Summary
Spray‐induced gene silencing (SIGS) is an innovative strategy for crop protection. However, the mechanism of SIGS is not known. Here, we first demonstrate that secondary small interfering RNA (siRNA) amplification limits the application of SIGS. A myosin5 gene (Myo5) was chosen as the target of SIGS in an agronomically important pathogen—Fusarium asiaticum. Five segments corresponding to the different regions of the Myo5 gene were found to efficiently silence Myo5, resulting in cell wall defects, life cycle disruption and virulence reduction. Myo5‐8 (one of the Myo5 segments) induced sequence‐specific RNA interference (RNAi) activity in F. asiaticum, F. graminearum, F. tricinctum and F. oxysporum, but not in other fungi, in vitro. Remarkably, the silencing of Myo5 lasted for only 9 h unless the double‐stranded RNA (dsRNA) was continuously supplied, because F. asiaticum is unable to maintain siRNA amplification. After spraying on plants, dsRNAs were more efficiently taken up via the wounded surface. The antifungal activity of dsRNAs taken up by plant cells was higher and longer lasting than that dried onto the plant surface. In contrast with dsRNAs in fungi, dsRNAs in plant cells could efficiently turn into substantial siRNAs via secondary amplification machinery. Our findings provide new implications to develop SIGS as a mainstream disease control strategy against Fusarium and other fungi.
Keywords: dsRNA in vitro, Fusarium, RNA interference, spray‐induced gene silencing
Introduction
Since the earliest days of agriculture, humans have had to protect crops against yield losses from diseases caused by pathogens. The arms race between crops and pathogens continues (Lamberth et al., 2013). Amongst the top 10 fungal plant pathogens, Fusarium species represent a highly destructive genus of pathogens (Dean et al., 2012), infecting a broad range of crops and causing various crop diseases, including Fusarium head blight and seedling blight of wheat and barley, Gibberella ear rot and stalk rot of maize, bakanae of rice, and vascular diseases and wilts of tomato and cotton (Bai and Shaner, 2004; Ma et al., 2013). Mycotoxins produced during the progression of these diseases threaten humans and animals (Woloshuk and Shim, 2013). Natural resistance against Fusarium pathogens is inadequate and current protective measures against Fusarium species mainly rely on chemical fungicides, alterations to cultural practices and crop rotations (Bai and Shaner, 2004; Lamberth et al., 2013). As noted earlier, however, fungicide resistance, environmental hazards and health side‐effects limit chemical fungicide applications (van den Bosch and Gilligan, 2008; Verweij et al., 2009).
RNA interference (RNAi) is a conserved regulatory mechanism of gene expression in many eukaryotes (Baulcombe, 2004; Fire et al., 1998). RNAi technology has been used to control insect pests and virus diseases of plants and cellular diseases of humans, termed ‘host‐induced gene silencing’ (HIGS) (Baulcombe, 2015; Baum et al., 2007; Pooggin et al., 2003). Subsequently, HIGS has been developed in multiple crop systems to effectively control diseases caused by fungal and oomycete pathogens (Nunes and Dean, 2012). Our recent research has shown that wheat can be protected against Fusarium graminearum by HIGS (Cheng et al., 2015). However, the broad application of HIGS has been limited by the lack of available transformation protocols, the transformability and genetic stability of engineered RNA silencing traits, and public acceptance of the production of genetically modified crops (Machado et al., 2017; Wang and Jin, 2017).
Recently, a new innovative strategy (spray‐induced gene silencing, SIGS), which simulates HIGS, without the need to develop stably transformed plants, has been demonstrated to be effective in the control of both F. graminearum and Botrytis cinerea (Aline et al., 2016; Wang et al., 2016). SIGS technology demonstrates a potential paradigm shift in crop protection. The double‐stranded DNAs (dsRNAs) sprayed onto plant surfaces have two possible pathways of entry into fungal cells: RNAs can be taken up by plant cells first and then transferred into the fungal cells, and/or RNAs can be taken up by the fungal cells directly (Aline et al., 2016; Wang et al., 2016). These RNAs subsequently work in two different ways: the RNAs taken up by plant cells induce the plant RNAi machinery and then the fungal RNAi machinery; the RNAs taken up by fungal cells induce the fungal RNAi machinery directly. To improve the application strategies of SIGS, the two different SIGS machineries need to be studied in detail. Here, a myosin5 gene (Myo5) was chosen as the target of SIGS in Fusarium. Myo5, a class I myosin, is a molecular motor that binds to actin and hydrolyses ATP to produce physical force (Hartman et al., 2011). Most eukaryotes rely on class I myosins for eukaryotic cytokinesis, organelle transport and cell polarization (Bloemink and Geeves, 2011; Hartman et al., 2011; Hofmann et al., 2009). In Fusarium, Myo5 is an essential gene and cannot be knocked out without causing mortality (Zhang et al., 2015; Zheng et al., 2015). By screening for the most effective segment sequences against Myo5 in Fusarium asiaticum, we identified one RNAi segment, which was then used to synthesize dsRNA for the study of SIGS machineries in both fungal cells and plant cells.
Results
Generation of RNAi constructs from Myo5
Eight segments of the Myo5 cDNA (3645 bp) from the F. asiaticum strain were obtained for their corresponding RNAi constructs (pRNAiMyo5‐1, pRNAiMyo5‐2, pRNAiMyo5‐3, pRNAiMyo5‐4, pRNAiMyo5‐5, pRNAiMyo5‐6, pRNAiMyo5‐7 and pRNAiMyo5‐8). The RNAi vector was constructed with a transcriptional unit for hairpin RNA expression spaced by a cutinase gene intron from Magnaporthe oryzae (Nakayashiki et al., 2005) (Fig. S1, see Supporting Information). The eight RNAi constructs were individually introduced into the PLS1 locus of the F. asiaticum strain by protoplast transformation. PLS1 is dispensable in F. asiaticum and has been used previously as a target for the introduction of genes at an ectopic locus (Cheng et al., 2015; Song et al., 2014). Molecular characterization confirmed that all of the Myo5RNAi transformants contained an appropriate integration in the PLS1 gene site. The transformants were designated Myo5RNAi‐1 to Myo5RNAi‐8 (Fig. S2, see Supporting Information).
Screening of RNAi segments
Effects of knockdown of Myo5 gene on F. asiaticum cell wall and transport
To identify how the Myo5 gene functions in F. asiaticum growth and hyphal development, we assessed the growth of the Myo5RNAi transformants and the wild‐type (WT) strain on potato dextrose agar (PDA) with or without various agents. On non‐amended PDA, the Myo5RNAi‐3, Myo5RNAi‐4, Myo5RNAi‐5, Myo5RNAi‐7 and Myo5RNAi‐8 transformants grew slowly and their aerial hyphae were thick and short (Fig. 1A). The transformants Myo5RNAi‐1, Myo5RNAi‐2 and Myo5RNAi‐6 grew almost normally under the same conditions, suggesting that the effects differ depending on which part of the Myo5 gene is silenced.
Figure 1.
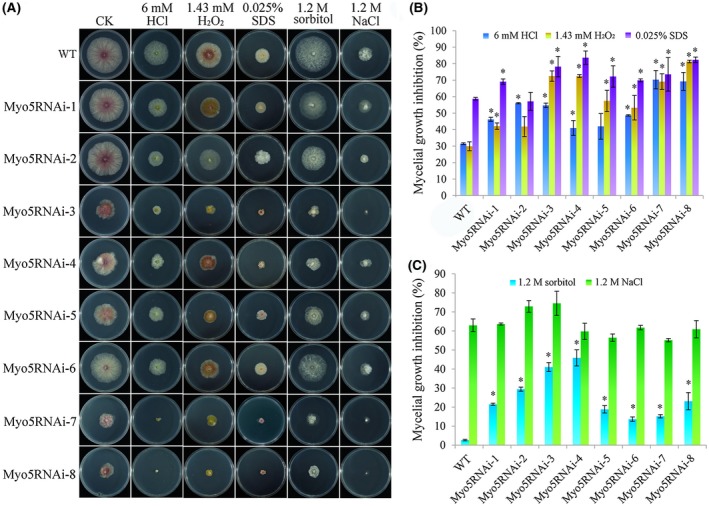
Sensitivity of wild‐type (WT) and Myo5RNAi [Myo5 gene RNA interference (RNAi) transformants] to osmotic and cell wall stress. (A) Growth of different strains on potato dextrose agar (PDA) plates or PDA amended with various compounds that generate osmotic and cell wall stress. Mycelial plugs were inoculated and cultured for 3 days at 25 °C in the dark. The experiment was repeated three times with similar results. CK, control; SDS, sodium dodecylsulfate. (B) Mycelial growth inhibition by compounds that generate cell wall stress. (C) Mycelial growth inhibition by compounds that generate osmotic stress. For (B) and (C), values are the means and standard errors of three experiments (*P < 0.05). [Colour figure can be viewed at wileyonlinelibrary.com]
On media containing solutes that increased the cell wall pressure [6 mm HCl, 1.43 mm H2O2 and 0.025% sodium dodecylsulfate (SDS)] (Song et al., 2014), the growth rate of Myo5RNAi transformants was strongly decreased (Fig. 1B). The inhibition of mycelial growth by HCl ranged from 41% to 70% for the Myo5RNAi transformants, but was 31% for the WT strain. Growth rates on H2O2‐ and SDS‐containing media were similar to those on HCl‐containing media. The morphology of the Myo5RNAi transformants suggested that interference with Myo5 caused defects in the cell wall that affected mycelial growth, and that the degree of defect differed amongst transformants.
The osmotic stress assays revealed that all eight Myo5RNAi transformants showed retarded growth in the presence of 1.2 m sorbitol (Fig. 1C); the growth inhibition caused by 1.2 m sorbitol ranged from 16% to 26% for the Myo5RNAi transformants, but was only 3% (±0.3%) for WT. The growth inhibition caused by 1.2 m NaCl did not differ significantly between the Myo5RNAi transformants and WT. This indicates that the growth inhibition caused by 1.2 m sorbitol could not be attributed only to osmotic stress. Unlike NaCl which induces osmotic stress, sorbitol is also a carbon source for fungi (Nakajima et al., 2016). We hypothesized that the transport of materials in the Myo5RNAi transformants was disturbed as a result of Myo5 interference.
To test our hypothesis, we examined the morphology of the conidia and hyphae of Myo5RNAi transformants using traditional microscopy and calcofluor white (CFW) staining (Fig. 2A). Microscopy revealed clear differences between the conidia of WT and those of Myo5RNAi‐2, Myo5RNAi‐3, Myo5RNAi‐4, Myo5RNAi‐5, Myo5RNAi‐7 and Myo5RNAi‐8 transformants. The WT conidia had four to five septa, whereas Myo5RNAi‐2, Myo5RNAi‐3, Myo5RNAi‐4, Myo5RNAi‐5, Myo5RNAi‐7 and Myo5RNAi‐8 transformants had only one to three septa, suggesting a possible defect in septum formation and distribution, as reported previously for an Myo2 mutant (Zheng et al., 2016). Myo5RNAi‐3, Myo5RNAi‐4, Myo5RNAi‐5, Myo5RNAi‐7 and Myo5RNAi‐8 displayed severely distorted and crooked mycelia with conglobate structures (Fig. 2A). It seemed that the cells of Myo5RNAi‐3, Myo5RNAi‐4, Myo5RNAi‐5, Myo5RNAi‐7 and Myo5RNAi‐8 transformants had expanded because of the abnormality of material transport and cell polarization caused by silencing of the Myo5 gene. Myo5 in Fusarium may also play a role in eukaryotic cytokinesis, organelle transport and cell polarization, as in most eukaryotes. More importantly, the hyphal cells of Myo5RNAi‐3, Myo5RNAi‐4, Myo5RNAi‐5, Myo5RNAi‐7 and Myo5RNAi‐8 transformants, but not WT, Myo5RNAi‐1, Myo5RNAi‐2 and Myo5RNAi‐6, contained many vacuoles (Fig. 2B), which might interfere with the transport of materials. To further confirm that chitin metabolism was disturbed in the Myo5RNAi transformants, the chitin content was measured. The results showed that the Myo5RNAi transformants produced 29.36%–40.67% chitin relative to its hyphal dry weight, which is significantly decreased compared with 59.26% for WT (Fig. 2C). Together, these data show that chitin metabolism is disturbed in the Myo5RNAi transformants, especially in Myo5RNAi‐3, Myo5RNAi‐4, Myo5RNAi‐5, Myo5RNAi‐7 and Myo5RNAi‐8, and results in abnormal cell structure, such as crooked mycelia and conglobate structures, caused by Myo5 silencing.
Figure 2.
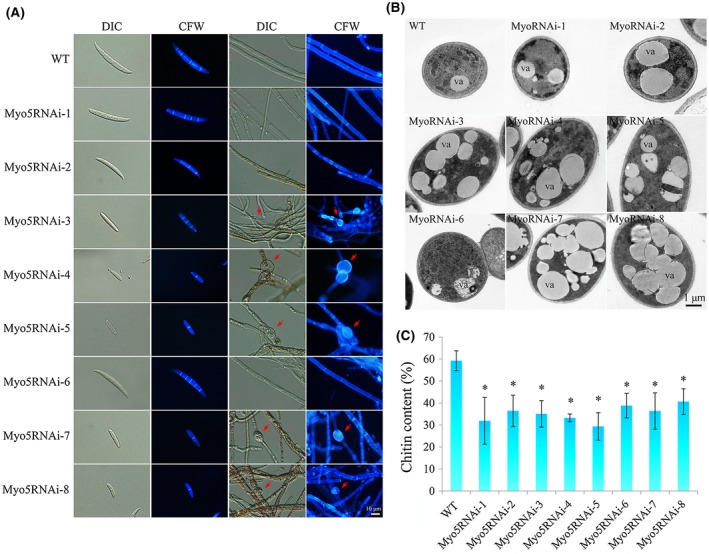
Analysis of spore and hyphal morphology, chitin distribution and chitin content. (A) Spore and hyphal morphology and chitin distribution. Macroconidia were collected from cultures that were maintained in carboxymethylcellulose (CMC) for 5 days (25 °C, 200 rpm) under constant fluorescent light. Hyphae of the wild‐type (WT) and Myo5RNAi strains were cultured in potato dextrose agar (PDA) for 3 days. Macroconidia and hyphae were stained with 10 μg/mL calcofluor white (CFW) and observed with an inverted fluorescence microscope (scale bar, 10 µm). The red arrows indicate the typical conglobate structures. DIC, Differential Interference Contrast. (B) Transmission electron microscopy of hyphae of the WT and Myo5RNAi transformants. Hyphae (3 days) were fixed with 2.5% (v/v) glutaraldehyde and post‐fixed with 1% (v/v) osmium tetroxide. va, vacuole. (C) Chitin content of fungal mycelia. Chitin content was measured at 72 h post‐inoculation with macroconidia and was expressed as a percentage of N‐acetylglucosamine (GlcNAc) relative to dry mycelial weight. The experiment was performed in triplicate. Values are the means and standard errors of three experiments (*P < 0.05). [Colour figure can be viewed at wileyonlinelibrary.com]
The life cycle and virulence of Myo5RNAi transformants
To further screen for Myo5RNAi segments that affect the life cycle of F. asiaticum, we assayed both asexual and sexual reproduction. Conidial production in carboxymethylcellulose (CMC) medium was similar for WT and the Myo5RNAi‐1, Myo5RNAi‐2 and Myo5RNAi‐6 transformants (Fig. 3A). In contrast, conidial production was significantly lower for Myo5RNAi‐3, Myo5RNAi‐4, Myo5RNAi‐5, Myo5RNAi‐7 and Myo5RNAi‐8 than for WT (Fig. 3A). Moreover, the conidia were shorter for the Myo5RNAi transformants than for WT (Fig. 3B). Sexual reproduction was also inhibited for the Myo5RNAi transformants. After 7 days on carrot agar, Myo5RNAi‐3, Myo5RNAi‐4, Myo5RNAi‐5, Myo5RNAi‐7 and Myo5RNAi‐8 failed to produce perithecia, but the other transformants and WT produced perithecia that were filled with ascospores (data not shown). After a further 14 days of cultivation, Myo5RNAi‐3, Myo5RNAi‐4, Myo5RNAi‐5, Myo5RNAi‐7 and Myo5RNAi‐8 produced some perithecia (Fig. 3C), but 46%–99% of the perithecia did not contain asci (Fig. 3D).
Figure 3.
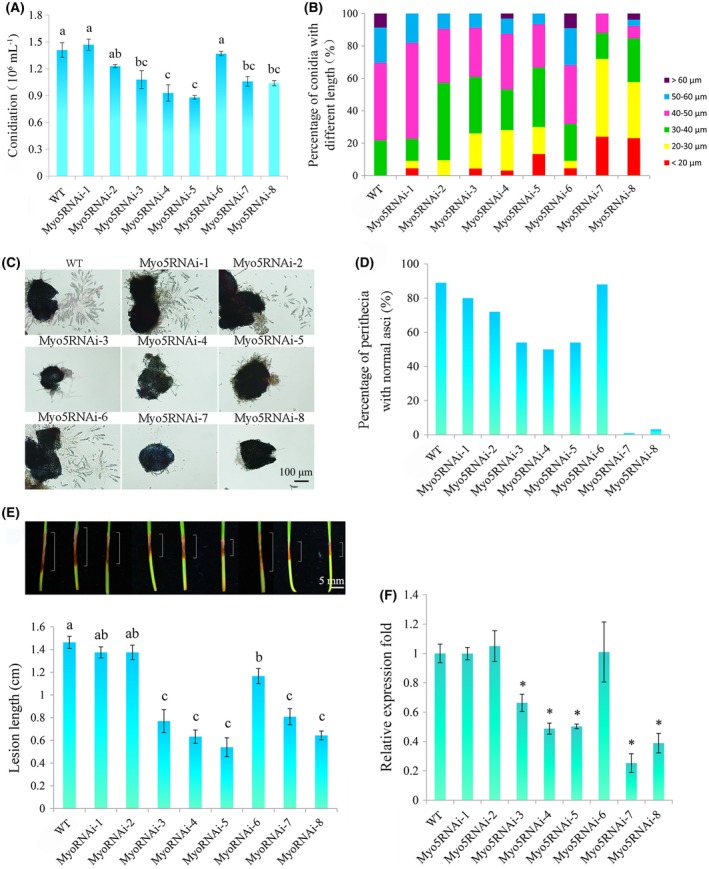
Conidiation, sexual reproduction, virulence and Myo5 gene expression of wild‐type (WT) and Myo5RNAi transformants. (A) Numbers of conidia produced in carboxymethylcellulose (CMC) medium after 5 days. Experiments were performed in triplicate. Different letters represent a significant difference at P < 0.05. (B) Lengths of conidia produced in CMC medium after 5 days. Two hundred conidia were examined for each strain. (C) Perithecia and asci rosettes. Images were captured 21 days after sexual induction. Scale bar, 100 µm. (D) Numbers of perithecia with normal asci at 21 days after sexual induction. One hundred perithecia were examined for each strain. (E) Virulence of WT and Myo5RNAi transformants on wheat seedlings as indicated by lesion length (see white lines in the photographs). Lesions were measured, and those of representative seedlings were photographed at 7 days post‐inoculation (dpi); the photographs are aligned with the bar graph. Values are means (± standard deviation, SD) of 30 replicate seedlings, and means with different letters are significantly different at P < 0.05. Scale bar, 5 mm. (F) Expression of the Myo5 gene in WT and Myo5RNAi strains. Total RNAs were extracted from 3‐day‐old mycelia. Gene expressional levels were normalized to the ubiquitin C‐terminal hydrolase gene. Values are means (± SD) of three independent biological duplicates. An asterisk indicates that the expression level was significantly lower (P < 0.05) in a transformant than in WT. [Colour figure can be viewed at wileyonlinelibrary.com]
The virulence of the Myo5RNAi transformants was determined by inoculation of seedlings of wheat cultivar Huaimai33. Lesion lengths did not differ significantly amongst WT and Myo5RNAi‐1 and Myo5RNAi‐2 at 7 days post‐inoculation (dpi) (Fig. 3E). In contrast, lesions were 20%–63% shorter for Myo5RNAi‐3, Myo5RNAi‐4, Myo5RNAi‐5, Myo5RNAi‐6, Myo5RNAi‐7 and Myo5RNAi‐8 than for WT (Fig. 3E). Quantitative real‐time polymerase chain reaction (qRT‐PCR) analyses confirmed that the expression of the Myo5 gene was significantly lower in Myo5RNAi‐3, Myo5RNAi‐4, Myo5RNAi‐5, Myo5RNAi‐7 and Myo5RNAi‐8 than in WT (Fig. 3F). These results demonstrate that silencing of certain parts of Myo5 interferes with the life cycle and virulence of F. asiaticum.
Effects of FaMyo5‐8 dsRNA on various Fusarium species and other fungi
Combining the above results, the Myo5‐8 segment sequence was chosen as the template of Myo5 dsRNA synthesis used in vitro. We subsequently assessed the in vitro RNAi effect of FaMyo5‐8 dsRNA against F. asiaticum (Fa), F. graminearum (Fg), F. tricinctum (Ft), F. oxysporum f. sp. lycopersici (Fo), F. verticillioides (Fv), Magnaporthe oryzae (Mo) and Botrytis cinerea (Bc). These fungi (Table S1, see Supporting Information) were closely related to F. asiaticum based on the phylogenetic analysis of the entire Myo5 gene (Fig. 4A). The similarities between the Myo5 gene of F. asiaticum and its homologues in the other fungi were as follows: 99% for F. graminearum, 82% for F. tricinctum, 84% for F. oxysporum f. sp. lycopersici, 82% for F. verticillioides, 76% for M. oryzae (91% query coverage) and 71% for B. cinerea (75% query coverage) (Fig. 4A). Microscopic analysis of fungal conidia treated with FaMyo5‐8 dsRNA revealed clear growth inhibition and abnormal mycelium with similar conglobate structures in F. asiaticum, F. graminearum, F. tricinctum and F. oxysporum f. sp. lycopersici (Fig. 4B–E). In addition, the sporulation of F. oxysporum f. sp. lycopersici was significantly inhibited by FaMyo5‐8 dsRNA. In contrast, the FaMyo5‐8 dsRNA had no effect on F. verticillioides, M. oryzae or B. cinerea (Fig. 4F–H). As expected, the expression level of Myo5 was highly inhibited in F. graminearum, F. tricinctum and F. oxysporum f. sp. lycopersici, but was not changed in F. verticillioides, M. oryzae or B. cinerea (Fig. 4I). We then determined the sequence similarity of the Myo5‐8 segment among the seven fungi. Consistent with the in vitro RNAi effect of FaMyo5‐8 dsRNA, the Myo5‐8 sequence of F. asiaticum had high similarity with that of F. graminearum, F. tricinctum and F. oxysporum f. sp. lycopersici (82%–99% identities), but lower similarity with that of F. verticillioides, M. oryzae and B. cinerea (68%–76% identities) (Fig. 4A). These results indicate that, although the sequence of the Myo5 gene is conserved in fungi, the sequence of the Myo5‐8 segment is more similar in F. asiaticum, F. graminearum, F. tricinctum and F. oxysporum f. sp. lycopersici than in the other fungi tested. Thus, Myo5‐8 dsRNA induced sequence‐specific RNAi activity in various Fusarium species, but not in F. verticillioides and other fungi.
Figure 4.
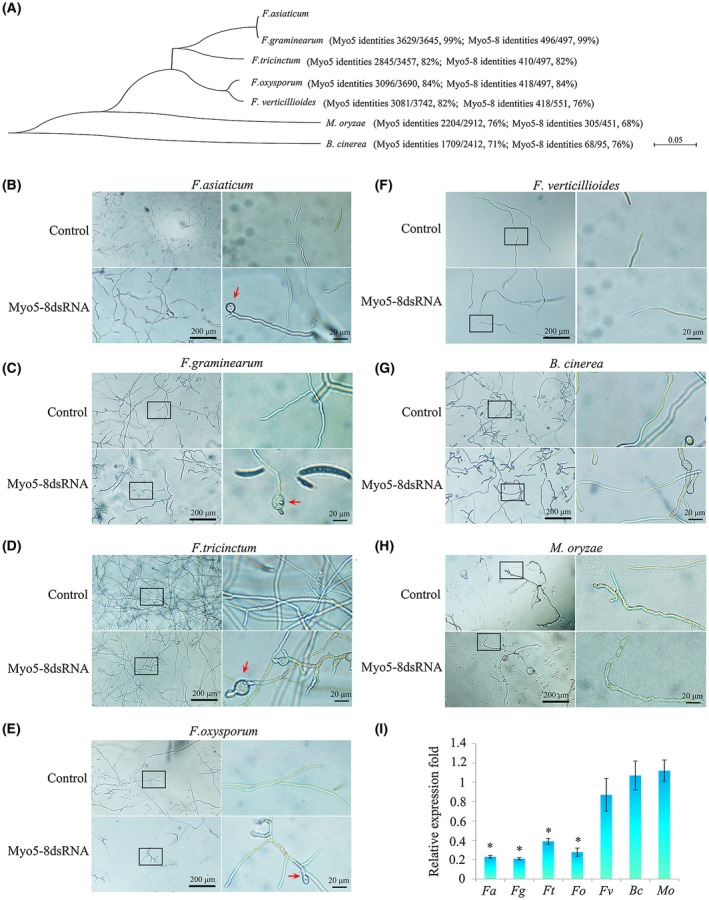
The effects of FaMyo5 double‐stranded RNA (dsRNA) on morphology and myosin‐5 gene expression of the different Fusarium species and other fungi. (A) Multiple sequence alignment of FaMyo5 with its homologues from F. graminearum, F. tricinctum, F. oxysporum, F. verticillioides, Magnaporthe oryzae and Botrytis cinerea. The identities of the full‐length Myo5 gene and of the Myo5‐8 segment are listed in parentheses. (B–H) The in vitro effects of FaMyo5‐8 dsRNAs on different Fusarium species, M. oryzae and B. cinerea. Hyphae were photographed after 24 h of cultivation. Each right panel is an enlarged view of the area in the black box in the left panel. The red arrows indicate the conglobate structures. (I) Expression of the myosin‐5 gene in different Fusarium species, M. oryzae and B. cinerea after treatment with FaMyo5 dsRNA. Total RNAs were extracted from 24‐h mycelia treated with FaMyo5 dsRNA. Gene expression levels were normalized to the ubiquitin C‐terminal hydrolase gene, and three independent biological duplicates were performed (* P < 0.05). [Colour figure can be viewed at wileyonlinelibrary.com]
Duration of the effect of Myo5‐8 dsRNA on F. asiaticum
To determine the duration of the RNAi triggered by Myo5‐8 dsRNA in F. asiaticum cells, we measured the rate of mycelial extension after Myo5‐8 dsRNA had been removed. As shown in Fig. 5A, the inhibition of mycelial extension lasted for 9 h after Myo5‐8 dsRNA had been removed from the SNA medium (0.1% KH2PO4, 0.1% KNO3, 0.05% MgSO4.7H2O, 0.05% KCl, 0.02% glucose and 0.02% sucrose). Within the first 5 h after dsRNA removal, the mycelium exhibited a comparable extension rate with that in medium continuously supplied with dsRNA, indicating effective RNAi in F. asiaticum cells within the first 5 h. However, the effect was reduced during the growth course from 5 to 9 h, and was lost after 9 h of growth (Fig. 5B). However, as shown in Fig. 5C, RNAi activity could last for 7 days with a continuous supply of dsRNA. Therefore, these observations indicate that the effects of FaMyo5‐8 dsRNA are short lived in F. asiaticum cells unless dsRNA is continuously supplied.
Figure 5.
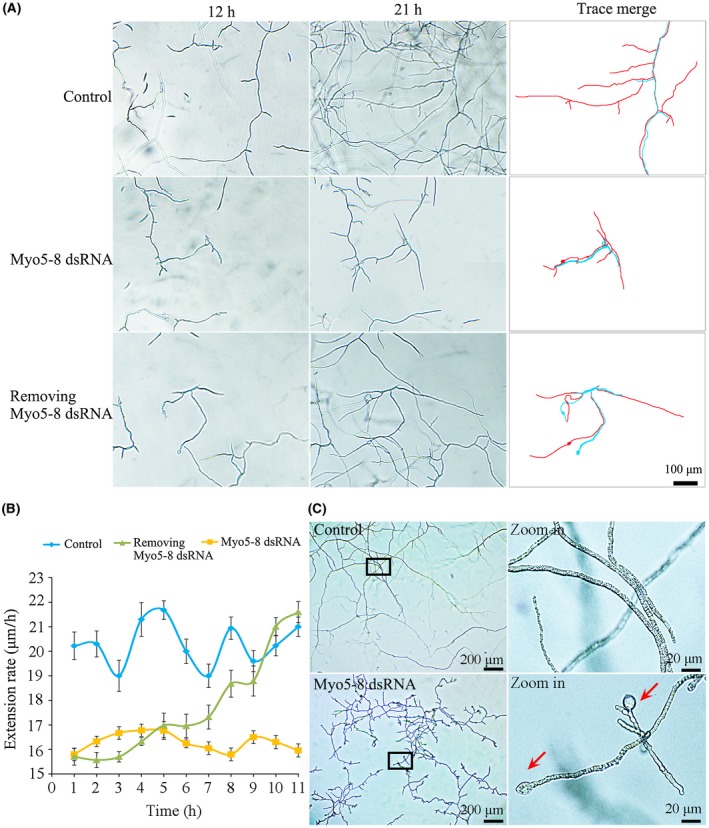
The duration of the effect of RNA interference (RNAi) in Fusarium asiaticum cells. Fusarium asiaticum was grown in SNA medium (0.1% KH2PO4, 0.1% KNO3, 0.05% MgSO4.7H2O, 0.05% KCl, 0.02% glucose and 0.02% sucrose) with or without 0.03 pm Myo5‐8 double‐stranded RNA (dsRNA). After 12 h, Myo5‐8 dsRNA was removed from some of the cultures, leaving three treatments: control (no Myo5‐8 dsRNA), continuous Myo5‐8 dsRNA or Myo5‐8 dsRNA for 12 h before removal. (A) Appearance of hyphae in the three treatments at 21 h (9 h after Myo5‐8 dsRNA had been removed from one of the treatments). In the panels on the right, blue lines indicate Fusarium growth traces at 12 h, and red lines indicate Fusarium growth traces at 21 h. (B) Extension rates of cultures from 12 h (when Myo5‐8 dsRNA had been removed from one of the treatments) to 23 h. Values are means from 10 independent replicates and are expressed as mycelium growth length per hour. (C) Appearance of hyphae in two treatments (control and continuous Myo5‐8 dsRNA) at 7 days. The panels on the right are larger versions of those on the left in the black boxes. The red arrows indicate conglobate structures. [Colour figure can be viewed at wileyonlinelibrary.com]
Generation and amplification of small interfering RNAs (siRNAs) in F. asiaticum cells
To investigate the mechanism causing FaMyo5‐8 dsRNAs to be short lived in F. asiaticum cells, we analysed the small RNAs (sRNAs) generated in F. asiaticum cells after Myo5‐8 dsRNA had been removed. The amount of total sRNAs in F. asiaticum cells was strongly reduced 5 h after Myo5‐8 dsRNA removal compared with that 1 h after Myo5‐8 dsRNA removal (Fig. 6A). Interestingly, the majority of siRNA species mapped to the Myo5‐8 segment. No difference was observed between the control (without dsRNA) and the Myo5‐8 dsRNA‐treated group (dsRNA treated for 12 h and then removed) with regard to the number of siRNAs that mapped to sites of Myo5‐1 to Myo5‐7. These results reveal that the majority of siRNA species in F. asiaticum are generated from exogenous dsRNA in the primary steps of RNAi. Therefore, secondary siRNA amplification using the target mRNA as a template was insufficient in F. asiaticum cells.
Figure 6.
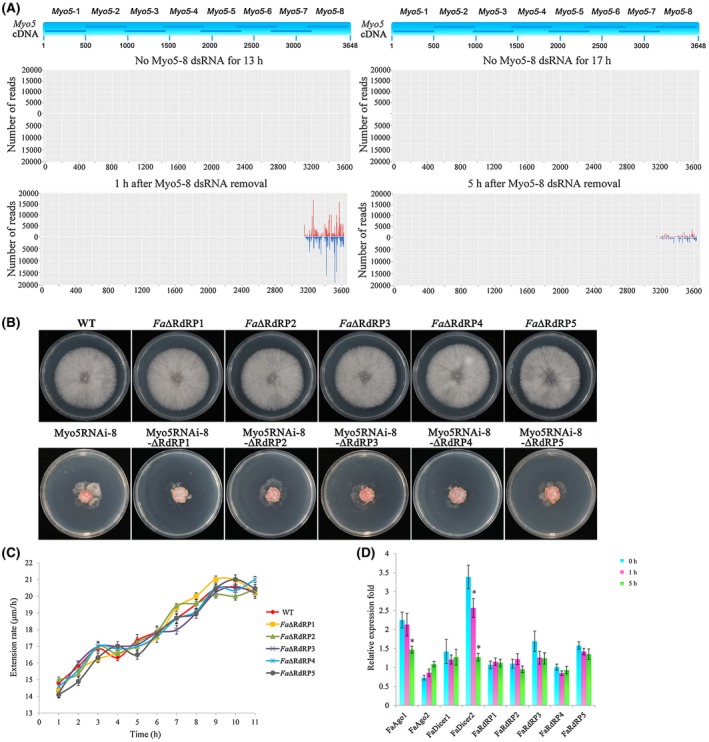
Secondary small interfering RNA (siRNA) amplification in Fusarium asiaticum .(A) Profiling of Myo5‐8 double‐stranded RNA (dsRNA)‐derived small RNAs (sRNAs) after Myo5‐8 dsRNA removal. Top panel is a diagram of eight fragments derived from Myo5 cDNA. Total sRNAs were isolated from four treatments: control‐1 (F. asiaticum was cultured for 13 h without Myo5‐8 dsRNA, no Myo5‐8 dsRNA for 13 h), control‐2 (F. asiaticum was cultured for 17 h without Myo5‐8 dsRNA, no Myo5‐8 dsRNA for 17 h), treatment‐1 (F. asiaticum was cultured for 12 h before removal, and cultured for a further 1 h after Myo5‐8 dsRNA removal), and treatment‐2 (F. asiaticum was cultured for 12 h before removal, and cultured for a further 5 h after Myo5‐8 dsRNA removal). sRNA reads were mapped to the sequence of Myo5 cDNA. (B) Growth pattern of RdRP gene mutants in the F. asiaticum wild‐type (WT) background (Fa∆RdRP1 to Fa∆RdRP5) and in the Myo5RNAi‐8 background (Myo5RNAi‐8‐∆RdRP1 to Myo5RNAi‐8‐∆RdRP5) on potato dextrose agar (PDA). Photographs were taken after 3 days of incubation at 25 °C. (C) Extension rates of F. asiaticum and five RdRP mutants after Myo5‐8 dsRNA removal. WT and five RdRP mutants were grown in SNA medium (0.1% KH2PO4, 0.1% KNO3, 0.05% MgSO4.7H2O, 0.05% KCl, 0.02% glucose and 0.02% sucrose) with 0.03 pm Myo5‐8 dsRNA. After 12 h, the Myo5‐8 dsRNA was removed from the cultures. Extension rates since Myo5‐8 dsRNA removal were calculated and expressed as mycelium growth length per hour. Values are means from 10 independent replicates. (D) The relative mRNA expression levels of RNA interference (RNAi)‐related genes in F. asiaticum after Myo5‐8 dsRNA removal for 0, 1 and 5 h. Expression of the ubiquitin C‐terminal hydrolase gene was used as the reference, and three independent biological duplicates were performed (* P < 0.05). [Colour figure can be viewed at wileyonlinelibrary.com]
To further substantiate the siRNA amplification machinery in F. asiaticum, we generated 10 RNA‐dependent RNA polymerase (RdRP) mutants (Fa∆RdRP1 to Fa∆RdRP5, Myo5RNAi‐8‐∆RdRP1 to Myo5RNAi‐8‐∆RdRP5) deficient for the critical enzyme of siRNA amplification. All 10 mutants showed no detectable change in mycelial growth or colony morphology on PDA compared with the WT or Myo5RNAi‐8 transformants (Fig. 6B). Meanwhile, these mutants exhibited normal asexual and sexual development (data not shown). Fa∆RdRP1 to Fa∆RdRP5 mutants displayed a similar duration of RNAi effect to WT (Fig. 6C), indicating that the siRNA amplification machinery had little or a short‐lived effect on the duration of RNAi in F. asiaticum.
We analysed the expression of nine RNAi‐related genes (two Dicer genes, FaDicer1 and FaDicer2, two Argonaute genes, FaAgo1 and FaAgo2, and five RdRP genes, FaRdRP1 to FaRdRP5) via qRT‐PCR after Myo5‐8 dsRNA had been removed from the medium. As shown in Fig. 6D, the relative amounts of transcripts of FaDicer1, FaAgo2 and the five FaRdRP genes remained substantially unchanged after Myo5‐8 dsRNA had been removed from the medium (from 0 to 5 h). In contrast, the relative amounts of transcripts of FaDicer2 and FaAgo1 were increased at 0 and 1 h, and were reduced at 5 h, after Myo5‐8 dsRNA had been removed from the medium (Fig. 6D). These results suggest that FaDicer2 and FaAgo1, but not FaDicer1, FaAgo2 or FaRdRPs, are critical for the RNAi pathway in F. asiaticum.
Effects of different methods of Myo5‐8 dsRNA spraying on plant resistance
Previous studies by Aline et al. (2016) have shown that dsRNA can be taken up by plant cells. To further explore whether dsRNA is absorbed via plant cells directly or via the wounded surface of the plant, we analysed the difference between intact coleoptiles and tip cut coleoptiles with regard to the uptake of dsRNA labelled with fluorescein‐12. Fluorescein‐dsRNAs were treated with DNase I/RNase and purified to remove the DNA, single‐stranded RNA (ssRNA) and excess fluorescein‐12‐UTP. As shown in Fig. S3A (see Supporting Information), after purification, the fluorescein‐12‐UTP and DNA template were removed from fluorescein‐dsRNAs, so that the effects of fluorescein‐12‐UTP and the DNA templet on the fluorescence signal were excluded (Fig. S3A). A strong fluorescence signal spread from the cut surface cells to distal cells after dsRNAs were sprayed onto the cut coleoptile. However, only a rare fluorescence signal could be detected in cells of intact coleoptiles under the same conditions (Fig. 7A), indicating that dsRNA was absorbed more efficiently via the wounded surface than the intact surface. To further investigate the way in which dsRNA spreads in coleoptiles, we separated the coleoptile from the seedling 3 days after spraying with fluorescein‐dsRNAs. Microscopic analyses revealed that the brightest fluorescence was located in the cut surface (in local tissue) and the lignified tracheary element (in distal tissue). The fluorescence signal could also be detected in the parenchyma cell around the tracheary element, indicating that the fluorescein‐dsRNA entered the damaged cells of the wounded surface and was transferred via the tracheary element (Fig. 7B). In addition, the signal generated by fluorescein‐dsRNAs (either fluorescein‐Myo5 dsRNAs or fluorescein‐GFP dsRNAs) was larger than the signal generated by fluorescein‐12‐UTP, which was concentrated in the region of spraying (Fig. S3B). As shown above, F. asiaticum was unable to maintain the silencing effect unless dsRNA was continuously supplied (Figs 5 and 6). In plant cells, the fluorescence signal could be detected even at 8 dpi, suggesting that Myo5‐8 dsRNAs are stable in the plant cells for at least 8 days.
Figure 7.
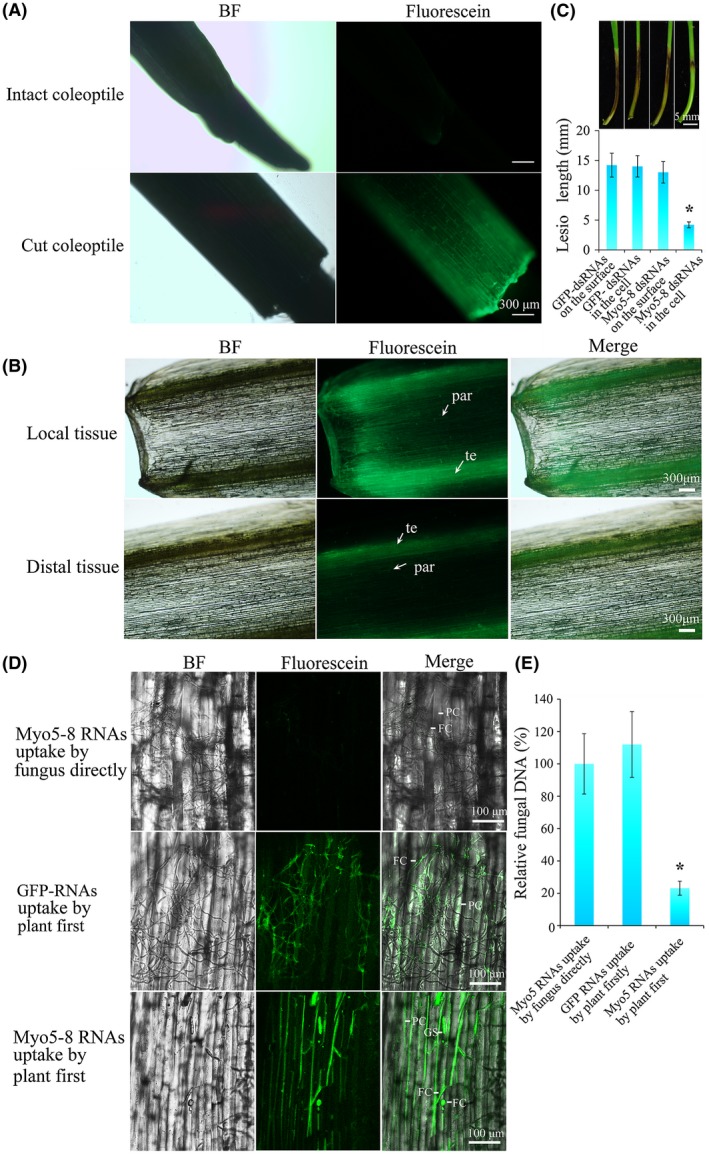
Double‐stranded RNA (dsRNA) uptake by the plant and effects on the fungal virulence of different pathways of Fusarium asiaticum uptake of dsRNA. (A) Microscopy of fluorescein‐Myo5‐8 dsRNA uptake by intact coleoptile and tip cut coleoptile. Detection of fluorescein‐Myo5‐8 dsRNA in intact coleoptile and tip cut coleoptile 24 h after spraying. Scale bar, 300 µm. BF, bright. (B) The spread of fluorescein‐Myo5‐8 dsRNA in the coleoptile. The fluorescence signal is located in the tracheary element (te) and parenchyma cell (par). Scale bar, 300 µm. (C) Resistance to F. asiaticum of wheat seedling sprayed with dsRNA onto the coleoptile surface or into the coleoptile cell. Lesions were measured and photographed at 7 days post‐inoculation (dpi). Values are means and standard errors of 30 replicate seedlings (*P < 0.05). Scale bar, 5 mm. GFP, green fluorescent protein. (D) Virulence of F. asiaticum that takes up Myo5‐8 dsRNA directly or from the plant cell. In one group, fluorescein‐Myo5‐8 dsRNAs or fluorescein‐GFP dsRNAs were inoculated onto the cut coleoptile and then into the macroconidial culture. Meanwhile, in the other group, SNA medium (0.1% KH2PO4, 0.1% KNO3, 0.05% MgSO4.7H2O, 0.05% KCl, 0.02% glucose and 0.02% sucrose) was inoculated onto the cut coleoptile and then into the macroconidial culture. After 12 h of cultivation, macroconidia from one group were inoculated onto the coleoptiles from another group. Fungal growth and fluorescence signals were visualized at 24 h post‐inoculation using confocal microscopy. The green signal reveals the uptake of fluorescein‐Myo5‐8 dsRNAs by plant cells (PC), germinating spores (GS) and fungal cells (FC). Scale bar, 100 µm. (E) Relative fungal biomass in coleoptiles. Control (RNA uptake by fungus directly): 10 000 macroconidia in SNA medium were mixed with 1 μg of fluorescein‐Myo5‐8 dsRNA and incubated at room temperature for 12 h; this was followed by inoculation on the cut coleoptile. Treatment (RNA uptake by plant first): 1 μg of fluorescein‐Myo5‐8 dsRNA was inoculated onto the cut coleoptile for 12 h; meanwhile, 10 000 macroconidia in SNA medium were incubated at room temperature for 12 h and were then inoculated onto the location of fluorescein‐Myo5‐8 dsRNA inoculation. After a further 24 h of cultivation, DNA was isolated from the coleoptiles. The real‐time quantitative polymerase chain reaction was performed using an equal quantity (30 ng) of corresponding DNA from control and treatment groups. The Fusarium‐specific gene Tri6 and wheat‐specific gene actin were used to calculate the fungal biomass. Data are represented as means ± standard deviation (SD), n = 10. *P < 0.05. [Colour figure can be viewed at wileyonlinelibrary.com]
The dsRNAs sprayed on the plants can be divided into two groups: either dried on the surface or absorbed by plant cells. We detected the effect of the two individual groups of dsRNAs on plant resistance against F. asiaticum. The lesion length of infected seedlings at 7 dpi in the group whose Myo5‐8 dsRNAs were dried on the surface was 13 mm, similar to that of two control groups (GFP dsRNA dried on the surface, 14.2 mm; GFP dsRNA absorbed by plant cells, 14 mm). However, in the group whose Myo5‐8 dsRNAs were absorbed by plant cells, the lesion length was 4.2 mm, displaying a significant disease reduction of 70% compared with the control group (Fig. 7C).
Once the plant is infected by the fungus, the dsRNAs dried on the surface should be taken up by the fungal cells directly, whereas the dsRNAs absorbed by the plant cells could be processed by plant cells and/or be transferred into fungal cells. To further investigate the effects of the two dsRNA transfer pathways on plant resistance against Fusarium, an infection experiment was designed to analyse the different effects. As shown in Fig. 7D, the fluorescence of Myo5‐8 dsRNA taken up by the fungal cells directly could not be detected at 12 h post‐inoculation. The fungal hyphae were long and grew vigorously on coleoptiles. In contrast, the fluorescence of the Myo5‐8 dsRNA transferred into fungal cells from plant cells was strongly detected at 12 h post‐inoculation. The fungal hyphae were short and crooked with conglobate structures. The fungal biomass was 76.84% lower in the treated group (coleoptiles were inoculated with fluorescein‐Myo5 dsRNAs first and then inoculated with macroconidia) than in the control (fluorescein‐Myo5 dsRNAs and macroconidia were mixed first and then were inoculated onto coleoptiles) (Fig. 7E). In another control treatment (coleoptiles were inoculated with fluorescein‐GFP dsRNAs first and then inoculated with macroconidia), the fluorescence of fluorescein‐GFP dsRNA transferred into fungal cells from plant cells was strongly detected at 12 h post‐inoculation and the fungal hyphae were long and grew vigorously on coleoptiles (Fig. 7D,E). These results suggest that the dsRNAs taken up by the plant cells first and then transferred into fungal cells will have a longer lasting effect on plant protection, compared with those taken up by fungal cells directly.
The SIGS mechanism in plant cells
To further investigate whether Myo5‐8 dsRNAs can be digested into siRNAs by the plant siRNA generation and amplification mechanism, we profiled Myo5‐8 dsRNA‐derived siRNAs in Myo5‐8 dsRNA‐sprayed and Myo5‐8 siRNA (obtained from the same amount of Myo5‐8 dsRNA via digestion with RNase III)‐sprayed coleoptiles at 12 h and 72 h after spraying. sRNA sequencing analysis revealed that the Myo5‐8 dsRNA‐derived reads from the Myo5‐8 dsRNA‐sprayed coleoptile at 72 h after spraying were increased substantially compared with those at 12 h after spraying (from 176 257 total reads to 448 426 total reads) (Table S2, see Supporting Information). In contrast, the reads from the Myo5‐8 siRNA‐sprayed coleoptile at 72 h after spraying were decreased compared with those at 12 h after spraying (from 804 169 total reads to 590 080 total reads) [Figs 8A, S4 (see Supporting Information) and Table S2]. Among the 18–27‐nucleotide (nt) reads, the increasing fold of 21‐nt and 24‐nt size classes was much greater than that of the other size classes, indicating that these classes were the main sRNA classes that were generated and amplified within plant cells (Fig. 8A).
Figure 8.
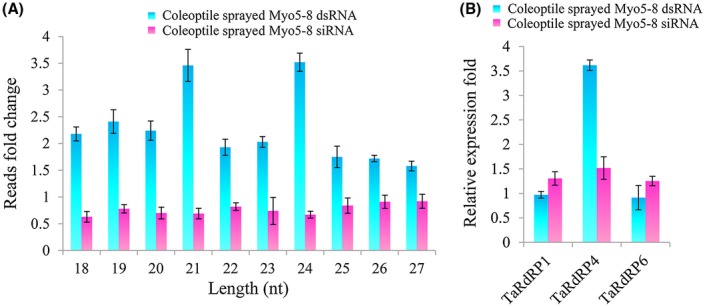
Small RNA sequencing (sRNA‐Seq) analysis of Myo5‐8 double‐stranded RNA (dsRNA)‐derived small interfering RNAs (siRNAs) and quantitative real‐time polymerase chain reaction (qRT‐PCR) analysis of RNA‐dependent RNA polymerase (RdRP) gene expression in Myo5‐8 dsRNA‐sprayed and Myo5‐8 siRNA‐sprayed coleoptiles. (A) Fold change of 18–27‐nucleotide (nt) reads of Myo5‐8 dsRNA‐derived siRNAs. The fold change was calculated as the quantity change of 18–27‐nt total sRNA reads with perfect matches to the Myo5‐8 dsRNA sequence on going from the coleoptiles at 12 h after spraying to the coleoptiles at 72 h after spraying. The bars represent the mean and standard deviation from three biological replicates. (B) Expression of three RdRP genes in Myo5‐8 dsRNA‐sprayed and Myo5‐8 siRNA‐sprayed coleoptiles. Total RNAs were extracted from 72‐h mycelia treated with Myo5‐8 dsRNA, Myo5‐8 siRNA and sterilized water (as control). Gene expression levels were normalized to the actin gene, and three independent biological duplicates were performed (* P < 0.05). [Colour figure can be viewed at wileyonlinelibrary.com]
It is noteworthy that the amplification of siRNA (to synthesize secondary siRNAs) usually uses the target mRNA as a template. However, the target mRNA of Myo5‐8 dsRNA‐derived siRNAs was absent in the wheat genome. To test whether the amplification of Myo5‐8 siRNA was generated by plant post‐transcriptional gene silencing, we additionally analysed the expression of RdRP genes in wheat. As a result of a lack of complete Triticum aestivum protein data, only three RdRPs were found in the T. aestivum database in the National Center for Biotechnology Information (NCBI) via BLASTp search with six Arabidopsis thaliana RdRP genes as query with default settings. The results showed that the T. aestivum proteins CAA09895.1 (TaRdRP1), CDM86506.1 (TaRdRP6) and CDM82152.1 (TaRdRP4) were homologous with the Arabidopsis thaliana proteins AtRdRP1, AtRdRP6 and AtRdRP4, respectively (Fig. S5, see Supporting Information). The qRT‐PCR results showed that the relative amounts of TaRdRP4 transcripts increased 3.6‐fold in the coleoptile sprayed with Myo5‐8 dsRNA compared with that sprayed with Myo5‐8 siRNA (Fig. 8B). These data suggest that the Myo5‐8 dsRNAs can be digested into siRNAs and then amplified by RdRP in T. aestivum. Further work should investigate whether the abundant secondary Myo5‐8 siRNAs can be amplified using the sense/antisense strand of Myo5‐8 dsRNA as the template.
Discussion
In the decade since its discovery, RNAi has been demonstrated to play diverse roles in a multitude of processes, including gene regulation, defence against viral infection and tumour suppression. For crop protection, a plant transgene‐based approach that expresses antisense or hairpin RNAi constructs directed against the target transcripts of pests or viruses (Baum et al., 2007; Pooggin et al., 2003), referred to as ‘host‐induced gene silencing’ (HIGS) (Cheng et al., 2015; Nowara et al., 2010; Tinoco et al., 2010), has been developed. Very recent reports have shown that a non‐transgenic RNAi strategy, SIGS, can replace HIGS without the need to develop stably transformed plants and available transformation protocols (Aline et al., 2016; Wang et al., 2016). However, the mechanism of SIGS in both plants and fungi is unclear (Machado et al., 2017; Wang and Jin, 2017). The objective of this study was to investigate the detailed mechanism of SIGS and to provide new insights into the application of SIGS.
We first identified and assayed the RNAi efficacy of different segments of the F. asiaticum Myo5 gene when such segments were applied to fungi. FaMyo5‐8 (one of the eight Myo5 segments) was highly conserved amongst F. asiaticum, F. graminearum, F. tricinctum and F. oxysporum, and FaMyo5‐8 dsRNAs induced sequence‐specific RNAi activity in these Fusarium species, but not in other fungi, in vitro. Interestingly, as F. asiaticum was unable to maintain the amplification of the siRNA machinery, the effects of FaMyo5‐8 dsRNAs were short lived in F. asiaticum cells unless dsRNA was continuously supplied. After spraying on plants, dsRNA was stable for 8 days. A wounded surface increased the uptake of dsRNA by plant cells. sRNA sequencing and qRT‐PCR analysis revealed that Myo5‐8 dsRNAs could be digested into siRNAs and then amplified by RdRP in T. aestivum. dsRNAs that were taken up by plant cells first and then transferred into fungal cells induced longer lasting RNAi effects to protect the plant against F. asiaticum when compared with dsRNAs taken up by the fungal cells directly.
The Myo5 gene was selected as the target sequence for silencing because it is essential for F. asiaticum survival (Zheng et al., 2015). Fusarium asiaticum mycelial growth on PDA was clearly retarded by the expression of the Myo5‐3, Myo5‐4, Myo5‐5, Myo5‐7 and Myo5‐8 RNAi constructs. The inhibition was substantially increased by the addition of an excess carbon supply (1.2 m sorbitol), indicating that the transport of materials in Myo5RNAi transformants was disturbed. Consistent with this finding, the chitin content in Myo5 RNAi transformants was lower. As a result, the cell wall was defective in Myo5‐3, Myo5‐4, Myo5‐5, Myo5‐7 and Myo5‐8 RNAi transformants. The cell wall is critical for fungal activity and survival (Gooday, 1995) and a defective cell wall may increase the extent to which dsRNAs enter the cells. Once the Myo5 dsRNA enters the cell through hyphal tips (the active site of cell wall metabolism), the subsequent dsRNAs will traverse the cell wall more easily because of cell wall defects.
The haploid period of the Fusarium life cycle, during which conidia are produced, lasts for a relatively long time. The haploid hyphae then form the second dikaryotic hyphae, which produce perithecium initials (Trail, 2009). To be effective, control measures must target a critical phase in the fungal life cycle (Trail et al., 2005). Myo5‐3, Myo5‐4, Myo5‐5, Myo5‐7 and Myo5‐8 dsRNAs reduced both the asexual and sexual reproduction of F. asiaticum, indicating that they have substantial potential to arrest critical phases in the life cycle of the fungus. Interestingly, the effects of different Myo5 segments on the Fusarium life cycle did not perfectly correlate with their effects on F. asiaticum development. Amongst these segments, Myo5‐8 was effective and stable in reducing both the life cycle and pathogenicity of F. asiaticum. These results suggest that RNAi‐triggering molecules derived from different regions of a given gene may have very different or even opposite effects on different biological processes. It seems that the levels of RNAi efficiency rely on the secondary or tertiary structure of target mRNAs. Therefore, screening for segments is essential for the identification of efficient dsRNAs for subsequent use in SIGS.
RNAi has been considered as a technology for the development of gene‐specific drugs for specific organisms (Mohr and Perrimon, 2012). The specificity, however, may also be a disadvantage, given that the genus Fusarium includes more than 90 species or varieties and that most Fusarium diseases are caused by two or more species (Bai and Shaner, 2004; Gerlach and Nirenberg, 1983). For example, Fusarium head blight is primarily caused by F. asiaticum and F. graminearum, but can also be caused by F. culmorum and F. avenaceum (Mesterházy, 1995). Comparative genomics has shown that the genomes of Fusarium species vary greatly in repeat content and in size, from the 36‐Mb genome of F. graminearum to the 61‐Mb genome of F. oxysporum (Ma et al., 2013). Thus, a sequence‐specific RNAi method is unlikely to control all Fusarium spp. Fortunately, the Myo5 gene is relatively conserved amongst Fusarium species, but is still different between Fusarium species and other fungi. The selected dsRNA segment, Myo5‐8, inhibited the growth of F. asiaticum, F. graminearum, F. tricinctum and F. oxysporum f. sp. lycopersici, but had no effect on F. verticillioides, M. oryzae or B. cinerea. These results suggest that Myo5‐8 dsRNA could be specific and effective for the control of many Fusarium species, but would not inhibit other fungi.
A vast amount of studies have shown that the RNAi pathway plays a role in the development and physiology of several fungi (Chang et al., 2012; Nicolas et al., 2013). However, a recent study by Chen et al. (2015) has shown that the RNAi machinery is not involved in vegetative development and pathogenesis in F. graminearum, indicating that the functions of RNAi components vary amongst different fungal species. The core RNAi machinery, including Dicer, Argonaute and RdRPs, appears to be largely conserved in fungi (Li et al., 2010), but some differences do exist (Drinnenberg et al., 2009). In some fungi, additional genes involved in RNAi have been identified which produce siRNAs by Dicer‐independent pathways, for example Neurospora crassa and Mucor circinelloides (Billmyre et al., 2013). In addition, there are some fungal species that lack RNAi machinery, such as Saccharomyces cerevisiae (Drinnenberg et al., 2011). In plants and Caenorhabditis elegans, two distinct populations of sRNAs have been proposed to participate in RNAi: ‘primary siRNAs’ (derived from Dicer nuclease‐mediated cleavage of the original trigger) and ‘secondary siRNAs’ (additional sRNAs whose synthesis requires an RdRP) (Pak and Fire, 2007). Although F. asiaticum possesses five RdRP genes, the silencing process lasts for a short time in F. asiaticum cells, because the degree of RNAi amplification is insufficient to cause continuous mRNA degradation. A long‐lasting RNAi effect is maintained by secondary siRNA production through RdRP activity in most eukaryotic organisms (Marker et al., 2010; Tang et al., 2003), whereas RdRP has not been identified in insects, which possess a systemic and long‐lasting RNAi process (Carthew and Sontheimer, 2009). In F. asiaticum, one reason why the silencing process lasts for only a short time may be that the RdRPs are non‐functional or transiently functional enzymes, different from RdRPs in plants, C. elegans, N. crassa and Dictyostelium discoideum (Agrawal et al., 2003).
To avoid being taken up by F. asiaticum directly, sprays need to be efficiently taken up by plant cells. Recently, Aline et al. (2016) have shown that dsRNAs labelled with the green fluorescent dye ATTO 488 are taken up by barley cells when sprayed onto barley leaf segments. However, it is not clear whether dsRNA is absorbed via plant cells directly or via the vascular bundle of the leaf cut surface. In wheat, the cell types of the tip and the shaft of the coleoptile are different. The vascular bundle is only present in the shaft of the coleoptile (Nikus and Jonsson, 1999; O'Brien and Thimann, 1965). Therefore, we used living wheat seedlings to analyse the dsRNA uptake by plant cells. We provided convincing evidence to show that dsRNA is absorbed more efficiently via the wounded surface than the intact surface. After spraying on the plant, dsRNA dries on the surface and/or is absorbed by plant cells. When the plant is infected by the fungus, the dsRNA that dries on the surface cannot be transferred to the infected tissue from the sprayed tissue through the vascular bundle (Fig. 7) and cannot lead to the spread of RNAi signalling in the plant. Although it may be taken up by fungal cells directly, dsRNA that dries on the surface works for only a short duration and has little or only short‐lived effects on plant resistance (Fig. 7).
Once sprayed on the wheat coleoptile, the dsRNA molecules are stable for 8 days. However, this is not sufficient to serve as an efficient disease control method, which requires a reasonable duration of efficacy. One of the most striking features of RNAi in plants, such as Arabidopsis, is its ability to amplify the initial response by the activities of RdRPs which synthesize secondary siRNAs (Tang et al., 2003). Secondary siRNAs are much more abundant than primary siRNAs and strongly repress the target gene expression level (Zhang and Ruvkun, 2012). Encouragingly, we observed a massive Myo5‐8 dsRNA‐derived siRNA increase when dsRNA was sprayed on wheat coleoptiles. Moreover, the expression level of TaRdRP4 was significantly induced. The synthesis of secondary siRNAs seems to depend on the target mRNA of siRNA. However, the target of the exogenous dsRNA does not exist in plants. Therefore, further study is required to determine whether the secondary Myo5‐8 siRNAs are amplified using the sense/antisense stand of Myo5‐8 dsRNA as the template.
In summary, our results demonstrate that the selected dsRNA, Myo5‐8, can be used to control a variety of Fusarium species. The effective way to utilize Myo5‐8 dsRNA to control Fusarium disease is to spray dsRNA on the wounded surface of the plant. The duration of SIGS efficacy was limited by the amplification of siRNA in Fusarium, but promoted by RdRP‐dependent secondary siRNA synthesis in wheat (Fig. S6, see Supporting Information). Our study provides new implications to develop SIGS as a mainstream disease control strategy.
Experimental procedures
Fungi, plants and culture conditions
The fungal species and strains used in this study are listed in Table S1. The F. asiaticum strain was isolated from a field in a region experiencing a Fusarium head blight epidemic in Jiangsu Province, China (Chen et al., 2007). Fusarium verticillioides ZJ11 and F. tricinctum ZJ32 were isolated from different fields experiencing maize ear rot epidemics in Zhejiang Province, China. Fusarium oxysporum f. sp. lycopersici JS51 was isolated from a diseased tomato grown in Jiangsu Province, China. Wheat variety Huaimai33 was maintained in our laboratory. Potato dextrose broth (PDB) and CMC broth were used for the assessment of mycelial growth and conidial production, respectively.
RNAi constructs, DNA and RNA isolation, Southern blot and qRT‐PCR
An Myo5 cDNA (3645 nt) from F. asiaticum was divided into eight fragments, which were designated as Myo5‐1 (nt 1–473), Myo5‐2 (nt 452–945), Myo5‐3 (nt 939–1454), Myo5‐4 (nt 1381–1915), Myo5‐5 (nt 1829–2314), Myo5‐6 (nt 2254–2740), Myo5‐7 (nt 2650–3207) and Myo5‐8 (nt 3149–3645) (Fig. S1A). These fragments were used to construct eight fungal RNAi constructs (pMyo5RNAi‐1 to pMyo5RNAi‐8). Each construct carried a G418 resistance cassette and a transcriptional unit for hairpin RNA expression with a cutinase gene intron spacer from M. oryzae (Nakayashiki et al., 2005) (Fig. S1B). All primers used in this study are listed in Table S3 (see Supporting Information) and are described in Fig. S1B.
Fungal DNA was extracted from mycelia as described previously (Nicholson et al., 1997). Fusarium strains were grown on PDA for 3 days, and total RNA was extracted from the mycelia with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions, and was transcribed into cDNA as described previously (Song et al., 2014). The full‐sized cDNA of Myo5 was amplified with the primers Myo5P1 and Myo5P2. PCRs were carried out in a TaKaRa PCR thermal cycler (TaKaRa TP600, Dalian, China) with gene‐specific PCR primers. Southern blot analyses of Myo5RNAi transformants was performed with the digoxigenin (DIG) labelling kit (Roche, Mannheim, Germany) (Song et al., 2014).
qRT‐PCR was performed with an ABI 7500 real‐time detection system (Applied Biosystems, Foster City, CA, USA). The fungal housekeeping gene ubiquitin C‐terminal hydrolase UBH (Gene ID: FGSG_01231) was used as an internal control (Min et al., 2014). Data from three biological replicates were used to calculate the mean and standard deviation.
Fungal transformation and assessments of stress sensitivity, mycelial growth, cell structure, chitin content, asexual and sexual reproduction, and pathogenicity
Fusarium asiaticum transformation, transformant identification, stress sensitivity, mycelial growth and chitin content were assessed as described previously (Song et al., 2014). Pathogenicity was assessed following seedling inoculation (Cheng et al., 2015).
Cell structure observation was performed using an inverted fluorescence microscope (Olympus IX71, Olympus Canada, Markham, ON, Canada). For the observation of septum and chitin distribution, conidia or hyphae were stained by CFW. Images were captured and analysed by Image‐Pro Plus software (Media Cybernetics, Silver Spring, MD, USA). Transmission electron microscopy (TEM) was carried out as described by Song et al. (2014).
For the determination of conidial production, 10 μL of macroconidia (5 × 105 spores/mL) were transferred into 20 mL of CMC medium; after the culture had been incubated for 5 days at 25 °C on a rotary shaker (200 rpm), the conidia were counted. For sexual development assays, mycelia were grown in Petri plates containing carrot agar for 5 days before the aerial mycelia were removed (with a glass spreader and a 2.5% sterilized Tween‐60 solution) to induce sexual reproduction by the mycelia remaining on the plates. After this treatment, all cultures were incubated under UV light (365 nm; HKiv Import & Export Co., Ltd., Xiamen, China) at 25 °C. The cultures were examined for perithecia and ascospores after 7 and 21 days of incubation.
dsRNA and siRNA synthesis and silencing in vitro
Myo5 dsRNA and control dsRNA [derived from the green fluorescent protein (GFP) coding sequence; sequence ID: HF675000.1] were generated using a MEGAscript RNAi Kit (Invitrogen) following MEGAscript protocols. Fluorescein‐Myo5 dsRNAs were labelled using a Fluorescein RNA Labeling Mix Kit, following the manufacturer's instructions (Sigma, St. Louis, MO, USA). The Myo5‐8 siRNAs were generated using Myo5‐8 dsRNAs, following the ShortCut RNaseIII (NEB, ipswich, MA, USA) manufacturer's instructions. Conidia were diluted to 1 × 104 mL–1 using SNA medium (0.1% KH2PO4, 0.1% KNO3, 0.05% MgSO4.7H2O, 0.05% KCl, 0.02% glucose and 0.02% sucrose) and were added to 0.1 pm Myo5‐8 dsRNA or 0.1 pm GFP dsRNA. Mixtures were placed in a 96‐well microtitre plate and observed with an inverted microscope.
For the determination of the duration of the RNAi effect, F. asiaticum was grown in SNA medium with or without 0.03 pm Myo5‐8 dsRNA. After 12 h, Myo5‐8 dsRNA was removed from some of the cultures by washing three times with SNA medium, leaving three treatments: control (no Myo5‐8 dsRNA, but with 0.03 pm GFP dsRNA), continuous Myo5‐8 dsRNA or Myo5‐8 dsRNA for 12 h before removal. The mycelial extension was monitored by capturing images every hour at 25 °C. The mycelial extension rate was calculated as described previously (Trinci, 1973). Fusarium asiaticum treated with continuous Myo5‐8 dsRNA was cultured for 7 days and photographed.
sRNA library construction and sequencing
For fungal sRNA library construction, total RNAs isolated from F. asiaticum were pooled and prepared [control group, cultured in SNA for 13 h and 17 h; treated group, cultured in SNA with 0.03 pm Myo5‐8 dsRNA for 12 h, then removed dsRNA and cultured for a further 1 h and 5 h (as described above)]. Mycelia were collected and washed three times with hyperpure water for sRNA extraction. For wheat coleoptile sRNA library construction, the uppermost 4 mm of a coleoptile of a 3‐day‐old Huaimai33 seedling was cut off (without damaging the wheat bud) and sprayed with 400 ng of Myo5‐8 dsRNA or Myo5‐8 siRNA. Total RNAs isolated from the uppermost 7 mm of 10 coleoptiles were extracted at 12 and 72 h after spraying.
Samples were ground to a powder with liquid nitrogen and total RNA was isolated using Trizol reagent (Invitrogen). Total RNA quality and quantity were assessed on an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and a Nanodrop instrument (Thermo®, Waltham, MA, USA) (Fig. S7, see Supporting Information). sRNA fractions were extracted from 2 μg of total RNA using 15% denaturing polyacrylamide gel (Kasschau et al., 2007).
sRNA libraries were constructed by the addition of 5′ adaptor (sequence GTTCAGAGTTCTACAGTCCGACGATC) and 3′ adaptor (sequence TGGAATTCTCGGGTGCCAAGG) using TruSeq Small RNA Sample Prep Kits (Illumina, San Diego, CA, USA) and sequenced using an Illumina Hiseq 2000/2500 by the Mega Genomics Company (Beijing, China). sRNA‐Seq was repeated three times.
Quality assessment was performed using CASAVA 1.8 (Illumina) (Fig. S8, see Supporting Information). After removal of the adaptor contaminants from the raw reads, the remaining sRNA sequences ranging from 18 to 27 bp were filtered and mapped to the FaMyo5 mRNA sequence using Bowtie (Langmead and Salzberg, 2012; Langmead et al., 2009).
dsRNA spray and effect measurement
For the determination of dsRNA uptake in plants, 3‐day‐old Huaimai33 seedlings with intact coleoptiles or wounded coleoptiles (the uppermost 4 mm of the coleoptile was cut off) were sprayed with 400 ng of fluorescein‐Myo5‐8 dsRNA. After 12 h, the seedlings were washed with sterile water and imaged using an inverted fluorescence microscope (Olympus IX71). To assay the spread of dsRNA in the coleoptiles, 400 ng of fluorescein‐Myo5‐8 dsRNAs were sprayed onto the wounded coleoptile. After 3 days, the seedlings were washed twice with sterile water and the uppermost 10 mm of the coleoptile was cross cut and observed using an inverted fluorescence microscope.
For the determination of dsRNA application on plants, 3‐day‐old Huaimai33 seedlings with intact coleoptiles or wounded coleoptiles (the uppermost 4 mm of the coleoptile was cut off) were sprayed with 0.1 pm of Myo5‐8 dsRNA or 0.1 pm of GFP‐specific 560‐nt dsRNA (control). After dsRNAs had dried on the surface or been absorbed by the plant cells (12 h after spraying), seedlings with intact coleoptiles were also cut (the uppermost 4 mm of the coleoptile was cut off), whereas seedlings with wounded coleoptiles were washed with sterile water. All the seedlings were inoculated with 3 µL of macroconidial suspension (5 × 105 spores/ml), and seedling inoculations were carried out in growth chambers. Thirty seedlings were inoculated, and the brown lesions of diseased seedlings were measured at 7 dpi.
To investigate the effect of different pathways by which dsRNA enters into fungal cells, an experiment was performed to detect the infection of F. asiaticum that takes up dsRNA directly or absorbs dsRNA from plant cells. For the observation of the infection of F. asiaticum that takes up dsRNA directly, 10 000 macroconidia in SNA medium were mixed with 1 μg of fluorescein‐Myo5‐8 dsRNA, incubated at room temperature for 12 h and then inoculated onto cut coleoptiles. For the observation of the infection of F. asiaticum that absorbs dsRNA from plant cells, 1 μg of fluorescein‐Myo5‐8 dsRNA was inoculated onto cut coleoptiles for 12 h; meanwhile, 10 000 macroconidia in SNA medium were incubated at room temperature for 12 h and were then inoculated onto the location of fluorescein‐Myo5‐8 dsRNA inoculation. Each treatment contained 10 coleoptiles. After a further 24 h of cultivation, the fungal biomass was assayed as described previously (Kumar et al., 2015). The coleoptiles were imaged using a Leica TCS SP8 (Leica Microsystems, Tokyo, Japan) equipped with a 10× objective. Images of fluorescing dsRNA were obtained using a 488‐nm excitation laser and a hybrid detector set to 480–520 nm.
Statistical analysis
All the data were analysed using SAS release 6.12 (SAS Institute, Cary, NC, USA) at a significance level of 0.05.
Supporting information
Fig. S1 A diagram of the eight Myo5 gene fragments used for making RNAi constructs, and the integration of the Myo5RNAi constructs into F. asiaticum. (a) The Myo5 gene sequence was divided into eight segments (Myo5‐1 to ‐8). A short overlap was located at the ends of the junction regions. (b) A schematic diagram for homologous recombination between the replacement vectors pMyo5RNAi and the PLS gene locus of F. asiaticum. The PCR primers and probe for detection of transformants are indicated.
Fig. S2 PCR and Southern blot analyses of Myo5RNAi transformants. (a) PCR products from the 5′ and 3′ regions of the disrupted PLS gene were amplified with the primers TetP9/PgpdP5 and Pgpd1P3/TetP10, respectively. The 1751‐ (U) and 1777‐bp (D) fragments were amplified from the 5′ and 3′ regions of the disrupted PLS gene, respectively. M: λDNA/PstI DNA Marker. (b) Southern blot analysis of Myo5RNAi transformants. The genomic DNAs were digested with NcoI and hybridized with a fragment of a neomycin resistance gene amplified with the primers G418ProbeP1/ G418ProbeP2. The sizes of hybridization fragments for the strains Myo5RNAi‐1 to Myo5RNAi‐8 were 6738/4225, 6758/4225, 6824/4225, 6862/4225, 6764/4225, 6766/4225, 6908/4225, and 6786/4225 bp, respectively.
Fig. S3 The labelling of Myo5‐8 dsRNA with fluorescein‐12‐UTP and spraying of fluorescein‐Myo5‐8 dsRNA and fluorescein‐UTP on cut coleoptile. (A) Fluorescein‐Myo5 dsRNAs were labeled using the fluorescein RNA Labeling Mix Kit and purified using the MEGAscript RNAi Kit (Invitrogen) following MEGAscript protocols. The products were detected using GelDoc XR and Quantity One 4.5.2 software (Bio‐Rad, Hercules, CA) with or without ethidium bromide (EB). (B) Microscopy of fluorescein‐Myo5‐8 dsRNA, fluorescein‐GFP dsRNA and fluorescein‐UTP uptake by tip cut coleoptile 24 h after spraying. Scale bar = 300 µm.
Fig. S4 Profiling of Myo5‐8 dsRNA derived sRNAs in coleoptiles spraying Myo5‐8 siRNA and Myo5‐8 dsRNA. Top panel is a diagram of eight fragments derived from Myo5 cDNA. The uppermost 4 mm of coleoptile of 3‐d‐old Huaimai33 seeding was cut off and sprayed with 400 ng Myo5‐8 dsRNA or Myo5‐8 siRNA. Small RNAs were isolated from the uppermost 7 mm of 10 coleoptiles and sequenced 12 hours and 72 hours post spraying.
Fig. S5 Phylogenetic analysis of the relationship between the six Arabidopsis RdRP proteins and the three wheat RdRP proteins. GenBank accession numbers are as follows: AtRdRP1, AY148431; AtRdRP2, At4g11130; AtRdRP3, At2g19910; AtRdRP4, At2g19920; AtRdRP5, At2g19930 and AtRdRP6, AF268093; Sequence alignment, distance calculation, and tree construction were performed with the use of the Clustal W program.
Fig. S6 A proposed model for Fusarium‐plant interaction after dsRNA sprayed. Fungal cell showing the dsRNA (on the plant surface) taken up by the fungal cells directly and the siRNA transferred from plant cell leading to target mRNA degradation. Fusarium is unable to maintain siRNA amplification by RdRPs machinery leading to short‐lasting SIGS effect. However, the plant cell could provide substantial siRNAs via secondary amplification machinery and determined the duration of SIGS.
Fig. S7 The quality of total RNA using for small RNA sequencing.
Fig. S8 The quality assessment of small RNA data.
Table S1 Strains used in this study.
Table S2 A list of small RNA that mapped to myosin 5 mRNA.
Table S3 Primers and sequences used in this study.
Acknowledgements
This research was supported by the National Natural Science Foundation of China (31701805), the Fundamental Research Funds for the Central Universities (KJQN201808), the National Natural Science Foundation of China (31772191 and 31730072) and a general financial grant from the China Postdoctoral Science Foundation (2016M601835).
References
- Agrawal, N. , Dasaradhi, P.V. , Mohmmed, A. , Malhotra, P. , Bhatnagar, R.K. and Mukherjee, S.K. (2003) RNA interference: biology, mechanism, and applications. Microbiol. Mol. Biol. Rev. 67, 657–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aline, K. , Biedenkopf, D. , Furch, A. , Weber, L. , Rossbach, O. , Abdellatef, E. , Linicus, L. , Johannsmeier, J. , Jelonek, L. and Goesmann, A. (2016) An RNAi‐based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathog. 12, e1005901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, G. and Shaner, G. (2004) Management and resistance in wheat and barley to Fusarium head blight. Annu. Rev. Phytopathol. 42, 135–161. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D. (2004) RNA silencing in plants. Nature, 431, 356–363. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D.C. (2015) VIGS, HIGS and FIGS: small RNA silencing in the interactions of viruses or filamentous organisms with their plant hosts. Curr. Opin. Plant Biol. 26, 141–146. [DOI] [PubMed] [Google Scholar]
- Baum, J.A. , Bogaert, T. , Clinton, W. , Heck, G.R. , Feldmann, P. , Ilagan, O. , Johnson, S. , Plaetinck, G. , Munyikwa, T. and Pleau, M. (2007) Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 25, 1322–1326. [DOI] [PubMed] [Google Scholar]
- Billmyre, R.B. , Calo, S. , Feretzaki, M. , Wang, X. and Heitman, J. (2013) RNAi function, diversity, and loss in the fungal kingdom. Chromosome Res. 21, 561–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloemink, M.J. and Geeves, M.A. (2011) Shaking the myosin family tree: biochemical kinetics defines four types of myosin motor. Semin. Cell Dev. Biol. 22, 961–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bosch, F. and Gilligan, C.A. (2008) Models of fungicide resistance dynamics. Annu. Rev. Phytopathol. 46, 123–147. [DOI] [PubMed] [Google Scholar]
- Carthew, R.W. and Sontheimer, E.J. (2009) Origins and mechanisms of miRNAs and siRNAs. Cell, 136, 642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, S.S. , Zhang, Z. and Liu, Y. (2012) RNA interference pathways in fungi: mechanisms and functions. Annu. Rev. Microbiol. 66, 305–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Chen, C. , Wang, J. , Jin, L. and Zhou, M. (2007) Genetic study on JS399‐19 resistance in hyphal fusion of Fusarium graminearum by using nitrate nonutilizing mutants as genetic markers. Acta Genet. Sin. 34, 469–476. [DOI] [PubMed] [Google Scholar]
- Chen, Y. , Gao, Q. , Huang, M. , Liu, Y. , Liu, Z. , Liu, X. and Ma, Z. (2015) Characterization of RNA silencing components in the plant pathogenic fungus Fusarium graminearum . Sci. Rep. 5, 12 500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, W. , Song, X.S. , Li, H.P. , Cao, L.H. , Sun, K. , Qiu, X.L. , Xu, Y.B. , Yang, P. , Huang, T. , Zhang, J.B. , Qu, B. and Liao, Y.C. (2015) Host‐induced gene silencing of an essential chitin synthase gene confers durable resistance to Fusarium head blight and seedling blight in wheat. Plant Biotechnol. J. 13, 1335–1345. [DOI] [PubMed] [Google Scholar]
- Dean, R. , Van Kan, J.A. , Pretorius, Z.A. , Hammond‐Kosack, K.E. , Di Pietro, A. , Spanu, P.D. , Rudd, J.J. , Dickman, M. , Kahmann, R. , Ellis, J. and Foster, G.D. (2012) The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinnenberg, I.A. , Fink, G.R. and Bartel, D.P. (2011) Compatibility with killer explains the rise of RNAi‐deficient fungi. Science, 333, 1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinnenberg, I.A. , Weinberg, D.E. , Xie, K.T. , Mower, J.P. , Wolfe, K.H. , Fink, G.R. and Bartel, D.P. (2009) RNAi in budding yeast. Science, 326, 544–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire, A. , Xu, S. , Montgomery, M.K. , Kostas, S.A. , Driver, S.E. and Mello, C.C. (1998) Potent and specific genetic interference by double‐stranded RNA in Caenorhabditis elegans . Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Gerlach, W. and Nirenberg, H.I. (1983) The genus Fusarium: a pictorial atlas. Mycologia, 75, 1110. [Google Scholar]
- Gooday, G.W. (1995) Cell walls. Growing Fungus, 20, 43–62. [Google Scholar]
- Hartman, M.A. , Finan, D. , Sivaramakrishnan, S. and Spudich, J.A. (2011) Principles of unconventional myosin function and targeting. Annu. Rev. Cell Dev. Biol. 27, 133–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann, W.A. , Richards, T.A. and de Lanerolle, P. (2009) Ancient animal ancestry for nuclear myosin. J. Cell Sci. 122, 636–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau, K.D. , Fahlgren, N. , Chapman, E.J. , Sullivan, C.M. , Cumbie, J.S. , Givan, S.A. and Carrington, J.C. (2007) Genome‐wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 5, e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, A. , Karre, S. , Dhokane, D. , Kage, U. , Hukkeri, S. and Kushalappa, A.C. (2015) Real‐time quantitative PCR based method for the quantification of fungal biomass to discriminate quantitative resistance in barley and wheat genotypes to fusarium head blight. J. Cereal. Sci. 64, 16–22. [Google Scholar]
- Lamberth, C. , Jeanmart, S. , Luksch, T. and Plant, A. (2013) Current challenges and trends in the discovery of agrochemicals. Science, 341, 742–746. [DOI] [PubMed] [Google Scholar]
- Langmead, B. and Salzberg, S.L. (2012) Fast gapped‐read alignment with Bowtie 2. Nat. Methods, 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B. , Trapnell, C. , Pop, M. and Salzberg, S.L. (2009) Ultrafast and memory‐efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L. , Chang, S.S. and Liu, Y. (2010) RNA interference pathways in filamentous fungi. Cell Mol. Life Sci. 67, 3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L.J. , Geiser, D.M. , Proctor, R.H. , Rooney, A.P. , O'Donnell, K. , Trail, F. , Gardiner, D.M. , Manners, J.M. and Kazan, K. (2013) Fusarium pathogenomics. Annu. Rev. Microbiol. 67, 399–416. [DOI] [PubMed] [Google Scholar]
- Machado, A.K. , Brown, N.A. , Urban, M. , Kanyuka, K. and Hammond‐Kosack, K. (2017) RNAi as an emerging approach to control Fusarium head blight disease and mycotoxin contamination in cereals. Pest Manag. Sci. 74, 790–799. 10.1002/ps.4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marker, S. , Le Mouel, A. , Meyer, E. and Simon, M. (2010) Distinct RNA‐dependent RNA polymerases are required for RNAi triggered by double‐stranded RNA versus truncated transgenes in Paramecium tetraurelia . Nucleic Acids Res. 38, 4092–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesterházy, A. (1995) Types and components of resistance to Fusarium head blight of wheat. Plant Breeding, 114, 377–386. [Google Scholar]
- Min, K. , Son, H. , Lim, J.Y. , Choi, G.J. , Kim, J.C. , Harris, S.D. and Lee, Y.W. (2014) Transcription factor RFX1 is crucial for maintenance of genome integrity in Fusarium graminearum . Eukaryot. Cell, 13, 427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr, S.E. and Perrimon, N. (2012) RNAi screening: new approaches, understandings, and organisms. Wiley Interdiscip. Rev. RNA, 3, 145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayashiki, H. , Hanada, S. , Nguyen, B.Q. , Kadotani, N. , Tosa, Y. and Mayama, S. (2005) RNA silencing as a tool for exploring gene function in ascomycete fungi. Fungal Genet Biol 42, 275. [DOI] [PubMed] [Google Scholar]
- Nakajima, Y. , Maeda, K. , Jin, Q. , Takahashiando, N. , Kanamaru, K. , Kobayashi, T. and Kimura, M. (2016) Oligosaccharides containing an α‐(1→2) (glucosyl/xylosyl)‐fructosyl linkage as inducer molecules of trichothecene biosynthesis for Fusarium graminearum . Int. J. Food Microbiol. 238, 215–221. [DOI] [PubMed] [Google Scholar]
- Nicholson, P. , Rezanoor, H.N. , Simpson, D.R. and Joyce, D. (1997) Differentiation and quantification of the cereal eyespot fungi Tapesia yallundae and Tapesia acuformis using a PCR assay. Plant Pathol. 46, 842–856. [Google Scholar]
- Nicolas, F.E. , Torres‐Martinez, S. and Ruiz‐Vazquez, R.M. (2013) Loss and retention of RNA interference in fungi and parasites. PLoS Pathog. 9, e1003089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikus, J. and Jonsson, L.M.V. (1999) Tissue localization of β‐glucosidase in rye, maize and wheat seedlings. Physiol. Plant. 107, 373–378. [DOI] [PubMed] [Google Scholar]
- Nowara, D. , Gay, A. , Lacomme, C. , Shaw, J. , Ridout, C. , Douchkov, D. , Hensel, G. , Kumlehn, J. and Schweizer, P. (2010) HIGS: host‐induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis . Plant Cell, 22, 3130–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes, C.C. and Dean, R.A. (2012) Host‐induced gene silencing: a tool for understanding fungal host interaction and for developing novel disease control strategies. Mol. Plant Pathol. 13, 519–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien, T.P. and Thimann, K.V. (1965) Histological studies on the coleoptile. I. Tissue and cell types in the coleoptile tip. Am. J. Bot. 52, 910–918. [Google Scholar]
- Pak, J. and Fire, A. (2007) Distinct populations of primary and secondary effectors during RNAi in C. elegans . Science, 315, 241–244. [DOI] [PubMed] [Google Scholar]
- Pooggin, M. , Shivaprasad, P.V. , Veluthambi, K. and Hohn, T. (2003) RNAi targeting of DNA virus in plants. Nat. Biotechnol. 21, 131–132. [DOI] [PubMed] [Google Scholar]
- Song, X.S. , Li, H.P. , Zhang, J.B. , Song, B. , Huang, T. , Du, X.M. , Gong, A.D. , Liu, Y.K. , Feng, Y.N. and Agboola, R.S. (2014) Trehalose 6‐phosphate phosphatase is required for development, virulence and mycotoxin biosynthesis apart from trehalose biosynthesis in Fusarium graminearum . Fungal Genet. Biol. 63, 24–41. [DOI] [PubMed] [Google Scholar]
- Tang, G. , Reinhart, B.J. , Bartel, D.P. and Zamore, P.D. (2003) A biochemical framework for RNA silencing in plants. Gene Dev. 17, 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco, M.L. , Dias, B.B. , Dall'Astta, R.C. , Pamphile, J.A. and Aragao, F.J. (2010) In vivo trans‐specific gene silencing in fungal cells by in planta expression of a double‐stranded RNA. BMC Biol. 8, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trail, F. (2009) For blighted waves of grain: Fusarium graminearum in the postgenomics era. Plant Physiol. 149, 103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trail, F. , Gaffoor, I. , Guenther, J.C. and Hallen, H.E. (2005) Using genomics to understand the disease cycle of the Fusarium head blight fungus, Gibberella zeae (anamorph Fusarium graminearum). Can. J. Plant Pathol. 27, 486–498. [Google Scholar]
- Trinci, A.P.J. (1973) The hyphal growth unit of wild type and spreading colonial mutants of Neurospora crassa . Arch. Mikrobiol. 91, 127–136. [DOI] [PubMed] [Google Scholar]
- Verweij, P.E. , Snelders, E. , Kema, G.H. , Mellado, E. and Melchers, W.J. (2009) Azole resistance in Aspergillus fumigatus: a side‐effect of environmental fungicide use? Lancet Infect. Dis. 9, 789–795. [DOI] [PubMed] [Google Scholar]
- Wang, M. and Jin, H. (2017) Spray‐induced gene silencing: a powerful innovative strategy for crop protection. Trends Microbiol. 25, 4–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. , Weiberg, A. , Lin, F.M. , Thomma, B.P. , Huang, H.D. and Jin, H. (2016) Bidirectional cross‐kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants, 2, 16 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woloshuk, C.P. and Shim, W.B. (2013) Aflatoxins, fumonisins, and trichothecenes: a convergence of knowledge. FEMS Microbiol. Rev. 37, 94–109. [DOI] [PubMed] [Google Scholar]
- Zhang, C.Q. , Chen, Y. , Yin, Y.N. , Ji, H.H. , Shim, W.B. , Hou, Y.P. , Zhou, M.G. , Li, X.D. and Ma, Z.H. (2015) A small molecule species specifically inhibits Fusarium myosin I. Environ. Microbiol. 17, 2735–2746. [DOI] [PubMed] [Google Scholar]
- Zhang, C. and Ruvkun, G. (2012) New insights into siRNA amplification and RNAi. RNA Biol. 9, 1045–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Z.T. , Hou, Y.P. , Cai, Y.Q. , Zhang, Y. , Li, Y.J. and Zhou, M.G. (2015) Whole‐genome sequencing reveals that mutations in myosin‐5 confer resistance to the fungicide phenamacril in Fusarium graminearum . Sci. Rep. 5, 8248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Z.T. , Liu, X.M. , Li, B. , Cai, Y.Q. , Zhu, Y.Y. and Zhou, M.G. (2016) Myosins FaMyo2B and Famyo2 affect asexual and sexual development, reduce pathogenicity, and FaMyo2B acts jointly with the myosin passenger protein FaSmy1 to affect resistance to phenamacril in Fusarium asiaticum . PLoS One, 11, e0154058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 A diagram of the eight Myo5 gene fragments used for making RNAi constructs, and the integration of the Myo5RNAi constructs into F. asiaticum. (a) The Myo5 gene sequence was divided into eight segments (Myo5‐1 to ‐8). A short overlap was located at the ends of the junction regions. (b) A schematic diagram for homologous recombination between the replacement vectors pMyo5RNAi and the PLS gene locus of F. asiaticum. The PCR primers and probe for detection of transformants are indicated.
Fig. S2 PCR and Southern blot analyses of Myo5RNAi transformants. (a) PCR products from the 5′ and 3′ regions of the disrupted PLS gene were amplified with the primers TetP9/PgpdP5 and Pgpd1P3/TetP10, respectively. The 1751‐ (U) and 1777‐bp (D) fragments were amplified from the 5′ and 3′ regions of the disrupted PLS gene, respectively. M: λDNA/PstI DNA Marker. (b) Southern blot analysis of Myo5RNAi transformants. The genomic DNAs were digested with NcoI and hybridized with a fragment of a neomycin resistance gene amplified with the primers G418ProbeP1/ G418ProbeP2. The sizes of hybridization fragments for the strains Myo5RNAi‐1 to Myo5RNAi‐8 were 6738/4225, 6758/4225, 6824/4225, 6862/4225, 6764/4225, 6766/4225, 6908/4225, and 6786/4225 bp, respectively.
Fig. S3 The labelling of Myo5‐8 dsRNA with fluorescein‐12‐UTP and spraying of fluorescein‐Myo5‐8 dsRNA and fluorescein‐UTP on cut coleoptile. (A) Fluorescein‐Myo5 dsRNAs were labeled using the fluorescein RNA Labeling Mix Kit and purified using the MEGAscript RNAi Kit (Invitrogen) following MEGAscript protocols. The products were detected using GelDoc XR and Quantity One 4.5.2 software (Bio‐Rad, Hercules, CA) with or without ethidium bromide (EB). (B) Microscopy of fluorescein‐Myo5‐8 dsRNA, fluorescein‐GFP dsRNA and fluorescein‐UTP uptake by tip cut coleoptile 24 h after spraying. Scale bar = 300 µm.
Fig. S4 Profiling of Myo5‐8 dsRNA derived sRNAs in coleoptiles spraying Myo5‐8 siRNA and Myo5‐8 dsRNA. Top panel is a diagram of eight fragments derived from Myo5 cDNA. The uppermost 4 mm of coleoptile of 3‐d‐old Huaimai33 seeding was cut off and sprayed with 400 ng Myo5‐8 dsRNA or Myo5‐8 siRNA. Small RNAs were isolated from the uppermost 7 mm of 10 coleoptiles and sequenced 12 hours and 72 hours post spraying.
Fig. S5 Phylogenetic analysis of the relationship between the six Arabidopsis RdRP proteins and the three wheat RdRP proteins. GenBank accession numbers are as follows: AtRdRP1, AY148431; AtRdRP2, At4g11130; AtRdRP3, At2g19910; AtRdRP4, At2g19920; AtRdRP5, At2g19930 and AtRdRP6, AF268093; Sequence alignment, distance calculation, and tree construction were performed with the use of the Clustal W program.
Fig. S6 A proposed model for Fusarium‐plant interaction after dsRNA sprayed. Fungal cell showing the dsRNA (on the plant surface) taken up by the fungal cells directly and the siRNA transferred from plant cell leading to target mRNA degradation. Fusarium is unable to maintain siRNA amplification by RdRPs machinery leading to short‐lasting SIGS effect. However, the plant cell could provide substantial siRNAs via secondary amplification machinery and determined the duration of SIGS.
Fig. S7 The quality of total RNA using for small RNA sequencing.
Fig. S8 The quality assessment of small RNA data.
Table S1 Strains used in this study.
Table S2 A list of small RNA that mapped to myosin 5 mRNA.
Table S3 Primers and sequences used in this study.


