Summary
Oxylipins are a newly emerging group of signals that serve defence roles or promote virulence. To identify specific host and fungal genes and oxylipins governing the interactions between maize and Fusarium verticillioides, maize wild‐type and lipoxygenase3 (lox3) mutant were inoculated with either F. verticillioides wild‐type or linoleate‐diol‐synthase 1‐deleted mutant (ΔFvlds1D). The results showed that lox3 mutants were more resistant to F. verticillioides. The reduced colonization on lox3 was associated with reduced fumonisin production and with a stronger and earlier induction of ZmLOX4, ZmLOX5 and ZmLOX12. In addition to the reported defence function of ZmLOX12, we showed that lox4 and lox5 mutants were more susceptible to F. verticillioides and possessed decreased jasmonate levels during infection, suggesting that these genes are essential for jasmonic acid (JA)‐mediated defence. Oxylipin profiling revealed a dramatic reduction in fungal linoleate diol synthase 1 (LDS1)‐derived oxylipins, especially 8‐HpODE (8‐hydroperoxyoctadecenoic acid), in infected lox3 kernels, indicating the importance of this molecule in virulence. Collectively, we make the following conclusions: (1) LOX3 is a major susceptibility factor induced by fungal LDS1‐derived oxylipins to suppress JA‐stimulating 9‐LOXs; (2) LOX3‐mediated signalling promotes the biosynthesis of virulence‐promoting oxylipins in the fungus; and (3) both fungal LDS1‐ and host LOX3‐produced oxylipins are essential for the normal infection and colonization processes of maize seed by F. verticillioides.
Keywords: Fusarium, jasmonic acid, linoleate diol synthase, maize, oxylipin cross‐talk, susceptibility genes
Introduction
Seed contamination by mycotoxigenic fungi threatens both human food and animal feed. Fusarium verticillioides (Sacc.) Nirenberg (teleomorph, Gibberella moniliformis Wineland) causes severe stalk and ear rot of maize and is found in plant debris in nearly all maize fields during harvest (Battilani et al., 2008). In the southern regions of Europe, F. verticillioides is the prevailing species in maize fields (Folcher et al., 2009; Logrieco et al., 2002) and can colonize diverse plant organs at all developmental stages. Symptomless infection, producing no visible damage, can exist systemically and causes the presence of the fungus to be overlooked (Battilani et al., 2003). Recently, infection of maize by F. verticillioides has gained scrutiny because the fungus produces a variety of toxic secondary metabolites, including fumonisins, bikaverin, fusaric acid, fusarin C and moniliformin (Bacon et al., 2004; Nelson et al., 1993). Fumonisins possess carcinogenic properties (Gelderblom et al., 1988), and are included in group 2B of toxins (possibly carcinogenic to humans) by the International Agency for Research on Cancer (IARC) (International Agency for Research on Cancer, 2002).
An understanding of the molecular signalling occurring during plant–pathogen interactions paves the way for the creation and utilization of better forms of disease resistance in staple crops. Recent studies have demonstrated the role of a subset of oxidized lipid molecules, collectively termed oxylipins, produced by the plant and the pathogen, proposed to serve as signals in plant–pathogen ecosystems (Borrego and Kolomiets, 2016; Christensen and Kolomiets, 2011; Gao and Kolomiets, 2009; Maschietto et al., 2015; Reverberi et al., 2010; Scarpari et al., 2014; Tsitsigiannis and Keller, 2007). These molecules are primarily produced via the oxidation of polyunsaturated fatty acids (PUFAs) through the action of dioxygenases, lipoxygenases (LOXs) in plants and fungi, and linoleate diol synthases (LDSs) in fungi (Andreou et al., 2009; Brodhun and Feussner, 2011; Camera et al., 2004; Feussner and Wasternack, 2002; Mosblech et al., 2009). Plant‐derived oxylipins can mimic the physiological role of endogenous fungal oxylipins that regulate growth, development, sporogenesis and mycotoxin biosynthesis, favouring or inhibiting these processes. In particular, plant oxylipins can modulate the production of conidia (Brodhagen and Keller, 2006; Tsitsigiannis and Keller, 2006) and mycotoxins (Burow et al., 1997; Gao et al., 2009; Reverberi et al., 2010; Roze et al., 2007), regulate the ratio of sexual/asexual spores (Tsitsigiannis et al., 2004a, 2004b) and be used for quorum sensing (Affeldt et al., 2012).
Although the functional and physiological characterization of LOXs was first addressed in dicots, only recently have studies aimed to unravel the role of oxylipins in monocot–pathogen interactions with the use of maize mutants (Christensen et al., 2014; Gao et al., 2007) and F. verticillioides mutants (Scala et al., 2014). The analysis of the maize knock‐out lox3 mutant provided convincing genetic evidence that the functional host LOX3 isoform and its 9‐oxylipin products are required for normal biosynthesis of fumonisin B1 and conidiation by F. verticillioides (Gao et al., 2007). Interestingly, susceptible hybrid varieties also produced the highest levels of the 9‐LOX product, 9‐hydroxyoctadecenoic acid (9‐HODE) (Dall'Asta et al., 2015), and its fatty acid precursor, linoleic acid (Dall'Asta et al., 2012). These results indicated that host‐derived oxylipins act as signals to regulate fungal secondary metabolism and reproduction.
The role of F. verticillioides‐derived oxylipins in endogenous processes was established by the inactivation of the LDS1 gene (LDS1), where the deletion mutant (hereafter referred to as ΔFvlds1D) was perturbed in development, secondary metabolism and virulence (Scala et al., 2014). It was uncovered that LDS1‐derived oxylipins act as negative regulators of fumonisin synthesis. Unexpectedly, the results suggested that LDS1‐dependent oxylipins, 8,13‐dihydroxyoctadecenoic acid (8,13‐diHODE) and 8‐hydroperoxyoctadecenoic acid (8‐HpODE) (the oxylipins most perturbed in ΔFvlds1D), may act as histone‐modifying compounds. Notably, the lack of these oxylipins in the ΔFvlds1D mutant correlated with an increase in the acetylation of H4 in the promoter of the fumonisin biosynthesis gene FUM1 (Scala et al., 2014). Thus, oxylipins may act as signals to reprogram gene expression by affecting promoter availability for transcription factors, as suggested in mammalian systems (Ravindra et al., 2012).
In this study, kernel fungal colonization bioassays developed by Christensen et al. (2012) were performed on the maize B73 inbred line and its near‐isogenic lox3 mutant (Gao et al., 2007) inoculated with either F. verticillioides wild‐type (hereafter referred to as FvWT) or ΔFvlds1D mutant to elucidate oxylipin‐mediated cross‐talk between maize and F. verticillioides. Through an integrated molecular and lipidomic approach, this study revealed the relative contributions of both host and fungal oxylipins to the outcome of the plant–pathogen interaction. The biosynthesis of selected oxylipins is presented in Fig. S1 (see Supporting Information). Fungal pathogenicity processes were monitored by measurement of the fungal biomass through the accumulation of the fungal‐specific membrane lipid, ergosterol, for colonization, enumeration of conidia and fumonisin accumulation. The expression of several selected LOX genes of maize and fungal fatty acid dioxygenases, including a LOX and two LDS genes, and extensive oxylipin profiling revealed that oxylipins of both plant and fungal origin coordinately control the fate of host–pathogen interaction by acting as defence‐ or virulence‐response modifiers. Functional characterization of the knock‐out mutants of ZmLOX4 and ZmLOX5 revealed that these two closely related 9‐LOX genes in maize are required for jasmonate‐mediated defence against F. verticillioides.
Results
Fusarium verticillioides colonization progresses linearly during the early stages of disease while the infected maize seed is alive, after which sporulation and mycotoxin biosynthesis saturate at advanced stages
This study tested the hypothesis that oxylipin cross‐talk is a mechanism governing the interaction between maize seed and F. verticillioides. To explore this interaction, it was necessary to understand the dynamics of disease development and to identify the time frame after infection when pathogenic processes are relevant. It was also crucial to identify the timing during which the host remains viable as this study relied on two interacting organisms for cross‐talk to occur. Previous work has uncovered that seed colonization by F. verticillioides varies dramatically when cultured on living kernels vs. dead kernels (Mukherjee et al., 2011).
To understand the dynamics of F. verticillioides disease progression leading to complete seed maceration, a kernel biological assay was performed on B73 wild‐type (WT) maize seeds. Colonization, conidiation, fumonisin accumulation and gene expression were measured at 3, 6, 9, 12 and 15 days post‐inoculation (dpi). Figure 1 shows that colonization, as measured by the content of the fungus‐specific membrane lipid, ergosterol, increased steadily from 3 to 9 dpi, and decreased at 12 and 15 dpi. Levels of conidia and fumonisin accumulation increased dramatically between 6 and 9 dpi, before peaking at 12 dpi and holding steady for the remainder of the time course. When sporulation and fumonisin accumulation were normalized by fungal biomass, it was observed that, per fungal unit, 12 and 15 dpi showed the greatest production of conidia and fumonisin, followed by 9 dpi. Gene expression analysis was unable to detect host transcripts at 12 or 15 dpi of F. verticillioides‐infected kernels compared with mock‐treated kernels at the same time points (data not shown), suggesting that seed was completely macerated and no longer viable, precluding any analyses of host–pathogen interactions at these time points. Based on these results, 6 and 9 dpi were selected for subsequent experimentation as time points representative of early and late stages of disease development, whilst maintaining the interaction between viable plant and fungus.
Figure 1.
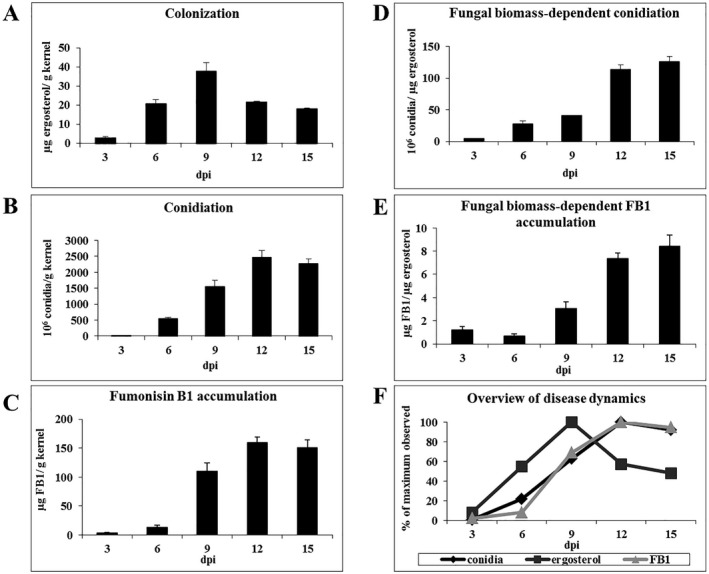
Dynamics of disease progression of Fusarium verticillioides (FvWT)‐infected B73 maize kernels. (A) Colonization. (B) Conidiation. (C) Fumonisin B1 accumulation. (D) Fungal biomass‐dependent conidiation. (E) Fungal biomass‐dependent fumonisin B1 accumulation. (F) Overview of disease progression. These factors were determined for B73 seed infected by the FvWT strain over a time course of 15 days post‐inoculation (dpi). Error bars indicate ± standard error (SE).
Oxylipin biosynthetic enzymes from both the seed and F. verticillioides govern the levels of colonization, fumonisin production and sporulation
Fungal parameters of colonization, fumonisin accumulation and conidia production were monitored (Fig. 2). At 6 and 9 dpi, both B73 and lox3 mutant kernels supported greater colonization of the FvWT strain compared with the ΔFvlds1D fungal mutant, as demonstrated by an increased amount of ergosterol (Fig. 2A). This was more evident at 9 dpi, where B73 kernels inoculated with FvWT showed the highest content of ergosterol compared with the other host–pathogen combinations. This finding confirmed the results of the previous study showing that a functional LDS1 gene is essential for fungal virulence (Scala et al., 2014). Interestingly, lox3 mutants were more resistant to colonization by FvWT at 9 dpi, as evidenced by the three‐fold reduced fungal biomass compared with the B73 line. This result suggests that LOX3‐derived 9‐oxylipins facilitate virulence by this pathogen. Furthermore, the lack of functional LOX3 in the host and the resulting increased resistance did not alter the reduced virulence of the ΔFvlds1D fungal mutant, which remained equally low regardless of the host genotype.
Figure 2.
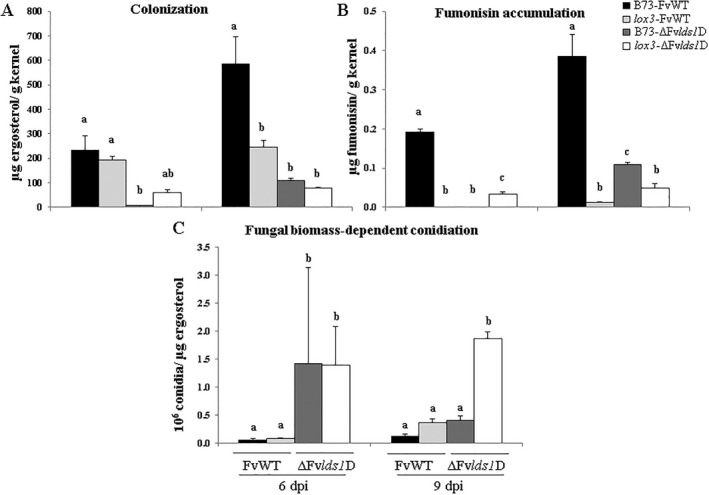
Colonization (based on ergosterol) (A), fumonisin accumulation (B) and conidiation as a function of fungal biomass as measured through ergosterol (C) on wild‐type (B73) and lox3 mutant maize kernels inoculated with wild‐type (FvWT) and oxylipin‐deficient (ΔFvlds1D) Fusarium verticillioides strains at 6 and 9 days post‐inoculation (dpi). Error bars indicate ± standard error (SE). The same letters above the histograms denote insignificant differences between means of the treatments within the same time point, resulting from Tukey's honestly significant difference test (P < 0.05).
Greater colonization of the B73 kernels by FvWT was also associated with significantly increased fumonisin levels at both time points (0.19 and 0.38 µg/g kernel at 6 and 9 dpi, respectively) (Fig. 2B). The ΔFvlds1D fungal mutant colonized seed and produced fumonisin to a lower extent than the FvWT strain in both maize lines. As shown in Fig. 2B, very low levels of ergosterol and no measurable toxin were recorded at 6 dpi in B73 kernels. At 9 dpi, a slight increase in fumonisin was observed for the ΔFvlds1D mutant in both maize genotypes, with greater accumulation in the WT host compared with the lox3 mutant. Overall, the ability of the ΔFvlds1D mutant strain to colonize seed and produce fumonisin remained significantly impaired compared with the FvWT strain. Notably, LDS1‐derived oxylipins appeared to have a greater effect on the fungal ability to colonize WT kernels, as evidenced by a six‐fold reduced ΔFvlds1D fungal biomass at day 9 compared with only a four‐fold reduction in fumonisin. These results suggest that, although both host LOX3‐ and fungal LDS1‐derived oxylipins contribute positively to seed colonization, LOX3‐derived metabolites have a greater impact on fumonisin biosynthesis compared with LDS1‐derived products.
Unexpectedly, although B73 kernels inoculated with FvWT showed significantly higher levels of colonization and fumonisin production compared with the lox3 mutant inoculated with the same strain, the number of ΔFvlds1D conidia was notably higher on the lox3 mutant at 9 dpi (Fig. 2C). These results suggest that oxylipins produced by both LOX3 and LDS1 suppress conidiation, but promote colonization and fumonisin synthesis.
FvLDS1 induces ZmLOX3 expression to suppress defence‐related maize 9‐LOXs
The expression levels of four maize 9‐LOX genes, ZmLOX3, ZmLOX4, ZmLOX5 and ZmLOX12, and two dual positional‐specific LOX isoforms, ZmLOX1 and ZmLOX2, were measured by real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) in WT and lox3 mutant infected by FvWT or ΔFvlds1D mutant strain. The expression profiles are reported in Fig. 3A–F as the relative expression of inoculated kernels over mock‐inoculated kernels. Of the genes tested, ZmLOX3, ZmLOX4, ZmLOX5 and ZmLOX12 showed strong changes in gene expression. In WT seed, throughout the time course, ZmLOX3 was significantly up‐regulated only in the kernels infected by FvWT, whereas the ΔFvlds1D mutant strain triggered lower gene expression levels (Fig. 3C). Infection by both F. verticillioides strains (FvWT and ΔFvlds1D) modulated ZmLOX4 expression in a trend intriguingly similar to that of ZmLOX5 (Fig. 3D,E). Both ZmLOX4 and ZmLOX5 were up‐regulated in both maize genotypes after fungal inoculations with both fungal strains. In infected B73 kernels, ZmLOX4 and ZmLOX5 transcripts accumulated to the greatest levels on inoculation with FvWT at 9 dpi, showing expression values of 13.47 and 5.29, respectively (P < 0.05). In contrast with WT kernels, earlier and significantly greater induction was observed for both genes in lox3 mutant kernels in response to FvWT at 6 dpi (10.11‐ and 7.52‐fold induction for ZmLOX4 and ZmLOX5, respectively), followed by a marked decrease thereafter. Earlier and greater induction of these two closely related paralogues in lox3 suggests their relevance to increased resistance to F. verticillioides observed in this mutant.
Figure 3.
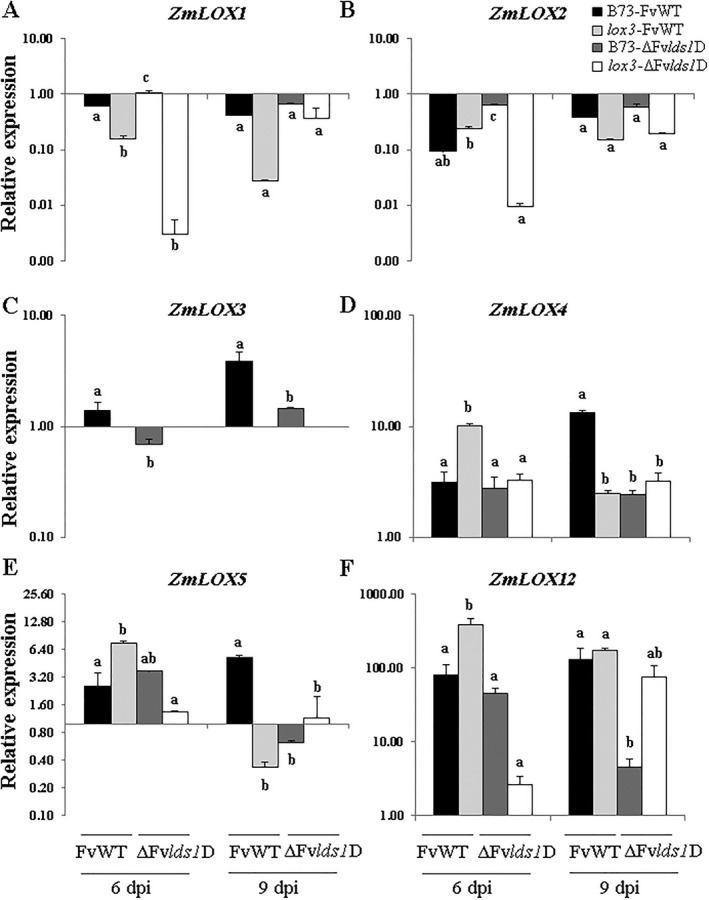
Relative expression of ZmLOX1 (A), ZmLOX2 (B), ZmLOX3 (C), ZmLOX4 (D), ZmLOX5 (E) and ZmLOX12 (F) in wild‐type (B73) and lox3 mutant maize kernels inoculated with wild‐type (FvWT) and oxylipin‐deficient (ΔFvlds1D) Fusarium verticillioides strains at 6 and 9 days post‐inoculation (dpi). Error bars indicate ± standard error (SE). The same letters above the histograms denote insignificant differences between means of the treatments within the same time point, resulting from Tukey's honestly significant difference test (P < 0.05).
ZmLOX12 showed the highest induction values with a remarkable 387‐fold induction of expression in response to infection by FvWT compared with mock‐treated kernels in the lox3 mutant kernels at 6 dpi. This value declined at 9 dpi, reaching similar levels of expression to B73 kernels inoculated with FvWT. Interestingly, ZmLOX12 also exhibited enhanced transcript levels in the lox3 mutant in response to ΔFvlds1D at 9 dpi, confirming the major role of this gene in maize resistance against F. verticillioides (Christensen et al., 2014).
In contrast with the induction of these 9‐LOXs, transcripts of ZmLOX1 and ZmLOX2 were decreased in both maize lines colonized by FvWT or ΔFvlds1D mutant strain (Fig. 3A,B), suggesting that these two closely related genes are not involved in seed defence against F. verticillioides. Unfortunately, to date, the function of ZmLOX1 and ZmLOX2 genes in the interactions with F. verticillioides is unknown, preventing any speculation.
Overall, LDS1‐produced oxylipins appear to modulate the expression of maize 9‐LOXs in this study. Although suppression of ZmLOX1 and ZmLOX2 by ΔFvlds1D is greater than that by FvWT, induction of the defence‐related ZmLOX4, ZmLOX5 and ZmLOX12 is not as pronounced in response to FvWT (Fig. 3). Taken together, the results support the notion of oxylipin‐mediated cross‐talk between fungi and their plant hosts (Borrego and Kolomiets, 2012; Christensen and Kolomiets, 2011) and, more specifically, that fungal oxylipins modulate the expression of plant oxylipin biosynthesis genes (Brodhagen and Keller, 2006; Reverberi et al., 2010).
ZmLOX4, ZmLOX5 and ZmLOX12 are required for normal jasmonic acid (JA) biosynthesis during maize defence against F. verticillioides seed infection
The overexpression of the two closely related paralogues, ZmLOX4 and ZmLOX5, in the infected lox3 mutant seed echoed the expression pattern observed for the known defence‐related gene, ZmLOX12 (Christensen et al., 2014), which prompted the hypothesis that ZmLOX4 and ZmLOX5 also contribute to seed defence against F. verticillioides.
To test this hypothesis, knock‐out mutants of ZmLOX4 and ZmLOX5 were infected with the FvWT strain, fungal growth was observed and spores were enumerated (Park, 2012). Two knock‐out alleles for ZmLOX4 (Fig. 4A,B) and three knock‐out alleles for ZmLOX5 (Fig. 4C,D) displayed dramatically increased fungal growth and conidia production compared with their respective near‐isogenic WT lines. These results provide genetic evidence that the ZmLOX4 and ZmLOX5 isoforms are essential for seed defence against F. verticillioides. Because ZmLOX4, ZmLOX5 and ZmLOX12 are overexpressed in lox3 kernels, we suggest that their enhanced induction in response to infection is one of the mechanisms explaining the increased resistance of the lox3 mutant.
Figure 4.
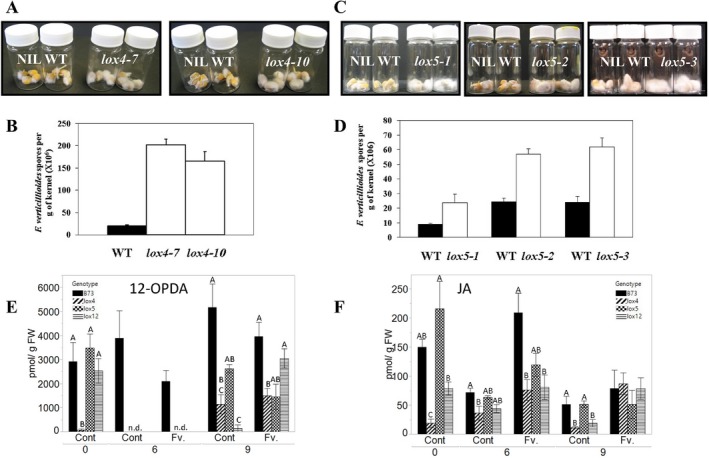
Production of conidia of Fusarium verticillioides (FvWT) was increased when grown on kernels of the lox4 and lox5 mutants compared to the wild type (B73). (A, C) Visual observation of fungal mycelial growth on lox4 and lox5 mutants, and near‐isogenic wild type kernels (NIL WT) 13 days post inoculation with F. verticillioides. The fungus easily colonized lox4 and lox5 mutant kernels more aggressively compared with wild type kernels. (B, D) Number of conidia produced on infected seed of maize lox4 and lox5 mutants, and NIL WTs, at 13 days post inoculation. (E, F) 12‐OPDA and JA content of lox4, lox5, and lox12 mutant kernels compared to WT. Vertical bars indicate ± se. Unconnected letters represent statistical differences within day post infection (DPI) by Tukey's HSD on log‐transformed data.
As 9‐LOXs, such as ZmLOX3, have been shown to negatively regulate JA biosynthesis in diverse maize organs (Gao et al., 2008, 2009), we tested the role of non‐JA‐producing 9‐LOXs, LOX4, LOX5 (Park et al., 2010) and LOX12 (Christensen et al., 2014), on JA production during F. verticillioides infection of maize seed; mutant lines were inoculated and jasmonates were profiled by liquid chromatography‐tandem mass spectrometry (LC‐MS/MS). Without a functional LOX4, seed did not accumulate normal levels of the JA precursor, 12‐oxo‐phytodienoic acid (12‐OPDA) or JA at basal levels. Remarkably, at 6 dpi, lox4, lox5 and lox12 mutants were unable to accumulate any detectable level of 12‐OPDA and less than half the normal level of JA compared with WT. Taken together, these data suggest that 9‐oxylipin products from LOX4, LOX5 and LOX12 are required to induce normal JA biosynthesis on F. verticillioides infection of maize seed.
Maize LOX3‐derived oxylipins modulate the expression of F. verticillioides oxylipin biosynthetic genes during infection
The relative expression of FvLOX1, FvLDS1, FvLDS2 and FvFAO genes was measured in B73 and lox3 mutant kernels at 6 and 9 dpi with FvWT and ΔFvlds1D strains (Fig. 5A–D). These genes are crucial in the synthesis of different oxylipins and in the pathogenic behaviour of F. verticillioides on Zea mays (Christensen and Kolomiets, 2011; Scala et al., 2013, 2014). Notably, FvLOX1 and FvLDS1 were the most strongly induced of the oxylipin biosynthesis genes, with significantly high transcript levels for the FvWT strain in lox3 mutant kernels at both days (Fig. 5A,B). Transcript levels of the FvLDS2 gene were similar in all host–pathogen combinations at 6 dpi. At 9 dpi, a significantly different expression pattern was measured for both fungi in both maize lines, with the highest induction for the ΔFvlds1D strain in B73 kernels, followed by FvWT in lox3 mutant kernels (Fig. 5C). It is also worth noting the significantly enhanced up‐regulation of the FvFAO gene observed in the FvWT strain in both maize lines at 6 dpi compared with the ΔFvlds1D strain, whereas gene expression was similarly affected at 9 dpi with both strains, as shown in Fig. 5D. Taken together, these results show that, during infection, expression levels of FvLOX1, FvLDS1 and FvLDS2 genes are modulated by host LOX genes.
Figure 5.
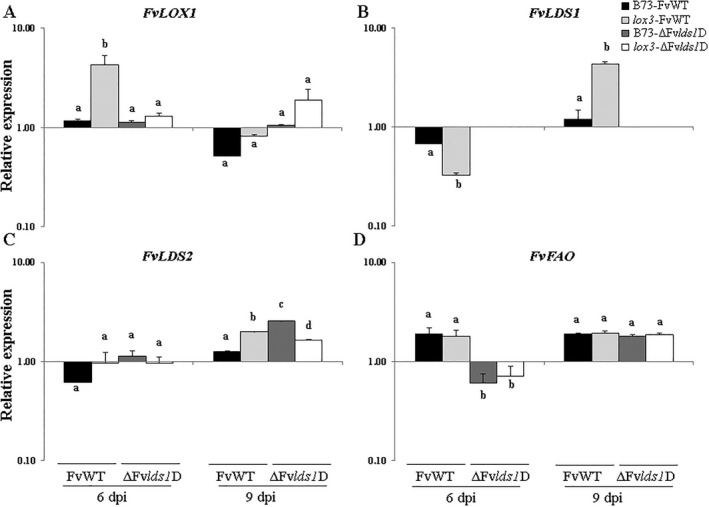
Relative expression of FvLOX1 (A), FvLDS1 (B), FvLDS2 (C) and FvFAO (D) genes in wild‐type (B73) and lox3 mutant maize kernels inoculated with wild‐type (FvWT) and oxylipin‐deficient (ΔFvlds1D) Fusarium verticillioides strains at 6 and 9 days post‐inoculation (dpi). Error bars indicate ± standard error (SE). The same letters above the histograms denote insignificant differences between means of the treatments within the same time point, resulting from Tukey's honestly significant difference test (P < 0.05).
Plant and fungal oxylipins transiently accumulate in infected kernels in a genotype‐dependent manner
Recent studies have shown that plants and fungi produce similar oxylipins, albeit through distinct biochemical pathways, whereas other oxylipins are unique to either kingdom (Christensen and Kolomiets, 2011). To identify the oxylipins altered on deletion of either the FvLDS1 or ZmLOX3 genes during seed colonization, fungal‐derived (Fig. 6A–F) and plant‐derived (Fig. 7A–F) oxylipins were quantified at 6 and 9 dpi. Overall, substantial fungal and host genotype‐dependent differences in oxylipin profiles were observed. With regard to fungal‐specific oxylipins, significant differences emerged amongst treatments and days of inoculation. Notably, at 6 dpi, the maize kernels (B73 as well as lox3) inoculated with the ΔFvlds1D mutant displayed a lower content of fungal oxylipins, suggesting a major role of FvLDS1 in the biosynthesis of 8‐HpODE and 8,13‐diHODE during the early stages of disease progression. Interestingly, both of these metabolites were increased at 9 dpi in a LOX3‐dependent manner, suggesting that LOX3‐mediated signalling is required to induce other fungal dioxygenases involved in 8‐HpODE and 8,13‐diHODE production during the later stages of disease development.
Figure 6.
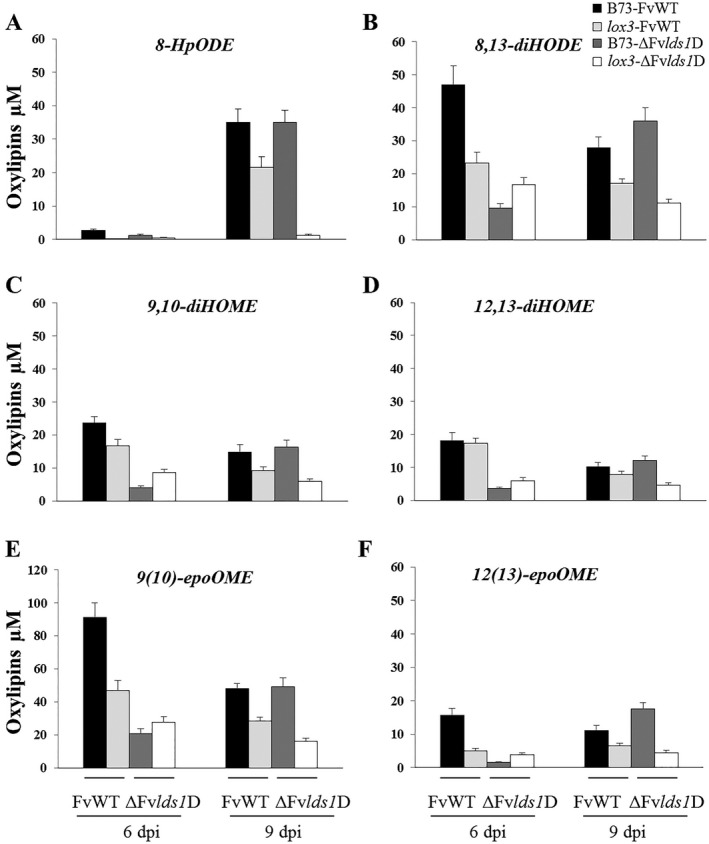
Multiple reaction monitoring (MRM) quantification of linoleate diol synthase (LDS)‐derived oxylipins [8‐hydroperoxyoctadecenoic acid, 8‐HpODE (A); 8,13‐dihydroxyoctadecenoic acid, 8,13‐diHODE (B); 9,10‐dihydroxymonoenoic acid, 9,10‐diHOME (C); 12,13‐dihydroxymonoenoic acid, 12,13‐diHOME (D); 9(10)‐epoxymonoenoic acid, 9(10)‐epoOME (E); 12(13)‐epoxymonoenoic acid, 12(13)‐epoOME (F)] in wild‐type (B73) and lox3 mutant maize kernels inoculated with wild‐type (FvWT) and oxylipin‐deficient (ΔFvlds1D) Fusarium verticillioides strains at 6 and 9 days post‐inoculation (dpi). Error bars indicate ± standard error (SE).
Figure 7.
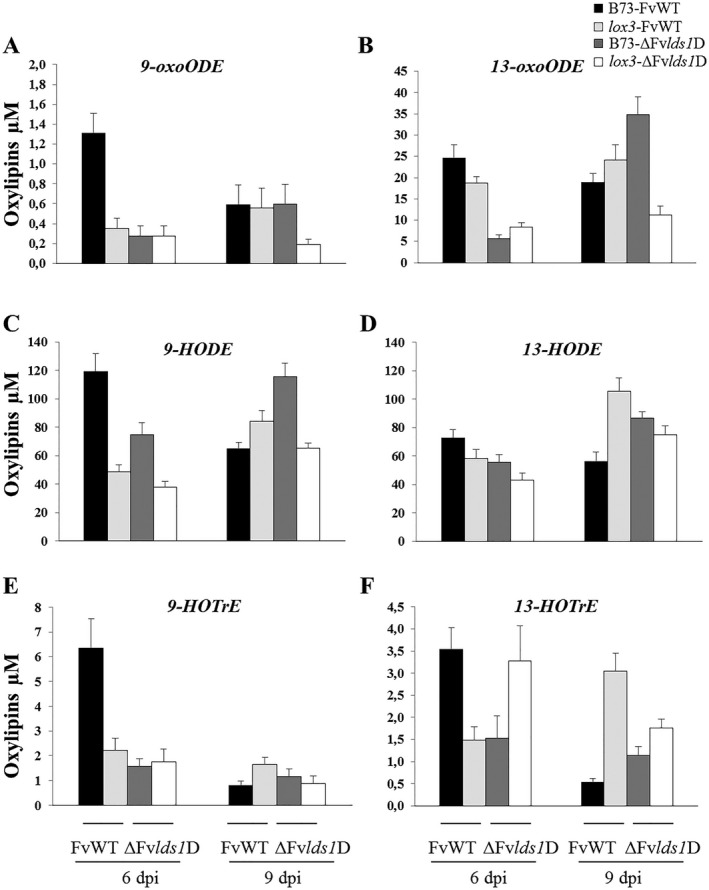
Multiple reaction monitoring (MRM) quantification of lipoxygenase (LOX)‐derived oxylipins [9‐oxo‐octadecenoic acid, 9‐oxoODE (A); 13‐oxo‐octadecenoic acid, 13‐oxoODE (B); 9‐hydroxyoctadecenoic acid, 9‐HODE (C); 13‐hydroxyoctadecenoic acid, 13‐HODE (D); 9‐hydroxyoctatrienoic acid, 9‐HOTrE (E); 13‐hydroxyoctatrienoic acid, 13‐HOTrE (F)] in wild‐type (B73) and lox3 mutant maize kernels inoculated with wild‐type (FvWT) and oxylipin‐deficient (ΔFvlds1D) Fusarium verticillioides strains at 6 and 9 days post‐inoculation (dpi). Error bars indicate ± standard error (SE).
Similar to the fungal‐specific oxylipins, the plant‐specific oxylipins 9‐hydroxyoctatrienoic acid (9‐HOTrE) and 13‐hydroxyoctatrienoic acid (13‐HOTrE) also displayed a host and pathogen genotype‐dependent accumulation pattern. 9‐HOTrE is a major product of LOX3 in seed (Gao et al., 2007) and was reduced in all genotype combinations compared with WT seed infected by FvWT at 6 dpi, suggesting that LOX3 is the major isoform involved in the biosynthesis of 9‐HOTrE in F. verticillioides‐infected seed as well. Interestingly, lox3 mutant seed accumulated significantly greater levels of 13‐HOTrE in response to FvWT at 9 dpi (Fig. 7F). These results are in agreement with previously published data suggesting that the lox3 mutant overexpresses the 13‐LOX genes, ZmLOX8 and ZmLOX10 (Gao et al., 2009), which may explain the increased levels of 13‐oxylipins.
The oxylipins potentially produced by both organisms included 9‐oxo‐octadecenoic acid (9‐oxoODE), 13‐oxo‐octadecenoic acid (13‐oxoODE), 9‐HODE, 13‐hydroxyoctadecenoic acid (13‐HODE), 9(10)‐epoxymonoenoic acid (9(10)‐epoOME), 12(13)‐epoxymonoenoic acid (12(13)‐epoOME), 9,10‐dihydroxymonoenoic acid (9,10‐diHOME) and 12,13‐dihydroxymonoenoic acid (12,13‐diHOME). In general, the stage of disease progression determines the organism whose genotype impacts the pattern of specific oxylipin accumulation to the greatest levels. At earlier time points, the fungal genotype is the major factor in determining the levels of 12,13‐diHOME, 9,10‐diHOME and 13‐oxoODE. At this stage of infection, the levels of 9(10)‐epoOME, 12(13)‐epoOME and 9‐oxoODE depend on both a functional LOX3 and LDS1 to reach the levels seen in WT. At advanced stages of disease development, the host genotype is the major factor in determining the levels of 9,10‐diHOME, 9(10)‐epoOME and 12(13)‐epoOME. Interestingly, both LDS1 and LOX3 are required for suppression of 13‐oxoODE, 9‐HOD and 13‐HOD; however, the levels of these same oxylipins are either reduced or return to WT levels in seeds of lox3 infected by ΔFvlds1D. As FvLDS1 is overexpressed during the colonization of lox3 seed, this may explain the increased accumulation pattern of these oxylipins during these treatments. Alternatively, the accumulation pattern is reminiscent of the expression profile for FvLDS2, which indicates that additional fungal oxygenases may serve as biosynthetic enzymes for these oxylipins during advanced disease progression.
Discussion
This is the first study to explore the role of oxylipin‐mediated cross‐talk in the plant–fungal interaction of maize seed and F. verticillioides. To gain insights into the role of host and pathogen oxylipins as unexplored mechanisms of virulence during seed infection processes, a functional analysis was performed with oxygenase mutants from F. verticillioides and maize reported to be essential for normal disease development. ZmLOX3 is a 9‐LOX required for normal susceptibility to F. verticillioides, as knock‐out mutants are resistant to fumonisin accumulation and are unable to support normal sporulation (Gao et al., 2007). FvLDS1 from F. verticillioides clusters with 9S‐DOX‐AOS from F. oxysporum (Fig. S2, see Supporting Information) and appears to serve as a negative regulator of growth and fumonisin production in vitro (Scala et al., 2014); however, its role in seed infections remains to be tested. To test the interaction of these two oxygenases in the context of oxylipin‐mediated cross‐talk, all permutations of host–pathogen combinations (B73–FvWT; lox3–FvWT; B73–ΔFvlds1D; lox3–ΔFvlds1D) were examined for disease development and mycotoxin production, transcriptional reprogramming and oxylipin accumulation.
Oxylipins are a family of oxidized fatty acids with potent activity as molecular signals for intra‐ and intercellular communications in animals, plants and fungi (Tsitsigiannis and Keller, 2006). Fungi respond to specific plant oxylipins with distinct alterations in sporulation (Dagenais et al., 2008; Horowitz Brown et al., 2008; Tsitsigiannis and Keller, 2006; Tsitsigiannis et al., 2004b) and secondary metabolism, depending on the oxylipin molecular species (Mita et al., 2007; Reverberi et al., 2010, Scala et al., 2014; Scarpari et al., 2014). The biochemical and structural similarities of plant and fungal oxylipins have been proposed to facilitate the reciprocal cross‐kingdom perception of these molecules which mimic the function of the endogenously produced oxylipins (Borrego and Kolomiets, 2012; Christensen and Kolomiets, 2011; Gao and Kolomiets, 2009). To date, information is scarce for any pathosystem for the impact of fungal‐derived oxylipins in mediating the responses of the plant. The functional characterization of the maize and F. verticillioides oxylipin synthesis pathways provides the opportunity to elucidate this complex and biologically meaningful mechanism of signalling across kingdoms in an agronomically important interaction.
Kernel bioassays are universally performed in many laboratories to study mycotoxin‐producing fungal pathogens and their interactions with host seeds (Christensen et al., 2012). However, to explore the interaction between seed and fungi at the molecular and biochemical level, it is fundamental that the host remains alive throughout the process to maintain the intimate signal exchange. To understand the timing of host viability and dynamics of the pathogenic process, a kernel biological assay was performed with seed of the B73 inbred line infected with FvWT. In our hands, beyond 9 dpi, the host was no longer alive as no expression of maize genes was detected in infected seed compared with mock controls at 12 or 15 dpi. Within this time frame, the levels of colonization, conidiation and fumonisin accumulation were observed to increase incrementally from 6 and 9 dpi at sufficient concentrations. Thus, 6 and 9 dpi were chosen for subsequent experiments utilizing all host and pathogen genotype combinations. It is interesting to note that, during the late stages of disease development, ergosterol levels decreased compared with those at 9 dpi. The exhaustion of kernel nutrients and the resource demands of conidiation are likely to contribute to death of the fungal mycelia and subsequent degradation of ergosterol (Charcosset and Chauvet, 2001).
The lox3 mutants were more resistant to F. verticillioides WT strain infection, as measured by the inability of lox3 mutant seed to support WT levels of fungal biomass, as determined through ergosterol quantification. The reduced colonization on lox3 kernels was associated with a striking reduction in fumonisin accumulation. These results confirmed the previous report of increased fumonisin resistance in lox3 mutant seed (Gao et al., 2007). The LDS1 knock‐out mutant displayed a dramatic reduction in colonization and fumonisin accumulation at all days tested compared with FvWT, which provides strong genetic evidence that both oxygenases are required to allow normal disease development. These observations contrast with the conclusions from a previous report of FvLDS1, which described a modest increase in colonization of ΔFvlds1D on maize cobs and increased fumonisin production in vitro (Scala et al., 2014). The different tissues and infection methods used may explain the differences between the observations. Similarly, the dramatic difference in fumonisin production between Scala et al. (2014) and this study can be attributed to the effect of a living host as substrate. Mukherjee et al. (2011) found that, compared with WT, F. verticillioides knock‐out strains of regulators of G‐protein signalling produced increased fumonisin on non‐viable maize kernels, but decreased fumonisin when colonizing living kernels.
Increased expression of ZmLOX3 was observed with the progression of infection, echoing a previous report on the expression pattern of ZmLOX3 in F. verticillioides‐infected cobs (Scala et al., 2014). The increased expression of ZmLOX3 correlated with the timing of the greatest difference between WT and lox3 mutant in terms of both colonization and fumonisin accumulation. Notably, ΔFvlds1D was unable to induce ZmLOX3 accumulation when transcript levels remained similar to mock‐infected seeds. The increased resistance of lox3 mutant seed correlated with increased expression of the defence‐related gene, ZmLOX12, and two additional 9‐LOXs, ZmLOX4 and ZmLOX5.
In a recent study, ZmLOX12 was implicated in maize defence against F. verticillioides, as the knock‐out disruption of this gene resulted in a dramatic increase in susceptibility to F. verticillioides not only in kernels, but in all below‐ and above‐ground organs tested (Christensen et al., 2014). ZmLOX12 is required for normal accumulation of the defensive phytohormone JA following F. verticillioides infection. In support of the conclusion that JA‐mediated signalling is the major mechanism behind defence against this pathogen, the JA‐deficient opr7opr8 double mutant displayed extreme susceptibility to F. verticillioides infection (Christensen et al., 2014). Here, a very strong induction of ZmLOX12 was observed in the lox3 mutant kernels, prompting the hypothesis that the overexpression of this gene in the lox3 mutant is an underlying mechanism behind the resistance of lox3 kernels to fumonisin contamination and Fusarium ear rot.
Concurrent with the expression pattern of ZmLOX12, segmentally duplicated paralogues, ZmLOX4 and ZmLOX5, were also strongly induced by FvWT in the infected lox3 mutant seed compared with mock. The lox4 and lox5 knock‐out mutants were found to be substantially more susceptible to F. verticillioides, suggesting that these two genes are required for normal defence against this pathogen (Park et al., 2010). The reduced colonization and fumonisin production characteristic of the lox3 mutation could be ascribed not only to the enhanced expression of ZmLOX12, but also to the stronger and earlier induction of ZmLOX4 and ZmLOX5 during the early stages of F. verticillioides seed infection. These two isoforms are tonoplast‐localized (Tolley et al., 2017) paralogues and, despite sharing over 95% identity at the amino acid level (Park et al., 2010), it appears that neither is capable of compensating for the absence of the other during F. verticillioides infection. All three non‐JA‐producing 9‐LOXs appear to be indispensable for the induction of normal jasmonate biosynthesis during infection, suggesting that each produces either a unique 9‐oxylipin required for activation of the JA pathway or acts in a sequential manner to eventually produce a sufficient amount of one or several oxylipins to reach a threshold for initiation of JA production. The role of JA as the major defence hormone required for immunity against F. verticillioides has been most convincingly shown by the analysis of the JA‐deficient opr7opr8 double mutant which displays complete loss of resistance to this pathogen (Christensen et al., 2014).
Oxylipin profiling of infected kernels revealed that the major 9‐oxylipin that was significantly reduced in the lox3 mutant was linolenic acid‐derived, 9‐HOD(T)E, confirming the results of previous oxylipin analyses of germinating seed (Gao et al., 2007). The increased substrate utilization of linolenic acid by ZmLOX3 may be a consequence of its subcellular localization. Recently, it has been reported that the rice orthologue of ZmLOX3 is plastid‐localized (Zhou et al., 2014), the membranes of which are rich in linolenic acid. These results suggest that LOX3‐derived oxylipins, e.g. 9‐HOTE, not only facilitate fungal colonization, but also strongly induce fumonisin biosynthesis, through the suppression of defence‐related genes, such as ZmLOX4, ZmLOX5 and ZmLOX12.
The most striking observation in terms of fungal oxylipins was the dramatic reduction in 8‐HpODE production by the ΔFvlds1D mutant grown on lox3 mutant kernels. This is in spite of similarly reduced colonization levels by ΔFvlds1D regardless of the host genotype (Fig. 2A). These results suggest that, although both FvLDS1 and ZmLOX3‐derived oxylipins are required to signal for normal production of 8‐HpODE, ZmLOX3 is capable of compensating for the lack of FvLDS1 potentially through the production of similar 9S‐oxylipin species. Given that the levels of 8‐HpODE are nearly identical in WT seed colonized by either FvWT or ΔFvlds1D, 8‐HpODE is probably dispensable during the infection process. During the early stages of disease development, these observations are in agreement with a previous report showing that ΔFvlds1D is unable to accumulate normal levels of 8‐HpODE; however, at later stages of growth, Scala et al. (2014) were unable to observe normal levels of 8‐HpODE or 8,13‐diHODE in vitro. The difference between the two studies may be a result of the effect of the living host on secondary metabolism (Mukherjee et al., 2011). Scala et al. (2013) found a dramatic induction of F. verticillioides oxylipin biosynthetic genes during the colonization of maize cobs compared with axenic cultures. The direct effect of fungal oxylipins with regard to their ability to induce ZmLOX3 for increased pathogenicity is currently under investigation.
With regard to the expression study of fungal oxylipin biosynthesis genes, transcript accumulation of one LOX (LOX1) and three LDS (LDS1, LDS2 and FAO) genes was investigated. In Aspergillus and other fungal genera, the LDS genes are named Ppo (PSI‐producing oxygenase) as they produce a mixture of oxidized molecules called Psi (Precocious sexual inducer) factors. The change in the ratio between these oxidized lipids affects the production of spores, increasing the production of sexual or asexual spores, depending on the specific oxylipin composition, or regulating secondary metabolism, such as mycotoxin synthesis. Silencing of specific Ppo or sole LOX genes reduced the ability of Aspergillus species to colonize tissues and led to an altered ratio between sexual/asexual sporulation (Horowitz Brown et al., 2008; Tsitsigiannis and Keller, 2006, 2007). Intriguingly, we observed a very strong induction of expression of FvLOX1 and FvLDS1 in the WT strain of F. verticillioides, in particular when grown in lox3 kernels at different times post‐inoculation. Although the physiological relevance of this induction is not clear, it is likely that F. verticillioides utilizes different oxylipin pathways to promote disease development. As each of the F. verticillioides LDS isoforms clusters into distinct clades (S1), it is expected that their oxylipin biosynthetic activities will be unique, and it becomes evident that higher order mutants are required to untangle the complex cross‐talk between the fungal oxylipin branches.
Figure 8 schematically depicts the current hypothetical model of the oxylipin‐mediated signal communication between maize seed and F. verticillioides. This model is based on current literature and the results described above. As a part of its pathogenesis strategy, on infection of a susceptible host, F. verticillioides produces an array of LDS1‐derived oxylipins that serve dual functions: (1) to signal changes in developmental programs, including colonization and secondary metabolism, e.g. mycotoxin production; and (2) to modulate the expression of host LOXs, the metabolic products of which facilitate the pathogenesis processes. Fungal oxylipins are probably perceived by the host cell receptor‐like complexes, such as the recently identified jasmonate and coronatine (JA structural mimic of bacterial origin) JAZ–COI1 receptor complex (Katsir et al., 2008), or by putative G protein/x96coupled receptors (GPCRs) of the host (Affeldt et al., 2012; Gookin et al., 2008). This perception of fungal oxylipins results in the activation of the pathogenicity‐related ZmLOX3 in the host, whose activity represses the expression of the defence‐related ZmLOX4, ZmLOX5 and ZmLOX12. In support of this hypothesized ability of fungal oxylipins to affect the expression of host LOX genes, Brodhagen et al. (2008) showed that oxylipin‐deficient mutants of Aspergillus nidulans were unable to normally induce peanut LOX.
Figure 8.
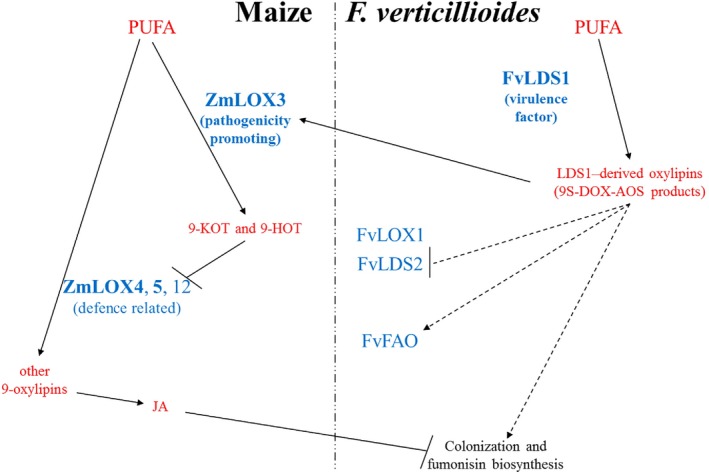
Model depicting the oxylipin‐mediated cross‐talk between maize and Fusarium verticillioides uncovered in this study. Polyunsaturated fatty acids (PUFAs) are substrates for maize or F. verticillioides oxygenases. In the host, the pathogenicity‐promoting ZmLOX3 is the primary isoform responsible for the biosynthesis of 9‐KOT and 9‐HOT in seed. In the fungus, the virulence factor, FvLDS1, appears to be responsible for the production of 9‐oxylipins that induce the expression of ZmLOX3 in infected seed. The increased production of ZmLOX3‐derived 9‐KOT, 9‐HOT or their downstream products suppresses the transcription of the defence‐related LOX isoforms, ZmLOX4, ZmLOX5 and ZmLOX12, which would normally catalyse the production of additional 9‐oxylipins to positively promote JA‐mediated defences to suppress F. verticillioides seed colonization. Symbols in red and blue represent metabolites and enzymes, respectively. The enzymes functionally characterized by this study are shown in bold. AOS, allene oxide synthase; DOX, dioxygenase; FAO, fatty acid oxygenase; 9‐HOT, 9‐hydroxyoctadecatrienoic acid; JA, jasmonic acid; 9‐KOT, 9‐ketooctadecatrienoic acid; LDS, linoleate diol synthase; LOX, lipoxygenase.
Taken together, these results suggest that FvLDS1‐derived oxylipins promote maize vulnerability to colonization through induction of the susceptibility gene, ZmLOX3, to suppress defence‐related branches of the oxylipin pathway. The down‐regulation of ZmLOX3 was recently reported as a mechanism behind Trichoderma virens‐mediated induced systemic resistance (Constantino et al., 2013). This prompts the intriguing hypothesis that 9‐LOXs are the targets for microbial effectors, such as small peptides (Djonović et al., 2006) or oxylipins (Brodhagen et al., 2008), in diverse plant–microbe interactions that can lead to transcriptional reprogramming for increased disease resistance or susceptibility.
Experimental Procedures
Plant materials and fungal strains
Two maize inbred lines were used in this study: the B73 inbred line, susceptible to F. verticillioides infection and fumonisin contamination, and its near‐isogenic line (NIL) mutant lox3–4, back‐crossed to B73 seven times, which is more resistant than WT. The generation of maize lox3, lox4, lox5 and lox12 knock‐out mutants by Mutator‐transposable element‐insertional mutagenesis has been described previously (Christensen et al., 2014; Gao et al., 2007; Park, 2012; Y. Park, unpublished results).
The fungal wild‐type strain (FvWT) used in this study was F. verticillioides ITEM 10027 (MPVP 294), which was isolated from maize in south Tuscany by the Department of Sustainable Crop Production, Piacenza, Italy. The oxylipin‐deficient ΔFvlds1D was obtained from the WT strain as described by Scala et al. (2014).
The strain ITEM 10027 (MPVP 294) is stored in the fungal collection of the Department of Sustainable Crop Production, Piacenza, Italy and the Institute of Sciences and Food Production, National Research Council, Bari, Italy. The oxylipin‐deficient ΔFvlds1D is stored in the F. verticillioides oxylipin‐deficient collection of Sapienza University, Rome, Italy. In both collections, strains are cryo‐conserved in glycerol (18%) and maintained at −80 °C.
Inoculum production
Working fungal cultures were maintained on Petri plates (9 cm Ø) on potato dextrose agar (PDA; HiMedia, Mumbai, India) and incubated at 25 °C with a 12‐h photoperiod for 14 days. A conidial suspension was obtained by scraping plates with 5 mL of autoclaved 0.1% Tween‐20 solution and filtering the conidial suspension through four layers of autoclaved cheesecloth. The spore suspension was adjusted to a final concentration of 106 conidia/mL based on a microscopic count using a Bürker chamber.
Kernel selection and inoculation assay
Kernels of similar shape and size, preferably flat, were selected for both the B73 line and the lox3 mutant. Kernel inoculation assay was essentially carried out as described by Christensen et al. (2012). Briefly, kernels were placed in a 200‐mL flask, surface sterilized with 70% ethanol by shaking at room temperature for 5 min, 1 min with sterile water and 10 min with 6% sodium hypochlorite. Finally, they were rinsed three times with sterile distilled water by shaking for 5 min each time, and dried at room temperature on autoclaved towels in sterile conditions.
To facilitate fungal infection, the embryo side was wounded at a depth of 0.5 mm using a sterilized razor blade. Groups of three kernels were placed in autoclaved 20‐mL glass scintillation vials. The kernels were inoculated with 200 µL of spore suspension (106 spores/mL), capped and mixed by Vortex® for 10 s. Control seeds (mock) received an equal amount of 0.1% Tween‐20. A humidity chamber was prepared by placing five autoclaved sheets of paper towel in a plastic container, to which 100 mL of distilled sterile water was added. All kernels were placed in the vials with the embryo side up to avoid differences in growth caused by the differential colonization of diverse kernel tissues. The caps were loosened to allow air exchange and the vials were placed in a humidity chamber. The chamber was not air‐ or water‐tight and an additional 50 µL of sterile distilled water was added daily to each vial throughout the experiment to preserve a high moisture level during the experiment time course and to avoid volatile accumulation inside the vials. The samples were incubated under a 12‐h light photoperiod at 28 °C for 6 and 9 dpi. At every time point, samples were frozen at −80 °C until analysis.
RNA extraction
Samples were ground under liquid nitrogen to obtain a very fine powder and 100 mg of powder was used for RNA extraction following the sodium dodecylsulfate (SDS) extraction protocol based on Prescott and Martin (1986).
The extracted RNA was purified with the RNA Cleanup protocol (Qiagen, Valencia, CA, USA), according to the manufacturer's instructions, including the optional on‐column DNase digestion (Qiagen), and stored at −80 °C. The amount and quality of total RNA were estimated by fluorometric assay (Qubit Invitrogen, ThermoFischer, Waltham, MA, USA) as well as by agarose gel electrophoresis.
Real‐time RT‐PCR expression analysis
A two‐step protocol was performed to study the expression of six plant LOX genes and four fungal genes involved in oxylipin synthesis (LOXs and LDSs). cDNA was synthesized from a 1‐µg sample of total RNA, following a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, San Mateo County, CA, USA) and stored at −20 °C.
Twenty nanograms of single‐stranded cDNA determined by fluorometric assay (Qubit Invitrogen) were used for quantitative RT‐PCR. Fungal gene‐specific primers were taken from the literature (Scala et al., 2013) or were designed using Primer3Plus software in accordance with the criteria required for quantitative PCR primer design (Udvardi et al., 2008). Their sequences are reported in Table S1 (see Supporting Information).
Primer gene specificity was tested with one of the extracted RNA samples using an amplification reaction and the melt curve to avoid any non‐specific amplification. The total reaction volume for the analysis was 25 µL: 7.5 µL of purified diluted cDNA (20 ng total), 5 µL of primer (4 µm) and 12.5 µL of FluoCycle™ II SYBR® Green master mix (EuroClone S.p.a., Milan, Italy). Relative quantitative analysis was performed using the MiniOpticon device (Bio‐Rad, Hercules, CA, USA) under the following conditions: an initial step at 95 °C for 3 min, followed by 40 cycles at 95 °C for 10 s and 60 °C (or 54 °C for LOX1 primers of both maize and Fusarium) for 30 s. Three replicates were employed for each tested sample and template‐free negative controls. Melting curve analysis, ranging from 60 to 95 °C with a 0.5 °C increment for 5 s, was used to identify different amplicons and to confirm primer pair specificity; primers for the reference genes showed high specificity at both temperatures.
Relative quantification was normalized to the reference gene (cullin for maize plant and β‐tubulin for Fusarium strains); the expression ratio and fold change (FC) were calculated using the 2−ΔΔCt method (Schmittgen and Livak, 2008) and calibrated on the mock‐inoculated kernels.
Conidial enumeration
Four vials with kernels were used as replicates for conidial enumeration. Five millilitres of methanol (Carlo Erba Reagents S.r.l., Milan, Italy) were added to each vial and mixed thoroughly using a Vortex®; 100 µL of spore suspension were extracted twice as technical replicates and diluted in a tube (Eppendorf, Hamburg, Germany) with 100 µL of sterile distilled water. Each 200‐µL aliquot was enumerated twice with a haemocytometer.
A separate set of biologically replicated vials was frozen at −80 °C for ergosterol analysis.
Ergosterol analysis
Four vials with kernels were used as replicates for ergosterol analysis. To extract ergosterol, 5 mL of methanol and 10 mL of chloroform (1 : 2, v/v) (Carlo Erba Reagents S.r.l.) were added to each vial and mixed for 10 s, followed by incubation in the dark for 24 h at room temperature.
Afterwards, 1 mL of extract was filtered and dried under a nitrogen stream. The solute was dissolved in 1 mL of n‐hexane and injected into a high‐performance liquid chromatography (HPLC) system: a Thermo Separation Product (TSP) instrument equipped with a TSP‐2000 pump, an AS‐3000 sampling system and a Spectra Focus UV‐Vis detector (San Jose, CA, USA). The system was controlled by TSP PCI1000 software. A Superspher Si‐60 column (particle size, 4 µm; 125 mm × 4 mm i.d.; Merck, Burlington, MA, USA) was used at ambient temperature, with a mobile phase of n‐hexane–isoamyl alcohol (98 : 2) at 1.0 mL/min; the injection volume was 30 µL. The UV detector was set at 280 nm. Ergosterol standards between 20 and 200 ng were injected (Pietri et al., 2004).
Fumonisin analysis
Free and total fumonisins were analysed as described by Dall'Asta et al. (2012). Samples were analysed on a 2695 Alliance separation system (Waters Co., Milford, MA, USA) equipped with a Quattro API triple quadrupole mass spectrometer with an electrospray source (Micromass, Waters, Manchester, UK). Experiments in multiple reaction monitoring (MRM) in positive ion mode were performed according to our previous work (Dall'Asta et al., 2012; Giorni et al., 2015).
Oxylipin analysis
Oxylipins were extracted as described by Scala et al. (2014). Samples were analysed by liquid chromatography (HPLC 1200 series rapid resolution; Agilent Technologies, Santa Clara, CA, USA) coupled to a mass spectrometer (G6410A series triple quadrupole, QqQ; Agilent Technologies) equipped with electrospray ionization (ESI). Experiments in MRM in negative ion mode [M‐H]− were performed and data processed as reported by Ludovici et al. (2014). Jasmonates were measured as described previously (Christensen et al., 2014)
Statistical analyses
The reported values for ergosterol, fumonisin, conidia, and plant and fungal gene expression are the average of three biological replicates. One‐factor analysis of variance (ANOVA) was performed on the observed means of the different parameters evaluated, and the significance of the different treatments (B73–FvWT, lox3–FvWT, B73–ΔFvlds1D and lox3–ΔFvlds1D) within each time point was evaluated by Tukey's honestly significant difference (HSD) test (P < 0.05).
Author Contributions
P.B. performed conception and design, data analysis, paper revision and coordination of contributions. A.L. performed data management and paper preparation. V.S. performed mutant strain delivery, conception of fungal oxylipin genes, data acquisition (oxilipins), data interpretation (Fv × maize: oxylipin gene expression and metabolites) and paper preparation. M.R. performed data analysis and interpretation (Fv × maize: oxylipin gene expression and metabolites), and paper revision. R.G. performed experiment management and data acquisition (F. verticillioides disease; Fv × maize: gene expression). C.F. performed data acquisition (fumonisin). C.D. performed data acquisition (fumonisin), data analysis and paper revision. Y.‐S.P. performed experiment management, and creation and functional analyses of lox4 and lox5 mutants (Fv × maize). J.B. performed experiments on hormone response to Fusarium infection. E.J.B. performed experiment management, data acquisition and elaboration, paper preparation and revision, and data interpretation. M.V.K. performed conception and design, creation of maize mutants, data analysis, paper preparation and revision, and data interpretation.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 The biosynthesis of selected oxylipins measured in this study based on current literature. No 8‐hydroperoxy fatty acid‐producing enzymes were detected in the genome of Fusarium verticillioides. FvLOX1 (FVEG 09897) and LOX2 (FVEG 03347) cluster with 13S‐LOX of Fusarium oxysporum (data not shown). Colours represent maize (green) or F. verticillioides (brown). HpODE, hydroperoxyoctadecadienoic acid; diHOME, dihydroxyoctadecenoic acid; HODE, hydroxyoctadecadienoic acid; oxo‐ODE, oxo‐octadecadienoic acid; epoOME, epoxyoctadecenoic acid; HOTrE, hydroxyoctadecatrienoic acid.
Fig. S2 Phylogenetic analysis of the Fusarium verticillioides fatty acid oxygenases. The evolutionary history was inferred using the maximum likelihood method based on the Poisson correction model (Zuckerkandl and Pauling, 1965). The bootstrap consensus tree inferred from 500 replicates (Felsenstein, 1985) is taken to represent the evolutionary history of the taxa analysed (Felsenstein, 1985). Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches (Felsenstein, 1985). Initial tree(s) for the heuristic search were obtained automatically by application of the Neighbour‐Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, followed by selection of the topology with the superior log likelihood value. The analysis involved 20 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 837 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 (Kumar et al., 2016). Nomenclature follows Sooman, Linda, and Ernst H. Oliw. “Discovery of a novel linoleate dioxygenase of Fusarium oxysporum and linoleate diol synthase of Colletotrichum graminicola.” Lipids 50.12 (2015): 1243–1252. https://onlinelibrary.wiley.com/doi/full/10.1007/s11745‐015‐4078‐9.
Table S1 Primers used in this study.
Acknowledgements
R.G. carried out this work within the PhD school ‘Agrisystem’ of Università Cattolica del Sacro Cuore (Italy). The work performed in this study was supported by a National Science Foundation Grant (IOS‐0544428) and United States Department of Agriculture – National Institute of Food and Agriculture USDA‐NIFA (2017–67013‐ 26524) to M.V.K.
References
- Affeldt, K.J. , Brodhagen, M. and Keller, N.P. (2012) Aspergillus oxylipin signaling and quorum sensing pathways depend on G protein‐coupled receptors. Toxins, 4, 695–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreou, A. , Brodhun, F. and Feussner, I. (2009) Biosynthesis of oxylipins in non‐mammals. Prog. Lipid Res. 48, 148–170. [DOI] [PubMed] [Google Scholar]
- Bacon, C.W. , Hinton, D.M. , Porter, J.K. , Glenn, A.E. and Kuldau, G. (2004) Fusaric acid, a Fusarium verticillioides metabolite, antagonistic to the endophytic biocontrol bacterium Bacillus mojavensis . Can. J. Bot. 82, 878–885. [Google Scholar]
- Battilani, P. , Rossi, V. and Pietri, A. (2003) Modelling Fusarium verticillioides infection and fumonisin synthesis in maize ears. Asp. Appl. Biol. 68, 91–100. [Google Scholar]
- Battilani, P. , Pietri, A. , Barbano, C. , Scandolara, A. , Bertuzzi, T. and Marocco, A. (2008) Logistic regression modeling of cropping systems to predict fumonisin contamination in maize. J. Agric. Food Chem. 56, 10 433–10 438. [DOI] [PubMed] [Google Scholar]
- Borrego, E.J. and Kolomiets, M.V. (2012) Lipid‐mediated signaling between fungi and plants In: Biocommunication of Fungi (Witzany G., ed), pp. 249–260. Town: Springer Dordrecht, Netherlands. [Google Scholar]
- Borrego, E.J. and Kolomiets, M.V. (2016) Synthesis and functions of jasmonates in maize. Plants, 5, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodhagen, M. and Keller, N.P. (2006) Signalling pathways connecting mycotoxin production and sporulation. Mol. Plant Pathol. 7, 285–301. [DOI] [PubMed] [Google Scholar]
- Brodhagen, M. , Tsitsigiannis, D.I. , Hornung, E. , Goebel, C. , Feussner, I. and Keller, N.P. (2008) Reciprocal oxylipin mediated cross‐talk in the Aspergillus–seed pathosystem. Mol. Microbiol. 67, 378–391. [DOI] [PubMed] [Google Scholar]
- Brodhun, F. and Feussner, I. (2011) Oxylipins in fungi. FEBS J. 278, 1047–1063. [DOI] [PubMed] [Google Scholar]
- Burow, G.B. , Nesbitt, T.C. , Dunlap, J. and Keller, N.P. (1997) Seed lipoxygenase products modulate Aspergillus mycotoxin biosynthesis. Mol. Plant–Microbe Interact. 10, 380–387. [Google Scholar]
- Camera, S.L. , Gouzerh, G. , Dhondt, S. , Hoffmann, L. , Fritig, B. , Legrand, M. and Heitz, T. (2004) Metabolic reprogramming in plant innate immunity: the contributions of phenylpropanoid and oxylipin pathways. Immunol. Rev. 198, 267–284. [DOI] [PubMed] [Google Scholar]
- Charcosset, J.Y. and Chauvet, E. (2001) Effect of culture conditions on ergosterol as an indicator of biomass in the aquatic hyphomycetes. Appl. Environ. Microbiol. 67, 2051–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, S.A. and Kolomiets, M.V. (2011) The lipid language of plant–fungal interactions. Fungal Genet. Biol. 48, 4–14. [DOI] [PubMed] [Google Scholar]
- Christensen, S.A. , Borrego, E. , Shim, W. , Isakeit, T. and Kolomiets, M. (2012) Quantification of fungal colonization, sporogenesis, and production of mycotoxins using kernel bioassays. J. Vis. Exp. 23, 3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, S.A. , Nemchenko, A. , Park, Y. , Borrego, E. , Huang, P. , Schmelz, E.A. , Kunze, S. , Feussner, I. , Yalpani, N. , Meeley, R. and Kolomiets, M.V. (2014) The novel monocot‐specific 9‐lipoxygenase ZmLOX12 is required to mount an effective jasmonate‐mediated defense against Fusarium verticillioides in maize. Mol. Plant–Microbe Interact. 27, 1263–1276. [DOI] [PubMed] [Google Scholar]
- Constantino, N. , Mastouri, F. , Damarwinasis, R. , Borrego, E. , Moran‐Diez, M.E. , Kenerley, C.M. , Gao, X. and Kolomiets, M.V. (2013) Root‐expressed maize lipoxygenase 3 negatively regulates induced systemic resistance to Colletotrichum graminicola in shoots. Front. Plant Sci. 4, 510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagenais, T.R.T. , Chung, D.W. , Giles, S.S. , Hull, C.M. , Andes, D. and Keller, N. (2008) Defects in conidiophore development and conidium–macrophage interactions in a dioxygenase mutant of Aspergillus fumigatus . Infect. Immun. 76, 3214–3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall'Asta, C. , Falavigna, C. , Galaverna, G. and Battilani, P. (2012) Role of maize hybrids and their chemical composition in Fusarium infection, fumonisin production and masking. J. Agric. Food Chem. 60, 3800–3808. [DOI] [PubMed] [Google Scholar]
- Dall'Asta, C. , Giorni, P. , Cirlini, M. , Reverberi, M. , Gregori, R. , Ludovici, M. , Camera, E. , Fanelli, C. , Battilani, P. and Scala, V. (2015) Maize lipids play a pivotal role in the fumonisin accumulation. World Mycotox. J. 8, 87–97. [Google Scholar]
- Djonović, S. , Pozo, M.J. , Dangott, L.J. , Howell, C.R. and Kenerley, C.M. (2006) Sm1, a proteinaceous elicitor secreted by the biocontrol fungus Trichoderma virens induces plant defense responses and systemic resistance. Mol. Plant–Microbe Interact. 19, 838–853. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution, 39, 783–791. [DOI] [PubMed] [Google Scholar]
- Feussner, I. and Wasternack, C. (2002) The lipoxygenase pathway. Annu. Rev. Plant Biol. 53, 275–297. [DOI] [PubMed] [Google Scholar]
- Folcher, L. , Jarry, M. , Weissenberger, A. , Gérault, F. , Eychenne, N. , Delos, M. and Regnault‐Roger, C. (2009) Comparative activity of agrochemical treatments on mycotoxin levels with regard to corn borers and Fusarium mycoflora in maize (Zea mays L.) fields. Crop Protoc. 28, 302–308. [Google Scholar]
- Gao, X. and Kolomiets, M.V. (2009) Host‐derived lipids and oxylipins are crucial signals in modulating mycotoxin production by fungi. Toxin Rev. 28, 79–88. [Google Scholar]
- Gao, X.Q. , Shim, W.B. , Gobel, C. , Kunze, S. , Feussner, I. , Meeley, R. , Balint‐Kurti, P. and Kolomiets, M. (2007) Disruption of a maize 9‐lipoxygenase results in increased resistance to fungal pathogens and reduced levels of contamination with mycotoxin fumonisin. Mol. Plant–Microbe Interact. 20, 922–933. [DOI] [PubMed] [Google Scholar]
- Gao, X.Q. , Brodhagen, M. , Isakeit, T. , Brown, S.H. , Gobel, C. , Betran, J. , Feussner, I. , Keller, N.P. and Kolomiets, M. (2009) Inactivation of the lipoxygenase ZmLOX3 increases susceptibility of maize to Aspergillus spp. Mol. Plant–Microbe Interact. 22, 222–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelderblom, W.C.A. , Jaskiewicz, J. , Marasas, W.F.O. , Thiel, P.G. , Horak, R.M. , Vleggar, R. and Kriek, N.P.J. (1988) Fumonisins: novel mycotoxins with cancer promoting activity produced by Fusarium moniliforme . Appl. Environ. Microbiol. 54, 1806–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorni, P. , Dall'Asta, C. , Reverberi, M. , Scala, V. , Ludovici, M. , Cirlini, M. , Galaverna, G. , Fanelli, C. and Battilani, P. (2015) Open field study of some Zea mays hybrids, lipid compounds and fumonisins accumulation. Toxins, 7, 3657–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gookin, T.E. , Kim, J. and Assmann, S.M. (2008) Whole proteome identification of plant candidate G‐protein coupled receptors in Arabidopsis, rice, and poplar: computational prediction and in‐vivo protein coupling. Genome Biol. 9, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz Brown, S. , Zarnowski, R. , Sharpee, W.C. and Keller, N.P. (2008) Morphological transition governed by density dependent and lipoxygenase activity in Aspergillus flavus . Appl. Environ. Microbiol. 74, 5674–5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Agency for Research on Cancer (IARC) . (2002) Summaries & Evaluations. Fumonisin B1 (Group 2B), 82, 301.
- Katsir, L. , Chung, H.S. , Koo, A.J. and Howe, G.A. (2008) Jasmonate signaling: a conserved mechanism of hormone sensing. Curr. Opin. Plant Biol. 11, 428–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S. , Stecher, G. and Tamura, K. (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logrieco, A. , Mulè, G. , Moretti, A. and Bottalico, A. (2002) Toxigenic Fusarium species and mycotoxins associated with maize ear rot in Europe. Eur. J. Plant Pathol. 108, 597–609. [Google Scholar]
- Ludovici, M. , Ialongo, C. , Reverberi, M. , Beccaccioli, M. , Scarpari, M. and Scala, V. (2014) Quantitative profiling of oxylipins through comprehensive LC‐MS/MS analysis of Fusarium verticillioides and maize kernels. Food Addit. Contam. 31, 2026–2033. [DOI] [PubMed] [Google Scholar]
- Maschietto, V. , Marocco, A. , Malachova, A. and Lanubile, A. (2015) Resistance to Fusarium verticillioides and fumonisin accumulation in maize inbred lines involves an earlier and enhanced expression of lipoxygenase (LOX) genes. J. Plant Physiol. 188, 9–18. [DOI] [PubMed] [Google Scholar]
- Mita, G. , Fasano, P. , De Domenico, S. , Perrone, G. , Epifani, F. , Iannacone, R. , Casey, R. and Santino, A. (2007) 9‐lipoxygenase metabolism is involved in the almond/Aspergillus carbonarius interaction. J. Exp. Bot. 58, 1803–1811. [DOI] [PubMed] [Google Scholar]
- Mosblech, A. , Feussner, I. and Heilmann, I. (2009) Oxylipins: structurally diverse metabolites from fatty acid oxidation. Plant Physiol. Biochem. 47, 511–517. [DOI] [PubMed] [Google Scholar]
- Mukherjee, M. , Kim, J.E. , Park, Y.S. , Kolomiets, M.V. and Shim, W.B. (2011) Regulators of G‐protein signalling in Fusarium verticillioides mediate differential host–pathogen responses on non‐viable versus viable maize kernels. Mol. Plant Pathol. 12, 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, P.E. , Desjardins, A.E. and Plattner, R.D. (1993) Fumonisins, mycotoxins produced by Fusarium species: biology, chemistry, and significance. Annu. Rev. Phytopathol. 31, 233–252. [DOI] [PubMed] [Google Scholar]
- Park, Y.S. (2012) Diverse Functions of the Two Segmentally Duplicated 9‐Lipoxygenases ZmLOX4 and ZmLOX5 of Maize (Doctoral dissertation). College Station, TX: Texas A & M University.
- Park, Y.S. , Kunze, S. , Ni, X. , Feussner, I. and Kolomiets, M.V. (2010) Comparative molecular and biochemical characterization of segmentally duplicated 9‐lipoxygenase genes ZmLOX4 and ZmLOX5 of maize. Planta, 231, 1425–1437. [DOI] [PubMed] [Google Scholar]
- Pietri, A. , Bertuzzi, T. , Pallaroni, L. and Piva, G. (2004) Occurrence of mycotoxins and ergosterol in maize harvested over 5 years in northern Italy. Food Addit. Contam. 21, 479–487. [DOI] [PubMed] [Google Scholar]
- Prescott, A. and Martin, C. (1986) A rapid method for the quantitative assessment of levels of specific mRNAs in plants. Plant Mol. Biol. Rep. 4, 219–224. [Google Scholar]
- Ravindra, K.C. , Narayan, V. , Lushington, G.H. , Peterson, B.R. and Prabhu, K.S. (2012) Targeting of histone acetyltransferase by cyclopentenone prostaglandin (12)‐PGJ(2) through covalent binding to Cys (1438). Chem. Res. Toxicol. 25, 337–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverberi, M. , Punelli, F. , Scarpari, M. , Camera, E. , Zjalic, S. , Ricelli, A. , Fanelli, C. and Fabbri, A.A. (2010) Lipoperoxidation affects ochratoxin A biosynthesis in Aspergillus ochraceus and its interaction with wheat seeds. Appl. Microbiol. Biotechnol. 85, 1935–1946. [DOI] [PubMed] [Google Scholar]
- Roze, L.V. , Beaudry, R.M. , Arthur, A.E. , Calvo, A.M. and Linz, J.E. (2007) Aspergillus volatiles regulate aflatoxin synthesis and asexual sporulation in Aspergillus parasiticus . Appl. Environ. Microbiol. 73, 7268–7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scala, V. , Camera, E. , Ludovici, M. , Dall'Asta, C. , Cirlini, M. , Giorni, P. , Battilani, P. , Bello, C. , Fabbri, A.A. , Fanelli, C. and Reverberi, M. (2013) Fusarium verticillioides and maize interaction in vitro: relationship between oxylipin cross‐talk and fumonisin synthesis. World Mycotox. J. 6, 343–351. [Google Scholar]
- Scala, V. , Giorni, P. , Cirlini, M. , Ludovici, M. , Visentin, I. , Cardinale, F. , Fabbri, A.A. , Fanelli, C. , Reverberi, M. , Battilani, P. , Galaverna, G. and Dall'Asta, C. (2014) LDS1 produced oxylipins are negative regulators of growth, conidiation and fumonisin synthesis in the fungal maize pathogen Fusarium verticillioides . Front. Microbiol. 5, 669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpari, M. , Punelli, M. , Scala, V. , Zaccaria, M. , Nobili, C. , Ludovici, M. , Camera, E. , Fabbri, A.A. , Reverberi, M. and Fanelli, C. (2014) Lipids in Aspergillus flavus–maize interaction. Front. Microbiol. 5, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen, T.D. and Livak, K.J. (2008) Analyzing real‐time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1118. [DOI] [PubMed] [Google Scholar]
- Tolley, J. , Nagashima, Y. , Gorman, Z. , Kolomiets, M. and Koiwa, H. (2017) Isoform‐specific subcellular localization of Zea mays lipoxygenases and oxo‐phytodienoate reductase 2. Plant Gene, 13, 36–41. [Google Scholar]
- Tsitsigiannis, D.I. and Keller, N.P. (2006) Oxylipins act as determinants of natural product biosynthesis and seed colonization in Aspergillus nidulans . Mol. Microbiol. 59, 882–892. [DOI] [PubMed] [Google Scholar]
- Tsitsigiannis, D.I. and Keller, N.P. (2007) Oxylipins as developmental and host–fungal communication signals. Trends Microbiol. 15, 109–118. [DOI] [PubMed] [Google Scholar]
- Tsitsigiannis, D.I. , Kowieski, T.M. , Zarnowski, R. and Keller, N.P. (2004a) Endogenous lipogenic regulators of spore balance in Aspergillus nidulans . Eukaryot. Cell, 3, 1398–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsitsigiannis, D.I. , Zarnowski, R. and Keller, N.P. (2004b) The lipid body protein, PpoA, coordinates sexual and asexual sporulation in Aspergillus nidulans . J. Biol. Chem. 279, 11 344–11 353. [DOI] [PubMed] [Google Scholar]
- Udvardi, M.K. , Czechowski, T. and Scheible, W.R. (2008) Eleven golden rules of quantitative RT‐PCR. Plant Cell, 20, 1736–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, G. , Ren, N. , Qi, J. , Lu, J. , Xiang, C. , Ju, H. , Cheng, J. and Lou, Y. (2014) The 9‐lipoxygenase Osr9‐LOX1 interacts with the 13‐lipoxygenase‐mediated pathway to regulate resistance to chewing and piercing–sucking herbivores in rice. Physiol. Plant. 152, 59–69. [DOI] [PubMed] [Google Scholar]
- Zuckerkandl, E. and Pauling, L. (1965) Evolutionary divergence and convergence in proteins In: Evolving Genes and Proteins (Bryson V. and Vogel H.J., eds), pp. 97–166. New York: Academic Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 The biosynthesis of selected oxylipins measured in this study based on current literature. No 8‐hydroperoxy fatty acid‐producing enzymes were detected in the genome of Fusarium verticillioides. FvLOX1 (FVEG 09897) and LOX2 (FVEG 03347) cluster with 13S‐LOX of Fusarium oxysporum (data not shown). Colours represent maize (green) or F. verticillioides (brown). HpODE, hydroperoxyoctadecadienoic acid; diHOME, dihydroxyoctadecenoic acid; HODE, hydroxyoctadecadienoic acid; oxo‐ODE, oxo‐octadecadienoic acid; epoOME, epoxyoctadecenoic acid; HOTrE, hydroxyoctadecatrienoic acid.
Fig. S2 Phylogenetic analysis of the Fusarium verticillioides fatty acid oxygenases. The evolutionary history was inferred using the maximum likelihood method based on the Poisson correction model (Zuckerkandl and Pauling, 1965). The bootstrap consensus tree inferred from 500 replicates (Felsenstein, 1985) is taken to represent the evolutionary history of the taxa analysed (Felsenstein, 1985). Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches (Felsenstein, 1985). Initial tree(s) for the heuristic search were obtained automatically by application of the Neighbour‐Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, followed by selection of the topology with the superior log likelihood value. The analysis involved 20 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 837 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 (Kumar et al., 2016). Nomenclature follows Sooman, Linda, and Ernst H. Oliw. “Discovery of a novel linoleate dioxygenase of Fusarium oxysporum and linoleate diol synthase of Colletotrichum graminicola.” Lipids 50.12 (2015): 1243–1252. https://onlinelibrary.wiley.com/doi/full/10.1007/s11745‐015‐4078‐9.
Table S1 Primers used in this study.


