Summary
Phytoplasmas are insect‐transmitted phytopathogenic bacteria, which secrete effector proteins that are often responsible for altering the plant morphology and behaviours of their vectors. Phytoplasma multifunctional effector proteins TENGU and SAP11 induce typical witches’ broom symptoms, but their mode of action remains unknown. Previously, we have identified a SAP11‐like effector from wheat blue dwarf phytoplasma, SWP1, which induces witches’ broom symptoms in Nicotiana benthamiana. In this study, we observed that SWP1‐expressing transgenic Arabidopsis thaliana plants showed typical witches’ broom symptoms. On overexpression of SWP1 truncation mutants in N. benthamiana, we identified that the coiled‐coil domain and nuclear localization were responsible for the induction of witches’ broom symptoms. In addition, using yeast two‐hybrid and bimolecular fluorescence complementation assays, we demonstrated that SWP1 interacts with A. thaliana transcription factor TCP18 (BRC1), the key negative regulator of branching signals in various plant species. Moreover, in planta co‐expression analysis showed that SWP1 promotes the degradation of BRC1 via a proteasome system. These findings suggest that the phytoplasma effector SWP1 induces witches’ broom symptoms through targeting of BRC1 and promoting its degradation.
Keywords: BRC1, coiled‐coil domain, proteasome TCP18, protein degradation, wheat blue dwarf
Introduction
Phytoplasmas [class Mollicutes, genus ‘Candidatus (Ca.) Phytoplasma’] are cell wall‐lacking bacterial pathogens that cause enormous losses in hundreds of plant species, including food crops, vegetables, fruit trees, shade trees and lumber (Hogenhout et al., 2008; Strauss, 2009). Plants infected with these bacteria exhibit an array of symptoms, including phyllody (the production of leaf‐like structures in place of floral parts), witches’ broom (the proliferation of axillary shoots) and virescence (the loss of normal flower pigments and the development of green flowers) (Lee et al., 2000). Phytoplasmas are restricted to the phloem sieve cells of host plants and are transmitted between plants by their sap‐sucking insect vectors, such as leafhoppers, plant hoppers and psyllids (Sugio et al., 2011b). Because efficient means to culture phytoplasmas outside their hosts are lacking, studies on phytoplasmas, particularly the molecular mechanisms of pathogenicity, are very laborious and difficult (Strauss, 2009). To date, the complete genomic sequences of only six phytoplasmas have been determined (Andersen et al., 2013; Bai et al., 2006; Kube et al., 2008; Orlovskis et al., 2017; Oshima et al., 2004; Tran‐Nguyen et al., 2008). Although several secreted proteins have been predicted as candidate effectors that are directly delivered into the cytoplasm of host cells through a Sec‐dependent protein translocation pathway (Bai et al., 2009; Sugio et al., 2011b), only three of these proteins have been studied in detail.
Unlike many effectors secreted by other plant‐associated microbes that directly target specific processes of the plant immune system, these three effectors primarily function in modulating plant development. TENGU, a small protein (38 residues) identified from onion yellows phytoplasma strain M (OY‐M; Ca. Phialophora asteris), and its homologues from several phytoplasma strains in the aster yellows group (group 16SrI) dramatically induce dwarfism and witches’ broom symptoms when overexpressed in Nicotiana benthamiana and/or Arabidopsis thaliana plants (Hoshi et al., 2009; Sugawara et al., 2013). In addition, TENGU induces the sterility of A. thaliana (Minato et al., 2014). SAP11, secreted by aster yellows phytoplasma strain witches’ broom (AY‐WB; Ca. P. asteris), causes a series of morphological changes in plants, including smaller rosettes, severely crinkled leaves, crinkled siliques and witches’ broom (Sugio et al., 2011a). Another AY‐WB effector, SAP54, transforms flowers of A. thaliana into leaf‐like vegetative tissues (Maclean et al., 2011). Notably, TENGU and SAP11 are responsible for the typical witches’ broom phenotype of phytoplasmas. TENGU inhibits an auxin‐related pathway, thereby leading to witches’ broom symptoms (Hoshi et al., 2009). However, the plant targets of TENGU have not been described to date. SAP11 binds and destabilizes Arabidopsis CIN (CINCINNATA)‐TCP (TEOSINTE‐BRANCHED, CYCLOIDEA, PROLIFERATION FACTOR 1 AND 2) transcription factors, resulting in a leaf morphogenesis change. However, no visible witches’ broom symptoms have been observed in CIN‐TCP‐silenced lines of Arabidopsis (Efroni et al., 2008; Sugio et al., 2011a). Thus, the mechanism of witches’ broom symptoms caused by SAP11 has not been well described to date. Therefore, direct molecular evidence for the mechanism of witches’ broom symptoms induced by phytoplasma effectors remains to be discovered.
The process that leads to axillary bud growth to produce a branch or to remain dormant in the axils of leaves is highly plastic and is frequently regulated by endogenous and environmental stimuli (Aguilar‐Martinez et al., 2007). Two models have been proposed to describe this process (Domagalska and Leyser, 2011; Ongaro and Leyser, 2008). One is the auxin transport canalization‐based model in which the growth of the lather branches is dependent on the establishment of auxin export from the axillary buds, and this auxin export is negatively regulated by strictly basipetal transport of auxin in the primary stem. The other model is the second messenger model in which strigolactones and cytokinins are two potential candidates regulated by auxin to translocate directly into axillary buds to modulate bud activity (Domagalska and Leyser, 2011; Dun et al., 2012). In Arabidopsis, BRC1 and its paralogue BRC2 are considered to be key integrators of these different pathways that negatively control shoot branching within axillary buds (Aguilar‐Martinez et al., 2007; Rameau et al., 2014). BRC1 and BRC2 belong to the TB1/CYC subclass of TCP transcription factors, which are plant specific and have profound effects on plant developmental processes (Nicolas and Cubas, 2016). In Arabidopsis, the TCP family comprises 24 members, which are classified into two classes (class I and class II), according to the alignment of the TCP domain. Class II is further divided into TB1/CYC and CIN subclasses based on additional diagnostic residues (Nicolas and Cubas, 2016).
SAP11CaPM, the homologue of SAP11 secreted by apple proliferation phytoplasma (AP; Ca. P. mali), targets and/or destabilizes at least three CIN‐TCP proteins (Janik et al., 2017; Tan et al., 2016). Nevertheless, an initial yeast two‐hybrid (Y2H) screen indicated interaction between SAP11CaPM and TCP18‐like (BRC1) of Malus × domestica, although this was not confirmed by further analyses (Janik et al., 2017). Notably, SAP11CaPM‐transgenic N. benthamiana plants did not exhibit a clear witches’ broom phenotype (Tan et al., 2016). It is possible that SAP11CaPM does not target and destabilize BRC1. SAP11 interacts with and destabilizes members of the CIN‐TCP subfamily. However, whether SAP11 targets TB1/CYC‐TCP proteins remains unknown (Sugio et al., 2011a). The increase in stem number caused by SAP11‐like proteins is most probably the result of the destabilization of BRC1 and BRC2. Different interactor ranges of SAP11 homologues may reflect their functional diversity. Thus, whether witches’ broom‐inducing proteins (SAP11‐like) interact with and destabilize TB1/CYC subclass proteins, particularly BRC1, must be determined.
Wheat blue dwarf (WBD) phytoplasma (group 16SrI) causes dwarfism, witches’ broom symptoms and sterility, leading to severe yield losses in wheat production in northwestern China (Chen et al., 2014). WBD is transmitted by Psammotettix striatus to diverse plant species, including members of the families Gramineae, Brassicaceae and Solanaceae (Gu et al., 2007). Previously, we have reported that SWP1 (SAP11‐like) induces typical witches’ broom symptoms when transiently expressed in N. benthamiana (Wang et al., 2018). In this study, we found that SWP1‐transgenic Arabidopsis also exhibits the typical proliferation phenotype. On overexpression of SWP1 deletion mutants in N. benthamiana, we identified that the coiled‐coil (CC) domain and nuclear localization are responsible for the witches’ broom induction of SWP1. In addition, we demonstrated that SWP1 interacts with BRC1 and promotes the degradation of BRC1 through a proteasome system. Finally, we discuss how the phytoplasma effector induces witches’ broom symptoms.
Results
Overexpression of SWP1 induces witches’ broom symptoms in Arabidopsis
Previously, we have found that transient expression of SWP1 induces a witches’ broom phenotype in N. benthamiana (Wang et al., 2018). To further analyse the effect of SWP1 on plant development, transgenic Arabidopsis lines expressing the mature SWP1 protein (without signal peptide) were generated. Three T3 homozygous lines (lines 8, 13 and 15) were selected for further study. Four weeks after flowering, these SWP1‐transgenic plants exhibited a significantly larger number of primary rosette (RI) branches than observed in wild‐type Col‐0 plants (Fig. 1a,b). Nevertheless, no apparent differences were observed with regard to the size of the rosettes, the shapes of the leaves, siliques and flowers, and the length of stems between SWP1‐transgenic and WT plants (Fig. S1, see Supporting Information). Reverse transcription‐polymerase chain reaction (RT‐PCR) assays showed that SWP1 was expressed (Fig. 1c). In addition, SWP1‐transgenic plants showed less jasmonic acid (JA) production, but no significant difference in the expression of LIPOXYGENASE2 (LOX2), compared with Col‐0 plants (Fig. S2, see Supporting Information). These results indicate that SWP1 is responsible for witches’ broom symptoms, but not for the dwarfism and sterility phenotypes.
Figure 1.
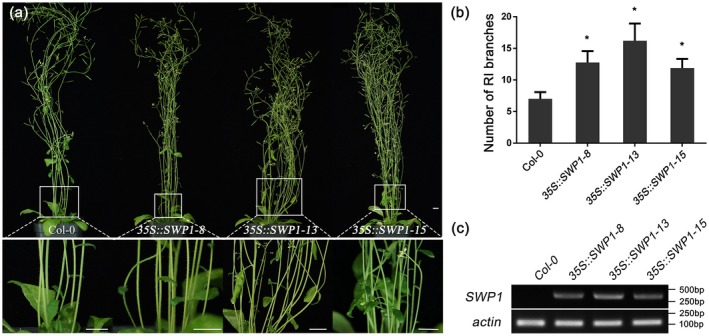
Overexpression of SWP1 induces witches’ broom symptoms in Arabidopsis. (a) Top: phenotypes of T3 homozygous 35S::SWP1 lines 8, 13 and 15. Bottom: close‐up of shoot phenotypes from the same plants. The plants were photographed at 4 weeks after flowering. Bars, 1 cm. (b) Number of primary rosette (RI) branches of the plants shown in (a). Bars represent the mean ± standard deviation of at least 40 plants. Asterisks indicate significant differences compared with Col‐0 [P < 0.05; one‐way analysis of variance (ANOVA)]. (c) The expression of SWP1 in transgenic plants was examined by semi‐reverse transcription‐polymerase chain reaction (semi‐RT‐PCR). The actin gene was used as internal reference. [Colour figure can be viewed at wileyonlinelibrary.com]
The CC domain and nuclear localization are required for the induction of witches’ broom symptoms
The three identified phytoplasma virulence effectors TENGU, SAP54 and SAP11 all contain a predicted CC structure, which is required for the targeting of host proteins, at least for SAP11 (Sugio et al., 2014). In addition, the nuclear localization signal (NLS) of SAP11 is required for the destabilization of CIN‐TCPs and for the induction of crinkled leaves in Arabidopsis (Sugio et al., 2014). SWP1 was also predicted to possess a CC domain and a C‐terminal monopartite‐type NLS (Fig. 2a) (Chen et al., 2014). To identify the functions of these domains of SWP1, a series of deletion mutants were transiently expressed in N. benthamiana using a viral vector‐mediated expression system. As shown in Figs 2c and S3a (see Supporting Information), N. benthamiana plants expressing SWP1ΔNLSCC and SWP1ΔCC did not show apparent morphological changes, similar to the plants expressing green fluorescent protein (GFP). However, deletion of the C‐terminal‐predicted NLS did not affect the witches’ broom‐inducing activity of SWP1 (Fig. 2c). The expression of SWP1 and its mutants was confirmed by western blot analysis with an antibody against SWP1, except for SWP1ΔNLSCC, which was too small to detect (Fig. S3b). Then, to test whether SWP1 accumulates in the plant nucleus, fusion proteins of GFP with mutants of SWP1 were transiently expressed in N. benthamiana leaves and subsequently examined to determine the subcellular localization. Unexpectedly, GFP‐SWP1 and GFP‐SWP1ΔNLS were observed in both nucleus and cytoplasm, similar to GFP (Fig. 2d). Thus, the predicted C‐terminal NLS of SWP1 was not apparently associated with the location of SWP1. To further determine the location of SWP1 function, an NLS of SV40 Large T‐antigen (NLSSV40) (Kalderon et al., 1984), a nuclear export signal (NES) of HIV‐Rev (Fischer et al., 1995) and a non‐functional NES (NESKO) (Sugio et al., 2014) were fused to the C‐terminus of SWP1ΔNLS. As expected, GFP‐SWP1‐NLS and GFP‐SWP1‐NES were located in the nucleus and cytoplasm, respectively (Fig. 2d). However, the witches’ broom‐inducing function was abolished when SWP1 was excluded from the plant cell nucleus by NES (Figs 2c,d and S3b). SWP1 and derivatives were expressed in each leaf sample used for microscopy (Fig. S3c). Collectively, these results indicate that the CC domain and nuclear localization are required for the function of SWP1 in inducing witches’ broom symptoms.
Figure 2.
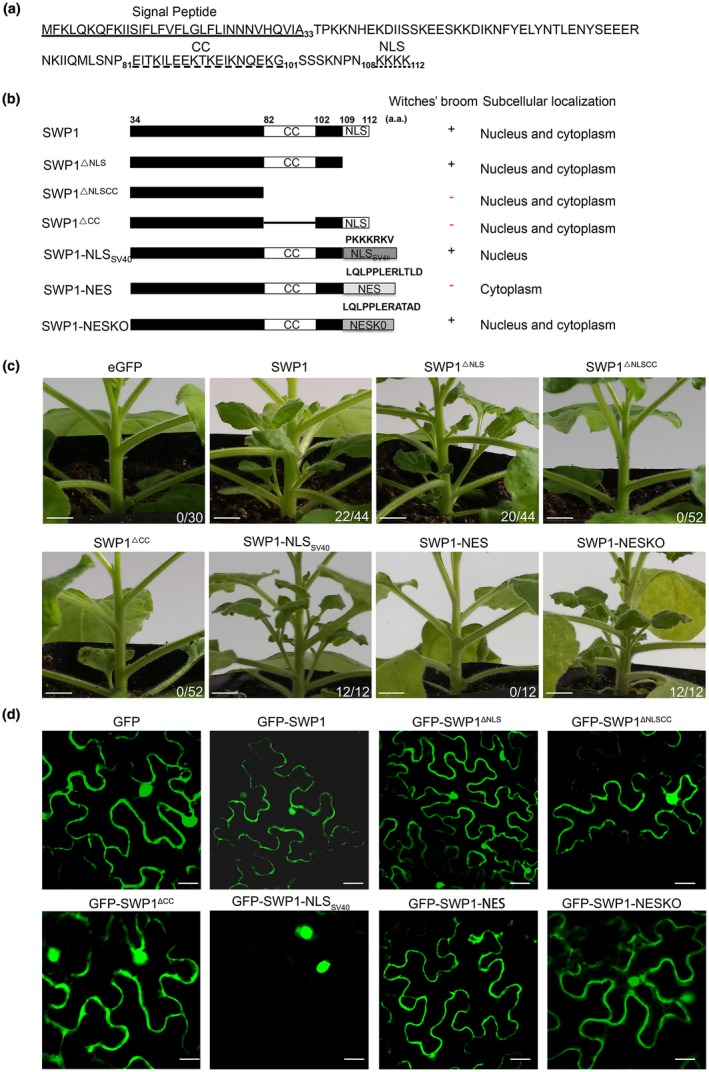
Coiled‐coil (CC) domain and nuclear localization of SWP1 are required for the induction of witches’ broom symptoms. (a) The amino acid (aa) sequence of SWP1. The signal peptide of SWP1 is underlined with a single line. The predicted CC domain and nuclear localization signal (NLS) are marked with a broken underline and a dotted underline, respectively. (b) Schematic diagram, phenotype and subcellular localization summary of SWP1 mutants. SWP1 was fused with a nuclear export signal (NES), a defective NES (NESKO) or the nuclear localization sequence of SV40 Large T‐antigen (NLSSV40). The presence and absence of witches’ broom symptoms are indicated by + and –, respectively. (c) Six‐week‐old Nicotiana benthamiana plants expressing SWP1 mutants. SWP1 and green fluorescent protein (GFP) were used as controls. Photographs were taken at 2 weeks after agro‐infiltration. Fractions indicate the number of plants exhibiting witches’ broom symptoms and the total number of plants tested. Bars, 0.5 cm. (d) Subcellular localization of N‐terminal GFP‐tagged SWP1 mutants in N. benthamiana leaves. Photographs were taken by an LSM 510 META confocal microscopy system with a wavelength of 488 nm. Bars, 75 μm. [Colour figure can be viewed at wileyonlinelibrary.com]
SWP1 targets shoot branching integrators BRC1 and BRC2
SAP11 and its homologue SAP11CaPM interact with different members of the CIN‐TCP family (Janik et al., 2017; Sugio et al., 2011a; Tan et al., 2016). To determine whether SWP1 targets TCPs, particularly CYC/TB1‐TCPs, we screened all members of the class II TCP family in Arabidopsis by Y2H assays. Eleven class II TCPs were fused to the transcription activation domain (AD) of the yeast transcriptional regulatory protein GAL4, and the mature SWP1 sequence was fused to the DNA‐binding domain (BD). Yeast cultures with BD‐SWP1 and AD‐TCP1, AD‐TCP2, AD‐TCP12 (BRC2), AD‐TCP18 (BRC1) or AD‐TCP24 were grown on SD/‐Trp‐Leu‐His‐Ade containing 40 μg/mL X‐α‐Gal (‐LWHA+X‐α‐Gal) to select for HIS3, ADE2, and MEL1 expression. Because of the potential autoactivation activity of TCPs (when expressed in the BD construct), we did not test the interaction by reciprocal arrangement. Notably, the interaction of SWP1 with BRC1 and BRC2 was further confirmed by repeating the Y2H assay (Fig. 3a). Moreover, we further determined this interaction in planta by bimolecular fluorescence complementation (BiFC) assays. A clear fluorescence signal was observed in the cell nucleus when NYFP (N‐terminal yellow fluorescent protein fragment)‐SWP1 was co‐expressed with CYFP (C‐terminal yellow fluorescent protein fragment)‐BRC1 and CYFP‐BRC2 in N. benthamiana leaves, but these signals were not detected in control samples (NYFP and CYFP were used as negative controls) (Fig. 3b). Furthermore, co‐expression of GFP‐SWP1 with mCherry‐BRC1 and mCherry‐BRC2 in N. benthamiana leaves indicated that SWP1 co‐localized with BRC1 and BRC2 in the plant cell nucleus (Fig. S4, see Supporting Information), which is consistent with the results of BiFC. Therefore, these results indicate that SWP1 interacts with the shoot branching integrators BRC1 and BRC2.
Figure 3.
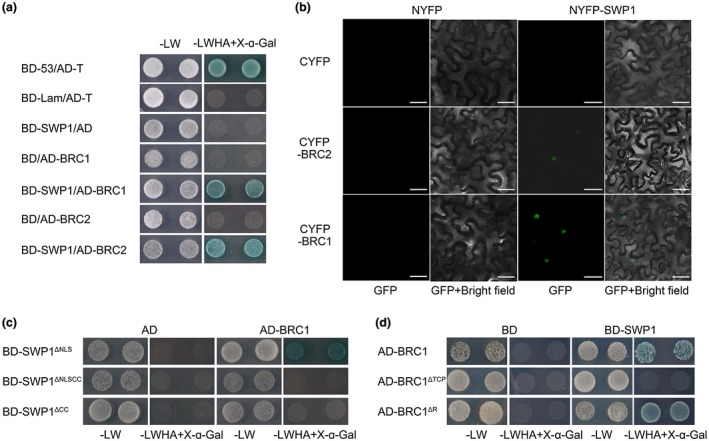
SWP1 targets the shoot branching integrators BRC1 and BRC2. (a) Yeast two‐hybrid (Y2H) assay was used to examine the interaction of BD‐SWP1 with AD‐BRC1 and AD‐BRC2. Blue yeast colonies on SD/‐Trp‐Leu‐His‐Ade medium containing 40 μg/mL X‐α‐Gal (‐LWHA+X‐α‐Gal) indicate positive interaction between the two proteins. BD‐53/AD‐T, positive control; BD‐Lam/AD‐T, negative control; BD‐SWP1/AD, BD/AD‐BRC1 and BD/AD‐BRC2, vector controls. (b) Bimolecular fluorescence complementation (BiFC) assays confirm the interaction of SWP1 with BRC1 and BRC2 in Nicotiana benthamiana. Fluorescence signals were observed at 60 h after agro‐infiltration using a confocal laser scanning microscope. The plasmid combinations NYFP/CYFP, NYFP‐SWP1/CYFP, NYFP/CYFP‐BRC1 and NYFP/CYFP‐BRC2 were used as negative controls. Bars, 30 μm. (c, d) Y2H assays of SWP1 mutants (c) and BRC1 mutants (d). [Colour figure can be viewed at wileyonlinelibrary.com]
In Arabidopsis, BRC1 is considered to be a key integrator of different pathways to negatively control shoot branching, whereas its paralogue BRC2 may play a minor and partly redundant role in this process (Aguilar‐Martinez et al., 2007; Rameau et al., 2014). Thus, BRC1 was chosen for further studies. The CC domain is required for witches’ broom symptom induction of SWP1; moreover, the CC domain is required for SAP11 interaction with its target TCP13 (Sugio et al., 2014). To determine the possible role of the SWP1 CC domain in the interaction between SWP1 and BRC1, we fused SWP1ΔNLS, SWP1ΔNLSCC and SWP1ΔCC to BD, followed by co‐expression with AD‐BRC1 in yeast cells (AD was used as a negative control). We observed that BD‐SWP1ΔNLSCC and BD‐SWP1ΔCC did not interact with BRC1 (Fig. 3c). Thus, similar to SAP11, the CC domain of SWP1 is required for its interaction with the target.
The TCP domain is shared in the TCP family, and is a 59‐amino‐acid, non‐canonical, basic helix‐loop‐helix motif that allows DNA binding and protein–protein interaction (Nicolas and Cubas, 2016). In addition, an 18–20‐residue arginine‐rich motif (the R domain) is present in five members of class II proteins (Martin‐Trillo and Cubas, 2010). Notably, these five proteins were all targeted by SWP1 (Figs 3a and S5, see Supporting Information). To determine whether the TCP domain and R domain function within the BRC1 interaction with SWP1, we co‐expressed the AD‐fused BRC1 deletion mutants (Fig. S6, see Supporting Information) with BD‐SWP1 in yeast cells. Y2H assay showed that the TCP domain is required for interaction with SWP1, but the R domain may be dispensable (Fig. 3d).
SWP1 targets BRC1 homologues from various plant species
WBD infects many monocot plants, such as wheat (Triticum aestivum, family Gramineae) and wild oat (Avena fatua L., family Gramineae), in the field and dicot plants, such as Madagascar periwinkle (Catharanthus roseus, family Apocynaceae) and tomato (Solanum lycopersicum, family Solanaceae), under laboratory conditions (Gu et al., 2007). Most host plants, such as wheat, wild oat and Madagascar periwinkle, exhibit a typical witches’ broom phenotype. However, a few host plants do not, such as tomato. To test whether this phenomenon is associated with the interaction of SWP1 and BRC1s, we cloned BRC1‐like genes of a monocotyledonous model plant Brachypodium distachyon (BdTB1_like, XP_003559513; BdTCP12_like, XP_014756189), Madagascar periwinkle (CrBRC1‐like, cra_locus_10370_iso_1_len_1494_ver_3) and tomato (SlBRC1b, NP_001234572.2) into pGADT7 and examined their interaction with SWP1 by Y2H assays. Yeast cells co‐expressing SWP1 and BRC1s of Brachypodium distachyon and Madagascar periwinkle grew on high‐stringency selection medium (‐LWHA+X‐α‐Gal), whereas those co‐expressing SWP1 and BRC1b from tomato did not (Fig. 4). Therefore, these findings suggest that witches’ broom symptoms caused by WBD phytoplasma most probably depend on the interaction of SWP1 with BRC1.
Figure 4.
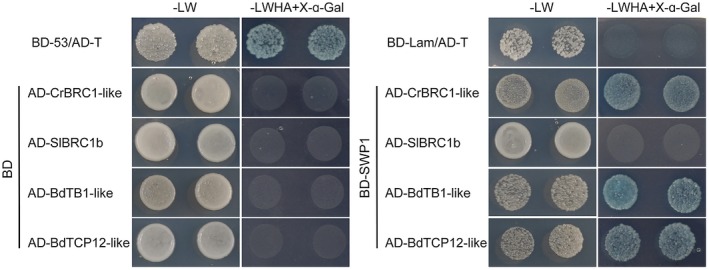
SWP1 targets BRC1 homologues of various plant species. Yeast two‐hybrid (Y2H) assays were used to examine the interaction between SWP1 and homologues of BRC1. The experimental details are described in Fig. 3a. [Colour figure can be viewed at wileyonlinelibrary.com]
SWP1 destabilizes BRC1 through a proteasome system
The shoot architecture of plants expressing SWP1 resembled the phenotype of the Arabidopsis brc1 mutant. Thus, we speculated that SWP1 most probably induced witches’ broom symptoms by destabilizing BRC1. To test this hypothesis, we co‐expressed BRC1 with GFP‐SWP1, GFP‐SWP1ΔCC and a negative control GFP in N. benthamiana leaves. Western blot analyses revealed that the protein abundance of BRC1 co‐expressed with GFP‐SWP1 was remarkably lower than that co‐expressed with GFP and GFP‐SWP1ΔCC (Fig. 5a, left blots). However, when mCherry was co‐expressed with these GFP fusions, the protein level of mCherry was not affected (Fig. 5a, right blots). These findings confirm that SWP1 and SAP11 promote the degradation of BRC1 in vivo. Furthermore, when BRC1 was co‐expressed with GFP‐SAP11, the protein abundance of BRC1 was decreased (Fig. S7, see Supporting Information). In contrast, the protein abundance of BRC1 was not decreased by GFP‐SAP11CaPM (Fig. S7). These results confirm that SAP11 may promote the degradation of A. thaliana BRC1, but SAP11CaPM does not.
Figure 5.
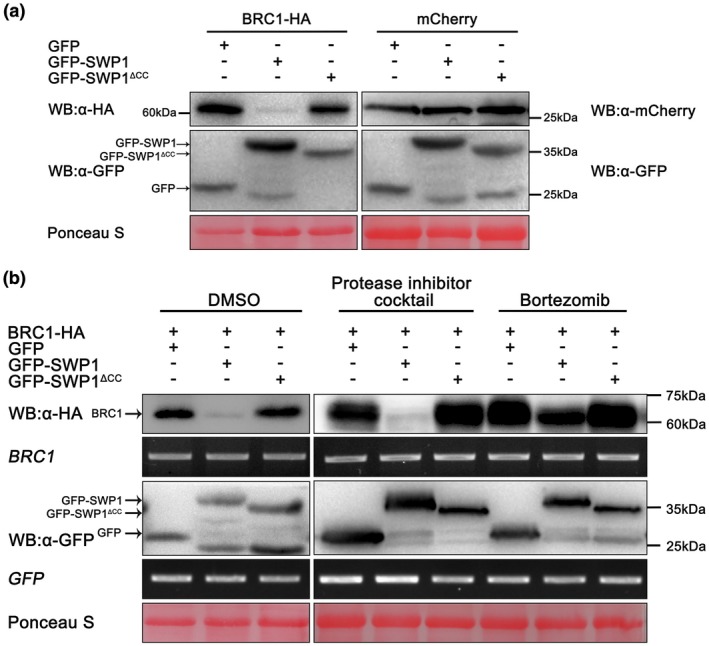
SWP1 destabilizes BRC1 through a proteasome system. (a) Green fluorescent protein (GFP), GFP‐SWP1 and GFP‐ SWP1ΔCC were co‐expressed with BRC1‐HA or mCherry in leaves of Nicotiana benthamiana. Samples were collected at 60 h after agro‐infiltration and subjected to western blot (WB). (b) Infiltrated tissues in (a, left panel) were treated with 50 μm bortezomib [dissolved in 5% dimethylsulfoxide (DMSO)], 1% protease inhibitor cocktail (dissolved in 5% DMSO) or 5% DMSO 4 h before harvest. The protein abundance and transcript level of the samples were examined by western blot and reverse transcription‐polymerase chain reaction (RT‐PCR), respectively. Similar results were observed in at least three independent repeats. Membranes were stained with Ponceau S to confirm equal loading. [Colour figure can be viewed at wileyonlinelibrary.com]
Proteasomes and proteases are the two major classes of cytoplasmic proteolysis system in eukaryotes. To determine which of these two systems was responsible for the degradation of BRC1, we treated N. benthamiana leaves co‐expressing BRC1 and GFP‐SWP1 with the proteasome inhibitor bortezomib (PS‐341) and protease inhibitor cocktail. Western blot analyses showed that PS‐341 treatment greatly inhibited the degradation of BRC1, whereas the protease inhibitor cocktail and dimethylsulfoxide (DMSO) treatments had no apparent effect on the degradation (Fig. 5b). RT‐PCR assay showed that the transcript levels of these genes were not obviously different (Fig. 5b). To further confirm whether the accumulation of BRC1 was mediated by a proteasome, two other proteasome inhibitors, epoxomicin and MG132, were also examined. The degradation of BRC1 was greatly inhibited by epoxomicin, whereas MG132 had no apparent effect (Fig. S8, see Supporting Information), possibly because of the rapid reversibility of action and the lower potency of combination, selectivity and metabolic stability of MG132 than of PS341 and epoxomicin (Kisselev and Goldberg, 2001). Collectively, these results suggest that SWP1 promotes the degradation of BRC1 through the proteasome system.
Discussion
As biotrophic pathogens, phytoplasmas establish a long‐term feeding relationship with living hosts to obtain nutrients. To achieve this relation, dozens of candidate effectors are predicted to be delivered into host cells (Bai et al., 2009), with some demonstrated to suppress plant immune responses, regulate plant development, increase insect vector reproduction and promote colonization (Hoshi et al., 2009; Lu et al., 2014; MacLean et al., 2014; Maejima et al., 2014; Minato et al., 2014; Sugio et al., 2011a; Tan et al., 2016; Wang et al., 2018). Witches’ broom symptoms, characterized by the production of many axillary shoots, are induced by most phytoplasmas, making witches’ broom one of the typical symptoms for the discovery of new phytoplasma candidates. Thus, this common symptom may play a vital role in the pathogenesis of phytoplasmas. Witches’ broom leads to more vegetative plant tissues, which increases the phloem network for phytoplasma replication and feeds more insect vectors. To date, two virulence effectors (TENGU and SAP11) and their homologues are known to induce this symptom. However, their mode of action remains largely unknown. In this study, we found that a SAP11‐like effector of WBD phytoplasma (SWP1) induced witches’ broom by direct interaction with and promotion of degradation of the plant key negative regulator of branching signals (BRC1), thereby elucidating the molecular mechanism of the induction of witches’ broom symptoms by a phytoplasma effector (Fig. 6).
Figure 6.
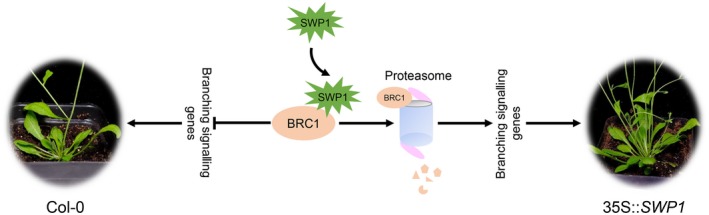
Scheme of SWP1 function in inducing witches’ broom symptoms by mediating the degradation of BRC1 through a proteasome system. [Colour figure can be viewed at wileyonlinelibrary.com]
TENGU was the first reported witches’ broom‐inducing effector. The amino acid sequence and symptom‐inducing function are highly conserved amongst homologues (Hoshi et al., 2009; Sugawara et al., 2013). However, TENGU is apparently an aster yellows (16SrI) group‐specific effector (Wang et al., 2014). Although the protein targets of TENGU have not been identified to date, microarray analysis of tengu‐transgenic Arabidopsis suggests that the effector suppresses the auxin signalling and biosynthesis pathways (Hoshi et al., 2009). Auxin is produced at the shoot apex and transported basipetally down the stem in the polar auxin transport stream to inhibit shoot branching (Ongaro and Leyser, 2008). Because auxin does not enter the axillary bud, this inhibition is indirect and systemic. However, BRC1 and its orthologues are well‐described, bud‐specific regulators and play a direct regulatory role in axillary bud activation (Domagalska and Leyser, 2011). Thus, the mechanisms of TENGU‐ and SWP1‐mediated shoot branching are most probably different, which is also consistent with their distinct intracellular localization. TENGU is localized in the cytoplasm (Hoshi et al., 2009), whereas SWP1 primarily functions in the nucleus. Only three phytoplasma effectors have been identified, and two induce witches’ broom with different mechanisms, suggesting the importance of witches’ broom for phytoplasmas.
Unlike TENGU, the second witches’ broom‐inducing effector SAP11 has been identified in diverse groups of phytoplasma; in addition, homologues show diversity in subcellular location, protein targets and symptom phenotypes (Janik et al., 2017; Sugio et al., 2011a, 2014; Tan et al., 2016; Wang et al., 2014). For example, SAP11 only localizes in plant nuclei (Bai et al., 2009), but SWP1 is localized in both the nuclei and cytoplasm (Fig. 2d). In addition, some phenotypes of SAP11‐transgenic Arabidopsis plants, such as smaller rosettes, severely crinkled leaves and crinkled siliques, are not observed in SWP1‐transgenic plants (Fig. S1) (Sugio et al., 2011a). However, SAP11CaPM‐transgenic N. benthamiana lines exhibit crinkled leaves and dwarf phenotypes, but no witches’ broom (Tan et al., 2016). Moreover, SWP1 and SAP11 promote the degradation of A. thaliana BRC1, but SAP11CaPM does not (Fig. S7). Furthermore, the expression of LOX2 and JA synthesis are down‐regulated in SAP11‐transgenic plants (Sugio et al., 2011a), whereas SWP1‐transgenic plants show less JA production, but no significant reduction in LOX2 expression, when compared with that of Col‐0 plants (Fig. S2). Different phenotypes induced by SAP11 homologues most probably result from their different plant targets. SAP11 targets all class II CIN‐TCPs, but TB1/CYC‐TCPs were not tested (Sugio et al., 2011a). SAP11CaPM interacts with at least three CIN‐TCPs, but not with TCP18‐like (BRC1) of apple (Janik et al., 2017), which possibly explains why SAP11CaPM does not induce witches’ broom. By contrast, SWP1 targets two CIN‐TCPs and all three TB1/CYC‐TCPs, including BRC1. A possible domain responsible for the diversity of protein targets is the CC domain, which is the least conserved region and is required for binding of both SAP11 and SWP1 to TCPs (Fig. 3c) (Sugio et al., 2014). However, how SAP11‐like proteins discriminate between different TCPs requires further study.
Shoot branching or tillering patterns are a fundamental component of plant architecture, which determines many aspects of plant morphology and adaptation to resource availability (Gonzalez‐Grandio et al., 2017). Excessive production of axillary branches will compete for limited resources and has a negative effect on plant growth (Dong et al., 2017). However, shoot branching is a favourable trait for crop breeding. For example, the modern maize (Zea mays ssp. mays), which has strong apical dominance with one primary axis of growth and produces only two or three shorter lateral branches on the main stem, was domesticated from its wild ancestor teosinte (Z. mays ssp. parviglumis), which is highly branched with long axillary branches (Doebley et al., 1995). This striking evolutionary change was largely affected by a natural variation, which led to the increased expression of the teosinte branched 1 (tb1) allele (homologous gene of BRC1) in maize and the subsequent inhibition of the growth of axillary buds (Burnham and Yagyu, 1961; Clark et al., 2006; Doebley et al., 1997). The maize tb1 mutant produces more tillers, leading to a ‘bushy’ profile, similar to its progenitor (teosinte) (Doebley et al., 1995; Hubbard et al., 2002). Thus, TB1 and BRC1 are further confirmed as major contributors to repress axillary bud growth.
Post‐translational modification is a tool employed by cells to regulate protein activity (Pogány et al., 2015). All three phytoplasma effectors characterized to date are associated with post‐translational proteolytic processing. SAP54 destabilizes plant MADS‐domain transcription factors in a ubiquitin/26S proteasome‐dependent manner (MacLean et al., 2014). Likewise, SAP11 destabilizes plant TCP transcription factors via an unknown pathway (Sugio et al., 2011a, 2014). In addition, TENGU and SAP11 themselves undergo proteolytic processing to generate small peptides (Lu et al., 2014; Sugawara et al., 2013). SWP1 promotes the degradation of BRC1 and undergoes post‐translational cleavage (Fig. S3b). Thus, post‐translational proteolysis of virulence effectors and/or their targets is apparently a common mechanism of phytoplasmas, although whether TENGU can also promote the degradation of its targets warrants further investigation. Notably, both SAP11 and SWP1 destabilize TCPs, but most probably via different pathways, and how SWP1 mediates BRC1 degradation requires further study.
Experimental Procedures
Plasmid construction
Plasmids of pBin‐NYFP, pBin‐CYFP, pBin61, pBin‐GFP and pBin‐P19 (Sun et al., 2013a, 2013b) were a kind gift from Professor Liying Sun (Northwest A&F University, Shaanxi, China). pGR107‐SWP1 and pGR107‐GFP have been described previously (Wang et al., 2018).
All constructs were generated using a ClonExpress II One‐Step Cloning Kit (Vazyme, Nanjing, Jiangsu, China) according to the manufacturer’s instructions. Primers used for plasmid construction contained the gene‐specific primer and at least 15 bp of a vector sequence available on each side of the cloning site. For the transgene assay, SWP1 was cloned into a binary plant expression vector pBI121 with a Cauliflower mosaic virus (CaMV) 35S promoter to generate the pBI121‐SWP1 construct (35S::SWP1). For viral vector‐mediated transient expression, a series of mutants of SWP1, including SWP1ΔNLS, SWP1ΔNLSΔCC, SWP1ΔCC, SWP1‐NES, SWP1‐NESKO and SWP1‐NLSSV40, were amplified using PrimeSTAR® Max DNA Polymerase (TAKARA, Beijing, China). These amplicons were then cloned into the Potato virus X vector pGR107. For the subcellular localization assay, SWP1 and its mutants were cloned to the C‐terminus of GFP into the plant expression vector pBin‐GFP; TCP12 and TCP18 were fused to the C‐terminus of mCherry into pBin‐mCherry. For Y2H, we cloned SWP1 and its mutants into pGBKT7 (Clontech, Mountain View, CA, USA). The coding sequences of class II TCP genes were amplified from the cDNA of A. thaliana. These amplicons were then ligated into pGADT7 (Clontech) as above. For BiFC, SWP1 was cloned into pBin‐NYFP, and TCP genes were cloned into pBin61‐CYFP. For co‐expression, TCP12 and TCP18 were amplified with a haemagglutinin (HA) tag at the C‐terminus and ligated into pBin61. All constructs were sequenced to verify the sequences of the inserts and are listed in Table S1 (see Supporting Information). Primer sequences are listed in Table S2 (see Supporting Information).
Transient expression in N. benthamiana
Nicotiana benthamiana plants were grown in a growth room at 23 °C and 60% humidity with a 12‐h/12‐h light/dark photoperiod. Agrobacterium tumefaciens strain GV3101 was used for transformation of the respective constructs. Transformants were cultured in Luria broth medium supplemented with appropriate antibiotics and 10 mm MES (pH 5.6) at 28 °C overnight. Cultures were then harvested and resuspended in infiltration buffer [10 mm MgCl2, 10 mm MES (pH 5.6) and 200 mm acetosyringone] to a final optical density at 600 nm (OD600) of 0.5. The resuspended cultures were then incubated at room temperature for 3 h and infiltrated into leaves of 4–6‐week‐old N benthamiana plants using a syringe without a needle.
For viral vector‐mediated transient expression, cultures were infiltrated into two leaves of 4‐week‐old N. benthamiana plants. Each strain was assayed in four to nine replicated plants. Systemic symptoms were scored and photographed at 14 days post‐inoculation. For subcellular localization, A. tumefaciens cells carrying GFP fusions or mCherry fusions were infiltrated into 6‐week‐old plants as described above, but the leaf samples were collected at 60 h after infiltration for confocal imaging. For BiFC assays, three Agrobacterium cultures carrying pBin‐NYFP fusions, pBin‐cYFP fusions and pBin‐P19 were mixed in a 1 : 5 : 1 ratio to a final OD600 = 1; samples were collected 60 h after infiltration for confocal imaging. For co‐expression assays, three Agrobacterium cultures carrying GFP fusions, BRC1‐HA and pBin‐P19 were mixed in a 1 : 1 : 1 ratio to OD600 = 0.3 for each. The proteasome inhibitors bortezomib/PS‐341 (50 μm) (MCE, Monmouth Junction, NJ, USA), MG132 (50 μm) (MCE) and epoxomicin (50 μm) (MCE), the protease inhibitor cocktail (1%) (Sigma‐Aldrich, St Louis, MO, USA) and 0.5% DMSO (a negative control) were infiltrated into the relevant area 56 h after agro‐infiltration; samples were then harvested after 4 h.
Western blot
Proteins were extracted using RIPA Lysis Buffer [50 mm Tris‐HCl, pH 7.4, 150 mm NaCl, 1% Triton X‐100, 1% sodium deoxycholate and 0.1% sodium dodecylsulfate (SDS)] with 1% protease inhibitor cocktail (Sigma‐Aldrich). After centrifugation at 15 294 g for 15 min, samples were mixed with 5 × sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) Sample Loading Buffer (Beyotime, Shanghai, China), boiled for 5 min and separated using 12%–15% SDS‐PAGE according to the protein size. Gels were electroblotted onto 0.22‐μm polyvinylidene difluoride (PVDF) membranes at 200 mA for 1–2 h. Membranes were blocked in 1 × TBST with 5% non‐fat dry milk [TBST buffer is TBS buffer (see below) with 0.05% Tween‐20] for 2 h at room temperature. Then, membranes were incubated in the primary antibodies at a 1 : 2000 dilution for 2 h at room temperature and washed three times for 10 min each with 1 × TBST. After incubating with an appropriate secondary antibody at a dilution of 1 : 2000 for 2 h at room temperature, membranes were washed as above, but with a final wash in TBS (150 mm NaCl, 20 mm Tris‐HCl, pH 7.4). Proteins were detected using an eECL Western Blot Kit (CWBIO, Beijing, China) according to the manufacturer’s instructions, with a SuperSignal ChemiDoc XRS+ system (Bio‐Rad, Hercules, CA, USA).
RT‐PCR
Total RNA was extracted following the protocol of a Biospin Plant Total RNA Extraction Kit (DNA‐free) (Bioeer, Beijing, China). First‐strand cDNA was synthesized from 1 μg of total RNA using a PrimeScript™ RT Reagent Kit with gDNA Eraser (Perfect Real Time) (TAKARA) according to the user manual. The primers used for RT‐PCR are listed in Table S2.
Y2H screen
Y2H assays were conducted by co‐transformation of Saccharomyces cerevisiae strain AH109 with two appropriate constructs using the GAL4 system (Clontech). The co‐transformed yeast cells were plated onto synthetic dropout minus tryptophan and leucine [SD/‐Trp‐Leu (‐LW)] plates for the assay of the presence of both plasmids. Interactions were determined by dropping 2 μL of yeast suspension (OD600 = 0.3) onto SD/‐Trp‐Leu‐His‐Ade plates containing 40 μg/mL X‐α‐Gal (Clontech) (‐LWHA+X‐α‐Gal).
Confocal microscopy
Leaves of N. benthamiana were harvested at 60 h after agro‐infiltration and examined under a Zeiss LSM 510 META confocal laser scanning microscope (Zeiss, Promenade, Jena, Germany). GFP, YFP and mCherry fluorescence were observed at excitation wavelengths of 488, 488 and 594 nm, respectively.
Generation of transgenic Arabidopsis lines
35S::SWP1 was introduced into Col‐0 by the floral dip method as described previously (Zhang et al., 2006) with A. tumefaciens strain GV3101. To select putative transgenic plants, sterilized T0 seeds were plated on Murashige and Skoog salt (MS) (QDRS BIOTEC, Qingdao, Shangdong, China) plates with 50 μg/mL kanamycin. The expression of SWP1 was confirmed by RT‐PCR. Seeds of the T1 generation were plated on the same selection plates. Single insertion lines were selected depending on a 3 : 1 ratio of resistant plants. Then, seeds (≥50, T2 generation) of these lines were collected and plated on the selected plates. The lines with 100% resistance were T3 homozygous plants with the single insertion, and these homozygous T3 lines were used for further studies.
Phenotypic analysis
Arabidopsis seeds, including the selected T3 homozygous SWP1‐transgenic lines and wild‐type Col‐0, were cold‐treated (4 °C) for 3 days and then sown on commercial soil in a growth room at 23 °C/20 °C in 60% humidity with a 14‐h/10‐h light/dark photoperiod. When plants had grown for 4 weeks after the main inflorescence became visible, branches (shoots longer than 0.5 cm) were counted and heights were measured from at least 40 individuals of each line.
Supporting information
Fig. S1 The 35S::SWP1 lines showed no apparent differences in the size of rosettes, shape of leaves, siliques and flowers, and length of main stems. (a) 26‐d‐old Arabidopsis plants of wild‐type (Col‐0) and three 35S::SWP1 lines. Bar = 1 cm. (b) Inflorescence stems of 8‐wk‐old Col‐0 and three 35S::SWP1 lines. Bars = 0.8 cm. (c) Length of main stems of plants in (b). Columns show the mean and standard errors (bars) calculated from 8 plants per line.
Fig. S2 SWP1 transgenic plants produce less JA. (a) RNA levels of the LOX2 gene in 35S::SWP1 and Col‐0 plants. RT‐qPCR results are normalized with the U‐box housekeeping gene AT5G15400 transcript. RNA was prepared from 4‐wk‐old Arabidopsis plants. Error bars represent the SEs of three replicates. Similar results were obtained from three independent biological experiments. (b) 35S::SWP1 transgenic plants produce less JA. Columns show mean of HPLC results of 4‐wk‐old 35S::SWP1 and Col‐0 plants from three biological replicates. (Scale bars = SE.) *P < 0.01 compared with Col‐0 (one‐way ANOVA).
Fig. S3 Western blot analyses of SWP1 and SWP1 mutants. (a) Six‐wk‐old N. benthamiana plants expressing SWP1 mutants. SWP1 and GFP were used as controls. Photographs were taken at two wk after agro‐infiltration. Bars = 1.5 cm. (b) Western blot with α‐SWP1 antibody showed the presence of SWP1 and SWP1 mutants in Figure 2c. Samples were collected 7 d after agro‐infiltration. *Indicates the objective bands. (c) Expression of GFP‐SWP1 and GFP‐SWP1 mutants in Figure 2d were confirmed by western blot using α‐GFP antibody. Ponceau S‐stained RuBisCo was used as the loading control.
Fig. S4 SWP1 co‐localized with BRC1 and BRC2 at the cell nucleus in N. benthamiana. Fluorescence signals were visualized at 60 h after agro‐infiltration by confocal microscopy. The boxed areas are shown at higher magnification. Bars = 20 μm.
Fig. S5 Yeast two‐hybrid analyses of the interaction between SWP1 and members of the class II TCPs. Experimental details are described in Figure 3a. The experiment was repeated three times with the same results.
Fig. S6 Schematic diagram of Arabidopsis BRC1 and its truncated versions. TCP, TCP domain; R, R domain.
Fig. S7 SWP1 homolog, SAP11, destabilizes Arabidopsis BRC1. BRC1‐HA was co‐expressed with GFP‐SAP11,GFP‐SAP11CaPM, or GFP in leaves of N. benthamiana. Samples were collected at 60 h after agro‐infiltration and subjected to western blot (WB). Membranes were stained with Ponceau S to confirm equal loading.
Fig. S8 Destabilization of BRC1 mediated by SWP1 was inhibited by proteasome inhibitors epoxomicin and MG132. N. benthamiana leaves co‐expressing BRC1 with GFP‐SWP1 or GFP were treated with 5% DMSO, 50 μM proteasome inhibitors epoxomicin, or 50 μM MG132 4 h before harvest, as indicated. The protein abundance was examined by western blot. Membranes were stained with Ponceau S to confirm equal loading. Similar results were observed in three independent experiments.
Table S1 List of plasmids used in this study.
Table S2 List of primers used in this study.
Acknowledgements
We thank Dr Jiajun Nie (Northwest A&F University) for editing the manuscript and Mr Ruofei Xu (Northwest A&F University) for assistance with gene transformation. The National Natural Science Foundation of China (31371913, 31570144) and the 111 Project from the Education Ministry of China (B07049) provided financial support for this work.
References
- Aguilar‐Martinez, J.A. , Poza‐Carrion, C. and Cubas, P. (2007) Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell, 19, 458–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, M.T. , Liefting, L.W. , Havukkala, I. and Beever, R.E. (2013) Comparison of the complete genome sequence of two closely related isolates of 'Candidatus Phytoplasma australiense' reveals genome plasticity. BMC Genomics, 14, 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, X. , Correa, V.R. , Toruño, T.Y. , Ammar, E. , Kamoun, S. and Hogenhout, S.A. (2009) AY‐WB phytoplasma secretes a protein that targets plant cell nuclei. Mol. Plant–Microbe Interact. 22, 18–30. [DOI] [PubMed] [Google Scholar]
- Bai, X. , Zhang, J. , Ewing, A. , Miller, S.A. , Jancso Radek, A. , Shevchenko, D.V. , Tsukerman, K. , Walunas, T. , Lapidus, A. , Campbel, J.W. and Hogenhout, S.A. (2006) Living with genome instability: the adaptation of phytoplasmas to diverse environments of their insect and plant hosts. J. Bacteriol. 188, 3682–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham, C.R. and Yagyu, P. (1961) Linkage relations of teosinte branched. Maize Genet. Coop. News Lett. 35, 87. [Google Scholar]
- Chen, W. , Li, Y. , Wang, Q. , Wang, N. and Wu, Y. (2014) Comparative genome analysis of wheat blue dwarf phytoplasma, an obligate pathogen that causes wheat blue dwarf disease in China. PLoS One, 9, e96436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, R.M. , Wagler, T.N. , Quijada, P. and Doebley, J. (2006) A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nat. Genet. 38, 594–597. [DOI] [PubMed] [Google Scholar]
- Doebley, J. , Stec, A. and Gustus, C. (1995) Teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics, 141, 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J. , Stec, A. and Hubbard, L. (1997) The evolution of apical dominance in maize. Nature, 386, 485. [DOI] [PubMed] [Google Scholar]
- Domagalska, M.A. and Leyser, O. (2011) Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell. Biol. 12, 211–221. [DOI] [PubMed] [Google Scholar]
- Dong, Z. , Li, W. , Unger‐Wallace, E. , Yang, J. , Vollbrecht, E. and Chuck, G. (2017) Ideal crop plant architecture is mediated by tassels replace upper ears1, a BTB/POZ ankyrin repeat gene directly targeted by TEOSINTE BRANCHED1. Proc. Natl. Acad. Sci. USA, 114, E8656–E8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun, E.A. , de Saint Germain, A. , Rameau, C. and Beveridge, C.A. (2012) Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiol. 158, 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni, I. , Blum, E. , Goldshmidt, A. and Eshed, Y. (2008) A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell, 20, 2293–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, U. , Huber, J. , Boelens, W.C. , Mattajt, L.W. and Lührmann, R. (1995) The HIV‐1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell, 82, 475–483. [DOI] [PubMed] [Google Scholar]
- Gonzalez‐Grandio, E. , Pajoro, A. , Franco‐Zorrilla, J.M. , Tarancon, C. , Immink, R.G. and Cubas, P. (2017) Abscisic acid signaling is controlled by a BRANCHED1/HD‐ZIP I cascade in Arabidopsis axillary buds. Proc. Natl. Acad. Sci. USA, 114, E245–E254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, P. , Wu, Y. and An, F. (2007) Host range testing and RFLP analysis of wheat blue dwarf phytoplasma. Acta Phytopathol. Sin. 37, 390–397. [Google Scholar]
- Hogenhout, S.A. , Oshima, K. , el Ammar, D. , Kakizawa, S. , Kingdom, H.N. and Namba, S. (2008) Phytoplasmas: bacteria that manipulate plants and insects. Mol. Plant. Pathol. 9, 403–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshi, A. , Oshima, K. , Kakizawa, S. , Ishii, Y. , Ozeki, J. , Hashimoto, M. , Komatsu, K. , Kagiwada, S. , Yamaji, Y. and Namba, S. (2009) A unique virulence factor for proliferation and dwarfism in plants identified from a phytopathogenic bacterium. Proc. Natl. Acad. Sci. USA, 106, 6416–6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard, L. , Mcsteen, P. , Doebley, J. and Hake, S. (2002) Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics, 162, 1927–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janik, K. , Mithofer, A. , Raffeiner, M. , Stellmach, H. , Hause, B. and Schlink, K. (2017) An effector of apple proliferation phytoplasma targets TCP transcription factors—a generalized virulence strategy of phytoplasma? Mol. Plant. Pathol. 18, 435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon, D. , Roberts, B.L. , Richardson, W.D. and Smith, A.E. (1984) A short amino acid sequence able to specify nuclear location. Cell, 39, 499–509. [DOI] [PubMed] [Google Scholar]
- Kisselev, A.F. and Goldberg, A.L. (2001) Proteasome inhibitors: from research tools to drug candidates. Chem. Biol. 8, 739. [DOI] [PubMed] [Google Scholar]
- Kube, M. , Schneider, B. , Kuhl, H. , Dandekar, T. , Heitmann, K. , Migdoll, A.M. , Reinhardt, R. and Seemüller, E. (2008) The linear chromosome of the plant‐pathogenic mycoplasma ‘Candidatus Phytoplasma mali’. BMC Genomics, 9, 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, I. , Davis, R.E. and Gundersen‐Rindal, D.E. (2000) Phytoplasma: phytopathogenic mollicutes. Annu. Rev. Microbiol. 54, 221–255. [DOI] [PubMed] [Google Scholar]
- Lu, Y. , Cheng, K. , Jiang, S. and Yang, J. (2014) Post‐translational cleavage and self‐interaction of the phytoplasma effector SAP11. Plant Signal. Behav. 9, e28991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean, A.M. , Sugio, A. , Makarova, O.V. , Findlay, K.C. , Grieve, V.M. , Tóth, R. , Nicolaisen, M. and Hogenhout, S.A. (2011) Phytoplasma effector sap54 induces indeterminate leaf‐like flower development in Arabidopsis plants. Plant Physiol. 157, 831–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean, A.M. , Orlovskis, Z. , Kowitwanich, K. , Zdziarska, A.M. , Angenent, G.C. , Immink, R.G. and Hogenhout, S.A. (2014) Phytoplasma effector SAP54 hijacks plant reproduction by degrading MADS‐box proteins and promotes insect colonization in a RAD23‐dependent manner. PLoS Biol. 12, e1001835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maejima, K. , Iwai, R. , Himeno, M. , Komatsu, K. , Kitazawa, Y. , Fujita, N. , Ishikawa, K. , Fukuoka, M. , Minato, N. , Yamaji, Y. , Oshima, K. and Namba, S. (2014) Recognition of floral homeotic MADS domain transcription factors by a phytoplasmal effector, phyllogen, induces phyllody. Plant J. 78, 541–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin‐Trillo, M. and Cubas, P. (2010) TCP genes: a family snapshot ten years later. Trends Plant. Sci. 15, 31–39. [DOI] [PubMed] [Google Scholar]
- Minato, N. , Himeno, M. , Hoshi, A. , Maejima, K. , Komatsu, K. , Takebayashi, Y. , Kasahara, H. , Yusa, A. , Yamaji, Y. , Oshima, K. , Kamiya, Y. and Namba, S. (2014) The phytoplasmal virulence factor TENGU causes plant sterility by downregulating of the jasmonic acid and auxin pathways. Sci. Rep. 4, 7399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas, M. and Cubas, P. (2016) TCP factors: new kids on the signaling block. Curr. Opin. Plant Biol. 33, 33–41. [DOI] [PubMed] [Google Scholar]
- Ongaro, V. and Leyser, O. (2008) Hormonal control of shoot branching. J. Exp. Bot. 59, 67–74. [DOI] [PubMed] [Google Scholar]
- Orlovskis, Z. , Canale, M.C. , Haryono, M. , Lopes, J.R.S. , Kuo, C.H. and Hogenhout, S.A. (2017) A few sequence polymorphisms among isolates of Maize bushy stunt phytoplasma associate with organ proliferation symptoms of infected maize plants. Ann. Bot. 119, 869–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima, K. , Kakizawa, S. , Nishigawa, H. , Jung, H.Y. , Wei, W. , Suzuki, S. , Arashida, R. , Nakata, D. , Miyata, S. , Ugaki, M. and Namba, S. (2004) Reductive evolution suggested from the complete genome sequence of a plant‐pathogenic phytoplasma. Nat. Genet. 36, 27–29. [DOI] [PubMed] [Google Scholar]
- Pogány, M. , Dankó, T. , Kámán‐Tóth, E. , Schwarczinger, I. and Bozsó, Z. (2015) Regulatory proteolysis in Arabidopsis–pathogen interactions. Int. J. Mol. Sci. 16, 23 177–23 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rameau, C. , Bertheloot, J. , Leduc, N. , Andrieu, B. , Foucher, F. and Sakr, S. (2014) Multiple pathways regulate shoot branching. Front. Plant Sci. 5, 741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss, E. (2009) Phytoplasma research begins to bloom. Science, 325, 388–390. [DOI] [PubMed] [Google Scholar]
- Sugawara, K. , Honma, Y. , Komatsu, K. , Himeno, M. , Oshima, K. and Namba, S. (2013) The alteration of plant morphology by small peptides released from the proteolytic processing of the bacterial peptide TENGU. Plant Physiol. 162, 2005–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio, A. , Kingdom, H.N. , MacLean, A.M. , Grieve, V.M. and Hogenhout, S.A. (2011a) Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc. Natl. Acad. Sci. USA, 108, E1254–E1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio, A. , MacLean, A.M. and Hogenhout, S.A. (2014) The small phytoplasma virulence effector SAP11 contains distinct domains required for nuclear targeting and CIN‐TCP binding and destabilization. New Phytol. 202, 838–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio, A. , MacLean, A.M. , Kingdom, H.N. , Grieve, V.M. , Manimekalai, R. and Hogenhout, S.A. (2011b) Diverse targets of phytoplasma effectors: from plant development to defense against insects. Annu. Rev. Phytopathol. 49, 175–195. [DOI] [PubMed] [Google Scholar]
- Sun, L. , Andika, I.B. , Kondo, H. and Chen, J. (2013a) Identification of the amino acid residues and domains in the cysteine‐rich protein of Chinese wheat mosaic virus that are important for RNA silencing suppression and subcellular localization. Mol. Plant Pathol. 14, 265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L. , Jing, B. , Andika, I.B. , Hu, Y. , Sun, B. , Xiang, R. , Kondo, H. and Chen, J. (2013b) Nucleo‐cytoplasmic shuttling of VPg encoded by Wheat yellow mosaic virus requires association with the coat protein. J. Gen. Virol. 94, 2790–2802. [DOI] [PubMed] [Google Scholar]
- Tan, C.M. , Li, C.H. , Tsao, N.W. , Su, L.W. , Lu, Y.T. , Chang, S.H. , Lin, Y.Y. , Liou, J.C. , Hsieh, L.C. , Yu, J.Z. , Sheue, C.R. , Wang, S.Y. , Lee, C.F. and Yang, J.Y. (2016) Phytoplasma SAP11 alters 3‐isobutyl‐2‐methoxypyrazine biosynthesis in Nicotiana benthamiana by suppressing NbOMT1. J. Exp. Bot. 67, 4415–4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran‐Nguyen, L.T. , Kube, M. , Schneider, B. , Reinhardt, R. and Gibb, K.S. (2008) Comparative genome analysis of ‘Candidatus Phytoplasma australiense’ (subgroup tuf‐Australia I; rp‐A) and ‘Ca. Phytoplasma asteris’ strains OY‐M and AY‐WB. J. Bacteriol. 190, 3979–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, N. , Li, Y. , Chen, W. , Yang, H. , Zhang, P. and Wu, Y. (2018) Identification of wheat blue dwarf phytoplasma effectors targeting plant proliferation and defence responses. Plant Pathol. 67, 603–609. [Google Scholar]
- Wang, Q. , Guo, Y. , Wang, N. , Li, Y. , Chen, W. , Chen, W. and Wu, Y. (2014) Identification of a conserved core genome with group‐specific genes from comparative genomics of ten different Candidatus Phytoplasma strains. J. Phytopathol. 162, 650–659. [Google Scholar]
- Zhang, X. , Henriques, R. , Lin, S.S. , Niu, Q.W. and Chua, N.H. (2006) Agrobacterium‐mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 The 35S::SWP1 lines showed no apparent differences in the size of rosettes, shape of leaves, siliques and flowers, and length of main stems. (a) 26‐d‐old Arabidopsis plants of wild‐type (Col‐0) and three 35S::SWP1 lines. Bar = 1 cm. (b) Inflorescence stems of 8‐wk‐old Col‐0 and three 35S::SWP1 lines. Bars = 0.8 cm. (c) Length of main stems of plants in (b). Columns show the mean and standard errors (bars) calculated from 8 plants per line.
Fig. S2 SWP1 transgenic plants produce less JA. (a) RNA levels of the LOX2 gene in 35S::SWP1 and Col‐0 plants. RT‐qPCR results are normalized with the U‐box housekeeping gene AT5G15400 transcript. RNA was prepared from 4‐wk‐old Arabidopsis plants. Error bars represent the SEs of three replicates. Similar results were obtained from three independent biological experiments. (b) 35S::SWP1 transgenic plants produce less JA. Columns show mean of HPLC results of 4‐wk‐old 35S::SWP1 and Col‐0 plants from three biological replicates. (Scale bars = SE.) *P < 0.01 compared with Col‐0 (one‐way ANOVA).
Fig. S3 Western blot analyses of SWP1 and SWP1 mutants. (a) Six‐wk‐old N. benthamiana plants expressing SWP1 mutants. SWP1 and GFP were used as controls. Photographs were taken at two wk after agro‐infiltration. Bars = 1.5 cm. (b) Western blot with α‐SWP1 antibody showed the presence of SWP1 and SWP1 mutants in Figure 2c. Samples were collected 7 d after agro‐infiltration. *Indicates the objective bands. (c) Expression of GFP‐SWP1 and GFP‐SWP1 mutants in Figure 2d were confirmed by western blot using α‐GFP antibody. Ponceau S‐stained RuBisCo was used as the loading control.
Fig. S4 SWP1 co‐localized with BRC1 and BRC2 at the cell nucleus in N. benthamiana. Fluorescence signals were visualized at 60 h after agro‐infiltration by confocal microscopy. The boxed areas are shown at higher magnification. Bars = 20 μm.
Fig. S5 Yeast two‐hybrid analyses of the interaction between SWP1 and members of the class II TCPs. Experimental details are described in Figure 3a. The experiment was repeated three times with the same results.
Fig. S6 Schematic diagram of Arabidopsis BRC1 and its truncated versions. TCP, TCP domain; R, R domain.
Fig. S7 SWP1 homolog, SAP11, destabilizes Arabidopsis BRC1. BRC1‐HA was co‐expressed with GFP‐SAP11,GFP‐SAP11CaPM, or GFP in leaves of N. benthamiana. Samples were collected at 60 h after agro‐infiltration and subjected to western blot (WB). Membranes were stained with Ponceau S to confirm equal loading.
Fig. S8 Destabilization of BRC1 mediated by SWP1 was inhibited by proteasome inhibitors epoxomicin and MG132. N. benthamiana leaves co‐expressing BRC1 with GFP‐SWP1 or GFP were treated with 5% DMSO, 50 μM proteasome inhibitors epoxomicin, or 50 μM MG132 4 h before harvest, as indicated. The protein abundance was examined by western blot. Membranes were stained with Ponceau S to confirm equal loading. Similar results were observed in three independent experiments.
Table S1 List of plasmids used in this study.
Table S2 List of primers used in this study.


