Summary
Ferredoxins, the major distributors for electrons to various acceptor systems in plastids, contribute to redox regulation and antioxidant defence in plants. However, their function in plant immunity is not fully understood. In this study, we show that the expression of the major leaf ferredoxin gene Fd2 is suppressed by Pseudomonas syringae pv. tomato (Pst) DC3000 infection, and that knockout of Fd2 (Fd2‐KO) in Arabidopsis increases the plant's susceptibility to both Pst DC3000 and Golovinomyces cichoracearum. On Pst DC3000 infection, the Fd2‐KO mutant accumulates increased levels of jasmonic acid and displays compromised salicylic acid‐related immune responses. Fd2‐KO also shows defects in the accumulation of reactive oxygen species induced by pathogen‐associated molecular pattern‐triggered immunity. However, Fd2‐KO shows enhanced R‐protein‐mediated resistance to Pst DC3000/AvrRpt2 infection, suggesting that Fd2 plays a negative role in effector‐triggered immunity. Furthermore, Fd2 interacts with FIBRILLIN4 (FIB4), a harpin‐binding protein localized in chloroplasts. Interestingly, Fd2, but not FIB4, localizes to stromules that extend from chloroplasts. Taken together, our results demonstrate that Fd2 plays an important role in plant immunity.
Keywords: ferredoxin, fibrillin, harpin, jasmonic acid, reactive oxygen species, salicylic acid, stromule
Introduction
To defend against a wide range of pathogenic microorganisms in the environment, plants have evolved a sophisticated innate immune system (Jones and Dangl, 2006; Schwessinger and Ronald, 2012). In addition to using preformed physical and chemical barriers, plants mount active responses to protect themselves. One branch of plant immunity, which is activated on recognition of conserved pathogen‐associated molecular patterns (PAMPs) by cell surface transmembrane proteins known as pattern recognition receptors, is referred to as PAMP‐triggered immunity (PTI) (Yamaguchi et al., 2013). To overcome PTI, pathogens deploy defence‐suppressing effector proteins that are secreted into the apoplastic space or are translocated into the host cytoplasm (Chisholm et al., 2006). To counter the activity of the pathogen effectors, plants use resistance (R) proteins that act as intracellular receptors. R proteins perceive pathogen effectors and thereby activate effector‐triggered immunity (ETI), which is a rapid and robust response often associated with a hypersensitive response (HR) (Akamatsu et al., 2013; Liu W et al., 2013).
Increasing numbers of studies have indicated that the chloroplast plays an important role in plant immunity (Nomura et al., 2012; de Torres Zabala et al., 2015). Various defence signalling molecules, including reactive oxygen species (ROS), salicylic acid (SA) and jasmonic acid (JA), are synthesized in the chloroplast (Serrano et al., 2016). In addition, the organelle is required for the integration of multiple environmental stimuli and for the active communication of the signals to other organelles (Padmanabhan and Dinesh‐Kumar, 2010).
Ferredoxins (Fds), a group of small iron–sulfur [2Fe–2S] cluster‐containing proteins, are soluble electron carriers that distribute electrons from photosystem I (PSI) to the various acceptor systems of metabolic processes in the chloroplast stroma (Hanke and Mulo, 2013). Fd1 and Fd2 are the photosynthetic and leaf‐localized Fds in Arabidopsis (Hanke and Hase, 2008). Fd2, which constitutes about 90% of the total leaf Fd complement, functions in linear photosynthetic electron transport; the less abundant Fd1 has been implicated in cyclic electron flow (Hanke et al., 2004). A previous study has found that the Fd2‐knockout mutant (Fd2‐KO) of Arabidopsis accumulates increased levels of ROS in the chloroplast, possibly because of the excess electrons transferred from PSI to O2 (Voss et al., 2008). Another study has reported that Fd2‐KO plants exhibit greater adaptation than wild‐type (WT) plants to long‐term high‐light conditions through the up‐regulation of photoprotection‐related genes (Liu J et al., 2013). In addition, transplastomic tobacco expressing Fd2 in its chloroplasts shows increased tolerance to abiotic stresses, and especially to low‐light conditions (Yamamoto et al., 2006).
FIBRILLIN4 (FIB4) is a thylakoid membrane‐ and plastoglobule‐localized protein that associates with the photosystem II (PSII) light‐harvesting complex (Friso et al., 2004; Galetskiy et al., 2008; Peltier et al., 2004; Vidi et al., 2006; Ytterberg et al., 2006). The fib4 knockdown apple tree mutant and Arabidopsis fib4 mutant are more susceptible than WT plants to bacterial pathogens (Singh et al., 2010). Recently, FIB4 has been found to be required for a high level of accumulation of plastoquinone in the plastoglobule (Singh et al., 2012). In addition, FIB4 interacts with the harpin protein, HrpN (Song et al., 2002). Harpins are glycine‐rich, cysteine‐lacking and heat‐stable proteins secreted via Type III secretion by Gram‐negative plant‐pathogenic bacteria (Choi et al., 2013; Wei et al., 1992). HrpN, the founder protein of the harpin family, is secreted by Erwinia amylovora and was the first pathogen‐independent HR elicitor characterized in plants (Wei et al., 1992).
Fds can enhance harpin‐mediated HR in plants. For example, PFLP, which is the Fd from sweet pepper, increases ROS generation and enhances the HR in tobacco in response to harpin treatments (Dayakar et al., 2003). Furthermore, overexpression of the sweet pepper PFLP gene in transgenic Arabidopsis activates HR‐associated events and increases resistance to Erwinia carotovora ssp. carotovora (ECC), but not to its harpin mutant strain. These results suggest that high levels of ectopic PFLP may lead to the recognition of the harpin and to the activation of the HR and other defence responses (Ger et al., 2014). However, the exact function of Fd in plant immune responses is still unclear.
In this study, we found that Arabidopsis Fd2 plays an important role in immune resistance to Pseudomonas syringae pv. tomato (Pst) DC3000 and Golovinomyces cichoracearum via the suppression of innate JA accumulation and regulation of PTI. Fd2 interacts with FIB4, and the expression of both genes increases in response to harpin treatment. In addition, Fd2 is localized in the stromules, which suggests that it might function in retrograde signal transduction. These results provide new insights into the mechanisms of Fd2‐mediated innate immunity in plants.
Results
Knockout of Fd2 increases susceptibility to the pathogens
To investigate the role of the major leaf Fd in plant innate immunity, we first quantified the expression level of Fd2 in Arabidopsis following infection by the virulent hemibiotrophic bacterium Pst DC3000. Within 12 h post‐infection (hpi), transcription of Fd2 was not significantly different between plants inoculated with buffer alone (control) and the bacterium (Fig. 1a, left). At 24 and 48 hpi, however, Fd2 transcript levels in bacterium‐inoculated plants were markedly lower than in control plants and also significantly lower than at 0 and 12 hpi (Fig. 1a, left). Unlike Fd2, FIB4 transcripts were not significantly affected by Pst DC3000 infection (Fig. 1a, right). We also performed immunoblot analysis using protein isolated from Col‐0 before and after infection with the polyclonal antibody generated against plant Fd (FDX1). Consistent with the transcription data, the Fd2 protein level was reduced at 2 days post‐infection (dpi) in Col‐0 (Fig. S1a,b, see Supporting Information). Taken together, these results demonstrate that Pst DC3000 infection reduces the expression of Fd2, but not FIB4, in Arabidopsis.
Figure 1.
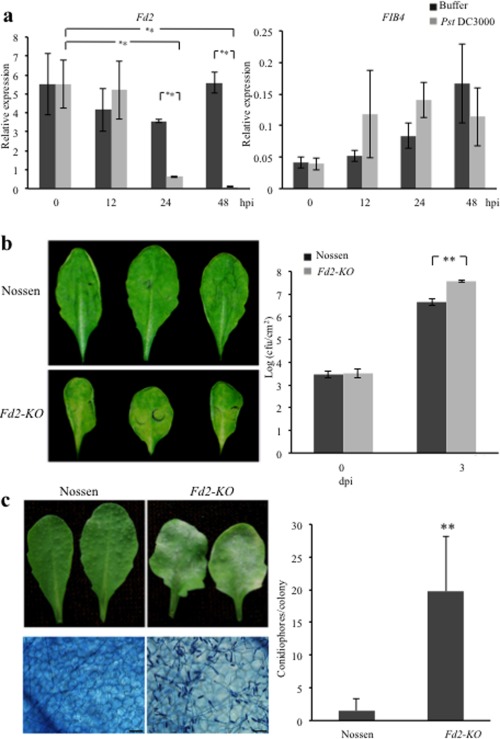
The Fd2‐knockout mutant (Fd2‐KO) exhibits increased susceptibility to pathogen infection. (a) After infiltration inoculation with Pseudomonas syringae pv. tomato (Pst) DC3000 [optical density at 600 nm (OD600) = 0.0002] or 10 mm MgCl2 as the control, total RNA was extracted from the inoculated leaves of 4‐week‐old Col‐0 plants at the indicated time points. The relative transcriptional levels of FIB4 and Fd2 were determined by quantitative real‐time polymerase chain reaction (qRT‐PCR). The transcription of Fd2 (left), but not FIB4 (right), was down‐regulated by Pst DC3000 at 24 and 48 h post‐infection (hpi). All qRT‐PCR assays were performed with Actin as the endogenous control. Error bars represent the standard deviation (SD) (n = 3) from three biological repeats, and the significance was determined at **P < 0.01 with Student's t‐test. (b) Four‐week‐old Nossen and Fd2‐KO plants were infected with Pst DC3000 (OD600 = 0.0002) by infiltration. Inoculated leaves at 4 days post‐infection (dpi) are shown on the left, and bacterial numbers at 0 and 3 dpi are indicated on the right. cfu, colony‐forming units. Error bars represent the SD (n = 3) from three biological repeats, and the significance was determined at **P < 0.01 with Student's t‐test. (c) Four‐week‐old Nossen and Fd2‐KO plants were inoculated with Golovinomyces cichoracearum. The infected leaves were photographed at 8 dpi (left top). Fungal growth, as indicated by staining with trypan blue, is shown on the left bottom. Bar, 50 μm. Fungal growth in plants at 5 dpi, as indicated by the number of conidiophores per colony, is shown on the right. The bars represent the mean and SD; significance was determined at **P < 0.01 with a one‐way analysis of variance. All results in this figure are from one of three independent experiments performed with similar results.
Next, we inoculated an Fd2‐KO mutant (Voss et al., 2008) and its WT background, Nossen, with Pst DC3000 by infiltration. At 4 dpi, bacterial speck disease symptoms were more severe on Fd2‐KO than on WT plants (Fig. 1b, left panel). Bacterial counts indicated that Fd2‐KO supported significantly more Pst DC3000 growth than WT (Fig. 1b, right panel). We also challenged Fd2‐KO mutant and WT plants with a virulent biotrophic powdery mildew G. cichoracearum strain (UCSC1), followed by quantification of conidiophore formation. The inoculation analysis showed that the Fd2‐KO mutant also displayed more susceptibility than the WT (Fig. 1c). Therefore, our results demonstrate that Fd2 is required for resistance against both the hemibiotrophic pathogen Pst DC3000 and the biotrophic pathogen G. cichoracearum.
The Fd2‐KO mutant displays reduced PTI‐induced ROS accumulation
To determine why Fd2‐KO is more susceptible to pathogen infection, we assessed whether the PTI pathway is affected by the absence of Fd2. We first measured the ROS burst in response to treatments with the PAMPs, flg22 and chitin, in Fd2‐KO and WT. Although ROS accumulation was detected in both Fd2‐KO and WT, ROS levels were lower in Fd2‐KO than in WT at all time points (Fig. 2a). Next, we analysed the transcriptional levels of two plasma membrane‐bound NADPH oxidases, AtrbohD and AtrbohF, that are required for apoplastic ROS accumulation (Torres et al., 2002). Consistent with the reduced ROS accumulation in response to PAMP treatments, Fd2‐KO displayed lower levels of both AtrbohD and AtrbohF transcripts than WT in the absence of any treatment (Fig. 2b). We also determined the transcriptional levels of FLG22‐INDUCED RECEPTOR‐LIKE KINASE 1 (FRK1), a marker gene of late PTI responses (Asai et al., 2002). As shown in Fig. 2c, FRK1 transcription did not differ significantly between Fd2‐KO and WT before inoculation. After inoculation with Pst DC3000, FRK1 transcription was strongly induced in both Fd2‐KO and WT; however, transcript levels were lower in Fd2‐KO than in WT, particularly at 2 dpi, when FRK1 transcript levels continued to increase in WT, but began to decline in Fd2‐KO (Fig. 2c). Based on these results, we conclude that Fd2 is involved in the regulation of PTI responses.
Figure 2.
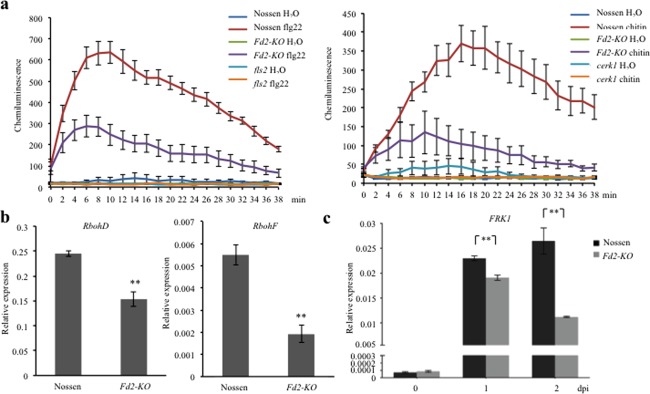
The Fd2‐knockout mutant (Fd2‐KO) is defective in pathogen‐associated molecular pattern (PAMP)‐triggered immunity (PTI)‐induced reactive oxygen species (ROS) accumulation. (a) To detect ROS, leaf strips of Nossen and Fd2‐KO plants were treated with 100 nm flg22 (left) or 200 μg/mL chitin (right) in buffer containing luminol and horseradish peroxidase. Luminescence was recorded at the indicated times. fls2 and cerk1 mutants were used as negative controls for flg22 and chitin treatment, respectively. Error bars represent the standard error (SE) (n = 8). (b) The relative transcriptional levels of RbohD (left) and RbohF (right) in 4‐week‐old non‐treated Nossen and Fd2‐KO plants as determined by quantitative real‐time polymerase chain reaction (qRT‐PCR). (c) The induction of FRK1 as indicated by qRT‐PCR in Nossen and Fd2‐KO plants before and after Pseudomonas syringae pv. tomato (Pst) DC3000 [optical density at 600 nm (OD600) = 0.0002] inoculation. In (b) and (c), Actin was used as the endogenous control; significance was determined at **P < 0.01 with Student's t‐test, and error bars represent SE (n = 3). Experiments were conducted in triplicate with similar results. dpi, days post‐infection.
The Fd2‐KO mutant accumulates high levels of JA in response to Pst DC3000 infection
To determine whether the biosynthesis of defence‐related hormones is affected by the absence of Fd2, we quantified the levels of SA, JA and JA‐isoleucine/leucine (JA‐Ile/Leu) in Pst DC3000‐infected Fd2‐KO and WT plants with buffer infiltration as a control via high‐performance liquid chromatography‐tandem mass spectroscopy (LC‐MS/MS). Because of the transient nature of increases in phytohormone concentrations following stress stimuli, these experiments were repeated three times, with each study containing three to five biological replicates. In all cases, the Fd2‐KO mutant accumulated significantly higher levels of JA and JA‐Ile/Leu than WT following inoculation with Pst DC3000 (2 dpi; Fig. 3a,b). The increase in JA levels in Fd2‐KO post‐infection was dramatic (over 90‐fold increases in most experiments conducted), particularly because WT showed no significant increase in JA accumulation at 2 dpi (Fig. 3a). A similar trend was observed with JA‐Ile/Leu, whose levels post‐infection were significantly higher in Fd2‐KO than in WT (Fig. 3b). As with JA levels, JA‐Ile/Leu levels in WT plants showed no significant increase at 2 dpi. Interestingly, loss of Fd2 might also result in increased resting levels of JA and JA‐Ile/Leu, although this trend was not consistent across all experiments, perhaps because of their low levels in the leaves of control plants. In addition, infection increased levels of SA in both Fd2‐KO and WT plants, but SA levels did not differ significantly between the mutant and WT post‐infection (Fig. 3c). Together, these results demonstrate that a loss of Fd2 leads to the generation of high levels of JA and JA‐Ile/Leu in response to Pst DC3000 infection, but does not affect SA accumulation.
Figure 3.
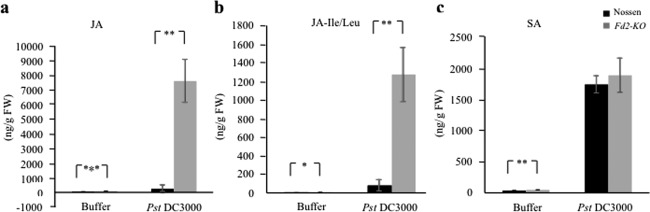
The Fd2‐knockout mutant (Fd2‐KO) accumulates high levels of jasmonic acid (JA) and JA‐isoleucine/leucine (JA‐Leu/Ile) in response to pathogen infection. Four‐week‐old Nossen and Fd2‐KO plants were inoculated with Pseudomonas syringae pv. tomato (Pst) DC3000 [optical density at 600 nm (OD600) = 0.001] or buffer (10 mm MgCl2) by leaf infiltration. At 2 days post‐infection (dpi), samples were collected for JA (a), JA‐Ile/Leu (b) and salicylic acid (SA) (c) quantification. Significance was determined using Student's t‐test (two‐tailed, type III, unequal variance); *P < 0.05, **P < 0.01 and ***P < 0.001. Error bars represent the standard deviation (SD) (n = 5 for all samples, except that n = 4 for Pst DC3000‐treated Fd2‐KO). The experiment was repeated three times, and representative results are shown. FW, fresh weight.
The induction of SA‐responsive defence genes is defective in Fd2‐KO in response to Pst DC3000 infection
Based on the differences in JA and JA‐Ile/Leu levels in Fd2‐KO vs. WT plants and on the potential interactions between the JA and SA signalling pathways, we measured the expression level of genes in both JA‐ and SA‐mediated immunity pathways. First, we analysed the transcription of PDF1.2, a JA‐responsive gene involved in defence against necrotrophic pathogens (Penninckx et al., 1998). In WT, inoculation with Pst DC3000 increased PDF1.2 transcript levels at 1 dpi and, to a lesser extent, at 2 dpi. Consistent with the high JA and JA‐Ile/Leu levels in Fd2‐KO after infection, PDF1.2 transcripts accumulated to a significantly higher level in Fd2‐KO than in WT at both 1 and 2 dpi (Fig. 4a). MYC2 is a JA‐responsive transcription factor that plays a key role in the regulation of cross‐talk between SA and JA (Lorenzo et al., 2004; Thaler et al., 2012). SID2 (also known as ICS1) encodes an isochorismate synthase that is required for the synthesis of SA from chorismate for plant defence (Wildermuth et al., 2001). MYC2 can activate the expression of the NAC transcription factor genes, which, in turn, reduce the expression of SID2 (Zheng et al., 2012). We found that MYC2 transcript levels were significantly higher in Fd2‐KO than in WT before and at 1 dpi with Pst DC3000, and were similar in both genotypes at 2 dpi (Fig. 4b). SID2 transcript levels were significantly lower in Fd2‐KO than in WT before and at 1 dpi with Pst DC3000, and were similar in both genotypes at 2 dpi (Fig. 4c). This inverse relationship between the levels of MYC2 and SID2 transcripts before and after Pst DC3000 infection correlates with the MYC2‐dependent reduction of SID2 expression.
Figure 4.
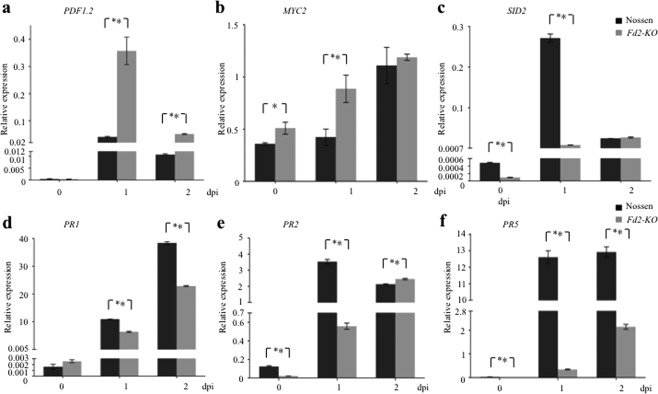
The Fd2‐knockout mutant (Fd2‐KO) displays enhanced expression of jasmonic acid (JA)‐responsive genes and compromised induction of salicylic acid (SA)‐responsive genes on pathogen infection. After inoculation with Pseudomonas syringae pv. tomato (Pst) DC3000 [optical density at 600 nm (OD600) = 0.0002] by infiltration, total RNA was extracted from the inoculated leaves of Nossen and Fd2‐KO plants at the indicated times. The relative transcriptional levels of PDF1.2 (a), MYC2 (b), SID2 (c), PR1 (d), PR2 (e) and PR5 (f) were determined by quantitative real‐time polymerase chain reaction (qRT‐PCR) with Actin as the endogenous control. Error bars represent the standard error (SE) (n = 3), and significance was determined at *P < 0.05 and **P < 0.01 with Student's t‐test. Experiments were conducted in triplicate with similar results. dpi, days post‐infection.
We also quantified the transcripts of SA‐responsive genes in Fd2‐KO and WT following Pst DC3000 infection. Interestingly, although SA accumulation was not affected, the transcript levels of PR1, PR2 and PR5 were reduced before and after infiltration with Pst DC3000 in Fd2‐KO plants (Fig. 4d–f). NPR1 encodes a transcription cofactor and is required for SA‐induced transcription of PR genes (Zhang et al., 1999). EDS5 localizes to the chloroplast envelope and exports SA from chloroplast to cytoplasm (Serrano et al., 2013). Consistent with the patterns of hormone accumulation and gene expression, the transcript levels of both NPR1 and EDS5 were significantly lower in Fd2‐KO than in WT before and after Pst DC3000 infection (Fig. S2a,b, see Supporting Information). Taken together, our data indicate that the higher accumulation of JA and JA‐Ile/Leu in the absence of Fd2 activates the JA signalling pathway, which may, in turn, suppress the SA signalling pathway after pathogen infection.
Fd2 negatively regulates Rps2‐mediated ETI
To investigate the role of Fd2 in ETI, we inoculated Fd2‐KO and WT plants with Pst DC3000 containing avrRpt2 and avrRpm1, whose products are recognized by the corresponding nucleotide‐binding oligomerization domain‐ and leucine‐rich repeat‐containing proteins, RPS2 and RPM1, respectively (Axtell and Staskawicz, 2003; Mackey et al., 2002). The non‐pathogenic Pst DC3000 strain with deletion of the hrcC gene (Pst DC3000/ΔhrcC), which is required for effector secretion, was used as the control. The inoculation assay showed that Fd2‐KO displayed enhanced resistance against Pst DC3000/avrRpt2, because the bacterial growth was less in Fd2‐KO than in WT (Fig. 5a). However, the resistance to Pst DC3000/avrRpm1 or Pst DC3000/ΔhrcC did not differ significantly between Fd2‐KO and WT (Fig. 5a). We then used 3,3‐diaminobenzidine tetrahydrochloride (DAB) staining to quantify H2O2 accumulation in Fd2‐KO and WT on Pst DC3000/avrRpt2 infection. Consistent with its enhanced resistance, Fd2‐KO accumulated higher H2O2 levels than WT in response to Pst DC3000/avrRpt2 inoculation (Fig. 5b). These results suggest that the Fd2 mutation enhances Rps2‐mediated resistance to Pst DC3000/avrRpt2 and causes increased H2O2 accumulation.
Figure 5.
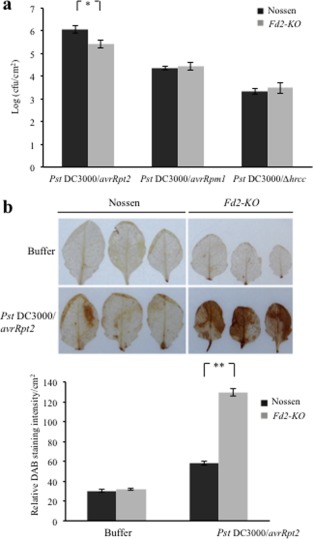
The Fd2‐knockout mutant (Fd2‐KO) is more resistant than Nossen to Pseudomonas syringae pv. tomato (Pst) DC3000/avrRpt2. (a) Four‐week‐old Nossen and Fd2‐KO plants were infiltration inoculated with Pst DC3000/avrRpt2, Pst DC3000/avrRpm1 or Pst DC3000/Δhrcc [optical density at 600 nm (OD600) = 0.0002]. Bacteria were counted at 3 days post‐infection (dpi). cfu, colony‐forming units. Error bars represent the standard deviation (SD) (n = 3), and significance was determined at *P < 0.05 with Student's t‐test. Experiments were conducted in triplicate with similar results. (b) Four‐week‐old Nossen and Fd2‐KO plants were spray inoculated with Pst DC3000/avrRpt2 (OD600 = 0.2) or with 10 mm MgCl2 buffer as the control. H2O2 production at 10 h after spraying was visualized by 3,3‐diaminobenzidine tetrahydrochloride (DAB) staining (top). The relative DAB staining intensity/cm2 is shown at the bottom. Error bars represent the standard error (SE) (n = 3).
Fd2 interacts with the hrpN‐binding protein FIB4
Because Fd is related to harpin‐mediated responses in plants and FIB4 is a harpin‐binding protein localized in chloroplasts (Dayakar et al., 2003; Song et al., 2002), we determined the relationship between Fd and FIB4. To investigate whether Fd interacts with FIB4, we first fused the two leaf Fds (Fd1 and Fd2) and FIB4 with glutathione S‐transferase (GST) and maltose‐binding protein (MBP), respectively. The GST‐Fd2 protein, but not GST‐Fd1 or GST, strongly bound to MBP‐FIB4, as revealed by anti‐MBP immunoblotting in an in vitro pull‐down assay (Fig. 6a). Next, we performed a co‐immunoprecipitation (co‐IP) assay following transient expression of Fd2‐GFP and FIB4‐HA in Nicotiana benthamiana leaves with the green fluorescent protein gene (GFP) and FIB4‐HA co‐expression as a negative control. After total protein had been extracted, samples were immunoprecipitated with anti‐GFP antibody. Anti‐haemagglutinin (anti‐HA) immunoblotting indicated that FIB4‐HA was co‐immunoprecipitated from the leaves co‐expressing both Fd2‐GFP and FIB4‐HA, but not from the leaves co‐expressing GFP and FIB4‐HA (Fig. 6b), indicating that Fd2 and FIB4 can form a protein complex in planta. We then used the split‐luciferase complementation assay to verify the interaction between Fd2 and FIB4 in planta (Chen et al., 2008). After the C‐termini of Fd2 and FIB4 had been fused with the N‐terminal (nLuc) and C‐terminal (cLuc) halves of luciferase, respectively, the constructs were transiently co‐expressed in N. benthamiana leaves. The complemented luciferase activity was detected with the combination of Fd2‐nLuc and FIB4‐cLuc, but not with the negative controls, nLuc and FIB4‐cLuc or Fd2‐nLuc and cLuc (Fig. 6c). To determine protein expression, we performed an immunoblotting assay with anti‐cLuc antibody to quantify the protein levels of FIB4‐cLuc in the leaves co‐expressing Fd2‐nLuc and FIB4‐cLuc or nLuc and FIB4‐cLuc (Fig. S3, see Supporting Information). In addition, the binding between FIB4 and HrpN was confirmed by pull‐down and co‐IP assays (Fig. S4a,b, see Supporting Information). Together, these results indicate that Fd2 interacts with the HrpN‐binding protein FIB4 in vitro and in vivo.
Figure 6.
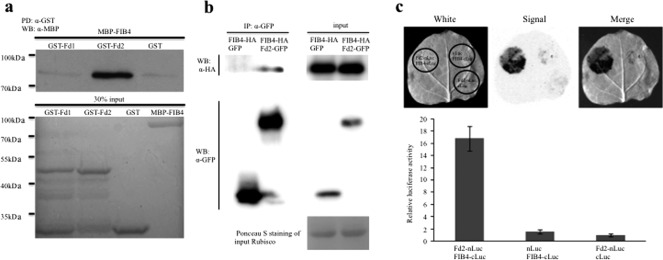
Fd2 interacts with the HrpN‐binding protein FIBRILLIN4 (FIB4). (a) Fd2, but not Fd1, interacts with FIB4 in vitro. Purified GST‐Fd1, GST‐Fd2 and glutathione S‐transferase (GST) were bound to glutathione resin beads and incubated with purified maltose‐binding protein (MBP)‐FIB4. After the preparation had been washed, the in vitro interaction of MBP‐FIB4 with GST‐Fd2 was detected by western blotting with α‐MBP antibody (top); 30% of the input was loaded and the sample amounts were shown by Coomassie blue staining (bottom). (b) Fd2 also interacts with FIB4 in vivo. FIB4‐HA and Fd2‐GFP were co‐expressed in Nicotiana benthamiana by agroinfiltration, with FIB4‐haemagglutinin (HA) and green fluorescent protein (GFP) co‐expression as the control. Proteins were extracted 3 days after infiltration, and immunoprecipitation was carried out with anti‐GFP antibody (left). The input is shown on the right. (c) Split‐luciferase complementation assays verified the interaction of Fd2 and FIB4. The left half of the N. benthamiana leaf was co‐infiltrated with Fd2‐nLuc/FIB4‐cLuc. The right half of the leaf was co‐infiltrated with nLuc/FIB4‐cLuc and Fd2‐nLuc/cLuc as a control. After 2 days of incubation, the inoculated leaves were sprayed with luciferin and chemiluminescence images were captured; the images are shown at the top. The relative luciferase activity values are indicated at the bottom; error bars represent the standard error (SE) (n = 10).
To determine the regulatory relationship between Fd2 and FIB4, we quantified the transcript levels of FIB4 and Fd2 in Fd2‐KO and fib4‐1, respectively. The analysis showed that levels of FIB4 transcripts were significantly lower in Fd2‐KO than in WT Nossen (Fig. S5a, see Supporting Information), suggesting that Fd2 positively regulates FIB4 expression. However, the levels of Fd2 transcript were significantly higher in fib4‐1 than in WT Col‐0 (Fig. S5b), suggesting that plants may compensate for the FIB4 mutation by inducing Fd2.
To study the expression pattern of FIB4 and Fd2 in plants after HrpN treatment, we sprayed 5 μg/mL GST‐HrpN onto 4‐week‐old Col‐0 plants, with GST as the control, and determined the transcript levels of FIB4 and Fd2 before and at 12 and 24 h after treatment. The analysis indicated that, relative to treatment with GST, GST‐HrpN increased Fd2 expression at 12 and 24 h after treatment and slightly increased FIB4 transcription at 12 h after treatment (Fig. S6a,b, see Supporting Information). These results demonstrate that HrpN causes accumulation of both FIB4 and Fd2 transcripts.
Fd2 is localized in stromules that extend from the chloroplast
We confirmed the chloroplast localization of Fd2 and FIB4 in the protoplasts isolated from N. benthamiana leaves that transiently expressed Fd2‐GFP or FIB4‐GFP (Fig. S7, see Supporting Information). Interestingly, Fd2‐GFP localized to stromules extending from the chloroplast (Fig. 7a, top). Some stromules with Fd2‐GFP signals were projected to the nuclei, in which weak signals of Fd2‐GFP were observed (Fig. 7a, top). As indicated by staining with the nuclear marker 4′,6‐diamidino‐2‐phenylindole (DAPI), Fd2‐GFP was in the nucleus that was associated with stromules (Fig. 7b). To exclude the possibility that the observed Fd2‐GFP signals were generated by free GFP (i.e. GFP tag cleaved from the Fd2‐GFP fusion protein), immunoblotting with anti‐GFP antibody was carried out using total protein extracted from the N. benthamiana leaf tissues transiently expressing Fd2‐GFP. As shown in Fig. S8 (see Supporting Information), no free GFP was detected. It follows that the signals in stromules and nuclei were from the Fd2‐GFP fusion protein. In contrast with Fd2‐GFP, FIB4‐GFP was not detected in the stromules or the nucleus (Fig. 7a, bottom). Therefore, our results demonstrate that Fd2 localizes to stromules of the chloroplast and possibly enters into the nucleus via stromules.
Figure 7.
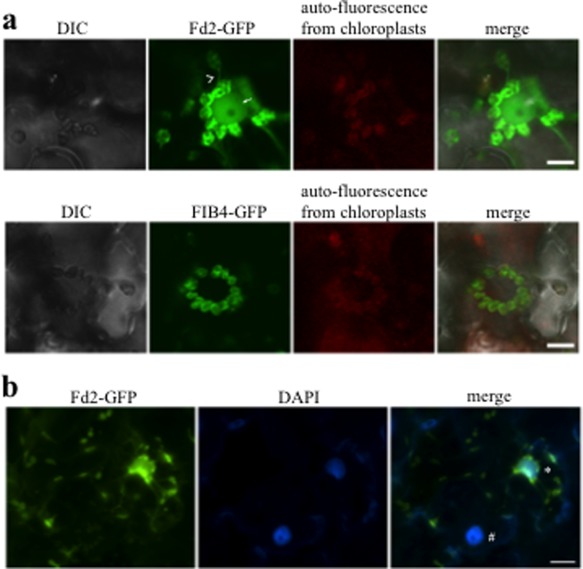
Fd2 localizes in stromules that extend from chloroplasts. (a) Confocal images of chloroplasts around the nuclei in Nicotiana benthamiana cells transiently expressing Fd2‐GFP (top) and FIB4‐GFP (bottom). Fd2, but not FIBRILLIN4 (FIB4), was found in stromules (top, indicated by arrowheads). A weak Fd2‐GFP signal was detected in the nucleus that was associated with the stromules (indicated by arrow). GFP, green fluorescent protein. (b) 4′,6‐Diamidino‐2‐phenylindole (DAPI) was used to visualize the nuclei. The nuclei that were connected with stromules showed strong Fd2‐GFP signals (indicated by *), whereas the nuclei without stromules attached showed very weak Fd2‐GFP signals (indicated by #). Bars, 10 μm (a) and 20 μm (b). DIC, Differential Interference Contrast.
Discussion
In this study, we analysed the functions of the primary Fd in Arabidopsis leaves, Fd2, in plant innate immunity. We found that loss of Fd2 causes strong JA and JA‐Ile/Leu accumulation on Pst DC3000 infection and reduces the expression of the genes in the SA signalling pathway, leading to enhanced susceptibility to Pst DC3000 and G. cichoracearum. Fd2 also plays a positive role in PTI‐mediated ROS accumulation, but negatively regulates the Rps2‐mediated ETI response. In addition, Fd2 interacts with the HrpN‐binding protein, FIB4, and expression of both Fd2 and FIB4 is induced by HrpN treatment. Furthermore, we found that Fd2 is a stromule‐localized protein and may be involved in the retrograde signalling pathway. Together, our results provide new insights into a role of Fd2, and more generally of the chloroplast, in the regulation of the plant immune responses.
Fd2 inhibits the accumulation of JA and its amino acid conjugate in response to infection
JA is associated with defence responses against necrotrophic pathogens and herbivorous insects (Glazebrook, 2005), whereas SA contributes to immune responses against biotrophic and hemibiotrophic pathogens (Vlot et al., 2009). The SA and JA defence pathways are often antagonistic to one another (Bari and Jones, 2009). The biosynthesis of JA, a lipid‐derived hormone, is initiated in the chloroplasts, but is completed in the peroxisome, and the final chloroplastic intermediate in JA biosynthesis is cis‐(+)‐12‐oxophytodienoic acid (OPDA) (Wasternack and Hause, 2013). OPDA is derived from oxidatively modified polyunsaturated fatty acids which, together with JA and methyl JA (MeJA), are collectively termed oxylipins (Howe and Schilmiller, 2002). A previous study has shown that knockout of the key components of the chloroplast photoprotection system in Arabidopsis produces high levels of JA and its precursor OPDA (Demmig‐Adams et al., 2013). Similarly, in the absence of Fd2, excess electrons generated from PSI are transferred to O2, leading to the formation of superoxide ( ), which could also cause lipid peroxidation. The formation of oxylipin hormones resulting from elevated lipid peroxides in the chloroplast may explain why the Fd2‐KO mutant accumulates more JA and JA‐Ile/Leu than WT following Pst DC3000 infection. Furthermore, although SA accumulates to the same level in WT and Fd2‐KO in the absence of pathogen attack, the induction of SA‐related genes is compromised in the Fd2‐KO mutant relative to WT on Pst DC3000 infection, which may result from the increased JA and JA‐Ile/Leu levels in the mutant. Researchers have recently reported that knockout of the gene encoding Arabidopsis NADPH‐dependent thioredoxin reductase C (NTRC) which, working together with peroxiredoxin, plays an important role in the chloroplast redox detoxification system, causes a strong increase in JA and JA‐Ile content and enhances disease susceptibility (Ishiga et al., 2016). Like the Fd2‐KO mutant in our study, the ntrc mutant showed no defects in SA accumulation, but reduced SID2 and PR1 transcription on infection.
RPS2‐mediated ETI is compromised in plants unable to accumulate SA (Clarke et al., 2000; Tao et al., 2003). Given our finding that Fd2‐KO plants show elevated JA accumulation and signalling and reduced SA signalling, it was surprising to find enhanced RPS2‐mediated ETI in Fd2‐KO. However, a recent study has shown that JA enhances RPS2‐mediated ETI, although the exact mechanism is unclear (Liu et al., 2016). Therefore, we propose that the resistance of Fd2‐KO to Pst DC3000/avrRpt2 is indirectly mediated by the high accumulation of JA in the absence of Fd2.
The oxidative burst, a hallmark of PTI responses, is accompanied by a biphasic accumulation of ROS. The first phase, occurring within 1 h following infection, is mostly an apoplastic accumulation that is tightly linked to NADPH oxidase activity; the second ROS burst, which occurs several hours after infection, is specific to pathogen attack and correlates with ETI and the HR (Shapiguzov et al., 2012; Stael et al., 2015). Growing evidence has indicated that chloroplasts play a crucial role in the second phase of ROS accumulation (Serrano et al., 2016). Increased ROS levels derived from ETI were found to compromise PSII efficiency, which leads to the progressive reduction in the plastoquinone pools involved in the transfer of electrons from PSII to PSI (Mur et al., 2010). In this case, Fd2 could be the reservoir to receive and distribute the extra electrons from PSI. When Fd2 is absent, these overproduced electrons will be transferred to O2, forming H2O2. This may explain why the Fd2‐KO mutant displays enhanced ROS accumulation on ETI activation.
Functions of Fd2 in the mediation of the harpin‐induced responses
Although the harpin protein was discovered over 20 years ago (Wei et al., 1992), the mechanism underlying harpin‐induced resistance and the HR is still not clear. In tobacco leaves, harpin infiltration causes the redistribution of chloroplasts in the palisade mesophyll cells and modification of the thylakoid membrane structure, which are early hallmarks of plant HR (Boccara et al., 2007). Thus, chloroplasts may be important in harpin‐induced plant responses. In addition, small amounts of HrpN are translocated into plant cells infected by E. amylovora (Bocsanczy et al., 2008). We found that the transient expression of HrpN in N. benthamiana by agroinoculation induced a weak, HR‐like cell death (Fig. S9, see Supporting Information), although whether this is the effect of HrpN located in plant cells is unclear.
Three harpin‐associated proteins have been reported in plants: AtHIPM, AtPIP1;4 and FIB4 (Li et al., 2015; Oh and Beer, 2007; Song et al., 2002). Among them, AtHIPM and AtPIP1;4 are involved in the mediation of enhanced plant growth on harpin treatment (Li et al., 2015; Oh and Beer, 2007), whereas the function of FIB4 in harpin‐mediated responses is ambiguous. FIB4, as a thylakoid membrane‐localized protein, increases plant immunity and detoxifies redox in the chloroplast (Singh et al., 2010). When treated with methyl viologen, which competes with Fd to accept electrons from PSI, the FIB4‐knockdown apple mutant accumulates higher superoxide levels than WT (Singh et al., 2010). The latter results, together with the current finding that Fd2 interacts with FIB4, suggest that FIB4 assists electron transport from photosynthesis to Fd. We therefore speculate that, when harpin is delivered into plant cells, it interacts with FIB4 and consequently disrupts Fd2 to receive electrons from photosynthesis, which is mediated by FIB4, ultimately leading to ROS accumulation in chloroplasts and cell death. However, more research is needed to clarify how Fd2 and FIB4 mediate plant responses to harpin.
Potential roles of Fd2 in the regulation of retrograde signalling
Activation of plant immunity depends on dramatic changes in the cellular redox status, which result in the reprogramming of the transcriptome and the establishment of both local and systemic defence (Spoel and Loake, 2011). In the stroma, Fd is the most upstream acceptor of electrons generated by photosynthesis, and Fd therefore determines the redox status of various downstream reductants, such as NADPH and thioredoxin (TRX) (Hanke and Mulo, 2013). To balance the redox outside of the chloroplast, NADPH‐dependent thioredoxin reductases (NTRA and NTRB) are required to maintain the pool of reduced TRX in the cytoplasm (Sweat and Wolpert, 2007). By transferring electrons generated by PSI to NADP+ via Fd, the chloroplast is the main NADPH‐producing organelle. Therefore, deletion of Fd2 may disrupt the reduction of cytoplasmic TRX by decreasing the amount of available NADPH, which can further affect nuclear gene transcription (Tada et al., 2008).
In addition, some plant transcription factors, such as TGA1 and TGA4 in Arabidopsis, rely on their oxidation state to regulate gene transcription (Després et al., 2003). The reductant which regulates the redox status in the nucleus is still unknown. Stromules, emanated from plastid bodies, have been found recently to be involved in the chloroplast to nuclear transport of immunity signals (Caplan et al., 2015). It is still unclear, however, how a protein can move from stromules to the nucleus across the inner and outer chloroplast membranes and to the nuclear envelope. According to our observations, Fd2 may be delivered from chloroplasts to the nucleus via stromules. This raises the possibility that Fd2 may be involved in retrograde chloroplast to nucleus signalling by reducing transcription factors in the nucleus. If this is correct, it is not surprising to find that the expression of many genes involved in the immune response, including genes regulating SA signalling and PTI, are regulated by Fd2.
Experimental Procedures
Plant growth conditions
The Arabidopsis Fd2–KO mutant (Voss et al., 2008) and the corresponding WT, Nossen, were kindly provided by Renate Scheibe's laboratory (University of Osnabrück, Germany) and by Hong‐Bin Wang's laboratory (Sun Yat‐sen University, China). Arabidopsis T‐DNA insertion mutant fib4–1 (SALK_014831) (Singh et al., 2010) was obtained from the Arabidopsis Biological Resource Center (http://www.biosci.ohio-state.edu/pcmb/Facilities/abrc/abrchome.htm) (Alonso et al., 2003). Arabidopsis plants were grown in a growth room at 20–22°C under a 10‐h light/14‐h dark cycle.
Pathogen infections
The powdery mildew pathogen G. cichoracearum (strain UCSC1) was maintained on the Arabidopsis pad4‐1 mutant. The inoculation method has been described previously (Shi et al., 2013; Wang et al., 2011). Trypan blue staining was used to monitor fungal growth in the leaves at 8 dpi. Fungal growth and conidiation were quantified as described previously (Consonni et al., 2006). Pst DC3000 infection by infiltration and a bacterial growth assay were performed as described previously (Geng et al., 2012) with minor differences: 4‐week‐old plants were used for infiltration, and leaf discs were harvested at 3 dpi for analysis of bacterial growth.
ROS measurement
An oxidative burst assay was carried out as described previously (Zhang et al., 2010) with minor modifications. The leaf strips were treated with 100 nm flg22 or 200 μg/mL chitin in 200 mL of buffer containing 20 μm luminol and 1 μg/mL horseradish peroxidase to immediately test the ROS burst. Each data point represented eight replicates.
DAB staining
To determine the accumulation of H2O2 after pathogen challenge, we stained the infected leaves with DAB. Four‐week‐old Nossen and Fd2‐KO plants were spray inoculated with an avirulent isolate containing avrRpt2. Ten hours after inoculation, the infected leaves were cut and briefly washed with water. After the leaves had been stained with 0.1% (w/v) DAB overnight in the dark, they were destained with 95% ethanol and preserved in 50% ethanol. H2O2 production was indicated by a reddish‐brown colour, and the relative DAB staining intensity was analysed by Image J.
Phytohormone quantification
Arabidopsis leaf samples were harvested at 2 dpi with Pst DC3000 or buffer and were flash frozen in liquid nitrogen. For phytohormone quantification, approximately 50 mg per sample was triple‐ground in liquid nitrogen and placed in a tube. A 1‐mL volume of 50 mm sodium phosphate buffer (pH 7.0) was added to each tube, and samples were extracted as described previously (Blakeslee and Murphy, 2016). The following were added to each tube as internal standards (ISTD): 12.5 ng of [2H6] (+)‐cis,trans‐abscisic acid (d6‐ABA, OlchemIm Ltd., Olomouc, Czech Republic, part #0342722); 5 ng of jasmonic‐d5 acid (2,4,4‐d3; acetyl‐2,2‐d2) (d5‐JA, CDN Isotopes, Pointe‐Claire, QC, Canada, part #D‐6936); and 12.5 ng of 2‐hydroxybenzoic acid‐d6 (d6‐SA, CDN Isotopes, part #D‐1156). Samples were mixed with a vortex apparatus, extracted for 20 min at 4°C with gentle shaking on a nutator, and centrifuged at 12 000 g for 15 min at 4°C. Samples were separated using an Agilent Poroshell 120EC‐C18 (3.5 × 50 mm2, 2.7 µm) column and an acidified water–methanol buffer system (buffer A, 0.1% acetate, 5% methanol in water; buffer B, 0.1% acetate in methanol). Gradient conditions were as follows: 3 min 2%–50% B, 2 min 50%–98% B, hold at 98% B for 2 min, and then back to 2% B for 1 min. Eluted compounds were further separated and quantified using an Agilent 6460 triple quadrupole dual mass spectrometer equipped with an electrospray ionization source (ESI) (Agilent Technologies, Santa Clara, CA, USA). Compounds were quantified in negative ion mode, and mass transitions for each compound are provided in Table S3 (see Supporting Information). Compound identity was confirmed by comparing retention times and mass transitions with the authentic standards described above. Because of the transient nature of increases in phytohormone concentrations following stress stimuli, these experiments were repeated three times, with each experiment containing three to five biological replicates.
Quantitative real‐time polymerase chain reaction (qRT‐PCR)
Total RNAs were extracted from 100 mg of Arabidopsis leaf tissue using TRIzol reagent (Thermo Fisher Scientific, 15596, Waltham, MA , USA) following the manufacturer's instructions. A 3‐μg quantity of total RNA was treated with DNase I (Invitrogen, 18068‐015, Carlsbad, CA, USA) to remove DNA contamination, according to the manufacturer's instructions. The RNA samples were then subjected to first‐strand cDNA synthesis using the Promega reverse transcription kit (Promega, A3500, Fitchburg, Wisconsin, USA) following the manufacturer's instructions. qRT‐PCR was carried out using the CFX96 Touch Real‐Time PCR Detection System (Bio‐Rad, Hercules, California, USA). The qRT‐PCR data were subjected to standard statistical analysis. Primers of the marker genes used in this study are listed in Table S2 (see Supporting Information).
Agrobacterium‐mediated transient expression in N. benthamiana, plant protein extraction and immunoblotting
Agrobacterium GV3101 carrying the indicated plasmids was infiltrated into 5‐week‐old N. benthamiana leaves. All primers used for plasmid construction in this study are listed in Table S1 (see Supporting Information). The agroinfiltration assay in N. benthamiana was carried out as described previously (Park et al., 2012). Total plant protein was extracted by grinding tissue in liquid nitrogen and immediately transferring the homogenate to 1 mL of extraction buffer containing 150 mm NaCl, 50 mm Tris‐HCl, pH 7.6, 1 mm ethylenediaminetetraacetic acid (EDTA), 0.5% Triton X‐100, 2 mm dithiothreitol (DTT) and plant protease inhibitor cocktail (Sigma‐Aldrich, St. Louis, Missouri, USA). The samples were centrifuged at 16200 g for 15 min at 4°C, and the supernatant was used for immunoblotting as described previously (Park et al., 2012).
Co‐IP assay in N. benthamiana
For co‐IP, 50 μL of a 50% slurry (v/v) of protein G‐agarose beads (Roche, 11719416001, Pleasanton, CA, USA) was washed with 1 × phosphate‐buffered saline (PBS) and then incubated with 5 μL of α‐GFP antibody (Roche, 11814460001) at 4°C overnight with gentle rotation. Following centrifugation and removal of the buffer, protein samples (500 μL) were added to the beads with shaking for 6 h at 4°C. After the beads had been washed four times with washing buffer (150 mm NaCl, 50 mm Tris‐HCl, pH 7.6, 1 mm EDTA, 0.3% Triton X‐100 and 2 mm DTT), 50 μL of 1 × sodium dodecylsulfate (SDS) loading buffer was added, and the sample was heated to 95°C for 5 min. Then, 15 μL of each sample was loaded for gel running and immunoblotting with α‐HA antibody (Roche, 1867423) or α‐Myc antibody (Sigma‐Aldrich, M4439).
GST pull‐down
The target genes were cloned into pGEX 6p‐1 and pMAL‐c2x vectors to express proteins in Escherichia coli with N‐terminus‐fused GST and MBP tags, respectively. The fusion proteins were prepared according to the manufacturer's instructions. The concentration of proteins in the supernatant was determined with the Bio‐Rad protein assay reagent (500‐0006). For the GST pull‐down assay, about 5 µg of purified GST‐fused proteins and GST were incubated with 40 μL of glutathione resin beads (GoldBio, G‐250‐100, St. Louis, MO, USA) for 4 h at 4°C with gentle shaking. After the supernatant had been removed and the pellet had been washed three times with PBS, 5 µg of purified MBP‐fused proteins (purified with amylose resin, NEB E8021S, Ipswich, Massachusetts, UK, following the manufacturer's instructions) was added to the beads with end‐over‐end mixing for 6 h at 4°C. The beads were then washed four times with PBST buffer (1 × PBS with 0.5% Triton X‐100 and 1 mm EDTA). Following centrifugation and removal of the supernatant, SDS loading buffer was added to the beads. The preparation was heated for 5 min before 15 μL of each sample was loaded for gel running. α‐MBP (NEB, E8032S) and α‐GST (Invitrogen, 71–7500) were used for immunoblot analysis.
Split‐luciferase complementation assay
Fd2 was cloned into the pCAMBIA NLuc vector with the N‐terminal half of luciferase (nLuc) using Gateway technology, resulting in Fd2‐nLuc; FIB4 was cloned into the pCAMBIA CLuc vector with the C‐terminal half of luciferase (cLuc), resulting in FIB4‐cLuc (Chen et al., 2008; Lee et al., 2015). After individual transformation into Agrobacterium GV3101, they were transiently co‐expressed in N. benthamiana leaves by infiltration with a final concentration of OD600 = 0.6 (OD600, optical density at 600 nm) for each construct. On the same leaves, GV3101 carrying the empty nLuc vector and the FIB4‐cLuc construct or the empty cLuc vector and the Fd2‐nLuc construct was co‐infiltrated as a control. Two days after inoculation, the inoculated leaves were sprayed with 1 mM luciferin, and chemiluminescence images were captured by ChemiDoc XRS+ (Bio‐Rad). The fluorescence intensity was quantified with Image Lab 5.2 software.
Microscopic analysis
Protoplasts were isolated as described previously (Breuers et al., 2012). Protoplasts were observed with a Nikon (Shinagawa, Tokyo, Japan) Eclipse 80i microscope and photographed with a Nikon DS‐Qi1 camera. Plant tissues were observed with a Nikon A1 confocal microscope. Images were processed with NIS‐Elements (Shinagawa, Tokyo, Japan) imaging software. To detect the GFP signal, 488 nm excitation and 507 nm detection wavelengths were used; to detect the chloroplast autofluorescence, 535 nm excitation and 620 nm detection wavelengths were used. For nuclear staining, 10 μg/mL of DAPI (Invitrogen, NL5995050) was used.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 The expression of Fd2 is reduced by Pseudomonas syringae pv. tomato (Pst) DC3000. (a) Total protein was extracted from Col‐0 before and at 2 days after Pst DC3000 infiltration. The protein level of Fd2 was determined by western blot using antiserum against ferredoxin in Spinacia oleracea (FDX1, top line) with Ponceau S staining of input Rubisco as the loading control (bottom line). (b) The relative intensity of immunoblotting bands is shown. Error bars represent the standard error (SE) (n = 3) from three biological repeats, and significance was determined at *P < 0.05 with Student's t‐test. dpi, days post‐infection.
Fig. S2 The transcriptional levels of NPR1 (a) and EDS5 (b) in Nossen and Fd2‐KO before and after Pseudomonas syringae pv. tomato (Pst) DC3000 [optical density at 600 nm (OD600) = 0.0002] infection. Quantitative real‐time polymerase chain reaction (qRT‐PCR) was carried out with Actin as the endogenous control. Significance was determined at *P < 0.05 and **P < 0.01 with Student's t‐test. Error bars represent the standard error (SE) (n = 3). Experiments were conducted in triplicate with similar results. dpi, days post‐infection.
Fig. S3 Protein levels of FIB4‐cLuc in Nicotiana benthamiana leaves transiently expressing split‐luciferase complementation vectors. Immunoblotting was carried out with the anti‐cLuc antibody.
Fig. S4 FIBRILLIN4 (FIB4) binds to HrpN. (a) FIB4 interacts with HrpN as indicated by a pull‐down assay. Purified GST‐HrpN and glutathione S‐transferase (GST) were bound to glutathione resin beads and incubated with purified maltose‐binding protein (MBP)‐FIB4. After the preparation had been washed, the in vitro interaction of MBP‐FIB4 with GST‐HrpN was detected by western blotting with α‐MBP antibody (top); 30% of the input was loaded and the sample amounts were shown by Coomassie blue staining (bottom). (b) FIB4 interacts with HrpN as indicated by co‐immunoprecipitation (co‐IP). FIB4‐GFP and Myc‐HrpN were co‐expressed in Nicotiana benthamiana by agroinfiltration, with Myc‐HrpN expressed alone as the control. Proteins were extracted 3 days after infiltration, and immunoprecipitation was carried out with anti‐green fluorescent protein (anti‐GFP) antibody (top). The input is shown at the bottom.
Fig. S5 The transcript analysis of FIB4 (a) and Fd2 (b) in Fd2‐KO and fib4‐1, respectively, relative to the wild‐types. FIB4 transcription was suppressed in Fd2‐KO, whereas Fd2 was induced in fib4‐1. Actin was used as the endogenous control. Significance was determined at **P < 0.01 with Student's t‐test. Error bars represent the standard error (SE) (n = 3). Experiments were conducted in triplicate with similar results.
Fig. S6 Both Fd2 and FIB4 are harpin‐responsive genes. Four‐week‐old Col‐0 plants were sprayed with 5 μg/mL glutathione S‐transferase (GST) and GST‐HrpN. Leaf tissues were harvested at the indicated times for analysis of the relative transcript levels of Fd2 (a) and FIB4 (b). Compared with the GST control, both Fd2 and FIB4 were up‐regulated by HrpN treatment. Actin was used as the endogenous control; error bars represent the standard error (SE) (n = 3); significance was determined at **P < 0.01 with Student's t‐test. Experiments were conducted in triplicate with similar results.
Fig. S7 Both FIBRILLIN4 (FIB4) and Fd2 are localized to chloroplasts. Protoplasts were prepared from Nicotiana benthamiana leaves transiently expressing FIB4‐GFP or Fd2‐GFP by agroinfiltration. Both Fd2‐GFP and FIB4‐GFP (green) localize to chloroplasts (red, autofluorescence). Bars, 10 μm.
Fig. S8 Immunoblotting to detect Fd2‐GFP transiently expressed in Nicotiana benthamiana leaves. Fd2‐GFP was transiently expressed in N. benthamiana by agroinfiltration. Total protein was extracted 3 days after infiltration, and western blot was carried out with anti‐green fluorescent protein (anti‐GFP) antibody.
Fig. S9 Transient expression of HrpN in Nicotiana benthamiana caused weak cell death. At 3 days after transient expression of HrpN‐GFP by agroinfiltration, a weak, hypersensitive response (HR)‐like cell death was observed on the leaf. The leaf expressing Fd2‐GFP was used as a control. Bars, 5 mm.
Table S1 Primers used for plasmid construction.
Table S2 Primers used for real‐time polymerase chain reaction (PCR) analysis.
Table S3 Tandem mass spectroscopy (MS/MS) settings and mass transition used to quantify salicylic acid (SA), jasmonic acid (JA) and JA‐isoleucine/leucine (JA‐Ile/Leu).
Acknowledgements
We thank R. Scheibe and H.‐B. Wang for providing Nossen and Fd2‐KO seeds. This work was supported by a grant from Plant Health Care Inc. (Grant No. #1000).
References
- Akamatsu, A. , Wong, H.L. , Fujiwara, M. , Okuda, J. , Nishide, K. , Uno, K. , Imai, K. , Umemura, K. , Kawasaki, T. , Kawano, Y. and Shimamoto, K. (2013) An OsCEBiP/OsCERK1‐OsRacGEF1‐OsRac1 module is an essential early component of chitin‐induced rice immunity. Cell Host Microbe, 13, 465–476. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M. , Stepanova, A.N. , Leisse, T.J. , Kim, C.J. , Chen, H. and Shinn, P. (2003) Genome‐wide insertional mutagenesis of Arabidopsis thaliana . Science, 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Asai, T. , Tena, G. , Plotnikova, J. , Willmann, M.R. , Chiu, W.‐L. , Gomez‐Gomez, L. , Boller, T. , Ausubel, F.M. and Sheen, J. (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature, 415, 977–983. [DOI] [PubMed] [Google Scholar]
- Axtell, M.J. and Staskawicz, B.J. (2003) Initiation of RPS2‐specified disease resistance in Arabidopsis is coupled to the AvrRpt2‐directed elimination of RIN4. Cell, 112, 369–377. [DOI] [PubMed] [Google Scholar]
- Bari, R. and Jones, J.D.G. (2009) Role of plant hormones in plant defence responses. Plant Mol. Biol. 69, 473–488. [DOI] [PubMed] [Google Scholar]
- Blakeslee, J.J. and Murphy, A.S. (2016) Microscopic and biochemical visualization of auxins in plant tissues In: Environmental Responses in Plants: Methods and Protocols (Duque P., ed.), pp. 37–53. New York, NY: Springer. [DOI] [PubMed] [Google Scholar]
- Boccara, M. , Schwartz, W. , Guiot, E. , Vidal, G. , De Paepe, R. , Dubois, A. and Boccara, A.‐C. (2007) Early chloroplastic alterations analysed by optical coherence tomography during a harpin‐induced hypersensitive response. Plant J. 50, 338–346. [DOI] [PubMed] [Google Scholar]
- Bocsanczy, A.M. , Nissinen, R.M. , Oh, C. and Beer, S.V. (2008) HrpN of Erwinia amylovora functions in the translocation of DspA/E into plant cells. Mol. Plant Pathol. 9, 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuers, F.K. , Bräutigam, A. , Geimer, S. , Welzel, U.Y. , Stefano, G. , Renna, L. , Brandizzi, F. and Weber, A.P. (2012) Dynamic remodeling of the plastid envelope membranes—a tool for chloroplast envelope in vivo localizations. Front. Plant Sci. 3, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan, J.L. , Kumar, A.S. , Park, E. , Padmanabhan, M.S. , Hoban, K. , Modla, S. , Czymmek, K. and Dinesh‐Kumar, S.P. (2015) Chloroplast stromules function during innate immunity. Dev. Cell, 34, 45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. , Zou, Y. , Shang, Y. , Lin, H. , Wang, Y. , Cai, R. , Tang, X. and Zhou, J.‐M. (2008) Firefly luciferase complementation imaging assay for protein–protein interactions in plants. Plant Physiol. 146, 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm, S.T. , Coaker, G. , Day, B. and Staskawicz, B.J. (2006) Host–microbe interactions: shaping the evolution of the plant immune response. Cell, 124, 803–814. [DOI] [PubMed] [Google Scholar]
- Choi, M.‐S. , Kim, W. , Lee, C. and Oh, C.‐S. (2013) Harpins, multifunctional proteins secreted by Gram‐negative plant‐pathogenic bacteria. Mol. Plant–Microbe Interact. 26, 1115–1122. [DOI] [PubMed] [Google Scholar]
- Clarke, J.D. , Volko, S.M. , Ledford, H. , Ausubel, F.M. and Dong, X. (2000) Roles of salicylic acid, jasmonic acid, and ethylene in cpr‐induced resistance in Arabidopsis. Plant Cell, 12, 2175–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consonni, C. , Humphry, M.E. , Hartmann, H.A. , Livaja, M. , Durner, J. , Westphal, L. , Vogel, J. , Lipka, V. , Kemmerling, B. , Schulze‐Lefert, P. , Somerville, S.C. and Panstruga, R. (2006) Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat. Genet. 38, 716–720. [DOI] [PubMed] [Google Scholar]
- Dayakar, B.V. , Lin, H.‐J. , Chen, C.‐H. , Ger, M.‐J. , Lee, B.‐H. , Pai, C.‐H. , Chow, D. , Huang, H.E. , Hwang, S.Y. , Chung, M.C. and Feng, T.Y. (2003) Ferredoxin from sweet pepper (Capsicum annuum L.) intensifying harpin pss‐mediated hypersensitive response shows an enhanced production of active oxygen species (AOS). Plant Mol. Biol. 51, 913–924. [DOI] [PubMed] [Google Scholar]
- Demmig‐Adams, B. , Cohu, C.M. , Amiard, V. , van Zadelhoff, G. , Veldink, G.A. , Muller, O. and Adams, W.W. (2013) Emerging trade‐offs—impact of photoprotectants (PsbS, xanthophylls, and vitamin E) on oxylipins as regulators of development and defense. New Phytol. 197, 720–729. [DOI] [PubMed] [Google Scholar]
- Després, C. , Chubak, C. , Rochon, A. , Clark, R. , Bethune, T. , Desveaux, D. and Fobert, P.R. (2003) The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell, 15, 2181–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso, G. , Giacomelli, L. , Ytterberg, A.J. , Peltier, J.‐B. , Rudella, A. , Sun, Q. and Wijk, K.J. (2004) In‐depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell, 16, 478–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galetskiy, D. , Susnea, I. , Reiser, V. , Adamska, I. and Przybylski, M. (2008) Structure and dynamics of photosystem II light‐harvesting complex revealed by high‐resolution FTICR mass spectrometric proteome analysis. J. Am. Soc. Mass Spectrom. 19, 1004–1013. [DOI] [PubMed] [Google Scholar]
- Geng, X. , Cheng, J. , Gangadharan, A. and Mackey, D. (2012) The coronatine toxin of Pseudomonas syringae is a multifunctional suppressor of Arabidopsis defense. Plant Cell, 24, 4763–4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ger, M.‐J. , Louh, G.‐Y. , Lin, Y.‐H. , Feng, T.‐Y. and Huang, H.‐E. (2014) Ectopically expressed sweet pepper ferredoxin PFLP enhances disease resistance to Pectobacterium carotovorum subsp. carotovorum affected by harpin and protease‐mediated hypersensitive response in Arabidopsis. Mol. Plant Pathol. 15, 892–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Hanke, G.T. and Hase, T. (2008) Variable photosynthetic roles of two leaf‐type ferredoxins in Arabidopsis, as revealed by RNA interference. Photochem Photobiol. 84, 1302–1309. [DOI] [PubMed] [Google Scholar]
- Hanke, G.T. and Mulo, P. (2013) Plant type ferredoxins and ferredoxin‐dependent metabolism. Plant Cell Environ. 36, 1071–1084. [DOI] [PubMed] [Google Scholar]
- Hanke, G.T. , Kimata‐Ariga, Y. , Taniguchi, I. and Hase, T. (2004) A post genomic characterization of Arabidopsis ferredoxins. Plant Physiol. 134, 255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, G.A. and Schilmiller, A.L. (2002) Oxylipin metabolism in response to stress. Curr. Opin. Plant Biol. 5, 230–236. [DOI] [PubMed] [Google Scholar]
- Ishiga, Y. , Ishiga, T. , Ikeda, Y. , Matsuura, T. and Mysore, K.S. (2016) NADPH‐dependent thioredoxin reductase C plays a role in nonhost disease resistance against Pseudomonas syringae pathogens by regulating chloroplast‐generated reactive oxygen species. PeerJ. 4, e1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Lee, D. , Bourdais, G. , Yu, G. , Robatzek, S. and Coaker, G. (2015) Phosphorylation of the plant immune regulator RPM1‐INTERACTING PROTEIN4 enhances plant plasma membrane H+‐ATPase activity and inhibits flagellin‐triggered immune responses in Arabidopsis. Plant Cell, 27, 2042–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L. , Wang, H. , Gago, J. , Cui, H. , Qian, Z. , Kodama, N. , Ji, H. , Tian, S. , Shen, D. , Chen, Y. , Sun, F. , Xia, Z. , Ye, Q. , Sun, W. , Flexas, J. and Dong, H. (2015) Harpin Hpa1 interacts with aquaporin PIP1;4 to promote the substrate transport and photosynthesis in Arabidopsis. Sci. Rep. 5, 17 207. doi: 10.1038/srep17207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Wang, P. , Liu, B. , Feng, D. , Zhang, J. , Su, J. , Zhang, Y. , Wang, J.F. and Wang, H.B. (2013) A deficiency in chloroplastic ferredoxin 2 facilitates effective photosynthetic capacity during long‐term high light acclimation in Arabidopsis thaliana . Plant J. 76, 861–874. [DOI] [PubMed] [Google Scholar]
- Liu, L. , Sonbol, F.‐M. , Huot, B. , Gu, Y. , Withers, J. , Mwimba, M. , Yao, J. , He, S.Y. and Dong, X. (2016) Salicylic acid receptors activate jasmonic acid signalling through a non‐canonical pathway to promote effector‐triggered immunity. Nat. Commun. 7, 13 099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W. , Liu, J. , Ning, Y. , Ding, B. , Wang, X. , Wang, Z. and Wang, G.L. (2013) Recent progress in understanding PAMP‐ and effector‐triggered immunity against the rice blast fungus Magnaporthe oryzae . Mol. Plant. 6, 605–620. [DOI] [PubMed] [Google Scholar]
- Lorenzo, O. , Chico, J.M. , Sánchez‐Serrano, J.J. and Solano, R. (2004) JASMONATE‐INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate‐regulated defense responses in Arabidopsis. Plant Cell, 16, 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey, D. , Holt, B.F. , Wiig, A. and Dangl, L. , J. (2002) RIN4 interacts with type III effector molecules and is required for RPM1‐mediated resistance in Arabidopsis. Curr. Biol. 108, 743–754. [DOI] [PubMed] [Google Scholar]
- Mur, L.A.J. , Aubry, S. , Mondhe, M. , Kingston‐Smith, A. , Gallagher, J. , Timms‐Taravella, E. , James, C. , Papp, I. , Hörtensteiner, S. , Thomas, H. and Ougham, H. (2010) Accumulation of chlorophyll catabolites photosensitizes the hypersensitive response elicited by Pseudomonas syringae in Arabidopsis. New Phytol. 188, 161–174. [DOI] [PubMed] [Google Scholar]
- Nomura, H. , Komori, T. , Uemura, S. , Kanda, Y. , Shimotani, K. , Nakai, K. , Furuichi, T. , Takebayashi, K. , Sugimoto, T. , Sano, S. , Suwastika, I.N , Fukusaki, E. , Yoshioka, H. , Nakahira, Y. and Shiina, T. (2012) Chloroplast‐mediated activation of plant immune signalling in Arabidopsis. Nat. Commun. 3, 926. [DOI] [PubMed] [Google Scholar]
- Oh, C.‐S. and Beer, S.V. (2007) AtHIPM, an ortholog of the apple HrpN‐interacting protein, is a negative regulator of plant growth and mediates the growth‐enhancing effect of HrpN in Arabidopsis. Plant Physiol. 145, 426–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan, M.S. and Dinesh‐Kumar, S.P. (2010) All hands on deck—the role of chloroplasts, endoplasmic reticulum, and the nucleus in driving plant innate immunity. Mol. Plant–Microbe Interact. 23, 1368–1380. [DOI] [PubMed] [Google Scholar]
- Park, C.‐H. , Chen, S. , Shirsekar, G. , Zhou, B. , Khang, C.H. , Songkumarn, P. , Afzal, A.J. , Ning, Y. , Wang, R. , Bellizzi, M. , Valent, B. and Wang, G.‐L. (2012) The Magnaporthe oryzae effector AvrPiz‐t targets the RING E3 Ubiquitin Ligase APIP6 to suppress pathogen‐associated molecular pattern‐triggered immunity in rice. Plant Cell, 24, 4748–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier, J.‐B. , Ytterberg, A.J. , Sun, Q. and van Wijk, K.J. (2004) New functions of the thylakoid membrane proteome of Arabidopsis thaliana revealed by a simple, fast, and versatile fractionation strategy. J. Biol. Chem. 279, 49 367–49 383. [DOI] [PubMed] [Google Scholar]
- Penninckx, I.A. , Thomma, B.P. , Buchala, A. , Métraux, J.P. and Broekaert, W.F. (1998) Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell, 10, 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwessinger, B. and Ronald, P.C. (2012) Plant innate immunity: perception of conserved microbial signatures. Annu. Rev. Plant Biol. 63, 451–482. [DOI] [PubMed] [Google Scholar]
- Serrano, I. , Audran, C. and Rivas, S. (2016) Chloroplasts at work during plant innate immunity. J Exp Bot. 67, 3845–3854. [DOI] [PubMed] [Google Scholar]
- Serrano, M. , Wang, B. , Aryal, B. , Garcion, C. , Abou‐Mansour, E. , Heck, S. , Geisler, M. , Mauch, F. , Nawrath, C. and Métraux, J.‐P. (2013) Export of salicylic acid from the chloroplast requires the multidrug and toxin extrusion‐like transporter EDS5. Plant Physiol. 162, 1815–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiguzov, A. , Vainonen, J.P. , Wrzaczek, M. and Kangasjärvi, J. (2012) ROS‐talk – how the apoplast, the chloroplast, and the nucleus get the message through. Front. Plant Sci. 3, 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H. , Shen, Q. , Qi, Y. , Yan, H. , Nie, H. , Chen, Y. , Zhao, T. , Katagiri, F. and Tang, D. (2013) BR‐SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell, 25, 1143–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, D.K. , Maximova, S.N. , Jensen, P.J. , Lehman, B.L. , Ngugi, H.K. and McNellis, T.W. (2010) FIBRILLIN4 is required for plastoglobule development and stress resistance in apple and Arabidopsis. Plant Physiol. 154, 1281–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, D.K. , Laremore, T.N. , Smith, P.B. , Maximova, S.N. and McNellis, T.W. (2012) Knockdown of FIBRILLIN4 gene expression in apple decreases plastoglobule plastoquinone content. PLoS One, 7, e47547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, X. , Bariola, P. , Linderoth, N. , Fan, H. and Wei, Z.‐M. (2002) Receptors for hypersensitive response elicitors and uses thereof. US Patent Application No. 20020007501.
- Spoel, S.H. and Loake, G.J. (2011) Redox‐based protein modifications: the missing link in plant immune signalling. Curr. Opin. Plant Biol. 14, 358–364. [DOI] [PubMed] [Google Scholar]
- Stael, S. , Kmiecik, P. , Willems, P. , Van Der Kelen, K. , Coll, N.S. , Teige, M. and Van Breusegem, F. (2015) Plant innate immunity–sunny side up? Trends Plant Sci. 20, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweat, T.A. and Wolpert, T.J. (2007) Thioredoxin h5 is required for victorin sensitivity mediated by a CC‐NBS‐LRR gene in Arabidopsis. Plant Cell, 19, 673–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada, Y. , Spoel, S.H. , Pajerowska‐Mukhtar, K. , Mou, Z. , Song, J. , Wang, C. , Zuo, J. and Dong, X. (2008) Plant immunity requires conformational charges of NPR1 via S‐nitrosylation and thioredoxins. Science, 321, 952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, Y. , Xie, Z. , Chen, W. , Glazebrook, J. , Chang, H.‐S. and Han, B. (2003) Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae . Plant Cell, 15, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler, J.S. , Humphrey, P.T. and Whiteman, N.K. (2012) Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 17, 260–270. [DOI] [PubMed] [Google Scholar]
- Torres, M.A. , Dangl, J.L. and Jones, J.D.G. (2002) Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA, 99, 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torres Zabala, M. , Littlejohn, G. , Jayaraman, S. , Studholme, D. , Bailey, T. , Lawson, T. , Tillich, M. , Licht, D. , Bölter, B. , Delfino, L. , Truman, W. , Mansfield, J. , Smirnoff, N. and Grant, M. (2015) Chloroplasts play a central role in plant defence and are targeted by pathogen effectors. Nat. Plants, 1, 15 074. [DOI] [PubMed] [Google Scholar]
- Vidi, P.‐A. , Kanwischer, M. , Baginsky, S. , Austin, J.R. , Csucs, G. , Dörmann, P. , Kessler, F. and Bréhélin, C. (2006) Tocopherol cyclase (VTE1) localization and vitamin E accumulation in chloroplast plastoglobule lipoprotein particles. J. Biol. Chem. 281, 11 225–11 234. [DOI] [PubMed] [Google Scholar]
- Vlot, A.C. , Dempsey, D.M.A. and Klessig, D.F. (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. [DOI] [PubMed] [Google Scholar]
- Voss, I. , Koelmann, M. , Wojtera, J. , Holtgrefe, S. , Kitzmann, C. , Backhausen, J.E. and Scheibe, R. (2008) Knockout of major leaf ferredoxin reveals new redox‐regulatory adaptations in Arabidopsis thaliana . Physiol. Plant. 133, 584–598. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Nishimura, M.T. , Zhao, T. and Tang, D. (2011) ATG2, an autophagy‐related protein, negatively affects powdery mildew resistance and mildew‐induced cell death in Arabidopsis. Plant J. 68, 74–87. [DOI] [PubMed] [Google Scholar]
- Wasternack, C. and Hause, B. (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111, 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Z. , Laby, R. , Zumoff, C. , Bauer, D. , He, S. , Collmer, A. and Beer, S. (1992) Harpin, elicitor of the hypersensitive response produced by the plant pathogen Erwinia amylovora . Science, 257, 85–88. [DOI] [PubMed] [Google Scholar]
- Wildermuth, M.C. , Dewdney, J. , Wu, G. and Ausubel, F.M. (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature, 414, 562–565. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, K. , Yamada, K. , Ishikawa, K. , Yoshimura, S. , Hayashi, N. , Uchihashi, K. , Ishihama, N. , Kishi‐Kaboshi, M. , Takahashi, A. , Tsuge, S. , Ochiai, H. , Tada, Y. , Shimamoto, K. , Yoshioka, H. and Kawasaki, T. (2013) A receptor‐like cytoplasmic kinase targeted by a plant pathogen effector is directly phosphorylated by the chitin receptor and mediates rice immunity. Cell Host Microbe, 13, 347–357. [DOI] [PubMed] [Google Scholar]
- Yamamoto, H. , Kato, H. , Shinzaki, Y. , Horiguchi, S. , Shikanai, T. , Hase, T. , Endo, T. , Nishioka, M. , Makino, A. , Tomizawa, K. and Miyake, C. (2006) Ferredoxin limits cyclic electron flow around PSI (CEF‐PSI) in higher plants—stimulation of CEF‐PSI enhances non‐photochemical quenching of Chl fluorescence in transplastomic tobacco. Plant Cell Physiol. 47, 1355–1371. [DOI] [PubMed] [Google Scholar]
- Ytterberg, A.J. , Peltier, J.‐B. and Van Wijk, K.J. (2006) Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiol. 140, 984–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Li, W. , Xiang, T. , Liu, Z. , Laluk, K. , Ding, X. , Zou, Y. , Gao, M. , Zhang, X. , Chen, S. , Mengiste, T. , Zhang, Y. and Zhou, J.‐M. (2010) Receptor‐like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe, 7, 290–301. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Fan, W. , Kinkema, M. , Li, X. and Dong, X. (1999) Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR‐1 gene. Proc. Natl. Acad. Sci. USA, 96, 6523–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, X.‐Y , Spivey, N.W. , Zeng, W. , Liu, P.‐P. , Fu, Z.Q. , Klessig, D.F. , He, S.Y. and Dong, X. (2012) Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe, 11, 587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 The expression of Fd2 is reduced by Pseudomonas syringae pv. tomato (Pst) DC3000. (a) Total protein was extracted from Col‐0 before and at 2 days after Pst DC3000 infiltration. The protein level of Fd2 was determined by western blot using antiserum against ferredoxin in Spinacia oleracea (FDX1, top line) with Ponceau S staining of input Rubisco as the loading control (bottom line). (b) The relative intensity of immunoblotting bands is shown. Error bars represent the standard error (SE) (n = 3) from three biological repeats, and significance was determined at *P < 0.05 with Student's t‐test. dpi, days post‐infection.
Fig. S2 The transcriptional levels of NPR1 (a) and EDS5 (b) in Nossen and Fd2‐KO before and after Pseudomonas syringae pv. tomato (Pst) DC3000 [optical density at 600 nm (OD600) = 0.0002] infection. Quantitative real‐time polymerase chain reaction (qRT‐PCR) was carried out with Actin as the endogenous control. Significance was determined at *P < 0.05 and **P < 0.01 with Student's t‐test. Error bars represent the standard error (SE) (n = 3). Experiments were conducted in triplicate with similar results. dpi, days post‐infection.
Fig. S3 Protein levels of FIB4‐cLuc in Nicotiana benthamiana leaves transiently expressing split‐luciferase complementation vectors. Immunoblotting was carried out with the anti‐cLuc antibody.
Fig. S4 FIBRILLIN4 (FIB4) binds to HrpN. (a) FIB4 interacts with HrpN as indicated by a pull‐down assay. Purified GST‐HrpN and glutathione S‐transferase (GST) were bound to glutathione resin beads and incubated with purified maltose‐binding protein (MBP)‐FIB4. After the preparation had been washed, the in vitro interaction of MBP‐FIB4 with GST‐HrpN was detected by western blotting with α‐MBP antibody (top); 30% of the input was loaded and the sample amounts were shown by Coomassie blue staining (bottom). (b) FIB4 interacts with HrpN as indicated by co‐immunoprecipitation (co‐IP). FIB4‐GFP and Myc‐HrpN were co‐expressed in Nicotiana benthamiana by agroinfiltration, with Myc‐HrpN expressed alone as the control. Proteins were extracted 3 days after infiltration, and immunoprecipitation was carried out with anti‐green fluorescent protein (anti‐GFP) antibody (top). The input is shown at the bottom.
Fig. S5 The transcript analysis of FIB4 (a) and Fd2 (b) in Fd2‐KO and fib4‐1, respectively, relative to the wild‐types. FIB4 transcription was suppressed in Fd2‐KO, whereas Fd2 was induced in fib4‐1. Actin was used as the endogenous control. Significance was determined at **P < 0.01 with Student's t‐test. Error bars represent the standard error (SE) (n = 3). Experiments were conducted in triplicate with similar results.
Fig. S6 Both Fd2 and FIB4 are harpin‐responsive genes. Four‐week‐old Col‐0 plants were sprayed with 5 μg/mL glutathione S‐transferase (GST) and GST‐HrpN. Leaf tissues were harvested at the indicated times for analysis of the relative transcript levels of Fd2 (a) and FIB4 (b). Compared with the GST control, both Fd2 and FIB4 were up‐regulated by HrpN treatment. Actin was used as the endogenous control; error bars represent the standard error (SE) (n = 3); significance was determined at **P < 0.01 with Student's t‐test. Experiments were conducted in triplicate with similar results.
Fig. S7 Both FIBRILLIN4 (FIB4) and Fd2 are localized to chloroplasts. Protoplasts were prepared from Nicotiana benthamiana leaves transiently expressing FIB4‐GFP or Fd2‐GFP by agroinfiltration. Both Fd2‐GFP and FIB4‐GFP (green) localize to chloroplasts (red, autofluorescence). Bars, 10 μm.
Fig. S8 Immunoblotting to detect Fd2‐GFP transiently expressed in Nicotiana benthamiana leaves. Fd2‐GFP was transiently expressed in N. benthamiana by agroinfiltration. Total protein was extracted 3 days after infiltration, and western blot was carried out with anti‐green fluorescent protein (anti‐GFP) antibody.
Fig. S9 Transient expression of HrpN in Nicotiana benthamiana caused weak cell death. At 3 days after transient expression of HrpN‐GFP by agroinfiltration, a weak, hypersensitive response (HR)‐like cell death was observed on the leaf. The leaf expressing Fd2‐GFP was used as a control. Bars, 5 mm.
Table S1 Primers used for plasmid construction.
Table S2 Primers used for real‐time polymerase chain reaction (PCR) analysis.
Table S3 Tandem mass spectroscopy (MS/MS) settings and mass transition used to quantify salicylic acid (SA), jasmonic acid (JA) and JA‐isoleucine/leucine (JA‐Ile/Leu).


