Summary
The plant membrane‐localized NADPH oxidases, also known as respiratory burst oxidase homologues (RBOHs), play crucial roles in various cellular activities, including plant disease responses, and are a major source of reactive oxygen species (ROS). Sclerotinia sclerotiorum is a cosmopolitan fungal pathogen that causes Sclerotinia stem rot (SSR) in soybean. Via a key virulence factor, oxalic acid, it induces programmed cell death (PCD) in the host plant, a process that is reliant on ROS generation. In this study, using protein sequence similarity searches, we identified 17 soybean RBOHs (GmRBOHs) and studied their contribution to SSR disease development, drought tolerance and nodulation. We clustered the soybean RBOH genes into six groups of orthologues based on phylogenetic analysis with their Arabidopsis counterparts. Transcript analysis of all 17 GmRBOHs revealed that, of the six identified groups, group VI (GmRBOH‐VI) was specifically and drastically induced following S. sclerotiorum challenge. Virus‐induced gene silencing (VIGS) of GmRBOH‐VI using Bean pod mottle virus (BPMV) resulted in enhanced resistance to S. sclerotiorum and markedly reduced ROS levels during disease development. Coincidently, GmRBOH‐VI‐silenced plants were also found to be drought tolerant, but showed a reduced capacity to form nodules. Our results indicate that the pathogenic development of S. sclerotiorum in soybean requires the active participation of specific host RBOHs, to induce ROS and cell death, thus leading to the establishment of disease.
Keywords: drought, NADPH oxidases, ROS, Sclerotinia sclerotiorum, Sclerotinia stem rot, soybean
Introduction
Plants continuously produce reactive oxygen species (ROS) as byproducts of different metabolic pathways, such as respiration and photosynthesis. In turn, these small molecules are constantly scavenged by the redox machinery of the cell. Therefore, a steady state is maintained under normal physiological conditions (Alscher et al., 1997; Apel and Hirt, 2004; Tripathy and Oelmüller, 2012). ROS can be toxic to various cell components, affecting proteins, lipids and nucleic acids, when levels reach a certain threshold (Sharma et al., 2012). Thus, many studies have focused on the detrimental effect of ROS. However, increasing evidence suggests a more intricate role for these molecules that may function upstream or downstream of various signalling events (Baxter et al., 2014). ROS can serve as secondary messengers as part of both inter‐ and intracellular signalling, regulating key cellular processes (Mittler et al., 2011). In biotic stress responses, the regulation of the cellular redox state is now an important area of research, because of the strong correlation between ROS signalling and stress responses (Apel and Hirt, 2004; Marino et al., 2012). The hypersensitive response (HR), a form of programmed cell death (PCD), is perhaps one of the most studied forms of resistance responses mounted by plant tissues against invading pathogens. This response is accompanied by the release of superoxide anion ( ) and hydrogen peroxide (H2O2) at the site of pathogen challenge, which is required for pathogen arrest and incompatibility. Although the timing and magnitude may differ, ROS are also produced during compatible interactions, contributing to successful infections by some pathogens (Gilbert and Wolpert, 2013; Kabbage et al., 2013; Williams et al., 2011). Overall, it is clear that ROS play an important role in stress responses, and contribute to the outcome of many plant–microbe interactions.
One of the major sources of ROS in plants are plasma membrane‐bound NADPH oxidases. They catalyse the conversion of O2 to , which is further converted into other ROS, such as hydroxyl radicals and H2O2 (Sagi and Fluhr, 2001). NADPH oxidases, also known as respiratory burst oxidase homologues (RBOHs), in the plant and animal kingdoms possess cytosolic FAD‐ and NADPH‐binding domains at their C‐terminal region, and six conserved transmembrane helices. The third and fifth helices, via key histidine residues, support two haem groups that are required for electron transfer across the plasma membrane (Lambeth, 2004; Sagi and Fluhr, 2006). The N‐terminal region contains variable numbers of calcium‐binding EF‐hand motifs and phosphorylation target sites that are important for their activity (Glyan'ko and Ischenko, 2010; Kimura et al., 2012; Kobayashi et al., 2007; Oda et al., 2010).
RBOHs have been identified in various plant species, including tomato, tobacco, Arabidopsis, Medicago truncatula, common bean, rice and maize (Arthikala et al., 2014; Li et al., 2015; Marino et al., 2011; Simon‐Plas et al., 2002; Wang et al., 2013). In Arabidopsis, they form a multigenic family comprising 10 genes (AtRBOHA–AtRBOHJ), and their activities have been implicated in various physiological events, including response to stress (Torres and Dangl, 2005). AtRBOHD, the most highly expressed Arabidopsis RBOH, mediates many processes, such as pathogen response, stomatal closure and systemic signalling in response to both abiotic and biotic stresses (Kwak et al., 2003; Miller et al., 2009; Torres et al., 2002). AtRBOHD is also regulated by both Ca2+‐dependent and Ca2+‐independent pathways during immune responses (Dubiella et al., 2013; Kadota et al., 2014, 2015). AtRBOHF has been shown to participate in abscisic acid (ABA) signal transduction and plays a key role in the interplay between intracellular oxidative stress and immune response to pathogens (Chaouch et al., 2012; Kwak et al., 2003; Marino et al., 2012), and has been implicated in non‐host resistance to Magnaporthe oryzae in Arabidopsis (Nozaki et al., 2013). AtRBOHD and AtRBOHF are considered to be the main Arabidopsis isoforms associated with responses to pathogens. Other studies have noted the involvement of Arabidopsis RBOHs in developmental processes. AtRBOHC has been shown to regulate root hair formation (Foreman et al., 2003), whereas AtRBOHB is essential for seed ripening and germination (Müller et al., 2009). AtRBOHH and AtRBOHJ modulate pollen tube growth and seed development (Kaya et al., 2014; Lassig et al., 2014). Interestingly, a role for these proteins was also noted in connection with mutualistic interactions. In the model legume Medicago truncatula, MtRBOHA has been shown to be important for nodule functioning; silencing of MtRBOHA decreases nitrogen fixation activity in nodules and the modulation of genes encoding the microsymbiont nitrogenase (Marino et al., 2011). In Phaseolus vulgaris, Arthikala et al. (2014) showed that the overexpression of PvRBOHB, a common bean NADPH oxidase gene, enhances symbiosome number, bacteroid size and nitrogen fixation in nodules. Therefore, several functional studies have placed RBOHs at the centre of ROS network regulation and associated biological processes in cells, thus demonstrating their importance to key metabolic functions in plants, including the pathogen response.
Sclerotinia sclerotiorum is a cosmopolitan fungal pathogen that infects virtually all dicotyledonous plants (Bolton et al., 2006; Kabbage et al., 2015). It has been traditionally viewed as a prototypical necrotroph, but recent findings have suggested that its pathogenic development may involve a brief biotrophic phase (Kabbage et al., 2013, 2015; Williams et al., 2011). Sclerotinia sclerotiorum can cause considerable damage to crop plants and has been proven to be difficult to control, with host resistance being inadequate. In soybean, this fungus causes Sclerotinia stem rot (SSR), also known as white mould disease. SSR can be a significant yield‐limiting disease, and yield losses greater than 10 million bushels (270 million kg) per year are common (Peltier et al., 2012).
Sclerotinia sclerotiorum is a prolific producer of cell wall‐degrading enzymes (CWDEs, e.g. pectinases, cellulases, hemicellulases), which facilitate plant cell wall degradation and host colonization (Amselem et al., 2011). In addition to its lytic repertoire, an important factor governing the pathogenic success of S. sclerotiorum is the secretion of the key virulence factor oxalic acid (OA). Mutants defective in OA production are poorly pathogenic and unable to overcome host defences (Kabbage et al., 2013; Liang et al., 2015; Williams et al., 2011). OA has been shown to contribute to pathogenesis in ways that facilitate the colonization of the host plant, including the inhibition of host defences (Williams et al., 2011), pH‐mediated activation of CWDEs and the inhibition of autophagy (Kabbage et al., 2013). Importantly, OA induces apoptotic‐like PCD, a process that is largely reliant on ROS (Kim et al., 2008). Thus, the regulation of ROS plays a critical role in the pathogenic success of S. sclerotiorum, particularly at the later stages of the infection process, when ROS generation and tissue cell death culminate in the establishment of disease (Williams et al., 2011).
As a result of the importance of RBOHs in ROS generation, we postulate that the up‐regulation of ROS and the ensuing cell death imposed by S. sclerotiorum require host NADPH oxidases in soybean. Using a combination of bioinformatics tools, expression studies and reverse genetic approaches, we show the key requirement of four soybean RBOHs (GmRBOHs), designated GmRBOH‐VI, for SSR development. The silencing of this group resulted in decreased ROS levels, which coincided with enhanced resistance to S. sclerotiorum. Remarkably, these plants were also found to be drought tolerant, but the silencing of GmRBOH‐VI affected root nodulation. Our results indicate that the pathogenic development of S. sclerotiorum in soybean requires the active participation of specific host RBOHs, to induce ROS and cell death, thus leading to the establishment of disease.
Results
Identification of the soybean RBOH family
The Arabidopsis genome contains 10 RBOHs (AtRBOHs) which have been widely studied and characterized (Marino et al., 2012). We conducted blastp searches against the soybean Joint Genome Institute (JGI) Phytozome (Wm82.a2.v1) (https://phytozome.jgi.doe.gov/pz/portal.html) and National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/protein ) databases using Arabidopsis protein sequences as reference queries, and identified 17 soybean RBOHs (GmRBOHs). The identified GmRBOHs were named GmRBOHA–GmRBOHQ (Table 1), depending on the location in the soybean genome and the widely accepted nomenclature (Torres and Dangl, 2005), and varied in size from 820 to 941 amino acids. Protein domain composition was analysed using the SMART alignment tool (http://smart.embl-heidelberg.de/smart/set_mode.cgi?GENOMIC=1), and revealed that all the GmRBOHs have conserved NADPH oxidase, ferric reductase, FAD‐ and NAD‐binding domains (Fig. 1A). They also contain a variable number (0–2) of EF‐hand motifs (Fig. 1A), which are known to play a key role in the calcium‐dependent regulation of RBOHs (Wong et al., 2007). We clustered the soybean RBOH genes into six groups of orthologues based on phylogenetic analysis with their Arabidopsis counterparts (Fig. 1B). AtRBOHs were distributed amongst all groups, except group I (Fig. 1B). The soybean genes GmRBOHD and GmRBOHO belong to group I; GmRBOHN and GmRBOHG belong to group II; GmRBOHA, GmRBOHF, GmRBOHJ and GmRBOHM belong to group III; GmRBOHH belongs to group IV; GmRBOHC, GmRBOHE, GmRBOHG and GmRBOHI belong to group V; and GmRBOHB, GmRBOHL, GmRBOHP and GmRBOHQ belong to group VI. Our analysis predicts an expanded family of at least 17 genes in the soybean genome that encode RBOH proteins, none of which have been examined previously.
Table 1.
Soybean respiratory burst oxidase homologue (GmRBOH) genes.
| Name of gene | Locus ID in JGI Phytozome (Wm82.a2.v1)* | NCBI accession number† | Protein size (predicted, amino acids) | Molecular weight (kDa) |
|---|---|---|---|---|
| GmRBOHA | Glyma.01G222700 | XP_003517484 | 927 | 105.94 |
| GmRBOHB | Glyma.03G236300 | XP_003521697 | 885 | 100.71 |
| GmRBOHC | Glyma.04G203200 | XP_003522455 | 928 | 104.86 |
| GmRBOHD | Glyma.05G021100 | XP_006579505 | 820 | 92.99 |
| GmRBOHE | Glyma.05G198700 | XP_014631288 | 898 | 100.98 |
| GmRBOHF | Glyma.05G212500 | XP_003525369 | 941 | 106.50 |
| GmRBOHG | Glyma.06G162300 | XP_003526909 | 941 | 105.56 |
| GmRBOHH | Glyma.07G130800 | XP_006583585 | 859 | 98.1 |
| GmRBOHI | Glyma.08G005900 | XP_003532261 | 888 | 100.49 |
| GmRBOHJ | Glyma.08G018900 | XP_003532995 | 941 | 106.70 |
| GmRBOHK | Glyma.09G073200 | XP_006587062 | 928 | 105.17 |
| GmRBOHL | Glyma.10G152200 | XP_003536070 | 825 | 93.72 |
| GmRBOHM | Glyma.11G020700 | XP_003538264 | 927 | 105.88 |
| GmRBOHN | Glyma.15G182000 | XP_014622948 | 935 | 105.90 |
| GmRBOHO | Glyma.17G078300 | XP_006600576 | 821 | 93.07 |
| GmRBOHP | Glyma.19G233900 | XP_003554649 | 887 | 101.12 |
| GmRBOHQ | Glyma.20G236200 | XP_003556516 | 889 | 101.23 |
*Joint Genome Institute (JGI) Phytozome (Wm82.a2.v1) (https://phytozome.jgi.doe.gov/pz/portal.html).
†National Center for Biotechnology Information (NCBI) accession number (http://www.ncbi.nlm.nih.gov/protein).
Figure 1.
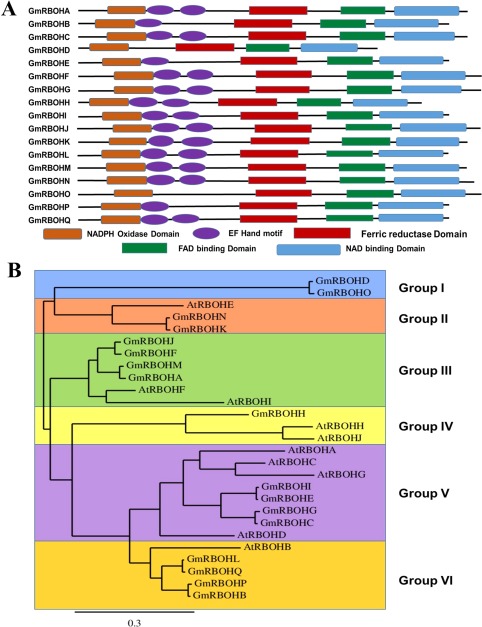
Domain organization and phylogenetic tree of soybean respiratory burst oxidase homologues (GmRBOHs). (A) Domain organization of 17 putative GmRBOHs. The domain organization is based on the SMART alignment tool (http://smart.embl-heidelberg.de/smart/set_mode.cgi?GENOMIC=1). (B) Phylogenetic relationship analysis of 17 GmRBOHs and 10 Arabidopsis RBOHs (AtRBOHs). The phylogenetic tree was constructed using PhyML 3.0 based on the maximum likelihood method (http://www.phylogeny.fr/advanced.cgi). Six groups of GmRBOHs were identified. Branch lengths are proportional to the number of substitutions per site (see scale bars). Only bootstrap values >50% were used to resolve branching.
Spatial expression profile of soybean RBOHs
RBOH genes have been reported to show tissue‐specific expression patterns in plants, including Arabidopsis, tomato and rice (Marino et al., 2011; Sagi and Fluhr, 2006; Wang et al., 2013). For example, AtRBOHA–AtRBOHG and AtRBOHI are expressed in roots, AtRBOHH and AtRBOHJ are pollen specific, whereas AtRBOHD and AtRBOHF are expressed throughout the plant (Sagi and Fluhr, 2006). To determine the tissue‐ and organ‐specific expression patterns of GmRBOHs, total RNA was extracted from roots, stems, flowers and leaves of 4‐week‐old soybean plants. Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) was performed using gene‐specific primers designed for each of the GmRBOHs (Table S1, see Supporting Information), and relative expression levels were calculated using Cons15, a calcium dependent protein kinase (CDPK)‐related protein, as internal control (Libault et al., 2008). Our analysis revealed that GmRBOHA is expressed at low levels in all tissues, whereas GmRBOHE and GmRBOHM are strongly and ubiquitously expressed throughout the plant (Fig. 2). GmRBOHB and GmRBOHL are specifically expressed in roots, whereas GmRBOHK and GmRBOHN appear to be mostly expressed in stems and roots. No flower‐ or leaf‐specific expression was detected, and the remainder of the GmRBOHs did not show any obvious organ‐specific expression (Fig. 2). In accordance with the results reported previously in other plant species, a variable expression pattern of GmRBOHs was detected depending on the tissue tested. The biological significance of such expression profiles requires further investigation.
Figure 2.
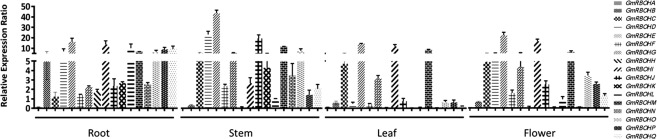
Expression profile of soybean respiratory burst oxidase homologue genes (GmRBOHs) in different tissues. The mRNA transcript levels of all 17 GmRBOHs were determined by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) in the root, stem, leaf and flower tissues. GmCons15 was used as an internal control. All experiments were performed with three independent biological repeats. Error bars represent the standard error (SE; n = 3).
Group VI GmRBOHs were specifically induced following S. sclerotiorum challenge
ROS regulation plays a key role in the pathogenic development of S. sclerotiorum. One of the major sources of ROS in plants are plasma membrane‐bound NADPH oxidases. Accordingly, we examined the expression pattern of GmRBOHs following S. sclerotiorum challenge in a time course experiment at 6, 12, 24, 48, 72 and 96 h post‐inoculation (hpi). Non‐infected stem tissue served as a control. Four‐week‐old soybean plants, ‘Williams 82’, were inoculated using the cut petiole inoculation technique (Peltier et al., 2009), in which an agar plug containing actively growing mycelia of S. sclerotiorum is inserted at the base of a cut petiole. This inoculation method is designed to mimic field conditions, in which fungal hyphae progress from germinating ascospores on the flower to the main stem of the soybean plant to cause typical SSR symptoms. Disease symptoms first appeared at 48 hpi; by 96 hpi, significant cell death could be seen on the inoculated stem (Fig. 3A). Our expression analysis of all 17 GmRBOHs (Fig. S1, see Supporting Information) revealed that, of the six groups of GmRBOHs (Fig. 1B), group VI (GmRBOH‐VI) was specifically and drastically induced during the time course (Fig. 3B). Although GmRBOHB transcript abundance increased by more than 20‐fold as early as 6 hpi, peak expression of all four members of this group coincided with the later stages of infection (48–96 hpi) and the development of disease symptoms (Fig. 3A). GmRBOHL (100‐fold increase) and GmRBOHP (50‐fold increase) were the most highly expressed at 96 hpi compared with uninfected controls. The expression of other GmRBOH members was either unaffected or down‐regulated during the same time course (Fig. S1). Our results suggest that GmRBOH‐VI members may be required by S. sclerotiorum for successful host colonization and SSR disease development.
Figure 3.
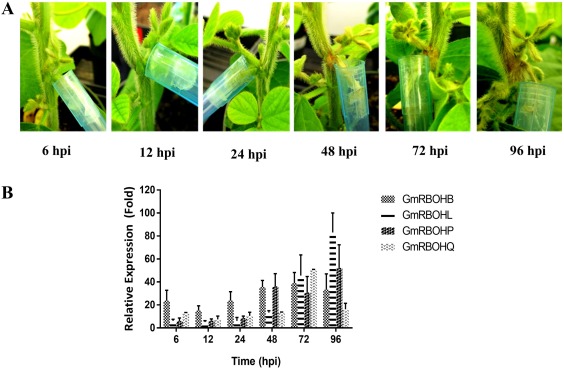
Disease progression and expression profiles of GmRBOH‐VI following infection with Sclerotinia sclerotiorum. (A) Disease symptoms observed following petiole inoculation with an agar plug containing actively growing mycelia of S. sclerotiorum at 6, 12, 24, 48, 72 and 96 h post‐inoculation (hpi). (B) RNAs isolated from non‐infected and infected soybean stems were employed to analyse the expression of GmRBOH‐VI using quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). The relative expression values were calculated by comparing the expression values of genes in inoculated vs. non‐inoculated soybean stem tissues using the 2−ΔΔ Ct method. GmCons15 was used as an endogenous control. Data are presented as means ± standard error (SE) from three independent experiments.
OA is considered to be a key pathogenicity factor for S. sclerotiorum. Via OA secretion, this fungus can provoke an increase in ROS levels within the host, leading to apoptotic‐like cell death and disease development (Kim et al., 2008; Williams et al., 2011). OA‐deficient mutants are unable to up‐regulate host ROS levels and are largely non‐pathogenic (Kabbage et al., 2013; Liang et al., 2015; Williams et al., 2011). Accordingly, we questioned whether the previously studied OA‐deficient mutant strain (A2) could alter the expression profile of GmRBOH‐VI in a similar manner to the wild‐type strain. We examined the expression pattern of GmRBOH‐VI following A2 challenge using the same time course as described for the wild‐type strain (Fig. 4A). Expression analysis revealed that this OA‐deficient mutant was unable to induce the expression of GmRBOH‐VI to wild‐type levels, and the contrast between the two strains was particularly evident in the later stages of the infection process (48–96 hpi, Fig. 4B). Thus, our results suggest that, in the absence of OA, S. sclerotiorum is unable to induce the expression of host RBOHs, to increase ROS levels or to trigger the cell death that is required for disease establishment.
Figure 4.
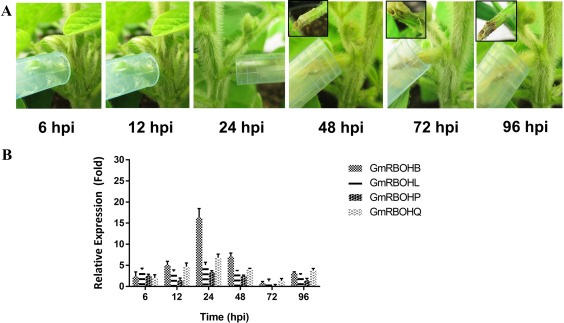
Disease progression and expression profile of GmRBOH‐VI following inoculation with an oxalic acid (OA)‐deficient (A2) strain of Sclerotinia sclerotiorum. (A) Disease symptoms observed at 6, 12, 24, 48, 72 and 96 h post‐inoculation (hpi) following A2 inoculation. (B) RNAs isolated from non‐infected and infected soybean stems were employed to analyse the expression of GmRBOH‐VI using quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). The relative expression values were calculated by comparing the expression value of genes in inoculated vs. non‐inoculated soybean stem tissues using the 2−ΔΔ Ct method. GmCons15 was used as an endogenous control. Data are presented as means ± standard error (SE) from three independent experiments.
Silencing of GmRBOH‐VI leads to enhanced resistance to S. sclerotiorum in an ROS‐dependent manner
Our expression analysis showed that soybean RBOH‐VI expression is significantly induced during the pathogenic development of S. sclerotiorum. We propose that these host genes may be required by the fungus for successful tissue colonization. Virus‐induced gene silencing (VIGS) using Bean pod mottle virus (BPMV) (Zhang et al., 2010, 2013) was employed to knock down the expression of GmRBOH‐VI. This BPMV VIGS system was originally developed using the soybean variety Williams 82 because of its susceptibility to this virus. However, BPMV‐infected Williams 82 plants showed strong resistance to S. sclerotiorum, making this variety unsuitable for our VIGS studies. We screened a large pool of soybean varieties and identified the variety Traff, which shows better tolerance to BPMV, but maintains a predictable response to S. sclerotiorum (data not shown). To evaluate the efficacy of our VIGS system in Traff, we silenced the soybean phytoene desaturase (GmPDS), a gene involved in carotenoid biosynthesis, and obtained consistent photobleaching of the host plants (Fig. S2, see Supporting Information).
As a result of the strong sequence similarities among RBOH‐VI group members, we were unable to silence these genes individually, despite numerous attempts. Thus, a single BPMV silencing construct (pBPMV‐GmRBOH‐VI) was designed to target all four members. The silencing efficiency of pBPMV‐GmRBOH‐VI was determined in Traff by qRT‐PCR and compared with an empty vector control (pBPMV‐0). The expression of the target genes (GmRBOHB, GmRBOHL, GmRBOHP and GmRBOHQ) was decreased significantly, and we were able to achieve a 45%–65% reduction in transcript levels compared with the expression of these genes in the empty vector control (Fig. 5A). GmRBOH‐VI‐silenced soybean plants were then evaluated for their response to S. sclerotiorum challenge; three biological replicates with eight plants each were used. The cut petiole inoculation method was employed as described previously. Five days following S. sclerotiorum inoculation, BPMV‐0 soybean plants showed typical SSR symptoms and began to wilt. In contrast, GmRBOH‐VI‐silenced plants did not show any wilting symptoms (Fig. 5). In GmRBOH‐VI‐silenced plants, lesion development was arrested shortly after reaching the main stem, and a red/dark discoloration was apparent at the edge of the lesion (Fig. 5B). The lesion length was quantified in both the empty vector control and GmRBOH‐VI‐silenced plants (Fig. 5C). Overall, these results suggest that silencing of GmRBOH‐VI genes leads to enhanced resistance in soybean against S. sclerotiorum infection, and indicate that this pathogen requires their activity to achieve pathogenic success.
Figure 5.
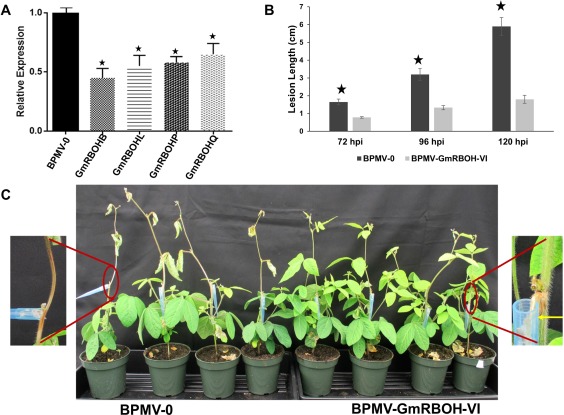
Silencing of GmRBOH‐VI leads to enhanced resistance to Sclerotinia sclerotiorum. (A) Silencing efficiency of GmRBOH‐VI. The first true leaves of 10‐day‐old soybean plants were used for the biolistic delivery of the Bean pod mottle virus (BPMV) constructs BPMV‐0 (empty vector control) and BPMV‐GmRBOH‐VI. The silencing efficiency was calculated by comparing the transcript levels of each GmRBOH‐VI gene in BPMV‐GmRBOH‐VI virus‐induced gene‐silenced plants with the corresponding levels in BPMV‐0‐infected plants. Lesion length (B) and disease symptoms (C) following petiole inoculation with S. sclerotiorum. Lesion lengths were measured from 72 to 120 h post‐inoculation (hpi) (B). At 120 hpi, the control plants were completely wilted in contrast with BPMV‐GmRBOH‐VI‐inoculated plants (C). Eight plants were used for each of the three biological repeats. Data are presented (A, B) as the mean ± standard deviation (SD) from three independent experiments. ⋆, Significant difference at the P < 0.05 level. Yellow arrow shows red discoloration at the edge of the lesion.
RBOHs catalyse the conversion of O2 to , which is further converted into other reactive oxygen molecules, including H2O2. We determined the H2O2 levels in GmRBOH‐VI‐silenced and empty vector control plants challenged with S. sclerotiorum, using the potassium iodide (KI) method, as described previously (Alexieva et al., 2001). Three biological replications and four plants per replication were evaluated in a time course experiment (6, 12, 24, 48, 72 and 96 hpi). Our data indicate that GmRBOH‐VI‐silenced plants produce significantly less H2O2 than empty vector control plants (Fig. 6). In BPMV‐0 control plants, H2O2 production increases in two phases. In the first phase, an increase in H2O2 levels is seen as early as 6 hpi. This is followed by a decrease until 24 hpi, where H2O2 levels once again increase continuously until 96 hpi as disease symptoms develop. At 96 hpi, as much as three times more H2O2 is produced in BPMV‐0 relative to GmRBOH‐VI‐silenced plants (Fig. 6). Overall, our results show that S. sclerotiorum induces ROS levels in soybean as part of its pathogenic development, a process that is reliant on host RBOHs.
Figure 6.
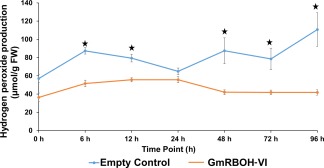
Silencing of GmRBOH‐VI coincides with reduced hydrogen peroxide (H2O2) production. H2O2 was quantified in infected and non‐infected soybean stem tissue using the potassium iodide (KI) spectrophotometric method. The mean and standard error of the mean (SEM) are shown (n = 6) and expressed on the basis of stem fresh weight (FW). ⋆, Significant difference at P < 0.05.
GmRBOH‐VI‐silenced soybean plants are drought tolerant
A role of RBOH genes in response to ROS‐inducing insults has been reported, including in response to drought and salinity treatments (Cheng et al., 2013; Lin et al., 2009; Wang et al., 2013, 2016). Drought is an important yield‐limiting stress in soybean production; therefore, we analysed the effect of GmRBOH‐VI silencing under water stress conditions. GmRBOH‐VI‐silenced plants and BPMV‐0‐inoculated plants were subjected to drought by depriving plants of water for 10 days, after which watering was resumed. After a water deprivation period of 7 days, BPMV‐0‐inoculated plants showed severe wilting symptoms, whereas GmRRBOH‐VI‐silenced plants maintained turgor (Fig. 7B). At 10 days, GmRBOH‐VI‐silenced plants also started to wilt (Fig. 7C). However, after watering was resumed, we observed that GmRBOH‐VI‐silenced plants recovered, whereas BPMV‐0‐inoculated plants did not. These results suggest that knocking down the expression of GmRBOH‐VI leads to increased drought tolerance, possibly by limiting oxidative damage and, ultimately, death of the plant imposed by elevated ROS levels during this stress.
Figure 7.
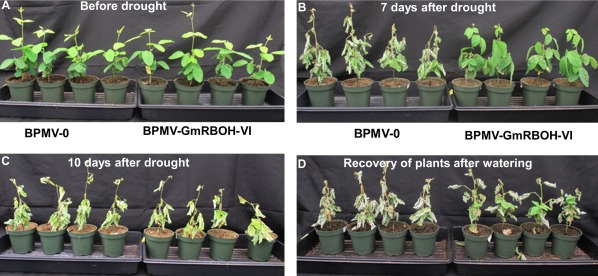
Knocking down the expression of GmRBOH‐VI leads to increased drought tolerance. Plants are shown before drought stress (A), and 7 days (B) and 10 days (C) after water deprivation. (D) Recovery of plants after watering was resumed. In each panel, the BPMV‐0 empty vector plants (left) and GmRBOH‐VI‐silenced plants (right) are shown. Eight plants were used for each of the three biological repeats.
Silencing of GmRBOH‐VI affects soybean nodulation
Previous studies have indicated the role of RBOHs in plant–legume symbioses. Knocking down the expression of MtRBOHA negatively affects nodule formation in Medicago truncatula (Marino et al., 2011), and Arthikala et al. (2014) have shown that the overexpression of PvRBOHB in P. vulgaris enhances symbiosome number, bacteroid size and nitrogen fixation in nodules.
To determine the effect of GmRBOH‐VI silencing on nodulation, we conducted nodulation assays in GmRBOH‐VI‐silenced and BPMV‐0 control plants. Ten‐day‐old soybean plants were inoculated with pBPMV‐GmRBOH‐VI and the control empty vector pBPMV‐0. Control and GmRBOH‐VI‐silenced plants were then inoculated with Bradyrhizobium diazoefficiens USDA110, and nodules were counted at 12 days post‐inoculation. A significant reduction in nodule number (P = 0.04) was observed in GmRBOH‐VI‐silenced plants compared with controls. GmRBOH‐VI‐silenced plants produced, on average, 69 nodules/plant, whereas the control produced 123 nodules/plant (Fig. 8B), representing an approximately 50% reduction in nodule formation. We did not find any differences in the structure or shape of the nodules between the treatments. This result indicates that knocking down the expression of GmRBOH‐VI leads to a significant decrease in soybean nodulation.
Figure 8.
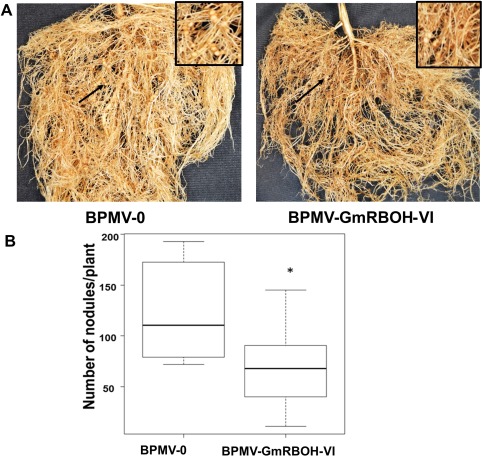
Silencing of GmRBOH‐VI reduces nodulation. (A) Nodule formation in empty vector control (BPMV‐0) and GmRBOH‐VI‐silenced plants (BPMV‐GmRBOH‐VI). (B) Number of nodules per plant. Plants were inoculated with 3 mL of Bradyrhizobium diazoefficiens USDA110 at an optical density of 0.15. Eighteen days after B. diazoefficiens inoculation, the number of nodules on each plant was counted manually. Nineteen plants for each treatment were used for the nodulation study, ∗Significant difference at P < 0.05.
Transient overexpression of GmRBOH‐VI in Nicotiana benthamiana leads to increased susceptibility to S. sclerotiorum
Considering our enhanced resistance phenotype observed in GmRBOH‐VI‐silenced soybean in response to S. sclerotiorum, we reasoned that the overexpression of these genes might facilitate fungal growth and colonization. Transient assays are difficult to perform in soybean, and so we opted to use N. benthamiana leaves to perform transient overexpression. Human influenza haemagglutinin (HA)‐tagged GmRRBOH‐VI was cloned into an Agrobacterium compatible vector downstream of a 35S promoter, and bacterial cells were infiltrated into N. benthamiana leaves. The presence of RBOH proteins was detected via immunoblots using anti‐HA antibody. At 24 h post‐agroinfiltration, detached leaves of N. benthamiana were challenged with agar plugs containing actively growing mycelia of S. sclerotiorum. The overexpression of GmRBOHB, GmRBOHL, GmRBOHP and GmRBOHQ in N. benthamiana enhanced disease development to varying levels and resulted in an approximately 40%–60% increase in lesion area compared with empty vector control leaves (Fig. 9B,C). These data suggest that the overexpression of GmRRBOH‐VI leads to increased susceptibility to S. sclerotiorum infection in N. benthamiana, and further confirms its positive role in the pathogenic development of S. sclerotiorum.
Figure 9.
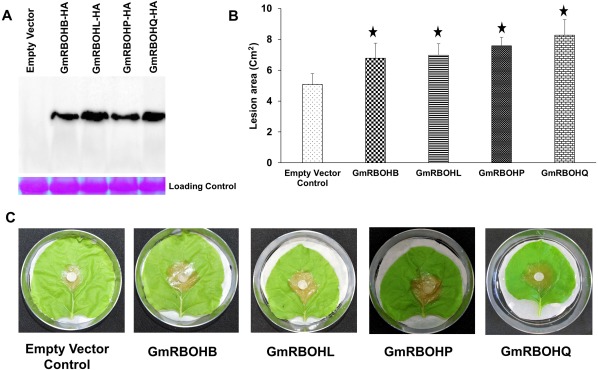
Transient overexpression of GmRBOH‐VI in Nicotiana benthamiana leads to enhanced susceptibility to Sclerotinia sclerotiorum. (A) Detection of GmRBOHB‐HA, GmRBOHL‐HA, GmRBOHP‐HA and GmRBOHQ‐HA from infiltrated N. benthamiana leaves. The pGWB414‐GmRBOHB‐HA, pGWB414‐GmRBOHL‐HA, pGWB414‐GmRBOHP‐HA, pGWB414‐GmRBOHQ‐HA and pGWB414‐eHA (empty vector) constructs were expressed in leaves by Agrobacterium infiltration, and samples were collected at 48 h post‐infiltration. Total soluble protein extracts were prepared and separated using sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and tagged GmRBOH proteins were detected using a haemagglutinin (HA)‐specific antibody. Equal loading of protein samples was confirmed by Ponceau staining. (B) Lesion area. pGWB414‐GmRBOHB‐HA, pGWB414‐GmRBOHL‐HA, pGWB414‐GmRBOHP‐HA, pGWB414‐GmRBOHQ‐HA and pGWB414‐eHA were expressed in N. benthamiana leaves using Agrobacterium. At 24 h post‐infiltration, leaves were detached and challenged with S. sclerotiorum. The lesion diameter was measured at 24 h post‐inoculation (hpi). (C) Lesion development in representative leaves. Mean lesion area ± standard deviation (SD) from three independent experiments was measured; each experiment contained five leaves. ⋆, Significant difference at P < 0.05.
Discussion
The cosmopolitan fungal pathogen S. sclerotiorum can modulate host defences and subvert plant PCD pathways to achieve pathogenic success. Indeed, S. sclerotiorum induces a cell death regime in the host plant which displays apoptotic features (e.g. DNA laddering), and the expression of anti‐apoptotic genes in plants prevents disease development (Kabbage et al., 2013; Kim et al., 2008). This pathogen makes efficient use of a simple dicarboxylic acid, OA, to commandeer a range of host processes, including the elicitation of PCD. It is believed that the timely induction of cell death during host colonization provides nutrients that are for the benefit of the pathogen. Emerging evidence suggests that ROS play a key role in this process (Kim et al., 2008; Williams et al., 2011). ROS are known intermediaries of PCD responses, and function as signalling molecules during pathogen development and pathogen–host interactions (Erental et al., 2008; Torres et al., 2006). We examined the underlying mechanisms of ROS generation in soybean (Glycine max) in response to S. sclerotiorum by identifying the soybean RBOH (GmRBOH) family and by characterizing its role in this pathogenic system. This study was prompted by previous observations indicating that one of the major sources of ROS in plants under pathogen attack is plasma membrane‐bound RBOH proteins, and that host redox regulation is important to S. sclerotiorum pathogenicity (Williams et al., 2011). Several lines of evidence are consistent with the following conclusions: (i) a group of GmRBOH (GmRBOH‐VI) genes is specifically induced following S. sclerotiorum challenge in soybean; (ii) GmRBOH‐VI induction may be reliant on the presence of the fungal secreted OA in the infection court, as OA‐deficient mutants are unable to induce GmRBOH expression and are non‐pathogenic; (iii) the silencing of GmRBOH‐VI leads to enhanced resistance to S. sclerotiorum and other ROS‐inducing insults; and (iv) GmRBOH‐VI silencing and disease resistance coincide with a marked decrease in ROS levels in the host plant. Therefore, S. sclerotiorum appears to co‐opt the soybean ROS machinery to its benefit by modulating the expression of host RBOHs. These genes provide a potential target for the generation of SSR‐resistant soybean lines.
Several studies have demonstrated the role of ROS production in plant immunity and other plant processes, including abiotic stress responses, growth and development. RBOHs play a key role in ROS generation, and different RBOHs may control different plant processes, as reported previously (Kadota et al., 2015; Torres and Dangl, 2005). In plant immunity, ROS are proposed to function as antimicrobial molecules, in plant cell wall reinforcement and as secondary messengers to activate additional defence responses. The implication of host RBOHs is well documented in defence responses, including HR‐PCD and pathogen‐associated molecular pattern (PAMP)‐triggered defences following pathogen recognition. In Arabidopsis, the two principal isoforms associated with the pathogen response are AtRBOHD and AtRBOHF. AtRBOHD affects many processes, including lignification, cell death control, stomatal closure, and systemic signalling in response to both abiotic and biotic stresses (Kwak et al., 2003; Miller et al., 2009; Torres et al., 2002). AtRBOHD is also regulated by both Ca2+‐dependent and Ca2+‐independent pathways during immune responses (Dubiella et al., 2013; Kadota et al., 2014, 2015). AtRBOHF and AtRBOHD have been shown to have redundant functions, as many of the observed phenotypes are enhanced in the Atrbohd and Atrbohf double mutants (Chaouch et al., 2012; Kwak et al., 2003; Marino et al., 2012). Although significant progress has been made in our understanding of RBOH function in response to pathogens, many of these studies have largely focused on biotrophic or hemibiotrophic pathogens.
Although NADPH oxidase activity and ROS production typically correlate with successful disease resistance responses against invading biotrophic pathogens, ROS may be advantageous to pathogens with predominantly necrotrophic lifestyles, such as S. sclerotiorum, which require dead host tissue. As stated above, PCD is essential for S. sclerotiorum pathogenicity, a process that requires ROS generation. Our results show that a group of soybean RBOH genes (GmRBOH‐VI) is specifically induced following S. sclerotiorum challenge, with peak expression at the later stages of the infection process. Silencing of GmRBOH‐VI leads to markedly decreased ROS production and enhanced resistance to this pathogen. Thus, S. sclerotiorum may induce ROS production to its advantage by increasing RBOH activity. In accordance, necrotrophs have been proposed to stimulate ROS production in host tissue to induce cell death and to facilitate infection (Marino et al., 2012). This was further supported by results in Arabidopsis showing that ROS levels correlate positively with the growth of Botrytis cinerea, a close relative of S. sclerotiorum, but negatively with the growth of the hemibiotrophic pathogen Pseudomonas syringae (Govrin and Levine, 2000). Increased resistance to another necrotrophic fungus, Alternaria brassicicola, was also observed in rbohD mutants in Arabidopsis (Pogány et al., 2009). Surprisingly, the silencing of RBOHB (SlRBOHB) in tomato led to increased susceptibility to B. cinerea, and its overexpression in N. benthamiana enhanced resistance to the same necrotrophic pathogen (Li et al., 2015). Although it is difficult to explain these contradictory results, it is, however, conceivable that similar pathogens may trigger different responses in a particular host. For example, S. sclerotiorum and B. cinerea are taxonomically closely related pathogens, but important differences in developmental and pathogenic features have been noted (Amselem et al., 2011). One of these differences is OA production, the requirment of which differs for the two pathogens depending on the host (Stefanato et al., 2008; Xue et al., 2015). Thus, such disparities may provoke different host responses. Alternatively, the involvement of different RBOH genes, and the timing of RBOH activity and ROS generation, may also be key to the outcome of a given host–microbe interaction. It should also be noted that RBOH activity is regulated by complex signalling events involving Ca2+‐based regulation, pattern recognition receptor (PRR) complexes and Rac GTPase (Kadota et al., 2015). Therefore, despite the common mechanism by which ROS are produced, RBOHs are at the crossroads of a complex network of signals, thus explaining the variable outcomes observed in different situations.
How S. sclerotiorum co‐opts the host's ROS/RBOH machinery is an important question. It is reasonable to speculate that the key pathogenicity factor OA plays a role in this interaction. In this study, we have shown that GmRBOH‐VI induction requires OA in the infection court, and that OA‐deficient mutants are unable to up‐regulate GmRBOH‐VI expression and are non‐pathogenic. We have also noted that the lack of GmRBOH‐VI transcript induction may be a result of the inability of the fungus to colonize host tissues. However, OA has been shown to have opposing functions, including the dampening of ROS in the initial stages of host colonization, but later promoting ROS production (Williams et al., 2011). Using a redox‐sensitive GFP system, Williams et al. (2011) showed that OA induces a reducing environment at the onset of infection to impede host defences, but, once the infection is initiated, an oxidative state persists leading to PCD of host tissue. Our results suggest that the later surge of ROS may be caused by the up‐regulation of RBOH activity in the host by S. sclerotiorum and that the timing of this activity and ROS production appear to be key to the pathogenic success of S. sclerotiorum. It is currently unclear whether the initial reductive state imposed by OA involves the dampening of RBOH gene expression. Our results show that the expression of other GmRBOHs is decreased during disease development. However, this down‐regulation occurs at the later stages of the infection process.
The involvement of RBOH genes in abiotic stress responses is well documented (Cheng et al., 2013; Lin et al., 2009; Wang et al., 2013, 2016). Drought, in particular, is an important yield‐limiting stress in soybean production. Soybean plants are most affected by drought during the reproductive growth phase, causing flower abortion, lower pod number and reduced seed per pod. We considered the effect of the silencing of GmRBOH‐VI on drought tolerance in soybean. Remarkably, the silencing of these genes delayed wilting and cell death imposed by water stress. Once watering was resumed, silenced plants were able to recover more quickly than control plants following prolonged exposure to drought conditions. During water deprivation, plant cell homeostasis is affected, causing elevated levels of ROS, a process that is probably mediated by RBOHs. High levels of ROS induce oxidative damage and, ultimately, death of the plant. The silencing of GmRBOH‐VI markedly reduced ROS levels and delayed cell death associated with water stress. Under field conditions, this could afford the plant valuable time to cope with extreme drought conditions and to improve recovery. However, ROS also act as important signalling molecules that communicate with phytohormone pathways, redox‐sensitive molecules and other ROS‐responsive processes to mediate acclimatization to various abiotic stresses (Bhattacharjee, 2005; Kaur et al., 2014; Marino et al., 2012). This is supported by results in rice (Wang et al., 2016) and tomato (Li et al., 2015), where osrbohA knockout and SlRBOHB‐silenced plants, respectively, were found to be more sensitive to drought stress. We speculate that, under our experimental conditions, silencing of GmRBOH‐VI maintains ROS at sublethal levels without impeding signalling events, thus limiting the accumulation of excessive ROS during prolonged drought stress, which is detrimental to recovery and survivability. It is important to note the expanded RBOH family in soybean, and other members may also be involved in abiotic stress signalling, including drought.
Whilst considering the potential utilization of GmRBOH‐VI‐silenced plants to confer resistance to S. sclerotiorum in soybean, we examined the effect of silencing on nodulation in this legume. A role for RBOH proteins has been reported in the symbiosis between legumes and nitrogen‐fixing rhizobia. In Medicago truncatula, MtRBOHA has been shown to be important for nodule functioning; silencing of MtRBOHA decreases nitrogen fixation activity in nodules (Marino et al., 2011). In P. vulgaris, the over‐expression of PvRBOHB enhances nodule nitrogen‐fixing activity and delays nodule senescence; however, it impedes the colonization of arbuscular mycorrhizal fungi (AMF) (Arthikala et al., 2014). Thus, RBOH genes can both inhibit and stimulate symbiotic interactions. In this study, we quantified nodules in control and GmRBOH‐VI‐silenced plants, and found that a significant reduction in nodule formation occurred in GmRBOH‐VI‐silenced soybean. This suggests that these genes may contribute to the establishment of symbiotic associations between soybean and rhizobia. However, further studies are required to establish whether the decrease in nodule number has a significant impact on the plant's overall nitrogen‐fixing capacity. It will also be interesting to determine whether GmRBOH‐VI silencing has a positive impact on mycorrhization, as observed in common bean (Arthikala et al., 2014). The generation of stable transgenic plants is underway to address these questions and to further assess tolerance to other biotic and abiotic stresses.
Numerous studies have discussed the importance of RBOH family members as adapter molecules orchestrating plant responses to developmental cues, environmental insults and microbes. In the case of S. sclerotiorum, it appears that this fungus can manipulate RBOH signalling to its advantage in soybean. We propose that the targeting of specific GmRBOH genes for silencing may constitute a viable strategy to limit SSR development and confer tolerance to other environmental insults.
Experimental Procedures
Plant material
Two varieties of soybean (Glycine max), Williams 82 and Traff, were used in this study. Traff was used for VIGS assays, whereas the gene expression study was performed on Williams 82. Soybean seedlings and plants were maintained in a growth chamber at 24 °C with a 16‐h light/8‐h dark photoperiod cycle. Fertilization was applied using standard practices.
Identification, domain search and phylogenetic analysis of soybean RBOHs (GmRBOHs)
Arabidopsis RBOH protein sequences were used to perform sequence similarity searches in JGI Phytozome (Wm82.a2.v1) (https://phytozome.jgi.doe.gov/pz/portal.html) (Schmutz et al., 2010) using a stringent cut‐off (E‐value = 0.0). We identified 17 GmRBOHs and searched their protein sequences for conserved domains using the SMART alignment tool (http://smart.embl-heidelberg.de/smart/set_mode.cgi?GENOMIC=1) (Letunic et al., 2015) and PFAM (http://www.sanger.ac.uk/science/tools/pfam) (Finn et al., 2016). The protein sequences of GmRBOH and AtRBOH were used in PhyML 3.0 to construct a maximum likelihood phylogenetic tree (http://www.phylogeny.fr/advanced.cgi) (Dereeper et al., 2008, 2010). Bootstrap values >50% were used to resolve branching.
Construction of BPMV VIGS and overexpression constructs
To make the GmRBOH‐VI silencing construct, the forward primer GmRbohSGVIF (5′‐AAGGGATCCTGCGAGCGATTACTTCGTGCT‐3′) and reverse primer GmRbohSGVIR (5′‐TTGGGTACC CACTCTGGTCACTACTTGCTG‐3′) were used to amplify a 307‐bp fragment. Restriction sites BamHI and KpnI (italic) were added to forward and reverse primers, respectively. An extra nucleotide in the reverse primer, shown in bold type, was added to maintain the viral open reading frame. The amplified fragment was ligated into the DNA‐based BPMV VIGS vector pBPMV‐IA‐D35 (Liu et al., 2011). Biolistic delivery of BPMV constructs was performed as described previously (Zhang et al., 2013). Silencing was monitored using the construct pBPMV‐IA‐PDS‐3R, which targets the soybean phytoene desaturase (PDS), leading to photobleaching of the plants (Zhang et al., 2010). For transient overexpression, GmRBOHB, GmRBOHL, GmRBOHP and GmRBOHQ coding sequences were amplified using their corresponding primers (Table S1) from soybean cDNAs. The coding regions were then cloned into the Gateway™ entry vector pDONR/Zeo (Life Technologies, Carlsbad, CA, USA) to produce pENTR/Zeo:GmRBOHB, pENTR/Zeo:GmRBOHL, pENTR/Zeo:GmRBOHP and pENTR/Zeo:GmRBOHQ by performing the BP clonase reaction following the manufacturer's protocol. pENTR/Zeo:GmRBOHB, pENTR/Zeo:GmRBOHL, pENTR/Zeo:GmRBOHP and pENTR/Zeo:GmRBOHQ were recombined into the binary vector pGWB414 upstream of a human influenza HA tag (Nakagawa et al., 2007), resulting in pGWB414:GmRBOHB–HA, pGWB414:GmRBOHL–HA, pGWB414:GmRBOHP–HA and pGWB414:GmRBOHQ–HA, respectively. The binary plasmids were transferred into the Agrobacterium strain GV3101 for further experiments.
Sclerotinia sclerotiorum infection and drought treatment
Disease assays were performed using the wild‐type isolate of S. sclerotiorum 1980 or the OA‐deficient mutant (A2) derived from this strain (Williams et al., 2011). Strains were grown at room temperature on potato dextrose agar (PDA). Soybean plants were infected with S. sclerotiorum using the cut petiole inoculation method (Hoffman et al., 2002). Actively growing S. sclerotiorum agar plugs were inserted into a cut petiole of the soybean plants using a 1‐mL pipette tip. VIGS plants were challenged with S. sclerotiorum 18 days after BPMV construct inoculation. In drought studies, plants were subjected to water stress over a period of 10 days. Before starting the stress, we ensured that all pots had equal weight, and received equal amounts of soil and water. After 10 days of continuous water stress, watering was resumed to assess the recovery of plants.
Immunoblotting
Total proteins were extracted from N. benthamiana leaves 48 h after agroinfiltration in lysis buffer [3× per fresh weight of tissue, 5% β‐mercaptoethanol, 1× complete protease inhibitor cocktail, 94% of 2× Laemmli buffer (Bio‐Rad, Hercules, CA, USA)]. Extracts were centrifuged at 10000g for 10 min. Supernatant (30 µL) was separated on an 8% sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) gel and transferred to nitrocellulose membrane using a trans‐blot semidry cell (Bio‐Rad) following the manufacturer's protocol. Ponceau staining [0.1% (w/v) Ponceau S in 1% (v/v) acetic acid] was performed to check for efficient protein transfer and equal loading. Skimmed milk powder (5%) was used as a blocking agent. A 1 : 1000 dilution of rabbit anti‐HA antibody (Cell Signaling Technology, Danvers, MA, USA) was used as primary antibody. The goat anti‐rabbit immunoglobulin G (IgG) horseradish peroxidase (HRP)‐linked antibody (Cell Signaling Technology) was used as secondary antibody. The luminescent signal was visualized using Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare Bio‐Sciences, Pittsburgh, PA, USA) and the ChemiDoc™ MP System (Bio‐Rad).
RNA isolation, reverse transcription and gene expression analysis
The internodal region at the infection site was used for RNA isolation, which included both symptomatic and non‐symptomatic tissue. Stem tissues were harvested and immediately frozen in liquid N2. RNA was isolated using Trizol reagent (Ambion Life Technologies, Carlsbad, CA, USA), and then treated with RNase‐free DNaseI (NEB Inc., Ipswich, MA, USA). The RNA was reverse transcribed using the AMV First‐Strand cDNA Synthesis Kit (NEB Inc.) and oligo‐dT primer according to the manufacturer's instructions. The cDNA was used as template for gene expression analysis using qRT‐PCR. qRT‐PCR was performed using a Sensi FAST SYBR® No‐ROX Kit (Bioline USA Inc., Taunton, MA, USA). Each reaction consisted of 5 μL of 2× SensiFAST SYBR No‐ROX Mix, 1 μL of 1 : 10‐fold diluted cDNAs, 0.4 μL of each 10 μm gene‐specific forward primer and reverse primer in a final volume of 10 μL. The primer pairs used for qRT‐PCR are shown in Table S1. qRT‐PCR was performed on a CFX96 real‐time PCR system (Bio‐Rad). The protocol was as follows: 2 min of initial denaturation at 95 °C; the samples were then subjected to cycling parameters of 95 °C for 5 s, 58 °C for 10 s and 72 °C for 20 s (for 40 cycles). The relative expression of the gene was calculated using the 2−ΔΔ Ct method (Livak and Schmittgen, 2001) with soybean GmCon15S as an endogenous control. Three biological repeats were performed for each sample.
H2O2 measurement
H2O2 determination in infected and non‐infected soybean stem tissue was performed using a modified KI method as described previously (Alexieva et al., 2001). In brief, plant tissues were harvested, immediately frozen in liquid N2, ground and stored at −80 ºC until H2O2 quantification. Frozen powder (1.5 g) was directly homogenized with 10 mL of a solution containing 0.1% w/v trichloroacetic acid (TCA) at 4 °C. The homogenized sample was centrifuged at 11,250g for 15 min at 4 °C. The reaction mixture consisted of 0.5 mL of 0.1% TCA, plant tissue extract supernatant, 0.5 mL of 100 mm potassium phosphate buffer and 2 mL of reagent mix (1 m KI w/v in fresh double‐distilled water). Care was taken to protect samples and solutions from light. The reaction was developed for 1 h in darkness and the absorbance was measured at 390 nm. Quantification was calculated using a standard curve prepared with known concentrations of H2O2.
Transient assay in N. benthamiana and symptom quantification
For Agrobacterium‐mediated transient overexpression of candidate genes in N. benthamiana, bacterial cultures (Agrobacterium tumefaciens GV3101) were grown overnight (28 °C, 200 rpm), pelleted by centrifugation and then resuspended in infiltration medium [9 mm MES (2‐(N‐morpholino)ethanesulfonic acid), 10 mm MgSO4, 10 mm MgCl2, pH 5.6, 300 µm acetosyringone]. Cell densities were adjusted to 0.9 (optical density at 600 nm, OD600). Leaves of 4–5‐week‐old N. benthamiana plants were infiltrated using a needleless syringe. Twenty‐four hours post‐agroinfiltration, detached leaves of N. benthamiana were challenged with agar plugs containing actively growing mycelia of S. sclerotiorum. Leaves were photograhed at 24 h post‐challenge, and the lesion area was calculated using the image analysis software ImageJ (Abramoff et al., 2004; Glozer, 2008).
Nodulation assay
Ten‐day‐old soybean seedlings were inoculated with pBPMV‐GmRBOH‐VI and the control empty vector pBPMV‐0 (Kandoth et al., 2013). Twenty‐one days following VIGS construct inoculation, control and GmRBOH‐VI‐silenced plants were inoculated with a 3‐mL culture of Bradyrhizobium diazoefficiens USDA110 at an optical density of 0.15. Whole plants were harvested after 12 days, the roots were cleaned and the number of nodules in each plant was counted manually.
Statistical analysis
All experiments consisted of three independent biological replicates. For statistical analysis, Student's t‐test was performed and P < 0.05 was considered to be significant. For nodulation data analysis, one‐way analysis of variance (ANOVA) was performed and P < 0.05 was considered to be significant.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Expression profile of soybean respiratory burst oxidase homologues (GmRBOHs) following infection with Sclerotinia sclerotiorum. hpi, hours post‐inoculation.
Fig. S2 Leaf phenotypes on the Traff soybean cultivar induced by Bean pod mottle virus (BPMV), GmRBOH‐VI and GmPDS silencing vectors.
Table S1 List of primers used for real‐time polymerase chain reaction (PCR) analysis and overexpression construct.
Acknowledgements
We wish to thank Ryan Kessens for technical assistance. This work was supported by the Wisconsin Soybean Marketing Board (#193446 to MK and DLS), the North Central Soybean Research Program (#187452 to MK and DLS), the United Soybean Board (#1420‐532‐5604 to MK, SAW, JHH) and the National Science Foundation (#1546742 to J‐MA).
References
- Abramoff, M.D. , Magalhães, P.J. and Ram, S.J. (2004) Image processing with ImageJ. Biophoton. Int. 11, 36–42. [Google Scholar]
- Alexieva, V. , Sergiev, I. , Mapelli, S. and Karanov, E. (2001) The effect of drought and ultraviolet radiation on growth and stress markers in pea and wheat. Plant Cell Environ. 24, 1337–1344. [Google Scholar]
- Alscher, R.G. , Donahue, J.L. and Cramer, C.L. (1997) Reactive oxygen species and antioxidants: relationships in green cells. Physiol. Plant. 100, 224–233. [Google Scholar]
- Amselem, J. , Cuomo, C.A. , van Kan, J.A. , Viaud, M. , Benito, E.P. , Couloux, A. , Coutinho, P.M. , de Vries, R.P. , Dyer, P.S. , Fillinger, S. , Fournier, E. , Gout, L. , Hahn, M. , Kohn, L. , Lapalu, N. , Plummer, K.M. , Pradier, J.M. , Quévillon, E. , Sharon, A. , Simon, A. , ten Have, A. , Tudzynski, B. , Tudzynski, P. , Wincker, P. , Andrew, M. , Anthouard, V. , Beever, R.E. , Beffa, R. , Benoit, I. , Bouzid, O. , Brault, B. , Chen, Z. , Choquer, M. , Collémare, J. , Cotton, P. , Danchin, E.G. , Da Silva, C. , Gautier, A. , Giraud, C. , Giraud, T. , Gonzalez, C. , Grossetete, S. , Güldener, U. , Henrissat, B. , Howlett, B.J. , Kodira, C. , Kretschmer, M. , Lappartient, A. , Leroch, M. , Levis, C. , Mauceli, E. , Neuvéglise, C. , Oeser, B. , Pearson, M. , Poulain, J. , Poussereau, N. , Quesneville, H. , Rascle, C. , Schumacher, J. , Ségurens, B. , Sexton, A. , Silva, E. , Sirven, C. , Soanes, D.M. , Talbot, N.J. , Templeton, M. , Yandava, C. , Yarden, O. , Zeng, Q. , Rollins, J.A. , Lebrun, M.H. and Dickman, M. (2011) Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea . PLoS Genet. 7, e1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel, K. and Hirt, H. (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. [DOI] [PubMed] [Google Scholar]
- Arthikala, M.‐K. , Sánchez‐López, R. , Nava, N. , Santana, O. , Cárdenas, L. and Quinto C. (2014) RbohB, a Phaseolus vulgaris NADPH oxidase gene, enhances symbiosome number, bacteroid size, and nitrogen fixation in nodules and impairs mycorrhizal colonization. New Phytol. 202, 886–900. [DOI] [PubMed] [Google Scholar]
- Baxter, A. , Mittler, R. and Suzuki, N. (2014) ROS as key players in plant stress signalling. J. Exp. Bot. 65, 1229–1240. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee, S. (2005) Reactive oxygen species and oxidative burst: roles in stress, senescence and signal transduction in plants. Curr. Sci. 89, 1113–1121. [Google Scholar]
- Bolton, M.D. , Thomma, B.P. and Nelson, B.D. (2006) Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 7, 1–16. [DOI] [PubMed] [Google Scholar]
- Chaouch, S. , Queval, G. and Noctor, G. (2012) AtRbohF is a crucial modulator of defence‐associated metabolism and a key actor in the interplay between intracellular oxidative stress and pathogenesis responses in Arabidopsis. Plant J. 69, 613–627. [DOI] [PubMed] [Google Scholar]
- Cheng, C. , Xu, X. , Gao, M. , Li, J. , Guo, C. , Song, J. and Wang, X. (2013) Genome‐wide analysis of respiratory burst oxidase homologs in grape (Vitis vinifera L.). Int. J. Mol. Sci. 14, 24 169–24 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper, A. , Guignon, V. , Blanc, G. , Audic, S. , Buffet, S. , Chevenet, F. , Dufayard, J.F. , Guindon, S. , Lefort, V. , Lescot, M. , Claverie, J.M. and Gascuel, O. (2008) Phylogeny.fr: robust phylogenetic analysis for the non‐specialist. Nucleic Acids Res. 36, W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper, A. , Audic, S. , Claverie, J.M. and Blanc, G. (2010) BLAST‐EXPLORER helps you build datasets for phylogenetic analysis. BMC Evol. Biol. 10, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubiella, U. , Seybold, H. , Durian, G. , Komander, E. , Lassig, R. , Witte, C.P. , Schulze, W.X. and Romeis, T. (2013) Calcium‐dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. USA, 110, 8744–8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erental, A. , Dickman, M.B. and Yarden, O. (2008) Sclerotial development in Sclerotinia sclerotiorum: awakening molecular analysis of a “Dormant” structure. Fungal Biol. Rev. 22, 6–16. [Google Scholar]
- Finn, R.D. , Coggill, P. , Eberhardt, R.Y. , Eddy, S.R. , Mistry, J. , Mitchell, A.L. , Potter, S.C. , Punta, M. , Qureshi, M. , Sangrador‐Vegas, A. , Salazar, G.A. , Tate, J. and Bateman, A. (2016) The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman, J. , Demidchik, V. , Bothwell, J.H. , Mylona, P. , Miedema, H. , Torres, M.A. , Linstead, P. , Costa, S. , Brownlee, C. , Jones, J.D. , Davies, J.M. and Dolan, L. (2003) Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature, 422, 442–446. [DOI] [PubMed] [Google Scholar]
- Gilbert, B.M. and Wolpert, T.J. (2013) Characterization of the LOV1‐mediated, victorin‐induced, cell‐death response with virus‐induced gene silencing. Mol. Plant–Microbe Interact. 26, 903–917. [DOI] [PubMed] [Google Scholar]
- Glozer, K. (2008) Protocol for Leaf Image Analysis‐Surface Area. Davis, CA: University of California. [Google Scholar]
- Glyan'ko, A.K. and Ischenko, A.A. (2010) Structural and functional characteristics of plant NADPH oxidase: a review. Appl. Biochem. Microbiol. 46, 463–471. [PubMed] [Google Scholar]
- Govrin, E.M. and Levine, A. (2000) The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea . Curr. Biol. 10, 751–757. [DOI] [PubMed] [Google Scholar]
- Hoffman, D.D. , Diers, B.W. , Hartman, G.L. , Nickell, C.D. , Nelson, R.L. , Pedersen, W.L. , Cober, E.R. , Graef, G.L. , Steadman, J.R. , Nelson, B.D. , Del Rio, L.E. , Grau, C.R. , Helms, T. , Anderson, T. , Poysa, V. , Rajcan, I. and Stienstra, W.C. (2002) Selected soybean plant introductions with partial resistance to Sclerotinia sclerotiorum . Plant Dis. 86, 971–980. [DOI] [PubMed] [Google Scholar]
- Kabbage, M. , Williams, B. and Dickman, M.B. (2013) Cell death control: the interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum . PLoS Pathog. 9, e1003287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabbage, M. , Yarden, O. and Dickman, M.B. (2015) Pathogenic attributes of Sclerotinia sclerotiorum: switching from a biotrophic to necrotrophic lifestyle. Plant Sci. 233, 53–60. [DOI] [PubMed] [Google Scholar]
- Kadota, Y. , Sklenar, J. , Derbyshire, P. , Stransfeld, L. , Asai, S. , Ntoukakis, V. , Jones, J.D. , Shirasu, K. , Menke, F. , Jones, A. and Zipfel, C. (2014) Direct regulation of the NADPH oxidase RBOHD by the PRR‐associated kinase BIK1 during plant immunity. Mol. Cell, 54, 43–55. [DOI] [PubMed] [Google Scholar]
- Kadota, Y. , Shirasu, K. and Zipfel, C. (2015) Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol. 56, 1472–1480. [DOI] [PubMed] [Google Scholar]
- Kandoth, P.K. , Heinz, R. , Yeckel, G. , Gross, N.W. , Juvale, P.S. , Hill, J. , Whitham, S.A. , Baum, T.J. and Mitchum, M.G. (2013) A virus‐induced gene silencing method to study soybean cyst nematode parasitism in Glycine max . BMC Res. Notes, 6, 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur, C. , Ghosh, A. , Pareek, A. , Sopory, S.K. and Singla‐Pareek, S.L. (2014) Glyoxalases and stress tolerance in plants. Biochem. Soc. Trans. 42, 485. [DOI] [PubMed] [Google Scholar]
- Kaya, H. , Nakajima, R. , Iwano, M. , Kanaoka, M.M. , Kimura, S. , Takeda, S. , Kawarazaki, T. , Senzaki, E. , Hamamura, Y. , Higashiyama, T. , Takayama, S. , Abe, M. and Kuchitsu, K. (2014) Ca2+‐activated reactive oxygen species production by Arabidopsis RbohH and RbohJ is essential for proper pollen tube tip growth. Plant Cell, 26, 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K.S. , Min, J.Y. and Dickman, M.B. (2008) Oxalic acid Is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Mol. Plant‐Microbe Interact. 21, 605–612. [DOI] [PubMed] [Google Scholar]
- Kimura, S. , Kaya, H. , Kawarazaki, T. , Hiraoka, G. , Senzaki, E. , Michikawa, M. and Kuchitsu, K. (2012) Protein phosphorylation is a prerequisite for the Ca2+‐dependent activation of Arabidopsis NADPH oxidases and may function as a trigger for the positive feedback regulation of Ca2+ and reactive oxygen species. Biochim. Biophys. Acta, 1823, 398–405. [DOI] [PubMed] [Google Scholar]
- Kobayashi, M. , Ohura, I. , Kawakita, K. , Yokota, N. , Fujiwara, M. , Shimamoto, K. , Doke, N. and Yoshioka, H. (2007) Calcium‐dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell, 19, 1065–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak, J.M. , Mori, I.C. , Pei, Z.M. , Leonhardt, N. , Torres, M.A. , Dangl, J.L. , Bloom, R.E. , Bodde, S. , Jones, J.D. and Schroeder, J.I. (2003) NADPH oxidase AtrbohD and AtrbohF genes function in ROS‐dependent ABA signaling in Arabidopsis. EMBO J. 22, 2623–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambeth, J.D. (2004) NOX enzymes and the biology of reactive oxygen. Nat. Rev. Immunol. 4, 181–189. [DOI] [PubMed] [Google Scholar]
- Lassig, R. , Gutermuth, T. , Bey, T.D. , Konrad, K.R. and Romeis, T. (2014) Pollen tube NAD(P)H oxidases act as a speed control to dampen growth rate oscillations during polarized cell growth. Plant J. 78, 94–106. [DOI] [PubMed] [Google Scholar]
- Letunic, I. , Doerks, T. and Bork, P. (2015) SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 43, D257–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Zhang, H. , Tian, L. , Huang, L. , Liu, S. , Li, D , and Song, F. (2015) Tomato SlRbohB, a member of the NADPH oxidase family, is required for disease resistance against Botrytis cinerea and tolerance to drought stress. Front. Plant Sci. 6, 463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, X. , Liberti, D. , Li, M. , Kim, Y.T. , Hutchens, A. , Wilson, R. and Rollins, J.A. (2015) Oxaloacetate acetylhydrolase gene mutants of Sclerotinia sclerotiorum do not accumulate oxalic acid, but do produce limited lesions on host plants. Mol. Plant Pathol. 16, 559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault, M. , Thibivilliers, S. , Bilgin, D.D. , Radwan, O. , Benitez, M. , Clough, S.J. and Stacey G. (2008) Identification of four soybean reference genes for gene expression normalization. Plant Genome, 1, 44–54. [Google Scholar]
- Lin, F. , Zhang, Y. and Jiang, M.Y. (2009) Alternative splicing and differential expression of two transcripts of nicotine adenine dinucleotide phosphate oxidase B gene from Zea mays . J. Integr. Plant Biol. 51, 287–298. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔCT method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Liu, J.‐Z. , Horstman, H.D. , Braun, E. , Graham, M.A. , Zhang, C. , Navarrem, D. , Qiu, W.L. , Lee, Y. , Nettleton, D. , Hill, J.H. and Whitham, S.A. (2011) Soybean homologs of MPK4 negatively regulate defense responses and positively regulate growth and development. Plant Physiol. 157, 1363–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino, D. , Andrio, E. , Danchin, E.G. , Oger, E. , Gucciardo, S. , Lambert, A. , Puppo, A. and Pauly, N. (2011) A Medicago truncatula NADPH oxidase is involved in symbiotic nodule functioning. New Phytol. 189, 580–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino, D. , Dunand, C. , Puppo, A. and Pauly, N. (2012) A burst of plant NADPH oxidases. Trends Plant Sci. 17, 9–15. [DOI] [PubMed] [Google Scholar]
- Miller, G. , Schlauch, K. , Tam, R. , Cortes, D. , Torres, M.A. , Shulaev, V. , Dangl, J.L. and Mittler, R. (2009) The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci. Signal. 2, ra45. [DOI] [PubMed] [Google Scholar]
- Mittler, R. , Vanderauwera, S. , Suzuki, N. , Miller, G. , Tognetti, V.B. , Vandepoele, K. , Gollery, M. , Shulaev, V. and Van Breusegem, F. (2011) ROS signaling: the new wave? Trends Plant Sci. 16, 300–309. [DOI] [PubMed] [Google Scholar]
- Müller, K. , Carstens, A.C. , Linkies, A. , Torres, M.A. and Leubner‐Metzger, G. (2009) The NADPH‐oxidase AtrbohB plays a role in Arabidopsis seed after‐ripening. New Phytol. 184, 885–897. [DOI] [PubMed] [Google Scholar]
- Nakagawa, T. , Suzuki, T. , Murata, S. , Nakamura, S. , Hino, T. , Maeo, K. , Tabata, R. , Kawai, T. , Tanaka, K. , Niwa, Y. , Watanabe, Y. , Nakamura, K. , Kimura, T. and Ishiguro, S. (2007) Improved gateway binary vectors: high‐performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71, 2095–2100. [DOI] [PubMed] [Google Scholar]
- Nozaki, M. , Kita, K. , Kodaira, T. and Ishikawa, A. (2013) AtRbohF contributes to non‐host resistance to Magnaporthe oryzae in Arabidopsis. Biosci. Biotechnol. Biochem. 77, 1323–1325. [DOI] [PubMed] [Google Scholar]
- Oda, T. , Hashimoto, H. , Kuwabara, N. , Akashi, S. , Hayashi, K. , Kojima, C. , Wong, H.L. , Kawasaki, T. , Shimamoto, K. , Sato, M. and Shimizu, T. (2010) Structure of the N‐terminal regulatory domain of a plant NADPH oxidase and its functional implications. J. Biol. Chem. 285, 1435–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier, A.J. , Hatfield, R.D. and Grau, C.R. (2009) Soybean stem lignin concentration relates to resistance to Sclerotinia sclerotiorum . Plant Dis. 93, 149–154. [DOI] [PubMed] [Google Scholar]
- Peltier, A.J. , Bradley, C.A. , Chilvers, M.I. , Malvick, D.K. , Mueller, D.S. , Wise, K.A. and Esker, P.D. (2012) Biology, yield loss and control of Sclerotinia stem rot of soybean. J. Integr. Pest Manag. 3, B1–B7. [Google Scholar]
- Pogány, M. , von Rad, U. , Grün, S. , Dongó, A. , Pintye, A. , Simoneau, P. , Bahnweg, G. , Kiss, L. , Barna, B. and Durner, J. (2009) Dual roles of reactive oxygen species and NADPH oxidase RBOHD in an arabidopsis‐alternaria pathosystem. Plant Physiol. 151, 1459–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi, M. and Fluhr, R. (2001) Superoxide production by plant homologues of the gp91(phox) NADPH oxidase. Modulation of activity by calcium and by tobacco mosaic virus infection. Plant Physiol. 126, 1281–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi, M. and Fluhr, R. (2006) Production of reactive oxygen species by plant NADPH oxidases. Plant Physiol. 141, 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz, J. , Cannon, S.B. , Schlueter, J. , Ma, J. , Mitros, T. , Nelson, W. , Hyten, D.L. , Song, Q. , Thelen, J.J. , Cheng, J. , Xu, D. , Hellsten, U. , May, G.D. , Yu, Y. , Sakurai, T. , Umezawa, T. , Bhattacharyya, M.K. , Sandhu, D. , Valliyodan, B. , Lindquist, E. , Peto, M. , Grant, D. , Shu, S. , Goodstein, D. , Barry, K. , Futrell‐Griggs, M. , Abernathy, B. , Du, J. , Tian, Z. , Zhu, L. , Gill, N. , Joshi, T. , Libault, M. , Sethuraman, A. , Zhang, X.C. , Shinozaki, K. , Nguyen, H.T. , Wing, R.A. , Cregan, P. , Specht, J. , Grimwood, J. , Rokhsar, D. , Stacey, G. , Shoemaker, R.C. and Jackson, S.A. (2010) Genome sequence of the palaeopolyploid soybean. Nature, 463, 178–183. [DOI] [PubMed] [Google Scholar]
- Sharma, P. , Jha, A.B. , Dubey, R.S. and Pessarakli, M. (2012) Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 1–26. [Google Scholar]
- Simon‐Plas, F. , Elmayan, T. and Blein, J.P. (2002) The plasma membrane oxidase NtrbohD is responsible for AOS production in elicited tobacco cells. Plant J. 31, 137–147. [DOI] [PubMed] [Google Scholar]
- Stefanato, L.F. , Abou‐Mansour, E. , van Kan, J. , Métraux, J.P. and Schoonbeek, H.J. (2008). Oxaloacetate acetyl hydrolase is responsible for oxalic acid production in Botrytis cinerea and required for lesion expansion on some, but not on most host plants In: Third Botrytis Genome Workshop, Tenerife, Spain. [Google Scholar]
- Torres, M.A. and Dangl, J.L. (2005) Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr. Opin. Plant Biol. 8, 397–403. [DOI] [PubMed] [Google Scholar]
- Torres, M.A. , Dangl, J.L. and Jones, J.D. (2002) Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. 99, 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathy, B.C. and Oelmüller, R. (2012) Reactive oxygen species generation and signaling in plants. Plant Signal. Behav. 7, 1621–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G.‐F. , Li, W.Q. , Li, W.Y. , Wu, G.L. , Zhou, C.Y. and Chen, K.M. (2013) Characterization of rice NADPH oxidase genes and their expression under various environmental conditions. Int. J. Mol. Sci. 14, 9440–9458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Zhang, M.M. , Wang, Y.J. , Gao, Y.T. , Li, R. , Wang, G.F. , Li, W.Q. , Liu, W.T. and Chen, K.M. (2016) The plasma membrane NADPH oxidase OsRbohA plays a crucial role in developmental regulation and drought‐stress response in rice. Physiol. Plant. 156, 421–443. [DOI] [PubMed] [Google Scholar]
- Williams, B. , Kabbage, M. , Kim, H.J. , Britt, R. and Dickman, M.B. (2011) Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog. 7, e1002107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, H.L. , Pinontoan, R. , Hayashi, K. , Tabata, R. , Yaeno, T. , Hasegawa, K. , Kojima, C. , Yoshioka, H. , Iba, K. , Kawasaki, T. and Shimamoto, K. (2007) Regulation of rice NADPH oxidase by binding of Rac GTPase to its N‐terminal extension. Plant Cell, 19, 4022–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, L. , Xiang, M. , White, D. and Chen, W. (2015) pH dependency of sclerotial development and pathogenicity revealed by using genetically defined oxalate‐minus mutants of Sclerotinia sclerotiorum . Environ. Microbiol. 17, 2896–2909. [DOI] [PubMed] [Google Scholar]
- Zhang, C. , Bradshaw, J.D. , Whitham, S.A. and Hill, J.H. (2010) The development of an efficient multipurpose bean pod mottle virus viral vector set for foreign gene expression and RNA silencing. Plant Physiol. 153, 52–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. , Whitham, S.A. and Hill, J.H. (2013). Virus‐induced gene silencing in soybean and common bean In: Virus‐Induced Gene Silencing: Methods and Protocols (Becker A., ed.),pp. 149–156. Totowa, NJ: Humana Press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Expression profile of soybean respiratory burst oxidase homologues (GmRBOHs) following infection with Sclerotinia sclerotiorum. hpi, hours post‐inoculation.
Fig. S2 Leaf phenotypes on the Traff soybean cultivar induced by Bean pod mottle virus (BPMV), GmRBOH‐VI and GmPDS silencing vectors.
Table S1 List of primers used for real‐time polymerase chain reaction (PCR) analysis and overexpression construct.


