Summary
Plant viruses often harm their hosts, which have developed mechanisms to prevent or minimize the effects of virus infection. Resistance and tolerance are the two main plant defences to pathogens. Although resistance to plant viruses has been studied extensively, tolerance has received much less attention. Theory predicts that tolerance to low‐virulent parasites would be achieved through resource reallocation from growth to reproduction, whereas tolerance to high‐virulent parasites would be attained through shortening of the pre‐reproductive period. We have shown previously that the tolerance of Arabidopsis thaliana to Cucumber mosaic virus (CMV), a relatively low‐virulent virus in this host, accords to these predictions. However, whether other viruses trigger the same response, and how A. thaliana copes with highly virulent virus infections remains unexplored. To address these questions, we challenged six A. thaliana wild genotypes with five viruses with different genomic structures, life histories and transmission modes. In these plants, we quantified virus multiplication, virulence, and the effects of infection on plant growth and reproduction, and on the developmental schedule. Our results indicate that virus multiplication varies according to the virus × host genotype interaction. Conversely, effective tolerance is observed only on CMV infection, and is associated with resource reallocation from growth to reproduction. Tolerance to the other viruses is observed only in specific host–virus combinations and, at odds with theoretical predictions, is linked to longer pre‐reproductive periods. These findings only partially agree with theoretical predictions, and contribute to a better understanding of pathogenic processes in plant–virus interactions.
Keywords: Arabidopsis thaliana, life history traits, resistance, resource reallocation, tolerance
Introduction
Viruses are often virulent parasites of plants. As such, viruses may cause important losses in crops, reduce the fitness of wild plants and therefore modulate ecosystem composition and dynamics (Anderson et al., 2004; Fraile and García‐Arenal, 2016). In the context of host–parasite co‐evolution, virulence is defined as the negative impact of parasite infection on host fitness (Little et al., 2010; Read, 1994). As virulence is the result of pathology of the host, it is determined by both host and parasite traits (Little et al., 2010); because it reduces host fitness, hosts have developed various defence strategies to avoid pathogen infection, reduce pathogen multiplication or reduce its virulence (Agnew et al., 2000). Two major defences of plants against viruses are resistance, which decreases within‐host virus multiplication and results in lower virus loads, and tolerance, which specifically decreases virulence regardless of the level of virus multiplication (Little et al., 2010; Råberg, 2014). The mechanisms of plant resistance to viruses and the role of resistance in plant–virus interactions have been analysed extensively (Csorba et al., 2009; de Ronde et al., 2014; Truniger and Aranda, 2009), whereas the study of plant tolerance to viruses has received comparatively much less attention (Jeger et al., 2006).
Our group has analysed the genetic variation of virulence and plant defence in the interaction between the model plant Arabidopsis thaliana L. Heynh. (Brassicaceae) and Cucumber mosaic virus (CMV, Cucumovirus, Bromoviridae). The infection of 21 wild genotypes of A. thaliana (from here on, Arabidopsis), representing the genetic variation of the species in Eurasia, with three CMV strains, showed that virus load was dependent on the host genotype × virus strain interaction, and that quantitative resistance to CMV was a host trait with moderate to high broad‐sense heritability (Pagán et al., 2007). Our previous work also indicated that virulence, estimated as the effect of CMV infection on viable seed production (progeny production), did not correlate with virus load as a result of host genotype × virus strain‐specific tolerance, and again tolerance was a host trait with moderate to high heritability (Pagán et al., 2007). Interestingly, tolerance was positively correlated with the length of post‐embryonic development (life span, LP) of the Arabidopsis genotypes (Pagán et al., 2007). This relationship was explained by the differential effect of CMV infection on host life history, resulting in differential resource allocation between life history traits: although all host genotypes delayed flowering on infection, only long life cycle genotypes modified resource allocation from vegetative growth to reproduction, thus decreasing the effects of infection on progeny production, i.e. attaining tolerance (Pagán et al., 2008).
There is ample evidence that major‐gene qualitative and quantitative resistance of plants to viruses depends on the plant genotype and is virus specific (Csorba et al., 2009; de Ronde et al., 2014; Pagán et al., 2016; Truniger and Aranda, 2009). This evidence may lead to the hypothesis that Arabidopsis genotypes will differ in their resistance to different viruses. However, life history theory predicts that parasitized hosts may modify optimal resource allocation, by increasing the reproductive effort and/or altering temporal life history schedules by reducing the pre‐reproductive period, to maximize fitness (Forbes, 1993; Minchella, 1985; Perrin et al., 1996). These general responses would be modulated according to the parasite's virulence (Gandon et al., 2002; Hochberg et al., 1992). Given that our previous results agree with such predictions, it could be hypothesized that infection by viruses other than CMV will also result in an increase in the reproductive effort of Arabidopsis, long life cycle genotypes being more tolerant to different viruses than short life cycle genotypes.
To test these hypotheses, we challenged six Arabidopsis genotypes with five viruses. The Arabidopsis genotypes had a short or long life cycle, and had been shown to be non‐tolerant or tolerant, respectively, to different strains of CMV (Pagán et al., 2007, 2008). The five viruses were CMV, Turnip crinkle virus (TCV, Carmovirus, Tombusviridae), Turnip mosaic virus (TuMV, Potyvirus, Potyviridae), Youcai mosaic virus (YoMV, Tobamovirus, Virgaviridae) and Cauliflower mosaic virus (CaMV, Caulimovirus, Caulimoviridae). These five virus species belong to different genera and families, so that they exemplify different genome expression strategies and, presumably, different modes of molecular interactions with the host plant. In addition, these five virus species exemplify different life histories, which may determine their ecology and evolution, as well as the patterns of host defence evolution. CMV is a typical generalist, with the broadest host range for a plant virus, as it infects more than 1200 species in more than 100 families of monocotyledonous and dicotyledonous plants. TuMV has a moderate host range limited to a few dicotyledonous families. TCV, YoMV and CaMV have natural host ranges restricted only to the Brassicaceae. CMV and TuMV are transmitted by aphids in a non‐persistent manner. CaMV is aphid transmitted in a bimodal manner, both non‐persistently and semi‐persistently. TCV is transmitted by chrysomelid beetles and YoMV by plant‐to‐plant contact. Last, only CMV is efficiently seed transmitted in different species of Brassicaceae, including A. thaliana (Hily et al., 2014; Pagán et al., 2014). The biology of these viruses has been analysed extensively (for reviews, see Carrington et al., 1987, 1989; Haas et al., 2002; Hollings and Stone, 1972; Jacquemond, 2012; Walsh and Jenner 2002). More significant, four of these virus species, CMV, TuMV, TCV and CaMV, have been found to infect field populations of Arabidopsis, with incidences of up to 70% according to year and host population site (Pagán et al., 2010), indicating that their interaction with Arabidopsis is significant in nature, and could have resulted in the evolution of host defences. YoMV has also been demonstrated to infect several wild species in experimental conditions (Cai et al., 2009), including Arabidopsis (Aguilar et al., 1996), and has also been detected in wild plant populations (Park et al., 2016).
Results show that resistance is a general response of Arabidopsis to virus infection displayed in a genotype‐specific manner. However, effective tolerance attained through modification of plant life history traits has a much narrower spectrum, as it is observed only in response to one of the five viruses tested. These observations may help to clarify the role of resistance and tolerance in plant–virus co‐evolution.
Results
To test the hypotheses that the defence responses of Arabidopsis to virus infection are genotype specific and are associated with the length of the life cycle, three Arabidopsis genotypes with a long life cycle and three with a short life cycle were challenged with five viruses. The experiment involved eight replicated plants per treatment (genotype × virus or mock‐inoculated; see Experimental Procedures for details) in a fully randomized design.
Infection symptoms in Arabidopsis genotypes
The six assayed Arabidopsis wild genotypes were systemically infected with the five assayed virus species; no immune or hypersensitive resistance responses were observed. The type and severity of the symptoms were dependent on the specific interaction between virus species and host genotype, and varied from asymptomatic (e.g. CMV in Cum‐0) to leaf distortion and extreme reduction of rosette growth, with no inflorescence production (e.g. TuMV in Cum‐0), with different degrees of leaf mottle and chlorosis, and growth reduction, in between these two extremes. Across host genotypes, CMV induced the mildest symptoms, whereas TCV and, in particular, TuMV induced the most severe symptoms (Fig. 1). The effect of infection on plant development varied largely according to the virus–host genotype interaction, sometimes resulting in plant sterilization: although all CMV‐, CaMV‐ and YoMV‐infected plants flowered and produced seed (except two of eight YoMV‐infected Kas‐0 plants, which flowered but did not produce seed), all TCV‐infected plants flowered, but only 78.3% produced seed, and 61.8% of TuMV‐infected plants flowered, but only 17.6% produced seed. The effect of TCV and TuMV infection on flower and seed production was more severe in the host genotypes with a long life cycle. Indeed, TuMV infection resulted in flowering in 100% of plants with a short life cycle genotype, with 31.6% producing seed (all of the Col‐1 genotype), whereas 13.3% of plants with a long life cycle genotype flowered and none produced seed (χ 2 = 3.78, P = 0.051). Also, 87.5% of TCV‐infected plants with a short life cycle genotype produced seed vs. 68.2% of those with a long life cycle, although this difference was not significant (χ 2 = 2.78, P = 0.095).
Figure 1.

Symptoms of virus infection in Arabidopsis. Symptoms at 15 days post‐inoculation (dpi) for Col‐1 (A–F) and Kas‐0 (G–L). Plants were mock‐inoculated (A, G) or infected with Cucumber mosaic virus (CMV) (B, H), Turnip crinkle virus (TCV) (C, I), Turnip mosaic virus (TuMV) (D, J), Youcai mosaic virus (YoMV) (E, K) or Cauliflower mosaic virus (CaMV) (F, L). Symptoms were as follows: mild growth reduction (B, H); chlorosis, severe stunting and leaf lamina reduction (C, D, I, J); or chlorosis, moderate stunting and leaf lamina reduction (E, F, K, L). All scale bars represent 1 cm.
Virus multiplication in Arabidopsis genotypes
For all assayed virus species, multiplication was estimated by quantitative real‐time reverse transcription‐polymerase chain reaction (RT‐qPCR) quantification of viral RNA, or qPCR quantification of DNA, accumulation in systemically infected leaves at 15 days post‐inoculation (dpi). The RNA/DNA accumulation of the different viruses varied over four orders of magnitude, being highest for TCV and lowest for CaMV (Table 1).
Table 1.
Multiplication of five viruses in six Arabidopsis wild genotypes. *
| host Genotype | CMV | TCV | TuMV | YoMV | CaMV |
|---|---|---|---|---|---|
| Col‐1 | 10.20 ± 3.42 | 320.91 ± 56.48 | 1.63 ± 0.43 | 3.61 ± 1.21 | 0.15 ± 0.06 |
| Ler | 5.06 ± 1.75 | 377.60 ± 70.37 | 2.50 ± 0.32 | 8.44 ± 1.25 | 0.24 ± 0.11 |
| Shak | 7.73 ± 3.56 | 494.64 ± 81.61 | 0.50 ± 0.058 | 3.86 ± 0.70 | 0.19 ± 0.10 |
| Ll‐0 | 14.25 ± 3.63 | 283.41 ± 42.18 | 0.58 ± 0.05 | 3.43 ± 0.59 | 0.18 ± 0.04 |
| Cum‐0 | 11.73 ± 5.21 | 181.31 ± 48.62 | 2.70 ± 1.07 | 5.41 ± 0.94 | 0.52 ± 0.12 |
| Kas‐0 | 6.75 ± 2.66 | 163.98 ± 31.21 | 2.92 ± 0.82 | 2.26 ± 0.59 | 1.42 ± 0.57 |
CaMV, Cauliflower mosaic virus; CMV, Cucumber mosaic virus; TCV, Turnip crinkle virus; TuMV, Turnip mosaic virus; YoMV, Youcai mosaic virus.
*Viral multiplication is quantified as nanograms of viral RNA/DNA per microgram of total RNA/DNA. Data values are means ± standard error of at least four replicate plants.
Generalized linear model (GzLM) analyses considering virus species and host life cycle length (short or long) as fixed factors, and host genotype nested to life cycle length as a random factor, in a full factorial model, showed that virus multiplication was dependent on all factors and their interactions (Wald χ 2 ≥ 19.01, P < 10−3). The effect of host genotype and life cycle length was solely a result of TCV multiplication, which was highest in the genotypes with a short life cycle. Removal of TCV from this analysis resulted in a non‐significant effect of host life cycle length (not shown). When multiplication across host genotypes was analysed for each virus separately, RNA/DNA accumulation varied significantly for all viruses (Wald χ 2 ≥ 14.37, P ≤ 0.013), except CMV (Wald χ 2 (5,27) = 8.105, P = 0.151) (Table 1). For each of the four viruses with significant differences, the six Arabidopsis genotypes ranked differently according to RNA/DNA accumulation, indicating that they differed in resistance to each of them.
Effects of virus infection on Arabidopsis seed production
The effect of virus infection on progeny production was quantified as the ratio of viable seeds produced by infected plants (SW i) relative to mock‐inoculated controls (SW m) (Table S1, see Supporting Information). The ratio SW i /SW m is an estimation of tolerance, which varies between zero and unity, with ratios > 1 indicating overcompensation. Importantly, this ratio is also an estimation of virulence V, which is usually measured as its reciprocal (V = 1 – SW i /SW m), with negative values indicating positive effects of infection on host fitness. For the estimation of tolerance/virulence, the actual weight of seeds produced per plant was converted into weight of viable seeds by multiplying by the viability of the seeds. The viability of the seeds from infected plants, estimated as the percentage germination, did not differ for mock‐inoculated and CMV‐, TuMV‐, YoMV‐ or CaMV‐infected plants. However, the viability of the seeds from Col‐1, Ler, Shak and Ll‐0 TCV‐infected plants was between 76% and 91% lower than that of the corresponding mock‐inoculated controls (χ 2 ≥ 11.45, P ≤ 10−3; Table S1, see Supporting Information). Thus, in all the analyses below, the values of SW i and SW m always refer to the weight of viable seeds.
The ratio SW i /SW m varied largely according to the virus species–host genotype interaction (Fig. 2). It was not significantly different from unity in the interactions of Kas‐0, Cum‐0 and Ll‐0 with CMV (complete tolerance), and was zero in the interaction of most host genotypes (except Col‐1) with TuMV (maximum virulence). GzLM analyses, considering virus species and life cycle length of the host genotype as fixed factors, and host genotype nested to life cycle length as a random factor, in a full factorial model, showed that SW i /SW m was dependent on all factors and their interactions (Wald χ 2 ≥ 31.23, P < 10−3). The effect of CMV and TCV infection on seed production was higher in the genotypes with a short life cycle than in those with a long life cycle (Wald χ 2 (1,34) = 33.10, P = 10−3 for CMV; Wald χ 2 (1,46) = 4.11, P = 0.043 for TCV). The effect of TuMV infection on seed production was higher in the genotypes with a long life cycle than in those with a short life cycle (Wald χ 2 (1,32) = 4.51, P = 0.034), which has limited meaning, as only TuMV‐infected Col‐1 plants produced seeds. The effects of YoMV and CaMV on seed production were similar for the genotypes with short and long life cycles (Wald χ 2 ≤ 3.39, P ≥ 0.066). It should be noted that, according to the hypotheses to be tested, these analyses focused on the factor ‘life cycle length’, and that the effect of infection on each individual host genotype did not always follow the above patterns (Fig. 2 and Table S1). Indeed, we detected significant effects of the interaction between the fixed factor ‘virus species’ and the random factor ‘host genotype nested to life cycle length’ on the SW i /SW m values.
Figure 2.
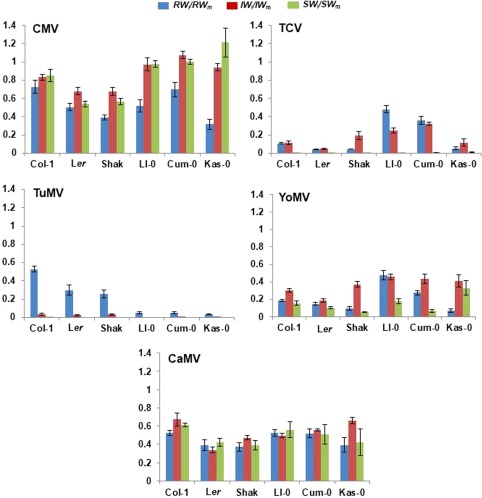
Effect of virus infection on Arabidopsis growth and reproduction. The effects of infection by Cucumber mosaic virus (CMV), Turnip crinkle virus (TCV), Turnip mosaic virus (TuMV), Youcai mosaic virus (YoMV) and Cauliflower mosaic virus (CaMV) on the growth and reproduction of six Arabidopsis wild genotypes, shown as the ratio of the weight in infected to mock‐inoculated plants of rosettes (RW i/RW m, blue bars), inflorescences (IW i/IW m, red bars) and viable seeds (SW i/SW m, green bars). Data are means ± standard error of at least four replicate plants.
When the effect of virus infection on seed production was analysed separately for each virus over the six host genotypes, significant differences amongst host genotypes were found for all viruses (Wald χ 2 ≥ 6.71, P ≤ 0.010). For TuMV, no comparison was possible (Fig. 2). The ratio SW i /SW m ranked differently for CMV, YoMV and TCV across host genotypes, indicating marked differences in virulence. Indeed, virulence was lowest for CMV (highest SW i /SW m ratio and thus highest tolerance), intermediate for YoMV and CaMV, and extremely high (i.e. lowest and ineffective tolerance) for TCV and TuMV, which nearly prevented seed production.
Effects of virus infection on resource allocation to growth and reproduction
The effect of virus infection on vegetative growth effort (growth effort) was quantified as the ratio of rosette weight of the infected to the mock‐inoculated plants: RW i /RW m. Infection by all five viruses resulted in a reduction in rosette weight (Fig. 2). The value of the ratio RW i /RW m was dependent on the virus species, the host genotype nested to life cycle length and the interactions virus × life cycle length and virus × host genotype nested to life cycle length (Wald χ 2 ≥ 187.55, P ≤ 10−3), but not on life cycle length (Wald χ 2 (1,147) = 0.82, P = 0.345). The effect of TCV and YoMV on growth effort was higher in the genotypes with a short life cycle (Wald χ 2 ≥ 11.49, P ≤ 0.001), and the opposite was observed for TuMV (Wald χ 2 (1,32) = 88.60, P ≤ 10−3). No effect of life cycle length was detected for CMV and CaMV (Wald χ 2 ≥ 0.36, P ≥ 0.223). When the effect of virus infection on growth effort was analysed separately for each virus over the six host genotypes, it differed significantly amongst host genotypes for all viruses (Wald χ 2 ≥ 16.95, P ≤ 0.005) (Fig. 2). The ratio RW i /RW m ranked differently for each virus across host genotypes, indicating differential effects of virus infection on growth effort.
The effect of virus infection on the development of reproductive structures was quantified as the ratio of inflorescence weight of infected to mock‐inoculated plants: IW i /IW m. Infection by all five viruses resulted in a reduction in inflorescence weight (Fig. 2). The value of the ratio IW i /IW m was dependent on the virus species, life cycle length, host genotype nested to life cycle length and all the interactions (Wald χ 2 ≥ 75.30, P ≤ 10−3). For CMV and YoMV, the effect of infection on reproductive effort was higher in the genotypes with a short life cycle (Wald χ 2 ≥ 10.57, P ≤ 10−3); again the opposite occurred for TuMV (Wald χ 2 (1,32) = 26.11, P = 10−3). TCV and CaMV infection had similar effects on reproductive structures in genotypes of both life cycle length groups (Wald χ 2 ≥ 0.76, P ≥ 0.064). When the effect of virus infection on reproductive effort was analysed separately for each virus over the six host genotypes, it differed significantly amongst host genotypes for all viruses (Wald χ 2 ≥ 54.56, P < 10−3) (Fig. 2). The ranking of the IW i /IW m values for each virus across host genotypes indicated differential effects of virus infection on IW, as for SW and RW.
As tolerance to CMV is attained in Arabidopsis by resource reallocation from growth to progeny production (Pagán et al., 2008), we analysed the effect of virus infection on the ratio (SW/RW)i/(SW/RW)m. This ratio was dependent on the virus species, life cycle length, host genotype nested to life cycle length and their interactions (Wald χ 2 ≥ 14.43, P ≤ 0.006). The value of the (SW/RW)i/(SW/RW)m ratio was higher in CMV‐infected genotypes with a long life cycle than in those with a short life cycle (Wald χ 2 (1,34) = 8.04, P = 0.005), indicating resource reallocation from growth to reproduction on infection. This ratio was similar for long and short life cycle genotypes infected with TCV, YoMV and CaMV (Wald χ 2 ≤ 2.29, P ≥ 0.130), although it differed for individual genotypes infected with YoMV (Fig. 3). Resource reallocation was not a host response to infection by these viruses, with the exception of Kas‐0 infected with YoMV (Fig. 3). Plants infected with TuMV were not included in the analyses, as only infected Col‐1 plants produced seed.
Figure 3.
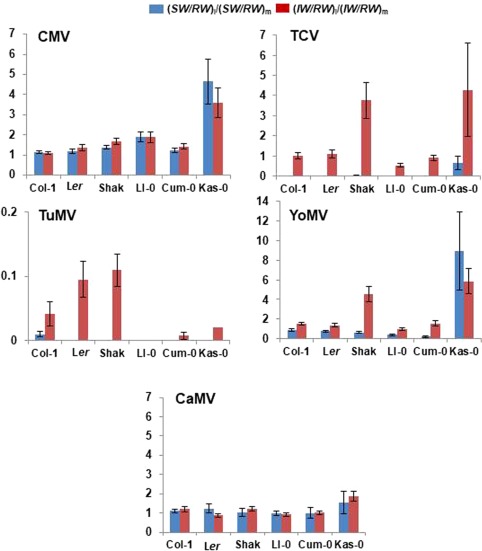
Effect of virus infection on resource allocation to growth and reproduction in Arabidopsis. The effects of infection by Cucumber mosaic virus (CMV), Turnip crinkle virus (TCV), Turnip mosaic virus (TuMV), Youcai mosaic virus (YoMV) and Cauliflower mosaic virus (CaMV) on resource allocation in six Arabidopsis wild genotypes, shown as the ratio of the weights of viable seeds to rosettes in infected vs. mock‐inoculated plants [(SW/RW)i/(SW/RW)m, blue bars] and as the ratio of the weights of inflorescences to rosettes in infected vs. mock‐inoculated plants [(IW/RW)i/(IW/RW)m, red bars]. Data are means ± standard error of at least four replicate plants. Note the different scale in each panel.
It could be possible that resource reallocation from growth to reproduction occurred, but did not yield differences in seed production. To test whether this was the case, we analysed whether there was resource reallocation from growth to the production of reproductive structures, as indicated by the ratio (IW/RW)i/(IW/RW)m. This ratio was dependent on the virus species, host genotype nested to life cycle length and the interactions virus species × life cycle length and virus species × host genotype nested to life cycle length (Wald χ 2 ≥ 22.47, P ≤ 10−3), but was not dependent on the life cycle length (Wald χ 2 (1,165) = 4.29, P = 0.117). The value of the (IW/RW)i/(IW/RW)m ratio was higher in CMV‐infected genotypes with a long life cycle than in those with a short life cycle (Wald χ 2 (1,34) = 7.49, P = 0.006), indicating resource reallocation from growth to reproduction on infection. This ratio was similar for long and short life cycle genotypes infected with TCV, YoMV and CaMV (Wald χ 2 ≤ 0.87, P ≥ 0.351), although it differed for individual host genotypes infected with all viruses (Fig. 3). Resource reallocation from growth to reproduction was not a host response to infection by these viruses, with the exception of Kas‐0 infected with YoMV and CaMV, and of Shak infected with TCV and YoMV. The ratio (IW/RW)i/(IW/RW)m was significantly lower than unity in all genotypes infected with TuMV (Fig. 3).
It should be noted that, as for SW i /SW m, analyses of all traits reported in this section particularly focused on the factor ‘life cycle length’, and the effect of infection on each individual host genotype did not always follow the above‐described patterns (Figs 2 and 3 and Table S1), as indicated by the significant effects of the interaction between factors (virus species, host life cycle length and host genotype nested to life cycle length) detected in most analysed traits.
In summary, the reallocation of resources from vegetative growth to reproduction, both to the formation of reproductive structures and progeny, is a specific response of plants from long life cycle genotypes to CMV infection, but is not a general response to virus infection.
Effects of virus infection on host developmental schedule
To analyse whether virus infection affected the temporal schedule of Arabidopsis development, we measured the total life span (LP), as well as the span of the growth and reproductive periods (GP and RP, respectively). LP is longer than GP + RP, as from the time the first silique shatters (end of RP) to complete plant senescence (end of LP) there is a period of silique maturation and progressive plant senescence that is not analysed here independently.
The effect of infection on host life span (LP i/LP m) (Fig. 4) was dependent on the virus species, life cycle length of the host genotype, host genotype nested to life cycle length and the interactions between these factors (Wald χ 2 ≥ 148.61, P < 10−3). The effect of infection on LP differed for each assayed virus. In CMV‐infected plants, LP i/LP m was significantly different for genotypes with long and short life cycles (Wald χ 2 (1,4) = 1.00, P = 0.016). LP was longer on infection in genotypes with a long life cycle and did not vary in those with a short life cycle, when compared with mock‐inoculated controls. In CaMV‐infected plants, LP i/LP m was also significantly different for genotypes with long and short life cycles (Wald χ 2 (1,4) = 6.87, P = 0.009). LP was shorter in genotypes with a long life cycle, but was not altered in those with a short life cycle. In TuMV‐infected plants, LP was shorter than in the corresponding controls in all genotypes, this reduction being more severe for genotypes with a long life cycle than in those with a short life cycle (Wald χ 2 (1,4) = 49.47, P < 10−3). Infection by TCV and YoMV did not have a life cycle length differential effect on LP (Wald χ 2 ≤ 2.57, P ≥ 0.109). LP was observed to decrease only in some genotypes (e.g. Ler and Kas‐0 for TCV, Ler for ToMV) and increased in the rest.
Figure 4.
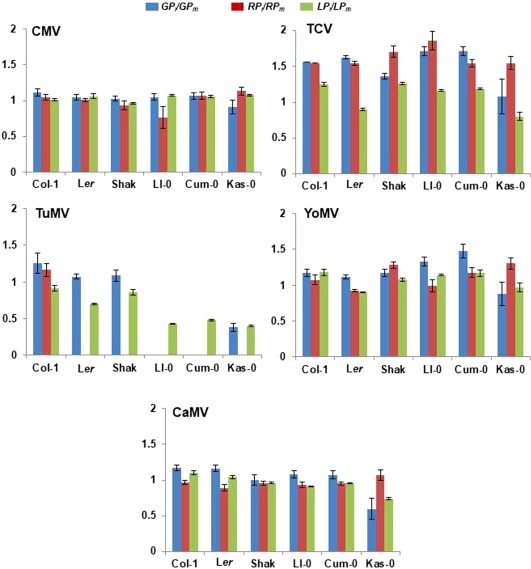
Effect of viral infection on the temporal schedule of Arabidopsis development. The effects of infection by Cucumber mosaic virus (CMV), Turnip crinkle virus (TCV), Turnip mosaic virus (TuMV), Youcai mosaic virus (YoMV) and Cauliflower mosaic virus (CaMV) on the temporal schedule of development of six Arabidopsis wild genotypes, shown as the ratio of the length in infected to mock‐inoculated plants of the pre‐reproductive growth period (GP i/GP m, blue bars), reproductive period (RP i/RP m, red bars) and life span (LP i/LP m, green bars). Data are means ± standard error of at least four replicate plants.
The effect of infection on the vegetative growth period (GP i/GP m) was dependent on the virus species, life cycle length of the host genotype, host genotype nested to life cycle length and the interactions between these factors (Wald χ 2 ≥ 24.31, P < 10−3) (Fig. 4). In TuMV‐infected plants, GP i/GP m was unaffected in genotypes with a short life cycle (Wald χ 2 ≤ 2.09, P ≥ 0.091), but most plant genotypes with a long life cycle never flowered. Infection by CMV, TCV, YoMV and CaMV did not have a differential effect on GP according to genotype life cycle length (Wald χ 2 ≤ 2.04, P ≥ 0.153). CMV infection did not affect GP in any genotype. TCV and YoMV infection resulted in an increase in GP in all genotypes, except Kas‐0, and CaMV infection resulted in an increase in GP of Col‐1 and Ler, but did not affect the other genotypes.
Lastly, the effect of infection on the reproductive period (RP i/RP m) (Fig. 4) was dependent on the virus species, the life cycle length of the host genotype, the host genotype nested to life cycle length and the interaction virus species × host genotype nested to life cycle length (Wald χ 2 ≥ 5.77, P ≤ 0.016), but not on the interaction virus species × life cycle length (Wald χ 2 (3,122) = 0.46, P = 0.928). Infection by CMV, CaMV and TuMV (in the only host genotype that flowered after infection, Col‐1) did not affect RP in any genotype. TCV infection resulted in an increase in RP in all genotypes, and YoMV infection resulted in an increase in RP in Shak, Cum‐0 and Kas‐0, a decrease in Ler and no effect in Col‐1 or Ll‐0.
In conclusion, the effect of infection on GP and LP, but not RP, was dependent on the life cycle length of the host genotype, but the sense and magnitude of the effect varied for each virus species. The effect of infection on RP varied according to host genotype and virus species.
Discussion
Understanding the mechanisms of host defence to parasites is a central question in biology. The analysis of plant defences to pathogens, including viruses, has focused on resistance, i.e. on the mechanisms that decrease the probability of infection and/or the multiplication of the pathogen within the infected hosts (Råberg, 2014). The literature on the genetic basis and molecular mechanisms of plant resistance to viruses is extensive (for reviews, see de Ronde et al., 2014; Truniger and Aranda, 2009). One major conclusion of the accumulated evidence is that the level of resistance is specific for each host × virus interaction, often at the level of host and virus genotypes, and thus resistance is a major factor in plant–virus co‐evolution (Fraile and García‐Arenal, 2010; Pagán et al., 2016). Other defences of plants to pathogens have received considerably less attention (Jeger et al., 2006), which is certainly the case for tolerance. Tolerance is defined as a process that reduces the harm of parasite infection (i.e. virulence), regardless of the level of parasite multiplication (Little et al., 2010; Råberg, 2014), and its mechanisms and role in host–pathogen co‐evolution remain underexplored (Best et al., 2014; Little et al., 2010).
We examined the response of six wild genotypes of Arabidopsis with long or short life cycle lengths to infection by five virus species with different life styles, genome organization and expression, host ranges and transmission mechanisms. Our purpose was to analyse whether both resistance and tolerance vary according to the host genotype × virus species interaction, and whether tolerance to other viruses is associated with life cycle length, as reported for tolerance to CMV (Pagán et al., 2007, 2008). The six assayed Arabidopsis genotypes differed in resistance to the five assayed virus species, as indicated by significant differences in virus multiplication, which were dependent on the host genotype × virus species interaction. The results also showed that the assayed viruses were virulent pathogens that significantly reduced plant growth and progeny production. The effect of virus infection on progeny production, measured by the ratio SW i/SW m, which we take as an estimate of virulence, was also dependent on the virus species × host genotype interaction. Interestingly, for each virus species, the effect of infection on seed production was unrelated to the level of virus multiplication in each host genotype (Fig. 2 and Table 1). A positive correlation between parasite multiplication and virulence is assumed in most models of virulence evolution (Alizon et al., 2009) and has been shown for many animal and plant pathogens (Bull and Lauring, 2014; Lipsitch and Moxon, 1997; Sacristán and García‐Arenal, 2008), but not always (Davies et al., 2001; Levin and Bull, 1994). The lack of correlation between these two traits has been explained by non‐linear tolerance to infection (Miller et al., 2006). Indeed, our group has reported previously that, in the Arabidopsis–CMV interaction, the observed lack of correlation between virus multiplication and virulence can be explained by host genotype‐specific tolerance (Pagán et al., 2007). Hence, these results indicate that, in Arabidopsis, tolerance to virus infection is a genotype‐specific response.
Here, genotype‐specific tolerance was also observed in response to CMV, but effective genotype‐specific tolerance to the other assayed viruses was not observed. The results of this study show that, when the assayed Arabidopsis genotypes are clustered according to their life cycle length, genotypes with a long life cycle (Cum‐0, Ll‐0 and Kas‐0; Group I genotypes in Pagán et al., 2007) are tolerant to CMV infection, which does not affect seed production. In the genotypes with a short life cycle (Col‐1, Ler, Shak; Group II genotypes in Pagán et al., 2007), however, infection reduces seed production by about 40% (Fig. 3). These results fully agree with previous observations (Hily et al., 2015; Pagán et al., 2008, 2009), which showed that long‐lived Arabidopsis genotypes were more tolerant than short‐lived genotypes to infection by different CMV genotypes. The observed host genotype differential responses are unrelated to rates of CMV multiplication, which do not differ for genotypes with long and short life cycles (Fig. 2). Indeed, tolerance to CMV showed a high broad‐sense heritability ( = 0.63), even higher than in our previous reports (Pagán et al., 2007, 2008). This indicates that the observed variation in the effect of CMV infection on seed production across plant genotypes is largely host dependent and not a by‐product of virus infection.
It has been proposed that host tolerance to parasites can be achieved by resource reallocation to different fitness components. Life history theory predicts that resource investment by organisms will be conditioned by trade‐offs between resource allocation to different fitness components, such as growth, reproduction and survival, and the optimal resource allocation will depend on the environmental conditions (Stearns, 1976). Theory proposes that, under conditions that increase mortality rates, such as parasitism, hosts will reduce the allocation of resources to growth, and will allocate more resources to reproduction, to maximize fitness under these unfavourable conditions (Forbes, 1993; Leventhal et al., 2014; Minchella, 1985; Perrin et al., 1996; Williams, 1966). Experimental support for this hypothesis derives mostly from studies of invertebrate animals and their parasites (e.g. Chadwick and Little, 2005; Fredensborg and Poulin, 2006; Michalakis and Hochberg, 1994; Polak and Starmer, 1998). Evidence from plants is scarcer, but it has been shown that tolerance to parasitism or herbivory may be explained by resource reallocation to reproduction (Agrawal, 2000; Fellous and Salvaudon, 2009; Strauss and Agrawal, 1999). Our results show that, on CMV infection, tolerant genotypes reallocate resources from growth to reproduction. This is consistent with previous data from our group indicating that, although phenotypic plasticity in response to CMV infection is modulated by environmental conditions, such as infection time relative to host development (Pagán et al., 2007, 2008), host plant density and virus incidence (Pagán et al., 2009), or light and temperature conditions (Hily et al., 2015), resource reallocation from growth to reproduction is a consistent and specific response of tolerant genotypes.
Life history theory also predicts that infection by highly virulent parasites, resulting in high host mortality rates, will result in shorter host pre‐reproductive periods, so that reproduction is achieved before resource depletion and death, whereas infection by low‐virulent parasites will delay host reproduction, allowing for compensation of parasite damage later in life (Gandon et al., 2002; Hochberg et al., 1992). In agreement with these predictions, the results show that, on infection, tolerance to CMV is associated with an increase in both life span and vegetative growth period, LP and GP, which does not occur in the genotypes in which CMV virulence is higher. Thus, our present and past (Pagán et al., 2008, 2009; Hily et al., 2015) results also agree with theoretical predictions on the modification of life history traits in response to parasitism.
Given the robustness of tolerance to CMV regardless of environmental conditions, it could be hypothesized that resource reallocation‐based tolerance would also operate to decrease harm caused by infection with other viruses. However, the effect of infection by TCV, TuMV, YoMV and CaMV on seed production was always as severe as in the CMV‐non‐tolerant genotypes (for YoMV and CaMV) or was orders of magnitude more severe (for TCV and TuMV), so that no expression of tolerance comparable with that of the long life cycle genotypes to CMV was attained. Indeed, no life cycle length‐associated, or genotype‐associated, resource reallocation from growth to reproduction was observed after infection by any of these viruses, with the notable exception of Kas‐0 after infection by YoMV and CaMV, the two less virulent viruses after CMV. A possible explanation for this absence of resource reallocation‐based tolerance is that a defence response based on resource reallocation was prevented by a strong reduction in vegetative growth (RW) as a result of infection with these four viruses. This could be the case for infection by TCV in genotypes with a short life cycle and by TuMV in genotypes with a long life cycle (RW i/RW m values of 0.07 and 0.04, respectively) or, to a lesser degree, for infection with YoMV in genotypes with both short and long life cycles (RW i/RW m values of 0.14 and 0.28, respectively). However, it would not apply to infection with CaMV of genotypes with a long life cycle, which results in a reduction in growth similar to that caused by CMV (RW i/RW m values of 0.49 for CaMV and 0.48 for CMV). These results strongly suggest that tolerance associated with resource reallocation from growth to reproduction is a specific response of some Arabidopsis genotypes to CMV infection, and that changes in resource allocation after infection by TCV, TuMV, YoMV and CaMV can be attributed to virus use of host resources, resulting in pathogenic effects of infection, rather than to responses that compensate for the negative effects of parasitism (Forbes, 1993; Hochberg et al., 1992; Perrin et al., 1996). In support of this interpretation, it is worth mentioning that the broad‐sense heritability in Arabidopsis genotypes of SW i/SW m and of (SW/RW)i/(SW/RW)m is much higher after infection by CMV ( of 0.63 and 0.51, respectively) than after infection by any other virus (0.12 < < 0.32 for SW i/SW m and 0.08 < < 0.31 for (SW/RW)i/(SW/RW)m).
A further interesting observation is that, contrary to theoretical predictions (Gandon et al., 2002; Hochberg et al., 1992), the length of the pre‐reproductive period of infected plants was unrelated to the virulence of the infecting virus, as it was unaffected by the less virulent viruses CMV and CaMV, and was increased by the more virulent viruses YoMV and TCV. The longer GP in the most virulent viruses may be explained by the complex regulation of the transition from growth to reproduction in Arabidopsis, which is determined by a variety of factors, including the attainment of a minimum rosette size (Irish, 2010; Smyth et al., 1990). This is best exemplified in the case of TuMV‐infected plants, as most infected genotypes suffered great reductions in their rosette size and never flowered, the virus acting as a castrating parasite.
In conclusion, genotype‐specific resistance to virus infection seems to be a general defence of Arabidopsis to a variety of virus species. Conversely, tolerance to virus infection associated with life history trait modifications seems to be a narrow‐spectrum response. This is an unexpected result at odds with theory and with observations of plant responses to cellular pathogens and herbivores. These findings contribute to a better understanding of plant–virus co‐evolutionary dynamics, and suggest that some aspects of life history theory should be revisited in the case of plant–virus interactions.
Experimental Procedures
Arabidopsis thaliana genotypes and virus isolates
The six wild genotypes of Arabidopsis used in this study, namely Columbia glabrata1 (Col‐1), Landsberg erecta (Ler), Shakdara (Shak), Llagostera (Ll‐0), Kashmir (Kas‐0) and Cumbres Mayores (Cum‐0), were originally provided by Dr Carlos Alonso‐Blanco, CNB, CSIC, Madrid, Spain. All genotypes were initially multiplied simultaneously under the same glasshouse conditions to reduce maternal effects. For plant growth, seeds were stratified at 4 °C for 4 days in 96‐well trays containing a 3 : 1 peat–vermiculite mix. Trays were then transferred to a glasshouse at 22 °C under long‐day conditions (16 h light, 70% relative humidity). Ten days later, seedlings were transplanted into individual 10‐cm‐diameter pots containing the same substrate.
Cucumber mosaic virus strain LS (LS‐CMV) was multiplied in Nicotiana clevelandii Gray. plants inoculated with transcripts from biologically active cDNA clones (Zhang et al., 1994). Turnip crinkle virus strain Massachusetts (TCV‐M) was multiplied in Nicotiana benthamiana Domin. plants from transcripts of biologically active cDNA clones provided by Professor Anne Simon (University of Maryland, College Park, MD, USA). Turnip mosaic virus isolate UK1 (TuMV‐UK1) was provided by Professor Fernando Ponz (CBGP, UPM‐INIA, Madrid, Spain) and multiplied in N. benthamiana plants. Youcai mosaic virus isolate Oilseed rape mosaic virus (YoMV‐ORSMV), originally provided by Professor Adrian Gibbs (Australian National University, Canberra, Australia), was multiplied in Nicotiana tabacum cv. Samsun. Cauliflower mosaic virus isolate Cabbage John Innes BJI (CaMV‐BJI) was multiplied in N. clevelandii from infectious clones provided by Dr Stéphane Blanc (INRA, Montpellier, France).
Arabidopsis plants were inoculated at the four‐leaf stage (stage 1.04 as in Boyes et al., 2001) with sap from young developing leaves of the virus propagation hosts in 0.1 m phosphate buffer pH 7.0 + 0.2% sodium diethylditiocarbamate (DIECA). Fifteen microlitres of sap were applied per plant and 15 µL of phosphate buffer were applied to mock‐inoculated controls. Eight plants of each genotype were inoculated with each virus or mock‐inoculated. Infection rates ranged between 60% (for CaMV) and 100% (for TCV and YoMV), but there were always at least four infected plants per treatment.
Quantification of virus multiplication
Virus multiplication was estimated as viral RNA/DNA accumulation at 15 dpi using RT‐qPCR or qPCR. At 15 dpi, three leaf discs of 4 mm in diameter were randomly harvested from different systemically infected rosette leaves of each plant, and total plant RNA was extracted using Trizol® reagent (Life Technologies, Carlsbad, CA, USA) or, for CaMV‐inoculated plants, total plant DNA was extracted in 50 mm NaCl, 100 mm Tris‐HCl, pH 7.5, 50 mm ethylenediaminetetraacetic acid (EDTA), pH 8.0, 10 mg/mL RNAse A (10 mg/mL). For each RT‐qPCR, 0.5–4 ng of total RNA/DNA were added to Brilliant III Ultra‐Fast SYBR Green qRT‐PCR Master Mix according to the manufacturer's recommendations (Agilent Technologies, Santa Clara, CA, USA) in a final volume of 10 µL. In each plate, purified viral RNA, in vitro transcripts from the cDNA clones or the DNA from the clone (for CaMV) was serially diluted in a 1 : 10 ratio to obtain a standard curve from 100 ng/µL to 0.1 fg/µL. Each plant sample was assayed in triplicate on a Light Cycler 480 II real‐time PCR system (Roche Diagnostics, Basel, Switzerland). Dissociation curves were generated to ascertain that only one single product was produced and detected in each case.
For the detection of LS‐CMV, the forward and reverse primers 5′TAAGAAGCTTGTTTCGCGCATTC3′ and 5′CGGAAAGATCGGATGATGAAGG3′ were used to amplify a region of 106 nucleotides (nt) from the coat protein gene (nucleotide positions 1951–1626 in Acc. No. AF127976.1), as described in Hily et al. (2014). For the detection of TCV‐M, primers 5′CACTCAGATTTAGGTACTCCCC3′ and 5′CACGCTAGATACACAACCCTC3′ were used to amplify 140 nt from the coat protein gene (positions 397–537 in Acc. No. HQ589261.1). For the detection of TuMV‐UK1, primers 5′TGTTCGGCTTGGATGGAA3′ and 5′TTAACGTCCTCGGTCGTATGC3′ were used to amplify 70 nt from the coat protein gene (positions 9515–9585 in Acc. No. AB194802.1), as described in Lunello et al. (2007). For the detection of YoMV, primers 5′CCTTCTGAGTGCGATTGTGA3′ and 5′GATGGACGCGACTCTTCTTC3′ were used to amplify 159 nt from the coat protein gene (positions 5746–5905 in Acc. No. NC_004422.1). For the detection of CaMV‐BJI, primers 5′TGAAATCCTCAGTGACCAAAAATC3′ and 5′TACAGGGGCAATCATTGATGAGC3′ were used to amplify 152 nt from the ORF III gene (positions 1865–2017 in Acc. No. KJ716236.1) as described in Chaouachi et al. (2008).
Quantification of Arabidopsis resource allocation and temporal life history traits
We focused on the life history traits related to the allocation of resources to growth and reproduction and to the temporal schedule of development. Plants were harvested on complete senescence and maintained at 65 °C in an oven until constant weight. Following Thompson and Stewart (1981), the rosette weight (RW) was taken as a measure of the vegetative growth effort, the weight of the reproductive structures, that is, the inflorescence (IW), was taken as a measure of the reproductive effort, and the weight of viable seeds (SW) was taken as a measure of progeny production. All weights are presented in grams.
The temporal schedule of Arabidopsis development was also quantified. Following Boyes et al. (2001), the span of the vegetative growth period (GP) was defined as the time from the end of stratification to the opening of the first flower. The span of the reproductive period (RP) was defined as the time from the opening of the first flower until the first silique shattered. Finally, the life span (LP) was defined as the period from seed germination until complete senescence. All of these periods were measured in days.
To assess seed viability, a germination assay was performed in which 200 seeds per treatment (host genotype × virus) were plated in Petri dishes on moist filter paper, stratified for 4 days in the dark at 4 °C and incubated for 4 days at 21 °C in a 16‐h light cycle. Then, germinated seeds were counted and the germination percentage was calculated.
Statistical analyses
For each trait (variable), the normality of data distribution was evaluated by a Shapiro–Wilk test and homoscedasticity was checked according to Levene's test for equality of error variances. As all variables were not normally distributed, virus multiplication, life history traits and the effects of infection on life history traits were analysed via full factorial GzLMs. Significance of differences amongst classes within each factor was determined by Fisher's least‐significant difference test (LSD). The frequency of viable seeds, plants producing flowers or seeds was compared by contingency tests. Analyses were performed using the software package SPSS v. 22 (SPSS Inc., Chicago, IL, USA).
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Values of life history traits for six Arabidopsis wild genotypes mock‐inoculated or infected by five viruses.
Acknowledgements
This research was funded by Plan Estatal de I+D+i, Secretaría de Estado de Investigación, MINECO, Spain (BFU2015–64018‐R). TuMV‐UK1 was provided by Professor Fernando Ponz (CBGP, UPM‐INIA, Madrid, Spain). The TCV‐M infectious clone was provided by Professor Anne Simon (University of Maryland, College Park, MD, USA). The CaMV‐BJI infectious clone was provided by Dr Stéphane Blanc (INRA, Montpellier, France). Antolín López Quirós provided excellent technical support. I.P. was supported by a Ramón y Cajal grant from MINECO, Spain (RYC‐2011–08574). A.S. was supported by a scholarship of an Erasmus Mundus EU programme (BRAVE, agreement number 2013‐2536/001‐001).
References
- Agnew, P. , Koella, J.C. and Michalakis, Y. (2000) Host life history responses to parasitism. Microbes Infect. 2, 891–896. [DOI] [PubMed] [Google Scholar]
- Agrawal, A.A. (2000) Overcompensation of plants in response to herbivory and the by‐product benefits of mutualism. Trends Plant Sci. 5, 309–313. [DOI] [PubMed] [Google Scholar]
- Aguilar, I. , Sánchez, F. , Martin Martin, A. , Martinez‐Herrera, D. and Ponz, F. (1996) Nucleotide sequence of Chinese rape mosaic virus (oilseed rape mosaic virus), a crucifer tobamovirus infectious on Arabidopsis thaliana . Plant Mol. Biol. 30, 191–197. [DOI] [PubMed] [Google Scholar]
- Alizon, S. , Hurford, A. , Mideo, N. and Van Baalen, M. (2009) Virulence evolution and the trade‐off hypothesis: history, current state of affairs and the future. J. Evol. Biol. 22, 245–259. [DOI] [PubMed] [Google Scholar]
- Anderson, P.K. , Cunningham, A.A. , Patel, N.G. , Morales, F.J. , Epstein, P.R. and Daszak, P. (2004) Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol. 19, 535–544. [DOI] [PubMed] [Google Scholar]
- Best, A. , White, A. and Boots, M. (2014) The coevolutionary implications of host tolerance. Evolution, 5, 1426–1435. [DOI] [PubMed] [Google Scholar]
- Boyes, D.C. , Zayed, A.M. , Ascenzi, R. , McCaskill, A.J. , Hoffman, N.E. , Davis, K.R. and Görlach, J. (2001) Growth stage‐based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell, 13, 1499–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull, J.J. and Lauring, A.S. (2014) Theory and empiricism in virulence evolution. PLoS Pathog. 10, e1004387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, L. , Chen, K. , Zhang, X. , Yan, L. , Hou, M. and Xu, Z. (2009) Biological and molecular characterization of a crucifer Tobamovirus infecting oilseed rape. Biochem. Genet. 47, 451–461. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C. , Morris, T.J. , Stockley, P.G. and Harrison, S.C. (1987) Structure and assembly of Turnip crinkle virus. IV. Analysis of the coat protein gene and implications of the subunit primary structure. J. Mol. Biol. 194, 265–276. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C. , Heaton, L.A. , Zuidema, D. , Hillman, B.I. and Morris, T.J. (1989) The genome structure of Turnip crinkle virus. Virology, 170, 219–226. [DOI] [PubMed] [Google Scholar]
- Chadwick, W. and Little, T. (2005) A parasite‐mediated life‐history shift in Daphnia magna . Proc. R. Soc. Lond. B: Biol. Sci. 272, 505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaouachi, M. , Fortabat, M.N. , Geldreich, A. , Yot, P. , Kerlan, C. , Kebdani, N. , Audeon, C. , Romaniuk, M. and Bertheau, Y. (2008) An accurate real‐time PCR test for the detection and quantification of cauliflower mosaic virus (CaMV): applicable in GMO screening. Eur. Food. Res. Technol. 3, 789–798. [Google Scholar]
- Csorba, T. , Pantaleo, V. and Burgyán, J. (2009) RNA silencing: an antiviral mechanism. Adv. Virus. Res. 75, 35–230. [DOI] [PubMed] [Google Scholar]
- Davies, C.M. , Webster, J.P. and Woolhouse, M.J. (2001) Trade‐offs in the evolution of virulence in an indirectly transmitted macroparasite. Proc. R. Soc. Lond. B: Biol. Sci. 268, 251–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellous, S. and Salvaudon, L. (2009) How can your parasites become your allies? Trends Parasitol. 25, 62–66. [DOI] [PubMed] [Google Scholar]
- Forbes, M.R.L. (1993) Parasitism and host reproductive effort. Oikos, 67, 444–450. [Google Scholar]
- Fraile, A. and García‐Arenal, F. (2010) The coevolution of plants and viruses: resistance and pathogenicity. Adv. Virus Res. 76, 1–32. [DOI] [PubMed] [Google Scholar]
- Fraile, A. and García‐Arenal, F. (2016) Environment and evolution modulate plant virus pathogenesis. Curr. Opin. Virol. 17, 50–56. [DOI] [PubMed] [Google Scholar]
- Fredensborg, B.L. and Poulin, R. (2006) Parasitism shaping host life‐history evolution: adaptive responses in a marine gastropod to infection by trematodes. J. Anim. Ecol. 75, 44–53. [DOI] [PubMed] [Google Scholar]
- Gandon, S. , Agnew, P. and Michalakis, Y. (2002) Coevolution between parasite virulence and host life‐history traits. Am. Nat. 160, 374–388. [DOI] [PubMed] [Google Scholar]
- Haas, M. , Bureau, M. , Geldreich, A. , Yot, P. and Keller, M. (2002) Cauliflower mosaic virus: still in the news. Mol. Plant Pathol. 3, 419–429. [DOI] [PubMed] [Google Scholar]
- Hily, J.M. , García, A. , Moreno, A. , Plaza, M. , Wilkinson, M.D. , Fereres, A. , Fraile, A. and García‐Arenal, F. (2014) The relationship between host lifespan and pathogen reservoir potential: an analysis in the system Arabidopsis thaliana–Cucumber mosaic virus . PLoS. Pathog. 10, e1004492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hily, J.M. , Poulicard, N. , Mora, M.Á. , Pagán, I. and García‐Arenal, F. (2015) Environment and host genotype determine the outcome of a plant–virus interaction: from antagonism to mutualism. New Phytol. 209, 812–822. [DOI] [PubMed] [Google Scholar]
- Hochberg, M.E. , Michalakis, Y. and De Meeus, T. (1992) Parasitism as a constraint on the rate of life‐history evolution. J. Evol. Biol. 5, 491–504. [Google Scholar]
- Hollings, M. and Stone, O.M. (1972) Turnip Crinkle Virus. C.M.I/A.A.B. Descriptions of Plant Viruses, No. 109. Warwick: Commonwealth Mycological Institute, Kew, Surrey, and Association of Applied Biologists.
- Irish, V.F. (2010) The flowering of Arabidopsis flower development. Plant J. 6, 1014–1028. [DOI] [PubMed] [Google Scholar]
- Jacquemond, M. (2012) Cucumber mosaic virus. Adv. Virus. Res. 84, 439–504. [DOI] [PubMed] [Google Scholar]
- Jeger, M.J. , Seal, S.E. and Van den Bosch, F. (2006) Evolutionary epidemiology of plant virus disease. Adv. Virus. Res. 67, 163–203. [DOI] [PubMed] [Google Scholar]
- Leventhal, G.E. , Dünner, R.P. and Barribeau, S.M. (2014) Delayed virulence and limited costs promote fecundity compensation upon infection. Am. Nat. 4, 480–493. [DOI] [PubMed] [Google Scholar]
- Levin, B.R. and Bull, J.J. (1994) Short‐sighted evolution and the virulence of pathogenic microorganisms. Trends Microbiol. 2, 76–81. [DOI] [PubMed] [Google Scholar]
- Lipsitch, M. and Moxon, R. (1997) Virulence and transmissibility of pathogens: what is the relationship? Trends Microbiol. 5, 31–37. [DOI] [PubMed] [Google Scholar]
- Little, T.J. , Shuker, D.M. , Colegrave, N. , Day, T. and Graham, A.L. (2010) The coevolution of virulence: tolerance in perspective. PLoS Pathog. 6, e1001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunello, P. , Mansilla, C. , Sánchez, F. and Ponz, F. (2007) A developmentally linked, dramatic, and transient loss of virus from roots of Arabidopsis thaliana plants infected by either of two RNA viruses. Mol. Plant–Microbe Interact. 12, 1589–1595. [DOI] [PubMed] [Google Scholar]
- Michalakis, Y. and Hochberg, M.E. (1994) Parasitic effects on host life‐history traits: a review of recent studies. Parasite, 1, 291–294. [DOI] [PubMed] [Google Scholar]
- Miller, M.R. , White, A. and Boots, M. (2006) The evolution of parasites in response to tolerance in their hosts: the good, the bad, and apparent commensalism. Evolution, 5, 945–956. [PubMed] [Google Scholar]
- Minchella, D.J. (1985) Host life‐history variation in response to parasitism. Parasitology, 90, 205–216. [Google Scholar]
- Pagán, I. , Alonso‐Blanco, C. and García‐Arenal, F. (2007) The relationship of within‐host multiplication and virulence in a plant–virus system. PLoS One, 2, e786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagán, I. , Alonso‐Blanco, C. and García‐Arenal, F. (2008) Host responses in life‐history traits and tolerance to virus infection in Arabidopsis thaliana . PLoS Pathog. 4, e1000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagán, I. , Alonso‐Blanco, C. and García‐Arenal, F. (2009) Differential tolerance to direct and indirect density‐dependent costs of viral infection in Arabidopsis thaliana . PLoS Pathog. 5, e1000531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagán, I. , Fraile, A. , Fernández‐Fueyo, E. , Montes, N. , Alonso‐Blanco, C. and García‐Arenal, F. (2010) Arabidopsis thaliana as a model for the study of plant–virus co‐evolution. Philos. Trans. R. Soc. B: Biol. Sci. 365, 1983–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagán, I. , Montes, N. , Milgroom, M.G. and García‐Arenal, F. (2014) Vertical transmission selects for reduced virulence in a plant virus and for increased resistance in the host. PLoS Pathog. 10, e1004293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagán, I. , Fraile, A. and García‐Arenal, F. (2016) Evolution of the interactions of viruses with their plant hosts In: Virus Evolution Current Research and Future Directions (Weaver S.C., Denison M., Roossinck M. and Vignuzzi M., eds), pp. 127–154. Wymondham, Norfolk: Caister Academic Press. [Google Scholar]
- Park, C.Y. , Lee, M.A. , Lee, S.H. , Kim, J.S. and Kim, H.G. (2016) First report of Youcai mosaic virus in Daisy Fleabane (Erigeron annuus). Plant Dis. 100, 1250. [Google Scholar]
- Perrin, N. , Christe, P. and Richner, H. (1996) On host life‐history response to parasitism. Oikos, 75, 317–320. [Google Scholar]
- Polak, M. and Starmer, W.T. (1998) Parasite‐induced risk of mortality elevates reproductive effort in male Drosophila. Proc. R. Soc. Lond. B: Biol. Sci. 265, 2197–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Råberg, L. (2014) How to live with the enemy: understanding tolerance to parasites. PLoS Biol. 12, e1001989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read, A.F. (1994) The evolution of virulence. Trends Microbiol. 2, 73–76. [DOI] [PubMed] [Google Scholar]
- de Ronde, D. , Butterbach, P. and Kormelink, R. (2014) Dominant resistance against plant viruses. Front. Plant Sci. 5, 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacristán, S. and García‐Arenal, F. (2008) The evolution of virulence and pathogenicity in plant pathogen populations. Mol. Plant Pathol. 9, 369–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, D.R. , Bowman, J.L. and Meyerowitz, E.M. (1990) Early flower development in Arabidopsis. Plant Cell, 8, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns, S.C. (1976) Life‐history tactics: a review of the ideas. Q. Rev. Biol. 51, 3–47. [DOI] [PubMed] [Google Scholar]
- Strauss, S.Y. and Agrawal, A.A. (1999) The ecology and evolution of plant tolerance to herbivory. Trends Ecol. Evol. 14, 179–185. [DOI] [PubMed] [Google Scholar]
- Thompson, K. and Stewart, A.J.A. (1981) The measurement and meaning of reproductive effort in plants. Am. Nat. 2, 205–211. [Google Scholar]
- Truniger, V. and Aranda, M.A. (2009) Recessive resistance to plant viruses. Adv. Virus Res. 75, 119–231. [DOI] [PubMed] [Google Scholar]
- Walsh, J.A. and Jenner, C.E. (2002) Turnip mosaic virus and the quest for durable resistance. Mol. Plant Pathol. 3, 289–300. [DOI] [PubMed] [Google Scholar]
- Williams, G.C. (1966) Adaptation and Natural Selection. Princeton, NJ: Princeton University Press. [Google Scholar]
- Zhang, L. , Hanada, K. and Palukaitis, P. (1994) Mapping local and systemic symptom determinants of cucumber mosaic cucumovirus in tobacco. J. Gen. Virol. 75, 3185–3191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Values of life history traits for six Arabidopsis wild genotypes mock‐inoculated or infected by five viruses.


