Summary
In plants, the mitogen‐activated protein kinase (MAPK) cascades are the central signaling pathways of the complicated defense network triggered by the perception of pathogen‐associated molecular patterns to repel pathogens. The Arabidopsis thaliana MAPK phosphatase 1 (AtMKP1) negatively regulates the activation of MAPKs. Recently, the AtMKP1 homolog of Nicotiana benthamiana (NbMKP1) was found in association with the Bamboo mosaic virus (BaMV) replication complex. This study aimed to investigate the role of NbMKP1 in BaMV multiplication in N. benthamiana. Silencing of NbMKP1 increased accumulations of the BaMV‐encoded proteins and the viral genomic RNA, although the same condition reduced the infectivity of Pseudomonas syringae pv. tomato DC3000 in N. benthamiana. On the other hand, overexpression of NbMKP1 decreased the BaMV coat protein accumulation in a phosphatase activity‐dependent manner in protoplasts. NbMKP1 also negatively affected the in vitro RNA polymerase activity of the BaMV replication complex. Collectively, the activity of NbMKP1 seems to reduce BaMV multiplication, inconsistent with the negatively regulatory role of MKP1 in MAPK cascades in terms of warding off fungal and bacterial invasion. In addition, silencing of NbMKP1 increased the accumulation of Foxtail mosaic virus but decreased Potato virus X. The discrepant effects exerted by NbMKP1 on different pathogens foresee the difficulty to develop plants with broad‐spectrum resistance through genetically manipulating a single player in MAPK cascades.
Keywords: Bamboo mosaic virus, MAPK cascade, MAPK phosphatase, PAMP‐triggered immunity, Potexvirus, RNA virus replication
Introduction
Dynamic interplay between viruses and their hosts will determine the outcomes of viral invasion. Because of the limited genome sizes, viruses must take advantage of cellular machinery to complete their infection cycle, including entry, trafficking, translation, replication, viral particle assembly, and exit of the viruses from infected cells. On the other hand, hosts have evolved defense mechanisms to counteract the viral infection. In plants, RNA interference is a major antiviral defense to eliminate or attenuate the viral infection (Obbard et al., 2009). In addition, innate immunity triggered by the perception of virus‐encoded proteins may induce hypersensitive and systemic acquired resistance responses to limit the virus to infected cells and impart resistance to the non‐infected tissues (Mandadi and Scholthof, 2013).
Recognition of pathogen‐associated molecular patterns (PAMPs), generally defined as conserved molecular signatures existing in the entire classes of microbes and with an essential function for these microbes, by plasma membrane‐localized pathogen recognition receptors (PRRs) is a basic component of plant defense (Zipfel, 2009). PAMP perception by PRRs triggers an array of signaling pathways such as the production of reactive oxygen species (ROS) and the activation of mitogen‐activated protein kinases (MAPKs), which in turn activate target proteins, e.g. transcription factors and enzymes, through phosphorylation and, eventually, lead to the activation of defense‐related genes, the synthesis of camalexin, and the ROS‐mediated hypersensitive response‐like cell death, among other defense responses (Meng and Zhang, 2013; Rodriguez et al., 2010). The MAPK cascades, consisting of a series of protein phosphorylation events, attribute to three tiers of kinases. In plants, MAPKs are activated during PAMP responses by upstream MAPK kinases (MAPKKs) through the phosphorylation at the conserved Thr and Tyr residues in the T‐E/D‐Y activation motif of MAPKs (Hamel et al., 2006). MAPKKs, in turn, are activated by MAPKK kinases (MAPKKKs) through the phosphorylation of two Ser/Thr residues in the S/T‐x(3 − 5)‐S/T motif of the MAPKK activation loop (Hamel et al., 2006). MAPK3, MAPK4, and MAPK6 are known to be activated in response to PAMP elicitation in Arabidopsis thaliana. MAPK4 was reported to be a negative regulator of defense‐related signaling pathways as evidenced by the constitutive defense responses in mpk4 mutants (Petersen et al., 2000). In contrast, MAPK3 and MAPK6 regulate positively the plant defense responses (Asai et al., 2002; Ren et al., 2002, 2008 ).
To avoid deleterious effects on the cells due to excessive defense responses, the activation of MAPKs must be negatively regulated. Protein phosphatases are important components to modulate the magnitude, duration, and physical outcome of PAMP signaling through dephosphorylation of the activated MAPKs. In plants, the phosphatases implicated in PAMP responses include Ser/Thr phosphatases (PP2C family), protein Tyr phosphatases (PTPs), and MAPK phosphatases (MKPs). MKPs are dual specificity phosphatases (DSPs) that de‐phosphorylate both Thr and Tyr of the T‐E/D‐Y motif (Wang et al., 2007). Disruption of the gene encoding A. thaliana MAPK phosphatase 1 (AtMKP1) conferred a hypersensitivity phenotype in response to genotoxic stress treatments (UV‐C and methyl methanesulphonate) upon the mutant plant (Ulm et al., 2001). Furthermore, the mkp1 mutant A. thaliana showed constitutive defense responses, including elevated levels of salicylic acid, camalexin, pathogenesis‐related gene expression, and resistance to the bacterial pathogen Pseudomonas syringae pv. tomato (Pto) DC3000 (Bartels et al., 2009). Genetic analysis revealed that MAPK6, but not MAPK3, is required for the mkp1‐dependent enhanced resistance to Pto DC3000 and PAMP‐induced growth inhibition observed in mkp1 seedlings, suggesting that AtMKP1 regulates negatively the MAPK6‐mediated PAMP responses (Anderson et al., 2011).
Bamboo mosaic virus (BaMV) is one of the members of the genus Potexvirus. The ~6.4 kb (+)‐strand genome of BaMV contains five open reading frames (ORFs) plus a cap structure and a poly(A) tail at the 5’ and 3’ ends, respectively (Lin et al., 1994). ORF1 directs the synthesis of the 155‐kDa viral replication protein (REPBaMV), principally responsible for transcription/replication of the viral genome and the synthesis of the 5’ cap structure (Huang et al., 2005; Li et al., 1998, 2001a, 2001b ). Organization of the functional domains and the domain‐associated enzymatic activity of REPBaMV in terms of 5’ cap formation and replication were summarized in a recent review (Meng and Lee, 2017). ORF2, 3 and 4 are overlapped, collectively referred to as triple gene block (TGB), and their translated proteins, TGBp1, TGBp2 and TGBp3, respectively, are essential for the virus movement in plants (Lin et al., 2004, 2006 ). OFR5 encodes the viral coat protein (CP), the only structural protein required for virion assembly. The interaction between CP and REPBaMV also plays a critical role in cell‐to‐cell movement of the viral genome (Lee et al., 2011). An 836‐nt satellite RNA of BaMV (satBaMV) was found more than two decades ago (Lin and Hsu, 1994). Many satBaMV variants have hitherto been isolated from fields, all featured with a conserved apical hairpin stem loop structure critical for efficient replication of satBaMV by REPBaMV (Annamalai et al., 2003). The barely discernable REPBaMV in BaMV‐infected plants had undermined the efforts in search of host factors interacting with REPBaMV by using biochemical approaches. Thus, it was an important finding that the presence of satBaMV SF4 greatly enhanced the accumulation level of REPBaMV in N. benthamiana leaves through Agrobacterium tumefaciens mediated REPBaMV‐encoding gene transformation (Lee et al., 2011).
The replication efficiency of plant RNA viruses is modulated by a variety of host proteins, which may target the viral replication‐associated proteins or RNAs, modify the microenvironment of the viral replication complex (VRC), and others (Ahlquist et al., 2003; Hyodo and Okuno, 2014). In speaking of BaMV, a methyltransferase (PNbMTS1) and a heat shock 90 homolog (NbHsp90) have been evidenced to interact with REPBaMV by the yeast two‐hybrid assay. PNbMTS1 has an AdoMet‐dependent inhibitory effect on BaMV accumulation (Cheng et al., 2009), while NbHsp90 promotes BaMV accumulation presumably by facilitating the maturation of REPBaMV or assembly of the functional VRC (Huang et al., 2012). Chloroplast phosphoglycerate kinase (NbPGK), cytoplasmic glyceraldehyde 3‐phosphate dehydrogenase (NbGAPDH), and NbHsp90 are able to bind the 3’ untranslated region (UTR) of BaMV. NbPGK increases BaMV accumulation (Lin et al., 2007), whereas NbGAPDH decreases the production efficiency of the viral negative‐strand RNA presumably due to its strong occupation on the poly(A) tail of the 3’ UTR (Prasanth et al., 2011). The N. benthamiana glutathione S‐transferase 4 (NbGST4) enhances BaMV replication efficiency, particularly for the viral negative‐strand RNA synthesis, by creating a more reduced microenvironment in favor of the activity of VCR (Chen et al., 2013).
To search for more host proteins probably associated with the VRC of BaMV, a protocol was developed recently to obtain the putative VRC from N. benthamiana leaves, and the host proteins selectively associated with REPBaMV after immunoprecipitation were identified by tandem mass spectrometry preceded by polyacrylamide gel electrophoresis (Lee et al., 2016). Involvement of each of the identified proteins in BaMV replication was then assessed by challenging the target gene‐silenced N. benthamiana with the genetically modified BaMV, which carries a CP promoter‐driven green fluorescent protein (GFP) gene. Several proteins were considered to be host factors because silencing of the protein‐encoding genes changed the GFP imaging in the virus‐inoculated leaves. Among them, MAPK phosphatase (MKP)‐like protein (named NbMKP1 thereafter) raised our attention because it has a surprisingly negative effect on the accumulation of BaMV‐encoded GFP. Therefore, we set out to elaborate the function of NbMKP1 in multiplication of BaMV and two other potexviruses in N. benthamiana. The results suggest that BaMV and Foxtail mosaic virus (FoMV), but not Potato virus X (PVX), could benefit from the activation of MAPK signaling pathway, which is generally considered to be a defense response against pathogenic fungi and bacteria.
Results
Increase of BaMV in N. benthamiana by NbMKP1 silencing
The Tobacco rattle virus (TRV)‐based method (Ratcliff et al., 2001) was employed in this study for gene silencing experiments. To determine the effect of NbMKP1 on BaMV accumulation in N. benthamiana, the plant was agroinfiltrated with pTRV1/pTRV2‐NbMKP1 followed by BaMV inoculation as described in Experimental Procedures. The plant treated with pTRV1/pTRV2‐Luc served as a mock control while that with pTRV1/pTRV2‐PDS was used to monitor the gene silencing progress (Luc and PDS stand for luciferase and phytoene desaturase genes, respectively). Apparently, the NbMKP1‐silenced plant looked as normal as the Luc‐silenced plant at the time when the PDS‐silenced plant showed vast white spots on the newly developed leaves (Fig. 1A). Real‐time quantitative PCR (qPCR) analysis indicates that the average amount of NbMKP1 transcript in leaves of the NbMKP1‐silenced plants was about one third of that in the Luc‐silenced plants (Fig. 1B, left panel), confirming the silencing status of the NbMKP1‐silenced plants. qPCR also indicates that the accumulation of BaMV genome, at 4 days after BaMV inoculation, in the NbMKP1‐silenced plants was about 2 folds of that accumulated in the Luc‐silenced plants (Fig. 1B, right panel). Accumulation of BaMV in N. benthamiana in this study could also be assessed by the expression of GFP, because the genome of the virus had been engineered to contain a CP promoter‐driven GFP gene. The fluorescent images of the inoculated leaves taken off from the Luc‐ and NbMKP1‐silenced plants were scanned and the pixels obtained were compared with each other. The leaves from the NbMKP1‐silenced plants had 90% more pixels than that from the control plants (Fig. 2A). Furthermore, the accumulation of BaMV CP was determined by western blotting. The result indicates that the average amount of CP in the NbMKP1‐silenced plants was about 30 fold more than that in the Luc‐silenced plants (Fig. 2B). The much greater difference in the accumulation of CP than GFP between the NbMKP1‐silenced and the control plants may be attributed to the presumably slower turnover rate of CP in vivo. To confirm the involvement of NbMKP1 in the MAPK pathways, the phosphorylation status of MAPKs in leaves was probed by Phospho‐p44/42 MAPK (Erk1/2) (Thr202/Tyr204) rabbit monoclonal antibody, which has been used to detect A. thaliana MAPK3, MAPK6, MAPK4/11 (Bethke et al., 2012; González Besteiro and Ulm, 2013; González Besteiro et al., 2011). Indeed, the phosphorylated MAPKs were accumulated more in the NbMKP1‐silenced than the control plants, suggesting that NbMKP1 should have a regulatory role in the MAPK pathways, although the identity of those substrate proteins remains to be confirmed.
Figure 1.
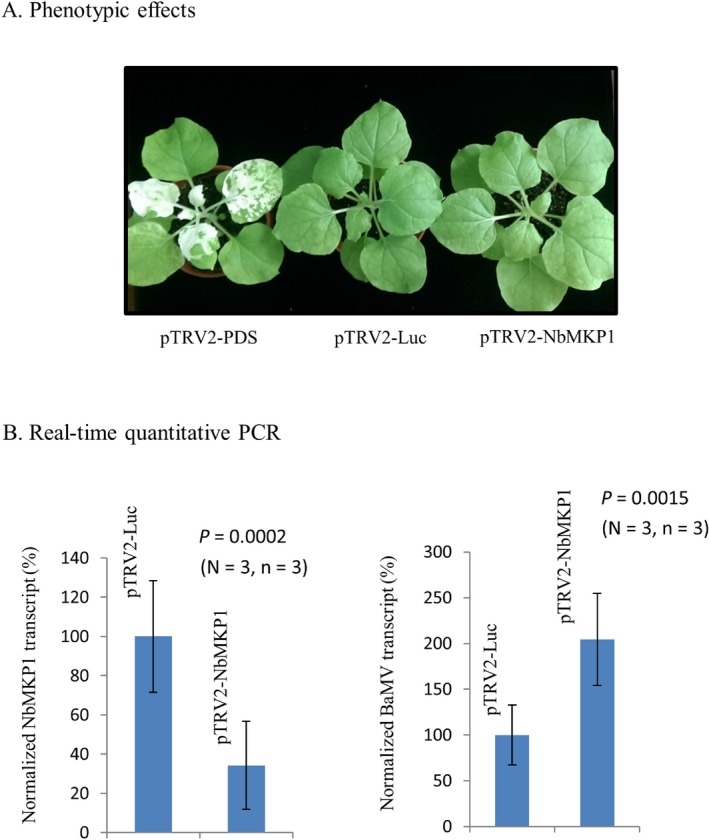
Increase of BaMV gRNA in the NbMKP1‐silenced N. benthamiana. Leaves of 4‐week‐old N. benthamiana were agroinfiltrated with pTRV1/pTRV2‐PDS, pTRV1/pTRV2‐Luc, or pTRV1/pTRV2‐NbMKP1 as described in Experimental Procedures. (A) Appearances of the silencing plants were photographed when the silencing effect of PDS was obvious. (B) The Luc‐ and NbMKP1‐silenced leaves were further inoculated with GFP‐carrying BaMV virions and the relative amounts of NbMKP1, BaMV genomic RNA, and β‐actin transcripts in the inoculated leaves were analyzed 4 days post‐inoculation by real‐time qPCR as described in Experimental Procedures. The statistical data points are the means of three independent experiments (N), each with three biological replicates (n).
Figure 2.
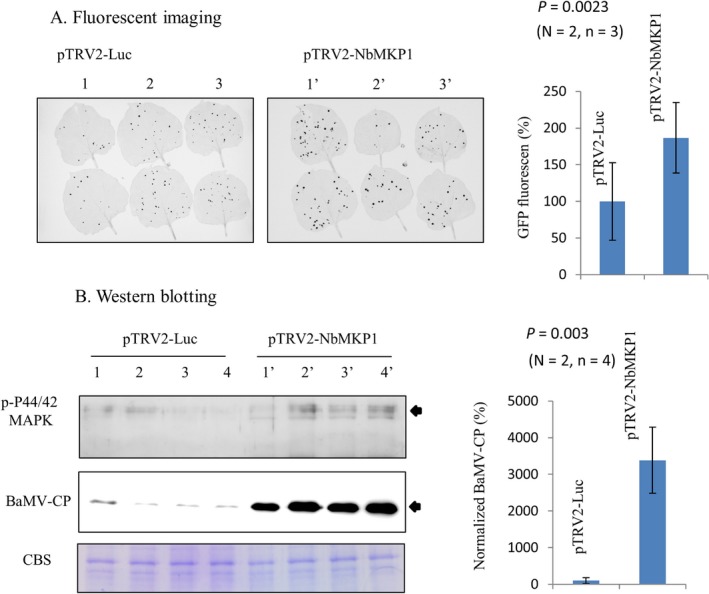
Increase of the BaMV‐encoded proteins in the NbMKP1‐silenced N. benthamiana. (A) Fluorescent images of BaMV‐inoculated leaves of N. benthamiana that had been agroinfiltrated with pTRV1/pTRV2‐Luc or pTRV1/pTRV2‐NbMKP1 prior to virus inoculation. Two leaves were inoculated per individual plant, with a total of six plants per independent experiment. The statistical data are the means of two independent experiments (N), with 3 plantlets (n) for each trial condition. (B) The relative accumulations of CP due to BaMV infection in the Luc‐ and NbMKP1‐silenced plants. The presence of activated MAPKs and the BaMV CP were detected by western blot using specific antibodies described in Experimental Procedures. The statistical data are the means of two independent experiments (N), each with four plantlets (n) per trial condition.
The NbMKP1‐silenced N. benthamiana was also inoculated with FoMV and PVX to determine if NbMKP1 exerts a common effect on other members of the genus Potexvirus. Accumulation of the FoMV CP was about 2.5 folds higher in the NbMKP1‐silenced plants than in the control (Fig. 3A). By contrast, NbMKP1 silencing had an opposite effect on PVX, an approximately 70% drop in the viral CP accumulation due to NbMKP1 silencing (Fig. 3B). The relative accumulation of PVX genomes in the inoculated leaves was further analyzed by Northern blotting to assure the suppression of PVX replication in the NbMKP1‐silenced leaves. The results indicate a significant drop, > 85%, in terms of PVX genomic RNA accumulation due to NbMKP1 silencing (Fig. 3C). This disagreement indicates that the effect exerted by NbMKP1 is different depending on the pathogen.
Figure 3.
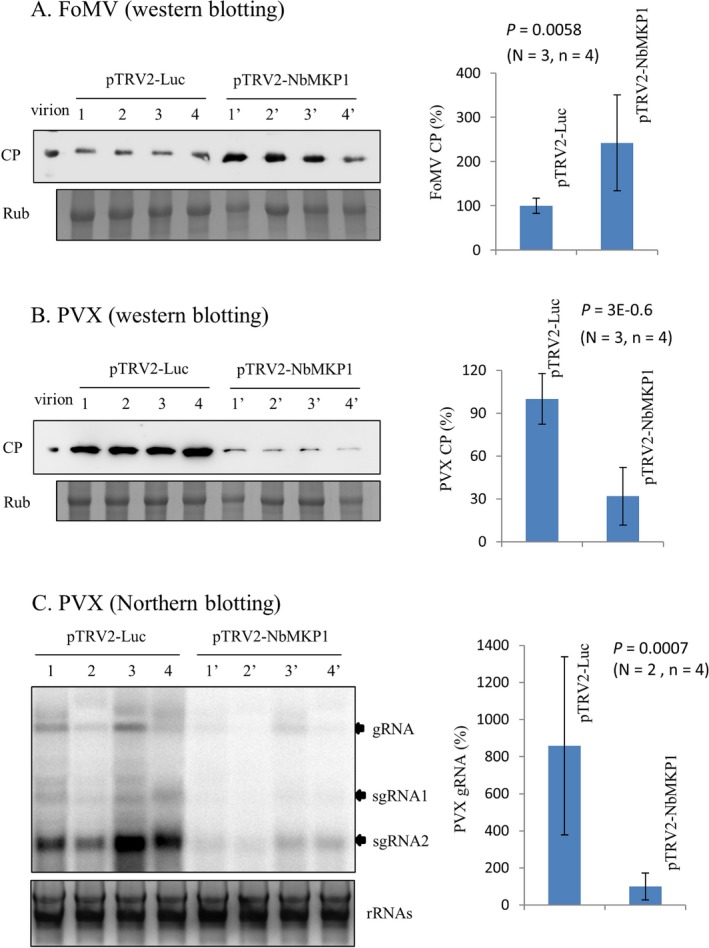
Differential effects of NbMKP1 on the accumulation of different potexviruses. The Luc‐ and NbMKP1‐silenced N. benthamiana plants were inoculated with Foxtail mosaic virus (FoMV) or Potato virus X (PVX). The relative accumulations of the viral CP in the inoculated leaves were analyzed by western blotting at 4 days after inoculation (Panel A, FoMV; Panel B, PVX), while that of the PVX genomes in the inoculated leaves were assayed by northern blotting at 2 days after inoculation (Panel C). The statistical data shown in the western and northern blotting assays are the means of three and two independent experiments (N), respectively. There were four plantlets (n) per trial condition.
Decrease of the BaMV CP in N. benthamiana protoplasts by NbMKP1 overexpression
The cDNA copy of NbMKP1 was cloned in our laboratory according to the information released by N. benthamiana Genome and Transcriptome Sequencing Consortium. This clone (GenBank accession number KX923320) contains 2580 base pairs encoding an 859 amino acid polypeptide as expected. It is notable that several nucleotide substitutions exist between the clone and the one released by the Consortium, denoted respectively as NbMKP1 and NbMKP1(QUT), with an identity of 98%. The substitutions result in 25 changes out of the 859 amino acid residues (Fig. 4). Nonetheless, about a half of the changes involve functionally conservative residues. As with other plant MKP1‐like proteins, Nicotiana tabacum MKP1 (NtMKP1), AtMKP1 and NbMKP1 belong to the dual specificity phosphatase family. The homologies between NbMKP1, NtMKP1 and AtMKP1 are about 94% and 52%, respectively. The phosphatase domain is located in the N terminus of the proteins featured with a catalytic motif H‐C‐x‐x‐G‐x‐x‐R.
Figure 4.

Comparison in amino acid sequences among NbMKP1, NbMKP1(QUT), NtMKP1, and AtMKP1. NbMKP1 was cloned in this study. NbMKP1(QUT) was retrieved from the database in Queensland University of Technology with the accession name Nbv5.1tr6400042, while NtMKP1 and AtMKP1 were retrieved from NCBI with accession numbers NP_001312232 and OAP02192, respectively. The residues in red are conserved among the proteins. Dots on top of the first sequence denote the residues that are different between NbMKP1 and NbMKP1(QUT). The residues underlined constitute the signature catalytic motif, H‐C‐x‐x‐G‐x‐x‐R, of the phosphatase domain, and the dagger symbol indicates the cysteine within the catalytic motif.
To examine the effect of NbMKP1 overexpression on BaMV accumulation, the NbMKP1 clone was inserted into plasmid pBI221, positioned downstream of the CaMV 35S promoter. The resulting plasmid pBI‐NbMKP1 was co‐introduced with the BaMV infectious clone pCBG into protoplasts prepared from healthy N. benthamiana leaves. Protoplasts co‐transfected with pBI221 and pCBG served as a control. The viral CP accumulated in to protoplasts was assayed 20 h post‐transfection by western blotting. The average accumulation of BaMV CP dropped 63% in protoplasts treated with pBI‐NbMKP1 in comparison with the control (Fig. 5A). Next, pBI‐NbMKP1 and pBI221 at different ratios (6:0, 1:5, and 0:6 g) were co‐transfected with pCBG into protoplasts to assure the NbMKP1 dependence of the accumulation of BaMV CP. The results show that the smallest amount of CP was found in protoplasts when the highest amount of pBI‐NbMKP1 was used (Fig. 5B).
Figure 5.
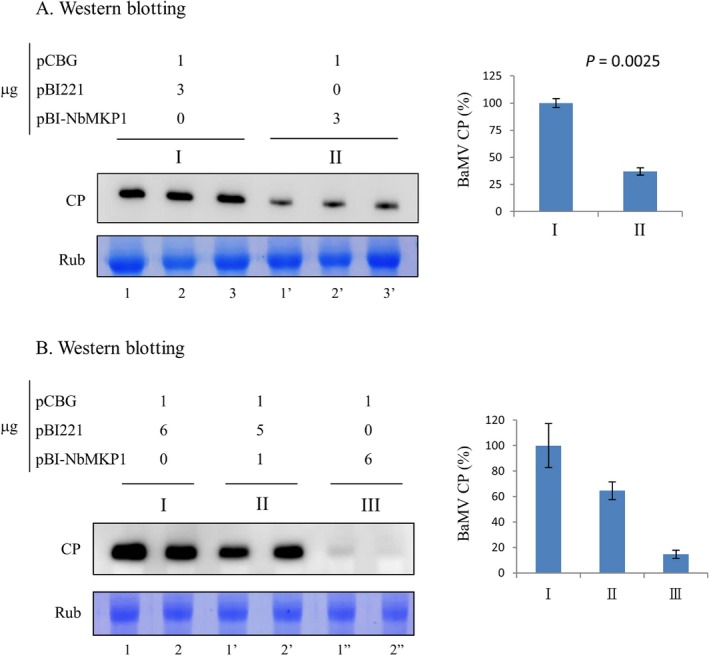
Decrease of the BaMV CP in the NbMKP1‐overexpressed protoplasts. (A) The BaMV infectious clone pCBG (1 μg) and the NbMKP1 expression plasmid pBI‐NbMKP1 at the indicated amounts were co‐transfected into 1 × 105 N. benthamiana protoplasts. The viral CP in the protoplasts was assayed 20 h post‐transfection. pBI221 was used to replace pBI‐NbMKP1 in the control. The experiment was performed in triplicate. (B) Indicated amounts of pCBG, pBI‐NbMKP1, and pBI221 were co‐transfected into protoplasts and the viral CP was assayed 20 h post‐transfection. The experiment was assayed in duplicate.
To assure that additional NbMKP1 was actually expressed from the exogenously transfected plasmid, a monoclonal antibody specifically recognizing the hemagglutinin tag (HA) was used in western blot to detect the presence of the C‐terminally HA‐fused NbMKP1 in pBI‐NbMKP1‐HA‐transfected protoplasts (Fig. 6). The result confirms the expression of NbMKP1‐HA, and shows that the BaMV CP dropped more than 70% under this condition in comparison with the pBI221‐transfected control. The catalytic cysteine (Cys235) in the phosphatase domain has been shown to be essential for AtMKP1 to relief MAPK activation (Ulm et al., 2002). To learn the relation of the phosphatase activity of NbMKP1 to BaMV accumulation, the corresponding cysteine residue (Cys201) was changed to serine and the mutational effect on the accumulation of BaMV CP was assayed. The C201S mutation not only abolished the inhibitory function of NbMKP1 in BaMV CP accumulation; it actually doubled the BaMV CP accumulation in comparison with the pBI221‐transfected control (Fig. 6). This result suggests that NbMKP1(C201S) was dominant over the endogenous NbMKP1. A competition between the inactive NbMKP1(C201S) and the endogenous NbMKP1 to bind the phosphoamino acid residues of their common activated MAPK substrates may cause this dominant negative effect.
Figure 6.
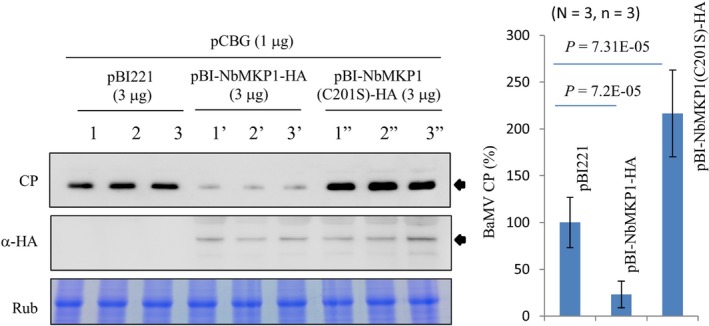
The effect of mutation in the catalytic cysteine residue on BaMV accumulation. Protoplasts (1 × 105) prepared from N. benthamiana leaves were co‐transfected with pCBG and pBI221, pBI‐NbMKP1‐HA, or pBI‐NbMKP1(C201S)‐HA as indicated. The BaMV CP in the protoplasts was assayed by western blotting 20 h post‐transfection. The expression of NbMKP1‐HA owing to the exogenously transfected plasmids was detected by western blotting using anti‐HA antibody. The statistical data are means of three independent experiments, each with three biological replicates.
Regulation of the RdRp activity of REPBaMV by NbMKP1
Results of both the silencing and overexpression experiments aforementioned indicate that the phosphatase activity of NbMKP1 decreased accumulation of the BaMV genome as well as the BaMV‐encoded proteins. Thus, it was interesting to determine whether the BaMV replication efficiency was actually affected by the phosphatase activity of NbMKP1. The REPBaMV‐enriched membrane fraction P30 was isolated from N. benthamiana at 2 days after the plant had been agroinfiltrated with pERep‐HA, pKSF4, and pEpNbMKP1 or pEpNbMKP1(C201S). pERep‐HA and pKSF4 were to express a C‐terminally HA‐fused REPBaMV and the satellite RNA SF4, respectively. Western blotting analysis indicates that agroinfiltration with pERep‐HA and pKSF4 was sufficient for plants to express REPBaMV‐HA in P30 and co‐expression of NbMKP1, either wild type or C201S mutant, did not considerably change the expression levels of REPBaMV (Fig. 7A). The inherent RNA‐dependent RNA polymerase (RdRp) activity of P30 using the endogenous SF4 RNA as the template was then initiated by adding NTP and [α‐32P]UTP into P30 suspension as the previous description (Lee et al., 2016). Apparently, the amount of the de novo synthesized α‐32P‐labeled SF4 decreased to ~ 40% compared with the control if the plant had also been agroinfiltrated with pEpNbMKP1 (Fig. 7B). Nonetheless, 50% more SF4 was produced if the plant had received pEpNbMKP1(C201S) compared with the control. These results confirm that the phosphatase activity of NbMKP1 has a negative impact on the RdRp activity of REPBaMV.
Figure 7.
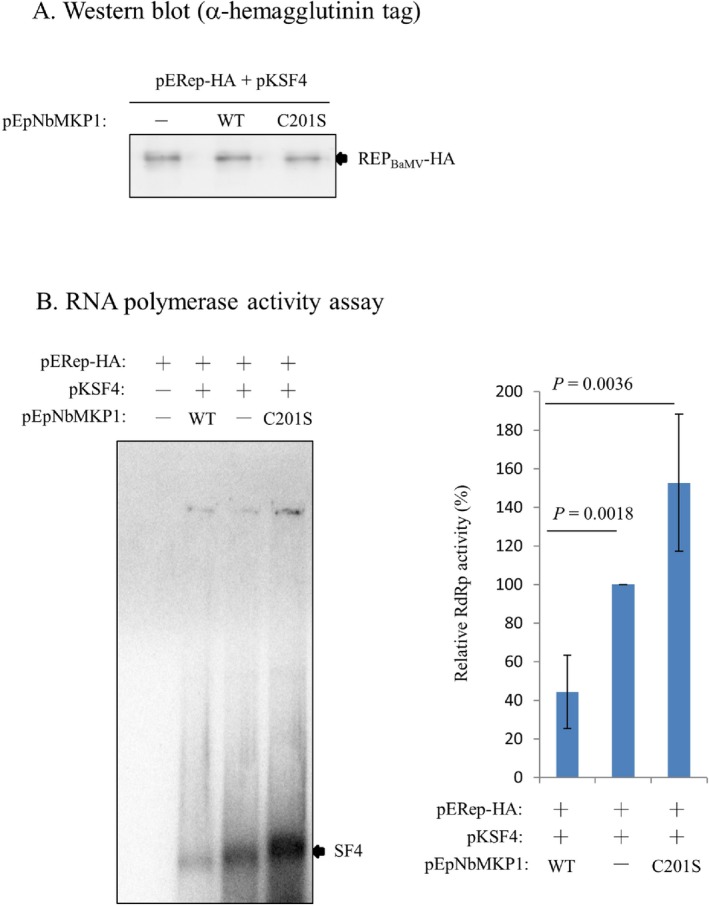
Regulation of the REPBaMV activity by NbMKP1. Cell extracts of N. benthamiana leaves, harvested on day 2 after agroinfiltration with binary plasmids pERep‐HA and pKSF4, in combination without or with plasmid pEpNbMKP1 or pEpNbMKP1(C201S), were subjected to centrifugation at 30000×g. The pellet (P30) after centrifugation were analyzed by (A) western blotting for the detection of REPBaMV‐HA and (B) in vitro RNA‐dependent RNA polymerase activity assay using the endogenous satellite RNA SF4 as the template. The statistical data are the means of three independent experiments.
Enhanced resistance to P. syringae by NbMKP1 silencing
AtMKP1 was reported to be a negative regulator of diverse PAMP responses such as activation of MAPK3 and MAPK6 and transient production of extracellular ROS (Anderson et al., 2011). Those authors also revealed that A. thaliana mkp1 seedlings and adult plants are more resistant to the virulent bacterial pathogen Pto DC3000. To determine whether NbMKP1 is functionally analogous to AtMKP1, the NbMKP1‐ and Luc‐silenced plants were infected with Pto DC3000 by infiltration at upper leaves. Necrosis zones, an indication of hypersensitive response, were visible only in the Luc‐silenced leaf by 18 h post‐infiltration (hpi), and a longer period was required for the NbMKP1‐silenced leaf to show cell necrosis (Fig. 8A). A delay of hypersensitive response indicates that the NbMKP1‐silenced plant displays a stronger basal defense against Pto DC3000. This notion is supported by the reports that plant innate immunity triggered by flagellin suppresses the hypersensitive response in non‐host tobacco leaves elicited by Pto DC3000 (Wei et al., 2012) and that N. benthamiana is considered to be a non‐host of Pto DC3000 (Mysore and Ryu, 2004). The bacterial numbers in the infected leaves were counted by plating the diluted leaf extract on agar plates selective for the growth of Pto DC3000. The average cell density at 18 hpi in the NbMKP1‐silenced leaves was only about 16% of that in the control leaves, further suggesting a stronger basal defense in the NbMKP1‐silenced plant (Fig. 8B). Collectively, NbMKP1 should have a similar role as AtMKP1 in PAMP responses.
Figure 8.
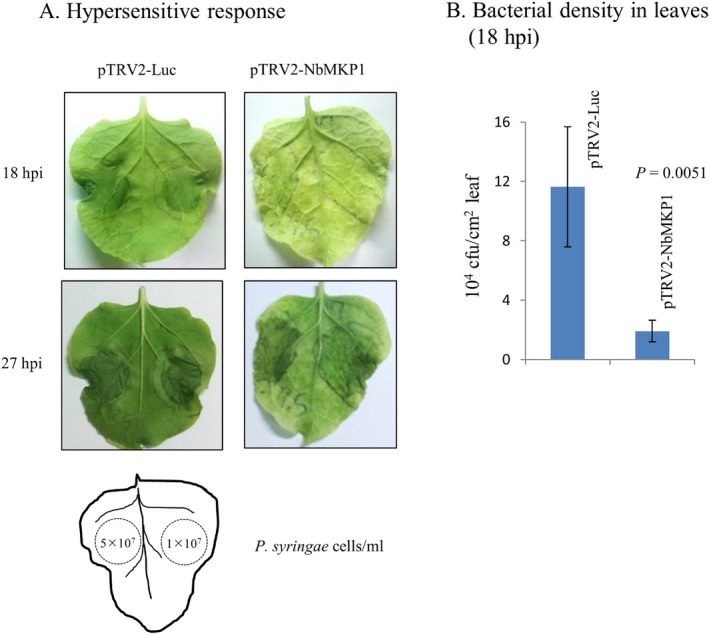
Increase in resistance to Pseudomonas syringae pv. tomato (Pto) DC3000 by NbMKP1 silencing. The Luc‐ and NbMKP1‐silenced N. benthamiana plants were infected with Pto DC3000 at concentrations of 5 × 107 and 1 × 107 cells/mL (left and right sides of the leaf, respectively) via leaf infiltration. (A) Emergence of the necrosis zones on leaves was photographed at 18 and 27 hours post‐infection (hpi). (B) The infiltration areas that had received Pto DC3000 at 1 × 107 cells/mL were cut off at 18 hpi and the cell density within was counted (colony forming units/cm2 leaf surface) as described in Experimental Procedures. The data are the means of those counted from four plantlets per trail condition.
Discussion
In plants, PAMP‐triggered immunity (PTI) and effector‐triggered immunity (ETI) deploy a multi‐layered defense against the invasion of fungi and bacteria. As the uncertainty about whether plant viruses possess molecular features meeting the definition of PAMPs, viruses have not always been included in the context of plant innate immunity models. Nonetheless, the concept of viruses being able to induce PTI/ETI in plants is increasingly generalized in recent review articles (Mandadi and Scholthof, 2013; Meng and Zhang, 2013; Nicaise, 2014), given that many important components of plant innate immunity are involved in antiviral defense mechanisms. These components include, but not limited to, (i) salicylic acid, induced by the recognition of plant R protein to pathogen effector, plays an important role in plant–virus interactions (Carr et al., 2010), (ii) MAPK4, a negatively regulatory player in plant PTI mechanisms, represses soybean defense against Bean pod mottle virus (Liu et al., 2011), and (iii) the receptor‐like protein kinases BAK1 and BKK1 (BAK1‐like kinase1), key activators of plant PRRs after PAMP perception, are required for A. thaliana resistance to plant viruses (Kørner et al., 2013; Yang et al., 2010). Nonetheless, a potential player of the innate immunity may have a positive role favorable for virus invasion as an elicitor‐inducible leucine‐rich repeat receptor‐like protein was found capable of assisting the movement of BaMV from cell to cell in N. benthamiana (Chen et al., 2017).
Plausibly, NbMKP1 is a negative regulator in PTI/ETI in N. benthamiana, because downregulation of the protein enhances the plant's basal defense against Pto DC3000 infection (Fig. 8). However, downregulation of NbMKP1 increases BaMV accumulation (Figs 1 and 2), while overexpression of the protein exerts an opposite effect (Figs 5 and 6). More specifically, the activity of NbMKP1 may result in a situation unfavorable for the RdRp activity of REPBaMV (Fig. 7). To our knowledge, this is the first demonstration of the involvement of MAPK phosphatase proteins in plant virus multiplication.
Based on the western blotting analysis using p44/42 MAPK antibody, a couple of phosphorylated MAPKs appeared when NbMKP1 was silenced (Fig. 2). This observation confirms the involvement of NbMKP1 in MAPKs cascades. Although it is unclear about what the phosphorylated MAPKs are and how NbMKP1 affects BaMV replication, possible explanations are discussed here according to the established models of plant MAPK cascades. AtMKP1 participates in the regulation of PTI/ETI signaling in A. thaliana through dephosphorylation of the activated AtMAPK3 and AtMAPK6; hence it is logical to assume a similar role of NbMKP1 in the innate immune systems of N. benthamiana. If this is the case, activation of the molecular targets of NbMKP1, presumably NbMAPK3/NbMAPK6, would benefit BaMV replication through MAPK‐activated downstream events, of which the generation of ROS may take part in what we have observed in this study. In N. benthamiana, a respiratory burst oxidase homolog B (RbohB) produces ROS in response to PAMPs to inhibit pathogen infection (Segonzac et al., 2011; Yoshioka et al., 2003). Recently, this RbohB‐derived ROS was found to be required for robust genome replication of Red clover necrotic mosaic virus (RCNMV) and Brome mosaic virus (Hyodo et al., 2017). The positive role of ROS in viral RNA replication was also observed in animal RNA viruses. For instance, mitochondrial ROS generation is required for efficient replication of enterovirus 71 (Cheng et al., 2014) and oxidative stress positively affects RNA genome replication of flavivirus and alphavirus (Gullberg et al., 2015). These findings prompt us to speculate that a similar ROS‐promoted RNA replication also occurs in BaMV. Another possibility is that the downstream events of activated MAPKs promote the expression or activation of some BaMV replication‐supporting host proteins. Although elucidating the detailed mechanism regarding how NbMKP1 regulates BaMV replication remains a challenging task, it should be an interesting issue for future research.
It is noteworthy that the silencing effect of NbMKP1 on viral RNA replication is not a universal phenomenon in the genus Potexvirus. Similarly to BaMV, the FoMV CP was accumulated more in the NbMKP1‐silenced N. benthamiana. By contrast, NbMKP1 silencing had an opposite effect on PVX. The mechanism underlying this discrepancy is still elusive, despite it is known that BaMV is more close to FoMV phylogenetically. From an application viewpoint, this discrepancy suggests that development of plants with broad‐spectrum resistance against different pathogens through manipulating PTI/ETI innate immunity systems may not be an easy task.
Experimental Procedures
Plasmids
According to the nucleotide (nt) information of NbMKP1 (Nbv5.1tr6400042) provided by N. benthamiana Genome and Transcriptome Sequencing Consortium (https://benthgenome.qut.edu.au/), the cDNA of NbMKP1 was amplified from a cDNA preparation of 6‐week‐old N. benthamiana leaves by PCR using primers 5'‐AACATCCCGGGATGTTGGGGGTAGATGAGAAGGACAGGGTA and 5'‐CATATGAGCTCCTAATATGTCTTCTCAAGTGAGAAACAAGGCAGATA. The amplified cDNA fragment was used to replace the β‐glucuronidase gene within the transient protein expression vector pBI221 using SmaI and SacI sites. The resulting plasmid pBI‐NbMKP1 was used to express NbMKP1 in N. benthamiana protoplasts. Substitution of serine for Cys201 of NbMKP1 produced by pBI‐NbMKP1 was performed according to the QuikChange site‐directed mutagenesis protocol using a pair of divergent primers 5’‐AGCTACCAGGGAGTGTCCCGA and 5’‐GTGCACAAAAACTCTCCCACCTTG. The HA‐coding sequence was added in frame to the 3’ end of NbMKP1 within pBI‐NbMKP1 or pBI‐NbMKP1(C201S) by the similar PCR‐based technique using a pair of divergent primers 5'‐CCAGATTACGCATAGGAGCTCGAATTTCCCCG and 5’‐AACGTCGTATGGATAATATGTCTTCTCAAGTGA. The cDNA copies of NbMKP1, both wild type and the C201S mutant, were also inserted into the binary plasmid pEpyon‐32K to become pEpNbMKP1 and pEpNbMKP1(C201S), which were used to overexpress NbMKP1 in plant leaves via agroinfiltration. For NbMKP1‐silencing experiments, a 212‐nt cDNA fragment of NbMKP1 (592‐803 nts) was inserted into pTRV2 via restriction sites EcoRI and XhoI (Lee et al., 2016). The BaMV infectious clone pCBG was used to direct the constitutive synthesis of a GFP‐carrying BaMV transcript in plant cells (Lin et al., 2004). The binary plasmids pERep‐HA and pKSF4 were used to produce REPBaMV‐HA and the BaMV satellite RNA SF4, respectively, in N. benthamiana leaves via agroinfiltration as described previously (Lee et al., 2016).
TRV‐induced gene silencing
A. tumefaciens C58C1 carrying pTRV1 or pTRV2 derivative (pTRV2‐NbMKP1, pTRV2‐PDS, or pTRV2‐Luc) was cultivated in LB medium supplemented with 50 μg/ml kanamycin at 28C, 200 rpm, for 24 h. The harvested cells after centrifugation were suspended in 10 mM MES [pH5.5] buffer supplemented with 10 mM MgCl2 to an OD600 of 1. Cells that carry pTRV1 and the specified pTRV2 derivative, respectively, were mixed at equal volumes and infiltrated into the undersides of leaves of 4‐week‐old N. benthamiana. In general, the silencing effect appeared in 4 to 5 weeks as evidenced by the emergence of white spotted regions on leaves of the plants that had received pTRV1 and pTRV2‐PDS.
Virus inoculation
Virions (0.5 μg) of BaMV, FoMV, or PVX were inoculated mechanically into the leaves of the silencing plants as indicated by the aid of carborundum dust. The virus‐inoculated leaves were harvested 4–5 days post‐inoculation for viral protein and RNA analyses. The expression of GFP in leaves due to the replication of GFP‐carrying BaMV was recorded with the Fujifilm LAS‐4000 imager and the resulting pixels were quantified using the Multi Gauge Version 3.0 software (Fuji Photo Film Co., LTD).
Protoplast transfection
Polyethylene glycol 4000‐mediated transfection was used to introduce 1 μg pCBG plus the indicated amounts of plasmids (pBI221, pBI‐NbMKP1, or pBI‐NbMKP1(C201S)) into 1 × 105 protoplasts prepared from 5‐week‐old N. benthamiana leaves according to a protocol described previously (Cheng et al., 2009). The transfected protoplasts were cultivated at room temperature in growth buffer (0.55 M mannitol‐MES [pH5.7], 1 μM CuSO4, 1 μM KI, 1 mM MgSO4, 0.2 mM K2HPO4, 1 mM KNO3, 10 mM CaCl2, and 30 μg/ml cefatoxime) for 16–24 h under a constant light.
Protein analysis
Leaves (∼0.1 g) frozen in liquid nitrogen or protoplasts (1.5 × 105 cells) collected by centrifugation were homogenized in 200 μl cold extraction buffer (50 mM Tris [pH 7.9], 100 mM KCl, 1 mM EDTA, 20% (v/v) glycerol, and 1 mM DTT) with a hand held polytron. The clarified solution after centrifugation at 21000 ×g for 5 min was used for protein analysis. Proteins such as the specified viral CP and the HA‐fused REPBaMV and NbMKP1 were detected by western blot analysis using anti‐CP and anti‐HA antibodies. The appearance of phosphorylated MAPKs in the leaf extract was detected by western blot using Phospho‐p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (197G2) rabbit monoclonal antibody #4377 (Cell Signaling Technology). The large subunit of ribulose‐1,5‐bisphosphate carboxylase (Rub) in samples, shown in the Coomassie blue‐stained 12% SDS‐PAGE, was used as an internal control for normalization. Imaging data of western blot and PAGE was processed with the Fujifilm LAS‐4000 imager and the resulting pixels were quantified using the Multi Gauge software.
RNA analysis
To prepare real‐time qPCR analysis, the total RNA of the collected leaves was isolated using TriPure Isolation Reagent and primed to synthesize cDNA using an oligo(dT)15 primer and MMLV Reverse Transcriptase (Promega, Fitchburg, WI). For each qPCR, 4 l of 4‐fold‐diluted cDNA was used with 7.5 l Kapa Sybr Fast qPCR Master Mix (Kapa Biosystems, Wilmington, MA), and the following paired primers (each 0.13 M) in a final 15 l solution: 5’‐CGTATGCACGGTCTGTTTCC and 5’‐TAGGCCATATGCCAAGATCA for NbMKP1 transcript (50–223 nts), 5’‐CACATCCGGCACTTACCA and 5’‐ATGTATCACGGAAATAAGAGTT for BaMV genome (1765–2001 nts), and 5’‐GATGAAGATACTCACAGAAAGA and 5’‐GTGGTTTCATGAATGCCAGCA for β‐actin transcript. The latter was used as an internal control for normalization of the expression of NbMKP1 and BaMV genome. The cycling condition began with an initial hold at 95°C for 3 min, followed by 30 cycles of 3 s at 95°C, 20 s at 58°C, and 20 s at 72°C. Reactions, including data collection, were carried out with the TOptical Gradient 96 Real‐Time Thermocycler (Biometra GmbH, Germany). For Northern blotting analysis, total RNA was extracted from PVX‐inoculated leaves on day 2 post inoculation. The RNA samples were denatured in buffer containing 10 mM phosphate, pH 6.3, and 1.2 M glyoxal, electrophoresed on agarose gels, and transferred onto nylon membrane. The membrane was then hybridized with a 32P‐labeled RNA probe specifically recognizing the PVX (+) 3’‐terminal sequence (Huang et al., 2012).
RdRp activity assay
The REPBaMV‐enriched membrane fraction (P30) was prepared from N. benthamiana leaves on day 2 after agroinfiltration with binary plasmids pERep‐HA, pKSF4, and pEpNbMKP1 or pEpNbMKP1(C201S) as indicated according to the method described previously (Lee et al., 2016). The RdRp activity assay using the satellite RNA SF4, endogenously present in P30, as the template was carried out by incubating an aliquot of P30 suspension in a solution that contained 30 mM Tris [pH8.8], 10 mM MgCl2, 50 mM NaCl, 20 mM dithiothreitol, 2 mM ATP, 2 mM CTP, 2 mM GTP, 2 μM UTP, 1.5 μl [α‐32P]UTP (6000 Ci/mmol, Perkin Elmer), and 1.5 μl RNase inhibitor. After incubation at 26C for 3 h, the reaction solution was extracted twice with an equal volume of phenol/chloroform (pH 4.5), and the nucleic acids within were precipitated with ethanol. The radiolabeled RNA products, generated by the RdRp activity of REPBaMV, were separated on a 1% agarose gel and analyzed with the Fujifilm BAS‐2500 phosphoimager.
Pseudomonas syringae infection
Pseudomonas syringae pv. tomato DC3000 was cultivated on King's B medium agar plate (20 g/L peptone, 1.5 g/L K2HPO4, 10 g/L glycerol, 5 mM MgSO4, and 1.5% agar, pH 7.2) at 30C. The cells, collected from the agar plates, were suspended in 10 mM MgCl2 solution to 5 × 107 and 1 × 107 cells/ml, and infiltrated into the undersides of the tested leaves of N. benthamiana using a blunt syringe. The appearance of cell necrosis zone on leaves was monitored over time after infiltration. The cell density of Pto DC3000 in leaves was determined by cutting leaf disks with a boring tool (inner diameter 0.7 cm) and placing the plant material in 500 µl of 10 mM MgCl2. The disks were homogenized and the resulting suspension was diluted and plated on King's B medium plates supplemented with 50 µg/ml rifampicin, 2 µg/ml cycloheximide, and 20 µg/ml nalidixic acid. Bacterial colony forming units (CFU) on the selective agar plates were counted 2 days after incubation at 30°C.
Statistical analysis
The significance of comparative data was analyzed statistically by Student's t‐test (tail = 1 type = 1).
Acknowledgements
This work was supported by grants MOST 103‐2321‐B‐005‐003 from the Ministry of Science and Technology, Taiwan, ROC. There is no conflict of interest regarding this study.
References
- Ahlquist, P. , Noueiry, A.O. , Lee, W.‐M. , Kushner, D.B. and Dye, B.T. (2003) Host factors in positive‐strand RNA virus genome replication. J. Virol. 77, 8181–8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J.C. , Bartels, S. , González Besteiro, M.A. , Shahollari, B. , Ulm, R. and Peck, S.C. (2011) Arabidopsis MAP Kinase Phosphatase 1 (AtMKP1) negatively regulates MPK6‐mediated PAMP responses and resistance against bacteria. Plant J. 67, 258–268. [DOI] [PubMed] [Google Scholar]
- Annamalai, P. , Hsu, Y.‐H. , Liu, Y.‐P. , Tsai, C.‐H. and Lin, N.‐S. (2003) Structural and mutational analyses of cis‐acting sequences in the 5’‐untranslated region of satellite RNA of bamboo mosaic potexvirus. Virology 311, 229–239. [DOI] [PubMed] [Google Scholar]
- Asai, T. , Tena, G. , Plotnikova, J. , Willmann, M.R. , Chiu, W.L. , Gomez‐Gomez, L. , Boller, T. , Ausubel, F.M. and Sheen, J. (2002) MAP kinase signaling cascade in Arabidopsis innate immunity. Nature 415, 977–983. [DOI] [PubMed] [Google Scholar]
- Bartels, S. , Anderson, J.C. , González Besteiro, M.A. , Carreri, A. , Hirt, H. , Buchala, A. , Métraux, J.P. , Peck, S.C. and Ulm, R. (2009) MAP kinase phosphatase1 and protein tyrosine phosphatase1 are repressors of salicylic acid synthesis and SNC1‐mediated responses in Arabidopsis. Plant Cell, 21, 2884–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke, G. , Pecher, P. , Eschen‐Lippold, L. , Tsuda, K. , Katagiri, F. , Glazebrook, J. , Scheel, D. and Lee, J. (2012) Activation of the Arabidopsis thaliana mitogen‐activated protein kinase MPK11 by the flagellin‐derived elicitor peptide, flg22. Mol. Plant Microbe Interact. 25, 471–480. [DOI] [PubMed] [Google Scholar]
- Carr, J.P. , Lewsey, M.G. and Palukaitis, P. (2010) Signaling in induced resistance. Adv. Virus Res. 76, 57–121. [DOI] [PubMed] [Google Scholar]
- Chen, I.‐H. , Chiu, M.‐H. , Cheng, S.‐F. , Hsu, Y.‐H. and Tsai, C.‐H. (2013) The glutathione transferase of Nicotiana benthamiana (NbGSTU4) plays a role in regulating the early replication of Bamboo mosaic virus . New Phytol. 199, 749–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, I.‐H. , Huang, Y.‐P. , Tseng, C.‐H. , Ni, J.‐T. , Tsai, C.‐H. , Hsu, Y.‐H. and Tsai, C.‐H. (2017) Nicotiana benthamiana elicitor‐inducible leucine‐rich repeat receptor‐like protein assists Bamboo mosaic virus cell‐to‐cell movement. Front. Plant Sci. 8, 1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, C.‐W. , Hsiao, Y.‐Y. , Wu, H.‐C. , Chuang, C.‐M. , Chen, J.‐S. , Tsai, C.‐H. , Hsu, Y.‐H. , Wu, Y.‐C. , Lee, C.‐C. and Meng, M. (2009) Suppression of Bamboo mosaic virus accumulation by a putative methyltransferase in Nicotiana benthamiana . J. Virol. 83, 5796–5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, M.‐L. , Weng, S.‐F. , Kuo, C.‐H. and Ho, H.‐Y. (2014) Enterovirus 71 induces mitochondrial reactive oxygen species generation that is required for efficient replication. PLoS One, 9, e113234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González Besteiro, M.A. and Ulm, R. (2013) Phosphorylation and stabilization of Arabidopsis MAP kinase phosphatase 1 in response to UV‐B stress. J. Biol. Chem. 288, 480–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González Besteiro, M.A. , Bartels, S. , Albert, A. and Ulm, R. (2011) Arabidopsis MAP kinase phosphatase 1 and its target MAP kinases 3 and 6 antagonistically determine UV‐B stress tolerance, independent of the UVR8 photoreceptor pathway. Plant J. 68, 727–737. [DOI] [PubMed] [Google Scholar]
- Gullberg, R.C. , Jordan Steel, J. , Moon, S.L. , Soltani, E. and Geiss, B.J. (2015) Oxidative stress influences positive strand RNA virus genome synthesis and capping. Virology 475, 219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel, L.P. , Nicole, M.C. , Sritubtim, S. , Morency, M.J. , Ellis, M. , Ehlting, J. , Beaudoin, N. , Barbazuk, B. , Klessig, D. , Lee, J. , Martin, G. , Mundy, J. , Ohashi, Y. , Scheel, D. , Sheen, J. , Xing, T. , Zhang, S. , Seguin, A. and Ellis, B.E. (2006) Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 11, 192–198. [DOI] [PubMed] [Google Scholar]
- Huang, Y.‐L. , Hsu, Y.‐H. , Han, Y.‐T. and Meng, M. (2005) mRNA guanylation catalyzed by the S‐adenosylmethionine‐dependent guanylyltransferase of Bamboo mosaic virus . J. Biol. Chem. 280, 13153–13162. [DOI] [PubMed] [Google Scholar]
- Huang, Y.‐W. , Hu, C.‐C. , Liou, M.‐R. , Chang, B.‐Y. , Tsai, C.‐H. , Meng, M. , Lin, N.‐S. and Hsu, Y.‐H. (2012) Hsp90 interacts specifically with viral RNA and differentially regulates replication initiation of Bamboo mosaic virus and associated satellite RNA. PLoS Pathog. 8, e1002726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyodo, K. , Hashimoto, K. , Kuchitsu, K. , Suzuki, N. and Okuno, T. (2017) Harnessing host ROS‐generating machinery for the robust genome replication of a plant RNA virus. Proc. Natl. Acad. Sci. USA, 114, E1282–E1290. USA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyodo, K. and Okuno, T. (2014) Host factors used by positive‐strand RNA plant viruses for genome replication. J. Gen. Plant Pathol. 80, 123–135. [Google Scholar]
- Kørner, C.J. , Klauser, D. , Niehl, A. , Domínguez‐Ferreras, A. , Chinchilla, D. , Boller, T. , Heinlein, M. and Hann, D.R. (2013) The immunity regulator BAK1 contributes to resistance against diverse RNA viruses. Mol. Plant Microbe Interact. 26, 1271–1280. [DOI] [PubMed] [Google Scholar]
- Lee, C.‐C. , Ho, Y.‐N. , Hu, R.‐H. , Yen, Y.‐T. , Wang, Z.‐C. , Lee, Y.‐C. , Hsu, Y.‐H. and Meng, M. (2011) The interaction between Bamboo mosaic virus replication protein and coat protein is critical for virus movement in plant hosts. J. Virol. 85, 12022–12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C.‐C. , Lin, T.‐L. , Lin, J.‐W. , Han, Y.‐T. , Huang, Y.‐T. , Hsu, Y.‐H. and Meng, M. (2016) Promotion of Bamboo mosaic virus accumulation in Nicotiana benthamiana by 5’3’ exonuclease NbXRN4. Front. Microbiol. 6, 1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y.‐I. , Cheng, Y.‐M. , Huang, Y.‐L. , Tsai, C.‐H. , Hsu, Y.‐H. and Meng, M. (1998) Identification and characterization of the E. coli‐expressed RNA‐dependent RNA polymerase of Bamboo mosaic virus . J. Virol. 72, 10093–10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y.‐I. , Chen, Y.‐J. , Hsu, Y.‐H. and Meng, M. (2001a) Characterization of the AdoMet‐dependent guanylyltransferase activity that is associated with the N terminus of Bamboo mosaic virus replicase. J. Virol. 75, 782–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y.‐I. , Shih, T.‐W. , Hsu, Y.‐H. , Han, Y.‐T. , Huang, Y.‐L. and Meng, M. (2001b) The helicase‐like domain of plant potexvirus replicase participates in the formation of 5’ cap structure of RNA by exhibiting an RNA 5’‐triphosphatase activity. J. Virol. 75, 12114–12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, M.‐K. , Chang, B.‐Y. , Liao, J.‐T. , Lin, N.‐S. and Hsu, Y.‐H. (2004) Arg‐16 and Arg‐21 in the N‐terminal region of the triple‐gene‐block protein 1 of Bamboo mosaic virus are essential for virus movement. J. Gen. Virol. 85, 251–259. [DOI] [PubMed] [Google Scholar]
- Lin, J.‐W. , Ding, M.‐P. , Hsu, Y.‐H. and Tsai, C.‐H. (2007) Chloroplast phosphoglycerate kinase, a gluconeogenetic enzyme, is required for efficient accumulation of Bamboo mosaic virus . Nucleic Acids Res. 35, 424–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, N.‐S. and Hsu, Y.‐H. (1994) A satellite RNA associated with bamboo mosaic potexvirus. Virology 202, 707–714. [DOI] [PubMed] [Google Scholar]
- Lin, M.‐K. , Hu, C.‐C. , Lin, N.‐S. , Chang, B.‐Y. and Hsu, Y.‐H. (2006) Movement of potexviruses requires species specific interactions among the cognate triple gene block proteins, as revealed by a trans‐complementation assay based on the Bamboo mosaic virus satellite RNA‐mediated expression system. J. Gen. Virol. 87, 1357–1367. [DOI] [PubMed] [Google Scholar]
- Lin, N.‐S. , Lin, B.‐Y. , Lo, N.‐W. , Hu, C.‐C. , Chow, T.‐Y. and Hsu, Y.‐H. (1994) Nucleotide sequence of the genomic RNA of bamboo mosaic potexvirus. J. Gen. Virol. 75, 2513–2518. [DOI] [PubMed] [Google Scholar]
- Liu, J.‐Z. , Horstman, H.D. , Braun, E. , Graham, M.A. , Zhang, C. , Navarre, D. , Qiu, W.L. , Lee, Y. , Nettleton, D. , Hill, J.H. and Whitham, S.A. (2011) Soybean homologs of MPK4 negatively regulate defense responses and positively regulate growth and development. Plant Physiol. 157, 1363–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandadi, K.K. and Scholthof, K.B. (2013) Plant immune responses against viruses: how does a virus cause disease? Plant Cell 25, 1489–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, M. and Lee, C.‐C. (2017) Function and structural organization of the replication protein of Bamboo mosaic virus . Front. Microbiol. 8, 522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, X. and Zhang, S. (2013) MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51, 245–266. [DOI] [PubMed] [Google Scholar]
- Mysore, K.S. and Ryu, C.‐M. (2004) Nonhost resistance: how much do we know? Trends Plant Sci. 9, 97–104. [DOI] [PubMed] [Google Scholar]
- Nicaise, V. (2014) Crop immunity against viruses: outcomes and future challenges. Front Plant Sci. 5, 660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard, D.J. , Gordon, K.H. , Buck, A.H. and Jiggins, F.M. (2009) The evolution of RNAi as a defense against viruses and transposable elements. Philos.Trans. R. Soc. Lond B Biol. Sci. 364, 99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, M. , Brodersen, P. , Naested, H. , Andreasson, E. , Lindhart, U. , Johansen, B. , Nielsen, H.B. , Lacy, M. , Austin, M.J. , Parker, J.E. , Sharma, S.B. , Klessig, D.F. , Martienssen, R. , Mattsson, O. , Jensen, A.B. and Mundy, J. (2000) Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 103, 1111–1120. [DOI] [PubMed] [Google Scholar]
- Prasanth, K.R. , Huang, Y.‐W. , Liou, M.‐R. , Wang, Y.‐L. , Hu, C.‐C. , Tsai, C.‐H. , Meng, M. , Lin, N.‐S. and Hsu, Y.‐H. (2011) Glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) negatively regulates the replication of Bamboo mosaic virus and its associated satellite RNA. J. Virol. 85, 8829–8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff, F. , Martin‐Hernandez, A.M. and Baulcombe, D.C. (2001) Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25, 237–245. [DOI] [PubMed] [Google Scholar]
- Ren, D. , Yang, H. and Zhang, S. (2002) Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J. Biol. Chem. 277, 559–565. [DOI] [PubMed] [Google Scholar]
- Ren, D. , Liu, Y. , Yang, K.Y. , Han, L. , Mao, G. , Glazebrook, J. and Zhang, S. (2008) A fungal‐responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA, 105, 5638–5643. USA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, M.C. , Petersen, M. and Mundy, J. (2010) Mitogen‐activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61, 621–649. [DOI] [PubMed] [Google Scholar]
- Segonzac, C. , Feike, D. , Gimenez‐Ibanez, S. , Hann, D.R. , Zipfel, C. and Rathjen, J.P. (2011) Hierarchy and roles of pathogen‐associated molecular pattern‐induced responses in Nicotiana benthamiana . Plant Physiol. 156, 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm, R. , Revenkova, E. , di Sansebastiano, G.P. , Bechtold, N. and Paszkowski, J. (2001) Mitogen‐activated protein kinase phosphatase is required for genotoxic stress relief in Arabidopsis. Genes Dev. 15, 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm, R. , Ichimura, K. , Mizoguchi, T. , Peck, S.C. , Zhu, T. , Wang, X. , Shinozaki, K. and Paszkowski, J. (2002) Distinct regulation of salinity and genotoxic stress responses by Arabidopsis MAP kinase phosphatase 1. EMBO J. 21, 6483–6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Chevalier, D. , Larue, C. , Ki Cho, S. and Walker, J.C. (2007) The protein phosphatases and protein kinases of Arabidopsis thaliana . Arabidopsis Book, 5, e0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, C.‐F. , Hsu, S.‐T. , Deng, W.‐L. , Wen, Y.‐D. and Huang, H.‐C. (2012) Plant innate immunity induced by flagellin suppresses the hypersensitive response in non‐host plants elicited by Pseudomonas syringae pv. averrhoi . PLoS One, 7, e41056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H. , Gou, X. , He, K. , Xi, D. , Du, J. , Lin, H. and Li, J. (2010) BAK1 and BKK1 in Arabidopsis thaliana confer reduced susceptibility to turnip crinkle virus. Eur. J. Plant Pathol. 127, 149–156. [Google Scholar]
- Yoshioka, H. , Numata, N. , Nakajima, K. , Katou, S. , Kawakita, K. , Rowland, O. , Jones, J.D. and Doke, N. (2003) Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans . Plant Cell, 15, 706–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C. (2009) Early molecular events in PAMP‐triggered immunity. Curr. Opin. Plant Biol. 12, 414–420. [DOI] [PubMed] [Google Scholar]


