During the past decade, many genomes have been sequenced from fungal and oomycete pathogens that interact biotrophically with plants, i.e. they thrive at least initially on living plant tissue. This has revealed genomes that often encode hundreds of proteins predicted to be secreted on the basis of N‐terminal signal peptides. Most of these proteins are unique or found only within restricted phylogenetic clades (Franceschetti et al., 2017). They are predicted to be ‘effectors’, i.e. proteins which, in some way, contribute to the virulence of the pathogen (see below). The fact that these filamentous microbes have hundreds of candidate effector genes is in stark contrast with bacterial pathogens, which typically have an order of magnitude fewer effector candidate genes. Although most of these hundreds of effectors currently lack evidence for significant roles in virulence, it is still striking that many of them appear to contribute measurably to virulence and that several of them seem to physically interact with numerous host proteins. In this Opinion Piece, we discuss these observations and attempt to address the apparent need for hundreds of effector candidate genes in these species. We suggest that this requirement reflects, in part, the need for effectors to target defence‐unrelated susceptibility components. Many of these, in turn, may be monitored (‘guarded’) by resistance‐triggering immune sensors. Potentially, pathogen success depends on additional sets of effectors dedicated to suppress this kind of surveillance.
Plant immunity is complex and organized into layers described by the so‐called ‘zig–zag model’ (Jones and Dangl, 2006). Plants exploit the fact that pathogens display indispensable pathogen‐associated molecular patterns (PAMPs). These molecules are generally recognized by plant plasma membrane‐resident pattern recognition receptors (PRRs), which activate pattern‐triggered immunity (PTI). To suppress PTI, pathogens secrete and deliver effectors to the host. Some effectors are thought to be transferred to the host cytosol, where they may be recognized by nucleotide‐binding leucine‐rich repeat‐type receptors, also referred to as NOD‐like receptor (NLR) proteins. This recognition leads to effector‐triggered immunity (ETI), which can include localized host programmed cell death, also called the hypersensitive response. When genetic variants of receptor genes determine whether ETI is triggered, they are referred to as resistance (R) genes. When variants of effector genes induce ETI, they are termed avirulence (Avr) genes. NLR proteins can recognize effectors directly or indirectly. Indirect recognition, based on the monitoring of an effector's action on the host target, provides the potential for NLR proteins to detect many effectors from the same or different pathogens. Some effectors serve to suppress ETI. An example is bacterial AvrRpt2, which suppresses RPM1‐mediated immunity triggered by AvrRPM1. It does this by cleaving plant RIN4 which otherwise is phosphorylated by AvrRPM1 (Jones and Dangl, 2006). An example in filamentous fungi is the effector SvrPm3a1/f1, recently identified in the wheat powdery mildew pathogen. SvrPm3a1/f1 suppresses immunity mediated by the R protein, Pm3a/f, which, in turn, recognizes the effector AvrPm3a2/f2 (Bourras et al., 2016). As discussed below, we anticipate that ETI‐suppressing effectors will be common. Recent functional analyses of effector repertoires in various filamentous pathogens have produced some remarkable findings, which have consequently raised two important aspects that are discussed below.
Genomes of filamentous pathogens often encode hundreds of candidate effectors. So why does the loss of function of some individual effectors have a severe impact on virulence? Investigations into the potential roles of these genes have included measurement of the effects of the experimental impairment of individual effectors through gene silencing or deletion. Such genetic approaches provide first evidence for a putative role in pathogenesis, which can be followed by functional studies unravelling the effector's molecular mechanisms. It has thus been straightforward to identify effector candidates that appear to contribute to virulence. For example, the expression of genes from the barley powdery mildew fungus (Blumeria graminis f.sp. hordei) can be suppressed through the transient expression of hairpin constructs of the gene sequence in the host, a process called host‐induced gene silencing (HIGS). This experimental approach was used to screen over 80 effector candidate genes. Most of these showed no significant contribution to virulence, potentially as a result of functional redundancy or the fact that they operate at a stage in the disease not included in the study. Yet, we successfully identified 21 that seemingly contributed to virulence (e.g. Pliego et al., 2013). The smut pathogen of maize, Ustilago maydis, also encodes hundreds of effector candidates, many of which are shared with other smut fungi. Although functional redundancy is clearly observed, knock‐outs of individual effectors can attenuate virulence (reviewed in Lanver et al., 2017). Likewise, the rice blast fungus Magnaporthe oryzae encodes many effector candidates, and yet single knock‐outs can compromise virulence (e.g. Zhang and Xu, 2014). Moreover, although the genome of the oomycete pathogen, Phytophthora infestans, potentially encodes >500 RXLR‐type effectors, silencing of AVR3a alone leads to a reduction in virulence (Whisson et al., 2016). In contrast with these examples from filamentous organisms, the bacterial pathogen Pseudomonas syringae pv. tomato (Pst) possesses an order of magnitude fewer effectors (approximately 30), and yet the simultaneous loss of multiple effectors is required to impact pathogenic efficiency measurably, indicating a high level of functional redundancy in the effector repertoire (Kvitko et al., 2009).
Based on these facts, a key question arises: why does the loss of so many individual effectors in filamentous plant pathogens lead to significant reductions in virulence? There are several possible explanations, including the following.
Many filamentous pathogens undergo complex infection stage transitions, involving the development of specialized structures for cell wall penetration, such as appressoria, or for host cell manipulation, such as haustoria. These structures are in intimate interaction with individual host cells for extended periods. Each stage may require the action of several factors that facilitate distinct structural changes in the host to accommodate the developmental changes in the pathogen. An example is the stimulation of the host cell to form the extrahaustorial membrane with specific host proteins associated with it; this is likely to be orchestrated by effector activity (e.g. reviewed in Whisson et al., 2016). Effectors may also be involved in the facilitation or promotion of nutrient acquisition, the requirements of which may be more complex than for bacteria – from the manipulation of biosynthetic pathways to the transport of metabolites. This may be particularly so for obligate biotrophic fungal pathogens. These microbial pathogens are utterly dependent on the living host for their food, and it is not difficult to see why several effectors are needed to manipulate host metabolism for the purposes of nutrition. The numbers of metabolic pathways and the numbers of potential effector targets in each pathway are potentially large.
There are examples of ETI‐suppressing effectors. Given the relatively extended interaction time with the host cell, plus the potential need to manipulate complex processes for nutrition and nutrient exchange, there are more opportunities for host immunity to be triggered. Consequently, additional effectors may be required, for example, to suppress specific ETI events resulting from NLR‐mediated monitoring of nutrition‐related processes. In this case, the silencing of any of these effectors would release and trigger a successful immune response, resulting in decreased virulence (see also below).
Filamentous plant pathogens can have broad host ranges. It is possible that some effectors are host‐specific, explaining the expansions in the overall effector repertoire. Moreover, rust fungi alternate between two distinct hosts, each expected to require separate sets of effectors for their colonization. Nevertheless, bacterial pathogens with lower effector numbers may also have broad host ranges, suggesting that this alone cannot explain the need for large numbers of effectors. In addition to distinct hosts, filamentous pathogens may colonize distinct host tissues, with separate effector requirements. This is the case for U. maydis, which deploys different effectors to colonize and manipulate the vegetative and floral parts of maize (Redkar et al., 2017).
An additional explanation could be that the silencing of one candidate effector actually results in an alteration in activity, biosynthesis and/or delivery of others. To our knowledge, this has not been investigated systematically in filamentous pathogen effectors, but has been observed for the expression of other fungal genes.
A more prosaic reason why so many effectors give a clear and severe phenotype when silenced could be that the effectors selected for characterization are the most highly expressed, i.e. are more easily detected by transcriptome and proteome studies. This was the case for those effector candidates studied by Pliego et al. (2013), and it is reasonable to expect that highly expressed effectors are also those that contribute most to virulence.
The methods used to measure the significance of effectors in virulence may provide an alternative set of answers to the conundrum of why so many filamentous effectors appear to be essential. For instance, HIGS may not just target an individual gene/transcript, but may affect several ‘off‐target’ genes simultaneously. One technical solution, which has been adopted to address this issue, is to complement the silencing phenotype by the transient expression of ‘silencing‐immune’ transgenes, i.e. genes in which synonymous codons are systematically replaced throughout the coding sequence (Pliego et al., 2013). Where this has been achieved, the complementation assays also address the criticism that expression of a transgene in the host may induce non‐specific resistance in the affected cells, which would show the same phenotype. Thus, the assay helps to reduce the likelihood of false positives.
The large numbers of effectors and the fact that individual effectors often make a substantial contribution to virulence are discoveries that are challenging to understand, as discussed above. Two recent studies conducted to obtain an insight into the plant molecular processes influenced by effectors have revealed yet more complexity (Mukhtar et al., 2011; Weßling et al., 2014). Effector candidates from the phylogenetically distinct pathogens Pst, Golovinomyces orontii (Go) and Hyaloperonospora arabidopsidis (Hpa) (representing three kingdoms of life: bacteria, fungi and oomycetes, respectively) have a very low number of host targets, many of which are shared by multiple effectors. The investigations revealed that single pathogen effector proteins in some cases can interact with multiple host proteins, and that certain host proteins are seemingly targeted by a multitude of effectors. These host proteins are themselves typically highly interconnected within the host protein interactome, suggesting that they are multifunctional. Notably, it was found that several of these host proteins are interaction partners of effectors from either a single pathogen (‘intraspecies convergence’) or different pathogen species (‘interspecies convergence’), revealing that some pathogen effectors from different kingdoms of life are aimed at a limited number of highly interconnected host proteins (designated as ‘hubs’; Fig. 1). Perhaps surprisingly, the majority of these ‘hub’ proteins are functionally not related to plant immunity, but implicated in a broad number of plant physiological processes. It thus seems that a major task of the respective effectors is not to directly undermine plant immunity, but to target plant proteins whose activity may in other ways be beneficial to the pathogen. The respective host proteins may thus include ‘susceptibility factors’, proteins whose activity is beneficial to disease development. This view is supported by the genetic analysis carried out in the study of Weßling et al. (2014). Approximately 25% of the mutant lines deficient in 124 candidate effector targets showed enhanced resistance to Hpa and/or Go, indicating that the encoded proteins are required for susceptibility per se, some of which may be negative regulators of immunity.
Figure 1.
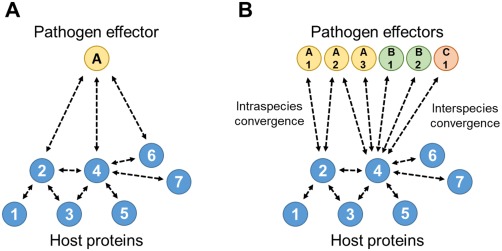
Scheme visualizing pathogen effector–host effector target promiscuity. The blue circles at the bottom represent host proteins; the yellow, green and red circles at the top represent pathogen effectors. Broken lines denote protein–protein interactions. (A) One pathogen effector interacts with multiple host proteins. (B) Intraspecies (left) and interspecies (right) convergence of pathogen effectors on host proteins. Differently coloured effector proteins originate from different pathogen species. Host proteins 2 and 4 can be considered as ‘hubs’.
Taken together, these findings raise a number of intriguing questions. First, why do many pathogen effectors appear to have multiple host targets. Second, why do multiple effectors apparently target the same host proteins? Again, several explanations are conceivable.
There may be structural reasons, as a host target or pathogen effector protein may have multiple binding sites on its surface, possibly allowing interaction with several interaction partners. Conversely, numerous target or effector proteins that converge on a single interaction partner might share a similar structural motif (three‐dimensional module), serving as a conserved protein–protein interaction interface. Such structural properties could result in functional redundancy of pathogen effector proteins that is independent of amino acid sequence similarity. This phenomenon may explain why several sequence‐diversified effectors target the same host components. However, what might be the biological reasons for such presumed operative redundancies? Multiple effectors targeting a single host protein could buffer against the loss of individual effector genes in the usually highly plastic pathogen genomes, driven by evolutionary pressure on recognition of individual effectors by host NLR proteins. It is well known that effectors that are avirulence determinants are typically dispensable for the pathogen without any obvious fitness penalty. A number of experimental studies support the functional redundancy of effector activities; for example, effectors AVR2, AVR3a and Pi02860 of P. infestans suppress INF1‐triggered immunity via different mechanisms, and several RXLR‐type effectors are able to inhibit the flg22‐triggered immune response (reviewed in Whisson et al., 2016).
Another possible explanation for the observed protein–protein interaction pattern might be the sequential delivery of effector proteins targeting the same host proteins, i.e. some effectors may target a given host protein early, whereas others do so later in the infection process. Indeed, transcriptome studies have revealed that pathogen effector genes may be expressed in consecutive ‘waves’ during plant colonization (O'Connell et al., 2012), which can be seen in the context of the stage transitions referred to above. Successive alternation of interacting effectors at host targets could also help to escape recognition by host NLR proteins. Effectors might also be cooperative: interaction with multiple effector proteins could be necessary to modulate effectively the function of a given host protein.
Some of the proposed host ‘hub’ proteins could play multiple roles in cellular homeostasis and, accordingly, be present in distinct multi‐protein complexes of unique composition. Several pathogen effectors might in fact be required for the adequate modulation of certain complexes involving these host targets in space and time, whereas it may not be beneficial to target other complexes. Examples of this might be the COP9 signalosome subunit CSN5 and certain members of the TCP transcription factor family (TCP13, TCP14, TCP15 and TCP19), which interact with numerous effector proteins from different pathogen species (Mukhtar et al., 2011; Weßling et al., 2014). CSN5 is a COP9 signalosome component possibly involved in the regulation of the activity of hundreds of cullin‐RING‐type E3 ubiquitin ligases (CRLs) through direct interaction. Some of these CRLs (e.g. CRL1 and CRL3) are required for efficient plant immunity via salicylic acid‐induced protein ubiquitination and subsequent degradation, whereas others act as negative regulators of immunity, such as NRL1, the target of Pi02860 (Whisson et al., 2016). Likewise, TCP transcription factors heterodimerize and interact with a number of transcriptional co‐repressors, thereby forming discrete protein–protein complexes, which could explain the requirement of apparent effector redundancy.
It is possible, in cases in which an effector interacts with more than one host protein, that these proteins interact with each other either directly or indirectly as part of a complex. Interaction with several host proteins may thus be required for correct positioning of an effector within the complex to disrupt, modulate or redirect the activity of the complex.
Although the scenarios outlined above provide plausible explanations for effector–target promiscuity, we cannot fully disregard the possibility that the findings to some extent reflect experimental artefacts, e.g. as a result of the ‘stickiness’ of the respective proteins in the yeast‐based assays. Although in planta validation of the protein–protein interactions by bimolecular fluorescence complementation (BiFC) and the altered pathogen infection phenotypes obtained with T‐DNA insertion mutants of the predicted host targets largely argue against such a possibility, additional experimental analyses will be necessary to substantiate further the current view. For example, it will be interesting to determine whether effectors targeting the same host protein have similar or diverse three‐dimensional protein structures. Furthermore, it remains to be seen whether the pathogen effectors activate or inhibit target proteins.
The studies of Mukhtar et al. (2011) and Weßling et al. (2014) may provide experimental support for the answers to the question why filamentous pathogen genomes encode hundreds of secreted effectors and why several of them, despite this, still appear to contribute significantly to virulence. Many of the effector targets identified appear to have no direct role in immunity, in turn suggesting that they are susceptibility components providing additional benefits to the pathogen. The fact that the knock‐out of some targets activates immunity suggests that they may include negative regulators of immunity, or that they may be monitored by NLRs, and that adapted pathogens secrete additional effectors to suppress this immunity activation. As alluded to by Weßling et al. (2014), this would call for yet another set of effectors with a role similar to those that suppress NLR monitoring of the PTI component, RIN4 (Jones and Dangl, 2006). Judging by the number of genes causing resistance phenotypes when mutated, the set of effectors suppressing NLR‐based monitoring may be large (Weßling et al., 2014). This kind of one‐by‐one study of effectors and their targets has the potential to uncover such relationships in ways not necessarily possible by classical R gene and Avr gene segregations, either because gene variants are not available or because of gene redundancy.
Acknowledgements
This paper was fostered by our participation in the COST Action FA1208 ‘Pathogen‐informed strategies for sustainable broad‐spectrum crop resistance’. H.T.‐C. was supported by the Villum Foundation (VKR023502) and the Independent Research Fund Denmark/Technology and Production (DFF‐6111‐00524). P.R.J.B. was supported by the Biotechnology and Biological Sciences Research Council grant BB/N009967/1. P.D.S. and R.P. were supported by the ERA‐CAPS project DURESTrit (BB/M004929/1 and PA 861/13). R.P. was additionally supported by grant PA 861/14 within the Priority Programme SPP1819, ‘Rapid evolutionary adaptation: Potential and constraints’, funded by the Deutsche Forschungsgemeinschaft (DFG).
References
- Bourras, S. , McNally, K.E. , Müller, M.C. , Wicker, T. and Keller, B. (2016) Avirulence genes in cereal powdery mildews: the gene‐for‐gene hypothesis 2.0. Front. Plant Sci. 7, 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschetti, M. , Maqbool, A. , Jiménez‐Dalmaroni, M.J. , Pennington, H.G. , Kamoun, S. and Banfield, M.J. (2017) Effectors of filamentous plant pathogens: commonalities amid diversity. Microbiol. Mol. Biol. Rev. 81, e00066–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kvitko, B.H. , Park, D.H. , Velásquez, A.C. , Wei, C.‐F. , Russell, A.B. , Martin, G.B. , Schneider, D.J. and Collmer, A. (2009) Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog. 5, e1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanver, D. , Tollot, M. , Schweizer, G. , Lo Presti, L. , Reissmann, S. , Ma, L.S. , Schuster, M. , Tanaka, S. , Liang, L. , Ludwig, N. and Kahmann, R. (2017) Ustilago maydis effectors and their impact on virulence. Nat. Rev. Microbiol. 15, 409–421. [DOI] [PubMed] [Google Scholar]
- Mukhtar, M.S. , Carvunis, A.‐R. , Dreze, M. , Epple, P. , Steinbrenner, J. , Moore, J. , Tasan, M. , Galli, M. , Hao, T. , Nishimura, M.T. , Pevzner, S.J. , Donovan, S.E. , Ghamsari, L. , Santhanam, B. , Romero, V. , Poulin, M.M. , Gebreab, F. , Gutierrez, B.J. , Tam, S. , Monachello, D. , Boxem, M. , Harbort, C.J. , McDonald, N. , Gai, L. , Chen, H. , He, Y. , European Union Effectoromics Consortium , Vandenhaute, J. , Roth, F.P. , Hill, D.E. , Ecker, J.R. , Vidal, M. , Beynon, J. , Braun, P. and Dangl, J.L. (2011) Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science, 333, 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell, R.J. , Thon, M.R. , Hacquard, S. , Amyotte, S.G. , Kleemann, J. , Torres, M.F. , Damm, U. , Buiate, E.A. , Epstein, L. , Alkan, N. , Altmueller, J. , Alvarado‐Balderrama, L. , Bauser, C.A. , Becker, C. , Birren, B.W. , Chen, Z. , Choi, J. , Crouch, J.A. , Duvick, J.P. , Farman, M.A. , Gan, P. , Heiman, D. , Henrissat, B. , Howard, R.J. , Kabbage, M. , Koch, C. , Kracher, B. , Kubo, Y. , Law, A.D. , Lebrun, M.H. , Lee, Y.H. , Miyara, I. , Moore, N. , Neumann, U. , Nordstroem, K. , Panaccione, D.G. , Panstruga, R. , Place, M. , Proctor, R.H. , Prusky, D. , Rech, G. , Reinhardt, R. , Rollins, J.A. , Rounsley, S. , Schardl, C.L. , Schwartz, D.C. , Shenoy, N. , Shirasu, K. , Sikhakolli, U.R. , Stueber, K. , Sukno, S.A. , Sweigard, J.A. , Takano, Y. , Takahara, H. , Trail, F. , van der Does, H.C. , Voll, L.M. , Will, I. , Young, S. , Zeng, Q. , Zhang, J. , Zhou, S. , Dickman, M.B. , Schulze‐Lefert, P. , van Themaat, E.V.L. , Ma, L.J. and Vaillancourt, L.J. (2012) Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat. Genet. 44, 1060–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pliego, C. , Nowara, D. , Bonciani, G. , Gheorghe, D.M. , Xu, R. , Surana, P. , Whigham, E. , Nettleton, D. , Bogdanove, A.J. , Wise, R.P. , Schweizer, P. , Bindschedler, L.V. and Spanu, P.D. (2013) Host‐induced gene silencing in barley powdery mildew reveals a class of ribonuclease‐like effectors. Mol. Plant–Microbe Interact. 26, 633–642. [DOI] [PubMed] [Google Scholar]
- Redkar, A. , Matei, A. and Doehlemann, G. (2017) Insights into host cell modulation and induction of new cells by the corn smut Ustilago maydis . Front. Plant Sci. 8, 899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weßling, R. , Epple, P. , Altmann, S. , He, Y. , Yang, L. , Henz, S.R. , McDonald, N. , Wiley, K. , Bader, K.C. , Gläßer, C. , Mukhtar, M.S. , Haigis, S. , Ghamsari, L. , Stephens, A.E. , Ecker, J.R. , Vidal, M. , Jones, J.D. , Mayer, K.F. , Ver Loren van Themaat, E. , Weigel, D. , Schulze‐Lefert, P. , Dangl, J.L. , Panstruga, R. and Braun, P. (2014) Convergent targeting of a common host protein‐network by pathogen effectors from three kingdoms of life. Cell Host Microbe, 16, 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisson, S.C. , Boevink, P.C. , Wang, S. and Birch, P.R. (2016) The cell biology of late blight disease. Curr. Opin. Microbiol. 34, 127–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S. and Xu, J.R. (2014) Effectors and effector delivery in Magnaporthe oryzae . PLoS Pathog. 10, e1003826. [DOI] [PMC free article] [PubMed] [Google Scholar]


