Summary
In the present study, we investigated the role of Trichoderma virens (TriV_JSB100) spores or cell‐free culture filtrate in the regulation of growth and activation of the defence responses of tomato (Solanum lycopersicum) plants against Fusarium oxysporum f. sp. lycopersici by the development of a biocontrol–plant–pathogen interaction system. Two‐week‐old tomato seedlings primed with TriV_JSB100 spores cultured on barley grains (BGS) or with cell‐free culture filtrate (CF) were inoculated with Fusarium pathogen under glasshouse conditions; this resulted in significantly lower disease incidence in tomato Oogata‐Fukuju plants treated with BGS than in those treated with CF. To dissect the pathways associated with this response, jasmonic acid (JA) and salicylic acid (SA) signalling in BGS‐ and CF‐induced resistance was evaluated using JA‐ and SA‐impaired tomato lines. We observed that JA‐deficient mutant def1 plants were susceptible to Fusarium pathogen when they were treated with BGS. However, wild‐type (WT) BGS‐treated tomato plants showed a higher JA level and significantly lower disease incidence. SA‐deficient mutant NahG plants treated with CF were also found to be susceptible to Fusarium pathogen and displayed low SA levels, whereas WT CF‐treated tomato plants exhibited moderately lower disease levels and substantially higher SA levels. Expression of the JA‐responsive defensin gene PDF1 was induced in WT tomato plants treated with BGS, whereas the SA‐inducible pathogenesis‐related protein 1 acidic (PR1a) gene was up‐regulated in WT tomato plants treated with CF. These results suggest that TriV_JSB100 BGS and CF differentially induce JA and SA signalling cascades for the elicitation of Fusarium oxysporum resistance in tomato.
Keywords: defence marker genes, induced systemic resistance, jasmonic acid, salicylic acid, tomato Fusarium wilt, Trichoderma virens
Introduction
Tomato (Solanum lycopersicum Mill.) plants are continually exposed to diverse biotic stresses from a wide variety of organisms, including bacteria, viruses and fungi, as well as damage from insects and herbivores. Fusarium wilt of tomato caused by Fusarium oxysporum f. sp. lycopersici Snyder and Hans (FOL) is a detrimental soil‐borne fungal disease occurring worldwide that has the potential to reduce the productivity of tomato crops in glasshouse and field conditions (Harikrushana et al., 2014; Sartaj et al., 2011). The fungus may cause severe yield losses in tomato depending on the resistance level of the cultivar and the environmental conditions. The application of soil fumigants, such as methyl bromide, is an effective method for disease management. However, the use of such pesticides can have negative impacts on the environment and human health (Sivan and Chet, 1993; Xie et al., 2015). Although the breeding of tomato cultivars resistant to Fusarium wilt is both cost‐effective and environmentally acceptable, new strains of FOL continue to emerge that counteract the resistance of these cultivars. These limitations in the control of Fusarium wilt disease have led researchers to focus on diverse alternative systems. Amongst these alternatives, beneficial microorganisms have gained considerable attention as an eco‐friendly and cost‐effective platform for the stimulation of disease resistance through induced systemic resistance (ISR), and for the promotion of growth in plants for sustainable crop production (Abdelrahman et al., 2016; Elsharkawy et al., 2012; Harel et al., 2014; Jogaiah et al., 2013, 2016; Murali et al., 2013; Nagaraju et al., 2012; Pel and Pieterse, 2013; Pieterse et al., 2014).
Amongst the growth‐promoting fungi, Trichoderma species are potentially the most commonly used organisms for agricultural crop improvement (Abdelrahman et al., 2016; Jogaiah et al., 2013; Nagaraju et al., 2012). The application of these species in the form of seed, root or soil treatments improves the uptake of soil nutrients by crop plants, increases soil fertility and enhances the production of growth‐promoting substances and bioactive metabolites that act as biopesticides (Jogaiah et al., 2013; Shoresh et al., 2010). Furthermore, Trichoderma isolates have strong antagonistic and mycoparasitic effects against phytopathogens, and are therefore able to reduce disease severity in plants (Elsharkawy et al., 2013; Viterbo and Horwitz, 2010). Trichoderma virens, which employs these mechanisms effectively, has been recognized as an aggressive mycoparasite that is capable of competing with pathogens at the site of infection (Djonovic et al., 2007). Priming of plants with Trichoderma spores or Trichoderma secondary metabolites has also been shown to produce rapid and effective defence responses against pathogen attack by acting as immunity stimulants (Contreras‐Cornejo et al., 2011; Elsharkawy et al., 2012; Hafez et al., 2013; Tucci et al., 2011; Yoshioka et al., 2012). Taken together, these reports suggest that the use of Trichoderma species for plant protection is a promising approach to reduce the use of fungicides, growth regulators and manpower; this will eventually lower the production costs and environmental impacts associated with pathogen defence. Furthermore, a greater knowledge of the defence responses triggered in plants by pathogens opens up new biotechnological avenues for crop protection.
Previous studies have shown that the priming of various host plants with Trichoderma species results in the activation of multiple pathways regulated by several signalling molecules. Simultaneous induction of the salicylic acid (SA), jasmonic acid (JA) and ethylene (ET) pathways by Trichoderma species has been reported following pathogen attack in Arabidopsis thaliana (Mathys et al., 2012), grape (Vitis vinifera) (Perazzolli et al., 2012), tomato (Jogaiah et al., 2013; Leonetti et al., 2017; Martínez‐Medina et al., 2013, 2017) and melon (Cucumis melo) (Martínez‐Medina et al., 2014). However, extensive studies on the plant signalling mechanisms triggered by T. virens strains or its metabolites in tomato against FOL are lacking. In the present study, we tested various treatment methods using T. virens (TriV_JSB100) spores cultured on barley grains (BGS) or cell‐free culture filtrate (CF) to determine their efficacy in inducing resistance to Fusarium wilt in tomato plants. In addition, the effect of BGS and CF treatment on tomato seed germination and plant growth parameters was evaluated. To dissect the possible signalling pathways responsible for the action of the BGS and CF inoculants of TriV_JSB100 in FOL disease resistance, JA and SA signalling in Trichoderma‐induced resistance was evaluated using phytohormonal and gene expression analyses of JA‐ and SA‐impaired tomato lines.
Results
Effects of TriV_JSB100 treatments on tomato seed germination
Tomato Oogata‐Fukuju seeds treated with barley grain TriV_JSB100 spores (BGS), TriV_JSB100 cell‐free culture filtrate (CF), BGS followed by FOL inoculation (BGS + FOL), CF followed by FOL inoculation (CF + FOL), FOL or sterile distilled water (mock) were grown on damp paper towel in a growth chamber (75 mol/m2/s photosynthetically active radiation) under a 12‐h light/12‐h dark cycle at 23 ± 2 °C. The seed germination percentage was recorded after 3 and 7 days for each treatment (Fig. 1). Tomato seed treated with BGS and BGS + FOL showed significant (P < 0.05) early seed germination of 98.75% and 94.50%, respectively, at 7 days post‐treatment relative to mock seed (Fig. 1). However, seed treated with CF and CF + FOL showed no significant differences in seed germination in comparison with mock seed (Fig. 1), whereas FOL‐treated seed exhibited a significant (P < 0.001) reduction in seed germination relative to mock seed (Fig. 1).
Figure 1.
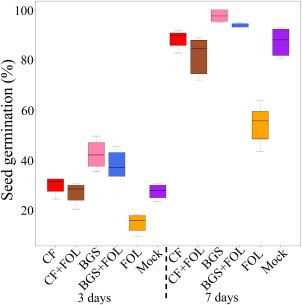
Box plot of the seed germination percentage of Oogata‐Fukuju tomato seeds at 3 and 7 days post‐treatment with different combinations of Trichoderma virens (TriV_JSB100). Tomato seeds were treated with TriV_JSB100 spores embedded on barley grains (BGS), with TriV_JSB100 cell‐free culture filtrate (CF), with BGS and challenge inoculated with Fusarium oxysporum f. sp. lycopersici (BGS + FOL), with CF and challenge inoculated with FOL (CF + FOL), with FOL (FOL) and with sterile distilled water (Mock). The box signifies the upper and lower quartiles, and the median is represented by a short black line within the box for each treatment.
Colonization and population density of TriV_JSB100 on roots of tomato
The in planta colonization of TriV_JSB100 in BGS‐treated, 1‐month‐old Oogata‐Fukuju tomato roots was examined under a light microscope and on potato dextrose agar (PDA) plates. The microscopic examination showed extensive blue‐stained mycelial growth of TriV_JSB100 in all three root segments (2, 4 and 6 cm) treated with BGS (Fig. 2a). In another set of samples, root segments were surface sterilized with 0.5% sodium hypochlorite and incubated on PDA plates for 5 days at 23 ± 2 °C. As a result of fungal colonization, significant (P = 0.000) production of TriV_JSB100 spores [9.16 × 105 colony‐forming units (CFU)/mL] was recorded in the 6‐cm root segment of BGS‐treated plants (Fig. 2b). Conversely, no Trichoderma mycelial growth was detected in the root segments treated with non‐Trichoderma (CF) inoculated plants.
Figure 2.
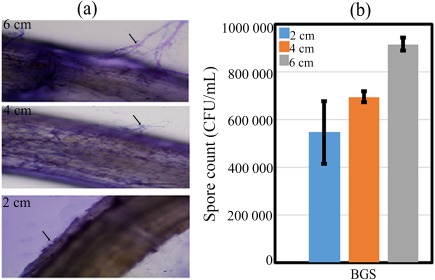
Root colonization and population density of 1‐month‐old Oogata‐Fukuju tomato plants treated with Trichoderma virens (TriV_JSB100). (a) Root segments treated with TriV_JSB100 spores cultured on barley grain (BGS) were stained with lactophenol and observed under a light microscope. (b) TriV_JSB100 spore population density of root segments treated with BGS grown on potato dextrose agar. Values are the means of four independent replications (n = 4). Bars represent standard error (SE).
Enhancement of vegetative growth and nutrient uptake in TriV_JSB100‐treated tomato plants
To understand the effects of root colonization by TriV_JSB100 BGS and TriV_JSB100 CF on plant growth and development, plant growth parameters were measured in treated 1‐month‐old tomato Oogata‐Fukuju plants under glasshouse conditions. Tomato plants treated with BGS, BGS + FOL and CF exhibited a significant (P < 0.01) increase in plant height relative to mock plants (Table 1). No significant differences in plant height were detected in tomato plants treated with CF + FOL in comparison with mock plants (Table 1). In contrast, a significant reduction (P < 0.001) in plant height was detected in tomato plants treated with FOL relative to mock plants (Table 1). The total number of leaves in BGS‐ and BGS + FOL‐treated plants was significantly higher (P < 0.01 and P < 0.05, respectively), with 21.75 and 20.75 leaves per plant, respectively, than that in mock plants (Table 1). In addition, no significant differences were detected in the total number of leaves between CF‐ and CF + FOL‐treated plants and mock plants (Table 1). With 11.25 leaves per plant, FOL‐treated plants displayed a significantly lower (P < 0.001) total number of leaves than that in mock plants (Table 1).
Table 1.
Effect of different Trichoderma virens (TriV_JSB100) treatments on 1‐month‐old Oogata‐Fukuju tomato growth parameters.
| Treatment | Plant height (cm) | No. of leaves | Shoot FW (g) | Root FW (g) | Shoot DW (g) | Root DW (g) |
|---|---|---|---|---|---|---|
| CF | 116.57 † | 17.50ns | 14.65ns | 8.30ns | 5.05ns | 1.80ns |
| CF + FOL | 109.95ns | 16.75ns | 12.57ns | 8.87ns | 4.50ns | 1.67ns |
| BGS | 138.87 ‡ | 21.75 † | 24.75 ‡ | 11.87 ‡ | 7.58 † | 2.73 ‡ |
| BGS + FOL | 120.10 † | 20.75* | 20.25 † | 10.97 † | 6.95 † | 1.97ns |
| FOL | 73.60 ‡ | 11.25 ‡ | 10.76* | 4.70 † | 1.87 † | 0.82 ‡ |
| Mock | 105.25 | 17.00 | 13.52 | 7.90 | 4.62 | 1.62 |
TriV_JSB100 spores cultured on barley grain (BGS), TriV_JSB100 cell‐free culture filtrate (CF), BGS followed by Fusarium oxysporum f. sp. lycopersici inoculation (BGS + FOL), CF followed by FOL inoculation (CF + FOL), FOL and sterile distilled water (Mock). Values are means of four independent replicates (n = 4). Significant levels are given as: *P < 0.05, † P < 0.01 and ‡ P < 0.001, according to Tukey's honestly significant difference test. ns, not significant.
DW, dry weight; FW, fresh weight.
The highest shoot and root fresh weight (FW) values were detected in BGS‐treated plants, whereas the lowest shoot and root FW values were detected in tomato plants treated with FOL (Table 1). Moreover, the greatest increase (P < 0.01) in shoot and root dry weight (DW) was observed in BGS‐treated plants (7.58 and 2.73 g DW, respectively) when compared with mock plants (Table 1). Conversely, significantly lower (P < 0.001) shoot and root DW was detected in tomato plants treated with FOL than in mock plants (Table 1). In addition, no significant differences in shoot and root FW or DW were detected between CF‐ and CF + FOL‐treated plants and mock plants (Table 1).
Enhanced nutrient uptake in plants generally results in increased plant growth and fitness. Hence, we analysed the uptake of major nutrients, such as nitrogen (N), phosphorus (P) and potassium (K). One‐month‐old tomato Oogata‐Fukuju plants treated with BGS and BGS + FOL exhibited a significant (P < 0.001 and P < 0.05, respectively) increase in N (6.95 and 6.50 mg/g FW, respectively), P (2.75 and 2.67 mg/g FW, respectively) and K (4.20 and 4.07 mg/g FW, respectively) content relative to mock plants (Fig. 3). Conversely, significantly lower (P < 0.01) N, P and K (3.40, 1.37 and 2.90 mg/g FW, respectively) content was detected in FOL‐treated plants than in mock plants (Fig. 3). In addition, no significant differences in N, P and K content were observed between CF‐ and CF + FOL‐treated plants and mock plants (Fig. 3).
Figure 3.
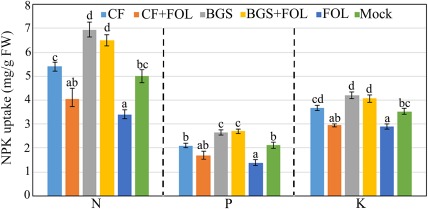
Changes in nitrogen (N), phosphorus (P) and potassium (K) levels in 1‐month‐old Oogata‐Fukuju tomato plants treated with different combinations of Trichoderma virens (TriV_JSB100). Tomato plants were treated with TriV_JSB100 spores embedded on barley grains (BGS), with BGS followed by Fusarium oxysporum f. sp. lycopersici inoculation (BGS + FOL), with TriV_JSB100 cell‐free culture filtrate (CF), with CF and challenge inoculated with FOL (CF + FOL), with FOL (FOL) and with sterile distilled water (Mock). Values are the means of four independent replicates (n = 4) ± standard error (SE). Treatment means followed by the same letter(s) within the column are not significantly different according to Tukey's honestly significant difference post hoc test. The data presented are from representative experiments that were repeated at least four times with similar results. FW, fresh weight.
Effect of TriV_JSB100 treatments on susceptibility of tomato plants to F. oxysporum
None of the TriV_JSB100 treatments were found to be phytotoxic to wild‐type (WT) and mutant tomato plants (Fig. 4a,b). Two‐week‐old tomato seedlings raised from seeds treated with BGS, CF or mock were either root dip inoculated with FOL or left untreated before being transplanted into clay pots (one plant/pot) pretreated with BGS or CF. All the TriV_JSB100 and pathogen‐inoculated seedlings (BGS, BGS + FOL, CF, CF + FOL, FOL and mock) were grown for another 15 days under glasshouse conditions before disease progression was recorded. One‐month‐old Castlemart, Moneymaker and Oogata‐Fukuju WT plants treated with BGS + FOL displayed significantly lower disease levels (33.4%, 34.07% and 35.8%, respectively) than those of FOL‐inoculated control plants (Fig. 4a,b). In addition, Castlemart, Moneymaker and Oogata‐Fukuju WT plants treated with CF + FOL displayed a moderate reduction in disease level (43.65%, 44.1% and 47.0%, respectively) relative to FOL‐inoculated control plants (Fig. 4a,b). The def1 mutant plants treated with CF + FOL showed a significantly lower disease incidence of 45.15% relative to FOL‐inoculated control plants (Fig. 4a). In contrast, def1 mutant plants treated with BGS + FOL showed no significant differences in disease level relative to FOL‐inoculated control plants (Fig. 4a). Moreover, NahG mutant plants treated with CF + FOL were highly susceptible to FOL, exhibiting 68.75% disease incidence (Fig. 4b). Conversely, NahG mutant plants treated with BGS + FOL showed a disease level that was significantly lower by 41.7% than that in FOL‐inoculated control plants (Fig. 4b). These results were highly reproducible using four independent replicates, confirming that the induction of resistance mechanisms by TriV_JSB100 BGS and CF required not only JA signalling, but also SA signalling.
Figure 4.
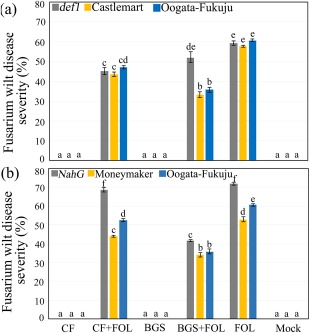
Effect of different Trichoderma virens (TriV_JSB100) treatments on Fusarium wilt disease severity in 1‐month‐old wild‐type and mutant tomato plants. (a) Fusarium wilt disease severity percentage in jasmonic acid‐deficient mutant def1, Castlemart and Oogata‐Fukuju tomato plants treated with TriV_JSB100 spores embedded on barley grain (BGS), BGS followed by F. oxysporum f. sp. lycopersici inoculation (BGS + FOL), TriV_JSB100 cell‐free culture filtrate (CF), CF and challenge inoculated with FOL (CF + FOL), FOL (FOL) or sterile distilled water (Mock). (b) Fusarium wilt disease severity percentage in salicylic acid‐deficient mutant NahG, Moneymaker and Oogata‐Fukuju tomato plants treated with BGS, BGS + FOL, CF, CF + FOL, FOL and mock. Values are the means of four independent replicates (n = 4) ± standard error (SE).Treatment means followed by the same letter(s) within the column are not significantly different according to Tukey's honestly significant difference post hoc test. The data presented are from representative experiments that were repeated at least four times with similar results.
Differential induction of JA and SA levels observed in TriV_JSB100‐primed tomato plants
To determine the phytohormonal changes in 2‐week‐old tomato seedlings treated with BGS, BGS + FOL, CF, CF + FOL, FOL and mock at 0, 4 and 24 h post‐treatment (hpt) with Fusarium pathogen, JA and SA contents were measured by liquid chromatography‐tandem mass spectrometry (LC‐MS/MS). The heatmap clustering of the JA level showed two major clades based on the plant genotype and hpt. Clade I contained the genotypes with low JA levels, whereas clade II contained the genotypes with higher JA levels (Fig. 5a). No JA was detected in the def1 mutant at 0 and 4 hpt, whereas only a slight non‐significant increase in JA was detected in BGS‐ and BGS + FOL‐treated plants at 24 hpt. Similarly, low levels of JA were detected in treated Castlemart and Oogata‐Fukuju WT plants at 0 hpt. In contrast, a substantial increase in JA level was observed in treated Castlemart and Oogata‐Fukuju WT plants at 4 and 24 hpt. Among the treatments, the greatest (P < 0.001) increases in JA level were observed in BGS‐ and BGS + FOL‐treated Castlemart (4.65 and 9.07 ng/mg FW, respectively) and Oogata‐Fukuju (3.02 and 7.05 ng/mg FW, respectively) WT plants at 24 hpt in comparison with levels in the mock plants (Fig. 5a). However, CF‐, CF + FOL‐ and FOL‐treated Castlemart and Oogata‐Fukuju WT plants showed a small increase in the JA level at 4 and 24 hpt relative to mock plants (Fig. 5a).
Figure 5.
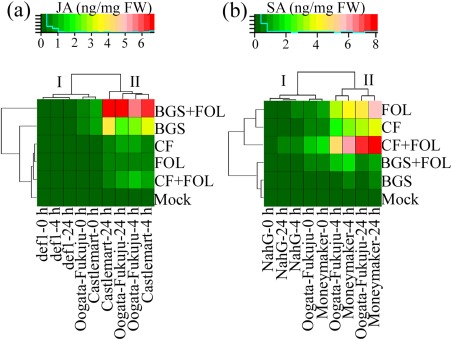
Heatmap clustering of jasmonic acid (JA) and salicylic acid (SA) levels (ng/mg FW) in 2‐week‐old wild‐type and mutant tomato seedlings treated with different combinations of Trichoderma virens (TriV_JSB100) after different time intervals (0, 4 and 24 h). (a) Changes in the level of JA in wild‐type Castlemart and Oogata‐Fukuju tomato seedlings and JA‐deficient mutant def1 seedlings treated with TriV_JSB100 spores embedded on barley grains (BGS), BGS followed by Fusarium oxysporum f. sp. lycopersici inoculation (BGS + FOL), TriV_JSB100 cell‐free culture filtrate (CF), CF and challenge inoculated with FOL (CF + FOL), FOL (FOL) or sterile distilled water (Mock). (b) Changes in the level of SA in wild‐type Moneymaker and Oogata‐Fukuju tomato seedlings and SA‐deficient mutant NahG seedlings treated with BGS, BGS + FOL, CF, CF + FOL, FOL and mock. The average mean of four independent replicates (n = 4) from each treatment was used to generate the heatmap employing the R package 3.2.2. The colour scale indicates the data matrix range. The data presented are from representative experiments that were repeated at least four times with similar results.
The heatmap clustering of the SA level also showed two major clades based on genotype and hpt (Fig. 5b). No SA was detected in the NahG mutant at 0 and 4 hpt, whereas only a slight increase was detected in CF‐, FOL‐ and CF + FOL‐treated plants at 24 hpt. Likewise, a low level of SA was detected in treated Moneymaker and Oogata‐Fukuju WT plants at 0 hpt (Fig. 5b). In contrast, a considerable increase in SA levels was observed in treated Moneymaker and Oogata‐Fukuju WT plants at 4 and 24 hpt. Among the treatments, the greatest (P < 0.001) increases in SA levels were found in FOL‐, CF‐ and CF + FOL‐treated Castlemart (3.60, 4.40 and 8.10 ng/mg FW, respectively) and Oogata‐Fukuju (2.77, 2.52 and 6.47 ng/mg FW, respectively) WT plants at 24 hpt relative to those in mock plants (Fig. 5b). However, BGS‐ and BGS + FOL‐treated Moneymaker and Oogata‐Fukuju WT plants showed only a slightly higher SA level at 4 and 24 hpt than that in mock plants (Fig. 5b).
Real‐time reverse transcriptase‐quantitative polymerase chain reaction (RT‐qPCR) of JA‐ and SA‐inducible gene expression in TriV_JSB100‐primed tomato plants
We were then interested in determining whether the differences in SA and JA levels had an impact on the responsive gene expression level in the tripartite interaction (tomato–TriV_JSB100–FOL). Thus, we used RT‐qPCR to examine the expression of the JA‐inducible defensin gene PDF1 and SA‐inducible pathogenesis‐related protein 1 acidic (PR1a) gene. Differential expression of PDF1 and PR1a was observed across the different treatments (Fig. 6a,b). Significantly higher (P < 0.001) PDF1 expression was detected in BGS‐ and BGS + FOL‐treated Castlemart (7.75‐ and 15.12‐fold, respectively) and Oogata‐Fukuju (6.05‐ and 14.10‐fold, respectively) WT plants at 24 hpt than in mock plants (Fig. 6a). In addition, FOL‐treated Castlemart and Oogata‐Fukuju WT plants displayed 2.08‐ and 2.10‐fold increases at 24 hpt relative to mock plants (Fig. 6a). However, no significant differences were detected in CF‐treated plants and mock plants (Fig. 6a). The expression level of PR1a was significantly (P < 0.001) up‐regulated in CF‐, CF + FOL‐ and FOL‐treated Moneymaker (19.55‐, 36.0‐ and 16.0‐fold, respectively) and Oogata‐Fukuju (10.10‐, 25.90‐ and 11.10‐fold, respectively) WT plants at 24 hpt in comparison with expression levels in mock plants (Fig. 6b). However, no significant differences were detected between BGS and mock plants (Fig. 6b). These results were fully reproducible across four biological replicate experiments and demonstrated that the elicitation of ISR induced by BGS is centred on the JA‐regulated priming mechanism, whereas treatment with CF triggers an activation of systemic acquired resistance (SAR) through the SA‐dependent signalling pathway.
Figure 6.
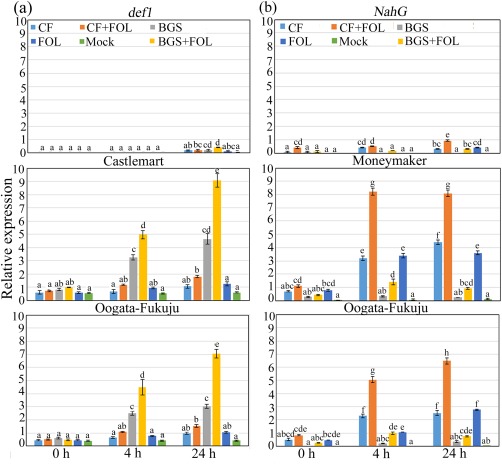
Bar plot of quantitative real‐time reverse transcriptase‐polymerase chain reaction (RT‐qPCR) of the jasmonic acid (JA)‐responsive defensin gene PDF1 and pathogenesis‐related protein 1 acidic (PR1a) gene in 2‐week‐old wild‐type and mutant tomato seedlings treated with different combinations of Trichoderma virens (TriV_JSB100) at different time intervals (0, 4 and 24 h). (a) Relative expression of PDF1 in wild‐type Castlemart and Oogata‐Fukuju seedlings and in JA‐deficient mutant def1 seedlings treated with TriV_JSB100 spores embedded on barley grains (BGS), BGS followed by Fusarium oxysporum f. sp. lycopersici inoculation (BGS + FOL), TriV_JSB100 cell‐free culture filtrate (CF), CF and challenge inoculated with FOL (CF + FOL), FOL (FOL) or sterile distilled water (Mock). (b) Relative expression of PR1a in wild‐type Moneymaker and Oogata‐Fukuju seedlings and in salicylic acid (SA)‐deficient mutant NahG seedlings treated with BGS, BGS + FOL, CF, CF + FOL, FOL and mock. Values are means of four independent replicates (n = 4) ± standard error (SE).Treatment means followed by the same letter(s) within the column are not significantly different according to Tukey's honestly significant difference post hoc test. The β‐actin gene was used as reference. The data presented are from representative experiments that were repeated at least four times with similar results.
Principal component analysis
Principal component analysis (PCA) was performed in order to gain further insight into the contribution of various TriV_JSB100 treatments to FOL disease reduction and growth promotion in tomato plants (Fig. 7). TriV_JSB100 treatments were significantly loaded into two major principal components (PC1 and PC2) explaining 83.24% of the variance. Early seed germination, NPK, FW, DW, disease level, SA and PR1a variables were loaded into PC1, explaining 64.75% of the variance, whereas disease level, JA and PDF1 variables were loaded into PC2, explaining 18.49% of the variance. The PCA plot reveals a clear segregation of the six different treatments (BGS, BGS + FOL, CF, CF + FOL, FOL and mock) with four biological replicates for each treatment; this indicates the high reproducibility and differential effect of the different treatments on the disease level and growth promotion characteristics in tomato plants (Fig. 7).
Figure 7.
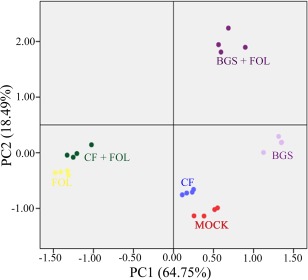
Principal component analysis (PCA) of seed germination, growth parameters, nutrient uptake, disease severity, phytohormone level and gene expression variables in tomato plants treated with different combinations of Trichoderma virens (TriV_JSB100). Data variables of tomato plants treated with TriV_JSB100 spores embedded on barley grains (BGS), BGS followed by Fusarium oxysporum f. sp. lycopersici inoculation (BGS + FOL), TriV_JSB100 cell‐free culture filtrate (CF), CF and challenge inoculated with FOL (CF + FOL), FOL (FOL) or sterile distilled water (MOCK) were clustered and loaded into two principal components PC1 and PC2.
Discussion
We have observed that tomato seeds treated with BGS and BGS + FOL germinate significantly (P < 0.05) earlier, at 7 days post‐treatment, relative to mock seeds (Fig. 1). However, no significant differences were observed between cell‐free culture filtrate (CF)‐, CF + FOL‐ and mock‐treated seeds (Fig. 1). A similar correlation was observed in our previous research (Abdelrahman et al., 2016), where the treatment of onion (Allium cepa) seed with a conidial suspension of T. longibrachiatum isolates produced remarkable changes in terms of early seed germination and improved seedling vigour when compared with the untreated control. Velazques‐Robledo et al. (2011) reported that tomato seeds susceptible to Rhizoctonia solani treated with two strains of T. virens showed improved seed germination relative to the untreated control. These results collectively indicate that Trichoderma spp. have a positive effect on seed germination in several cropping systems. In the present study, we have demonstrated that the TriV_JSB100 treatments result in improved growth parameters in 1‐month‐old tomato plants (Table 1). The current findings are consistent with a recent study by Colla et al. (2015), which showed that coating maize (Zea mays) seeds with T. atroviride resulted in a higher seed germination rate, and coating wheat (Triticum aestivum) seeds resulted in improved seedling growth parameters through higher shoot and root DW and grain yield than that of uncoated controls. Likewise, T. longibrachiatum treatment of bulb onions improved growth parameters such as seedling height and FW (Abdelrahman et al., 2016). The enhanced seed germination rates and plant growth parameters observed in this study might be caused by successful colonization by TriV_JSB100, leading to an improvement in nutrient uptake and induction of indole acetic acid (Abdelrahman et al., 2016; Jogaiah et al., 2013; Shoresh et al., 2010; Yadav et al., 2011).
To confirm the positive effect of TriV_JSB100 treatments on nutrient uptake, the levels of N, P and K were measured in 1‐month‐old tomato Oogata‐Fukuju WT plants treated with BGS, BGS + FOL, CF, CF + FOL, FOL and mock treatments. We observed a significant increase in the levels of N, P and K in the BGS‐ and BGS + FOL‐treated plants relative to the mock plants (Fig. 3); no significant differences were observed between CF‐, CF + FOL‐ and mock‐treated plants (Fig. 3). Nutrients are important factors in disease control and can affect plant fitness, pathogens and plant–pathogen interactions (Dordas, 2008). For example, the severity of root rot and crown disease in tomato plants was reduced by increases in the uptake of nitrate‐nitrogen and copper (Duffy and Défag, 1999). Similarly, increased P uptake improved the growth and yield of infected citrus (Citrus limon) trees (Zhao et al., 2013). Our results indicate that T. virens is required to mobilize and increase nutrient uptake by the plant; T. virens is more efficient and competitive than many other soil microbes in promoting growth and improving plant fitness (Hermosa et al., 2012; Singh et al., 2008). Therefore, we can hypothesize that the energy required to induce TriV_JSB100‐mediated FOL resistance does not affect tomato plant development. Conversely, the significant reduction in nutrient levels on FOL treatment might be caused by the fact that N assimilation requires the reduction of nitrate to ammonium; this is affected by environmental factors, such as pathogen attack or heavy metals (Cao et al., 2015; Masclaux‐Daubresse et al., 2010).
Many studies on growth‐promoting fungi have shown that Trichoderma spp. induce direct and indirect resistance in plants against pathogen attacks; these species are therefore recognized as key regulators of the hormone signalling pathways involved in pathogen‐associated disease resistance (Ferrigo et al., 2014; Leonetti et al., 2017; Martínez‐Medina et al., 2017; Shoresh et al., 2010; Tucci et al., 2011). In the present study, TriV_JSB100 BGS treatments significantly reduced the disease level, significantly increased the JA level and up‐regulated the expression of PDF1 in WT tomato plants relative to FOL‐treated control plants (Figs 4a, 5a and 6a). Conversely, JA‐deficient def1 mutants treated with BGS + FOL showed a high disease level, lower JA levels and no PDF1 expression (Figs 4a, 5a and 6a). This result suggests that treatments with BGS are largely dependent on JA biosynthesis to trigger the plant defence responses that are essential to decrease disease incidence. Our results are consistent with a previous study by Contreras‐Cornejo et al. (2011), who suggested that the defence responses elicited by Trichoderma in Arabidopsis against Botrytis cinerea involved the defence hormone JA as an important factor in boosting plant immunity. Similar correlations have been established by Hafez et al. (2013), wherein the soil application of T. viride to tomato plants at 24 hpt with F. oxysporum or Rhizoctonia solani resulted in an increase in the expression of JA‐related PDF1 and PDF2 genes. Similarly, Mathys et al. (2012) have shown that induced resistance in Arabidopsis roots treated with T. hamatum is regulated by JA‐ and ET‐related genes. Moreover, the JA‐inducible genes lipoxygenase (Lox1) and phenylalanine ammonia‐lyase (Pal1) and the ET‐inducible genes ethylene receptor (ETR1) and constitutive triple response 1 (CTR1) were found to be induced both locally and systemically on treatment with T. asperellum T‐203 spores alone. However, induction with T. asperellum T‐203 and subsequent challenge with the bacterial pathogen Pseudomonas syringae pv. lachrymans increased the expression of peroxidase (POX), glucanase (GLU) and other pathogenesis‐related proteins in cucumber (Cucumis sativus), with these observations providing evidence of cross‐talk between tripartite interactions (Shoresh et al., 2005).
In CF + FOL‐treated WT plants, a moderate reduction in the disease level, a significant increase in SA content and the up‐regulation of the PR1a SA marker gene were observed relative to FOL‐treated control plants (Figs 4b, 5b and 6b). SA‐deficient NahG plants treated with CF + FOL, however, showed an increase in disease incidence to a level similar to that of FOL‐treated control plants and exhibited lower SA levels and no expression of PR1a (Figs 4b, 5b and 6b). These results indicate that the CF‐induced defence mechanism is heavily dependent on SA signalling. Likewise, Arabidopsis treated with T. asperellum CF showed up‐regulation of SA‐related genes associated with resistance to Cucumber mosaic virus; this suggests that the SA pathway plays an important role in Trichoderma CF systemic resistance (Elsharkawy et al., 2013). A similar observation was reported in Arabidopsis plants treated with secondary metabolites of T. virens; this resulted in the enhanced accumulation of components of the SA‐dependent plant defence response (Velazques‐Robledo et al., 2011). In addition, Tucci et al. (2011) observed that Trichoderma CF treatment triggered ISR through SA‐dependent gene expression. The expression of defence genes induced by JA transduction increased the already activated ISR response over time, indicating that this phenomenon of overlapping expression occurs against biotrophic and necrotrophic pathogens both locally and systemically. There have also been several reports of the involvement of JA and SA in defence responses in several crops treated with Trichoderma and challenged with pathogens (Contreras‐Cornejo et al., 2011; Hermosa et al., 2012; Korolev et al., 2008; Martínez‐Medina et al., 2013, 2017). The balance between JA and SA homeostasis is known to be important for the interaction between beneficial microbe, pathogen and host plant that results in the activation of defence signalling pathways (Contreras‐Cornejo et al., 2011; Leonetti et al., 2017; Martínez‐Medina et al., 2017; Pieterse et al., 2009). The present study revealed that BGS treatment boosted plant defences more strongly than CF treatment. Such differentiation between BGS and CF treatments has been reported previously for T. asperellum in association with Arabidopsis (Elsharkawy et al., 2013). We hypothesize that the differential induction of plant defences with BGS and CF treatments could be related to variation in the abundance and type of elicitor secreted by T. virens. For example, the expression and secretion of the eliciting protein small protein 1 (SM1) by T. virens was much higher in the presence of the host plant than in T. virens CF alone (Djonovic et al., 2006, 2007). Moreover, the application of T. virens Gv29–8 to maize roots triggered ISR against Colletotrichum graminicola, and the Sm1 elicitor protein secreted by T. virens was shown to be necessarily required for the induction of ISR in maize, rice and cotton (Djonovic et al., 2006, 2007). A recent study by Salas‐Marina et al. (2015) reported that Sm1 and eliciting plant response‐like protein 1 (Epl1), secreted by T. virens and T. atroviride, induced resistance against Alternaria solani, B. cinerea and P. syringae pv. tomato in tomato. They also showed that T. virens was more effective than T. atroviride in promoting tomato plant growth. Similar induction of plant ISR by different species of Trichoderma has also been reported (Elsharkawy et al., 2012, 2013; Hafez et al., 2013; Harman et al., 2004; Hermosa et al., 2012).
In our study, we found that priming tomato plants with TriV_JSB100 both reduces the incidence of Fusarium wilt disease and produces several added benefits to the host plants. The associated beneficial responses include a significant increase in plant growth and JA and SA phytohormone levels, and the up‐regulation of defence marker genes. Notably, these beneficial responses occurred both with and without pathogen infection. Therefore, it can be inferred that the priming of host plants by TriV‐JSB100 conveys the ability to respond strongly when exposed to pathogens. This suggests that plants are equipped to respond to pathogen infection by priming with TriV_JSB100 or such other beneficial organisms (Conrath et al., 2006; Nagaraju et al., 2012; Shoresh et al., 2010; Van Wees et al., 2008; Walters and Heil, 2007). Furthermore, even when the host plant was infected and the corresponding defence responses were triggered, the growth of the plant was not affected in TriV_JSB100‐primed plants. Hence, it is clear that TriV_JSB100 priming of tomato has an overwhelmingly positive influence in terms of reducing disease incidence, and that it does not adversely affect the host plant in terms of energy costs.
Conclusions
This study examined the effectiveness of two different T. virens inoculants, TriV_JSB100 BGS and TriV_JSB100 CF, in tomato plants infected with the FOL pathogen. Our results indicated significantly lower Fusarium wilt disease incidence in tomato plants treated with TriV_JSB100 BGS than TriV_JSB100 CF. Furthermore, examination of disease incidence, phytohormone levels and defence marker gene expression following TriV_JSB100 BGS and TriV_JSB100 CF treatments in JA‐ and SA‐impaired mutants (def1 and NahG, respectively) demonstrated that TriV_JSB100 BGS‐mediated ISR is centred on the JA‐regulated priming mechanism, whereas the CF‐induced mechanism is centred on the SA priming mechanism. We have also reported that root colonization by TriV_JSB100 resulted in significantly higher nutrient uptake, which may have increased crop productivity. Thus, our results indicate that TriV_JSB100 can be exploited for both commercial disease protection and increased crop productivity, and that different TriV_JSB100 treatments can be used to obtain durable resistance.
Experimental Procedures
Plant materials
Seeds of the Japanese tomato cultivar Oogata‐Fukuju, which is highly susceptible to Fusarium wilt, were purchased from Takii Seed Co. Ltd., Kyoto, Japan, and were used throughout the study. Seeds of the SA‐deficient mutant NahG and its parental WT Moneymaker were kindly provided by the Tomato Genetic Resource Center at the University of California at Davis, CA, USA. Seeds of the JA‐deficient mutant def1 and its parental WT Castlemart were obtained from the Leibniz‐Institute of Vegetable and Ornamental Crops, Großbeeren, Germany. Seeds were sterilized with 0.5% sodium hypochlorite for 5 min and washed thoroughly in sterile distilled water for 10 min before use.
Pathogen inoculum
FOL CK3–1 (Namiki et al., 1994), the pathogen responsible for Fusarium wilt in tomato, was obtained from Dr Tsuge at Nagoya University, Nagoya, Japan. A pure culture of FOL was maintained on PDA plates and slants at 22 ± 2 °C for 7 days. After incubation, fungal hyphal tips were taken from the growing edge with a sterile sharp needle and transferred to 250 mL of potato dextrose broth. The broth was incubated at 22 ± 2 °C under 12 h/12 h alternate cycles of near‐ultraviolet light and darkness for 7 days. Samples were then centrifuged at 11,952 g for 10 min and the pellets were washed three times in 25 mL of sterile distilled water. Subsequently, the washed fungal pellets were suspended in 25 mL of sterile distilled water. The conidial spore load was calculated using a haemocytometer and adjusted to 1 × 107 conidial spores/mL for inoculation.
Plant growth‐promoting fungus T. virens isolate
The bio‐agent T. virens (TriV_JSB100; NCBI accession number KC569356) was isolated from tomato rhizosphere soil from a farmer's field, Jodhpur (26°18′N; 73°04′E), Rajasthan State, India.
Barley grain spore (BGS) production
Hayadori 2 barley grains procured from Kaneko Seeds, Tokyo, Japan and autoclaved (250 g in 150 mL distilled water) on two consecutive days were used as a substrate. The substrate was later inoculated with TriV_JSB100 (106 spores/mL) under sterile conditions and incubated for 15 days. Completely colonized and air‐dried (in ambient conditions) fungal substrate samples were ground to a particle size of 1 mm using a hand coffee grinder. The fungal‐colonized barley particles were then tested for TriV_JSB100 spore production and checked for contamination by incubation on PDA plates at 23 ± 2 °C for 5 days. After incubation, the spores were scraped from the PDA plates and suspended in sterile distilled water before the resultant spore suspensions were diluted with sterile distilled water to final concentrations of 2%, 5% and 10%. Finally, the conidial spore loads from all the dilutions were calculated using a haemocytometer and adjusted to 1 × 106 conidial spores/mL for further analyses.
Cell‐free culture filtrate (CF) production
Ten mycelial discs of 6‐day‐old TriV_JSB100 culture were grown in 500‐mL Erlenmeyer conical flasks containing 150 mL of potato dextrose broth. The fungus was cultured without shaking at room temperature (23 ± 2 °C) for 5 days. The crude culture filtrate was separated from mycelia and filtered through two layers of a Whatman (Sigma‐Aldrich, St. Louis, MO, USA) No. 2 filter; 100 mL of filtrate were centrifuged at 1,328 g and the resultant culture filtrate was diluted to final concentrations of 5%, 10% and 15% in sterile distilled water for further analyses.
Screening for beneficial traits
Isolates derived from different concentrations of TriV_JSB100 BGS (2%, 5% and 10%) and CF (5%, 10% and 15%) were tested individually for certain characteristics associated with beneficial traits, such as root colonization, phosphate solubilization and indole‐3‐acetic acid content, according to the procedure of Jogaiah et al. (2013). The best results were obtained with isolates of 10% BGS, carrying 106 spores/mL, and 15% CF; these were used as the optimized doses for seed treatments in this study (Table S1, see Supporting Information).
Assay of TriV_JSB100 effects on Oogata‐Fukuju tomato seed germination
Two hundred grams of Oogata‐Fukuju tomato seeds were surface sterilized as described previously; 100 g of sterilized seeds were then treated with 100 mL of BGS suspension in which 0.5 g of BGS powder was suspended in sterile distilled water carrying conidial spores (106 spores/mL) at room temperature. A further 50 g of sterilized seeds were soaked in 50 mL of 15% CF suspension, and the remaining 50 g of sterilized seeds were soaked in 50 mL of sterilized distilled water. The following experiments were conducted with four biological replicates (20 seeds/replicate):
Sterilized tomato seeds were treated with BGS powder at room temperature for 6 h (BGS).
Sterilized tomato seeds were treated with BGS as above and then soaked in 50 mL of FOL spore suspension (1 × 107) for 30 min (BGS + FOL).
Sterilized tomato seeds were soaked in 50 mL of CF at room temperature for 6 h (CF).
Sterilized tomato seeds were soaked in 50 mL of CF as above and then soaked in 50 mL of FOL spore suspension (1 × 107) for 30 min (CF + FOL).
Sterilized tomato seeds were soaked in 50 mL of FOL spore suspension (1 × 107) for 30 min (FOL); or sterilized tomato seeds were soaked in 50 mL of sterile distilled water for 30 min (mock).
The treated and pathogen‐inoculated seeds (BGS, BGS + FOL, CF, CF + FOL, FOL and mock) were then placed equidistant on moistened blotter paper towel according to the method of Sudisha et al. (2005), and incubated in a growth chamber at 23 ± 2 °C with a 12‐h light/12‐h dark photoperiod. Seedling emergence was observed daily and the germination percentage was calculated after 2 weeks of incubation using the following formula:
Colonization and population density of TriV_JSB100 in Oogata‐Fukuju tomato roots
To examine the colonization and population density of TriV_JSB100 in tomato, 2‐week‐old Oogata‐Fukuju tomato seedlings, raised in growth chambers from BGS‐treated seeds with and without FOL treatment as described above, were transplanted into clay pots [17.5 cm in diameter × 11.2 cm in depth; filled with peat moss and vermiculite (1 : 1, v/v) containing 50 mg N/kg, 500 mg P/kg and 100 mg K/kg, pH 6–6.5, water‐holding capacity ∼70% (Napura Yodo, Yanmar Agricultural Equipment, Tokyo, Japan)], pretreated with BGS as a root‐supporting medium and maintained under glasshouse conditions (23 ± 2 °C, 90% relative humidity) for another 15 days. One‐month‐old BGS‐treated plants were harvested separately. Only the root portion was retained after soil remnants had been completely removed by washing in running tap water for 30 min. Root portions were then surface sterilized (NaOCl, 1.5% for 3 min), washed three times in sterile distilled water, air dried and then cut into three parts: root base (portion closest to the stem), middle section and root tip. Each of the sterilized root parts was further reduced into 2‐cm segments and placed on PDA plates supplemented with 200 mg/L of chloramphenicol. The morphology of the fungal mycelia grown from the root segments was compared with the original isolate on the same medium. To study the colonization of T. virens, the root segments were heated with 10% potassium hydroxide for 15 min at 60 °C, treated with 2.5% hydrochloric acid for 10 min, stained with 0.2% trypan blue and observed under a light microscope. The spore load was counted using a haemocytometer following the procedure of Murali et al. (2013).
Assay of TriV_JSB100 treatments on tomato Oogata‐Fukuju vegetative growth and nutrient uptake
In a separate experiment, 2‐week‐old tomato Oogata‐Fukuju seedlings, raised from CF‐, BGS‐ or sterile distilled water‐treated seeds grown in a growth chamber as described above, were subjected to root dip inoculation; treated 2‐week‐old tomato plants were uprooted completely and the root portion only of the seedlings was soaked with 50 mL of FOL spore suspension (1 × 107) for 30 min. Roots of the mock plants were immersed in 50 mL of sterile distilled water for 30 min. Treated and pathogen‐inoculated plant seedlings (BGS, BGS + FOL, CF, CF + FOL, FOL and mock) in four biological replicates (one plant/replica/pot) were transplanted into clay pots containing the supporting mixture as mentioned above, pretreated with BGS or CF for BGS‐ and CF‐treated samples, respectively, and maintained under glasshouse conditions as mentioned above for another 15 days. One‐month‐old plant samples from each respective treatment were selected and the plant height, number of leaves, and shoot and root FW were measured. Both shoot and root were labelled and oven dried at 80 °C for 24 h, and the shoot and root DW were recorded. In a further experiment, plant materials treated as described above were harvested, digested and the total N and P contents of each were determined using the vanadomolybdate phosphoric acid yellow colour method (Kitson and Mellon, 1944) and Kjeldahl method (Jackson, 1962), respectively. Total K content was determined by flame photometry (Jackson, 1973).
Analysis of disease incidence
To determine the effectiveness of TriV_JSB100 treatment in protecting tomato plants from Fusarium wilt disease, 2‐week‐old tomato seedlings (def1 mutant, Castlemart, NahG mutant, Moneymaker and Oogata‐Fukuju), raised from seeds treated with BGS, CF or sterile distilled water as described above, were root dip inoculated with 50 mL FOL spore suspension (1 × 107) for 30 min at room temperature. Two‐week‐old treated and pathogen‐inoculated seedlings (BGS, BGS + FOL, CF, CF + FOL, FOL and mock) were transplanted into clay pots (as described above) pretreated with BGS or CF for each respective condition. The plants were watered at regular intervals, and 0.1% NPK (Hyponex 6 : 10 : 5, Osaka, Japan) dissolved in water was provided twice. Each tomato treatment consisted of 16 plants (one plant/pot with four plants per replicate and four replicates); the experiment was repeated twice. Plants were observed to determine disease progression based on observed symptoms, such as dark‐brown vascular discoloration, slight vein clearing on outer leaflets, stunting, wilting associated with the presence of foliar chlorosis and seedling death. The disease incidence was recorded 30 days after pathogen inoculation and calculated using the formula:
Extraction and quantification of SA and JA contents in treated tomato and challenge inoculated plants
For JA and SA quantification, 2‐week‐old seedlings (def1 mutant, Castlemart, NahG mutant, Moneymaker and Oogata‐Fukuju), raised from seed treated with BGS, CF or sterile distilled water in a growth chamber as described above, were root dip inoculated with 50 mL of FOL. Treated‐ and pathogen‐inoculated 2‐week‐old whole seedlings (BGS, BGS + FOL, CF, CF + FOL, FOL and mock) were harvested separately in four replicates at 0, 4 and 24 hpt, frozen in liquid nitrogen and vacuum dried. For each sample, 200 mg of frozen sample were then placed in a 2‐mL microfuge tube and ground in a bead beater (Qiagen or equivalent, Qiagen, Tokyo, Japan) with 3‐mm tungsten beads at 25 Hz/s for 2 min. A 1‐mL aliquot of ethyl acetate spiked with internal standards prepared in methanol (10 ng of d2JA/µL and 20 ng of d4SA/µL) was then added to each sample. Each treatment also included an extraction control containing no plant material. A 3‐mm tungsten bead was placed in each microfuge tube and samples were extracted in the bead beater for 2 min at 25 Hz/s, placed on ice for 10 min and then centrifuged at 13 000 g for 20 min at 4 °C. The supernatant was carefully removed to a new 2‐mL microfuge tube and the pellets were re‐extracted with 1 mL of ethyl acetate without standards; both supernatants were then pooled. Following a further 10‐min incubation on ice, the extract was dried in a vacuum concentrator to near dryness (∼90 min), mixed with 500 µL of 70% methanol (v/v) and centrifuged at 1000 rpm for 10 min at 4 °C. A 4‐µL aliquot of the resultant extract was transferred to LC‐MS vials and analysed by LC/MS‐MS.
Samples were analysed using an AB SCIEX 3200 QTRAP LC‐MS/MS system (AB Sciex, Framingham, MA, USA), coupled with a hybrid triple quadrupole/linear ion trap mass spectrometer with a turbo V™ ion source, and analysed using software version 1.5.1 (Shimadzu Corporation, Kyoto, Japan). Chromatographic separation was carried out by injecting 4 µL of each sample onto a Mightysil RP‐18 GP column (5 mm, Aqua, Kanto Chemical, Co., Inc., Tokyo, Japan) at 40 °C. A mobile phase composed of 100% solvent A (90% ultrapure water : 10% acetonitrile : 0.1% formic acid) to 100% solvent B (5% ultrapure water : 95% acetonitrile : 0.1% formic acid) was obtained over 20 min. Solvent B was held at 100% for 5 min and then returned to 100% solvent A for 15 min of equilibration prior to the next injection with a flow rate of 200 µL/min.
A multiple reaction monitoring ion with a specific mass‐to‐charge ratio generated from each endogenous JA, SA or internal standard (the parent ion) was selected and fragmented to obtain its daughter ions; a specific daughter ion was used for the generation of the corresponding compound's chromatogram. The parent ions, daughter ions and collision energies used in these analyses are summarized in Table S2 (see Supporting Information). SA and JA levels of treated‐ and non‐treated tomato plants with or without pathogen challenge were quantified by comparing their sample peak area with the peak area of their respective internal standards.
Expression analysis of JA and SA marker gene expression in TriV_JSB100‐primed tomato seedlings
The SA‐inducible gene PR1a (Accession No: M69247) (F, 5′‐GAGGGCAGCCGTGCAA‐3′; R, 5′‐CACATTTTTCCACCAACACATTG‐3′), JA‐inducible plant defensin gene PDF1 (Hafez et al., 2013) (F, 5′‐ CAATGTAACTTAAAGTGCCTAATTATG‐3′; R, 5′‐CTTATCAGATCTCAATGGAGAAATC‐3′) and reference gene β‐actin (F, 5′‐TTGCCGCATGCCATTCT‐3′; R, 5′‐TCGGTGAGGATATTCATCAGGTT‐3′) (Jogaiah et al., 2013) primer sequences were used for gene expression analysis. For RNA extraction, 2‐week‐old whole seedlings (def1 mutant, Castlemart, NahG mutant, Moneymaker and Oogata‐Fukuju), raised from seeds treated with BGS, CF or sterile distilled water as described above, were challenge inoculated with 50 mL of FOL. Treated‐ and pathogen‐inoculated samples (BGS, BGS + FOL, CF, CF + FOL, FOL and mock) were harvested at set time points (0, 4 and 24 hpt). The experiment was conducted in quadruplicate and repeated twice. Total RNA extraction, reverse transcription and RT‐qPCR were performed as described previously (Jogaiah et al., 2013).
Data analysis
All experiments were conducted with four biological replicates. The data obtained were subjected to one‐way analysis of variance. Significant (P < 0.05) differences among treatments were obtained by Tukey's honestly significant difference post hoc test using SPSS software (SPSS 20.0, SPSS Inc., Chicago, IL, USA). PCA was generated by SPSS software. Box plot analysis of seed germination percentage and heatmap clustering analysis of JA and SA content with the different treatments were generated using R package version 3.2.2 (www.r-project.org).
Disclosure
The authors have no conflicts of interest related to the work described in this article.
Author Contributions
SJ and S‐iI conceived and designed the experiments. SJ and MA performed the experiments. SJ, MA and S‐iI analysed the data. SJ and S‐iI contributed reagents/materials/analysis tools. SJ, MA, L‐SPT and S‐iI wrote the manuscript. SJ and MA prepared the figures/graphs. SJ, MA and S‐iI revised the manuscript. All the authors read and approved the final manuscript.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Biochemical characterizations of Trichoderma virens TriV_JSB100.
Table S2 Compound mass optimizing parameters using triple quadrupole (Q) mass spectrometry. De‐clustering potential (DP), entrance potential (EP), collision cell entrance potential (CEP), collision cell exit potential (CXP) and collision energy (CE).
Acknowledgements
All the authors thank Yamaguchi University, Japan and Karnatak University, India for providing the laboratory facilities to perform and execute the experiments. This work was supported by the Japan Society for the Promotion of Science [Grant in Aid for Postdoctoral Research (No. P 11086)] and the Department of Science and Technology, New Delhi, India [Grant in Aid for Scientific Research SB/YS/LS‐92/2014].
Contributor Information
Sudisha Jogaiah, Email: jsudish@kud.ac.in.
Shin‐Ichi Ito, Email: shinsan@yamaguchi-u.ac.jp.
References
- Abdelrahman, M. , Abdel‐Motaal, F. , El‐Sayed, M. , Jogaiah, S. , Shigyo, M. , Ito, S‐I. and Tran, L.S. (2016) Dissection of Trichoderma longibrachiatum‐induced defense in onion (Allium cepa L.) against Fusarium oxysporum f. sp. cepa by target metabolite profiling. Plant Sci. 246, 128–138. [DOI] [PubMed] [Google Scholar]
- Cao, J. , Cheng, C. , Yang, J. and Wang, Q. (2015) Pathogen infection drives patterns of nutrient resorption in citrus plants. Sci. Rep. 5, 14 675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colla, G. , Rouphael, Y. , Bonini, P. and Cardarelli, M. (2015) Coating seeds with endophytic fungi enhances growth, nutrient uptake, yield and grain quality of winter wheat. Int. J. Plant Prod. 9, 171–190. [Google Scholar]
- Conrath, U. , Beckers, G.J.M. , Flors, V. , Garcia‐Agustin, P. , Jakab, G. , Mauch, F. , Newman, M.A. , Pieterse, C.M.J. , Poinssot, B. , Pozo, M.J. , Pugin, A. , Schaffrath, U. , Ton, J. , Wendehenne, D. , Zimmerli, L. and Mauch‐Mani, B. (2006) Priming: getting ready for battle. Mol. Plant–Microbe Interact. 19, 1062–1071. [DOI] [PubMed] [Google Scholar]
- Contreras‐Cornejo, H.A. , Macias‐Rodriguez, L. , Beltran‐Pena, E. , Herrera‐Estrella, E. and Lopez‐Bucio, J. (2011) Trichoderma‐induced plant immunity likely involves both hormonal‐ and camalex independent mechanisms in Arabidopsis thaliana and confers resistance against necrotrophic fungus Botrytis cinerea . Plant Signal. Behav. 6, 1554–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djonovic, S. , Pozo, M.J. , Dangott, L.J. , Howell, C.R. and Kenerley, C.M. (2006) Sm1, a proteinaceous elicitor secreted by the biocontrol fungus Trichoderma virens induces plant defense responses and systemic resistance. Mol. Plant–Microbe Interact. 19, 838–853. [DOI] [PubMed] [Google Scholar]
- Djonovic, S. , Vargas, W.A. , Kolomiets, M.V. , Horndeski, M. , Wiest, A. and Kenerley, C.M. (2007) A proteinaceous elicitor Sm1 from the beneficial fungus Trichoderma virens is required for induced systemic resistance in maize. Plant Physiol. 145, 875–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dordas, C. (2008) Role of nutrients in controlling plant diseases in sustainable agriculture. A review. Agron. Sustain. Dev. 28, 33–46. [Google Scholar]
- Duffy, B.K. and Défag, G. (1999) Macro‐ and microelement fertilizers influence the severity of Fusarium crown and root rot of tomato in a soilless production system. Hortscience, 34, 287–291. [Google Scholar]
- Elsharkawy, M.M. , Shimizu, M. , Takahashi, H. and Hyakumachi, M. (2012) The plant growth‐promoting fungus Fusarium equiseti and the arbuscular mycorrhizal fungus Glomus mosseae induce systemic resistance against Cucumber mosaic virus in cucumber plants. Plant Soil, 361, 397–409. [Google Scholar]
- Elsharkawy, M.M. , Shimizu, M. , Takahashi, H. , Ozaki, K. and Hyakumachi, M. (2013) Induction of systemic resistance against cucumber mosaic virus in Arabidopsis thaliana by Trichoderma asperellum SKT‐1. Plant Pathol. J. 29, 193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrigo, D. , Raiola, A. , Piccolo, E. , Scopel, C. and Causin, R. (2014) Trichoderma harzianum T22 induces in maize systemic resistance against Fusarium verticillioides . J. Plant Pathol. 96, 133–142. [Google Scholar]
- Hafez, E.E. , Hashem, M. , Mahmoud, M.B. , El‐Saadani, M.A. and Seham, A.A. (2013) Induction of new defensin genes in tomato plants via pathogens—biocontrol agent interaction. J. Plant Pathol. Microbiol. 4, 167. [Google Scholar]
- Harel, Y.M. , Mehari, Z.H. , Rav‐David, D. and Elad, Y. (2014) Systemic resistance to gray mold induced in tomato by benzothiadiazole and Trichoderma harzianum T39. Phytopathology, 104, 150–157. [DOI] [PubMed] [Google Scholar]
- Harikrushana, P. , Ramchandra, S. and Shah, K.R. (2014) Study of wilt producing Fusarium sp. from tomato (Lycopersicon esculentum Mill). Int. J. Curr. Microbiol. Appl. Sci. 3, 854–858. [Google Scholar]
- Harman, G.E. , Howell, C.R. , Viterbo, A. , Chet, I. and Lorito, M. (2004) Trichoderma species – opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2, 43–56. [DOI] [PubMed] [Google Scholar]
- Hermosa, R. , Viterbo, A. , Chet, I. and Monte, E. (2012) Plant‐beneficial effects of Trichoderma and of its genes. Microbiology, 158, 17–25. [DOI] [PubMed] [Google Scholar]
- Jackson, M.L. (1962) Soil Chemical Analysis. Eaglewood Cliffs, NY: Prentice Hall, Inc. [Google Scholar]
- Jackson, M.L. (1973) Soil Chemical Analysis. New Delhi: Prentice Hall of India Pvt. Ltd. [Google Scholar]
- Jogaiah, S. , Abdelrahman, M. , Tran, L.S. and Ito, S‐I. (2013) Characterization of rhizosphere fungi that mediate resistance in tomato against bacterial wilt disease. J. Exp. Bot. 64, 3829–3842. [DOI] [PubMed] [Google Scholar]
- Jogaiah, S. , Mahantesh, K. , Sharathchnadra, R.G. , Shetty, H.S. , Vedamurthy, A.B. and Tran, L.S. (2016) Isolation and evaluation of proteolytic actinomycete isolates as novel inducers of pearl millet downy mildew disease protection. Sci. Rep. 6, 30 789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitson, R.E. and Mellon, M.G. (1944) Colorimetric determination of phosphorus as molybdivanadophosphoric acid. Ind. Eng. Chem. 16, 379–382. [Google Scholar]
- Korolev, N. , Rav‐David, D. and Elad, Y. (2008) The role of phytohormones in basal resistance and Trichoderma‐induced systemic resistance to Botrytis cinerea in Arabidopsis thaliana . BioControl, 53, 667–683. [Google Scholar]
- Leonetti, P. , Zonno, M.C. , Molinari, S. and Altomare, C. (2017) Induction of SA‐signaling pathway and ethylene biosynthesis in Trichoderma harzianum treated tomato plants after infection of the root‐knot nematode Meloidogyne incognita . Plant Cell Rep. 36, 621–631. [DOI] [PubMed] [Google Scholar]
- Martínez‐Medina, A. , Fernandez, I. , Sanchez‐Guzman, M. , Jung, S.C. , Pascual, J.A. and Poz, M.J. (2013) Deciphering the hormonal signalling network behind the systemic resistance induced by Trichoderma harzianum in tomato. Front. Plant Sci. 4, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez‐Medina, A. , Mar Alguacil, M.D. , Pascual, J.A. and Van Wees, S.C.M. (2014) Phytohormone profiles induced by Trichoderma isolates correspond with their biocontrol and plant growth‐promoting activity on melon plants. J. Chem. Ecol. 40, 804–815. [DOI] [PubMed] [Google Scholar]
- Martínez‐Medina, A. , Fernandez, I. , Lok, G.B. , Pozo, M.J. , Pieterse, C.M. and Van Wees, S.C. (2017) Shifting from priming of salicylic acid‐ to jasmonic acid‐regulated defences by Trichoderma protects tomato against the root knot nematode Meloidogyne incognita . New Phytol. 213, 1363–1377. [DOI] [PubMed] [Google Scholar]
- Masclaux‐Daubresse, C. , Daneial‐Vedele, F. , Dechorgnat, J. , Chardon, F. , Gaufichon, L. and Suzuki, A. (2010) Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann. Bot. 105, 1141–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathys, J. , De Cremer, K. , Timmermans, P. , Van Kerckhove, S. , Lievens, B. , Vanhaecke, M. , Cammue, B.P. and De Coninck, B. (2012) Genome‐wide characterization of ISR induced in Arabidopsis thaliana by Trichoderma hamatum T382 against Botrytis cinerea infection. Front. Plant Sci. 3, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murali, M. , Sudisha, J. , Amruthesh, K.N. , Ito, S‐I. and Shekar Shetty, H. (2013) Rhizosphere fungus Penicillium chrysogenum promotes growth and induces defence‐related genes and downy mildew disease resistance in pearl millet. Plant Biol. 15, 111–118. [DOI] [PubMed] [Google Scholar]
- Nagaraju, A. , Sudisha, J. , Mahadevamurthy, S. and Ito, S‐I. (2012) Seed priming with Trichoderma harzianum isolates enhances plant growth and induces resistance against Plasmopara halstedii, an incitant of sunflower downy mildew disease. Aust. J. Plant Pathol. 41, 609–620. [Google Scholar]
- Namiki, F. , Shiomi, T. , Kayamura, T. and Tsuge, T. (1994) Characterization of the formae speciales of Fusarium oxysporum causing wilt of cucurbits by DNA fingerprinting with nuclear repetitive DNA sequences. Appl. Environ. Microbiol. 60, 2684–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pel, M.J.C. and Pieterse, C.M.J. (2013) Microbial recognition and evasion of host immunity. J . Exp. Bot. 64, 1237–1248. [DOI] [PubMed] [Google Scholar]
- Perazzolli, M. , Moretto, M. , Fontana, P. , Ferrarini, A. , Velasco, R. , Moser, C. , Delledonne, M. and Pertot, I. (2012) Downy mildew resistance induced by Trichoderma harzianum T39 in susceptible grapevines partially mimics transcriptional changes of resistant genotypes. BMC Genomics, 13, 660–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M.J. , Leon‐Reyes, A. , Van der Ent, S. and Van Wees, S.C.M. (2009) Networking by small‐molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M.J. , Zamioudis, C. , Berendsen, R.L. , Weller, D.M. , Van Wees, S.C.M. and Bakker, P.A.H.M. (2014) Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375. [DOI] [PubMed] [Google Scholar]
- Salas‐Marina, M.A. , Isordia‐Jasso, M.I. , Islas‐Osuna, M.A. , Delgado‐Sánchez, P. , Jimenez‐Bremont, J.F. , Rodríguez‐Kessler, M. , Rosales‐Saavedra, M.T. , Herrera‐Estrella, A. and Casas‐Flores, S. (2015) The Epl1 and Sm1 proteins from Trichoderma atroviride and Trichoderma virens differentially modulate systemic disease resistance against different life style pathogens in Solanum lycopersicum . Front. Plant Sci. 6, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartaj, A.S. , Sakamoto, K. and Inubushi, K. (2011) Effect of Penicillium sp. EU0013 inoculation on tomato growth and Fusarium wilt. Hortic. Res. 65, 69–73. [Google Scholar]
- Shoresh, M. , Yedidia, I. and Chet, I. (2005) Involvement of jasmonic acid/ethylene signaling pathway in the systemic resistance induced in cucumber by Trichoderma asperellum T203. Phytopathology, 95, 76–84. [DOI] [PubMed] [Google Scholar]
- Shoresh, M. , Harman, G.E. and Mastouri, F. (2010) Induced systemic resistance and plant responses to fungal biocontrol agents. Annu. Rev. Phytopathol. 48, 21–43. [DOI] [PubMed] [Google Scholar]
- Singh, V. , Joshi, B.B. , Awasthi, S.K. and Srivastava, S.N. (2008) Ecofriendly management of red rot disease of sugarcane with Trichoderma strains. Sugar Tech., 10, 158–161. [Google Scholar]
- Sivan, A. and Chet, I. (1993) Integrated control of Fusarium crown and root rot of tomato with Trichoderma harzianum in combination with methyl bromide or soil solarization. Crop Prot. 12, 380–386. [Google Scholar]
- Sudisha, J. , Amruthesh, K.N. , Deepak, S.A. , Shetty, N.P. , Sarosh, B.R. and Shekar Shetty, H. (2005) Comparative efficacy of strobilurin fungicides against downy mildew disease of pearl millet. Pest Biochem. Physiol. 81, 188–197. [Google Scholar]
- Tucci, M. , Ruocco, M. , Masi, L.D. , Palma, M.D. and Lorito, M. (2011) The beneficial effect of Trichoderma spp. on tomato is modulated by the plant genotype. Mol. Plant Pathol. 12, 341–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wees, S.C.M. , Van der Ent, S. and Pieterse, C.M.J. (2008) Plant immune responses triggered by beneficial microbes. Curr. Opin. Plant Biol. 11, 443–448. [DOI] [PubMed] [Google Scholar]
- Velazques‐Robledo, R. , Contreras‐Cornejo, H.A. , Macias‐Rodriguez, L. , Hernandez‐Morales, A. , Aguirre, J. , Casas‐Flores, S. , Lopez‐Bucio, J. and Herrera‐Estrella, A. (2011) Role of the 4‐phosphopantetheinyl transferase of Trichoderma virens in secondary metabolism and induction of plant defense responses. Mol. Plant–Microbe Interact. 24, 1459–1471. [DOI] [PubMed] [Google Scholar]
- Viterbo, A. and Horwitz, B.A. (2010) Mycoparasitism In: Cellular and Molecular Biology of Filamentous Fungi (Borkovich K.A. and Ebbole D.J., eds), pp. 676–693. Washington: American Society of Microbiology. [Google Scholar]
- Walters, D.R. and Heil, M. (2007) Costs and trade‐offs associated with induced resistance. Physiol. Mol. Plant Pathol. 71, 3–17. [Google Scholar]
- Xie, H. , Yan, D. , Mao, L. , Wang, Q. , Li, Y. , Ouyang, C. , Guo, M. and Cao, A. (2015) Evaluation of methyl bromide alternatives efficacy against soil‐borne pathogens, nematodes and soil microbial community. PLoS One, 10, e0117980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav, J. , Verma, J.P. and Tiwari, K.N. (2011) Plant growth promoting activities of fungi and their effect on chickpea plant growth. Asian J. Biol. Sci. 4, 291–299. [Google Scholar]
- Yoshioka, Y. , Ichikawa, H. , Naznin, H.A. , Kogure, A. and Hyakumachi, M. (2012) Systemic resistance induced in Arabidopsis thaliana by Trichoderma asperellum SKT‐1, a microbial pesticide of seed borne diseases of rice. Pest Manag. Sci. 68, 60–66. [DOI] [PubMed] [Google Scholar]
- Zhao, H. , Sun, R. , Albrecht, U. , Padmanabhan, C. , Wang, A. , Coffey, M.D. , Girke, T. , Wang, Z. , Close, T.J. , Roose, M. , Yokomi, R.K. , Folimonova, S. , Vidalakis, G. , Rouse, R. , Bowman, K.D. and Jin, H. (2013) Small RNA profiling reveals phosphorus deficiency as a contributing factor in symptom expression for citrus huanglongbing disease. Mol. Plant. 6, 301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Biochemical characterizations of Trichoderma virens TriV_JSB100.
Table S2 Compound mass optimizing parameters using triple quadrupole (Q) mass spectrometry. De‐clustering potential (DP), entrance potential (EP), collision cell entrance potential (CEP), collision cell exit potential (CXP) and collision energy (CE).


