Summary
The subversion of plant cellular functions is essential for bacterial pathogens to proliferate in host plants and cause disease. Most bacterial plant pathogens employ a type III secretion system to inject type III effector (T3E) proteins inside plant cells, where they contribute to the pathogen‐induced alteration of plant physiology. In this work, we found that the Ralstonia solanacearum T3E RipAY suppresses plant immune responses triggered by bacterial elicitors and by the phytohormone salicylic acid. Further biochemical analysis indicated that RipAY associates in planta with thioredoxins from Nicotiana benthamiana and Arabidopsis. Interestingly, RipAY displays γ‐glutamyl cyclotransferase (GGCT) activity to degrade glutathione in plant cells, which is required for the reported suppression of immune responses. Given the importance of thioredoxins and glutathione as major redox regulators in eukaryotic cells, RipAY activity may constitute a novel and powerful virulence strategy employed by R. solanacearum to suppress immune responses and potentially alter general redox signalling in host cells.
Keywords: glutathione, plant immunity, Ralstonia, redox, thioredoxin, type III effector
Introduction
To infect a plant, pathogens need to manipulate host cells to turn an otherwise hostile environment into a niche suitable for pathogen proliferation. For Gram‐negative bacterial pathogens of plants and animals, an essential means to achieve host manipulation is the type III secretion system (T3SS), a molecular syringe that injects proteins, termed type III effectors (T3Es), directly inside host cells (Galan et al., 2014).
Plants have developed a complex immune system capable of perceiving multiple kinds of microbial molecules, which are interpreted as invasion patterns (IPs; Cook et al., 2015). Some bacterial proteins can be perceived directly at the plant cell surface by receptors localized at the plasma membrane, termed pattern recognition receptors (PRRs; Zipfel, 2014). Bacterial proteins delivered inside plant cells can also be perceived, in a direct or indirect manner, by intracellular receptors containing nucleotide‐binding and leucine‐rich repeat (NB‐LRR) domains (NLRs; Khan et al., 2016). The activation of these IP receptors leads to an extensive reprogramming of host cells in order to strengthen defences and initiate immune responses (Bigeard et al., 2015; Boller and Felix, 2009; Tsuda and Katagiri, 2010). This immune activation is expected to prevent the proliferation of the perceived bacteria, and to prepare plant cells for an efficient defence response against further invading bacteria. To neutralize this plant surveillance system and the subsequent immune responses, successful bacterial pathogens have evolved T3Es that are able to suppress plant immune signalling at multiple levels (Deslandes and Rivas, 2012; Macho and Zipfel, 2015). This immune suppression is supported by the subversion of additional plant functions (Macho, 2016). T3E manipulation of plant cellular functions is key to allow bacterial proliferation and, ultimately, the development of disease, but presents the additional risk of T3E perception by NLRs (Khan et al., 2016). The definition of opposite forces that determine the outcome of a plant–bacteria interaction (bacterial virulence activities versus plant immune responses) has led to the proposition of a model presenting a complex evolutionary arms race between effectors and plant defence responses (Jones and Dangl, 2006; Win et al., 2012). The complex outcome of this interaction will influence the perception and response to further bacteria at the site of infection and distal tissues.
Plants rely on a balanced cellular environment to maintain basal levels of immune regulators and to mount defence responses after pathogen perception. Redox regulators efficiently buffer the redox status of plant cells, and play important roles in the activation of immune responses after pathogen perception (Spoel and Loake, 2011). However, our knowledge on whether and how pathogens manipulate redox regulation to cause disease is still scarce.
Ralstonia solanacearum is often considered to be one of the most destructive bacterial pathogens, causing bacterial wilt disease in more than 250 plant species worldwide (Mansfield et al., 2012). Ralstonia solanacearum can live for long periods in water or soil, and is able to penetrate plants through the roots. After plant invasion, R. solanacearum reaches the vascular system and uses the xylem vessels to colonize the whole plant. Massive bacterial replication and production of exopolysaccharide eventually lead to the collapse of the vascular system, therefore causing severe wilting and plant death (Peeters et al., 2013b). It has been shown that R. solanacearum is able to express genes related to the T3SS throughout different stages of the infection process (Monteiro et al., 2012). Interestingly, the versatile lifestyle of R. solanacearum and the colonization of different plant organs correlate with a larger number of T3Es compared with other bacterial pathogens: bacteria from a single R. solanacearum strain can inject up to 70 T3Es (termed Rips for Ralstonia injected proteins) inside plant cells (Peeters et al., 2013a). Although the role of several of these effectors has been studied (Deslandes and Genin, 2014), the relevance and function of most remain unknown, and therefore the R. solanacearum T3E repertoire constitutes a powerful tool for the identification of novel bacterial virulence activities and for the investigation of the pathogen manipulation of plant functions. One of these T3Es, RipAY, has been shown to contribute to R. solanacearum virulence in eggplant leaves (Macho et al., 2010). Interestingly, RipAY is widely conserved amongst the R. solanacearum strains sequenced to date (Clarke et al., 2015; Peeters et al., 2013a), and is one of the few Rips that does not trigger hypersensitive response (HR)‐like responses in any of the plant genotypes tested (Clarke et al., 2015).
In this work, we studied the potential basis of the virulence activity of RipAY, and found that RipAY suppresses immune responses triggered by bacterial elicitors and the defence‐related phytohormone salicylic acid (SA). Further biochemical analysis indicated that RipAY associates with several redox regulators and that the manipulation of plant redox regulation is essential for the observed suppression of immune responses.
Results
RipAY can suppress immune responses
Several T3Es from different bacterial pathogens contribute to the suppression of plant defence responses (Deslandes and Rivas, 2012; Macho and Zipfel, 2015). The activation of plant PRRs by microbial elicitors leads to a plethora of signalling events and immune responses, including an early burst of reactive oxygen species (ROS) and the activation of a cascade of mitogen‐activated protein kinases (MAPKs) (Boller and Felix, 2009; Macho and Zipfel, 2014). The peptide flg22 (representative of one of the elicitor domains of bacterial flagellin) is perceived in most plants by a PRR complex formed by the leucine‐rich repeat receptor kinases FLS2 and BAK1 (Gómez‐Gómez and Boller, 2000; Sun et al., 2013), and is commonly employed to elicit and probe PRR‐dependent responses in laboratory conditions. To determine whether RipAY has the potential to suppress elicitor‐induced responses in plant cells, we used an Agrobacterium tumefaciens‐mediated transient expression system in Nicotiana benthamiana leaf tissues. Two days after infiltration with A. tumefaciens, we observed a strong reduction in the flg22‐triggered ROS burst in half‐leaves expressing RipAY, fused to a green fluorescent protein (GFP) tag or a haemagglutinin‐StrepII (HA‐StrepII) tag (Figs 1a,b and S1, see Supporting Information). As controls, we used A. tumefaciens carrying constructs to trigger the expression of GFP or GFP‐HA‐StrepII, infiltrated in the other half of the leaf, allowing a direct comparison with RipAY‐expressing tissues (Figs 1a,b and S1). Interestingly, RipAY expression did not cause a clear and reproducible suppression of flg22‐triggered MAPK activation (Fig. S2, see Supporting Information). This may indicate that RipAY differentially affects responses to flg22, although an influence of the differential sensitivity of the assays for the determination of the ROS burst and the activation of MAPKs cannot be excluded.
Figure 1.
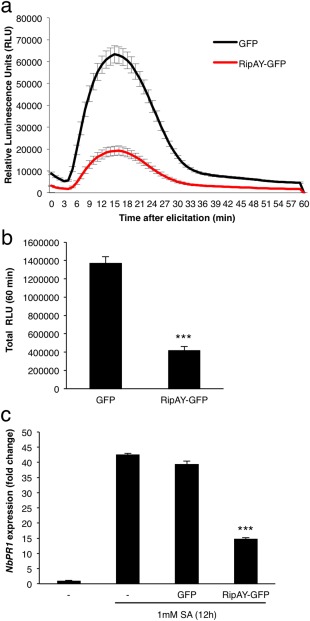
RipAY‐GFP suppresses immune responses in Nicotiana benthamiana leaves. Agrobacterium tumefaciens was used to induce the transient expression of RipAY‐GFP in one half of the leaf and green fluorescent protein (GFP) in the other half. (a, b) Oxidative burst triggered by 50 nm flg22 in N. benthamiana tissues at 2.5 days post‐inoculation (dpi) with A. tumefaciens, and measured in a luminol‐based assay as relative luminescence units (RLU). Values are the average ± standard error (SE) (n = 24). (c) Quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of NbPR1 transcripts in N. benthamiana leaves, either untreated or treated with 1 mm salicylic acid (SA) for 12 h, at 2.5 dpi with A. tumefaciens. Gene expression values are relative to the NbEF1α housekeeping gene and are normalized to untreated N. benthamiana tissues. Values are the average ± SE (n = 3). Asterisks indicate significant differences compared with the corresponding GFP control at P < 0.001. The experiments were repeated at least three times with similar results.
SA plays a major role in the activation of immune responses after the perception of different types of IPs (Vlot et al., 2009). To determine whether RipAY interferes with SA‐dependent responses, we measured the expression of the N. benthamiana orthologue of the Arabidopsis gene PATHOGENESIS‐RELATED‐1 (PR1), which is a hallmark of SA‐dependent responses (Vlot et al., 2009; Ward et al., 1991). Expression of RipAY‐GFP significantly reduced the SA‐induced accumulation of NbPR1 transcripts (Fig. 1c), suggesting that RipAY is able to suppress SA‐dependent gene expression.
RipAY‐GFP localizes in the nucleus and cytoplasm in N. benthamiana cells
To determine the subcellular localization of RipAY in plant cells, we transiently expressed RipAY fused to a monomeric red fluorescent protein (mRFP) tag in epidermal cells of N. benthamiana and observed the RFP fluorescence using confocal microscopy. As a control, we co‐expressed a nuclear addressed Nuclear Localisation Signal (NLS)‐GFP. Agrobacterium‐mediated transient expression indicated a cytoplasmic and nuclear localization of RipAY‐mRFP (Figs 2 and S3, see Supporting Information). Western blot analysis confirmed that most of the signal is associated with the full‐length RipAY‐mRFP fusion protein present in the observed tissues (Fig. S4, see Supporting Information).
Figure 2.
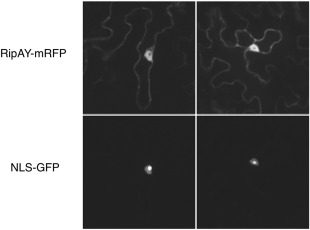
RipAY‐mRFP localizes to cytoplasmic and nuclear plant cell compartments. Two representative images from the observation of RipAY‐mRFP and NLS‐GFP co‐transformed Nicotiana benthamiana leaf cells. The top images are taken in the red channel, showing the localization of RipAY‐mRFP, with a cytoplasmic and nuclear localization. The bottom images, taken in the green channel, show the nuclear localization of NLS‐GFP, with strong accumulation in the nucleolus. Each square has a real‐life size of 164 μm. GFP, green fluorescent protein; mRFP, monomeric red fluorescent protein.
RipAY associates with plant h‐type thioredoxins
In order to identify plant proteins that interact with RipAY, we expressed RipAY‐GFP in N. benthamiana and purified the fusion protein, together with associated proteins, by immunoprecipitation using agarose beads coupled to an anti‐GFP nanobody. To increase the robustness of our analysis, we also expressed RipAY‐HA‐StrepII and purified the fusion protein by affinity pull‐down using a Superflow resin bound to Strep‐Tactin. As controls, we expressed and purified GFP and GFP‐HA‐StrepII, respectively. The purified RipAY and associated proteins were then digested with trypsin and processed by liquid chromatography‐tandem mass spectrometry (LC‐MS/MS). In several biological replicates, LC‐MS/MS analysis of RipAY purifications with both tags showed abundant peptides from several members of the thioredoxin h (TRX‐h) family (Table 1), whereas no TRX‐h peptides were identified after purification of GFP or GFP‐HA‐StrepII. Thioredoxins are small enzymes that reduce disulfide bonds of target proteins, and have been found to regulate a variety of biological processes in plants (Gelhaye et al., 2005), including immune responses (Kneeshaw et al., 2014; Tada et al., 2008). Members of the TRX‐h family are mostly considered to be cytosolic proteins, although membrane and nuclear localizations have also been demonstrated in specific cases (Delorme‐Hinoux et al., 2016; Gelhaye et al., 2005). To confirm this finding, we selected the four NbTRX‐h proteins identified by LC‐MS/MS and their closest paralogues (Fig. S5, see Supporting Information) and co‐expressed them, fused to a C‐terminal FLAG tag, together with RipAY‐GFP. GFP immunoprecipitation assays indicated that all NbTRX‐h tested associated in planta with RipAY‐GFP, but not with GFP alone used as a control (Fig. 3a).
Table 1.
Thioredoxin peptides identified in liquid chromatography‐tandem mass spectrometry (LC‐MS/MS) analysis after purification of RipAY‐GFP and RipAY‐HA‐StrepII from Nicotiana benthamiana leaves.
| Gene name | Gene number | Exclusive unique peptide count | Protein coverage (%) | Best mascot ion score | |
|---|---|---|---|---|---|
| RipAY‐GFP purification | |||||
| Thioredoxin h type | NbS00049748g0003.1 | 5 | 56 | 120 | |
| Thioredoxin h type 1 | NbS00034448g0009.1 | 14 | 82 | 119 | |
| Thioredoxin h type 1 | NbS00026639g0011.1 | 5 | 81 | 124.8 | |
| Thioredoxin h type 2 | NbS00012766g0011.1 | 7 | 76 | 95.1 | |
| RipAY‐HA‐StrepII purification | |||||
| Thioredoxin h type | NbS00049748g0003.1 | 5 | 51 | 91.2 | |
| Thioredoxin h type 1 | NbS00034448g0009.1 | 14 | 83 | 113 | |
| Thioredoxin h type 1 | NbS00026639g0011.1 | 5 | 82 | 125.5 | |
| Thioredoxin h type 2 | NbS00012766g0011.1 | 5 | 59 | 95.1 | |
Figure 3.
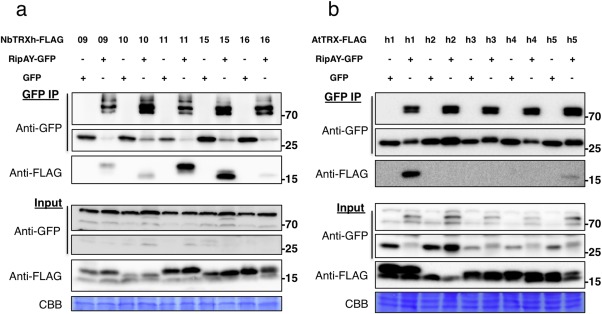
RipAY associates with h‐type thioredoxins in plant cells. GFP or RipAY‐GFP was co‐expressed with plant h‐type thioredoxins in Nicotiana benthamiana before immunoprecipitation using green fluorescent protein (GFP)‐trap beads. (a) RipAY association with the indicated h‐type thioredoxins from N. benthamiana (NbTRX‐h). (b) RipAY association with the indicated h‐type thioredoxins from Arabidopsis (AtTRX‐h). Immunoblots were analysed using anti‐GFP or anti‐FLAG antibody. CBB, Coomassie brilliant blue. Molecular weight (kDa) marker bands are indicated for reference. The experiments were repeated three times with similar results.
To determine whether RipAY interaction is extended to TRX‐h from other plant species, we co‐expressed RipAY‐GFP together with five different TRX‐h proteins from Arabidopsis (AtTRX‐h1–5) in N. benthamiana. GFP immunoprecipitation assays indicated that AtTRX‐h1 and AtTRX‐h5 interact with RipAY‐GFP (Fig. 3b). AtTRX‐h1 was most similar to the NbTRX‐h proteins tested (Fig. S5), and showed a stronger interaction with RipAY relative to AtTRX‐h5 (Fig. 3b). It is noteworthy that we occasionally detected weaker interactions with AtTRX‐h3 and AtTRX‐h4 (data not shown). Interestingly, AtTRX‐h5 plays an important role in the regulation of immune responses, predominantly through the redox regulation of the SA‐dependent transcriptional co‐activator NONEXPRESSOR OF PATHOGENESIS‐RELATED GENES 1 (NPR1) (Kneeshaw et al., 2014; Tada et al., 2008). The expression of TRX‐h5 is up‐regulated by flg22, SA and pathogen infection (Laloi et al., 2004; Reichheld et al., 2002; Tada et al., 2008), and trxh5 mutants show compromised PR1 expression after SA treatment and compromised systemic acquired resistance (SAR) (Tada et al., 2008).
RipAY cysteine (Cys) is not a target of thioredoxin redox regulation
Plant proteins that associate with pathogen effectors are usually considered to be virulence targets, but they can also contribute to the activation of effector activities by different mechanisms (Win et al., 2012). Considering that TRXs generally modulate the redox status of target proteins by reducing disulfide bonds in target Cys residues, we explored whether this is the case for the interaction between TRX‐h proteins and RipAY. Preliminary sequence analysis suggested that RipAY is not susceptible to the reduction of intramolecular disulfide bonds, as it has only one Cys residue (C333; Fig. S6, see Supporting Information). Accordingly, mutation of C333 to a serine (C333S) did not affect the RipAY suppression of the flg22‐triggered ROS burst (Fig. 4a), and RipAY C333S retained the ability to associate with TRXs in co‐immunoprecipitation assays (Fig. 4b). These results suggest that the interaction between RipAY and TRXs is not based on intramolecular or intermolecular disulfide bond reduction.
Figure 4.
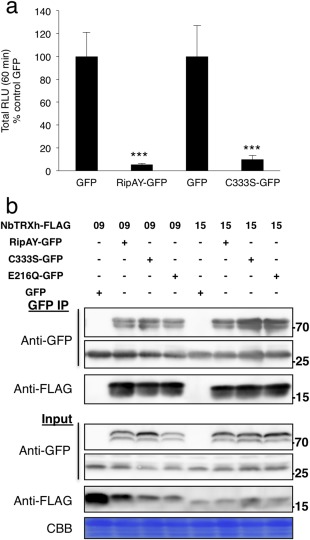
RipAY cysteine 333 is not critical for RipAY functions in Nicotiana benthamiana. (a) Suppression of flg22‐triggered reactive oxygen species (ROS) burst by RipAY‐GFP and RipAY‐C333S‐GFP variant transiently expressed in N. benthamiana leaves. Agrobacterium tumefaciens was used to induce the transient expression of RipAY‐GFP (or RipAY‐C333S‐GFP) in one half of the leaf and green fluorescent protein (GFP) in the other half. The oxidative burst was triggered by 50 nm flg22 in N. benthamiana tissues at 2.5 days post‐inoculation (dpi) with A. tumefaciens, and measured in a luminol‐based assay as relative luminescence units (RLU). Values are the average ± standard error (SE) (n = 24), and are represented as a percentage of the corresponding GFP control. Asterisks indicate significant differences compared with the corresponding GFP control at P < 0.001. (b) Association between RipAY‐C333S or RipAY‐E216Q variants and N. benthamiana h‐type thioredoxins. GFP, RipAY‐GFP, RipAY‐C333S‐GFP or RipAY‐E216Q‐GFP was co‐expressed with NbTRX‐h‐09‐FLAG and NbTRX‐h‐15‐FLAG before immunoprecipitation using GFP‐trap beads. Immunoblots were analysed using anti‐GFP or anti‐FLAG antibody. CBB, Coomassie brilliant blue. Molecular weight (kDa) marker bands are indicated for reference. The experiments were repeated three times with similar results.
Thioredoxins as potential targets of RipAY virulence activities
To investigate whether h‐type TRXs regulate immune responses in N. benthamiana, we employed a genetic approach based on virus‐induced gene silencing (VIGS) or the overexpression of different NbTRX‐h genes. We designed two different silencing constructs, which targeted different groups of genes for silencing: the NbTRX‐h9si construct is designed to target NbTRX‐h9 and NbTRX‐h11, whereas the NbTRX‐h15si construct is designed to target NbTRX‐h15, NbTRX‐h10 and NbTRX‐h16 (Fig. S7a, see Supporting Information). Transient expression of the mentioned silencing constructs in 3‐week‐old N. benthamiana plants led to a reduction in mRNA levels of the corresponding NbTRX‐h genes in younger leaves at 20 days post‐infiltration (dpi) (Fig. S7b). Strikingly, although plants expressing the TRX‐h15si construct showed a slight decrease in flg22‐triggered ROS, plants expressing the TRX‐h9si construct displayed an enhanced ROS burst after elicitor treatment (Fig. S7c). These results may be caused by the different roles of the different TRX‐h proteins, although RipAY associated with all of them (Fig. 3a), or to pleiotropic effects on additional TRXs or other redox regulators caused by the silencing of the selected TRX‐h genes.
Overexpression of the different NbTRX‐h genes from a cauliflower mosaic virus 35S promoter (generating NbTRX‐h‐FLAG fusion proteins) did not cause significant differences in flg22‐triggered ROS (Fig. S8a, see Supporting Information). However, the overexpression of several NbTRX‐h genes partially rescued the inhibition of flg22‐triggered ROS caused by RipAY (Fig. S8b), suggesting that h‐type TRXs are associated with the virulence activity of RipAY.
As RipAY associates with AtTRX‐h5 (Fig. 3b) and this protein is required for several immune responses (including PR1 expression and SAR; Tada et al., 2008), we tested whether an AtTRX‐h5 loss‐of‐function mutant (Attrxh5−4; Sweat and Wolpert, 2007) was also impaired in the flg22‐triggered ROS burst, as this response is clearly inhibited by RipAY (Fig. 1). However, flg22‐triggered ROS in Attrxh5−4 showed a pattern similar to that in Col‐0 wild‐type plants (Fig. S9, see Supporting Information). This suggests that additional TRXs may contribute to the regulation of the flg22‐triggered ROS burst, or that RipAY may additionally target other plant proteins to suppress elicitor‐induced responses.
RipAY displays γ‐glutamyl cyclotransferase (GGCT) activity to degrade glutathione in planta
RipAY has no overall sequence similarity with other proteins from any organism, including plants or other plant pathogens. However, a search in the National Center for Biotechnology Information (NCBI) conserved domain database (Marchler‐Bauer et al., 2015) indicated that the RipAY central region (amino acids 126–252; E value = 1.65e‐05; Fig. S6) is similar to proteins from the GGCT‐like superfamily, associated with the degradation of glutathione in a multitude of organisms, including plants (Kumar et al., 2015; Oakley et al., 2008; Paulose et al., 2013). Therefore, we decided to test whether RipAY has an impact on the glutathione content of plant cells. At 1 dpi with A. tumefaciens, the glutathione content of N. benthamiana tissues expressing RipAY‐GFP was decreased to 30% of that of control tissues expressing GFP, despite the fact that RipAY‐GFP accumulation was still very low (Fig. 5a). The glutathione content decreased progressively in the following days, reaching 7.3% of that of control tissues at 3 dpi (Fig. 5a). The key catalytic glutamic acid residue for GGCT activity in human and plant GGCTs is conserved in RipAY (E216; Fig. S6), and mutation of this residue to glutamine has been reported to abolish GGCT activity without altering the general protein structure (Oakley et al., 2008). Mutation of RipAY E216 to glutamine (E216Q) did not affect RipAY accumulation or the interaction with plant thioredoxins (Fig. 4b), but completely abolished the RipAY‐mediated decrease in plant glutathione levels (Fig. 5a), indicating that the observed glutathione degradation is a product of RipAY GGCT activity.
Figure 5.
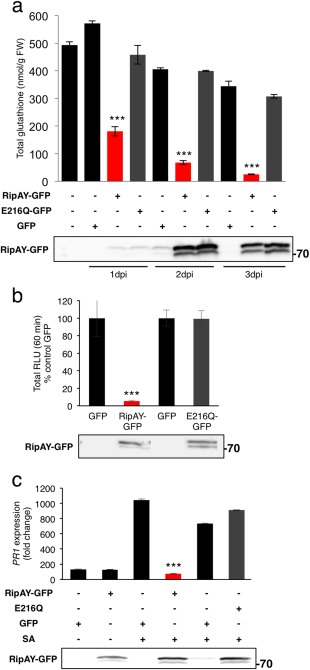
RipAY displays γ‐glutamyl cyclotransferase (GGCT) activity in plant cells, which is required to degrade glutathione and suppress immune responses. (a) Total glutathione content in Nicotiana benthamiana tissues at 1, 2 or 3 days post‐inoculation with Agrobacterium tumefaciens inducing the expression of green fluorescent protein (GFP), RipAY‐GFP or RipAY‐E216Q‐GFP. Values are the average ± standard error (SE) (n = 3). (b) Agrobacterium tumefaciens was used to induce the transient expression of RipAY‐GFP (or RipAY‐E216Q‐GFP) in one half of the leaf and GFP in the other half. The oxidative burst was triggered by 50 nm flg22 in N. benthamiana tissues at 2.5 days post‐inoculation with A. tumefaciens, and measured in a luminol‐based assay as relative luminescence units (RLU). Values are the average ± SE (n = 24), and are represented as a percentage of the corresponding GFP control. (c) Quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of NbPR1 transcripts in N. benthamiana leaves, either untreated or treated with 1 mm salicylic acid (SA) for 12 h, at 2.5 days post‐inoculation with A. tumefaciens. Gene expression values are relative to the NbEF1α housekeeping gene and are normalized to untreated N. benthamiana tissues. Values are the average ± SE (n = 3). Immunoblots were analysed using anti‐GFP antibody to verify protein accumulation. Asterisks indicate significant differences compared with the corresponding GFP control at P < 0.001. The experiments were repeated three times with similar results.
Intriguingly, we were unable to generate Arabidopsis transgenic lines expressing RipAY, probably because of the toxicity of sustained RipAY expression. Even the basal expression detected using a dexamethasone (DEX)‐inducible construct (data not shown) prevented us from selecting RipAY‐expressing plants. To determine whether RipAY is able to degrade glutathione in Arabidopsis cells, we isolated mesophyll protoplasts from Arabidopsis leaves and transfected them with a construct driving the expression of RipAY‐HA‐StrepII. Protoplasts expressing RipAY‐HA‐StrepII showed a depletion of glutathione, which required an intact E216 residue (Fig. S10a, see Supporting Information). The RipAY‐mediated degradation of glutathione correlated with a suppression of SA‐induced accumulation of AtPR1 transcripts (Fig. S10c), indicating that RipAY also acts as a GGCT and suppresses immune‐related signalling in Arabidopsis cells.
GGCT activity is required for the suppression of immune responses
Glutathione is a master redox buffer in prokaryotes and eukaryotes, and plays a major role in plant redox homeostasis (Noctor et al., 2011, 2012). Interestingly, glutathione is essential for the regulation of plant responses to environmental stresses, including immune responses (Frendo et al., 2013; Ghanta and Chattopadhyay, 2011). Considering this, we speculated that the GGCT activity could contribute to RipAY immune suppression activities. Indeed, the E216Q mutation suppressed the ability of RipAY to inhibit the flg22‐triggered ROS burst (Fig. 5b) and SA‐induced PR1 expression (Figs 5c and S10c), indicating that RipAY GGCT activity is essential for RipAY suppression of the tested immune responses. Interestingly, the E216Q mutation did not affect RipAY interaction with plant thioredoxins (Fig. 4b).
Considering the dramatic impact of RipAY in plant glutathione content and the flg22‐triggered ROS burst (Figs 1 and 5), we decided to test whether a plant mutant with low glutathione content displays a similar inhibition of flg22‐triggered ROS. The pad2‐1 mutant accumulates approximately 20% of glutathione compared with wild‐type plants, and is affected in several immune responses (Dubreuil‐Maurizi and Poinssot, 2012). Strikingly, we did not detect differences in the flg22‐triggered ROS burst in pad2‐1 mutants compared with Col‐0 wild‐type plants (Fig. S11a, see Supporting Information). Moreover, the pad2‐1 mutation did not cause a robust increase in the development of disease symptoms caused by R. solanacearum infection (Fig. S11b). As an alternative chemical approach to detect the effect of glutathione depletion in immune responses, we treated N. benthamiana tissues with 1‐chloro‐2,4‐dinitrobenzene (CDNB), which has been shown to decrease glutathione contents in plant cells (Okuma et al., 2011). Treatment with increasing CDNB concentrations resulted in a reduction in glutathione content, which correlated with a reduction in the flg22‐triggered ROS burst (Fig. S12), supporting the notion that a decrease in glutathione concentration causes an inhibition of the plant response to elicitors.
Discussion
Redox regulation plays an important role in plant responses to biotic and abiotic stress (Spoel and Loake, 2011; Suzuki et al., 2011). Therefore, the manipulation of redox regulators may constitute a powerful tool for pathogens to subvert plant immune responses and have a broad impact on plant stress signalling. In this work, we found that the expression of the R. solanacearum T3E RipAY in plant cells leads to a suppression of elicitor and SA‐triggered immune responses. Further biochemical analysis showed that RipAY degrades glutathione in plant cells. Glutathione is a master redox buffer (Noctor et al., 2011, 2012), which contributes to the activation of diverse immune responses (Frendo et al., 2013; Ghanta and Chattopadhyay, 2011). RipAY immune suppression activities are abolished in a mutant affected on the catalytic core of GGCT activity, which cannot degrade glutathione. This points to the degradation of glutathione as an important virulence activity of RipAY towards the suppression of immune responses. Similarly, we showed that chemical depletion of glutathione in plant cells leads to the suppression of elicitor‐triggered immune responses. This is consistent with previous reports showing a major contribution of glutathione to plant defence responses. For instance, the pad2‐1 mutant (deficient in glutathione biosynthesis) is impaired in the accumulation of camalexin, glucosinolates and SA, and shows deficient SA‐dependent responses, including PR1 expression (Dubreuil‐Maurizi and Poinssot, 2012). In addition, pad2 mutant plants are impaired in flg22‐triggered callose deposition (Clay et al., 2009) and are more susceptible to a wide range of pathogens and pests (Dubreuil‐Maurizi and Poinssot, 2012). The association between glutathione content and plant responses to biotic stress is also supported by other reports using different glutathione‐deficient mutants (Ball et al., 2004). Surprisingly, in our experimental conditions, the pad2‐1 mutant was not affected in the flg22‐triggered ROS burst and did not show a clearly increased susceptibility to R. solanacearum. Considering that GGCT activity is required for RipAY suppression of the tested immune responses, these results suggest different possible scenarios: (i) RipAY GGCT activity may have a stronger impact on glutathione content relative to the effect of the pad2‐1 mutant; (ii) RipAY degradation of other γ‐glutamyl compounds, in addition to glutathione, may contribute to the observed inhibition of immune responses; or (iii) a synergistic effect between glutathione degradation and the targeting of other plant proteins may be required for the full virulence activity of RipAY. It is also possible that additional effects of the pad2‐1 mutation counteract the effect of glutathione deficiency, such as the reported up‐regulation of RBOHD (Dubreuil‐Maurizi et al., 2011), which encodes the main NADPH oxidase responsible for the flg22‐triggered ROS burst (Nühse et al., 2007; Zhang et al., 2007).
RipAY associates with several h‐type thioredoxins from N. benthamiana and Arabidopsis. The h‐type thioredoxins play important roles in immune responses, especially AtTRX‐h5 (Kneeshaw et al., 2014; Tada et al., 2008), which we found to associate with RipAY in plant cells. Plant interactors can be effector targets, but may also contribute to effector activation as helpers (Win et al., 2012). Therefore, we considered the possibility of TRXs acting as chaperones/helpers for RipAY, and/or contributing to activate its function. The most predominant activity described for thioredoxins is the reduction of disulfide bonds in target proteins. Targets of cytosolic thioredoxins usually have numerous Cys residues suitable for redox regulation (Ueoka‐Nakanishi et al., 2013). However, RipAY has only one Cys residue (C333; Fig. S6), and we found that this Cys is not required for the interaction with thioredoxins in planta or for the RipAY‐mediated suppression of the flg22‐triggered ROS burst. Moreover, the interaction between RipAY and the different TRXs tested is very strong and stable. This is in contrast with the transient interaction between TRXs and their targets, which usually can only be detected by stabilizing the interaction with a mutation in the second Cys of the TRX catalytic domain (Hisabori et al., 2005). Therefore, RipAY sequence analysis and our experimental data suggest that the interaction between RipAY and thioredoxins is not based on the usual thioredoxin‐mediated reduction of disulfide bonds. In addition, we found that the overexpression of several TRXs inhibits the immune suppression activity of RipAY instead of contributing to it. This suggests that TRX overexpression could compensate for the RipAY targeting of endogenous thioredoxins, which would point to a scenario in which thioredoxins are virulence targets of RipAY, or in which additional redox buffering by TRXs counteracts RipAY activity.
TRXs have been postulated as effector targets based on the presence of TRX‐like domains in NB‐LRR proteins (integrated decoy model; Cesari et al., 2014; Nishimura et al., 2015). Indeed, the victorin toxin from the fungus Cochliobolus victoriae takes advantage of the guarding of plant TRXs by NB‐LRR proteins to induce cell death in plants, which promotes disease development (Lorang et al., 2012; Sweat and Wolpert, 2007). Recently, an effector from the plant‐parasitic nematode Meloidogyne javanica has been found to manipulate the ferredoxin–thioredoxin system in root plastids, which activates ROS scavenging and suppresses immune responses, enhancing susceptibility to the nematode (Lin et al., 2016). Arabidopsis TRX‐h5 and TRX‐h3 have also been found to interact with an effector from the plant‐pathogenic oomycete Hyaloperonospora arabidopsidis (Wessling et al., 2014). This suggests that pathogens from different kingdoms may have evolved to associate with plant TRXs as a general strategy to promote plant infection. Our results suggest that RipAY may affect different targets to collectively manipulate redox regulation in plant cells, leading to the suppression of immune responses (Fig. S13, see Supporting Information). The reducing activities of TRXs seem to be associated with glutathione and glutathione reductases (Gelhaye et al., 2004; Meyer et al., 2012; Reichheld et al., 2007), and TRXs and glutathione have been found to play partially overlapping roles in yeast and plants (Kovacs et al., 2015; Sharma et al., 2000). Therefore, it may be difficult to uncouple a potential effect of RipAY on TRXs and glutathione.
In a recent report, Fujiwara et al. (2016) showed that RipAY expression inhibits yeast growth, degrades glutathione and interacts with TRXs in yeast. A detailed biochemical analysis indicated that RipAY displays a remarkably strong GGCT activity, which is required for the reported inhibition of yeast growth (Fujiwara et al., 2016). Moreover, infiltration of R. solanacearum in eggplant leaves causes a RipAY GGCT‐dependent depletion of glutathione (Fujiwara et al., 2016). Interestingly, this report shows that RipAY requires the presence of eukaryotic TRXs to degrade glutathione in vitro and in yeast cells, and suggests that TRXs function as activators for RipAY GGCT activity. Consistent with our data in planta, Fujiwara et al. (2016) showed that a redox‐inactive mutant of AtTRX‐h5 (C39S C42S) still interacts with RipAY, and a RipAY C333S mutant still interacts with TRXs in yeast, pointing to a redox‐independent interaction between RipAY and TRXs. In the light of these results, and considering that GGCT activity is essential for RipAY suppression of immune responses, we cannot rule out the possibility that the interaction with TRXs somehow contributes to the function of RipAY via an as yet unknown mechanism not based on redox regulation.
The mechanism of RipAY‐mediated suppression of immunity after association with TRXs and degradation of glutathione could be related to the transcriptional co‐activator NPR1. The activation and turnover of NPR1 rely on post‐translational modifications, and are both required for the activation and maintenance of a subset of SA‐dependent immune responses (Pajerowska‐Mukhtar et al., 2013; Saleh et al., 2015). SA activates the reduction of the oligomeric cytosolic form of NPR1, catalysed by glutathione and TRX through disulfide reduction and denitrosylation (Kneeshaw et al., 2014; Kovacs et al., 2015; Mou et al., 2003; Tada et al., 2008). Then, the reduced monomeric form of NPR1 translocates to the nucleus to activate the transcription of defence‐related genes. RipAY targeting of redox regulators could impact NPR1, among other regulators of immune responses. Consistently, silencing of NPR1 in tomato enhances the infection by R. solanacearum (Chen et al., 2009). Further work is required to decipher whether the molecular mechanism of RipAY suppression of immunity is related to the redox regulation of NPR1 or other regulators of SA signalling.
The RipAY targeting of redox signalling and the suppression of immune responses could contribute to the virulence activity of RipAY in natural infections. It is important to note that our results showing plant immune suppression by RipAY were obtained after transient expression of RipAY in plant tissues. In natural infections, bacteria are expected to translocate small amounts of most effector proteins, in comparison with the abundance of effector proteins accumulated in plant tissues after transient expression. However, Fujiwara et al. (2016) have recently shown that RipAY has an extremely high catalytic activity to degrade glutathione compared with other eukaryotic GGCTs, which supports the idea that RipAY could exert a similar function at lower concentrations within plant cells. Consistent with this hypothesis, we failed to obtain Arabidopsis transgenic lines expressing RipAY, probably because of the toxicity of sustained RipAY expression, even using a plasmid containing a DEX‐inducible promoter driving the expression of RipAY. This is most probably a result of the residual low expression of this promoter in basal conditions, which was observed in N. benthamiana (data not shown). It is noteworthy, however, that we did not detect any visible cell death during our transient expression assays (2–3 dpi) or even much later, up to 6–8 dpi, when both RipAY‐GFP and the GFP‐expressing control started to show signs of chlorosis. This suggests that our results, obtained at 2–3 dpi, are not a product of a general cell death phenomenon.
TRXs and glutathione have multiple targets in plant cells and regulate multiple processes in addition to biotic stress, such as abiotic stress and plant development (Frendo et al., 2013; Gelhaye et al., 2005; Ghanta and Chattopadhyay, 2011; Kneeshaw et al., 2014; Noctor et al., 2011, 2012; Tada et al., 2008). Therefore, by targeting TRXs and glutathione, R. solanacearum could globally perturb plant signalling to benefit pathogen proliferation (Fig. S13). Given the general roles of TRXs and glutathione as antioxidants (Gelhaye et al., 2005; Szalai et al., 2009), their manipulation could have a broader impact in the context of natural infections, including, for example, a deficiency of ROS detoxification. Therefore, it will be important to decipher the complex activity of RipAY in the intricate infection process by R. solanacearum, where the colonization of different plant organs in different stages of the infection is expected to have specific requirements in terms of manipulation of plant processes. This may also allow us to reinterpret the relevance of different plant targets for RipAY virulence activities.
When this article was in preparation, an additional report showed that RipAY is activated by thioredoxins, degrades glutathione and suppresses elicitor‐triggered responses in N. benthamiana (Mukaihara et al., 2016). Collectively, these results, together with those published by Fujiwara et al. (2016), and our findings reported in this article, robustly indicate that RipAY targets redox regulators to suppress immune responses during R. solanacearum infection.
Experimental Procedures
Plant material and growth conditions
Nicotiana benthamiana wild‐type plants were grown on soil with one plant per pot in an environmentally controlled growth room at 22 ºC with a 16‐h photoperiod and a light intensity of 100–150 mE/m2/s. Arabidopsis thaliana wild‐type (Col‐0) and Arabidopsis mutants pad2‐1 (Glazebrook and Ausubel, 1994) and Attrxh5‐4 (Sweat and Wolpert, 2007) were grown in a growth chamber at 22 °C with a 10‐h photoperiod and a light intensity of 100–150 mE/m2/s.
Chemicals
The flg22 peptide (TRLSSGLKINSAKDDAAGLQIA) was purchased from Abclonal, Woburn, MA, USA. Sequencing‐grade modified trypsin was purchased from Promega (Madison, WI, USA). The Nanosep centrifugal filter units with Omega membrane (MWCO 10 kDa) were purchased from Pall Incorporation (New York, NY, USA). All other chemicals were purchased from Sigma‐Aldrich (St. Louis, MO, USA) unless otherwise stated.
Bacterial strains and cultivation conditions
Escherichia coli DH5α was grown overnight at 37 °C and 220 rpm in Luria–Bertani (LB) medium supplemented with appropriate antibiotics (rifampicin, 50 mg/L; kanamycin, 25 mg/L; carbenicillin, 50 mg/L; gentamycin, 25 mg/L; spectinomycin, 50 mg/L). Agrobacterium tumefaciens strains GV3101 and GV3101(pMP90RK) were grown at 29 °C and 220 rpm in LB medium supplemented with appropriate antibiotics (rifampicin, 50 mg/L; kanamycin, 25 mg/L; carbenicillin, 50 mg/L; gentamycin, 25 mg/L; spectinomycin, 50 mg/L). Ralstonia solanacearum GMI1000 strain was cultivated overnight in rich medium (Plener et al., 2010).
RNA isolation, cDNA amplification and quantitative RT‐PCR
Total RNA was extracted using the E.Z.N.A. Plant RNA kit with DNA digestion on‐column (Biotek, Winooski, VT, USA) according to the manufacturer's instructions. RNA samples were quantified with a Nanodrop spectrophotometer (Thermo Scientific, Waltham, MA, USA). First‐strand cDNA was synthesized using the iScript™ cDNA synthesis kit (Biorad, Hercules, CA, USA). The open reading frames (ORFs) of thioredoxins were amplified from the first‐strand cDNA using Q5 Hot Start High‐Fidelity DNA polymerase (New England Biolabs). Quantitative RT‐PCR was performed using iTaq™ Universal SYBR Green Supermix (Biorad) and a CFX96 Real‐time system (Biorad), and quantitative PCR data were analysed as described by Livak and Schmittgen (2001). Primers used for cDNA amplification and quantitative PCR are listed in Table S1 (see Supporting Information).
Plasmids
The gene fragments amplified were cloned into the pENTR‐D‐TOPO or pDONR207 vector (Thermo Scientific, Waltham, MA, USA) and then subcloned into different gateway binary vectors via attL‐attR recombinant (LR) reactions (Thermo Scientific, Waltham, MA, USA). The RipAY (Rsp1022) gene from the R. solanacearum GMI1000 strain (pACC384, pDONR207 base) and RipAY mutants were subcloned into pGWB505 (Nakagawa et al., 2007) to generate a C‐terminal fusion with an enhanced green fluorescent protein (eGFP) tag, and pXCSG‐HAStrep (Witte et al., 2004) to generate a C‐terminal fusion with an HA‐StrepII tag. The amplified ORFs of the thioredoxins were subcloned into pGWB511 (Nakagawa et al., 2007) carrying a 3‐terminal sequence encoding the FLAG tag. pGWB constructs were transformed into A. tumefaciens GV3101, whereas pXCSG‐HAStrep constructs were transformed into A. tumefaciens GV3101 (pMP90RK). The RipAY‐mRFP construct was generated by LR recombination of the RipAY ENTRY clone (pACC384, pDONR207 base), with the pDEST binary vector pGR0029RFP2 (Deslandes et al., 2003). The NLS‐GFP construct was generated by LR recombination of an NLS SV40 pENTRY clone (pNP50, pENTR sd/d/topo base) into the pDEST binary vector pMDC83 (Curtis and Grossniklaus, 2003).
Agrobacterium‐mediated transient expression
Agrobacterium‐mediated transient expression in N. benthamiana was performed as described previously (Li, 2011). Before infiltration, the bacterial suspension was adjusted to a final optical density at 600 nm (OD600) of 0.5. Samples were taken at 1–3 dpi for analysis based on experimental requirements.
Measurement of ROS generation
Oxidative burst measurements were performed as described previously (Gimenez‐Ibanez et al., 2009), with some modifications. ROS were elicited with 50 nm flg22, and luminescence was measured over 60 min using a Microplate luminescence reader (Varioskan flash, Thermo Scientific). The luminescence was recorded in each well during 400 ms every minute over 60 min. The data obtained from the ROS burst assay were analysed and represented in two different ways, showing the relative luminescence units (RLU) produced per minute (representing the kinetics of ROS production in different samples over 60 min) and the total accumulative values of RLU for each sample (representing the total ROS production in different samples over the duration of the assay).
MAPK activation
MAPK activation assays were performed using 4–5‐week‐old N. benthamiana, as described previously (Segonzac et al., 2011), with several modifications. Two days after Agrobacterium infiltration, four 8‐mm‐diameter leaf discs were taken from infiltrated leaves and frozen in liquid nitrogen as a negative control. The rest of the leaves were then elicited for 15 min after vacuum infiltration of 100 nm flg22, and samples were taken and frozen in liquid nitrogen. MAPK activation was monitored by Western blot with Phospho‐p44/42 MAPK (Erk1/2; Thr‐202/Tyr‐204) antibodies from Cell Signaling (Danvers, MA, USA), according to the manufacturer's protocol. Blots were stained with Coomassie brilliant blue to verify equal loading.
Confocal microscopy
Confocal laser scanning microscopy was performed using a Leica AOBS confocal microscope (Wetzlar, Germany) employing the abaxial side of Agrobacterium‐infiltrated N. benthamiana leaves. Leaves were observed from 24 to 48 h post‐infiltration. After co‐transformation of plant cells with RipAY‐mRFP and NLS‐GFP, both red and green channels were recorded, signal saturation was avoided and several fields of view were inspected to obtain a global overview of the putative localization of RipAY‐mRFP.
Measurements of total cellular glutathione in N. benthamiana leaves
Total cellular glutathione was measured using a Glutathione Assay Kit (Sigma‐Aldrich), according to the manufacturer's instructions. Briefly, 100 mg of leaf tissue were collected and frozen in liquid nitrogen at 1, 2 and 3 dpi. The frozen tissues were ground with a tissuelyser (Qiagen, Hilden, Germany) and 1.0 mL of 5% 5‐sulfosalicylic acid was added to the powder to extract total glutathione. After centrifugation, 10 μL of the supernatant was used for glutathione measurement, which was then normalized to the weight of the original sample used.
Site‐directed mutagenesis
RipAY‐E216Q and RipAY‐C333S mutant variants were generated using the QuickChange lightning Site‐Directed Mutagenesis Kit (Life Technologies) following the manufacturer's instructions. RipAY/pDONR207 plasmid was used as template and the primers used for mutagenesis are listed in Table S1.
Infections with R. solanacearum
Four‐week‐old A. thaliana plants, grown in Jiffy pods, were inoculated, without wounding, by soil drenching. An overnight‐grown bacterial suspension was diluted to obtain an inoculum of 5 × 107 colony‐forming units/mL. Typically, 2 L of inoculum was used to soak up to 50 A. thaliana‐containing Jiffy pods. Once the Jiffy pods had been drenched completely (after approximately 10 min), the plants were removed from the bacterial solution and placed on a bed of potting mixture soil in the same original inoculation tray. The genotypes to be tested (here Col‐0 versus pad2‐1) were placed in a predefined random order in order to allow an unbiased analysis of wilting. Three real biological repetitions were performed (each inoculated with a separate GMI1000 inoculum), with each repetition containing 17 Col‐0 and 16 pad2‐1 plants. Daily scoring of visible wilting on a scale of 0–4 (0–100% leaf wilting) was evaluated using Kaplan–Meier survival analysis, log‐rank test and hazard ratio calculation, as described previously (Remigi et al., 2011; Wang et al., 2016).
Large‐scale immunoprecipitation assays for LC‐MS/MS analysis
Large‐scale immunoprecipitation assays for LC‐MS/MS analysis were performed as described previously (Kadota et al., 2016), with several modifications. Three to five grams of N. benthamiana leaf materials at 2 dpi were frozen and ground in liquid nitrogen. Total proteins were extracted using protein extraction buffer [100 mm Tris‐HCl, pH 7.5, 150 mm NaCl, 10% glycerol, 5 mm Ethylene diamine tetraacetic acid (EDTA), 10 mm Dithiothreitol (DTT), 2 mm Phenylmethylsulfonyl fluoride (PMSF), 10 mm NaF, 10 mm Na2MoO4, 2 mm NaVO3, 0.5% (v/v) IGEPAL (IGEPAL CA‐630), 1% (v/v) Plant Protease Inhibitor cocktail (Sigma, St. Louis, MO, USA). To use Strep‐Tactin beads, 100 µg/mL avidin was included in the protein extraction buffer. Extracts were centrifuged twice at 15 000 g for 15 min at 4 °C to completely remove debris. GFP‐trap beads (ChromoTek, Martinsried, Germany) or Strep‐Tactin beads (IBA‐lifesciences, Goettingen, Germany) were added to the supernatant and incubated for 1 h at 4 °C with slow but constant rotation. Conjugated beads were washed three times with 1 mL cold wash buffer [100 mm Tris‐HCl, pH 7.5, 150 mm NaCl, 10% glycerol, 2 mm DTT, 10 mm NaF, 10 mm Na2MoO4, 2 mm NaVO3, 0.5% (v/v) IGEPAL, 1% (v/v) Plant Protease Inhibitor cocktail (Sigma)] and twice with wash buffer without IGEPAL before stripping interacting proteins from the beads by boiling in 50 µL Laemmli sample buffer (Biorad) for 10 min. Immunoprecipitated proteins were separated on precast sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) gels (Biorad). Gels were washed five times with MilliQ water (Merck Millipore, Darmstadt, Germany) and stained with Coomassie brilliant blue G250 (Biorad) for 1 h. The stained gels were destained twice with MilliQ water and cut into pieces for protein identification.
Co‐immunoprecipitation
Nicotiana benthamiana leaves were co‐infiltrated with A. tumefaciens GV3101 carrying plasmids to induce the expression of RipAY‐GFP (pGWB505) and TRX‐h‐FLAG (pGWB511). Leaves infiltrated with GV3101 carrying GFP (pGWB505) and TRX‐h‐FLAG (pGWB511) were used as negative controls. After 2 dpi, total proteins were extracted and immunoprecipitation was performed with GFP‐trap beads as described above, except that the conjugated beads were washed five times with 1 mL cold wash buffer with IGEPAL before stripping interacting proteins from the beads by boiling in 50 µL SDS loading buffer for 10 min. The immunoprecipitated proteins were separated on SDS‐PAGE gels and Western blot was performed using the anti‐FLAG (Sigma) and anti‐GFP (Abiocode, Agoura Hills, CA, USA) primary antibodies, respectively. Blots were stained with Coomassie brilliant blue to verify equal loading.
Protein digestion, LC‐MS/MS analysis and protein identification
Detailed information on protein digestion, LC‐MS/MS and protein identification can be found in Methods S1.
VIGS of thioredoxins in N. benthamiana
VIGS was performed using Tobacco rattle virus (TRV)‐based VIGS vectors as described previously (Senthil‐Kumar and Mysore, 2014). Silencing fragments of NbTrx09/11 (NbTRX‐h9si) and NbTrx10/15/16 (NbTRX‐h15si) were amplified using the primers listed in Table 1, and cloned into pEASY vectors (Transgene, Beijing, China). After digestion with EcoRI and XhoI (New England Biolabs), the silencing fragments were subcloned into TRV2 vectors to yield the TRV2:NbTRX‐h9 and TRV2:NbTRX‐h15 silencing constructs. These constructs were transformed into A. tumefaciens GV3101 for agroinfiltration in the lower leaves of 3‐week‐old N. benthamiana plants.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Methods S1 Protein digestion, liquid chromatography‐tandem mass spectrometry (LC‐MS/MS) and protein identification.
Fig. S1 RipAY‐HA‐StrepII suppresses flg22‐triggered reactive oxygen species (ROS) in Nicotiana benthamiana leaves. Agrobacterium tumefaciens was used to induce the transient expression of RipAY‐HA‐StrepII in one half of the leaf and GFP‐HA‐StrepII in the other half. (a, b) Oxidative burst triggered by 50 nm flg22 in N. benthamiana tissues at 2.5 days post‐inoculation (dpi) with A. tumefaciens, and measured in a luminol‐based assay as relative luminescence units (RLU). Values are the average ± standard error (SE) (n = 24). Asterisks indicate significant differences compared with the corresponding GFP control at P < 0.001. The experiment was repeated at least three times with similar results. GFP, green fluorescent protein; HA, haemagglutinin.
Fig. S2 RipAY does not cause a clear and reproducible suppression of flg22‐triggered mitogen‐activated protein kinase (MAPK) activation. Agrobacterium tumefaciens was used to induce the transient expression of RipAY‐GFP in one half of the leaf and GFP in the other half. MAPK activation was induced with 100 nm flg22 and analysed 15 min after flg22 treatment using anti‐phosphorylated MAPK antibody (anti‐pMAPK). Immunoblots were also analysed using anti‐GFP antibody to verify protein accumulation. CBB, Coomassie brilliant blue; GFP, green fluorescent protein. Molecular weight (kDa) marker bands are indicated for reference. The experiments were repeated three times with similar results.
Fig. S3 RipAY‐mRFP localizes to cytoplasmic and nuclear plant cell compartments. Four additional independent confocal microscopy images of Nicotiana benthamiana epidermal cells expressing RipAY‐mRFP. mRFP, monomeric red fluorescent protein; N, nucleus; arrows, cytoplasmic strands.
Fig. S4 Western blot analysis of RipAY‐mRFP. Western blot analysis of the Nicotiana benthamiana tissues used to determine the subcellular localization of RipAY‐mRFP (lanes marked in red). The expected sizes of the RipAY‐mRFP fusion protein and free mRFP are indicated for reference. These representative experiments indicate that most of the signal observed under the confocal microscope is associated with the full‐length RipAY‐mRFP. A faint band with the expected size of free monomeric red fluorescent protein (mRFP) was occasionally observed, and could correspond to the cleavage of a small proportion of the mRFP tag in planta or during the protein extraction process.
Fig. S5 Nicotiana benthamiana thioredoxins studied in this work. (a) The h‐type thioredoxins identified to be associated with RipAY, and their equivalent nomenclature in the N. benthamiana genome versions (www.solgenomics.org) and this work. (b) Phylogenetic tree of the N. benthamiana and Arabidopsis thaliana h‐type thioredoxins studied in this work.
Fig. S6 RipAY sequence analysis. Amino acid sequence of RipAY from Ralstonia solanacearum GMI1000, used in this work. The GGCT domain is underlined in red. The conserved catalytic glutamate residue (E216) is marked with a green box. The only Cys residue (C333) is marked with a blue box.
Fig. S7 Genetic analysis by silencing of Nicotiana benthamiana thioredoxins. (a) Phylogenetic tree of the N. benthamiana h‐type thioredoxins studied in this work. Two silencing constructs were generated to silence the two different groups marked with red boxes. (b) Quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of NbTRX‐h transcripts in N. benthamiana leaves at 20 days post‐inoculation (dpi) of the lower leaves with Agrobacterium tumefaciens expressing a control construct or the different silencing constructs, as detailed in Experimental procedures. Gene expression values are relative to the NbEF1α housekeeping gene and are normalized to tissues expressing the silencing control. (c) Oxidative burst was triggered by 50 nm flg22 in N. benthamiana tissues silenced for the different NbTRX‐h genes, and measured in a luminol‐based assay as relative luminescence units (RLU) over 60 min. Values are the average ± standard error (SE) (n = 24). Asterisks indicate significant differences compared with the corresponding virus‐induced gene silencing (VIGS) control at P < 0.05. The experiments were repeated at least three times with similar results.
Fig. S8 Analysis of the impact of overexpression of NbTRX‐h genes on the flg22‐triggered reactive oxygen species (ROS) burst. (a) Agrobacterium tumefaciens was used to induce the transient expression of different NbTRX‐h‐FLAG fusion proteins in one half of the leaf and GFP in the other half. (b) Agrobacterium tumefaciens was used to induce the transient expression of RipAY‐GFP in one half of the leaf and GFP in the other half, co‐inoculated with A. tumefaciens with no plasmid or triggering the transient expression of different NbTRX‐h‐FLAG fusion proteins. The oxidative burst was triggered by 50 nm flg22 in Nicotiana benthamiana tissues at 2.5 days post‐inoculation (dpi) with A. tumefaciens, and measured in a luminol‐based assay as relative luminescence units (RLU). Values are the average ± standard error (SE) (n = 24). Immunoblots were analysed using anti‐GFP and anti‐FLAG antibodies to verify protein accumulation. CBB, Coomassie brilliant blue. Asterisks indicate significant differences compared with the value obtained in control RipAY‐expressing tissues at P < 0.001. The experiments were repeated three times with similar results.
Fig. S9 The trxh5‐4 mutation does not affect the flg22‐triggered reactive oxygen species (ROS) burst. The oxidative burst was triggered by 50 nm flg22 in wild‐type Col‐0 and trxh5‐4 mutant plants, and measured in a luminol‐based assay as relative light units (RLU). Values are the average ± standard error (SE) (n = 24). The experiments were repeated three times with similar results.
Fig. S10 RipAY degrades glutathione and suppresses salicylic acid (SA)‐triggered PR1 induction in Arabidopsis mesophyll protoplasts. (a) Total glutathione content in Arabidopsis mesophyll protoplasts expressing GFP‐HA‐Strep, RipAY‐HA‐Strep or RipAY‐E216Q‐HA‐Strep. Values are the average ± standard error (SE) (n = 3). Approximately 15 000 protoplasts were used per sample. (b) Immunoblot to confirm protein accumulation in transfected protoplasts, using anti‐haemagglutinin (anti‐HA). CBB, Coomassie brilliant blue. (c) Quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of AtPR1 transcripts in Arabidopsis mesophyll protoplasts either untreated or treated with 0.1 mM SA for 2 h. Gene expression values are relative to the AtActin2 housekeeping gene and are normalized to untreated protoplasts. Values are the average ± SE (n = 3). Asterisks indicate significant differences compared with the corresponding green fluorescent protein (GFP) control at P < 0.001. The experiments were repeated twice with similar results.
Fig. S11 Analysis of flg22‐triggered reactive oxygen species (ROS) burst and Ralstonia solanacearum infection in the pad2‐1 mutant. (a) The pad2‐1 mutation does not affect the flg22‐triggered ROS burst. The oxidative burst triggered by 50 nm flg22 in wild‐type Col‐0 and pad2‐1 mutant plants was measured in a luminol‐based assay as relative light units (RLU). Values are the average ± standard error (SE) (n = 24). The experiments were repeated three times with similar results. (b) The bacterial wilt disease provoked by R. solanacearum GMI1000 is not distinguishable between Col‐0 and pad2‐1. Hazard ratio representation of three biological independent inoculation experiments. The median of the three data points is represented. Using a one‐tailed paired t‐test (P = 0.11), the pad2‐1 hazard ratio is not distinguishable from the baseline [hazard ratio of Col‐0 inoculated line against itself = 1; log10(hazard ratio) = 0].
Fig. S12 1‐Chloro‐2,4‐dinitrobenzene (CDNB) application causes a reduction in plant cellular glutathione and suppresses the flg22‐triggered reactive oxygen species (ROS) burst. (a) Total glutathione content in Nicotiana benthamiana tissues after an overnight treatment with different concentrations of CDNB. The values are the average ± standard error (SE) (n = 3). (b) The oxidative burst was triggered by 50 nm flg22 in N. benthamiana tissues after an overnight treatment with different concentrations of CDNB, and measured in a luminol‐based assay as relative luminescence units (RLU) over 60 min. Values are the average ± SE (n = 24). Asterisks indicate significant differences compared with the mock control at P < 0.001. The experiments were repeated at least three times with similar results.
Fig. S13 Model suggesting the impact of RipAY on redox activities regulated by glutathione and h‐type thioredoxins. NPR1, NONEXPRESSOR OF PATHOGENESIS‐RELATED GENES 1; PR, pathogenesis‐related; PRR, pattern recognition receptor; SA, salicylic acid; TRX‐h, thioredoxin h.
Table S1 Primers used in this study for the amplification of thioredoxin genes from Nicotiana benthamiana and Arabidopsis thaliana cDNA and quantitative reverse transcription‐polymerase chain reaction (RT‐PCR), as indicated.
Acknowledgements
We thank Rosa Lozano‐Durán for critical reading and suggestions to the manuscript. We also thank members of the Macho and Lozano‐Durán laboratories for helpful discussions, and Xinyu Jian for technical and administrative assistance during this work. We thank Laurent Noël, Thomas Wolpert and Benoît Poinssot for sharing biological materials. We are grateful to Cécile Pouzet and Aurélie Le Ru of the T.R.I. GENOTOUL microscopy platform (FRAIB, Toulouse, France) for performing confocal imaging. Research in the Macho laboratory was supported by the Shanghai Center for Plant Stress Biology (Chinese Academy of Sciences), National Natural Science Foundation of China (grant 31571973) and the Chinese 1000 Talents Program. A.‐C.C. and N.P. were supported by LABEX TULIP (ANR‐10‐LABX‐41 and ANR‐11‐IDEX‐0002‐02).
References
- Ball, L. , Accotto, G.P. , Bechtold, U. , Creissen, G. , Funck, D. , Jimenez, A. , Kular, B. , Leyland, N. , Mejia‐Carranza, J. , Reynolds, H. , Karpinski, S. and Mullineaux, P.M. (2004) Evidence for a direct link between glutathione biosynthesis and stress defense gene expression in Arabidopsis. Plant Cell, 16, 2448–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigeard, J. , Colcombet, J. and Hirt, H. (2015) Signaling mechanisms in pattern‐triggered immunity (PTI). Mol. Plant, 8, 521–539. [DOI] [PubMed] [Google Scholar]
- Boller, T. and Felix, G. (2009) A renaissance of elicitors: perception of microbe‐associated molecular patterns and danger signals by pattern‐recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. [DOI] [PubMed] [Google Scholar]
- Cesari, S. , Bernoux, M. , Moncuquet, P. , Kroj, T. and Dodds, P.N. (2014) A novel conserved mechanism for plant NLR protein pairs: the “integrated decoy” hypothesis. Front. Plant Sci. 5, 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.Y. , Lin, Y.M. , Chao, T.C. , Wang, J.F. , Liu, A.C. , Ho, F.I. and Cheng, C.P. (2009) Virus‐induced gene silencing reveals the involvement of ethylene‐, salicylic acid‐ and mitogen‐activated protein kinase‐related defense pathways in the resistance of tomato to bacterial wilt. Physiol. Plant. 136, 324–335. [DOI] [PubMed] [Google Scholar]
- Clarke, C.R. , Studholme, D.J. , Hayes, B. , Runde, B. , Weisberg, A. , Cai, R. , Wroblewski, T. , Daunay, M.C. , Wicker, E. , Castillo, J.A. and Vinatzer, B.A. (2015) Genome‐enabled phylogeographic investigation of the quarantine pathogen Ralstonia solanacearum Race 3 Biovar 2 and screening for sources of resistance against its core effectors. Phytopathology, 105, 597–607. [DOI] [PubMed] [Google Scholar]
- Clay, N.K. , Adio, A.M. , Denoux, C. , Jander, G. and Ausubel, F.M. (2009) Glucosinolate metabolites required for an Arabidopsis innate immune response. Science, 323, 95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, D.E. , Mesarich, C.H. and Thomma, B.P. (2015) Understanding plant immunity as a surveillance system to detect invasion. Annu. Rev. Phytopathol. 53, 541–563. [DOI] [PubMed] [Google Scholar]
- Curtis, M.D. and Grossniklaus, U. (2003) A gateway cloning vector set for high‐throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme‐Hinoux, V. , Bangash, S.A. , Meyer, A.J. and Reichheld, J.P. (2016) Nuclear thiol redox systems in plants. Plant Sci. 243, 84–95. [DOI] [PubMed] [Google Scholar]
- Deslandes, L. and Genin, S. (2014) Opening the Ralstonia solanacearum type III effector tool box: insights into host cell subversion mechanisms. Curr. Opin. Plant Biol. 20, 110–117. [DOI] [PubMed] [Google Scholar]
- Deslandes, L. and Rivas, S. (2012) Catch me if you can: bacterial effectors and plant targets. Trends Plant Sci. 17, 644–655. [DOI] [PubMed] [Google Scholar]
- Deslandes, L. , Olivier, J. , Peeters, N. , Feng, D.X. , Khounlotham, M. , Boucher, C. , Somssich, I. , Genin, S. and Marco, Y. (2003) Physical interaction between RRS1‐R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. USA, 100, 8024–8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil‐Maurizi, C. and Poinssot, B. (2012) Role of glutathione in plant signaling under biotic stress. Plant Signal Behav. 7, 210–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil‐Maurizi, C. , Vitecek, J. , Marty, L. , Branciard, L. , Frettinger, P. , Wendehenne, D. , Meyer, A.J. , Mauch, F. and Poinssot, B. (2011) Glutathione deficiency of the Arabidopsis mutant pad2‐1 affects oxidative stress‐related events, defense gene expression, and the hypersensitive response. Plant Physiol. 157, 2000–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frendo, P. , Baldacci‐Cresp, F. , Benyamina, S.M. and Puppo, A. (2013) Glutathione and plant response to the biotic environment. Free Radic. Biol. Med. 65, 724–730. [DOI] [PubMed] [Google Scholar]
- Fujiwara, S. , Kawazoe, T. , Ohnishi, K. , Kitagawa, T. , Popa, C. , Valls, M. , Genin, S. , Nakamura, K. , Kuramitsu, Y. , Tanaka, N. and Tabuchi, M. (2016) RipAY, a plant pathogen effector protein, exhibits robust gamma‐glutamyl cyclotransferase activity when stimulated by eukaryotic thioredoxins. J. Biol. Chem. 291, 6813–6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan, J.E. , Lara‐Tejero, M. , Marlovits, T.C. and Wagner, S. (2014) Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu. Rev. Microbiol. 68, 415–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelhaye, E. , Rouhier, N. and Jacquot, J.P. (2004) The thioredoxin h system of higher plants. Plant Physiol. Biochem. 42, 265–271. [DOI] [PubMed] [Google Scholar]
- Gelhaye, E. , Rouhier, N. , Navrot, N. and Jacquot, J.P. (2005) The plant thioredoxin system. Cell. Mol. Life Sci. 62, 24–35. [DOI] [PubMed] [Google Scholar]
- Ghanta, S. and Chattopadhyay, S. (2011) Glutathione as a signaling molecule: another challenge to pathogens. Plant Signal. Behav. 6, 783–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez‐Ibanez, S. , Hann, D.R. , Ntoukakis, V. , Petutschnig, E. , Lipka, V. and Rathjen, J.P. (2009) AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr. Biol. 19, 423–429. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. and Ausubel, F.M. (1994) Isolation of phytoalexin‐deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc. Natl. Acad. Sci. USA, 91, 8955–8959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez‐Gómez, L. and Boller, T. (2000) FLS2: an LRR receptor‐like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell, 5, 1003–1011. [DOI] [PubMed] [Google Scholar]
- Hisabori, T. , Hara, S. , Fujii, T. , Yamazaki, D. , Hosoya‐Matsuda, N. and Motohashi, K. (2005) Thioredoxin affinity chromatography: a useful method for further understanding the thioredoxin network. J. Exp. Bot. 56, 1463–1468. [DOI] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kadota, Y. , Macho, A.P. and Zipfel, C. (2016) Immunoprecipitation of plasma membrane receptor‐like kinases for identification of phosphorylation sites and associated proteins. Methods Mol. Biol. 1363, 133–144. [DOI] [PubMed] [Google Scholar]
- Khan, M. , Subramaniam, R. and Desveaux, D. (2016) Of guards, decoys, baits and traps: pathogen perception in plants by type III effector sensors. Curr. Opin. Microbiol. 29, 49–55. [DOI] [PubMed] [Google Scholar]
- Kneeshaw, S. , Gelineau, S. , Tada, Y. , Loake, G.J. and Spoel, S.H. (2014) Selective protein denitrosylation activity of thioredoxin‐h5 modulates plant Immunity. Mol. Cell, 56, 153–162. [DOI] [PubMed] [Google Scholar]
- Kovacs, I. , Durner, J. and Lindermayr, C. (2015) Crosstalk between nitric oxide and glutathione is required for NONEXPRESSOR OF PATHOGENESIS‐RELATED GENES 1 (NPR1)‐dependent defense signaling in Arabidopsis thaliana . New Phytol. 208, 860–872. [DOI] [PubMed] [Google Scholar]
- Kumar, S. , Kaur, A. , Chattopadhyay, B. and Bachhawat, A.K. (2015) Defining the cytosolic pathway of glutathione degradation in Arabidopsis thaliana: role of the ChaC/GCG family of gamma‐glutamyl cyclotransferases as glutathione‐degrading enzymes and AtLAP1 as the Cys–Gly peptidase. Biochem. J. 468, 73–85. [DOI] [PubMed] [Google Scholar]
- Laloi, C. , Mestres‐Ortega, D. , Marco, Y. , Meyer, Y. and Reichheld, J.P. (2004) The Arabidopsis cytosolic thioredoxin h5 gene induction by oxidative stress and its W‐box‐mediated response to pathogen elicitor. Plant Physiol. 134, 1006–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. (2011) Infiltration of Nicotiana benthamiana protocol for transient expression via Agrobacterium. Bio‐protocol Bio101: e95. [Google Scholar]
- Lin, B. , Zhuo, K. , Chen, S. , Hu, L. , Sun, L. , Wang, X. , Zhang, L.H. and Liao, J. (2016) A novel nematode effector suppresses plant immunity by activating host reactive oxygen species‐scavenging system. New Phytol. 209, 1159–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(T)(‐Delta Delta C) method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lorang, J. , Kidarsa, T. , Bradford, C.S. , Gilbert, B. , Curtis, M. , Tzeng, S.C. , Maier, C.S. and Wolpert, T.J. (2012) Tricking the guard: exploiting plant defense for disease susceptibility. Science, 338, 659–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho, A.P. (2016) Subversion of plant cellular functions by bacterial type‐III effectors: beyond suppression of immunity. New Phytol. 210, 51–57. [DOI] [PubMed] [Google Scholar]
- Macho, A.P. and Zipfel, C. (2014) Plant PRRs and the activation of innate immune signaling. Mol. Cell, 54, 263–272. [DOI] [PubMed] [Google Scholar]
- Macho, A.P. and Zipfel, C. (2015) Targeting of plant pattern recognition receptor‐triggered immunity by bacterial type‐III secretion system effectors. Curr. Opin. Microbiol. 23C, 14–22. [DOI] [PubMed] [Google Scholar]
- Macho, A.P. , Guidot, A. , Barberis, P. , Beuzon, C.R. and Genin, S. (2010) A competitive index assay identifies several Ralstonia solanacearum type III effector mutant strains with reduced fitness in host plants. Mol. Plant–Microbe Interact. 23, 1197–1205. [DOI] [PubMed] [Google Scholar]
- Mansfield, J. , Genin, S. , Magori, S. , Citovsky, V. , Sriariyanum, M. , Ronald, P. , Dow, M. , Verdier, V. , Beer, S.V. , Machado, M.A. , Toth, I. , Salmond, G. and Foster, G.D. (2012) Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler‐Bauer, A. , Derbyshire, M.K. , Gonzales, N.R. , Lu, S. , Chitsaz, F. , Geer, L.Y. , Geer, R.C. , He, J. , Gwadz, M. , Hurwitz, D.I. , Lanczycki, C.J. , Lu, F. , Marchler, G.H. , Song, J.S. , Thanki, N. , Wang, Z. , Yamashita, R.A. , Zhang, D. , Zheng, C. and Bryant, S.H. (2015) CDD: NCBI's conserved domain database. Nucleic Acids Res. 43, D222–D226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, Y. , Belin, C. , Delorme‐Hinoux, V. , Reichheld, J.P. and Riondet, C. (2012) Thioredoxin and glutaredoxin systems in plants: molecular mechanisms, crosstalks, and functional significance. Antioxid. Redox Signal. 17, 1124–1160. [DOI] [PubMed] [Google Scholar]
- Monteiro, F. , Genin, S. , van Dijk, I. and Valls, M. (2012) A luminescent reporter evidences active expression of Ralstonia solanacearum type III secretion system genes throughout plant infection. Microbiology, 158, 2107–2116. [DOI] [PubMed] [Google Scholar]
- Mou, Z. , Fan, W. and Dong, X. (2003) Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell, 113, 935–944. [DOI] [PubMed] [Google Scholar]
- Mukaihara, T. , Hatanaka, T. , Nakano, M. and Oda, K. (2016) Ralstonia solanacearum type III effector RipAY is a glutathione‐degrading enzyme that is activated by plant cytosolic thioredoxins and suppresses plant immunity. mBio. 7, e00359‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, T. , Suzuki, T. , Murata, S. , Nakamura, S. , Hino, T. , Maeo, K. , Tabata, R. , Kawai, T. , Tanaka, K. , Niwa, Y. , Watanabe, Y. , Nakamura, K. , Kimura, T. and Ishiguro S. (2007) Improved Gateway binary vectors: high‐performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71, 2095–2100. [DOI] [PubMed] [Google Scholar]
- Nishimura, M.T. , Monteiro, F. and Dangl, J.L. (2015) Treasure your exceptions: unusual domains in immune receptors reveal host virulence targets. Cell, 161, 957–960. [DOI] [PubMed] [Google Scholar]
- Noctor, G. , Queval, G. , Mhamdi, A. , Chaouch, S. and Foyer, C.H. (2011) Glutathione. Arabidopsis Book, 9, e0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor, G. , Mhamdi, A. , Chaouch, S. , Han, Y. , Neukermans, J. , Marquez‐Garcia, B. , Queval, G. and Foyer, C.H. (2012) Glutathione in plants: an integrated overview. Plant Cell Environ. 35, 454–484. [DOI] [PubMed] [Google Scholar]
- Nühse, T.S. , Bottrill, A.R. , Jones, A.M.E. and Peck, S.C. (2007) Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J. 51, 931–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley, A.J. , Yamada, T. , Liu, D. , Coggan, M. , Clark, A.G. and Board, P.G. (2008) The identification and structural characterization of C7orf24 as gamma‐glutamyl cyclotransferase. An essential enzyme in the gamma‐glutamyl cycle. J. Biol. Chem. 283, 22 031–22 042. [DOI] [PubMed] [Google Scholar]
- Okuma, E. , Sarwar‐Jahan, M.D. , Munemasa, S. , Anowar‐Hossain, M. , Muroyama, D. , Mahbub‐Islam, M. , Ogawa, K. , Watanabe‐Sugimoto, M. , Nakamura, Y. , Shimoishi, Y. , Mori, I.C. and Murata, Y. (2011) Negative regulation of abscisic acid‐induced stomatal closure by glutathione in Arabidopsis. J. Plant Physiol. 168, 2048–2055. [DOI] [PubMed] [Google Scholar]
- Pajerowska‐Mukhtar, K.M. , Emerine, D.K. and Mukhtar, M.S. (2013) Tell me more: roles of NPRs in plant immunity. Trends Plant Sci. 18, 402–411. [DOI] [PubMed] [Google Scholar]
- Paulose, B. , Chhikara, S. , Coomey, J. , Jung, H.I. , Vatamaniuk, O. and Dhankher, O.P. (2013) A gamma‐glutamyl cyclotransferase protects Arabidopsis plants from heavy metal toxicity by recycling glutamate to maintain glutathione homeostasis. Plant Cell, 25, 4580–4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters, N. , Carrere, S. , Anisimova, M. , Plener, L. , Cazale, A.C. and Genin, S. (2013a) Repertoire, unified nomenclature and evolution of the Type III effector gene set in the Ralstonia solanacearum species complex. BMC Genomics, 14, 859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters, N. , Guidot, A. , Vailleau, F. and Valls, M. (2013b) Ralstonia solanacearum, a widespread bacterial plant pathogen in the post‐genomic era. Mol. Plant Pathol. 14, 651–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plener, L. , Manfredi, P. , Valls, M. and Genin, S. (2010) PrhG, a transcriptional regulator responding to growth conditions, is involved in the control of the type III secretion system regulon in Ralstonia solanacearum . J. Bacteriol. 192, 1011–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichheld, J.P. , Khafif, M. , Riondet, C. , Droux, M. , Bonnard, G. and Meyer, Y. (2007) Inactivation of thioredoxin reductases reveals a complex interplay between thioredoxin and glutathione pathways in Arabidopsis development. Plant Cell, 19, 1851–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichheld, J.P. , Mestres‐Ortega, D. , Laloi, C. and Meyer, Y. (2002) The multigenic family of thioredoxin h in Arabidopsis thaliana: specific expression and stress response. Plant Physiol. Biochem. 40, 685–690. [Google Scholar]
- Remigi, P. , Anisimova, M. , Guidot, A. , Genin, S. and Peeters, N. (2011) Functional diversification of the GALA type III effector family contributes to Ralstonia solanacearum adaptation on different plant hosts. New Phytol. 192, 976–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh, A. , Withers, J. , Mohan, R. , Marques, J. , Gu, Y. , Yan, S. , Zavaliev, R. , Nomoto, M. , Tada, Y. and Dong, X. (2015) Posttranslational modifications of the master transcriptional regulator NPR1 enable dynamic but tight control of plant immune responses. Cell Host Microbe, 18, 169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segonzac, C. , Feike, D. , Gimenez‐Ibanez, S. , Hann, D.R. , Zipfel, C. and Rathjen, J.P. (2011) Hierarchy and roles of pathogen‐associated molecular pattern‐induced responses in Nicotiana benthamiana . Plant Physiol. 156, 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthil‐Kumar, M. and Mysore, K.S. (2014) Tobacco rattle virus‐based virus‐induced gene silencing in Nicotiana benthamiana . Nat. Protoc. 9, 1549–1562. [DOI] [PubMed] [Google Scholar]
- Sharma, K.G. , Sharma, V. , Bourbouloux, A. , Delrot, S. and Bachhawat, A.K. (2000) Glutathione depletion leads to delayed growth stasis in Saccharomyces cerevisiae: evidence of a partially overlapping role for thioredoxin. Curr. Genet. 38, 71–77. [DOI] [PubMed] [Google Scholar]
- Spoel, S.H. and Loake, G.J. (2011) Redox‐based protein modifications: the missing link in plant immune signalling. Curr. Opin. Plant Biol. 14, 358–364. [DOI] [PubMed] [Google Scholar]
- Sun, Y. , Li, L. , Macho, A.P. , Han, Z. , Hu, Z. , Zipfel, C. , Zhou, J.M. and Chai, J. (2013) Structural basis for flg22‐induced activation of the Arabidopsis FLS2‐BAK1 immune complex. Science, 342, 624–628. [DOI] [PubMed] [Google Scholar]
- Suzuki, N. , Koussevitzky, S. , Mittler, R. and Miller, G. (2011) ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 35, 259–270. [DOI] [PubMed] [Google Scholar]
- Sweat, T.A. and Wolpert, T.J. (2007) Thioredoxin h5 is required for victorin sensitivity mediated by a CC‐NBS‐LRR gene in Arabidopsis. Plant Cell, 19, 673–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalai, G. , Kellos, T. , Galiba, G. and Kocsy, G. (2009) Glutathione as an antioxidant and regulatory molecule in plants under abiotic stress conditions. J. Plant Growth Regul. 28, 66–80. [Google Scholar]
- Tada, Y. , Spoel, S.H. , Pajerowska‐Mukhtar, K. , Mou, Z. , Song, J. , Wang, C. , Zuo, J. and Dong, X. (2008) Plant immunity requires conformational changes [corrected] of NPR1 via S‐nitrosylation and thioredoxins. Science, 321, 952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda, K. and Katagiri, F. (2010) Comparing signaling mechanisms engaged in pattern‐triggered and effector‐triggered immunity. Curr. Opin. Plant Biol. 13, 459–465. [DOI] [PubMed] [Google Scholar]
- Ueoka‐Nakanishi, H. , Sazuka, T. , Nakanishi, Y. , Maeshima, M. , Mori, H. and Hisabori, T. (2013) Thioredoxin h regulates calcium dependent protein kinases in plasma membranes. FEBS J. 280, 3220–3231. [DOI] [PubMed] [Google Scholar]
- Vlot, A.C. , Dempsey, D.A. and Klessig, D.F. (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. [DOI] [PubMed] [Google Scholar]
- Wang, K. , Remigi, P. , Anisimova, M. , Lonjon, F. , Kars, I. , Kajava, A. , Li, C.H. , Cheng, C.P. , Vailleau, F. , Genin, S. and Peeters, N. (2016) Functional assignment to positively selected sites in the core type III effector RipG7 from Ralstonia solanacearum . Mol. Plant Pathol. 17, 553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, E.R. , Uknes, S.J. , Williams, S.C. , Dincher, S.S. , Wiederhold, D.L. , Alexander, D.C. , Ahl‐Goy, P. , Metraux, J.P. and Ryals, J.A. (1991) Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell, 3, 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessling, R. , Epple, P. , Altmann, S. , He, Y. , Yang, L. , Henz, S.R. , McDonald, N. , Wiley, K. , Bader, K.C. , Gläßer, C. , Mukhtar, M.S. , Haigis, S. , Ghamsari, L. , Stephens, A.E. , Ecker, J.R. , Vidal, M. , Jones, J.D. , Mayer, K.F. , Ver Loren van Themaat, E. , Weigel, D. , Schulze‐Lefert, P. , Dangl, J.L. , Panstruga, R. and Braun, P. (2014) Convergent targeting of a common host protein‐network by pathogen effectors from three kingdoms of life. Cell Host Microbe, 16, 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win, J. , Chaparro‐Garcia, A. , Belhaj, K. , Saunders, D.G. , Yoshida, K. , Dong, S. , Schornack, S. , Zipfel, C. , Robatzek, S. , Hogenhout, S.A. and Kamoun, S. (2012) Effector biology of plant‐associated organisms: concepts and perspectives. Cold Spring Harb. Symp. Q. Biol. 77, 235–247. [DOI] [PubMed] [Google Scholar]
- Witte, C.P. , Noel, L.D. , Gielbert, J. , Parker, J.E. and Romeis, T. (2004) Rapid one‐step protein purification from plant material using the eight‐amino acid StrepII epitope. Plant Mol. Biol. 55, 135–147. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Shao, F. , Li, Y. , Cui, H. , Chen, L. , Li, H. , Zou, Y. , Long, C. , Lan, L. , Chai, J. , Chen, S. , Tang, X. and Zhou, J.M. (2007) A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP‐induced immunity in plants. Cell Host Microbe, 1, 175–185. [DOI] [PubMed] [Google Scholar]
- Zipfel, C. (2014) Plant pattern recognition receptors. Trends Immunol. 35, 345–351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Methods S1 Protein digestion, liquid chromatography‐tandem mass spectrometry (LC‐MS/MS) and protein identification.
Fig. S1 RipAY‐HA‐StrepII suppresses flg22‐triggered reactive oxygen species (ROS) in Nicotiana benthamiana leaves. Agrobacterium tumefaciens was used to induce the transient expression of RipAY‐HA‐StrepII in one half of the leaf and GFP‐HA‐StrepII in the other half. (a, b) Oxidative burst triggered by 50 nm flg22 in N. benthamiana tissues at 2.5 days post‐inoculation (dpi) with A. tumefaciens, and measured in a luminol‐based assay as relative luminescence units (RLU). Values are the average ± standard error (SE) (n = 24). Asterisks indicate significant differences compared with the corresponding GFP control at P < 0.001. The experiment was repeated at least three times with similar results. GFP, green fluorescent protein; HA, haemagglutinin.
Fig. S2 RipAY does not cause a clear and reproducible suppression of flg22‐triggered mitogen‐activated protein kinase (MAPK) activation. Agrobacterium tumefaciens was used to induce the transient expression of RipAY‐GFP in one half of the leaf and GFP in the other half. MAPK activation was induced with 100 nm flg22 and analysed 15 min after flg22 treatment using anti‐phosphorylated MAPK antibody (anti‐pMAPK). Immunoblots were also analysed using anti‐GFP antibody to verify protein accumulation. CBB, Coomassie brilliant blue; GFP, green fluorescent protein. Molecular weight (kDa) marker bands are indicated for reference. The experiments were repeated three times with similar results.
Fig. S3 RipAY‐mRFP localizes to cytoplasmic and nuclear plant cell compartments. Four additional independent confocal microscopy images of Nicotiana benthamiana epidermal cells expressing RipAY‐mRFP. mRFP, monomeric red fluorescent protein; N, nucleus; arrows, cytoplasmic strands.
Fig. S4 Western blot analysis of RipAY‐mRFP. Western blot analysis of the Nicotiana benthamiana tissues used to determine the subcellular localization of RipAY‐mRFP (lanes marked in red). The expected sizes of the RipAY‐mRFP fusion protein and free mRFP are indicated for reference. These representative experiments indicate that most of the signal observed under the confocal microscope is associated with the full‐length RipAY‐mRFP. A faint band with the expected size of free monomeric red fluorescent protein (mRFP) was occasionally observed, and could correspond to the cleavage of a small proportion of the mRFP tag in planta or during the protein extraction process.
Fig. S5 Nicotiana benthamiana thioredoxins studied in this work. (a) The h‐type thioredoxins identified to be associated with RipAY, and their equivalent nomenclature in the N. benthamiana genome versions (www.solgenomics.org) and this work. (b) Phylogenetic tree of the N. benthamiana and Arabidopsis thaliana h‐type thioredoxins studied in this work.
Fig. S6 RipAY sequence analysis. Amino acid sequence of RipAY from Ralstonia solanacearum GMI1000, used in this work. The GGCT domain is underlined in red. The conserved catalytic glutamate residue (E216) is marked with a green box. The only Cys residue (C333) is marked with a blue box.
Fig. S7 Genetic analysis by silencing of Nicotiana benthamiana thioredoxins. (a) Phylogenetic tree of the N. benthamiana h‐type thioredoxins studied in this work. Two silencing constructs were generated to silence the two different groups marked with red boxes. (b) Quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of NbTRX‐h transcripts in N. benthamiana leaves at 20 days post‐inoculation (dpi) of the lower leaves with Agrobacterium tumefaciens expressing a control construct or the different silencing constructs, as detailed in Experimental procedures. Gene expression values are relative to the NbEF1α housekeeping gene and are normalized to tissues expressing the silencing control. (c) Oxidative burst was triggered by 50 nm flg22 in N. benthamiana tissues silenced for the different NbTRX‐h genes, and measured in a luminol‐based assay as relative luminescence units (RLU) over 60 min. Values are the average ± standard error (SE) (n = 24). Asterisks indicate significant differences compared with the corresponding virus‐induced gene silencing (VIGS) control at P < 0.05. The experiments were repeated at least three times with similar results.
Fig. S8 Analysis of the impact of overexpression of NbTRX‐h genes on the flg22‐triggered reactive oxygen species (ROS) burst. (a) Agrobacterium tumefaciens was used to induce the transient expression of different NbTRX‐h‐FLAG fusion proteins in one half of the leaf and GFP in the other half. (b) Agrobacterium tumefaciens was used to induce the transient expression of RipAY‐GFP in one half of the leaf and GFP in the other half, co‐inoculated with A. tumefaciens with no plasmid or triggering the transient expression of different NbTRX‐h‐FLAG fusion proteins. The oxidative burst was triggered by 50 nm flg22 in Nicotiana benthamiana tissues at 2.5 days post‐inoculation (dpi) with A. tumefaciens, and measured in a luminol‐based assay as relative luminescence units (RLU). Values are the average ± standard error (SE) (n = 24). Immunoblots were analysed using anti‐GFP and anti‐FLAG antibodies to verify protein accumulation. CBB, Coomassie brilliant blue. Asterisks indicate significant differences compared with the value obtained in control RipAY‐expressing tissues at P < 0.001. The experiments were repeated three times with similar results.
Fig. S9 The trxh5‐4 mutation does not affect the flg22‐triggered reactive oxygen species (ROS) burst. The oxidative burst was triggered by 50 nm flg22 in wild‐type Col‐0 and trxh5‐4 mutant plants, and measured in a luminol‐based assay as relative light units (RLU). Values are the average ± standard error (SE) (n = 24). The experiments were repeated three times with similar results.
Fig. S10 RipAY degrades glutathione and suppresses salicylic acid (SA)‐triggered PR1 induction in Arabidopsis mesophyll protoplasts. (a) Total glutathione content in Arabidopsis mesophyll protoplasts expressing GFP‐HA‐Strep, RipAY‐HA‐Strep or RipAY‐E216Q‐HA‐Strep. Values are the average ± standard error (SE) (n = 3). Approximately 15 000 protoplasts were used per sample. (b) Immunoblot to confirm protein accumulation in transfected protoplasts, using anti‐haemagglutinin (anti‐HA). CBB, Coomassie brilliant blue. (c) Quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of AtPR1 transcripts in Arabidopsis mesophyll protoplasts either untreated or treated with 0.1 mM SA for 2 h. Gene expression values are relative to the AtActin2 housekeeping gene and are normalized to untreated protoplasts. Values are the average ± SE (n = 3). Asterisks indicate significant differences compared with the corresponding green fluorescent protein (GFP) control at P < 0.001. The experiments were repeated twice with similar results.
Fig. S11 Analysis of flg22‐triggered reactive oxygen species (ROS) burst and Ralstonia solanacearum infection in the pad2‐1 mutant. (a) The pad2‐1 mutation does not affect the flg22‐triggered ROS burst. The oxidative burst triggered by 50 nm flg22 in wild‐type Col‐0 and pad2‐1 mutant plants was measured in a luminol‐based assay as relative light units (RLU). Values are the average ± standard error (SE) (n = 24). The experiments were repeated three times with similar results. (b) The bacterial wilt disease provoked by R. solanacearum GMI1000 is not distinguishable between Col‐0 and pad2‐1. Hazard ratio representation of three biological independent inoculation experiments. The median of the three data points is represented. Using a one‐tailed paired t‐test (P = 0.11), the pad2‐1 hazard ratio is not distinguishable from the baseline [hazard ratio of Col‐0 inoculated line against itself = 1; log10(hazard ratio) = 0].
Fig. S12 1‐Chloro‐2,4‐dinitrobenzene (CDNB) application causes a reduction in plant cellular glutathione and suppresses the flg22‐triggered reactive oxygen species (ROS) burst. (a) Total glutathione content in Nicotiana benthamiana tissues after an overnight treatment with different concentrations of CDNB. The values are the average ± standard error (SE) (n = 3). (b) The oxidative burst was triggered by 50 nm flg22 in N. benthamiana tissues after an overnight treatment with different concentrations of CDNB, and measured in a luminol‐based assay as relative luminescence units (RLU) over 60 min. Values are the average ± SE (n = 24). Asterisks indicate significant differences compared with the mock control at P < 0.001. The experiments were repeated at least three times with similar results.
Fig. S13 Model suggesting the impact of RipAY on redox activities regulated by glutathione and h‐type thioredoxins. NPR1, NONEXPRESSOR OF PATHOGENESIS‐RELATED GENES 1; PR, pathogenesis‐related; PRR, pattern recognition receptor; SA, salicylic acid; TRX‐h, thioredoxin h.
Table S1 Primers used in this study for the amplification of thioredoxin genes from Nicotiana benthamiana and Arabidopsis thaliana cDNA and quantitative reverse transcription‐polymerase chain reaction (RT‐PCR), as indicated.


