Summary
The identification of phytopathogen proteins that are differentially expressed during the course of the establishment of an infection is important to better understand the infection process. In vitro approaches, using plant extracts added to culture medium, have been used to identify such proteins, but the biological relevance of these findings for in planta infection are often uncertain until confirmed by in vivo studies. Here, we compared the proteins of Pectobacterium carotovorum ssp. carotovorum strain PccS1 differentially expressed in Luria–Bertani medium supplemented with extracts of the ornamental plant Zantedeschia elliotiana cultivar ‘Black Magic’ (in vitro) and in plant tissues (in vivo) by two‐dimensional electrophoresis coupled with mass spectrometry. A total of 53 differentially expressed proteins (>1.5‐fold) were identified (up‐regulated or down‐regulated in vitro, in vivo or both). Proteins that exhibited increased expression in vivo but not in vitro, or in both conditions, were identified, and deletions were made in a number of genes encoding these proteins, four of which (clpP, mreB, flgK and eda) led to a loss of virulence on Z. elliotiana, although clpP and mreB were later also shown to be reduced in growth in rich and minimal media. Although clpP, flgK and mreB have previously been reported as playing a role in virulence in plants, this is the first report of such a role for eda, which encodes 2‐keto‐3‐deoxy‐6‐phosphogluconate (KDPG) aldolase, a key enzyme in Entner–Doudoroff metabolism. The results highlight the value of undertaking in vivo as well as in vitro approaches for the identification of new bacterial virulence factors.
Keywords: calla lily, Entner–Doudoroff, in vivo, Pectobacterium, proteomics, Zantedeschia
Introduction
Summer‐flowering calla lily (Zantedeschia spp.) cultivars play an important role in cut‐flower and bulb production worldwide (Snijder et al., 2004; Wright, 1998). Soft rot disease caused by Pectobacterium carotovorum ssp. carotovorum (Pcc; formerly Erwinia carotovora ssp. carotovora) leads to substantial economic losses in calla lily, as well as in other crops and ornamental monocots (Ma et al., 2007; Mansfield et al., 2012; Ni et al., 2010; Snijder et al., 2004; Toth et al., 2003; Wright, 1998; Yishay et al., 2008). As there are no effective measures to control the disease, it is a major limiting factor in horticultural production, although strict agronomic measures and the selection of resistant cultivars have alleviated disease losses to some extent (Luzzatto‐Knaan et al., 2014; Ni et al., 2010).
The main pathogenicity determinants of Pcc are plant cell wall‐degrading enzymes (PCWDEs), which include pectate lyase (Pel), polygalacturonase (Peh), cellulase (Cel) and protease (Prt) (Bortoli‐German et al., 1994; Heikinheimo et al., 1995; Hinton et al., 1990; Mae et al., 1995; Pirhonen et al., 1991; Toth et al., 2003). The availability of genomic sequences for Pectobacterium strains (Bell et al., 2004; Koskinen et al., 2012; Park et al., 2012) has facilitated detailed studies on these and other virulence determinants (Grinter et al., 2012; Ma et al., 2014; Yap et al., 2004). However, the protein expression patterns of Pcc in a host plant during the establishment of infection remain poorly understood.
Proteomic approaches have been widely used in the study of plant–bacterial interactions. Examples include the identification and functional analysis of Xanthomonas campestris pv. campestris proteins expressed in culture medium (Chung et al., 2007), X. axonopodis pv. citri proteins differentially expressed in response to different growth conditions (Mehta and Rosato, 2001), comparison of the secretomes of Pectobacterium atrosepticum induced by host extracts (Mattinen et al., 2007) and impacts induced by host plant extracts or ions on Dickeya dadantii grown in minimal medium (MM) (Babujee et al., 2007). Most of these studies have been performed with plant extracts to mimic in planta conditions. However, there is no evidence to show whether findings from these studies are directly relevant to what happens in planta during infection.
Mehta and Rosato (2003) developed a simple and rapid protocol for obtaining microorganisms directly from inoculated leaves. Five new X. axonopodis pv. citri proteins differentially regulated by the host plant in vivo were reported in this study. Using this protocol and the corresponding proteomics approaches, Andrade et al. (2008) analysed the in vivo proteome of X. campestris pv. campestris in different cultivars of infected Brassica oleracea. Despite such technical progress, comparison of data from plant–bacterial interactions in vitro and in vivo is in its infancy, hindering our understanding of pathogen responses to living plants in the establishment of infection.
Recently, the in vitro and in vivo profiles of outer membrane (OM) proteins in the rice pathogen Acidovorax avenae ssp. avenae have been reported (Ibrahim et al., 2012). Combining liquid chromatography‐tandem mass spectrometry (LC‐MS/MS) and in silico OM proteome and in silico genome‐wide analyses, it was anticipated that a high number of OM proteins were differentially expressed in the two conditions, with those expressed in vivo possibly indicating a role in virulence.
In this study, protein profiles from in vitro and in vivo conditions were generated for PccS1 following its interaction with the Zantedeschia elliotiana cultivar ‘Black Magic’. With two‐dimensional electrophoresis (2‐DE) coupled with MS, we identified 53 proteins that revealed differential expression in vitro, in vivo or in both compared with the control. On testing mutant strains affected in genes encoding 14 of these proteins (ClpP, MreB, FlgK, KdgR, PrmA, AceE, Pgk, AspA, AtpD, HslO, Upp, NusA, SspA and Zwf), three (ClpP, MreB and FlgK) were found to be affected in virulence. In addition, following further analysis of a mutant in gene zwf, an additional protein, Eda, was further examined and found to be involved in virulence. Although ClpP, FlgK and MreB have previously been reported to play a role in the virulence of plant pathogens, to the best of our knowledge, this is the first report in which the Entner–Doudoroff pathway central enzyme Eda has been shown to play such a role.
Results
Bacterial cells recovered from the plant‐pathogenic bacterium infiltrated in vivo
An examination of bacterial populations in petioles of the potted plants (shown in Fig. S1, see Supporting Information) revealed that there was a clear increase in the population at 8 h after inoculation (HAI), and the initial symptoms of disease could be observed at this time. However, the rotting of plant tissue was not visualized clearly until 16 HAI, at which time there were sufficient quantities of proteins from extracts for 2‐DE, and there was no bacterial protein in the negative controls (data not shown).
Comparative proteomic profiling analyses
The protein expression patterns of PccS1‐LB [PccS1 cultured in Luria–Bertani broth (LB)], PccS1‐LB‐Ze (PccS1 cultured in LB + 0.4% petiole extract) and PccS1‐V‐Ze (PccS1 grown in planta) were analysed by 2‐DE, and resulted in a total of 386, 366 and 369 protein spots, respectively, on the 2‐DE maps (Fig. 1). When comparing the differential protein spots obtained from PccS1‐LB‐Ze or PccS1‐V‐Ze with the control (PccS1‐LB), PccS1‐LB‐Ze showed 47 proteins with increased expression and 23 with decreased expression (70 proteins in total), whereas PccS1‐V‐Ze showed 50 proteins with increased expression and 45 with decreased expression (95 proteins in total) (P ≤ 0.05, expression ratio ≥ 1.5). When comparing the 2‐DE protein profiles between PccS1‐V‐Ze and PccS1‐LB‐Ze, there was a total of 133 proteins showing significant differential expression between PccS1‐V‐Ze and PccS1‐LB‐Ze, 68 showing increased and 65 showing decreased expression in PccS1‐V‐Ze relative to PccS1‐LB‐Ze.
Figure 1.
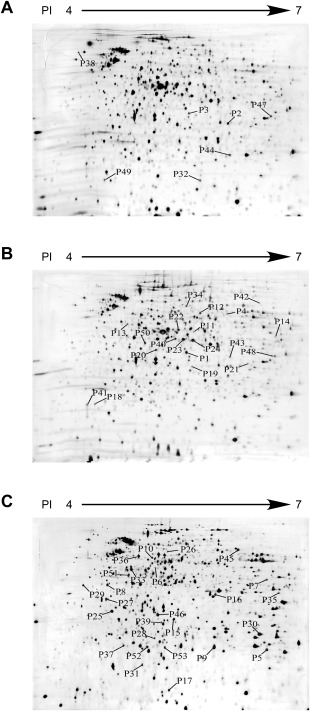
Two‐dimensional electrophoresis (2‐DE) images of proteins extracted from the bacterial cells of Pectobacterium carotovorum ssp. carotovorum strain PccS1 cultured in Luria–Bertani broth (LB) (PccS1‐LB) (A), in LB supplemented with 0.4% petiole extract of Zantedeschia elliotiana (PccS1‐LB‐Ze, PccS1 interaction with the host plant in vitro) (B) and recovered from petioles of potted Z. elliotiana plants (PccS1‐V‐Ze, PccS1 interaction with the host in vivo) (C). The differentially expressed protein spots were numbered above the threshold ratio of 1.5. Protein spots identified by tandem mass spectrometry (MS/MS) are listed in Table 1.
Differentially expressed proteins analysed by MS
Of the 165 proteins showing differential expression in the in vitro condition [PccS1‐LB‐Ze (70 proteins)] and/or in vivo condition [PccS1‐V‐Ze (95 proteins)] compared with the control (PccS1‐LB), 74 proteins were excised from silver‐stained gels and their tryptic fragments were analysed by MS/MS. Of these 74 proteins, 53 were successfully identified and divided into eight groups based on functional annotation (Fig. 2), and the degrees of differential expression of the individual proteins within these groups were examined (Fig. 3). Table 1 shows details of the identification of differentially expressed proteins using MS analysis. Among the proteins identified, there were 37 proteins in PccS1‐LB‐Ze and 50 in PccS1‐V‐Ze showing differential expression when compared with the control (PccS1‐LB). Of the 37 proteins in PccS1‐LB‐Ze, 23 were up‐regulated and 14 were down‐regulated, with nine of these proteins showing greater than a five‐fold change in expression when compared with the control. In PccS1‐V‐Ze, 32 of the 50 proteins were up‐regulated and 18 down‐regulated, with 30 of the proteins showing greater than a five‐fold change in expression when compared with the control.
Figure 2.
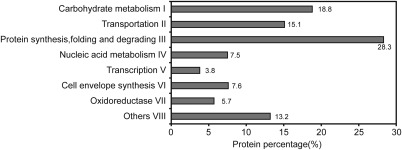
Distribution of 53 differentially expressed proteins following proteomic analyses in vivo and in vitro based on functional annotation according to the COG (Clusters of Orthologous Groups) database of the National Center for Biotechnology Information (NCBI).
Figure 3.
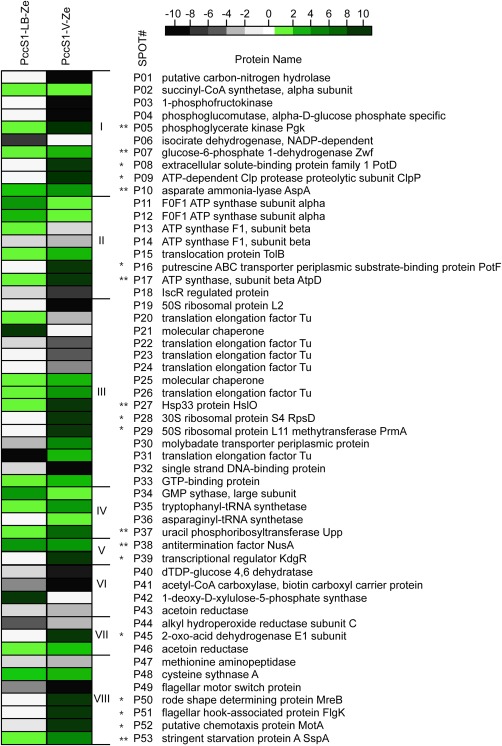
Differentially expressed protein profiles of PccS1‐LB‐Ze [Pectobacterium carotovorum ssp. carotovorum strain PccS1 cultured in Luria–Bertani broth (LB) supplemented with 0.4% petiole extract of Zantedeschia elliotiana] (left column) and PccS1‐V‐Ze (recovered from petioles of potted Z. elliotiana plants) (right column) compared with those of PccS1‐LB (PccS1 cultured in LB); the degrees of differential expression are presented according to the colour scale at the top left; the green boxes indicate up‐regulation, the black boxes indicate down‐regulation and the white boxes represent proteins with no significant difference from those of PccS1‐LB. *Proteins significantly up‐regulated only in vivo. **Proteins significantly up‐regulated both in vitro and in vivo.
Table 1.
Differentially expressed proteins identified by tandem mass spectrometry.
| Spot | Accession number | Organism | Score | M r (cal) | p i (cal) | Sequence coverage (%) | No. of peptides matched |
|---|---|---|---|---|---|---|---|
| Carbohydrate metabolism | |||||||
| P01 | gi|50123194 | Pectobacterium atrosepticum SCRI1043 | 228 | 32.8 | 5.53 | 18 | 4 |
| P02 | gi|253687633 | P. carotovorum PC1 | 268 | 30.1 | 5.56 | 23 | 6 |
| P03 | gi|50121652 | P. atrosepticum SCRI1043 | 342 | 33.8 | 5.46 | 29 | 7 |
| P04 | gi|253687606 | P. carotovorum PC1 | 459 | 58.9 | 5.70 | 28 | 14 |
| P05 | gi|50122833 | P. atrosepticum SCRI1043 | 107 | 41.3 | 5.24 | 8 | 3 |
| P06 | gi|253688259 | P. carotovorum PC1 | 543 | 46.2 | 5.09 | 45 | 18 |
| P07 | gi|253688219 | P. carotovorum PC1 | 503 | 56.2 | 6.07 | 38 | 16 |
| P08 | gi|253688246 | P. carotovorum PC1 | 102 | 38.7 | 5.01 | 16 | 3 |
| P09 | gi|227328849 | P. carotovorum WPP14 | 339 | 23.2 | 5.57 | 19 | 4 |
| P10 | gi|253686801 | P. carotovorum PC1 | 306 | 53.3 | 5.13 | 16 | 7 |
| Transportation | |||||||
| P11 | gi|50123430 | P. atrosepticum SCRI1043 | 103 | 55.4 | 5.49 | 21 | 8 |
| P12 | gi|227113925 | P. brasiliensis PBR1692 | 524 | 55.4 | 5.49 | 34 | 11 |
| P13 | gi|253690617 | P. carotovorum PC1 | 531 | 50.3 | 4.96 | 36 | 11 |
| P14 | gi|227327556 | P. carotovorum WPP14 | 309 | 45.8 | 6.97 | 28 | 8 |
| P15 | gi|254421831 | Synechococcus sp. PCC 7335 | 274 | 52.3 | 4.56 | 12 | 6 |
| P16 | gi| 253688080 | P. carotovorum PC1 | 240 | 44.3 | 5.72 | 20 | 7 |
| P17 | gi|112791384 | P. odoriferum | 77 | 46.2 | 4.71 | 14 | 2 |
| P18 | gi|253690287 | P. carotovorum PC1 | 286 | 21.4 | 4.63 | 29 | 6 |
| Protein synthesis, folding and degradation | |||||||
| P19 | gi|224585232 | Salmonella enterica ssp. enterica serovar Paratyphi C str. RKS4594 | 375 | 27.9 | 10.82 | 39 | 12 |
| P20 | gi|253686604 | P. carotovorum PC1 | 329 | 43.4 | 5.23 | 32 | 9 |
| P21 | gi|50122803 | P. atrosepticum SCRI1043 | 293 | 68.9 | 4.83 | 10 | 4 |
| P22 | gi|253686604 | P. carotovorum PC1 | 486 | 43.4 | 5.29 | 47 | 17 |
| P23 | gi|253686604 | P. carotovorum PC1 | 531 | 43.4 | 5.29 | 48 | 19 |
| P24 | gi|253686604 | P. carotovorum PC1 | 117 | 43.4 | 5.23 | 36 | 10 |
| P25 | gi|50122803 | P. atrosepticum SCRI1043 | 421 | 68.9 | 4.83 | 16 | 7 |
| P26 | gi|253686604 | P. carotovorum PC1 | 259 | 43.4 | 5.23 | 31 | 11 |
| P27 | gi|253690256 | P. carotovorum PC1 | 413 | 32.7 | 4.68 | 21 | 6 |
| P28 | gi|50122927 | P. atrosepticum SCRI1043 | 240 | 23.5 | 10.08 | 59 | 12 |
| P29 | gi|253686645 | P. carotovorum PC1 | 504 | 32.3 | 4.40 | 44 | 11 |
| P30 | gi|253687663 | P. carotovorum PC1 | 160 | 27.8 | 6.12 | 7 | 2 |
| P31 | gi|253686604 | P. carotovorum PC1 | 113 | 43.4 | 5.23 | 13 | 5 |
| P32 | gi|227326029 | P. carotovorum WPP14 | 476 | 19.2 | 5.46 | 29 | 7 |
| P33 | gi|253686962 | P. carotovorum PC1 | 140 | 43.1 | 4.79 | 16 | 5 |
| Nucleic acid metabolism | |||||||
| P34 | gi|253689372 | P. carotovorum PC1 | 269 | 58.9 | 5.37 | 20 | 9 |
| P35 | gi|253690238 | P. carotovorum PC1 | 339 | 37.6 | 5.94 | 20 | 11 |
| P36 | gi|253688172 | P. carotovorum PC1 | 369 | 52.6 | 5.00 | 29 | 14 |
| P37 | gi|253687524 | P. carotovorum PC1 | 148 | 22.6 | 5.74 | 29 | 5 |
| Transcription | |||||||
| P38 | gi|253686985 | P. carotovorum PC1 | 313 | 56.5 | 4.53 | 27 | 15 |
| P39 | gi|253688272 | P. carotovorum PC1 | 218 | 29.7 | 5.25 | 52 | 11 |
| Cell envelope synthesis | |||||||
| P40 | gi|145298428 | Aeromonas salmonicida ssp. salmonicida A449 | 133 | 41.4 | 5.78 | 19 | 4 |
| P41 | gi|253686650 | P. carotovorum PC1 | 230 | 16.6 | 4.71 | 20 | 3 |
| P42 | gi|50120070 | P. atrosepticum SCRI1043 | 338 | 67.9 | 6.04 | 19 | 10 |
| P43 | gi|253689156 | P. carotovorum PC1 | 323 | 43.9 | 5.74 | 24 | 8 |
| Oxidoreductase | |||||||
| P44 | gi|123443378 | Yersinia enterocolitica ssp. enterocolitica 8081 | 179 | 22.4 | 5.80 | 18 | 3 |
| P45 | gi| 251756507 | P. carotovorum PC1 | 135 | 99.8 | 5.46 | 11 | 11 |
| P46 | gi|253686701 | P. carotovorum PC1 | 463 | 27.1 | 5.14 | 37 | 12 |
| Others | |||||||
| P47 | gi|253687336 | P. carotovorum PC1 | 239 | 29.6 | 5.88 | 43 | 10 |
| P48 | gi|50119836 | P. atrosepticum SCRI1043 | 546 | 34.5 | 5.51 | 34 | 9 |
| P49 | gi|253688960 | P. carotovorum PC1 | 117 | 14.9 | 4.62 | 29 | 3 |
| P50 | gi|253686657 | P. carotovorum PC1 | 254 | 37.1 | 5.06 | 32 | 9 |
| P51 | gi| 253688966 | P. carotovorum PC1 | 101 | 60.1 | 4.74 | 3 | 6 |
| P52 | gi|148254924 | Bradyrhizobium sp. BTAil | 79 | 28.2 | 5.17 | 3 | 4 |
| P53 | gi| 49609794 | P. atrosepticum SCRI1043 | 52 | 24.5 | 5.11 | 9 | 2 |
Almost one‐half (25 proteins) of the 53 identified proteins belonged to groups I and III (Fig. 2): ‘carbohydrate metabolism’ and ‘protein synthesis, folding and degradation’, respectively. The majority of these proteins were from PccS1‐V‐Ze, which in almost all cases showed greater changes in expression than proteins from PccS1‐LB‐Ze. However, where changes in expression were observed, there was a good correlation with those exhibiting up‐ or down‐regulation between the two conditions (Fig. 3). In the other groups, almost all of the proteins were significantly up‐regulated in both PccS1‐LB‐Ze and PccS1‐V‐Ze, again with the majority of proteins from PccS1‐V‐Ze tending to exhibit greater changes in expression than those from PccS1‐LB‐Ze. All of the proteins in groups IV and V, involved in ‘nucleic acid metabolism’ and ‘transcription’, were up‐regulated in PccS1‐V‐Ze from at least 1.8‐ to over 10‐fold. From all of the above proteins, 10 (PotD [P08], ClpP [P09], PotF [P16], RpsD [P28], PrmA [P29], KdgR [P39], AceE [P45], MreB [P50], FlgK [P51] and MotA [P52]) were selected for further study from PccS1‐V‐Ze, and showed expression levels over 10‐fold up‐regulated, but with little or no differential expression in PccS1‐LB‐Ze (when compared with the PccS1‐LB control). In addition, eight proteins (Pgk [P05], Zwf [P07], AspA [P10], AtpD [P17], HslO [P27], Upp [P37], NusA [P38] and SspA [P53]) were selected showing up to 10‐fold up‐regulation in both PccS1‐V‐Ze and PccS1‐LB‐Ze compared with the control (in the majority of cases being more highly up‐regulated in PccS1‐V‐Ze than in PccS1‐LB‐Ze).
Changes in the transcriptional levels of genes encoding significantly up‐regulated proteins in the interaction of PccS1 with Z. elliotiana in vivo
To relate gene and protein expression levels, the transcription levels of genes encoding proteins with an eight‐ to more than 10‐fold increase in expression level following the interaction of PccS1 with Z. elliotiana in vivo (PccS1‐V‐Ze) were assessed by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) (Fig. 4), and compared with that of PccS1 cultured in LB (PccS1‐LB). All of these genes showed transcript levels up‐regulated in PccS1‐V‐Ze with increased expression in their respective proteins following proteomics, e.g. P29 (prmA), P39 (kdgR), P45 (aceE) and P50 (mreB) had transcriptional levels up‐regulated between four‐ and 11‐fold compared with the control (Fig. 4). In all cases, the transcript levels of these genes in PccS1‐LB‐Ze were similar to those of the control.
Figure 4.
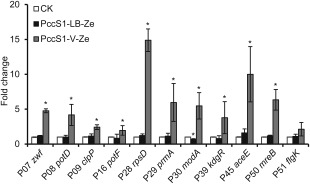
Relative mRNA levels of genes in Pectobacterium carotovorum ssp. carotovorum strain PccS1 in different conditions determined by quantitative reverse transcription‐polymerase chain reaction. The data were analysed using 16s rDNA as an endogenous control. The white bar represents the relative mRNA levels of genes from bacteria (PccS1‐LB) grown to stationary phase in Luria–Bertani broth (LB) used as a control, the black bar the relative mRNA levels of genes from bacteria (PccS1‐LB‐Ze) grown to stationary phase in LB + 0.4% petiole extract of Zantedeschia elliotiana, and the grey bar the relative mRNA levels of genes from bacteria following interaction with Z. elliotiana in vivo (PccS1‐V‐Ze) for 16 h. *Significant difference in mRNA level of the bacterial cells compared with that cultured in LB (P < 0.05, Student's t‐test).
Pathogenicity assay of mutant strains
Experiments were set up to investigate the role of the 10 differentially expressed proteins from PccS1‐V‐Ze in the virulence of PccS1. These experiments involved biparental mating to produce single gene deletion mutants of clpP [P09], prmA [P29], kdgR [P39], aceE [P45], mreB [P50] and flgK [P51]. Diagnostic PCR for individual genes confirmed that they were disrupted in the mutant strains (data not shown). No deletion mutants were obtained for potD [P08], potF [P16], rpsD [P28] and motA [P52], and these were not investigated further.
Pathogenicity tests of the six mutant strains compared with that of the wild‐type were performed by inoculation into detached leaves from whole plants of Z. elliotiana (Fig. 5A). The maceration assay revealed that there was no significant difference in virulence between the prmA, kdgR and aceE mutants and the wild‐type strain, whereas clpP, mreB and flgK mutants exhibited reduced virulence and a number of other phenotypic effects. Details on changes in virulence and other phenotypes, together with the complementation of mutants clpP, flgK and mreB, have been reported elsewhere (Deng et al., 2014; Li et al., 2013; Yang et al., 2012). It should be noted that both clpP and mreB mutants were also affected in growth in both rich and minimal media, which appears to explain their reduced virulence phenotype. ClpP and FlgK have been reported previously to play a role in the virulence of plant pathogens, including FlgK in Xanthomonas axonopodis pv. glycines on soybean (Athinuwat et al., 2009) and ClpP in D. dadantii on Chinese cabbage (Li et al., 2010). The roles of MreB and FlgK in virulence carried out as part of this study have also been reported previously (Deng et al., 2014; Yang et al., 2012).
Figure 5.
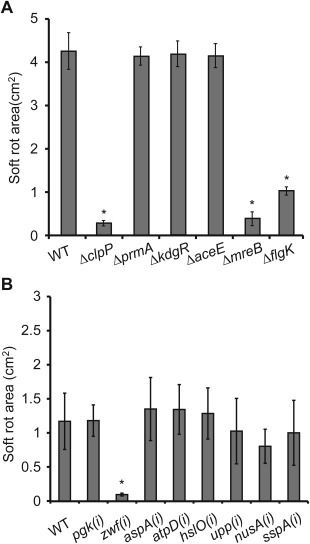
Disease lesions caused by Pectobacterium carotovorum ssp. carotovorum strain PccS1 (wild‐type) and deletion mutants ΔclpP, ΔmreB, ΔflgK, ΔprmA, ΔkdgR and ΔaceE in leaves of Zantedeschia elliotiana (A) and PccS1 (wild‐type) and insertion mutants pgk(i), zwf(i), aspA(i), atpD(i), hslO(i), upp(i), nusA(i) and sspA(i) in Chinese cabbage leaves (B). The leaves were inoculated at small wound sites with a 10‐μL bacterial suspension at an optical density at 600 nm (OD600) of 1.2. Three replicates were used for each treatment, and the experiment was repeated three times. Vertical bars represent standard errors. *Significant difference between the wild‐type strain and mutant (P < 0.05; t‐test).
The in vivo assay conducted uncovered proteins involved in virulence that were not identified in the in vitro screen, suggesting that a combination of both in vitro and in vivo analyses adds value to the search for virulence. However, the proteins identified from the in vivo screen have previously been associated with reduced virulence (see above). We therefore extended the study to include an additional set of proteins (Pgk [P05], Zwf [P07], AspA [P10], AtpD [P17], HslO [P27], Upp [P37], NusA [P38] and SspA [P53]), which showed increased expression both in vivo and in vitro, in an attempt to identify novel virulence determinants. To accelerate virulence testing for this group of proteins, we first generated insertion mutants and tested their virulence (Fig. 5B). Of these mutants, only zwf(i), affected in zwf and encoding an enzyme in the pentose phosphate pathway involved in metabolism, was reduced in virulence, and was taken forward for deletion mutagenesis in line with the mutants produced following the in vivo screen. However, when the zwf deletion mutant (Δzwf) was tested in planta, no reduction in virulence was observed.
In D. dadantii 3937, zwf lies upstream of gene eda in a two‐gene operon (Conway et al., 1991; Hugouvieux‐Cotte‐Pattat and Robert‐Baudouy, 1994). eda encodes 2‐keto‐3‐deoxy‐6‐phosphogluconate aldolase, an enzyme in the Entner–Doudoroff pathway involved in the catalysis of glucose to pyruvic acid as an alternative pathway to glycolysis in the production of ATP (Conway et al., 1991). Based on the incomplete draft genome sequence of PccS1, a similar operon structure was also observed in this strain (Fig. 6A). PCR, using primers zwf‐F and eda‐R on cDNA, confirmed that zwf and eda are co‐transcribed in PccS1 (Fig. 6A,B). To determine whether the insertion mutant in zwf affected virulence through its actions on the expression of eda, a deletion mutant in eda was generated and was shown to be reduced in virulence, whereas complementation of the eda mutant restored virulence (Fig. 6C). The mutation had no effect on growth in both LB and MM compared with the wild‐type and complemented strains (Fig. S2, see Supporting Information). An alignment of amino acids from Eda using BioEdit software (Hall, 1999) against six other Eda proteins, including D. dadantii 3937 KdgA, showed 88% identity with Escherichia coli K‐12 and 89% identity with D. dadantii 3937 (Fig. 7A). In line with the findings in D. dadantii 3937, which suggest that KdgA (an equivalent to Eda) is involved in the utilization of pectin ((Hugouvieux‐Cotte‐Pattat and Robert‐Baudouy, 1994), growth of the PccS1 eda mutant on pectinase agar was reduced compared with that of the wild‐type strain (Fig. 7B), whereas complementation of the mutant restored the wild‐type phenotype.
Figure 6.
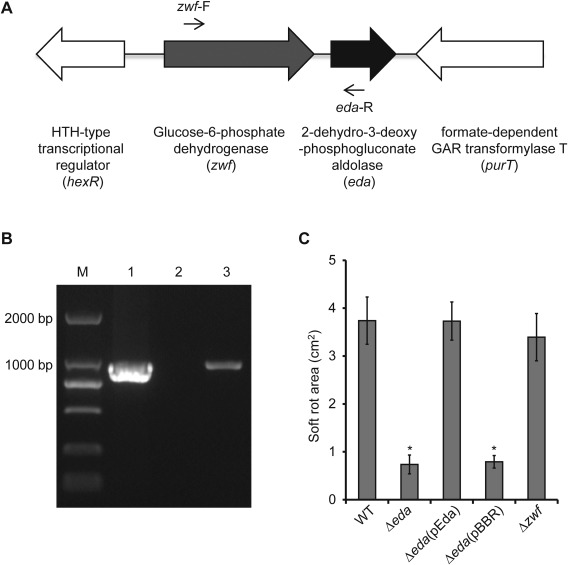
(A) Genetic structure of the zwf‐eda operon in the draft genome sequence of Pectobacterium carotovorum ssp. carotovorum strain PccS1. (B) Polymerase chain reaction (PCR) analysis showing the co‐transcription of zwf‐eda using the primers zwf‐F and eda‐R which span across both genes: 1, genomic DNA; 2, RNA (no reverse transcript); 3, cDNA. The product size is 1000 bp. (C) Pathogenicity assay on Zantedeschia elliotiana leaves showing the reduction in virulence of the Δeda deletion mutant and its subsequent complementation, but no reduction in virulence for Δzwf. *Significant difference between the wild‐type strain and mutant (P < 0.05; t‐test).
Figure 7.
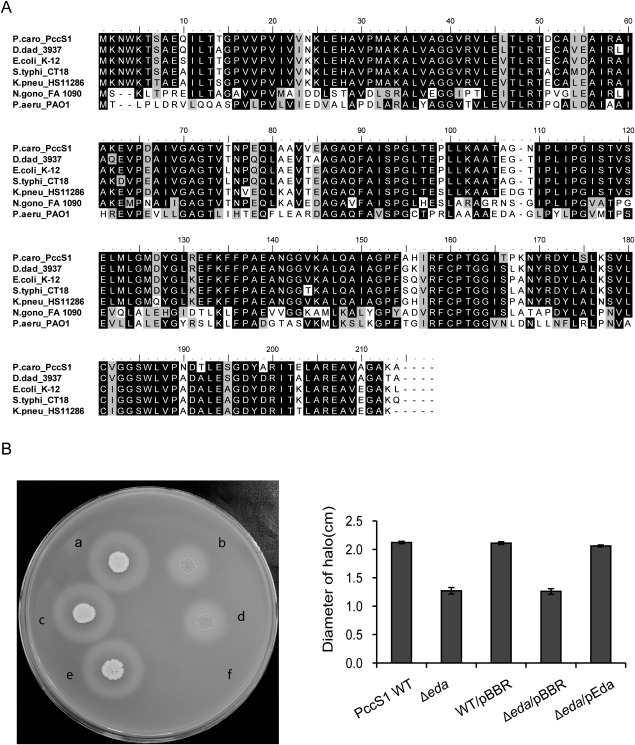
Protein alignment of Eda and growth of the eda mutant on pectinase agar. (A) Alignment of deduced amino acid sequences of Pectobacterium carotovorum PccS1 protein Eda with homologues from other bacteria. Identical residues are shaded in black, conservative substitutions (similarity > 70%) are shaded in grey and non‐conservative substitutions are shaded in white. The multiple sequence alignment was generated using BioEdit (Version 7.2.5) and the alignment output was produced using Adobe Ilustrator (CS5). Sequence IDs are abbreviated as follows: Pectobacterium carotovorum ssp. carotovorum PccS1, ‘P.caro_PccS1’; Dickeya dadantii 3937, ‘D.dad_3937’; Escherichia coli K‐12 substr. MG1655 (NC_000913.3 ), ‘E.coli_K12’; Salmonella enterica ssp. enterica serovar Typhi str. CT18 (NC_003198.1), ‘S.typhi_CT18’; Klebsiella pneumoniae ssp. pneumoniae (NC_016845.1), ‘K.pneu_HS11286’; Neisseria gonorrhoeae FA 1090 (NC_002946.2), ‘N. gono_FA 1090’; Pseudomonas aeruginosa PAO1 (NC_002516.2), ‘P.aeru_PAO1’. (B) Growth of PccS1 strains on pectinase agar at 27°C for 72 h followed by flooding with 7.5% (w/v) copper acetate for 1–2 h: a, PccS1 WT; b, Δeda; c, WT/pBBR; d, Δeda/pBBR; e, Δeda/pEda (complemented strain); f, MgSO4 buffer control.
Discussion
Recently, functional genomic strategies, including proteomics and transcriptomics, have contributed to our understanding of plant–pathogen interactions (Delaunois et al., 2014; Mark et al., 2005). Proteomics has improved dramatically to assist in the large‐scale functional assignment of candidate proteins involved in pathogenicity (Mehta et al., 2008). Such studies include the identification of important proteins involved in pathogenesis, e.g. Svx, a quorum‐sensing‐controlled virulence factor secreted via the type II secretion pathway (Corbett et al., 2005), and the porin OmpA, involved in binding to specific host cell receptor molecules (Babujee et al., 2007). As a result of the difficulties of extracting bacterial nucleic acids and proteins from plant material, most studies are performed by the growth of bacteria in medium supplemented with plant extracts to mimic in planta conditions, and this has revealed many important bacterial proteins involved in both pathogenic and symbiotic interactions with plants (Cheng et al., 2010). However, the major drawback of this approach is the lack of a true interaction between the pathogen and its host (Mark et al., 2005). Although an increasing number of researchers are studying the differential expression of nucleic acids in plants during both pathogenic and beneficial interactions, only a very few have addressed changes in the bacterial proteome during these interactions, e.g. proteomic analysis of Xanthomonas campestris pv. campestris during its interaction with the host plant in vivo (Andrade et al., 2008; Villeth et al., 2009). It is clear that differences are certain to occur between bacterial interactions with the host plant in vitro and in vivo, with many processes occurring in vivo that would not occur in vitro, e.g. the onset and targeting of host defences and the way in which a pathogen responds to these defences, such as altering one or more of its metabolic pathways or switching ion channels. Such complex interactions could not be replicated using plant extracts alone (Haider and Pal, 2013; Nie et al., 2007).
It is highly likely that the proteins found in our 2‐DE maps do not represent all of the proteins expressed during the infection process, but rather result in only a partial view of this aspect of the PccS1 proteome. The gel electrophoresis and subsequent MS identification steps are both biased largely towards highly abundant proteins (Lubec and Afjehi‐Sadat, 2007; Mehta et al., 2008). However, even with this proviso, there were still a large number of differentially expressed proteins (133 proteins) between the in vivo (PccS1‐V‐Ze) and in vitro (PccS1‐LB‐Ze) conditions, and between these conditions and the control PccS1‐LB (165 proteins). Among the 53 proteins successfully identified from these 165 proteins, the expression levels of 16 proteins in PccS1‐LB‐Ze were similar to those in PccS1‐LB, but only three proteins in PccS1‐V‐Ze showed the same expression level as in PccS1‐LB, suggesting that a more diverse range of proteins is differentially expressed in the in vivo condition (Fig. 3).
The 53 proteins were successfully identified and divided into eight groups depending on their functional annotation (Fig. 2). The most abundant proteins present in the total fraction were involved in protein synthesis, folding, degradation and carbohydrate metabolism. Many of the proteins that were up‐regulated in PccS1‐LB‐Ze in response to petiole exudates were involved in the utilization of nutrients. This observation is consistent with a transcriptomic study of bacterial responses to plant root exudates (Mark et al., 2005). However, in PccS1‐V‐Ze, expression of these proteins showed some differences with PccS1‐LB‐Ze, i.e. proteins including P09 (ClpP), P16 (PotF), P29 (PrmA), P39 (KdgR), P45 (AceE), P50 (MreB) and P51 (FlgK) were highly up‐regulated, whereas others, including P1 (Cpa), P3 (FruK) and P4 (Pgm), were highly down‐regulated. When the bacteria were cultured in medium supplemented with plant extracts, the metabolic pathways of the bacteria were affected, and the expression of nutrient‐related genes involved in pathogenesis was induced differentially. However, when the pathogen infects plants, these plants will produce defence factors, e.g. signal molecules, proteins and hormones, to defend against invasion by the pathogen, and pathogenic bacteria will also change their gene expression to influence the plant accordingly, complex reactions that cannot take place when using plant extracts alone. Perhaps for this reason, proteins involved in protein synthesis, folding and degradation from the in vivo conditions accounted for 28.3% of the eight protein functions. Another group of proteins that were up‐regulated belonged to nucleic acid metabolism and transportation, including ATP synthase, translocation protein and GTP‐binding protein, which have all been shown previously to be required during the interaction of bacteria with their plant hosts (Caldelari et al., 2006).
Among the genes encoding the 10 proteins with expression levels over 10‐fold in PccS1 when interacting with Z. elliotiana in vivo compared with in vitro, six genes were successfully deleted in PccS1 to generate mutants. The remaining four could not be deleted, and may therefore be necessary for PccS1 viability. The pathogenicity of the six mutants was tested on Z. elliotiana, and the results revealed that mutants ΔprmA, ΔaceE and ΔkdgR showed no significant difference in virulence compared with the wild‐type strain, and were therefore not studied further. However, mutants ΔclpP, ΔflgK and ΔmreB showed a significant reduction in virulence on the leaves or petioles of Z. elliotiana. ClpP is a serine‐type Prt and is required for many important functions, including growth at high temperature, motility, synthesis of degradative enzymes and sporulation in E. coli (Porankiewicz et al., 1999), the rapid adaptive response of intracellular pathogens, such as Listeria monocytogenes, during their infectious process (Gaillot et al., 2000), and the mediation of biofilm formation on implanted medical devices in Staphylococcus epidermidis (Wang et al., 2007). ClpP proteolytic activity is essential for the virulence of many animal‐pathogenic bacteria, including Salmonella enterica serovar typhimurium, Streptococcus pneumoniae, L. monocytogenes, E. coli and Staphylococcus aureus (Frees et al., 2012; Gaillot et al., 2000; Iyoda and Watanabe, 2005; Knudsen et al., 2013; Michel et al., 2006; Park et al., 2010; Tomoyasu et al., 2005), but there are fewer studies on ClpP associated with the virulence of plant‐pathogenic bacteria, e.g. only in D. dadantii 3937 (Li et al., 2010). FlgK is a component of the hook‐filament junction of flagella in bacteria, which is an organelle used for motility in liquids and on surfaces. Bacterial motility has been described as a pathogenicity determinant in pectobacteria (Perombelon, 2002). MreB is a rod shape‐determining and cytoskeletal protein, and has been implicated in peptidoglycan biosynthesis and cell stabilization (Garner et al., 2011). The results from these single gene deletion mutants of PccS1 suggest that all three genes are important to Pcc infection of the living host.
Although this study identified three proteins (ΔclpP, ΔflgK and ΔmreB) from the in vivo‐only condition that play a role in virulence, only MreB was identified for the first time as part of this study, and this finding has been published elsewhere (Deng et al., 2014). We therefore investigated proteins that were increased in expression in both in vivo and in vitro conditions in order to identify further virulence‐related proteins. Of the eight further genes tested, insertion mutagenesis identified only zwf as a potential virulence candidate. However, after further investigation, a mutation in the eda gene, which is adjacent to and downstream of zwf in the PccS1 genome, was found to be responsible for the reduced virulence phenotype. Although zwf is part of the pentose phosphate pathway that operates alongside glycolysis in primary bacterial metabolism, eda is part of the alternative Entner–Doudoroff pathway (Conway et al., 1991). In D. dadantii strain 3937 (formerly Erwinia chrysanthemi), the KDPG adolase enzyme encoded by kdgA (which is an alternative name used for the eda gene in this bacterium) is involved in the catabolism of pectins following breakdown of 2‐keto‐3‐deoxygluconate (KDG) to pyruvate and glyceraldehyde phosphate (Hugouvieux‐Cotte‐Pattat and Robert‐Baudouy, 1994). This is in line with our findings that the eda gene in PccS1 is highly similar to that of D. dadantii 3937 (Fig. 7A), and that the mutant is less able to grow on pectinase medium than the wild‐type and complemented strains (Fig. 7B). The role of eda, a key enzyme in the Entner–Doudoroff pathway, in virulence may thus be associated with the breakdown of pectin during the infection process.
Although this study was limited to the capture of proteins inside the cells of PccS1 only, and at only a single time point during the establishment of infection, we identified a number of proteins that showed increased expression in vivo compared with in vitro conditions, as well as in vivo and in vitro compared with the control. Based on these expression profiles, 14 mutants were produced in a number of selected genes, with four of these demonstrating a role in virulence of PccS1. Further studies are now required to use this and other approaches to identify further proteins/genes involved in the virulence of PccS1 on Z. elliotiana, and to further elucidate the role of Eda in the virulence of Pectobacterium.
Experimental Procedures
Bacterial strains, plasmids and culture conditions
The bacterial strains and plasmids used in this work are listed in Table S1 (see Supporting Information). All bacterial strains were cultured in Luria–Bertani broth (LB) or on LB agar plates (LA) (1% tryptone, 0.5% yeast extract, 1% NaCl and 1.5% agar for LA plate, w/v). The growth temperature for PccS1 and its mutant strains was 28°C and for E. coli strains was 37°C. When required, antibiotics were added to the medium at the following final concentrations: rifampicin (Rif), 100 µg/mL; kanamycin (Kan), 50 µg/mL; gentamicin (Gm), 25 µg/mL.
Plant extract preparation and bacterial growth
Plant extracts were prepared according to Mattinen et al. (2007) and Mehta and Rosato (2001). Briefly, 100 g of fresh petioles from Z. elliotiana ‘Black Magic’, following 45 days of growth in the glasshouse, were ground with 1000 mL of water in a blender. The mixture was then filtered through micro‐cloth and centrifuged at 4 000 g for 20 min. The supernatant was passed through a 0.2‐µm filter, followed by ultrafiltration (Millipore, Red Bank, NJ, USA) twice to eliminate proteins larger than 5 kDa. Finally, the extract was filter sterilized again and stored at −20°C.
Bacteria were grown in LB supplemented with 0.4% petiole extracts of Z. elliotiana (hereafter, PccS1‐LB‐Ze (in vitro)] until the optical density at 600 nm (OD600) reached 1.2, and were then used for protein extraction. Protein extracted from the bacteria grown to stationary phase in LB was used as a control (PccS1‐LB).
Infiltration and recovery of bacterial cells from the host plant
To determine the optimum period for maximum bacterial growth and cell recovery from inoculated plants, to enable sufficient total protein for analysis, cells were extracted just prior to collapse of the cell walls at the surface of necrosis in the petioles. Testing of the bacterial population dynamics in the host after inoculation was performed according to Andrade et al. (2008). Briefly, a single colony of PccS1 was inoculated into 20 mL of LB and grown at 28°C until stationary phase (OD600 = 1.2). After centrifugation at 3 000 g for 10 min at 4°C, the cell pellets were washed three times with distilled water and finally suspended in distilled water at a cell density of OD600 = 1.2. Potted plants of Z. elliotiana ‘Black Magic’ were grown in the glasshouse, and 45‐day‐old petioles were chosen for inoculation with 100 µL of PccS1 suspension. Water‐infiltrated petioles were used as controls. The infiltrated petioles were collected at 0, 4, 8, 12, 16 and 20 HAI, sterilized with 70% alcohol and then cut into smaller pieces with a sterile scalpel and maintained for 1 h in sterile glass plates containing 20 mL of distilled water to allow the bacteria to move from the petioles into the water. After this time, 100 µL of the water was plated onto LB and incubated at 28°C for 16 h, and the number of resulting colonies was counted. The tests were repeated three times with three to five replicate petioles at each time point, and the results were then subjected to a t‐test.
Pathogen cells taken directly from plant material [PccS1‐V‐Ze (in vivo)] were recovered at 16 HAI (population dynamics and protein concentration tests confirmed that this was the optimum period for cell recovery) and harvested by centrifugation at 4 000 g for 20 min. The cell pellets were used for protein extraction and total RNA isolation.
Extraction of proteins
Proteins were extracted from three replicate bacterial cell pellets of PccS1‐V‐Ze, PccS1‐LB and PccS1‐LB‐Ze for each condition. Briefly, the cell pellets were washed twice with 20 mL of washing buffer (50 m Tris‐HCI, pH 7.2), centrifuged at 14 000 g for 30 min at 4°C, and then disrupted by an Qproteome Bacterial Protein Prep Kit (Qiagen, Hilden, Germany) according to the Qproteome Bacterial Protein Preparation Handbook. The cell pellets were thawed on ice for 15 min and resuspended in 10 mL of native lysis buffer (with lysozyme and nuclease added). The samples were incubated on ice for 30 min and then gently mixed two or three times by swirling the solution before centrifugation at 14 000 g for 30 min at 4°C to pellet the cellular debris. The cell lysate supernatant was retained and then 100 mL of acetone were added and the samples were precipitated for 12 h at −20°C. The proteins were pelleted by centrifugation at 14 000 g for 20 min and the pellet was resuspended in rehydration buffer (RB). Protein samples were stored at −20°C and protein quantification was performed according to Bradford (1976). The concentration of the extracted protein was detected using a 2‐D Quant Kit (GE Healthcare, Chicago, IL, USA).
2‐DE, gel staining and image generation
Isoelectric focusing (IEF, first dimension) was performed with an IPGphor 3 system a gel system of immobilized pH gradients (GE Healthcare). Dry IPG strips (24 cm, pH 4–7, GE Healthcare) containing 1.2 mg of protein were rehydrated for 12 h at 20°C in rehydration buffer. IEF was performed in five steps: 300 V for 0.5 h, 700 V for 0.5 h, 1500 V for 1.5 h, linear gradient increase to 9000 V for 3 h and 9000 V for 4 h. The focused strips were equilibrated first in sodium dodecylsulfate (SDS) equilibration buffer (50 mm Tris, 6 m urea, 30% glycerol, 2% SDS, trace amount of bromophenol blue) containing 1% w/v dithiothreitol for 15 min, and then in equilibration solution with 4% w/v iodoacetamide for 15 min. For the second dimension, proteins were separated according to their molecular weights on 12.5% polyacrylamide gels. After being stained with colloidal Coomassie blue G‐250 (CBB), the wet gels were scanned using an ImageScanner (UMAX Powerlook1100, Taiwan, China). Gel images were analysed with ImageMaster 2D platinum 5.0 software (GE Healthcare). At least three gels were analysed for each of the different conditions. Proteins were considered to be differentially expressed when the average values exceeded the threshold of a two‐fold difference (P < 0.05) (Cheng et al., 2009; Zhao et al., 2011).
Trypsin digestion and MS analysis
The protein profile of PccS1 interaction with Z. elliotiana [PccS1‐V‐Ze (in vivo]] was compared with that from the cells of PccS1 cultured in LB [PccS1‐LB (control)] or LB + 0.4% petiole extract of Z. elliotiana [PccS1‐LB‐Ze (in vitro)]. Protein spots of interest were excised from the 2‐D gels and destained in 120 µL of 50% methanol. The pieces of gel were shrunk with 100 µL acetonitrile and then rehydrated with 50 µL of 100 mm NH4HCO3 three times, and the samples were dried by vacuum centrifugation in a vacuum heater. Trypsin solution was added to the dried gel spots and incubated at 4°C for 30 min. In‐gel tryptic digestion was performed overnight at 37°C. Samples were frozen at −20°C before spotting on a matrix‐assisted laser desorption/ionization (MALDI) plate. Aliquots of each hydrolysed sample were mixed with 1.5 µL of matrix solution of 0.5% α‐cyano‐4‐hydroxycinnamic acid (CHCA, w/v) in 70% acetonitrile and 30% trifluoroacetic acid (0.1% TFA, w/v), 800 µL of which were immediately spotted and air dried before MS analysis (Mattinen et al., 2007; Zhao et al., 2011). The samples were analysed on a 4700 Proteomics Analyzer MALDI‐TOF/TOF system (ABI, Carlsbad, CA, USA). All spectra were obtained in positive reflector mode, using an accelerating voltage of about 20 kV for the MS mode. Molecular ions displaying sufficient signal were submitted to MS/MS analyses, carried out in the positive precursor ion fragmentation mode at a laser frequency of 200 Hz. The MS/MS spectra were acquired in the reflector‐positive mode after collision‐induced dissociation (CID) or LIFT™ fragmentation with external calibration. The MS/MS spectra were interpreted automatically and manually (de novo sequencing) using Mascot distiller software. The peptide masses obtained were employed for identification with the MASCOT program (Matrix Science, London, UK) using the National Center for Biotechnology Information (NCBI) and Pfam (http://pfam.sanger.ac.uk/), and only the matches to Pectobacterium species were considered as positive identifications. Peptide sequences obtained by de novo sequencing were analysed using blast (Mattinen et al., 2007; Shevchenko et al., 1996; Villeth et al., 2009).
RNA extraction and qRT‐PCR analysis
To confirm the differential expression of proteins in PccS1 in vitro and following interaction with the host plant (in vivo), the transcriptional levels of the selected genes in PccS1 were measured by qRT‐PCR. Bacterial total RNA was extracted using an OMEGA Kit (OMEGA, Norcross, GA, USA) according to the manufacturer's instructions. The purity and concentration of RNA were determined with a microspectrophotometer (Nanodrop 1000, Thermo TaKaRa, Dalian, China) and gel electrophoresis. Two micrograms of RNA were used to synthesize cDNA according to the manufacturer's instructions for the PrimeScript™ RT Reagent (Perfect Real Time) Kit (TaKaRa). The 16S rDNA was used as a normalized control. The primers (Table S2, see Supporting Information) used for qRT‐PCR were designed using primer 3.0 (http://primer3.ut.ee/). qRT‐PCR was performed with an ABI 7500 Fast Real‐Time PCR System (Applied Biosystems, Carlsbad, CA, USA) using SYBR Premix Ex Taq™ (Perfect Real Time) (TaKaRa). Primer specificity was assessed using the dissociation curve protocol. The conditions for PCR amplification were as follows: initial denaturation of 95°C for 15 s, followed by 40 cycles of 95°C for 5 s and 64°C for 34 s. The data were analysed by 7500 software (v.2.0.6) and standard deviations were calculated. The expression of the genes was calculated by the comparative C T method described by Schmittgen and Livak (2008) using the following formula: fold change = 2–ΔΔ C T, where ΔΔC T i for gene of interest i = (C Tgene i – C T16SrDNA)mutant/(C Tgene i – C T16SrDNA)wild‐type. For the analysis of co‐transcribed genes, PCR with primers (Table S3, see Supporting Information) flanking zwf (zwf‐F) and eda (eda‐R) was conducted as described previously (Pacheco et al., 2012). Two micrograms of RNA (no reverse transcript) and cDNA were used as PCR templates, with genomic DNA used as the positive control. There were three independent biological experiments with sampling carried out in duplicate.
Gene modification in PccS1
Both gene insertion and gene deletion mutants were used in this study. Gene insertion mutants were constructed according to Yu et al. (2007). In short, the internal fragments from the target genes were amplified by the gene‐specific primers (Table S3), and released by KpnІ and HindIII digestion from the pMD19‐T (simple) subclones. These fragments were cloned into the same sites of pEX18 (Table S1) to produce gene insertion constructs. The constructs were conjugally transferred to PccS1 by E. coli S17‐1 (Table S1). The conjugant cells were spread onto LB plates supplemented with Rif and Gm. The plates were incubated at 28°C for 24–48 h for selection. The colonies that developed on the selection medium were candidates for target gene insertion mutants. To confirm the insertion mutants, in addition to the above gene‐specific primers, Gm‐specific primers and PccS1 strain‐specific primers were also used.
The gene deletion mutants were constructed and complemented according to Li et al. (2010). Briefly, the upstream and downstream regions of the target genes were amplified by PCR using the relevant primers (Table S3). A Kan cassette, amplified from pET30 (Table S1), was ligated with these two fragments and cloned into pMD19‐T, verified by sequencing, excised with appropriate restriction enzymes (as indicated in Table S3) and cloned into pEX18 (Table S1) to yield a gene insertion plasmid vector. This vector was transferred into PccS1 by conjugation using E. coli S17‐1 (Table S1). To select strains with chromosomal deletions, transconjugants were plated on LB containing 10% sucrose (Solarbio, Beijing, China) and Kan. Colonies having resistance to sucrose and Kan were isolated and confirmed by PCR using primers outside the gene of interest. To construct plasmids for complementation, target genes and their promoter region were amplified and cloned into pBBR1MCS5 (Table S1). Complementation vectors (Table S1) were introduced into the mutants by conjugation, as described above, and complemented strains were selected by resistance to Gm. All of the constructs and mutants were verified by PCR and DNA sequencing.
Pathogenicity assay
Pathogenicity assays were performed as described previously (Gu et al., 2009; Murata et al., 1991). PccS1 and the mutant strains were grown in LB at 28°C overnight. Cells were harvested and resuspended in distilled water and adjusted to an OD600 of 1.2 for inoculation. For all deletion mutants, petioles of leaves detached from approximately 45‐day‐old Z. elliotiana plants were used for the pathogenicity assays. For insertion mutants, as a screen prior to retesting of deletion mutants on Z. elliotiana, leaves of Chinese cabbage were used. Before inoculation, the leaves were washed thoroughly with distilled water. For each site, the surface of the leaves was punctured with a sterile toothpick and 10 μL of the wild‐type or mutant strains of PccS1 were inoculated in parallel into the tissue. Distilled water was inoculated into leaves as a control. The leaves were placed in a moist container and incubated in a moist chamber at 28°C. The area (cm2) of maceration was measured at 16 HAI. The pathogenicity assays were repeated three times with three replicates for each treatment. Growth of the eda mutant in both LB and MM (with 0.2% glucose) at 28°C was compared with that of the wild‐type and complemented strains over a 12‐h period following inoculation of a 100‐fold dilution of overnight culture of the same medium. Where relevant, the growth of other mutants in rich and minimal media was tested and published elsewhere.
Exo‐enzyme activity assays
To examine the role of the PccS1 eda gene in pectin utilization, the exo‐enzyme activity of the wild‐type and mutant strains was assayed. Briefly, 10 μL of an overnight culture (normalized to 104 cells/mL in 10 mm MgSO4) were spotted onto pectinase plates as described in Andro et al. (1984) and incubated at 27°C for 72 h, followed by flooding with 7.5% (w/v) copper acetate for 1–2 h. Colonies positive for Pel activity produce creamy double haloes on a translucent light blue background. Three repetitions were performed for each assay.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Bacterial population dynamics curve during the establishment of infection. A Pectobacterium carotovorum ssp. carotovorum strain PccS1 double‐distilled water suspension (100 µL) [optical density at 600 nm (OD600) = 1.2] was inoculated in 45‐day‐old petioles of potted plants of Zantedeschia elliotiana. The infiltrated petioles were collected at 0, 4, 8, 12, 16 and 20 h after inoculation (HAI), and the number of colonies were counted. The tests were repeated three times with three to five plants each time and the results were subjected to t‐test.
Fig. S2 Growth of Pectobacterium carotovorum ssp. carotovorum strain PccS1 wild‐type (WT), Δeda, WT with empty vector pBBR, Δeda with empty vector pBBR and Δeda with complementing vector pBBR (pEda). Luria–Bertani broth (LB) (A) and minimal medium (MM) plus 0.2% glucose (B) over a 12‐h period at 28°C.
Table S1 Bacterial strains and plasmids used in this study.
Table S2 Primers used in this study for quantitative real‐time polymerase chain reaction.
Table S3 Oligonucleotide sequences for gene modification in this study.
Acknowledgements
This work was supported by the Special Fund for Agro‐Scientific Research in the Public Interest of China (Grant No. 201303015) and the China Scholarship Council (No. 201406850042). It was supported by a grant from the Scottish Government's Rural and Environmental Sciences and Analytical Services (RESAS) Division. The authors declare no conflicts of interest.
Contributor Information
Ian Toth, Email: ian.toth@hutton.ac.uk.
Jiaqin Fan, Email: fanjq@njau.edu.cn.
References
- Andrade, A.E. , Silva, L.P. , Pereira, J.L. , Noronha, E.F. , Reis, F.B. , Bloch, C. , dos Santos, M.F. , Domont, G.B. , Franco, O.L. and Mehta, A. (2008) In vivo proteome analysis of Xanthomonas campestris pv. campestris in the interaction with the host plant Brassica oleracea . FEMS Microbiol. Lett. 281, 167–174. [DOI] [PubMed] [Google Scholar]
- Andro, T. , Chambost, J.P. , Kotoujansky, A. , Cattaneo, J. , Bertheau, Y. , Barras, F. , Van Gijsegem, F. and Coleno, A. (1984) Mutants of Erwinia chrysanthemi defective in secretion of pectinase and cellulase. J. Bacteriol. 160, 1199–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athinuwat, D. , Prathuangwong, S. , Cursino, L. and Burr, T. (2009) Xanthomonas axonopodis pv. glycines soybean cultivar virulence specificity is determined by avrBs3 homolog avrXg1 . Phytopathology, 99, 996–1004. [DOI] [PubMed] [Google Scholar]
- Babujee, L. , Venkatesh, B. , Yamazaki, A. and Tsuyumu, S. (2007) Proteomic analysis of the carbonate insoluble outer membrane fraction of the soft‐rot pathogen Dickeya dadantii (syn. Erwinia chrysanthemi) strain 3937. J. Proteome Res. 6, 62–69. [DOI] [PubMed] [Google Scholar]
- Bell, K.S. , Sebaihia, M. , Pritchard, L. , Holden, M.T.G. , Hyman, L.J. , Holeva, M.C. , Thomson, N.R. , Bentley, S.D. , Churcher, L.J. , Mungall, K. , Atkin, R. , Bason, N. , Brooks, K. , Chillingworth, T. , Clark, K. , Doggett, J. , Fraser, A. , Hance, Z. , Hauser, H. , Jagels, K. , Moule, S. , Norbertczak, H. , Ormond, D. , Price, C. , Quail, M.A. , Sanders, M. , Walker, D. , Whitehead, S. , Salmond, G.P. , Birch, P.R. , Parkhill, J. and Toth, I.K. (2004) Genome sequence of the enterobacterial phytopathogen Erwinia carotovora subsp atroseptica and characterization of virulence factors. Proc. Natl. Acad. Sci. USA, 101, 11 105–11 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortoli‐German, I. , Brun, E. , Py, B. , Chippaux, M. and Barras, F. (1994) Periplasmic disulphide bond formation is essential for cellulase secretion by the plant pathogen Erwinia chrysanthemi . Mol. Microbiol. 11, 545–553. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Caldelari, I. , Mann, S. , Crooks, C. and Palmer, T. (2006) The tat pathway of the plant pathogen Pseudomonas syringae is required for optimal virulence. Mol. Plant–Microbe Interact. 19, 200–212. [DOI] [PubMed] [Google Scholar]
- Cheng, Z.Y. , Duan, J. , Hao, Y.A. , McConkey, B.J. and Glick, B.R. (2009) Identification of bacterial proteins mediating the interactions between Pseudomonas putida UW4 and Brassica napus (Canola). Mol. Plant–Microbe Interact. 22, 686–694. [DOI] [PubMed] [Google Scholar]
- Cheng, Z.Y. , McConkey, B.J. and Glick, B.R. (2010) Proteomic studies of plant–bacterial interactions. Soil Biol. Biochem. 42, 1673–1684. [Google Scholar]
- Chung, W.J. , Shu, H.Y. , Lu, C.Y. , Wu, C.Y. , Tseng, Y.H. , Tsai, S.F. and Lin, C.H. (2007) Qualitative and comparative proteomic analysis of Xanthomonas campestris pv. campestris 17. Proteomics, 7, 2047–2058. [DOI] [PubMed] [Google Scholar]
- Conway, T. , Yi, K.C. , Egan, S.E. , Wolf, R.E. Jr. and Rowley, D.L. (1991) Locations of the zwf, edd, and eda genes on the Escherichia coli physical map. J. Bacteriol. 173, 5247–5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett, M. , Virtue, S. , Bell, K. , Birch, P. , Burr, T. , Hyman, L. , Lilley, K. , Poock, S. , Toth, I. and Salmond, G. (2005) Identification of a new quorum‐sensing‐controlled virulence factor in Erwinia carotovora subsp. atroseptica secreted via the type II targeting pathway. Mol. Plant–Microbe Interact. 18, 334–342. [DOI] [PubMed] [Google Scholar]
- Delaunois, B. , Jeandet, P. , Clement, C. , Baillieul, F. , Dorey, S. and Cordelier, S. (2014) Uncovering plant–pathogen crosstalk through apoplastic proteomic studies. Front Plant Sci. 5, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, Y.M. , Du, S. , Wang, H. , Yang, Z.L. , Liu, F.Q. and Fan, J.Q. (2014) Functional characterization of mreB gene from Pectobacterium carotovorum subsp. carotovorum . J. Nanjing Agric. Univ. 37, 45–52. [Google Scholar]
- Frees, D. , Andersen, J.H. , Hemmingsen, L. , Koskenniemi, K. , Baek, K.T. , Muhammed, M.K. , Gudeta, D.D. , Nyman, T.A. , Sukura, A. , Varmanen, P. and Savijoki, K. (2012) New insights into Staphylococcus aureus stress tolerance and virulence regulation from an analysis of the role of the ClpP protease in the strains Newman, COL, and SA564. J. Proteome Res. 11, 95–108. [DOI] [PubMed] [Google Scholar]
- Gaillot, O. , Pellegrini, E. , Bregenholt, S. , Nair, S. and Berche, P. (2000) The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes . Mol. Microbiol. 35, 1286–1294. [DOI] [PubMed] [Google Scholar]
- Garner, E.C. , Bernard, R. , Wang, W.Q. , Zhuang, X.W. , Rudner, D.Z. and Mitchison, T. (2011) Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis . Science, 333, 222–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinter, R. , Milner, J. and Walker, D. (2012) Ferredoxin containing bacteriocins suggest a novel mechanism of iron uptake in Pectobacterium spp. PLoS One, 7, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, C.Y. , Fan, J.Q. , Yang, X. , Hu, B.S. , Liu, F.Q. and Zhang, Y.C. (2009) Identification of the pathogen and quorum quenching study on bacterial soft rot of colored calla lily. J. Nanjing Agric. Univ. 32, 71–77. [Google Scholar]
- Haider, S. and Pal, R. (2013) Integrated analysis of transcriptomic and proteomic data. Curr. Genomics, 14, 91–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, T.A. (1999) BioEdit: a user‐friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. [Google Scholar]
- Heikinheimo, R. , Flego, D. , Pirhonen, M. , Karlsson, M.B. , Eriksson, A. , Mae, A. , Kõiv, V. and Palva, E.T. (1995) Characterization of a novel pectate lyase from Erwinia carotovora sbsp carotovora . Mol. Plant–Microbe Interact. 8, 207–217. [DOI] [PubMed] [Google Scholar]
- Hinton, J.C. , Gill, D.R. , Lalo, D. , Plastow, G.S. and Salmond, G.P. (1990) Sequence of the peh gene of Erwinia carotovora: homology between Erwinia and plant enzymes. Mol. Microbiol. 4, 1029–1036. [DOI] [PubMed] [Google Scholar]
- Hugouvieux‐Cotte‐Pattat, N. and Robert‐Baudouy, J. (1994) Molecular analysis of the Erwinia chrysanthemi region containing the kdgA and zwf genes. Mol. Microbiol. 11, 67–75. [DOI] [PubMed] [Google Scholar]
- Ibrahim, M. , Shi, Y. , Qiu, H. , Li, B. , Jabeen, A. , Li, L.P. , Liu, H. , Kube, M. , Xie, G. , Wang, Y. , Blondel, C. , Santiviago, C.A. , Contreras, I. and Sun, G. (2012) Differential expression of in vivo and in vitro protein profile of outer membrane of Acidovorax avenae subsp avenae . PloS One, 7, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyoda, S. and Watanabe, H. (2005) ClpXP protease controls expression of the type III protein secretion system through regulation of RpoS and GrlR levels in enterohemorrhagic Escherichia coli . J. Bacteriol. 187, 4086–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen, G.M. , Olsen, J.E. , Aabo, S. , Barrow, P. , Rychlik, I. and Thomsen, L.E. (2013) ClpP deletion causes attenuation of Salmonella typhimurium virulence through mis‐regulation of RpoS and indirect control of CsrA and the SPI genes. Microbiology, 159, 1497–1509. [DOI] [PubMed] [Google Scholar]
- Koskinen, J.P. , Laine, P. , Niemi, O. , Nykyri, J. , Harjunpaa, H. , Auvinen, P. , Paulin, L. , Pirhonen, M. , Palva, T. and Holm, L. (2012) Genome sequence of Pectobacterium sp. strain SCC3193. J. Bacteriol. 194, 6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T. , Jiang, H. , Yang, Z.L. , Deng, Y.M. , Du, S. , Liu, F.Q. , Wang, H. and Fan, J. (2013) Functional characterization of clpP gene from Pectobacterium carotovorum subsp. carotovorum . J. Nanjing Agric. Univ. 36, 58–64. [Google Scholar]
- Li, Y. , Yamazaki, A. , Zou, L.F. , Biddle, E. , Zeng, Q.A. , Wang, Y.J. , Lin, H. , Wang, Q. and Yang, C.H. (2010) ClpXP protease regulates the type III secretion system of Dickeya dadantii 3937 and is essential for the bacterial virulence. Mol. Plant–Microbe Interact. 23, 871–878. [DOI] [PubMed] [Google Scholar]
- Lubec, G. and Afjehi‐Sadat, L. (2007) Limitations and pitfalls in protein identification by mass spectrometry. Chem. Rev. 107, 3568–3584. [DOI] [PubMed] [Google Scholar]
- Luzzatto‐Knaan, T. , Kerem, Z. , Doron‐Faigenboim, A. and Yedidia, I. (2014) Priming of protein expression in the defence response of Zantedeschia aethiopica to Pectobacterium carotovorum . Mol. Plant Pathol. 15, 364–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, B. , Hibbing, M.E. , Kim, H.S. , Reedy, R.M. , Yedidia, I. , Breuer, J. , Glasner, J.D. , Perna, N.T. , Kelman, A. and Charkowski, A.O. (2007) Host range and molecular phylogenies of the soft rot enterobacterial genera Pectobacterium and Dickeya . Phytopathology, 97, 1150–1163. [DOI] [PubMed] [Google Scholar]
- Ma, B. , Charkowski, A.O. , Glasner, J.D. and Perna, N.T. (2014) Identification of host–microbe interaction factors in the genomes of soft rot‐associated pathogens Dickeya dadantii 3937 and Pectobacterium carotovorum WPP14 with supervised machine learning. BMC Genomics, 15, 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mae, A. , Heikinheimo, R. and Palva, E.T. (1995) Structure and regulation of the Erwinia carotovora subspecies carotovora SCC3193 cellulase gene celV1 and the role of cellulase in phytopathogenicity. Mol. Gen. Genet. 247, 17–26. [DOI] [PubMed] [Google Scholar]
- Mansfield, J. , Genin, S. , Magori, S. , Citovsky, V. , Sriariyanum, M. , Ronald, P. , Dow, M. , Verdier, V. , Beer, S.V. , Machado, M.A. , Toth, I. , Salmond, G. and Foster, G.D. (2012) Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark, G.L. , Dow, J.M. , Kiely, P.D. , Higgins, H. , Haynes, J. , Baysse, C. , Abbas, A. , Foley, T. , Franks, A. , Morrissey, J. and O'Gara, F. (2005) Transcriptome profiling of bacterial responses to root exudates identifies genes involved in microbe–plant interactions. Proc. Natl. Acad. Sci. USA, 102, 17 454–17 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattinen, L. , Nissinen, R. , Riipi, T. , Kalkkinen, N. and Pirhonen, M. (2007) Host‐extract induced changes in the secretome of the plant pathogenic bacterium Pectobacterium atrosepticum . Proteomics, 7, 3527–3537. [DOI] [PubMed] [Google Scholar]
- Mehta, A. and Rosato, Y.B. (2001) Differentially expressed proteins in the interaction of Xanthomonas axonopodis pv. citri with leaf extract of the host plant. Proteomics, 1, 1111–1118. [DOI] [PubMed] [Google Scholar]
- Mehta, A. and Rosato, Y.B. (2003) A simple method for in vivo expression studies of Xanthomonas axonopodis pv. citri . Curr. Microbiol. 47, 400–403. [DOI] [PubMed] [Google Scholar]
- Mehta, A. , Brasileiro, A.C.M. , Souza, D.S.L. , Romano, E. , Campos, M.A. , Grossi‐De‐Sa, M.F. , Silva, M.S. , Franco, O.L. , Fragoso, R.R. , Bevitori, R. and Rocha, T.L. (2008) Plant–pathogen interactions: what is proteomics telling us?. FEBS J. 275, 3731–3746. [DOI] [PubMed] [Google Scholar]
- Michel, A. , Agerer, F. , Hauck, C.R. , Herrmann, M. , Ullrich, J. , Hacker, J. and Ohlsen, K. (2006) Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J. Bacteriol. 188, 5783–5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata, H. , Mcevoy, J.L. , Chatterjee, A. , Collmer, A. and Chatterjee, A.K. (1991) Molecular cloning of an aepa gene that activates production of extracellular pectolytic, cellulolytic, and proteolytic enzymes in Erwinia carotovora subsp carotovora . Mol. Plant–Microbe Interact. 4, 239–246. [Google Scholar]
- Ni, L. , Guo, L. , Custers, J.B.M. and Zhang, L. (2010) Characterization of calla lily soft rot caused by Pectobacterium carotovorum subsp carotovorum ZT0505: bacterial growth and pectate lyase activity under different conditions. J. Plant Pathol. 92, 421–428. [Google Scholar]
- Nie, L. , Wu, G. , Culley, D.E. , Scholten, J.C.M. and Zhang, W. (2007) Integrative analysis of transcriptomic and proteomic data: challenges, solutions and applications. Crit. Rev. Biotechnol. 27, 63–75. [DOI] [PubMed] [Google Scholar]
- Pacheco, A.R. , Curtis, M.M. , Ritchie, J.M. , Munera, D. , Waldor, M.K. , Moreira, C.G. and Sperandio, V. (2012) Fucose sensing regulates bacterial intestinal colonization. Nature, 492, 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, C.Y. , Kim, E.H. , Choi, S.Y. , Tran, T.D.H. , Kim, I.H. , Kim, S.N. , Pyo, S. and Rhee, D.K. (2010) Virulence attenuation of Streptococcus pneumoniae clpP mutant by sensitivity to oxidative stress in macrophages via an NO‐mediated pathway. J. Microbiol. 48, 229–235. [DOI] [PubMed] [Google Scholar]
- Park, T.H. , Choi, B.S. , Choi, A.Y. , Choi, I.Y. , Heu, S. and Park, B.S. (2012) Genome sequence of Pectobacterium carotovorum subsp. carotovorum strain PCC21, a pathogen causing soft rot in Chinese cabbage. J. Bacteriol. 194, 6345–6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perombelon, M.C.M. (2002) Potato diseases caused by soft rot erwinias: an overview of pathogenesis. Plant Pathol. 51, 1–12. [Google Scholar]
- Pirhonen, M. , Saarilahti, H. , Karlsson, M.B. and Palva, E.T. (1991) Identification of pathogenicity determinants of Erwinia carotovora subsp carotovora by transposon mutagenesis. Mol. Plant–Microbe Interact. 4, 276–283. [Google Scholar]
- Porankiewicz, J. , Wang, J.M. and Clarke, A.K. (1999) New insights into the ATP‐dependent Clp protease: Escherichia coli and beyond. Mol. Microbiol. 32, 449–458. [DOI] [PubMed] [Google Scholar]
- Schmittgen, T.D. and Livak, K.J. (2008) Analyzing real‐time PCR data by the comparative C‐T method. Nat. Protoc. 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Shevchenko, A. , Wilm, M. , Vorm, O. and Mann, M. (1996) Mass spectrometric sequencing of proteins from silver stained polyacrylamide gels. Anal. Chem. 68, 850–858. [DOI] [PubMed] [Google Scholar]
- Snijder, R.C. , Cho, H.R. , Hendriks, M.M.W.B. , Lindhout, P. and van Tuyl, J.M. (2004) Genetic variation in Zantedeschia spp. (Araceae) for resistance to soft rot caused by Erwinia carotovora subsp carotovora . Euphytica, 135, 119–128. [Google Scholar]
- Tomoyasu, T. , Takaya, A. , Handa, Y. , Karata, K. and Yamamoto, T. (2005) ClpXP controls the expression of LEE genes in enterohaemorrhagic Escherichia coli . FEMS Microbiol. Lett. 253, 59–66. [DOI] [PubMed] [Google Scholar]
- Toth, I.K. , Bell, K.S. , Holeva, M.C. and Birch, P.R.J. (2003) Soft rot erwiniae: from genes to genomes. Mol. Plant Pathol. 4, 17–30. [DOI] [PubMed] [Google Scholar]
- Villeth, G.R. , Reis, F.B. , Tonietto, A. , Huergo, L. , de Souza, E.M. , Pedrosa, F.O. , Franco, O.L. and Mehta, A. (2009) Comparative proteome analysis of Xanthomonas campestris pv. campestris in the interaction with the susceptible and the resistant cultivars of Brassica oleracea . FEMS Microbiol. Lett. 298, 260–266. [DOI] [PubMed] [Google Scholar]
- Wang, C.Z. , Li, M. , Dong, D. , Wang, J.P. , Ren, J. , Otto, M. and Gao, Q. (2007) Role of ClpP in biofilm formation and virulence of Staphylococcus epidermidis . Microbes Infect. 9, 1376–1383. [DOI] [PubMed] [Google Scholar]
- Wright, P.J. (1998) A soft rot of calla (Zantedeschia spp.) caused by Erwinia carotovora subspecies carotovora . NZ J. Crop Hort. 26, 331–334. [Google Scholar]
- Yang, Z.L. , Deng, Y.M. , Du, S. , Li, T. , Jiang, H. , Liu, F.Q. and Fan, J. (2012) Function of flgK gene in Pectobacterium carotovorum subsp. carotovorum . Wei sheng wu xue bao (Acta Microbiol. Sin.) 52, 703–709. [PubMed] [Google Scholar]
- Yap, M.N. , Barak, J.D. and Charkowski, A.O. (2004) Genomic diversity of Erwinia carotovora subsp carotovora and its correlation with virulence. Appl. Environ. Microbiol. 70, 3013–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yishay, M. , Burdman, S. , Valverde, A. , Luzzatto, T. , Ophir, R. and Yedidia, I. (2008) Differential pathogenicity and genetic diversity among Pectobacterium carotovorum ssp carotovorum isolates from monocot and dicot hosts support early genomic divergence within this taxon. Environ. Microbiol. 10, 2746–2759. [DOI] [PubMed] [Google Scholar]
- Yu, F. , Zaleta‐Rivera, K. , Zhu, X. , Huffman, J. , Millet, J.C. , Harris, S.D. , Yuen, G. , Li, X.C. and Du, L. (2007) Structure and biosynthesis of heat‐stable antifungal factor (HSAF), a broad‐spectrum antimycotic with a novel mode of action. Antimicrob. Agents Chemother. 51, 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Qian, G. , Yin, F. , Fan, J. , Zhai, Z. , Liu, C. , Hu, B. and Liu, F. (2011) Proteomic analysis of the regulatory function of DSF‐dependent quorum sensing in Xanthomonas oryzae pv. oryzicola . Microb. Pathog. 50, 48–55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Bacterial population dynamics curve during the establishment of infection. A Pectobacterium carotovorum ssp. carotovorum strain PccS1 double‐distilled water suspension (100 µL) [optical density at 600 nm (OD600) = 1.2] was inoculated in 45‐day‐old petioles of potted plants of Zantedeschia elliotiana. The infiltrated petioles were collected at 0, 4, 8, 12, 16 and 20 h after inoculation (HAI), and the number of colonies were counted. The tests were repeated three times with three to five plants each time and the results were subjected to t‐test.
Fig. S2 Growth of Pectobacterium carotovorum ssp. carotovorum strain PccS1 wild‐type (WT), Δeda, WT with empty vector pBBR, Δeda with empty vector pBBR and Δeda with complementing vector pBBR (pEda). Luria–Bertani broth (LB) (A) and minimal medium (MM) plus 0.2% glucose (B) over a 12‐h period at 28°C.
Table S1 Bacterial strains and plasmids used in this study.
Table S2 Primers used in this study for quantitative real‐time polymerase chain reaction.
Table S3 Oligonucleotide sequences for gene modification in this study.


