Summary
Nitrogen (N) availability can impact plant resistance to pathogens by the regulation of plant immunity. To better understand the links between N nutrition and plant defence, we analysed the impact of N availability on Medicago truncatula resistance to the root pathogen Aphanomyces euteiches. This oomycete is considered to be the most limiting factor for legume production. Ten plant genotypes were tested in vitro for their resistance to A. euteiches in either complete or nitrate‐deficient medium. N deficiency led to enhanced or reduced susceptibility depending on the plant genotype. Focusing on four genotypes displaying contrasting responses, we determined the impact of N deficiency on plant growth and shoot N concentration, and performed expression analyses on N‐ and defence‐related genes, as well as the quantification of soluble phenolics and different amino acids in roots. Our analyses suggest that N modulation of plant resistance is not linked to plant response to N deprivation or to mechanisms previously identified to be involved in plant resistance. Furthermore, our studies highlight a role of glutamine in mediating the susceptibility to A. euteiches in M. truncatula.
Keywords: amino acids, Aphanomyces euteiches; glutamine; Medicago truncatula; nitrate; nitrogen; plant defence
Introduction
Nutrients are essential for the growth and development of plants and microorganisms, and are also recognized as important factors in the fate of plant interactions with beneficial microbes and pathogens (Bonneau et al., 2013; Dordas, 2008; Fagard et al., 2014; Hacquard et al., 2016; Mur et al., 2017). Among the different nutrients, nitrogen (N) plays a particularly important role. Indeed, N constitutes approximately 2%–5% of dry matter, making it quantitatively the most important element in plants. Agronomical data indicate that, in addition to the impact on yield, the quantity and quality of N supply can impact a plant's ability to cope with pathogen attacks, leading to enhanced or decreased resistance. There is, however, no general rule on the outcome of N fertilization on disease severity. For example, in potato, N supply leads to opposite results, depending on the pathogen model (Mittelstrass et al., 2006). These differences could be correlated, in part, with the pathogenic lifestyle (biotrophic or necrotrophic). Other factors could be involved in the modulation by N of plant resistance to pathogens, such as the plant capacity to mobilize N and/or synthesize defence compounds (Ballini et al., 2013; Dietrich et al., 2004). Moreover, a recent study has indicated that the form of N (nitrate or ammonium) affects the defence response and resistance to bacterial infection in tobacco (Gupta et al., 2013). Finally, N can quantitatively and qualitatively affect pathogen virulence and pathogen nutrition in planta (Snoeijers et al., 2000). Taken together, these data indicate that N supply can affect plant resistance. However, in general, the underlying mechanisms remain unclear.
The different steps involved in N metabolism have long been studied, and many of the corresponding proteins have been characterized (Masclaux‐Daubresse et al., 2010). Interestingly, the contribution of several of these proteins to plant defence has been highlighted recently. Arabidopsis mutants affected in different steps of N transport and assimilation, such as the nitrate reductase (NR) double mutant (nia1 nia2) (Modolo et al., 2006; Oliveira et al., 2009), the high‐affinity nitrate transporter NRT2.1 mutant (Camañes et al., 2012) and the putative nitrate transporter NRT2.6 (Dechorgnat et al., 2012) mutant, were all found to display an altered sensitivity to pathogens. Conversely, pathogens modulate the expression of genes or the activity of enzymes involved in N transport, N assimilation, N remobilisation and amino acid metabolism (Fagard et al., 2014).
Amino acid contents and their relative concentrations are modified during plant–pathogen interactions (Fagard et al., 2014; Zeier, 2013). Different explanations can be proposed to interpret these modifications. Amino acids (per se or as precursors of defence molecules) represent an important resource for setting up induced resistance in plants. Amino acids also constitute a nutrient source that is crucial for pathogen survival and propagation (Pellier et al., 2003; Solomon et al., 2003). The modification of plant amino acid composition could therefore represent a plant defence strategy which could lead to N deprivation for pathogens through, for instance, N remobilisation from diseased tissues to healthy parts (Olea et al., 2004; Tavernier et al., 2007). Alternatively pathogens, through co‐evolution, could adapt their preference to amino acids accumulated in infected plants or could manipulate plant N metabolism, inducing the accumulation of favourable amino acids at the infection site (Solomon and Oliver, 2002). The links between amino acid metabolism and plant resistance were reinforced by results showing that Arabidopsis thaliana lines impaired in the LHT1 amino acid transporter (Liu et al., 2010) or overexpressing the pepper ASPARAGINE SYNTHETASE1 gene (CaAS1) (Hwang et al., 2011) exhibited enhanced resistance to Pseudomonas syringae.
Finally, modulation of N metabolism could impact molecular defence responses. For instance, nitric oxide and reactive oxygen species levels are altered in response to N deprivation (Schachtman and Shin, 2007; Thalineau et al., 2016). Salicylic acid (SA) levels and SA‐mediated defences, mainly linked to the redox state, are also dependent on N nutrition (Gupta et al., 2013; Yaeno and Iba, 2008). Transcriptome analyses have further highlighted that the expression of genes involved in the response to biotic stress [such as mitogen‐activated protein kinase 3 (MPK3), MPK7, peroxidase‐ and R protein‐encoding genes] is affected by N limitation in Arabidopsis (Peng et al., 2007).
Parasitic oomycetes cause severe diseases on many crop plants. Aphanomyces euteiches is a pathogenic oomycete responsible for root rot in many legume species (Gaulin et al., 2007), and is considered to be the most limiting factor for legume production. Resistance to A. euteiches has been investigated in legumes and, particularly, in the model plant Medicago truncatula. The reaction of M. truncatula lines to A. euteiches infection varies from susceptibility to resistance (Djébali et al., 2012; Vandemark and Grünwald, 2004). Comparative analyses of susceptible (F83005.5) and tolerant (A17) genotypes have indicated that resistance mechanisms include the protection of the central cylinder against pathogen invasion, associated with frequent pericycle cell divisions, lignin deposition and the accumulation of soluble phenolic compounds (Djébali et al., 2009, 2011). These results have been corroborated by genetic studies. A major locus associated with resistance to A. euteiches, namely AER1, has been identified (Pilet‐Nayel et al., 2009) and confirmed by a genome‐wide association study, which identified two major independent loci on top of chromosome 3, one of which co‐localized with prAe1 (formerly named AER1) (Bonhomme et al., 2014). The functional role of the prAe1 locus has been investigated recently (Badis et al., 2015) using two near‐isogenic lines, NR and NS, partially resistant and susceptible, respectively, differing in the allelic state of prAe1. Transcriptomic and biochemical analyses demonstrated the significant role of secondary metabolites, particularly of flavonoids, in resistance, and identified a chalcone‐derived compound as a potent inhibitor of A. euteiches.
The aim of this work was to study the effect of N nutrition on M. truncatula resistance to A. euteiches. Ten genotypes of M. truncatula were grown in vitro on complete or N‐deficient (NØ) medium. Parameters related to plant growth and root development, as well as relevant indicators of M. truncatula resistance to A. euteiches, were scored. N deficiency led to contrasting responses of the different genotypes to A. euteiches, resulting in either enhanced or decreased resistance of plants to the pathogen. To understand how NØ conditions lead to contrasting behaviours between M. truncatula genotypes, we focused our investigations on four genotypes to perform biochemical (quantification of soluble phenolics and amino acids) and molecular (gene expression related to N metabolism or involved in defence responses) analyses. Our analyses suggest that N modulation of plant resistance is not linked to plant response to N deprivation nor to mechanisms previously identified to be involved in plant resistance. Furthermore, our studies highlight the importance of glutamine (Gln) in enhancing plant susceptibility to A. euteiches.
Results
Nitrogen deficiency enhances the resistance of the A17 genotype to A. euteiches
To assess the effects of N nutrition on the interaction between M. truncatula and A. euteiches, the A17 genotype was grown in vitro for 10–21 days on two media: complete medium (containing 3.3 mm nitrate as sole N source) and nitrate deficient NØ medium. On NØ medium, the first symptoms of N deficiency (plant growth parameters) were observed after 10 days of culture compared with plants cultivated on complete medium. Significant differences in plant N content appeared later, after 14 days of culture (Fig. S1, see Supporting Information).
The A17 genotype was less affected by A. euteiches on NØ medium relative to complete medium, as it displayed a lower percentage of symptomatic tissues (stem necrosis and root browning) (Fig. 1A), and plant root development was less affected by inoculation (Fig. S1). Moreover, a significantly lower level of pathogen was detected in roots by enzyme‐linked immunosorbent assay (ELISA) (Fig. 1B) using a polyclonal antibody raised against A. euteiches (Slezack et al., 1999). This was correlated with an eight‐fold decrease (ΔΔCt = 3) in the transcript levels of A. euteiches tubulin (AeTUB) gene (Fig. 1C). Thus, A17 was more resistant to A. euteiches on NØ medium relative to complete medium.
Figure 1.
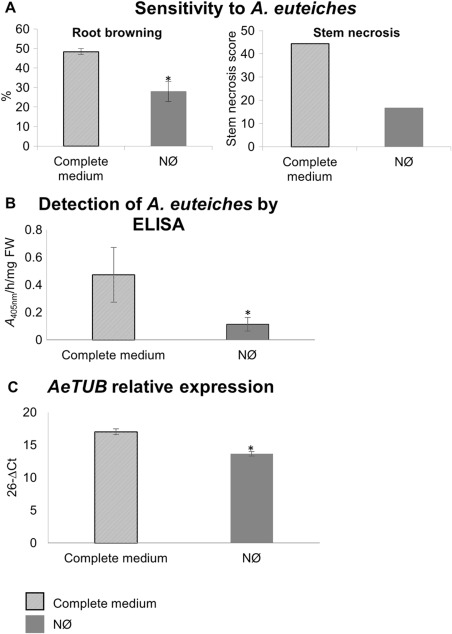
The susceptibility of the A17 genotype to Aphanomyces euteiches is decreased on nitrate‐deficient (NØ) medium. (A) Susceptibility to A. euteiches as scored by the percentage of symptomatic tissues [stem necrosis at 21 days of culture and root browning at 10 days post‐inoculation (dpi), see Experimental Procedures]. (B) Quantification by enzyme‐linked immunosorbent assay (ELISA) of A. euteiches in root extracts. FW, fresh weight. (C) Relative expression of AeTUB using MtEF1 as reference gene. Plants were cultivated in vitro for 3 weeks on complete medium or NØ medium and inoculated with A. euteiches. For ELISA, the background signal from control non‐inoculated roots was subtracted from the signal detected in inoculated roots. Error bars indicate standard errors (n = 16–20 for symptoms; n = 4–6 biological replicates for ELISAs and AeTUB expression) and asterisks indicate significant differences (P < 0.05). Data from a representative experiment out of four independent experiments are shown.
We verified whether this NØ‐induced resistance (NØ‐IR) was a result of the fact that the virulence of A. euteiches was affected by N deficiency. For this purpose, we measured AeCRN13 and AeCRN5 expression, two CRN candidate effectors identified through a transcriptomic approach on A. euteiches (Gaulin et al., 2008; Ramirez‐Garcés et al., 2016). No difference in CRN expression between complete and NØ media was observed when AeCRN13 and AeCRN5 gene expression levels were normalized to AeTUB gene expression, a constitutively expressed gene frequently used to quantify root colonisation levels (Fig. 2). Thus, these results indicate that N deficiency does not affect the expression of the CRN13 and CRN5 effectors, and therefore possibly does not affect the virulence of A. euteiches.
Figure 2.
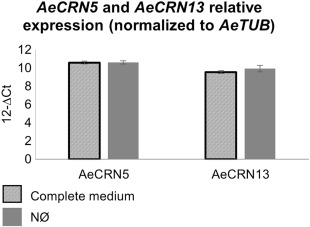
Increased resistance of A17 on nitrate‐deficient (NØ) medium is not related to changes in AeCRN5 and AeCRN13 expression. Transcripts levels of AeCRN5 and AeCRN13 normalized to AeTUB expression. One‐day‐old seedlings were inoculated with Aphanomyces euteiches and cultivated in vitro for 10 days on complete medium or NØ medium. No expression of AeCRN5 and AeCRN13 was detected in control non‐inoculated plants. Error bars indicate standard errors (n = 4 biological replicates).
Effects of nitrogen deficiency on the M. truncatula–A. euteiches interaction are dependent on plant genotype
To study whether N deficiency impacted similarly the behaviour of other genotypes of M. truncatula to A. euteiches, we extended our initial experiments to nine other genotypes.
As for A17, stem necrosis and root browning were assessed and root infection was measured by ELISA. Plant growth and root architectural parameters were also determined as relevant indicators of M. truncatula resistance to A. euteiches. Principal component analysis (PCA) was performed using these different parameters (Bonhomme et al., 2014).
A representative PCA of three independent experiments is displayed in Fig. S2 (see Supporting Information). ELISA, root browning and stem necrosis were positively correlated with each other and negatively correlated with growth parameters, such as shoot biomass. PC1, which captured 44% of the variance, represents a ‘synthetic’ measurement of plant resistance to A. euteiches. Thus, the more a genotype lies on the negative side of this axis, the more susceptible it is to the pathogen. First, the results show that, on complete medium, the different genotypes display different behaviours: F83005.5 and SA028064 represent the most susceptible and most resistant genotypes, respectively, whereas the other genotypes display intermediate levels of resistance. This classification is similar to that found by others (Bonhomme et al., 2014), confirming the relevance of our assessment of resistance to A. euteiches. Second, interestingly, N deficiency affects plant susceptibility to A. euteiches differently according to genotype (Fig. S2).
We focused on four genotypes which showed, reproducibly, a significantly different response to N deficiency (Fig. 3, Table 1): the genotypes F83005.5 and SA022322, which were more susceptible on NØ (NØ‐IS for NØ‐induced susceptibility), and the genotypes A17 and DZA045‐6, which displayed enhanced resistance on NØ (NØ‐IR) (Fig. 3, Table 1). The DZA315‐16 genotype, which displayed a very strong increase in resistance on NØ medium (Fig. S2), was not chosen as its response to N deficiency was too marked compared with the other genotypes, and could therefore not adequately represent the processes involved in the N modulation of plant immunity in the majority of the other genotypes. To understand why NØ medium impacted differently on the resistance of the four genotypes to A. euteiches, we quantified the soluble phenolics and amino acids, and analysed the expression of the genes involved in N metabolism and defence in roots. These analyses were performed at 10 days of culture, a time point at which N deficiency is already effective (Fig. S1).
Figure 3.
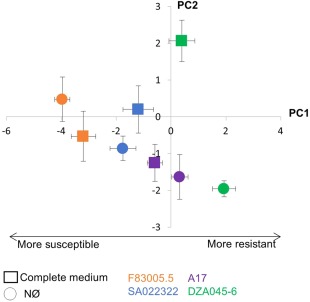
Close‐up of the principal component analysis (PCA) of plant susceptibility to Aphanomyces euteiches (Fig. S2, see Supporting Information), highlighting the behaviour of the four selected genotypes cultivated in complete or nitrate‐deficient (NØ) medium. Susceptibility of the Medicago truncatula genotypes A17, F83005.5, DZA045‐6 and SA022322 under contrasting N availability (complete medium, squares; NØ medium, circles) was compared by PCA (PC1 explains 44% of the variance). The negative side of the PC1 axis is highly correlated with the genotype susceptibility to A. euteiches (ELISA, root browning and stem necrosis, see Fig. S2). Bars indicate standard error (n = 4 biological replicates). Data from a representative experiment out of three independent experiments are shown.
Table 1.
Classification of the four selected genotypes according to the modulation of their resistance to Aphanomyces euteiches by nitrate deficiency.
| Genotypes | PC1NØ – PC1complete * | Behaviour |
|---|---|---|
| DZA045‐6 | 1.7a | NØ‐IR |
| A17 | 1.0a | |
| SA022322 | −0.71b | NØ‐IS |
| F83005.5 | −0.76b |
*Statistical analyses were performed on the difference between the PC1 value obtained on nitrate‐deficient (NØ) medium and the PC1 value obtained on complete medium in three independent experiments. Letters indicate significant differences (P < 0.05).
Nitrogen deficiency leads to changes in secondary metabolism and defence gene expression
We first determined the concentrations of soluble phenolic compounds in roots (Fig. 4). Inoculation by A. euteiches led to a significant increase in soluble phenolic concentrations for all genotypes. Similarly, N deficiency increased the basal concentration of soluble phenolics, and the combination of both N deficiency and inoculation by A. euteiches led to the highest concentrations of these compounds (Fig. 4).
Figure 4.
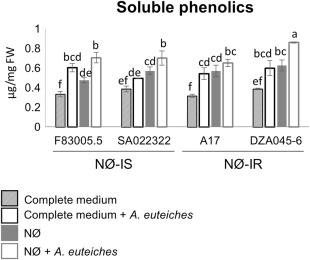
Infection by Aphanomyces euteiches and nitrogen (N) deficiency increase phenolic compounds in Medicago truncatula roots. The concentrations of root‐soluble phenolics of the four M. truncatula genotypes F83005.5, SA022322, A17 and DZA045‐6 were determined in plants cultivated in vitro for 10 days on complete medium or nitrate‐deficient (NØ) medium and inoculated or not with A. euteiches. FW, fresh weight. Error bars indicate standard errors (n = 4 biological replicates) and letters indicate significant differences (P < 0.05). Data from a representative experiment out of three independent experiments are shown. NØ‐IS, genotype displaying NØ‐induced susceptibility; NØ‐IR, genotype displaying NØ‐induced resistance.
To study further the contribution of secondary metabolism to plant resistance, we analysed, by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR), the expression of genes involved in secondary metabolism and, more broadly, in plant defence. Among the numerous genes studied (above 20 genes, Table S1, see Supporting Information), many were identified as more strongly expressed in partially resistant (NR) than in partially susceptible (NS) near‐isogenic lines, and could contribute to plant resistance (Badis et al., 2015). Expression analyses showed that many of these genes were differentially expressed depending on inoculation by A. euteiches, N nutrition or plant genotype (data not shown). A small group of genes displayed interesting profiles (Fig. 5). Three genes (MtDRP, encoding a disease resistance protein; MtBGL2, encoding a β‐1,3‐glucanase; and MtERF1, encoding an ethylene‐responsive transcription factor) were induced significantly by A. euteiches on complete medium in all or almost all genotypes. As they did not discriminate between the genotypes depending on their resistance level, these genes do not appear to contribute to plant resistance in these conditions. On NØ medium, the expression levels of these genes were not correlated with the degree of resistance of the different genotypes. Three other genes (MtCOMT1, MtPAL2, MtCHS2), involved in secondary metabolism, were induced significantly by the pathogen in most genotypes and displayed, interestingly, a basal expression level (e.g. in the absence of pathogen) that was correlated with plant resistance to A. euteiches on complete medium: the most sensitive genotype (F83005.5, Fig. 3) displayed the lowest and the most resistant genotype (DZA045‐6, Fig. 3) displayed the highest expression levels of these genes (Fig. 5). These genes could therefore contribute to plant resistance in complete medium. On NØ medium, the NØ‐IS genotypes displayed a significantly increased basal expression of these genes (except for MtCHS2 for F83005.5), although they were more susceptible to A. euteiches in these conditions (Fig. 5). Conversely the NØ‐IR genotypes displayed no significant change in the basal expression of these genes on NØ medium, despite being more resistant when experiencing N limitation (Fig. 5). Altogether, these expression analyses indicate that these genes possibly contributing to resistance to A. euteiches in complete medium appear not to determine resistance under N deficiency.
Figure 5.
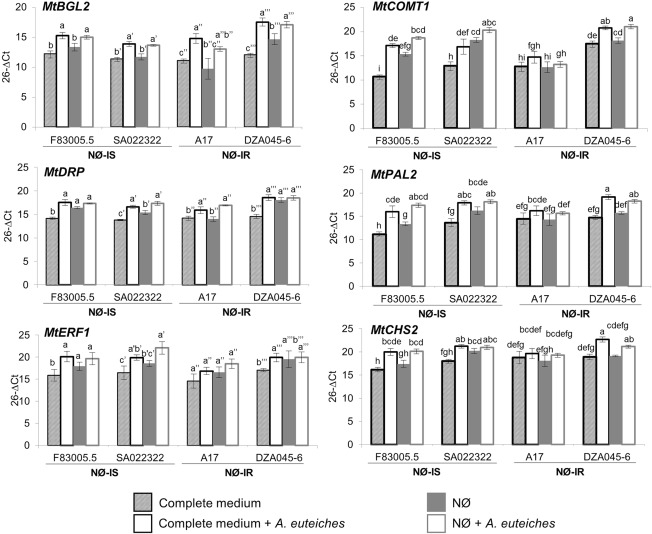
Expression analysis of secondary metabolism and defence genes in the four selected genotypes. Gene expression levels were determined using MtERF1 as reference gene in roots of the four Medicago truncatula genotypes, F83005.5, SA022322, A17 and DZA045‐6, after 10 days of culture on complete medium or nitrate‐deficient (NØ) medium and inoculated or not with Aphanomyces euteiches. Error bars indicate standard errors (n = 4 biological replicates) and letters indicate significant differences (P < 0.05). NØ‐IS, genotype displaying NØ‐induced susceptibility; NØ‐IR, genotype displaying NØ‐induced resistance. MtBGL2, β‐1,3‐glucanase gene; MtDRP, disease resistance protein gene; MtCOMT1, chalcone‐O‐methyltransferase gene; MtPAL2, phenyl ammonia‐lyase gene; MtCHS2, chalcone synthase gene.
Nitrogen deficiency leads to genotype‐specific responses and highlights the low N responsiveness of DZA045‐6
To assess whether NØ‐IR could be linked to an induction of plant defence as a result of N stress, we analysed the response of the four genotypes to N deficiency at the growth and gene expression levels. As shown in Fig. S3 (see Supporting Information), N deficiency affected all genotypes, resulting in lower shoot biomass than in complete medium at 10 and 21 days of culture (Fig. S3A,B) and lower shoot N concentrations at 21 days (Fig. S3C). No effect of genotype could be seen on N concentration. Shoot biomass, however, responded to N deficiency differently depending on the plant genotype (Fig. S3). At 21 days of culture, the highest decrease in shoot biomass on N deficiency was observed for SA022322, an intermediate decrease for A17 and the lowest decrease for DZA045‐6 and F83005.5. This highlighted that the SA022322 and F83005.5 genotypes responded differently to N deficiency despite both displaying NØ‐IS.
We analysed the expression of genes involved in N metabolism: MtNIA1 and MtNIA2, which encode two isoforms of NR; MtNRT2.1 and MtNRT2.3, which encode nitrate transporters (Pellizzaro et al., 2015); MtGS1a, MtGS1b and MtGS2a, which encode the two cytosolic and major plastid‐located Gln synthetase enzymes, respectively (Seabra et al., 2013).
qRT‐PCR analyses were performed in M. truncatula roots after 10 days of culture to assess whether the four genotypes differentially expressed these genes. Two main conclusions can be drawn from the data displayed in Fig. 6. First, the F83005.5, SA022322 and A17 genotypes can be discriminated in complete medium from the DZA045‐6 genotype based on the expression profiles of the MtNRT2.1 and MtNIA1 genes. These nitrate‐regulated genes display a strongly reduced basal expression level (ΔΔCt = 5, that is at least 30‐fold lower) in DZA045‐6 (Fig. 6). This altered response to nitrate of DZA045‐6 was confirmed by its reduced sensitivity to chlorate, a toxic analogue of nitrate, compared with the three other genotypes (Fig. S4, see Supporting Information). Second, on complete medium, for the NØ‐IS genotypes, inoculation led to a significant increase in MtGS1b (and of MtGS1a for F83005.5) mRNA levels, which was not observed in NØ‐IR genotypes in complete medium. Therefore, our developmental and gene analyses did not highlight N responses specific to NØ‐IS or NØ‐IR genotypes, apart from MtGS1b induction in inoculated roots of NØ‐IS genotypes on complete medium.
Figure 6.
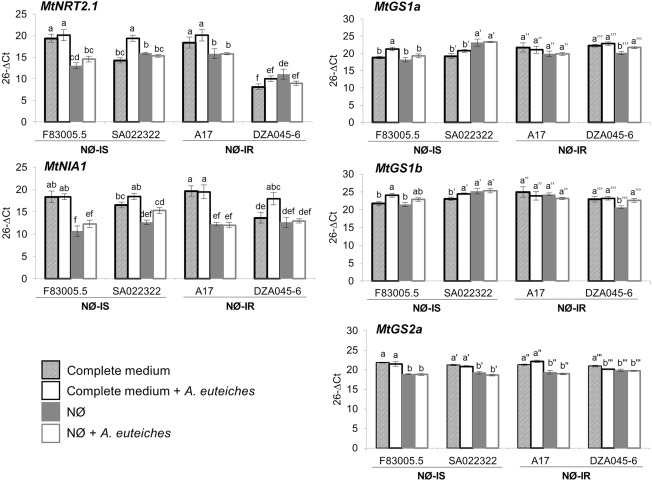
Expression analysis of nitrogen‐linked genes in the four selected genotypes. Gene expression levels were determined using MtERF1 as reference gene in roots of the four Medicago truncatula genotypes, F83005.5, SA022322, A17 and DZA045‐6, after 10 days of culture on complete medium or nitrate‐deficient (NØ) medium and inoculated or not with Aphanomyces euteiches. Error bars indicate standard errors (n = 4 biological replicates) and letters indicate significant differences (P < 0.05). NØ‐IS, genotype displaying NØ‐induced susceptibility; NØ‐IR, genotype displaying NØ‐induced resistance. MtNRT2.1, nitrate transporter gene; MtNIA1, nitrate reductase gene; MtGS1a, MtGS1b, cytosolic glutamine synthetase genes; MtGS2a, chloroplastic glutamine synthetase gene.
Aphanomyces euteiches infection induces higher concentrations of specific amino acids in NØ‐IS genotypes than in NØ‐IR genotypes
To analyse more precisely whether the four selected genotypes differ in their N economy, the concentrations of different amino acids in the roots were determined in the different genotypes after 10 days of culture on complete medium or NØ medium in the presence or not of A. euteiches (Table S2, see Supporting Information). In non‐inoculated roots, cultivation on NØ medium led to marked changes in amino acid concentrations. We observed a significant decrease in the concentration of total amino acids (Fig. S5A, see Supporting Information) on NØ medium in A17 and DZA045‐6, the two genotypes showing NØ‐IR, as well as a significant increase in the concentration of minor amino acids (Fig. S5B) in all genotypes, except DZA045‐6. The enhanced basal levels of these amino acids on NØ medium reflect changes in minor amino acid homeostasis, already highlighted during N starvation in Arabidopsis roots (Krapp et al., 2011). These latter analyses confirmed the altered N response of DZA045‐6.
Six amino acids [Gln, arginine (Arg), lysine (Lys), tyrosine (Tyr), methionine (Met), tryptophan (Trp)] showed interesting profiles, as they responded to A. euteiches differently according to genotype (Fig. 7). In complete medium, on inoculation, a significant increase in these six amino acids was observed in F83005.5 and SA022322 (NØ‐IS genotypes), whereas a smaller increase or no significant change was observed in the NØ‐IR genotypes (A17 and DZA045‐6), resulting in about two‐ to three‐fold lower contents of these amino acids in the latter genotypes when inoculated (Fig. 7A, Table S2). In NØ medium (Fig. 7B, Table S2), a significantly lower concentration in inoculated NØ‐IR roots compared with inoculated NØ‐IS roots was maintained only for Gln (1.5–3‐fold lower; Figs 7B and S6, see Supporting Information). Thus, only Gln levels discriminated between inoculated NØ‐IR and NØ‐IS genotypes on both complete and NØ media, with high levels of Gln correlated with lower levels of resistance. Based on these results and literature reporting a specific role of Gln in plant–pathogen interactions (Liu et al., 2010; Tavernier et al., 2007), we decided to investigate the involvement of this amino acid in A. euteiches infection and plant resistance.
Figure 7.
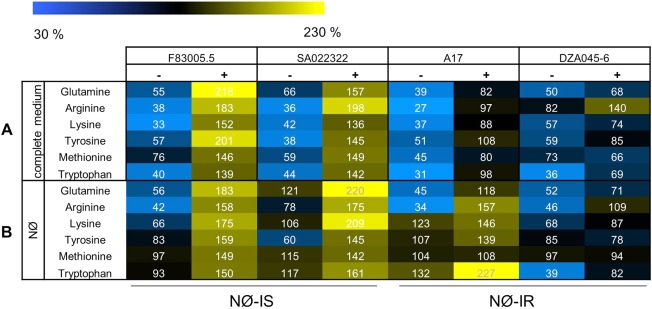
Root contents on complete medium (A) and nitrate‐deficient (NØ) medium (B) for the six specific amino acids whose accumulation is significantly induced by Aphanomyces euteiches in NØ‐IS genotypes on complete medium. Amino acid quantification was performed on four genotypes (F83005.5, SA022322, A17 and DZA045‐6). Plants were cultivated in vitro for 10 days on complete medium or NØ medium and inoculated (+) or not (–) with A. euteiches. Amino acids were measured on three biological replicates. The percentage of the average level of each amino acid is displayed for the four selected genotypes in each condition. For each amino acid, the average levels were determined as the mean levels of the amino acid in all conditions, and were equal to 7.5 µmol/g dry weight for glutamine (Gln), 1.6 µmol/g dry weight for arginine (Arg) and lysine (Lys), 0.8 µmol/g dry weight for tyrosine (Tyr), 0.3 µmol/g dry weight for methionine (Met) and 1.3 µmol/g dry weight for tryptophan (Trp). The colour scale limits were 30% (blue) and 230% (yellow) of the root amino acid average content. Absolute values for amino acid contents are displayed in Table S2 (see Supporting Information). NØ‐IS, genotype displaying NØ‐induced susceptibility; NØ‐IR, genotype displaying NØ‐induced resistance.
Gln is a mediator of the M. truncatula–A. euteiches interaction
To test the effect of Gln on plant susceptibility to A. euteiches, we tested whether the addition of Gln to plant culture medium would induce susceptibility in a condition in which the plant is normally resistant. For this, we cultivated the NØ‐IR genotypes A17 and DZA045‐6 on NØ medium supplemented with Gln as the sole N source. We observed a significant increase in root infection of A17 and DZA045‐6 in this case compared with NØ medium (Fig. 8A), but also compared with complete medium containing nitrate (data not shown).
Figure 8.
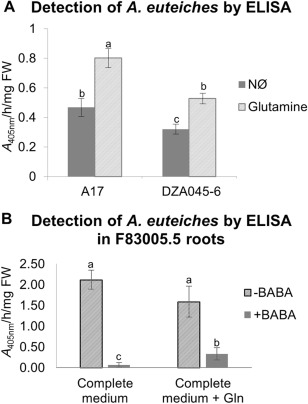
Effect of glutamine (Gln) on susceptibility to Aphanomyces euteiches. Quantification by enzyme‐linked immunosorbent assay (ELISA) of A. euteiches in root extracts. (A) The A17 and DZA045‐6 genotypes were cultivated in vitro for 3 weeks on nitrate‐deficient (NØ) medium or Gln medium and inoculated with A. euteiches. (B) The F83005.5 genotype was cultivated in vitro for 3 weeks on complete medium supplemented or not with 200 µm of β‐aminobutyric acid (BABA), or on complete medium supplemented or not with 200 µm of Gln and 200 µm of BABA. The background signal from control non‐inoculated roots was subtracted from the signal detected in inoculated roots. Error bars indicate standard error (n = 6 biological replicates for ELISAs) and letters indicate significant differences (P < 0.05). Data from a representative experiment out of three (A) and two (B) independent experiments are shown. FW, fresh weight.
Conversely, we tested whether the reduction in M. truncatula susceptibility to A. euteiches was correlated with a decrease in Gln levels. For this, we treated the NØ‐IS genotype (F83005.5) with β‐aminobutyric acid (BABA), a non‐protein amino acid known to prime the plant response to biotic stress (Conrath et al., 2002). We observed that BABA treatment decreased the susceptibility to A. euteiches (Fig. 8B), concomitant with a significant decrease in endogenous Gln content (–50%) in the plant infected by A. euteiches. Interestingly, as reported previously in another biological model (Wu et al., 2010), the addition of Gln partially reversed the protective effect of BABA (Fig. 8B). Therefore, these experiments highlight a role of Gln in the M. truncatula–A. euteiches interaction.
Discussion
In our study, we observed that N availability affects M. truncatula susceptibility to the hemibiotrophic oomycete A. euteiches. This result was not linked, however, to the pathogen lifestyle, but to plant genotype. Indeed, we observed: (1) genotypes that become more susceptible to A. euteiches on NØ medium (NØ‐IS genotypes) and (2) genotypes that become more resistant to A. euteiches on NØ medium (NØ‐IR genotypes).
No correlation could be made, however, between genotype resistance on complete medium and genotype behaviour on NØ medium (NØ‐IS or NØ‐IR). For example, R108 and DZA045‐6 showed a similar resistance to A. euteiches in complete medium (Fig. S2), yet the former genotype displayed NØ‐IS, whereas the latter displayed NØ‐IR.
We propose different hypotheses to explain the genotypic variability of N modulation of M. truncatula resistance to A. euteiches.
First, we tested whether the N modulation of plant resistance could be linked to a genetic variability of the regulation by N of the plant defence response. We analysed the contribution of soluble phenolic compounds, which play an important role in resistance to A. euteiches. Indeed, Djébali et al. (2011) showed that the difference in resistance between A17 and F83005.5 genotypes could be explained, in part, by an increase in soluble phenolics at 3 days post‐inoculation (dpi) in infected A17 roots, which does not occur in the sensitive F83005.5 genotype. Furthermore, secondary metabolites, in particular phenylpropanoids and flavonoids, accumulate during low N nutrition (Fritz et al., 2006) and could thus play a role in NØ‐IR. The quantification of soluble phenolics did not highlight significant differences between genotypes. It is possible that, at the stage at which N deficiency occurs (10 days of culture), the differences in the levels of soluble phenolics occurring at an earlier stage (3–6 dpi; Djébali et al., 2011) between tolerant and susceptible genotypes are overridden by developmentally and N deficiency‐induced increases in soluble phenolics. Alternatively, the lack of specificity of the Folin–Ciocalteu reagent could prevent the accurate determination of soluble phenolics because of the interference with compounds accumulating in these conditions (Everette et al., 2010). The analysis of the expression of the genes involved in secondary metabolism or in defence (Fig. 5) highlighted that the expression levels of a small set of genes were correlated with the plant resistance level on complete medium, and confirmed the potential importance of these genes for plant resistance, as proposed in the analysis of NR and NS near‐isogenic lines (Badis et al., 2015). Our data show, in addition, that the expression of some of these genes is modulated by N deficiency, but no correlation was observed between gene expression and genotype resistance on NØ medium, also suggesting that mechanisms leading to NØ‐IR and NØ‐IS do not involve the same genes. Whether this is indeed the case awaits functional studies assessing the impact of knock‐outs in these genes on plant resistance on complete and NØ medium. In contrast, a recent transcriptomic study on the N modulation of tomato susceptibility to Botrytis cinerea highlighted the repression by the pathogen of a subset of defence response genes in N‐limiting conditions and their induction in N‐sufficient conditions (Vega et al., 2015). It would be interesting to study whether these genes contribute to genotypic variability in the resistance of wild Solanum species to this pathogen.
Second, the contrasting behaviours in the response to A. euteiches in NØ medium could be caused by differences in the plant capacity to use N efficiently from the culture medium and/or to mobilize its N resources during N deficiency. Only a few studies have reported the effect of plant genetic diversity on the modulation of resistance by N. In rice, Ballini et al. (2013) have shown that high N input increases susceptibility to Magnaporthe oryzae and have suggested that high N use efficiency (NUE) is linked to high N‐induced susceptibility. In order to assess this link, we measured shoot biomass and N concentration, monitored the expression of the genes involved in nitrate transport and assimilation, and determined amino acid levels and composition in the four selected genotypes cultivated in different conditions. Although all genotypes responded to N starvation, the modalities of plant adaptation to this nutritional stress varied. Indeed, these different analyses clearly set the DZA045‐6 genotype apart from the other genotypes, as it displayed lower expression of nitrate‐related genes on complete medium (Fig. 6) and strongly reduced amino acid changes in response to N deficiency (Fig. 7); this suggests an altered response to nitrate and N deprivation compared with the other genotypes. A more thorough characterization of the DZA045‐6 genotype would be of interest to assess the importance of altered N response in different processes, including the setting up and regulation of symbioses.
NØ medium affected the different N‐related parameters, but no significant differences discriminating between NØ‐IR and NØ‐IS genotypes were observed. In particular, as mentioned above, although DZA045‐6 was NØ‐IR, like the A17 genotype, it displayed an altered response to nitrate compared with A17 (Figs 6 and S4). Likewise, the two NØ‐IS genotypes, F83005.5 and SA022322, displayed the lowest and highest decrease in shoot biomass on N deficiency, respectively (Fig. S3). These results suggest that N modulation of plant resistance to A. euteiches is not correlated with plant response to N availability.
The analysis of amino acid composition highlighted six amino acids (Gln, Arg, Lys, Tyr, Met, Trp) which were highly induced by infection in NØ‐IS genotypes, but much less or not at all in NØ‐IR genotypes in complete medium. In NØ medium, the differences in these amino acid levels between inoculated NØ‐IS genotypes and the inoculated NØ‐IR genotype A17 were attenuated for most minor amino acids. Gln levels, however, discriminated between NØ‐IS and NØ‐IR genotypes in both N‐replete and N‐deficient conditions, with high Gln levels associated with enhanced plant susceptibility.
This positive correlation was further sustained by two other experiments: (i) Gln provision prevented NØ‐IR and even led to enhanced susceptibility compared with complete medium (containing nitrate as sole N source), stressing the specific role of Gln in favouring A. euteiches infection (Fig. 8A); (ii) conversely, a treatment (BABA) inducing resistance in a NØ‐IS genotype was correlated with lower levels of Gln and was partially reversed by the addition of Gln (Fig. 8B).
Gln is known to play a role in plant–pathogen interactions. On the pathogen side, Gln is well known to be a favourable N source for fungal development in vitro (Tudzynski, 2014) and high levels of Gln in planta could therefore favour pathogen growth. Conversely, Gln has been shown to repress fungal virulence in the hemibiotrophic fungus Fusarium oxysporum and to inhibit the formation of appressoria, highly specialized infection structures, in the rice blast fungus Magnaporthe oryzae (López‐Berges et al., 2010; Marroquin‐Guzman and Wilson, 2015). On the plant side, reports show that the plant metabolic pathways activated during the necrotrophic stage are similar to those induced during senescence (Swartzberg et al., 2007; Tavernier et al., 2007). The accumulation of Gln linked to N remobilization by cytosolic Gln synthetase in the plant host has been proposed to be involved in plant defence in response to Colletotrichum lindemuthianum (Tavernier et al., 2007), and is correlated with the onset of the necrotrophic phase of the pathogen. Gln accumulation has been proposed to signal to the pathogenic fungus that the plant is translocating N and to possibly trigger the onset of the necrotrophic stage. In complete medium, we observed a higher accumulation of transcripts of the MtGS1b gene on root inoculation in the lines displaying NØ‐IS (Fig. 6), suggestive of the induction of N remobilization during pathogen infection, but this was not observed on NØ medium (Fig. 6). Therefore, it would be interesting to test whether other types of regulation affect Gln synthetase, as Tyr nitration of GS1a has been proposed to control plant–microbe interactions in M. truncatula (Melo et al., 2011). More recent studies have identified Gln as an important mediator of plant–pathogen interactions through redox modulation. Liu et al. (2010) have shown that knocking out a single amino acid transporter gene (LHT1), which uses Gln as a main substrate, is sufficient to confer the resistance of Arabidopsis against a large spectrum of pathogens. These studies highlight the specific role of Gln, rather than plant N status, in plant defence, with Gln deficiency leading to the activation of plant defence responses by enhancing cellular redox stress. Our data are consistent with this role of Gln in plant immunity, as the more susceptible genotypes displayed higher Gln contents on infection, and conditions favouring Gln accumulation increased A. euteiches infection.
The effect of Gln on plant immunity appears to be broad, because it also decreases the protection against A. euteiches conferred by BABA in the susceptible F83005.5 genotype. Gln has been reported to reverse a wide range of phenotypes induced by BABA, including reduced plant growth, anthocyanin accumulation, resistance to heat stress and resistance to a bacterial pathogen (Wu et al., 2010). This reversal has been attributed to the alleviation of an amino acid stress induced by BABA, as it is a non‐proteinogenic amino acid which cannot be metabolized by plants. BABA has been reported to alter amino acid balance (Luna et al., 2014; Singh et al., 2010), but whether BABA affects Gln levels has not been reported.
In conclusion, in this work, we show that N deficiency affects the resistance of M. truncatula to A. euteiches and this is dependent on plant genotype. Mechanisms leading to NØ‐IR and NØ‐IS seem to be independent of those involved in quantitative resistance (Badis et al., 2015; Djébali et al., 2011) and in plant perception of N deficiency. Finally, we have identified Gln as an N source important for A. euteiches infection. This suggests the role of Gln in a cross‐talk between plant and pathogen, but the exact mechanisms involving Gln production and its effects on A. euteiches infection remain unknown.
Experimental Procedures
Plant growth and inoculation by A. euteiches
Ten M. truncatula genotypes, belonging to the core collection CC8 (INRA, Montpellier, France) (Jemalong‐A17, F83005.5, DZA045‐6, SA022322, SA028064, SALSES71B, ESP105.L, DZA315‐16 and DZA012‐J) and R108, were used in this study. The seeds of M. truncatula were scarified according to Djébali et al. (2009). After overnight at 4 °C, seeds were germinated in phytochambers with 16 h light at 350 µmol/m2/s at 23 °C/8 h night cycle at 21 °C. One day after germination, seedlings were transferred to square plates (12 cm × 12 cm) containing a modified M medium (Bécard and Fortin, 1988). This modified medium was enriched in phosphate (final concentration, 1.3 mm) without sugar and contained either 3.3 mm nitrate (complete medium) or no nitrate (NØ medium). After sealing with parafilm, plates were placed vertically in the culture chamber (16 h light at 350 µmol/m2/s at 23 °C/8 h night cycle at 21 °C) for 7–21 days. Altogether, 20 seedlings per condition (defined as a given genotype cultivated on complete or NØ medium and inoculated or not) were analysed. Four biological replicates per condition were made by pooling the roots from five seedlings for each replicate at harvest.
The strain Aphanomyces euteiches Drechs ATCC 201684 was used to inoculate seeds 1 day after germination. Zoospores were produced as described in Rey et al. (2013) and roots were inoculated with 500 zoospores.
Symptom annotations and root architectural analyses
The effects of inoculation and N deficiency were scored after 10 or 21 days of culture by determining plant growth and developmental parameters [root and shoot biomass, primary root (RI) length, lateral root (LR) number and length] and disease symptoms (Bonhomme et al., 2014; Djébali et al., 2009), including the percentage of dead plants, percentage of yellow cotyledons, stem necrosis, percentage of primary root browning and ELISA. Stem necrosis was scored semi‐quantitatively as ‘0’ (no symptom), ‘1’ (partial necrosis) or ‘2’ (total necrosis of stem) for each living plant, and the total stem necrosis score for a given condition is expressed as a percentage of the maximal possible level of stem necrosis, that is: 100 × sum of necrosis scores of scored plants/(2 × number of scored plants).
Root system architecture was analysed on scans of plates containing seedlings grown for 10 days on complete medium or NØ medium in the presence or not of A. euteiches, using ImageJ software (Schneider et al., 2012) to measure the RI and LR lengths and LR density (which is the average number of LRs per centimetre of RI). For each experiment, about 20 seedlings per condition were analysed.
Assessment of infection by ELISA
Assessment of A. euteiches development in roots was performed by ELISA using a rabbit polyclonal serum raised against A. euteiches and a mouse anti‐rabbit IgG alkaline phosphatase conjugate, as described by Slezack et al. (1999), on protein extracts obtained from roots from pooled plants after 21 days of culture. Alkaline phosphatase activity was monitored by following the increase in absorbance at 405 nm, and is expressed as the slope of the obtained curve per milligram of root fresh weight.
RNA extraction, reverse transcription and quantitative PCR on transformed roots
Total RNA was extracted from transformed roots using TRIzol® Reagent (Fisher Scientific, Illkirch, France) according to the recommendations of the manufacturer. For quantitative PCR, RNAs (0.5–1 µg) were reverse transcribed in a final volume of 20 µL in the presence of RNasin (Promega, Charbonnières, France) and oligo(dT)15, with MMLV reverse transcriptase (Promega), as recommended by the manufacturer.
Quantitative PCR was performed on reverse‐transcribed RNAs on four independent biological replicates per condition. For the analysis of the four selected genotypes, 16 conditions [4 genotypes × 2 nutritions × 2 inoculation conditions (mock/inoculated)] were analysed and two independent plant cultures were performed. Technical replicates showed a much lower variance than biological replicates. Quantitative PCRs were performed in an ABI PRISM 7900 apparatus (Applied Biosystems®, Saint‐Aubin, France) in a final volume of 15 µL using Absolute SYBR green ROX Mix (Thermo Scientific, Camberley, Surrey, UK), 0.3 µm of gene‐specific primers and 5 µL of cDNA template diluted 60‐fold. The reference genes used for normalization were MtEF1α and MtUBQ. Expression levels were calculated by determining ΔCt between test and reference genes (ΔCttest gene – reference gene). As both reference genes led to similar results, only data obtained with MtEF1 are shown. For MtEF1, the average Ct in all experiments was equal to 14 cycles. As 40 cycles were performed for qPCR analysis, a gene was considered not to be expressed if ΔCttest gene – reference gene ≥ 26. Therefore, gene expression was quantified by the parameter 26 – ΔCttest gene – reference gene, which is higher when the test gene is more highly expressed (Bari et al., 2006; Truong et al., 2015). When normalizing AeCRN5 and AeCRN13 expression to AeTUB expression, as the average Ct of AeTUB was 28, the value 12 – ΔCttest gene – AeTUBgene was used for the quantification of gene expression. Primers used for qPCR all displayed a high amplification efficiency (90%–100%) and their sequences are listed in Table S3 (see Supporting Information).
Soluble phenolics and amino acid quantification
Soluble phenolic quantification was performed on 100 mg root fresh weight according to Singleton and Rossi (1965) using the Folin–Ciocalteu reagent and ferulic acid as a reference standard.
Profiling of the amino acid content in plant samples was performed using the extraction and chromatographic procedures described in Jubault et al. (2008).
Statistical analysis
Statistical analyses were performed using one‐ or two‐way analysis of variance (ANOVA), followed by Fisher's test. Data were considered to be significantly different for P < 0.05. The PCA and Monte Carlo tests were performed using the statistical ade4TkGUI software to integrate the different measurements of plant development and susceptibility to A. euteiches.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 The A17 genotype experiences nitrogen (N) deficiency and is less affected by Aphanomyces euteiches on nitrate‐deficient (NØ) medium. content in shoot at 10 days (A), total biomass at 10 days (B), shoot to root ratio at 10 days (C) and percentage of total nitrogen at 14 days (D) in A17 genotype cultivated on complete medium (hatched bars) or NØ medium (grey bars). Data from a representative experiment are shown. Error bars indicate standard error (n = 4 biological samples for content and total biomass; n = 16–20 for shoot to root ratio; n = 4–6 for percentage total nitrogen). Asterisks denote significant differences (P < 0.05). FW, fresh weight. (E) A17 seedlings 10 days after culture on complete or NØ medium and inoculated or not with A. euteiches. Scale bar: 2 cm.
Fig. S2 Nitrogen (N) deficiency affects plant susceptibility to Aphanomyces euteiches differently according to genotype. Principal component analysis (PCA) of plant susceptibility to A. euteiches in the 10 different genotypes cultivated in complete or nitrate‐deficient (NØ) medium. (A) Correlation circle of the measured variables. The negative side of the PC1 axis is highly correlated with the genotype susceptibility to A. euteiches [enzyme‐linked immunosorbent assay (ELISA), root and stem browning]. FW, fresh weight. (B) PCA representation of the susceptibility of different genotypes of M. truncatula (A17, F83005.5, DZA045‐6, SA022322, SA028064, SALSES71B, ESP105.L, R108, DZA315.16 and DZA012.J) under contrasting N availability (complete medium, squares; NØ medium, circles). Bars indicate standard error (n = 4 biological replicates). Data from a representative experiment out of three independent experiments are shown.
Fig. S3 Analysis of the response to nitrogen (N) deficiency (NØ) of the four selected genotypes. (A) Shoot biomass at 21 days. (B) Shoot biomass at 10 days. (C) N concentration in shoots at 21 days. Error bars indicate standard errors (n = 4–6 biological samples) and letters indicate significant differences (P < 0.05). NØ‐IS, genotype displaying NØ‐induced susceptibility; NØ‐IR, genotype displaying NØ‐induced resistance.
Fig. S4 Enhanced resistance to chlorate of the DZA045‐6 genotype. The four genotypes (F83005.5, SA022322, A17 and DZA045‐6) were cultivated in vitro for 10 days on complete medium in the presence of different concentrations of chlorate, and the primary root length was determined. Error bars indicate standard error (n = 4 biological replicates).
Fig. S5 Quantification of total and minor amino acid levels in roots of the four selected genotypes. (A) Total amino acid levels. (B) Minor amino acid levels. DW, dry weight. Amino acid quantification was performed on four genotypes (F83005.5, SA022322, A17 and DZA045‐6). Plants were cultivated in vitro for 10 days on complete medium or nitrate‐deficient (NØ) medium and inoculated or not with Aphanomyces euteiches. Error bars indicate standard errors (n = 3 biological replicates) and letters indicate significant differences (P < 0.05).
Fig. S6 Glutamine levels in the four selected genotypes cultivated in vitro for 10 days on complete or nitrate‐deficient (NØ) medium in the presence or not of Aphanomyces euteiches. Error bars indicate standard error (n = 3 biological replicates) and letters indicate significant differences (P < 0.05).
Table S1 Defence‐related/secondary metabolism genes analysed by RT‐qPCR in the four selected genotypes cultivated in complete or N‐deficient NØ medium and inoculated or not with A. euteiches.
Table S2 Root amino acid concentrations (µmol/g dry weight) in the four selected genotypes. Letters indicate significant differences (n = 3 biological replicates, P < 0.05).
Table S3 Primers used for gene expression analyses by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR).
Acknowledgements
We would like to thank E. Gaulin, A. Nars and C. Jacquet for providing the A. euteiches strain Drechs ATCC 201684 and for advice on the pathosystem, E. Dumas‐Gaudot for the polyclonal antibody directed against A. euteiches and J.‐M. Prospéri for seeds of the CC8 collection.
E. Thalineau was funded by a PhD contract (2013‐32) from the Ministère de l'Enseignement Supérieur et de la Recherche. This work was supported by a grant from AgroSup Dijon and the Conseil Régional de Bourgogne (PARI8).
References
- Badis, Y. , Bonhomme, M. , Lafitte, C. , Huguet, S. , Balzergue, S. , Dumas, B. and Jacquet, C. (2015) Transcriptome analysis highlights preformed defences and signalling pathways controlled by the prAe1 quantitative trait locus (QTL), conferring partial resistance to Aphanomyces euteiches in Medicago truncatula . Mol. Plant Pathol. 16, 973–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballini, E. , Nguyen, T.T. and Morel, J.‐B. (2013) Diversity and genetics of nitrogen‐induced susceptibility to the blast fungus in rice and wheat. Rice, 6, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari, R. , Datt Pant, B. , Stitt, M. and Scheible, W.R. (2006) PHO2, microRNA399, and PHR1 define a phosphate‐signaling pathway in plants. Plant Physiol. 141, 988–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bécard, G. and Fortin, J.A. (1988) Early events of vesicular–arbuscular mycorrhiza formation on Ri T‐DNA transformed roots. New Phytol. 108, 211–218. [DOI] [PubMed] [Google Scholar]
- Bonhomme, M. , Andre, O. , Badis, Y. , Ronfort, J. , Burgarella, C. , Chantret, N. , Prosperi, J.M. , Briskine, R. , Mudge, J. , Debelle, F. , Navier, H. , Miteul, H. , Hajri, A. , Baranger, A. , Tiffin, P. , Dumas, B. , Pilet‐Nayel, M.L. , Young, N.D. and Jacquet, C. (2014) High‐density genome‐wide association mapping implicates an F‐box encoding gene in Medicago truncatula resistance to Aphanomyces euteiches . New Phytol. 201, 1328–1342. [DOI] [PubMed] [Google Scholar]
- Bonneau, L. , Huguet, S. , Wipf, D. , Pauly, N. and Truong, H.N. (2013) Combined phosphate and nitrogen limitation generates a nutrient stress transcriptome favorable for arbuscular mycorrhizal symbiosis in Medicago truncatula . New Phytol. 199, 188–202. [DOI] [PubMed] [Google Scholar]
- Camañes, G. , Pastor, V. , Cerezo, M. , García‐Andrade, J. , Vicedo, B. , García‐Agustín, P. and Flors, V. (2012) A deletion in NRT2.1 attenuates Pseudomonas syringae‐induced hormonal perturbation, resulting in primed plant defenses. Plant Physiol. 158, 1054–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrath, U. , Pieterse, C.M. and Mauch‐Mani, B. (2002) Priming in plant–pathogen interactions. Trends Plant Sci. 7, 210–216. [DOI] [PubMed] [Google Scholar]
- Dechorgnat, J. , Patrit, O. , Krapp, A. , Fagard, M. and Daniel‐Vedele, F. (2012) Characterization of the Nrt2.6 gene in Arabidopsis thaliana: a link with plant response to biotic and abiotic stress. PLoS One, 7, e42491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich, R. , Ploss, K. and Heil, M. (2004) Constitutive and induced resistance to pathogens in Arabidopsis thaliana depends on nitrogen supply. Plant Cell Environ. 27, 896–906. [Google Scholar]
- Djébali, N. , Jauneau, A. , Ameline‐Torregrosa, C. , Chardon, F. , Jaulneau, V. , Mathe, C. , Bottin, A. , Cazaux, M. , Pilet‐Nayel, M.L. , Baranger, A. , Aouani, M.E. , Esquerre‐Tugaye, M.T. , Dumas, B. , Huguet, T. and Jacquet, C. (2009) Partial resistance of Medicago truncatula to Aphanomyces euteiches is associated with protection of the root stele and is controlled by a major QTL rich in proteasome‐related genes. Mol. Plant–Microbe Interact. 22, 1043–1055. [DOI] [PubMed] [Google Scholar]
- Djébali, N. , Mhadhbi, H. , Lafitte, C. , Dumas, B. , Esquerré‐Tugayé, M.‐T. , Aouani, M.E. and Jacquet, C. (2011) Hydrogen peroxide scavenging mechanisms are components of Medicago truncatula partial resistance to Aphanomyces euteiches . Eur. J. Plant Pathol. 131, 559–571. [Google Scholar]
- Djébali, N. , Aribi, S. , Taamalli, W. , Arraouadi, S. , Aouani, M.E. and Badri, M. (2012) Natural variation of Medicago truncatula resistance to Aphanomyces euteiches . Eur. J. Plant Pathol. 135, 831–843. [Google Scholar]
- Dordas, C. (2008) Role of nutrients in controlling plant diseases in sustainable agriculture. A review. Agron. Sustain. Dev. 28, 33–46. [Google Scholar]
- Everette, J.D. , Bryant, Q.M. , Green, A.M. , Abbey, Y.A. , Wangila, G.W. and Walker, R.B. (2010) Thorough study of reactivity of various compound classes toward the Folin−Ciocalteu reagent. J. Agric. Food Chem. 58, 8139–8144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard, M. , Launay, A. , Clément, G. , Courtial, J. , Dellagi, A. , Farjad, M. , Krapp, A. , Soulié, M.‐C. and Masclaux‐Daubresse, C. (2014) Nitrogen metabolism meets phytopathology. J. Exp. Bot. 65, 5643–5656. [DOI] [PubMed] [Google Scholar]
- Fritz, C. , Palacios‐Rojas, N. , Feil, R. and Stitt, M. (2006) Regulation of secondary metabolism by the carbon-nitrogen status in tobacco: nitrate inhibits large sectors of phenylpropanoid metabolism. Plant J. 46, 533–548. [DOI] [PubMed] [Google Scholar]
- Gaulin, E. , Jacquet, C. , Bottin, A. and Dumas, B. (2007) Root rot disease of legumes caused by Aphanomyces euteiches . Mol. Plant Pathol. 8, 539–548. [DOI] [PubMed] [Google Scholar]
- Gaulin, E. , Madoui, M.‐A. , Bottin, A. , Jacquet, C. , Mathé, C. , Couloux, A. , Wincker, P. and Dumas, B. (2008) Transcriptome of Aphanomyces euteiches: new oomycete putative pathogenicity factors and metabolic pathways. PLoS One, 3, e1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, K.J. , Brotman, Y. , Segu, S. , Zeier, T. , Zeier, J. , Persijn, S.T. , Cristescu, S.M. , Harren, F.J. , Bauwe, H. , Fernie, A.R. , Kaiser, W.M. and Mur, L.A. (2013) The form of nitrogen nutrition affects resistance against Pseudomonas syringae pv. phaseolicola in tobacco. J. Exp. Bot. 64, 553–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacquard, S. , Kracher, B. , Hiruma, K. , Münch, P.C. , Garrido‐Oter, R. , Thon, M.R. , Weimann, A. , Damm, U. , Dallery, J.F. , Hainaut, M. , Henrissat, B. , Lespinet, O. , Sacristán, S. , Ver Loren van Themaat, E. , Kemen, E. , McHardy, A.C. , Schulze‐Lefert, P. and O'Connell, R.J. (2016) Survival trade‐offs in plant roots during colonization by closely related beneficial and pathogenic fungi. Nat. Commun. 7, 11 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, I.S. , An, S.H. and Hwang, B.K. (2011) Pepper asparagine synthetase 1 (CaAS1) is required for plant nitrogen assimilation and defense responses to microbial pathogens. Plant J. 67, 749–762. [DOI] [PubMed] [Google Scholar]
- Jubault, M. , Hamon, C. , Gravot, A. , Lariagon, C. , Delourme, R. , Bouchereau, A. and Manzanares‐Dauleux, M.J. (2008) Differential regulation of root arginine catabolism and polyamine metabolism in clubroot‐susceptible and partially resistant Arabidopsis genotypes. Plant Physiol. 146, 2008–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp, A. , Berthomé, R. , Orsel, M. , Mercey‐Boutet, S. , Yu, A. , Castaings, L. , Elftieh, S. , Major, H. , Renou, J.P. and Daniel‐Vedele, F. (2011) Arabidopsis roots and shoots show distinct temporal adaptation patterns toward nitrogen starvation. Plant Physiol. 157, 1255–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, G. , Ji, Y. , Bhuiyan, N.H. , Pilot, G. , Selvaraj, G. , Zou, J. and Wei, Y. (2010) Amino acid homeostasis modulates salicylic acid‐associated redox status and defense responses in Arabidopsis. Plant Cell, 22, 3845–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López‐Berges, M.S. , Rispail, N. , Prados‐Rosales, R.C. and Pietro, A.D. (2010) A nitrogen response pathway regulates virulence functions in Fusarium oxysporum via the protein kinase TOR and the bZIP Protein MeaB. Plant Cell, 22, 2459–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna, E. , van Hulten, M. , Zhang, Y. , Berkowitz, O. , Lopez, A. , Petriacq, P. , Sellwood, M.A. , Chen, B. , Burrell, M. , van de Meene, A. , Pieterse, C.M. , Flors, V. and Ton, J. (2014) Plant perception of β‐aminobutyric acid is mediated by an aspartyl‐tRNA synthetase. Nat. Chem. Biol. 10, 450–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marroquin‐Guzman, M. and Wilson, R.A. (2015) GATA‐dependent glutaminolysis drives appressorium formation in Magnaporthe oryzae by suppressing TOR inhibition of cAMP/PKA signaling. PLoS Pathog. 11, e1004851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux‐Daubresse, C. , Daniel‐Vedele, F. , Dechorgnat, J. , Chardon, F. , Gaufichon, L. and Suzuki, A. (2010) Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann. Bot. 105, 1141–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo, P.M. , Silva, L.S. , Ribeiro, I. , Seabra, A.R. and Carvalho, H.G. (2011) Glutamine synthetase is a molecular target of nitric oxide in root nodules of Medicago truncatula and is regulated by tyrosine nitration. Plant Physiol. 157, 1505–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelstrass, K. , Treutter, D. , Plessl, M. , Heller, W. , Elstner, E.F. and Heiser I. (2006) Modification of primary and secondary metabolism of potato plants by nitrogen application differentially affects resistance to Phytophthora infestans and Alternaria solani . Plant Biol. 8, 653–661. [DOI] [PubMed] [Google Scholar]
- Modolo, L.V. , Augusto, O. , Almeida, I.M.G. , Pinto‐Maglio, C.A.F. , Oliveira, H.C. , Seligman, K. and Salgado, I. (2006) Decreased arginine and nitrite levels in nitrate reductase‐deficient Arabidopsis thaliana plants impair nitric oxide synthesis and the hypersensitive response to Pseudomonas syringae . Plant Sci. 171, 34–40. [Google Scholar]
- Mur, L.A.J. , Simpson, C. , Kumari, A. , Gupta, A.K. and Gupta, K.J. (2017) Moving nitrogen to the centre of plant defence against pathogens. Ann. Bot. 119, 703–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olea, F. , Pérez‐García, A. , Cantón, F.R. , Rivera, M.E. , Cañas, R. , Avila, C. , Cazorla, F.M. , Cánovas, F.M. and de Vicente, A. (2004) Up‐regulation and localization of asparagine synthetase in tomato leaves infected by the bacterial pathogen Pseudomonas syringae . Plant Cell Physiol. 45, 770–780. [DOI] [PubMed] [Google Scholar]
- Oliveira, H.C. , Justino, G.C. , Sodek, L. and Salgado, I. (2009) Amino acid recovery does not prevent susceptibility to Pseudomonas syringae in nitrate reductase double‐deficient Arabidopsis thaliana plants. Plant Sci. 176, 105–111. [Google Scholar]
- Pellier, A.‐L. , Laugé, R. , Veneault‐Fourrey, C. and Langin, T. (2003) CLNR1, the AREA/NIT2‐like global nitrogen regulator of the plant fungal pathogen Colletotrichum lindemuthianum is required for the infection cycle. Mol. Microbiol. 48, 639–655. [DOI] [PubMed] [Google Scholar]
- Pellizzaro, A. , Clochard, T. , Planchet, E. , Limami, A.M. and Morère‐Le Paven, M.‐C. (2015) Identification and molecular characterization of Medicago truncatula NRT2 and NAR2 families. Physiol. Plant. 154, 256–269. [DOI] [PubMed] [Google Scholar]
- Peng, M. , Bi, Y.‐M. , Zhu, T. and Rothstein, S.J. (2007) Genome‐wide analysis of Arabidopsis responsive transcriptome to nitrogen limitation and its regulation by the ubiquitin ligase gene NLA. Plant Mol. Biol. 65, 775–797. [DOI] [PubMed] [Google Scholar]
- Pilet‐Nayel, M.L. , Prosperi, J.M. , Hamon, C. , Lesne, A. , Lecointe, R. , Le Goff, I. , Herve, M. , Deniot, G. , Delalande, M. , Huguet, T. , Jacquet, C. and Baranger, A. (2009) AER1, a major gene conferring resistance to Aphanomyces euteiches in Medicago truncatula . Phytopathology, 99, 203–208. [DOI] [PubMed] [Google Scholar]
- Ramirez‐Garcés, D. , Camborde, L. , Pel, M.J.C. , Jauneau, A. , Martinez, Y. , Néant, I. , Leclerc, C. , Moreau, M. , Dumas, B. and Gaulin, E. (2016) CRN13 candidate effectors from plant and animal eukaryotic pathogens are DNA‐binding proteins which trigger host DNA damage response. New Phytol. 210, 602–617. [DOI] [PubMed] [Google Scholar]
- Rey, T. , Nars, A. , Bonhomme, M. , Bottin, A. , Huguet, S. , Balzergue, S. , Jardinaud, M.F. , Bono, J.J. , Cullimore, J. , Dumas, B. , Gough, C. and Jacquet, C. (2013) NFP, a LysM protein controlling Nod factor perception, also intervenes in Medicago truncatula resistance to pathogens. New Phytol. 198, 875–886. [DOI] [PubMed] [Google Scholar]
- Schachtman, D.P. and Shin, R. (2007) Nutrient sensing and signaling: NPKS. Annu. Rev. Plant Biol. 58, 47–69. [DOI] [PubMed] [Google Scholar]
- Schneider, C.A. , Rasband, W.S. and Eliceiri, K.W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabra, A.R. , Silva, L.S. and Carvalho, H.G. (2013) Novel aspects of glutamine synthetase (GS) regulation revealed by a detailed expression analysis of the entire GS gene family of Medicago truncatula under different physiological conditions. BMC Plant Biol. 13, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, P. , Wu, C.‐C. and Zimmerli, L. (2010) Beta‐aminobutyric acid priming by stress imprinting. Plant Signal. Behav. 5, 878–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton, V.L. and Rossi, J.A. (1965) Colorimetry of total phenolics with phosphomolybdic‐phosphotungstic acid reagents. Am. J. Enol. Vitic. 16, 144–158. [Google Scholar]
- Slezack, S. , Dumas‐Gaudot, E. , Rosendahl, S. , Kjøller, R. , Paynot, M. , Negrel, J. and Gianinazzi, S. (1999) Endoproteolytic activities in pea roots inoculated with the arbuscular mycorrhizal fungus Glomus mosseae and/or Aphanomyces euteiches in relation to bioprotection. New Phytol. 142, 517–529. [Google Scholar]
- Snoeijers, S.S. , Pérez‐García, A. , Joosten, M.H.A.J. and Wit, P.J.G.M.D. (2000) The effect of nitrogen on disease development and gene expression in bacterial and fungal plant pathogens. Eur. J. Plant Pathol. 106, 493–506. [Google Scholar]
- Solomon, P.S. and Oliver, R.P. (2002) Evidence that gamma‐aminobutyric acid is a major nitrogen source during Cladosporium fulvum infection of tomato. Planta, 214, 414–420. [DOI] [PubMed] [Google Scholar]
- Solomon, P.S. , Tan, K.‐C. and Oliver, R.P. (2003) The nutrient supply of pathogenic fungi; a fertile field for study. Mol. Plant Pathol. 4, 203–210. [DOI] [PubMed] [Google Scholar]
- Swartzberg, D. , Kirshner, B. , Rav‐David, D. , Elad, Y. and Granot, D. (2007) Botrytis cinerea induces senescence and is inhibited by autoregulated expression of the IPT gene. Eur. J. Plant Pathol. 120, 289–297. [Google Scholar]
- Tavernier, V. , Cadiou, S. , Pageau, K. , Laugé, R. , Reisdorf‐Cren, M. , Langin, T. and Masclaux‐Daubresse, C. (2007) The plant nitrogen mobilization promoted by Colletotrichum lindemuthianum in Phaseolus leaves depends on fungus pathogenicity. J. Exp. Bot. 58, 3351–3360. [DOI] [PubMed] [Google Scholar]
- Thalineau, E. , Truong, H.‐N. , Berger, A. , Fournier, C. , Boscari, A. , Wendehenne, D. and Jeandroz, S. (2016) Cross‐regulation between N metabolism and nitric oxide (NO) signaling during plant immunity. Front. Plant Sci. 7, 472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong, H.N. , Thalineau, E. , Bonneau, L. , Fournier, C. , Potin, S. , Balzergue, S. , van Tuinen, D. , Jeandroz, S. and Morandi, D. (2015) The Medicago truncatula hypermycorrhizal B9 mutant displays an altered response to phosphate and is more susceptible to Aphanomyces euteiches . Plant Cell Environ. 38, 73–88. [DOI] [PubMed] [Google Scholar]
- Tudzynski, B. (2014) Nitrogen regulation of fungal secondary metabolism in fungi. Front. Microbiol. 5, 656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandemark, G.J. and Grünwald, N.J. (2004) Reaction of Medicago truncatula to Aphanomyces Euteiches race 2. Arch. Phytopathol. Plant Prot. 37, 59–67. [Google Scholar]
- Vega, A. , Canessa, P. , Hoppe, G. , Retamal, I. , Moyano, T.C. , Canales, J. , Gutiérrez, R.A. and Rubilar, J. (2015) Transcriptome analysis reveals regulatory networks underlying differential susceptibility to Botrytis cinerea in response to nitrogen availability in Solanum lycopersicum . Front Plant Sci. 6, 911–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C.‐C. , Singh, P. , Chen, M.‐C. and Zimmerli, L. (2010) L‐Glutamine inhibits beta‐aminobutyric acid‐induced stress resistance and priming in Arabidopsis. J. Exp. Bot. 61, 995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaeno, T. and Iba, K. (2008) BAH1/NLA, a RING‐type ubiquitin E3 ligase, regulates the accumulation of salicylic acid and immune responses to Pseudomonas syringae DC3000. Plant Physiol. 148, 1032–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeier, J. (2013) New insights into the regulation of plant immunity by amino acid metabolic pathways. Plant Cell Environ. 36, 2085–2103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 The A17 genotype experiences nitrogen (N) deficiency and is less affected by Aphanomyces euteiches on nitrate‐deficient (NØ) medium. content in shoot at 10 days (A), total biomass at 10 days (B), shoot to root ratio at 10 days (C) and percentage of total nitrogen at 14 days (D) in A17 genotype cultivated on complete medium (hatched bars) or NØ medium (grey bars). Data from a representative experiment are shown. Error bars indicate standard error (n = 4 biological samples for content and total biomass; n = 16–20 for shoot to root ratio; n = 4–6 for percentage total nitrogen). Asterisks denote significant differences (P < 0.05). FW, fresh weight. (E) A17 seedlings 10 days after culture on complete or NØ medium and inoculated or not with A. euteiches. Scale bar: 2 cm.
Fig. S2 Nitrogen (N) deficiency affects plant susceptibility to Aphanomyces euteiches differently according to genotype. Principal component analysis (PCA) of plant susceptibility to A. euteiches in the 10 different genotypes cultivated in complete or nitrate‐deficient (NØ) medium. (A) Correlation circle of the measured variables. The negative side of the PC1 axis is highly correlated with the genotype susceptibility to A. euteiches [enzyme‐linked immunosorbent assay (ELISA), root and stem browning]. FW, fresh weight. (B) PCA representation of the susceptibility of different genotypes of M. truncatula (A17, F83005.5, DZA045‐6, SA022322, SA028064, SALSES71B, ESP105.L, R108, DZA315.16 and DZA012.J) under contrasting N availability (complete medium, squares; NØ medium, circles). Bars indicate standard error (n = 4 biological replicates). Data from a representative experiment out of three independent experiments are shown.
Fig. S3 Analysis of the response to nitrogen (N) deficiency (NØ) of the four selected genotypes. (A) Shoot biomass at 21 days. (B) Shoot biomass at 10 days. (C) N concentration in shoots at 21 days. Error bars indicate standard errors (n = 4–6 biological samples) and letters indicate significant differences (P < 0.05). NØ‐IS, genotype displaying NØ‐induced susceptibility; NØ‐IR, genotype displaying NØ‐induced resistance.
Fig. S4 Enhanced resistance to chlorate of the DZA045‐6 genotype. The four genotypes (F83005.5, SA022322, A17 and DZA045‐6) were cultivated in vitro for 10 days on complete medium in the presence of different concentrations of chlorate, and the primary root length was determined. Error bars indicate standard error (n = 4 biological replicates).
Fig. S5 Quantification of total and minor amino acid levels in roots of the four selected genotypes. (A) Total amino acid levels. (B) Minor amino acid levels. DW, dry weight. Amino acid quantification was performed on four genotypes (F83005.5, SA022322, A17 and DZA045‐6). Plants were cultivated in vitro for 10 days on complete medium or nitrate‐deficient (NØ) medium and inoculated or not with Aphanomyces euteiches. Error bars indicate standard errors (n = 3 biological replicates) and letters indicate significant differences (P < 0.05).
Fig. S6 Glutamine levels in the four selected genotypes cultivated in vitro for 10 days on complete or nitrate‐deficient (NØ) medium in the presence or not of Aphanomyces euteiches. Error bars indicate standard error (n = 3 biological replicates) and letters indicate significant differences (P < 0.05).
Table S1 Defence‐related/secondary metabolism genes analysed by RT‐qPCR in the four selected genotypes cultivated in complete or N‐deficient NØ medium and inoculated or not with A. euteiches.
Table S2 Root amino acid concentrations (µmol/g dry weight) in the four selected genotypes. Letters indicate significant differences (n = 3 biological replicates, P < 0.05).
Table S3 Primers used for gene expression analyses by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR).


