Summary
Puccinia coronata f. sp. avenae (Pca) causes crown rust disease in cultivated and wild oat (Avena spp.). The significant yield losses inflicted by this pathogen make crown rust the most devastating disease in the oat industry. Pca is a basidiomycete fungus with an obligate biotrophic lifestyle, and is classified as a typical macrocyclic and heteroecious fungus. The asexual phase in the life cycle of Pca occurs in oat, whereas the sexual phase takes place primarily in Rhamnus species as the alternative host. Epidemics of crown rust happens in areas with warm temperatures (20–25 °C) and high humidity. Infection by the pathogen leads to plant lodging and shrivelled grain of poor quality.
Disease symptoms: Infection of susceptible oat varieties gives rise to orange–yellow round to oblong uredinia (pustules) containing newly formed urediniospores. Pustules vary in size and can be larger than 5 mm in length. Infection occurs primarily on the surfaces of leaves, although occasional symptoms develop in the oat leaf sheaths and/or floral structures, such as awns. Symptoms in resistant oat varieties vary from flecks to small pustules, typically accompanied by chlorotic halos and/or necrosis. The pycnial and aecial stages are mostly present in the leaves of Rhamnus species, but occasionally symptoms can also be observed in petioles, young stems and floral structures. Aecial structures display a characteristic hypertrophy and can differ in size, occasionally reaching more than 5 mm in diameter.
Taxonomy: Pca belongs to the kingdom Fungi, phylum Basidiomycota, class Pucciniomycetes, order Pucciniales and family Pucciniaceae.
Host range: Puccinia coronata sensu lato can infect 290 species of grass hosts. Pca is prevalent in all oat‐growing regions and, compared with other cereal rusts, displays a broad telial host range. The most common grass hosts of Pca include cultivated hexaploid oat (Avena sativa) and wild relatives, such as bluejoint grass, perennial ryegrass and fescue. Alternative hosts include several species of Rhamnus, with R. cathartica (common buckthorn) as the most important alternative host in Europe and North America.
Control: Most crown rust management strategies involve the use of rust‐resistant crop varieties and the application of fungicides. The attainment of the durability of resistance against Pca is difficult as it is a highly variable pathogen with a great propensity to overcome the genetic resistance of varieties. Thus, adult plant resistance is often exploited in oat breeding programmes to develop new crown rust‐resistant varieties.
Useful website: https://www.ars.usda.gov/midwest-area/st-paul-mn/cereal-disease-lab/docs/cereal-rusts/race-surveys/.
Keywords: avirulence factors, buckthorn, crown rust, fungicides, genetic resistance, oat
Introduction
Oat (Avena sativa) is the sixth largest cereal crop based on worldwide production [Food and Agriculture Organization (FAO), 2014], with Russia, Canada, the European Union, Australia and the USA as the major producers [US Department of Agriculture, Foreign Agricultural Service (USDA‐FAS), 2017]. Although oat is primarily grown as a livestock feed crop, A. sativa ranks fourth amongst the cereals after wheat, rice and corn based on human consumption. Wholegrain oat products are usually attractive to consumers because of their impact on cholesterol levels in humans and their preservative‐free properties, as well as high nutritional and fibre content (van den Broeck et al., 2016; Burnette et al., 1992).
Unfortunately, the global prevalence of Puccinia coronata f. sp. avenae (Pca) hinders oat production and reduces the economic value of the grain, as infection affects the grain yield, kernel weight and groat percentage (Doehlert et al., 2001; Holland and Munkvold, 2001; Humphreys and Mather, 1996). Moreover, crown rust infection weakens straw production and causes oat plants to lodge (Endo and Boewe, 1958). The earliest documented account of P. coronata was made in 1767, as described by Tozzetti (1952), and, since then, several subdivisions have been suggested, including splitting into physiological forms (formae speciales), varieties and subspecies. Numerous classifications based on spore morphology, as well as telial and aecial hosts, have been described to reflect variations within P. coronata sensu lato (Brown, 1937; Cummins, 1971; Eriksson, 1908; Eriksson and Henning, 1894; Peturson, 1954; Urban and Markova, 1994); however, such assignments fail to represent genetic differences between isolates or true host ranges (Anikster et al., 2003; Eshed and Dinoor, 1980). Nonetheless, some of these designations remain in use to indicate the major grass hosts, as in the case of Pca. Most recently, the implementation of molecular analysis has shown that P. coronata encompasses multiple species (Szabo, 2006) and suggests the division of the complex into seven species (Liu and Hambleton, 2013). For the purpose of this review, we refer to Pca as a pathogen affecting primarily cultivated and wild oat, as well as some grasses, such as Lolium, and which, in previous publications, has been denominated as P. coronata var. avenae f. sp. avenae (Liu and Hambleton, 2013) or P. coronata var. avenae, PcSP2 (Szabo, 2006). In this review, we summarize the current knowledge on this pathogen, including the economic importance and life cycle, as well as perspectives on rust virulence, disease management and future research directions.
Economic Importance
Historical reports of damage to oat crops caused by Pca first appeared in the late 1800s. Crop failures as a result of crown rust infection were first reported in Europe (Cornu, 1880) and the Baltics (Sivers, 1887), just shortly before the pathogen was noted in the USA (Thaxter, 1890). Since then, oat crown rust epidemics have continued to affect oat production, resulting in 10%–40% yield losses (Behnken et al., 2009; Martinelli et al., 1994; Simons, 1970). Throughout the 20th century, crown rust epidemics have occurred intermittently in different regions of the world. Severe damage caused by this pathogen has been reported in South America (Gassner, 1916), Portugal (D'Oliveira, 1942), Australia (Waterhouse, 1952), Israel (Wahl and Schreiter, 1953), southeastern Europe (Kostic, 1959) and the USA (Sherf, 1954). Since the 1990s, epidemics of crown rust have occurred almost annually in Brazil and Uruguay (Leonard and Martinelli, 2005; Wahl and Schreiter, 1953). More recently, Pca has been reported to pose a serious risk to oat production in Tunisia (Hammami et al., 2010) and Canada (Chong et al., 2008).
In years in which the weather is not conducive to infection or inoculum pressure is not high, yield losses caused by crown rust can be as low as 5%. According to records at the US Department of Agriculture, Agricultural Research Service, Cereal Disease Laboratory (USDA‐ARS CDL), oat crown rust epidemics remained relatively mild (less than 10% losses) in the USA from 1981 to 2013, with the exception of 1991 and 1993, when losses reached 15% in North Dakota, and, in 1997, when Minnesota and Louisiana reported 20% losses (Long and Hughes, 2000). Severe losses occurred in the USA in 2014, when over 13 million bushels, equivalent to 18.7% of the country's oat production, were lost as a result of damage by Pca. During this epidemic, yield losses in the two major oat‐producing states, Minnesota and South Dakota, were 50% and 35%, respectively (USDA‐ARS CDL, 2014).
Life Cycle and Infection Process
Puccinia coronata f. sp. avenae possesses five infectious stages that are associated with either sexual or asexual reproductive phases in its life cycle (Fig. 1) (Simons, 1970). In the Middle East, Europe and North America, where alternative hosts grow in close association with oat, both sexual and asexual stages of Pca exist (Dinoor, 1977; Simons, 1985; Wahl, 1970). In contrast, alternative hosts are uncommon or absent in East Africa, South America, Australia and New Zealand, and the pathogen is probably limited to a repetitive asexual stage in these regions (Harder and Haber, 1992; Simons, 1985). The asexual infection phase (telial stage) occurs entirely in oat, whereas sexual reproduction (aecial stage) takes place in alternative hosts (Dietz, 1926; Simons, 1985). The asexual phase involves repeated cycles of infection and sporulation mediated by urediniospores that can repeat as quickly as every 2 weeks (Fig. 2). At this stage, Pca is dikaryotic, with each single‐celled urediniospore containing two haploid nuclei. The urediniospores germinate on the adaxial and abaxial leaf surfaces under suitable conditions (i.e. mild temperatures, adequate moisture and short exposure to light). Once germinated, these spores form appressoria and, subsequently, a penetration peg, which allows the fungus to penetrate the stoma and gain access to the mesophyll space of the leaf. A substomatal vesicle is formed in the stomatal cavity, from which infection hyphae originate, and hyphal tips elongate to produce specialized haustorial mother cells that develop into haustoria—structures essential for nutrient uptake (Staples and Macko, 1984). The intercellular branching of the infection hyphae proceeds until a fungal colony is formed in the surrounding leaf tissue, which, after 7–10 days, gives rise to sporulating uredinia that produce a new set of urediniospores. The uredinia emerge as bright orange–yellow oblong pustules (Fig. 2) that constitute the characteristic symptom of infection (Harder and Haber, 1992; Jackson et al., 2008; Simons, 1970; Staples and Macko, 1984).
Figure 1.
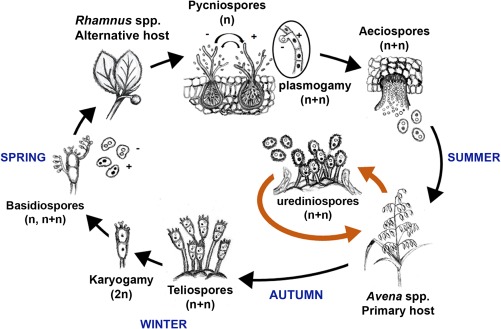
Life cycle of Puccinia coronata f. sp. avenae. The asexual phase of the cycle (orange arrows) is completed in domestic and wild oat species during summer as multiple rounds of infection. The sexual phase of the cycle begins with the differentiation of teliospores in late summer or autumn to withstand cold winter temperatures. Teliospores undergo karyogamy and complete one meiotic event to produce basidiospores in the spring. Basidiospores carry either the (–) or (+) mating type and probably undergo mitosis prior to germination and infection of buckthorn (Rhamnus species). In buckthorn, the fungus differentiates pycniospores, which come into contact with neighbour hyphae from the opposite mating type, enabling plasmogamy. Aeciospores then form and infect oat to re‐initiate the asexual cycle. Drawing by M. Figueroa.
Figure 2.
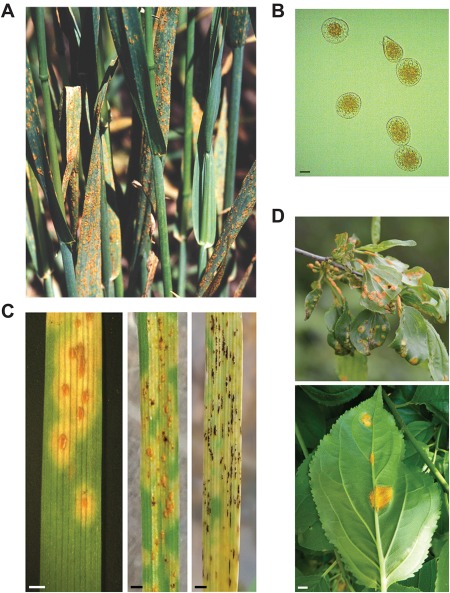
Symptoms of oat crown rust infection. (A) Infection of oat by Puccinia coronata f. sp. avenae in the field. A high density of pustules (clusters of urediniospores) is shown in the leaves. Photograph by R. Caspers. (B) Micrograph of urediniospores; scale, 10 µm. (C) Close‐up image of pustules in susceptible oat variety Marvelous (left) and two stages of teliospore formation (middle and right); scale, 2 mm. (D) Photographs of crown rust in common buckthorn (Rhamnus cathartica). Symptoms are shown in the leaves and petioles. Pycnia are present on the adaxial surface of the leaf (top), whereas aecia form on the abaxial surface (top and bottom); scale, 2 mm. Courtesy photographs by R. Caspers and E. Byamukama.
The sexual phase of Pca involves both oat and the alternative host, common buckthorn (Rhamnus spp.) (Fig. 1). Late in the cropping season, as the plant starts to senesce, rust infection sites differentiate teliospores (Fig. 2). These dikaryotic, thick‐walled, survival structures germinate in the spring and undergo meiosis to produce haploid basidiospores, which subsequently infect growing buckthorn leaves (Mendgen, 1984; USDA‐ARS CDL, 2017). Once in buckthorn, Pca completes additional developmental processes, resulting in the spermatial stage. At this stage, pycnia are formed on the adaxial surface of the leaf and produce pycniospores, which act as gametes to fuse with receptive hyphae and re‐establish a dikaryotic stage in the fungus (Fig. 2) (Harder and Haber, 1992; Simons, 1985). After plasmogamy, aecium formation occurs on the abaxial surface, and these cylindrical‐shaped structures produce aeciospores that re‐infect the grass host (Figs 1 and 2) (Harder and Haber, 1992). Each of the haploid nuclei contributed by the two gametes remains in the aeciospores (dikaryotic), and therefore represents a complete haplotype genome inherited from a single parent.
Urediniospores and aeciospores are wind transmitted and can travel long distances (Jackson et al., 2008). For instance, during early summer in North America, urediniospores from infected oat in Mexico and the southern USA may be blown northward to the north‐central region of the USA and the provinces of Manitoba and Saskatchewan in Canada, a route known as the Puccinia pathway (Harder and Haber, 1992). During autumn, the wind may carry urediniospores south to infect winter oat (Forbes, 1939). However, in the northern USA, where buckthorn is abundant, inoculum from the alternative host probably plays a more important role in crown rust outbreaks than do urediniospores carried through the Puccinia pathway (USDA‐ARS CDL, 2017). The northward movement of crown rust inoculum also occurs in Europe (Klenová‐Jiráková et al., 2010; Simons, 1985). Moreover, migrating birds have been shown to play a role in both the northward and southward dispersal of spores during spring and autumn, respectively, and even across continents (da Silva et al., 2016; Warner and French, 1970).
Pathogenicity and Population Biology of Pca
The development of rust diseases results from a failure of the host plant's immune system to recognize the pathogen and activate defence responses. As a first line of defence, plant immunity involves the recognition of conserved pathogen components, such as chitin in fungi, known as pathogen‐associated molecular patterns (PAMPs) (Dangl et al., 2013; Dodds and Rathjen, 2010; Hogenhout et al., 2009; Toruño et al., 2016). However, adapted pathogens have evolved mechanisms to suppress or prevent the activation of these plant basal defences, often through the action of secreted effector proteins. Thus, as a second line of defence, plants have also evolved mechanisms to detect such effectors (Dangl et al., 2013; Dodds and Rathjen, 2010; Petre and Kamoun, 2014; Toruño et al., 2016). This latter plant recognition system (refers to Effector‐Triggered Immunity, ETI) was initially characterized at the genetic level by H. H. Flor, who developed the gene‐for‐gene hypothesis based on interactions in the flax rust disease system (Flor, 1971). In this model, dominant resistance (R) genes in the host plant confer recognition of specific avirulence (Avr) genes from the pathogen. This model has now been verified at the molecular level, with plant R genes found to encode intracellular immune receptor proteins mostly belonging to the nucleotide‐binding leucine‐rich repeat (NB‐LRR) class (Dodds and Rathjen, 2010; Jones and Dangl, 2006). The Avr proteins recognized by these receptors are typically effector proteins that are delivered into host cells. Work in the flax rust system demonstrated that rust haustoria can deliver effector proteins to the host plant cell, which, if recognized by the corresponding R proteins, trigger immune responses and resistance (Dodds et al., 2004, 2006; Garnica et al., 2014). The relationship between crown rust resistance in oat and Pca virulence displays typical gene‐for‐gene characteristics, with resistance conferred by multiple single dominant resistance genes, each displaying different specificities towards pathogen isolates (Chong et al., 2000, 2008). To date, more than 100 crown rust resistance (Pc) genes have been described in oat (Table S1, see Supporting Information) (Gnanesh et al., 2014; Graichen et al., 2010; Tan and Carson, 2013; USDA‐ARS CDL, 2016), although the lack of molecular information makes it difficult to determine whether some of these genes may be the same or are alleles at a common locus. In contrast with this wealth of R genes, no effectors have yet been identified in Pca, and the knowledge of rust pathogen Avr proteins remains limited to Melampsora lini, the causal agent of flax rust, and Hemileia vastatrix, the causal agent of coffee leaf rust (Maia et al., 2017; Ravensdale et al., 2011). Nonetheless, it is evident that virulence profiles can vary greatly amongst isolates of Pca, and this can only be explained by the existence of distinct effector/Avr gene repertoires in the pathogen.
As documented by national and international surveys, Pca is one of the most pathogenically diverse cereal rust fungi, exhibiting the rapid emergence of new virulent isolates (Carson, 2011; Chong and Kolmer, 1993; Klenová‐Jiráková et al., 2010; Park, 2008; Simons, 1985). Pathogen characterization is conducted on a set of oat genotypes carrying one or more Pc genes in various genotypic backgrounds to define pathogenicity based on compatible (virulent) or incompatible (avirulent) interactions, each assigned to a physiological race (Chong et al., 2000; Leonard et al., 2004; van Niekerk et al., 2001). The response of each differential genotype to an isolate is recorded using a 0–4 infection type (IT) scale (Fig. 3), with the cut‐off between compatible and incompatible usually at either IT 3 or >3. Simons and Michel (1964) reported about 400 physiological races of Pca worldwide (Harder and Haber, 1992), which clearly demonstrates the pathogenic variability of the pathogen. The number of Pc genes represented in the differential sets used to identify pathotypes (races) in surveys varies among countries, and has been updated over time in response to the rise of new virulences (Chong et al., 2000). For instance, in the USA, the USDA‐ARS CDL utilizes a set of 40 oat differentials. There is no universal set of differentials used across research institutions, although there is some overlap of both Pc genes and oat differentials (Table 1).
Figure 3.
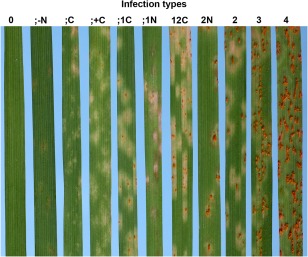
Examples of infection types (ITs) on the disease rating scale as developed by Murphy (1935). ITs are used to score seedling resistance and to conduct physiological race assignments of Puccinia coronata f. sp. avenae. Incompatible reactions include the following: 0, no urediniospores; fleck (;), presence of flecks; 1, few small pustules; 2, small pustules, presence of green islands; all of these may be accompanied by necrosis (N) and/or chlorosis (C). Compatible reactions include the following: 3, large pustules surrounded by chlorotic halos; 4, large pustules, often coalescing. Those illustrated here (photographs by R. F. Park) were scored as, left to right: 0, ;–N, ;C, ;+C, ;1C, ;1N, 12C, 2N, 2, 3 and 4.
Table 1.
Oat differential genotypes used in the USA, Australia, Canada and Brazil to assign pathotypes of Puccinia coronata f. sp. avenae.
| Gene(s) | Australia | USA | Brazil * | Canada † |
|---|---|---|---|---|
| Pc1 | X | |||
| Pc3c, Pc4c, Pc6c, Pc9 | X | |||
| Pc4, Pc5 | X | |||
| Pc6, Pc7, Pc8, Pc21 | X | |||
| Pc6d | X | |||
| Pc14 | X | X | ||
| Pc15, Pc16, Pc17 | X | |||
| Pc35 | X | X | ||
| Pc36 | X | X | X | |
| Pc38 | X | X | X | X |
| Pc38, Pc39, Pc52 | X | |||
| Pc39 | X | X | X | X |
| Pc39, Pc61, PcBettong | X | |||
| Pc40 | X | X | X | |
| Pc45 | X | X | X | X |
| Pc46 | X | X | X | X |
| Pc48 | X | X | X | X |
| Pc48+ | X | |||
| Pc48, Pc56 | X | |||
| Pc50 | X | X | X | X |
| Pc50+ | X | |||
| Pc51 | X | X | X | X |
| Pc52 | X | X | X | X |
| Pc53 | X | X | ||
| Pc54 | X | X | X | |
| Pc55 | X | X | X | |
| Pc56 | X | X | X | X |
| Pc56+ | X | |||
| Pc57 | X | X | ||
| Pc58 | X | X | X | X |
| Pc59 | X | X | X | X |
| Pc60 | X | X | X | |
| Pc60, Pc61 | X | |||
| Pc61 | X | X | ||
| Pc61+ | X | |||
| Pc61, PcBettong | X | |||
| Pc62 | X | X | X | X |
| Pc63 | X | X | X | |
| Pc64 | X | X | X | X |
| Pc67 | X | X | ||
| Pc68 | X | X | X | X |
| Pc70 | X | X | ||
| Pc71 | X | X | ||
| Pc91 | X | X | X | |
| Pc92 | X | |||
| Pc94 | X | X | X | |
| Pc96 | X | X | ||
| Pc97 | X | |||
| Pc98 | X | |||
| Pc101 | X | |||
| Pc103‐1 | X | |||
| Pc104 | X | |||
| PcH546 | X | |||
| PcH548 | X | |||
| PcMortlock, PcCulgoa | X | |||
| PcWIX1, PcWIX2 | X | |||
| Unknown (Bondvic) | X | |||
| Unknown (X716) | X | |||
| Unknown (Marvelous) | X | |||
| Unknown (H548) | X | |||
| Unknown (IA B605X sel.) | X | |||
| Unknown (WI X4361‐9) | X | |||
| Unknown (TAM‐O‐405) | X | |||
| Unknown (Belle) | X | |||
| Unknown (HiFi) | X | |||
| Unknown (Leggett) | X | |||
| Unknown (Stainless) | X | |||
| Total | 42 | 40 | 28 | 24 |
The deployment of race‐specific seedling genes has been a method of choice to mitigate oat crown rust damage in the field. Unfortunately, this strategy has not delivered resistance durability and has resulted in boom‐and‐bust cycles as pathogen populations evolve virulence to previously resistant varieties over time (Chong and Seaman, 1997; Chong and Zegeye, 2004; McCallum et al., 2007; McDonald and Linde, 2002; Park et al., 2000). These shifts occur as a result of the selective pressure exerted by the limited genetic diversity in the crop on the rust population, leading to a change in virulence frequencies within populations. Thus, the evolutionary capacity of the pathogen should be considered in disease management strategies. Alternative hosts play an important role in the epidemiology and evolution of pathogenicity in some rust species, as they act as a reservoir for the pathogen and allow sexual recombination, which leads to a more diverse population (Zhao et al., 2016). Given that buckthorn is prevalent in North America, high rates of sexual recombination probably account for the extremely polymorphic Pca populations and the rapid evolution of new virulent pathotypes (Leonard, 2002).
Studies in North America have shown that race diversity is greater amongst isolates collected on Rhamnus carthartica or in oat found in close proximity to R. carthartica (Fleischmann, 1965; Murphy, 1935), suggesting that the alternative host may play a role in generating the genetic diversity of Pca. Nevertheless, high levels of pathogenic variation in Pca are not exclusive to sexual populations, as asexual populations in regions in which the alternative host is absent or rare, such as Australia, are also highly polymorphic (Park, 2013; Simons, 1985). In Australia, surveys from 1998 to 2010 identified more than 100 Pca races, much higher than found for the same period of time in other cereal rust species that are also not sexually active (Park, 2008; Park and Wellings, 2012). These observations raise questions about the role of buckthorn in the pathogenic diversity of Pca and whether other factors, such as repeated introductions, high mutation rates or somatic hybridization and recombination, are also important contributors to the diversity and virulence in Pca populations (Bartos et al., 1969; Klenová‐Jiráková et al., 2010; Park and Wellings, 2012; Simons, 1970). It is possible that the genetic diversity of Pca, resulting from the re‐assortment of pathogenicity factors through sexual reproduction, can be further enhanced by multiple molecular mechanisms that introduce variability into asexual populations (Simons, 1985).
The characterization of the genetic diversity of Pca at the molecular level is limited. A preliminary analysis of 12 Australian Pca isolates using DNA amplification fingerprinting markers detected two pathogen subpopulations, indicating two distinctive exotic rust isolate introductions (Brake et al., 2001). Klenová‐Jiráková et al. (2010) used an amplified fragment length polymorphism (AFLP) analysis to evaluate the genetic diversity of 40 Pca isolates collected from seven different countries (Israel, Czech Republic, Belarus, Estonia, Hungary, Austria and Serbia), which confirmed the high genetic diversity of the species, as each isolate presented a unique AFLP molecular pattern, although genetic similarity was found for those isolates from the Czech Republic, Austria and Serbia. To our knowledge, simple sequence repeat‐expressed sequence tag (SSR‐EST) markers have only been developed to study the genetic diversity of Puccinia coronata f. sp. lolii (pathogen of ryegrass) (Dracatos et al., 2006), but not for Pca. Similarly, the development of single nucleotide polymorphism (SNP) markers to conduct genetic studies in Pca has not been implemented, as EST or genomic sequences are not yet available for this pathogen.
Disease Management Approaches
Management measures to prevent crown rust outbreaks include the use of biocontrol agents, removal of the alternative host, development of varieties with resistance and application of fungicides (Fig. 4) (Hoffman et al., 2006; McCallum et al., 2007; Simons, 1970).
Figure 4.

Crown rust management strategies. (A) Effect of fungicide treatment in an oat field. Right section of the field depicts damage of the pathogen in the absence of fungicide treatment on a susceptible variety, in contrast with the left side of the field that was treated with fungicide. (B) Effect of genetic resistance in the field. Photograph illustrates the positive effect of using the oat variety Deon versus a variety that does not contain genetic resistance. Deon was selected from the cross Sesqui × 2/Bettong//MN02108 and was released by the Minnesota Agricultural Experiment Station in 2012. Courtesy photographs by R. Caspers and E. Byamukama.
Common buckthorn is a shrub native to Europe, northwest Africa and western Asia, and was first introduced to the USA in the mid‐1800s as an ornamental plant and windbreak. The species quickly spread through the upper midwestern and northeastern regions of the USA and has now reached Saskatchewan and the Maritime provinces of Canada (Knight et al., 2007). The invasiveness of buckthorn in forests, prairies and savannas has led to its classification as a restricted noxious weed in parts of both the USA and Canada [US Department of Agriculture, Natural Resources Conservation Service (USDA‐NRCS), 2017], and has awakened interest in the launch of stringent eradication programmes to reduce its ecological impact. However, a significant challenge to these measures is to successfully prevent the re‐emergence of buckthorn, particularly in remote areas. The USA, together with other countries, including Canada, Kenya, Latvia, Estonia and Russia, where buckthorn has become a problem, have passed legislation to prevent the dissemination and encourage the eradication of this invasive weed (Sherf et al., 1956), which has resulted in a great reduction in buckthorn in places such as Iowa (Simons, 1985). Currently, the biological control of buckthorn using insects is also being explored in North America and Europe [Centre for Agriculture and Bioscience International (CABI), 2015].
Oat crown rust can reduce grain yield by as much as one‐half in susceptible varieties, which may be mitigated by fungicide application (Martinelli et al., 1994; Picinini and Fernandes, 1994). May et al. (2014) demonstrated that fungicide application improved both the yield and grain quality in crown rust‐susceptible varieties, when sprayed after the flag leaf had fully emerged, but the more resistant variety, ‘Leggett’, showed no yield response to fungicide application. The decision to apply fungicide is often influenced by cost–benefit analysis, dictated by grain value and fungicide and application costs. For instance, in Minnesota, where oat is largely grown for feed, the grain has a lower value compared with that used for food and milling, most often making chemical intervention uneconomic. Nevertheless, in 2015, oat crown rust infections in Minnesota became so severe that, in the following growing season, many farmers applied fungicide to the crop for the first time. The number of fungicides that control crown rust is limited, not just for lack of effective chemistry, but because some fungicides are not labelled for use in oat in some regions. Products containing pyraclostrobin or azoxystrobin (alone or in combination), or a combination of propiconazole and tryfloxystrobin can control the pathogen (Behnken et al., 2009). However, some of these products have restrictions that prevent application 30–40 days prior to harvest, making it difficult to time the application for disease control without encroaching on the mandated labelling regulations.
The high evolutionary capacity of Pca and the increasing use of fungicides in oat raise the prospect of fungicide insensitivity in this pathogen. A recent evaluation by Oliver (2014) argues for the classification of rust fungi within the high‐risk group of pathogens to evolve fungicide insensitivity. The polycyclic and airborne nature of rust fungi strongly resembles that of pathogens which have developed fungicide insensitivity in the past. Reduced sensitivity to demethylation inhibitor and quinone outside inhibitor fungicides has been detected in Puccinia triticina, the wheat leaf rust pathogen (Arduim et al., 2012). Similarly, the monitoring of fungicide effectiveness for more than a decade in Phakopsora pachyrhizi, the causal agent of Asian soybean rust, demonstrated a reduction in fungicide sensitivity over time (Godoy et al., 2016).
It is worth noting that, although fungicides can reduce the impact of rust, their use comes with environmental and economic costs, as well as risks to human health. Research on the impact of fungicide applications on yield, grain quality, milling characteristics and plant development is still required. Finally, oat occupies an important niche in organic production systems, where chemical treatments are not permitted, and thus there is increasing market pressure to avoid the use of fungicides and to employ alternative approaches.
Genetic Resistance to Oat Crown Rust
Oat crown rust resistance may be mediated by race‐specific or non‐race‐specific mechanisms (Carson, 2011; Leonard, 2002; Periyannan et al., 2017). Race‐specific resistance is underpinned by the concept of the gene‐for‐gene interaction, which has been fundamental in the breeding of disease‐resistant crop varieties (Dodds and Rathjen, 2010; Flor, 1971), and has been extensively exploited in oat breeding programmes to control crown rust since the early 1900s (Ohm and Shaner, 1992). This type of resistance depends on highly heritable single genes that typically elicit a hypersensitive response, which either completely or partially inhibits rust sporulation (Ohm and Shaner, 1992). The use of seedling resistance against Pca was intensified in the USA in the late 1920s with the introduction of the highly resistant oat varieties ‘Victoria’ from Uruguay and ‘Bond’ from Australia (Ohm and Shaner, 1992; Simons, 1985). As oat with the Victoria‐type resistance (Pc2 and Pc11) showed strong protection against crown rust (Coffman, 1977; Simons, 1970), breeding efforts were rapidly undertaken to transfer the resistance from Victoria to US‐adapted oat germplasm. The first varieties carrying the Victoria‐derived resistance were released in 1940. Unfortunately, such success was short lived as these varieties were quickly devastated by Victoria blight caused by Helminthosporium victoriae in 1946 (Murphy and Meehan, 1946), as Pc2 had the pleiotropic effect of making lines with this gene highly susceptible to Victoria blight (Lorang et al., 2007). Varieties carrying the Bond‐derived resistance (Pc3 and Pc4), as well as other varieties, such as ‘Landhafer’, ‘Ukraine’ and ‘Santa Fe’, replaced Victoria‐derived varieties after the epidemic of H. victoriae. The resistance of these varieties was eventually overcome with the emergence of new crown rust races (Simons, 1985). Learning from these lessons, oat breeding programmes have been reluctant to use single sources of resistance since the mid‐1960s (Simons, 1985). Nevertheless, the field durability of crown rust‐resistant varieties continues to be challenged in the USA (Carson, 2011), Canada (McCallum et al., 2007), Australia (Park, 2008), and Brazil and Uruguay (Leonard and Martinelli, 2005).
There are many examples that illustrate the limited durability of Pc genes. For instance, Pc38 and Pc39 were released in the early 1980s and were used together; however, by the end of the decade, virulence to these genes, individually and in combination, was reported (Chong and Seaman, 1997; McCallum et al., 2007). Similarly, Pc48, which was first deployed in the 1990s, was defeated in 2001 (Chong and Zegeye, 2004). Two R genes that have been fairly effective are Pc68 and Pc91 (McCallum et al., 2007; McCartney et al., 2011; Rooney et al., 1994). Varieties carrying Pc68 were released in the early 1990s in Canada and remained effective until 2001. Later, Pc68 was combined with Pc94 in the variety ‘Leggett’, which was released in 2004 (McCallum et al., 2007). However, virulence to Pc94 has now been reported (Chong et al., 2011; R. F. Park, unpublished). Only a few races have been identified in the USA that overcome Pc91 and, for this reason, there is interest in the use of this gene in combination with other Pc genes (Chong et al., 2008). However, Pc91 lacked durability when deployed in Australia, with virulence detected 6 years after its release in the variety Drover (Park, 2013). Two years later, two further virulent pathotypes had been detected and approximately 35% of isolates analysed from eastern Australia were virulent for Pc91 (R. F. Park, unpublished). Notwithstanding, gene pyramiding is seen as a valid strategy for the use of race‐specific genes to more effectively extend the durability of R genes. The effectiveness of this approach will probably depend on the genes, the number of genes in use and whether or not such virulence combinations already exist in the pathogen population. For example, virulence on the variety ‘AC Assiniboia’, which contains three genes (Pc38, Pc39 and Pc68), was detected only 4 years after its release in North America (Leonard, 2007; McCallum et al., 2007). The same variety was released in Australia in 1999, under the name ‘Graza 68’, and virulence was detected just 2 years later (R. F. Park, unpublished). Thus, a priori knowledge of the pathogenic diversity of the pathogen population may be helpful to better inform decisions about gene pyramiding schemes to maximize resistance durability.
In addition to race‐specific resistance, oat breeding programmes have benefited from non‐specific quantitative resistance, often known as adult plant resistance (APR) or partial resistance (Ohm and Shaner, 1992). APR does not typically manifest at the seedling stage, and crop protection comes from the reduction in fungal sporulation or delay of symptom appearance (Díaz‐Lago et al., 2003; Jones, 1978; Lin et al., 2014; Portyanko et al., 2005). As such, APR is usually effective against all or at least a wide range of rust genotypes, a characteristic that is highly attractive in cases such as crown rust where so many pathotypes exist. The identification and characterization of APR genes in oat against Pca has been difficult because of the complexity of inheritance or the presence of race‐specific resistance within a single oat variety (e.g. Victoria, Santa Fe and Ukraine) (Klos et al., 2017; Loarce et al., 2016; Upadhyaya and Baker, 1960). In other instances, inheritance of APR can be less complex; for example, Victoria/Garry oat contain only two genes, Pc27 and Pc28, contributing to the overall APR phenotype (Simons et al., 1978; Upadhyaya and Baker, 1960). The Canadian A. sterilis accession 1387 is a case in which a single partially dominant gene, Pc69, controls APR (Harder et al., 1984). Currently, six genes have been designated to be APR‐conditioning genes in oat, namely Pc27, Pc28, Pc69, Pc72, Pc73 and Pc74 (Table S1) (USDA‐ARS CDL, 2016), and more than 25 quantitative trait loci (QTLs) associated with APR have been mapped in oat (Acevedo et al., 2010; Babiker et al., 2015; Barbosa et al., 2006; Jackson et al., 2007; Lin et al., 2014; Portyanko et al., 2005; Zhu and Kaeppler, 2003).
In contrast with race‐specific resistance, which often does not remain effective for more than 5 years in the field (Carson, 2011), APR can provide rust protection for extended periods of time. One example that illustrates such resistance durability is the variety ‘Red Rustproof’ (A. byzantina K. Koch), an oat introduced into the USA in the 1860s, which served as a progenitor of most winter oat in the country (Stanton, 1955). Red Rustproof oat have several strains that were widely used in the field for many years as some carried consistently effective crown rust resistance for more than 100 years (Luke et al., 1972). The resistance in Red Rustproof oat is thought to be primarily associated with APR because both late‐rusting (delayed symptom development) and slow‐rusting (low disease severity) traits are manifested. According to genetic studies of the variety Red Rustproof‐14, these resistance traits are controlled by a few genes with high heritability (Luke et al., 1972). However, Red Rustproof oat also carry race‐specific resistance, as hypersensitive‐like responses to some races of Pca occurs (Luke et al., 1972). Such race specificity could perhaps be explained by the presence of uncharacterized race‐specific genes, in addition to the gene Pc1, which was isolated in Red Rustproof oat and shown to be linked to Pc2 (Davies and Jones, 1927; Dietz and Murphy, 1930) (Table S1).
A second example of durability is provided by the oat line MN841801, which exhibits APR and has remained partially resistant to crown rust infection for over 30 years (Leonard, 2002). MN841801 was developed by the oat breeding programme at the University of Minnesota in the 1960s, and was the result of a cross between 65B663 (a selection from Florad/58‐7, originally from Coker Pedigreed Seed Company) and 65B1362, both highly resistant lines (Leonard, 2002). Over the years, the durable resistance of MN84801 has attracted much attention and several studies conducted by independent groups to map the QTLs associated with the resistance phenotype reached different conclusions. The inheritance of APR in MN841801 was examined by Chong (2000), who used recombinant inbred lines (RILs) from the cross AC Assiniboia/MN841801, and found two complementary APR genes effective against a specific pathotype of Pca. In contrast, in a cross between MN841801‐1 (a reselection of MN841801) and ‘Noble‐2’ (a reselection of ‘Noble’), molecular markers allowed the identification of four major and three minor QTLs that control crown rust resistance (Acevedo et al., 2010; Portyanko et al., 2005), whereas another study using 6K oat SNPs and Kompetitive allele‐specific PCR (KASP) found only one major QTL in the Assiniboia/MN841801 RIL population (Lin et al., 2014). Efforts to determine the underlying mechanism of APR in MN841801 still continue and have extended to comparative transcriptome analysis of MN841801 and Noble‐2 (Loarce et al., 2016). Such transcriptome profiling comparisons reveal an interesting list of defence‐related genes that are up‐regulated during crown rust infection, including those for signal perception and transduction, hormone production and cell wall modification, amongst others.
Another source of APR that has been studied is ‘TAM O‐301’, a variety developed from a cross between a Texas‐adapted A. sativa and the A. sterilis accession PI295919 (Hoffman et al., 2006). TAM O‐301 carries the R gene Pc58 (Simons et al., 1978) and, according to results using restriction fragment length polymorphism (RLFP) and AFLP markers in a ‘Ogle’/TAM O‐301 RIL mapping population, the resistance from TAM O‐301 is also controlled by two additional genes, probably responsible for the APR phenotype (Jackson et al., 2007, 2008; Portyanko et al., 2001). However, TAM O‐301 could also contain more than one seedling resistance gene (R. F. Park, unpublished). As exemplified by QTL identification in the oat lines MN841801 and TAM O‐301, the detection of genes for quantitative resistance is expedited by new marker technologies that are proving advantageous in the alleviation of mapping complexities. Newly developed high‐throughput genetic marker assay systems, such as SNP, KASP and diversity arrays technology (DArT), are of benefit in oat breeding, with the construction of dense genetic maps enabling the discovery of additional resistance QTLs against crown rust in different oat crosses and RILs (Babiker et al., 2015; Barbosa et al., 2006; Chaffin et al., 2016; Jackson et al., 2007; Zhu and Kaeppler, 2003). In parallel, such systems are also used for genome‐wide association study (GWAS) as an alternative route to locate crown rust resistance QTLs (Klos et al., 2017; Montilla‐Bascón et al., 2015). Apart from their application in marker‐assisted selection, molecular markers are essential for map‐based cloning of resistance genes (Cabral et al., 2014) as a basic step towards the determination of the mechanisms by which these genes exert their function. In this context, recent advances in genotyping by sequencing (GBS) and RNA sequencing (RNA‐seq) in oat (Gutierrez‐Gonzalez et al., 2013; Huang et al., 2014) have provided essential resources to enable functional studies of candidate genes.
Finding New Sources of Resistance Against Oat Crown Rust
The discovery of diverse resistance genes in A. sterilis certainly made important contributions towards the exploitation of multiple gene resistance in breeding programmes (Leonard, 2007; McCallum et al., 2007; Simons, 1985). Indeed, more than 45 effective Pc genes were obtained from A. sterilis, including Pc38, Pc39, Pc68 and the majority of the APR genes. Several other wild oat species have been screened and used to identify new resistance genes (Aung et al., 2010; Cabral and Park, 2016; Cabral et al., 2011; Mitchell Fetch et al., 2007; Rooney et al., 1994). Recent sources of both race‐specific and APR include A. strigosa, A. glabrota, A. trichophylla, A. longiglumus, A. magna and A. murphyi (Cabral and Park, 2014; Sowa et al., 2016; USDA‐ARS CDL, 2016); some of these sources originated from Moroccan and Israeli collections (Dinoor, 1970; Tan and Carson, 2013; Wahl, 1970). Carson (2010) screened accessions of A. barbata from the Mediterranean region and found some additional sources of resistance for both seedling and adult stages, including a new type of ‘blotchy’ reaction in adult plants. Thus, it is unlikely that resistance resources from wild species have been exhausted, and systematic evaluations of seed banks could enhance current management approaches. In a similar framework, non‐host resistance holds promise to provide quantitative and durable disease resistance against a broad spectrum of pathogens (Bettgenhaeuser et al., 2014; Figueroa et al., 2015; Heath, 2000). Brachypodium distachyon, a temperate grass that belongs to the Pooideae subfamily, is closely related to oat (Gutierrez‐Gonzalez and Garvin, 2011; Kellogg, 2001) and has been shown to harbour resistance against multiple wheat cereal rusts (Ayliffe et al., 2013; Dawson et al., 2015; Figueroa et al., 2013). Thus, it is worthwhile to examine its potential against crown rust to unravel genes or loci that are associated with either disease resistance or susceptibility using modern biological techniques such as genome editing.
Future Research Directions and Perspectives
To date, no seedling or APR resistance gene has been cloned from oat. However, current efforts to assemble genome references for A. sativa and A. strigosa will deliver improvements in molecular and genetic tools that will facilitate the identification and mapping of known resistance genes or QTLs. Furthermore, new approaches, such as mutagenesis resistance gene enrichment (MutRenSeq), developed to rapidly clone disease resistance genes in wheat (Steuernagel et al., 2016), offer promise for the cloning of oat resistance genes. Such efforts will lead to valuable tools for the introgression and/or pyramiding of resistance genes into elite oat germplasm and help to develop new varieties with enhanced resistance against crown rust.
Wild relatives are a known reservoir of genetic diversity that has been extremely advantageous to oat breeding programmes in the past, and there should be further efforts to screen germplasm collections. Nevertheless, we must keep in mind that the introgression of traits from alien germplasm can be difficult and extremely laborious. Lack of synteny between species could translate into suppressed recombination, failure to achieve chromosome pairing and hybridization incompatibility, amongst other challenges (Wulff and Moscou, 2014). However, these can be overcome by the employment of special techniques, such as embryo rescue, the development of synthetic hexaploids and colchicine treatment coupled with backcrossing to improve fertility (Loskutov and Rines, 2011; Rines et al., 2007). The generation of resistance gene cassettes is one option to stack alien resistance genes, but this would require the development of genetically modified (GM) oat varieties. Like other GM crops, such an approach would face significant hurdles in terms of public acceptability, especially given that oat occupies a health food niche in the market. Thus, as we conceive viable paths towards sustainable oat production and health, gene editing should be considered as a powerful tool to access untapped genetic sources and make advances in the management of this devastating disease. The identification of susceptibility factors may provide opportunities to engineer broad‐spectrum resistance by the modification of gene alleles in oat. Three naturally occurring examples that support the feasibility of this approach are the mlo resistance in barley against powdery mildew (Humphry et al., 2006), as well as Lr34‐ and Lr67‐mediated resistance in wheat against rust and powdery mildew (Krattinger et al., 2009, 2011; Moore et al., 2015). In the case of mlo resistance, recessive mlo resistance alleles of the Mlo locus are responsible for non‐race specific resistance, and the simultaneous modification of all three Mlo homoeoalleles in wheat resulted in broad‐spectrum resistance to Blumeria graminis f. sp. tritici (Wang et al., 2014). Likewise, Lr34 and Lr67 illustrate how simple amino acid changes in a putative adenosine triphosphate‐binding cassette transporter and a hexose transporter, respectively, can confer broad‐spectrum resistance (Dodds and Lagudah, 2016; Krattinger et al., 2011). Current work to make precise genome edits using Clustered Regularly Interspaced Short Palindromic Repeat/CRISPR Associated protein 9 (CRISPR/Cas9), coupled with studies to determine susceptibility factors in oat, will open up doors to control crown rust at a global scale.
Little is known about the mechanisms that drive genetic variability in Pca; however, efforts are underway to generate high‐quality reference genomes for this species to enable comparative genomics and effector gene discovery. Once these resources are in place, population genomics studies will shed light into the genetic diversity of Pca. Such studies may determine the contribution of sexual recombination and other factors to the rapid evolution of this pathogen. An understanding of the diversity of Pca is essential in staying ahead of the pathogen's ability to overcome R genes and in the prevention of large‐scale epidemics, which will negatively impact oat growers. Thus, monitoring of the population dynamics and virulence shifts of Pca should be an important component to design strategies to control crown rust.
Conclusions
In the past few years, the management of oat crown rust has become more difficult as the pathogen has evolved virulence to most of the R genes deployed in the field (Carson, 2008, 2011; Park, 2008). The future release of a genome reference for A. sativa and other relatives will be key to the acceleration of the identification of novel disease resistance genes and will allow us to design more effective disease management strategies. Although there might not be a silver bullet to achieve durable resistance, we must consider the importance of protecting current research and development investments by avoiding the deployment of single R genes in the field. This will require strategic planning of effective gene combinations, and tailored deployment of resistant varieties amongst geographical regions. Ongoing developments in molecular genetics and genomics tools are expected to have a tremendous impact in improving our understanding of the genetic basis of resistance to crown rust in oat, pathogenicity in Pca and the host–pathogen interaction. The high genetic variability of Pca makes this pathogen an ideal model to investigate virulence evolution in rust fungi.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Oat resistance genes against Puccinia coronata f. sp. avenae.
Acknowledgements
We acknowledge support by the University of Minnesota Experimental Station USDA‐NIFA Hatch/Figueroa project MIN‐22–058, Smith project MIN‐01–023, USDA‐ARS Kianian and the Australian Grains Research Development Corporation. We thank Howard Rines and P. N. Dodds for discussions and revisions during manuscript preparation, as well as E. Byamukama and R. Caspers for sharing photographs used to construct the figures. The authors declare no conflicts of interest.
References
- Acevedo, M. , Jackson, E.W. , Chong, J. , Rines, H.W. , Harrison, S. and Bonman, J.M. (2010) Identification and validation of quantitative trait loci for partial resistance to crown rust in oat. Phytopathology, 100, 511–521. [DOI] [PubMed] [Google Scholar]
- Anikster, A.Y. , Eilam, T. , Manisterski, J. and Leonard, K.J. (2003) Self‐fertility and other distinguishing characteristics of a new morphotype of Puccinia coronata pathogenic on smooth brome grass. Mycologia, 95, 87–97. [DOI] [PubMed] [Google Scholar]
- Arduim, G.D.S. , Reis, E.M. , Barcellos, A.L. and Turra, C. (2012) In vivo sensitivity reduction of Puccinia triticina races, causal agent of wheat leaf rust, to DMI and QoI fungicides. Summa Phytopathol. 38, 306–311. [Google Scholar]
- Aung, T. , Zwer, P. , Park, R. , Davies, P. , Sidhu, P. and Dundas, I. (2010) Hybrids of Avena sativa with two diploid wild oats (CIav6956) and (CIav7233) resistant to crown rust. Euphytica, 174, 189–198. [Google Scholar]
- Ayliffe, M. , Singh, D. , Park, R. , Moscou, M. and Pryor, T. (2013) Infection of Brachypodium distachyon with selected grass rust pathogens. Mol. Plant–Microbe Interact. 26, 946–957. [DOI] [PubMed] [Google Scholar]
- Babiker, E.M. , Gordon, T.C. , Jackson, E.W. , Chao, S. , Harrison, S.A. , Carson, M.L. , Obert, D.E. and Bonman, J.M. (2015) Quantitative trait loci from two genotypes of oat (Avena sativa) conditioning resistance to Puccinia coronata . Phytopathology, 105, 239–245. [DOI] [PubMed] [Google Scholar]
- Barbosa, M.M. , Federizzi, L.C. , Milach, S.C.K. , Martinelli, J.A. and Thomé, G.C. (2006) Molecular mapping and identification of QTLs associated to oat crown rust partial resistance. Euphytica, 150, 257–269. [Google Scholar]
- Bartos, P. , Dyck, P.L. and Samborski, D.J. (1969) Adult‐plant leaf rust resistance in Thatcher and Marquis wheat: a genetic analysis of the host–parasite interaction. Can. J. Bot. 47, 267–269. [Google Scholar]
- Behnken, L.M. , Breitenbach, F.R. and Miller, R.P. (2009) Impact of Foliar Fungicide to Control Crown Rust in Oats in 2009 Available at: https://www.extension.umn.edu/agriculture/forages/pest/docs/umn-ext-impact-of-foliar-fungicide-to-control-crown-rusts-in-oats.pdf [accessed 21 April 2017].
- Bettgenhaeuser, J. , Gilbert, B. , Ayliffe, M. and Moscou, M.J. (2014) Nonhost resistance to rust pathogens – a continuation of continua. Front. Plant Sci. 5, 664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brake, V.M. , Irwin, J.A.G. and Park, R.F. (2001) Genetic variability in Australian isolates of Puccinia coronata f. sp. avenae assessed with molecular and pathogenicity markers. Australas. Plant Pathol. 30, 259–266. [Google Scholar]
- van den Broeck, C.H. , Londono, M.D. , Timmer, R. , Smulders, J.M. , Gilissen, J.L. , and van der Meer, M.I. (2016) Profiling of nutritional and health‐related compounds in oat varieties. Foods, 5, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, M.R. (1937) A study of crown rust, Puccinia coronata Corda, in Great Britain: physiologic specialization in the uredospore stage. Ann. Appl. Biol. 24, 504–527. [Google Scholar]
- Burnette, D. , Lenz, M. , Sisson, P.F. , Sutherland, S. and Weaver, S.H. (1992) Marketing, processing, and uses of oat for food In: Oat Science and Technology (Marshall H. and Sorrells M., eds), pp. 247–263. Madison, WI: ASA/CSSA. [Google Scholar]
- Cabral, A.L. and Park, R.F. (2014) Seedling resistance to Puccinia coronata f. sp. avenae in Avena strigosa, A. barbata and A. sativa . Euphytica, 196, 385–395. [Google Scholar]
- Cabral, A. and Park, R. (2016) Genetic analysis of seedling resistance to crown rust in five diploid oat (Avena strigosa) accessions. J. Appl. Genet. 56, 27–36. [DOI] [PubMed] [Google Scholar]
- Cabral, A.L. , Singh, D. and Park, R.F. (2011) Identification and genetic characterisation of adult plant resistance to crown rust in diploid and tetraploid accessions of Avena . Ann. Appl. Biol. 159, 220–228. [Google Scholar]
- Cabral, A.L. , Ganesh, B.N. , Fetch, J.M. , McCartney, C. , Fetch, T. , Park, R.F. , Menzies, J.G. , McCallum, B. , Nanaiah, G.K. and Goyal, A. (2014) Oat fungal diseases and the application of molecular marker technology for their control In: Future Challenges in Crop Protection against Fungal Pathogens (Goyal A. and Manoharachary C., eds), pp. 343–358. New York: Springer. [Google Scholar]
- Carson, M.L. (2008) Virulence frequencies in oat crown rust in the United States from 2001 through 2005. Plant Dis. 92, 379–384. [DOI] [PubMed] [Google Scholar]
- Carson, M.L. (2010) Additional sources of broad‐spectrum resistance to Puccinia coronata f. sp. avenae from Canadian accessions of Avena barbata . Plant Dis. 94, 1405–1410. [DOI] [PubMed] [Google Scholar]
- Carson, M.L. (2011) Virulence in oat crown rust (Puccinia coronata f. sp. avenae) in the United States from 2006 through 2009. Plant Dis. 95, 1528–1534. [DOI] [PubMed] [Google Scholar]
- Centre for Agriculture and Bioscience International (CABI) (2015) Rhamnus cathartica (buckthorn) In: Invasive Species Compendium. Wallingford, Oxfordshire: CABI; Available at: www.cabi.org/isc. [Google Scholar]
- Chaffin, A.S. , Huang, Y.‐F. , Smith, S. , Bekele, W.A. , Babiker, E. , Gnanesh, B.N. , Foresman, B.J. , Blanchard, S.G. , Jay, J.J. , Reid, R.W. , Wight, C.P. , Chao, S. , Oliver, R. , Islamovic, E. , Kolb, F.L. , McCartney, C. , Mitchell Fetch, J.W. , Beattie, A.D. , Bjørnstad, Å. , Bonman, J.M. , Langdon, T. , Howarth, C.J. , Brouwer, C.R. , Jellen, E.N. , Klos, K.E. , Poland, J.A. , Hsieh, T.‐F. , Brown, R. , Jackson, E. , Schlueter, J.A. and Tinker, N.A. (2016) A consensus map in cultivated hexaploid oat reveals conserved grass synteny with substantial subgenome rearrangement. Plant Genome, 9, 1–21. [DOI] [PubMed] [Google Scholar]
- Chong, J. (2000) Inheritance of resistance to two Puccinia coronata isolates in a partial resistant oat line MN841801. Acta Phytopathol. Entomol. Hung. 35, 37–40. [Google Scholar]
- Chong, J. and Kolmer, J.A. (1993) Virulence dynamics and phenotypic diversity of Puccinia coronata f.sp. avenae in Canada from 1974 to 1990. Can. J. Bot. 71, 248–255. [Google Scholar]
- Chong, J. and Seaman, W.L. (1997) Incidence and virulence of Puccinia coronata f. sp. avenae in Canada in 1995. Can. J. Plant Pathol. 19, 176–180. [Google Scholar]
- Chong, J. and Zegeye, T. (2004) Physiologic specialization of Puccinia coronata f. sp. avenae, the cause of oat crown rust, in Canada from 1999 to 2001. Can. J. Plant Pathol. 26, 97–108. [Google Scholar]
- Chong, J. , Leonard, K.J. and Salmeron, J.J. (2000) A North American system of nomenclature for Puccinia coronata f. sp. avenae . Plant Dis. 84, 580–585. [DOI] [PubMed] [Google Scholar]
- Chong, J. , Gruenke, J. , Dueck, R. , Mayert, W. and Woods, S. (2008) Virulence of oat crown rust [Puccinia coronata f. sp. avenae] in Canada during 2002–2006. Can. J. Plant Pathol. 30, 115–123. [Google Scholar]
- Chong, J. , Gruenke, J. , Dueck, R. , Mayert, W. , Fetch, J.M. and McCartney, C. (2011) Virulence of Puccinia coronata f. sp. avenae in the Eastern Prairie Region of Canada during 2007–2009. Can. J. Plant Pathol. 33, 77–87. [Google Scholar]
- Coffman, F. (1977) Oat history, identification and classification. US Dep. Agric. Agric. Res. Serv. Tech. Bull. 1516, 356. [Google Scholar]
- Cornu, M.M. (1880) Notes sur quelques parasites des plantes vivantes: générations alternantes; pezizes a sclérotes. Bull. Soc. Bot. Fr. 27, 209–210. [Google Scholar]
- Cummins, G.B. (1971) Key to the genera of rust fungi In: The Rust Fungi of Cereals, Grasses and Bamboos, p. 1 Berlin Heidelberg: Springer. [Google Scholar]
- Dangl, J.L. , Horvath, D.M. and Staskawicz, B.J. (2013) Pivoting the plant immune system from dissection to deployment. Science, 341, 746–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, D.W. and Jones, E.T. (1927) Further studies on the inheritance of resistance to crown rust (P. coronata, Corda) in fs segregates of a cross between Red Rustproof (A. sterilis) and Scotch potato oats (A. sativa). Welsh J. Agric. 3, 232–235. [Google Scholar]
- Dawson, A.M. , Bettgenhaeuser, J. , Gardiner, M. , Green, P. , Hernández‐Pinzón, I. , Hubbard, A. and Moscou, M.J. (2015) The development of quick, robust, quantitative phenotypic assays for describing the host–nonhost landscape to stripe rust. Front. Plant Sci. 6, 876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz‐Lago, J.E. , Stuthman, D.D. and Leonard, K.J. (2003) Evaluation of components of partial resistance to oat crown rust using digital image analysis. Plant Dis. 87, 667–674. [DOI] [PubMed] [Google Scholar]
- Dietz, S. (1926) The alternate hosts of crown rust, Puccinia coronata Corda. J. Agric. Res. 33, 953–970. [Google Scholar]
- Dietz, S.M. and Murphy, H.C. (1930) Inheritance of resistance to Puccinia coronata avenae, p. f. III. Phytopathology, 20, 120. [Google Scholar]
- Dinoor, A. (1970) Sources of oat crown rust resistance in hexaploid and tetraploid wild oats in Israel. Can. J. Bot. 48, 153–161. [Google Scholar]
- Dinoor, A. (1977) Oat crown rust resistance in Israel. Ann. N. Y. Acad. Sci. 287, 357–366. [Google Scholar]
- Dodds, P.N. and Lagudah, E.S. (2016) Starving the enemy. Science, 354, 1377–1378. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N. and Rathjen, J.P. (2010) Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N. , Lawrence, G.J. , Catanzariti, A.‐M. , Ayliffe, M.A. and Ellis, J.G. (2004) The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell, 16, 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N. , Lawrence, G.J. , Catanzariti, A.‐M. , Teh, T. , Wang, C.‐I.A. , Ayliffe, M.A. , Kobe, B. and Ellis, J.G. (2006) Direct protein interaction underlies gene‐for‐gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci USA, 103, 8888–8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehlert, D.C. , McMullen, M.S. and Hammond, J.J. (2001) Genotypic and environmental effects on grain yield and quality of oat grown in North Dakota. Crop Sci. 41, 1066–1072. [Google Scholar]
- D'Oliveira, B. (1942) A estacao agronomica e os problemas nacionais de fitopatologia. Rev. Agron. 30, 414–438. [Google Scholar]
- Dracatos, P.M. , Dumsday, J.L. , Olle, R.S. Cogan, N.O.I. , Dobrowolski, M.P. , Fujimori, M. , Roderick, H. , Stewart, A.V. , Smith, K.F. and Forster, J.W. (2006) Development and characterization of EST‐SSR markers for the crown rust pathogen of ryegrass (Puccinia coronata f.sp. lolii). Genome, 49, 572–583. [DOI] [PubMed] [Google Scholar]
- Endo, R.M. and Boewe, G. (1958) Losses caused by crown rust of oats in 1956 and 1957. Plant Dis. Rep. 42, 1126–1128. [Google Scholar]
- Eriksson, J. (1908) Neue studien über die spezielisierung der grasbewohnenden kronenrostarten. Arch. Bot. 8, 1–26. [Google Scholar]
- Eriksson, J. and Henning, E. (1894) Die hauptresultate einer neuen untersuchung über die getreideroste. Z. Pflanzenkr. 4, 66–73. [Google Scholar]
- Eshed, N. and Dinoor, A. (1980) Genetics of pathogenicity in Puccinia coronata: pathogenic specialization at the host genus level. Phytopathology, 70, 1042–1046. [Google Scholar]
- Figueroa, M. , Alderman, S. , Garvin, D.F. and Pfender, W.F. (2013) Infection of Brachypodium distachyon by formae speciales of Puccinia graminis: early infection events and host–pathogen incompatibility. PLoS One, 8, e56857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa, M. , Castell‐Miller, C.V. , Li, F. , Hulbert, S.H. and Bradeen, J.M. (2015) Pushing the boundaries of resistance: insights from Brachypodium–rust interactions. Front. Plant Sci. 6, 558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann, G. (1965) Variability in the physiologic race populations of oat crown rust isolated from aecia and uredia. Plant Dis. Rep. 49, 132–133. [Google Scholar]
- Flor, H.H. (1971) Current status of the gene‐for‐gene concept. Annu. Rev. Phytopathol. 9, 275–296. [Google Scholar]
- Food and Agriculture Organization (FAO). (2014) FAOSTAT Statistical Database, Rome: FAO; Available at: http://www.fao.org/faostat/en/#data [accessed 1 April 2017]. [Google Scholar]
- Forbes, I. (1939) Factors affecting the development of Puccinia coronata in Louisiana. Phytopathology, 29, 659–684. [Google Scholar]
- Garnica, D.P. , Nemri, A. , Upadhyaya, N.M. , Rathjen, J.P. and Dodds, P.N. (2014) The ins and outs of rust haustoria. PLoS Pathog. 10, e1004329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassner, G. (1916) Die geteriedroste und ihr aufreten im subtropischedn ostlichen sudamerika. Zentralbl. Bakteriol. Parassitenkd. Infekt. Hyg. Abt. 2, 305–381. [Google Scholar]
- Gnanesh, B. , Fetch, J. , Zegeye, T. , McCartney, C. and Fetch, T. (2014) Oat In: Alien Gene Transfer in Crop Plants, Volume 2: Achievements and Impacts (Pratap A. and Kumar J., eds), pp. 51–73. New York, NY: Springer‐Verlag. [Google Scholar]
- Godoy, C.V. , Seixas, C.D.S. , Soares, R.M. , Marcelino‐Guimarães, F.C. , Meyer, M.C. and Costamilan, L.M. (2016) Asian soybean rust in Brazil: past, present, and future. Pesqui. Agropecuária Bras. 51, 407–421. [Google Scholar]
- Graichen, F.A.S. , Martinelli, J.A. , Federizzi, L.C. , Pacheco, M.T. , Chaves, M.S. and Wesp, C.L. (2010) Inheritance of resistance to oat crown rust in recombinant inbred lines. Sci. Agric. 67, 435–440. [Google Scholar]
- Gutierrez‐Gonzalez, J.J. and Garvin, D.F. (2011) Reference genome‐directed resolution of homologous and homeologous relationships within and between different oat linkage maps. Plant Genome J. 4, 178. [Google Scholar]
- Gutierrez‐Gonzalez, J.J. , Tu, Z.J. and Garvin, D.F. (2013) Analysis and annotation of the hexaploid oat seed transcriptome. BMC Genomics, 14, 471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammami, I. , Allagui, M.B. , Chakroun, M. and EL‐Gazzeh, M. (2010) Natural population of oat crown rust in Tunisia. Phytopathol. Mediterr. 49, 35–41. [Google Scholar]
- Harder, D.E. and Haber, S. (1992) Oat diseases and pathologic techniques In: Oat Science and Technology (Marshall H. and Sorells M., eds), pp. 307–425. Madison, WI: ASA/CSA. [Google Scholar]
- Harder, D.E. , McKenzie, R.I.H. and Martens, J.W. (1984) Inheritance of adult plant resistance to crown rust in an accession of Avena sterilis . Phytopathology, 74, 352–353. [Google Scholar]
- Heath, M.C. (2000) Nonhost resistance and nonspecific plant defenses. Curr. Opin. Plant Biol. 3, 315–319. [DOI] [PubMed] [Google Scholar]
- Hoffman, D.L. , Chong, J. , Jackson, E.W. and Obert, D.E. (2006) Characterization and mapping of a crown rust resistance gene complex (Pc58) in TAM O‐301. Crop Sci. 46, 2630–2635. [Google Scholar]
- Hogenhout, S.A. , Van der Hoorn, R.A.L. , Terauchi, R. and Kamoun, S. (2009) Emerging concepts in effector biology of plant‐associated organisms. Mol. Plant–Microbe Interact. 22, 115–122. [DOI] [PubMed] [Google Scholar]
- Holland, J. and Munkvold, G. (2001) Genetic relationships of crown rust resistance, grain yield, test weight, and seed weight in oat. Crop Sci. 41, 1041–1050. [Google Scholar]
- Huang, Y.‐F. , Poland, J.A. , Wight, C.P. , Jackson, E.W. and Tinker, N.A. (2014) Using genotyping‐by‐sequencing (GBS) for genomic discovery in cultivated oat. PLoS One, 9, e102448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys, D.G. and Mather, D.E. (1996) Heritability of β‐glucan, groat percentage, and crown rust resistance in two oat crosses. Euphytica, 91, 359–364. [Google Scholar]
- Humphry, M. , Consonni, C. and Panstruga, R. (2006) mlo‐based powdery mildew immunity: silver bullet or simply non‐host resistance? Mol. Plant Pathol. 7, 605–610. [DOI] [PubMed] [Google Scholar]
- Jackson, E.W. , Obert, D.E. , Menz, M. , Hu, G. , Avant, J.B. , Chong, J. and Bonman, J.M. (2007) Characterization and mapping of oat crown rust resistance genes using three assessment methods. Phytopathology, 97, 1063–1070. [DOI] [PubMed] [Google Scholar]
- Jackson, E.W. , Obert, D.E. , Chong, J. , Avant, J. and Bonman, J. (2008) Detached‐leaf method for propagating Puccinia coronata and assessing crown rust resistance in oat. Plant Dis. 92, 1400–1406. [DOI] [PubMed] [Google Scholar]
- Jones, I.T. (1978) Components of adult plant resistance to powdery mildew (Erysiphe graminis f. sp. avenae) in oats. Ann. Appl. Biol. 90, 233–239. [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kellogg, E.A. (2001) Evolutionary history of the grasses. Plant Physiol. 125, 1198–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenová‐Jiráková, H. , Leišová‐Svobodova, L. , Hanzalová, A. and Kučera, L. (2010) Diversity of oat crown rust (Puccinia coronata f.sp. avenae) isolates detected by virulence and AFLP analyses. Plant Prot. Sci. 46, 98–106. [Google Scholar]
- Klos, K.E. , Yimer, B.A. , Babiker, E.M. Beattie, A.D. , Bonman, J.M. , Carson, M.L. , Chong, J. , Harrison, S.A. , Ibrahim, A.M.H. , Kolb, F.L. , McCartney, C.A. , McMullen, M. , Fetch, J.M. , Mohammadi, M. , Murphy, J.P. and Tinker, N.A. (2017) Genome‐wide association mapping of crown rust resistance in oat elite germplasm. Plant Genome, 10, 1–13. [DOI] [PubMed] [Google Scholar]
- Knight, K.S. , Kurylo, J.S. , Endress, A.G. , Stewart, J.R. and Reich, P.B. (2007) Ecology and ecosystem impacts of common buckthorn (Rhamnus cathartica): a review. Biol. Invasions, 9, 925–937. [Google Scholar]
- Kostic, B. (1959) The cereal rusts in the south‐eastern part of Yugoslavia in 1958 and 1959. Robigo, 9, 8–12. [Google Scholar]
- Krattinger, S.G. , Lagudah, E.S. , Spielmeyer, W. , Singh, R.P. , Huerta‐Espino, J. , McFadden, H. , Bossolini, E. , Selter, L.L. and Keller, B. (2009) A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science, 323, 1360–1363. [DOI] [PubMed] [Google Scholar]
- Krattinger, S.G. , Lagudah, E.S. , Wicker, T. , Risk, J.M. , Ashton, A.R. , Selter, L.L. , Matsumoto, T. and Keller, B. (2011) Lr34 multi‐pathogen resistance ABC transporter: molecular analysis of homoeologous and orthologous genes in hexaploid wheat and other grass species. Plant J. 65, 392–403. [DOI] [PubMed] [Google Scholar]
- Leonard, K.J. (2002) Oat lines with effective adult plant resistance to crown rust. Plant Dis. 86, 593–598. [DOI] [PubMed] [Google Scholar]
- Leonard, K.J. (2007) Persistent virulence associations in sexual populations of Puccinia coronata . Plant Pathol. 56, 35–45. [Google Scholar]
- Leonard, K.J. and Martinelli, J.A. (2005) Virulence of oat crown rust in Brazil and Uruguay. Plant Dis. 89, 802–808. [DOI] [PubMed] [Google Scholar]
- Leonard, K.J. , Anikster, Y. and Manisterski, J. (2004) Patterns of virulence in natural populations of Puccinia coronata on wild oat in Israel and in agricultural populations on cultivated oat in the United States. Phytopathology, 94, 505–514. [DOI] [PubMed] [Google Scholar]
- Lin, Y. , Gnanesh, B.N. , Chong, J. Chen, G. , Beattie, A.D. , Mitchell Fetch, J.W. , Kutcher, H.R. , Eckstein, P.E. , Menzies, J.G. , Jackson, E.W. and McCartney, C.A. (2014) A major quantitative trait locus conferring adult plant partial resistance to crown rust in oat. BMC Plant Biol. 14, 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, M. and Hambleton, S. (2013) Laying the foundation for a taxonomic review of Puccinia coronata s.l. in a phylogenetic context. Mycol. Prog. 12, 63–89. [Google Scholar]
- Loarce, Y. , Navas, E. , Paniagua, C. , Fominaya, A. , Manjón, J.L. and Ferrer, E. (2016) Identification of genes in a partially resistant genotype of Avena sativa expressed in response to Puccinia coronata infection. Front. Plant Sci. 7, 731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, D. L. , and Hughes, M.E. (2000) Small Grain Losses due to Rust US Department of Agriculture Research Survey, St. Paul, MN: Cereal Disease Laboratory online publication CDL–EP#007 Available at: https://www.ars.usda.gov/midwest-area/stpaul/cereal-disease-lab/docs/small-grain-losses-due-to-rust/small-grain-losses-due-to-rust/ [accessed 24 April 2017].
- Lorang, J.M. , Sweat, T.A. and Wolpert, T.J. (2007) Plant disease susceptibility conferred by a “resistance” gene. Proc. Natl. Acad. Sci. USA, 104, 14 861–14 866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loskutov, I.G. and Rines, H.W. (2011) Avena In: Wild Crop Relatives: Genomic and Breeding Resources Cereals (Kole C., ed), pp. 109–183. Berlin Heidelberg: Springer‐Verlag. [Google Scholar]
- Luke, H.H. , Chapman, W.H. and Barnett, R.D. (1972) Horizontal resistance of Red Rustproof oats to crown rust. Phytopathology, 62, 414–417. [Google Scholar]
- Maia, T. , Badel, J.L. , Marin‐Ramirez, G. , de Rocha, C.M. , Fernandes, M.B. , da Silva, J.C.F. , de Azevedo‐Junior, G.M. and Brommonschenkel, S.H. (2017) The Hemileia vastatrix effector HvEC‐016 suppresses bacterial blight symptoms in coffee genotypes with the SH1 rust resistance gene. New Phytol. 213, 1315–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli, J.A. , Federizzi, L.C. and Bennedetti, A.C. (1994) Effect of oat cultivars mixtures and seed treatments on the restriction of leaf rust disease progress. Summa Phytopathol. 20, 113–115. [Google Scholar]
- May, W.E. , Ames, N. , Irvine, R.B. , Kutcher, H.R. , Lafond, G.P. and Shirtliffe, S.J. (2014) Are fungicide applications to control crown rust of oat beneficial? Can. J. Plant Sci. 94, 911–922. [Google Scholar]
- McCallum, B.D. , Fetch, T. and Chong, J. (2007) Cereal rust control in Canada. Aust. J. Agric. Res. 58, 639–647. [Google Scholar]
- McCartney, C.A. , Stonehouse, R.G. , Rossnagel, B.G. , Eckstein, P.E. , Scoles, G.J. , Zatorski, T. , Beattie, A.D. and Chong, J. (2011) Mapping of the oat crown rust resistance gene Pc91 . Theor. Appl. Genet. 122, 317–325. [DOI] [PubMed] [Google Scholar]
- McDonald, B.A. and Linde, C. (2002) Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 40, 349–379. [DOI] [PubMed] [Google Scholar]
- Mendgen, K. (1984) Development and physiology of teliospores In: The Cereal Rust, Vol. 1 (Roelfs A. and Bushnell W., eds), pp. 375–398. Orlando, FL: Academic Press, Inc. [Google Scholar]
- Menzies, J. , Xue, A. , Dueck, R. and Gruenke, J. (2015) Virulence of Puccinia coronata f. sp. avenae in Canada; 2010 to 2013. In: Proceedings of the 14th International Cereal Rusts and Powdery Mildews Conference, Copenhagen, Denmark, p. 95 Helsingør, Denmark: The European and Mediterranean Cereal Rusts Foundation. [Google Scholar]
- Mitchell Fetch, J.W. , Duguid, S.D. , Brown, P.D. Chong, J. , Fetch, T.G. , Haber, S.M. , Menzies, J.G. , Ames, N. , Noll, J. , Aung, T. and Stadnyk, K.D. (2007) Leggett oat. Can. J. Plant Sci. 87, 509–512. [Google Scholar]
- Montilla‐Bascón, G. , Rispail, N. , Sánchez‐Martín, J. , Rubiales, D. , Mur, L.A.J. , Langdon, T. , Howarth, C.J. and Prats, E. (2015) Genome‐wide association study for crown rust (Puccinia coronata f. sp. avenae) and powdery mildew (Blumeria graminis f. sp. avenae) resistance in an oat (Avena sativa) collection of commercial varieties and landraces. Front. Plant Sci. 6, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, J. , Herrera‐Foessel, S. , Lan, C. , Schnippenkoetter, W. , Ayliffe, M. , Huerta‐Espino, J. , Lillemo, M. , Viccars, L. , Milne, R. , Periyannan, S. , Kong, X. , Spielmeyer, W. , Talbot, M. , Bariana, H. , Patrick, J.W. , Dodds, P. , Singh, R. and Lagudah, E. (2015) A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 47, 1494–1498. [DOI] [PubMed] [Google Scholar]
- Murphy, H.C. (1935) Physiologic specialization in Puccinia coronata avenae . US Dep. Agric. Tech. Bull. 433, 48. [Google Scholar]
- Murphy, H.C. and Meehan, F. (1946) Reaction of oat varieties to a new species of Helminthosporium . Phytopathology, 36, 407. [Google Scholar]
- van Niekerk, B.D. , Pretorius, Z.A. and Boshoff, W.H.P. (2001) Pathogenic variability of Puccinia coronata f. sp. avenae and P. graminis f. sp. avenae on oat in South Africa. Plant Dis. 85, 1085–1090. [DOI] [PubMed] [Google Scholar]
- Ohm, H. and Shaner, G. (1992) Breeding oat for resistance to diseases In: Oat Science and Technology (Marshall H. and Sorells M., eds), pp. 657–698. Madison, WI: ASA/CSA. [Google Scholar]
- Oliver, R.P. (2014) A reassessment of the risk of rust fungi developing resistance to fungicides. Pest Manag. Sci. 70, 1641–1645. [DOI] [PubMed] [Google Scholar]
- Park, R.F. (2008) Breeding cereals for rust resistance in Australia. Plant Pathol. 57, 591–602. [Google Scholar]
- Park, R.F. (2013) New oat crown rust pathotype with virulence for Pc91 . Cereal Rust Rep. 11, 8–10. [Google Scholar]
- Park, R.F. and Wellings, C.R. (2012) Somatic hybridization in the Uredinales. Annu. Rev. Phytopathol. 50, 219–239. [DOI] [PubMed] [Google Scholar]
- Park, R.F. , Meldrum, S.M. and Oates, J.D. (2000) Recent pathogenic changes in the leaf (brown) rust pathogen of wheat and the crown rust pathogen of oats in Australia in relation to host resistance. Acta Phytopathol. Entomol. Hung. 35, 387–394. [Google Scholar]
- Periyannan, S. , Milne, R. , Figueroa, M. , Lagudah, E. and Dodds, P. (2017) An overview of genetic rust resistance: from broad to specific mechanisms. PLoS Pathog. 13, e1006380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petre, B. and Kamoun, S. (2014) How do filamentous pathogens deliver effector proteins into plant cells? PLoS Biol. 12, e1001801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peturson, B. (1954) The relative prevalence of specialized forms of Puccinia coronata that occur on Rhamnus cathartica in Canada. Can. J. Bot. 32, 40–47. [Google Scholar]
- Picinini, E.C. and Fernandes, J.M.C. (1994) Efficacy of fungicides for controlling crown rust of oats. Fitopatol. Bras. 19, 74–78. [Google Scholar]
- Portyanko, V.A. , Hoffman, D.L. , Lee, M. and Holland, J.B. (2001) A linkage map of hexaploid oat based on grass anchor DNA clones and its relationship to other oat maps. Genome, 44, 249–265. [PubMed] [Google Scholar]
- Portyanko, V.A. , Chen, G. , Rines, H.W. , Phillips, R.L. , Leonard, K.J. , Ochocki, G.E. and Stuthman, D.D. (2005) Quantitative trait loci for partial resistance to crown rust, Puccinia coronata, in cultivated oat, Avena sativa L. Theor. Appl. Genet. 111, 313–324. [DOI] [PubMed] [Google Scholar]
- Ravensdale, M. , Nemri, A. , Thrall, P.H. , Ellis, J.G. and Dodds, P.N. (2011) Co‐evolutionary interactions between host resistance and pathogen effector genes in flax rust disease. Mol. Plant Pathol. 12, 93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rines, H.W. , Porter, H.L. , Carson, M.L. and Ochocki, G. (2007) Introgression of crown rust resistance from diploid oat Avena strigosa into hexaploid cultivated oat A. sativa by two methods: direct crosses and through an initial 2x+4x synthetic hexaploid. Euphytica, 158, 67–79. [Google Scholar]
- Rooney, W.L. , Rines, H.W. and Phillips, R.L. (1994) Identification of RFLP markers linked to crown rust resistance genes Pc 91 and Pc92 in oat. Crop Sci. 34, 940–944. [Google Scholar]
- Sherf, A. , Bragonier, W. , Shurtleff, M. and Browning, J. (1956) A Collection of Miscellaneous Material Relating to Eradication and Control of Buckthorn (Rhamnus sp.) as the Alternate Host of the Organism (Puccinia coronata) Causing Crown Rust of Oats. Ames, IA: Department of Botany and Plant Pathology, Iowa State College.
- Sherf, A.F. (1954) The 1953 crown and stem rust epidemic of oats in Iowa. Proc. Iowa Acad. Sci. 61, 161–169. [Google Scholar]
- da Silva, L.P. , Pereira Coutinho, A. , Heleno, R.H. , Tenreiro, P.Q. and Ramos, J.A. (2016) Dispersal of fungi spores by non‐specialized flower‐visiting birds. J. Avian Biol. 47, 438–442. [Google Scholar]
- Simons, M.D. (1970) Crown Rust of Oats and Grasses Monograph. Worcester, MA: APS at The Hefferman Press, Inc. [Google Scholar]
- Simons, M.D. (1985) Crown rust In: The Cereal Rusts, Vol. 2 (Roelfs A. and Bushnell W., eds), pp. 131–172. Orlando, FL: Academic Press, Inc. [Google Scholar]
- Simons, M.D. and Michel, L. (1964) International register of pathogenic races of Puccinia coronata var. avenae . Plant Dis. Rep. 48, 763–766. [Google Scholar]
- Simons, M.D. , Martens, J.W. , McKenzie, R.I.H. , Nishiyama, I. , Sadanaga, K. , Sebesta, J. , and Thomas, H. (1978) Oats: a standardized system of nomenclature for genes and chromosomes and catalog of genes governing characters US Department of Agricultural Handbook, pp. 1–40. Madison, WI: US Department of Agriculture. [Google Scholar]
- Sivers, M.N. (1887) Ein probeanbau verschiedener hafersorten. Balt. Wochenschr. Landwirt. Sch. Gewerbefl. Handel. Dorpat. 39, 390–391. [Google Scholar]
- Sowa, S. , Paczos‐Grzęda, E. , Koroluk, A. , Okoń, S. , Ostrowska, A. , Ociepa, T. , Chrząstek, M. and Kowalczyk, K. (2016) Resistance to Puccinia coronata f. sp. avenae in Avena magna, A. murphyi, and A. insularis . Plant Dis. 100, 1184–1191. [DOI] [PubMed] [Google Scholar]
- Stanton, P. (1955) Oat identification and classification. US Dept. Agr. Tech. Bull. 1100, 206. [Google Scholar]
- Staples, R. and Macko, V. (1984) Germination of urediospores and differentiation of infection structures In: The Cereal Rusts, Vol. 1 (Roelfs A. and Bushnell W., eds), pp. 255–289. Orlando, FL: Academic Press, Inc. [Google Scholar]
- Steuernagel, B. , Periyannan, S.K. , Hernández‐Pinzón, I. , Witek, K. , Rouse, M.N. , Yu, G. , Hatta, A. , Ayliffe, M. , Bariana, H. , Jones, J.D.G. , Lagudah, E.S. and Wulff, B.B.H. (2016) Rapid cloning of disease‐resistance genes in plants using mutagenesis and sequence capture. Nat. Biotechnol. 34, 652–655. [DOI] [PubMed] [Google Scholar]
- Szabo, L.J. (2006) Deciphering species complexes: Puccinia andropogonis and Puccinia coronata, examples of differing modes of speciation. Mycoscience, 47, 130–136. [Google Scholar]
- Tan, M.Y.A. and Carson, M.L. (2013) Screening wild oat accessions from Morocco for resistance to Puccinia coronata . Plant Dis. 97, 1544–1548. [DOI] [PubMed] [Google Scholar]
- Thaxter, R. (1890) Report of Mycologist, Fourteenth Annual Report. New Haven, CT: Connecticut Agricultural Experiment Station.
- Toruño, T.Y. , Stergiopoulos, I. and Coaker, G. (2016) Plant–pathogen effectors: cellular probes interfering with plant defenses in spatial and temporal manners. Annu. Rev. Phytopathol. 54, 419–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tozzetti, G. (1952) True nature, causes and sad effects of the rust, the bunt, the smut, and other maladies of wheat, and of oats in the field (L. R. Tehon, transl.). In: Phytopathol, Classic, No. 9. American Phytopathological Society, Washington, DC (originally published, 1767).
- Upadhyaya, Y.M. and Baker, E.P. (1960) Studies on the mode of inheritance of Hajira type stem rust resistance and Victoria type crown rust resistance as exhibited in crosses involving the oat variety Garry. Proc. Linn. Soc. New South Wales, 85, 157–179. [Google Scholar]
- Urban, Z. and Markova, J. (1994) The rust fungi of grasses in Europe. 1. Puccinia coronata Corda. Acta Univ. Carolinae Biol. 37, 93–147. [Google Scholar]
- USDA‐ARS CDL (2014) Oat Loss to Rust USDA‐ARS Cereal Disease Laboratory. Available at: https://www.ars.usda.gov/ARSUserFiles/50620500/Smallgrainlossesduetorust/2014loss/2014oatloss.pdf [accessed 1 April 2017].
- USDA‐ARS CDL (2016) Oat Crown Rust Resistance Genes USDA‐ARS Cereal Disease Laboratory. Available at: http://www.ars.usda.gov/Main/docs.htm?docid=10342 [accessed 18 March 2017].
- USDA‐ARS CDL (2017) Oat Crown Rust USDA‐ARS Cereal Disease Laboratory. Available at: https://www.ars.usda.gov/midwest-area/st-paul-mn/cereal-disease-lab/docs/cereal-rusts/oat-crown-rust/ [accessed 1 April 2017].
- US Department of Agriculture, Foreign Agricultural Service (USDA‐FAS) (2017) World Agricultural Production Washington, DC: USDA Foreign Agricultural Service. Available at: https://www.fas.usda.gov/data/world-agricultural-production [accessed 10 March 2017].
- US Department of Agriculture, Natural Resources Conservation Service (USDA‐NRCS) (2017) Rhamnus catharthica L., Common Buckthorn Greensboro, NC: USDA National Resources Conservation Service, National Plant Data Team. Available at: https://plants.usda.gov/core/profile?symbol=RHCA3 [accessed 7 July 2017].
- Vieira, E.A. , Carvalho, F.I.F. , Chaves, M.S. , de Oliveira, A.C. , Benin, G. , Hartwig, I. , Silva, J.A.G. , Bertan, I. , Martins, A.F. and Martins, L.F. (2007) Virulence variability of Puccinia coronata f. sp. avenae isolates collected in three counties from Rio Grande do Sul State, Brazil. Plant Dis. 91, 66–70. [DOI] [PubMed] [Google Scholar]
- Wahl, I. (1970) Prevalence and geographic distribution of resistance to crown rust in Avena sterilis . Phytopathology, 60, 746–749. [Google Scholar]
- Wahl, I. and Schreiter, S. (1953) A highly virulent physiological race of crown rust on oats in Israel. Bull. Isr. Res. Counc. 3, 256–257. [Google Scholar]
- Wang, Y. , Cheng, X. , Shan, Q. , Zhang, Y. , Liu, J. , Gao, C. and Qiu, J.L. (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. [DOI] [PubMed] [Google Scholar]
- Warner, G. and French, D. (1970) Dissemination of fungi by migratory birds: survival and recovery of fungi from birds. Can. J. Bot. 48, 907–910. [Google Scholar]
- Waterhouse, P.M. (1952) Australian rust studies. IX. Physiologic rust determinations and surveys of cereal rusts. Proc. Linn. Soc. New South Wales. 77, 209–258. [Google Scholar]
- Wulff, B.B.H. and Moscou, M.J. (2014) Strategies for transferring resistance into wheat: from wide crosses to GM cassettes. Front. Plant Sci. 5, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J. , Wang, M. , Chen, X. and Kang, Z. (2016) Role of alternate hosts in epidemiology and pathogen variation of cereal rusts. Annu. Rev. Phytopathol. 54, 207–228. [DOI] [PubMed] [Google Scholar]
- Zhu, S. and Kaeppler, H. (2003) Identification of quantitative trait loci for crown rust in oat line MAM17‐5. Crop Sci. 43, 358–366. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Oat resistance genes against Puccinia coronata f. sp. avenae.


