Summary
Phytophthora cinnamomi is one of the most devastating plant pathogens in the world. It infects close to 5000 species of plants, including many of importance in agriculture, forestry and horticulture. The inadvertent introduction of P. cinnamomi into natural ecosystems, including a number of recognized Global Biodiversity Hotspots, has had disastrous consequences for the environment and the biodiversity of flora and fauna.
The genus Phytophthora belongs to the Class Oomycetes, a group of fungus‐like organisms that initiate plant disease through the production of motile zoospores. Disease control is difficult in agricultural and forestry situations and even more challenging in natural ecosystems as a result of the scale of the problem and the limited range of effective chemical inhibitors. The development of sustainable control measures for the future management of P. cinnamomi requires a comprehensive understanding of the cellular and molecular basis of pathogen development and pathogenicity. The application of next‐generation sequencing technologies to generate genomic and transcriptomic data promises to underpin a new era in P. cinnamomi research and discovery. The aim of this review is to integrate bioinformatic analyses of P. cinnamomi sequence data with current knowledge of the cellular and molecular basis of P. cinnamomi growth, development and plant infection. The goal is to provide a framework for future research by highlighting potential pathogenicity genes, shedding light on their possible functions and identifying suitable targets for future control measures.
Taxonomy
Phytophthora cinnamomi Rands; Kingdom Chromista; Phylum Oomycota or Pseudofungi; Class Oomycetes; Order Peronosporales; Family Peronosporaceae; genus Phytophthora.
Host range
Infects about 5000 species of plants, including 4000 Australian native species. Host plants important for agriculture and forestry include avocado, chestnut, macadamia, oak, peach and pineapple.
Disease symptoms
A root pathogen which causes rotting of fine and fibrous roots, but which can also cause stem cankers. Root damage may inhibit water movement from roots to shoots, leading to dieback of young shoots.
Useful websites
http://fungidb.org/fungidb/; http://genome.jgi.doe.gov/Phyci1/Phyci1.home.html; http://www.ncbi.nlm.nih.gov/assembly/GCA_001314365.1; http://www.ncbi.nlm.nih.gov/assembly/GCA_001314505.1
Keywords: dieback disease, Oomycetes, Phytophthora cinnamomi, root pathogen, soil‐borne pathogen
Introduction
Phytophthora cinnamomi is one of the most devastating plant pathogens known. It has a world‐wide distribution and a host range approaching 5000 species (Cahill et al., 2008; Jung et al., 2013). In addition to causing substantial economic losses in agriculture, forestry and horticulture, the inadvertent introduction of P. cinnamomi has had disastrous consequences for natural ecosystems and biodiversity. Prime examples include the impact of P. cinnamomi on chestnut and holm oak forests in Europe (Serrazina et al., 2015; Sghaier‐Hammami et al., 2013), on avocado and macadamia worldwide (Akinsanmi et al., 2017; Reeksting et al., 2016) and on natural vegetation in south‐east and south‐west Australia (Cahill et al., 2008; Jung et al., 2013) (Fig. 1). About 4000 Australian native species, sometimes close to 50% of species present, are susceptible in forests and heathlands in Western Australia (Shearer et al., 2004; Weste, 2003). The recognition of the magnitude of the environmental problems caused by P. cinnamomi has led to its inclusion in the list of Key Threatening Processes in the Commonwealth Environmental Protection & Biodiversity Conservation Act 1999 and to the development of a National Threat Abatement Plan aimed at the management and control of P. cinnamomi diseases (Australian Government, 2014).
Figure 1.

Phytophthora cinnamomi kills thousands of plant species in natural ecosystems in Western Australia, threatening the environment and biodiversity. (A) An uninfected area within a Eucalypt forest south of Perth dominated by Eucalyptus marginata (Jarrah), Banksia grandis and Xanthorea species (grasstrees). (B) Xanthorea species and many proteaceous plants in Western Australia are highly susceptible to P. cinnamomi. When present, P. cinnamomi dramatically changes the floral composition of the region, with more resistant species, such as acacias, rushes and sedges, replacing the plants that have been killed.
Phytophthora is a genus in the Oomycetes, a Class in Phylum Pseudofungi within the Kingdom Chromista (Beakes et al., 2012; Cavalier‐Smith and Chao, 2006). The defining character of the Chromista is the production of motile asexual spores possessing a flagellum adorned by tubular hairs which are responsible for forward movement (Beakes et al., 2012; Cahill et al., 1996). For many species of Phytophthora, the motile zoospores are the main infective agent that initiates plant disease. Phytophthora cinnamomi has been ranked in the Top 10 Oomycete plant pathogens based on scientific and economic importance (Kamoun et al., 2015). It has been estimated that, in California, losses in avocado crops caused by P. cinnamomi exceed US$40 million annually (Ploetz, 2013).
A number of key questions relating to the management of P. cinnamomi diseases urgently need answers. What factors are responsible for P. cinnamomi's extremely extensive host range? Many of these factors must be species specific because other Phytophthora species, such as P. infestans and P. sojae, have narrow host ranges (Cooke and Andersson, 2013; Dorrance, 2013). Is P. cinnamomi's success in establishing disease a result of its ability to avoid triggering host defence and/or to suppress or overcome host defences? What aspects of P. cinnamomi molecular or cellular make‐up would be good targets for novel, specific and sustainable control measures? The answers to these questions will be informed by a better understanding of the cellular and molecular biology of P. cinnamomi development and infection strategies.
The advent and application of next‐generation sequencing technologies herald a new era in P. cinnamomi research and discovery. The genomes (78 Mb) of three isolates of P. cinnamomi have now been sequenced and are publicly available (http://fungidb.org/fungidb/; http://genome.jgi.doe.gov/Phyci1/Phyci1.home.html; http://www.ncbi.nlm.nih.gov/assembly/GCA_001314365.1; http://www.ncbi.nlm.nih.gov/assembly/GCA_001314505.1). The first sequencing project was conducted at the Joint Genome Institute (JGI). The sequence data are in draft form, with only preliminary annotation, and are accessible on JGI and FungiDB websites. Sequence data for two further isolates were released in 2016 [available on the National Center for Biotechnology Information (NCBI) website]. These unannotated data have been assembled into about 4000 contigs. At the time of writing, there were two published analyses of P. cinnamomi transcriptomes available (Meyer et al., 2016; Reitmann et al., 2016). The first reports data from Eucalyptus nitens, 5 days after inoculation with P. cinnamomi; the second reports data from a library of cysts and germinated cysts.
This present Pathogen Profile focuses on information that has become available since the last Pathogen Profile on P. cinnamomi was published in 2005 (Hardham, 2005). The goal of the article is to integrate bioinformatic analyses of P. cinnamomi sequence data with our current understanding of the cellular and molecular basis of growth, development and plant infection strategies employed by P. cinnamomi and other Phytophthora species. It is hoped that this will provide a framework for future research by highlighting pathogenicity genes, shedding light on their possible functions and identifying potential targets for future control measures.
The P. cinnamomi Lifecycle
Phytophthora cinnamomi is a soil‐borne pathogen with sexual and asexual phases in its lifecycle (Zentmyer, 1980). It can grow saprophytically on dead organic matter or parasitically on susceptible hosts. Typically, P. cinnamomi infects fine, feeder roots, but it can also invade woody stems, especially through wounds or natural breaks in the peridermal layer (O'Gara et al., 2015). Growth within the root system causes root rotting and interferes with water uptake and transport to the shoot, resulting in wilting and chlorosis of the foliage. Plants may die rapidly, or may survive, often without showing disease symptoms, for many years. The ability of P. cinnamomi to grow saprophytically in the soil or symptomlessly in infected plants is a major contributing factor to the long‐term survival of the pathogen. Sexual oospores, asexual chlamydospores and intracellular hyphal aggregates are thought to enable the pathogen to survive for long periods under adverse conditions and make complete eradication of the disease extremely difficult (Jung et al., 2013).
Gene silencing experiments in P. sojae have shown that the signalling pathways involved in oospore development include PsGK5, a G‐protein‐coupled receptor (GPCR) that has a phosphatidylinositol phosphate (PIP) kinase domain at its C‐terminus (Yang et al., 2013), and PsMPK7, a stress‐associated mitogen‐activated protein kinase (MAPK) (Gao et al., 2015). The P. cinnamomi genome contains a homologue for both of these genes (Table 1).
Table 1.
Phytophthora cinnamomi proteins involved in sporulation.
| P. cinnamomi gene name* | Homologues used for blast query | Proposed protein function | References |
|---|---|---|---|
| Oospore development | |||
| PHYCI_277748 |
P. sojae PsGK5
PHYSO_335695 |
G‐protein‐coupled receptor with C‐terminal phosphatidylinositol phosphate (PIP) kinase domain Silencing > inhibits oospore development |
Yang et al. (2013) |
| PHYCI_112968 |
P. sojae PsMPK7
PHYSO_355777 |
Stress‐associated mitogen‐activated protein kinase (MAPK). Gene expression is up‐regulated in zoospores, cysts and germinating cysts Silencing > inhibits oospore development; reduces virulence |
Gao et al. (2015) |
| Asexual sporulation and zoosporogenesis | |||
|
PHYCI_89449 PHYCI_233998 PHYCI_111102 † PHYCI_199460 † |
P. cinnamomi α‐tubulin | α‐tubulin subunit of microtubule. Microtubules are required for shape and spacing of nuclei within sporangia and form the framework of the flagella axoneme | Hardham (1987); Hyde and Hardham (1993) |
| PHYCI_88710 | P. cinnamomi β‐tubulin | β‐tubulin subunit of microtubule. Microtubules are required for shape and spacing of nuclei within sporangia and form the framework of the flagella axoneme | Weerakoon et al. (1998) |
| PHYCI_93292 |
P. infestans Pigpb1
PITG_06376 |
G‐protein β‐subunit Silencing > inhibits sporangium formation |
Latijnhouwers and Govers (2003) |
| PHYCI_90010 |
P. sojae PsMPK1
ACJ09359 |
MAPK. Gene expression is up‐regulated in sporulating hyphae and early infection Silencing > inhibits sporangium formation; reduces virulence |
Li et al. (2014) |
| PHYCI_80900 |
P. cinnamomi PcLpv
AF315065 |
Small family of high‐molecular‐weight glycoproteins contained in zoospore large peripheral vesicles. Gene expression peaks 6–8 h after induction of sporulation | Marshall et al. (2001b) |
| PHYCI_92681 |
P. infestans PiCdc14
PITG_18578 |
Phosphatase involved in cell cycle regulation and localized near the basal bodies at the nuclear apex Silencing > inhibits sporangium formation Overexpression > inhibits sporangial cleavage |
Ah‐Fong and Judelson (2011) |
|
PHYCI_97894 PHYCI_290439 |
P. infestans actin PITG_15078 |
Actin microfilaments are reorganized during sporangial cleavage | Jackson and Hardham (1998) |
| PHYCI_91218 |
P. infestans PiGK4
PITG_05519 |
G‐protein‐coupled receptor with a C‐terminal PIP kinase domain Silencing > inhibits sporangial cleavage |
Hua et al. (2013) |
| PHYCI_243332 |
P. sojae PsMYB1
PHYSO_351786 |
Myb transcription factor. Transcription increases in sporulating hyphae, germinating cysts and early infection Silencing > inhibits sporangial cleavage and zoospore release; causes direct germination of sporangia; reduces virulence |
Zhang et al. (2012) |
| PHYCI_95800 |
P. sojae PsGPR11
PHYSO_352568 |
G‐protein‐coupled receptor which does not interact with PsGPA1 Silencing > cleavage is normal, but zoospore release from sporangia is inhibited; reduces virulence |
Wang et al. (2010) |
*Accession number in FungiDB.
†Genome assembly shows these as truncated α‐tubulin sequences, but this is likely to be an assembly error.
Asexual Sporulation and Zoosporogenesis
Sporangiogenesis: sporangium development
Reduction in nutrient availability triggers the development of asexual, multinucleate sporangia (Fig. 2) (Hardham, 2005). The shape and distribution of 20–30 nuclei within the sporangia are maintained by arrays of microtubules (Hyde and Hardham, 1992, 1993). Microtubules are composed of tubulin dimers. The P. cinnamomi genome is likely to contain four α‐tubulin genes, although the current assembly shows only truncated versions for two of them (Table 1). There is a single β‐tubulin gene in the P. cinnamomi genome, as reported previously (Weerakoon et al., 1998).
Figure 2.
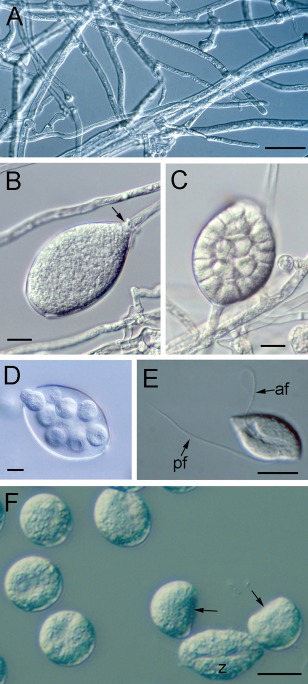
The asexual lifecycle of Phytophthora cinnamomi. (A) Vegetative hyphae. (B) Mature, uncleaved sporangium which is sealed off from the subtending hypha by a septum (arrow). (C) Cleaved sporangium in which uninucleate domains have been formed through the development of a system of cleavage membranes that will become the plasma membranes of the future zoospores. (D) Zoospore release from a sporangium through an apical pore. (E) A biflagellate zoospore. The two flagella emerge from the groove along the ventral surface of the zoospore. The anterior flagellum (af) projects forward and pulls the cell along. The posterior flagellum (pf) trails behind the zoospore and functions in changing the direction of swimming. (F) A zoospore (z) and six young cysts, two of which still retain the initially flat region (arrows) corresponding to the zoospore ventral surface. Bars, 10 µm. (B) and (F) Reproduced with permission from Hee et al. (2013).
The analysis of P. cinnamomi transcriptomes during sporangiogenesis has revealed that changes in gene expression begin rapidly after the induction of sporulation. Screening of 5280 cDNA clones from a library constructed from mycelial mRNA isolated 4 h after the induction of sporulation identified 226 genes whose expression was up‐regulated compared with levels in vegetative hyphae (Narayan, 2004). Many of the proteins encoded by these genes are involved in protein synthesis, signal transduction, general metabolism and cell structure. The quantification of RNA blots reveals a range of distinct expression patterns during sporangiogenesis. Transcript levels for genes encoding translation elongation factor 1α (PcEF1a), calmodulin (PcCaM), S‐adenosylhomocysteinase (PcSAc), annexin VII (PcAnx) and the SET1 transcription factor, PcSet1, rise transiently 2–6 h after the induction of sporulation (Fig. S1, see Supporting Information) (Marshall et al., 2001a, 2001b; Narayan, 2004). The expression of genes encoding glyceraldehyde‐3‐phosphate dehydrogenase (PcGDH), pyruvate phosphate dikinase (PcPdk) and lactate dehydrogenase (PcLDH) decreases initially, before transcript levels peak 4 h after induction. The expression of the gene encoding ubiquitin, PcUbq, increases within the first 30 min and elevated transcript levels are maintained for the ensuing 6 h. By contrast, the expression of PcCen, a P. cinnamomi centrin gene, changes little over the 24‐h time course. Transient elevation of gene expression has also been documented during P. infestans sporangiogenesis from microarrays (Judelson et al., 2009). In P. infestans and P. sojae, sporangial development is inhibited by silencing the expression of genes encoding the G‐protein β‐subunit Pigpb1, the MAPK PsMPK1 and the cell cycle phosphatase PiCdc14 (Ah‐Fong and Judelson, 2011; Latijnhouwers and Govers, 2003; Li et al., 2014). Phytophthora cinnamomi has homologues to each of these three proteins (Table 1). Future studies using RNA‐sequencing (RNA‐Seq) to analyse sporulation transcriptomes will provide a comprehensive understanding of the global changes in gene expression associated with sporangiogenesis.
Zoosporogenesis: sporangial cleavage and zoospore release
Subdivision of the multinucleate sporangia into uninucleate zoospores (Figs 2, 3) is induced by a decrease in temperature, and a ‘cold box’ motif involved in the ensuing changes in gene expression has been identified in the promoters of zoosporogenesis‐specific genes in P. infestans (Tani and Judelson, 2006). During sporangial cytokinesis (zoosporogenesis), organelles are apportioned and arranged within the cytoplasm of the future zoospores (Hardham, 2005). Three types of storage vesicle, for example, become distributed in specific zones of the zoospore cortical cytoplasm, mitochondria adopt a subcortical localization, actin microfilaments associate with the zoospore plasma membrane and the flagella are assembled. Actin microfilaments play a range of essential roles in eukaryotic cells and actin genes are expressed throughout the Phytophthora lifecycle (Yan and Liou, 2006). The P. cinnamomi genome includes two actin genes (Table 1).
Figure 3.
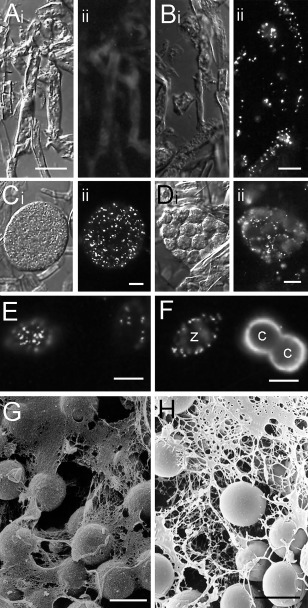
Synthesis of secretory proteins during Phytophthora cinnamomi sporulation and their secretion during encystment. These processes are exemplified in (A)–(F) by immunolabelling with monoclonal antibody (mAb) PcCpa2 and in (G) and (H) by mucin‐like material visualized by scanning electron microscopy (SEM). Micrographs in (Ai)–(Di) are bright field images of the same field of view as shown in the fluorescence images in (Aii)–(Dii). (A) Cryosection of vegetative hyphae. No components in vegetative hyphae react with PcCpa2 mAb after immunofluorescence labelling. (B) Cryosection of sporulating hyphae immunolabelled with PcCpa2 mAb. PcCpa2 reacts with three high‐molecular‐weight polypeptides that are synthesized after the induction of sporulation and packaged into zoospore dorsal vesicles (Gubler and Hardham, 1988). (C) Mature sporangium immunolabelled with PcCpa2 mAb. Dorsal vesicles containing PcCpa2 proteins are randomly distributed throughout the sporangial cytoplasm. (D) During sporangial cleavage, the dorsal vesicles labelled by PcCpa2 mAb become distributed near cleavage membranes that will form the dorsal surface of the future zoospores. (E) PcCpa2‐containing vesicles next to the zoospore dorsal surface. (F) PcCpa2‐containing vesicles in the zoospore (z) cortical cytoplasm and on the surface of two young cysts (c). The absence of immunolabelling in the region of contact between the two cysts may be because this was the ventral surface of both cells or because the antibody did not have access to this region. (G) Mucin‐like material secreted during zoospore encystment on a root surface visualized by SEM after critical point drying. (H) Mucin‐like material secreted during zoospore encystment on a root surface visualized by cryo‐SEM. Images in (A)–(F) are courtesy of Dr Michele Cope. The image in (H) is reproduced with permission from Hardham et al. (1994). Bars, 10 µm.
Increases in the concentration of cytoplasmic Ca2+ and pH are required for the induction of sporangial cleavage in P. cinnamomi (Jackson and Hardham, 1996; Suzaki et al., 1996), and the involvement of Ca2+ in signal transduction during zoosporogenesis has been substantiated by experimental studies using inhibitors of phospholipase C and Ca2+ channels (Tani et al., 2004). In P. infestans and P. sojae, silencing of the expression of genes encoding a GPCR, PiGK4 and the Myb transcription factor, PsMYB1, also inhibits sporangial cleavage (Hua et al., 2013; Zhang et al., 2012). Silencing of PsMYB1 in P. sojae causes a switch from indirect to direct sporangial germination. Phytophthora cinnamomi possesses homologues of both of these genes (Table 1).
After sporangial cytokinesis, zoospores are released into an evanescent vesicle originating from material in the apical papilla of the sporangium. Zoospore expulsion is thought to be driven by hydrostatic pressure within the sporangium because the process still occurs in a P. parasitica transformant that makes flagella‐less zoospores (Narayan et al., 2010). However, silencing of the gene encoding the PsGPR11 GPCR in P. sojae inhibits zoospore release, although sporangial cleavage apparently occurs normally (Wang et al., 2010).
Cellular and Molecular Basis of P. cinnamomi Infection Strategies
Active movement to a potential host plant
Dissemination of P. cinnamomi via hyphal growth may be especially important for the uphill spread of the pathogen (O'Gara et al., 2005) but, in most cases, infection is initiated through the active movement of biflagellate zoospores. Phytophthora flagella have the 9 + 2 microtubular substructure typical of eukaryotic flagella (Hardham, 1987) and the P. cinnamomi genome contains genes encoding eukaryotic flagellar proteins. Two of these, namely dynein light chain 1 (PcDLC1) and radial spoke protein 6 (PcRSP6) (Table 2), have been cloned (Narayan, 2004; Narayan et al., 2010). Their expression increases soon after the induction of asexual sporulation, consistent with flagella assembly during zoosporogenesis (Cope and Hardham, 1994). Silencing of the expression of a PcDLC1 homologue in P. parasitica (PnDLC1) leads to the formation of zoospores that lack flagella, suggesting that interference in the production of this outer dynein arm component aborts flagellar assembly (Narayan et al., 2010).
Table 2.
Phytophthora cinnamomi proteins involved in the initiation of plant infection.
| P. cinnamomi gene name* | Homologues used for blast query | Protein description and proposed function | References |
|---|---|---|---|
| Zoospore motility, chemotaxis and encystment | |||
| PHYCI_551329 |
P. cinnamomi PcDLC1
ADI77080.1 |
Dynein light chain 1 is a leucine‐rich repeat protein that is a component of the flagellar outer dynein arm Silencing > inhibits flagella assembly |
Narayan et al. (2010) |
| PHYCI_78591 | P. cinnamomi PcRSP6 | Flagellar radial spoke protein 6. Occurs along the length of the flagella | Narayan (2004) |
|
PHYCI_79928 PHYCI_71401 |
P. parasitica PnCen1
PPTG_05358 PnCen2 PPTG_12273 |
Centrin is a 20‐kDa protein that occurs within the flagella and a connecting band between the two basal bodies and in an anterior flagellar root | Harper et al. (1995) |
| PHYCI_207361 |
P. infestans Pigpa1
PITG_03162 P. sojae PsGPA1 PHYSO_323786 |
G‐protein α‐subunit Silencing > disrupts zoospore motility, inhibits chemotaxis, negative geotaxis and auto‐aggregation and induces encystment; reduces virulence |
Hua et al. (2008); Latijnhouwers et al. (2004) |
| PHYCI_232701 |
P. sojae PsHint1
PHYSO_494520 |
Histidine triad domain‐containing protein that interacts with PsGPA1 Silencing > inhibits chemotaxis and induces encystment; reduces virulence |
Zhang et al. (2016) |
| PHYCI_91218 † |
P. infestans PiGK4
PITG_05519 |
G‐protein‐coupled receptor with a C‐terminal phosphatidylinositol phosphate (PIP) kinase domain Silencing of P. sojae PsGK4 (PHYSO_286453) > inhibits chemotaxis and induces encystment |
Yang et al. (2013) |
| PHYCI_95800 |
P. sojae PsGPR11
PHYSO_352568 |
G‐protein‐coupled receptor which does not interact with PsGPA1 Silencing > no effect on chemotaxis, but induces encystment; reduces virulence |
Wang et al. (2010) |
| PHYCI_92931 |
P. sojae PsSAK1
PHYSO_545105 |
Mitogen‐activated protein kinase (MAPK) Silencing > induces encystment; reduces virulence |
Li et al. (2010) |
| Cyst adhesion, protection and germination; germling growth; appressorium formation | |||
|
PHYCI_1255 PHYCI_208606 |
P. cinnamomi PcVsv1
AAX84973.1 |
220‐kDa protein stored in small ventral vesicles in zoospores and secreted during encystment. PcVsv1 contains 47 copies of a thrombospondin Type I repeat motif, a domain occurring in adhesive proteins. PcVsv1 expression is induced during asexual sporulation | Robold and Hardham (2005) |
|
PHYCI_310810 PHYCI_80900 PHYCI_7831 PHYCI_300556 PHYCI_85845 |
P. cinnamomi 1PcLpv, partial AAK27342.1 AAK27345.1 |
DNA, RNA and immunoblots show the presence of three PcLpv genes that encode three 11–14‐kb transcripts and three 500–600‐kDa proteins. Five annotated genes in the P. cinnamomi genome with homology to cloned PcLpv genes encode proteins <54 kDa; the two largest contain long regions of undetermined sequence. PcLpv expression is up‐regulated during sporulation. PcLpv is stored in large peripheral vesicles, is not secreted during encystment and is degraded during germling growth | Gubler and Hardham (1988, 1990); Marshall et al. (2001a) |
| 3PHYCI_95662 |
P. parasitica PnCcp
PPTG_01661 |
12‐kDa protein stored within large peripheral vesicles in zoospores and secreted during encystment. Gene expression is up‐regulated in sporulating hyphae and zoospores | Škalamera and Hardham (2006); Zhang et al. (2013) |
|
PHYCI_91218 PHYCI_481282 PHYCI_376712 PHYCI_277748 PHYCI_269807 PHYCI_212799 PHYCI_105008 PHYCI_285878 PHYCI_291554 PHYCI_260267 PHYCI_97820 PHYCI_3455 PHYCI_105811 |
P. infestans PiGK4
PITG_05519 P. sojae PsGK4 PHYSO_286453 |
G‐protein‐coupled receptor with C‐terminal PIP kinase domain Silencing of PsGK4 and PiGK4 > inhibits cyst germination |
Hua et al. (2013); Yang et al. (2013) |
|
PHYCI_95800 PHYCI_89253 PHYCI_91364 |
P. sojae PsGPR11
PHYSO_352568 |
G‐protein‐coupled receptor which does not interact with PsGPA1 Silencing > inhibits cyst germination; reduces virulence |
Wang et al. (2010) |
| PHYCI_207361 |
P. sojae PsGPA1
PHYSO_323786 |
G‐protein α‐subunit. Single copy gene Silencing > inhibits cyst germination; reduces virulence |
Hua et al. (2008) |
| PHYCI_232701 |
P. sojae PsHint1
PHYSO_494520 |
Histidine triad domain‐containing protein that interacts with PsGPA1 Silencing > inhibits cyst germination; causes branched germ tubes; inhibits hyphal extension |
Zhang et al. (2016) |
| PHYCI_92931 |
P. sojae PsSAK1
ACJ09358 |
MAPK Silencing > inhibits cyst germination and appressorium formation; reduces virulence |
Li et al. (2010) |
| PHYCI_243332 |
P. sojae PsMYB1
PHYSO_351786 |
Myb transcription factor whose transcript levels are decreased in PsSAK1‐silenced mutants. Transcription of PsMYB1 increased in germinating cysts and early infection Silencing > inhibits cyst germination; reduces virulence |
Zhang et al. (2012) |
|
PHYCI_257808 PHYCI_461512 |
P. infestans PiNIFC1–3
PITG_11238, PITG_11237, PITG_11239 |
NIF proteins interact with nuclear LIM transcription factors to regulate gene expression. Three of four PiNIF genes are expressed during zoosporogenesis Silencing > inhibits cyst germination by 60% |
Judelson and Tani (2007); Tani et al. (2005) |
| PHYCI_112968 |
P. sojae PsMPK7
PHYSO_355777 |
Stress‐associated MAPK which is up‐regulated in zoospores, cysts and germinating cysts Silencing > abnormal germ tubes; swelling of the hyphal apex; reduces virulence |
Gao et al. (2015) |
| PHYCI_91649 |
P. sojae PsVPS1
PHYSO_562318 |
Dynamin‐related vacuolar sorting protein that mediates budding of clathrin‐coated vesicles from the late Golgi apparatus Silencing > induces apical swelling and branching of germ tubes and reduces hyphal growth; reduces virulence |
Li et al. (2013) |
|
PHYCI_327508 PHYCI_253325 PHYCI_253304 ‡ |
P. parasitica PPMUCL1
PPTG_17796 |
Mucin‐like proteins. High‐molecular‐weight secreted glycoproteins found in biofilms produced by germinated cysts. Biofilm mucins may provide protection against desiccation | Larousse et al. (2014) |
| PHYCI_196387 § |
P. infestans PITG_23049 P. parasitica PPTG_13017 |
P. infestans Car90 (cyst germination‐specific acidic repeat) protein which is expressed in germinating cysts and during appressorium formation. Car proteins contain an octapeptide tandem repeat found in mammalian mucins | Görnhardt et al. (2000) |
| PHYCI_232811 |
P. infestans Pi‐ts1
PITG_13139 |
Threonine synthase. Increased gene expression in germinated cysts with appressoria | Grenville‐Briggs et al. (2005) |
| PHYCI_306811 |
P. infestans Pi‐met1
PITG_01072 |
Methionine synthase. Increased gene expression in germinated cysts with appressoria | Grenville‐Briggs et al. (2005) |
| PHYCI_96428 |
P. infestans Pi‐kari1
PITG_02925 |
Ketol‐acid reductoisomerase. Increased gene expression in germinated cysts with appressoria | Grenville‐Briggs et al. (2005) |
| PHYCI_111395 |
P. infestans Pi‐als1
PITG_03410 |
Acetolactate synthase. Increased gene expression in germinated cysts with appressoria | Grenville‐Briggs et al. (2005) |
| PHYCI_95137 |
P. infestans Pi‐trp1
PITG_00221 |
Tryptophan synthase. Increased gene expression in germinated cysts with appressoria | Grenville‐Briggs et al. (2005) |
| Plant colonization – hyphal growth, haustoria formation | |||
| PHYCI_137521 |
P. ramorum PSURA_75613.1 P. parasitica PPTG_16290.1 |
Mucin or mucin‐like protein isolated from P. ramorum mycelial cell walls. It has homology to the P. infestans Car mucins. PSURA_75613.1 has a transmembrane (TM) domain | Larousse et al. (2014); Meijer et al. (2006) |
|
PHYCI_111888 PHYCI_202815 |
P. ramorum PSURA_83136 P. parasitica PPTG_17896.1 |
Mucin or mucin‐like protein isolated from P. ramorum mycelial cell walls with homology to the P. infestans Car mucins | Larousse et al. (2014); Meijer et al. (2006) |
| PHYCI_115477 |
P. ramorum PSURA_80868.1 P. parasitica PPTG_13138.1 |
Mucin or mucin‐like protein isolated from P. ramorum mycelial cell walls with homology to the P. infestans Car mucins | Larousse et al. (2014); Meijer et al. (2006) |
|
PHYCI_218680 PHYCI_271751 |
P. ramorum PSURA_75750 P. parasitica PPTG_01865.1 |
Mucin or mucin‐like protein isolated from P. ramorum mycelial cell walls. It has homology to the P. infestans Car mucins. PSURA_75750.1 has a TM domain | Larousse et al. (2014); Meijer et al. (2006); Reitmann et al. (2016) |
| PHYCI_88279 |
P. parasitica PpPDI1
PPTG_18309 |
Secreted protein disulfide isomerase that induces cell death Over‐expression of GFP‐PpPDI1 > enhances haustoria formation; increases virulence |
Meng et al. (2015) |
*Accession number in FungiDB.
†Additional PiGK4 homologues are listed in the cyst germination section.
‡These are the top three P. cinnamomi genes homologous to PPMUCL1 in table S3 in Larousse et al. (2014).
§The P. cinnamomi gene sequence with the closest homology to PITG_23049. The sequence is only 88 amino acids in length and the comparison yields an E‐value of 2e−34. PHYCI_196387 is not included in the list of 60 P. cinnamomi mucin genes with homology to PPMUCL1 in table S3 in Larousse et al. (2014).
Zoospore chemotaxis is a vital aspect of disease initiation and the P. cinnamomi genome contains homologues of three genes implicated in this process in P. infestans and P. sojae, namely a G‐protein α‐subunit (GPA1), a GPA1‐interacting protein (Hint1) and a GPCR (GK4) (Table 2). Silencing of these three genes in P. infestans or P. sojae reduces the period of zoospore motility before encystment and inhibits chemotaxis, negative geotaxis and auto‐aggregation (Hua et al., 2008; Latijnhouwers et al., 2004; Yang et al., 2013). By contrast, silencing of two other GPCRs, PsGPR11 and PsSAK1, in P. sojae does not inhibit zoospore chemotaxis, although it does induces encystment (Li et al., 2010; Wang et al., 2010; Yang et al., 2013). There are homologues of these two genes in the P. cinnamomi genome (Table 2).
GPCRs involved in zoospore motility are thought to reside in the zoospore plasma membrane, but, as yet, there is no evidence for this. However, antibody binding to a flagellar membrane protein in P. cinnamomi rapidly induces zoospore encystment (Hardham and Suzaki, 1986). This flagella surface antigen is thus conserved across the Phytophthora genus and it would be fascinating to determine whether it is a GPCR involved in the regulation of zoospore motility.
Attachment and protection at the plant surface
Phytophthora cinnamomi zoospores swim towards the elongation zone of plant roots, often settling preferentially in the grooves above epidermal anticlinal walls (Hardham, 2005). At high densities, they may exhibit auto‐aggregation and cluster at certain sites on the plant root surface (Fig. 4), a phenomenon requiring both chemotaxis and bioconvection (Savory et al., 2014). Encysting zoospores attach to the root surface through the secretion of a 250‐kDa adhesin protein, PcVsv1, stored in small ventrally located vesicles (Hardham and Gubler, 1990; Robold and Hardham, 2005). Expression of the PcVsv1 gene is up‐regulated during asexual sporulation, coincident with the appearance of ventral vesicles. Homologues of the Vsv1 adhesin occur throughout the Oomycetes.
Figure 4.
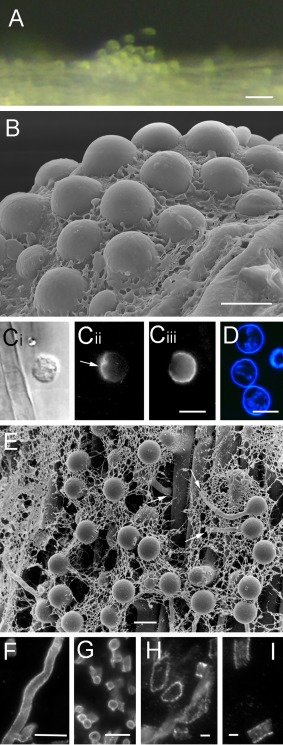
Adhesion and penetration by Phytophthora cinnamomi on the plant surface. (A) Aggregation of motile zoospores on the surface of a plant root. (B) A cluster of cysts embedded in mucin‐like material secreted during encystment on a root surface. (C) A cyst on the surface of a plant root labelled with PcVsv1 monoclonal antibody (mAb) (Cii) or soybean agglutinin (SBA) (Ciii). The same cell is shown in bright field in (Ci). Proteins secreted from zoospore ventral vesicles form an adhesive pad between the cyst and the root (arrow in Cii). SBA binds to N‐acetylgalactosyl and galactosyl residues in PcCpa2 glycoproteins that are secreted from dorsal vesicles onto the zoospore dorsal surface which faces away from the root (Ciii). (D) The cell wall that is rapidly formed on the surface of young cysts is stained by calcofluor. (E) Cysts on the surface of a plant root germinate and the germ tube often penetrates the root along the periclinal wall between adjacent epidermal cells (arrows). (F–I) Secreted cell wall‐degrading enzymes coat the surface of P. cinnamomi hyphae. Polygalacturonases (F–H) and endoglucanases (I) are immunolabelled by polyclonal antibodies raised against Sclerotinia sclerotiorum (Martel et al., 1996), Fusarium moniliforme (De Lorenzo et al., 1987), Colletotrichum lindemuthianum (Hugouvieux et al., 1995) and Macrophomina phaseolina (Jones and Wang, 1997) enzymes, respectively. Bars: (A) 50 µm; (B–G) 10 µm; (H, I) 2 µm.
Studies in P. parasitica have shown that a 12‐kDa complement control protein is also secreted during zoospore encystment (Škalamera and Hardham, 2006). There are single Ccp genes in both P. parasitica and P. cinnamomi genomes (Table 2). Ccp proteins are stored in an outer shell surrounding an inner core of Lpv glycoproteins within large peripheral vesicles (Gubler and Hardham, 1990; Zhang et al., 2013). Remarkably, during encystment, the small Ccp proteins are secreted, whereas the 500–600‐kDa Lpv proteins are retained, an example of selective protein secretion. In mammals, multiple complement control domains facilitate a function in cell adhesion but, as Phytophthora Ccp proteins contain only a single complement control module, an adhesive role seems unlikely. In P. cinnamomi, Lpv proteins apparently serve as a protein store utilized during germling growth (Gubler and Hardham, 1990). DNA and RNA blots show the presence of three PcLpv genes which give rise to three transcripts of 11–14 kb in size (Marshall et al., 2001b). None of the Phytophthora genomes in FungiDB include Lpv genes that would give rise to transcripts of this length. Of the five predicted P. cinnamomi transcripts with homology to cloned partial Lpv genes, the two largest include long regions of undetermined nucleotides (Table 2). Sequencing of the cloned, partial PcLpv genes has shown that the C‐terminal half of the Lpv proteins consists of 12–18 copies of almost identical 178‐amino‐acid repeats (Marshall et al., 2001b), a factor that has so far confounded the identification and sequence determination of the complete length of an Lpv gene.
A set of three large PcCpa glycoproteins of >330 kDa in molecular weight is secreted from small dorsal vesicles to form a mucilage‐like coating over the cyst surface (Gubler and Hardham, 1988; Hardham, 2005). The appearance of the secreted PcCpa material (Fig. 4) is similar to that of the biofilm formed by mucin‐like (MUCL) proteins secreted by P. parasitica germinated cysts onto the host surface (Larousse et al., 2014). According to Larousse et al. (2014, Table S3), the P. cinnamomi genome contains 60 genes with similarity to P. parasitica PPMUCL1. Other P. cinnamomi genes are homologous to P. infestans cyst‐germination‐specific acidic repeat (Car) mucins (Görnhardt et al., 2000) and to six cell wall mucins in P. ramorum (Meijer et al., 2006) (Table 2). In the absence of PcCpa sequence information, there are currently no molecular data linking these proteins with the cyst or hyphal mucins described in these other Phytophthora species. Not all P. parasitica PPMUCL genes are expressed during biofilm formation (Larousse et al., 2014), and thus it is possible that some mucin gene homologues in P. cinnamomi might encode the PcCpa glycoproteins synthesized during sporulation, stored in dorsal vesicles and secreted during zoospore encystment. The confirmation or negation of this hypothesis awaits the identification of the PcCpa genes, but immunoblots show that secreted PPMUCL glycoproteins are of a similar size to the PcCpa glycoproteins. Mucins secreted by epithelial cells in animals form a highly hydrated barrier that protects the underlying cells and tissues against pathogen invasion. Biofilms formed by pathogenic organisms are thought to enhance virulence by contributing to host attachment and protecting against desiccation. It is thus possible that mucins secreted by Phytophthora zoospores and germinated cysts serve similar protective functions during the establishment of plant infection.
Plant penetration and colonization
Cyst germination and chemotropic growth
Phytophthora cinnamomi cysts typically germinate 20–30 min after zoospore encystment. Germination can occur in distilled water, suggesting that it might be programmed to follow zoospore encystment in the absence of external chemical signals. Consistent with the expectation that signalling proteins will be involved in directing changes in gene expression, protein synthesis and the switch from cell motility to secretion as the dominant cellular activity, silencing of the GPCRs, PiGK4 and PsGPR11, not only curtails zoospore motility, but also inhibits cyst germination (Hua et al., 2013; Wang et al., 2010; Yang et al., 2013). Phytophthora cinnamomi has 13 GPCR genes with homology to PiGK4 and three with homology to PsGPR11 (Table 2). Phytophthora cinnamomi also has close homologues to three other classes of genes implicated by gene silencing studies in cyst germination, namely a MAPK PsSAK1 (Li et al., 2010), a Myb transcription factor PsMYB1 (Zhang et al., 2012) and three NIF proteins that interact with nuclear LIM transcription factors (Judelson and Tani, 2007; Tani et al., 2005) (Table 2). Silencing of the expression of PsSAK1 also inhibits appressorium formation and leads to the production of longer germ tubes (Li et al., 2010). Phytophthora cinnamomi also has homologues to two P. sojae proteins, the MAPK PsMPK7 and a dynamin‐related vacuolar sorting protein PsVPS1, which function in regulating the polarity of germ tube extension (Gao et al., 2015; Li et al., 2013), and to five genes encoding enzymes involved in amino acid synthesis whose expression is up‐regulated during appressorium formation (Grenville‐Briggs et al., 2005) (Table 2).
Cell wall‐degrading enzymes (CWDEs)
Initial penetration and subsequent colonization of the plant are made possible by the action of a wide range of pathogen degradative enzymes that digest components of the plant cell wall. Phytophthora genomes contain large multigene families encoding CWDEs that contain one or more Carbohydrate‐Active enZyme (CAZyme) modules (Blackman et al., 2014; Götesson et al., 2002; Larroque et al., 2012; Ospina‐Giraldo et al., 2010).
Gene‐by‐gene analysis of predicted P. cinnamomi transcripts included in the FungiDB database using dbCAN (http://csbl.bmb.uga.edu/dbCAN/) indicates that there are 438 CWDE genes in the P. cinnamomi genome (Tables 3, 4, 5), a number similar to the 431 and 423 CWDE genes identified in P. parasitica and P. infestans, respectively, using the same approach (Blackman et al., 2014) (Table S1, see Supporting Information). The 438 CWDEs in P. cinnamomi include 271 proteins containing a glycoside hydrolase (GH) module (Table 3), 17 proteins containing an auxiliary activity (AA) module (Table 4), 53 proteins containing a carbohydrate esterase (CE) module (Table 4), 42 proteins containing a polysaccharide lyase (PL) module (Table 4) and 64 proteins containing only a non‐catalytic carbohydrate‐binding module (CBM) (Table 5). Proteins from these five classes of CAZymes degrade the main polysaccharides in the plant cell wall, namely cellulose, hemicelluloses, pectins and β‐1,3‐glucans (Tables 3, 4, 5). Cellulose is attacked by CWDEs containing modules from nine GH, five CBM and three AA families. Hemicelluloses are attacked by CWDEs containing modules from 12 GH, four CE and six CBM families. Pectins are attacked by CWDEs containing modules from 10 GH, three PL, three CE and one CBM family. The identification of 26 GH28 polygalacturonase genes in FungiDB (15 full length and 11 partial) confirms the earlier report of more than 17 P. cinnamomi polygalacturonase genes based on Southern DNA blots (Götesson et al., 2002).
Table 3.
Phytophthora cinnamomi proteins involved in plant cell wall degradation: enzymes containing glycosyl hydrolase (GH) modules.
| CAZyme family | Potential substrates | Potential enzyme activities | Number of genes |
|---|---|---|---|
| GH1 | Cellulose, hemicellulose (XG), pectin (RGI) | β‐Glucosidase, β‐galactosidase, β‐mannosidase, exo‐β‐1,4‐glucanase | 14 |
| GH2 | Hemicellulose (mannans), glycoproteins | β‐Mannosidase | 1 |
| GH3 | Cellulose, hemicellulose (XG), pectin (RGI), AGPs | β‐Glucosidase, exo‐β‐1,4‐glucosidase, β‐1,4‐xylosidase, β‐1,3‐glucosidase, α‐l‐arabinofuranosidase | 26 |
| GH5 | Cellulose, hemicellulose (xylans, galactomannans), β‐1,3‐glucans | Endo‐β‐1,4‐glucanase, β‐1,4‐cellobiosidase, endo‐β‐1,4‐xylanase, endo‐β‐1,4‐mannosidase, β‐1,3‐glucosidase | 30 |
| GH6 | Cellulose | Endo‐β‐1,4‐glucanase, cellobiohydrolase | 7 |
| GH7 | Cellulose | Endo‐β‐1,4‐glucanase, cellobiohydrolase | 7 |
| GH10 | Hemicellulose (xylans) | Endo‐β‐1,4‐β‐xylanase | 6 |
| GH12 | Cellulose, hemicellulose (XG) | Endo‐β‐1,4‐glucanase, XG endo‐β‐1,4‐glucanase | 10 |
| GH13 | Starch | α‐Amylase, α‐glucosidase | 2 |
| GH16 | Hemicellulose (XG), β‐1,3‐glucans | XG endo‐β‐1,4‐glucanase, β‐1,3‐glucosidase | 21 |
| GH17 | β‐1,3‐Glucans | Endo‐β‐1,3‐glucosidase | 21 |
| GH18 | N‐linked oligosaccharides | Endo‐β‐N‐acetylglucosaminidase | 3 |
| GH19 | N‐linked oligosaccharides | Endo‐β‐N‐acetylglucosaminidase | 1 |
| GH28 | Pectin (HG) | Polygalacturonase | 26 |
| GH30 | Cellulose, hemicellulose (XG), pectin (RGI), AGPs | β‐Glucosidase, endo‐β‐1,4‐xylanase, xylan β‐1,4‐xylosidase, endo‐β‐1,6‐galactanase, β‐1,6‐glucanase | 17 |
| GH31 | Starch, hemicellulose (XG) | α‐Glucosidase, α‐xylosidase | 6 |
| GH32 | Sucrose | Invertase | 3 |
| GH35 | Hemicellulose (XG), pectin (HG), AGPs | β‐Galactosidase, exo‐β‐1,4‐galactanase | 1 |
| GH37 | Trehalose (α,α‐1,1‐glucans) | α,α‐Trehalase | 2 |
| GH38 | N‐linked oligosaccharides | α‐Mannosidase | 1 |
| GH43 | Hemicellulose (xylans), pectin (RGI), AGPs | α‐l‐Arabinofuranosidase | 7 |
| GH45 | Cellulose, β‐glucans | Endo‐glucanase | 1 |
| GH47 | N‐linked oligosaccharides | α‐Mannosidase | 5 |
| GH53 | Pectin (RGI) | Endo‐β‐1,4‐galactanase | 6 |
| GH54 | Pectin (RGI) | α‐l‐Arabinofuranosidase | 2 |
| GH63 | N‐linked oligosaccharides | α‐1,3‐Glucosidase, α‐glucosidase | 1 |
| GH72 | β‐1,3‐Glucans | β‐1,3‐Glucanosyl‐transglycosylase | 10 |
| GH78 | Pectin (RGI) | α‐l‐Rhamnosidase | 6 |
| GH81 | β‐1,3‐Glucans | Endo‐β‐1,3‐glucanase | 12 |
| GH85 | AGPs | Endo‐β‐N‐acetylglucosaminidase | 1 |
| GH89 | N‐linked oligosaccharides | α‐N‐Acetylglucosaminidase | 1 |
| GH105 | Pectin (RGI) | Unsaturated rhamnogalacturonyl hydrolase | 1 |
| GH109 | O‐linked oligosaccharides | α‐N‐Acetylgalactosaminidase | 8 |
| GH114 | Glycoproteins | Endo‐α‐1,4‐polygalactosaminidase | 1 |
| GH123 | O‐linked oligosaccharides | Glycosphingolipid β‐N‐acetylgalactosaminidase | 1 |
| GH131 | Cellulose, hemicellulose (β‐1,4‐glucans) | Exo‐β‐1,3/1,6‐glucanase, endo‐β‐1,4‐glucanase | 3 |
| Total | 271 |
AGP, arabinogalactan protein; CAZyme, Carbohydrate‐Active enZyme; HG, homogalacturonan; RGI, rhamnogalacturonan I; XG, xyloglucan.
Table 4.
Phytophthora cinnamomi proteins involved in plant cell wall degradation: enzymes containing auxiliary activity (AA), carbohydrate esterase (CE) and polysaccharide lyase (PL) modules.
| CAZyme family | Potential substrates | Potential enzyme activities | Number of genes |
|---|---|---|---|
| Auxiliary activity modules | |||
| AA1 | Benzenediol, catechol | Laccase, oxidoreductase, ferroxidase | 1 |
| AA7 | Cellobiose, chitin, glycoproteins | Glucooligosaccharide oxidase, chitooligosaccharide oxidase | 8 |
| AA8 | Cellulose | Iron reductase domain | 5 |
| AA10 | Cellulose | Copper‐dependent monooxygenase | 3 |
| Total | 17 | ||
| Carbohydrate esterase modules | |||
| CE1 | Hemicellulose | Feruloyl esterase | 5 |
| CE2 | Hemicellulose | Acetyl xylan esterase | 1 |
| CE4 | Hemicellulose, N‐linked oligosaccharides | Acetyl xylan esterase, peptidoglycan GlcNAc deacetylase | 2 |
| CE5 | Hemicellulose | Acetyl xylan esterase, cutinase | 9 |
| CE8 | Pectin (HG) | Pectin methylesterase | 17 |
| CE12 | Pectin (HG, RGI) | Pectin and RGI acetylesterase, acetyl xylan esterase | 12 |
| CE13 | Pectin (HG) | Pectin acetylesterase | 7 |
| Total | 53 | ||
| Polysaccharide lyase modules | |||
| PL1 | Pectin (HG) | Pectate lyase, pectin lyase | 21 |
| PL3 | Pectin (HG, RGI) | Pectate lyase | 22 |
| PL4 | Pectin (RGI) | Rhamnogalacturonan lyase | 6 |
| Total | 42 | ||
CAZyme, Carbohydrate‐Active enZyme; GlcNAc, N‐acetylglucosamine; HG, homogalacturonan; RGI, rhamnogalacturonan I.
Table 5.
Phytophthora cinnamomi proteins involved in plant cell wall degradation: enzymes containing carbohydrate‐binding (CBM) modules.
| CAZyme family | Potential substrates | Number of genes |
|---|---|---|
| CBM1 | Cellulose | 14 |
| CBM9 | Hemicellulose (xylans) | 1 |
| CBM13 | Hemicellulose (xylans) | 9 |
| CBM14 | Chitin | 1 |
| CBM16 | Cellulose, hemicellulose (glucomannans) | 1 |
| CBM17 | Cellulose | 2 |
| CBM20 | Starch, fucose | 3 |
| CBM23 | Hemicellulose (mannans) | 1 |
| CBM25 | Starch | 1 |
| CBM32 | Pectin (galactose, PGA and β‐galactosyl‐β‐1,4‐GlcNAc) | 2 |
| CBM37 | Cellulose, hemicellulose (xylans) | 2 |
| CBM38 | Inulin | 1 |
| CBM40 | Sialides | 1 |
| CBM47 | Fucose | 4 |
| CBM48 | Glycogen (starch) | 1 |
| CBM50 | Chitin, glycoproteins | 2 |
| CBM60 | Hemicellulose (xylans) | 1 |
| CBM63 | Cellulose | 16 |
| Total | 63 |
CAZyme, Carbohydrate‐Active enZyme; GlcNAc, N‐acetylglucosamine; PGA, polygalacturonic acid.
Phytophthora cinnamomi polygalacturonases are synthesized and secreted during an early phase of growth after subculturing in vitro. Although the total protein concentration in the culture medium continues to increase over a 15‐day growth period, polygalacturonase activity peaks 5–6 days after inoculation of the medium (J. Schick et al., unpublished observations). Secreted polygalacturonase proteins can be immunolocalized on the hyphal surface (Fig. 4) and recombinant, single‐chain, anti‐polygalacturonase antibodies can inhibit the activity of polygalacturonases secreted by P. cinnamomi (Manatunga et al., 2005).
The measurement of P. cinnamomi polygalacturonase transcript levels using quantitative real‐time polymerase chain reaction (qPCR) shows that the GH28 genes are differentially expressed both in vitro and in planta (E. Landgren et al., unpublished observations). In in vitro assays using 5% V8 broth or Ribeiro's minimal medium (Ribeiro, 1978) supplemented with defined carbon sources, some P. cinnamomi GH28 genes are expressed regardless of the type of carbohydrate present, some are expressed most highly in the presence of glucose and others are expressed most highly in the presence of pectin (Fig. S2, see Supporting Information). These results provide evidence of the operation of carbon catabolite repression mechanisms in the regulation of GH28 gene expression. qPCR measurement of P. cinnamomi GH28 transcript levels during the infection of lupin roots by P. cinnamomi also indicates that, as in the lupin–P. parasitica interaction (Blackman et al., 2015), some GH28 genes are expressed early in plant infection, with transcript levels decreasing as infection proceeds (Fig. S3, see Supporting Information).
Hyphal growth
Having penetrated the plant surface, P. cinnamomi hyphae grow intracellularly or intercellularly through the root cortex and into the central vascular bundle (Fig. 5). Blockage of the xylem through hyphal obstruction and deposition of material by the plant inhibits water movement from the roots to the shoots, resulting in water stress (Ruiz Gómez et al., 2015). Necrosis of infected fine feeder roots exacerbates the problem and can lead to rapid plant death (McConnell and Balci, 2015; Oßwald et al., 2014).
Figure 5.
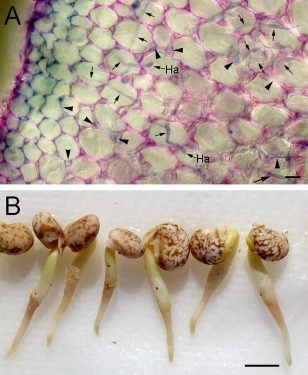
Colonization and lesion development in lupin (Lupinus augustifolius) roots 20 h after inoculation with Phytophthora cinnamomi zoospores. (A) A transverse hand‐section of an infected lupin root. During initial colonization of the root, hyphae grow from the epidermis, through the cortex and into the vascular cylinder. Hyphal growth may be intracellular (arrows) or intercellular (arrowheads). Two putative haustoria (Ha) are indicated. (B) Lesions develop on the lupin roots just below the surface of the zoospore suspension whose approximate position is marked by black ink spots. Bars, 10 µm.
Phytophthora cinnamomi hyphae, like other tip‐growing cells, extend through the fusion of small transport vesicles at the hyphal apex and the secretion of new membrane and wall material. Phytophthora and other Oomycete cell walls are based on a cellulosic, rather than chitinous, microfibrillar framework. According to a recent study, P. cinnamomi and other Phytophthora and Peronosporales species have Type I cell walls, distinguished mainly by their lack of N‐acetylglucosamine and their cellulose content (Mélida et al., 2013).
Pathogen CWDEs are also secreted through vesicle fusion at the hyphal apex, and the consequent hydrolysis of plant cell walls is indicated by changes in wall morphology, cell separation and tissue maceration (Redondo et al., 2015; Ruiz Gómez et al., 2015). There is some evidence that these changes are accompanied by de‐esterification of plant cell wall pectins during the infection of holm oak (Quercus ilex) by P. cinnamomi (Ruiz Gómez et al., 2015); however, the causal relationship between these features and details of the molecular mechanisms underlying plant cell wall degradation await future research.
Haustoria
Although previously considered to be a necrotrophic pathogen, recent observations of putative P. cinnamomi haustoria within cortical and vascular tissues in a number of host plants, including Lupinus augustifolius (Fig. 5A), have suggested that P. cinnamomi can grow as a hemibiotroph (Crone et al., 2013b; Redondo et al., 2015). In Arabidopsis, the extrahaustorial membrane surrounding haustoria of the Oomycete, Hyaloperonospora arabidopsidis, contains the plasmodesmata‐located protein1 (PDL1) (Caillaud et al., 2014). The demonstration of the presence of homologues of PDL1 around the P. cinnamomi haustoria‐like structures could confirm their identification. Molecular details of Phytophthora haustorial function are yet to be elucidated, although a protein disulfide isomerase PpPDI1 has been associated with haustoria and enhanced pathogenicity in P. parasitica (Meng et al., 2015). The P. cinnamomi genome contains a homologue of the PpPDI1 gene (Table 2) and characterization of its encoded protein may provide important information on P. cinnamomi haustorium function.
The timing of the transition from an initially biotrophic to a subsequent necrotrophic phase of plant infection by P. cinnamomi is influenced by the plant species and environmental conditions. Maintenance of the biotrophic phase means that P. cinnamomi can be present in infected plants in the absence of obvious disease symptoms (Crone et al., 2013a, 2013b). Long‐term survival in infected plants can be achieved through the formation of chlamydospores, stromata, hyphal aggregates within lignitubers and oospores (Crone et al., 2013b; Jung et al., 2013).
Plant–Pathogen Interactions
Effectors and elicitors
During infection, P. cinnamomi, like other plant pathogens, secretes a diverse range of effector molecules into the plant apoplast. From the pathogen's point of view, the intended function of these effectors is to facilitate the establishment of disease. From the plant's point of view, the goal is to recognize the effectors and trigger a defence response that will inhibit the development of disease. When an intended effector is recognized by the plant and elicits a defence response, it is termed an avirulence factor or elicitor.
Effector proteins produce metabolic or structural changes in host cells that aid pathogen growth and favour the development of disease. They are secreted by the pathogen and act either in the plant apoplast or symplast. Apoplastic effectors include CWDEs, elicitins, toxins and inhibitors of plant enzymes. Symplastic effectors are translocated across the plant plasma membrane by an as yet unknown mechanism and act within the plant cell cytoplasm. The function of the majority of known or putative cytoplasmic effectors is still unclear, but some have been shown to suppress host defence responses, such as callose deposition or hypersensitive cell death (Dou et al., 2008a; Du et al., 2015). Effectors have sequence variants that may or may not be recognized by a potential host plant. In many cases, it was the recognized avirulence protein form that triggers host defence that was first identified.
A great diversity of avirulence proteins and other elicitors have been described and studied. Elicitors may be proteins, carbohydrates or lipids, and may be of pathogen or plant origin. In the latter case, an elicitor can arise, for example, as a result of the digestion of a plant cell wall component by a pathogen enzyme. Many elicitor molecules are highly conserved across broad taxonomic groups and are often referred to as pathogen‐ or microbe‐associated molecular patterns (PAMPS or MAMPs). PAMPs trigger basal defence responses, also referred to as PAMP‐triggered immunity (PTI), including localized cell wall reinforcement and callose deposition, production of antimicrobial compounds, such as phytoalexins and reactive oxygen species, and, in some cases, cell death (Chang et al., 2015). Some elicitors are conserved, but occur in only one or a few genera. Small cysteine‐rich proteins, called elicitins, produced by species of Phytophthora, including P. cinnamomi, are examples of this latter class of elicitor. Elicitins function in plant sterol uptake, and thus, when they are not recognized by the plant, Phytophthora elicitins are likely to serve as pathogenicity factors and can be considered to be effectors.
The potential role of many P. cinnamomi effectors in pathogenicity has been presented already as part of the discussion of the infection strategies employed, especially during the early stages of disease establishment, such as adhesion and penetration. However, full disease development involves a great diversity of complex plant–pathogen interactions and will depend on many putative effectors not yet discussed. Some effectors will facilitate attack and colonization so that the pathogen can access the nutrients it needs for growth and reproduction. Other effectors will be central to pathogen defence mechanisms that provide protection against plant defences.
Putative effectors of pathogen attack strategies
P. cinnamomi elicitins
Elicitins are 10‐kDa globular proteins produced by all Phytophthora and some Pythium species (Duclos et al., 1998; Jiang et al., 2006). They are not recognized by most plants, but, in some solanaceous species (notably Nicotiana species) and some cultivars of radish and turnip, they trigger plant hypersensitive cell death and can induce plant resistance to a variety of bacterial and fungal pathogens (Oßwald et al., 2014). Phytophthora species typically contain 10–20 elicitin genes (Jiang et al., 2006). blast analysis of the P. cinnamomi genome, using the PHYCI_98389 β‐cinnamomin gene as query, resulted in the identification of 32 putative elicitin sequences (Table S2, see Supporting Information), considerably more than the single α‐cinnamomin and β‐cinnamomin and two highly acidic elicitin (HAE) genes previously reported in P. cinnamomi (Duclos et al., 1998).
Phytophthora elicitins bind dehydroergosterol and, because Phytophthora species cannot synthesize sterols, they are believed to serve an essential role in Phytophthora development and pathogenicity through their ability to transfer sterols from plant membranes to the pathogen (Osman et al., 2001; Rodrigues et al., 2006). qPCR measurements have indicated that all four previously reported cinnamomin genes are expressed in vitro (Horta et al., 2008). In cork oak (Quercus suber) roots, transcript levels of α‐cinnamomin increased about three‐fold, whereas those for β‐cinnamomin and α‐HAE decreased by about 20% and 90%, respectively, during a 24‐h period after inoculation with P. cinnamomi. Expression of the β‐HAE gene was detected only at 36 h post‐inoculation (hpi). These results may indicate that β‐cinnamomin mainly functions during the initial establishment of infection, whereas α‐cinnamomin functions throughout colonization and sporulation (Horta et al., 2008). The reported silencing of the expression of the β‐cinnamomin gene reduces the pathogen's ability to penetrate and colonize the roots of Q. suber and chestnut (Castanea sativa) seedlings compared with wild‐type controls, leading to decreased symptom severity in the inoculated plants (Horta et al., 2010; Maia et al., 2012). As these studies are based on only one silenced transformant line, they must be viewed as being preliminary; nevertheless, they represent an exciting advance in the demonstration of P. cinnamomi elicitin function. The establishment of a reproducible and stable transformation strategy for P. cinnamomi would greatly facilitate the elucidation of P. cinnamomi gene function and the molecular basis of plant infection.
Plant recognition of cinnamomin contributes to the induction of host defence responses. Pretreatment of oak (Q. suber or Q. ilex) or chestnut (C. sativa) roots with α‐ or β‐cinnamomin results in a significant reduction in root colonization by P. cinnamomi (Ebadzad et al., 2015; Maia et al., 2008; Medeira et al., 2012). Further studies of the role of cinnamomins in Phytophthora–plant interactions may be aided by the development of an improved method for the production and purification of β‐cinnamomin (Hofzumahaus and Schallmey, 2013).
Small cysteine‐rich (SCR) proteins
Phytophthora species produce a second category of small cysteine‐rich (SCR) effector proteins. The first representative, P. cactorum PcF, induces the expression of plant pathogenicity genes and programmed cell death (Orsomando et al., 2001). Previously analysed Phytophthora species contain 3–19 SCR genes (Chen et al., 2015; Orsomando et al., 2011). A search using P. cactorum SCR genes has revealed that the P. cinnamomi genome contains one homologue of PcF, eight homologues of Scr96, 15 homologues of Scr99 and one homologue of Scr121 (Table 6), and thus appears to have the largest PcF/SCR gene family reported so far. Silencing of P. cactorum Scr96 reduces pathogen virulence on Nicotiana benthamiana leaves and makes it more sensitive to oxidative stress (Chen et al., 2015), indicating that Phytophthora SCR proteins are effectors that function as virulence factors under the same circumstances in which they trigger plant cell death.
Table 6.
Phytophthora cinnamomi proteins involved in plant–pathogen interactions: putative effectors involved in plant colonization.
| P. cinnamomi gene name (in FungiDB) | Homologues used for blast query | Proposed protein function | References |
|---|---|---|---|
| Elicitins * | |||
| PHYCI_127817 |
P. cinnamomi
α‐cinnamomin |
Sterol carrier protein believed to be required for virulence | Duclos et al. (1998); Horta et al. (2008) |
| PHYCI_98389 |
P. cinnamomi
β‐cinnamomin |
Sterol carrier protein Silencing > reduces virulence |
Duclos et al. (1998); Horta et al. (2010); Maia et al. (2012) |
| PHYCI_251414 |
P. cinnamomi
Highly acidic elicitin‐α (HAE‐α) |
Sterol carrier protein believed to be required for virulence | Duclos et al. (1998); Osman et al. (2001) |
| PHYCI_98390 |
P. cinnamomi
HAE‐β |
Sterol carrier protein believed to be required for virulence | Duclos et al. (1998); Osman et al. (2001) |
| Small cysteine‐rich (SCR) toxins | |||
| PHYCI_83830 |
P. cactorum PcF
AAK63068 |
PcF and PcF‐like toxin proteins, small cysteine‐rich proteins from P. cactorum | Nicastro et al. (2009); Orsomando et al. (2001) |
|
PHYCI_93258 PHYCI_93260 PHYCI_93259 PHYCI_92597 PHYCI_323321 PHYCI_85664 PHYCI_85660 PHYCI_97296 |
P. cactorum Scr96
ALC04448 |
PcF toxin family of SCR proteins in P. infestans, P. sojae, P. ramorum and P. cactorum
In planta, P. cactorum Scr96 transcript levels are high in early infection (12 hpi) and then decline. Silencing > reduces virulence and increases sensitivity to oxidative stress |
Chen et al. (2015) |
|
PHYCI_90211 PHYCI_96480 PHYCI_20145 PHYCI_97226 PHYCI_330581 PHYCI_234114 PHYCI_90122 PHYCI_259880 PHYCI_254679 PHYCI_252576 PHYCI_269098 PHYCI_237327 PHYCI_194590 PHYCI_256642 PHYCI_241899 |
P. cactorum Scr99
ALC04449 |
PcF toxin family of SCR proteins In planta, Scr99 transcript levels increase during the first 48 hpi |
Chen et al. (2015) |
| PHYCI_97299 |
P. cactorum Scr121
ALC04450 |
PcF toxin family of SCR proteins In planta, Scr121 transcript levels increase during the first 48 hpi |
Chen et al. (2015) |
| Transglutaminases | |||
|
PHYCI_89523 PHYCI_208557 PHYCI_471842 PHYCI_89525 PHYCI_89524 PHYCI_209063 PHYCI_471855 PHYCI_288438 PHYCI_1375 PHYCI_227786 PHYCI_240555 PHYCI_1278 PHYCI_15457 PHYCI_261219 PHYCI_181539 PHYCI_13742 PHYCI_12440 PHYCI_178520 PHYCI_134840 PHYCI_198929 PHYCI_97108 |
P. ramorum GP42
PSURA_53744, PSURA_83169 |
Two GP42 transglutaminases have been found in hyphal cell wall proteomes in P. infestans and P. ramorum. They catalyse peptide bond formation that can increase protein stability. GP42 transglutaminases induce plant defence responses. They are expressed early in infection, but their function in pathogenicity is unknown | Brunner et al. (2002); Grenville‐Briggs et al. (2010); Martins et al. (2014b); Meijer et al. (2006) |
| Crinkler (CRN) effectors | |||
|
PHYCI_99711
†
PHYCI_98916 PHYCI_93895 PHYCI_82897 PHYCI_75608 PHYCI_68121 PHYCI_557374 PHYCI_251494 PHYCI_213501 PHYCI_148758 PHYCI_115716 PHYCI_141767 PHYCI_105597 PHYCI_111648 |
P. infestans
45 CRN proteins representative of those reported in table S10 in Haas et al. (2009) |
CRN effectors are secreted proteins that may cause crinkling and necrosis. The conserved N‐terminus includes LFLAK and DWL motifs that function in protein uptake into the plant cytoplasm. Variable C‐terminal regions are associated with a range of effector functions, including translocation into the plant nucleus. Many CRN proteins suppress plant defence, including hypersensitive cell death, possibly through interaction with plant catalases In P. sojae, silencing > reduces virulence |
Chen et al. (2013); Haas et al. (2009); Liu et al. (2011); Mafurah et al. (2015); Stam et al. (2013); Van Damme et al. (2012); Zhang et al. (2015) |
| NLPs (Nep1‐like proteins) | |||
|
72 NLP homologues in P. cinnamomi
[see Table S4 (Supporting Information) for full list] |
P. sojae NLPs
PHYSO_562453 PHYSO_509399 PHYSO_249691 |
NLPs are Nep1‐like proteins that have an NPP necrosis‐inducing Phytophthora protein (PFAM PF05630) domain. Secreted and believed to act within the plant cytoplasm. Induce a range of defence responses, including changes in gene expression, callose deposition, production of ethylene, nitric oxide, reactive oxygen species (ROS) and phytoalexins, and cell death. They promote pathogen growth and pathogenicity | Bailey et al. (2005); Feng and Li (2013); Kanneganti et al. (2006); Oome and Van Den Ackerveken (2014); Qutob et al. (2006); Santhanam et al. (2013) |
| RXLR effectors | |||
| 171 P. cinnamomi proteins have an RxLR or RxL(E/D/Q) motif and an N‐terminal SP. Most also have the EER motif |
P. infestans RxLR 122 RxLR‐containing genes described in Haas et al. (2009) |
Small secreted proteins containing an RxLR motif believed to be responsible for protein translocation across the plant plasma membrane RxLR effectors have diverse sequences and are likely to have diverse functions |
Haas et al. (2009) |
| PHYCI_87160 |
P. sojae Avr3b
PHYSO_286971 P. infestans Avr3b PITG_15732 PITG_15679 |
Effector containing RxLR, EER and Nudix domains. Nudix hydrolases are ADP‐ribose/NADH pyrophosphorylases In planta expression increases susceptibility to Phytophthora |
Dong et al. (2011) |
|
PHYCI_324245
‡
PHYCI_129006 PHYCI_568683 |
P. sojae
PsPSR2 PHYSO_290752 |
RxLR effectors that suppress gene silencing in plants and promote infection | Qiao et al. (2013); Xiong et al. (2014) |
|
PHYCI_24296 PHYCI_297058 |
P. infestans PiSNE1
PITG_13157 |
An RxLx effector that is targeted to the plant nucleus and that suppresses plant cell necrosis | Kelley et al. (2010) |
*Accession numbers of the two elicitin and two HAE genes described by Duclos et al. (1998) are listed. There are 32 homologous sequences in the P. cinnamomi genome with E‐values of <E‐05.
†The 14 P. cinnamomi sequences that contain an N‐terminal secretion signal and conserved LFLAK and DWL domains. These sequences are a subset of 280 P. cinnamomi sequences with homology to 45 representative P. infestans CRN proteins listed in Haas et al. (2009). The full list of 42 genes containing LFLAK and DWL domains is presented in Table S2 (see Supporting Information) with information on secretion motifs and possible N‐terminal truncation.
‡The top three sequences identified in the blast analysis.
GP42 transglutaminase
A 42‐kDa transglutaminase, GP42, was initially identified in P. sojae because of its elicitor activity (Nürnberger et al., 1994). Transglutaminases occur in multiple Phytophthora species and catalyse an acyl transfer reaction that renders peptide bonds more resistant to proteolytic degradation. A single P. cinnamomi transglutaminase sequence has been described (Martins et al., 2014b), but blast analysis with the two P. ramorum GP42 genes shows that there are 21 putative P. cinnamomi sequences (Table 6). GP42 transglutaminase homologues are expressed during early infection, suggesting that they may function during the establishment of Phytophthora disease (Brunner et al., 2002).
CRN crinklers
Members of the Crinkler (CRN) group of effectors were first identified amongst P. infestans genes that caused crinkling and necrosis when transiently expressed in N. benthamiana cells (Torto et al., 2003), although more recent studies have shown that the majority of CRN effectors suppress plant defence (Chen et al., 2013; Haas et al., 2009; Stam et al., 2013). CRN proteins have a conserved N‐terminal domain that includes LFLAK and DWL motifs that function in directing translocation of the CRN proteins from the apoplast into the plant cytoplasm (Schornack et al., 2010). Variable C‐terminal domains endow CRN proteins with a range of functions, including nuclear localization required for effector function (Liu et al., 2011; Mafurah et al., 2015; Van Damme et al., 2012).
All Phytophthora species examined so far have large multigene families of CRN genes (Haas et al., 2009; Tyler et al., 2006). pblast searches of the P. cinnamomi genome with 45 representative P. infestans CRN proteins from the list in Table S10 in Haas et al. (2009) identified 280 P. cinnamomi homologous sequences. Of these, 42 contain conserved LFLAK and DWL domains (Tables 6 and S3, see Supporting Information). Of the 42 proteins, 14 have an N‐terminal secretion signal and 11 have sequences associated with non‐classical secretion pathways. If the N‐terminal truncation of four additional proteins is caused by assembly errors, these proteins may also be secreted, giving a total of 29 putative P. cinnamomi CRN effectors. A P. cinnamomi CRN homologue, shown in the current genome assembly to be truncated at the N‐terminus, is the most highly expressed P. cinnamomi gene 5 days after inoculation of Eucalyptus nitens (Meyer et al., 2016). Thirteen of the 42 P. cinnamomi CRN homologues have a nuclear localization signal (NLS) in the C‐terminal region (Table S3). Further characterization of the cohort of 29 putative P. cinnamomi CRN proteins is likely to be an important area for future research. The CRN proteins may be critical factors in P. cinnamomi's success as a pathogen and its ability to infect such a wide range of plant species.
NLPs: Nep1‐like protein
A second group of secreted effectors believed to act within the plant cell cytoplasm and to cause plant cell necrosis is the Nep1‐like proteins (NLPs). NLPs occur widely among bacteria, fungi and Oomycetes (Gijzen and Nürnberger, 2006; Oome and Van Den Ackerveken, 2014). They contain a conserved NPP1 (necrosis‐inducing Phytophthora protein) domain. The NLP gene family is especially large in the Oomycetes. Phytophthora sojae and P. ramorum have been reported to possess 50–60 members, although more than one‐half are predicted to be pseudogenes (Gijzen and Nürnberger, 2006). Phytophthora cinnamomi has 72 putative NLP genes (Tables 6 and S4, see Supporting Information). NLPs may contribute to pathogen virulence by eliciting a range of plant defence responses or by acting as toxins (Bailey et al., 2005; Oome and Van Den Ackerveken, 2014; Qutob et al., 2002; Santhanam et al., 2013).
RxLR proteins
Phytophthora genomes include hundreds of genes that encode putative effectors containing an RxLR motif, believed to direct the passage of the effector from the plant apoplast into the plant cell cytoplasm (Dou et al., 2008b). Analysis of the genomes of P. infestans, P. ramorum and P. sojae has shown the presence of approximately 560, 370 and 390 RxLR‐containing, small secreted proteins, respectively (Haas et al., 2009; Jiang et al., 2008; Wang et al., 2011). The use of 122 of the P. infestans RxLR genes in pblast searches of the P. cinnamomi genome produced hits with 340 proteins which also have an N‐terminal signal peptide. Manual screening for the presence of RxLR and EER motifs resulted in the identification of 171 RxLR genes in the P. cinnamomi genome (Table 6). Despite the potential importance of RxLR proteins in the infection of plants by Phytophthora pathogens, to date, the molecular function of only a few RxLR effectors has been determined or their precise role in plant pathogenicity established. Of 10 Phytophthora RxLR effectors that have been functionally characterized, only three have homologues in the P. cinnamomi genome (Table 6).
Avr3b Nudix hydrolase
Seven RxLR genes in the Avr3b family in P. sojae contain a C‐terminal Nudix motif found in Nudix hydrolases, which catalyse the hydrolysis of nucleoside diphosphate derivatives (Dong et al., 2011). pblast searches showed that the P. cinnamomi genome contains a number of potential Avr3b homologues, but, in the current assembly, only sequence PHYCI_87160 has all four domains typical of Nudix hydrolases, i.e. a signal peptide, RxLR, EER and Nudix domains (Table 6). Three (PHYCI_257012, PHYCI_95983 and PHYCI_305326) of six other homologous P. cinnamomi sequences are missing 5′ data which might have included secretion signal and RxLR domains; one (PHYCI_194603) of the six sequences is missing 3′ data which might have included a Nudix domain; and two (PHYCI_130927 and PHYCI_552641) of the six sequences are missing 5′ and 3′ data. It seems likely that the absence of domains characteristic of Avr3b effectors from some or all of these six sequences is a result of assembly errors, meaning that there could be up to seven Avr3b homologues in P. cinnamomi.
In Arabidopsis, synthesis of a Nudix hydrolase, AtNUDT7, is induced by pathogen attack and acts as a negative regulator of plant defence against bacterial and Oomycete pathogens (Bartsch et al., 2006). It is suggested that Phytophthora Avr3b effectors may down‐regulate plant defence by mimicking endogenous plant Nudix hydrolases (Dong et al., 2011).
PSR suppressors of RNA silencing
A study of P. sojae RxLR effectors has identified two genes whose encoded proteins are able to suppress RNA silencing in plants (Xiong et al., 2014). The gene family, designated PSR (Phytophthora suppressor of RNA silencing), is reported to be conserved, with at least one gene present in a number of Phytophthora species, including P. cinnamomi (Xiong et al., 2014). pblast analysis in FungiDB, comparing one of the P. sojae PsPSR2 proteins (PHYSO_290752) with sequences in P. cinnamomi genomes, yielded a number of proteins with 30%–40% identity. It is thus surprising that the P. cinnamomi (var. robiniae) sequence listed in Table S1 in Xiong et al. (2014) is 94% identical to the P. sojae PHYSO_290752 sequence, but only 41% identical to the top P. cinnamomi (var. cinnamomi) hit. Nevertheless, there is evidence of a family of PSR2‐like proteins in P. cinnamomi (Table 6). Transient expression of PSR or PSR‐like genes in N. benthamiana increases their susceptibility to P. infestans infection, although details of the mechanism(s) involved are yet to be elucidated (Qiao et al., 2013).
PiSNE1 suppressor
A P. infestans protein with an RxLR‐like motif, PiSNE1 (P. infestans, suppressor of necrosis 1), is transcribed during the biotrophic phase of infection in tomato and suppresses the induction of plant cell necrosis and programmed cell death in response to pathogen attack (Kelley et al., 2010). The PiSNE1 protein has an NLS and is translocated to the plant nucleus. There are two putative homologues of PiSNE1 in the P. cinnamomi genome, both of which have an NLS according to the Plant‐PLoc algorithm (Table 6).
Putative effectors of pathogen defence strategies
Pathogens may attempt to avoid the triggering of host defences by masking or changing the sequence of elicitors recognized by the plant (Dodds et al., 2006; Fujikawa et al., 2012). If these strategies are unsuccessful, the pathogen will need to deal with multifaceted plant defences, possibly through a variety of mechanisms, including suppression, deactivation and tolerance.
Protection against reactive oxygen species (ROS)
To establish disease, pathogens must be able to survive an oxidative burst that may occur as part of the plant's defence response. The oxidative burst involves the production of reactive oxygen species, such as the superoxide anion ( ) and H2O2. To detoxify these molecules, phytopathogens produce superoxide dismutases (SODs) which convert to H2O2, and catalases and peroxidases that catalyse the breakdown of H2O2 to water and oxygen.
The P. cinnamomi genome contains three SOD genes (Table 7). Two SODs use manganese as a cofactor and the third uses copper/zinc ions. During plant–pathogen interactions, pathogen SOD gene expression may be up‐regulated, and encoded SODs act as pathogenicity factors (Rolke et al., 2004; Veluchamy et al., 2012). Plants, in turn, may be able to inhibit the function of pathogen SODs, as the activity of P. cinnamomi MnSODs has been reported to be lower in the presence of avocado root or cell wall components (Guzmán‐Deara et al., 2013).
Table 7.
Phytophthora cinnamomi proteins involved in plant–pathogen interactions: putative effectors involved in combating plant defences.
| P. cinnamomi gene name (in FungiDB) | Homologues used for blast query | Proposed protein function | References |
|---|---|---|---|
| Defence against reactive oxygen species (ROS) | |||
| PHYCI_89802 |
P. parasitica PnMnSOD2
PPTG_04295 |
Mitochondrial manganese superoxide dismutase | Blackman et al. (2005) |
| PHYCI_213407 |
P. parasitica PnMnSOD1a
PPTG_04112 |
Cytosolic manganese superoxide dismutase | Blackman et al. (2005) |
| PHYCI_74368 |
P. parasitica
PPTG_06915 |
Copper/zinc superoxide dismutase | Blackman et al. (2005) |
| PHYCI_316206 |
P. parasitica PnCat1
PPTG_06664 |
Catalase that is predicted to be cytosolic | Blackman and Hardham (2008) |
| PHYCI_94911 |
P. parasitica PnCat2
PPTG_06713 |
Catalase that is predicted to be targeted to peroxisomes | Blackman and Hardham (2008) |
| PHYCI_97994 |
P. parasitica PnCat3
PPTG_06866 |
Catalase that is predicted to be cytosolic | Blackman and Hardham (2008) |
| PHYCI_238395 |
P. parasitica PPTG_02738 PPTG_04280 |
Catalase‐peroxidases that are predicted to be secreted | Blackman and Hardham (2008) |
| PHYCI_112968 |
P. sojae PsMPK7
PHYSO_355777 |
Stress‐associated mitogen‐activated protein kinase (MAPK) Silencing > reduces ability to suppress plant ROS; reduces virulence |
Gao et al. (2015); Li et al. (2010) |
| Glucanase inhibitor proteins (GIPs) | |||
|
PHYCI_99638 PHYCI_585554 PHYCI_245084 PHYCI_322876 PHYCI_430555 PHYCI_99743 PHYCI_217079 |
P. parasitica GIPs PPTG_10494 PPTG_13057 PPTG_10494 PPTG_13156 PPTG_13062 |
Glucanase inhibitor proteins (GIPs). Chymotrypsin serine protease homologues that lack the catalytic triad of amino acids, His‐Asp‐Ser, required for protease activity. Differential expression during plant infection and evidence for co‐evolution with host endoglucanases | Damasceno et al. (2008); Martins et al. (2014a); Rose et al. (2002) |
| Kazal‐like protease inhibitors: EPI1–4 | |||
|
PHYCI_116215 PHYCI_313058 PHYCI_237404 PHYCI_150461 |
P. infestans EPI1
PITG_22681 |
Kazal‐like protease inhibitor During infection, P. infestans EPI1 inhibits the tomato P69B subtilisin (PR7). EPI1 and P69B genes have similar expression profiles |
Sels et al. (2008); Tian et al. (2004) |
|
PHYCI_15239 PHYCI_230761 PHYCI_247340 PHYCI_279459 PHYCI_296195 |
P. infestans EpI10
PITG_12129 |
Kazal‐like protease inhibitor that inhibits plant subtilisin A proteases | Chinnapun et al. (2009); Tian et al. (2005) |
| Cystatin‐like protease inhibitors: EPIC1–4 | |||
|
PHYCI_95628 PHYCI_313378 PHYCI_16414 PHYCI_247204 PHYCI_13396 PHYCI_97582 |
P. infestans EPIC2A
PITG_09175 P. infestans EPIC1 PITG_09169 |
Cystatin‐like protease inhibitors. Inhibit tomato papain‐like protease Rcr3 and potato papain‐like cysteine protease, C14 | Kaschani and van der Hoorn (2011); Song et al. (2009); Tian et al. (2007) |
The P. cinnamomi genome contains three catalase genes, each homologous to one of three PnCat genes in P. parasitica (Blackman and Hardham, 2008) (Table 7). In the current version of the P. cinnamomi genome, there are also one full‐length (PHYCI_238395) (Table 7) and two truncated (PHYCI_12861 and PHYCI_12862) catalase‐peroxidase genes, all of which have signal peptides directing their secretion.
The sensing of oxidative stress is typically followed by the activation of signal transduction pathways involving MAPKs. The expression of genes encoding two P. sojae MAPKs, PsSAK1 and PsMPK7, is induced by H2O2 (Gao et al., 2015; Li et al., 2010), and P. cinnamomi has homologues of both genes (Tables 1 and 7). Silencing of PsSAK1 impairs plant colonization (Li et al., 2010). Silencing of PsMPK7 leads to increased sensitivity to oxidative stress and reduced pathogenicity (Gao et al., 2015).
Inhibitors of plant endoglucanases (GIPs)
β‐1,3‐Glucans are key components of cell walls of Oomycetes and fungi. As part of their defence response, plants may produce endoglucanases that target pathogen wall β‐1,3‐glucans, not only weakening the cell wall, but also releasing glucan fragments that are potent elicitors of the defence response (Sels et al., 2008). Phytophthora species have evolved a counter‐defence strategy by producing β‐1,3‐glucanase inhibitor proteins (GIPs) that inhibit the activity of plant endo‐β‐1,3‐glucanases (York et al., 2004).
Interrogation of the P. cinnamomi genome with five P. parasitica GIP genes indicates that P. cinnamomi contains seven putative full‐length GIP genes (Table 7), as well as one partial sequence (PHYCI_92737). All lack the catalytic triad His–Asp–Ser. The full‐length genes have a secretion signal peptide at the N‐terminus. The partial gene is truncated at the N‐terminus, possibly as a result of sequencing or assembly errors. The expression of one of the full‐length genes, PHYCI_217079, increases during the first 24 h after inoculation of roots of chestnut before decreasing over the next 12 h (Martins et al., 2014a).
Protease inhibitors
Plant proteases play important roles in plant immunity. They are involved in pathogen detection, activation of defence responses and degradation of pathogen proteases (Bae et al., 2013; Figueiredo et al., 2014). Early recognition and proteolysis of pathogen proteins can release elicitors and induce other defence strategies. However, as part of the continuing arms race between hosts and pathogens, Phytophthora and other microorganisms have evolved proteinaceous inhibitors of plant proteases.
The first identified extracellular protease inhibitor (EPI) produced by a plant‐pathogenic microorganism was P. infestans EPI1 (Tian et al., 2004). EPI1 contains two Kazal‐like domains found in some serine protease inhibitor proteins. EPI1 specifically inhibits members of the subtilisin A class of serine proteases, including the tomato pathogenesis‐related protein P69B subtilase (Sels et al., 2008). The P. cinnamomi genome contains three full‐length (Table 7) and one truncated (PHYCI_150461) EPI1 homologue. The protein encoded by a second member of the P. infestans EPI multigene family, EPI10, also inhibits P69B activity (Tian et al., 2005). A homologue in P. palmivora, Ppepi10, inhibits subtilisin A and an uncharacterized rubber plant (Hevea brasiliensis) protease (Chinnapun et al., 2009). The P. cinnamomi genome contains five EPI10 homologues (Table 7).
A second family of P. infestans inhibitors, the cystatin‐like protease inhibitors (EPIC), contain cystatin‐like protease inhibitor domains (Song et al., 2009; Tian et al., 2007). Phytophthora infestans EPIC1 and EPIC2B inhibit the papain‐like tomato proteases PIP1 and Rcr3, and the potato papain‐like cysteine protease, C14 (Kaschani and van der Hoorn, 2011; Song et al., 2009). Phytophthora cinnamomi contains six full‐length (Table 7) and three truncated (PHYCI_237355, PHYCI_126736 and PHYCI_237330) EPIC homologues.
Geographical and Host Range
Improvements in the sensitivity and accuracy of detection and in identification techniques have confirmed that P. cinnamomi has spread throughout the world from its centre of origin in South‐east Asia, with the number of susceptible plant species now approaching 5000 (Jung et al., 2013). Phytophthora cinnamomi has been detected in 15 biodiversity hotspots, regions in which ‘exceptional concentrations of endemic species are undergoing exceptional loss of habitat’ (Myers et al., 2000), including the Mediterranean Basin, south‐west Western Australia and the fynbos of South Africa (Bezuidenhout et al., 2010).
The Mediterranean Basin contains more than 25 000 species of plant, about 50% of which are endemic to the region (Myers et al., 2000). Much of the area is populated by the evergreen oaks, Quercus ilex (holm oak) and Q. suber (cork oak), which are highly susceptible to infection by P. cinnamomi. Dehesa forests of Q. ilex and Q. suber within herbaceous pastures are the most widespread agroforestry land‐use system in Europe (Martín‐García et al., 2015), and dense Q. ilex forests comprise much of the maquis vegetation of the Mediterranean islands and archipelagos (Scanu et al., 2015). Over the last 30 years, extensive Mediterranean oak decline has been attributed primarily to P. cinnamomi (Camilo‐Alves et al., 2013; Linaldeddu et al., 2014; Martín‐García et al., 2015). P. cinnamomi is also one of two Phytophthora species responsible for the destructive ink disease of sweet chestnut (C. sativa), with serious economic consequences in Europe for timber and nut production and soil stability (Dal Maso and Montecchio, 2015). In south‐west Western Australia, over 3500 plant species, many of which are endemic, are susceptible to P. cinnamomi. P. cinnamomi is responsible for dramatic changes to the composition of plant communities over vast areas (Shearer et al., 2007) and threatens macrofungal and bird biodiversity in this region (Anderson et al., 2010; Davis et al., 2014). In South Africa, sclerophyllous shrubs in the Agathosma genus are part of the Cape Floral Kingdom, a small region in terms of land area, but an area which has the greatest known species richness (Cowling et al., 1996). Agathosma species are also grown for commercial use and their decline has been associated with infection by a number of soil‐borne Phytophthora species, including P. cinnamomi (Bezuidenhout et al., 2010).
It is predicted that global warming and climate change will increase P. cinnamomi disease problems in agriculture and forestry, and will exacerbate problems in biodiversity hotspots, including increasing the rate of extinction of endemic species (Burgess et al., 2017; Malcolm et al., 2006). The Mediterranean Basin and south‐west Western Australia are considered to be especially vulnerable. Experimental simulations of the effects of potential weather extremes on infection of holm oak by P. cinnamomi indicate that periods of drought following waterlogging will accelerate disease development and the rate of oak seedling mortality (Corcobado et al., 2014). Increased temperatures and soil water content will favour P. cinnamomi growth and dispersal, but accurate predictions of the effects of changing climatic conditions on disease development also need to take into account a range of other factors, such as effects on plant hosts. Mathematical models based on data from P. cinnamomi that utilize multiple parameters associated with climatic change and pathogen and plant responses are being developed (Dal Maso and Montecchio, 2015; Thompson et al., 2013). Globalization and increased movement of plant material throughout the world also provide enhanced opportunities, via genetic recombination, for the development of novel isolates with altered virulence and/or host range.
Integrated Disease Management Strategies
Key requirements for successful prediction, control and management of diseases include a thorough knowledge of the cellular and molecular basis of pathogen biology and pathogenicity; methods for rapid and sensitive detection and identification; multifaceted control procedures; and access to resistant plant cultivars. Our current understanding of P. cinnamomi biology and infection strategies has been described above. An overview of detection and control methods follows.
Detection and identification
The development of immunodiagnostic techniques based on antibodies that recognize pathogen molecules (antigens) is a major improvement over traditional methods which assess morphological and physiological characteristics (Hardham, 2005). However, the fact that the targeted antigen may not be present in all viable samples can lead to false negative results. DNA, by contrast, is present throughout an organism's lifecycle, and DNA‐based assays utilizing qPCR to amplify DNA have a number of advantages over immunodiagnostic techniques (Elliot et al., 2015; Engelbrecht et al., 2013; Eshraghi et al., 2011b; Vincelli and Tisserat, 2008). It should, however, be remembered that assays targeting DNA do not discriminate between viable and non‐viable material (O'Brien et al., 2009).
The sensitivity and specificity of qPCR‐based assays are influenced by the selection of the DNA sequence that is targeted and the design of the oligonucleotide primers that are used (O'Brien et al., 2009). Nested PCR assays which incorporate two rounds of amplification utilizing different primers may improve taxonomic discrimination and increase assay sensitivity (Engelbrecht et al., 2013; Schena et al., 2008). In multiplex assays, differentially tagged primer pairs have been used to simultaneously test for the presence of P. cinnamomi and other pathogen species in the same soil, water or plant samples (Kostov et al., 2016). Specific recognition of DNA sequences has also been used to detect P. cinnamomi in planta through the application of fluorescence in situ hybridization (Li et al., 2014).
Control strategies
As a result of the phylogenetic distance between Phytophthora and true fungi, most fungicides are ineffective and the choice of chemicals available for Phytophthora management is limited. The two chemicals most frequently used to control P. cinnamomi are metalaxyl (a phenylamide) and phosphite (a phosphonate and the active ingredient in fosetyl‐Al); however, their prolonged use has led to decreased sensitivity and the development of resistance (Dobrowolski et al., 2008; Hu et al., 2010).
Phosphite can control P. cinnamomi diseases directly through the inhibition of pathogen growth or indirectly through enhancement of plant defence. In vitro, phosphite induces cell wall lysis and hyphal abnormalities, and reduces hyphal growth and spore production or germination (King et al., 2010; McCarren et al., 2009a, 2009b). Initial investigations have documented the differential expression of 44 P. cinnamomi genes after phosphite treatment, including the down‐regulation of nine genes encoding putative cellulose synthases, a feature consistent with increased hyphal wall lysis (King et al., 2010; Wong et al., 2009).
Phosphite induction of plant defence varies depending on the plant species, but often allows normally susceptible plants to resist infection (Anderson et al., 2012; Daniel and Guest, 2006; Eshraghi et al., 2011a; Lim et al., 2013; Pilbeam et al., 2011; Shearer and Crane, 2012; Suddaby et al., 2008). Phosphite can be phytotoxic, and the mode and rate of its application need to be monitored and the most suitable methods used (Akinsanmi and Drenth, 2013; Anderson et al., 2012; Crane and Shearer, 2014; Gentile et al., 2009; Groves et al., 2015; Pilbeam et al., 2011; Scott et al., 2015; Shearer and Fairman, 2007; Stasikowski et al., 2014). Changes in the plant transcriptome and proteome induced by phosphite are now beginning to be explored (Burra et al., 2014; Eshraghi et al., 2011a, 2014; Lim et al., 2013).
The identification of biological control agents for P. cinnamomi has been limited (but see Bosso et al., 2016), although the application of mulches, composts or animal manure can inhibit P. cinnamomi growth, perhaps through the secretion of cellulase enzymes by microorganisms proliferating in the mulch (Aryantha and Guest, 2006; Evidente et al., 2009; Richter et al., 2011). The scale of P. cinnamomi distribution, its ability to survive for years in soil or symptomless plants, and the extent of plant susceptibility make the management of P. cinnamomi diseases challenging and difficult. The benefits of implementing integrated strategies have been demonstrated for the management of avocado root rot, the most important disease of avocado worldwide (Ramírez‐Gil et al., 2017). A combination of drenching in metalaxyl, mancozeb and silicate, with phosphite injection, addition of compost to the soil and mulching with organic material reduce disease occurrence and increase the production of first class fruit by almost 70% and 44%, respectively, relative to untreated controls.
Concluding Remarks
In the years leading up to the publication of the previous Pathogen Profile on P. cinnamomi in Molecular Plant Pathology in 2005, a major contributor to the then current state of knowledge was the application of immunological techniques to studies of P. cinnamomi cellular and molecular biology and the development of novel diagnostic assays. Immunocytochemistry has continued to expand our understanding but, over the last decade, additional approaches have helped to elucidate key aspects of P. cinnamomi biology and pathogenicity, such as those addressed by the questions posed at the beginning of this review article, relating to P. cinnamomi's extensive host range, infection strategies and potential targets for novel controls.
In the years leading up to the present Pathogen Profile on P. cinnamomi, a major component of our increased knowledge has been the release of the P. cinnamomi genome sequence. A central goal of this review article has been to place new molecular data obtained through bioinformatic analysis of the genomic information within the context of our understanding of the cellular biology of P. cinnamomi growth and infection. In so doing, a number of potentially important candidates for future research have been identified. For example, P. cinnamomi has the largest family of genes encoding SCR virulence factors reported to date. How critical is the role of SCR proteins in plant infection? Do they contribute to P. cinnamomi's wide host range? It is also clear that P. cinnamomi is well equipped to deal with induced plant defence, containing, as it does, abundant CRN, Avr3b and PsSNE1 homologues, all of which have been shown to suppress host defence in other Phytophthora species.
In the next phase of research on P. cinnamomi, it will be crucial to identify key pathogenicity genes and to determine the role of their encoded proteins in plant infection. One approach that will be central to this endeavour will be the application of next‐generation sequencing technologies, such as RNA‐Seq, to obtain comprehensive information on P. cinnamomi transcriptomes during development and plant infection.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Relative expression of selected Phytophthora cinnamomi genes before and after the induction of sporulation by transfer of mycelia to mineral salts solution. The graphs show data derived by the quantification of bands on RNA blots (Narayan, 2004) and are indicative of a number of different expression profiles during sporulation. Anx, annexin; CaM, calmodulin; Cen, centrin; EF1α, elongation factor 1α; GDH, glyceraldehyde‐3‐phosphate dehydrogenase; LDH, d‐lactate dehydrogenase; Pc, P. cinnamomi; Pdk, pyruvate phosphate dikinase; SAc, S‐adenosylhomocysteinase; Set1, SET1 transcription factor; Ubq, ubiquitin; STk, serine/threonine protein kinase.
Fig. S2 Transcript levels of three Phytophthora cinnamomi polygalacturonase genes in cultures growing in 5% V8 broth and defined media. Mycelia were bulked in media containing 2% glucose as the sole carbon source and then transferred to 5% V8 broth or one of six defined media. Transcript levels were measured using quantitative polymerase chain reaction (qPCR) (Dandipat, 2009).
Fig. S3 Transcript levels of three Phytophthora cinnamomi polygalacturonase genes in roots of lupin (Lupinus augustifolius) after inoculation. Transcript levels were measured using quantitative polymerase chain reaction (qPCR) (Dandipat, 2009).
Table S1 The number of genes in Carbohydrate‐Active enZyme (CAZyme) families in the genomes of Phytophthora cinnamomi (Pcin) and P. parasitica (Ppar).
Table S2 Putative Phytophthora cinnamomi elicitin genes. An initial group of 32 P. cinnamomi sequences with E‐values of <E‐05 was obtained by blast analysis of the P. cinnamomi genome in FungiDB with the β‐cinnamomin gene, PHYCI_98389. Subsequently, two additional P. cinnamomi sequences with high homology to PITG_06908 and PHYSO_30815 were identified. The table shows the P. infestans (or, in one case, P. sojae) elicitins to which the 34 P. cinnamomi sequences are most similar.
Table S3 Putative Crinkler (CRN) homologues in the Phytophthora cinnamomi genome. A pblast search using 45 CRN proteins representative of those reported in table S10 in Haas et al. (2009) resulted in the identification of 280 homologous sequences. These 280 sequences were analysed for the presence of conserved LFLAK and DWE motifs. This yielded a list of 42 putative P. cinnamomi CRN genes. These sequences were then analysed for the presence of a classical secretion signal (SP) or for evidence of secretion via a non‐classical pathway, as indicated by an NN score of >0.6. The list of 42 putative CRN genes includes six genes that are truncated at the N‐terminus in the current P. cinnamomi genome assembly. It also includes 13 genes that are not truncated at the N‐terminus, but for which there is no evidence for secretion. Genes were also analysed for the presence of a nuclear localization signal (NLS).
Table S4 Putative Phytophthora cinnamomi Nep1‐like protein (NLP) genes. Three P. sojae NLP genes, PHYSO_562453, PHYSO_509399 and PHYSO_249691, were used to blast the P. cinnamomi genome in FungiDB. This resulted in the identification of the 72 putative P. cinnamomi NLP genes listed here. The degree of homology of each of the P. cinnamomi genes to each of the three P. sojae genes is indicated by the scores and E‐values in the table. Genes shown with light or dark blue shading lack homology to one or two, respectively, of the three P. sojae genes. Only sequences with E‐values of <E‐05 are included.
References
- Ah‐Fong, A.M.V. and Judelson, H.S. (2011) New role for Cdc14 phosphatase: localization to basal bodies in the oomycete Phytophthora and its evolutionary coinheritance with eukaryotic flagella. PLoS One, 6, e16725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinsanmi, O.A. and Drenth, A. (2013) Phosphite and metalaxyl rejuvenate macadamia trees in decline caused by Phytophthora cinnamomi . Crop Protect. 53, 29–36. [Google Scholar]
- Akinsanmi, O.A. , Neal, J. , Drenth, A. and Topp, B. (2017) Characterization of accessions and species of macadamia to stem infection by Phytophthora cinnamomi . Plant Pathol. 66, 186–193. [Google Scholar]
- Anderson, J.M. , Pegg, K.G. , Scott, C. and Drenth, A. (2012) Phosphonate applied as a pre‐plant dip controls Phytophthora cinnamomi root and heart rot in susceptible pineapple hybrids. Aust. Plant Pathol. 41, 59–68. [Google Scholar]
- Anderson, P. , Brundrett, M. , Grierson, P. and Robinson, R. (2010) Impact of severe forest dieback caused by Phytophthora cinnamomi on macrofungal diversity in the northern jarrah forest of Western Australia. For. Ecol. Manage. 259, 1033–1040. [Google Scholar]
- Aryantha, N.P. and Guest, D.I. (2006) Mycoparasitic and antagonistic inhibition on Phytophthora cinnamomi Rands by microbial agents isolated from manure composts. Plant Pathol. J. 5, 291–298. [Google Scholar]
- Australian Government (2014) Threat Abatement Plan for Disease in Natural Ecosystems Caused by Phytophthora cinnamomi. Canberra: Commonwealth of Australia. [Google Scholar]
- Bae, C. , Kim, S.M. , Lee, D.J. and Choi, D. (2013) Multiple classes of immune‐related proteases associated with the cell death response in pepper plants. PLoS One, 8, e63533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, B.A. , Bae, H. , Strem, M.D. , Antúnez De Mayolo, G. , Guiltinan, M.J. , Verica, J.A. , Maximova, S.N. and Bowers, J.H. (2005) Developmental expression of stress response genes in Theobroma cacao leaves and their response to Nep1 treatment and a compatible infection by Phytophthora megakarya . Plant Physiol. Biochem. 43, 611–622. [DOI] [PubMed] [Google Scholar]
- Bartsch, M. , Gobbato, E. , Bednarek, P. , Debey, S. , Schultze, J.L. , Bautor, J. and Parker, J.E. (2006) Salicylic acid‐independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7 . Plant Cell, 18, 1038–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beakes, G.W. , Glockling, S.L. and Sekimoto, S. (2012) The evolutionary phylogeny of the oomycete “fungi”. Protoplasma, 249, 3–19. [DOI] [PubMed] [Google Scholar]
- Bezuidenhout, C.M. , Denman, S. , Kirk, S.A. , Botha, W.J. , Mostert, L. and McLeod, A. (2010) Phytophthora taxa associated with cultivated Agathosma, with emphasis on the P. citricola complex and P. capensis sp. nov. Persoonia: Mol. Phylogen. Evol. Fung. 25, 32–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman, L.M. and Hardham, A.R. (2008) Regulation of catalase activity and gene expression during Phytophthora nicotianae development and infection of tobacco. Mol. Plant Pathol. 9, 495–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman, L.M. , Mitchell, H.J. and Hardham, A.R. (2005) Characterisation of manganese superoxide dismutase from Phytophthora nicotianae . Mycol. Res. 109, 1171–1183. [DOI] [PubMed] [Google Scholar]
- Blackman, L.M. , Cullerne, D.P. and Hardham, A.R. (2014) Bioinformatic characterisation of genes encoding cell wall degrading enzymes in the Phytophthora parasitica genome. BMC Genomics, 15, 785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman, L.M. , Cullerne, D.P. , Torreña, P. , Taylor, J. and Hardham, A.R. (2015) RNA‐Seq analysis of the expression of genes encoding cell wall degrading enzymes during infection of lupin (Lupinus angustifolius) by Phytophthora parasitica . PLoS One, 10, e0136899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosso, L. , Scelza, R. , Varlese, R. , Meca, G. , Testa, A. , Rao, M.A. and Cristinzio, G. (2016) Assessing the effectiveness of Byssochlamys nivea and Scopulariopsis brumptii in pentachlorophenol removal and biological control of two Phytophthora species. Fungal Biol. 120, 645–653. [DOI] [PubMed] [Google Scholar]
- Brunner, F. , Rosahl, S. , Lee, J. , Rudd, J.J. , Geiler, C. , Kauppinen, S. , Rasmussen, Scheel, D. and Rnberger, N.T. (2002) Pep‐13, a plant defense‐inducing pathogen‐associated pattern from Phytophthora transglutaminases. EMBO J. 21, 6681–6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess, T.I. , Scott, J.K. , McDougall, K.L. , Stukely, M.J.C. , Crane, C. , Dunstan, W.A. , Brigg, F. , Andjic, V. , White, D. , Rudman, T. , Arentz, F. , Ota, N. and Hardy, G.E.S.J. (2017) Current and projected global distribution of Phytophthora cinnamomi, one of the world's worst plant pathogens. Global Change Biol. 23, 1661–1674. [DOI] [PubMed] [Google Scholar]
- Burra, D.D. , Berkowitz, O. , Hedley, P.E. , Morris, J. , Resjö, S. , Levander, F. , Liljeroth, E. , Andreasson, E. and Alexandersson, E. (2014) Phosphite‐induced changes of the transcriptome and secretome in Solanum tuberosum leading to resistance against Phytophthora infestans . BMC Plant Biol. 14, 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill, D.M. , Cope, M. and Hardham, A.R. (1996) Thrust reversal by tubular mastigonemes: immunological evidence for a role of mastigonemes in forward motion of zoospores of Phytophthora cinnamomi . Protoplasma, 194, 18–28. [Google Scholar]
- Cahill, D.M. , Rookes, J.E. , Wilson, B.A. , Gibson, L. and McDougall, K.L. (2008) Phytophthora cinnamomi and Australia's biodiversity: impacts, predictions and progress towards control. Aust. J. Bot. 56, 279–310. [Google Scholar]
- Caillaud, M.C. , Wirthmueller, L. , Sklenar, J. , Findlay, K. , Piquerez, S.J. , Jones, A.M. , Robatzek, S. , Jones, J.D. and Faulkner, C. (2014) The plasmodesmal protein PDLP1 localises to haustoria‐associated membranes during downy mildew infection and regulates callose deposition. PLoS Pathog. 10, e1004496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilo‐Alves, C.S. , da Clara, M.I.E. and de Almeida Ribeiro, N.M.C. (2013) Decline of Mediterranean oak trees and its association with Phytophthora cinnamomi: a review. Eur. J. For. Res. 132, 411–432. [Google Scholar]
- Cavalier‐Smith, T. and Chao, E.E.Y. (2006) Phylogeny and megasystematics of phagotrophic heterokonts (kingdom Chromista). J. Mol. Evol. 62, 388–420. [DOI] [PubMed] [Google Scholar]
- Chang, Y.H. , Yan, H.Z. and Liou, R.F. (2015) A novel elicitor protein from Phytophthora parasitica induces plant basal immunity and systemic acquired resistance. Mol. Plant Pathol. 16, 123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X.R. , Xing, Y.P. , Li, Y.P. , Tong, Y.H. and Xu, J.Y. (2013) RNA‐Seq reveals infection‐related gene expression changes in Phytophthora capsici . PLoS One, 8, e74588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X.‐R. , Li, Y.‐P. , Li, Q.‐Y. , Xing, Y.‐P. , Liu, B.‐B. , Tong, Y.‐H. and Xu, J.‐Y. (2015) SCR96, a small cysteine‐rich secretory protein of Phytophthora cactorum, can trigger cell death in the Solanaceae and is important for pathogenicity and oxidative stress tolerance. Mol. Plant Pathol. 17, 577–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnapun, D. , Tian, M. , Day, B. and Churngchow, N. (2009) Inhibition of a Hevea brasiliensis protease by a Kazal‐like serine protease inhibitor from Phytophthora palmivora . Physiol. Mol. Plant Pathol. 74, 27–33. [Google Scholar]
- Cooke, D.E.L. and Andersson, B. (2013) Phytophthora infestans and potato late blight in Europe In Phytophthora: A Global Perspective (Lamour K., ed.). pp. 59–67. Wallingford, Oxfordshire: CAB International. [Google Scholar]
- Cope, M. and Hardham, A.R. (1994) Synthesis and assembly of flagellar surface antigens during zoosporogenesis in Phytophthora cinnamomi . Protoplasma, 180, 158–168. [Google Scholar]
- Corcobado, T. , Cubera, E. , Juárez, E. , Moreno, G. and Solla, A. (2014) Drought events determine performance of Quercus ilex seedlings and increase their susceptibility to Phytophthora cinnamomi . Agr. Forest Meteorol. 192–193, 1–8. [Google Scholar]
- Cowling, R.M. , MacDonald, I.A.W. and Simmons, M.T. (1996) The Cape Peninsula, South Africa: physiographical, biological and historical background to an extraordinary hot‐spot of biodiversity. Biodivers. Conserv. 5, 527–550. [Google Scholar]
- Crane, C.E. and Shearer, B.L. (2014) Comparison of phosphite application methods for control of Phytophthora cinnamomi in threatened communities. Aust. Plant Pathol. 43, 143–149. [Google Scholar]
- Crone, M. , McComb, J.A. , O'Brien, P.A. and Hardy, G.E.S.J. (2013a) Annual and herbaceous perennial native Australian plant species are symptomless hosts of Phytophthora cinnamomi in the Eucalyptus marginata (jarrah) forest of Western Australia. Plant Pathol. 62, 1057–1062. [Google Scholar]
- Crone, M. , McComb, J.A. , O'Brien, P.A. and Hardy, G.E.S.J. (2013b) Survival of Phytophthora cinnamomi as oospores, stromata, and thick‐walled chlamydospores in roots of symptomatic and asymptomatic annual and herbaceous perennial plant species. Fungal Biol. 117, 112–123. [DOI] [PubMed] [Google Scholar]
- Damasceno, C.M.B. , Bishop, J.G. , Ripoll, D.R. , Win, J. , Kamoun, S. and Rose, J.K.C. (2008) Structure of the glucanase inhibitor protein (GIP) family from Phytophthora species suggests coevolution with plant endo‐β‐1,3‐glucanases. Mol. Plant–Microbe Interact. 21, 820–830. [DOI] [PubMed] [Google Scholar]
- Dal Maso, E. and Montecchio, L. (2015) Large‐scale fuzzy rule‐based prediction for suitable chestnut ink disease sites: a case study in north‐east Italy. Forest Pathol. 45, 311–323. [Google Scholar]
- Dandipat, N. (2009) Elucidating the role of polygalacturonases in the establishment of Phytophthora diseases. BSc Honours. Canberra, ACT: The Australian National University.
- Daniel, R. and Guest, D. (2006) Defence responses induced by potassium phosphonate in Phytophthora palmivora‐challenged Arabidopsis thaliana . Physiol. Mol. Plant Pathol. 67, 194–201. [Google Scholar]
- Davis, R.A. , Valentine, L.E. , Craig, M.D. , Wilson, B. , Bancroft, W.J. and Mallie, M. (2014) Impact of Phytophthora‐dieback on birds in Banksia woodlands in south west Western Australia. Biol. Conserv. 171, 136–144. [Google Scholar]
- De Lorenzo, G. , Salvi, G. , Degra, L. , D'Ovidio, R. and Cervone, F. (1987) Induction of extracellular polygalacturonase and its mRNA in the phytopathogenic fungus Fusarium moniliforme . J. Gen. Microbiol. 133, 3365–3373. [Google Scholar]
- Dobrowolski, M.P. , Shearer, B.L. , Colquhoun, I.J. , O'Brien, P.A. and Hardy, G.E.S. (2008) Selection for decreased sensitivity to phosphite in Phytophthora cinnamomi with prolonged use of fungicide. Plant Pathol. 57, 928–936. [Google Scholar]
- Dodds, P.N. , Lawrence, G.J. , Catanzariti, A.M. , Teh, T. , Wang, C.I.A. , Ayliffe, M.A. , Kobe, B. and Ellis, J.G. (2006) Direct protein interaction underlies gene‐for‐gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA, 103, 8888–8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, S. , Yin, W. , Kong, G. , Yang, X. , Qutob, D. , Chen, Q. , Kale, S.D. , Sui, Y. , Zhang, Z. , Dou, D. , Zheng, X. , Gijzen, M. , Tyler, B. and Wang, Y. (2011) Phytophthora sojae avirulence effector Avr3b is a secreted NADH and ADP‐ribose pyrophosphorylase that modulates plant immunity. PLoS Pathog. 7, e1002353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrance, A.E. (2013) Phytophthora sojae on soybean In Phytophthora: A Global Perspective (Lamour K., ed.). pp. 79–86. Wallingford, Oxfordshire: CAB International. [Google Scholar]
- Dou, D. , Kale, S.D. , Wang, X. , Chen, Y. , Wang, Q. , Wang, X. , Jiang, R.H.Y. , Arredondo, F.D. , Anderson, R.G. , Thakur, P.B. , McDowell, J.M. , Wang, Y. and Tyler, B.M. (2008a) Conserved C‐terminal motifs required for avirulence and suppression of cell death by Phytophthora sojae effector Avr1b. Plant Cell, 20, 1118–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou, D. , Kale, S.D. , Wang, X. , Jiang, R.H.Y. , Bruce, N.A. , Arredondo, F.D. , Zhang, X. and Tyler, B.M. (2008b) RXLR‐mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen‐encoded machinery. Plant Cell, 20, 1930–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, Y. , Mpina, M.H. , Birch, P.R.J. , Bouwmeester, K. and Govers, F. (2015) Phytophthora infestans RXLR effector AVR1 interacts with exocyst component Sec5 to manipulate plant immunity. Plant Physiol. 169, 1975–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclos, J. , Fauconnier, A. , Coelho, A.C. , Bollen, A. , Cravador, A. and Godfroid, E. (1998) Identification of an elicitin gene cluster in Phytophthora cinnamomi . Mitochon. DNA, 9, 231–237. [DOI] [PubMed] [Google Scholar]
- Ebadzad, G. , Medeira, C. , Maia, I. , Martins, J. and Cravador, A. (2015) Induction of defence responses by cinnamomins against Phytophthora cinnamomi in Quercus suber and Quercus ilex subs. rotundifolia . Eur. J. Plant Pathol. 143, 705–723. [Google Scholar]
- Elliot, M. , Schlenzig, A. , Harris, C.M. , Meagher, T.R. and Green, S. (2015) An improved method for qPCR detection of three Phytophthora spp. in forest and woodland soils in northern Britain. Forest Pathol. 45, 537–539. [Google Scholar]
- Engelbrecht, J. , Duong, T.A. and van den Berg, N. (2013) Development of a nested quantitative real‐time PCR for detecting Phytophthora cinnamomi in Persea americana rootstocks. Plant Dis. 97, 1012–1017. [DOI] [PubMed] [Google Scholar]
- Eshraghi, L. , Anderson, J. , Aryamanesh, N. , Shearer, B. , McComb, J. , Hardy, G.E.S. and O'Brien, P.A. (2011a) Phosphite primed defence responses and enhanced expression of defence genes in Arabidopsis thaliana infected with Phytophthora cinnamomi . Plant Pathol. 60, 1086–1095. [Google Scholar]
- Eshraghi, L. , Aryamanesh, N. , Anderson, J.P. , Shearer, B. , McComb, J.A. , Hardy, G.E.S.J. and O'Brien, P.A. (2011b) A quantitative PCR assay for accurate in planta quantification of the necrotrophic pathogen Phytophthora cinnamomi . Eur. J. Plant Pathol. 131, 419–430. [Google Scholar]
- Eshraghi, L. , Anderson, J.P. , Aryamanesh, N. , McComb, J.A. , Shearer, B. and Hardy, J. (2014) Defence signalling pathways involved in plant resistance and phosphite‐mediated control of Phytophthora cinnamomi . Plant Mol. Biol. Rep. 32, 342–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evidente, A. , Cristinzio, G. , Punzo, B. , Andolfi, A. , Testa, A. and Melck, D. (2009) Flufuran, an antifungal 3,5‐disubstituted furan produced by Aspergillus flavus Link. Chem. Biodivers. 6, 328–334. [DOI] [PubMed] [Google Scholar]
- Feng, B.Z. and Li, P.Q. (2013) Molecular characterization and functional analysis of the Nep1‐like protein‐encoding gene from Phytophthora capsici . Genet. Mol. Res. 12, 1468–1478. [DOI] [PubMed] [Google Scholar]
- Figueiredo, A. , Monteiro, F. and Sebastiana, M. (2014) Subtilisin‐like proteases in plant–pathogen recognition and immune priming: a perspective. Front. Plant Sci. 5, 739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikawa, T. , Sakaguchi, A. , Nishizawa, Y. , Kouzai, Y. , Minami, E. , Yano, S. , Koga, H. , Meshi, T. and Nishimura, M. (2012) Surface α‐1,3‐glucan facilitates fungal stealth infection by interfering with innate immunity in plants. PLoS Pathog. 8, e1002882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, J. , Cao, M. , Ye, W. , Li, H. , Kong, L. , Zheng, X. and Wang, Y. (2015) PsMPK7, a stress‐associated mitogen‐activated protein kinase (MAPK) in Phytophthora sojae, is required for stress tolerance, reactive oxygenated species detoxification, cyst germination, sexual reproduction and infection of soybean. Mol. Plant Pathol. 16, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile, S. , Valentino, D. and Tamietti, G. (2009) Control of ink disease by trunk injection of potassium phosphite. J. Plant Pathol. 91, 565–571. [Google Scholar]
- Gijzen, M. and Nürnberger, T. (2006) Nep1‐like proteins from plant pathogens: recruitment and diversification of the NPP1 domain across taxa. Phytochemistry, 67, 1800–1807. [DOI] [PubMed] [Google Scholar]
- Görnhardt, B. , Rouhara, I. and Schmelzer, E. (2000) Cyst germination proteins of the potato pathogen Phytophthora infestans share homology with human mucins. Mol. Plant–Microbe Interact. 13, 32–42. [DOI] [PubMed] [Google Scholar]
- Götesson, A. , Marshall, J.S. , Jones, D.A. and Hardham, A.R. (2002) Characterization and evolutionary analysis of a large polygalacturonase gene family in the oomycete plant pathogen Phytophthora cinnamomi . Mol. Plant–Microbe Interact. 15, 907–921. [DOI] [PubMed] [Google Scholar]
- Grenville‐Briggs, L.J. , Avrova, A.O. , Bruce, C.R. , Williams, A. , Whisson, S.C. , Birch, P.R.J. and Van West, P. (2005) Elevated amino acid biosynthesis in Phytophthora infestans during appressorium formation and potato infection. Fungal Genet. Biol. 42, 244–256. [DOI] [PubMed] [Google Scholar]
- Grenville‐Briggs, L.J. , Avrova, A.O. , Hay, R.J. , Bruce, C.R. , Whisson, S.C. and Van West, P. (2010) Identification of appressorial and mycelial cell wall proteins and a survey of the membrane proteome of Phytophthora infestans . Fungal Biol. 114, 702–723. [DOI] [PubMed] [Google Scholar]
- Groves, E. , Howard, K. , Hardy, G. and Burgess, T. (2015) Role of salicylic acid in phosphite‐induced protection against Oomycetes; a Phytophthora cinnamomi–Lupinus augustifolius model system. Eur. J. Plant Pathol. 141, 559–569. [Google Scholar]
- Gubler, F. and Hardham, A.R. (1988) Secretion of adhesive material during encystment of Phytophthora cinnamomi zoospores, characterized by immunogold labelling with monoclonal antibodies to components of peripheral vesicles. J. Cell Sci. 90, 225–235. [Google Scholar]
- Gubler, F. and Hardham, A.R. (1990) Protein storage in large peripheral vesicles in Phytophthora zoospores and its breakdown after cyst germination. Exp. Mycol. 14, 393–404. [Google Scholar]
- Guzmán‐Deara, J. , Reyes‐De la Cruz, H. , Beltrán‐Peña, E.M. , Castro‐Mercado, E. and García‐Pineda, E. (2013) Identification and characterization of superoxide dismutase in Phytophthora cinnamomi . Protoplasma, 250, 779–785. [DOI] [PubMed] [Google Scholar]
- Haas, B.J. , Kamoun, S. , Zody, M.C. , Jiang, R.H.Y. , Handsaker, R.E. , Cano, L.M. , Grabherr, M. , Kodira, C.D. , Raffaele, S. , Torto‐Alalibo, T. , Bozkurt, T.O. , Ah‐Fong, A.M.V. , Alvarado, L. , Anderson, V.L. , Armstrong, M.R. , Avrova, A. , Baxter, L. , Beynon, J. , Boevink, P.C. , Bollmann, S.R. , Bos, J.I.B. , Bulone, V. , Cai, G.H. , Cakir, C. , Carrington, J.C. , Chawner, M. , Conti, L. , Costanzo, S. , Ewan, R. , Fahlgren, N. , Fischbach, M.A. , Fugelstad, J. , Gilroy, E.M. , Gnerre, S. , Green, P.J. , Grenville‐Briggs, L.J. , Griffith, J. , Grunwald, N.J. , Horn, K. , Horner, N.R. , Hu, C.H. , Huitema, E. , Jeong, D.H. , Jones, A.M.E. , Jones, J.D.G. , Jones, R.W. , Karlsson, E.K. , Kunjeti, S.G. , Lamour, K. , Liu, Z.Y. , Ma, L.J. , Maclean, D. , Chibucos, M.C. , McDonald, H. , McWalters, J. , Meijer, H.J.G. , Morgan, W. , Morris, P.F. , Munro, C.A. , O'Neill, K. , Ospina‐Giraldo, M. , Pinzon, A. , Pritchard, L. , Ramsahoye, B. , Ren, Q.H. , Restrepo, S. , Roy, S. , Sadanandom, A. , Savidor, A. , Schornack, S. , Schwartz, D.C. , Schumann, U.D. , Schwessinger, B. , Seyer, L. , Sharpe, T. , Silvar, C. , Song, J. , Studholme, D.J. , Sykes, S. , Thines, M. , van de Vondervoort, P.J.I. , Phuntumart, V. , Wawra, S. , Weide, R. , Win, J. , Young, C. , Zhou, S.G. , Fry, W. , Meyers, B.C. , Van West, P. , Ristaino, J. , Govers, F. , Birch, P.R.J. , Whisson, S.C. , Judelson, H.S. and Nusbaum, C. (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans . Nature, 461, 393–398. [DOI] [PubMed] [Google Scholar]
- Hardham, A.R. (1987) Microtubules and the flagellar apparatus in zoospores and cysts of the fungus Phytophthora cinnamomi . Protoplasma, 137, 109–124. [Google Scholar]
- Hardham, A.R. (2005) Phytophthora cinnamomi . Mol. Plant Pathol. 6, 589–604. [DOI] [PubMed] [Google Scholar]
- Hardham, A.R. and Suzaki, E. (1986) Encystment of zoospores of the fungus, Phytophthora cinnamomi, is induced by specific lectin and monoclonal antibody binding to the cell surface. Protoplasma, 133, 165–173. [Google Scholar]
- Hardham, A.R. and Gubler, F. (1990) Polarity of attachment of zoospores of a root pathogen and pre‐alignment of the emerging germ tube. Cell Biol. Int. Rep. 14, 947–956. [Google Scholar]
- Hardham, A.R. , Cahill, D.M. , Cope, M. , Gabor, B.K. , Gubler, F. and Hyde, G.J. (1994) Cell surface antigens of Phytophthora spores: biological and taxonomic characterization. Protoplasma, 181, 213–232. [Google Scholar]
- Harper, J.D.I. , Gubler, F. , Salisbury, J.L. and Hardham, A.R. (1995) Centrin association with the flagellar apparatus in spores of Phytophthora cinnamomi . Protoplasma, 188, 225–235. [Google Scholar]
- Hee, W.Y. , Torrena, P.S. , Blackman, L.M. and Hardham, A.R. (2013) Phytophthora cinnamomi in Australia In Phytophthora: A Global Perspective (Lamour K., ed.). pp. 124–134. Wallingford, UK: CAB International. [Google Scholar]
- Hofzumahaus, S. and Schallmey, A. (2013) Escherichia coli‐based expression system for the heterologous expression and purification of the elicitin β‐cinnamomin from Phytophthora cinnamomi . Protein Expr. Purif. 90, 117–123. [DOI] [PubMed] [Google Scholar]
- Horta, M. , Sousa, N. , Coelho, A.C. , Neves, D. and Cravador, A. (2008) In vitro and in vivo quantification of elicitin expression in Phytophthora cinnamomi . Physiol. Mol. Plant Pathol. 73, 48–57. [Google Scholar]
- Horta, M. , Caetano, P. , Medeira, C. , Maia, I. and Cravador, A. (2010) Involvement of the β‐cinnamomin elicitin in infection and colonisation of cork oak roots by Phytophthora cinnamomi . Eur. J. Plant Pathol. 127, 427–436. [Google Scholar]
- Hu, J. , Hong, C. and Strmoberg, E.L. (2010) Mefenoxam sensitivity in Phytophthora cinnamomi isolates. Plant Dis. 94, 39–44. [DOI] [PubMed] [Google Scholar]
- Hua, C. , Wang, Y. , Zheng, X. , Dou, D. , Zhang, Z. , Govers, F. and Wang, Y. (2008) A Phytophthora sojae G‐protein α subunit is involved in chemotaxis to soybean isoflavones. Eukaryot. Cell, 7, 2133–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, C. , Meijer, H.J.G. , de Keijzer, J. , Zhao, W. , Wang, Y. and Govers, F. (2013) GK4, a G‐protein‐coupled receptor with a phosphatidylinositol phosphate kinase domain in Phytophthora infestans, is involved in sporangia development and virulence. Mol. Microbiol. 88, 352–370. [DOI] [PubMed] [Google Scholar]
- Hugouvieux, V. , Centis, S. , Lafitte, C. and Esquerré‐Tugayé, M.T. (1995) Characterization of Colletotrichum lindemuthianum endopolygalacturonase with molecular probes. CR Acad. Sci. III‐VIE, 318, 113–120. [Google Scholar]
- Hyde, G.L. and Hardham, A.R. (1992) Confocal microscopy of microtubule arrays in cryosectioned sporangia of Phytophthora cinnamomi . Exp. Mycol. 16 207–218. [Google Scholar]
- Hyde, G.J. and Hardham, A.R. (1993) Microtubules regulate the generation of polarity in zoospores of Phytophthora cinnamomi . Eur. J. Cell Biol. 62, 75–85. [PubMed] [Google Scholar]
- Jackson, S.L. and Hardham, A.R. (1996) A transient rise in cytoplasmic free calcium is required for the induction of cytokinesis in zoosporangia of Phytophthora cinnamomi . Eur. J. Cell Biol. 69, 180–188. [PubMed] [Google Scholar]
- Jackson, S.L. and Hardham, A.R. (1998) Dynamic rearrangement of the filamentous actin network occurs during zoosporogenesis and encystment in the oomycete Phytophthora cinnamomi . Fungal Genet. Biol. 24, 24–33. [DOI] [PubMed] [Google Scholar]
- Jiang, R.H.Y. , Tyler, B.M. , Whisson, S.C. , Hardham, A.R. and Govers, F. (2006) Ancient origin of elicitin gene clusters in Phytophthora genomes. Mol. Biol. Evol. 23, 338–351. [DOI] [PubMed] [Google Scholar]
- Jiang, R.H.Y. , Tripathy, S. , Govers, F. and Tyler, B.M. (2008) RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc. Natl. Acad. Sci. USA, 105, 4874–4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, R.W. and Wang, H. (1997) Immunolocalization of a α‐1,4‐endoglucanase from Macrophomina phaseolina expressed in planta. Can. J. Microbiol. 43, 491–495. [Google Scholar]
- Judelson, H.S. and Tani, S. (2007) Transgene‐induced silencing of the zoosporogenesis‐specific NIFC gene cluster of Phytophthora infestans involves chromatin alterations. Eukaryot. Cell, 6, 1200–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judelson, H.S. , Narayan, R. , Ah‐Fong, A.M.V. and Kim, K.S. (2009) Gene expression changes during asexual sporulation by the late blight agent Phytophthora infestans occur in discrete temporal stages. Mol. Genet. Genomics, 281, 193–206. [DOI] [PubMed] [Google Scholar]
- Jung, T. , Colquhoun, I.J. and Hardy, G.E.S.J. (2013) New insights into the survival strategy of the invasive soilborne pathogen Phytophthora cinnamomi in different natural ecosystems in Western Australia. Forest Pathol. 43, 266–288. [Google Scholar]
- Kamoun, S. , Furzer, O. , Jones, J.D.G. , Judelson, H.S. , Ali, G.S. , Dalio, R.J.D. , Roy, S.G. , Schena, L. , Zambounis, A. , Panabières, F. , Cahill, D. , Ruocco, M. , Figueiredo, A. , Chen, X.R. , Hulvey, J. , Stam, R. , Lamour, K. , Gijzen, M. , Tyler, B.M. , Grünwald, N.J. , Mukhtar, M.S. , Tomé, D.F.A. , Tör, M. , Van Den Ackerveken, G. , McDowell, J. , Daayf, F. , Fry, W.E. , Lindqvist‐Kreuze, H. , Meijer, H.J.G. , Petre, B. , Ristaino, J. , Yoshida, K. , Birch, P.R.J. and Govers, F. (2015) The Top 10 oomycete pathogens in molecular plant pathology. Mol. Plant Pathol. 16, 413–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanneganti, T.D. , Huitema, E. , Cakir, C. and Kamoun, S. (2006) Synergistic interactions of the plant cell death pathways induced by Phytophthora infestans Nep1‐like protein PiNPP1.1 and INF1 elicitin. Mol. Plant–Microbe Interact. 19, 854–863. [DOI] [PubMed] [Google Scholar]
- Kaschani, F. and van der Hoorn, R.A.L. (2011) A model of the C14‐EPIC complex indicates hotspots for a protease‐inhibitor arms race in the oomycete–potato interaction. Plant Signal. Behav. 6, 109–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, B.S. , Lee, S.J. , Damasceno, C.M.B. , Chakravarthy, S. , Kim, B.D. , Martin, G.B. and Rose, J.K.C. (2010) A secreted effector protein (SNE1) from Phytophthora infestans is a broadly acting suppressor of programmed cell death. Plant J. 62, 357–366. [DOI] [PubMed] [Google Scholar]
- King, M. , Reeve, W. , Van der Hoek, M.B. , Williams, N. , McComb, J. , O'Brien, P.A. and Hardy, G.E.S.J. (2010) Defining the phosphite‐regulated transcriptome of the plant pathogen Phytophthora cinnamomi . Mol. Genet. Genomics, 284, 425–435. [DOI] [PubMed] [Google Scholar]
- Kostov, K. , Verstappen, E.C.P. , Bergervoet, J.H.W. , de Weerdt, M. , Schoen, C.D. , Slavov, S. and Bonants, P.J.M. (2016) Multiplex detection and identification of Phytophthora spp. using target‐specific primer extension and Luminex xTAG technology. Plant Pathol. 65, 1008–1021. [Google Scholar]
- Larousse, M. , Govetto, B. , Séassau, A. , Etienne, C. , Industri, B. , Theodorakopoulos, N. , Deleury, E. , Ponchet, M. , Panabières, F. and Galiana, E. (2014) Characterization of PPMUCL1/2/3, three members of a new Oomycete‐specific mucin‐like protein family residing in Phytophthora parasitica biofilm. Protist, 165, 275–292. [DOI] [PubMed] [Google Scholar]
- Larroque, M. , Barriot, R. , Bottin, A. , Barre, A. , Rougé, P. , Dumas, B. and Gaulin, E. (2012) The unique architecture and function of cellulose‐interacting proteins in oomycetes revealed by genomic and structural analyses. BMC Genomics, 13, 605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latijnhouwers, M. and Govers, F. (2003) A Phytophthora infestans G‐protein β‐subunit is involved in sporangium formation. Eukaryot. Cell, 2, 971–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latijnhouwers, M. , Ligterink, W. , Vleeshouwers, V.G.A.A. , van West, P. and Govers, F. (2004) A Gα subunit controls zoospore motility and virulence in the potato late blight pathogen Phytophthora infestans . Mol. Microbiol. 51, 925–936. [DOI] [PubMed] [Google Scholar]
- Li, A. , Wang, Y. , Tao, K. , Dong, S. , Huang, Q. , Dai, T. , Zheng, X. and Wang, Y. (2010) PsSAK1, a stress‐activated MAP kinase of Phytophthora sojae, is required for zoospores viability and infection of soybean. Mol. Plant–Microbe Interact. 23, 1022–1031. [DOI] [PubMed] [Google Scholar]
- Li, A. , Zhang, M. , Wang, Y. , Li, D. , Liu, X. , Tao, K. , Ye, W. and Wang, Y. (2014) PsMPK1, an SLT2‐type mitogen‐activated protein kinase, is required for hyphal growth, zoosporogenesis, cell wall integrity, and pathogenicity in Phytophthora sojae . Fungal Genet. Biol. 65, 14–24. [DOI] [PubMed] [Google Scholar]
- Li, D. , Zhao, Z. , Huang, Y. , Lu, Z. , Yao, M. , Hao, Y. , Zhai, C. and Wang, Y. (2013) PsVPS1, a dynamin‐related protein, is involved in cyst germination and soybean infection of Phytophthora sojae . PLoS One, 8, e58623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, S. , Borza, T. , Peters, R.D. , Coffin, R.H. , Al‐Mughrabi, K.I. , Pinto, D.M. and Wang‐Pruski, G. (2013) Proteomics analysis suggests broad functional changes in potato leaves triggered by phosphites and a complex indirect mode of action against Phytophthora infestans . J. Proteomics, 93, 207–223. [DOI] [PubMed] [Google Scholar]
- Linaldeddu, B.T. , Scanu, B. , Maddau, L. and Franceschini, A. (2014) Diplodia corticola and Phytophthora cinnamomi: the main pathogens involved in holm oak decline on Caprera Island (Italy). Forest Pathol. 44, 191–200. [Google Scholar]
- Liu, T. , Ye, W. , Ru, Y. , Yang, X. , Gu, B. , Tao, K. , Lu, S. , Dong, S. , Zheng, X. , Shan, W. , Wang, Y. and Dou, D. (2011) Two host cytoplasmic effectors are required for pathogenesis of Phytophthora sojae by suppression of host defences. Plant Physiol. 155, 490–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mafurah, J.J. , Ma, H. , Zhang, M. , Xu, J. , He, F. , Ye, T. , Shen, D. , Chen, Y. , Rajput, N.A. and Dou, D. (2015) A virulence essential CRN effector of Phytophthora capsici suppresses host defense and induces cell death in plant nucleus. PLoS One, 10, e0127965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia, I. , Medeira, C. , Melo, E. and Cravador, A. (2008) Quercus suber infected by Phytophthora cinnamomi. Effects at cellular level of cinnamomin on roots, stem and leaves. Microsc. Microanal. 14, 146–147. [Google Scholar]
- Maia, I. , Horta, M. , Cravador, A. and Medeira, C. (2012) Loss of aggressiveness of Phytophthora cinnamomi (beta‐cinnamomin silenced strain) in the infection of Castanea sativa . Microsc. Microanal. 18, 17–18. [Google Scholar]
- Malcolm, J.R. , Liu, C. , Neilson, R.P. , Hansen, L. and Hannah, L. (2006) Global warming and extinctions of endemic species from biodiversity hotspots. Conserv. Biol. 20, 538–548. [DOI] [PubMed] [Google Scholar]
- Manatunga, V. , Sanati, H. , Tan, P. and O'Brien, P. (2005) Maceration of plant tissue by fungi is inhibited by recombinant antipectinase antibodies. Eur. J. Plant Pathol. 112, 211–220. [Google Scholar]
- Marshall, J.S. , Ashton, A.R. , Govers, F. and Hardham, A.R. (2001a) Isolation and characterization of four genes encoding pyruvate, phosphate dikinase in the oomycete plant pathogen Phytophthora cinnamomi . Curr. Genet. 40, 73–81. [DOI] [PubMed] [Google Scholar]
- Marshall, J.S. , Wilkinson, J.M. , Moore, T. and Hardham, A.R. (2001b) Structure and expression of the genes encoding proteins resident in large peripheral vesicles of Phytophthora cinnamomi zoospores. Protoplasma, 215, 226–239. [DOI] [PubMed] [Google Scholar]
- Martel, M.B. , Létoublon, R. and Fěvre, M. (1996) Purification of endo polygalacturonases from Sclerotinia sclerotiorum: multiplicity of the complex enzyme system. Curr. Microbiol. 33, 243–248. [DOI] [PubMed] [Google Scholar]
- Martín‐García, J. , Solla, A. , Corcobado, T. , Siasou, E. and Woodward, S. (2015) Influence of temperature on germination of Quercus ilex in Phytophthora cinnamomi, P. gonapodyides, P. quercina and P. psychrophila infested soils. Forest Pathol. 45, 215–223. [Google Scholar]
- Martins, I.M. , Martins, F. , Belo, H. , Vaz, M. , Carvalho, M. , Cravador, A. and Choupina, A. (2014a) Cloning, characterization and in vitro and in planta expression of a glucanase inhibitor protein (GIP) of Phytophthora cinnamomi . Mol. Biol. Rep. 41, 2453–2462. [DOI] [PubMed] [Google Scholar]
- Martins, I.M. , Matos, M. , Costa, R. , Silva, F. , Pascoal, A. , Estevinho, L.M. and Choupina, A.B. (2014b) Transglutaminases: recent achievements and new sources. Appl. Microbiol. Biotechnol. 98, 6957–6964. [DOI] [PubMed] [Google Scholar]
- McCarren, K.L. , McComb, J.A. , Shearer, B.L. and Hardy, G.E.S.J. (2009a) In vitro influence of phosphite on chlamydospore production and viability of Phytophthora cinnamomi . Forest Pathol. 39, 210–216. [Google Scholar]
- McCarren, K.L. , McComb, J.A. , Shearer, B.L. and Hardy, G.E.S.J. (2009b) Phosphite impact on the in vitro production and viability of selfed oospores by Phytophthora cinnamomi . Forest Pathol. 39, 124–132. [Google Scholar]
- McConnell, M.E. and Balci, Y. (2015) Fine root dynamics of oak saplings in response to Phytophthora cinnamomi infection under different temperatures and durations. Forest Pathol. 45, 155–164. [Google Scholar]
- Medeira, C. , Maia, I. , Ribeiro, C. , Candeias, I. , Melo, E. , Sousa, N. and Cravador, A. (2012) Alpha cinnamomin elicits a defence response against Phytophthora cinnamomi in Castanea sativa . Acta Hortic. 940, 315–322. [Google Scholar]
- Meijer, H.J.G. , van de Vondervoort, P.J.I. , Yin, Q.Y. , de Koster, C.G. , Klis, F.M. , Govers, F. and de Groot, P.W.J. (2006) Identification of cell wall‐associated proteins from Phytophthora ramorum . Mol. Plant–Microbe Interact. 19, 1348–1358. [DOI] [PubMed] [Google Scholar]
- Mélida, H. , Sandoval‐Sierra, J.V. , Diéguez‐Uribeondo, J. and Bulone, V. (2013) Analyses of extracellular carbohydrates in oomycetes unveil the existence of three different cell wall types. Eukaryot. Cell, 12, 194–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, Y. , Zhang, Q. , Zhang, M. , Gu, B. , Huang, G. , Wang, Q. and Shan, W. (2015) The protein disulfide isomerase 1 of Phytophthora parasitica (PpPDI1) is associated with the haustoria‐like structures and contributes to plant infection. Front. Plant Sci. 6, 632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, F.E. , Shuey, L.S. , Naidoo, S. , Mamni, T. , Berger, D.K. , Myburg, A.A. , van den Berg, N. and Naidoo, S. (2016) Dual RNA‐sequencing of Eucalyptus nitens during Phytophthora cinnamomi challenge reveals pathogen and host factors influencing compatibility. Front. Plant Sci. 7, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, N. , Mittermeler, R.A. , Mittermeler, C.G. , Da Fonseca, G.A.B. and Kent, J. (2000) Biodiversity hotspots for conservation priorities. Nature, 403, 853–858. [DOI] [PubMed] [Google Scholar]
- Narayan, R. (2004) Characterization of pre‐sporangium stage sporulation genes in the Oomycete plant pathogen Phytophthora cinnamomi. PhD. Canberra, ACT: The Australian National University.
- Narayan, R.D. , Blackman, L.M. , Shan, W.X. and Hardham, A.R. (2010) Phytophthora nicotianae transformants lacking dynein light chain 1 produce non‐flagellate zoospores. Fungal Genet. Biol. 47, 663–671. [DOI] [PubMed] [Google Scholar]
- Nicastro, G. , Orsomando, G. , Ferrari, E. , Manconi, L. , Desario, F. , Amici, A. , Naso, A. , Carpaneto, A. , Pertinhez, T.A. , Ruggieri, S. and Spisni, A. (2009) Solution structure of the phytotoxic protein PcF: the first characterized member of the Phytophthora PcF toxin family. Protein Sci. 18, 1786–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nürnberger, T. , Nennstiel, D. , Jabs, T. , Sacks, W.R. , Hahlbrock, K. and Scheel, D. (1994) High affinity binding of a fungal oligopeptide elicitor to parsley plasma membranes triggers multiple defense responses. Cell, 78, 449–460. [DOI] [PubMed] [Google Scholar]
- O'Brien, P.A. , Williams, N. and Hardy, G.E.S. (2009) Detecting Phytophthora . Crit. Rev. Microbiol. 35, 169–181. [DOI] [PubMed] [Google Scholar]
- O'Gara, E. , Howard, K. , Wilson, B. and Hardy, G.E.S. (2005) Management of Phytophthora cinnamomi for biodiversity conservation in Australia: Part 1 – A review of current management. A report funded by the Commonwealth Government Department of the Environment and Heritage by the Centre for Phytophthora Science and Management, Murdoch University, Western Australia.
- O'Gara, E. , Howard, K. , McComb, J. , Colquhoun, I.J. and Hardy, G.E.S.J. (2015) Penetration of suberized periderm of a woody host by Phytophthora cinnamomi . Plant Pathol. 64, 207–215. [Google Scholar]
- Oome, S. and Van Den Ackerveken, G. (2014) Comparative and functional analysis of the widely occurring family of Nep1‐like proteins. Mol. Plant–Microbe Interact. 27, 1081–1094. [DOI] [PubMed] [Google Scholar]
- Orsomando, G. , Lorenzi, M. , Raffaelli, N. , Rizza, M.D. , Mezzetti, B. and Ruggieri, S. (2001) Phytotoxic protein PcF, purification, characterization, and cDNA sequencing of a novel hydroxyproline‐containing factor secreted by the strawberry pathogen Phytophthora cactorum . J. Biol. Chem. 276, 21 578–21 584. [DOI] [PubMed] [Google Scholar]
- Orsomando, G. , Brunetti, L. , Pucci, K. , Ruggeri, B. and Ruggieri, S. (2011) Comparative structural and functional characterization of putative protein effectors belonging to the PcF toxin family from Phytophthora spp. Protein Sci. 20, 2047–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman, H. , Vauthrin, S. , Mikes, V. , Milat, M.L. , Panabières, F. , Marais, A. , Brunie, S. , Maume, B. , Ponchet, M. and Blein, J.P. (2001) Mediation of elicitin activity on tobacco is assumed by elicitin‐sterol complexes. Mol. Biol. Cell, 12, 2825–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ospina‐Giraldo, M.D. , Griffith, J.G. , Laird, E.W. and Mingora, C. (2010) The CAZyome of Phytophthora spp.: a comprehensive analysis of the gene complement coding for carbohydrate‐active enzymes in species of the genus Phytophthora . BMC Genomics, 11, 525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oßwald, W. , Fleischmann, F. , Rigling, D. , Coelho, A.C. , Cravador, A. , Diez, J. , Dalio, R.J. , Horta Jung, M. , Pfanz, H. , Robin, C. , Sipos, G. , Solla, A. , Cech, T. , Chambery, A. , Diamandis, S. , Hansen, E. , Jung, T. , Orlikowski, L.B. , Parke, J. , Prospero, S. and Werres, S. (2014) Strategies of attack and defence in woody plant–Phytophthora interactions. Forest Pathol. 44, 169–190. [Google Scholar]
- Pilbeam, R.A. , Howard, K. , Shearer, B.L. and Hardy, G.E.S.J. (2011) Phosphite stimulated histological responses of Eucalyptus marginata to infection by Phytophthora cinnamomi . Trees Struct. Funct. 25, 1121–1131. [Google Scholar]
- Ploetz, R.C. (2013) Phytophthora root rot of Avocado In: Phytophthora: A Global Perspective (Lamour K., ed.). pp. 197–203. Wallingford, Oxfordshire: CAB International. [Google Scholar]
- Qiao, Y. , Liu, L. , Xiong, Q. , Flores, C. , Wong, J. , Shi, J. , Wang, X. , Liu, X. , Xiang, Q. , Jiang, S. , Zhang, F. , Wang, Y. , Judelson, H.S. , Chen, X. and Ma, W. (2013) Oomycete pathogens encode RNA silencing suppressors. Nat. Genet. 45, 330–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qutob, D. , Kamoun, S. and Gijzen, M. (2002) Expression of a Phytophthora sojae necrosis‐inducing protein occurs during transition from biotrophy to necrotrophy. Plant J. 32, 361–373. [DOI] [PubMed] [Google Scholar]
- Qutob, D. , Kemmerling, B. , Brunner, F. , Kufner, I. , Engelhardt, S. , Gust, A.A. , Luberacki, B. , Seitz, H.U. , Stahl, D. , Rauhut, T. , Glawischnig, E. , Schween, G. , Lacombe, B. , Watanabe, N. , Lam, E. , Schlichting, R. , Scheel, D. , Nau, K. , Dodt, G. , Hubert, D. , Gijzen, M. and Nürnberger, T. (2006) Phytotoxicity and innate immune responses induced by Nep1‐like proteins. Plant Cell, 18, 3721–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez‐Gil, J.G. , Castañeda‐Sánchez, D.A. and Morales‐Osorio, J.G. (2017) Production of avocado trees infected with Phytophthora cinnamomi under different management regimes. Plant Pathol. 66, 623–632. [Google Scholar]
- Redondo, M.Á. , Pérez‐Sierra, A. , Abad‐Campos, P. , Torres, L. , Solla, A. , Reig‐Armiñana, J. and García‐Breijo, F. (2015) Histology of Quercus ilex roots during infection by Phytophthora cinnamomi . Trees Struct. Funct. 29, 1943–1957. [Google Scholar]
- Reeksting, B.J. , Olivier, N.A. and van den Berg, N. (2016) Transcriptome responses of an ungrafted Phytophthora root rot tolerant avocado (Persea americana) rootstock to flooding and Phytophthora cinnamomi . BMC Plant Biol. 16, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitmann, A. , Berger, D.K. and van Den Berg, N. (2016) Putative pathogenicity genes of Phytophthora cinnamomi identified via RNA‐Seq analysis of pre‐infection structures. Eur. J. Plant Pathol. 147, 211–228. [Google Scholar]
- Ribeiro, O.K. (1978) A Source Book of the Genus Phytophthora. Cornell University: J. Cramer. [Google Scholar]
- Richter, B.S. , Ivors, K. , Shi, W. and Benson, D.M. (2011) Cellulase activity as a mechanism for suppression of Phytophthora root rot in mulches. Phytopathology, 101, 223–230. [DOI] [PubMed] [Google Scholar]
- Robold, A.V. and Hardham, A.R. (2005) During attachment Phytophthora spores secrete proteins containing thrombospondin type 1 repeats. Curr. Genet. 47, 307–315. [DOI] [PubMed] [Google Scholar]
- Rodrigues, M.L. , Archer, M. , Martel, P. , Miranda, S. , Thomaz, M. , Enguita, F.J. , Baptista, R.P. , Pinho e Melo, E. , Sousa, N. , Cravador, A. and Carrondo, M.A. (2006) Crystal structures of the free and sterol‐bound forms of β‐cinnamomin. Biochim. Biophys. Acta, Proteins Proteomics, 1764, 110–121. [DOI] [PubMed] [Google Scholar]
- Rolke, Y. , Liu, S. , Quidde, T. , Williamson, B. , Schouten, A. , Weltring, K.M. , Siewers, V. , Tenberge, K.B. , Tudzynski, B. and Tudzynski, P. (2004) Functional analysis of H2O2‐generating systems in Botyris cinerea: the major Cu–Zn‐superoxide dismutase (BCSOD1) contributes to virulence on French bean, whereas a glucose oxidase (BCGOD1) is dispensible. Mol. Plant Pathol. 5, 17–27. [DOI] [PubMed] [Google Scholar]
- Rose, J.K.C. , Ham, K.S. , Darvill, A.G. and Albersheim, P. (2002) Molecular cloning and characterization of glucanase inhibitor proteins: coevolution of a counterdefence mechanism by plant pathogens. Plant Cell 14, 1329–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz Gómez, F.J. , Navarro‐Cerrillo, R.M. , Sánchez‐Cuesta, R. and Pérez‐de‐Luque, A. (2015) Histopathology of infection and colonization of Quercus ilex fine roots by Phytophthora cinnamomi . Plant Pathol. 64, 605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhanam, P. , Van Esse, H.P. , Albert, I. , Faino, L. , Nürnberger, T. and Thomma, B.P.H.J. (2013) Evidence for functional diversification within a fungal Nep1‐like protein family. Mol. Plant–Microbe Interact. 26, 278–286. [DOI] [PubMed] [Google Scholar]
- Savory, A.I.M. , Grenville‐Briggs, L.J. , Wawra, S. , Van West, P. and Davidson, F.A. (2014) Auto‐aggregation in zoospores of Phytophthora infestans: the cooperative roles of bioconvection and chemotaxis. J. R. Soc. Interface, 11, 20140017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanu, B. , Linaldeddu, B.T. , Deidda, A. and Jung, T. (2015) Diversity of Phytophthora species from declining Mediterranean maquis vegetation, including two new species, Phytophthora crassamura and P. ornamentata sp. nov. PLoS One, 10, e0143234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena, L. , Duncan, J.M. and Cooke, D.E.L. (2008) Development and application of a PCR‐based 'molecular tool box' for the identification of Phytophthora species damaging forests and natural ecosystems. Plant Pathol. 57, 64–75. [Google Scholar]
- Schornack, S. , Van Damme, M. , Bozhurt, T.O. , Cano, L.M. , Smoker, M. , Thines, M. , Gaulin, E. , Kamoun, S. and Huitema, E. (2010) Ancient class of translocated oomycete effectors targets the host nucleus. Proc. Natl. Acad. Sci. USA, 107, 17 421–17 426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, P.M. , Barber, P.A. and Hardy, G.E.S.J. (2015) Novel phosphite and nutrient application to control Phytophthora cinnamomi disease. Aust. Plant Pathol. 44, 431–436. [Google Scholar]
- Sels, J. , Mathys, J. , De Coninck, B.M.A. , Cammue, B.P.A. and De Bolle, M.F.C. (2008) Plant pathogenesis‐related (PR) proteins: a focus on PR peptides. Plant Physiol. Biochem. 46, 941–950. [DOI] [PubMed] [Google Scholar]
- Serrazina, S. , Santos, C. , Machado, H. , Pesquita, C. , Vicentini, R. , Pais, M.S. , Sebastiana, M. and Costa, R. (2015) Castanea root transcriptome in response to Phytophthora cinnamomi challenge. Tree Genet. Genomes, 11, 19. [Google Scholar]
- Sghaier‐Hammami, B. , Valero‐Galvàn, J. , Romero‐Rodríguez, M.C. , Navarro‐Cerrillo, R.M. , Abdelly, C. and Jorrín‐Novo, J. (2013) Physiological and proteomics analyses of Holm oak (Quercus ilex subsp. ballota [Desf.] Samp.) responses to Phytophthora cinnamomi . Plant Physiol. Biochem. 71, 191–202. [DOI] [PubMed] [Google Scholar]
- Shearer, B.L. and Crane, C.E. (2012) Variation within the genus Lambertia in efficacy of low‐volume aerial phosphite spray for control of Phytophthora cinnamomi . Aust. Plant Pathol. 41, 47–57. [Google Scholar]
- Shearer, B.L. and Fairman, R.G. (2007) Application of phosphite in a high‐volume foliar spray delays and reduces the rate of mortality of four Banksia species infected with Phytophthora cinnamomi . Aust. Plant Pathol. 36, 358–368. [Google Scholar]
- Shearer, B.L. , Crane, C.E. and Cochrane, A. (2004) Quantification of the susceptibility of the native flora of the south‐west botanical province, Western Australia, to Phytophthora cinnamomi . Aust. J. Bot. 52, 435–443. [Google Scholar]
- Shearer, B.L. , Crane, C.E. , Barrett, S. and Cochrane, A. (2007) Phytophthora cinnamomi Sinvasion, a major threatening process to conservation of flora diversity in the South‐west Botanical Province of Western Australia. Aust. J. Bot. 55, 225–238. [Google Scholar]
- Škalamera, D. and Hardham, A.R. (2006) PnCcp, a Phytophthora nicotianae protein containing a single complement control protein module, is sorted into large peripheral vesicles in zoospores. Aust. Plant Pathol. 35, 593–603. [Google Scholar]
- Song, J. , Win, J. , Tian, M.Y. , Schornack, S. , Kaschani, F. , Ilyas, M. , Van der Hoorn, R.A.L. and Kamoun, S. (2009) Apoplastic effectors secreted by two unrelated eukaryotic plant pathogens target the tomato defense protease Rcr3. Proc. Natl. Acad. Sci. USA, 106, 1654–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam, R. , Jupe, J. , Howden, A.J.M. , Morris, J.A. , Boevink, P.C. , Hedley, P.E. and Huitema, E. (2013) Identification and characterisation of CRN effectors in Phytophthora capsici shows modularity and functional diversity. PLoS One, 8, e59517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasikowski, P.M. , McComb, J.A. , Scott, P. , Paap, T. , O'Brien, P.A. and Hardy, G.E.S.J. (2014) Calcium sulphate soil treatments augment the survival of phosphite‐sprayed Banksia leptophylla infected with Phytophthora cinnamomi . Aust. Plant Pathol. 43, 369–379. [Google Scholar]
- Suddaby, T. , Alhussaen, K. , Daniel, R. and Guest, D. (2008) Phosphonate alters the defence responses of Lambertia species challenged by Phytophthora cinnamomi . Aust. J. Bot. 56, 550–556. [Google Scholar]
- Suzaki, E. , Suzaki, T. , Jackson, S.L. and Hardham, A.R. (1996) Changes in intracellular pH during zoosporogenesis in Phytophthora cinnamomi . Protoplasma, 191, 79–83. [Google Scholar]
- Tani, S. and Judelson, H.S. (2006) Activation of zoosporogenesis‐specific genes in Phytophthora infestans involves a 7‐nucleotide promoter motif and cold‐induced membrane rigidity. Eukaryot. Cell, 5, 745–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani, S. , Yatzkan, E. and Judelson, H.S. (2004) Multiple pathways regulate the induction of genes during zoosporogenesis in Phytophthora infestans . Mol. Plant–Microbe Interact. 17, 330–337. [DOI] [PubMed] [Google Scholar]
- Tani, S. , Kim, K.S. and Judelson, H.S. (2005) A cluster of NIF transcriptional regulators with divergent patterns of spore‐specific expression in Phytophthora infestans . Fungal Genet. Biol. 42, 42–50. [DOI] [PubMed] [Google Scholar]
- Thompson, S. , Levin, S. and Rodriguez‐Iturbe, I. (2013) Linking plant disease risk and precipitation drivers: a dynamical systems framework. Am. Nat. 181, E1–E16. [DOI] [PubMed] [Google Scholar]
- Tian, M. , Huitema, E. , da Cunha, L. , Torto‐Alalibo, T. and Kamoun, S. (2004) A Kazal‐like extracellular serine protease inhibitor from Phytophthora infestans targets the tomato pathogenesis‐related protease P69B. J. Biol. Chem. 279, 26 370–26 377. [DOI] [PubMed] [Google Scholar]
- Tian, M. , Benedetti, B. and Kamoun, S. (2005) A second kazal‐like protease inhibitor from Phytophthora infestans inhibits and interacts with the apoplastic pathogenesis‐related protease P69B of tomato. Plant Physiol. 138, 1785–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, M. , Win, J. , Song, J. , Van Der Hoorn, R. , Van Der Knaap, E. and Kamoun, S. (2007) A Phytophthora infestans cystatin‐like protein targets a novel tomato papain‐like apoplastic protease. Plant Physiol. 143, 364–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torto, T.A. , Li, S. , Styer, A. , Huitema, E. , Testa, A. , Gow, N.A.R. , Van West, P. and Kamoun, S. (2003) EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora . Genome Res. 13, 1675–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler, B.M. , Tripathy, S. , Zhang, X.M. , Dehal, P. , Jiang, R.H.Y. , Aerts, A. , Arredondo, F.D. , Baxter, L. , Bensasson, D. , Beynon, J.L. , Chapman, J. , Damasceno, C.M.B. , Dorrance, A.E. , Dou, D.L. , Dickerman, A.W. , Dubchak, I.L. , Garbelotto, M. , Gijzen, M. , Gordon, S.G. , Govers, F. , Grunwald, N.J. , Huang, W. , Ivors, K.L. , Jones, R.W. and Kamoun, S. (2006) Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science, 313, 1261–1266. [DOI] [PubMed] [Google Scholar]
- Van Damme, M. , Bozkurt, T.O. , Cakir, C. , Schornack, S. , Sklenar, J. , Jones, A.M.E. and Kamoun, S. (2012) The Irish potato famine pathogen Phytophthora infestans translocates the CRN8 kinase into host plant cells. PLoS Pathog. 8, e1002875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veluchamy, S. , Williams, B. , Kim, K. and Dickman, M.B. (2012) The CuZn superoxide dismutase from Sclerotinia sclerotiorum is involved with oxidative stress tolerance, virulence, and oxalate production. Physiol. Mol. Plant Pathol. 78, 14–23. [Google Scholar]
- Vincelli, P. and Tisserat, N. (2008) Nucleic acid‐based pathogen detection in applied plant pathology. Plant Dis. 92, 660–669. [DOI] [PubMed] [Google Scholar]
- Wang, Q. , Han, C. , Ferreira, A.O. , Yu, X. , Ye, W. , Tripathy, S. , Kale, S.D. , Gu, B. , Sheng, Y. , Sui, Y. , Wang, X. , Zhang, Z. , Cheng, B. , Dong, S. , Shan, W. , Zheng, X. , Dou, D. , Tyler, B.M. and Wang, Y. (2011) Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell, 23, 2064–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y.L. , Li, A.N. , Wang, X.L. , Zhang, X. , Zhao, W. , Dou, D.L. , Zheng, X.B. and Wang, Y.C. (2010) GPR11, a putative seven‐transmembrane G protein‐coupled receptor, controls zoospore development and virulence of Phytophthora sojae . Eukaryot. Cell, 9, 242–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerakoon, N.D. , Roberts, J.K. , Lehnen, L.P. , Wilkinson, J.M. , Marshall, J.S. and Hardham, A.R. (1998) Isolation and characterization of the single α‐tubulin gene in Phytophthora cinnamomi . Mycologia, 90, 85–95. [Google Scholar]
- Weste, G. (2003) The dieback cycle in Victorian forests: a 30‐year study of changes caused by Phytophthora cinnamomi in Victorian open forests, woodlands and heathlands. Aust. Plant Pathol. 32, 247–256. [Google Scholar]
- Wong, M.H. , McComb, J. , Hardy, G.E.S. and O'Brien, P.A. (2009) Phosphite induces expression of a putative proteophosphoglycan gene in Phytophthora cinnamomi . Aust. Plant Pathol. 38, 235–241. [Google Scholar]
- Xiong, Q. , Ye, W. , Choi, D. , Wong, J. , Qiao, Y. , Tao, K. , Wang, Y. and Ma, W. (2014) Phytophthora suppressor of RNA silencing 2 is a conserved RxLR effector that promotes infection in soybean and Arabidopsis thaliana . Mol. Plant–Microbe Interact. 27, 1379–1389. [DOI] [PubMed] [Google Scholar]
- Yan, H.Z. and Liou, R.F. (2006) Selection of internal control genes for real‐time quantitative RT‐PCR assays in the oomycete plant pathogen Phytophthora parasitica . Fungal Genet. Biol. 43, 430–438. [DOI] [PubMed] [Google Scholar]
- Yang, X. , Zhao, W. , Hua, C. , Zheng, X. , Jing, M. , Li, D. , Govers, F. , Meijer, H.J.G. and Wang, Y. (2013) Chemotaxis and oospore formation in Phytophthora sojae are controlled by G‐protein‐coupled receptors with a phosphatidylinositol phosphate kinase domain. Mol. Microbiol. 88, 382–394. [DOI] [PubMed] [Google Scholar]
- York, W.S. , Qin, Q. and Rose, J.K.C. (2004) Proteinaceous inhibitors of endo‐β‐glucanases. Biochim. Biophys. Acta, Proteins Proteomics, 1696, 223–233. [DOI] [PubMed] [Google Scholar]
- Zentmyer, G.A. (1980) Phytophthora Cinnamomi and the Diseases It Causes. St Paul, MN: The American Phytopathological Society. [Google Scholar]
- Zhang, M. , Lu, J. , Tao, K. , Ye, W. , Li, A. , Liu, X. , Kong, L. , Dong, S. , Zheng, X. and Wang, Y. (2012) A myb transcription factor of Phytophthora sojae, regulated by MAP kinase PsSAK1, is required for zoospore development. PLoS One, 7, e40246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M. , Li, Q. , Liu, T. , Liu, L. , Shen, D. , Zhu, Y. , Liu, P. , Zhou, J.M. and Dou, D. (2015) Two cytoplasmic effectors of Phytophthora sojae regulate plant cell death via interactions with plant catalases. Plant Physiol. 167, 164–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W. , Blackman, L.M. and Hardham, A.R. (2013) Transient fusion and selective secretion of vesicle proteins in Phytophthora nicotianae zoospores. PeerJ, 2013, e221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Zhai, C. , Hua, C. , Qiu, M. , Hao, Y. , Nie, P. , Ye, W. and Wang, Y. (2016) PsHint1, associated with the G‐protein α subunit PsGPA1, is required for the chemotaxis and pathogenicity of Phytophthora sojae . Mol. Plant Pathol. 17, 272–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Relative expression of selected Phytophthora cinnamomi genes before and after the induction of sporulation by transfer of mycelia to mineral salts solution. The graphs show data derived by the quantification of bands on RNA blots (Narayan, 2004) and are indicative of a number of different expression profiles during sporulation. Anx, annexin; CaM, calmodulin; Cen, centrin; EF1α, elongation factor 1α; GDH, glyceraldehyde‐3‐phosphate dehydrogenase; LDH, d‐lactate dehydrogenase; Pc, P. cinnamomi; Pdk, pyruvate phosphate dikinase; SAc, S‐adenosylhomocysteinase; Set1, SET1 transcription factor; Ubq, ubiquitin; STk, serine/threonine protein kinase.
Fig. S2 Transcript levels of three Phytophthora cinnamomi polygalacturonase genes in cultures growing in 5% V8 broth and defined media. Mycelia were bulked in media containing 2% glucose as the sole carbon source and then transferred to 5% V8 broth or one of six defined media. Transcript levels were measured using quantitative polymerase chain reaction (qPCR) (Dandipat, 2009).
Fig. S3 Transcript levels of three Phytophthora cinnamomi polygalacturonase genes in roots of lupin (Lupinus augustifolius) after inoculation. Transcript levels were measured using quantitative polymerase chain reaction (qPCR) (Dandipat, 2009).
Table S1 The number of genes in Carbohydrate‐Active enZyme (CAZyme) families in the genomes of Phytophthora cinnamomi (Pcin) and P. parasitica (Ppar).
Table S2 Putative Phytophthora cinnamomi elicitin genes. An initial group of 32 P. cinnamomi sequences with E‐values of <E‐05 was obtained by blast analysis of the P. cinnamomi genome in FungiDB with the β‐cinnamomin gene, PHYCI_98389. Subsequently, two additional P. cinnamomi sequences with high homology to PITG_06908 and PHYSO_30815 were identified. The table shows the P. infestans (or, in one case, P. sojae) elicitins to which the 34 P. cinnamomi sequences are most similar.
Table S3 Putative Crinkler (CRN) homologues in the Phytophthora cinnamomi genome. A pblast search using 45 CRN proteins representative of those reported in table S10 in Haas et al. (2009) resulted in the identification of 280 homologous sequences. These 280 sequences were analysed for the presence of conserved LFLAK and DWE motifs. This yielded a list of 42 putative P. cinnamomi CRN genes. These sequences were then analysed for the presence of a classical secretion signal (SP) or for evidence of secretion via a non‐classical pathway, as indicated by an NN score of >0.6. The list of 42 putative CRN genes includes six genes that are truncated at the N‐terminus in the current P. cinnamomi genome assembly. It also includes 13 genes that are not truncated at the N‐terminus, but for which there is no evidence for secretion. Genes were also analysed for the presence of a nuclear localization signal (NLS).
Table S4 Putative Phytophthora cinnamomi Nep1‐like protein (NLP) genes. Three P. sojae NLP genes, PHYSO_562453, PHYSO_509399 and PHYSO_249691, were used to blast the P. cinnamomi genome in FungiDB. This resulted in the identification of the 72 putative P. cinnamomi NLP genes listed here. The degree of homology of each of the P. cinnamomi genes to each of the three P. sojae genes is indicated by the scores and E‐values in the table. Genes shown with light or dark blue shading lack homology to one or two, respectively, of the three P. sojae genes. Only sequences with E‐values of <E‐05 are included.


