Summary
The Potyviridae family is a major group of plant viruses that includes c. 200 species, most of which have narrow host ranges. The potyvirid P1 leader proteinase self‐cleaves from the remainder of the viral polyprotein and shows large sequence variability linked to host adaptation. P1 proteins can be classified as Type A or Type B on the basis, amongst other things, of their dependence or not on a host factor to develop their protease activity. In this work, we studied Type A proteases from the Potyviridae family, characterizing their host factor requirements. Our in vitro cleavage analyses of potyvirid P1 proteases showed that the N‐terminal domain is relevant for host factor interaction and suggested that the C‐terminal domain is also involved. In the absence of plant factors, the N‐terminal end of Plum pox virus P1 antagonizes protease self‐processing. We performed extended deletion mutagenesis analysis to define the N‐terminal antagonistic domain of P1. In viral infections, removal of the P1 protease antagonistic domain led to a gain‐of‐function phenotype, strongly increasing local infection in a non‐permissive host. Altogether, our results shed new insights into the adaptation and evolution of potyvirids.
Keywords: host adaptation, host factor, P1 proteases, polyprotein processing, Potyviridae, RNA silencing suppression
Several pathogens code for proteases that participate in the promotion of infectivity and virulence. They regulate pathogen replication, target specific host factors and enhance vector performance to increase pathogen fitness and transmission (Bak et al., 2017; Büttner, 2016; Krausslich and Wimmer, 1988; Tong, 2002).
Polyprotein processing for the release of mature subunits is a common strategy used by viruses of the picorna‐like supergroup, which includes the Potyviridae family (Koonin et al., 2008). The Potyviridae family is a major group of plant viruses including nearly 200 species (Wylie et al., 2017). The viral RNA genome is translated into large polyproteins that are further processed into functional peptides by virus‐encoded endopeptidases (Adams et al., 2005; Revers and García, 2015). Amongst these are P1 proteins, chymotrypsin‐like serine proteases located at the beginning of viral polyproteins. Generally present in one copy, some potyvirids code for two P1 proteins with different proteolytic specificities (Rodamilans et al., 2013; Valli et al., 2007).
P1 is the most divergent of potyviral proteins (Mengual‐Chuliá et al., 2016; Shukla et al., 1991; Valli et al., 2007). Despite this significant variability, the P1 C‐terminal region is relatively well conserved. It harbours a serine protease domain responsible for cis‐cleavage at the P1–HC junction, and thus P1 self‐releases from the polyprotein (Adams et al., 2005). P1 acts as a non‐essential accessory factor (Verchot and Carrington, 1995b), but its detachment from the polyprotein is indispensable for viral viability, as the lack of self‐cleavage impairs the functionality of the downstream silencing suppressor HC (Pasin et al., 2014; Shan et al., 2015; Verchot and Carrington, 1995a).
In the Potyviridae, polyprotein processing has been shown to be an important regulatory mechanism (Ivanov et al., 2014). Non‐lethal mutations in the trans‐acting protease NIaPro or its recognition sites are critical for host adaptation (Calvo et al., 2014; Chen et al., 2008). Moreover, recent work with the P1 protein of Plum pox virus (PPV) provides evidence that the N‐terminal region acts as a negative regulator of P1 self‐cleavage, modulating viral replication during infection (Pasin et al., 2014).
Potyvirid P1 proteins can be classified as Type A or Type B on the basis of the phylogenetic relationship, shared isoelectric point, RNA silencing suppression activity and plant factor requirements (Fig. 1A). The protease activity of Type B proteins, such as P1b from Cucumber vein yellowing virus (CVYV), has no host factor requirements (Rodamilans et al., 2013). Type A proteins, which include PPV P1, depend on as yet unknown host factor(s) for processing, as full‐length P1s self‐cleave in the wheat germ extract (WGE) translation system, but show no activity in the rabbit reticulocyte lysate (RRL) system. In contrast with full‐length Type A proteins, a truncated PPV P1 protease with the N‐end at amino acid V164 (P1Pro) is active in both WGE and RRL systems (Pasin et al., 2014). Considering that PPV P1 is able to function independently of plant factors when devoid of the antagonist N‐terminal region, it is reasonable to anticipate that other Type A P1s would exhibit similar autonomous protease domains. We tested this hypothesis in two Type A P1 proteins: P1 from Turnip mosaic virus (TuMV) and P1a from CVYV. We first evaluated the in vitro cleavage activity of these full‐length proteins and included P1, amplified from an infectious clone of Ugandan cassava brown streak virus (UCBSV) (Pasin et al., 2017), as a theoretical Type B P1 protein (Methods S1, see Supporting Information). UCBSV is an atypical potyvirid that lacks the silencing suppressor HC and encodes a single P1 (Mbanzibwa et al., 2009; Patil et al., 2015). Analysis of in vitro cleavage assays confirmed the classification of TuMV P1 and CVYV P1a as Type A proteins, as they were able to perform auto‐cleavage only in WGE, but not in RRL. It also categorized P1 UCBSV as a Type B protein, as its self‐cleavage processing was independent of the translation system used (Fig. S1, see Supporting Information). To define core protease domains in CVYV P1a and TuMV P1, we set up a series of deletion mutants based on an alignment between PPV and TuMV P1s and CVYV P1a, and using as reference the VELI motif (Valli et al., 2007) (Fig. 1A, Methods S1). In vitro results showed that neither CVYV P1a nor TuMV P1 presented a protease domain active in both WGE and RRL (Fig. 1B,C). For both proteins, the largest tested deletions impaired protease activity in WGE and RRL (R404 and L406 of CVYV P1a, Q223 and R230 of TuMV P1). Smaller deletions (S402 for CVYV, C218 for TuMV) allowed processing in WGE, but prevented cleavage in RRL. These results show that CVYV and TuMV Type A protease domains are functional, but their cofactor requirements differ from the autonomous protease domain found in PPV P1.
Figure 1.
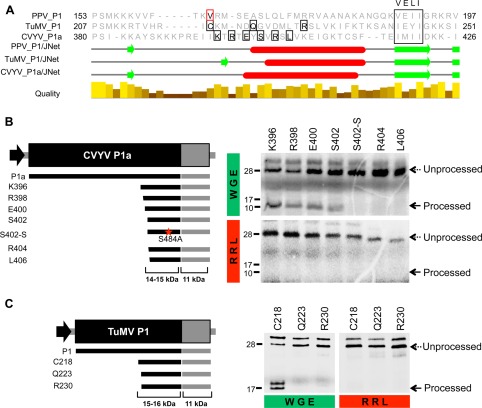
Proteolytic activity of Cucumber vein yellowing virus (CVYV) P1a and Turnip mosaic virus (TuMV) P1 minimal protease domains in wheat germ extract (WGE) and rabbit reticulocyte lysate (RRL) in vitro translation systems. (A) Partial alignment of Plum pox virus (PPV) P1, TuMV P1 and CVYV P1a. The N‐terminal amino acids of PPV P1Pro (V164) and of each construct tested for proteolytic activity are boxed in red and black, respectively. The VELI motif is marked. Secondary structures were predicted using the JNet algorithm included in the Jalview package (Waterhouse et al., 2009): α‐helices, red ovals; β‐sheets, green arrows; below, alignment conservation bars. (B, C) Diagrams showing DNA constructs with progressive deletions of CVYV P1a (B) and TuMV P1 (C) coding sequences, amplified by polymerase chain reaction (PCR) and subjected to in vitro transcription and in vitro translation. Expected translation products and their molecular weights are displayed below. Truncation products are named according to the first amino acid (in addition to the initial methionine) that is maintained. Mutation in the serine of CVYV P1a (S484A) is marked with a red star. Self‐cleavage activity was evaluated in WGE and RRL in vitro translation systems. 35S‐labelled translation products were resolved by sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and detected by autoradiography. Processed and unprocessed products are marked (right); left, molecular weight markers. Unaccounted extra bands might originate from additional initiation events of translation.
Previous data have suggested that small N‐terminal modifications of PPV P1Pro render the protease domain partially dependent on host factors (Pasin et al., 2014). To verify this, the first two amino acids of PPV P1Pro were mutated to alanine (P1ProAA); catalytic mutants were included as negative controls (Fig. S2, Methods S1, see Supporting Information). PPV P1ProAA cleavage capacity was reduced by more than 70% with respect to that of P1Pro when assayed in RRL, whereas its activity in WGE remained almost unaltered (Fig. S2B,C). Our results suggest that plant factors recognize the C‐terminal part of Type A P1, and indicate that, when the N‐terminus of PPV P1Pro is disturbed, it adopts a plant cofactor‐dependent configuration similar to core protease domains of Type A proteins from TuMV and CVYV.
The boundaries of the C‐terminal protease domain of PPV P1 were mapped between amino acids 162–170 (Pasin et al., 2014). To define the limits of the N‐terminal sequences that interfere with the protease activity of PPV P1 in the absence of plant cofactors, we prepared a new set of P1 N‐terminal deletion mutants fused to an HC fragment. Catalytic mutants were included as negative controls (Fig. 2A, Methods S1). As reported, full‐length P1 presented high protease activity in WGE (>50%), and showed no detectable processing in RRL. In contrast, P1Pro showed high processing capacity in both WGE and RRL, with more than 50% of P1Pro detached from the HC part (Fig. 2B). Compared with P1Pro, P1 constructs with shorter N‐terminal truncations showed high processing ratios in WGE (>50%); however, their activity in RRL gradually declined as the N‐terminus length increased (Fig. 2B). These findings show that there is no well‐defined antagonistic module at the N‐terminal region of PPV P1, but sequences of this region progressively contribute to prevent cleavage activity in the absence of plant cofactors.
Figure 2.
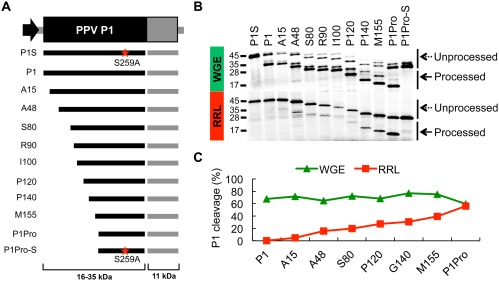
Proteolytic activities of Plum pox virus (PPV) P1 N‐terminal deletion constructs in wheat germ extract (WGE) and rabbit reticulocyte lysate (RRL) in vitro translation systems. (A) Diagram showing DNA constructs with progressive deletions of PPV P1 coding sequences, amplified by polymerase chain reaction (PCR) and subjected to in vitro transcription and in vitro translation. Expected translation products and their molecular weights are displayed below. Truncation products are named according to the first amino acid (in addition to the initial methionine) that is maintained. Mutations in the catalytic serines of PPV P1 and PPV P1Pro (S259A) are marked with a red star. (B) Self‐cleavage activity was evaluated in RRL and WGE in vitro translation mixtures. 35S‐labelled products were resolved by sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and detected by autoradiography. Processed and unprocessed products are marked (right); left, molecular weight markers. (C) Quantification analysis of the protease activity of the truncated P1 proteins. The plot was built on the basis of P1 cleavage efficiency estimated by the program Quantity One. One hundred was assigned as the maximum translation value, which includes processed and unprocessed products of the P1 construct.
Inefficient P1 processing impairs HC silencing suppressor activity and viral viability (Pasin et al., 2014; Shan et al., 2015). After in vitro translation experiments, we sought to further characterize PPV P1Pro in planta, assessing how its self‐cleavage might affect HC functions. Transient RNA silencing assays were performed in Nicotiana benthamiana and Cucumis sativus, hosts that allow efficient and suboptimal PPV P1 processing, respectively (Fig. 3, Methods S1). Green fluorescent protein (GFP) was used as the inducer of RNA silencing and as a reporter of HC silencing suppressor activity in plants expressing P1‐, P1S‐ (catalytic mutant of P1 protease), P1Pro‐HC or a control empty vector. GFP and HC accumulation were assessed by immunoblot analysis at 6 days post‐agroinfiltration (dpa). Both proteins were detected when P1‐HC was expressed in N. benthamiana, but not in C. sativus plants. In contrast, P1Pro released functional HC and allowed high levels of GFP accumulation in both N. benthamiana and C. sativus. Unprocessed P1‐HC could not be detected in the conditions tested.
Figure 3.
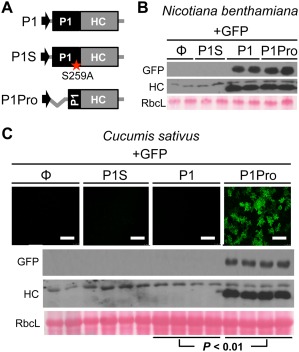
Effects of upstream P1 sequences on Plum pox virus (PPV) HC silencing suppressor activity in Nicotiana benthamiana and Cucumis sativus. (A) Diagram of the agroinfiltrated constructs. Mutation in the catalytic serine of PPV P1 (S259A) is marked with a red star. (B, C) Transient expression by agroinfiltration was performed in N. benthamiana (B) and cucumber (C) leaves. A green fluorescent protein (GFP)‐expressing construct was co‐infiltrated with the empty vector (Φ) or with P1‐, P1S‐ and P1Pro‐HC constructs. Samples were analysed at 6 days post‐agroinfiltration (dpa). Images of C. sativus samples were taken under a confocal microscope; scale bars, 100 µm. GFP and HC accumulation were assessed by anti‐GFP and anti‐HC immunoblot assay; RbcL, Ponceau red‐stained blots showing the large subunit of Rubisco protein as loading control. The difference in GFP signal intensity between the indicated samples is statistically significant (P < 0.01) by Student's t‐test (n = 4).
One of the aspects limiting host–potyvirid compatibility may be the absence of an appropriate plant cofactor for P1 release from the rest of the polyprotein. In agreement with this view, replacement of P1 by CVYV P1a facilitated local infection of PPV in cucumber (Carbonell et al., 2012; Shan et al., 2015). To address the capacity of P1Pro to overcome host‐specific restrictions to viral infection, C. sativus plants were agroinoculated with wild‐type PPV or the viral clone PPV‐P1Pro, in which the N‐terminal antagonistic domain of P1 was deleted (Fig. 4A, Methods S1). The silencing suppressor p19 was included in some of the infiltration mixtures to rescue HC defects that might derive from limited P1 self‐cleavage. Confocal microscopy analysis of inoculated cucumber leaves showed the presence of GFP in all viral samples co‐infiltrated with p19. When p19 was absent from the infiltration mixtures, GFP could only be observed in PPV‐P1Pro samples (Fig. 4B). This result suggests that, in cucumber, free HC able to support PPV infection is liberated by P1Pro, but not by full‐length P1. This assumption was confirmed by immunoblot analysis using anti‐coat protein (anti‐CP) and anti‐HC antibodies. CP was observed in all samples co‐agroinfiltrated with p19, but released HC could only be detected in PPV‐P1Pro samples, independent of the presence or absence of p19. No systemic infection could be detected in any of the inoculated cucumber plants at 21 dpa (not shown). Nicotiana benthamiana plants were used as positive controls of unrestricted infection and, in this host, CP and HC could be detected in all viral clone samples (Fig. 4C). The potyvirid NIb cistron codes for the viral RNA‐dependent RNA polymerase, whose activity is abolished by the removal of the GDD catalytic motif. To demonstrate PPV‐P1Pro replication in inoculated cucumber leaves, an independent experiment was performed including a PPV‐P1Pro replication‐defective mutant (PPV‐P1ProΔGDD). Reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) analysis of positive and negative viral genome strands confirms PPV‐P1Pro replication in these leaves (Fig. S3, Methods S1, see Supporting Information).
Figure 4.
Effects of N‐terminal truncation of P1 on Plum pox virus (PPV) infection in cucumber leaves. (A) PPV and PPV‐P1Pro infectious clones were delivered to Cucumis sativus and Nicotiana benthamiana plants by co‐agroinfiltration with empty vector (Φ) or p19 silencing suppressor; Φ and p19 constructs were co‐agroinfiltrated together as a negative control. (B) At 7 days post‐agroinfiltration (dpa), confocal micrographs of inoculated cucumber leaves were taken; scale bars, 100 µm. (C) In inoculated cucumber (top panel) and N. benthamiana leaves (bottom), viral accumulation and release of HC were assessed by anti‐coat protein (anti‐CP) and anti‐HC immunoblot assays. Each lane corresponds to a pool of four agroinfiltrated plants; RbcL, Ponceau red‐stained blots showing the large subunit of Rubisco protein as loading control. (D) Model summarizing the results obtained with PPV and PPV‐P1Pro. P1 cleavage and RNA silencing suppressor activity (RSS) are marked in each case. Hosts that promote or inhibit P1 cleavage, RSS activity and viral infections are indicated by arrow‐headed or bar‐headed lines, respectively. Nb, N. benthamiana; Cs, C. sativus.
The data presented in this article support a view in which P1 leader proteases are, at least in part, responsible for the narrow host ranges of potyvirid species and the driving forces of Potyviridae speciation, as suggested previously (Desbiez and Lecoq, 2004; Maliogka et al., 2012; Rodamilans et al., 2013; Salvador et al., 2008b; Shan et al., 2015; Valli et al., 2007). We propose that this role is directly dependent on cofactor requirements of P1 processing and thus the release of functional silencing suppressors to counteract host antiviral defences (Fig. 4D). P1 self‐cleavage defines viral viability and infection onset, and balances viral amplification rates and immune response severity (Pasin et al., 2014; Verchot and Carrington, 1995a, 1995b).
N‐terminal deletions of PPV P1 allowed the identification of P1Pro, a minimal cofactor‐independent protease domain of P1 (Pasin et al., 2014). Here, we show that P1Pro is the most efficient of the constructs tested, and N‐terminal extensions progressively restore the domain that antagonizes P1 self‐cleavage in the RRL system (Fig. 2). Moreover, the fact that point mutations of P1Pro that do not disturb its intrinsic protease activity make it dependent on the plant cofactor(s) (Fig. S2) highlights the strict requirements for P1 autonomous cleavage. Compatible with this view, our efforts to find plant cofactor‐independent regions similar to PPV P1Pro in representative Type A proteins, TuMV P1 and CVYV P1a, resulted in core protease domains that still require plant cofactor(s) for their activation (Fig. 1). Further analysis is needed to elucidate whether PPV P1 represents a unique link between Type A and Type B P1 proteases, or whether its cleavage kinetics are recapitulated in any of the ∼200 members of the Potyviridae family.
The lack of dependence on a specific cofactor suggests that P1Pro might self‐cleave in plants unsuitable for full‐length P1 processing, and thus expand viral host ranges. In agreement with this assumption, in vivo transient expression experiments showed efficient P1Pro release from HC in cucumber plants, a non‐permissive host of PPV (Fig. 3). Moreover, the mutant virus PPV‐P1Pro is able to boost local replication in C. sativus leaves, indicating that part of the host restriction found by PPV is caused by P1–host cofactor(s) incompatibility and thus silencing suppressor defects (Fig. 4). These results not only highlight the relevance of P1 leader proteinases as host range determinants, but also pose interesting questions about the role of the P1 protein in viral adaptation. PPV P1 acts as a regulator of viral infection (Pasin et al., 2014), and deletions of the N‐terminal part of PPV P1 gradually alter the self‐cleavage ability of the core protease (Fig. 2). It is therefore reasonable to envisage that different deletions on this non‐essential gene could modulate viral replication rates and facilitate viral adaptation in specific environments, in the same way that PPV‐P1Pro is able to overcome host incompatibilities and promote local viral amplification in C. sativus (Fig. 4). Certainly, a role for this type of deletion in the potyvirid adaptive process would be in agreement with the large diversity of Type A P1 proteins. Of note, although this is the first report of P1 deletions facilitating the infection of a particular host, genome deletions involved in adaptation to the host have been described for the highly variable N‐terminal part of potyviral CP, and have also been reported for other viruses (Carbonell et al., 2013; Ojosnegros et al., 2011; Salvador et al., 2008a; Sorrell et al., 2010; Tatineni and Dawson, 2012; Tromas et al., 2014; Willcocks et al., 1994).
This work stresses the relevance of endopeptidases in viral infections and fosters the idea that virus‐encoded proteases are important antiviral targets (Gutierrez‐Campos et al., 1999; Kim et al., 2016; Krausslich and Wimmer, 1988). It is imperative to obtain a full understanding of how P1 proteins work, and comprehend their role in viral infections, host range definition and evolution. This work contributes to all of this and sets the lead for future studies into the further characterization of P1–host factor(s) interaction.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Methods S1 Experimental procedures.
Fig. S1 Phylogenetic tree and in vitro proteolytic activity of representative P1 proteins from Type A and Type B groups. (A) P1 proteins were aligned and the phylogenetic tree was built. Bootstrap percentages are shown next to the branches. Type A and B proteins are indicated. GenBank accessions are as follows: Plum pox virus (PPV), NP734339; Turnip mosaic virus (TuMV), NP734213; Cucumber vein yellowing virus (CVYV), YP308878; Ugandan cassava brown streak virus (UCBSV), CBA13048. (B) Self‐cleavage activity of Type A proteins from TuMV and CVYV, and a Type B protein from UCBSV, was evaluated in wheat germ extract (WGE) and rabbit reticulocyte lysate (RRL) in vitro translation systems. Diagrams show DNA constructs of the polyprotein coding sequences, amplified by polymerase chain reaction (PCR) and subjected to in vitro transcription and in vitro translation. Catalytic mutations are indicated with red stars. 35S‐labelled products were resolved by sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and detected by autoradiography. Processed and unprocessed products are marked (left); right, molecular weight markers.
Fig. S2 Proteolytic activity of Plum pox virus (PPV) P1Pro and P1ProAA truncated proteins in rabbit reticulocyte lysate (RRL) and wheat germ extract (WGE) in vitro translation systems. (A) Diagram of DNA constructs of coding sequences of different PPV variants amplified by polymerase chain reaction (PCR) and subjected to in vitro transcription and further in vitro translation. Expected translation products and their molecular weights are shown below. Mutations in the serine of the P1 active centre (S259A) and in two residues at the N‐terminus of the P1Pro domain (V164A and R165A) are indicated with a red star and a red triangle, respectively. (B) Self‐cleavage activity was evaluated in WGE and RRL in vitro translation mixtures. 35S‐labelled translation products were resolved by sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and detected by autoradiography. Processed and unprocessed polyprotein products are marked with solid and dotted arrows, respectively (right); left, molecular weight markers. (C) Quantification analysis of the proteolytic activity of P1 variants, estimated by the program Quantity One. One hundred was assigned as the maximum translation value, which includes processed and unprocessed products of the P1 construct.
Fig. S3 Analysis of viral replication in the inoculated leaves of Cucumis sativus. (A) Diagram of the agroinfiltrated constructs. In the replication‐defective mutant, NIb deleted amino acids that render replicase inactive are marked with a red line. (B) At 7 days post‐agroinfiltration (dpa), local viral accumulation was assessed by anti‐coat protein (anti‐CP) immunoblot assay. Each lane corresponds to one plant sample; RbcL, Ponceau red‐stained blot showing the large subunit of Rubisco protein as loading control. (C) Reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) analysis of the viral accumulation of the positive (left) and negative (right) genome strands. All data were quantified relative to the average value of PPV‐P1Pro for the positive and negative strands, respectively. Bars show mean ± standard deviation (SD) (n = 4); letters indicate P < 0.01, one‐way analysis of variance (ANOVA) and Bonferroni post‐hoc test.
Acknowledgements
We are grateful to B. García for technical assistance, to F. Ponz and D. Baulcombe for supply of materials and to the Rijk Zwaan Company for providing cucumber seeds. H.S. is supported by the China Scholarship Council. This work was funded by grants BIO2013–49053‐R, BIO2016–80572‐R and Plant KBBE PCIN‐2013‐056 from the Spanish Government and the FEDER program. The authors have no conflicts of interest to declare.
Contributor Information
Fabio Pasin, Email: fpasin@gate.sinica.edu.tw.
Juan Antonio García, Email: jagarcia@cnb.csic.es.
Bernardo Rodamilans, Email: brodamilans@cnb.csic.es.
References
- Adams, M.J. , Antoniw, J.F. and Beaudoin, F. (2005) Overview and analysis of the polyprotein cleavage sites in the family Potyviridae . Mol. Plant Pathol. 6, 471–487. [DOI] [PubMed] [Google Scholar]
- Bak, A. , Cheung, A.L. , Yang, C. , Whitham, S.A. and Casteel, C.L. (2017) A viral protease relocalizes in the presence of the vector to promote vector performance. Nat. Commun. 8, 14 493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner, D. (2016) Behind the lines—actions of bacterial type III effector proteins in plant cells. FEMS Microbiol. Rev. 40, 894–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo, M. , Malinowski, T. and García, J.A. (2014) Single amino acid changes in the 6K1‐CI region can promote the alternative adaptation of Prunus‐ and Nicotiana‐propagated Plum pox virus C isolates to either host. Mol. Plant–Microbe Interact. 27, 136–149. [DOI] [PubMed] [Google Scholar]
- Carbonell, A. , Dujovny, G. , García, J.A. and Valli, A. (2012) The Cucumber vein yellowing virus silencing suppressor P1b can functionally replace HCPro in Plum pox virus infection in a host‐specific manner. Mol. Plant–Microbe Interact. 25, 151–164. [DOI] [PubMed] [Google Scholar]
- Carbonell, A. , Maliogka, V.I. , Pérez, J. D J. , Salvador, B. , León, D.S. , García, J.A. and Simón‐Mateo, C. (2013) Diverse amino acid changes at specific positions in the N‐terminal region of the coat protein allow Plum pox virus to adapt to new hosts. Mol. Plant–Microbe Interact. 26, 1211–1224. [DOI] [PubMed] [Google Scholar]
- Chen, K.C. , Chiang, C.H. , Raja, J.A. , Liu, F.L. , Tai, C.H. and Yeh, S.D. (2008) A single amino acid of NIaPro of Papaya ringspot virus determines host specificity for infection of papaya. Mol. Plant–Microbe Interact. 21, 1046–1057. [DOI] [PubMed] [Google Scholar]
- Desbiez, C. and Lecoq, H. (2004) The nucleotide sequence of Watermelon mosaic virus (WMV, Potyvirus) reveals interspecific recombination between two related potyviruses in the 5' part of the genome. Arch. Virol. 149, 1619–1632. [DOI] [PubMed] [Google Scholar]
- Gutierrez‐Campos, R. , Torres‐Acosta, J.A. , Saucedo‐Arias, L.J. and Gomez‐Lim, M.A. (1999) The use of cysteine proteinase inhibitors to engineer resistance against potyviruses in transgenic tobacco plants. Nat. Biotechnol. 17, 1223–1226. [DOI] [PubMed] [Google Scholar]
- Ivanov, K.I. , Eskelin, K. , Löhmus, A. and Mäkinen, K. (2014) Molecular and cellular mechanisms underlying potyvirus infection. J. Gen. Virol. 95, 1415–1429. [DOI] [PubMed] [Google Scholar]
- Kim, S.H. , Qi, D. , Ashfield, T. , Helm, M. and Innes, R.W. (2016) Using decoys to expand the recognition specificity of a plant disease resistance protein. Science, 351, 684–687. [DOI] [PubMed] [Google Scholar]
- Koonin, E.V. , Wolf, Y.I. , Nagasaki, K. and Dolja, V.V. (2008) The Big Bang of picorna‐like virus evolution antedates the radiation of eukaryotic supergroups. Nat. Rev. Microbiol. 6, 925–939. [DOI] [PubMed] [Google Scholar]
- Krausslich, H.G. and Wimmer, E. (1988) Viral proteinases. Annu. Rev. Biochem. 57, 701–754. [DOI] [PubMed] [Google Scholar]
- Maliogka, V.I. , Salvador, B. , Carbonell, A. , Sáenz, P. , León, D. , Oliveros, J. , Delgadillo, M. , García, J. and Simón‐Mateo, C. (2012) Virus variants with differences in the P1 protein coexist in a Plum pox virus population and display particular host‐dependent pathogenicity features. Mol. Plant Pathol. 13, 877–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbanzibwa, D.R. , Tian, Y. , Mukasa, S.B. and Valkonen, J.P.T. (2009) Cassava brown streak virus (Potyviridae) encodes a putative Maf/HAM1 pyrophosphatase implicated in reduction of mutations and a P1 proteinase that suppresses RNA silencing but contains no HC‐Pro. J. Virol. 83, 6934–6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengual‐Chuliá, B. , Bedhomme, S. , Lafforgue, G. , Elena, S.F. and Bravo, I.G. (2016) Assessing parallel gene histories in viral genomes. BMC Evol. Biol. 16, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojosnegros, S. , García‐Arriaza, J. , Escarmís, C. , Manrubia, S.C. , Perales, C. , Arias, A. , Mateu, M.G. , Domingo, E. and Malik, H.S. (2011) Viral genome segmentation can result from a trade‐off between genetic content and particle stability. PLoS Genet. 7, e1001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasin, F. , Simón‐Mateo, C. and García, J.A. (2014) The hypervariable amino‐terminus of P1 protease modulates potyviral replication and host defense responses. PLoS Pathog. 10, e1003985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasin, F. , Bedoya, L.C. , Bernabé‐Orts, J.M. , Gallo, A. , Simón‐Mateo, C. , Orzaez, D. and García, J.A. (2017) Multiple T‐DNA delivery to plants using novel mini binary vectors with compatible replication origins. ACS Synth. Biol. 6, 1962–1968. [DOI] [PubMed] [Google Scholar]
- Patil, B.L. , Legg, J.P. , Kanju, E. and Fauquet, C.M. (2015) Cassava brown streak disease: a threat to food security in Africa. J. Gen. Virol. 96, 956–968. [DOI] [PubMed] [Google Scholar]
- Revers, F. and García, J.A. (2015) Molecular biology of potyviruses. Adv. Virus Res. 92, 101–199. [DOI] [PubMed] [Google Scholar]
- Rodamilans, B. , Valli, A. and García, J.A. (2013) Mechanistic divergence between P1 proteases of the family Potyviridae . J. Gen. Virol. 94, 1407–1414. [DOI] [PubMed] [Google Scholar]
- Salvador, B. , Delgadillo, M.O. , Sáenz, P. , García, J.A. and Simón‐Mateo, C. (2008a) Identification of Plum pox virus pathogenicity determinants in herbaceous and woody hosts. Mol. Plant–Microbe Interact. 21, 20–29. [DOI] [PubMed] [Google Scholar]
- Salvador, B. , Sáenz, P. , Yángüez, E. , Quiot, J.B. , Quiot, L. , Delgadillo, M.O , García, J.A. and Simón‐Mateo, C. (2008b) Host‐specific effect of P1 exchange between two potyviruses. Mol. Plant Pathol. 9, 147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, H. , Pasin, F. , Valli, A. , Castillo, C. , Rajulu, C. , Carbonell, A. , Simón‐Mateo, C. , García, J.A. and Rodamilans, B. (2015) The Potyviridae P1a leader protease contributes to host range specificity. Virology, 476, 264–270. [DOI] [PubMed] [Google Scholar]
- Shukla, D.D. , Frcnkel, M.J. and Ward, C.W. (1991) Structure and function of the potyvirus genome with special reference to the coat protein coding region. Can. J. Plant Pathol. 13, 178–191. [Google Scholar]
- Sorrell, E.M. , Song, H. , Pena, L. and Perez, D.R. (2010) A 27‐amino‐acid deletion in the neuraminidase stalk supports replication of an avian H2N2 influenza A virus in the respiratory tract of chickens. J. Virol. 84, 11 831–11 840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatineni, S. and Dawson, W.O. (2012) Enhancement or attenuation of disease by deletion of genes from Citrus tristeza virus. J. Virol. 86, 7850–7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, L. (2002) Viral proteases. Chem. Rev. 102, 4609–4626. [DOI] [PubMed] [Google Scholar]
- Tromas, N. , Zwart, M.P. , Forment, J. and Elena, S.F. (2014) Shrinkage of genome size in a plant RNA virus upon transfer of an essential viral gene into the host genome. Genome Biol. Evol. 6, 538–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valli, A. , López‐Moya, J.J. and García, J.A. (2007) Recombination and gene duplication in the evolutionary diversification of P1 proteins in the family Potyviridae . J. Gen. Virol. 88, 1016–1028. [DOI] [PubMed] [Google Scholar]
- Verchot, J. and Carrington, J.C. (1995a) Debilitation of plant potyvirus infectivity by P1 proteinase‐inactivating mutations and restoration by second‐site modifications. J. Virol. 69, 1582–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot, J. and Carrington, J.C. (1995b) Evidence that the potyvirus P1 proteinase functions in trans as an accessory factor for genome amplification. J. Virol. 69, 3668–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse, A.M. , Procter, J.B. , Martin, D.M. , Clamp, M. and Barton, G.J. (2009) Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics, 25, 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcocks, M.M. , Ashton, N. , Kurtz, J.B. , Cubitt, W.D. and Carter, M.J. (1994) Cell culture adaptation of astrovirus involves a deletion. J. Virol. 68, 6057–6058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie, S.J. , Adams, M. , Chalam, C. , Kreuze, J. , López‐Moya, J.J. , Ohshima, K. , Praveen, S. , Rabenstein, F. , Stenger, D. , Wang, A. and Murilo Zerbini, F. (2017) ICTV virus taxonomy profile: Potyviridae . J. Gen. Virol. 98, 352–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Methods S1 Experimental procedures.
Fig. S1 Phylogenetic tree and in vitro proteolytic activity of representative P1 proteins from Type A and Type B groups. (A) P1 proteins were aligned and the phylogenetic tree was built. Bootstrap percentages are shown next to the branches. Type A and B proteins are indicated. GenBank accessions are as follows: Plum pox virus (PPV), NP734339; Turnip mosaic virus (TuMV), NP734213; Cucumber vein yellowing virus (CVYV), YP308878; Ugandan cassava brown streak virus (UCBSV), CBA13048. (B) Self‐cleavage activity of Type A proteins from TuMV and CVYV, and a Type B protein from UCBSV, was evaluated in wheat germ extract (WGE) and rabbit reticulocyte lysate (RRL) in vitro translation systems. Diagrams show DNA constructs of the polyprotein coding sequences, amplified by polymerase chain reaction (PCR) and subjected to in vitro transcription and in vitro translation. Catalytic mutations are indicated with red stars. 35S‐labelled products were resolved by sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and detected by autoradiography. Processed and unprocessed products are marked (left); right, molecular weight markers.
Fig. S2 Proteolytic activity of Plum pox virus (PPV) P1Pro and P1ProAA truncated proteins in rabbit reticulocyte lysate (RRL) and wheat germ extract (WGE) in vitro translation systems. (A) Diagram of DNA constructs of coding sequences of different PPV variants amplified by polymerase chain reaction (PCR) and subjected to in vitro transcription and further in vitro translation. Expected translation products and their molecular weights are shown below. Mutations in the serine of the P1 active centre (S259A) and in two residues at the N‐terminus of the P1Pro domain (V164A and R165A) are indicated with a red star and a red triangle, respectively. (B) Self‐cleavage activity was evaluated in WGE and RRL in vitro translation mixtures. 35S‐labelled translation products were resolved by sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and detected by autoradiography. Processed and unprocessed polyprotein products are marked with solid and dotted arrows, respectively (right); left, molecular weight markers. (C) Quantification analysis of the proteolytic activity of P1 variants, estimated by the program Quantity One. One hundred was assigned as the maximum translation value, which includes processed and unprocessed products of the P1 construct.
Fig. S3 Analysis of viral replication in the inoculated leaves of Cucumis sativus. (A) Diagram of the agroinfiltrated constructs. In the replication‐defective mutant, NIb deleted amino acids that render replicase inactive are marked with a red line. (B) At 7 days post‐agroinfiltration (dpa), local viral accumulation was assessed by anti‐coat protein (anti‐CP) immunoblot assay. Each lane corresponds to one plant sample; RbcL, Ponceau red‐stained blot showing the large subunit of Rubisco protein as loading control. (C) Reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) analysis of the viral accumulation of the positive (left) and negative (right) genome strands. All data were quantified relative to the average value of PPV‐P1Pro for the positive and negative strands, respectively. Bars show mean ± standard deviation (SD) (n = 4); letters indicate P < 0.01, one‐way analysis of variance (ANOVA) and Bonferroni post‐hoc test.


