Summary
Bacterial canker disease is considered to be one of the most destructive diseases of tomato (Solanum lycopersicum), and is caused by the seed‐borne Gram‐positive bacterium Clavibacter michiganensis ssp. michiganensis (Cmm). This vascular pathogen generally invades and proliferates in the xylem through natural openings or wounds, causing wilt and canker symptoms. The incidence of symptomless latent infections and the invasion of tomato seeds by Cmm are widespread. Pathogenicity is mediated by virulence factors and transcriptional regulators encoded by the chromosome and two natural plasmids. The virulence factors include serine proteases, cell wall‐degrading enzymes (cellulases, xylanases, pectinases) and others. Mutational analyses of these genes and gene expression profiling (via quantitative reverse transcription‐polymerase chain reaction, transcriptomics and proteomics) have begun to shed light on their roles in colonization and virulence, whereas the expression of tomato genes in response to Cmm infection suggests plant factors involved in the defence response. These findings may aid in the generation of target‐specific bactericides or new resistant varieties of tomato. Meanwhile, various chemical and biological controls have been researched to control Cmm. This review presents a detailed investigation regarding the pathogen Cmm, bacterial canker infection, molecular interactions between Cmm and tomato, and current perspectives on improved disease management.
Keywords: crop disease, foliar infection, genome, Microbacteriaceae, molecular biology, plant–microbe interactions, systemic infection
Introduction
Tomato is the world's largest vegetable crop with an annual production at 169 million tons, representing 16% of total vegetable production and 58 billion US dollars [Food and Agricultural Organization (FAO), 2016]. Bacterial canker of tomato, caused by Clavibacter michiganensis ssp. michiganensis (Cmm), is considered to be a serious threat to the processing and fresh market tomato industries, having caused catastrophic epidemics in most tomato‐growing areas of the world (Blank et al., 2016; Kleitman et al., 2008; Smith, 1910; Volcani, 1985).
Cmm is a non‐motile, Gram‐positive actinomycete, and the only recognized species of the genus Clavibacter. It has been grouped into several subspecies which cause devastating diseases in agricultural crops (Table 1; Davis et al., 1984), although digital DNA–DNA hybridization (dDDH) and average nucleotide identity (ANI) suggest that the subspecies fit the criteria for separate species (Tambong, 2017).
Table 1.
Clavibacter michiganensis subspecies and hosts
| Subspecies | Common name or symptoms | Major hosts | Reference |
|---|---|---|---|
| californiensis | Asymptomatic | Tomato | Yasuhara‐Bell and Alvarez ( 2015) |
| capsici | Bacterial canker disease | Bell pepper; sweet pepper | Oh et al. ( 2016) |
| chilensis | Asymptomatic | Tomato | Yasuhara‐Bell and Alvarez ( 2015) |
| insidiosus | Wilting and stunting | Alfalfa | McCulloch ( 1925) |
| michiganensis | Bacterial wilt and canker | Tomato | Davis et al. ( 1984); Strider ( 1969) |
| nebraskensis | Wilt and blight disease | Maize | Vidaver and Mandel ( 1974) |
| phaseoli | Leaf yellowing | Common bean | Gonzalez and Trapiello ( 2014) |
| sepedonicus | Ring rot disease | Potato | Manzer and Genereux ( 1981) |
| tessellarius | Leaf freckles and leaf spot | Wheat | Carlson and Vidaver ( 1982) |
Plants infected with Cmm show various symptoms depending on the age of the host plant, cultivar susceptibility and virulence of Cmm (Gleason et al., 1993), together with certain environmental conditions, including temperature and humidity (de León et al., 2011). When plants are infected at early stages of their life (as seeds or young seedlings), they develop systemic infections (also called primary infections) that affect fruit quality and yield, and typically lead to plant death. In contrast, older plants usually develop foliar infections (also called secondary infections), which cause chlorosis of leaves, but may or may not affect the quality and yield of the current crop.
During systemic infection, plants can at first appear asymptomatic, with symptoms showing after 6–8 weeks as the bacterium multiplies to a high titre (Gitaitis et al., 1991), reaching 109–1010 per gram of plant tissue homogenate (Meletzus et al., 1993). Cmm invades and proliferates in xylem vessels, causing browning of the internal vasculature, with the gradual degradation of vascular tissues (Fig. 1). This interferes with water transport and leads to wilting during early stages of infection (Eichenlaub and Gartemann, 2011). Tomato plants that are infected during late developmental stages may be asymptomatic or have a slow wilting process (Gitaitis et al., 1991; Sharabani et al., 2013). They can survive infection and may produce marketable fruit, but can become an efficient source of infection for the successive growing season (Sharabani et al., 2013).
Figure 1.
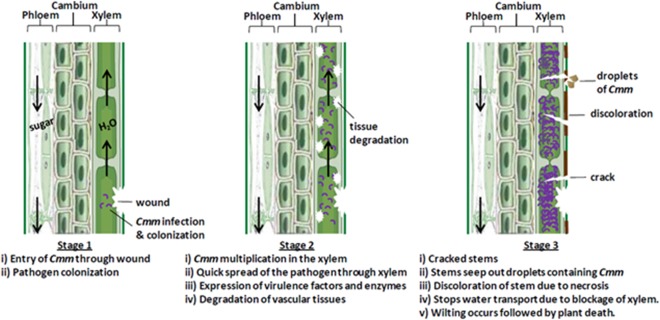
Schematic diagram showing the stages of bacterial canker disease progression in tomato stem following Clavibacter michiganensis ssp. michiganensis (Cmm) infection.
The effects of Cmm, including failure of tomato production and premature death of the entire plant, can result in substantial economic losses (Chang et al., 1992). Field experiments have recorded yield losses of up to 84% in commercial fields of Ontario, Canada (Poysa, 1993), 20%–30% in France (Rat et al., 1991) and 46% in Illinois, USA (Chang et al., 1992), although the occurrence of this disease is sporadic and losses caused by outbreaks can also be minor.
Few Gram‐positive phytopathogens have been studied in detail to explore the mechanisms utilized by bacteria to sense the host plant, colonize and counteract plant defence responses. However, the publication of Cmm genome sequences has provided an excellent platform for molecular investigations into disease induction and host–pathogen interactions (Gartemann et al., 2008), providing insights into the mechanisms of Cmm virulence and the defence responses of tomato.
This review illustrates the current knowledge of bacterial canker disease, the global analysis approaches used to dissect Cmm–tomato molecular interactions, together with disease management.
Cmm Transmission
Infected seeds are responsible for the long‐distance transmission and dissemination of Cmm, enabling its introduction to previously disease‐free regions (Fatmi et al., 1991). Genetic diversity amongst Cmm isolates from narrow geographical ranges (Kleitman et al., 2008; Milijašević‐Marčić et al., 2012; Tancos et al., 2015; Thapa et al., 2017; Wassermann et al., 2017) suggests that the introduction of foreign strains is common, as is their ability to adapt to the new habitat (Jacques et al., 2012).
Seed infection occurs when Cmm enters the seed coat and endosperm through the vascular system of an infected female parent (Fatmi and Schaad, 1988). Seeds that are contaminated externally with Cmm, through contact with other sources of the bacterium, can also serve as the initial source of inoculum for systemic infections (Quesada‐Ocampo et al., 2012). Contaminated soil and plant debris are also important sources of systemic infections (Hadas et al., 2005). Cmm can survive in plant debris under natural field conditions in the USA for more than 10 months (Fatmi and Schaad, 2002), and may contribute to soil‐borne infections for up to 4 years (Ciccarone and Carilli, 1948, as cited in Fatmi and Schaad, 2002).
Cmm enters leaves, stems and roots through wounds or natural openings, including stomata and hydathodes (Carlton et al., 1998; Tancos et al., 2013), allowing the disease to spread laterally via splashing or dripping water or from contaminated tools (Carlton et al., 1998; Chang et al., 1991; Gitaitis et al., 1991; Ricker and Riedel, 1993; Xu et al., 2010).
As C. michiganensis subspecies that are pathogenic in a particular plant host can endophytically colonize non‐host plants, the disease may also be disseminated as endophytes associated with non‐host plants (Thapa et al., 2017). Previous reports of bacterial and fungal endophytes have demonstrated similar patterns (Darrasse et al., 2010; Ploch and Thines, 2011; Sowley et al., 2010).
Molecular Biology of Cmm Pathogenesis
Genomic features of Cmm
Sequenced genomes of Cmm tomato pathogen strains include a chromosome of 3.3–3.6 Mbp and a plasmid, pCM1, of 31–59 kb. Most of these strains have a second plasmid, pCM2, of 64–109 kb, which, compared with pCM1, is less conserved amongst tomato pathogenic strains in both size and gene content. By contrast, both plasmids are often absent in tomato endophytic strains of C. michiganensis, some of which cause disease in other plant species (Thapa et al., 2017). Although much genetic research has been conducted with the wild‐type Cmm strain NCPPB382 (Cmm382), other strains have been sequenced more recently (Gartemann et al., 2008; Thapa et al., 2017).
Cmm belongs to the phylum Actinobacteria which is characterized as having genomes with high GC content. However, there are about 20 regions identified in the Cmm382 genome with low GC content (Gartemann et al., 2008), many of which are involved in pathogenesis. In particular, a 129‐kb low GC region near the origin of replication is recognized as a part of the chromosomal chp/tomA pathogenicity island (PAI) (Gartemann et al., 2008). The chp/tomA PAI is the only genomic island present in all 12 sequenced Cmm strains that are pathogenic in tomato, whereas it is absent in the five sequenced tomato endophyte strains of C. michiganensis (Thapa et al., 2017). Within this PAI, the chp subregion contains putative protease‐encoding genes, and other genes known to be involved in virulence, whereas the tomA subregion contains tomA, encoding tomatinase, which is involved in the degradation of tomatin and provides basal defence to tomato (Kaup et al., 2005; Stork et al., 2008).
Virulence genes
Although the precise functions and interactions of Cmm virulence genes remain unknown, expression studies and the effects of mutations on disease progression have begun to reveal the general roles of some genes. As a number of putative virulence factors are present on the two plasmids, pCM1 and pCM2, and on the chp/tomA PAI, several studies have utilized natural variants or derivative strains that lack these specific loci. Strain Cmm27, a deletion mutant of Cmm382 that lacks the chp/tomA PAI, exhibits reduced colonization of tomato compared with Cmm382, and does not produce disease symptoms (Chalupowicz et al., 2010). Other derivatives of Cmm382 include Cmm101, Cmm102 and Cmm100, which lack the plasmids pCM2, pCM1 or both, respectively. Cmm100 will colonize plants without producing a wilting phenotype (although it does induce localized leaf blisters after foliar infection), and is therefore considered to be non‐virulent. Cmm101 and Cmm102 both exhibit reduced virulence compared with Cmm382, but remain pathogenic (Chalupowicz et al., 2017).
Similar to Cmm100 (which lacks pCM1 and pCM2), a plasmid‐free derivative of strain CmmCASJ002 colonizes plants with titres similar to its parent strain, but does not elicit tomato canker symptoms. Similar to Cmm101 (which lacks pCM2), a derivative of CmmCASJ002 that lacks pCM2, but retains pCM1, exhibits reduced wilting and weak pathogenicity, but colonizes plants with titres similar to the parent strain. However, CmmCASJ001 and CmmCASJ007 are both wild‐type isolates that naturally lack pCM2 and yet retain pathogenicity, with virulence no lower than that of Cmm382. Removing pCM1 from CmmCASJ001 does, however, eliminate tomato canker symptoms and produce five‐fold lower bacterial titres compared with the parent strain (Thapa et al., 2017).
It seems therefore that pCM1 is a major contributing factor to virulence, whereas the role of pCM2 is strain dependent (Thapa et al., 2017). Nevertheless, most tomato‐pathogenic Cmm strains harbour both plasmids (e.g. nine of 11 Californian isolates; Thapa et al., 2017) or genes normally associated with both plasmids, as determined by polymerase chain reaction (PCR) (e.g. 46 of 51 New York isolates, and 12 of 12 Argentinian isolates; Tancos et al., 2015; Wassermann et al., 2017). It is therefore possible that the requirement for pCM2 in the pathogenicity of some Cmm strains, but not others, stems from the integration of pCM2‐associated genes into the chromosome. In support of this concept, C. michiganensis ssp. nebraskensis, which does not harbour plasmids, encodes on its chromosome homologues of 28 genes which are present on plasmids in other C. michiganensis subspecies (Tambong, 2017). Although PCR‐based detection failed to find plasmid‐associated genes in some isolates of Cmm, this method may be limited by overly specific primers that are unable to hybridize with more divergent sequences.
Of the specific genes associated with virulence that are found on each plasmid, on the chp/tomA PAI and at other chromosomal locations (Table 2), the majority encode proteases and other plant cell wall‐degrading enzymes (Burger et al., 2005; Savidor et al., 2012). Tomatinase, encoded on the chp/tomA PAI, is also an important contributor to virulence.
Table 2.
Locations and summary of mutant and expression studies for putative virulence genes of Clavibacter michiganensis ssp. michiganensis.
| Time of expression (hpi) | ||||||
|---|---|---|---|---|---|---|
| Gene | Location | Expression in minimal medium† | Systemic infection | Foliar infection | Results from genetic studies | Suspected role |
| Chp family proteases | ||||||
| chpA* | PAI | Yes | ||||
| chpB* | PAI | |||||
| chpC | PAI | Yes | 72‡ | 8‡ | Mutant has reduced incidence of plants with foliar blisters, reduced incidence of wilting (Chalupowicz et al., 2017), reduction of in planta titre and weak disease symptoms (Stork et al., 2008) | Colonization, virulence |
| chpD* | PAI | |||||
| chpE | PAI | 192§ | ||||
| chpF | PAI | Yes | 192§ | |||
| chpG | PAI | Yes | 192§ | No effect on systemic colonization or virulence, abolished HR in non‐host plants (Lu et al., 2015; Stork et al., 2008) | ||
| pat‐1 | pCM2 | 24, 48‡, 12, 24, 72, 96, 168¶, 192§ | Induces virulence when cloned into Cmm100 (Meletzus et al., 1993). | Virulence | ||
| phpA | pCM2 | Yes | Does not induce virulence when cloned into Cmm100 (Burger et al., 2005) | |||
| phpB | pCM2 | No | Does not induce virulence when cloned into Cmm100 (Burger et al., 2005) | |||
| Chymotrypsin‐related serine proteases | ||||||
| ppaA | PAI | Yes | ||||
| ppaB1 | PAI | Yes | 192[Link], [Link] | |||
| ppaB2 | PAI | Yes | 192[Link], [Link] | |||
| ppaC | PAI | Yes | 192§ | |||
| ppaD | PAI | Yes | 192§ | |||
| ppaE | PAI | No | ||||
| ppaF | Other | No | ||||
| ppaG | Other | No | 192§ | |||
| ppaH | Other | Yes | 192§ | |||
| ppaI | Other | No | 192§ | |||
| ppaJ | pCM1 | Yes | ||||
| Subtilase proteases | ||||||
| sbtA | PAI | No | 16‡ | Mutant has reduced incidence of foliar blisters, less wilting (Chalupowicz et al., 2017) | Virulence | |
| sbtB | Other | No | Early†† | |||
| sbtC | Other | No | 192§ | |||
| Cellulases | ||||||
| celA | pCM1 | Yes | 8, 16, 24, 28[Link], [Link], 12, 24, 72, 96, 168¶, 192§ | Not detected at ≥ four‐fold | Mutant does not induce wilting in Cmm101; gene induces wilting when cloned into Cmm100 (Jahr et al., 2000) | Virulence |
| celB* | Other | No | ||||
| Xylanases | ||||||
| xysA | Other | Yes | ||||
| xysB | Other | No | Early†† | |||
| Pectinases | ||||||
| pgaA | Other | Yes | 192§ | 8, 16‡ | Mutant has reduced incidence of foliar blisters (Chalupowicz et al., 2017) | Virulence (foliar) |
| pelA1 | PAI | 192§ | ||||
| pelA2 | PAI | |||||
| Endoglucanases | ||||||
| endX/Y | Other | 8, 16‡ | Mutant has reduced incidence of foliar blisters (Chalupowicz et al., 2017) | Virulence (foliar) | ||
| Transcriptional regulators | ||||||
| vatr1 | Other | Reduced wilting symptoms during systemic infection (Chalupowicz et al., 2017; Savidor et al., 2014) | Virulence | |||
| vatr2 | Other | 24, 48, 72‡ | 8, 16‡ | Reduced incidence of foliar blisters, less wilting (Chalupowicz et al., 2017) | Virulence | |
| wcoP | 240‡‡ | |||||
| Other | ||||||
| gmdA | Other | No | ||||
| manB | No | |||||
| perF | 8, 16, 24, 48‡ | Reduced incidence of foliar blisters (Chalupowicz et al., 2017) | Virulence (foliar) | |||
| srtA | Reduced incidence of foliar blisters (Chalupowicz et al., 2017) | Virulence (foliar) | ||||
| tomA | PAI | |||||
*Presumed pseudogene.
†Transcript levels significantly greater than in medium supplemented with plant tissue homogenate for at least one time point (Flügel et al., 2012).
‡Transcript levels ≥ four‐fold relative to that at time zero (Chalupowicz et al., 2017).
§Protein detected in planta at 8 days post‐infection, and significantly up‐regulated in medium supplemented with xylem sap vs. rich defined medium (Savidor et al., 2012).
¶Transcript levels significantly greater than in minimal medium (Chalupowicz et al., 2010).
**Proteomic analysis could not differentiate between ppaB1 and ppaB2, which only differ by one amino acid (Savidor et al., 2012).
††Transcript levels significantly greater in xylem mimicking medium vs. minimal medium (Hiery et al., 2013).
‡‡Transcript levels significantly greater at 10 days post‐infection vs. in minimal medium (Flügel et al., 2012).
Tomatinase
Encoded on the chp/tomA PAI, tomatinase (Tom) is amongst 13 predicted secreted proteins that are common to all Cmm tomato pathogens (Tancos et al., 2015; Thapa et al., 2017), with the possible exception of CmmNCPPB3264 (by Southern blot; Kaup et al., 2005). TomA degrades the tomato alkaloid α‐tomatine, which is implicated in plant host defence against microbial pathogens. However, a tomA insertional mutant of Cmm101 retained similar virulence and bacterial titres to Cmm101, suggesting that tomA is not a virulence factor (Kaup et al., 2005). Nevertheless, as the comparison was made using Cmm101, a pCM2‐free derivative which already shows reduced virulence compared with the parent strain Cmm382, it is possible that tomA may contribute to the virulence of Cmm. A study of Fusarium oxysporum f. sp. lycopersici, a fungal pathogen of tomato, found that tomatinase is required for full virulence, although it is not essential for pathogenicity (Pareja‐Jaime et al., 2008).
Serine proteases
Putative virulence‐associated Cmm proteases are encoded by the plasmids, the chp/tomA PAI and other chromosomal loci (Table 2; Fig. 2), and are members of three distinct families. They are secreted proteins whose substrates are not yet known. Nonetheless, these degradative enzymes might facilitate bacterial colonization via the degradation and alteration of host cell physiology to ultimately acquire nutrients and manipulate the host defence responses.
Figure 2.
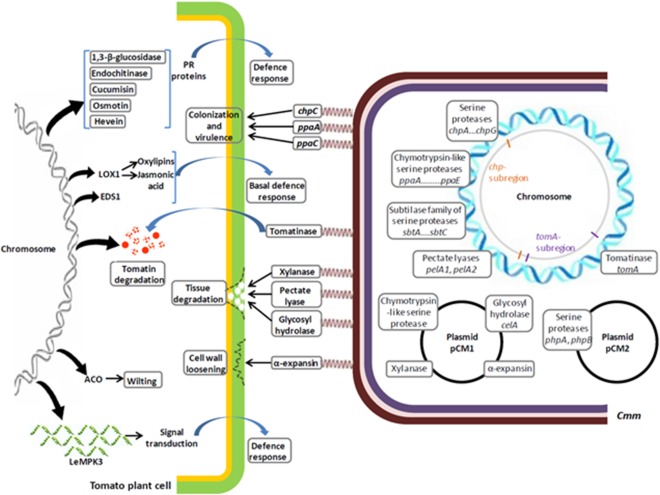
Schematic diagram showing putative roles of select pathogen and host genes during Clavibacter michiganensis ssp. michiganensis (Cmm) infection.
Proteases are implicated in various plant–microbe interactions. An extracellular protease (ecpAXoc) from Xanthomonas oryzae pv. oryzicola, a bacterial pathogen of rice, induces chlorosis‐ and necrosis‐like symptoms when injected into rice leaves, indicating that it is a virulence factor (Zou et al., 2012). In Fusarium oxysporum f. sp. lycopersici, a causative agent of Fusarium wilt in tomato, the secreted serine protease FoSep1 cleaves extracellular tomato chitinases, which are host defensive enzymes that target fungal cell walls (Jashni et al., 2015). In contrast, the secreted serine protease PrtA of the Gram‐negative bacterial pathogen of grapevines, Xylella fastidiosa strain Temecula1, reduces biofilm formation and is classified as an antivirulence factor, which, when disrupted, results in a hypervirulent phenotype (Gouran et al., 2016). Antivirulent factors may include proteins whose expression is advantageous outside of the host, but detrimental within the host, and therefore its expression is eventually lost or differentially regulated via evolution (Bliven and Maurelli, 2012). Thus, although not all proteases involved in plant–microbe interactions are necessarily virulence factors, those which are antivirulence factors are expected to be less common or expressed at lower levels in pathogenic Cmm.
Pat‐1 family
The Chp or Pat‐1 family of serine proteases is encoded by three genes on plasmid pCM2 (pat‐1, phpA and phpB) and seven genes on the chp/tomA PAI (chpA–chpG; Burger et al., 2005). Most of the tested Chp family genes (chpA, chpC, chpF, chpG and phpA, but not phpB) produce up‐regulated transcripts in minimal medium compared with medium supplemented with plant tissue homogenate (Flügel et al., 2012). Expression of pat‐1, chpC, chpE, chpF and chpG is detected at the transcript and/or protein level in systemically infected plants, and the expression of chpC is detected during foliar infection (Chalupowicz et al., 2010, 2017; Savidor et al., 2012).
Introduction of plasmid‐encoded pat‐1 into Cmm100, a derivative of strain Cmm382 that lacks both plasmids, restores some level of virulence and results in the wilting of infected plants (Meletzus et al., 1993), whereas simultaneous introduction of both phpA and phpB into Cmm100 does not produce wilting symptoms in tomato (Burger et al., 2005). Therefore, pat‐1 appears to be the most and possibly only impactful virulence gene on the pCM2 plasmid. Although some pathogenic Cmm naturally lack pCM2 (Thapa et al., 2017), it would be interesting to determine whether such strains possess a pat‐1 orthologue elsewhere in the genome.
The remaining Chp family proteases are clustered in a region of about 50 kb on the PAI of Cmm382 (Stork et al., 2008). All of these tested chromosomal genes (chpC, chpE, chpF and chpG), when mutated, show a lower severity of leaf blisters during foliar infection (Chalupowicz et al., 2017). Mutation of chpC also leads to a dramatic reduction of in planta titre, followed by weak disease symptoms (Stork et al., 2008), including a significantly reduced incidence of leaf blisters during foliar infection and significantly reduced incidence of wilting during systemic infection (Chalupowicz et al., 2017). It is therefore implicated in both colonization and virulence.
Plant gene expression is also affected by chpC. Compared with plants infected with the Cmm382 wild‐type strain, tomato plants infected with a ΔchpC mutant strain show increased levels of transcripts encoding 1‐aminocyclopropane‐1‐carboxylic acid oxidase 1 (ACO1) and pathogenesis‐related protein 4 (PR4) during blister formation (Chalupowicz et al., 2017). ACO1 is an enzyme involved in the biosynthesis of ethylene, a plant hormone with roles in development and defence, which is normally up‐regulated in Cmm‐infected plants (Savidor et al., 2012). Tomato plants that are insensitive to ethylene or deficient in its production experience increased wilting symptoms when infected with Cmm, compared with wild‐type plants (Balaji et al., 2008). Together, these results suggest that ethylene plays a role in defence against Cmm pathogenesis, and that ChpC is involved in suppression of this host defence. Reduced disease symptoms during infection with the ΔchpC mutant strain may be caused in part by impaired suppression of ACO1 expression by the plant host, and therefore impaired suppression of the host's defence response (Chalupowicz et al., 2017).
In contrast with ΔchpC, mutation of chpG in the Cmm101 strain has no effect on either virulence or colonization in tomato (Stork et al., 2008), but abolishes an apparent hypersensitive response (HR) in non‐host plants (Lu et al., 2015; Stork et al., 2008). HR is a plant‐induced death of cells surrounding an infection, which can prevent the spread of microbial pathogens. Cmm causes localized necrosis consistent with HR when infiltrated into the non‐host plants tobacco and Mirabilis jalapa. Evidence suggesting that the observed necrosis is caused by HR rather than toxicity include a distribution of Cmm that is limited to the infiltrated area in the non‐host plants, and the absence of necrosis when these plants are simultaneously infiltrated with inhibitors of plant metabolism (Alarcón et al., 1998).
In the ΔchpG mutant of Cmm101, the abolished HR could be rescued by complementation with intact chpG, but not with pat‐1, phpA or phpB, indicating that the pCM2‐encoded proteases do not contribute to HR. Introduction of chpG (but not chpC) into tobacco (Nicotiana tabacum) leaves via Agrobacterium tumefaciens infiltration leads to HR, which demonstrates that chpG is sufficient to induce this response. The study authors hypothesized that ChpG can cleave a plant‐derived target, leading to conformational changes in the target, which then activate an extracellular leucine‐rich repeat (eLRR) domain‐containing R protein (Lu et al., 2015). Ultimately, this approach might be helpful to identify the substrate(s) of ChpG through testing of the cognate receptor protein in N. tabacum.
The genes chpA, chpB and chpD are believed to be pseudogenes as they have frame shifts and/or in‐frame stop codons (Stork et al., 2008), although chpA is known to produce transcripts (Flügel et al., 2012).
Ppa family
Chymotrypsin‐like serine proteases belonging to the Ppa family are encoded by 11 genes: six on the PAI (ppaA–ppaE, including ppaB1 and ppaB2), four at a pair of other chromosomal locations (ppaF–ppaI) and one on the pCM1 plasmid (ppaJ; Gartemann et al., 2008). Like the pat‐1 family genes, many of these (ppaA, ppaB1, ppaB2, ppaC, ppaD, ppaH and ppaJ, but not ppaE, ppaF, ppaG and ppaI) produce up‐regulated transcripts in minimal medium compared with medium supplemented with plant tissue homogenate (Flügel et al., 2012). In addition, the expression of ppaB1, ppaB2, ppaC, ppaD, ppaG, ppaH and ppaI is detected in systemically infected plants by proteomic analysis (Savidor et al., 2012). Mutation of ppaA or ppaC results in lower severity of symptoms in infected plants, although the results are not statistically significant (Chalupowicz et al., 2017).
Subtilase proteases
The third group of secreted serine proteases is the subtilase proteases. Of these, sbtA is encoded by the PAI and sbtB and sbtC are encoded elsewhere on the chromosome (Gartemann et al., 2008). Intriguingly, they have high similarity to the SBT1, SBT2 and P69 subtilases of tomato (Gartemann et al., 2008; Janzik et al., 2000; Jorda et al., 1999). Although none of the sbt genes are expressed in minimal medium (Flügel et al., 2012), they are each detected at the transcript and/or protein level in foliar or systemically infected plants (Chalupowicz et al., 2017; Hiery et al., 2013; Savidor et al., 2012), suggesting that they play a role after perception of the host. Mutation of each sbt results in lower severity of leaf blisters during foliar infection, but only mutation of sbtA significantly reduces the incidence of symptoms during both foliar and systemic infection (Chalupowicz et al., 2017).
Cell wall‐degrading enzymes
In addition to proteases, Cmm is able to secrete hydrolytic carbohydrate‐active enzymes (CAZymes) that are involved in metabolism and degradation of plant cell wall components including polysaccharides, oligosaccharides and glycoconjugates (Cantarel et al., 2009). These enzymes include cellulases, xylanases and pectate lyases, which participate in the breakdown of the plant cell wall components cellulose, xylan and pectin, respectively, to facilitate bacterial colonization and nutrient acquisition (Cantarel et al., 2009). Glycome profiling of tomato plants inoculated with CmmCASJ002 shows reduced arabinogalactan epitopes, and loosening of xyloglucan epitopes, which indicate severe modification of plant cell wall integrity. This modification is consistent with the extensive tissue damage during the later stages of Cmm infection (Thapa et al., 2017).
Based on secretome analysis, Cmm strains are predicted to possess more CAZymes than other pathogenic C. michiganensis subspecies, as well as other Gram‐positive and xylem‐limited bacterial phytopathogens (Thapa et al., 2017). On the contrary, tomato endophytic Clavibacter strains have fewer total CAZymes relative to pathogenic Cmm (Thapa et al., 2017). Overall, the high number of CAZymes in Cmm suggests hydrolysis potential for a greater range of polysaccharides, and may be an indication of host specificity.
Cellulases
Located on the pCM1 plasmid, celA encodes a protein with high similarity to the endo‐β‐1,4‐glucanases of family A1 cellulases (Jahr et al., 2000). Its homologue on the chromosome outside the PAI, celB, is believed to be a pseudogene due to a truncated C‐terminal domain (Gartemann et al., 2008). Although one study found that mutation of celB reduced the severity of leaf blisters during foliar infection (Chalupowicz et al., 2017), the results were not statistically significant.
celA transcripts are abundant in minimal medium (Flügel et al., 2012). Its transcripts and proteins are detected during systemic infection of tomato plants (Chalupowicz et al., 2010, 2017; Savidor et al., 2012), whereas transcript abundance is lower during foliar infection (Chalupowicz et al., 2017). Consistent with these expression patterns, celA is considered to be a key player for pathogenicity in systemic infection. Like pat‐1, introduction of celA into Cmm100, a derivative of strain Cmm382 that lacks both plasmids, restores virulence and results in wilting of infected plants (Meletzus et al., 1993). Similarly, introduction of celA into a plasmid‐free CmmCASJ002 derivative or CmmCASJ001 derivative partially restores pathogenicity (Thapa et al., 2017). Furthermore, disruption of celA in Cmm101 (a derivative of Cmm382 which contains the celA‐bearing pCM1, but not pCM2) abolishes wilting during infection. These results demonstrate that celA is a major pathogenicity gene on pCM1, and is critical to bacterial canker disease, although other pCM1 genes may affect colonization or the utilization of nutrients (Jahr et al., 2000).
The 78‐kDa CelA (Jahr et al., 2000) is predicted to be a secreted protein, and is amongst 13 core secreted proteins present in all tomato‐pathogenic Cmm strains (Thapa et al., 2017). It consists of three domains: an N‐terminal catalytic domain, a cellulose binding domain and a C‐terminal domain with homology to α‐expansin from plants. Removal of any one of these domains abolishes wilt induction, indicating that all domains are required for CelA‐associated pathogenicity (Jahr et al., 2000). The C‐terminal α‐expansin domain is probably involved in non‐enzymatic loosening of the plant cell wall (Georgelis et al., 2014) by removing hydrogen bonds between carbohydrate polymers (Cosgrove, 1998), which may improve access to cellulose for hydrolysis. An additional α‐expansin located on the chromosome (Gartemann et al., 2008) may also participate in pathogenesis.
Two endoglucanases encoded by the endX/Y gene may also participate in cellulose degradation. Transcripts from this gene are abundant during foliar infection of tomato, and mutation of endX/Y significantly decreases the incidence of leaf blisters during foliar infection, whereas symptoms during systemic infection are largely unaltered (Chalupowicz et al., 2017). Two endoglucanases are also amongst 13 core secreted proteins present in all tomato‐pathogenic Cmm (Thapa et al., 2017).
Xylanases
Two β‐1,4‐xylanase encoding genes, xysA and xysB, are located on the chromosome outside of the PAI. Transcripts of xysA are significantly up‐regulated in minimal medium relative to xylem‐mimicking medium (Hiery et al., 2013), and transcripts of both xysA and xysB are detected at low levels in systemically infected plants (Chalupowicz et al., 2010).
Pectinases
The Cmm genome encodes a putative polygalacturonase, which depolymerizes pectin, and two pectate lyases which act on pectate, a product of pectin degradation in plants.
Transcripts from the polygalacturonase (pga), located on the chromosome outside the chp/tomA PAI, are up‐regulated during the growth of Cmm382 in minimal medium (Flügel et al., 2012) and during the early stages of foliar infection (Chalupowicz et al., 2017). The protein is also detected during systemic infection, and is significantly up‐regulated in medium supplemented with xylem sap vs. rich defined medium (Savidor et al., 2012). Mutation of pga results in a significantly reduced incidence of leaf blisters during foliar infection (Chalupowicz et al., 2017).
The pectate lyases (PelA1 and PelA2) are encoded on the chp/tomA PAI of Cmm and share 90%–92% identity (Strider, 1969; Thapa et al., 2017). During systemic infection, transcripts of pelA1 are abundant during the early stages of systemic infection (Chalupowicz et al., 2010), and the protein is present in planta. It is also significantly up‐regulated in medium supplemented with xylem sap compared with rich defined medium (Savidor et al., 2012). In minimal medium, the transcript abundance of pelA1 is greater than that of pelA2 in strains Cmm382, CmmCASJ002 and CmmCA00002. Perhaps because of its higher expression levels, deletion of pelA1 from CmmCASJ002 leads to a reduction in wilting symptoms relative to the wild‐type, whereas deletion of pelA2 does not affect symptoms. Neither deletion affects bacterial titres in planta (Thapa et al., 2017).
Other proteins
Other proteins found to play a role in pathogenesis include perforin and sortase. Mutation of genes for perforin (perF) or sortase (srtA) results in a significantly lower incidence of symptoms during foliar infection. The incidence and severity of systemic infection also appear to be lower in the mutants, although results in this case were not significant (Chalupowicz et al., 2017).
Transcriptional regulators
Plasmid‐encoded factors
Because xylanase activity is not detected in Cmm strains that lack pCM1, it was previously thought that the xylanase genes resided on this plasmid (Meletzus et al., 1993). However, it is now known that the genes xysA and xysB are located on the chromosome, suggesting a potential role of plasmid‐borne transcriptional regulators. In fact, transcripts of xysA are present, but significantly reduced, in Cmm100, which lacks plasmids, whereas their levels are not significantly altered in Cmm27, which lacks the PAI (both xysA and xysB are located outside of the PAI). In contrast, xysB transcript levels appear to be altered (up‐ or down‐regulated at various time points) in both Cmm100 and Cmm27 (Chalupowicz et al., 2010).
Other chromosomal genes whose expression is altered by the removal of plasmids include those encoding the serine proteases ChpC and PpaA, and the cell wall‐degrading enzymes CelB and PelA1. Transcripts of both chpC and ppaA are significantly reduced in Cmm strains that lack either or both of the plasmids (Cmm100, Cmm101 and Cmm102). Because this reduction in expression does not appear to be additive (Chalupowicz et al., 2010), transcriptional regulators on the two plasmids may participate at different positions in the same hierarchical pathway(s) that ultimately activates the transcription of chpC and ppaA.
Transcripts of celB and pelA1 are up‐regulated in Cmm100, suggesting that at least one plasmid participates in the transcriptional repression of these genes. The cellulase‐encoding celB is also up‐regulated in Cmm102, but not Cmm101 or Cmm27 (Chalupowicz et al., 2010). These results indicate that the plasmid pCM1 is involved in the transcriptional repression of celB, whereas genes located on pCM2 and on the PAI are not involved.
PAI‐encoded factors
As mentioned above, xysB transcript levels appear to be altered not only in Cmm100, but also in Cmm27, which lacks the chp/tomA PAI (Chalupowicz et al., 2010). Transcript levels of celA (located on pCM1) and pat‐1 (located on pCM2) are also significantly reduced in Cmm27 (Chalupowicz et al., 2017). Together, these results indicate that the PAI may encode transcriptional regulators that contribute to the expression of both chromosomal (xysB) and plasmid‐encoded (celA and pat‐1) virulence genes.
Vatr1 and Vatr2
Chromosomal genes outside of the chp/tomA PAI also play a role in the regulation of Cmm virulence factors. The mutation of vatr1, a member of the TetR family of transcriptional repressors, results in significantly higher levels of transcripts of 26 genes, including xysB and the subtilase protease sbtB, whereas the mutation of vatr2 from the GntR family of transcriptional regulators (members of which can be repressors or activators) significantly increases the expression of 16 genes (Savidor et al., 2014).
Both vatr1 and vatr2 also participate in positive regulatory pathways for virulence factors. Mutation analyses show that vatr1 positively regulates 34 genes and vatr2 positively regulates 36 genes, with 25 of these commonly regulated by both vatr1 and vatr2. Amongst the genes whose transcripts are down‐regulated in response to these mutations are pat‐1, sbtC and celA for Δvatr1 mutants, and pat‐1 and phpA for Δvatr2 mutants (Savidor et al., 2014).
Some of these regulated genes (including pat‐1, phpA and celA) are located on the pCM1 and pCM2 plasmids. This, together with the findings that plasmids affect the regulation of chromosomal virulence genes (above), highlights the interdependence between chromosomal and plasmid genes in virulence expression. Plasmid and chromosomal genes may share cis regulatory elements that are affected by the same trans factors, and may cooperate in regulatory pathways.
Indeed, both vatr1 and vatr2 regulate other transcriptional regulators, indicating that they occupy high positions in the hierarchy of virulence gene regulation, and vatr1 is a positive regulator of vatr2 (Savidor et al., 2014). All 10 of the vatr‐regulated genes identified as transcription factors are located on the chromosome (and outside of the chp/tomA PAI).
Plants infected with the vatr mutants show a significant reduction in wilting symptoms during systemic infection (Chalupowicz et al., 2017; Savidor et al., 2014), and this reduction is much more pronounced with Δvatr2 mutants than Δvatr1 mutants (Savidor et al., 2014). Mutation of vatr2 also leads to a significant reduction in the incidence of leaf blisters during foliar infection (Chalupowicz et al., 2017). Given that vatr1 is a positive regulator of vatr2, the greater reduction in systemic virulence observed with the Δvatr2 mutant is probably the result of complete loss of its expression, compared with partial loss of vatr2 expression in the Δvatr1 mutant. Neither mutation (Δvatr1 or Δvatr2) affects the growth of Cmm in rich medium, demonstrating that their effect on virulence is a result of their effects on virulence genes and not on general housekeeping processes (Savidor et al., 2014). Consistent with their role in virulence, plants infected with Δvatr1 or Δvatr2 mutants of Cmm produce significantly reduced levels of ACO and ethylene vs. plants infected with wild‐type Cmm (Savidor et al., 2014).
Host responses to infection
A number of tomato genes that are involved in defence against pathogenesis are up‐regulated during Cmm infection, including genes involved in the production and scavenging of free oxygen radicals, enhanced protein turnover and hormone (including ethylene) synthesis and response (Balaji et al., 2008).
To counteract pathogen invasion, plants can initiate signal transduction cascades involving various phosphatases and kinases (Balaji et al., 2008; Beckers et al., 2009; Mayrose et al., 2004). In addition to known phosphatases and kinases, two phospholipase D signal‐transducing proteins of tomato were up‐regulated during Cmm infection (Savidor et al., 2012).
To combat pathogen infection, host plants activate their basal defence responses via pathogenesis‐related (PR) proteins on recognition of extracellular pathogen‐associated molecular patterns (PAMPs) (Nurnberger et al., 2004; Zipfel and Felix, 2005). Generally, these PR proteins have antimicrobial properties and are involved in cellular activities, such as defence signalling, cell wall hydrolysis, production of reactive oxygen species, contact toxicity and alkalinization of the medium (Asai et al., 2002; van Loon et al., 2006). The proteome of Cmm‐infected tomato reveals a cluster of differentially expressed PR proteins compared with mock‐infected controls, including 1,3‐β‐glucosidase, endochitinase, cucumisin‐like serine protease, and osmotin‐ and hevein‐like proteins (Savidor et al., 2012).In addition, several proteins related to specific plant defence responses are induced in the infected plant (Savidor et al., 2012): lipoxygenase‐1 (LOX1), which may be involved in the synthesis of oxylipins or jasmonic acid (Bohland et al., 1997; Gardner, 1991; Melan et al., 1993; Vick and Zimmerman, 1983); enhanced disease susceptibility 1 (EDS1), which is crucial for the basal defence response against pathogens (Parker et al., 1996; Wiermer et al., 2005), and proteins similar to Phytophthora‐inhibited protease 1 (PIP1) and PepEST, which are involved in the response to potential virulence factors (Ko et al., 2005; Tian et al., 2007).
Conversely, various tomato proteins are suppressed in Cmm‐infected stems, including developmentally regulated plasma membrane polypeptide (DREPP), threonine or serine kinase for signal transduction, and four secreted class III peroxidases (Savidor et al., 2012). Class III peroxidases are produced during the course of lignin production, wound healing and defence, and their expression is normally up‐regulated in plants on bacterial, fungal or viral infection (Harrison et al., 1995; Hiraga et al., 2001; van Loon and Geelen, 1971; Reimers et al., 1992). The down‐regulation of these plant proteins in Cmm‐infected tomato (Savidor et al., 2012) suggests that Cmm interferes with this plant defence response.
Overall, research related to the response of tomato towards Cmm infection and the mechanisms associated with symptom development is still in progress. The availability of the completed tomato genome sequence (Tomato Genome Consortium, 2012) will enable further investigations of gene functions and new strategies for disease prevention.
Control of Cmm
Four subspecies of C. michiganensis, including Cmm, are classified as quarantine organisms by the European and Mediterranean Plant Protection Organization (EPPO) because of the serious economic threat that they pose (EPPO, 2017). Because dissemination occurs through the use of contaminated seeds and infected transplants, as well as through Cmm‐infested soil, equipment and tools, prevention of harmful cultural practices is extremely important. Reducing plant stress by maintaining an optimum population, nutrition, weed management and fertility will also make plants less likely to become diseased.
Because of its prominent impact, research has been undertaken to develop additional efficient tools for the detection and management of Cmm (Fig. 3). Various reliable and sensitive assays for Cmm diagnosis have been discussed in earlier reviews (de León et al., 2011; Sen et al., 2015). Long‐distance transmission through infected seeds means that seed testing is essential. However, although several bioassays are being developed and employed for the detection of Cmm in seeds, they are mostly used as presumptive tests because of their imprecise nature. Thus, the generation of a fast, sensitive and cost‐efficient detection method is crucial to expedite the diagnosis at an early stage.
Figure 3.
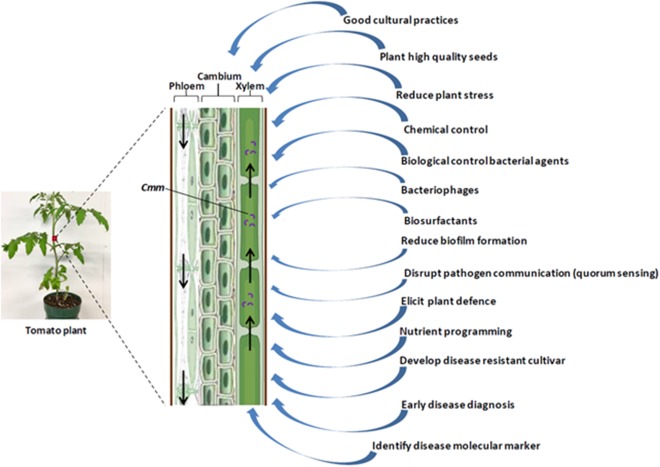
Schematic diagram summarizing the strategies to manage bacterial canker disease of tomato.
Chemical control of Cmm
Several chemical treatments for seeds, plants or soil have been studied for Cmm control. The most popular of these include the copper compounds, copper hydroxide and copper sulfate, and bactericides such as streptomycin, mancozeb and their combinations (Hausbeck et al., 2000; Kasselaki et al., 2011; de León et al., 2008; Werner et al., 2002). Although these antimicrobial compounds result in efficient reduction of bacterial titres, they are ultimately inadequate to protect the plant, and some are phytotoxic or promote resistance (de León et al., 2008; Yang et al., 2002). Other phytosanitary methods, such as seed treatments with acidified nitrite or 1% hydrochloric acid, and soil treatment with formaldehyde, reduce both the bacterial titre and symptom development, but are only partially effective against Cmm (Kasselaki et al., 2011; Pradhanang and Colier, 2009; Sharma and Kumar, 2000). A study using thermotherapy (hot water) for seed disinfection found that temperatures of 48 and 52 °C were effective for some seed cultivars, whereas higher temperatures (56 and 60 °C) were detrimental to seed germination and vigour (Divsalar et al., 2014). Therefore, the application of this eco‐friendly approach will require the identification of the optimum temperature and time for particular seed varieties.
Other less conventional antimicrobials are also being studied for use against Cmm. For example, the hexapeptide KCM21 elicits strong bactericidal activity against Cmm by rupturing the bacterial cells (Choi et al., 2014). Twelve potent small molecule inhibitors of Cmm have also been found, which are various piperidines, benzimidazoles, phenols, phenoxy isopropanolamines and pyrrolidones (Xu et al., 2015). Using single‐step and sequential passage resistance assays, no resistant Cmm colonies were observed following incubation at lethal and sublethal concentrations of each compound (Xu et al., 2015), suggesting that resistance to these compounds is not likely to be an immediate problem. However, the application of various agricultural chemicals or antibiotics faces various challenges, including the emergence of resistant pathogens, environmental health concerns and regulatory constraints. The detailed characterization of these compounds is therefore crucial.
Plant‐derived organic substances, such as extracts and essential oils, also have potential for the control of Cmm. Fragarin, purified from the soluble fraction of strawberry leaves, quickly inhibits the growth of C. michiganensis ssp. sepedonicus (ring rot disease of potato) via dissipation of the membrane potential (Filippone et al., 2001). Essential oils of thyme, oregano, dictamnus and marjoram are capable of inhibiting Cmm growth in vitro (Daferera et al., 2003). Related to oregano, Origanum onites L. provides essential oil, extracts and pure metabolites (such as carvacrol) which exhibit potent antibacterial activity against Cmm, and substantially decrease symptom development without affecting the germination or growth of tomato seedlings (Kotan et al., 2014). Moreover, the suppression of bacterial canker using tomato‐ or pepper‐based composts in combination with chicken or cattle manure is quite effective (Yogev et al., 2009), and treatment with lysozyme, an antimicrobial enzyme produced by animals, significantly reduces the incidence of bacterial canker on glasshouse tomatoes (Utkhede and Koch, 2004).
In contrast with these biocidal agents, other chemicals can be used to enhance the plant's own defences towards Cmm (Soylu et al., 2003; Werner et al., 2002). For example, pretreatment of tomato seedlings with acibenzolar‐S‐methyl (ASM) significantly reduces Cmm growth and disease severity during the course of infection (Baysal et al., 2003). This ASM‐mediated enhanced resistance is associated with increased activities of plant peroxidase and chitinase (Soylu et al., 2003). In addition, DL‐β‐aminobutyric acid (BABA) treatment remarkably suppresses symptom development caused by Cmm via enhanced peroxidase and phenylalanine ammonia‐lyase activities of the host plant (Baysal et al., 2005). These findings suggest that plant defence activators, particularly ASM and BABA, are linked to the induction of tomato plant resistance to bacterial canker.
Biological control
Competing microorganisms or viruses can help to limit the growth of phytopathogens by producing biocidal substances, inducing the plant's own resistance mechanisms (Grady et al., 2016), or directly parasitizing the pathogen. A large proportion of such biological control agents consists of rhizobacteria, including many genera that are able to efficiently colonize plant roots and suppress the invading soil‐borne pathogens (Schroth and Hancock, 1981; Weller, 1988).
Biocontrol agents having antagonistic activity towards Cmm have been reported, including seed and root treatments with fluorescent pseudomonads under glasshouse conditions (Amkraz et al., 2010; Boudyach et al., 2010). Seed treatment with both Pseudomonas and Bacillus strains improved the quality of tomato seeds and immensely decreased the incidence of bacterial canker in field studies (Kasselaki et al., 2011; Umesha, 2006).
Pseudomonads are well recognized for antibiosis, one of the key biocontrol mechanisms, whereby they can produce different antimicrobial metabolites, including phenazines, pyrrolnitrin and hydrogen cyanide (HCN), together with various degradative enzymes, for disease suppression (Haas and Defago, 2005; Weller and Thomashow, 1993). Accordingly, the reduction of canker disease development and Cmm population by Pseudomonas sp. LBUM300 is attributed to the production of the antibiotics 2,4‐diacetylphloroglucinol (DAPG) and HCN (Lanteigne et al., 2012; Paulin et al., 2017).
Other than pseudomonads, microorganisms with Cmm‐inhibiting activity under glasshouse conditions include Streptomyces sp. strain HL‐12 (Yuan et al., 2009), Bacillus subtilis, Trichoderma harzianum and Rhodosporidium diobovatum (Utkhede and Koch, 2004).
Phage‐based biological control is also being studied as a promising alternative because of its specificity, ease of preparation and inexpensive production (Jones et al., 2007). Bacteriophage CMP1, originally isolated from Cmm‐infected, overwintering tomato stems (Echandi and Sun, 1973), is a member of the Siphoviridae family of viruses and specifically infects Cmm. Its genome encodes an endolysin with peptidase activity (Wittmann et al., 2011) which specifically lyses Cmm, but not any other bacteria (Wittmann et al., 2010). The phage itself may have potential in preventing Cmm, but, for more efficient control, its endolysin was produced recombinantly in tomato plants, giving them complete resistance to Cmm (Wittmann et al., 2016).
Resistant cultivars
As for more traditional genetic improvement, attempts to breed a Cmm‐resistant tomato variety have been modest, and no commercial tomato cultivar possesses total resistance (Gleason et al., 1993; Liedl et al., 2013; Poysa, 1993). However, as mentioned above, Cmm may be present as an endophyte on some non‐host plants (Thapa et al., 2017) and can elicit HR in others via the ChpG protein (Lu et al., 2015). Further research into the mechanisms that reduce pathogenicity in non‐hosts may expedite the development of Cmm‐resistant cultivars, either through traditional breeding or genetic engineering. In addition, resistance may be engineered through the expression of recombinant antibodies that interfere with key virulence factors (Safarnejad et al., 2011).
Conclusions
Bacterial canker disease caused by Cmm is a serious threat in the majority of tomato‐growing areas, and is considered to be one of the most devastating plant diseases.
Current knowledge of Cmm implicates various virulence factors encoded by the plasmids, PAI and other chromosomal regions, yet, for many of these factors, their precise role in virulence remains unknown. The identification and study of these factors is crucial to understand the mechanism of pathogenicity, and to serve as a basic platform for the generation of target‐specific bactericides. Similarly, an understanding of the molecular basis of host–pathogen interactions may identify key host factors for susceptibility and resistance, which may aid in the creation of new resistant tomato varieties.
Acknowledgements
This work was largely funded by Mitacs‐Accelerate Fund (IT07941‐Yuan_OGVG) and Ontario Greenhouse Vegetable Growers (OGVG). We also appreciate the support and fund from Agriculture and Agri‐Food Canada, Growing Forward‐II (project J‐001332 and project J‐001589) and conducted by the authors as a part of their duties. The project was also partially funded by the Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery grant RGPIN‐2015‐06052 awarded to Z.‐C.Y. We thank Niki Bennett very much for useful discussions. The authors declare no conflict of interest.
References
- Alarcón, C. , Castro, J. , Muñoz, F. , Arce‐Johnson, P. and Delgado, J. (1998) Protein(s) from the Gram‐positive bacterium Clavibacter michiganensis subsp. michiganensis induces a hypersensitive response in plants. Phytopathology, 88, 306–310. [DOI] [PubMed] [Google Scholar]
- Amkraz, N. , Boudyach, E. , Boubaker, H. , Bouizgarne, B. and Aoumar, A.A.B. (2010) Screening for fluorescent pseudomonads, isolated from the rhizosphere of tomato, for antagonistic activity toward Clavibacter michiganensis subsp. michiganensis . World J. Microbiol. Biotechnol. 26, 1059–1065. [Google Scholar]
- Asai, T. , Tena, G. , Plotnikova, J. , Willmann, M.R. , Chiu, W.L. , Gomez‐Gomez, L. , Boller, T. , Ausubel, F.M. and Sheen, J. (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature, 415, 977–983. [DOI] [PubMed] [Google Scholar]
- Balaji, V. , Mayrose, M. , Sherf, O. , Jacob‐Hirsch, J. , Eichenlaub, R. , Iraki, N. , Manulis‐Sasson, S. , Rechavi, G. , Barash, I. and Sessa, G. (2008) Tomato transcriptional changes in response to Clavibacter michiganensis subsp. michiganensis reveal a role for ethylene in disease development. Plant Physiol. 146, 1797–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baysal, O. , Soylu, E.M. and Soylu, S. (2003) Induction of defence‐related enzymes and resistance by the plant activator acibenzolar‐S‐methyl in tomato seedlings against bacterial canker caused by Clavibacter michiganensis ssp. michiganensis . Plant Pathol. 52, 747–753. [Google Scholar]
- Baysal, O. , Gursoy, Y. , Ornek, H. and Duru, A. (2005) Induction of oxidants in tomato leaves treated with DL‐beta‐amino butyric acid (BABA) and infected with Clavibacter michiganensis ssp. michiganensis . Eur. J. Plant. Pathol. 112, 361–369. [Google Scholar]
- Beckers, G.J. , Jaskiewicz, M. , Liu, Y. , Underwood, W.R. , He, S.Y. , Zhang, S. and Conrath, U. (2009) Mitogen‐activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana . Plant Cell, 21, 944–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank, L. , Cohen, Y. , Borenstein, M. , Shulhani, R. , Lofthouse, M. , Sofer, M. and Shtienberg, D. (2016) Variables associated with severity of bacterial canker and wilt caused by Clavibacter michiganensis subsp. michiganensis in tomato greenhouses. Phytopathology, 106, 254–261. [DOI] [PubMed] [Google Scholar]
- Bliven, K.A. and Maurelli, A.T. (2012) Antivirulence genes: insights into pathogen evolution through gene loss. Infect. Immun. 80, 4061–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohland, C. , Balkenhohl, T. , Loers, G. , Feussner, I. and Grambow, H.J. (1997) Differential induction of lipoxygenase isoforms in wheat upon treatment with rust fungus elicitor, chitin oligosaccharides, chitosan, and methyl jasmonate. Plant Physiol. 114, 679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudyach, E.H. , Fatmi, M. , Akhayat, O. , Benizri, E. and Aoumar, A.A.B. (2010) Selection of antagonistic bacteria of Clavibacter michiganensis subsp. michiganensis and evaluation of their efficiency against bacterial canker of tomato. Biocont. Sci. Tech. 11, 141–149. [Google Scholar]
- Burger, A. , Grafen, I. , Engemann, J. , Niermann, E. , Pieper, M. , Kirchner, O. , Gartemann, K.‐H. and Eichenlaub, R. (2005) Identification of homologues to the pathogenicity factor Pat‐1, a putative serine protease of Clavibacter michiganensis subsp. michiganensis . Microbiol. Res. 160, 417–427. [DOI] [PubMed] [Google Scholar]
- Cantarel, B.L. , Coutinho, P.M. , Rancurel, C. , Bernard, T. , Lombard, V. and Henrissat, B. (2009) The carbohydrate‐active enzymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, R.R. and Vidaver, A.K. (1982) Taxonomy of Corynebacterium plant pathogens, including a new pathogen of wheat, based on polyacrylamide gel electrophoresis of cellular proteins. Int. J. Syst. Evol. Microbiol. 32, 315–326. [Google Scholar]
- Carlton, W.M. , Braun, E.J. and Gleason, M.L. (1998) Ingress of Clavibacter michiganensis subsp. michiganensis into tomato leaves through hydathodes. Phytopathology, 88, 525–529. [DOI] [PubMed] [Google Scholar]
- Chalupowicz, L. , Cohen‐Kandli, M. , Dror, O. , Eichenlaub, R. , Gartemann, K.H. , Sessa, G. , Barash, I. and Manulis‐Sasson, S. (2010) Sequential expression of bacterial virulence and plant defense genes during infection of tomato with Clavibacter michiganensis subsp. michiganensis . Phytopathology, 100, 252–261. [DOI] [PubMed] [Google Scholar]
- Chalupowicz, L. , Barash, I. , Reuven, M. , Dror, O. , Sharabani, G. , Gartemann, K.H. , Eichenlaub, R. , Sessa, G. and Manulis‐Sasson, S. (2017) Differential contribution of Clavibacter michiganensis ssp. michiganensis virulence factors to systemic and local infection in tomato. Mol. Plant Pathol. 18, 336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, R. , Ries, S. and Pataky, J. (1991) Dissemination of Clavibacter michiganensis subsp. michiganensis by practices used to produce tomato transplants. Phytopathology, 81, 1276–1281. [Google Scholar]
- Chang, R.J. , Ries, S.M. and Pataky, J.K. (1992) Reductions in yield of processing tomatoes and incidence of bacterial canker. Plant Dis. 76, 805–809. [Google Scholar]
- Choi, J. , Baek, K.‐H. and Moon, E. (2014) Antimicrobial effects of a hexapeptide KCM21 against Pseudomonas syringae pv. tomato DC3000 and Clavibacter michiganensis subsp. michiganensis . Plant Pathol. J. 30, 245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarone, A. and Carilli, A. (1948) Osservazion di campo su Corynebacterium michiganense (Smith) Jensen e considerazioni su un possible cao di sua supravvivenza nel terreno. Boll. Staz. Patol. Vegetal. 6, 277–280. [Google Scholar]
- Cosgrove, D.J. (1998) Cell wall loosening by expansins. Plant Physiol. 118, 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daferera, D.J. , Ziogas, B.N. and Polissiou, M.G. (2003) The effectiveness of plant essential oils on the growth of Botrytis cinerea, Fusarium sp. and Clavibacter michiganensis subsp. michiganensis . Crop Prot. 22, 39–44. [Google Scholar]
- Darrasse, A. , Darsonval, A. , Boureau, T. , Brisset, M.N. , Durand, K. and Jacques, M.A. (2010) Transmission of plant‐pathogenic bacteria by nonhost seeds without induction of an associated defense reaction at emergence. Appl. Environ. Microbiol. 76, 6787–6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, M.J. , Gillaspie, A.G. , Vidaver, A.K. and Harris, R.W. (1984) Clavibacter: a new genus containing some phytopathogenic coryneform bacteria, including Clavibacter xyli subsp. xyli sp. nov., subsp. nov. and Clavibacter xyli subsp. cynodontis subsp. nov., pathogens that cause ratoon stunting disease of sugarcane and bermudagrass stunting disease. Int. J. Syst. Bacteriol. 34, 107–117. [Google Scholar]
- Divsalar, M. , Shakeri, M. and Khandan, A. (2014) Study on thermotherapy treatment effects on seed germination and vigor of tomato cultivars. Int. J. Plant Soil Sci. 3, 799–809. [Google Scholar]
- Echandi, E. and Sun, M. (1973) Isolation and characterization of a bacteriophage for identification of Corynebacterium michiganense . Phytopathology, 63, 1398–1401. [Google Scholar]
- Eichenlaub, R. and Gartemann, K.H. (2011) The Clavibacter michiganensis subspecies: molecular investigation of gram‐positive bacterial plant pathogens. Annu. Rev. Phytopathol. 49, 445–464. [DOI] [PubMed] [Google Scholar]
- EPPO . (2017) EPPO A2 List of pests recommended for regulation as quarantine pests. https://www.eppo.int/QUARANTINE/listA2.htm [Google Scholar]
- Fatmi, M. and Schaad, N. (1988) Semiselective agar medium for isolation of Clavibacter michiganense subsp. michiganense from tomato seed. Phytopathology, 78, 121–126. [Google Scholar]
- Fatmi, M. and Schaad, N. (2002) Survival of Clavibacter michiganensis ssp. michiganensis in infected tomato stems under natural field conditions in California, Ohio and Morocco. Plant Pathol. 51, 149–154. [Google Scholar]
- Fatmi, M. , Schaad, N. and Bolkan, H. (1991) Seed treatments for eradicating Clavibacter michiganensis subsp. michiganensis from naturally infected tomato seeds. Plant Dis. 75, 383–385. [Google Scholar]
- Filippone, M.P. , Diaz‐Ricci, J.C. , Castagnaro, A.P. and Farías, R.N. (2001) Effect of fragarin on the cytoplasmic membrane of the phytopathogen Clavibacter michiganensis . Mol. Plant–Microbe Interact. 14, 925–928. [DOI] [PubMed] [Google Scholar]
- Flügel, M. , Becker, A. , Gartemann, K.H. and Eichenlaub, R. (2012) Analysis of the interaction of Clavibacter michiganensis subsp. michiganensis with its host plant tomato by genome‐wide expression profiling. J. Biotechnol. 160, 42–54. [DOI] [PubMed] [Google Scholar]
- Food and Agricultural Organization (FAO) . (2016) Data Available at http://faostat.fao.org/site/339/default.aspx [accessed on 1 October 2016].
- Gardner, H.W. (1991) Recent investigations into the lipoxygenase pathway of plants. Biochem. Biophys. Acta, 1084, 221–239. [DOI] [PubMed] [Google Scholar]
- Gartemann, K.‐H. , Abt, B. , Bekel, T. , Burger, A. , Engemann, J. , Flügel, M. , Gaigalat, L. , Goesmann, A. , Gräfen, I. , Kalinowski, J. , Kaup, O. , Kirchner, O. , Krause, L. , Linke, B. , McHardy, A. , Meyer, F. , Pohle, S. , Rückert, C. , Schneiker, S. , Zellermann, E.M. , Pühler, A. , Eichenlaub, R. , Kaiser, O. and Bartels, D. (2008) The genome sequence of the tomato‐pathogenic actinomycete Clavibacter michiganensis subsp. michiganensis NCPPB382 reveals a large island involved in pathogenicity. J. Bacteriol. 190, 2138–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgelis, N. , Nikolaidis, N. and Cosgrove, D.J. (2014) Biochemical analysis of expansin‐like proteins from microbes. Carbohydr. Polym. 100, 17–23. [DOI] [PubMed] [Google Scholar]
- Gitaitis, R. , Beaver, R. and Voloudakis, A. (1991) Detection of Clavibacter michiganensis subsp. michiganensis in symptomless tomato transplants. Plant Dis. 75, 834–838. [Google Scholar]
- Gleason, M.L. , Gitaitis, R.D. and Ricker, M.D. (1993) Recent progress in understanding and controlling bacterial canker of tomato in eastern North America. Plant Dis. 77, 1069–1076. [Google Scholar]
- Gonzalez, A.J. and Trapiello, E. (2014) Clavibacter michiganensis subsp. phaseoli subsp. nov., pathogenic in bean. Int. J. Syst. Evol. Microbiol. 64, 1752–1755. [DOI] [PubMed] [Google Scholar]
- Gouran, H. , Gillespie, H. , Nascimento, R. , Chakraborty, S. , Zaini, P.A. , Jacobson, A. , Phinney, B.S. , Dolan, D. , Durbin‐Johnson, B.P. , Antonova, E.S. , Lindow, S.E. , Mellema, M.S. , Goulart, L.R. and Dandekar, A.M. (2016) The secreted protease PrtA controls cell growth, biofilm formation and pathogenicity in Xylella fastidiosa . Sci. Rep. 6, 31 098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady, E.N. , MacDonald, J. , Liu, L. , Richman, A. and Yuan, Z.C. (2016) Current knowledge and perspectives of Paenibacillus: a review. Microb. Cell Fact. 15, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, D. and Defago, G. (2005) Biological control of soil‐borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3, 307–319. [DOI] [PubMed] [Google Scholar]
- Hadas, R. , Kritzman, G. , Klietman, F. , Gefen, T. and Manulis, S. (2005) Comparison of extraction procedures and determination of the detection threshold for Clavibacter michiganensis ssp. michiganensis in tomato seeds. Plant Pathol. 54, 643–649. [Google Scholar]
- Harrison, S.J. , Curtis, M.D. , McIntyre, C.L. , Maclean, D.J. and Manners, J.M. (1995) Differential expression of peroxidase isogenes during the early stages of infection of the tropical forage legume Stylosanthes humilis by Colletotrichum gloeosporioides . Mol. Plant–Microbe Interact. 8, 398–406. [DOI] [PubMed] [Google Scholar]
- Hausbeck, M. , Bell, J. , Medina‐Mora, C. , Podolsky, R. and Fulbright, D. (2000) Effect of bactericides on population sizes and spread of Clavibacter michiganensis subsp. michiganensis on tomatoes in the greenhouse and on disease development and crop yield in the field. Phytopathology, 90, 38–44. [DOI] [PubMed] [Google Scholar]
- Hiery, E. , Adam, S. , Reid, S. , Hofmann, J. , Sonnewald, S. and Burkovski, A. (2013) Genome‐wide transcriptome analysis of Clavibacter michiganensis subsp. michiganensis grown in xylem mimicking medium. J. Biotechnol. 168, 348–354. [DOI] [PubMed] [Google Scholar]
- Hiraga, S. , Sasaki, K. , Ito, H. , Ohashi, Y. and Matsui, H. (2001) A large family of class III plant peroxidases. Plant Cell Physiol. 42, 462–468. [DOI] [PubMed] [Google Scholar]
- Jacques, M.A. , Durand, K. , Orgeur, G. , Balidas, S. , Fricot, C. , Bonneau, S. , Quillévéré, A. , Audusseau, C. , Olivier, V. , Grimault, V. and Mathis, R. (2012) Phylogenetic analysis and polyphasic characterization of Clavibacter michiganensis strains isolated from tomato seeds reveal that nonpathogenic strains are distinct from C. michiganensis subsp. michiganensis . Appl. Environ. Microbiol. 78, 8388–8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahr, H. , Dreier, J. , Meletzus, D. , Bahro, R. and Eichenlaub, R. (2000) The endo‐beta‐1,4‐glucanase CelA of Clavibacter michiganensis subsp. michiganensis is a pathogenicity determinant required for induction of bacterial wilt of tomato. Mol. Plant–Microbe Interact. 13, 703–714. [DOI] [PubMed] [Google Scholar]
- Janzik, I. , Macheroux, P. , Amrhein, N. and Schaller, A. (2000) LeSBT1, a subtilase from tomato plants – overexpression in insect cells, purification, and characterization. J. Biol. Chem. 275, 5193–5199. [DOI] [PubMed] [Google Scholar]
- Jashni, M.K. , Dols, I.H. , Iida, Y. , Boeren, S. , Beenen, H.G. , Mehrabi, R. , Collemare, J. and de Wit, P.J. (2015) Synergistic action of a metalloprotease and a serine protease from Fusarium oxysporum f. sp. lycopersici cleaves chitin‐binding tomato chitinases, reduces their antifungal activity, and enhances fungal virulence. Mol. Plant–Microbe Interact. 28, 996–1008. [DOI] [PubMed] [Google Scholar]
- Jones, J. , Jackson, L. , Balogh, B. , Obradovic, A. , Iriarte, F. and Momol, M. (2007) Bacteriophages for plant disease control. Annu. Rev. Phytopathol. 45, 245–262. [DOI] [PubMed] [Google Scholar]
- Jorda, L. , Coego, A. , Conejero, V. and Vera, P. (1999) A genomic cluster containing four differentially regulated subtilisin‐like processing protease genes in tomato plants. J. Biol. Chem. 274, 2360–2365. [DOI] [PubMed] [Google Scholar]
- Kasselaki, A.‐M. , Goumas, D. , Tamm, L. , Fuchs, J. , Cooper, J. and Leifert, C. (2011) Effect of alternative strategies for the disinfection of tomato seed infected with bacterial canker (Clavibacter michiganensis subsp. michiganensis). NJAS – Wageningen J. Life Sci. 58, 145–147. [Google Scholar]
- Kaup, O. , Grafen, I. , Zellermann, E.M. , Eichenlaub, R. and Gartemann, K.H. (2005) Identification of a tomatinase in the tomato‐pathogenic actinomycete Clavibacter michiganensis subsp. michiganensis NCPPB382. Mol. Plant–Microbe Interact. 18, 1090–1098. [DOI] [PubMed] [Google Scholar]
- Kleitman, F. , Barash, I. , Burger, A. , Iraki, N. , Falah, Y. , Sessa, G. , Weinthal, D. , Chalupowicz, L. , Gartemann, K.‐H. , Eichenlaub, R. and Manulis‐Sasson, S. (2008) Characterization of a Clavibacter michiganensis subsp. michiganensis population in Israel. Eur. J. Plant Pathol. 121, 463–475. [Google Scholar]
- Ko, M.K. , Jeon, W.B. , Kim, K.S. , Lee, H.H. , Seo, H.H. , Kim, Y.S. and Oh, B.J. (2005) A Colletotrichum gloeosporioides‐induced esterase gene of nonclimacteric pepper (Capsicum annuum) fruit during ripening plays a role in resistance against fungal infection. Plant Mol. Biol. 58, 529–541. [DOI] [PubMed] [Google Scholar]
- Kotan, R. , Cakir, A. , Ozer, H. , Kordali, S. , Cakmakci, R. , Dadasoglu, F. , Dikbas, N. , Aydin, T. and Kazaz, C. (2014) Antibacterial effects of Origanum onites against phytopathogenic bacteria: possible use of the extracts from protection of disease caused by some phytopathogenic bacteria. Sci. Hortic. 172, 210–220. [Google Scholar]
- Lanteigne, C. , Gadkar, V.J. , Wallon, T. , Novinscak, A. and Filion, M. (2012) Production of DAPG and HCN by Pseudomonas sp. LBUM300 contributes to the biological control of bacterial canker of tomato. Phytopathology, 102, 967–973. [DOI] [PubMed] [Google Scholar]
- de León, L. , Siverio, F. , López, M.M. and Rodríguez, A. (2008) Comparative efficiency of chemical compounds for in vitro and in vivo activity against Clavibacter michiganensis subsp. michiganensis, the causal agent of tomato bacterial canker. Crop Prot. 27, 1277–1283. [Google Scholar]
- de León, L. , Siverio, F. , López, M.M. and Rodríguez, A. (2011) Clavibacter michiganensis subsp. michiganensis, a seedborne tomato pathogen: healthy seeds are still the goal. Plant Dis. 95, 1328–1338. [DOI] [PubMed] [Google Scholar]
- Liedl, B.E. , Labate, J.A. , Stommel, J.R. , Slade, A. , and Kole, C. (2013) Genetics, Genomics, and Breeding of Tomato. Boca Raton, FL: CRC Press. [Google Scholar]
- van Loon, L.C. and Geelen, J.L.M.C. (1971) The relation of polyphenoloxidase and peroxidase to symptom expression in tobacco var. “Samsun NN” after infection with tobacco mosaic virus. Acta Phytopathol. Acad. Sci. Hung. 6, 9–20. [Google Scholar]
- van Loon, L.C. , Rep, M. and Pieterse, C.M. (2006) Significance of inducible defense‐related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. [DOI] [PubMed] [Google Scholar]
- Lu, Y. , Hatsugai, N. , Katagiri, F. , Ishimaru, C.A. and Glazebrook, J. (2015) Putative serine protease effectors of Clavibacter michiganensis induce a hypersensitive response in the apoplast of Nicotiana species. Mol. Plant–Microbe Interact. 28, 1216–1226. [DOI] [PubMed] [Google Scholar]
- Manzer, F. and Genereux, H. (1981) Ring rot In: Compendium of Potato Disease (Hooker W.J. ed.), pp. 31–32. St Paul, MN: American Phytopathological Society Press. [Google Scholar]
- Mayrose, M. , Bonshtien, A. and Sessa, G. (2004) LeMPK3 is a mitogen activated protein kinase with dual specificity induced during tomato defense and wounding responses. J. Biol. Chem. 279, 14 819–14 827. [DOI] [PubMed] [Google Scholar]
- McCulloch, L. (1925) Aplanobacter insidiosum n. sp., the cause of an Alfalfa disease. Phytopathology, 15, 496–497. [Google Scholar]
- Melan, M.A. , Dong, X. , Endara, M.E. , Davis, K.R. , Ausubel, F.M. and Peterman, T.K. (1993) An Arabidopsis thaliana lipoxygenase gene can be induced by pathogens, abscisic acid, and methyl jasmonate. Plant Physiol. 101, 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meletzus, D. , Bermphol, A. , Dreier, J. and Eichenlaub, R. (1993) Evidence for plasmid‐encoded virulence factors in the phytopathogenic bacterium Clavibacter michiganensis subsp. michiganensis NCPPB382. J. Bacteriol. 175, 2131–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milijašević‐Marčić, S. , Gartemann, K.‐H. , Frohwitter, J. , Eichenlaub, R. , Todorović, B. , Rekanović, E. and Potočnik, I. (2012) Characterization of Clavibacter michiganensis subsp. michiganensis strains from recent outbreaks of bacterial wilt and canker in Serbia. Eur. J. Plant Pathol. 134, 697–711. [Google Scholar]
- Nurnberger, T. , Brunner, F. , Kemmerling, B. and Piater, L. (2004) Innate immunity in plants and animals: striking similarities and obvious differences. Immunol. Rev. 198, 249–266. [DOI] [PubMed] [Google Scholar]
- Oh, E.J. , Bae, C. , Lee, H.B. , Hwang, I.S. , Lee, H.I. , Yea, M.C. , Yim, K.O. , Lee, S. , Heu, S. , Cha, J.S. and Oh, C.S. (2016) Clavibacter michiganensis subsp. capsici subsp. nov., causing bacterial canker disease in pepper. Int. J. Syst. Evol. Microbiol. 66, 4065–4070. [DOI] [PubMed] [Google Scholar]
- Pareja‐Jaime, Y. , Roncero, M.I.G. and Ruiz‐Roldán, M.C. (2008) Tomatinase from Fusarium oxysporum f. sp. lycopersici is required for full virulence on tomato plants. Mol. Plant–Microbe Interact. 21, 728–736. [DOI] [PubMed] [Google Scholar]
- Parker, J.E. , Holub, E.B. , Frost, L.N. , Falk, A. , Gunn, N.D. and Daniels, M.J. (1996) Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell, 8, 2033–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulin, M.M. , Novinscak, A. , Lanteigne, C. , Gadkar, V.J. and Filion, M. (2017) Interaction between 2,4‐diacetylphloroglucinol‐ and hydrogen cyanide‐producing Pseudomonas brassicacearum LBUM300 and Clavibacter michiganensis subsp. michiganensis in the tomato rhizosphere. Appl. Environ. Microbiol. 83, e00073–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploch, S. and Thines, M. (2011) Obligate biotrophic pathogens of the genus Albugo are widespread as asymptomatic endophytes in natural populations of Brassicaceae. Mol. Ecol. 20, 3692–3699. [DOI] [PubMed] [Google Scholar]
- Poysa, V. (1993) Evaluation of tomato breeding lines resistant to bacterial canker. Can. J. Plant Pathol. 15, 301–304. [Google Scholar]
- Pradhanang, P.M. and Colier, G. (2009) How effective is hydrochloric acid treatment to control Clavibacter michiganensis subsp. michiganensis contamination in tomato seed? Acta Hortic. 808, 81–85. [Google Scholar]
- Quesada‐Ocampo, L. , Landers, N. , Lebeis, A. , Fulbright, D. and Hausbeck, M. (2012) Genetic structure of Clavibacter michiganensis subsp. michiganensis populations in Michigan commercial tomato fields. Plant Dis. 96, 788–796. [DOI] [PubMed] [Google Scholar]
- Rat, B. , Poissonnier, J. , Goisque, M.J. and Burgaud, A. (1991) Le point sur le chancre bactérien. Fruit Légu. 86, 38–40. [Google Scholar]
- Reimers, P.J. , Guo, A. and Leach, J.E. (1992) Increased activity of a cationic peroxidase associated with an incompatible interaction between Xanthomonas oryzae pv oryzae and rice (Oryza sativa). Plant Physiol. 99, 1044–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricker, M. and Riedel, R. (1993) Effect of secondary spread of Clavibacter michiganensis subsp. michiganensis on yield of northern processing tomatoes. Plant Dis. 77, 364–366. [Google Scholar]
- Safarnejad, M.R. , Jouzani, G.S. , Tabatabaie, M. , Twyman, R.M. and Schillberg, S. (2011) Antibody‐mediated resistance against plant pathogens. Biotechnol. Adv. 29, 961–971. [DOI] [PubMed] [Google Scholar]
- Savidor, A. , Teper, D. , Gartemann, K.‐H. , Eichenlaub, R. , Chalupowicz, L. , Manulis‐Sasson, S. , Barash, I. , Tews, H. , Mayer, K. , Giannone, R.J. , Hettich, R.L. and Sessa, G. (2012) The Clavibacter michiganensis subsp. michiganensis–tomato interactome reveals the perception of pathogen by the host and suggests mechanisms of infection. J. Proteome Res. 11, 736–750. [DOI] [PubMed] [Google Scholar]
- Savidor, A. , Chalupowicz, L. , Teper, D. , Gartemann, K.H. , Eichenlaub, R. , Manulis‐Sasson, S. , Barash, I. and Sessa, G. (2014) Clavibacter michiganensis subsp. michiganensis Vatr1 and Vatr2 transcriptional regulators are required for virulence in tomato. Mol. Plant–Microbe Interact. 27, 1035–1047. [DOI] [PubMed] [Google Scholar]
- Schroth, M.N. and Hancock, J.C. (1981) Selected topics in biological control. Annu. Rev. Microbiol. 35, 453–476. [DOI] [PubMed] [Google Scholar]
- Sen, Y. , van der Wolf, J. , Visser, R.G. and van Heusden, S. (2015) Bacterial canker of tomato: current knowledge of detection, management, resistance, and interactions. Plant Dis. 99, 4–13. [DOI] [PubMed] [Google Scholar]
- Sharabani, G. , Shtienberg, D. , Borenstein, M. , Shulhani, R. , Lofthouse, M. , Sofer, M. , Chalupowicz, L. , Barel, V. and Manulis‐Sasson, S. (2013) Effects of plant age on disease development and virulence of Clavibacter michiganensis subsp. michiganensis on tomato. Plant Pathol. 62, 1114–1122. [Google Scholar]
- Sharma, J.P. and Kumar, S. (2000) Management of Ralstonia wilt through soil disinfectant, mulch, lime and cakes in tomato (Lycopersicon esculentum). Indian J. Agric. Sci. 70, 17–19. [Google Scholar]
- Smith, E. (1910) A new tomato disease of economic importance. Science, 31, 794–796. [Google Scholar]
- Sowley, E.N.K. , Dewey, F.M. and Shaw, M.W. (2010) Persistent, symptomless, systemic, and seed‐borne infection of lettuce by Botrytis cinerea . Eur. J. Plant Pathol. 126, 61–71. [Google Scholar]
- Soylu, S. , Baysal, O. and Soylu, E.M. (2003) Induction of disease resistance by the plant activator, acibenzolar‐S‐methyl (ASM), against bacterial canker (Clavibacter michiganensis subsp. michiganensis) in tomato seedlings. Plant Sci. 165, 1069–1075. [Google Scholar]
- Stork, I. , Gartemann, K.H. , Burger, A. and Eichenlaub, R. (2008) A family of serine proteases of Clavibacter michiganensis subsp. michiganensis: chpC plays a role in colonization of the host plant tomato. Mol. Plant Pathol. 9, 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strider, D.L. (1969) Bacterial canker of tomato caused by Corynebacterium michiganense: a literature review and bibliography In: Technical Bulletin No. 193. Raleigh, NC: North Carolina Agricultural Experiment Station. [Google Scholar]
- Tambong, J.T. (2017) Comparative genomics of Clavibacter michiganensis subspecies, pathogens of important agricultural crops. PLoS One, 12, e0172295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tancos, M.A. , Chalupowicz, L. , Barash, I. , Manulis‐Sasson, S. and Smart, C.D. (2013) Tomato fruit and seed colonization by Clavibacter michiganensis subsp. michiganensis through external and internal routes. Appl. Environ. Microbiol. 79, 6948–6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tancos, M.A. , Lange, H.W. and Smart, C.D. (2015) Characterizing the genetic diversity of the Clavibacter michiganensis subsp. michiganensis population in New York. Bacteriology, 105, 169–179. [DOI] [PubMed] [Google Scholar]
- Thapa, S.P. , Pattathil, S. , Hahn, M.G. , Jacques, M.A. , Gilbertson, R.L. and Coaker, G. (2017) Genomic analysis of Clavibacter michiganensis reveals insight into virulence strategies and genetic diversity of a Gram‐positive bacterial pathogen. Mol. Plant–Microbe Interact. 30, 786–802. [DOI] [PubMed] [Google Scholar]
- Tian, M. , Win, J. , Song, J. , van der Hoorn, R. , van der Knaap, E. and Kamoun, S. (2007) A Phytophthora infestans cystatin‐like protein targets a novel tomato papain‐like apoplastic protease. Plant Physiol. 143, 364–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Corsortium . (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesha, S. (2006) Occurrence of bacterial canker in tomato fields of Karnataka and effect of biological seed treatment on disease incidence. Crop Prot. 25, 375–381. [Google Scholar]
- Utkhede, R. and Koch, C. (2004) Biological treatments to control bacterial canker of greenhouse tomatoes. Biocontrol, 49, 305–313. [Google Scholar]
- Vick, B.A. and Zimmerman, D.C. (1983) The biosynthesis of jasmonic acid: a physiological role for plant lipoxygenase. Biochem. Biophys. Res. Commun. 111, 470–477. [DOI] [PubMed] [Google Scholar]
- Vidaver, A.K. and Mandel, M. (1974) Corynebacterium nebraskense, a new, orange‐pigmented phytopathogenic species. Int. J. Syst. Evol. Microbiol. 24, 482–485. [Google Scholar]
- Volcani, Z. (1985) Bacterial Diseases of Plants in Israel. Bet Dagan: Agricultural Research Organization.
- Wassermann, E. , Montecchia, M.S. , Correa, O.S. , Damián, V. and Romero, A.M. (2017) Clavibacter michiganensis subsp. michiganensis strains virulence and genetic diversity. A first study in Argentina. Eur. J. Plant Pathol. 149, 35–42. [Google Scholar]
- Weller, D.M. (1988) Biological control of soilborne plant pathogens in the rhizosphere with bacteria. Annu. Rev. Phytopathol. 26, 379–407. [Google Scholar]
- Weller, D.M. and Thomashow, L.S. (1993) Use of rhizobacteria for biocontrol. Curr. Opin. Biotechnol. 4, 306–311. [Google Scholar]
- Werner, N.A. , Fulbright, D.W. , Podolsky, R. , Bell, J. and Hausbeck, M.K. (2002) Limiting populations and spread of Clavibacter michiganensis subsp. michiganensis on seedling tomatoes in the greenhouse. Plant Dis. 86, 535–542. [DOI] [PubMed] [Google Scholar]
- Wiermer, M. , Feys, B.J. and Parker, J.E. (2005) Plant immunity: the EDS1 regulatory node. Curr. Opin. Plant Biol. 8, 383–389. [DOI] [PubMed] [Google Scholar]
- Wittmann, J. , Eichenlaub, R. and Dreiseikelmann, B. (2010) The endolysins of bacteriophages CMP1 and CN77 are specific for the lysis of Clavibacter michiganensis strains. Microbiology, 156, 2366–2373. [DOI] [PubMed] [Google Scholar]
- Wittmann, J. , Gartemann, K.H. , Eichenlaub, R. and Dreiseikelmann, B. (2011) Genomic and molecular analysis of phage CMP1 from Clavibacter michiganensis subspecies michiganensis . Bacteriophage, 1, 6–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann, J. , Brancato, C. , Berendzen, K.W. and Dreiseikelmann, B. (2016) Development of a tomato plant resistant to Clavibacter michiganensis using the endolysin gene of bacteriophage CMP1 as a transgene. Plant Pathol. 65, 496–502. [Google Scholar]
- Xu, X. , Miller, S.A. , Baysal‐Gurel, F. , Gartemann, K.H. , Eichenlaub, R. and Rajashekara, G. (2010) Bioluminescence imaging of Clavibacter michiganensis subsp. michiganensis infection of tomato seeds and plants. Appl. Environ. Microbiol. 76, 3978–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X. , Kumar, A. , Deblais, L. , Pina‐Mimbela, R. , Nislow, C. , Fuchs, J.R. , Miller, S.A. and Rajashekara, G. (2015) Discovery of novel small molecule modulators of Clavibacter michiganensis subsp. michiganensis . Front. Microbiol. 6, 1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X.‐E. , Long, X.‐X. , Ni, W.‐Z. , Ye, Z.‐Q. , He, Z.‐L. , Stoffella, P.J. and Calvert, D.V. (2002) Assessing copper thresholds for phytotoxicity and potential dietary toxicity in selected vegetable crops. J. Environ. Sci. Health B, 37, 625–635. [DOI] [PubMed] [Google Scholar]
- Yasuhara‐Bell, J. and Alvarez, A.M. (2015) Seed‐associated subspecies of the genus Clavibacter are clearly distinguishable from Clavibacter michiganensis subsp. michiganensis . Int. J. Syst. Evol. Microbiol. 65, 811–826. [DOI] [PubMed] [Google Scholar]
- Yogev, A. , Raviv, M. , Kritzman, G. , Hadar, Y. , Cohen, R. , Kirshner, B. and Katan, J. (2009) Suppression of bacterial canker of tomato by composts. Crop Prot. 28, 97–103. [Google Scholar]
- Yuan, H‐P. , Min, H. , Lv, Z‐M. and Li, Z‐M. (2009) Antimicrobial activity of isolate HL‐12 against Clavibacter michiganensis subsp. michiganensis in the presence of cadmium. Ecotoxicology, 18, 447–454. [DOI] [PubMed] [Google Scholar]
- Zipfel, C. and Felix, G. (2005) Plants and animals: a different taste for microbes? Curr. Opin. Plant Biol. 8, 353–360. [DOI] [PubMed] [Google Scholar]
- Zou, H.S. , Song, X. , Zou, L.F. , Yuan, L. , Li, Y.R. , Guo, W. , Che, Y.Z. , Zhao, W.X. , Duan, Y.P. and Chen, G.Y. (2012) EcpA, an extracellular protease, is a specific virulence factor required by Xanthomonas oryzae pv. oryzicola but not by X. oryzae pv. oryzae in rice. Microbiology, 158, 2372–2383. [DOI] [PubMed] [Google Scholar]


