Summary
RNA viruses have very compact genomes and so provide a unique opportunity to study how evolution works to optimize the use of very limited genomic information. A widespread viral strategy to solve this issue concerning the coding space relies on the expression of proteins with multiple functions. Members of the family Potyviridae, the most abundant group of RNA viruses in plants, offer several attractive examples of viral factors which play roles in diverse infection‐related pathways. The Helper Component Proteinase (HCPro) is an essential and well‐characterized multitasking protein for which at least three independent functions have been described: (i) viral plant‐to‐plant transmission; (ii) polyprotein maturation; and (iii) RNA silencing suppression. Moreover, multitudes of host factors have been found to interact with HCPro. Intriguingly, most of these partners have not been ascribed to any of the HCPro roles during the infectious cycle, supporting the idea that this protein might play even more roles than those already established. In this comprehensive review, we attempt to summarize our current knowledge about HCPro and its already attributed and putative novel roles, and to discuss the similarities and differences regarding this factor in members of this important viral family.
Keywords: multifunctional proteins, proteinase, RNA silencing suppressor, transmission
Introduction
Members of the family Potyviridae are the most abundant and socio‐economically relevant RNA viruses infecting plants (Scholthof et al., 2011; Valli et al., 2015); therefore, they have been the subject of intense studies worldwide. This family is formed by eight genera (Brambyvirus, Bymovirus, Ipomovirus, Macluravirus, Poacevirus, Potyvirus, Rymovirus and Tritimovirus) which are differentiated by their genome composition and structure, RNA sequence and transmission vectors (Revers and García, 2015). Most potyvirids (i.e. viruses belonging to the Potyviridae family) have monopartite, single‐stranded and positive‐sense genomes of around 10 000 nucleotides that are encapsidated by multiple units of a single coat protein (CP) in flexuous and filamentous virus particles of 680–900 nm in length and 11–14 nm in diameter (Kendall et al., 2008). Exceptionally, bymoviruses are peculiar in this regard, as they have a bipartite genome that is encapsidated separately. Inside the infected cells, the viral RNA of potyvirids is uncoated and translated into polyproteins which are proteolytically processed by viral‐encoded proteinases producing, in most cases, the following mature viral gene products: P1, the helper component proteinase (HCPro), P3, 6K1, CI, 6K2, NIa (VPg + Pro), NIb and CP. As mentioned above, bymoviruses have two genomic RNA segments that are independently translated. In addition to the large polyproteins, transframe products, named P3N‐PIPO and P3N‐ALT, which share the N‐terminal region of P3, are produced from RNA variants generated via transcriptional slippage during viral replication (Hagiwara‐Komoda et al., 2016; Olspert et al., 2015; Rodamilans et al., 2015). Furthermore, the same mechanism is also used during the replication of some sweet potato potyviruses to produce an additional transframe product, termed P1N‐PISPO, which overlaps with the P1 cistron (Mingot et al., 2016; Untiveros et al., 2016).
RNA viruses in general are known to have small and condensed genomes which, at least in part, might be a result of: (i) intrinsic structural restrictions (e.g. topology and stability) of the RNA molecule (Gorbalenya et al., 2006); (ii) the need to minimize the negative impact of the error‐prone viral replication (Holmes, 2003); or (iii) the need to protect themselves from the action of antiviral host defence mechanisms (Eusebio‐Cope and Suzuki, 2015). As a consequence, RNA viruses are under intense selective pressure to optimize the use of their genomic information. To cope with this restriction, they exploit diverse strategies in order to produce/recruit all the required components to ensure infection success (Ahlquist et al., 2003; Atkins et al., 2016; Firth and Brierley, 2012; Sztuba‐Solinska et al., 2011). One of these strategies relies on the expression of viral proteins with several functions. In particular, the well‐characterized RNA viruses of the family Potyviridae provide fascinating examples of multitasking proteins (e.g. Sorel et al., 2014; Weber and Bujarski, 2015). Here, we present a comprehensive review concerning the potyvirid HCPro, with particular emphasis on members of the genus Potyvirus, in which at least three clearly independent functions have been described.
Transmission—A Historical Overview of HCPro Discovery
Potyviruses are transmitted by aphids by a mode of transmission that is described as non‐persistent, as it occurs rapidly, with the duration of acquisition and inoculation phases in the range of seconds to minutes without retention periods (Bradley, 1952; Day and Irzykiewicz, 1954; Kassanis, 1941). This fast and usually efficient mode of transmission was recognized as a serious caveat in the adoption of control measures against pathogenic virus dissemination, because it leaves virtually no time available for effective insecticide treatment aimed to target their vectors. Therefore, intense research efforts took place to better understand potyviral transmission. In this context, the role of HCPro in this process was found even before it was known that it was a viral protein. The name ‘Helper Component’ was coined to describe the existence of a ‘component’ of unknown source, but present in infected plants, which ‘helped’ the transmission of potyviruses mediated by aphid vectors. How this function was identified is an extraordinary story that reveals the resources, skills and imagination of those researchers involved in its discovery (Pirone and Thornbury, 1984). Chronologically, the finding of certain natural virus isolates with altered transmission properties was the first indication that this function was genetically regulated (Kamm, 1969; Simons, 1976). The use of aphid artificial feeding systems, based on stretched parafilm membranes, was instrumental in the verification that insects often failed to transmit the disease when purified virions were used for the transmission assay (Pirone and Megahed, 1966). Hence, this result indicated that the viral particle alone is not sufficient for efficient transmission. Taking advantage of UV radiation treatments to inactivate viral RNAs, it was shown that a UV‐resistant component (probably a protein) must be acquired by aphids simultaneously (or prior) to virions in order to transmit the virus (Govier and Kassanis, 1974a, 1974b; Kassanis and Govier, 1971a, 1971b). Later, equipped with very simple experimental tools, the purification of the active factor was achieved and allowed the generation of specific antisera (Govier et al., 1977; Thornbury et al., 1985), which was certainly crucial to establish its origin as part of the viral polyprotein (Carrington et al., 1989a; Dougherty and Hiebert, 1980; Hiebert et al., 1984). Indeed, antibodies against HCPro have been very useful to establish the presence of this viral factor in amorphous inclusions of cells infected with some potyviruses (De Mejia et al., 1985), as well as to unravel other aspects of HCPro that will be described in diverse sections of this article.
Based on results from the experiments described above, a molecular mechanism by which HCPro participates in the transmission process was suggested long ago by Govier and Kassanis (1974b). The so‐called ‘bridge hypothesis’ proposes that the helper component acts as a reversible link between the viral particle and the vector mouthparts (Fig. 1). Over the years, accumulative evidence has provided ample support for this hypothesis, whereas alternative models, such as the proposition of a direct interaction between CP and aphid receptors with HCPro acting to expose CP binding sites (Salomon and Bernardi, 1995), failed to reach generalization. Among the most remarkable outcomes of these efforts were the identification and validation of conserved domains in CP and HCPro that are involved in vector transmission. In CP, a highly conserved ‘DAG’ motif had been earlier predicted to play a role in transmission (Harrison and Robinson, 1988; Laín et al., 1988), which was further confirmed by mutagenesis analyses (Atreya et al., 1990, 1991, 1995). With regard to the identification of relevant domains in HCPro, the characterization of transmission‐defective isolates in different viruses (Huet et al., 1994; Peng et al., 1998; Thornbury et al., 1990) led to the identification of at least two separate motifs required for the bridge hypothesis to occur: a PTK amino acid triad that interacts with CP (Huet et al., 1994; Peng et al., 1998) and a KITC motif that participates in retention to an unknown structure in the aphid mouthparts (Blanc et al., 1998; Huet et al., 1994) (Fig. 1). The presence of these amino acids might not be sufficient for function and, indeed, other regions in HCPro were later proposed to affect transmissibility (Canto et al., 1995; Llave et al., 2002; Seo et al., 2010). Importantly, predictions based on the bridge hypothesis have been confirmed, and they include: (i) the identification of HCPro retention sites in aphid stylets (Moreno et al., 2012; Wang et al., 1998); (ii) the direct interaction between CP and HCPro (Blanc et al., 1997; Roudet‐Tavert et al., 2002; Seo et al., 2010); and (iii) the location of HCPro in a protruding tip at one end of the viral particle (Torrance et al., 2006).
Figure 1.
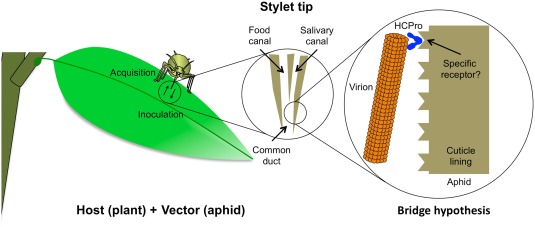
‘Bridge hypothesis’ for aphid transmission of potyviruses. Left: an aphid is feeding from an infected plant. Centre: longitudinal section of the mandibular stylet (the external flanking maxillae have been omitted to simplify the figure), including the two parallel channels (the food canal that connects to the digestive system and the salivary canal that allows secretions during feeding) joining at the common duct. Right: a helper component proteinase (HCPro) complex (depicted in a dimeric form) is bound at one end of the viral particle and allows a reversible interaction with potential receptors located over the cuticle lining (internal side of the stylet tip). It should be noted that this figure is a predictive representation of the viral transmission process based on very limited available experimental data on interactions and the consequent role of HCPro during this process (see text for details). Hence, it cannot be ruled out, for instance, that the HCPro–CP interaction might occur all along the viral particle and non‐dimeric forms of HCPro may play a role in viral transmission.
An intriguing observation linked to the discovery of HCPro is the unusual aphid‐mediated transmission of the potexvirus Potato aucuba mosaic virus (PaMV), which only takes place when PaMV‐infected plants are co‐infected with a potyvirus (Kassanis, 1961). The further finding of an equivalent DAG motif at the N‐terminus of the PaMV CP provided a putative explanation for the observed trans‐complementation. In the same study, an elegant demonstration of the relevance of the DAG amino acid triad was obtained by engineering this motif in the CP of Potato virus X, a non‐DAG, non‐aphid‐borne, potexvirus, as this modification rendered the aphid transmission of this virus dependent on HCPro (Baulcombe et al., 1993). It is worth mentioning that the compatibility of different HCPros to support the transmission of other potyviruses has also been confirmed (Flasinski and Cassidy, 1998; Lecoq and Pitrat, 1985; López‐Moya et al., 1995; Sako and Ogata, 1981). Indeed, this trans‐complementation property of HCPro is believed to play an important ecological role by driving the evolution of the helper strategy as a way to avoid the negative impact of genetic bottlenecks associated with non‐persistent virus transmission (Pirone and Blanc, 1996).
The purification of an HCPro still active during transmission was useful for the study of diverse features of this protein. Although the insertion of a 6 × His tag facilitated HCPro purification in the context of a viral infection using an Ni2+‐charged resin (Blanc et al., 1999), the same purification protocol was successfully applied in other viruses without attaching the 6 × His tag to HCPro (Wang and Pirone, 1999). These results suggest that intrinsic biochemical properties of the protein require the interaction with metallic ions, an observation that agrees with previous studies mentioning the relevance of divalent cations in the buffer (in particular Mg2+) during transmission assays (Thornbury and Pirone, 1983; Thornbury et al., 1985).
The expression of functional HCPro in heterologous systems has provided a useful methodology to speed up research on potyvirus transmission. Hence, the correct activity of HCPro was maintained when the protein was expressed in transgenic plants (Berger et al., 1989), in insect cells using a baculovirus‐based system (Thornbury et al., 1993) or in yeast (Ruiz‐Ferrer et al., 2004). In addition, transient expression systems in plants, using either viral vectors (Sasaya et al., 2000) or agro‐infiltration (Goytia et al., 2006), also succeeded in producing transmission‐active HCPro.
Remarkably, it was also shown that HCPro plays a key role in the semi‐persistent dispersion of Wheat streak mosaic virus (WSMV), a member of the Tritimovirus genus transmitted by eriophyid mites (Stenger et al., 2005b). Moreover, despite the low overall sequence similarity between the tritimovirus and potyvirus HCPros, mutations in conserved cysteine residues affected the transmission process in viruses belonging to these two genera (Atreya and Pirone, 1993; Llave et al., 1999; Young et al., 2007). As a detailed characterization of the role of HCPro in transmission mediated by vectors other than aphids awaits to be addressed, it is not currently clear whether this function has been acquired independently in different Potyviridae genera (convergent evolution), or has been derived from a common ancestral virus that was transmitted by an ancestral arthropod (adaptation).
Finally, other aspects that remain to be determined in order to better understand the role of HCPro in transmission include the stoichiometry and geometry of the reversible virion–HCPro–vector interactions, which seem to involve multimers of HCPro (Plisson et al., 2003; Ruiz‐Ferrer et al., 2005), and the location of this factor at one end of the viral particles (Torrance et al., 2006). Curiously, the visualization of virions within insect stylets has only been attempted and achieved with potyviruses in a very reduced number of studies (Wang et al., 1996), and only a few attempts to discover the vector receptors have been pursued and communicated (Dombrovsky et al., 2007; Fernández‐Calvino et al., 2010). Thus, at this point, it is still uncertain whether the potyvirus‐specific aphid receptor co‐localizes or shares properties with the putative receptors of viruses from other families (Blanc et al., 2014; Uzest et al., 2007).
RNA Silencing Suppression—Fight for Survival
RNA silencing is a highly conserved, sequence‐specific, regulatory mechanism that shuts down the expression of target genes at the transcriptional and post‐transcriptional level. The entire silencing machinery is formed by partially overlapping modules, which are accordingly activated in the presence of diverse double‐stranded (ds) RNA molecules and have different roles during development (some good reviews on RNA silencing have been published recently: Bologna and Voinnet, 2014; Castel and Martienssen, 2013; Chang et al., 2012). As part of its many tasks, RNA silencing plays a key antiviral role in organisms from different kingdoms (Bronkhorst and van Rij, 2014; Chang et al., 2012; Ding, 2010; Huang et al., 2014; Li et al., 2002; Szittya and Burgyan, 2013; Zhang et al., 2015). In the case of plants, for instance, it is well established that viruses generate viral‐derived dsRNAs as a consequence of: (i) viral replication; (ii) the tendency of RNA to fold in hairpin‐like structures; and/or (iii) the transcription of bidirectional mRNAs. These dsRNAs are first recognized and processed by RNase III‐like enzymes belonging to the Dicer family, which cut them into viral‐derived short interfering RNA (vsiRNA) duplexes of 21–24 nucleotides in length. Analogously, another batch of these vsiRNAs derives from newly synthesized dsRNAs generated by the action of RNA‐dependent RNA polymerases (RDRs). After stabilization via HUA enhancer 1 (HEN1)‐mediated methylation of their 3′ ends, vsiRNA duplexes are recruited by Argonaute (AGO)‐containing complexes, where only the so‐called ‘guide strand’ is retained to further direct the complex towards complementary RNA/DNA sequences in order to promote silencing (Zhang et al., 2015). A basic description of the antiviral silencing pathway against plant RNA viruses is illustrated in Fig. 2.
Figure 2.
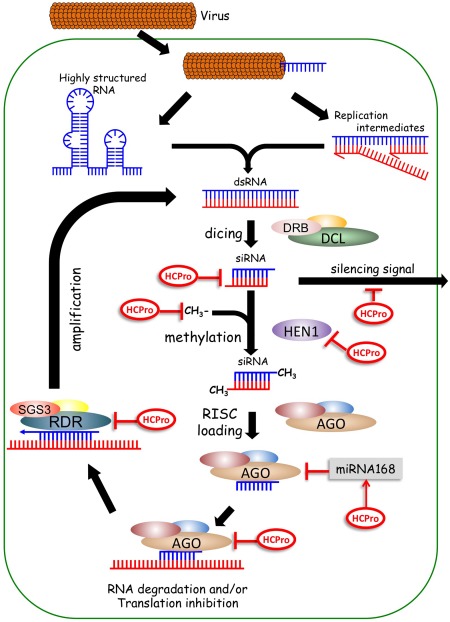
Potential targets of helper component proteinase (HCPro) in the antiviral RNA silencing pathway. Simplified schematic representation of the RNA silencing‐mediated defences in plants that are deployed against RNA viruses. Steps of the cascade at which HCPro from different potyvirids may be acting in order to block this defensive response are indicated. AGO, Argonaute protein; DCL, Dicer‐like protein; DRB, double‐stranded RNA‐binding protein; HEN1, HUA enhancer 1; RDR, RNA‐dependent RNA polymerase; RISC, RNA‐induced silencing complex; SGS3, supressor of gene silencing 3.
During their evolution, viruses have had to develop ways to fight back against RNA silencing in order to survive. The most effective strategy appears to be that based on the expression of viral proteins, called RNA silencing suppressors (RSSs), with the capacity to block or interfere with antiviral silencing. The HCPro protein from members of the genus Potyvirus was indeed the first RSS to be described (Anandalakshmi et al., 1998; Kasschau and Carrington, 1998). Many studies since then have revealed that HCPro can counteract the silencing‐based defensive barrier by targeting multiple steps of the cascade (Fig. 2 and Table 1). Interestingly, some of these studies have also shown that only HCPro from members of the genera Potyvirus and Rymovirus appears to have RNA silencing suppression activity, whereas this function relies on another protein in members of the remaining genera (Giner et al., 2010; Mingot et al., 2016; Tatineni et al., 2012; Untiveros et al., 2016; Young et al., 2012).
Table 1.
Helper component proteinase (HCPro)‐targeted steps of the antiviral RNA silencing pathway.
| Targeted step | Molecular mechanism | Potyvirus | References |
|---|---|---|---|
| vsiRNA uploading | Sequestration of vsiRNAs | TEV, PPV, PRSV, ZYMV, TuMV | Garcia‐Ruiz et al. (2015); Lakatos et al. (2006); Sahana et al. (2014); Shiboleth et al. (2007); Valli et al. (2015) |
| vsiRNA methylation | Inhibition of production | PVY, PVA | Cañizares et al. (2013); Ivanov et al. (2016); Soitamo et al. (2011) |
| Binding and inactivation of HEN1 | ZYMV | Jamous et al. (2011) | |
| Effector | Down‐regulation of AGO1 | TEV | Varallyay and Havelda (2013) |
| Interaction with AGO1 | PVA | Ivanov et al. (2016) | |
| Amplification | Down‐regulation of RDR6 | SCMV | Zhang et al. (2008) |
| Movement of silencing signal | Sequestration of siRNAs? | PVY, TEV | Delgadillo et al. (2004); Hamilton et al. (2002); Pfeffer et al. (2002) |
| Induction of endogenous silencing suppressors? | Interaction with rgs‐CaM and RAV2 | TEV, TuMV | Anandalakshmi et al. (2000); Endres et al. (2010) |
AGO1, Argonaute protein 1; HEN1, HUA enhancer 1; PPV, Plum pox virus; PRSV, Papaya ringspot virus; PVA, Potato virus A; PVY, Potato virus Y; RDR6, RNA‐dependent RNA polymerase 6; rgs‐CaM, calmodulin‐related protein; SCMV, Sugarcane mosaic virus; TEV, Tobacco etch virus; TuMV, Turnip mosaic virus; vsiRNA, viral‐derived short interfering RNA; ZYMV, Zucchini yellow mosaic virus.
The molecular mechanism by which HCPro interferes with antiviral silencing remained elusive until Lakatos et al. (2006) found that, similar to the well‐characterized tombusviral RSS P19, the Tobacco etch virus (TEV) HCPro prevents the loading of vsiRNAs into the silencing effector complexes by direct binding to these molecules in a size‐specific manner. Although vsiRNA sequestration seems to be a quite common anti‐silencing mechanism for the HCPros of diverse potyviruses, other non‐mutually exclusive alternatives have been proposed (Table 1). For instance, HCPro has been found to interfere with the methylation of the vsiRNA 3′ end either by the inhibition of the production of the methyl group through disturbance of the methionine cycle (Ivanov et al., 2016; Soitamo et al., 2011), or by direct interaction with and inhibition of HEN1 (Jamous et al., 2011). Interference with AGO‐containing effector complexes was also described for the HCPro expressed by TEV and Potato virus A (PVA). In the first case, TEV HCPro takes advantage of the homeostatic self‐regulation properties of the host RNA silencing pathway (Mallory and Vaucheret, 2010) and enhances the expression of miR168 with the consequent down‐regulation of its endogenous targets, which include the mRNA of the antiviral AGO1 (Varallyay and Havelda, 2013). In the second case, PVA HCPro interacts directly with AGO1 in ribosomal complexes, supporting the idea that this RSS is somehow able to alleviate the putative translational repression of the potyviral genome mediated by RNA silencing (Ivanov et al., 2016). Furthermore, HCPro can interfere with the RDR‐mediated amplification step, as in the case of the Sugarcane mosaic virus (SCMV) HCPro, which down‐regulates RDR6 by interfering with the transcription of RDR6 mRNA (Zhang et al., 2008). Finally, it has also been observed that HCPro blocks a long‐distance silencing signal that moves ahead of the viral infection (Delgadillo et al., 2004; Hamilton et al., 2002; Pfeffer et al., 2002). Based on previous results (Lewsey et al., 2016; Melnyk et al., 2011; Molnar et al., 2010), it is reasonable to hypothesize that vsiRNAs move through the whole plant via the vascular system, and that HCPro‐mediated blockage of the long‐distance silencing signal relies on direct vsiRNA interaction and sequestration at the infected tissues.
Host factors are also relevant for HCPro‐mediated silencing suppression. Such is the case for the tobacco rgs‐CaM, a calmodulin‐related protein which interacts directly with TEV HCPro and works as an endogenous RSS (eRSS) (Anandalakshmi et al., 2000). Moreover, Endres et al. (2010) found that the Related to ABI3/VP1 2 (RAV2) ethylene‐induced transcription factor from Arabidopsis thaliana is required for the anti‐silencing activity of the Turnip mosaic virus (TuMV) HCPro. They observed that HCPro interacts with RAV2 and induces the transcription of certain putative eRSSs, including the calmodulin‐related protein CML38, which seems to be the A. thaliana homologue of the above‐mentioned tobacco rgs‐CaM. Altogether, these results raise the possibility that HCPro recruits eRSSs in a direct (protein–protein interaction) and/or indirect (by RAV2‐mediated transcriptional activation) fashion in order to interfere with host defence mechanisms mediated by RNA silencing. Intriguingly, results from other experiments, which are discussed below, indicate that HCPro–rgs‐CaM interaction certainly targets this viral protein for degradation (Nakahara et al., 2012).
As the different RNA silencing modules in plants partially overlap, viral RSSs usually interfere not only with the antiviral part, but also with those modules controlling plant developmental programmes. Indeed, the presence of pleiotropic developmental defects, associated with disturbances in miRNA function, in transgenic plants constitutively expressing HCPro, supports this assumption (Chapman et al., 2004; Kasschau et al., 2003; Mallory et al., 2002) and renders feasible the idea that the silencing suppression activity of HCPro causes some of the observed potyviral‐induced disease symptoms in infected plants. Mlotshwa et al. (2005) observed that overexpression of Dicer‐like protein 1, the enzyme responsible for miRNA synthesis, rescued the developmental anomalies caused by HCPro, but did not correct defects in miRNA pathways. This suggests that disturbance in one or a few miRNA‐controlled factors, rather than general impairments in miRNA function, underlies the HCPro‐associated developmental disorders. In agreement with this suggestion, misregulation of AUXIN RESPONSE FACTOR 8 by miR167 was concluded to be the main cause of developmental abnormalities induced by HCPro and other viral silencing suppressors (Jay et al., 2011). However, more recent results challenge this conclusion (Mlotshwa et al., 2016).
Whether HCPro interference with diverse RNA silencing modules is a collateral effect of silencing suppression or a deliberate viral strategy to favour the infection process is still a matter of debate. In this regard, synthetic evolution experiments offer an attractive opportunity to analyse these two options. Torres‐Barceló et al. (2008), for instance, introduced several mutations on TEV HCPro and tested not only the effects of these changes on RNA silencing suppression activity, but also, later, on the infection of tobacco plants (Torres‐Barceló et al., 2010), the natural TEV host. Hence, they found that HCPro hypersuppressor variants rapidly evolved towards variants with moderate, wild‐type‐like, anti‐silencing capacity, suggesting that this HCPro activity is indeed fine tuned during TEV infection to minimize the unwanted side‐effects of silencing blockage on normal plant developmental patterns (Torres‐Barceló et al., 2010).
Structure Versus Function—HCPro is a Multidomain Viral Protein
After the discovery of its contribution to aphid‐mediated plant‐to‐plant transmission, another function was ascribed to HCPro: the maturation of viral factors by releasing itself from the rest of the polyprotein. Bacterial and in vitro studies provided evidence that HCPro is a cis‐acting proteinase that functions co‐translationally and independently of a plant factor, with the cleavage site between a glycine dipeptide at its C‐terminus (Carrington et al., 1989a, 1989b). Genetic analyses by site‐directed mutagenesis further characterized two residues, one cysteine and one histidine, as the catalytic diad for proteolytic activity, categorizing HCPro in the cysteine‐type proteinase family (Oh and Carrington, 1989). Further analyses defined the consensus cleavage sequence surrounding the glycine dipeptide at the HCPro C‐terminus to be YXVGG (positions P4 to P1′) (Carrington and Herndon, 1992). HCPro is currently classified in the C6 peptidase superfamily (Rawlings et al., 2016).
Together with the characterization of the protease domain, amino acids and motifs relevant for aphid transmission, movement, RNA binding and RNA silencing suppression were also examined. Schematically, HCPro can be divided into three domains (the indicated positions correspond to TEV HCPro): an N‐terminal part (amino acids 1–100) required for aphid transmission; a central region (amino acids 101–299) in charge of RNA silencing suppression and other functions; and a C‐terminal domain (amino acids 300–459) harbouring the proteolytic activity of HCPro (Hasiów‐Jaroszewska et al., 2014) (Fig. 3A). As mentioned above, a zinc finger‐like domain located at the N‐terminus of HCPro, which includes the KITC motif, is associated with potyviral aphid‐mediated transmission (Atreya and Pirone, 1993; Atreya et al., 1992) (Fig. 3A). The specific involvement in helping the transmission of the N‐terminal part was also supported by the emergence of spontaneous TEV, Lettuce mosaic virus (LMV) and Onion yellow dwarf virus deletion mutants which, even lacking the first 89, 108 or 92 amino acids of HCPro, respectively, were able to complete the whole viral infection cycle, except for propagation by aphids (Dolja et al., 1993; German‐Retana et al., 2000; Takaki et al., 2006).
Figure 3.
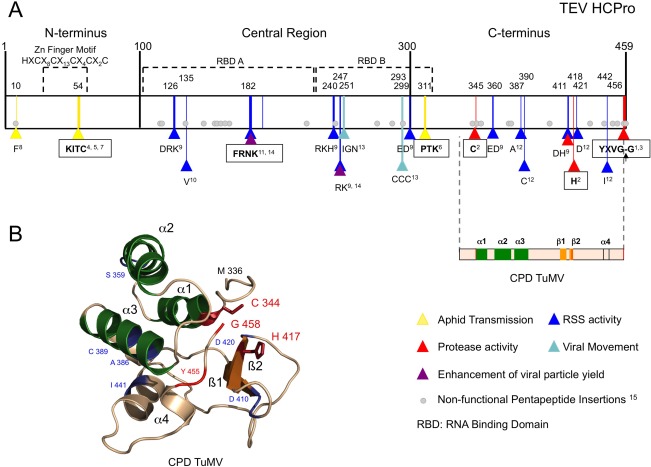
Helper component proteinase (HCPro) structural and functional features. (A) Schematic representation of a representative potyviral HCPro (from Tobacco etch virus, TEV) divided into three main regions. The best‐characterized motifs are shown in squares. Amino acids relevant for a given function, which are conserved at least among the Potyvirus genus, are marked with triangles at their corresponding positions. Amino acids relevant for viral movement (marked in light blue) were described before the characterization of HCPro RNA silencing suppression activity; therefore, their real role might be misassigned. Pentapeptide insertions that render Plum pox virus (PPV) HCPro poorly functional or non‐functional as an RNA silencing suppressor (RSS) are depicted as grey circles at the equivalent TEV HCPro positions. A two‐dimensional representation of the Turnip mosaic virus HCPro structure solved by Guo et al. (2011) encompasses the equivalent C‐terminal region of TEV HCPro. Superscript numbers indicate the following references: 1Carrington et al. (1989a); 2Oh and Carrington (1989); 3Carrington and Herndon (1992); 4Atreya et al. (1992); 5Atreya and Pirone (1993); 6Huet et al. (1994); 7Dolja et al. (1993); 8Blanc et al. (1998); 9Kasschau and Carrington (2001); 10González‐Jara et al. (2005); 11Shiboleth et al. (2007); 12Torres‐Barceló et al. (2008); 13Cronin et al. (1995); 14Valli et al. (2014); 15Varrelmann et al. (2007). (B) Crystal structure of the cysteine protease domain of Turnip mosaic virus HCPro (Guo et al., 2011; PDB code 3RNV). The corresponding L and R domains of papain‐like proteases would be represented by the α‐helices shown in green and the β‐sheets shown in orange, respectively. Those amino acids highlighted in (A) are also indicated in (B).
Cronin et al. (1995) described two motifs in the central region of HCPro relevant for viral movement. Two years later, using a series of alanine‐scanning mutants built in a TEV‐GUS chimeric virus background, Kasschau et al. (1997) described several amino acids relevant for genome amplification and long‐distance movement that were located mainly in the central region of HCPro. In 2001, and after HCPro was characterized as an RSS, the same group found a strong correlation between silencing suppression and the genome amplification and movement defects they had observed in the alanine‐scanning mutants (Kasschau and Carrington, 2001). They also showed that proteinase and anti‐silencing activities worked independently in most studied cases. This indicates that the proteinase function per se is not needed for RNA silencing suppression. However, there was a mutation located at the C‐terminal part of the protein which disturbed both proteolytic activity and RNA silencing suppression, which demonstrates that the protease domain is also required for silencing suppression activity, for instance, to provide the protein with appropriate folding. Furthermore, experiments of scanning mutagenesis via pentapeptide insertions in the Plum pox virus (PPV) HCPro (Varrelmann et al., 2007) or point amino acid substitutions in TEV HCPro (Torres‐Barceló et al., 2008) also support a key role of the protein central domain for RNA silencing suppression, and corroborate the idea of inter‐domain interactions. On the other hand, a study on Papaya ringspot virus (PRSV) showed that the amino terminal part of HCPro is involved in the systemic infection of zucchini (Yap et al., 2009) (Fig. 3A), which is in agreement with the results obtained previously by Atreya and Pirone. (1993) in Tobacco vein mottling virus. All of these findings suggest that HCPro from distinct viruses may have different inter‐domain interactions, and such interplay between domains may be relevant from structural and functional points of view.
Some early studies attributed to HCPro the ability to bind nucleic acids in a sequence‐non‐specific manner (Maia and Bernardi, 1996; Merits et al., 1998). The involvement of the central region of HCPro in RNA binding was further described using different deletion mutants (Urcuqui‐Inchima et al., 2000). This study divided the central region of the Potato virus Y (PVY) HCPro into domains A and B, which bind RNA in vitro independently (Fig. 3A). Remarkably, Lakatos et al. (2006) showed that the RNA silencing suppression activity of TEV HCPro involved siRNA binding (see above) and, later, the conserved FRNK motif, which overlaps with the RNA‐binding domain A, was shown to be relevant for HCPro–siRNA interaction (Shiboleth et al., 2007; Wu et al., 2010). Moreover, a study based on HCPro from PPV described that this protein also works as an enhancer of viral particle yield (see below). Mutational analyses located the relevant amino acids for this novel activity also in the central region of HCPro (Valli et al., 2014) (Fig. 3A).
Even in the early reports on HCPro, it was proposed that this viral protein normally adopts a complex quaternary structure (Thornbury et al., 1985). This idea was later supported by diverse studies on the self‐interaction of HCPro from PVA, PVY and LMV, in which crucial motifs for oligomerization were found by yeast two‐hybrid assay at both the N‐terminal and C‐terminal parts of the protein (Guo et al., 1999; Urcuqui‐Inchima et al., 1999a, 1999b). Similar results were obtained years later for TuMV HCPro using bimolecular fluorescence complementation assays (Zheng et al., 2011). Plisson et al. (2003) studied this matter more precisely via protein purification from infected plants and the characterization of both wild‐type and an N‐terminal deletion mutant of LMV HCPro, which lacks its first 100 amino acids. As the full‐length protein and the shorter version were observed in size exclusion analysis to behave as a dimer or trimer in solution, the authors concluded that the N‐terminus of LMV HCPro is not involved in self‐interaction. Furthermore, chemical crosslinking confirmed the presence of dimers, tetramers and higher order oligomers in solution, whereas the observation of two‐dimensional crystals by electron microscopy showed the appearance of dimers that bound to form tetramers. In agreement with earlier observations regarding the role of cations in HCPro stabilization, crystal formation occurred only in the presence of Mg2+. Additional structural studies, which were conducted with TEV HCPro purified from infected plants and observed by electron microscopy, confirmed the oligomerization states mentioned above. Although dimers, tetramers and hexamers of HCPro were indeed observed in solution, an adjusted model proposed that, at least in the particular case of TEV, the self‐interaction between monomers occurs on a V‐shaped conformation with HCPro located in an antiparallel orientation (Ruiz‐Ferrer et al., 2005).
The most recent data regarding the structural features of HCPro come from a three‐dimensional crystal structure solved by Guo et al. (2011) corresponding to 158 amino acids, including the protease domain, from TuMV HCPro (Fig. 3B). This peptide was produced in bacteria, and the formation of oligomers was actively avoided in order to facilitate crystal formation, so that structural questions regarding dimerization are still unanswered. In any case, the atomic structure of amino acids 336–458 showed several features of high interest. First, it confirmed the identity of the previously proposed protease catalytic diad and established the presence of the C‐terminal glycine tightly bound to the enzymatic cleft. This observation might indeed explain the exclusive cis‐acting mode of HCPro, as the terminal glycine would occupy the space needed for the catalytic site to remain active. Unfortunately, attempts by Guo et al. (2011) to remove this amino acid in order to make the proteinase active in trans, as was later performed for the CP serine proteinase of alphaviruses (Aggarwal et al., 2014), were unsuccessful. Second, the overall structure of this domain allowed for accurate comparisons with existing structures of other cysteine‐like proteinases, such as papain, indicating that the HCPro atomic arrangement differs significantly from the distinctive papain‐like folding. It presents a highly reduced four‐helical domain that harbours the catalytic cysteine and in which helices α1–α3 roughly cover the L domain of papain (Fig. 3B, in green); it also contains a small β‐barrel that carries the catalytic histidine in which strands β1–β2 would match the R domain of papain (Fig. 3B, in orange). Intriguingly, comparison with other cysteine proteinases revealed clear similarities between HCPro and the alphavirus nsP2 protein, as both have a compact fold with similar secondary structural topology. All in all, the atomic model of this domain represents the perfect opportunity to become more fully acquainted with its proteinase activity. Previous studies using high doses of human cystatin C (García et al., 1993) and phytocystatins and human stefin A (Wen et al., 2004) have shown inhibition of the HCPro proteolytic activity in vitro, and genetically modified plants expressing oryzacystatin I have been proven to be resistant to TEV and PVY infection (Gutierrez‐Campos et al., 1999). Now, with a molecular structure of the protease domain available, it should be possible to design novel chemicals aiming to disturb HCPro self‐cleavage as an effective antiviral strategy.
Bacterially expressed HCPro is also useful to raise antisera, allowing the study of protein subcellular localization. For instance, antiserum to PPV HCPro recognized not only this protein, but also the HCPro from 10 other potyviral species, and was able to: (i) label amorphous inclusions in the cytoplasm of plant cells infected with PPV, PRSV, Pepper mottle virus and Tobacco vein mottling virus; (ii) label pinwheels in cells infected with Bean yellow mosaic virus and Clover yellow vein virus (ClYVV); (iii) give scattered signals in the cytoplasm of cells infected with Bidens mottle virus; and (iv) highlight nuclear inclusions in cells infected with TEV and Beet mosaic virus (Riedel et al., 1998). Similarly, HCPro from Cowpea aphid‐borne mosaic virus was used to prepare antiserum for immunofluorescence assays, which showed diffuse distribution of the protein in the cytoplasm of naturally infected cells (Mlotshwa et al., 2002). Bimolecular fluorescence complementation assays located transiently expressed TuMV HCPro oligomers diffused in the cytoplasm of plant cells and/or associated in granules along the endoplasmic reticulum (Zheng et al., 2011; Zilian and Maiss, 2011). The most recent and thorough examination of subcellular localization comes from a study performed by del Toro et al. (2014) with the PVY HCPro fused to diverse fluorophores. In addition to a diffuse presence of this viral protein in the cytoplasm, they also observed distinct protein distributions (e.g. amorphous cytoplasmic inclusions containing α‐tubulin, dot‐like inclusions distributed regularly throughout the cytoplasm and associated with the endoplasmic reticulum and the microtubule cytoskeleton, and all over the microtubules) that were influenced by the environmental conditions. Altogether, these results suggest that HCPro might not be attached to one single region inside infected cells; instead, its location may change during the infection cycle in order to cope with its multiple functions and/or as a response to external changes. The spatial/temporal distribution of HCPro as well as the putative link between this potentially dynamic subcellular localization and diverse HCPro functions deserve further study.
Additional Roles of HCPro—The Advantage of Being Promiscuous
HCPro interacts with several host and viral proteins and, because most of these interactions appear to be unrelated to the three well‐known roles of this viral factor, namely aphid transmission, viral polyprotein processing and suppression of host antiviral RNA silencing, it has been proposed that such interactions are part of additional, much less characterized, functions of HCPro during potyvirid infections (Table 2 summarizes these interactions and the hypothetical role that they play during the infection cycle). For instance, it has been shown that HCPro from several potyviruses interacts and modulates the activity of the host proteasome. Ballut et al. (2005), who proposed this role for the first time, found that LMV HCPro binds to and inhibits the activity of the 20S proteasome. Surprisingly, the presence of HCPro only inhibits the RNase activity of this multicatalytic complex, which targets in vitro the viral RNA genome for degradation, whereas the proteolytic activity of the 20S proteasome is either unchanged or even slightly stimulated. Later, it was described that PVY HCPro interacts with the PAA, PBB and PBE subunits of the A. thaliana 20S proteasome, but not with the PAE subunit, which certainly carries ribonuclease activity (Jin et al., 2007a). However, Dielen et al. (2011) were later able to detect the interaction between LMV HCPro and PAE in diverse systems, even in the context of an LMV infection in lettuce. Similar studies with PRSV demonstrated that the proteasome inhibitor MG132 has a positive effect on PRSV accumulation in papaya, and that PRSV HCPro, similar to PVY HCPro, interacts with PAA, but not with the PAE subunit of the papaya 20S proteasome (Sahana et al., 2012). Moreover, additional experiments by Sahana et al. (2012) indicated that the PAA and PAE subunits interact with each other. Thus, these authors mitigated discrepancies with the HCPro–PAE interaction and its consequences by proposing that: (i) binding between HCPro and PAA may be sufficient to disturb the RNase activity of PAE or may prevent the interaction of the PAA and PAE subunits; and (ii) HCPro from different potyviruses might interact with different components of the 20S proteasome, depending on the specific plant–virus combination (Sahana et al., 2012). All in all, results from the above‐mentioned studies suggest that the 20S proteasome works as another defence layer against members of the Potyviridae family, and that HCPro interferes with the proteasome activity as a viral counteractive measure.
Table 2.
Diverse activities of the helper component proteinase (HCPro) whose roles in viral infection have not been fully characterized.
| Activity | Function | Hypothetical aims | Virus | References |
|---|---|---|---|---|
| Interaction with PAA, PBB, PBE or PAE proteasome subunits | Inhibition of the 20S proteasome | Counter‐action of a proteasome‐based plant defence mechanism |
LMV PVY PRSV |
Ballut et al. (2005); Dielen et al. (2011); Jin et al. (2007b); Sahana et al. (2012) |
|
Interaction with NtMinD, NtDXS, CF1β‐subunit of chloroplast ATP synthase, Ferrodoxin‐5 |
Reduction of the photosynthesis rate | General weakening of the host |
PVY SCMV |
Cheng et al. (2008); Gunasinghe and Berger (1991); Jin et al. (2007a); Li et al. (2015); Tu et al. (2015a, 2015b) |
| Interaction with PaCRT | Disturbance of Ca2+ binding to PaCRT | Blocking of the Ca2+‐mediated activation of host defences | PRSV | Shen et al. (2010) |
| Interaction with HIP2 | Blocking of HIP2 activity | Disturbance of certain signalling networks of defence responses |
PVA PVY TEV |
Haikonen et al. (2013a, 2013b) |
| Formation of cytoplasmic granules | Recruitment of both host and viral factors |
Overcoming of RNA silencing‐based defences Attainment of optimal viral translation |
PVA TuMV |
Hafrén et al. (2015) |
| Interaction with eIF4E/eIF(iso)4E | Recruitment of translation initiation factors | Attainment of optimal viral translation | PVA, PVY, TEV | Ala‐Poikela et al. (2011) |
| Interaction with VPg and CI | Protein allocation at the tip of virions |
Transmission or movement Attainment of optimal viral translation |
ClYVV WSMV PSbMV LMV PVA PPV |
Choi et al. (2000); Guo et al. (2001); Ivanov et al. (2016); Roudet‐Tavert et al. (2007); Yambao et al. (2003); Zilian and Maiss (2011) |
| Interaction with CP? | Proper formation of viral particles | Coordination of different stages of the viral infection cycle | PPV | Valli et al. (2014) |
ClYVV, Clover yellow vein virus; CP, coat protein; eIF4E, eukaryotic translation initiation factor 4E; eIF(iso)4E, eukaryotic translation initiation factor isoform 4E; HIP2, HCPro interacting protein 2; LMV, Lettuce mosaic virus; NtDXS, tobacco chloroplast protein 1‐deoxy‐d‐xylulose‐5‐phosphate synthase; PaCRT, papaya calreticulin; PPV, Plum pox virus; PRSV, Papaya ringspot virus; PSbMV, Pea seed‐borne mosaic virus; PVA, Potato virus A; PVY, Potato virus Y; SCMV, Sugarcane mosaic virus; TEV, Tobacco etch virus; TuMV, Turnip mosaic virus; WSMV, Wheat streak mosaic virus; ZYMV, Zucchini yellow mosaic virus.
Potyvirid infections frequently alter chloroplast number and morphology, leading to a decreased level of photosynthesis in infected tissue (Pompe‐Novak et al., 2001). Indeed, HCPro has been found previously to accumulate in chloroplasts of PVY‐infected tobacco cells (Gunasinghe and Berger, 1991), and further analyses have reported an interaction between a chloroplast protein, NtMinD, and PVY HCPro (Jin et al., 2007b). Given that homodimers of NtMinD participate in chloroplast division, PVY HCPro might prevent NtMinD self‐interaction, with a consequent alteration in chloroplast number (Jin et al., 2007b). Moreover, a recent study not only confirmed the presence of PVY HCPro in the chloroplast, but also showed that the ATPase activity of NtMinD is reduced in the presence of this viral protein (Tu et al., 2015b). Such observations allowed these authors to provide an explanation for the commonly observed abnormal morphology of chloroplasts in the presence of PVY. In a parallel study, Tu et al. (2015a) also found that PVY HCPro interacts in tobacco with the CF1β‐subunit of the chloroplast ATP synthase. Such an interaction leads to a decreased number of active enzymatic complexes, with a consequent overall reduction in ATP synthesis in the chloroplast of both HCPro transgenic and PVY‐infected tobacco plants, which, in the end, reduces the net photosynthetic rate. The interaction between HCPro and the tobacco chloroplast protein 1‐deoxy‐d‐xylulose‐5‐phosphate synthase (NtDXS) has been described recently (Li et al., 2015). As NtDXS is a limiting enzyme for plastidic isoprenoid biosynthesis in plants (Estévez et al., 2001), an effect of this interaction in the production of diverse isoprenoids, such as chlorophylls, tocopherols, carotenoids or abscisic acid (ABA), is expected. Certainly, PVY HCPro enhances the activity of NtDXS, thereby boosting the isoprenoid biosynthesis pathway, with a consequent increase in the level of certain pigments, ABA and ABA‐responsive genes (Li et al., 2015). Moreover, Cheng et al. (2008) showed that SCMV HCPro interacts with the maize chloroplast precursor, but not with the mature form, of ferrodoxin‐5. Therefore, this interaction might disturb the post‐translational import of ferrodoxin‐5 into maize chloroplasts, which would then lead to the perturbation of chloroplast structure and function. However, although evidence for the implication of HCPro in chloroplast distortion, photosynthesis reduction and alteration of isoprenoid metabolism in infected plants is very strong, the meanings of these HCPro‐mediated effects for virus infection remain unclear.
PRSV HCPro binds to the papaya calreticulin (PaCRT) protein, in particular with its calcium‐binding domain located at the protein C‐terminus, whereas PRSV infection enhances PaCRT transcription in the early days post‐infection (Shen et al., 2010). Given that Ca2+ is considered to be an essential second messenger that participates in many plant signal pathways, including defence signalling (Zhang et al., 2014), HCPro might disturb the calcium‐binding capacity of PaCRT and thereby mitigate the activation of downstream pathways (Shen et al., 2010).
PVA HCPro has been found to interact with the HCPro interacting protein 2 (HIP2) from Solanum tuberosum and Nicotiana tabacum, two natural hosts of PVA (Haikonen et al., 2013b). Moreover, as a positive interaction was also observed for HCPro from PVY and TEV, which have a similar host range to PVA, but not for HCPro from Pea seed‐borne mosaic virus (PSbMV), which infects just a few species in the Solanaceae family, a role of this interaction in virus–host specificity was proposed (Haikonen et al., 2013a). HIP2 is a microtubule‐associated protein similar to A. thaliana SPR2 and, as evidence of the importance of the HCPro–HIP2 interaction for viral infection, depletion of this host factor or mutations in HCPro abolishing HIP2 binding reduced the PVA titre in different hosts. Although the precise functional role of this interaction is currently unknown, SPR2 interacts with: (i) many receptor‐like kinases associated with plant innate immunity and (ii) two transcription factors related to immune responses (Mukhtar et al., 2011). This led Haikonen et al. (2013a, 2013b) to hypothesize that HIP2 controls certain signalling networks of defence responses, and that HCPro might subvert this controller, via protein–protein interaction, to the benefit of the virus.
PVA induces the formation of small aggregates containing the acidic ribosomal protein P0 in the cytoplasm of infected cells, referred to as PVA‐induced granules (PGs) (Hafrén et al., 2015). The formation of PGs is specifically triggered by HCPro and, in addition to P0, they contain HCPro, the RNA silencing effector AGO1, the oligouridylate‐binding protein 1, varicose, an isoform of translation initiation factor 4E [eIF(iso)4E] and even the viral RNA genome (Hafrén et al., 2015). Notably, only anti‐silencing‐proficient HCPro variants have been shown to promote the formation of PGs, as observed by direct mutagenesis. Based on these results, and the known link between host proteins located in PGs and the viral VPg, the authors proposed that the formation of these granules is required to overcome RNA silencing‐based defences via the relocation of AGO1 towards PGs, and to achieve optimal viral expression mediated by VPg (Hafrén et al., 2015).
Ala‐Poikela et al. (2011) found clear evidence of direct interaction between the HCPro from three different potyviruses (PVA, PVY and TEV) and the translation initiation factors eIF(iso)4E and eIF4E from potato and tobacco. Moreover, a putative eIF4E‐binding motif was identified at the C‐terminal part of PVA HCPro, which showed a high degree of conservation among other potyviruses. Certainly, the disruption of this motif by direct mutagenesis had a negative impact on HCPro–eIF4E binding and was detrimental to the virulence of PVA, supporting the idea that such an interaction plays an important, yet unknown, role during viral infection (Ala‐Poikela et al., 2011). However, this inference should be taken with some caution, as a further study showed that this mutation strongly reduced the RNA silencing suppression activity of PVA HCPro (Hafrén et al., 2015).
HCPro interacts with itself (discussed above) and with some of the other viral proteins. Physical interaction between HCPro and VPg has been described for different potyviruses (Ivanov et al., 2016; Roudet‐Tavert et al., 2007; Yambao et al., 2003), suggesting that the joint action of these two proteins might play a general role during potyviral infections. Intriguingly, as already mentioned, Torrance et al. (2006) showed the presence of a protruding tip at one end in a fraction of potyviral virions, which was suggested to be formed by HCPro in association with VPg. The authors discussed that this interaction might play a role in aphid‐mediated, plant‐to‐plant transmission or even in cell‐to‐cell movement. Moreover, different lines of evidence have shown that interaction of HCPro with VPg involves the same central domain of the latter protein that interacts with eIF4E (Roudet‐Tavert et al., 2007; Yambao et al., 2003) Indeed, HCPro and eIF4E from LMV and lettuce, respectively, compete for VPg binding (Roudet‐Tavert et al., 2007). The outstanding relevance of the VPg–eIF4E (in its two isoforms) interaction for potyvirid infections has been extensively studied as a model of plant recessive resistance (Robaglia and Caranta, 2006; Truniger and Aranda, 2009; Wang and Krishnaswamy, 2012). The most accepted, but not yet demonstrated, model proposes that VPg works as a pseudo‐cap structure that recruits translation complexes for viral use. As already mentioned, HCPro and eIF4E also interact with each other (Ala‐Poikela et al., 2011), so that the deduction of the actual role of HCPro in this protein trio seems complicated. HCPro might be part of the translational complex that is recruited by VPg at the 5′ end of the viral genome to either carry out an unknown function or, in line with the silencing suppression activities of both viral proteins (Rajamäki and Valkonen, 2009), to interfere with the hypothetical inhibition of virus translation mediated by host‐deployed RNA silencing defences, as proposed recently (Ivanov et al., 2016).
Interaction of HCPro with the CI protein of quite a large number of potyvirids has also been detected using different experimental systems (Choi et al., 2000; Guo et al., 2001; Ivanov et al., 2016; Zilian and Maiss, 2011). CI is a multifunctional RNA helicase that participates in viral replication and cell‐to‐cell movement (Sorel et al., 2014) and, as for HCPro, it is attached to the tip at one end of a fraction of viral particles, at least in the case of PVA (Gabrenaite‐Verkhovskaya et al., 2008). It is possible to envisage a scenario in which HCPro somehow collaborates with CI in virus cell‐to‐cell movement, or even that HCPro moves between adjacent cells, as part of a ribonucleic complex, to exert any of its multiple functions in a newly infected neighbour cell, as suggested previously (Rojas et al., 1997).
Given the well‐established role of HCPro in viral plant‐to‐plant transmission, at least for members of the genera Potyvirus and Tritimovirus, the interaction between HCPro and CP is the most evident among potyvirid proteins. As expected, such binding has been detected in diverse viruses by different methods (Blanc et al., 1997; Guo et al., 2001; Kang et al., 2004; Lin et al., 2009; Peng et al., 1998; Roudet‐Tavert et al., 2002; Seo et al., 2010). Intriguingly, the HCPro–CP interaction has also been detected in aphid non‐transmissible potyviruses (Manoussopoulos et al., 2000; Roudet‐Tavert et al., 2002), suggesting the existence of a functional role for this protein–protein complex different from aphid‐mediated transmission. In agreement with this hypothesis, Valli et al. (2014) found that HCPro plays a key role in PPV infection by enhancing the yield of full‐length viral particles. This novel function of HCPro is not linked to its other main activities, as observed by direct mutagenesis. Furthermore, this activity appears to be highly specific, meaning that HCPro would act only on its cognate CP. Although the exact molecular mechanism by which HCPro enhances the yield of intact virions is currently unknown, the authors proposed two non‐mutually exclusive possibilities, both agreeing with the known localization of HCPro at the end of the viral particles: (i) HCPro is involved in the initial steps of the assembly of CP subunits; and/or (ii) HCPro stabilizes viral particles once they are fully assembled. They also speculated about how the spatiotemporal availability of HCPro might function as a device that coordinates different stages of the viral cycle, namely translation, replication and encapsidation, in the infected cell (Valli et al., 2014). Indeed, a recent report has located HCPro in 6K2‐induced replication vesicles in PVA‐infected plants (Lõhmus et al., 2016).
HCPro as a Trigger and Target of Plant Defence Responses—Defence, Counter‐Defence, Counter‐Counter‐Defence
Given the outstanding importance of HCPro in multiple steps of viral infection, it is not surprising that its recognition by the host might induce mechanisms to counteract its action and trigger other defence responses. Moreover, as with the proviral activities of HCPro, the antiviral reactions elicited by HCPro can also contribute to the development of disease symptoms (García and Pallás, 2015).
Defence responses triggered by HCPro can be non‐specific. For instance, Pruss et al. (2004) showed that, whereas TEV HCPro suppresses RNA silencing‐related antiviral defences, it confers enhanced broad‐spectrum resistance against multiple pathogens, including heterologous viruses, via both salicylic acid (SA)‐dependent and SA‐independent mechanisms. Evidence for the alteration of SA‐mediated defences as a consequence of TEV HCPro expression in transgenic lines was also provided by Alamillo et al. (2006). The enhancement of host defence responses induced by potyviral HCPro appears to be temperature dependent (Shams‐Bakhsh et al., 2007). More recent results have suggested that HCPro might enhance the expression of defence‐related genes in the SA pathway by reducing the DNA methylation at their promoter regions, which is associated with a drastic reduction of siRNAs derived from these sequences (Yang et al., 2016).
HCPro also induces more specific defence responses. Namely, this viral protein can act as an elicitor of R gene‐driven effector‐triggered immunity. This is the case for some strains of PVY, which induce a hypersensitive response (HR) that restricts the virus in necrotic local lesions in potato cultivars harbouring the dominant resistance genes Nctbr and Nytbr (Moury et al., 2011; Tian and Valkonen, 2015). These resistance genes appear to recognize similar structural determinants in the central region of HCPro of PVY° (Nytbr) and PVYC (Nctbr) strains (Tian and Valkonen, 2013, 2015). PVY isolates overcoming Nytbr often cause veinal necrosis in tobacco, and some determinants of this phenotype have been identified in HCPro (Faurez et al., 2012; Tribodet et al., 2005). However, avirulence determinants of Nytbr are different from those responsible for the induction of veinal necrosis (Tian and Valkonen, 2015).
Some PVY isolates induce necrotic symptoms in potato tubers and a mutation in HCPro linked to the ability to induce tuber necrosis is also involved in the induction of veinal necrosis in tobacco (Glais et al., 2015; Tribodet et al., 2005). There is no evidence that veinal necrosis in tobacco and potato tuber necrosis are HR‐like responses to specific interactions between avirulence factors. The fitness decrease caused by point mutations associated with the acquisition of necrosis properties in tobacco may suggest that the necrotic reaction is associated with a defensive response (Rolland et al., 2009). However, the fact that these mutations also have a fitness cost in a host that does not show necrotic symptoms questions this conclusion.
Necrotic symptoms have also been observed in tobacco plants infected with PVA modified by mutations in a highly variable region of the central part of the HCPro protein (Haikonen et al., 2013a). These mutations, which affect interactions with a microtubule‐associated protein (see above for HCPro–HIP2 interaction) and have been suggested to cause conformational changes in adjacent regions of the protein, were associated with a reduction in viral accumulation and the induction of many defence‐related genes, including ethylene‐ and jasmonic acid‐inducible genes, at the onset of necrosis (Haikonen et al., 2013a). Taken together, these data suggest a scenario in which alterations in HCPro conformation by mutations that overcome R gene‐mediated specific resistance affect functional interactions with other host factors and induce alternative defence responses (Tian and Valkonen, 2015).
Another example of a resistance gene elicited by HCPro of a potyvirus is a gene located at the complex Rsv1 locus of soybean, probably belonging to the nucleotide‐binding site‐leucine‐rich repeat (NBS‐LRR) class, which recognizes the HCPro of Soybean mosaic virus (SMV) (Eggenberger et al., 2008; Hajimorad et al., 2008; Wen et al., 2013). The precise mechanism involved in the induction of resistance by SMV HCPro is still unclear. The HCPro‐responsive resistance gene alone allows limited replication at the inoculation site. However, the complete Rsv1 cluster, which includes at least one additional, P3‐responsive, SMV resistance gene, confers extreme resistance against avirulent SMV variants (Wen et al., 2013). The first identified SMV isolate able to overcome the resistance conferred by the Rsv1 locus caused a lethal systemic HR phenotype, probably as a result of a weak interaction of the viral avirulence factors and the host resistance genes (Hajimorad et al., 2005). HCPro probably contributes to this phenotype, as some SMV isolates carrying mutations at HCPro also provoked systemic HR in soybean plants only containing the HCPro‐responsive gene of the Rsv1 cluster (Wen et al., 2013), and a single amino acid substitution in this viral protein allowed virulent SMV to cause severe rugosity and local necrotic lesions, instead of lethal systemic HR, in soybean expressing the complete Rsv1 cluster (Seo et al., 2011). Interestingly, the gain of virulence of SMV on the Rsv1 soybean genotype had a fitness penalty in susceptible rsv1 plants, and this trade‐off was a consequence of the mutations introduced in HCPro during the adaptation to the resistance selective pressure (Khatabi et al., 2013). This observation emphasizes the convenience for the host of triggering antiviral defences against important multifunctional proteins, as this strategy might cause a high global fitness cost, even extinction, for the escaping viruses.
Some of the HCPro contributions to the induction of host defence responses may be indirect. It has been reported that ClYVV activates SA signalling and HR‐related pathways, causing systemic necrosis and plant death, in pea containing Cyn1, a gene mapped in a genomic region that corresponds to an R‐gene‐analogue gene cluster in the genome of Medicago truncatula (Ravelo et al., 2007). Indeed, point mutations in ClYVV HCPro that attenuate RNA silencing suppression activity and symptom expression in broad bean (Yambao et al., 2008) reduce the ability of ClYVV to activate the SA signalling pathway and to induce cell death in Cyn1‐containing plants (Atsumi et al., 2009). Although these results might suggest that ClYVV HCPro itself is the elicitor of the Cyn1‐controlled response, the authors consider that it is more likely that the reduced activity of the mutated HCPro limits viral amplification and, subsequently, the accumulation of the host factor(s) triggering the defence response (Atsumi et al., 2009). A similar scenario, in which reduced HCPro activity maintains viral amplification below the levels that induce host detrimental effects, has been proposed to explain why a PPV mutant with unrestricted P1–HCPro processing causes more severe symptoms with lower accumulation levels than the wild‐type virus (Pasin et al., 2014).
The destruction of HCPro is another defence response deployed by the plant to counteract RNA silencing suppression and other activities of this important virulence factor. As mentioned above, the calmodulin‐related protein rgs‐CaM from tobacco was identified as a host factor that interacts with TEV HCPro and contributes by itself to suppress RNA silencing (Anandalakshmi et al., 2000). More recently, it has been observed that binding of rgs‐CaM to the dsRNA‐binding domains of different viral RSSs, including HCPro from ClYVV, directs them to the autophagy‐like pathway for degradation (Nakahara et al., 2012). Therefore, although HCPro–rgs‐CaM interaction is soundly supported by all the experimental evidence available, the integration of both positive and negative effects of this interaction on the suppression of RNA silencing in a comprehensive model is still missing.
The Diversity of HCPro and HCPro‐Like Proteins—Similar but Different
The Potyviridae family comprises viruses from eight different genera. Most of the studies presented here have been carried out with HCPro from species of the Potyvirus genus, which is by far the most abundant. In members of this genus, the N‐terminal part of viral polyproteins follows the same pattern: a P1 leader serine proteinase that processes itself to separate from HCPro, which, in turn, cleaves at its C‐terminus to be released from the rest of the polyprotein (Fig. 4A). In potyviruses, as well as in members of the genus Rymovirus, HCPro has a molecular weight of around 50 kDa, and those motifs described in this review are predominantly conserved. As described above, the most outstanding feature of HCPro from poty‐ and rymoviruses is its ability to suppress RNA silencing. To date, the only discovered exceptions to this rule are the sweet potato‐infecting potyviruses, which express an apparently normal HCPro variant that has no evident anti‐silencing activity. Even more surprising is the fact that all of these viruses express an atypically long P1 with a viral polymerase slippage site that generates an extra open reading frame (ORF), termed PISPO. This new ORF gives rise to a transframe protein, named P1N‐PISPO, with RNA silencing suppression activity (Clark et al., 2012; Li et al., 2012; Mingot et al., 2016; Untiveros et al., 2016).
Figure 4.
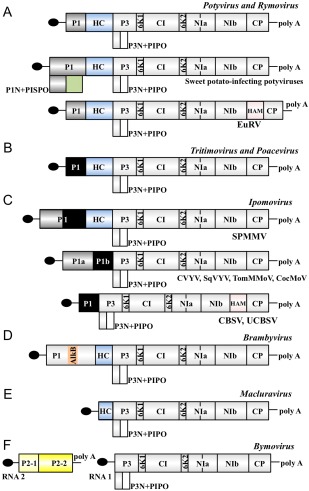
Schematic representation of genomic organization in viruses from different genera of the family Potyviridae. The long open reading frame is shown as a box divided into mature viral products. PIPO open reading frame is indicated as the box below P3. The terminal protein VPg is depicted as a black ellipse. P1a and P1a‐like proteins are represented by grey boxes, whereas P1b and P1b‐like proteins are represented by black boxes. Features that are not shared by all potyvirids are highlighted in different colours. (A) Potyvirus and Rymovirus genera. The PISPO open reading frame in sweet potato‐infecting potyviruses is indicated as a pale green box below P1. The extra protein HAM between NIb and CP in Euphorbia ringspot virus (Knierim et al., 2017) is highlighted in pink. (B) Tritimovirus and Poacevirus. (C) Ipomovirus. The diversity among members of this genus has been reviewed (Dombrovsky et al., 2014). A HAM extra protein (in pink) was also present in a subset of ipomoviruses. (D) Brambyvirus. The AlkB domain in P1 from Blackberry virus Y is highlighted in pale orange. (E) Macluravirus. (F) Bymovirus. RNA2, unique in the Potyviridae family, is highlighted in yellow.
Poacevirus and Tritimovirus are two related genera bearing HCPro of similar or slightly reduced size compared with poty‐ and rymoviruses (Fig. 4A and B). Although these four genera share the same genome organization, the RNA silencing suppression activity is exerted by P1, instead of HCPro, in poace‐ and tritimoviruses, and the HCPro of the tritimovirus WSMV is not needed for virus viability (Stenger et al., 2005a, 2007; Tatineni et al., 2012; Young et al., 2012). These observations are in perfect agreement with sequence comparison data, showing that strong similarities among HCPro variants from viruses of these four genera are only displayed at the protease domain (C‐terminal region) (Guo et al., 2011). In contrast, the central region of poace‐ and tritimoviral HCPros, where the anti‐silencing activity mainly maps in potyviruses, is very different and does not contain the typical FRNK motif.
The most diverse potyvirids with regard to genome organization at the 5′ end are those belonging to the genus Ipomovirus, which can be divided into two groups based on the presence or absence of HCPro (Fig. 4C). The first ipomovirus species to be described was Sweet potato mild mottle virus (SPMMV) (Colinet et al., 1998), a virus that encodes an unusually large P1 protein that works as an RSS (Giner et al., 2010). Interestingly, this virus codes for an HCPro that is similar in size to that of potyviruses, but contains no RNA silencing suppression activity. Phylogenetic analyses aligned SPMMV closer to tritimoviruses than to other potyvirids (Stenger et al., 1998) and, as expected, SPMMV HCPro lacks sequence similarity with potyviral HCPros outside the protease domain. Ipomoviruses without HCPro have one P1 copy (Mbanzibwa et al., 2009) or two divergent P1 copies in tandem (Desbiez et al., 2016; Janssen et al., 2005; Li et al., 2008; Valli et al., 2006) at the N‐terminal part of the viral polyprotein. Remarkably, as in the case of SPMMV, all ipomoviruses lacking HCPro use P1 as an RSS.
Susaimuthu et al. (2008) identified and fully sequenced Blackberry virus Y, which was classified as the founder member of a new potyvirid genus, named Brambyvirus (Fig. 4D). Downstream of an unusual P1, the Blackberry virus Y genome codes for an atypical HCPro, reduced in size (36 kDa) and bearing in common with HCPro from other potyvirids only the cysteine protease domain. It is still unknown which protein from this virus, if any, blocks the RNA silencing‐based defences deployed by the host.
Bymovirus is the only bipartite genus of the Potyviridae family (Fig. 4F). Bymovirus RNA1 codes for a polyprotein that starts at a protein homologous to the potyviral P3 and follows the Potyviridae genomic pattern until the 3′ untranslated region (UTR) (Kashiwazaki et al., 1990). The bymovirus RNA2 codes for two proteins: the second is not related to any of the potyvirid proteins, but the first is described as HCPro‐like because of its cysteine proteinase domain (Kashiwazaki et al., 1991). This protein (P2‐1) is very small (28 kDa) and has no other motifs that relate it to other potyvirid HCPros.
The first member of the genus Macluravirus to be fully sequenced was Chinese yam necrotic mosaic virus (Kondo and Fujita, 2012). This virus presents the smallest monopartite genome in the family Potyviridae (Fig. 4E). It lacks a P1 leader proteinase and codes for an HCPro of just 29 kDa. Whether or not this protein has RSS activity is still unknown. Macluraviral HCPro appears to be more similar to bymoviral P2‐1 than to other potyvirid HCPro.
The closest relative of HCPro outside the Potyviridae family can be found in the picorna‐like, fungal‐infecting, hypoviruses. The sequence similarity, putative active site and cleavage site composition relate HCPro to p29 and p48 cysteine proteinases of Cryphonectria hypoviruses (Choi et al., 1991a, 1991b; Shapira and Nuss, 1991). A study performed by Suzuki et al. (1999) mapped the p29 symptom determinants outside the protease domain, in a region within the N‐terminus of the protein. This domain contains four cysteine residues, similar to the conserved residues in the zinc finger domain of HCPro, which are essential for virus viability. Moreover, both p29 and HCPro proteins alter host developmental processes when expressed in the absence of virus infection (Suzuki et al., 2003). Even more important is the fact that p29 has synergistic effects with other fungal viruses (Sun et al., 2006), probably linked, as in the case of HCPro, to the RNA silencing suppression activity that is displayed by p29 in the natural fungal host and in plants (Segers et al., 2006).
Unrelated plant viruses encoding proteins similar to HCPro can be found in the Closteroviridae family. They belong to the Sindbis virus‐like supergroup and share in common with potyvirids the presence of leader proteinases with C‐terminal papain‐like domains, which are also multifunctional factors with apparent crucial domain interplay. Unlike poty‐ and rymoviral HCPro, these leader proteinases seem to lack RNA silencing suppression activity, but are certainly involved in genome amplification and participate in cell‐to‐cell movement (Peng et al., 2001). Proteins related to HCPro are also found in animal viruses. Such is the case of alphaviruses, which, similar to closteroviruses, also belong to the Sindbis virus‐like supergroup, and encode leader proteinases sharing remarkable structural homology with HCPro at the level of its cysteine protease domain (Guo et al., 2011).
Future Perspectives—Looking Forward
The genome organization of viruses belonging to the Potyviridae family is highly conserved in a large core region that starts at the P3 cistron. Coincidentally, mature proteins encoded at this viral segment are all released from polyprotein precursors by proteolytic processing conducted by the NIapro protease (Valli et al., 2015). In contrast, the upstream genomic region is highly variable, even among members of the same genus, and encodes proteins that are liberated from the polyprotein precursors by self‐cleavage. Thus, it is tempting to speculate that an ancient potyvirid precursor had a simplified genome that only coded for the NIapro‐processed module. Although a sound and confident prediction of the evolutionary history of Potyviridae is beyond the scope of this review, we dare to speculate that the first step towards contemporary potyvirids would have been the acquisition of a second genome element in a bymovirus, including what would have been the first HCPro‐related protein: the P2‐1 cysteine proteinase. Either as a subsequent step from a bymovirus, or as a parallel event from the proposed potyvirid ancestor, an HCPro‐like gene would have been incorporated into the viral genome to give rise to the simplest modern monopartite potyvirid: a macluravirus. Then, further evolution would have boosted HCPro size, diversity and functional complexity.
However, what was the primordial function of the proto‐HCPro? We do not know, but it is unlikely that such a small protein was able to suppress silencing or to help transmission by aphids or other vectors. We do not even know whether or not this function is still conserved by the currently large HCPros from different potyvirid genera. Research in the barely studied HCPro from macluraviruses and brambyviruses, as well as in P2‐1 from bymoviruses, could certainly shed some light on the evolutionary path, not only of these multifunctional viral proteins, but also of the entire viral family.
HCPro is quite a well‐conserved protein in members of the genus Potyvirus for which the nucleotide sequences of this factor have been determined so far; therefore, the large diversity of this factor within the entire Potyviridae family is surprising. This could be explained by the assumption that the primordial HCPro was a recently acquired accessory factor, having some flexibility to evolve and incorporate new functions. In this scenario, HCPro could adopt diverse activities in the different evolutionary lineages from which the potyvirid genera originated. Moreover, several new activities could pyramid in a single protein, as occurred with the HCPro of potyviruses, although the coupling among different protein functions might restrict its ability to evolve (Hasiów‐Jaroszewska et al., 2014). As HCPro has been shown to be the elicitor and target of different plant defence responses the escape from these responses should also limit its potential to evolve.
The fact that engineered members of the Potyvirus genus depleted of HCPro are unable to infect wild‐type plants, but can infect RNA silencing‐deficient plants, and that unrelated RSSs are able to functionally replace HCPro, indicates that the main function of the present potyviral HCPro is the suppression of RNA silencing‐mediated antiviral defences (Carbonell et al., 2012; Garcia‐Ruiz et al., 2010; Maliogka et al., 2012). However, further studies using systems biology approaches will be required to decipher the contribution to the overall silencing suppression of the HCPro activities somehow related to this function (Table 1).
Although the silencing suppression‐unrelated activities of HCPro are not absolutely essential, they have been shown to be relevant for viral infection. Further characterization of these activities to understand how they are integrated into the infection process also needs to be the target of future studies. Indeed, the development of appropriate real‐time imaging techniques that allow the unveiling of HCPro localization dynamics in the infected cell would be especially helpful for this aim.
In spite of being the first HCPro function identified, very little is known about how HCPro plays its role as a bridge during aphid transmission. The identification of the HCPro receptor in the aphid stylet and the characterization of the dynamics of virion–HCPro–aphid interactions that govern both the acquisition and release of viral particles by insects are among the most interesting future challenges of HCPro research.
Finally, whereas the crystal structure of the protease domain of a potyviral HCPro has been solved at 2.0 Å resolution (Guo et al., 2011), no high‐resolution structure of the complete HCPro is currently available. Solving the structure of HCPro alone and bound to viral and host co‐factors, or even bound to nucleic acids (e.g. siRNAs, miRNAs), would be of great value to understand the multiple functions of this amazing protein.
Acknowledgements
We are very grateful to the two anonymous reviewers for their invaluable advice during manuscript editing. Work in the authors’ laboratories was funded by grants from the Ministerio de Economía, Industria y Competitividad (BIO2013‐49053‐R, BIO2016‐80572‐R and Plant KBBE PCIN‐2013‐056 to J.A.G., AGL2013‐42537‐R and AGL2016‐75529‐R to J.J.L.‐M., and BIO2015‐73900‐JIN to A.A.V.). We acknowledge the support of the “Severo Ochoa Programme for Centres of Excellence in R&D” of the Ministerio de Economía, Industria y Competitividad to CNB (SEV‐2013‐0347) and CRAG (SEV‐2015‐0533). Research in CRAG is also supported by CERCA institution (Generalitat de Catalunya).
Contributor Information
Adrián A. Valli, Email: avalli@cnb.csic.es
Juan Antonio García, Email: jagarcia@cnb.csic.es.
References
- Aggarwal, M. , Dhindwal, S. , Kumar, P. , Kuhn, R.J. and Tomar, S. (2014) Trans‐protease activity and structural insights into the active form of the alphavirus capsid protease. J. Virol. 88, 12 242–12 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlquist, P. , Noueiry, A.O. , Lee, W.M. , Kushner, D.B. and Dye, B.T. (2003) Host factors in positive‐strand RNA virus genome replication. J. Virol. 77, 8181–8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamillo, J.M. , Sáenz, P. and García, J.A. (2006) Salicylic acid‐mediated and RNA‐silencing defense mechanisms cooperate in the restriction of systemic spread of plum pox virus in tobacco. Plant J. 48, 217–227. [DOI] [PubMed] [Google Scholar]
- Ala‐Poikela, M. , Goytia, E. , Haikonen, T. , Rajamaki, M.‐L. and Valkonen, J.P.T. (2011) Helper component proteinase of genus Potyvirus is an interaction partner of translation initiation factors eIF(iso)4E and eIF4E that contains a 4E binding motif. J. Virol. 85, 6784–6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anandalakshmi, R. , Pruss, G.J. , Ge, X. , Marathe, R. , Mallory, A.C. , Smith, T.H. and Vance, V.B. (1998) A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA, 95, 13 079–13 084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anandalakshmi, R. , Marathe, R. , Ge, X. , Herr, J.M. Jr , Mau, C. , Mallory, A. , Pruss, G. , Bowman, L. and Vance, V.B. (2000) A calmodulin‐related protein that suppresses posttranscriptional gene silencing in plants. Science, 290, 142–144. [DOI] [PubMed] [Google Scholar]
- Atkins, J.F. , Loughran, G. , Bhatt, P.R. , Firth, A.E. and Baranov, P.V. (2016) Ribosomal frameshifting and transcriptional slippage: from genetic steganography and cryptography to adventitious use. Nucleic Acids Res. 44, 7007–7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atreya, C.D. and Pirone, T.P. (1993) Mutational analysis of the helper component‐proteinase gene of a potyvirus: effects of amino acid substitutions, deletions, and gene replacement on virulence and aphid transmissibility. Proc. Natl. Acad. Sci. USA, 90, 11 919–11 923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atreya, C.D. , Raccah, B. and Pirone, T.P. (1990) A point mutation in the coat protein abolishes aphid transmissibility of a potyvirus. Virology, 178, 161–165. [DOI] [PubMed] [Google Scholar]
- Atreya, C.D. , Atreya, P.L. , Thornbury, D.W. and Pirone, T.P. (1992) Site‐directed mutations in the potyvirus HC‐Pro gene affect helper component activity, virus accumulation, and symptom expression in infected tobacco plants. Virology, 191, 106–111. [DOI] [PubMed] [Google Scholar]
- Atreya, P.L. , Atreya, C.D. and Pirone, T.P. (1991) Amino acid substitutions in the coat protein result in loss of insect transmissibility of a plant virus. Proc. Natl. Acad. Sci. USA, 88, 7887–7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atreya, P.L. , Lopez‐Moya, J.J. , Atreya, C.D. and Pirone, T.P. (1995) Mutational analysis of the coat protein N‐terminal amino acids involved in potyvirus transmission by aphids. J. Gen. Virol. 76, 265–270. [DOI] [PubMed] [Google Scholar]
- Atsumi, G. , Kagaya, U. , Kitazawa, H. , Nakahara, K.S. and Uyeda, I. (2009) Activation of the salicylic acid signaling pathway enhances Clover yellow vein virus virulence in susceptible pea cultivars. Mol. Plant–Microbe Interact. 22, 166–175. [DOI] [PubMed] [Google Scholar]
- Ballut, L. , Drucker, M. , Pugnière, M. , Cambon, F. , Blanc, S. , Roquet, F. , Candresse, T. , Schmid, H.P. , Nicolas, P. , Le Gall, O. and Badaoui, S. (2005) HcPro, a multifunctional protein encoded by a plant RNA virus, targets the 20S proteasome and affects its enzymic activities. J. Gen. Virol. 86, 2595–2603. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D.C. , Lloyd, J. , Manoussopoulos, I.N. , Roberts, I.M. and Harrison, B.D. (1993) Signal for potyvirus‐dependent aphid transmission of potato aucuba mosaic virus and the effect of its transfer to potato virus X. J. Gen. Virol. 74, 1245–1253. [DOI] [PubMed] [Google Scholar]
- Berger, P.H. , Hunt, A.G. , Domier, G.M. , Hellman, G.M. , Stram, Y. , Thornbury, D.W. and Pirone, T.P. (1989) Expression in transgenic plants of a viral gene product that mediates insect transmission of potyviruses. Proc. Natl. Acad. Sci. USA, 86, 8402–8406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc, S. , López‐Moya, J.J. , Wang, R.Y. , García‐Lampasona, S. , Thornbury, D.W. and Pirone, T.P. (1997) A specific interaction between coat protein and helper component correlates with aphid transmission of a potyvirus. Virology, 231, 141–147. [DOI] [PubMed] [Google Scholar]
- Blanc, S. , Ammar, E.D. , Garcia‐Lampasona, S. , Dolja, V.V. , Llave, C. , Baker, J. and Pirone, T.P. (1998) Mutations in the potyvirus helper component protein: effects on interactions with virions and aphid stylets. J. Gen. Virol. 79, 3119–3122. [DOI] [PubMed] [Google Scholar]
- Blanc, S. , Dolja, V.V. , Llave, C. and Pirone, T.P. (1999) Histidine‐tagging and purification of tobacco etch potyvirus helper component protein. J. Virol. Methods, 77, 11–15. [DOI] [PubMed] [Google Scholar]
- Blanc, S. , Drucker, M. and Uzest, M. (2014) Localizing viruses in their insect vectors. Annu. Rev. Phytopathol. 52, 403–425. [DOI] [PubMed] [Google Scholar]
- Bologna, N.G. and Voinnet, O. (2014) The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis . Annu. Rev. Plant Biol. 65, 473–503. [DOI] [PubMed] [Google Scholar]
- Bradley, R.H.E. (1952) Studies on the aphid transmission of a strain of henbane mosaic virus. Ann. Appl. Biol. 39, 78–97. [Google Scholar]
- Bronkhorst, A.W. and van Rij, R.P. (2014) The long and short of antiviral defense: small RNA‐based immunity in insects. Curr. Opin. Virol. 7, 19–28. [DOI] [PubMed] [Google Scholar]
- Canto, T. , López‐Moya, J.J. , Serra‐Yoldi, M.T. , Díaz‐Ruiz, J.R. and López‐Abella, D. (1995) Different helper component mutations associated with lack of aphid transmissibility in two isolates of potato virus Y. Phytopathology, 85, 1519–1524. [Google Scholar]
- Cañizares, M.C. , Lozano‐Durán, R. , Canto, T. , Bejarano, E.R. , Bisaro, D.M. , Navas‐Castillo, J. and Moriones, E. (2013) Effects of the crinivirus coat protein‐interacting plant protein SAHH on post‐transcriptional RNA silencing and its suppression. Mol. Plant–Microbe Interact. 26, 1004–1015. [DOI] [PubMed] [Google Scholar]
- Carbonell, A. , Dujovny, G. , Garcia, J.A. and Valli, A. (2012) The Cucumber vein yellowing virus silencing suppressor P1b can functionally replace HCPro in Plum pox virus infection in a host‐specific manner. Mol. Plant–Microbe Interact. 25, 151–164. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C. and Herndon, K.L. (1992) Characterization of the potyviral HC‐Pro autoproteolytic cleavage site. Virology, 187, 308–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington, J.C. , Cary, S.M. , Parks, T.D. and Dougherty, W.G. (1989a) A second proteinase encoded by a plant potyvirus genome. EMBO J. 8, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington, J.C. , Freed, D.D. and Sanders, T.C. (1989b) Autocatalytic processing of the potyvirus helper component proteinase in Escherichia coli and in vitro . J. Virol. 63, 4459–4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel, S.E. and Martienssen, R.A. (2013) RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 14, 100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, S.S. , Zhang, Z. and Liu, Y. (2012) RNA interference pathways in fungi: mechanisms and functions. Annu. Rev. Microbiol. 66, 305–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, E.J. , Prokhnevsky, A.I. , Gopinath, K. , Dolja, V.V. and Carrington, J.C. (2004) Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 18, 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y.Q. , Liu, Z.M. , Xu, J. , Zhou, T. , Wang, M. , Chen, Y.T. , Li, H.F. and Fan, Z.F. (2008) HC‐Pro protein of sugar cane mosaic virus interacts specifically with maize ferredoxin‐5 in vitro and in planta. J. Gen. Virol. 89, 2046–2054. [DOI] [PubMed] [Google Scholar]
- Choi, G.H. , Pawlyk, D.M. and Nuss, D.L. (1991a) The autocatalytic protease p29 encoded by a hypovirulence‐associated virus of the chesnut blight fungus resembles the potyvirus‐encoded protease HC‐Pro. Virology, 183, 747–752. [DOI] [PubMed] [Google Scholar]
- Choi, G.H. , Shapira, R. and Nuss, D.L. (1991b) Cotranslational autoproteolysis involved in gene expression from a double‐stranded RNA genetic element associated with hypovirulence of the chesnut blight fungus. Proc. Natl. Acad. Sci. USA, 88, 1167–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, I.R. , Stenger, D.C. and French, R. (2000) Multiple interactions among proteins encoded by the mite‐transmitted wheat streak mosaic tritimovirus. Virology, 267, 185–198. [DOI] [PubMed] [Google Scholar]
- Clark, C.A. , Davis, J.A. , Abad, J.A. , Cuellar, W.J. , Fuentes, S. , Kreuze, J.F. , Gibson, R.W. , Mukasa, S.B. , Tugume, A.K. , Tairo, F.D. and Valkonen, J.P.T. (2012) Sweetpotato viruses: 15 years of progress on understanding and managing complex diseases. Plant Dis. 96, 168–185. [DOI] [PubMed] [Google Scholar]
- Colinet, D. , Kummert, J. and Lepoivre, P. (1998) The nucleotide sequence and genome organization of the whitefly transmitted sweetpotato mild mottle virus: a close relationship with members of the family Potyviridae . Virus Res. 53, 187–196. [DOI] [PubMed] [Google Scholar]
- Cronin, S. , Verchot, J. , Haldeman‐Cahill, R. , Schaad, M.C. and Carrington, J.C. (1995) Long‐distance movement factor: a transport function of the potyvirus helper component proteinase. Plant Cell, 7, 549–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day, M.F. and Irzykiewicz, H. (1954) On the mechanism of transmission of non‐persistent phytopathogenic viruses by aphids. Aust. J. Biol. Sci. 7, 251–273. [DOI] [PubMed] [Google Scholar]
- Delgadillo, M.O. , Sáenz, P. , Salvador, B. , García, J.A. and Simón‐Mateo, C. (2004) Human influenza virus NS1 protein enhances viral pathogenicity and acts as an RNA silencing suppressor in plants. J. Gen. Virol. 85, 993–999. [DOI] [PubMed] [Google Scholar]
- del Toro, F. , Tena Fernández, F. , Tilsner, J. , Wright, K.M. , Tenllado, F. , Chung, B.N. , Praveen, S. and Canto, T. (2014) Potato virus Y HCPro localization at distinct, dynamically related and environment‐influenced structures in the cell cytoplasm. Mol. Plant–Microbe Interact. 27, 1331–1343. [DOI] [PubMed] [Google Scholar]
- De Mejia, M.V.G. , Hiebert, E. , Purcifull, D.E. , Thornbury, D.W. and Pirone, T.P. (1985) Identification of potyviral amorphous inclusion protein as a nonstructural virus‐specific protein related to helper component. Virology, 142, 34–43. [DOI] [PubMed] [Google Scholar]
- Desbiez, C. , Verdin, E. , Tepfer, M. , Wipf‐Scheibel, C. , Millot, P. , Dafalla, G. and Lecoq, H. (2016) Characterization of a new cucurbit‐infecting ipomovirus from Sudan. Arch. Virol. 161, 2913–2915. [DOI] [PubMed] [Google Scholar]
- Dielen, A.S. , Sassaki, F.T. , Walter, J. , Michon, T. , Menard, G. , Pagny, G. , Krause‐Sakate, R. , Maia Ide, G. , Badaoui, S. , Le Gall, O. , Candresse, T. and German‐Retana, S. (2011) The 20S proteasome alpha5 subunit of Arabidopsis thaliana carries an RNase activity and interacts in planta with the Lettuce mosaic potyvirus HcPro protein. Mol. Plant Pathol. 12, 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, S.W. (2010) RNA‐based antiviral immunity. Nat. Rev. Immunol. 10, 632–644. [DOI] [PubMed] [Google Scholar]
- Dolja, V.V. , Herndon, K.L. , Pirone, T.P. and Carrington, J.C. (1993) Spontaneous mutagenesis of a plant potyvirus genome after insertion of a foreign gene. J. Virol. 67, 5968–5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrovsky, A. , Gollop, N. , Chen, S.B. , Chejanovsky, N. and Raccah, B. (2007) In vitro association between the helper component‐proteinase of zucchini yellow mosaic virus and cuticle proteins of Myzus persicae . J. Gen. Virol. 88, 1602–1610. [DOI] [PubMed] [Google Scholar]
- Dombrovsky, A. , Reingold, V. and Antignus, Y. (2014) Ipomovirus – an atypical genus in the family Potyviridae transmitted by whiteflies. Pest Manag. Sci. 70, 1553–1567. [DOI] [PubMed] [Google Scholar]
- Dougherty, W.G. and Hiebert, E. (1980) Translation of potyvirus RNA in a rabbit reticulocyte lysate: identification of nuclear inclusion proteins as products of tobacco etch virus RNA translation and cylindrical inclusion protein as a product of the potyvirus genome. Virology, 104, 174–182. [DOI] [PubMed] [Google Scholar]
- Eggenberger, A.L. , Hajimorad, M.R. and Hill, J.H. (2008) Gain of virulence on Rsv1‐genotype soybean by an avirulent Soybean mosaic virus requires concurrent mutations in both P3 and HC‐Pro. Mol. Plant–Microbe Interact. 21, 931–936. [DOI] [PubMed] [Google Scholar]
- Endres, M.W. , Gregory, B.D. , Gao, Z. , Foreman, A.W. , Mlotshwa, S. , Ge, X. , Pruss, G.J. , Ecker, J.R. , Bowman, L.H. and Vance, V. (2010) Two plant viral suppressors of silencing require the ethylene‐inducible host transcription factor RAV2 to block RNA silencing. PLoS Pathog. 6, e1000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estévez, J.M. , Cantero, A. , Reindl, A. , Reichler, S. and Leon, P. (2001) 1‐Deoxy‐D‐xylulose‐5‐phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. J. Biol. Chem. 276, 22 901–22 909. [DOI] [PubMed] [Google Scholar]
- Eusebio‐Cope, A. and Suzuki, N. (2015) Mycoreovirus genome rearrangements associated with RNA silencing deficiency. Nucleic Acids Res. 43, 3802–3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faurez, F. , Baldwin, T. , Tribodet, M. and Jacquot, E. (2012) Identification of new Potato virus Y (PVY) molecular determinants for the induction of vein necrosis in tobacco. Mol. Plant Pathol. 13, 948–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández‐Calvino, L. , Goytia, E. , López‐Abella, D. , Giner, A. , Urizarna, M. , Vilaplana, L. and López‐Moya, J.J. (2010) The helper‐component protease transmission factor of tobacco etch potyvirus binds specifically to an aphid ribosomal protein homologous to the laminin receptor precursor. J. Gen. Virol. 91, 2862–2873. [DOI] [PubMed] [Google Scholar]
- Firth, A.E. and Brierley, I. (2012) Non‐canonical translation in RNA viruses. J. Gen. Virol. 93, 1385–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flasinski, S. and Cassidy, B.G. (1998) Potyvirus aphid transmission requires helper component and homologous coat protein for maximal efficiency. Arch. Virol. 143, 2159–2172. [DOI] [PubMed] [Google Scholar]
- Gabrenaite‐Verkhovskaya, R. , Andreev, I.A. , Kalinina, N.O. , Torrance, L. , Taliansky, M.E. and Mäkinen, K. (2008) Cylindrical inclusion protein of potato virus A is associated with a subpopulation of particles isolated from infected plants. J. Gen. Virol. 89, 829–838. [DOI] [PubMed] [Google Scholar]
- García, J.A. and Pallás, V. (2015) Viral factors involved in plant pathogenesis. Curr. Opin. Virol. 11, 21–30. [DOI] [PubMed] [Google Scholar]
- García, J.A. , Cervera, M.T. , Riechmann, J.L. and López‐Otín, C. (1993) Inhibitory effects of human cystatin C on plum pox potyvirus proteases. Plant Mol. Biol. 22, 697–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Ruiz, H. , Takeda, A. , Chapman, E.J. , Sullivan, C.M. , Fahlgren, N. , Brempelis, K.J. and Carrington, J.C. (2010) Arabidopsis RNA‐dependent RNA polymerases and dicer‐like proteins in antiviral defense and small interfering RNA biogenesis during Turnip mosaic virus infection. Plant Cell, 22, 481–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Ruiz, H. , Carbonell, A. , Hoyer, J.S. , Fahlgren, N. , Gilbert, K.B. , Takeda, A. , Giampetruzzi, A. , Garcia Ruiz, M.T. , McGinn, M.G. , Lowery, N. , Martinez Baladejo, M.T. and Carrington, J.C. (2015) Roles and programming of Arabidopsis ARGONAUTE proteins during Turnip mosaic virus infection. PLoS Pathog. 11, e1004755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German‐Retana, S. , Candresse, T. , Alias, E. , Delbos, R.P. and Le Gall, O. (2000) Effects of green fluorescent protein or β‐glucuronidase tagging on the accumulation and pathogenicity of a resistance‐breaking Lettuce mosaic virus isolate in susceptible and resistant lettuce cultivars. Mol. Plant–Microbe Interact. 13, 316–324. [DOI] [PubMed] [Google Scholar]
- Giner, A. , Lakatos, L. , García‐Chapa, M. , López‐Moya, J.J. and Burgyán, J. (2010) Viral protein inhibits RISC activity by argonaute binding through conserved WG/GW motifs. PLoS Pathog. 6, e1000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glais, L. , Faurez, F. , Tribodet, M. , Boulard, F. and Jacquot, E. (2015) The amino acid 419 in HC‐Pro is involved in the ability of PVY isolate N605 to induce necrotic symptoms on potato tubers. Virus Res. 208, 110–119. [DOI] [PubMed] [Google Scholar]
- González‐Jara, P. , Atencio, F.A. , Martínez‐García, B. , Barajas, D. , Tenllado, F. and Díaz‐Ruíz, J.R. (2005) A single amino acid mutation in the plum pox virus helper component‐proteinase gene abolishes both synergistic and RNA silencing suppression activities. Phytopathology, 95, 894–901. [DOI] [PubMed] [Google Scholar]
- Gorbalenya, A.E. , Enjuanes, L. , Ziebuhr, J. and Snijder, E.J. (2006) Nidovirales: evolving the largest RNA virus genome. Virus Res. 117, 17–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govier, D.A. and Kassanis, B. (1974a) Evidence that a component other than the virus particle is needed for aphid transmission of potato virus Y. Virology, 57, 285–286. [DOI] [PubMed] [Google Scholar]
- Govier, D.A. and Kassanis, B. (1974b) A virus‐induced component of plant sap needed when aphids acquire potato virus Y from purified preparations. Virology, 61, 420–426. [DOI] [PubMed] [Google Scholar]
- Govier, D.A. , Kassanis, B. and Pirone, T.P. (1977) Partial purification and characterization of the potato virus Y helper component. Virology, 78, 306–314. [DOI] [PubMed] [Google Scholar]
- Goytia, E. , Fernandez‐Calvino, L. , Martinez‐Garcia, B. , Lopez‐Abella, D. and Lopez‐Moya, J.J. (2006) Production of plum pox virus HC‐Pro functionally active for aphid transmission in a transient‐expression system. J. Gen. Virol. 87, 3413–3423. [DOI] [PubMed] [Google Scholar]
- Gunasinghe, U.B. and Berger, P.H. (1991) Association of potato virus Y gene products with chlorolasts in tobacco. Mol. Plant–Microbe Interact. 4, 452–457. [Google Scholar]
- Guo, D.Y. , Merits, A. and Saarma, M. (1999) Self‐association and mapping of interaction domains of helper component‐proteinase of potato A potyvirus. J. Gen. Virol. 80, 1127–1131. [DOI] [PubMed] [Google Scholar]
- Guo, D.Y. , Rajamaki, M.L. , Saarma, M. and Valkonen, J.P.T. (2001) Towards a protein interaction map of potyviruses: protein interaction matrixes of two potyviruses based on the yeast two‐hybrid system. J. Gen. Virol. 82, 935–939. [DOI] [PubMed] [Google Scholar]
- Guo, B. , Lin, J. and Ye, K. (2011) Structure of the autocatalytic cysteine protease domain of potyvirus helper‐component proteinase. J. Biol. Chem. 286, 21 937–21 943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez‐Campos, R. , Torres‐Acosta, J.A. , Saucedo‐Arias, L.J. and Gomez‐Lim, M.A. (1999) The use of cysteine proteinase inhibitors to engineer resistance against potyviruses in transgenic tobacco plants. Nat. Biotechnol. 17, 1223–1226. [DOI] [PubMed] [Google Scholar]
- Hafrén, A. , Lohmus, A. and Mäkinen, K. (2015) Formation of Potato virus A‐induced RNA granules and viral translation are interrelated processes required for optimal virus accumulation. PLoS Pathog. 11, e1005314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara‐Komoda, Y. , Choi, S.H. , Sato, M. , Atsumi, G. , Abe, J. , Fukuda, J. , Honjo, M.N. , Nagano, A.J. , Komoda, K. , Nakahara, K.S. , Uyeda, I. and Naito, S. (2016) Truncated yet functional viral protein produced via RNA polymerase slippage implies underestimated coding capacity of RNA viruses. Sci. Rep. 6, 21 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haikonen, T. , Rajamaki, M.L. , Tian, Y.P. and Valkonen, J.P. (2013a) Mutation of a short variable region in HCpro protein of Potato virus A affects interactions with a microtubule‐associated protein and induces necrotic responses in tobacco. Mol. Plant–Microbe Interact. 26, 721–733. [DOI] [PubMed] [Google Scholar]
- Haikonen, T. , Rajamäki, M.L. and Valkonen, J.P. (2013b) Interaction of the microtubule‐associated host protein HIP2 with viral helper component proteinase is important in infection with potato virus A. Mol. Plant–Microbe Interact. 26, 734–744. [DOI] [PubMed] [Google Scholar]
- Hajimorad, M.R. , Eggenberger, A.L. and Hill, J.H. (2005) Loss and gain of elicitor function of Soybean mosaic virus G7 provoking Rsv1‐mediated lethal systemic hypersensitive response maps to P3. J. Virol. 79, 1215–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajimorad, M.R. , Eggenberger, A.L. and Hill, J.H. (2008) Adaptation of Soybean mosaic virus avirulent chimeras containing P3 sequences from virulent strains to Rsv1‐genotype soybeans is mediated by mutations in HC‐Pro. Mol. Plant–Microbe Interact. 21, 937–946. [DOI] [PubMed] [Google Scholar]
- Hamilton, A. , Voinnet, O. , Chappell, L. and Baulcombe, D. (2002) Two classes of short interfering RNA in RNA silencing. EMBO J. 21, 4671–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, B.D. and Robinson, D.J. (1988) Molecular variation in vector‐borne plant viruses: epidemiological significance. Philos. Trans. R. Soc. London Ser. B: Biol. Sci. 321, 447–462. [DOI] [PubMed] [Google Scholar]
- Hasiów‐Jaroszewska, B. , Fares, M.A. and Elena, S.F. (2014) Molecular evolution of viral multifunctional proteins: the case of Potyvirus HC‐Pro. J. Mol. Evol. 78, 75–86. [DOI] [PubMed] [Google Scholar]
- Hiebert, E. , Thornbury, D.W. and Pironet, T.P. (1984) Immunoprecipitation analysis of potyviral in vitro translation products using antisera to helper component of tobacco vein mottling virus and potato virus Y. Virology, 135, 1–9. [DOI] [PubMed] [Google Scholar]
- Holmes, E.C. (2003) Error thresholds and the constraints to RNA virus evolution. Trends Microbiol. 11, 543–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, T. , Cui, Y. and Zhang, X. (2014) Involvement of viral microRNA in the regulation of antiviral apoptosis in shrimp. J. Virol. 88, 2544–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet, H. , Gal‐On, A. , Meir, E. , Lecoq, H. and Raccah, B. (1994) Mutations in the helper component protease gene of zucchini yellow mosaic virus affect its ability to mediate aphid transmissibility. J. Gen. Virol. 75, 1407–1414. [DOI] [PubMed] [Google Scholar]
- Ivanov, K.I. , Eskelin, K. , Basic, M. , De, S. , Lohmus, A. , Varjosalo, M. and Makinen, K. (2016) Molecular insights into the function of the viral RNA silencing suppressor HCPro. Plant J. 85, 30–45. [DOI] [PubMed] [Google Scholar]
- Jamous, R.M. , Boonrod, K. , Fuellgrabe, M.W. , Ali‐Shtayeh, M.S. , Krczal, G. and Wassenegger, M. (2011) The helper component‐proteinase of the Zucchini yellow mosaic virus inhibits the Hua Enhancer 1 methyltransferase activity in vitro . J. Gen. Virol. 92, 2222–2226. [DOI] [PubMed] [Google Scholar]
- Janssen, D. , Martín, G. , Velasco, L. , Gómez, P. , Segundo, E. , Ruiz, L. and Cuadrado, I.M. (2005) Absence of a coding region for the helper component‐proteinase in the genome of cucumber vein yellowing virus, a whitefly‐transmitted member of the Potyviridae . Arch. Virol. 150, 1439–1447. [DOI] [PubMed] [Google Scholar]
- Jay, F. , Wang, Y. , Yu, A. , Taconnat, L. , Pelletier, S. , Colot, V. , Renou, J.P. and Voinnet, O. (2011) Misregulation of AUXIN RESPONSE FACTOR 8 underlies the developmental abnormalities caused by three distinct viral silencing suppressors in Arabidopsis. PLoS Pathog. 7, e1002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Y.S. , Ma, D.Y. , Dong, J.L. , Jin, J.C. , Li, D.F. , Deng, C.W. and Wang, T. (2007a) HC‐Pro protein of Potato Virus Y can interact with three Arabidopsis 20S proteasome subunits in planta. J. Virol. 81, 12 881–12 888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Y.S. , Ma, D.Y. , Dong, J.L. , Li, D.F. , Deng, C.W. , Jin, J.C. and Wang, T. (2007b) The HC‐Pro protein of Potato virus Y interacts with NtMinD of tobacco. Mol. Plant–Microbe Interact. 20, 1505–1511. [DOI] [PubMed] [Google Scholar]
- Kamm, J.A. (1969) Change in transmissibility of Bean yellow mosaic virus by aphids. Ann. Entomol. Soc. Am. 62, 47–50. [Google Scholar]
- Kang, S.H. , Lim, W.S. and Kim, K.H. (2004) A protein interaction map of soybean mosaic virus strain G7H based on the yeast two‐hybrid system. Mol. Cells, 18, 122–126. [PubMed] [Google Scholar]
- Kashiwazaki, S. , Minobe, Y. , Omura, T. and Hibino, H. (1990) Nucleotide sequence of barley yellow mosaic virus RNA 1: a close evolutionary relationship with potyviruses. J. Gen. Virol. 71, 2781–2790. [DOI] [PubMed] [Google Scholar]
- Kashiwazaki, S. , Minobe, Y. and Hibino, H. (1991) Nucleotide sequence of barley yellow mosaic virus RNA2. J. Gen. Virol. 72, 995–999. [DOI] [PubMed] [Google Scholar]
- Kassanis, B. (1941) Transmission of tobacco etch viruses by aphides. Ann. Appl. Biol. 28, 238–243. [Google Scholar]
- Kassanis, B. (1961) The transmission of potato aucuba mosaic virus by aphids from plants also infected by potato viruses A or Y. Virology, 13, 93–97. [DOI] [PubMed] [Google Scholar]
- Kassanis, B. and Govier, D.A. (1971a) New evidence on the mechanism of aphid transmission of potato C and potato aucuba mosaic viruses. J. Gen. Virol. 10, 99–101. [DOI] [PubMed] [Google Scholar]
- Kassanis, B. and Govier, D.A. (1971b) The role of the helper virus in aphid transmission of potato aucuba mosaic virus and potato virus C. J. Gen. Virol. 13, 221–228. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D. and Carrington, J.C. (1998) A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell, 95, 461–470. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D. and Carrington, J.C. (2001) Long‐distance movement and replication maintenance functions correlate with silencing suppression activity of potyviral HC‐Pro. Virology, 285, 71–81. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D. , Cronin, S. and Carrington, J.C. (1997) Genome amplification and long‐distance movement functions associated with the central domain of tobacco etch potyvirus helper component‐proteinase. Virology, 228, 251–262. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D. , Xie, Z.X. , Allen, E. , Llave, C. , Chapman, E.J. , Krizan, K.A. and Carrington, J.C. (2003) P1/HC‐Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell, 4, 205–217. [DOI] [PubMed] [Google Scholar]
- Kendall, A. , McDonald, M. , Bian, W. , Bowles, T. , Baumgarten, S.C. , Shi, J. , Stewart, P.L. , Bullitt, E. , Gore, D. , Irving, T.C. , Havens, W.M. , Ghabrial, S.A. , Wall, J.S. and Stubbs, G. (2008) Structure of flexible filamentous plant viruses. J. Virol. 82, 9546–9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatabi, B. , Wen, R.H. and Hajimorad, M.R. (2013) Fitness penalty in susceptible host is associated with virulence of Soybean mosaic virus on Rsv1‐genotype soybean: a consequence of perturbation of HC‐Pro and not P3. Mol. Plant Pathol. 14, 885–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knierim, D. , Menzel, W. and Winter, S. (2017) Analysis of the complete genome sequence of euphorbia ringspot virus, an atypical member of the genus Potyvirus . Arch. Virol. 162, 291–293. [DOI] [PubMed] [Google Scholar]
- Kondo, T. and Fujita, T. (2012) Complete nucleotide sequence and construction of an infectious clone of Chinese yam necrotic mosaic virus suggest that macluraviruses have the smallest genome among members of the family Potyviridae . Arch. Virol. 157, 2299–2307. [DOI] [PubMed] [Google Scholar]
- Laín, S. , Riechmann, J.L. , Méndez, E. and García, J.A. (1988) Nucleotide sequence of the 3′ terminal region of plum pox potyvirus RNA. Virus Res. 10, 325–342. [DOI] [PubMed] [Google Scholar]
- Lakatos, L. , Csorba, T. , Pantaleo, V. , Chapman, E.J. , Carrington, J.C. , Liu, Y.P. , Dolja, V.V. , Calvino, L.F. , López‐Moya, J.J. and Burgyán, J. (2006) Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 25, 2768–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecoq, H. and Pitrat, M. (1985) Specificity of the helper‐component‐mediated aphid transmission of three potyviruses infecting muskmelon. Phytopathology, 75, 890–893. [Google Scholar]
- Lewsey, M.G. , Hardcastle, T.J. , Melnyk, C.W. , Molnar, A. , Valli, A. , Urich, M.A. , Nery, J.R. , Baulcombe, D.C. and Ecker, J.R. (2016) Mobile small RNAs regulate genome‐wide DNA methylation. Proc. Natl. Acad. Sci. USA, 113, E801–E810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F. , Xu, D. , Abad, J. and Li, R. (2012) Phylogenetic relationships of closely related potyviruses infecting sweet potato determined by genomic characterization of Sweet potato virus G and Sweet potato virus 2 . Virus Genes, 45, 118–125. [DOI] [PubMed] [Google Scholar]
- Li, H. , Ma, D. , Jin, Y. , Tu, Y. , Liu, L. , Leng, C. , Dong, J. and Wang, T. (2015) Helper component‐proteinase enhances the activity of 1‐deoxy‐D‐xylulose‐5‐phosphate synthase and promotes the biosynthesis of plastidic isoprenoids in Potato virus Y‐infected tobacco. Plant Cell Environ. 38, 2023–2034. [DOI] [PubMed] [Google Scholar]
- Li, H.W. , Li, W.X. and Ding, S.W. (2002) Induction and suppression of RNA silencing by an animal virus. Science, 296, 1319–1321. [DOI] [PubMed] [Google Scholar]
- Li, W.M. , Hilf, M.E. , Webb, S.E. , Baker, C.A. and Adkins, S. (2008) Presence of P1b and absence of HC‐Pro in Squash vein yellowing virus suggests a general feature of the genus Ipomovirus in the family Potyviridae . Virus Res. 135, 213–219. [DOI] [PubMed] [Google Scholar]
- Lin, L. , Shi, Y. , Luo, Z. , Lu, Y. , Zheng, H. , Yan, F. , Chen, J. , Chen, J. , Adams, M.J. and Wu, Y. (2009) Protein–protein interactions in two potyviruses using the yeast two‐hybrid system. Virus Res. 142, 36–40. [DOI] [PubMed] [Google Scholar]
- Llave, C. , Martínez, B. , Díaz‐Ruiz, J.R. and López‐Abella, D. (1999) Helper component mutations in nonconserved residues associated with aphid transmission efficiency of a pepper isolate of potato virus Y. Phytopathology, 89, 1176–1181. [DOI] [PubMed] [Google Scholar]
- Llave, C. , Martínez, B. , Díaz‐Ruiz, J.R. and López‐Abella, D. (2002) Amino acid substitutions within the Cys‐rich domain of the tobacco etch potyvirus HC‐Pro result in loss of transmissibility by aphids. Arch. Virol. 147, 2365–2375. [DOI] [PubMed] [Google Scholar]
- Lõhmus, A. , Varjosalo, M. and Mäkinen, K. (2016) Protein composition of 6K2‐induced membrane structures formed during Potato virus A infection. Mol. Plant Pathol. 17, 943–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López‐Moya, J.J. , Canto, T. , Díaz‐Ruíz, J.R. and López‐Abella, D. (1995) Transmission by aphids of a naturally non‐transmissible plum pox virus isolate with the aid of potato virus Y helper component. J. Gen. Virol. 76, 2293–2297. [DOI] [PubMed] [Google Scholar]
- Maia, I.G. and Bernardi, F. (1996) Nucleic acid‐binding properties of a bacterially expressed potato virus Y helper component‐proteinase. J. Gen. Virol. 77, 869–877. [DOI] [PubMed] [Google Scholar]
- Maliogka, V.I. , Calvo, M. , Carbonell, A. , García, J.A. and Valli, A. (2012) Heterologous RNA‐silencing suppressors from both plant‐ and animal‐infecting viruses support plum pox virus infection. J. Gen. Virol. 93, 1601–1611. [DOI] [PubMed] [Google Scholar]
- Mallory, A. and Vaucheret, H. (2010) Form, function, and regulation of ARGONAUTE proteins. Plant Cell, 22, 3879–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A.C. , Reinhart, B.J. , Bartel, D. , Vance, V.B. and Bowman, L.H. (2002) A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro‐RNAs in tobacco. Proc. Natl. Acad. Sci. USA, 99, 15 228–15 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoussopoulos, I.N. , Maiss, E. and Tsagris, M. (2000) Native electrophoresis and Western blot analysis (NEWeB): a method for characterization of different forms of potyvirus particles and similar nucleoprotein complexes in extracts of infected plant tissues. J. Gen. Virol. 81, 2295–2298. [DOI] [PubMed] [Google Scholar]
- Mbanzibwa, D.R. , Tian, Y.P. , Mukasa, S.B. and Valkonen, J.P.T. (2009) Cassava brown streak virus (Potyviridae) encodes a putative Maf/HAM1 pyrophosphatase implicated in reduction of mutations and a P1 proteinase that suppresses RNA silencing but contains no HC‐Pro. J. Virol. 83, 6934–6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnyk, C.W. , Molnar, A. , Bassett, A. and Baulcombe, D.C. (2011) Mobile 24 nt small RNAs direct transcriptional gene silencing in the root meristems of Arabidopsis thaliana . Curr. Biol. 21, 1678–1683. [DOI] [PubMed] [Google Scholar]
- Merits, A. , Guo, D.Y. and Saarma, M. (1998) VPg, coat protein and five non‐structural proteins of potato A potyvirus bind RNA in a sequence‐unspecific manner. J. Gen. Virol. 79, 3123–3127. [DOI] [PubMed] [Google Scholar]
- Mingot, A. , Valli, A. , Rodamilans, B. , San León, D. , Baulcombe, D.C. , García, J.A. and López‐Moya, J.J. (2016) The P1N‐PISPO trans‐frame gene of sweet potato feathery mottle potyvirus is produced during virus infection and functions as an RNA silencing suppressor. J. Virol. 90, 3543–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlotshwa, S. , Verver, J. , Sithole‐Niang, I. , Gopinath, K. , Carette, J. , Van Kammen, A.B. and Wellink, J. (2002) Subcellular location of the helper component‐proteinase of Cowpea aphid‐borne mosaic virus . Virus Genes, 25, 207–216. [DOI] [PubMed] [Google Scholar]
- Mlotshwa, S. , Schauer, S.E. , Smith, T.H. , Mallory, A.C. , Herr, J.M. , Roth, B. , Merchant, D.S. , Ray, A. , Bowman, L.H. and Vance, V.B. (2005) Ectopic DICER‐LIKE1 expression in P1/HC‐Pro Arabidopsis rescues phenotypic anomalies but not defects in microRNA and silencing pathways. Plant Cell, 17, 2873–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlotshwa, S. , Pruss, G.J. , MacArthur, J.L. , Reed, J.W. and Vance, V.S. (2016) Developmental defects of the P1/HC‐Pro potyviral suppressor are not due to misregulation of AUXIN RESPONSE FACTOR 8. Plant Physiol. 172, 1853–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar, A. , Melnyk, C.W. , Bassett, A. , Hardcastle, T.J. , Dunn, R. and Baulcombe, D.C. (2010) Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science, 328, 872–875. [DOI] [PubMed] [Google Scholar]
- Moreno, A. , Tjallingii, W.F. , Fernandez‐Mata, G. and Fereres, A. (2012) Differences in the mechanism of inoculation between a semi‐persistent and a non‐persistent aphid‐transmitted plant virus. J. Gen. Virol. 93, 662–667. [DOI] [PubMed] [Google Scholar]
- Moury, B. , Caromel, B. , Johansen, E. , Simon, V. , Chauvin, L. , Jacquot, E. , Kerlan, C. and Lefebvre, V. (2011) The helper component proteinase cistron of Potato virus Y induces hypersensitivity and resistance in potato genotypes carrying dominant resistance genes on chromosome IV. Mol. Plant–Microbe Interact. 24, 787–797. [DOI] [PubMed] [Google Scholar]
- Mukhtar, M.S. , Carvunis, A.R. , Dreze, M. , Epple, P. , Steinbrenner, J. , Moore, J. , Tasan, M. , Galli, M. , Hao, T. , Nishimura, M.T. , Pevzner, S.J. , Donovan, S.E. , Ghamsari, L. , Santhanam, B. , Romero, V. , Poulin, M.M. , Gebreab, F. , Gutierrez, B.J. , Tam, S. , Monachello, D. , Boxem, M. , Harbort, C.J. , McDonald, N. , Gai, L. , Chen, H. , He, Y. , European Union Effectoromics Consortium, Vandenhaute, J. , Roth, F.P. , Hill, D.E. , Ecker, J.R. , Vidal, M. , Beynon, J. , Braun, P. and Dangl, J.L. (2011) Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science, 333, 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara, K.S. , Masuta, C. , Yamada, S. , Shimura, H. , Kashihara, Y. , Wada, T.S. , Meguro, A. , Goto, K. , Tadamura, K. , Sueda, K. , Sekiguchi, T. , Shao, J. , Itchoda, N. , Matsumura, T. , Igarashi, M. , Ito, K. , Carthew, R.W. and Uyeda, I. (2012) Tobacco calmodulin‐like protein provides secondary defense by binding to and directing degradation of virus RNA silencing suppressors. Proc. Natl. Acad. Sci. USA, 109, 10 113–10 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, C.S. and Carrington, J.C. (1989) Identification of essential residues in potyvirus proteinase HC‐Pro by site‐directed mutagenesis. Virology, 173, 692–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olspert, A. , Chung, B.Y. , Atkins, J.F. , Carr, J.P. and Firth, A.E. (2015) Transcriptional slippage in the positive‐sense RNA virus family Potyviridae . EMBO Rep. 16, 995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasin, F. , Simón‐Mateo, C. and García, J.A. (2014) The hypervariable amino‐terminus of P1 protease modulates potyviral replication and host defense responses. PLoS Pathog. 10, e1003985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, C.W. , Peremyslov, V.V. , Mushegian, A.R. , Dawson, W.O. and Dolja, V.V. (2001) Functional specialization and evolution of leader proteinases in the family Closteroviridae . J. Virol. 75, 12 153–12 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Y.H. , Kadoury, D. , Gal‐On, A. , Wang, Y. and Raccah, B. (1998) Mutations in the HC‐Pro gene of zucchini yellow mosaic potyvirus: effects on aphid transmission and binding to purified virions. J. Gen. Virol. 79, 897–904. [DOI] [PubMed] [Google Scholar]
- Pfeffer, S. , Dunoyer, P. , Heim, F. , Richards, K.E. , Jonard, G. and Ziegler‐Graff, V. (2002) P0 of beet western yellows virus is a suppressor of posttranscriptional gene silencing. J. Virol. 76, 6815–6824. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pirone, T.P. and Blanc, S. (1996) Helper‐dependent vector transmission of plant viruses. Annu. Rev. Phytopathol. 34, 227–247. [DOI] [PubMed] [Google Scholar]
- Pirone, T.P. and Megahed, S. (1966) Aphid transmissibility of some purified viruses and viral RNAs. Virology, 30, 631–637. [DOI] [PubMed] [Google Scholar]
- Pirone, T.P. and Thornbury, D.W. (1984) The involvement of a helper component in non‐persistent transmission of plant viruses by aphids. Microbiol. Sci. 1, 191–193. [PubMed] [Google Scholar]
- Plisson, C. , Drucker, M. , Blanc, S. , German‐Retana, S. , Le Gall, O. , Thomas, D. and Bron, P. (2003) Structural characterization of HC‐Pro, a plant virus multifunctional protein. J. Biol. Chem. 278, 23 753–23 761. [DOI] [PubMed] [Google Scholar]
- Pompe‐Novak, M. , Wrischer, M. and Ravnikar, M. (2001) Ultrastructure of chloroplasts in leaves of potato plants infected by potato virus YNTN . Phyton, 41, 215–226. [Google Scholar]
- Pruss, G.J. , Lawrence, C.B. , Bass, T. , Li, Q.Q. , Bowman, L.H. and Vance, V. (2004) The potyviral suppressor of RNA silencing confers enhanced resistance to multiple pathogens. Virology, 320, 107–120. [DOI] [PubMed] [Google Scholar]
- Rajamäki, M.L. and Valkonen, J.P.T. (2009) Control of nuclear and nucleolar localization of nuclear inclusion protein A of picorna‐like Potato virus A in Nicotiana species. Plant Cell, 21, 2485–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravelo, G. , Kagaya, U. , Inukai, T. , Sato, M. and Uyeda, I. (2007) Genetic analysis of lethal tip necrosis induced by Clover yellow vein virus infection in pea. J. Gen. Plant Pathol. 73, 59–65. [Google Scholar]
- Rawlings, N.D. , Barrett, A.J. and Finn, R. (2016) Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 44, D343–D350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revers, F. and García, J.A. (2015) Molecular biology of potyviruses. Adv. Virus Res. 92, 101–199. [DOI] [PubMed] [Google Scholar]
- Riedel, D. , Lesemann, D.E. and Maiss, E. (1998) Ultrastructural localization of nonstructural and coat proteins of 19 potyviruses using antisera to bacterially expressed proteins of plum pox potyvirus. Arch. Virol. 143, 2133–2158. [DOI] [PubMed] [Google Scholar]
- Robaglia, C. and Caranta, C. (2006) Translation initiation factors: a weak link in plant RNA virus infection. Trends Plant Sci. 11, 40–45. [DOI] [PubMed] [Google Scholar]
- Rodamilans, B. , Valli, A. , Mingot, A. , San León, D. , Baulcombe, D. , López‐Moya, J.J. and García, J.A. (2015) RNA polymerase slippage as a mechanism for the production of frameshift gene products in plant viruses of the Potyviridae family. J. Virol. 89, 6965–6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas, M.R. , Zerbini, F.M. , Allison, R.F. , Gilbertson, R.L. and Lucas, W.J. (1997) Capsid protein and helper component proteinase function as potyvirus cell‐to‐cell movement proteins. Virology, 237, 283–295. [DOI] [PubMed] [Google Scholar]
- Rolland, M. , Kerlan, C. and Jacquot, E. (2009) The acquisition of molecular determinants involved in potato virus Y necrosis capacity leads to fitness reduction in tobacco plants. J. Gen. Virol. 90, 244–252. [DOI] [PubMed] [Google Scholar]
- Roudet‐Tavert, G. , German‐Retana, S. , Delaunay, T. , Delécolle, B. , Candresse, T. and Le Gall, O. (2002) Interaction between potyvirus helper component‐proteinase and capsid protein in infected plants. J. Gen. Virol. 83, 1765–1770. [DOI] [PubMed] [Google Scholar]
- Roudet‐Tavert, G. , Michon, T. , Walter, J. , Delaunay, T. , Redondo, E. and Le Gall, O. (2007) Central domain of a potyvirus VPg is involved in the interaction with the host translation initiation factor eIF4E and the viral protein HcPro. J. Gen. Virol. 88, 1029–1033. [DOI] [PubMed] [Google Scholar]
- Ruiz‐Ferrer, V. , Goytia, E. , Martínez‐Garcia, B. , López‐Abella, D. and López‐Moya, J.J. (2004) Expression of functionally active helper component protein of Tobacco etch potyvirus in the yeast Pichia pastoris . J. Gen. Virol. 85, 241–249. [DOI] [PubMed] [Google Scholar]
- Ruiz‐Ferrer, V. , Boskovic, J. , Alfonso, C. , Rivas, G. , Llorca, O. , López‐Abella, D. and López‐Moya, J.J. (2005) Structural analysis of tobacco etch potyvirus HC‐Pro oligomers involved in aphid transmission. J. Virol. 79, 3758–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahana, N. , Kaur, H. , Basavaraj, Tena, F. , Jain, R.K. , Palukaitis, P. , Canto, T. and Praveen, S. (2012) Inhibition of the host proteasome facilitates papaya ringspot virus accumulation and proteosomal catalytic activity is modulated by viral factor HcPro. PLoS One, 7, e52546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahana, N. , Kaur, H. , Jain, R.K. , Palukaitis, P. , Canto, T. and Praveen, S. (2014) The asparagine residue in the FRNK box of potyviral helper‐component protease is critical for its sRNA binding and subcellular localization. J. Gen. Virol. 95, 1167–1177. [DOI] [PubMed] [Google Scholar]
- Sako, N. and Ogata, K. (1981) Different helper factors associated with aphid transmission of some potyviruses. Virology, 112, 762–765. [DOI] [PubMed] [Google Scholar]
- Salomon, R. and Bernardi, F. (1995) Inhibition of viral aphid transmission by the N‐terminus of the maize dwarf mosaic virus coat protein. Virology, 213, 676–679. [DOI] [PubMed] [Google Scholar]
- Sasaya, T. , Torrance, L. , Cowan, G. and Ziegler, A. (2000) Aphid transmission studies using helper component proteins of Potato virus Y expressed from a vector derived from Potato virus X . J. Gen. Virol. 81, 1115–1119. [DOI] [PubMed] [Google Scholar]
- Scholthof, K.‐B.G. , Adkins, S. , Czosnek, H. , Palukaitis, P. , Jacquot, E. , Hohn, T. , Hohn, B. , Saunders, K. , Candresse, T. , Ahlquist, P. , Hemenway, C. and Foster, G.D. (2011) Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 12, 938–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segers, G.C. , van Wezel, R. , Zhang, X. , Hong, Y. and Nuss, D.L. (2006) Hypovirus papain‐like protease p29 suppresses RNA silencing in the natural fungal host and in a heterologous plant system. Eukaryot. Cell, 5, 896–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, J.K. , Kang, S.H. , Seo, B.Y. , Jung, J.K. and Kim, K.H. (2010) Mutational analysis of interaction between coat protein and helper component‐proteinase of Soybean mosaic virus involved in aphid transmission. Mol. Plant Pathol. 11, 265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, J.K. , Sohn, S.H. and Kim, K.H. (2011) A single amino acid change in HC‐Pro of soybean mosaic virus alters symptom expression in a soybean cultivar carrying Rsv1 and Rsv3 . Arch. Virol. 156, 135–141. [DOI] [PubMed] [Google Scholar]
- Shams‐Bakhsh, M. , Canto, T. and Palukaitis, P. (2007) Enhanced resistance and neutralization of defense responses by suppressors of RNA silencing. Virus Res. 130, 103–109. [DOI] [PubMed] [Google Scholar]
- Shapira, R. and Nuss, D.L. (1991) Gene expression by a hypovirulence‐associated virus of the chesnut blight fungus involves two papain‐like protease activities. J. Biol. Chem. 266, 19 419–19 425. [PubMed] [Google Scholar]
- Shen, W. , Yan, P. , Gao, L. , Pan, X. , Wu, J. and Zhou, P. (2010) Helper component‐proteinase (HC‐Pro) protein of Papaya ringspot virus interacts with papaya calreticulin. Mol. Plant Pathol. 11, 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiboleth, Y.M. , Haronsky, E. , Leibman, D. , Arazi, T. , Wassenegger, M. , Whitham, S.A. , Gaba, V. and Gal‐On, A. (2007) The conserved FRNK box in HC‐Pro, a plant viral suppressor of gene silencing, is required for small RNA binding and mediates symptom development. J. Virol. 81, 13 135–13 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, J.N. (1976) Aphid transmission of a nonaphid‐transmissible strain of tobacco etch virus. Phytopathology, 66, 652–654. [Google Scholar]
- Soitamo, A.J. , Jada, B. and Lehto, K. (2011) HC‐Pro silencing suppressor significantly alters the gene expression profile in tobacco leaves and flowers. BMC Plant Biol. 11, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorel, M. , Garcia, J.A. and German‐Retana, S. (2014) The Potyviridae cylindrical inclusion helicase: a key multipartner and multifunctional protein. Mol. Plant–Microbe Interact. 27, 215–226. [DOI] [PubMed] [Google Scholar]
- Stenger, D.C. , Hall, J.S. , Choi, I.R. and French, R. (1998) Phylogenetic relationships within the family Potyviridae: wheat streak mosaic virus and brome streak mosaic virus are not members of the genus Rymovirus . Phytopathology, 88, 782–787. [DOI] [PubMed] [Google Scholar]
- Stenger, D.C. , French, R. and Gildow, F.E. (2005a) Complete deletion of Wheat streak mosaic virus HC‐Pro: a null mutant is viable for systemic infection. J. Virol. 79, 12 077–12 080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenger, D.C. , Hein, G.L. , Gildow, F.E. , Horken, K.M. and French, R. (2005b) Plant virus HC‐Pro is a determinant of eriophyid mite transmission. J. Virol. 79, 9054–9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenger, D.C. , Young, B.A. , Qu, F. , Morris, T.J. and French, R. (2007) Wheat streak mosaic virus lacking helper component‐proteinase is competent to produce disease synergism in double infections with Maize chlorotic mottle virus . Phytopathology, 97, 1213–1221. [DOI] [PubMed] [Google Scholar]
- Sun, L.Y. , Nuss, D.L. and Suzuki, N. (2006) Synergism between a mycoreovirus and a hypovirus mediated by the papain‐like protease p29 of the prototypic hypovirus CHV1‐EP713. J. Gen. Virol. 87, 3703–3714. [DOI] [PubMed] [Google Scholar]
- Susaimuthu, J. , Tzanetakis, I.E. , Gergerich, R.C. and Martin, R.R. (2008) A member of a new genus in the Potyviridae infects Rubus . Virus Res. 131, 145–151. [DOI] [PubMed] [Google Scholar]
- Suzuki, N. , Chen, B.S. and Nuss, D.L. (1999) Mapping of a hypovirus p29 protease symptom determinant domain with sequence similarity to potyvirus HC‐Pro protease. J. Virol. 73, 9478–9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, N. , Maruyama, K. , Moriyama, M. and Nuss, D.L. (2003) Hypovirus papain‐like protease p29 functions in trans to enhance viral double‐stranded RNA accumulation and vertical transmission. J. Virol. 77, 11 697–11 707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szittya, G. and Burgyan, J. (2013) RNA interference‐mediated intrinsic antiviral immunity in plants. Curr. Top Microbiol. Immunol. 371, 153–181. [DOI] [PubMed] [Google Scholar]
- Sztuba‐Solinska, J. , Stollar, V. and Bujarski, J.J. (2011) Subgenomic messenger RNAs: mastering regulation of (+)‐strand RNA virus life cycle. Virology, 412, 245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaki, F. , Sano, T. and Yamashita, K. (2006) The complete nucleotide sequence of attenuated Onion yellow dwarf virus: a natural potyvirus deletion mutant lacking the N‐terminal 92 amino acids of HC‐Pro. Arch. Virol. 151, 1439–1445. [DOI] [PubMed] [Google Scholar]
- Tatineni, S. , Qu, F. , Li, R. , Morris, T.J. and French, R. (2012) Triticum mosaic poacevirus enlists P1 rather than HC‐Pro to suppress RNA silencing‐mediated host defense. Virology, 433, 104–115. [DOI] [PubMed] [Google Scholar]
- Thornbury, D.W. and Pirone, T.P. (1983) Helper components of two potyviruses are serologically distinct. Virology, 125, 487–490. [DOI] [PubMed] [Google Scholar]
- Thornbury, D.W. , Hellmann, G.M. , Rhoads, R.E. and Pirone, T.P. (1985) Purification and characterization of potyvirus helper component. Virology, 144, 260–267. [DOI] [PubMed] [Google Scholar]
- Thornbury, D.W. , Patterson, C.A. , Dessens, J.T. and Pirone, T.P. (1990) Comparative sequence of the helper component (HC) region of potato virus Y and a HC‐defective strain, potato virus C. Virology, 178, 573–578. [DOI] [PubMed] [Google Scholar]
- Thornbury, D.W. , van den Heuvel, J.F.J.M. , Lesnaw, J.A. and Pirone, T.P. (1993) Expression of potyvirus proteins in insect cells infected with a recombinant baculovirus. J. Gen. Virol. 74, 2731–2735. [DOI] [PubMed] [Google Scholar]
- Tian, Y.P. and Valkonen, J.P. (2013) Genetic determinants of Potato virus Y required to overcome or trigger hypersensitive resistance to PVY strain group O controlled by the gene Ny in potato. Mol. Plant–Microbe Interact. 26, 297–305. [DOI] [PubMed] [Google Scholar]
- Tian, Y.P. and Valkonen, J.P. (2015) Recombination of strain O segments to HCpro‐encoding sequence of strain N of Potato virus Y modulates necrosis induced in tobacco and in potatoes carrying resistance genes Ny or Nc . Mol. Plant Pathol. 16, 735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrance, L. , Andreev, I.A. , Gabrenaite‐Verhovskaya, R. , Cowan, G. , Makinen, K. and Taliansky, M.E. (2006) An unusual structure at one end of potato potyvirus particles. J. Mol. Biol. 357, 1–8. [DOI] [PubMed] [Google Scholar]
- Torres‐Barceló, C. , Martín, S. , Daròs, J.A. and Elena, S.F. (2008) From hypo‐ to hypersuppression: effect of amino acid substitutions on the RNA‐silencing suppressor activity of the Tobacco etch potyvirus HC‐Pro. Genetics, 180, 1039–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres‐Barceló, C. , Daròs, J.A. and Elena, S.F. (2010) Compensatory molecular evolution of HC‐Pro, an RNA‐silencing suppressor from a plant RNA virus. Mol. Biol. Evol. 27, 543–551. [DOI] [PubMed] [Google Scholar]
- Tribodet, M. , Glais, L. , Kerlan, C. and Jacquot, E. (2005) Characterization of Potato virus Y (PVY) molecular determinants involved in the vein necrosis symptom induced by PVYN isolates in infected Nicotiana tabacum cv. Xanthi. J. Gen. Virol. 86, 2101–2105. [DOI] [PubMed] [Google Scholar]
- Truniger, V. and Aranda, M.A. (2009) Recessive resistance to plant viruses. Adv. Virus Res. 75, 119–159. [DOI] [PubMed] [Google Scholar]
- Tu, Y. , Jin, Y. , Ma, D. , Li, H. , Zhang, Z. , Dong, J. and Wang, T. (2015a) Interaction between PVY HC‐Pro and the NtCF1bβ‐subunit reduces the amount of chloroplast ATP synthase in virus‐infected tobacco. Sci. Rep. 5, 15 605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, Y. , Zhang, Z. , Li, D. , Li, H. , Dong, J. and Wang, T. (2015b) Potato virus Y HC‐Pro reduces the ATPase activity of NtMinD, which results in enlarged chloroplasts in HC‐Pro transgenic tobacco. PLoS One, 10, e0136210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untiveros, M. , Olspert, A. , Artola, K. , Firth, A.E. , Kreuze, J.F. and Valkonen, J.P. (2016) A novel sweet potato potyvirus ORF is expressed via polymerase slippage and suppresses RNA silencing. Mol. Plant Pathol. 17, 1111–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urcuqui‐Inchima, S. , Maya, I.G. , Drugeon, G. , Haenni, A.L. and Bernardi, F. (1999a) Effect of mutations within the Cys‐rich motif of potyvirus helper component‐proteinase on self‐interaction. J. Gen. Virol. 80, 2809–2812. [DOI] [PubMed] [Google Scholar]
- Urcuqui‐Inchima, S. , Walter, J. , Drugeon, G. , German‐Retana, S. , Haenni, A.L. , Candresse, T. , Bernardi, F. , L. and Gall, O. (1999b) Potyvirus helper component‐proteinase self‐interaction in the yeast two‐hybrid system and delineation of the interaction domain involved. Virology, 258, 95–99. [DOI] [PubMed] [Google Scholar]
- Urcuqui‐Inchima, S. , Maia, I.G. , Arruda, P. , Haenni, A.L. and Bernardi, F. (2000) Deletion mapping of the potyviral helper component‐proteinase reveals two regions involved in RNA binding. Virology, 268, 104–111. [DOI] [PubMed] [Google Scholar]
- Uzest, M. , Gargani, D. , Drucker, M. , Hebrard, E. , Garzo, E. , Candresse, T. , Fereres, A. and Blanc, S. (2007) A protein key to plant virus transmission at the tip of the insect vector stylet. Proc. Natl. Acad. Sci. USA, 104, 17 959–17 964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valli, A. , Martín‐Hernández, A.M. , López‐Moya, J.J. and García, J.A. (2006) RNA silencing suppression by a second copy of the P1 serine protease of Cucumber vein yellowing ipomovirus (CVYV), a member of the family Potyviridae that lacks the cysteine protease HCPro. J. Virol. 80, 10 055–10 063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valli, A. , Oliveros, J.C. , Molnar, A. , Baulcombe, D. and García, J.A. (2011) The specific binding to 21‐nt double‐stranded RNAs is crucial for the anti‐silencing activity of Cucumber vein yellowing virus P1b and perturbs endogenous small RNA populations. RNA, 17, 1148–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valli, A. , Gallo, A. , Calvo, M. , Pérez, J.J. and García, J.A. (2014) A novel role of the potyviral helper component proteinase contributes to enhance the yield of viral particles. J. Virol. 88, 9808–9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valli, A. , García, J.A. and López‐Moya, J.J. (2015) Potyviridae In: Encyclopedia of Life Sciences (eLS). Chichester: Wiley; DOI: 10.1002/9780470015902.a0000755.pub3. [DOI] [Google Scholar]
- Varallyay, E. and Havelda, Z. (2013) Unrelated viral suppressors of RNA silencing mediate the control of ARGONAUTE1 level. Mol. Plant Pathol. 14, 567–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varrelmann, M. , Maiss, E. , Pilot, R. and Palkovics, L. (2007) Use of pentapeptide‐insertion scanning mutagenesis for functional mapping of the plum pox virus helper component proteinase suppressor of gene silencing. J. Gen. Virol. 88, 1005–1015. [DOI] [PubMed] [Google Scholar]
- Wang, A. and Krishnaswamy, S. (2012) Eukaryotic translation initiation factor 4E‐mediated recessive resistance to plant viruses and its utility in crop improvement. Mol. Plant Pathol. 13, 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R.Y. and Pirone, T.P. (1999) Purification and characterization of turnip mosaic virus helper component protein. Phytopathology, 89, 564–567. [DOI] [PubMed] [Google Scholar]
- Wang, R.Y. , Ammuar, E.D. , Thornbury, D.W. , Lopez‐Moya, J.J. and Pirone, T.P. (1996) Loss of potyvirus transmissibility and helper‐component activity correlate with non‐retention of virions in aphid stylets. J. Gen. Virol. 77, 861–867. [DOI] [PubMed] [Google Scholar]
- Wang, R.Y. , Powell, G. , Hardie, J. and Pirone, T.P. (1998) Role of the helper component in vector‐specific transmission of potyviruses. J. Gen. Virol. 79, 1519–1524. [DOI] [PubMed] [Google Scholar]
- Weber, P.H. and Bujarski, J.J. (2015) Multiple functions of capsid proteins in (+) stranded RNA viruses during plant–virus interactions. Virus Res. 196, 140–149. [DOI] [PubMed] [Google Scholar]
- Wen, R. , Zhang, S.C. , Michaud, D. and Sanfaçon, H.N. (2004) Inhibitory effects of cystatins on proteolytic activities of the Plum pox potyvirus cysteine proteinases. Virus Res. 105, 175–182. [DOI] [PubMed] [Google Scholar]
- Wen, R.H. , Khatabi, B. , Ashfield, T. , Saghai Maroof, M.A. and Hajimorad, M.R. (2013) The HC‐Pro and P3 cistrons of an avirulent Soybean mosaic virus are recognized by different resistance genes at the complex Rsv1 locus. Mol. Plant–Microbe Interact. 26, 203–215. [DOI] [PubMed] [Google Scholar]
- Wu, H.W. , Lin, S.S. , Chen, K.C. , Yeh, S.D. and Chua, N.H. (2010) Discriminating mutations of HC‐Pro of Zucchini yellow mosaic virus with differential effects on small RNA pathways involved in viral pathogenicity and symptom development. Mol. Plant–Microbe Interact. 23, 17–28. [DOI] [PubMed] [Google Scholar]
- Yambao, M.L. , Masuta, C. , Nakahara, K. and Uyeda, I. (2003) The central and C‐terminal domains of VPg of Clover yellow vein virus are important for VPg–HCPro and VPg–VPg interactions. J. Gen. Virol. 84, 2861–2869. [DOI] [PubMed] [Google Scholar]
- Yambao, M.L. , Yagihashi, H. , Sekiguchi, H. , Sekiguchi, T. , Sasaki, T. , Sato, M. , Atsumi, G. , Tacahashi, Y. , Nakahara, K.S. and Uyeda, I. (2008) Point mutations in helper component protease of clover yellow vein virus are associated with the attenuation of RNA‐silencing suppression activity and symptom expression in broad bean. Arch. Virol. 153, 105–115. [DOI] [PubMed] [Google Scholar]
- Yang, L. , Xu, Y. , Liu, Y. , Meng, D. , Jin, T. and Zhou, X. (2016) HC‐Pro viral suppressor from Tobacco vein banding mosaic virus interferes with DNA methylation and activates the salicylic acid pathway. Virology, 497, 244–250. [DOI] [PubMed] [Google Scholar]
- Yap, Y.K. , Duangjit, J. and Panyim, S. (2009) N‐terminal of Papaya ringspot virus type‐W (PRSV‐W) helper component proteinase (HC‐Pro) is essential for PRSV systemic infection in zucchini. Virus Genes, 38, 461–467. [DOI] [PubMed] [Google Scholar]
- Young, B.A. , Hein, G.L. , French, R. and Stenger, D.C. (2007) Substitution of conserved cysteine residues in wheat streak mosaic virus HC‐Pro abolishes virus transmission by the wheat curl mite. Arch. Virol. 152, 2107–2111. [DOI] [PubMed] [Google Scholar]
- Young, B.A. , Stenger, D.C. , Qu, F. , Morris, T.J. , Tatineni, S. and French, R. (2012) Tritimovirus P1 functions as a suppressor of RNA silencing and an enhancer of disease symptoms. Virus Res. 163, 672–677. [DOI] [PubMed] [Google Scholar]
- Zhang, C. , Wu, Z. , Li, Y. and Wu, J. (2015) Biogenesis, function, and applications of virus‐derived small RNAs in plants. Front. Microbiol. 6, 1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Du, L. and Poovaiah, B.W. (2014) Calcium signaling and biotic defense responses in plants. Plant Signal. Behav. 9, e973818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Du, P. , Lu, L. , Xiao, Q. , Wang, W. , Cao, X. , Ren, B. , Wei, C. and Li, Y. (2008) Contrasting effects of HC‐Pro and 2b viral suppressors from Sugarcane mosaic virus and Tomato aspermy cucumovirus on the accumulation of siRNAs. Virology, 374, 351–360. [DOI] [PubMed] [Google Scholar]
- Zheng, H. , Yan, F. , Lu, Y. , Sun, L. , Lin, L. , Cai, L. , Hou, M. and Chen, J. (2011) Mapping the self‐interacting domains of TuMV HC‐Pro and the subcellular localization of the protein. Virus Genes, 42, 110–116. [DOI] [PubMed] [Google Scholar]
- Zilian, E. and Maiss, E. (2011) Detection of plum pox potyviral protein–protein interactions in planta using an optimized mRFP‐based bimolecular fluorescence complementation system. J. Gen. Virol. 92, 2711–2723. [DOI] [PubMed] [Google Scholar]


