Summary
Wheat streak mosaic virus (WSMV) causes wheat streak mosaic, a disease of cereals and grasses that threatens wheat production worldwide. It is a monopartite, positive‐sense, single‐stranded RNA virus and the type member of the genus Tritimovirus in the family Potyviridae. The only known vector is the wheat curl mite (WCM, Aceria tosichella), recently identified as a species complex of biotypes differing in virus transmission. Low rates of seed transmission have been reported. Infected plants are stunted and have a yellow mosaic of parallel discontinuous streaks on the leaves. In the autumn, WCMs move from WSMV‐infected volunteer wheat and other grass hosts to newly emerged wheat and transmit the virus which survives the winter within the plant, and the mites survive as eggs, larvae, nymphs or adults in the crown and leaf sheaths. In the spring/summer, the mites move from the maturing wheat crop to volunteer wheat and other grass hosts and transmit WSMV, and onto newly emerged wheat in the fall to which they transmit the virus, completing the disease cycle. WSMV detection is by enzyme‐linked immunosorbent assay (ELISA), reverse transcription‐polymerase chain reaction (RT‐PCR) or quantitative RT‐PCR (RT‐qPCR). Three types of WSMV are recognized: A (Mexico), B (Europe, Russia, Asia) and D (USA, Argentina, Brazil, Australia, Turkey, Canada). Resistance genes Wsm1, Wsm2 and Wsm3 have been identified. The most effective, Wsm2, has been introduced into several wheat cultivars. Mitigation of losses caused by WSMV will require enhanced knowledge of the biology of WCM biotypes and WSMV, new or improved virus detection techniques, the development of resistance through traditional and molecular breeding, and the adaptation of cultural management tactics to account for climate change.
Keywords: Aceria tosichella, cereal crops, Tritimovirus, wheat curl mite, WSMV
Introduction
Wheat streak mosaic virus (WSMV) was first observed by Peltier in Nebraska in the Central Great Plains of the USA in 1922 and described as ‘yellow mosaic’ (McKinney, 1937; Staples and Allington, 1956). WSMV was formerly placed in the genus Rymovirus together with mite‐transmitted viruses of the family Potyviridae. Later, the complete genome sequence and evolutionary analysis established WSMV with the whitefly‐transmitted Sweet potato mild mottle virus and not with Ryegrass mosaic virus, the type member of the genus Rymovirus (Stenger et al., 1998). The finding thus proposed a new genus known as ‘Tritimovirus’ within the family Potyviridae, for which WSMV is the type member (Rabenstein et al., 2002). It is transmitted by the wheat curl mite (WCM, Aceria tosichella Keifer). The virus is widely distributed in most wheat‐growing regions of the world, including the USA, Canada, Mexico, Brazil, Argentina, Europe, Turkey, Iran, Australia and New Zealand (Hadi et al., 2011; Navia et al., 2013).
WSMV is hosted by many plant species of the family Poaceae, including wheat (Triticum aestivum L.), oat (Avena sativa L.), barley (Hordeum vulgare L.), maize (Zea mays L.), millet (Panicum), Setaria and Echinochloa spp., and several other grasses (Chalupníková et al., 2017; Dráb et al., 2014; French and Stenger, 2002). Given the potentially devastating impact of WSMV on affected cereal crops, the occurrence of this disease in wheat has been a cause for concern because losses can range from minimal to complete crop failure (French and Stenger, 2003). Improvement in WSMV resistance is an important aspect of wheat production, and the development of resistant cultivars has helped to increase production (Price et al., 2010a).
Because of the devastating economic impact caused by WSMV in wheat‐growing countries around the globe, and its significance in the plant pathology community, a comprehensive report updating the knowledge on WSMV is warranted. Therefore, this review examines current knowledge on WSMV including virus biology, genome architecture, mechanism of transmission, host range, disease symptoms and cycle, diagnostic tools, genetic diversity, host resistance, and management strategies and tactics.
Causal Agent of the Disease
WSMV is the causal agent of wheat streak mosaic disease. WSMV is a non‐enveloped, flexible, filamentous, rod‐shaped virus composed of a monopartite, positive‐sense, single‐stranded RNA genome (ssRNA+). The WSMV genome size is ∼9.3–9.4 kb and expands into a single open reading frame (ORF), which is transcribed into a large polyprotein (Fig. 1). This polyprotein is cleaved into at least 10 mature proteins: P1 (P1 protein: 40 kDa); HC‐Pro (helper component protease: 44 kDa); P3 (P3 protein: 32 kDa); 6K1 and 6K2 (6 kDa protein); CI (cytoplasmic inclusion protein: 73 kDa); VPg (viral protein genome‐linked proteinase: 23 kDa); NIa (nuclear Inclusion putative protease: 26 kDa); NIb (Nuclear Inclusion putative polymerase: 57 kDa) CP (coat protein: 37 kDa) (Choi et al., 2002; Chung et al., 2008; Stenger et al., 1998). The recently described short ORF (PIPO) is expressed as a fusion protein with the N‐terminal half of P3 (P3N‐PIPO) (Chung et al., 2008). The role of these distinct proteins has been deciphered in various processes, including suppressor of RNA silencing (Young et al., 2012), genome amplification, protein–protein interactions, RNA binding and amplification of the virus genome, cell‐to‐cell and systemic transport, virion assembly (Rojas et al., 1997; Tatineni and French, 2014; Tatineni et al., 2017) and proteolytic processing (Schaad et al., 1996). The 5′‐terminus has a VPg and the 3′‐terminus has a poly (A) tail. The RNA is infectious and serves as both the genome and viral messenger.
Figure 1.
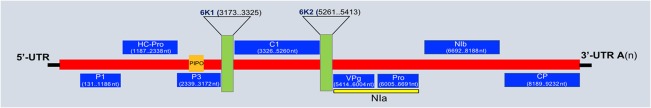
Genome architecture of Wheat streak mosaic virus (WSMV). The genome size of WSMV is 9.3–9.4 kb and has a single open reading frame, which is transcribed into a large polyprotein. This polyprotein is composed of 10 proteins: P1 (P1 protein: 40 kDa); HC‐Pro (helper component protease: 44 kDa); P3 (P3 protein: 32 kDa); 6K1 and 6K2 (6 kDa protein); CI (cytoplasmic inclusion protein: 73 kDa); VPg (viral protein genome‐linked proteinase: 23 kDa); NIa (nuclear Inclusion putative protease: 26 kDa); NIb (Nuclear Inclusion putative polymerase: 57 kDa) CP (coat protein: 37 kDa). nt, nucleotides; UTR, untranslated region.
Protein functions
P1
The P1 protein of WSMV has serine proteinase activity, is known to mediate suppression of RNA silencing and plays a role in the enhancement of disease symptoms in WSMV‐P1‐expressing transgenic plants infected with Potato virus X (PVX) (Young et al., 2012).
HC‐Pro
Mutations of HC‐Pro in potyviruses affect multiple functions, including disruption of polyprotein processing, aphid transmission, long‐distance movement, maintenance of replication and suppression of post‐transcriptional gene silencing (PTGS) (Carrington et al., 1996; Llave et al., 2002; Ruiz‐Ferrer et al., 2005). However, WSMV HC‐Pro shares two functions in common with potyviruses: mediation of vector transmission and cysteine proteinase activity (Stenger et al., 2005a; Young et al., 2007). Moreover, deletion of the HC‐Pro coding region shows no effect on WSMV virulence in wheat, oats and corn (Stenger et al., 2005b). Mutation analyses of the WSMV HC‐Pro protein suggest that it plays a role in replication and is dispensable for systemic movement (Stenger et al., 2006). WSMV HC‐Pro does not mediate suppression of RNA silencing when tested in Nicotiana benthamiana (Young et al., 2012). WSMV HC‐Pro exhibits no effect on disease synergism in maize co‐infected with a WSMV HC‐Pro complete deletion mutant and Maize chlorotic mottle virus (MCMV) (Stenger et al., 2007).
CP
WSMV CP is 349 amino acids long. The C‐terminal aspartic acid residues (D216, D289, D290, D326, D333 and D334) of CP are involved in host‐specific virus movement and play a role in efficient cell‐to‐cell movement in wheat and long‐distance transport in maize (Tatineni and French, 2014). WSMV CP contains three flexible linker motifs: SGSGS‐1 (36–40 amino acids), SGSGS‐2 (43–47 amino acids) and SGSGS‐3 (53–57 amino acids). Deletion of these motifs, either individually or jointly, elicits symptoms similar to the wild‐type (Tatineni et al., 2017). The CP amino acids 6–27 and 85–100 are required for efficient virion assembly and/or systemic infection and cell‐to‐cell movement (Tatineni and French, 2014). Deletions in the N‐terminal region (58–84 amino acids) of the CP enhance the accumulation of CP and genomic RNA, alter CP‐specific protein profiles and cause severe symptom phenotypes in multiple cereal hosts, including wheat, maize, rye and barley (Tatineni et al., 2017). The N‐terminal region of WSMV CP is a host‐ and strain‐specific long‐distance transport factor (LTF) in maize. The differing amino acids (AS to EP at position 20/21; Q to L at position 30; AG to VE at position 50/52) in the N‐terminus of CP between the WSMV‐S81 and WSMV‐T isolates are crucial for interactions with the maize inbred line SDp2 (Tatineni et al., 2011). Recently, it has been reported that amino mutations of aspartic acid residues at amino acid positions 289 or 326 (D289A or D326A) at the carboxy‐proximal region of CP significantly reduce mite transmission (Tatineni et al., 2018).
Virus Transmission by WCM
The only known vector of WSMV is an obligatory phytophagous WCM, which is amongst the most important eriophyid mite pests of agricultural crops (Navia et al., 2013; Oldfield and Proeseler, 1996). This microscopic mite (Fig. 2) inhabits sheltered sites on the plant which protect it from desiccation (Navia et al., 2013), and the haplodiploid single unfertilized female is capable of initiating a population (Miller et al., 2012), which increases its ability to successfully spread the viruses it transmits. In addition to wheat, WCMs can transmit WSMV to barley, oats, corn, rye and many wild annual kinds of grass (Table 1). As a result of its wind‐borne dispersal, the mite is widely distributed in cereal fields and grasslands, which boosts the ability of WSMV to spread within cereal‐producing regions worldwide. The capability of WCMs to successfully colonize new plants is remarkable. After landing on new plants, WCMs are able to multiply very rapidly and attain, after two generations (14 days), a population density 25% higher than that on the plant from which they dispersed (Kiedrowicz et al., 2017a). This confirms the great dispersal and colonization potential of WCMs, which influences the spread of wheat streak mosaic.
Figure 2.

Scanning electron microscopy (SEM) image of wheat curl mite (Aceria tosichella) specimens on a wheat leaf.
Table 1.
Host range of Wheat streak mosaic virus.
| Host | Common name | Reference | |
|---|---|---|---|
| Cereals | |||
| Avena barbata | Bearded oat | Coutts et al. ( 2014) | |
| Avena sativa | Oat | Brakke ( 1971) | |
| Hordeum vulgare | Barley | Brakke ( 1971) | |
| Panicum millaceum | Broomcorn millet | Sill and Agusiobo ( 1955); Vacke et al. ( 1986); Ellis et al. ( 2004) | |
| Pennisetum glaucum | Pearl millet | Seifers et al. ( 1996) | |
| Secale cereale | Cereal rye | Vacke et al. ( 1986); Ito et al. ( 2012) | |
| Setaria italica | Foxtail millet | Truol et al. ( 2010) | |
| Sorghum bicolor | Sorgum | Seifers et al. ( 1996) | |
| Triticum aestivum | Wheat | Brakke ( 1971) | |
| Zea mays | Maize | Brakke ( 1971) | |
| Wild grasses | |||
| Aegilops cylindrica | Jointed goatgrass | Sill and Connin (1953) | |
| Agropyron repens | Couch grass | Dráb et al. ( 2014); Singh and Kundu ( 2017) | |
| Agrostis capillaris | Common bent | Chalupníková et al. ( 2017) | |
| Alopecurus pratensis | Meadow foxtail | Dráb et al. ( 2014) | |
| Anthoxanthum odoratum | Sweet vernal‐grass | Chalupníková et al. ( 2017) | |
| Arrhenatherum elatius | False oat‐grass | Dráb et al. ( 2014) | |
| Austrostipa compressa | Speargrass | Vincent et al. ( 2014) | |
| Avena fatua | Wild oat | Vacke et al. ( 1986) | |
| Avena strigesa | Wild oats | Vacke et al. ( 1986) | |
| Brachypodium distachyon | Purple false brome | Mandadi et al., (2014) | |
| Briza maxima | Blowfly grass | Coutts et al. ( 2014) | |
| Bromus arvenis | Field brome | Sill and Connin (1953) | |
| Bromus japonicus | Japanese brome | Wegulo et al. ( 2008) | |
| Bromus rigidus | Brome grass | Coutts et al. ( 2014) | |
| Bromus secalinus | Cheat grass | Sill and Connin (1953) | |
| Bromus tectorum | Downy brome | Sill and Connin (1953) | |
| Cenchrus longispinus | Mat sandbur | Connin ( 1956) | |
| Cenchrus pauciflours | Sandbur | Wegulo et al. ( 2008) | |
| Cynodon dactylon | Couch grass | Ellis et al. ( 2004) | |
| Digitaria sanguinalis | Hairy crab grass | Vacke et al. ( 1986) | |
| Echinochloa crus‐galli | Barnyardgrass | Sill and Connin (1953) | |
| Echinochloa colonum | Junglerice | Khadivar and Nasrolahnejad ( 2009) | |
| Elymus repens | Quackgrass | Ito et al. ( 2012) | |
| Eragrostis cilianensis | Stink grass | Connin ( 1956) | |
| Eragrostis curvula | African lovegrass | Ellis et al. ( 2004) | |
| Eriochloa acuminata | Tapertip cupgrass | Seifers et al. ( 2010) | |
| Eriochloa contracta | Prairie cupgrass | Christian and Willis ( 1993) | |
| Eleusine tristachya | Spike goosegrass | Ellis et al. ( 2004) | |
| Elymus canadensis | Canada wild rye | Ito et al. ( 2012) | |
| Holcus lanatus | Soft‐grass | Chalupníková et al. ( 2017) | |
| Holcus mollis | Creeping soft‐grass | Chalupníková et al. ( 2017) | |
| Hordeum leporinum | Barley grass | Coutts et al. ( 2014) | |
| Lagurus ovatus | Hare's‐tail | Vacke et al. ( 1986) | |
| Lolium mitiflorum | Annual ryegrass | Vacke et al. ( 1986); Ellis et al. ( 2004) | |
| Lolium rigidum | Ryegrass | Coutts et al. ( 2014) | |
| Panicum dichotomiflorum | Fall panicgrass | Sill and Connin (1953) | |
| Panicum capillare | Witch grass | Coutts et al. ( 2008a, 2008b) | |
| Phalaris aquatica | Phalaris | Ellis et al. ( 2004) | |
| Phleum pratense | Timothy‐grass | Dráb et al. ( 2014) | |
| Poa pratensis | Bluegrass | Ito et al. ( 2012); Dráb et al. (2014) | |
| Setaria viridis | Green bristlegrass | Sill and Connin (1953) | |
| Tragus australianus | Small burr grass | Coutts et al. ( 2008a, 2008b) |
The WCM was identified as the agent transmitting WSMV by Slykhuis (1955), and the recent use of DNA sequence data and experimental host bioassays has shown that the WCM is, in fact, a species complex consisting of several divergent genetic lineages (probably cryptic species) (Miller et al., 2013; Skoracka et al., 2012, 2013). Some lineages are highly host specific to single wild‐growing grass species, whereas others are less host specialized and feed on several plant species, including cereals (Skoracka et al., 2013, 2017). The genetic and host range variability within the WCM complex corresponds to the virus vectoring ability amongst WCM lineages (Hein et al., 2012; Schiffer et al., 2009). Up to now, it has been shown that only two lineages within the complex can transmit plant viruses in wheat (Hein et al., 2012).
These lineages have been designated as type 1 and type 2 in Australia (Carew et al., 2009); they match the genotypes found in North America (Hein et al., 2012), and correspond to European and South American MT‐8 and MT‐1 lineages designated by Skoracka et al. (2013, 2014). Laboratory‐based transmission trials using these two types collected in Australia have indicated that only type 2 (MT‐1) is able to transmit WSMV (Schiffer et al., 2009). However, both genotypes collected in North America have been found to effectively transmit WSMV, although at varying rates (Seifers et al., 2002). WCM type 2 (MT‐1) transmits WSMV at an average rate of 43%–68%, depending on the vector's phenological stage, and also reproduces more rapidly in the presence of WSMV relative to type 1 (MT‐8) (Siriwetwiwat, 2006). This result may suggest that a specific symbiotic relationship between WCM type 2 (MT‐1) and WSMV exists, which enables higher success for both the mite and virus, e.g. better reproductive rates for the mites and therefore better chance of virus dispersal. Some arthropod‐borne plant viruses exhibit close relationships with their vector, and vector fitness is often higher on infected host plants (e.g. Belliure et al., 2005).
In Poland, these two WCM biotypes also differ in colonization strategy, and biotype 1 (MT‐8) has a uniform distribution, whereas biotype 2 (MT‐1) occurs unexpectedly in only a few localities within the country, but attains very high densities there (about 30% higher than MT‐8) (Skoracka et al., 2017). All results obtained to date now suggest that biotype 2 (MT‐1) is able to multiply more rapidly and transmit WSMV more efficiently than biotype 1 (MT‐8). These differences in virus transmission efficiency therefore indicate that these two biotypes may require different control and management strategies. As they are divergent phenotypes, they may respond differently to control measures. The next steps directed towards WCM management should focus on genotyping methods to enable straightforward and rapid identification of the biotype in the field.
Virus transmission rates may be determined not only by mite genotype, but also by virus genetic strain. It has been shown that virus isolate and mite genotype, but not source location or WCM colony age, have a significant influence on WSMV transmission, and the existence of cryptic species within WCMs and numerous genotypes of WSMV complicates the epidemiology and poses a challenge to the management of this virus (Wosula et al., 2016).
Undoubtedly, the existence of divergent WCM lineages has implications not only for the management of the WCM and WSMV, but also for the study of the biology and genetics of virus transmission. All further research on the relationships between the WCM and WSMV should be based on the molecular identification of WCM lineages and should focus on particular lineages instead of WCM sensu lato.
However, to date, the fundamental knowledge about relationships between the WCM and WSMV has been based solely on WCM sensu lato. The WCM acquires WSMV during feeding, when it penetrates the epidermal cells using thin, dagger‐like chelicerae. The mites are subsequently infective for up to 9 days at 20–25 ºC after they have been removed from an infected plant or after moulting to the next developmental stage (Navia et al., 2013; Orlob, 1966). Mites can remain infective for up to 2 months at 3 °C, which indicates that overwintering specimens can be a source of WSMV inoculum (Navia et al., 2013). All mobile stages of the WCM (larva, nymph and adult) can be infective. However, virus transmission efficiency differs amongst stages, with immature stages having a higher efficiency than adults. Moreover, for adults to be effectively infective, they must acquire the virus as an immature stage (del Rosario and Sill, 1965; Orlob, 1966; Siriwetwiwat, 2006; Slykhuis, 1955) and, to acquire the virus, the mite requires 15–30 min of feeding on the plant (Orlob, 1966). It has been suggested that WSMV circulates, but does not multiply, in its vector (Paliwal, 1980).
WCMs can transmit other viruses apart from WSMV, such as Triticum mosaic virus (TriMV) and Wheat mosaic virus (WMoV), and can cause mixed infections (Byamukama et al., 2014; Seifers et al., 2011; de Wolf and Seifers, 2008). Such double or even triple infections have been found more frequently (47%) than single infections of winter wheat by WSMV (5%) in the Central Great Plains of the USA (Byamukama et al., 2016). In another experiment, yield loss was 96% when a susceptible wheat cultivar was co‐inoculated with WSMV and TriMV, compared with single inoculation (yield losses of 53% and 50% caused by single inoculation of wheat by TriMV and WSMV, respectively) (Byamukama et al., 2014). Oliveira‐Hofman et al. (2015) found that transmission of WSMV by WCM genotype 2 (MT‐1) was higher from singly infected source plants than from those co‐infected with TriMV.
The high level of infection of a wheat crop with WSMV is associated with the presence of abundant grasses and volunteer wheat plants which serve as hosts for WCMs and WSMV and provide an effective ‘green bridge’ refuge for WCMs between harvesting of the current season's crop and planting of the next season's crop (Somsen and Sill, 1970). When the quality of green bridge food decreases because of host maturity or overcrowding, WCMs start their aerial movement by wind currents into wheat fields from nearby grass vegetation or fields with volunteer wheat that harbour viruliferous mites (Kiedrowicz et al., 2017b; Somsen and Sill, 1970). For example, in Australia, a 40% WSMV incidence and about 5000 WCMs per spike were found at the margin of a wheat crop associated with abundant grasses and volunteer wheat plants in an adjacent pasture (so‐called ‘edge effect’) (Coutts et al., 2008a, 2008b). However, Byamukama et al. (2016) have shown that viruliferous WCMs can be found in any part of the field by the end of the growing season, not only at the edges of wheat fields. Hunger et al. (1992) and Somsen and Sill (1970) found that, as plants mature, they become more resistant to virus infection and develop fewer and milder symptoms. WCMs usually attain high population densities at the end of the wheat growing season, which ensures the infestation and subsequent virus infection of various green bridge hosts, including volunteer wheat and grasses. If conditions allow the survival of these hosts until autumn‐planted winter wheat emerges, the probability of WSMV transmission to autumn‐planted wheat increases, resulting in some level of disease and yield loss every year (Byamukama et al., 2016). The control of grasses and volunteer cereals before the planting of winter wheat and the use of resistant cultivars have been suggested as effective strategies for WCM and WSMV management (Coutts et al., 2008a, 2008b).
Apart from green bridges, climate and weather conditions may influence the levels of WCM infestation and WSMV infection. It has been suggested that high temperatures are the most preferable for MT‐1 and MT‐8 WCM lineages (Kuczyński et al., 2016). According to Orlob (1966), dry and hot conditions favour the development of WCM populations. Indeed, in Nebraska, USA, the drier western regions are more conducive for WCM population build up than are the less dry eastern regions. In addition, in a year with dry and warm conditions, considerably more WCMs were trapped during a field experiment (Byamukama et al., 2016). Conversely, in Australia, wet summers and autumns, as well as westerly frontal winds, provide good conditions for WCM development and spread, which increases the probability of virus outbreaks (Coutts et al., 2008a, 2008b).
Virus–vector interactions may also be altered by nutrient availability. It has been shown that enrichment of CO2 concentration has no observable effects on WCM populations, which suggests that increases in atmospheric CO2 may not directly alter WCM populations and WSMV spread (Miller et al., 2015). Interestingly, nitrogen fertilization increased WCM population growth rates when mites were WSMV infested, but had the opposite effect on non‐viruliferous mites. This outcome was interpreted as a virus–vector mutualism that is conditional on nitrogen limitation. Although, at high nitrogen rates, the interaction between virus and vector was mutually beneficial, at low nitrogen rates the transmission was beneficial for the virus, but detrimental for the vector (as the vector is expected to be nitrogen limited). Therefore, the increase in population growth rate of a viruliferous vector associated with nitrogen may result in virus outbreaks. From a disease management perspective, these results provide a recommendation about the timing and amount of fertilization, suggesting that fertilization should be avoided at the time of year at which the WCM disperses to green bridge plants (Miller et al., 2015).
It has also been suggested that there might be a host‐dependent trade‐off in virus transmission capability by the WCM. Mites reared on western wheatgrass (Agropyron smithii Rydb.) transmit WSMV at significantly lower rates than mites reared on wheat. Once these mites have adapted to wheat, they transmit WSMV at rates comparable with those of colonies that have always been reared on wheat (del Rosario and Sill, 1965). Undoubtedly, given the increasing prevalence and spread of WSMV in many continents, there is still a demand to better understand the biology, ecology and genetics of the WCM complex in order to design effective management strategies for the WCM and associated viruses.
Virus Transmission by Seeds
WSMV transmission by seed was first described in maize in seed production fields in Iowa, and a very low percentage of seed transmission (0.1%) of the virus was found (Hill et al., 1974). Jones et al. (2005) identified WSMV seed‐borne infection in eight wheat genotypes by testing for the virus in seedlings. They found 0.2%–0.5% seed transmission across genotypes and up to 1.5% transmission in individual genotypes, indicating that the rate of transmission was lower across the wheat breeding collection tested and higher in individual genotypes. Such a low seed transmission rate is likely to have little significance epidemiologically in an individual field. However, the epidemiological significance is amplified when one considers the increased probability of global spread of the virus through local, regional and international exchange of germplasm.
Host Range of WSMV
WSMV has a wide host range, including cereals and other grass species. Wheat (Triticum aestivum) is a major host for the virus and the preferred host for the vector mite, A. tosichella biotypes 1 and 2 (lineages MT‐8 and MT‐1), which are known to vector the virus. Other cereal hosts include oats (Avena sativa), barley (Hordeum vulgare), rye (Secale cereale), maize (Zea mays), foxtail millet (Setaria italica), broom‐corn millet or millet (Panicum miliaceum) (Table 1) (Brakke, 1971; Coutts et al., 2014; Vacke et al., 1986), and the mite also feeds and reproduces on these cereals. However, some cereals are susceptible to the virus, but are not good hosts for mites, for example barley (Hordeum vulgare) and rye (Secale cereale). Various annual and perennial grasses serve as hosts of WSMV, including Agropyron repens, Agrostis capillaris, Avena fatua, Bromus japonicus, Brachypodium distachyon and Holcus mollis (Table 1) (Chalupníková et al., 2017; Dráb et al., 2014; Mandadi et al., 2014; Singh and Kundu, 2017; Wegulo et al., 2008).
Disease Symptoms
WSMV on young leaves starts as light green streaks which elongate to form discontinuous yellow to pale green stripes, forming a mosaic pattern running parallel to the leaf veins as symptoms progress in spring (Vacke et al., 1986) (Fig. 3A). These symptoms are often difficult to diagnose as they can be easily confused with nutritional disorders, environmental effects or chemical damage. Plants in field margins closest to the source of WCMs are often the first, and may be the only ones, to show symptoms. With low to moderate levels of infection, a gradation of the intensity of symptoms may be seen across a field, with the most severe symptoms at the edge of the field closest to the WCM source. In severe epidemics, plants in entire fields can become symptomatic (Fig. 3B,C). In winter wheat, infections that cause serious yield losses occur in the autumn. However, symptoms usually appear the following spring, except when there are prolonged warm temperatures late into the autumn, in which case symptoms can appear in the autumn. The appearance of symptoms in the autumn is an indication that severe epidemics may develop in the following spring. In the spring, plants infected in the autumn appear stunted, yellow, less upright than healthy plants and poorly tillered if infections occur early in the autumn. Yellowing intensifies as the temperatures become warmer. Spikes may not develop in severely infected plants or may be poorly filled with shrivelled kernels in less severely infected plants. The effects of spring infections on symptom development and yield are usually subtle (Somsen and Sill, 1970). Recent studies by Tatineni et al. (2017) have shown that deletion of CP amino acids 58–84 leads to the development of serious chlorotic streaks and spots, followed by acute chlorosis in wheat, maize, barley and rye, compared with mild to moderate chlorotic streaks and mosaic symptoms caused by wild‐type WSMV.
Figure 3.

Wheat streak mosaic virus (WSMV) disease symptoms on hosts. (A) WSMV‐infected wheat cv. Cubus showing advanced symptoms with linear streaks coalescing into almost solid yellow areas. (B) WSMV‐infected wheat cv. Vlada mechanically inoculated with WSMV isolate (CZlab, accession no. FJ216408). (C) A section of a wheat field affected by a severe epidemic of wheat streak mosaic in western Nebraska, USA in May 2017. Note the intense yellowing and stunting of the wheat crop.
Disease Cycle
The only known vector of WSMV is the WCM, biotypes 1 and 2 (lineages MT‐8 and MT‐1) (Slykhuis, 1955). The preferred host for these lineages is wheat. However, several other cereal crops (e.g. cereal rye, maize, barley and oat) and wild grasses (e.g. couch grass, false oat‐grass), which are WSMV hosts, are also hosts to the mite (Table 1). In winter wheat, initial infections occur during the autumn when viruliferous mites move from WSMV‐infected volunteer wheat and other cereal and grass hosts, aided by wind, to the newly emerged wheat on which they feed and, during this process, transmit WSMV (Fig. 4).
Figure 4.
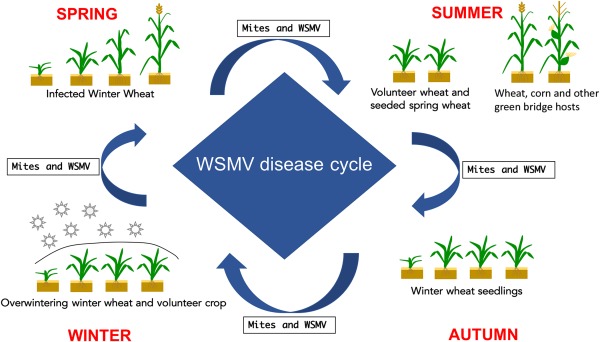
The life cycle of Wheat streak mosaic virus (WSMV).
Infections that occur in the autumn cause the most significant yield losses. The amount of yield loss is determined by the following factors: the presence of volunteer wheat and other mite and virus hosts proximal to wheat fields during planting, the density of mite populations, time of infection in the autumn, prevailing temperatures during the autumn and cultivar susceptibility. The higher the population densities of mites and mite and virus hosts near a wheat field during planting, the earlier infections occur in the autumn. The milder and more prolonged the temperatures remain in the autumn, and the higher the susceptibility of the wheat cultivar planted, the greater the yield loss (Hunger et al., 1992; Slykhuis et al., 1957). The mites overwinter as eggs, larvae, nymphs and adults in the crown and WSMV overwinters in the live tissues of wheat plants and other hosts.
In the spring, when temperatures warm up, mites become active and are spread by wind within and between fields. They feed and transmit the virus to healthy plants. During and after heading, mites move from the leaves and other above‐ground parts of the wheat plants to sites within the spikes, in which they feed and are protected. Their populations build up to high levels during spike development. When the wheat crop matures and starts to dry down, the mites must find new hosts with green tissue on which they can feed and survive during the summer. Hence, they move to volunteer wheat and other grass hosts, which serve as a green bridge for the mites and virus between harvesting and planting in the autumn. Following planting in the autumn, the mites move onto the newly emerged wheat and transmit WSMV, completing the disease cycle.
The WSMV disease cycle in spring wheat is similar to that in winter wheat, except that initial infections occur in spring after wheat emergence and the disease cycle is completed in the following spring, when mites move onto the newly emerged wheat and transmit the virus. Because of the timing of planting, the risk for significant losses as a result of WSMV in spring wheat is less than that in winter wheat. However, depending on the environmental conditions and proximity to spring wheat of infected winter wheat and other virus hosts with high mite populations, losses can be as significant in spring wheat as in winter wheat.
WSMV Diagnosis and Quantification
WSMV infection has historically been detected by means of symptoms on leaves. However, symptoms on leaves are not a reliable method for the confirmation of WSMV because other viruses can cause similar symptoms. Two near‐identical serological methods are available for the detection of WSMV which are based on enzyme‐linked immunosorbent assay (ELISA): double antibody sandwich‐ELISA (DAS‐ELISA) and triple antibody sandwich‐ELISA (TAS‐ELISA). ELISA is the most established method for the monitoring of viruses, but is less effective than methods based on cDNA amplification (polymerase chain reaction, PCR) because of its low sensitivity (Izzo et al., 2012), its inability to recognize all related viral strains (Coutts et al., 2011) and its inefficiency to interpret viral accumulations (Schubert et al., 2015).
WSMV has been detected by molecular methods, such as reverse transcription‐polymerase chain reaction (RT‐PCR) or quantitative RT‐PCR (RT‐qPCR) (Dráb et al., 2014; Gadiou et al., 2009; Schubert et al., 2015). Most of the PCR‐based detection protocols have targeted the viral CP gene (Gadiou et al., 2009; Singh and Kundu, 2017). European isolate WSMV‐ΔE has been detected by polymerase chain reaction‐restriction fragment length polymorphism (PCR‐RFLP) targeting the conserved ClaI restriction site in the core CP gene sequence (Gadiou et al., 2009). Multiplex RT‐PCR is being used not only for the detection of viral pathogens, but also for strain identification of viral pathogens. RT‐PCR and multiplex RT‐PCR provide indications of the presence or absence of WSMV, rather than the virus titre in a sample using RT‐qPCR (Chalupníková et al., 2017; Price et al., 2010b).
In contrast, RT‐qPCR has enabled the quantification of the virus concentration of several plant RNA viruses, including WSMV (Chalupníková et al., 2017; Dráb et al., 2014). The method is preferred for absolute virus quantification to study virus biology, virus gene expression, and virus–host and virus–vector interactions. Using RT‐qPCR, Tatineni et al. (2010) quantified WSMV concentrations in wheat with single and double infections by WSMV and TriMV, and revealed that the two viruses induced cultivar‐specific disease synergism in wheat. Using FAM (Fluorescein) and ATTO‐labelled (bright fluorophores) sequence‐specific probes in RT‐qPCR, Schubert et al. (2015) revealed a higher accumulation of RNA in the USA PV57 strain compared with European isolates. Overall, RT‐qPCR is preferred for absolute virus quantification to study virus biology, virus gene expression, and virus–host and virus–vector interactions.
Genetic Diversity of WSMV
WSMV is widely distributed in the wheat‐growing regions of the world, including North and South America, Australia, Asia, Europe and Russia (Table S1, see Supporting Information). The extent of the genetic diversity of WSMV has been evaluated between various isolates with different origins. Variability based on the whole genome divided WSMV isolates into three major clades, namely clade A, clade B and clade D (Schubert et al., 2015) (Fig. 5A). Clade A represents isolates from Mexico, known as El‐Batán. Clade B contains isolates from Europe, Russia and Turkey (Gadiou et al., 2009) (Table S1). Clade B isolates from Europe, also known as WSMV‐ΔE, are characterized by a deletion of triplet codon GCA at nucleotide position 8412 to 8414, resulting in deletion of the glycine amino acid at position 2761 in the sequence of the CP (Gadiou et al., 2009). Whole‐genome comparative analyses of clade B isolates revealed differences in the putative protein P1/HC‐Pro cleavage site in addition to the CP gene between European, American and Asian isolates (Choi et al., 2002; Schubert et al., 2015). The P1/HCPro cleavage site for clade A isolates is HGLRWY/GDS, clade B isolates contain the motif HGLRWY/C(G)EP(S) and isolates from America and Asia possess the motif HGL(F)RWY/GDQ (Schubert et al., 2015).
Figure 5.
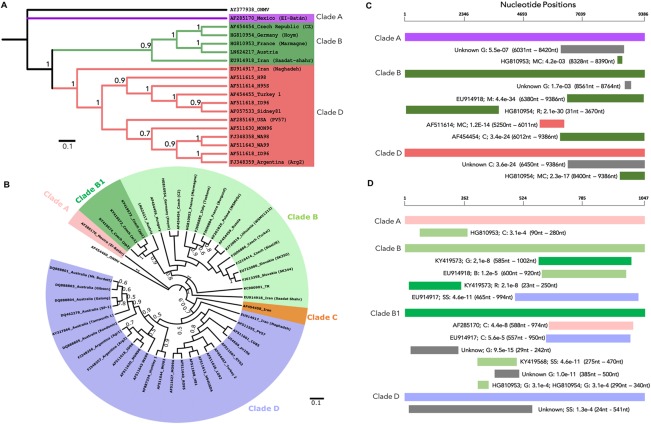
Genetic diversity of various types of Wheat streak mosaic virus (WSMV). (A) Based on the whole genome of WSMV. (B) Based on the coat protein gene sequence of WSMV. An isolate from Mexico (El‐Batán) represents clade A; isolates from Europe represent clade B and include an Asian isolate from Iran (Saadat‐shahr); WSMV grass isolates from the Czech Republic are classified into type B1; an isolate from Iran (AF454458) represents clade C; isolates from the USA, Argentina, Turkey and Australia represent clade D. Oat necrotic mottle virus (ONMV) (AY377938_ONMV) was used as an outgroup. For the generation of the tree, nucleotide sequences were aligned using ClustalX2 (Larkin et al., 2007) and the tree was constructed using MEGA 7 (Kumar et al., 2016) as described previously (Singh et al., 2018). The tree was viewed using ITOL (https://itol.embl.de/). The neighbour‐joining method was used for the construction of the tree and the reliability of the branches was inferred from a bootstrap analysis of 1000 replicates. The dataset supporting the results for the study has been submitted to the TreeBASE repository (http://treebase.org/treebase-web/home.html) and is publicly accessible at http://purl.org/phylo/treebase/phylows/study/TB2:S22140. (C, D) Recombinant analysis of WSMV based on the full genome and coat protein nucleotide sequences. Analyses were performed using various algorithms included in the RDP software package (Martin et al., 2015) as described previously (Singh and Kundu, 2017). The type strain AF285169_PV57 (USA) was used as a reference. The order of the designation of the recombination events is as follows: accession number, algorithm used (R, RDP; G, GENECONV; C, Chimaera; MaxChi; B, Bootscan; SS, SiScan; 3seq; LARD), P‐value and nucleotide position. Only recombination events with P < 0.05 detected by at least three different algorithms are shown. Numbers of events with P > 0.05 are given in parentheses. Recombination events were observed in five algorithms: RDP, GENECONV, Bootscan, Chimaera and SiScan. Algorithms MaxChi, 3seq and LARD did not detect significant recombination events.
Clade D includes isolates from North and South America, Australia, Canada and Turkey (Dwyer et al., 2007; Robinson and Murray, 2013). Clade D isolates of American origin are divided into four subclades: D1 contains isolates from the American Pacific Northwest (APNW); D2 contains isolates from Kansas and Colorado; D3 contains isolates from Kansas, Kentucky, Ohio and Missouri; and D4 contains isolates from Kansas and Nebraska, including Sidney 81 (French and Stenger, 2002). The characteristic triplet deletion in the CP, similar to WSMV‐ΔE isolates from Europe, was later identified in clade D isolates originating from North America (Robinson and Murray, 2013). Earlier phylogenetic analysis based on the CP gene showed the existence of clade C, in addition to clade A, clade B and clade D (Robinson and Murray, 2013; Stenger and French, 2009) (Fig. 5B). Clade C comprises isolates from Iran (Dwyer et al., 2007) (Table S1). More recent analysis of the WSMV whole genome from Iran revealed that one isolate (Iran_Saadat) clustered with clade B, and another isolate (Iran_Naghadeh) aligned together with clade D, resulting in the hypothesis of three distinct genotypes coexisting in Iran (Schubert et al., 2015).
WSMV has a diverse host range, and grasses serve as one of the important natural reservoirs of the virus. It has been revealed recently that WSMV which infects grasses from the Czech Republic shares high similarity with clade B isolates from other countries in Europe. Therefore, a new clade has been introduced, and is known as clade B1 (Singh and Kundu, 2017). Based on the CP gene sequence, clade A isolates share ∼79% (nucleotides) and 83% (amino acids) with clade B isolates, 73% (nucleotides) and 76% (amino acids) with clade B1 isolates (grasses) and 78% (nucleotides) and 84% (amino acids) with clade D isolates. Clade B isolates share a high similarity of ∼92% (nucleotides) and 94% (amino acids) with clade B1 isolates and ∼90% (nucleotides) and 95% (amino acids) with clade D isolates. Clade B1 isolates, represented by grasses from the Czech Republic, show a similarity of 85% (nucleotides) and 88% (amino acids) to clade D isolates (Singh and Kundu, 2017).
Whole‐genome recombinant analysis of various WSMV clades has shown that recombination mainly occurs at the 3′ end of the sequence (Schubert et al., 2015). Clade A isolates recombine with clade B isolate, HG810953_Marmagne, from France (8328–8390 nucleotides) (Fig. 5C). Clade B isolates (Europe/Asia) recombine only with isolates from within this cluster, as well as with clade D isolates (5250–6011 nucleotides) (Fig. 5C). Clade D isolates show recombination events with clade B isolate HG810954_Hoym from Germany (8400–9386 nucleotides). In addition, recombination prediction based on a partial CP gene sequence has revealed that clade A isolates recombine with isolates from clade B (isolate HG810953_Marmagne from France at 90–280 nucleotides) (Singh and Kundu, 2017) (Fig. 5D). Clade B isolates recombine within this cluster, with clade B1 (isolate KY419573_P. pretense at 585–1002 nucleotides; 23–250 nucleotides) and clade D (Fig. 5D). Clade B1 isolates are predicted to recombine with all three clades: clade A, clade B and clade D (Singh and Kundu, 2017) (Fig. 5D). There is an unknown recombination that occurs in clade D. However, further experimental investigations are required to decipher the potential outcome of recombinant analysis to expand our understanding of the various clades of WSMV.
Plant Resistance to WSMV
Resistance to WSMV was first reported in perennial Triticeae relatives, such as Thinopyrum intermedium and Thinopyrum ponticum (Chen et al., 2003; Friebe et al., 1993; Harvey et al., 1999). Three resistance genes have been identified: Wsm1, Wsm2 and Wsm3 (Fahim et al., 2012b; Friebe et al., 2009; Haley et al., 2002; Lu et al., 2012). These resistance genes have been introduced into cultivated wheat lines. The resistance gene Wsm1 is associated with chromosome 4D and has led to the release of the winter wheat cultivar Mace (Graybosch et al., 2009). The resistance gene Wsm2 is associated with chromosome arm 3BS and has led to the release of several cultivars of wheat, including RonL (Seifers et al., 2006), Snowmass (Haley et al., 2011), Clara CL (Martin et al., 2014) and Oakley CL (Zhang et al., 2015). However, both Wsm1 and Wsm2 are ineffective at higher temperatures (Seifers et al., 2013). The third true resistance gene, Wsm3, has recently been identified and has been proven to be effective at higher temperatures than Wsm1 and Wsm2 (Fahim et al., 2012b). However, Wsm3 is not yet available in any commercial wheat cultivars (Richardson et al., 2014). The commercially available WSMV‐resistant wheat cultivars were developed in the USA and there are no reports of WSMV‐resistant cultivars or other cereal species in Europe.
Resistance genes to the WCM vector have been identified in grass species: Aegilops tauschii (2n = 2x = 14, DD), Thinopyrum ponticum, (2n = 10x = 70, JJJJsJs) and Th. intermedium (2n = 6x = 42, JJsS) (Fahim et al., 2011; Fedak and Han, 2005; Qi et al., 1979). The grass genes intercross to hexaploid wheat, but very few wheat cultivars possess effective resistance against the WCM because of virulent WCM populations (Hakizimana et al., 2004; Martin et al., 1976; Murugan et al., 2011). However, WCM resistance remains a compelling approach to reduce losses caused by WSMV. Two distant hybrids between spring wheat and the grass Agropyron glaucum, Zhong1 and Zhong2, show effective resistance towards both WSMV and its WCM vector (Chen et al., 2003; Han et al., 2003; Qi et al., 1979).
Disease Management
The management of WSMV is aimed at the minimization or elimination of the risks of infection of wheat. The highest risk is volunteer wheat which emerges in a wheat field just before harvest following a hailstorm. Other risks include: volunteer wheat in summer crops other than wheat; crops or grassy weeds that are hosts of WCMs or WSMV, e.g. maize, that are allowed to grow past autumn wheat emergence; a cool, wet summer which favours the growth of volunteer wheat and other hosts, as well as the survival and reproduction of WCMs, and also prolongs the period of growth of summer host crops; a prolonged autumn with above normal temperatures; and early planting of wheat.
Because WSMV cannot be controlled by chemicals and the chemical control of WCMs is ineffective (Fritts et al., 1999), the most effective strategy for the management of WSMV is to use cultural practices. Pre‐harvest volunteer wheat, especially volunteer wheat that emerges in a wheat field as a result of a hailstorm, should be controlled with herbicides or tillage. Post‐harvest volunteer wheat should also be controlled. To be effective, volunteer wheat should be completely dead at least 2 weeks before planting. Grassy weeds in and close to fields in which wheat will be planted in the autumn should be controlled with tillage or herbicides. Early planting of wheat should be avoided. The combined effects of mites and virus when wheat is planted early include heavy and widespread infections in the autumn, leading to severe epidemics the following spring that result in substantial yield losses.
Wheat should not be planted next to late‐maturing summer crops that are hosts to WCMs or WSMV, such as maize, foxtail millet, sorghum or small grain cover crops. When available, wheat cultivars with greater resistance or tolerance to WSMV that are adapted to the local area or region should be planted. High‐risk wheat fields should be planted last. These are the fields adjacent to grassy weeds and late‐maturing host crops. An integrated disease management approach that combines as many as possible of these strategies and tactics will most effectively reduce losses caused by WSMV, as illustrated in McMechan and Hein (2016), who showed that cultivar resistance and delayed planting improved the yields of three winter wheat cultivars under high WSMV intensity.
Conclusions and Future Direction
WSMV continues to be a threat to wheat production worldwide. Research is needed that will provide information to enhance our understanding of the biology, ecology and epidemiology of the disease and its WCM vector, including the knowledge that the WCM constitutes a species complex. Improved techniques for rapid detection and diagnosis will be essential to growers in making timely and informed management decisions. These techniques include the use of molecular tools, such as RT‐PCR and RT‐qPCR. In addition, the increasingly common whole‐genome sequencing approach provides the opportunity to search for signatures of polyphagy, detoxification and WSMV vectoring abilities in different WCM biotypes, which offers new possibilities for the development of wheat protection strategies. The genetic engineering of resistance to WSMV in wheat, for example through the expression of artificial polycistronic microRNA (Fahim et al., 2012a) and gene silencing (Li et al., 2005), will complement traditional resistance breeding strategies to achieve higher and more effective levels of resistance. One recent addition to genetic engineering is the development of the characteristic clustered regularly interspaced short palindromic repeats/CRISPR‐associated 9 (CRISPR/Cas9) protein that has emerged as a potent genome‐editing tool to confer resistance against geminiviruses (Baltes et al., 2015). Therefore, it will be intriguing to implement CRISPR/Cas9 to modify WSMV/WCM genes in order to develop effective resistance against virus and vector.
WSMV mutants with deletion in amino acids in the CP region are capable of systemic infection, although with delayed and milder symptoms (Tatineni and French, 2014). Therefore, the availability of a series of viable CP deletion mutants of WSMV will greatly facilitate our understanding of the complexity of WSMV–host interactions. Furthermore, it will be interesting to identify the different amino acids in different strains of WSMV that are vital for the interactions with the vector and hosts using molecular approaches. The interaction of viruses with their hosts is a rather complex and dynamic process, involving numerous interactions amongst viral proteins and host proteins. An improved understanding of the complex interactions of WSMV‐derived proteins that alter the host cellular machinery, as well as the identification of host genes, will contribute to the development of novel sources of resistance and other control measures. New technology, such as next‐generation sequencing (RNA‐sequencing) of hosts infected with WSMV, will provide valuable insights into host factors that differentially interact with the virus, thus enhancing our understanding of the mechanisms of host–virus interaction, as well as the nature and mechanisms of long‐distance transport of viruses in monocot plants.
Climate change poses new challenges because of its influence on the biology, ecology and epidemiology of WSMV and its WCM vector. The current trend in climate change is towards warmer temperatures globally. The implications of this trend are that there will be more frequent outbreaks of severe WSMV epidemics over larger areas or regions. The increased frequency of outbreaks of severe epidemics, coupled with an increased probability of long‐distance dispersal through the exchange of infected germplasm amongst researchers locally, regionally and globally, means that greater yield losses will be expected. Concerted efforts will be needed to mitigate these losses. These will include breeding for resistance to WSMV and the WCM using traditional methods, as well as molecular tools; vigilance in implementing management tactics, especially the control of volunteer wheat and other crop and grass hosts of WSMV and the WCM; modification or adaptation of management tactics to account for climate change and differences in the biology and ecology of WCM biotypes; and educating growers, crop consultants, extension educators and the public about the disease and how to manage it to protect yields.
Supporting information
Table S1 Diversity of Wheat streak mosaic virus isolates.
Acknowledgements
Work in J.K.K.'s laboratory was supported by grants QJ1530373 and RO0417 from the Ministry of Agriculture, Czech Republic. A.S. was supported by the National Science Centre in Poland (grant no. UMO‐2016/21/B/NZ8/00786) (results on WCM MT‐1 and MT‐8 dispersal and colonization abilities were obtained during the course of this project).
References
- Baltes, N.J. , Hummel, A.W. , Konecna, E. , Cegan, R. , Bruns, A.N. , Bisaro, D.M. and Voytas, D.F. (2015) Conferring resistance to geminiviruses with the CRISPR‐Cas prokaryotic immune system. Nat. Plants. 1, 15 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belliure, B. , Janssen, A. , Maris, P.C. , Peters, D. and Sabelis, M.W. (2005) Herbivore arthropods benefit from vectoring plant viruses. Ecol. Lett. 8, 70–79. [Google Scholar]
- Brakke, M.K. (1971) Wheat Streak Mosaic Virus. CMI/AAB Descriptions of Plant Viruses No. 48. Wellesbourne, Warwickshire: Association of Applied Biologists; Available at: http://www.dpvweb.net/dpv/showdpv.php?dpvno=048 [accessed on 15 October 2017]. [Google Scholar]
- Byamukama, E. , Tatineni, S. , Hein, G. , McMechan, J. and Wegulo, S.N. (2016) Incidence of Wheat streak mosaic virus, Triticum mosaic virus, and Wheat mosaic virus in wheat curl mites recovered from maturing winter wheat spikes. Plant Dis. 100, 318–323. [DOI] [PubMed] [Google Scholar]
- Byamukama, E. , Wegulo, S.N. , Tatineni, S. , Hein, G.L. , Graybosch, R.A. , Baenziger, P.S. and French, R. (2014) Quantification of yield loss caused by Triticum mosaic virus and Wheat streak mosaic virus in winter wheat under field conditions. Plant Dis. 98, 127–133. [DOI] [PubMed] [Google Scholar]
- Carew, M. , Schiffer, M. , Umina, P. , Weeks, A. and Hoffmann, A. (2009) Molecular markers indicate that the wheat curl mite, Aceria tosichella Keifer, may represent a species complex in Australia. Bull. Entomol. Res. 99, 479–486. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C. , Kasschau, K.D. , Mahajan, S.K. and Schaad, M.C. (1996) Cell‐to‐cell and long‐distance transport of viruses in plants. Plant Cell, 8, 1669–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalupníková, J. , Kundu, J.K. , Singh, K. , Bartaková, P. and Beoni, E. (2017) Wheat streak mosaic virus: incidence in field crops, potential reservoir within grass species and uptake in winter wheat cultivars. J. Integr. Agric. 16, 60 345–60 357. [Google Scholar]
- Chen, Q. , Conner, R.L. , Li, H.J. , Sun, S.C. , Ahmad, F. , Laroche, A. and Graf, R.J. (2003) Molecular cytogenetic discrimination and reaction to Wheat streak mosaic virus and the wheat curl mite in Zhong series of wheat–Thinopyrum intermedium partial amphiploids. Genome, 46, 135–145. [DOI] [PubMed] [Google Scholar]
- Choi, I.R. , Horken, K.M. , Stenger, D.C. and French, R. (2002) Mapping of the P1 proteinase cleavage site in the polyprotein of Wheat streak mosaic virus (genus Tritimovirus). J. Gen. Virol. 83, 443–450. [DOI] [PubMed] [Google Scholar]
- Christian, M.L. and Willis, W.G. (1993) Survival of Wheat streak mosaic virus in grass hosts in Kansas from wheat harvest to fall wheat emergence. Plant Dis. 77, 239–242. [Google Scholar]
- Chung, B.Y. , Miller, W.A. , Atkins, J.F. and Firth, A.E. (2008) An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. USA, 105, 5897–5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connin, R.V. (1956) The host range of the wheat curl mite, vector of Wheat streak mosaic . J. Econ. Entomol. 48, 1–4. [Google Scholar]
- Coutts, B.A. , Banovic, M. , Kehoe, M.A. , Severtson, D.L. and Jones, R.A.C. (2014) Epidemiology of Wheat streak mosaic virus in wheat in a Mediterranean‐type environment. Eur. J. Plant Pathol. 140, 797–813. [Google Scholar]
- Coutts, B.A. , Hammond, N.E.B. , Kehoe, M.A. and Jones, R.A.C. (2008a) Finding Wheat streak mosaic virus in southwest Australia. Aust. J. Agric. Res. 59, 836–843. [Google Scholar]
- Coutts, B.A. , Kehoe, M.A. , Webster, C.G. , Wylie, S.J. and Jones, R.A.C. (2011) Zucchini yellow mosaic virus: biological properties, detection procedures and comparison of coat protein gene sequences. Arch. Virol. 156, 2119–2131. [DOI] [PubMed] [Google Scholar]
- Coutts, B.A. , Strickland, G.R. , Kehoe, M.A. , Severtson, D.L. and Jones, R.A.C. (2008b) The epidemiology of Wheat streak mosaic virus in Australia: case histories, gradients, mite vectors, and alternative hosts. Aust. J. Agric. Res. 59, 844–853. [Google Scholar]
- de Wolf, E. and Seifers, D. (2008) Triticum mosaic: a new wheat disease in Kansas. Kansas State University, Agricultural Experiment Station and Cooperative Extension Service (EP‐145). Available at: https://www.bookstore.ksre.ksu.edu/pubs/EP145.pdf [accessed on 20 November 2017].
- del Rosario, M.S. and Sill, W.H. Jr. (1965) Physiological strains of Aceria tulipae and their relationships to the transmission of Wheat streak mosaic virus . Phytopathology, 55, 1168–1175. [Google Scholar]
- Dráb, T. , Svobodová, E. , Ripl, J. , Jarošová, J. , Rabenstein, F. , Melcher, U. and Kundu, J.K. (2014) SYBR Green I based RT‐qPCR assays for the detection of RNA viruses of cereals and grasses. Crop Pasture Sci. 65, 1323–1328. [Google Scholar]
- Dwyer, G.I. , Gibbs, M.J. , Gibbs, A.J. and Jones, R.A.C. (2007) Wheat streak mosaic virus in Australia: relationship to isolates from the Pacific Northwest of the USA and its dispersion via seed transmission. Plant Dis. 91, 164–170. [DOI] [PubMed] [Google Scholar]
- Ellis, M.H. , Rebetzke, G.J. , Kelman, W.M. , Moore, C.S. and Hyles, J.E. (2004) Detection of Wheat streak mosaic virus in four pasture grass species in Australia. Plant Pathol. 53, 239. [Google Scholar]
- Fahim, M. , Larkin, P.J. , Haber, S. , Shorter, S. , Lonergan, P.F. and Rosewarne, G.M. (2012b) Effectiveness of three potential sources of resistance in wheat against Wheat streak mosaic virus under field conditions. Australas. Plant Pathol. 41, 301–309. [Google Scholar]
- Fahim, M. , Mechanicos, A. , Ayala‐Navarrete, L. , Haber, S. and Larkin, P.J. (2011) Resistance to Wheat streak mosaic virus – a survey of resources and development of molecular markers. Plant Pathol. 61, 425–440. [Google Scholar]
- Fahim, M. , Millar, A.A. , Wood, C.C. and Larkin, P.J. (2012a) Resistance to Wheat streak mosaic virus generated by expression of an artificial polycistronic microRNA in wheat. Plant Biotechnol. J. 10, 150–163. [DOI] [PubMed] [Google Scholar]
- Fedak, G. and Han, F. (2005) Characterization of derivatives from wheat–Thinopyrum wide crosses. Cytogenet. Genome Res. 109, 360–367. [DOI] [PubMed] [Google Scholar]
- French, R. and Stenger, D.C. (2002) Wheat Streak Mosaic Virus. CMI/AAB Descriptions of Plant Viruses No. 398. Wellesbourne, Warwickshire: Association of Applied Biologists; Available at: http://www.dpvweb.net/dpv/showdpv.php?dpvno=393 [accessed on 2 September 2017]. [Google Scholar]
- French, R. and Stenger, D.C. (2003) Evolution of Wheat streak mosaic virus: dynamics of population growth within plants may explain limited variation. Annu. Rev. Phytopathol. 41, 199–214. [DOI] [PubMed] [Google Scholar]
- Friebe, B. , Jiang, J. , Gill, B.S. and Dyck, P.L. (1993) Radiation‐induced nonhomologous wheat Agropyron intermedium chromosomal translocations conferring resistance to leaf rust. Theor. Appl. Genet. 86, 141–149. [DOI] [PubMed] [Google Scholar]
- Friebe, B. , Qi, L.L. , Wilson, D.L. , Chang, Z.J. , Seifers, D.L. , Martin, T.J. , Fritz, A.K. and Gill, B.S. (2009) Wheat–Thinopyrum intermedium recombinants resistant to Wheat streak mosaic virus and Triticum mosaic virus . Crop Sci. 49, 1221–1226. [Google Scholar]
- Fritts, D.A. , Michels, G.J., Jr. and Rush, C.M. (1999) The effects of planting date and insecticide treatments on the incidence of High Plains Disease in corn. Plant Dis. 83, 1125–1128. [DOI] [PubMed] [Google Scholar]
- Gadiou, S. , Kúdela, O. , Ripl, J. , Rabenstein, F. , Kundu, J.K. and Glasa, M. (2009) An amino acid deletion in Wheat streak mosaic virus capsid protein distinguishes a homogeneous group of European isolates and facilitates their specific detection. Plant Dis. 93, 1209–1213. [DOI] [PubMed] [Google Scholar]
- Graybosch, R.A. , Peterson, C.J. , Baenziger, P.S. , Baltensperger, D.D. , Nelson, L.A. , Jin, Y. , Kolmer, J. , Seabourn, B. , French, R. , Hein, G. , Martin, T.J. , Beecher, B. , Schwarzacher, T. and Heslop‐Harrison, P. (2009) Registration of ‘Mace’ hard red winter wheat. J. Plant Regist. 3, 51–56. [Google Scholar]
- Hadi, B.A.R. , Langham, M.A.C. , Osborne, L. and Tilmon, K.J. (2011) Wheat streak mosaic virus on wheat: biology and management. J. Integr. Pest Manag. 2, 1–5. [Google Scholar]
- Hakizimana, F. , Ibrahim, A.M.H. , Langham, M.A.C. , Rudd, J.C. and Haley, S.D. (2004) Generation means analysis of Wheat streak mosaic virus resistance in winter wheat. Euphytica, 139, 133–139. [Google Scholar]
- Haley, S.D. , Johnson, J.J. , Peairs, F.B. , Stromberger, J.A. , Heaton, E.E. , Seifert, S.A. , Kottke, R.A. , Rudolph, J.B. , Martin, T.J. , Bai, G. , Chen, X. , Bowden, R.L. , Jin, Y. , Kolmer, J.A. , Seifers, D.L. , Chen, M.‐S. and Seabourn, B.W. (2011) Registration of ‘Snowmass’ wheat. J. Plant Regist. 5, 87–90. [Google Scholar]
- Haley, S.D. , Martin, T.J. , Quick, J.S. , Seifers, D.L. , Stromberger, J.A. , Clayshulte, S.R. , Clifford, B.L. , Peairs, F.B. , Rudolph, J.B. , Johnson, J.J. , Gill, B.S. and Friebe, B. (2002) Registration of CO960293‐2 wheat germplasm resistant to Wheat streak mosaic virus and Russian wheat aphid. Crop Sci. 42, 1381–1382. [Google Scholar]
- Han, F.P. , Fedak, G. , Benabdelmouna, A. , Armstrong, K. and Ouellet, T. (2003) Characterization of six wheat × Thinopyrum intermedium derivatives by GISH, RFLP, and multicolor GISH. Genome, 46, 490–495. [DOI] [PubMed] [Google Scholar]
- Harvey, T.L. , Seifers, D.L. , Martin, T.J. , Brown‐Guedira, G. and Gill, B.S. (1999) Survival of wheat curl mites on different sources of resistance in wheat. Crop Sci. 39, 1887–1889. [Google Scholar]
- Hein, G.L. , French, R. , Siriwetwiwat, B. and Amrine, J.W. (2012) Genetic characterization of North American populations of wheat curl mite and dry bulb mite. J. Econ. Entomol. 105, 1801–1808. [DOI] [PubMed] [Google Scholar]
- Hill, J.H. , Martinson, C.A. and Russell, W.A. (1974) Seed transmission of Maize dwarf mosaic and Wheat streak mosaic viruses in maize and response of inbred lines. Crop Sci. 14, 232–235. [Google Scholar]
- Hunger, R.M. , Sherwood, J.L. , Evans, C.K. and Montana, J.R. (1992) Effects of planting date and inoculation date on severity of Wheat streak mosaic in hard red winter wheat cultivars. Plant Dis. 76, 1056–1060. [Google Scholar]
- Ito, D. , Miller, Z. , Menalled, F. , Moffet, M. and Burrows, M. (2012) Relative susceptibility among alternative host species prevalent in the Great Plains to Wheat streak mosaic virus . Plant Dis. 96, 1185–1192. [DOI] [PubMed] [Google Scholar]
- Izzo, M.M. , Kirkland, P.D. , Gu, X. , Lele, Y. , Gunn, A.A. and House, J.K. (2012) Comparison of three diagnostic techniques for detection of rotavirus and coronavirus in calf faeces in Australia. Aust. Vet. J. 90, 122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, R.A.C. , Coutts, B.A. , Mackie, A.E. and Dwyer, G.I. (2005) Seed transmission of Wheat streak mosaic virus shown unequivocally in wheat. Plant Dis. 89, 1048–1050. [DOI] [PubMed] [Google Scholar]
- Khadivar, R.S. and Nasrolahnejad, S. (2009) Serological and molecular detection of Wheat streak mosaic virus (WSMV) in cereal fields of Golestan province, Northern Iran. Int. J. Plant Prod. 16, 4. [Google Scholar]
- Kiedrowicz, A. , Kuczyński, L. , Laska, A. , Lewandowski, M. , Proctor, H. and Skoracka, A. (2017a) Dispersal strategies in passively spreading phytophagous mites. In: ASAB Easter Conference 2017, Liverpool, 5–7 April 2017 Abstract book, p. 10. The Association for the Study of Animal Behaviour, University of Liverpool, Liverpool.
- Kiedrowicz, A. , Kuczyński, L. , Lewandowski, M. , Proctor, H. and Skoracka, A. (2017b) Behavioural responses to potential dispersal cues in two economically important species of cereal‐feeding eriophyid mites. Sci. Rep. 7, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczyński, L. , Rector, B.G. , Kiedrowicz, A. , Lewandowski, M. , Szydło, W. and Skoracka, A. (2016) Thermal niches of two invasive genotypes of the wheat curl mite Aceria tosichella: congruence between physiological and geographical distribution data. PLoS One, 11, e0154600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S. , Stecher, G. and Tamura, K. (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, M.A. , Blackshields, G. , Brown, N.P. , Chenna, R. , McGettigan, P.A. , McWilliam, H. , Valentin, F. , Wallace, I.M. , Wilm, A. , Lopez, R. , Thompson, J.D. , Gibson, T.J. and Higgins, D.G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics, 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- Li, Z. , Liu, Y. and Berger, P.H. (2005) Transgene silencing in wheat transformed with the WSMV‐CP gene. Biotechnology, 4, 62–68. [Google Scholar]
- Llave, C. , Martinez, B. , Diaz‐Ruiz, J.R. and Lopez‐Abella, D. (2002) Amino acid substitutions witin the cys‐rich domain of the tobacco etch potyvirus HC‐Pro result in loss of transmissibility by aphids. Arch. Virol. 147, 2365–2375. [DOI] [PubMed] [Google Scholar]
- Lu, H. , Kottke, R. , Devkota, R. , Amand, P.S. , Bernardo, A. , Bai, G. , Byrne, P. , Martin, T.J. , Haley, S.D. and Rudd, J. (2012) Consensus mapping and identification of markers for marker assisted selection in wheat. Crop Sci. 52, 720–728. [Google Scholar]
- Mandadi, K.K. , Pyle, J.D. and Scholthof, K.B.G. (2014) Comparative analysis of antiviral responses in Brachypodium distachyon and Setaria viridis reveal conserved and unique outcomes among C3 and C4 plant defenses. Mol. Plant Microbe Interact. 27, 1277–1290. [DOI] [PubMed] [Google Scholar]
- Martin, T.J. , Harvey, T.L. and Livers, R.W. (1976) Resistance to Wheat streak mosaic virus and its vector Aceria tulipae . Phytopathology, 66, 346–349. [Google Scholar]
- Martin, D.P. , Murrell, B. , Golden, M. , Khoosal, A. and Muhire, B. (2015) RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. 1, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, T.J. , Zhang, G. , Fritz, A.K. , Miller, R. and Chen, M. (2014) Registration of ‘Clara CL’ wheat. J. Plant Regist. 8, 38–42. [Google Scholar]
- McKinney, H.H. (1937) Mosaic Diseases of Wheat and Related Cereals US Department of Agriculture Circular No. 442, 1–23. Available at: https://archive.org/stream/mosaicdiseasesof442mcki/mosaicdiseasesof442mcki_djvu.txt [accessed on 1 October 2017].
- McMechan, A.J. and Hein, G.L. (2016) Planting date and variety selection for management of viruses transmitted by the wheat curl mite (Acari: Eriophyidae). J. Econ. Entomol. 109, 70–77. [DOI] [PubMed] [Google Scholar]
- Miller, Z.J. , Lehnhoff, E.A. , Menalled, F.D. and Burrows, M. (2015) Effects of soil nitrogen and atmospheric carbon dioxide on Wheat streak mosaic virus and its vector (Aceria tosichella Kiefer). Plant Dis. 99, 1803–1807. [DOI] [PubMed] [Google Scholar]
- Miller, A.D. , Skoracka, A. , Navia, D. , Mendonca, R.S. , Szydło, W. , Schultz, M.B. , Michael Smith, C. , Truol, G. and Hoffmann, A.A. (2013) Phylogenetic analyses reveal extensive cryptic speciation and host specialization in an economically important mite taxon. Mol. Phylogenet. Evol. 66, 928–940. [DOI] [PubMed] [Google Scholar]
- Miller, A.D. , Umina, P.A. , Weeks, A.R. and Hoffmann, A.A. (2012) Population genetics of the wheat curl mite (Aceria tosichella Keifer) in Australia: implications for the management of wheat pathogens. Bull. Entomol. Res. 102, 199–212. [DOI] [PubMed] [Google Scholar]
- Murugan, M. , Sotelo Cardona, P. , Duraimurugan, P. , Whitfield, A.E. , Schneweis, D. , Starkey, S. and Smith, C.M. (2011) Wheat curl mite resistance: interactions of mite feeding with wheat streak mosaic virus infection. J. Econ. Entomol. 104, 1406–1414. [DOI] [PubMed] [Google Scholar]
- Navia, D. , de Mendonca, R.S. , Skoracka, A. , Szydło, W. , Knihinicki, D. , Hein, G.L. , da Silva Pereira, P.R. , Truol, G. and Lau, D. (2013) Wheat curl mite, Aceria tosichella, and transmitted viruses: an expanding pest complex affecting cereal crops. Exp. Appl. Acarol. 59, 95–143. [DOI] [PubMed] [Google Scholar]
- Oldfield, G.N. and Proeseler, G. (1996) Eriophyoid mites as vectors of plant pathogens In: Eriophyoid Mites: Their Biology, Natural Enemies and Control (Lindquist E.E., Sabelis M.W., Bruin J., eds), pp. 259–273. Amsterdam: Elsevier Science BV. [Google Scholar]
- Oliveira‐Hofman, C. , Wegulo, S.N. , Tatineni, S. and Hein, G.L. (2015) Impact of Wheat streak mosaic virus and Triticum mosaic virus coinfection of wheat on transmission rates by wheat curl mites. Plant Dis. 99, 1170–1174. [DOI] [PubMed] [Google Scholar]
- Orlob, G.B. (1966) Feeding and transmission characteristics of Aceria tulipae Keifer as vector of Wheat streak mosaic virus . J. Phytopathol. 55, 218–238. [Google Scholar]
- Paliwal, Y.C. (1980) Relationship of Wheat streak mosaic and Barley stripe mosaic viruses to vector and nonvector eriophyid mites. Arch. Virol. 63, 129–132. [DOI] [PubMed] [Google Scholar]
- Price, J.A. , Smith, J. , Simmons, A. , Fellers, J. and Rush, C.M. (2010b) Multiplex real‐time RT‐PCR for detection of Wheat streak mosaic virus and Tritcum mosaic virus . J. Virol. Methods, 165, 198–201. [DOI] [PubMed] [Google Scholar]
- Price, J. , Workneh, F. , Evett, S. , Jones, D. , Arthur, J. and Rush, C.M. (2010a) Effects of Wheat streak mosaic virus on root development and water‐use efficiency of hard red winter wheat. Plant Dis. 94, 766–770. [DOI] [PubMed] [Google Scholar]
- Qi, S.Y. , Yu, S. , Zhang, X.Y.H. , Yu, G.H. and Song, F.Y. (1979) Studies on distant hybridization between spring wheat and Agropyron glaucum . Sci. Agric. Sinica, 2, 1–11. [Google Scholar]
- Rabenstein, F. , Seifers, D.L. , Schubert, J. , French, R. and Stenger, D.C. (2002) Phylogenetic relationships, strain diversity and biogeography of tritimoviruses. J. Gen. Virol. 83, 895–906. [DOI] [PubMed] [Google Scholar]
- Richardson, K. , Miller, A.D. , Hoffmann, A.A. and Larkin, P. (2014) Potential new sources of wheat curl mite resistance in wheat to prevent the spread of yield‐reducing pathogens. Exp. Appl. Acarol. 64, 1–19. [DOI] [PubMed] [Google Scholar]
- Robinson, M.D. and Murray, T.D. (2013) Genetic variation of Wheat streak mosaic virus in the United States Pacific Northwest. Phytopathology, 103, 98–104. [DOI] [PubMed] [Google Scholar]
- Rojas, M.R. , Zerbini, F.M. , Allison, R.F. , Gilbertson, R.L. and Lucas, W.J. (1997) Capsid protein and helper component‐proteinase function as Potyvirus cell‐to‐cell movement proteins. Virology, 237, 283–295. [DOI] [PubMed] [Google Scholar]
- Ruiz‐Ferrer, V. , Boskovic, J. , Alfonso, C. , Rivas, G. , Llorca, O. , Lopez‐Abella, D. and Lopez‐Moya, J.J. (2005) Structural analysis of tobacco etch potyvirus HC‐Pro oligomers involved in aphid transmission. J. Virol. 79, 3758–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaad, M.C. , Haldeman‐Cahill, R. , Cronin, S. and Carrington, J.C. (1996) Analysis of the VPg‐proteinase (NIa) encoded by tobacco etch potyvirus: effects of mutations on subcellular transport, proteolytic processing, and genome amplification. J. Virol. 70, 7039–7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer, M. , Umina, P. , Carew, M. , Hoffmann, A. , Rodoni, B. and Miller, A. (2009) The distribution of wheat curl mite (Aceria tosichella) lineages in Australia and their potential to transmit Wheat streak mosaic virus . Ann. Appl. Biol. 155, 371–379. [Google Scholar]
- Schubert, J. , Ziegler, A. and Rabenstein, F. (2015) First detection of Wheat streak mosaic virus in Germany: molecular and biological characteristics. Arch. Virol. 160, 1761–1766. [DOI] [PubMed] [Google Scholar]
- Seifers, D.L. , Haber, S. , Martin, T.J. and Zhang, G. (2013) New sources of temperature sensitive resistance to Wheat streak mosaic virus in wheat. Plant Dis. 97, 1051–1056. [DOI] [PubMed] [Google Scholar]
- Seifers, D.L. , Harvey, T.L. , Kofoid, K.D. and Stegmeier, W.D. (1996) Natural infection of pearl millet and sorghum by Wheat streak mosaic virus in Kansas. Plant Dis. 80, 179–185. [Google Scholar]
- Seifers, D.L. , Harvey, T.L. , Louie, R. , Gordon, D.T. and Martin, T.J. (2002) Differential transmission of isolates of the High Plains virus by different sources of wheat curl mites. Plant Dis. 86, 138–142. [DOI] [PubMed] [Google Scholar]
- Seifers, D.L. , Martin, T.J. and Fellers, J.P. (2010) An experimental host range for Triticum mosaic virus . Plant Dis. 94, 1125–1131. [DOI] [PubMed] [Google Scholar]
- Seifers, D.L. , Martin, T.J. and Fellers, J.P. (2011) Occurrence and yield effects of wheat infected with Triticum mosaic virus in Kansas. Plant Dis. 95, 183–188. [DOI] [PubMed] [Google Scholar]
- Seifers, D.L. , Martin, T.J. , Harvey, T.L. , Haber, S. and Haley, S.D. (2006) Temperature sensitive and efficacy of Wheat streak mosaic virus resistance derived from CO960293 wheat. Plant Dis. 90, 623–628. [DOI] [PubMed] [Google Scholar]
- Sill, W.H., Jr. and Agusiobo, P.C. (1955) Host range studies of the Wheat streak mosaic virus. Plant Dis. Rep. 39, 633–642. [Google Scholar]
- Sill, W.H., Jr. and Connin, R.V. (1953) Summary of the known host range of Wheat streak mosaic virus. Trans. Kans. Acad. Sci. 56, 411–417. [Google Scholar]
- Singh, K. and Kundu, J.K. (2017) Variations in Wheat streak mosaic virus coat protein sequence among crop and non‐crop hosts. Crop and Pasture Sci. 68, 328–336. [Google Scholar]
- Singh, K. , Winter, M. , Zouhar, M. and Ryšánek, P. (2018) Cyclophilins: less studied proteins with critical roles in pathogenesis. Phytopathology, 108, 6–14. [DOI] [PubMed] [Google Scholar]
- Siriwetwiwat, B. (2006) Interactions between the wheat curl mite Aceria tosichella Keifer (Eriophyidae), and Wheat streak mosaic virus and distribution of wheat curl mite biotypes in the field. PhD dissertation, University of Nebraska‐Lincoln.
- Skoracka, A. , Kuczyński, L. , Santos de Mendonça, R. , Dabert, M. , Szydło, W. , Knihinicki, D. , Truol, G. and Navia, D. (2012) Cryptic species within the wheat curl mite Aceria tosichella (Keifer) (Acari: Eriophyoidea), revealed by mitochondrial, nuclear and morphometric data. Invertebr. Syst. 26, 417–433. [Google Scholar]
- Skoracka, A. , Kuczyński, L. , Szydło, W. and Rector, B. (2013) The wheat curl mite Aceria tosichella (Acari: Eriophyoidea) is a complex of cryptic lineages with divergent host ranges: evidence from molecular and plant bioassay data. Biol. J. Linnean. Soc. 109, 165–180. [Google Scholar]
- Skoracka, A. , Lewandowski, M. , Rector, B.G. , Szydło, W. and Kuczyński, L. (2017) Spatial and host‐related variation in prevalence and population density of wheat curl mite (Aceria tosichella) cryptic genotypes in agricultural landscapes. PLoS One, 12, e0169874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoracka, A. , Rector, B. , Kuczyński, L. , Szydło, W. , Hein, G. and French, R. (2014) Global spread of wheat curl mite by its most polyphagous and pestiferous lineages. Ann. Appl. Biol. 165, 222–235. [Google Scholar]
- Slykhuis, J.T. (1955) Aceria tulipae Keifer (Acarina: Eriophyidae) in relation to the spread of Wheat streak mosaic . Phytopathology, 45, 116–128. [Google Scholar]
- Slykhuis, J.T. , Andrews, J.E. and Pittman, U.J. (1957) Relation of date of seeding winter wheat in southern Alberta to losses from Wheat streak mosaic, root rot, and rust. Can. J. Plant Sci. 37, 113–127. [Google Scholar]
- Somsen, H.W. and Sill, W.H. (1970) The wheat curl mite, Aceria tulipae Keifer, in relation to epidemiology and control of wheat streak mosaic. Available at: https://www.ksre.k-state.edu/historicpublications/pubs/STB162.pdf [accessed on 20 September 2017].
- Staples, R. and Allington, W.B. (1956) Streak Mosaic of Wheat in Nebraska and its Control, Research Bulletin No. 178. Lincoln, NE: University of Nebraska‐Lincoln College of Agriculture, Agricultural Experiment Station.
- Stenger, D.C. and French, R. (2009) Wheat streak mosaic virus genotypes introduced to Argentina are closely related to isolates from the American Pacific Northwest and Australia. Arch. Virol. 154, 331–336. [DOI] [PubMed] [Google Scholar]
- Stenger, D.C. , French, R. and Gildow, F.E. (2005b) Complete deletion of Wheat streak mosaic virus HC‐Pro: a null mutant is viable for systemic infection. J. Virol. 79, 12 077–12 080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenger, D.C. , Hall, J.S. , Choi, I. and French, R. (1998) Phylogenetic relationships within the family Potyviridae: Wheat streak mosaic virus and Brome streak mosaic virus are not members of the genus Rymovirus. Phytopathology, 88, 782–787. [DOI] [PubMed] [Google Scholar]
- Stenger, D.C. , Hein, G.L. , Gildow, F.E. , Horken, K.M. and French, R. (2005a) Plant virus HC‐Pro is a determinant of eriophyid mite transmission. J. Virol. 79, 9054–9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenger, D.C. , Young, B.A. and French, R. (2006) Random mutagenesis of Wheat streak mosaic virus HC‐Pro: noninfectious interfering mutations in a gene dispensable for systemic infection of plants. J. Gen. Virol. 87, 2741–2747. [DOI] [PubMed] [Google Scholar]
- Stenger, D.C. , Young, B.A. , Qu, F. , Morris, T.J. and French, R. (2007) Wheat streak mosaic virus lacking HC‐Pro is competent to produce disease synergism in double infections with Maize chlorotic mottle virus. Phytopathology, 97, 1213–1221. [DOI] [PubMed] [Google Scholar]
- Tatineni, S. , Elowsky, C. and Graybosch, R.A. (2017) Wheat streak mosaic virus coat protein deletion mutants elicit more severe symptoms than wild‐type virus in multiple cereal hosts. Mol. Plant–Microbe Interact. 30, 974–983. [DOI] [PubMed] [Google Scholar]
- Tatineni, S. and French, R. (2014) The C‐terminus of Wheat streak mosaic virus coat protein is involved in differential infection of wheat and maize through host‐specific long‐distance transport. Mol. Plant–Microbe Interact. 27, 150–162. [DOI] [PubMed] [Google Scholar]
- Tatineni, S. , Graybosch, R.A. , Hein, G.L. , Wegulo, S.N. and French, R. (2010) Wheat cultivar‐specific disease synergism and alteration of virus accumulation during co‐infection with Wheat streak mosaic virus and Triticum mosaic virus . Phytopathology, 100, 230–238. [DOI] [PubMed] [Google Scholar]
- Tatineni, S. , McMechan, A.J. and Hein, G.L. (2018) Wheat streak mosaic virus coat protein is a determinant for vector transmission by the wheat curl mite. Virology, 514, 42–49. [DOI] [PubMed] [Google Scholar]
- Tatineni, S. , Van Winkle, D.H. and French, R. (2011) The N‐terminal region of Wheat streak mosaic virus coat protein is a host‐ and strain‐specific long‐distance transport factor. J. Virol. 85, 1718–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truol, G. , Sagadin, M. and Rodriguez, M. (2010) Fox tail millet (Setaria italica L.): a new reservoir species of the Wheat streak mosaic virus (WSMV) in the province of Buenos Aires. Biocell, 34, A135. [Google Scholar]
- Vacke, J. , Zacha, V. and Jokeš, M. (1986) Identification of virus in wheat new to Czechoslovakia. In: Proceedings of the Xth Czechoslovak Plant Protection Conference, Brno. 209–210.
- Vincent, S.J. , Coutts, B.A. and Jones, R.A.C. (2014) Effects of introduced and indigenous viruses on native plants: exploring their disease causing potential at the agro‐ecological interface. PLoS One, 9, e91224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegulo, S.N. , Hein, G.L. , Klein, R.N. and French, R.C. (2008) Managing Wheat Streak Mosaic Nebraska Cooperative Extension EC1871. Lincoln, NE: University of Nebraska. Available at: http://extensionpublications.unl.edu/assets/pdf/ec1871.pdf [accessed on 20 August 2017].
- Wosula, E.N. , McMechan, A.J. , Oliveira‐Hofman, C. , Wegulo, S.N. and Hein, G.L. (2016) Differential transmission of two isolates of Wheat streak mosaic virus by five wheat curl mite populations. Plant Dis. 100, 154–158. [DOI] [PubMed] [Google Scholar]
- Young, B.A. , Hein, G.L. , French, R. and Stenger, D.C. (2007) Substitution of conserved cysteine residues in Wheat streak mosaic virus HC‐Pro abolishes virus transmission by the wheat curl mite. Arch. Virol. 152, 2107–2111. [DOI] [PubMed] [Google Scholar]
- Young, B.A. , Stenger, D.C. , Qu, F. , Morris, T.J. , Tatineni, S. and French, R. (2012) Tritimovirus P1 functions as a suppressor of RNA silencing and an enhancer of disease symptoms. Virus Res. 163, 672–677. [DOI] [PubMed] [Google Scholar]
- Zhang, G. , Martin, T.J. , Fritz, A.K. , Miller, R. , Chen, M.‐S. , Bowden, R.L. and Johnson, J.J. (2015) Registration of ‘Oakley CL’ wheat. J. Plant Regist. 9, 190–195. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Diversity of Wheat streak mosaic virus isolates.


