Summary
Pantoea agglomerans, a widespread epiphytic bacterium, has evolved into a hypersensitive response and pathogenicity (hrp)‐dependent and host‐specific gall‐forming pathogen by the acquisition of a pathogenicity plasmid containing a type III secretion system (T3SS) and its effectors (T3Es). Pantoea agglomerans pv. betae (Pab) elicits galls on beet (Beta vulgaris) and gypsophila (Gypsophila paniculata), whereas P. agglomerans pv. gypsophilae (Pag) incites galls on gypsophila and a hypersensitive response (HR) on beet. Draft genome sequences were generated and employed in combination with a machine‐learning approach and a translocation assay into beet roots to identify the pools of T3Es in the two pathovars. The genomes of the sequenced Pab4188 and Pag824‐1 strains have a similar size (∼5 MB) and GC content (∼55%). Mutational analysis revealed that, in Pab4188, eight T3Es (HsvB, HsvG, PseB, DspA/E, HopAY1, HopX2, HopAF1 and HrpK) contribute to pathogenicity on beet and gypsophila. In Pag824‐1, nine T3Es (HsvG, HsvB, PthG, DspA/E, HopAY1, HopD1, HopX2, HopAF1 and HrpK) contribute to pathogenicity on gypsophila, whereas the PthG effector triggers HR on beet. HsvB, HsvG, PthG and PseB appear to endow pathovar specificities to Pab and Pag, and no homologous T3Es were identified for these proteins in other phytopathogenic bacteria. Conversely, the remaining T3Es contribute to the virulence of both pathovars, and homologous T3Es were found in other phytopathogenic bacteria. Remarkably, HsvG and HsvB, which act as host‐specific transcription factors, displayed the largest contribution to disease development.
Keywords: galls, gypsophila, Pantoea agglomerans pv. betae, Pantoea agglomerans pv. gypsophilae, pathoadaptive, pseB, table beet
Introduction
Many Gram‐negative plant‐ and animal‐pathogenic bacteria employ a type III secretion system (T3SS) to secrete effectors (T3Es) and colonize their host organisms (Buttner and He, 2009). In phytopathogenic bacteria, T3SSs are encoded by the hrp/hrc (hypersensitive response and pathogenicity/hypersensitive response and conserved) gene cluster, which is required for bacteria to cause disease in susceptible plants and to elicit the hypersensitive response (HR) in resistant plants (Lindgren, 1997). The T3SS delivers T3Es directly into the cytosol of eukaryotic cells, and thus allows the manipulation of host cellular activities to the benefit of the pathogen. T3Es can subvert host signalling pathways, suppress the immune response, modify the cytoskeleton structure and interfere with cellular trafficking (Dou and Zhou, 2012; Lindeberg et al., 2012). Most known T3Es belong to conserved protein families that are present in evolutionary distant bacteria encoding the T3SS (McCann and Guttman, 2008). However, a given bacterial pathogen species or pathovar may encode T3Es which have emerged during the evolution of its pathogenicity and may be linked to host specificity (Nissan et al., 2006; Sokurenko et al., 1999).
Pantoea agglomerans is widespread in diverse natural habitats; in particular, it is associated with many plants as a common epiphyte and endophyte (Kobayashi and Palumbo, 2000; Lindow and Brandl, 2003). In addition, it is the most commonly opportunistic bacterial pathogen isolated from humans (Cruz et al., 2007). Pantoea agglomerans pv. gypsophilae (Pag) (formerly Erwinia herbicola pv. gypsophilae) was first reported as a gall‐forming bacterium on gypsophila that limits plant propagation (Cooksey, 1986), whereas its related pathovar Pantoea agglomerans pv. betae (Pab) was later described as a gall‐forming pathogen on beet (Burr et al., 1991). Pag induces galls on gypsophila and HR on beet, whereas Pab is tumorigenic on beet and gypsophila. However, on gypsophila, galls caused by Pab are morphologically distinguishable from galls produced by Pag (Burr et al., 1991). The virulence of both pathovars evolved by the acquisition of a plasmid containing a pathogenicity island (PAI), which was distributed among genetically diverse populations of P. agglomerans (Weinthal et al., 2007). The pathogenicity plasmids of Pag and Pab, designated as pPATHPag and pPATHPab, respectively, may vary in size (Manulis et al., 1991) and their curing results in a loss of pathogenicity (Weinthal et al., 2007).
pPATHPag of the Pag strain 824‐1 has been investigated more extensively. It has a size of approximately 135 kb that accommodates a PAI of ∼75 kb (Barash and Manulis‐Sasson, 2009). It harbours an intact hrp/hrc cluster containing a functional T3SS (Mor et al., 2001), eight T3Es, six of which significantly reduce gall formation on mutagenesis (Barash and Manulis‐Sasson, 2007), a cluster of genes encoding the biosynthetic enzymes of the plant hormones auxin and cytokinins (Clark et al., 1993; Lichter et al., 1995), multiple and diverse insertion sequences (Guo et al., 2002), and partial sequences of known bacterial genes (Manulis and Barash, 2003). The structure of the PAI and its plasmid location support the premise of a recent evolution of pathogenesis (Barash and Manulis‐Sasson, 2009). Although it is not yet clear how host specificity at the pathovar level is determined, many reports support the hypothesis that differential repertoires of T3Es may be involved (e.g., Hajri et al., 2009; Lindeberg et al., 2009). Comparative analysis of the T3E inventories of Pag and Pab could shed new light on the role of effectors in the host specificity of these pathovars.
Advances in genome sequencing have allowed the identification of many novel T3Es in various phytopathogenic bacteria (Baltrus et al., 2012; Kvitko et al., 2009; O'Brien et al., 2011; Peeters et al., 2013). Predictions of T3Es have generally been based on sequence homology to known effectors from other bacterial pathogens or the presence of eukaryotic characteristics indicative of a function within the host cell (Collmer et al., 2002; Da Silva et al., 2002). Effectors have also been predicted based on the presence of conserved cis‐acting regulatory motifs in the promoter region of the gene or translocation signals at the protein N‐terminus (Jiang et al., 2009; Petnicki‐Ocwieja et al., 2002). Recently, a machine‐learning approach has been utilized for the prediction of T3Es (Burstein et al., 2009, 2015; Lifshitz et al., 2014; Teper et al., 2016). The machine‐learning approach extracts features that distinguish effector from non‐effector proteins and uses them to train a diverse arsenal of machine‐learning classification algorithms. Teper et al. (2016) used a machine‐learning approach based on 79 features, including homology to known T3Es of plant‐ and animal‐pathogenic bacteria, genomic proximity to other effectors, GC content, sequence homology to genes encoded in bacteria that do not encode a T3SS (a negative feature), amino acid composition at the N‐terminus and in the entire protein, and the presence of specific regulatory elements. After validation of candidate effectors identified in the first round of machine learning, new information is incorporated for the second round of prediction (Teper et al., 2016).
In contrast with the information on T3Es residing on pPATHPag, the information on the T3Es of pPATHPab is quite scanty (reviewed in Barash and Manulis‐Sasson, 2009). Furthermore, the possibility of chromosomal T3Es has not been investigated previously. The major goal of this study was to progress towards the identification of the complete inventories of T3Es in Pag and Pab, and to analyse their relationship to the differential host specificities between the two pathovars. To this aim, we first generated draft genome sequences of the two pathovars, and then employed a machine‐learning approach to predict their T3Es. A translocation assay into cells of beet roots was devised based on features of the Pag effector PthG and employed for experimental validation. Finally, the contribution of each effector to gall formation was quantitatively assessed and its possible involvement in host specificity was evaluated.
Results
Generation of draft genome sequences of Pag and Pab
Genome sequencing of Pag strain 824‐1 and Pab strain 4188 was performed using the Illumina MiSeq platform. Raw reads (250‐bp paired‐end) were pre‐processed using Trimmomatic (Bolger et al., 2014) and assembled using Velvet (Zerbino and Birney, 2008). The genomes of both pathovars were found to be almost identical in size (∼5 MB), had the same GC content (∼55%) and contained ∼4600 predicted open reading frames (ORFs) (Table 1). Similar characteristics have been reported for other genomes of P. agglomerans (e.g. Lim et al., 2014; Matsuzawa et al., 2012; Smith et al., 2013). Genome sequence assemblies of Pab4188 and Pag824‐1 were deposited in the GenBank database under the accession numbers GCA_001662025.1 and GCA_001661985.1, respectively.
Table 1.
Characteristics of the draft genomes of Pantoea agglomerans pvs. betae and gypsophilae.
| Genome characteristics | Pab4188 | Pag824‐1 |
|---|---|---|
| Genome size (MB) | 5.02 | 5.01 |
| Number of contigs (99% of the genome) | 34 | 19 |
| N50 | 499.008 | 499.033 |
| GC content (%) | 54.9 | 54.8 |
| Number of open reading frames | 4599 | 4598 |
N50 is a statistical measure of an average length of a set of sequences.
Prediction of T3Es of Pab4188 and Pag824‐1
To identify novel T3Es of the Pantoea pathovars, we employed a machine‐learning approach similar to that used previously to identify T3Es in Xanthomonas euvesicatoria (Teper et al., 2016) and Pseudomonas aeruginosa (Burstein et al., 2015), and type IV effectors (T4Es) in Legionella pneumophila and Coxiella burnetii (Burstein et al., 2009; Lifshitz et al., 2014). The machine‐learning algorithm was based on 49 features that potentially differentiate T3Es from non‐effectors. The performance scores of the top 15 features in Pab4188 and Pag824‐1 are reported in Tables S1 and S2 (see Supporting Information), respectively. Feature performance was estimated by area under curve‐receiver operating characteristic (AUC‐ROC) and area under the precision‐recall curve (AUPRC), and was carried out on 10‐fold cross‐validation. The AUC‐ROC and AUPRC scores achieved on the Pab dataset were 0.816 ± 0.105 and 0.942 ± 0.036, respectively, and on the Pag dataset were 1.0 ± 0 and 1.0 ± 0, respectively. The T3E prediction scores for ORFs of Pab4188 and Pag824‐1, as determined by the machine‐learning approach, are given in Tables S3 and S4 (see Supporting Information), respectively.
Translocation assay for predicted T3Es
To examine the translocation of the predicted effectors, we devised a translocation assay based on the Pag effector PthG (Ezra et al., 2000). As indicated earlier, Pag elicits HR on beet. This HR is mediated by recognition of the PthG effector (Ezra et al., 2000). Deletion analysis of PthG identified a 225‐amino‐acid domain at the protein C‐terminus (amino acids 247 to 472), which is sufficient to trigger HR on beet (HR domain), and a 43‐amino‐acid domain at the protein N‐terminus, which is sufficient for translocation (translocation signal; Nissan et al., 2003). A PthG translocation cassette (Fig. 1) composed of a multiple cloning site and the PthG HR domain fused to a histidine (His) tag was constructed and cloned under the Plac promoter into the pVSP61 plasmid to yield pVSP‐PtHis (Fig. 1). The HR domain on pVSP‐PtHis could not be translocated into beet cells unless fused to the N‐terminal translocation signal of a T3E. For each predicted effector, a gene fragment containing the N‐terminus (∼70 amino acids) of the ORF, together with a 20‐bp upstream sequence containing the ribosome binding site, was polymerase chain reaction (PCR) amplified and fused in frame to the PthG HR domain within the translocation cassette. The translocation cassette with or without the N‐terminus of the predicted effector was transformed into Pab4188 and into a Pab mutant lacking a functional T3SS. The expression of all the fusion constructs in the obtained strains was validated by dot blot using anti‐His antibodies (Fig. S1, see Supporting Information). Each of the strains was inoculated [106 colony‐forming units (cfu)/mL] into beet cubes, as described in Experimental Procedures, and monitored for the elicitation of HR (appearing within 24–48 h and indicating translocation), gall formation (appearing after 4–5 days and indicating lack of translocation) or no symptoms (expected for the T3SS mutant used as negative control) (Fig. 2).
Figure 1.
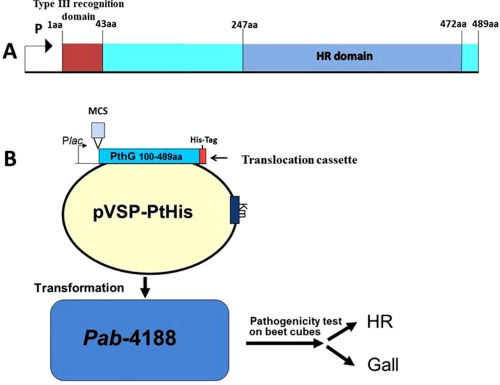
(A) Schematic structure of PthG. (B) Translocation cassette for the translocation assay of type III effectors (T3Es) into beet cubes. The translocation cassette consists of multiple cloning sites (MCSs) and the C‐terminus of PthG containing the hypersensitive response (HR) domain fused to a His‐Tag. The translocation cassette was cloned into pVSP61 under the lac promoter to yield kanamycin (Km)‐resistant pVSP‐PtHis. aa, amino acid. Details of the assay procedure are described in the Results section.
Figure 2.

Translocation assay on beet cubes. 1, water control; 2, Pab4188 (gall formation); 3, Pab4188 with the translocation vector, pVSP‐PtHis, containing the N‐terminus of HsvG (hypersensitive response, HR); 4, Pab4188 with the empty vector, pVSP‐PtHis (gall formation).
The ORFs to be tested for translocation were selected on the basis of the prediction scores obtained by the machine‐learning analysis, homology to known T3Es in other bacteria and the presence of a hrp box upstream to the ORF. The hrp box is the recognition sequence that activates the HrpL regulon in P. agglomerans (Nizan‐Koren et al., 2003). Highly ranked ORFs showing homology to non‐effector genes were excluded. As most ORFs with high prediction scores were located on contigs originating from plasmids, chromosomal ORFs with high scores were selected for the translocation assay in a search for chromosomal T3Es. The results illustrated in Tables 2 and 3 describe the translocation of Pab and Pag effectors, respectively. Translocation was demonstrated for nine of 19 ORFs that were examined for the Pab4188 strain, and for eight of 17 that were examined for the Pag824‐1 strain. All the translocated ORFs in both pathovars harboured an hrp box. Despite the fact that five ORFs in Pab and three in Pag contained an hrp box, they were not translocated (Tables 2 and 3). In both pathovars, these included HopAK1 and HrpN, which were defined as harpins (Choi et al., 2013), and HopVI, whose Pseudomonas syringae homologue was shown to be a translocated effector (Lindeberg et al., 2005). Three highly ranked chromosomal ORFs from each pathovar were not translocated. The latter finding, together with the observation that all the identified T3Es resided on plasmid contigs, suggests that the T3Es in both pathovars were restricted to the pPATH plasmids.
Table 2.
Translocation assay for predicted Pantoea agglomerans pv. betae type III effectors.*
| Gene/ORF* | Prediction score † | hrp box ‡ | Translocation |
|---|---|---|---|
| HsvG | 0.997 | + | + |
| HsvB | 0.997 | + | + |
| HopAF1 | 0.996 | + | + |
| HopD1 | 0.996 | + | + |
| HopR1 | 0.996 | – | – |
| HopX2 | 0.995 | + | + |
| HopAY1 | 0.994 | + | + |
| 1611 | 0.993 | + | – |
| PthG | 0.992 | + | + |
| PseB | 0.991 | + | + |
| 1595 | 0.99 | – | – |
| HrpN | 0.989 | + | – |
| HrpK | 0.989 | + | + |
| HopV1 | 0.973 | + | – |
| HopAK1 | 0.971 | – | – |
| 2716 § | 0.683 | + | – |
| 2223 § | 0.62 | – | – |
| 585 § | 0.567 | + | – |
| 1337 § | 0.236 | – | – |
Table 3.
Translocation assay for predicted Pantoea agglomerans pv. gypsophilae type III effectors.*
| Gene/ORF* | Prediction score † | hrp box ‡ | Translocation |
|---|---|---|---|
| HopAF1 | 0.987 | + | + |
| 2073 | 0.987 | – | – |
| HopAY1 | 0.986 | + | + |
| HsvG | 0.986 | + | + |
| HopV1 | 0.985 | + | – |
| HopX2 | 0.984 | + | + |
| HopR1 | 0.983 | – | – |
| 2097 | 0.983 | – | – |
| HrpK | 0.98 | + | + |
| HsvB | 0.97 | + | + |
| HopD1 | 0.946 | + | + |
| HopAK1 | 0.94 | + | – |
| PthG | 0.90 | + | + |
| HrpN | 0.858 | + | – |
| 2878 § | 0.843 | – | – |
| 3721 § | 0.842 | – | – |
| 2591 § | 0.822 | – | – |
*Translocation was performed as in Table 2.
†Prediction score was determined by the machine‐learning approach.
‡Presence of hrp box downstream to the open reading frame (ORF).
§Located on a chromosomal contig.
The inventories of the T3Es in Pab and Pag
Based on this and previous studies, and as summarized in Table 4 and Fig. S2 (see Supporting Information), there are eight functional T3Es and two harpins in Pab, and nine functional T3Es and two harpins in Pag. Four functional effectors (i.e. PseB, HopAY1, HopAF1 and HrpK) were uncovered in P. agglomerans during this study, whereas the remaining effectors have been identified previously (Barash and Manulis‐Sasson, 2007). Effectors present in both pathovars showed high identity at the amino acid level (75%–100%). HsvB, HsvG and PthG have been reported previously as unique P. agglomerans T3Es (Ezra et al., 2000; Nissan et al., 2006) and homologous proteins have not been reported as T3Es in any other bacteria. HopV1 and HopR1 which, in other bacteria, were defined as T3Es were present in both pathovars, but were not translocated, presumably as a result of modification in their N‐terminal sequences, which constitute the T3E translocation signal (Buttner and He, 2009). PthG and HopD1 appeared to be specific to Pag because only partial sequences encoding these effectors were found in Pab. Conversely, PseB is a novel T3E that is exclusively present in Pab and was submitted to the GenBank database under the accession number KX 083348. With the exception of HsvG, HsvB, PthG and PseB, which were unique to P. agglomerans, the remaining effectors were homologues (66%–97% identity) of T3Es in various Pseudomonas syringae pathovars or Erwinia amylovora, and could have been acquired by horizontal gene transfer. The two harpins HopAK1 and HrpN were present in both pathovars and, as expected, were not translocated.
Table 4.
The inventory of type III effectors (T3Es) and harpins of P. agglomerans pvs. betae and gypsophilae.
| Virulence of Pab4188* | Virulence of Pag824‐1* | ||||
|---|---|---|---|---|---|
| ORF | Presence in Pab | Presence in Pag | Gypsophila | Beet | Gypsophila |
| T3Es | |||||
| HsvG † | + | + | H | None | H |
| HsvB † | + | + | None | H | None |
| PthG | + (truncated) | + | None | None ‡ | H |
| PseB | + | ‐ | H | M | None ‡ |
| DspA/E | + | + | H | M | H |
| HopAY1 | + | + | M | M | L |
| HopD1 | + (truncated) | + | None ‡ | None ‡ | L |
| HopX2 | + | + | M | M | L |
| HopAF1 | + | + | M | M | L |
| HrpK | + | + | M | M | L |
| Harpins | |||||
| HopAK1 | + | + | None | None | None |
| HrpN | + | + | None | None | None |
*Virulence of the corresponding type III effector mutant expressed as the percentage reduction in gall fresh weight relative to the wild‐type. Degrees of contribution to virulence are as follows: high (H), above 70% reduction in gall size; medium (M), between 21% and 69% reduction; low (L), below 20% reduction; None, no effect on virulence. Results are the average of three independent experiments. Average gall size for Pab4188 or Pag824‐1 in gypsophila is 202 ± 12 mg, and for Pab4188 in beet is 171 ± 10 mg.
†Gall reduction in mutants was 90% or above.
‡Partial sequence (PthG and HopD1 in Pab) or completely deleted (PseB in Pag).
To quantitatively assess the contribution of individual effectors to Pag and Pab pathogenicity, each effector gene was individually mutagenized in Pag and/or Pab, and the resulting mutants were used to infect beet cubes and/or gypsophila cuttings. Gall formation was monitored in the infected tissues and the percentage reduction in gall fresh weight was measured relative to the wild‐type. The effectors that showed the most prominent contribution to virulence were HsvB in the beet pathovar and HsvG in the gypsophila pathovar, as their mutants could cause above 90% reduction in gall development (Table 4). These two paralogous effectors have been reported previously as host‐specific transcription factors (TFs) essential for pathogenicity (Nissan et al., 2006). Next in rank were PseB of the beet pathovar and PthG of the gypsophila pathovar, whose mutagenesis caused 75%–90% reduction in gall formation in gypsophila, although the effect of PseB on beet was significantly lower than on gypsophila. DspA/E was also rated high and medium in the gypsophila and beet pathovars, respectively (Table 4). The third group of effectors included HopAY1, HopX2, HopAF1 and HrpK, whose mutagenesis caused 40%–75% reduction in gall development. HopD1, which was partially deleted in the betae pathovar, exhibited a slight effect on gall formation in gypsophila by the gypsophilae pathovar. Mutation in the harpins HopAK1 and HrpN did not affect pathogenicity.
Discussion
Most Gram‐negative phytopathogenic bacteria translocate large repertoires of T3Es into the host cell to defeat plant defence and mediate pathogenesis (Gohre and Robatzek, 2008). The present study indicates that Pab and Pag harbour only eight and nine functional effectors, respectively. In contrast, all other reported plant‐pathogenic bacteria possess numerous functional effectors. For example, Pseudomonas syringae pv. tomato (Pst) DC3000 translocates 28 T3Es into the plant cell (Kvitko et al., 2009), whereas Xanthomonas euvesicatoria translocates 30 (Hann et al., 2010; Teper et al., 2016). A large‐scale identification, annotation and phylogenetic analysis of T3Es from Ralstonia solanacearum species complex RSCC yielded a total of more than 94 T3Es (Genin and Denny, 2012; Peeters et al., 2013). These comparisons emphasize the high diversification of T3Es present in phytopathogenic bacteria. The development of a host defence and surveillance system imposes intense selective pressure on T3Es, which act as key components in bacterial virulence (Dodds, 2010; Jones and Dangl, 2006). The T3Es may enlarge the host range of a given bacterium by suppressing host defence reactions that are induced after the recognition of pathogen‐associated molecular patterns, resulting in effector‐triggered susceptibility. In other cases, individual T3Es may be specifically recognized by the host to trigger HR, resulting in an effector‐induced resistance. These events have led to the concept of a co‐evolutionary arms race that holds a central paradigm in the interpretation of pathogen–host interactions (Pitman et al., 2005; Starvinides et al., 2008). Accordingly, the reciprocal selective pressures imposed by host–pathogen interactions drive hosts to evolve mechanisms to detect and eliminate invading microbes, whilst leading to the selection of pathogen variants that can suppress or avoid these mechanisms. Consequently, the repertoire of T3E variants increases with time and seems to be proportional to the duration of host–pathogen interactions.
The finding that P. agglomerans pvs. betae and gypsophilae harbour only a few functional effectors that are sufficient to trigger galls in their respective hosts (Table 4) is in accordance with the hypothesis that these two pathovars have evolved recently (Barash and Manulis‐Sasson, 2009). Notably, no resistant cultivars of either gypsophila or beet to P. agglomerans that could provoke and enhance the evolution of new effectors have been reported. It is noteworthy that, of the 28 T3Es identified in Pst DC3000, a minimal repertoire of eight effectors could restore near wild‐type growth and symptoms in the model plant Nicotiana benthamiana, which became a host to this pathogen following deletion of the hopQ1‐1 effector gene (Cunnac et al., 2011). The latter study emphasizes the functional redundancy present in the pool of T3Es that evolved during the evolution of phytopathogenic bacteria (Alfano and Collmer, 2004).
The quantification of gall fresh weight following the infection of beet and gypsophila with P. agglomerans strains mutagenized in individual T3Es allowed the assessment of the contribution of each effector to virulence. The highest contribution was provided by HsvB in beet and HsvG in gypsophila, whose mutants exhibited a reduction in gall formation by more than 90% relative to wild‐type bacteria (Table 4). These two paralogous effectors have been reported previously as host‐specific TFs that are present in both pathovars (Nissan et al., 2006; Valinsky et al., 2002). The activation domain of HsvG has been reported to have two direct repeats (71 and 74 amino acids), whereas that of HsvB has only one repeat (Nissan et al., 2006). Exchanging the repeat domains between HsvG and HsvB resulted in a switch in host specificity (Nissan et al., 2006). A target gene of HsvG, namely HSVGT, was isolated from the host plant Gypsophila paniculata (Nissan et al., 2012). HSVGT encodes a predicted acidic protein that harbours characteristic motifs of eukaryotic TFs, such as a bipartite nuclear localization signal, a zinc finger and a leucine zipper. Further analysis has demonstrated that HsvG binds to the HSVGT promoter, indicating that HSVGT is a direct target of HsvG (Nissan et al., 2012). It can be hypothesized that HsvG targets host TFs to effectively repress the transcription of defence‐associated or other major plant genes that promote infection (Canonne and Rivas, 2012). It is also possible that HsvG may directly affect gall symptoms, similar to the TAL effector AvrBs3 of Xanthomonas campestris pv. euvesicatoria (Kay and Bonas, 2009). AvrBs3 binds directly to the promoter of UPA20, which encodes a master regulator TF of the basic helix–loop–helix class, and presumably causes cell enlargement through the activation of UPA7, an α‐expansin encoding gene. HsvB is a paralogous effector of HsvG that differs mainly in the absence of one of the two direct amino acid repeats of HsvG (Nissan et al., 2006). Similar to HsvG, it localizes to the plant nucleus, contains a helix–turn–helix‐rich region and activates transcription in yeast. Therefore, a similar mode of action to HsvG might be postulated for HsvB. The recent availability of the beet genome (Dohm, et al., 2014), as opposed to the absence of the gypsophila genome, should facilitate the characterization of the HsvB target genes in beet.
Mutations in PthG, PseB and DspA/E had a very strong effect on P. agglomerans‐induced gall formation (Table 4). PthG and PseB have not been reported as T3Es in any bacteria other than P. agglomerans and, like HsvG and HsvB, presumably evolved by a pathoadaptive mechanism (Sokurenko et al., 1999). PthG, which triggers HR on multiple beet species, has been characterized previously as a genus‐specific rather than cultivar‐specific avirulence factor (Ezra et al., 2004). Hypothetical homologous proteins of PthG are widespread among phytopathogenic bacteria, including Pseudomonas syringae, Xanthomonas sp. and Erwinia. When blasted by HHpred (Söding et al., 2005), a remote homology to Bartonella and Legionella effectors was detected, possibly as a result of the presence of a common FIC domain, which is involved in reversible adenylylation events. PthG shares 35% identity with the phosphocholine transferase AnkX of Legionella pneumophila which participates in phosphocholine transfer by the FIC domain (Campanacci et al., 2013). Transgenic tobacco plants (Nicotiana tabacum var. Samsun NN) with PthG show a two‐ to four‐fold increase in free indole acetic acid and a 1.5‐ to two‐fold increase in ethylene (Weinthal et al., 2010). In addition, various phenotypic features of transgenic lines, such as leaf deformation, dwarf stature, leaf vein branching, loss of apical dominance and other suggested pathological hormone imbalances, are presumably initiated by the increase in auxin and ethylene. PseB of the betae pathovar has no homologous genes in any other bacteria and its function is not yet known. DspA/E belongs to the AvrE family of T3Es and has been reported previously in the gall‐forming P. agglomerans (Mor et al., 2001). T3Es of the AvrE family are widespread in plant‐pathogenic bacteria and are found in the genera Pantoea, Dickeya and Pectobacterium (Bogdanove et al., 1998a). AvrE of Pantoea stewartii and Pseudomonas syringae has been shown recently to target protein phosphatase 2A (PP2A) complexes in susceptible hosts (Jin et al., 2016). Effectors of the AvrE family are generally encoded by genes adjacent to the T3SS cluster, which has led to the suggestion that they have been acquired by bacteria together with T3SS (Bogdanove et al., 1998b).
Mutants of HopAY1, HopX2 and HopAF1 exhibit a medial reduction in gall development. HopAY1 has been reported in Pseudomonas as an effector with cysteine‐type endopeptidase activity (Baltrus et al., 2011). HopX2 of the HopX family was first isolated from Pseudomonas syringae pv. maculicola (Guttman et al., 2002) and contains a putative catalytic triad characteristic of cysteine proteases (Nimchuk et al., 2007). HopAF1 has been detected in several sequenced strains of Pseudomonas syringae. Its protein is structurally related to bacterial deamidases and inhibits ethylene biosynthesis (Washington, 2013). HopD1 has been shown recently to target the Arabidopsis TF NTL9, resulting in the suppression of effector‐triggered immunity responses (Block et al., 2014). HrpK was identified as a putative type III translocator in Pst DC3000 (Petnicki‐Ocwieja et al., 2005). The observation that the homologous effectors acquired from other phytopathogenic bacteria were aimed at different plant targets is in accordance with the observed lack of functional redundancy in the gall‐forming P. agglomerans (Barash and Manulis‐Sasson, 2009).
The gall‐forming P. agglomerans serves as a unique model for the identification of the course of events that leads to the emergence of new plant bacterial pathogens. Pathogenicity was acquired through the evolution of a plasmid‐borne PAI that harbours the T3SS and all the T3Es required for gall development (Barash and Manulis‐Sasson, 2007; and present work). The discovery of the complete repertoires of T3Es in the two pathovars of P. agglomerans allows an interpretation of their relevance to pathovar specificity. As pointed out earlier, two groups of T3Es could be distinguished (Table 4): the first group includes unique effectors of P. agglomerans (i.e. HsvG, HsvB, PthG and PseB), which presumably were acquired by a pathoadaptive mechanism. The second group includes ORFs with homology to T3Es in other bacteria, which could have been acquired by horizontal gene transfer. The first group appears to endow pathovar specificity, namely HsvG and PthG induce galls by Pag on gypsophila and HR on beet, whereas HsvB, HsvG and PseB elicit galls by Pab on beet and gypsophila. It is noteworthy that the functional contribution of T3Es to host specificity is not necessarily related to their distribution between the two pathovars. Thus, HsvG and HsvB are present in both pathovars; however, HsvG is recognized only by gypsophila, whereas HsvB is recognized only by beet (Nissan et al., 2006). PthG incites galls on gypsophila, but elicits HR on beet, thus preventing development on this host (Ezra et al., 2000). PseB is exclusively located and functional in Pab, but is recognized by the two hosts. In contrast with the effectors that evolved by a pathoadaptive mechanism, those acquired by horizontal gene transfer are equally distributed and functional in both pathovars. An exception is HopD1, whose sequence is truncated in the gypsophilae pathovar and is functional only in the betae pathovar (Table 4).
Experimental Procedures
Bacterial strains, growth conditions and pathogenicity tests
The bacterial strains and plasmids used in this study are listed in Table S5 (see Supporting Information). Wild‐type and mutant strains of P. agglomerans were grown in Luria–Bertani (LB) broth or agar at 28 ºC, whereas Escherichia coli strains were cultured on the same medium at 37 ºC. Antibiotics were used at the following concentrations (µg/mL): ampicillin (Amp), 100; kanamycin (Km), 50; rifampicin (Rif), 150; spectinomycin (Spec), 50; tetracycline (Tet), 15. All strains were maintained as glycerol stocks at −80 ºC.
Pathogenicity tests on gypsophila were performed on cuttings of Gypsophila paniculata var. Golan (Danziger Ltd., Bet Dagan, Israel) essentially according to Lichter et al. (1995). After removal of an approximately 2‐mm section from the bottom of the stem, the cuttings (10 for each treatment) were inoculated by dipping into bacterial suspension of 106 cells/mL for 30 min and placed in vermiculite‐filled trays for symptom visualization. The glasshouse temperature was maintained at 22–25 ºC and high humidity was generated by computer‐controlled mist sprinklers that were activated every 20 min for 10 s. Pathogenicity was scored 15–20 days after inoculation. The degree of virulence was determined by removal of the galls from the infected cuttings and measurement of their fresh weight. Pathogenicity tests on table beet cubes were performed according to Ezra et al., (2000). Whole matured beets (Beta vulgaris, Egyptian Red Beet) were soaked in 1% hypochlorite for 10 min and then washed twice in sterile water. They were then cut into 0.5 × 0.7 × 0.7 cm cubes under sterile conditions and placed on sterile 1.5% water agar in a Petri dish. Inoculation was carried out with culture grown overnight on LB agar by puncturing the top of the cube and inserting the bacteria with a sterile toothpick. The dishes were incubated for 5 days at 28 ºC prior to testing for gall formation. Virulence was expressed by measuring the fresh weight of the removed galls.
DNA manipulation
Isolation of genomic or plasmid DNA from P. agglomerans and E. coli, cloning, ligation, transformation and other DNA manipulations were performed according to standard procedures (Ausubel et al., 1995) or as recommended by the supplier. The cloning vectors used in this study are listed in Table S5. DNA fragments were amplified by PCR with Tac (Super‐Therm polymerase JMR‐801; Roche, Mannheim, Germany). Primers were synthesized by Hylabs Laboratories (Rehovot, Israel) and are described in Table S6 (see Supporting Information). PCR was performed directly on bacterial colonies. Amplification was carried out in a Biometra thermocycler (Bio‐Rad Laboratories, Hercules, CA, USA), as described previously (Chalupowicz et al., 2008). Escherichia coli and P. agglomerans strains were transformed by electroporation with a Gene Pulser apparatus (Bio‐Rad Laboratories) according to the manufacturer's instructions. Triparental mating was performed with the E. coli helper plasmid pRK2073 (Specr) on LB agar plates, as described elsewhere (Ditta et al., 1980; Manulis et al., 1998). DNA sequencing was carried out in the Instrumentation and Services Center at Tel‐Aviv University.
DNA sequence homology searches were carried out with the National Center for Biotechnology Information blast programs blastn, blastx and PSI blast. The alignment of the amino acid sequences was carried out with the programs clustalX and genedoc. Restrictions maps, ORF searches and additional molecular analyses were carried out with Sci Ed Central for Windows 95, (Scientific and Educational Software, Cary, NC, USA).
Dot blot analysis
Bacterial cells of the tested strain were grown overnight in 3 mL of LB medium, followed by transfer of 0.6 mL of cell suspension into 3 mL of fresh LB medium, and grown further for approximately 4 h, until an optical density at 600 nm (OD600) of 0.5 was reached. A sample of 50 μL of bacterial cells was then mixed with 10 μL of 6 × sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) sample buffer (375 mm Tris‐HCl, pH 6.8, 6% SDS, 48% glycerol, 9% mercaptoethanol) and 5 μL were spotted onto a nitrocellulose strip which was allowed to dry at room temperature. The strips were incubated for 1 h at room temperature in 5% skimmed milk in TBST (10 mm Tris, pH 8.0, 150 mm NaCl, 0.5% Tween 20). Rabbit anti‐6 × His‐tag antibodies (1 : 10 000, abcam, San Francisco, CA, USA) were then added and incubated for 1 h. Membranes were washed three times for 10 min with TBST and incubated with a 1 : 8000 dilution of alkaline phosphatase‐conjugated anti‐rabbit antibodies (abcam) for 1 h. The blots were washed three times with TBST and developed using BCIP/NBT (5‐bromo‐4‐chloro‐3‐indolyl‐phosphate/nitro blue tetrazolium) phosphatase substrate (KPL, Gaithersburg, MD, USA) according to the manufacturer's protocol.
Generation of mutants
Mutations of the tested genes were constructed by gene disruption using a suicidal vector as described by Kalogeraki and Winans (1997). For the generation of a mutation in pseB, as an example, a 289‐bp internal fragment of pseB was amplified with primers PseBM‐F and PseBM‐R (Table S6), digested with KpnI and SacI, and cloned into pVIK165 (KmR) predigested with the same enzymes to obtain pVIK165::pseB (Table S5). The resulting plasmid, which carried the vegetative replication origin of plasmid R6K without its pir gene, was replicated in E. coli S17‐1/λpir. Mutation was verified by PCR with the primers PseBM_ConF and PseBM_ConR (Table S6), followed by digestion with KpnI and SacI. The KmR mutant was mobilized into Pab4188 (RifR) by triparental mating and selected on LB plates amended with Km and Rif. Mutation was verified again by employing PCR with the corresponding primers (Table S6). The latter procedure was employed for the generation of marker exchange mutants in all other effector candidates described in Table 4 and Fig. S3 (see Supporting Information), except HsvG, HsvB, PthG, DspA/E and HopD1 (formerly AvrPphD) in Pag, which have been reported previously (Ezra et al., 2000; Guo et al. 2002; Mor et al., 2001; Nissan et al., 2006). Mutants were complemented by triparental mating with the intact ORF of the disrupted gene using pVSP61 as a vector.
Genome assembly and ORF prediction
Raw reads were preprocessed by removing low‐quality start and end sequences using Trimmomatic‐0.3 (Bolger et al., 2014) with the following parameters: LEADING: 30 TRAILING: 30. Reads were then assembled using Velvet‐1.2.10 (Zerbino and Birney, 2008) with the following parameters: kmer: 97(Pab), 135(Pag); exp_cov: 450; ins_length:600; cov_cutoff=auto. ORF prediction was carried out on the assembled contigs using Prodigal (Hyatt et al., 2010) with default parameters. Genomic annotations were obtained by blasting each ORF against the genome of Pantoea anannatis (NC_013956).
Machine‐learning scheme
A machine‐learning approach was employed as described previously (Burstein et al., 2009; Lifshitz et al., 2014). Briefly, an ensemble classification algorithm consisting of four different classification algorithms was used. The algorithms that were used included: naïve Bayes (Langley et al., 1992), Bayesian networks (Heckerman et al., 1995), support vector machine (SVM) (Burges, 1998), and tutorial of support vector machines for pattern recognition (Vapnik, 1999) and random forest (Breiman, 2001). For each classifier, except for random forest, which performs feature selection internally, a ‘Wrapper’ procedure was applied for feature selection (Kohavi and John, 1997). The performance of each classifier was estimated by using the mean AUPRC over 10‐fold cross‐validation, as described in Lifshitz et al. (2014). The latter was calculated using AUC calculator 0.2 (Davis and Goadrich, 2006). The weight of each classifier in the final ensemble classifier was the score it received after being trained on the full training dataset. The final classification score was computed for each ORF as a weighted mean of its scores from the four different classifiers, according to their weights. For random forest, we used the Random Forest R package (Liaw and Matthew, 2002) based on the original Fortran implementation as described by Breiman (2001), and, for the other classifiers, the WEKA Java library was used with default parameters (Hall et al., 2009).
Databases used in machine learning
The bacterial protein database was created by loading the RefSeq database (ftp://ftp.ncbi.nlm.nih.gov/refseq/release/bacteria) and following down‐sampling by taking every 10th sequence. The plants protein database was downloaded from RefSeq (ftp://ftp.ncbi.nlm.nih.gov/refseq/release/plant/).
The positive dataset was built by blasting the predicted ORFs in the genomes of Pag and Pab against known effectors in Pag and Pab, respectively, as well as against known effectors in P. stewartii, P. ananatis and Pseudomonas syringae. ORFs that had a hit with an E‐value lower than 10e−50 were taken as the positive dataset. The negative dataset was built by taking all the ORFs with an E‐value of zero when blasting them against the genome of E. coli K‐12.
Feature extraction
For each predicted ORF in the genomes of Pag and Pab, 49 features were extracted and used for training of the machine‐learning algorithm. These features can be divided into five general groups.
(1) Homology features (six features). The predicted protein sequence of each ORF was blasted against tailored databases of plant proteins, bacterial proteins and effector proteins using blast‐p. Two features were extracted from each blast result: (i) the BIT score of the best hit in the database; and (ii) the number of hits with an E‐value below 0.01.
(2) Physical features (33 features). The length and GC content of each predicted ORF was measured. Another Boolean feature, indicating whether the predicted ORF is less than 100 nucleotides, was extracted. The frequency of each amino acid was calculated in the 25 amino acids which comprise the ORF N‐terminus. In addition, each ORF was analysed by predicting coiled‐coil motifs using the pipeline that was created in the Russell Laboratories according to Lupas et al. (1991). These predictions were made for the N‐terminal part of the ORF (first quarter of the predicted protein), the C‐terminal part (last quarter of the predicted protein) and the middle of the protein. The total length and number of predicted coiled‐coils were calculated for the entire ORF. The predicted ORFs were also analysed for transmembrane domains using HMMTOP (Tusnády and Simon, 2001). The analysis was carried out as in the coiled‐coils features on the N‐terminus, C‐terminus and middle of the protein. In addition, the total length and number of transmembrane domains were calculated for the entire protein.
(3) Genome organization (seven features). For each predicted ORF, the number of known effectors that reside in proximity to 5, 10, 15, 20, 25 and 30 ORFs upstream and downstream was measured. In addition, the distance to the closest ORF was measured by counting the number of predicted ORFs that reside between them.
(4) Amino acid similarity to effectors (one feature). The similarity of the amino acid composition of each ORF was compared with known effectors using a log‐likelihood score. The likelihood function for an effector was calculated by measuring the frequency of each amino acid among effectors and then multiplying according to the amino acid sequence of the ORF. As a background, the frequencies of amino acids among all ORFs were measured and multiplied according to the amino acid sequence of the ORF.
(5) Regulatory elements (two features) in 300 base pairs upstream to the start codon of each predicted ORF were searched for the presence of the hrp‐box bipartite motif described by Nissan et al. (2005). The motif was searched as an exact match, or allowing one mismatch in sequence or in the gap between the two parts of the motif.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Dot blot of type III effector (T3E) fusion constructs used for translocation assay. Expression of the fusion constructs was validated by dot blot using anti‐His antibodies (abcam, San Francisco, CA, USA). Pab4188: 1, HsvG; 2, HsvB; 3, HopAF1; 4, HopD1; 5, HopR1; 6, HopX2; 7, HopAY1; 8, 1611; 9, PthG; 10, PseB; 11, 1595; 12, HrpN; 13, HrpK; 14, HopV1; 15, 2716; 16, 2223; 17, 585; 18, 1337. Pag824‐1: 1, HopAF1; 2, 2073; 3, HopAY1; 4, HsvG; 5, HopV1; 6, HopX2; 7, HopR1; 8, 2097; 9, HrpK; 10, HsvB; 11, HopD1; 12, HopAK1; 13, PthG; 14, HrpN; 15, 2878; 16, 3721; 17, 2591.
Fig. S2 Reduction in gall formation by mutants of Pab4188 in beet or gypsophila (a) and by mutants of Pag824‐1 in gypsophila (b). Results are expressed as the percentage reduction in gall fresh weight relative to the wild‐type. Average gall size for Pab4188 or Pag824‐1 in gypsophila is 202 ± 12 mg, and for Pab4188 in beet is 171 ± 10 mg. Results are the average of three independent experiments.
Fig. S3 Confirmation of a mutation in type III effectors (T3Es) of Pab4188 (a) and Pag824‐1 (b) by polymerase chain reaction (PCR). Primers for each T3E are given in Table S6 (see Supporting Information). (a) C, wild‐type Pab4188; M, molecular weight marker 100–500‐bp ladder; 1, PseB; 2, HopAY1; 3, HopX2; 4, HopAF1; 5, HopV1; 6, HopR1; 7, HrpK; 8, HopAK1; 9, HopD1. (b) C, wild‐type Pag824‐1; M, molecular weight marker 100–500‐bp ladder; 1, HopAY1; 2, HopX2; 3, HopAF1; 4, HopV1; 5, HopR1; 6, HrpK; 7, HopAK1.
Table S1 Performance scores of the top 15 features in Pantoea agglomerans pv. betae (Pab).
Table S2 Performance scores of the top 15 features in Pantoea agglomerans pv. gypsophilae (Pag).
Table S3 Prediction scores of open reading frames (ORFs) in Pantoea agglomerans pv. betae (Pab).
Table S4 Prediction scores of open reading frames (ORFs) in Pantoea agglomerans pv. gypsophilae (Pag).
Table S5 Bacterial strains and plasmids used in this study.
Table S6 Sequence of primers used in this study.
Acknowledgements
This research was supported by the Israel Science Foundation under grant number 1404/13.
Contributor Information
Isaac Barash, Email: isaaci@post.tau.ac.il.
Tal Pupko, Email: talp@post.tau.ac.il.
References
- Alfano, J.R. and Collmer, A. (2004) Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42, 385–414. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M. , Brent, R. , Kingston, R.E. , Moore, D.D. , Seidman, J.G. , Smith, J.A. and Struhl, K. (eds) (1995) Current Protocols in Molecular Biology. New York: Wiley. [Google Scholar]
- Baltrus, D.A. , Nishimura, M.T. , Romanchuk, A. , Chang, J.H. , Mukhtar, M.S. , Cherkis, K.J. , Roach, J. , Grant, S.R. , Jones, C.D. and Dangl, J.F. (2011) Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathogens, 7, e1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltrus, D.A. , Nishimura, M.T. , Dougherty, K.M. , Biswas, S. , Mukhtar, M.S. , Vicente, J. , Holub, E.B. and Dangl, J.F. (2012) The molecular basis of host specialization in bean pathovars of Pseudomonas syringae . Mol. Plant–Microbe Interact. 25, 877–888. [DOI] [PubMed] [Google Scholar]
- Barash, I. and Manulis‐Sasson, S. (2007) Virulence mechanisms and host specificity of gall‐forming Pantoea agglomerans . Trends Microbiol. 15, 538–545. [DOI] [PubMed] [Google Scholar]
- Barash, I. and Manulis‐Sasson, S. (2009) Recent evolution of bacterial pathogens: the gall‐forming Pantoea agglomerans case. Annu. Rev. Phytopathol. 47, 133–152. [DOI] [PubMed] [Google Scholar]
- Block, A. , Toruno, T.Y. , Elowsky, C.G. , Zhang, C. , Steinbrenner, J. , Beynon, J. and Alfano, J.R. (2014) The Pseudomonas syringae type III effector HopD1 suppresses effector‐triggered immunity, localizes to the endoplasmic reticulum, and targets the Arabidopsis transcription factor NTL9. New Phytol. 201, 1358–1370. [DOI] [PubMed] [Google Scholar]
- Bogdanove, A. , Bauer, D.W. and Beer, S.V. (1998a) Erwinia amylovora secretes DspE, a pathogenicity factor and functional AvrE homolog through the Hrp (type III secretion) pathway. J. Bacteriol. 180, 2244–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanove, A.J. , Kim, J.F. , Wei, Z.M. , Kolchinsky, P. , Charkowski, A.O. , Conlin, A.K. , Collmer, A. and Beer, S.V. (1998b) Homology and functional similarity of an hrp‐linked pathogenicity locus, dspEF, of Erwinia amylovora and the avirulence locus avrE of Pseudomonas syringae pathovar tomato . Proc. Natl. Acad. Sci. USA, 95, 1325–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger, A.M. , Lohse, M. and Usadel, B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman, L. (2001) Random forest. Mach. Learn. 45, 5–32. [Google Scholar]
- Burges, C.J.C. (1998) A tutorial on support vector machines for pattern recognition. Data Min. Knowl. Discov. 2, 121–167. [Google Scholar]
- Burr, T.J. , Katz, B.H. , Abawi, G.S. and Crosier, D.C. (1991) Comparison of tumorigenic strains of Erwinia herbicola isolated from table beet with Erwinia herbicola pv. gypsophilae . Plant Dis. 75, 855–858. [Google Scholar]
- Burstein, D. , Zusman, T. , Degtyar, E. , Viner, R. , Segal, G. and Pupko, T. (2009) Genome‐scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog. 5, e1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein, D. , Satanower, S. , Simovitch, M. , Belnik, Y. , Zehavi, M. , Yerushalmi, G. , Ben‐Aroya, S. , Pupko, T. and Banin, F. (2015) Novel type III effectors in Pseudomonas aeruginosa . MBio, 6, e0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner, D. and He, S.Y. (2009) Type III protein secretion in plant pathogenic bacteria. Plant Physiol. 150, 1656–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanacci, V. , Mukherjee, S. , Roy, C.R. and Cherfils, J. (2013) Structure of the Legionella effector AnkX reveals the mechanism of phosphocholine transfer by the FIC domain. Embo J. 32, 1469–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canonne, J. and Rivas, S. (2012) Bacterial effectors target the plant cell nucleus to subvert host transcription. Plant Sig. Behav. 7, 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalupowicz, L. , Manulis‐Sasson, S. , Itkin, M. , Sacher, A. , Sessa, G. and Barash, I. (2008) Quorum sensing system affects gall development incited by Pantoea agglomerans pv. gypsophilae . Mol. Plant–Microbe Interact. 21, 1094–1105. [DOI] [PubMed] [Google Scholar]
- Choi, M.S. , Kim, W. , Lee, C. and Oh, C.S. (2013) Harpins, multifunctional proteins secreted by gram‐negative plant‐pathogenic bacteria. Mol. Plant–Microbe Interact. 26, 1115–1122. [DOI] [PubMed] [Google Scholar]
- Clark, E. , Manulis, S. , Ophir, Y. , Barash, I. and Gafni, Y. (1993) Cloning and characterization of iaaM and iaaH from Erwinia herbicola pathovar gypsophilae . Phytopathology, 83, 234–240. [Google Scholar]
- Collmer, A. , Lindeberg, M. , Petnicki‐Ocwieja, T. , Schneider, D.J. and Alfano, J.R. (2002) Genomic mining type III secretion system effectors in Pseudomonas syringae yields new picks for all TTSS prospectors. Trends Microbiol. 10, 462–469. [DOI] [PubMed] [Google Scholar]
- Cooksey, D.A. (1986) Galls of Gypsophila paniculata caused by Erwinia herbicola . Plant Dis. 70, 464–468. [Google Scholar]
- Cruz, A.T. , Cazacu, A.C. and Al, C.H. (2007) Pantoea agglomerans, a plant pathogen causing human disease. J. Clin. Microbiol. 45, 1989–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunnac, S. , Chakravarthy, S. , Kvitkoa, B.H. , Russell, A.B. , Gregory, B. , Martin, G.B. and Collmer, A. (2011) Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae. Proc . Natl. Acad. Sci. USA, 108, 2975–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva, A.C.R. , Ferro, J.A. , Reinach, F.C. , Farah, C.S. , Furlan, L.R. Quaggio, R.B. , Monteiro‐Vitorello, C.B. , Van Sluys, M.A. , Almeida, N.F. , Alves, L.M. , do Amaral, A.M. , Bertolini, M.C. , Camargo, L.E. , Camarotte, G. , Cannavan, F. , Cardozo, J. , Chambergo, F. , Ciapina, L.P. , Cicarelli, R.M. , Coutinho, L.L. , Cursino‐Santos, J.R. , El‐Dorry, H. , Faria, J.B. , Ferreira, A.J. , Ferreira, R.C. , Ferro, M.I. , Formighieri, E.F. , Franco, M.C. , Greggio, C.C. , Gruber, A. , Katsuyama, A.M. , Kishi, L.T. , Leite, R.P. , Lemos, E.G. , Lemos, M.V. , Locali, E.C. , Machado, M.A. , Madeira, A.M. , Martinez‐Rossi, N.M. , Martins, E.C. , Meidanis, J. , Menck, C.F. , Miyaki, C.Y. , Moon, D.H. , Moreira, L.M. , Novo, M.T. , Okura, V.K. , Oliveira, M.C. , Oliveira, V.R. , Pereira, H.A. , Rossi, A. , Sena, J.A. , Silva, C. , de Souza, R.F. , Spinola, L.A. , Takita, M.A. , Tamura, R.E. , Teixeira, E.C. , Tezza, R.I. , Trindade dos Santos, M. , Truffi, D. , Tsai, S.M. , White, F.F. , Setubal, J.C. and Kitajima, J.P. (2002) Comparison of genomes of two Xanthomonas pathogens with differing host specificities. Nature, 417, 453–459. [DOI] [PubMed] [Google Scholar]
- Davis, J. and Goadrich, M. (2006) The relationship between Precision‐Recall and ROC curves. In: Proceedings of the 23rd International Conference on Machine Learning, pp. 233–240. New York: Association for Computing Machinery.
- Ditta, G. , Stanfield, S. , Corbin, D. and Helinsky, D.R. (1980) Broad host range DNA cloning system for gram‐negative bacteria: construction of a gene bank of Rhizobium meliloti . Proc. Natl. Acad. Sci. USA, 77, 7343–7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N. (2010) Genome evolution in plant pathogens. Science, 330, 1486–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm, J.C. , Minoche, A.E. , Holtgräwe, D. , Capella‐Gutiérrez, S. , Zakrzewski, F. , Tafer, H. , Rupp, O. , Sörensen, T.R. , Stracke, R. , Reinhardt, R. , Goesmann, A. , Kraft, T. , Schulz, B. , Stadler, P.F. , Schmidt, T. , Gabaldón, T. , Lehrach, H. , Weisshaar, B. and Himmelbauer, H. (2014) The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature, 505, 546–549. [DOI] [PubMed] [Google Scholar]
- Dou, D.J.M. and Zhou, J.M. (2012) Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe, 12, 484–495. [DOI] [PubMed] [Google Scholar]
- Ezra, D. , Barash, I. , Valinsky, L. and Manulis, S. (2000) The dual function in virulence and host range restriction of a gene isolated from the pPATHEhg plasmid of Erwinia herbicola pv. gypsophilae . Mol. Plant–Microbe Interact. 13, 693–698. [DOI] [PubMed] [Google Scholar]
- Ezra, D. , Barash, I. , Weinthal, D.M. , Gaba, V. and Manulis, S. (2004) pthG from Pantoea agglomerans pv. gypsophilae encodes an avirulence effector that determines incompatibility in multiple beet species. Mol. Plant Pathol. 5, 105–114. [DOI] [PubMed] [Google Scholar]
- Genin, S. and Denny, T.P. (2012) Pathogenomics of the Ralstonia solanacearum species complex. Annu. Rev. Phytopathol. 50, 67–89. [DOI] [PubMed] [Google Scholar]
- Gohre, V. and Robatzek, S. (2008) Breaking the barriers: microbial effector molecules subvert plant immunity. Annu. Rev. Phytopathol. 46, 189–215. [DOI] [PubMed] [Google Scholar]
- Guo, M. , Manulis, S. , Mor, H. and Barash, I. (2002) The presence of diverse IS elements and an avrPphD homologue that acts as a virulence factor on the pathogenicity plasmid of Erwinia herbicola pv. gypsophilae . Mol. Plant–Microbe Interact. 15, 709–716. [DOI] [PubMed] [Google Scholar]
- Guttman, D.S. , Vinatzer, B.A. , Sarkar, S.F. , Ranall, M.V. , Kettler, G. and Greenberg, J.T. (2002) A functional screen for type III (Hrp) secretome of the plant pathogen Pseudomonas syringae . Science, 295, 1722–1726. [DOI] [PubMed] [Google Scholar]
- Hajri, A. , Brin, C. , Hunault, G. , Lardeux, F. , Lemaire, C. , Manceau, C. , Boureau, T. and Poussier, S. (2009) A “repertoire for repertoire” hypothesis: repertoires of type three effectors are candidate determinants of host specificity in Xanthomonas . PLoS One, 4, e6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, M. , Frank, E. , Holmes, G. , Pfahringer, B. , Reutemann, P. and Witten, I.H. (2009) The WEKA data mining software: an update. ACM SIGKDD Explor. 11, 10–18. [Google Scholar]
- Hann, D.R. , Gimenez‐Ibanez, S. and Rathjen, J.P. (2010) Bacterial virulence effectors and their activities. Curr. Opin. Plant Biol. 13, 388–393. [DOI] [PubMed] [Google Scholar]
- Heckerman, D. , Geiger, D. and Chickering, D. (1995) Learning Bayesian networks: the combination of knowledge and statistical data. Mach. Learn. 20, 197–243. [Google Scholar]
- Hyatt, D. , Chen, G.L. , LoCascio, P.F. , Land, M.L. , Larimer, F. and Hauser, L.J. (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics, 11, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W. , Jiang, B.L. , Xu, R.Q. , Huang, J.D. , We, H.Y. , Jiang, G.F. , Cen, W.J., Liu, J., Ge, Y.Y., Li, G.H., Su, L.L., Hang, X.H., Tang, D.J., Lu, G.T., Feng, J.X., He, Y.Q. and Tang, J.L. (2009) Identification of six type III effector genes with the PIP box in Xanthomonas campestris pv. campestris and five of them contribute individually to full pathogenicity. Mol. Plant–Microbe Interact. 22, 1401–1411. [DOI] [PubMed] [Google Scholar]
- Jin, L. , Ham, J.H. , Hage, R. , Zhao, W. , Soto‐Hernández, J. , Lee, S.Y. , Paek, S.M. , Kim, M.G. , Boone, C. , Coplin, D.L. and Mackey, D. (2016) Direct and indirect targeting of PP2A by conserved bacterial type‐III effector proteins. PLoS Pathog. 12, e1005609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kalogeraki, V. and Winans, S.C. (1997) Suicide plasmids containing promoterless reporter genes can simultaneously disrupt and create fusions to target genes of diverse bacteria. Gene, 188, 69–75. [DOI] [PubMed] [Google Scholar]
- Kay, S. and Bonas, U. (2009) How Xanthomonas type III effectors manipulate the host plant. Curr. Opin. Microbiol. 12, 37–43. [DOI] [PubMed] [Google Scholar]
- Kobayashi, D.Y. and Palumbo, J.D. (2000) Bacterial endophytes and their effects on plants and uses in agriculture In: Microbial Endophytes (Bacon C.W. and White J.F., Jr., eds), pp. 199–233. Basel: Marcel Dekker Inc. [Google Scholar]
- Kohavi, R. and John, G.H. (1997) Wrappers for feature subset selection. Artif. Intellig. 97, 273–324. [Google Scholar]
- Kvitko, B.H. , Park, D.H. , Velásquez, A.C. , Wei, C.F. , Russell, A.B. , Martin, G.B. , Schneider, D.J. and Collmer, A. (2009) Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PloS Pathog. 5, e1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley, P. , Iba, W. and Thompson, K. (1992) An analysis of Bayesian classifiers. In: Aaai‐92, Proceedings of the Tenth National Conference on Artificial Intelligence, pp. 223–238. AAAI Press, San Jose.
- Liaw, A. and Matthew, W. (2002) Classification and regression by randomForest. R News, 2, 18–22. [Google Scholar]
- Lichter, A. , Barash, I. , Valinsky, L. and Manulis, S. (1995) The genes involved in cytokinin biosynthesis in Erwinia herbicola pv. gypsophilae: characterization and role in gall formation. J. Bacteriol. 177, 4457–4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifshitz, Z. , Burstein, D. , Schwartz, K. , Shuman, H.A. , Pupko, T. and Segal, G. (2014) Identification of novel Coxiella burnetii Icm/Dot effectors and genetic analysis of their involvement in modulating a mitogen‐activated protein kinase pathway. Infect. Immun. 82, 3740–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, J.A. , Lee, D.H. , Kim, B.Y. and Heu, S. (2014) Draft genome sequence of Pantoea agglomerans R190, a producer of antibiotics against phytopathogens and foodborne pathogens. J. Biotechnol. 188, 7–8. [DOI] [PubMed] [Google Scholar]
- Lindeberg, M. , Stavrinides, J.H.C.J. , Alfano, J.R. , Collmer, A. , Dangl, J.L. , Greenberg, J.T. , Mansfield, J.W. and Guttman, D.S. (2005) Proposed guidelines for unified nomenclature and phylogenetic analysis of type III hop effector proteins in the plant pathogen Pseudomonas syringae . Mol. Plant–Microbe Interact. 18, 275–282. [DOI] [PubMed] [Google Scholar]
- Lindeberg, M. , Cunnac, S. and Collmer, A. (2009) The evolution of Pseudomonas syringe host specificity and type III effectors. Mol. Plant Pathol. 10, 767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeberg, M. , Cunnac, S. and Collmer, A. (2012) Pseudomonas syringae type III effector repertoires: last words in endless arguments. Trends Microbiol. 20, 199–208. [DOI] [PubMed] [Google Scholar]
- Lindgren, P.B. (1997) The role of hrp genes during plant–bacterial interactions. Annu. Rev. Phytopathol. 35, 129–152. [DOI] [PubMed] [Google Scholar]
- Lindow, S.E. and Brandl, M. (2003) Microbiology of the phyllosphere. Appl. Environ. Microbiol. 69, 1875–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas, A. , Van Dyke, M. and Stock, J. (1991) Predicting coiled coils from protein sequences. Science, 252, 1162–1164. [DOI] [PubMed] [Google Scholar]
- Manulis, S. and Barash, I. (2003) Pantoea agglomerans pvs. gypsophilae and betae, a recently evolved pathogen?. Mol. Plant Pathol. 4, 307–314. [DOI] [PubMed] [Google Scholar]
- Manulis, S. , Gafni, Y. , Clark, E. , Zutra, D. , Ophir, Y. and Barash, I. (1991) Identification of a plasmid DNA probe for detection of Erwinia herbicola pathogenic on Gypsophila paniculata . Phytopathology, 81, 54–57. [Google Scholar]
- Manulis, S. , Haviv‐Chesner, A. , Brandl, M.T. , Lindow, S.E. and Barash, I. (1998) Differential involvement of indole‐3‐acetic acid biosynthetic pathways in pathogenicity and epiphytic fitness of Erwinia herbicola pv. gypsophilae . Mol. Plant–Microbe Interact. 11, 634–642. [DOI] [PubMed] [Google Scholar]
- Matsuzawa, T. , Moria, K. , Kadowaki, T. , Shimada, M. , Tashiro, K. , Kuhara, S. , Inagawa, H. , Soma, G. and Takegawa, K. (2012) Genome sequence of Pantoea agglomerans strain IG1. J. Bacteriol. 194, 1258–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann, H.C. and Guttman, D.S. (2008) Evolution of the type III secretion system and its effectors in plantmicrobe interactions. New Phytologist, 177, 33–47. [DOI] [PubMed] [Google Scholar]
- Mor, H. , Manulis, S. , Zuc, M. , Nizan, R. , Coplin, D.L. and Barash, I. (2001) Genetic organization of the hrp gene cluster and dspAE/BF operon in Erwinia herbicola pv. gypsophilae . Mol. Plant–Microbe Interact. 14, 431–436. [DOI] [PubMed] [Google Scholar]
- Nimchuk, Z.L. , Fisher, E.J. , Desveaux, D. , Chang, J.H. and Dangl, J.L. (2007) The HopX (AvrPphE) family of Pseudomonas syringae type III effectors require a catalytic triad and a novel N‐terminal domain for function. Mol Plant–Microbe Interact. 20, 346–357. [DOI] [PubMed] [Google Scholar]
- Nissan, G. , Manulis, S. and Barash, I. (2003) Molecular determinants affecting activity of the virulence effectors, PthG and HsvG of Erwinia herbicola pv. gypsophilae In: 11th International Congress of Molecular Plant–Microbe Interactions, Petersburg, Russia, p. 152.
- Nissan, G. , Manulis, S. , Weinthal, D. , Sessa, G. and Barash, I. (2005) Analysis of promoters recognized by HrpL, an alternative sigma‐factor protein from Pantoea agglomerans pv. gypsophilae . Mol. Plant–Microbe Interact. 18, 634–643. [DOI] [PubMed] [Google Scholar]
- Nissan, G. , Manulis‐Sasson, S. , Weinthal, D.M. , Mor, H. , Sessa, G. and Barash, I. (2006) The type III effectors HsvG and HsvB of gall‐forming Pantoea determine host specificity and function as transcriptional activators. Mol. Microbiol. 61, 1118–1131. [DOI] [PubMed] [Google Scholar]
- Nissan, G. , Manulis‐Sasson, S. , Chalupowicz, L. , Teper, D. , Yeheskel, A. , Pasmanik‐Chor, M. , Sessa, G. and Barash, I. (2012) The type III effector HsvG of the gall‐forming Pantoea agglomerans mediates expression of the host gene HSVGT . Mol. Plant–Microbe Interact. 25, 231–240. [DOI] [PubMed] [Google Scholar]
- Nizan‐Koren, R. , Manulis, S. , Mor, H. , Iraki, N.M. and Barash, I. (2003) The regulatory cascade that activates the Hrp regulon in Erwinia herbicola pv. gypsophilae . Mol. Plant–Microbe Interact. 16, 249–260. [DOI] [PubMed] [Google Scholar]
- O'Brien, H.F. , Thakur, S. and Guttman, D.S. (2011) Evolution of plant pathogenesis in Pseudomonas syringe: a genomic perspective. Annu. Rev. Phytopathol. 49, 269–289. [DOI] [PubMed] [Google Scholar]
- Peeters, N. , Carrère, S. , Anisimova, M. , Plener, L. , Cazalé, A.C. and Genin, S. (2013) Repertoire, unified nomenclature and evolution of the Type III effector gene set in the Ralstonia solanacearum species complex. BMC Genomics, 14, 859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petnicki‐Ocwieja, T. , Schneider, D.J. , Tam, V.C. , Chancey, S.T. , Shan, L. , Jamir, Y. , Schechter, L.M. , Janes, M.D. , Buell, C.R. , Tang, X. , Collmer, A. and Alfano, J.R. (2002) Genomewide identification of proteins secreted by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA, 99, 7652–7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petnicki‐Ocwieja, T. , van Dijk, K. and Alfano, J.R. (2005) The hrpK operon of Pseudomonas syringae pv. tomato DC3000 encodes two proteins secreted by the type III (Hrp) protein secretion system: HopB1 and HrpK, a putative type III translocator. J. Bacteriol. 187, 649–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman, A.R. , Jackson, R.W. , Mansfield, J.W. , Kaitell, V. , Thwaites, R. and Arnold, D.L. (2005) Exposure to host resistance mechanisms drives evolution of bacterial virulence in plants. Curr. Biol. 15, 2230–2235. [DOI] [PubMed] [Google Scholar]
- Smith, D.D.N. , Kirzinger, M.W.B. and Stavrinides, J. (2013) Draft genome sequence of the antibiotic‐producing cystic fibrosis isolate Pantoea agglomerans Tx10. Genome, 1, e00904–e00913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söding, J. , Biegert, A. and Lupas, A.N. (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokurenko, E.V. , Hasty, D.L. and Dykhuizen, D.E. (1999) Pathoadaptive mutations: gene loss and variation in bacterial pathogens. Trends Microbiol. 7, 191–195. [DOI] [PubMed] [Google Scholar]
- Starvinides, J. , McCann, H.C. and Guttman, D.S. (2008) Host–pathogen interplay and the evolution of bacterial effectors. Cell. Microbiol. 10, 285–292. [DOI] [PubMed] [Google Scholar]
- Teper, D. , Burstein, D. , Salomon, D. , Gershovitz, M. , Pupko, T. and Sessa, G. (2016) Identification of novel Xanthomonas euvesicatoria type III effector proteins by a machine‐learning approach. Mol. Plant Pathol. 17, 398–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnády, G.E. and Simon, I. (2001) The HMMTOP transmembrane topology prediction server. Bioinformatics, 17, 849–850. [DOI] [PubMed] [Google Scholar]
- Valinsky, L. , Barash, I. , Chalupowicz, L. , Ezra, D. and Manulis, S. (2002) Regulation of hsvG, a host specific virulence gene from Erwinia herbicola pv. gypsophilae . Physiol. Mol. Plant Pathol. 60, 19–29. [Google Scholar]
- Vapnik, V. (1999) The Nature of Statistical Learning Theory. New York: Springer. [Google Scholar]
- Washington, E.J. (2013) Charaterization of the role of Pseudomonas syringe type III effector HopAF1 in virulence. PhD dissertation, North Carolina at Chapel Hill, NC, USA.
- Weinthal, D.M. , Barash, I. , Panijel, M. , Valinsky, L. , Gaba, V. and Manulis‐Sasson, S. (2007) Distribution and replication of the pathogenicity plasmid pPATH in diverse populations of the gall‐forming Pantoea agglomerans . Appl. Environ. Microbiol. 73, 7552–7561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinthal, D.M. , Yablonski, S. , Singer, S. , Barash, I. , Manulis‐Sasson, S. and Gaba, V. (2010) The type III effector PthG of Pantoea agglomerans pv. gypsophilae modifies host plant responses to auxin, cytokinin and light. Eur. J. Plant Pathol. 128, 289–302. [Google Scholar]
- Zerbino, D.R. and Birney, E. (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Dot blot of type III effector (T3E) fusion constructs used for translocation assay. Expression of the fusion constructs was validated by dot blot using anti‐His antibodies (abcam, San Francisco, CA, USA). Pab4188: 1, HsvG; 2, HsvB; 3, HopAF1; 4, HopD1; 5, HopR1; 6, HopX2; 7, HopAY1; 8, 1611; 9, PthG; 10, PseB; 11, 1595; 12, HrpN; 13, HrpK; 14, HopV1; 15, 2716; 16, 2223; 17, 585; 18, 1337. Pag824‐1: 1, HopAF1; 2, 2073; 3, HopAY1; 4, HsvG; 5, HopV1; 6, HopX2; 7, HopR1; 8, 2097; 9, HrpK; 10, HsvB; 11, HopD1; 12, HopAK1; 13, PthG; 14, HrpN; 15, 2878; 16, 3721; 17, 2591.
Fig. S2 Reduction in gall formation by mutants of Pab4188 in beet or gypsophila (a) and by mutants of Pag824‐1 in gypsophila (b). Results are expressed as the percentage reduction in gall fresh weight relative to the wild‐type. Average gall size for Pab4188 or Pag824‐1 in gypsophila is 202 ± 12 mg, and for Pab4188 in beet is 171 ± 10 mg. Results are the average of three independent experiments.
Fig. S3 Confirmation of a mutation in type III effectors (T3Es) of Pab4188 (a) and Pag824‐1 (b) by polymerase chain reaction (PCR). Primers for each T3E are given in Table S6 (see Supporting Information). (a) C, wild‐type Pab4188; M, molecular weight marker 100–500‐bp ladder; 1, PseB; 2, HopAY1; 3, HopX2; 4, HopAF1; 5, HopV1; 6, HopR1; 7, HrpK; 8, HopAK1; 9, HopD1. (b) C, wild‐type Pag824‐1; M, molecular weight marker 100–500‐bp ladder; 1, HopAY1; 2, HopX2; 3, HopAF1; 4, HopV1; 5, HopR1; 6, HrpK; 7, HopAK1.
Table S1 Performance scores of the top 15 features in Pantoea agglomerans pv. betae (Pab).
Table S2 Performance scores of the top 15 features in Pantoea agglomerans pv. gypsophilae (Pag).
Table S3 Prediction scores of open reading frames (ORFs) in Pantoea agglomerans pv. betae (Pab).
Table S4 Prediction scores of open reading frames (ORFs) in Pantoea agglomerans pv. gypsophilae (Pag).
Table S5 Bacterial strains and plasmids used in this study.
Table S6 Sequence of primers used in this study.


