Summary
Xanthomonas oryzae pv. oryzae (Xoo) causes bacterial blight, a serious disease of rice. Xoo secretes a repertoire of cell wall‐degrading enzymes, including cellulases, xylanases and pectinases, to degrade various polysaccharide components of the rice cell wall. A secreted Xoo cellulase, CbsA, is not only a key virulence factor of Xoo, but is also a potent inducer of innate immune responses of rice. In this study, we solved the crystal structure of the catalytic domain of the CbsA protein to a resolution of 1.86 Å. The core structure of CbsA shows a central distorted TIM barrel made up of eight β strands with N‐ and C‐terminal loops enclosing the active site, which is a characteristic structural feature of an exoglucanase. The aspartic acid at the 131st position of CbsA was predicted to be important for catalysis and was therefore mutated to alanine to study its role in the catalysis and biological functions of CbsA. Intriguingly, the D131A CbsA mutant protein displayed the enzymatic activity of a typical endoglucanase. D131A CbsA was as proficient as wild‐type (Wt) CbsA in inducing rice immune responses, but was deficient in virulence‐promoting activity. This indicates that the specific exoglucanase activity of the Wt CbsA protein is required for this protein to promote the growth of Xoo in rice.
Keywords: cellulases, endoglucanase, exoglucanase, rice, Xanthomonas oryzae pv. oryzae
Introduction
In order to gain access to the nutrients inside host cells, an invading plant pathogen must be able to degrade the complex cell wall. Cellulose, hemicellulose and pectin form the major components of the plant cell wall. Cellulose is a linear polysaccharide composed of glucose residues linked to each other by β‐1,4 glycosidic linkages. Although it is chemically a simple molecule, as it possesses only one type of monomeric sugar, it is a complex structural entity (Hon, 1994). Cellulases break down cellulose into simple monosaccharides and/or oligosaccharides. Cellulases are broadly classified into exoglucanases/cellobiohydrolases (CBHs) and endoglucanases, which differ in their architecture, substrate preference and mode of action. Exoglucanases prefer crystalline substrates, have active sites in a closed tunnel and processively remove cellobiose units from the chain ends (Teeri, 1997; Teeri et al., 1998). On the other hand, endoglucanases prefer amorphous regions, have their active sites in an open cleft and make random internal cuts in the polymer (Teeri, 1997).
A number of bacterial plant pathogens belonging to the genera Xanthomonas, Erwinia, Ralstonia, etc., and fungal pathogens of the genera Rhizoctonia, Magnaporthe, etc., secrete cellulases. The major secreted cellulases produced by these plant pathogens have been demonstrated to be required for complete virulence of the organism (Gough et al., 1988; Roberts et al., 1988; Xia et al., 2016). Cellulases are also potent inducers of defence responses in host tissues. For instance, cellulase treatment induces the production of salicylic acid, a defence hormone, and confers resistance to mild mottle virus attack in pepper (Sato et al., 2011).
Xanthomonas oryzae pv. oryzae (Xoo) causes a serious bacterial blight disease of rice. It secretes a variety of cell wall‐degrading enzymes (CWDEs), including an endoglucanase (ClsA), predicted CBH (CbsA), xylanase (XynB), lipase/esterase (LipA), pectin methyl esterase (Pmt) and polygalacturonase (PglA), to degrade the rice cell wall. Purified preparations of CbsA, ClsA and LipA are potent inducers of rice defence responses, such as callose deposition and programmed cell death. Prior treatment of rice leaves with any one of these enzymes provides enhanced resistance against subsequent infection by Xoo (Jha et al., 2007). Mutations in the clsA, cbsA, lipA and xynB genes have been shown to affect virulence on rice, whereas mutations in the pmt and pglA genes have minimal effects on virulence (Jha et al., 2007; Rajeshwari et al., 2005; Tayi et al., 2016b). A mutation in the cbsA gene has a much greater effect on virulence than mutations in any of the other genes, suggesting that the CbsA protein is much more critical for virulence than the other Xoo secreted CWDEs. A correlation has been observed between the tissue specificity of bacterial pathogens of plants and the presence of a functional CbsA‐like protein. In general, functional CbsA‐like proteins are encoded in the genomes of xylem‐dwelling pathogens, such as Xoo, but are either absent from, or are predicted to be non‐functional in, the genomes of bacteria that multiply in the intercellular spaces of the palisade parenchymatous tissues (Ryan et al., 2011). Because of the importance of this protein in the rice–Xoo interaction, we have chosen to solve its crystal structure with the objective of performing structure–function studies.
The crystal structures of many exoglucanases and endoglucanases of both bacterial and fungal origin have been solved. According to the CAZy classification of glycoside hydrolases (GHs), exoglucanases belong to GH families GH5, GH6, GH7, GH9 and GH48 (Cantarel et al., 2009; Henrissat, 1991). The structures of GH6 exoglucanases, five from fungi and one from bacteria, have been solved. These include the Cel6A proteins of Trichoderma reesei, Humicola insolens and Chaetomium thermophyllum, Cel6A and Cel6C of Coprinopsis cineria, and Cel6B of Thermobifida fusca (Liu et al., 2010; Rouvinen et al., 1990; Sandgren et al., 2013; Tamura et al., 2012; Thompson et al., 2012; Varrot et al., 1999a). The structures of all of these exoglucanases revealed a distorted seven‐stranded α/β barrel and showed a tunnel enclosing the active site which allows for the processive style of activity shown by CBHs. The GH6 family of exoglucanases catalyses reactions via a general acid–base mechanism. In these enzymes, the catalytic acid has been identified to be an aspartic acid, but the existence of a single residue functioning as a catalytic base is unclear. It is now well accepted that, in GH6 family exoglucanases, a proton‐transferring network, involving an aspartate, a serine and two water molecules, performs the function of a catalytic base via a Grotthuss mechanism (Thompson et al., 2012; Vuong and Wilson, 2009).
In this study, we have solved the crystal structure of the catalytic domain of CbsA. The structural features and biochemical activities shown by CbsA confirm that it is an exoglucanase as predicted. We performed biochemical and functional characterization of a mutant CbsA protein in which the active site aspartic acid at the 131st position, which is predicted to function as a catalytic base, is converted to alanine. Interestingly, we found that this mutation (D131A) converted wild‐type (Wt) CbsA, a cellulase which is an exoglucanase by structure, into an endoglucanase by activity. This gain of function by the mutant protein did not affect its ability to induce rice defence responses. However, it did not support the virulence‐promoting activity of CbsA, thus indicating the essentiality of a specific exoglucanase activity in the pathogenesis of Xoo.
Results
Domain architecture of CbsA
Bioinformatics analysis of the sequence of the CbsA protein (∼56 kDa) revealed the presence of an N‐terminal signal peptide of 32 amino acids, followed by a catalytic domain containing 425 amino acids with predicted exoglucanase activity and a C‐terminal fibronectin type 3 domain of 109 amino acids. The catalytic domain belongs to an α/β protein fold, whereas the C‐terminal domain belongs to a fibronectin fold. Based on the CAZy classification, CbsA could be classified into the GH6 family of enzymes. The secreted form of CbsA is ∼45 kDa and has only the catalytic domain without the C‐terminal fibronectin domain. The crystallization and preliminary crystallographic studies of the catalytic domain of 425 residues have been reported previously (Kumar et al., 2012).
Overall structure of CbsA
The crystal structure of the catalytic domain of CbsA (PDB ID: 5XYH) was solved to a resolution of 1.86 Å with R work/R free (%) values of 18.89/23.77 by the molecular replacement method using the catalytic domain of a CBH Cel6B (E3) from T. fusca (PDB ID: 4B4H) as the search model. The refinement statistics are presented in Table 1. The core structure of the catalytic domain of CbsA shows a central distorted β‐barrel made up of eight strands. The structure shows two distinct loops: an N‐terminal (Y125 to Q145) loop and a C‐terminal (P406 to L428) loop, which form a tunnel enclosing the active site (Fig. 1A,B). Four conserved tryptophan residues were found to line the active site pocket of CbsA (Fig. S1, see Supporting Information). These tryptophans lining the active site pocket help to anchor the substrate, possibly by virtue of a π–π interaction with the sugar rings (Koivula et al., 1996; Varrot et al., 1999b). The structure also reveals the presence of two additional loops, P278–S291 and G335–G355, specific to bacterial exoglucanases, which are not found in any of the fungal CBHs (Fig. 2A).
Table 1.
Data collection and refinement statistics
| PDB ID | 5XYH |
|---|---|
| Data collection | |
| Diffraction source | Rigaku MicroMax‐007 HF Cu |
| Wavelength (Å) | 1.5418 |
| Space group | P212121 |
| Cell parameters (Å) | a = 46.14, b = 90.72, c = 99.78 |
| Resolution range (Å) * | 25–1.86 (1.93–1.86) |
| Overall B factor from Wilson plot (Å2) | 17.3 |
| Total observations | 225 473 |
| Unique reflections | 33 848 (3169) |
| Completeness (%) | 93.9 (89.4) |
| Redundancy | 6.7 (5.6) |
| <I/σI> | 31.69 (7.75) |
| R merge (%) | 4.9 (18.3) |
| Refinement statistics | |
| Resolution (Å) | 1.86 |
| No. of reflections | 33 638 |
| R work/R free (%) | 18.89/23.77 |
| No. of residues | 424 |
| No. of atoms | |
| Protein | 3212 |
| Water | 474 |
| B‐factors (Å2) | |
| Protein | 16.15 |
| Water | 19.36 |
| Root‐mean‐square deviations | |
| Bond lengths (Å) | 0.014 |
| Bond angles (deg) | 0.87 |
| Ramachandran plot | |
| Most favoured region (%) | 97.16 |
| Additionally allowed region (%) | 2.84 |
| Disallowed region (%) | 0 |
*Values in parentheses are for the highest resolution shell.
Figure 1.
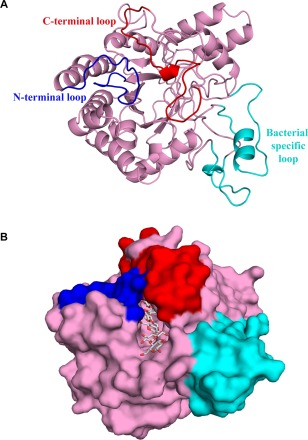
Crystal structure of the catalytic domain of CbsA. (A) CbsA has a central distorted β‐barrel (pink) made up of eight strands. The N‐ and C‐terminal loops are shown in blue and red, respectively, whereas the unique loops present towards the C‐terminal region are shown in cyan. (B) The CbsA structure with the modelled substrate. The surface representation of the enzyme represents a well‐defined substrate‐binding tunnel responsible for processive hydrolysis of the single‐chain cellulose polymer. Three molecules of cellobiose were modelled from PDB ID: 4B4F.
Figure 2.
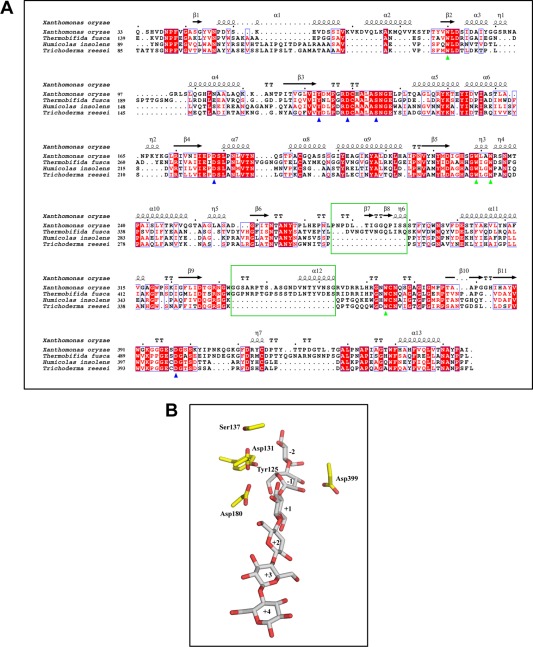
Multiple sequence alignment of the CbsA catalytic domain and putative catalytic residues. (A) Multiple sequence alignment of the CbsA catalytic domain with homologous regions from other bacteria and fungi. Residues marked with a green triangle are conserved tryptophans lining the tunnel and blue triangles indicate the putative catalytic residues. The sequences of the bacterial‐specific loops are enclosed in green boxes. (B) Putative catalytic residues present in close proximity to the modelled substrate. The subsites of the substrate‐binding tunnel are numbered from the non‐reducing end to the reducing end as −2, −1, +1, +2, +3 and +4. Residues around the active site are shown with sticks.
Active site of CbsA
In CbsA, the presence of loops results in the formation of an active site tunnel spanning a length of ∼47.7 Å and classifies CbsA as an exoglucanase, wherein its putative catalytic activity could be to perform processive hydrolysis of crystalline substrates. In order to obtain a better understanding of the active site tunnel and to decipher the putative catalytic residues, the crystal structure of CbsA was superimposed on that of CBH Cel6B (E3) from T. fusca complexed with cellobiose with a root‐mean‐square deviation (rmsd) of 0.959 Å (PDB ID 4B4F). Subsequently, this non‐hydrolysable moiety was modelled on the CbsA structure (Fig. 2B). In Cel6B of T. fusca, the D274 residue functions as a catalytic acid, whereas the D226 and S232 residues were found to be part of a water network which performs the role of a catalytic base (Vuong and Wilson, 2009). In the apo structure of Xoo CbsA, the D180, D131 and S137 residues are analogous to the D274, D226 and S232 residues of T. fusca, and are similarly positioned in the active site tunnel, suggesting identical roles for these residues in catalysis. Apart from the above‐mentioned residues which are involved directly in acid–base catalysis, two other amino acids, which were highly conserved in all CBHs and were demonstrated to play a role in catalysis, are Y125 and D399 of Xoo CbsA (Fig. 2B). In T. reesei, the Y169 residue (the Y125 analogue) is postulated to play a role in the distortion of the glucose entering the tunnel and its conversion into a more active conformation. In addition, it has been proposed to play a role in ensuring the protonation states of acidic aspartates D175 and D221 (residues analogous to D131 and D180 of Xoo CbsA, respectively) which are part of the tunnel (Koivula et al., 1996). Previously, the D405 residue of H. insolens exoglucanase Cel6A (analogous to D399 of Xoo CbsA) has been proposed to function as a potential catalytic base (Varrot et al., 1999b). However, the role of this residue as a catalytic base is questionable on the basis of its position and environment. This particular aspartate of the active site forms a conserved salt bridge with arginine and suggests a structure‐stabilizing function to this residue (Koivula et al., 2002; Sandgren et al., 2013). Both Y125 and D399 of Xoo CbsA were found to occupy identical positions and possess similar interactions with other amino acids as identified in T. fusca Cel6B.
D131A CbsA displays an endo mode of activity
We initiated structure–function studies of the CbsA protein by making mutations that are expected to be critical for biochemical activity. As indicated above, the D131 residue is expected to be part of a water network that functions as a catalytic base. We made a D131A mutant of the CbsA protein and assessed the effect of the mutation on protein function. The activity of purified D131A CbsA was tested on the polysaccharide substrate carboxymethylcellulose (CMC) and on the soluble oligosaccharides cellobiose, cellotriose, cellotetraose, cellopentaose and cellohexaose. CMC is considered to be a hallmark substrate for endoglucanases because of its amorphous nature. In a CMC plate assay, Xoo ClsA (endoglucanase) and D131A CbsA showed a much larger halo than Wt CbsA, suggesting that CMC is cleaved better by D131A CbsA than by Wt CbsA (Fig. 3A). The specific activities of Wt CbsA and D131A CbsA on CMC were also calculated by measuring the reducing sugars released from CMC on enzyme activity (Table S1, see Supporting Information). The hydrolysed products of CMC obtained by the activity of Wt CbsA, D131A CbsA and ClsA were analysed by thin layer chromatography (TLC) (Fig. 3B). Wt CbsA showed very limited activity on CMC, releasing cellobiose and cellotriose in minor quantities, indicating an exo mode of activity (Fig. 3B, band 2). ClsA displayed an endo mode of activity by producing a smear emerging from the loading spot on the TLC sheet (indicative of the release of oligosaccharides of various lengths by making random cuts) (Fig. 3B, band 3). On the other hand, D131A CbsA, unlike Wt CbsA, but very similar to ClsA, released a range of oligosaccharides from CMC, as evident from the smear on the TLC sheet, suggesting random internal cleavages being made by this enzyme, similar to a typical endoglucanase (Fig. 3B, band 4). This endo mode of activity shown by D131A CbsA was further confirmed by viscometric analysis (Fig. 3C). Enzymes which make internal cuts in the substrate are expected to decrease its viscosity much more rapidly than enzymes which act from the ends of substrate molecules. ClsA reduced the viscosity of the CMC solution drastically. Wt CbsA did not decrease the viscosity of the CMC solution, even on prolonged incubation, whereas D131A CbsA decreased the viscosity at a much faster rate than Wt CbsA, but not as efficiently as ClsA (Fig. 3C). The activity of D131A CbsA on soluble oligosaccharides is also identical to ClsA (Fig. 3D). Under the conditions tested, the substrate cellobiose (G2) was not acted upon by any of the three enzymes Wt CbsA, D131A CbsA or ClsA. Wt CbsA could not act on cellotriose (G3), whereas D131A CbsA and ClsA released cellobiose (G2) from this substrate. Wt CbsA converts cellotetraose (G4) to cellobiose (G2), whereas D131A CbsA and ClsA release cellobiose (G2) and cellotriose (G3) from this substrate. Cellopentaose (G5) and cellohexaose (G6) are converted by Wt CbsA, D131A CbsA and ClsA to cellobiose (G2) and cellotriose (G3) (Fig. 3D).
Figure 3.
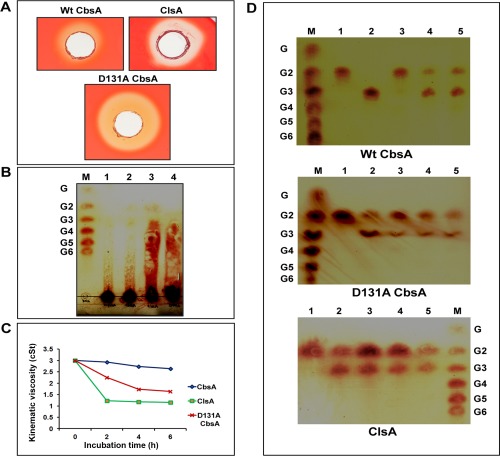
D131A CbsA displays an endo mode of activity. (A) Equal amounts of purified preparations of wild‐type (Wt) CbsA, D131A CbsA and ClsA proteins were added to wells in carboxymethylcellulose (CMC) plates and stained with Congo red after 36–48 h of incubation, as described in Experimental procedures. The presence of a halo around the well indicates the activity of the enzyme on CMC. (B) Separation of hydrolysed products by thin layer chromatography (TLC). The CMC substrate was incubated with buffer, Wt CbsA, D131A CbsA and ClsA, and the complete reaction mixture was spotted on a TLC sheet in the following lanes: 1, CMC + buffer; 2, CMC + Wt CbsA; 3, CMC + ClsA; 4, CMC + D131A CbsA. A mix of the oligosaccharides glucose to cellohexaose (G–G6) was loaded as a ladder in the lane labelled as M. Sugars present in the reaction mixture were separated using butanol, acetic acid and water in a 2 : 1 : 1 ratio as the mobile phase, and were detected using orcinol and sulfuric acid reagent, as described in Experimental procedures. (C) Viscometric analysis of CMC solution on activity of Wt CbsA, D131A CbsA and ClsA. The flow time of the reaction mixture containing CMC together with buffer, Wt CbsA, D131A CbsA and ClsA was measured at regular intervals using a viscosity bath maintained at a constant temperature of 37 οC. As described in Experimental procedures, the kinematic viscosity of the samples was calculated and plotted against the incubation time. (D) Activity of Wt CbsA, D131A CbsA and ClsA on soluble oligosaccharides. The oligosaccharide substrates cellobiose G2 (1), cellotriose G3 (2), cellotetraose G4 (3), cellopentaose G5 (4) and cellohexaose G6 (5) were incubated with Wt CbsA, D131A CbsA and ClsA, and the complete reaction mixture was loaded onto the TLC sheet. The sugars released were separated and detected as described in Experimental procedures. A mix of oligosaccharides (G–G6) is shown in the lane labelled as M.
D131A mutant CbsA does not support the virulence of Xoo on rice
The CbsA protein is an important virulence factor for Xoo. A cbsA– mutant of Xoo is virulence deficient (Jha et al., 2007). We wanted to determine whether the change in activity from the exo mode to the endo mode for D131A CbsA can still support the in planta virulence function of CbsA. Rice leaves were inoculated with Wt Xoo (BXO43), the cbsA– mutant, the cbsA– mutant expressing Wt CbsA and the cbsA– mutant expressing D131A CbsA. The cbsA– mutant strain expressing Wt CbsA (complemented strain) partially complemented the virulence deficiency of the mutant, whereas the cbsA– mutant strain expressing D131A CbsA exhibited severe virulence deficiency (Fig. 4A). The lesions caused by the cbsA– mutant strain expressing D131A were significantly shorter in comparison with those of the cbsA– mutant strain expressing Wt CbsA, as well as the cbsA– mutant strain itself (Fig. 4B). Overall, these results suggest that the altered activity caused by the D131A mutation affects the ability of this protein to promote Xoo virulence.
Figure 4.
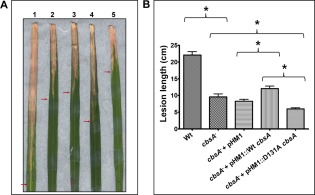
Virulence phenotype of D131A mutant CbsA. (A) Rice leaves were inoculated with wild‐type (Wt) Xanthomonas oryzae pv. oryzae (Xoo) (BXO43) (1), the cbsA– mutant (2), the cbsA– mutant + pHM1 (3), the cbsA– mutant + pHM1::Wt cbsA (4) and the cbsA– mutant + pHM1::D131A cbsA (5). At 20 days post‐inoculation, the images of infected rice leaves were captured. (B) Lesion lengths were measured after 20 days. Error bars indicate the standard deviation of readings from at least 10 inoculated leaves. Similar results were obtained in independent experiments. A Student's two‐tailed t‐test for independent means was performed for the following groups: wild‐type and cbsA mutant, mutant with empty vector (control strain), mutant expressing Wt CbsA (complemented strain) and mutant expressing D131A CbsA. *All the compared values were significantly different at the P < 0.05 level.
D131A CbsA is proficient in the induction of immune responses in rice
Purified preparations of Xoo‐secreted CWDEs, such as LipA, CbsA and ClsA, have been shown to be elicitors of immune responses, e.g. callose deposition and programmed cell death, in rice tissues (Jha et al., 2007). Callose deposition is a hallmark defence response shown by the host, wherein β‐1,3‐glucan, a polymeric substance, is deposited at the site of pathogen entry as a way of strengthening the cell wall. Programmed cell death is a host defence response in which infected cells undergo localized cell death to prevent further spread of the pathogen. We investigated whether D131A CbsA is compromised in its ability to induce plant defence responses. Infiltration of D131A CbsA into rice leaves induced callose deposits as efficiently as Wt CbsA (Fig. 5A). Similarly, treatment of rice roots with D131A CbsA induced programmed cell death as efficiently as Wt CbsA, as shown by extensive propidium iodide (PI) internalization and dispersal within the cells (Fig. 5B).
Figure 5.
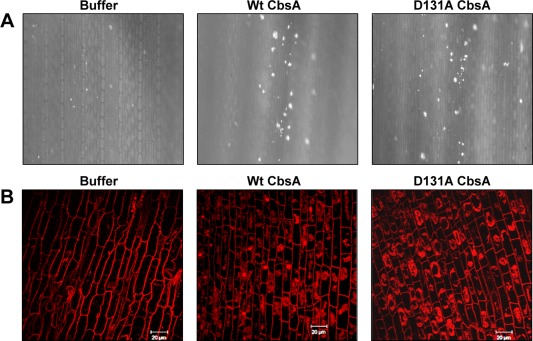
D131A mutation does not affect the ability of CbsA to induce defence responses in rice tissues. (A) Callose deposition in rice leaves: 10–15‐day‐old rice leaves were infiltrated with 100 µL of buffer, Wt CbsA or D131A CbsA proteins of concentration 0.1 mg/mL. The leaves were subsequently treated to remove chlorophyll, stained with aniline blue and visualized under an epifluorescence microscope. Bright spots in the images are the callose deposits. (B) Programmed cell death in rice roots: roots of 2–3‐day‐old rice seedlings were excised and treated with buffer, Wt CbsA or D131A CbsA proteins (0.5 mg/mL) for 16 h, stained with propidium iodide (PI) and examined under a confocal microscope. Extensive internalization of PI and dispersal within the cell are indicative of programmed cell death (PCD). Scale bar measures 20 µm. The concentrations of protein used are the lowest effective concentrations for the wild type protein to elicit callose deposition and programmed cell death respectively.
In a previous study, we have shown that the CbsA protein of Xoo is required for the ability of the bacterium to induce rice defence responses during infection (Tayi et al., 2016a). A mutant of Xoo that is defective in the Type 3 secretion system (T3SS) is an inducer of rice defence responses (Jha et al., 2007). A T3SS– cbsA– double mutant strain is deficient in inducing callose deposition in rice leaves, and a complemented clone expressing Wt CbsA restores to the T3SS– cbsA– mutant the ability to induce callose deposits (Tayi et al., 2016a). To determine whether, in vivo, the altered biochemical activity of the D131A mutant protein can substitute for the activity of the Wt protein in eliciting rice innate immune responses during infection, a T3SS– cbsA– strain expressing D131A CbsA was generated and tested for its ability to induce callose deposition in rice. The T3SS– cbsA– strain expressing the D131A mutant is as efficient as the Wt complemented strain (i.e. T3SS– cbsA– expressing Wt CbsA) in the elicitation of callose deposition in rice leaves (Fig. 6A). The numbers of callose deposits induced by the T3SS– cbsA– strain expressing D131A CbsA are similar to those induced by the T3SS– cbsA– strain expressing Wt CbsA (Fig. 6B).
Figure 6.
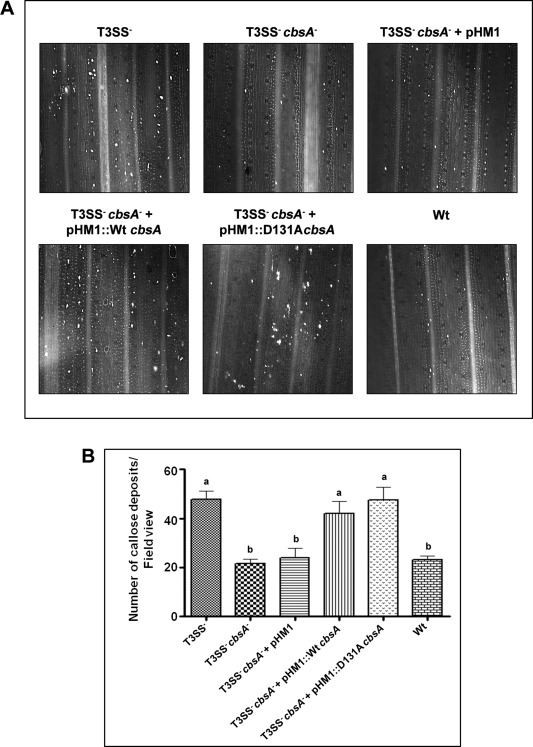
D131A CbsA is as efficient as the wild‐type (Wt) protein in the induction of callose deposition in rice leaves in vivo. (A) Ten‐ to 15‐day‐old rice leaves were infiltrated with one of the following strains of Xanthomonas oryzae pv. oryzae: T3SS– mutant, T3SS– cbsA – double mutant, T3SS– cbsA – + pHM1 (empty vector control), T3SS– cbsA – + pHM1::cbsA (Wt complemented clone), T3SS– cbsA – + pHM1::D131AcbsA and BXO43 (Wt). The leaves were subsequently treated to remove chlorophyll, stained with aniline blue and visualized under an epifluorescence microscope. Bright spots in the images are the callose deposits. (B) The average numbers of callose spots from at least four leaves and three to four different viewing areas in each experiment were plotted. Error bars represent standard deviation (SD). A Student's two‐tailed t‐test for independent means was performed in pairwise combinations for all the values. Values with the same letter (either a or b) are not significantly different at P < 0.05. Similar results were obtained in independent experiments.
Discussion
As part of their virulence repertoire, plant pathogens utilize a battery of enzymes, including endoglucanases and CBHs, to systematically hydrolyse the plant cell wall. Xoo causes the serious bacterial blight disease of rice. The CbsA protein is an important virulence factor of this bacterium. In the present study, the crystal structure of the Xoo CbsA protein was solved to a resolution of 1.86 Å. The presence of the N‐ and C‐terminal loops enclosing the active site confirms that this protein is an exoglucanase and that it could possibly perform processive hydrolysis of crystalline cellulose (Rouvinen et al., 1990; Varrot et al., 2003). In the absence of such loops, an open active site cleft would be displayed, as typically present in endoglucanases (Davies et al., 2000). There are increasing lines of evidence to suggest that the substrate‐enclosing loops display conformational changes on binding of the substrates and, occasionally, may facilitate endo‐cleavage; thus explaining the catalytic facets of both exo‐ and endo‐hydrolysis shown by CBHs (Varrot et al., 1999b; Zou et al., 1999).
Mutational studies of several CBHs have demonstrated a very important role for the aspartic acids of the active site in catalysis. Amongst these various aspartic acid residues, the analogues of Xoo CbsA D131 have been reported to be important for catalysis in these enzymes (Koivula et al., 2002, Vuong and Wilson, 2009). Although the role of this particular aspartic acid as a catalytic base via the Grotthuss mechanism in exoglucanases is increasingly accepted, mutation of this residue to alanine still retains the enzymatic activity. Recent simulation work on T. reesei Cel6A has suggested that proton hopping along a short water wire is responsible for rescuing the catalytic activity of its D175A mutant protein (which is analogous to the Xoo D131A CbsA mutant protein) (Mayes et al., 2016).
The inability of Wt CbsA or any true exoglucanase to act as efficiently as an endoglucanase on CMC is a result of their tunnel‐shaped active sites and the bulky carboxymethyl substitutions on the glucose molecules of CMC which hinder the movement of the processive enzyme along the CMC molecule. The D131A CbsA enzyme showed greater activity than the Wt CbsA on CMC, as revealed by the CMC plate assay as well as specific activity calculations. The mutant enzyme also decreased the viscosity of CMC solution much more quickly than Wt CbsA, although not as efficiently as the typical endoglucanase, ClsA. These results clearly indicate that the mutant enzyme has attained the endo mode of activity. Vuong and Wilson (2009) have made similar observations regarding activity on CMC for the analogous mutant protein D226A from T. fusca Cel6B. Vuong and Wilson (2009) explained that the internal cleavage of CMC by D226A may be caused by the local changes brought about by replacing aspartic acid with alanine. A smaller side chain of alanine may allow the modified glucose residues of CMC to fit into the active site, enabling the mutant enzyme to move along a CMC molecule smoothly until a group of unmodified glucose residues is found to carry out internal cleavage (Vuong and Wilson, 2009; Wu et al., 2013). Interestingly, the crystal structure of the analogous mutant protein D175A Cel6A of T. reesei showed the presence of an open active site, which is a characteristic feature of an endoglucanase, as a result of the large conformational change of the N‐terminal loop (172–182) harbouring the residue (Koivula et al., 2002). A similar loop conformational change can be expected even in the D131A CbsA mutant of Xoo, generating a wider active site, which can explain its endo mode of activity.
The introduction of the D131A cbsA clone into the cbsA mutant background does not restore, even partially, the virulence deficiency of the cbsA mutant. This is in contrast with the observation that the introduction of the Wt cbsA gene into the cbsA mutant background results in partial restoration of the virulence deficiency of the mutant. Full complementation is not observed, we believe, because the complementing plasmid is unstable during growth within rice leaves (Tayi and Sonti, unpublished results). The endoglucanase activity of the D131A CbsA mutant does not appear to be a substitute for the Wt activity of the protein in promoting virulence on rice. It seems likely that efficient hydrolysis of host cell walls during bacterial growth within rice xylem vessels (the tissue compartment in which the pathogen multiplies) is not possible without the exoglucanase activity of the CbsA protein. Intriguingly, the D131A mutant shows weaker virulence than the cbsA null mutant. One possible explanation for this observation is that Xoo‐secreted plant CWDEs (such as cellulases, xylanases, esterases, pectinases, etc.) may act in a coordinated manner, although we do not have specific evidence for this. We believe that D131A CbsA may be competing with ClsA for substrate and that, in vivo, the products of D131A CbsA action are not as good substrates for the other CWDEs as those produced by the action of ClsA. Thus, the presence of D131A CbsA would inhibit the effectiveness of the other CWDEs, making the cbsA mutant strain carrying D131A show weaker virulence than the mutant alone.
Interestingly, the purified D131A CbsA protein is as efficient in inducing defence responses (callose deposition and programmed cell death) in rice tissues as Wt CbsA. In addition, the D131A CbsA protein is as good as the Wt CbsA protein at rescuing the inability of the T3SS– cbsA– mutant to induce rice innate immune responses. This indicates that, in vivo, the D131A CbsA mutant protein can substitute for the Wt protein in eliciting rice innate immune responses during infection. In the majority of cases, the ability of a CWDE to elicit host immune responses comes from its activity on the host cell wall and the release of cell wall degradation products which are recognized as DAMPs (damage‐associated molecular patterns) by the host immune system. For instance, in the case of a secreted esterase LipA of Xoo, the biochemically inactive enzyme S95A LipA is deficient in inducing rice defence responses (Aparna et al., 2009). However, there are examples wherein some surface structural motif of the CWDE is recognized as a PAMP (pathogen‐associated molecular pattern) for the induction of immune responses in the host, and the enzymatic activity of the protein is not required for this purpose. For example, ethylene‐inducing xylanase from T. reesei and cellulase from Rhizoctonia solani have been shown to be inducers of host defence responses without the requirement of any enzymatic activity (Enkerli et al., 1999; Ma et al., 2015; Ron and Avni, 2004).
We have mutated a few other predicted key catalytic residues, such as D180, S137 and Y125, to alanine, but none of these mutations result in a complete loss of biochemical activity. The D226A S232A Cel6B mutant of T. fusca exhibits complete loss of activity, but the analogous mutant of CbsA (D131A S137A) is not expressed in Xoo. Therefore, at the moment, we are unable to determine whether CbsA is a PAMP or a DAMP.
CbsA also shows a distinct structural feature in the form of two additional loops found only in bacterial CBHs. As these loops are not present in proximity to the active sites, their role in catalytic activity can be ruled out. CbsA also has a fibronectin domain at its C‐terminal end that is not required for biochemical activity as it is missing from the secreted protein. The carbohydrate‐binding domains of GHs are the counterparts of the fibronectin domain, and have a well‐established role in binding to crystalline substrates and increasing the efficiency of catalytic domains (Kataeva et al., 2002; Nakamura et al., 2016). In addition, a nine‐residue loop region from the fibronectin‐like domain of a pectate lyase PelL of Dickeya dadantii has been reported to be involved in the control of the secretion of the protein via the Type 2 secretion system by interacting with the secretion system components (Pineau et al., 2014). The specific roles of the fibronectin domain and bacterial‐specific loops of CbsA in the virulence of Xoo are being investigated.
Experimental Procedures
Bacterial strains, plasmids, primers and culture media used
The bacterial strains and plasmids used in this study are listed in Table 2. The primers used in this study are listed in Table S2 (see Supporting Information). Xoo strains were grown at 28 οC in peptone sucrose (PS) medium. Escherichia coli strains were grown in Luria–Bertani (LB) medium at 37 οC. The Xoo single crossovers obtained in the process of generating the deletion mutant of Xoo cbsA were selected and grown on nutrient agar and nutrient broth medium. The concentrations of antibiotics used were as follows: rifampicin (Rf), 50 μg/mL; ampicillin (Ap), 50 μg/mL; spectinomycin (Sp), 50 μg/mL; kanamycin (Km), 25 μg/mL for E. coli and 15 μg/mL for Xoo.
Table 2.
List of bacterial strains and plasmids used in this study.
| Bacterial strain/plasmid | Relevant characteristic(s) | Reference/source |
|---|---|---|
| Escherichia coli strains | ||
| DH5α | λ– f80d lacZDM15 D(lacZYA‐argF) U169 recA1 endA hsdR17 ( ) supE44 thi‐1 gyrA relA1 | Invitrogen, Carlsbad, CA, US |
| S17‐1 | RP4‐2 Tc::Mu‐Km::Tn7 pro hsdR recA Tra+ used as mobilizing strain | Simon et al. (1983) |
| Xanthomonas oryzae pv. oryzae strains | ||
| BXO1 | Wild‐type; Indian isolate | Laboratory collection |
| BXO43 | rif‐2; derivative of BXO1 | Laboratory collection |
| ΔcbsA | rif‐2; derivative of BXO43 | This study |
| ΔcbsA/pHM1 | ΔcbsA/pHM1; rif‐2; Spr derivative of ΔcbsA | This study |
| ΔcbsA/pHM1::cbsA | ΔcbsA/pHM1::cbsA; rif‐2; Spr derivative of ΔcbsA | This study |
| ΔcbsA/pHM1::D131AcbsA | ΔcbsA/pHM1::D131AcbsA; rif‐2; Spr derivative of ΔcbsA | This study |
| T3SS– | hrpB6::bla rif‐2; HR–, Apr derivative of BXO43 | Jha et al. (2007) |
| T3SS– cbsA– | ΔcbsA:: rif‐2; Apr derivative of T3SS– | Tayi et al. (2016a) |
| T3SS– cbsA–/pHM1 | T3SS– cbsA–/pHM1; rif‐2; Apr, Spr; derivative of T3SS– cbsA– | Tayi et al. (2016a, b) |
| T3SS– cbsA–/pHM1::cbsA | T3SS– cbsA–/pHM1::cbsA; rif‐2; Apr, Spr; derivative of T3SS– cbsA– | This study |
| T3SS– cbsA–/pHM1::D131AcbsA | T3SS– cbsA–/pHM1::D131AcbsA; rif‐2; Apr, Spr; derivative of T3SS– cbsA– | This study |
| Plasmids | ||
| pK18mobsacB | Allelic exchange suicide vector SacB derivative of pK18mob; sacB Tra–Mob+, Kmr does not replicate in X. oryzae pv. oryzae | Schäfer, Tauch et al. (1994) |
| pTL1 | pK18mobsacB+1373 bp of A+C fragment of cbsA gene | Tayi et al. (2016a, b) |
| pBSKS | Cloning vector; Apr | Stratagene |
| pBSKS Wt CbsA | pBSKS + 1701 bp cbsA | This study |
| pBSKS D131A CbsA | pBSKS + 1701 bp D131AcbsA | This study |
| pHM1 | Broad‐host‐range cosmid vector (13.3 kb); Spr | Innes, Hirose et al. (1988) |
| pHM1 Wt CbsA | pHM1 + 1701 bp cbsA gene | This study |
| pHM1 D131A CbsA | pHM1 + 1701 bp D131AcbsA gene | This study |
Molecular biology and microbiology techniques
Genomic DNA isolation was performed as described in Leach et al. (1990). For the amplification of the Wt cbsA gene and the D131A version of cbsA, high‐fidelity Phusion polymerase (Finnzymes, Waltham, MA, USA) was used, and Taq polymerase from KAPA Biosystems (Boston, MA, US) was used for all screening purposes. Restriction digestions were carried out using Thermo Fischer Scientific (Waltham, MA, US) Fast Digest enzymes. Ligation reactions were carried out using T4 DNA ligase. Plasmids were purified using either a Macherey Nagel (Duren, Germany) plasmid purification kit or by the alkaline lysis method. Gel extraction and polymerase chain reaction (PCR) purifications were carried out using Macherey Nagel gel extraction and PCR clean‐up kits. Agarose gel electrophoresis, transformation of E. coli, biparental matings and electroporation of plasmids into Xoo were performed as described previously (Ray et al., 2000; Subramoni and Sonti, 2005).
Sequencing and analysis
An ABI Prism 3700 automated DNA sequencer (Perkin‐Elmer, Foster City, CA, USA) was used to carry out sequencing of DNA and the sequences obtained were analysed using the blast algorithm at the National Center for Biotechnology Information database.
CbsA structure solution
The crystal structure of the catalytic domain of CBH (CbsA) from Xoo was determined by the molecular replacement method using Cel6B (E3) from T. fusca (PDB ID: 4B4H) as the search model. Initial phasing was performed using MOLREP‐AUTO MR from the CCP4 suite (Collaborative Computational Project, 1994). The program autobuild from PHENIX (Adams et al., 2010) was used for initial model building, followed by iterative cycles of manual model building and refinement using COOT (Emsley and Cowtan, 2004) and PHENIX. A strong electron density could be seen in the active site pocket in which we could not fit any sugar molecule unambiguously and hence was not accounted for. Water molecules were included using COOT, guided by 2Fo–Fc and Fo–Fc maps. The final model contains 424 amino acids and 474 water molecules. The data collection and refinement statistics are given in Table 1.
Generation of in‐frame deletion cbsA mutant of Xoo
To generate an in‐frame deletion in the cbsA gene of Xoo, a recombinant plasmid pTL1 (pk18mobsacB with in frame‐deleted cbsA fragment), generated in an earlier study (Tayi et al., 2016a), was used. The plasmid pTL1 was transformed into S17‐1 (Simon et al., 1983). Biparental matings were then set up between the S17‐1 E. coli cells containing pTL1 and Wt Xoo (BXO43). BXO43 cells which have integrated the plasmid at the correct locus in the genome were selected by kanamycin resistance and confirmed by PCR using a pair of primers flanking the cbsA gene (Table S2). Confirmed single recombinants were grown in nutrient broth without kanamycin and plated on 1% potato sucrose agar (PSA) plates to isolate cells which had undergone a second recombination event. Double recombinants which include both mutants and Wt cells were screened and the in frame deletion of the cbsA gene in the mutants was confirmed as described previously (Tayi et al., 2016a).
Generation of D131A CbsA mutant
The cbsA gene was PCR amplified from the genome of the BXO43 strain of Xoo (Wt), with primers having HindIII and EcoRI restriction sites at the 5′ and 3′ ends, respectively (Table S2). The amplified gene was restriction digested with these enzymes to obtain a product with sticky ends. We used the pBSKS (Stratagene, San Diego, CA, US) plasmid as a shuttle vector for gene amplification and site‐directed mutagenesis studies. Plasmid pBSKS was double digested (with the same enzymes) and used for ligation with the doubly digested cbsA gene product. The ligation product was transformed into DH5α cells and the vector–gene construct was used for site‐directed mutagenesis studies. The D131A mutant was generated by PCR amplification using the primers listed in Table S2, followed by 30 min of digestion with DpnI to digest the Wt cbsA‐pBSKS constructs. The mutations were then confirmed by DNA sequencing. The positive constructs were then again digested with HindIII and EcoRI, and ligated in HindIII–EcoRI‐digested pHM1 vector. These pHM1‐CbsA mutant constructs were transformed into DH5α cells. The plasmids were purified from these cultures and used for electroporation into a Xoo strain which has an in‐frame deletion of the cbsA gene. This strain was used further for purification of the D131A CbsA mutant protein.
Enzyme purification
Wt Xoo (BXO43) was used for purification of the CbsA and ClsA proteins. D131A CbsA protein was purified from a cbsA – mutant strain expressing D131A CbsA from the pHM1 cosmid. The strains were grown until saturation. The extracellular culture supernatant was obtained by pelleting the cells at 4°C, 9.8 Kg, for 20 min. Secreted proteins in the culture supernatant were precipitated using ammonium sulfate (55% saturation) and the pellet was dissolved in 0.01 m potassium phosphate buffer, pH 6.0. Excess salt was removed by extensive dialysis against the same buffer. Proteins in the dialysed sample were separated into various fractions by cation exchange chromatography using a MONO S column with a linear gradient of 0.01 m potassium phosphate buffer, pH 6.0 (Buffer A), to 0.01 m potassium phosphate buffer, pH 6.0, and 1 m NaCl (Buffer B). Subsequently, using gel filtration chromatography (Superdex 200, GE Healthcare, Chicago, Illinois, US), the Wt CbsA and D131A CbsA proteins were purified to homogeneity in 0.02 m Tris‐Cl, pH 8.0, and 0.02 m NaCl. The purity of the proteins was confirmed by sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE).
CMC plate assay
A 1.2% agarose suspension containing 0.1% of CMC was poured into Petri plates. After solidification, wells were cut out and filled with either the enzyme or buffer. Following incubation at 28°C for 36–48 h, the plates were stained with 1% Congo red solution and then destained with 1 m NaCl. A zone of clearance around the well indicates cellulase activity.
Thin layer chromatography
Ten micrograms of the oligomeric substrates cellobiose, cellotriose, cellotetraose, cellopentaose and cellohexaose were hydrolysed to completion by incubation with 1 µm of any one of the following: Wt CbsA, D131A CbsA or ClsA enzyme. Similarly, 100 µg of CMC was treated with 1 µm of enzyme for 16 h at 37°C. Hydrolysis products were separated on 0.2‐mm aluminium sheet silica gel 60 plates (Merck, Kenilworth, NJ, US) with a butanol, acetic acid and water mixture (2 : 1 : 1) as eluent. Sugars were detected by dipping the plates in a freshly prepared solution containing 10% sulfuric acid and 0.1% methanolic orcinol (1 : 1), followed by heating the plates at 80°C until the colour developed.
Estimation of reducing sugars
One millilitre of a reaction mixture containing 0.5 mL of 2% CMC solution and 0.2 µm of either Wt CbsA or D131A CbsA was incubated for 4 h at 37°C. To stop the reaction, 3 mL of 3,5‐dinitro salicylic acid (DNS) solution was added to the complete reaction mixture, followed by boiling. The absorbance of the samples and glucose standards was measured at 540 nm in a spectrophotometer and the specific activity of the enzyme was calculated.
Viscosity measurements
Five millilitres of 2% (w/v) solution of CMC were hydrolysed with 0.2 µm of ClsA, Wt CbsA or D131A CbsA in a 10‐mL reaction. The flow time of the reaction mixture was determined at intervals of 2, 4 and 6 h in a kinematic viscosity bath (Cannon in USA, State College, PA, US) at 37°C. The kinematic viscosity was calculated by multiplying the correction value specific for the vessel with the flow time of the sample.
Virulence assay
Wt Xoo (BXO43), cbsA– mutant, cbsA– mutant expressing either the Wt CbsA protein or D131A CbsA protein from pHM1 vector and cbsA– mutant carrying the empty pHM1 vector were grown in PS broth with appropriate antibiotics. The bacterial cells were pelleted by centrifugation at 2.4 Kg for 5 min at room temperature, washed and resuspended in sterile Milli Q (MQ) water. Forty to 45‐day‐old Taichung Native‐1 (TN‐1) rice leaf tips were cut with surgical scissors dipped in a bacterial suspension with an optical density at 600 nm (OD600) of 1. Lesion lengths were measured at 20 days post‐inoculation.
Callose deposition assay
TN‐1 rice seedlings (about 10–15 days old) were infiltrated with buffer (10 mm phosphate buffer, pH 6.0), Wt CbsA or D131A CbsA proteins (of 0.1 mg/mL) or cultures of the Xoo strains Wt (BXO43), T3SS–, T3SS– cbsA–, T3SS– cbsA– + pHM1 (empty vector control strain), T3SS– cbsA– + pHM1::WtcbsA and T3SS– cbsA– + pHM1::D131AcbsA with OD600 = 1.0 using a needleless syringe. Sixteen hours later, the infiltrated zones (approximately 1 cm in length) were cut from the leaves and pigments were removed by heating at 60°C with alcohol. Subsequently, the samples were stained with 0.5% aniline blue solution prepared in 150 mm K2HPO4 and analysed under an epifluorescence microscope using a blue filter and ×10 objective.
Cell death assay
TN‐1 rice seeds were surface sterilized and germinated on sterile filter paper overlaid on 0.5% agar in Petri dishes; 1–2‐cm‐long root tips were excised from the seedlings and treated with buffer (10 mm phosphate buffer, pH 6.0), Wt CbsA or D131A CbsA protein (500 µL of 0.5 mg/mL). After incubation for 16–18 h, the roots were washed with sterile MQ water and stained with PI. The PI‐stained roots were mounted in 50% glycerol on glass slides. The PI internalization in root cells was further detected in an LSM‐510 Meta confocal microscope (Carl Zeiss, Jena, Germany) under a 63× oil immersion objective using an He–Ne laser excited at 543 nm. One‐micrometre‐thick longitudinal optical sections were captured and further projected to obtain an image of 2–3 μm total thickness. All images were analysed using LSM software.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 The conserved tryptophans lining the active site tunnel. Tryptophans around the active site tunnel which help to anchor the substrate by virtue of π–π interactions with the sugar rings are shown with sticks. (Ligand was modelled from PDB ID: 4B4F.)
Table S1 Specific activity of wild‐type (Wt) CbsA and D131A CbsA.
Table S2 List of primers used in this study.
Acknowledgements
We acknowledge the help received from Amit Rajak, Dr Prabhavathi Devi and Dr R. B. N. Prasad of the Lipid Research Facility of the CSIR‐Indian Institute of Chemical Technology in carrying out the viscosity experiments. LT, SK and RN acknowledge fellowships from the Indian Council of Medical Research, the University Grants Commission and the Council of Scientific and Industrial Research, respectively. This work was supported by the XIIth five year plan project, Plant–Microbe and Soil Interactions (BSC0117), of the Council of Scientific and Industrial Research. RVS and RS were also supported by a J. C. Bose Fellowship from the Department of Science and Technology, Government of India. The authors have no conflicts of interest to declare.
Contributor Information
Rajan Sankaranarayanan, Email: sankar@ccmb.res.in.
Ramesh V. Sonti, Email: sonti@ccmb.res.in
References
- Adams, P.D. , Afonine, P.V. , Bunkóczi, G. , Chen, V.B. , Davis, I.W. , Echols, N. , Headd, J.J. , Hung, L.‐W. , Kapral, G.J. , Grosse‐Kunstleve, R.W. , McCoy, A.J. , Moriarty, N.W. , Oeffner, R. , Read, R.J. , Richardson, D.C. , Richardson, J.S. , Terwilliger, T.C. and Zwart, P.H. (2010) PHENIX: a comprehensive Python‐based system for macromolecular structure solution. Acta Crystallogr. D: Biol. Crystallogr. 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparna, G. , Chatterjee, A. , Sonti, R.V. and Sankaranarayanan, R. (2009) A cell wall‐degrading esterase of Xanthomonas oryzae requires a unique substrate recognition module for pathogenesis on rice. Plant Cell, 21, 1860–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel, B.L. , Coutinho, P.M. , Rancurel, C. , Bernard, T. , Lombard, V. and Henrissat, B. (2009) The Carbohydrate‐Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project . (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D: Biol. Crystallogr. 50, 760. [DOI] [PubMed] [Google Scholar]
- Davies, G.J. , Brzozowski, A.M. , Dauter, M. , Varrot, A. and Schülein, M. (2000) Structure and function of Humicola insolens family 6 cellulases: structure of the endoglucanase, Cel6B, at 1.6 Å resolution. Biochem. J. 348, 201–207. [PMC free article] [PubMed] [Google Scholar]
- Emsley, P. and Cowtan, K. (2004) Coot: model‐building tools for molecular graphics. Acta Crystallogr. D: Biol. Crystallogr. 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Enkerli, J. , Felix, G. and Boller, T. (1999) The enzymatic activity of fungal xylanase is not necessary for its elicitor activity. Plant Physiol. 121, 391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough, C.L. , Dow, J.M. , Barber, C.E. and Daniels, M.J. (1988) Cloning of two endoglucanase genes of Xanthomonas campestris pv. campestris: analysis of the role of the major endoglucanase in pathogenesis. Mol. Plant–Microbe Interact. 1, 275–281. [Google Scholar]
- Henrissat, B. (1991) A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280, 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon, D.N.‐S. (1994) Cellulose: a random walk along its historical path. Cellulose, 1, 1–25. [Google Scholar]
- Innes, R.W. , Hirose, M.A. and Kuempel, P.L. (1988) Induction of nitrogen‐fixing nodules on clover requires only 32 kilobase pairs of DNA from the Rhizobium trifolii symbiosis plasmid. J. Bacteriol. 170, 3793–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha, G. , Rajeshwari, R. and Sonti, R.V. (2007) Functional interplay between two Xanthomonas oryzae pv. oryzae secretion systems in modulating virulence on rice. Mol. Plant–Microbe Interact. 20, 31–40. [DOI] [PubMed] [Google Scholar]
- Kataeva, I.A. , Seidel, R.D. , Shah, A. , West, L.T. , Li, X.‐L. and Ljungdahl, L.G. (2002) The fibronectin type 3‐like repeat from the Clostridium thermocellum cellobiohydrolase CbhA promotes hydrolysis of cellulose by modifying its surface. Appl. Environ. Microbiol. 68, 4292–4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivula, A. , Reinikainen, T. , Ruohonen, L. , Valkeajärvi, A. , Claeyssens, M. , Teleman, O. , Kleywegt, G.J. , Szardenings, M. , Rouvinen, J. , Jones, T.A. and Teeri, T.T. (1996) The active site of Trichoderma reesei cellobiohydrolase II: the role of tyrosine 169. Protein Eng. 9, 691–699. [DOI] [PubMed] [Google Scholar]
- Koivula, A. , Ruohonen, L. , Wohlfahrt, G. , Reinikainen, T. , Teeri, T.T. , Piens, K. , Claeyssens, M. , Weber, M. , Vasella, A. , Becker, D. , Sinnott, M.L. , Zou, J.‐Y. , Kleywegt, G.J. , Szardenings, M. , Ståhlberg, J. and Jones, T.A. (2002) The active site of cellobiohydrolase Cel6A from Trichoderma reesei: the roles of aspartic acids D221 and D175. J. Am. Chem. Soc. 124, 10 015–10 024. [DOI] [PubMed] [Google Scholar]
- Kumar, S. , Haque, A.S. , Jha, G. , Sonti, R.V. and Sankaranarayanan, R. (2012) Crystallization and preliminary crystallographic studies of CbsA, a secretory exoglucanase from Xanthomonas oryzae pv. oryzae . Acta Crystallogr. Sect. F: Struct. Biol. Cryst. Commun. 68, 1191–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach, J.E. , White, F.F. , Rhoads, M.L. , and Leung, H. (1990) A repetitive DNA sequence differentiates Xanthomonas campestris pv. oryzae from other pathovars of X. campestris. Mol. Plant-Microbe Interact. 3, 238–246. [Google Scholar]
- Liu, Y. , Yoshida, M. , Kurakata, Y. , Miyazaki, T. , Igarashi, K. , Samejima, M. , Fukuda, K. , Nishikawa, A. and Tonozuka, T. (2010) Crystal structure of a glycoside hydrolase family 6 enzyme, CcCel6C, a cellulase constitutively produced by Coprinopsis cinerea . FEBS J. 277, 1532–1542. [DOI] [PubMed] [Google Scholar]
- Ma, Y. , Han, C. , Chen, J. , Li, H. , He, K. , Liu, A. and Li, D. (2015) Fungal cellulase is an elicitor but its enzymatic activity is not required for its elicitor activity. Mol. Plant Pathol. 16, 14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayes, H.B. , Knott, B.C. , Crowley, M.F. , Broadbelt, L.J. , Ståhlberg, J. and Beckham, G.T. (2016) Who's on base? Revealing the catalytic mechanism of inverting family 6 glycoside hydrolases. Chem. Sci. 7, 5955–5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, A. , Tasaki, T. , Ishiwata, D. , Yamamoto, M. , Okuni, Y. , Visootsat, A. , Maximilien, M. , Noji, H. , Uchiyama, T. , Samejima, M. , Igarashi, K. and Iino, R. (2016) Single‐molecule imaging analysis of binding, processive movement, and dissociation of cellobiohydrolase Trichoderma reesei Cel6A and its domains on crystalline cellulose. J. Biol. Chem. 291, 22 404–22 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineau, C. , Guschinskaya, N. , Robert, X. , Gouet, P. , Ballut, L. and Shevchik, V.E. (2014) Substrate recognition by the bacterial type II secretion system: more than a simple interaction. Mol. Microbiol. 94, 126–140. [DOI] [PubMed] [Google Scholar]
- Rajeshwari, R. , Jha, G. and Sonti, R.V. (2005) Role of an in planta‐expressed xylanase of Xanthomonas oryzae pv. oryzae in promoting virulence on rice. Mol. Plant–Microbe Interact. 18, 830–837. [DOI] [PubMed] [Google Scholar]
- Ray, S.K. , Rajeshwari, R. and Sonti, R.V. (2000) Mutants of Xanthomonas oryzae pv. oryzae deficient in general secretory pathway are virulence deficient and unable to secrete xylanase. Mol. Plant–Microbe Interact. 13, 394–401. [DOI] [PubMed] [Google Scholar]
- Roberts, D. , Denny, T. and Schell, M. (1988) Cloning of the egl gene of Pseudomonas solanacearum and analysis of its role in phytopathogenicity. J. Bacteriol. 170, 1445–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron, M. and Avni, A. (2004) The receptor for the fungal elicitor ethylene‐inducing xylanase is a member of a resistance‐like gene family in tomato. Plant Cell, 16, 1604–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouvinen, J. , Bergfors, T. , Teeri, T. , Knowles, J. and Jones, T. (1990) Three‐dimensional structure of cellobiohydrolase II from Trichoderma reesei . Science, 249, 380–386. [DOI] [PubMed] [Google Scholar]
- Ryan, R.P. , Vorhölter, F.‐J. , Potnis, N. , Jones, J.B. , Van Sluys, M.‐A. , Bogdanove, A.J. and Dow, J.M. (2011) Pathogenomics of Xanthomonas: understanding bacterium–plant interactions. Nat. Rev. Microbiol. 9, 344–355. [DOI] [PubMed] [Google Scholar]
- Sandgren, M. , Wu, M. , Karkehabadi, S. , Mitchinson, C. , Kelemen, B.R. , Larenas, E.A. , Ståhlberg, J. and Hansson, H. (2013) The structure of a bacterial cellobiohydrolase: the catalytic core of the Thermobifida fusca family GH6 cellobiohydrolase Cel6B. J. Mol. Biol. 425, 622–635. [DOI] [PubMed] [Google Scholar]
- Sato, C. , Oka, N. , Nabeta, K. and Matsuura, H. (2011) Cellulase applied to the leaves of sweet pepper (Capsicum annuum L. var. grossum) upregulates the production of salicylic and azelaic acids. Biosci. Biotechnol. Biochem. 75, 761–763. [DOI] [PubMed] [Google Scholar]
- Schafer, A. , Tauch, A. , Jager, W. , Kalinowski, J. , Thierbach, G. and Puhler, A. (1994) Small mobilizable multi‐purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 145, 69–73. [DOI] [PubMed] [Google Scholar]
- Simon, R. , Priefer, U. and Pühler, A. (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1, 784–791. [Google Scholar]
- Subramoni, S. and Sonti, R.V. (2005) Growth deficiency of a Xanthomonas oryzae pv. oryzae fur mutant in rice leaves is rescued by ascorbic acid supplementation. Mol. Plant–Microbe Interact. 18, 644–651. [DOI] [PubMed] [Google Scholar]
- Tamura, M. , Miyazaki, T. , Tanaka, Y. , Yoshida, M. , Nishikawa, A. and Tonozuka, T. (2012) Comparison of the structural changes in two cellobiohydrolases, CcCel6A and CcCel6C, from Coprinopsis cinerea–a tweezer‐like motion in the structure of CcCel6C. FEBS J. 279, 1871–1882. [DOI] [PubMed] [Google Scholar]
- Tayi, L. , Maku, R. , Patel, H.K. and Sonti, R. (2016a) Action of multiple cell wall degrading enzymes is required for elicitation of innate immune responses during Xanthomonas oryzae pv. oryzae infection in rice. Mol. Plant–Microbe Interact. 29, 599–608. [DOI] [PubMed] [Google Scholar]
- Tayi, L. , Maku, R.V. , Patel, H.K. and Sonti, R.V. (2016b) Identification of pectin degrading enzymes secreted by Xanthomonas oryzae pv. oryzae and determination of their role in virulence on rice. PLoS One, 11, e0166396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeri, T.T. (1997) Crystalline cellulose degradation: new insight into the function of cellobiohydrolases. Trends Biotechnol. 15, 160–167. [Google Scholar]
- Teeri, T. , Koivula, A. , Linder, M. , Wohlfahrt, G. , Divne, C. and Jones, T. (1998) Trichoderma reesei cellobiohydrolases: why so efficient on crystalline cellulose? Biochem. Soc. Trans. 26, 173–178. [DOI] [PubMed] [Google Scholar]
- Thompson, A.J. , Heu, T. , Shaghasi, T. , Benyamino, R. , Jones, A. , Friis, E.P. , Wilson, K.S. and Davies, G.J. (2012) Structure of the catalytic core module of the Chaetomium thermophilum family GH6 cellobiohydrolase Cel6A. Acta Crystallogr. D: Biol. Crystallogr. 68, 875–882. [DOI] [PubMed] [Google Scholar]
- Varrot, A. , Hastrup, S. , Schülein, M. and Davies, G.J. (1999a) Crystal structure of the catalytic core domain of the family 6 cellobiohydrolase II, Cel6A, from Humicola insolens, at 1.92 Å resolution. Biochem. J. 337, 297–304. [PMC free article] [PubMed] [Google Scholar]
- Varrot, A. , Schülein, M. and Davies, G.J. (1999b) Structural changes of the active site tunnel of Humicola insolens cellobiohydrolase, Cel6A, upon oligosaccharide binding. Biochemistry, 38, 8884–8891. [DOI] [PubMed] [Google Scholar]
- Varrot, A. , Frandsen, T.P. , von Ossowski, I. , Boyer, V. , Cottaz, S. , Driguez, H. , Schülein, M. and Davies, G.J. (2003) Structural basis for ligand binding and processivity in cellobiohydrolase Cel6A from Humicola insolens . Structure, 11, 855–864. [DOI] [PubMed] [Google Scholar]
- Vuong, T.V. and Wilson, D.B. (2009) The absence of an identifiable single catalytic base residue in Thermobifida fusca exocellulase Cel6B. FEBS J. 276, 3837–3845. [DOI] [PubMed] [Google Scholar]
- Wu, M. , Bu, L. , Vuong, T.V. , Wilson, D.B. , Crowley, M.F. , Sandgren, M. , Ståhlberg, J. , Beckham, G.T. and Hansson, H. (2013) Loop motions important to product expulsion in the Thermobifida fusca glycoside hydrolase family 6 cellobiohydrolase from structural and computational studies. J. Biol. Chem. 288, 33 107–33 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, T. , Li, Y. , Sun, D. , Zhuo, T. , Fan, X. and Zou, H. (2016) Identification of an extracellular endoglucanase that is required for full virulence in Xanthomonas citri subsp. citri . PLoS One, 11, e0151017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, J‐Y. , Kleywegt, G.J. , Ståhlberg, J. , Driguez, H. , Nerinckx, W. , Claeyssens, M. , Koivula, A. , Teeri, T.T. and Jones, T.A. (1999) Crystallographic evidence for substrate ring distortion and protein conformational changes during catalysis in cellobiohydrolase Ce16A from Trichoderma reesei . Structure, 7, 1035–1045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 The conserved tryptophans lining the active site tunnel. Tryptophans around the active site tunnel which help to anchor the substrate by virtue of π–π interactions with the sugar rings are shown with sticks. (Ligand was modelled from PDB ID: 4B4F.)
Table S1 Specific activity of wild‐type (Wt) CbsA and D131A CbsA.
Table S2 List of primers used in this study.


