Summary
Pathogen–host interaction is a complicated process; pathogens mainly infect host plants to acquire nutrients, especially sugars. Rhizoctonia solani, the causative agent of sheath blight disease, is a major pathogen of rice. However, it is not known how this pathogen obtains sugar from rice plants. In this study, we found that the rice sugar transporter OsSWEET11 is involved in the pathogenesis of sheath blight disease. Quantitative real‐time polymerase chain reaction (qRT‐PCR) and β‐d‐glucuronidase expression analyses showed that R. solani infection significantly enhanced OsSWEET11 expression in leaves amongst the clade III SWEET members. The analyses of transgenic plants revealed that Ossweet11 mutants were less susceptible, whereas plants overexpressing OsSWEET11 were more susceptible, to sheath blight compared with wild‐type controls, but the yield of OsSWEET11 mutants and overexpressors was reduced. SWEETs become active on oligomerization. Split‐ubiquitin yeast two‐hybrid, bimolecular fluorescence complementation and co‐immunoprecipitation assays showed that mutated OsSWEET11 interacted with normal OsSWEET11. In addition, expression of conserved residue mutated AtSWEET1 inhibited normal AtSWEET1 activity. To analyse whether inhibition of OsSWEET11 function in mesophyll cells is related to defence against this disease, mutated OsSWEET11 was expressed under the control of the Rubisco promoter, which is specific for green tissues. The resistance of transgenic plants to sheath blight disease, but not other disease, was improved, whereas yield production was not obviously affected. Overall, these results suggest that R. solani might acquire sugar from rice leaves by the activation of OsSWEET11 expression. The plants can be protected from infection by manipulation of the expression of OsSWEET11 without affecting the crop yield.
Keywords: mesophyll cell, OsSWEET11, resistance, rice, sheath blight disease
Introduction
Sheath blight, which is one of the three major diseases of rice, is caused by infection with Rhizoctonia solani AGI‐1A and is responsible for severe yield losses (Slaton et al., 2003). This disease affects rice throughout its life cycle, from the seedling to heading stage, and causes lesions on leaves, sheaths and even panicles. It causes withering of leaves and sheaths, and reduction in the rate of seed set. At later stages of infection, the entire plant may wither and lodge. Sheath blight can reduce the yield of rice from 8% to 50%, depending on the severity of the disease, stage of the crop at which it was infected by the fungus and overall environmental conditions (Savary et al., 2000).
The use of fungicides remains the mainstay of strategies for the control of sheath blight (Rajesh et al., 2016). However, extensive use of chemicals poses health risks, causes financial strain on farmers and is damaging to the environment. Thus, research aimed at the identification of pathogen‐resistant rice and deciphering of the mechanisms underlying pathogen–plant interaction is urgently needed. Previous studies have shown that the overexpression of chitinase and β‐1,3‐glucanase in rice enhances the resistance to different anastomosis groups of R. solani (Mao et al., 2014; Sripriya et al., 2017). However, the overexpression of OsACS2, which encodes an enzyme involved in ethylene biosynthesis, increases the endogenous production of ethylene and enhances the resistance to rice sheath blight and rice blast (Helliwell et al., 2013). In addition, the overexpression of OsPGIP1, which codes for polygalacturonase‐inhibiting protein, significantly improves the resistance to sheath blight in rice (Wang et al., 2015). Recent evidence has suggested that transgenic lines with inducible production of ethylene exhibit resistance to R. solani, without any yield penalty (Helliwell et al., 2013). The overexpression of OsOSM1, which encodes a protein belonging to the pathogenesis‐related protein 5 family, has been shown to enhance resistance to sheath blight in rice in field trials (Xue et al., 2016). Pyramided transgenic plant lines which co‐express both the genes (OsCHI11 and AtNPR1) were found to have more improved performance against sheath blight tolerance than single gene transformants. (Karmakar et al., 2017). However, to date, no gene that confers significant resistance has been identified in rice.
The process through which R. solani obtains nutrients, such as sugar required for its proliferation, from plants remains unknown. The elucidation of the process for the acquisition of nutrients by a pathogen is often helpful to devise effective strategies for the generation of pathogen‐resistant plants. The recently identified SWEETs constitute a family of sugar transporters (Chen et al., 2010). On average, 20 SWEET genes have been identified to be present in many higher plant species (Chen LQ et al., 2015). In terms of fungi, during Botrytis cinerea infection, AtSWEET4, AtSWEET15 and AtSWEET17 are induced (Ferrari et al., 2007). Golovinomyces cichoracearum infection induces several AtSWEETs, most prominently AtSWEET12 (Chen et al., 2010). OsSWEET11/Os8N3 is involved in the bacterial blight disease response in rice (Yang et al., 2006). However, the role of OsSWEET11 in other types of pathogen infections has not been reported. Recent studies have shown that SWEETs are responsible for susceptibility to disease in many plant species; they act as the targets of effector proteins secreted by pathogens during host–microbe interactions. For example, Xanthomonas oryzae pv. oryzae (Xoo) strain PXO99A can produce the transcription activator‐like (TAL) effector, PthXo1, which binds directly to the OsSWEET11 promoter (Chen et al., 2010; Yang et al., 2006). The TAL effectors are delivered to the cytoplasm of plant cells through the type III secretion system and enter the nucleus to induce the expression of specific SWEET genes, ensuring the delivery of sucrose to the apoplasts of the colonized cells (Chen et al., 2010). For example, OsSWEET11/Xa13, OsSWEET13/Xa25 and OsSWEET14 have been identified as targets of Xoo effectors in rice (Antony et al., 2010; Hutin et al., 2015; Liu et al., 2011; Yang et al., 2006). In cotton, the expression of a sucrose transporter, GhSWEET10, can be activated by Avrb6, a TAL effector of X. citri ssp. malvacearum, during its invasion (Cox et al., 2017).
Despite the progress in the elucidation of plant–pathogen interactions, two critical issues regarding the nutrient acquisition process of pathogens need to be addressed to modify pathogen resistance in rice. The first issue that needs clarification is how R. solani acquires nutrients, particularly sugar, from infected plants to support its propagation. The second issue that needs to be resolved is how the balance between the expense on pathogen resistance and that on plant growth and development is maintained.
The phylogenetic analysis of plant SWEET genes has revealed that they can be grouped into four clades, as first defined in Arabidopsis (Chen et al., 2010). SWEETs grouped into clades I and II function in the transport of hexoses, whereas those included in clades III and IV are sucrose and fructose transporters, respectively (Chen LQ et al., 2010, 2015). Previous studies have shown that SWEETs play diverse functions in plant development and production, and in the senescence of leaves (Quirino et al., 1999), sugar loading in phloem (Chen et al., 2012), nectar production (Lin et al., 2014), pollen viability (Yang et al., 2006) and grain filling (Ma et al., 2017; Sosso et al., 2015). All of these processes are important for the improvement of the rice yield. Thus, the manipulation of nutrient partitioning in specific tissues is a critical factor that needs to be considered to generate pathogen‐resistant rice. However, the role of SWEETs in rice sheath blight has not yet been identified.
In this study, we analysed the R. solani AG1‐1A‐mediated induction of OsSWEET11 (which belongs to clade III) and investigated the resistance of the OsSWEET11 mutant and overexpression rice lines to sheath blight. We found that OsSWEET11 has a negative effect on sheath blight resistance. Interestingly, similar to proton–sucrose symporters, which tend to form dimers for normal functioning (Krügel et al., 2008, 2013), SWEETs have been reported to be activated on oligomerization (Xuan et al., 2013). To avoid interference with the essential role of OsSWEET11 during grain filling in rice (Ma et al., 2017), we used the Rubisco promoter to express the mutated form of OsSWEET11 (mSWEET11) which can inhibit OsSWEET11 sugar transport activity by forming a trimer with wild‐type OsSWEET11 to inhibit OsSWEET11‐mediated sugar transport in leaf cells during pathogen infection. The results showed that the inhibition of OsSWEET11 in leaf cells improved the resistance of transgenic rice to sheath blight without affecting the yield.
Results
Rhizoctonia solani AG1‐1A infection induces the expression of OsSWEET11
OsSWEET11 and OsSWEET14 are targets of the type III secretion effector in bacterial blight disease; however, whether OsSWEETs are involved in sheath blight disease is not known. To analyse this question, clade III OsSWEET gene expression was analysed. The expression levels of OsSWEET12–OsSWEET15 were not altered by infection (Fig. S1, see Supporting Information). However, the expression of OsSWEET11 in the leaves and sheaths of mock (medium)‐ and R. solani AG1‐1A‐infected 1‐month‐old plants was induced by R. solani AG1‐1A after 48 h, but not by mock treatment (Fig. 1A). After 72 h of infection, OsSWEET11 was induced five‐ and three‐fold in the leaves and sheaths, respectively (Fig. 1A). The transcriptional activation of OsSWEET11 by R. solani AG1‐1A infection was further verified using transgenic plants expressing the pOsSWEET11::GUS construct (Fig. 1B) (Ma et al., 2017). In the mock treatment, the expression of β‐d‐glucuronidase (GUS) was barely detectable in both leaves and sheaths. However, higher GUS staining was observed in the leaves and sheaths after 48 h of R. solani AG1‐1A infection (Fig. 1C). These results indicate that R. solani AG1‐1A infection induces the expression of OsSWEET11, but not OsSWEET12–OsSWEET15.
Figure 1.

Expression patterns of OsSWEET11 induced by Rhizoctonia solani AG1‐1A infection. (A) Expression levels of OsSWEET11 were analysed after 0, 24, 48 and 72 h of R. solani infection. Solid potato dextrose agar (PDA) medium was used as a control (Mock), and R. solani AG1‐1A cultured on the surface of PDA was used for the infection of rice leaves and sheaths. Ten leaves from each time point were sampled for RNA extraction. The experiments were repeated three times, and the data represent the means ± standard error (SE) (n = 3). Significant differences at P < 0.05 are indicated by different letters. (B) Schematic representation of the plasmid construct used for β‐d‐glucuronidase (GUS) expression in the transgenic plants, in which the OsSWEET11 promoter drives GUS expression. (C) Analysis of GUS expression in leaf and sheath after 48 h of mock and AG1‐1A infection.
Expression of OsSWEET11 is positively correlated with the lesion caused by sheath blight
Because OsSWEET11 expression was induced by R. solani AG1‐1A, the causative agent of sheath blight disease, we performed genetic studies on plants in which OsSWEET11 was knocked out or overexpressed. The response of two OsSWEET11 knock‐out mutants (Ossweet11‐1 and Ossweet11‐2) and two overexpression (OX) lines (OX2 and OX4) to R. solani AG1‐1A infection was assessed. Before testing the R. solani response, the OsSWEET11 expression level was examined in four independent OsSWEET11 OX lines. The data showed that OsSWEET11 was more highly expressed in OX lines than in wild‐type plants (Fig. S2, see Supporting Information). The fungal pathogen was inoculated on detached leaves, and the lesion area was evaluated after 72 h (Fig. 2A). After 72 h, the lesion area with respect to the total leaf area was compared amongst the wild‐type, knock‐out mutants and OX lines (Fig. 2B); the lesion areas were around 40%, 30% and 60%, respectively (Fig. 2B), indicating that the Ossweet11 mutants were less susceptible, whereas the OX lines were more susceptible, to R. solani AG1‐1A compared with the wild‐type plants.
Figure 2.
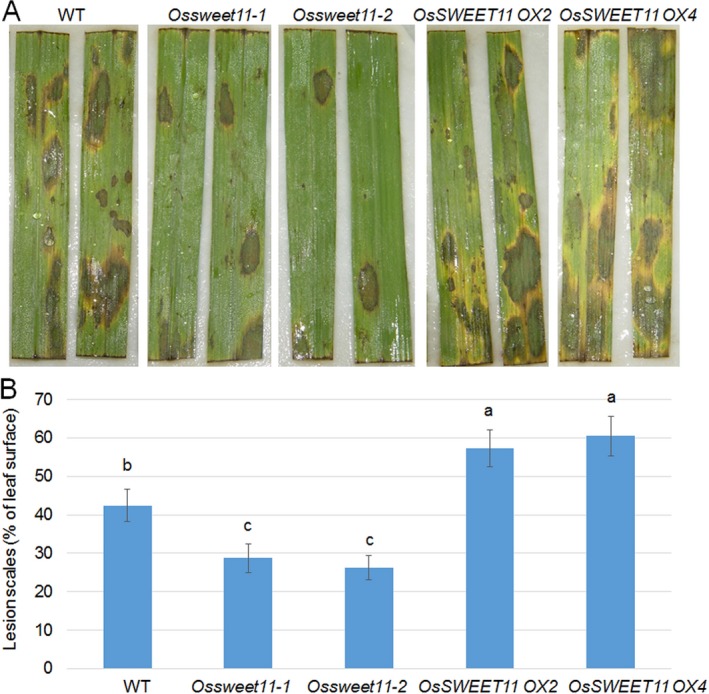
Response of OsSWEET11 mutants and rice overexpression lines to Rhizoctonia solani AG1‐1A. (A) Leaves from wild‐type (WT) plants, OsSWEET11 mutants (Ossweet11‐1 and Ossweet11‐2) and OsSWEET11 rice overexpression lines (OsSWEET11 OX2 and OsSWEET11 OX4) were inoculated with R. solani AG1‐1A and photographed after infection. Six leaves from each line were analysed, and the experiments were repeated three times. (B) The lesion scales were analysed for R. solani AG1‐1A‐infected leaves by determining the lesion area on the leaf surface. Data represent the means ± standard error (SE) (n > 10). Significant differences at P < 0.05 are indicated by different letters.
The OsSWEET11‐silenced lines were insensitive to bacterial blight caused by PXO99A (Yang et al., 2006). In addition, the OsSWEET11 mutants were insensitive, whereas the OsSWEET11 OX plants were sensitive, to sheath blight disease. Therefore, the response of OsSWEET11 OX plants to PXO99A infection was analysed. The leaves of 2‐month‐old wild‐type and OsSWEET11 OX plants (OX2 and OX4) were inoculated with PXO99A; the lesion length in wild‐type plants was around 11 cm, whereas, in OX2 and OX4, it was around 17 and 15 cm, respectively (Fig. S3, see Supporting Information), indicating that OsSWEET11 OX plants were more susceptible to bacterial blight disease caused by the PXO99A strain.
Mutation at conserved residues inhibits the activity of AtSWEET1
Although OsSWEET11 knock‐out mutants showed reduced sheath blight disease index, grain filling and seed set were severely affected (Fig. S4, see Supporting Information) (Ma et al., 2017). In a previous study, the mutation of conserved residues, Y57 and G58, was found to inhibit the glucose transport activity of AtSWEET1, and co‐expression of wild‐type and mutated AtSWEET1 significantly reduced the glucose transport activity of AtSWEET1 in the EBY4000 yeast strain, which indicates that the oligomerization of SWEET is necessary for its function (Xuan et al., 2013). Therefore, mSWEET11 was expressed using a cell type‐specific promoter to inhibit OsSWEET11‐mediated sugar efflux in the cells, and thus to protect plants from sheath blight disease whilst maintaining normal grain filling. Mutated SWEET11 was generated by collecting and aligning the sequences of AtSWEET1, OsSWEET2b, OsSWEET11 and OsSWEET11 homologues from different model plants (Fig. 3A). The results showed that OsSWEET11 and its homologues contained a relatively longer cytosolic tail compared with that in AtSWEET1 and OsSWEET2b, and many residues were highly conserved amongst these SWEET proteins (Fig. 3A). The SUSY7 yeast strain is able to analyse sucrose transport activity (Lalonde et al., 2003), but OsSWEET11 did not show activity in SUSY7 (Fig. S5, see Supporting Information). Therefore, AtSWEET1 was used to analyse conserved residue function. Five residues (P23, G76, P162, P191 and Q202) of AtSWEET1 with conserved residues (P31, G82, P169, P198 and Q209) in OsSWEET11 were selected for mutation. Empty vector, AtSWEET1 and mutated AtSWEET1 were expressed in the EBY4000 yeast strain deficient in hexose transport, and 2% glucose and maltose were used as carbon sources for the glucose transport activity test of AtSWEET1. The expression of AtSWEET1 and mutated AtSWEET1 in yeast EBY4000 revealed that AtSWEET1 successfully complemented the growth of yeast cells, whereas the five mutated AtSWEET1 proteins failed to complement the growth in glucose‐containing medium (Fig. 3B).
Figure 3.
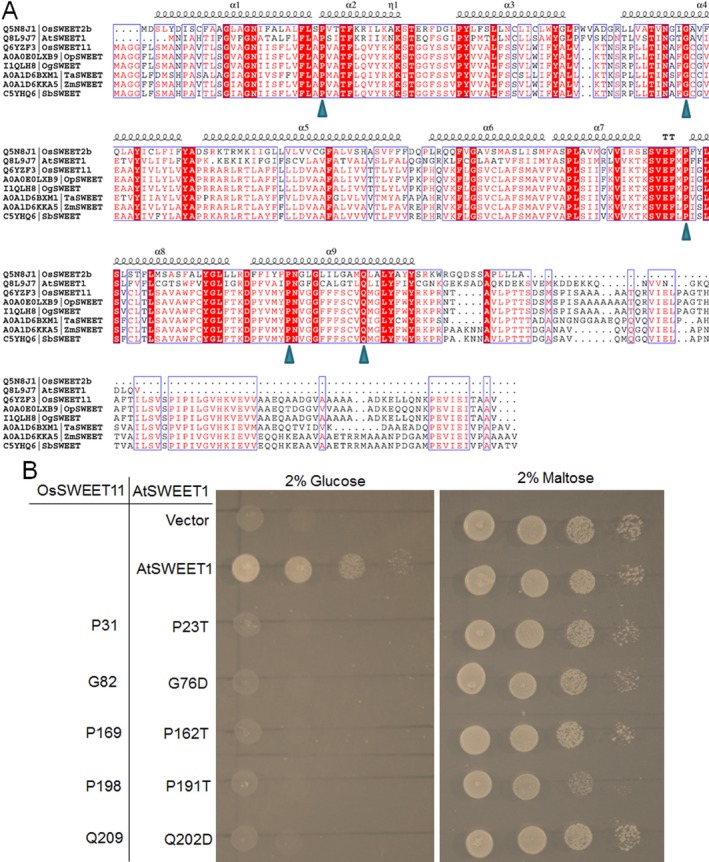
Alignment of amino acid sequences of the selected SWEETs and functional analysis of the conserved residues in glucose transport. (A) Sequence alignment of OsSWEET2b, AtSWEET1a, OsSWEET11, OpSWEET, OgSWEET, TaSWEET, ZmSWEET and SbSWEET. The conserved residues are shown in red boxes. The triangle indicates the residues tested for glucose transport function. (B) Growth assays of mutant AtSWEET1 proteins expressed in EBY4000 yeast strain were performed on YNB (Yeast Nitrogen Base without Amino Acids) medium containing 2% glucose or maltose. AtSWEET1 mutants carrying P23A, G76D, P162A, P191T and Q202D led to the loss of glucose transport activity. Empty (pDRf1) vector and AtSWEET1 were used as negative and positive controls, respectively. The positions of the conserved residues in OsSWEET11 are shown in the left panel. The yeast cells were grown at 28 °C for 3 days.
Mutated OsSWEET11 interacts with wild‐type OsSWEET11
Mutated OsSWEET11 (mSWEET11) was constructed by mutating three conserved residues (P31, G82 and P169) amongst the five described above. Whether there was any interaction between mSWEET11 and OsSWEET11 was determined by performing a mating‐based split‐ubiquitin assay. OsSWEET11 and mSWEET11 were cloned into Nub vector pXN25_GW, and OsSWEET11 was cloned into Cub vector pMETYC_GW; NubWT and NubG were used as positive and negative controls, respectively. The yeast growth assay showed that OsSWEET11 interacted with itself or mSWEET11 (Fig. 4A). Furthermore, a split green fluorescent protein (GFP) system was used to confirm their interaction in tobacco leaves. The open reading frames (ORFs) of OsSWEET11 and mSWEET11 were cloned into the pXNGW vector, in which the N‐terminal half of the yellow fluorescent protein (YFP) sequence was C‐terminally fused to OsSWEET11, whereas the ORF of mSWEET11 was cloned into the pXCGW vector, in which the C‐terminal half of the cyan fluorescent protein (CFP) sequence was C‐terminally fused to mSWEET11. The interaction of the fusion proteins transiently co‐expressed in Nicotiana benthamiana was checked by observing fluorescence using a confocal microscope. The fusion proteins were found to be localized to the plasma membrane (Fig. 4B). The interaction between OsSWEET11 and mSWEET11 was confirmed by performing a co‐immunoprecipitation (Co‐IP) assay in a tobacco transient expression system. OsSWEET11‐Myc alone or OsSWEET11‐Myc together with mSWEET11‐GFP was expressed in N. benthamiana leaves, and the expression of OsSWEET11‐Myc and mSWEET11‐GFP was detected using Western blot analysis with anti‐Myc or anti‐GFP antibodies. Before IP, OsSWEET11‐Myc and mSWEET11‐GFP were successfully expressed in tobacco leaves, and anti‐Myc antibody was used for IP. After IP, the anti‐GFP antibody was used to detect the presence of mSWEET11‐GFP (Fig. 4C). The results indicated that OsSWEET11‐Myc interacted with mSWEET11‐GFP. Next, we inferred whether mSWEET11 can affect OsSWEET11 by co‐expression of mutated AtSWEET1 (mAtSWEET1; mutation at P23, G76 and P162 which correspond to the residues in mSWEET11) and AtSWEET1 in the EBY4000 yeast strain. AtSWEET1 was driven by a strong ADH promoter, whereas mAtSWEET1 was driven by a strong PMA1 promoter. The results indicated that OsSWEET11 interacts with mSWEET11, and mAtSWEET1 significantly inhibits the glucose transport activity of AtSWEET1 (Fig. 4D).
Figure 4.
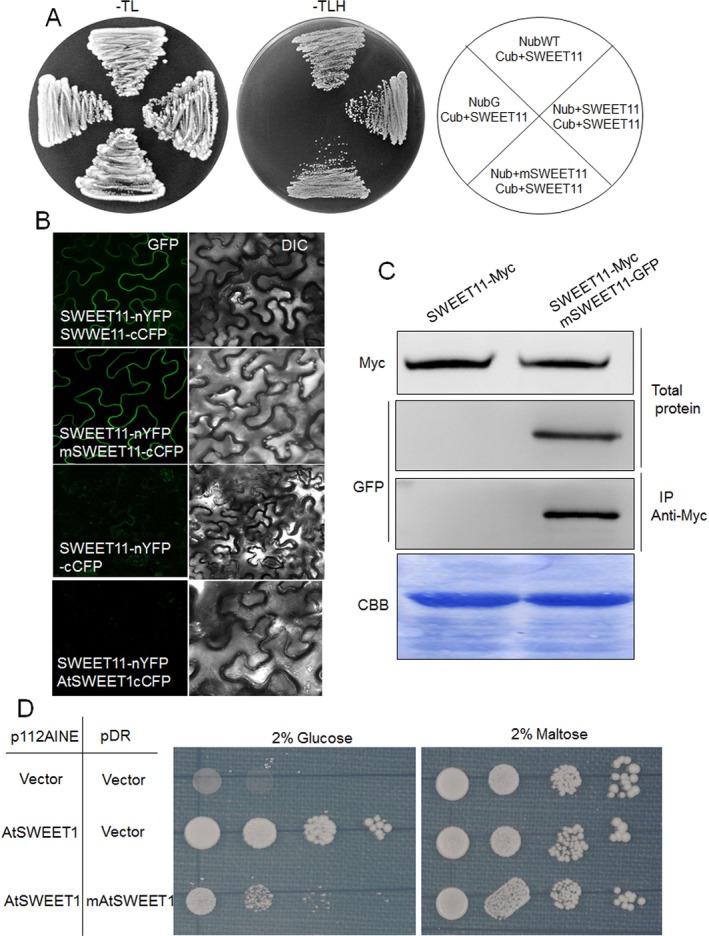
Assessment of the interaction between wild‐type (WT) and mutated OsSWEET11 and inhibition of glucose transport by co‐expression of WT and mutant AtSWEET1 proteins in yeast EBY4000. (A) Interaction between OsSWEET11 and mOsSWEET11 was analysed using a split‐ubiquitin yeast two‐hybrid system. mSWEET11 was cloned into the Nub vector, whereas OsSWEET11 was cloned into the Cub (C‐terminal ubiquitin domain driven by methionine‐repressible MET25 promoter and fused to the artificial PLV transcription factor) or Nub vector. NubWT (Nub) and NubG (N‐terminal ubiquitin domain carrying a glycine mutation) were used as positive and negative controls, respectively. (B) Observation of the yellow fluorescent protein (YFP) fluorescence. Reconstitution of the YFP fluorescence from OsSWEET11‐nYFP + OsSWEET11‐cCFP or OsSWEET11‐nYFP + mOsSWEET11‐cCFP (left, fluorescence channel; right, bright field). Co‐expression of OsSWEET11‐nYFP + cCFP or OsSWEET11‐nYFP + AtSWEET1‐cCFP was used as negative control. Bars, 20 μm. (C) Co‐immunoprecipitation (Co‐IP) was performed to analyse the interaction between OsSWEET11 and mOsSWEET11 in tobacco leaves. OsSWEET11‐Myc or OsSWEET11‐Myc + mOsSWEET11‐GFP was transformed into tobacco leaves using Agrobacterium‐mediated transformation. The total protein and Myc antibody‐immunoprecipitated proteins were analysed using Western blot analysis with either Myc or green fluorescent protein (GFP) antibodies. CBB (Coomassie Brilliant Blue) staining was used as a loading control. (D) The p112AINE empty vector or p112AINE‐SWEET1 construct was co‐transformed with pDRf1 empty vector or pDR‐mAtSWEET1 construct into the yeast strain, EBY4000. Growth assays of yeast cells co‐expressing WT and mutant AtSWEET1 protein, mAtSWEET1, were performed on YNB (Yeast Nitrogen Base without Amino Acids) medium containing 2% glucose or maltose. The yeast cells were grown at 28 °C for 3 days.
Expression of mSWEET11, driven by the Rubisco promoter, in rice mitigates the lesions of sheath blight disease
The photosynthetic production of sugar occurs in mesophyll cells. Rubisco is the major protein expressed during the photosynthesis process; therefore, a 2.0‐kb fragment of the Rubisco small subunit promoter was used to express mSWEET11 (Fig. 5A). More than 10 individual transgenic plants were produced. The tissue‐specific expression of OsSWEET11 was analysed in the roots, leaves, sheaths and flowers of wild‐type and three independent pRubisco‐mSWEET11 lines (#1, #2 and #6). The results showed that OsSWEET11 was highly expressed in the leaves and sheaths of the three pRubisco‐mSWEET11 lines, but exhibited similar levels in the roots and flowers of wild‐type and pRubisco‐mSWEET11 lines (Fig. 5B). The expression of OsSWEET11 was around 20–23‐fold higher in the leaves, whereas it was around six‐fold higher in the sheaths of pRubisco‐mSWEET11 lines compared with the expression in the corresponding tissues of the wild‐type plants (Fig. 5B). We also analysed the response of pRubisco‐mSWEET11 lines to R. solani AG1‐1A infection. After 72 h of infection, the manifestations of the infection on leaves were photographed (Fig. 5C). The results indicated that the lesion area was around 42% in wild‐type plants, whereas it was around 25.5–29% in the leaves of pRubisco‐mSWEET11 plants (Fig. 5D). These results indicated that mSWEET11 was highly expressed in the leaves and sheaths of pRubisco‐mSWEET11 transgenic plants, and that the transgenic plants were less susceptible to R. solani AG1‐1A infection.
Figure 5.
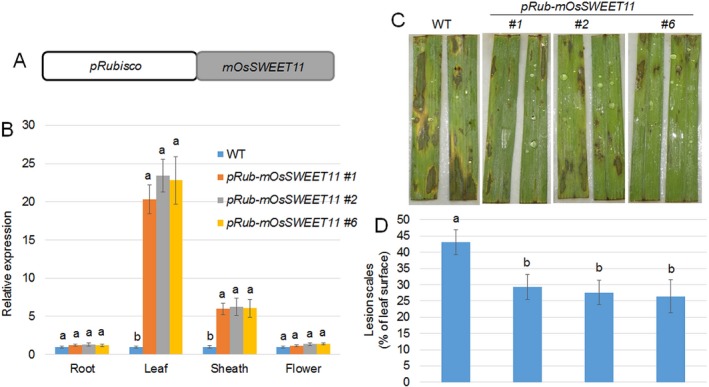
Expression levels of OsSWEET11 and disease symptoms in the transgenic plants expressing mOsSWEET11 under the control of the Rubisco promoter. (A) Schematic diagram showing the construct used for the generation of transgenic plants. The Rubisco promoter was used to drive mOsSWEET11, a mutated form of OsSWEET11. (B) The expression levels of OsSWEET11 were analysed in the roots, leaves, sheaths and flowers of wild‐type and three independent transgenic plants (pRub‐mOsSWEET11 #1, #2 and #6) by quantitative real‐time polymerase chain reaction (qRT‐PCR). The experiments were repeated three times. Significant differences at P < 0.05 are indicated by different letters. (C) The leaves of wild‐type and three independent transgenic plants (pRub‐mOsSWEET11 #1, #2 and #6) were infected with Rhizoctonia solani AG1‐1A and photographed after 3 days of infection. Six leaves from each line were analysed, and the experiments were repeated three times. (D) The lesion scales were analysed for the R. solani AG1‐1A‐infected leaves shown in (C) by determination of the lesion area on the leaf surface. Data represent means ± standard error (SE) (n > 10). Significant differences at P < 0.05 are indicated by different letters.
Furthermore, the response of pRubisco‐mSWEET11 plants to PXO99A‐mediated bacterial blight disease was assessed. The leaves of 2‐month‐old wild‐type and pRubisco‐mSWEET11 plants (#1 and #2) were inoculated with PXO99A; the lesion lengths in the wild‐type plants were around 11 cm, whereas those in the #1 and #2 pRubisco‐mSWEET11 plants were around 16 and 17 cm, respectively (Fig. S6, see Supporting Information), indicating that pRubisco‐mSWEET11 plants were susceptible to bacterial blight disease caused by strain PXO99A. In addition, Magnaporthe oryzae Guy11 was inoculated into pRubisco‐mSWEET11 plants. The pRubisco‐mSWEET11 plants exhibited wild‐type‐like response to M. oryzae (Fig. S7, see Supporting Information).
Transgenic rice plants expressing mSWEET11 under the control of the Rubisco promoter maintain normal yield
The Ossweet11 mutants were resistant to R. solani AG1‐1A (Fig. 2), but the grain filling of mutant plants was affected (Fig. S4) (Ma et al., 2017). In the pRubisco‐mSWEET11 transgenic plants, the sugar efflux activity of OsSWEET11 might be specifically inhibited in the mesophyll cells, and thus provide resistance to R. solani AG1‐1A when challenged by infection. Therefore, the plant morphology was analysed. The OX lines exhibited a severe dwarf phenotype, whereas the pRubisco‐mSWEET11 plants were similar to the wild‐type plants (Fig. 6A). Furthermore, we compared the 1000‐grain weight of wild‐type, OsSWEET11 mutant, OX and pRubisco‐mSWEET11 plants. The results showed that OsSWEET11 mutants had the lowest grain weight, and this value was higher for the OX lines than for the mutants, but was lower than that of the wild‐type and pRubisco‐mSWEET11 plants. The pRubisco‐mSWEET11 plants developed normal grains, which were similar to those of the wild‐type plants (Fig. 6B). In addition, the seed size, tiller number and grain number of pRubisco‐mSWEET11 plants were similar to those of the wild‐type plants (Fig. 6C–F).
Figure 6.
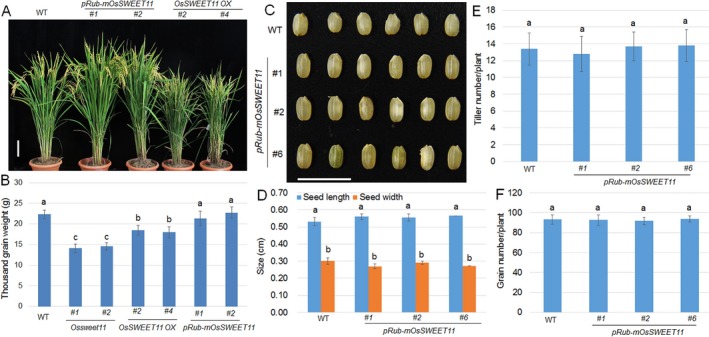
Morphology and grain weight of wild‐type plants, OsSWEET11 mutants, overexpressors and pRub‐mOsSWEET11 transgenic plants. (A) Four‐month‐old wild‐type plants, OsSWEET11 overexpressors and pRub‐mOsSWEET11 transgenic plants were photographed. Scale bar, 10 cm. (B) The 1000‐grain weight was analysed using the seeds from the different plants. (C) Seeds harvested from wild‐type and pRub‐mOsSWEET11 transgenic plants were photographed. Scale bar, 1 cm. (D) The seed length and width in wild‐type and pRub‐mOsSWEET11 transgenic plants were analysed. More than 100 seeds were analysed. (E) The tiller number in wild‐type and pRub‐mOsSWEET11 transgenic plants was analysed. More than 20 plants were analysed. (F) The grain number in wild‐type and pRub‐mOsSWEET11 transgenic plants was analysed. More than 20 plants were analysed. Significant differences at P < 0.05 are indicated by different letters.
Discussion
Rice sheath blight, bacterial blight and blast disease are the three major diseases of rice, which severely affect yields. The defence mechanism of rice against R. solani is not yet known. OsSWEET11 is known to be a target of the type III effector secreted from Xoo PXO99A. It can be highly induced on PXO99A infection and has been shown to be negatively related to resistance to bacterial blight (Yang et al., 2006). In this study, we found that OsSWEET11 was induced in rice infected with R. solani AG1‐1A, a major causative agent of rice sheath blight disease. However, other clade III OsSWEET genes (OsSWEET12–OsSWEET15) were not altered by infection with R. solani AG1‐1A. The transcript levels of the gene and pOsSWEET11::GUS analysis consistently revealed that R. solani AG1‐1A infection significantly induced the expression of OsSWEET11 in the leaves and sheaths of rice, suggesting a specific role of OsSWEET11 in response to sheath blight disease. Based on these results, we conducted further genetic studies using the CRISPR‐Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR‐associated)‐based knock‐out mutants and transgenic plants in which the ubiquitin promoter was used to drive the overexpression of OsSWEET11. In the OsSWEET11 OX lines, the transcript levels of OsSWEET11 were considerably higher than those in the wild‐type control. The pathogen infection experiments showed that the OsSWEET11 knock‐out mutants were less susceptible, whereas the OsSWEET11 overexpressing plants were more susceptible, to sheath blight disease compared with the wild‐type plants, suggesting that OsSWEET11 is a gene responsible for the susceptibility of rice to sheath blight disease.
Ossweet11 mutants were resistant to both sheath blight and bacterial blight disease, and thus OsSWEET11 may be a potential target for the breeding of resistant rice. However, the OsSWEET11 mutants exhibited severe defects in grain filling and seed quality (Ma et al., 2017). Previously, TAL effectors of Xoo have been used to generate TALENs for editing the OsSWEET14/Os11N3 promoter region, to which the type III effector binds. The modified plants showed remarkable resistance to bacterial blight disease without causing any change in the normal expression of OsSWEET14 (Li et al., 2012). In a previous study, we identified that the co‐expression of AtSWEET1 with the non‐functional mutant form of AtSWEET1 significantly inhibited the glucose transport activity in the yeast EBY4000 strain (Xuan et al., 2013). OsSWEET11 can be induced by R. solani AG1‐1A. Therefore, we speculated that the specific expression of the mutated form of OsSWEET11 in certain tissues or cell types could inhibit sucrose transport in the target cells in which R. solani AG1‐1A hijacks sugar, and without affecting the grain filling process. Before testing this, we selected conserved residues (P23A, G76D, P162A, P191T and Q202D) in AtSWEET1, OsSWEET2b, OsSWEET11 and their homologues from other plants via sequence alignment and evaluated their importance in the glucose transport activity of AtSWEET1, because OsSWEET11 failed to transport sucrose in the SUSY7 yeast strain (Fig. S5). It might be that OsSWEET11 does not localize at the plasma membrane or that OsSWEET11 sucrose transport affinity is much lower than that of AtSUC2. The results showed that the mutation of any one of the five residues abolished the glucose transport activity of AtSWEET1 in the EBY4000 yeast strain. Furthermore, transgenic rice plants expressing mutated OsSWEET11 were generated, with mutations at three positions, P31, G82 and P169; these are conserved positions corresponding to P23, G76D and P162A of AtSWEET1, respectively. Further, split‐ubiquitin‐based yeast two‐hybrid, split GFP and Co‐IP assays showed that OsSWEET11 interacts with mSWEET11 in yeast and plants, and their interaction was observed in the plasma membrane of plant cells. In addition, the co‐expression of AtSWEET1 with mutated AtSWEET1 (with mutations at P23, G76D and P162) significantly inhibited the glucose transport activity of AtSWEET1. Our results suggest that the expression of mSWEET11 could very likely inhibit the normal function of OsSWEET11. It would be interesting to perform further experiments to test the activity of OsSWEET11 sucrose transport activity, as well as the inhibition of OsSWEET11, by the co‐expression of mSWEET11 in other possible systems.
Mesophyll cells are known to be the main sites of photosynthesis, which leads to the production of sugar. The Rubisco protein is mainly expressed in mesophyll cells. Therefore, the Rubisco small subunit promoter was used to express mSWEET11 to determine whether the inhibition of OsSWEET11 function in mesophyll cells can improve the resistance of rice to sheath blight disease, because leaves of rice are vulnerable to this pathogen. Unlike in wild‐type plants, the expression level of OsSWEET11 was higher in the leaves and sheaths, but was similar in the roots and flowers, in pRub‐mSWEET11 plants. In addition, pRub‐mSWEET11 plants exhibited resistance to sheath blight disease, suggesting that the sugar efflux activity of intrinsic OsSWEET11, induced by R. solani AG1‐1A, was counteracted by mSWEET11 expressed in the mesophyll. However, the grain weight of pRub‐mSWEET11 plants was similar to that of wild‐type plants, whereas it was affected in all the OsSWEET11 mutants and overexpressors (Fig. 6). However, whether OsSWEET11 induced in mesophyll cells by R. solani AG1‐1A or Rubisco promoter activity is restricted to mesophyll cells needs to be evaluated further. pRub‐mSWEET11 plants exhibited susceptibility to bacterial blight disease caused by Xoo PXO99A, suggesting that mSWEET11 expression does not interfere with Xoo susceptibility. PXO99A strictly relies on the transactivation of OsSWEET11 by its major TALE PthXo1. PXO99A does not transactivate other SWEETs which could complement the loss of OsSWEET11 transactivation (Yang et al., 2006). It could also be induced in the vasculature in which X. oryzae is prevalent, or mSWEET11 expression under the Rubisco promoter may activate other SWEETs, which may efflux sugar to X. oryzae. Nevertheless, atsweet2 mutants were more susceptible to Pythium infection (Chen HY et al., 2015), suggesting a diverse role of SWEETs in the disease response. Expressing of mSWEET11 under control of the vasculature specific promoter may provide resistant lines against PXO99A. pRub‐mSWEET11 plants exhibited a similar response to M. oryzea Guy11, suggesting that this mutation is not associated with rice blast disease. Not surprisingly, the plants overexpressing OsSWEET11 were susceptible to PXO99A. To determine whether OsSWEET11 expression is tightly connected with defence, the PBZ1 level was analysed in the OsSWEET11 mutant, overexpressors and pRubisco‐mSWEET11 plants. The data showed that the expression level of PBZ1 was similar in these lines, indicating that mutation and higher expression of OsSWEET11 did not alter plant defence in non‐inoculated plants (Fig. S8, see Supporting Information).
Rhizoctonia solani AG1‐1A activates OsSWEET11 via an unknown mechanism to efflux sugar from the cytosol to the apoplast for its own use. This is a typical strategy of pathogens in microbe–plant interactions. Given the importance of sugar acquisition from the host plant to the pathogen, microbes other than R. solani AG1‐1A might also take advantage of the sugar efflux transport activity of OsSWEET11, or other SWEETs in rice. Therefore, SWEETs, as sugar efflux transporters, play critical roles during microbe–plant interactions. In this study, the expression of OsSWEET11 was activated by Rhizoctonia solani to efflux sucrose to the apoplasm, which may be further hydrolysed to glucose via cell wall invertase, and taken up by R. solani (Fig. 7). More importantly, seed set and grain filling in rice were not significantly affected by this inhibition, and the SWEET11 level was similar in wild‐type and pRubisco‐mSWEET11 plants in flower tissue, indicating that the Rubisco promoter is not active in flower tissue. Taken together, our findings suggest that the sugar transporter, OsSWEET11, plays an important role in the infection of sheath blight disease, and that manipulation of the expression of susceptible genes may be an efficient way to protect plants from the attack of pathogens without affecting their yield.
Figure 7.
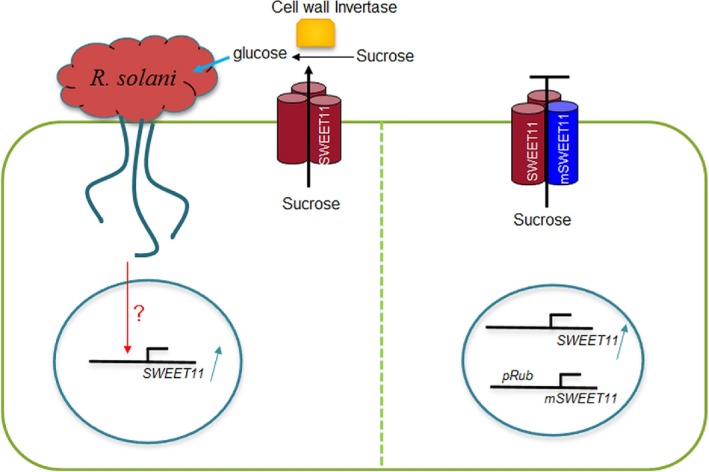
Schematic representation of the hypothesized function of OsSWEET11 during Rhizoctonia solani infection. Rhizoctonia solani infection activates the expression of OsSWEET11 via an unknown mechanism to efflux sucrose to the apoplasm, and cell wall invertase catabolizes sucrose to glucose and fructose. Further, glucose can be taken up into R. solani cells. The expression of mutated OsSWEET11 via the Rubisco promoter may generate a non‐functional OsSWEET11 complex. The wild‐type (WT) OsSWEET11 induced by R. solani may form heterotrimers consisting of WT OsSWEET11 and mutated OsSWEET11, which may be expressed under the Rubisco promoter, and the non‐functional OsSWEET11 complex may inhibit sugar efflux.
Experimental Procedures
Plant growth and pathogen inoculation
Rice plants were grown under glasshouse conditions at Shenyang Agricultural University, Shenyang, China. The daily high and low temperatures of the glasshouse were typically 30 and 23 °C, respectively. The plants were propagated by selfing.
For inoculation of the pathogen R. solani AGI‐1A, rice plants (Nipponbare) were grown in the glasshouse for 1 month. The second youngest leaf from the main tiller was cut into a 10‐cm piece, placed on moistened filter paper and kept in a Petri dish (36 cm × 36 cm × 2.5 cm). For each entry, four leaves were used as one replicate with four replications per entry in a completely randomized design. Colonized potato dextrose agar (PDA) medium (diameter, 7 mm) was excised using a circular cutter from the PDA plate and placed on the abaxial surface of each leaf piece. The leaves were incubated in a chamber with continuous light at 25 °C for 72 h. The filter paper was kept moist with sterile water. After 72 h, the lesion length on each cut leaf piece was measured, and each leaf was visually rated on a scale of 0–9, with ‘0’ for the absence of lesion and ‘9’ for the lesion that covered 90%–100% of the leaf surface. Visual scores of 1–8 represented 10%–80% diseased leaf area (Prasad and Eizenga, 2008).
For PXO99A infection, the PXO99A strain (Hopkins et al., 1992) was cultured on solid peptone sucrose agar (PSA) medium at 28 °C for 3 days (Yu et al., 2011). After full growth, the bacteria were suspended in distilled (dH2O) to an optical density (OD) of 1.0. The leaves were infected with bacteria after cutting them 1 cm away from the tip using a pair of scissors. For the PXO99A infection assay, 2‐month‐old plants were used, and the lesion length was measured after 2 weeks of infection.
The M. oryzae isolate Guy11 was cultivated on oatmeal medium under weak light for 2 weeks to generate spores. For spray inoculation, 3‐week‐old seedlings were sprayed with 1 × 105/mL spores of M. oryzae, as described previously (Ning et al., 2015). The number of typical susceptible lesions in each seedling was counted to evaluate the infection level.
Gene expression constructs and generation of transgenic plants
For the generation of the gene expression construct, a 2.0‐kb fragment from the promoter region of Rubisco ribulose bisphosphate carboxylase small chain (Os12g17600) was amplified using rice Nipponbare genomic DNA with a primer pair: Rubisco F and R. The polymerase chain reaction (PCR) product was cloned into the pEASY‐Blunt vector (Transgen Biotech, Beijing, China). The plasmid containing the promoter sequence and a shuttle vector, pTCK303 (Du et al., 2016), were digested with Hind III and Spe I, and the promoter fragment was integrated into the pTCK303 vector.
The SWEET mutant plasmids were prepared using overlap extension PCR. Fragments containing three point mutations, OsSWEET11M‐1 (P31T: CCA‐ACA), OsSWEET11M‐2 (G82D: GGC‐GAC) and OsSWEET11M‐3 (P169T: CCG‐ACG), based on the OsSWEET11 cDNA sequence (XM_015792937), were amplified using the overlap extension PCR method. The PCR products were cloned and sequenced. The sequences of PCR primers are listed in Table S1 (see Supporting Information). The mutated OsSWEET11 coding sequences were amplified from the plasmids containing the point mutations using the primer pair, 11_303F and 11_303 R, which introduced Spe I and Sac I sites into the amplified fragments. The PCR products were cloned into the Rubisco promoter‐containing plasmid at the Spe I and Sac I sites. The cloning was confirmed by sequencing before the recombinant plasmid was introduced into Agrobacterium tumefaciens EHA‐105 for transformation of rice callus, as described earlier (Hiei et al., 1994).
The OsSWEET11 OX construct was generated by amplifying the coding sequence of OsSWEET11 using the primer pair, 11 F and 11 R, and rice Nipponbare cDNA as the template. The PCR product was integrated into the pEASY‐blunt vector and sequenced. The plasmid was digested with BamH I and Sac I. The fragment containing the coding sequence of OsSWEET11 was integrated into pTCK303. The plasmid was confirmed by sequencing before it was introduced into A. tumefaciens EHA‐105 for transformation of rice callus.
RNA extraction and quantitative real‐time polymerase chain reaction (qRT‐PCR) analysis
Total RNA was isolated using an RNeasy Plant Mini Kit (QIAGEN, Duesseldorf, Germany) or TRIzol reagent (Takara, Dalian, Liaoning, China), and was treated with RQ‐RNase free DNase (Promega, Madison, WI, USA) for removal of genomic DNA contamination. For cDNA synthesis, reverse transcriptase RNaseH (Toyobo, http://www.toyobo-global.com/) or a GoScript Reverse Transcription Kit was used, according to the manufacturer's instructions (Promega). The products obtained in qRT‐PCR were quantified using Illumina Research Quantity software, Illumina Eco 3.0 (Illumina, San Diego, CA, USA), and the values were normalized against the Ubiquitin levels in the same samples. The primers used for qRT‐PCR are listed in Table S1.
Retrieval of SWEET homologue sequences and their alignment
The sequences of AtSWEET1, OsSWEET2b, OsSWEET11 and OsSWEET11 homologues from different model plants (Table S2, see Supporting Information) were retrieved from the UniProt database (http://www.uniprot.org/). These sequences were analysed by multiple sequence alignment using the ClustalW (version 2) program (Larkin et al., 2007).
Assay of sugar transport activity of mutated AtSWEET1 in yeast
The yeast hexose transporter mutant strain, EBY4000 [hxt1–17D::loxPgal2D::loxP stl1D::loxP agt1D::loxP ydl247wD::loxP yjr160cD::loxP], was used to test the hexose transport activity (Wieczorke et al., 1999). Wild‐type and mutant AtSWEET1 were expressed in EBY4000 using the pDRf1‐GW vector. The transformants were selected on solid SD–Ura(Synthetic Dropout Medium–Uracil) with 2% maltose as the carbon source at 28 °C for 3 days. Subsequently, the growth of yeast cells on SD medium containing 2% glucose or maltose was monitored. AtSUC2 and OsSWEET11 were expressed in the SUSY7 yeast strain using the pDRf1‐GW vector to test sucrose transport activity.
Mating‐based split‐ubiquitin system
For mating‐based split‐ubiquitin assays, OsSWEET11 and mutated OsSWEET11 (mSWEET11) ORFs were cloned into the mating‐based split‐ubiquitin Nub vectors, pXN22_GW and pXN25_GW, and the Cub vector, pMETYC_GW. The assays were performed as described previously (Lalonde et al., 2010).
Split GFP assay
The nYFP and cCFP sequences were fused to the C‐terminal sequences of OsSWEET11 and mutated OsSWEET11 (mSWEET11) in pXNGW and pXCGW vectors, respectively (Xuan et al., 2013). The fusion proteins were introduced into N. benthamiana leaves using the Agrobacterium‐mediated transient expression method (Kim et al., 2009a). The interactions of the co‐expressed proteins were monitored by detection of YFP fluorescence under a confocal microscope (SP5; Leica, Solms, Germany). All the constructs were verified by DNA sequencing.
Co‐IP and Western blot analyses
OsSWEET11‐Myc or mSWEET11‐GFP was co‐transformed with OsSWEET11‐Myc in N. benthamiana leaves by Agrobacterium‐mediated transformation for transient expression. The expressed proteins were extracted and Co‐IP assays were performed, as described previously (Kim et al., 2009b). The microsomal fraction was prepared by centrifugation at 20 000 and 100 000 g for 1 h. The resulting pellet was resuspended in extraction buffer containing 1% Triton X‐100. After centrifugation at 100 000 g for 10 min, the solubilized proteins were incubated with anti‐Myc antibody bound to protein A beads for 2 h. The beads were washed four times with extraction buffer containing 0.1% Triton X‐100 and eluted with sodium dodecylsulfate (SDS) sample buffer.
From each sample, 20 µg of protein was separated by sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and electrotransferred onto Immobilon‐P transfer membrane (MILLIPORE JAPAN, Tokyo, Japan). The membranes were incubated in TBS (tris buffered saline) containing 5% skimmed milk and 0.05% Tween‐20 for 60 min and blotted with primary antibodies at 4 °C for 2 h. The anti‐Myc (1 : 2000; Abcam, Cambridge, MA, USA) and anti‐GFP (1 : 2000; Abcam) antibodies were used as primary antibodies. The membranes were incubated for 1 h with an anti‐mouse or anti‐rabbit horseradish peroxidase (HRP)‐conjugated secondary antibody (1 : 2000; Cell Signaling Technology Boston, MA, USA). The blots were visualized by chemiluminescence using an ECL Western Blotting Detection System (GE Healthcare, Piscataway, NJ, USA).
Statistical analysis
Statistical calculations were performed using Prism 5 (GraphPad, San Diego, CA, USA). All the data are expressed as means ± standard error (SE). Comparisons between different groups were performed using one‐way analysis of variance (ANOVA), followed by Bonferroni's multiple comparison test. P < 0.05 was considered to be statistically significant.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Primer sequences used in this study.
Table S2 Information on SWEET11 homology.
Fig. S1 Expression of OsSWEET12–OsSWEET15 genes induced by Rhizoctonia solani infection. The expression levels of OsSWEETs were analysed after 0, 24, 48 and 72 h of R. solani infection. Ten leaves were sampled at each time point for RNA extraction. The experiments were repeated three times, and the data represent means ± standard error (SE) (n = 3). Significant differences at P < 0.05 are indicated by different letters.
Fig. S2 Expression level of OsSWEET11 in the rice overexpression lines. The expression level of OsSWEET11 was analysed using RNA extracted from the leaves of 2‐week‐old wild‐type and OsSWEET11 overexpression lines (OsSWEET11 OX1, OX2, OX4 and OX5). The expression level in the wild‐type plant was defined as ‘1’, and the relative expression levels are shown. Significant differences at P < 0.05 are indicated by different letters.
Fig. S3 Sensitivity of OsSWEET11 overexpression (OX) plants to PXO99A. The leaves of 2‐month‐old wild‐type and OsSWEET11 OX plants (#2 and #4) were inoculated with PXO99A. The lesion length was measured after 2 weeks of infection. Significant differences at P < 0.05 are indicated by different letters.
Fig. S4 The phenotypes of wild‐type and ossweet11‐1 seeds. The wild‐type and ossweet11‐1 mutant seeds were photographed. A comparison of wild‐type and ossweet11‐3 mutant seeds shows poor filling in the latter.
Fig. S5 Sucrose transport activity test in the SUSY7 yeast strain. Growth assays of AtSUC2 and OsSWEET11 proteins expressed in the SUSY7 yeast strain were performed on YNB (Yeast Nitrogen Base without Amino Acids) medium containing 2% sucrose or glucose. Empty (pDRf1) vector and AtSUC2 were used as negative and positive controls, respectively. The yeast cells were grown at 28 °C for 3 days.
Fig. S6 Sensitivity of pRub‐mOsSWEET11 plants to PXO99A infection. The leaves of 2‐month‐old wild‐type and pRub‐mOsSWEET11 plants (#1 and #2) were inoculated with PXO99A. The lesion length was measured after 2 weeks of infection. Significant differences at P < 0.05 are indicated by different letters.
Fig. S7 Disease symptoms in plants expressing OsSWEET11 mutants on infection with Magnaporthe oryzae. (A) The leaves of wild‐type and two independent transgenic plants (pRub‐mOsSWEET11 #1 and #2) were infected with M. oryzae Guy11. After 3 days of infection, the leaves were photographed. Ten leaves from each line were analysed, and the experiments were repeated three times. (B) The lesion scales were analysed for the M. oryzae Guy11‐infected leaves shown in (A) by determination of the lesion area on the leaf surface. The data represent means ± standard error (SE) (n > 10). Significant differences at P < 0.05 are indicated by different letters.
Fig. S8 Expression levels of PBZ1 in plants expressing OsSWEET11 mutants. The levels of PBZ1 were analysed in the leaves of wild‐type plants (WT), Ossweet11 mutants, OsSWEET11 overexpressors and pRub‐mOsSWEET11 lines. More than six leaves from each line were used for RNA extraction. Data represent the means ± standard error (SE) (n = 3), and the experiments were repeated at least three times. Significant differences at P < 0.05 are indicated by different letters.
Acknowledgements
This work was supported by Ministry of Science and Technology of China, National Key R&D Program of China (2016YFD0100101), an initiative grant (880416008) from Shenyang Agricultural University and the projects of the Agricultural Science Institute of Wenzhou (2016C02050‐4), Key Laboratory of Crop Breeding in South Zhejiang (2017SZCB01) and the Natural Science Foundation of Jiangsu Province, China (BK20151424). EBY4000 and SUSY7 yeast strains were obtained from Wolf Frommer's laboratory.
References
- Antony, G. , Zhou, J. , Huang, S. , Li, T. , Liu, B. , White, F. and Yang, B. (2010) Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os‐11N3 . Plant Cell, 22, 3864–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H.Y. , Huh, J. , Yu, Y. , Ho, L. , Chen, L. , Tholl, D. , Frommer, W.B. and Guo, W.J. (2015) The Arabidopsis vacuolar sugar transporter SWEET2 limits carbon sequestration from roots and restricts Pythium infection. Plant J. 83, 1046–1058. [DOI] [PubMed] [Google Scholar]
- Chen, L.Q. , Hou, B.H. , Lalonde, S. , Takanaga, H. , Hartung, M.L. , Qu, X.Q. , Guo, W.J. , Kim, J.G. , Underwood, W. , Chaudhuri, B. , Chermak, D. , Antony, G. , White, F.F. , Somerville, S.C. , Mudgett, M.B. and Frommer, W.B. (2010) Sugar transporters for intercellular exchange and nutrition of pathogens. Nature, 468, 527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L.Q. , Qu, X.Q. , Hou, B.H. , Sosso, D. , Osorio, S. , Fernie, A.R. and Frommer, W.B. (2012) Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science, 335, 207–211. [DOI] [PubMed] [Google Scholar]
- Chen, L.Q. , Cheung, L.S. , Feng, L. , Tanner, W. and Frommer, W.B. (2015) Transport of sugars. Annu. Rev. Biochem. 84, 865–894. [DOI] [PubMed] [Google Scholar]
- Cox, K.L. , Meng, F. , Wilkins, K.E. , Li, F. , Wang, P. , Booher, N.J. , Carpenter, S.C.D. , Chen, L.Q. , Zheng, H. , Gao, X. , Zheng, Y. , Fei, Z. , Yu, J.Z. , Isakeit, T. , Wheeler, T. , Frommer, W.B. , He, P. , Bogdanove, A.J. and Shan, L. (2017) TAL effector driven induction of a SWEET gene confers susceptibility to bacterial blight of cotton. Nat. Commun. 8, 15 588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, Y. , He, W. , Deng, C. , Chen, X. , Gou, L. , Zhu, F. , Guo, W. , Zhang, J. and Wang, T. (2016) Flowering‐related RING Protein 1 (FRRP1) regulates flowering time and yield potential by affecting histone H2B monoubiquitination in rice (Oryza sativa). PLoS One, 11, e0150458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari, S. , Galletti, R. , Denoux, C. , Lorenzo, G.D. , Ausubel, F.M. and Dewdney, J. (2007) Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is independent of salicylic acid, ethylene, or jasmonate signaling but requires phytoalexin deficient3 . Plant Physiol. 144, 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell, E.E. , Wang, Q. and Yang, Y. (2013) Transgenic rice with inducible ethylene production exhibits broad‐spectrum disease resistance to the fungal pathogens Magnaporthe oryzae and Rhizoctonia solani . Plant Biotechnol. J. 11, 33–42. [DOI] [PubMed] [Google Scholar]
- Hiei, Y. , Ohta, S. , Komari, T. and Kumashiro, T. (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T‐DNA. Plant J. 6, 271–282. [DOI] [PubMed] [Google Scholar]
- Hopkins, C.M. , White, F.F. , Choi, S.H. , Guo, A. and Leach, J.E. (1992) Identification of a family of avirulence genes from Xanthomonas oryzae pv. oryzae . Mol. Plant–Microbe Interact. 5, 451–459. [DOI] [PubMed] [Google Scholar]
- Hutin, M. , Sabot, F. , Ghesquiere, A. , Koebnik, R. and Szurek, B. (2015) A knowledge‐based molecular screen uncovers a broad‐spectrum OsSWEET14 resistance allele to bacterial blight from wild rice. Plant J. 84, 694–703. [DOI] [PubMed] [Google Scholar]
- Karmakar, S. , Molla, K.A. , Das, K. , Sarkar, S.N. , Datta, S.K. and Datta, K. (2017) Dual gene expression cassette is superior than single gene cassette for enhancing sheath blight tolerance in transgenic rice. Sci. Rep. 7, 7900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.G. , Li, X. , Roden, J.A. , Taylor, K.W. , Aakre, C.D. , Su, B. , Lalonde, S. , Kirik, A. , Chen, Y. , Baranage, G. , McLane, H. , Martin, G.B. and Mudgett, M.B. (2009. a) Xanthomonas T3S effector XopN suppresses PAMP‐triggered immunity and interacts with a tomato atypical receptor‐like kinase and TFT1. Plant Cell, 21, 1305–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T.W. , Guan, S. , Sun, Y. , Deng, Z. , Tang, W. , Shang, J.X. , Sun, Y. , Burlingame, A.L. and Wang, Z.Y. (2009. b) Brassinosteroid signal transduction from cell‐surface receptor kinases to nuclear transcription factors. Nat. Cell. Biol. 11, 1254–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krügel, U. , Veenhoff, L.M. , Langbein, J. , Wiederhold, E. , Liesche, J. , Friedrich, T. , Grimm, B. , Martinoia, E. , Poolman, B. and Kühn, C. (2008) Transport and sorting of the Solanum tuberosum sucrose transporter SUT1 is affected by posttranslational modification. Plant Cell, 20, 2497–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krügel, U. , Wiederhold, E. , Pustogowa, J. , Hackel, A. , Grimm, B. and Kühn, C. (2013) Site directed mutagenesis of StSUT1 reveals target amino acids of regulation and stability. Biochimie, 95, 2132–2144. [DOI] [PubMed] [Google Scholar]
- Lalonde, S. , Weise, A. , Walsh, R.P. , Ward, J.M. and Frommer, W.B. (2003) Fusion to GFP blocks intercellular trafficking of the sucrose transporter SUT1 leading to accumulation in companion cells. BMC Plant Biol. 3, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde, S. , Sero, A. , Pratelli, R. , Pilot, G. , Chen, J. , Sardi, M.I. , Parsa, S.A. , Kim, D.Y. , Acharya, B.R. , Stein, E.V. , Hu, H.C. , Villiers, F. , Takeda, K. , Yang, Y. , Han, Y.S. , Schwacke, R. , Chiang, W. , Kato, N. , Loqué, D. , Assmann, S.M. , Kwak, J.M. , Schroeder, J.I. , Rhee, S.Y. and Frommer, W.B. (2010) A membrane protein/signaling protein interaction network for Arabidopsis version AMPv2. Front. Physiol. 1, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, M.A. , Blackshields, G. , Brown, N.P. , Chenna, R. , McGettigan, P.A. , McWilliam, H. , Valentin, F. , Wallace, I.M. , Wilm, A. , Lopez, R. , Thompson, J.D. , Gibson, T.J. and Higgins, D.G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics, 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- Li, T. , Liu, B. , Spalding, M.H. , Weeks, D.P. and Yang, B. (2012) High‐efficiency TALEN‐based gene editing produces disease‐resistant rice. Nat. Biotechnol. 30, 390–392. [DOI] [PubMed] [Google Scholar]
- Lin, I.W. , Sosso, D. , Chen, L.Q. , Gase, K. , Kim, S.G. , Kessler, D. , Klinkenberg, P.M. , Gorder, M.K. , Hou, B.H. , Qu, X.Q. , Carter, C.J. , Baldwin, I.T. and Frommer, W.B. (2014) Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9. Nature, 508, 546–549. [DOI] [PubMed] [Google Scholar]
- Liu, Q. , Yuan, M. , Zhou, Y. , Li, X. , Xiao, J. and Wang, S. (2011) A paralog of the MtN3/saliva family recessively confers race‐specific resistance to Xanthomonas oryzae in rice. Plant Cell Environ. 34, 1958–1969. [DOI] [PubMed] [Google Scholar]
- Ma, L. , Zhang, D. , Miao, Q. , Yang, J. , Xuan, Y. and Hu, Y. (2017) Essential role of sugar transporter OsSWEET11 during the early stage of rice grain filling. Plant Cell Physiol. 58, 863–873. [DOI] [PubMed] [Google Scholar]
- Mao, B. , Liu, X. , Hu, D. and Li, D. (2014) Co‐expression of RCH10 and AGLU1 confers rice resistance to fungal sheath blight Rhizoctonia solani and blast Magnaporthe oryzae and reveals impact on seed germination. World. J. Microbiol. Biotechnol. 30, 1229–1238. [DOI] [PubMed] [Google Scholar]
- Ning, Y. , Shi, X. , Wang, R. , Fan, J. , Park, C.H. , Zhang, C. , Zhang, T. , Ouyang, X. , Li, S. and Wang, G.L. (2015) OsELF3‐2, an ortholog of Arabidopsis ELF3, interacts with the E3 ligase APIP6 and negatively regulates immunity against Magnaporthe oryzae in rice. Mol. Plant. 8, 1679–1682. [DOI] [PubMed] [Google Scholar]
- Prasad, B. and Eizenga, G.C. (2008) Rice sheath blight disease resistance identified in Oryza spp. accessions. Plant Dis. 92, 1503–1509. [DOI] [PubMed] [Google Scholar]
- Quirino, B.F. , Normanly, J. and Amasino, R.M. (1999) Diverse range of gene activity during Arabidopsis thaliana leaf senescence includes pathogen‐independent induction of defense‐related genes. Plant Mol. Biol. 40, 267–278. [DOI] [PubMed] [Google Scholar]
- Savary, S. , Willocquet, L. , Elazegui, F.A. , Castilla, N.P. and Teng, P.S. (2000) Rice pest constraints in tropical Asia: quantification of yield losses due to rice pests in a range of production situations. Plant Dis. 84, 357–369. [DOI] [PubMed] [Google Scholar]
- Slaton, N.A. , Cartwright, R.D. , Meng, J. , Gbur, E.E. and Norman, R.J. (2003) Sheath blight severity and rice yield as affected by nitrogen fertilizer rate, application method, and fungicide. Agron. J. 95, 1489–1496. [Google Scholar]
- Sosso, D. , Luo, D. , Li, Q.B. , Sasse, J. , Yang, J. , Gendrot, G. , Suzuki, M. , Koch, K.E. , McCarty, D.R. , Chourey, P.S. , Rogowsky, P.M. , Ross‐Ibarra, J. , Yang, B. and Frommer, W.B. (2015) Seed filling in domesticated maize and rice depends on SWEET‐mediated hexose transport. Nat. Genet. 47, 1489–1493. [DOI] [PubMed] [Google Scholar]
- Sripriya, R. , Parameswari, C. and Veluthambi, K. (2017) Enhancement of sheath blight tolerance in transgenic rice by combined expression of tobacco osmotin (ap24) and rice chitinase (chi11) genes. In Vitro Cell Dev. Plant, 53, 12–21. [Google Scholar]
- Wang, R. , Lu, L. , Pan, X. , Hu, Z. , Ling, F. , Yan, Y. , Liu, Y. and Lin, Y. (2015) Functional analysis of OsPGIP1 in rice sheath blight resistance. Plant Mol. Biol. 87, 181–191. [DOI] [PubMed] [Google Scholar]
- Wieczorke, R. , Krampe, S. , Weierstall, T. , Freidel, K. , Hollenberg, C.P. and Boles, E. (1999) Concurrent knock‐out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae . FEBS. Lett. 464, 123–128. [DOI] [PubMed] [Google Scholar]
- Xuan, Y.H. , Hu, Y.B. , Chen, L.Q. , Sosso, D. , Ducat, D.C. , Hou, B.H. and Frommer, W.B. (2013) Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. Proc. Natl. Acad. Sci. USA, 110, E3685–E3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, X. , Cao, Z. , Zhang, X.T. , Wang, Y. , Zhang, Y.F. , Chen, Z.X. , Pan, X.B. and Zuo, S.M. (2016) Overexpression of OsOSM1 enhances resistance to rice sheath blight. Plant Dis. 100, 1634–1642. [DOI] [PubMed] [Google Scholar]
- Yang, B. , Sugio, A. and White, F.F. (2006) Os8N3 is a host disease‐susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. USA, 103, 10 503–10 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y. , Streubel, J. , Balzergue, S. , Champion, A. , Boch, J. , Koebnik, R. , Feng, J. , Verdier, V. and Szurek, B. (2011) Colonization of rice leaf blades by an African strain of Xanthomonas oryzae pv. oryzae depends on a new TAL effector that induces the rice nodulin‐3 Os11N3 gene. Mol. Plant–Microbe Interact. 24, 1102–1113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Primer sequences used in this study.
Table S2 Information on SWEET11 homology.
Fig. S1 Expression of OsSWEET12–OsSWEET15 genes induced by Rhizoctonia solani infection. The expression levels of OsSWEETs were analysed after 0, 24, 48 and 72 h of R. solani infection. Ten leaves were sampled at each time point for RNA extraction. The experiments were repeated three times, and the data represent means ± standard error (SE) (n = 3). Significant differences at P < 0.05 are indicated by different letters.
Fig. S2 Expression level of OsSWEET11 in the rice overexpression lines. The expression level of OsSWEET11 was analysed using RNA extracted from the leaves of 2‐week‐old wild‐type and OsSWEET11 overexpression lines (OsSWEET11 OX1, OX2, OX4 and OX5). The expression level in the wild‐type plant was defined as ‘1’, and the relative expression levels are shown. Significant differences at P < 0.05 are indicated by different letters.
Fig. S3 Sensitivity of OsSWEET11 overexpression (OX) plants to PXO99A. The leaves of 2‐month‐old wild‐type and OsSWEET11 OX plants (#2 and #4) were inoculated with PXO99A. The lesion length was measured after 2 weeks of infection. Significant differences at P < 0.05 are indicated by different letters.
Fig. S4 The phenotypes of wild‐type and ossweet11‐1 seeds. The wild‐type and ossweet11‐1 mutant seeds were photographed. A comparison of wild‐type and ossweet11‐3 mutant seeds shows poor filling in the latter.
Fig. S5 Sucrose transport activity test in the SUSY7 yeast strain. Growth assays of AtSUC2 and OsSWEET11 proteins expressed in the SUSY7 yeast strain were performed on YNB (Yeast Nitrogen Base without Amino Acids) medium containing 2% sucrose or glucose. Empty (pDRf1) vector and AtSUC2 were used as negative and positive controls, respectively. The yeast cells were grown at 28 °C for 3 days.
Fig. S6 Sensitivity of pRub‐mOsSWEET11 plants to PXO99A infection. The leaves of 2‐month‐old wild‐type and pRub‐mOsSWEET11 plants (#1 and #2) were inoculated with PXO99A. The lesion length was measured after 2 weeks of infection. Significant differences at P < 0.05 are indicated by different letters.
Fig. S7 Disease symptoms in plants expressing OsSWEET11 mutants on infection with Magnaporthe oryzae. (A) The leaves of wild‐type and two independent transgenic plants (pRub‐mOsSWEET11 #1 and #2) were infected with M. oryzae Guy11. After 3 days of infection, the leaves were photographed. Ten leaves from each line were analysed, and the experiments were repeated three times. (B) The lesion scales were analysed for the M. oryzae Guy11‐infected leaves shown in (A) by determination of the lesion area on the leaf surface. The data represent means ± standard error (SE) (n > 10). Significant differences at P < 0.05 are indicated by different letters.
Fig. S8 Expression levels of PBZ1 in plants expressing OsSWEET11 mutants. The levels of PBZ1 were analysed in the leaves of wild‐type plants (WT), Ossweet11 mutants, OsSWEET11 overexpressors and pRub‐mOsSWEET11 lines. More than six leaves from each line were used for RNA extraction. Data represent the means ± standard error (SE) (n = 3), and the experiments were repeated at least three times. Significant differences at P < 0.05 are indicated by different letters.


