Summary
Xanthomonas citri ssp. citri (Xcc) is an important plant‐pathogenic bacterium that causes citrus canker disease worldwide. PthA, a transcriptional activator‐like (TAL) effector, directs the expression of the canker susceptibility gene CsLOB1. Here, we report our recent progress in the functional characterization of CsLOB1. Subcellular localization analysis of CsLOB1 protein in citrus protoplast revealed that CsLOB1 is primarily localized in the nucleus. We showed that CsLOB1 expression driven by dexamethasone (DEX) in CsLOB1‐GR transgenic plants is associated with pustule formation following treatment with DEX. Pustule formation was not observed in DEX‐treated wild‐type plants and in non‐treated CsLOB1‐GR transgenic plants. Water soaking is typically associated with symptoms of citrus canker. Weaker water soaking was observed with pustule formation in CsLOB1‐GR transgenic plants following DEX treatment. When CsLOB1‐GR‐transgenic Duncan grapefruit leaves were inoculated with Xcc306ΔpthA4 and treated with DEX, typical canker symptoms, including hypertrophy, hyperplasia and water soaking symptoms, were observed on DEX‐treated transgenic plant leaves, but not on mock‐treated plants. Twelve citrus genes that are induced by PthA4 are also stimulated by the DEX‐induced expression of CsLOB1. As CsLOB1 acts as a transcriptional factor, we identified putative targets of CsLOB1 via bioinformatic and electrophoretic mobility shift assays. Cs2g20600, which encodes a zinc finger C3HC4‐type RING finger protein, has been identified to be a direct target of CsLOB1. This study advances our understanding of the function of CsLOB1 and the molecular mechanism of how Xcc causes canker symptoms via CsLOB1.
Keywords: citrus canker, C3HC4‐type RING finger protein, CsLOB1, susceptibility gene, Xanthomonas
Introduction
Citrus canker caused by Xanthomonas citri ssp. citri (Xcc) is an important disease of citrus worldwide. Citrus canker is characterized by the formation of necrotic, raised lesions on leaves, stems and fruit with raised, brown, water‐soaked margins, usually with a yellow halo around the lesion. On heavily infected trees, citrus canker causes severe defoliation, twig dieback, general tree decline, blemished fruit and premature fruit drop (Gottwald et al., 2001). The Asiatic form, or A type, of citrus canker affects a wide range of hosts, including Citrus spp. and many closely related rutaceous plants (Graham et al., 2004).
Although many genes of Xcc contribute to pathogenicity, PthA, a type III secretion system (T3SS) effector, is a critical pathogenicity determinant (Swarup et al., 1992; Yan and Wang, 2012). Variants of the PthA effector are widely present in Xanthomonas spp. that cause citrus canker disease (Al‐Saadi et al., 2007; Cubero and Graham, 2002; Jalan et al., 2013; Swarup et al., 1992). Representative strains of the five different pathotypes responsible for citrus canker, XccA, XccA*, XccAw, X. fuscans ssp. aurantifolii (Xfa) B and C, contain at least one pthA‐like gene, which are designated pthA, pthA*, pthAw, pthB and pthC, respectively, and are essential for pustule formation on citrus. Loss of PthA leads to loss of pustule symptoms and reduced bacterial growth (Swarup et al., 1992; Yan and Wang, 2012). Amongst all the citrus canker‐related bacteria, members of the XccA clade are the most virulent and widespread (Zhang et al., 2015). Notably, Xcc strain 306 (Xcc306), a member of the XccA group, contains four pthA‐related genes, named pthA1, pthA2, pthA3 and pthA4, of which only pthA4 is competent for pustule formation (da Silva et al., 2002; Yan and Wang, 2012). Pth effectors are members of the transcriptional activator‐like (TAL) effector family, which direct the expression of specific disease susceptibility genes during infection (Bogdanove et al., 2010). The effectors bind to effector‐binding elements (EBEs) within the promoter regions via a series of amino acid repeats in the central coding portion (Boch et al., 2009; Hann et al., 2010; Moscou and Bogdanove, 2009). A target of the TAL effector AvrBs3 from X. campestris pv. vesicatoria is upa20, which encodes a bHLH family transcriptional factor and acts as a regulator of cell enlargement (Kay et al., 2007). In rice, three major susceptibility genes, Os8N3 (OsSWEET11), Os11N3 (OsSWEET14) and Os12N3 (OsSWEET13), are targets of the TAL effectors from the bacterial blight pathogen X. oryzae pv. oryzae (Antony et al., 2010; Chu et al., 2006; Yang et al., 2006).
In citrus, PthA4 targets CsLOB1, a plant‐specific transcriptional factor in the lateral organ boundaries (LOB) domain family (Hu et al., 2014). PthA4 and its functionally equivalent Pth effectors recognize EBEs in the promoter of CsLOB1 (Hu et al., 2014). All PthA variants are associated with an increase in CsLOB1 expression on infection. Designed TAL effectors (dTALEs) targeting specific binding sites within the CsLOB1 promoter, but not CsSWEET1, were able to restore pustule formation and enhance bacterial growth when expressed in the pthA4 mutant of Xcc (XccΔpthA4). CsSWEET1 is a homologue of the SWEET sugar transporter and rice disease susceptibility genes OsSWEET11 and OsSWEET14 in citrus (Antony et al., 2010; Yang et al., 2006). Despite the significant progress in the characterization of CsLOB1, the detailed molecular mechanism of how CsLOB1 is involved in canker symptom development remains unknown. In this study, we conducted the functional characterization of CsLOB1, including its subcellular localization, involvement in canker symptom development and the identification of downstream targets of CsLOB1.
Results
Tissue expression pattern and subcellular localization analyses of the CsLOB1 gene
To explore the expression pattern of CsLOB1 in citrus, we tested its expression level in flowers, roots, stems and leaves of Duncan grapefruit using quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis. The expression level of CsLOB1 was higher in leaves and stems than in flowers and roots (Fig. 1A).
Figure 1.
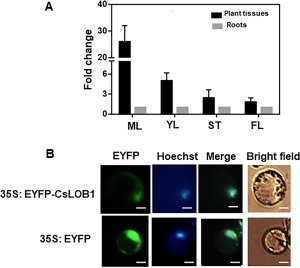
Tissue expression pattern and subcellular localization analyses of the CsLOB1 gene. (A) Expression pattern of the CsLOB1 gene in different tissues of Duncan grapefruit. The endogenous housekeeping gene used was GAPDH (Cs2g14940, glyceraldehyde 3‐phosphate dehydrogenase). Each value represents the mean ± standard deviation (SD) of three replicates. YL, young leaves, 15 days after flushing; ML, mature leaves, over 30 days after flushing; FL, folded flowers; ST, stems. Roots, fresh lateral root. All tissues in each replicate are from the same plant. (B) Subcellular localization of CsLOB1. 35S:EYFP‐CsLOB1 was transformed into citrus protoplast via polyethylene glycol (PEG)‐calcium‐mediated transfection. CsLOB1 localization to the nucleus was confirmed by co‐localization with Hoechst 33342. The fluorescence figures were taken 10 min after Hoechst dye addition to the protoplast at the rate of 1 : 600. The green colour indicates the fluorescence of enhanced yellow fluorescent protein (EYFP). The blue colour indicates the nuclear localization stained by Hoechst. Scale bar represents 10 μm.
As a putative transcription factor in citrus, it is predicted that CsLOB1 modulates downstream target genes inside the nucleus. To test the subcellular localization of CsLOB1 in citrus cells, we transiently expressed 35S:EYFP‐CsLOB1 (Fig. S1A, see Supporting Information) in citrus protoplast (Yoo et al., 2007). Fluorescence microscopy analyses showed that EYFP‐CsLOB1 co‐localized with the nuclear stain Hoechst 33342, indicating that CsLOB1 was primarily localized in the nucleus (Fig. 1B). To further confirm the localization of CsLOB1, we co‐transformed citrus protoplast with 35S:EYFP‐CsLOB1 and mCherry‐NLS (Fig. S1B) via polyethylene glycol (PEG)‐calcium‐mediated transfection (Yoo et al., 2007). Nuclear localization was indicated by the red fluorescence signal of the mCherry‐NLS protein, in which the mCherry gene is fused with the nuclear localization signal (NLS) sequence at the carboxyl terminus. Transient expression of mCherry‐NLS, which localizes to the nucleus, overlapped with 35S:EYFP‐CsLOB1 when co‐transformed into citrus protoplast (Fig. S1B), further confirming that CsLOB1 localizes to the nucleus.
Ectopic CsLOB1 expression in citrus induces pustule formation
To test a direct causal relationship between canker symptoms and the expression of CsLOB1, we took advantage of the dexamethasone (DEX, a synthetic glucocorticoid)‐induced nuclear targeting of reporter construct containing the glucocorticoid receptor (GR). In the absence of DEX, the fusion protein is present in the cytoplasm in a complex with heat shock protein 90 (HSP90), which inhibits the function of the fusion protein by preventing its nuclear localization (Galigniana et al., 1998; Picard et al., 1988). Transgenic Duncan grapefruit (Citrus paradisi Macf.) plants expressing 35S:CsLOB1‐GR were generated. CsLOB1 was fused to the hormone‐binding domain of GR and under the control of the ubiquitously expressed 35S promoter (Figs 2A and S2, see Supporting Information). The35S:CsLOB1‐GR construct also contains green fluorescent protein (GFP) and neomycin phosphotransferase II (NptII) to facilitate the screening of transgenic plants (Fig. 2A). The expression of CsLOB1 in transgenic plants was further confirmed using qRT‐PCR (Fig. 2B). Interestingly, DEX (100 μm)‐treated leaves of CsLOB1 transgenic plants exhibited pustule formation (Fig. 2C). Green fluorescence was observed in CsLOB1 transgenic plants as a result of the presence of GFP in the CsLOB1 expression construct, but not in wild‐type plants (Fig. 2D). Pustule symptoms were not observed in DEX‐treated wild‐type plants and in mock‐treated CsLOB1 transgenic plants (Fig. 2C). Only weak water soaking was observed with pustule symptoms in DEX‐treated CsLOB1 transgenic plants (Fig. 2C).
Figure 2.
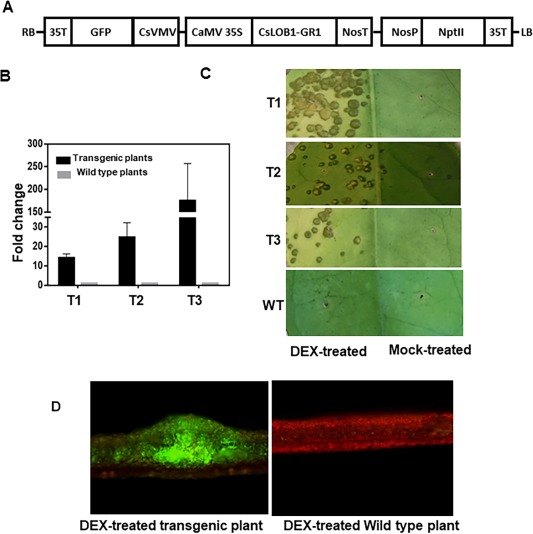
Ectopic CsLOB1 expression in citrus induces pustule formation. (A) Diagram of the 35S:CsLOB1‐GR plasmid used to construct CsLOB1 transgenic plants. The 35S:CsLOB1‐GR construct contains green fluoresent protein (GFP) and neomycin phosphotransferase II (NptII) to facilitate the screening of transgenic plants. (B) Validation of CsLOB1 gene expression in different CsLOB1 transgenic lines (T1, T2 and T3) without dexamethasone (DEX) treatment by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). Each value represents the mean ± standard deviation (SD) of three replicates. The housekeeping gene GAPDH (glyceraldehyde 3‐phosphate dehydrogenase) was used as the endogenous control. Each value represents the fold change (transgenic plants vs. wild‐type plants) ± SD. T1, T2, T3, three different transgenic lines. (C) CsLOB1 transgenic plant leaves show canker‐like symptoms at about 1 month post‐treatment with DEX. The leaves were injected with DEX (100 μm) or mock solution (equivalent volume of solvent without DEX). The photograph was taken at 1 month post‐inoculation. T1, T2, T3, different transgenic plant lines; WT, wild‐type plant. (D) Pustules on CsLOB1 transgenic plant leaves at about 1 month post‐treatment with DEX. Left: pustules on CsLOB1 transgenic plant leaves show green fluorescence under a fluorescence microscope. Right: non‐transgenic plants observed under a fluorescence microscope; Scale bar represents 200 μm.
Ectopic CsLOB1 expression in citrus restores Xcc306ΔpthA4 canker symptoms
Water soaking is a known characteristic of canker symptoms (Brunings and Gabriel, 2003). CsLOB1 expression alone induced weak water soaking symptoms (Fig. 2C), indicating that other virulence factors in addition to PthA4 of Xcc are required to induce water soaking. When CsLOB1 transgenic Duncan grapefruit leaves were inoculated with Xcc306ΔpthA4, and then treated with DEX solution or mock, water soaking and pustule symptoms were observed on DEX‐treated transgenic plant leaves, but not on mock‐treated plants, at 7 days post‐inoculation (Fig. 3A).
Figure 3.
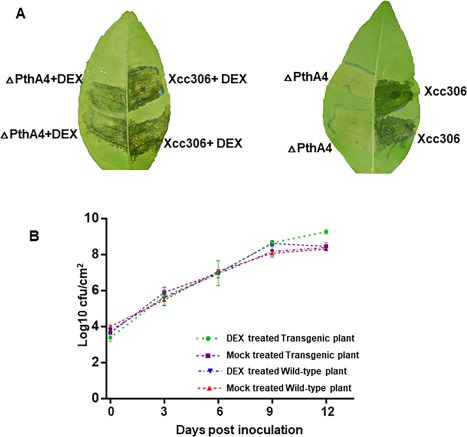
Ectopic CsLOB1 expression in citrus restores Xcc306ΔpthA4 canker symptoms. (A) Transgenic plant leaves were inoculated with Xcc306ΔpthA4 at 5 × 108 colony‐forming units (cfu)/mL, and then with 20 μL of dexamethasone (DEX, 100 μm) or mock solution at 24 h after bacterial inoculation. The photograph was taken at 7 days post‐inoculation. (B) Overexpression of CsLOB1 promotes the growth of Xcc306ΔpthA4. Leaves were inoculated with Xcc306ΔpthA4 at a concentration of 5 × 105 cfu/mL. The bacterial population was measured at the time points indicated. Error bar indicates the standard deviation for two replicates.
The bacterial population was higher in DEX‐treated CsLOB1 transgenic plant leaves than in mock‐treated transgenic plant leaves inoculated with Xcc306ΔpthA4 at 9 days post‐inoculation, and also higher than in DEX‐ and mock‐treated wild‐type plant leaves inoculated with Xcc306ΔpthA4 (Fig. 3B).
Gene expression induced by CsLOB1
Our previous study has suggested that Xcc secretes PthA4, which translocates into the plant nucleus to induce the expression of the susceptibility gene CsLOB1, which, in turn, regulates downstream genes to cause pustule symptoms (Hu et al., 2014). We reasoned that the ectopic expression of CsLOB1 would induce many genes induced by PthA4. To test this hypothesis, we selected 12 genes which were induced by both PthA4 and dTALE targeting CsLOB1 (Hu et al., 2014; Zhang et al., 2016) to test their induction by CsLOB1 with qRT‐PCR. The selected genes include expansin genes (orange1.1t00187, Cs7g32410, Cs9g15100.1), pectate lyase genes (Cs2g23970 and orange1.1t00910), an endoglucanase gene (Cs2g20750), a polygalacturonase gene (Cs7g01690) and a gibberellin‐regulated gene (Cs6g17190). CsLOB1 transgenic plant leaves were treated with DEX solution and then used for RNA extraction at 36 h post‐inoculation. Most of the 12 selected genes which were induced by PthA4 were also induced by CsLOB1 (Fig. 4).
Figure 4.

Citrus genes induced by CsLOB1. mRNA samples were extracted from CsLOB1 transgenic plant leaves treated with dexamethasone (DEX) solution and mock solution at 36 h post‐inoculation. The housekeeping gene GAPDH (glyceraldehyde 3‐phosphate dehydrogenase) was used as an endogenous control. Each value represents the mean ± standard deviation (SD) of three replicates.
Targets of CsLOB1
CsLOB1 is a transcriptional factor and probably exerts its effect by binding to the promoter region of target genes. It has been shown that LOB domain proteins bind to a 6‐bp LBD motif (GCGGCG) (Bell et al., 2012; Husbands et al., 2007). To identify the targets of CsLOB1, we searched the LBD motif in the promoter regions of 218 PthA4‐activated genes identified in our previous study (Hu et al., 2014, 2016). In total, the LBD motif has been identified in the promoter region of 26 PthA‐activated genes (Table S1, see Supporting Information).
We then tested the interactions between purified CsLOB1 and the LBD motif‐containing probes. Of the 26 LBD motif‐containing probes, only the Cs2g20600 probe interacted with CsLOB1 (Fig. 5A). The addition of 30 × unlabelled Cs2g20600 probe reduced the binding of CsLOB1 with the biotin‐labelled probe (Fig. 5B). Mutation of the LBD motif to ATAATA abolished the interaction between the probe and CsLOB1 (Fig. 5C). The qRT‐PCR result indicates that the gene expression of Cs2g20600 is up‐regulated on DEX‐treated CsLOB1‐GR transgenic plant leaves compared with the control at 8 h post‐inoculation (Fig. S3, see Supporting Information).
Figure 5.
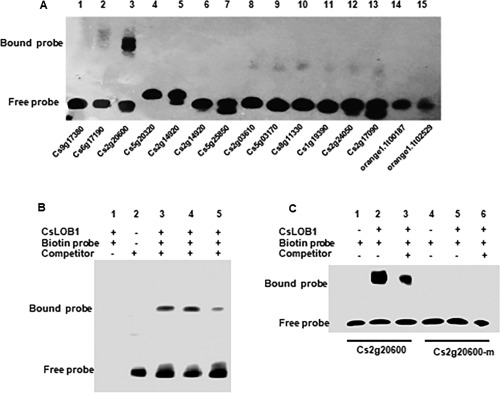
Electrophoretic mobility shift assay (EMSA) of CsLOB1 interaction with putative targets. (A) EMSA results revealed the interaction between biotin‐labelled LBD motif‐containing probes and CsLOB1 protein. (B) Competition for Cs2g20600 binding with the unlabelled probe was performed at 5× and 30× of the labelled probe. (C) Binding of the Cs2g20600 probe and mutant Cs2g20600 probe with CsLOB1. Unlabelled probe was added at 50× of the labelled probe. Cs2g20600‐m, mutant probe.
Discussion
In this study, we have demonstrated that CsLOB1 expression in citrus results in pustule formation. We used CsLOB1 fused with GR, which resides in the cytoplasm as part of a heteromeric complex with HSP90 before DEX treatment. Unliganded GR binds to the HSP complex primarily via the hormone‐binding domain that associates with HSP90 (Aoyama and Chua, 1997). After treatment, DEX binds to the hormone‐binding domain of GR, thus releasing CsLOB1‐GR to localize to the nucleus to exert its functions. This is indicated by pustule development and the induction of downstream genes in CsLOB1 transgenic grapefruit after DEX treatment. DEX treatment of wild‐type plants does not induce pustule development and the expression of the selected genes. The data presented here, previous data on canker symptom induction by dTALEs targeting EBECsLOB1 (Hu et al., 2014) and the requirement of CsLOB1 for canker symptoms (Jia et al., 2016, 2017) provide strong support that CsLOB1 is the canker susceptibility gene (Hu et al., 2014). Xcc infects citrus and causes pustule symptoms via the dominant pathogenicity factor PthA4, which activates the susceptibility gene CsLOB1, which, in turn, regulates downstream genes required for pustule symptom development. Importantly, citrus genes induced by PthA4 (Hu et al., 2014) were also induced by CsLOB1, confirming that PthA4 exerts its effect through CsLOB1. In addition, CsLOB1 shows higher expression in leaves and stems compared with roots, probably indicating that CsLOB1 expression is tissue specific and partially explaining why Xcc usually causes typical pustule lesions on certain tissues, including leaves and stems.
Our results suggest that CsLOB1 is the direct cause of pustule development, and the induction of CsLOB1 is sufficient to cause pustule development; however, water soaking requires other factors in addition to the induction of CsLOB1. Weak water soaking was observed in CsLOB1 transgenic grapefruit leaves after DEX treatment, but was obvious in CsLOB1 transgenic grapefruit leaves after XccΔpthA4 infection and DEX treatment. Water soaking is an important step in bacterial infection of the phyllosphere (Xin et al., 2016). PthA and related genes have been reported to be required for water soaking symptoms of citrus canker (Swarup et al., 1991). Interestingly, both PthA4 and CsLOB1 induce the expression of a pectate lyase gene (Hu et al., 2014). A pectate lyase gene has been suggested to be an indirect S gene target of the TAL effector AvrHah1 in Xanthomonas gardneri and contributes to water soaking in bacterial spot of tomato. dTALEs for pectate lyase complement water soaking when delivered by the avrHah1 mutant of X. gardneri (Schwartz et al., 2017). Induction of the pectate lyase gene might be responsible for the weak water soaking observed in DEX‐treated CsLOB1‐GR transgenic citrus. Previously, water soaking caused by Xcc has been hypothesized to result from increased water uptake from the xylem through capillary action as a result of the loss of free intercellular space because of cell swelling and cell division, which hydrates and swells xanthan gum (Brunings and Gabriel, 2003). Our data support this model. In this process, the activation of pectate lyase may also contribute to water soaking.
With CsLOB1 as the major canker susceptibility gene, it has been suggested that canker‐resistant citrus varieties could be generated via recessive resistance strategies by modification of EBE in the promoter region or coding region of CsLOB1 (Hu et al., 2014; Jia et al., 2016, 2017). This is similar to the generation of disease‐resistant rice by modification of EBEs of rice S gene promoters or coding regions (Antony et al., 2010; Li et al., 2012; Zhou et al., 2015). To understand the potential side effect of such a strategy, it is critical to understand the function of CsLOB1. CsLOB1 belongs to the plant‐specific LOB domain family, which consists of a conserved DNA‐binding Cys repeat motif (CX2CX6CX3C), an invariant glycine residue and a coiled‐coil Leu‐zipper‐like motif (LX6LX3LX6L) involved in protein–protein interactions (Shuai et al., 2002). LBD proteins are transcriptional factors involved in the regulation of lateral organ development, anthocyanin and nitrogen metabolism, and respond to hormones and environmental stimuli (Gendron et al., 2012; Majer and Hochholdinger, 2011). As expected, CsLOB1 contains multiple NLSs and localizes to the cell nucleus. Interestingly, citrus contains 36 LBD proteins with CsLOB2 and CsLOB3, which share 67.9% and 71.0% identity, respectively, to CsLOB1, and have similar functions to CsLOB1. Canker pustule symptoms are restored by dTALEs targeting CsLOB2 and CsLOB3 expressed in XccΔpthA4 (Zhang et al., 2016). RNA sequencing (RNA‐seq) analysis has shown that CsLOB1, CsLOB2 and CsLOB3 all regulate a set of cell wall metabolic genes, which might be involved in canker symptom development. However, neither CsLOB2 nor CsLOB3 contains the same EBE sequence in its promoter as CsLOB1, thus avoiding elicitation by PthA4. The redundancy of CsLOB1, CsLOB2 and CsLOB3 potentially explains the lack of side effect of the mutation of CsLOB1 alone (Jia et al., 2016).
Electrophoretic mobility shift assay (EMSA) results revealed that CsLOB1 shows strong and specific interaction with the promoter of Cs2g20600. Cs2g20600 encodes a zinc finger C3HC4‐type RING finger protein. C3HC4‐type RING finger proteins are known to be involved in numerous cellular processes, such as transcription, signal transduction through protein–protein interactions and ubiquitination, as most C3HC4‐type RING finger proteins are E3 ubiquitin ligases (Wu et al., 2014). However, how Cs2g20600 contributes to canker symptom development remains unknown. Future work will investigate the function of Cs2g20600 and its role in canker symptom development.
Experimental Procedures
Plant materials
All citrus plants used for inoculation were grown in a glasshouse at temperatures ranging from 25 to 30 °C at the Citrus Research and Education Center, University of Florida, Lake Alfred, FL, USA. Bacterial inoculation of plants was conducted in a quarantine glasshouse at 28 °C, 80% humidity, 16 h of daylight and 8 h of darkness.
Bacterial inoculations and population growth calculation
Xcc strains, including Xcc306 and Xcc306ΔpthA4, were kept in 25% glycerol and preserved in a freezer at −80 ºC. The Xcc strains were recovered and cultured on nutrient agar (NA) plates at 28 ºC. The Xcc strain used for inoculation was a bacterial suspension solution, which was diluted to the required concentration with double‐distilled water. An optical density at 600 nm (OD600) of 0.5 is equivalent to about 5 × 108 colony‐forming units (cfu)/mL. Bacterial suspensions of 5 × 105 cfu/mL were injected into the abaxial surface of citrus leaves with a 5‐mL needleless syringe. To calculate the bacterial population, inoculated leaves were rinsed with sterile water to remove dust from the leaf surface. Three discs (1 cm2) of the inoculated region were taken by a clean hole puncher and ground in 1 mL of sterile tap water. After a series of dilutions, 50 µL of bacterial suspensions were dropped onto NA medium by pipettes and incubated at 28 °C for 2 days. The colony counts were recorded to calculate the bacterial populations. Each experiment was repeated three times. The symptoms on the inoculation region were observed and captured at different time points with a digital camera.
RNA extraction and reverse transcription
Leaf samples were collected, frozen in liquid nitrogen and stored at −80 °C until RNA extraction. RNA extraction was performed for all plant tissues using an RNeasy Plant Mini Kit (QIAGEN, Valencia, CA, USA). RNA quality and concentration were determined with an ND‐8000 Nanodrop spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) based on the absorbance ratio. RNA samples with OD260/280 between 1.8 and 2.0 and OD260/230 > 2.0 were used for further analysis. The RNA was then processed via DNase I treatment and first‐strand cDNA synthesis using an RNA reverse transcription kit (QIAGEN, Valencia, CA, USA) according to the manufacturer's instructions.
qRT‐PCR analyses
All primers used for qRT‐PCR in various experiments were designed on the website (https://www.idtdna.com/Primerquest/Home/Index). Primers were designed with melting temperatures between 60 and 64 °C, oligo lengths of 18–30 bp, GC contents of 40%–60% and amplicon sizes between 70 and 150 bp. qRT‐PCR was performed using KiCqStart® SYBR Green qPCR ReadyMix™ (Applied Biosystems, Foster City, CA, USA). The gene‐specific primer sequences are listed in Table S2 (see Supporting Information). qRT‐PCR was performed using an Applied Biosystems 7500 Fast Real‐time PCR system (Applied Biosystems, Foster City, CA, USA). The endogenous housekeeping gene used was Cs2g14940 (GAPDH, glyceraldehyde 3‐phosphate dehydrogenase). The experiments were repeated twice, and each experiment contained three replicates. Each reaction was run in a 20‐μL volume reaction system containing 5 μL of template cDNA, 10 μL of KiCqStart SYBR Green qPCR Readymix and a final primer concentration of 300 nm. All reactions were performed under the following conditions: 30 s at 95 °C, and 40 cycles of 5 s at 95 °C and 30 s at 60 °C, in 96‐well optical reaction plates (Applied Biosystems, Foster City, CA, USA). A melting curve was generated from 60 °C to 95 °C at the end of the reaction to verify the specificity of the amplicon for each primer pair. The gene expression levels in all samples were determined by the number of cycles (Ct) required for the amplification‐related fluorescence to reach a specific threshold level of detection. The raw Ct value was analysed using QuantStudio™ design and analysis software v1.4.1. The 2−ΔΔCt method was used for relative quantification.
PEG‐mediated transient expression in citrus protoplasts
Grapefruit seedlings germinated from seeds were cultured in a dark glasshouse for 15 days. Small stem segments from the seedlings were obtained using a fresh sharp blade and digested in an enzyme solution [20 mm MES (pH 5.7) containing 1.5% (w/v) cellulase R10, 0.4% (w/v) macerozyme R10, 0.4 m mannitol and 20 mm KCl; the solution was kept at 55 °C for 10 min, and then cooled to room temperature (25 °C), and 10 mm CaCl2, 1–5 mm β‐mercaptoethanol and 0.1% bovine serum albumin (BSA) were added] to obtain protoplasts. The concentration and quality of the collected protoplasts were checked under a microscope. Approximately 10 μL of DNA (20 μg of plasmid DNA) were mixed with 100 μL of protoplasts (approximately 2 × 104 protoplasts). Then, 110 μL of PEG solution [20%–40% (w/v) PEG4000 in double‐distilled H2O containing 0.2 m mannitol and 100 mm CaCl2) were added and mixed. The transfection mixture was incubated at room temperature for 15 min. The transfection mixture was diluted with 400 μL of W5 solution. The supernatant was removed after centrifugation at 100 g for 2 min at room temperature. The protoplasts were resuspended gently and then cultured in a six‐well tissue culture plate overnight. All solutions and detailed protocols processed in this research were followed by PEG‐calcium mediated transfection was conducted as described previously (Yoo et al., 2007).
Generation of transgenic Duncan grapefruit plants expressing 35S:CsLOB1‐GR
The coding region of CsLOB1 of 711 bp without start and stop codons was amplified with primers containing XhoI/EcoRI restriction sites, and subcloned into the pCAMBIA‐1380 plasmid to generate the 35S:CsLOB1‐GR construct with GR fused in‐frame to the C‐terminus of CsLOB1. The GR1 domain was amplified from BIΔGR, which was kindly provided by Dr R. W. Davis (Lloyd et al., 1994). To facilitate screening, GFP and NptII sequences were also included in the final construct (Fig. 2). The 35S:CsLOB1‐GR plasmid was transferred into Agrobacterium strain EHA105 and used for plant transformation. Duncan grapefruit epicotyl segments were used as explants for transformation, and the Agrobacterium‐mediated citrus transformation was performed as described previously (Orbović and Grosser, 2015). GFP fluorescence in putative transgenic lines was evaluated using an epifluorescence stereomicroscope. The transgenic plants were confirmed by PCR analysis (Table S3, see Supporting Information). Transgenic grapefruit shoots were micrografted in vitro onto 1‐month‐old Carrizo citrange [C. sinensis (L.) Osbeck × Poncirus trifoliata (L.) Raf.] rootstock seedlings. The grafted shoots were potted on to grow in the glasshouse after 1 month.
Protein purification
The coding sequence of CsLOB1 of 714 bp was amplified with primers containing EcoRI/SalI restriction sites, and subcloned into the protein expression vector pGEX‐4T‐1, named pGEX‐4T‐1‐CsLOB1, using Q5® High‐Fidelity 2× Master Mix (New England Biolabs, Ipswich, MA, USA). The expression vector pGEX‐4T‐1‐CsLOB1 was confirmed by sequencing, and transformed into BL21 competent Escherichia coli (New England Biolabs, Ipswich, MA, USA). Escherichia coli BL21 was cultured in 10 mL of Luria–Bertani (LB) medium at 37 °C overnight; 0.1 mL of the overnight culture was transferred into 50 mL of LB medium containing 50 µm kanamycin and incubated at 37 °C until A 600 nm reached OD = 0.6. A final concentration of 0.25 mm of isopropylthiogalactopyranoside (IPTG) was added to the LB medium. The incubation was continued for 10 h at 16 °C. Bacterial cells were harvested by centrifuging the culture at 4000 g for 30 min. The pellet was resuspended in phosphate‐buffered saline (PBS) and sonicated on ice. The supernatant was then purified using Pierce™ glutathione agarose according to the instructions of the manufacturer (Thermo Scientific, Waltham, MA, USA). The fractions and purified GST‐CsLOB1 protein were analysed by sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS–PAGE) on a 12% polyacrylamide gel, and determined using a protein molecular standard (New England Biolabs, Ipswich, MA, USA).
EMSA
CsLOB1 protein (1 µg/µL) was used for EMSAs. Twenty‐six LBD motif‐containing probes were synthesized and biotin labelled for EMSA (50‐bp promoter sequences containing the LBD motif in the centre) using the Biotin 3′ End DNA Labelling Kit (Thermo Scientific, Waltham, MA, USA). EMSA was then performed using a LightShift™ Chemiluminescent EMSA Kit according to the manufacturer's instructions (Thermo Scientific, Waltham, MA, USA).
Author Contributions
N.W. conceived and supervised the project. H.J., S.D. and N.W. designed the experiment. S.D., H.J. and D.T. conducted the experiments. N.W. and S.D. wrote the manuscript. All authors read, revised and approved the final manuscript.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 (A) Diagram of the vector 35S:EYFP‐CsLOB1. This vector is based on the vector pSAT6‐EYFP‐C1 and was used for subcellular localization. (B) Confirmation of CsLOB1 subcellular localization. The vectors 35S:CsLOB1‐EYFP and mCherry‐NLS were co‐transformed into citrus protoplast. The fluorescence signal was detected with fluorescence microscopy. The green colour indicates the fluorescence of enhanced yellow fluorescent protein (EYFP), whereas the red colour indicates the nuclear localization due to red fluorescent protein. The scale bar represents 10 μm.
Fig. S2 (A) CsLOB1 transgenic plant shoots show green fluorescence under a fluorescence microscope: 1, transgenic plant shoot [green fluorescent protein (GFP) signal]; 2, non‐transgenic shoot without GFP signal. (B) CsLOB1 transgenic plants show normal growth when compared with a wild‐type plant.
Fig. S3 Validation of Cs2g20600 gene expression in CsLOB1 transgenic plant leaves treated with DEX (100 μM) or mock solution at 8 hours post inoculation by qRT‐PCR. The housekeeping gene GAPDH was used as the endogenous control. Each value represents the fold change (DEX‐treatment vs mock‐treatment) ± SD of three replicates. Transgenic: DEX‐treated transgenic plant leaves; Wild type: DEX‐treated wild type plant leaves.
Table S1 Probes used in electrophoretic mobility shift assay (EMSA).
Table S2 Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) primer sequences used in this study.
Table S3 Primer sequences used in this study.
Acknowledgements
This project was supported by the US Department of Agriculture‐National Institute of Food and Agriculture (USDA‐NIFA) Plant Biotic Interactions Program 2017‐67013‐26527 and the Chinese Scholarship Council. We thank Dr R. W. Davis for providing BIΔGR.
Contributor Information
Changyong Zhou, Email: zhoucy@cric.cn.
Nian Wang, Email: nianwang@ufl.edu.
References
- Al‐Saadi, A. , Reddy, J.D. , Duan, Y.P. , Brunings, A.M. , Yuan, Q. and Gabriel, D.W. (2007) All five host‐range variants of Xanthomonas citri carry one pthA homolog with 17.5 repeats that determines pathogenicity on citrus, but none determine host‐range variation. Mol. Plant–Microbe Interact. 20, 934–943. [DOI] [PubMed] [Google Scholar]
- Antony, G. , Zhou, J. , Huang, S. , Li, T. , Liu, B. , White, F. and Yang, B. (2010) Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os‐11N3 . Plant Cell, 22, 3864–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama, T. and Chua, N.H. (1997) A glucocorticoid‐mediated transcriptional induction system in transgenic plants. Plant J. 11, 605–612. [DOI] [PubMed] [Google Scholar]
- Bell, E.M. , Lin, W‐C. , Husbands, A.Y. , Yu, L. , Jaganatha, V. , Jablonska, B. , Mangeon, A. , Neff, M.M. , Girke, T. and Springer, P.S. (2012) Arabidopsis lateral organ boundaries negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proc. Natl. Acad. Sci. USA, 109, 21 146–21 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch, J. , Scholze, H. , Schornack, S. , Landgraf, A. , Hahn, S. , Kay, S. , Lahaye, T. , Nickstadt, A. and Bonas, U. (2009) Breaking the code of DNA binding specificity of TAL‐type III effectors. Science, 326, 1509–1512. [DOI] [PubMed] [Google Scholar]
- Bogdanove, A.J. , Schornack, S. and Lahaye, T. (2010) TAL effectors: finding plant genes for disease and defense. Curr. Opin. Plant Biol. 13, 394–401. [DOI] [PubMed] [Google Scholar]
- Brunings, A.M. and Gabriel, D.W. (2003) Xanthomonas citri: breaking the surface. Mol. Plant Pathol. 4, 141–157. [DOI] [PubMed] [Google Scholar]
- Chu, Z. , Fu, B. , Yang, H. , Xu, C. , Li, Z. , Sanchez, A. , Park, Y.J. , Bennetzen, J.L. , Zhang, Q. and Wang, S. (2006) Targeting xa13, a recessive gene for bacterial blight resistance in rice. Theor. Appl. Genet. 112, 455–461. [DOI] [PubMed] [Google Scholar]
- Cubero, J. and Graham, J.H. (2002) Genetic relationship among worldwide strains of Xanthomonas causing canker in citrus species and design of new primers for their identification by PCR. Appl. Environ. Microbiol. 68, 1257–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galigniana, M.D. , Scruggs, J.L. , Herrington, J. , Welsh, M.J. , Carter‐Su, C. , Housley, P.R. and Pratt, W.B. (1998) Heat shock protein 90‐dependent (geldanamycin‐inhibited) movement of the glucocorticoid receptor through the cytoplasm to the nucleus requires intact cytoskeleton. Mol. Endocrinol. 12, 1903–1913. [DOI] [PubMed] [Google Scholar]
- Gendron, J.M. , Liu, J.‐S. , Fan, M. , Bai, M.‐Y. , Wenkel, S. , Springer, P.S. , Barton, M.K. and Wang, Z.‐Y. (2012) Brassinosteroids regulate organ boundary formation in the shoot apical meristem of Arabidopsis . Proc. Natl. Acad. Sci. USA, 109, 21 152–21 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwald, T. , Hughes, G. , Graham, J. , Sun, X. and Riley, T. (2001) The citrus canker epidemic in Florida: the scientific basis of regulatory eradication policy for an invasive species. Phytopathology, 91, 30–34. [DOI] [PubMed] [Google Scholar]
- Graham, J.H. , Gottwald, T. , Cubero, J. and Achor, D.S. (2004) Xanthomonas axonopodis pv. citri: factors affecting successful eradication of citrus canker. Mol. Plant Pathol. 5, 1–15. [DOI] [PubMed] [Google Scholar]
- Hann, D. , Gimenez‐Ibanez, S. and Rathjen, J.P. (2010) Bacterial virulence effectors and their activities. Curr. Opin. Plant Biol. 13, 388–393. [DOI] [PubMed] [Google Scholar]
- Hu, Y. , Zhang, J. , Jia, H. , Sosso, D. , Li, T. , Frommer, W.B. , Yang, B. , White, F.F. , Wang, N. and Jones, J.B. (2014) Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. USA, 111, E521–E529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y. , Duan, S. , Zhang, Y. , Shantharaj, D. , Jones, J.B. and Wang, N. (2016) Temporal transcription profiling of sweet orange in response to PthA4‐mediated Xanthomonas citri subsp. citri infection. Phytopathology, 106, 442–451. [DOI] [PubMed] [Google Scholar]
- Husbands, A. , Bell, E.M. , Shuai, B. , Smith, H.M. and Springer, P.S. (2007) LATERAL ORGAN BOUNDARIES defines a new family of DNA‐binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res. 35, 6663–6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalan, N. , Kumar, D. , Andrade, M.O. , Yu, F. , Jones, J.B. , Graham, J.H. , White, F.F. , Setubal, J.C. and Wang, N. (2013) Comparative genomic and transcriptome analyses of pathotypes of Xanthomonas citri subsp. citri provide insights into mechanisms of bacterial virulence and host range. BMC Genomics, 14, 551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, H. , Orbovic, V. , Jones, J.B. and Wang, N. (2016) Modification of the PthA4 effector binding elements in Type I CsLOB1 promoter using Cas9/sgRNA to produce transgenic Duncan grapefruit alleviating XccΔpthA4:dCsLOB1.3 infection. Plant Biotechnol. J. 14, 1291–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, H. , Zhang, Y. , Orbović, V. , Xu, J. , White, F.F. , Jones, J.B. and Wang, N. (2017) Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol. J. 15, 817–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay, S. , Hahn, S. , Marois, E. , Hause, G. and Bonas, U. (2007) A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science, 318, 648–651. [DOI] [PubMed] [Google Scholar]
- Li, T. , Liu, B. , Spalding, M.H. , Weeks, D.P. and Yang, B. (2012) High‐efficiency TALEN‐based gene editing produces disease‐resistant rice. Nat. Biotechnol. 30, 390–392. [DOI] [PubMed] [Google Scholar]
- Lloyd, A.M. , Schena, M. , Walbot, V. and Davis, R.W. (1994) Epidermal cell fate determination in Arabidopsis: patterns defined by a steroid‐inducible regulator. Science, 266, 436–439. [DOI] [PubMed] [Google Scholar]
- Majer, C. and Hochholdinger, F. (2011) Defining the boundaries: structure and function of LOB domain proteins. Trends Plant Sci. 16, 47–52. [DOI] [PubMed] [Google Scholar]
- Moscou, M.J. and Bogdanove, A.J. (2009) A simple cipher governs DNA recognition by TAL effectors. Science, 326, 1501. [DOI] [PubMed] [Google Scholar]
- Orbović, V. and Grosser, J. (2015) Citrus transformation using juvenile tissue explants. Methods Mol Biol. 1224, 245–257. [DOI] [PubMed] [Google Scholar]
- Picard, D. , Salser, S. and Yamamoto, K.R (1988) A movable and regulable inactivation function within the steroid binding domain of the glucocorticoid receptor. Cell, 54, 1073–1080. [DOI] [PubMed] [Google Scholar]
- Schwartz, A. , Morbitzer, R. , Lahaye, T. and Staskawicz, B.J. (2017) TALE‐induced bHLH transcription factors that activate a pectate lyase contribute to water soaking in bacterial spot of tomato. Proc. Natl. Acad. Sci. USA, 114, E897–E903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai, B. , Reynaga‐Peña, C.G. and Springer, P.S. (2002) The lateral organ boundaries gene defines a novel, plant‐specific gene family. Plant Physiol. 129, 747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva, A.C.R. , Ferro, J.A. , Reinach, F.C. , Farah, C.S. , Furlan, L.R. , Quaggio, R.B. , Monteiro‐Vitorello, C.B. , Van Sluys, M.A. , Almeida, N.F. , Alves, L.M.C. , do Amaral, A.M. , Bertolini, M.C. , Camargo, L.E.A. , Camarotte, G. , Cannavan, F. , Cardozo, J. , Chambergo, F. , Ciapina, L.P. , Cicarelli, R.M.B. , Coutinho, L.L. , Cursino‐Santos, J.R. , El‐Dorry, H. , Faria, J.B. , Ferreira, A.J.S. , Ferreira, R.C.C. , Ferro, M.I.T. , Formighieri, E.F. , Franco, M.C. , Greggio, C.C. , Gruber, A. , Katsuyama, A.M. , Kishi, L.T. , Leite, R.P. , Lemos, E.G.M. , Lemos, M.V.F. , Locali, E.C. , Machado, M.A. , Madeira, A.M.B.N. , Martinez‐Rossi, N.M. , Martins, E.C. , Meidanis, J. , Menck, C.F.M. , Miyaki, C.Y. , Moon, D.H. , Moreira, L.M. , Novo, M.T.M. , Okura, V.K. , Oliveira, M.C. , Oliveira, V.R. , Pereira, H.A. , Rossi, A. , Sena, J.A.D. , Silva, C. , de Souza, R.F. , Spinola, L.A.F. , Takita, M.A. , Tamura, R.E. , Teixeira, E.C. , Tezza, R.I.D. , Trindade dos Santos, M. , Truffi, D. , Tsai, S.M. , White, F.F. , Setubal, J.C. and Kitajima, J.P. (2002) Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature, 417, 459–463. [DOI] [PubMed] [Google Scholar]
- Swarup, S. , Defeyter, R. , Brlansky, R. and Gabriel, D. (1991) A pathogenicity locus from Xanthomonas citri enables strains from several pathovars of X. campestris to elicit cankerlike lesions on citrus. Phytopathology, 81, 802–809. [Google Scholar]
- Swarup, S. , Yang, Y. , Kingsley, M. and Gabriel, D. (1992) An Xanthomonas citri pathogenicity gene, pthA, pleiotropically encodes gratuitous avirulence on nonhosts. Mol. Plant–Microbe Interact. 5, 204–213. [DOI] [PubMed] [Google Scholar]
- Wu, W. , Cheng, Z. , Liu, M. , Yang, X. and Qiu, D. (2014) C3HC4‐type RING finger protein NbZFP1 is involved in growth and fruit development in Nicotiana benthamiana . PLoS One, 9, e99352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin, X. , Nomura, K. , Aung, K. , Velásquez, A.C. , Yao, J. , Boutrot, F. , Chang, J.H. , Zipfel, C. and He, S.Y. (2016) Bacteria establish an aqueous living space in plants crucial for virulence. Nature, 539, 524–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Q. and Wang, N. (2012) High‐throughput screening and analysis of genes of Xanthomonas citri subsp. citri involved in citrus canker symptom development. Mol. Plant–Microbe Interact. 25, 69–84. [DOI] [PubMed] [Google Scholar]
- Yang, B. , Sugio, A. and White, F.F. (2006) Os8N3 is a host disease‐susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. USA, 103, 10 503–10 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, S.‐D. , Cho, Y.‐H. and Sheen, J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Huguet‐Tapia, J.C. , Hu, Y. , Jones, J. , Wang, N. , Liu, S. and White, F.F. (2016) Homologs of CsLOB1 in citrus function as disease susceptibility genes in citrus canker. Mol. Plant Pathol. 18, 798–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Jalan, N. , Zhou, X. , Goss, E. , Jones, J.B. , Setubal, J.C. , Deng, X. and Wang, N. (2015) Positive selection is the main driving force for evolution of citrus canker‐causing Xanthomonas . ISME J. 9, 2128–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Peng, Z. , Long, J. , Sosso, D. , Liu, B. , Eom, J.‐S. , Huang, S. , Liu, S. , Vera Cruz, C. , Frommer, W.B. , White, F.F. and Yang, B. (2015) Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 82, 632–643. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 (A) Diagram of the vector 35S:EYFP‐CsLOB1. This vector is based on the vector pSAT6‐EYFP‐C1 and was used for subcellular localization. (B) Confirmation of CsLOB1 subcellular localization. The vectors 35S:CsLOB1‐EYFP and mCherry‐NLS were co‐transformed into citrus protoplast. The fluorescence signal was detected with fluorescence microscopy. The green colour indicates the fluorescence of enhanced yellow fluorescent protein (EYFP), whereas the red colour indicates the nuclear localization due to red fluorescent protein. The scale bar represents 10 μm.
Fig. S2 (A) CsLOB1 transgenic plant shoots show green fluorescence under a fluorescence microscope: 1, transgenic plant shoot [green fluorescent protein (GFP) signal]; 2, non‐transgenic shoot without GFP signal. (B) CsLOB1 transgenic plants show normal growth when compared with a wild‐type plant.
Fig. S3 Validation of Cs2g20600 gene expression in CsLOB1 transgenic plant leaves treated with DEX (100 μM) or mock solution at 8 hours post inoculation by qRT‐PCR. The housekeeping gene GAPDH was used as the endogenous control. Each value represents the fold change (DEX‐treatment vs mock‐treatment) ± SD of three replicates. Transgenic: DEX‐treated transgenic plant leaves; Wild type: DEX‐treated wild type plant leaves.
Table S1 Probes used in electrophoretic mobility shift assay (EMSA).
Table S2 Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) primer sequences used in this study.
Table S3 Primer sequences used in this study.


