Summary
Plant viruses in the genus Carlavirus include more than 65 members. Plants infected with carlaviruses exhibit various symptoms, including leaf malformation and plant stunting. Cysteine‐rich protein (CRP) encoded by carlaviruses has been reported to be a pathogenicity determinant. Carlavirus CRPs contain two motifs in their central part: a nuclear localization signal (NLS) and a zinc finger motif (ZF). In addition to these two conserved motifs, carlavirus CRPs possess highly divergent, N‐terminal, 34 amino acid residues with unknown function. In this study, to analyse the role of these distinct domains, we tested six carlavirus CRPs for their RNA silencing suppressor activity, ability to enhance the pathogenicity of a heterologous virus and effects on virus accumulation levels. Although all six tested carlavirus CRPs showed RNA silencing suppressor activity at similar levels, symptoms induced by the Potato virus X (PVX) heterogeneous system exhibited two different patterns: leaf malformation and whole‐plant stunting. The expression of each carlavirus CRP enhanced PVX accumulation levels, which were not correlated with symptom patterns. PVX‐expressing CRP with mutations in either NLS or ZF did not induce symptoms, suggesting that both motifs play critical roles in symptom expression. Further analysis using chimeric CRPs, in which the N‐terminal region was replaced with the corresponding region of another CRP, suggested that the N‐terminal region of carlavirus CRPs determined the exhibited symptom types. The up‐regulation of a plant gene upp‐L, which has been reported in a previous study, was also observed in this study; however, the expression level was not responsible for symptom types.
Keywords: carlavirus, cysteine‐rich protein, pathogenicity determinant, RNA silencing suppressor, symptom
Introduction
Plants infected with viruses exhibit various symptoms, including necrosis, chlorosis and symptoms associated with developmental defects. Various viral proteins have been reported to be involved in these defects. One of these viral pathogenicity determinants is a viral protein called cysteine‐rich protein (CRP), which is encoded by several plant viruses in diverse genera, including hordei‐, tobra‐, furo‐, peclu‐, beny‐, allexi‐, viti‐, mandari‐, tricho and carlaviruses (Andika et al., 2012; Liu et al., 2002; Lukhovitskaya et al., 2005; Senshu et al., 2011; Te et al., 2005; Yelina et al., 2002). Although CRPs of viruses in different genera do not exhibit significant amino acid similarity between them, they are generally characterized by the presence of multiple cysteine residues, which are also found in zinc finger proteins and some transcriptional regulatory proteins (Auld, 2001; Berg and Godwin, 1997; Lukhovitskaya et al., 2013). CRPs encoded by plant viruses can be classified into two groups based on their domain structures. CRPs encoded by hordei‐, tobra‐, furo‐, and pecluviruses contain a conserved CGxxH motif (C, cysteine; G, glycine; H, histidine; x, any amino acid residue) (Sun et al., 2013), whereas CRPs encoded by beny‐, allexi‐, viti‐ and carlaviruses have an arginine‐rich nuclear localization signal (NLS) in the central part of the protein. Carlavirus CRPs have a characteristic zinc finger motif (ZF) located adjacent to the NLS. Both NLS and ZF are highly conserved in carlavirus CRPs, implying a role of carlavirus CRPs as nucleic acid‐binding proteins. Indeed, the CRPs encoded by Chrysanthemum virus B (CVB) and Potato virus M (PVM) have been shown to bind RNA and DNA in vitro (Gramstat et al., 1990; Lukhovitskaya et al., 2009).
The genus Carlavirus, which belongs to the family Betaflexiviridae, includes more than 65 members that have a negative impact on the production of agricultural crops and ornamentals (Adams et al., 2011; Martelli et al., 2007). Carlaviruses have a monopartite, single‐stranded and positive‐sense RNA genome. Their RNA genome contains six open reading frames (ORFs). ORF1 encodes a viral replicase. Three overlapping ORFs, ORF2–ORF4, encode movement proteins (MPs), named triple gene block proteins 1, 2 and 3 (TGBp1, TGBp2 and TGBp3), ORF5 encodes a coat protein (CP) and the product of ORF6 at the 3′ proximal end is CRP. To date, CRPs of four carlaviruses have been analysed for their pathogenicity: PVM (Senshu et al., 2011), CVB (Lukhovitskaya et al., 2005, 2013), Potato virus H (PVH, Li et al., 2013) and Sweet potato chlorotic fleck virus (SPCFV, Deng et al., 2015). All of these four CRPs were demonstrated to be pathogenicity determinants in a separate experiment using similar, heterologous expression analyses with a Potato virus X (PVX) vector in tobacco plants, but showed significantly different effects on the pathogenicity of the vector. Infection of PVX expressing CVB CRP (PVX‐CVB CRP) induced rapid systemic necrosis, whereas infection of PVX‐PVM CRP or PVX‐PVH CRP induced only leaf curling, neither of which was observed in control PVX vector‐infected plants (Li et al., 2013; Lukhovitskaya et al., 2005; Senshu et al., 2011). A recent report has suggested that the CRP encoded by CVB acts as a eukaryotic transcription factor in the host plant nucleus to modulate the morphogenesis of plant cells, which is one of the most persuasive mechanisms of CRP‐mediated symptom expression (Lukhovitskaya et al., 2013). However, the molecular mechanisms underlying the different effects of CRPs of carlaviruses on plants remain elusive.
Together with their function as pathogenicity determinants, CRPs of many plant viruses serve as RNA silencing suppressors (RSSs) (Benyvirus, Chiba et al., 2013; Tobravirus, Martin‐Hernández and Baulcombe, 2008; Furovirus, Sun et al., 2013; Hordeivirus, Te et al., 2005). RSS activity has also been reported in CRPs of carlaviruses, including SPCFV (Deng et al., 2015), CVB (Lukhovitskaya et al., 2014), PVH (Li et al., 2013) and PVM (Senshu et al., 2011). However, the characteristics of these CRPs as RSSs are considerably different. First, they have notably different RSS activities; SPCFV CRP possesses strong RSS activity, similar to that of Cucumber mosaic virus (CMV) 2b, a well‐known efficient RSS, whereas CRPs of PVM and PVH exhibit weaker RSS activity compared with SPCFV CRP (Deng et al., 2015; Li et al., 2013; Senshu et al., 2011). Second, it is dependent on the carlavirus species as to whether or not the NLS of each CRP is essential for RSS activity; NLS in SPCFV CRP plays a critical role in RSS activity, whereas CVB CRP acts as an RSS independent of its nuclear localization (Deng et al., 2015; Lukhovitskaya et al., 2014). In addition, as mentioned above, the symptoms induced by CRPs of different carlavirus species under heterologous expression by a PVX vector are quite variable (Deng et al., 2015; Li et al., 2013; Lukhovitskaya et al., 2005; Senshu et al., 2011). It is unknown whether the different characteristics of CRPs as RSSs contribute to these symptomatic varieties of PVX CRP‐infected plants.
In this study, we compared the functions of CRPs encoded by six different carlaviruses. Our results suggest that all CRPs tested possess similar RSS activity, albeit at low levels, whereas the phenotype in plants induced by each CRP on expression by a heterologous virus varies considerably. The symptom pattern changed according to the N‐terminal region, which was shown by analysis using chimeric CRPs. These results demonstrated that the N‐terminal region of CRP in carlaviruses plays a role in the determination of symptom types.
Results
CRPs of six different carlavirus species possess RNA silencing suppression activity
To determine the characteristic differences in CRPs, we investigated the RSS activity of six carlavirus CRPs: PVM, Daphne virus S (DVS), CVB, Lily symptomless virus (LSV), Helleborus net necrosis virus (HeNNV) and Narcissus common latent virus (NLV). Our preliminary attempts to demonstrate the RSS activity of these carlavirus CRPs with the Agrobacterium‐mediated green fluorescent protein (GFP) transient co‐expression assay (agro‐patch assay; Johansen and Carrington, 2001) did not clearly indicate RSS activity, with the exception of PVM and HeNNV CRPs that infrequently recovered weak GFP fluorescence (data not shown). This is consistent with previous reports showing that CRPs of carlaviruses, as well as those of other viruses, have relatively weak RSS activity (Chiba et al., 2013). Indeed, CVB CRP did not show any RSS activity with the agro‐patch assay (Lukhovitskaya et al., 2009). Thus, we next applied the virus movement complement assay (Bayne et al., 2005; Senshu et al., 2011) to determine whether CRPs of carlaviruses have weak RSS activity undetectable with the agro‐patch assay. p25 encoded by PVX functions as both an MP and RSS (Verchot‐Lubicz, 2005). Using a GFP‐tagged and p25‐deficient PVX (PVXΔp25‐GFP), in combination with two p25 mutants, p25T117A and p25A104V, which have a single amino acid mutation that disrupts the function as an RSS or MP, respectively (Bayne et al., 2005), we tested the PVX movement‐complementing activity of six carlavirus CRPs, with p19, a known RSS encoded by Tomato bushy stunt virus (TBSV), as a positive control. When co‐expressed with p25T117A, PVXΔp25‐GFP requires an additional RSS to move to neighbouring cells because of the induction of antiviral silencing in host plants. Indeed, when co‐expressed with a negative control β‐glucuronidase (GUS), PVXΔp25‐GFP was confined to single cells, whereas fluorescent foci consisting of multiple cells were observed when RSS p19 was co‐expressed. To examine whether carlavirus CRPs possess RSS activity in this system, we co‐infiltrated PVXΔp25‐GFP and p25T117A or p25A104V with each of the carlavirus CRPs, GUS or p19. On co‐infiltration of PVXΔp25‐GFP, p25T117A and the carlavirus CRP, PVX‐GFP fluorescent foci consisted of multiple cells, in contrast with a negative control which consisted of mostly single cells, indicating that the carlavirus CRPs suppress RNA silencing. However, in contrast, the fluorescent foci were confined to single cells on co‐infiltration of PVXΔp25‐GFP, p25A104V and the carlavirus CRPs (Fig. 1A). Hence, these results clearly suggest that the carlavirus CRPs in this study function as RSSs, but not as MPs that can promote PVX movement.
Figure 1.
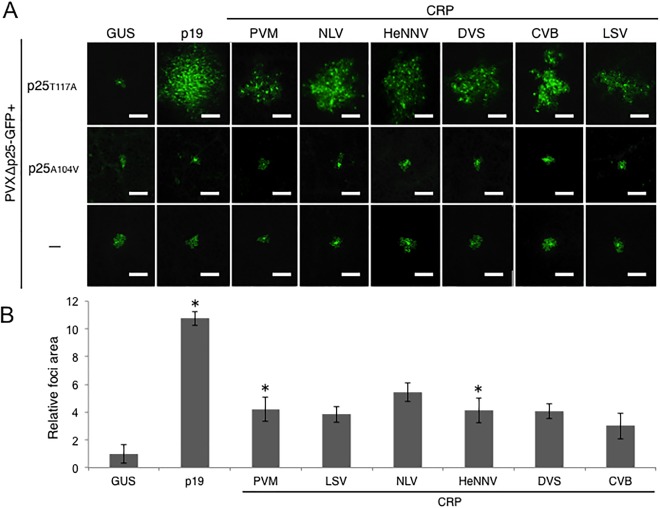
(A) Evaluation of RNA silencing suppressor (RSS) activity by viral movement complementation assay in Nicotiana benthamiana leaves. Carlavirus cysteine‐rich proteins (CRPs), β‐glucuronidase (GUS, negative control) and p19 (RSS but not movement protein) were tested for their ability to complement the movement of PVXΔp25‐GFP only (designed as ‘−’, bottom row), PVXΔp25‐GFP co‐expressed with p25T117A (RSS‐deficient p25, top row) or p25A104V (movement‐deficient p25, middle row). Cells expressing green fluorescent protein (GFP) were visualized using fluorescence microscopy and photographs were taken at 5 days post‐inoculation (dpi). Representative images of 20 fluorescence foci are shown. Scale bars, 50 µm. (B) Quantification of the area of the GFP fluorescence derived from PVXΔp25‐GFP in Fig. 1A. Twenty foci were selected randomly from at least three leaves of two independent replicates for each expression construct (GUS, p19 and carlavirus CRPs), and the fluorescent areas were measured using the BZ‐II Image analysis application (KEYENCE, Osaka, Japan). The relative areas of foci normalized to GUS (n = 20) are shown. Error bars represent the standard deviations. Asterisk (*) indicates a significant (P < 0.001; Student's t‐test) difference between GUS and each tested sample. CVB, Chrysanthemum virus B; DVS, Daphne virus S; HeNNV, Helleborus net necrosis virus; LSV, Lily symptomless virus; NLV, Narcissus common latent virus; PVM, Potato virus M.
Subsequently, the RSS activity of carlavirus CRPs was confirmed by the measurement of the fluorescent area at 5 days post‐inoculation (dpi). The mean area of each complementation assay was normalized with that of GUS. The area measurements were larger in all co‐expressions of carlavirus CRPs relative to that of GUS, although they were relatively smaller than that of p19 (Fig. 1B). All observed fluorescent foci produced by the co‐expression of carlavirus CRPs consisted of multiple cells, resulting in larger area measurements. Significant increases in the florescent area were confirmed in the co‐expression of PVM and HeNNV CRPs (P < 0.001, Student's t‐test), and those of other CRPs repeatedly consisted of multiple cells, indicating that the carlavirus CRPs act as weak RSSs.
Symptoms induced by CRP differ between carlavirus species
To analyse the function of each CRP as a pathogenicity determinant, we expressed carlavirus CRPs from a PVX vector, whose infection was launched by an Agrobacterium tumefaciens infiltration into Nicotiana occidentalis plants. At 5 dpi, necrosis appeared on inoculated leaves of all N. occidentalis plants infected with the PVX vector harbouring each CRP (PVX CRP), whereas only mild chlorosis appeared in plants infected with the PVX empty vector. In contrast, subsequent systemic infection of each CRP‐expressing PVX caused different symptoms at 20 dpi: PVX‐PVM_CRP, PVX‐LSV_CRP and PVX‐NLV_CRP caused severe stunting with rugosity, whereas PVX‐HeNNV_CRP, PVX‐DVS_CRP and PVX‐CVB_CRP caused leaf malformations, and the PVX empty vector induced mosaic and very mild malformation on apical leaves (Fig. 2). Thus, expression of the carlavirus CRPs in N. occidentalis enhances the virulence of PVX.
Figure 2.
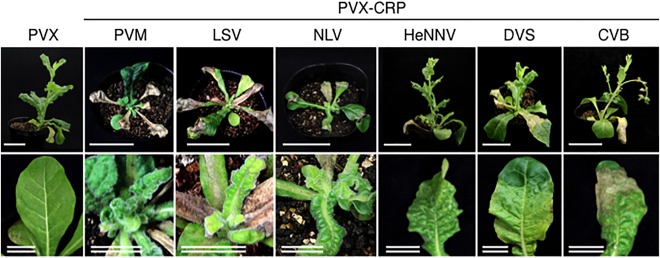
Symptoms on Nicotiana occidentalis plants infected with a Potato virus X (PVX) vector expressing carlavirus cysteine‐rich proteins (CRPs). Representative images of 12 plants for each carlavirus CRP are shown. Photographs were taken at 20 days post‐inoculation (dpi). Scale bars, 5 cm; 1cm (double line). CVB, Chrysanthemum virus B; DVS, Daphne virus S; HeNNV, Helleborus net necrosis virus; LSV, Lily symptomless virus; NLV, Narcissus common latent virus; PVM, Potato virus M.
PVX RNA accumulation was increased significantly in all N. occidentalis apical leaves infected with the six PVX CRPs compared with those infected with a control PVX vector (P < 0.05, F‐test). The smallest increase was approximately two‐fold in PVX‐PVM_CRP‐infected plants and the highest increase was about 12‐fold in PVX‐CVB_CRP‐infected plants (Fig. 3). No significant relationships were found between enhanced viral RNA accumulation and the RSS activities of the six tested carlavirus CRPs shown in Fig. 1, as reported in many other RSSs of plant viruses (Ruiz‐Ruiz et al., 2013; Tsuda et al., 2007). Moreover, these enhanced virus accumulation levels could not explain the different symptoms exhibited by infection with PVX harbouring different carlavirus CRPs. For example, PVX‐NLV_CRP and PVX‐HeNNV_CRP accumulated approximately at the same level (by approximately eight‐fold compared with the control), but infection with PVX‐NLV_CRP and PVX‐HeNNV_CRP resulted in stunting of the plants and leaf malformations, respectively (Figs 2 and 3).
Figure 3.
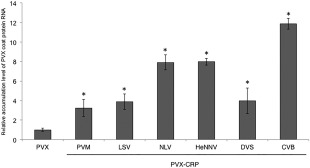
Virus RNA accumulation levels in systemic leaves of Nicotiana occidentalis plants infected with a Potato virus X (PVX) vector expressing carlavirus cysteine‐rich proteins (CRPs). Viral RNAs were detected by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis at 14 days post‐inoculation (dpi). Values presented are relative to those of systemic leaves of plants infected with a control PVX vector. Four biological replicates were used, and two technical replicates were performed for each biological replicate. The error bars represent the standard deviations. Asterisk (*) indicates a significant (P < 0.05, F‐test) difference between PVX and each PVX‐carlavirus CRP. CVB, Chrysanthemum virus B; DVS, Daphne virus S; HeNNV, Helleborus net necrosis virus; LSV, Lily symptomless virus; NLV, Narcissus common latent virus; PVM, Potato virus M.
NLS and ZF are involved in the enhancement of pathogenicity, but NLS is independent of RNA silencing suppression
To analyse the effects of NLS and ZF on CRP‐mediated symptom development, we made use of the previously analysed PVM CRP mutants, mNLS and mZF (Senshu et al., 2011). mNLS and mZF carry alanine mutations in the arginine residues of NLS and the cysteine residues of ZF that are presumed to form the zinc finger structure, respectively. These two CRP mutants were expressed in N. occidentalis leaves using the PVX vector (namely PVX‐PVM_CRPmNLS and PVX‐PVM_CRPmZF). PVX‐PVM_CRP induced necrosis on the infiltrated area of the leaves at 6 dpi (Fig. 4). In contrast, necrosis was not observed on leaves inoculated with either PVX‐PVM_CRPmZF or PVX‐PVM_CRPmNLS (Fig. 4); faint chlorosis was observed in the infiltrated area, similar to that observed with the control PVX vector. Moreover, subsequent systemic infection of PVX‐PVM_CRP caused stunting of the whole plant at 14 dpi, whereas infection of either PVX‐PVM_CRPmNLS or PVX‐PVM_CRPmZF only induced mosaic symptoms on apical leaves, typical of those caused by control PVX infection (Fig. 4). These results prompted us to investigate whether PVX‐PVM_CRPmNLS or PVX‐PVM_CRPmZF retained its mutated CRP sequence on systemic infection. Reverse transcription‐polymerase chain reaction (RT‐PCR) and subsequent direct sequencing detected the CRP sequence from PVX‐PVM_CRP‐infected systemic leaves, but not from PVX‐PVM_CRPmNLS‐ or PVX‐PVM_CRPmZF‐infected leaves. Neither mutated nor reverted CRP was detected in the upper systemic leaves of N. occidentalis plants infected with these mutants (data not shown), indicating that PVX carrying mutated CRP failed to infect systemically N. occidentalis plants.
Figure 4.
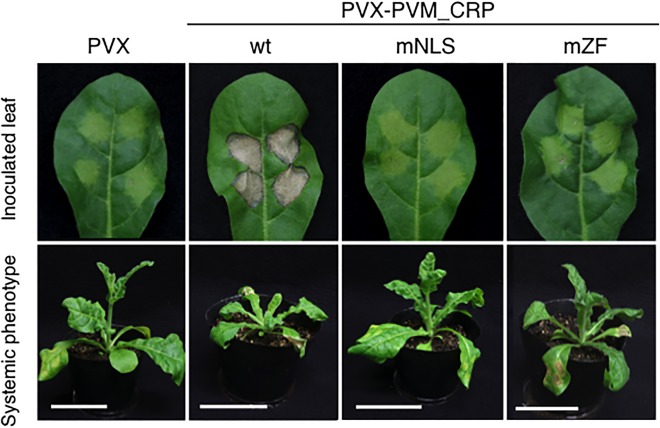
Effects of Potato virus M (PVM) cysteine‐rich protein (CRP) and its mutants on the pathogenicity of a Potato virus X (PVX) vector. Top panels represent Nicotiana occidentalis leaves inoculated with Potato virus X (PVX), PVX‐PVM_CRP (wt), PVX‐PVM_CRPmNLS (mNLS) and PVX‐PVM_CRPmZF (mZF). The systemic phenotypes that arose later are shown in the bottom panels. Photographs of infected leaves (top panels) and whole plants (bottom panels) were taken at 6 and 14 days post‐inoculation (dpi), respectively. Representative images of 12 plants for each mutant test are shown. Scale bars, 5 cm. NLS, nuclear localization signal; ZF, zinc finger motif.
We assumed that the inability of PVX vectors expressing NLS‐ and ZF‐mutated CRPs to infect whole plants could be explained by the lack of RSS activity of these mutants, which significantly reduced the accumulation efficiency of the vectors. To test this idea, we repeated the virus movement complementation assay with PVM CRPmNLS and mZF mutants. As shown in Fig. 1, co‐expression of wild‐type PVM CRP and p25T117A complemented the movement of PVXΔp25‐GFP, which resulted in fluorescent foci consisting of multiple cells. This RSS activity of PVM CRP was not influenced by mutation into NLS, as the fluorescent foci consisted of multiple cells, similar to those in PVM CRP, when PVM CRPmNLS was co‐expressed (Fig. 5A). In contrast, PVM CRPmZF in combination with p25T117A did not complement the movement of PVXΔp25‐GFP, as indicated by the GFP fluorescence confined to single cells. These results were confirmed by the measurement of the area of fluorescent foci, which showed that co‐expression with PVM CRP and PVM CRPmNLS promoted the movement of PVXΔp25‐GFP by about 4.2‐fold compared with that with GUS, whereas PVM CRPmZF did not increase the relative fluorescent focal area of PVXΔp25‐GFP (Fig. 5B). No complementation of the movement of PVXΔp25‐GFP was observed when NLS and ZF mutants were co‐expressed with p25A104V (data not shown). These data indicate that ZF, but not NLS, is required for RSS activity, although both motifs are involved in the increase in pathogenicity of the PVX vector.
Figure 5.
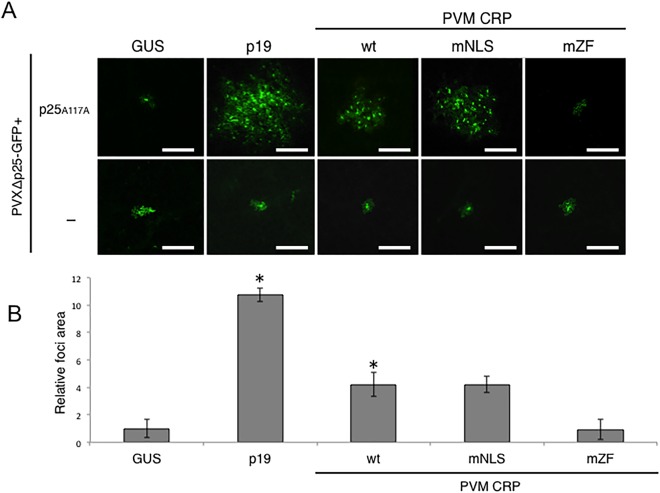
(A) Influence of nuclear localization signal (NLS) and zinc finger motif (ZF) mutations on RNA silencing suppressor (RSS) activity of Potato virus M (PVM) cysteine‐rich protein (CRP). Constructs expressing PVM CRP (wt), PVM CRPmNLS (mNLS) or PVM CRPmZF (mZF), as indicated, were co‐expressed with PVXΔp25‐GFP and p25T117A or negative control (buffer only, −) in Nicotiana benthamiana leaves. Cells expressing green fluorescent protein (GFP) were visualized using fluorescence microscopy and photographs were taken at 5 days post‐inoculation (dpi). Representative images of 20 fluorescence foci are shown. Scale bars, 50 µm. (B) Quantification of the area of the GFP fluorescence foci derived from PVX‐GFP in Fig. 5A. Twenty foci were selected randomly from at least three leaves of two independent replicates for each expression construct: wt, mNLS, mZF, GUS and p19. Images and measurements of PVM shown here are the same as shown in Fig. 1B. The fluorescent areas were measured using the BZ‐II Image analysis application (KEYENCE, Osaka, Japan). The relative sizes normalized to β‐glucuronidase (GUS) (n = 20) are shown. Error bars represent the standard deviations. Asterisk (*) indicates a significant (P < 0.001, Student's t‐test) difference between GUS and each carlavirus CRP. PVX, Potato virus X.
Symptom type is determined by the N‐terminal region of carlavirus CRPs
Although the NLS and ZF motifs of carlavirus CRPs are well conserved, the amino acid sequences of their N‐terminal region are highly divergent (Fig. 6A). To examine the possibility that the N‐terminal region of CRP is responsible for the different symptoms caused by a PVX vector, we engineered chimeric CRPs, designated CVBN(34)PVMC(73)_CRP and PVMN(34)CVBC(73)_CRP, in which the N‐terminal 34 amino acid residues of PVM CRP and CVB CRP were reciprocally exchanged (Fig. 6B). As shown in Fig. 2, on expression from the PVX vector, CVB CRP and PVM CRP exhibited different effects on symptoms of a PVX vector in N. occidentalis plants; CVB CRP induced leaf malformations, whereas PVM CRP induced whole‐plant stunting (Figs 2 and 6C). These symptoms were dramatically changed by the replacement of the N‐terminal regions; infection with PVX‐PVMN(34)CVBC(73)_CRP induced a phenotype similar to that exhibited by PVX‐PVM_CRP. To the contrary, infection by PVX‐CVBN(34)PVMC(73)CRP induced symptoms typical of PVX‐CVB_CRP (Fig. 6C). Collectively, these data suggest that the N‐terminal region of carlavirus CRP is involved in symptom type determination.
Figure 6.
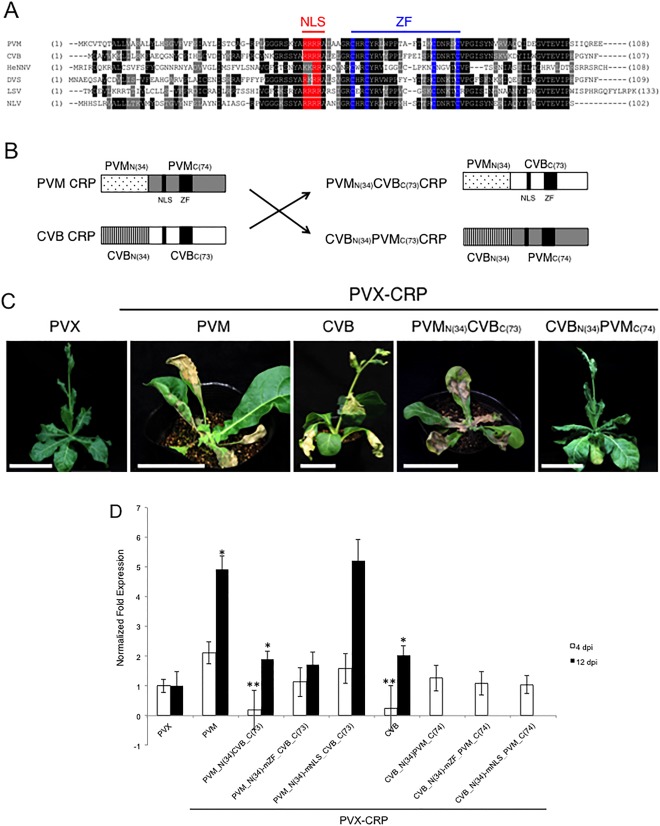
(A) Sequence alignment of cysteine‐rich proteins (CRPs) of six carlaviruses. The nuclear localization signal (NLS) (shown in red) and zinc finger motif (ZF) (blue) are conserved in the central region of each protein. Identical and similar amino acid residues are boxed and shaded in grey, respectively. The numbers of amino acid residues upstream and downstream of the aligned regions are shown in parentheses. Note that the sequence similarity of the N‐terminal region is relatively low compared with the central region containing NLS and ZF motifs. (B) Schematic representation of chimeric CRPs. The N‐terminal 34 amino acid residues of Potato virus M (PVM) CRP or Chrysanthemum virus B (CVB) CRP were exchanged, resulting in the chimeric CRP constructs PVMN(34)CVBC(73)CRP and CVBN(34)PVMC(74)CRP, respectively. (C) Systemic symptoms of Nicotiana occidentalis plants infected with Potato virus X (PVX), PVX‐CVB_CRP, PVX‐PVM_CRP, PVX‐PVMN(34)CVBC(73)CRP and PVX‐CVBN(34)PVMC(74)CRP. Photographs were taken at 13 days post‐inoculation (dpi). Representative images of 12 plants for each PVX CRP are shown. Scale bars, 5 cm. (D) Effect of the chimeric CRPs and motif mutations on the expression of the upp‐L gene responsible for leaf malformation in N. occidentalis. The expression levels of upp‐L were analysed by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) using the specific primer set. Total RNA was extracted from inoculated leaves at 4 dpi and upper leaves at 12 dpi. The values presented are relative to those of leaves of plants infected with a control PVX vector. Three biological replicates were used, and two technical replicates were performed for each biological replicate. The error bars represent the standard deviations. Asterisk (*) indicates significant (*P < 0.05 and **P < 0.001, Student's t‐test) differences between PVX and PVX vectors expressing CRPs.
A recent study has shown that CVB CRP activates the expression of a plant gene upp‐L which regulates cell size and proliferation, resulting in the induction of leaf malformation in plants (Lukhovitskaya et al., 2013). This activation occurs through the specific interaction between the regulatory region in the upp‐L promoter and CVB CRP acting as a plant transcription factor (Lukhovitskaya et al., 2013). A question that arises is whether the ability to up‐regulate upp‐L gene expression is shared among carlavirus CRPs. We thus analysed the expression level of upp‐L by quantitative RT‐PCR (qRT‐PCR). Eight constructs were analysed: wild‐type PVX‐PVM_CRP and PVX‐CVB_CRP, PVX vectors expressing two chimeric CRPs between PVM and CVB, which were used in Fig. 6C, and four PVX vectors expressing chimeric CRPs with mZF or mNLS mutation introduced to examine the role of motifs conserved in carlavirus CRPs. To monitor detailed expression changes of upp‐L, we extracted total RNA from inoculated leaves at 4 dpi and from upper leaves at 12 dpi. Our qRT‐PCR results demonstrated no up‐regulation of upp‐L at 4 dpi in N. occidentalis plants infected with all of the above viruses, but upp‐L expression was significantly increased at 12 dpi in those infected with PVX‐PVM_CRP, PVX‐ PVMN(34)CVBC(73)_CRP and PVX‐CVB_CRP (Fig. 6D), suggesting that both PVM CRP and CVB CRP have the ability to up‐regulate upp‐L gene expression, as shown with CVB_CRP of another strain in a previous report. In contrast, at 4 dpi, there was a significant decrease in upp‐L expression in plants infected with PVX‐CVB_CRP and PVX‐PVMN(34)CVBC(73)_CRP, which implies that the C‐terminal region of CVB CRP contributes to the down‐regulation of upp‐L. Moreover, considering that there were no significant changes in upp‐L expression in plants infected with PVX‐PVMN(34)‐mZF_CVBC(73)_CRP and PVX‐PVMN(34)‐mNLS_CVBC(73)_CRP, it is suggested that ZF and NLS motifs of CVB CRP participate in the down‐regulation of upp‐L at 4 dpi. The data from plants infected with PVX vectors expressing chimeric CRPs whose C‐terminal region was derived from PVM CRP (CVBN(34)PVMC(73)CRP, CVBN(34)‐mZF_PVMC(73)CRP and CVBN(34)‐mNLS_PVMC(73)CRP) at 12 dpi were excluded, because our RT‐PCR analyses revealed that they lost the inserted chimeric CRP sequences in the upper leaves, suggesting that these viruses failed to infect N. occidentalis plants systemically. In contrast, other viruses were confirmed to retain the inserted CRP sequence at 12 dpi. Collectively, our results demonstrate that CRPs of at least two different carlaviruses (CVB and PVM) have the potential to up‐regulate upp‐L expression. However, the expression level is unlikely to contribute to the symptom pattern, as seen in N. occidentalis plants infected with PVX‐PVMN(34)CVBC(73)_CRP and PVX‐CVB_CRP, which yield similar upp‐L expression levels at both 4 and 12 dpi, but exhibit different symptoms (Fig. 6C). Taken together, our results demonstrate that carlavirus CRPs share the potential to up‐regulate upp‐L expression, although some may decrease its expression at early time points, suggesting that the different symptom types are not determined by the upp‐L expression level alone.
Discussion
All carlavirus CRPs used in this study were shown to act as RSSs, suggesting that CRPs encoded by carlaviruses are RSSs. The RSS activities of different CRPs were weak, and were detected by the virus movement complement assay, but not by the Agrobacterium patch assay. This weak RSS activity has led to speculation that CRPs are not necessarily required for successful infections, at least in some plant–carlavirus interactions. Indeed, sweet potato C6 virus, a virus belonging to the genus Carlavirus, lacks CRP in its genome (De Souza et al., 2013). However, another carlavirus CRP encoded by SPCFV has been shown to act as a strong RSS, whose activity is detectable by the Agrobacterium patch assay, which suggests that it plays an important role in virus infection (Deng et al., 2015). Notable differences between SPCFV CRP and the six tested CRPs in this study not only include the RSS activity level, but also the need for NLS motifs for RSS activity, in which the NLS motif is essential for the RSS activity of SPCFV CRP (Deng et al., 2015), but not for that of PVM and CVB CRP (Fig. 5; Lukhovitskaya et al., 2014), although the biological significance of these differences remains elusive. Given the high conservation of NLSs in carlavirus CRPs, it is possible that the six carlaviruses tested in this study require NLSs for other functions of CRPs in their life cycle, such as vector transmission, as well as the enhancement of pathogenicity independent of RSS activity.
In addition to carlavirus CRPs, there are several viral proteins that contain NLSs consisting of basic residues, such as Rice stripe virus (RSV) p3 (Xiong et al., 2009), Potato virus A (PVA) Nla (VPg) (Rajamäki and Valkonen, 2009), Citrus tristeza virus (CTV) p23 (Ruiz‐Ruiz et al., 2013), Beet necrotic yellow vein virus (BNYVV) and Beet soil‐borne mosaic virus (BSBMV) cysteine‐rich p14 (Chiba et al., 2013), and CMV 2b protein (González et al., 2012). RSV p3 and CTV p23 are RSSs whose activity requires NLS (Ruiz‐Ruiz et al., 2013; Xiong et al., 2009). Carlavirus CRPs tested in this study have parallels with BNYVV and BSBMV CRP and CMV 2b protein, in that NLS plays a role in the enhancement of virus pathogenicity, but not in RSS activity (Chiba et al., 2013; Du et al., 2014; González et al., 2012). As carlaviruses are positive‐stranded RNA viruses that replicate in the cytoplasm, most of their encoded proteins, including CRP, do not function in the nucleus in principle. However, it has been reported that the accumulation of 2b of CMV, which is also a positive‐stranded RNA virus, in the nucleus enhances virus virulence (Du et al., 2014). In addition, CMV 2b has been shown to be a transcriptional activator in yeast (Ham et al., 1999). Based on these findings, we could argue that the function of CRP in the host cell nucleus partially contributes to the virulence of carlaviruses.
One of the most convincing functions of nuclear‐localized CRPs is a transcriptional activator. A study of CVB CRP demonstrated that it acts as a plant transcription factor in the nucleus and modulates host cell morphogenesis, which consequently induces leaf malformation symptoms (Lukhovitskaya et al., 2013). If we assume that carlavirus CRPs share transcription factor‐like activity, we can explain the fact that both NLS and ZF are highly conserved and crucial for the enhancement of viral pathogenicity. In addition, it is tempting to suggest that the transcription factor‐like activity contributes to the efficient virus propagation in infected plants, because we have shown that PVX‐PVM_CRPmNLS and PVX‐PVM_CRPmZF probably have low fitness attributed to low virus accumulation levels, hence resulting in unsuccessful infections. Future experiments will investigate how CRP‐mediated transcriptional activation provides benefits to carlavirus infections.
In accordance with the idea that CRP's function as a transcription factor contributes to symptoms, increased virus RNA accumulation levels, which are caused by CRP, are not correlated with the development of symptoms. A similar observation has also been reported in previous studies on CTV p23, a pathogenicity determinant containing both NLS and ZF motifs, which induces symptoms without an increase in viral accumulation (Ruiz‐Ruiz et al., 2013). Similarly, symptoms induced by Pepper mild mottle virus are controlled by RSS activity, but not by viral accumulation (Tsuda et al., 2007). Taken together, our results suggest that the development of symptoms is associated with the transcription factor‐like activity that requires NLS and ZF, but the symptom patterns are not correlated with the virus RNA accumulation levels.
Therefore, a key remaining question is how different symptoms are determined and exhibited by carlavirus CRPs. Our results using PVX expressing chimeric CRPs between PVM and CVB clearly demonstrated that the N‐terminal region of CRP determines the symptom patterns (Fig. 6C). A previous study has suggested that a plant gene upp‐L, whose expression is increased in plants infected with PVX‐CVB_CRP, underlies the mechanism of symptom expression by CRPs (Lukhovitskaya et al., 2013). In agreement with this previous study, upp‐L was up‐regulated in plants infected with both PVX‐PVM_CRP and PVX‐CVB_CRP at 12 dpi, suggesting that the transcription factor‐like activity found in CVB CRP is shared amongst at least two carlavirus CRPs. However, the symptom patterns were not correlated with the upp‐L expression level. Although plants infected with PVX‐PVMN(34)CVBC(73)_CRP and PVX‐PVM_CRP, which both expressed CRP containing the N‐terminal region from PVM CRP, induced similar symptoms, upp‐L expression was decreased in the plant infected with PVX‐PVMN(34)CVBC(73)_CRP, but not with PVX‐PVM_CRP, at 4 dpi, which was also observed in plants infected with PVX‐CVB_CRP at 4 dpi. We suspect that the decrease in upp‐L expression at 4 dpi can be attributed to a resistance response in inoculated leaves by PVX expressing CRP containing the C‐terminal region from CVB CRP, which is manifested as necrosis in the leaf vein (data not shown). Supporting this assumption, the expression of plant resistance genes was strongly increased in plants infected with PVX‐CVB_CRP compared with PVX‐PVM_CRP (data not shown). Therefore, we assumed that PVX‐CVB_CRP could similarly induce the up‐regulation of upp‐L as PVX‐PVM_CRP; however, it induced strong plant resistance responses in the early phases of infection, which resulted in decreased upp‐L expression through an unknown mechanism. Collectively, our data suggest that different symptoms exhibited by carlavirus CRPs are determined by the N‐terminal region, but not by upp‐L expression, although its role in plants remains unclear.
The N‐terminus of CRPs may determine their subcellular and tissue localization, which leads to different symptoms. In our preliminary observations, PVM CRP localized not only to the nucleus but also to the cytoplasm in leaf epidermal cells of N. benthamiana plants (data not shown). As reported in the CMV 2b protein, which partitions between the nucleus and cytoplasm (Du et al., 2014), PVM CRP is probably a nucleus‐ and cytoplasm‐localized protein. CVB CRP was observed mostly in the nucleus, but also in the cytoplasm (Lukhovitskaya et al., 2009). It is possible that the N‐terminal region of CRP determines the balance of localization between the nucleus and cytoplasm, as well as detailed localization patterns. Indeed, a previous study has demonstrated that the subcellular localization of CRP encoded by Chinese wheat mosaic virus (CWMV, genus Furovirus) was determined by its C‐terminal region containing variable amino acids (Sun et al., 2013). It is interesting to note that, using the SOSUI program (Mitaku et al., 2002), a transmembrane region is predicted in the N‐terminal 10–32 amino acids of PVM CRP, but not predicted in CVB CRP. This could explain the difference between CRP of PVM and CVB, although a transmembrane region is not predicted in the other four CRPs. Several previous reports have suggested that interactions between viruses and cellular compartments are crucial for virus long‐distance movements and systemic infection (Fagoaga et al., 2011; Kim et al., 2007). In our preliminary attempts, in N. occidentalis plants infected with PVX‐PVM CRP, expression of PVM CRP was not detected in the shoot apical meristem by in situ hybridization using PVM CRP as a probe, where the upp‐L gene is expressed and PVX‐CVB_CRP is predicted to be localized (Lukhovitskaya et al., 2013) (data not shown). The data imply that tissue tropism of the PVX vector harbouring CRPs of CVB and PVM is different. Further studies are needed to confirm this preliminary observation and to further understand the mechanisms of the subcellular localization and tissue tropism of carlavirus CRPs.
This article has demonstrated the conservation and divergence of functions of carlavirus CRPs. Carlavirus CRPs share functions as RSSs, and commonly enhance viral RNA accumulation. Moreover, PVX expressing carlavirus CRP induces enhanced symptoms in plants, suggesting that carlavirus CRPs are pathogenicity determinants. However, the symptoms induced by carlavirus CRPs are notably different, indicating that there are fundamental differences among homologous proteins encoded by carlaviruses. For future elucidation of the underlying mechanisms of these differences among carlavirus CRPs, the N‐terminal region might shed light on the question.
Experimental Procedures
Construction of binary plasmids and PVX recombinants
The binary plasmids for the expression of GUS and p19 have been described previously (Senshu et al., 2011). Six CRPs used in this study were obtained from the following templates: the plasmid pBI121‐pvmCRP (Senshu et al., 2011) for PVM; the synthetic gene (Eurofins Genomics, Tokyo, Japan) based on the nucleotide sequence accession number NC008266 for NLV; the cDNA clone coding for the CRP region of HeNNV (accession number AB623047; Shiraishi et al., 2011); the cDNA clone coding for the CRP region of DVS (AB889483; Fujita et al., 2015); extracted total RNA from infected leaves of CVB (ATCC PV‐349); and extracted RNA from infected leaves of LSV in Japan. They were PCR amplified and cloned into the binary PVX vector pCAMV using SmaI and SalI sites (Hoshi et al., 2009). The pvmCRP mutants have been described previously (Senshu et al., 2011). The chimeric CRPs between PVM and CVB were generated by recombinant PCR with the specific primers listed in Table S1 (see Supporting Information). The chimeric CRPs with mutated ZF or NLS were generated by recombinant PCR with primers containing corresponding mutations in either of the motifs. The primers used for the construction of these chimeric CRPs with mutated ZF/NLS are also listed in Table S1. The plasmids for the movement complementation assay have been described previously (Senshu et al., 2011).
Plant materials and agroinfiltration
Plants were maintained in growth chambers at 20–25°C throughout the assays. The methods used for A. tumefaciens infiltration have been described previously (Senshu et al., 2011). Agrobacterium cultures carrying appropriate plasmids were resuspended in infiltration buffer to a final optical density at 600 nm of 1.0. For the movement complementation assay, an Agrobacterium culture carrying the PVXΔp25‐GFP binary plasmid was diluted 10 000‐fold and mixed 1 : 1 : 1 with two Agrobacterium cultures carrying the binary plasmid of the p25 mutant and of each CRP. For the upp‐L expression analysis, suspensions were diluted to a final optical density at 600 nm of 0.1.
RNA isolation and analysis
Total RNA isolation and qRT‐PCR were carried out as described previously (Senshu et al., 2011). For the analysis of viral RNA accumulation (Fig. 3), total RNA was extracted from apical leaves at 20 dpi and treated with RNase‐free recombinant DNase I (TaKaRa, Shiga, Japan). cDNA was obtained with the high‐capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions. Primers amplifying the CP gene of PVX (PVXCP159F and PVXCP239R; Senshu et al., 2011) were used to quantify PVX RNA accumulation. To quantify the Nh upp‐L gene, specific primers (Table S1) were used. Primers amplifying the N. benthamiana elongation factor 1α gene, which was used as a reference gene for both PVX RNA and Nh upp‐L, were EF1U and EF1D (Komatsu et al., 2010). Nh upp‐L expression was measured by qRT‐PCR only for total RNA samples from plants infected with a PVX vector retaining a CRP insert, which was checked by conventional one‐step RT‐PCR using the SuperScript III One‐Step RT‐PCR System with Platinum Taq (Life Technologies, Carlsbad, CA, USA) with PVC_F and PVC_R primers in Table S1, which flank the multiple cloning site of a PVX vector. Three biological replicates were used for the analysis of both genes, and two technical replicates were performed within each biological replicate.
GFP florescence analysis
GFP fluorescence of infiltrated leaves was observed using a fluorescence microscope (BZ9000; KEYENCE, Osaka, Japan). The fluorescent areas were measured using the BZ‐II Image analysis application (KEYENCE). All images were edited using Adobe Photoshop CS6 software (Adobe Systems, San Jose, CA, USA).
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Primers used in this study.
Acknowledgements
We thank D. C. Baulcombe (University of Cambridge, Cambridge, UK) for providing the cDNA clone. We also thank K. Nohara (Graduate School of Agricultural and Life Sciences, University of Tokyo, Tokyo, Japan) for technical assistance. This study was supported by a grant‐in‐aid for JSPS Research Fellow Grant Number JP256903.
References
- Adams, M.J. , Candresse, T. , Hammond, J. , Kreuze, J.F. , Martelli, G.P. , Namba, S. , Pearson, M.N. , Ryu, K.H. , Saldarelli, P. and Yoshikawa, N. (2011) Family betaflexiviridae In: Virus Taxonomy. Ninth Report of the International Committee on Taxonomy of Viruses, (King A.M.Q., Adams M.J., Carstens E.B. and Lefkowitz E.J., eds), pp. 920–941. London: Elsevier Academic Press. [Google Scholar]
- Andika, I.B. , Kondo, H. , Nishiguchi, M. and Tamada, T. (2012) The cysteine‐rich proteins of beet necrotic yellow vein virus and tobacco rattle virus contribute to efficient suppression of silencing in roots. J. Gen. Virol. 93, 1841–1850. [DOI] [PubMed] [Google Scholar]
- Auld, D.S. (2001) Zinc coordination sphere in biochemical zinc sites. Biometals, 14, 271–313. [DOI] [PubMed] [Google Scholar]
- Bayne, E.H. , Rakitina, D.V. , Morozov, S.Y. and Baulcombe, D.C. (2005) Cell‐to‐cell movement of potato potexvirus X is dependent on suppression of RNA silencing. Plant J. 44, 471–482. [DOI] [PubMed] [Google Scholar]
- Berg, J.M. and Godwin, H.A. (1997) Lessons from zinc‐binding peptides. Annu. Rev. Biophys. Biomol. Struct. 26, 357–371. [DOI] [PubMed] [Google Scholar]
- Chiba, S. , Hleibieh, K. , Delbianco, A. , Klein, E. , Ratti, C. , Ziegler‐Graff, V. , Bouzoubaa, S. and Gilmer, D. (2013) The Benyvirus RNA silencing suppressor is essential for long‐distance movement, requires both zinc‐finger and NoLS basic residues but not a nucleolar localization for its silencing‐suppression activity. Mol. Plant–Microbe Interact. 26, 168–181. [DOI] [PubMed] [Google Scholar]
- Deng, X.G. , Peng, X.J. , Zhu, F. , Chen, Y.J. , Zhu, T. , Qin, S.B. , Xi, D.H. and Lin, H.H. (2015) A critical domain of Sweet potato chlorotic fleck virus nucleotide‐binding protein (NaBp) for RNA silencing suppression, nuclear localization and viral pathogenesis. Mol. Plant Pathol. 16, 365–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza, J. , Fuentes, S. , Savenkov, E.I. , Cuellar, W. and Kreuze, J.F. (2013) The complete nucleotide sequence of sweet potato C6 virus: a carlavirus lacking a cysteine‐rich protein. Arch. Virol. 158, 1393–1396. [DOI] [PubMed] [Google Scholar]
- Du, Z. , Chen, A. , Chen, W. , Liao, O. , Zhang, H. , Bao, Y. , Roossinck, M.J. and Carr, J.P. (2014) Nuclear‐cytoplasmic partitioning of cucumber mosaic virus protein 2b determines the balance between its roles as a virulence determinant and an RNA‐silencing suppressor. J. Virol. 88, 5228–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagoaga, C. , Pensabene, G. , Moreno, P. , Navarro, L. , Flores, R. and Peña, L. (2011) Ectopic expression of the p23 silencing suppressor of Citrus tristeza virus differentially modifies viral accumulation and tropism in two transgenic woody hosts. Mol. Plant Pathol. 12, 898–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, N. , Komatsu, K. , Neriya, Y. , Kagiwada, S. , Hara, S. , Miyazaki, A. , Netsu, O. , Hashimoto, M. , Yamaji, Y. and Namba, S. (2015) Complete genome sequence of a Japanese isolate of Daphne virus S. Cytologia, 80, 327–330. [Google Scholar]
- González, I. , Rakitina, D. , Semashko, M. , Taliansky, M. , Praveen, S. , Palukaitis, P. , Carr, J.P. , Kalinina, N. and Canto, T. (2012) RNA binding is more critical to the suppression of silencing function of Cucumber mosaic virus 2b protein than nuclear localization. RNA, 18, 771–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramstat, A. , Courtpozanis, A. and Rohde, W. (1990) The 12 kDa protein of potato virus M displays properties of a nucleic acid‐binding regulatory protein. FEBS Lett. 276, 34–38. [DOI] [PubMed] [Google Scholar]
- Ham, B.K. , Lee, T.H. , You, J.S. , Nam, Y.W. , Kim, J.K. and Paek, K.H. (1999) Isolation of a putative tobacco host factor interacting with cucumber mosaic virus‐encoded 2b protein by yeast two‐hybrid screening. Mol. Cells, 9, 548–555. [PubMed] [Google Scholar]
- Hoshi, A. , Oshima, K. , Kakizawa, S. , Ishii, Y. , Ozeki, J. , Hashimoto, M. , Komatsu, K. , Kagiwada, S. , Yamaji, Y. and Namba, S. (2009) A unique virulence factor for proliferation and dwarfism in plants identified from a phytopathogenic bacterium. Proc. Natl. Acad. Sci. USA, 106, 6416–6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen, L.K. and Carrington, J.C. (2001) Silencing on the spot. Induction and suppression of RNA silencing in the Agrobacterium‐mediated transient expression system. Plant Physiol. 126, 930–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.H. , Ryabov, E.V. , Kalinina, N.O. , Rakitina, D.V. , Gillespie, T. , MacFarlane, S. , Haupt, S. , Brown, J.W. and Taliansky, M. (2007) Cajal bodies and the nucleolus are required for a plant virus systemic infection. EMBO J. 26, 2169–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu, K. , Hashimoto, M. , Ozeki, J. , Yamaji, Y. , Maejima, K. , Senshu, H. , Himeno, M. , Okano, Y. , Kagiwada, S. and Namba, S. (2010) Viral‐induced systemic necrosis in plants involves both programmed cell death and the inhibition of viral multiplication, which are regulated by independent pathways. Mol. Plant–Microbe Interact. 23, 283–293. [DOI] [PubMed] [Google Scholar]
- Li, Y.Y. , Zhang, R.N. , Ziang, H.Y. , Abouelnasr, H. , Li, D.W. , Yu, J.L. , McBeath, G.H. and Han, C.G. (2013) Discovery and characterization of a novel carlavirus infecting potatoes in China. PLoS One, 8, e69255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , Reavy, B. , Swanson, M. and MacFarlane, S. (2002) Functional replacement of the tobacco rattle virus cysteine‐rich protein by pathogenicity proteins from unrelated plant viruses. Virology, 298, 232–239. [DOI] [PubMed] [Google Scholar]
- Lukhovitskaya, N. , Yelina, N. , Zamyatnin, A.J. , Schepetilnikov, M. , Solovyev, A. , Sandgren, M. , Morozov, S.Y. , Valkonen, J.P. and Savenkov, E.L. (2005) Expression, localization and effects on virulence of the cysteine‐rich 8 kDa protein of Potato mop‐top virus. J. Gen. Virol. 86, 2879–2889. [DOI] [PubMed] [Google Scholar]
- Lukhovitskaya, N.I. , Ignatovich, I.V. , Savenkov, E.I. , Schiemann, J. , Morozov, S.Y. and Solovyev, A.G. (2009) Role of the zinc‐finger and basic motifs of chrysanthemum virus B p12 protein in nucleic acid binding, protein localization and induction of a hypersensitive response upon expression from a viral vector. J. Gen. Virol. 90, 723–733. [DOI] [PubMed] [Google Scholar]
- Lukhovitskaya, N.I. , Solovieva, A.D. , Boddeti, S.K. , Thaduri, S. , Solovyev, A.G. and Savenkov, E.I. (2013) An RNA virus‐encoded zinc‐finger protein acts as a plant transcription factor and induces a regulator of cell size and proliferation in two tobacco species. Plant Cell, 25, 960–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukhovitskaya, N.I. , Vetukuri, R.R. , Sama, I. , Thaduri, S. , Solovyev, A.G. and Savenkov, E.I. (2014) A viral transcription factor exhibits antiviral RNA silencing suppression activity independent of its nuclear localization. J. Gen. Virol. 95, 2831–2837. [DOI] [PubMed] [Google Scholar]
- Martelli, G.P. , Adams, M.J. , Kreuze, J.F. and Dolja, V.V. (2007) Family Flexiviridae: a case study in virion and genome plasticity. Annu. Rev. Phytopathol. 45, 73–100. [DOI] [PubMed] [Google Scholar]
- Martin‐Hernández, A.M. and Baulcombe, D.C. (2008) Tobacco rattle virus 16‐kilodalton protein encodes a suppressor of RNA silencing that allows transient viral entry in meristems. J. Virol. 82, 4064–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitaku, S. , Hirokawa, T. and Tsuji, T. (2002) Amphiphilicity index of polar amino acids as an aid in the characterization of amino acid preference at membrane–water interfaces. Bioinformatics, 18, 608–616. [DOI] [PubMed] [Google Scholar]
- Rajamäki, M.L. and Valkonen, J.P. (2009) Control of nuclear and nucleolar localization of nuclear inclusion protein a of picorna‐like Potato virus A in Nicotiana species. Plant Cell, 21, 2485–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz‐Ruiz, S. , Soler, N. , Sanchez‐Navarro, J. , Fagoaga, C. , Lopez, C. , Navarro, L. , Mareno, P. , Pena, L. and Flores, R. (2013) Citrus tristeza virus p23: determinants for nucleolar localization and their influence on suppression of RNA silencing and pathogenesis. Mol. Plant–Microbe Interact. 26, 306–318. [DOI] [PubMed] [Google Scholar]
- Senshu, H. , Yamaji, Y. , Minato, N. , Shiraishi, T. , Maejima, K. , Hashimoto, M. , Miura, C. , Neriya, Y. and Namba, S. (2011) A dual strategy for the suppression of host antiviral silencing: two distinct suppressors for viral replication and viral movement encoded by potato virus M. J. Virol. 85, 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi, T. , Hoshi, H. , Eimori, K. , Kawanishi, T. , Komatsu, K. , Hashimoto, M. , Maejima, K. , Yamaji, Y. and Namba, S. (2011) First report of Helleborus net necrosis virus isolated from hellebores with black death syndrome in Japan. J. Gen. Plant Pathol. 77, 269–272. [Google Scholar]
- Sun, L. , Andika, I.B. , Kondo, H. and Chen, J. (2013) Identification of the amino acid residues and domains in the cysteine‐rich protein of Chinese wheat mosaic virus that are important for RNA silencing suppression and subcellular localization. Mol. Plant Pathol. 14, 265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Te, J. , Melcher, U. , Howard, A. and Verchot‐Lubicz, J. (2005) Soilborne wheat mosaic virus (SBWMV) 19K protein belongs to a class of cysteine rich proteins that suppress RNA silencing. Virol. J. 2, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda, S. , Kubota, K. , Kanda, A. , Ohki, T. and Meshi, T. (2007) Pathogenicity of pepper mild mottle virus is controlled by the RNA silencing suppression activity of its replication protein but not the viral accumulation. Phytopathology, 976, 412–420. [DOI] [PubMed] [Google Scholar]
- Verchot‐Lubicz, J. (2005) A new cell‐to‐cell transport model for potexviruses. Mol. Plant–Microbe Interact. 18, 283–290. [DOI] [PubMed] [Google Scholar]
- Xiong, R. , Wu, J. , Zhou, Y. and Zhou, X. (2009) Characterization and subcellular localization of an RNA silencing suppressor encoded by Rice stripe tenuivirus. Virology, 25, 29–40. [DOI] [PubMed] [Google Scholar]
- Yelina, N. , Savenkov, E. , Solovyev, A. , Morozov, S. and Valkonen, J. (2002) Long‐distance movement, virulence, and RNA silencing suppression controlled by a single protein in hordei‐ and potyviruses: complementary functions between virus families. J. Virol. 76, 981–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Primers used in this study.


