Summary
Soft‐rot diseases of plants attributed to Dickeya dadantii result from lysis of the plant cell wall caused by pectic enzymes released by the bacterial cell by a type II secretion system (T2SS). Arabidopsis thaliana can express several lines of defence against this bacterium. We employed bacterial mutants with defective envelope structures or secreted proteins to examine early plant defence reactions. We focused on the production of AtrbohD‐dependent reactive oxygen species (ROS), callose deposition and cell death as indicators of these reactions. We observed a significant reduction in ROS and callose formation with a bacterial mutant in which genes encoding five pectate lyases (Pels) were disrupted. Treatment of plant leaves with bacterial culture filtrates containing Pels resulted in ROS and callose production, and both reactions were dependent on a functional AtrbohD gene. ROS and callose were produced in response to treatment with a cellular fraction of a T2SS‐negative mutant grown in a Pels‐inducing medium. Finally, ROS and callose were produced in leaves treated with purified Pels that had also been shown to induce the expression of jasmonic acid‐dependent defence genes. Pel catalytic activity is required for the induction of ROS accumulation. In contrast, cell death observed in leaves infected with the wild‐type strain appeared to be independent of a functional AtrbohD gene. It was also independent of the bacterial production of pectic enzymes and the type III secretion system (T3SS). In conclusion, the work presented here shows that D. dadantii is recognized by the A. thaliana innate immune system through the action of pectic enzymes secreted by bacteria at the site of infection. This recognition leads to AtrbohD‐dependent ROS and callose accumulation, but not cell death.
Keywords: Arabidopsis, Dickeya, innate immunity, oligogalacturonides, pectate lyases
Introduction
Confronted by a variety of pathogenic organisms, plants have evolved several lines of defence which are expressed via a large array of genes at the site of infection and systemically (Moore et al., 2011; Spoel and Dong, 2012). A first line of defence, resembling the innate immune system in animals (Nürnberger et al., 2004), resides in the recognition of the pathogen by special membrane receptors via pathogen‐associated molecular patterns (PAMPs). These PAMPS are conserved microbial components, such as bacterial flagellar proteins, peptidoglycans, elongation factor‐Tu or fungal chitin (for reviews, see Boller and Felix, 2009; Brunner and Nürnberger, 2012; Newman et al., 2013). Pathogen recognition can also take place via damage‐associated molecular patterns (DAMPs), such as fragments of plant cell wall released by the action of plant cell wall‐degrading enzymes secreted by some pathogenic microorganisms (Denoux et al., 2008; Ridley et al., 2001). The receptors activate signalling cascades involving complex cellular and molecular events that collectively contribute to limit pathogen multiplication and the appearance of disease symptoms. Hallmark events of PAMP‐triggered immunity are a rapid activation of mitogen‐activated protein kinase (MAPK) cascades, a burst of reactive oxygen species (ROS) and a transient influx of calcium ions from the apoplast. PAMP‐stimulated immunity leads to the production of many secondary metabolites, antimicrobial peptides and cell wall modifications, including callose deposition (Aslam et al., 2009; Boller and Felix, 2009; Boudsocq and Sheen, 2013; Lecourieux et al., 2006; Monaghan and Zipfel, 2012; Torres, 2010; Segonzac et al., 2011). Phytohormones, including ethylene (ET), salicylic acid (SA), jasmonate and abscisic acid (ABA), are central to the modulation of innate immunity responses (Pieterse et al., 2012). Successful pathogens have acquired the ability to counteract PAMP‐triggered immunity (PTI). Many pathogens have the potential to inject effector proteins into the host cell and disrupt cellular homeostasis; however, effectors may be recognized by specific plant receptor proteins resulting in the hypersensitive response (HR) and elevated host resistance, designated as effector‐triggered immunity (ETI) (Coll et al., 2011; Dodds and Rathjen, 2010; Jones and Dangl, 2006). Pathogens that are necrotrophs produce plant cell wall depolymerases and/or toxins that kill cells at the site of the infection. Thus, necrotrophs are able to attack a wide range of plants. The pathogenicity of necrotrophs is multifactorial and often dependent on environmental conditions. As a result, immune reactions induced by these pathogens may vary greatly according to the infection conditions, and therefore are often difficult to analyse (Mengiste, 2012).
The soft‐rot Enterobacteriaceae, which belong to the genera Pectobacterium and Dickeya (formerly Erwinia carotovora and E. chrysanthemi), are typical broad‐host‐range necrotrophs causing wilt, soft‐rot and blackleg diseases of various plants. Although the modes of action of the virulence factors produced by Pectobacterium and Dickeya have been characterized (Charkowski et al., 2012; Davidsson et al., 2013; Reverchon and Nasser, 2013; Toth et al., 2003, 2011), less is known about the microbial components that elicit a host response. Initially, Pectobacterium wasabiae was used to study the plant defence response and, in Arabidopsis thaliana, the bacterium induced signalling cascades mainly mediated by ET and jasmonic acid (JA) (Palva et al., 1993). The induction of signalling activities occurred when plants were treated with plant cell wall‐degrading enzymes present in the bacterial culture filtrates (Norman et al., 1999; Norman‐Setterblad et al., 2000). These culture filtrates conferred resistance against the bacterium, suggesting that oligogalacturonides (OGs) released through the action of pectinolytic enzymes were the elicitors of this reaction. Pectobacterium carotovorum also produces type III effectors, including a harpin (HrpN) which contributes to virulence and DspE which induces an HR‐like cell death on Nicotiana benthamiana (Kim et al., 2011). HrpN has been shown to induce lesion formation and systemic resistance in A. thaliana (Kariola et al., 2003) with the expression of SA‐dependent and ET/JA‐dependent marker genes. Treatment of plants with both HrpN and a polygalacturonase (PehA) led to an amplified expression of defence marker genes and increased lesion formation, revealing a cooperative action between the two types of elicitor. In addition, a necrosis‐inducing protein (Nip) causing HR‐like cell death on tobacco leaf tissue was identified in harpin‐less P. carotovorum natural isolates (Mattinen et al., 2004), which showed sequence homology to necrosis‐ and ethylene‐inducing elicitors of fungi and oomycetes.
The A. thaliana immune response triggered by D. dadantii seems to be another story. The three SA‐, ET‐ and JA‐mediated signalling pathways activated by the bacterium are also induced by treatment with purified pectate lyases (Pels: PelB, PelI and PelL), representing the different families of isoenzymes secreted by bacterial cells during pathogenesis. The SA pathway is activated by treatment with any of these enzymes, whereas the ET and JA pathways respond only to PelB and PelI (Fagard et al., 2007). Induction of the SA pathway by D. dadantii also takes place via the siderophores produced by this bacterium, achromobactin and chrysobactin, a response attributed to the capacity of these compounds to mobilize iron in leaf tissues (Dellagi et al., 2009). However, like P. carotovorum and other necrotrophs (Glazebrook, 2005; Norman‐Setterblad et al., 2000), only ET and JA are the mediators for a certain level of resistance against the bacterium. Activation of the SA pathway by D. dadantii has been viewed as a method to modulate the plant immune response through the known antagonistic mechanism existing between the SA and JA signalling cascades (Dellagi et al., 2009; Glazebrook, 2005). In addition, changes in the cell wall structure and production of ROS generated through the NADPH‐oxidase AtrbohD are early reactions that contribute to the formation of a resistance barrier against D. dadantii (Fagard et al., 2007). Dickeya dadantii can also cause the death of plant cells surrounding the maceration symptoms (Kraepiel et al., 2011). This cell death appears to be an efficient mechanism against the spread and survival of D. dadantii (Kraepiel et al., 2011).
To determine whether there is a causal link between the diverse defence responses induced by D. dadantii in A. thaliana, we investigated the mode of recognition of this bacterium by the plant. Using the Col‐0 wild‐type ecotype and T‐DNA insertion lines harbouring either single or double atrbohD and atrbohF mutations (Torres et al., 2002), we ascertained that the ROS production triggered by D. dadantii is dependent on NADPH oxidase. We then analysed a set of bacterial mutants with potential changes in the ability to induce early reactions triggered by the wild‐type, including the production of ROS, callose deposition and cell death. The main conclusion was that D. dadantii is recognized by the A. thaliana innate immune system through the action of pectic enzymes released by the bacterium in plant cell walls. Plant cell death induced by the bacterium is a reaction independent of the action of pectinases and the type III secretion system (T3SS).
Results
ROS accumulation elicited by D. dadantii is affected in a pel‐negative mutant
Infection of A. thaliana by D. dadantii 3937 triggers the production of ROS, mainly H2O2, which accumulate in walls and intercellular spaces at the sites of bacterial colonization (Fagard et al., 2007). Using T‐DNA insertion lines harbouring either single or double atrbohD and atrbohF mutations, Fagard et al. (2007) identified AtrbohD as the main respiratory burst oxidase isoform involved in this extracellular ROS production. Because ROS may also be produced intracellularly in defence to pathogen attack (Kraepiel et al., 2011; Rojas and Mysore, 2012), in this study we monitored ROS accumulation using two imaging probes: the water‐soluble compound 3,3′‐diaminobenzidine (DAB) to check the extracellular accumulation of ROS and the membrane‐permeable dihydrodichlorofluorescein diacetate (DCFH‐DA) (Swanson et al., 2011) to detect intracellular ROS. In Col‐0 leaves infected with the wild‐type strain, a DAB reaction increasing over time was visible at 9 h post‐infiltration (hpi) (Fig. 1A). As expected, there was no DAB reaction in leaves of the atrbohD mutant. With the DCFH‐DA probe, we detected a fluorescent signal in Col‐0 leaves at the 16‐hpi time point. This signal was strongly reduced in leaves of the atrbohD/atrbohF double mutant line and in the atrbohD single mutant (Fig. 1B). In leaves of the atrbohF mutant, the signal was also reduced, but much less so than that observed for the atrbohD single mutant. From these data, we conclude that ROS production triggered by D. dadantii infection in A. thaliana essentially depends on a functional AtrbohD NADPH oxidase. Because AtrbohD is a critical enzyme in PAMP immune signalling (Benschop et al., 2007; Nühse et al., 2007), we examined whether this ROS burst was induced by PAMP.
Figure 1.
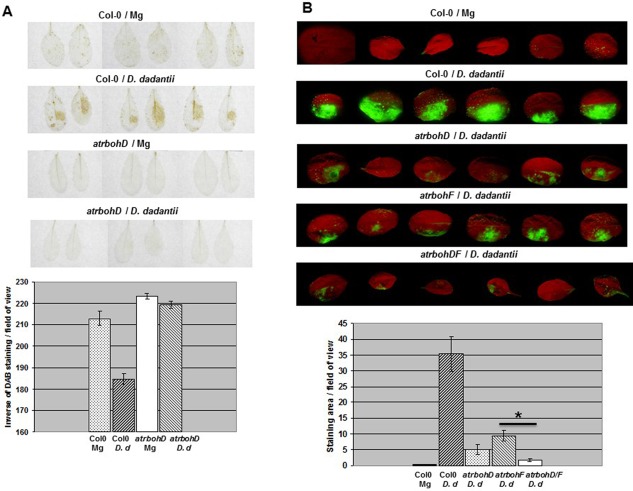
Reactive oxygen species (ROS) accumulation triggered by Dickeya dadantii wild‐type strain in Arabidopsis thaliana. Leaves of Col‐0 wild‐type and atrbohD/F single and double mutant lines, as indicated, were infiltrated with a D. dadantii suspension or an MgSO4 solution (Mg) according to the method described in Experimental Procedures. (A) 3,3′‐Diaminobenzidine (DAB) staining: photographs show representative reactions observed in leaves harvested at 12 h post‐infiltration (hpi); ROS detection by DAB staining on atrbohF and atrbohDF mutant lines, described by Fagard et al. (2007), was not carried out in this study; quantification (see Experimental Procedures) is expressed as the inverse of grey pixels/leaf counted in a field of 100 mm2; n = 8 leaves and bars represent standard error. (B) Dihydrodichlorofluorescein diacetate (DCFH‐DA) fluorescence: photographs show representative reactions observed in leaves harvested at 16 hpi; quantification of the fluorescence corresponds to the index of grey pixels/leaf; n = 8 leaves and bars represent standard error. Asterisk on graph in (B) indicates a statistically significant difference between atrbohF single and double mutant lines in response to D. dadantii (Mann–Whitney, P < 0.01). Experiments were carried out three times with similar results.
To investigate the bacterial component(s) triggering the oxidative burst, we heat inactivated the bacteria prior to inoculation and checked their eliciting activity. Boiling of D. dadantii cells for 15 min resulted in reduced ROS production (Fig. 2), indicating that thermostable components, such as lipopolysaccharides (LPSs) and peptidoglycans, were unlikely to be major triggers of the reaction. We made the assumption that D. dadantii mutants impaired in the biogenesis of potential elicitors of basal immunity could have lost the capacity to induce ROS production. We used a set of isogenic mutants (described in Experimental Procedures) with mutations of the flagellin structural gene fliC, the hrcC gene from the T3SS locus, the outC/D genes from the type II secretion system (T2SS) operon (called out), the five ‘major’ endo‐pectate lyase‐encoding genes pelA to pelE (called pel) and the achromobactin and chrysobactin siderophore biosynthetic genes acsA and cbsE. We also surveyed deep‐rough mutants (RH6065 and R1456) altered in the core oligosaccharide structure of the LPS. All these mutants, examined by DAB staining, were able to induce an oxidative burst (Fig. S1, see Supporting Information). However, the DAB coloration observed with the pel strain was weaker than that conferred by the wild‐type, unlike the out mutants, for which the coloration did not differ significantly. The use of the DCFH‐DA probe allowed similar observations. Only the pel mutant showed a reduced fluorescence (Fig. 3B and, for details, Fig. S2, see Supporting Information). Interestingly, we found that the outC mutant defective in the secretion of pectin‐degrading enzymes (Fig. 3D) was still able to macerate leaf tissues, although the symptoms remained confined to the inoculated area (Fig. 3C), indicating the leakiness of this mutant within the plant. In contrast, the pel mutant did not produce any visible maceration symptoms (Fig. 3C). We tested the eliciting activity of cellular fractions (CFs) prepared from the outC mutant. The CF obtained from the outC mutant induced more ROS accumulation than the CF prepared from the wild‐type strain, suggesting that the non‐secreted enzymes accumulated in the outC mutant were responsible for ROS induction (Fig. 3A and, for details, Fig. S2). In addition, no ROS accumulation was found after treatment with a CF from Escherichia coli (Fig. S2). Collectively, these data indicate that ROS accumulation elicited by D. dadantii is caused by the capability of this microorganism to produce pectin‐degrading enzymes.
Figure 2.
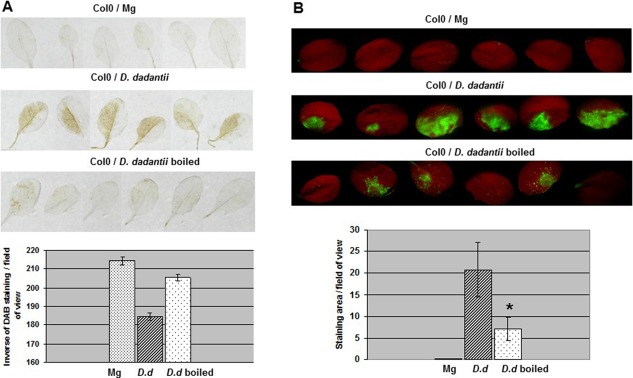
Thermo‐lability of the Dickeya dadantii inducing factor(s) of reactive oxygen species (ROS) accumulation. Leaves of Col‐0 wild‐type were inoculated with a boiled or control bacterial suspension, or an MgSO4 solution (Mg), as indicated, according to the method described in Experimental Procedures. 3,3′‐Diaminobenzidine (DAB) (A) and dihydrodichlorofluorescein diacetate (DCFH‐DA) (B) staining and quantification were performed as reported in Fig. 1. Asterisk on graph in (B) indicates a statistically significant difference between D. dadantii boiled and control samples (Mann–Whitney, P < 0.01). Experiments were carried out twice with similar results.
Figure 3.
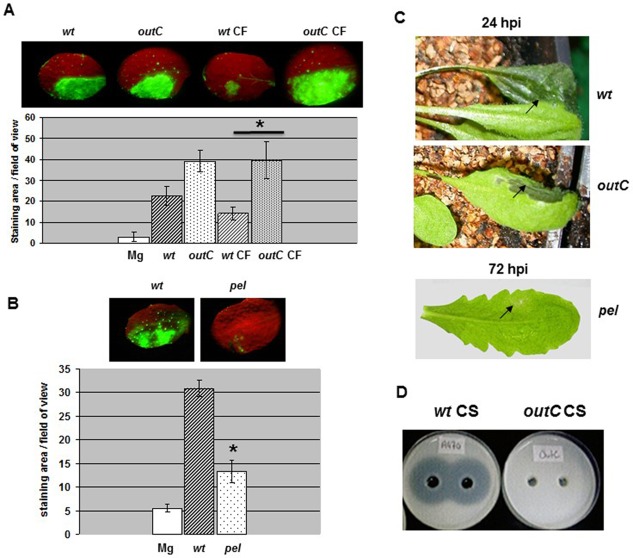
Reactive oxygen species (ROS) accumulation triggered by the Dickeya dadantii outC and pel mutants in Arabidopsis thaliana. Leaves of Col‐0 wild‐type (wt) were inoculated with a bacterial suspension of the strain to be tested, or with a sample of the corresponding soluble cellular fraction (CF), or an MgSO4 solution (Mg) (as indicated in the figure), according to the method described in Experimental Procedures. (A, B) Dihydrodichlorofluorescein diacetate (DCFH‐DA) staining and quantification were performed as reported in Fig. 1. Photographs show one representative reaction for each condition. In (A), asterisk on the graph indicates the difference between wt CF and outC CF (Mann–Whitney, P < 0.05). In (B), asterisk on the graph indicates the difference between wt and pel strain (Mann–Whitney, P < 0.01). (C) Photographs show representative symptoms caused by the strains tested observed at the time point indicated. The strains tested are indicated by their genotype. Experiments were carried out three times with similar results. (D) Cup‐plate assay for the detection of pectate lyase activity in bacterial culture supernatant fluids (CS) from wt and outC strains. The presence of translucent halos surrounding holes indicates that the substrate (polygalacturonate, PGA) in the agar medium was degraded after deposition of the CSs.
Dickeya dadantii Pels trigger AtrbohD‐dependent ROS accumulation
In D. dadantii strain 3937, eight calcium‐dependent Pels (PelA, PelB, PelC, PelD, PelE, PelI, PelL and PelZ), controlled at the transcriptional level by several regulators responding to various stimuli, have been characterized (Hugouvieux‐Cotte‐Pattat et al. 2014; Reverchon and Nasser, 2013). They are secreted via the Out system (Bouley et al. 2001; Pineau et al. 2014). Notably, the five isoforms PelA to PelE, which are the major macerating enzymes (Tardy et al., 1997), are inducible by polygalacturonate (PGA) and their secretion takes place during the late exponential phase of growth. Therefore, to investigate the inducing effect of Pels on the oxidative burst, we infiltrated leaves with a filter‐sterilized culture supernatant fluid prepared from wild‐type bacteria (wt CS) grown in a minimal medium with glycerol as the carbon source and PGA as the pel gene inducer (M9 PGA). To avoid a rapid degradation of the tissues, wt CS was diluted 10‐fold before use (see Experimental Procedures). We analysed the time course of ROS accumulation after infiltration using the DCFH‐DA staining method (Fig. 4). In Col‐0, we observed the presence of fluorescent zones corresponding to the infiltrated areas at the 3‐hpi time point. These reactions were always stronger in intensity than those observed in leaves treated with a sample of M9 PGA (M in Fig. 4A). Heat inactivation of wt CS prior to infiltration resulted in a significant reduction in fluorescence (Fig. 4A). There was no reaction with CSs prepared from bacterial cultures grown in the absence of PGA (data not shown). Leaves of the atrbohDF double mutant were not reactive to wt CS infiltration (Fig. 4A). A CS prepared from the pel mutant did not give any relevant DCFH‐DA reaction (Fig. 4B). Using the DAB staining method, we made similar observations (Fig. S3, see Supporting Information). We conclude that the ability of wild‐type CSs to induce an oxidative burst is a result of the enzymatic activity of Pels in the plant, rather than to PGA catabolites possibly present in M9 PGA medium or generated during bacterial growth in vitro. Therefore, all or part of the Pels (PelA to PelE) can be viewed as the causal factor(s) of the oxidative burst triggered by D. dadantii in A. thaliana.
Figure 4.
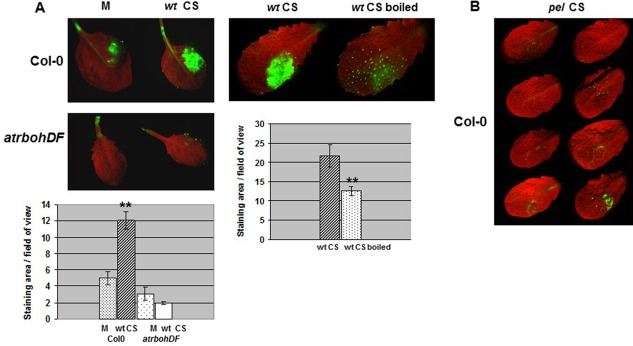
Role of Dickeya dadantii pectic enzymes in reactive oxygen species (ROS) induction by a dihydrodichlorofluorescein diacetate (DCFH‐DA) staining analysis. (A) Leaves of Col‐0 wild‐type and atrbohDF mutant line were inoculated with a sample of culture supernatant fluid from the wild‐type strain (wt CS), or a sample of M9 polygalacturonate (PGA) medium (M) (as indicated on the figure), according to the method described in Experimental Procedures. The wt CS was boiled for 15 min (as indicated) prior to infiltration in Col‐0 leaves. DCFH‐DA staining and quantification were performed as reported in Fig. 1. Photographs show one representative reaction for each condition. Asterisks on the graph corresponding to wt CS indicate the difference between M and wt CS, on Col‐0 (Mann–Whitney, P < 0.01). Asterisks on the graph corresponding to boiled wt CS indicate the difference between wt CS and wt CS boiled, on Col‐0 (Mann–Whitney, P < 0.05). Experiments were carried out three times with similar results. (B) Leaves of Col‐0 were inoculated with a sample of culture supernatant fluid from the pel mutant strain (pel CS) as reported in (A). The results presented correspond to one experiment. This experiment was carried out twice with similar results.
Dickeya dadantii Pels induce AtrbohD‐dependent callose deposition in A. thaliana
The formation of callose is a reaction of A. thaliana plants in response to D. dadantii infection. Callose deposits could be observed at 8 hpi in both the extracellular compartment of parenchyma and along vascular tissues (Fagard et al., 2007). Callose deposition is a PAMP‐triggered reaction, but can also occur in response to a variety of signals depending on the challenging interaction (Luna et al., 2011). Therefore, we investigated the origin of this response in the D. dadantii–A. thaliana interaction. We examined whether the formation of callose was dependent on a functional AtrbohD gene. Callose was detected with aniline blue and quantified from the photographs. We found that the reaction was significantly reduced in D. dadantii‐inoculated leaves from the atrbohD mutant (Fig. 5A). It was also drastically reduced in Col‐0 leaves when D. dadantii cells were heat inactivated prior to inoculation (Fig. 5B). Because these data were reminiscent of those observed in the induction of the oxidative burst, we investigated the bacterial mutants for their capacity to induce callose formation. The hrcC and acsA cbsE mutations, as well as those impairing the LPS structure, did not affect the ability of bacteria to induce callose (Fig. S4, see Supporting Information). The outC and fliC mutations resulted in a slight reduction in the reaction (Fig. S4). However, callose deposition was strongly reduced in leaves inoculated with the pel mutant, suggesting a role of Pel(s) in the elicitation of this response (Fig. S5A, see Supporting Information). Accordingly, we detected the presence of callose in Col‐0 leaves after infiltration of a CS prepared from wild‐type bacteria grown in M9 PGA (Fig. S5B). Deposits were visible (3 hpi) in the limb and along the vessels. There was no reaction in leaves treated with M9 PGA. No callose deposition was observed after infiltration of a heat‐inactivated CS (Fig. S5B), indicating that the bacterial Pel activity produced in planta was probably at the origin of the reaction. Consistent with this, the eliciting activity of CFs from outC cells was stronger than that prepared from the wild‐type (Fig. S6, see Supporting Information).
Figure 5.
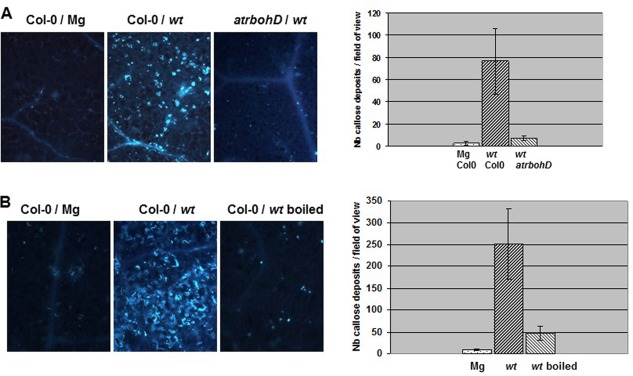
Callose deposition induced by Dickeya dadantii in Arabidopsis thaliana. (A) Leaves of the Col‐0 wild‐type and atrbohD mutant line were inoculated with a bacterial suspension from the wild‐type strain (wt) or an MgSO4 solution (Mg) (as indicated in the figure), according to the method described in Experimental Procedures. (B) Heat inactivation was achieved by boiling the suspension (as indicated in the figure) for 15 min prior to infiltration in Col‐0 leaves. Callose deposits were detected with aniline blue staining in leaves harvested at the 8‐h post‐infiltration (hpi) time point. Photographs show one representative reaction for each condition. The corresponding graphs show a quantification of the reactions (see Experimental Procedures) given as the number of callose spots per leaf counted in a field of 0.6 mm2; n = 8 leaves and bars represent standard error. Experiments were carried out three times with similar results. Nb, number.
Individual Pels induce ROS accumulation, callose deposition and expression of JA‐dependent marker genes
To ascertain the role of D. dadantii Pels in the induction of ROS accumulation and callose deposition, we examined the effect of purified preparations of PelB and PelI at 3 hpi of Col‐0 plants. PelB and PelI are representative members of the two families of endo‐pectate lyases produced by D. dadantii, PL1 and PL3, respectively (www.cazy.org), and both are very active in the maceration of plant tissues (Shevchik et al., 1997a, b). We observed the presence of a DAB reaction in infiltrated tissues with PelB and PelI. The two enzymes gave similar reactions (Fig. 6A). In order to determine whether Pel catalytic activity was necessary for the induction of ROS accumulation, we compared the action of PelI with that of the enzymatically inactive PelIQ227A mutant protein, in which a glutamine residue in the catalytic site was replaced by an alanine residue (Creze et al., 2008; C. Creze and V. E. Shevchik, unpublished data). PelIQ227A did not cause tissue maceration or induce ROS accumulation (Fig. 6B), indicating the need for enzymatic activity for these reactions.
Figure 6.
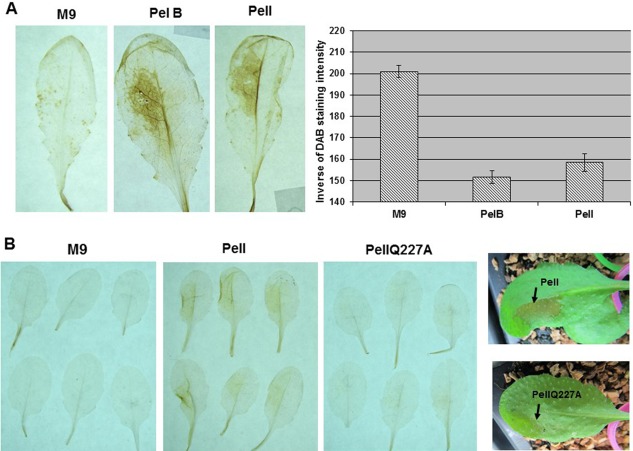
Role of individual pectate lyases in reactive oxygen species (ROS) induction. (A) Leaves of Col‐0 wild‐type were infiltrated with a sample of purified Dickeya dadantii pectate lyase, PelB or PelI, or with M9 medium (as indicated in the figure). Details are presented in Experimental Procedures. Photographs labelled PelB, PelI and M9 show representative reactions detected by 3,3′‐diaminobenzidine (DAB) staining in leaves harvested at the 3‐h post‐infiltration (hpi) time point; quantifications were performed as reported in Fig. 1. Experiments were carried out twice with similar results. (B) Photographs show the DAB reactions obtained with PelI compared with the PelIQ227A mutant as indicated. On the right, photographs show the maceration symptoms caused by PelI at 18 hpi, which were not observed with PelIQ227A. Experiments were carried out twice with similar results.
The infiltration of PelB and PelI caused the accumulation of callose deposits particularly visible in zones bordering the lesions. We found no significant difference between PelB and PelI (Fig. 7). To further determine the role of these enzymes in the induction of the A. thaliana basal immune response, we monitored, by reverse transcription‐polymerase chain reaction (RT‐PCR), the expression of defence marker genes representative of the JA‐mediated signalling pathway (Fig. S7, see Supporting Information). As PelI can be subject to proteolytic cleavage with consequences on protein activity (Shevchik et al., 1998; see Discussion), we tested native PelI and the cleaved form, which consists of an isolated catalytic domain (designated as PelI Cat in Fig. S7). The accumulation of AOX1a gene transcripts, apparent at 6 hpi, in response to PelI and PelB (with a good dose effect shown for PelI Cat diluted 10 times before infiltration) increased by 24 hpi. We also observed an accumulation of PDF1 and CHI‐B gene transcripts in response to PelI, PelI Cat and PelB at 24 hpi. The VSP2 gene appeared to be slightly induced at 6 hpi by PelI and PelI Cat, and an accumulation of VSP2 transcripts was visible at 24 hpi. The induction of the VSP2 gene by PelB, visible at 6 hpi, decreased at 24 hpi.
Figure 7.
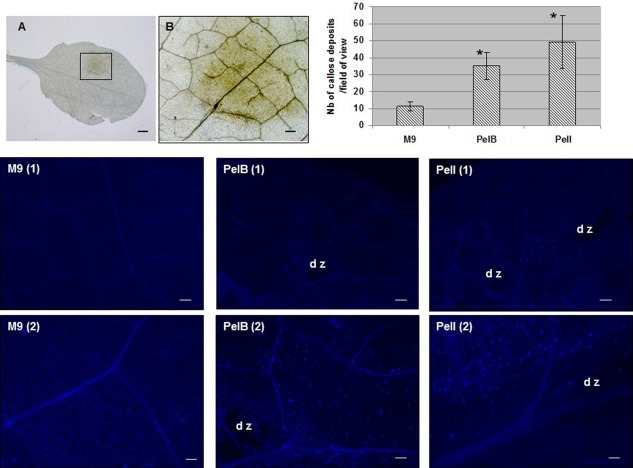
Role of individual pectate lyases in callose deposition. Leaves of Col‐0 wild‐type were infiltrated with individual pectate lyases as reported in Fig. 6. They were collected at the 3‐h post‐infiltration (hpi) time point to be stained by aniline blue for microscopic observations of callose deposits. Photographs in (A) and (B) show a browning (3 hpi) caused by PelB visualized by light microscopy. Bar, 1 mm (A); bar, 250 µm (B). Photographs labelled as M9, PelB and PelI show representative reactions of callose deposition observed at two magnifications for each condition: bar, 250 µm in (1); bar, 100 µm in (2); dz, degraded zone. For callose deposit quantification, n = 6 leaves and bars on the graph represent standard error. Asterisks indicate a statistically significant difference between PelB or PelI and control samples (Mann–Whitney, P = 0.01). Experiments were carried out twice with similar results.
Pectic enzymes do not cause the cell death reaction triggered by D. dadantii in A. thaliana
In D. dadantii‐infected A. thaliana plants, cell death was described as a reaction appearing around the macerated area (Kraepiel et al., 2011). As there are several mechanisms that trigger cell death (Dickman and Fluhr, 2013), we asked whether, on the basis of the data presented above, there was a link between cell death, ROS production and Pel activity. We followed the appearance of cell death in leaves of Col‐0 plants infected with the wild‐type strain using trypan blue staining. We visualized a dark blue coloration corresponding to the inoculated zone at 12 hpi. A similar reaction was induced after inoculation of the outC and pel mutants (Fig. 8). We tested whether this reaction was still visible after symptom appearance. With the wild‐type strain, the coloration appeared to be confined to a region surrounding the macerated tissue (Fig. S8A, see Supporting Information). With the outC and pel mutants, the tissues were not macerated at 16 hpi, and the coloration remained visible throughout the inoculated zone. We found no difference in the induction of this reaction between the wild‐type ecotype and the atrbohD mutant line (Fig. S8A), indicating that ROS accumulation elicited by the bacterium was not essential for the expression of this type of cell death. To verify that cell death was not caused by pectic enzymes, we tested the effect of a CS from the wild‐type strain grown in M9 PGA, after infiltration in Col‐0 leaves: there was no reaction (Fig. S8B). There was also no reaction after infiltration of a CS prepared from the pel mutant, indicating that the absence of reaction with a wild‐type CS was not caused by a negative effect of Pels on the expression of a potential cell death elicitor. In Col‐0 and atrbohD plants, D. dadantii‐induced cell death was not affected by the hrcC mutation (Fig. S9, see Supporting Information), which led us to conclude that the T3SS or cognate effectors did not induce the reaction. Independent of pectic enzymes and ROS and generally independent of type II and type III secretion, this cell death remains to be mechanistically and functionally explained.
Figure 8.
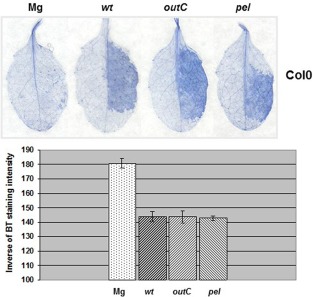
Cell death induced by Dickeya dadantii wild‐type, outC and pel strains in Arabidopsis thaliana. Leaves of Col‐0 plants were inoculated with a bacterial suspension of the strain to be tested, or an MgSO4 solution (Mg) (as indicated in the figure), according to the method described in Experimental Procedures. Trypan blue (BT) staining was performed on leaves harvested at the 12‐h post‐infiltration (hpi) time point. Photographs show one representative reaction for each condition. The graph shows a quantification of the staining intensity (see Experimental Procedures) expressed as the inverse of grey pixels/leaf counted in a field of 100 mm2; n = 12 leaves and bars represent standard error. Experiments were carried out three times with similar results.
Discussion
Dickeya dadantii causes a systemic symptom of maceration on A. thaliana resulting from the action of a set of pectin‐degrading enzymes produced by the bacterium in the plant tissue causing cell wall degradation. In a first report, Fagard et al. (2007) showed that the early phase of infection by D. dadantii is accompanied by an extracellular production of ROS and callose deposits visible until the tissues are degraded. These authors also showed that the A. thaliana atrbohD single mutant, as well as the atrbohD/F double mutant, shows a clear susceptibility phenotype, revealing that ROS produced by the NADPH oxidase isoform AtrbohD are involved in plant resistance to this bacterium. In the present study, we found the existence of a causal link between ROS production and callose deposition reminiscent of the PTI pathway in response to typical PAMPs (Daudi et al., 2012; Galletti et al., 2011; Zhang et al., 2007). By investigating the aptitude of D. dadantii mutants with defects in envelope structure or secreted proteins to induce these plant reactions, we discovered the role of D. dadantii Pels in the recognition process between the bacterium and the plant.
Using DCFH‐DA as an imaging probe, we determined that the oxidative burst induced by D. dadantii is accompanied by the intracellular accumulation of ROS. In the absence of a functional atrbohD gene, this ROS accumulation does not take place, which confirms the major role of AtrbohD in the interaction between D. dadantii and A. thaliana. Fagard et al. (2007) observed a high accumulation of atrbohD gene transcripts, relative to those of the atrbohF gene, in D. dadantii‐infected Col‐0 plants, indicating that transcriptional control of the AtrbohD gene regulates ROS production. However, a post‐translational activation of the enzyme cannot be excluded, as AtrbohD activity can be subject to several steps of control during the innate immune response (Dubiella et al., 2013; Kadota et al., 2014; Li et al., 2014; Rasul et al., 2012). Rather intriguing is the link between AtrbohD‐dependent ROS production and the intracellular accumulation of ROS. AtrbohD‐dependent intracellular ROS accumulation during the non‐host resistance response of A. thaliana towards the bacterium Erwinia amylovora has also been described (Launay et al., 2016). Cross‐talk between different pools of ROS has been reported. A study by Davletova et al. (2005) proposed the existence of cross‐talk between pools of ROS in the apoplast and in the chloroplast during light stress. Therefore, it seems possible that a high or sustained ROS production via a transmembrane NADPH oxidase could result in the elevation of the intracellular ROS content. The recent finding that an A. thaliana aquaporin is a facilitator of H2O2 transport across the plasma membrane, which mechanistically links the induction of apoplastic H2O2 to the activation of PTI pathways (Tian et al., 2016), provides an attractive explanation for our observation.
Our study shows that AtrbohD‐mediated ROS production is required for callose accumulation. We did not observe significant changes in these plant responses between D. dadantii wild‐type and mutant strains, except with the mutant defective in the production of the five Pels, PelA to PelE, suggesting a role of these enzymes in the triggering process. However, the fact that an out‐deficient mutant lacking a functional T2SS triggers a ROS burst comparable with that induced by the wild‐type strain may seem contradictory (Fig. 3A, left panel; Fagard et al., 2007). This discrepancy could be caused by the leakiness of the outC mutation. Another explanation could be that bacterial cell death of the outC mutant in planta releases Pel enzymes which, as shown here, can trigger ROS and callose accumulation. Indeed, the soluble cell fraction of the outC mutant contains larger amounts of pectinases than that of the wild‐type strain because the enzymes normally produced by bacteria remain accumulated in the periplasm. Thus, the CF of the outC mutant causes a higher ROS production than that of the wild‐type strain (Fig. 3A, right panel). The role of pectinases in the triggering process is further supported by the analysis of ROS and callose production in response to CSs from the wild‐type and pel‐deficient strains. Direct evidence is provided by the study of the effect of purified PelB and PelI enzymes, which are representatives of two families of Pel secreted by D. dadantii, PL1 and PL3, respectively (www.cazy.org; Shevchik et al., 1997a, b). Leaf infiltration of these Pels triggers ROS formation and callose deposition. In the same way, these enzymes are able to induce the expression of Chi‐B and PDF1, two defence genes representative of the JA‐ and JA/ET‐signalling pathways, respectively, as well as that of the AOX1 gene, known to respond to changes in the cellular oxidative status. The VPS2 gene, a marker of the JA/ABA‐mediated pathway, appeared to be inducible by PelI and the isolated catalytic domain of this enzyme, PelI Cat (see next paragraph). The VSP2 gene induction by PelB at 6 hpi which, however, decreased at 24 hpi suggests that variations in the timing and intensity of the JA‐mediated defence response may occur depending on the enzyme. These results corroborate the data of Fagard et al. (2007), who showed that the induction of Chi‐B and AOX1 genes by D. dadantii takes place in response to individual Pels.
The question raised by these data concerns the mechanism by which D. dadantii Pels induce a plant immune response reminiscent of PTI. A previous study by Zhang et al. (2014) has demonstrated that a polygalacturonase from the fungal pathogen Botrytis cinerea is recognized as a microbe‐associated molecular pattern (MAMP) by an A. thaliana leucine‐rich repeat receptor‐like protein encoded by the RBPG1 gene, which is essential for the expression of defence‐inducing activity. In this case, the defence‐inducing activity of the enzyme appeared to be independent of its catalytic activity. The PelI protein could also be viewed as a direct elicitor of the basal defence system. Indeed, proteins called harpins, such as HrpW from E. amylovora and Pseudomonas syringae, are well‐known elicitors of plant defence and their C‐terminal domain shares homology with Pels of family PL3, including PelI (Charkowski et al., 1998; Kim and Beer, 1998). However, harpins are heat stable and have no catalytic activity. A study by Shevchik et al. (1998) showed that the PelI catalytic domain (PelI Cat), deleted from its N‐terminal fibronectin‐like type III domain, is able to induce necrosis on tobacco leaves and the formation of capsidiol in cell culture. However, experimental evidence indicates that the capacity of PelI Cat to trigger these plant defence reactions is strictly dependent on its catalytic activity (V. S. Shevchik, unpublished data). It is worth remembering that the harpin of E. amylovora was detected in leaves of apple seedlings by immunolabelling (Périno et al., 1999), whereas Pels could not be identified in planta by tissue printing, probably because these enzymes, which are cell wall destructive, lead to a rapid disorganization of the tissues. Therefore, it seems unlikely that Pels act as direct elicitors of the defences observed. Their action is probably connected to their enzymatic activity. The finding that a catalytically inactive Pel (PelIQ227A) was also inefficient in inducing ROS accumulation strongly supports this hypothesis.
The enzymatic characteristics of D. dadantii endo‐pectate lyases, including PelB and PelI, have been reviewed in detail (Hugouvieux‐Cotte‐Pattat et al., 2014). Endo‐pectate lyases attack randomly inside the PGA chain and generate multiple products of various sizes. The processivity and final end products of these enzymes depend on the structure of their active site, and therefore the presence of several isozymes in a plant tissue can lead to a mixture of small and large oligomers. Therefore, the common aptitude of PelB and PelI to generate the ROS burst and callose deposition could result from the release of large amounts of OGs. The observation of callose deposits in Pel‐treated tissues around the lesions (Fig. 7) suggests that cell wall degradation is required for induction of the reaction. Thus, the formation of ROS and callose, as well as the activation of the JA‐mediated signalling pathway, can be hypothesized to be part of a PTI‐like response to OGs released from plant cell wall pectin. It is acknowledged that OGs function as DAMPs recognized by plant receptors involved in the first phase of the immune response (Denoux et al., 2008; Galletti et al., 2009). The signalling role of these molecules in PTI was studied by treating plants with purified preparations of OGs with a diverse degree of polymerization, ranging from 10 to 15 on average. Purified OGs obtained with a fungal endopolygalacturonase proved to be able to induce an AtrbohD‐mediated ROS production and callose accumulation in A. thaliana, as well as an array of defences conferring resistance to B. cinerea (Galletti et al., 2008).
We found that D. dadantii is able to cause necrosis in A. thaliana, a symptom of cell death sometimes observed in the absence of maceration symptoms or at the front of maceration (Kraepiel et al., 2011). Interestingly, the cell death described previously on N. benthamiana in response to P. carotovorum appeared to be a reaction preceding maceration. In that case, it was dependent on the expression of the bacterial T3SS (Kim et al., 2011). The cell death induced by D. dadantii can also occur before maceration, and without the involvement of ROS production mediated by AtrbohD. None of the bacterial mutants tested in this study were affected in this response. In particular, integrity of the T3SS was dispensable, indicating that the D. dadantii causal agent of cell death is different from that of P. carotovorum. We could also rule out a potential role of flagellin. Thus, the underlying mechanism remains to be elucidated. However, it is interesting to note that cell death induced by D. dadantii in A. thaliana is independent of the pectinolytic activity and of AtrbohD‐dependent ROS production. One hypothesis could be that this cell death is beneficial to the bacterium to obtain access to nutrients without needing to degrade pectin. Another hypothesis is that the death of plant cells is a defence reaction working in the plant against the bacterium, as suggested by Kraepiel et al. (2011).
In conclusion, the present study shows that D. dadantii can induce in the host A. thaliana a PTI‐like response, including the accumulation of ROS and callose deposits dependent on the action of the NADPH oxidase isoform AtrbohD, as well as activation of the JA‐mediated signalling pathway. This response requires the production of bacterial Pels known to act as virulence determinants. The Pel catalytic activity is required for ROS induction, which indicates that the catabolic products (OGs) released by the enzymes are probably involved in the induction of the immune response. The D. dadantii–A. thaliana pathosystem provides a model in which OGs, which represent a carbon source for bacterial growth and are co‐regulators of the virulence genes, are also susceptible to influence the infectious process by triggering the plant immune system. In this interaction, the role of plant cell death, either beneficial or detrimental to D. dadantii, remains to be elucidated.
Experimental Procedures
Plant material and growth conditions
Arabidopsis thaliana seeds from the Col‐0 ecotype were obtained from the INRA Versailles collection (Versailles, France). Single and double atrbohD and atrbohF mutants (Torres et al., 2002) were kindly provided by J. Dangl (Chapel Hill, NC, USA). Plants were grown on soil as described previously (Fagard et al., 2007).
Bacterial strains, bacterial material and growth conditions
The wild‐type strain D. dadantii (previously named Erwinia chrysanthemi 3937) (Samson et al., 2005) was isolated from Saintpaulia ionantha H. Wendl. (African violet). The isogenic mutant strains contain the following null mutations: outC::uidA‐Km (Kazemi‐Pour et al., 2004), outD::uidA‐Cm (G. Condemine, Villeurbanne, France), ΔpelADE ΔpelBC (pel) (Beaulieu et al., 1993), fliC::uidA‐Km (V. E. Shevchik), hrcC::uidA‐Km (Kraepiel et al., 2011) and acsA::Cm cbsE::Spec (Franza et al., 2005). The LPS‐defective mutants RH6065 and R1456, isolated as phage phiEC2‐resistant derivatives from strain 3937, have been described previously (Schoonejans et al., 1987). For plant inoculations, bacterial growth conditions were as described in Fagard et al. (2007). CSs and soluble CFs were obtained from bacterial cultures grown in M9 minimal medium (Sambrook et al., 1989) at 30°C with shaking to the late exponential phase (optical density at 600 nm of 0.8). M9 medium was supplemented with glycerol (2 g/L) as a carbon source and PGA (2 g/L) as a Pel inducer. CSs were filter sterilized and stored at −20°C until use. For CF preparations, the cultures were centrifuged and washed in M9 medium at 4°C before a sonication step on ice. Lysis mixtures were centrifuged for 45 min at 10 000 g at 4°C and the supernatant fluids were harvested. They were kept on ice before infiltration into plant leaves.
Determination of Pel activity
Pel activity was detected by the cup‐plate technique (Andro et al., 1984). When necessary, Pel activity was assessed in CSs as reported previously (Franza et al., 1999). Pel activity is expressed as micromoles of unsaturated products liberated per minute and per milligram (dry weight) of bacteria.
Plant inoculations and treatments
We used a syringe without a needle to infiltrate approximately one‐half of a leaf with the bacterial suspension or the sample of CS to be tested. Bacterial suspensions for inoculation were performed in 10 mm MgSO4 at a concentration of 107 colony‐forming units (cfu)/mL; a 10 mm MgSO4 solution (Mg) was used for control leaves. CSs were diluted in M9 medium before infiltration to achieve a Pel activity (specific activity of about 0.02) similar to that produced by a bacterial culture corresponding to 107 cfu/mL. Routinely, we used five plants and two leaves per plant per condition. The Pels PelB, PelI and PelIQ227A were purified to homogeneity from recombinant E. coli BL21 strains (Shevchik et al. 1997a, 1997b). For histochemical assays, they were diluted in M9 medium to reach a concentration of 200 µg/mL. For RT‐PCR analysis, Pels were diluted in 10 mm Tris‐HCl (pH 8). Samples of 100 µL were used to infiltrate approximately one‐quarter of a leaf.
Histochemical assays
To detect H2O2 in leaves, we used the DAB coloration method described by Torres et al. (2002). DAB reacts with H2O2, resulting in the formation of a dark brown precipitate. To monitor intracellular H2O2 accumulation, we used the fluorogenic probe DCFH‐DA, according to the procedure described previously (Kieu et al., 2012). Callose deposition was detected by aniline blue staining, as described previously (Fagard et al., 2007). Leaves inoculated with bacterial suspensions or CSs were observed with a Zeiss (Jena, Germany) fluorescence microscope. Leaves treated with individual Pels were examined by stereofluorescence microscopy (Azio Zoom V.16, Carl Zeiss Inc., Jena, Germany). Cell death was visualized using trypan blue staining, as described by Kraepiel et al. (2011). For image analysis, we used Image J software (National Institutes of Health, Bethesda, MD, USA). Quantifications were carried out as described previously (Kieu et al., 2012). For DAB, aniline blue and trypan blue staining, the colour image was transformed into an eight‐bit grey scale picture before quantification. For DCFH‐DA fluorescence, green was converted into grey and mean grey values were calculated. All assays were realized three times (except when specified) with similar results.
RNA extraction and RT‐PCR analysis
For RNA extraction, infiltrated leaves were cut off at the indicated times, immediately frozen in liquid nitrogen and stored at −70 ºC. Total RNA preparations and RT‐PCR analysis were carried out according to the procedure described previously (Fagard et al., 2007). The gene‐specific primers used in this analysis were as follows: AOX1a‐F (At3g22370), 5′‐ACATCTGCTTGGATATGGACTAGAG‐3′; AOX1a‐R, 5′‐TAAGTAACCAAGGAAATAAGCGTTG‐3′; PDF1‐F (At5g44420), 5′‐TCATGGCTAAGTTTGCTTCCATCATCACCC‐3′; PDF1‐R, 5′‐GTAGATTTAACATGGGACG‐3′; CHI‐B‐F, 5′‐TTGTCCTGCTAGAGGTTTCTACACT‐3′; CHI‐B‐R, 5′‐ATCAAGATTACCACCAGGATTAACA‐3′; VSP2‐F (At5g24770), 5′‐TGCATCTTTACCAAAACATCATAGA‐3′; VSP2‐R, 5′‐ATGTTGTATCCTTTCTTCACGAGAC‐3′; EF1α‐F (At1g07940), 5′‐TTGGCGGCACCCTTAGCTGGATCA‐3′; EF1α‐R, 5′‐ATGCCCCAGGACATCGTGATTTCAT‐3′.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Reactive oxygen species (ROS) burst triggered by Dickeya dadantii mutants in Arabidopsis thaliana. Leaves of Col‐0 wild‐type were inoculated with a suspension of the bacterial strain to be tested or MgSO4 solution (Mg), according to the method described in Experimental Procedures. 3,3′‐Diaminobenzidine (DAB) staining and quantification were performed as reported in Fig. 1. Photographs show one representative reaction for each condition observed in leaves harvested at 12 h post‐infiltration (hpi). Data are representative of three independent experiments. Bars, standard error; n = 16 leaves. Asterisk indicates statistically significant difference between D. dadantii wild‐type and pel mutant (Mann–Whitney, P < 0.01). Bacterial strains tested are indicated by their genotype or their name [for the lipopolysaccharide (LPS) mutant] as described in the text.
Fig. S2 Photographs showing reactive oxygen species (ROS) accumulation detected by dihydrodichlorofluorescein diacetate (DCFH‐DA) staining used for quantification. The photographs correspond to experiments described in the text and shown in Fig. 3. CF, cellular fraction.
Fig. S3 Role of Dickeya dadantii pectic enzymes in reactive oxygen species (ROS) burst induction in Arabidopsis thaliana by a 3,3′‐diaminobenzidine (DAB) staining analysis. (A) Leaves of Col‐0 wild‐type were infiltrated with a sample of culture supernatant fluid from wild‐type strain (wt CS), or a sample of M9 polygalacturonate (PGA) medium (M) (as indicated in the figure), according to the method described in Experimental Procedures. (B) The atrbohD mutant line and wt CS boiled for 15 min prior to leaf infiltration were tested. DAB staining and quantification were performed as reported in Fig. 1. Photographs show one representative reaction for each condition. In (A, left), asterisk indicates the difference between M and wt CS, on Col‐0 (Mann–Whitney, P < 0.01). In (A, right), asterisk indicates the difference between wt CS and wt CS boiled, on Col‐0 (Mann–Whitney, P < 0.05). Experiments were carried out twice with similar results. (B) Leaves of Col‐0 wild‐type and atrbohD mutant line were infiltrated with a sample of culture supernatant fluid from the pel mutant strain (pel CS) as indicated in the figure. The experiment was carried out twice with similar results.
Fig. S4 Callose deposition induced by Dickeya dadantii mutants in Arabidopsis thaliana. Leaves of Col‐0 plants were inoculated with a suspension from wild‐type or mutant strains, or an MgSO4 solution (Mg) (as indicated in the figure), according to the method described in Experimental Procedures. Callose detection and quantification were performed as described in Fig. 5. Only the quantification is shown. Bacterial strains are referred to by their genotype or their name [for the lipopolysaccharide (LPS) mutant] as indicated in the text. Experiments were carried out three times with similar results. Nb, number.
Fig. S5 Callose deposition induced by Dickeya dadantii pectic enzymes in Arabidopsis thaliana. (A) Leaves of Col‐0 wild‐type were inoculated with a bacterial suspension from the strain to be tested or an MgSO4 solution (Mg) (as indicated by their genotypes in the figure), according to the method described in Experimental Procedures. (B) Col‐0 leaves were infiltrated with a sample of culture supernatant fluid from the wild‐type strain (wt CS), or a sample of M9 polygalacturonate (PGA) medium (M), or a sample of wt CS boiled for 15 min prior to infiltration. Callose detection and quantification were performed as described in Fig. 5. Experiments were carried out three times with similar results. Nb, number.
Fig. S6 Callose deposition induced by the Dickeya dadantii outC mutant in Arabidopsis thaliana. Leaves of Col‐0 plants were inoculated with a bacterial suspension of the strain to be tested, a sample of the corresponding cellular fraction (CF), or MgSO4 solution (Mg), according to the method described in Experimental Procedures. Callose detection and quantification were performed as described in Fig. 5. Experiments were carried out three times with similar results. Nb, number.
Fig. S7 Expression of jasmonic acid (JA)‐controlled marker genes in Col‐0 plants in response to purified Dickeya dadantii pectate lyases. The expression of defence genes was monitored by reverse transcription‐polymerase chain reaction after leaf infiltration with pectate lyases PelB, PelI and PelI Cat or Tris buffer, as indicated. Details are presented in the text and in Experimental Procedures. The constitutively expressed EF1α gene was used as an internal control. Experiments were carried out twice with similar results. The figure was rearranged for clarity.
Fig. S8 Cell death phenotype: independence of AtrbohD‐mediated reactive oxygen species (ROS) accumulation and of Dickeya dadantii pectic enzymes. (A) Leaves of Col‐0 wild‐type and atrbohD mutant line were inoculated with a bacterial suspension from the strains to be tested (as indicated in the figure), according to the method described in Experimental Procedures. (B) Col‐0 leaves were infiltrated with culture supernatant fluids (CSs) corresponding to the wt and pel strains (as indicated) grown in M9 polygalacturonate (PGA) medium. Trypan blue staining was performed on eight leaves harvested at the 16‐h post‐infiltration (hpi) (A) and 3‐hpi (B) time points. Photographs show representative reactions for each condition. Experiments were carried out three times with similar results.
Fig. S9 Cell death phenotype caused by a Dickeya dadantii hrcC mutant in Arabidopsis thaliana. (A) Leaves of Col‐0 wild‐type were inoculated with a bacterial suspension from the strains to be tested, or an MgSO4 solution (Mg) (as indicated in the figure), according to the method described in Experimental Procedures. (B) Inoculations were carried out on Col‐0 and atrbohD leaves. Trypan blue (BT) staining was performed on eight leaves harvested at the 12‐h post‐infiltration (hpi) (A) and 16‐hpi (B) time points. Photographs show representative reactions for each condition. Experiments were carried out three times with similar results.
Supporting Information
Acknowledgements
Research in the laboratory was supported by the Institut National de la Recherche Agronomique and the Université Pierre et Marie Curie. We thank Guy Condemine, Sylvie Reverchon and William Nasser for providing D. dadantii outC mutant strains and their encouragement in this work. We are grateful to the reviewers for helpful comments to improve the manuscript. D.E. thanks Professor Larry Barton for reading the initial English version of the manuscript. We thank Hervé Ferry for technical support.
References
- Andro, T. , Chambost, J.‐P. , Kotoujansky, A. , Cattaneo, J. , Bertheau, Y. , Barras, F. , VanGijsegem, F. and Coleno, A. (1984) Mutants of Erwinia chrysanthemi defective in secretion of pectinase and cellulase. J. Bacteriol. 160, 1199–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam, S.Z. , Erbs, G. , Morrissey, K.L. , Newman, M.‐A. , Chinchilla, D. , Boller, T. , Molinaro, A. , Jackson, R.W. and Cooper, R.M. (2009) Microbe‐associated molecular pattern (MAMP) signatures, synergy, size and charge: influences on perception or mobility and host defence responses. Mol. Plant Pathol. 10, 375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu, C. , Boccara, M. and Van Gijsegem, F. (1993) Pathogenic behaviour of pectinase‐defective Erwinia chrysanthemi mutants on different plants. Mol. Plant–Microbe Interact. 6, 197–202. [Google Scholar]
- Benschop, J.J. , Mohammed, S. , O'Flaherty, M. , Heck, A.J. , Slijper, M. and Menke, F.L. (2007) Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell. Proteomics, 6, 1198–1214. [DOI] [PubMed] [Google Scholar]
- Boller, T. and Felix, G. (2009) A renaissance of elicitors: perception of microbe‐associated molecular patterns and danger signals by pattern‐recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. [DOI] [PubMed] [Google Scholar]
- Boudsocq, M. and Sheen, J. (2013) CDPKs in immune and stress signalling. Trends Plant Sci. 18, 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouley, J. , Condemine, G. and Shevchik, V.E. (2001) The PDZ domain of OutC and the N‐terminal region of OutD determine the secretion specificity of the type II out pathway of Erwinia chrysanthemi . J. Mol. Biol. 208, 205–219. [DOI] [PubMed] [Google Scholar]
- Brunner, F. and Nürnberger, T. (2012) Identification of immunogenic microbial patterns takes the fast lane. Proc. Natl. Acad. Sci. USA, 109, 4029–4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charkowski, A.O. , Alfano, J.R. , Preston, G. , Yuan, J. , He, S.Y. and Collmer, A. (1998) The Pseudomonas syringae pv. tomato HrpW protein has domains similar to harpins and pectate lyases and can elicit the plant hypersensitive response and bind to pectate. J.Bacteriol. 180, 5211–5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charkowski, A. , Blanco, C. , Condemine, G. , Expert, D. , Franza, T. , Hayes, C. , Hugouvieux‐Cotte‐Pattat, N. , López Solanilla, E. , Low, D. , Moleleki, L. , Pirhonen, M. , Pitman, A. , Perna, N. , Reverchon, S. , Rodríguez Palenzuela, P. , San Francisco, M. , Toth, I. , Tsuyumu, S. , van der Waals, J. , van der Wolf, J. , Van Gijsegem, F. , Yang, C.H. and Yedidia, I. (2012) The role of secretion systems and small molecules in soft‐rot enterobacteriaceae pathogenicity. Annu. Rev. Phytopathol. 50, 425–449. [DOI] [PubMed] [Google Scholar]
- Coll, N.S. , Epple, P. and Dangl, J.L. (2011) Programmed cell death in the plant immune system. Cell Death Differ. 18, 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creze, C. , Castang, S. , Derivery, E. , Haser, R. , Hugouvieux‐Cotte‐Pattat, N. , Shevchik, V.E. and Gouet, P. (2008) The crystal structure of pectate lyase peli from soft rot pathogen Erwinia chrysanthemi in complex with its substrate. J Biol. Chem. 283, 8260–8268. [DOI] [PubMed] [Google Scholar]
- Daudi, A. , Cheng, Z. , O'Brien, J.A. , Mammarella, N. , Khan, S. , Ausubel, F.M. and Bolwell, G.P. (2012) The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern‐triggered immunity. Plant Cell, 24, 275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova, S. , Rizhsky, L. , Liang, H. , Shengqiang, Z. , Oliver, D.J. , Coutu, J. , Shulaev, V. , Schlauch, K. , and Mittler, R. (2005) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell. 17, 268–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidsson, P.R. , Kariola, T. , Niemi, O. and Palva, E.T. (2013) Pathogenicity of and plant immunity to soft rot bacteria. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellagi, A. , Segond, D. , Rigault, M. , Fagard, M. , Simon, C. , Saindrenan, P. and Expert, D. (2009) Microbial siderophores exert a subtle role in Arabidopsis during infection by manipulating the immune response and the iron status. Plant Physiol. 150, 1687–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoux, C. , Galletti, R. , Mammarella, N. , Gopalan, S. , Werck, G. , De Lorenzo, G. , Ferrari, S. , Ausubel, F.M. and Dewdney, J. (2008) Activation of defense response pathways by OGs and flg22 elicitors in Arabidopsis seedlings. Mol. Plant. 1, 423–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickman, M.B. and Fluhr, R. (2013) Centrality of host cell death in plant–microbe interactions. Annu. Rev. Phytopathol. 51, 543–570. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N. and Rathjen, J.P. (2010) Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Dubiella, U. , Seybold, H. , Durian, G. , Komander, E. , Lassig, R. , Witte, C.P. , Schulze, W.X. and Romeis, T. (2013) Calcium‐dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc . Natl. Acad. Sci. USA, 110, 8744–8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard, M. , Dellagi, A. , Roux, C. , Périno, C. , Rigault, M. , Boucher, V. , Shevchik, V.E. and Expert, D. (2007) Arabidopsis thaliana expresses multiple lines of defense to counterattack Erwinia chrysanthemi . Mol. Plant–Microbe Interact. 20, 794–805. [DOI] [PubMed] [Google Scholar]
- Franza, T. , Sauvage, C. and Expert, D. (1999) Iron regulation and pathogenicity in Erwinia chrysanthemi 3937: role of the Fur repressor protein. Mol. Plant–Microbe Interact. 12, 119–128. [DOI] [PubMed] [Google Scholar]
- Franza, T. , Mahé, B. and Expert, D. (2005) Erwinia chrysanthemi requires a second iron transport route dependent on the siderophore achromobactin for extracellular growth and plant infection. Mol. Microbiol. 55, 261–275. [DOI] [PubMed] [Google Scholar]
- Galletti, R. , Denoux, C. , Gambetta, S. , Dewdney, J. , Ausubel, F.M. , De Lorenzo, G. and Ferrari, S. (2008) The AtrbohD‐mediated oxidative burst elicited by oligogalacturonides in Arabidopsis is dispensable for the activation of defense responses effective against Botrytis cinerea . Plant Physiol. 148, 1695–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletti, R. , Ferrari, S. and De Lorenzo, G. (2001) Arabidopsis MPK3 and MPK6 play different roles in basal and oligogalacturonide‐ or flagellin‐induced resistance against Botrytis cinerea. Plant Physiol. 157, 804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletti, R. , De Lorenzo, G. and Ferrari, S. (2009) Host‐derived signals activate plant innate immunity. Plant Signal. Behav. 4, 33–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Hugouvieux‐Cotte‐Pattat, N. , Condemine, G. and Shevchik, V. (2014) Bacterial pectate lyases, structural and functional diversity. Environ. Microbiol. Rep. 6, 427–440. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nat. Rev. 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kadota, Y. , Sklenar, J. , Derbyshire, P. , Stransfeld, L. , Asai, S. , Ntoukakis, V. , Jones, J.D. , Shirasu, K. , Menke, F. , Jones, A. and Zipfel, C. (2014) Direct regulation of the NADPH oxidase RBOHD by the PRR‐associated kinase BIK1 during plant immunity. Mol. Cell, 54, 43–55. [DOI] [PubMed] [Google Scholar]
- Kariola, T. , Palomäki, T.A. , Brader, G. and Palva, E.T. (2003) Erwinia carotovora sbsp. carotovora and Erwinia‐derived elicitors HrpN and PehA trigger distinct but interacting defense responses and cell death in Arabidopsis. Mol. Plant–Microbe Interact. 16, 179–187. [DOI] [PubMed] [Google Scholar]
- Kazemi‐Pour, N. , Condemine, G. and Hugouvieux‐Cotte‐Pattat, N. (2004) The secretome of the plant pathogenic bacterium Erwinia chrysanthemi . Proteomics, 4, 3177–3188. [DOI] [PubMed] [Google Scholar]
- Kieu, N.P. , Aznar, A. , Segond, D. , Rigault, M. , Simond‐Côte, E. , Kunz, C. , Soulié, M.‐C. , Expert, D. and Dellagi, A. (2012) Iron deficiency affects plant defence responses and confers resistance to Dickeya dadantii and Botrytis cinerea . Mol. Plant Pathol. 13, 807–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.S. , Thammarat, P. , Lommel, S.A. , Hogan, C.S. and Charkowski, A.O. (2011) Pectobacterium carotovorum elicits plant cell death with DspE/F but the P. carotovorum DspE does not suppress callose or induce expression of plant genes early in plant–microbe interactions. Mol. Plant–Microbe Interact. 24, 773–786. [DOI] [PubMed] [Google Scholar]
- Kim, J.F. and Beer, S.V. (1998) HrpW of Erwinia amylovora, a new harpin that contains a domain homologous to pectate lyases of a distinct class. J. Bacteriol. 180, 5203–5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraepiel, Y. , Pédron, J. , Patrit, O. , Simond‐Côte, E. , Hermand, V. and van Gijsegem, F. (2011) Analysis of the plant bos1 mutant highlights necrosis as an efficient defence mechanism during D. dadantii/Arabidopsis thaliana interaction. PLoS One, 6, e18991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Launay, A. , Patrit, O. , Wenes, E. and Fagard, M. (2016) DspA/E contributes to apoplastic accumulation of ROS in nonhost A. thaliana . Front. Plant Sci. 7, 00545 10.3389/fpls.2016.00545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecourieux, D. , Ranjeva, R. and Pugin, A. (2006) Calcium in plant defence‐signalling pathways. New Phytol. 171, 249–269. [DOI] [PubMed] [Google Scholar]
- Li, L. , Li, M. , Yu, L. , Zhou, Z. , Liang, X. , Liu, Z. , Cai, G. , Gao, L. , Zhang, X. , Wang, Y. , Chen, S. and Zhou, J.M. (2014) The FLS2‐associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe, 15, 329–338. [DOI] [PubMed] [Google Scholar]
- Luna, E. , Pastor, V. , Robert, J. , Flors, V. , Mauch‐Mani, B. and Ton, J. (2011) Callose deposition: a multifaceted plant defense response. Mol. Plant–Microbe Interact. 24, 183–193. [DOI] [PubMed] [Google Scholar]
- Mattinen, L. , Tshuikina, M. , Mäe, A. and Pirhonen, M. (2004) Identification and characterization of Nip, a necrosis‐inducing virulence protein of Erwinia carotovora sbsp. carotovora . Mol. Plant–Microbe Interact. 17, 1366–1375. [DOI] [PubMed] [Google Scholar]
- Mengiste, T. (2012) Plant immunity to necrotrophs. Annu. Rev. Phytopathol. 50, 267–294. [DOI] [PubMed] [Google Scholar]
- Monaghan, J. and Zipfel, F. (2012) Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 15, 349–357. [DOI] [PubMed] [Google Scholar]
- Moore, J.W. , Loake, G.L. and Spoel, S.H. (2011) Transcription dynamics in plant immunity. Plant Cell, 23, 2809–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, M.A. , Sundelin, T. , Nielsen, J.T. and Erbs, G. (2013) MAMP (microbe‐associated molecular pattern) triggered immunity in plants. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman, C. , Vidal, S. and Palva, E.T. (1999) Oligogalacturonide‐mediated induction of a gene involved in jasmonic acid synthesis in response to the cell‐wall‐degrading enzymes of the plant pathogen Erwinia carotovora . Mol. Plant–Microbe Interact. 12, 640–644. [DOI] [PubMed] [Google Scholar]
- Norman‐Setterblad, C. , Vidal, S. and Palva, E.T. (2000) Interacting signal pathways control defense gene expression in Arabidopsis in response to cell‐wall degrading enzymes from Erwinia carotovora . Mol. Plant–Microbe Interact. 13, 430–438. [DOI] [PubMed] [Google Scholar]
- Nühse, T.S. , Bottrill, A.R. , Jones, A.M. and Peck, S.C. (2007) Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J. 51, 931–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nürnberger, T. , Brunner, F. , Kemmerling, B. and Piater, L. (2004) Innate immunity in plants and animals: striking similarities and obvious differences. Immunol. Rev. 198, 249–266. [DOI] [PubMed] [Google Scholar]
- Palva, T.K. , Holmström, K.O. , Heino, P. and Palva, E.T. (1993) Induction of plant defense response by exoenzymes of Erwinia carotovora subsp. carotovora . Mol. Plant–Microbe Interact. 6, 190–196. [Google Scholar]
- Périno, C. , Gaudriault, S. , Vian, B. and Barny, M.A. (1999) Visualization of harpin secretion in planta during infection of apple seedlings by Erwinia amylovora . Cell. Microbiol. 1, 131–141. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M.J. , Van der Does, D. , Zamioudis, C. , Leon‐Reyes, A. and Van Wees, S.C.M. (2012) Hormonal modulation of plant immunity. Annu. Rev. Cell. Dev. 28, 489–521. [DOI] [PubMed] [Google Scholar]
- Pineau, C. , Gushinskaya, N. , Robert, X. , Gouet, P. , Ballut, L. and Shevchik, V.E. (2014) Substrate recognition by the bacterial type II secretion system: more than a simple interaction. Mol. Microbiol. 94, 126–140. [DOI] [PubMed] [Google Scholar]
- Rasul, S. , Dubreuil‐Maurizi, C. , Lamotte, O. , Koen, E. , Poinssot, B. , Alcaraz, G. , Wendehenne, D. and Jeandroz, S. (2012) Nitric oxide production mediates oligogalacturonide‐triggered immunity and resistance to Botrytis cinerea in Arabidopsis thaliana . Plant Cell Environ. 35, 1483–1499. [DOI] [PubMed] [Google Scholar]
- Reverchon, S. and Nasser, W. (2013) Dickeya ecology: environment sensing and regulation of virulence programme. Environ. Microbiol. Rep. 5, 622–636. [DOI] [PubMed] [Google Scholar]
- Ridley, B.L. , O’Neill, M.A. and Mohnen, D. (2001) Pectins: structure, biosynthesis, and oligogalacturonide‐related signaling. Phytochemistry 57, 929–967. [DOI] [PubMed] [Google Scholar]
- Rojas, C. and Mysore, K.S. (2012) Glycolate oxidase is an alternative source for H2O2 production during plant defense responses and functions independently from NADPH oxidase. Plant Signal. Behav. 7, 752–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. , Fritsch, E.F. and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn Cold Spring Harbor, NY: Cold Spring Harbor Press. [Google Scholar]
- Samson, R. , Legendre, J.B. , Christen, R. , Fischer‐Le Saux, M. , Achouak, W. and Gardan, L. (2005) Transfer of Pectobacterium chrysanthemi (Burkholder et al. 1953) Brenner et al. 1973 and Brenneria paradisiaca to the genus Dickeya gen. nov. as Dickeya chrysanthemi comb. nov. and Dickeya paradisiaca comb. nov. and delineation of four novel species, Dickeya dadantii sp. nov., Dickeya dianthicola sp. nov., Dickeya dieffenbachiae sp. nov. and Dickeya zeae sp. nov. Int. J. Syst. Evol. Microbiol. 55, 1415–1427. [DOI] [PubMed] [Google Scholar]
- Schoonejans, E. , Expert, D. and Toussaint, A. (1987) Characterization and virulence properties of Erwinia chrysanthemi ϕEC2‐resistant mutants. J. Bacteriol. 169, 4011–4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segonzac, C. , Feike, D. , Gimenez‐Ibanez, S. , Hann, D.R. , Zipfel, C. and Rathjen, J.P. (2011) Hierarchy and roles of pathogen‐associated molecular pattern‐induced responses in Nicotiana benthamiana . Plant Physiol. 156, 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchik, V.E. , Robert‐Baudouy, J. and Condemine, G. (1997a) Specific interaction between OutD, an Erwinia chrysanthemi outer membrane protein of the general secretory pathway, and secreted proteins. EMBO J. 16, 3007–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchik, V.E. , Robert‐Baudouy, J. and Hugouvieux‐Cotte‐Pattat, N. (1997b) The pectate lyase PelI of Erwinia chrysanthemi belongs to a new family. J. Bacteriol. 179, 7321–7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchik, V.E. , Boccara, M. , Vedel, R. and Hugouvieux‐Cotte‐Pattat, N. (1998) Processing of the pectate lyase PelI by extracellular proteases of Erwinia chrysanthemi 3937. Mol. Microbiol. 29, 1459–1469. [DOI] [PubMed] [Google Scholar]
- Spoel, S.H. and Dong, X. (2012) How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 12, 89–100. [DOI] [PubMed] [Google Scholar]
- Swanson, S.J. , Choi, W.G. , Chanoca, A. and Gilroy, S. (2011) In vivo imaging of Ca2+, pH, and reactive oxygen species using fluorescent probes in plants. Annu. Rev. Plant Biol. 62, 273–297. [DOI] [PubMed] [Google Scholar]
- Tardy, F. , Nasser, W. , Robert‐Baudouy, J. and Hugouvieux‐Cotte‐Pattat, N. (1997) Comparative analysis of the five major Erwinia chrysanthemi pectate lyases: enzyme characteristics and potential inhibitors. J. Bacteriol. 179, 2503–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, S. , Wang, X. , Li, P. , Wang, H. , Ji, H. , Xie, J. , Qiu, Q. , Shen, D. and Dong, H. (2016) Plant aquaporin AtPIP1;4 links apoplastic H2O2 induction to disease immunity pathways. Plant Physiol. 171, 1635–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, M.A. (2010) ROS in biotic interactions. Physiol Plant. 138, 414–429. [DOI] [PubMed] [Google Scholar]
- Torres, M.A. , Dangl, J.L. and Jones, J.D. (2002) A. thaliana gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc . Natl. Acad. Sci. USA, 99, 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth, I.K. , Bell, K.S. , Holeva, M.C. and Birch, P.R.J. (2003) Soft rot erwiniae: from genes to genomes. Mol. Plant Pathol. 4, 17–30. [DOI] [PubMed] [Google Scholar]
- Toth, I.K. , van der Wolf, J.M. , Saddler, G. , Lojkowska, E. , Hélias, V. , Pirhonen, M. , Tsor (Lahkim), L. and Elphinstone, J.G. (2011) Dickeya species: an emerging problem for potato production in Europe. Plant Pathol. 60, 385–399. [Google Scholar]
- Zhang, J. , Shao, F. , Li, Y. , Cui, H. , Chen, L. , Li, H. , Zou, Y. , Long, C. , Lan, L. , Chai, I. , Chen, S. , Tang, X. and Zhou, J.‐M. (2007) A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP‐induced immunity in plants. Cell Host Microbe, 1, 175–185. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Kars, I. , Essenstam, B. , Liebrand, T.W.H. , Wagemakers, L. , Elberse, J. , Tagkalaki, P. , Tjoitang, D. , van den Ackerveken, G. and van Kan, J.A.L. (2014) Fungal endopolygalacturonases are recognized as microbe‐associated molecular patterns by the Arabidopsis receptor‐like protein RESPONSIVENESS TO BOTRYTIS POLYGALACTURONASES1. Plant Physiol. 164, 352–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Reactive oxygen species (ROS) burst triggered by Dickeya dadantii mutants in Arabidopsis thaliana. Leaves of Col‐0 wild‐type were inoculated with a suspension of the bacterial strain to be tested or MgSO4 solution (Mg), according to the method described in Experimental Procedures. 3,3′‐Diaminobenzidine (DAB) staining and quantification were performed as reported in Fig. 1. Photographs show one representative reaction for each condition observed in leaves harvested at 12 h post‐infiltration (hpi). Data are representative of three independent experiments. Bars, standard error; n = 16 leaves. Asterisk indicates statistically significant difference between D. dadantii wild‐type and pel mutant (Mann–Whitney, P < 0.01). Bacterial strains tested are indicated by their genotype or their name [for the lipopolysaccharide (LPS) mutant] as described in the text.
Fig. S2 Photographs showing reactive oxygen species (ROS) accumulation detected by dihydrodichlorofluorescein diacetate (DCFH‐DA) staining used for quantification. The photographs correspond to experiments described in the text and shown in Fig. 3. CF, cellular fraction.
Fig. S3 Role of Dickeya dadantii pectic enzymes in reactive oxygen species (ROS) burst induction in Arabidopsis thaliana by a 3,3′‐diaminobenzidine (DAB) staining analysis. (A) Leaves of Col‐0 wild‐type were infiltrated with a sample of culture supernatant fluid from wild‐type strain (wt CS), or a sample of M9 polygalacturonate (PGA) medium (M) (as indicated in the figure), according to the method described in Experimental Procedures. (B) The atrbohD mutant line and wt CS boiled for 15 min prior to leaf infiltration were tested. DAB staining and quantification were performed as reported in Fig. 1. Photographs show one representative reaction for each condition. In (A, left), asterisk indicates the difference between M and wt CS, on Col‐0 (Mann–Whitney, P < 0.01). In (A, right), asterisk indicates the difference between wt CS and wt CS boiled, on Col‐0 (Mann–Whitney, P < 0.05). Experiments were carried out twice with similar results. (B) Leaves of Col‐0 wild‐type and atrbohD mutant line were infiltrated with a sample of culture supernatant fluid from the pel mutant strain (pel CS) as indicated in the figure. The experiment was carried out twice with similar results.
Fig. S4 Callose deposition induced by Dickeya dadantii mutants in Arabidopsis thaliana. Leaves of Col‐0 plants were inoculated with a suspension from wild‐type or mutant strains, or an MgSO4 solution (Mg) (as indicated in the figure), according to the method described in Experimental Procedures. Callose detection and quantification were performed as described in Fig. 5. Only the quantification is shown. Bacterial strains are referred to by their genotype or their name [for the lipopolysaccharide (LPS) mutant] as indicated in the text. Experiments were carried out three times with similar results. Nb, number.
Fig. S5 Callose deposition induced by Dickeya dadantii pectic enzymes in Arabidopsis thaliana. (A) Leaves of Col‐0 wild‐type were inoculated with a bacterial suspension from the strain to be tested or an MgSO4 solution (Mg) (as indicated by their genotypes in the figure), according to the method described in Experimental Procedures. (B) Col‐0 leaves were infiltrated with a sample of culture supernatant fluid from the wild‐type strain (wt CS), or a sample of M9 polygalacturonate (PGA) medium (M), or a sample of wt CS boiled for 15 min prior to infiltration. Callose detection and quantification were performed as described in Fig. 5. Experiments were carried out three times with similar results. Nb, number.
Fig. S6 Callose deposition induced by the Dickeya dadantii outC mutant in Arabidopsis thaliana. Leaves of Col‐0 plants were inoculated with a bacterial suspension of the strain to be tested, a sample of the corresponding cellular fraction (CF), or MgSO4 solution (Mg), according to the method described in Experimental Procedures. Callose detection and quantification were performed as described in Fig. 5. Experiments were carried out three times with similar results. Nb, number.
Fig. S7 Expression of jasmonic acid (JA)‐controlled marker genes in Col‐0 plants in response to purified Dickeya dadantii pectate lyases. The expression of defence genes was monitored by reverse transcription‐polymerase chain reaction after leaf infiltration with pectate lyases PelB, PelI and PelI Cat or Tris buffer, as indicated. Details are presented in the text and in Experimental Procedures. The constitutively expressed EF1α gene was used as an internal control. Experiments were carried out twice with similar results. The figure was rearranged for clarity.
Fig. S8 Cell death phenotype: independence of AtrbohD‐mediated reactive oxygen species (ROS) accumulation and of Dickeya dadantii pectic enzymes. (A) Leaves of Col‐0 wild‐type and atrbohD mutant line were inoculated with a bacterial suspension from the strains to be tested (as indicated in the figure), according to the method described in Experimental Procedures. (B) Col‐0 leaves were infiltrated with culture supernatant fluids (CSs) corresponding to the wt and pel strains (as indicated) grown in M9 polygalacturonate (PGA) medium. Trypan blue staining was performed on eight leaves harvested at the 16‐h post‐infiltration (hpi) (A) and 3‐hpi (B) time points. Photographs show representative reactions for each condition. Experiments were carried out three times with similar results.
Fig. S9 Cell death phenotype caused by a Dickeya dadantii hrcC mutant in Arabidopsis thaliana. (A) Leaves of Col‐0 wild‐type were inoculated with a bacterial suspension from the strains to be tested, or an MgSO4 solution (Mg) (as indicated in the figure), according to the method described in Experimental Procedures. (B) Inoculations were carried out on Col‐0 and atrbohD leaves. Trypan blue (BT) staining was performed on eight leaves harvested at the 12‐h post‐infiltration (hpi) (A) and 16‐hpi (B) time points. Photographs show representative reactions for each condition. Experiments were carried out three times with similar results.
Supporting Information


