Summary
Vascular wilt caused by Verticillium dahliae is a destructive disease that represents a chronic economic problem for crop production worldwide. In this work, we characterized two new regulators of pathogenicity in this species. Vph1 (VDAG_06555) was identified in a candidate gene approach as a putative homologue of the transcription factor Ste12. Vhb1 (VDAG_08786), identified in a forward genetics approach, is similar to the homeobox transcription factor Htf1, reported as a regulator of conidiogenesis in several fungi. Deletion of vph1 did not affect vegetative growth, whereas deletion of vhb1 greatly reduced sporulation rates in liquid medium. Both mutants failed to induce Verticillium wilt symptoms. However, unlike Δvph1, Δvhb1 could be re‐isolated from the vascular system of some asymptomatic plants. Confocal microscopy further indicated that Δvph1 and Δvhb1 differed in their behaviour in planta; Δvph1 could not penetrate the root cortex, whereas Δvhb1 was impaired in its ability to colonize the xylem. In agreement with these observations, only Δvhb1 could penetrate cellophane paper. On cellophane, wild‐type and Δvhb1 strains produced numerous short branches with swollen tips, resembling the hyphopodia formed on root surfaces, contrasting with Δvph1, which generated unbranched long filaments without swollen tips. A microarray analysis showed that these differences in growth were associated with differences in global transcription patterns, and allowed us to identify a large set of novel genes potentially involved in virulence in V. dahliae. Ste12 homologues are known regulators of invasive growth, but Vhb1 is the first putative Htf1 homologue identified with a critical role in virulence.
Keywords: host colonization, transcriptional regulators, Verticillium wilt, virulence factors
Introduction
The soil‐borne fungus Verticillium dahliae, the main causal agent of Verticillium wilt, affects a wide variety of plants, including agronomically important crops, ornamental flowers, trees and shrubs (Klosterman et al., 2009; Pegg and Brady, 2002). The main features of the V. dahliae disease cycle have been elucidated. In the presence of host root exudates, melanized resting structures, the microsclerotia (MS), germinate, produce hyphae that penetrate the epidermis and cortex, and grow inter‐ and intracellularly until reaching the xylem (Eynck et al., 2007; Vallad and Subbarao, 2008). In the xylem, a combination of hyphal growth and sporulation is thought to contribute to vascular colonization, eventually obstructing the xylem fluid. This, together with the secretion of toxins, leads to the appearance of disease symptoms (Luo et al., 2014). Large amounts of MS are produced in wilted tissue at late stages of infection, which can remain in the soil as dormant viable structures for long periods (Fradin and Thomma, 2006; Klimes et al., 2015). Verticillium dahliae is particularly difficult to control largely because of its lack of host specificity and vascular lifestyle combined with the high durability of MS.
An understanding of the genetic basis underlying each major event in the disease cycle of V. dahliae may contribute to the development of novel strategies to control Verticillium wilt. Transcription factors (TFs) are key targets of the signalling pathways that govern development and virulence in fungi. These proteins bind specific DNA sequences in the promoter regions, thus controlling the expression of genes whose activation/repression is required to induce changes in the cell in response to environmental stimuli, including those coming from the host (Hughes, 2011). Our understanding of the molecular mechanisms that control V. dahliae development and virulence is rapidly increasing (Klimes et al., 2015). However, only a handful of TF genes have been functionally characterized to date in this species. TFs are classified into different types based on their conserved DNA‐binding domains. Zinc finger proteins are amongst the major types of transcriptional regulator. The V. dahliae genome is predicted to encode 937 zinc finger TFs, only three of which have been functionally characterized to date: the C2H2 zinc finger proteins Vta2 and VdCrz1, and Vdpf1, containing the fungal‐specific Zn(II)2‐Cys6 domain (Luo et al., 2016; Tran et al., 2014; Xiong et al., 2015). Proteins containing the DNA‐binding motif MADS‐box are another major group of TFs. Recently, the functional characterization of a MADS‐box protein from V. dahliae, VdMcm1, has been reported (Xiong et al., 2016). These TFs appear to perform pleiotropic functions regulating aspects of vegetative growth, such as MS production and melanization of cultures, sporulation rates or hyphal growth, as well as virulence (Luo et al., 2016; Tran et al., 2014; Xiong et al., 2015, 2016). A putative homologue of Sge1, a TF involved in the regulation of effector gene expression, has also been shown to play roles in virulence, hyphal growth and conidiation (Santhanam and Thomma, 2013). The APSES domain represents a type of basic helix–loop–helix DNA‐binding domain which is specific to fungi. In contrast with other TFs characterized in V. dahliae, the APSES protein Vst1 is a critical regulator of development, but dispensable for host invasion (Sarmiento‐Villamil et al., 2016).
The determination of the signalling pathway(s) in which different TFs perform their functions is a critical element to fully understand the molecular mechanisms underlying cellular processes. For many TFs, these aspects are not yet known. Moreover, although some TFs are clearly connected with particular signalling pathways, the general picture can vary considerably amongst species. VdCrz1 is a putative homologue of the calcium signalling pathway target Crz1, and there is evidence of its link to this pathway in V. dahliae (Xiong et al., 2015). APSES proteins, however, have been associated with cyclic adenosine monophosphate (cAMP) signalling (Bockmühl and Ernst, 2001; Sonneborn et al., 1999; Tebarth et al., 2003), one of the major pathways controlling development and virulence in fungi. Some key components of cAMP signalling have been characterized in V. dahliae, including the protein kinase A gene VdPKAC1 (Tzima et al., 2010) and the G‐protein β subunit gene VGB (Tzima et al., 2012). In V. dahliae, G‐protein/cAMP signalling is a negative regulator of MS biogenesis and a positive regulator of virulence (Tzima et al., 2012), and the APSES TF Vst1 seems to negatively interact with this pathway (Sarmiento‐Villamil et al., 2016). Mitogen‐activated protein kinase (MAPK) cascades, together with G‐protein/cAMP signalling, are major regulators of morphogenesis and virulence. A number of components of these pathways have been studied in V. dahliae, including MAPK genes from the so‐called Pathogenicity MAPK (PMK) cascade and from the stress‐activated high osmolarity glycerol (HOG) cascade (Rauyaree et al., 2005; Tian et al., 2016; Wang et al., 2016). Putative homologues of the transmembrane proteins Mbs2 and Sho1, upstream components of these cascades, have also been characterized in this species (Qi et al., 2016; Tian et al., 2014). These studies have revealed that MAPK signalling plays a role in the regulation of virulence and different aspects of growth, most remarkably MS formation, again indicating that development and plant infection may fall under the same regulatory pathways. However, TFs that might function as major downstream targets of MAPK signalling have not been reported to date in V. dahliae. In yeast, the TF Ste12 is a key target of MAPK signalling (Elion et al., 2005). In pathogenic fungi, Ste12 and Ste12‐like proteins regulate invasive growth, functioning as downstream targets of PMK (Rispail and Di Pietro, 2010). Ste12 TFs are characterized by the presence of a homeodomain‐like DNA‐binding motif, the Ste motif, located at the N‐terminus and which shares residues involved in DNA binding with the homeodomains of the widespread homeobox family of regulatory proteins (Yuan and Fields, 1991). However, the Ste motif is distinct from typical homeodomains and Ste12 proteins form a group of transcriptional regulators specific to fungi.
TFs with typical homeodomains are also present in fungi and appear to play diverse roles. A well‐characterized case of homeodomain proteins in plant pathogens are those encoded by the Ustilago maydis b locus, which control pathogenic development (Schulz et al., 1990). In Magnaporthe oryzae, Kim et al. (2009) functionally characterized all homeodomain TF genes identified in this species, Mohox1 to Mohox7. Mohox2 and Mohox6 were involved in sporulation, whereas the absence of Mohox1 and Mohox4 resulted in greatly reduced hyphal growth. Mohox7, however, was critical for appressorium development, but not for invasive growth. Liu et al. (2010) further confirmed that Mohox2, which they designated HTF1, was essential for conidiogenesis. Zheng et al. (2012) characterized HTF1 homologues in three Fusarium species, providing evidence that the role of this homeobox TF in the regulation of conidiogenesis is conserved. Neither in Magnaporthe nor in Fusarium did Htf1 play a role in invasive growth (Liu et al., 2010; Zheng et al., 2012). In Botrytis cinerea, the homeodomain TF BcHOX8, identified as up‐regulated during Arabidopsis thaliana infection, plays a role in hyphal growth, conidiogenesis and in the production of the finger‐like‐tip infectious hyphae (Antal et al., 2012). Although BcHOX8 was not essential for plant infection, its absence had a negative impact on the invasion of certain hosts and tissues.
In this work, we report the functional characterization in V. dahliae of vph1, a putative orthologue of ste12 identified in a candidate gene approach, and vhb1, identified in a forward genetics approach, which encodes a protein exhibiting a typical homeodomain and similar to the homeobox TF Htf1 involved in sporulation in several fungi. We found that vph1 and vhb1 are both critically required for virulence. However, we also provide evidence that their absence blocks host invasion at different stages of the disease cycle. In addition, transcription pattern profiling analyses in a condition that mimics the penetration phase further suggest different functions for Vph1 and Vhb1.
Results
Identification of a putative Ste12 homologue in V. dahliae
TFs of the Ste12 family are characterized by the presence of a highly conserved homeodomain‐like motif, the Ste domain, in the N‐terminal region (Wong Sak Hoi and Dumas, 2010). To identify putative Ste12 encoding genes in the genome of V. dahliae strain VdLs.17, we performed a protein–protein blast search using the Ste domain of Saccharomyces cerevisiae Ste12. This search detected a single significant match, VDAG_06555. We named this putative orthologue of ste12 in V. dahliae vph1. A distinctive feature of Ste12 proteins from filamentous fungi is that they contain two zinc finger C2H2‐type domains in the C‐terminus (Rispail and Di Pietro, 2010; Wong Sak Hoi and Dumas, 2010). Based on the Broad Institute annotation (Klosterman et al., 2011), VDAG_06555 encoded a 685‐amino‐acid protein having a single zinc finger motif (C2H2‐type). In Botrytis cinerea and Colletotrichum lindemuthianum, alternative splicing mechanisms generating two transcript variants that encoded full‐sized and truncated versions of Ste12 were observed (Schamber et al., 2010; Wong Sak Hoi et al., 2007). Analysis of the V. dahliae genomic region encoding the Ste12 homologue by blastx searches, and the determination of all putative open reading frames (ORFs), showed that alternative splicing could also encode a larger variant of Vph1, a 713‐amino‐acid protein which contains two zinc finger domains (Fig. 1).
Figure 1.

Organization of vph1, showing alternative splicing of transcripts. (a) Organization of exons (E) and introns (I) in the two transcripts. (b) Predicted structure of the full‐length Vph1 protein containing two zinc finger C2H2‐type domains (Vph1–2ZF) and the truncated version containing only one (Vph1–1ZF). Amino acid positions of the Ste motif and zinc fingers are indicated. (c) Amino acid alignment of the Ste motif (black bar) and zinc finger domains. The two zinc fingers are indicated by the square boxes, whereas the grey bar indicates the single zinc finger of Vph1–1ZF. Domains included are from the following proteins: Verticillium. dahliae Vph1–2ZF and Vph1–1ZF; V. nonalfalfae VDBG_08894; Colletotrichum lagenarium Cst1; Magnaporthe grisea Mst12; Fusarium oxysporum Fost12; Aspergillus nidulans SteA; Penicillium marneffei StlA; Saccharomyces cerevisiae Ste12; Candida albicans Cph1; Rhizophagus irregularis GintSTE. Black shading indicates identical or similar amino acid residues in all proteins, dark grey shading indicates a conservation of residues below 100% and above 80%, and light grey shading indicates a conservation of residues below 80% and above 60%.
Identification of a homeobox TF as a putative regulator of morphogenesis and virulence in V. dahliae by random insertional mutagenesis
In parallel with the candidate gene approach, we used forward genetics to identify novel regulators of virulence and/or development in V. dahliae. To generate deletion mutants, we used constructs in binary vectors compatible with Agrobacterium tumefaciens‐mediated transformation (ATMT) (García‐Pedrajas et al., 2013; Paz et al., 2011). Transformants that did not exhibit deletion of the target gene, but ectopic integration of the T‐DNA, were not discarded, but were used to generate a collection of mutants with random insertions. These random mutants were initially screened for the presence of visible phenotypic alterations in in vitro culture. Mutant D‐10–4F, generated in this way, exhibited alterations in MS production and reduced sporulation, and was selected for further analysis. When inoculated in tomato plants, D‐10–4F failed to induce Verticillium wilt symptoms.
An inverse polymerase chain reaction (PCR) approach was then performed (see Experimental Procedures) to characterize T‐DNA insertions in mutant D‐10–4F. We identified two T‐DNA integration points, one located between genes VDAG_05485 and VDAG_05486, and the other 512 bp upstream of the VDAG_08786 start codon. Although these insertions did not affect the ORFs, they may nevertheless affect the expression of these genes, and thereby alter their function(s). As our aim was to identify genes with regulatory functions, we focused on VDAG_08786 encoding a 592‐amino‐acid protein with a homeodomain in the N‐terminal region (Fig. 2). We named this gene vhb1 (Verticillium dahliae homeobox protein 1). Within the homeobox family, vhb1 exhibited the highest similarity to htf1/hox2 (Fig. 2), characterized as a regulator of conidiogenesis in M. oryzae and several Fusarium species (Kim et al., 2009; Liu et al., 2010; Zheng et al., 2012).
Figure 2.

Amino acid alignment of the homeodomains of Vhb1 and similar proteins. The homeodomains included are from the following proteins: Verticillium dahliae Vhb1; Colletotrichum gloeosporoides CGGC5_1994; Fusarium graminearum FgHTF1; Fusarium oxysporum FoHTF1; Fusarium verticillioides FvHTF1; Magnaporthe oryzae MoHOX2/MoHTF1; Magnaporthe poae MAPG_03141; Sclerotinia sclerotiorum SS1G_01567; V. dahliae VDAG_03007 and VDAG_05070. Black shading indicates identical or similar amino acid residues in all homeodomains included, dark grey shading indicates a conservation of residues below 100% and above 80%, and light grey shading indicates a conservation of residues below 80% and above 60%.
Targeted deletion of vph1 and vhb1 reveals their critical roles in virulence
To assess the involvement of the ste12 orthologue vph1 and the uncharacterized homeobox gene vhb1 in the regulation of morphogenesis and/or pathogenicity in V. dahliae, deletion mutants were generated using the OSCAR (One Step Construction of Agrobacterium‐Recombination‐ready‐plasmids) method (Paz et al., 2011). A modification was introduced to this method so that transformed strains constitutively expressed a synthetic SGFP‐TYG gene (Maor et al., 1998) (Fig. S1, see Supporting Information). A PCR analysis indicated deletion of vph1 and vhb1 in two and four independent transformants, respectively. For each gene, a deletion mutant was selected to confirm deletion through a Southern blot analysis with probes derived from their respective ORFs. A second set of Southern blots with a hygR probe revealed that no secondary T‐DNA integration events occurred in these mutants (Fig. S2, see Supporting Information), confirming that no other genes were affected. In addition, a transformant in which the T‐DNA had integrated ectopically and with no phenotypic alterations was selected to be used as a green fluorescent protein (GFP)‐tagged control strain representative of wild‐type (WT) behaviour in our fluorescence microscopy experiments.
Strains deleted for vph1 did not exhibit any phenotypic alterations when grown on solid or liquid media. Mutants lacking vhb1 displayed normal growth on solid medium, but showed a drastic reduction in sporulation rates in liquid medium (Figs 3 and S3, see Supporting Information). Infection assays using tomato as a host were then performed to test the virulence of Δvph1 and Δvhb1 strains. Characteristic symptoms of Verticillium wilt were observed between 3 and 7 weeks post‐inoculation on inoculation with the WT and GFP‐tagged control strain (Fig. 4a). The severity of symptoms caused by both strains did not differ significantly (Fig. 4b), confirming that the GFP‐tagged ectopic transformant that was selected behaved as the parental strain, and could therefore be used as a control of WT infection. By contrast, Δvph1 and Δvhb1 did not generate any Verticillium wilt symptoms (Fig. 4a,b). Verticillium wilt symptoms induced in tomato by the parental WT Dvd‐T5 are restricted to wilting of the lower leaves (Fig. 4a, arrows) and plant stunting. To characterize in more detail the effect of vph1 and vhb1 deletion on virulence, Nicotiana benthamiana and lettuce were also used as hosts in virulence assays. Unlike tomato, N. benthamiana plants inoculated with WT strain Dvd‐T5 exhibited severe Verticillium wilt symptoms and plant stunting (Fig. 5a). In contrast with the WT and GFP‐tagged control strain, N. benthamiana plants inoculated with either Δvph1 or Δvhb1 were indistinguishable from those mock inoculated with water (Fig. 5a,b). The results of lettuce virulence assays further confirmed that vph1 and vhb1 are critical virulence factors (Fig. S4, see Supporting Information).
Figure 3.
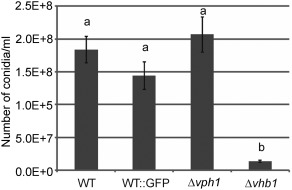
Deletion of vhb1 results in reduced sporulation rates in liquid medium. Conidia from 4‐day‐old cultures, initially inoculated with 106 conidia/mL of each strain, were counted in a Fuchs–Rosenthal haemocytometer. Three replicates of each strain were included in each experiment and five independent experiments were performed. Different letters indicate statistically significant differences (P < 0.05) according to a Bonferroni–Dunn test. Bars indicate standard error.
Figure 4.
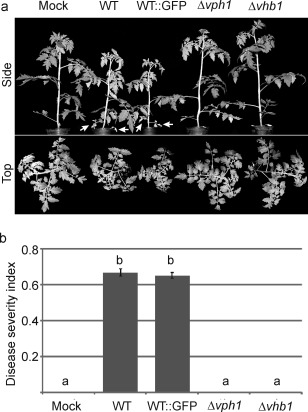
Deletion of vph1 or vhb1 renders the Verticillium dahliae strains avirulent towards tomato plants. (a) Solanum lycopersicum L. variety Moneymaker plants inoculated with wild‐type (WT) and WT green fluorescent protein (GFP)‐tagged strains generated characteristic symptoms, including wilting of the lower leaves (white arrows) and plant stunting, whereas plants inoculated with Δvph1 or Δvhb1 were indistinguishable from those mock inoculated with water. (b) Average of disease severity at 3 weeks post‐inoculation, calculated according to a 0–3 scale: 0, healthy leaf; 1, chlorotic leaf; 2, leaf with wilt symptoms; 3, fully necrotic leaf. Different letters indicate statistically significant differences (P < 0.05) according to a Bonferroni–Dunn test. Bars indicate standard error.
Figure 5.
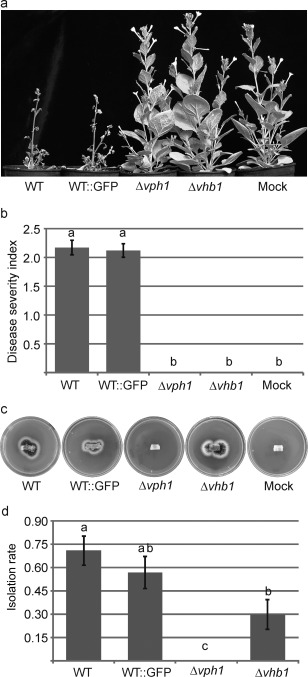
Deletion of vph1 or vhb1 renders the Verticillium dahliae strains avirulent towards Nicotiana benthamiana. (a) Nicotiana benthamiana plants inoculated with wild‐type (WT) and WT green fluorescent protein (GFP)‐tagged strains generated characteristic Verticillium wilt symptoms, whereas plants inoculated with Δvph1 or Δvhb1 were indistinguishable from those mock inoculated with water. (b) Average of disease severity at 7 weeks post‐inoculation, calculated according to a 0–3 scale: 0, healthy leaf; 1, chlorotic leaf; 2, leaf with wilt symptoms; 3, fully necrotic leaf. Different letters indicate statistically significant differences (P < 0.05) according to a Bonferroni–Dunn test. Bars indicate standard error. (c) Re‐isolation of V. dahliae strains from the vascular system of inoculated N. benthamiana plants. Images were taken after 15 days of incubation of the stem sections on potato dextrose agar (PDA) at 24 ºC. (d) Quantification of the frequency of re‐isolation from the vascular system. Eight plants were assayed per strain and the experiment was repeated three times. The re‐isolation rate was analysed by the application of generalized linear models (GzLMs) using Logit as the link function and Binomial as the underlying distribution. Different letters indicate statistically significant differences (P < 0.05) according to the sequential Bonferroni method for error correction. Bars represent the standard error.
To assay whether mutant strains were able to reach the vascular vessels, stem sections below the first internode of N. benthamiana plants were collected at 7 weeks post‐inoculation, and placed onto potato dextrose agar (PDA) plates. In plants inoculated with Δvph1, mycelium outgrowth from the vascular system was never observed. Unlike Δvph1, Δvhb1 could be re‐isolated from vascular tissue (Fig. 5c), although the rate of isolation was significantly lower in the Δvhb1 strain than in the WT (Fig. 5d).
Deletion of vph1 impairs penetration of the root cortex, whereas deletion of vhb1 affects proliferation in the xylem
The colonization of tomato root tissue by GFP‐tagged strains Δvph1 and Δvhb1 was visualized, and compared with that observed during WT development (WT::GFP) using confocal laser scanning microscopy (CLSM). We found that the combined use of CLSM and GFP‐tagged V. dahliae derivatives enabled the in situ localization of the fungus on/in tomato roots without any tissue manipulation, as reported previously for olive (Prieto et al., 2009). CLSM imagery showed that tomato roots were rapidly colonized by both mutant strains soon after inoculation [1 day post‐inoculation (dpi)], similar to the WT (Fig. 6a,e,i). The colonization of the tomato root surface by V. dahliae was not uniform; some root regions showed an abundance of hyphal colonization, whereas others were completely devoid of the fungus. In general, V. dahliae hyphae were more frequently found near root hairs (Fig. 6a,e,i). However, at the beginning of the infection process, we observed differences in host colonization between the two mutants, and between the mutants and WT. We found that the cortical tissue of tomato roots was internally colonized by Δvhb1 at early stages (4 dpi), similar to the observed colonization pattern of the WT strain (Fig. 6c,d,g,h). However, although we also observed that the WT internally colonized the vascular tissue at this time point (4 dpi) (Fig. 6c,d), Δvhb1 was not observed to proliferate in the vascular tissue at any time point (Fig. 6g,h). By contrast with both WT and Δvhb1, Δvph1 was only detected on the tomato root surface (Fig. 6k,l). Internal colonization of the cortical root tissue by this mutant was never observed at any time point up to the end of the experiment. The colonization pattern of each V. dahliae strain was similar at 4–11 dpi, when both Δvhb1 and Δvph1 were barely detectable in tomato roots, similar to the WT strain (data not shown). No significant changes in the sampled plants were observed from 11 dpi until the end of the experiment.
Figure 6.
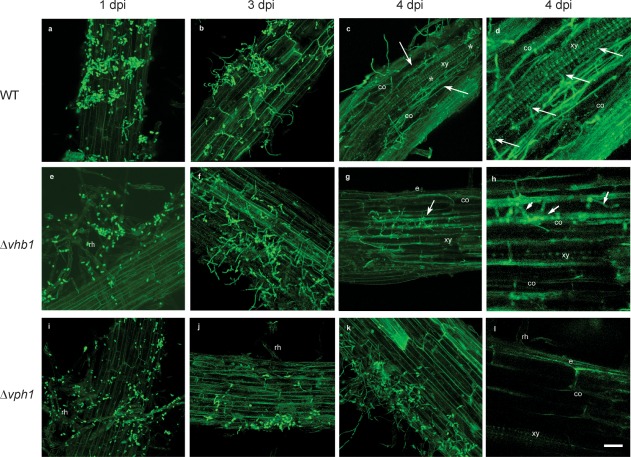
Confocal laser scanning microscopy (CLSM) images of the time course of the colonization process of tomato roots by Verticillium dahliae wild‐type (WT) and mutant green fluorescent protein (GFP)‐tagged strains (in green). Confocal analysis was performed on 3–4‐cm‐long roots to show surface and inner V. dahliae colonization. Images are projections of 25 adjacent confocal optical sections. The focal step between confocal optical sections was 0.5 μm. (a) Surface colonization of tomato roots by WT at 1 day post‐inoculation (dpi). (b) Surface colonization of tomato roots by WT at 3 dpi. Most of the conidia have germinated and proliferation of hyphae can be observed on the root surface. (c, d) Confocal optical sections of internal colonization (arrowed) and intercellular colonization of the vascular tissue (*) by WT at 4 dpi. (e) Surface colonization of tomato roots by Δvhb1 at 1 dpi. Root hairs are externally colonized by Δvhb1 conidia. (f) Surface colonization of tomato roots by Δvhb1 at 3 dpi. Proliferation of hyphae on the root surface is observed. (g, h) Intercellular colonization of the cortical tissue (arrowed) of the tomato root by Δvhb1 at 4 dpi. The vascular tissue was never invaded by Δvhb1. (i) Surface colonization of tomato roots by Δvph1 at 1 dpi. Root hairs are externally colonized by Δvph1 conidia. (j) Surface colonization of tomato roots by Δvph1 at 3 dpi. Only some hyphae were observed on the root surface. (k, l) Superficial colonization of the tomato root by Δvph1 at 4 dpi. The cortical tissue was never invaded by Δvph1. Scale bar represents 20 μm in all panels. co, cortical cells; e, epidermis; rh, root hairs; xy, xylem vessel cells.
Re‐introduction of the WT gene in the deletion mutants restores pathogenicity
To fully establish that the loss of pathogenicity observed in Δvph1 and Δvhb1 was a result of the deletion of the corresponding gene, we transformed each mutant with a construct containing the entire target gene ORF plus 1 kb of the flanking sequences to capture putative regulatory elements. A PCR analysis confirmed the re‐introduction of the corresponding target gene by random integration of the complementation constructs in all transformants tested (data not shown). Complemented transformed strains were selected for each of the genes, and designated as Δvph1C and Δvhb1C. Virulence assays using N. benthamiana as a host were performed with strains Δvph1C and Δvhb1C alongside the parental strain Dvd‐T5 and deletion mutants Δvph1 and Δvhb1. The results of these assays confirmed that, in the Δvph1 strain complemented with vph1, and in the Δvhb1 strain complemented with vhb1, pathogenicity was restored. The Δvph1C and Δvhb1C strains were as virulent as the WT strain Dvd‐T5 (Fig. S5a, see Supporting Information). Complementation of Δvhb1 with vhb1 also reversed the defect in spore production in liquid medium that was observed in the vhb1 deletion mutant (Fig. S5b).
The global transcription pattern of Δvph1 greatly differs from those of Δvhb1 and WT in a condition that mimics the penetration phase
To identify putative regulated targets of the Vph1 and Vhb1 TFs, we compared the global gene expression patterns in the mutants with those of the WT strain using microarray technology. As it is technically difficult to obtain V. dahliae gene expression profiles that are not strongly biased towards plant RNA during the initial stages of infection because of the small amount of fungus present, we investigated whether growth on plates covered with cellophane could be used as a condition to mimic the penetration phase in vitro. Interestingly, microscopic observations showed that the WT V. dahliae strain grown on cellophane produced numerous short branches with swollen tips, from which the penetration hyphae emerged (Fig. S6, see Supporting Information). These structures closely resembled those observed in planta (Vallad and Subbarao, 2008; Zhang et al., 2013; Zhao et al., 2014), which have been described recently as hyphopodia (Reusche et al., 2014; Zhao et al., 2016). Consistent with the differential behaviour in planta of Δvhb1 and Δvhp1 revealed by the re‐isolation from the vascular system assay and CLSM images, the former was able to penetrate cellophane paper, whereas the Δvph1 strain was not (Fig. 7a,b). Furthermore, the morphology of Δvhb1 grown on cellophane paper was indistinguishable from that of WT (Fig. 7c). By contrast, Δvph1 produced long filaments which were not swollen at the tips. The re‐introduction of the vph1 gene in Δvph1 restored the ability of the strain to penetrate cellophane paper and produce hyphopodium‐like structures (Fig. S7, see Supporting Information). We considered that growth on plates covered with cellophane could provide a good indication in vitro of the initial stages of infection, and extracted RNA from the WT and the deletion mutant strains grown in this condition for microarray analysis.
Figure 7.
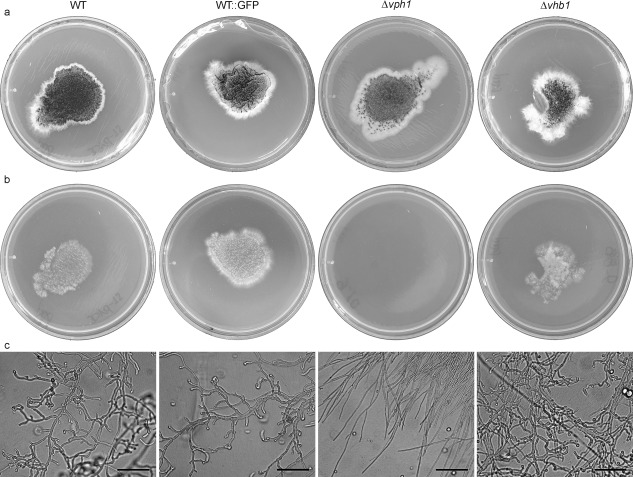
Growth of wild‐type (WT) and mutant Verticillium dahliae strains on potato dextrose agar (PDA) medium covered with cellophane membranes. (a) Strains grown on PDA covered with cellophane for 4 days at 24 °C. (b) Plates incubated at 24 °C for 24 h after removal of the cellophane membrane with the fungal colony. (c) Microscopic observation of strains grown on PDA covered with cellophane for 2 days. Scale bar, 50 µm.
We found that the global expression patterns of Δvph1 and WT grown on cellophane differed considerably: 434 genes had a differential expression of at least two‐fold in both strains (File S1, see Supporting Information). By contrast, WT and Δvhb1 showed very similar global transcription patterns when grown on cellophane; only 39 genes were differentially expressed by at least two‐fold in both strains, 23 of which were also identified as differentially expressed in WT vs. Δvph1 (Table 1 and File S2, see Supporting Information). The 434 genes differentially expressed in WT vs. Δvph1 were assigned to different biological and molecular functional categories (Fig. 8 and Files S3 and S4, see Supporting Information). Roughly one‐half of the differential genes identified could not be assigned to any functional category (Fig. 8). To validate microarray data, real‐time reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) was conducted to confirm differential expression in eight selected genes. The gene expression profiles determined by both microarray and RT‐qPCR show the same trend, with some variation in the magnitude of differential expression in six of the eight genes (Table S1, see Supporting Information).
Table 1.
Genes differentially expressed in the wild‐type (WT) vs. Δvhb1.
| Gene ID | Functional annotation | log2 FC WT vs. Δvhb1 | log2 FC WT vs. Δvph1 |
|---|---|---|---|
| VDAG_00107 | Guanyl‐specific ribonuclease F1 | −1.19 | −1.18 |
| VDAG_00169 | Conserved hypothetical protein | −1.72 | – |
| VDAG_00194 | CMR1 | −1.14 | – |
| VDAG_00195 | Pig1 | −1.85 | – |
| VDAG_00798 | Calphotin | −1.05 | – |
| VDAG_00900 | Predicted protein | −1.45 | −2.77 |
| VDAG_00901 | Killer toxin subunits α/β | −1.23 | −2.33 |
| VDAG_01659 | Ent‐kaurene oxidase | −1.77 | −1.64 |
| VDAG_01660 | FAD‐binding domain‐containing protein | −2.21 | −2.22 |
| VDAG_01661 | Conserved hypothetical protein | −2.40 | −2.33 |
| VDAG_01662 | Ent‐kaurene oxidase | −2.52 | −2.67 |
| VDAG_01663 | Ent‐kaurene oxidase | −2.33 | −2.34 |
| VDAG_02082 | β‐Glucanase | −1.60 | 2.38 |
| VDAG_02103 | Hypothetical protein | −3.38 | −4.51 |
| VDAG_02503 | Hypothetical protein | −1.24 | −1.63 |
| VDAG_02917 | Conserved hypothetical protein | 1.23 | 1.19 |
| VDAG_03477 | Predicted protein | −1.67 | −1.12 |
| VDAG_03551 | Pectate lyase | −1.71 | – |
| VDAG_03653 | Conserved hypothetical protein | −1.22 | −1.26 |
| VDAG_04535 | Aflatoxin biosynthesis ketoreductase nor‐1 | −1.12 | −1.13 |
| VDAG_04550 | Secreted protein | −2.18 | −2.26 |
| VDAG_04979 | Secreted aspartic proteinase | 2.11 | – |
| VDAG_05094 | FAD‐binding domain‐containing protein | −1.18 | – |
| VDAG_05303 | Conserved hypothetical protein | −1.18 | −1.26 |
| VDAG_06236 | Conserved hypothetical protein | −1.25 | −1.50 |
| VDAG_06930 | Conserved hypothetical protein | −1.54 | −2.04 |
| VDAG_07075 | 7‐Dehydrocholesterol reductase | −1.87 | – |
| VDAG_07145 | Vanillyl‐alcohol oxidase | −1.44 | – |
| VDAG_07608 | Exopolygalacturonase | −1.68 | – |
| VDAG_07682 | Hypothetical protein | −1.14 | – |
| VDAG_07817 | β‐Xylosidase | −1.06 | −1.26 |
| VDAG_08101 | Aldehyde dehydrogenase | −1.24 | – |
| VDAG_08741 | Endochitinase | 2.26 | – |
| VDAG_08956 | Hypothetical protein | −1.76 | – |
| VDAG_09137 | Conserved hypothetical protein | −1.37 | – |
| VDAG_09296 | Predicted protein | −1.28 | −1.68 |
| VDAG_09517 | Conserved hypothetical protein | −1.11 | – |
| VDAG_09977 | Conserved hypothetical protein | −1.27 | −1.94 |
| VDAG_09980 | Conserved hypothetical protein | −2.48 | −2.12 |
| VDAG_10270 | Conserved hypothetical protein | −1.65 | – |
FC, fold change.
Figure 8.
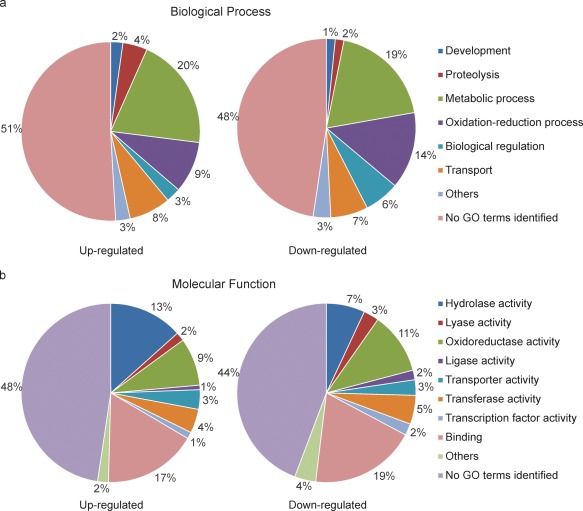
Functional categories of the genes under Vph1 regulation. The 207 genes induced in the presence of Vph1, i.e. up‐regulated in the wild‐type (WT) (left), and the 227 genes induced in the absence of Vph1, i.e. down‐regulated in WT (right), were assigned to several biological process categories (a) and molecular functional categories (b) according to the Gene Ontology Consortium database.
Interestingly, considering the behaviour in planta of Δvph1, a number of genes identified as differentially expressed in WT vs. Δvph1 coded for proteins potentially involved in the initial interaction with the host. The onset of pathogenic development is associated, for instance, with increased expression of secreted proteins (Ranganathan and Garg, 2009; de Sain and Rep, 2015). In V. dahliae, 7.4% of the genome is predicted to encode secreted proteins (Klosterman et al., 2011). We observed that 102 genes encoding secreted proteins were differentially expressed in WT vs. Δvph1, meaning that 13% of the secretome was differentially expressed in these two strains. A total of 29 carbohydrate‐active enzyme (CAZY) genes were also differentially expressed, 16 of which potentially encoded cell wall‐degrading enzymes (CWDEs); roughly two‐thirds of these genes were up‐regulated in WT compared with their expression in Δvph1. These CAZYs could play diverse roles associated with the infection process, including degradation of the plant cell walls, in addition to modifications of the fungal cell wall. Proteolysis activity was also induced in WT (Fig. 8). Proteases from fungal pathogens are associated with the degradation of host proteins during some interactions (Jashni et al., 2015). The WT condition was also enriched in genes involved in morphological development, suggesting that Vph1 might also be involved in the regulation of morphological changes that might be required at the onset of pathogenic development. The blast2go analysis assigned several hypothetical protein genes to this functional category, among them VDAG_05055, the gene with the highest level of induction in WT vs. Δvph1 (Table 2). We also identified five genes up‐regulated in WT highly similar to appressorium development‐associated genes. These included VDAG_08233 and VDAG_07904, putative homologues of Magnaporthe grisea GAS1 and GAS2 (Xue et al., 2002), respectively, VDAG_03883 also encoding a Gas1‐like protein, and VDAG_07677 and VDAG_08575, similar to Colletotrichum gloeosporioides CAP22 and CHIP3, respectively (Hwang and Kolattukudy, 1995; Kim et al., 2000).
Table 2.
Top 30 up‐regulated genes in the wild‐type (WT) vs. Δvph1.
| Gene ID | Functional annotation | Normalized intensity value | log2 FC | FC | |
|---|---|---|---|---|---|
| WT | Δvph1 | ||||
| VDAG_05055 | Conserved hypothetical protein | 1205.4 | 62.39 | 4.44 | 21.7 |
| VDAG_10293 | Predicted protein | 4384.1 | 307.24 | 4.14 | 17.7 |
| VDAG_04552 | Conserved hypothetical protein | 3389.2 | 260.78 | 3.97 | 15.7 |
| VDAG_07675 | Conserved hypothetical protein | 2442.9 | 221.5 | 3.57 | 11.9 |
| VDAG_01820 | Conserved hypothetical protein | 7526.4 | 887.55 | 3.48 | 11.2 |
| VDAG_07904 | MAS1 protein | 1171.2 | 117.29 | 3.41 | 10.6 |
| VDAG_02463 | Conserved hypothetical protein | 18141 | 2322 | 3.34 | 10.1 |
| VDAG_09643 | Predicted protein | 994.35 | 105.99 | 3.18 | 9.06 |
| VDAG_02618 | Conserved hypothetical protein | 533.19 | 59.6 | 3.15 | 8.86 |
| VDAG_06039 | Conserved hypothetical protein | 6935.3 | 1218.3 | 3.14 | 8.80 |
| VDAG_07668 | Conserved hypothetical protein | 1820.9 | 220.06 | 3.06 | 8.33 |
| VDAG_04811 | Conserved hypothetical protein | 1468.2 | 252.08 | 2.89 | 7.42 |
| VDAG_08840 | Conserved hypothetical protein | 4133.9 | 636.17 | 2.87 | 7.29 |
| VDAG_08181 | l‐Fucose transporter | 40273 | 6900.2 | 2.85 | 7.23 |
| VDAG_08476 | Cytochrome P450 61 | 1544.6 | 437.45 | 2.83 | 7.12 |
| VDAG_09229 | Conserved hypothetical protein | 509.63 | 132.45 | 2.81 | 7.01 |
| VDAG_05655 | Conserved hypothetical protein | 8104.2 | 1440.4 | 2.81 | 7.00 |
| VDAG_00625 | Sphingoid long‐chain base transporter RSB1 | 1844.9 | 405.36 | 2.79 | 6.93 |
| VDAG_07991 | Predicted protein | 430.81 | 62.23 | 2.78 | 6.89 |
| VDAG_05508 | Predicted protein | 462.59 | 94.79 | 2.68 | 6.42 |
| VDAG_09086 | Conserved hypothetical protein | 578.92 | 95.84 | 2.66 | 6.32 |
| VDAG_00445 | Triacylglycerol lipase | 210.1 | 55.01 | 2.59 | 6.02 |
| VDAG_08233 | CAS1 | 1122.8 | 192 | 2.58 | 5.98 |
| VDAG_08448 | Lovastatin nonaketide synthase | 1605 | 265.79 | 2.57 | 5.96 |
| VDAG_03556 | Laccase‐1 | 726.2 | 121.27 | 2.56 | 5.9 |
| VDAG_02697 | Conserved hypothetical protein | 1084.4 | 205.81 | 2.54 | 5.81 |
| VDAG_09655 | Conserved hypothetical protein | 166.61 | 39.35 | 2.41 | 5.31 |
| VDAG_07221 | Endoglucanase‐4 | 11351 | 2128.6 | 2.39 | 5.22 |
| VDAG_05448 | Metalloprotease | 2582.7 | 614.48 | 2.38 | 5.2 |
| VDAG_02082 | β‐Glucanase | 220.66 | 40.65 | 2.38 | 5.2 |
FC, fold change.
Several genes previously characterized in V. dahliae were also differentially expressed in WT vs. Δvph1. For example, the WT up‐regulated gene VDAG_06010 encodes an nucleotide‐rhamnose synthase/epimerase‐reductase (NRS/ER) protein involved in the synthesis of rhamnose and found to be required for virulence in V. dahliae (Santhanam et al., 2016). Similarly, VDAG_05890, encoding a cytochrome P450 monooxygenase (VdCYP1) with a role in virulence (Zhang et al., 2016), was also induced in WT in our analysis. Intriguingly, two necrosis and ethylene‐inducing protein (NLP) genes, VDAG_1995 (NLP2), previously characterized as induced in planta and involved in virulence (Santhanam et al., 2013; Zhou et al., 2012), and VDAG_04550 (NPL4), were down‐regulated in WT vs. Δvph1.
Genes encoding proteins potentially involved in oxidation–reduction processes were more abundant in the population of genes down‐regulated in WT; 9% of the genes identified as up‐regulated in WT and 14% of the genes identified as down‐regulated in WT fell into this category (Fig. 8). Repression of secondary metabolism in WT appeared to account in part for these differences. Thus, a putative cluster containing four putative cytochrome p450 genes, VDAG_01659, VDAG_01662, VDAG_01661 and VDAG_01663, was down‐regulated in WT. Genes from VDAG_08284 to VDAG_08291 comprised another cluster repressed in WT. Genes from VDAG_00900 to VDAG_00904 were also down‐regulated in WT; three of these five genes encode representatives of the secretome. Several of the genes from these clusters were amongst those with the highest levels of down‐regulation in WT (Table 3), which may further indicate that their repression plays a role in the early infection process.
Table 3.
Top 30 down‐regulated genes in the wild‐type (WT) vs. Δvph1.
| Gene ID | Functional annotation | Normalized intensity value | log2 FC | FC | |
|---|---|---|---|---|---|
| WT | Δvph1 | ||||
| VDAG_02103 | Hypothetical protein | 1031.77 | 18844.21 | 4.51 | 22.81 |
| VDAG_05420 | Ent‐kaurene oxidase | 215.91 | 6993.33 | 4.32 | 19.99 |
| VDAG_04019 | Secretory phospholipase A2 | 388.68 | 1956.21 | 3.95 | 15.44 |
| VDAG_03342 | Phosphate‐repressible phosphate permease | 72.20 | 1092.86 | 3.63 | 12.41 |
| VDAG_03624 | Hypothetical protein | 267.02 | 1780.19 | 3.19 | 9.14 |
| VDAG_07384 | Predicted protein | 94.34 | 745.50 | 3.03 | 8.17 |
| VDAG_04530 | UVI‐1 | 335.42 | 4737.90 | 2.97 | 7.81 |
| VDAG_01167 | Multidrug resistance protein CDR1 | 60.47 | 414.80 | 2.92 | 7.55 |
| VDAG_08032 | Conserved hypothetical protein | 269.44 | 1786.48 | 2.81 | 7.01 |
| VDAG_07494 | Secreted protein | 2694.99 | 9754.51 | 2.79 | 6.93 |
| VDAG_00900 | Predicted protein | 2694.99 | 9754.51 | 2.77 | 6.83 |
| VDAG_02908 | Secretory phospholipase A2 | 321.46 | 1778.62 | 2.74 | 6.68 |
| VDAG_10133 | Reticulon‐4‐interacting protein | 1358.43 | 6852.54 | 2.74 | 6.67 |
| VDAG_01662 | Ent‐kaurene oxidase | 177.57 | 893.70 | 2.67 | 6.37 |
| VDAG_02238 | Predicted protein | 5965.96 | 22152.21 | 2.62 | 6.13 |
| VDAG_01054 | Conserved hypothetical protein | 87.05 | 498.31 | 2.60 | 6.08 |
| VDAG_02468 | Hypothetical protein | 2841.28 | 8030.44 | 2.57 | 5.95 |
| VDAG_08870 | YcaC | 319.38 | 1846.23 | 2.54 | 5.81 |
| VDAG_03732 | Hypothetical protein | 869.13 | 3521.20 | 2.48 | 5.59 |
| VDAG_05374 | MFS hexose transporter | 40.81 | 164.74 | 2.46 | 5.49 |
| VDAG_08288 | Catechol O‐methyltransferase | 686.96 | 1726.44 | 2.45 | 5.47 |
| VDAG_03684 | Predicted protein | 120.19 | 467.66 | 2.45 | 5.46 |
| VDAG_07138 | Conserved hypothetical protein | 35.98 | 132.94 | 2.42 | 5.35 |
| VDAG_05122 | Hypothetical protein | 1609.74 | 7685.12 | 2.35 | 5.12 |
| VDAG_01663 | Ent‐kaurene oxidase | 2062.62 | 10816.07 | 2.34 | 5.07 |
| VDAG_06028 | Rds1 | 1545.35 | 3797.40 | 2.33 | 5.04 |
| VDAG_01661 | Conserved hypothetical protein | 283.00 | 1363.72 | 2.33 | 5.03 |
| VDAG_00901 | Killer toxin subunits α/β | 161.28 | 854.43 | 2.33 | 5.02 |
| VDAG_00902 | LysM domain‐containing protein | 176.61 | 834.47 | 2.33 | 5.01 |
| VDAG_07616 | Conserved hypothetical protein | 2166.41 | 8621.91 | 2.32 | 4.98 |
FC, fold change.
In yeast, Ste12 can bind specific DNA sequences and also relies on other TFs to bind DNA and regulate the expression of different sets of target genes. The pheromone response element (PRE) sequence TAGAAACA has been identified as being directly bound by Ste12; at least two PREs are required for efficient binding (reviewed in Wong Sak Hoi and Dumas, 2010). In filamentous fungi, very few studies have focused on the identification of the DNA sequences recognized by Ste12‐like proteins, and little is known about how they regulate their target genes. It has been suggested that the highly conserved Ste motif could bind similar DNA sequences in different species. Indeed, Clste12, a Ste12 protein from C. lindemuthianum, recognizes PRE sequences (Wong Sak Hoi and Dumas, 2010; Wong Sak Hoi et al., 2007). In this work, we used a bioinformatics approach to determine whether the promoters of the genes differentially expressed in WT vs. Δvph1 were enriched in PREs. We analysed 1000 bp upstream of the start codon of the 434 differentially expressed genes, a region likely to capture the cis‐regulatory sites, identifying a single PRE in 24 of them and two PREs in only one gene. When the same approach was applied to the entire V. dahliae strain VdLs.17 annotated genome, a single PRE was identified in the putative promoter of 413 genes and two PREs were identified in 11 genes. This means that there is a slight enrichment in PREs in the promoter region of the differentially expressed genes: 5.8% of them contained this element vs. 4% in the general population. However, this slight enrichment was not considered to be significant as the frequency of identification of PREs in the differentially expressed genes was not higher than the expected frequency of random occurrence, and usually a single PRE was present.
Discussion
Regulators of transcription are key downstream elements in the signalling pathways that control cellular processes. In plant‐pathogenic fungi, the functional characterization of these regulatory proteins, the identification of their target genes and the analysis of how they interact with different signalling pathways are critical for an understanding of the molecular mechanisms controlling virulence. In this work, using forward and reverse genetics approaches, we have identified two previously uncharacterized TFs in the vascular pathogen V. dahliae, designated Vph1 and Vhb1, and have shown that they are both critical for virulence, but act at different stages of pathogenic development.
Vph1 is a putative Ste12 homologue in V. dahliae. Ste12 proteins contain a homeodomain‐like motif in the N‐terminus which is divergent from typical homeodomains (Wong Sak Hoi and Dumas, 2010). The Ste12 proteins examined in filamentous fungi additionally contain zinc finger C2H2‐type domains in the C‐terminus (Rispail and Di Pietro, 2010; Wong Sak Hoi and Dumas, 2010). We found that the Vph1 protein possessed these hallmark features of Ste12 homologues in filamentous fungi, and that, as in other species, alternative splicing could generate truncated and full‐sized versions of Vph1 containing one and two zinc fingers, respectively.
In yeast, Ste12 is a key downstream target of the Fus1/Kss1 MAPK pathway controlling mating and filamentation (Elion et al., 2005). A similar MAPK cascade containing a single Fus1/Kss1 homologue, usually referred to as Pmk1, appears to play a conserved role in the regulation of invasive growth in filamentous pathogenic fungi (Rispail and Di Pietro, 2010; Wong Sak Hoi and Dumas, 2010). Mutants lacking Ste12 activity and those lacking Pmk1 are both affected in penetration and invasive growth (Rispail and Di Pietro, 2010). This provides indirect evidence that, in these species, Ste12 might also function as a downstream target of MAPK. Nevertheless, Ste12 is thought to regulate a smaller subset of functions under MAPK regulation as the phenotypes of ste12 mutants are less pleiotropic than those of MAPK genes (Rispail and Di Pietro, 2010). In V. dahliae, a putative homologue of Pmk1, designated Vmk1, was functionally characterized (Rauyaree et al., 2005). Mutants lacking vmk1 were severely impaired in their ability to cause disease and were also affected in microsclerotia production and sporulation. We found that Δvph1 strains were not virulent, but were not affected in microsclerotium and spore production. This result suggests that Vph1 could be a downstream target of Vmk1 in the regulation of invasive growth, but not of developmental processes associated with vegetative growth. However, we found that deletion of vph1 had a greater impact on virulence than that reported previously in mutants disrupted for vmk1. The latter were severely reduced in virulence (Rauyaree et al., 2005), whereas Δvph1 strains were non‐pathogenic in three different hosts examined in this study and were never re‐isolated from the vascular system of inoculated plants. In vivo CLSM analysis confirmed that Δvph1 was affected in the penetration phase. Furthermore, when grown in vitro on cellophane, Δvph1 strains lacked the structures resembling hyphopodia characteristic of the WT grown in this condition. Other components of MAPK signalling have been functionally characterized in V. dahliae, including homologues of the transmembrane mucin Msb2 and the tetraspan transmembrane protein Sho1 (Qi et al., 2016; Tian et al., 2014). Msb2 and Sho1 are known upstream components of the HOG MAPK pathways, but a link with Pmk1 signalling has also been established. For instance, in F. oxysporum, Sho1 promotes the phosphorylation of Fmk1 and cooperates with Mbs2 to regulate invasive growth (Pérez‐Nadales and Di Pietro, 2015). In V. dahliae, VdMsb regulates MS development, stress responses and virulence (Tian et al., 2014), sharing roles with Vmk1 (Rauyaree et al., 2005) and MAPK proteins from the HOG pathway (Qi et al., 2016; Tian et al., 2016). The absence of each of these MAPK signalling components played a role in reducing virulence in V. dahliae. However, this effect was not as pronounced as in Δvph1, where pathogenic development was abolished. Different pathways controlling the onset of pathogenic development in V. dahliae could therefore function upstream of Vph1.
To identify potential targets of Vph1 during the penetration phase, we performed a microarray analysis of WT and Δvph1 grown on cellophane paper. This analysis showed that the remarkable growth differences of WT and Δvph1 observed in this condition were associated with clear differences in transcription pattern profiles. A number of the genes identified as differentially expressed in both strains encoded proteins for which a potential role in the onset of pathogenic development could be proposed. For instance, the WT condition was enriched in genes encoding secreted proteins. Three of these, VDAG_08265, VDAG_09614 and VDAG_05848, corresponded to secreted aspartic proteases (SAPs), which, in several pathogenic fungi, are known to be critical virulence factors. VDAG_08265 is highly similar to aspergillopepsin F, involved in host invasion in Aspergillus fumigatus (Lee and Kolattukudy, 1995), whereas VDAG_09614 and VDAG_05848 exhibit the highest similarities to Candida albicans candidopepsins Sap2 and Sap3. These proteases play roles in adhesion and host invasion and, for Sap2, a direct role in the hydrolysis of proteins of the host immune system was confirmed (Naglik et al., 2004). Interestingly, the Ste12 homologue Cph1 is amongst the transcriptional activators of SAP genes in C. albicans (Naglik et al., 2004). VDAG_05448, related to Mep1 extracellular metalloproteases, was also induced in WT. Cocciodioides posadasii Mep1, which exhibits significant similarity to VDAG_05448, is secreted by endospores and digests the cell surface antigen SOWgp, thus avoiding detection by the host cell defences (Hung et al., 2005).
The WT condition was also enriched in genes potentially encoding CAZYs; 19 of the 29 differentially expressed CAZY genes were up‐regulated in WT. Moreover, 11 of the CAZY genes which were up‐regulated in WT encoded putative CWDEs, whereas only four genes which were down‐regulated in WT corresponded to this category. The homologies exhibited by a number of CAZYs clearly pointed to a role in the degradation of the plant cell walls. Thus, Vph1 regulates certain different activities controlling the initiation of infection, such as the production of secreted proteinases potentially involved in adhesion, host tissue damage and degradation of host proteins to avoid recognition, and also the production of CAZYs involved in the degradation of plant cell walls to facilitate host tissue penetration.
However, a few of the CAZY genes up‐regulated in WT could play a role in fungal cell wall dynamics associated with the infection process, rather than in plant cell wall degradation. For example, VDAG_07727 is highly similar to chitin deacetylases which deacetylate chitin to chitosan, a change in the cell wall that might allow certain fungal pathogens to escape recognition by the host. The fungal pathogen C. lindemuthianum secretes an endo‐chitin de‐N‐acetylase (ClCDA) to modify exposed hyphal chitin during penetration and infection of plants (Blair et al., 2006). The cell wall of Cryptococcus neoformans predominantly contains chitosan. Upadhya et al. (2016) showed that deletion of the three chitin deacetylase genes of C. neoformans renders strains avirulent. VDAG_09238 was highly similar to dfg5 which encodes a glycosylphosphatidylinositol (GPI)‐anchored cell wall α‐1,6‐mannanase required for the incorporation of glycoproteins into the cell wall (Ao et al., 2015; Maddi et al., 2012).
In a number of air‐borne pathogens, MAPK‐deficient mutants are not able to differentiate appressoria, whereas strains lacking Ste12 are still able to produce appressoria, although not functional (Rispail and Di Pietro, 2010). Microscopic analyses using GFP‐tagged V. dahliae showed that this species produces structures not highly differentiated, but similar to appressoria swellings on root surfaces (Vallad and Subbarao, 2008; Zhang et al., 2013; Zhao et al., 2014), although the contribution of these structures to the penetration of host tissue remains unclear. We observed that V. dahliae WT, when grown on cellophane paper, produces similar hyphal swellings, and that the Δvph1 strains were clearly impaired in their ability to generate them. Furthermore, a number of genes related to appressoria development were identified in the microarray analysis, all exhibiting induced levels of expression in WT. These included VDAG_07904 and VDAG_08233, which were 10 and six times more strongly expressed in WT than in the mutant, respectively, and which appear to be homologues of M. grisea GAS2 and GAS1, specifically expressed in appressoria and under Pmk1 regulation (Xue et al., 2002). Mutants lacking GAS1 or GAS2 are defective in penetration. These results suggest that the Ste12 homologue Vph1 might control, amongst other aspects related to virulence, the differentiation of infection structures, potentially under MAPK regulation.
An increasing number of studies have focused on the characterization of V. dahliae TFs. However, there are still very few reports on the mechanisms by which they regulate gene expression. The absence of Vdpf affects the expression of genes encoding components of cAMP/G‐protein signalling, amongst other genes involved in morphogenesis and virulence (Luo et al., 2016). This de‐regulation of cAMP/G‐protein signalling could contribute to the reduced virulence displayed by mutants lacking Vdpf (Luo et al., 2016). The potential sequences recognized by Vdpf in the promoter of its target genes were found to correspond to the binding sites of GAL4 domain‐containing TFs (Luo et al., 2016). In the Δvph1 strain, in which pathogenic development is abolished, global transcription profiling determined that genes encoding components of major signalling pathways, including MAPK, were not differentially expressed compared with WT. Our results indicate that Vph1 might function mainly through the regulation of genes directly involved in the establishment of pathogenic development, rather than the regulation of signalling associated with virulence. Little is known about the DNA sequences recognized by Ste12 homologues in filamentous fungi. However, the DNA‐binding Ste motif is highly conserved and, in C. lindemuthianum, ClSte12 has been shown to be able to bind PRE sequences, identified as direct binding sites of Ste12 in yeast (Wong Sak Hoi et al., 2007). We used a bioinformatics approach to investigate the presence of PREs in V. dahliae putative promoter sequences. We found that PREs were present at a frequency similar to the expected frequency of random occurrence in both the differentially expressed genes and the entire V. dahliae annotated genome. This result strongly suggests that the mechanism of action of the Ste12 homologue in V. dahliae does not involve binding to PREs in the promoter region of its target genes. Putative DNA sequences recognized by Vph1 need to be determined experimentally.
As it has been documented in yeast, Vph1 might rely on other TFs to bind DNA. The identification of Vph1‐interacting partners is also of great interest to characterize how this TF regulates different target genes. In Sordaria macroscopa, the Ste12 homologue interacts with the MADS‐box TF Mcm1, an interaction which is mediated by the Ste motif (Nolting and Pöggeler, 2006). In V. dahliae, functional characterization of an Mcm1 homologue, VdMcm1, has been reported recently, and, in addition, RNA‐sequencing (RNA‐seq) was used to identify its putative targets (Xiong et al., 2016). The phenotype displayed by VdMcm1 deletion mutants is not similar to that of Δvph1; unlike the latter, ΔVdMcm1 exhibits clear defects in vegetative growth. With regard to virulence, the impact of vph1 deletion was higher than that of VdMcm1 deletion. Moreover, comparison of their transcription profiles revealed little overlap between VdMcm1‐ and Vph1‐regulated genes. For example, although both TFs appeared to regulate the expression of secreted protein genes, only 12 of the 102 secreted protein genes identified as repressed in ΔVdMcm1 (Xiong et al., 2016) were also differentially expressed in Δvph1 vs. WT. Intriguingly, 11 of these genes were up‐regulated rather than down‐regulated in Δvph1 compared with WT. Taken together, these results indicate that, in V. dahliae, the MADS‐box VdMcm1 is not a major interacting partner of the Ste12 homologue Vph1.
Strains lacking the homeobox TF Vhb1, the other regulatory protein characterized in this work, were also avirulent when tested on N. benthamiana, tomato and lettuce. However, in contrast with Δvph1, Δvhb1 was not affected in the penetration phase, but later on in its ability to colonize the vascular system. CLSM of GFP‐tagged strains clearly showed in vivo the ability of the mutant lacking vhb1 to penetrate the root cortex, similar to WT. However, in contrast with WT, Δvhb1 was not detected in the xylem. In Verticillium species, it has been proposed that, on reaching the xylem, the fungus undergoes a sporulation process to generate a unicellular form, which is carried upwards in the transpiration stream, playing an important role in vascular colonization (Puhalla and Bell, 1981). Within the homeobox family of TFs, Vhb1 showed the highest similarities to M. oryzae MoHox2/MoHtf1 (Kim et al., 2009; Liu et al., 2010) and Htf1 homologues in Fusarium species (Zheng et al., 2012), which all play important roles in sporulation. We found that V. dahliae vhb1 also plays a role in sporulation; although Δvhb1 strains produced normal verticillate conidiophores on solid medium, they exhibited significantly reduced sporulation rates in liquid medium. In M. oryzae and F. graminearum, Htf1 plays a key role in sporulation, but is dispensable for virulence (Liu et al., 2010; Zheng et al., 2012). By contrast, our characterization of Vhb1 in the vascular pathogen V. dahliae provides the first report of a putative homologue of Htf1 proteins with a critical role in host proliferation and virulence. Interestingly, as mentioned above, Δvhb1 was specifically affected in a phase of pathogenic development, proliferation in the xylem, which has been associated with sporulation. Further experimentation will be required to determine whether the role of vhb1 in the colonization of the vascular vessels is associated with a role in the regulation of sporulation mechanisms associated with virulence.
The results of an in vitro penetration assay were in agreement with the observation in vivo that Δvhb1 was not impaired in its ability to penetrate the root cortex. Thus, Δvhb1 was able to penetrate cellophane paper and reach the underlying medium, and grew on this surface generating numerous short branches with swollen tips that resembled penetration pegs; this behaviour was indistinguishable from that of WT. In addition, the transcription pattern profiles of Δvhb1 and WT were very similar when grown in a condition that mimicked the penetration phase. This result indicates that Vhb1 may control the expression of genes whose induction/repression is required at later stages of pathogenic development. Interestingly, 36 of 39 genes identified as differentially expressed in WT vs. Δvhb1 were induced in the absence of vhb1 (i.e. up‐regulated in the mutant). When the stringency was lowered to consider additional genes as differentially expressed, most genes were still identified as repressed in WT (data not shown). These results suggest that Vhb1 might function as a repressor of gene expression. Further investigation is required to confirm this point and to identify the targets of Vhb1 during the colonization of the xylem.
The identification of key regulators of the different stages of pathogenic development is of great interest to dissect the progression of the disease cycle in V. dahliae at the molecular level. In this work, we have characterized two TFs, Vph1 and Vhb1, which are critical for root cortex penetration and proliferation in the vascular system, respectively. Similar to other homeodomain‐like Ste12 homologues in plant pathogens, Vph1 is critical for invasive growth. A transcriptomic analysis determined that Vph1 might regulate diverse activities required to establish infection, which are worth exploring in future work. The analysis of Vph1 regulation, including the elucidation of the putative role of alternative splicing, will also be of great interest. Vhb1, containing a typical homeodomain, was required for fungal proliferation in the xylem and its absence also resulted in the absence of Verticillium wilt symptoms. Strains with altered Vhb1 function might therefore be used as tools to identify activities required for vascular colonization. In this sense, it will be of interest not only to identify putative targets of Vhb1 during initial stages of vascular colonization, but also to determine whether the plant responses potentially associated with the development of symptoms are affected in infections with Δvhb1.
Experimental Procedures
Fungal strains and growth conditions
Verticillium dahliae strain DvdT5 (Dobinson et al., 1996) was used as parental strain to generate deletion mutants. Strains were maintained on PDA (Difco, Le Pont de Claix, France). Transformants were propagated on PDA supplemented with hygromycin B (50 µg/mL) or nourseothricin (50 µg/mL), according to the selection marker used in the transformation construct. Plates were incubated at 24 °C. Potato dextrose broth (PDB, Difco) or YEPS (1% yeast extract, 2% bactopeptone, 2% sucrose) were used for liquid cultures, which were incubated at 24 °C and 250 rpm.
Isolation and analysis of the genomic sequences flanking T‐DNA insertion(s)
To identify the gene(s) whose mutation gave rise to the observed phenotypic alterations in the random mutant D‐10–4F, the inverse PCR approach developed by Ochman et al. (1988) was employed. For the digestion of genomic DNA, enzymes HindIII and BamHI were used. Inverse PCR of the circularized DNA molecules was performed with the primer combinations Dvps3F_7751/Dvps3R_‐880 and Dvps3F_7751/Hyg‐R(210) (Table S2, see Supporting Information) for HindIII and BamHI digestions respectively. A second round of amplification was then performed with an inner set of primers: primer combinations attL3F/attR2R and attL3F/Hyg‐R(210). PCR products were sequenced and sequences were aligned with the V. dahliae genome to identify the point of insertion(s) of the T‐DNA.
Deletion of vph1 and vhb1 and complementation of the deletion mutants with the corresponding deleted gene
Deletion constructs were generated using the OSCAR method (Paz et al., 2011), but replacing the vector pA‐Hyg‐OSCAR by pA‐Hyg‐GFP‐OSCAR containing a synthetic SGFP‐TYG cassette (Maor et al., 1998), so that the transformants obtained constitutively expressed GFP (see Fig. S1). Primers used to amplify 1 kb of the sequences flanking vph1 (VDAG_06555) and vhb1 (VDAG_08786) are listed in Table S2. ATMT of V. dahliae was performed as described by Dobinson et al. (2004) with minor modifications. Individual transformants were analysed for gene replacement by PCR and Southern blot analysis. PCR analysis was performed with the primer combinations Vph1F_1676/Vph1R_2147 and Vhb1_670F/Vhb1_1114R (Table S2), which amplified roughly 400 bp of vph1 and vhb1 ORFs, respectively. Deletion of vph1 or vhb1 in transformants that did not amplify the ORF band was further confirmed by Southern blot analysis. A deletion mutant from each gene was selected for a Southern blot analysis study with probes derived from the ORFs and the hygR marker and genomic DNAs digested with several different restriction enzymes (see Fig. S2). To confirm deletion, genomic DNAs were digested with either HindIII or XhoI and simultaneously hybridized with probes derived from vph1 and vhb1 ORFs, amplified using the PCR DIG DNA labelling kit (Roche, Basel, Switzerland). The HindIII digestion generated hybridization bands of about 1.4 and 11 kb for the WT allele of vhb1 and vph1, respectively; the presence of only the 1.4‐kb band confirmed the deletion of vph1, whereas the presence of only the 11‐kb band confirmed the deletion of vhb1, in the transformants obtained with the respective deletion constructs. Restriction digestion with XhoI generated hybridization bands of approximately 1.5 and 1.3 kb for the WT allele of vhb1 and vph1, respectively. The presence of only the 1.5‐ or 1.3‐kb band also confirmed deletion in Δvph1 and Δvhb1, respectively (Fig. S2). To determine whether more than one integration event of the T‐DNA occurred in these deletion mutants, the probes were stripped from the membranes and these were re‐hybridized with a probe corresponding to the hygR marker, amplified with primers HygF_Oscar and HygR_Oscar (Table S2). The presence of single hybridization bands, a 4.3‐kb band when DNAs were digested with HindIII and a 5.8‐kb band when DNAs were digested with XhoI, confirmed the absence of additional integration events in Δvph1 (Fig. S2). These digestions could not distinguish single from multiple integration events in vhb1 as the vhb1 deletion construct contained two internal restriction sites for each enzyme. Two additional Southern blot hybridizations were therefore performed, digesting genomic DNAs with either EcoRV or XbaI, to confirm the absence of secondary integration events in Δvhb1. In both cases, the single expected bands indicative of gene replacement, a band of 7.9 kb for EcoRV digestion and a band of 9.7 kb for XbaI digestion, were detected (Fig. S2).
To re‐introduce vph1 and vhb1 in the corresponding deletion mutant, thus generating complemented strains Δvph1C and Δvhb1C, a construct was generated for each gene containing the entire ORF plus approximately 1 kb of the flanking sequences, and was ectopically integrated into the genome. To amplify the vph1 and vhb1 fragments containing the gene ORF plus flanking sequences, primer combinations Vph1_‐1200F_attB2r/Vph1_3232R_attB1r and Vhb1_‐1031F_attB2r/Vhb1_2950R_attB1r (Table S2), respectively, were used, together with Thermo Scientific Phusion High Fidelity DNA Polymerase. The amplified 4432‐bp vph1 and 3981‐bp vhb1 bands were gel purified and cloned into the binary vector pOSCAR (Paz et al., 2011). As deletion mutants were hygromycin resistant, nourseothricin (NTC) resistance was used for selection in these complementation constructs. With this purpose, pA‐NTC‐OSCAR was used as the marker donor vector in the OSCAR BP clonase reaction (Paz et al., 2011). On ATMT of V. dahliae Δvph1 and Δvhb1 strains with the corresponding construct, the presence of the WT vph1 and vhb1 genes was confirmed by PCR using primer combinations Vph1F_1676/Vph1R_2147 and Vhb1_670F/Vhb1_1114R (Table S2), respectively.
Plant infection assays
Pathogenicity assays with N. benthamiana and Solanum lycopersicum L. variety Moneymaker were performed following a root dip method (Wellman, 1939), with minor modifications as described in Sarmiento‐Villamil et al. (2016). Pathogenicity tests were also performed with Lactuca sativa as a host using a soil‐based root drench method (Klosterman and Hayes, 2009). Three independent experiments were conducted with each of the three hosts.
To test the ability of strains to reach the vascular system in N. benthamiana infections, at 7 weeks post‐inoculation, stem sections around the first internode were collected and placed on PDA medium. PDA plates were incubated at 24 ºC for 15 days observing fungal outgrowth. Eight plants per treatment were assayed in this way and the experiment was repeated three times.
Assessment of V. dahliae root colonization by CLSM
Colonization of tomato roots by WT, Δvph1 and Δvhb1 strains was visualized in S. lycopersicum L. variety Moneymaker using CLSM. Tomato seedlings were inoculated with WT and mutant GFP‐tagged strains, as described previously for the pathogenicity assays. For microscopic observation, tomato plants were carefully uprooted and the root systems were washed by dipping them in tap water to remove attached sand particles. Roots from plants inoculated with each strain were thoroughly analysed by CLSM, as described previously (Prieto et al., 2009). Several root segments (length, 4–5 cm) representative of the whole root system of each tomato treatment were analysed using confocal microscopy during a period of over 20 dpi to monitor V. dahliae colonization. Inoculated plants were sequentially taken. Three plants per day were taken from 1 to 5 dpi, whereas two plants were taken every 2 days from 5 to 15 dpi and another two plants at 21 dpi from each treatment.
Colonization of tomato roots by V. dahliae was analysed from three‐dimensional (3D) confocal optical stacks collected using an Axioskop 2 MOT microscope (Carl Zeiss Inc., Jena, Germany) equipped with an argon laser, controlled by Carl Zeiss Laser Scanning System LSM5 PASCAL software (Carl Zeiss Inc.). GFP‐tagged fungal cells were excited with the 488‐nm argon laser line and detected in the 500–520‐nm window. CLSM data were recorded and then transferred for analysis to Zeiss LSM Image Browser version 4.0 (Carl Zeiss Inc.). Projections from adjacent confocal optical sections were made to build up the images finally selected for presentation in this study. Figures were processed with PHOTOSHOP 4.0 software (Adobe Systems Inc., San Jose, CA, USA).
Cellophane penetration assay
Cellophane penetration assays were carried out according to Prados‐Rosales and Di Pietro (2008) with minor modifications. Briefly, 105 conidia of each strain were plated on PDA covered with cellophane sheets. Plates were incubated at 24 °C and samples were taken for observations using light microscopy (Nikon Optiphot microscope, Tokyo, Japan) at regular intervals: 24, 48, 72 and 96 h. After 96 h, the cellophane sheet with the fungal colony was removed and the ability of the different strains to penetrate the cellophane membrane and reach the underlying medium was assessed by incubating the plates for an additional 24 h. Three replicates of each strain were included in each assay and five independent assays were performed.
RNA sample preparation and microarray analysis
For the microarray analysis, 105 conidia of each strain were plated on PDA covered with cellophane membranes. After 48 h of growth at 24 ºC in the dark, mycelia from three independent plates of each strain were scraped off the cellophane membranes, mixed and ground to a fine powder under liquid nitrogen. Total RNAs were extracted using a Spectrum™ Plant Total RNA Kit (Sigma‐Aldrich, Saint Louis, MO, USA). RNA integrity was confirmed by Tape Station using the R6K Screen Tape (Agilent Technologies, Santa Clara, CA, USA).
Verticillium dahliae custom array design, probe preparation, hybridizations and scanning and data processing were performed by Bioarray (Elche, Spain) as described in Sarmiento‐Villamil et al. (2016). Genes were designated as differentially expressed if their differential expression ratios were at least two‐fold, with P < 0.05 (moderated t‐tests) according to a parametric test. For identification, the genes were assigned the same ID as in the annotated strain VdLs.17 genome (Klosterman et al., 2011). Differentially expressed genes were classified into molecular function and biological process categories according to the Gene Ontology (GO) Consortium database (http://www.geneontology.org/) using blast2go® software (version 3.0) (http://www.blast2go.de) with hypergeometric test P < 0.001.
RT‐qPCR
For RT‐qPCR, fungal samples were prepared and total RNAs were extracted as described previously for the microarray analysis. The mRNAs were transcribed to cDNA using MultiScribe Reverse Transcriptase (Life Technologies). The qPCRs were carried out using 5 × HOT FIREPol EvaGreen qPCR Mix Plus (Solis BioDyne, Tartu, Estonia), following the manufacturer's instructions, in a StepOne Thermal Cycler (Applied Biosystems, Foster City, CA, USA) obtaining the quantification cycle (Cq) values from OneStep Software®. Relative expression levels were determined by the ΔΔCt method based on two technical replicates and three biological replicates per sample and using actin and GADPH (glyceraldehyde 3‐phosphate dehydrogenase) as housekeeping genes. The primers used for RT‐qPCR are listed in Table S2.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Modified version of the One Step Construction of Agrobacterium‐Recombination‐ready‐plasmids (OSCAR) method used to generate deletion mutants in this work. We used OSCAR (Paz et al., 2011), but replaced vector pA‐Hyg‐OSCAR with a new version containing a synthetic SGFP‐TYG gene under the regulation of the Aspergillus nidulans Pgdp promoter. To construct the new vector, the SGFP‐TYG cassette was excised from vector gGFP (Maor et al., 1998) by digestion with ClaI and XbaI, blunt ended using T4 DNA polymerase and ligated into pA‐Hyg‐OSCAR (Paz et al., 2011), digested with BamHI and also blunt ended. Two vectors were generated in this way, pA‐Hyg‐GFP‐OSCAR‐5 (shown in figure) and pA‐Hyg‐GFP‐OSCAR‐6, which differ only in the orientation of the green fluorescent protein (GFP) cassette. Deletion constructs generated with pA‐Hyg‐GFP‐OSCAR vectors generate deletion mutants in which the target gene open reading frame (ORF) is replaced by a hygR marker and the SGFP‐TYG cassette.
Fig. S2 Confirmation of vhb1 and vph1 deletion and absence of secondary integration events in the mutants by Southern blot analyses. The samples analysed included: parental strain Dvd‐T5 (WT), a transformant with a random insertion of the T‐DNA harbouring a hygR marker and the green fluorescent protein (GFP) cassette (WT GFP), a strain deleted for vhb1 according to polymerase chain reaction (PCR) analysis (Δvhb1), and a strain deleted for vph1 according to PCR analysis (Δvph1). (a, b) Confirmation of vhb1 and vph1 deletion. Genomic DNAs were singly digested with HindIII (a) and XhoI (b), and membranes were simultaneously hybridized with probes derived from vhb1 and vph1 open reading frames (ORFs). (c, d) Confirmation of absence of secondary integration events in Δvph1. After confirmation of deletion, membranes with the HindIII‐ and XhoI‐digested DNAs were stripped from the probes and re‐hybridized with hygR; the vph1 deletion construct contains a single HindIII and XhoI restriction site and these digestions therefore allow discrimination between single and multiple integration events in Δvph1. (e, f) Confirmation of absence of secondary integration events in Δvhb1. Genomic DNAs were singly digested with EcoRV and XbaI, and hybridized with hygR; the vhb1 deletion construct contains a single EcoRV and XbaI restriction site and these digestions therefore allow discrimination between single and multiple integration events in Δvhb1. For each Southern blot, a schematic representation of the vhb1 and vph1 WT and mutant alleles, indicating the position of the corresponding restriction sites, is presented on the left.
Fig. S3 Deletion of vph1 or vhb1 does not affect growth on solid medium. Images were taken after 11 days of growth on potato dextrose agar (PDA) at 24 °C.
Fig. S4 Strains deleted for vph1 or vhb1 do not induce Verticillium wilt symptoms in Lactuca sativa. Average of disease severity at 3 weeks post‐inoculation; different letters indicate statistically significant differences (P < 0.05) according to a Bonferroni–Dunn test. Bars indicate standard error.
Fig. S5 Complemented strains Δvph1C and Δvhb1C exhibit wild‐type (WT) phenotypes. (a) Δvph1C and Δvhb1C display WT levels of virulence towards Nicotiana benthamiana. Left: N. benthamiana plants inoculated with the complemented strains Δvph1C and Δvhb1C and the WT parental strain, exhibiting characteristic Verticillium wilt symptoms, compared with plants inoculated with Δvph1 or Δvhb1, which are indistinguishable from those mock inoculated with water. Right: average of disease severity at 7 weeks post‐inoculation, calculated according to a 0–3 scale: 0, healthy leaf; 1, chlorotic leaf; 2, leaf with wilt symptoms; 3, fully necrotic leaf. Different letters indicate statistically significant differences (P < 0.05) according to a Bonferroni–Dunn test. Bars indicate standard error. (b) Complementation with the WT gene reverses the sporulation defect of Δvhb1. Left: liquid cultures of strains grown for 4 days in YEPS (1% yeast extract, 2% bactopeptone, 2% sucrose). Right: sporulation rates in 4‐day‐old liquid cultures, initially inoculated with 106 conidia/mL of each strain. Conidia were counted in a Fuchs–Rosenthal haemocytometer. Three replicates of each strain were included in each experiment and four independent experiments were performed. Different letters indicate statistically significant differences (P < 0.05) according to a Bonferroni–Dunn test. Bars indicate standard error.
Fig. S6 Verticillium dahliae grown on cellophane sheets develops hyphopodium‐like structures. Images of wild‐type (WT) strain Dvd‐T5 grown on potato dextrose agar (PDA) medium covered with cellophane sheets for 48 h at 24 ºC and captured by light microscopy. Arrows point to hyphal tip structures that resemble penetration pegs. Scale bar, 50 µm.
Fig. S7 Complementation of Δvph1 with the wild‐type (WT) gene restores the ability to penetrate cellophane paper. (a) Strains grown on potato dextrose agar (PDA) covered with cellophane for 4 days at 24 °C. (b) Plates incubated at 24 °C for 24 h after removing the cellophane membrane with the fungal colony. (c) Microscopic observation of strains grown on PDA covered with cellophane for 2 days. Scale bar, 20 µm.
File S1 List of the 434 genes differentially expressed in the wild‐type (WT) vs. Δvph1 showing differential expression values.
File S2 List of the 39 genes differentially expressed in the wild‐type (WT) vs. Δvhb1 showing differential expression values.
File S3 Genes differentially expressed in the wild‐type (WT) vs. Δvph1 corresponding to the different biological process categories according to the gene ontology (GO) analysis.
File S4 Genes differentially expressed in the wild‐type (WT) vs. Δvph1 corresponding to the different molecular function categories according to the gene ontology (GO) analysis.
Table S1 Real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) validation of microarray data in selected genes.
Table S2 Specific primers used in this study.
Acknowledgements
This research was supported by Grants AGL2009–13445 from the Spanish Ministry of Science and Innovation and AGL2013–48980‐R from the Spanish Ministry of Economy and Competitiveness, co‐funded by the European Union (FEDER funds). J.L.S.‐V. was a recipient of an F.P.I. fellowship (BES‐2010–033084) from the Spanish Ministry of Science and Innovation. The authors thank Professor Antonio Martín for the use of the confocal microscopy facilities.
References
- Antal, Z. , Rascle, C. , Cimerman, A. , Viaud, M. , Billon‐Grand, G. , Choquer, M. and Bruel, C. (2012) The homeobox BcHOX8 gene in Botrytis cinerea regulates vegetative growth and morphology. PLoS One, 7, e48134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao, J. , Chinnici, J.L. , Maddi, A. and Free, S.J. (2015) The N‐linked outer chain mannans and the Dfg5p and Dcw1p endo‐α‐1,6‐mannanases are needed for incorporation of Candida albicans glycoproteins into the cell wall. Eukaryot. Cell, 14, 792–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair, D.E. , Hekmat, O. , Schüttelkopf, A.W. , Shrestha, B. , Tokuyasu, K. , Withers, S.G. and van Aalten, D.M.F. (2006) Structure and mechanism of chitin deacetylase from the fungal pathogen Colletotrichum lindemuthianum . Biochemistry, 45, 9416–9426. [DOI] [PubMed] [Google Scholar]
- Bockmühl, D.P. and Ernst, J.F. (2001) A potential phosphorylation site for an A‐type kinase in the Efg1 regulator protein contributes to hyphal morphogenesis of Candida albicans . Genetics, 157, 1523–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobinson, K. , Grant, S. and Kang, S. (2004) Cloning and targeted disruption, via Agrobacterium tumefaciens‐mediated transformation, of a trypsin protease gene from the vascular wilt fungus Verticillium dahliae . Curr. Genet. 45, 104–110. [DOI] [PubMed] [Google Scholar]
- Dobinson, K.F. , Tenuta, G.K. and Lazarovits, G. (1996) Occurrence of race 2 of Verticillium dahliae in processing tomato fields in southwestern Ontario. Can. J. Plant Pathol. 18, 55–58. [Google Scholar]
- Elion, E.A. , Qi, M. and Chen, W. (2005) Signaling specificity in yeast. Science, 307, 687–688. [DOI] [PubMed] [Google Scholar]
- Eynck, C. , Koopmann, B. , Grunewaldt‐Stoecker, G. , Karlovsky, P. and Von Tiedemann, A. (2007) Differential interactions of Verticillium longisporum and V. dahliae with Brassica napus detected with molecular and histological techniques. Eur. J. Plant Pathol. 118, 259–274. [Google Scholar]
- Fradin, E.F. and Thomma, B.P.H.J. (2006) Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo‐atrum . Mol. Plant Pathol. 7, 71–86. [DOI] [PubMed] [Google Scholar]
- García‐Pedrajas, M. , Paz, Z. , Andrews, D. , Baeza‐Montañez, L. and Gold, S. (2013) Rapid deletion plasmid construction methods for protoplast and Agrobacterium‐based fungal transformation systems In: Laboratory Protocols in Fungal Biology (Gupta V.K., Tuohy M.G., Ayyachamy M., Turner K.M. and O'Donovan A., eds), pp. 375–393. New York: Springer. [Google Scholar]
- Hughes, T. (2011) Introduction to “a handbook of transcription factors” In: A Handbook of Transcription Factors (Hughes T., ed), pp. 1–6. New York: Springer. [DOI] [PubMed] [Google Scholar]
- Hung, C.‐Y. , Seshan, K.R. , Yu, J.‐J. , Schaller, R. , Xue, J. , Basrur, V. , Gardner, M.J. and Cole, G. (2005) A metalloproteinase of Coccidioides posadasii contributes to evasion of host detection. Infect. Immun. 73, 6689–6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, C.‐S. and Kolattukudy, P.E. (1995) Isolation and characterization of genes expressed uniquely during appressorium formation by Colletotrichum gloeosporioides conidia induced by the host surface wax. Mol. Gen. Genet. 247, 282–294. [DOI] [PubMed] [Google Scholar]
- Jashni, M.K. , Mehrabi, R. , Collemare, J. , Mesarich, C.H. and de Wit, P.J.G.M. (2015) The battle in the apoplast: further insights into the roles of proteases and their inhibitors in plant–pathogen interactions. Front. Plant Sci. 6, 584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. , Park, S.‐Y. , Kim, K.S. , Rho, H.‐S. , Chi, M.‐H. , Choi, J. , Park, J. , Kong, S. , Park, J. , Goh, J. and Lee, Y.‐H. (2009) Homeobox transcription factors are required for conidiation and appressorium development in the rice blast fungus Magnaporthe oryzae . PLoS Genet. 5, e1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.‐K. , Liu, Z.‐M. , Li, D. and Kolattukudy, P.E. (2000) Two novel genes induced by hard‐surface contact of Colletotrichum gloeosporioides conidia. J. Bacteriol. 182, 4688–4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimes, A. , Dobinson, K.F. , Thomma, B.P.H.J. and Klosterman, S.J. (2015) Genomics spurs rapid advances in our understanding of the biology of vascular wilt pathogens in the genus Verticillium . Annu. Rev. Phytopathol. 53, 181–198. [DOI] [PubMed] [Google Scholar]
- Klosterman, S.J. and Hayes, R.J. (2009) A soilless Verticillium wilt assay using an early flowering lettuce line. Plant Dis. 93, 691–698. [DOI] [PubMed] [Google Scholar]
- Klosterman, S.J. , Atallah, Z.K. , Vallad, G.E. and Subbarao, K.V. (2009) Diversity, pathogenicity, and management of Verticillium species. Annu. Rev. Phytopathol. 47, 39–62. [DOI] [PubMed] [Google Scholar]
- Klosterman, S.J. , Subbarao, K.V. , Kang, S. , Veronese, P. , Gold, S.E. , Thomma, B.P.H.J. , Chen, Z. , Henrissat, B. , Lee, Y.‐H. , Park, J. , García‐Pedrajas, M.D. , Barbara, D.J. , Anchieta, A. , de Jonge, R. , Santhanam, P. , Maruthachalam, K. , Atallah, Z. , Amyotte, S.G. , Paz, Z. , Inderbitzin, P. , Hayes, R.J. , Heiman, D.I. , Young, S. , Zeng, Q. , Engels, R. , Galagan, J. , Cuomo, C.A. , Dobinson, K.F. and Ma, L.‐J. (2011) Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens. PLoS Pathog. 7, e1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.D. and Kolattukudy, P.E. (1995) Molecular cloning of the cDNA and gene for an elastinolytic aspartic proteinase from Aspergillus fumigatus and evidence of its secretion by the fungus during invasion of the host lung. Infect. Immun. 63, 3796–3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W. , Xie, S. , Zhao, X. , Chen, X. , Zheng, W. , Lu, G. , Xu, J.‐R. and Wang, Z. (2010) A Homeobox gene is essential for conidiogenesis of the rice blast fungus Magnaporthe oryzae . Mol. Plant–Microbe Interact. 23, 366–375. [DOI] [PubMed] [Google Scholar]
- Luo, X. , Xie, C. , Dong, J. , Yang, X. and Sui, A. (2014) Interactions between Verticillium dahliae and its host: vegetative growth, pathogenicity, plant immunity. Appl. Microbiol. Biotechnol. 98, 6921–6932. [DOI] [PubMed] [Google Scholar]
- Luo, X. , Mao, H. , Wei, Y. , Cai, J. , Xie, C. , Sui, A. , Yang, X. and Dong, J. (2016) The fungal‐specific transcription factor Vdpf influences conidia production, melanized microsclerotia formation and pathogenicity in Verticillium dahliae . Mol. Plant Pathol. 17, 1364–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddi, A. , Fu, C. and Free, S.J. (2012) The Neurospora crassa dfg5 and dcw1 genes encode α‐1,6‐mannanases that function in the incorporation of glycoproteins into the cell wall. PLoS One, 7, e38872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maor, R. , Puyesky, M. , Horwitz, B.A. and Sharon, A. (1998) Use of green fluorescent protein (GFP) for studying development and fungal–plant interaction in Cochliobolus heterostrophus . Mycol. Res. 102, 491–496. [Google Scholar]
- Naglik, J. , Albrecht, A. , Bader, O. and Hube, B. (2004) Candida albicans proteinases and host/pathogen interactions. Cell Microbiol. 6, 915–926. [DOI] [PubMed] [Google Scholar]
- Nolting, N. and Pöggeler, S. (2006) A STE12 homologue of the homothallic ascomycete Sordaria macrospora interacts with the MADS box protein MCM1 and is required for ascosporogenesis. Mol. Microbiol. 62, 853–868. [DOI] [PubMed] [Google Scholar]
- Ochman, H. , Gerber, A.S. and Hartl, D.L. (1988) Genetic applications of an inverse polymerase chain reaction. Genetics, 120, 621–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz, Z. , García‐Pedrajas, M.D. , Andrews, D.L. , Klosterman, S.J. , Baeza‐Montañez, L. and Gold, S.E. (2011) One step construction of Agrobacterium‐recombination‐ready‐plasmids (OSCAR), an efficient and robust tool for ATMT based gene deletion construction in fungi. Fungal Genet. Biol. 48, 677–684. [DOI] [PubMed] [Google Scholar]
- Pegg, G.F. and Brady, B.L. (2002) Verticillium Wilts. New York: CABI Pub.[WorldCat] [Google Scholar]
- Pérez‐Nadales, E. and Di Pietro, A. (2015) The transmembrane protein Sho1 cooperates with the mucin Msb2 to regulate invasive growth and plant infection in Fusarium oxysporum . Mol. Plant Pathol. 16, 593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prados‐Rosales, R.C. and Di Pietro, A. (2008) Vegetative hyphal fusion is not essential for plant infection by Fusarium oxysporum . Eukaryot. Cell, 7, 162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto, P. , Navarro‐Raya, C. , Valverde‐Corredor, A. , Amyotte, S.G. , Dobinson, K.F. and Mercado‐Blanco, J. (2009) Colonization process of olive tissues by Verticillium dahliae and its in planta interaction with the biocontrol root endophyte Pseudomonas fluorescens PICF7. Microb. Biotechnol. 2, 499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhalla, J.E. and Bell, A.A. (1981) Genetics and biochemistry of wilt pathogens In: Fungal Wilt Diseases of Plants (Mace M.E., Bell A.A. and Beckman C.H., eds.), pp. 146–192. New York: Academic Press. [Google Scholar]
- Qi, X. , Zhou, S. , Shang, X. and Wang, X. (2016) VdSho1 regulates growth, oxidant adaptation and virulence in Verticillium dahliae . J. Phytopathol. 164, 1064–1074. [Google Scholar]
- Ranganathan, S. and Garg, G. (2009) Secretome: clues into pathogen infection and clinical applications. Genome Med. 1, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauyaree, P. , Ospina‐Giraldo, M. , Kang, S. , Bhat, R. , Subbarao, K. , Grant, S. and Dobinson, K.F. (2005) Mutations in VMK1, a mitogen‐activated protein kinase gene, affect microsclerotia formation and pathogenicity in Verticillium dahliae . Curr. Genet. 48, 109–116. [DOI] [PubMed] [Google Scholar]
- Reusche, M. , Truskina, J. , Thole, K. , Nagel, L. , Rindfleisch, S. , Tran, V.T. , Braus‐Stromeyer, S.A. , Braus, G.H. , Teichmann, T. and Lipka, V. (2014) Infections with the vascular pathogens Verticillium longisporum and Verticillium dahliae induce distinct disease symptoms and differentially affect drought stress tolerance of Arabidopsis thaliana . Environ. Exp. Bot. 108, 23–37. [Google Scholar]
- Rispail, N. and Di Pietro, A. (2010) The homeodomain transcription factor Ste12: connecting fungal MAPK signalling to plant pathogenicity. Commun. Integr. Biol. 3, 327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sain, M. and Rep, M. (2015) The role of pathogen‐secreted proteins in fungal vascular wilt diseases. Int. J. Mol. Sci. 16, 23 970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhanam, P. and Thomma, B.P.H.J. (2013) Verticillium dahliae Sge1 differentially regulates expression of candidate effector genes. Mol. Plant–Microbe Interact. 26, 249–256. [DOI] [PubMed] [Google Scholar]
- Santhanam, P. , van Esse, H.P. , Albert, I. , Faino, L. , Nürnberger, T. and Thomma, B.P.H.J. (2013) Evidence for functional diversification within a fungal NEP1‐like protein family. Mol. Plant–Microbe Interact. 26, 278–286. [DOI] [PubMed] [Google Scholar]
- Santhanam, P. , Boshoven, J.C. , Salas, O. , Bowler, K. , Islam, M.T. , Saber, M.K. , van den Berg, G.C.M. , Bar‐Peled, M. and Thomma, B.P.H.J. (2016) Rhamnose synthase activity is required for pathogenicity of the vascular wilt fungus Verticillium dahliae . Mol. Plant Pathol. 18, 347–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmiento‐Villamil, J.L. , García‐Pedrajas, N.E. , Baeza‐Montañez, L. and García‐Pedrajas, M.D. (2016) The APSES transcription factor Vst1 is a key regulator of development in microsclerotium and resting mycelium producing Verticillium species. Mol. Plant Pathol. doi: 10.1111/mpp.12496 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schamber, A. , Leroch, M. , Diwo, J. , Mendgen, K. and Hahn, M. (2010) The role of mitogen‐activated protein (MAP) kinase signalling components and the Ste12 transcription factor in germination and pathogenicity of Botrytis cinerea . Mol. Plant Pathol. 11, 105–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz, B. , Banuett, F. , Dahl, M. , Schlesinger, R. , Schäfer, W. , Martin, T. , Herskowitz, I. and Kahmann, R. (1990) The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain‐related motif. Cell, 60, 295–306. [DOI] [PubMed] [Google Scholar]
- Sonneborn, A. , Bockmühl, D.P. and Ernst, J.F. (1999) Chlamydospore formation in Candida albicans requires the Efg1p morphogenetic regulator. Infect. Immun. 67, 5514–5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebarth, B. , Doedt, T. , Krishnamurthy, S. , Weide, M. , Monterola, F. , Dominguez, A. and Ernst, J.F. (2003) Adaptation of the Efg1p morphogenetic pathway in Candida albicans by negative autoregulation and PKA‐dependent repression of the EFG1 Gene. J. Mol. Biol. 329, 949–962. [DOI] [PubMed] [Google Scholar]
- Tian, L. , Xu, J. , Zhou, L. and Guo, W. (2014) VdMsb regulates virulence and microsclerotia production in the fungal plant pathogen Verticillium dahliae . Gene, 550, 238–244. [DOI] [PubMed] [Google Scholar]
- Tian, L. , Wang, Y. , Yu, J. , Xiong, D. , Zhao, H. and Tian, C. (2016) The mitogen‐activated protein kinase kinase VdPbs2 of Verticillium dahliae regulates microsclerotia formation, stress response, and plant infection. Front. Microbiol. 7, 1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, V.‐T. , Braus‐Stromeyer, S.A. , Kusch, H. , Reusche, M. , Kaever, A. , Kühn, A. , Valerius, O. , Landesfeind, M. , Aßhauer, K. , Tech, M. , Hoff, K. , Pena‐Centeno, T. , Stanke, M. , Lipka, V. and Braus, G.H. (2014) Verticillium transcription activator of adhesion Vta2 suppresses microsclerotia formation and is required for systemic infection of plant roots. New Phytol. 202, 565–581. [DOI] [PubMed] [Google Scholar]
- Tzima, A. , Paplomatas, E.J. , Rauyaree, P. and Kang, S. (2010) Roles of the catalytic subunit of cAMP‐dependent protein kinase A in virulence and development of the soilborne plant pathogen Verticillium dahliae . Fungal Genet. Biol. 47, 406–415. [DOI] [PubMed] [Google Scholar]
- Tzima, A.K. , Paplomatas, E.J. , Tsitsigiannis, D.I. and Kang, S. (2012) The G protein β subunit controls virulence and multiple growth‐ and development‐related traits in Verticillium dahliae . Fungal Genet. Biol. 49, 271–283. [DOI] [PubMed] [Google Scholar]
- Upadhya, R. , Lam, W.C. , Maybruck, B. , Specht, C.A. , Levitz, S.M. and Lodge, J.K. (2016) Induction of protective immunity to cryptococcal infection in mice by a heat‐killed, chitosan‐deficient strain of Cryptococcus neoformans . MBio, 7, e00547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallad, G.E. and Subbarao, K.V. (2008) Colonization of resistant and susceptible lettuce cultivars by a green fluorescent protein‐tagged isolate of Verticillium dahliae . Phytopathology, 98, 871–885. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Tian, L. , Xiong, D. , Klosterman, S.J. , Xiao, S. and Tian, C. (2016) The mitogen‐activated protein kinase gene, VdHog1, regulates osmotic stress response, microsclerotia formation and virulence in Verticillium dahliae . Fungal Genet. Biol. 88, 13–23. [DOI] [PubMed] [Google Scholar]
- Wellman, F.L. (1939) A technique for studying host resistance and pathogenicity in tomato fusarium wilt. Phytopathology, 29, 945–956. [Google Scholar]
- Wong Sak Hoi, J. and Dumas, B. (2010) Ste12 and Ste12‐like proteins, fungal transcription factors regulating development and pathogenicity. Eukaryot. Cell, 9, 480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong Sak Hoi, J. , Herbert, C. , Bacha, N. , O'Connell, R. , Lafitte, C. , Borderies, G. , Rossingnol, M. , Rougé, P. and Dumas, B. (2007) Regulation and role of a STE12‐like transcription factor from the plant pathogen Colletotrichum lindemuthianum . Mol. Microbiol. 64, 68–82. [DOI] [PubMed] [Google Scholar]
- Xiong, D. , Wang, Y. , Tang, C. , Fang, Y. , Zou, J. and Tian, C. (2015) VdCrz1 is involved in microsclerotia formation and required for full virulence in Verticillium dahliae . Fungal Genet. Biol. 82, 201–212. [DOI] [PubMed] [Google Scholar]
- Xiong, D. , Wang, Y. , Tian, L. and Tian, C. (2016) MADS‐Box transcription factor VdMcm1 regulates conidiation, microsclerotia formation, pathogenicity, and secondary metabolism of Verticillium dahliae . Front. Microbiol. 7, 1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, C. , Park, G. , Choi, W. , Zheng, L. , Dean, R.A. and Xu, J.‐R. (2002) Two novel fungal virulence genes specifically expressed in appressoria of the rice blast fungus. Plant Cell, 14, 2107–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Y.L. and Fields, S. (1991) Properties of the DNA‐binding domain of the Saccharomyces cerevisiae STE12 protein. Mol. Cell Biol. 11, 5910–5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D.‐D. , Wang, X.‐Y. , Chen, J.‐Y. , Kong, Z.‐Q. , Gui, Y.‐J. , Li, N.‐Y. , Bao, Y.‐M. and Dai, X.‐F. (2016) Identification and characterization of a pathogenicity‐related gene VdCYP1 from Verticillium dahliae . Sci. Rep. 6, 27 979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W.‐W. , Jiang, T.‐F. , Cui, X. , Qi, F.‐J. and Jian, G.‐L. (2013) Colonization in cotton plants by a green fluorescent protein labelled strain of Verticillium dahliae . Eur. J. Plant Pathol. 135, 867–876. [Google Scholar]
- Zhao, P. , Zhao, Y.‐L. , Jin, Y. , Zhang, T. and Guo, H.‐S. (2014) Colonization process of Arabidopsis thaliana roots by a green fluorescent protein‐tagged isolate of Verticillium dahliae . Protein Cell, 5, 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y.‐L. , Zhou, T.‐T. and Guo, H.‐S. (2016) Hyphopodium‐specific VdNoxB/VdPls1‐dependent ROS‐Ca2+ signaling is required for plant infection by Verticillium dahliae . PLoS Pathog. 12, e1005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, W. , Zhao, X. , Xie, Q. , Huang, Q. , Zhang, C. , Zhai, H. , Xu, L. , Lu, G. , Shim, W.‐B. and Wang, Z. (2012) A conserved homeobox transcription factor Htf1 is required for phialide development and conidiogenesis in Fusarium species. PLoS One, 7, e45432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, B.‐J. , Jia, P.‐S. , Gao, F. and Guo, H.‐S. (2012) Molecular characterization and functional analysis of a necrosis‐ and ethylene‐inducing, protein‐encoding gene family from Verticillium dahliae . Mol. Plant–Microbe Interact. 25, 964–975. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Modified version of the One Step Construction of Agrobacterium‐Recombination‐ready‐plasmids (OSCAR) method used to generate deletion mutants in this work. We used OSCAR (Paz et al., 2011), but replaced vector pA‐Hyg‐OSCAR with a new version containing a synthetic SGFP‐TYG gene under the regulation of the Aspergillus nidulans Pgdp promoter. To construct the new vector, the SGFP‐TYG cassette was excised from vector gGFP (Maor et al., 1998) by digestion with ClaI and XbaI, blunt ended using T4 DNA polymerase and ligated into pA‐Hyg‐OSCAR (Paz et al., 2011), digested with BamHI and also blunt ended. Two vectors were generated in this way, pA‐Hyg‐GFP‐OSCAR‐5 (shown in figure) and pA‐Hyg‐GFP‐OSCAR‐6, which differ only in the orientation of the green fluorescent protein (GFP) cassette. Deletion constructs generated with pA‐Hyg‐GFP‐OSCAR vectors generate deletion mutants in which the target gene open reading frame (ORF) is replaced by a hygR marker and the SGFP‐TYG cassette.
Fig. S2 Confirmation of vhb1 and vph1 deletion and absence of secondary integration events in the mutants by Southern blot analyses. The samples analysed included: parental strain Dvd‐T5 (WT), a transformant with a random insertion of the T‐DNA harbouring a hygR marker and the green fluorescent protein (GFP) cassette (WT GFP), a strain deleted for vhb1 according to polymerase chain reaction (PCR) analysis (Δvhb1), and a strain deleted for vph1 according to PCR analysis (Δvph1). (a, b) Confirmation of vhb1 and vph1 deletion. Genomic DNAs were singly digested with HindIII (a) and XhoI (b), and membranes were simultaneously hybridized with probes derived from vhb1 and vph1 open reading frames (ORFs). (c, d) Confirmation of absence of secondary integration events in Δvph1. After confirmation of deletion, membranes with the HindIII‐ and XhoI‐digested DNAs were stripped from the probes and re‐hybridized with hygR; the vph1 deletion construct contains a single HindIII and XhoI restriction site and these digestions therefore allow discrimination between single and multiple integration events in Δvph1. (e, f) Confirmation of absence of secondary integration events in Δvhb1. Genomic DNAs were singly digested with EcoRV and XbaI, and hybridized with hygR; the vhb1 deletion construct contains a single EcoRV and XbaI restriction site and these digestions therefore allow discrimination between single and multiple integration events in Δvhb1. For each Southern blot, a schematic representation of the vhb1 and vph1 WT and mutant alleles, indicating the position of the corresponding restriction sites, is presented on the left.
Fig. S3 Deletion of vph1 or vhb1 does not affect growth on solid medium. Images were taken after 11 days of growth on potato dextrose agar (PDA) at 24 °C.
Fig. S4 Strains deleted for vph1 or vhb1 do not induce Verticillium wilt symptoms in Lactuca sativa. Average of disease severity at 3 weeks post‐inoculation; different letters indicate statistically significant differences (P < 0.05) according to a Bonferroni–Dunn test. Bars indicate standard error.
Fig. S5 Complemented strains Δvph1C and Δvhb1C exhibit wild‐type (WT) phenotypes. (a) Δvph1C and Δvhb1C display WT levels of virulence towards Nicotiana benthamiana. Left: N. benthamiana plants inoculated with the complemented strains Δvph1C and Δvhb1C and the WT parental strain, exhibiting characteristic Verticillium wilt symptoms, compared with plants inoculated with Δvph1 or Δvhb1, which are indistinguishable from those mock inoculated with water. Right: average of disease severity at 7 weeks post‐inoculation, calculated according to a 0–3 scale: 0, healthy leaf; 1, chlorotic leaf; 2, leaf with wilt symptoms; 3, fully necrotic leaf. Different letters indicate statistically significant differences (P < 0.05) according to a Bonferroni–Dunn test. Bars indicate standard error. (b) Complementation with the WT gene reverses the sporulation defect of Δvhb1. Left: liquid cultures of strains grown for 4 days in YEPS (1% yeast extract, 2% bactopeptone, 2% sucrose). Right: sporulation rates in 4‐day‐old liquid cultures, initially inoculated with 106 conidia/mL of each strain. Conidia were counted in a Fuchs–Rosenthal haemocytometer. Three replicates of each strain were included in each experiment and four independent experiments were performed. Different letters indicate statistically significant differences (P < 0.05) according to a Bonferroni–Dunn test. Bars indicate standard error.
Fig. S6 Verticillium dahliae grown on cellophane sheets develops hyphopodium‐like structures. Images of wild‐type (WT) strain Dvd‐T5 grown on potato dextrose agar (PDA) medium covered with cellophane sheets for 48 h at 24 ºC and captured by light microscopy. Arrows point to hyphal tip structures that resemble penetration pegs. Scale bar, 50 µm.
Fig. S7 Complementation of Δvph1 with the wild‐type (WT) gene restores the ability to penetrate cellophane paper. (a) Strains grown on potato dextrose agar (PDA) covered with cellophane for 4 days at 24 °C. (b) Plates incubated at 24 °C for 24 h after removing the cellophane membrane with the fungal colony. (c) Microscopic observation of strains grown on PDA covered with cellophane for 2 days. Scale bar, 20 µm.
File S1 List of the 434 genes differentially expressed in the wild‐type (WT) vs. Δvph1 showing differential expression values.
File S2 List of the 39 genes differentially expressed in the wild‐type (WT) vs. Δvhb1 showing differential expression values.
File S3 Genes differentially expressed in the wild‐type (WT) vs. Δvph1 corresponding to the different biological process categories according to the gene ontology (GO) analysis.
File S4 Genes differentially expressed in the wild‐type (WT) vs. Δvph1 corresponding to the different molecular function categories according to the gene ontology (GO) analysis.
Table S1 Real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) validation of microarray data in selected genes.
Table S2 Specific primers used in this study.


