Summary
DEFORMED ROOT AND LEAVES1 (DRL1) is an Arabidopsis homologue of the yeast TOXIN TARGET4 (TOT4)/KILLER TOXIN‐INSENSITIVE12 (KTI12) protein that is physically associated with the RNA polymerase II‐interacting protein complex named Elongator. Mutations in DRL1 and Elongator lead to similar morphological and molecular phenotypes, suggesting that DRL1 and Elongator may functionally overlap in Arabidopsis. We have shown previously that Elongator plays an important role in both salicylic acid (SA)‐ and jasmonic acid (JA)/ethylene (ET)‐mediated defence responses. Here, we tested whether DRL1 also plays a similar role as Elongator in plant immune responses. Our results show that, although DRL1 partially contributes to SA‐induced cytotoxicity, it does not play a significant role in SA‐mediated expression of PATHOGENESIS‐RELATED genes and resistance to the virulent bacterial pathogen Pseudomonas syringae pv. maculicola ES4326. In contrast, DRL1 is required for JA/ET‐ and necrotrophic fungal pathogen Botrytis cinerea‐induced defence gene expression and for resistance to B. cinerea and Alternaria brassicicola. Furthermore, unlike the TOT4/KTI12 gene which, when overexpressed in yeast, confers zymocin resistance, a phenotype of the tot4/kti12 mutant, overexpression of DRL1 does not change B. cinerea‐induced defence gene expression and resistance to this pathogen. Finally, DRL1 contains an N‐terminal P‐loop and a C‐terminal calmodulin (CaM)‐binding domain and is a CaM‐binding protein. We demonstrate that both the P‐loop and the CaM‐binding domain are essential for the function of DRL1 in B. cinerea‐induced expression of PDF1.2 and ORA59, and in resistance to B. cinerea, suggesting that the function of DRL1 in plant immunity may be regulated by ATP/GTP and CaM binding.
Keywords: Arabidopsis, Botrytis cinerea, DRL1, Elongator, ethylene, jasmonic acid, salicylic acid
Introduction
As a result of their sessile nature, plants have to endure and withstand a wide variety of microbial pathogens with diverse modes of attack and, accordingly, have evolved a highly intricate immune system involving pathogen recognition, signal transduction and the activation of appropriate transcriptional changes. Depending on the lifestyle of the invading pathogen, different signal molecules are synthesized to activate respective defence signalling pathways that are the most effective for resisting the invader (Loake and Grant, 2007; van Loon et al., 2006; Pozo et al., 2005). Salicylic acid (SA), jasmonic acid (JA) and ethylene (ET) are the primary defence signal molecules that play central roles in the regulation of plant immune responses (Pieterse et al., 2009). SA‐activated defence signalling is generally effective against biotrophic and hemibiotrophic pathogens, whereas JA/ET‐mediated defence responses confer resistance to necrotrophic pathogens (Glazebrook, 2005; Thomma et al., 2001). Although SA‐ and JA/ET‐mediated defence pathways have been well characterized (Broekaert et al., 2006; Kazan and Manners, 2008; Vlot et al., 2009), the signalling components of these pathways still remain to be fully uncovered.
Elongator is an RNA polymerase II‐interacting protein complex, which was first identified in yeast and was later purified from human and Arabidopsis cells (Hawkes et al., 2002; Nelissen et al., 2010; Otero et al., 1999; Wittschieben et al., 1999). This complex consists of six subunits and its structure is highly conserved among yeast, humans and Arabidopsis. The six Elongator (ELP) subunits in Arabidopsis were named ELONGATA2 (ELO2)/ELP1, ELP2, ELO3/ELP3, ELO1/ELP4, ELP5 and ELP6 (Nelissen et al., 2010). It should be noted that the acronym ‘ELP’ has been used previously to describe ‘EDM2‐lik proteins’ in Arabidopsis (Eulgem et al., 2007). A large body of evidence in yeast and humans has shown that Elongator plays important roles in diverse cellular and molecular processes, including histone modification, tRNA modification, exocytosis, α‐tubulin acetylation and zygotic paternal genome demethylation (Creppe et al., 2009; Hawkes et al., 2002; Huang et al., 2005; Okada et al., 2010; Rahl et al., 2005; Winkler et al., 2002). In plants, Elongator has been shown to function in meristem and leaf development, cell cycle progression and response to abiotic stresses (Chen et al., 2006; Nelissen et al., 2005; Xu et al., 2012; Zhou et al., 2009). Recently, we have demonstrated that Elongator also plays an important role in SA‐ and JA/ET‐mediated plant defence responses (Defraia et al., 2010, 2013; Wang et al., 2015). Arabidopsis elo/elp mutants exhibit delayed and/or reduced induction of defence genes, including the SA pathway marker gene PATHOGENESIS‐RELATED GENE1 (PR1) and the JA/ET defence pathway marker gene PLANT DEFENSIN1.2 (PDF1.2), and display enhanced suceptibility to the hemibiotrophic bacterial pathogen Pseudomonas syringae and the necrotrophic fungal pathogens Botrytis cinerea and Alternaria brassicicola (Defraia et al., 2010, 2013; Wang et al., 2015). Although the molecular mechanisms by which Elongator modulates plant immunity still require further investigation (Wang et al., 2013), these results indicate that Elongator is required for full activation of the plant immune system.
DEFORMED ROOT AND LEAVES1 (DRL1) is an Arabidopsis homologue of the yeast TOXIN TARGET4 (TOT4)/KILLER TOXIN‐INSENSITIVE12 (KTI12) protein (Butler et al., 1994; Frohloff et al., 2001), which is physically associated with the Elongator complex (Petrakis et al., 2005). TOT4/KTI12 does not specify an Elongator subunit (Fichtner et al., 2002), but yeast cells lacking TOT4/KTI12 are similar to elp mutants in resistance to the Kluyveromyces lactis toxin zymocin and also share other general phenotypes with elp mutants (Petrakis et al., 2005), indicating a close functional relationship between TOT4/KTI12 and Elongator. DRL1 is also physically associated with Elongator, and the Arabidopsis drl1 and elo/elp mutants display similar morphological phenotypes, including narrow leaves, disorganized shoot apical meristem and short roots, and molecular phenotypes, such as transcriptome changes (Nelissen et al., 2003, 2005; Xu et al., 2012), suggesting that DRL1 and Elongator may also functionally overlap in plants. Although the role of Elongator in SA‐ and JA/ET‐mediated defence signalling pathways has been defined (Defraia et al., 2010, 2013; Wang et al., 2015), whether or not DRL1 also functions in these pathways has not been tested.
DRL1 contains several highly conserved regions present in adenosine triphosphate/guanosine triphosphate (ATP/GTP)‐binding or ‐utilizing proteins (Kaziro et al., 1991; Nelissen et al., 2003). The first is a P‐loop [G‐X(4)‐G‐K‐S/T] spanning amino acids 8 to 15 in the N‐terminus (Nelissen et al., 2003). This P‐loop is highly conserved among TOT4/KTI12 homologues (Fichtner et al., 2002). An allele in which this motif is deleted fails to complement the zymocin‐resistant phenotype of the yeast tot4/kti12 mutant, indicating that the P‐loop is essential for the function of TOT4/KTI12 (Fichtner et al., 2002). Whether this motif is important for the function of DRL1 in plants is unknown. The second is the G region (amino acids 148–152, N‐K/R‐X‐D) of GTP‐binding proteins, which interacts directly with the guanine ring of GTP. The third is P‐X(2)‐A/S‐T (amino acids 194–199), which has been found in many ATP/GTP‐utilizing enzymes. However, DRL1 lacks other highly conserved regions found in GTP‐binding proteins; it may therefore not be a bona fide GTP‐bining protein and may instead exert its function on ATP/GTP binding or transfer ATP/GTP to other proteins (Nelissen et al., 2003). DRL1 is also a calmodulin (CaM)‐binding protein. It has been shown that the C‐terminal 100 amino acids of DRL1 bind CaM in a calcium‐dependent manner (Nelissen et al., 2003). A motif L‐X(3)‐F‐X(2)‐L‐X(5)‐L (amino acids 260–273) within the C‐terminal 100 amino acids was considered as the calmodulin‐binding domain (CBD) (Nelissen et al., 2003). However, it is currently unclear whether the CBD is necessary for the function of DRL1.
Here, we show that DRL1 is required for JA/ET‐mediated, but not SA‐mediated, defence responses. Mutations in DRL1 delay and/or decrease JA/ET‐ and B. cinerea‐induced defence gene expression and compromise resistance to B. cinerea and A. brassicicola. We further demonstrate that both the N‐terminal P‐loop and the C‐terminal CBD of DRL1 are essential for its function in plant immune responses.
Results
Mutations in DRL1 partially restore SA tolerance to npr1 and reduce JA/ET‐induced, but not SA‐induced, defence gene expression
NONEXPRESSOR OF PR GENES1 (NPR1) is a master transcription coactivator of the SA signalling pathway (Dong, 2004). Mutations in the NPR1 gene not only block SA signalling, but also make the mutant plants hypersensitive to SA‐induced cytotoxicity (Cao et al., 1994, 1997; Delaney et al., 1995; Kinkema et al., 2000; Shah et al., 1997). On half‐strength Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) supplemented with high concentrations of SA, npr1 seedlings are chlorotic and fail to develop beyond the cotyledon stage, whereas the wild‐type exhibits tolerance to SA cytotoxicity. We have shown previously that mutations in the Elongator subunits ELP2 and ELO3/ELP3 restore SA tolerance to npr1 (Defraia et al., 2010, 2013). As DRL1 is functionally associated with Elongator (Nelissen et al., 2003; Xu et al., 2012), mutations in the DRL1 gene might also restore SA tolerance. To test this hypothesis, we identified two transfer DNA (T‐DNA) insertion homozygous lines, SALK_056915 and SALK_140551, which exhibited significantly reduced expression of DRL1 and displayed a morphology similar to that of the previously charaterized elp2 mutant (Fig. S1a,b, see Supporting Information). As SALK lines are in the Col‐0 genetic background (Alonso et al., 2003), SALK_056915 and SALK_140551 were named drl1‐C1 and drl1‐C2, respectively. We generated double mutants drl1‐C1 npr1 and drl1‐C2 npr1 by crossing with the npr1‐3 mutant and germinated the double mutant seeds on half‐strength MS medium containing 0.3 mm of SA. As reported previously (Cao et al., 1997; Kinkema et al., 2000), the npr1 seedlings turned chlorotic because of SA cytotoxicity, whereas the wild‐type seedlings remained green (Fig. 1a). The drl1‐C1 npr1 and drl1‐C2 npr1 seedlings exhibited an intermediate phenotype (Fig. 1a), indicating partial restoration of SA tolerance in the double mutants. However, the drl1‐C1 npr1 and drl1‐C2 npr1 mutant plants were as susceptible as npr1 to the hemibiotrophic bacterial pathogen P. syringae pv. maculicola (Psm) ES4326 (Fig. S2, see Supporting Information), suggesting that mutations in DRL1 might not affect resistance to bacterial pathogens.
Figure 1.
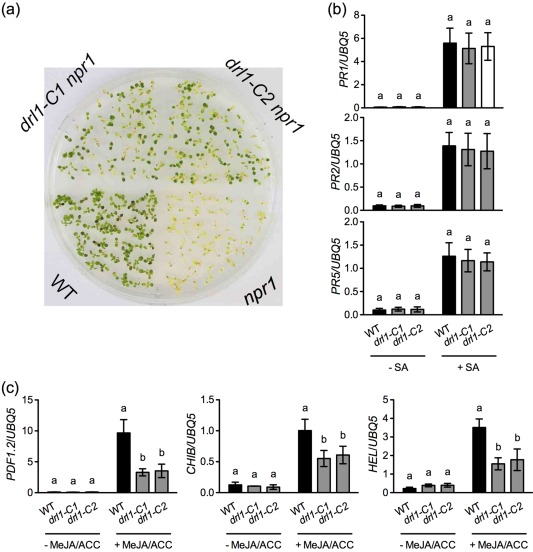
Tolerance to salicylic acid (SA)‐induced cytotoxicity in drl1 npr1 double mutants, and SA‐ and jasmonic acid/ethylene (JA/ET)‐induced defence gene expression in drl1 mutants. (a) Seeds were plated on half‐strength Murashige and Skoog (MS) agar medium containing 0.3 mm SA. After 3 days of stratification, the plate was transferred to a growth chamber and photographed 10 days later. WT, wild‐type. (b, c) Two‐week‐old seedlings grown on half‐strength MS medium were treated with or without 0.5 mm SA (b) and with or without 0.1 mm methyl jasmonate (MeJA) plus 0.1 mm 1‐aminocyclopropane‐1‐carboxylic acid (ACC) (MeJA/ACC) (c). Total RNA was extracted from plant tissues, except roots, collected 24 h later and subjected to quantitative polymerase chain reaction (qPCR) analysis. Expression was normalized against the constitutively expressed UBQ5. Data represent the mean of three biological replicates with standard deviation (SD). Different letters above the bars indicate significant differences [P < 0.05, one‐way analysis of variance (ANOVA)]. Comparison was made separately among the wild‐type, drl1‐C1 and drl1‐C2 for each treatment. The experiments were repeated three times with similar trends.
As mutations in Elongator genes inhibit SA‐ and JA/ET‐mediated defence signalling (Defraia et al., 2010, 2013; Wang et al., 2015), we tested whether the drl1 mutations affect SA‐ and JA/ET‐induced defence gene expression. To this end, we treated the drl1 mutants with SA or the JA derivative methyl jasmonate (MeJA) plus the ET precursor 1‐aminocyclopropane‐1‐carboxylic acid (ACC) by soaking 2‐week‐old seedlings grown on half‐strength MS plates with 0.5 mm SA or 0.1 mm MeJA plus 0.1 mm ACC. Twenty‐four hours later, the induction of three SA pathway genes, PR1, PR2 and PR5, as well as three JA/ET‐inducible genes, PDF1.2, BASIC CHITINASE (CHIB) and HEVEIN‐LIKE (HEL), was examined by real‐time quantitative PCR (qPCR). As shown in Fig. 1b, expression of the three PR genes was induced to similar levels after SA treatment in the wild‐type and the drl1 mutants, suggesting that DRL1 is not essential for SA‐induced PR gene expression. However, the drl1 mutations inhibited JA/ET‐induced expression of PDF1.2, CHIB and HEL (Fig. 1c), indicating that DRL1 is required for full induction of the three JA/ET‐inducible defence genes.
Mutations in DRL1 inhibit B. cinerea‐induced, but not Psm ES4326‐induced, defence gene expression
To test whether DRL1 is required for pathogen‐induced defence gene expression, we inoculated 4‐week‐old soil‐grown drl1 and wild‐type plants with Psm ES4326 or the necrotrophic fungal pathogen B. cinerea and monitored Psm ES4326‐induced expression of PR1, PR2 and PR5, as well as B. cinerea‐induced expression of PDF1.2, CHIB, HEL and OCTADECANOID ‐ RESPONSIVE ARABIDOPSIS AP2/ERF59 (ORA59). The previously characterized elp2 mutant was included as a control (Defraia et al., 2010). Expression of these defence genes, except ORA59, was examined at 0 and 24 h post‐inoculation (hpi). Expression of ORA59 was examined at 0 and 6 hpi, as this gene is induced early after B. cinerea infection (Pré et al., 2008). In the wild‐type plants, the three PR genes were significantly induced by Psm ES4326, and PDF1.2, CHIB, HEL and ORA59 were significantly induced by B. cinerea (Fig. 2a–g). In the elp2 mutant, the induction of these defence genes was significantly reduced (Fig. 2a–g), confirming the previous results (Defraia et al., 2010; Wang et al., 2015). In contrast, compared with the wild‐type, Psm ES4326‐induced expression of PR genes was not significantly altered in the drl1 mutants, whereas B. cinerea‐induced expression of PDF1.2, HEL and ORA59 was significantly reduced in these mutants (Fig. 2a–g). These results indicate that DRL1 is differentially required for Psm ES4326‐ and B. cinerea‐induced defence gene expression.
Figure 2.
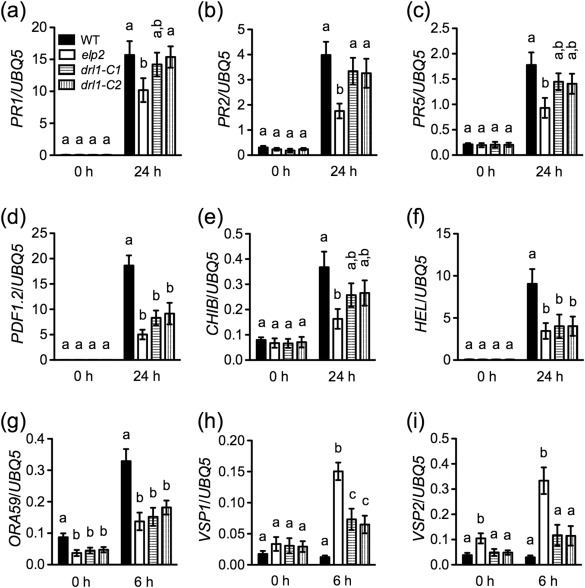
Pathogen‐induced defence gene expression in drl1 mutants. (a–c) Wild‐type (WT), elp2, drl1‐C1 and drl1‐C2 plants were inoculated with Pseudomonas syringae pv. maculicola (Psm) ES4326 [optical density at 600 nm (OD600) = 0.001]. Total RNA was extracted from the inoculated leaves collected at the indicated time points and analysed for the expression of PR1 (a), PR2 (b) and PR5 (c) using quantitative polymerase chain reaction (qPCR). (d–i) Wild‐type, elp2, drl1‐C1 and drl1‐C2 plants were inoculated with Botrytis cinerea spores. Total RNA was extracted from the inoculated leaves collected at the indicated time points and analysed for the expression of PDF1.2 (d), CHIB (e), HEL (f), ORA59 (g), VSP1 (h) and VSP2 (i) using qPCR. Expression was normalized against the constitutively expressed UBQ5. Data represent the mean of three biological replicates with standard deviation (SD). Different letters above the bars indicate significant differences [P < 0.05, one‐way analysis of variance (ANOVA)]. The comparison was made separately among the genotypes for each time point. All experiments were repeated three times with similar trends.
We have shown previously that ELP2 suppresses the expression of wound‐responsive genes (Wang et al., 2015). During B. cinerea infection, expression of the wound‐responsive genes VEGETATIVE STORAGE PROTEIN1 (VSP1), VSP2 and JASMONATE RESPONSIVE1 (JR1) are significantly enhanced in the elp2 mutant. To test whether DRL1 plays a similar role in the modulation of wound‐responsive gene expression, we examined the expression levels of VSP1 and VSP2 in the samples used for the analysis of ORA59 expression. As reported previously (Wang et al., 2015), VSP1 and VSP2 were significantly up‐regulated in the elp2 mutant (Fig. 2h, i). VSP1 was also significantly up‐regulated in the drl1 mutants, but the expression levels of VSP1 and VSP2 were significantly lower in the drl1 mutants than in the elp2 mutant (Fig. 2h, i). These results suggest that DRL1 plays a less important role than ELP2 in suppressing the expression of wound‐responsive genes during B. cinerea infection.
Mutations in DRL1 compromise resistance to the necrotrophic fungal pathogens B. cinerea and A. brassicicola, but not the hemibiotrophic bacterial pathogen Psm ES4326
We have shown previously that mutations in Elongator genes compromise resistance to the hemibiotrophic bacterial pathogen Psm ES4326 and the necrotrophic fungal pathogens B. cinerea and A. brassicicola (Defraia et al., 2010; Wang et al., 2015). To test whether DRL1 plays a similar role in the mediation of disease resistance, we tested the susceptibility of drl1‐C1 and drl1‐C2 to Psm ES4326, B. cinerea and A. brassicicola. The elp2 mutant was again included in the experiment as a control. As reported previously (Defraia et al., 2010; Wang et al., 2015), the elp2 mutant was significantly more susceptible than the wild‐type to all three pathogens (Fig. 3). In contrast, the drl1 mutants were as susceptible as the wild‐type to the bacterial pathogen Psm ES4326 (Fig. 3a), but were more susceptible than the wild‐type to the two necrotrophic fungal pathogens (Fig. 3b–e). Although the susceptibility of the drl1 mutants to B. cinerea and A. brassicicola was not as strong as that of the elp2 mutant (Fig. 3b–e), these results demonstrate that DRL1 is also an important player in the mediation of resistance to necrotrophic fungal pathogens.
Figure 3.
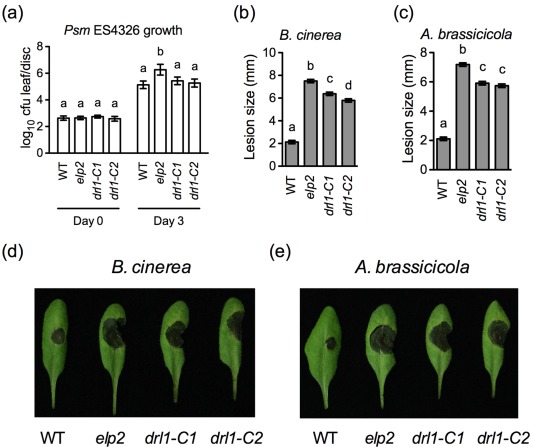
Disease susceptibility of drl1 mutants. (a) Leaves of 4‐week‐old wild‐type (WT), elp2, drl1‐C1 and drl1‐C2 plants were inoculated with Pseudomonas syringae pv. maculicola (Psm) ES4326 [optical density at 600 nm (OD600) = 0.0001]. The in planta bacterial titres were determined immediately and at 3 days post‐inoculation. Data represent the mean of eight biological replicates with standard deviation (SD). Different letters above the bars indicate significant differences [P < 0.05, one‐way analysis of variance (ANOVA)]. The comparison was made separately among the genotypes for each time point. The experiment was repeated three times with similar trends. cfu, colony‐forming unit. (b, c) Sizes of the necrotic lesions formed on Botrytis cinerea‐ (b) and Alternaria brassicicola‐infected (c) wild‐type, elp2, drl1‐C1 and drl1‐C2 plants. Lesion sizes on 72 leaves measured in three independent experiments were combined and analysed as a one‐way ANOVA, blocked by experiment. The resulting mean and standard error (SE) are presented. Different letters above the bars indicate significant differences (P < 0.05). (d, e) Symptoms of the necrotic lesions formed on B. cinerea‐ (d) and A. brassicicola‐infected (e) wild‐type, elp2, drl1‐C1 and drl1‐C2 plants. Photographs were taken at 4 days post‐inoculation.
Overexpression of DRL1 does not affect defence gene expression and disease resistance
To further confirm that the defence phenotypes of the drl1 mutants are caused by mutations in the DRL1 gene, we generated transgenic drl1‐C1 plants expressing a wild‐type DRL1 driven by its native promoter or a modified, constitutive 35S promoter (Mindrinos et al., 1994). Multiple transgenic lines were generated and analysed for both constructs (Fig. S3, see Supporting Information), and one representative transgenic line for each construct is shown in Fig. 4. Expression of DRL1 was restored to wild‐type levels in the DRL1::DRL1 drl1‐C1 transgenic plants (Fig. 4a). On B. cinerea infection, the expression levels of DRL1 were not altered in the wild‐type and DRL1::DRL1 drl1‐C1 plants (Fig. 4a), indicating that DRL1 may not be regulated at the transcriptional level. Compared with the wild‐type, B. cinerea‐induced expression of the defence genes ORA59 and PDF1.2 was significantly inhibited in drl1‐C1, but was induced to wild‐type levels in DRL1::DRL1 drl1‐C1 plants (Fig. 4b, c). Furthermore, although drl1‐C1 was significantly more susceptible than the wild‐type to B. cinerea, DRL1::DRL1 drl1‐C1 plants were as susceptible as the wild‐type (Fig. 4d, e). These results demonstrate that the DRL1::DRL1 transgene complements the defence phenotypes of the drl1‐C1 mutant.
Figure 4.
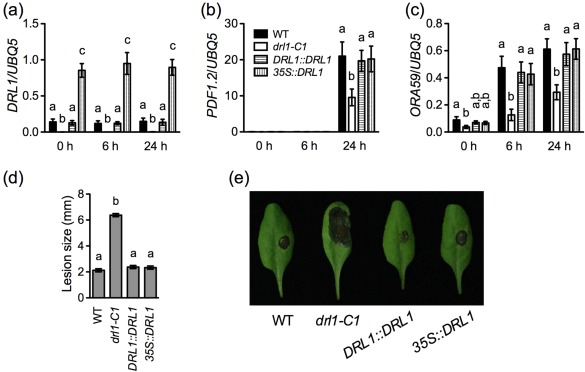
Transgenic expression of DRL1 in drl1 plants. (a–c) Wild‐type (WT), drl1‐C1, DRL1::DRL1 and 35S::DRL1 plants were inoculated with Botrytis cinerea spores. Total RNA was extracted from the inoculated leaves collected at the indicated time points and analysed for the expression of DRL1 (a), PDF1.2 (b) and ORA59 (c) using quantitative polymerase chain reaction (qPCR). Expression was normalized against the constitutively expressed UBQ5. Data represent the mean of three biological replicates with standard deviation (SD). Different letters above the bars indicate significant differences [P < 0.05, one‐way analysis of variance (ANOVA)]. The comparison was made separately among the genotypes for each time point. The experiments were repeated three times with similar trends. (d) Sizes of the necrotic lesions formed on B. cinerea‐infected wild‐type, drl1‐C1, DRL1::DRL1 and 35S::DRL1 plants. Lesion sizes on 72 leaves measured in three independent experiments were combined and analysed as a one‐way ANOVA, blocked by experiment. The resulting mean and standard error (SE) are presented. Different letters above the bars indicate significant differences (P < 0.05). (e) Symptoms of the necrotic lesions formed on B. cinerea‐infected wild‐type, drl1‐C1, DRL1::DRL1 and 35S::DRL1 plants. Photographs were taken at 4 days post‐inoculation.
In 35S::DRL1 drl1‐C1 transgenic plants, expression of DRL1 was significantly elevated (Figs 4a and S3a). However, B. cinerea‐induced expression of ORA59 and PDF1.2 in 35S::DRL1 drl1‐C1 plants was comparable with that in wild‐type plants (Figs 4b, c and S3b). In line with the gene expression results, 35S::DRL1 drl1‐C1 transgenic plants were as susceptible as the wild‐type to B. cinerea (Figs 4d, e and S3c). These results indicate that overexpression of the DRL1 gene does not change defence gene induction and B. cinerea resistance.
The drl1 mutation alters B. cinerea‐induced transcriptome reprogramming
To define how DRL1 modulates fungal pathogen‐induced transcriptional changes at the genome level, we performed a microarray experiment on B. cinerea‐infected drl1‐C1 and wild‐type plants (NCBI GEO Series number GSE79961). Triplicate experiments were performed independently, and the data were analysed to identify genes that showed a two‐fold or higher induction or suppression with a low q value (≤0.05) in drl1‐C1 and the wild‐type. Similar to multiple previous microarray analyses (AbuQamar et al., 2006; Rowe et al., 2010; Wang et al., 2015; Windram et al., 2012), dramatic transcriptome changes were detected in the wild‐type plants after B. cinerea infection. The numbers of genes that were up‐ and down‐regulated by two‐fold or more at 6 hpi (2238 and 1887, respectively) were close to those (1812 and 1531, respectively) reported previously (Wang et al., 2015), confirming the transcriptional changes induced by B. cinerea infection. Interestingly, although profound transcriptional changes also occurred in the drl1‐C1 mutant, the numbers of genes that were up‐ and down‐regulated by two‐fold or more (1400 and 998, respectively) were much lower than those (2238 and 1887, respectively) in the wild‐type (Fig. 5a). The drl1‐C1 mutation also shifted the transcriptome profiles of the mutant plants. Although most of the genes up‐ or down‐regulated two‐fold or more in drl1‐C1 were also found in the wild‐type, approximately 10.6% and 11.6% of the genes that were up‐ and down‐regulated in drl1‐C1, respectively, were not found in the wild‐type (Fig. 5a). Taken together, these results indicate that the drl1 mutation has a significant impact on B. cinerea infection‐induced transcriptome reprogramming.
Figure 5.
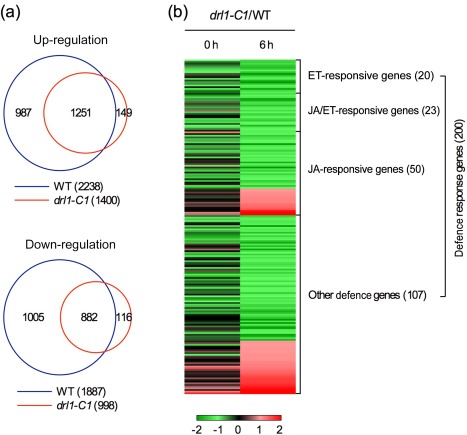
Botrytis cinerea‐induced transcriptome changes in drl1 plants. (a) Overlaps among the genes that were up‐ or down‐regulated at 6 h post‐inoculation (hpi) in drl1‐C1 and the wild‐type (WT). (b) Heatmap analysis of the 200 defence genes that were differentially expressed between drl1‐C1 and the wild‐type at 6 hpi. A clear down‐regulation (green) of the majority of the 200 genes was observed.
To identify genes whose expression is modulated by DRL1, we analysed the genes that were differentially expressed between drl1‐C1 and the wild‐type. Genes that showed a two‐fold or larger difference in their expression levels with a low q value (≤0.05) were chosen for further analysis. A total of 354 (115 up‐ and 239 down‐regulated) and 803 (404 up‐ and 399 down‐regulated) genes were differentially expressed between drl1‐C1 and the wild‐type at 0 and 6 hpi, respectively. Gene ontology (GO) analysis of the genes differentially expressed at 6 hpi revealed that genes involved in plant defence, such as response to chitin, defence response and innate immune response, were significantly enriched in the down‐regulated genes (Fig. S4a, see Supporting Information), whereas genes involved in metabolic reactions, such as sulfur compound biosynthesis, photosynthesis and amino acid metabolism, were significantly enriched in the up‐regulated genes (Fig. S4b). A total of 200 defence response genes, which were differentially expressed between drl1‐C1 and the wild‐type at 6 hpi, are displayed as a heat map (Fig. 5b and Table S3, see Supporting Information). These genes include 20 ET‐responsive genes, 23 JA/ET‐responsive genes, 50 JA‐responsive genes and 107 other defence genes. All ET‐ and JA/ET‐responsive genes and the majority of JA‐responsive and other defence genes were down‐regulated in the drl1‐C1 mutant. Among the down‐regulated genes are a group of well‐characterized defence genes, including ORA59, ETHYLENE RESPONSE FACTOR1 (ERF1), WRKY DNA BINDING PROTEIN 33 (WRKY33), SIGMA FACTOR BINDING PROTEIN 1 (SIB1), PHYTOALEXIN DEFICIENT 3 (PAD3), GDSL LIPASE‐LIKE 1 (GLIP1) and CYP79B2 (Table 1), which have been demonstrated to function in resistance to necrotrophic fungal pathogens (Ferrari et al., 2007; Kliebenstein et al., 2005; Lai et al., 2011; Lorenzo et al., 2003; Oh et al., 2005; Pré et al., 2008; Zheng et al., 2006). Therefore, DLR1 may contribute to B. cinerea resistance by modulating the expression of these defence genes.
Table 1.
Partial list of the defence genes that were differentially expressed between drl1‐C1 and the wild‐type during Botrytis cinerea infection.
| AGI locus | Gene name | drl1‐C1/WT | AGI description | |||
|---|---|---|---|---|---|---|
| 0 h | 6 h | |||||
| log2(FC) | q value | log2(FC) | q value | |||
| Defence‐response genes | ||||||
| At1g06160 | ORA59 | −1.818 | 0 | −1.091 | 0 | OCTADECANOID‐RESPONSIVE ARABIDOPSIS AP2/ERF 59 |
| At3g23240 | ERF1 | −1.249 | 0 | ETHYLENE‐RESPONSIVE TRANSCRIPTION FACTOR 1 | ||
| At2g38470 | WRKY33 | −1.081 | 0.000012 | WRKY DNA‐BINDING PROTEIN 33 | ||
| At3g56710 | SIB1 | −1.056 | 0.000862 | SIGMA FACTOR BINDING PROTEIN 1 | ||
| At3g26830 | PAD3 | −1.252 | 0.000001 | −1.375 | 0.000005 | BIFUNCTIONAL DIHYDROCAMALEXATE SYNTHASE/CAMALEXIN SYNTHASE |
| At5g40990 | GLIP | −1.222 | 0 | GDSL LIPASE 1 | ||
| At4g39950 | CYP79B2 | −1.735 | 0 | CYTOCHROME P450, FAMILY 79, SUBFAMILY B, POLYPEPTIDE 2 | ||
| At2g26020 | PDF1.2b | −1.514 | 0 | PLANT DEFENSIN 1.2b | ||
| At5g44430 | PDF1.2c | −1.692 | 0 | PLANT DEFENSIN 1.2c | ||
| At3g04720 | HEL | −1.052 | 0.00026 | −1.53 | 0.00003 | HEVEIN‐LIKE |
| Wound response genes | ||||||
| At3g16470 | JR1 | 1.198 | 0.000043 | JA‐RESPONSIVE PROTEIN 1 | ||
| At5g24780 | VSP1 | 1.082 | 0.00033 | VEGETATIVE STORAGE PROTEIN 1 | ||
| At5g24770 | VSP2 | 3.368 | 0 | VEGETATIVE STORAGE PROTEIN 2 | ||
Both the N‐terminal P‐loop and the C‐terminal CBD of DRL1 are required for its function in plant immunity
DRL1 contains an N‐terminal P‐loop and a C‐terminal CBD (Nelissen et al., 2003). The P‐loop is required for the function of TOT4/KTI12 in yeast (Fichtner et al., 2002). To test whether the P‐loop and the CBD of DRL1 are required for the function of DRL1 in plant immunity, we introduced point mutations into the DRL1 cDNA to generate a P‐loop mutant, in which a conserved lysine residue (K14) was changed to arginine, and a CBD mutant, in which two conserved amino acid residues, a phenylalanine and a leucine (F264 and L267), were changed to aspartate and glutamate, respectively (Fig. 6a). In addition, a valine residue (V265) was also changed to aspartate to further modify the net charge of the domain (Fig. 6a). Mutated drl1 cDNAs driven by the DRL1 native promoter were transformed into drl1‐C1 to test the functionality of the P‐loop and the CBD mutant. Multiple transgenic lines were produced and characterized for both constructs (Fig. S5, see Supporting Information) and one representative transgenic line for each construct is shown in Fig. 6. Neither the P‐loop nor the CBD mutant complemented the defence phenotypes of drl1‐C1, including decreased induction of the defence genes PDF1.2 and ORA59 by B. cinerea infection and enhanced susceptibility to B. cinerea (Fig. 6b–e). These results clearly indicate that both the P‐loop and the CBD of DRL1 are important for its function in the plant immune response.
Figure 6.
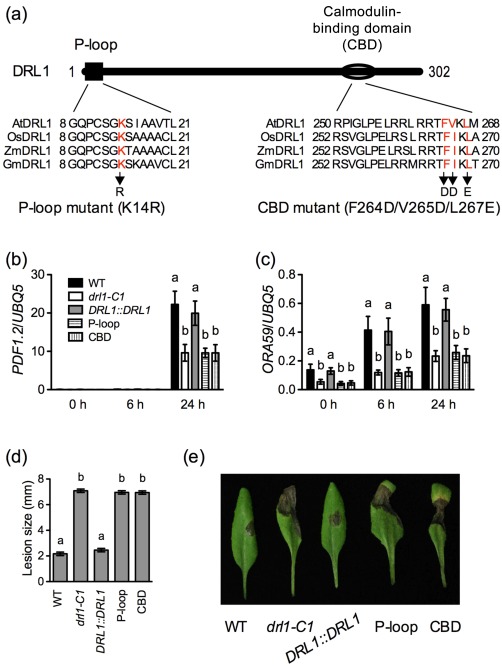
Characterization of the P‐loop and C‐terminal calmodulin‐binding domain (CBD) mutants of DRL1. (a) Schematic representation of the P‐loop and CBD mutations. The P‐loop and the C‐terminal CBD of Arabidopsis DRL1 (AtDRL1) are aligned with those of rice (OsDRL1), maize (ZmDRL1) and soybean (GmDRL1) DRL1. Only sequences that are part of the alignment are shown. The mutated amino acid residues are labelled in red. Arrows indicate the mutations created in the P‐loop and CBD mutants. (b, c) Wild‐type (WT), drl1‐C1, DRL1::DRL1, P‐loop mutant and CBD mutant plants were inoculated with Botrytis cinerea spores. Total RNA was extracted from the inoculated leaves collected at the indicated time points and analysed for the expression of PDF1.2 (b) and ORA59 (c) using quantitative polymerase chain reaction (qPCR). Expression was normalized against the constitutively expressed UBQ5. Data represent the mean of three biological replicates with standard deviation (SD). Different letters above the bars indicate significant differences [P < 0.05, one‐way analysis of variance (ANOVA)]. The comparison was made separately among the genotypes for each time point. The experiments were repeated three times with similar trends. (d) Sizes of the necrotic lesions formed on B. cinerea‐infected wild‐type, drl1‐C1, DRL1::DRL1, P‐loop mutant and CBD mutant plants. Lesion sizes on 72 leaves measured in three independent experiments were combined and analysed as a one‐way ANOVA, blocked by experiment. The resulting mean and standard error (SE) are presented. Different letters above the bars indicate significant differences (P < 0.05). (e) Symptoms of the necrotic lesions formed on B cinerea‐infected wild‐type, drl1‐C1, DRL1::DRL1, P‐loop mutant and CBD mutant plants. Photographs were taken at 4 days post‐inoculation.
Discussion
In this study, we analysed the function of DRL1 in plant immune responses. Our results show that: (i) DRL1 makes a partial contribution to SA‐induced cytotoxicity, but does not play a significant role in SA‐mediated PR gene expression and Psm ES4326 resistance; (ii) DRL1 is required for JA/ET‐induced expression of PDF1.2, CHIB and HEL, B. cinerea‐induced expression of a large group of defence genes, and resistance to B. cinerea and A. brassicicola; (iii) overexpression of the DRL1 gene does not alter the basal expression and induction of PDF1.2 and ORA59 and resistance to B. cinerea; and (iv) both the N‐terminal P‐loop and the C‐terminal CBD of DRL1 are critical for the basal expression and induction of PDF1.2 and ORA59 and for resistance to B. cinerea.
It has been shown that yeast cells lacking TOT4/KTI12 or an Elongator subunit display indistinguishable pehnotypes, including resistance to the K. lactis toxin zymocin and hypersensitivity to high temperature (above 38 °C) and drugs, such as caffeine and calcofluor white (Fichtner et al., 2002; Frohloff et al., 2001). Similarly, Arabidopsis drl1 and elo/elp mutants exhibit almost identical morphological phenotypes, such as narrow leaves, disorganized shoot apical meristem, and short roots, and similar transcriptome changes (Nelissen et al., 2003, 2005; Xu et al., 2012). These results suggest significant functional overlap between TOT4/KTI12 or DRL1 and the Elongator complex. However, our results indicate that DRL1 and Elongator do not play largely overlapping roles in plant immune responses. Whilst Elongator is required for both SA‐ and JA/ET‐mediated defence responses, DRL1 only contributes to JA/ET‐mediated defence signalling (Figs 1, 2, 3), indicating that Elongator has a broader role than DRL1 in plant immune responses. Furthermore, although both drl1 and elo/elp mutants are suspectible to B. cinerea and A. brassicicola, drl1 mutants are significantly less susceptible than the elp2 mutant (Fig. 3b, c), suggesting that DRL1 plays a less important function than Elongator, even in JA/ET‐mediated resistance to necrotrophic fungal pathogens.
DRL1 is required for JA/ET‐induced expression of the defence marker genes PDF1.2, CHIB and HEL (Fig. 1c), indicating that it positively contributes to JA/ET‐mediated defence responses. Indeed, mutations in DRL1 inhibit B. cinerea‐induced expression of many JA/ET‐responsive defence genes, including ORA59 and ERF1 (Fig. 5b and Table 1), which encode two well‐documented AP2/ERF domain transcription factors that have been shown to directly regulate PDF1.2 expression and B. cinerea resistance (Lorenzo et al., 2003; Pré et al., 2008). DRL1 is also required for transcription factor WRKY33‐mediated defence signalling (Zheng et al., 2006). In drl1, basal expression of the WRKY33 gene and B. cinerea‐induced expression of several WRKY33 target genes, including PAD3 and GLIP1, are reduced (Table 1). Both PAD3 and GLIP1 have been demonstrated to play positive roles in resistance to B. cinerea (Ferrari et al., 2007; Oh et al., 2005). In addition, the induction of the SIGMA FACTOR BINDING PROTEIN1 (SIB1) gene, which encodes a WRKY33‐interacting protein required for B. cinerea resistance (Lai et al., 2011), is also decreased (Table 1). Therefore, the reduced defence gene expression and enhanced susceptibility to B. cinerea in drl1 are probably mainly attributed to the attenuated JA/ET‐ and WRKY33‐mediated defence signalling.
In yeast, loss of TOT4/KTI12 function, elevation of TOT4/KTI12 copy number and overexpression of TOT4/KTI12 all lead to resistance to the K. lactis toxin zymocin (Fichtner et al., 2002; Frohloff et al., 2001), implying that the cellular level of TOT4/KTI12 may play an important role in the regulation of zymocin sensitivity. Interestingly, although ectopic expression of the DRL1 gene rescues the growth retardation phenotype of yeast cells lacking TOT4/KTI12, overexpression of DRL1 in wild‐type yeast does not elicit zymocin resistance (Jun et al., 2015). Similarly, overexpression of DRL1 in Arabidopsis does not confer any detectable morphological and defence phenotypes (Figs S6, see Supporting Information, and 4). These results suggest that cellular DRL1 protein levels may not be vigorously regulated and that the basal level of DRL1 may be sufficient for its function in plant development and immunity. In support of these hypotheses, expression of the DRL1 gene is not altered by B. cinerea infection (Fig. 4a), implying that DRL1 may not be regulated at the transcriptional level.
The DRL1 protein contains two (P‐loop and N‐K/R‐X‐D box) of the five highly conserved sequence motifs that are required for GTP binding and hydrolysis in GTP‐binding proteins (Kaziro et al., 1991; Nelissen et al., 2003). The missing motifs are critical for GTP hydrolysis (Kaziro et al., 1991), suggesting that DRL1 might not be a bona fide GTP‐binding protein. In addition, previous work has shown that deletion of the P‐loop renders TOT4/KTI12 inactive (Fichtner et al., 2002), and we found that substitution of an arginine for the conserved lysine residue (K14R) in the P‐loop destroys the function of DRL1 (Fig. 6). These results clearly demonstrate that the P‐loop is important for the function of TOT4/KTI12 and DRL1. As DRL1 contains another conserved motif (P‐X‐X‐A/S‐T), which exists in many ATP/GTP‐utilizing proteins, and the P‐loop is also found in many nucleotide triphosphate‐utilizing enzymes, it has been proposed that DRL1 may exert its function on ATP/GTP binding or transfer ATP/GTP to another protein (Nelissen et al., 2003). However, the precise relationship between DRL1 and ATP/GTP requires further investigation.
It has been demonstrated that DRL1 is a CaM‐binding protein. Nelissen et al. (2003) reported that the C‐terminal 100 amino acids of DRL1 bind CaM in a calcium‐dependent manner, and identified a motif L‐X(3)‐F‐X(2)‐L‐X(5)‐L (amino acids 260–273) within this region as the CBD. Very recently, uisng a bioinformatic approach, Jun et al. (2015) have identified another CBD in the N‐terminus of the protein, but whether it binds CaM has not been tested. CaM‐binding proteins have been implicated in plant immune responses (Poovaiah et al., 2013). For instance, the CaM‐binding transcription factor SIGNAL RESPONSIVE1 (SR1)/CALMODULIN BINDING TRANSCRIPTION ACTIVATOR3 (CAMAT3) interacts with the CGCG box motifs in the promoters of ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1) and NON‐RACE‐SPECIFIC DISEASE RESISTANCE1 (NDR1), which encode two key componenets of the SA‐ and resistance gene‐mediated defence pathways, to negatively regulate EDS1 and NDR1 expression (Du et al., 2009; Nie et al., 2012). Another CaM‐binding transcription factor CALMODULIN‐BINDING PROTEIN 60‐LIKE.g (CBP60g) is recruited to the promoter of the SA biosynthesis gene ISOCHORISMATE SYNTHASE1 (ICS1)/SALICYLIC ACID INDUCTION DEFICIENT2 (SID2) to activate its transcription (Wang et al., 2009). Here, we demonstrate that DRL1 is another CaM‐binding protein functioning in plant immune responses. Substituions of three amino acids in the C‐terminal CBD of DRL1 made the protein unable to complement the drl1 defence phenotype (Fig. 6), suggesting that CaM binding at this motif may be important for the function of DRL1 in plant immunity. Whether the putative N‐terminal CBD is also required for the function of DRL1 remains to be tested (Jun et al., 2015). Neverthelesss, as it is well known that CaM binding is a highly conserved mechanism modulating target protein functions in eukaryotic cells (Poovaiah et al., 2013), it will be interesting to determine how CaM binding affects the function of DRL1.
In yeast, deletion of TOT4/KTI12 does not affect the structural integrity of the Elongator complex (Fichtner et al., 2002), indicating that TOT4/KTI12 is not a stable structural component of the complex. However, TOT4/KTI12 and DRL1 are physically associated with the Elongator complex in yeast and Arabidopsis, respectively (Petrakis et al., 2005; Xu et al., 2012), suggesting that they may function through Elongator. Indeed, deletion of TOT4/KTI12 leads to a decrease in histone acetylation levels in chromatin, a molecular phenotype also seen in yeast cells lacking ELP3 (Petrakis et al., 2005). As TOT4/KTI12 does not have histone acetyltransferase (HAT) activity, it is probably required for the normal HAT activity of the Elongator complex. Whether TOT4/KTI12 or DRL1 is also required for Elongator's other molecular functions, such as tRNA modification and DNA demethylation/methylation, is not clear (Esberg et al., 2006; Jia et al., 2015; Okada et al., 2010; Wang et al., 2013). Nevertheless, so far, the available evidence suggests that TOT4/KTI12 and DRL1 are modifiers of Elongator in yeast and Arabidopsis, respectively (Nelissen et al., 2003; Petrakis et al., 2005). The results presented here indicate that DRL1 is involved in most, but not all, of Elongator's functions. As the N‐terminal P‐loop and the C‐terminal CBD are required for the function of DRL1 in plant immunity, how ATP/GTP and/or CaM binding regulates the activities of DRL1 and Elongator deserves further investigation.
Experimental Procedures
Plant materials and growth conditions
The wild‐type used was the Arabidopsis thaliana (L.) Heynh. Columbia (Col‐0) ecotype and the mutant alleles used were Atelp2‐5 (Defraia et al., 2010), drl1‐C1 (SALK_056915) and drl1‐C2 (SALK_140551). The T‐DNA insertion lines were obtained from the Arabidopsis Biological Resource Center at Ohio State University. Homozygous mutant plants of the T‐DNA insertion lines were confirmed with a pair of primers flanking the T‐DNA insertions (Table S1, see Supporting Information) and the left border primer LBa1 (Alonso et al., 2003). Arabidopsis seeds were sown on autoclaved soil (Metro‐Mix200, Grace‐Sierra, http://www.sungro.com) and vernalized at 4 °C for 3 days. Plants were germinated and grown at approximately 23 °C under a 16‐h light/8‐h dark regime.
Pathogen infection
Inoculation of plants with Psm ES4326 was performed by pressure infiltration with a 1‐mL needleless syringe, as described previously (Zhang et al., 2012). After inoculation, eight infected leaves, one from each plant, were collected for each genotype or time point to examine the growth of the pathogen. The B. cinerea strain B05 and A. brassicicola strain MUCL 20297 were used in this study. Botrytis cinerea and A. brassicicola inoculation were performed as described previously (Wang et al., 2013). Briefly, the fungal pathogens were grown on BD Difco Potato Dextrose Agar (Becton, Dickinson and Company, Sparks, MD, USA) for about 10 days at 24 °C. Spores were harvested, resuspended in BD Difco Potato Dextrose Broth (Becton, Dickinson and Company) at a density of (1–5) × 105 spores/mL, and incubated for 2 h prior to inoculation. Five‐microliter spore suspensions were dropped on the adaxial surface of rosette leaves (one drop on each leaf), where the leaves had been gently wounded with a needle. Symptoms were monitored for 3–4 days (4–5 days for A. brassicicola). The lengths and widths of the lesions were measured with a caliper before disease symptoms expanded beyond inoculated leaves and the average of the length and the width was used to represent the size of a lesion. In each experiment, 24 plants per genotype were used for three sub‐experiments. Each sub‐experiment was performed in the same flat under the same clear plastic dome. In each flat, plants from different genotypes (eight plants per genotype) were randomly arranged. One leaf on each plant was inoculated and all leaves were inoculated with the same spore suspension. A total of 24 lesions (one on each plant) were measured and used for statistical analysis.
RNA analysis
RNA extraction, reverse transcription and real‐time qPCR analysis were carried out as described by Defraia et al. (2010). In each experiment, three independent biological samples were collected at each time point per genotype and analysed. UBQ5 was used as the reference gene for qPCR normalization, because it is one of the most stably expressed genes (Gutierrez et al., 2008). The primers used for qPCR in this study have either been reported previously or are shown in Table S2 (see Supporting Information).
Microarray analysis
Four‐week‐old soil‐grown plants were inoculated with the B. cinerea strain B05. Total RNA samples extracted from leaf tissues collected at the indicated time points after B. cinerea inoculation were subjected to microarray analysis as described previously (Wang et al., 2013).
The mean signals obtained from Agilent Feature Extraction software were background corrected using the normexp + offset method, in which a small positive offset (k = 50) was added to move the corrected intensities away from zero (Ritchie et al., 2007). The resulting data were log transformed (using 2 as the base) and normalized between individual samples by scaling the individual log‐transformed signal intensities so that all datasets had comparable lower quartile, median and upper quartile values (Smyth, 2004). After normalization, the empirical Bayes moderated t‐statistics, which are implemented in the limma Bioconductor package (Smyth, 2004), were performed for differential expression detection. In each comparison, a P value and fold change were computed for each gene locus. The gene expression fold changes were computed based on the normalized log‐transformed signal intensity data. To control the false discovery rate and correct multiple hypothesis testing, a q value was calculated and used to assess the significance of each test employing the approach of Benjamini and Hochberg (1995). The comparison results were further explored to obtain numbers of overlapping genes between different comparisons.
Plasmid construction and plant transformation
A pair of primers (SmaI‐DRL1F, 5′‐TCCCCCGGGCTCTTGAACATCTTCAGCGTC‐3′; SacI‐StrepII‐DRL1R, 5′‐CGAGCTCATTTTTCAAATTGAGGATGAGACCATGCACTAGCGTTATTACCTCCAAACTC‐3′) was used to amplify the promoter and coding regions of DRL1 from genomic DNA. The PCR products were digested with SmaI and SacI, and then ligated into the corresponding sites of the vector pBI101, resulting in the plasmid pBI101‐DRL1::DRL1. Site‐directed mutagenesis of the conserved amino acid residues in the P‐loop and the CBD of DRL1 was performed in the pBI101‐DRL1::DRL1 construct using a PCR‐based Quick‐Change site‐directed mutagenesis kit (Stratagene, LaJolla, CA, USA). The primers used for the site‐directed mutagenesis of the P‐loop were DRL1(K14R)F (5′‐CCTTGTAGTGGTAGATCAATAGCTGCAG‐3′) and DRL1(K14R)R (5′‐CTGCAGCTATTGATCTACCACTACAAGG‐3′), and, for the CBD mutagenesis, the primers were DRL1(FVKL/DDKE)F (5′‐GAGGCTTCGAAGAACGGATGATAAAGAGATGGGTCAATCGAG‐3′) and DRL1(FVKL/DDKE)R (5′‐CTCGATTGACCCATCTCTTTATCATCCGTTCTTCGAAGCCTC‐3′). The presence of the expected mutations was confirmed by DNA sequencing. For the overexpression of DRL1, a pair of primers (SalI‐DRL1F, 5′‐GCGTCGACATGGCGCTAGTTGTGATTTGTG‐3′; SacI‐StrepII‐DRL1R) was used to amplify the coding region of DRL1 from genomic DNA. The PCR products were digested with SalI and SacI, and then ligated into the corresponding sites of the vector pBI1.4T, resulting in the plasmid pBI1.4T‐35S::DRL1. The plasmids were introduced into the Agrobacterium tumefaciens strain GV3101 (pMP90) by electroporation and transformed into the drl1‐C1 mutant following the floral dip method (Clough and Bent, 1998).
Chemical treatment
Two‐week‐old seedlings grown on half‐strength MS medium were treated with 0.5 mm SA or a combination of 0.1 mm MeJA and 0.1 mm ACC. Seedlings for negative controls were treated with water. The aerial parts of the seedlings were collected and subjected to total RNA extraction.
Statistical methods
Except for those used in microarray analysis, statistical analyses were performed using the one‐way analysis of variance (ANOVA) in Prism 5.0b (GraphPad Software, La Jolla, CA, USA). Lesion sizes measured in three independent experiments were combined and analysed as a one‐way ANOVA, blocked by experiment, using JMP 11 (JMP Software, Cary, NC, USA). Other experiments were conducted three times with similar trends, and results from a representative experiment are presented.
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: DRL1 (At1g13870); PR1 (At2g14610); PR2 (At3g57260); PR5 (At1g75040); PDF1.2 (At5g44420); CHIB (At3g12500); HEL (At3g04720); WRKY33 (At2g38470); ORA59 (At1g06160); VSP1 (At5g24780); VSP2 (At5g24770); UBQ5 (At3g62250); and NCBI Gene Expression Omnibus Series number GSE79961 (microarray data).
Conflict of Interest
The authors declare no conflicts of interest.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website.
Fig. S1 Characterization of drl1 T‐DNA insertion mutants.
Fig. S2 Growth of Pseudomonas syringae pv. maculicola (Psm) ES4326 in drl1 npr1 double mutants.
Fig. S3 Characterization of 35S::DRL1 drl1 transgenic plants.
Fig. S4 Gene ontology (GO) analysis of the genes differentially expressed between drl1 and the wild‐type.
Fig. S5 Characterization of multiple independent transgenic lines expressing the P‐loop and C‐terminal calmodulin‐binding domain (CBD) mutants of DRL1.
Fig. S6 Morphology of the DRL1::DRL1 drl1 and 35S::DRL1 drl1 transgenic plants.
Table S1 Primers used for the identification of homozygous T‐DNA insertion mutant plants.
Table S2 Primers used for quantitative polymerase chain reaction (qPCR) analysis of gene expression.
Table S3 Defence genes that were differentially expressed between drl1‐C1 and the wild‐type during Botrytis cinerea infection.
Acknowledgements
We thank Dr Jeffery A. Rollins (University of Florida, FL, USA) for the B. cinerea strain B05, Dr Xinnian Dong (Duke University, NC, USA) for the A. brassicicola strain MUCL 20297 and the Arabidopsis Biological Resource Center at Ohio State University (Columbus, OH, USA) for SALK_056915 and SALK_140551 seeds. This work was partially supported by a grant from the National Science Foundation (IOS‐0842716) awarded to Z.M.
References
- AbuQamar, S. , Chen, X. , Dhawan, R. , Bluhm, B. , Salmeron, J. , Lam, S. , Dietrich, R.A. and Mengiste, T. (2006) Expression profiling and mutant analysis reveals complex regulatory networks involved in Arabidopsis response to Botrytis infection. Plant J. 48, 28–44. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M. , Stepanova, A.N. , Leisse, T.J. , Kim, C.J. , Chen, H. , Shinn, P. , Stevenson, D.K. , Zimmerman, J. , Barajas, P. , Cheuk, R. , Gadrinab, C. , Heller, C. , Jeske, A. , Koesema, E. , Meyers, C.C. , Parker, H. , Prednis, L. , Ansari, Y. , Choy, N. , Deen, H. , Geralt, M. , Hazari, N. , Hom, E. , Karnes, M. , Mulholland, C. , Ndubaku, R. , Schmidt, I. , Guzman, P. , Aguilar‐Henonin, L. , Schmid, M. , Weigel, D. , Carter, D.E. , Marchand, T. , Risseeuw, E. , Brogden, D. , Zeko, A. , Crosby, W.L. , Berry, C.C. and Ecker, J.R. (2003) Genome‐wide insertional mutagenesis of Arabidopsis thaliana . Science, 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Benjamini, Y. and Hochberg, Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B, 57, 289–300. [Google Scholar]
- Broekaert, W.F. , Delaure, S.L. , De Bolle, M.F. and Cammue, B.P. (2006) The role of ethylene in host–pathogen interactions. Annu. Rev. Phytopathol. 44, 393–416. [DOI] [PubMed] [Google Scholar]
- Butler, A.R. , White, J.H. , Folawiyo, Y. , Edlin, A. , Gardiner, D. and Stark, M.J. (1994) Two Saccharomyces cerevisiae genes which control sensitivity to G1 arrest induced by Kluyveromyces lactis toxin. Mol. Cell. Biol. 14, 6306–6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H. , Bowling, S.A. , Gordon, S. and Dong, X. (1994) Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell, 6, 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H. , Glazebrook, J. , Clark, J.D. , Volko, S. and Dong, X. (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell, 88, 57–63. [DOI] [PubMed] [Google Scholar]
- Chen, Z. , Zhang, H. , Jablonowski, D. , Zhou, X. , Ren, X. , Hong, X. , Schaffrath, R. , Zhu, J.K. and Gong, Z. (2006) Mutations in ABO1/ELO2, a subunit of holo‐Elongator, increase abscisic acid sensitivity and drought tolerance in Arabidopsis thaliana . Mol. Cell. Biol. 26, 6902–6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Creppe, C. , Malinouskaya, L. , Volvert, M.L. , Gillard, M. , Close, P. , Malaise, O. , Laguesse, S. , Cornez, I. , Rahmouni, S. , Ormenese, S. , Belachew, S. , Malgrange, B. , Chapelle, J.P. , Siebenlist, U. , Moonen, G. , Chariot, A. and Nguyen, L. (2009) Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha‐tubulin. Cell, 136, 551–564. [DOI] [PubMed] [Google Scholar]
- Defraia, C.T. , Zhang, X. and Mou, Z. (2010) Elongator subunit 2 is an accelerator of immune responses in Arabidopsis thaliana . Plant J. 64, 511–523. [DOI] [PubMed] [Google Scholar]
- Defraia, C.T. , Wang, Y. , Yao, J. and Mou, Z. (2013) Elongator subunit 3 positively regulates plant immunity through its histone acetyltransferase and radical S‐adenosylmethionine domains. BMC Plant Biol. 13, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney, T.P. , Friedrich, L. and Ryals, J.A. (1995) Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA, 92, 6602–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, X. (2004) NPR1, all things considered. Curr. Opin. Plant Biol. 7, 547–552. [DOI] [PubMed] [Google Scholar]
- Du, L. , Ali, G.S. , Simons, K.A. , Hou, J. , Yang, T. , Reddy, A.S. and Poovaiah, B.W. (2009) Ca(2+)/calmodulin regulates salicylic‐acid‐mediated plant immunity. Nature, 457, 1154–1158. [DOI] [PubMed] [Google Scholar]
- Esberg, A. , Huang, B. , Johansson, M.J. and Bystrom, A.S. (2006) Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol. Cell, 24, 139–148. [DOI] [PubMed] [Google Scholar]
- Eulgem, T. , Tsuchiya, T. , Wang, X.J. , Beasley, B. , Cuzick, A. , Tor, M. , Zhu, T. , McDowell, J.M. , Holub, E. and Dangl, J.L. (2007) EDM2 is required for RPP7‐dependent disease resistance in Arabidopsis and affects RPP7 transcript levels. Plant J. 49, 829–839. [DOI] [PubMed] [Google Scholar]
- Ferrari, S. , Galletti, R. , Denoux, C. , De Lorenzo, G. , Ausubel, F.M. and Dewdney, J. (2007) Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is independent of salicylic acid, ethylene, or jasmonate signaling but requires PHYTOALEXIN DEFICIENT3 . Plant Physiol. 144, 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichtner, L. , Frohloff, F. , Burkner, K. , Larsen, M. , Breunig, K.D. and Schaffrath, R. (2002) Molecular analysis of KTI12/TOT4, a Saccharomyces cerevisiae gene required for Kluyveromyces lactis zymocin action. Mol. Microbiol. 43, 783–791. [DOI] [PubMed] [Google Scholar]
- Frohloff, F. , Fichtner, L. , Jablonowski, D. , Breunig, K.D. and Schaffrath, R. (2001) Saccharomyces cerevisiae Elongator mutations confer resistance to the Kluyveromyces lactis zymocin. EMBO J. 20, 1993–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Gutierrez, L. , Mauriat, M. , Guenin, S. , Pelloux, J. , Lefebvre, J.F. , Louvet, R. , Rusterucci, C. , Moritz, T. , Guerineau, F. , Bellini, C. and Van Wuytswinkel, O. (2008) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription‐polymerase chain reaction (RT‐PCR) analysis in plants. Plant Biotechnol. J. 6, 609–618. [DOI] [PubMed] [Google Scholar]
- Hawkes, N.A. , Otero, G. , Winkler, G.S. , Marshall, N. , Dahmus, M.E. , Krappmann, D. , Scheidereit, C. , Thomas, C.L. , Schiavo, G. , Erdjument‐Bromage, H. , Tempst, P. and Svejstrup, J.Q. (2002) Purification and characterization of the human elongator complex. J. Biol. Chem. 277, 3047–3052. [DOI] [PubMed] [Google Scholar]
- Huang, B. , Johansson, M.J. and Bystrom, A.S. (2005) An early step in wobble uridine tRNA modification requires the Elongator complex. RNA, 11, 424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, Y. , Tian, H. , Li, H. , Yu, Q. , Wang, L. , Friml, J. and Ding, Z. (2015) The Arabidopsis thaliana elongator complex subunit 2 epigenetically affects root development. J. Exp. Bot. 66, 4631–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun, S.E. , Cho, K.H. , Hwang, J.Y. , Abdel‐Fattah, W. , Hammermeister, A. , Schaffrath, R. , Bowman, J.L. and Kim, G.T. (2015) Comparative analysis of the conserved functions of Arabidopsis DRL1 and yeast KTI12. Mol. Cells, 38, 243–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan, K. and Manners, J.M. (2008) Jasmonate signaling: toward an integrated view. Plant Physiol. 146, 1459–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaziro, Y. , Itoh, H. , Kozasa, T. , Nakafuku, M. and Satoh, T. (1991) Structure and function of signal‐transducing GTP‐binding proteins. Annu. Rev. Biochem. 60, 349–400. [DOI] [PubMed] [Google Scholar]
- Kinkema, M. , Fan, W. and Dong, X. (2000) Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell, 12, 2339–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein, D.J. , Rowe, H.C. and Denby, K.J. (2005) Secondary metabolites influence Arabidopsis/Botrytis interactions: variation in host production and pathogen sensitivity. Plant J. 44, 25–36. [DOI] [PubMed] [Google Scholar]
- Lai, Z. , Li, Y. , Wang, F. , Cheng, Y. , Fan, B. , Yu, J.Q. and Chen, Z. (2011) Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell, 23, 3824–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loake, G. and Grant, M. (2007) Salicylic acid in plant defence–the players and protagonists. Curr. Opin. Plant Biol. 10, 466–472. [DOI] [PubMed] [Google Scholar]
- van Loon, L.C. , Geraats, B.P. and Linthorst, H.J. (2006) Ethylene as a modulator of disease resistance in plants. Trends Plant Sci. 11, 184–191. [DOI] [PubMed] [Google Scholar]
- Lorenzo, O. , Piqueras, R. , Sanchez‐Serrano, J.J. and Solano, R. (2003) ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell, 15, 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindrinos, M. , Katagiri, F. , Yu, G.L. and Ausubel, F.M. (1994) The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide‐binding site and leucine‐rich repeats. Cell, 78, 1089–1099. [DOI] [PubMed] [Google Scholar]
- Murashige, T. and Skoog, F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nelissen, H. , Clarke, J.H. , de Block, M. , de Block, S. , Vanderhaeghen, R. , Zielinski, R.E. , Dyer, T. , Lust, S. , Inze, D. and van Lijsebettens, M. (2003) DRL1, a homolog of the yeast TOT4/KTI12 protein, has a function in meristem activity and organ growth in plants. Plant Cell, 15, 639–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen, H. , Fleury, D. , Bruno, L. , Robles, P. , de Veylder, L. , Traas, J. , Micol, J.L. , van Montagu, M. , Inze, D. and van Lijsebettens, M. (2005) The elongata mutants identify a functional Elongator complex in plants with a role in cell proliferation during organ growth. Proc. Natl. Acad. Sci. USA, 102, 7754–7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen, H. , de Groeve, S. , Fleury, D. , Neyt, P. , Bruno, L. , Bitonti, M.B. , Vandenbussche, F. , van der Straeten, D. , Yamaguchi, T. , Tsukaya, H. , Witters, E. , De Jaeger, G. , Houben, A. and Van Lijsebettens, M. (2010) Plant Elongator regulates auxin‐related genes during RNA polymerase II transcription elongation. Proc. Natl. Acad. Sci. USA, 107, 1678–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie, H. , Zhao, C. , Wu, G. , Wu, Y. , Chen, Y. and Tang, D. (2012) SR1, a calmodulin‐binding transcription factor, modulates plant defense and ethylene‐induced senescence by directly regulating NDR1 and EIN3 . Plant Physiol. 158, 1847–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, I.S. , Park, A.R. , Bae, M.S. , Kwon, S.J. , Kim, Y.S. , Lee, J.E. , Kang, N.Y. , Lee, S. , Cheong, H. and Park, O.K. (2005) Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola . Plant Cell, 17, 2832–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, Y. , Yamagata, K. , Hong, K. , Wakayama, T. and Zhang, Y. (2010) A role for the elongator complex in zygotic paternal genome demethylation. Nature, 463, 554–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero, G. , Fellows, J. , Li, Y. , de Bizemont, T. , Dirac, A.M. , Gustafsson, C.M. , Erdjument‐Bromage, H. , Tempst, P. and Svejstrup, J.Q. (1999) Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell. 3, 109–118. [DOI] [PubMed] [Google Scholar]
- Petrakis, T.G. , Sogaard, T.M. , Erdjument‐Bromage, H. , Tempst, P. and Svejstrup, J.Q. (2005) Physical and functional interaction between Elongator and the chromatin‐associated Kti12 protein. J. Biol. Chem. 280, 19 454–19 460. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M. , Leon‐Reyes, A. , van der Ent, S. and van Wees, S.C. (2009) Networking by small‐molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316. [DOI] [PubMed] [Google Scholar]
- Poovaiah, B.W. , Du, L. , Wang, H. and Yang, T. (2013) Recent advances in calcium/calmodulin‐mediated signaling with an emphasis on plant–microbe interactions. Plant Physiol. 163, 531–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo, M.J. , van Loon, L.C. and Pieterse, C.M.J. (2005) Jasmonate‐signals in plant–microbe interactions. J. Plant Growth Regul. 23, 211–222. [Google Scholar]
- Pré, M. , Atallah, M. , Champion, A. , de Vos, M. , Pieterse, C.M. and Memelink, J. (2008) The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 147, 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl, P.B. , Chen, C.Z. and Collins, R.N. (2005) Elp1p, the yeast homolog of the FD disease syndrome protein, negatively regulates exocytosis independently of transcriptional elongation. Mol. Cell, 17, 841–853. [DOI] [PubMed] [Google Scholar]
- Ritchie, M.E. , Silver, J. , Oshlack, A. , Holmes, M. , Diyagama, D. , Holloway, A. and Smyth, G.K. (2007) A comparison of background correction methods for two‐colour microarrays. Bioinformatics, 23, 2700–2707. [DOI] [PubMed] [Google Scholar]
- Rowe, H.C. , Walley, J.W. , Corwin, J. , Chan, E.K. , Dehesh, K. and Kliebenstein, D.J. (2010) Deficiencies in jasmonate‐mediated plant defense reveal quantitative variation in Botrytis cinerea pathogenesis. PLoS Pathog. 6, e1000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, J. , Tsui, F. and Klessig, D.F. (1997) Characterization of a salicylic acid‐insensitive mutant (sai1) of Arabidopsis thaliana identified in a selective screen utilizing the SA‐inducible expression of the tms2 gene. Mol. Plant–Microbe Interact. 10, 69–78. [DOI] [PubMed] [Google Scholar]
- Smyth, G.K. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3: Article 3, [DOI] [PubMed] [Google Scholar]
- Thomma, B.P. , Penninckx, I.A. , Broekaert, W.F. and Cammue, B.P. (2001) The complexity of disease signaling in Arabidopsis. Curr. Opin. Immunol. 13, 63–68. [DOI] [PubMed] [Google Scholar]
- Vlot, A.C. , Dempsey, D.A. and Klessig, D.F. (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. [DOI] [PubMed] [Google Scholar]
- Wang, C. , Ding, Y. , Yao, J. , Zhang, Y. , Sun, Y. , Colee, J. and Mou, Z. (2015) Arabidopsis Elongator subunit 2 positively contributes to resistance to the necrotrophic fungal pathogens Botrytis cinerea and Alternaria brassicicola . Plant J. 83, 1019–1033. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Tsuda, K. , Sato, M. , Cohen, J.D. , Katagiri, F. and Glazebrook, J. (2009) Arabidopsis CaM binding protein CBP60g contributes to MAMP‐induced SA accumulation and is involved in disease resistance against Pseudomonas syringae . PLoS Pathog. 5, e1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , An, C. , Zhang, X. , Yao, J. , Zhang, Y. , Sun, Y. , Yu, F. , Amador, D.M. and Mou, Z. (2013) The Arabidopsis Elongator complex subunit2 epigenetically regulates plant immune responses. Plant Cell, 25, 762–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windram, O. , Madhou, P. , McHattie, S. , Hill, C. , Hickman, R. , Cooke, E. , Jenkins, D.J. , Penfold, C.A. , Baxter, L. , Breeze, E. , Kiddle, S.J. , Rhodes, J. , Atwell, S. , Kliebenstein, D.J. , Kim, Y.S. , Stegle, O. , Borgwardt, K. , Zhang, C. , Tabrett, A. , Legaie, R. , Moore, J. , Finkenstadt, B. , Wild, D.L. , Mead, A. , Rand, D. , Beynon, J. , Ott, S. , Buchanan‐Wollaston, V. and Denby, K.J. (2012) Arabidopsis defense against Botrytis cinerea: chronology and regulation deciphered by high‐resolution temporal transcriptomic analysis. Plant Cell, 24, 3530–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, G.S. , Kristjuhan, A. , Erdjument‐Bromage, H. , Tempst, P. and Svejstrup, J.Q. (2002) Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc. Natl. Acad. Sci. USA, 99, 3517–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittschieben, B.O. , Otero, G. , de Bizemont, T. , Fellows, J. , Erdjument‐Bromage, H. , Ohba, R. , Li, Y. , Allis, C.D. , Tempst, P. and Svejstrup, J.Q. (1999) A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell, 4, 123–128. [DOI] [PubMed] [Google Scholar]
- Xu, D. , Huang, W. , Li, Y. , Wang, H. , Huang, H. and Cui, X. (2012) Elongator complex is critical for cell cycle progression and leaf patterning in Arabidopsis . Plant J. 69, 792–808. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Wang, C. , Zhang, Y. , Sun, Y. and Mou, Z. (2012) The Arabidopsis mediator complex subunit16 positively regulates salicylate‐mediated systemic acquired resistance and jasmonate/ethylene‐induced defense pathways. Plant Cell, 24, 4294–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Z. , Qamar, S.A. , Chen, Z. and Mengiste, T. (2006) Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48, 592–605. [DOI] [PubMed] [Google Scholar]
- Zhou, X. , Hua, D. , Chen, Z. , Zhou, Z. and Gong, Z. (2009) Elongator mediates ABA responses, oxidative stress resistance and anthocyanin biosynthesis in Arabidopsis. Plant J. 60, 79–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website.
Fig. S1 Characterization of drl1 T‐DNA insertion mutants.
Fig. S2 Growth of Pseudomonas syringae pv. maculicola (Psm) ES4326 in drl1 npr1 double mutants.
Fig. S3 Characterization of 35S::DRL1 drl1 transgenic plants.
Fig. S4 Gene ontology (GO) analysis of the genes differentially expressed between drl1 and the wild‐type.
Fig. S5 Characterization of multiple independent transgenic lines expressing the P‐loop and C‐terminal calmodulin‐binding domain (CBD) mutants of DRL1.
Fig. S6 Morphology of the DRL1::DRL1 drl1 and 35S::DRL1 drl1 transgenic plants.
Table S1 Primers used for the identification of homozygous T‐DNA insertion mutant plants.
Table S2 Primers used for quantitative polymerase chain reaction (qPCR) analysis of gene expression.
Table S3 Defence genes that were differentially expressed between drl1‐C1 and the wild‐type during Botrytis cinerea infection.


