Summary
Plant NLR genes encode sensitive immune receptors that can mediate the specific recognition of pathogen avirulence effectors and activate a strong defence response, termed effector‐triggered immunity. The expression of NLRs requires strict regulation, as their ability to trigger immunity is dependent on their dose, and overexpression of NLRs results in autoimmunity and massive fitness costs. An elaborate interplay of different mechanisms controlling NLR transcript levels allows plants to maximize their defence capacity, whilst limiting negative impact on their fitness. Global suppression of NLR transcripts may be a prerequisite for the fast evolution of new NLR variants and the expansion of this gene family. Here, we summarize recent progress made towards a comprehensive understanding of NLR transcript‐level expression control. Multiple mechanistic steps, including transcription as well as co‐/post‐transcriptional processing and transcript turn‐over, contribute to balanced base levels of NLR transcripts and allow for dynamic adjustments to defence situations.
Keywords: alternative polyadenylation, alternative splicing, nonsense‐mediated decay, plant disease resistance genes, post‐transcriptional regulation, small RNAs, transcriptional regulation
Introduction
Like other multicellular organisms, plants have evolved immune receptors that allow them to recognize pathogenic microorganisms or pests (Jacob et al., 2013). Particularly efficient are disease resistance (R) gene‐encoded receptors, which typically mediate very strong immunity (Hammond‐Kosack and Jones, 1996). On specific interactions with pathogen avirulence (avr) gene products, R proteins can induce the hypersensitive response (HR), a form of programmed cell death which, together with other defence reactions, provides efficient protection against biotrophic or hemibiotrophic pathogens (Coll et al., 2010). Most known R genes encode immune receptors with a central ARC (APAF‐1, disease resistance proteins, CED‐4)‐type nucleotide‐binding site (NB) and a C‐terminal leucine‐rich repeat (LRR) domain (Jacob et al., 2013; Jones et al., 2016). In Arabidopsis thaliana (Arabidopsis), most of these NB‐LRRs, or NLRs, contain at their N‐terminus a coiled coil (CC) or Toll Interleukin‐1 Receptor (TIR) domain (Meyers et al., 2003). Examples of other domains present at the N‐terminus of NLRs are Solanaceae domains (SDs) in some Solanaceae NLRs, BED‐DNA‐binding zinc‐finger domains of Populus trichocarpa NLRs or the WRKY domains in the Arabidopsis proteins WRKY19/MEKK4 and RRS1. In some cases, additional domains occur in other parts of NLR proteins. Generally, CC‐NB‐LRR genes are present in monocot and dicot genomes, whereas TIR‐NB‐LRRs are entirely absent in monocots, but strongly represented in dicots (Jacob et al., 2013). Although NLR repertoires are relatively compact in ancestral plant lineages (∼25 NLRs in the bryophyte Physcomitrella patens and ∼2 NLRs in the lycophyte Selaginella mollendorffii), the presence of these proteins massively expanded to one of the largest and most variable families in advanced plant clades, reaching more than 400 members in rice (Oryza sativa) or grape (Vitis vinifera) (Clark et al., 2007; Jacob et al., 2013; Ossowski et al., 2008). The number of NLR genes seems generally to correlate with the total number of genes in a genome. However, there are some exceptions; with papaya, watermelon and cucumber featuring relatively low numbers of NLRs (Zhang et al., 2016).
Pathogen avr genes are fast evolving and encode polymorphic effector proteins that are often transmitted into plant cells, where they interact with host proteins to either attenuate host immune responses or otherwise enhance pathogen fitness (virulence) in the host environment (Abramovitch et al., 2006; Chisholm et al., 2006; Dangl and McDowell, 2006). Effector recognition by NLR immune receptors triggers a set of defence reactions referred to as effector‐triggered immunity (ETI). How NLRs induce ETI is poorly understood. At this point, no general pathway of NLR‐mediated defence induction has been identified, and it seems that many different mechanisms have evolved that link NLR‐facilitated effector recognition to downstream defence responses (Dodds and Rathjen, 2010; Li et al., 2015). In addition to serving as specific receptors for pathogen effectors, several widely conserved members of this receptor family have been proposed to function as helper NLRs, that have a function in the transduction or amplification of defence signals (Bonardi et al., 2011; Li et al., 2015).
Appropriate homeostasis of NLR activity is critical for their function (Li et al., 2015). NLR protein levels must be above a certain threshold to sufficiently activate defence signalling (Bieri et al., 2004; Holt et al., 2005). However, mutations that either constitutively activate NLR immune receptors or result in elevated NLR expression levels can lead to embryo lethality, spontaneous cell death and/or retarded plant growth (Mackey et al., 2003; Palma et al., 2010; Shirano et al., 2002; Xiao et al., 2003). NLR‐mediated resistance can be associated with a substantial reduction in plant fitness (Tian et al., 2003). The phenomenon of hybrid necrosis can be caused by aberrant genetic interactions involving NLR genes in hybrid genomes. Imbalanced activity of NLR genes in such hybrid contexts can result in autoimmunity‐related phenomena, such as spontaneous HR (Bomblies and Weigel, 2007; Bomblies et al., 2007). Similar effects are often observed in transgenic plant lines overexpressing NLRs (Li et al., 2015; Stokes et al., 2002).
Post‐translational mechanisms involving the HSP90 chaperone, the co‐chaperones RAR1 and SGT1, as well as other components of the SKP1‐Cullin‐F‐box (SCF)‐dependent protein degradation pathway, are known to control NLR protein levels and activity (Bieri et al., 2004; Cheng et al., 2011; Holt et al., 2005; Hubert et al., 2003; Schulze‐Lefert, 2004). In addition, control of NLR transcript levels can be important for their function (Mohr et al., 2010). For example, subtle increases in constitutive mRNA levels (of less than two‐fold) of the Arabidopsis NLR gene SNC1 convert normal‐looking plants into dwarfed individuals with strong constitutively activated immune responses (Li et al., 2007; Stokes et al., 2002).
Transcripts of many NLR genes are known to accumulate in response to defence induction or related stimuli. Of 124 Arabidopsis NLR genes represented on the widely used ATH1 microarray, 75 were found to exhibit at least two‐fold higher transcript levels in response to one or more of 15 examined defence‐related treatments (Mohr et al., 2010). A recent RNA‐sequencing (RNA‐seq) analysis uncovered 55 up‐regulated NLR transcripts in response to treatment with the microbe‐associated molecular pattern (MAMP) flg22 in Arabidopsis (Yu et al., 2013). Massive up‐regulation of NLR transcripts after defence induction has also been observed in other plant species, such as Brassica rapa, soybean and rice (Brechenmacher et al., 2015; Chen et al., 2015; Ribot et al., 2008). Experimental detection of elevated transcript levels may reflect enhanced rates of transcription, but also changes in alternative transcript splicing or polyadenylation and/or altered transcript stability.
Transcript up‐regulation of NLRs may locally enhance their potency in pathogen recognition and defence induction (Mohr et al., 2010). Transiently enhanced expression of NLRs in response to cues hinting at the presence of pathogens may increase sensitivity towards pathogen effectors, allowing for stronger and faster immune reactions. As the overexpression of NLR genes is known, in many cases, to lead to constitutive defence activation, broad‐spectrum pathogen resistance as well as spontaneous HR cell death, the defence‐associated up‐regulation of these genes may also generally contribute to inducible immune responses, such as pattern‐triggered immunity (PTI) or ETI.
The immunity of plants can be developmentally regulated (McDowell et al., 2005) and NLR genes can be expressed in a tissue/organ‐specific manner. A recent meta‐analysis suggested that global NLR expression patterns of a plant species can reflect organ‐specific effector challenges encountered by this organism (Much et al., 2017). For example, Arabidopsis exhibits a high preference for NLR expression in shoots relative to roots, whereas the legume, Lotus, shows the opposite trend. Both species show low levels of NLR expression in reproductive tissues. Generally, NLRs seem to be preferentially expressed in tissues in which exposure to effectors is most anticipated. Circadian rhythms may play a similar role in preparing plants for possible pathogen attacks by transient up‐regulation of NLR transcripts at certain times of the day (Bhardwaj et al., 2011; Wang et al., 2011)
During the past 10 years, a large body of literature has accumulated showing that regulatory mechanisms can target NLR transcripts at every step in their expression process (Fig. 1). Constitutive levels of NLR transcripts often seem to be set to a very narrowly defined ground state. In addition, many NLR genes exhibit differential expression of their full‐length transcripts and/or specific transcript isoforms in response to defence induction. Here, we review transcript‐level expression control processes of NLRs. Post‐translational mechanisms of NLR regulation have been covered in several other reviews (Belkhadir et al., 2004b; Li et al., 2015; Schulze‐Lefert, 2004; Shirasu, 2009).
Figure 1.
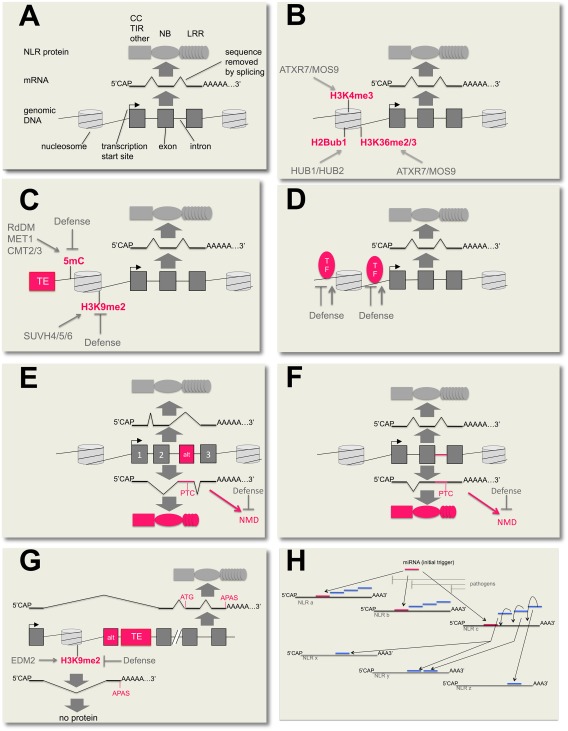
General mechanisms of NLR transcript regulation described in this review. (A) General flow of genetic information from chromatin‐associated genomic DNA through transcription and transcript processing to mature mRNAs, and from mRNAs via translation to NLR proteins. CC, coiled coil; LRR, leucine‐rich repeat; NB, nucleotide‐binding site; TIR, Toll Interleukin‐1 Receptor domain. (B) Post‐translational histone modifications associated with active transcription. H3K4me3, H3K36me2, H3K36me3 and H2ub1 levels positively correlate with transcript levels of some NLRs. In Arabidopsis, the H3K4 and H3K36 methyltransferase ATXR7/MOS9 and the H2 ubiquitylases HUB1 and HUB2 are known to promote high levels of these epigenetic marks. Although not shown for NLR loci so far, some post‐translational histone modifications, such as H3K4me3, can be inducible during pathogen defence. Post‐translational modifications shown in this figure are not meant to be present at the exact location indicated, but rather in the genomic area of the NLR gene. (C) Transposon‐associated silencing marks. Transposon insertions can recruit epigenetic marks silencing their expression into the context of NLR genes, where these marks can have suppressive effects on NLR transcription (or other expression‐related processes; see G). At some NLR loci, the levels of these silencing marks, such as 5mC or H3K9me2, decrease after defence induction. In Arabidopsis, H3K9me2 is established by the SUVH (Suppressor of variegation homologue) 4, 5 and 6 H3 methyltransferases, whereas 5mC is established by the RNA‐directed DNA methylation (RdDM) pathway and maintained by the MET1, CMT2 or CMT3 DNA methyltransferases. How these marks are removed during defence induction is not known. (D) Regulatory transcription factors (TFs), such as WRKYs, can affect the transcript levels of certain NLRs. A broad role of WRKYs in NLR transcription is likely, as their binding sites are enriched in the promoters of Arabidopsis NLRs. Many WRKY TFs are known to be controlled by defence signals and can play activating or suppressing roles in defence gene expression. (E, F) Expression of a large number of NLRs is known to be affected by alternative splicing. This process can result in the inclusion of alternative exons (E) or retained introns (F) in the respective mRNAs. Either one of these processes can lead to the inclusion of in‐frame premature termination codons (PTCs), which may result in truncated NLR proteins and/or recruit the respective transcripts to the nonsense‐mediated decay (NMD) RNA degradation pathway. In at least some cases, truncated NLRs generated by alternative splicing are probably functional by associating with full‐length NLRs or acting as downstream signalling components. Defence‐associated suppression of NMD can lead to increased abundance of some NLR isoforms, probably resulting in heightened states of immunity. (G) An increasing number of NLRs are found to be regulated by alternative polyadenylation. In the case of Arabidopsis RPP7, a transposon recruited the silencing mark H3K9me2 into the first intron of this NLR gene. High levels of H3K9me2 in this area suppress the use of a proximal polyadenylation site and promote the production of full‐length RPP7 coding mRNAs. Reduced levels of H3K9me2 caused by defence induction or mutations in the EDM2 gene lead to enhanced use of the proximal polyadenylation site, increased levels of a prematurely terminated non‐coding transcript and, consequently, reduced levels of full‐length RPP7 coding transcripts. Alternative polyadenylation can be combined with alternative splicing by the differential inclusion of alternative terminal exons, as in RPP7. (A–G) Basic graphical elements used in these panels are defined in (A). Features specifically important or unique for the respective mechanisms are shown in red. TE, transposable element; alt, alternative exon; Defence, defence‐inducing signals; thin pointed arrows, activating interactions; thin blunt arrows, suppressing interactions. (H) MicroRNA (miRNA)‐triggered cascades of secondary phased small interfering RNAs (phasiRNAs). In several cases, miRNAs targeting conserved regions in NLR transcripts were shown to serve as initial triggers (short red horizontal lines) of secondary phasiRNA (short blue horizontal lines) cascades, which comprehensively mediate post‐transcriptional co‐silencing of NLR gene families. On association with primary target NLR transcripts (e.g. NLRs a, b and c in the figure), these miRNAs trigger the production of phasiRNAs which amplify and diversify the initial trigger and reinforce the silencing of the primary NLR targets (e.g. NLRs a, b and c), but also secondary NLR targets (e.g. NLRs x, y and z). Consequently, the secondary phasiRNAs target a wider set of NLR transcripts than the initial miRNA trigger. Such NLR silencing cascades can be suppressed by microbial RNA silencing suppressors resulting in enhanced immunity.
Defence Signals Regulating NLR Transcript Levels
In addition to R gene‐encoded NLRs as specific receptors of pathogen effectors, a second type of receptor, termed pattern recognition receptor (PRR), is involved in the recognition of pathogen‐derived molecules. Molecular recognition of widely conserved MAMPs by PRRs triggers a broad set of defence reactions, jointly referred to as PTI (Couto and Zipfel, 2016). In contrast with NLR‐triggered signalling processes, which are only peripherally understood, a variety of signalling mechanisms have been implicated in the regulation of PTI. These involve numerous regulatory proteins, such as protein kinases, ion channels, G‐proteins, NADPH oxidases, lipase‐like proteins and transcription factors (TFs). Details of such PTI‐associated signalling processes have been summarized recently in several excellent reviews (Bigeard et al., 2015; Couto and Zipfel, 2016; Tsuda and Katagiri, 2010).
Changes in NLR transcript levels can be controlled by unique stimuli, but may also respond to a wide variety of different defence signals. Some NLR genes, such as barley Mla6 or Mla13 (Halterman et al., 2003), only seem to specifically respond to the recognition of their respective cognate avr effector, whereas others are less limited in the spectrum of stimuli to which they respond and exhibit transcript‐level changes during a variety of different defence situations (Mohr et al., 2010). In many cases, the universal defence hormone salicylic acid (SA) seems to be a critical regulator of NLR expression (Shirano et al., 2002; Xiao et al., 2003; Yang and Hua, 2004).
Chromatin‐Associated Mechanisms of NLR Transcript Regulation
Studies on the RPP5 NLR cluster in the Arabidopsis accession Col‐0 have contributed substantially to our understanding of NLR expression control. Suppressor screens for mutations inhibiting autoimmunity in genetic backgrounds with enhanced activity of the RPP5 cluster member SNC1 have uncovered several MOS (modifiers of snc1) loci encoding regulators of NLR expression (Li et al., 2015; van Wersch et al., 2016). The Col‐0 RPP5 cluster is a complex 90‐kb locus harbouring eight closely related NLR genes and two transposons, including the haplotype‐specific RPP4 and SNC1, two NLR genes which have been shown to be functional and mediate pathogen resistance (van der Biezen et al., 2002; Noël et al., 1999; Parker et al., 1993, 1997; Stokes et al., 2002). A first hint at the possible epigenetic regulation of NLR expression stems from the analysis of SNC1, as ectopic insertions of transgenes containing the promoter and transcribed region of this gene resulted in much higher transcript levels than the endogenous SNC1 copy, which seems to be present in a repressive chromatin environment (Li et al., 2007).
Trimethylation of lysine 4 of histone H3 (H3K4me3) has been identified as one possible epigenetic mark required for the appropriate expression of the adjacent RPP5 cluster members RPP4 and SNC1 (Xia et al., 2013). The SET (Su(var)3–9, Enhancer‐of‐zeste, Trithorax) type H3K4 methyltransferase ATXR7 is responsible for establishing this mark at the RPP4 and SNC1 loci. Transcript levels of both NLR genes are reduced in an atxr7 mutant, as are the defence responses triggered by them.
Similar phenotypes are observed in the mos9 mutant, which has been identified as a suppressor of the autoimmunity phenotype of snc1‐1 (Xia et al., 2013). MOS9, a nuclear plant‐specific protein, which bears no discernible motifs or domains, seems to be physically associated with ATXR7, assisting this histone methyltransferase in its catalytic function. H3K4me3 is a mark universally associated with actively transcribed genes (Kouzarides, 2007). A similar situation was observed for LAZ5, a TIR‐NB‐LRR‐encoding gene unlinked to the RPP5 cluster, which, for correct expression and function, requires the SET domain group 8 (SDG8) histone methyltransferase (Palma et al., 2010). SDG8 is responsible for di‐ or trimethylation of H3K36. Like methylation of H3K4, these marks are generally associated with active transcription. Whether the levels of any of these histone modifications change at their target NLR loci in response to defence induction, or whether they play any specific roles in defence‐related regulation of other NLRs, has not been reported at this point. However, defence‐inducing treatments have been found to increase H3K4me3 levels at other Arabidopsis loci (Jaskiewicz et al., 2011).
In addition to methylation, ubiquitylation of histones is known to affect transcription. Two E3 ubiquitin ligases, HUB1 and HUB2, are known to mono‐ubiquitylate histone H2B to H2Bub1 in Arabidopsis. Both of these enzymes control H2Bub1 levels at SNC1 and RPP4, and positively contribute to the transcription of these NLR genes (Zou et al., 2014). In response to defence induction, H2Bub1 levels at SNC1 and RPP4 exhibit a moderate increase, which is associated with elevated transcript levels of these genes.
Another example of a post‐translational histone modification exhibiting differential levels at an NLR gene after defence induction is the canonical transposon silencing mark H3K9me2 (Tsuchiya and Eulgem, 2013). After induction of an ETI response, H3K9me2 levels transiently decrease within the first intron of RPP7, affecting alternative polyadenylation (APA) of this NLR gene (see below).
The histone mark H3K9me2 is functionally linked to cytosine methylation, another type of epigenetic signal found to have an impact on the expression of NLR genes. The complex interplay of de novo methylation, maintenance methylation and de‐methylation mechanisms controls the methylation of the nucleobase cytosine at position 5. Methylated cytosine (5mC) occurs in plants at the symmetrical GC and CHG motifs or asymmetrical CHH sites (H = any nucleobase, except G). De novo methylation seems to be globally controlled in Arabidopsis by the RNA‐directed DNA methylation (RdDM) pathway (Law and Jacobsen, 2010). Well‐characterized Arabidopsis maintenance methyltransferases include MET1, which methylates cytosines at hemimethylated GC, and CMT2 and CMT3, which catalyse cytosine methylation at CHH or CHG sites associated with H3K9me2. Cytosine demethylation requires active base excision followed by DNA repair. As in the case of H3K9me2, 5mC is primarily involved in the silencing of transposons. In the promoters of genes, 5mC typically has a repressive effect on transcription.
Transcript levels of several NLR genes have been found to be controlled by cytosine methylation. Generally, it seems that this mark is often recruited to NLR genes by transposon insertions. The Arabidopsis NLR gene RMG1 (Resistance Methylated Gene 1), encoding a TIR‐NB‐LRR protein, is expressed at high levels in response to the MAMP flg22. RMG1 transcripts are suppressed by cytosine‐methylated helitron transposon regions in this gene's promoter and flg22‐induced processes seem to counteract this suppressive mechanism (Yu et al., 2013). Although its details are unknown, this regulatory mechanism may be linked to the flg22‐induced decrease of several transcripts encoding components of the RdDM and 5mC maintenance pathways (Yu et al., 2013). Defence induction is known to globally affect 5mC levels in Arabidopsis (Deleris et al., 2016; Dowen et al., 2012; Pavet et al., 2006; Yu et al., 2013). For example, avr effectors of the bacterial pathogen Pseudomonas syringae pv tomato DC3000 induce hypomethylation and chromatin de‐condensation at centromeric and pericentromeric repeats (Pavet et al., 2006). SA, as well as various P. syringae strains, induce comprehensive changes in 5mC patterns in genic regions and transposons of the Arabidopsis genome (Dowen et al., 2012). The MAMP flg22 has also been shown to induce globally differential cytosine methylation in transposons and transposon‐associated defence genes (Yu et al., 2013).
Regulation of NLR Gene Expression by TFs
Major types of TF known to control transcript levels of defence‐related genes are members of the WRKY, ERF and TGA‐bZIP families (Eulgem, 2005). WRKY TFs typically bind to pathogen/elicitor‐responsive W box cis‐elements (hexameric consensus sequence: TTGACC/T) (Eulgem and Somssich, 2007; Eulgem et al., 2000; Rushton et al., 2010). Mohr et al. (2010) demonstrated a key role of W boxes in controlling constitutive levels and the defence‐associated transient accumulation of RPP8 transcripts. RPP8 encodes a CC‐NB‐LRR in the Arabidopsis accession Ler, which mediates strong race‐specific resistance against isolates of the pathogenic oomycete Hyaloperonospora arabidopsidis. RPP8 exhibits low basal transcript levels, which are transiently elevated in response to various defence‐inducing stimuli. In contrast with a wild‐type RPP8 transgene, an RPP8 construct containing mutated W boxes in its promoter failed to confer RPP8‐mediated immunity against H. arabidopsidis in a susceptible Arabidopsis background. W boxes were further found to be generally enriched in promoter regions of Arabidopsis NLR genes (Mohr et al., 2010), supporting a general role of these promoter elements and, possibly, WRKY TFs in the transcriptional regulation of NLR genes. Although, at this point, no other types of TF seem to be known to be involved in transcriptional regulation of NLR genes, it is unlikely that such roles are exclusively executed by WRKYs.
Alternative Splicing in NLR Expression Control
Alternative splicing (AS) has been recognized as a main contributor to transcriptome and proteome diversity. Dynamic AS processes, which are responsive to environmental or developmental stimuli, can regulate the abundance of defined functional mRNA isoforms, and thereby serve as a mechanism of quantitative gene expression control.
In Arabidopsis, AS is known to affect approximately 60% of its multi‐exon genes (Marquez et al., 2012). Studies in other dicot or monocot angiosperm species have suggested that AS affects 30%–50% of their multi‐exon genes (Mandadi et al., 2014; Shen et al., 2014; Thatcher et al., 2014).
Multiple reports have linked dynamic AS events to the activation of plant immune responses (Dinesh‐Kumar and Baker, 2000; Howard et al., 2013; Mandadi et al., 2014; Yang et al., 2014; Zhang and Gassmann, 2007). Many examples of alternative NLR transcript splicing have been described, some of which seem to be constitutive and not affected by defence signalling, and some of which are dynamic and responsive to defence‐related signals (Table 1) (Gassmann, 2008; Jordan et al., 2002; Yang et al., 2014). Although many cases of AS in plants result in a changed composition of exonic coding sequences and, consequently, in changed protein sequences, the retention of intronic sequences seems to be predominant in plants (Reddy et al., 2013). This form of AS can introduce premature in‐frame stop codons, which can destabilize transcripts by subjecting them to nonsense‐mediated decay, a pathway of RNA degradation (see below). However, at least in some cases of NLR expression control, such alternative transcript isoforms seem to remain stable and to encode truncated protein versions that contribute to successful immunity (see below).
Table 1.
Examples of alternative splicing in NLR gene expression.
| Species/NLR | Known variants | Comments | Reference |
|---|---|---|---|
| Tobacco N | 2 | Alternative exon in intron 3; both isoforms are required for full immunity | Whitham et al. (1994); Dinesh‐Kumar and Baker (2000) |
| Tomato Bs4 | 2 | Alternative splice donor sites | Schornack et al. (2013) |
| Potato Y‐1 | 4 | Differential use of cryptic intron combined with alternative polyadenylation | Vidal et al. (2002) |
| Medicago truncatula RCT1 | >4 | Combined alternative splicing/polyadenylation leading to numerous different transcript isoforms representing at least four distinct coding sequence variants | Tang et al. (2013) |
| Common bean JA1tr | 7 | Result suggest the existence of seven transcripts generated by alternative use splice donor–acceptor sites and polyadenylation | Ferrier‐Cana et al. (2005) |
| Soybean Rj2 | 2 | Intron 4 retention; regular transcript is sufficient for function | Tang et al. (2016) |
| Flax L6 | 4 | Intron 3 retention; alternative splice donor or acceptor sites | Lawrence et al. (1995) |
| Flax M | 12 | Intron retention, exon skipping and use of multiple splice acceptor sites | Schmidt et al. (2007) |
| Arabidopsis RPS4 | 3 | Intron 2 and/or intron 3 retention; all isoforms are required for full immunity | Gassmann et al. (1999); Zhang and Gassmann (2003) |
| Arabidopsis RAC1 | 2 | Intron 1 retention | Borhan et al. (2004) |
| Barley Mla6‐2 | 2 | Intron retention | Halterman et al. (2001) |
| Barley Mla13 | 5 | Alternative spliced introns in 5′ untranslated region (5′ UTR) | Halterman et al. (2003) |
| Rice Pi‐ta | 12 | 12 different transcript isoforms generated by combined alternative splicing and polyadenylation with some forms also having different 5′ ends | Costanzo and Jia (2009) |
| Rice RGA5 | 2 | Intron 3 retention; isoform with retained intron does not confer resistance to Magnaporthe oryzae | Cesari et al. (2013) |
| Wheat LR10 | 2 | Intron retention | Sela et al. (2012) |
| Wheat Sr35 | 2 | Intron 3 retention (second intron in 3′ UTR) | Saintenac et al. (2013) |
One of the first reports on AS in NLR regulation demonstrated the critical importance of two splice isoforms of the tobacco TIR‐NB‐LRR gene N, which mediates resistance against Tobacco mosaic virus (TMV) (Dinesh‐Kumar and Baker, 2000; Whitham et al., 1994). Both isoforms are required for full immunity: the full‐length protein encoding the shorter isoform Ns and the longer Nl version, which encodes a truncated protein via a retained alternative exon leading to reading‐frame shift and a premature translational termination codon. Interestingly, N‐mediated TMV recognition in tobacco triggers a transient change in the Ns/Nl transcript ratio, which is essential for a successful defence against TMV. A 3′ untranslated region (3′ UTR) downstream of the N coding sequence seems to be critically important for the control of this ratio. The mechanistic details of this dynamic AS process are not known.
AS has also been described for the barley CC‐NB‐LRR encoding Mla6 and Mla13 genes (Halterman and Wise, 2004; Halterman et al., 2001, 2003; Shen et al., 2003). Mla13 is a particularly interesting example as the ratio of its five different splice isoforms changes after defence induction by its cognate powdery mildew strain (Halterman et al., 2003). Even more complex is the situation for the rice CC‐NB‐LRR gene Pi‐ta, which expresses 12 different transcript splice forms, 11 of which are predicted to encode proteins and some of which are differentially expressed after defence induction by rice blast (Costanzo and Jia, 2009).
Additional examples of alternative NLR transcript splicing include wheat LR10 and Sr35 (Saintenac et al., 2013; Sela et al., 2012), rice RGA5 (Cesari et al., 2013), flax L6 and M (Anderson et al., 1997; Lawrence et al., 1995), tomato Bs4 (Schornack et al., 2004), potato Y‐1 (Vidal et al., 2002), Medicago truncatula RCT1 (Tang et al., 2013), common bean JA1tr (Ferrier‐Cana et al., 2005), soybean Rj2 (Tang et al., 2016) and the Arabidopsis genes RPS4 (Gassmann et al., 1999) and RAC1 (Borhan et al., 2004). As in the case of N, MLA13 and Pi‐ta, the relative abundance of its six splice forms changes for RPS4 (Zhang and Gassmann, 2007) and, similar to N, AS of RPS4 was found to be critical for its function (Zhang and Gassmann, 2003). This also applies to RCT1, for which the presence or absence of alternative transcripts affects the outcome of pathogenic interactions of M. truncatula and races of the fungus Colletotrichum trifolii (Tang et al., 2013). Strikingly, the AS pattern of RCT1 orthologues is conserved between M. truncatula and its close relative M. sativa. A recent RNA‐seq study found nine NLR genes to exhibit differential accumulation of alternative splice isoforms after viral infections in the monocot model system Brachypodium distachyon (Mandadi et al., 2014).
Interestingly, most TIR‐NB‐LRR genes share a common gene structure, with the first exon encoding the TIR domain, the second exon encoding the NB domain and the remaining exons encoding the LRR region as well as additional C‐terminal parts (Yang et al., 2014). Common to several NLR genes encoding TIR‐NB‐LRR proteins, such as N or RPS4, AS leads to the expression of truncated proteins partially or fully lacking their LRR, NB‐LRR or other C‐terminal portions (Gassmann, 2008; Yang et al., 2014). Typically, this is a consequence of the retention of intronic sequences introducing premature in‐frame stop codons into the mature mRNAs. The resulting truncated protein variants may serve critical functions during defence activation, as the transcript isoforms encoding them seem to be needed for full immunity in many cases. Multiple studies have shown intramolecular or intermolecular interactions between individual NLR protein domains to have important roles in the regulation of their activity (Belkhadir et al., 2004a; Bendahmane et al., 2002; Hwang et al., 2000; Moffett et al., 2002; Rairdan and Moffett, 2006; Takken and Goverse, 2012). Thus, by engaging in protein–protein interactions with defined NLR partners, accumulating TIR‐NB‐LRR protein fragments resulting from induced splicing processes may serve to regulate NLR activity (Gassmann, 2008). Systematic protein–protein interaction studies support such a role of truncated NLRs (Nandety et al., 2013). Alternatively or additionally, truncated NLRs may also play a role in downstream defence signalling processes, as has been demonstrated for the mammalian TIR domain protein MyD88 (Kenny and O'Neill, 2008). This possibility is supported by the fact that the overexpression of isolated TIR or CC domains of some plant NLRs can constitutively induce defence activation (Bernoux et al., 2011; Collier et al., 2011; Maekawa et al., 2011; Swiderski et al., 2009; Zhang et al., 2004).
Molecular processes controlling alternative NLR transcript splicing do not seem to be understood in great detail at this point. Some splicing factors, such as serine and arginine‐rich (SR) proteins, are known to be stress regulated (Ali and Reddy, 2008; Palusa et al., 2007). In B. distachyon, multiple genes encoding putative splicing regulators have been found to exhibit differential splicing after viral infections (Mandadi and Scholthof, 2015). Thus, in some cases, differential activity of regulatory proteins may contribute to dynamic NLR transcript splicing after defence induction. A partial loss‐of‐function mutation of MOS12 encoding an arginine‐rich protein related to the human SR‐type splicing regulator cyclin 1 has been reported to affect the splicing patterns of the Arabidopsis NLR genes SNC1 and RPS4 (Xu et al., 2012). Components of the evolutionarily conserved nuclear spliceosome‐associated MOS4‐associated complex (MAC) (Monaghan et al., 2009, 2010; Palma et al., 2007) have been found to physically interact with the MOS12 protein (Xu et al., 2012). Mutations in genes encoding other MAC components have also been shown to affect splicing‐related processes controlled by MOS12 (Xu et al., 2012). Roles of SR proteins, like MOS12, in SNC1 and RPS4 transcript splicing are further supported by splicing defects of these NLRs in a mutant of the SR importin‐encoding MOS4 gene (Xu et al., 2012). SR importins are known to function as nuclear import receptors for SR proteins (Ström and Weis, 2001). Although these results strongly link MAC components to alternative SNC1 splicing, direct physical interactions of these proteins with SNC1 transcripts have not been reported so far.
In addition to direct involvement of splicing regulators, AS can be affected by the rate of Pol II elongation during transcription (de la Mata et al., 2010). Low rates of Pol II elongation can favour the use of upstream splice sites over their downstream alternatives. Such kinetic coupling between transcription and splicing can theoretically be controlled by defence‐associated changes in chromatin states. Epigenetic marks can also serve the recruitment of splicing factors to sites engaged in co‐transcriptional transcript processing. It will be interesting to see whether future studies can establish functional links between defence‐associated changes in cytosine methylation or histone modifications and dynamic alternative splicing processes affecting the expression of defence genes in general or, specifically, NLR genes.
Alternative Polyadenylation as a Mode of NLR Regulation
In contrast with AS, the significance of APA for global gene expression has been realized only very recently (Proudfoot, 2011). At least 70% of all Arabidopsis genes are known to be subject to APA (Sherstnev et al., 2012; Wu et al., 2011). APA can result in distinct transcript isoforms that differ in their coding sequences, but also their 3′ UTRs, which are typically unique for each transcript and important for their stability (Proudfoot, 2011).
Mechanistically, the processes of AS and APA are distinct, although they can also act together (see Fig. 1G). Cleavage of native mRNAs at their 3′ end and the addition of a poly A tail can be controlled by a set of cis‐elements at the 3′ end of the pre‐mRNA, referred to as polyadenylation signals (Proudfoot, 2011; Tian and Graber, 2012). A highly conserved protein complex of more than 50 subunits is typically recruited to polyadenylation sites (Hunt et al., 2008; Lutz, 2008).
Several recently published studies have indicated that APA is also involved in the regulation of plant immune responses (Lyons et al., 2013; Shen et al., 2011; Thomas et al., 2012, Tsuchiya and Eulgem, 2013). For example, the Arabidopsis RNA‐binding protein FPA affects APA of the ERF4 TF gene in response to the MAMP flg22 (Lyons et al., 2013). This mechanism seems to limit the extent of MAMP‐induced defence responses. Furthermore, the Arabidopsis polyadenylation complex component CPSF30 controls APA site use in introns and 3′ UTRs of a large number of defence‐associated genes, and positively contributes to SA‐dependent defence responses and HR‐like cell death (Bruggeman et al., 2014; Thomas et al., 2012).
Early evidence for an important role of APA in NLR regulation stems from a massive parallel signature sequencing analysis, which showed that transcripts of 72 Arabidopsis NLR genes are possibly alternatively polyadenylated (Tan et al., 2007). The only in‐depth analysis of APA in NLR regulation has been reported for the Arabidopsis NLR gene RPP7. This CC‐NB‐LRR gene mediates strong ETI of the Arabidopsis accession Col against the Hiks1 isolate of H. arabidopsidis (McDowell et al., 2000). The gene structure of RPP7 is unusually complex and features three non‐coding upstream exons, three central coding exons and three non‐coding downstream exons. The COPIA‐R7 retrotransposon, which is inserted in the first RPP7 intron, recruits high levels of the transposon silencing mark H3K9me2 into this region of RPP7, together with a polyadenylation signal in its 5′ long terminal repeat (Tsuchiya and Eulgem, 2013). Loss of RPP7‐mediated resistance in edm2 (enhanced downy mildew 2) (Eulgem et al., 2007) mutants is associated with a strong shift in RPP7 polyadenylation site use, leading to strong accumulation of proximally polyadenylated non‐coding transcripts and, consequently, reduced production of full‐length RPP7 coding transcripts (Lei et al., 2013; Tsuchiya and Eulgem, 2013). Similar effects were observed in mutants of ASI1/IBM2 (Saze et al., 2013; Wang et al., 2013). EDM2 encodes a histone H3‐binding PHD‐finger protein (Eulgem et al., 2007; Lei et al., 2013; Tsuchiya and Eulgem, 2014), whereas IBM2 encodes an RNA‐binding protein (Saze et al., 2013; Wang et al., 2013). Thus, an unconventional mechanism may link histone marks to the RNA processing machinery.
EDM2 promotes high levels of H3K9me2 in the first RPP7 intron and suppresses the use of the proximal polyadenylation site in this area. H3K9me2 is critical for this process, as the suvh4/5/6 triple mutant, which is deficient in three Arabidopsis H3K9 methyltransferases (Ebbs and Bender, 2006), phenocopies the effects of EDM2 on RPP7 expression and function. The EDM2‐ and H3K9me2‐dependent polyadenylation mechanism is responsive to pathogen recognition and dynamically adjusts RPP7 expression levels during the induction of immune responses (Tsuchiya and Eulgem, 2013).
Post‐Transcriptional Regulation of NLR Transcripts by Small RNAs
In addition to silencing at the transcriptional level by the mediation of cytosine methylation, plant small RNAs can also suppress the expression of target genes at the post‐transcriptional level. MicroRNAs (miRNAs) and some types of small interfering RNAs (siRNAs), such as secondary cis‐ or trans‐acting siRNAs (casiRNAs; tasiRNAs), can bind to complementary sequences in target mRNAs and suppress their expression, either by endonucleolytic cleavage followed by degradation or translational suppression (Axtell, 2013).
The first evidence for a role of small RNAs in the suppression of NLR genes was provided by Yi and Richards (2007), who found small RNAs to co‐suppress several members of the Arabidopsis Col‐0 RPP5 cluster, including SNC1 and RPP4. Although the mechanistic details of this process are not known, this process appears to involve 21‐nucleotide siRNAs generated from overlapping sense and antisense transcripts within the RPP5 cluster, as well as the Dicer‐like protein DCL4 and the Argonaute member AGO1. Additional studies have suggested roles of miRNAs in the cleavage of NLR transcripts in Brassica rapa, Vitis vinifera, Pinus taeda and Citrus trifoliata (Carra et al., 2009; He et al., 2008; Lu et al., 2007; Park and Shin, 2015; Song et al., 2010).
Recent studies have provided substantial evidence for the use of phased siRNAs (phasiRNAs) in the broad silencing of NLR family members in legumes and Solanaceae (Park and Shin, 2015). This type of secondary siRNA can be triggered in Arabidopsis by 22‐nucleotide miRNAs (Allen and Howell, 2010). By targeting conserved regions in 74 NLR transcripts of the legume M. truncatula, the miR1507, miR2109 and miR2118 families of highly abundant 22‐nucleotide miRNAs trigger a cascade of phasiRNAs that probably serve as casiRNAs or tasiRNAs, suppressing the expression of at least 60% of the 540 NLR genes of this organism (Zhai et al., 2011). One of these miRNA families, miR2118, is closely related to the miR482 family of Solanaceae, which can also serve as a miRNA trigger of a regulatory cascade suppressing NLR expression (Shivaprasad et al., 2012). Although variable in sequence, members of the widely conserved miR482/miR2118 superfamily target transcript sequences encoding the P‐loop motif of NLR nucleotide‐binding sites, as well as LRR motifs of these immune receptors. Transient expression in Nicotiana benthamiana leaves supported a role of miR482 in NLR transcript decay associated with RDR6‐dependent production of secondary phasiRNAs (Shivaprasad et al., 2012). Intriguingly, this silencing cascade can be suppressed in plants infected with various viral or bacterial pathogens, resulting in enhanced levels of miR482‐targeted NLR transcripts and, as a likely consequence, enhanced basal defence states, as NLR overexpression is known to trigger defence reactions even in the absence of the respective cognate avr ligands.
In addition to miR482‐type miRNAs, six additional miRNA families from the three Solanaceae species tobacco, tomato and potato were also found to cleave NLR transcripts and to trigger secondary phasiRNAs (Li et al., 2012). These include two miRNAs (nta‐miR6019 and nta‐miR6020) that guide the cleavage of the tobacco TMV resistance gene N. Transient overexpression of these two miRNAs in N. benthamiana leaves attenuated N‐mediated TMV resistance, demonstrating the significance of these miRNAs for plant immunity (Li et al., 2012). Evidence for similar phasiRNA networks suppressing NLR expression was also found in many other plant species, including gymnosperms (Howell et al., 2007; Johnson et al., 2009; Klevebring et al., 2009; Zhai et al., 2011), suggesting that this type of control mechanism evolved early in land plants. The ancient gymnosperm Norway spruce features 19 miRNA families targeting over 750 NLR genes to generate phasiRNAs. This also includes members of the miR482/miR2118 superfamily (Xia et al., 2015). Based on an extensive analysis of the NLR contingents of 70 land plants, coupled with extensive small RNA data, Zhang et al. (2016) observed a tight association between the diversity of NLRs and the emergence of miRNAs targeting them. They proposed a co‐evolutionary model according to which miRNAs targeting NLRs are frequently generated de novo from highly duplicated NLR genes.
Consistent with the involvement of RDR6 in the generation of secondary siRNAs and the roles of these small RNAs in NLR suppression, an Arabidopsis rdr6 mutant exhibits enhanced resistance against virulent and avirulent strains of P. syringae (Boccara et al., 2014). Enhanced resistance against P. syringae was also observed by knock down of the miR482‐related Arabidopsis miR472, whereas overexpression of this NLR‐targeting miRNA reduced pathogen resistance. In RDR6‐ and miR472‐deficient lines, numerous NLR transcripts exhibit altered constitutive levels and a stronger defence‐associated induction compared with wild‐type Arabidopsis.
Taken together, all these observations indicate that miRNAs can serve as master regulators triggering cascades of secondary phasiRNAs, which collectively post‐transcriptionally suppress the expression of a wide set of NLRs sharing conserved target motifs. This type of mechanism may serve one or several important purposes (see below).
1. Enhanced NLR evolvability by global NLR transcript dosage control
The necessity for fast‐paced NLR evolution inevitably must lead to the occasional emergence of highly expressed variants causing a substantial fitness penalty for the plant. Even if expression levels of individual NLRs are limited, expansion of this gene family may result in intolerably high levels of NLR transcripts and, consequently, NLR proteins. Thus, miRNA‐controlled global NLR suppression may keep collective levels of NLR transcripts under a critical threshold and provide a background allowing for safe diversification and expansion within NLR gene families (Shivaprasad et al., 2012). An miRNA‐triggered phasiRNA cascade globally suppressing transcripts of the pentatricopeptide repeat gene family in Arabidopsis has been proposed to serve a similar purpose and to quench rapid expansion of this fast‐evolving and diversifying gene family (Allen and Howell, 2010; Lurin et al., 2004; Saha et al., 2007).
2. Counter‐counter defence by pathogen‐induced transient overexpression of NLR genes
As described above, the miR482‐triggered silencing cascade can be suppressed by viral or bacterial pathogens (Shivaprasad et al., 2012). This effect may be caused by microbial RNA silencing suppressors, which attenuate miR482 levels. Various types of pathogen seem to be able to suppress the host RNA silencing machinery, which originally evolved as a mechanism enhancing pathogen virulence (Li and Ding, 2006; Navarro et al., 2008; Pumplin and Voinnet, 2013; Qiao et al., 2013). By non‐specifically counteracting miRNA‐dependent defence responses, pathogens may also counteract the miR482/miR2118‐mediated NLR suppression mechanism and transiently elicit high NLR transcript levels and, consequently, a broad activation of defence reactions, as observed in NLR‐overexpressing plant lines (Park and Shin, 2015; Shivaprasad et al., 2012).
3. Immune suppression to enable symbioses with microbes
In legumes, highly abundant miRNAs and associated phasiRNAs regulate NLR expression en masse (Zhai et al., 2011). The extent of this NLR suppression system may imply a function in symbiosis with nitrogen‐fixing rhizobia bacteria, a process specific to legumes. One of the NLR‐targeting miRNAs, miR482, triggers hypernodulation in soybean and is transiently up‐regulated in this host in response to inoculation with rhizobia (Li et al., 2010). Another NLR‐targeting miRNA, miR1507, is up‐regulated in a hypernodulating soybean mutant (Li et al., 2010). Thus, global suppression of NLR transcripts associated with rhizobial colonization may enable interactions with these beneficial symbionts and support the nodulation process. As some NLR‐associated miRNAs are also conserved in Solanaceae and other plant clades, they may have a wider role in supporting plant–microbial symbioses, such as the mycorrhizal colonization of roots (Zhai et al., 2011).
Nonsense‐Mediated Decay of NLR Transcripts
Originally, nonsense‐mediated mRNA decay (NMD) was identified as a cytoplasmic surveillance mechanism degrading aberrant mRNAs (Maquat, 2004; Schweingruber et al., 2013). In many cases, the NMD machinery recognizes premature translational termination codons (PTCs) in conjunction with downstream exon junction complexes. NMD‐targeted transcripts enter general mRNA degradation processes (Nicholson and Mühlemann, 2010). An increasing body of literature supports a wide role of NMD in eukaryotic gene expression control, and NMD seems to regulate the transcript levels of specific genes in response to a wide variety of biological signals (Bruno et al., 2011; Drechsel et al., 2013; Gloggnitzer et al., 2014). Interestingly, impairment of NMD by mutations in Arabidopsis leads to constitutive immunity (Jeong et al., 2011; Rayson et al., 2012; Riehs‐Kearnan et al., 2012). This effect can be rescued by mutations in defence signalling components (Riehs‐Kearnan et al., 2012). A recent RNA‐seq analysis found that transcripts of 50 Arabidopsis NLR genes were up‐regulated in an NMD‐deficient background, and were therefore probably suppressed by the NMD machinery (Gloggnitzer et al., 2014). Splice variants of a subset of these, all of the TIR‐NB‐LRR‐type, have typical features of NMD targets, such as PTCs, and were found to exhibit increased half‐lives in the NMD‐ and defence‐deficient smg7/pad4 background. The Ler allele of one of these NLRs, RPS6Ler, was found to be causal for the strong constitutive immunity phenotype observed in an NMD‐deficient mixed Col‐0/Ler ecotype background. As proposed for NLR‐targeting miRNA/phasiRNA cascades, NMD may also enhance plant fitness and NLR evolvability by controlling global NLR transcript dosage and preventing inappropriately high NLR activity levels. Intriguingly, NMD appears to play a role in defence induction similar to the miRNA/phasiRNA cascades described above. The perception of MAMPs can trigger transient suppression of NMD in Arabidopsis and, consequently, a temporary increase in NMD target transcripts, including those of NLRs, which is associated with enhanced disease resistance against the virulent DC3000 strain of P. syringae (Gloggnitzer et al., 2014).
AS can generate NLR transcripts with NMD‐eliciting features. At least in some cases, excision of cryptic introns results in NMD‐susceptible NLR transcripts, whereas transcripts containing retained introns, the most prevalent form of alternative splice products in plants, are often not targeted by NMD, even if featuring PTCs (Gloggnitzer et al., 2014; Gohring et al., 2014). Instead, such transcripts can be retained in the nucleus, where they are inaccessible to the NMD machinery (Gohring et al., 2014).
Roles of Transposons in the Evolution of NLR Regulatory Mechanisms
NLR genes are one of the fastest evolving gene families in plants. Their often‐found organization in complex gene clusters is believed to be critical for the fast pace of their structural and functional diversification (Meyers et al., 2003; Michelmore and Meyers, 1998). Frequent recombination events at such loci, followed by diversifying selection, may generate the structural diversity needed to match high avr effector evolution rates in the microbial world (Jacob et al., 2013; McDowell and Simon, 2006). The need for well‐balanced expression probably forces on new NLR varieties a maturation process to evolve suitable regulatory mechanisms. High enrichment of transposons in NLR clusters is often observed (Miyao et al., 2003; Wei et al., 2002). In the recent past, transposons have emerged as a major source of the ‘raw material’ needed for the fast evolution of nuanced NLR expression control. For example, in Solanaceae, the insertion of miniature inverted‐repeat transposable elements (MITEs) within NLR regions appears to be conserved, as numerous families of these transposons were found to reside within six syntenic NLR regions in tomato, potato and tobacco (Kuang et al., 2009). These insertions are suspected to affect gene function and regulation in many cases. One of them, MiS1‐1, is critical for the function and regulation of the tobacco TIR‐NB‐LRR gene N, by providing the alternative exon, which is retained in the alternatively spliced Nl transcript (see above) (Dinesh‐Kumar and Baker, 2000; Kuang et al., 2009).
The insertion of the LTR retrotransposon Renovator upstream of the rice NLR gene Pit provided promoter sequences directing sufficiently high transcript levels of this gene for effective disease resistance against the fungal pathogen Magnaporthe oryzae (Hayashi and Yoshida, 2009). Likewise, insertion of the COPIA‐R7 LTR retrotransposon into the first intron of the 5′ UTR of the Arabidopsis NLR gene RPP7 recruited an H3K9me2‐dependent APA mechanism critical for the function of this gene (Tsuchiya and Eulgem, 2013). In addition to COPIA‐R7, additional transposon sequences affect the complex regulation of RPP7. Several transposon and other repeat sequences in its 5′ UTR seem to attract massive cytosine methylation to a region directly upstream of the transcription start‐site (Le et al., 2014). However, cytosine methylation in this region is suppressed by demethylases to a level that allows for sufficient levels of transcription of this gene. Furthermore, electrophoretic mobility shift experiments with Arabidopsis nuclear protein extracts have shown that the Simplehat transposon in the distal promoter of RPP7 provides a docking site for an unknown DNA‐binding activity and serves as a cis‐element mediating elevated GUS reporter gene expression in response to the recognition of H. arabidopsidis (Y.‐H. Wei and T. Eulgem, unpublished).
Conclusions
The need for tightly regulated and balanced expression of NLR genes has led to the evolution of multiple mechanisms of NLR transcript‐level expression control (Fig. 1). Several themes in this context reoccur.
The involvement of AS in NLR expression control is widespread (Table 1). Defence‐associated changes in the ratios between different splice forms often seem to be critical for immunity. In some cases, AS seems to result in splice forms encoding truncated NLRs, which can be functionally important for defence induction.
Numerous examples of NLR suppression by miRNA‐induced phasiRNA cascades as well as NMD are emerging. A probably common role of these mechanisms is global NLR transcript dosage control, which may be a critical prerequisite for the evolution of new NLRs and the expansion of this gene family. Pathogen‐triggered down‐regulation of these NLR suppressive mechanisms allows for a transient defence induction.
Transposons are likely to play an important role in the fast evolution of NLR genes by providing the ‘raw material’ for regulatory sequences and chromatin states serving their expression control.
Defence induction can trigger a transient global reduction of transposon silencing marks, such as 5mC and H3K9me2. At least in some cases, these epigenetic signals affect NLR expression and function.
In addition to NLR genes, the expression of other components of the plant immune system must be tightly controlled. The overexpression and misregulation of numerous defence regulators are known to result in autoimmunity and growth retardation, similar to the effects observed in plant lines with enhanced NLR activity. Thus, the regulatory mechanisms discussed here are unlikely to be limited to NLR genes. For example, the involvement of WRKY TFs and W box promoter elements is clearly widespread amongst many genes encoding components of the plant immune system. However, the tolerance of plants to a relaxation of NLR control mechanisms is low, and mutations affecting these are associated with reduced plant fitness.
A comprehensive understanding of NLR expression control is not only of academic interest. Efforts to increase pathogen and pest resistance of crops are strongly dependent on breeding‐based transfer of NLR genes into different genetic backgrounds. The design of genetically engineered crops expressing new NLR derivatives is a promising approach for the near future. An accurate knowledge of the gene regulatory requirements for optimal expression of the respective NLR genes is likely to increase the success of breeding‐ and gene transfer‐based approaches targeting enhanced pathogen resistance of crop cultivars.
Acknowledgements
The authors wish to apologize to all colleagues whose work was not cited here because of space limitations. We thank Dr Yu‐Hung Wei for sharing unpublished results. RPP7/EDM2‐related work in the Eulgem laboratory was funded by National Science Foundation grants IOB‐6890764, IOS‐1052556, MCB‐1330905 and IOS‐1457329. YL received additional support from the China Scholarship Council and the National Natural Science Foundation of China (grant #31401312).
References
- Abramovitch, R.B. , Anderson, J.C. and Martin, G.B. (2006) Bacterial elicitation and evasion of plant innate immunity. Nat. Rev. Mol. Cell Biol. 7, 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, G.S. and Reddy, A.S. (2008) Regulation of alternative splicing of pre‐mRNAs by stresses. Curr. Top. Microbiol. Immunol. 326, 257–275. [DOI] [PubMed] [Google Scholar]
- Allen, E. and Howell, M.D. (2010) miRNAs in the biogenesis of trans‐acting siRNAs in higher plants. Semin. Cell Dev. Biol. 21, 798–804. [DOI] [PubMed] [Google Scholar]
- Anderson, P.A. , Lawrence, G.J. , Morrish, B.C. , Ayliffe, M.A. , Finnegan, E.J. and Ellis, J.G. (1997) Inactivation of the Flax rust resistance gene M associated with loss of a repeated unit within the leucine‐rich repeat coding region. Plant Cell, 9, 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell, M.J. (2013) Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 64, 137–159. [DOI] [PubMed] [Google Scholar]
- Belkhadir, Y. , Nimchuk, Z. , Hubert, D.A. , Mackey, D. and Dangl, J.L. (2004a) Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell, 16, 2822–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir, Y. , Subramaniam, R. and Dangl, J.L. (2004b) Plant disease resistance protein signaling: NBS‐LRR proteins and their partners. Curr. Opin. Plant Biol. 7, 391–399. [DOI] [PubMed] [Google Scholar]
- Bendahmane, A. , Farnham, G. , Moffett, P. and Baulcombe, D.C. (2002) Constitutive gain‐of‐function mutants in a nucleotide binding site‐leucine rich repeat protein encoded at the Rx locus of potato. Plant J. 32, 195–204. [DOI] [PubMed] [Google Scholar]
- Bernoux, M. , Ve, T. , Williams, S. , Warren, C. , Hatters, D. , Valkov, E. , Zhang, X. , Ellis, J.G. , Kobe, B. and Dodds, P.N. (2011) Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self‐association, signaling, and autoregulation. Cell Host Microbe, 9, 200–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj, V. , Meier, S. , Petersen, L.N. , Ingle, R.A. and Roden, L.C. (2011) Defence responses of Arabidopsis thaliana to infection by Pseudomonas syringae are regulated by the circadian clock. PLoS One, 6, e26968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieri, S. , Mauch, S. , Shen, Q.H. , Peart, J. , Devoto, A. , Casais, C. , Ceron, F. , Schulze, S. , Steinbiß, H.H. , Shirasu, K. and Schulze‐Lefert, P. (2004) RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell, 16, 3480–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Biezen, E.A. , Freddie, C.T. , Kahn, K. , Parker, J.E. and Jones, J.D.G. (2002) Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR‐NB‐LRR genes and confers downy mildew resistance through multiple signaling components. Plant J. 29, 439–451. [DOI] [PubMed] [Google Scholar]
- Bigeard, J. , Colcombet, J. and Hirt, H. (2015) Signaling mechanisms in pattern‐triggered immunity (PTI). Mol. Plant. 8, 521–539. [DOI] [PubMed] [Google Scholar]
- Boccara, M. , Sarazin, A. , Thiebeauld, O. , Jay, F. , Voinnet, O. , Navarro, L. and Colot, V. (2014) The Arabidopsis miR472‐RDR6 silencing pathway modulates PAMP‐ and effector‐triggered immunity through the post‐transcriptional control of disease resistance genes. PLoS Pathog. 10, e1003883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies, K. and Weigel, D. (2007) Hybrid necrosis: autoimmunity as a potential gene‐flow barrier in plant species. Nat. Rev. Genet. 8, 382–393. [DOI] [PubMed] [Google Scholar]
- Bomblies, K. , Lempe, J. , Epple, P. , Warthmann, N. , Lanz, C. , Dangl, J.L. and Weigel, D. (2007) Autoimmune response as a mechanism for a Dobzhansky–Muller‐type incompatibility syndrome in plants. PLoS Biol. 5, e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonardi, V. , Tang, S. , Stallmann, A. , Roberts, M. , Cherkis, K. and Dangl, J.L. (2011) Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc. Natl. Acad. Sci. USA, 108, 16 463–16 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borhan, M.H. , Holub, E.B. , Beynon, J.L. , Rozwadowski, K. and Rimmer, S.R. (2004) The Arabidopsis TIR‐NB‐LRR gene RAC1 confers resistance to Albugo candida (white rust) and is dependent on EDS1 but not PAD4 . Mol. Plant–Microbe Interact. 17, 711–719. [DOI] [PubMed] [Google Scholar]
- Brechenmacher, L. , Nguyen, T.H.N. , Zhang, N. , Jun, T.H. , Xu, D. , Mian, M.R. and Stacey, G. (2015) Identification of soybean proteins and genes differentially regulated in near isogenic lines differing in resistance to aphid infestation. J. Proteome Res. 14, 4137–4146. [DOI] [PubMed] [Google Scholar]
- Bruggeman, Q. , Garmier, M. , de Bont, L. , Soubigou‐Taconnat, L. , Mazubert, C. , Benhamed, M. , Raynaud, C. , Bergounioux, C. and Delarue, M. (2014) The polyadenylation factor subunit CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR30: a key factor of programmed cell death and a regulator of immunity in Arabidopsis. Plant Physiol. 165, 732–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno, I.G. , Karam, R. , Huang, L. , Bhardwaj, A. , Lou, C.H. , Shum, E.Y. , Song, H.W. , Corbett, M.A. , Gifford, W.D. , Gecz, J. and Pfaff, S.L. (2011) Identification of a microRNA that activates gene expression by repressing nonsense‐mediated RNA decay. Mol. Cell, 42, 500–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carra, A. , Mica, E. , Gambino, G. , Pindo, M. , Moser, C. , Pe, M.E. and Schubert, A. (2009) Cloning and characterization of small non‐coding RNAs from grape. Plant J. 59, 750–763. [DOI] [PubMed] [Google Scholar]
- Cesari, S. , Thilliez, G. , Ribot, C. , Chalvon, V. , Michel, C. , Jauneau, A. , Rivas, S. , Alaux, L. , Kanzaki, H. , Okuyama, Y. , Morel, J.B. , Fournier, E. , Tharreau, D. , Terauchi, R. and Kroj, T. (2013) The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR‐Pia and AVR1‐CO39 by direct binding. Plant Cell, 25, 1463–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. , Pang, W. , Chen, B. , Zhang, C. and Piao, Z. (2015) Transcriptome analysis of Brassica rapa near‐isogenic lines carrying clubroot‐resistant and ‐susceptible alleles in response to Plasmodiophora brassicae during early infection. Front Plant Sci. 6, 1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y.T. , Li, Y. , Huang, S. , Huang, Y. , Dong, X. , Zhang, Y. and Li, X. (2011) Stability of plant immune‐receptor resistance proteins is controlled by SKP1‐CULLIN1‐F‐box (SCF)‐mediated protein degradation. Proc. Natl. Acad. Sci. USA, 108, 14 694–14 699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm, S.T. , Coaker, G. , Day, B. and Staskawicz, B.J. (2006) Host–microbe interactions: shaping the evolution of the plant immune response. Cell, 124, 803–814. [DOI] [PubMed] [Google Scholar]
- Clark, R.M. , Schweikert, G. , Toomajian, C. , Ossowski, S. , Zeller, G. , Shinn, P. , Warthmann, N. , Hu, T.T. , Fu, G. , Hinds, D.A. , Chen, H. , Frazer, K.A. , Huson, D.H. , Schölkopf, B. , Nordborg, M. , Rätsch, G. , Ecker, J.R. and Weigel, D. (2007) Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana . Science, 317, 338–342. [DOI] [PubMed] [Google Scholar]
- Coll, N.S. , Vercammen, D. , Smidler, A. , Clover, C. , Van Breusegem, F. , Dangl, J.L. and Epple, P. (2010) Arabidopsis type I metacaspases control cell death. Science, 330, 1393–1397. [DOI] [PubMed] [Google Scholar]
- Collier, S.M. , Hamel, L.P. and Moffett, P. (2011) Cell death mediated by the N‐terminal domains of a unique and highly conserved class of NB‐LRR protein. Mol. Plant–Microbe Interact. 24, 918–931. [DOI] [PubMed] [Google Scholar]
- Costanzo, S.C. and Jia, Y.L. (2009) Alternatively spliced transcripts of Pi‐ta blast resistance gene in Oryza sativa . Plant Sci. 177, 468–478. [Google Scholar]
- Couto, D. and Zipfel, C. (2016) Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16, 537–552. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L. and McDowell, J.M. (2006) Two modes of pathogen recognition by plants. Proc. Natl. Acad. Sci. USA, 103, 8575–8576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleris, A. , Halter, T. and Navarro, L. (2016) DNA methylation and demethylation in plant immunity. Annu. Rev. Phytopathol. 54, 579–603. [DOI] [PubMed] [Google Scholar]
- Dinesh‐Kumar, S.P. and Baker, B.J. (2000) Alternatively spliced N resistance gene transcripts: their possible role in tobacco mosaic virus resistance. Proc. Natl. Acad. Sci. USA, 97, 1908–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N. and Rathjen, J.P. (2010) Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Dowen, R.H. , Pelizzola, M. , Schmitz, R.J. , Lister, R. , Dowen, J.M. , Nery, J.R. , Dixon, J.E. and Ecker, J.R. (2012) Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. USA, 109, 2183–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsel, G. , Kahles, A. , Kesarwani, A.K. , Stauffer, E. , Behr, J. , Drewe, P. , Rätsch, G. and Wachter, A. (2013) Nonsense‐mediated decay of alternative precursor mRNA splicing variants is a major determinant of the Arabidopsis steady state transcriptome. Plant Cell, 25, 3726–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbs, M.L. and Bender, J. (2006) Locus‐specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell, 18, 1166–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem, T. (2005) Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci. 10, 71–78. [DOI] [PubMed] [Google Scholar]
- Eulgem, T. and Somssich, I.E. (2007) Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10, 366–371. [DOI] [PubMed] [Google Scholar]
- Eulgem, T. , Rushton, P.J. , Robatzek, S. and Somssich, I.E. (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. [DOI] [PubMed] [Google Scholar]
- Eulgem, T. , Tsuchiya, T. , Wang, X. , Beasley, B. , Cuzick, A. , Tör, M. , Zhu, T. , McDowell, J.M. , Holub, E. and Dangl, J.L. (2007) EDM2 is required for RPP7‐dependent disease resistance in Arabidopsis and affects RPP7 transcript levels. Plant J. 49, 829–839. [DOI] [PubMed] [Google Scholar]
- Ferrier‐Cana, E. , Macadré, C. , Sévignac, M. , David, P. , Langin, T. and Geffroy, V. (2005) Distinct post‐transcriptional modifications result into seven alternative transcripts of the CC‐NBS‐LRR gene JA1tr of Phaseolus vulgaris . Theor. Appl. Genet. 110, 895–905. [DOI] [PubMed] [Google Scholar]
- Gassmann, W. (2008) Alternative splicing in plant defense. Curr. Top. Microbiol. Immunol. 326, 219–233. [DOI] [PubMed] [Google Scholar]
- Gassmann, W. , Hinsch, M.E. and Staskawicz, B.J. (1999) The Arabidopsis RPS4 bacterial‐resistance gene is a member of the TIR‐NBS‐LRR family of disease‐resistance genes. Plant J. 20, 265–277. [DOI] [PubMed] [Google Scholar]
- Gloggnitzer, J. , Akimcheva, S. , Srinivasan, A. , Kusenda, B. , Riehs, N. , Stampfl, H. , Bautor, J. , Dekrout, B. , Jonak, C. , Jiménez‐Gómez, J.M. , Parker, J.E. and Riha, K. (2014) Nonsense‐mediated mRNA decay modulates immune receptor levels to regulate plant antibacterial defense. Cell Host Microbe, 16, 376–390. [DOI] [PubMed] [Google Scholar]
- Gohring, J. , Jacak, J. and Barta, A. (2014) Imaging of endogenous messenger RNA splice variants in living cells reveals nuclear retention of transcripts inaccessible to nonsense‐mediated decay in Arabidopsis. Plant Cell, 26, 754–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halterman, D.A. and Wise, R.P. (2004) A single‐amino acid substitution in the sixth leucine‐rich repeat of barley MLA6 and MLA13 alleviates dependence on RAR1 for disease resistance signaling. Plant J. 38, 215–226. [DOI] [PubMed] [Google Scholar]
- Halterman, D.A. , Zhou, F. , Wei, F. , Wise, R.P. and Schulze‐Lefert, P. (2001) The MLA6 coiled‐coil, NBS‐LRR protein confers AvrMla6‐dependent resistance specificity to Blumeria graminis f. sp. hordei. in barley and wheat. Plant J. 25, 335–348. [DOI] [PubMed] [Google Scholar]
- Halterman, D.A. , Wei, F. and Wise, R.P. (2003) Powdery mildew‐induced Mla mRNAs are alternatively spliced and contain multiple upstream open reading frames. Plant Physiol. 131, 558–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond‐Kosack, K.E. and Jones, J.D.G. (1996) Plant disease resistance genes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 575–607. [DOI] [PubMed] [Google Scholar]
- Hayashi, K. and Yoshida, H. (2009) Refunctionalization of the ancient rice blast disease resistance gene Pit by the recruitment of a retrotransposon as a promoter. Plant J. 57, 413–425. [DOI] [PubMed] [Google Scholar]
- He, X.F. , Fang, Y.Y. , Feng, L. and Guo, H.S. (2008) Characterization of conserved and novel microRNAs and their targets, including a TuMV‐induced TIR‐NBS‐LRR class R gene‐derived novel miRNA in Brassica. FEBS Lett. 582, 2445–2452. [DOI] [PubMed] [Google Scholar]
- Holt, B.F. 3rd , Belkhadir, Y. and Dangl, J.L. (2005) Antagonistic control of disease resistance protein stability in the plant immune system. Science, 309, 929–932. [DOI] [PubMed] [Google Scholar]
- Howard, B.E. , Hu, Q. , Babaoglu, A.C. , Chandra, M. , Borghi, M. , Tan, X. , He, L. , Winter‐Sederoff, H. , Gassmann, W. , Veronese, P. and Heber, S. (2013) High‐throughput RNA sequencing of Pseudomonas‐infected Arabidopsis reveals hidden transcriptome complexity and novel splice variants. PLoS One, 8, e74183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, M.D. , Fahlgren, N. , Chapman, E.J. , Cumbie, J.S. , Sullivan, C.M. , Givan, S.A. , Kasschau, K.D. and Carrington, J.C. (2007) Genome‐wide analysis of the RNA‐DEPENDENT RNA POLYMERASE6/DICER‐LIKE4 pathway in Arabidopsis reveals dependency on miRNA‐ and tasiRNA‐directed targeting. Plant Cell, 19, 926–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert, D.A. , Tornero, P. , Belkhadir, Y. , Krishna, P. , Takahashi, A. , Shirasu, K. and Dangl, J.L. (2003) Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J. 22, 5679–5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, A.G. , Xu, R.Q. , Addepalli, B. , Rao, S. , Forbes, K.P. , Meeks, L.R. , Xing, D. , Mo, M. , Zhao, H. , Bandyopadhyay, A. , Dampanaboina, L. , Marion, A. , Lanken, C.V. and Li, Q.Q. (2008) Arabidopsis mRNA polyadenylation machinery: comprehensive analysis of protein–protein interactions and gene expression profiling. BMC Genomics, 9, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, C.‐F. , Bhakta, A.V. , Truesdell, G.M. , Pudlo, W.M. and Williamson, V.M. (2000) Evidence for a role of the N terminus and leucine‐rich repeat region of the Mi gene product in regulation of localized cell death. Plant Cell, 12, 1319–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob, F. , Vernaldi, S. and Maekawa, T. (2013) Evolution and conservation of plant NLR functions. Front Immunol. 4, 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskiewicz, M. , Conrath, U. and Peterhänsel, C. (2011) Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep. 12, 50–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, H.J. , Kim, Y.J. , Kim, S.H. , Kim, Y.H. , Lee, I.J. , Kim, Y.K. and Shin, J.S. (2011) Nonsense‐mediated mRNA decay factors, UPF1 and UPF3, contribute to plant defense. Plant Cell Physiol. 52, 2147–2156. [DOI] [PubMed] [Google Scholar]
- Johnson, C. , Kasprzewska, A. , Tennessen, K. , Fernandes, J. , Nan, G.L. , Walbot, V. , Sundaresan, V. , Vance, V. and Bowman, L.H. (2009) Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res. 19, 1429–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D. , Vance, R.E. and Dangl, J.L. (2016) Intracellular innate immune surveillance devices in plants and animals. Science, 354, aaf6395. [DOI] [PubMed] [Google Scholar]
- Jordan, T. , Schornack, S. and Lahaye, T. (2002) Alternative splicing of transcripts encoding Toll‐like plant resistance proteins–what's the functional relevance to innate immunity? Trends Plant Sci. 7, 392–398. [DOI] [PubMed] [Google Scholar]
- Kenny, F.E. and O'Neill, L.A.J. (2008) Signalling adaptors used by Toll‐like receptors: an update. Cytokine, 43, 342–349. [DOI] [PubMed] [Google Scholar]
- Klevebring, D. , Street, N.R. , Fahlgren, N. , Kasschau, K.D. , Carrington, J.C. , Lundeberg, J. and Jansson, S. (2009) Genome‐wide profiling of populus small RNAs. BMC Genomics, 10, 620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides, T. (2007) Chromatin modifications and their function. Cell, 128, 693–705. [DOI] [PubMed] [Google Scholar]
- Kuang, H. , Padmanabhan, C. , Li, F. , Kamei, A. , Bhaskar, P.B. , Ouyang, S. , Jiang, J. , Buell, C.R. and Baker, B. (2009) Identification of miniature inverted‐repeat transposable elements (MITEs) and biogenesis of their siRNAs in the Solanaceae: new functional implications for MITEs. Genome Res. 19, 42–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law, J.A. and Jacobsen, S.E. (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, G.J. , Finnegan, E.J. , Ayliffe, M.A. and Ellis, J.G. (1995) The L6 gene for flax rust resistance is related to the Arabidopsis bacterial resistance gene RPS2 and the tobacco viral resistance gene N . Plant Cell, 7, 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, T.N. , Schumann, U. , Smith, N.A. , Tiwari, S. , Au, P.C.K. , Zhu, Q.H. , Taylor, J.M. , Kazan, K. , Llewellyn, D.J. , Zhang, R. , Dennis, E.S. and Wang, M.B. (2014) DNA demethylases target promoter transposable elements to positively regulate stress responsive genes in Arabidopsis. Genome Biol. 15, 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, M. , La, H. , Lu, K. , Wang, P. , Miki, D. , Ren, Z. , Duan, C.G. , Wang, X. , Tang, K. , Zeng, L. , Yang, L. , Zhang, H. , Nie, W. , Liu, P. , Zhou, J. , Liu, R. , Zhong, Y. , Liu, D. and Zhu, J.K. (2013) Arabidopsis EDM2 promotes IBM1 distal polyadenylation and regulates genome DNA methylation patterns. Proc. Natl. Acad. Sci. USA, 111, 527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F. and Ding, S.W. (2006) Virus counterdefense: diverse strategies for evading the RNA‐silencing immunity. Annu. Rev. Microbiol. 60, 503–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F. , Pignatta, D. , Bendix, C. , Brunkard, J.O. , Cohn, M.M. , Tung, J. , Sun, H. , Kumar, P. and Baker, B. (2012) MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. USA, 109, 1790–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Deng, Y. , Wu, T. , Subramanian, S. and Yu, O. (2010) Misexpression of miR482, miR1512, and miR1515 increases soybean nodulation. Plant Physiol. 153, 1759–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Kapos, P. and Zhang, Y. (2015) NLRs in plants. Curr. Opin. Immunol. 32, 114–121. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Yang, S. , Yang, H. and Hua, J. (2007) The TIR‐NB‐LRR gene SNC1 is regulated at the transcript level by multiple factors. Mol. Plant–Microbe Interact. 20, 1449–1456. [DOI] [PubMed] [Google Scholar]
- Lu, S. , Sun, Y.H. , Amerson, H. and Chiang, V.L. (2007) MicroRNAs in loblolly pine (Pinus taeda L.) and their association with fusiform rust gall development. Plant J. 51, 1077–1098. [DOI] [PubMed] [Google Scholar]
- Lurin, C. , Andrés, C. , Aubourg, S. , Bellaoui, M. , Bitton, F. , Bruyère, C. , Caboche, M. , Debast, C. , Gualberto, J. , Hoffmann, B. , Lecharny, A. , Ret, M.L. , Martin‐Magniette, M.L. , Mireau, H. , Peeters, N. , Renou, J.P. , Szurek, B. , Taconnat, L. and Small, I. (2004) Genome‐wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell, 16, 2089–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz, C.S. (2008) Alternative polyadenylation: a twist on mRNA 3′ end formation. ACS Chem. Biol. 3, 609–617. [DOI] [PubMed] [Google Scholar]
- Lyons, R. , Iwase, A. , Gänsewig, T. , Sherstnev, A. , Duc, C. , Barton, G.J. , Hanada, K. , Higuchi‐Takeuchi, M. , Matsui, M. , Sugimoto, K. , Kazan, K. , Simpson, G.G. and Shirasu, K. (2013) The RNA‐binding protein FPA regulates flg22‐triggered defense responses and transcription factor activity by alternative polyadenylation. Sci. Rep. 3, 2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey, D. , Belkhadir, Y. , Alonso, J.M. , Ecker, J.R. and Dangl, J.L. (2003) Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2‐mediated resistance. Cell, 112, 379–389. [DOI] [PubMed] [Google Scholar]
- Maekawa, T. , Cheng, W. , Spiridon, L.N. , Töller, A. , Lukasik, E. , Saijo, Y. , Liu, P. , Shen, Q.H. , Micluta, M.A. , Somssich, I.E. , Takken, F.L. , Petrescu, A.J. , Chai, J. and Schulze‐Lefert, P. (2011) Coiled‐coil domain‐dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe, 9, 187–199. [DOI] [PubMed] [Google Scholar]
- Mandadi, K.K. and Scholthof, K.B. (2015) Genome‐wide analysis of alternative splicing landscapes modulated during plant–virus interactions in Brachypodium distachyon . Plant Cell, 27, 71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandadi, K.K. , Pyle, J.D. and Scholthof, K.B. (2014) Comparative analysis of antiviral responses in Brachypodium distachyon and Setaria viridis reveals conserved and unique outcomes among C3 and C4 plant defenses. Mol. Plant–Microbe Interact. 27, 1277–1290. [DOI] [PubMed] [Google Scholar]
- Maquat, L.E. (2004) Nonsense‐mediated mRNA decay: splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol. 5, 89–99. [DOI] [PubMed] [Google Scholar]
- Marquez, Y. , Brown, J.W. , Simpson, C. , Barta, A. and Kalyna, M. (2012) Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 22, 1184–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mata, M. , Lafaille, C. and Kornblihtt, A.R. (2010) First come, first served revisited: factors affecting the same alternative splicing event have different effects on the relative rates of intron removal. RNA, 16, 904–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell, J.M. and Simon, S.A. (2006) Recent insights into R gene evolution. Mol. Plant Pathol. 7, 437–448. [DOI] [PubMed] [Google Scholar]
- McDowell, J.M. , Cuzick, A. , Can, C. , Beynon, J. , Dangl, J.L. and Holub, E.B. (2000) Downy mildew (Peronospora parasitica) resistance genes in Arabidopsis vary in functional requirements for NDR1, EDS1, NPR1, and salicylic acid accumulation. Plant J. 22, 523–530. [DOI] [PubMed] [Google Scholar]
- McDowell, J.M. , Williams, S.G. , Funderburg, N.T. , Eulgem, T. and Dangl, J.L. (2005) Genetic analysis of developmentally regulated resistance to downy mildew (Hyaloperonospora parasitica) in Arabidopsis thaliana . Mol. Plant–Microbe Interact. 18, 1226–1234. [DOI] [PubMed] [Google Scholar]
- Meyers, B.C. , Kozik, A. , Griego, A. , Kuang, H. and Michelmore, R.W. (2003) Genome‐wide analysis of NBS‐LRR‐encoding genes in Arabidopsis. Plant Cell, 15, 809–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore, R.W. and Meyers, B.C. (1998) Clusters of resistance genes in plants evolve by divergent selection and a birth‐and‐death process. Genome Res. 8, 1113–1130. [DOI] [PubMed] [Google Scholar]
- Miyao, A. , Tanaka, K. , Murata, K. , Sawaki, H. , Takeda, S. , Abe, K. , Shinozuka, Y. , Onosato, K. and Hirochika, H. (2003) Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon‐rich regions of the genome. Plant Cell, 15, 1771–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett, P. , Farnham, G. , Peart, J. and Baulcombe, D.C. (2002) Interaction between domains of a plant NBS‐LRR protein in disease resistance‐related cell death. EMBO J. 21, 4511–4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr, T.J. , Mammarella, N.D. , Hoff, T. , Woffenden, B.J. , Jelesko, J.G. and McDowell, J.M. (2010) The Arabidopsis downy mildew resistance gene RPP8 is induced by pathogens and salicylic acid and is regulated by W box cis elements. Mol. Plant–Microbe Interact. 23, 1303–1315. [DOI] [PubMed] [Google Scholar]
- Monaghan, J. , Xu, F. , Gao, M. , Zhao, Q. , Palma, K. , Long, C. , Chen, S. , Zhang, Y. and Li, X. (2009) Two Prp19‐like U‐box proteins in the MOS4‐associated complex play redundant roles in plant innate immunity. PLoS Pathog. 5, e1000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan, J. , Xu, F. , Xu, S. , Zhang, Y. and Li, X. (2010) Two putative RNA‐binding proteins function with unequal genetic redundancy in the MOS4‐associated complex. Plant Physiol. 154, 1783–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Much, D. , Gupta, V. , Bachmann, A. , Busch, W. , Kelly, S. , Mun, T. and Andersen, S.U. (2017) Organ‐specific NLR resistance gene expression varies with plant symbiotic status. bioRxiv. doi: 10.1101/135764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandety, R.S. , Caplan, J.L. , Cavanaugh, K. , Perroud, B. , Wroblewski, T. , Michelmore, R.W. and Meyers, B.C. (2013) The role of TIR‐NBS and TIR‐X proteins in plant basal defense responses. Plant Physiol. 162, 1459–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, L. , Jay, F. , Nomura, K. , He, S.Y. and Voinnet, O. (2008) Suppression of the microRNA pathway by bacterial effector proteins. Science, 321, 964–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson, P. and Mühlemann, O. (2010) Cutting the nonsense: the degradation of PTC‐containing mRNAs. Biochem. Soc. Trans. 38, 1615–1620. [DOI] [PubMed] [Google Scholar]
- Noël, L. , Moores, T.L. , van der Biezen, E.A. , Parniske, M. , Daniels, M.J. , Parker, J.E. and Jones, J.D. (1999) Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell, 11, 2099–2111. [PMC free article] [PubMed] [Google Scholar]
- Ossowski, S. , Schneeberger, K. , Clark, R.M. , Lanz, C. , Warthmann, N. and Weigel, D. (2008) Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 18, 2024–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma, K. , Zhao, Q. , Cheng, Y.T. , Bi, D. , Monaghan, J. , Cheng, W. , Zhang, Y. and Li, X. (2007) Regulation of plant innate immunity by three proteins in a complex conserved across the plant and animal kingdoms. Genes Dev. 21, 1484–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma, K. , Thorgrimsen, S. , Malinovsky, F.G. , Fiil, B.K. , Nielsen, H.B. , Brodersen, P. , Hofius, D. , Petersen, M. and Mundy, J. (2010) Autoimmunity in Arabidopsis acd11 is mediated by epigenetic regulation of an immune receptor. PLoS Pathog. 6, e1001137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palusa, S.G. , Ali, G.S. and Reddy, A.S. (2007) Alternative splicing of pre‐mRNAs of Arabidopsis serine/arginine‐rich proteins: regulation by hormones and stresses. Plant J. 49, 1091–1107. [DOI] [PubMed] [Google Scholar]
- Park, J.H. and Shin, C. (2015) The role of plant small RNAs in NB‐LRR regulation. Brief Funct. Genomics, 14, 268–274. [DOI] [PubMed] [Google Scholar]
- Parker, J.E. , Szabò, V. , Staskawicz, B.J. , Lister, C. , Dean, C. , Daniels, M.J. and Jones, J.D.G. (1993) Phenotypic characterization and molecular mapping of the Arabidopsis thaliana locus RPP5, determining resistance to Peronospora parasitica . Plant J. 4, 821–833. [Google Scholar]
- Parker, J.E. , Coleman, M.J. , Szabò, V. , Frost, L.N. , Schmidt, R. , van der Biezen, E.A. , Moores, T. , Dean, C. , Daniels, M.J. and Jones, J.D. (1997) The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the Toll and Interleukin‐1 receptors with N and L6 . Plant Cell, 9, 879–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavet, V. , Quintero, C. , Cecchini, N.M. , Rosa, A.L. and Alvarez, M.E. (2006) Arabidopsis displays centromeric DNA hypomethylation and cytological alterations of heterochromatin upon attack by Pseudomonas syringae . Mol. Plant–Microbe Interact. 19, 577–587. [DOI] [PubMed] [Google Scholar]
- Proudfoot, N.J. (2011) Ending the message: poly(A) signals then and now. Genes Dev. 25, 1770–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumplin, N. and Voinnet, O. (2013) RNA silencing suppression by plant pathogens: defence, counter‐defence and counter‐counter‐defence. Nat. Rev. Microbiol. 11, 745–760. [DOI] [PubMed] [Google Scholar]
- Qiao, Y. , Liu, L. , Xiong, Q. , Flores, C. , Wong, J. , Shi, J. , Wang, X. , Liu, X. , Xiang, Q. , Jiang, S. , Zhang, F. , Wang, Y. , Judelson, H.S. , Chen, X. and Ma, W. (2013) Oomycete pathogens encode RNA silencing suppressors. Nat. Genet. 45, 330–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rairdan, G.J. and Moffett, P. (2006) Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell, 18, 2082–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayson, S. , Arciga‐Reyes, L. , Wootton, L. , De Torres Zabala, M. , Truman, W. , Graham, N. , Grant, M. and Davies, B. (2012) A role for nonsense‐mediated mRNA decay in plants: pathogen responses are induced in Arabidopsis thaliana NMD mutants. PLoS One, 7, e31917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, A.S. , Marquez, Y. , Kalyna, M. and Barta, A. (2013) Complexity of the alternative splicing landscape in plants. Plant Cell, 25, 3657–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribot, C. , Hirsch, J. , Balzergue, S. , Tharreau, D. , Nottéghem, J.L. , Lebrun, M.H. and Morel, J.B. (2008) Susceptibility of rice to the blast fungus, Magnaporthe grisea . J. Plant Physiol. 165, 114–124. [DOI] [PubMed] [Google Scholar]
- Riehs‐Kearnan, N. , Gloggnitzer, J. , Dekrout, B. , Jonak, C. and Riha, K. (2012) Aberrant growth and lethality of Arabidopsis deficient in nonsense‐mediated RNA decay factors is caused by autoimmune‐like response. Nucleic Acids Res. 40, 5615–5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton, P.J. , Somssich, I.E. , Ringler, P. and Shen, Q.J. (2010) WRKY transcription factors. Trends Plant Sci. 15, 247–258. [DOI] [PubMed] [Google Scholar]
- Saha, D. , Prasad, A.M. and Srinivasan, R. (2007) Pentatricopeptide repeat proteins and their emerging roles in plants. Plant Physiol. Biochem. 45, 521–534. [DOI] [PubMed] [Google Scholar]
- Saintenac, C. , Zhang, W. , Salcedo, A. , Rouse, M.N. , Trick, H.N. , Akhunov, E. and Dubcovsky, J. (2013) Identification of wheat gene Sr35 that confers resistance to Ug99 stem rust race group. Science, 341, 783–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze, H. , Kitayama, J. , Takashima, K. , Miura, S. , Harukawa, Y. , Ito, T. and Kakutani, T. (2013) Mechanism for full‐length RNA processing of Arabidopsis genes containing intragenic heterochromatin. Nat. Commun. 4, 2301. [DOI] [PubMed] [Google Scholar]
- Schmidt, S. , Lombardi, M. , Gardiner, D.M. , Ayliffe, M. and Anderson, P.A. (2007) The M flax rust resistance pre‐mRNA is alternatively spliced and contains a complex upstream untranslated region. Theor Appl Genet. 115, 373–382. [DOI] [PubMed] [Google Scholar]
- Schornack, S. , Ballvora, A. , Gürlebeck, D. , Peart, J. , Ganal, M. , Baker, B. , Bonas, U. and Lahaye, T. (2004) The tomato resistance protein Bs4 is a predicted non‐nuclear TIR‐NB‐LRR protein that mediates defense responses to severely truncated derivatives of AvrBs4 and overexpressed AvrBs3. Plant J. 37, 46–60. [DOI] [PubMed] [Google Scholar]
- Schulze‐Lefert, P. (2004) Plant immunity: the origami of receptor activation. Curr. Biol. 14, R22–R24. [PubMed] [Google Scholar]
- Schweingruber, C. , Rufener, S.C. , Zünd, D. , Yamashita, A. and Mühlemann, O. (2013) Nonsense‐mediated mRNA decay – mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim. Biophys. Acta, 1829, 612–623. [DOI] [PubMed] [Google Scholar]
- Sela, H. , Spiridon, L.N. , Petrescu, A.J. , Akerman, M. , Mandel‐Gutfreund, Y. , Nevo, E. , Loutre, C. , Keller, B. , Schulman, A.H. and Fahima, T. (2012) Ancient diversity of splicing motifs and protein surfaces in the wild emmer wheat (Triticum dicoccoides) LR10 coiled coil (CC) and leucine‐rich repeat (LRR) domains. Mol. Plant Pathol. 13, 276–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Q.H. , Zhou, F. , Bieri, S. , Haizel, T. , Shirasu, K. and Schulze‐Lefert, P. (2003) Recognition specificity and RAR1/SGT1 dependence in barley Mla disease resistance genes to the powdery mildew fungus. Plant Cell, 15, 732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Y. , Venu, R.C. , Nobuta, K. , Wu, X. , Notibala, V. , Demirci, C. , Meyers, B.C. , Wang, G.L. , Ji, G. and Li, Q.Q. (2011) Transcriptome dynamics through alternative polyadenylation in developmental and environmental responses in plants revealed by deep sequencing. Genome Res. 21, 1478–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Y. , Zhou, Z. , Wang, Z. , Li, W. , Fang, C. , Wu, M. , Ma, Y. , Liu, T. , Kong, L.A. , Peng, D.L. and Tian, Z. (2014) Global dissection of alternative splicing in paleopolyploid soybean. Plant Cell, 26, 996–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherstnev, A. , Duc, C. , Cole, C. , Zacharaki, V. , Hornyik, C. , Ozsolak, F. , Milos, P.M. , Barton, G.J. and Simpson, G.G. (2012) Direct sequencing of Arabidopsis thaliana RNA reveals patterns of cleavage and polyadenylation. Nat. Struct. Mol. Biol. 19, 845–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirano, Y. , Kachroo, P. , Shah, J. and Klessig, D.F. (2002) A gain‐of‐function mutation in an Arabidopsis Toll Interleukin1 receptor‐nucleotide binding site‐leucine‐rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell, 14, 3149–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu, K. (2009) The HSP90‐SGT1 chaperone complex for NLR immune sensors. Annu. Rev. Plant Biol. 60, 139–164. [DOI] [PubMed] [Google Scholar]
- Shivaprasad, P.V. , Chen, H.M. , Patel, K. , Bond, D.M. , Santos, B.A. and Baulcombe, D.C. (2012) A microRNA superfamily regulates nucleotide binding site‐leucine‐rich repeats and other mRNAs. Plant Cell, 24, 859–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, C. , Wang, C. , Zhang, C. , Korir, N.K. , Yu, H. , Ma, Z. and Fang, J. (2010) Deep sequencing discovery of novel and conserved microRNAs in trifoliate orange (Citrus trifoliata). BMC Genomics, 11, 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes, T.L. , Kunkel, B.N. and Richards, E.J. (2002) Epigenetic variation in Arabidopsis disease resistance. Genes Dev. 16, 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ström, A.C. and Weis, K. (2001) Importin‐beta‐like nuclear transport receptors. Genome Biol. 2, REVIEWS3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiderski, M.R. , Birker, D. and Jones, J.D. (2009) The TIR domain of TIR‐NB‐LRR resistance proteins is a signaling domain involved in cell death induction. Mol. Plant–Microbe Interact. 22, 157–165. [DOI] [PubMed] [Google Scholar]
- Takken, F.L. and Goverse, A. (2012) How to build a pathogen detector: structural basis of NB‐LRR function. Curr. Opin. Plant Biol. 15, 375–384. [DOI] [PubMed] [Google Scholar]
- Tan, X. , Meyers, B.C. , Kozik, A. , West, M.A. , Morgante, M. , St Clair, D.A. , Bent, A.F. and Michelmore, R.W. (2007) Global expression analysis of nucleotide binding site‐leucine rich repeat‐encoding and related genes in Arabidopsis. BMC Plant Biol. 7, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, F. , Yang, S. , Gao, M. and Zhu, H. (2013) Alternative splicing is required for RCT1‐mediated disease resistance in Medicago truncatula . Plant Mol. Biol. 82, 367–374. [DOI] [PubMed] [Google Scholar]
- Tang, F. , Yang, S. and Zhu, H. (2016) Functional analysis of alternative transcripts of the soybean Rj2 gene that restricts nodulation with specific rhizobial strains. Plant Biol. 18, 537–541. [DOI] [PubMed] [Google Scholar]
- Thatcher, S.R. , Zhou, W. , Leonard, A. , Wang, B.B. , Beatty, M. , Zastrow‐Hayes, G. , Zhao, X. , Baumgarten, A. and Li, B. (2014) Genome‐wide analysis of alternative splicing in Zea mays: landscape and genetic regulation. Plant Cell, 26, 3472–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, P.E. , Wu, X. , Liu, M. , Gaffney, B. , Ji, G. , Li, Q.Q. and Hunt, A.G. (2012) Genome‐wide control of polyadenylation site choice by CPSF30 in Arabidopsis. Plant Cell, 24, 4376–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, B. and Graber, J.H. (2012) Signals for pre‐mRNA cleavage and polyadenylation. Wiley Interdiscip. Rev. RNA. 3, 385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, D. , Traw, M.B. , Chen, J.Q. , Kreitman, M. and Bergelson, J. (2003) Fitness costs of R‐gene‐mediated resistance in Arabidopsis thaliana . Nature, 423, 74–77. [DOI] [PubMed] [Google Scholar]
- Tsuchiya, T. and Eulgem, T. (2013) An alternative polyadenylation mechanism coopted to the Arabidopsis RPP7 gene through intronic retrotransposon domestication. Proc. Natl. Acad. Sci. USA, 110, E3535–E3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya, T. and Eulgem, T. (2014) The PHD‐finger module of the Arabidopsis thaliana defense regulator EDM2 can recognize triply modified histone H3 peptides. Plant Signal. Behav. 9, e29202‐1–e29202‐4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda, K. and Katagiri, F. (2010) Comparing signaling mechanisms engaged in triggered and effector‐triggered immunity. Curr. Opin. Plant Biol. 13, 459–465. [DOI] [PubMed] [Google Scholar]
- Vidal, S. , Cabrera, H. , Andersson, R.A. , Fredriksson, A. and Valkonen, J.P. (2002) Potato gene Y‐1 is an N gene homolog that confers cell death upon infection with potato virus Y . Mol. Plant–Microbe Interact. 15, 717–727. [DOI] [PubMed] [Google Scholar]
- Wang, W. , Barnaby, J.Y. , Tada, Y. , Li, H. , Tör, M. , Caldelari, D. , Lee, D.U. , Fu, X.D. and Dong, X. (2011) Timing of plant immune responses by a central circadian regulator. Nature, 470, 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Duan, C.G. , Tang, K. , Wang, B. , Zhang, H. , Lei, M. , Lu, K. , Mangrauthia, S.K. , Wang, P. , Zhu, G. , Zhao, Y. and Zhu, J.K. (2013) RNA‐binding protein regulates plant DNA methylation by controlling mRNA processing at the intronic heterochromatin‐containing gene IBM1 . Proc. Natl. Acad. Sci. USA, 110, 15 467–15 472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, F. , Wing, R.A. and Wise, R.P. (2002) Genome dynamics and evolution of the Mla (powdery mildew) resistance locus in barley. Plant Cell, 14, 1903–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wersch, R. , Li, X. and Zhang, Y. (2016) Mighty dwarfs: Arabidopsis autoimmune mutants and their usages in genetic dissection of plant immunity. Front Plant Sci. 7, 1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham, S. , Dinesh‐Kumar, S.P. , Choi, D. , Hehl, R. , Corr, C. and Baker, B. (1994) The product of the Tobacco Mosaic Virus resistance gene N: similarity to Toll and the Interleukin‐1 receptor. Cell, 78, 1101–1115. [DOI] [PubMed] [Google Scholar]
- Wu, X. , Liu, M. , Downie, B. , Liang, C. , Ji, G. , Li, Q.Q. and Hunt, A.G. (2011) Genome‐wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc. Natl. Acad. Sci. USA, 108, 12 533–12 538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, R. , Xu, J. , Arikit, S. and Meyers, B.C. (2015) Extensive families of miRNAs and PHAS loci in Norway spruce demonstrate the origins of complex phasiRNA networks in seed plants. Mol. Biol. Evol. 32, 2905–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, S. , Cheng, Y.T. , Huang, S. , Win, J. , Soards, A. , Jinn, T.L. , Jones, J.D. , Kamoun, S. , Chen, S. , Zhang, Y. and Li, X. (2013) Regulation of transcription of nucleotide‐binding leucine‐rich repeat‐encoding genes SNC1 and RPP4 via H3K4 trimethylation. Plant Physiol. 162, 1694–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, S. , Brown, S. , Patrick, E. , Brearley, C. and Turner, J.G. (2003) Enhanced transcription of the Arabidopsis disease resistance genes RPW8.1 and RPW8.2 via a salicylic acid‐dependent amplification circuit is required for hypersensitive cell death. Plant Cell, 15, 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, F. , Xu, S. , Wiermer, M. , Zhang, Y. and Li, X. (2012) The cyclin L homolog MOS12 and the MOS4‐associated complex are required for the proper splicing of plant resistance genes. Plant J. 70, 916–928. [DOI] [PubMed] [Google Scholar]
- Yang, S. and Hua, J. (2004) A haplotype‐specific Resistance gene regulated by BONZAI1 mediates temperature‐dependent growth control in Arabidopsis. Plant Cell, 16, 1060–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S. , Tang, F. and Zhu, H. (2014) Alternative splicing in plant immunity. Int. J. Mol. Sci. 15, 10 424–10 445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, H. and Richards, E.J. (2007) A cluster of disease resistance genes in Arabidopsis is coordinately regulated by transcriptional activation and RNA silencing. Plant Cell, 19, 2929–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, A. , Lepere, G. , Jay, F. , Wang, J. , Bapaume, L. , Wang, Y. , Abraham, A.L. , Penterman, J. , Fischer, R.L. , Voinnet, O. and Navarro, L. (2013) Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc. Natl. Acad. Sci. USA, 110, 2389–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, J. , Jeong, D.H. , De Paoli, E. , Park, S. , Rosen, B.D. , Li, Y. , González, A.J. , Yan, Z. , Kitto, S.L. , Grusak, M.A. , Jackson, S.A. , Stacey, G. , Cook, D.R. , Green, P.J. , Sherrier, D.J. and Meyers, B.C. (2011) MicroRNAs as master regulators of the plant NB‐LRR defense gene family via the production of phased, trans‐acting siRNAs. Genes Dev. 25, 2540–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X.C. and Gassmann, W. (2003) RPS4‐mediated disease resistance requires the combined presence of RPS4 transcripts with full‐length and truncated open reading frames. Plant Cell, 15, 2333–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X.C. and Gassmann, W. (2007) Alternative splicing and mRNA levels of the disease resistance gene RPS4 are induced during defense responses. Plant Physiol. 145, 1577–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Dorey, S. , Swiderski, M. and Jones, J.D. (2004) Expression of RPS4 in tobacco induces an AvrRps4‐independent HR that requires EDS1, SGT1 and HSP90. Plant J. 40, 213–224. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Xia, R. , Kuang, H. and Meyers, B.C. (2016) The diversification of plant NBS‐LRR defense genes directs the evolution of microRNAs that target them. Mol. Biol. Evol. 33, 2692–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, B. , Yang, D.L. , Shi, Z. , Dong, H. and Hua, J. (2014) Monoubiquitination of histone 2B at the disease resistance gene locus regulates its expression and impacts immune responses in Arabidopsis. Plant Physiol. 165, 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]


