Center of Nanoscience & Nanotechnology (National Chung‐Hsing University, Taichung, Taiwan). This research was supported by the Ministry of Science and Technology Grant 99‐2313‐B‐005‐019‐MY3.
References
- Atabekov, J.G. , Rodionova, N.P. , Karpova, O.V. , Kozlovsky, S.V. and Poljakov, V.Y. (2000) The movement protein‐triggered in situ conversion of potato virus X virion RNA from a nontranslatable into a translatable form. Virology, 271, 259–263. [DOI] [PubMed] [Google Scholar]
- Atkins, D. , Hull, R. , Wells, B. , Roberts, K. , Moore, P. and Beachy, R.N. (1991) The Tobacco mosaic virus 30K movement protein in transgenic tobacco plants is localized to plasmodesmata. J. Gen. Virol. 72, 209–211. [DOI] [PubMed] [Google Scholar]
- Balmer, Y. , Vensel, W.H. , Tanaka, C.K. , Hurkman, W.J. , Gelhaye, E. , Rouhier, N. , Jacquot, J.P. , Manieri, W. , Schürmann, P. , Droux, M. and Buchanan, B.B. (2004) Thioredoxin links redox to the regulation of fundamental processes of plant mitochondria. Proc. Natl. Acad. Sci. USA, 101, 2642–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamunusinghe, D. , Hemenway, C.L. , Nelson, R.S. , Sanderfoot, A.A. , Ye, C.M. , Silva, M.A. , Payton, M. and Verchot‐Lubicz, J. (2009) Analysis of Potato virus X replicase and TGBp3 subcellular locations. Virology, 393, 272–285. [DOI] [PubMed] [Google Scholar]
- Bayne, E.H. , Rakitina, D.V. , Morozov, S.Y. and Baulcombe, D.C. (2005) Cell‐to‐cell movement of Potato potexvirus X is dependent on suppression of RNA silencing. Plant J. 44, 471–482. [DOI] [PubMed] [Google Scholar]
- Bhat, S. , Folimonova, S.Y. , Cole, A.B. , Ballard, K.D. , Lei, Z. , Watson, B.S. , Sumner, L.W. and Nelson, R.S. (2013) Influence of host chloroplast proteins on Tobacco mosaic virus accumulation and intercellular movement. Plant Physiol. 161, 134–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, I.H. , Chou, W.J. , Lee, P.Y. , Hsu, Y.H. and Tsai, C.H. (2005) The AAUAAA motif of Bamboo mosaic virus RNA is involved in minus‐strand RNA synthesis and plus‐strand RNA polyadenylation. J. Virol. 79, 14 555–14 561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, C.W. , Hsiao, Y.Y. , Wu, H.C. , Chuang, C.M. , Chen, J.S. , Tsai, C.H. , Hsu, Y.H. , Wu, Y.C. , Lee, C.C. and Meng, M. (2009) Suppression of Bamboo mosaic virus accumulation by a putative methyltransferase in Nicotiana benthamiana . J. Virol. 83, 5796–5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, S.F. , Huang, Y.P. , Wu, Z.R. , Hu, C.C. , Hsu, Y.H. and Tsai, C.H. (2010) Identification of differentially expressed genes induced by Bamboo mosaic virus infection in Nicotiana benthamiana by cDNA‐amplified fragment length polymorphism. BMC Plant Biol. 10, 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, S.F. , Huang, Y.P. , Chen, L.H. , Hsu, Y.H. and Tsai, C.H. (2013a) Chloroplast phosphoglycerate kinase is involved in the targeting of Bamboo mosaic virus to chloroplasts in Nicotiana benthamiana plants. Plant Physiol. 163, 1598–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, S.F. , Tsai, M.S. , Huang, C.L. , Huang, Y.P. , Chen, I.H. , Lin, N.S. , Hsu, Y.H. , Tsai, C.H. and Cheng, C.P. (2013b) Ser/Thr kinase‐like protein of Nicotiana benthamiana is involved in the cell‐to‐cell movement of Bamboo mosaic virus . PLoS One, 8, e62907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, M.H. , Chen, I.H. , Baulcombe, D.C. and Tsai, C.H. (2010) The silencing suppressor P25 of Potato virus X interacts with Argonaute1 and mediates its degradation through the proteasome pathway. Mol. Plant Pathol. 11, 641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, Y.L. , Hung, Y.J. , Tseng, Y.H. , Hsu, H.T. , Yang, J.Y. , Wung, C.H. , Lin, N.S. , Meng, M. , Hsu, Y.H. and Chang, B.Y. (2013) The stable association of virion with the triple‐gene‐block protein 3‐based complex of Bamboo mosaic virus . PLoS Pathog. 9, e1003405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky, V. , Knorr, D. , Schuster, G. and Zambryski, P. (1990) The P30 movement protein of Tobacco mosaic virus is a single‐strand nucleic acid binding protein. Cell, 60, 637–647. [DOI] [PubMed] [Google Scholar]
- Collet, J.F. and Messens, J. (2010) Structure, function, and mechanism of thioredoxin proteins. Antioxid. Redox Signal. 13, 1205–1216. [DOI] [PubMed] [Google Scholar]
- DiMaio, F. , Chen, C.C. , Yu, X. , Frenz, B. , Hsu, Y.H. , Lin, N.S. and Egelman, E.H. (2015) The molecular basis for flexibility in the flexible filamentous plant viruses. Nat. Struct. Mol. Biol. 22, 642–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridborg, I. , Grainger, J. , Page, A. , Coleman, M. , Findlay, K. and Angell, S. (2003) TIP, a novel host factor linking callose degradation with the cell‐to‐cell movement of Potato virus X . Mol. Plant–Microbe Interact. 16, 132–140. [DOI] [PubMed] [Google Scholar]
- Gelhaye, E. , Rouhier, N. and Jacquot, J.P. (2003) Evidence for a subgroup of thioredoxin h that requires GSH/Grx for its reduction. FEBS Lett. 555, 443–448. [DOI] [PubMed] [Google Scholar]
- Gelhaye, E. , Rouhier, N. and Jacquot, J.P. (2004) The thioredoxin h system of higher plants. Plant Physiol. Biochem. 42, 265–271. [DOI] [PubMed] [Google Scholar]
- Heinlein, M. (2015) Plant virus replication and movement. Virology, 479‐480, 657–671. [DOI] [PubMed] [Google Scholar]
- Howard, A.R. , Heppler, M.L. , Ju, H.J. , Krishnamurthy, K. , Payton, M.E. and Verchot‐Lubicz, J. (2004) Potato virus X TGBp1 induces plasmodesmata gating and moves between cells in several host species whereas CP moves only in N. benthamiana leaves. Virology, 328, 185–197. [DOI] [PubMed] [Google Scholar]
- Hsu, H.T. , Chou, Y.L. , Tseng, Y.H. , Lin, Y.H. , Lin, T.M. , Lin, N.S. , Hsu, Y.H. and Chang, B.Y. (2008) Topological properties of the triple gene block protein 2 of Bamboo mosaic virus . Virology, 379, 1–9. [DOI] [PubMed] [Google Scholar]
- Hsu, H.T. , Tseng, Y.H. , Chou, Y.L. , Su, S.H. , Hsu, Y.H. and Chang, B.Y. (2009) Characterization of the RNA‐binding properties of the triple‐gene‐block protein 2 of Bamboo mosaic virus . Virol. J. 6, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y.L. , Han, Y.T. , Chang, Y.T. , Hsu, Y.H. and Meng, M. (2004) Critical residues for GTP methylation and formation of the covalent m7GMP‐enzyme intermediate in the capping enzyme domain of bamboo mosaic virus. J. Virol. 78, 1271–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y.P. , Chen, J.S. , Hsu, Y.H. and Tsai, C.H. (2013) A putative Rab‐GTPase activation protein from Nicotiana benthamiana is important for Bamboo mosaic virus intercellular movement. Virology, 447, 292–299. [DOI] [PubMed] [Google Scholar]
- Huh, S.U. and Paek, K.H. (2013) Plant RNA binding proteins for control of RNA virus infection. Front. Physiol. 4, 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi, K. , Nishikiori, M. and Ishikawa, M. (2010) Interactions between tobamovirus replication proteins and cellular factors: their impacts on virus multiplication. Mol. Plant–Microbe Interact. 23, 1413–1419. [DOI] [PubMed] [Google Scholar]
- Ishiwatari, Y. , Fujiwara, T. , McFarland, K.C. , Nemoto, K. , Hayashi, H. , Chino, M. and Lucas, W.J. (1998) Rice phloem thioredoxin h has the capacity to mediate its own cell‐to‐cell transport through plasmodesmata. Planta, 205, 12–22. [DOI] [PubMed] [Google Scholar]
- Joudrier, P. , Gautier, M.F. , de Lamotte, F. and Kobrehel, K. (2005) The thioredoxin h system: potential applications. Biotechnol. Adv. 23, 81–85. [DOI] [PubMed] [Google Scholar]
- Ju, H.J. , Samuels, T.D. , Wang, Y.S. , Blancaflor, E. , Payton, M. , Mitra, R. , Krishnamurthy, K. , Nelson, R.S. and Verchot‐Lubicz, J. (2005) The Potato virus X TGBp2 movement protein associates with endoplasmic reticulum‐derived vesicles during virus infection. Plant Physiol. 138, 1877–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy, K. , Heppler, M. , Mitra, R. , Blancaflor, E. , Payton, M. , Nelson, R.S. and Verchot‐Lubicz, J. (2003) The Potato virus X TGBp3 protein associates with the ER network for virus cell‐to‐cell movement. Virology, 309, 135–151. [DOI] [PubMed] [Google Scholar]
- Kumar, D. , Kumar, R. , Hyun, T.K. and Kim, J.Y. (2015) Cell‐to‐cell movement of viruses via plasmodesmata. J. Plant Res. 128, 37–47. [DOI] [PubMed] [Google Scholar]
- Laliberte, J.F. , Beauchemin, C. and Boutet, N. (2007) Visualization of the interaction between the precursors of VPg, the viral protein linked to the genome of Turnip mosaic virus, and the translation eukaryotic initiation factor iso 4E in planta. J. Virol. 81, 775–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, P. , Yeh, W.B. , Tsai, C.W. and Lin, N.S. (2010) A unique glycine‐rich motif at the N‐terminal region of Bamboo mosaic virus coat protein is required for symptom expression. Mol. Plant–Microbe Interact. 23, 903–914. [DOI] [PubMed] [Google Scholar]
- Lee, C.C. , Ho, Y.N. , Hu, R.H. , Yen, Y.T. , Wang, Z.C. , Lee, Y.C. , Hsu, Y.H. and Meng, M. (2011) The interaction between Bamboo mosaic virus replication protein and coat protein is critical for virus movement in plant hosts. J. Virol. 85, 12 022–12 031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y.I. , Cheng, Y.M. , Huang, Y.L. , Tsai, C.H. , Hsu, Y.H. and Meng, M. (1998) Identification and characterization of the Escherichia coli‐expressed RNA‐dependent RNA polymerase of Bamboo mosaic virus . J. Virol. 72, 10 093–10 099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y.I. , Chen, Y.J. , Hsu, Y.H. and Meng, M. (2001a) Characterization of the AdoMet‐dependent guanylyltransferase activity that is associated with the N terminus of Bamboo mosaic virus replicase. J. Virol. 75, 782–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y.I. , Shih, T.W. , Hsu, Y.H. , Han, Y.T. , Huang, Y.L. and Meng, M. (2001b) The helicase‐like domain of plant potexvirus replicase participates in formation of RNA 5' cap structure by exhibiting RNA 5'‐triphosphatase activity. J. Virol. 75, 12 114–12 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J.Y. , Mendu, V. , Pogany, J. , Qin, J. and Nagy, P.D. (2012) The TPR domain in the host Cyp40‐like cyclophilin binds to the viral replication protein and inhibits the assembly of the tombusviral replicase. PLoS Pathog. 8, e1002491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, L. , Luo, Z.P. , Yan, F. , Lu, Y.W. , Zheng, H.Y. and Chen, J.P. (2011) Interaction between potyvirus P3 and ribulose‐1,5‐bisphosphate carboxylase/oxygenase (RubisCO) of host plants. Virus Genes, 43, 90–92. [DOI] [PubMed] [Google Scholar]
- Lin, M.K. , Chang, B.Y. , Liao, J.T. , Lin, N.S. and Hsu, Y.H. (2004) Arg‐16 and Arg‐21 in the N‐terminal region of the triple‐gene‐block protein 1 of Bamboo mosaic virus are essential for virus movement. J. Gen. Virol. 85, 251–259. [DOI] [PubMed] [Google Scholar]
- Lin, M.K. , Hu, C.C. , Lin, N.S. , Chang, B.Y. and Hsu, Y.H. (2006) Movement of potexviruses requires species‐specific interactions among the cognate triple gene block proteins, as revealed by a trans‐complementation assay based on the Bamboo mosaic virus satellite RNA‐mediated expression system. J. Gen. Virol. 87, 1357–1367. [DOI] [PubMed] [Google Scholar]
- Lin, N.S. , Lin, F.Z. , Huang, T.Y. and Hsu, Y.H. (1992) Genome properties of Bamboo mosaic virus . Mol. Plant Pathol. 82, 731–734. [Google Scholar]
- Lin, N.S. , Lin, B.Y. , Lo, N.W. , Hu, C.C. , Chow, T.Y. and Hsu, Y.H. (1994) Nucleotide sequence of the genomic RNA of Bamboo mosaic potexvirus . J. Gen. Virol. 75, 2513–2518. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Schiff, M. and Dinesh‐Kumar, S.P. (2002) Virus‐induced gene silencing in tomato. Plant J. 31, 777–786. [DOI] [PubMed] [Google Scholar]
- Lough, T.J. , Shash, K. , Xoconostle‐Cázares, B. , Hofstra, K.R. , Beck, D.L. , Balmori, E. , Forster, R.L.S. and Lucas, W.J. (1998) Molecular dissection of the mechanism by which potexvirus triple gene block proteins mediate cell‐to‐cell transport of infectious RNA. Mol. Plant–Microbe Interact. 11, 801–814. [Google Scholar]
- Lough, T.J. , Netzler, N.E. , Emerson, S.J. , Sutherland, P. , Carr, F. , Beck, D.L. , Lucas, W.J. and Forster, R.L. (2000) Cell‐to‐cell movement of potexviruses: evidence for a ribonucleoprotein complex involving the coat protein and first triple gene block protein. Mol. Plant–Microbe Interact. 13, 962–974. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J. and Lee, J.Y. (2004) Plasmodesmata as a supracellular control network in plants. Nat. Rev. Mol. Cell Biol. 5, 712–726. [DOI] [PubMed] [Google Scholar]
- Maurer‐Stroh, S. , Eisenhaber, B. and Eisenhaber, F. (2002) N‐terminal N‐myristoylation of proteins: refinement of the sequence motif and its taxon‐specific differences. J. Mol. Biol. 317, 523–540. [DOI] [PubMed] [Google Scholar]
- Melcher, U. (2000) The '30K' superfamily of viral movement proteins. J. Gen. Virol. 81, 257–266. [DOI] [PubMed] [Google Scholar]
- Meng, L. , Wong, J.H. , Feldman, L.J. , Lemaux, P.G. and Buchanan, B.B. (2010) A membrane‐associated thioredoxin required for plant growth moves from cell to cell, suggestive of a role in intercellular communication. Proc. Natl. Acad. Sci. USA, 107, 3900–3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, Y. , Vignols, F. and Reichheld, J.P. (2002) Classification of plant thioredoxins by sequence similarity and intron position. Methods Enzymol. 347, 394–402. [DOI] [PubMed] [Google Scholar]
- Meyer, Y. , Reichheld, J.P. and Vignols, F. (2005) Thioredoxins in Arabidopsis and other plants. Photosynth. Res. 86, 419–433. [DOI] [PubMed] [Google Scholar]
- Mitra, R. , Krishnamurthy, K. , Blancaflor, E. , Payton, M. , Nelson, R.S. and Verchot‐Lubicz, J. (2003) The Potato virus X TGBp2 protein association with the endoplasmic reticulum plays a role in but is not sufficient for viral cell‐to‐cell movement. Virology, 312, 35–48. [DOI] [PubMed] [Google Scholar]
- Morozov, S.Y. and Solovyev, A.G. (2003) Triple gene block: modular design of a multifunctional machine for plant virus movement. J. Gen. Virol. 84, 1351–1366. [DOI] [PubMed] [Google Scholar]
- Morozov, S.Y. , Solovyev, A.G. , Kalinina, N.O. , Fedorkin, O.N. , Samuilova, O.V. , Schiemann, J. and Atabekov, J.G. (1999) Evidence for two nonoverlapping functional domains in the Potato virus X 25K movement protein. Virology, 260, 55–63. [DOI] [PubMed] [Google Scholar]
- Nakasugi, K. , Crowhurst, R.N. , Bally, J. , Wood, C.C. , Hellens, R.P. and Waterhouse, P.M. (2013) De novo transcriptome sequence assembly and analysis of RNA silencing genes of Nicotiana benthamiana . PLoS One. 8, e59534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oparka, K.J. , Prior, D.A. , Santa Cruz, S. , Padgett, H.S. and Beachy, R.N. (1997) Gating of epidermal plasmodesmata is restricted to the leading edge of expanding infection sites of Tobacco mosaic virus (TMV). Plant J. 12, 781–789. [DOI] [PubMed] [Google Scholar]
- Park, M.R. , Seo, J.K. and Kim, K.H. (2013) Viral and nonviral elements in potexvirus replication and movement and in antiviral responses. Adv. Virus Res. 87, 75–112. [DOI] [PubMed] [Google Scholar]
- Park, M.R. , Jeong, R.D. and Kim, K.H. (2014) Understanding the intracellular trafficking and intercellular transport of potexviruses in their host plants. Front. Plant Sci. 5, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichheld, J.P. , Mestres‐Ortega, D. , Laloi, C. and Meyer, Y. (2002) The multigenic family of thioredoxin h in Arabidopsis thaliana: specific expression and stress response. Plant Physiol. Biochem. 40, 685–690. [Google Scholar]
- Rodionova, N.P. , Karpova, O.V. , Kozlovsky, S.V. , Zayakina, O.V. , Arkhipenko, M.V. and Atabekov, J.G. (2003) Linear remodeling of helical virus by movement protein binding. J. Mol. Biol. 333, 565–572. [DOI] [PubMed] [Google Scholar]
- Ruggenthaler, P. , Fichtenbauer, D. , Krasensky, J. , Jonak, C. and Waigmann, E. (2009) Microtubule‐associated protein AtMPB2C plays a role in organization of cortical microtubules, stomata patterning, and tobamovirus infectivity. Plant Physiol. 149, 1354–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz, M.T. , Voinnet, O. and Baulcombe, D.C. (1998) Initiation and maintenance of virus‐induced gene silencing. Plant Cell, 10, 937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smotrys, J.E. and Linder, M.E. (2004) Palmitoylation of intracellular signaling proteins: regulation and function. Annu. Rev. Biochem. 73, 559–587. [DOI] [PubMed] [Google Scholar]
- Sun, L. , Ren, H. , Liu, R. , Li, B. , Wu, T. , Sun, F. , Liu, H. , Wang, X. and Dong, H. (2010) An h‐type thioredoxin functions in tobacco defense responses to two species of viruses and an abiotic oxidative stress. Mol. Plant–Microbe Interact. 23, 1470–1485. [DOI] [PubMed] [Google Scholar]
- Tada, Y. , Spoel, S.H. , Pajerowska‐Mukhtar, K. , Mou, Z. , Song, J. , Wang, C. , Zuo, J. and Dong, X. (2008) Plant immunity requires conformational changes [corrected] of NPR1 via S‐nitrosylation and thioredoxins. Science, 321, 952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarrago, L. , Laugier, E. , Zaffagnini, M. , Marchand, C.H. , Le Marechal, P. , Lemaire, S.D. and Rey, P. (2010) The plant thioredoxin CDSP32 regenerates 1‐cys methionine sulfoxide reductase B activity through the direct reduction of sulfenic acid. J. Biol. Chem. 285, 14 964–14 972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomenius, K. , Clapham, D. and Meshi, T. (1987) Localization by immunogold cytochemistry of the virus‐coded 30K protein in plasmodesmata of leaves infected with tobacco mosaic virus. Virology, 160, 363–371. [DOI] [PubMed] [Google Scholar]
- Tsai, C.H. , Cheng, C.P. , Peng, C.W. , Lin, B.Y. , Lin, N.S. and Hsu, Y.H. (1999) Sufficient length of a poly(A) tail for the formation of a potential pseudoknot is required for efficient replication of Bamboo mosaic potexvirus RNA. J. Virol. 73, 2703–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng, Y.H. , Hsu, H.T. , Chou, Y.L. , Hu, C.C. , Lin, N.S. , Hsu, Y.H. and Chang, B.Y. (2009) The two conserved cysteine residues of the triple gene block protein 2 are critical for both cell‐to‐cell and systemic movement of Bamboo mosaic virus . Mol. Plant–Microbe Interact. 22, 1379–1388. [DOI] [PubMed] [Google Scholar]
- Verchot‐Lubicz, J. (2005) A new cell‐to‐cell transport model for Potexviruses. Mol. Plant–Microbe Interact. 18, 283–290. [DOI] [PubMed] [Google Scholar]
- Verchot‐Lubicz, J. , Torrance, L. , Solovyev, A.G. , Morozov, S.Y. , Jackson, A.O. and Gilmer, D. (2010) Varied movement strategies employed by triple gene block‐encoding viruses. Mol. Plant–Microbe Interact. 23, 1231–1247. [DOI] [PubMed] [Google Scholar]
- Vijayapalani, P. , Chen, J.C. , Liou, M.R. , Chen, H.C. , Hsu, Y.H. and Lin, N.S. (2012) Phosphorylation of Bamboo mosaic virus satellite RNA (satBaMV)‐encoded protein P20 downregulates the formation of satBaMV‐P20 ribonucleoprotein complex. Nucleic Acids Res. 40, 638–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, S. , Deom, C.M. , Beachy, R.N. and Lucas, W.J. (1989) Movement protein of Tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science, 246, 377–379. [DOI] [PubMed] [Google Scholar]
- Wu, C.H. , Lee, S.C. and Wang, C.W. (2011) Viral protein targeting to the cortical endoplasmic reticulum is required for cell–cell spreading in plants. J. Cell Biol. 193, 521–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J.P. , Liu, Q. , Zhang, H.L. , Jia, Q. , Hong, Y.G. and Liu, Y.L. (2013) The rubisco small subunit is involved in tobamovirus movement and Tm‐2(2)‐mediated extreme resistance. Plant Physiol. 161, 374–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 The morphology of NbTRXh2‐knockdown and control (Luc‐knockdown) plants. No significant differences were observed between NbTRXh2‐knockdown and control plants. The positive control is the phytoene desaturase‐knockdown plant indicated as PDS. The negative control is the luciferase‐knockdown plant indicated as Luc.
Fig. S2 The expression profiles of NbTRXh2 and viral protein accumulation in Nicotiana benthamiana plants after virus inoculation. Four‐week‐old plants were inoculated with 500 ng of Barley mosaic virus (BaMV), Potato virus X (PVX) or Cucumber mosaic virus (CMV). The total RNAs and proteins were extracted from inoculated leaves at 1, 3, 5 and 7 days post‐inoculation (dpi) and used to determine the expression levels of NbTRXh2 by real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) in (A) and the accumulation levels of the viral coat protein (CP) by Western blotting in (B). The numbers in the table shown under the statistic bars are the average and standard deviation (n = 3) of the relative expression of NbTRXh2. M, mock‐inoculated plants; B, BaMV‐inoculated plants; P, PVX‐inoculated plants; C, CMV‐inoculated plants. The expression level of the mock‐inoculated plant (M1) was set as 100%. Asterisks indicate statistically significant differences of the indicated group analysed by Student's t‐test (*P < 0.05; ***P < 0.001).
Fig. S3 Cell‐to‐cell movement of Barley mosaic virus (BaMV) in Luc‐ and NbTRXh2‐knockdown plants. (A) Areas of green fluorescent protein (GFP) foci on pCBG (500 ng of the infectious BaMV cDNA viral vector which can express GFP)‐inoculated Nicotiana benthamiana leaves (about 4‐week‐old plants) at 1.5 and 3 days post‐inoculation, measured by fluorescence microscopy. Bar length, 0.2 mm. (B) Statistical analysis of the results obtained from (A); y‐axis is the GFP focus size (mm2). The numbers shown above the bars are the average and standard deviation of 10 foci from Luc‐ and NbTRXh2‐knockdown plants. Asterisks indicate statistically significant differences of the indicated group analysed by Student's t‐test (***P < 0.001).


