Summary
The effector protein AvrP is secreted by the flax rust fungal pathogen (Melampsora lini) and recognized specifically by the flax (Linum usitatissimum) P disease resistance protein, leading to effector‐triggered immunity. To investigate the biological function of this effector and the mechanisms of specific recognition by the P resistance protein, we determined the crystal structure of AvrP. The structure reveals an elongated zinc‐finger‐like structure with a novel interleaved zinc‐binding topology. The residues responsible for zinc binding are conserved in AvrP effector variants and mutations of these motifs result in a loss of P‐mediated recognition. The first zinc‐coordinating region of the structure displays a positively charged surface and shows some limited similarities to nucleic acid‐binding and chromatin‐associated proteins. We show that the majority of the AvrP protein accumulates in the plant nucleus when transiently expressed in Nicotiana benthamiana cells, suggesting a nuclear pathogenic function. Polymorphic residues in AvrP and its allelic variants map to the protein surface and could be associated with differences in recognition specificity. Several point mutations of residues on the non‐conserved surface patch result in a loss of recognition by P, suggesting that these residues are required for recognition.
Keywords: crystal structure, effector‐triggered immunity, flax rust (Melampsora lini) effector, NLR [nucleotide‐binding and oligomerization domain (NOD)‐like receptor, nucleotide‐binding/leucine‐rich repeat receptor], nuclear localization, plant disease resistance, zinc finger
Introduction
During infection, many plant pathogens deliver proteins known as effectors into host cells to aid colonization. To counteract this infection strategy, plants have evolved intracellular receptors, known as resistance (R) proteins, which recognize effectors and initiate defence responses, collectively known as effector‐triggered immunity (ETI) (Dodds and Rathjen, 2010; Jones and Dangl, 2006). ETI is typically defined by the hypersensitive response (HR), which results in death of the infected cell. Most R proteins contain nucleotide‐binding (NB) and leucine‐rich repeat (LRR) domains and are related to mammalian nucleotide‐binding and oligomerization domain (NOD)‐like receptors (NLRs) (Bonardi et al., 2012). Effectors are often highly variable, both within and between species, and the recognition of effectors is highly specific. Pathogen effector–plant R protein interactions were described genetically in the 1950s by the gene‐for‐gene theory (Flor, 1956). Effectors recognized by R proteins are termed avirulence (Avr) proteins. The recognition of Avr proteins by R proteins is accomplished by direct association, indirect association through accessory proteins or the recognition of host proteins modified by Avr proteins (Dodds and Rathjen, 2010).
Effectors from filamentous eukaryotic plant pathogens, such as fungi and oomycetes, have a high level of genetic diversity, and few features have been identified in their protein sequences to help predict biological functions (Bozkurt et al., 2012; Oliveira‐Garcia and Valent, 2015; Rafiqi et al., 2012). Fungal and oomycete intracellular effectors have been found to suppress immunity by interaction with host proteins to modulate plant metabolism, including hormone signalling and transcription; however, only a few have been assigned a specific function (Djamei et al., 2011; McLellan et al., 2013; Okmen and Doehlemann, 2014). Increasing numbers of fungal and oomycete effectors have been reported to move into the host nucleus, suggesting that these effectors may interfere with host nuclear processes. For instance, the effector domains of oomycete Crinkler (CRN) effectors target the host nucleus and accumulate in specific subnuclear compartments, leading to necrosis (Ramirez‐Garces et al., 2016; Schornack et al., 2010; Stam et al., 2013a, 2013b). The Phytophthora PSR (suppressor of RNA silencing) effector binds to the Arabidopsis PSR1‐interacting protein 1 (PINP1), which regulates small RNA accumulation in the host nucleus, to promote infection (Qiao et al., 2015). The Ustilago maydis effector See1 (seedling efficient effector 1) interacts with a maize homologue of yeast suppressor of the G2 allele of skp1 (SGT1) in both the cytosol and nucleus of leaf cells, resulting in re‐activation of plant DNA synthesis and tumour formation (Redkar et al., 2015). A putative Colletotrichum graminicola effector, CgEP1 (Colletotrichum graminicola effector protein 1), targets the host nucleus and binds directly to maize genomic DNA (Vargas et al., 2016).
Genetic studies of the interaction between flax (Linum usitatissimum) and its rust pathogen (Melampsora lini) have identified a number of R genes in the host and Avr genes in the rust pathogen. R genes at the L, M, N and P loci have been cloned and encode NLR proteins with an N‐terminal Toll/interleukin‐1 receptor (TIR) domain (Ravensdale et al., 2011). In flax rust, Avr genes have been cloned from six loci, AvrL567, AvrL2, AvrM, AvrM14, AvrP123 and AvrP4, and all encode secreted proteins (Anderson et al., 2016; Barrett et al., 2009; Catanzariti et al., 2006; Dodds et al., 2004). Apart from AvrM14, which shows homology to nudix hydrolases, these Avr proteins have no homologues with defined functions in current databases. Because these Avr proteins are recognized by intracellular host R proteins, they have been proposed to enter plant cells during infection, a process that has been observed for AvrM by confocal microscopy (Rafiqi et al., 2010). A physical interaction between Avr and R proteins has been shown using yeast two‐hybrid (Y2H) assays for the AvrL567:L5/L6/L7 pairs and the AvrM:M pair (Catanzariti et al., 2010; Dodds et al., 2006). Structure‐guided mutagenesis studies have revealed that specific recognition of AvrL567 effectors results from multiple amino acid contacts between Avr and R proteins (Wang et al., 2007). In the case of AvrM, the C‐terminal domain of AvrM is required for M‐dependent cell death and is directly involved in the interaction (Catanzariti et al., 2010; Ve et al., 2013).
The screening of an M. lini haustorium‐specific cDNA library revealed a haustorially expressed secreted protein gene cosegregating with a gene at the AvrP123 locus (Catanzariti et al., 2006). So far, 10 AvrP123 alleles have been identified from rust isolates collected from flax species L. usitatissimum and Linum marginale, including AvrP and AvrP123 (Barrett et al., 2009; Catanzariti et al., 2006; Lawrence et al., 1981b). In flax, at least six rust resistance genes (P, P1–P5) occur at the complex P locus (Islam and Mayo, 1990). The P and P1–P3 resistance specificities are defined by the recognition of allelic variants of AvrP123 (Barrett et al., 2009; Catanzariti et al., 2006; Dodds and Thrall, 2009). Transient expression of AvrP123 induces cell death activities in flax plants containing the P1, P2 or P3 resistance genes, and AvrP induces P‐dependent cell death activity (Catanzariti et al., 2006; Dodds and Thrall, 2009). AvrP123 and its allelic variants, including AvrP, are cysteine (Cys)‐rich proteins. They contain a sequence motif resembling that found in Kazal family serine protease inhibitors, where the Cys residues form disulfide bonds (Barrett et al., 2009; Catanzariti et al., 2006). However, recent biochemical studies of purified AvrP protein have suggested that the Cys residues may instead bind metal ions, and the arrangement of Cys residues in AvrP is similar to that in the plant homeodomain (PHD) motif, which binds zinc (Zn) ions (Zhang et al., 2017).
To further investigate fungal effector function and recognition by R proteins, we determined the crystal structure of AvrP. Structural comparisons reveal similarities to proteins involved in gene regulation, and microscopic analysis indicates nuclear enrichment of AvrP when expressed in planta. Structure‐guided mutagenesis and transient expression assays reveal surface residues of AvrP that are required for recognition by the P resistance protein.
Results
Crystal structure of AvrP
The AvrP and AvrP123 genes are alleles at the AvrP123 locus, encoding proteins with a 23‐amino‐acid N‐terminal signal peptide (Barrett et al., 2009; Catanzariti et al., 2006). The predicted mature forms of the AvrP and AvrP123 proteins, consisting of residues 24–111 and 24–117, respectively, were expressed in Escherichia coli and purified as described previously (Zhang et al., 2017). AvrP was crystallized using ZnCl2 as an additive; however, AvrP123 failed to crystallize (Zhang et al., 2017). The structure of AvrP was solved using the multi‐wavelength anomalous diffraction (MAD) approach (Table S1, see Supporting Information). The final model of AvrP contains two molecules in the asymmetric unit with a root‐mean‐square deviation of 0.5 Å. The refined model of the AvrP structure encompasses residues 26–102; the N‐terminal residues 24–25 and the C‐terminal residues 103–111 have no interpretable electron density, suggesting that they are flexible within the crystal. The structure adopts an elongated shape, containing four β‐strands (β1–β4) and a C‐terminal α‐helix (α1) connected by loops (Fig. 1a). The structure shows three Zn ions bound in each protein molecule. Each Zn ion has a tetrahedral coordination with either four Cys or three Cys and one histidine (His) residue (Fig. 1b–d). The four β‐strands form an elongated and twisted β‐sheet, and coordinate Zn1 by residues C36, C38, C78 and C81 (Fig. 1a,b,e). Zn2 is coordinated by residues C53 and C67 in the elongated β2β3 loop, C89 in the β4 strand and H93 in the β4α1 loop. Zn3 is coordinated by residues C59 and C61 in the β2β3 loop, C97 in the α1 helix and H101 in the C‐terminal loop.
Figure 1.
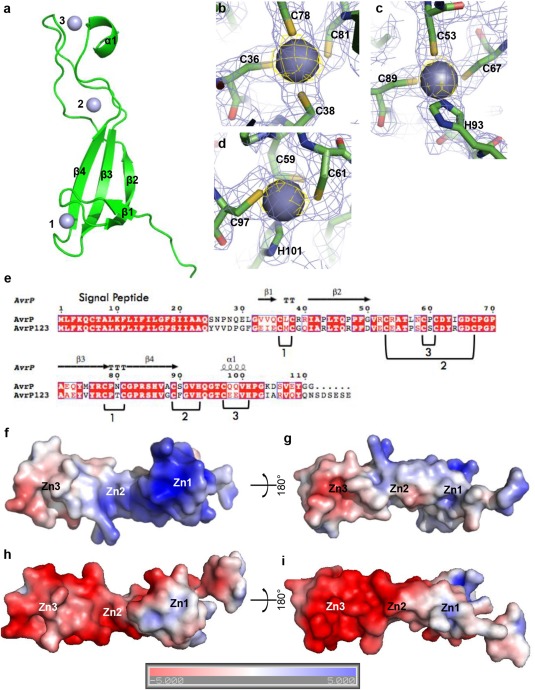
The crystal structure of AvrP. (a) Ribbon diagram showing the overall structure of AvrP. Zinc (Zn) ions are shown as numbered spheres. (b–d) Tetrahedral coordination of Zn ions with CCCC (b) and CCCH (c, d) residue combinations. The images show the electron density map (blue mesh) of AvrP contoured at 1.0 σ and the anomalous difference map (yellow mesh) for the Zn ions contoured at 4.0 σ. The AvrP residues are shown as sticks and the Zn ions are shown as spheres. (e) Sequence alignment of AvrP and AvrP123. The secondary structure elements of AvrP are displayed above the alignment. ‘T’ corresponds to a β‐turn. The Zn‐binding motifs are numbered and the Zn‐binding residues are indicated under the alignment. (f–i) Surface electrostatic potentials of AvrP (f, g) and AvrP123 (h, i). Positive potential is shown in blue and negative potential is shown in red (coloured continuously between −5 and 5 kT/e). Zn1, Zn2 and Zn3 indicate the relative positions of the Zn ions.
The interface between the two molecules within the asymmetric unit of AvrP crystals features few side‐chain interactions and does not appear to be biologically relevant. Size‐exclusion chromatography coupled with multi‐angle light scattering (SEC‐MALS) showed that both AvrP and AvrP123 are monomers in solution (experimental and theoretical molecular weights correspond to 10.5 and 9.8 kDa for AvrP, and 10.7 and 10.6 kDa for AvrP123, respectively) (Fig. S1a, see Supporting Information).
Examination of the surface electrostatic potential of AvrP reveals an uneven distribution. The Zn1‐binding region forms a positively charged head (Fig. 1f). The C‐terminal α‐helix and the β2β3 loop within the Zn3‐binding motif feature a negatively charged surface patch (Fig. 1g). Modelling the AvrP123 structure using AvrP as a template (sequence identity, 60%) reveals an extended negatively charged surface area in the Zn3‐binding region of AvrP123 (Fig. 1h,i), whereas the positively charged surface patch in the Zn1‐binding region is much smaller than that of AvrP (Fig. 1h). The flax rust AvrM effectors contain positively charged surface patches that mediate binding to head‐groups of negatively charged phospholipids, such as phosphatidylinositol phosphates (PIPs), but this binding ability is not essential for plant intracellular accumulation of secreted AvrM (Ve et al., 2013). AvrP and AvrP123 did not bind to PIPs in a dot‐blot lipid‐binding assay (Fig. S1b), indicating that they are unlikely to enter the plant cells through a PIP‐binding pathway. Both AvrP and AvrP123 showed weak binding to phosphatidic acid at protein concentrations of 0.3 μg/mL. However, this is likely to be non‐specific, given the significantly lower affinity compared with the positive control AvrM.
Zn‐binding motifs in AvrP have features of Zn‐finger domains
The Zn‐binding topology of AvrP is similar to the treble‐clef Zn‐finger domain family, which includes PHD, RING (really interesting new gene), B‐box and FYVE (domain shared by Fab1, YOTB, Vac1 and EEA1) domains. These domains each bind to two Zn ions through interleaved Zn‐binding motifs, known as the ‘cross‐brace’ topology, in which the first and third C/C or C/H pairs bind to the first Zn ion and the second and fourth C/C or C/H pairs bind to the second Zn ion (Grishin, 2001) (Fig. S2a, see Supporting Information). Each subgroup of the treble‐clef Zn‐finger family is characterized by distinct arrangements of the Zn ion‐coordinating residues. However, they have similar protein folds consisting of two orthogonally positioned β‐hairpins and C‐terminal α‐helices (ββα) (Fig. S2a,b). Each β‐turn of the β‐hairpins contains a pair of residues that are involved in binding two different Zn ions. The AvrP structure contains a similar, but distinct, ‘cross‐brace’ topology, with six C/C or C/H pairs bound to three Zn ions (Fig. 1e and S2c,d). The secondary structure of AvrP has a ββα order similar to the treble‐clef Zn‐finger family proteins with a Cys pair located in each β‐turn. The second β‐hairpin of AvrP (in the Zn1 region) contains the sequence motif CPXCG, which is common in treble‐clef Zn‐finger domains (Wang et al., 1998). However, in AvrP, the two β‐hairpins are separated by a long loop and are involved in the binding of one Zn ion.
Zn‐binding motifs are required for AvrP‐triggered cell death
The addition of ZnCl2 to the growth medium enhanced the production of AvrP protein in E. coli and Zn was required for AvrP crystallization (Zhang et al., 2017). Therefore, we tested the effect of site‐directed mutations in Zn‐binding motifs on the ability of AvrP to trigger P‐dependent cell death in planta. Three mutants were made, targeting each of the Zn‐binding sites separately by replacing all four coordinating Cys and His residues of Zn1, Zn2 or Zn3 with alanine. Mature wild‐type AvrP protein and its mutant variants were expressed in transgenic Nicotiana tabacum (tobacco) leaves containing the flax P resistance gene by Agrobacterium infiltration. The AvrP constructs, either with or without a haemagglutinin (HA) tag, triggered cell death in P‐containing tobacco leaves, whereas the Zn‐binding motif mutants (HA‐tagged) failed to trigger cell death (Fig. 2a). All AvrP proteins are detectable by immunoblotting, although the Zn1 and Zn2 mutant variants accumulated to lower levels than the wild‐type and Zn3 mutant proteins (Fig. 2b), indicating that the Zn1 and Zn2 mutations may affect protein stability. The low protein accumulation of the Zn1 and Zn2 mutants may be insufficient to trigger P‐dependent cell death. Collectively, these data suggest that the integrity of each of the three Zn‐binding sites is important for AvrP recognition by P, probably because they are required for the adoption of the correct folded structure of the protein.
Figure 2.
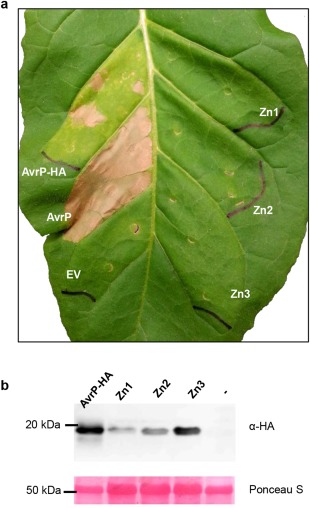
Agrobacterium‐mediated expression of zinc (Zn)‐binding motif mutants of AvrP. AvrP protein (lacking the signal peptide) fused with a haemagglutinin (HA) tag (AvrP‐HA) or without a tag (AvrP), and HA‐tagged AvrP containing mutations in the Zn‐binding residues (Zn1, Zn2 and Zn3), were expressed in a transgenic tobacco leaf containing the P resistance gene. (a) Leaf photographed at 5 days post‐infiltration (dpi). Empty vector (EV) was used as a negative control. (b) Protein extracts from infiltrated leaf tissue expressing HA‐tagged AvrP or Zn mutant proteins as in (a) or from non‐infiltrated leaf tissue (–) were size fractionated by gel electrophoresis and analysed by immunoblotting with antibodies against HA. Samples were collected 2 days after infiltration. The positions and sizes (kDa) of protein molecular mass standards are indicated. The lower panel shows Ponceau S staining of ribulose‐1,5‐bisphosphate carboxylase/oxygenase (Rubisco) on the blotted nitrocellulose membrane to indicate relative protein loading. The western blot shows representative images from two independent experiments.
Structural comparisons and functional implications
To investigate the similarity of AvrP to other known structures in the Protein Data Bank (PDB), structure‐based similarity searches were performed using the Dali server (Holm and Rosenstrom, 2010). Dali reports structural similarities based on a Z‐score, where proteins with Z‐scores > 2 are considered to have similar folds (Holm and Rosenstrom, 2010). Although no high‐scoring structural similarities were found that encompassed the entire AvrP structure, a region involving both hairpin β‐strands and the Zn1‐binding site in AvrP showed structural similarities with several proteins (Fig. 3a–e). The structural similarities in this region are unlikely to have been detected based on sequence alone, as the protein sequence identities with AvrP are low (∼10%) (Fig. 3f). Of the four most closely related structures, ZPR1 is the only protein shown to bind Zn (Fig. 3d) (Mishra et al., 2007). The proteins that showed similarity to AvrP in this region are over‐represented by proteins involved in transcription and translation, suggesting that AvrP may possess a similar function. We therefore performed nucleic acid‐binding assays with AvrP and AvrP123, but no significant nucleic acid‐binding activity was observed under the conditions used (Fig. S3, References S1, see Supporting Information). AvrP showed weak single‐stranded RNA (ssRNA) binding at high (15 µm) protein concentrations but, given the concentrations used, this appears unlikely to be of biological significance.
Figure 3.
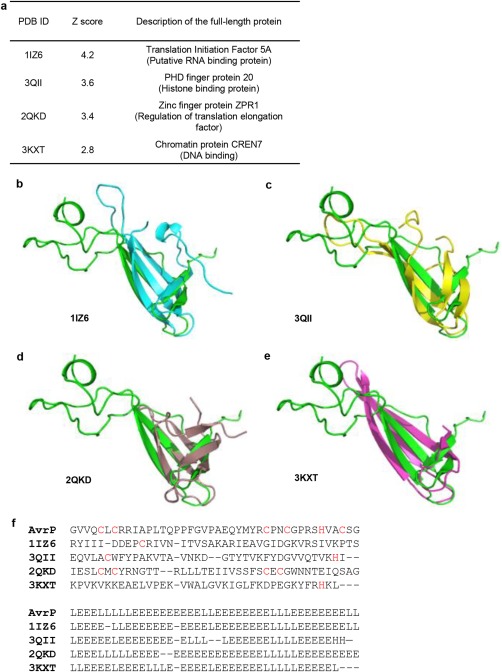
Structural alignment of AvrP and Dali hits with low sequence similarity. (a) Representative hits from Dali structure search. (b–e) Structure superimposition of AvrP and the hits, showing the similarities in the Zn1‐binding region. The AvrP structure is shown in green. The Protein Data Bank (PDB) IDs of the superimposed structures are indicated below each image. The residue regions aligned with AvrP and their corresponding domains are 16–63 (SH3‐like β‐barrel), 26–66 (Tudor domain), 11–58 (zinc finger) and 6–49 (not assigned) for LIZ6 (Yao et al., 2003), 3QII (Adams‐Cioaba et al., 2012), 2QKD (Mishra et al., 2007) and 3KXT (Feng et al., 2010), respectively. (f) Structure‐based sequence alignment (top) generated using Dali, showing that AvrP has low sequence identities with the Dali hits. Only sequences of the superimposed structures are shown. Cysteine and histidine residues are highlighted in red. The bottom panel shows the alignment of elements of secondary structure; helices, β‐strands and loops are shown as H, E and L, respectively.
AvrP and AvrP123 localize preferentially to the nuclei of plant cells
The localization of AvrP and AvrP123 inside plant cells was investigated by Agrobacterium‐mediated expression of C‐terminal citrine (CTR) fusion proteins in N. benthamiana leaves (Fig. 4a). Compared with CTR alone, which showed a nuclear–cytoplasmic distribution, both AvrP‐CTR and AvrP123‐CTR fusion proteins showed nuclear accumulation, although some cytosolic fluorescence was also detected. To quantify the nuclear enrichment of AvrP and AvrP123, the ratio of nuclear to cytosolic fluorescence intensity was calculated. Compared with CTR alone, AvrP‐CTR and AvrP123‐CTR showed more than a two‐fold and six‐fold increase in nuclear relative to cytosolic fluorescence, respectively (Fig. 4b; Table S2, see Supporting Information). The expected molecular masses of AvrP and AvrP123 fused with the CTR tag are 36.9 and 37.7 kDa, respectively, and so these proteins may be small enough to diffuse into the plant nucleus (Wang and Brattain, 2007). However, the enhanced accumulation of AvrP and AvrP123 in the nucleus, compared with the cytosol, suggests that they are retained in the nucleus. No nuclear localization motifs were evident in these protein sequences, and so the mechanism of nuclear targeting is unknown. CTR‐tagged AvrP and AvrP123 were able to trigger cell death in transgenic flax containing P and P2, respectively, indicating that they remained recognizable by their corresponding R proteins (Fig. S4, see Supporting Information).
Figure 4.
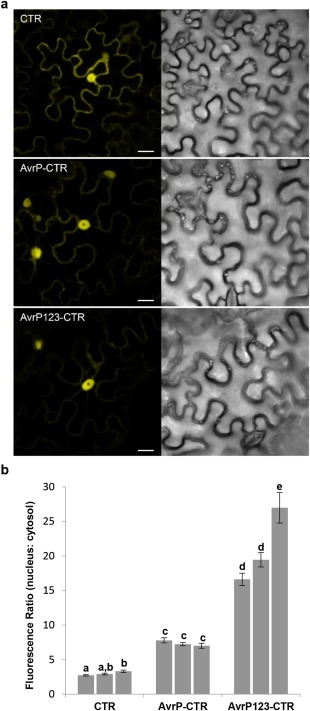
Subcellular localization of citrine (CTR)‐tagged AvrP and AvrP123. (a) Confocal images of Nicotiana benthamiana cells showing an increased ratio of fluorescence in the nucleus relative to the cytosol in leaves expressing AvrP‐CTR and AvrP123‐CTR, compared with CTR alone. The fluorescence image (left) and corresponding transmitted light image (right) were taken at 2 days post‐infiltration (dpi). Bars, 20 µm. (b) Ratio of average fluorescence intensity of nucleus to cytosol in transformed cells. Column values represent mean ratios ± standard error (SE) (n = 30) from three independent experiments for each protein. Different letters denote a significant difference, whereas the same letters denote no significant difference (two‐tailed t‐test, P < 0.05).
Non‐conserved positions are enriched on one side of the AvrP structure
A number of different AvrP123 alleles have been identified from rust isolates collected from flax, L. marginale (Lm) or L. usitatissimum (Lu), including an intra‐allelic recombinant between AvrP and AvrP123, designated as bs25 (Barrett et al., 2009; Catanzariti et al., 2006; Lawrence et al., 1981b). They show varying recognition specificities by the corresponding P, P1, P2 and P3 resistance proteins (Fig 5a and S5, see Supporting Information) (Dodds and Thrall, 2009). A high level of polymorphism is observed amongst these seven AvrP123 variants (Fig. 5b), with 36 differences found between the mature AvrP (Lu4) and AvrP123 (Lu1) proteins. However, the 10 Cys and two His residues involved in Zn binding are strictly conserved amongst all the variants. Two sequence‐related subgroups can be defined. The first, designated here as the AvrP group, includes Lu2, Lu3, Lu6 and AvrP, which share sequence identities of 86%–98%. The second group, designated here as the AvrP123 group, includes AvrP123 and Lm1, which share an identity of 89% and contain a six‐amino‐acid extension at the C‐terminus compared with the AvrP group. The sequence identity between the two groups is ∼60%. The bs25 protein is identical to AvrP for the first 60 amino acids and to AvrP123 for the remainder of the protein, and is recognized exclusively by the P2 resistance protein (Dodds and Thrall, 2009). Therefore, the C‐terminal region of the AvrP protein most probably influences recognition by the P resistance protein, and the N‐terminal region may influence recognition by P1 and P3. The Lu2 protein has eight amino acid differences compared with AvrP and is not detected by any of the P protein variants. Most of the residues that differ between Lu2 and AvrP localize to the C‐terminal region, which further supports the role of this region in the recognition by P.
Figure 5.
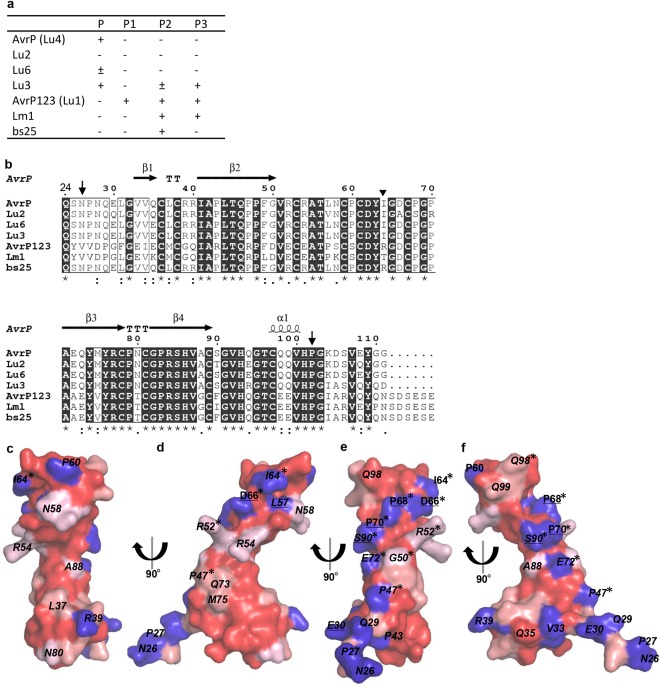
Polymorphic residues on the surface of AvrP variants. (a) The recognition spectra of AvrP variants by P family resistance proteins (Dodds and Thrall, 2009). The AvrP variants were expressed in flax leaves containing the flax P family resistance genes by Agrobacterium infiltration. Plants were scored 12 days after infiltration. ‘+’ indicates that cell death was induced, and ‘–’ indicates that no response was induced. (b) Sequence alignment of AvrP variants. The sequences of mature proteins were aligned using Clustal Omega (Sievers et al., 2011). Residue conservation is indicated below the alignment by an asterisk (fully conserved), a colon (strongly similar) and a period (weakly similar), whereas blank means that there is no similarity. The secondary structure elements of AvrP are shown on top of the alignment and the region included in the structure is located between the downward arrows. ‘T’ corresponds to a β‐turn. The inverted triangle indicates the point at which the bs25 sequence changes from AvrP to the AvrP123 sequence. (c–f) Conservation mapping on the surface of the AvrP structure. The degree of conservation, from strong to weak, is indicated by dark to light red. Non‐conserved residues are shown in blue. Surface residues that are polymorphic between AvrP and Lu2, and AvrP and AvrP123, are underlined and italicized, respectively. Residues targeted for mutation are marked with an asterisk.
The polymorphic residues of the AvrP123 variants are spread over the entire protein sequence. To investigate their structural distribution, we scored residue conservation using Clustal Omega (Sievers et al., 2011) and mapped the non‐conserved residues onto the surface of the AvrP structure. This analysis showed conserved surface patches on the concave side of the structure and the surface of the β3 and β4 strands (Fig. 5c,d), whereas non‐conserved residues are enriched on the opposite side of the protein (designated the variable surface) (Fig. 5e,f). On the variable surface, eight residues at non‐conserved positions occur close to one another, including residues I64, D66, P68 and P70 in the β2β3 loop, and residue S90 in the β4α1 loop (using AvrP numbering), which form a line threaded across the surface of the structure. In addition to these five residues, P47 in the β2 strand and E72 in the β3 strand are located on the same relatively flat surface. The non‐conserved N‐terminal residues N26, P27, Q29 and E30 further extend the variable surface. The non‐conserved residues in the C‐terminal region (103–111) could not be mapped as this region is missing from the structure. Amongst the eight residues polymorphic between the AvrP and Lu2 proteins, four are in the variable surface at positions 66, 68, 70 and 90. Notably, residue 66 changes from aspartate (containing a negatively charged side‐chain) in AvrP to alanine in Lu2, and residue 70 changes from proline to arginine (containing a positively charged side‐chain). These observations suggest that residues 66, 68, 70 and 90 may be involved in P recognition. However, residues D66, P68 and P70 do not differ between AvrP and AvrP123, and their roles in P1, P2 or P3 recognition are currently unknown.
Surface polymorphic residues of AvrP are involved in R protein recognition
To further investigate the variable surface, we tested whether mutations of non‐conserved residues in the variable surface of AvrP affect the P‐mediated cell death response. Mutations were introduced into the non‐conserved residues of AvrP, including P47R, I64R, E72A, D66A, P68S, P70R and S90T. As the C‐terminal region of AvrP may be responsible for P recognition, we also tested Q94E and E108Q, which differ between AvrP and Lu2, although Q94 is not in the variable surface, and E108 is not modelled in the structure of AvrP, as well as residues G50 and R52, which differ between AvrP and AvrP123 and occur at the periphery of the variable surface. When expressed in P‐containing flax as yellow fluorescent protein (YFP) fusions, the P47R, I64R and P70R mutants of AvrP did not trigger cell death, whereas the other mutants triggered cell death responses comparable with that of the wild‐type protein (Fig. 6a). Although protein expression of the YFP‐tagged AvrP and mutants could not be detected in flax (Fig. S6a, see Supporting Information), the same constructs were transiently expressed in N. benthamiana and stable protein overexpression was detected for all of them, except for the P47R and P70R mutants (Fig. S6b), which seemed to accumulate at a lower level, indicating that these proteins may be unstable. When expressed in P‐containing transgenic tobacco as HA fusions, the P47R, I64R and D66A mutants abolished P‐mediated cell death activity, whereas the P68S, P70R, S90T, Q94E and E108Q mutations led to weaker cell death activity compared with the wild‐type protein (Figs 6b and S7, see Supporting Information). All proteins were detectable in N. tabacum, except for the P47R mutant (Fig. 6c). Collectively, these data show that mutations of several residues in the variable surface and the C‐terminal Q94 and E108 residues of AvrP affect P‐mediated cell death activity.
Figure 6.
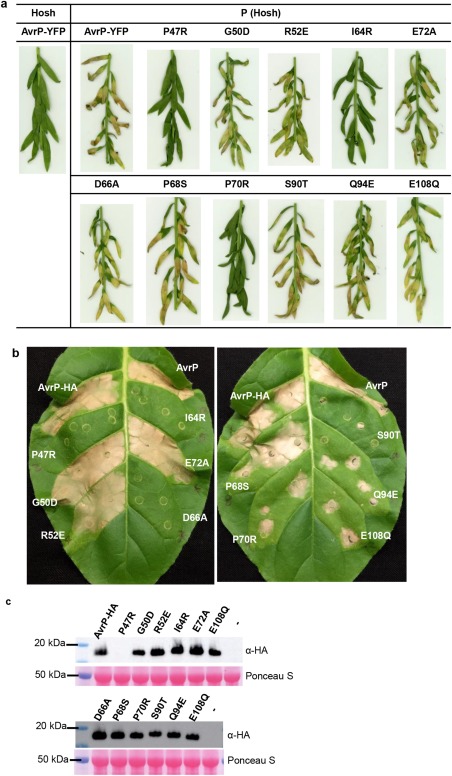
Mutational analysis of AvrP residues. (a) Cell death assay of yellow fluorescent protein (YFP)‐tagged AvrP or AvrP mutants transiently expressed in flax lines with [P (Hosh)] or without (Hosh) the P gene. (b) Transgenic tobacco leaf containing the P resistance gene from flax was infiltrated with Agrobacterium cultures containing plasmids expressing AvrP, or haemagglutinin (HA)‐tagged AvrP (AvrP‐HA), and corresponding HA‐tagged mutants. (c) Protein extracts from (b) and non‐infiltrated (–) leaf tissue were size fractionated by gel electrophoresis and analysed by immunoblotting with anti‐HA antibodies. Samples were taken at 2 days post‐infiltration (dpi). The bottom panel shows Ponceau S staining of Rubisco on the blotted nitrocellulose membrane to indicate relative protein loading. The western blot shows representative images from two independent experiments.
Testing for direct interaction of AvrP and AvrP123 with P and P2
Recognition by direct interaction between effector and R proteins has been shown for the AvrM:M and AvrL567:L6 pairs (Catanzariti et al., 2010; Dodds et al., 2006), with polymorphic surface residues of AvrL567 and AvrM determining recognition specificity (Ravensdale et al., 2012; Ve et al., 2013; Wang et al., 2007). The highly specific pattern of recognition of AvrP123 variants by the cognate R proteins is consistent with a direct recognition model (Dodds and Thrall, 2009). We tested for direct interaction of AvrP and AvrP123 with P and P2 using Y2H assays, but did not observe any positive interactions (Fig. 7a and S8a, see Supporting Information). Stable protein expression was detected in yeast (Figs S6c and S8c). However, we did note that AvrP123 fused to the GAL4 DNA‐binding domain (BD) led to the autoactivation of the reporter gene, even without the activation domain (AD) fusion partners (Figs 7b and S8b). Interaction was also not detected using co‐immunoprecipitation in planta under the conditions tested (Fig. S8d).
Figure 7.
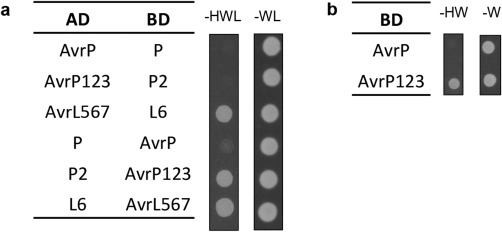
AvrP123 shows transcriptional activation in yeast. (a) AvrP/AvrP123 and P/P2 interaction assays in yeast. Avr and R gene constructs were fused to either a GAL4 activation domain (AD) or a binding domain (BD) in vectors that enable yeast transformants to grow in minimal medium lacking tryptophan and leucine (–WL), respectively. As a result of the interaction between AD and BD fusion proteins, the HIS3 reporter gene is activated, enabling the yeast to grow in medium lacking histidine, tryptophan and leucine (–HWL). L6 and AvrL567 constructs were used as positive controls. (b) BD‐AvrP123 construct activates yeast growth in the absence of AD constructs in medium lacking histidine and tryptophan (–HW).
Discussion
We report the crystal structure of the flax rust effector protein AvrP, revealing a novel elongated Zn‐binding protein with an unusual Zn‐binding topology. The Zn‐binding region of AvrP shows limited similarities to DNA‐binding and chromatin‐associated proteins. Localization studies show that AvrP and AvrP123 fused to the CTR fluorescence tag have a nucleo‐cytosolic localization, with an enhanced nuclear accumulation relative to CTR. The structure of AvrP shows that polymorphic residues in the AvrP family that influence R protein specificity map to the surface of the structure. Analysis of the surface conservation amongst AvrP variants, combined with mutational studies, reveals a variable surface that may play a role in specific recognition by the P family of R proteins.
AvrP is a Cys‐rich effector protein that binds to Zn ions
Distinct structures of fungal and oomycete effectors have been reported previously. Most structures of the oomycete RXLR effectors that have been published to date comprise a disordered RXLR uptake region and a three‐α‐helix fold effector domain, termed the WY domain (Boutemy et al., 2011; Win et al., 2012; Yaeno et al., 2011). Despite divergent sequences, the WY domains maintain a conserved structural fold across different oomycete species (Win et al., 2012). Similar to the oomycete RXLR effectors, sequence‐divergent effectors identified from the rice blast fungus Magnaporthe oryzae contain a common six‐stranded β‐sandwich structure related to the structure of the wheat pathogen Pyrenophora tritici‐repentis effector ToxB (de Guillen et al., 2015). These observations suggest that, despite very different sequences amongst fungal and oomycete effectors, structure‐based homology searches will contribute significantly to our understanding of effector functions.
Two other M. lini effector structures have been reported: AvrL567 (Wang et al., 2007) and AvrM (Ve et al., 2013). Both structures show very low similarity with other known structures, giving no clues to their biological functions. The structure of AvrP is unrelated to either of these effector proteins and, although the entire AvrP structure has no structural homologues, the Zn1‐binding region of AvrP shares similarities with Zn‐finger proteins. The Zn‐binding residues are strictly conserved amongst AvrP variants, suggesting that they probably maintain similar overall structures. A distant homologue of AvrP123, Mlp 124530, has been identified in Melampsora larici‐populina (poplar leaf rust) (Duplessis et al., 2011; Hacquard et al., 2012). Mlp 124530 has low sequence identity with the AvrP variants (∼25%). However, all 12 residues that are involved in Zn‐ion binding in AvrP are conserved in Mlp 124530, suggesting that it may have a similar structure to AvrP and that the binding of Zn ions may be a common feature in the AvrP‐like effector family. This family can be represented by the C‐X1–2‐C‐X13–14‐C‐X5‐C‐X‐C‐X5–7‐C‐X10‐C‐X6‐C‐X3‐H‐X7‐H motif, a novel form of Zn‐binding motif that can be used for the future identification of AvrP‐like effector candidates.
Cys‐rich proteins are over‐represented amongst fungal effectors and, in most cases, it is expected that these Cys residues form disulfide bonds (Bozkurt et al., 2012; Stergiopoulos and de Wit, 2009), which may enhance protein stability, especially as they are secreted in the protease‐rich apoplastic space (van den Burg et al., 2003). Disruption of disulfide bonds can result in the loss of pathogenic function (van den Burg et al., 2003; van den Hooven et al., 2001; Kooman‐Gersmann et al., 1997). We show here that the Cys residues in AvrP instead bind to Zn ions. Similar to disulfides, Zn binding may be important for protein stability and may promote correct protein folding. Accordingly, in this study, we showed that mutations of the Zn‐binding motifs in AvrP reduce protein accumulation and disrupt effector‐triggered cell death in planta. Although an expanded family of Zn‐finger proteins has been identified in the search for fungal effector candidates (Duplessis et al., 2011; Guyon et al., 2014), no molecular details of Zn binding have been revealed previously.
Enriched nuclear localization of AvrP suggests a nuclear function
The structure of AvrP shows some structural similarities to several Zn‐finger proteins, many of which are transcription factors that bind nucleic acids or histones and affect gene expression. Microscopic analysis of CTR‐tagged AvrP indicated that it shows enhanced accumulation in the host nucleus compared with the CTR protein when expressed in plant cells. These two properties suggest that AvrP might function inside the nucleus, possibly as a regulator of host cell transcription. However, we observed no significant nucleic acid‐binding activity for AvrP or AvrP123. AvrP showed weak ssRNA‐binding activity at a high protein concentration, but AvrP123 did not, suggesting that this property may not be biologically significant, assuming that these two proteins share a common pathogenicity mechanism. The ssRNA‐binding activity of AvrP may result from the positively charged surface of AvrP. Structural similarities with nucleic acid‐binding proteins have been identified for several fungal effectors; however, as found for AvrP, the follow‐up biochemical assays did not support a nucleic acid‐binding function (Blondeau et al., 2015; Pedersen et al., 2012; Wang et al., 2007). It is possible that nucleic acid‐binding domains were used as a structural template for effector diversification during evolution of the pathogen, leading to diverse effector functions (Pedersen et al., 2012).
There are a number of effectors secreted by bacteria that have been shown to influence host gene transcription. For example, the bacterial transcription activator‐like (TAL) effector proteins, which contain a nuclear localization signal (NLS) and an acidic transcriptional activation domain (AD), hijack host gene transcription by binding directly to DNA in the host (Zhang et al., 2015). However, AvrP does not have an identifiable NLS. The C‐terminal regions of AvrP variants share some similatities with AD domains, which contain an excess of negatively charged (D and E) residues and are structurally disordered (Ptashne and Gann, 1997). In our Y2H assays, BD‐fused AvrP123 strongly activated the HIS3 reporter gene (upstream of GAL4) in the absence of the AD fusion (Fig. 7b), suggesting that AvrP123 can directly influence transcriptional activation in yeast. Further investigation is required to test whether AvrP variants can act as transcriptional regulators in flax, possibly through interaction with host‐derived nuclear proteins, and whether nuclear localization is required to trigger R protein‐specific defence.
Mutational analysis of AvrP suggests that surface residues control recognition specificity by the P resistance protein
We show that polymorphic residues are located on the surface of the AvrP structure and that non‐conserved residues are enriched on one side of the structure. Different cell death activities were observed in flax and tobacco for several AvrP mutants, which may represent differences in protein stability in the two expression systems. The P70R mutant, which was stably expressed in tobacco but not in flax, showed a weak cell death activity in tobacco, but this activity was abolished in flax. The P47R mutant, however, which lost cell death activity in both systems, showed lower protein accumulation in N. benthamiana compared with the wild‐type protein and no expression in tobacco. Collectively, several point mutations of non‐conserved residues in the variable surface of AvrP reduced AvrP‐triggered cell death in P‐containing flax lines and transgenic tobacco. Amongst these, residues 66, 68, 70 and 90 differ between AvrP and the Lu2 protein, which is not recognized by P. Mutations of these residues in AvrP to Lu2 residues reduced cell death caused by P recognition in tobacco (Figs 6b and S7). When expressed in P‐containing flax, mutation of D66 also suppressed cell death, although such suppression was not evident for the P68 and S90 mutants (Fig. 6a). Although residues I64 and E72 are conserved between AvrP and Lu2, mutation of I64 abolished AvrP‐triggered necrosis, suggesting that this residue is involved in the recognition of AvrP by P. In AvrP123, I64 and E72 residues are replaced by arginine and alanine, respectively (Fig. 5b). The changes in side‐chain charge at these positions may prevent recognition between AvrP123 and P. Mutations of the C‐terminal polymorphic residue Q94 and E108 also affect P recognition, even though they are not part of the variable surface. We lack structural information for the C‐terminal nine residues of AvrP, including E108. Nevertheless, it is possible that these residues may affect P recognition and the function of adjacent residues, such as Q94.
Like the structures of the AvrL567 and AvrM flax rust effectors, the structure of AvrP revealed a novel protein fold with limited structural similarity to proteins of known function. Although we speculate that AvrP is targeted to the host nucleus during infection, the role of AvrP in pathogen virulence remains unknown. The coordination of Zn ions by AvrP appears to be important for correct folding of the protein, and therefore for recognition by the P resistance protein. Although a direct interaction between AvrP and P could not be demonstrated here, non‐conserved residues clustered on a surface patch of AvrP appear to be involved in specific recognition by P.
Experimental Procedures
Vectors and gene constructs
For transient expression in tobacco, the cDNAs of the AvrP and AvrP123 proteins (Catanzariti et al., 2006), lacking the predicted signal peptides, were cloned into the pL and/or pLH vector, which contain an HA epitope sequence, using ligation‐independent cloning (LIC) (Eschenfeldt et al., 2009; Stols et al., 2002). For transient expression in flax and Y2H assays, the same gene fragments were cloned into pAM‐PATpro35S:GWY‐YFPv (Bernoux et al., 2008) and Gateway‐compatible Y2H vectors (Bernoux et al., 2011), respectively, using Gateway cloning. AvrP mutations were introduced into these constructs using the Phusion site‐directed mutagenesis kit (Thermo Fisher Scientific, http://www.thermofisher.com/, Waltham, Massachusetts, USA) and primers containing the corresponding mutation. To express proteins in N. benthamiana for fluorescence microscopy, gene splicing by polymerase chain reaction (PCR)‐driven overlap extension was used to generate constructs containing the AvrP and AvrP123 coding sequences, lacking the predicted signal peptide and stop codon, fused to CTR with an in‐frame linker encoding GPGP. The PCR products were cloned as SpeI‐PvuI fragments into the binary vector pCAMBIA3301 between NOS promoter and terminator sequences (De Block et al., 1984; Horsch et al., 1984). The CTR gene sequence was cloned as an XbaI‐PvuI fragment into the same vector to generate a CTR‐only expression construct. The oligonucleotides used in this study are summarized in Table S3 (see Supporting Information).
Plant material
A transgenic W38 tobacco (N. tabacum) line containing the P resistance gene was made by transformation with the P2/P‐RVX construct (Dodds et al., 2001). The flax (L. usitatissimum) lines Hoshangabad, Bison, Koto (containing P), Akmolinsk (P1), Abyssinian (P2) and Leona (P3), and the Bison backcross derivatives containing these resistance genes, have been described previously (Islam and Mayo, 1990; Lawrence et al., 1981a). A line containing the P resistance gene in Hoshangabad, P (Hosh), was generated by three generations of backcrossing.
Structure determination
AvrP was expressed, purified and crystallized as described by Zhang et al. (2017). Data were collected at the Australian Synchrotron MX1 beamline (Cowieson et al., 2015) using Blu‐Ice software (McPhillips et al., 2002). The diffraction data were used to a resolution of 2.52 Å and the structure was solved using the MAD technique. The structure was refined to final R work and R free values of 22% and 26%, respectively. The AvrP structure and data used to derive the structure have been deposited with the Protein Data Bank (www.pdb.org.) with PDB ID 5VJJ. Details are described in Methods S1 (see Supporting Information) and Table S1.
Transient in planta expression assays
Agrobacterium cultures containing the corresponding vectors were grown overnight at 28 °C in lysogeny broth (LB; 10 g/L tryptone, 5 g/L yeast extract and 10 g/L NaCl) medium with appropriate antibiotic selections. The cells were pelleted and resuspended in infiltration mix (10 mm MgCl2, 200 μm acetosyringone) to an optical density at 600 nm (OD600) of 0.5 or 1.0, followed by incubation at room temperature for 2 h. Cultures were infiltrated into leaves of 4‐week‐old tobacco or flax with a 1‐mL syringe. The infiltrated tobacco plants were incubated in growth chambers at 24 °C with 200 µmol/m2/s light intensity and an 8‐h light and 16‐h dark cycle. The flax plants were kept in a glasshouse at 24 °C with natural light and day/night cycle.
Confocal microscopy
Constructs were introduced into N. benthamiana leaves via agroinfiltration. Samples were collected at 2 days post‐infiltration (dpi) and viewed on a Zeiss (Oberkochen, Germany) LSM 780 confocal microscope using an LD C‐Apochromat 40×/1.1 W Korr M27 water immersion objective, with an excitation wavelength of 514 nm and a collection window of 520–600 nm for citrine and 637–759 nm for chlorophyll autofluorescence. Both fluorescence and transmitted light images were collected using the following settings: size, 212 × 212 µm2; 2048 × 2048 pixels; scan time, 30 s. Fluorescent images of cell nuclei were brought to just below saturation to enable comparison between samples. Cytosolic and nuclear fluorescence intensity values were measured using ImageJ (Schneider et al., 2012) in the Fiji package (Schindelin et al., 2012). Average fluorescence intensities were calculated from 30 cells. Cytosolic samples were taken from regions of the cytosol that were perpendicular to the plane of imaging and free of chloroplasts. Nuclear samples were taken from a central plane avoiding the nucleolus. The experiment was repeated three times. P values were calculated using Student's t‐test. The results were tabulated and graphed in Microsoft Excel.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Protein solution property and protein–lipid binding assays.
Fig. S2 The cross‐brace zinc‐binding topology.
Fig. S3 Electrophoretic mobility shift assay (EMSA) for nucleic acid binding using the Pentaprobe approach.
Fig. S4 Cell death assay of AvrP and AvrP123 citrine (CTR) fusions.
Fig. S5 Specific recognition of AvrP variants by P family resistance proteins.
Fig. S6 Immunoblotting of AvrP and P.
Fig. S7 Cell death scoring of AvrP mutants.
Fig. S8 Test for interaction of AvrP and AvrP123 with P and P2.
Table S1 X‐Ray data collection, structure solution and refinement statistics.
Table S2 Ratios of average fluorescence intensity in the nucleus and the cytosol (Av Nuc:Cyt) for AvrP‐CTR, AvrP123‐CTR and citrine (CTR).
Table S3 Oligonucleotides used in cloning.
Methods S1 Supplemental experimental procedures.
References S1 Supplemental references.
Acknowledgements
This research was supported by Australian Research Council (ARC) Discovery Projects DP120100685, DP130104098 and DP160102244. XZ was a recipient of an ANZ Trustees PhD Scholarship for Medical Research in Queensland. BK is a National Health and Medical Research Council (NHMRC) Principal Research Fellow (1003325 and 1110971). MB was a recipient of an ARC Discovery Early Career Research Award (DE130101292). We acknowledge the use of the University of Queensland Remote Operation Crystallization and X‐ray Diffraction Facility (UQ ROCX), and the assistance of Karl Byriel and Gordon King. X‐Ray diffraction data collection was undertaken on MX beamlines at the Australian Synchrotron. We thank Kim Newell for providing technical assistance.
Contributor Information
Xiaoxiao Zhang, Email: xiaoxiao.zhang@csiro.au.
Bostjan Kobe, Email: b.kobe@uq.edu.au.
References
- Adams‐Cioaba, M.A. , Li, Z. , Tempel, W. , Guo, Y. , Bian, C. , Li, Y. , Lam, R. and Min, J. (2012) Crystal structures of the Tudor domains of human PHF20 reveal novel structural variations on the Royal Family of proteins. FEBS Lett. 586, 859–865. [DOI] [PubMed] [Google Scholar]
- Anderson, C. , Khan, M.A. , Catanzariti, A.M. , Jack, C.A. , Nemri, A. , Lawrence, G.J. , Upadhyaya, N.M. , Hardham, A.R. , Ellis, J.G. , Dodds, P.N. and Jones, D.A. (2016) Genome analysis and avirulence gene cloning using a high‐density RADseq linkage map of the flax rust fungus, Melampsora lini . BMC Genomics, 17, 667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett, L.G. , Thrall, P.H. , Dodds, P.N. , van der Merwe, M. , Linde, C.C. , Lawrence, G.J. and Burdon, J.J. (2009) Diversity and evolution of effector loci in natural populations of the plant pathogen Melampsora lini . Mol. Biol. Evol. 26, 2499–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernoux, M. , Timmers, T. , Jauneau, A. , Briere, C. , de Wit, P.J.G.M. , Marco, Y. and Deslandes, L. (2008) RD19, an Arabidopsis cysteine protease required for RRS1‐R‐mediated resistance, is relocalized to the nucleus by the Ralstonia solanacearum PopP2 effector. Plant Cell, 20, 2252–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernoux, M. , Ve, T. , Williams, S. , Warren, C. , Hatters, D. , Valkov, E. , Zhang, X. , Ellis, J.G. , Kobe, B. and Dodds, P.N. (2011) Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self‐association, signaling, and autoregulation. Cell Host Microbe, 9, 200–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondeau, K. , Blaise, F. , Graille, M. , Kale, S.D. , Linglin, J. , Ollivier, B. , Labarde, A. , Lazar, N. , Daverdin, G. , Balesdent, M.H. and Choi, D.H. (2015) Crystal structure of the effector AvrLm4–7 of Leptosphaeria maculans reveals insights into its translocation into plant cells and recognition by resistance proteins. Plant J. 83, 610–624. [DOI] [PubMed] [Google Scholar]
- Bonardi, V. , Cherkis, K. , Nishimura, M.T. and Dangl, J.L. (2012) A new eye on NLR proteins: focused on clarity or diffused by complexity? Curr. Opin. Immunol. 24, 41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutemy, L.S. , King, S.R.F. , Win, J. , Hughes, R.K. , Clarke, T.A. , Blumenschein, T.M.A. , Kamoun, S. and Banfield, M.J. (2011) Structures of Phytophthora RXLR effector proteins: a conserved but adaptable fold underpins functional diversity. J. Biol. Chem. 286, 35 834–35 842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkurt, T.O. , Schornack, S. , Banfield, M.J. and Kamoun, S. (2012) Oomycetes, effectors, and all that jazz. Curr. Opin. Plant Biol. 15, 483–492. [DOI] [PubMed] [Google Scholar]
- van den Burg, H.A. , Westerink, N. , Francoijs, K.J. , Roth, R. , Woestenenk, E. , Boeren, S. , de Wit, P.J. , Joosten, M.H. and Vervoort, J. (2003) Natural disulfide bond‐disrupted mutants of AVR4 of the tomato pathogen Cladosporium fulvum are sensitive to proteolysis, circumvent Cf‐4‐mediated resistance, but retain their chitin binding ability. J. Biol. Chem. 278, 27 340–27 346. [DOI] [PubMed] [Google Scholar]
- Catanzariti, A.M. , Dodds, P.N. , Lawrence, G.J. , Ayliffe, M.A. and Ellis, J.G. (2006) Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell, 18, 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzariti, A.M. , Dodds, P.N. , Ve, T. , Kobe, B. , Ellis, J.G. and Staskawicz, B.J. (2010) The AvrM effector from flax rust has a structured C‐terminal domain and interacts directly with the M resistance protein. Mol. Plant–Microbe Interact. 23, 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowieson, N.P. , Aragao, D. , Clift, M. , Ericsson, D.J. , Gee, C. , Harrop, S.J. , Mudie, N. , Panjikar, S. , Price, J.R. , Riboldi‐Tunnicliffe, A. and Williamson, R. (2015) MX1: a bending‐magnet crystallography beamline serving both chemical and macromolecular crystallography communities at the Australian Synchrotron. J Synchrotron Radiat. 22, 187–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Block, M. , Herrera‐Estrella, L. , Van Montagu, M. , Schell, J. and Zambryski, P. (1984) Expression of foreign genes in regenerated plants and in their progeny. EMBO J. 3, 1681–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djamei, A. , Schipper, K. , Rabe, F. , Ghosh, A. , Vincon, V. , Kahnt, J. , Osorio, S. , Tohge, T. , Fernie, A.R. , Feussner, I. and Feussner, K. (2011) Metabolic priming by a secreted fungal effector. Nature, 478, 395–398. [DOI] [PubMed] [Google Scholar]
- Dodds, P. and Thrall, P. (2009) Recognition events and host–pathogen co‐evolution in gene‐for‐gene resistance to flax rust. Funct. Plant Biol. 36, 395–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N. and Rathjen, J.P. (2010) Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N. , Lawrence, G.J. and Ellis, J.G. (2001) Six amino acid changes confined to the leucine‐rich repeat beta‐strand/beta‐turn motif determine the difference between the P and P2 rust resistance specificities in flax. Plant Cell, 13, 163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N. , Lawrence, G.J. , Catanzariti, A.M. , Ayliffe, M.A. and Ellis, J.G. (2004) The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell, 16, 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N. , Lawrence, G.J. , Catanzariti, A.M. , Teh, T. , Wang, C.I. , Ayliffe, M.A. , Kobe, B. and Ellis, J.G. (2006) Direct protein interaction underlies gene‐for‐gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA, 103, 8888–8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplessis, S. , Cuomo, C.A. , Lin, Y.C. , Aerts, A. , Tisserant, E. , Veneault‐Fourrey, C. , Joly, D.L. , Hacquard, S. , Amselem, J. , Cantarel, B.L. and Chiu, R. (2011) Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc. Natl. Acad. Sci. USA, 108, 9166–9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschenfeldt, W.H. , Lucy, S. , Millard, C.S. , Joachimiak, A. and Mark, I.D. (2009) A family of LIC vectors for high‐throughput cloning and purification of proteins. Methods Mol. Biol. 498, 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Y. , Yao, H. and Wang, J. (2010) Crystal structure of the crenarchaeal conserved chromatin protein Cren7 and double‐stranded DNA complex. Protein Sci. 19, 1253–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor, H.H. (1956) The complementary genic systems in flax and flax rust. Adv. Genet. 8, 29–54. [Google Scholar]
- Grishin, N.V. (2001) Treble clef finger – a functionally diverse zinc‐binding structural motif. Nucleic Acids Res. 29, 1703–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Guillen, K. , Ortiz‐Vallejo, D. , Gracy, J. , Fournier, E. , Kroj, T. and Padilla, A. (2015) Structure analysis uncovers a highly diverse but structurally conserved effector family in phytopathogenic fungi. PLoS Pathog. 11, e1005228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyon, K. , Balague, C. , Roby, D. and Raffaele, S. (2014) Secretome analysis reveals effector candidates associated with broad host range necrotrophy in the fungal plant pathogen Sclerotinia sclerotiorum . BMC Genomics, 15, 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacquard, S. , Joly, D.L. , Lin, Y.C. , Tisserant, E. , Feau, N. , Delaruelle, C. , Legué, V. , Kohler, A. , Tanguay, P. , Petre, B. and Frey, P. (2012) A comprehensive analysis of genes encoding small secreted proteins identifies candidate effectors in Melampsora larici‐populina (poplar leaf rust). Mol. Plant–Microbe Interact. 25, 279–293. [DOI] [PubMed] [Google Scholar]
- Holm, L. and Rosenstrom, P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hooven, H.W. , van den Burg, H.A. , Vossen, P. , Boeren, S. , de Wit, P.J.G.M. and Vervoort, J. (2001) Disulfide bond structure of the AVR9 elicitor of the fungal tomato pathogen Cladosporium fulvum: evidence for a cystine knot. Biochemistry, 40, 3458–3466. [DOI] [PubMed] [Google Scholar]
- Horsch, R.B. , Fraley, R.T. , Rogers, S.G. , Sanders, P.R. , Lloyd, A. and Hoffmann, N. (1984) Inheritance of functional foreign genes in plants. Science, 223, 496–498. [DOI] [PubMed] [Google Scholar]
- Islam, M.R. and Mayo, G.M.E. (1990) A compendium on host genes in flax conferring resistance to flax rust. Plant Breed. 104, 89–100. [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kooman‐Gersmann, M. , Vogelsang, R. , Hoogendijk, E.C.M. and De Wit, P.J.G.M. (1997) Assignment of amino acid residues of the AVR9 peptide of Cladosporium fulvum that determine elicitor activity. Mol. Plant–Microbe Interact. 10, 821–829. [DOI] [PubMed] [Google Scholar]
- Lawrence, G.J. , Mayo, G.M.E. and Shepherd, K.W. (1981a) Interactions between genes controlling pathogenicity in the flax rust fungus. Phytopathology, 71, 12–19. [Google Scholar]
- Lawrence, G.J. , Shepherd, K.W. and Mayo, G.M.E. (1981b) Fine‐structure of genes controlling pathogenicity in flax rust, Melampsora‐lini . Heredity, 46, 297–313. [Google Scholar]
- McLellan, H. , Boevink, P.C. , Armstrong, M.R. , Pritchard, L. , Gomez, S. , Morales, J. , Whisson, S.C. , Beynon, J.L. and Birch, P.R. (2013) An RxLR effector from Phytophthora infestans prevents re‐localisation of two plant NAC transcription factors from the endoplasmic reticulum to the nucleus. PLoS Pathog. 9, e1003670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhillips, T.M. , McPhillips, S.E. , Chiu, H.J. , Cohen, A.E. , Deacon, A.M. , Ellis, P.J. , Garman, E. , Gonzalez, A. , Sauter, N.K. , Phizackerley, R.P. and Soltis, S.M. (2002) Blu‐Ice and the distributed control system: software for data acquisition and instrument control at macromolecular crystallography beamlines. J. Synchrotron Radiat. 9, 401–406. [DOI] [PubMed] [Google Scholar]
- Mishra, A.K. , Gangwani, L. , Davis, R.J. and Lambright, D.G. (2007) Structural insights into the interaction of the evolutionarily conserved ZPR1 domain tandem with eukaryotic EF1A, receptors, and SMN complexes. Proc. Natl. Acad. Sci. USA, 104, 13 930–13 935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okmen, B. and Doehlemann, G. (2014) Inside plant: biotrophic strategies to modulate host immunity and metabolism. Curr. Opin. Plant Biol. 20, 19–25. [DOI] [PubMed] [Google Scholar]
- Oliveira‐Garcia, E. and Valent, B. (2015) How eukaryotic filamentous pathogens evade plant recognition. Curr. Opin. Microbiol. 26, 92–101. [DOI] [PubMed] [Google Scholar]
- Pedersen, C. , Ver Loren van Themaat, E. , McGuffin, L.J. , Abbott, J.C. , Burgis, T.A. , Barton, G. , Bindschedler, L.V. , Lu, X. , Maekawa, T. , Weßling, R. and Cramer, R. (2012) Structure and evolution of barley powdery mildew effector candidates. BMC Genomics, 13, 694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne, M. and Gann, A. (1997) Transcriptional activation by recruitment. Nature, 386, 569–577. [DOI] [PubMed] [Google Scholar]
- Qiao, Y. , Shi, J. , Zhai, Y. , Hou, Y. and Ma, W. (2015) Phytophthora effector targets a novel component of small RNA pathway in plants to promote infection. Proc. Natl. Acad. Sci. USA, 112, 5850–5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiqi, M. , Gan, P.H. , Ravensdale, M. , Lawrence, G.J. , Ellis, J.G. , Jones, D.A. , Hardham, A.R. and Dodds, P.N. (2010) Internalization of flax rust avirulence proteins into flax and tobacco cells can occur in the absence of the pathogen. Plant Cell, 22, 2017–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiqi, M. , Ellis, J.G. , Ludowici, V.A. , Hardham, A.R. and Dodds, P.N. (2012) Challenges and progress towards understanding the role of effectors in plant–fungal interactions. Curr. Opin. Plant Biol. 15, 477–482. [DOI] [PubMed] [Google Scholar]
- Ramirez‐Garces, D. , Camborde, L. , Pel, M.J. , Jauneau, A. , Martinez, Y. , Neant, I. , Leclerc, C. , Moreau, M. , Dumas, B. and Gaulin, E. (2016) CRN13 candidate effectors from plant and animal eukaryotic pathogens are DNA‐binding proteins which trigger host DNA damage response. New Phytol. 210, 602–617. [DOI] [PubMed] [Google Scholar]
- Ravensdale, M. , Nemri, A. , Thrall, P.H. , Ellis, J.G. and Dodds, P.N. (2011) Co‐evolutionary interactions between host resistance and pathogen effector genes in flax rust disease. Mol. Plant Pathol. 12, 93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravensdale, M. , Bernoux, M. , Ve, T. , Kobe, B. , Thrall, P.H. , Ellis, J.G. and Dodds, P.N. (2012) Intramolecular interaction influences binding of the flax L5 and L6 resistance proteins to their AvrL567 ligands. PLoS Pathog. 8, e1003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redkar, A. , Hoser, R. , Schilling, L. , Zechmann, B. , Krzymowska, M. , Walbot, V. and Doehlemann, G. (2015) A secreted effector protein of Ustilago maydis guides maize leaf cells to form tumors. Plant Cell, 27, 1332–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin, J. , Arganda‐Carreras, I. , Frise, E. , Kaynig, V. , Longair, M. , Pietzsch, T. , Preibisch, S. , Rueden, C. , Saalfeld, S. , Schmid, B. and Tinevez, J.Y. (2012) Fiji: an open‐source platform for biological‐image analysis. Nat Methods, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, C.A. , Rasband, W.S. and Eliceiri, K.W. (2012) NIH image to ImageJ: 25 years of image analysis. Nat. Methods, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schornack, S. , van Damme, M. , Bozkurt, T.O. , Cano, L.M. , Smoker, M. , Thines, M. , Gaulin, E. , Kamoun, S. and Huitema, E. (2010) Ancient class of translocated oomycete effectors targets the host nucleus. Proc. Natl. Acad. Sci. USA, 107, 17 421–17 426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers, F. , Wilm, A. , Dineen, D. , Gibson, T.J. , Karplus, K. , Li, W. , Lopez, R. , McWilliam, H. , Remmert, M. , Söding, J. and Thompson, J.D. (2011) Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam, R. , Howden, A.J.M. , Delgado‐Cerezo, M. , Amaro, T.M.M.M. , Motion, G.B. , Pham, J. and Huitema, E. (2013a) Characterization of cell death inducing Phytophthora capsici CRN effectors suggests diverse activities in the host nucleus. Front Plant Sci. 4, 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam, R. , Jupe, J. , Howden, A.J.M. , Morris, J.A. , Boevink, P.C. , Hedley, P.E. and Huitema, E. (2013b) Identification and characterisation of CRN effectors in Phytophthora capsici shows modularity and functional diversity. PLoS One, 8, e59517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergiopoulos, I. and de Wit, P.J.G.M. (2009) Fungal effector proteins. Annu. Rev. Phytopathol. 47, 233–263. [DOI] [PubMed] [Google Scholar]
- Stols, L. , Gu, M. , Dieckman, L. , Raffen, R. , Collart, F.R. and Donnelly, M.I. (2002) A new vector for high‐throughput, ligation‐independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr. Purif. 25, 8–15. [DOI] [PubMed] [Google Scholar]
- Vargas, W.A. , Sanz‐Martin, J.M. , Rech, G.E. , Armijos‐Jaramillo, V.D. , Rivera, L.P. , Echeverria, M.M. , Díaz‐Mínguez, J.M. , Thon, M.R. and Sukno, S.A. (2016) A fungal effector with host nuclear localization and DNA‐binding properties is required for maize anthracnose development. Mol. Plant–Microbe Interact. 29, 83–95. [DOI] [PubMed] [Google Scholar]
- Ve, T. , Williams, S.J. , Catanzariti, A.M. , Rafiqi, M. , Rahman, M. , Ellis, J.G. , Hardham, A.R. , Jones, D.A. , Anderson, P.A. , Dodds, P.N. and Kobe, B. (2013) Structures of the flax‐rust effector AvrM reveal insights into the molecular basis of plant‐cell entry and effector‐triggered immunity. Proc. Natl. Acad. Sci. USA, 110, 17 594–17 599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B. , Jones, D.N. , Kaine, B.P. and Weiss, M.A. (1998) High‐resolution structure of an archaeal zinc ribbon defines a general architectural motif in eukaryotic RNA polymerases. Structure, 6, 555–569. [DOI] [PubMed] [Google Scholar]
- Wang, C.I. , Guncar, G. , Forwood, J.K. , Teh, T. , Catanzariti, A.M. , Lawrence, G.J. , Loughlin, F.E. , Mackay, J.P. , Schirra, H.J. , Anderson, P.A. and Ellis, J.G. (2007) Crystal structures of flax rust avirulence proteins AvrL567‐A and ‐D reveal details of the structural basis for flax disease resistance specificity. Plant Cell, 19, 2898–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R.W. and Brattain, M.G. (2007) The maximal size of protein to diffuse through the nuclear pore is larger than 60 kDa. FEBS Lett. 581, 3164–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win, J. , Krasileva, K.V. , Kamoun, S. , Shirasu, K. , Staskawicz, B.J. and Banfield, M.J. (2012) Sequence divergent RXLR effectors share a structural fold conserved across plant pathogenic oomycete species. PLoS Pathog. 8, e1002400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaeno, T. , Li, H. , Chaparro‐Garcia, A. , Schornack, S. , Koshiba, S. , Watanabe, S. , Kigawa, T. , Kamoun, S. and Shirasu, K. (2011) Phosphatidylinositol monophosphate‐binding interface in the oomycete RXLR effector AVR3a is required for its stability in host cells to modulate plant immunity. Proc. Natl. Acad. Sci. USA, 108, 14 682–14 687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, M. , Ohsawa, A. , Kikukawa, S. , Tanaka, I. and Kimura, M. (2003) Crystal structure of hyperthermophilic archaeal initiation factor 5A: a homologue of eukaryotic initiation factor 5A (eIF‐5A). J. Biochem. 133, 75–81. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Yin, Z. and White, F. (2015) TAL effectors and the executor R genes. Front Plant Sci. 6, 641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Nguyen, N. , Breen, S. , Outram, M.A. , Dodds, P.N. , Kobe, B. , Solomon, P.S. and Williams, S.J. (2017) Production of small cysteine‐rich effector proteins in Escherichia coli for structural and functional studies. Mol. Plant Pathol. 18, 141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Protein solution property and protein–lipid binding assays.
Fig. S2 The cross‐brace zinc‐binding topology.
Fig. S3 Electrophoretic mobility shift assay (EMSA) for nucleic acid binding using the Pentaprobe approach.
Fig. S4 Cell death assay of AvrP and AvrP123 citrine (CTR) fusions.
Fig. S5 Specific recognition of AvrP variants by P family resistance proteins.
Fig. S6 Immunoblotting of AvrP and P.
Fig. S7 Cell death scoring of AvrP mutants.
Fig. S8 Test for interaction of AvrP and AvrP123 with P and P2.
Table S1 X‐Ray data collection, structure solution and refinement statistics.
Table S2 Ratios of average fluorescence intensity in the nucleus and the cytosol (Av Nuc:Cyt) for AvrP‐CTR, AvrP123‐CTR and citrine (CTR).
Table S3 Oligonucleotides used in cloning.
Methods S1 Supplemental experimental procedures.
References S1 Supplemental references.


