Summary
Infection caused by the synergistic interaction of two plant viruses is typically manifested by severe symptoms and increased accumulation of either virus. In potex–potyviral synergism, the potyviral RNA silencing suppressor helper component proteinase (HCPro) is known to enhance the pathogenicity of the potexvirus counterpart. In line with this, Potato virus X (PVX; genus Potexvirus) genomic RNA (gRNA) accumulation and gene expression from subgenomic RNA (sgRNA) are increased in Nicotiana benthamiana by Potato virus A (PVA; genus Potyvirus) HCPro expression. Recently, we have demonstrated that PVA HCPro interferes with the host cell methionine cycle by interacting with its key enzymes S‐adenosyl‐l‐methionine synthetase (SAMS) and S‐adenosyl‐l‐homocysteine hydrolase (SAHH). To study the involvement of methionine cycle enzymes in PVX infection, we knocked down SAMS and SAHH. Increased PVX sgRNA expression between 3 and 9 days post‐infiltration (dpi) and upregulation of (–)‐strand gRNA accumulation at 9 dpi were observed in the SAHH‐silenced background. We found that SAMS and SAHH silencing also caused a significant reduction in glutathione (GSH) concentration, specifically in PVX‐infected plants between 2 and 9 dpi. Interestingly, HCPro expression in PVX‐infected plants caused an even stronger reduction in GSH levels than did SAMS + SAHH silencing and a similar level of reduction was also achieved by knocking down GSH synthetase. PVX sgRNA expression was increased in the GSH synthetase‐silenced background. GSH is a major antioxidant of plant cells and therefore GSH shortage may explain the strong oxidative stress and severe symptoms observed during potex–potyvirus mixed infection.
Keywords: glutathione, HCPro, methionine cycle, potexvirus, potyvirus, S‐adenosyl‐l‐homocysteine hydrolase, silencing suppressor
Introduction
Mixed infections are common under field conditions and the interaction between viruses can be either synergistic or antagonistic (Syller, 2012). In synergism, one of the infecting viruses enhances the accumulation and/or pathogenicity of the other. This situation often leads to the expression of more severe disease than that caused by either virus alone (Senanayake and Mandal, 2014; Wang et al., 2009). Enhanced replication, movement to otherwise non‐invaded tissues and breakdown of host resistance by suppression of post‐transcriptional gene silencing are amongst the several causes deemed to be responsible for the higher virus titre and greater severity of symptoms (Untiveros et al., 2007). Mixed infections between potex‐ and potyviruses commonly lead to synergistic interactions, but the relationship can also be antagonistic (Ross, 1950). A recent study on mixed infection by Papaya ringspot virus (PSRV, a Potyvirus) and Papaya mosaic virus (PapMV, a Potexvirus) showed order‐dependent variation in their interaction. Synergistic interaction develops when PRSV infects the host simultaneously or before PapMV, whereas antagonism occurs if the infection sequence is reversed (Chávez‐Calvillo et al., 2016).
Potato virus X (PVX; genus Potexvirus) is ranked by several plant virologists to be amongst the top ten scientifically and economically important plant viruses (Scholthof et al., 2011). It has a monopartite positive‐sense RNA of 6.4 kb in size. Of the five proteins it encodes, the 165K replicase is translated directly from the genomic RNA (gRNA) open reading frame (ORF) 1. ORFs 2, 3 and 4, cumulatively known as triple gene block (TGB) genes, code for a multifunctional 25K protein (p25), having roles in movement and RNA silencing suppression, and two endoplasmic reticulum‐associated movement proteins, a 12K protein (p12) and an 8K protein (p8). TGB encoded proteins are translated from subgenomic (sg) transcripts. Another sgRNA hosting ORF 5 encodes the coat protein (CP). PVX generally produces mild mosaic symptoms in potato and mottling or necrotic spotting in tobacco. However, symptoms can increase drastically when it infects in combination with some of the potyvirus group members (Senanayake and Mandal, 2014). Several viruses belonging to the genus potyvirus, e.g. Potato virus Y (PVY) and Potato virus A (PVA), have been reported to undergo synergistic interaction with PVX. PVA causes up to 40% yield loss on co‐infection with PVX and Potato virus S (PVS) (Dedic, 1975; Hameed et al., 2014). In the current investigation, we have used PVX and PVA as model viruses to study the molecular mechanism behind the synergistic PVA–PVX infection in Nicotiana benthamiana. Aguilar et al. (2015) reported both systemic necrosis and increased PVX sgRNA accumulation during PVX–potyvirus interactions in N. benthamiana. They identified PVX P25 as the main pathogenicity determinant for systemic necrosis.
Potyviral helper component proteinase (HCPro) has been identified as an essential factor in potex–potyviral synergism (Shi et al., 1997). In order to induce synergism, replication of the whole potyviral genome is not necessary; rather, the expression of only the P1‐HCPro segment of the genome is sufficient to produce a similar degree of effect (Pruss et al., 1997; Vance et al., 1995). Moreover, the role of P1‐HCPro in the pathogenicity enhancement of unrelated viruses, such as Cucumber mosaic virus and Tobacco mosaic virus, has led to the formulation of the existing hypothesis on potyvirus‐mediated synergism in which the antiviral RNA silencing suppression property of HCPro is hailed to be responsible. HCPro is thought to break down a plant's antiviral defence, which is taken advantage of by other co‐infecting viruses to accumulate beyond their normal host limits. In addition, mutations made to the central domain of HCPro, impairing its silencing suppression activity, render the protein unable to induce synergism (Shi et al., 1997, Kasschau and Carrington, 1998, Marathe et al., 2000, Kasschau and Carrington, 2001, Voinnet, 2001, González‐Jara et al., 2005).
Potyviral HCPro binds small interfering RNAs (siRNAs), which is a function that has been proposed to sequester siRNAs away from the programming of RNA‐induced silencing complexes (RISCs) (Garcia‐Ruiz et al., 2015; Del Toro et al., 2017). Another proposed mechanism for the suppression of RNA silencing by HCPro is the destabilization of siRNAs (Mallory et al., 2001; Yu et al., 2005). For efficient silencing, double‐stranded siRNAs are stabilized by 2′‐O‐methylation at their 3′ ends catalysed by Hua enhancer 1 (HEN1) (Yu et al., 2005). Recent studies have indicated that HCPro interacts with two enzymes of the host methionine cycle, S‐adenosyl‐l‐methionine synthetase (SAMS) and S‐adenosyl‐l‐homocysteine hydrolase (SAHH) (Cañizares et al., 2013; Ivanov et al., 2016). Inhibition of SAMS activity by HCPro has been proposed to deprive methyl transferase HEN1 of its methyl donor S‐adenosyl‐l‐methionine (SAM; Ivanov et al., 2016). In addition, the knockdown of SAHH has been found to decrease siRNA accumulation and suppress local RNA silencing (Cañizares et al., 2013). Our current study indicates that the HCPro‐mediated local disruption of the methionine cycle and the reduction of glutathione (GSH) concentration as a result of synergistic interactions potentially have a crucial role to play in PVX–PVA synergism.
Results
Selection of a marker pair for the simultaneous quantification of PVX and PVA
Owing to their stability and ease of quantification, fluorescent protein‐based detection methods have garnered wide acceptance in plant studies. They can be imaged as well as quantified accurately from intact leaf tissues without any need for cell lysis or sample preparation (Dhillon et al., 2009; Richards et al., 2003; Stephan et al., 2011). The expression of fluorescent proteins from viral genomes within the host cell can accurately reflect the viral gene expression level. Pasin et al. (2014) have recently developed a semi‐high‐throughput method for fluorescence intensity quantification of potyviruses from intact infected leaf discs. Following a similar strategy, we developed a rapid dual fluorescence marker‐based method for the simultaneous detection of PVX and PVA during mixed infection.
Simultaneous detection of PVX and PVA in the system required the selection of a suitable pair of fluorescence markers with adequate sensitivity, yet non‐overlapping excitation/emission (Ex/Em) spectra. We screened for a suitable marker to be used with PVX tagged with green fluorescent protein (GFP) (PVX‐GFP). Nicotiana benthamiana leaves were infiltrated with Agrobacterium [optical density at 600 nm (OD600) = 0.5] carrying PVA infectious complementary DNAs (icDNAs) tagged with red fluorescent protein (RFP) (PVA‐RFP), yellow fluorescent protein (YFP) (PVA‐YFP) or cyan fluorescent protein (CFP) (PVA‐CFP) (constructs presented in Table 1). Fluorescence signals were measured under several recommended Ex/Em ultraviolet–visible (UV–vis) wavelengths at 4 days post‐infiltration (dpi). Samples infiltrated with β‐glucuronidase (GUS) were used as negative controls. Table 2 compiles the highest fold change achieved in comparison with the background fluorescence of mock plants for each marker, together with the corresponding Ex/Em wavelengths. RFP and YFP showed the highest 263‐fold difference, whereas CFP gave only a weak three‐fold difference. As the Ex/Em spectra of RFP and GFP do not overlap, they were considered to be the best pair for dual virus quantification.
Table 1.
List of constructs used in this study.
| Construct name | Gene cassette | Vector | Description | Reference |
|---|---|---|---|---|
| PVX | 35S‐PVX::GFP/RLUC‐nos | pGreen | Full‐length, wild‐type PVX icDNA tagged with GFP and RLUC | This study |
| PVA‐(RFP/YFP/CFP) | 35S‐PVAwt::RFP/YFP/CFP‐nos | pRD400 | Full‐length, wild‐type PVA icDNA tagged with either RFP/YFP or CFP | Ivanov et al. ( 2016) and this study |
| pHG‐SAMS | 35S‐SAMS(hp)‐ocs | pHG12 | Plasmid expressing hairpin RNA targeting the SAMS gene family | Ivanov et al. ( 2003) |
| pHG‐SAHH | 35S‐SAHH(hp)‐ocs | pHG12 | Plasmid expressing hairpin RNA targeting the SAHH gene family | Ivanov et al. ( 2003) |
| pHG‐HEN1 | 35S‐HEN1(hp)‐ocs | pHG12 | Plasmid expressing hairpin RNA targeting the HEN1 gene family | Ivanov et al. ( 2003) |
| pHG‐GSHS | 35S‐GSHS(hp)‐ocs | pHG8 | Plasmid expressing hairpin RNA targeting the GSHS gene family | This study |
| HCPro | 35S‐HCPro‐nos | pRD400 | Plasmid expressing PVA HCPro | Hafrén et al. ( 2015) |
| HCProSDM | 35S‐HCProSDM‐RFP‐nos | pSITEII‐6C1 | Plasmid expressing silencing suppression‐deficient mutant of HCPro fused to RFP | Hafrén et al. ( 2015) |
| HCPro4EBM | 35S‐HCPro4EBM‐RFP‐nos | pSITEII‐6C1 | Plasmid expressing eIF4E binding‐deficient mutant of HCPro fused to RFP | Hafrén et al. ( 2015) |
| GUS | 35S‐GUS‐nos | pRD400 | Plasmid expressing uidA gene encoding β‐glucuronidase (GUS) | Eskelin et al. ( 2011) |
| FLUC | 35S‐FLUC‐nos | pRD400 | Plasmid expressing intron‐spliced FLUC | Hafrén et al. ( 2015) |
CFP, cyan fluorescent protein; eIF4E, eukaryotic translation initiation factor 4E; GFP, green fluorescent protein; HCPro, helper component proteinase; HEN1, Hua enhancer 1; icDNA, infectious complementary DNA; PVA, Potato virus A; PVX, Potato virus X; RFP, red fluorescent protein; RLUC, Renilla luciferase; SAMS, S‐adenosyl‐l‐methionine synthetase; SAHH, S‐adenosyl‐l‐homocysteine hydrolase; YFP, yellow fluorescent protein.
Table 2.
Selected excitation/emission (Ex/Em) wavelengths giving maximum fold difference for individual fluorescence markers
| Construct | Ex/Em (nm) | Maximum fold change * |
|---|---|---|
| PVX::GFP | 400/509 | 13 |
| PVA::RFP | 555/584 | 263 |
| PVA::YFP | 500/530 | 263 |
| PVA::CFP | 433/475 | 3 |
*Fold change was calculated by dividing the fluorescence intensity of the samples containing the fluorescent protein in question by the background signal level of the β‐glucuronidase (GUS)‐containing mock samples. Both test and mock plants were Agrobacterium infiltrated at an optical density of OD600 = 0.1.
Validation of markers
In order to test the sensitivity of the selected markers in the quantification of PVA and PVX, we infiltrated N. benthamiana leaves with series of Agrobacterium dilutions (OD600 = 0.5, 0.1, 0.02 and 0.004) carrying PVA‐RFP and PVX‐GFP icDNA (Fig. 1A,B). Leaf samples were collected at 3 dpi and the fluorescent proteins were measured under selected Ex/Em wavelengths. The fluorescence intensity was found to increase with increasing amount of Agrobacterium infiltration (Fig. 1A,B). A high r 2 coefficient of determination in linear regression for both markers ( = 0.90 and = 0.91) suggested their suitability for the quantitative estimation of GFP derived from PVX sgRNA and RFP produced as a part and subsequently processed from PVA polyprotein.
Figure 1.
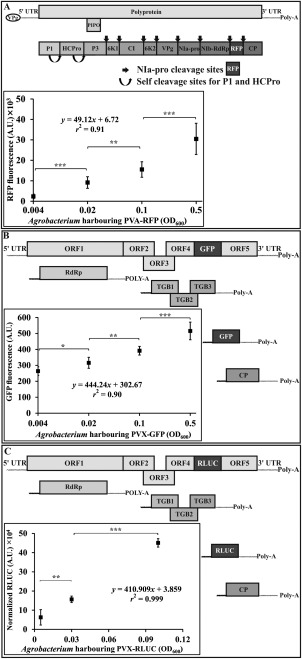
Quantifiability of Potato virus A‐red fluorescent protein (PVA‐RFP), Potato virus X‐green fluorescent protein (PVX‐GFP) and Potato virus A‐Renilla luciferase (PVX‐RLUC). Each panel (A–C) depicts a schematic presentation of the viral construct used (not drawn to scale). Although RFP is expressed as part of the PVA polyprotein, GFP and RLUC are products of the PVX subgenomic RNAs (sgRNAs). Plants were infiltrated with increasing amounts of Agrobacterium carrying an infectious complementary DNA (icDNA) to express PVA‐RFP (A), PVX‐GFP (B) and PVX‐RLUC (C). Sampling of PVA‐RFP (n = 8) and PVX‐GFP (n = 8) expressing plants was performed from local leaves at 5 days post‐infiltration (dpi) and from PVX‐RLUC expressing plants at 3 dpi (n = 4). 35S‐Firefly luciferase (35S‐FLUC) [optical density at 600 nm (OD600) = 0.01] was used to normalize RLUC amongst the sets. Student's t‐test was carried out to estimate the significance of the differences between various Agrobacterium concentrations (*P < 0.05; **P < 0.01; ***P < 0.001).
Renilla luciferase (RLUC)‐based assay has proven to be an outstanding method for the measurement of the viral gene expression level, owing to its high sensitivity and linearity over several orders of magnitude (Eskelin et al., 2010). As the other PVX construct used herein harbours RLUC (Table 1), another set of experiments was carried out to correlate the RLUC expression level with different dilutions (0.1, 0.03 and 0.005) of Agrobacterium carrying PVX‐RLUC (Fig. 1C). From the graphs, it is evident that the variation in the level of RLUC expression and GFP fluorescence with varying Agrobacterium OD does not follow the same scale. That is, a two‐fold increase in RLUC expression level is not reflected as a two‐fold increase in GFP fluorescence. This essentially indicates the lower sensitivity of GFP when compared with RLUC; moreover, the high coefficient of determination ( = 0.99) advocates a linear correlation between infiltration OD and RLUC expression level.
PVX and PVA infection dynamics during single and mixed infections
Next, we measured the variation in the respective GFP and RFP accumulation during the synergistic interaction between PVX and PVA, and compared this with their accumulation during single infections. Agrobacterium carrying PVX‐GFP and PVA‐RFP (OD600 = 0.01) was infiltrated both individually and as a mixture, and the fluorescence intensities of the respective markers were measured from the local leaves on the third, fifth, seventh and 10th days (Fig. 2A,C), and from the systemic leaves on the fifth, seventh, 10th and 14th days (Fig. 2B,D). As an independent method for the validation of the accumulation of the corresponding viruses, we also measured PVX (Fig. 2E,F) and PVA (Fig. 2G,H) gRNA levels from the same samples. PVX and PVA accumulation patterns in both local and systemic leaves, as well as in single and mixed infection scenarios, were distinctly different. PVA expression in local leaves kept on increasing (for PVA single infection) or reached a plateau (for PVX–PVA mixed infection) during the studied period. In the systemic leaves, PVA expression reached a maximum in both cases by the 10th day, followed by a slight fall in the fluorescence level on the 14th day. PVA expression levels were lower in mixed infection sets than in singly infected sets in both local and systemic leaves. The quantification of the PVA RNA level also corroborated this observation. PVX, however, accumulated at a high level in local leaves in both single and mixed infection sets. Although fluorescence as a result of PVX sgRNA expression and the accumulation of PVX gRNA were boosted significantly in the local leaves of mixed infection sets, the fold difference was modest. The situation was entirely different in systemic leaves, where PVX initially started to spread briskly in both sets but, after reaching a maximum at 7 dpi, the singly infected sets showed a drastic decrease in PVX titre at 10 dpi and, by 14 dpi, the systemic leaves showed almost complete recovery from PVX infection. Interestingly, PVX in the mixed infection sets did not show any sign of a decrease in titre, but continued to increase in terms of fluorescence and RNA level during the study period. In spite of collecting samples as late as 10 dpi from the local leaves, the Agrobacterium‐infiltrated leaves retained a fairly good shape with no significant signs of senescence. A systemic necrosis response developed in plants with mixed PVX and PVA infection by 14 dpi, as visualized in Fig. S1 (see Supporting Information). The retarded growth phenotype of these plants was also evident by that time. GFP and RFP fluorescence patterns from PVX and PVA, respectively, corroborated well with their individual RNA accumulation patterns, validating the robustness of the detection technique. Figure S2 (see Supporting Information) demonstrates the absence of interference of the fluorescence intensity of one marker by the fluorescence of the other at the given Ex/Em parameters used during the course of this experiment. Moreover, representative plots for correlation between individual RNA level and corresponding fluorescence level have been provided to demonstrate the linearity between them.
Figure 2.
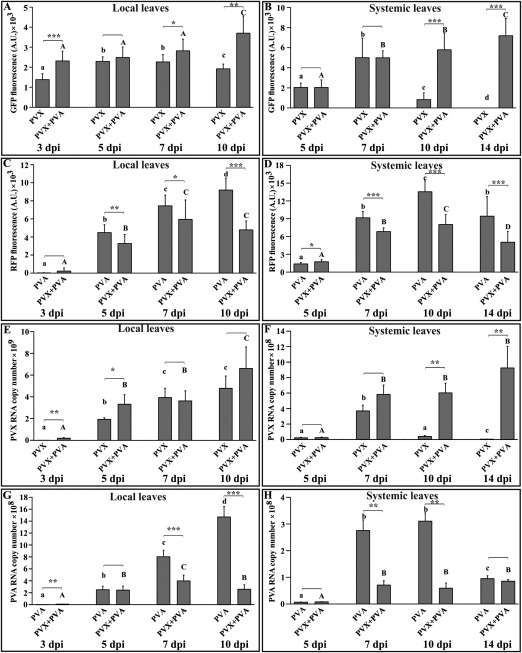
Potato virus X (PVX)‐derived green fluorescent protein (GFP) expression is significantly upregulated in systemic leaves as a result of mixed infection with Potato virus A‐red fluorescent protein (PVA‐RFP). (A, B) GFP fluorescence at designated time points on single PVX‐GFP infection and co‐infection with PVA‐RFP in local and systemic leaves, respectively. (C, D) RFP fluorescence at the same time points as in (A) and (B) on single PVA‐RFP infection and co‐infection with PVX‐GFP from local and systemic leaves, respectively. (E, F) PVX genomic RNA (gRNA) level corresponding to the fluorescence intensities presented in (A) and (B), respectively. (G, H) PVA RNA level corresponding to the fluorescence intensities presented in (C) and (D), respectively. Agrobacterium carrying the viral infectious complementary DNAs (icDNAs) was infiltrated at an optical density at 600 nm (OD600) = 0.01, whereas mock plants were infiltrated with Agrobacterium carrying the 35S‐β‐glucuronidase (35S‐GUS) construct at OD600 = 0.01. The third, fifth, seventh and 10th day samples were obtained from local leaves, and the fifth, seventh, 10th and 14th day samples were obtained from young systemic leaves. The fluorescence results are presented after subtraction of the background fluorescence level (mock) for that particular marker within each set. Each infection set comprises 15 individual plants further divided into subsets of three designated for individual time points. Each plant was sampled at four different infiltration points for local samples and four different systemic leaves for systemic samples, making a total of n = 12 replicates per reading. The RNA copy number was normalized using the housekeeping gene protein phosphatase 2A (PP2A). Student's t‐test was carried out to estimate the significance of the differences between single versus mixed infection (*P < 0.05; **P < 0.01; ***P < 0.001). Different letters above the bars indicate significant differences (P < 0.05) between time points in single infections (lowercase letters) and mixed infections (uppercase letters).
Role of HCPro in PVX and PVA synergism
The role of HCPro in the synergistic enhancement of the accumulation of several non‐related viruses (including PVX) has been known for two decades (Shi et al., 1997). Studies showing that mutations in the central domain of HCPro render it unable to suppress silencing, as well as inducing synergism, have strengthened the idea that these two functions of HCPro are interrelated (González‐Jara et al., 2005). In this context, we employed PVA HCPro and two mutants: a silencing‐deficient mutant (HCProSDM) and a eukaryotic translation initiation factor 4E (eIF4E) binding‐deficient mutant (HCPro4EBM) (Hafrén et al., 2015). We infiltrated young leaves with Agrobacterium harbouring PVX‐RLUC (OD600 = 0.01) together with one of the following constructs: 35S‐HCPro (OD600 = 0.3); 35S‐HCProSDM (OD600 = 1) and 35S‐HCPro4EBM (OD600 = 1). Higher OD600 of infiltration was used for the mutants to achieve a similar expression level to the wild‐type HCPro, as in Hafrén et al. (2015). Agrobacterium carrying 35S‐GUS was used as a control. Wild‐type HCPro led to a significant increase in RLUC expression by 5 dpi, whereas synergism was lost with HCProSDM and HCPro4EBM (Fig. 3A). The silencing suppression capacity of PVA HCPro4EBM was debilitated (Hafrén et al., 2015). Therefore, the results with both mutants confirm that antiviral RNA silencing suppression of HCPro is an essential requirement for the synergistic response.
Figure 3.
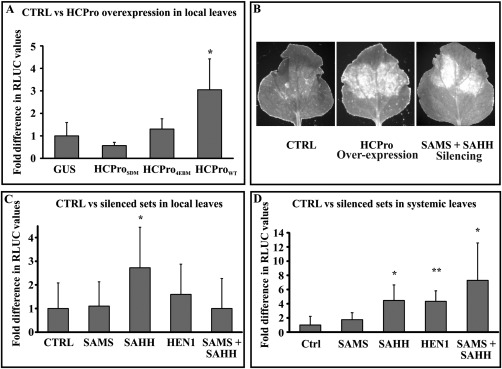
The silencing suppression activity of HCPro (helper component proteinase) is essential and disruption of the host cell methionine cycle contributes to Potato virus X–Potato virus A (PVX–PVA) synergism. (A) Relative variation in RLUC (Renilla luciferase) activities (n = 5) from PVX‐RLUC when co‐expressed with HCPro and its mutants. Agrobacterium carrying PVX‐RLUC was infiltrated at an optical density at 600 nm (OD600) = 0.01, wild‐type HCPro (HCProWT) at OD600 = 0.3, silencing suppression‐deficient (HCProSDM) and eukaryotic translation initiation factor 4E (eIF4E) binding‐deficient (HCPro4EBM) mutants of HCPro at OD600 = 1 and β‐glucuronidase (GUS) as negative control at OD600 = 0.3. Samples were collected from local leaves at 9 days post‐infiltration (dpi). Normalization of the RLUC levels was performed using 35S‐Firefly luciferase (35S‐FLUC) Agrobacterium infiltrated at OD600 = 0.01. Results are presented in terms of the fold difference compared with the control (GUS) set at a value of unity. Based on Student's t‐test, only the wild‐type HCPro was able to enhance PVX‐RLUC expression significantly over the GUS control (*P < 0.05). CTRL, control. (B) Visual examination of PVX‐derived GFP revealed enhanced GFP accumulation in HCPro‐overexpressing leaves, but also in leaves in which both of the methionine cycle enzymes, S‐adenosyl‐l‐methionine synthetase (SAMS) and S‐adenosyl‐l‐homocysteine hydrolase (SAHH), were silenced simultaneously. In this experiment, PVX‐GFP was infiltrated in the lower leaves on day zero at OD600 = 0.05 and allowed to spread systemically. The silencing and overexpression constructs were infiltrated into the top leaves on the 10th day. GFP fluorescence from the silenced/overexpressed spots was visualized on the 13th day under UV light. (C) Fold difference in PVX‐derived RLUC activities between control and silenced sets in local leaves. All sample sets (biological replicates within a set n = 5) were Agrobacterium infiltrated with PVX‐RLUC infectious complementary DNA (icDNA) at OD600 = 0.0001 accompanied by the silencing constructs, each at OD600 = 0.4. Empty pHG12 plasmid (CTRL) was used as a negative control. Sampling was performed at 6 dpi. Student's t‐test was carried out to calculate the significance of the differences between CTRL vs. silenced samples. (D) Fold difference in PVX‐derived RLUC activities (n = 5 or more), similar to (C), but from systemic leaves. The lower leaves were infiltrated with PVX‐RLUC (OD600 = 0.05) and infection was allowed to spread systemically. At 5 days after initiation of the infection, the top leaves were infiltrated with the respective silencing constructs at OD600 = 0.4. Samples were collected at 10 dpi from the top leaf areas into which the silencing constructs had been infiltrated. 35S‐FLUC (OD600 = 0.01) was used to normalize the RLUC values within each set. Statistical significances were calculated between the silenced sets against empty pHG12 (CTRL) (*P < 0.05; **P < 0.01).
Involvement of the methionine cycle in PVX–PVA synergism
In our earlier study, we have shown that one putative mechanism by which HCPro carries out silencing suppression is through the local disruption of the methionine cycle (Ivanov et al., 2016). HCPro reduces SAMS activity and interacts with SAHH during PVA infection in planta. In the light of the knowledge on these molecular interactions, we sought to elucidate the events taking place during the synergistic interaction between PVX and PVA. We monitored PVX accumulation in leaves in which SAMS and SAHH had simultaneously been knocked down to determine whether PVX gene expression was affected under these circumstances. The lower leaves of the plants were infiltrated with PVX‐GFP (OD600 = 0.05; day 0) and the infection was left to spread systemically. The top leaves were infiltrated with the silencing constructs pHG‐SAMS and pHG‐SAHH (OD600 = 0.4) or the 35S‐HCPro expression construct (OD600 = 0.15) at 10 dpi. Control for silencing was Agrobacterium containing empty Hellsgate plasmid (pHG‐CTRL). Visual examination of GFP fluorescence from the top leaves showed the strong accumulation of PVX‐derived GFP in HCPro‐overexpressing leaves at 13 dpi (Fig. 3B). A comparable accumulation was also seen in SAMS + SAHH‐silenced leaves (Fig. 3B), but not in control leaves. These results prompted us to probe further into the possible role of methionine cycle enzymes in HCPro‐mediated synergism.
In the next set of experiments, we quantified PVX‐RLUC expression in the SAMS‐, SAHH‐ and HEN1‐silenced backgrounds. RLUC activity was assayed from both local and systemic leaves (Fig. 3C,D). Interestingly, only SAHH silencing showed a consistent increase in PVX sgRNA expression in both local leaves at 6 dpi and systemic leaves at 10 dpi. HEN1 and simultaneous SAMS + SAHH silencing also boosted PVX expression significantly, as measured from systemic leaves at 10 dpi, whereas SAMS silencing did not show any effect on PVX expression in either of these cases.
Relationship between the methionine cycle and PVX accumulation
In our previous article (Ivanov et al., 2016), we showed that the downregulation of SAMS, SAHH and HEN1 enhanced the accumulation of HCPro‐less PVA (PVAΔHCPro), and hypothesized that, under these circumstances, HEN1 is deprived of its substrate SAM and, consequently, small RNA (sRNA) stabilization via methylation is inhibited. We hypothesized that PVX may benefit from less stable siRNAs, similar to PVA. In order to better understand the PVX gene expression pattern in SAMS‐, SAHH‐, SAMS + SAHH‐ and HEN1‐silenced conditions, we performed an infection dynamics study between 3 and 9 dpi (Fig. 4A–E).
Figure 4.
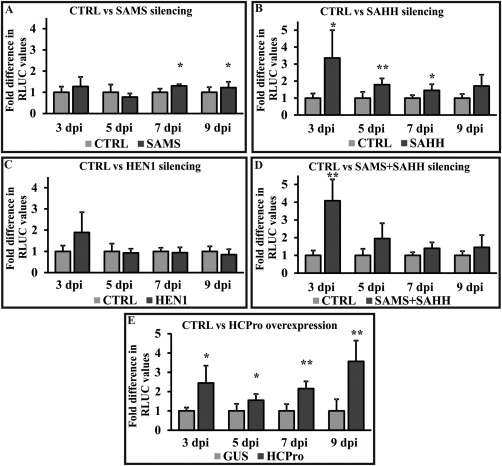
HCPro (helper component proteinase) overexpression and S‐adenosyl‐l‐homocysteine hydrolase (SAHH) silencing upregulate Potato virus X (PVX) gene expression. The PVX infection profile in terms of RLUC (Renilla luciferase) expression in the S‐adenosyl‐l‐methionine synthetase (SAMS)‐, SAHH‐, Hua enhancer 1 (HEN1)‐silenced and HCPro overexpression background was determined between 3 and 9 days post‐infiltration (dpi). The fold differences in PVX‐derived RLUC values between controls [for silencing, empty pHG12 plasmid; for overexpression, β‐glucuronidase (GUS)] and silenced/overexpressed sets are given at the designated time points. All samples were infiltrated with PVX‐RLUC at an optical density at 600 nm (OD600) = 0.01. Individual silencing and overexpression constructs were infiltrated at OD600 = 0.4. 35S‐Firefly luciferase (35S‐FLUC) (OD600 = 0.01) was used for normalization of the RLUC values. (A–D) Fold differences between RLUC values from control (CTRL) (set to unity) and SAMS‐/SAHH‐/HEN1‐/SAMS + SAHH‐silenced samples, respectively. (E) Fold difference in PVX‐derived RLUC values in GUS‐ and HCPro‐overexpressing sample sets. Statistical significances were calculated between the marked sample and its corresponding control (*P < 0.05; **P < 0.01).
SAMS and HEN1 silencing did not convincingly boost PVX‐RLUC sgRNA expression at most of the time points. Although, during the later days (7 and 9 dpi), PVX sgRNA expression in SAMS‐silenced sets showed significant enhancement compared with the control sets, the fold difference was too low (1.2–1.3‐fold) to draw any firm conclusions. SAHH‐silenced sets, however, demonstrated a consistent pattern of significant enhancement of RLUC expression from sgRNA between 3 and 7 dpi, and this trend continued at 9 dpi in local leaves (Fig. 4B). Interestingly, the combined effect of SAMS + SAHH silencing (Fig. 4D) followed the pattern of SAHH silencing alone. The PVX sgRNA expression level, however, when co‐expressed with HCPro, was approximately three‐fold enhanced during the whole period of time (Fig. 4E). HEN1, SAMS and SAHH silencing, as well as the expression of HCPro, between 3 and 9 dpi was confirmed by reverse transcription‐polymerase chain reaction (RT‐PCR) (Fig. S3, see Supporting Information).
We next wanted to determine whether the transient knockdown of methionine cycle components had any effect on (+)‐ and (–)‐gRNA accumulation during synergism. We extracted total RNA from the representative sets on the third, fifth and ninth days, and prepared (+)‐ and (–)‐strand‐specific cDNA using primers from the PVX RNA polymerase‐encoding region (RdRP). (+)‐ and (–)‐RNA‐specific products were quantified using quantitative PCR (qPCR). For most of the silencing conditions (SAMS, SAHH and HEN1), (+)‐ and (–)‐gRNA accumulation was slightly lower than the control during the initial phase of the infection. As the infection progressed, this difference was reduced, as seen in the fifth day samples, and, on the ninth day, both (+)‐ and (–)‐strand gRNA accumulation in the silenced sets marginally bypassed that of the control (Fig. 5A–H). HCPro expression in PVX‐infected leaves, however, demonstrated higher accumulation of PVX gRNAs throughout the whole studied period (Fig. 5G,H). Calculation from qPCR results showed a significant increase in (–)‐strand accumulation in the case of SAMS‐silenced (Fig. 5B) and SAHH‐silenced (Fig. 5D) sets (on the ninth day) compared with non‐silenced plants, an aspect of synergism previously described in Nicotiana tabacum (Pruss et al., 1997; Vance et al., 1995). However, the fold differences observed here were lower than those presented in those reports. The report from González‐Jara et al. (2005) also indicated a similar phenomenon in N. benthamiana infected with PVX + PVY/Tobacco etch virus (TEV), but to a lesser extent, which is much in line with our observations herein.
Figure 5.
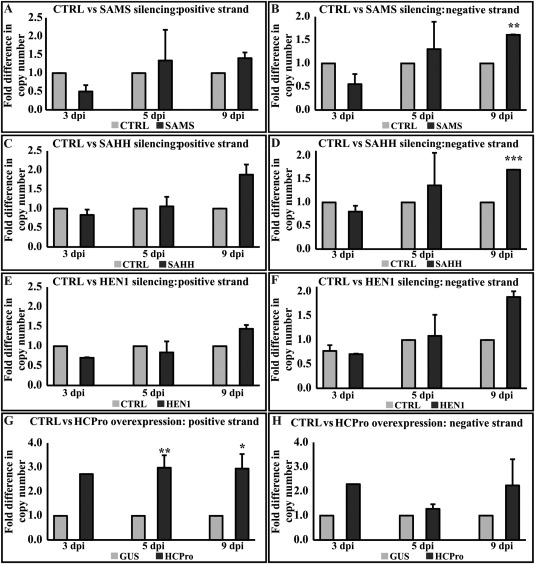
Potato virus X (PVX) (+)‐ and (–)‐strand RNA accumulation is enhanced in HCPro (helper component proteinase)‐overexpressing plants and in Hua enhancer 1 (HEN1)‐, S‐adenosyl‐l‐homocysteine hydrolase (SAHH)‐ and S‐adenosyl‐l‐methionine synthetase (SAMS)‐silenced plants. Variation in accumulation of PVX (+)‐ and (–)‐RNAs in the SAMS‐, SAHH‐, HEN1‐silenced and HCPro‐overexpressing backgrounds was determined at different time points. Experimental conditions are the same as those in Fig. 4. (+)‐ and (–)‐strand‐specific primers binding to the RNA polymerase‐encoding region (RdRP) were used to prepare cDNA from the purified RNA samples and for polymerase chain reaction (PCR) amplification. (A, C, E, G) Fold difference in accumulation of PVX (+)‐RNA when compared with control [CTRL: empty pHG12 plasmid or β‐glucuronidase (GUS)]. (B, D, F, H) Fold difference in accumulation of PVX (–)‐RNA when compared with CTRL (empty pHG12 plasmid or GUS). Copy numbers of the respective strands were calculated from a standard curve (*P < 0.05; **P < 0.01; ***P < 0.001).
Role of GSH in PVX–PVA synergism
As the methionine cycle shares a common metabolite, homocysteine, with the trans‐sulfuration pathway to produce methionine from cysteine (Wilson et al., 1976), reduction in homocysteine generation via a blockage in S‐adenosylhomocysteine (SAH) hydrolysis has been hypothesized to be replenished by pulling cysteine flux more towards methionine synthesis upstream rather than GSH production downstream. As a result of this intimate coupling between the methionine cycle and the trans‐sulfuration pathway, we decided to investigate whether HCPro‐mediated disruption of the methionine cycle could affect GSH biosynthesis and whether this could have any role in synergism. The variation in the GSH levels in the test samples PVX + HCPro, pHG‐SAMS + PVX and pHG‐SAHH + PVX, taking place between 2 and 9 dpi, was compared with that of a number of controls (PVX alone, PVX + GUS, PVX + pHG‐CTRL, PVX + pHG‐HEN1, PVA alone, HCPro alone and non‐infected plants) to eliminate effects other than those specifically related to synergism. As a positive control, we used the glutathione synthetase (GSHS) silencing construct (pHG‐GSHS + PVX). In the controls PVX alone, PVX + GUS, PVX + pHG‐CTRL, PVX + pHG‐HEN1, PVA alone and HCPro alone, and non‐infected plants, the GSH level either stayed the same or increased between days 2 and 9 (Fig. 6A). Although HCPro interferes with the methionine cycle, it could not alone reduce GSH levels, whereas HCPro together with PVX could. GSH also decreased in the SAMS‐, SAHH‐, SAMS + SAHH‐ and GSHS‐silenced plants infected with PVX (Fig. 6A). Partial disruption of the methionine cycle by silencing either SAMS or SAHH showed only approximately 10% reduction in GSH level in the presence of PVX, whereas silencing of both together resulted in a stronger reduction in the GSH level. In the HEN1‐silenced and PVX‐infected set, the GSH level did not decrease. HEN1 is a methyl transferase, which exploits SAM as a methyl donor and produces SAH as a byproduct, but the methionine cycle is not dependent on HEN1 activity. Therefore, it is logical that the silencing of HEN1 does not lead to the disruption of the methionine cycle. The positive control, i.e. GSHS silencing + PVX, resulted in approximately 50% reduction in GSH level. HCPro expression produced a very strong response, similar to GSHS silencing, but only in PVX‐infected plants. This led to the question of whether the decrease in GSH level is simply a consequence of PVX–PVA interaction or a pro‐PVX event that can boost PVX expression. To determine whether a reduction in GSH level actually boosts PVX accumulation, we monitored PVX‐RLUC expression in the GSHS‐silenced background. Interestingly, GSHS silencing showed significant enhancement in RLUC accumulation from 2 dpi, and approximately two‐fold higher PVX sgRNA expression was also noted at 5 and 7 dpi (Fig. 6B–D). These data suggest that reduction of GSH in cells in the presence of PVX and HCPro is an important factor in PVX–PVA synergism.
Figure 6.
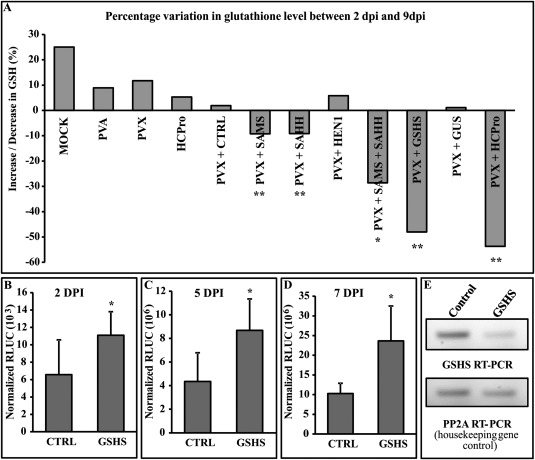
Decrease in glutathione (GSH) concentration occurs in S‐adenosyl‐l‐methionine synthetase (SAMS)‐, S‐adenosyl‐l‐homocysteine hydrolase (SAHH)‐ and glutathione synthetase (GSHS)‐silenced and HCPro (helper component proteinase)‐overexpressing plants. (A) The change in GSH concentration between 2 days post‐infiltration (dpi) (n = 6) and 9 dpi (n = 6) is shown as a percentage. The experimental conditions were essentially the same as in Fig. 4. Potato virus A (PVA) and HCPro alone were infiltrated at an optical density at 600 nm (OD600) of 0.01 and 0.3, respectively, and mock sets were infiltrated with induction buffer (devoid of Agrobacterium). For measurement of the GSH level, reduced glutathione was derivatized to N‐ethylmaleimide (NEM)‐GSH and quantified via liquid chromatography‐mass spectrometry (LC‐MS). The GSH concentration from each sample was calculated from the LC‐MS data using a standard curve plotted with known dilutions of derivatized GSH. A stable isotope of GSH was used to normalize technical variation in GSH detection across all the sets. (B–D) Potato virus X (PVX)‐derived RLUC (Renilla luciferase) values measured from empty pHG12 (CTRL) and GSHS‐silenced background at 2, 5 and 7 dpi. The GSHS silencing construct was infiltrated at OD600 = 0.4. 35S‐Firefly luciferase (35S‐FLUC) (Agrobacterium infiltrated at OD600 = 0.01) was used to normalize RLUC values. (E) Visual validation of knockdown of GSHS gene. Semi‐quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) with 25 amplification cycles was used to demonstrate differential band intensities in an agarose gel. PP2A, protein phosphatase 2A. Statistical significance for the silenced sets was calculated against the empty pHG12 (CTRL) control, whereas significance for the HCPro overexpression sets was calculated against the β‐glucuronidase (GUS) control (*P < 0.05; **P < 0.01).
Discussion
In this study, we set out to investigate the molecular mechanism behind the synergistic interaction between potex‐ and potyviruses during co‐infection. As evident from this and previous studies (Shi et al., 1997), the silencing suppressor activity of potyviral HCPro is essential for enhanced PVX accumulation. In a recent paper, we proposed that part of the HCPro silencing suppression activity can be attributed to its capacity to interfere with the host cell methionine cycle (Ivanov et al., 2016). We hypothesized that the interactions of HCPro with the methionine cycle enzymes SAMS and SAHH could also play a role in PVX–PVA synergism. The data obtained during the course of this study indicate that disruption of the methionine cycle indeed leads to enhanced PVX gene expression. In addition, reduced cellular GSH amounts were observed during synergistic PVA and PVX infection and when the methionine cycle had been disrupted. Based on these results, we propose that the GSH biosynthesis pathway, which is linked with the methionine cycle, is inhibited in these PVX‐infected plants, which express HCPro. We also propose that a reduction in GSH biosynthesis activity may create a cellular redox environment advantageous for enhanced PVX gene expression, but detrimental for the plant during potex–potyvirus co‐infection.
To date, a wide range of techniques have been employed to quantify virus accumulation. Both RLUC and fluorescent proteins have been shown to be reliable biomarkers for fast and easy quantification of potyviral gene expression (Eskelin et al., 2010; Pasin et al., 2014). In the current study, we used two fluorescent proteins, GFP and RFP, expressed from PVX and PVA icDNAs, respectively, for the simultaneous quantification of PVX and PVA gene expression. We validated the applicability of this dual marker system in the mixed infection context by comparing the viral expression patterns with information from previous studies. PVX has been reported to accumulate in three‐ to five‐fold greater amounts in mixed infection with PVY than alone in tobacco (Vance et al., 1995), whereas no such variation in potyvirus titre has been observed (Vance, 1991). In N. benthamiana, synergism is associated with the enhanced accumulation of PVX sgRNAs (Aguilar et al., 2015). Our results reiterate these observations, supporting the validity of simultaneous GFP‐ and RFP‐based quantification for mixed infection. RLUC, as a more sensitive marker, was, however, used for the quantification of PVX sgRNA gene expression when applicable.
RNA silencing is a robust antiviral defence mechanism in plants (Tomari and Zamore, 2005). The RNA silencing suppressor HCPro has developed several strategies to counter host defence systems. During viral RNA silencing, the methionine cycle, together with HEN1, a methyl transferase responsible for siRNA methylation, plays a pivotal role in the stabilization of siRNAs prior to their loading onto the RISC (Yu et al., 2005). HEN1 uses SAM, the product of SAMS, as the methyl group donor to methylate siRNAs, with the simultaneous production of SAH as the byproduct. SAH is a potent feedback inhibitor for the methyltransferases and SAHH efficiently removes it from the system to produce adenosine and homocysteine. We have proposed (Ivanov et al., 2016) that SAMS activity and, consequently, siRNA methylation, as well as SAHH activity, are inhibited by HCPro during PVA infection. In this study, we silenced SAMS, SAHH and HEN1, one at a time as well as in combination, and observed the changes in PVX sgRNA expression at different time points. Although SAHH silencing consistently enhanced RLUC accumulation from PVX sgRNA, no reproducible pattern of enhancement was induced by silencing of SAMS. Moreover, silencing of SAMS + SAHH together did not enhance PVX accumulation any better than silencing SAHH alone. As PVX carries a 7‐methylguanylate cap at the 5′ end of its genome, the dependence of PVX infection on SAM as a donor for cap methylation cannot be ignored as a factor that could be affected by SAMS silencing.
Seminal work on potex–potyvirus synergism by Pruss et al. (1997) established that, during single infection by PVX, accumulation continues to increase during the initial phase of infection. However, after a certain time point, the rate of increase diminishes and, subsequently, a gradual fall in both viral RNA and protein expression level can be observed. During the synergistic interaction, the increasing trend in PVX accumulation is prolonged. We observed that, for SAHH‐silenced sets, major enhancement in PVX‐derived RLUC values was observed during the initial phase of infection. In contrast, during HCPro overexpression, PVX accumulation was low during the earlier stages and increased as the infection continued. In addition, the fold difference in PVX RNA accumulation and sgRNA gene expression in HCPro‐overexpressing plants was clearly higher than that in any of the silenced backgrounds. The reason behind this could be that HCPro‐mediated interference with the methionine cycle activity may result in a more complete local loss of function than achieved by general silencing of the methionine cycle enzymes. Another reason may be that HCPro, being highly multifunctional in nature, may undergo other simultaneous interactions within the cells, which may also contribute cumulatively to PVX accumulation. A differential increase in (–)‐strand accumulation during synergism has been detected in several studies conducted on N. tabacum (Andika et al., 2012; González‐Jara et al., 2004; Pruss et al., 1997; Vance et al., 1995), and it has been presented as a hallmark for potex–potyviral synergism. In this study, we showed greater accumulation of (–)‐strand PVX RNA in SAMS‐ and SAHH‐silenced sets during later days (9 dpi), although the same was not detected in HEN1‐silenced or HCPro‐overexpressing plants. However, this is a common observation in the case of mixed infection in N. benthamiana. Only a slight increase in PVX (–)‐strand RNA accumulation has been reported in the case of co‐infections with PVY and TEV (González‐Jara et al., 2004), and a similar level of accumulation has been shown with several PPV HCPro variants independent of whether or not they induce synergism (González‐Jara et al., 2005).
The methionine cycle shares common metabolites with several metabolic pathways, such as the methionine salvage, polyamine, ethylene and GSH biosynthesis pathways. The roles of several intermediates of these closely associated pathways have been reported to be involved in many plant–pathogen interactions, including viral interactions (Alazem and Lin, 2015; Ball et al., 2004; Höller et al., 2010; Jiménez‐Bremont et al., 2014; Zechmann et al., 2007b). Disruption of the methionine cycle via selective downregulation of its central components may alter the flow of metabolites to these various pathways. GSH biosynthesis in this context deserves special inspection, as its level within the cells has been predicted to affect virus infection in many ways. An elevated GSH level has been correlated with reduced development of symptoms of virus infections (Gullner et al., 1999; Zechmann et al., 2007b). Enhanced activity of GSH metabolism has also been correlated with sulfur‐induced resistance in Tobacco mosaic virus (TMV)‐infected N. tabacum plants (Höller et al., 2010). As cysteine is a rate‐limiting substrate for GSH biosynthesis (Courtney‐Martin et al., 2008; Zechmann et al., 2008), it seems logical to believe that disruption of the methionine cycle could affect the GSH level downstream by altering the balance between homocysteine and cysteine. Migration of cysteine towards methionine synthesis probably deprives the GSH production pathway from its fair share of cysteine. This ultimately leads to a reduction in GSH accumulation. To examine the relationship between the GSH level in the host and PVX–PVA synergism, we knocked down GSHS, which we anticipated would block GSH biosynthesis. Similar to the SAHH‐silenced background, in the GSHS‐silenced background, PVX sgRNA expression was enhanced. We employed liquid chromatography‐mass spectrometry (LC‐MS) to quantify GSH in the infected leaves at two time points. PVX, together with HCPro expression, methionine cycle disruption or GSHS knockdown, led to a reduction in GSH level as the infection progressed. Samples expressing HCPro, PVA or PVX alone did not have any effect on the GSH level. This essentially shows that, in spite of its interactions with the methionine cycle, the enzyme HCPro alone cannot influence GSH levels, and that the reduction in GSH is truly a synergism‐specific event.
Variation in the GSH level by altering the levels of its precursors has been hypothesized previously as a potential virus‐induced phenomenon in the case of Zucchini yellow mosaic virus (ZYMV, also a potyvirus) infecting Cucurbita pepo (Zechmann et al., 2007a). In addition, transcriptional profiling has shown that PVX–PVY synergism leads to pronounced oxidative stress in the host compared with single infection by PVX or PVY (García‐Marcos et al., 2009). GSH is the main antioxidant in cells maintaining the homeostasis of the cellular redox potential via quenching of oxidative radicals, whereas it is itself converted into its oxidized form (GSSG). Integrating our current findings with this information, it is possible to speculate that, during synergism, HCPro‐mediated disruption of the methionine cycle, together with a PVX‐specific unknown factor, leads to a reduction in GSH production, the effect of which results in increased oxidative stress. This scenario is supported by the results showing that GSH reduction takes place when the methionine cycle is disrupted by silencing of SAMS and SAHH in PVX‐infected cells.
In conclusion, we would like to propose a model for further testing (see Fig. 7) in which the methionine cycle, together with the trans‐sulfuration pathway, plays a crucial role in PVX–PVA synergism. Disruption of the methionine cycle can affect both PVX RNA accumulation and expression. However, all the components of the methionine cycle may not be equally important for synergism. SAMS may have other roles to play in the expression of the cap‐dependent PVX genome; hence, its downregulation by HCPro did not seem to be very effective in boosting PVX. Interaction between HCPro and SAHH, however, seemed to be more crucial for PVX–PVA synergism. SAHH is positioned at a pivotal point in the methionine cycle. Reduced SAHH activity, via enhanced SAH accumulation, leads to reduced HEN1 activity and siRNA instability, which may benefit PVX to some extent. Second, as SAH is not hydrolysed, the supply of homocysteine via this route may be progressively reduced. When the methionine cycle is deprived of homocysteine precursor for methionine production, plant cells solve this problem by channelling cysteine flux towards homocysteine production via an alternative route, the trans‐sulfuration pathway. As cysteine is also a precursor of GSH, its use in homocysteine production is likely to be reflected as a reduction in GSH biosynthesis, which we propose could be an important synergism‐related phenomenon. As HCPro, together with PVX infection, probably causes many types of alteration in host cells, we cannot exclude the involvement of other metabolic or signalling pathways as a cause of the reduced GSH amounts, which is a subject for further investigations. It also remains to be studied why PVX sgRNA accumulation is enhanced during GSH depletion and oxidative stress, in spite of the damage caused to the host cell.
Figure 7.
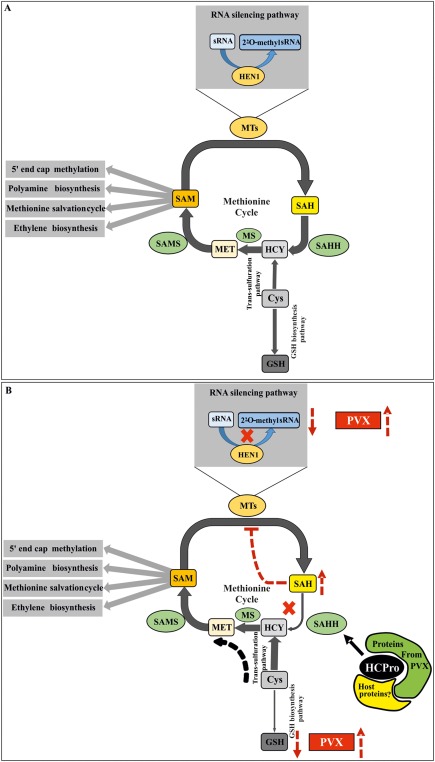
A hypothetical model for the mechanism behind HCPro (helper component proteinase)‐mediated enhancement of Potato virus X (PVX) gene expression. (A) Schematic representation of the methionine cycle and how it is connected with the trans‐sulfuration and glutathione (GSH) biosynthesis pathways. S‐Adenosyl‐l‐methionine synthetase (SAMS) and S‐adenosyl‐l‐homocysteine hydrolase (SAHH), the major players of the methionine cycle, are at the heart of the model proposed. The role of SAMS is to convert methionine (MET) into S‐adenosyl‐l‐methionine (SAM). A methyl transferase (MT) [Hua enhancer 1 (HEN1) in this case] then uses SAM as its substrate to methylate its subject [small RNA (sRNA) in this case]. In this reaction, S‐Adenosyl‐l‐homocysteine (SAH) is produced as a byproduct. SAH is a potential feedback inhibitor for most of the MTs, including HEN1, and hence it needs to be hydrolysed by SAHH. As a result, adenosine and homocysteine (HCY) are produced. Methionine synthase (MS) further catalyses the conversion of HCY to MET, thus closing the circuit. However, there is another route for HCY to enter the methionine cycle. This is via the trans‐sulfuration pathway, where cysteine (Cys) acts as a precursor to HCY. Cys is also a precursor for GSH biosynthesis further downstream. (B) Putative alterations in the flow of metabolites in the concerned pathways on PVX–Potato virus A (PVA) mixed infection. In the context of potyvirus infection, HCPro has already been shown to interact with SAMS and to reduce its activity. The product of SAMS, SAM, is a universal methyl donor and an important metabolite, which takes part in various cellular pathways. PVX, being a capped virus, also depends on SAM for cap methylation. Interaction of HCPro with SAHH causes the overaccumulation of SAH in the system, HEN1 inhibition and the destabilization of small interfering RNAs (siRNAs), which is reflected as a higher PVX titre. Another direct effect of SAHH downregulation is debilitated conversion of SAH to HCY, which is predicted to pull Cys flux towards the methionine cycle via the trans‐sulfuration pathway, depriving GSH of its fair share of Cys. This is reflected in lower GSH production downstream, leading to a remarkably reduced GSH concentration. In addition, this condition upregulates PVX gene expression. Moreover, GSH reduction changes the cellular redox homeostasis, which may be reflected as the increased pathogenicity of PVX during synergistic PVX–PVA interaction.
Experimental Procedures
Plants
Nicotiana benthamiana plants were grown in soil at 22 °C, 50% relative humidity and under a 16‐h light/8‐h dark photoperiod in an environmentally controlled glasshouse.
Viruses and expression constructs
All the constructs used in this study are listed in Table 1. The PVX construct was based on pgR106/7 (http://www.plantsci.cam.ac.uk/research/davidbaulcombe/methods/vigs). 35S‐PVX::RLUC‐nos was constructed by PCR amplification of the intron containing the rluc‐RBC‐1I gene from the 35S‐PVA::rlucint‐nos construct (Eskelin et al., 2011). The primer sequences were as follows: forward primer, 5′‐AGGCGCGCCATGACTTCGAAAGTTTATGATC‐3′ (AscI cloning site in italic); reverse primer, 5′‐CACGTCGACTTATTGTTCATTTTTGAGAAC‐3′ (SalI cloning site in italic). The amplified fragment was inserted into pgR107 via the corresponding cloning sites. PVA constructs were based on the full‐length infectious cDNA clone of PVA strain B11 (GenBank accession number AJ296311). Cloning of the full‐length, wild‐type PVA icDNA tagged with GFP has been described in Ivanov et al. (2003) and, in this study, GFP was replaced with RFP, YFP or CFP genes.
Most of the constructs for silencing experiments have been described elsewhere (see Table 1 for references). For GSHS knockdown experiments, a conserved region within GSHS gene family members (two in number) was identified and PCR fragments corresponding to that region were generated from N. benthamiana cDNA using appropriate primers. The primer sequences were as follows: forward, 5′‐ATTAGATCATCTCATCTCAG‐3′; reverse, 5′‐ATAAGTTCCATATATCCCAA‐3′. The PCR fragments obtained were inserted into pGEM‐T Easy vector (Promega, Madison, USA) and then recombined into pHellsgate 8 silencing vector (CSIRO Plant Industry, Australian capital territory, Australia) via an intermediate vector pDONR/Zeo (Life Technologies, Thermo Fisher Scientific, California, USA) using standard molecular cloning techniques. The monocistronic overexpression constructs used in this study have been described elsewhere (Table 1).
Agroinfiltration
Agrobacterium tumefaciens carrying selected constructs was grown overnight at 28 °C in Luria‐Bertani (LB) medium supplemented with 10 mm 2‐(N‐morpholino)ethanesulfonic acid (MES) (pH 6.3), 20 µm acetosyringone and appropriate antibiotics. On the next day, the overnight cultures were diluted 1 : 10 in the same medium and allowed to grow for at least 5 h (OD600 ∼ 1.5). Cells were harvested by centrifugation at 3500 g for 10 min at room temperature, followed by gentle re‐suspension of the pellets in the induction buffer [10 mm MES (pH 6.3), 10 mm MgCl2 and 150 µm acetosyringone]. This step was repeated twice more to ensure thorough washing of the cells. OD600 of the cell suspension was measured using an Eppendorf BioPhotometer (Germany) to estimate the cell density, which was then subsequently adjusted to an appropriate level for infiltration by diluting with the induction buffer. Infiltration mix was left to rest at room temperature for 1–2 h. Nicotiana benthamiana plants at the four‐leaf stage were selected for infiltration. At least four to six plants of uniform size (biological replicates) comprised each experiment. Young leaves were infiltrated by gently pressing infiltration mix through a syringe (without needle) into the abaxial surface.
Agrobacterium carrying the silencing constructs (see Table 1) or control empty vector was infiltrated at OD600 = 0.4, together with Agrobacterium strains harbouring PVX constructs (OD600 = 0.01) and Firefly luciferase (FLUC) (OD600 = 0.01), into N. benthamiana leaves. Silencing of the host genes was verified by comparing the mRNA levels with semi‐quantitative PCR amplification of isolated cDNA. cDNA synthesis from the control and silenced sets was carried out using a RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Scientific, Vilnius, Lithuania), primed with random hexamer. Subsequent PCR amplification for 25 cycles was carried out using the following primers: SAMS: forward, 5′‐GGGTTTGGATGCTGACAACT‐3′; reverse, 5′‐GTCACCTGCACCAATCTCCT‐3′; SAHH: forward, 5′‐GTCGAAATGCCTGGTCTTAT‐3′; reverse, 5′‐ACCATCTAACTTCAGCGCCT‐3′; HEN1: forward, 5′‐GCAATTTTGGCATCTGTAGGA‐3′; reverse, 5′‐AAAGAACCCCTCCAATTGCT‐3′; GSHS: forward, 5′‐GTTTCTTGAAAACAAAGATGAC‐3′; reverse, 5′‐CGTTGTGGTTTCATTACGTA‐3′. To verify silencing, the band intensities were compared in the agarose gel. However, the HCPro RNA level was visualized in the agarose gel on normal PCR amplification (35 cycles) using the following primers: HCPro forward, 5′‐CAACTCTGATTGATGTGGCA‐3′; HCPro reverse, 5′‐AAATTGGATCAATTGCCGAA‐3′.
Luciferase assay
The plants were sampled at various time points post‐infiltration by cutting 5‐mm leaf discs with a cork borer. All the samples were collected from the infiltrated area, approximately equidistant from the point of infiltration. The discs were frozen immediately in liquid nitrogen and stored at −80 °C. Dual luciferase assay of virus‐derived RLUC and control FLUC was carried out following Eskelin et al. (2010).
Fluorescence intensity measurement
Black, 96‐well, flat‐bottomed plates were used for the study. Infected leaves were sampled using a cork borer with an internal diameter of 5 mm. Similar to the luciferase assay, leaf discs for the study were collected at the same distance from the infiltration point and placed upside down in the wells; 100 μL water were provided for each well to prevent the sample from drying out. Fluorescence intensity quantification was carried out in a Tecan Infinite M200 monochromator‐based plate reader (Switzerland). During the standardization of the method, the top readings were used to acquire the best Ex and Em spectra for each fluorescent protein and to calculate the fold difference over the background. Subsequently virus quantification with the aid of fluorescent proteins was carried out using the selected Ex/Em pair (UV–vis wavelength).
Western blotting
The standard western blotting procedure was followed. A polyclonal rabbit anti‐HCPro antibody (at 1 : 15 000 dilution) was used for HCPro detection. The antibody was produced in‐house through the immunization of rabbits with purified recombinant proteins expressed in Escherichia coli.
qPCR and RT‐PCR experiments
Total RNA was extracted from 100 mg of leaf tissue using a standard Trizol reagent‐based extraction technique; 1000 ng of RNA from each set were used for DNase treatment (Promega), followed by cDNA preparation using Superscript III following the protocol provided by the manufacturer. Reverse primer (5′‐AACTGCCCAAAATTGCAAAC‐3′) and forward primer (5′‐TTTGGGAGGAAGATCTGTGG‐3′) from the RdRP of the PVX genome were used to prepare (+)‐ and (–)‐strand‐specific cDNA, respectively. qPCR was carried out using Maxima SYBR Green qPCR Master mix (Thermo Fisher Scientific) and a CFX96/384 Touch™ Real‐Time PCR Detection System (BioRad, California, USA) to calculate the copy number of the target gene in the individual samples. For quantification of the PVA RNA level, primers from its CP region [PVA CP forward primer (5′‐CATGCCCAGGTATGGTCTTC‐3′) and PVA CP reverse primer (5′‐ATCGGAGTGGTTGCAGTGAT‐3′)] were used to amplify that segment from the total cDNA. The protein phosphatase 2A (PP2A) gene [forward primer (5′‐GACCCTGATGTTGATGTTCG‐3′) and reverse primer (5′‐GAGGGATTTGAAGAGAGATTTC‐3′)] and F‐box gene [forward primer (5′‐GGCACTCACAAACGTCTATTTC‐3′); reverse primer (5′‐TGGGAGGCATCCTGCTTAT‐3′)] were used as housekeeping gene controls (Lõhmus et al., 2017). The standard curve for PVX copy number calculation was obtained using the known dilution of pGR107 carrying the PVX‐RLUC insert.
GSH derivatization for LC‐MS studies
A method for GSH quantification was developed. Auto‐oxidation is a major challenge in GSH quantification, resulting in its highly artefactual underestimation. In order to counter this issue, the thiol alkylating agent N‐ethylmaleimide (NEM) was used to derivatize GSH to produce stable GS‐NEM, which was subsequently detected via LC‐MS. Sample preparation was performed essentially as described by Giustarini et al. (2013) with certain modifications. Six leaf discs (5 mm in diameter each) were collected in a tube, frozen immediately in liquid nitrogen and stored at −80 °C until the point of sample preparation. The discs were crushed in liquid nitrogen; 200 μL of Tris‐NEM buffer (50 mm Tris buffer, 31 mm NEM) were added immediately to the frozen tissue and mixed thoroughly to ensure adequate contact between sample and buffer. As the tissues thawed in the buffer, the reaction mix was shortly vortexed again and incubated at room temperature for 2 min. Subsequently 33 μL of 60% Trichloroacetic acid (TCA) were added to the reaction mixture and centrifuged at 15700 g for 2 min. The supernatant was collected and used for LC‐MS directly or stored at −80 °C until analysis. Each set was analysed with at least three biological replicates and, for internal standardization purposes, each alkylation reaction was supplemented with an equal amount of GSH stable isotope [glutathione (glycine‐13C2,15N) disodium salt, Toronto Research Chemicals (Canada)]. GSH quantification was carried out using the regression equation of a standard curve obtained by plotting the acquired signals against the known concentrations of GS‐NEM.
LC‐MS methodology for NEM‐GSH quantification
Samples were injected into an Acquity UPLC system (Waters, Manchester, UK), equipped with an Acquity UPLC® HSS T3 1.8 µm, LC Column 50 × 2.1 mm. The UPLC was operated with a flow rate of 0.3 mL/min in gradient mode at 40 °C. Solvents used in the gradient were A (0.1% formic acid in water) and B [0.1% formic acid in acetonitrile–isopropanol (1 : 1)]. The initial conditions of the linear gradient were A (100%) and B (0%); the conditions were changed to A (70%) and B (30%) for 6 min and to A (0%) and B (100%) for the following 2 min, after which the initial conditions were re‐imposed for 3 min. The injection volume was 1.0 μL. Mass spectra were recorded with a Waters Synapt G2‐Si mass spectrometer (Waters). Measurements were performed using negative electrospray ionization (ESI) in resolution mode. Ions were scanned in the range 50–2000 m/z. MS analyses were performed with scan times of 0.1 s. The following conditions were used: capillary voltage, 2.0 kV; source temperature, 120 °C; sampling cone, 40.0; source offset, 80.0; desolvation temperature, 600 °C; cone gas flow, 100 L/h; desolvation gas flow, 1000 L/h; nebulizer gas flow, 6.5 bar. Leucine–encephalin was used as a lock mass and calibration was performed with sodium formate and Ultramark 1621.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Mixed infection by Potato virus X (PVX) and Potato virus A (PVA) induces a strong necrosis response and retards the growth of Nicotiana benthamiana plants. Top row: leaves of mock plants, PVA‐infected plants, PVX‐infected plants and PVA–PVX co‐infected plants from above at 14 days post‐infiltration (dpi). Bottom row: retarded growth caused by PVA–PVX co‐infection. When compared with mock plants, single infections by PVA and PVX also appeared to reduce growth slightly.
Fig. S2 Robustness of the method. (A) Mean of mock and Potato virus X‐green fluorescent protein (PVX‐GFP) detected from all samples under excitation/emission (Ex/Em) parameters for red fluorescent protein (RFP) measurement (n = 96). (B) Mean of mock and Potato virus A‐red fluorescent protein (PVA‐RFP) detected from all samples under Ex/Em parameters for GFP measurement (n = 96). PVA‐RFP (C) and PVX‐GFP (E) are shown to demonstrate how the actual signal appears side‐by‐side against the virtually insignificant measurement artefacts which may be incorporated into the results as a result of possible signal leakage. (D, F) Representative evidence for linear correlation between RFP fluorescence/PVA‐RFP RNA copy number and GFP fluorescence/PVX‐GFP RNA copy number, respectively.
Fig. S3 Validation of the silencing of methionine cycle components and HCPro (helper component proteinase) overexpression throughout the course of the infection experiment. (A–C) Knockdown of the indicated genes at 3, 5 and 9 days post‐infiltration (dpi). Semi‐quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) (25 cycles) was used to amplify the genes of interest. A less intense band in the agarose gel demonstrates that silencing of the corresponding mRNAs had occurred. (D) HCPro RNA level in the overexpression sets on the third and ninth days, and a western blot of a 5‐dpi sample probed with α‐HCPro antibody to confirm the overexpression of HCPro. GUS, β‐glucuronidase; HEN1, Hua enhancer 1; SAMS, S‐adenosyl‐l‐methionine synthetase; SAHH, S‐adenosyl‐l‐homocysteine hydrolase.
Acknowledgements
We thank Säde Virkki for growing the plants for this study, and Professors David Baulcombe and Lesley Torrance for the generous gift of PVX clones, and CSIRO Plant Industry, Australia for the pHELLSGATE 12 silencing vector. This work was supported by the Jane and Aatos Erkko Foundation and the Academy of Finland grant (1298254) to K.M. S.D. was supported by the Erasmus Mundus Action 2 program BRAVE and G.C‐C. by the Centre for International Mobility (KM‐14–9111).
References
- Aguilar, E. , Almendral, D. , Allende, L. , Pacheco, R. , Chung, B.N. , Canto, T. and Tenllado, F. (2015) The P25 protein of potato virus X (PVX) is the main pathogenicity determinant responsible for systemic necrosis in PVX‐associated synergisms. J Virol. 89, 2090–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alazem, M. and Lin, N.‐S. (2015) Roles of plant hormones in the regulation of host–virus interactions. Mol. Plant Pathol. 16, 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andika, I.B. , Kondo, H. , Nishiguchi, M. and Tamada, T. (2012) The cysteine‐rich proteins of beet necrotic yellow vein virus and tobacco rattle virus contribute to efficient suppression of silencing in roots. J. Gen. Virol. 93, 1841–1850. [DOI] [PubMed] [Google Scholar]
- Ball, L. , Accotto, G.‐P. , Bechtold, U. , Creissen, G. , Funck, D. , Jimenez, A. , Kular, B. , Leyland, N. , Mejia‐Carranza, J. , Reynolds, H. , Karpinski, S. and Mullineaux, P.M. (2004) Evidence for a direct link between glutathione biosynthesis and stress defense gene expression in Arabidopsis. Plant Cell, 16, 2448–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañizares, M.C. , Lozano‐Durán, R. , Canto, T. , Bejarano, E.R. , Bisaro, D.M. , Navas‐Castillo, J. and Moriones, E. (2013) Effects of the crinivirus coat protein–interacting plant protein SAHH on post‐transcriptional RNA silencing and its suppression. Mol. Plant–Microbe Interact. 26, 1004–1015. [DOI] [PubMed] [Google Scholar]
- Chávez‐Calvillo, G. , Contreras‐Paredes, C.A. , Mora‐Macias, J. , Noa‐Carrazana, J.C. , Serrano‐Rubio, A.A. , Dinkova, T.D. , Carrillo‐Tripp, M. and Silva‐Rosales, L. (2016) Antagonism or synergism between papaya ringspot virus and papaya mosaic virus in Carica papaya is determined by their order of infection. Virology, 489, 179–191. [DOI] [PubMed] [Google Scholar]
- Chen, I.H. , Chiu, M.‐H. , Cheng, S.‐F. , Hsu, Y.‐H. and Tsai, C.‐H. (2013) The glutathione transferase of Nicotiana benthamiana NbGSTU4 plays a role in regulating the early replication of Bamboo mosaic virus. New Phytol. 199, 749–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney‐Martin, G. , Rafii, M. , Wykes, L.J. , Ball, R.O. and Pencharz, P.B. (2008) Methionine‐adequate cysteine‐free diet does not limit erythrocyte glutathione synthesis in young healthy adult men. J. Nutr. 138, 2172–2178. [DOI] [PubMed] [Google Scholar]
- Dedic, P. (1975) The effect of Potato virus A (PVA) on yield in some potato varieties. Ochr. Rostl. 11, 127–133. [Google Scholar]
- Del Toro, F.J. , Donaire, L. , Aguilar, E. , Chung, B.N. , Tenllado, F. and Canto, T. (2017) Potato virus Y HCPro suppression of antiviral silencing in Nicotiana benthamiana plants correlates with its ability to bind in vivo to 21‐ and 22‐nucleotide small RNAs of viral sequence. J. Virol. 91, e00367–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon, T. , Chiera, J.M. , Lindbo, J.A. and Finer, J.J. (2009) Quantitative evaluation of six different viral suppressors of silencing using image analysis of transient GFP expression. Plant Cell Rep. 28, 639–647. [DOI] [PubMed] [Google Scholar]
- Eskelin, K. , Suntio, T. , Hyvärinen, S. , Hafren, A. and Mäkinen, K. (2010) Renilla luciferase‐based quantitation of Potato virus A infection initiated with Agrobacterium infiltration of N. benthamiana leaves. J. Virol. Methods, 164, 101–110. [DOI] [PubMed] [Google Scholar]
- Eskelin, K. , Hafrén, A. , Rantalainen, K.I. and Mäkinen, K. (2011) Potyviral VPg enhances viral RNA translation and inhibits reporter mRNA translation in planta. J. Virol. 85, 9210–9221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García‐Marcos, A. , Pacheco, R. , Martiáñez, J. , González‐Jara, P. , Díaz‐Ruíz, J.R. and Tenllado, F. (2009) Transcriptional changes and oxidative stress associated with the synergistic interaction between Potato virus X and Potato virus Y and their relationship with symptom expression. Mol. Plant–Microbe Interact. 22, 1431–1444. [DOI] [PubMed] [Google Scholar]
- Garcia‐Ruiz, H. , Carbonell, A. , Hoyer, J.S. , Fahlgren, N. , Gilbert, K.B. , Takeda, A. , Giampetruzzi, A. , Garcia Ruiz, M.T. , McGinn, M.G. , Lowery, N. , Martinez Baladejo, M.T. , Carrington, J.C. and Ding, B. (2015) Roles and programming of Arabidopsis ARGONAUTE proteins during Turnip mosaic virus infection. PLoS Pathog. 11, e1004755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giustarini, D. , Dalle‐Donne, I. , Milzani, A. , Fanti, P. and Rossi, R. (2013) Analysis of GSH and GSSG after derivatization with N‐ethylmaleimide. Nat. Protoc, 8, 1660–1669. [DOI] [PubMed] [Google Scholar]
- González‐Jara, P. , Tenllado, F. , Martinez‐Garcia, B. , Atencio, F.A. , Barajas, D. , Vargas, M. , Diaz‐Ruiz, J. and Diaz‐Ruiz, J.R. (2004) Host‐dependent differences during synergistic infection by Potyviruses with Potato virus X . Mol. Plant Pathol. 5, 29–35. [DOI] [PubMed] [Google Scholar]
- González‐Jara, P. , Atencio, F.A. , Martínez‐García, B. , Barajas, D. , Tenllado, F. and Díaz‐Ruíz, J.R. (2005) A single amino acid mutation in the plum pox virus helper component‐proteinase gene abolishes both synergistic and RNA silencing suppression activities. Phytopathology, 98, 894–901. [DOI] [PubMed] [Google Scholar]
- Gullner, G. , Tóbiás, I. , Fodor, J. and Kömives, T. (1999) Elevation of glutathione level and activation of glutathione‐related enzymes affect virus infection in tobacco. Free Radic Res. 31, S155–S161. [DOI] [PubMed] [Google Scholar]
- Hafrén, A. , Lõhmus, A. and Mäkinen, K. (2015) Formation of Potato virus A‐induced RNA granules and viral translation are interrelated processes required for optimal virus accumulation. PLoS Pathog. 11, e1005314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hameed, A. , Iqbal, Z. , Asad, S. and Mansoor, S. (2014) Detection of multiple potato viruses in the field suggests synergistic interactions among potato viruses in Pakistan. Plant Pathol. J. 30, 407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höller, K. , Király, L. , Künstler, A. , Müller, M. , Gullner, G. , Fattinger, M. and Zechmann, B. (2010) Enhanced glutathione metabolism is correlated with sulfur‐induced resistance in Tobacco mosaic virus‐infected genetically susceptible Nicotiana tabacum plants. Mol. Plant–Microbe Interact. 23, 1448–1459. [DOI] [PubMed] [Google Scholar]
- Ivanov, K.I. , Puustinen, P. , Gabrenaite, R. , Vihinen, H. , Rönnstrand, L. , Valmu, L. , Kalkkinen, N. and Mäkinen, K. (2003) Phosphorylation of the potyvirus capsid protein by protein kinase CK2 and its relevance for virus infection. Plant Cell, 15, 2124–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, K.I. , Eskelin, K. , Bašić, M. , De, S. , Lõhmus, A. , Varjosalo, M. and Mäkinen, K. (2016) Molecular insights into the function of the viral RNA silencing suppressor HCPro. Plant J. 85, 30–45. [DOI] [PubMed] [Google Scholar]
- Jiménez‐Bremont, J.F. , Marina, M. , Guerrero‐González, M.D.L.L. , Rossi, F.R. , Sánchez‐Rangel, D. , Rodríguez‐Kessler, M. , Ruiz, O.A. and Gárriz, A. (2014) Physiological and molecular implications of plant polyamine metabolism during biotic interactions. Front. Plant Sci. 5, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau, K.D. and Carrington, J.C. (1998) A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell, 95, 461–470. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D. and Carrington, J.C. (2001) Long‐distance movement and replication maintenance functions correlate with silencing suppression activity of potyviral HC‐Pro. Virology, 285, 71–81. [DOI] [PubMed] [Google Scholar]
- Lõhmus, A. , Hafrén, A. and Mäkinen, K. (2017) Coat protein regulation by CK2, CPIP, HSP70, and CHIP is required for Potato virus A replication and coat protein accumulation. J. Virol. 91, e01316‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A.C. , Ely, L. , Smith, T.H. , Marathe, R. , Anandalakshmi, R. , Fagard, M. , Vaucheret, H. , Pruss, G. , Bowman, L. and Vance, V.B. (2001) HC‐Pro suppression of transgene silencing eliminates the small RNAs but not transgene methylation or the mobile signal. Plant Cell, 13, 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marathe, R. , Anandalakshmi, R. , Smith, T.H. , Pruss, G.J. and Vance, V.B. (2000) RNA viruses as inducers, suppressors and targets of post‐transcriptional gene silencing. Plant Mol. Biol. 43, 295–306. [DOI] [PubMed] [Google Scholar]
- Pasin, F. , Kulasekaran, S. , Natale, P. , Simón‐Mateo, C. and García, J.A. (2014) Rapid fluorescent reporter quantification by leaf disc analysis and its application in plant–virus studies. Plant Methods, 10, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss, G. , Ge, X. , Shi, X.M. , Carrington, J.C. and Vance, V.B. (1997) Plant viral synergism: the potyviral genome encodes a broad‐range pathogenicity enhancer that transactivates replication of heterologous viruses. Plant Cell, 9, 859–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, H.A. , Halfhill, M.D. , Millwood, R.J. and Stewart, C.N. (2003) Quantitative GFP fluorescence as an indicator of recombinant protein synthesis in transgenic plants. Plant Cell Rep. 22, 117–121. [DOI] [PubMed] [Google Scholar]
- Ross, A.F. (1950) Local lesion formation and virus production following simultaneous inoculation with Potato virus‐X and virus‐Y . Phytopathology, 40, 24. [Google Scholar]
- Scholthof, K.‐B.G. , Adkins, S. , Czosnek, H. , Palukaitis, P. , Jacquot, E. , Hohn, T. , Hohn, B. , Saunders, K. , Candresse, T. , Ahlquist, P. , Hemenway, C. and Foster, G.D. (2011) Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 12, 938–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senanayake, D.M.J.B. and Mandal, B. (2014) Expression of symptoms, viral coat protein and silencing suppressor gene during mixed infection of a N–Wi strain of Potato virus Y and an asymptomatic strain of Potato virus X . Virus Dis. 25, 314–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, X.M. , Miller, H. , Verchot, J. , Carrington, J.C. and Vance, V.B. (1997) Mutations in the region encoding the central domain of helper component‐proteinase (HC‐Pro) eliminate Potato virus X/potyviral synergism. Virology, 231, 35–42. [DOI] [PubMed] [Google Scholar]
- Stephan, D. , Slabber, C. , George, G. , Ninov, V. , Francis, K.P. and Burger, J.T. (2011) Visualization of plant viral suppressor silencing activity in intact leaf lamina by quantitative fluorescent imaging. Plant Methods, 7, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syller, J. (2012) Facilitative and antagonistic interactions between plant viruses in mixed infections. Mol. Plant Pathol. 13, 204–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari, Y. and Zamore, P.D. (2005) Perspective: machines for RNAi. Genes Dev. 19, 517–529. [DOI] [PubMed] [Google Scholar]
- Untiveros, M. , Fuentes, S. and Salazar, L.F. (2007) Synergistic interaction of sweet potato chlorotic stunt virus (Crinivirus) with Carla‐, Cucumo‐, Ipomo‐, and potyviruses infecting sweet potato. Plant Dis. 91, 669–676. [DOI] [PubMed] [Google Scholar]
- Vance, V.B. (1991) Replication of potato virus X RNA is altered in coinfections with Potato virus Y . Virology, 182, 486–494. [DOI] [PubMed] [Google Scholar]
- Vance, V.B. , Berger, P.H. , Carrington, J.C. , Hunt, A.G. and Shi, X.M. (1995) 5' proximal potyviral sequences mediate Potato virus X/potyviral synergistic disease in transgenic tobacco. Virology, 206, 583–590. [DOI] [PubMed] [Google Scholar]
- Voinnet, O. (2001) RNA silencing as a plant immune system against viruses. Trends Genet. 17, 449–459. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Turina, M. , Medina, V. and Falk, B.W. (2009) Synergistic interaction between the Potyvirus, Turnip mosaic virus and the Crinivirus, Lettuce infectious yellows virus in plants and protoplasts. Virus Res. 144, 163–170. [DOI] [PubMed] [Google Scholar]
- Wilson, L.G. , Reuveny, Z. and Varner, J.E. (1976) Chapter: 19 – Sulfate reduction A2 In: Plant Biochemistry, 3rd edn ( Bonner, J. , ed.), pp. 599–632. San Diego, CA: Academic Press. [Google Scholar]
- Yu, B. , Yang, Z. , Li, J. , Minakhina, S. , Yang, M , Padgett, R.W , Steward, R. and Chen, X. (2005) Methylation as a crucial step in plant microRNA biogenesis. Science (New York, NY), 307, 932–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechmann, B. , Zellnig, G. and Müller, M. (2007a) Virus‐induced changes in the subcellular distribution of glutathione precursors in Cucurbita pepo (L.). Plant Biol. 9, 427–434. [DOI] [PubMed] [Google Scholar]
- Zechmann, B. , Zellnig, G. , Urbanek‐Krajnc, A. and Müller, M. (2007b) Artificial elevation of glutathione affects symptom development in ZYMV‐infected Cucurbita pepo L. plants. Arch. Virol. 152, 747–762. [DOI] [PubMed] [Google Scholar]
- Zechmann, B. , Müller, M. and Zellnig, G. (2008) Modified levels of cysteine affect glutathione metabolism in plant cells In: Sulfur Assimilation and Abiotic Stress in Plants (Khan D.N.A., Singh D.S. and Umar D.S., eds.), pp. 193–206. Berlin, Heidelberg: Springer. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Mixed infection by Potato virus X (PVX) and Potato virus A (PVA) induces a strong necrosis response and retards the growth of Nicotiana benthamiana plants. Top row: leaves of mock plants, PVA‐infected plants, PVX‐infected plants and PVA–PVX co‐infected plants from above at 14 days post‐infiltration (dpi). Bottom row: retarded growth caused by PVA–PVX co‐infection. When compared with mock plants, single infections by PVA and PVX also appeared to reduce growth slightly.
Fig. S2 Robustness of the method. (A) Mean of mock and Potato virus X‐green fluorescent protein (PVX‐GFP) detected from all samples under excitation/emission (Ex/Em) parameters for red fluorescent protein (RFP) measurement (n = 96). (B) Mean of mock and Potato virus A‐red fluorescent protein (PVA‐RFP) detected from all samples under Ex/Em parameters for GFP measurement (n = 96). PVA‐RFP (C) and PVX‐GFP (E) are shown to demonstrate how the actual signal appears side‐by‐side against the virtually insignificant measurement artefacts which may be incorporated into the results as a result of possible signal leakage. (D, F) Representative evidence for linear correlation between RFP fluorescence/PVA‐RFP RNA copy number and GFP fluorescence/PVX‐GFP RNA copy number, respectively.
Fig. S3 Validation of the silencing of methionine cycle components and HCPro (helper component proteinase) overexpression throughout the course of the infection experiment. (A–C) Knockdown of the indicated genes at 3, 5 and 9 days post‐infiltration (dpi). Semi‐quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) (25 cycles) was used to amplify the genes of interest. A less intense band in the agarose gel demonstrates that silencing of the corresponding mRNAs had occurred. (D) HCPro RNA level in the overexpression sets on the third and ninth days, and a western blot of a 5‐dpi sample probed with α‐HCPro antibody to confirm the overexpression of HCPro. GUS, β‐glucuronidase; HEN1, Hua enhancer 1; SAMS, S‐adenosyl‐l‐methionine synthetase; SAHH, S‐adenosyl‐l‐homocysteine hydrolase.


