Summary
Erwinia amylovora is the causal agent of the fire blight disease in some plants of the Rosaceae family. The non‐host plant Arabidopsis serves as a powerful system for the dissection of mechanisms of resistance to E. amylovora. Although not yet known to mount gene‐for‐gene resistance to E. amylovora, we found that Arabidopsis activated strong defence signalling mediated by salicylic acid (SA), with kinetics and amplitude similar to that induced by the recognition of the bacterial effector avrRpm1 by the resistance protein RPM1. Genetic analysis further revealed that SA signalling, but not signalling mediated by ethylene (ET) and jasmonic acid (JA), is required for E. amylovora resistance. Erwinia amylovora induces massive callose deposition on infected leaves, which is independent of SA, ET and JA signalling and is necessary for E. amylovora resistance in Arabidopsis. We also observed tumour‐like growths on E. amylovora‐infected Arabidopsis leaves, which contain enlarged mesophyll cells with increased DNA content and are probably a result of endoreplication. The formation of such growths is largely independent of SA signalling and some E. amylovora effectors. Together, our data reveal signalling requirements for E. amylovora‐induced disease resistance, callose deposition and cell fate change in the non‐host plant Arabidopsis. Knowledge from this study could facilitate a better understanding of the mechanisms of host defence against E. amylovora and eventually improve host resistance to the pathogen.
Keywords: callose, cell growth, defence signalling, effectors, endoreplication, fire blight, programmed cell death
Introduction
The Gram‐negative bacterium Erwinia amylovora belongs to the enterobacterial family, which includes human and animal pathogens, such as Yersinia spp., Shigella spp. and Salmonella spp. Erwinia amylovora is the causative agent of the devastating fire blight disease challenging certain rosaceous plants, including apples, pears and some ornamental plants (Eastgate, 2000; Vanneste and Eden‐Green, 2001). Outbreaks of fire blight cause tremendous economic losses worldwide. The disease is usually managed through an integrated approach, consisting of horticultural practice, antibiotics and chemical applications, and cultivar breeding (Gusberti et al., 2015). Genetic engineering provides a potentially powerful approach to enhance plant resistance to E. amylovora, the success of which relies on a thorough understanding of the mechanisms governing plant resistance to E. amylovora. Through whole‐genome sequencing, transcriptomics and molecular studies, many genes with potential roles in the regulation of E. amylovora resistance have been identified in plants (Kamber et al., 2016; Malnoy et al., 2012). An understanding of the molecular mechanisms by which these genes act to enhance resistance against E. amylovora is important for the development of strategies to reduce and/or prevent fire blight and to mitigate losses in agriculture.
Erwinia amylovora employs various virulence factors to suppress plant innate immunity and promote bacterial virulence (Oh and Beer, 2005; Pique et al., 2015; Vrancken et al., 2013). One of the major virulence factors of E. amylovora is the type III secretion system (TTSS) that delivers effector proteins to plant cells to cause disease (Grant et al., 2006; Hueck, 1998). Erwinia amylovora and Pseudomonas syringae share similar TTSS genes that are clustered in a large pathogenicity island on the bacterial chromosome, a region also called the hypersensitive response and pathogenicity (hrp) and disease‐specific (dsp) gene cluster (Alfano and Collmer, 2004; Choi et al., 2013). Among the effector proteins delivered by the TTSS of E. amylovora, DspA/E is a functional homologue of AvrE of P. syringae and plays a critical role in the promotion of E. amylovora virulence and cell death in both host and non‐host plants (Bogdanove et al., 1998a; Boureau et al., 2006; Gaudriault et al., 1997; Oh et al., 2007). Either as a secreted effector of E. amylovora or as a cell‐free elicitor, HrpN has been shown to induce a hypersensitive response (HR), callose deposition, broad‐spectrum disease resistance and/or growth of treated plants (Boureau et al., 2011; Dong et al., 1999; Reboutier et al., 2007; Wei et al., 1992). Erwinia amylovora also produces and secretes two polysaccharides, amylovoran and levan, which are important for biofilm formation of the bacterium and presumably also play a role in the survival of the bacterium in host tissue. These compounds could block host vascular tissue, thereby causing the wilting of vegetative shoots and promoting bacterial virulence (Eastgate, 2000; Pique et al., 2015; Vrancken et al., 2013).
The molecular mechanisms by which plants resist E. amylovora are not well understood. Such resistance is generally considered to be quantitative, possibly involving multiple genetic loci in host genomes (Khan et al., 2013; Le Roux et al., 2010; Malnoy et al., 2012). No specific gene‐for‐gene interaction involving the recognition of E. amylovora effectors by specific plant resistance genes has been reported so far. In addition to host plants (e.g. apple and pear), non‐host plants (e.g. Arabidopsis and tobacco) have been used to elucidate the plant mechanisms of resistance to E. amylovora. Although host plants show varying susceptibility to E. amylovora, non‐host plants generally show durable resistance to the bacterium. Despite the differences in supporting the growth of the bacterium, both types of plant respond to E. amylovora infection by the activation of certain common defence pathways, including cell ion leakage, cell death, callose deposition, defence gene expression and defence signalling pathways (Bogdanove et al., 1998a; Boureau et al., 2006, 2011; DebRoy et al., 2004; Dong et al., 1999; Gaudriault et al., 1997; Launay et al., 2016; Oh et al., 2007; Reboutier et al., 2007; Venisse et al., 2001; Wei et al., 1992). A study with Arabidopsis has also indicated that de novo protein synthesis is required for non‐host resistance to E. amylovora (Moreau et al., 2012). Thus, non‐host plants provide a powerful system to elucidate the plant mechanisms of resistance to E. amylovora.
Although E. amylovora induces multifaceted defence responses in plants, whether some of these responses are required for plant resistance to the bacterium is not well understood. For instance, although exogenous jasmonic acid (JA) treatment protects apple plants against E. amylovora, some JA‐related genes are induced and others are suppressed by E. amylovora infection (Degrave et al., 2008; Duge De Bernonville et al., 2012). Thus, it is unclear whether JA is required for E. amylovora resistance. Salicylic acid (SA) is another defence molecule linked to E. amylovora resistance. The treatment of apple and pear with SA analogues protects the plants from fire blight disease (Brisset et al., 2000; Sparla et al., 2004). The expression of many SA regulatory genes is induced by E. amylovora infection in apple and Arabidopsis (Bonasera et al., 2006; Degrave et al., 2008; Moreau et al., 2012; Venisse et al., 2002). Overexpression of the apple homolog of Arabidopsis NONEXPRESSOR OF PR GENES 1 (NPR1; a key SA signalling gene), MpNPR1, confers increased disease resistance to E. amylovora in apple (Malnoy et al., 2007). In addition, the Arabidopsis NONEXPRESSOR OF PR GENES 1 (NPR1) gene is required for HrpN of E. amylovora‐induced disease resistance in Arabidopsis (Dong et al., 1999). Although these studies support the importance of SA in E. amylovora resistance, the lack of loss‐of‐function analysis of key SA genes, e.g. using mutants or gene silencing, makes it difficult to assess whether SA signalling is required for E. amylovora resistance in host plants. Indeed, some studies from the non‐host Arabidopsis challenge this role of SA. Loss of function in the SA regulatory gene, ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1), compromises E. amylovora resistance in Arabidopsis (Aarts et al., 1998; Degrave et al., 2008; Moreau et al., 2012). However, a mutant impaired in ISOCHORISMATE SYNTHASE 1 (ICS1) (also called SA INDUCTION‐DEFICIENT 2 and EDS16), encoding the major SA biosynthetic enzyme (Nawrath and Metraux, 1999; Ng et al., 2011; Wildermuth et al., 2001), did not alter E. amylovora‐induced cell death, compared with the wild‐type (Degrave et al., 2008). Therefore, whether or not SA is required for E. amylovora resistance is still in question.
In this report, we examined the defence signalling pathways required for resistance to E. amylovora. We confirmed that E. amylovora infection activates SA production in both the host plant apple and the non‐host plant Arabidopsis. Although not yet known to mount gene‐for‐gene resistance to E. amylovora in plants (Malnoy et al., 2012), Arabidopsis in the presence of E. amylovora activates SA signalling with a similar amplitude and kinetics to that induced by recognition of the P. syringae effector avrRpm1 by the Arabidopsis resistance protein RPM1. Genetic analysis revealed that E. amylovora resistance in Arabidopsis is dependent on SA, but independent of JA and ethylene (ET) pathways. We further showed that SA, ET and JA pathways are not required for callose synthesis induced by E. amylovora. In addition, we observed that E. amylovora induced tumour‐like growths in Arabidopsis, which contain enlarged cells with increased nuclear DNA content. The formation of such abnormal growths is largely independent of SA and E. amylovora effectors. Together, this study reveals signalling requirements for E. amylovora‐induced disease resistance, callose deposition and cell fate change in the non‐host plant Arabidopsis. Information obtained from this study could be used to better understand the defence mechanisms of host plants in response to E. amylovora, and subsequently aid in the development of strategies to improve plant resistance to the destructive fire blight disease.
Results
Arabidopsis activates strong SA‐mediated defence in response to E. amylovora
The model plant Arabidopsis shows non‐host resistance to E. amylovora and has been used as a powerful system to study the plant mechanisms of E. amylovora resistance. In order to further elucidate how Arabidopsis activates defence against E. amylovora, we compared the kinetics of SA accumulation and SA signalling activation (using the expression of the SA marker gene PR1) induced by E. amylovora with those induced by some P. syringae strains. We used two doses {optical density at 600 nm (OD600) = 0.1 [108 colony‐forming units (cfu)/mL] and 0.01 (107 cfu/mL)} of E. amylovora and collected the inoculated leaves at 0, 6, 12, 24 and 48 h post‐inoculation (hpi). In parallel, we also included three P. syringae pv. maculicola (Pma) strains. Pma HrcC– lacks the T3SS to deliver effectors to plant cells, and thus only activates basal defence in the host (Hauck et al., 2003; Tsuda et al., 2008a; Wang et al., 2009). Pma DG3 is a virulent strain that induces effector‐triggered susceptibility and Pma avrRpm1 (also called Pma DG34) expresses the avirulent effector avrRpm1, recognized by the R protein RPM1, and can induce effector‐triggered immunity (Alfano et al., 2000; Guttman et al., 2002). The data for the P. syringae strains have been reported previously (Hamdoun et al., 2013). Here, we only included the data for Pma HrcC– in Fig. 1 for a comparison. We found that SA accumulation induced by E. amylovora (0.1) was observed as early as 6 hpi and was much higher than that induced by Pma HrcC– (0.1) (Fig. 1A,B). Consistent with SA accumulation, PR1 transcripts were also much higher in plants infected with E. amylovora (0.1) than those infected with Pma HrcC– (0.1) (Fig. 1C). The degree of SA and PR1 induction by E. amylovora was dosage dependent, as much lower accumulation of SA and PR1 transcripts was observed with the infection at the lower dose of E. amylovora (0.01) (Fig. 1). Interestingly, the kinetics and amplitude of SA and PR1 transcripts induced by E. amylovora (0.1) were actually comparable with those induced by the avirulent strain Pma avrRpm1, and faster and higher than those induced by the virulent strain Pma DG3 (Fig. 1 and Hamdoun et al., 2013). Erwinia amylovora‐induced SA accumulation occurs largely through the major SA biosynthetic pathway catalysed by ICS1, because a loss‐of‐function mutation in ICS1, ics1–1, abolished SA accumulation on E. amylovora infection (Fig. S1, see Supporting Information).
Figure 1.
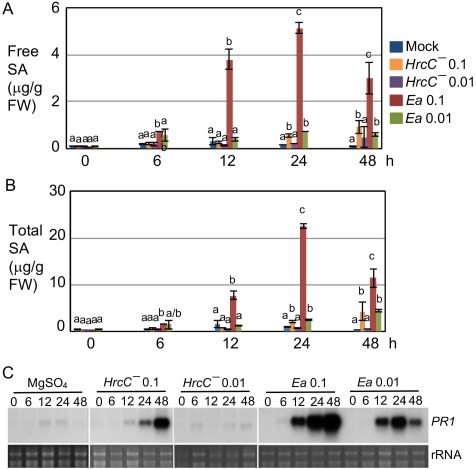
Erwinia amylovora induces dynamic changes in salicylic acid (SA) accumulation and PR1 expression in Arabidopsis. The fourth to sixth leaves of Col‐0 plants were infiltrated with E. amylovora (Ea) at an optical density at 600 nm (OD600) of 0.1 or 0.01, Pseudomonas syringae pv. maculicola strain HrcC– at OD600 = 0.1 or 0.01 or 10 mm MgSO4. The infected leaves were collected at 0, 6, 12, 24 and 48 h post‐inoculation (hpi) for SA and RNA analyses. (A) Free SA measurement. (B) Total SA measurement. (C) Northern blotting analysis of PR1 expression. Images of rRNA were used as loading controls. Statistical analysis was performed with one‐way analysis of variance (ANOVA) Fisher's partial least‐squares difference (PLSD) tests (StatView 5.0.1). Different letters in (A) and (B) indicate significant difference among the samples at each time point (P < 0.05). These experiments were repeated twice with similar results. FW, fresh weight.
Compared with the rapid and strong activation of the SA pathway in Arabidopsis on E. amylovora infection (Fig. 1), the host plant apple (Malus × domestica cv. ‘Royal Gala’) only produced modest levels of SA when challenged with E. amylovora (108 cfu/mL) at 48 hpi (Fig. S2, see Supporting Information). Such a modest activation of the SA pathway has also been reported previously (Milcevicová et al., 2010). The difference in SA pathway activation between the host plant apple and the non‐host plant Arabidopsis underscores the importance of SA signalling in non‐host resistance against E. amylovora.
Arabidopsis responds to Pma avrRpm1 via typical gene‐for‐gene interaction, using the plant R protein RPM1 to recognize the bacterial avirulence protein AvrRpm1. This recognition could result in an HR within hours of infection. Although E. amylovora‐induced SA signalling activation showed similar kinetics and amplitude to that induced by Pma avrRpm1 (Fig. 1 and Hamdoun et al., 2013), E. amylovora is not known to activate a rapid HR in Arabidopsis as shown with Pma avrRpm1 infection. Only limited leaf necrosis was observed in E. amylovora‐infected leaves several days post‐infection (Degrave et al., 2008 and our observation). The HR is usually correlated with massive ion leakage in the dying cells. Consistent with the lack of acute HR at the early infection stage, we found that E. amylovora induced much less ion leakage in Col‐0, compared with Pma avrRpm1 (Fig. 2). Thus, unlike Pma avrRpm1, E. amylovora‐induced SA signal activation is uncoupled from massive cell death.
Figure 2.
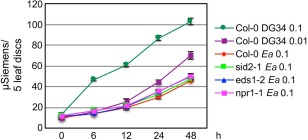
Erwinia amylovora‐induced ion leakage in Arabidopsis is independent of salicylic acid (SA) and less than that induced by RPM1–avrRpm1 recognition. Arabidopsis leaves were infiltrated with E. amylovora (Ea) or Pseudomonas syringae pv. maculicola (Pma DG34) avrRpm1 and collected after infection for ion leakage measurement. The data represent the average of triplicate samples ± standard deviation. These experiments were repeated twice with similar results.
Non‐host resistance to E. amylovora in Arabidopsis is SA dependent, but independent of ET‐ and JA‐mediated signalling
Although SA and some SA pathway‐related genes are known to be induced by E. amylovora in Arabidopsis and apple, whether or not SA is required for E. amylovora resistance has not been firmly established. The Arabidopsis mutant defective in the SA regulatory gene EDS1, but not in the major SA synthesis gene ICS1, showed greater cell death than wild‐type plants on E. amylovora infection (Degrave et al., 2008; Moreau et al., 2012). These observations suggest that EDS1, but not SA in general, is required for E. amylovora resistance in Arabidopsis.
To further clarify the role of SA in defence against E. amylovora in Arabidopsis, we infiltrated additional SA mutants (eds5–3, pad4–1, npr1–1 and nahG, in addition to ics1–1 and eds1–2) with E. amylovora. EDS5 encodes a chloroplast protein proposed to transport SA from the chloroplast to the cytoplasm in a cell (Nawrath et al., 2002; Serrano et al., 2013; Yamasaki et al., 2013). PHYTOALEXIN DEFICIENT 4 (PAD4) interacts with EDS1 and probably acts in the same pathway as EDS1 for defence regulation based on biochemical, genetic and microarray studies (Bartsch et al., 2006; Feys et al., 2001; Jirage et al., 1999; Ng et al., 2011; Sato et al., 2007). Both eds5–3 and pad4–1 mutants accumulate much reduced SA levels under defence conditions (Feys et al., 2001; Nawrath et al., 2002; Ng et al., 2011). NPR1 is a master regulator of SA signalling (Seyfferth and Tsuda, 2014; Yan and Dong, 2014). The nahG plant overexpresses the bacterial SA hydroxylase which metabolizes SA to catechol, leading to a greater susceptibility of plants to a range of pathogens (Delaney et al., 1994; Friedrich et al., 1995; Gaffney et al., 1993; Kachroo et al., 2000; Lu et al., 2001; Mur et al., 1997; van Wees and Glazebrook, 2003). To allow better growth of E. amylovora in Arabidopsis plants, we covered the infiltrated plants with a dome to maintain high humidity. In addition, we used an E. amylovora strain with streptomycin resistance for a more accurate quantification of E. amylovora replication on Arabidopsis leaves based on antibiotic selection. Under these conditions, we found that all SA mutants tested were able to support greater E. amylovora growth than Col‐0, and the nahG plant showed the highest bacterial population (Fig. 3). These data establish that SA is required for non‐host resistance to E. amylovora in Arabidopsis.
Figure 3.
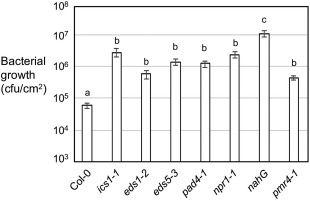
Salicylic acid (SA)‐defective plants and the pmr4–1 mutant disrupted in callose synthesis are more susceptible than Col‐0 to Erwinia amylovora. The fourth to sixth leaves of 30‐day‐old plants were infiltrated with E. amylovora at an optical density at 600 nm (OD600) of 0.1. Infected leaf discs (38 mm2) were taken for the measurement of bacterial growth at 3 days post‐inoculation (dpi). The initial inoculum (0 dpi) was about 8 × 103 colony‐forming units (cfu)/leaf disc for these plants. Each data point represents the average of 20–24 samples from four independent experiments. The error bars represent the standard error of the mean. Statistical analysis was performed with Student's t‐test (StatView 5.0.1). Different letters indicate significant difference among the samples (P < 0.05).
To further test how the SA pathway is related to cell death formation during the Arabidopsis–E. amylovora interaction, we measured ion leakage in several SA mutants. We found that the SA mutants showed a similar level of ion leakage to Col‐0 on E. amylovora infection (Fig. 2). These data further support that SA‐mediated defence is uncoupled from E. amylovora‐induced cell death.
To investigate whether additional defence signalling pathways are involved in E. amylovora resistance, we infected mutants impaired in ET or JA signalling. ein2–1 is an ET‐insensitive mutant (Alonso et al., 1999), whereas jar1–1 and jin1–7 are JA‐insensitive mutants (Lorenzo et al., 2004; Staswick et al., 1992). We found that these ET and JA mutants did not differ from Col‐0 in their ability to support E. amylovora growth (Fig. S3, see Supporting Information). Therefore, we conclude that ET and JA pathways are dispensable for resistance to E. amylovora in Arabidopsis.
Erwinia amylovora‐induced callose deposition is independent of SA, ET and JA pathways
Erwinia amylovora is known to induce massive callose deposition in apple and Arabidopsis plants (Boureau et al., 2011; DebRoy et al., 2004; Degrave et al., 2008). To better understand the signalling pathways required for E. amylovora‐induced cell wall defence in plants, we examined callose deposition on E. amylovora infection in Arabidopsis mutants disrupted in SA, ET and/or JA‐mediated defence. We found that, on E. amylovora infection, single mutants disrupted in SA, ET or JA pathways showed similar levels of callose deposition to Col‐0 (Figs S4 and S5, see Supporting Information). Further, disruption of two or more pathways mediated by SA, ET and/or JA did not affect E. amylovora‐induced callose deposition (Fig. 4). Thus, we conclude that E. amylovora‐induced callose deposition is independent of SA, ET and JA pathways. However, callose deposition was completely abolished in the powdery mildew resistant 4–1 (pmr4–1) mutant, which shows impaired callose synthesis (Fig. 4 and Moreau et al., 2012; Nishimura et al., 2003). Interestingly, we found that pmr4–1 supported greater E. amylovora growth (Fig. 3). Because the SA mutants were compromised in E. amylovora resistance, but not in callose deposition, and the callose‐deficient mutant pmr4–1 was compromised in E. amylovora resistance, we conclude that callose deposition is necessary, but not sufficient, for E. amylovora resistance.
Figure 4.
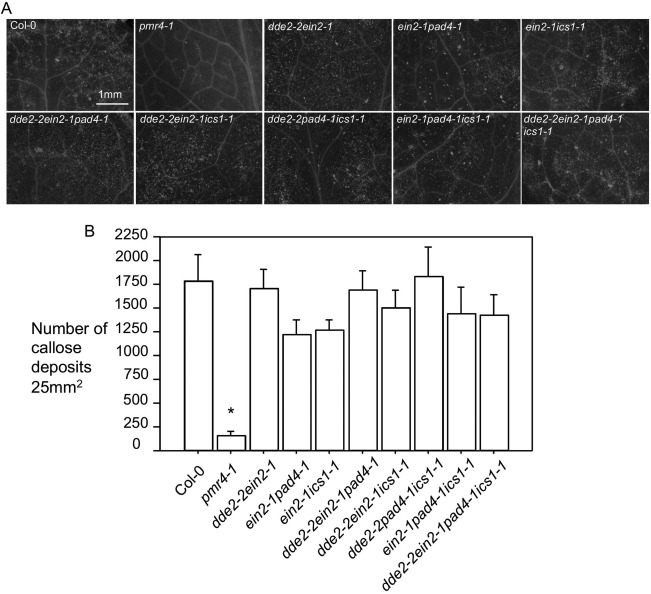
Erwinia amylovora‐induced callose deposition in Arabidopsis is independent of salicylic acid (SA), ethylene (ET) and jasmonic acid (JA) signalling. Erwinia amylovora [optical density at 600 nm (OD600) = 0.01]‐infiltrated leaves of each genotype were harvested for fixation and staining with aniline blue. (A) Images of callose deposits. Callose deposits appeared as fluorescent dots and were photographed with an AxioCam MRc5 camera connected to a fluorescence stereoscope microscope. (B) Quantification of callose deposits. The number of callose deposits of each genotype was quantified using ImageJ (Version 1.45s). Each data point was an average of at least six images from four different leaves ± standard deviation. Statistical analysis was performed with one‐way analysis of variance (ANOVA) Fisher's partial least‐squares difference (PLSD) tests (StatView 5.0.1). The asterisk indicates significant difference of pmr4–1 from other genotypes (P < 0.05). These experiments were repeated twice with similar results.
Erwinia amylovora infection induces cell growth in Arabidopsis
Bacteria, fungi and nematodes are known to induce abnormal cell growth in addition to promoting cell death in plants (de Almeida Engler et al., 1999; Chalupowicz et al., 2006; Chandran et al., 2009; Hamdoun et al., 2013; Hansen and Meins, 1986; Marois et al., 2002). Erwinia amylovora is known to induce cell death in both host and non‐host plants, but whether it affects cell growth has not been reported. We observed tumour‐like growths on E. amylovora‐infiltrated Arabidopsis leaves at 4 days post‐inoculation (dpi), which appeared to be transparent protrusions that varied in size (Fig. S6, see Supporting Information). For a closer observation of the abnormal growths, we fixed the growth regions and embedded them in LR white resin. With thin sections (1 µm), we found that the growth regions contained mostly enlarged mesophyll cells and the number of enlarged cells varied in different tumours (Fig. 5A). Quantification of abnormal growths by counting the clear protrusions on E. amylovora‐infiltrated leaves revealed that abnormal growths were induced in an E. amylovora dosage‐dependent manner (Fig. 5B).
Figure 5.
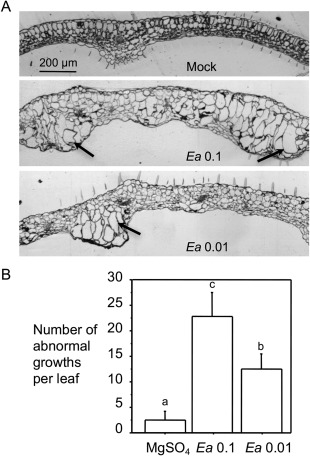
Erwinia amylovora induces abnormal cell growths in Arabidopsis. Erwinia amylovora‐infiltrated (Ea) and 10 mm MgSO4‐treated (Mock) leaves were observed for leaf morphology. (A) Leaf cross‐sections. The treated leaves were fixed, embedded in LR white resin and cut with an ultramicrotome into 1‐µm sections. Leaf cross‐sections were stained with 0.1% toluidine blue O and photographed using an AxioCam MRc5 camera (Zeiss, Inc., Göttingen, Germany) connected to a dissecting microscope. Arrows indicate enlarged cells in abnormal growth regions in leaves. The size bar represents 200 μm and applies to all images. (B) Quantification of the tumour‐like growths. The abnormal growths appeared to be transparent protrusions on the infected leaves and were counted at 5 days post‐inoculation (dpi) with the assistance of a dissecting microscope. At least 25 leaves were used for counting. Error bars represent standard deviation. Statistical analysis was performed with one‐way analysis of variance (ANOVA) Fisher's partial least‐squares difference (PLSD) tests (StatView 5.0.1). Different letters indicate significant difference amongst the samples (P < 0.05).
Enlarged cells are usually associated with increased nuclear content, which probably results from endoreplication, a process involving multiple rounds of DNA synthesis without cell mitosis (Bramsiepe et al., 2010; De Veylder et al., 2011). To quantify the nuclear content of the enlarged cells, we fixed the abnormal growth regions and embedded them in paraffin. Leaf cross‐sections, 15 µm thick, were stained with 4′,6‐diamidino‐2‐phenylindole (DAPI) to visualize nuclei. Consistent with their increased cell size, the enlarged cells had much larger nuclei (Fig. 6A–F). The stained nuclei were measured for relative fluorescence intensity using ImageJ (Version 1.45s). We arbitrarily set the average fluorescence value measured for guard cells as 2C. The relative nuclear DNA content of other cell types was calculated on the basis of that of the guard cells. We found that normal mesophyll cells had an average of about 20C nuclear DNA content. The enlarged cells, however, had an average nuclear DNA content of 60C, suggesting an activation of endoreplication in these cells (Fig. 6G).
Figure 6.
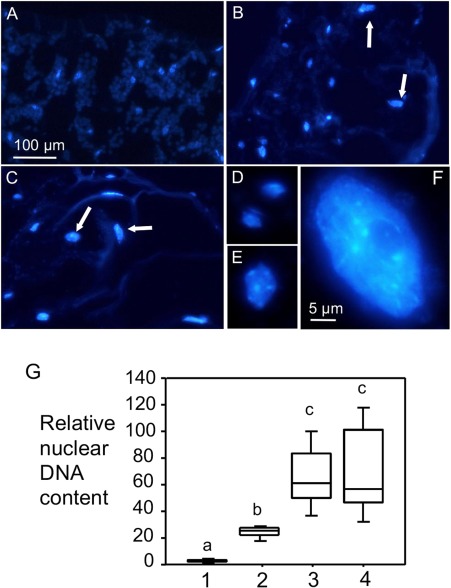
Erwinia amylovora infection induces the formation of enlarged mesophyll cells with increased nuclear content. Erwinia amylovora‐infiltrated or mock‐treated leaves were harvested at 5 days post‐inoculation (dpi), fixed and embedded in paraplast, and sectioned for 15‐µm slices with a microtome. Leaf cross‐sections were stained with 4′,6‐diamidino‐2‐phenylindole (DAPI) to visualize nuclei. Nuclei images (A–F) were captured using a Nikon DS cooled camera attached to a compound fluorescence microscope. The images are from the following samples: (A) 10 mm MgSO4; (B) E. amylovora [optical density at 600 nm (OD600) = 0.1]; (C) E. amylovora (OD600 = 0.01); (D) typical nuclei of guard cells from (A); (E) a typical nucleus of a normal mesophyll cell from (A); (F) a typical nucleus of an enlarged mesophyll cell induced by E. amylovora. Arrows indicate large nuclei (B, C). The size bar in (A) represents 100 µm and applies to (A–C), whereas the size bar in (F) represents 5 μm and applies to (D–F). (G) Quantification of relative nuclear DNA content. The numbers represent the relative nuclear DNA contents of the following types of cell: 1, guard cell nuclei from mock‐treated leaves; the value was set to 2C for the calculation of the relative nuclear contents of the other cell types; 2, normal mesophyll cells from mock‐treated leaves; 3, enlarged mesophyll cells induced by E. amylovora (OD600 = 0.1); 4, enlarged mesophyll cells induced by E. amylovora (OD600 = 0.01). At least 60 nuclei were used for each data point. Error bars represent standard deviation. Statistical analysis was performed with one‐way analysis of variance (ANOVA) Fisher's partial least‐squares difference (PLSD) tests (StatView 5.0.1). Different letters indicate significant difference amongst the samples (P < 0.05). The nuclear contents of guard cells and normal mesophyll cells from E. amylovora‐infected leaves were comparable with those from the corresponding cells in mock‐treated leaves (data not shown).
To elucidate the factors from Arabidopsis which affect E. amylovora‐induced cell growth, we examined several SA mutants. We found that ics1–1, pad4–1 and npr1–1 mutants had a similar number of tumour‐like growths to Col‐0, whereas nahG plants showed far fewer growths than Col‐0 (Fig. 7). It is worth noting that nahG plants were hypersusceptible to E. amylovora (Fig. 3) and most leaves had wilted by the time tumour growths were counted, preventing the assessment of cell growth (data not shown). Like the SA mutants, pmr4–1 had a similar number of tumour‐like growths to Col‐0 in the presence of E. amylovora. These results suggest that E. amylovora‐induced cell growth in Arabidopsis is largely independent of the SA pathway and callose formation.
Figure 7.
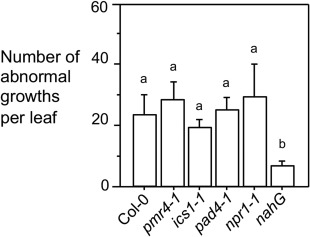
Erwinia amylovora‐induced tumour growths are largely independent of salicylic acid (SA). Plants were infiltrated with E. amylovora [optical density at 600 nm (OD600) = 0.1] or 10 mm MgSO4 as a control and observed for leaf morphology. The number of abnormal growths was counted at 5 days post‐inoculation (dpi) on the infected leaves with the assistance of a dissecting microscope. At least 25 leaves per genotype were used for counting. Error bars represent standard deviation. Different letters indicate significant difference amongst the samples (P < 0.05). These experiments were repeated twice with similar results.
We further tested whether E. amylovora effectors are required for cell growth control in Arabidopsis. Both dspE/A and hrpN are E. amylovora effectors delivered to the host via the TTSS, and these proteins play crucial roles in E. amylovora virulence (Bogdanove et al., 1998a; Boureau et al., 2006, 2011; Dong et al., 1999; Gaudriault et al., 1997; Oh et al., 2007; Wei et al., 1992). Erwinia amylovora strains lacking these effectors or the functional TTSS compromised the virulence of the bacterium (Fig. 8A and Boureau et al., 2006; Wei et al., 1992). We found that these three strains did not affect the number of abnormal growths in Arabidopsis, compared with the wild‐type E. amylovora strain (Fig. 8B). These data indicate that E. amylovora effectors are not necessary for cell growth in Arabidopsis.
Figure 8.
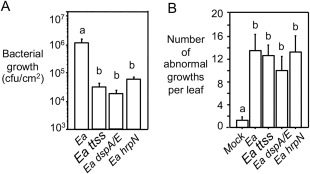
Erwinia amylovora effectors are not required for tumour growth in Arabidopsis. Plants were infiltrated with different E. amylovora strains [optical density at 600 nm (OD600) = 0.1] (Ea) or with 10 mm MgSO4 as a control (Mock). (A) Quantification of bacterial growth (cfu, colony‐forming unit). Each data point represents the average of 12 samples from two independent experiments ± standard error of the mean. (B) Quantification of abnormal growth. At least 25 leaves per genotype were used for each counting. Error bars represent standard deviation. Different letters indicate significant difference amongst the samples (P < 0.05). This experiment was repeated twice with similar results.
Discussion
Although not a natural host to E. amylovora, Arabidopsis activates strong defence against the bacterium and thus has been used as a powerful system for the dissection of the mechanisms of resistance to E. amylovora. Here, we report a study to elucidate the signalling requirements for some E. amylovora‐induced phenotypes in order to gain a better understanding of the non‐host resistance displayed by Arabidopsis to the bacterium. Our data showed that E. amylovora infection activates strong SA production and signalling in Arabidopsis with kinetics and amplitude similar to that induced by RPM1–avrRpm1 recognition during Arabidopsis–P. syringae interaction. An intact SA pathway, but not JA and ET pathways, is required for E. amylovora resistance in Arabidopsis. Callose deposition activated by E. amylovora can be uncoupled from defence signalling mediated by SA, JA and ET. Erwinia amylovora also induces cell growth in Arabidopsis, largely independent of plant SA signalling or effectors from the pathogen. Overall, this study has revealed the molecular components involved in the interaction between the non‐host plant Arabidopsis and E. amylovora. The knowledge obtained from this study could be employed to better understand the resistance mechanisms used by host plants, such as apple and pear, to defend against E. amylovora.
Non‐host resistance is the most common resistance against a broad range of pathogens in plants and can be grouped into two types (Gill et al., 2015; Senthil‐Kumar and Mysore, 2013; Thordal‐Christensen, 2003). Type I non‐host resistance induces no symptoms because the pathogens cannot overcome pre‐existing obstacles in plants, such as physical and chemical barriers. Type II non‐host resistance is associated with the development of cell death at the infection site. Erwinia amylovora activates type II non‐host resistance in Arabidopsis. However, we did not observe the typical HR symptoms at early infection stages as shown with Pma avrRpm1. Limited necrotic cell death was only observed at later stages of E. amylovora infection (Degrave et al., 2008 and data not shown). Consistent with these observations, E. amylovora elicited much less ion leakage than Pma avrRpm1 in Arabidopsis (Fig. 2). Nevertheless, E. amylovora induces rapid and strong accumulation of SA and PR1 transcripts in Arabidopsis, the kinetics and amplitude of which are similar to those induced by RPM1–avrRpm1 recognition (Fig. 1 and Hamdoun et al., 2013). Thus, non‐host resistance to E. amylovora observed with Arabidopsis shares similarities to, but is also different from, the resistance triggered by certain gene‐for‐gene interactions. The similarities could be explained by the activation of certain common downstream defence signalling pathways in plants. Indeed, both E. amylovora and P. syringae share the conserved TTSS and some effector genes (Alfano and Collmer, 2004; Choi et al., 2013; Oh and Beer, 2005), which could induce these downstream defence pathways in plants. However, Arabidopsis–P. syringae interaction involves classical gene‐for‐gene resistance, whereas it is unknown whether plants can use their specific resistance genes to recognize any cognate effectors of E. amylovora (Khan et al., 2013; Le Roux et al., 2010; Malnoy et al., 2012). In general, resistance to E. amylovora is considered as a quantitative trait which may involve multiple gene loci. Such differences in pathogen recognition by plants could explain the lack of acute HR in E. amylovora‐infected plants. However, we cannot rule out the possibility that Arabidopsis has evolved R proteins to recognize specifically certain E. amylovora effectors without invoking any HR.
Amongst the known defence signalling pathways, SA, JA and ET pathways have been linked to plant responses to E. amylovora. The data reported here demonstrate that SA signalling is required for E. amylovora resistance in Arabidopsis, whereas JA and ET pathways are dispensable. This role of SA in E. amylovora resistance is consistent with the known function of SA in regulating both host and non‐host resistances in several other plant pathosystems (Kachroo et al., 2000; Lu et al., 2001; Tsuda et al., 2008b; van Wees and Glazebrook, 2003), and could be applied to E. amylovora host plants. Indeed, SA has been shown to be important for E. amylovora in apple. SA accumulation and the expression of several SA regulatory genes were induced by E. amylovora infection in apple (Fig. 1; Bonasera et al., 2006; Moreau et al., 2012; Venisse et al., 2002). In addition, manipulation of the SA pathway in apple plants, either by exogenous SA analogue application or overexpression of the NPR1 gene, conferred increased disease resistance to E. amylovora (Brisset et al., 2000; Malnoy et al., 2007; Sparla et al., 2004). Interestingly, we only observed a modest increase in SA in apple plants susceptible to E. amylovora. On the contrary, the non‐host plant Arabidopsis activates quick and strong SA signalling in the presence of E. amylovora. It is possible that such a difference in SA signal activation can account for the different resistance to E. amylovora displayed by apple and Arabidopsis. Thus, increasing the SA pathway could be a powerful approach to enhance the resistance to E. amylovora in host plants such as apple and pear.
One striking feature of the plant response to E. amylovora infection is the formation of massive callose deposits in the infected area. The major callose synthase gene PMR4 is required for callose deposition in Arabidopsis (Moreau et al., 2012). However, it is unclear whether callose deposition is related to E. amylovora resistance and how the main defence signalling pathways affect callose deposition. In particular, the role of SA in E. amylovora‐induced callose deposition in Arabidopsis has not been completely defined. On the one hand, a study by Degrave et al. (2008) showed SA‐independent callose deposition induced by E. amylovora. On the other, another study by DebRoy et al. (2004) showed that the avrE effector expressed by P. syringae could suppress callose deposition in an SA‐dependent manner. They suggested that the E. amylovora effector dspA/E, a homologue of P. syringae avrE that functionally cross‐complements with dspA/E, could similarly target SA‐dependent cell wall defence. Here, using single or combinations of two or more mutants disrupting SA, JA and/or ET pathways, our data conclusively showed that callose deposition is independent of these three main defence signalling pathways in Arabidopsis. As the SA mutants were more susceptible to E. amylovora, yet accumulated wild‐type levels of callose, and the callose‐deficient mutant pmr4–1 was more susceptible to E. amylovora, we conclude that callose deposition is necessary, but not sufficient, for E. amylovora resistance in Arabidopsis. We also recognized the discrepancy in our E. amylovora growth data for the ics1 and pmr4–1 mutants (Fig. 3), compared with the observations reported earlier (Moreau et al., 2012). Such a difference could be explained by many factors, for instance, the different E. amylovora strains, sampling times and plant growth conditions used in the two laboratories. Nevertheless, our data suggest that the cell wall‐based resistance to E. amylovora is different from defence signalling mediated by SA, JA and ET in non‐host Arabidopsis. It would be interesting to further investigate whether cell wall modification induced by E. amylovora is important for resistance in the host plants. If so, manipulation of the cell wall could provide an additional strategy to SA signalling modification to enhance host resistance to fire blight disease.
We further reported here that E. amylovora infection activated tumour‐like growths in Arabidopsis. Many pathogens are known to induce cell fate change in the infected plants, including the activation of cell death, cell division and cell enlargement in the infected area. Although cell death has been widely recorded as a readout of disease symptoms (Greenberg and Yao, 2004), cell division and cell enlargement have been largely overlooked in studies of plant–pathogen interactions. Several pathogens have been reported to induce cell division and/or cell enlargement, in some cases leading to tumour‐like growths in plants (de Almeida Engler et al., 1999; Braun, 1956; Chalupowicz et al., 2006; Chandran et al., 2009; Hamdoun et al., 2013; Marois et al., 2002). We found that E. amylovora‐induced cell growth in Arabidopsis was independent of some effectors of the bacterium and SA signalling of Arabidopsis. Additional experiments are needed to identify molecular factors from E. amylovora and Arabidopsis that are important for abnormal growths in plants.
We speculate that such tumour‐like growths resemble cankers found in E. amylovora‐infected apple. It has been proposed that, in order to limit the spread of E. amylovora, apple plants produce extra cell layers to seal off diseased tissue, thus forming a canker. In turn, the canker provides an important structure in planta for E. amylovora to survive the winter, from which the bacterium emerges as ooze in the next spring for further infection (Eastgate, 2000; Malnoy et al., 2012). However, how E. amylovora induces canker formation in apple and the morphology of these cankers are not well understood. Further investigation of bacterial and/or plant factors contributing to tumour‐like growths in Arabidopsis could help to reveal the molecular mechanisms underlying canker formation in E. amylovora‐infected apples, leading to a better control of fire blight.
Erwinia amylovora is one of the top 10 plant‐pathogenic bacteria identified by the international scientific community. It causes the devastating fire blight disease in economically important pome fruit‐producing plants and some ornamental plants (Mansfield et al., 2012). The mechanisms of plant resistance to E. amylovora are not well understood. Here, we provide evidence to show that SA signalling and callose deposition are required for E. amylovora resistance in the non‐host plant Arabidopsis. Although it is difficult to directly test the requirements of SA signalling and callose deposition in host resistance to E. amylovora because of the inherent experimental restrictions with the host genetic system, the conservation of the genes involved in SA signalling and callose formation in Arabidopsis and host plants strongly suggests a similar requirement of these two processes for E. amylovora resistance in host plants. Thus, genes important for SA‐ and cell wall‐mediated defence provide potentially powerful molecular tools for genetic modification in order to improve the resistance of economically important crops to the destructive fire blight disease.
Experimental Procedures
Plant materials
The Arabidopsis plants used in this work were all in the Colombia‐0 background and were grown in growth chambers with a 12‐h light/12‐h dark cycle, light intensity of 200 µmol/m2/s, 60% humidity and 22 °C. The mutants ics1–1, eds1–2, eds5–3, pad4–1, npr1–1, jar1–1 and nahG have been described previously (Lu et al., 2009; Ng et al., 2011; Staswick et al., 1992; Wang et al., 2011). ein2–1 was provided by Caren Chang at University of Maryland College Park, jin1–7 by Barbara Kunkel at Washington University and pmr4–1 by Shunyuan Xiao at University of Maryland College Park. The multiple SA, ET and/or JA gene knockout mutants (dde2–2ein2–1, ein2–1pad4–1, ein2–1sid2–2, dde2–2ein2–1pad4–1, dde2–2ein2–1sid2–2, dde2–2pad4–1sid2–2, ein2–1pad4–1sid2–2 and dde2–2ein2–1pad4–1sid2–2) have been described previously (Tsuda et al., 2009) and were obtained from the Arabidopsis Biological Resource Center.
Malus × domestica cv. ‘Royal Gala’ was propagated in vitro as described previously (Ko et al., 2002; Norelli et al., 1988) and grown in a glasshouse under supplemental lighting to maintain a 16‐h day length and average temperature of 28 °C. Plants were watered daily and treated weekly with nutrient solution (MiracleGro) and Osmocote (Scott's Miracle‐Gro Products, Marysville, Ohio, USA).
Bacterial culture and infection assays
Erwinia amylovora strain Ea273 (Ea) is a wild‐type strain isolated from apple in New York, and the E. amylovora strain with streptomycin resistance was isolated from pear in Washington State. Ea dspA/E is a dspA/E deletion mutant and Ea hrpN is an hrpN deletion mutant. The Ea ttss mutant cannot export effectors to the host cell. These Ea mutants are in the Ea273 background and were created by homologous recombination (Bogdanove et al., 1998b; Wei and Beer, 1993). Erwinia amylovora strains were kindly provided by Steven V. Beer at Cornell University. All E. amylovora strains were grown at 30 ºC in Lysogeny broth (LB) medium with appropriate antibiotics for selection. For apple infection, actively growing shoots were sprayed with E. amylovora (108 cfu/mL) or the mock solution containing 10 mm phosphate buffer, pH 6.5, to run off using a hand‐pumped spray bottle, and then immediately wounded with a sterile florist frog as described previously (Bonasera et al., 2006). Treated leaves were harvested at 48 hpi for SA analysis.
For Arabidopsis infection, the fourth to sixth leaves of 30‐day‐old plants were infiltrated with E. amylovora strains at the indicated concentrations, using a 1‐mL needleless syringe. The 10 mm MgSO4 solution was used as mock treatment. All bacterial cultures and plant material inoculated with E. amylovora strains were maintained in isolation and autoclaved at the end of the experiment to prevent bacterial escape from the laboratory. The infected plants were covered with a clear dome to maintain 100% humidity. At 3 dpi, leaf discs of 7 mm in diameter from the infected leaves were excised using a core borer and ground in 10 mm MgSO4. The surface area of each leaf disc is 38 mm2. Serial dilutions of the ground mixture were made and plated on LB plates containing appropriate antibiotics. Each bacterial data point was an average of samples taken from at least eight different plants from two or more independent experiments ± standard error of the mean. Except for the bacterial growth assay, all other infection assays were performed with plants uncovered.
The culture of Pma ES4326 strains and the inoculation of Arabidopsis have been described previously (Wang et al., 2011).
Ion leakage assay
Arabidopsis leaves infected with E. amylovora or Pma avrRpm1 were collected immediately after inoculation and cut with a 7‐mm core borer. Five leaf discs from five different plants were washed in de‐ionized water twice and placed in 5 mL of de‐ionized water. Each time point had triplicate samples for each genotype and each treatment. Solution conductivity was measured using an EC meter (The London Company, Welwyn International Inc., Cleveland, OH, USA) at the indicated times after leaf collection.
SA measurement
Bacteria‐infected and mock (10 mm MgSO4)‐treated leaves from Arabidopsis or apple were extracted for free and total SA (glucosylated SA), and quantified with a high‐performance liquid chromatograph as described previously (Ng et al., 2011; Wang et al., 2011).
RNA analysis
Bacteria‐infected and mock‐treated leaves from 30‐day‐old Arabidopsis plants were collected for RNA extraction and northern blotting, as described previously (Lu et al., 2009). Radioactive probes were prepared by PCR, using an antisense primer specific for a gene fragment in the presence of [32P]dCTP. Primers for PR1 have been described previously (Lu et al., 2009).
Analysis of leaf morphology using light microscopy
Bacteria‐infected and mock‐treated leaves from 30‐day‐old Arabidopsis plants were examined for tumour‐like growths, which appeared to be transparent protrusions on leaves and could be observed with the assistance of a dissection microscope. At least 25 leaves from 12–15 plants were used for the quantification of tumour‐like growths for each treatment. To observe more detailed leaf morphology, the treated leaves were collected at 4 dpi and cut into 2 × 4‐mm2 sections. The sections were fixed, embedded in LR white resin and further sliced with an ultramicrotome into 1‐µm sections. Leaf cross‐sections were stained with 0.1% toluidine blue O and photographed using an AxioCam MRc5 camera (Zeiss, Inc., Göttingen, Germany) connected to a dissection microscope.
Nuclear DNA quantification by DAPI staining
Leaves of 30‐day‐old Arabidopsis plants treated with E. amylovora or 10 mm MgSO4 were collected and cut into 3 × 6‐mm2 sections, using at least six sections from six plants for each treatment. The sections were fixed, embedded in paraplast, cut into 15‐µm slices with a microtome, and stained with DAPI to visualize nuclei. Relative nuclear DNA quantification was analysed as described previously (Hamdoun et al., 2013).
Callose staining
Leaves of 30‐day‐old Arabidopsis plants treated with E. amylovora (0.01) or 10 mm MgSO4 were boiled in alcoholic lactophenol (phenol–glycerol–lactic acid–water: 1 : 1 : 1 : 1, v/v) for 2 min and rinsed with 50% ethanol. The transparent leaves were incubated for 1 h in 0.15 m phosphate buffer (pH 9.5) containing 0.01% aniline blue (Lu et al., 2003), prior to microscopic analysis. The stained leaves were visualized and photographed with a fluorescence dissection microscope (Leica M80, Leica Microsystems, Wetzlar, Germany) connected to a CCD camera (Leica IC80 HD). Callose deposition was quantified by measurement of the relative fluorescence intensity emitted by aniline blue stain using ImageJ (Version 1.45s).
Conflicts of Interest
The authors have no conflicts of interest to declare.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Erwinia amylovora‐induced salicylic acid (SA) accumulation is ISOCHORISMATE SYNTHASE 1 (ICS1) dependent in Arabidopsis. The fourth to sixth leaves of Col‐0 and ics1‐1 plants were infiltrated with E. amylovora at an optical density at 600 nm (OD600) of 0.1 and collected at 24 h post‐inoculation (hpi) for SA analyses. (A) Free SA measurement. (B) Total SA measurement. Statistical analysis was performed with one‐way analysis of variance (ANOVA) Fisher's partial least‐squares difference (PLSD) tests (StatView 5.0.1). Asterisks indicate significant difference between E. amylovora‐inoculated Col‐0 and ics1‐1 plants at the same time point (P < 0.05). These experiments were repeated twice with similar results. FW, fresh weight.
Fig. S2 Erwinia amylovora induces salicylic acid (SA) accumulation in apple. Erwinia amylovora (Ea; 0.1)‐inoculated and mock‐inoculated apple leaves were harvested at 48 h post‐inoculation (hpi) and extracted for total and free SA followed by high‐performance liquid chromatography (HPLC) measurement. Asterisks indicate significant difference between mock‐ and E. amylovora‐inoculated samples for free or total SA level (P < 0.05). These experiments were repeated twice with similar results. FW, fresh weight.
Fig. S3 Arabidopsis mutants disrupted in ethylene (ET) and jasmonic acid (JA) signalling are not compromised in resistance to Erwinia amylovora. The fourth to sixth leaves of 30‐day‐old plants were inoculated with E. amylovora at an optical density at 600 nm (OD600) = 0.1 [108 colony‐forming units (cfu)/mL]. Inoculated leaf discs (each 38 mm2) were taken for the measurement of bacterial growth at 3 days post‐inoculation (dpi). Each data point represents the average of 12 samples from two independent experiments ± standard error of the mean. Note that there is no difference between the samples. These experiments were repeated twice with similar results.
Fig. S4 Erwinia amylovora‐induced callose deposition in Arabidopsis is independent of salicylic acid (SA) signalling. Erwinia amylovora‐infected leaves of each genotype were harvested for fixation and staining with aniline blue. (A) Images of callose deposits. (B) Quantification of callose deposits. Each data point was an average of at least six images from four different leaves. The asterisk indicates significant difference of pmr4‐1 from the other genotypes (P < 0.05). These experiments were repeated twice with similar results.
Fig. S5 Erwinia amylovora‐induced callose deposition in Arabidopsis is independent of ethylene (ET) and jasmonic acid (JA) signalling. Erwinia amylovora‐inoculated leaves of each genotype were harvested for fixation and staining with aniline blue. (A) Images of callose deposits. Callose deposits appeared as fluorescent dots and were photographed with an AxioCam MRc5 camera connected to a fluorescence stereoscope microscope. (B) Quantification of callose deposits. The number of callose deposits of each genotype was quantified using ImageJ (Version 1.45s). Each data point was an average of at least six images from four different leaves. Note that there is no difference between the samples. These experiments were repeated twice with similar results.
Fig. S6 Erwinia amylovora induces tumour‐like growths in Arabidopsis leaves. Plants were infiltrated with E. amylovora at an optical density at 600 nm (OD600) of 0.1 [108 colony‐forming units (cfu)/mL] (Ea) or mock solution (10 mm MgSO4) (Mock). The abnormal growth phenotype usually appeared at about 4 days post‐inoculation (dpi). (A) Images of abnormal growths on the leaf abaxial side. (B) Leaf cross‐section. Leaves were sectioned into thin slices by hand sectioning and imaged with a camera connected to a dissection microscope. Arrows indicate abnormal growths as transparent protrusions on the infected leaves.
Acknowledgements
We thank members of the Lu laboratory and Wilbur Hershberger at USDA‐ARS in Kearneysville, WV, USA for assistance with this work. Images of leaf cross‐sections and nuclei were taken with microscopes at the Keith R. Porter Microscopy Facility at the University of Maryland Baltimore County (UMBC). This work was partially supported by a grant from the National Science Foundation (NSF 1456140) to H.L.
References
- Aarts, N. , Metz, M. , Holub, E. , Staskawicz, B.J. , Daniels, M.J. and Parker, J.E. (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene‐mediated signaling pathways in Arabidopsis . Proc. Natl. Acad. Sci. USA, 95, 10 306–10 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano, J.R. and Collmer, A. (2004) Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42, 385–414. [DOI] [PubMed] [Google Scholar]
- Alfano, J.R. , Charkowski, A.O. , Deng, W.L. , Badel, J.L. , Petnicki‐Ocwieja, T. , van Dijk, K. and Collmer, A. (2000) The Pseudomonas syringae Hrp pathogenicity island has a tripartite mosaic structure composed of a cluster of type III secretion genes bounded by exchangeable effector and conserved effector loci that contribute to parasitic fitness and pathogenicity in plants. Proc. Natl. Acad. Sci. USA, 97, 4856–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida Engler, J. , De Vleesschauwer, V. , Burssens, S. , Celenza, J.L. Jr. , Inze, D. , Van Montagu, M. , Engler, G. and Gheysen, G. (1999) Molecular markers and cell cycle inhibitors show the importance of cell cycle progression in nematode‐induced galls and syncytia. Plant Cell, 11, 793–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, J.M. , Hirayama, T. , Roman, G. , Nourizadeh, S. and Ecker, J.R. (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science, 284, 2148–2152. [DOI] [PubMed] [Google Scholar]
- Bartsch, M. , Gobbato, E. , Bednarek, P. , Debey, S. , Schultze, J.L. , Bautor, J. and Parker, J.E. (2006) Salicylic acid‐independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell, 18, 1038–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanove, A. , Kim, J. , Wei, Z. , Kolchinsky, P. , Charkowski, A. , Conlin, A. , Collmer, A. and Beer, S.V. (1998a) Homology and functional similarity of an hrp‐linked pathogenicity locus, dspEF, of Erwinia amylovora and the avirulence locus avrE of Pseudomonas syringae pathovar tomato . Proc. Natl. Acad. Sci. USA, 95, 1325–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanove, A.J. , Bauer, D.W. and Beer, S.V. (1998b) Erwinia amylovora secretes DspE, a pathogenicity factor and functional AvrE homolog, through the Hrp (type III secretion) pathway. J. Bacteriol. 180, 2244–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasera, J.M. , Kim, J.F. and Beer, S.V. (2006) PR genes of apple: identification and expression in response to elicitors and inoculation with Erwinia amylovora . BMC Plant Biol. 6, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boureau, T. , ElMaarouf‐Bouteau, H. , Garnier, A. , Brisset, M.N. , Perino, C. , Pucheu, I. and Barny, M.A. (2006) DspA/E, a type III effector essential for Erwinia amylovora pathogenicity and growth in planta, induces cell death in host apple and nonhost tobacco plants. Mol. Plant–Microbe Interact. 19, 16–24. [DOI] [PubMed] [Google Scholar]
- Boureau, T. , Siamer, S. , Perino, C. , Gaubert, S. , Patrit, O. , Degrave, A. , Fagard, M. , Chevreau, E. and Barny, M.A. (2011) The HrpN effector of Erwinia amylovora, which is involved in type III translocation, contributes directly or indirectly to callose elicitation on apple leaves. Mol. Plant–Microbe Interact. 24, 577–584. [DOI] [PubMed] [Google Scholar]
- Bramsiepe, J. , Wester, K. , Weinl, C. , Roodbarkelari, F. , Kasili, R. , Larkin, J.C. , Hülskamp, M. and Schnittger, A. (2010) Endoreplication controls cell fate maintenance. PLoS Genet. 6, e1000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, A.C. (1956) The activation of two growth‐substance systems accompanying the conversion of normal to tumor cells in crown gall. Cancer Res. 16, 53–56. [PubMed] [Google Scholar]
- Brisset, M. , Cesbron, S. , Thomson, S. and Paulin, J. (2000) Acibenzolar‐S‐methyl induces the accumulation of defense‐related enzymes in apple and protects from fire blight. Eur. J. Plant Pathol. 106, 529–536. [Google Scholar]
- Chalupowicz, L. , Barash, I. , Schwartz, M. , Aloni, R. and Manulis, S. (2006) Comparative anatomy of gall development on Gypsophila paniculata induced by bacteria with different mechanisms of pathogenicity. Planta, 224, 429–437. [DOI] [PubMed] [Google Scholar]
- Chandran, D. , Inada, N. , Hather, G. , Kleindt, C.K. and Wildermuth, M.C. (2009) Laser microdissection of Arabidopsis cells at the powdery mildew infection site reveals site‐specific processes and regulators. Proc. Natl. Acad. Sci. USA, 107, 460–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, M.S. , Kim, W. , Lee, C. and Oh, C.S. (2013) Harpins, multifunctional proteins secreted by gram‐negative plant‐pathogenic bacteria. Mol. Plant–Microbe Interact. 26, 1115–1122. [DOI] [PubMed] [Google Scholar]
- De Veylder, L. , Larkin, J.C. and Schnittger, A. (2011) Molecular control and function of endoreplication in development and physiology. Trends Plant Sci. 16, 624–634. [DOI] [PubMed] [Google Scholar]
- DebRoy, S. , Thilmony, R. , Kwack, Y.B. , Nomura, K. and He, S.Y. (2004) A family of conserved bacterial effectors inhibits salicylic acid‐mediated basal immunity and promotes disease necrosis in plants. Proc. Natl. Acad. Sci. USA, 101, 9927–9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degrave, A. , Fagard, M. , Perino, C. , Brisset, M.N. , Gaubert, S. , Laroche, S. , Patrit, O. and Barny, M.A. (2008) Erwinia amylovora type three‐secreted proteins trigger cell death and defense responses in Arabidopsis thaliana . Mol. Plant–Microbe Interact. 21, 1076–1086. [DOI] [PubMed] [Google Scholar]
- Delaney, T.P. , Uknes, S. , Vernooij, B. , Friedrich, L. , Weymann, K. , Negrotto, D. , Gaffney, T. , Gut‐Rella, M. , Kessmann, H. , Ward, E. and Ryals, J. (1994) A central role of salicylic acid in plant disease resistance. Science, 266, 1247–1250. [DOI] [PubMed] [Google Scholar]
- Dong, H. , Delaney, T.P. , Bauer, D.W. and Beer, S.V. (1999) Harpin induces disease resistance in Arabidopsis through the systemic acquired resistance pathway mediated by salicylic acid and the NIM1 gene. Plant J. 20, 207–215. [DOI] [PubMed] [Google Scholar]
- Duge De Bernonville, T. , Gaucher, M. , Flors, V. , Gaillard, S. , Paulin, J.P. , Dat, J.F. and Brisset, M.N. (2012) T3SS‐dependent differential modulations of the jasmonic acid pathway in susceptible and resistant genotypes of Malus spp. challenged with Erwinia amylovora . Plant Sci. 188, 1–9. [DOI] [PubMed] [Google Scholar]
- Eastgate, J.A. (2000) Erwinia amylovora: the molecular basis of fireblight disease. Mol. Plant Pathol. 1, 325–329. [DOI] [PubMed] [Google Scholar]
- Feys, B.J. , Moisan, L.J. , Newman, M.A. and Parker, J.E. (2001) Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20, 5400–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich, L. , Vernooij, B. , Gaffney, T. , Morse, A. and Ryals, J. (1995) Characterization of tobacco plants expressing a bacterial salicylate hydroxylase gene. Plant Mol. Biol. 29, 959–968. [DOI] [PubMed] [Google Scholar]
- Gaffney, T. , Friedrich, L. , Vernooij, B. , Negrotto, D. , Nye, G. , Uknes, S. , Ward, E. , Kessmann, H. and Ryals, J. (1993) Requirement of salicylic acid for the induction of systemic acquired resistance. Science, 261, 754–756. [DOI] [PubMed] [Google Scholar]
- Gaudriault, S. , Malandrin, L. , Paulin, J.P. and Barny, M.A. (1997) DspA, an essential pathogenicity factor of Erwinia amylovora showing homology with AvrE of Pseudomonas syringae, is secreted via the Hrp secretion pathway in a DspB‐dependent way. Mol. Microbiol. 26, 1057–1069. [DOI] [PubMed] [Google Scholar]
- Gill, U.S. , Lee, S. and Mysore, K.S. (2015) Host versus nonhost resistance: distinct wars with similar arsenals. Phytopathology, 105, 580–587. [DOI] [PubMed] [Google Scholar]
- Grant, S.R. , Fisher, E.J. , Chang, J.H. , Mole, B.M. and Dangl, J.L. (2006) Subterfuge and manipulation: type III effector proteins of phytopathogenic bacteria. Annu. Rev. Microbiol. 60, 425–449. [DOI] [PubMed] [Google Scholar]
- Greenberg, J. and Yao, N. (2004) The role and regulation of programmed cell death in plant–pathogen interactions. Cell. Microbiol. 6, 201–211. [DOI] [PubMed] [Google Scholar]
- Gusberti, M. , Klemm, U. , Meier, M.S. , Maurhofer, M. and Hunger‐Glaser, I. (2015) Fire blight control: the struggle goes on. A comparison of different fire blight control methods in Switzerland with respect to biosafety, efficacy and durability. Int. J. Environ. Res. Public Health, 12, 11 422–11 447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman, D.S. , Vinatzer, B.A. , Sarkar, S.F. , Ranall, M.V. , Kettler, G. and Greenberg, J.T. (2002) A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae . Science, 295, 1722–1726. [DOI] [PubMed] [Google Scholar]
- Hamdoun, S. , Liu, Z. , Gill, M. , Yao, N. and Lu, H. (2013) Dynamics of defense responses and cell fate change during Arabidopsis–Pseudomonas syringae interactions. PLoS One, 8, e83219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, C.E. and Meins, F. Jr. (1986) Evidence for a cellular gene with potential oncogenic activity in plants. Proc. Natl. Acad. Sci. USA, 83, 2492–2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck, P. , Thilmony, R. and He, S.Y. (2003) A Pseudomonas syringae type III effector suppresses cell wall‐based extracellular defense in susceptible Arabidopsis plants. Proc. Natl. Acad. Sci. USA, 100, 8577–8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueck, C.J. (1998) Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62, 379–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage, D. , Tootle, T.L. , Reuber, T.L. , Frost, L.N. , Feys, B.J. , Parker, J.E. , Ausubel, F.M. and Glazebrook, J. (1999) Arabidopsis thaliana PAD4 encodes a lipase‐like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA, 96, 13 583–13 588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo, P. , Yoshioka, K. , Shah, J. , Dooner, H.K. and Klessig, D.F. (2000) Resistance to turnip crinkle virus in Arabidopsis is regulated by two host genes and is salicylic acid dependent but NPR1, ethylene, and jasmonate independent. Plant Cell, 12, 677–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamber, T. , Buchmann, J.P. , Pothier, J.F. , Smits, T.H. , Wicker, T. and Duffy, B. (2016) Fire blight disease reactome: RNA‐seq transcriptional profile of apple host plant defense responses to Erwinia amylovora pathogen infection. Sci. Rep. 6, 21 600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, M.A. , Zhao, Y.F. and Korban, S.S. (2013) Identification of genetic loci associated with fire blight resistance in Malus through combined use of QTL and association mapping. Physiol. Plant. 148, 344–353. [DOI] [PubMed] [Google Scholar]
- Ko, K. , Norelli, J. , Reynoird, J. , Aldwinckle, H. and Brown, S. (2002) T4 lysozyme and attacin genes enhance resistance of transgenic ‘Galaxy’ apple against Erwinia amylovora . J. Am. Soc. Hort. Sci. 127, 515–519. [Google Scholar]
- Launay, A. , Patrit, O. , Wenes, E. and Fagard, M. (2016) DspA/E contributes to apoplastic accumulation of ROS in non‐host A. thaliana . Front. Plant Sci. 7, 545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roux, P.M. , Khan, M.A. , Broggini, G.A. , Duffy, B. , Gessler, C. and Patocchi, A. (2010) Mapping of quantitative trait loci for fire blight resistance in the apple cultivars 'Florina' and 'Nova Easygro'. Genome, 53, 710–722. [DOI] [PubMed] [Google Scholar]
- Lorenzo, O. , Chico, J.M. , Sanchez‐Serrano, J.J. and Solano, R. (2004) JASMONATE‐INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate‐regulated defense responses in Arabidopsis. Plant Cell, 16, 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, H. , Rate, D.N. , Song, J.T. and Greenberg, J.T. (2003) ACD6, a novel ankyrin protein, is a regulator and an effector of salicylic acid signaling in the Arabidopsis defense response. Plant Cell, 15, 2408–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, H. , Salimian, S. , Gamelin, E. , Wang, G. , Fedorowski, J. , LaCourse, W. and Greenberg, J.T. (2009) Genetic analysis of acd6–1 reveals complex defense networks and leads to identification of novel defense genes in Arabidopsis. Plant J. 58, 401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, M. , Tang, X. and Zhou, J.M. (2001) Arabidopsis NHO1 is required for general resistance against Pseudomonas bacteria. Plant Cell, 13, 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnoy, M. , Jin, Q. , Borejsza‐Wysocka, E.E. , He, S.Y. and Aldwinckle, H.S. (2007) Overexpression of the apple MpNPR1 gene confers increased disease resistance in Malus x domestica . Mol. Plant–Microbe Interact. 20, 1568–1580. [DOI] [PubMed] [Google Scholar]
- Malnoy, M. , Martens, S. , Norelli, J.L. , Barny, M.A. , Sundin, G.W. , Smits, T.H. and Duffy, B. (2012) Fire blight: applied genomic insights of the pathogen and host. Annu. Rev. Phytopathol. 50, 475–494. [DOI] [PubMed] [Google Scholar]
- Mansfield, J. , Genin, S. , Magori, S. , Citovsky, V. , Sriariyanum, M. , Ronald, P. , Dow, M.A.X. , Verdier, V. , Beer, S.V. , Machado, M.A. and Toth, I.A.N. (2012) Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marois, E. , Van den Ackerveken, G. and Bonas, U. (2002) The Xanthomonas type III effector protein AvrBs3 modulates plant gene expression and induces cell hypertrophy in the susceptible host. Mol. Plant–Microbe Interact. 15, 637–646. [DOI] [PubMed] [Google Scholar]
- Milcevicová, R. , Gosch, C. , Halbwirth, H. , Stich, K. , Hanke, M.V. , Peil, A. , Flachowsky, H. , Rozhon, W. , Jonak, C. , Oufir, M. and Hausman, J.F. (2010) Erwinia amylovora‐induced defense mechanisms of two apple species that differ in susceptibility to fire blight. Plant Sci. 179, 60–67. [Google Scholar]
- Moreau, M. , Degrave, A. , Vedel, R. , Bitton, F. , Patrit, O. , Renou, J.P. , Barny, M.A. and Fagard, M. (2012) EDS1 contributes to nonhost resistance of Arabidopsis thaliana against Erwinia amylovora . Mol. Plant–Microbe Interact. 25, 421–430. [DOI] [PubMed] [Google Scholar]
- Mur, L.A. , Bi, Y.M. , Darby, R.M. , Firek, S. and Draper, J. (1997) Compromising early salicylic acid accumulation delays the hypersensitive response and increases viral dispersal during lesion establishment in TMV‐infected tobacco. Plant J. 12, 1113–1126. [DOI] [PubMed] [Google Scholar]
- Nawrath, C. and Metraux, J.P. (1999) Salicylic acid induction‐deficient mutants of Arabidopsis express PR‐2 and PR‐5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell, 11, 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath, C. , Heck, S. , Parinthawong, N. and Metraux, J.P. (2002) EDS5, an essential component of salicylic acid‐dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell, 14, 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, G. , Seabolt, S. , Zhang, C. , Salimian, S. , Watkins, T.A. and Lu, H. (2011) Genetic dissection of salicylic acid‐mediated defense signaling networks in Arabidopsis. Genetics, 189, 851–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, M.T. , Stein, M. , Hou, B.H. , Vogel, J.P. , Edwards, H. and Somerville, S.C. (2003) Loss of a callose synthase results in salicylic acid‐dependent disease resistance. Science, 301, 969–972. [DOI] [PubMed] [Google Scholar]
- Norelli, J.L. , Aldwinckle, H.S. and Beer, S.V. (1988) Virulence of Erwinia amylovora strains to Malus sp. Novole plants grown in vitro and in the greenhouse. Phytopathology, 78, 1292–1297. [Google Scholar]
- Oh, C.S. and Beer, S.V. (2005) Molecular genetics of Erwinia amylovora involved in the development of fire blight. FEMS Microbiol. Lett. 253, 185–192. [DOI] [PubMed] [Google Scholar]
- Oh, C.S. , Martin, G.B. and Beer, S.V. (2007) DspA/E, a type III effector of Erwinia amylovora, is required for early rapid growth in Nicotiana benthamiana and causes NbSGT1‐dependent cell death. Mol. Plant Pathol. 8, 255–265. [DOI] [PubMed] [Google Scholar]
- Pique, N. , Minana‐Galbis, D. , Merino, S. and Tomas, J.M. (2015) Virulence factors of Erwinia amylovora: a review. Int. J. Mol. Sci. 16, 12 836–12 854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboutier, D. , Frankart, C. , Briand, J. , Biligui, B. , Laroche, S. , Rona, J.P. , Barny, M.A. and Bouteau, F. (2007) The HrpN(ea) harpin from Erwinia amylovora triggers differential responses on the nonhost Arabidopsis thaliana cells and on the host apple cells. Mol. Plant–Microbe Interact. 20, 94–100. [DOI] [PubMed] [Google Scholar]
- Sato, M. , Mitra, R.M. , Coller, J. , Wang, D. , Spivey, N.W. , Dewdney, J. , Denoux, C. , Glazebrook, J. and Katagiri, F. (2007) A high‐performance, small‐scale microarray for expression profiling of many samples in Arabidopsis–pathogen studies. Plant J. 49, 565–577. [DOI] [PubMed] [Google Scholar]
- Senthil‐Kumar, M. and Mysore, K.S. (2013) Nonhost resistance against bacterial pathogens: retrospectives and prospects. Annu. Rev. Phytopathol. 51, 407–427. [DOI] [PubMed] [Google Scholar]
- Serrano, M. , Wang, B. , Aryal, B. , Garcion, C. , Abou‐Mansour, E. , Heck, S. , Geisler, M. , Mauch, F. , Nawrath, C. and Métraux, J.P. (2013) Export of salicylic acid from the chloroplast requires the multidrug and toxin extrusion‐like transporter EDS5. Plant Physiol. 162, 1815–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfferth, C. and Tsuda, K. (2014) Salicylic acid signal transduction: the initiation of biosynthesis, perception and transcriptional reprogramming. Front. Plant Sci. 5, 697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparla, F. , Rotino, L. , Valgimigli, M. , Pupillo, P. and Trost, P. (2004) Systemic resistance induced by benzothiadiazole in pear inoculated with the agent of fire blight (Erwinia amylovora). Sci. Hortic. 101, 269–279. [Google Scholar]
- Staswick, P.E. , Su, W. and Howell, S.H. (1992) Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA, 89, 6837–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal‐Christensen, H. (2003) Fresh insights into processes of nonhost resistance. Curr. Opin. Plant Biol. 6, 351–357. [DOI] [PubMed] [Google Scholar]
- Tsuda, K. , Glazebrook, J. and Katagiri, F. (2008a) The interplay between MAMP and SA signaling. Plant Signal. Behav. 3, 359–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda, K. , Sato, M. , Glazebrook, J. , Cohen, J.D. and Katagiri, F. (2008b) Interplay between MAMP‐triggered and SA‐mediated defense responses. Plant J. 53, 763–775. [DOI] [PubMed] [Google Scholar]
- Tsuda, K. , Sato, M. , Stoddard, T. , Glazebrook, J. and Katagiri, F. (2009) Network properties of robust immunity in plants. PLoS Genet. 5, e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste, J.L. and Eden‐Green, S. (2001) Fire blight: the disease and its causative agent, Erwinia amylovora . Plant Pathol. 50, 417–418. [Google Scholar]
- Venisse, J.S. , Gullner, G. and Brisset, M.N. (2001) Evidence for the involvement of an oxidative stress in the initiation of infection of pear by Erwinia amylovora . Plant Physiol. 125, 2164–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venisse, J.S. , Malnoy, M. , Faize, M. , Paulin, J.P. and Brisset, M.N. (2002) Modulation of defense responses of Malus spp. during compatible and incompatible interactions with Erwinia amylovora . Mol. Plant–Microbe Interact. 15, 1204–1212. [DOI] [PubMed] [Google Scholar]
- Vrancken, K. , Holtappels, M. , Schoofs, H. , Deckers, T. and Valcke, R. (2013) Pathogenicity and infection strategies of the fire blight pathogen Erwinia amylovora in Rosaceae: state of the art. Microbiology, 159, 823–832. [DOI] [PubMed] [Google Scholar]
- Wang, G.F. , Seabolt, S. , Hamdoun, S. , Ng, G. , Park, J. and Lu, H. (2011) Multiple roles of WIN3 in regulating disease resistance, cell death, and flowering time in Arabidopsis. Plant Physiol. 156, 1508–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Tsuda, K. , Sato, M. , Cohen, J.D. , Katagiri, F. and Glazebrook, J. (2009) Arabidopsis CaM binding protein CBP60g contributes to MAMP‐induced SA accumulation and is involved in disease resistance against Pseudomonas syringae . PLoS Pathog. 5, e1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wees, S.C. and Glazebrook, J. (2003) Loss of non‐host resistance of Arabidopsis NahG to Pseudomonas syringae pv. phaseolicola is due to degradation products of salicylic acid. Plant J. 33, 733–742. [DOI] [PubMed] [Google Scholar]
- Wei, Z.M. and Beer, S.V. (1993) HrpI of Erwinia amylovora functions in secretion of harpin and is a member of a new protein family. J. Bacteriol. 175, 7958–7967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Z.M. , Laby, R.J. , Zumoff, C.H. , Bauer, D.W. , He, S.Y. , Collmer, A. and Beer, S.V. (1992) Harpin, elicitor of the hypersensitive response produced by the plant pathogen Erwinia amylovora . Science, 257, 85–88. [DOI] [PubMed] [Google Scholar]
- Wildermuth, M.C. , Dewdney, J. , Wu, G. and Ausubel, F.M. (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature, 414, 562–565. [DOI] [PubMed] [Google Scholar]
- Yamasaki, K. , Motomura, Y. , Yagi, Y. , Nomura, H. , Kikuchi, S. , Nakai, M. and Shiina, T. (2013) Chloroplast envelope localization of EDS5, an essential factor for salicylic acid biosynthesis in Arabidopsis thaliana . Plant Signal. Behav. 8, e23603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, S. and Dong, X. (2014) Perception of the plant immune signal salicylic acid. Curr. Opin. Plant Biol. 20C, 64–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Erwinia amylovora‐induced salicylic acid (SA) accumulation is ISOCHORISMATE SYNTHASE 1 (ICS1) dependent in Arabidopsis. The fourth to sixth leaves of Col‐0 and ics1‐1 plants were infiltrated with E. amylovora at an optical density at 600 nm (OD600) of 0.1 and collected at 24 h post‐inoculation (hpi) for SA analyses. (A) Free SA measurement. (B) Total SA measurement. Statistical analysis was performed with one‐way analysis of variance (ANOVA) Fisher's partial least‐squares difference (PLSD) tests (StatView 5.0.1). Asterisks indicate significant difference between E. amylovora‐inoculated Col‐0 and ics1‐1 plants at the same time point (P < 0.05). These experiments were repeated twice with similar results. FW, fresh weight.
Fig. S2 Erwinia amylovora induces salicylic acid (SA) accumulation in apple. Erwinia amylovora (Ea; 0.1)‐inoculated and mock‐inoculated apple leaves were harvested at 48 h post‐inoculation (hpi) and extracted for total and free SA followed by high‐performance liquid chromatography (HPLC) measurement. Asterisks indicate significant difference between mock‐ and E. amylovora‐inoculated samples for free or total SA level (P < 0.05). These experiments were repeated twice with similar results. FW, fresh weight.
Fig. S3 Arabidopsis mutants disrupted in ethylene (ET) and jasmonic acid (JA) signalling are not compromised in resistance to Erwinia amylovora. The fourth to sixth leaves of 30‐day‐old plants were inoculated with E. amylovora at an optical density at 600 nm (OD600) = 0.1 [108 colony‐forming units (cfu)/mL]. Inoculated leaf discs (each 38 mm2) were taken for the measurement of bacterial growth at 3 days post‐inoculation (dpi). Each data point represents the average of 12 samples from two independent experiments ± standard error of the mean. Note that there is no difference between the samples. These experiments were repeated twice with similar results.
Fig. S4 Erwinia amylovora‐induced callose deposition in Arabidopsis is independent of salicylic acid (SA) signalling. Erwinia amylovora‐infected leaves of each genotype were harvested for fixation and staining with aniline blue. (A) Images of callose deposits. (B) Quantification of callose deposits. Each data point was an average of at least six images from four different leaves. The asterisk indicates significant difference of pmr4‐1 from the other genotypes (P < 0.05). These experiments were repeated twice with similar results.
Fig. S5 Erwinia amylovora‐induced callose deposition in Arabidopsis is independent of ethylene (ET) and jasmonic acid (JA) signalling. Erwinia amylovora‐inoculated leaves of each genotype were harvested for fixation and staining with aniline blue. (A) Images of callose deposits. Callose deposits appeared as fluorescent dots and were photographed with an AxioCam MRc5 camera connected to a fluorescence stereoscope microscope. (B) Quantification of callose deposits. The number of callose deposits of each genotype was quantified using ImageJ (Version 1.45s). Each data point was an average of at least six images from four different leaves. Note that there is no difference between the samples. These experiments were repeated twice with similar results.
Fig. S6 Erwinia amylovora induces tumour‐like growths in Arabidopsis leaves. Plants were infiltrated with E. amylovora at an optical density at 600 nm (OD600) of 0.1 [108 colony‐forming units (cfu)/mL] (Ea) or mock solution (10 mm MgSO4) (Mock). The abnormal growth phenotype usually appeared at about 4 days post‐inoculation (dpi). (A) Images of abnormal growths on the leaf abaxial side. (B) Leaf cross‐section. Leaves were sectioned into thin slices by hand sectioning and imaged with a camera connected to a dissection microscope. Arrows indicate abnormal growths as transparent protrusions on the infected leaves.


