Summary
Reactive oxygen species (ROS) are associated with various developmental processes and host–pathogen interactions in pathogenic fungi. Peroxidases are a group of ROS‐detoxifying enzymes that are involved in the oxidative stress response and in a variety of physiological processes. In this study, we performed a genome‐wide functional characterization of putative peroxidase genes in Fusarium graminearum, a head blight pathogen of cereal crops. We identified 31 putative peroxidase genes and generated deletion mutants for these genes. Twenty‐six of the deletion mutants showed developmental phenotypes indistinguishable from that of the wild‐type, and five deletion mutants exhibited phenotypic changes in at least one phenotypic category. Four deletion mutants, fca6, fca7, fpx1 and fpx15, showed increased sensitivity to extracellular H2O2. Deletion mutants of FCA7 also exhibited reduced virulence and increased trichothecene production compared with those of the wild‐type strain, suggesting that Fca7 may play an important role in the host–pathogen interaction in F. graminearum. To identify the transcription factors (TFs) regulating FCA6, FCA7, FPX1 and FPX15 in response to oxidative stress, we screened an F. graminearum TF mutant library for growth in the presence of H2O2 and found that multiple TFs co‐regulated the expression of FCA7 under oxidative stress conditions. These results demonstrate that a complex network of transcriptional regulators of antioxidant genes is involved in oxidative stress responses in this fungus. Moreover, our study provides insights into the roles of peroxidases in developmental processes and host–pathogen interactions in plant‐pathogenic fungi.
Keywords: Fusarium graminearum, oxidative stress response, peroxidase
Introduction
Fusarium graminearum, one of the most economically important plant pathogens, causes Fusarium head blight (FHB) in wheat, barley and rice, as well as ear rot in maize (Leslie and Summerell, 2006). Epidemics of FHB cause serious yield losses in major cereal crops worldwide (Goswami and Kistler, 2004; Windels, 2000). In addition to yield losses, this fungus is responsible for the contamination of grains with mycotoxins, such as trichothecenes and zearalenone, which are harmful to humans and to livestock (Desjardins and Proctor, 2007). In particular, trichothecenes are potent inhibitors of protein synthesis (Arunachalam and Doohan, 2013) and are well‐studied virulence factors in F. graminearum (Proctor et al., 1995).
Reactive oxygen species (ROS), such as superoxides ( ), hydroxyl radicals (OH•) and hydrogen peroxide (H2O2), are generated as byproducts of aerobic respiration and metabolic pathways that primarily occur in mitochondria, peroxisomes and chloroplasts (Heller and Tudzynski, 2011). On the one hand, because excessive amounts of ROS can damage cellular components by oxidizing membrane lipids, cellular proteins and nucleic acids, living organisms possess efficient ROS‐degrading mechanisms (Camhi et al., 1995). On the other hand, some ROS, particularly H2O2, act as secondary messengers in important signal transduction pathways (Apel and Hirt, 2004). Therefore, the delicate balance between ROS generation and scavenging is expected to be tightly regulated by a complex antioxidant defence mechanism comprising both enzymatic and non‐enzymatic components.
ROS also play an important role in plant–pathogen interactions. During plant infections, phytopathogenic fungi are often exposed to oxidative stress conditions caused by the oxidative burst, a rapid and transient accumulation of ROS (Mehdy, 1994; Wojtaszek, 1997). The oxidative burst is an immediate and non‐specific plant defence response triggered by pathogen attack. The excessive accumulation of ROS induces other plant defence responses, such as the cross‐linking of cell walls and programmed cell death of plant cells at infection sites, and can also directly kill microbial pathogens (Lamb and Dixon, 1997; Levine et al., 1994; Torres et al., 2006). Therefore, plant‐pathogenic fungi have evolved effective ROS‐scavenging mechanisms to detoxify plant‐derived ROS and to successfully colonize host plants.
Peroxidases are major H2O2‐decomposing enzymes that catalyse the oxidation of various organic and inorganic compounds using H2O2 or organic hydroperoxides as electron acceptors (Heller and Tudzynski, 2011). Peroxidases are involved in the oxidative stress response as antioxidant enzymes, and many studies have attempted to define their roles in ROS detoxification during the initial plant infection process (Garre et al., 1998; Mir et al., 2015; Robbertse et al., 2003; Skamnioti et al., 2007). Moreover, peroxidases, such as catalase, catalase‐peroxidase and peroxiredoxin, are required as redox controllers in various physiological processes (Fourquet et al., 2008; König et al., 2012), demonstrating that they are also closely involved in a variety of biological processes.
Several important signal mediators, such as transcription factors (TFs), which orchestrate oxidative stress responses, have been identified in fungi. Yap1 and Skn7 are the best‐characterized TFs; they play crucial roles in the oxidative stress response by regulating the expression of genes encoding antioxidant enzymes in Saccharomyces cerevisiae (Lee et al., 1999). Several studies have revealed that Yap1 and Skn7 have a conserved function in the oxidative stress response in various plant‐pathogenic fungi, including Ustilago maydis, Magnaporthe oryzae, Alternaria alternata, Cochliobolus heterostrophus and Botrytis cinerea (Chen et al., 2012; Guo et al., 2011; Lev et al., 2005; Lin et al., 2009; Molina and Kahmann, 2007; Shalaby et al., 2014; Temme and Tudzynski, 2009). In F. graminearum, several known and novel TFs involved in the oxidative stress response have been characterized (Jiang et al., 2015; Lysøe et al., 2011; Montibus et al., 2013; Wang et al., 2011). These TFs have been reported to mediate the oxidative stress response by regulating the expression of genes encoding putative antioxidant enzymes (peroxidases). However, whether or not these putative antioxidant genes are indeed important for the oxidative stress response in F. graminearum remains unknown.
Recent studies have revealed that oxidative stress is also related to secondary metabolite biosynthesis in fungi (Hong et al., 2013a; Montibus et al., 2015). In Aspergillus species, aflatoxin biosynthesis is triggered in response to oxidative stress (Hong et al., 2013a; Reverberi et al., 2012). Accordingly an exogenous treatment with H2O2 leads to enhanced trichothecene production and induces the expression of trichothecene biosynthetic genes in F. graminearum (Ponts et al., 2006, 2007). Several studies have suggested that secondary metabolites may act in concert with antioxidant enzymes to protect cells against oxidative stress (Hong et al., 2013a, 2013b).
In this study, we functionally characterized 31 putative peroxidase genes through an extensive phenome analysis of F. graminearum. The aims of this study were as follows: (i) to characterize the function of putative peroxidases in various developmental processes, including vegetative growth, conidiation, sexual development, virulence, mycotoxin production and the oxidative stress response; and (ii) to identify TFs that regulate the expression of the genes encoding these putative peroxidases in response to oxidative stress. To our knowledge, this is the first study to investigate the function of genome‐wide putative peroxidase genes in the development and infection processes of phytopathogenic fungi. Our results provide insights into the roles of peroxidases in intracellular processes and host–pathogen interactions.
Results
Identification of putative peroxidase genes in F. graminearum
We used the previously constructed fungal peroxidase database (http://peroxidase.riceblast.snu.ac.kr) to identify all of the putative peroxidase genes in F. graminearum (Choi et al., 2014). The F. graminearum genome contains 23 haem peroxidases and eight non‐haem peroxidases, including five previously reported putative monofunctional catalase genes (FCA1, FCA2, FCA3, FCA4 and FCA5), two putative bifunctional catalase‐peroxidase genes (FCA6 and FCA7) (Lee et al., 2014) and three NADPH oxidase genes (NOXA, NOXB and NOXC) (Takemoto et al., 2007; Wang et al., 2014) (Table 1). We designated the remaining 21 peroxidase genes as FPX1 to FPX21 (F. graminearum peroxidase) for convenience. To investigate the phylogenetic relationships between the peroxidases of F. graminearum, we constructed a phylogenetic tree based on the predicted amino acid sequences of these peroxidases (Fig. 1A,B). Because there was no evolutionary relationship between haem (Fig. 1A) and non‐haem (Fig. 1B) peroxidases, a separate phylogenetic tree was constructed for each group. Peroxidases belonging to the same family generally clustered together, indicating the genetic similarity of their protein sequences. The relationships between peroxidase families were relatively weak, as indicated by the low bootstrap values.
Table 1.
Putative peroxidases in Fusarium graminearum.
| Types of peroxidase | Locus ID | Gene name | Reference | ||
|---|---|---|---|---|---|
| Haem peroxidase | Catalase superfamily | Catalase | FGSG_06554 | FCA1 | Lee et al. ( 2014) |
| FGSG_06733 | FCA2 | Lee et al. ( 2014) | |||
| FGSG_16526 | FCA3 | Lee et al. ( 2014) | |||
| FGSG_02881 | FCA4 | Lee et al. ( 2014) | |||
| FGSG_06596 | FCA5 | Lee et al. ( 2014) | |||
| Class I peroxidase | Catalase‐peroxidase | FGSG_02974 | FCA6 | Lee et al. ( 2014) | |
| FGSG_12369 | FCA7 | Lee et al. ( 2014) | |||
| Cytochrome c peroxidase | FGSG_01245 | FPX1 | This study | ||
| FGSG_10606 | FPX2 | This study | |||
| Hybrid ascorbate‐cytochrome c peroxidase | FGSG_04434 | FPX3 | This study | ||
| Class II peroxidase | Other class II peroxidase | FGSG_16013 | FPX4 | This study | |
| Haloperoxidase superfamily | Haloperoxidase | FGSG_03708 | FPX5 | This study | |
| FGSG_17448 | FPX6 | This study | |||
| FGSG_02341 | FPX7 | This study | |||
| FGSG_08911 | FPX8 | This study | |||
| FGSG_03436 | FPX9 | This study | |||
| Peroxidase‐cyclooxygenase superfamily | Prostaglandin H synthase (cyclooxygenase) | FGSG_17094 | FPX10 | This study | |
| Linoleate diol synthase (PGHS‐like) | FGSG_02668 | FPX11 | This study | ||
| FGSG_10960 | FPX12 | This study | |||
| FGSG_11146 | FPX13 | This study | |||
| NADPH oxidase superfamily | NoxA | FGSG_00739 | NOXA | Wang et al. ( 2014) | |
| NoxB | FGSG_10807 | NOXB | Wang et al. ( 2014) | ||
| NoxC | FGSG_11195 | NOXC | This study | ||
| Non‐haem peroxidase | Peroxiredoxin superfamily | 1‐cysteine peroxiredoxin | FGSG_07536 | FPX14 | This study |
| Typical 2‐cysteine peroxiredoxin | FGSG_03180 | FPX15 | This study | ||
| Atypical 2‐cysteine peroxiredoxin (type Q, BCP) | FGSG_10296 | FPX16 | This study | ||
| Atypical 2‐cysteine peroxiredoxin (type II, type V) | FGSG_08677 | FPX17 | This study | ||
| FGSG_00353 | FPX18 | This study | |||
| Alkylhydroperoxidase superfamily | Carboxymuconolactone decarboxylase (no peroxidase activity) | FGSG_01796 | FPX19 | This study | |
| FGSG_10039 | FPX20 | This study | |||
| Glutathione peroxidase | Fungal–bacterial glutathione peroxidase | FGSG_06150 | FPX21 | This study | |
Figure 1.
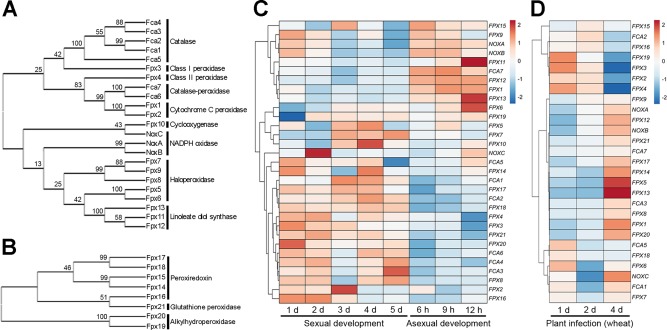
Phylogenetic and transcriptional analysis of peroxidase genes. Phylogenetic trees based on the amino acid sequences of 23 putative haem peroxidases (A) and eight putative non‐haem peroxidases (B) in Fusarium graminearum. The phylogenetic trees were constructed using the mega program (version 6.06) by the neighbour‐joining method with 2000 bootstrap replicates (Tamura et al., 2013). The numbers at the nodes represent the bootstrap percentages. (C, D) Heatmap visualization of the peroxidase gene transcriptional profiles during asexual and sexual development (C) and plant infection (D). The heatmap depicts peroxidase gene transcript abundances during various asexual and sexual developmental stages based on log2‐based relative transcript abundances compared with the 0‐day (sexual development), 3‐h (asexual development) and mock‐inoculated (plant infection) samples. Red and blue represent higher and lower expression, respectively. The rows represent transcriptional units. The expression data were obtained from previous studies (Harris et al., 2016; Sikhakolli et al., 2012; Son et al., 2013, 2016) and visualized using ClustVis (Metsalu and Vilo, 2015).
To assess the genetic requirements for peroxidases in fungal developmental processes, we analysed transcript profiles during sexual and asexual development of the fungus (Fig. 1C) and during plant infection (Fig. 1D). RNA‐sequencing (RNA‐seq) and microarray results were obtained from previous studies (Harris et al., 2016; Sikhakolli et al., 2012; Son et al., 2013, 2016), reanalysed and visualized using ClustVis (Metsalu and Vilo, 2015). Expression profiles during plant infection were additionally normalized to that of β‐tubulin (FGSG_09530). Approximately two‐thirds of the studied genes (FPX5–FPX16 in Fig. 1C) were up‐regulated during the initial and/or late stages of sexual reproduction, but most of the genes were down‐regulated during conidiation. The other 10 genes (FPX15–FPX19 in Fig. 1C) were up‐regulated during asexual reproduction, but showed maintained or reduced expression after sexual induction, with the exception of FPX9, NOXA and NOXB. During the early infection of wheat, approximately one‐half of the studied genes (FPX15–FPX4 and FCA5–FPX7 in Fig. 1D) were up‐regulated at the initial stage of infection (1 and 2 days after inoculation), whereas the other one‐half of the genes were up‐regulated during the later days of infection (4 days after inoculation). These results suggest that peroxidase‐mediated molecular processes are closely related to fungal development and infection in F. graminearum.
Targeted deletion of putative peroxidase genes in F. graminearum
To investigate the functions of the putative peroxidase genes, we performed targeted gene deletion by homologous recombination. Each peroxidase gene of the F. graminearum wild‐type strain Z‐3639 was replaced with the geneticin resistance gene (GEN) to create individual gene deletion mutants (Table S1, see Supporting Information). Successful disruption of 24 peroxidase genes was confirmed by Southern blot hybridization using a 5′ or 3′ flanking region as a probe (Fig. S1, see Supporting Information). The seven deletion mutants of the catalase and catalase‐peroxidase genes, fca1–fca7, were derived in a previous study (Lee et al., 2014).
We analysed 31 peroxidase deletion mutants for defects in various developmental processes, including vegetative growth, sexual and asexual development, trichothecene production, virulence and the oxidative stress response. Overall, we found that five peroxidase mutants were defective in at least one phenotypic category compared with the wild‐type (Table 2). None of the peroxidase deletion mutants exhibited defects in vegetative growth or conidiation (Table S2, see Supporting Information). When cultured on complete medium (CM) and minimal medium (MM), there was no significant difference in radial growth or colony morphology between the wild‐type and the peroxidase deletion mutant strains. Moreover, all of the 31 peroxidase deletion mutants were normal with regard to conidial production (Table S2). With respect to sexual development, only the noxA deletion mutants showed defects in perithecium production (Fig. 2A). The wild‐type and other peroxidase deletion mutants produced normal perithecia 7 days after sexual induction.
Table 2.
Summary of the phenotypes of the Fusarium graminearum peroxidase mutants.
| Developmental processes | Mutants showing defects |
|---|---|
| Vegetative growth* | None (same as wild‐type) |
| Conidiation† | None (same as wild‐type) |
| Sexual development‡ | noxA |
| Oxidative stress response§ | fca6, fca7, fpx1, fpx15 |
| Pathogenicity¶ | noxA, fca7 |
| Trichothecene production** | fca7 |
*Radial growth was measured after 5 days of incubation on complete medium (CM) and minimal medium (MM).
†The conidia were counted after 3 days of incubation in carboxymethyl cellulose (CMC) medium.
‡The formation of perithecia was observed 10 days after sexual induction on carrot agar.
§The sensitivity to oxidants (H2O2) was tested.
¶The disease index was measured 21 days after inoculation.
**Trichothecene production in minimal medium containing 5 mm agmatine (MMA) was analysed.
Figure 2.

Sexual development and virulence of NADPH oxidase deletion mutants. (A) Perithecium formation by the NADPH oxidase deletion mutants. Cirrhi (indicated by white arrows) were observed in the wild‐type (WT) and in the noxB and noxC deletion mutants 10 days after sexual induction. Scale bar, 500 µm. (B) Virulence on wheat heads. The centre spikelet of each wheat head was injected with 10 µL of a conidial suspension, and photographs were taken at 21 days after inoculation. Arrowheads indicate the inoculated spikelets. ‘Mock’ indicates wheat heads that were mock inoculated with 0.01% Tween 20.
NADPH oxidase genes in F. graminearum
The functions of two NADPH oxidase genes, NOXA and NOXB, in sexual development and pathogenicity have been characterized previously (Wang et al., 2014; Zhang et al., 2016). Here, we identified and characterized a novel NADPH oxidase gene, NOXC. Disruption of NOXA caused significant defects in sexual development, whereas deletion mutants of NOXB and NOXC showed no defects in perithecium production or maturation (Fig. 2A). Perithecia with cirrhi were observed in noxB and noxC mutants 10 days after sexual induction. In assays of fungal infection of wheat heads, the noxA deletion mutants showed significantly reduced virulence (P < 0.01, t‐test), whereas the deletion mutants of noxB and noxC caused typical head blight symptoms (Fig. 2B).
Peroxidases involved in the oxidative stress response
To investigate the sensitivity of the 31 peroxidase deletion mutants to oxidative stress, all of the knockout mutants were cultured in CM supplemented with 10 mm H2O2. Only four deletion mutants (fca6, fca7, fpx1 and fpx15) exhibited significantly altered sensitivity to oxidative stress mediated by H2O2 compared with that of the wild‐type strain (Fig. 3A). Of these four deletion mutants, fca7 and fpx15 were much more susceptible to H2O2 than were the other mutants.
Figure 3.
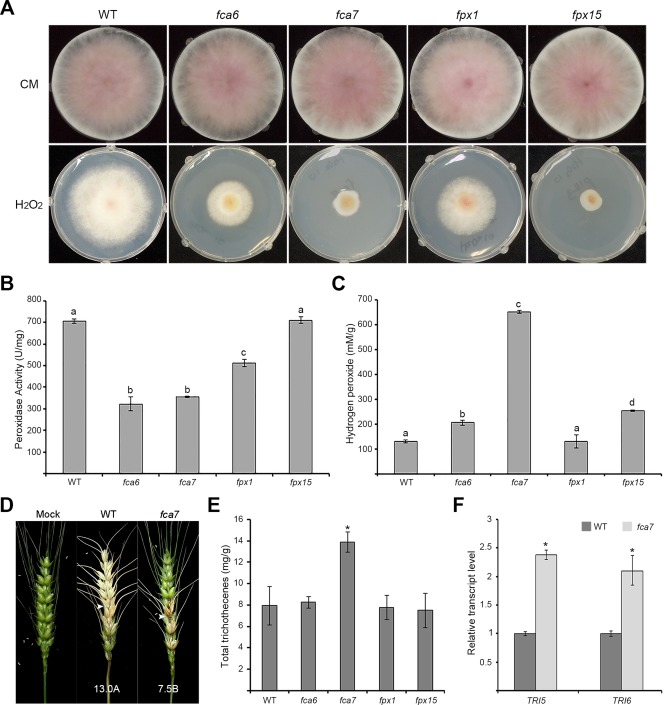
Peroxidases involved in the oxidative stress responses in Fusarium graminearum. (A) Oxidative stress sensitivity of F. graminearum strains. The mycelial growth of four peroxidase deletion mutants was evaluated on complete medium (CM) with and without supplementation with 10 mm H2O2. Photographs were taken 5 days after inoculation. (B) Peroxidase enzyme activities of the F. graminearum strains. (C) Hydrogen peroxide detection in mycelia of the F. graminearum strains. (D) Virulence on wheat heads. The centre spikelet of each wheat head was injected with 10 µL of a conidial suspension, and photographs were taken 21 days after inoculation. Arrowheads indicate the inoculated spikelets. ‘Mock’ indicates wheat heads that were mock inoculated with 0.01% Tween 20. (E) Total trichothecene production by the F. graminearum strains. Each strain was grown in minimal medium containing 5 mM agmatine (MMA) for 7 days. Trichothecenes were analysed by gas chromatography‐mass spectrometry (GC‐MS) and quantified based on the biomass of each strain. (F) Transcript levels of TRI5 and TRI6 in the wild‐type (WT) and fca7 deletion mutant strains. The transcript levels were analysed by quantitative real‐time polymerase chain reaction (qRT‐PCR) 4 days after inoculation in MMA. Asterisk indicates significant difference: P < 0.01, Tukey's test.
To determine whether the deletion of these four peroxidase genes leads to decreased peroxidase enzyme activity, we measured the peroxidase activity of the deletion mutants under oxidative conditions (Fig. 3B). The total peroxidase activities of the fca6, fca7 and fpx1 deletion mutants were significantly reduced compared with that of the wild‐type strain. Under our experimental conditions, the peroxidase activity of the fpx15 deletion mutant was similar to that of the wild‐type strain. These results suggest that the oxidative stress sensitivities of the fca6, fca7 and fpx1 deletion mutants are mainly caused by decreased peroxidase enzyme activity.
To investigate the H2O2 degradation capabilities of Fca6, Fca7, Fpx1 and Fpx15, we measured the concentration of H2O2 in mycelia after short‐term treatment of the cultures with H2O2 (Fig. 3C). We found that the H2O2 concentration in mycelia of the fca7 strain was significantly increased compared with that of the wild‐type. The concentration of H2O2 in the mycelia of the other deletion mutants was slightly higher than or similar to that of the wild‐type.
Peroxidases required for virulence and mycotoxin production
To determine whether the deletion of peroxidase genes affects the virulence of F. graminearum, we tested the pathogenicity of the peroxidase deletion mutants on flowering wheat heads. We found that the fca6, fpx1 and fpx15 deletion mutants exhibited normal virulence, despite being highly susceptible to H2O2. However, only the fca7 deletion mutant showed reduced (but not abolished) virulence compared with that of the wild‐type strain (Fig. 3D).
We also analysed total trichothecene production (deoxynivalenol and 15‐acetyl‐deoxynivalenol) by the peroxidase deletion mutants. Interestingly, the accumulation of trichothecenes by the fca7 deletion mutant was significantly enhanced compared with that of the wild‐type. The other peroxidase deletion mutants, including fca6, fpx1 and fpx15, produced amounts of trichothecenes similar to that of the wild‐type strain (Fig. 3E). The quantitative real‐time polymerase chain reaction (qRT‐PCR) results demonstrated that the expression of the trichothecene biosynthetic genes TRI5 and TRI6 was highly induced in the fca7 deletion mutant compared with the wild‐type strain (Fig. 3F). These results indicate that Fca7 has non‐redundant and crucial functions in virulence and trichothecene biosynthesis in F. graminearum. However, both the wild‐type and the FCA7 overexpression (FCA7oe) strains produced similar levels of trichothecenes, perhaps because the basal expression of FCA7 was sufficient to negatively regulate the production of trichothecenes under our experimental conditions (Fig. S2, see Supporting Information).
Identification of TFs involved in the oxidative stress response
To dissect the regulatory mechanisms of the major peroxidases, we first attempted to identify TFs involved in the oxidative stress response in F. graminearum among 657 TF mutants (Son et al., 2011b). We first screened the TF mutants using various oxidative stress‐inducing agents (10 mm H2O2, 0.1 mm menadione and 1 mm diamide). From the TF mutants showing altered sensitivity to oxidative stress‐inducing agents, we preferentially selected eight TF mutants that were highly sensitive to H2O2 for further study (Fig. 4A and Table 3). Of these, three TFs (FgAp1, FgSkn7 and Zif1) have been functionally characterized previously in F. graminearum (Jiang et al., 2015; Montibus et al., 2013; Wang et al., 2011). FGSG_05171 encodes a homologue of Neurospora crassa Cys‐3 and Aspergillus nidulans MetR involved in the regulation of the sulfur regulatory circuit (Fu et al., 1989; Natorff et al., 2003), and FGSG_01100 encodes a homologue of FoxO1 required for the oxidative stress response in mammalian cell systems (Furukawa‐Hibi et al., 2005). The three putative TFs containing a Zn(II)2Cys6 DNA‐binding domain (FGSG_08924 and FGSG_01293) and a C2H2 zinc finger domain (FGSG_01298) have not been functionally characterized in other fungi (Table 3).
Figure 4.
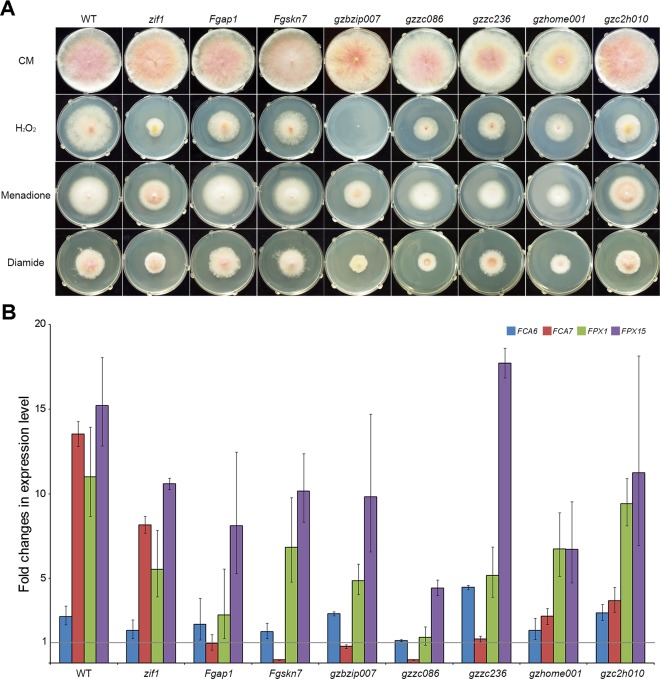
Characterization of transcription factors (TFs) involved in the oxidative stress response. (A) Oxidative stress sensitivity of eight TF mutants. The mycelial growth of the TF deletion mutants was evaluated on complete medium (CM) and on CM supplemented with 10 mm H2O2, 0.1 mm menadione or 1 mm diamide. Photographs were taken 5 days after inoculation. (B) Transcript abundances of peroxidase genes in the TF mutants. The transcript levels of FCA6, FCA7, FPX1 and FPX15 were analysed by quantitative real‐time polymerase chain reaction (qRT‐PCR). Total RNA was isolated from wild‐type (WT) and TF deletion mutant strains grown for 30 min in CM only or in CM supplemented with 5 mm H2O2.
Table 3.
Putative transcription factors involved in the oxidative stress response.
| Locus ID | Gene name | Description of the gene product | Species | Homologue | Reference |
|---|---|---|---|---|---|
| FGSG_01555 | ZIF1 | Related to bZIP transcription factor | Fusarium graminearum | ZIF1 | Wang et al. ( 2011) |
| FGSG_08800 | FgAP1 | Related to AP1‐like transcription factor | F. graminearum | FgAP1 | Montibus et al. ( 2013) |
| FGSG_06359 | FgSKN7 | Related to SKN7 | F. graminearum | FgSKN7 | Jiang et al. ( 2015) |
| FGSG_05171 | GzbZIP007 | Related to regulatory protein cys‐3 | Neurospora crassa | CYS‐3 | Fu et al. ( 1989) |
| FGSG_08924 | GzZC086 | Conserved hypothetical protein | N/A | N/A | Son et al. ( 2011b) |
| FGSG_01293 | GzZC236 | Related to Zn(II)2Cys6 transcriptional activator | N/A | N/A | Son et al. ( 2011b) |
| FGSG_01100 | GzHOME001 | Related to LIM homeobox protein | Human | FoxO1 | Furukawa‐Hibi et al. ( 2005) |
| FGSG_01298 | GzC2H010 | Conserved hypothetical protein | N/A | N/A | Son et al. ( 2011b) |
Expression of four peroxidase genes in TF deletion mutants
We hypothesized that the TFs involved in the oxidative stress response regulate the expression of the four peroxidase genes that are required for the oxidative stress response. We performed qRT‐PCR to measure the transcript levels of FCA6, FCA7, FPX1 and FPX15 in the wild‐type strain and in the eight selected TF deletion mutants under normal and oxidative stress conditions (Fig. 4B). The transcript levels of FCA6, FCA7, FPX1 and FPX15 were markedly up‐regulated in response to H2O2 in the wild‐type strain. In the TF deletion mutants, the expression levels of FCA6, FPX1 and FPX15 were increased by H2O2 treatment (similar to the wild‐type), whereas the transcript levels of FCA7 showed a different pattern. In seven TF deletion mutants (Fgap1, Fgskn7, gzbzip007, gzzc086, gzzc236, gzhome001 and gzc2h010), the expression of FCA7 was not highly induced or was even reduced by H2O2 treatment (Fig. 4B). In the zif1 mutants, the expression of FCA7 was relatively highly expressed following H2O2 treatment compared with the expression in the other deletion mutants, indicating that FCA7 expression is a key factor in the oxidative stress response in TF mutants.
Genetic relationship among TFs and FCA7
To investigate the genetic relationships between FCA7 and the eight TFs, we generated TF mutants with FCA7 overexpression by outcrosses (Table S1). Of the seven TF deletion mutants (Fgap1, Fgskn7, gzbzip007, gzzc086, gzzc236, gzhome001 and gzc2h010) that showed unchanged or reduced expression of FCA7 (Fig. 4B), the oxidative stress sensitivity of six (Fgap1, Fgskn7, gzzc086, gzzc236, gzhome001 and gzc2h010) was restored to some degree by the overexpression of FCA7 (Fig. 5). Of these, the gzzc236 mutants have been reported previously to show reduced virulence (Son et al., 2011b). Although the oxidative stress sensitivity of these mutants was restored by the overexpression of FCA7, virulence was not recovered (Fig. S4, see Supporting Information). Because the gzbzip007 deletion mutants did not grow at all on CM supplemented with 10 mm H2O2, we assayed the sensitivities of the gzbzip007 mutants to relatively low oxidative stress conditions (medium containing 3 mm H2O2). The vegetative growth of gzbzip007 under mild oxidative stress conditions was not restored by the overexpression of FCA7 (Fig. S3, see Supporting Information). The oxidative stress sensitivity of the zif1 mutant was slightly restored by FCA7 overexpression, consistent with the qRT‐PCR results. Taken together, these results demonstrate that the altered sensitivities of the TF mutants to oxidative stress are primarily caused by the repression of FCA7.
Figure 5.
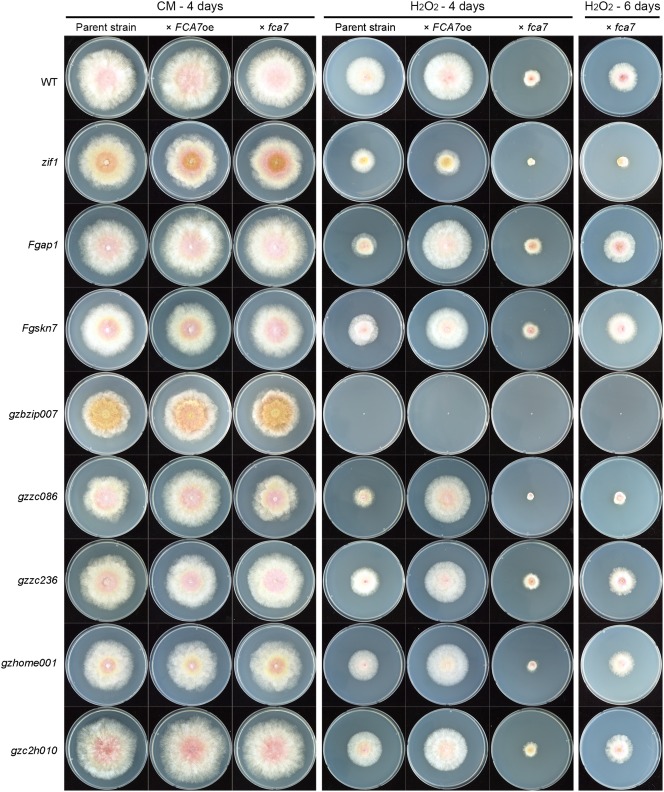
Oxidative stress sensitivity of eight transcription factor (TF) mutants carrying an FCA7 deletion or FCA7 overexpression (FCA7oe). The mycelial growth of TF mutant strains on complete medium (CM) supplemented with 10 mm H2O2 is shown. Photographs were taken 4 and 6 days after inoculation. WT, wild‐type.
We also outcrossed the TF deletion mutants to a heterothallic fca7 deletion strain, mat2 fca7. If these TFs are involved in a different antioxidant system, the double mutation would be expected to produce synergistic defects in the oxidative stress response. We found that two TF deletion mutants (zif1 and gzzc086) showed markedly increased sensitivity to oxidative stress when combined with the deletion of FCA7 (Fig. 5). Based on the phenotypes of the gzzc086 mutants carrying FCA7 deletion or overexpression, we suspected that GzZC086 has a regulatory function for FCA7 expression and that it is involved in multiple antioxidant pathways. Double deletion mutants of fca7 and TFs (Fgap1, Fgskn7, gzzc236, gzhome001 and gzc2h010) did not show synergistic effects on oxidative stress sensitivity compared with the corresponding single‐gene deletion mutants.
Genetic regulatory network of the TFs involved in oxidative stress responses
We further examined the transcript levels of the TF genes in the wild‐type strain under normal and oxidative stress conditions using qRT‐PCR (Fig. 6A). The expression of five TF genes (ZIF1, FgAP1, FgSKN7, GzbZIP007 and GzZC086) was significantly up‐regulated in response to H2O2, whereas the expression of the other three TF genes (GzZC236, GzHOME001 and GzC2H010) was not induced under our experimental conditions. To investigate the genetic regulatory network with which TF genes are associated, we examined the expression of the five TF genes showing increased expression under oxidative stress conditions in eight TF deletion mutants. We compared the fold change in expression of these five TF genes after H2O2 treatment in the eight TF deletion mutants with the fold change in the wild‐type strain (Fig. 6B). H2O2‐mediated induction of ZIF1, FgAP1 and FgSKN7 was nearly abolished in the gzzc086 mutants, and GzZC086 was not induced in gzhome001. We also found that the expression of ZIF1 and FgSKN7 did not increase in response to H2O2 in the gzc2h010 and gzzc236 mutants, respectively. The expression of GzbZIP007 was not reduced in any of the TF deletion mutants, indicating that GzbZIP007 is involved in a completely independent regulatory system for oxidative stress responses. Furthermore, it appeared that feedback regulatory mechanisms among the TFs, ZIF1–GzZC086, ZIF1–Gz236 and FgAP1–GzZC236, modulated the genetic regulatory networks. Based on our data, we propose a simplified genetic regulatory network that illustrates how TF genes are involved in the oxidative stress response in F. graminearum (Fig. 6C).
Figure 6.
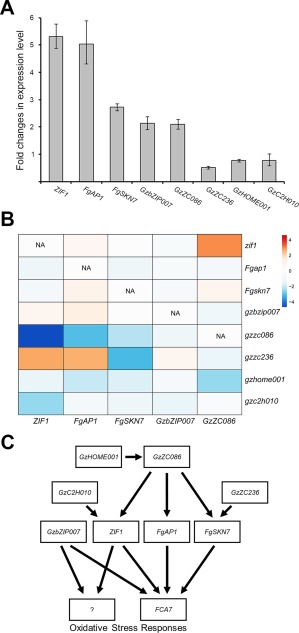
Genetic network of transcription factor (TF) genes involved in the oxidative stress response in Fusarium graminearum. (A) Fold change values in the expression of TF genes after H2O2 treatment in the wild‐type strain. The transcript levels of the TF genes were analysed by quantitative real‐time polymerase chain reaction (qRT‐PCR). Total RNA was isolated from the wild‐type strain grown for 30 min in complete medium (CM) only or in CM supplemented with 5 mm H2O2. (B) Heatmap of selected TF genes that were up‐regulated in the wild‐type strain in response to H2O2. The log base ratio of the fold change in gene expression in each deletion mutant compared with that in the wild‐type strain was converted to a heatmap using ClustVis (Metsalu and Vilo, 2015). (C) Proposed genetic network of TFs and FCA7 in the oxidative stress response. ?, unknown antioxidant components.
In conclusion, we found that, although the expression of FCA7 was not the only component of the oxidative stress response, Fca7 was the major antioxidant enzyme produced. We also concluded that at least six TFs (FgAP1, FgSKN7, GzZC086, GzZC236, GzHOME001 and GzC2H010) regulated the expression of FCA7 under oxidative stress conditions, demonstrating the existence of a complex network of transcriptional activators of antioxidant genes.
Discussion
Because plant‐pathogenic fungi encounter both internal and external oxidative stresses during development and during the process of plant infection, they have evolved effective ROS‐detoxifying mechanisms (Heller and Tudzynski, 2011). In particular, the rapid generation of H2O2 is one of the earliest plant defence responses to occur following the perception of signals associated with the presence of pathogens (Lamb and Dixon, 1997; Levine et al., 1994), and the roles of H2O2‐scavenging enzymes, such as catalases and catalase‐peroxidases, during plant invasion have been investigated in various phytopathogenic fungi (Garre et al., 1998; Robbertse et al., 2003; Schouten et al., 2002; Skamnioti et al., 2007; Tanabe et al., 2011). H2O2 also plays an important role as a signalling molecule in various developmental processes of fungi, such as cell differentiation and sexual development (Gessler et al., 2007; Hansberg et al., 1993; Lara‐Ortíz et al., 2003). Thus, cellular ROS levels should be extremely fine‐tuned by peroxidases during fungal development. In this study, we investigated the roles of putative peroxidases in F. graminearum. We identified 31 genes encoding putative peroxidases in F. graminearum and characterized their functions, not only in the oxidative stress response, but also in various developmental processes.
The F. graminearum genome contains 23 peroxidases belonging to the 11 haem peroxidase families and eight peroxidases belonging to the six non‐haem peroxidase families. In yeast, only two haem peroxidase families and five non‐haem peroxidase families have been found (Choi et al., 2014). A comparison of the predicted peroxidase genes in plant‐pathogenic fungi, including F. graminearum, M. oryzae, C. heterostrophus and yeast, revealed that the genomes of plant‐pathogenic fungi generally contain numerous haem peroxidase genes (Choi et al., 2014). Several studies have reported that certain haem peroxidase genes, such as haloperoxidase genes and catalase‐peroxidase genes, are mainly found in the genomes of phytopathogens (Gasselhuber et al., 2015; Zámocký and Obinger, 2010; Zámocký et al., 2012). These results imply a potential role of haem peroxidases in pathogenicity.
Phenome analysis of 31 peroxidase deletion mutants in F. graminearum revealed that only five peroxidase deletion mutants (fca6, fca7, fpx1, fpx15 and noxA) were involved in the various developmental processes examined. Other peroxidases may be able to functionally compensate for the loss of a single peroxidase, indicating overlapping and redundant functions of these peroxidases. The clustered peroxidase gene expression profiles observed during reproductive processes also support the overlapping functions of peroxidases in F. graminearum (Fig. 1C). Likewise, the deletion of multiple catalase genes in Cryptococcus neoformans did not cause visible phenotypic changes, because of the presence of a robust and redundant antioxidant defence system in this species (Giles et al., 2006). Moreover, in other filamentous fungi, such as B. cinerea and M. oryzae, deletion of the genes required for the oxidative stress response did not cause phenotypic changes in the fungal growth rate or conidia production (Huang et al., 2011; Temme and Tudzynski, 2009).
With respect to sexual development, only noxA deletion mutants showed defective perithecia production. Previous studies have demonstrated that NADPH oxidase‐dependent ROS signalling is important for cellular differentiation and development in fungi (Heller and Tudzynski, 2011; Takemoto et al., 2007). In this study, we identified three NADPH oxidase homologues in F. graminearum: NoxA, NoxB and NoxC. The function of NoxC has been poorly studied in filamentous fungi, whereas the functions of NoxA and NoxB in various cellular differentiation processes, including sexual development and pathogenicity, have been well characterized (Cano‐Domínguez et al., 2008; Egan et al., 2007; Giesbert et al., 2008; Lara‐Ortíz et al., 2003; Scott and Eaton, 2008; Segmüller et al., 2008; Takemoto et al., 2007; Wang et al., 2014; Zhang et al., 2016). A recent study has reported that NoxC exists in only seven Ascomycota and that most of these seven species are phytopathogenic fungi, suggesting that NoxC may have a specialized function in pathogenicity (Takemoto et al., 2007). Moreover, NOXC showed distinct and unique expression patterns during fungal reproduction compared with those of NOXA and NOXB (Fig. 1C). However, deletion of NOXC did not affect the pathogenicity of F. graminearum. Thus, the function of NOXC remains unclear.
Four peroxidase genes, FCA6, FCA7, FPX1 and FPX15, were closely involved in the oxidative stress response, but deletion of FPX15 did not affect peroxidase activity in F. graminearum (Fig. 3B). FPX15 is predicted to encode a non‐haem peroxidase belonging to the typical 2‐cysteine peroxiredoxin family. Peroxiredoxins possess both peroxidase and molecular chaperone activities (Jang et al., 2004), and the peroxidase‐to‐chaperone switch is triggered by oxidative stress (Jang et al., 2004; Kim et al., 2009). Based on previous and current results, we conclude that the increased sensitivity of fpx15 to oxidative stress is not directly related to its peroxidase activity, but to its chaperone activity.
Although the total peroxidase enzyme activity of fca6 was decreased to a level similar to that of fca7, the concentration of accumulated H2O2 in the mycelia of fca7 was much higher than that in those of fca6. FCA7 homologues have been predicted to encode an extracellular catalase‐peroxidase (Zámocký et al., 2009), and Fca7 contains a potential signal sequence for secretion predicted by SignalP 4.0 (Petersen et al., 2011). These results suggest that Fca7 may play a predominant role in the detoxification of external H2O2 in F. graminearum. Consistent with the fact that plant‐derived ROS scavenging by extracellular peroxidases is critical for successful infection, only fca7 mutants were reduced in virulence compared with that of the wild‐type strain.
Proteomic analysis of wheat spikelets during infection by F. graminearum revealed that proteins related to the oxidative burst pathway are induced at the early infection stage (Zhou et al., 2005, 2006). In this study, we discovered that, although deletion mutants of FCA6, FCA7, FPX1 and FPX15 showed increased sensitivity to oxidative stress mediated by H2O2, only fca7 deletion mutants exhibited reduced virulence compared with that of the wild‐type strain. In a number of phytopathogenic fungi, deletion of a gene that is essential for survival in the presence of high concentrations of H2O2 does not lessen virulence; this may be a result of the presence of alternative antioxidant systems (Robbertse et al., 2003; Temme and Tudzynski, 2009). Taken together, we conclude that Fca7 may function as a major H2O2‐scavenging enzyme and may play an irreplaceable role during the infection process in F. graminearum.
Recent studies have reported that oxidative stress and secondary metabolism are tightly linked in filamentous fungi, and have suggested that secondary metabolites play a protective role in the adaptation of plants to stress conditions (Hong et al., 2013a; Montibus et al., 2015). In F. graminearum, trichothecenes have been reported to be virulence factors for the fungal infection of wheat heads (Desjardins et al., 1996; Maier et al., 2006), and trichothecene biosynthesis has been shown to be triggered by exogenous H2O2 treatment associated with the increased expression of TRI genes (Ponts et al., 2006, 2007). In this study, we found that the fca7 deletion mutant produced more trichothecenes than the wild‐type strain, and that Fca7 is the major H2O2‐scavenging enzyme expressed in F. graminearum under oxidative stress conditions (Fig. 3C,E). Based on these findings, we conclude that Fca7‐mediated modulation of H2O2 may be one of the major determinants of trichothecene production in F. graminearum.
Eight TFs involved in the oxidative stress response were identified in F. graminearum. Previously, homologues of Yap1 and Skn7 were identified and functionally characterized in F. graminearum (Jiang et al., 2015; Montibus et al., 2013). Yap1 and Skn7 are central TFs that regulate the expression of oxidative stress‐related genes in S. cerevisiae (Lee et al., 1999). A novel bZIP TF, Zif1, has also been reported in F. graminearum (Wang et al., 2011). In this study, we identified five new TFs (GzbZIP007, GzZC086, GzZC236, GzHOME001 and GzC2H010) that are involved in the oxidative stress response in this fungus. We proposed a hypothetical simplified genetic network of TF genes in the oxidative stress response (Fig. 6C). Because studies of genetic networks governing the oxidative stress response have been limited because of the lack of TF mutants, further studies of these TFs in filamentous fungi, including F. graminearum, will expand our understanding of the molecular mechanisms underlying the oxidative stress response in this type of fungus.
We found that multiple TFs (FgAP1, FgSKN7, GzZC086, GzZC236, GzHOME001 and GzC2H010) co‐regulated the expression of FCA7 in response to oxidative stress, reflecting a complex network of transcriptional activators of antioxidant genes. A cooperative role of these TFs has been reported in previous studies (Calvo et al., 2012; Mulford and Fassler, 2011; Shalaby et al., 2014). In S. cerevisiae and C. heterostrophus, Yap1 and Skn7 cooperate to activate the expression of antioxidant genes in response to oxidative stress (Mulford and Fassler, 2011; Shalaby et al., 2014). In F. graminearum, the genetic relationship between orthologues of Skn7 and Atf1 has been investigated (Jiang et al., 2015). We further identified the putative TF binding sites in the upstream regions (–500 to −1 bp) of FCA7 using MatchTM software (Kel et al., 2003). The promoter of FCA7 was found to contain two C2H2 zinc finger protein‐binding regions (GCCCC and TTGGC) and several binding motifs (ACCTG, GCTGT, CCTGT, etc.) for homeobox proteins, suggesting that FCA7 might be under the direct regulation of GzC2H010 (C2H2 zinc finger) and/or GzHOME001 (homeobox).
We also found that three TFs (ZIF1, GzZC086 and GzbZIP007) may be involved in alternative antioxidant systems, such as the non‐enzymatic antioxidant response (Apel and Hirt, 2004). Non‐enzymatic antioxidants include low‐molecular‐weight compounds, such as glutathione, ascorbate and cysteine. Homologues of CYS‐3 have been reported to regulate sulfur metabolism, which is involved in the biosynthesis of amino acids, such as cysteine and methionine (Kong et al., 2015; Marzluf, 1997). The roles of cysteine and methionine residues in proteins in antioxidant functions have been reported in several studies (Fauchon et al., 2002; Levine et al., 2000; Pócsi et al., 2004). Thus, the increased H2O2 sensitivity of the gzbzip007 deletion mutant may be a result of defects in sulfur metabolism. In M. oryzae, deletion mutants of MoMETR, an orthologue of CYS‐3, showed methionine auxotrophy and hypersensitivity to H2O2 (Kong et al., 2015).
In conclusion, our work reveals that F. graminearum possesses a robust antioxidant system that is involved in the maintenance of the cellular ROS balance during cell differentiation and proliferation, and that major peroxidases are involved in the oxidative stress response in this species. We suggest that Fca7 is particularly important for pathogen–host interactions and that multiple TFs co‐regulate the expression of FCA7 under oxidative stress conditions.
Experimental Procedures
Fungal strains and culture media
The F. graminearum wild‐type strain Z‐3639 (Bowden and Leslie, 1999) and transgenic strains derived from this strain were used in this study (Table S1). The catalase deletion mutants (fca1–fca7) and TF deletion mutants used in this study have been derived previously (Lee et al., 2014; Son et al., 2011b). Putative peroxidase genes were identified from the Fungal Peroxidase Database (fPoxDB; http://peroxidase.riceblast.snu.ac.kr) (Choi et al., 2014). The protein sequences of these genes were obtained from the MIPS Fusarium graminearum database (FGDB; http://mips.helmholtz-muenchen.de/genre/proj/FGDB/) (Wong et al., 2011). All strains were stored as mycelial suspensions in 20% glycerol solution at −80 °C. The culture media used in this study were prepared following the Fusarium laboratory manual (Leslie and Summerell, 2006). To induce conidial production, carboxymethyl cellulose (CMC) and yeast malt agar (YMA) were used as described previously (Cappellini and Peterson, 1965; Harris, 2005). MM containing 5 mm agmatine (MMA) was used for trichothecene production (Gardiner et al., 2009).
Nucleic acid manipulations, Southern blotting and PCR
Genomic DNA was extracted from mycelial powder according to the Fusarium laboratory manual (Leslie and Summerell, 2006). Total RNA was extracted from mycelia ground in liquid nitrogen using the Easy‐Spin Total RNA Extraction Kit (iNtRON Biotech, Seongnam, South Korea). Standard protocols were used for restriction endonuclease digestion, agarose gel electrophoresis, Southern blotting and hybridization with 32P‐labelled probes (Sambrook and Russell, 2001). The primers for PCR and qRT‐PCR used in this study (Table S3, see Supporting Information) were synthesized by an oligonucleotide synthesis facility (Bionics, Seoul, South Korea). PCR procedures were performed according to the manufacturer's instructions (TaKaRa Bio, Inc., Ostu, Japan).
Targeted gene deletion
The double‐joint (DJ) PCR strategy was used to construct fusion PCR products for targeted gene deletion (Yu et al., 2004). To create deletion strains, the 5′ and 3′ flanking regions of the target genes were amplified from the genomic DNA of the wild‐type strain, and GEN was amplified from pII99 using the primer pair Gen‐for/Gen‐Rev. Three amplicons (5′ flanking region, 3′ flanking region and GEN) were fused in a second round of DJ PCR. Finally, fusion constructs were amplified using nested primers to generate split markers. The resulting constructs were transformed into the wild‐type strain as described previously (Son et al., 2011a). Southern hybridization was performed to confirm single‐copy integration.
Vegetative growth, conidiation and sexual development
Radial growth rates on CM and MM were measured 5 days after inoculation with freshly grown culture plugs from MM. Conidial production was measured by counting the number of conidia after incubating culture plugs from CM in 5 mL of CMC for 3 days at 25 °C on a rotary shaker (200 rpm).
For self‐fertilization, fungal strains were grown on carrot agar plates for 5 days. To induce sexual reproduction, aerial mycelia were removed with sterile 2.5% Tween 60 solution (Leslie and Summerell, 2006). For outcrosses, mycelia of heterothallic female strains grown on carrot agar plates for 5 days were fertilized with 1 mL of a conidial suspension from male strains. After sexual induction, all cultures were incubated under near‐UV light (wavelength, 365 nm; HKiv Import & Export Co., Ltd., Xiamen, China) at 25 °C.
Virulence assays and trichothecene analysis
For the virulence test, the point inoculation method was performed using the susceptible wheat cultivar Eunpamil as described previously (Son et al., 2011a). Ten microlitres of conidial suspension (106 conidia/mL) harvested from CMC were injected into the middle of the spikelet. After inoculation, the plants were incubated in a high‐humidity chamber for 3 days and transferred to a glasshouse. The number of spikelets displaying symptoms of FHB was determined 21 days after inoculation. More than five replicated inoculations per strain and two independent mutant strains were used in the experiment.
Total trichothecene production (deoxynivalenol and 15‐acetyl‐deoxynivalenol) was measured as described previously (Son et al., 2011a). Cultures grown in MMA were filtered through cheesecloth, and filtrates were extracted with an ethyl acetate–methanol solution. The dehydrated extracts were derivatized with Sylon BTZ (N,O‐bis(trimethylsilyl)acetamide + trimethylchlorosilane + trimethylsilylimidazole, 3 : 2 : 3; Supelco, Bellefonte, PA, USA), and the derivatized products were analysed using a Shimadzu QP‐5000 gas chromatograph‐mass spectrometer (Shimadzu, Kyoto, Japan). The total trichothecene concentration was quantified based on the biomass produced by each strain in MMA. The experiment was repeated five times.
Oxidative stress sensitivity assays
To evaluate the effects of oxidative stress on the mycelial growth of the peroxidase deletion mutants, 10 mm H2O2 was used. Agar plugs from actively growing cultures were transferred to CM with or without supplementation with H2O2, and the plates were incubated at 25 °C for 5 days. To screen 657 TF deletion mutants (Son et al., 2011b), CM supplemented with 10 mm H2O2 was used. At least two independent tests were performed for each assay, and each strain was tested in triplicate.
qRT‐PCR analysis
Conidial suspensions (106 conidia/mL) harvested from YMA were inoculated into 50 mL of liquid CM and incubated for 24 h at 25 °C on a rotary shaker (200 rpm). Total RNA was extracted from strains grown for an additional 30 min in CM only or in CM supplemented with 5 mm H2O2. cDNA was synthesized from total RNA using SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA). qRT‐PCR was performed using SYBR Green Super Mix (Bio‐Rad, Hercules, CA, USA) and a 7500 real‐time PCR system (Applied Biosystems, Foster City, CA, USA), using primer pairs specific for peroxidase genes (Table S3). The cyclophilin gene (CYP1; FGSG_07439) was used as a reference gene. qRT‐PCR was performed three times with two replicates per run, and the transcript level of each target gene was calculated as described previously (Livak and Schmittgen, 2001).
To measure the transcript levels of the trichothecene biosynthetic genes, TRI5 and TRI6, we incubated conidia from the wild‐type and peroxidase deletion mutant strains in MMA for 4 days, isolated total RNA from each strain and performed qRT‐PCR as described above.
Peroxidase enzyme activity assay and H2O2 assay
Fungal strains grown for 24 h in 50 mL liquid CM were incubated for an additional 30 min in CM supplemented with 5 mm H2O2. Crude proteins were extracted from harvested mycelia (ground in liquid nitrogen) of wild‐type and peroxidase deletion mutant strains using 1 mL of potassium phosphate buffer (250 mm, pH 7.0) supplemented with 1 mm phenylmethylsulfonyl fluoride, a protease inhibitor. The protein concentration was determined colorimetrically by the Bradford assay (Bio‐Rad). Total peroxidase enzyme activities were measured using the Quantichrom Peroxidase Assay (BioAssay Systems, Hayward, CA, USA) according to the manufacturer's instructions. The quantification of H2O2 in semi‐dried mycelia was determined using an Amplex Red hydrogen peroxide/peroxidase assay kit (Invitrogen).
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Fusarium graminearum strains used in this study.
Table S2 Summary of the peroxidase mutant phenotypes.
Table S3 Primers used in this study.
Fig. S1 Targeted gene deletion. Each peroxidase‐encoding gene (A–X) was deleted individually from the genome of the Fusarium graminearum wild‐type (WT) strain Z‐3639. GEN, geneticin resistance gene cassette. The sizes of the DNA standards (in kilobases) are indicated to the left of the blot.
Fig. S2 Total trichothecene production of wild‐type (WT) and FCA7 overexpression (FCA7oe) strains. Each strain was grown in agmatine (MMA) for 7 days. Trichothecenes were analysed by gas chromatography‐mass spectrometry (GC‐MS) and quantified based on the biomass of each strain.
Fig. S3 Mycelial growth on complete medium (CM) supplemented with 4 mm H2O2 of gzbzip007 mutant strains carrying an FCA7 deletion or with FCA7 overexpression (FCA7oe). The photographs were taken 4 days after inoculation. WT, wild‐type.
Fig. S4 Virulence on wheat heads. The centre spikelet of each wheat head was injected with a conidial suspension of one of the fungal strains. The photographs were taken 21 days after inoculation. The arrowheads indicate the inoculated spikelets. ‘Mock’ indicates wheat heads that were mock inoculated with 0.01% Tween 20. WT, wild‐type.
Acknowledgements
This work was supported by the National Research Foundation of Korea (2013R1A6A3A04059121) and the Strategic Initiative for Microbiomes in Agriculture and Food funded by the Ministry of Agriculture, Food and Rural Affairs (916006‐2), South Korea.
References
- Apel, K. and Hirt, H. (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. [DOI] [PubMed] [Google Scholar]
- Arunachalam, C. and Doohan, F.M. (2013) Trichothecene toxicity in eukaryotes: cellular and molecular mechanisms in plants and animals. Toxicol. Lett. 217, 149–158. [DOI] [PubMed] [Google Scholar]
- Bowden, R.L. and Leslie, J.F. (1999) Sexual recombination in Gibberella zeae . Phytopathology, 89, 182–188. [DOI] [PubMed] [Google Scholar]
- Calvo, I.A. , García, P. , Ayté, J. and Hidalgo, E. (2012) The transcription factors Pap1 and Prr1 collaborate to activate antioxidant, but not drug tolerance, genes in response to H2O2 . Nucleic Acids Res. 40, 4816–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camhi, S.L. , Lee, P. and Choi, A.M. (1995) The oxidative stress response. New Horiz. 3, 170–182. [PubMed] [Google Scholar]
- Cano‐Domínguez, N. , Álvarez‐Delfín, K. , Hansberg, W. and Aguirre, J. (2008) NADPH oxidases NOX‐1 and NOX‐2 require the regulatory subunit NOR‐1 to control cell differentiation and growth in Neurospora crassa . Eukaryot. Cell, 7, 1352–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappellini, R.A. and Peterson, J.L. (1965) Macroconidium formation in submerged cultures by a non‐sporulating strain of Gibberella zeae . Mycologia, 57, 962–966. [Google Scholar]
- Chen, L.‐H. , Lin, C.‐H. and Chung, K.‐R. (2012) Roles for SKN7 response regulator in stress resistance, conidiation and virulence in the citrus pathogen Alternaria alternata . Fungal Genet. Biol. 49, 802–813. [DOI] [PubMed] [Google Scholar]
- Choi, J. , Détry, N. , Kim, K.‐T. , Asiegbu, F.O. , Valkonen, J.P. and Lee, Y.‐H. (2014) fPoxDB: fungal peroxidase database for comparative genomics. BMC Microbiol. 14, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins, A.E. and Proctor, R.H. (2007) Molecular biology of Fusarium mycotoxins. Int. J. Food Microbiol. 119, 47–50. [DOI] [PubMed] [Google Scholar]
- Desjardins, A.E. , Proctor, R.H. , Bai, G. , McCormick, S.P. , Shaner, G. , Buechley, G. and Hohn, T.M. (1996) Reduced virulence of trichothecene‐nonproducing mutants of Gibberella zeae in wheat field tests. Mol. Plant–Microbe Interact. 9, 775–781. [Google Scholar]
- Egan, M.J. , Wang, Z.‐Y. , Jones, M.A. , Smirnoff, N. and Talbot, N.J. (2007) Generation of reactive oxygen species by fungal NADPH oxidases is required for rice blast disease. Proc. Natl. Acad. Sci. USA, 104, 11 772–11 777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauchon, M. , Lagniel, G. , Aude, J.‐C. , Lombardia, L. , Soularue, P. , Petat, C. , Marguerie, G. , Sentenac, A. , Werner, M. and Labarre, J. (2002) Sulfur sparing in the yeast proteome in response to sulfur demand. Mol. Cell, 9, 713–723. [DOI] [PubMed] [Google Scholar]
- Fourquet, S. , Huang, M.‐E. , D'Autreaux, B. and Toledano, M.B. (2008) The dual functions of thiol‐based peroxidases in H2O2 scavenging and signaling. Antioxid. Redox Signal. 10, 1565–1576. [DOI] [PubMed] [Google Scholar]
- Fu, Y.‐H. , Paietta, J.V. , Mannix, D.G. and Marzluf, G.A. (1989) cys‐3, the positive‐acting sulfur regulatory gene of Neurospora crassa, encodes a protein with a putative leucine zipper DNA‐binding element. Mol. Cell. Biol. 9, 1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa‐Hibi, Y. , Kobayashi, Y. , Chen, C. and Motoyama, N. (2005) FOXO transcription factors in cell‐cycle regulation and the response to oxidative stress. Antioxid. Redox Signal. 7, 752–760. [DOI] [PubMed] [Google Scholar]
- Gardiner, D.M. , Kazan, K. and Manners, J.M. (2009) Novel genes of Fusarium graminearum that negatively regulate deoxynivalenol production and virulence. Mol. Plant–Microbe Interact. 22, 1588–1600. [DOI] [PubMed] [Google Scholar]
- Garre, V. , Tenberge, K.B. and Eising, R. (1998) Secretion of a fungal extracellular catalase by Claviceps purpurea during infection of rye: putative role in pathogenicity and suppression of host defense. Phytopathology, 88, 744–753. [DOI] [PubMed] [Google Scholar]
- Gasselhuber, B. , Jakopitsch, C. , Zámocký, M. , Furtmüller, P.G. and Obinger, C. (2015) Mechanistic aspects of catalase‐peroxidase In: Heme Peroxidases (Raven E. and Dunford H.B., eds.), pp. 156–180. Cambridge, UK: Royal Society of Chemistry. [Google Scholar]
- Gessler, N.N. , Aver'yanov, A.A. and Belozerskaya, T.A. (2007) Reactive oxygen species in regulation of fungal development. Biochemistry, 72, 1091–1109. [DOI] [PubMed] [Google Scholar]
- Giesbert, S. , Schuerg, T. , Scheele, S. and Tudzynski, P. (2008) The NADPH oxidase Cpnox1 is required for full pathogenicity of the ergot fungus Claviceps purpurea . Mol. Plant Pathol. 9, 317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles, S.S. , Stajich, J.E. , Nichols, C. , Gerrald, Q.D. , Alspaugh, J.A. , Dietrich, F. and Perfect, J.R. (2006) The Cryptococcus neoformans catalase gene family and its role in antioxidant defense. Eukaryot. Cell, 5, 1447–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami, R.S. and Kistler, H.C. (2004) Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 5, 515–525. [DOI] [PubMed] [Google Scholar]
- Guo, M. , Chen, Y. , Du, Y. , Dong, Y. , Guo, W. , Zhai, S. , Zhang, H. , Dong, S. , Zhang, Z. , Wang, Y. , Wang, P. and Zheng, X. (2011) The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus Magnaporthe oryzae . PLoS Pathog. 7, e1001302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansberg, W. , De Groot, H. and Sies, H. (1993) Reactive oxygen species associated with cell differentiation in Neurospora crassa . Free Radic. Biol. Med. 14, 287–293. [DOI] [PubMed] [Google Scholar]
- Harris, L.J. , Balcerzak, M. , Johnston, A. , Schneiderman, D. and Ouellet, T. (2016) Host‐preferential Fusarium graminearum gene expression during infection of wheat, barley, and maize. Fungal Biol. 120, 111–123. [DOI] [PubMed] [Google Scholar]
- Harris, S.D. (2005) Morphogenesis in germinating Fusarium graminearum macroconidia. Mycologia, 97, 880–887. [DOI] [PubMed] [Google Scholar]
- Heller, J. and Tudzynski, P. (2011) Reactive oxygen species in phytopathogenic fungi: signaling, development, and disease. Annu. Rev. Phytopathol. 49, 369–390. [DOI] [PubMed] [Google Scholar]
- Hong, S.‐Y. , Roze, L.V. and Linz, J.E. (2013a) Oxidative stress‐related transcription factors in the regulation of secondary metabolism. Toxins, 5, 683–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, S.‐Y. , Roze, L.V. , Wee, J. and Linz, J.E. (2013b) Evidence that a transcription factor regulatory network coordinates oxidative stress response and secondary metabolism in aspergilli. Microbiologyopen, 2, 144–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, K. , Czymmek, K.J. , Caplan, J.L. , Sweigard, J.A. and Donofrio, N.M. (2011) HYR1‐mediated detoxification of reactive oxygen species is required for full virulence in the rice blast fungus. PLoS Pathog. 7, e1001335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, H.H. , Lee, K.O. , Chi, Y.H. , Jung, B.G. , Park, S.K. , Park, J.H. , Lee, J.R. , Lee, S.S. , Moon, J.C. , Yun, J.W. , Choi, Y.O. , Kim, W.Y. , Kang, J.S. , Cheong, G.W. , Yun, D.J. , Rhee, S.G. , Cho, M.J. and Lee, S.Y. (2004) Two enzymes in one: two yeast peroxiredoxins display oxidative stress‐dependent switching from a peroxidase to a molecular chaperone function. Cell, 117, 625–635. [DOI] [PubMed] [Google Scholar]
- Jiang, C. , Zhang, S. , Zhang, Q. , Tao, Y. , Wang, C. and Xu, J.R. (2015) FgSKN7 and FgATF1 have overlapping functions in ascosporogenesis, pathogenesis and stress responses in Fusarium graminearum . Environ. Microbiol. 17, 1245–1260. [DOI] [PubMed] [Google Scholar]
- Kel, A.E. , Gößling, E. , Reuter, I. , Cheremushkin, E. , Kel‐Margoulis, O.V. and Wingender, E. (2003) MATCHTM: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 31, 3576–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.Y. , Jang, H.H. , Lee, J.R. , Sung, N.R. , Lee, H.B. , Lee, D.H. , Park, D.J. , Kang, C.H. , Chung, W.S. , Lim, C.O. , Yun, D.J. , Kim, W.Y. , Lee, K.O. and Lee, S.Y. (2009) Oligomerization and chaperone activity of a plant 2‐Cys peroxiredoxin in response to oxidative stress. Plant Sci. 177, 227–232. [Google Scholar]
- Kong, S. , Park, S.Y. and Lee, Y.H. (2015) Systematic characterization of the bZIP transcription factor gene family in the rice blast fungus, Magnaporthe oryzae . Environ. Microbiol. 17, 1425–1443. [DOI] [PubMed] [Google Scholar]
- König, J. , Muthuramalingam, M. and Dietz, K.‐J. (2012) Mechanisms and dynamics in the thiol/disulfide redox regulatory network: transmitters, sensors and targets. Curr. Opin. Plant Biol. 15, 261–268. [DOI] [PubMed] [Google Scholar]
- Lamb, C. and Dixon, R.A. (1997) The oxidative burst in plant disease resistance. Annu. Rev. Plant Biol. 48, 251–275. [DOI] [PubMed] [Google Scholar]
- Lara‐Ortíz, T. , Riveros‐Rosas, H. and Aguirre, J. (2003) Reactive oxygen species generated by microbial NADPH oxidase NoxA regulate sexual development in Aspergillus nidulans . Mol. Microbiol. 50, 1241–1255. [DOI] [PubMed] [Google Scholar]
- Lee, J. , Godon, C. , Lagniel, G. , Spector, D. , Garin, J. , Labarre, J. and Toledano, M.B. (1999) Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 274, 16 040–16 046. [DOI] [PubMed] [Google Scholar]
- Lee, Y. , Min, K. , Son, H. , Park, A.R. , Kim, J.‐C. , Choi, G.J. and Lee Y.W. (2014) ELP3 is involved in sexual and asexual development, virulence, and the oxidative stress response in Fusarium graminearum . Mol. Plant–Microbe Interact. 27, 1344–1355. [DOI] [PubMed] [Google Scholar]
- Leslie, J.F. and Summerell, B.A. (2006) The Fusarium Laboratory Manual. Ames, IA: Blackwell Publishing. [Google Scholar]
- Lev, S. , Hadar, R. , Amedeo, P. , Baker, S.E. , Yoder, O. and Horwitz, B.A. (2005) Activation of an AP1‐like transcription factor of the maize pathogen Cochliobolus heterostrophus in response to oxidative stress and plant signals. Eukaryot. Cell, 4, 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, A. , Tenhaken, R. , Dixon, R. and Lamb, C. (1994) H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell, 79, 583–593. [DOI] [PubMed] [Google Scholar]
- Levine, R.L. , Moskovitz, J. and Stadtman, E.R. (2000) Oxidation of methionine in proteins: roles in antioxidant defense and cellular regulation. IUBMB Life, 50, 301–307. [DOI] [PubMed] [Google Scholar]
- Lin, C.‐H. , Yang, S.L. and Chung, K.‐R. (2009) The YAP1 homolog‐mediated oxidative stress tolerance is crucial for pathogenicity of the necrotrophic fungus Alternaria alternata in citrus. Mol. Plant–Microbe Interact. 22, 942–952. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2–ΔΔ C T method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lysøe, E. , Pasquali, M. , Breakspear, A. and Kistler, H.C. (2011) The transcription factor FgStuAp influences spore development, pathogenicity, and secondary metabolism in Fusarium graminearum . Mol. Plant–Microbe Interact. 24, 54–67. [DOI] [PubMed] [Google Scholar]
- Maier, F.J. , Miedaner, T. , Hadeler, B. , Felk, A. , Salomon, S. , Lemmens, M. , Kassner, H. and Schäfer, W. (2006) Involvement of trichothecenes in fusarioses of wheat, barley and maize evaluated by gene disruption of the trichodiene synthase (Tri5) gene in three field isolates of different chemotype and virulence. Mol. Plant Pathol. 7, 449–461. [DOI] [PubMed] [Google Scholar]
- Marzluf, G.A. (1997) Molecular genetics of sulfur assimilation in filamentous fungi and yeast. Annu. Rev. Microbiol. 51, 73–96. [DOI] [PubMed] [Google Scholar]
- Mehdy, M.C. (1994) Active oxygen species in plant defense against pathogens. Plant Physiol. 105, 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metsalu, T. and Vilo, J. (2015) ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 43, W566–W570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir, A.A. , Park, S.‐Y. , Sadat, M.A. , Kim, S. , Choi, J. , Jeon, J. and Lee, Y.H. (2015) Systematic characterization of the peroxidase gene family provides new insights into fungal pathogenicity in Magnaporthe oryzae . Sci. Rep. 5, 11 831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina, L. and Kahmann, R. (2007) An Ustilago maydis gene involved in H2O2 detoxification is required for virulence. Plant Cell, 19, 2293–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montibus, M. , Ducos, C. , Bonnin‐Verdal, M.‐N. , Bormann, J. , Ponts, N. , Richard‐Forget, F. and Barreau, C. (2013) The bZIP transcription factor Fgap1 mediates oxidative stress response and trichothecene biosynthesis but not virulence in Fusarium graminearum . PLoS One, 8, e83377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montibus, M. , Pinson‐Gadais, L. , Richard‐Forget, F. , Barreau, C. and Ponts, N. (2015) Coupling of transcriptional response to oxidative stress and secondary metabolism regulation in filamentous fungi. Crit. Rev. Microbiol. 41, 295–308. [DOI] [PubMed] [Google Scholar]
- Mulford, K.E. and Fassler, J.S. (2011) Association of the Skn7 and Yap1 transcription factors in the Saccharomyces cerevisiae oxidative stress response. Eukaryot. Cell, 10, 761–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natorff, R. , Sieńko, M. , Brzywczy, J. and Paszewski, A. (2003) The Aspergillus nidulans metR gene encodes a bZIP protein which activates transcription of sulphur metabolism genes. Mol. Microbiol. 49, 1081–1094. [DOI] [PubMed] [Google Scholar]
- Petersen, T.N. , Brunak, S. , von Heijne, G. and Nielsen, H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods, 8, 785–786. [DOI] [PubMed] [Google Scholar]
- Pócsi, I. , Prade, R.A. and Penninckx, M.J. (2004) Glutathione, altruistic metabolite in fungi. Adv. Microb. Physiol. 49, 1–76. [DOI] [PubMed] [Google Scholar]
- Ponts, N. , Pinson‐Gadais, L. , Verdal‐Bonnin, M.‐N. , Barreau, C. and Richard‐Forget, F. (2006) Accumulation of deoxynivalenol and its 15‐acetylated form is significantly modulated by oxidative stress in liquid cultures of Fusarium graminearum . FEMS Microbiol. Lett. 258, 102–107. [DOI] [PubMed] [Google Scholar]
- Ponts, N. , Pinson‐Gadais, L. , Barreau, C. , Richard‐Forget, F. and Ouellet, T. (2007) Exogenous H2O2 and catalase treatments interfere with Tri genes expression in liquid cultures of Fusarium graminearum . FEBS Lett. 581, 443–447. [DOI] [PubMed] [Google Scholar]
- Proctor, R.H. , Hohn, T.M. and McCormick, S.P. (1995) Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Mol. Plant–Microbe Interact. 8, 593–601. [DOI] [PubMed] [Google Scholar]
- Reverberi, M. , Gazzetti, K. , Punelli, F. , Scarpari, M. , Zjalic, S. , Ricelli, A. , Fabbri, A.A. and Fanelli, C. (2012) Aoyap1 regulates OTA synthesis by controlling cell redox balance in Aspergillus ochraceus . Appl. Microbiol. Biotechnol. 95, 1293–1304. [DOI] [PubMed] [Google Scholar]
- Robbertse, B. , Yoder, O.C. , Nguyen, A. , Schoch, C.L. and Turgeon, B.G. (2003) Deletion of all Cochliobolus heterostrophus monofunctional catalase‐encoding genes reveals a role for one in sensitivity to oxidative stress but none with a role in virulence. Mol. Plant–Microbe Interact. 16, 1013–1021. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. and Russell, D.W. (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Schouten, A. , Tenberge, K.B. , Vermeer, J. , Stewart, J. , Wagemakers, L. , Williamson, B. and van Kan, J.A. (2002) Functional analysis of an extracellular catalase of Botrytis cinerea . Mol. Plant Pathol. 3, 227–238. [DOI] [PubMed] [Google Scholar]
- Scott, B. and Eaton, C.J. (2008) Role of reactive oxygen species in fungal cellular differentiations. Curr. Opin. Microbiol. 11, 488–493. [DOI] [PubMed] [Google Scholar]
- Segmüller, N. , Kokkelink, L. , Giesbert, S. , Odinius, D. , van Kan, J. and Tudzynski, P. (2008) NADPH oxidases are involved in differentiation and pathogenicity in Botrytis cinerea . Mol. Plant–Microbe Interact. 21, 808–819. [DOI] [PubMed] [Google Scholar]
- Shalaby, S. , Larkov, O. , Lamdan, N.L. and Horwitz, B.A. (2014) Genetic interaction of the stress response factors ChAP1 and Skn7 in the maize pathogen Cochliobolus heterostrophus . FEMS Microbiol. Lett. 350, 83–89. [DOI] [PubMed] [Google Scholar]
- Sikhakolli, U.R. , López‐Giráldez, F. , Li, N. , Common, R. , Townsend, J.P. and Trail, F. (2012) Transcriptome analyses during fruiting body formation in Fusarium graminearum and Fusarium verticillioides reflect species life history and ecology. Fungal Genet. Biol. 49, 663–673. [DOI] [PubMed] [Google Scholar]
- Skamnioti, P. , Henderson, C. , Zhang, Z. , Robinson, Z. and Gurr, S.J. (2007) A novel role for catalase B in the maintenance of fungal cell‐wall integrity during host invasion in the rice blast fungus Magnaporthe grisea . Mol. Plant–Microbe Interact. 20, 568–580. [DOI] [PubMed] [Google Scholar]
- Son, H. , Lee, J. , Park, A.R. and Lee, Y.‐W. (2011a) ATP citrate lyase is required for normal sexual and asexual development in Gibberella zeae . Fungal Genet. Biol. 48, 408–417. [DOI] [PubMed] [Google Scholar]
- Son, H. , Seo, Y.‐S. , Min, K. , Park, A.R. , Lee, J. , Jin, J.‐M. , Lin, Y. , Cao, P. , Hong, S.Y. , Kim, E.K. , Lee, S.H. , Cho, A. , Lee, S. , Kim, M.G. , Kim, Y. , Kim, J.E. , Kim, J.C. , Choi, G.J. , Yun, S.H. , Lim, J.Y. , Kim, M. , Lee, Y.H. , Choi, Y.D. and Lee, Y.W. (2011b) A phenome‐based functional analysis of transcription factors in the cereal head blight fungus, Fusarium graminearum . PLoS Pathog. 7, e1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son, H. , Kim, M.‐G. , Min, K. , Seo, Y.‐S. , Lim, J.Y. , Choi, G.J. , Kim, J.C. , Chae, S.K. and Lee, Y.W. (2013) AbaA regulates conidiogenesis in the ascomycete fungus Fusarium graminearum . PLoS One, 8, e72915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son, H. , Lim, J.Y. , Lee, Y. and Lee, Y.‐W. (2016) Utilization of a conidia‐deficient mutant to study sexual development in Fusarium graminearum . PLoS One, 11, e0155671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto, D. , Tanaka, A. and Scott, B. (2007) NADPH oxidases in fungi: diverse roles of reactive oxygen species in fungal cellular differentiation. Fungal Genet. Biol. 44, 1065–1076. [DOI] [PubMed] [Google Scholar]
- Tamura, K. , Stecher, G. , Peterson, D. , Filipski, A. and Kumar, S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe, S. , Ishii‐Minami, N. , Saitoh, K.‐I. , Otake, Y. , Kaku, H. , Shibuya, N. , Nishizawa, Y. and Minami, E. (2011) The role of catalase‐peroxidase secreted by Magnaporthe oryzae during early infection of rice cells. Mol. Plant–Microbe Interact. 24, 163–171. [DOI] [PubMed] [Google Scholar]
- Temme, N. and Tudzynski, P. (2009) Does Botrytis cinerea ignore H2O2‐induced oxidative stress during infection? Characterization of Botrytis activator protein 1. Mol. Plant–Microbe Interact. 22, 987–998. [DOI] [PubMed] [Google Scholar]
- Torres, M.A. , Jones, J.D.G. and Dangl, J.L. (2006) Reactive oxygen species signaling in response to pathogens. Plant Physiol. 141, 373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Mogg, C. , Walkowiak, S. , Joshi, M. and Subramaniam, R. (2014) Characterization of NADPH oxidase genes NoxA and NoxB in Fusarium graminearum . Can. J. Plant Pathol. 36, 12–21. [Google Scholar]
- Wang, Y. , Liu, W. , Hou, Z. , Wang, C. , Zhou, X. , Jonkers, W. , Ding, S. , Kistler, H.C. and Xu, J.R. (2011) A novel transcriptional factor important for pathogenesis and ascosporogenesis in Fusarium graminearum . Mol. Plant–Microbe Interact. 24, 118–128. [DOI] [PubMed] [Google Scholar]
- Windels, C.E. (2000) Economic and social impacts of Fusarium head blight: changing farms and rural communities in the Northern Great Plains. Phytopathology, 90, 17–21. [DOI] [PubMed] [Google Scholar]
- Wojtaszek, P. (1997) Oxidative burst: an early plant response to pathogen infection. Biochem. J. 322, 681–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, P. , Walter, M. , Lee, W. , Mannhaupt, G. , Münsterkötter, M. , Mewes, H.‐W. , Adam, G. and Güldener, U. (2011) FGDB: revisiting the genome annotation of the plant pathogen Fusarium graminearum . Nucleic Acids Res. 39, D637–D639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J.‐H. , Hamari, Z. , Han, K.‐H. , Seo, J.‐A. , Reyes‐Dominguez, Y. and Scazzocchio, C. (2004) Double‐joint PCR: a PCR‐based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41, 973–981. [DOI] [PubMed] [Google Scholar]
- Zámocký, M. and Obinger, C. (2010) Molecular phylogeny of heme peroxidases In: Biocatalysis Based on Heme Peroxidases (Torres E. and Ayala M., eds.), pp. 7–35. Berlin: Springer‐Verlag. [Google Scholar]
- Zámocký, M. , Furtmüller, P.G. and Obinger, C. (2009) Two distinct groups of fungal catalase/peroxidases. Biochem. Soc. Trans. 37, 772–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zámocký, M. , Gasselhuber, B. , Furtmüller, P.G. and Obinger, C. (2012) Molecular evolution of hydrogen peroxide degrading enzymes. Arch. Biochem. Biophys. 525, 131–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. , Lin, Y. , Wang, J. , Wang, Y. , Chen, M. , Norvienyeku, J. , Li, G. , Yu, W. and Wang, Z. (2016) FgNoxR, a regulatory subunit of NADPH oxidases, is required for female fertility and pathogenicity in Fusarium graminearum . FEMS Microbiol. Lett. 363, fnv223. [DOI] [PubMed] [Google Scholar]
- Zhou, W. , Kolb, F.L. and Riechers, D.E. (2005) Identification of proteins induced or upregulated by Fusarium head blight infection in the spikes of hexaploid wheat (Triticum aestivum). Genome, 48, 770–780. [DOI] [PubMed] [Google Scholar]
- Zhou, W. , Eudes, F. and Laroche, A. (2006) Identification of differentially regulated proteins in response to a compatible interaction between the pathogen Fusarium graminearum and its host, Triticum aestivum . Proteomics, 6, 4599–4609. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Fusarium graminearum strains used in this study.
Table S2 Summary of the peroxidase mutant phenotypes.
Table S3 Primers used in this study.
Fig. S1 Targeted gene deletion. Each peroxidase‐encoding gene (A–X) was deleted individually from the genome of the Fusarium graminearum wild‐type (WT) strain Z‐3639. GEN, geneticin resistance gene cassette. The sizes of the DNA standards (in kilobases) are indicated to the left of the blot.
Fig. S2 Total trichothecene production of wild‐type (WT) and FCA7 overexpression (FCA7oe) strains. Each strain was grown in agmatine (MMA) for 7 days. Trichothecenes were analysed by gas chromatography‐mass spectrometry (GC‐MS) and quantified based on the biomass of each strain.
Fig. S3 Mycelial growth on complete medium (CM) supplemented with 4 mm H2O2 of gzbzip007 mutant strains carrying an FCA7 deletion or with FCA7 overexpression (FCA7oe). The photographs were taken 4 days after inoculation. WT, wild‐type.
Fig. S4 Virulence on wheat heads. The centre spikelet of each wheat head was injected with a conidial suspension of one of the fungal strains. The photographs were taken 21 days after inoculation. The arrowheads indicate the inoculated spikelets. ‘Mock’ indicates wheat heads that were mock inoculated with 0.01% Tween 20. WT, wild‐type.


