Summary
Fusaric acid (FA) is amongst the oldest identified secondary metabolites produced by Fusarium species, known for a long time to display strong phytotoxicity and moderate toxicity to animal cells; however, the cellular targets of FA and its function in fungal pathogenicity remain unknown. Here, we investigated the role of FA in Fusarium oxysporum, a soil‐borne cross‐kingdom pathogen that causes vascular wilt on more than 100 plant species and opportunistic infections in humans. Targeted deletion of fub1, encoding a predicted orthologue of the polyketide synthase involved in FA biosynthesis in F. verticillioides and F. fujikuroi, abolished the production of FA and its derivatives in F. oxysporum. We further showed that the expression of fub1 was positively controlled by the master regulator of secondary metabolism LaeA and the alkaline pH regulator PacC through the modulation of chromatin accessibility at the fub1 locus. FA exhibited strong phytotoxicity on tomato plants, which was rescued by the exogenous supply of copper, iron or zinc, suggesting a possible function of FA as a chelating agent of these metal ions. Importantly, the severity of vascular wilt symptoms on tomato plants and the mortality of immunosuppressed mice were significantly reduced in fub1Δ mutants and fully restored in the complemented strains. Collectively, these results provide new insights into the regulation and mode of action of FA, as well as on the function of this phytotoxin during the infection process of F. oxysporum.
Keywords: fungal pathogenicity, fusaric acid, Fusarium oxysporum, metal chelation, mycotoxins, phytotoxicity, virulence
Introduction
Fungi cause major plant diseases and destroy or contaminate each year a significant proportion of global agricultural production, making them by far the most damaging class of plant pathogen (Fisher et al., 2012; Strange and Scott, 2005). Moreover, opportunistic fungal pathogens of humans can provoke life‐threatening systemic infections, particularly in immunocompromised patients (Fridkin, 2005). The soil‐inhabiting fungus Fusarium oxysporum has been ranked amongst the top 10 fungal pathogens in molecular plant pathology based on scientific/economic importance, and causes vascular wilt disease in more than 100 different crops (Armstrong and Armstrong, 1981; Dean et al., 2012). In addition, F. oxysporum isolates can cause opportunistic infections in humans, ranging from superficial or locally invasive to disseminated, depending on the immune status of the host (Nucci and Anaissie, 2007). Fusarium oxysporum f. sp. lycopersici FGSC 9935 (FOL 4287) is a fully sequenced isolate (Ma et al., 2010) able to kill both tomato plants and immunosuppressed mice (Ortoneda et al., 2004). Therefore, this isolate represents an excellent model for the study of the genetic basis of cross‐kingdom pathogenicity in fungi.
Many fungi produce secondary metabolites that are toxic to plants or animals (Berthiller et al., 2013). Fusaric acid (FA), a picolinic acid derivative originally isolated from Fusarium heterosporium (Yabuta et al., 1937), was the first fungal phytotoxin isolated from infected host plants (Gäumann, 1957) and is known for its high phytotoxicity (Niehaus et al., 2014; Stipanovic et al., 2011). FA also exhibits toxicity towards animals, including notochord malformation in zebrafish (Yin et al., 2015), and neurotoxicity in mammals (Porter et al., 1995), and towards bacteria (Bacon et al., 2006; Ruiz et al., 2015). Although several studies on the mode of action of FA have been conducted, the cellular basis of its toxicity remains poorly understood. Suggested mechanisms include the modification of cell membrane potential, inhibition of ATP synthesis, chelation of metal ions or electrolyte leakage (D'Alton and Etherton, 1984; Marrè et al., 1993; Pavlovkin, 1998; Ruiz et al., 2015). Recently, chromatin condensation, cytochrome c release, DNA fragmentation and hydrogen peroxide accumulation have been reported in FA‐treated plant cell cultures, suggesting a possible involvement of programmed cell death in FA toxicity (Jiao et al., 2013; Samadi and Shahsavan Behboodi, 2006).
The polyketide synthase (PKS) Fub1 has been identified recently as the first enzyme of the FA biosynthetic pathway in Fusarium verticillioides (Brown et al., 2012). The fub1 gene is part of the FA gene cluster, and its inactivation is sufficient to completely block FA production (Brown et al., 2012; Niehaus et al., 2014). In the present work, we studied the role of Fub1 in F. oxysporum. We found that fub1 was essential for the production of FA and its derivatives in this fungus, and that its transcription was positively regulated by LaeA, a master regulator of secondary metabolism, and the alkaline pH regulator PacC. We further demonstrated that the loss of Fub1 and FA in F. oxysporum led to reduced virulence in tomato plants and immunodepressed mice. Finally, we showed that phytotoxicity of FA could be reduced by supplying copper, iron or zinc to the plants. Our results establish a functional role for FA in fungal virulence on plants and mammals.
Results
Inactivation of the PKS Fub1 abolishes FA production in F. oxysporum
A blastp search in the Fusarium Comparative Database (Broad Institute), using Fub1 from Fusarium fujikuroi (FFUJ_02105) as a bait, identified a single predicted Fub1 orthologue (FOXG_15248) displaying 89% overall identity with the query protein. Manual inspection of the F. oxysporum fub1 locus identified all other members of the FA gene cluster previously described in F. verticillioides (Brown et al., 2012) and F. fujikuroi (Niehaus et al., 2014) (Fig. S1A, see Supporting Information). Interestingly, two additional putative genes were present between fub3 and fub4 in different F. oxysporum isolates, including the reference strain FOL 4287 (Fig. S1A) (Brown et al., 2015). Both genes are neighbours in other Fusarium species, but not located in the FA gene cluster. For example, in F. fujikuroi, the orthologues of these two genes, FFUJ_11046 and FFUJ_11047, are located on chromosome 10, whereas the FA gene cluster is located on chromosome 3 (Fig. S1A). It is currently unknown whether the insertion of these two additional genes has any effect on the FA gene cluster.
Recently, additional components of the cluster, including two Zn(II)2Cys6 transcription factors, have been identified in different Fusarium species (Brown et al., 2015; Studt et al., 2016). To study the role of Fub1 in FA production by F. oxysporum, we replaced the entire FOXG_15248 coding sequence with the hygromycin B resistance gene (hphr), generating several fub1Δ strains (Fig. S1B and S1C). To determine whether FOXG_15248 was responsible for FA production in F. oxysporum, extracts from cultures of the different strains grown on potato dextrose agar (PDA) or Czapek‐Dox agar (CDA) were analysed by high‐performance liquid chromatography/electrospray ionization‐tandem mass spectrometry (HPLC/ESI‐MS/MS). This approach allows the reliable and sensitive quantification of several hundred fungal analytes, including almost all mycotoxins for which standards are commercially available (Malachova et al., 2014). FA and its derivative fusarinolic acid (FnA) were detected in wild‐type extracts, but not in those of the fub1Δ mutant (Fig. 1A). Interestingly, the total amount of mycotoxin (FA + FnA) was approximately 2.5 times higher in CDA than in PDA cultures, with an FA : FnA ratio of 29.5 : 1 in the former and 1 : 2.75 in the latter (Fig. 1A). These data are consistent with those reported in F. fujikuroi (Niehaus et al., 2014). In addition to FA and FnA, beauvericin (Bea) and bikaverin (Bik) were detected in all samples. Interestingly, both compounds were more abundant in the fub1Δ cultures compared with the wild‐type, especially in CDA (Fig. 1B). Reintroduction of the intact fub1 allele into fub1Δ, yielding the complemented fub1ΔC strain (Fig. S2, see Supporting Information), fully restored the wild‐type FA levels (Fig. 1). Thus, Fub1 is responsible for the production of FA and its derivatives in F. oxysporum.
Figure 1.
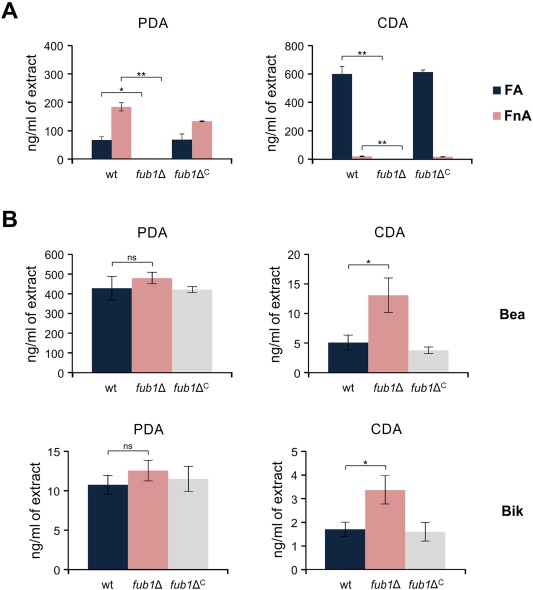
Fub1 is required for the production of fusaric acid (FA) and its derivatives in Fusarium oxysporum. (A) The amounts of FA and fusarinolic acid (FnA) in cultures of the indicated strains, grown for 3 days on potato dextrose agar (PDA) or Czapek‐Dox agar (CDA), were quantified by liquid chromatography/tandem mass spectrometry and expressed as nanograms per millilitre of extract. (B) The quantification of beauvericin (Bea) and bikaverin (Bik) was performed as in (A). wt, wild‐type strain. Bars represent standard errors from two independent fungal cultures. *P < 0.05; **P < 0.001; ns, not significant.
Next, we tested the potential toxicity of FA on F. oxysporum. When the wild‐type strain or fub1Δ were cultured on PDA supplemented with 0.25 or 0.5 mg/mL FA, both showed a significant and comparable reduction in radial growth, whereas no growth was detectable at 0.75 mg/mL FA (Fig. S3, see Supporting Information).
Effect of pH and nutrients on fub1 transcript levels and FA production
In F. fujikuroi, fub1 transcription is positively regulated by the pH response factor PacC at pH 8, but not at pH 4 (Niehaus et al., 2014). We noted that CDA has an initial pH of 6.8 ± 0.2, whereas PDA has an initial pH of about 5.6 ± 0.2. Moreover, the pH in CDA, in which NaNO3 is the sole nitrogen source, tended to increase during fungal growth (data not shown). To discriminate between the effects of medium composition and pH on FA biosynthesis, we germinated conidia of the wild‐type strain in potato dextrose broth (PDB) and transferred the germlings to Czapek‐Dox liquid (CDL) or fresh PDB buffered to either pH 5 or pH 7 (see Experimental procedures for details). Unexpectedly, in both media, fub1 transcription and FA production were much higher under moderate acidic conditions, although, as expected, CDL induced more FA (Fig. 2A, B). The effect of pH on FA production was stronger than that of the medium composition, as reflected by the finding that FA production was higher in PDB at pH 5 than in CDL at pH 7 (Fig. 2A, B). Our data indicate that both pH and nutrients are important factors in the regulation of FA biosynthesis, and that this regulation may differ between F. oxysporum and F. fujikuroi. We next tested the role of PacC in fub1 regulation using a pacC loss‐of‐function mutant (Caracuel et al., 2003). When germlings of the different strains were grown in glutamine minimal medium (GMM) buffered to either pH 5 or pH 7, fub1 transcript levels were 10 times lower in pacCΔ relative to the wild‐type at both pH values (Fig. 2C). Thus, PacC functions as a positive regulator of fub1 within this pH range.
Figure 2.
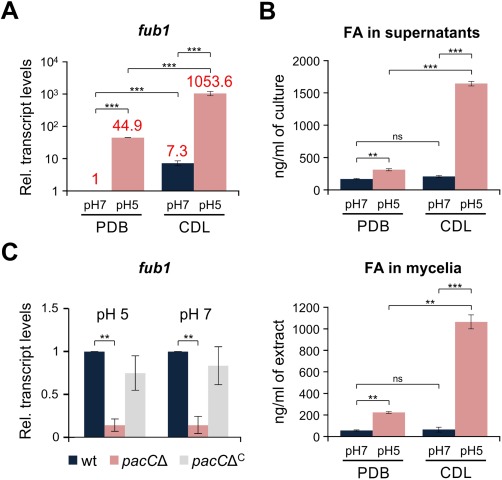
pH and medium composition regulate fub1 transcript levels and fusaric acid (FA) production. (A) Quantitative real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) was performed in the wild‐type strain germinated for 16 h in potato dextrose broth (PDB) and then transferred for 3 h to fresh PDB or Czapek‐Dox liquid (CDL) buffered at the indicated pH with 100 mm 2‐(N‐morpholino)ethanesulfonic acid (MES). Transcript levels of fub1 are expressed relative to those in PDB at pH 7 (see numbers above the columns for exact data). Bars represent standard errors from two independent biological experiments with three technical replicates each. (B) Quantification of FA in culture supernatants (top panel) and mycelia (bottom panel) of the wild‐type strain grown as in (A) performed by liquid chromatography/tandem mass spectrometry. Bars represent standard errors from two independent fungal cultures. (C) Quantitative real‐time RT‐PCR was performed in the indicated strains germinated for 16 h in glutamine minimal medium (GMM) and then transferred for 3 h to fresh GMM buffered at the indicated pH with 100 mm MES. Transcript levels of fub1 are expressed relative to those of the wild‐type at both pH values. Bars represent standard errors from two independent biological experiments with three technical replicates each. **P < 0.001; ***P < 0.0001; ns, not significant; wt, wild‐type.
Chromatin structure at the fub1 locus is controlled by the global regulator of secondary metabolism LaeA and the pH response factor PacC
Although the fub1Δ mutants did not show a detectable growth defect on PDA or CDA, we noted that their growth in the presence of hygromycin B was markedly reduced (Figs 3A and S4, see Supporting Information). Interestingly, the complemented fub1ΔC strains showed a similar growth defect on hygromycin, whereas the transformants carrying an ectopic insertion of the knockout construct (Ect) (Fig. S1C) did not (Figs 3A and S4). We hypothesized that this phenotype could be caused by a chromatin regulatory effect on transcription of the hph hygromycin resistance gene inserted at the fub1 locus. In line with this hypothesis, we found that hph transcript levels were between 30 and 50 times higher in Ect than in fub1Δ and fub1ΔC (Fig. 3B). LaeA is a global regulator of secondary metabolite gene clusters in different fungi (Bok and Keller, 2004; Butchko et al., 2012; Lopez‐Berges et al., 2013; Wiemann et al., 2010), and has been reported previously to regulate FA production in Fusarium (Lopez‐Berges et al., 2013; Niehaus et al., 2014). Transcript levels of fub1 in the wild‐type and the laeAΔC strains were between 300 and 500 times higher than in the laeAΔ mutant (Fig. 3C). We next examined the role of LaeA in chromatin remodelling and transcriptional regulation at the F. oxysporum fub1 locus, using real‐time quantitative polymerase chain reaction (PCR) with promoter‐ and gene‐specific primers (Fig. 4A) on genomic DNA (gDNA) obtained from mycelia treated with micrococcal nuclease (MNase) (Fig. S5, see Supporting Information). Relative chromatin accessibility, calculated as the ratio of amplification from untreated versus MNase‐treated mycelia, was about six times higher in wild‐type and laeAΔC compared with laeAΔ (Fig. 4B). Moreover, relative chromatin accessibility was significantly lower at pH 7 in comparison with pH 5, and in a pacCΔ strain at both pH values compared with the wild‐type (Fig. 4C, D), in line with the previous finding that fub1 transcript levels are lower at pH 7 and in pacCΔ (Fig. 2A, C). We conclude that chromatin accessibility and transcription at the fub1 locus, as well as the production of FA and its derivatives, are positively regulated by LaeA, moderate acidic pH and PacC.
Figure 3.
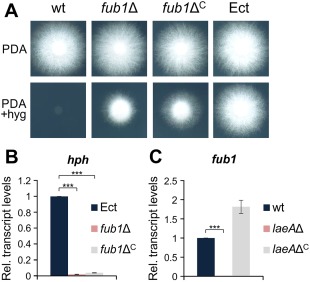
Transcript levels of fub1 are controlled by LaeA. (A) Colonies of the indicated strains grown on potato dextrose agar (PDA) with or without 50 mg/mL hygromycin B for 3 days at 28 ºC. (B, C) Quantitative real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) was performed in the indicated strains germinated for 16 h in potato dextrose broth (PDB) and then transferred to fresh PDB for 1 h. Transcript levels of the hph (B) and fub1 (C) genes are expressed relative to those of the ectopic transformant and the wild‐type strain, respectively. wt, wild‐type strain; Ect, ectopic transformant. Bars represent standard errors from two independent biological experiments with three technical replicates each. ***P < 0.0001.
Figure 4.

LaeA, pH and PacC regulate chromatin modifications at the fub1 locus. (A) Physical map of the promoter region of the fub1 gene located in the fusaric acid (FA) gene cluster. Primers used for chromatin analysis are indicated. (B–D) Real‐time quantitative polymerase chain reaction (PCR) performed on genomic DNA (gDNA) of the indicated strains grown as in Fig. 3B, C. Relative chromatin accessibility was calculated as the ratio of amplification levels obtained with gDNA from untreated mycelia versus gDNA from micrococcal nuclease (MNase)‐treated mycelia, and represented relative to that of the wild‐type (B), the wild‐type at pH 5 (C) or the wild‐type at different pH values (D), with each of the indicated primer pairs (see Table S1). wt, wild‐type strain. Bars represent standard errors from two independent biological experiments with three technical replicates each. *P < 0.05; **P < 0.001; ***P < 0.0001.
Fub1 and FA are not required for the growth of F. oxysporum under copper‐, iron‐ or zinc‐limiting conditions
The ability of FA to chelate metal ions, such as iron or copper, has been known for a long time (Lakshminarayanan and Subramanian, 1955; Malini, 1966; Pan et al., 2010; Tamari and Kaji, 1952). In a recent study, FA has been shown to chelate different metal ions, including Fe2+, Fe3+, Cu2+, Mn2+ and Zn2+ (Ruiz et al., 2015; Yin et al., 2015). We thus asked whether the production of FA is required for the growth of F. oxysporum under metal‐limiting conditions. The depletion of copper, iron or zinc was achieved by the addition of the specific chelators bathocuproinedisulfonic acid disodium salt (BCS), bathophenanthrolinedisulfonic acid disodium salt hydrate (BPS) and N,N,N′,N′‐tetrakis(2‐pyridylmethyl)ethylenediamine (TPEN), respectively (Fig. 5A). Unexpectedly, although copper is an essential micronutrient in most living organisms (Vulpe and Packman, 1995), we observed no detectable growth defect in F. oxysporum grown under copper limitation (Fig. 5A), even in the presence of BCS concentrations up to 1 mm (data not shown). A similar result has been reported previously in Aspergillus fumigatus (Park et al., 2014). By contrast, depletion of iron and zinc resulted in severe growth defects, as expected. In any case, inactivation of Fub1 had no additional effect on growth (Fig. 5A). Next, we asked whether Fub1 was required for fungal growth at toxic concentrations of copper, iron or zinc. When the wild‐type and fub1Δ strains were grown on GMM with up to 5 mm of the different metal ions, no significant differences were observed between strains (Fig. 5B). These results demonstrate that FA is not essential for growth under limiting or toxic concentrations of copper, iron or zinc. However, we noted that transcript levels of fub1 were significantly reduced in the presence of these three metal ions (Fig. 5C).
Figure 5.
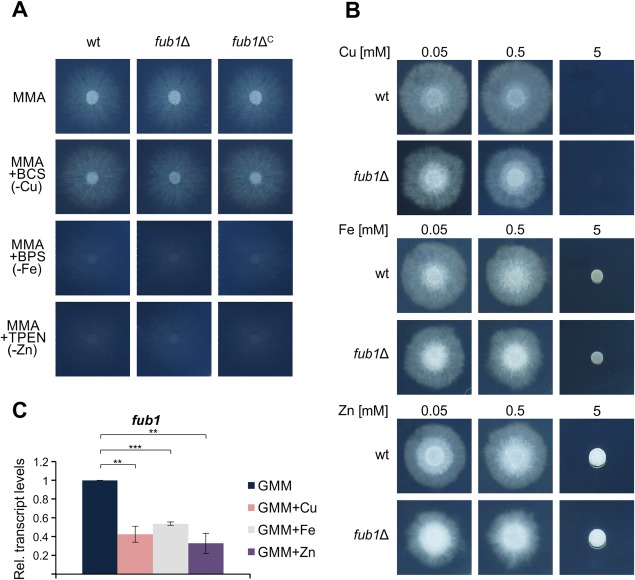
Expression of fub1 is repressed by copper, iron and zinc. (A) Growth of the indicated strains on minimal medium agar (MMA) with or without the indicated metal chelators. Plates were cultured for 3 days at 28 ºC. (B) Growth of the indicated strains on solid glutamine minimal medium (GMM) supplemented with the indicated concentrations of copper, iron or zinc. Plates were cultured for 3 days at 28 ºC. (C) Quantitative real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) was performed in the wild‐type strain germinated for 16 h in GMM without copper, iron or zinc and then supplemented, or not, with 50 µm CuSO4, FeSO4 or ZnSO4 for 2 h. Transcript levels of fub1 are expressed relative to those in GMM. wt, wild‐type strain; BCS, bathocuproinedisulfonic acid disodium salt; BPS, bathophenanthrolinedisulfonic acid disodium salt hydrate; TPEN, N,N,N′,N′‐tetrakis(2‐pyridylmethyl)ethylenediamine. Bars represent standard errors from two independent biological experiments with three technical replicates each. **P < 0.001; ***P < 0.0001.
FA toxicity in tomato plants is reversed by the exogenous addition of copper, iron and zinc
The production and phytotoxic properties of FA have been studied over the past 75 years (Bacon et al., 1996; Dong et al., 2014; Gäumann, 1957, 1958; Yabuta et al., 1937), and a number of mechanisms for FA toxicity have been suggested, most related to modifications in the plant cell membrane (D'Alton and Etherton, 1984). To test FA toxicity, roots of 3‐week‐old tomato plants were immersed in sterile water with or without FA. Plants maintained in the presence of 0.5–1 mm FA exhibited a progressive depigmentation of the stem, most probably a result of anthocyanin degradation, followed by a general loss of turgor and, finally, wilting of the entire plant (Fig. 6A). Importantly, the external addition of copper, iron or zinc, either to the FA solution or by foliar spraying, a process by which leaves can take up ions through the stomata and distribute them throughout the plant (Eddings and Brown, 1967; Neumann and Prinz, 1975), rendered plants more resistant to FA and significantly increased stem strength and pigmentation (Fig. 6B–D). Furthermore, the phytotoxic effect of FA was partially recapitulated by immersion of the roots in a solution containing the membrane‐permeable chelator TPEN (Fig. 7), but not the membrane‐impermeable chelators BPS or BCS (data not shown). Collectively, these results suggest that the phytotoxicity of FA is mediated by chelation of metal ions inside the plant.
Figure 6.
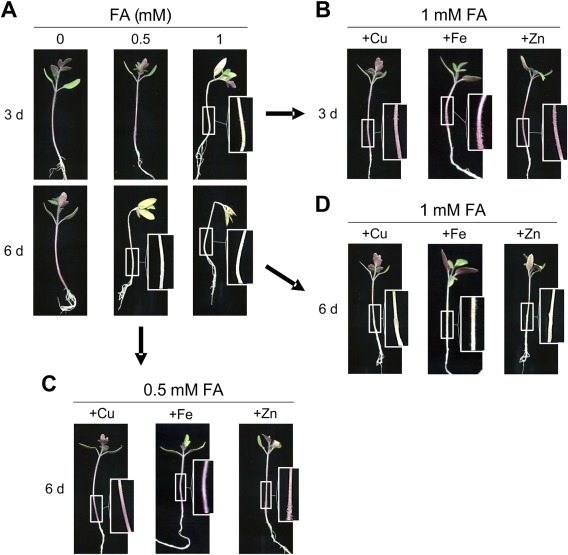
Phytotoxic effect of fusaric acid (FA) on tomato plants is remediated by exogenous copper, iron or zinc. (A) Roots of 3‐week‐old seedlings of tomato plants (cultivar Monika) were immersed in sterile water with the indicated concentrations of FA for 3 and 6 days. (B–D) Leaves of plants were sprayed with 0.025% CuSO4, FeSO4 or ZnSO4 solutions before immersing roots in FA solution. Boxed areas are shown at double magnification.
Figure 7.

The membrane‐permeable zinc chelator N,N,N′,N′‐tetrakis(2‐pyridylmethyl)ethylenediamine (TPEN) causes similar phytotoxicity symptoms in tomato plants to fusaric acid. Leaves of tomato plants were pretreated, or not, with a 0.025% ZnSO4 solution and roots were immersed in sterile water containing 4 µm of the zinc chelator TPEN for 3 and 6 days. Boxed areas are shown at double magnification.
FA is a virulence factor of F. oxysporum on tomato plants and immunodepressed mice
We noted that the expression of fub1 in F. oxysporum was markedly up‐regulated during the early stages of plant infection (Fig. 8A) and therefore tested the role of FA production in virulence. Tomato plants whose roots were inoculated with conidia of the F. oxysporum wild‐type or fub1ΔC strains showed progressive wilt symptoms and usually died before day 25 post‐inoculation (dpi) (Fig. 8B). In contrast, plants inoculated with the fub1Δ mutant displayed a significantly reduced mortality rate (Fig. 8B) and most survived the assay, developing only mild disease symptoms. Moreover, the amount of fungal biomass in roots and stems was markedly reduced in fub1Δ in comparison with the wild‐type and complemented strains (Fig. 8C). Thus, FA is required for full virulence of F. oxysporum in tomato plants.
Figure 8.
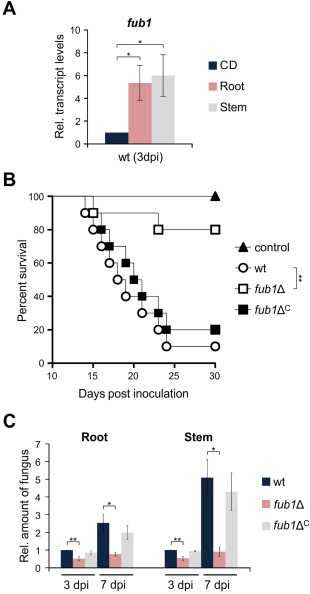
Fusaric acid (FA) production is required for full virulence of Fusarium oxysporum on tomato plants. (A) Quantitative real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) was performed in the wild‐type strain germinated for 16 h in potato dextrose broth (PDB) and then transferred to Czapek‐Dox liquid (CDL) for 3 h or for inoculated tomato roots and stems at 3 days post‐inoculation (dpi). Transcript levels of fub1 are expressed relative to those in CDL. (B) Groups of 10 tomato plants (cultivar Monika) were inoculated by dipping roots into a suspension of 5 × 106 freshly obtained microconidia/mL of the indicated fungal strains. Percentage survival was plotted for 30 days. Data shown are from one representative experiment. Experiments were performed three times with similar results. (C) Quantitative real‐time PCR was used to measure the relative amount of fungal DNA in total genomic DNA extracted from tomato roots and stems at 3 and 7 dpi with the indicated strains. Amplification levels are expressed relative to those of plants infected with the wild‐type strain. wt, wild‐type strain. Bars represent standard deviations from two independent biological experiments with three technical replicates each.*P < 0.05; **P < 0.001.
As the tomato pathogenic F. oxysporum strain can also infect and kill immunosuppressed mice (Ortoneda et al., 2004), we tested the role of FA production during infection of a mammalian host. Inoculation with 107 conidia of the wild‐type or fub1ΔC strain resulted in the killing of all animals before 15 dpi (Fig. 9A), and transcripts of fub1 were detected inside the host (Fig. S6, see Supporting Information). However, animals inoculated with two independent fub1Δ mutants showed significantly delayed mortality (Fig. 9A). In contrast with plant infection, the fungal burden in kidney, liver and lung of surviving mice did not differ significantly between the strains (Fig. 9B). These results suggest that FA contributes to the virulence of F. oxysporum in mammals, but is not required for dissemination in the host.
Figure 9.
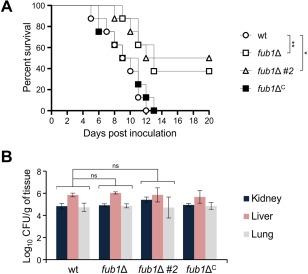
Fusaric acid (FA) is a virulence factor in mice. (A) Groups of 10 immunosuppressed Oncins France 1 male mice were inoculated with 107 microconidia of the indicated strains by lateral tail vein injection. Percentage survival was plotted for 20 days. Data shown are from one representative experiment. Experiments were performed three times with similar results. (B) Four randomly chosen surviving mice inoculated with 107 microconidia of the indicated strains were sacrificed at 5 days post‐inoculation (dpi) and homogenates obtained from the indicated organs were quantitatively cultured on potato dextrose agar (PDA). wt, wild‐type strain. *P < 0.05; **P < 0.001; ns, not significant.
Discussion
FA was discovered almost 80 years ago (Yabuta et al., 1937) and was the first fungal toxin whose production was detected in planta (Gäumann, 1957). Its strong phytotoxicity (Niehaus et al., 2014; Stipanovic et al., 2011), moderate toxicity in animals (Porter et al., 1995; Yin et al., 2015) and bacteria (Bacon et al., 2006; Ruiz et al., 2015), and its pharmacological properties (Song and Yee, 2001; Wang and Ng, 1999), make the study of FA biosynthesis and regulation of great interest. Moreover, FA inhibits the growth of fungi, including its producer Fusarium. However, FA‐producing strains use a variety of strategies, such as active export or enzymatic modification, to protect themselves from the toxin (Crutcher et al., 2015; Studt et al., 2016). Since the recent discovery of the FA biosynthetic gene cluster, different components of the cluster have been characterized (Brown et al., 2012, 2015; Niehaus et al., 2014; Studt et al., 2016). Inactivation of Fub1, the PKS acting in the first step of the FA biosynthetic pathway, completely abolishes the production of FA and its derivatives in different Fusarium species (Brown et al., 2012; Niehaus et al., 2014) (this work). Here, we used targeted deletion of fub1 to demonstrate, for the first time, a role of FA in the virulence of the cross‐kingdom pathogen F. oxysporum on plant and mammalian hosts.
Chromatin‐mediated regulation of FA production
The regulation of FA production has been studied for almost 80 years. Initially, it was proposed that FA was mainly produced under alkaline conditions (Yabuta et al., 1939), whereas later studies suggested that nitrogen sufficiency and slightly acidic media were optimal for FA production (Pitel and Vining, 1970). Here, we compared two different media, potato dextrose (PD) and Czapek‐Dox (CD), in both solid and liquid versions. Although PD is a richer and more complex medium, we found that the production of FA was higher in CD, a medium that has been known for a long time to promote FA production (Löffler and Mouris, 1992). By contrast, Bik and Bea were preferentially produced in PD. The exact reason for this difference is currently unknown. We hypothesized that pH could act as a key regulatory factor, and observed significantly higher fub1 expression and FA production at pH 5 relative to pH 7. Our results are in contrast with those reported in F. fujikuroi, showing a higher expression of fub1 at pH 8 relative to pH 4, requiring the alkaline pH regulator PacC (Niehaus et al., 2014). We also confirmed that PacC is required for full expression of fub1 at both pH 5 and pH 7. The seemingly contradictory results between F. oxysporum and F. fujikuroi could be explained by the different experimental conditions used in the two studies: 2‐(N‐morpholino)ethanesulfonic acid (MES)‐buffered versus unbuffered media, respectively (Niehaus et al., 2014). It is known that the pH of an unbuffered culture can change rapidly during fungal growth. However, the optimum pH for fub1 expression and FA production in F. oxysporum could be around pH 5 or higher, a range in which PacC is still active. In line with this hypothesis, pacC transcript levels were similar at pH 5 and pH 7, but almost undetectable at pH 4 (Caracuel et al., 2003). The global regulator of secondary metabolism LaeA (Bok and Keller, 2004; Butchko et al., 2012; Lopez‐Berges et al., 2013; Wiemann et al., 2010) has been shown previously to regulate FA production in Fusarium (Lopez‐Berges et al., 2013; Niehaus et al., 2014). LaeA contains a conserved S‐adenosylmethionine (SAM)‐binding site essential for its function, contributes to histone H3 lysine 9 trimethylation (Reyes‐Dominguez et al., 2010) and links transcriptional and epigenetic control of gene expression (Sarikaya‐Bayram et al., 2014). In line with previous reports suggesting a positive role of LaeA in FA biosynthesis (Lopez‐Berges et al., 2013; Niehaus et al., 2014), we showed here that the inactivation of LaeA leads to a significant decrease in chromatin accessibility at the FA gene cluster. These findings, together with the reduced expression of the hph gene when inserted at the site of fub1, suggest a major regulatory function of LaeA in remodelling chromatin structure at the F. oxysporum FA locus. In addition, we showed that moderate acidic pH and PacC contribute to an increase in chromatin accessibility at the fub1 locus. Although our data suggest that this contribution requires LaeA, this remains to be confirmed experimentally. The fact that inactivation of LaeA has, by far, the strongest effect on the expression of the FA gene cluster and FA production suggests that other stimuli, such as nutrients or pH, may converge on this master regulator of secondary metabolism to regulate expression of the gene cluster.
FA has long been known for its ability to chelate metal ions (Lakshminarayanan and Subramanian, 1955; Malini, 1966; Pan et al., 2010; Tamari and Kaji, 1952). However, the regulation of FA biosynthesis by metals has not been studied so far. Here, we showed that transcript levels of fub1 are negatively regulated by copper, iron or zinc. Similarly, transcript levels of sidC, a LaeA‐regulated gene functioning in the biosynthesis of the siderophore ferricrocin, are also down‐regulated in the presence of iron (Eisendle et al., 2004; Lopez‐Berges et al., 2013; Perrin et al., 2007). Although this suggests that FA might function in metal uptake, we found that fub1 was not essential for the growth of F. oxysporum in copper‐, iron‐ or zinc‐limiting conditions, most probably because more specific and efficient uptake mechanisms are present in filamentous fungi, such as high‐affinity copper and zinc transporters (Park et al., 2014; Vicentefranqueira et al., 2005) and siderophore‐assisted iron uptake (Schrettl and Haas, 2011). Alternatively, metal‐chelating FA might be used by Fusarium to inhibit microbial competitors in the soil or to improve growth at toxic metal concentrations. Indeed, FA is exported in F. fujikuroi and F. oxysporum f. sp. vasinfectum via the Major Facilitator Superfamily (MFS) transporters Fub11 and FubT, respectively (Crutcher et al., 2015, Studt et al., 2016). However, we found that a lack of FA production was not detrimental during fungal growth in toxic copper, iron or zinc conditions.
Mechanism of FA phytotoxicity and role in virulence
Early studies established the phytotoxic activity of FA and its role in the induction of wilt symptoms in plants (Gäumann, 1957, 1958; Yabuta et al., 1937). Our study confirmed that tomato seedlings develop typical wilt symptoms when their roots are exposed to FA. The fact that wilting was observed in the cotyledons and lower leaves suggests that FA is transported and distributed throughout the entire plant. Similar wilt symptoms have been reported in water melon seedlings (Hong‐Sheng et al., 2008).
The precise mechanism of phytotoxicity of FA remains unknown. A number of studies have suggested that it could be related to its ability to chelate different metal ions (Gäumann, 1958; Lakshminarayanan and Subramanian, 1955; Ruiz et al., 2015; Tamari and Kaji, 1952). Here, we showed that the addition of copper, iron or zinc to FA‐treated plants significantly reduces wilting. Importantly, the inhibition of FA toxicity was also functional when the metal ions and FA were applied to different parts of the plant (leaves and roots, respectively), indicating that the chelating mechanism occurs inside the plant. This is further supported by the fact that the membrane‐permeable metal chelator TPEN, but not the membrane‐impermeable chelators BPS or BCS, was able to exert a toxic effect similar to that of FA. Although additional mechanisms of FA toxicity cannot be ruled out, our results clearly support a causal link between FA phytotoxicity and metal chelation.
FA is one of the first fungal toxins for which a functional role in virulence has been proposed (Gäumann, 1957, 1958) and several studies have provided circumstantial evidence linking FA production to plant pathogenicity (Dong et al., 2014; Gapillout et al., 1996; Venter and Steyn, 1998). However, to date, no formal proof for such a role has been provided. Here, we demonstrated that mutants lacking fub1, which are unable to produce FA or its derivatives, are significantly reduced in their capacity to cause mortality in tomato plants. Interestingly, these mutants also caused less mortality in immunosuppressed mice, showing, for the first time, the relevance of FA production during fungal infection of mammals. Although the pH of mammalian blood is around pH 7.3, which is not favourable for FA production, small amounts of FA might be sufficient to promote fungal virulence in mammals. In addition, it cannot be ruled out that FA production is under positive regulation inside the host. Indeed, FA has been shown previously to be produced in blood cultures in an LaeA‐ and VeA‐dependent manner (Lopez‐Berges et al., 2013). Previously, the mycotoxin Bea has also been shown to contribute to infection of F. oxysporum in plants and mice. This suggests that the production of secondary metabolites, many of which are regulated by the Velvet complex and LaeA, could play a role in the capacity of F. oxysporum to attack both plant and animal hosts. In line with this idea, mutants lacking VeA or LaeA are significantly attenuated in virulence on tomato plants and mice (Lopez‐Berges et al., 2013), as are Velvet complex mutants in other human and plant pathogenic fungi (Bok et al., 2005; Jiang et al., 2011; Laskowski‐Peak et al., 2012; Lee et al., 2012; Lopez‐Berges et al., 2013; Merhej et al., 2012; Myung et al., 2009; Webster and Sil, 2008; Wiemann et al., 2010). Additional studies, including investigations on the combinatory/synergistic effects of co‐occurring mycotoxins, are required to fully understand the role of secondary metabolite production in the cross‐kingdom pathogenicity of F. oxysporum.
Experimental Procedures
Fungal isolates and culture conditions
Fusarium oxysporum f. sp. lycopersici race 2 wild‐type isolate 4287 (FGSC 9935) was used in all experiments. Fungal strains were stored as microconidial suspensions at −80 °C with 30% glycerol. For the extraction of gDNA and microconidia production, cultures were grown in PDB at 28 °C (Di Pietro and Roncero, 1998). For the analysis of gene expression and relative chromatin accessibility, freshly obtained microconidia were germinated for 14–16 h in PDB or GMM. Germlings were harvested by filtration, washed three times in sterile water and transferred to fresh PDB, CDL or GMM with or without 50 µm CuSO4, FeSO4 or ZnSO4 for the indicated time periods. pH 5 and pH 7 buffered conditions were achieved using 100 mm MES, when indicated. For the determination of colony growth, 2 × 104 microconidia were spotted onto PDA, CDA, minimal medium agar (MMA) or GMM with or without FA (0–0.75 mg/mL), with or without 200 µm BPS, 200 µm BCS or 4 µm TPEN, and with or without CuSO4, FeSO4 or ZnSO4 (0.05–5 mm). Plates were incubated at 28 °C for the indicated time periods. All experiments included two replicates and were performed at least three times with similar results.
Fungal strains
PCRs were routinely performed with VELOCITY™ DNA Polymerase (Bioline, London, UK) using an MJ Mini™ Personal Thermal Cycler (Bio‐Rad, Madrid, Spain) (see Table S1, Supporting Information, for a complete list of primer sequences used in this study). All fungal transformations and purification of the transformants by monoconidial isolation were performed as described previously (Di Pietro and Roncero, 1998). The cassette for targeted replacement of the entire coding region of the F. oxysporum fub1 gene with the hygromycin B resistance marker (Punt et al., 1987) was assembled by a fusion PCR method (Szewczyk et al., 2006). DNA fragments flanking the fub1 coding region were amplified from gDNA of F. oxysporum wild‐type with the primers fub1‐F1 + fub1‐R1 and fub1‐F2 + fub1‐R2, respectively, whereas the hygromycin B resistance marker, under the control of the Aspergillus nidulans gpdA promoter and trpC terminator, was amplified from the pAN7‐1 plasmid (Punt et al., 1987) with primers fub1‐hph‐F + fub1‐hph‐R. The three DNA fragments were then PCR fused with the primers fub1‐F1n + fub1‐R2n. The fub1Δ allele obtained was used to transform protoplasts of the F. oxysporum wild‐type strain to hygromycin B resistance (Fig. S1B). Transformants showing homologous insertion of the construct were genotyped by PCR of gDNA with primers fub1‐F1 + fub1‐R2 (not shown) and by Southern blot analysis (Fig. S1C). To generate a construct for complementation of the fub1Δ strain, a 9645‐bp fragment, spanning from 1077 bp upstream of the wild‐type F. oxysporum fub1 translation initiation codon to 1092 bp downstream of the translation termination codon, was amplified by PCR with the primers fub1‐F1 + fub1‐R2. The amplified fragment was used to co‐transform protoplasts of the fub1Δ strain with the phleomycin B resistance gene under the control of the A. nidulans gpdA promoter and trpC terminator, amplified from the pAN8‐1 plasmid (Mattern et al., 1988) with the primers gpdA−15b + trpC−8b (Fig. S2A). Several phleomycin‐resistant co‐transformants were analysed for the presence of a functional fub1 allele by PCR with the gene‐specific primers fub1‐F3 + fub1‐R3 (Fig. S2B). Among the different complemented strains, we selected one in which the production of FA and derivatives (Fig. 1) and fub1 transcript levels returned to wild‐type values (Fig. S2C, D).
Nucleic acid manipulations and quantitative real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) analysis
Total RNA and gDNA were extracted from F. oxysporum mycelia following previously reported protocols (Chomczynski and Sacchi, 1987; Raeder and Broda, 1985). The quality and quantity of extracted nucleic acids were determined by running aliquots in ethidium bromide‐stained agarose gels and by spectrophotometric analysis in a NanoDrop ND‐1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA), respectively. Routine nucleic acid manipulations were performed according to standard protocols (Sambrook and Russell, 2001). DNA and protein sequence databases were searched using the blast algorithm (Altschul et al., 1990). Quantitative RT‐PCR was performed as described previously (Lopez‐Berges et al., 2010, 2012) using FastStart Essential DNA Green Master (Roche Diagnostics SL, Barcelona, Spain) in a CFX Connect Real‐Time System (Bio‐Rad). Gene‐specific primers (see Table S1) were designed to flank an intron, if possible. Transcript levels were calculated by comparative ΔCt and normalized to act1.
Analysis of chromatin structure
Mycelia of F. oxysporum strains grown under the indicated conditions were harvested by filtration, lyophilized and ground to a fine powder in a Mini‐BeadBeater 8 (BioSpec Products, Bartlesville, OK, USA). Nuclease digestion was performed as described previously (Basheer et al., 2009; Gonzalez and Scazzocchio, 1997; Lopez‐Berges et al., 2013). Briefly, 20 mg of lyophilized mycelium was suspended in 1 mL of MNase buffer (250 mm sucrose, 60 mm KCl, 15 mm NaCl, 0.5 mm CaCl2, 3 mm MgCl2), and 300 mL of the suspension were treated for 5 min with 3 U of MNase (Sigma, Madrid, Spain) at 37 °C. The reaction was terminated by adding stop buffer [2% sodium dodecylsulfate (SDS), 40 mm ethylenediaminetetraacetic acid (EDTA)]. DNA was obtained by phenol–chloroform extraction, precipitated, washed with 70% ethanol, dissolved in water and treated with RNAse (see Fig. S5). Quantitative real‐time PCR was performed as described above using promoter‐ and gene‐specific primers (see Table S1). Chromatin accessibility was expressed by comparative ΔCt as the ratio between amplification levels from untreated gDNA relative to those obtained from MNase‐digested gDNA. Values were presented relative to those of the wild‐type strain.
Mycotoxin quantification
The quantification of FA and derivatives was performed as described previously (Lopez‐Berges et al., 2013). Samples were obtained from fungal colonies grown for 3 days at 28 °C on PDA or CDA, and from mycelia and supernatant of the wild‐type strain germinated in PDB for 16 h, and then transferred for 3 h to fresh PDB or CDL buffered to pH 5 or pH 7. Samples were homogenized in acetonitrile–water–glacial acetic acid (79 : 20 : 1, v/v/v) with a Homogenizer Workcenter T10 basic (IKA®, Wilmington, NC, USA) for 1 min at a rate of 4 mL solvent per gram of sample. The mix was re‐homogenized after 2 min of repose, filtered, centrifuged for 10 min at 12 000 g and the supernatant was lyophilized. Dry crude extracts were reconstituted in the solvent, and mycotoxin detection and quantification were performed with a QTrap 5000 LC‐MS/MS System (Applied Biosystems, Foster City, CA, USA) equipped with a TurboIonSpray electrospray ionization (ESI) source and a 1290 Series UPLC System (Agilent, Waldbronn, Germany), as described previously (Malachova et al., 2014). Supernatant samples were lyophilized directly and then reconstituted in the solvent for quantification.
Determination of FA toxicity on tomato plants
Three‐week‐old seedlings of tomato plants (cultivar Monika) were individually root immersed in inoculum tubes containing pH 6 sterile water with different FA concentrations or 200 µm BPS, 200 µm BCS or 4 µm TPEN, and placed in a glasshouse for the indicated time periods. Copper, iron and zinc foliar spraying was performed, when indicated, 2 h before root immersion. Briefly, plant roots were carefully covered with cling film and leaves were sprayed twice with 0.025% CuSO4, FeSO4 or ZnSO4 in 0.1% Tween‐20 solution. When the leaves were completely dry, the roots were washed three times in sterile water before immersion in the indicated solutions. Symptoms were monitored daily and scored 3 and 6 days after FA or chelator treatment.
Plant infection assays
Tomato root inoculation assays were performed as described previously (Di Pietro and Roncero, 1998) using 2‐week‐old tomato seedlings (cultivar Monika). The severity of disease symptoms and plant survival were recorded daily for 30 days. Ten plants were used for each treatment. Virulence experiments were performed at least three times with similar results. Plant survival was calculated by the Kaplan–Meier method and compared among groups using the log‐rank test. Data were analysed with the software GraphPad Prism 4. The quantification of fungal biomass in planta was performed as described previously (Pareja‐Jaime et al., 2010) using total gDNA extracted from tomato roots or stems infected with F. oxysporum strains at 3 or 7 dpi. Relative amounts of fungal gDNA were calculated by comparative ΔCt of the Fusarium act1 gene normalized to the tomato EFα1 gene (see Table S1).
Animal infection assays
Mice were cared for in accordance with the principles outlined by the European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes (European Treaty Series, No. 123; http://conventions.coe.int/Treaty/en/Treaties/Html/123.htm). Experimental conditions were approved by the Animal Welfare Committee of the Faculty of Medicine, Universitat Rovira i Virgili. Infection assays with immunodepressed mice were performed as described previously (Ortoneda et al., 2004). Briefly, groups of 10 Oncins France (OF) 1 male mice (Charles River, Criffa S.A., Barcelona, Spain) were immunosuppressed with an intraperitoneal 200 mg/kg dose of cyclophosphamide (Laboratorios Funk S.A., Barcelona, Spain) 2 days before inoculation, and then every 5 days, and infected by injecting 0.2 mL of an inoculum of 107 conidia into a lateral vein of the tail. Survival was recorded daily for the indicated time periods. Infection experiments with each individual strain were performed at least three times. Survival was estimated by the Kaplan–Meier method and compared among groups using the log‐rank test. To determine fungal tissue burden, randomly chosen surviving mice inoculated with 107 conidia were sacrificed at 5 dpi. Kidneys, livers and lungs were aseptically removed, weighed and homogenized in sterile saline, and 10‐fold serial dilutions were spread onto PDA. The plates were incubated at 28 °C, the colonies were counted after 3 days and the number of colony‐forming units (CFU) per gram of organ was calculated. Fungal colony counts were converted to log10 and compared using the analysis of variance test. Data were analysed with the software GraphPad Prism 4.
Accession numbers
Sequence data can be found in the GenBank/EMBL database or in the Fusarium Comparative Genome database under the following accession numbers: Fub1, FOXG_15248; Act1, FOXG_01569; EFα1, NC_015443; pAN7‐1 (PgpdA‐hygr‐TtrpC), Z32698; pAN8‐1 (PgpdA‐phleor‐TtrpC), Z32751.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website.
Fig. S1 Identification of the Fusarium oxysporum fusaric acid (FA) gene cluster and fub1 knockout strategy. (A) Conserved synteny between Fusarium fujikuroi and F. oxysporum FA gene clusters. Note that the two genes inserted between fub3 and fub4 in F. oxysporum are present in F. fujikuroi in another chromosome. (B) Fusarium oxysporum fub1 locus and targeted gene disruption construct. (C) Southern blot analysis. Genomic DNA of the indicated strains was treated with XhoI, separated on a 0.7% agarose gel, transferred to a nylon membrane and hybridized with the DNA probe indicated in (B). wt, wild‐type.
Fig. S2 Generation and selection of the fub1Δ complemented strain (fub1ΔC). (A) Strategy of fub1Δ complementation by co‐transformation with a fub1 wild‐type allele and the phleomycin resistance marker. The relative positions of the polymerase chain reaction (PCR) primers used for genotyping are indicated. phleor, phleomycin resistance gene. (B) PCR amplification of genomic DNA (gDNA) of the indicated strains using primers F3 and R3. The complemented strains, fub1ΔC and fub1ΔC #2, produce a banding pattern consistent with the integration of an intact fub1 allele. wt, wild‐type strain. (C) Quantitative real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) performed in the indicated strains germinated for 16 h in potato dextrose broth (PDB) and transferred for 3 h to Czapec‐Dox liquid (CDL). Transcript levels of fub1 are expressed relative to those in the wild‐type strain. Bars represent standard errors from two independent biological experiments with three technical replicates each. (D) The amounts of fusaric acid (FA) and fusarinolic acid (FnA) in cultures of the indicated strains, grown for 3 days on Czapek‐Dox agar (CDA), were quantified by liquid chromatography/tandem mass spectrometry and expressed as nanograms per millilitre of extract.
Fig. S3 Mycelial growth on potato dextrose agar (PDA) with or without fusaric acid (FA). Growth of the indicated strains cultured for 3 days at 28 ºC. wt, wild‐type.
Fig. S4 Mycelial growth in Czapek‐Dox agar (CDA) with or without hygromycin B. Growth of the indicated strains cultured for 3 days at 28 ºC. wt, wild‐type.
Fig. S5 Nucleosomal repeat length in Fusarium oxysporum. Genomic DNA was extracted from lyophilized mycelium of the indicated strains, treated with micrococcal nuclease (MNase) for 5 min at 37 ºC (T), separated in a 2% agarose gel, stained with ethidium bromide and visualized under UV light. DNA extracted from untreated mycelium was loaded as a control (UT). M, DNA marker. wt, wild‐type.
Fig. S6 Fusarium oxysporum fub1 is expressed during infection of mice. (A, B) Melt curves in quantitative real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) experiments of the indicated samples. Note the detection of a non‐specific amplicon in mice samples which makes quantification impossible.
Table S1 Primers used in this study.
Acknowledgements
We are grateful to Esther Martínez Aguilera (Universidad de Córdoba, Spain) for technical assistance. We thank Professor Hamid Reza Zamanizadeh (Islamic Azad University Tehran, Iran) and Dr Antonio Moretti (Institute of Sciences of Food Production, CNR, Bari, Italy) for helpful discussions. This research was supported by the following grants: BIO2013‐47870‐R from the Spanish Ministerio de Economía y Competitividad (MINECO); CVI‐7319 from the Junta de Andalucía; and Marie Curie FP7‐PEOPLE‐ITN‐607963 FUNGIBRAIN from the European Comission. C.L.‐D. has an FPI fellowship from MINECO. V.G. has an ERASMUS fellowship from the European Commission. The authors declare no conflicts of interest.
Contributor Information
Antonio Di Pietro, Email: ge2dipia@uco.es.
Manuel S. López‐Berges, Email: ge2snlpm@uco.es
References
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. and Lipman, D.J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Armstrong, G.M. and Armstrong, J.K. (1981) Formae speciales and races of Fusarium oxysporum causing wilt diseases In: Fusarium: Diseases, Biology and Taxonomy (Cook R. ed), pp. 391–399. University Park, PA: Penn State University Press. [Google Scholar]
- Bacon, C.W. , Porter, J.K. , Norred, W.P. and Leslie, J.F. (1996) Production of fusaric acid by Fusarium species. Appl. Environ. Microbiol. 62, 4039–4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon, C.W. , Hinton, D.M. and Hinton, A. Jr. (2006) Growth‐inhibiting effects of concentrations of fusaric acid on the growth of Bacillus mojavensis and other biocontrol Bacillus species. J. Appl. Microbiol. 100, 185–194. [DOI] [PubMed] [Google Scholar]
- Basheer, A. , Berger, H. , Reyes‐Dominguez, Y. , Gorfer, M. and Strauss, J. (2009) A library‐based method to rapidly analyse chromatin accessibility at multiple genomic regions. Nucleic Acids Res. 37, e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthiller, F. , Crews, C. , Dall'asta, C. , Saeger, S.D. , Haesaert, G. , Karlovsky, P. , Oswald, I.P. , Seefelder, W. , Speijers, G. and Stroka, J. (2013) Masked mycotoxins: a review. Mol. Nutr. Food Res. 57, 165–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok, J.W. and Keller, N.P. (2004) LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot Cell, 3, 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok, J.W. , Balajee, S.A. , Marr, K.A. , Andes, D. , Nielsen, K.F. , Frisvad, J.C. and Keller, N.P. (2005) LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryot. Cell, 4, 1574–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, D.W. , Butchko, R.A. , Busman, M. and Proctor, R.H. (2012) Identification of gene clusters associated with fusaric acid, fusarin, and perithecial pigment production in Fusarium verticillioides . Fungal Genet. Biol. 49, 521–532. [DOI] [PubMed] [Google Scholar]
- Brown, D.W. , Lee, S.H. , Kim, L.H. , Ryu, J.G. , Lee, S. , Seo, Y. , Kim, Y.H. , Busman, M. , Yun, S.H. , Proctor, R.H. and Lee, T. (2015) Identification of a 12‐gene fusaric acid biosynthetic gene cluster in Fusarium species through comparative and functional genomics. Mol. Plant–Microbe Interact. 28, 319–332. [DOI] [PubMed] [Google Scholar]
- Butchko, R.A. , Brown, D.W. , Busman, M. , Tudzynski, B. and Wiemann, P. (2012) Lae1 regulates expression of multiple secondary metabolite gene clusters in Fusarium verticillioides . Fungal Genet. Biol. 49, 602–612. [DOI] [PubMed] [Google Scholar]
- Caracuel, Z. , Roncero, M.I. , Espeso, E.A. , Gonzalez‐Verdejo, C.I. , Garcia‐Maceira, F.I. and Di Pietro, A. (2003) The pH signalling transcription factor PacC controls virulence in the plant pathogen Fusarium oxysporum . Mol. Microbiol. 48, 765–779. [DOI] [PubMed] [Google Scholar]
- Crutcher, F.K. , Liu, J. , Puckhaber, L.S. , Stipanovic, R.D. , Bell, A.A. and Nichols, R.L. (2015) FUBT, a putative MFS transporter, promotes secretion of fusaric acid in the cotton pathogen Fusarium oxysporum f. sp. vasinfectum . Microbiology, 161, 875–883. [DOI] [PubMed] [Google Scholar]
- Chomczynski, P. and Sacchi, N. (1987) Single‐step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem. 162, 156–159. [DOI] [PubMed] [Google Scholar]
- D'Alton, A. and Etherton, B. (1984) Effects of fusaric acid on tomato root hair membrane potentials and ATP levels. Plant Physiol. 74, 39–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, R. , Van Kan, J.A. , Pretorius, Z.A. , Hammond‐Kosack, K.E. , Di Pietro, A. , Spanu, P.D. , Rudd, J.J. , Dickman, M. , Kahmann, R. , Ellis. J. and Foster, G.D. (2012) The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro, A. and Roncero, M.I. (1998) Cloning, expression, and role in pathogenicity of pg1 encoding the major extracellular endopolygalacturonase of the vascular wilt pathogen Fusarium oxysporum . Mol. Plant–Microbe Interact. 11, 91–98. [DOI] [PubMed] [Google Scholar]
- Dong, X. , Xiong, Y. , Ling, N. , Shen, Q. and Guo, S. (2014) Fusaric acid accelerates the senescence of leaf in banana when infected by Fusarium . World J. Microbiol. Biotechnol. 30, 1399–1408. [DOI] [PubMed] [Google Scholar]
- Eddings, J.L. and Brown, A.L. (1967) Absorption and translocation of foliar‐applied iron. Plant Physiol. 42, 15–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisendle, M. , Oberegger, H. , Buttinger, R. , Illmer, P. and Haas, H. (2004) Biosynthesis and uptake of siderophores is controlled by the PacC‐mediated ambient‐pH regulatory system in Aspergillus nidulans . Eukaryot. Cell, 3, 561–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, M.C. , Henk, D.A. , Briggs, C.J. , Brownstein, J.S. , Madoff, L.C. , McCraw, S.L. and Gurr, S.J. (2012) Emerging fungal threats to animal, plant and ecosystem health. Nature, 484, 186–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridkin, S.K. (2005) The changing face of fungal infections in health care settings. Clin. Infect. Dis. 41, 1455–1460. [DOI] [PubMed] [Google Scholar]
- Gapillout, I. , Milat, M.L. and Blein, J.P. (1996) Effects of fusaric acid on cells from tomato cultivars resistant or susceptible to Fusarium oxysporum f. sp. lycopersici. Eur. J. Plant Pathol. 102, 127–132. [Google Scholar]
- Gäumann, E. (1957) Fusaric acid as a wilt toxin. Phytopathology, 47, 342–357. [Google Scholar]
- Gäumann, E. (1958) The mechanism of fusaric acid injury. Phytopathology, 48, 670–686. [Google Scholar]
- Gonzalez, R. and Scazzocchio, C. (1997) A rapid method for chromatin structure analysis in the filamentous fungus Aspergillus nidulans . Nucleic Acids Res. 25, 3955–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong‐Sheng, W. , Wei, B. , Dong‐Yang, L. , Ning, L. , Rong‐Rong, Y. , Waseem, R. , Shen, Q.R. (2008) Effect of fusaric acid on biomass and photosynthesis of watermelon seedlings leaves. Caryologia, 61, 258–268. [Google Scholar]
- Jiang, J. , Liu, X. , Yin, Y. and Ma, Z. (2011) Involvement of a velvet protein FgVeA in the regulation of asexual development, lipid and secondary metabolisms and virulence in Fusarium graminearum . PLoS One, 6, e28291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao, J. , Zhou, B. , Zhu, X. , Gao, Z. and Liang, Y. (2013) Fusaric acid induction of programmed cell death modulated through nitric oxide signalling in tobacco suspension cells. Planta, 238, 727–737. [DOI] [PubMed] [Google Scholar]
- Lakshminarayanan, K. and Subramanian, D. (1955) Is fusaric acid a vivotoxin? Nature, 176, 697–698. [Google Scholar]
- Laskowski‐Peak, M.C. , Calvo, A.M. , Rohrssen, J. and Smulian, A.G. (2012) VEA1 is required for cleistothecial formation and virulence in Histoplasma capsulatum . Fungal Genet. Biol. 49, 838–846. [DOI] [PubMed] [Google Scholar]
- Lee, J. , Myong, K. , Kim, J.E. , Kim, H.K. , Yun, S.H. and Lee, Y.W. (2012) FgVelB globally regulates sexual reproduction, mycotoxin production and pathogenicity in the cereal pathogen Fusarium graminearum . Microbiology, 158, 1723–1733. [DOI] [PubMed] [Google Scholar]
- Löffler, H.J.M. and Mouris, J.R. (1992) Fusaric acid: phytotoxicity and in vitro production by Fusarium oxysporum f.sp. lilii, the causal agent of basal rot in lilies. Neth. J. Plant Pathol. 98, 107–115. [Google Scholar]
- Lopez‐Berges, M.S. , Rispail, N. , Prados‐Rosales, R.C. and Di Pietro, A. (2010) A nitrogen response pathway regulates virulence functions in Fusarium oxysporum via the protein kinase TOR and the bZIP protein MeaB. Plant Cell, 22, 2459–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Berges, M.S. , Capilla, J. , Turra, D. , Schafferer, L. , Matthijs, S. , Jochl, C. , Cornelis, P. , Guarro, J. , Haas, H. and Di Pietro, A. (2012) HapX‐mediated iron homeostasis is essential for rhizosphere competence and virulence of the soilborne pathogen Fusarium oxysporum . Plant Cell, 24, 3805–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Berges, M.S. , Hera, C. , Sulyok, M. , Schafer, K. , Capilla, J. , Guarro, J. and Di Pietro, A. (2013) The velvet complex governs mycotoxin production and virulence of Fusarium oxysporum on plant and mammalian hosts. Mol. Microbiol. 87, 49–65. [DOI] [PubMed] [Google Scholar]
- Ma, L.J. , van der Does, H.C. , Borkovich, K.A. , Coleman, J.J. , Daboussi, M.J. , Di Pietro, A. , Dufresne, M. , Freitag, M. , Grabherr, M. , Henrissat, B. , Houterman, P.M. , Kang, S. , Shim, W.B. , Woloshuk, C. , Xie, X. , Xu, J.R. , Antoniw, J. , Baker, S.E. , Bluhm, B.H. , Breakspear, A. , Brown, D.W. , Butchko, R.A. , Chapman, S. , Coulson, R. , Coutinho, P.M. , Danchin, E.G. , Diener, A. , Gale, L.R. , Gardiner, D.M. , Goff, S. , Hammond‐Kosack, K.E. , Hilburn, K. , Hua‐Van, A. , Jonkers, W. , Kazan, K. , Kodira, C.D. , Koehrsen, M. , Kumar, L. , Lee, Y.H. , Li, L. , Manners, J.M. , Miranda‐Saavedra, D. , Mukherjee, M. , Park, G. , Park, J. , Park, S.Y. , Proctor, R.H. , Regev, A. , Ruiz‐Roldan, M.C. , Sain, D. , Sakthikumar, S. , Sykes, S. , Schwartz, D.C. , Turgeon, B.G. , Wapinski, I. , Yoder, O. , Young, S. , Zeng, Q. , Zhou, S. , Galagan, J. , Cuomo, C.A. , Kistler, H.C. and Rep, M. (2010) Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium . Nature, 464, 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malachova, A. , Sulyok, M. , Beltran, E. , Berthiller, F. and Krska, R. (2014) Optimization and validation of a quantitative liquid chromatography‐tandem mass spectrometric method covering 295 bacterial and fungal metabolites including all regulated mycotoxins in four model food matrices. J. Chromatogr. A, 1362, 145–156. [DOI] [PubMed] [Google Scholar]
- Malini, S. (1966) Heavy metal chelates of fusaric acid: in vitro spectrophotometry. J. Phytopathol. 57, 221–231. [Google Scholar]
- Marrè, M.T. , Vergani, P. and Albergoni, F.G. (1993) Relationship between fusaric acid uptake and its binding to cell structures by leaves of Egeria densa and its toxic effects on membrane permeability and respiration. Physiol. Mol. Plant Pathol. 42, 141–157. [Google Scholar]
- Mattern, I.E. , Punt, P.J. and van den Hondel, D.A. (1988) A vector of Aspergillus transformation conferring phleomycin resistance. Fungal Genet. News, 35, 25. [Google Scholar]
- Merhej, J. , Urban, M. , Dufresne, M. , Hammond‐Kosack, K.E. , Richard‐Forget, F. and Barreau, C. (2012) The velvet gene, FgVe1, affects fungal development and positively regulates trichothecene biosynthesis and pathogenicity in Fusarium graminearum . Mol. Plant Pathol. 13, 363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung, K. , Li, S. , Butchko, R.A. , Busman, M. , Proctor, R.H. , Abbas, H.K. and Calvo, A.M. (2009) FvVE1 regulates biosynthesis of the mycotoxins fumonisins and fusarins in Fusarium verticillioides . J. Agric. Food Chem. 57, 5089–5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann, P.M. and Prinz, R. (1975) Foliar iron spray potentiates growth of seedlings on iron‐free media. Plant Physiol. 55, 988–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehaus, E.M. , von Bargen, K.W. , Espino, J.J. , Pfannmuller, A. , Humpf, H.U. and Tudzynski, B. (2014) Characterization of the fusaric acid gene cluster in Fusarium fujikuroi . Appl. Microbiol. Biotechnol. 98, 1749–1762. [DOI] [PubMed] [Google Scholar]
- Nucci, M. and Anaissie, E. (2007) Fusarium infections in immunocompromised patients. Clin. Microbiol. Rev. 20, 695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortoneda, M. , Guarro, J. , Madrid, M.P. , Caracuel, Z. , Roncero, M.I. , Mayayo, E. and Di Pietro, A. (2004) Fusarium oxysporum as a multihost model for the genetic dissection of fungal virulence in plants and mammals. Infect. Immun. 72, 1760–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, J.H. , Lin, Y.C. , Tan, N. and Gu, Y.C. (2010) Cu(II): a “signaling molecule” of the mangrove endophyte Fusarium oxysporum ZZF51? Biometals, 23, 1053–1060. [DOI] [PubMed] [Google Scholar]
- Pareja‐Jaime, Y. , Martin‐Urdiroz, M. , Roncero, M.I. , Gonzalez‐Reyes, J.A. and Roldan Mdel, C. (2010) Chitin synthase‐deficient mutant of Fusarium oxysporum elicits tomato plant defence response and protects against wild‐type infection. Mol. Plant Pathol. 11, 479–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, Y.S. , Lian, H. , Chang, M. , Kang, C.M. and Yun, C.W. (2014) Identification of high‐affinity copper transporters in Aspergillus fumigatus . Fungal Genet. Biol. 73, 29–38. [DOI] [PubMed] [Google Scholar]
- Pavlovkin, J. (1998) Effect of fusaric acid on the electrical properties of maize root hairs plasmalemma. Agriculture, 44, 350–355. [Google Scholar]
- Perrin, R.M. , Fedorova, N.D. , Bok, J.W. , Cramer, R.A. , Wortman, J.R. , Kim, H.S. , Nierman, W.C. and Keller, N.P. (2007) Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 3, e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitel, D.W. and Vining, L.C. (1970) Accumulation of dehydrofusaric acid and its conversion to fusaric and 10‐hydroxyfusaric acids in cultures of Gibberella fujikuroi . Can. J. Biochem. 48, 623–630. [DOI] [PubMed] [Google Scholar]
- Porter, J.K. , Bacon, C.W. , Wray, E.M. and Hagler, W.M. Jr. (1995) Fusaric acid in Fusarium moniliforme cultures, corn, and feeds toxic to livestock and the neurochemical effects in the brain and pineal gland of rats. Nat. Toxins, 3, 91–100. [DOI] [PubMed] [Google Scholar]
- Punt, P.J. , Oliver, R.P. , Dingemanse, M.A. , Pouwels, P.H. and van den Hondel, C.A. (1987) Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli . Gene, 56, 117–124. [DOI] [PubMed] [Google Scholar]
- Raeder, U. and Broda, P. (1985) Rapid preparation of DNA from filamentous fungi. Lett. Appl. Microbiol. 1, 17–22. [Google Scholar]
- Reyes‐Dominguez, Y. , Bok, J.W. , Berger, H. , Shwab, E.K. , Basheer, A. , Gallmetzer, A. , Scazzocchio, C. , Keller, N. and Strauss, J. (2010) Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans . Mol. Microbiol. 76, 1376–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz, J.A. , Bernar, E.M. and Jung, K. (2015) Production of siderophores increases resistance to fusaric acid in Pseudomonas protegens Pf‐5. PLoS One, 10, e0117040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samadi, L. and Shahsavan Behboodi, B. (2006) Fusaric acid induces apoptosis in saffron root‐tip cells: roles of caspase‐like activity, cytochrome c, and H2O2 . Planta, 225, 223–234. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. , Russell, D. (2001) Molecular Cloning: A Laboratory Manual, 3th edn New York, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Sarikaya‐Bayram, O. , Bayram, O. , Feussner, K. , Kim, J.H. , Kim, H.S. , Kaever, A. , Feussner, I. , Chae, K.S. , Han, D.M. , Han, K.H. and Braus, G.H. (2014) Membrane‐bound methyltransferase complex VapA‐VipC‐VapB guides epigenetic control of fungal development. Dev. Cell, 29, 406–420. [DOI] [PubMed] [Google Scholar]
- Schrettl, M. and Haas, H. (2011) Iron homeostasis–Achilles' heel of Aspergillus fumigatus? Curr. Opin. Microbiol. 14, 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J.J. and Yee, N.K. (2001) A concise synthesis of fusaric acid and (S)‐(+)‐fusarinolic acid. J. Org. Chem. 66, 605–608. [DOI] [PubMed] [Google Scholar]
- Stipanovic, R.D. , Puckhaber, L.S. , Liu, J. and Bell, A.A. (2011) Phytotoxicity of fusaric acid and analogs to cotton. Toxicon, 57, 176–178. [DOI] [PubMed] [Google Scholar]
- Strange, R.N. and Scott, P.R. (2005) Plant disease: a threat to global food security. Ann. Rev. Phytopathol. 43, 83–116. [DOI] [PubMed] [Google Scholar]
- Studt, L. , Janevska, S. , Niehaus, E.M. , Burkhardt, I. , Arndt, B. , Sieber, C.M. , Humpf, H.U. , Dickschat, J.S. and Tudzynski, B. (2016) Two separate key enzymes and two pathway‐specific transcription factors are involved in fusaric acid biosynthesis in Fusarium fujikuroi . Environ. Microbiol. 18, 936–956. [DOI] [PubMed] [Google Scholar]
- Szewczyk, E. , Nayak, T. , Oakley, C.E. , Edgerton, H. , Xiong, Y. , Taheri‐Talesh, N. , Osmani, S.A. and Oakley, B.R. (2006) Fusion PCR and gene targeting in Aspergillus nidulans . Nat. Protoc. 1, 3111–3120. [DOI] [PubMed] [Google Scholar]
- Tamari, K. and Kaji, J. (1952) Studies on the mechanism of injurious action of fusarinic acid on plant‐growth. J. Agric. Chem. Soc. Jpn. 26, 345–349. [Google Scholar]
- Venter, S.L. and Steyn, P.J. (1998) Correlation between fusaric acid production and virulence of isolates of Fusarium oxysporum that causes potato dry rot in South Africa. Potato Res. 41, 289–294. [Google Scholar]
- Vicentefranqueira, R. , Moreno, M.A. , Leal, F. and Calera, J.A. (2005) The zrfA and zrfB genes of Aspergillus fumigatus encode the zinc transporter proteins of a zinc uptake system induced in an acid, zinc‐depleted environment. Eukaryot Cell, 4, 837–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulpe, C.D. and Packman, S. (1995) Cellular copper transport. Annu. Rev. Nutr. 15, 293–322. [DOI] [PubMed] [Google Scholar]
- Wang, H. and Ng, T.B. (1999) Pharmacological activities of fusaric acid (5‐butylpicolinic acid). Life Sci. 65, 849–856. [DOI] [PubMed] [Google Scholar]
- Webster, R.H. and Sil, A. (2008) Conserved factors Ryp2 and Ryp3 control cell morphology and infectious spore formation in the fungal pathogen Histoplasma capsulatum . Proc. Natl. Acad. Sci. USA, 105, 14 573–14 578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemann, P. , Brown, D.W. , Kleigrewe, K. , Bok, J.W. , Keller, N.P. , Humpf, H.U. and Tudzynski, B. (2010) FfVel1 and FfLae1, components of a velvet‐like complex in Fusarium fujikuroi, affect differentiation, secondary metabolism and virulence. Mol. Microbiol. 77, 972–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuta, T. , Kambe, K. and Hayashi, T. (1937) Biochemistry of the bakanae fungus. I. Fusarinic acid, a new product of the bakanae fungus. J. Agric. Chem. Soc. Jpn. 10, 1059–1068. [Google Scholar]
- Yabuta, T. , Sumiki, Y. , Aso, K. , Tamura, T. , Igarashi, H. and Tamari, K. (1939) Biochemical studies on the bakanae fungus. IV. The culture conditions for producing gibberellin or fusaric acid. J. Agric. Chem. Soc. Jpn. 15, 1209–1220. [Google Scholar]
- Yin, E.S. , Rakhmankulova, M. , Kucera, K. , de Sena Filho, J.G. , Portero, C.E. , Narvaez‐Trujillo, A. , Holley, S.A. and Strobel, S.A. (2015) Fusaric acid induces a notochord malformation in zebrafish via copper chelation. Biometals, 28, 783–789. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website.
Fig. S1 Identification of the Fusarium oxysporum fusaric acid (FA) gene cluster and fub1 knockout strategy. (A) Conserved synteny between Fusarium fujikuroi and F. oxysporum FA gene clusters. Note that the two genes inserted between fub3 and fub4 in F. oxysporum are present in F. fujikuroi in another chromosome. (B) Fusarium oxysporum fub1 locus and targeted gene disruption construct. (C) Southern blot analysis. Genomic DNA of the indicated strains was treated with XhoI, separated on a 0.7% agarose gel, transferred to a nylon membrane and hybridized with the DNA probe indicated in (B). wt, wild‐type.
Fig. S2 Generation and selection of the fub1Δ complemented strain (fub1ΔC). (A) Strategy of fub1Δ complementation by co‐transformation with a fub1 wild‐type allele and the phleomycin resistance marker. The relative positions of the polymerase chain reaction (PCR) primers used for genotyping are indicated. phleor, phleomycin resistance gene. (B) PCR amplification of genomic DNA (gDNA) of the indicated strains using primers F3 and R3. The complemented strains, fub1ΔC and fub1ΔC #2, produce a banding pattern consistent with the integration of an intact fub1 allele. wt, wild‐type strain. (C) Quantitative real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) performed in the indicated strains germinated for 16 h in potato dextrose broth (PDB) and transferred for 3 h to Czapec‐Dox liquid (CDL). Transcript levels of fub1 are expressed relative to those in the wild‐type strain. Bars represent standard errors from two independent biological experiments with three technical replicates each. (D) The amounts of fusaric acid (FA) and fusarinolic acid (FnA) in cultures of the indicated strains, grown for 3 days on Czapek‐Dox agar (CDA), were quantified by liquid chromatography/tandem mass spectrometry and expressed as nanograms per millilitre of extract.
Fig. S3 Mycelial growth on potato dextrose agar (PDA) with or without fusaric acid (FA). Growth of the indicated strains cultured for 3 days at 28 ºC. wt, wild‐type.
Fig. S4 Mycelial growth in Czapek‐Dox agar (CDA) with or without hygromycin B. Growth of the indicated strains cultured for 3 days at 28 ºC. wt, wild‐type.
Fig. S5 Nucleosomal repeat length in Fusarium oxysporum. Genomic DNA was extracted from lyophilized mycelium of the indicated strains, treated with micrococcal nuclease (MNase) for 5 min at 37 ºC (T), separated in a 2% agarose gel, stained with ethidium bromide and visualized under UV light. DNA extracted from untreated mycelium was loaded as a control (UT). M, DNA marker. wt, wild‐type.
Fig. S6 Fusarium oxysporum fub1 is expressed during infection of mice. (A, B) Melt curves in quantitative real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) experiments of the indicated samples. Note the detection of a non‐specific amplicon in mice samples which makes quantification impossible.
Table S1 Primers used in this study.


