Summary
Powdery mildew, caused by the biotrophic fungal pathogen Blumeria graminis f. sp. tritici (Bgt), is a major threat to the production of wheat (Triticum aestivum). It is of great importance to identify new resistance genes for the generation of Bgt‐resistant or Bgt‐tolerant wheat varieties. Here, we show that the wheat copine genes TaBON1 and TaBON3 negatively regulate wheat disease resistance to Bgt. Two copies of TaBON1 and three copies of TaBON3, located on chromosomes 6AS, 6BL, 1AL, 1BL and 1DL, respectively, were identified from the current common wheat genome sequences. The expression of TaBON1 and TaBON3 is responsive to both pathogen infection and temperature changes. Knocking down of TaBON1 or TaBON3 by virus‐induced gene silencing (VIGS) induces the up‐regulation of defence responses in wheat. These TaBON1‐ or TaBON3‐silenced plants exhibit enhanced wheat disease resistance to Bgt, accompanied by greater accumulation of hydrogen peroxide and heightened cell death. In addition, high temperature has little effect on the up‐regulation of defence response genes conferred by the silencing of TaBON1 or TaBON3. Our study shows a conserved function of plant copine genes in plant immunity and provides new genetic resources for the improvement of resistance to powdery mildew in wheat.
Keywords: copine, disease resistance, powdery mildew, temperature, Triticum aestivum
Introduction
Hexaploid wheat (Triticum aestivum L. AABBDD) is a major food crop that provides dietary carbohydrates for more than one‐third of the world's population. Its production is critical for global food security, but its grain yield and quality are negatively affected by a variety of pathogenic viruses, bacteria, fungi, oomycetes and nematodes. Amongst these, Blumeria graminis f. sp. tritici (Bgt) causes wheat powdery mildew which is one of the most destructive wheat diseases worldwide (Yao et al., 2007). The utilization of resistant wheat cultivars is the most effective and economic strategy for the control of wheat powdery mildew. It is therefore important to identify new powdery mildew resistance regulators from the wheat genome for traditional and molecular breeding.
As sessile organisms, plants cannot hide or escape when attacked by various pathogens. They have evolved at least two layers of innate immunity for defence against these would‐be invaders (Jones and Dangl, 2006). The first layer, known as pathogen‐associated molecular pattern (PAMP)‐triggered immunity (PTI), is achieved by a set of plant plasma membrane‐located pattern recognition receptors (PRRs), which confer basal defence by the perception of conserved PAMPs, such as flagellin, EF‐Tu and chitin (Bernoux et al., 2011; Jones and Dangl, 2006; Segonzac and Zipfel, 2011; Zhang and Zhou, 2010). The induced PTI defence responses, including reactive oxygen species accumulation, callose deposition and the activation of defence‐related genes, inhibit or prevent the colonization of plants by pathogens. Some pathogens can overcome PTI by producing effectors to suppress the defence response and therefore to enhance disease development. Plants have evolved a second layer of innate immunity, namely effector‐triggered immunity (ETI), which is activated on direct or indirect recognition of effectors by disease resistance (R) genes, mostly coding intracellular receptors, namely NLRs (Nod‐Like Receptors) (Jones and Dangl, 2006). The detection of pathogen effectors by plant NLR proteins often culminates in a hypersensitive response (HR) associated with locally induced cell death that blocks the spread of the pathogen (Hofius et al., 2007; Morel and Dangl, 1997). More than 58 R genes for powdery mildew have been designated in wheat (McIntosh et al., 2017). Most are race‐specific resistance genes based on a gene‐for‐gene system. This type of resistance is usually short lived because of the high evolutionary potential of the pathogen (McDonald and Linde, 2002).
Copines are calcium‐dependent phospholipid‐binding proteins, consisting of highly conserved N‐terminal C2 domains (named C2A and C2B) and a C‐terminal von Willebrand factor A (vWA) domain (Creutz et al., 1998). In Arabidopsis, the C2 domains and the vWA domain of the copine protein BON1 have been shown to play critical functions through their calcium‐binding and protein–protein interacting activities, respectively (Bennypaul et al., 2012). Copines are present in diverse species of plants, and can be grouped into type I and type III copines in each of the 16 species analysed (Zou et al., 2016). In the dicot plant Arabidopsis, all three copine genes, AtBON1 and AtBON2 in type I, and AtBON3 in type III, are involved in disease resistance regulation. AtBON1 in Arabidopsis negatively regulates the immune receptor NLR gene SNC1 (Suppressor of npr1‐1, constitutive 1), and the bon1 loss‐of‐function mutant has an enhanced disease resistance and, consequently, a temperature‐dependent growth defect (Yang and Hua, 2004). The Arabidopsis BON1 is also a positive regulator of the abscisic acid‐ and bacterial pathogen‐triggered stomatal closure response, indicating an opposing role of BON1 at pre‐invasion and post‐invasion defence responses (Gou et al., 2015). In addition, AtBON1 has overlapping functions with AtBON2 and AtBON3 in repressing programmed cell death and defence responses through multiple NLR‐like genes (Li et al., 2009), and bon1bon2bon3 triple loss‐of‐function mutants die from heightened defence responses (Yang et al., 2006). Although the functions of copine genes have been studied in Arabidopsis, the roles of these genes in monocots or crop plants, such as common wheat, remain unknown.
Virus‐induced gene silencing (VIGS) is a useful strategy for the study of gene function in cereals. It uses a Barley stripe mosaic virus (BSMV)‐based silencing system in monocot plants (Bennypaul et al., 2012; Holzberg et al., 2002), and has been successfully utilized in wheat gene function studies (Ahmed et al., 2017; Kage et al., 2017; Liu et al., 2016).
In this study, we investigated the function of copine genes in common wheat: TaBON1 and TaBON3. We found that the expression of TaBON1 and TaBON3 is altered by pathogen infection and temperature changes. Most importantly, the silencing of copine genes TaBON1 or TaBON3 up‐regulates defence response genes in the absence of pathogen invasion and enhances resistance against powdery mildew in wheat. Our results thus reveal a conserved function of copine genes in the regulation of disease resistance in dicots and monocots.
Results
Identification of copines in common wheat
Given that the copines play key roles in disease resistance in Arabidopsis, we were interested in elucidating the roles of copines in common wheat, especially in disease resistance. In Arabidopsis, the loss of AtBON1 function leads to enhanced disease resistance to the virulent hemibiotrophic bacterial pathogen Pseudomonas syringae pv. tomato DC3000 and the biotrophic oomycete pathogen Hyaloperonospora parasitica (Yang and Hua, 2004). Therefore, we hypothesized that its homologues in common wheat might also regulate resistance to obligate biotrophic fungi, such as powdery mildew. To identify copine genes in wheat, we used the cDNA sequences of the Arabidopsis AtBON1 gene to blast search the genome sequences of wheat at the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/). Two cDNAs (Sequence IDs: AK334076.1 and AK330585.1) were identified and their corresponding genes were named as TaBON1 and TaBON3 to indicate the type I and type III copines to which they belong (Zou et al., 2016). The deduced TaBON1 and TaBON3 protein sequences have 66%–68% sequence identity to AtBON1. Because common wheat is hexaploid and has A, B, D sets of genomes, we expected that there would be three copies of each of the TaBON1 and TaBON3 genes. We determined the chromosomal locations of these TaBON genes using their cDNA sequences to blast the chromosome‐based draft sequence of hexaploid wheat (https://phytozome.jgi.doe.gov/pz/portal.html#!search?show=BLAST&method=Org_Taestivum_er). Two genes corresponding to the TaBON1 cDNA sequences were found on chromosomes 6AS and 6BL, and were therefore designated as homeologous to TaBON‐A1 and TaBON‐B1. Three genes corresponding to TaBON3 cDNA were located on chromosomes 1AL, 1BL and 1DL, and were therefore designated as homeologous to TaBON‐A3, TaBON‐B3 and TaBON‐D3. Phylogenetic analysis of the copine proteins from Arabidopsis, rice and wheat revealed that TaBON1 is closely related to the rice OsBON1 (Os02g0521300) protein, with a sequence identity of 86%. TaBON3 is closely related to the OsBON3 protein (Os05g0373300) with a sequence identity of 82%–85% (Fig. 1A). Based on the previous classification of plant copine proteins (Zou et al., 2016), TaBON1 is a member of type I and TaBON3 is a member of type III copines. We further compared homeologous copies of the TaBON1 and TaBON3 genes based on the annotation of the draft genome sequences. All predicted TaBON proteins contain features of copine, including two C2 domains (C2A and C2B) at the N‐terminus and a vWA domain at the C‐terminus (Fig. 1B). These domains are almost identical in amino acid sequences amongst homeologous copies of TaBON1 or TaBON3 (Figs S1 and S2, see Supporting Information). All copies of TaBON1 and TaBON3 genes have 13–16 exons based on annotation (Fig. 1C). The exon/intron junction sites are conserved not only amongst homeologues, but also amongst all TaBON genes. The lengths of the corresponding introns are almost identical amongst homeologues, but less conserved between TaBON1 and TaBON3 genes. The major differences amongst homeologues lie in the 5′ and/or 3′ end of the gene. TaBON1‐B lacks two exons at the 5′ and one exon at the 3′ end compared with TaBON‐A1, whereas TaBON‐D3 lacks one exon at the 5′ end compared with TaBON‐A3 or TaBON‐B3. Whether or not these differences are a result of mis‐annotation has yet to be determined experimentally. Nevertheless, we conclude that common wheat has at least two homeologous TaBON1 genes and three homeologous TaBON3 genes.
Figure 1.
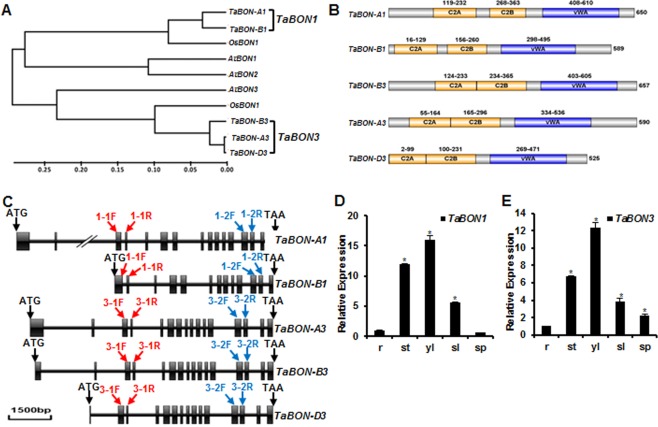
Phylogenetic tree, domain organization and tissue expression pattern of TaBON1 and TaBON3. (A) Neighbour‐joining tree of copine family members from Arabidopsis thaliana (At), Oryza sativa (Os) and Triticum aestivum (Ta) generated by MEGA5.02 software. The inferred phylogeny was tested by 1000 bootstrap replicates. The scale bar indicates the branch length. (B) Conserved domains identified in SMART and displayed on the IBS1.0.2 program (http://ibs.biocuckoo.org/download.php). Numbers represent the amino acid residues in the domain or the whole protein. (C) Diagram of gene structure of TaBON1 and TaBON3. Boxes and lines represent exons and introns, respectively. The red arrows indicate the primer sites for the amplification of fragments used in virus‐induced gene silencing (VIGS) constructs of TaBON1 and TaBON3. The blue arrows indicate the primer sites for quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) to check the expression of TaBON1 or TaBON3. 1‐1F stands for TaBON1‐1F, 3‐1F for TaBON3‐1F, and so on. (D, E) qRT‐PCR analysis of TaBON1 (D) and TaBON3 (E) expression levels in various tissues, including the root (r), stem (st), young leaf (yl) (McCouch et al., 2013), senescent leaf (sl) and spike (sp). The expression is normalized by the expression level in the root. TaActin was used as the internal control. Similar results were seen in all three biological replicates and one biological experiment with three technical repeats is shown. Asterisks indicate statistically significant differences in comparison with the root at P ≤ 0.01 (Student's t‐test).
We subsequently used quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) assay to assess the expression patterns of these genes in different tissues, including root, stem, young leaf, senescent leaf and spike. Specific primers were designed to differentiate between TaBON1 and TaBON3, but they were not able to differentiate homeologous copies. Expression of TaBON1 and TaBON3 was detectable in all tested wheat tissues and the tissue expression pattern was similar between TaBON1 and TaBON3. Both genes had the highest expression in young leaves and the lowest expression in roots (Fig. 1D,E).
Silencing of TaBON1 or TaBON3 induces defence response genes in wheat
To assess the function of TaBON1 and TaBON3 in wheat, we silenced these two genes via a BSMV‐based VIGS approach (Zhou et al., 2007). This approach has been shown to be effective for reverse genetics in wheat (Scofield et al., 2005). Because homeologous genes probably have redundant functions, we silenced all TaBON1 (TaBON‐A1 and TaBON‐B1) and TaBON3 (TaBON‐A3, TaBON‐B3 and TaBON‐D3) genes using regions of the cDNA sequences specific to TaBON1 or TaBON3 gene VIGS, respectively. The chlorosis phenotype was observed by 8 days post‐inoculation (dpi), indicating successful virus infection (Fig. 2A). The efficiency of VIGS was monitored using the wheat phytoene desaturase gene (PDS) as a control for VIGS, and leaf bleaching was observed by 10 dpi at 22 °C, indicating an efficient silencing of the PDS gene (Fig. S3A, see Supporting Information). The efficiency of VIGS of TaBON was analysed through qRT‐PCR using the fourth leaves of seedlings. The endogenous TaBON1 and TaBON3 transcripts were 80%–90% reduced in leaves inoculated with the BSMV virus containing the respective TaBON1 and TaBON3 fragments compared with control plants inoculated with the BSMV virus only (Fig. 2B,C). The specificity of silencing was also shown by RT‐PCR: the expression of TaBON3 was not altered in TaBON1‐silenced lines and the expression of TaBON1 was not altered in TaBON3‐silenced lines (Fig. 2D,E).
Figure 2.
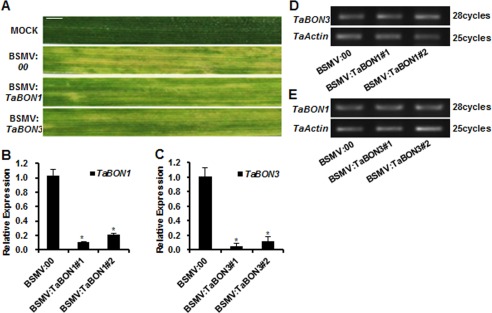
Knocking down of TaBON1 and TaBON3 expression by Barley stripe mosaic virus (BSMV)‐mediated gene silencing. (A) The leaf phenotype of successful virus infection. Photographs were captured at 8 days after virus inoculation. MOCK: wheat leaves treated with 2 × GKP buffer. Scale bars, 1 cm. (B, C) Relative transcript levels of TaBON1 (B) and TaBON3 (C) in plants at 22 °C inoculated with the BSMV‐based silencing system and assayed by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). Expression was normalized by the expression level in the control (BSMV:00). TaActin was used as the internal control. Similar results were seen in all three biological replicates and one biological set with three technical repeats is shown. Asterisks indicate statistically significant differences in comparison with the control (BSMV:00) at P ≤ 0.01 (Student's t‐test). (D, E) RNA expression levels of TaBON3 in TaBON1‐silenced (D) and TaBON1 in TaBON3‐silenced (E) plants before Blumeria graminis f. sp. tritici (Bgt) infection determined by semi‐quantitative RT‐PCR analysis.
Because knocking out of the Arabidopsis BON1 gene induces autoimmunity with the up‐regulation of defence genes (Yang and Hua, 2004), we tested whether or not the silencing of TaBON genes would also affect the transcript levels of defence response genes in the absence of pathogens. We chose TaPR2 and TaPR10 as marker genes for defence responses because they are highly induced during plant defence (Caruso et al., 1999; Mohammadi et al., 2012). By qRT‐PCR analysis, the transcripts of TaPR2 and TaPR10 were found to accumulate to higher levels in the TaBON1‐ and TaBON3‐silenced lines than in BSMV:00 control plants (Fig. 3A,B). We further analysed H2O2 accumulation and cell death in silenced plants, because such events occurred in the Arabidopsis bon1 mutant. Using 3,3′‐diaminobenzidine (DAB) and trypan blue staining to assess H2O2 and cell death, respectively, we did not find a difference between silenced plants and control plants (Fig. 3C,D). These results suggest that the knockdown of TaBON1 and TaBON3 causes the up‐regulation of defence response genes, but not significant cell death, in the absence of the pathogen in wheat.
Figure 3.
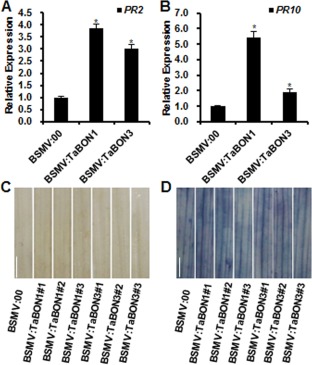
Up‐regulation of defence‐related features in silenced wheat plants without Blumeria graminis f. sp. tritici (Bgt) infection at 22 °C. (A, B) RNA expression of pathogenesis‐related genes TaPR2 (A) and TaPR10 (B) analysed by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) in BSMV:00‐, BSMV:TaBON1‐ and BSMV:TaBON3‐inoculated plants at 22 °C. RNA samples were isolated from the fourth leaves of wheat seedlings. Expression is normalized by the expression level in the control (BSMV:00). TaActin was used as the internal control. Similar results were seen in all three biological replicates and one biological replicate with three technical repeats is shown. Asterisks indicate statistically significant differences in comparison with the control (BSMV:00) at P ≤ 0.01 (Student's t‐test). (C, D) The fourth leaves at 14 days before challenge with Bgt were stained with 3,3′‐diaminobenzidine (DAB) (C) and trypan blue (D). Scale bars, 1 cm. Three plants for TaBON1 and TaBON3 silencing were analysed.
TaBON1 and TaBON3 negatively regulate wheat resistance against powdery mildew
To assess the roles of TaBON genes in resistance against the powdery mildew pathogen Bgt, we analysed their expression during Bgt infection. Plants of the common wheat variety Sumai 3 were infected with Bgt at the 2‐week‐old stage, and gene expression was analysed on infected leaves collected at different time points by qRT‐PCR (Fig. 4A,B). The expression of TaBON1 was increased by two‐ to nine‐fold at 1, 2 and 4 h post‐inoculation (hpi), whereas the expression of TaBON3 was up‐regulated by two‐ to 11‐fold at 1, 2 and 4 hpi. The induction of both genes reached a maximum at 4 hpi and decreased slightly at 8 and 12 hpi. At 24, 36 and 48 hpi, the expression of these two genes decreased to one‐ to two‐fold of the pre‐infection level. The induction of wheat PR genes TaPR2 and TaPR10 by Bgt in common wheat occurred at 2 hpi and reached a maximum at 12 hpi (Fig. S4, see Supporting Information). Therefore, TaBON1 and TaBON3 appear to show an early induction by Bgt, and maximum induction occurs earlier than that of the PR gene, suggesting a role of these genes in the early defence response to Bgt.
Figure 4.
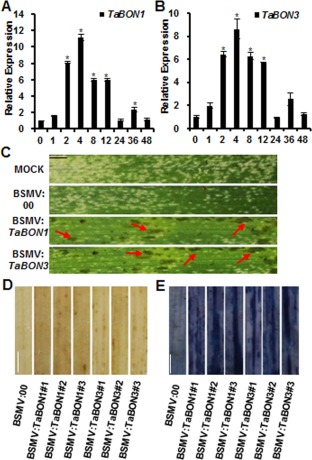
Silencing of TaBON1 or TaBON3 enhanced resistance to Blumeria graminis f. sp. tritici (Bgt). (A, B) Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis of the relative expression levels of TaBON1 (A) and TaBON3 (B) in Bgt‐infected common wheat leaves. RNA samples were isolated from the leaves of Bgt‐infected common wheat Sumai 3 at 0, 1, 2, 4, 8, 12, 24, 36 and 48 h post‐inoculation (hpi). Expression was normalized by the expression level at 0 hpi. TaActin was used as the internal control. Similar results were seen in all three biological replicates and one biological replicate with three technical repeats is shown. Asterisks indicate statistically significant differences in comparison with 0 hpi at P ≤ 0.01 (Student's t‐test). (C) The fourth leaves at 14 days after infection with spores of Bgt isolated from the field. Representative leaves were photographed at 5 days post‐inoculation (dpi). The red arrows indicate the location of the lesions. (D) The fourth leaves at 14 days were challenged with Bgt and sampled at 3 dpi. Three silenced plants for TaBON1 and TaBON3 were analysed. H2O2 accumulation at infection sites was stained using 3,3′‐diaminobenzidine (DAB). (E) The fourth leaves at 14 days were challenged with Bgt and sampled at 24 hpi. Three silenced plants for each of TaBON1 and TaBON3 were analysed. Scale bars, 1 cm. Cell death accumulation at infection sites was stained with trypan blue.
We subsequently assessed VIGS lines of TaBON1 or TaBON3 for their resistance to the virulent Bgt field‐isolated strain. Wheat plants were pre‐inoculated with buffer alone (MOCK), virus with the silencing vector BSMV:00 or TaBON1‐ and TaBON3‐silenced constructs on the second leaf. At 14 days after virus inoculation, the fourth leaf was detached and infected with fresh spores of virulent Bgt. Disease symptoms were analysed at 5 days after Bgt inoculation. Whereas leaves from plants pre‐inoculated with MOCK or BSMV:00 vector developed extensive powdery growth on the surface, leaves from both TaBON1‐ and TaBON3‐silenced lines showed significantly less powdery growth, indicating that the knockdown of TaBON1 and TaBON3 enhanced resistance to powdery mildew. In addition, we assessed resistance to Bgt using un‐detached leaves, and reduced powdery growth was also observed in TaBON1‐ and TaBON3‐silenced lines (Fig. S3D). Interestingly, extensive lesions developed on Bgt‐infected leaves in TaBON1‐ and TaBON3‐silenced lines, but not the control lines (Fig. 4C). DAB staining revealed that H2O2 accumulation was enhanced in TaBON1‐ and TaBON3‐silenced plants compared with BSMV:00‐infected control plants at 3 dpi of Bgt (Fig. 4D). Trypan blue staining showed that cell death was also significantly increased in TaBON1‐ and TaBON3‐silenced plants compared with BSMV:00‐infected control plants at 24 hpi (Fig. 4E). Therefore, TaBON1 and TaBON3 negatively regulate wheat disease resistance to Bgt and its associated H2O2 accumulation and cell death.
Silencing of TaBON1 or TaBON3 induces the up‐regulation of defence response genes in wheat at 28°C
Because the Arabidopsis BON1 gene shows a higher expression at lower temperature (Hua et al., 2001; Yang and Hua, 2004), we analysed the expression of the two wheat copine genes in leaves at three different temperatures using qRT‐PCR. The expression of TaBON1 and TaBON3 was significantly lower at higher temperatures of 25 and 32 °C than at the normal growth temperature of 22 °C, with the lowest expression at 32 °C (Fig. 5A,B).
Figure 5.
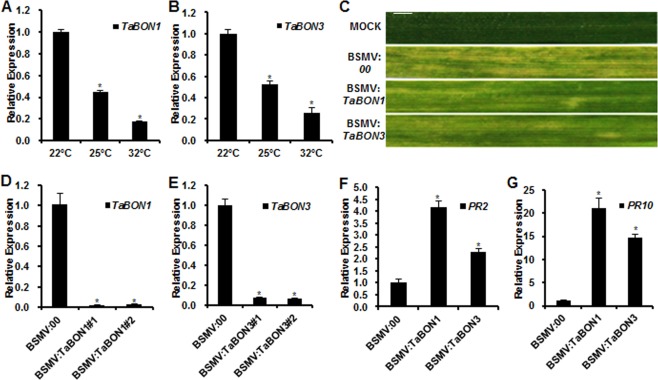
Temperature effects on the expression of TaBON1 and TaBON3, as well as defence induced by the silencing of TaBON1 and TaBON3. (A, B) Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis of TaBON1 (A) and TaBON3 (B) relative expression levels at different temperatures in common wheat leaves. RNA samples were isolated from the leaves of high‐temperature‐treated common wheat Sumai 3 at 22, 25 and 32 °C. Expression was normalized by the expression level at 22 °C. (C) The leaf phenotype of successful virus infection. Photographs were captured at 8 days after virus inoculation. MOCK: wheat leaves treated with 2 × GKP buffer. Scale bars, 1 cm. (D, E) Relative transcript levels of TaBON1 (D) and TaBON3 (E) in plants at 28 °C inoculated with a Barley stripe mosaic virus (BSMV)‐based silencing system and assayed by qRT‐PCR. (F, G) RNA expression of pathogenesis‐related genes TaPR2 (F) and TaPR10 (G) analysed by qRT‐PCR in BSMV:00‐, BSMV:TaBON1‐ and BSMV:TaBON3‐inoculated plants at 28 °C. Expression was normalized by the expression level in the control (BSMV:00). TaActin was used as the internal control. Similar results were seen in all three biological replicates, and one biological replicate with three technical repeats is shown. Asterisks indicate statistically significant differences in comparison with the control at P ≤ 0.01 (Student's t‐test).
Moderately high temperature was found to suppress the enhanced disease resistance in Arabidopsis BON1 knockout plants resulting from NLR up‐regulation. We therefore analysed the disease resistance phenotypes of the TaBON‐silenced plants at 28 °C, a moderately higher growth temperature for wheat. The VIGS system was efficient to knock down the TaBON1 and TaBON3 genes at 28 °C. Chlorosis was observed by 8 days after virus inoculation on the third leaves at 28 °C, indicating successful virus infection (Fig. 5C). Analysis by qRT‐PCR of the fourth leaves revealed a substantial reduction in endogenous TaBON1 and TaBON3 expression by VIGS (Fig. 5D,E). Because Bgt does not grow at 28 °C and therefore cannot be used to test disease resistance, we analysed the expression of defence genes by qRT‐PCR in TaBON1 and TaBON3 knockdown plants without Bgt infection. Marker genes TaPR2 and TaPR10 were both significantly up‐regulated in the TaBON1 and TaBON3 knockdown plants compared with the control plants (Fig. 5F,G), and the extent of up‐regulation was comparable between 28 and 22 °C (Fig. 3).
Discussion
The wheat powdery mildew fungus Bgt is an obligate biotrophic fungus and a causal agent of one of the most economically important diseases of wheat. As a result of its rapid evolution, novel virulent Bgt races often appear to circumvent race‐specific resistance genes in wheat. Thus, it is of great importance to identify new resistance genes from the wheat genome for the generation of Bgt‐tolerant wheat. In Arabidopsis, the evolutionarily conserved copine proteins have been shown to be negative regulators of plant disease resistance to both bacterial and fungal pathogens (Yang et al., 2006). In this study, we found a role of the wheat copine TaBON1 and TaBON3 genes in disease resistance to powdery mildew in wheat, which provides new gene resources for resistance to powdery mildew in wheat.
A previous study has found that copines in higher plants can be grouped into types I and III, where AtBON1 and AtBON2 belong to type I and AtBON3 belongs to type III (Zou et al., 2016). In monocot species, there is usually one gene of the type I group and one gene of the type III group. Common wheat is a hexaploid with an A, B and D genome, and we found A and B copies of the TaBON1 gene in the type I group, as well as A, B and D copies of TaBON3 in the type III group. Apparently, a D copy of TaBON1 is absent from the wheat genome. This could be a result of missing information from the current draft genome sequence, or a real loss of the copy from the D genome. Amongst the different copies of the TaBON1 or TaBON3 genes, the sequence identity is 97%–99% at the amino acid sequence level. The variations amongst gene copies often reside in the N‐terminal region which is important for protein localization. These variations might therefore confer functional diversity to different copies of the TaBON genes (Bennypaul et al., 2012). It will be interesting to investigate whether or not differences are indeed true (not a result of mis‐annotation) and, if so, the biological consequence of the differences.
In Arabidopsis, AtBON1 plays an important role in plant immunity and stomatal closure control (Gou et al., 2015; Hua et al., 2001; Yang and Hua, 2004), whereas AtBON2 and AtBON3 play a minor role in immunity as their single knockout mutants do not show a defect, but have a synergistic effect with the AtBON1 mutant in plant immunity (Yang et al., 2006). Unlike the Arabidopsis copines, silencing of TaBON1 or TaBON3 leads to enhanced disease resistance to Bgt. It appears that TaBON genes of the two types have an equal and non‐redundant function in plant immunity. It is likely that the two types of copine genes have distinct functions in plants, which enables the maintenance of copine genes in two groups in both monocots and dicots. The distinct function is not yet known from this study, but the results strongly indicate that copine genes have a conserved role in plant disease resistance from monocots to dicots.
In Arabidopsis, high temperature suppresses bon1 autoimmunity (Yang and Hua, 2004), Here, we found that high temperature has no drastic effect on the induction of defence response genes conferred by silencing of TaBON1 or TaBON3. This resembles the bon1bon3 and bon1bon2 double mutants, where high temperature does not suppress autoimmunity (Yang et al., 2006). It is possible that the temperature‐sensitive SNC1 is the major NLR gene activated in bon1, and that multiple NLR and NLR‐like genes are activated in bon1bon3 and bon1bon2 double mutants, and these NLR‐like genes are not all temperature sensitive. The involvement of NLR genes in the enhanced resistance of TaBON‐silenced wheat plants has not been studied, but it is possible that R genes could be up‐regulated in TaBON‐silenced plants and not all of them are temperature sensitive.
The production of ROS is one of the earliest events in plant cells after infection by pathogens including Bgt (Fig. S5, see Supporting Information). ROS accumulation in plant cells may induce callose deposition, defence gene activation and hypersensitive cell death, which is usually beneficial for plants for resisting biotrophic and obligate hemibiotrophic pathogens (Kim et al., 2012; Lehmann et al., 2015; Luna et al., 2011; Torres et al., 2006). Unlike the Arabidopsis BON1 knockout mutant, TaBON1 or TaBON3 knockdown plants do not exhibit over‐accumulation of ROS or spontaneous cell death in the absence of pathogen infection, Whether or not this difference between Arabidopsis and wheat copines is caused by residual function of TaBON1 and TaBON3 in knockdown plants is yet to be experimentally determined. Nevertheless, these plants produced more ROS and showed more extensive cell death after Bgt infection than control plants. This heightened response probably leads to the enhanced resistance phenotype of TaBON1 or TaBON3 knockdown plants. In summary, the wheat copine genes TaBON1 and TaBON3 negatively modulate disease resistance against wheat powdery mildew. This is probably through the modulation of H2O2 and cell death accumulation during the interaction of wheat and Bgt. This finding indicates a conserved function of plant copines in plant immunity and provides new genetic resources for the improvement of resistance to powdery mildew in wheat.
Experimental Procedures
Plant material and growth conditions
Common wheat variety Sumai 3, a highly susceptible variety to powdery mildew, was used throughout these experiments. Wheat plants were grown in a light climate box under long‐day conditions (16 h : 8 h, light : dark) and 60% relative humidity. For temperature treatments, plants were grown to 2 weeks at 22 °C and then transferred to 25 and 32 °C, respectively, for 5 days.
Sequence analysis, domain prediction and phylogenetic tree construction
Full‐length copine protein sequences were collected from NCBI (http://www.ncbi.nlm.nih.gov/), TAIR (http://www.arabidopsis.org/), Gramene (http://www.gramene.org/) and Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html). Conserved domains were predicted using tools at SMART (http://smart.embl-heidelberg.de/). Multiple sequence alignment of full‐length copine proteins was carried out using the MEGA5.02 program (http://www.megasoftware.net/history.php) with default parameters, and then displayed on GeneDoc (http://www.softpedia.com/get/Science-CAD/GeneDoc.shtml). MEGA5.02 software was also used to create the phylogenetic trees of the copine family members.
Bgt isolates and inoculation
The naturally occurring Bgt population was collected from the field in Nanjing, Jiangsu Province, China (latitude 31°14″N to 32°37″N, longitude 118°22″E to 119°14″E) and was propagated for disease resistance tests. Sumai 3 plants were used as inoculum to breed fungi under spore‐proof glasshouse conditions. The infected Sumai 3 plants were grown in a 16 h/8 h, 20 °C/18 °C, day/night cycle, 80% relative humidity, and used to inoculate the surface of the tested leaves by gently shaking conidia (Chen et al., 2007).
RNA extraction and qRT‐PCR
Total RNA was extracted using TRIzol reagent (TaKaRa, Dalian, China) according to the manufacturer's instructions. The first strand of complementary DNA (cDNA) was synthesized using the Transcript 1st strand cDNA synthesis kit (Vazyme, Nanjing, China). qRT‐PCR was performed in a CFX 96 Real Time PCR System (Bio‐Rad Laboratories, USA) in 20 µL of reaction mixture containing 2 × SYBR Premix Ex Taq Kit (TaKaRa, http://www.takara-bio.com/), 0.4 µm of each primer and 1 µL of tenfold‐diluted cDNA template. The PCR program used was as follows: 95 °C for 30 s, followed by 40 cycles of 5 s at 95 °C, 30 s at 59 °C and 30 s at 72 °C, and a final 10 min at 72 °C. Three technical replicates were run for each cDNA sample. The sequences of qRT‐PCR primers are listed in Table 1.
Table 1.
Information for the primers used in this study.
| Name of the primer | Primer sequence (5′–3′) | Note |
|---|---|---|
| TaBON1‐VIGS‐1F | CCTTAATTAAGCAATCCCATAGTTGTCGTG | VIGS vector construction |
| TaBON1‐VIGS‐1R | CCGCGGCCGCATGAAACTGCGGATCAATAT | |
| TaBON1–2F | GGTCCTCTACTTAGCACC | qRT‐PCR primers for TaBON1 |
| TaBON1–2R | TTTCCTTGAAGTCAGCCC | |
| TaBON3‐VIGS‐1F | CCTTAATTAAAGTGATCCTATGTTGGTGGT | VIGS vector construction |
| TaBON3‐VIGS‐1R | CCGCGGCCGCTTCAGTGGTGTGTTATGGTA | |
| TaBON3‐2F | AGTTGGAGTTGAAGGCAT | qRT‐PCR primers for TaBON3 |
| TaBON3‐2R | TCTTTTGTTTCTTGCTGG | |
| PR2‐F | GCGTGAAGGTGGTGATTT | qRT‐PCR primers for TaPR2 (DQ090946) |
| PR2‐R | GTGCCCGTTACACTTGGAT | |
| PR10‐F | ACGGAGCGGATGTGGAAG | qRT‐PCR primers for TaPR10 (CV778999) |
| PR10‐R | GCCACCTGCGACTTGAGC | |
| Actin ‐F | CCTTCGTTTGGACCTTGCTG | qRT‐PCR primers for TaActin |
| Actin‐R | AGCTGCTCCTAGCCGTTTCC |
BSMV‐mediated TaBON1 and TaBON3 gene silencing
To generate the BSMV:TaBON1 and BSMV:TaBON3 constructs, the 194‐bp and 224‐bp sequences of TaBON1 and TaBON3 were amplified from Sumai 3. These two fragments were then each inserted into the BSMV:00 vector in reverse orientation to form the recombinant vectors BSMV:TaBON1 and BSMV:TaBON3. The second fully expanded leaves of 2‐week‐old Sumai 3 seedlings were inoculated with virus containing BSMV:TaBON1 or BSMV:TaBON3 by gently sliding pinched fingers from the leaf base to the tip three times, with virus containing BSMV:TaPDS or BSMV:00 as the control. Three independent sets of plants were prepared for each treatment with 50 seedlings for each BSMV virus. Thirty seedlings were mock treated with 2 × GKP buffer (50 mm glycine, 30 mm K2HPO4, pH 9.2, 1% bentonite, 1% celite) as a negative control. Fourteen days after virus inoculation, the detached fourth leaves from 30 plants were placed on 6‐Benzylaminopurine (6‐BA) medium and inoculated with high densities of freshly collected Bgt. The phenotypes of inoculated leaves were observed and photographed at 5 dpi.
Observations of H2O2 and cell death accumulation
H2O2 production in plants was detected by an endogenous peroxidase‐dependent in situ histochemical staining procedure using DAB (Thordal‐Christensen et al., 1997). The fourth leaves from silenced plants at 0 and 3 dpi after Bgt infection were stained with 1 mg/mL DAB dissolved in 1 m Tris‐HCl (pH 7.5) for 6–8 h, discoloured in absolute ethyl alcohol and stored in 50% glycerol (Chen et al., 2016). Trypan blue staining was performed to reveal dead plant cells (Thordal‐Christensen et al., 1997). Tissues were immersed in 0.4% trypan blue solution for 6 h, bleached with a solution of acetic acid–ethanol (3 : 1) at 37 °C and then stored in 50% glycerol.
Disclosure
The authors have no conflicts of interest to declare.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Sequence alignment of three homeologues of TaBON1 in common wheat. Amino acid sequences of TaBON1. TaBON‐A1 and TaBON‐B1 represent TaBON1 proteins from the common wheat A and B genomes, respectively. The conserved C2A, C2B and von Willebrand factor A (vWA) domains are underlined separately in red, blue and orange, respectively.
Fig. S2 Sequence alignment of three homeologues of TaBON3 in common wheat. Amino acid sequences of TaBON3. TaBON‐A3, TaBON‐B3 and TaBON‐D3 represent TaBON3 proteins from the common wheat A, B and D genomes, respectively. The conserved C2A, C2B and von Willebrand factor A (vWA) domains are underlined separately in red, blue and orange, respectively.
Fig. S3 Silencing of TaBON1 or TaBON3 enhanced resistance to Blumeria graminis f. sp. tritici (Bgt) in living wheat leaves. (A) The leaf phenotype of successful virus infection. Photographs were captured at 10 days after virus inoculation. MOCK: wheat leaves treated with 2 × GKP buffer. Scale bars, 1 cm. (B, C) The expression levels of TaBON1 (B) and TaBON3 (C) in plants at 22 °C inoculated with a Barley stripe mosaic virus (BSMV)‐based silencing system and assayed by reverse transcription‐polymerase chain reaction (RT‐PCR). TaActin was used as the internal control. Similar results were seen in all three biological replicates and one biological repeat is shown. (D) The disease phenotype of living leaves at 14 days after infection with spores of Bgt isolated from the field. Representative leaves were photographed at 5 dpi.
Fig. S4 TaPR genes are induced by Blumeria graminis f. sp. tritici (Bgt) infection. Quantitative reverse‐transcription polymerase chain reaction (qRT‐PCR) analysis of the relative expression levels of TaPR2 and TaPR10 in Bgt‐infected common wheat leaves. RNA samples were isolated from the leaves of Bgt‐infected common wheat Sumai 3 at 0, 1, 2, 4, 8, 12, 24, 36 and 48 h post‐inoculation (hpi). Expression is normalized by the expression level at 0 hpi. TaActin was used as the internal control. One biological replicate with three technical repeats is shown. Asterisks indicate statistically significant differences in comparison with 0 hpi at P ≤ 0.01 (Student's t‐test).
Fig. S5 H2O2 accumulation and cell death in response to Blumeria graminis f. sp. tritici (Bgt) infection in BSMV:00. The fourth leaves of BSMV:00‐inoculated plants challenged with Bgt and stained with either 3,3′‐diaminobenzidine (DAB) for H2O2 accumulation sampled at 0 and 3 days post‐inoculation (dpi) (A) or with trypan blue for cell death at 0, 24, 48 and 72 hpi (B). Scale bars, 1 cm.
Acknowledgements
The authors would like to thank Dr Xiue Wang (Nanjing Agricultural University) for wheat materials and vectors. We thank Zongkuan Wang, Jia Zhao, Wentao Wan and Xiaobing Wang for technical assistance. This work was supported by grants from the Natural Science Foundation of Jiangsu Province (BK20150662), the Open Project Program of State Key Laboratory Breeding Base for Zhejiang Sustainable Pest and Disease Control (2010DS700124‐KF1509) and Jiangsu Collaborative Innovation Center for Modern Crop Production.
References
- Ahmed, S.M. , Liu, P. , Xue, Q. , Ji, C. , Qi, T. , Guo, J. , Guo, J. and Kang, Z. (2017) TaDIR1–2, a wheat ortholog of lipid transfer protein AtDIR1 contributes to negative regulation of wheat resistance against Puccinia striiformis f. sp tritici . Front. Plant Sci. 8, 521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennypaul, H.S. , Mutti, J.S. , Rustgi, S. , Kumar, N. , Okubara, P.A. and Gill, K.S. (2012) Virus‐induced gene silencing (VIGS) of genes expressed in root, leaf, and meiotic tissues of wheat. Funct. Integr. Genomics, 12, 143–156. [DOI] [PubMed] [Google Scholar]
- Bernoux, M. , Ellis, J.G. and Dodds, P.N. (2011) New insights in plant immunity signaling activation. Curr. Opin. Plant Biol. 14, 512–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso, C. , Chilosi, G. , Caporale, C. , Leonardi, L. , Bertini, L. , Magro, P. and Buonocore, V. (1999) Induction of pathogenesis‐related proteins in germinating wheat seeds infected with Fusarium culmorum . Plant Sci. 140, 87–97. [Google Scholar]
- Chen, T. , Xiao, J. , Xu, J. , Wan, W. , Qin, B. , Cao, A. , Chen, W. , Xing, L. , Du, C. , Gao, X. , Zhang, S. , Zhang, R. , Shen, W. , Wang, H. and Wang, X. (2016) Two members of TaRLK family confer powdery mildew resistance in common wheat. BMC Plant Biol. 16, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.‐P. , Xing, L.‐P. , Wu, G.‐J. , Wang, H.‐Z. , Wang, X.‐E. , Cao, A.‐Z. and Chen, P.‐D. (2007) Plastidial glutathione reductase from Haynaldia villosa is an enhancer of powdery mildew resistance in wheat (Triticum aestivum). Plant Cell Physiol. 48, 1702–1712. [DOI] [PubMed] [Google Scholar]
- Creutz, C.E. , Tomsig, J.L. , Snyder, S.L. , Gautier, M.‐C. , Skouri, F. , Beisson, J. and Cohen, J. (1998) The copines, a novel class of C2 domain‐containing, calcium‐dependent, phospholipid‐binding proteins conserved from Paramecium to humans. J. Biol. Chem. 273, 1393–1402. [DOI] [PubMed] [Google Scholar]
- Gou, M.Y. , Zhang, Z.M. , Zhang, N. , Huang, Q.S. , Monaghan, J. , Yang, H.J. , Shi, Z. , Zipfel, C. and Hua, J. (2015) Opposing effects on two phases of defense responses from concerted actions of HEAT SHOCK COGNATE70 and BONZAI1 in Arabidopsis. Plant Physiol. 169, 2304–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofius, D. , Tsitsigiannis, D.I. , Jones, J.D. and Mundy, J. (2007) Inducible cell death in plant immunity. Semin. Cancer Biol. 17, 166–187. [DOI] [PubMed] [Google Scholar]
- Holzberg, S. , Brosio, P. , Gross, C. and Pogue, G.P. (2002) Barley stripe mosaic virus‐induced gene silencing in a monocot plant. Plant J. 30, 315–327. [DOI] [PubMed] [Google Scholar]
- Hua, J. , Grisafi, P. , Cheng, S.H. and Fink, G.R. (2001) Plant growth homeostasis is controlled by the Arabidopsis BON1 and BAP1 genes. Gene Dev. 15, 2263–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kage, U. , Karre, S. , Kushalappa, A.C. and McCartney, C. (2017) Identification and characterization of a fusarium head blight resistance gene TaACT in wheat QTL‐2DL. Plant Biotechnol. J. 15, 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, C. , Meskauskiene, R. , Zhang, S.R. , Lee, K.P. , Ashok, M.L. , Blajecka, K. , Herrfurth, C. , Feussner, I. and Apel, K. (2012) Chloroplasts of Arabidopsis are the source and a primary target of a plant‐specific programmed cell death signaling pathway. Plant Cell, 24, 3026–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann, S. , Serrano, M. , L'Haridon, F. , Tjamos, S.E. and Metraux, J.P. (2015) Reactive oxygen species and plant resistance to fungal pathogens. Phytochemistry, 112, 54–62. [DOI] [PubMed] [Google Scholar]
- Li, Y.Q. , Pennington, B.O. and Hua, J. (2009) Multiple R‐like genes are negatively regulated by BON1 and BON3 in Arabidopsis. Mol. Plant–Microbe Interact. 22, 840–848. [DOI] [PubMed] [Google Scholar]
- Liu, J. , Zhang, T.R. , Jia, J.Z. and Sun, J.Q. (2016) The wheat mediator subunit TaMED25 interacts with the transcription factor TaEIL1 to negatively regulate disease resistance against powdery mildew. Plant Physiol. 170, 1799–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna, E. , Pastor, V. , Robert, J. , Flors, V. , Mauch‐Mani, B. and Ton, J. (2011) Callose deposition: a multifaceted plant defense response. Mol. Plant–Microbe Interact. 24, 183–193. [DOI] [PubMed] [Google Scholar]
- McCouch, S. , Baute, G.J. , Bradeen, J. , Bramel, P. , Bretting, P.K. , Buckler, E. , Burke, J.M. , Charest, D. , Cloutier, S. , Cole, G. , Dempewolf, H. , Dingkuhn, M. , Feuillet, C. , Gepts, P. , Grattapaglia, D. , Guarino, L. , Jackson, S. , Knapp, S. , Langridge, P. , Lawton‐Rauh, A. , Lijua, Q. , Lusty, C. , Michael, T. , Myles, S. , Naito, K. , Nelson, R.L. , Pontarollo, R. , Richards, C.M. , Rieseberg, L. , Ross‐Ibarra, J. , Rounsley, S. , Hamilton, R.S. , Schurr, U. , Stein, N. , Tomooka, N. , van der Knaap, E. , van Tassel, D. , Toll, J. , Valls, J. , Varshney, R.K. , Ward, J. , Waugh, R. , Wenzl, P. and Zamir, D. (2013) Agriculture: feeding the future. Nature, 499, 23–24. [DOI] [PubMed] [Google Scholar]
- McDonald, B.A. and Linde, C. (2002) The population genetics of plant pathogens and breeding strategies for durable resistance. Euphytica, 124, 163–180. [Google Scholar]
- McIntosh, R.A. , Dubcovsky, J. , Rogers, W.J. , Morris, C.F. and Xia, X.C. (2017) Catalogue of gene symbols for wheat: 2017 supplement. Annu. Wheat News Lett. [Google Scholar]
- Mohammadi, M. , Srivastava, S. , Hall, J.C. , Kav, N.N.V. and Deyholos, M.K. (2012) Two wheat (Triticum aestivum) pathogenesis‐related 10 (PR‐10) transcripts with distinct patterns of abundance in different organs. Mol. Biotechnol. 51, 103–108. [DOI] [PubMed] [Google Scholar]
- Morel, J.B. and Dangl, J.L. (1997) The hypersensitive response and the induction of cell death in plants. Cell Death Differ. 4, 671–683. [DOI] [PubMed] [Google Scholar]
- Scofield, S.R. , Huang, L. , Brandt, A.S. and Gill, B.S. (2005) Development of a virus‐induced gene‐silencing system for hexaploid wheat and its use in functional analysis of the Lr21‐mediated leaf rust resistance pathway. Plant Physiol. 138, 2165–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segonzac, C. and Zipfel, C. (2011) Activation of plant pattern‐recognition receptors by bacteria. Curr. Opin. Microbiol. 14, 54–61. [DOI] [PubMed] [Google Scholar]
- Thordal‐Christensen, H. , Zhang, Z. , Wei, Y. and Collinge, D.B. (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley–powdery mildew interaction. Plant J. 11, 1187–1194. [Google Scholar]
- Torres, M.A. , Jones, J.D.G. and Dangl, J.L. (2006) Reactive oxygen species signaling in response to pathogens. Plant Physiol. 141, 373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S. , Yang, H. , Grisafi, P. , Sanchatjate, S. , Fink, G.R. , Sun, Q. and Hua, J. (2006) The BON/CPN gene family represses cell death and promotes cell growth in Arabidopsis. Plant J. 45, 166–179. [DOI] [PubMed] [Google Scholar]
- Yang, S.H. and Hua, J. (2004) A haplotype‐specific resistance gene regulated by BONZAI1 mediates temperature‐dependent growth control in Arabidopsis. Plant Cell, 16, 1060–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, G. , Zhang, J. , Yang, L. , Xu, H. , Jiang, Y. , Xiong, L. , Zhang, C. , Zhang, Z. , Ma, Z. and Sorrells, M.E. (2007) Genetic mapping of two powdery mildew resistance genes in einkorn (Triticum monococcum L.) accessions. Theor. Appl. Genet. 114, 351–358. [DOI] [PubMed] [Google Scholar]
- Zhang, J. and Zhou, J.M. (2010) Plant immunity triggered by microbial molecular signatures. Mol. Plant, 3, 783–793. [DOI] [PubMed] [Google Scholar]
- Zhou, H. , Li, S. , Deng, Z. , Wang, X. , Chen, T. , Zhang, J. , Chen, S. , Ling, H. , Zhang, A. , Wang, D. and Zhang, X. (2007) Molecular analysis of three new receptor‐like kinase genes from hexaploid wheat and evidence for their participation in the wheat hypersensitive response to stripe rust fungus infection. Plant J. 52, 420–434. [DOI] [PubMed] [Google Scholar]
- Zou, B. , Hong, X. , Ding, Y. , Wang, X. , Liu, H. and Hua, J. (2016) Identification and analysis of copine/BONZAI proteins among evolutionarily diverse plant species. Genome, 59, 565–573. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Sequence alignment of three homeologues of TaBON1 in common wheat. Amino acid sequences of TaBON1. TaBON‐A1 and TaBON‐B1 represent TaBON1 proteins from the common wheat A and B genomes, respectively. The conserved C2A, C2B and von Willebrand factor A (vWA) domains are underlined separately in red, blue and orange, respectively.
Fig. S2 Sequence alignment of three homeologues of TaBON3 in common wheat. Amino acid sequences of TaBON3. TaBON‐A3, TaBON‐B3 and TaBON‐D3 represent TaBON3 proteins from the common wheat A, B and D genomes, respectively. The conserved C2A, C2B and von Willebrand factor A (vWA) domains are underlined separately in red, blue and orange, respectively.
Fig. S3 Silencing of TaBON1 or TaBON3 enhanced resistance to Blumeria graminis f. sp. tritici (Bgt) in living wheat leaves. (A) The leaf phenotype of successful virus infection. Photographs were captured at 10 days after virus inoculation. MOCK: wheat leaves treated with 2 × GKP buffer. Scale bars, 1 cm. (B, C) The expression levels of TaBON1 (B) and TaBON3 (C) in plants at 22 °C inoculated with a Barley stripe mosaic virus (BSMV)‐based silencing system and assayed by reverse transcription‐polymerase chain reaction (RT‐PCR). TaActin was used as the internal control. Similar results were seen in all three biological replicates and one biological repeat is shown. (D) The disease phenotype of living leaves at 14 days after infection with spores of Bgt isolated from the field. Representative leaves were photographed at 5 dpi.
Fig. S4 TaPR genes are induced by Blumeria graminis f. sp. tritici (Bgt) infection. Quantitative reverse‐transcription polymerase chain reaction (qRT‐PCR) analysis of the relative expression levels of TaPR2 and TaPR10 in Bgt‐infected common wheat leaves. RNA samples were isolated from the leaves of Bgt‐infected common wheat Sumai 3 at 0, 1, 2, 4, 8, 12, 24, 36 and 48 h post‐inoculation (hpi). Expression is normalized by the expression level at 0 hpi. TaActin was used as the internal control. One biological replicate with three technical repeats is shown. Asterisks indicate statistically significant differences in comparison with 0 hpi at P ≤ 0.01 (Student's t‐test).
Fig. S5 H2O2 accumulation and cell death in response to Blumeria graminis f. sp. tritici (Bgt) infection in BSMV:00. The fourth leaves of BSMV:00‐inoculated plants challenged with Bgt and stained with either 3,3′‐diaminobenzidine (DAB) for H2O2 accumulation sampled at 0 and 3 days post‐inoculation (dpi) (A) or with trypan blue for cell death at 0, 24, 48 and 72 hpi (B). Scale bars, 1 cm.


