Summary
Root‐knot nematodes (RKNs, Meloidogyne incognita) are economically important endoparasites with a wide host range. We used a comprehensive transcriptomic approach to investigate the expression of both tomato and RKN genes in tomato roots at five infection time intervals from susceptible plants and two infection time intervals from resistant plants, grown under soil conditions. Differentially expressed genes during susceptible (1827, tomato; 462, RKN) and resistance (25, tomato; 160, RKN) interactions were identified. In susceptible responses, tomato genes involved in cell wall structure, development, primary and secondary metabolite, and defence signalling pathways, together with RKN genes involved in host parasitism, development and defence, are discussed. In resistance responses, tomato genes involved in secondary metabolite and hormone‐mediated defence responses, together with RKN genes involved in starvation stress‐induced apoptosis, are discussed. In addition, 40 novel differentially expressed RKN genes encoding secretory proteins were identified. Our findings provide novel insights into the temporal regulation of genes involved in various biological processes from tomato and RKN simultaneously during susceptible and resistance responses, and reveal the involvement of a complex network of biosynthetic pathways during disease development.
Keywords: effectors, Meloidogyne incognita (RKN), metabolic networks, resistance, susceptible, tomato roots, transcriptome
Introduction
Root‐knot nematodes (RKNs; Meloidogyne spp.) are devastating polyphagous endoparasites that parasitize many cultivated plants worldwide and pose a serious threat to global food security. The estimated crop losses caused by RKN infections are over 100 billion dollars annually (Trudgill and Blok, 2001). RKNs are highly sophisticated parasites that hijack host machinery by secreting effector molecules to initiate and maintain feeding cells inside the host roots in order to complete their life cycle within 4–6 weeks (Abad and Williamson, 2010). The effector molecules consist of enzymes, peptides, small metabolites or other biomolecules, and have many roles in plant parasitism (Mitchum et al., 2013, Shukla et al., 2016). The majority of effector molecules are produced in the oesophageal glands (two subventral and one dorsal), whereas others are synthesized in the hypodermis and amphids. Effectors have been identified using various approaches, such as expressed sequence tag (EST) or genomic datasets and direct sequencing of gland cell mRNA (Bellafiore et al., 2008; Huang et al., 2003; Rutter et al., 2014a). A number of RKN effectors, including cellulases, xylanases, annexins, expansins, calreticulin, fatty acid retinol‐binding (FAR) protein and chorismate mutase, have been characterized previously (Haegeman et al., 2013; Huang et al., 2005; Jaouannet et al., 2013; Jaubert et al., 2005; Truong et al., 2015).
The zig‐zag model of the plant immune system proposes two branches of pathogen‐induced plant defence response: pattern‐triggered immunity (PTI) and effector‐triggered immunity (ETI) (Jones and Dangl, 2006). Recently, members of a conserved family of nematode pheromones (ascarosides) have been identified as nematode pathogen‐associated molecular patterns (PAMPs) (Manosalva et al., 2015). The resistance gene (R gene) from tomato, Mi‐1.2, which belongs to the nucleotide‐binding site–leucine‐rich repeat (NBS‐LRR) class, activates ETI and confers resistance against three species of Meloidogyne (Milligan et al., 1998). Direct or indirect interaction of the Mi gene with an as yet unknown avirulence (Avr) protein elicits a cascade of signal transduction pathways that activate defence responses (Hwang and Williamson, 2003; Williamson and Kumar, 2006). Previous studies have reported that Mi‐mediated resistance is characterized by localized cell death when a second‐stage juvenile (J2) attempts to establish a feeding site inside the host roots (Williamson and Hussey, 1996). As a result, the development of the feeding site is impaired and is unable to supply nutrition to nematodes. Previous studies have reported the involvement of mitogen‐activated protein kinase (MAPK) signalling cascades, resulting in the production of reactive oxygen species (ROS) and the participation of the Rme1 gene for the activation of defence responses, including salicylic acid (SA) and ethylene (ET) signalling pathways (Bhattarai et al., 2008; Branch et al., 2004; de Ilarduya et al., 2001; Li et al., 2006; Mantelin et al., 2013, Molinari et al., 2013).
Transcriptomic changes occurring in either susceptible or resistant plants during host–RKN interactions have been studied using various approaches, such as RNA blotting, differential cDNA library screening, EST sequencing, microarrays and, more recently, next‐generation sequencing (NGS) (Abad and Williamson, 2010). Microarray‐based transcriptomic studies have been conducted in Arabidopsis (Barcala et al., 2010; Fuller et al., 2007; Hammes et al., 2005; Jammes et al., 2005), tomato (Bar‐Or et al., 2005; Portillo et al., 2013; Schaff et al., 2007), a resistant soybean line (Ibrahim et al., 2011) and RKN‐tolerant aubergine Solanum torvum (Bagnaresi et al., 2013). To date, only a few studies have utilized NGS to investigate differential host gene expression patterns during RKN–host interactions, including rice galls and giant cells (GCs) (Ji et al., 2013; Kyndt et al., 2012), resistant soybean roots (Beneventi et al., 2013), resistant and susceptible alfalfa cultivars (Postnikova et al., 2015) and common bean roots (Santini et al., 2016). However, previous studies were either on hosts or dissected nematodes with a limited number of disease developmental stages. Only one study to date has reported both host and nematode genes from resistant and susceptible cultivars with RKN‐infected alfalfa roots grown in in vitro conditions and with a limited number of infective stages (Postnikova et al., 2015).
The availability of relatively well‐annotated genome reference sequences of both tomato and RKN have made the tomato–RKN system an excellent crop model for the study of host–pathogen interactions. In this study, we investigated the expression profiles of both tomato and RKN genes from in vivo infected tomato roots under soil‐grown conditions at five infection time intervals in a susceptible line and two infection time intervals in a resistant line. The study revealed complex changes in genes involved in cell wall architecture, development, hormonal signalling cascades and defence responses elicited by RKN in susceptible and resistant tomato roots. In addition, the repertoire of RKN genes that are most probably involved in parasitism, nematode growth, development and defence is presented. To our knowledge, this is the first comprehensive study to highlight simultaneously the expression profiles of tomato and RKN during susceptible and resistant responses in in vivo RKN‐infected tomato roots under soil‐grown conditions.
Results
Sequencing data and transcriptome mapping
Experiments were conducted to study the life cycle of RKN inside tomato roots and for a selection of disease developmental stages (Fig. 1). Transcriptome sequencing generated 1 154 560 291 paired‐end reads for replicate 1 and 537 461 341 single‐end reads for replicate 2 of 100 bp in length. Quality filtering of raw reads was performed for a mean phred quality score of 20 and any contaminating adapter sequences were removed (Table S1, see Supporting Information). For both replicates, 95%–99% good‐quality 35–88 bp reads were retained, which were used for mapping onto the reference genome employing TopHat2. In total, 72%–92% of good‐quality reads were mapped onto the Solanum lycopersicum genome (SL2.50 assembly) across all samples (Table 1). For infected samples, 0.1%–6.3% of reads were mapped to the Meloidogyne incognita genome (Table 1). All the sequencing data, raw as well as processed, have been submitted to the Gene Expression Omnibus (GEO) repository at the National Center for Biotechnology Information (NCBI) with accession no. GSE88763 and Sequence read archive (SRA) accession no. SRP091567.
Figure 1.
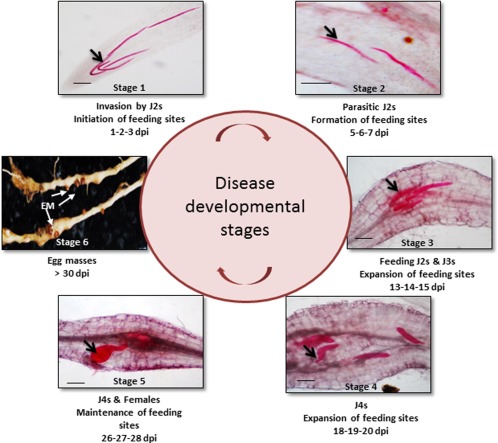
Meloidogyne incognita life cycle in roots of susceptible tomato cultivar Pusa Ruby (PR). Whole mounts (10 μm) of acid fuchsin‐stained roots with parasitic nematodes (black arrows) at different stages (stages 1–5) inside the roots. dpi, days post‐infection; EM, egg masses; J2s, second‐stage juveniles.
Table 1.
Statistics of good‐quality reads mapped onto the tomato and nematode reference genomes.
| Sample | Replicate 1 | Replicate 2 | ||||||
|---|---|---|---|---|---|---|---|---|
| No. of read pairs aligned onto Solanum lycopersicum genome | Percentage read pairs aligned (%) | No. of read pairs aligned onto Meloidogyne incognita genome | Percentage read pairs aligned (%) | No. of reads aligned onto Solanum lycopersicum genome | Percentage reads aligned (%) | No. of reads aligned onto Meloidogyne incognita genome | Percentage reads aligned (%) | |
| PR_stage_0 | 67 446 974 | 92.4 | 10 687 | 0.0 | 11 923 705 | 95.0 | 498 | 0.0 |
| PR_UI_stage_1 | 66 575 269 | 89.4 | 21 248 | 0.0 | 23 296 422 | 93.9 | 1777 | 0.0 |
| PR_UI_stage_2 | 48 847 742 | 91.6 | 107 | 0.0 | 36 674 766 | 95.4 | 1044 | 0.0 |
| PR_UI_stage_3 | 49 304 943 | 82.7 | 1169 | 0.0 | 8 761 649 | 94.0 | 822 | 0.0 |
| PR_UI_stage_4 | 58 326 864 | 78.4 | 1065 | 0.0 | 41 281 223 | 93.8 | 3273 | 0.0 |
| PR_UI_stage_5 | 78 560 558 | 90.2 | 1111 | 0.0 | 39 893 121 | 92.7 | 1672 | 0.0 |
| PR_I_stage_1 | 72 787 284 | 90.7 | 62 935 | 0.1 | 22 781 784 | 91.2 | 10 552 | 0.0 |
| PR_I_stage_2 | 54 697 086 | 84.8 | 102 658 | 0.2 | 11 485 953 | 94.6 | 8366 | 0.1 |
| PR_I_stage_3 | 76 668 601 | 90.9 | 663 329 | 0.8 | 29 797 293 | 91.0 | 252 817 | 0.8 |
| PR_I_stage_4 | 52 626 967 | 85.8 | 827 860 | 1.4 | 15 107 463 | 90.1 | 358 995 | 2.1 |
| PR_I_stage_5 | 63 581 675 | 74.9 | 5 377 656 | 6.3 | 11 078 964 | 81.2 | 1 026 425 | 7.5 |
| MM_stage_0 | 57 113 244 | 92.1 | 114 | 0.0 | 50 439 346 | 93.2 | 2658 | 0.0 |
| M36_stage_0 | 79 366 778 | 92.0 | 205 | 0.0 | 23 983 409 | 95.1 | 498 | 0.0 |
| M36_UI_stage_1 | 24 077 820 | 88.4 | 19 314 | 0.1 | 36 635 563 | 94.0 | 2657 | 0.0 |
| M36_UI_stage_2 | 20 417 489 | 74.6 | 39 740 | 0.1 | 34 522 313 | 95.4 | 779 | 0.0 |
| M36_I_stage_1 | 82 496 591 | 89.9 | 54 895 | 0.1 | 15 613 149 | 94.5 | 12 528 | 0.1 |
| M36_I_stage_2 | 78 517 115 | 90.5 | 89 928 | 0.1 | 32 888 657 | 94.7 | 47 024 | 0.1 |
Replicate 1, paired‐end reads; Replicate 2, single‐end reads; I, infected; UI, uninfected; PR, Pusa Ruby (susceptible cultivar); MM, Money Maker (susceptible cultivar); M36, transgenic MM (resistant line).
Identification of tomato genes from susceptible tomato (Pusa Ruby, PR) and resistant tomato (transgenic Money Maker, M36) plants
Differential gene expression analysis and annotations
Five different stages of infection and their corresponding uninfected controls were selected to study the susceptible response during RKN infection in tomato roots. A total of 24 411 tomato genes were identified across these five stages. Stage‐wise comparison of infected roots with their corresponding uninfected controls identified a total of 1827 significant differentially expressed genes (DEGs) (adjusted P < 0.05 and log2 fold change ≥ ±2) (Fig. 2a; Table S2, see Supporting Information). No significant DEGs were identified at stage 1. Eighteen significant DEGs were identified at stage 2 with more genes showing up‐regulation than down‐regulation. At stage 3, 905 significant DEGs were obtained. At stages 4 and 5, 1054 and 1308 DEGs, respectively, were identified with more genes showing down‐regulation (56.8% and 55.9%, respectively).
Figure 2.
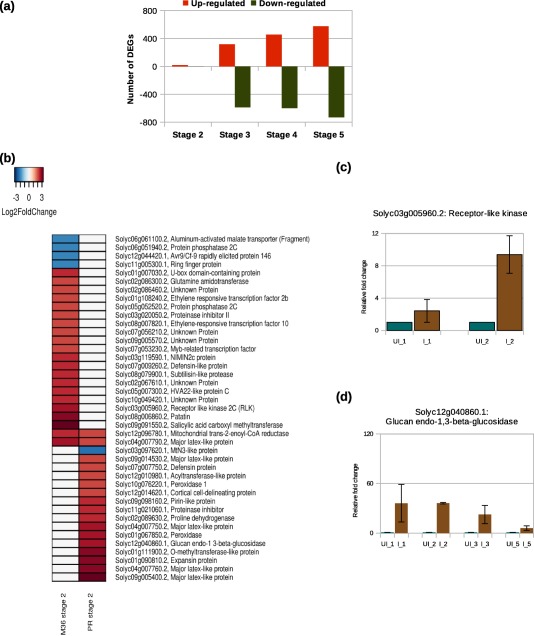
(a) Distribution of significant differentially expressed genes (DEGs) in tomato at different stages during the susceptible response. (b) Comparative expression profile of tomato DEGs detected during the susceptible (Pusa Ruby, PR) and resistance (transgenic Money Maker, M36) responses at stage 2. Differential expression was calculated with respect to the corresponding uninfected controls. Each row of the heatmap represents a gene and each column represents a stage of disease development. The colour key is given in the top‐left corner of the figure. (c, d) Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) validation of RNA sequencing (RNA‐seq) data for the tomato genes, receptor‐like kinase and glucan endo‐1,3‐β‐glucosidase, during resistance and susceptible responses, respectively. The y axes represent the relative fold change (calculated using the ΔΔCt method) in gene expression at various stages relative to the corresponding uninfected controls. The data are representative of two technical replicates and three independent biological replicates. Bars indicate standard errors. I, infected; UI, uninfected.
In resistant plants, only the first two stages of infection (stages 1 and 2) and corresponding uninfected controls were selected for study. A total of 23 393 tomato genes were identified across these two stages. Stage‐wise comparison of infected roots with their corresponding uninfected controls identified no significant DEGs at stage 1 and only 25 at stage 2, with 21 up‐regulated and four down‐regulated genes (Table S3, see Supporting Information). In addition, we compared the significant DEGs obtained from susceptible and resistance responses. As no significant DEG was observed at stage 1, DEGs obtained at stage 2 (18 in the susceptible response and 25 in the resistance response) were compared. Only two up‐regulated genes were common at stage 2 between susceptible and resistance responses: one encodes mitochondrial trans−2‐enoyl‐CoA reductase and the other encodes major latex‐like protein; the rest of the genes were distinct (Fig. 2b). In addition, quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) was performed to validate the expression profiles of tomato genes, including glucan endo‐1,3‐β‐glucosidase in the susceptible response and receptor‐like kinase in the resistance response (Fig. 2c,d).
Furthermore, sample‐to‐sample correlation was observed on the basis of the global gene expression profiles of the genes identified across all stages in the susceptible response (Fig. S1, see Supporting Information). Gene ontology and protein class annotation of the DEGs in both susceptible and resistance responses was performed (Figs S2–S4, see Supporting Information). Gene set enrichment analysis revealed significant results in molecular function category only at two stages (stages 4 and 5) during the susceptible response (Fig. S5, see Supporting Information).
Differential regulation of genes resulting in altered cell wall architecture
Our data reflect significant alteration in the expression of 85 tomato genes involved in cell wall degradation (43 genes), cell wall modification (29 genes), cell wall proteins (five genes) and cell wall synthesis (eight genes) during the susceptible response (Figs 3a, S6, see Supporting Information). By contrast, during resistance response, no significant alteration was observed in the expression of genes involved in the modulation of cell wall architecture.
Figure 3.

Schematic representation of the gene expression patterns of subsets of tomato differentially expressed genes (DEGs) involved in plant cell wall architecture (a), development (b), primary metabolism (c) and transporter genes (d) in the susceptible response. The heatmap represents only a subset of genes from each category (consult Supporting Information for the expression of all DEGs involved in each category). Each row represents a gene and each column represents a stage of disease development. Labels on the right side of the heatmap show the gene id, category to which it belongs, followed by the annotation. The colour key is given in the top‐left corner of the heatmap. RKN, root‐knot nematode.
Differential regulation of plant developmental genes and primary metabolism
During the susceptible response, 33 genes involved in the plant cell cycle (three genes), cytoskeletal organization (two genes), root cap proteins (seven genes), transcription factors controlling developmental processes (16 genes) and other developmental processes (5 genes) were differentially expressed (Figs 3b and S7, see Supporting Information). Furthermore, the differential expression of 102 genes, involved in photosynthesis (12 genes), major and minor carbohydrate metabolism (nine genes), fermentation (five genes), the tricarboxylic acid (TCA) cycle (four genes), mitochondrial electron transport (one gene), nitrogen metabolism (two genes), amino acid metabolism (17 genes), tetrapyrrole synthesis (four genes) nucleotide metabolism (12 genes) and lipid metabolism (36 genes) (Figs 3c and S8a–d, see Supporting Information), was observed. By contrast, during resistance response, a gene encoding patatin protein was up‐regulated at stage 2 and no alteration was observed in genes involved in primary metabolism.
Differential regulation of transporter genes
The expression of solute transporter genes was altered in the susceptible response, which included various carbohydrate and sugar transporters (19 genes), lipid transporters (seven genes), aquaporins (13 genes), and peptide, nitrate and amino acid transporters (18 genes) (Figs 3d and S9a–d, see Supporting Information). In addition, genes encoding multidrug and toxic compound extrusion (MATE) efflux transporters (15 genes), ATP‐binding cassette (ABC) transporters (eight genes) and various ion transporters (32 genes) were altered (Figs 3d and S9e,f). By contrast, no significant alteration was observed in the resistance response.
Suppression of plant defence responses by alteration of secondary and phytohormone metabolism
The secondary metabolite pathway analysis revealed alterations in biosynthetic pathways of various secondary metabolites including the phenylpropanoid pathway, flavonoid pathway, polyamine pathway and isoprenoid pathway (Figs 4 and S10a,b, see Supporting Information).
Figure 4.
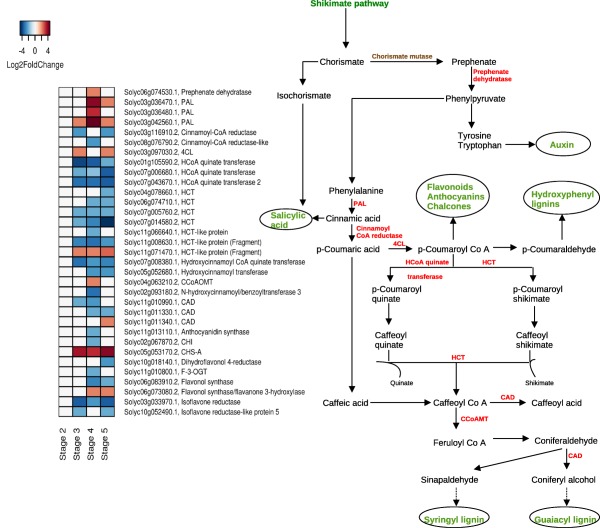
Schematic representation of the gene expression patterns (left) of tomato differentially expressed genes (DEGs) involved in the phenylpropanoid biosynthetic pathway (right). Pathway showing DEGs (in red) and end products of pathways (in green) during the susceptible response. In the heatmap, each row represents a gene and each column represents a stage of disease development. Labels on the right of the heatmap show the gene id followed by the annotation. The colour key is given in the top‐left corner of the figure. 4CL, 4‐hydroxycinnamoyl CoA ligase; CAD, cinnamoyl alcohol dehydrogenase; CCoAMT, caffeoyl CoA‐O‐methyltransferase; HCT, hydroxycinnamoyl shikimate/quinate hydroxycinnamoyl transferase; HCoA, hydroxycinnamoyl Co A; PAL, phenylalanine ammonia lyase.
In the phenylpropanoid biosynthetic pathway, genes encoding phenylalanine ammonia lyase (PAL) and 4‐hydroxycinnamoyl CoA ligase (4CL) were up‐regulated during disease development. Other downstream genes, including cinnamoyl alcohol dehydrogenase (CAD) and flavonol synthase, were down‐regulated (Fig. 4). We also found alterations in genes involved in isoprenoid and polyamine metabolism, together with the down‐regulation of four genes involved in gibberellin biosynthesis (Fig. S10a,b). In addition, the alteration in expression of genes involved in auxin signalling (16 genes) and cytokinin synthesis (four genes) was observed (Fig. S11a, see Supporting Information). Abscisic acid (ABA) synthesis and responsive genes were also altered, including the up‐regulation of lycopene β‐cyclase, FIP1 and HVA22‐like protein c, at stages 3–5 (Figs S10a and S11b).
Genes coding for enzymes involved in ET biosynthesis and signalling, including 1‐aminocyclopropane‐1‐carboxylic acid (ACC) synthase (ACS; two genes), ACC oxidase (ACO; 16 genes) and ET‐responsive transcription factors (ERFs; 32 genes), were differentially expressed (Figs S10b and S12, see Supporting Information). In addition, we found the differential expression of genes involved in jasmonic acid (JA) synthesis (eight genes) and SA‐responsive signalling (eight genes) during disease development (Fig. S13a,b, see Supporting Information). Genes related to oxidative stress, including glutathione S‐transferases (GSTs), peroxidases and thioredoxins, were largely down‐regulated on RKN infection during later stages (Fig. S13c).
During resistance responses, we found up‐regulation of genes encoding receptor‐like kinase and protein phosphatase 2C (regulator of ABA signalling). The genes encoding ABA‐responsive HVA22‐like protein c and an SA carboxyl methyltransferase were also up‐regulated at stage 2. In addition, two ERF genes were up‐regulated at stage 2, indicating active hormone‐mediated defence responses (Fig. 2b).
Expression profiles of tomato genes validated through quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR)
In total, 20 genes were selected (up‐regulation or down‐regulation) that showed a significant fold change (≤2 or ≥2) for further validation by qRT‐PCR (Table S4, see Supporting Information). A gene encoding an open reading frame (Solyc01g080500.2) and tubulin α chain (Solyc08g006890.2), showing uniform expression in all stages, were used as internal controls for tomato. The gene expression patterns obtained through qRT‐PCR were coincident with the RNA sequencing (RNA‐seq) expression profiles. The correlation coefficient (R 2) was 0.80 at P = 2.2e‐16 (Fig. S14, see Supporting Information).
Identification of RKN genes from infected root samples of susceptible and resistant tomato plants
Reads aligned onto RKN genome and differential expression analysis
Good‐quality reads from infected samples of susceptible plants were aligned onto the RKN genome (Table 1). Non‐tomato mapped reads led to the identification of 10 131 protein‐encoding RKN genes. Hierarchical clustering of the global expression profiles clustered the genes into two groups (Fig. S15, see Supporting Information).
In the susceptible response, we identified 462 significant DEGs (adjusted P < 0.05 and log2 fold change ≥ 2) relative to stage 1 (Figs 5a and S16a,b; Table S5, see Supporting Information). As the same population of RKN was used to study susceptible and resistance responses, RKN‐predicted genes of the same stage from both interactions were compared. In the resistance response, no significant alteration was found at stage 1 and 160 significant DEGs were identified at stage 2 relative to the susceptible response (Table S6, see Supporting Information).
Figure 5.
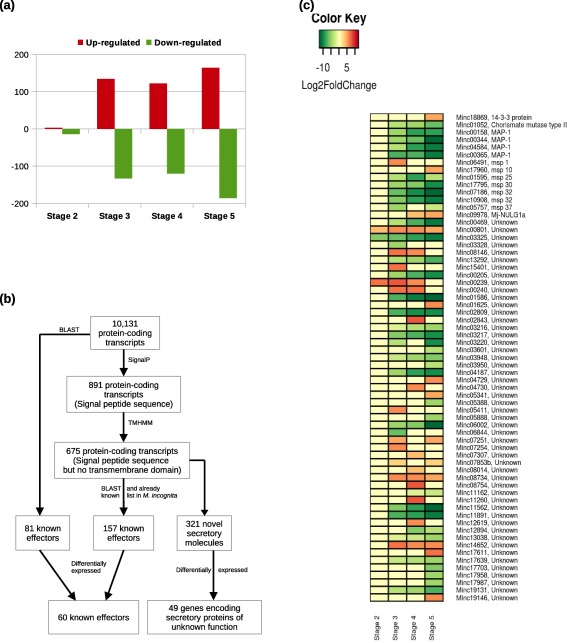
(a) Distribution of significant root‐knot nematode (RKN) differentially expressed genes (DEGs) detected at different stages of infection relative to stage 1 during the susceptible response. (b) Workflow summary of the bioinformatics strategy used to identify RKN candidate effector genes. (c) Schematic representation of the gene expression patterns of RKN DEGs reported to be involved in parasitism during the susceptible response. Each row represents a gene and each column represents a stage of disease development. Labels on the right show the gene id followed by the annotation. The colour key is given in the top‐left corner of the figure.
Effector identification
A search for candidate effectors amongst 10 131 RKN mapped translated proteins using SignalP 4.1, followed by TMHMM 2.0, identified 675 proteins that have a signal peptide sequence, but no transmembrane domain. Further, through manual screening based on reported putative effectors, we identified 157 putative effectors and 312 novel secretory molecules. In addition, blast results identified a further 81 known effectors, eight of which were found to have a signal peptide sequence only (Fig. 5b). We also looked for the differential expression of these 559 candidate effectors at different stages during disease development in the susceptible response. A total of 109 genes were differentially expressed (Fig. 5c), 60 of which have been reported previously as effectors and 49 could be classified as potentially secreted molecules with unknown functions.
Differential regulation of RKN genes encoding cell wall‐degrading enzymes and peptidases
We found significant expression of genes encoding 10 members of the glycosyl hydrolase (GH) and six members of the pectate lyase (PL) families at stages 1 and 2 during the susceptible response. Furthermore, expression of the UDP‐glucosyltransferase gene was significant at stages 1 and 2 and down‐regulated at stages 3–5, whereas the expression of the gene encoding carboxylesterase was up‐regulated at stages 3–5. The expression of a member of carbohydrate‐binding module family 20 was specifically down‐regulated at stages 3 and 5 (Fig. 6a). We also found stage‐specific differential expression of 30 genes encoding different peptidases, including aspartic, cysteine, metallo and serine peptidases (Fig. 6a). The expression of six genes encoding aspartic and serine peptidases was significant during stages 1 and 2, but later they were down‐regulated. However, the expression of eight genes coding for various peptidases was up‐regulated at later stages. In addition, the expression of five genes encoding metallo and serine peptidases was significant at stages 2–4, but down‐regulated at stage 5.
Figure 6.
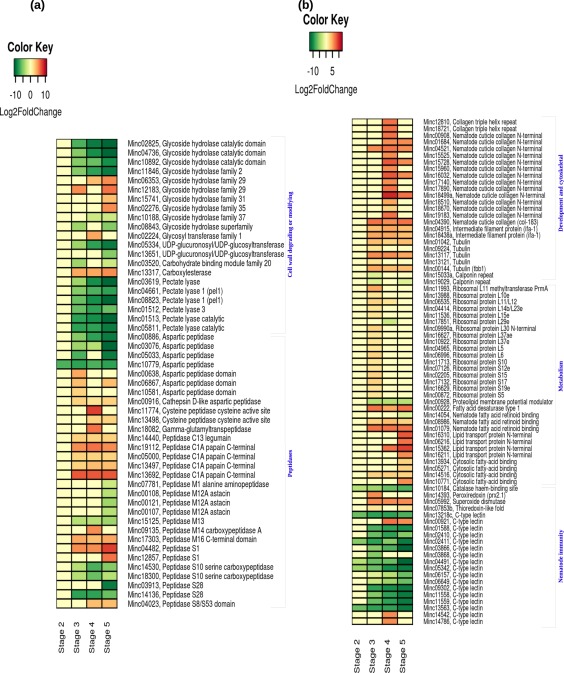
Schematic representation of the gene expression patterns of root‐knot nematode (RKN) differentially expressed genes (DEGs) involved in cell wall degradation and peptidase encoding (a) and development, metabolism and nematode immunity (b), detected during the susceptible response. Each row represents a gene and each column represents a stage of disease development. Labels on the right side of the heatmap show the gene id followed by the annotation. The colour key is given in the top‐left corner of the figure.
Interestingly, in the resistance response, we observed the up‐regulation of seven genes encoding members of the GH family and five genes encoding members of the PL family, together with seven genes encoding aspartic and cysteine peptidases, at stage 2 relative to the susceptible response (Table S6). The expression of two genes (of five) encoding aspartic peptidase and one gene (of two) encoding a cysteine peptidase was unique to the resistance response.
Differential regulation of RKN genes involved in nematode development and metabolism
Genes encoding two cuticle collagen and five tubulin proteins were up‐regulated at stage 3, whereas none were up‐regulated at stage 2. The expression of 16 cuticle collagen genes was induced at stage 4, indicating the dynamic transition and development of RKNs into J4 and adult worms. Two genes coding for intermediate filament protein 1 (IFA‐1) were up‐regulated during all stages of RKN development, whereas two genes coding for calponin protein were down‐regulated. In total, 17 genes encoding ribosomal proteins (RPs) were found to be significantly altered during disease development. RPL29e was found to be down‐regulated at stage 4, whereas another 16 genes were up‐regulated at stage 3 only (Fig. 6b). By contrast, in the resistance response, the expression of cuticle collagen genes was not significantly altered. In addition, we found the up‐regulation of one actin, two actin‐binding protein‐coding genes [abnormal nuclear anchorage (anc‐1) and levamisole resistant (lev‐11)], three myosin heavy chain structural genes, six unc genes and one plectin repeat‐containing gene.
We identified the differential regulation of 14 genes related to lipid metabolism and transport. A gene encoding fatty acid desaturase 1 was significantly induced during the sedentary stages (stages 3–5) of RKN, indicating the synthesis of fatty acids required for RKN development. We also observed significant up‐regulation of four genes encoding lipid transport proteins and three genes encoding cytosolic fatty acid‐binding protein at stage 5 (Fig. 6b). During the resistance response, we observed significant up‐regulation of genes encoding FAR protein and cytosolic fatty acid binding at stage 2 relative to the susceptible response.
Differential regulation of effector genes that suppress the plant defence response
In our study, we found significant expression of a catalase protein‐coding gene at stage 1 and down‐regulation at stages 2–5. In addition, we observed significant up‐regulation of a peroxiredoxin gene (PRX2.1) at stage 3 and superoxide dismutase gene (SOD) at stages 3–5. Apart from antioxidants, we identified the differential expression of 17 genes coding for C‐type lectins. Nine of the 16 genes were significantly down‐regulated during the later stages and five throughout disease development (Fig. 6b).
A gene encoding 14‐3‐3 protein was significantly induced at stage 5 (Fig. 5c). In addition, we observed the differential expression of seven genes coding for oesophageal gland cell secretory proteins (msp): msp1 was significantly induced at stage 3 and msp10 at stage 5, whereas msp25, msp30, msp32 and msp37 were significantly suppressed at later stages (Fig. 5c).
In the resistance response, we found significant up‐regulation of a gene encoding MAP‐1 protein relative to the susceptible response at stage 2 (Table S6). In addition, significant up‐regulation of two GSTs, one hydroperoxide reductase, one aldehyde dehydrogenase, one catalase protein, 15 C‐type lectins (five were unique to resistance responses) and two autophagy‐related proteins was observed.
Expression profiling of a few RKN genes by qRT‐PCR
The expression profiles of six RKN genes (Table S4) were determined in RKN‐infected tomato root tissues at stage 2 (early) and stage 5 (late) of disease development by qRT‐PCR. A significant differential expression amongst RKN genes was observed (Fig. S17, see Supporting Information), which was confirmed by Student's t‐test at P < 0.1 and P < 0.05. The expression of genes encoding EF1, collagen, MAP1, peptidase and C‐type lectin was down‐regulated at stage 5 relative to stage 2. However, the expression of the gene coding for Mj‐NULG1a was up‐regulated at stage 5 relative to stage 2.
Discussion
To the best of our knowledge, this is the first comprehensive analysis of global gene expression profiles of both tomato and RKN at five stages during the susceptible response and two stages during the resistance response from in vivo RKN‐infected tomato roots under soil‐grown conditions. DEGs were identified by comparing the expression profiles from infected roots with the equivalent developmental stages of uninfected roots. This approach identified a large set of DEGs, together with their temporal regulation during specific stages of disease development. In addition, we constructed a model to depict the dynamic changes in expression of the genes involved in biological processes from both tomato and RKN during susceptible and resistance responses based on the summary of transcriptome data (Fig. 7).
Figure 7.
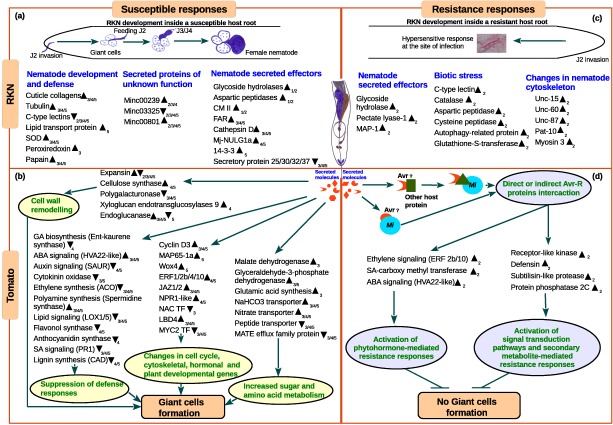
A model depicting the complex changes in the biochemical processes in both tomato and root‐knot nematode (RKN) during susceptible and resistant interactions based on the summary of transcriptome data. (a) Changes in the expression of RKN genes during the susceptible response. (b) Changes in the expression of tomato genes during the susceptible response. (c) Changes in the expression of RKN genes during the resistance response. (d) Changes in the expression of tomato genes during the resistance response. The up‐arrowhead indicates up‐regulated genes, down‐arrowhead indicates down‐regulated genes and numerical subscript in the arrowhead indicates the specific stage in which expression was observed. Host metabolic processes that are modulated on RKN infection are highlighted in coloured circles. ABA, abscisic acid; ACO, 1‐aminocyclopropane‐1‐carboxylic acid oxidase; Avr, avirulence protein; CAD, cinnamoyl alcohol dehydrogenase; CM II, chorismate mutase type II; ERF, ethylene‐responsive transcription factor; FAR, nematode fatty acid retinol‐binding protein; GA, gibberellins; LBD, lateral organ body domain‐containing transcription factor; LOX, lipoxygenase; MAP‐1, Meloidogyne avirulence protein; MATE, multidrug and toxic compound extrusion; NAC TF, no apical meristem‐containing transcription factor; MAP65‐1a, microtubule‐associated protein 65‐1a; pat, paralysed arrest two‐fold; PR1, pathogenesis‐related protein 1; SA, salicylic acid; SOD, superoxide dismutase; unc, uncoordinated; Wox4, WUS homeobox‐containing gene.
RNA‐seq data revealed 462 RKN DEGs during susceptible (Fig. 7a) and 160 RKN DEGs at stage 2 during resistance (Fig. 7c) responses. Here, for the first time, changes in RKN gene expression from in vivo infected resistant tomato roots is presented. Furthermore, RNA‐seq data revealed 1827 tomato DEGs during susceptible (Fig. 7b) and 25 tomato DEGs during resistance (Fig. 7d) responses.
RKNs use the mechanical force of the stylet and secrete cell wall‐degrading or modifying enzymes (CWD/MEs), including endoglucanases, PLs, polygalacturonases, expansins, etc., to macerate plant roots during invasion and migration (Mitchum et al., 2103). Accordingly, we observed the activation of RKN genes encoding CWD/MEs, belonging to GH and PL families, at stages 1–2 (Fig. 7a) during the susceptible response. When compared with the susceptible response, in the resistance response, the differential expression of four (of seven) genes encoding members of the GH family and two (of five) genes encoding members of the PL family was unique (Fig. 7c). The correlation between the reduction in invasion and increased expression, together with the different set of RKN CWD/MEs, is noteworthy and requires further investigation. After the initiation of feeding cells (stage 2 in our study), the expression of genes encoding CWD/MEs was reduced in nematodes and induced in plants to allow the remodelling of cell walls for the formation of feeding cells (reviewed in Sobczak et al., 2011). Moreover, the up‐regulation of tomato genes encoding expansin and glucan endo‐β‐glucosidase at stage 2 in our study indicates the initiation of dynamic changes in plant cell wall architecture (Fig. 7b). However, at later stages (stages 3–5), 53% of the plant genes encoding CWD/MEs (46 of 86) were down‐regulated (Fig. S6), indicating that CWD/MEs are tightly regulated in RKN‐infected tomato roots. By contrast, during the resistance response, a lack of differential expression of any plant CWD/ME suggests that these enzymes are not induced in the absence of GC and gall formation. Considering the expression of plant CWD/MEs during both susceptible and resistance responses, it is probable that plant CWD/MEs are largely involved in cell expansion and cell wall restructuring during gall formation, and that RKN effectors are involved in the modulation of their expression during disease development.
In susceptible plants, dramatic morphological changes occur in plant roots leading to a characteristic gall development. During this process, the crucial roles of genes encoding transcription factors from the no apical meristem (NAC) and lateral organ boundaries domain‐containing (LBD) families have been reported (Cabrera et al., 2014; Portillo et al., 2013). In our study, five genes encoding members of the NAC domain transcription factor family were specifically down‐regulated at stage 3, whereas LBD4 was up‐regulated. Furthermore, seven novel genes encoding members of the root cap protein were down‐regulated at stages 3–5 (Fig. S7). This indicates their role in transcriptional reprogramming to control root tip development and lateral root initiation. A number of amino acids are also synthesized in GCs as nutrients for parasitic nematodes. Biochemical assays of amino acids indicate that glutamine is essential for cyst nematode development (Betka et al., 1991). In our study, the down‐regulation of tomato genes encoding glutamate decarboxylase and tyrosine aminotransferase, which catabolize glutamate and tyrosine, was observed at stages 3–5. This inhibits the breakdown of amino acids and allows their availability for RKN development, as shown in cyst nematodes. With high demand for nutrient transport, we observed the differential regulation of tomato genes encoding peptide transporters, nodulin‐like sugar transporters, aquaporins, ion transporters and auxin efflux carriers. Furthermore, our study reports, for the first time, the differential expression of 15 genes encoding MATE efflux family proteins, which are known to play a role in plant growth, development and transport of xenobiotic organic cations (Fig. 7b; Eckardt, 2001). In contrast with the susceptible response, no alteration in expression of tomato genes involved in the plant cell cycle, cell division, primary or secondary metabolism was observed in the resistance response, confirming the cessation of GC formation.
In susceptible plants, feeding RKNs go through the transition from larval stages to adults, which is accompanied by the formation of a new cuticle. In our study, 16 cuticle biosynthetic genes were identified and their temporal gene expression patterns corresponding to RKN developmental stages indicated their role as structural proteins in the cuticle (Figs 6a and 7a). Earlier studies on proteases from both animal‐ and plant‐parasitic nematodes have suggested their roles in digestion, embryogenesis, cuticle remodelling and parasitism (references in Haegeman et al., 2012). Furthermore, we observed the specific up‐regulation of 16 (of 17) RKN genes encoding ribosomal proteins (not reported previously) during the susceptible response, which are known to play a role in the structural assembly of the ribosome. By contrast, the absence of feeding cell development in resistant plants leads to starvation‐induced stress in nematodes and, subsequently, cessation of their development. Accordingly, no significant change in the expression of genes encoding RKN cuticle collagens was observed in resistant plants. However, the up‐regulation of RKN genes that regulate actin and myosin filament dynamics was observed, possibly reflecting changes in the cytoskeleton and body wall muscles under starvation stress. For example, the unc‐87 gene, which is known to stabilize actin filaments for correct functioning of body wall muscles (Yamashiro et al., 2007), was up‐regulated at stage 2 (Fig. 7c).
Progress has been made to functionally characterize several RKN genes encoding a wide range of effectors (Mitchum et al., 2013), which have diverse roles during parasitism. For instance, the nematode FAR‐1 effector is known to interfere with lipid signalling, to alter cell wall organization and to suppress the plant defence response (Iberkleid et al., 2015; Prior et al., 2001). We found the up‐regulation of two RKN genes encoding FAR at stages 3–5 during susceptible interactions, which can be related to the suppression of plant defence responses during the later stages of disease development. Another effector gene Mj‐nulg1a, which is known to target GC nuclei, has been implicated in reducing the parasitism ability of nematodes at the early stages (2–5 days) of infection (Lin et al., 2013). In our study, the up‐regulation of the Mj‐nulg1a gene at later stages (stages 4–5) during the susceptible response suggests that it plays a role in the suppression of plant defence responses throughout disease development. Interestingly, during the resistance response, the up‐regulation of a FAR gene and Mj‐nulg1a gene at stage 2 was also observed (Fig. 7c). As Mi‐mediated resistance is not complete, their up‐regulation during the resistance response might suggest that a few parasitic juveniles (that have escaped the plant defence response) attempt to suppress the defence response. A well‐characterized RKN effector, CM II, was up‐regulated at stages 1–2 during the susceptible response, which is in agreement with the reported role of CM II during the early stages in the promotion of the metabolism of chorismate to prephenate and the suppression of SA synthesis (Huang et al., 2005). Another gene encoding a putative RKN avirulence (MAP‐1) protein belonging to a multigene family and expressed in amphids has been described previously to play a role during the early stages of RKN infection (Castagnone‐Sereno et al., 2009; Semblat et al., 2001; Vieira et al., 2011). More recently, all members of the map‐1 gene family from M. incognita have been shown to possess similar conserved CLE‐like motifs of at least 12 amino acids, but the number and arrangements of repeats are different (Rutter et al., 2014b). Here, we report, for the first time, the expression patterns of map‐1 genes during parasitic stages in both susceptible and resistance responses. We observed that four map‐1 genes were up‐regulated at stage 2 during the resistance response (one was unique to the resistance response), and were down‐regulated at stages 3–5 during the susceptible response (one was unique to the susceptible response). Based on their structural properties and expression patterns, it is probable that they play a role during the early stages of infection and may mimic host proteins to modulate plant developmental processes, similar to plant CLE‐like peptides (Leasure and He, 2012).
A number of candidate effector genes encode secretory proteins (msp) that are specific to M. incognita, but have not yet been annotated (Huang et al., 2003). However, the functional roles of msp16, msp9, msp12, msp18, msp20, msp24, msp33 and msp40 genes in nematode parasitism have been investigated using gene‐silencing approaches (Huang et al., 2006; Niu et al., 2016; Shivakumara et al., 2016; Xie et al., 2016; Xue et al., 2013). Our study showed, for the first time, that msp1, msp10, msp25, msp30, msp32 and msp37 are expressed throughout the parasitic cycle of RKN during the susceptible response (Fig. 7a), also indicating their crucial role in parasitism. Interestingly, the up‐regulation of msp37 and msp32 genes was observed at stage 2 during the resistance response (Fig. 7c) and needs to be investigated further. In addition, we identified 40 novel RKN DEGs encoding secretory proteins during the susceptible response based on the presence of a signal peptide sequence and absence of a transmembrane domain (Fig. 5a,b). The stage‐specific expression profile of these novel RKN genes indicates their potential function in nematode parasitism and/or development.
In the resistance response, the up‐regulation in expression of tomato genes encoding the defensin protein (Stotz et al., 2009) and subtilisin‐like protease, leading to the production of phytoalexins and stress‐induced proteolysis, was observed. We also found the up‐regulation of genes involved in the activation of signal transduction pathways, including receptor‐like kinase and protein phosphatase 2C. However, host defences are suppressed by parasitic nematodes during susceptible interactions (Favery et al., 2016). In our study, we detected the up‐regulation of tomato genes involved in polyamine and sterol biosynthesis and the suppression of genes involved in lignin, flavonoid, isoflavonoid and anthocyanin biosynthesis (Fig. 7b) during the susceptible response. Furthermore, the up‐regulation of three genes involved in the biosynthetic pathway of spermine and the down‐regulation of 13 genes encoding ACO indicate the suppression of ET biosynthesis (Hewezi et al., 2010; Kumar et al., 1997). In agreement with previous studies, the down‐regulation of commonly used SA markers (pathogenesis‐related genes, subtilisin‐like proteases and β‐1,3‐glucanase) and the up‐regulation of genes encoding ERFs 1, 2, 7, 9 and 10 was observed at stages 4–5 during the susceptible response (Fig. 7b). In addition, genes encoding JAZ proteins (JAZ 1‐3) were up‐regulated, whereas a downstream transcription factor gene, MYC2 (myelocytomatosis‐related proteins), was down‐regulated. This suggests that JAZ proteins are actively involved in the suppression of the JA‐regulated transcription of genes (Chini et al., 2007) during disease development.
In our study, the components of ET, ABA and SA signalling were differentially regulated during both the susceptible and resistance responses. For instance, the ABA‐responsive genes, HVA22‐like and MLP, together with the SA‐responsive gene, methyl carboxylase, were up‐regulated at stage 2 in the resistance response and at stages 3–5 in the susceptible response. Interestingly, HVA22‐like protein is known to be induced by ABA and acts downstream of GA‐Myb transcription factor to suppress gibberellin (GA)‐mediated apoptosis (Guo and Ho, 2008). The expression pattern of the HVA22 gene during tomato–RKN interactions has not been reported previously. Furthermore, the genes encoding ERF10 and ERF2b were up‐regulated at stage 2 in the resistance response and at stages 4 and 5 in the susceptible response, indicating an active role of ET signalling in plant–nematode interactions. The involvement of ABA‐, ET‐ and SA‐mediated defence responses during both susceptible and resistance responses suggests an overlap between biotic and abiotic stress signalling pathways in tomato–RKN interactions (Fig. 7d). Taken together, ABA and ET signalling pathways are induced, whereas SA and JA pathways are suppressed, in susceptible interactions (Fig. 7b); ABA and ET signalling pathways are induced at the early stages during resistance interactions.
To evade plant defence responses throughout disease development, RKNs produce an array of antioxidants, such as catalases, peroxidases, peroxiredoxin, thioredoxins, GSTs and SOD (Haegeman et al., 2012; Latina, 2015). In our study, an RKN gene encoding catalase was up‐regulated at stage 2 during the resistance response and down‐regulated at stages 2–5 during the susceptible response. In addition, we observed the up‐regulation of two genes encoding GST at stage 2 during the resistance response. Significant induction of these antioxidants indicates their role in scavenging host‐derived ROS. In addition, RKN C‐type lectins, which are known to suppress the production of ROS, were up‐regulated at stage 2 during the resistance response and down‐regulated at stages 3–5 during the susceptible response (Fig. 7a). Furthermore, during the resistance response, an increase in expression of seven RKN genes encoding peptidases and two RKN genes encoding autophagy‐related protein at stage 2 (reported for the first time in this study) indicates starvation‐induced proteolysis of proteins and apoptotic cell death, respectively (Fig. 7c).
In conclusion, this study provides much deeper and novel insights into the molecular mechanisms involved in plant–nematode interactions. This study has led to the identification of a large number of novel and known DEGs simultaneously from tomato and RKN, and their specific modulation during susceptible and/or resistance responses in in vivo infected tomato roots under soil‐grown conditions. The large repertoire of genes will greatly facilitate basic and applied research on plant–nematode interactions.
Experimental Procedures
Plant material and RKN infection
The two susceptible tomato cultivars used in this study were Pusa Ruby (PR) and Moneymaker (MM). In addition, a resistant transgenic tomato MM line containing the Mi gene (M36) was used. Seeds of the transgenic M36 line were obtained from Professor Valerie M. Williamson (University of California, Davis, CA, USA). Tomato seeds were germinated in trays (36 cm × 25 cm × 7 cm) in sterile soil : solrite (1 : 4) at 22 ± 2 ºC, 16‐h light/8‐h dark cycle, in a glasshouse culture room. Three‐week‐old seedlings were transferred to black trays, each containing 50 pots (each 4 cm in diameter) in soil : sand (1 : 1). Stocks of M. incognita were originally obtained from Professor Uma Rao (IARI, Delhi, India) and were maintained on susceptible tomato and aubergine plants in a glasshouse culture room. Infected roots were collected; egg masses were hand dissected and kept in sterile water for hatching at room temperature. Five‐week‐old tomato seedlings were inoculated with 1500 freshly hatched J2s per plant and water (control). Whole roots at different days post‐infection (dpi) were collected with two to five technical replicates, quickly washed with water and then frozen in liquid nitrogen to prevent RNA degradation. The experiment was conducted with two biological replicates. Whole root samples were stored at −80 ºC until RNA isolation.
Disease developmental stages
The selection of different stages of infection was performed independently using acid fuchsin staining before conducting the main experiments. RKN infection was monitored for 30 days after infection in PR and 7 days after infection in M36 by staining the roots with acid fuchsin (Fisher Scientific International, Inc., NH, USA) to determine the exact developmental stages of RKN inside the roots, as described by Byrd et al. (1983). Whole mounts of roots were prepared to count the number of nematodes at each stage for both susceptible and resistance tomato lines under the stereomicroscope (Discovery v.20, Zeiss). This study led to the selection of five different infected stages in the susceptible response, two stages in the resistance response and a stage 0 (0 dpi/uninfected) (Fig. 1). As nematode infection is not synchronous, tissue from three consecutive infected days was pooled as follows to enrich the tissue for that particular stage of nematode infection: stage 1 (1, 2, 3 dpi; invasion of J2s/initiation of feeding sites); stage 2 (5, 6, 7 dpi; parasitic J2s/formation of feeding sites); stage 3 (13, 14, 15 dpi; feeding J2s and J3s/expansion of feeding sites); stage 4 (18, 19, 20 dpi; J4s/maintenance of feeding sites); stage 5 (26, 27, 28 dpi; J4s and females/maintenance of feeding sites). In the resistance response, we considered only the first two stages for the study because it has been determined that Mi‐mediated resistance is an early and rapid response, which is characterized by localized cell death at the site of infection. As a result, the disease does not progress further (Williamson, 1998). The approximate numbers of parasitic nematodes inside the infected roots at each stage were as follows: stage 1 susceptible plants had 550 nematodes, whereas stage 1 resistant plants had 211 nematodes; stage 2 susceptible plants had 680 nematodes, whereas stage 2 resistant plants had 340 nematodes; stage 3, stage 4 and stage 5 susceptible plants had 430, 700 and 800 nematodes, respectively.
Library preparation and sequencing
Total RNA isolation of uninfected controls and different infected stages from whole roots was carried out independently using Trizol (Molecular Research Center, Inc., Cincinnati, USA) following the manufacturer's protocol. The quality and quantity of isolated RNA were checked on a 1.2% denaturing agarose gel and NanoVue (GE Healthcare, IL, USA), respectively. For each sample, 4 mg of total RNA were employed to construct transcriptome libraries using a TruSeq RNA sample preparation kit v2 (Illumina, San Diego, USA) according to the manufacturer's protocol. cDNA libraries were quantified using an Agilent BioAnalyzer 2100 (Agilent Technologies, CA, USA) loaded on a paired‐end (PE) read flow cell (TruSeq v3 kit, Illumina) for the first biological replicate and on a single‐end (SE) read flow cell (TruSeq v3 kit, Illumina) for the second biological replicate. Cluster generation was performed on cBot (TruSeq PE and SE cluster kit v3‐cBot‐HS, Illumina) and sequenced on a HiSeq 2000 platform (Illumina).
Transcriptome data analysis
The 100‐bp PE and SE reads were demultiplexed using the CASAVA tool (Illumina); the quality was assessed using FastQC (Andrews, 2010) and filtered using PrinSeq (Schmieder and Edwards, 2011) and Cutadapt (Martin, 2011). A flow summary of transcriptome data analysis is presented in Fig. 8. Filtered reads were aligned independently onto S. lycopersicum genome build SL2.50 (Tomato Genome Consortium, 2012) and M. incognita WB release 5 (Abad et al., 2008) using TopHat v2.0.14 (Kim et al., 2013) on default parameters. After alignments onto the two genomes individually, uniquely mapped reads were further used for quantification by HTSeq (Anders et al., 2015) based on gene annotations. A gene was considered to be expressed and included in the downstream analysis if at least 10 reads were mapped to it aggregated across all the samples. In addition, tomato genes were annotated using the online PANTHER database (Huaiyu et al., 2016) into protein classes and Gene Ontology (GO) was performed with S. lycopersicum as the reference set. To identify transcription factors, blastx was performed against the Plant Transcription Factors Database (PlnTFDB; Jin et al., 2014) at e‐value = 1e‐5 and percentage similarity ≥ 70%.
Figure 8.
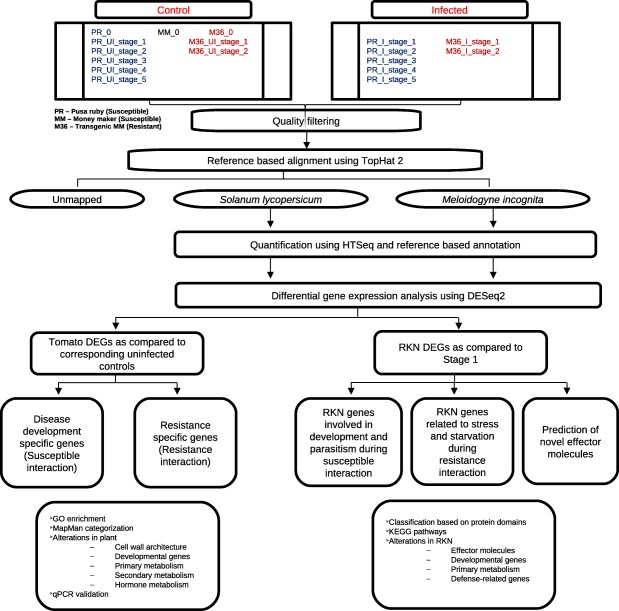
Workflow summary used for transcriptome data analysis.
Prediction of effector molecules
Protein sequences were assessed for presence of possible signal peptides using SignalP 4.1 (Petersen et al., 2011) and lack of trans‐membrane domain using TMHMM v.2.0 (Krogh et al., 2001). In addition, we carried out a blast search against the coding sequences of known effectors from Meloidogyne sp. and cyst nematodes at e‐value = 1e‐5 and percentage similarity ≥ 70%. Furthermore, ‘Annotate your protein’ tool was used on the dbCAN web server for automated annotation of carbohydrate‐active enzymes (Yin et al., 2012). Additional data mining was performed from the available literature.
Differential gene expression analysis and annotation
For both tomato and RKN, DEGs were analysed using DESeq2 (Love et al., 2014) in R (v 3.2.3, R Core Team, 2015). Further, to investigate susceptible and resistant tomato responses, the expression of different infected stages was compared with the corresponding uninfected controls to prevent bias caused by root development. A false discovery rate (FDR) cut‐off of 0.05 and log2 fold change ≥±2 was used throughout the study (unless stated otherwise). Parametric gene set enrichment analysis on tomato DEGs was performed using AgriGO v 1.2 (Du et al., 2010) with S. lycopersicum as reference set. Gene enrichment with a significant threshold of 0.05 after Hochberg FDR correction was used. Functional categorization of DEGs was performed using MapMan 3.6.0RC1 (Usadel et al., 2009). In order to highlight specific components and their functions in various pathways, we combined the annotations from gff, GO and MapMan. Redundancy in annotation was checked based on the gene locations on the tomato genome, as well as nucleotide and protein sequence similarity.
To investigate RKN responses in susceptible plants, RKN predicted genes from stage 1 were used as a baseline. In order to analyse differences in the RKN response to different host plants, RKN (same population of J2) predicted genes from resistant plants were compared with RKN predicted genes from susceptible plants of the same stage. Functional annotation of RKN DEGs from InterPro (IPR) protein domains was used for GO annotation and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used for pathway analysis. In addition, we performed blastx search against Meloidogyne sp. and cyst nematode proteins retrieved from the UniProt database. Furthermore, to improve the annotation, blastn search was performed against M. incognita stage‐specific transcriptome data (http://www6.inra.fr/meloidogyne_incognita) and transcript sequences of M. hapla (WormBase release 8) and M. floridensis (WormBase release 8). blast search was performed at e‐value = 1e‐5 and percentage similarity ≥ 70%. Redundancy in annotation at the amino acid sequence was checked. Figure S18 (see Supporting Information) shows a representative multiple sequence alignment and percentage identity matrix of the translated amino acid sequences of genes encoding PL 1.
Expression profiling by qRT‐PCR
For qRT‐PCR, the contaminating DNA was removed by treating 10 µg of total RNA with Dnase I enzyme (NEB, MA, USA). The quantity and quality of isolated RNA were checked on 1.2% denaturing agarose gel and NanoVue, respectively. Two micrograms of DNase‐treated RNA were reverse transcribed using Superscript III (Invitrogen, CA, USA) following the manufacturer's protocol. Quantitative real‐time PCR was performed with SYBR‐Green technology (Roche Holdings AG, Basel, Switzerland) on a CFX connect real‐time system (Biorad Inc., CA, USA). Specific qRT‐PCR primers were designed using the PrimerQuest tool provided by Integrated DNA Technologies (IDT Inc., IA, USA). The relative fold change was calculated using the ΔΔCt method.
The RNA‐seq data were validated for tomato genes using qRT‐PCR with the same set of tissues as used for transcriptome study. Specific primers for 20 tomato genes were designed for validation. All reactions were carried out with two technical replicates and three independent biological replicates.
To assess the expression profiles of RKN genes, independent experiments were conducted wherein total RNA from RKN J2 juveniles (approximately 10 000 juveniles), stage 2 knots (around 300 knots) and stage 5 knots (around 100 knots) was extracted using the Trizol method. The experiments were conducted using knots to enrich the tissue for the nematode stage. Specific primers for seven RKN genes were designed and the 18S rRNA gene was used as an internal control (Papolu et al., 2013). Stage 2 (early stage) was used as the control sample and the expression of different genes was quantified at stage 5 (late stage). A Student's t‐test was performed for statistical significance at P < 0.1 and P < 0.05. For all the M. incognita qRT‐PCR experiments, the specificity of the PCR amplification was tested using plant genes as negative controls. The uninfected root tissues and RKN‐infected root tissues were used to test the specificity of the amplification, and primers that showed amplification only in the RKN‐infected tissue were used for qRT‐PCR analysis. All reactions were carried out with two technical replicates and two independent biological replicates.
Author Contributions
A.K. conceived the project and designed the experiments. P.K. and N.S. set up the RKN infection in tomatoes and performed staining experiments. N.S. performed the wet experiments and the transcriptomic data analysis. Bioinformatics work was performed under the supervision of R.Y. and R.G., and R.Y. also carried out some of the bioinformatics analysis. N.S., R.Y. and A.K. wrote the manuscript and all the authors made changes to the initial and revised manuscripts, and approved the final version.
Conflicts of Interest
The authors have no conflicts of interest to declare.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website.
Fig. S1 Heatmap showing the correlation between various susceptible samples and replicates based on global expression profiles.
Fig. S2 Gene ontology (GO) annotation of significant differentially expressed genes (DEGs) of tomato detected during the susceptible response at various stages of disease development.
Fig. S3 Distribution into various protein classes of significant differentially expressed genes (DEGs) of tomato detected during the susceptible response.
Fig. S4 Distribution into various protein classes of significant differentially expressed genes (DEGs) of tomato detected at stage 2 during susceptible (a) and resistant (b) responses.
Fig. S5 Gene set enrichment analysis of molecular functions for differentially expressed genes (DEGs) of tomato detected during the susceptible response at stage 4 (a) and stage 5 (b).
Fig. S6 Schematic representation of the gene expression patterns of significant differentially expressed genes (DEGs) of tomato involved in plant cell wall architecture detected during the susceptible response.
Fig. S7 Schematic representation of the gene expression patterns of significant differentially expressed genes (DEGs) of tomato involved in plant root development detected during the susceptible response.
Fig. S8 Schematic representation of the gene expression patterns of significant differentially expressed genes (DEGs) of tomato involved in primary sugar metabolism (a), amino acid metabolism (b), lipid and fatty acid metabolism (c) and other primary metabolism categories (d) detected during the susceptible response.
Fig. S9 Schematic representation of the gene expression patterns of significant differentially expressed genes in tomato roots detected during the susceptible response: (a) sugar transporter genes; (b) lipid transporter genes; (c) aquaporin transporter genes; (d) amino acid and peptide transporter genes; (e) multidrug and toxic compound extrusion (MATE) transporter family genes; and (f) various inorganic ion transporter genes.
Fig. S10 Schematic representation of the gene expression patterns (left) of differentially expressed genes (DEGs) of tomato involved in the isoprenoid (a) and polyamine (b) biosynthetic pathways (right). Pathways showing DEGs (in red) and end products of pathways (in green) during the susceptible response.
Fig. S11 Schematic representation of the gene expression patterns of significant differentially expressed genes (DEGs) of tomato from auxin‐, cytokinin‐ and brassinosteroid‐responsive (a) and abscisic acid (ABA)‐responsive (b) categories detected during the susceptible response.
Fig. S12 Heatmap of log2 fold changes of significant differentially expressed ethylene‐responsive transcription factors detected at various stages of disease development in susceptible tomato roots.
Fig. S13 Schematic representation of the gene expression patterns of differentially expressed genes (DEGs) of tomato involved in jasmonic acid (a) and salicylic acid (b) biosynthesis and signalling, and oxidative stress‐related genes (c), detected during the susceptible response.
Fig. S14 Scatter plot showing the correlation between RNA sequencing (RNA‐seq) and quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) expression profiles.
Fig. S15 Heatmap showing the global gene expression profile of total root‐knot nematode (RKN) genes identified from infected tomato roots at five different infection time intervals.
Fig. S16 (a) Distribution of significant root‐knot nematode (RKN) differentially expressed genes (DEGs) into the top 30 InterPro (IPR) protein domains detected at different stages of infection from susceptible tomato roots relative to stage 1. (b) Distribution of significant RKN DEGs into the top 30 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways detected at different stages of infection from susceptible tomato roots relative to stage 1.
Fig. S17 Expression profiles of root‐knot nematode (RKN) genes determined by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis.
Fig. S18 (a) Multiple sequence alignment of translated amino acid sequences of root‐knot nematode (RKN) genes encoding pectate lyase 1. (b) Percentage identity matrix of translated amino acid sequences of RKN genes encoding pectate lyase 1.
Table S1 Statistics of raw reads and quality filtering of transcriptome libraries.
Table S2 Significant differentially expressed tomato genes detected at various stages of disease development during the susceptible response.
Table S3 Significant differentially expressed tomato genes detected at stage 2 during the resistance response.
Table S4 Primer sequences of tomato genes and root‐knot nematode (RKN) genes used for quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) expression analysis.
Table S5 Significant differentially expressed root‐knot nematode (RKN) genes detected at various stages of disease development during the susceptible response.
Table S6 Significant differentially expressed root‐knot nematode (RKN) genes detected at stage 2 during the resistance response.
Acknowledgements
A.K. thanks National Agricultural Science Fund – Indian Council of Agricultural Research (NASF‐ICAR), Delhi and University of Delhi – Research and Development (DU‐R&D) grant authorities for funding this research. RNA sequencing was carried out at Institute of Genomics and Integrative Biology (IGIB), Delhi, India and National Research Centre on Plant Biotechnology (NRCPB), Delhi, India. We also thank Dr Surekha‐Katiyar Agarwal (DU South Campus, Delhi, India) for her help during transcriptome library preparations. The computing facilities of Center for Biological Sequence Analysis – Technical University of Denmark (CBS‐DTU) and Computerome (both at Technical University of Denmark, Lyngby, Denmark) are gratefully acknowledged. We thank Ravi Shankar and Vandana Chawla (IHBT, Institute of Himalayan Bioresource Technology; Palampur, India) for initial help in bioinformatics analysis.
References
- Abad, P. and Williamson, V.M. (2010) Plant nematode interaction: a sophisticated dialogue In Advances in Botanical Research, Vol. 53 (Kader J.C., Delseny M., eds), pp. 147–192. Philadelphia, PA: Elsevier. [Google Scholar]
- Abad, P. , Gouzy, J. , Aury, J. , Castagnone‐Sereno, P. , Danchin, E. , Deleury, E. , Perfus‐Barbeoch, L. , Anthouard, V. , Artiguenave, F. , Blok, V.C. , Caillaud, M.C. , Coutinho, P.M. , Dasilva, C. , Luca, F. , Deau, F. , Esquibet, M. , Flutre, T. , Goldstone, J.V. , Hamamouch, N. , Hewezi, T. , Jaillon, O. , Jubin, C. , Leonetti, P. , Magliano, M. , Maier, T.R. , Markov, G.V. , McVeigh, P. , Pesole, G. , Poulain, J. , Robinson‐Rechavi, M. , Sallet, E. , Ségurens, B. , Steinbach, D. , Tytgat, T. , Ugarte, E. , Ghelder, C. , Veronico, P. , Baum, T.J. , Blaxter, M. , Bleve‐Zacheo, T. , Davis, E.L. , Ewbank, J.J. , Favery, B. , Grenier, E. , Henrissat, B. , Jones, J.T. , Laudet, V. , Maule, A.G. , Quesneville, H. , Rosso, M.‐N. , Schiex, T. , Smant, G. , Weissenbach, J. and Wincker, P. (2008) Genome sequence of the metazoan plant‐parasitic nematode Meloidogyne incognita . Nat. Biotechnol. 32, 909–915. [DOI] [PubMed] [Google Scholar]
- Anders, S. , Pyl, P.T. and Huber, W. (2015) HTSeq–A python framework to work with high‐throughput sequencing data. Bioinformatics, 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, S. (2010) FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc [accessed on April 2010].
- Bagnaresi, P. , Sala, T. , Irdani, T. , Scotto, C. , Lamontanara, A. , Beretta, M. , Rotino, G.L. , Sestili, S. , Cattivelli, L. and Sabatini, E. (2013) Solanum torvum responses to the root‐knot nematode Meloidogyne incognita . BMC Genomics, 14, 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcala, M. , Garcia, A. , Cabrera, J. , Casson, S. , Lindsey, K. , Favery, B. , García‐Casado, G. , Solano, R. , Fenoll, C. and Escobar, C. (2010) Early transcriptomic events in microdissected Arabidopsis nematode‐induced giant cells. Plant J. 61, 698–712. [DOI] [PubMed] [Google Scholar]
- Bar‐Or, C. , Kapulnik, Y. and Koltai, H. (2005) A broad characterization of the transcriptional profile of the compatible tomato response to the plant parasitic root knot nematode Meloidogyne javanica . Eur. J. Plant Pathol. 111, 181–192. [Google Scholar]
- Bellafiore, S. , Shen, Z. , Rosso, M.N. , Abad, P. , Shih, P. and Briggs, S.P. (2008) Direct identification of the Meloidogyne incognita secretome reveals proteins with host cell reprogramming potential. PLoS Pathog, 4, e1000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beneventi, M.A. , da Silva, O.B., Jr , de Sá, M.E.L. , Firmino, A.A.P. , de Amorim, R.M.S. , Albuquerque, E.V.S. , da Silva, M.C.M. , da Silva, J.P. , Campos, M.A. , Lopes, M.J.C. , Togawa, R.C. , Pappas, G.J., Jr . and de Sa, M.F.G. (2013) Transcription profile of soybean–root‐knot nematode interaction reveals a key role of phythormones in the resistance reaction. BMC Genomics, 14, 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betka, M. , Grundler, F. and Wyss, U. (1991) Influence of changes in the nurse cell system (syncytium) on the development of the cyst nematode Heterodera schachtii – single amino acids. Phytopathology, 81, 75–79. [Google Scholar]
- Bhattarai, K.K. , Xie, Q.G. , Mantelin, S. , Bishnoi, U. , Girke, T. , Navarre, D.A. and Kaloshian, I. (2008) Tomato susceptibility to root‐knot nematodes requires an intact jasmonic acid signalling pathway. Mol. Plant–Microbe Interact. 21, 1205–1214. [DOI] [PubMed] [Google Scholar]
- Branch, C. , Hwang, C.F. , Navarre, D.A. and Williamson, V.M. (2004) Salicylic acid is part of the Mi‐1‐mediated defense response to root‐knot nematode in tomato. Mol. Plant–Microbe Interact. 17, 351–356. [DOI] [PubMed] [Google Scholar]
- Byrd, D.W. , Kirkpatrick, T. and Barker, K.R. (1983) An improved technique for clearing and staining plant tissues for detection of nematodes. J. Nematol. 15, 142–143. [PMC free article] [PubMed] [Google Scholar]
- Cabrera, J. , Diaz‐Manzano, F.E. , Sanchez, M. , Rosso, M.N. , Melillo, T. , Goh, T. , Fukaki, H. , Cabello, S. , Hofmann, J. , Fenoll, C. and Escobar, C. (2014) A role for LATERAL ORGAN BOUNDARIES‐DOMAIN 16 during the interaction Arabidopsis–Meloidogyne spp. provides a molecular link between lateral root and root‐knot nematode feeding site development. New Phytol. 203, 632–645. [DOI] [PubMed] [Google Scholar]
- Castagnone‐Sereno, P. , Semblat, J.P. and Castagnone, C. (2009) Modular architecture and evolution of the map−1 gene family in the root‐knot nematode Meloidogyne incognita . Mol. Genet. Genomics, 282, 547–554. [DOI] [PubMed] [Google Scholar]
- Chini, A. , Fonseca, S. , Fernandez, G. , Adie, B. , Chico, J.M. , Lorenzo, O. , Garcia‐Casado, G. , Lopez‐Vidriero, I. , Lozano, F.M. , Ponce, M.R. , Micol, J.L. and Solano, R. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature, 448, 666–671. [DOI] [PubMed] [Google Scholar]
- Du, Z. , Zhou, X. , Ling, Y. , Zhang, Z. and Su, Z. (2010) AgriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38, 64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckardt, N.A. (2001) Move it on out with MATEs. Plant Cell, 13, 1477–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favery, B. , Quentin, M. , Jaubert‐Possamai, S. and Abad, P. (2016) Gall‐forming root‐knot nematodes hijack key plant cellular functions to induce multinucleate and hypertrophied feeding cells. J. Insect Physiol. 84, 60–69. [DOI] [PubMed] [Google Scholar]
- Fuller, V.L. , Lilley, C.J. , Atkinson, H.J. and Urwin, P.E. (2007) Differential gene expression in Arabidopsis following infection by plant‐parasitic nematodes Meloidogyne incognita and Heterodera schachtii . Mol. Plant Pathol. 8, 595–609. [DOI] [PubMed] [Google Scholar]
- Guo, W.J. and Ho, T.H.D. (2008) An abscisic acid‐induced protein, HVA22, inhibits gibberellin‐mediated programmed cell death in cereal aleurone cells. Plant Physiol. 147, 1710–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegeman, A. , Mantelin, S. , Jones, J.T. and Gheysen, G. (2012) Functional roles of effectors of plant‐parasitic nematodes. Gene, 492, 19–31. [DOI] [PubMed] [Google Scholar]
- Haegeman, A. , Bauters, L. , Kyndt, T. , Rahman, M.M. and Gheysen, G. (2013) Identification of candidate effector genes in the transcriptome of the rice root knot nematode Meloidogyne graminicola . Mol. Plant Pathol. 14, 379–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes, U.Z. , Schachtman, D.P. , Berg, R.H. , Nielsen, E. , Koch, W. , McIntyre, L.M. and Taylor, C.G. (2005) Nematode‐induced changes of transporter gene expression in Arabidopsis roots. Mol. Plant–Microbe Interact. 18, 1247–1257. [DOI] [PubMed] [Google Scholar]
- Hewezi, T. , Howe, P.J. , Maier, T.R. , Hussey, R.S. , Mitchum, M.G. , Davis, E.L. and Baum, T.J. (2010) Arabidopsis spermidine synthase is targeted by an effector protein of the cyst nematode Heterodera schachtii . Plant Physiol. 152, 968–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huaiyu, M.I. , Poudel, S. , Casagrande, A.M.J.T. and Thomas, P.D. (2016) PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 44, D336–D342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, G. , Gao, B. , Maier, T. , Allen, R. , Davis, E.L. , Baum, T.J. and Hussey, R.S. (2003) A profile of putative parasitism genes expressed in the oesophageal gland cells of the root‐knot nematode Meloidogyne incognita . Mol. Plant–Microbe Interact. 16, 376–381. [DOI] [PubMed] [Google Scholar]
- Huang, G. , Dong, R. , Allen, R. , Davis, E.L. , Baum, T.J. and Hussey, R.S. (2005) Two chorismate mutase genes from the root‐knot nematode Meloidogyne incognita . Mol. Plant Pathol. 6, 23–30. [DOI] [PubMed] [Google Scholar]
- Huang, G. , Dong, R. , Allen, R. , Davis, E.L. , Baum, T.J. and Hussey, R.S. (2006) A root‐knot nematode secretory peptide functions as a ligand for a plant transcription factor. Mol. Plant–Microbe Interact. 19, 463–470. [DOI] [PubMed] [Google Scholar]
- Hwang, C.F. and Williamson, V.M. (2003) Leucine‐rich repeat‐mediated intramolecular interactions in nematode recognition and cell death signalling by the tomato resistance protein Mi. Plant J. 34, 585–593. [DOI] [PubMed] [Google Scholar]
- Iberkleid, I. , Sela, N. and Miyara, S.B. (2015) Meloidogyne javanica fatty acid‐ and retinol‐binding protein (Mj‐FAR‐1) regulates expression of lipid‐, cell wall‐, stress‐ and phenylpropanoid‐related genes during nematode infection of tomato. BMC Genomics, 16, 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim, H.M.M. , Hosseini, P. , Alkharouf, N.W. , Hussein, E.H.A. , El‐Din, A.E.K.Y.G. , Aly, M.A.M. and Matthews, B.F. (2011) Analysis of gene expression in soybean (Glycine max) roots in response to the root‐knot nematode Meloidogyne incognita using microarrays and KEGG pathways. BMC Genomics, 12, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ilarduya, O.M. , Moore, A.E. and Kaloshian, I. (2001) The tomato Rme1 locus is required for Mi‐1‐mediated resistance to root‐knot nematodes and the potato aphid. Plant J. 27, 417–425. [DOI] [PubMed] [Google Scholar]
- Jammes, F. , Lecomte, P. , de Almeida‐Engler, J. , Bitton, F. , Martin‐Magniette, M.L. , Renou, J.P. , Abad, P. and Favery, B. (2005) Genome‐wide expression profiling of the host response to root‐knot nematode infection in Arabidopsis . Plant J. 44, 447–458. [DOI] [PubMed] [Google Scholar]
- Jaouannet, M. , Magliano, M. , Arguel, M.J. , Gourgues, M. , Evangelisti, E. , Abad, P. and Rosso, M.N. (2013) The root‐knot nematode calreticulin Mi‐CRT is a key effector in plant defense suppression. Mol. Plant–Microbe Interact. 26, 97–105. [DOI] [PubMed] [Google Scholar]
- Jaubert, S. , Milac, A.L. , Petrescu, A.J. , de Almeida‐Engler, J. , Abad, P. and Rosso, M.N. (2005) In planta secretion of a calreticulin by migratory and sedentary stages of root‐knot nematode. Mol. Plant–Microbe Interact. 18, 1277–1284. [DOI] [PubMed] [Google Scholar]
- Ji, H. , Gheysen, G. , Denil, S. , Lindsey, K. , Topping, J.F. , Nahar, K. , Haegeman, A. , de Vos, W.H. , Trooskens, G. , Criekinge, W.V. , Meyer, T.M. and Kyndt, T. (2013) Transcriptional analysis through RNA sequencing of giant cells induced by Meloidogyne graminicola in rice roots. J. Exp. Bot. 64, 3885–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, J.P. , Zhang, H. , Kong, L. , Gao, G. and Luo, J.C. (2014) PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 42, D1182–D1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 244, 323–329. [DOI] [PubMed] [Google Scholar]
- Kim, D. , Pertea, G. , Trapnell, C. , Pimentel, H. , Kelley, R. and Salzberg, S.L. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh, A. , Larsson, B. , Heijne, G. and Sonnhammer, E.L.L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. [DOI] [PubMed] [Google Scholar]
- Kumar, A. , Altabella, T. , Taylor, M.A. and Tiburcio, A.F. (1997) Recent advances in polyamine research. Trends Plant Sci. 2, 124–130. [Google Scholar]
- Kyndt, T. , Denil, S. , Haegeman, A. , Trooskens, G. , Bauters, L. , Van Criekinge, W. , De Meyer, T. and Gheysen, G. (2012) Transcriptional reprogramming by root knot and migratory nematode infection in rice. New Phytol. 196, 887–900. [DOI] [PubMed] [Google Scholar]
- Latina, R. (2015) Functional analysis of Meloidogyne graminicola C‐type lectins and their role in the nematode–rice interaction. Master's Thesis, University of Ghent, Ghent, Belgium.
- Leasure, C.D. and He, Z.H. (2012) CLE and RGF family peptide hormone signalling in plant development. Mol. Plant, 5, 1173–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q. , Xie, Q.G. , Smith‐Becker, J. , Navarre, D.A. and Kaloshian, I. (2006) Mi‐1‐mediated aphid resistance involves salicylic acid and mitogen‐activated protein kinase signalling cascades. Mol. Plant–Microbe Interact. 19, 655–664. [DOI] [PubMed] [Google Scholar]
- Lin, B. , Zhuo, K. , Wu, P. , Cui, R. , Zhang, L.H. and Liao, J. (2013) A novel effector protein, MJ‐NULG1a, targeted to giant cell nuclei plays a role in Meloidogyne javanica parasitism. Mol. Plant–Microbe Interact. 26, 55–66. [DOI] [PubMed] [Google Scholar]
- Love, M.I. , Huber, W. and Anders, S. (2014) Moderated estimation of fold change and dispersion for RNA‐Seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manosalva, P. , Manohar, M. , von Reuss, S.H. , Chen, S. , Koch, A. , Kaplan, F. , Choe, A. , Micikas, R.J. , Wang, X. , Kogel, K.H. , Sternberg, P.W. , Williamson, V.M. , Schroeder, F.C. and Klessig, D.F. (2015) Conserved nematode signalling molecules elicit plant defenses and pathogen resistance. Nat. Commun. 6, 7795. doi: 10.1038/ncomms8795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantelin, S. , Bhattarai, K.K. , Jhaveri, T.Z. and Kaloshian, I. (2013) Mi‐1‐mediated resistance to Meloidogyne incognita in tomato may not rely on ethylene but hormone perception through ETR3 participates in limiting nematode infection in a susceptible host. PLoS One, 8, e63281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M. (2011) Cutadapt removes adapter sequences from high‐throughput sequencing reads. EMBnet.journal, 17, 10–12. [Google Scholar]
- Milligan, S.B. , Bodeau, J. , Yaghoobi, J. , Kaloshian, I. , Zabel, P. and Williamson, V.M. (1998) The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine‐rich repeat family of plant genes. Plant Cell, 10, 1307–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchum, M.G. , Hussey, R.S. , Baum, T.J. , Wang, X. , Elling, A.A. , Wubben, M. and Davis, E.L. (2013) Nematode effector proteins: an emerging paradigm of parasitism. New Phytol. 199, 879–894. [DOI] [PubMed] [Google Scholar]
- Molinari, S. , Fanelli, E. and Leonetti, P. (2013) Expression of tomato SA‐responsive pathogenesis‐related genes in Mi‐1‐mediated and SA‐induced resistance to root‐knot nematodes. Mol. Plant Pathol. 15, 255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu, J.H. , Liu, P. , Liu, Q. , Chen, C. , Guo, Q. , Yin, J. , Yang, G. and Jian, H. (2016) Msp40 effector of root‐knot nematode manipulates plant immunity to facilitate parasitism. Sci. Rep. 6, 19 443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papolu, P.K. , Gantasala, N.P. , Kamaraju, D. , Banakar, P. , Sreevathsa, R. and Rao, U. (2013) Utility of host delivered RNAi of two FMRF amide like peptides, flp‐14 and flp‐18, for the management of root‐knot nematode, Meloidogyne incognita . PLoS One, 8, e80603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, T.N. , Brunak, S. , Heijne, G. and Nielsen, H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods, 8, 785–786. [DOI] [PubMed] [Google Scholar]
- Portillo, M. , Cabrera, J. , Lindsey, K. , Topping, J. , Andres, M.F. , Emiliozzi, M. , Oliveros, J.C. , Gracia‐Casado, G. , Solano, R. , Koltai, H. , Resnick, N. and Escobar, C. (2013) Distinct and conserved transcriptomic changes during nematode‐induced giant cell development in tomato compared with Arabidopsis: a functional role for gene repression. New Phytol. 197, 1276–1290. e [DOI] [PubMed] [Google Scholar]
- Postnikova, O.A. , Hult, M. , Shao, J. , Skantar, A. and Nemchinov, L.G. (2015) Transcriptome analysis of resistant and susceptible alfalfa cultivars infected with root‐knot nematode Meloidogyne incognita . PLoS One, 10, e0118269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior, A.E. , Jones, J.T. , Blok, V.C. , Beauchamp, J. , McDermott, L. , Cooper, A. and Kennedy, M.W. (2001) A surface‐associated retinol‐ and fatty acid‐binding protein (Gp‐FAR‐1) from the potato cyst nematode Globodera pallida: lipid binding activities, structural analysis and expression pattern. Biochem. J. 356, 387–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2015) R: A language and environment for statistical computing Vienna, Austria: R Foundation for Statistical Computing. Available online at: https://www.R-project.org.
- Rutter, W.B. , Hewezi, T. , Abubucker, S. , Maier, T.R. , Huang, G. , Mitreva, M. , Hussey, R.S. and Baum, T.J. (2014a) Mining novel effector proteins from the oesophageal gland cells of Meloidogyne incognita . Mol. Plant–Microbe Interact. 27, 965–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter, W.B. , Hewezi, T. , Maier, T.R. , Mitchum, M.G. , Davis, E.L. , Hussey, R.S. and Baum, T.J. (2014b) Members of the Meloidogyne avirulence protein family contain multiple plant ligand‐like motifs. Nematology, 104, 875–885. [DOI] [PubMed] [Google Scholar]
- Santini, L. , Munhoz, C.F. , Bonfim, M.F. Jr. , Brandão, M.M. , Inomoto, M.M. and Vieira, M.L.C. (2016) Host transcriptional profiling at early and later stages of the compatible interaction between Phaseolus vulgaris and Meloidogyne incognita . Nematology, 106, 282–294. [DOI] [PubMed] [Google Scholar]
- Schaff, J.E. , Nielsen, D.M. , Smith, C.P. , Scholl, E.H. and Bird, D.M. (2007) Comprehensive transcriptome profiling in tomato reveals a role for glycosyltransferase in Mi‐mediated nematode resistance. Plant Physiol. 144, 1079–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder, R. and Edwards, R. (2011) Quality control and preprocessing of metagenomic datasets. Bioinformatics, 27, 863–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semblat, J.P. , Rosso, M.N. , Hussey, R.S. , Abad, P. and Castagnone‐Sereno, P. (2001) Molecular cloning of a cDNA encoding an amphid‐secreted putative avirulence protein from the root‐knot nematode Meloidogyne incognita . Mol. Plant–Microbe Interact. 14, 72–79. [DOI] [PubMed] [Google Scholar]
- Shivakumara, T.N. , Papolu, P.K. , Dutta, T.K. , Kamaraju, D. , Chaudhary, S. and Rao, U. (2016) RNAi‐induced silencing of an effector confers transcriptional oscillation in another group of effectors in the root‐knot nematode, Meloidogyne incognita . Nematology, 18, 857–870. [Google Scholar]
- Shukla, N. , Kaur, P. and Kumar, A. (2016) Molecular aspects of plant–nematode interactions. Ind. J. Plant Physiol. 21, 477–488. DOI 10.1007/s40502‐016‐0263‐y. [Google Scholar]
- Sobczak, M. , Fudali, S. and Wieczorek, K. (2011) Cell wall modifications induced by nematodes In: Genomics and Molecular Genetics of Plant–Nematode Interactions (Jones J., Gheysen G. and Fenoll C., eds), pp. 395–422. Dordrecht: Springer. [Google Scholar]
- Stotz, H.U. , Spence, B. and Wang, Y. (2009) A defensin from tomato with dual function in defense and development. Plant Mol. Biol. 71, 131–143. [DOI] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature, 485, 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudgill, D.L. and Blok, V.C. (2001) Apomictic, polyphagous root‐knot nematodes, exceptionally successful and damaging biotrophic root pathogens. Annu. Rev. Phytopathol. 39, 53–77. [DOI] [PubMed] [Google Scholar]
- Truong, N.M. , Nguyen, C.N. , Abad, P. , Quentin, M. and Favery, B. (2015) Function of root‐knot nematode effectors and their targets in plant parasitism In: Advances in Botanical Research, Vol. 73 (Escobar C., Fenoll C., eds), pp. 293–324. Elsevier, USA. [Google Scholar]
- Usadel, B. , Poree, F. , Nagel, A. , Lohse, M. , Czedil‐Eysenberg, A. and Stitt, M. (2009) A guide to using MapMan to visualize and compare omics data in plants: a case study in the crop species, Maize. Plant Cell Environ. 32, 1211–1229. [DOI] [PubMed] [Google Scholar]
- Vieira, P. , Danchin, E.G. , Neveu, C. , Crozat, C. , Jaubert, S. , Hussey, R.S. , Engler, G. , Abad, P. , de Almeida‐Engler, J. , Castagnone‐Sereno, P. and Rosso, M.N. (2011) The plant apoplasm is an important recipient compartment for nematode secreted proteins. J. Exp. Bot. 62, 1241–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson, V.M. (1998) Root‐knot nematode resistance genes in tomato and their potential for future use. Annu. Rev. Phytopathol. 36, 277–293. [DOI] [PubMed] [Google Scholar]
- Williamson, V.M. and Hussey, R.S. (1996) Nematode pathogenesis and resistance in plants. Plant Cell, 8, 1735–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson, V.M. and Kumar, A. (2006) Nematode resistance in plants: the battle underground. Trends Genet. 22, 396–403. [DOI] [PubMed] [Google Scholar]
- Xie, J. , Li, S. , Mo, C. , Wang, G. , Xiao, X. and Xiao, Y. (2016) A novel Meloidogyne incognita effector Misp12 suppresses plant defense response at later stages of nematode parasitism. Front. Plant Sci. 7, 964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, B. , Hamamouch, N. , Li, C. , Huang, G. and Hussey, R.S. (2013) The 8D05 parasitism gene of Meloidogyne incognita is required for successful infection of host roots. Phytopathology, 103, 175–181. [DOI] [PubMed] [Google Scholar]
- Yamashiro, S. , Gimona, M. and Ono, S. (2007) UNC‐87, a calponin repeat protein in C. elegans, antagonizes ADF/cofilin‐mediated actin filament dynamics. J. Cell Sci. 120, 3022–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Y. , Mao, X. , Yang, J.C. , Chen, X. , Mao, F. and Xu, T. (2012) A web resource for automated carbohydrate‐active enzyme annotation. Nucleic Acids Res. 40, W445–W451. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website.
Fig. S1 Heatmap showing the correlation between various susceptible samples and replicates based on global expression profiles.
Fig. S2 Gene ontology (GO) annotation of significant differentially expressed genes (DEGs) of tomato detected during the susceptible response at various stages of disease development.
Fig. S3 Distribution into various protein classes of significant differentially expressed genes (DEGs) of tomato detected during the susceptible response.
Fig. S4 Distribution into various protein classes of significant differentially expressed genes (DEGs) of tomato detected at stage 2 during susceptible (a) and resistant (b) responses.
Fig. S5 Gene set enrichment analysis of molecular functions for differentially expressed genes (DEGs) of tomato detected during the susceptible response at stage 4 (a) and stage 5 (b).
Fig. S6 Schematic representation of the gene expression patterns of significant differentially expressed genes (DEGs) of tomato involved in plant cell wall architecture detected during the susceptible response.
Fig. S7 Schematic representation of the gene expression patterns of significant differentially expressed genes (DEGs) of tomato involved in plant root development detected during the susceptible response.
Fig. S8 Schematic representation of the gene expression patterns of significant differentially expressed genes (DEGs) of tomato involved in primary sugar metabolism (a), amino acid metabolism (b), lipid and fatty acid metabolism (c) and other primary metabolism categories (d) detected during the susceptible response.
Fig. S9 Schematic representation of the gene expression patterns of significant differentially expressed genes in tomato roots detected during the susceptible response: (a) sugar transporter genes; (b) lipid transporter genes; (c) aquaporin transporter genes; (d) amino acid and peptide transporter genes; (e) multidrug and toxic compound extrusion (MATE) transporter family genes; and (f) various inorganic ion transporter genes.
Fig. S10 Schematic representation of the gene expression patterns (left) of differentially expressed genes (DEGs) of tomato involved in the isoprenoid (a) and polyamine (b) biosynthetic pathways (right). Pathways showing DEGs (in red) and end products of pathways (in green) during the susceptible response.
Fig. S11 Schematic representation of the gene expression patterns of significant differentially expressed genes (DEGs) of tomato from auxin‐, cytokinin‐ and brassinosteroid‐responsive (a) and abscisic acid (ABA)‐responsive (b) categories detected during the susceptible response.
Fig. S12 Heatmap of log2 fold changes of significant differentially expressed ethylene‐responsive transcription factors detected at various stages of disease development in susceptible tomato roots.
Fig. S13 Schematic representation of the gene expression patterns of differentially expressed genes (DEGs) of tomato involved in jasmonic acid (a) and salicylic acid (b) biosynthesis and signalling, and oxidative stress‐related genes (c), detected during the susceptible response.
Fig. S14 Scatter plot showing the correlation between RNA sequencing (RNA‐seq) and quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) expression profiles.
Fig. S15 Heatmap showing the global gene expression profile of total root‐knot nematode (RKN) genes identified from infected tomato roots at five different infection time intervals.
Fig. S16 (a) Distribution of significant root‐knot nematode (RKN) differentially expressed genes (DEGs) into the top 30 InterPro (IPR) protein domains detected at different stages of infection from susceptible tomato roots relative to stage 1. (b) Distribution of significant RKN DEGs into the top 30 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways detected at different stages of infection from susceptible tomato roots relative to stage 1.
Fig. S17 Expression profiles of root‐knot nematode (RKN) genes determined by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis.
Fig. S18 (a) Multiple sequence alignment of translated amino acid sequences of root‐knot nematode (RKN) genes encoding pectate lyase 1. (b) Percentage identity matrix of translated amino acid sequences of RKN genes encoding pectate lyase 1.
Table S1 Statistics of raw reads and quality filtering of transcriptome libraries.
Table S2 Significant differentially expressed tomato genes detected at various stages of disease development during the susceptible response.
Table S3 Significant differentially expressed tomato genes detected at stage 2 during the resistance response.
Table S4 Primer sequences of tomato genes and root‐knot nematode (RKN) genes used for quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) expression analysis.
Table S5 Significant differentially expressed root‐knot nematode (RKN) genes detected at various stages of disease development during the susceptible response.
Table S6 Significant differentially expressed root‐knot nematode (RKN) genes detected at stage 2 during the resistance response.


