Summary
Fusarium wilt is one of the most serious diseases affecting cotton. However, the pathogenesis and mechanism by which Fusarium oxysporum overcomes plant defence responses are unclear. Here, a new group D mitogen‐activated protein kinase (MAPK) gene, GhMPK20, was identified and functionally analysed in cotton. GhMPK20 expression was significantly induced by F. oxysporum. Virus‐induced gene silencing (VIGS) of GhMPK20 in cotton increased the tolerance to F. oxysporum, whereas ectopic GhMPK20 overexpression in Nicotiana benthamiana reduced F. oxysporum resistance via disruption of the salicylic acid (SA)‐mediated defence pathway. More importantly, an F. oxysporum‐induced MAPK cascade pathway composed of GhMKK4, GhMPK20 and GhWRKY40 was identified. VIGS of GhMKK4 and GhWRKY40 also enhanced F. oxysporum resistance in cotton, and the function of GhMKK4–GhMPK20 was shown to be essential for F. oxysporum‐induced GhWRKY40 expression. Together, our results indicate that the GhMKK4–GhMPK20–GhWRKY40 cascade in cotton plays an important role in the pathogenesis of F. oxysporum. This research broadens our knowledge of the negative role of the MAPK cascade in disease resistance in cotton and provides an important scientific basis for the formulation of Fusarium wilt prevention strategies.
Keywords: cotton, Fusarium oxysporum, Gossypium hirsutum; MAPK cascade; negative regulation
Introduction
Plants have been exposed to many pathogens and have evolved multiple mechanisms of innate immunity (Jones and Takemoto, 2004). In general, the plant innate immunity system has two main branches: pathogen‐associated molecular pattern (PAMP)‐triggered immunity (PTI) and effector‐triggered immunity (ETI) (Jones and Dangl, 2006; Medzhitov and Janeway, 1997). Successful pathogens can invade and colonize plants by the suppression of host immunity systems. Bacterial pathogens can inhibit the defence signalling pathway by the secretion of effector molecules (Schechter et al., 2006). Similarly, fungi secrete a wide diversity of effectors into host cells to manipulate plant immunity (Yu X et al., 2012).
Plant immunity systems are tightly controlled by many phytohormones and a series of complex signal transduction processes. Salicylic acid (SA) and jasmonic acid (JA) are the most important hormones for plant immunity systems (Thaler et al., 2012). SA is an essential signalling molecule that induces systemic acquired resistance (SAR) and is primarily affected by responses to biotrophic pathogens (Glazebrook, 2005). The JA‐mediated pathway is primarily induced by resistance to herbivores and necrotrophic pathogens (Glazebrook, 2005). The SA pathway is typically prioritized over the JA pathway, and signalling crosstalk between SA and JA commonly manifests as reciprocal antagonism and may be adaptive (Thaler et al., 2012). Amongst the intricate signalling networks, mitogen‐activated protein kinase (MAPK) cascades are the major modules that identify and amplify external signals into intracellular components (Meng and Zhang, 2013; Rodriguez et al., 2010). MAPK cascade activation is one of the cellular responses induced following PAMP recognition in plants (Asai et al., 2002). Canonical MAPK cascades include three tiers of reversibly phosphorylated kinases: MAPK kinase (MAPKK) kinases (MAPKKKs), MAPKKs and MAPKs (Rodriguez et al., 2010). Previous studies have reported that many pathogens disrupt plant immunity by perturbing the plant MAPK pathways. AtMPK4 is a widely studied MAPK gene, and the activation of MAPK4 by flg22 negatively regulates the innate immune response in plants (Gao et al., 2008). Overexpression of a Pseudomonas syringae effector, HopAI1, inhibits MPK3 and MPK6 activation in response to flg22 in Arabidopsis (Zhang et al., 2007). In addition, HopF2, another P. syringae effector, blocks the kinase activity of MKK5 via its ADP‐ribosyltransferase activity (Feng and Zhou, 2012). In addition to bacterial pathogens, fungal pathogens attack plants by the disruption of plant MAPK cascades. As a model fungus, the mechanism of Phytophthora infestans infection has been widely studied. Phytophthora infestans is a member of a genus of pathogens destructive to members of the nightshade family. Phytophthora infestans secretes effectors to manipulate host innate immunity (Yang et al., 2017). For example, Avh238 and Avh241 are the RxLR effectors of P. infestans that cause cell death in various plant species (Yang et al., 2017; Yu X et al., 2012). Recent studies have reported that Avh238‐ and Avh241‐triggered cell death is required for the mitogen‐activated protein kinase kinase 2 (MEK2)–wound‐induced protein kinase (WIPK) cascade in Nicotiana benthamiana (Yang et al., 2017; Yu X et al., 2012). However, the infection mechanisms of other fungi and the mechanisms by which fungi disrupt plant MAPK cascades are unclear.
MAPK cascades affect plant innate immune responses by interacting with their substrates, and WRKY transcription factors (TFs) are essential MAPK substrates (Zhang T et al., 2016). WRKY TFs have been shown to bind W‐box [TTGACC(T)] in the promoter regions of target genes (Rushton et al., 2010), and many studies have reported that WRKY TFs are involved in the regulation of plant defence responses to pathogens (Rushton et al., 2010). In Arabidopsis, AtWRKY33 has been shown to regulate both SA‐ and JA‐mediated defence pathways (Birkenbihl et al., 2012), and OsWRKY30 overexpression has been shown to increase resistance to rice fungal pathogens by positively regulating the JA‐mediated pathway in rice (Peng et al., 2012). However, little is known about the function of WRKY TFs with regard to the defence response in non‐model plants, and potential mechanisms underlying the negative regulation of WRKY TFs on the plant defence response remain ambiguous.
Cotton (Gossypium hirsutum) is one of the most important fibre‐ and oil‐producing commercial crops (Sunilkumar et al., 2006). Fusarium wilt and Verticillium wilt are the fungal diseases that are the greatest threat to cotton. Fusarium oxysporum f. sp. vasinfectum is the main cause of Fusarium wilt disease and is present in almost all cotton‐growing regions worldwide (Gaspar et al., 2014). Fusarium wilt is a vascular disease that causes the vascular tissue to turn brown and results in substantial crop losses. Fusarium oxysporum infects the roots and then enters the vascular tissue, where it secretes many virulence determinants, such as small peptides or phytotoxins (Pietro et al., 2003). Many studies have reported that F. oxysporum, similar to other fungal pathogens, has evolved multiple strategies to counter plant immunity systems (Morrissey and Osbourn, 1999). The F. oxysporum SNF1 (FoSNF1) gene may encode a cell wall‐degrading enzyme. Mutations in FoSNF1 reduce the virulence of F. oxysporum by repressing the production of cell wall‐degrading enzymes (Ospina‐Giraldo et al., 2003). In addition, the Six1 protein is secreted by F. oxysporum during xylem colonization and is required for full virulence of F. oxysporum f. sp. lycopersici in susceptible tomato lines (Rep et al., 2005). However, few studies have examined the pathogenesis of F. oxysporum disruption of defence responses in cotton.
Upland cotton is an allotetraploid that most probably arose from interspecific hybridization (Page et al., 2013). Previously, 56 MAPKs and 102 WRKYs have been confirmed in upland cotton by genome‐wide analysis (Dou et al., 2014; Zhang X et al., 2016). Some MAPKs, such as GhMPK7 (Shi et al., 2010), GhMPK11 (Wang F et al., 2016) and GhMPK16 (Shi et al., 2011), and WRKYs, such as GhWRKY15 (Yu F et al., 2012), GhWRKY27a (Yan et al., 2015) and GhWRKY40 (Wang et al., 2014), have been shown to affect plant defence responses.
In this study, a new cotton group D MAPK gene, GhMPK20, was identified and characterized. Silencing of GhMPK20 in cotton enhanced the tolerance to F. oxysporum, and ectopic GhMPK20 overexpression in N. benthamiana reduced the resistance to F. oxysporum. More importantly, results from a series of genetic and biochemical analyses indicated that GhMPK20 plays an important role in the pathogenic process of F. oxysporum in cotton via the MKK4–MPK20–WRKY40 pathway. Together, these results broaden our knowledge regarding the biological roles of the MAPK cascade in disease resistance, and significantly improve our understanding of the pathogenesis of F. oxysporum.
Results
Characterization and subcellular localization of GhMPK20
Many recent studies have reported that MAPKs play important roles in plant disease resistance in Arabidopsis thaliana (MAPK Group, 2002). To identify new F. oxysporum‐induced MAPK genes in cotton, we detected the expression levels of MAPK genes in cotton after F. oxysporum infection, and expression analyses were performed in the cotton cotyledon. One MAPK gene, designated GhMPK20, was chosen for detailed characterization because it was significantly induced by F. oxysporum (more than eight‐fold) (Fig. S1A, see Supporting Information).
The full‐length GhMPK20 cDNA (GenBank accession number: HQ828072) is 2085 bp, with a 71‐bp 5′‐untranslated region (UTR), 193‐bp 3′‐UTR and 1821‐bp open reading fragment (ORF), encoding a 606‐amino‐acid peptide with a predicted molecular weight of 69.16 kDa and an isoelectric point of 9.22. Phylogenetic analysis showed that GhMPK20 can be clustered into the group D MAPK family (Fig. S2, see Supporting Information). Multiple sequence alignments demonstrated that GhMPK20 shares high homology with other group D members, such as A. thaliana AtMPK20 (73.49%), G. hirsutum GhMPK16 (60.79%) and Oryza sativa OsMPK20 (72.98%) (Fig. 1A). GhMPK20 also exhibits significant family similarity, with 11 conserved subdomains, a phosphorylation motif (TDY motif) in an activation loop and an extended C‐terminal region not present in group A, B and C MAPKs (Fig. 1A). Moreover, the 5172‐bp genomic sequence of GhMPK20 (GenBank accession number: JX470183) contains nine introns, similar to the structures of the other group D MAPKs, but greater in size than the other group D MAPKs (Fig. 1B).
Figure 1.

Sequence analysis and genomic DNA structures of GhMPK20. (A) Alignment of the amino acid sequences of GhMPK20 (HQ828072) with AtMPK20 (AEC10180), GhMPK15 (XP_016747991), GhMPK16 (ADI52623), GhMPK19 (NP_001314211) and OsMPK20 (XP_015621247). Identical amino acids are shaded in black. The conserved subdomains are shown with numerals (I–XI) and underlined at the top of the sequences. The TDY motif is marked by a dot. (B) Comparison of the genomic DNA structure of GhMPK20 with that of other group D mitogen‐activated protein kinases (MAPKs). The legend displays the pattern of exons, introns and untranslated regions (UTRs). The scale of the x‐axis indicates the length of the sequence. The abbreviations of the species names are as follows: At, Arabidopsis thaliana; Gh, Gossypium hirsutum; Os, Oryza sativa.
To establish the intracellular localization of GhMPK20, a reporter gene encoding green fluorescent protein (GFP) was fused to the C‐terminus of GhMPK20 under the control of the Cauliflower mosaic virus 35S (CaMV35S) promoter in pBI121‐GFP (Fig. 2A). The 35S::GhMPK20‐GFP plasmid and the control 35S::GFP plasmid were transiently expressed separately in N. benthamiana leaves using Agrobacterium infiltration (Fig. S3, see Supporting Information). As shown in Fig. 2B, fluorescent signals from 35S::GFP and 35S::GhMPK20‐GFP were distributed throughout the cytoplasm and nucleus. These results further confirm that GhMPK20 potentially functions in both the cytoplasm and nucleus.
Figure 2.
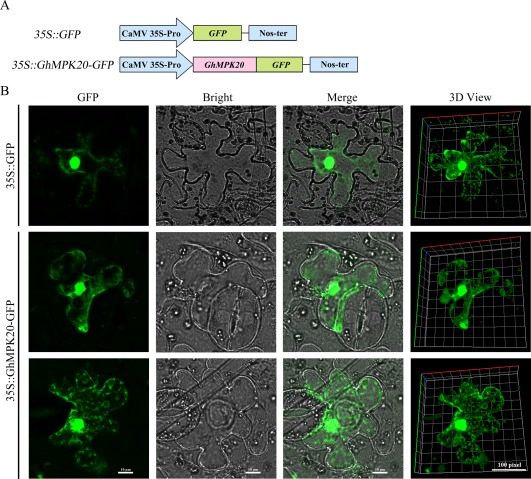
Subcellular localization of GhMPK20 in Nicotiana benthamiana leaves. (A) Schematic diagram of the 35S::GhMPK20‐GFP fusion construct and 35S::GFP construct. (B) Transient expression of the 35S::GhMPK20‐GFP fusion construct and the 35S::GFP construct in N. benthamiana leaves. Green fluorescence was observed with an LSM 880 META confocal microscope (Carl Zeiss). GFP, green fluorescent protein.
To determine the tissue‐specific expression patterns of GhMPK20, RNA was extracted from the roots, stems, leaves and cotyledons of 7‐day‐old cotton seedlings for quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). As shown in Fig. 3A, GhMPK20 was expressed in all tissues investigated in this study.
Figure 3.
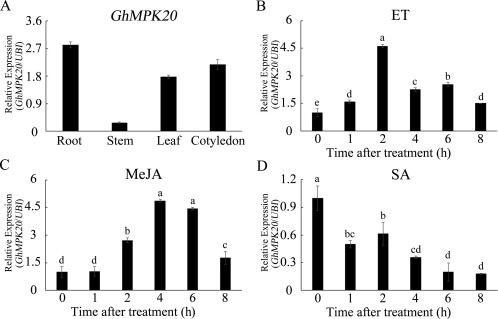
Expression patterns of GhMPK20 in different tissues and under different stress conditions. (A) Tissue‐specific expression of GhMPK20 was detected in the roots, stems, leaves and cotyledons. GhMPK20 expression levels were analysed after cotton seedlings were separately treated with 5 mm ethylene (ET) released from ethephon (B), 100 μm methyl jasmonate (MeJA) (C) and 10 mm salicylic acid (SA) (D). Data are presented as the mean ± standard error (SE) of three independent experiments (n = 6). The letters above the columns indicate significant differences (P < 0.05) according to Tukey's honestly significant difference (HSD) test.
GhMPK20‐silenced cotton displays enhanced resistance to F. oxysporum
To check whether GhMPK20 expression is induced by many disease resistance‐related hormones [such as ethylene (ET), JA and SA], cotton seedlings were separately exposed to various phytohormones. As shown in Fig. 3B,C, GhMPK20 expression was significantly enhanced after ET and JA treatments. SA treatment induced a significant decrease in the GhMPK20 expression level (Fig. 3D). These results indicate that GhMPK20 may play crucial roles in plant disease defence signal transduction pathways.
To examine whether GhMPK20 participates in cotton F. oxysporum resistance, virus‐induced gene silencing (VIGS) was used to silence GhMPK20 in cotton. By 21 days post‐infiltration (dpi), GhMPK20 mRNA levels were typically reduced (Fig. 4A). After 5 days of infection with F. oxysporum spore suspension, the degree of morbidity of F. oxysporum was determined. The degree of morbidity of F. oxysporum in GhMPK20‐silenced (CRV::20‐01 and CRV::20‐02) leaves was remarkably less than that in non‐silenced leaves (CRV::00) (Fig. 4B). The pathogen growth assay results showed that the F. oxysporum copy number in GhMPK20‐silenced cotton was less than that in non‐silenced cotton (Fig. 4C). In addition, the leaves of GhMPK20‐silenced cotton accumulated less brown staining than non‐silenced leaves, as demonstrated by diaminobenzidine (DAB) staining (Fig. 4B). To elucidate the mechanism underlying this enhanced resistance, the expression levels of SA‐mediated defence pathway genes and pathogenesis‐related (PR) genes were examined. The SA‐mediated genes and PR genes showed much higher expression levels in GhMPK20‐silenced cotton than in non‐silenced leaves after F. oxysporum infection (Fig. 4D). These results show that GhMPK20 silencing enhances F. oxysporum resistance in cotton.
Figure 4.
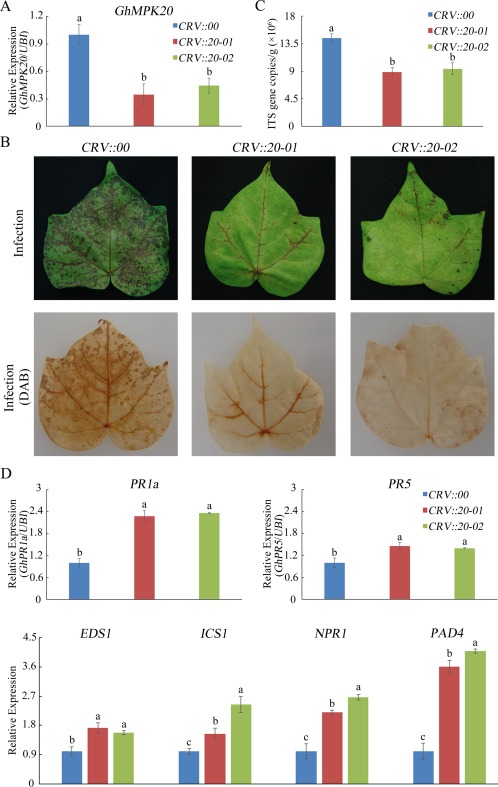
Silencing of GhMPK20 in cotton increases the tolerance to Fusarium oxysporum. (A) GhMPK20 expression levels in GhMPK20‐silenced cotton plants. (B) Representative phenotypes of GhMPK20‐silenced cotton after infection with F. oxysporum for 5 days. DAB, diaminobenzidine. (C) Pathogen accumulation level in GhMPK20‐silenced plants after infection with F. oxysporum for 5 days. ITS indicates the nuclear ribosomal DNA internal transcribed spacer (ITS) sequences of fungi. (D) Expression level of salicylic acid (SA)‐mediated defence pathway genes and pathogenesis‐related (PR) genes in GhMPK20‐silenced cotton plants after infection with F. oxysporum for 5 days. The error bars in (A), (C) and (D) indicate the mean ± standard error (SE) of three independent experiments (n = 9). The letters above the columns represent significant differences (P < 0.05) based on Tukey's honestly significant difference (HSD) test.
GhMPK20‐overexpressing plants display enhanced sensitivity to F. oxysporum
To further detect the biological function of GhMPK20 in disease resistance, transgenic N. benthamiana plants overexpressing GhMPK20 were generated, and three independent overexpressing lines (10#, 13# and 17#, named OE1, OE2 and OE3, respectively) were selected (Fig. S4, see Supporting Information). The T3 generations of these three transgenic plants were used for further functional study.
After plants were infected with F. oxysporum for 5 days, the leaves of overexpressing (OE) plants showed more serious wilting symptoms and chlorosis than did vector control (Vec) plants (Fig. 5A). Histochemical staining with trypan blue and DAB showed that OE leaves accumulated more blue and brown staining than did Vec leaves (Fig. 5A). The expression levels of SA‐mediated defence pathway genes and PR genes were also examined. As shown in Fig. 5B, the expression levels of all the tested genes were significantly decreased in OE leaves. These results indicate that GhMPK20‐overexpressing plants display enhanced susceptibility to F. oxysporum.
Figure 5.
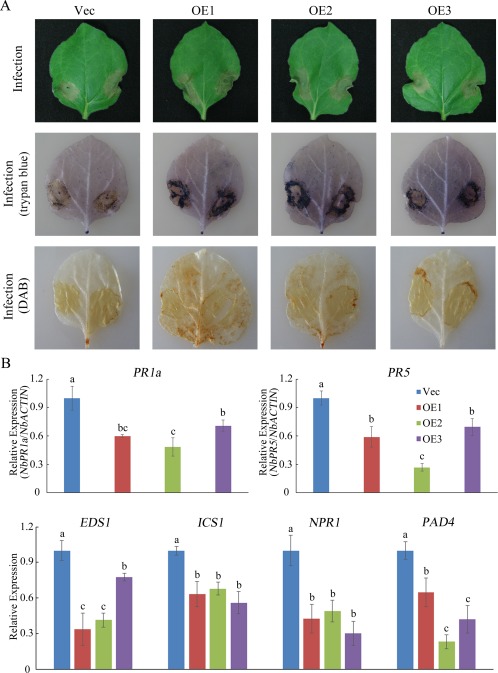
GhMPK20 overexpression reduces the tolerance to Fusarium oxysporum in transgenic Nicotiana benthamiana. (A) Representative phenotypes of GhMPK20‐overexpressing plants after infection with F. oxysporum for 5 days. DAB, diaminobenzidine. (B) Expression levels of salicylic acid (SA)‐mediated defence pathway genes and pathogenesis‐related (PR) genes in GhMPK20‐overexpressing N. benthamiana after infection with F. oxysporum for 5 days. Data are presented as the mean ± standard error (SE) of three independent experiments (n = 9). The letters above the columns indicate significant differences (P < 0.05) based on Tukey's honestly significant difference (HSD) test.
GhMPK20 interacts with GhMKK4 in vitro and in vivo
MAPKKs are potential upstream genes of MAPKs (MAPK Group, 2002). To investigate the upstream component of GhMPK20, based on our previous studies and the cotton genome, the interactions between GhMPK20 and some cotton MKKs [such as the A group MKK gene GhMKK6 (Gene ID: 107932005), B group MKK gene GhMKK3 (GenBank accession number: HQ828070), C group MKK genes GhMKK4 (GenBank accession number: FJ966886) and GhMKK5 (GenBank accession number: HQ637469), and D group MKK gene GhMKK9 (GenBank accession number: HQ651069)] were detected using yeast two‐hybrid assays. The yeast two‐hybrid assay results showed that only the positive control clone and the clone co‐transformed with GhMPK20 and GhMKK4 grew well on SD medium DDO and QDO/X (SD, synthetically defined medium; SD medium DDO, SD medium without Leu and Trp; SD medium QDO/X, SD medium with X‐a‐gal and without Ade, His, Leu, Trp) plates, indicating that GhMKK4 may be the upstream component of GhMPK20 (Fig. 6A). The interaction between GhMPK20 and GhMKK4 was further confirmed using the bimolecular fluorescence complementation (BiFC) system, in which GhMPK20‐yellow fluorescent protein (YFP)C and GhMKK4‐YFPN were co‐transformed into onion epidermal cells through particle bombardment. In accordance with the localization of GhMPK20 (Fig. 2) and GhMKK4 (Li et al., 2014), the fluorescent signal was detected in both the cytoplasm and nucleus (Fig. 6B). Furthermore, co‐immunoprecipitation (Co‐IP) assays verified the interaction between GhMPK20 and GhMKK4. The Co‐IP result showed that GhMKK4‐FLAG could be immunoprecipitated by GhMPK20‐MYC in protoplasts (Fig. 6C). These results demonstrate that GhMPK20 interacts with GhMKK4 in vitro and in vivo.
Figure 6.
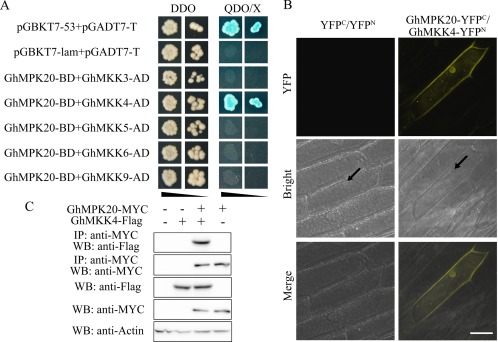
Interaction between GhMPK20 and GhMKK4. (A) GhMPK20 specifically interacts with GhMKK4 according to the yeast two‐hybrid (Y2H) system assay results. The indicated BD and AD fusion constructs were co‐transformed into the Y2H Gold yeast strain and grown on the SD media DDO and QDO/X. (B) The interaction between GhMPK20 and GhMKK4 was confirmed by bimolecular fluorescence complementation (BiFC) experiments. GhMPK20‐YFPC and GhMKK4‐YFPN fusion constructs were co‐transformed into onion epidermal cells using the particle bombardment method. Yellow fluorescence was observed with an LSM 510 META confocal microscope (Carl Zeiss). Arrowheads indicate nuclei. Bar, 50 μm. YFP, yellow fluorescent protein. (C) Co‐immunoprecipitation analysis was performed using MYC antibody in cotton protoplasts. GhMPK20‐MYC and GhMKK4‐FLAG fusion constructs were co‐transformed into cotton protoplasts. Western blot analysis shows protein expression levels. SD, synthetically defined medium; SD medium DDO, SD medium without Leu and Trp; SD medium QDO/X, SD medium with X‐a‐gal and without Ade, His, Leu, Trp.
GhMKK4‐silenced cotton displays enhanced resistance to F. oxysporum
To test whether GhMKK4 has functions similar to those of GhMPK20, we used VIGS to silence GhMKK4 in cotton. After cotton plants were infiltrated with Agrobacterium for 3 weeks, GhMKK4 expression was significantly reduced in CRV::GhMKK4 cotton plants (Fig. S5, see Supporting Information). Then, CRV::00 and CRV::GhMKK4 (CRV::4‐01 and CRV::4‐02) were root‐wounded and placed in an F. oxysporum spore suspension. After these plants were infected with F. oxysporum for 5 days, the CRV::GhMKK4 (CRV::4‐01, CRV::4‐02) plants showed resistance phenotypes and presented less brown staining than CRV::00 leaves, as determined by DAB staining (Fig. 7A). Furthermore, the SA‐mediated genes and PR genes showed the same expression profiles between CRV::GhMKK4 and CRV::GhMPK20 plants after F. oxysporum infection for 5 days (Fig. 7B).
Figure 7.
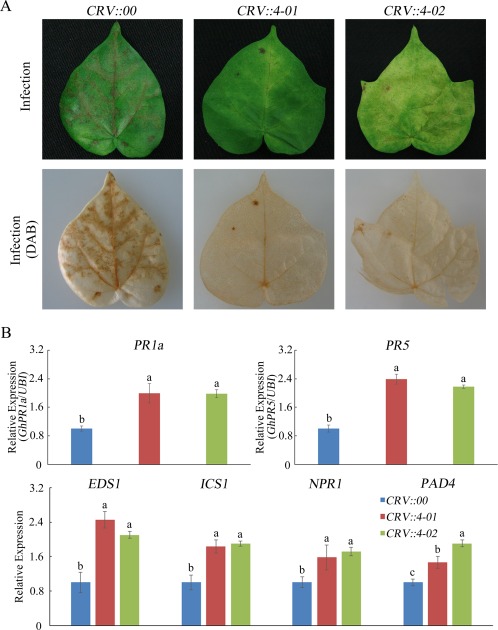
Silencing of GhMKK4 in cotton increases the tolerance to Fusarium oxysporum. (A) Representative phenotypes of GhMKK4‐silenced cotton after infection with F. oxysporum for 5 days. DAB, diaminobenzidine. (B) Expression levels of salicylic acid (SA)‐mediated defence pathway genes and pathogenesis‐related (PR) genes in GhMKK4‐silenced cotton after infection with F. oxysporum for 5 days. Data are presented as the mean ± standard error (SE) of three independent experiments (n = 9). The letters above the columns indicate significant differences (P < 0.05) based on Tukey's honestly significant difference (HSD) test.
GhMKK4–GhMPK20 function upstream of GhWRKY40 under F. oxysporum infection
Many studies have shown that WRKY TFs are amongst the most important substrates for MAPK cascades (Ishihama and Yoshioka, 2012). Previously, we have shown that GhWRKY40 is induced by fungal disease and GhWRKY40 interacts with GhMPK20 (Wang et al., 2014). In this study, the yeast two‐hybrid assay result showed that the positive control clone and the clone co‐transformed with GhMPK20 and GhWRKY40 grew well on SD medium DDO and QDO/X plates (Fig. 8A). Co‐IP also showed that GhWRKY40‐FLAG could be immunoprecipitated by GhMPK20‐MYC in protoplasts (Fig. 8B).
Figure 8.
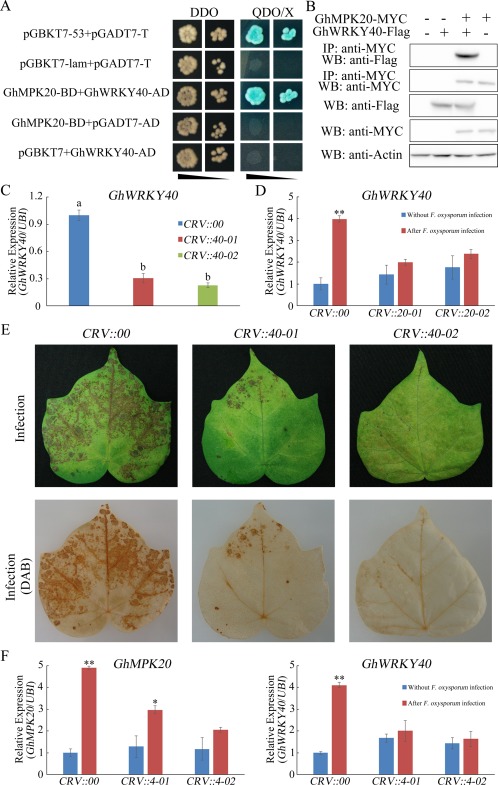
Function of GhMKK4–GhMPK20 is essential for Fusarium oxysporum‐induced GhWRKY40 expression. (A) GhMPK20 interacts with GhWRKY40 according to the yeast two‐hybrid (Y2H) system assay results. The indicated BD and AD fusion constructs were co‐transformed into the Y2H Gold yeast strain and grown on the SD media DDO and QDO/X. (B) Co‐immunoprecipitation (Co‐IP) analysis was performed using MYC antibody in cotton protoplasts. GhMPK20‐MYC and GhWRKY40‐FLAG fusion constructs were co‐transformed into cotton protoplasts. Western blot analysis shows protein expression levels. (C) GhWRKY40 expression levels in GhWRKY40‐silenced cotton plants. The data are presented as the mean ± standard error (SE) of three independent experiments (n = 9). The letters above the columns represent significant differences (P < 0.05) based on Tukey's honestly significant difference (HSD) test. (D) GhWRKY40 expression levels in GhMPK20‐silenced cotton plants with or without F. oxysporum infection. The error bars indicate the mean ± SE of three independent experiments (n = 9). Asterisks above the lines indicate significant differences (*P < 0.05, **P < 0.01) based on Tukey's HSD test. (E) Representative phenotypes of GhWRKY40‐silenced cotton after infection with F. oxysporum for 5 days. DAB, diaminobenzidine. (F) GhMPK20 and GhWRKY40 expression levels in GhMKK4‐silenced cotton plants with or without F. oxysporum infection. The error bars indicate the mean ± SE of three independent experiments (n = 9). Asterisks above the lines indicate significant differences (*P < 0.05, **P < 0.01) based on Tukey's HSD test. SD, synthetically defined medium; SD medium DDO, SD medium without Leu and Trp; SD medium QDO/X, SD medium with X‐a‐gal and without Ade, His, Leu, Trp.
To prove that these three signalling elements are indeed from the same cascade, a Phos‐tag‐based method was used in the phosphorylation assay. Phos‐tag, together with a chelated divalent cation, such as Mn2+, can bind phosphate groups (Longoni et al., 2015), and phosphorylated proteins can be resolved from their non‐phosphorylated forms based on differences in their migration rates (Bekešová et al., 2015). As shown in Fig. S6 (see Supporting Information), the phosphorylation levels of GhMPK20 were remarkably higher in cotton protoplasts co‐transformed with GhMKK4‐FLAG and GhMPK20‐MYC than in those transformed with only GhMPK20‐MYC (Fig. S6A). Furthermore, the phosphorylation levels of GhWRKY40 were higher in cotton protoplasts co‐transformed with GhMPK20‐MYC and GhWRKY40‐FLAG than in those transformed with only GhWRKY40‐FLAG (Fig. S6B).
To investigate whether GhWRKY40 expression is induced by F. oxysporum, cotton seedlings were exposed to F. oxysporum. GhWRKY40 transcript accumulation was typically elevated at the different time points following F. oxysporum infection (Fig. S7, see Supporting Information). To identify the function of GhWRKY40 on F. oxysporum infection, we used VIGS to silence GhWRKY40 in cotton (Fig. 8C). Five days after F. oxysporum infection, the leaves from CRV::GhWRKY40 exhibited milder wilting symptoms and chlorosis than did CRV::00 cotton (Fig. 8E). DAB staining showed that the CRV::GhWRKY40 leaves accumulated less brown staining than CRV::00 leaves (Fig. 8E).
To confirm that the function of GhMKK4–GhMPK20 is required for F. oxysporum‐induced GhWRKY40 expression. GhMKK4‐silenced and GhMPK20‐silenced leaves were infected with F. oxysporum. Compared with CRV::00 control plants, GhWRKY40 expression did not show obvious changes in GhMKK4‐silenced or GhMPK20‐silenced cotton plants after F. oxysporum infection (Fig. 8D,F). Furthermore, GhMPK20 expression was increased in the leaves of CRV::00 cotton plants, whereas that in CRV::GhMKK4 plants showed no obvious changes after F. oxysporum infection (Fig. 8F). These results suggest that the function of GhMKK4–GhMPK20 is essential for F. oxysporum‐induced GhWRKY40 expression.
Discussion
MAPK pathways are amongst the most important signal transduction pathways in plants, and many MAPK genes have been identified; however, information regarding the functions of group D MAPKs remains limited. In this study, GhMPK20, a new group D MAPK gene, was identified in G. hirsutum, and silencing of GhMPK20 in cotton enhanced resistance to F. oxysporum.
GhMPK20 possesses the typical features of group D MAPKs, such as the TDY activation motif, an extended C‐terminal region and the lack of a CD domain (Fig. 1) (MAPK group). The first identified function of group D MAPKs is enhanced resistance to fungal and bacterial infections by the overexpression of OsBWMK1 through constitutive PR gene expression (Cheong et al., 2003). Maize ZmMPK17 is involved in multiple stress responses, such as cold, osmotic stress and SA‐mediated pathogen defence (Pan et al., 2012). Moreover, active AtMPK18 can be dephosphorylated by PHS1 in the cytoplasm to regulate cortical microtubule functions (Walia et al., 2009). In cotton, GhMPK20 has many homologous genes, including GhMPK15 (GenBank accession number: XP_016747991), GhMPK16 (GenBank accession number: ADI52623) and GhMPK19 (GenBank accession number: NP_001314211) (Fig. 1). However, the functions of TDY MAPKs in cotton are unclear. In our previous studies, cotton GhMPK16 enhanced resistance to fungal (Colletotrichum nicotianae and Alternaria alternata) and bacterial (Pseudomonas solanacearum) pathogens in transgenic plants by rapidly inducing the expression of some PR genes (Shi et al., 2011). In this study, GhMPK20 was shown to differ from other group D MAPKs and conferred sensitivity to F. oxysporum stress in transgenic plants (Fig. 5).
Most of the plant MAPKs reported recently play important roles in pathogen resistance, whereas few MAPKs have been identified as negative effectors. AtMPK4, which can be activated by AtMKK1 and AtMKK2, is a negative regulator of the innate immune response in plants (Gao et al., 2008). Furthermore, functional analysis of Arabidopsis immune‐related MAPKs identified a negative role for MPK3 in the regulation of defence gene expression, SA accumulation and disease resistance to P. syringae (Frey et al., 2014). The present study revealed that GhMPK20 expression was highly induced in cotton after infection with F. oxysporum, suggesting that F. oxysporum might activate GhMPK20 to inhibit the defence response during infection. Furthermore, VIGS of GhMPK20 in cotton enhanced the tolerance to F. oxysporum by increasing the expression of some PR and SA‐related genes (Fig. 4). As a result of experimental condition limitations, cotton transformation is very difficult. Considering the easy transformation and convenience of studying F. oxysporum resistance, gain‐of‐function analysis was first performed by heterologous ectopic expression in N. benthamiana. Overexpression of GhMPK20 in N. benthamiana resulted in opposite phenotypes for GhMPK20‐silenced cotton. These data provide novel information and enrich our understanding of the functions of group D MAPKs in cotton.
In Arabidopsis, the AtMEKK1–AtMKK4/5–AtMPK3/6 cascade is involved in the regulation of the plant innate immune response, and the AtYODA–AtMKK4/5–AtMPK3/6 module plays critical roles in pathogen signalling and stomatal development (MAPK Group, 2002; Rodriguez et al., 2010). In addition to the model plant Arabidopsis, some cascades in other plants, such as tomato, tobacco, rice and apple, have also been identified (Kishi‐Kaboshi et al., 2010; Pedley and Martin, 2004; Wang et al., 2010; Yang et al., 2001). However, the components of these cascades require further study because they are largely incomplete. In our previous studies, we reported that GhWRKY40 is a potential downstream gene of GhMPK20 (Wang et al., 2014). Here, based on the yeast two‐hybrid, BiFC and Co‐IP analyses, a MAPK cascade composed of GhMKK4, GhMPK20 and GhWRKY40 was identified (Figs 6 and 8). GhMKK4 has been shown to enhance disease susceptibility by disrupting the SA signalling pathway (Li et al., 2014). GhWRKY40 overexpression also decreased the resistance of transgenic plants to pathogen infection and decreased the accumulation of PR1 transcripts (Wang et al., 2014). In this study, silencing of GhMKK4, GhMPK20 and GhWRKY40 in cotton enhanced the resistance to F. oxysporum. Furthermore, we found that the function of GhMKK4–GhMPK20 is essential for F. oxysporum‐induced GhWRKY40 expression (Fig. 8).
SA has been identified as one of the most crucial signalling molecules in the mediation of plant defence responses (Vlot et al., 2009). After pathogen infection, SA rapidly accumulates and activates the plant innate immunity response (Gao et al., 2017). PR1 is a key marker gene of SA signalling. High expression levels of PR1 indicate that SA signalling has been activated (He et al., 2007). NPR1 is another key marker of SA signalling, and an npr1 mutant was found to be SA insensitive, as it exhibited enhanced susceptibility to many bacterial and fungal pathogens (Delaney et al., 1995; Glazebrook et al., 1996). An eds5 mutant also enhanced disease susceptibility by inhibiting SA synthesis, and this phenotype could be rescued by the addition of SA (Rogers and Ausubel, 1997). In this study, we noticed that SA‐related genes, such as EDS1, ICS1, NPR1, PAD4 and PR1, were significantly induced after F. oxysporum infection in GhMKK4‐silenced (Fig. 7), GhMPK20‐silenced (Fig. 4) and GhWRKY40‐silenced (Fig. S8, see Supporting Information) cotton. Furthermore, as shown in Fig. S10 (see Supporting Information), after F. oxysporum infection for 5 days, the degree of F. oxysporum morbidity was remarkably lower after the external application of SA (10 mm), although the phenotype was not rescued completely. These results indicate that the GhMKK4–GhMPK20–GhWRKY40 cascade disrupts the SA signalling pathway and/or inhibits SA synthesis. Ding et al. (2011) have reported that, although SA‐ and JA‐mediated defence signalling pathways are known to have an antagonistic relationship, synergy between the two is coordinated in resistance‐related early molecular events. The sequential induction of SA‐ and JA‐mediated pathways would be an effective coordination mechanism of the two antagonistic pathways (Ding et al., 2011). Based on our results, we speculate that the GhMKK4–GhMPK20–GhWRKY40 cascade negatively regulates F. oxysporum resistance by disrupting this coordination.
Fusarium wilt disease has devastating effects on cotton, causing substantial crop losses in most of the major cotton‐producing areas worldwide (Wang et al., 2006). Many studies have reported that Fusarium wilt is a component of a disease complex with root‐knot nematodes and that F. oxysporum can secrete many enzymes to break the physical barriers and antifungal compounds of plants (Pietro et al., 2003; Wang et al., 2006). However, the pathogenic mechanism of F. oxysporum and the mechanism by which F. oxysporum overcomes plant defence responses are unclear. In this study, we report a new strategy by which F. oxysporum attacks plants. After root invasion and vascular colonization, the MAPK cascade, composed of GhMKK4, GhMPK20 and GhWRKY40, is affected by F. oxysporum. This MAPK cascade may disrupt the SA‐related defence pathway by decreasing the expression of some SA‐related genes and PR genes (Figure S9). Our results compensate for deficiency of knowledge in the pathogenic process of F. oxysporum in cotton and provide an important scientific basis for the formulation of Fusarium wilt prevention strategies.
Experimental Procedures
Plant materials, growth conditions and stress treatments
Cotton (G. hirsutum L. cv. lumian 22) seedlings were planted in hydroponic cultures and received 16 h of continuous light followed by 8 h of darkness per 24‐h period under glasshouse conditions at 26 ± 1 °C. For expression pattern analyses, uniform cotton seedlings were sprayed with ET released from ethephon (5 mm), JA (100 μm) and SA (10 mm) or inoculated with F. oxysporum conidial suspensions (106 conidia/mL) using the root‐dip method. Transgenic and vector control (Vec) N. benthamiana seedlings were planted in soil under a 16‐h light/8‐h dark photoperiod at 25 ± 1 °C. For F. oxysporum infections performed on cotton plants, the seedlings were inoculated with F. oxysporum conidial suspensions (106 conidia/mL) using the root‐dip method. In addition, the F. oxysporum conidial suspensions (106 conidia/mL) were injected into the leaves of GhMPK20‐overexpressing N. benthamiana using syringes. The treated materials were collected and frozen at −80 °C for RNA extraction. Each treatment was repeated at least three times.
GhMPK20 cloning, sequence analyses, vector construction and genetic transformation
The methods of cloning GhMPK20 cDNA and genomic sequences were performed as described previously (Zhang et al., 2012). Amino acid sequence alignments and the phylogenetic tree of GhMPK20 and other MAPKs were produced by DNAMAN and Molecular Evolutionary Genetics Analysis (mega) software 4.0, respectively. The ORF of GhMPK20 was cloned and inserted into the vector pRI 201‐AN (TaKaRa, Japan) under the control of a CaMV35S promoter. GhMPK20‐overexpressing N. benthamiana plants were obtained according to previously described procedures (Zhang et al., 2011). The primers used in this study are shown in Table S1 (see Supporting Information).
Subcellular localization of GhMPK20
To achieve transient expression, the recombinant plasmids 35S::GhMPK20‐GFP and 35S::GFP were transformed into Agrobacterium tumefaciens strain GV3101. After A. tumefaciens was cultured overnight, the cultures were pelleted and resuspended [optical density at 600 nm (OD600) = 1] in infiltration medium (10 mm MgCl2, 10 mm MES‐NaOH, pH 3.8, 200 μm acetosyringone). The leaves of 5‐week‐old N. benthamiana were used for transient expression. The fluorescent signals were observed with an LSM 880 META confocal microscope (Carl Zeiss, Germany).
RNA extraction and expression analyses of GhMPK20
The modified cetyl‐trimethyl‐ammonium bromide (CTAB) method (Wang et al., 2011) or TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was used to extract total RNA from all experimental samples. Subsequently, first‐strand cDNA was synthesized using EasyScript First‐Strand cDNA Synthesis SuperMix (Transgen, Beijing, China). The expression patterns of target genes for functional assays were analysed by qRT‐PCR. Reactions were performed in a 20‐µL reaction volume using a CFX96™ Real‐time Detection System (Bio‐Rad) and Bestar SybrGreen qPCR Mastermix (DBI, Germany). The following PCR program was used: pre‐denaturation at 95 °C for 2 min; 40 cycles of 95 °C for 10 s, 55 °C for 30 s and 72 °C for 30 s; and a final melt cycle from 55 to 98 °C. The G. hirsutum ubiquitin (UBI) or N. benthamiana β‐actin gene was used as the standard control.
Agrobacterium‐mediated VIGS
The method for VIGS was performed according to Gu et al. (2014). The fragments of GhMPK20, GhMKK4 and GhWRKY40 were inserted into the pCLCrV‐A vector. The primers used are shown in Table S1. The recombinant vectors pCLCrV‐A and pCLCrV‐B were transformed into A. tumefaciens strain EHA105. After A. tumefaciens was cultured overnight, the cultures were pelleted and resuspended (OD600 = 1) in infiltration medium as described above. The Agrobacterium suspensions containing pCLCrV‐A or pCLCrV‐B were mixed equally and inoculated into two fully expanded cotton cotyledons after incubation for 3 h. The leaves of the inoculated cotton plants were used for assays 3 weeks after inoculation. Each assay was performed using at least three independent biological replicates.
Yeast two‐hybrid, BiFC and Co‐IP assays
The interaction between GhMPK20 and cotton MKKs (GhMKK3, GhMKK4, GhMKK5, GhMKK6 or GhMKK9) or GhWRKY40 was detected with the Matchmaker Gold Yeast Two‐Hybrid System (Clontech, Japan) and the BiFC system as described previously (Li et al., 2013). For the yeast two‐hybrid system, the ORF region of GhMPK20 was cloned and fused to the GAL4 DNA‐binding domain in the bait plasmid pGBKT7. The ORF regions of GhWRKY40 or MKKs were separately cloned into the GAL4 activation domain in the prey vector pGADT7. The appropriate combinations of two plasmids amongst these plasmids were co‐transformed into the yeast two‐hybrid Gold strain and plated on DDO (SD medium without Leu and Trp [SD/–Leu/–Trp]) medium. Then, the positive clones were grown on the selective SD medium (QDO/X, SD medium with X‐a‐gal and without Ade, His, Leu, Trp [SD/–Ade/–His/–Leu/–Trp with X‐a‐gal]).
For the BiFC system, GhMPK20 and GhMKK4 were fused into pUC‐SPYCE‐35S and pUC‐SPYNE‐35S, respectively. Subsequently, these two recombinant plasmids were co‐transformed into onion epidermal cells using the particle bombardment method. The fluorescent signal was detected with a confocal microscope (LSM 510 META; Carl Zeiss).
For the Co‐IP assay, cotton seedlings were germinated under 16 h of continuous light followed by 8 h of darkness per 24‐h period. Cotton cotyledons were used to prepare protoplasts as described by Chen et al. (2017). Cotyledons were cut into 0.5‐mm strips and immediately transferred into 10 mL of enzyme solution [2.0% cellulase R10, 0.4% macerozyme R10, 0.6 m mannitol, 10 mm MES (pH 5.7), 20 mm KCl, 10 mm CaCl2 and 0.1% bovine serum albumin (BSA)] to digest at 25 °C after being vacuum prepared for 30 min. The following steps and protoplast transformation were performed as described by Yoo et al. (2007). GhMPK20 was inserted into the pGreenII 62‐MYC vector (GhMPK20‐MYC), and GhMKK4 or GhWKRY40 was fused into the pPZP211‐Flag vector (GhMKK4‐FLAG or GhWRKY40‐FLAG). The Co‐IP assay was performed using a ProFound c‐MYC Tag IP/Co‐IP kit (Thermo Fisher) according to previously described procedures (Wang C et al., 2016).
Phos‐tag sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE)
GhMKK4‐FLAG, GhMPK20‐MYC and GhWRKY40‐FLAG were transformed into cotton protoplasts as described above. Total proteins were extracted from leaves using extraction buffer [50 mm Tris/HCl (pH 7.5), 150 mm NaCl, 1% (v/v) Triton X‐100, 0.1% (w/v) SDS, phosphatase and protease inhibitors (PhosStop™ and EDTA‐free Complete™, Roche)]. The SDS‐PAGE separating gel consisted of 8% acrylamide, 50 μm Phos‐tag acrylamide (APExBIO, USA), 357 mm Tris buffer (pH 8.8) and 100 μm MnCl2. Phos‐tag SDS‐PAGE was performed using Mn2+‐Phos‐tag SDS‐PAGE according to the manufacturer's protocol.
Statistical analysis
All of the experiments in this study were performed at least three times. The error bars in each graph indicate the mean values ± standard error (SE) of replicates. Statistical significance between different measurements was assessed by Tukey's honestly significant difference (HSD) test using IBM Statistical Product and Service Solutions (SPSS) Statistics software version 19 (IBM, USA).
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Mitogen‐activated protein kinase (MAPK) expression patterns were analysed after cotton seedlings were infected with Fusarium oxysporum. (A) Expression pattern of GhMPK20 in cotyledon after F. oxysporum infection. (B) Expression pattern of GhMPK20 after root wounding. (C) Expression pattern of GhMPK6 after F. oxysporum infection. (D) Expression pattern of GhMPK7 after F. oxysporum infection. (E) Expression pattern of GhMPK11 after F. oxysporum infection. (F) Expression pattern of GhMPK16 after F. oxysporum infection. Data are presented as the mean ± standard error (SE) of three independent experiments (n = 6). The letters above the columns represent significant differences (P < 0.05) based on Tukey's honestly significant difference (HSD) test.
Fig. S2 Phylogenetic analysis of GhMPK20 and other plant mitogen‐activated protein kinase (MAPK) proteins. The neighbour‐joining phylogenetic tree was constructed using mega 4.0. The numbers above or below the branches indicate the bootstrap values (>50%) from 500 replicates. The GenBank IDs follow the protein names. The abbreviations of the species names are as follows: At, Arabidopsis thaliana; Gh, Gossypium hirsutum; Nt, Nicotiana tabacum; Zm, Zea mays; Le, Lycopersicon esculentum; Mt, Medicago truncatula.
Fig. S3 Sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) of transiently expressed proteins (35S::GFP and 35S::GhMPK20‐GFP) followed by western blotting with anti‐green fluorescent protein (anti‐GFP). 1, Nicotiana benthamiana leaves transiently expressing the 35S::GFP plasmid using Agrobacterium infiltration; 2, N. benthamiana leaves transiently expressing the 35S::GhMPK20‐GFP plasmid using Agrobacterium infiltration.
Fig. S4 GhMPK20 expression levels in nine independent transgenic lines obtained from T1 progeny. WT, wild‐type.
Fig. S5 GhMKK4 expression levels in GhMKK4‐silenced cotton plants. The error bars indicate the mean ± standard error (SE) of three independent experiments (n = 6). The letters above the columns represent significant differences (P < 0.05) based on Tukey's honestly significant difference (HSD) test.
Fig. S6 Phosphorylation assay using Phos‐tag polyacrylamide gel electrophoresis (PAGE). (A) Phosphorylation assay of GhMPK20 in cotton protoplasts transformed with GhMPK20‐MYC or co‐transformed with GhMKK4‐FLAG and GhMPK20‐MYC by western blotting. (B) Phosphorylation assay of GhWRKY40 in cotton protoplasts transformed with GhWRKY40‐FLAG or co‐transformed with GhMPK20‐MYC and GhWRKY40‐FLAG by western blotting. P and U indicate the phosphorylated and unphosphorylated forms, respectively.
Fig. S7 GhWRKY40 expression levels were analysed after cotton seedlings were infected with Fusarium oxysporum. Data are presented as the mean ± standard error (SE) of three independent experiments (n = 6). The letters above the columns represent significant differences (P < 0.05) based on Tukey's honestly significant difference (HSD) test.
Fig. S8 Expression levels of PDF1.2 and salicylic acid (SA)‐mediated defence pathway genes in GhWRKY40‐silenced cotton after Fusarium oxysporum infection. The error bars indicate the mean ± standard error (SE) of three independent experiments (n = 6). The letters above the columns represent significant differences (P < 0.05) based on Tukey's honestly significant difference (HSD) test.
Fig. S9 Expression levels of salicylic acid (SA)‐ and jasmonic acid (JA)‐mediated defence pathway genes in wild‐type cotton before infection or after Fusarium oxysporum infection for 5 days. The error bars indicate the mean ± standard error (SE) of three independent experiments (n = 6). The letters above the columns represent significant differences (P < 0.05) based on Tukey's honestly significant difference (HSD) test.
Fig. S10 Representative phenotypes of GhWRKY40‐silenced cotton after infection with Fusarium oxysporum for 5 days by the external application of salicylic acid (SA) (10 mm).
Table S1 Primers used in this study.
Acknowledgements
This work was financially supported by grants from the National Natural Science Foundation of China (Grant Nos. 31171837 and 31471424). The authors have no conflicts of interest to declare.
References
- Asai, T. , Tena, G. , Plotnikova, J. , Willmann, M.R. , Chiu, W.‐L. , Gomez‐Gomez, L. , Boller, T. , Ausubel, F.M. and Sheen, J. (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature, 415, 977–983. [DOI] [PubMed] [Google Scholar]
- Bekešová, S. , Komis, G. , Krenek, P. , Vyplelová, P. , Ovecka, M. , Luptovciak, I. , Illés, P. , Kucharová, A. and Šamaj, J. (2015) Monitoring protein phosphorylation by acrylamide pendant Phos‐Tag™ in various plants. Front Plant Sci. 6, 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl, R.P. , Diezel, C. and Somssich, I.E. (2012) Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol. 159, 266–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Lu, X. , Shu, N. , Wang, S. , Wang, J. , Wang, D. , Guo, L. and Ye, W. (2017) Targeted mutagenesis in cotton (Gossypium hirsutum L.) using the CRISPR/Cas9 system. Sci Rep. 7, 44 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong, Y.H. , Moon, B.C. , Kim, J.K. , Kim, C.Y. , Kim, M.C. , Kim, I.H. , Park, C.Y. , Kim, J.C. , Park, B.O. , Koo, S.C. , Yoon, H.W. , Chung, W.S. , Lim, C.O. , Lee, S.Y. and Cho, M.J. (2003) BWMK1, a rice mitogen‐activated protein kinase, locates in the nucleus and mediates pathogenesis‐related gene expression by activation of a transcription factor. Plant Physiol. 132, 1961–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney, T.P. , Friedrich, L. and Ryals, J.A. (1995) Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA, 92, 6602–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou, L. , Zhang, X. , Pang, C. , Song, M. , Wei, H. , Fan, S. and Yu, S. (2014) Genome‐wide analysis of the WRKY gene family in cotton. Mol. Genet. Genomics, 289, 1103–1121. [DOI] [PubMed] [Google Scholar]
- Ding, L. , Xu, H. , Yi, H. , Yang, L. , Kong, Z. , Zhang, L. , Xue, S. , Jia, H. and Ma, Z. (2011) Resistance to hemi‐biotrophic F. graminearum infection is associated with coordinated and ordered expression of diverse defense signaling pathways. PLoS One, 6, e19008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, F. and Zhou, J.M. (2012) Plant–bacterial pathogen interactions mediated by type III effectors. Curr. Opin. Plant Biol. 15, 469–476. [DOI] [PubMed] [Google Scholar]
- Frey, N.F. , Garcia, A.V. , Bigeard, J. , Zaag, R. , Bueso, E. , Garmier, M. , Pateyron, S. , de Tauzia‐Moreau, M.L. , Brunaud, V. , Balzergue, S. , Colcombet, J. , Aubourg, S. , Martin‐Magniette, M.L. and Hirt, H. (2014) Functional analysis of Arabidopsis immune‐related MAPKs uncovers a role for MPK3 as negative regulator of inducible defences. Genome Biol. 15, R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, M. , Liu, J. , Bi, D. , Zhang, Z. , Cheng, F. , Chen, S. and Zhang, Y. (2008) MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen‐activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 18, 1190–1198. [DOI] [PubMed] [Google Scholar]
- Gao, Y. , Wu, Y. , Du, J. , Zhan, Y. , Sun, D. , Zhao, J. , Zhang, S. , Li, J. and He, K. (2017) Both light‐induced SA accumulation and ETI mediators contribute to the cell death regulated by BAK1 and BKK1. Front. Plant Sci. 8, 622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar, Y.M. , McKenna, J.A. , McGinness, B.S. , Hinch, J. , Poon, S. , Connelly, A.A. , Anderson, M.A. and Heath, R.L. (2014) Field resistance to Fusarium oxysporum and Verticillium dahliae in transgenic cotton expressing the plant defensin NaD1. J. Exp. Bot. 65, 1541–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. , Rogers, E.E. and Ausubel, F.M. (1996) Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics, 143, 973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Z. , Huang, C. , Li, F. and Zhou, X. (2014) A versatile system for functional analysis of genes and microRNAs in cotton. Plant Biotechnol. J. 12, 638–649. [DOI] [PubMed] [Google Scholar]
- He, K. , Gou, X. , Yuan, T. , Lin, H. , Asami, T. , Yoshida, S. , Russell, S.D. and Li, J. (2007) BAK1 and BKK1 regulate brassinosteroid‐dependent growth and brassinosteroid‐independent cell‐death pathways. Curr. Biol. 17, 1109–1115. [DOI] [PubMed] [Google Scholar]
- Ishihama, N. and Yoshioka, H. (2012) Post‐translational regulation of WRKY transcription factors in plant immunity. Curr. Opin. Plant Biol. 15, 431–437. [DOI] [PubMed] [Google Scholar]
- Jones, D.A. and Takemoto, D. (2004) Plant innate immunity—direct and indirect recognition of general and specific pathogen‐associated molecules. Curr. Opin. Immunol. 16, 48–62. [DOI] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kishi‐Kaboshi, M. , Okada, K. , Kurimoto, L. , Murakami, S. , Umezawa, T. , Shibuya, N. , Yamane, H. , Miyao, A. , Takatsuji, H. , Takahashi, A. and Hirochika, H. (2010) A rice fungal MAMP‐responsive MAPK cascade regulates metabolic flow to antimicrobial metabolite synthesis. Plant J. 63, 599–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Zhang, L. , Wang, X. , Zhang, W. , Hao, L. , Chu, X. and Guo, X. (2013) Cotton GhMPK6a negatively regulates osmotic tolerance and bacterial infection in transgenic Nicotiana benthamiana, and plays a pivotal role in development. FEBS J. 280, 5128–5144. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Zhang, L. , Lu, W. , Wang, X. , Wu, C.A. and Guo, X. (2014) Overexpression of cotton GhMKK4 enhances disease susceptibility and affects abscisic acid, gibberellin and hydrogen peroxide signalling in transgenic Nicotiana benthamiana . Mol. Plant Pathol. 15, 94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longoni, P. , Douchi, D. , Cariti, F. , Fucile, G. and Goldschmidt‐Clermont, M. (2015) Phosphorylation of the light‐harvesting complex II isoform Lhcb2 is central to state transitions. Plant Physiol. 169, 2874–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAPK Group . (2002) Mitogen‐activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 7, 301–308. [DOI] [PubMed] [Google Scholar]
- Medzhitov, R. and Janeway, C.A Jr. (1997) Innate immunity: the virtues of a nonclonal system of recognition. Cell, 91, 295–298. [DOI] [PubMed] [Google Scholar]
- Meng, X. and Zhang, S. (2013) MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51, 245–266. [DOI] [PubMed] [Google Scholar]
- Morrissey, J.P. and Osbourn, A.E. (1999) Fungal resistance to plant antibiotics as a mechanism of pathogenesis. Microbiol. Mol. Biol. Rev. 63, 708–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ospina‐Giraldo, M.D. , Mullins, E. and Kang, S. (2003) Loss of function of the Fusarium oxysporum SNF1 gene reduces virulence on cabbage and Arabidopsis . Curr. Genet. 44, 49–57. [DOI] [PubMed] [Google Scholar]
- Pan, J. , Zhang, M. , Kong, X. , Xing, X. , Liu, Y. , Zhou, Y. , Liu, Y. , Sun, L. and Li, D. (2012) ZmMPK17, a novel maize group D MAP kinase gene, is involved in multiple stress responses. Planta, 235, 661–676. [DOI] [PubMed] [Google Scholar]
- Page, J.T. , Huynh, M.D. , Liechty, Z.S. , Grupp, K. , Stelly, D. , Hulse, A.M. , Ashrafi, H. , Van Deynze, A. , Wendel, J.F. and Udall, J.A. (2013) Insights into the evolution of cotton diploids and polyploids from whole‐genome re‐sequencing. G3 (Bethesda), 3, 1809–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedley, K.F. and Martin, G.B. (2004) Identification of MAPKs and their possible MAPK kinase activators involved in the Pto‐mediated defense response of tomato. J. Biol. Chem. 279, 49 229–49 235. [DOI] [PubMed] [Google Scholar]
- Peng, X. , Hu, Y. , Tang, X. , Zhou, P. , Deng, X. , Wang, H. and Guo, Z. (2012) Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta, 236, 1485–1498. [DOI] [PubMed] [Google Scholar]
- Pietro, A.D. , Madrid, M.P. , Caracuel, Z. , Delgado‐Jarana, J. and Roncero, M.I. (2003) Fusarium oxysporum: exploring the molecular arsenal of a vascular wilt fungus. Mol. Plant Pathol. 4, 315–325. [DOI] [PubMed] [Google Scholar]
- Rep, M. , Meijer, M. , Houterman, P.M. , van der Does, H.C. and Cornelissen, B.J. (2005) Fusarium oxysporum evades I‐3‐mediated resistance without altering the matching avirulence gene. Mol. Plant–Microbe Interact. 18, 15–23. [DOI] [PubMed] [Google Scholar]
- Rodriguez, M.C. , Petersen, M. and Mundy, J. (2010) Mitogen‐activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61, 621–649. [DOI] [PubMed] [Google Scholar]
- Rogers, E. and Ausubel, F.M. (1997) Arabidopsis enhanced disease susceptibility mutants exhibit enhanced susceptibility to several bacterial pathogens and alterations in PR‐1 gene expression. Plant Cell, 9, 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton, P.J. , Somssich, I.E. , Ringler, P. and Shen, Q.J. (2010) WRKY transcription factors. Trends Plant Sci. 15, 247–258. [DOI] [PubMed] [Google Scholar]
- Schechter, L.M. , Vencato, M. , Jordan, K.L. , Schneider, S.E. , Schneider, D.J. and Collmer, A. (2006) Multiple approaches to a complete inventory of Pseudomonas syringae pv. tomato DC3000 type III secretion system effector proteins. Mol. Plant–Microbe Interact. 19, 1180–1192. [DOI] [PubMed] [Google Scholar]
- Shi, J. , An, H.L. , Zhang, L. , Gao, Z. and Guo, X. (2010) GhMPK7, a novel multiple stress‐responsive cotton group C MAPK gene, has a role in broad spectrum disease resistance and plant development. Plant Mol. Biol. 74, 1–17. [DOI] [PubMed] [Google Scholar]
- Shi, J. , Zhang, L. , An, H. , Wu, C. and Guo, X. (2011) GhMPK16, a novel stress‐responsive group D MAPK gene from cotton, is involved in disease resistance and drought sensitivity. BMC Mol. Biol. 12, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunilkumar, G. , Campbell, L.M. , Puckhaber, L. , Stipanovic, R.D. and Rathore, K.S. (2006) Engineering cottonseed for use in human nutrition by tissue‐specific reduction of toxic gossypol. Proc. Natl. Acad. Sci. USA, 103, 18 054–18 059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler, J.S. , Humphrey, P.T. and Whiteman, N.K. (2012) Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 17, 260–270. [DOI] [PubMed] [Google Scholar]
- Vlot, A.C. , Dempsey, D.A. and Klessig, D.F. (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. [DOI] [PubMed] [Google Scholar]
- Walia, A. , Lee, J.S. , Wasteneys, G. and Ellis, B. (2009) Arabidopsis mitogen‐activated protein kinase MPK18 mediates cortical microtubule functions in plant cells. Plant J. 59, 565–575. [DOI] [PubMed] [Google Scholar]
- Wang, C. , Ulloa, M. and Roberts, P.A. (2006) Identification and mapping of microsatellite markers linked to a root‐knot nematode resistance gene (RKN1) in Acala NemX cotton (Gossypium hirsutum L.). Theor. Appl. Genet. 112, 770–777. [DOI] [PubMed] [Google Scholar]
- Wang, C. , Lu, W. , He, X. , Wang, F. , Zhou, Y. , Guo, X. and Guo, X. (2016) The cotton mitogen‐activated protein kinase 3 functions in drought tolerance by regulating stomatal responses and root growth. Plant Cell Physiol. 57, 1629–1642. [DOI] [PubMed] [Google Scholar]
- Wang, F. , Wang, C. , Yan, Y. , Jia, H. and Guo, X. (2016) Overexpression of cotton GhMPK11 decreases disease resistance through the gibberellin signaling pathway in transgenic Nicotiana benthamiana . Front Plant Sci. 7, 689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Xiao, H. , Chen, G. , Zhao, X. , Huang, C. , Chen, C. and Wang, F. (2011) Isolation of high‐quality RNA from Reaumuria soongorica, a desert plant rich in secondary metabolites. Mol. Biotechnol. 48, 165–172. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Yan, Y. , Li, Y. , Chu, X. , Wu, C. and Guo, X. (2014) GhWRKY40, a multiple stress‐responsive cotton WRKY gene, plays an important role in the wounding response and enhances susceptibility to Ralstonia solanacearum infection in transgenic Nicotiana benthamiana . PLoS One, 9, e93577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X.J. , Zhu, S.Y. , Lu, Y.F. , Zhao, R. , Xin, Q. , Wang, X.F. and Zhang, D.P. (2010) Two coupled components of the mitogen‐activated protein kinase cascade MdMPK1 and MdMKK1 from apple function in ABA signal transduction. Plant Cell Physiol. 51, 754–766. [DOI] [PubMed] [Google Scholar]
- Yan, Y. , Jia, H. , Wang, F. , Wang, C. , Liu, S. and Guo, X. (2015) Overexpression of GhWRKY27a reduces tolerance to drought stress and resistance to Rhizoctonia solani infection in transgenic Nicotiana benthamiana . Front Physiol. 6, 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, B. , Wang, Q. , Jing, M. , Guo, B. , Wu, J. , Wang, H. , Wang, Y. , Lin, L. , Wang, Y. , Ye, W. , Dong, S. and Wang, Y. (2017) Distinct regions of the Phytophthora essential effector Avh238 determine its function in cell death activation and plant immunity suppression. New Phytol. 214, 361–375. [DOI] [PubMed] [Google Scholar]
- Yang, K.Y. , Liu, Y. and Zhang, S. (2001) Activation of a mitogen‐activated protein kinase pathway is involved in disease resistance in tobacco. Proc. Natl. Acad. Sci. USA, 98, 741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, S.D. , Cho, Y.H. and Sheen, J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yu, F. , Huaxia, Y. , Lu, W. , Wu, C. , Cao, X. and Guo, X. (2012) GhWRKY15, a member of the WRKY transcription factor family identified from cotton (Gossypium hirsutum L.), is involved in disease resistance and plant development. BMC Plant Biol. 12, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X. , Tang, J. , Wang, Q. , Ye, W. , Tao, K. , Duan, S. , Lu, C. , Yang, X. , Dong, S. , Zheng, X. and Wang, Y. (2012) The RxLR effector Avh241 from Phytophthora sojae requires plasma membrane localization to induce plant cell death. New Phytol. 196, 247–260. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Shao, F. , Li, Y. , Cui, H. , Chen, L. , Li, H. , Zou, Y. , Long, C. , Lan, L. , Chai, J. , Chen, S. , Tang, X. and Zhou, J.M. (2007) A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP‐induced immunity in plants. Cell Host Microbe, 1, 175–185. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Xi, D. , Luo, L. , Meng, F. , Li, Y. , Wu, C.A. and Guo, X. (2011) Cotton GhMPK2 is involved in multiple signaling pathways and mediates defense responses to pathogen infection and oxidative stress. FEBS J. 278, 1367–1378. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Li, Y. , Lu, W. , Meng, F. , Wu, C.A. and Guo, X. (2012) Cotton GhMKK5 affects disease resistance, induces HR‐like cell death, and reduces the tolerance to salt and drought stress in transgenic Nicotiana benthamiana . J. Exp. Bot. 63, 3935–3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, T. , Chen, S. and Harmon, A.C. (2016) Protein–protein interactions in plant mitogen‐activated protein kinase cascades. J. Exp. Bot. 67, 607–618. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Xu, X. , Yu, Y. , Chen, C. , Wang, J. , Cai, C. and Guo, W. (2016) Integration analysis of MKK and MAPK family members highlights potential MAPK signaling modules in cotton. Sci Rep. 6, 29 781. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Mitogen‐activated protein kinase (MAPK) expression patterns were analysed after cotton seedlings were infected with Fusarium oxysporum. (A) Expression pattern of GhMPK20 in cotyledon after F. oxysporum infection. (B) Expression pattern of GhMPK20 after root wounding. (C) Expression pattern of GhMPK6 after F. oxysporum infection. (D) Expression pattern of GhMPK7 after F. oxysporum infection. (E) Expression pattern of GhMPK11 after F. oxysporum infection. (F) Expression pattern of GhMPK16 after F. oxysporum infection. Data are presented as the mean ± standard error (SE) of three independent experiments (n = 6). The letters above the columns represent significant differences (P < 0.05) based on Tukey's honestly significant difference (HSD) test.
Fig. S2 Phylogenetic analysis of GhMPK20 and other plant mitogen‐activated protein kinase (MAPK) proteins. The neighbour‐joining phylogenetic tree was constructed using mega 4.0. The numbers above or below the branches indicate the bootstrap values (>50%) from 500 replicates. The GenBank IDs follow the protein names. The abbreviations of the species names are as follows: At, Arabidopsis thaliana; Gh, Gossypium hirsutum; Nt, Nicotiana tabacum; Zm, Zea mays; Le, Lycopersicon esculentum; Mt, Medicago truncatula.
Fig. S3 Sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) of transiently expressed proteins (35S::GFP and 35S::GhMPK20‐GFP) followed by western blotting with anti‐green fluorescent protein (anti‐GFP). 1, Nicotiana benthamiana leaves transiently expressing the 35S::GFP plasmid using Agrobacterium infiltration; 2, N. benthamiana leaves transiently expressing the 35S::GhMPK20‐GFP plasmid using Agrobacterium infiltration.
Fig. S4 GhMPK20 expression levels in nine independent transgenic lines obtained from T1 progeny. WT, wild‐type.
Fig. S5 GhMKK4 expression levels in GhMKK4‐silenced cotton plants. The error bars indicate the mean ± standard error (SE) of three independent experiments (n = 6). The letters above the columns represent significant differences (P < 0.05) based on Tukey's honestly significant difference (HSD) test.
Fig. S6 Phosphorylation assay using Phos‐tag polyacrylamide gel electrophoresis (PAGE). (A) Phosphorylation assay of GhMPK20 in cotton protoplasts transformed with GhMPK20‐MYC or co‐transformed with GhMKK4‐FLAG and GhMPK20‐MYC by western blotting. (B) Phosphorylation assay of GhWRKY40 in cotton protoplasts transformed with GhWRKY40‐FLAG or co‐transformed with GhMPK20‐MYC and GhWRKY40‐FLAG by western blotting. P and U indicate the phosphorylated and unphosphorylated forms, respectively.
Fig. S7 GhWRKY40 expression levels were analysed after cotton seedlings were infected with Fusarium oxysporum. Data are presented as the mean ± standard error (SE) of three independent experiments (n = 6). The letters above the columns represent significant differences (P < 0.05) based on Tukey's honestly significant difference (HSD) test.
Fig. S8 Expression levels of PDF1.2 and salicylic acid (SA)‐mediated defence pathway genes in GhWRKY40‐silenced cotton after Fusarium oxysporum infection. The error bars indicate the mean ± standard error (SE) of three independent experiments (n = 6). The letters above the columns represent significant differences (P < 0.05) based on Tukey's honestly significant difference (HSD) test.
Fig. S9 Expression levels of salicylic acid (SA)‐ and jasmonic acid (JA)‐mediated defence pathway genes in wild‐type cotton before infection or after Fusarium oxysporum infection for 5 days. The error bars indicate the mean ± standard error (SE) of three independent experiments (n = 6). The letters above the columns represent significant differences (P < 0.05) based on Tukey's honestly significant difference (HSD) test.
Fig. S10 Representative phenotypes of GhWRKY40‐silenced cotton after infection with Fusarium oxysporum for 5 days by the external application of salicylic acid (SA) (10 mm).
Table S1 Primers used in this study.


