Summary
During plant–pathogen interactions, pathogenic bacteria have evolved multiple strategies to cope with the sophisticated defence systems of host plants. Proline iminopeptidase (PIP) is essential to Xanthomonas campestris pv. campestris (Xcc) virulence, and is conserved in many plant‐associated bacteria, but its pathogenic mechanism remains unclear. In this study, we found that disruption of pip in Xcc enhanced its flagella‐mediated bacterial motility by decreasing intracellular bis‐(3′,5′)‐cyclic dimeric guanosine monophosphate (c‐di‐GMP) levels, whereas overexpression of pip in Xcc restricted its bacterial motility by elevating c‐di‐GMP levels. We also found that PIP is a type III secretion system‐dependent effector capable of eliciting a hypersensitive response in non‐host, but not host plants. When we transformed pip into the host plant Arabidopsis, higher bacterial titres were observed in pip‐overexpressing plants relative to wild‐type plants after Xcc inoculation. The repressive function of PIP on plant immunity was dependent on PIP's enzymatic activity and acted through interference with the salicylic acid (SA) biosynthetic and regulatory genes. Thus, PIP simultaneously regulates two distinct regulatory networks during plant–microbe interactions, i.e. it affects intracellular c‐di‐GMP levels to coordinate bacterial behaviour, such as motility, and functions as a type III effector translocated into plant cells to suppress plant immunity. Both processes provide bacteria with the regulatory potential to rapidly adapt to complex environments, to utilize limited resources for growth and survival in a cost‐efficient manner and to improve the chances of bacterial survival by helping pathogens to inhabit the internal tissues of host plants.
Keywords: bacterial motility, pathogen–host interactions, proline iminopeptidase, repression of host immunity
Introduction
Xanthomonas campestris pv. campestris (Xcc) is the causal agent of black rot disease in cruciferous plants (Williams, 1980). We have shown previously that the xccR/pip locus, which encodes the LuxR homologue XccR, regulates the expression of the proline iminopeptidase (PIP, EC 3.4.11.5) gene pip, and disruption of either one of the two genes leads to significantly attenuated virulence in Xcc (Zhang et al., 2007). Notably, pip is genetically conserved in many plant‐associated bacteria, and is positioned adjacent to the luxR gene in the genome (González and Venturi, 2013). PIP was first identified and characterized in Escherichia coli (Sarid et al., 1959). It specifically catalyses the removal of the N‐terminal proline residue from peptides or proteins (Cunningham and O'Connor, 1997), and can also remove N‐terminal l‐alanine or d‐alanine from substrates, albeit at lower efficiency (Alonso and Garcia, 1996). In the food industry, PIP is used to debitterize the peptides in cheese processing (Leenhouts et al., 1998; Tan et al., 1993) and in the synthesis of proline‐containing dipeptides (Yamamoto et al., 2010, 2011). These studies have elaborated the biochemical properties and biotechnological applications of PIP. However, the molecular mechanism underlying how PIP modulates bacterial virulence is currently unknown.
Flagella and flagellum‐mediated cell swimming allow bacteria to approach nutrient sources, invade host cells and escape from host immune system recognition (Chaban et al., 2015; Duan et al., 2013; Josenhans and Suerbaum, 2002; Ottemann and Miller, 1997; Tian et al., 2015). It is believed that flagella‐mediated bacterial motility is important for the virulence of pathogens to their host organisms (Broek and Vanderleyden, 1995; Chaban et al., 2015; Duan et al., 2013; Hossain et al., 2005; Josenhans and Suerbaum, 2002). Flagella play important roles throughout the bacterial infection cycle: flagella‐mediated swimming motility enables bacteria to reach the host tissues and, once there, they play additional roles, including the promotion of surface adhesion and biofilm formation, which drives bacterial penetration between cell–cell junctions during infection (Chaban et al., 2015; Ottemann and Miller, 1997). Xcc possesses a single polar flagellum and more than 40 of its genes are predicted to be involved in flagellar biogenesis and motility (Yang et al., 2009). The flagellar genes are regulated in a three‐tier hierarchy by the RpoD (σ70) housekeeping protein and two alternative sigma factors, RpoN2 (σ54) and FliA (σ28) (Yang et al., 2009). At the top of the regulatory cascade, FleQ functions as a σ54‐cognate activator (Hu et al., 2005). During the invasion process, bacterial motility increases the chances of forming surface‐attached colonies for bacterial infection (Apel and Surette, 2008). Conversely, bacterial flagellins (FliC), the major structural proteins of Gram‐negative flagella, can also trigger defence responses in both plants and animals (Felix et al., 1999; Gewirtz et al., 2001). A flg22 oligopeptide derived from the most conserved region of Pseudomonas syringae flagellin, which acts as a pathogen‐associated molecular pattern (PAMP), is perceived by the host's FLS2 pattern recognition receptor to invoke PAMP‐triggered immunity in plant cells (Zipfel et al., 2004). The bacterial alkaline protease AprA has been found to degrade flagellin monomers for the evasion of host immunity (Pel et al., 2014). The above examples highlight how the fine tuning of flagellar gene expression is crucial for successful bacterial pathogen invasion.
The bacterial second messenger bis‐(3′,5′)‐cyclic dimeric guanosine monophosphate (c‐di‐GMP) regulates the transition from the motile planktonic state to sessile community‐based behaviour (Wolfe and Visick, 2008). High intracellular levels of c‐di‐GMP promote sessile lifestyles, such as a biofilm formation lifestyle, whereas low levels of c‐di‐GMP facilitate motility (Wolfe and Visick, 2008). Cellular c‐di‐GMP levels are controlled through the opposing activities of diguanylate cyclases with GGDEF domains and phosphodiesterases with EAL or HD‐GYP domains, with the former involved in c‐di‐GMP synthesis and the latter in c‐di‐GMP degradation (Hengge, 2009; Jenal and Malone, 2006; Römling and Amikam, 2006). Increasing evidence indicates that a range of cellular effectors, such as RpfG, PilZ domain proteins, transcription factors, RNA riboswitches and enzymatically inactive GGDEF and/or EAL domain proteins, interact directly with c‐di‐GMP and exert diverse influences on bacterial cells (Breaker, 2011; Hengge, 2009; Ryan et al., 2012). FleQ, as a master transcriptional regulator, controls flagella and exopolysaccharide (EPS) gene expression by responding to c‐di‐GMP (Hickman and Harwood, 2008). The Clp global regulator directly regulates the fliC flagellin gene of Xcc, and a large quantity of c‐di‐GMP binds specifically to Clp to abolish the interaction between Clp and the fliC promoter, thereby reducing bacterial motility (Lee et al., 2003; Tao et al., 2010).
The bacterial type III secretion system (T3SS) is an injectisome evolutionarily related to the bacterial flagellum (Notti et al., 2015). Many Gram‐negative plant‐ and animal‐pathogenic bacteria exploit the evolutionarily conserved T3SS to overcome host defences by the direct delivery of effectors to the host cell interior (He et al., 2004). It is interesting to note that c‐di‐GMP plays fundamental roles in the control of multiple important bacterial export pathways. High levels of c‐di‐GMP lead to the repression of Pseudomonas aeruginosa T3SS expression (Moscoso et al., 2011) and the inhibition of ATPase activity through direct allosteric control of the export of ATPase proteins by binding with c‐di‐GMP (Trampari et al., 2015). Type III effectors from plant‐pathogenic bacteria have multiple targets in plants and interfere with broad cellular pathways, including host immune responses, cytoskeletal dynamics, proteasome‐dependent protein degradation and phytohormone signalling (Buttner, 2016). About 40 T3SS effectors have been identified in xanthomonads, and a few have been found to interact with host‐specific factors to exert virulence functions or to trigger immune responses (Gürlebeck et al., 2006; White et al., 2009). Salicylic acid (SA) induction in plants is a general resistance mechanism used against pathogen infection (Vlot et al., 2009). The prevention of SA accumulation leads to disease susceptibility (Gaffney et al., 1993), whereas SA treatment confers resistance to a variety of biotrophic pathogens (Ryals et al., 1996; White, 1979). SA production in plants occurs mainly through the isochorismate pathway (Chen et al., 2009). In Arabidopsis, isochorismate synthase 1 (ICS1), a crucial enzyme in the SA biosynthetic pathway, is induced by biotic and abiotic stresses (Wildermuth et al., 2001). SA activates defence responses through its downstream component, non‐expressor of pathogenesis‐related genes 1 (NPR1), which functions as a key regulator of the SA signalling pathway (Dong, 2004). EDS1 (enhanced disease susceptibility 1) contributes to post‐invasive non‐host resistance, as well as to SA‐dependent and SA‐independent defence signalling, in incompatible host–pathogen interactions (Bartsch et al., 2006; Lipka et al., 2008; Wiermer et al., 2005). The EDS1/PAD4 (phytoalexin deficient 4) complex acts upstream of SA signalling, and contributes to the positive feedback loop for SA accumulation (Feys et al., 2001). To counteract plant resistance, the type III effector can suppress plant innate immunity by the up‐regulation of hormones that are antagonistic to SA, such as the P. syringae AvrPtoB effector, which triggers elevated foliar abscisic acid (ABA) levels to suppress callose deposition and increase plant susceptibility to pathogens (de Torres et al., 2006). The X. campestris pv. vesicatoria effector XopJ interacts with the RPT6 plant proteasomal subunit to interfere with SA‐dependent defence responses by the inhibition of proteasome activity (Ustun et al., 2013). The chloroplast‐localized J domain‐containing HopI1 causes chloroplast thylakoid structure remodelling by interaction with Hsp70, thereby suppressing SA accumulation (Jelenska et al., 2007).
Here, we show that the disruption of pip in Xcc results in increased bacterial motility and the activation of multiple flagellar genes. The reduced level of c‐di‐GMP observed in the pip mutant implies that PIP has a regulatory effect on c‐di‐GMP synthesis or catabolism, which may be advantageous for bacterial communication and group behaviour. We also found that, as a type III effector, PIP can modulate the host plant immune system by the repression of the SA signalling pathway. Our findings offer an insight into the role of PIP by showing that it not only modulates bacterial physiological processes, but also controls the environmental niches for successful establishment of infection by bacterial pathogens.
Results
Key amino acid residues for PIP enzymatic activity
We searched the MEROPS database, an integrated source of information on peptidases (Rawlings, 2016), and found that PIP belongs to the prolyl aminopeptidase S33 family, which preferentially releases an N‐terminal proline residue from peptides. Three residues (S106, D246 and H273) were predicted to comprise the enzymatic active site of PIP (Fig. 1A). To verify the contribution of these key residues to enzymatic activity, PIP variants, each with a single alanine amino acid substitution, were expressed from plasmid pET30a in E. coli BL21 (DE3). The enzyme activity assays showed that the alteration of any of these three residues almost completely abolished the enzyme's proline residue‐releasing ability from the substrate (Fig. 1B), demonstrating that these three amino acids are equally essential for the enzymatic activity of PIP.
Figure 1.
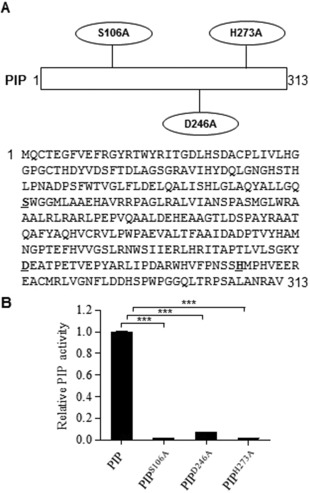
Key amino acid residues for proline iminopeptidase (PIP) enzymatic activity. (A) The amino acid sequence of PIP. Three amino acid residues at the PIP enzyme active site are shown with oval borders and with underlined bold letters in the sequence. The key residues, including a serine residue, a histidine residue and an aspartic acid residue, were mutated to an alanine residue separately. (B) Relative enzymatic activity of PIP and derived mutated proteins. The recombinant proteins were expressed and purified from Escherichia coli. The enzymatic activities of PIP were determined using l‐proline p‐nitroanilide trifluoroacetate salt (PPNA) as a substrate. Data are shown as the mean ± SEM (standard error of the mean) of three independent experiments. Statistical significance was determined using a one‐way analysis of variance (ANOVA; ***P < 0.0001).
Disruption of PIP leading to increased Xcc motility
Our previous work has shown that PIP is important for Xcc virulence and that pip expression is induced in planta (Zhang et al., 2007). However, how PIP affects the bacterial infection process remains unknown. Hence, we first tested the swimming phenotype in Xcc strains on XOLN medium (Fu and Tseng, 1990) (plant environment mimicking medium) motility plates (0.3% agar). The pip‐deficient mutant (pip –) strain showed significantly higher swimming motility, whereas the pip –/pLAFR3‐pip strain restored the motility to the wild‐type level (Fig. 2). Plasmids pLAFR3‐pip S106A, pLAFR3‐pip D246A and pLAFR3‐pip H273A were separately transformed into the pip‐deficient mutant, but the bacterial motility was not restored, indicating that PIP‐modulated bacterial swimming ability is dependent on PIP enzymatic activity (Fig. 2). We further examined the expression of the flagella‐associated genes listed in Table S1 (see Supporting Information). Real‐time quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) assays showed that a lack of pip led to an extensive induction of flagellum genes in XOLN medium. Of the three classes of flagella‐related genes, half of the tested genes were significantly increased in the pip‐deficient mutant. Class II genes, such as fleN, fliA (σ28) and flgH, were significantly increased by 5.42‐fold, 28.93‐fold and 84.21‐fold, respectively (Fig. 3A–C). The expression levels of fliQ and flgB genes were increased even more, by as much as 141.06‐fold and 130.84‐fold, respectively (Fig. 3D,E), whereas the levels of the other tested flagella‐related genes remained unchanged (Table S1). The levels of Class III genes, such as fliC and fliD, increased by 49.43‐fold and 9.45‐fold, respectively (Fig. 3F,G). The expression levels of the same genes in the pip –/pLAFR3‐pip strain were complemented or increased compared with that in wild‐type Xcc 8004, except for fleN (Fig. 3). In contrast, pip overexpression in Xcc 8004 restricted gene expression to even lower levels compared with the pip –/pLAFR3‐pip strain (Fig. 3). These results suggest that PIP is a repressor or negative regulator of flagellum‐mediated motility.
Figure 2.
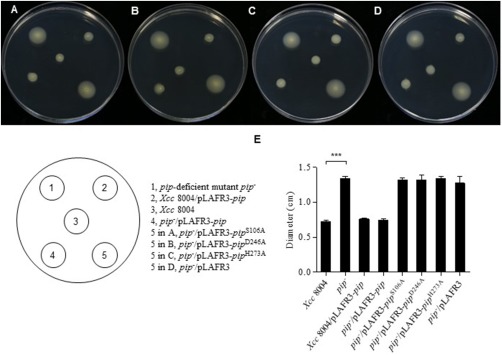
Regulation of Xanthomonas campestris pv. campestris (Xcc) motility by pip. (A–D) Motility phenotype of wild‐type Xcc 8004, pip –, Xcc 8004/pLAFR3‐pip and pip –/pLAFR3‐pip strains on XOLN swimming plates. Photographs were taken at 48 h after incubation at 28 °C. (E) Bars represent the mean diameters of the migration zones of each strain. Error bar represents SEM (standard error of the mean) for at least three replicates with statistical analysis using one‐way analysis of variance (ANOVA; ***P < 0.0001).
Figure 3.
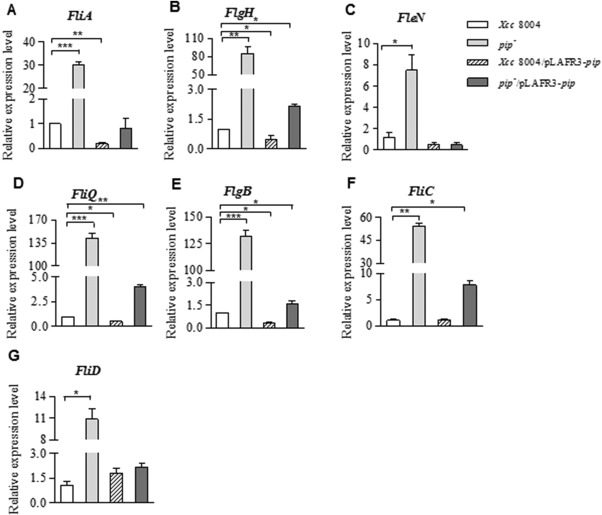
Proline iminopeptidase (PIP) affects flagellar gene expression. (A–G) Relative expression levels of flagellar genes in wild‐type Xcc 8004, pip –, Xcc 8004/pLAFR3‐pip and pip –/pLAFR3‐pip strains determined by quantitative reverse transcription‐polymerase chain reaction (RT‐PCR). The expression level of each gene in Xanthomonas campestris pv. campestris (Xcc) 8004 is indicated as 1.0. 16S rRNA was used as the internal reference. Error bar represents SEM (standard error of the mean) for at least three replicates with statistical analysis using one‐way analysis of variance (ANOVA; ***P < 0.0001, **P < 0.001, *P < 0.05).
Intracellular c‐di‐GMP level modulated by PIP
The important second messenger c‐di‐GMP has been shown to control a variety of biological processes, including motility, in many bacteria (Römling et al., 2013). To explore whether PIP‐controlled bacterial motility is correlated with c‐di‐GMP content, the intracellular levels of c‐di‐GMP were measured by high‐pressure liquid chromatography‐tandem mass spectrometry (HPLC‐MS/MS). As shown in Fig. 4, the pip‐deficient mutant had a lower level of c‐di‐GMP than the wild‐type Xcc 8004. The c‐di‐GMP level was markedly elevated by introducing pLAFR3‐pip into the pip‐deficient mutant (Fig. 4). Overexpression of pip in Xcc 8004 resulted in an even higher concentration of intracellular c‐di‐GMP, and it was 2.5‐fold higher than that of the wild‐type strain (Fig. 4). These results are consistent with those of previous studies reporting lower levels of c‐di‐GMP‐promoted bacterial motility (Jenal and Malone, 2006), and high levels of c‐di‐GMP repressing flagellar assembly by binding to FleQ (Ha and O'Toole, 2015). We propose that PIP may modulate bacterial motility via the c‐di‐GMP signalling pathway.
Figure 4.

Bacterial intracellular bis‐(3′,5′)‐cyclic dimeric guanosine monophosphate (c‐di‐GMP) levels are correlated with proline iminopeptidase (PIP) expression. c‐di‐GMP was extracted from the bacterial cells of each strain at an optical density at 600 nm (OD600) of 0.2, and quantified using high‐pressure liquid chromatography‐tandem mass spectrometry (HPLC‐MS/MS). Values were normalized by total protein contents measured by Bradford protein assay. The data represent the averages from three replicates; error bars indicate SEM (standard error of the mean). Statistical analysis was performed using one‐way analysis of variance (ANOVA; *P < 0.05, **P < 0.001).
PIP as a new type III effector
In plant‐pathogenic bacteria, the T3SS is encoded by hrp (hypersensitive response and pathogenicity) and hrc (hypersensitive response and conserved) gene clusters (Lindgren et al., 1986). HrcV, the conserved structural component of the T3SS apparatus, is involved in substrate docking and is responsible for the secretion of effector proteins (Hartmann and Büttner, 2013). We constructed an in‐frame deletion mutant of hrcV (ΔhrcV) impaired in its T3SS function. To investigate whether PIP can be delivered into the plant cytoplasm, we constructed C‐terminal translational calmodulin‐dependent adenylate cyclase (Cya) fusions to PIP (1–100 amino acids). The kinetics of cyclic adenosine monophosphate (cAMP) accumulation in plant leaves were examined following inoculation with Xcc 8004 or ΔhrcV strains expressing PIP(1–100)‐Cya. As additional controls, plants were inoculated with Xcc 8004 or ΔhrcV containing an empty vector (pLAFR3), Xcc 8004/pLAFR3‐eGFP(1–244)‐cya as a negative control and Xcc 8004/pLAFR3‐AvrBS2(1–98)‐cya as a positive control. As shown in Fig. 5A,B, although the Cya‐tagged fusion proteins were detected in the cell lysates of Xcc 8004/pLAFR3‐pip (1–100)‐cya, Xcc 8004/pLAFR3‐AvrBS2(1–98)‐cya, Xcc 8004/pLAFR3‐eGFP(1–244)‐cya and ΔhrcV/pLAFR3‐pip (1–100) ‐cya strains by western blotting, cAMP accumulation was detected only in the Xcc 8004/pLAFR3‐pip (1–100)‐cya and Xcc 8004/pLAFR3‐AvrBS2(1–98)‐cya inoculated plants, consistent with the western blotting results in the culture supernatants (Fig. 5B). These data indicate that PIP secretion and delivery into plant cells depend on the T3SS. Some bacterial effectors can lead to a hypersensitive reaction (HR) on non‐host plants (Mysore and Ryu, 2004). To further investigate the effect of PIP in different plants, the purified PIP protein was allowed to infiltrate plant leaves. The lesions caused by cell death were observed at 2 days post‐infiltration in non‐host tobacco leaves, and the HR lesion became more severe with greater amounts of PIP protein (Fig. 6A). In contrast, PIP protein infiltration did not induce HR lesions in the leaves of host cabbage plants (Fig. 6C,D). Similarly, when using different strains to inoculate tobacco leaves, we found that the pip‐overexpressing (Xcc 8004/pLAFR3‐pip) strain caused HR lesions more quickly than did the wild‐type Xcc 8004 and the pip‐deficient mutant (Fig. 6E). Interestingly, PIP enzymatic activity was dispensable for the elicitation of cell death in non‐host plants because the same phenotypes were observed when wild‐type PIP or the PIPS106A protein (one of the three enzymatic activity‐deficient proteins was selected to conduct the assays) was injected into tobacco leaves (Fig. 6A,B). Both PIP and PIPS106A failed to elicit cell death in host cabbage plant leaves (Fig. 6C,D), indicating that PIP of Xcc escaped recognition by the host plant immune system, and that this helped to boost bacterial virulence.
Figure 5.

Proline iminopeptidase (PIP) is a new type III effector. (A) The calmodulin‐dependent adenylate cyclase (Cya) activity was determined by testing cyclic adenosine monophosphate (cAMP) concentration after 10 h of infiltration of bacterial strains into Arabidopsis plant leaves. Values were normalized by the protein contents in each sample. ΔhrcV is a type III secretion‐deficient mutant. The data were repeated at least three times. (B) The expression assay of Cya‐tagged fusion protein in bacterial cells and culture supernatants through western blotting with anti‐Cya mouse monoclonal antibody. ND, not detected.
Figure 6.
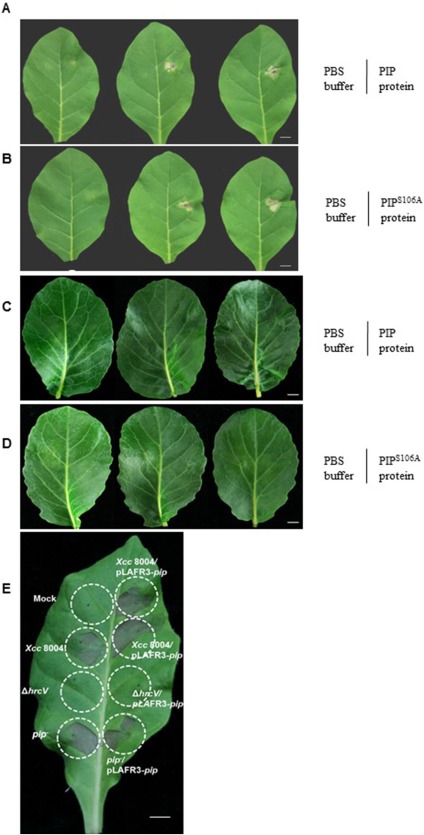
Cell death and hypersensitive response (HR) observation in non‐host plant tobacco and host plant cabbage leaves. Purified proline iminopeptidase (PIP) protein (A) and PIPS106A protein (B) were injected into non‐host leaves and led to cell death symptoms after 2 days. Left halves: phosphate‐buffered saline (PBS) treatment as the negative control; right halves: different concentrations of PIP protein (A) or PIPS106A protein (B) treatment for cell death observation. From left to right: 1 mg/mL, 5 mg/mL and 10 mg/mL of protein. PIP protein (C) and PIPS106A protein (D) could not induce cell death in cabbage leaves, and the protein inocula were the same as in (A) and (B). (E) HR assay of different bacterial strains with a bacterial titre of 109 colony‐forming units (CFU)/mL in tobacco leaves. Mock: 10 mm MgCl2 buffer. Scale bar, 1 cm.
PIP interferes with the SA signalling pathway to benefit bacterial growth
Transgenic Arabidopsis plants that express wild‐type PIP or PIPS106A were obtained to determine whether the enzymatic activity of PIP influences host plant resistance. Both the wild‐type and S106A mutated PIP protein were detectable in transgenic Arabidopsis plants on the western blots (Fig. 7A). The enzymatic activity of PIP in the leaves of pip transgenic plants was much higher than that in those of the wild‐type Col‐0 and pip S106A transgenic plants (Fig. 7B). To measure bacterial growth in planta, 4‐week‐old Arabidopsis seedlings were infiltrated by Xcc cells, and the number of colony‐forming units (CFUs) recovered from the infected leaves was counted at 72 h post‐inoculation (hpi) (Fig. 7C). The number of CFUs recovered from pip transgenic Arabidopsis was log 1.06‐fold higher than the number of CFUs recovered from wild‐type plants (Fig. 7C), whereas the number of CFUs in the pip S106A transgenic plants did not differ from that of the wild‐type plants. These data show that the pip transgenic Arabidopsis plants were more susceptible than their wild‐type counterparts to Xcc, and that PIP enzymatic activity played an important role in the repression of host resistance.
Figure 7.
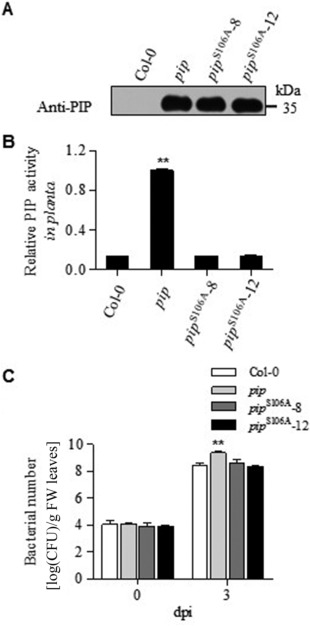
Proline iminopeptidase (PIP) activity is essential for Xanthomonas campestris pv. campestris (Xcc) pathogenicity. (A) Western blot assay of PIP expression in wild‐type Col‐0 and pip or pip S106A transgenic plants; PIPS106A‐8 and PIPS106A‐12 represent two independent transgenic lines. (B) Relative PIP enzymatic activities were determined by normalization to the level in pip transgenic plants (n = 9). (C) Bacterial populations in Col‐0 and different transgenic plants (n = 5). Statistical analysis was a one‐way analysis of variance (ANOVA) (**P < 0.001), and the error bar represents SEM (standard error of the mean). dpi, days post‐inoculation; FW, fresh weight.
Whether plant–microbial interactions are beneficial or detrimental to plants depends largely on the phytohormone homeostasis in the plant (Denance et al., 2013). To investigate further whether plant immunity is influenced by PIP, gene expression in the SA signalling pathway was measured by real‐time quantitative RT‐PCR. As shown in Fig. 8, the transcription levels of ICS1, NPR1, EDS1 and PAD4 in PIP‐expressing plants were 14.12‐, 2.8‐, 2.3‐ and 1.2‐fold lower, respectively, than those in wild‐type plants at 24 h after Xcc infection. The ICS1 and NPR1 expression levels in PIPS106A‐expressing plants were comparable with those in wild‐type plants, and the levels of EDS1 and PAD4 were 64% and 48% of the wild‐type levels, respectively. We also measured SA levels in Arabidopsis plants using HPLC‐MS/MS. After Xcc infection, the pip transgenic plants accumulated lower levels of free SA than the wild‐type plants (reduced to 67.2%), whereas the PIP activity‐deficient (pip S106A) plants showed unrestricted SA accumulation (Fig. 8E). These data indicate that bacterial PIP interferes with the SA signalling pathway of the host plant, which is beneficial for bacterial infection.
Figure 8.
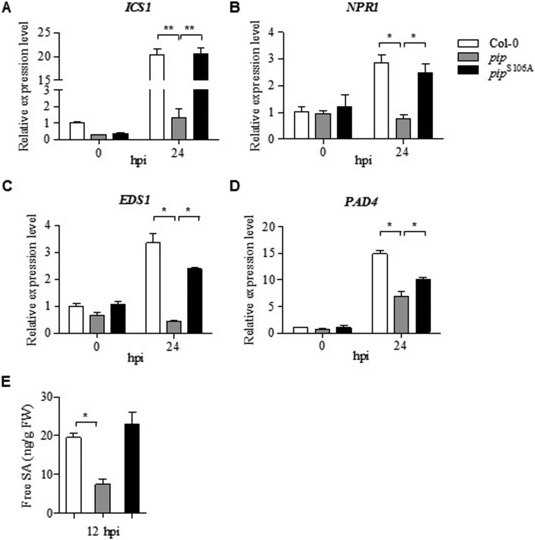
Proline iminopeptidase (PIP) interference with the salicylic acid (SA) signalling pathway in host Arabidopsis plants. Quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of SA biogenesis and signalling pathway gene expression in wild‐type Col‐0, transgenic pip and pip S106A plants, including (A) ICS1, (B) NPR1, (C) EDS1 and (D) PAD4, with actin2 as the internal reference. (E) Free SA levels in Xanthomonas campestris pv. campestris (Xcc)‐infected plants measured by liquid chromatography‐tandem mass spectrometry (LC‐MC/MS). Error bar represents SEM (standard error of the mean) for at least three replicates with statistical analysis using a one‐way analysis of variance (ANOVA; **P < 0.001, *P < 0.05). FW, fresh weight; hpi, hours post‐inoculation.
Discussion
During co‐evolutionary processes, plant pathogens have evolved various strategies to counteract plant immune systems. The role of the T3SS and bacterial flagella in pathogenesis is well established, but having one protein involved in both systems is uncommon in bacteria. Our study has revealed that PIP not only modulates bacterial motility, but also acts as a type III effector capable of inhibiting plant immunity. We propose that PIP may exert its effects during the early stages of infection with Xcc, but the mechanism underlying how the two systems are coordinated is still unknown.
PIPs are evolutionarily conserved in many plant‐associated bacteria and are involved in bacterial pathogenesis (González and Venturi, 2013; Zhang et al., 2007), but their respective mechanisms of action have not been established. Here, we observed that the pip‐deficient mutant showed much higher swimming ability than the wild‐type strain on XOLN medium motility plates (Fig. 2). XOLN is a basal salt medium that mimics a low‐nutrient environment in plants (Fu and Tseng, 1990). We found that the expression levels of nine flagellar genes in the three‐level hierarchical regulatory cascade, including the fliA (σ28) regulatory factor, for example, significantly increased in the pip‐deficient mutant (Fig. 3A). With regard to the flagellar genes, when compared with the wild‐type strain, the expression of fliQ and flgB increased 100‐fold (Fig. 3D,E). Conversely, the majority of the flagellar genes tested showed decreased expression levels in Xcc 8004/pLAFR3‐pip compared with the wild‐type strain (Figs 2 and 3), although bacterial motility showed no significant differences between them. One possible reason may be that the catalytic activity of PIP in the wild‐type strain is saturated for the control of bacterial movement, and so excessive PIP in the Xcc 8004/pLAFR3‐pip strain has no further effect on motility. Because pip expression was induced when Xcc 8004 infiltrated host plant leaves (Zhang et al., 2007), we speculate that this may cause reduced bacterial motility in the host plant. Co‐regulation of flagellar genes by PIP is helpful for the control of bacterial motility behaviour. Flagella‐mediated motility enables bacteria to reach optimal niches, but becomes less essential when the pathogen contacts the host tissues (Chaban et al., 2015; Josenhans and Suerbaum, 2002). Consequently, the motility apparatus is subject to strict energy‐saving control during pathogen–host interaction. We also noticed that the expression of fliC (encoding flagellin) was regulated by PIP (Fig. 3F). Flagellin molecules are the main structural proteins assembled into flagella and can be secreted into the culture medium (Komoriya et al., 1999). A highly conserved 22‐amino‐acid peptide (flg22) region of flagellin (which serves as a PAMP) was responsible for detectable elicitation of the Arabidopsis defence responses mediated by the plant's FLS2 (Sun et al., 2006). In some Xcc strains, the variable flg22 peptides elicit resistance responses in host plant leaves (Sun et al., 2006); thus, restriction of bacterial PAMP levels may circumvent dramatic defence responses in plants. In Xcc 8004, the flagellin flg22 domain amino acid sequence (QQLSSGKRITSASVDAAGLAIS) is identical to that of the Xcc B186 strain (Sun et al., 2006), which has no defence‐eliciting activity on Arabidopsis Col‐0.
c‐di‐GMP, a novel global bacterial second messenger, is involved in motility, EPS production, biofilm formation and virulence, and its metabolism is controlled by many proteins with GGDEF, HD‐GYP and EAL domains (Fouhy et al., 2006; Ryan and Dow, 2010). Consistent with previous findings, the lack of PIP and the increased intracellular c‐di‐GMP level in the reduced mobility pip‐overexpressing strain led to decreased c‐di‐GMP levels in bacterial cells with increased motility compared with the wild‐type strain (Figs 2 and 4).We propose that pip may directly or indirectly affect c‐di‐GMP metabolism and alter bacterial motility, but the interacting target of PIP is unclear. In our assay, the induction of flagella‐associated genes was negatively correlated with the intracellular content of c‐di‐GMP in the different strains (Figs 3 and 4), suggesting that PIP may affect flagellar gene expression via the c‐di‐GMP signalling pathway. The expression of multiple flagella genes is controlled by the c‐di‐GMP‐binding transcriptional regulator FleQ (Hickman and Harwood, 2008), and so it is also possible that the altered levels of c‐di‐GMP mediated by PIP may occur through FleQ to influence bacterial motility. We found that PIP exerts its modulating function on bacterial motility via its enzymatic activity (Figs 3 and 4), and it is highly likely that PIP modulates unidentified targets at the post‐translational level. An important feature of c‐di‐GMP regulation is its ability to control critical lifestyle transitions, such as motile–sessile transitions. We propose that PIP may contribute to the modulation of bacterial motility when Xcc contacts plant tissues.
Pathogenic bacteria deliver virulence effectors into the plant cytoplasm mainly through bacterial secretion systems to ensure their destructive function (Gerlach and Hensel, 2007). In Gram‐negative bacteria, type III effectors are the most important virulence factors for the reprogramming of host cellular functions for bacterial benefit (He, 1998). Surprisingly, PIP was verified as a new T3SS effector by the detection of PIP secretion in culture supernatants and when the PIP‐Cya fusion protein triggered cAMP accumulation in an hrcV‐dependent manner in the host plants (Fig. 5). HrpX is a global regulator of T3SS gene expression (Koebnik et al., 2006; Wengelnik and Bonas, 1996). Our previous results have shown that both HrpX and XccR are responsible for pip expression in host cabbage plants (Zhang et al., 2012, 2007), indicating that PIP may respond to different environmental signals to co‐ordinate bacterial behaviour. Intriguingly, purified PIP elicited cell death in non‐host tobacco plant leaves, but not in host cabbage leaves, and the phenotypes were not related to PIP enzymatic activity (Fig. 6). It is likely that the intact structure of the PIP protein, rather than its enzyme activity per se, is recognized by non‐host plants. Generally, type III effectors function primarily inside host cells when translocated by bacteria. The purified PIP protein triggers cell death on tobacco leaves, suggesting that it could also be recognized outside the plant cells. It is possible that the PIP protein could be released from the dead or broken bacterial cells when bacteria enter the plant apoplast.
Successful pathogens have evolved diverse strategies to subvert the multilayered plant immune system and exploit their respective host plants (Boller and He, 2009). The prevention of the accumulation of SA, a key immune hormone, is an important strategy employed by plant pathogens (Kumar, 2014), as reducing its levels in plants leads to disease susceptibility (Gaffney et al., 1993). We introduced pip and pip S106A into Arabidopsis plants by Agrobacterium‐mediated transformation (floral dipping). Plants with PIP enzymatic activity were more susceptible to Xcc infection, whereas pip S106A plants were not (Fig. 7C). We observed that PIP reduced the expression of SA biosynthesis‐related (ICS1) and resistance response‐related (EDS1, PAD4 and NPR1) transcripts in pip transgenic plants after Xcc infection, a finding consistent with lower SA accumulation (Fig. 8). These results show that Xcc PIP is involved in the repression of SA accumulation and interference with the SA signalling pathway to favour disease.
This work reveals the functionality of PIP at different locations in its host: by involvement in flagella‐associated motility regulation or as a virulence effector capable of overcoming the plant immune system by interfering with its SA signalling pathway. The fine tuning of pip gene expression at different stages of plant infection suggests the need for hierarchical and temporal control over its translocation. PIP can remove N‐terminal prolines from peptides or proteins (Cunningham and O'Connor, 1997), making such substrates potentially modifiable by PIP. However, PIP triggers HR in non‐host plant leaves in the absence of enzymatic activity, and so the possibility cannot be excluded that the targets of PIP are not its enzymatic substrates. Exploring these substrates in bacteria and plants will augment our understanding of the molecular mechanisms involved in plant–pathogen interactions. These results will provide a strategy with which to defeat pathogen invasion or enhance plant resistance to improve crop quality.
Experimental Procedures
Bacterial strains and growth conditions
The bacterial strains, plasmids and primers used in this study are shown in Table S2 (see Supporting Information). Xcc 8004 (Daniels et al., 1984) and derivative strains were cultured at 28 °C in NYG rich medium (Daniels et al., 1984) and XOLN medium (0.7 g/L K2HPO4, 0.2 g/L KH2PO4, 1.0 g/L (NH4)2SO4, 0.1 g/L MgCl2•6H2O, 0.01 g/L FeSO4•7H2O, 0.001 g/L MnCl2, 0.625 g/L yeast extract, 0.625 g/L tryptone, pH 7.5) (Fu and Tseng, 1990). Escherichia coli and Agrobacterium tumefaciens strain EHA105 were grown at 37 °C and 28 °C on Luria–Bertani (LB) agar plates or in LB broth with shaking (200 rpm), respectively. The antibiotics used were ampicillin (100 μg/mL), kanamycin (50 μg/mL), rifampicin (50 μg/mL), spectinomycin (50 μg/mL), tetracycline (10 μg/mL in liquid medium and 3 μg/mL in solid medium) and hygromycin B (20 μg/mL) for Arabidopsis transformant selection.
Protein expression, purification and antibody preparation
The pip coding sequence from the Xcc 8004 genome was PCR amplified with pip NdeI F and pip XhoI R primers and confirmed as correct by sequencing. The amplicon was ligated to the pET30a vector to generate the protein expression vector. Site‐directed mutagenesis of S106A, D246A and H273A active site residues in PIP was conducted on pET30a‐pip using the fast mutagenesis system (Transgen Biotech, Haidian District, China). The resultant expression vectors were transformed individually into E. coli BL21 (DE3) for the expression of His6‐tagged proteins. The early logarithmic phase [optical density at 600 nm (OD600) = 0.4] E. coli culture was induced overnight with 0.3 mm isopropyl‐β‐d‐thiogalactopyranoside at 16 °C with shaking (160 rpm). The pellet was harvested by centrifugation (5 min, 10 000 g, 4 °C), and then suspended in Ni‐NTA binding buffer (300 mm NaCl, 10 mm imidazole, 50 mm NaH2PO4, pH 8.0) containing 1 mm phenylmethanesulfonyl fluoride, lysed in a high‐pressure homogenizer and centrifuged at 10 000 g for 20 min. The supernatant was affinity purified using Ni‐NTA His•Bind Resin to obtain the His‐tagged fusion proteins according to the procedure recommended by the manufacturer (Novagen, Whitehouse Station, NJ, USA). The purified His‐tagged PIP protein was used to immunize and boost rabbits to obtain antibodies against PIP, and serum was collected after the fourth booster injection.
An Amicon® Ultra‐4 centrifugal filter device (molecular weight cut‐off, 10 kDa; Millipore, Whitehouse Station, NJ, USA) was used for protein concentration or buffer exchange. The protein purification was examined by 10% sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE), followed by Coomassie blue staining.
RNA extraction and quantitative RT‐PCR analyses
For the bacterial flagella‐associated gene expression assays, Xcc and its derivative strains were cultured to the mid‐logarithmic phase (OD600 = 1.0) in NYG medium and harvested by centrifugation. The cell pellet was resuspended in XOLN medium, the cells were cultured for 6 h at 28 °C and then harvested at 4 °C by centrifugation (10 000 g, 5 min). Total RNA was isolated using TRIzol® reagent (Invitrogen, Waltham, MA, USA).
For the SA signalling pathway‐related gene expression assay in plants, leaves (0.1 g) were collected and ground to a powder in liquid nitrogen, and 1 mL of TRIzol was added to isolate total RNA from the samples.
Total RNA (1 μg) from the bacteria or plants was used as a template to synthesize cDNA using a ReverTra Ace® qPCR RT Master Mix with the gDNA remover kit (Toyobo, Osaka, Japan). Quantitative real‐time RT‐PCR analyses were conducted using iQ SYBR Green Supermix (Bio‐Rad, Hercules, CA, USA) in a C1000 Touch Thermal Cycler (Bio‐Rad, Hercules, CA, USA) with bacterial 16S rRNA as a reference gene to quantify the transcript expression levels of flagella‐associated genes, or the Actin 2 reference gene to quantify plant SA‐related gene expression. The primer sequences are listed in Table S2.
Bacterial motility assays
XOLN medium soft‐agar motility plates (0.3% agar) were used to determine the swimming motility of the bacterial strains. After stabbing the bacterial colonies onto these plates, the diameter of the migration zone was measured after 48 h of incubation at 28 °C. One‐way analysis of variance (ANOVA) was used to compare the motility differences between the strains. The data reported are representative of three independent experiments, which all gave similar results.
Determination of intracellular c‐di‐GMP levels
c‐di‐GMP concentrations were measured in the Xcc strains. Each strain was inoculated from frozen stock into XOLN medium and cultured at 28 °C until the OD600 reached 0.2. Intracellular levels of c‐di‐GMP were determined by HPLC‐MS/MS according to a previously described procedure (Waters et al., 2008). Briefly, to extract c‐di‐GMP, 50 mL of each culture was harvested by centrifugation (5000 g, 10 min). The cell pellets were resuspended in 900 μL of cold extraction solution (40% acetonitrile, 40% methanol, 0.1% formic acid, 19.9% distilled water), vortexed for 30 s, incubated on ice for 15 min and then lysed by non‐contact ultrasonication (Bioruptor UCD‐200, Diagenode, Seraing (Ougrée), Belgium). The supernatants were extracted three times using extraction solution with centrifugation (16 100 g, 10 min) between each step; thereafter they were pooled and dried by lyophilization. The resulting samples were resuspended in 60 μL of HPLC‐grade water and analysed by HPLC‐MS/MS on an AB SCIEX QTRAP 4500 system (AB SCIEX, Foster City, CA, USA). Reverse‐phase liquid chromatography was performed using a Synergi Hydro‐RP 80A LC column (4 μm, 150 mm × 2 mm; Phenomenex, Torrance, CA, USA). MS was conducted using electrospray ionization, with the analyses in negative‐ion mode. The amount of c‐di‐GMP in the samples was estimated using a standard curve generated from pure c‐di‐GMP (Biolog Life Science Institute, Bremen, Germany). c‐di‐GMP levels were normalized against the total proteins. To determine protein amounts, 3 mL of each culture were pelleted by centrifugation, and the cells were lysed with 300 μL of phosphate‐buffered saline (PBS) using an ultrasonic oscillator. After centrifugation (10 000 g, 20 min), the protein concentrations in each supernatant were measured by the Bradford Protein Assay kit (Bio‐Rad, Hercules, CA, USA).
Plant inoculation and non‐host HR assay
Plasmid pRF419 (Zhang et al., 2007) (renamed here as pLAFR3‐pip) was transformed into Xcc 8004 to generate Xcc/pLAFR3‐pip. The pip S106A fragment in pET30a was PCR amplified with pip BamH I F and pip Hind III R primers. Its sequence was verified and then cloned into pLAFR3 to obtain pLAFR3‐pip S106A, and the resultant plasmid was transformed into Xcc 8004.
Xcc strains were cultured to OD600 = 1.0, the cell numbers were adjusted to 108 in 10 mm MgCl2 buffer and then used to inoculate 4‐week‐old SR1 tobacco or cabbage leaves. The leaves were inspected for HR lesion at 2 days after inoculation.
The PIP and PIPS106A proteins purified from E. coli cells were diluted to different concentrations (1, 5 and 10 mg/mL in PBS, using PBS as a control), injected with a needless syringe into tobacco SR1 and cabbage leaves for disease or HR lesion development, and then photographed at 48 h after protein infiltration.
Generation of bacterial in‐frame deletion mutants
Xcc 8004 was used as the parental strain for deletion mutant generation. In‐frame deletion of hrcV was conducted using the primers listed in Table S2 according to previously described methods (Slater et al., 2000). Briefly, two flanking hrcV fragments were PCR amplified with two primer pairs (LhrcV F‐EcoR I/LhrcV R‐Hind III and RhrcV F‐Hind III/RhrcV R‐BamH I). Both fragments were cleaved with Hind III, and then ligated to generate an intermediate fused fragment. The PCR‐amplified DNA fusion fragment from LhrcV primer F‐EcoR I and RhrcV primer R‐BamH I primers was cloned into a pK18mobsacB vector. The recombinant plasmid was transformed into Xcc 8004 by electroporation. Transformants were selected on LB medium supplemented with rifampicin and kanamycin. Positive colonies were plated onto NYG medium containing 5% (w/v) sucrose and rifampicin to select for the second cross‐over event resulting in the loss of the sacB gene. The hrcV in‐frame deletion mutant was verified by sequencing.
Bacterial growth in planta
Xcc was resuspended in 10 mm MgCl2 and inoculated at a cell density of 1 × 105 CFU/mL into the leaves of 4‐week‐old Arabidopsis plants with a needleless syringe. The bacterial population in each leaf was counted at 72 hpi using a Petri dish colony counting method. Each data point consisted of at least four replicates.
Adenylate cyclase assays
The first 100‐amino‐acid coding sequence fragment of PIP was amplified with the primers pip(1–100)‐Cya SacI F and pip(1–100)‐Cya BamH I R, verified by sequencing and ligated to pCPP3214 with the Cya coding sequence (Schechter et al., 2004). Next, the pip (1–100)‐cya fragment was digested with EcoR I and Hind III, and cloned into pLAFR3 to generate pLAFR3‐pip (1–100)‐cya. PCR‐amplified DNA fragments coding for the N‐terminal 98 amino acids of AvrBs2 (Casper‐Lindley et al., 2002) and the full length of enhanced green fluorescent protein (eGFP) (Genbank accession No. KM019171), the obtained fragments were ligated with the large fragment of BamH I/EcoR I‐digested pLAFR3‐pip (1–100)‐cya by a seamless assembly cloning kit (Trelief SoSoo Cloning Kit Ver. 2, TSINGKE, Changping District, China) to generate pLAFR3‐AvrBs2(1–98)‐cya and pLAFR3‐eGFP(1–244)‐cya. The plasmids were transformed into Xcc 8004 and ΔhrcV to generate Xcc 8004/pLAFR3‐pip (1–100)‐cya, ΔhrcV/pLAFR3‐pip (1–100)‐cya, Xcc 8004/pLAFR3‐AvrBS2(1–98)‐cya and Xcc 8004/pLAFR3‐eGFP(1–244)‐cya, respectively.
The bacterial cells and the culture supernatants were collected separately. The supernatants were added to an equal volume of 20% trichloroacetic acid (TCA) and incubated for 30 min on ice for protein precipitation; after centrifugation, the pellets were washed in cold acetone and then dissolved in PBS for western blot. The remaining cells were boiled in SDS lysis buffer containing 50 mm Tris‐HCl (pH 6.8), 2% (w/v) SDS, 10% (v/v) glycerol, 1% (v/v) β‐mercaptoethanol (2‐ME) and 0.1% (w/v) BPB (bromphenol blue). Protein samples were separated by electrophoresis on 10% SDS‐PAGE gels and transferred to PVDF (polyvinylidene fluoride) (Millipore, Whitehouse Station, NJ, USA) membranes using a semidry transfer system (Bio‐Rad, Hercules, CA, USA). Western blot working solution was prepared by mixing 100 µL of the stable peroxide solution and the luminol/enhancer solution (Promega, Madison, WI, USA). Cya fusion proteins were detected using primary anti‐Cya (3D1) mouse monoclonal IgG antibody (Santa Cruz Biotechnology, Dallas, TX, USA).
Adenylate cyclase activity assays in plant tissue were conducted as described previously (Schechter et al., 2004). Briefly, 0.1–0.2 g of leaves were collected at 10 hpi by Xcc strains (OD600 = 0.8), frozen in liquid nitrogen, ground to a powder and suspended in 100 μL of 0.1 m HCl. The concentrations of cAMP were measured in leaf samples using a cAMP complete enzyme‐linked immunosorbent assay (ELISA) kit (Enzo Life Sciences, Farmingdale, NY, USA) according to the manufacturer's instructions. The protein content of each sample was determined by the Bio‐Rad protein assay (Bio‐Rad, Hercules, CA, USA) for cAMP level normalization. The mean values obtained represent data triplicates from each separate experiment.
PIP enzymatic activity assays
PIP activity was assayed with l‐proline p‐nitroanilide trifluoroacetate salt (PPNA) as a substrate as described previously (Zhang et al., 2007). The reaction was performed according to the procedure of Sigma (http://www.sigmaaldrich.com/content/dam/sigma-aldrich/docs/Sigma/General_Information/proline_iminopeptidase.pdf) at 37 °C for 10 min. The purified PIP, PIPS106A, PIPD246A and PIPH273A proteins were extracted from E. coli cells using PBS; 5 µg of protein of each sample was used to assay the enzyme activity.
Transgenic Arabidopsis leaves (about 0.1 g) were frozen with liquid nitrogen and ground to a powder. PIP or PIPS106A protein was extracted using 500 μL of NP‐40 buffer (50 mm Tris‐HCl, 100 mm KCl, 2.5 mm MgCl2, 0.1% NP‐40) and centrifuged at 10 000 g for 30 min; the total protein in the supernatant was quantified by the Bio‐Rad protein assay system (Bio‐Rad, Hercules, CA, USA). For PIP enzymatic assay, 100 μg of total protein (or 5 µg of PIP protein from E. coli) were added to the reaction buffer [80 mm Tris‐HCl, pH 8.0, 4 mm dithiothreitol (DTT), 1.5 mm ethylenediaminetetraacetic acid (EDTA) and 0.17 mm PPNA] and incubated at 37 ºC for 10 min, using sodium acetate (pH 4.0) (final concentration, 250 mm) to stop the reaction. The absorbance of p‐nitroaniline released by PIP was measured at 410 nm.
The relative enzymatic activities of PIP variant proteins were calculated with respect to the established mean of wild‐type PIP activity. Four independent experiments were performed in triplicate.
Construction of transgenic plants
To create DNA encoding N‐terminal 3×FLAG‐PIP (or 3×FLAG‐PIPS106A) fusion proteins, PCR was conducted with the primers pip N‐flag BamH I F and pip N‐flag SacI R using pET30a‐pip (or pET30a‐pip S106A) as a template. The PCR products were cloned into the pGEM‐T vector (Promega, Madison, WI, USA) and verified by DNA sequencing, and the resulting BamH I‐SacI fragment was fused to the binary expression vector pCAMBIA1300, followed by transformation into the Agrobacterium tumefaciens strain EHA105. Arabidopsis thaliana (ecotype Col‐0) was transformed using the floral dip method by Agrobacterium tumefaciens (containing the transgenes pip or pip S106A) strains as described previously (Clough and Bent, 1998; Zhang et al., 2006). Treated plants were allowed to set seeds, which were then plated onto hygromycin B (20 μg/mL) medium for transformant selection. Total protein was extracted from the transgenic plants, and 50 μg was size fractionated on a 10% SDS‐PAGE gel. The PIP and PIPS106A expression levels in transgenic plants were verified by western blotting using anti‐PIP rabbit polyclonal antibody.
SA measurement using HPLC‐MS/MS
Plants were infected with Xcc 8004 using a needless syringe. One leaf on each plant was infiltrated with a suspension of Xcc (OD600 = 0.5). At 12 hpi, free SA measurement was performed with the leaf tissues from 4‐week‐old plants, as described previously (Bowling et al., 1994). Briefly, 0.1 g of leaves were ground into a powder in liquid nitrogen; 1 mL of 90% methanol was added, the sample was centrifuged at 14 000 g for 10 min and the supernatant was transferred to a new tube. The pellet was re‐extracted with 0.5 mL of 100% methanol, and the supernatant was combined with the first‐step supernatant, and dried in a speed vacuum to a final volume of ∼50 μL. The residue was resuspended in 500 μL of 0.1 m sodium acetate (pH 5.5), and an equal volume of 10% TCA was added to the tube. After centrifugation at 14 000 g for 10 min, the supernatant was transferred to a new tube and partitioned with 1 mL of ethylacetate–cyclohexane (1 : 1). After centrifugation at 14 000 g for 10 min, the organic phase was transferred to a new tube, dried in a speed vacuum, dissolved in 150 μL of 100% methanol and re‐centrifuged at 14 000 g for 10 min. The supernatant was used for HPLC‐MS/MS analysis on an AB SCIEX QTRAP 4500 system (AB SCIEX). Reverse‐phase liquid chromatography was performed with an Agilent Extend C18 column (3.5 μm, 2.1 mm × 100 mm; Agilent, Santa Clara, CA, USA). The sample was eluted with 0.8% acetic acid (pH 5.5) (solvent A), 100% acetonitrile (solvent B) and A : B (75 : 25) at a flow rate of 0.40 mL/min. The quantity of SA in each sample was estimated using a standard curve generated from pure SA (Sigma, Whitehouse Station, NJ, USA). SA levels were normalized against the fresh leaf weight.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Tested flagellar genes in Xanthomonas campestris pv. campestris (Xcc) 8004.
Table S2 Bacterial strains and plasmids used in this work.
Acknowledgements
This work was funded by the National Basic Research Program of China (973 Program 2015CB150600), the National Key Research and Development Plan (No. 2016YFD0100600) and the National Natural Science Foundation of China (No. 31370161). We have no conflicts of interest to declare.
Contributor Information
Rongxiang Fang, Email: fangrx@im.ac.cn.
Yantao Jia, Email: jiayt@im.ac.cn.
References
- Alonso, J. and Garcia, J.L. (1996) Proline iminopeptidase gene from Xanthomonas campestris pv. citri . Microbiology, 142, 2951–2957. [DOI] [PubMed] [Google Scholar]
- Apel, D. and Surette, M.G. (2008) Bringing order to a complex molecular machine: the assembly of the bacterial flagella. Biochim. Biophys. Acta, 1778, 1851–1858. [DOI] [PubMed] [Google Scholar]
- Bartsch, M. , Gobbato, E. , Bednarek, P. , Debey, S. , Schultze, J.L. , Bautor, J. and Parker, J.E. (2006) Salicylic acid‐independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the nudix hydrolase NUDT7 . Plant Cell, 18, 1038–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller, T. and He, S.Y. (2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science, 324, 742–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling, S.A. , Guo, A. , Cao, H. , Gordon, A. , Klessig, D.F. and Dong, X. (1994) A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell, 6, 1845–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker, R.R. (2011) Prospects for riboswitch discovery and analysis. Mol. Cell, 43, 867–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broek, A.V. and Vanderleyden, J. (1995) The role of bacterial motility, chemotaxis, and attachment in bacteria plant interactions. Mol. Plant–Microbe Interact. 8, 800–810. [Google Scholar]
- Buttner, D. (2016) Behind the lines—actions of bacterial type III effector proteins in plant cells. FEMS Microbiol. Rev. 40, 894–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper‐Lindley, C. , Dahlbeck, D. , Clark, E.T. and Staskawicz, B.J. (2002) Direct biochemical evidence for type III secretion‐dependent translocation of the AvrBs2 effector protein into plant cells. Proc. Natl. Acad. Sci. USA, 99, 8336–8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaban, B. , Hughes, H.V. and Beeby, M. (2015) The flagellum in bacterial pathogens: for motility and a whole lot more. Semin. Cell Dev. Biol. 46, 91–103. [DOI] [PubMed] [Google Scholar]
- Chen, Z. , Zheng, Z. , Huang, J. , Lai, Z. and Fan, B. (2009) Biosynthesis of salicylic acid in plants. Plant Signal. Behav. 4, 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cunningham, D.F. and O'Connor, B. (1997) Proline specific peptidases. Biochim. Biophys. Acta, 1343, 160–186. [DOI] [PubMed] [Google Scholar]
- Daniels, M.J. , Barber, C.E. , Turner, P.C. , Sawczyc, M.K. , Byrde, R.J.W. and Fielding, A.H. (1984) Cloning of genes involved in pathogenicity of Xanthomonas campestris pv. campestris using the broad host range cosmid pLAFR1. EMBO J. 3, 3323–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denance, N. , Sanchez‐Vallet, A. , Goffner, D. and Molina, A. (2013) Disease resistance or growth: the role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 4, 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, X.N. (2004) NPR1, all things considered. Curr. Opin. Plant Biol. 7, 547–552. [DOI] [PubMed] [Google Scholar]
- Duan, Q. , Zhou, M. , Zhu, L. and Zhu, G. (2013) Flagella and bacterial pathogenicity. J. Basic Microbiol. 53, 1–8. [DOI] [PubMed] [Google Scholar]
- Felix, G. , Duran, J.D. , Volko, S. and Boller, T. (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18, 265–276. [DOI] [PubMed] [Google Scholar]
- Feys, B.J. , Moisan, L.J. , Newman, M.A. and Parker, J.E. (2001) Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20, 5400–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouhy, Y. , Lucey, J.F. , Ryan, R.P. and Dow, J.M. (2006) Cell–cell signaling, cyclic di‐GMP turnover and regulation of virulence in Xanthomonas campestris . Res. Microbiol. 157, 899–904. [DOI] [PubMed] [Google Scholar]
- Fu, J.F. and Tseng, Y.H. (1990) Construction of lactose‐utilizing Xanthomonas campestris and production of xanthan gum from whey. Appl. Environ. Microbiol. 56, 919–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney, T. , Friedrich, L. , Vernooij, B. , Negrotto, D. , Nye, G. , Uknes, S. , Ward, E. , Kessmann, H. and Ryals, J. (1993) Requirement of salicylic acid for the induction of systemic acquired resistance. Science, 261, 754–756. [DOI] [PubMed] [Google Scholar]
- Gerlach, R.G. and Hensel, M. (2007) Protein secretion systems and adhesins: the molecular armory of gram‐negative pathogens. Int. J. Med. Microbiol. 297, 401–415. [DOI] [PubMed] [Google Scholar]
- Gewirtz, A.T. , Simon, P.O. , Schmitt, C.K. , Taylor, L.J. , Hagedorn, C.H. , O'Brien, A.D. , Neish, A.S. and Madara, J.L. (2001) Salmonella typhimurium translocates flagellin across intestinal epithelia, inducing a proinflammatory response. J. Clin. Invest. 107, 99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González, J.F. and Venturi, V. (2013) A novel widespread interkingdom signaling circuit. Trends Plant Sci. 18, 167–174. [DOI] [PubMed] [Google Scholar]
- Gürlebeck, D. , Thieme, F. and Bonas, U. (2006) Type III effector proteins from the plant pathogen Xanthomonas and their role in the interaction with the host plant. J. Plant Physiol. 163, 233–255. [DOI] [PubMed] [Google Scholar]
- Ha, D.G. and O'Toole, G.A. (2015) c‐di‐GMP and its effects on biofilm formation and dispersion: a Pseudomonas aeruginosa review. Microbiol Spectr. 3, MB‐0003–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann, N. and Büttner, D. (2013) The inner membrane protein HrcV from Xanthomonas spp. is involved in substrate docking during type III secretion. Mol. Plant–Microbe Interact. 26, 1176–1189. [DOI] [PubMed] [Google Scholar]
- He, S.Y. (1998) Type III protein secretion systems in plant and animal pathogenic bacteria. Annu. Rev. Phytopathol. 36, 363–392. [DOI] [PubMed] [Google Scholar]
- He, S.Y. , Nomura, K. and Whittam, T.S. (2004) Type III protein secretion mechanism in mammalian and plant pathogens. Biochim. Biophys. Acta, 1694, 181–206. [DOI] [PubMed] [Google Scholar]
- Hengge, R. (2009) Principles of c‐di‐GMP signalling in bacteria. Nat. Rev. Microbiol. 7, 263–273. [DOI] [PubMed] [Google Scholar]
- Hickman, J.W. and Harwood, C.S. (2008) Identification of FleQ from Pseudomonas aeruginosa as a c‐di‐GMP‐responsive transcription factor. Mol. Microbiol. 69, 376–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain, M.M. , Shibata, S. , Aizawa, S.I. and Tsuyumu, S. (2005) Motility is an important determinant for pathogenesis of Erwinia carotovora subsp carotovora . Physiol. Mol. Plant Pathol. 66, 134–143. [Google Scholar]
- Hu, R.M. , Yang, T.C. , Yang, S.H. and Tseng, Y.H. (2005) Deduction of upstream sequences of Xanthomonas campestris flagellar genes responding to transcription activation by FleQ. Biochim. Biophys. Acta, 335, 1035–1043. [DOI] [PubMed] [Google Scholar]
- Jelenska, J. , Yao, N. , Vinatzer, B.A. , Wright, C.M. , Brodsky, J.L. and Greenberg, J.T. (2007) A J domain virulence effector of Pseudomonas syringae remodels host chloroplasts and suppresses defenses. Curr. Biol. 17, 499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal, U. and Malone, J. (2006) Mechanisms of cyclic‐di‐GMP signaling in bacteria. Annu. Rev. Genet. 40, 385–407. [DOI] [PubMed] [Google Scholar]
- Josenhans, C. and Suerbaum, S. (2002) The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 291, 605–614. [DOI] [PubMed] [Google Scholar]
- Koebnik, R. , Kruger, A. , Thieme, F. , Urban, A. and Bonas, U. (2006) Specific binding of the Xanthomonas campestris pv. vesicatoria AraC‐type transcriptional activator HrpX to plant‐inducible promoter boxes. J. Bacteriol. 188, 7652–7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komoriya, K. , Shibano, N. , Higano, T. , Azuma, N. , Yamaguchi, S. and Aizawa, S. (1999) Flagellar proteins and type III‐exported virulence factors are the predominant proteins secreted into the culture media of Salmonella typhimurium . Mol. Microbiol. 34, 767–779. [DOI] [PubMed] [Google Scholar]
- Kumar, D. (2014) Salicylic acid signaling in disease resistance. Plant Sci. 228, 127–134. [DOI] [PubMed] [Google Scholar]
- Lee, M.C. , Weng, S.F. and Tseng, Y.H. (2003) Flagellin gene fliC of Xanthomonas campestris is upregulated by transcription factor Clp. Biochim. Biophys. Acta, 307, 647–652. [DOI] [PubMed] [Google Scholar]
- Leenhouts, K. , Bolhuis, A. , Boot, J. , Deutz, I. , Toonen, M. , Venema, G. , Kok, J. and Ledeboer, A. (1998) Cloning, expression, and chromosomal stabilization of the Propionibacterium shermanii proline iminopeptidase gene (pip) for food‐grade application in Lactococcus lactis . Appl. Environ. Microbiol. 64, 4736–4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren, P.B. , Peet, R.C. and Panopoulos, N.J. (1986) Gene cluster of Pseudomonas syringae pv. phaseolicola controls pathogenicity of bean plants and hypersensitivity of nonhost plants. J. Bacteriol. 168, 512–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka, U. , Fuchs, R. and Lipka, V. (2008) Arabidopsis non‐host resistance to powdery mildews. Curr. Opin. Plant Biol. 11, 404–411. [DOI] [PubMed] [Google Scholar]
- Moscoso, J.A. , Mikkelsen, H. , Heeb, S. , Williams, P. and Filloux, A. (2011) The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c‐di‐GMP signalling. Environ. Microbiol. 13, 3128–3138. [DOI] [PubMed] [Google Scholar]
- Mysore, K.S. and Ryu, C.M. (2004) Nonhost resistance: how much do we know? Trends Plant Sci. 9, 97–104. [DOI] [PubMed] [Google Scholar]
- Notti, R.Q. , Bhattacharya, S. , Lilic, M. and Stebbins, C.E. (2015) A common assembly module in injectisome and flagellar type III secretion sorting platforms. Nat. Commun. 6, 7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottemann, K.M. and Miller, J.F. (1997) Roles for motility in bacterial–host interactions. Mol. Microbiol. 24, 1109–1117. [DOI] [PubMed] [Google Scholar]
- Pel, M.J. , van Dijken, A.J. , Bardoel, B.W. , Seidl, M.F. , van der Ent, S. , van Strijp, J.A. and Pieterse, C.M. (2014) Pseudomonas syringae evades host immunity by degrading flagellin monomers with alkaline protease AprA. Mol. Plant–Microbe Interact. 27, 603–610. [DOI] [PubMed] [Google Scholar]
- Rawlings, N.D. (2016) Peptidase specificity from the substrate cleavage collection in the MEROPS database and a tool to measure cleavage site conservation. Biochimie, 122, 5–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling, U. and Amikam, D. (2006) Cyclic di‐GMP as a second messenger. Curr. Opin. Microbiol. 9, 218–228. [DOI] [PubMed] [Google Scholar]
- Römling, U. , Galperin, M.Y. and Gomelsky, M. (2013) Cyclic di‐GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77, 1–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals, J.A. , Neuenschwander, U.H. , Willits, M.G. , Molina, A. , Steiner, H.Y. and Hunt, M.D. (1996) Systemic acquired resistance. Plant Cell, 8, 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, R.P. and Dow, J.M. (2010) Intermolecular interactions between HD‐GYP and GGDEF domain proteins mediate virulence‐related signal transduction in Xanthomonas campestris . Virulence, 1, 404–408. [DOI] [PubMed] [Google Scholar]
- Ryan, R.P. , Tolker‐Nielsen, T. and Dow, J.M. (2012) When the PilZ don't work: effectors for cyclic di‐GMP action in bacteria. Trends Microbiol. 20, 235–242. [DOI] [PubMed] [Google Scholar]
- Sarid, S. , Berger, A. and Katchalski, E. (1959) Proline iminopeptidase. J. Biol. Chem. 234, 1740–1746. [PubMed] [Google Scholar]
- Schechter, L.M. , Roberts, K.A. , Jamir, Y. , Alfano, J.R. and Collmer, A. (2004) Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J. Bacteriol. 186, 543–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater, H. , Alvarez‐Morales, A. , Barber, C.E. , Daniels, M.J. and Dow, J.M. (2000) A two‐component system involving an HD‐GYP domain protein links cell–cell signalling to pathogenicity gene expression in Xanthomonas campestris . Mol. Microbiol. 38, 986–1003. [DOI] [PubMed] [Google Scholar]
- Sun, W.X. , Dunning, F.M. , Pfund, C. , Weingarten, R. and Bent, A.F. (2006) Within‐species flagellin polymorphism in Xanthomonas campestris pv campestris and its impact on elicitation of Arabidopsis FLAGELLIN SENSING2‐dependent defenses. Plant Cell, 18, 764–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, P. , Van Kessel, T. , Van de Veerdonk, F. , Zuurendonk, P. , Bruins, A. and Konings, W. (1993) Degradation and debittering of a tryptic digest from beta‐casein by aminopeptidase N from Lactococcus lactis subsp. cremoris WG2. Appl. Environ. Microbiol. 59, 1430–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, F. , He, Y.W. , Wu, D.H. , Swarup, S. and Zhang, L.H. (2010) The cyclic nucleotide monophosphate domain of Xanthomonas campestris global regulator Clp defines a new class of cyclic di‐GMP effectors. J. Bacteriol. 192, 1020–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, F. , Yu, C. , Li, H.Y. , Wu, X.L. , Li, B. , Chen, H.M. , Wu, M.S. and He, C.Y. (2015) Alternative sigma factor RpoN2 is required for flagellar motility and full virulence of Xanthomonas oryzae pv. oryzae . Microbiol Res. 170, 177–183. [DOI] [PubMed] [Google Scholar]
- de Torres, M. , Mansfield, J.W. , Grabov, N. , Brown, I.R. , Ammouneh, H. , Tsiamis, G. , Forsyth, A. , Robatzek, S. , Grant, M. and Boch, J. (2006) Pseudomonas syringae effector AvrPtoB suppresses basal defence in Arabidopsis . Plant J. 47, 368–382. [DOI] [PubMed] [Google Scholar]
- Trampari, E. , Stevenson, C.E. , Little, R.H. , Wilhelm, T. , Lawson, D.M. and Malone, J.G. (2015) Bacterial rotary export ATPases are allosterically regulated by the nucleotide second messenger cyclic‐di‐GMP. J. Biol. Chem. 290, 24 470–24 483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustun, S. , Bartetzko, V. and Bornke, F. (2013) The Xanthomonas campestris type III effector XopJ targets the host cell proteasome to suppress salicylic‐acid mediated plant defence. PLoS Pathog. 9, e1003427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot, A.C. , Dempsey, D.A. and Klessig, D.F. (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. [DOI] [PubMed] [Google Scholar]
- Waters, C.M. , Lu, W. , Rabinowitz, J.D. and Bassler, B.L. (2008) Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di‐GMP levels and repression of vpsT . J. Bacteriol. 190, 2527–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengelnik, K. and Bonas, U. (1996) HrpXv, an AraC‐type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria . J. Bacteriol. 178, 3462–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, F.F. , Potnis, N. , Jones, J.B. and Koebnik, R. (2009) The type III effectors of Xanthomonas . Mol. Plant Pathol. 10, 749–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, R.F. (1979) Acetylsalicylic acid (aspirin) induces resistance to tobacco mosaic virus in tobacco. Virology, 99, 410–412. [DOI] [PubMed] [Google Scholar]
- Wiermer, M. , Feys, B.J. and Parker, J.E. (2005) Plant immunity: the EDS1 regulatory node. Curr. Opin. Plant Biol. 8, 383–389. [DOI] [PubMed] [Google Scholar]
- Wildermuth, M.C. , Dewdney, J. , Wu, G. and Ausubel, F.M. (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature, 414, 562–565. [DOI] [PubMed] [Google Scholar]
- Williams, P. (1980) Black rot: a continuing threat to world crucifers. Plant Dis. 64, 736–742. [Google Scholar]
- Wolfe, A.J. and Visick, K.L. (2008) Get the message out: cyclic‐di‐GMP regulates multiple levels of flagellum‐based motility. J. Bacteriol. 190, 463–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, Y. , Usuki, H. , Iwabuchi, M. and Hatanaka, T. (2010) Prolyl aminopeptidase from Streptomyces thermoluteus subsp. fuscus strain NBRC14270 and synthesis of proline‐containing peptides by its S144C variant. Appl. Environ. Microbiol. 76, 6180–6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, Y. , Usuki, H. , Kumagai, Y. , Mukaihara, T. , Yamasato, A. and Hatanaka, T. (2011) Synthesis of prolyl‐hydroxyproline using prolyl aminopeptidase from Streptomyces aureofaciens TH‐3. Process Biochem. 46, 1560–1564. [Google Scholar]
- Yang, T.C. , Leu, Y.W. , Chang‐Chien, H.C. and Hu, R.M. (2009) Flagellar biogenesis of Xanthomonas campestris requires the alternative sigma factors RpoN2 and FliA and is temporally regulated by FlhA, FlhB, and FlgM. J. Bacteriol. 191, 2266–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J.X. , Kan, J.H. , Zhang, J.Q. , Guo, P. , Chen, X.Y. , Fang, R.X. and Jia, Y.T. (2012) Synergistic activation of the pathogenicity‐related proline iminopeptidase gene in Xanthomonas campestris pv. campestris by HrpX and a LuxR homolog. Appl. Environ. Microbiol. 78, 7069–7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Jia, Y. , Wang, L. and Fang, R. (2007) A proline iminopeptidase gene upregulated in planta by a LuxR homologue is essential for pathogenicity of Xanthomonas campestris pv. campestris . Mol. Microbiol. 65, 121–136. [DOI] [PubMed] [Google Scholar]
- Zhang, X.R. , Henriques, R. , Lin, S.S. , Niu, Q.W. and Chua, N.H. (2006) Agrobacterium‐mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. [DOI] [PubMed] [Google Scholar]
- Zipfel, C. , Robatzek, S. , Navarro, L. , Oakeley, E.J. , Jones, J.D.G. , Felix, G. and Boller, T. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature, 428, 764–767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Tested flagellar genes in Xanthomonas campestris pv. campestris (Xcc) 8004.
Table S2 Bacterial strains and plasmids used in this work.


