Summary
Hypothetical proteins without defined functions are largely distributed in all sequenced bacterial genomes. Understanding their potent functionalities is a basic demand for bacteriologists. Xanthomonas oryzae pv. oryzae (Xoo), the causal agent of bacterial leaf blight of rice, is one of the model systems for the study of molecular plant pathology. One‐quarter of proteins in the genome of this bacterium are defined as hypothetical proteins, but their roles in Xoo pathogenicity are unknown. Here, we generated in‐frame deletions for six hypothetical proteins selected from strain PXO99A and found that one of them (PXO_03177) is required for the full virulence of this strain. PXO_03177 is conserved in Xanthomonas, and is predicted to contain two domains relating to polysaccharide synthesis. However, we found that mutation of this gene did not affect the production or modification of extracellular polysaccharides (EPSs) and lipopolysaccharides (LPSs), two major polysaccharides produced by Xoo relating to its infection. Interestingly, we found that inactivation of PXO_03177 significantly impaired biofilm formation and tolerance to sodium dodecyl sulfate (SDS), both of which are considered to play key roles during Xoo infection in rice leaves. These findings thus enable us to define a function for PXO_03177 in the virulence of Xoo. Furthermore, we also found that the global regulator Clp controls the transcription of PXO_03177 by direct binding to its promoter region, presenting the first cellular regulatory pathway for the modulation of expression of this hypothetical protein gene. Our results provide reference information for PXO_03177 homologues in Xanthomonas.
Keywords: hypothetical protein, transcription, virulence, Xanthomonas oryzae pv. oryzae
Introduction
Xanthomonas oryzae pv. oryzae (Xoo), a Gram‐negative bacterium with a single polar flagellum, is the causal agent of bacterial leaf blight of rice (Swings et al., 1990). This disease causes severe losses and is found most predominantly in tropical Asian countries (Ferluga et al., 2007). Xoo infects rice leaves mainly through hydathodes or wounds at the leaf tip, multiplies in the intercellular spaces and enters into xylem vessels (Nguyen et al., 2016). Recently, this bacterium has been defined as one of the top 10 plant‐pathogenic bacteria in molecular plant pathology (Mansfield et al., 2012). A wide range of virulence factors have been reported to promote Xoo infection. They include, but are not limited to, xanthomonadin (a yellow soluble pigment), extracellular polysaccharides (EPSs), lipopolysaccharides (LPSs), adhesins, biofilms and cell motility (Li et al., 2015; Pradhan et al., 2012; Wang et al., 2013). In this bacterium, the production of these virulence factors is controlled by multiple regulators, including two‐component signal transduction systems (Yang et al., 2012; Zheng et al., 2016), the cyclic di‐GMP (guanosine 3',5'‐monophosphate) signalling pathway (Su et al., 2016), the quorum‐sensing signal (Ham, 2013), transcriptional regulators, i.e. GamR (Rashid et al., 2016) and Clp (Qian et al., 2013), and type II and III secretion systems (Chan and Goodwin, 1999; Hood et al., 2010; Ray et al., 2000). In most cases, these Xoo regulators show crosstalk in their functional regulations. For example, intracellular cyclic di‐GMP signalling serves as a multi‐connector responsible for the regulation of the quorum‐sensing signal (diffusible signal factor, DSF), the RpfC/RpfG two‐component system and the type III secretion system of this bacterium (Ham, 2013; Yang et al., 2016). Thus, these studies collectively demonstrate that Xoo employs complicated cellular signalling and regulatory pathways in crosstalk or non‐crosstalk to control the production of multiple virulence factors, promoting its infection of rice leaves. However, these identified regulators also reveal that only a limited number of genes in the Xoo genome have been experimentally confirmed as virulence modulators, and that a large number of genes, with or without predicted functions in the Xoo genome, are still unknown.
Proteins encoded by genes without predicted functions are always defined as hypothetical proteins (Qian et al., 2012). They may lack sequence homology to other species, and thus cannot be assigned a name. This type of protein constitutes a large proportion of genome annotations in all sequenced bacterial genomes (Salzberg et al., 2008). An understanding of their potent functionalities is an important consideration for bacteriologists. As an example, the Xoo PXO99A genome has a single circular chromosome of 5 240 075 bp and contains 5083 protein‐coding genes, one‐quarter of which are annotated as hypothetical proteins (Lei et al., 2013; Salzberg et al., 2008). A number of recent reports have revealed that multiple hypothetical proteins could function as previously uncharacterized virulence determinants in different Xanthomonas species. Representative examples include PXO_06181 in Xoo (Yu et al., 2017), Xoryp_02235, Xoryp_00885, Xoc_15235 and Xoryp_22910 in X. oryzae pv. oryzicola (Guo et al., 2012a) and XALc_0007 in X. albilineans (Rott et al., 2011). Collectively, these earlier reports not only point out that hypothetical proteins may serve as a rich source for the discovery of new Xanthomonas virulence determinants, but also suggest that virulence‐associated analyses could help to confer hypothetical proteins with defined biological functions in Xanthomonas. This could, in turn, further provide reference information on their corresponding homologues in other bacterial genomes.
The objectives of this study were to identify virulence‐associated hypothetical proteins and to dissect their roles in virulence‐associated functions, as well as their cellular transcriptional regulations, using strain PXO99A as the working model. Here, we report that one (PXO_03177) of the six hypothetical proteins randomly selected from strain PXO99A is involved in virulence. Bioinformatics analyses show that this protein is highly conserved in a wide range of Xanthomonas species. Further genetic and phenotypic evidence shows that this protein plays a key role in multiple Xoo virulence‐related functions, including biofilm formation and stress tolerance. Finally, we also show that the transcription of this gene is regulated by the global regulator Clp, where Clp binds to its promoter region. These findings not only identify the conserved hypothetical protein PXO_03177 as a new virulence determinant in Xoo, but also present its functionality and cellular regulation. Thus, our results bring a previously uncharacterized and conserved hypothetical protein into the Xanthomonas–host interaction arena, and provide corresponding reference information for its homologues in other bacterial genomes, i.e. Xanthomonas.
Results
PXO_03177 is a hypothetical protein that mediates Xoo virulence
The aim of our laboratory was to generate a mutant library comprising in‐frame deletions for each gene using Xoo PXO99A as a working model. Six mutants lacking hypothetical protein genes (PXO_01628, PXO_02766, PXO_03177, PXO_03326, PXO_03847 and PXO_04624) were generated in the laboratory (Tables 1 and S1, see Supporting Information). We thus selected these six available mutants to screen whether any of them were impaired in virulence on the susceptible host rice, as described in detail in Experimental procedures. The results showed that, compared with the wild‐type, the PXO_03177 mutant exhibited a two‐fold decrease in lesion length in rice leaves at 7 and 14 days post‐inoculation (dpi) (Fig. 1). Under similar test conditions, the other five mutants showed a lesion length on rice leaves similar to that of the wild‐type (Fig. S1, see Supporting Information). These findings suggest that only one of the six hypothetical proteins tested in this study was involved in Xoo virulence. To validate this point, we generated a complemented strain containing a plasmid‐borne PXO_03177, and found that this complemented strain retained the wild‐type ability to cause lesions in rice leaves (Fig. 1). Finally, we also showed that mutation of PXO_03177 did not alter the growth ability of the wild‐type in plant cell‐mimic medium (XOM3) and nutrient‐rich medium (nutrient broth, NB) (Procedures S1, Fig. S2, see Supporting Information). Taken together, these results reveal that the hypothetical protein PXO_03177 is not involved in growth, but serves as a new virulence determinant in Xoo.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristics* | Source |
|---|---|---|
| Strain | ||
| Escherichia coli | ||
| DH5a | F–, φ80dlacZΔM15, Δ(lacZYA‐argF)U169, deoR, recA1, endA1, hsdR17(rk–,mk+), phoA, supE44, λ‐, thi‐1, gyrA96 | Xu et al. (2016) |
| BL21 (DE3) | F –, ompT, gal, dcm, lon, hsdSB( ), λ(DE3) | Xu et al. (2016) |
| XL1‐Blue MRF′ Kan | D(mcrA)183, D(mcrCB‐hsdSMR‐mrr)173, endA1, supE44, thi‐1, recA1 gyrA96,relA1, lac, [F′proAB lacIqZDM15 Tn5 (KmR)] | Guo et al. (2009) |
| Xanthomonas oryzae pv. oryzae | ||
| PXO99A | Philippine race 6; wild‐type strain (WT) | Salzberg et al. (2008) |
| Δclp | clp in‐frame deletion mutant of strain PXO99A | This study |
| Cclp | Δclp harbouring plasmid pUFR047‐clp (complemented strain); GmR, AmpR | This study |
| ΔPXO_03177 | PXO_03177 in‐frame deletion mutant of strain PXO99A | This study |
| CPXO_03177 | ΔPXO_03177 harbouring plasmid pUFR047‐PXO_03177 (complemented strain); GmR, AmpR | This study |
| PXO99A(pUFZ75) | PXO99A harbouring plasmid pUFZ75 (GFP‐labelled strain); KmR | This study |
| ΔPXO_03177(pUFZ75) | ΔPXO_02671 harbouring plasmid pUFZ75 (GFP‐labelled strain); KmR | This study |
| Plasmid | ||
| pK18mobsacB | Allelic exchange suicide vector, sacB oriT(RP4); KmR | Zhao et al. (2012) |
| pK18‐PXO_3177 | pK18mobsacB with two PXO_03177 flanking fragments; KmR | This study |
| pK18‐clp | pK18mobsacB with two clp flanking fragments; KmR | This study |
| pUFR047 | Broad‐host‐range expression vector; IncW, GmR, AmpR, Mob+, Mob(p), lacZα+, Par+ | Andrade et al. (2014) |
| pUFR047‐clp | pUFR047 carrying clp gene with its native promoter region; GmR, AmpR | This study |
| pUFR047‐PXO_03177 | pUFR047 carrying PXO_03177 gene with its native promoter region; GmR, AmpR | This study |
| pUFR047‐PXO_03177‐Flag | pUFR047 carrying PXO_03177 gene with its native promoter region and Flag label; GmR, AmpR | This study |
| pUFZ75 | Ptrp‐TIR‐gfp cassette in pUFR034, KmR | Guo et al. (2012b) |
| pGEX‐6p‐1 | Vector for protein expression, AmpR | Deng et al. (2012) |
| pGEX‐6p‐1‐clp | pGEX‐6p‐1 with the coding region of Clp, AmpR | This study |
| pTRG | Plasmid used for protein expression in bacterial one‐hybrid assay, TetR | Guo et al. (2009) |
| pTRG‐Clp | pTRG with the coding region of Clp, TetR | This study |
| pBXcmT | Plasmid used for DNA cloning in bacterial one‐hybridization assay, ChloR | Guo et al. (2009) |
| pBXcmT‐PXO_03177 | pBXcmT with the PXO_03177 promoter region, ChloR | This study |
*GFP, green fluorescent protein; KmR, GmR, AmpR, TetR, ChloR: kanamycin, gentamicin, ampicillin, tetracycline, chloramphenicol resistance, respectively.
Figure 1.
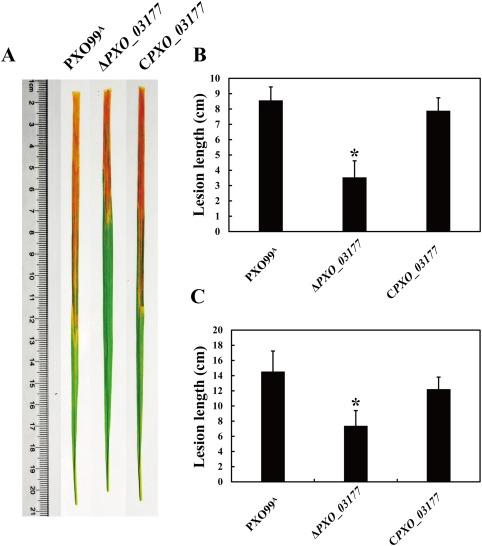
PXO_03177 is involved in Xanthomonas oryzae pv. oryzae virulence on susceptible rice plants. (A) Representative images of susceptible IR24 rice leaves inoculated with the various strains at 14 days post‐inoculation (dpi). (B, C) Lesion length caused by test strains on IR24 rice leaves at 7 dpi (B) and 14 dpi (C). Bars are means ± standard deviation (SD) (n = 20). PXO99A is wild‐type X. oryzae pv. oryzae; ΔPXO_03177 is the PXO_03177 deletion mutant; CPXO_03177 is the complemented strain of ΔPXO_03177 containing a plasmid‐borne PXO_03177. Data from triplicate experiments are shown. *P < 0.01.
PXO_03177 is conserved in multiple Xanthomonas species, but with different genomic annotations
To better understand PXO_03177, we further performed detailed bioinformatics analyses. PXO_03177 is 695 amino acids in size and has a predicted pI = 6.81 and molecular weight of 78.97 kDa (http://web.expasy.org/compute_pi/). Although PXO_03177 was a hypothetical protein in strain PXO99A, it was predicted to possess two domains, i.e. Glyco_tran_WbsX (PF14307) (Tao et al., 2004) and RgpF (PF05045) (Shibata et al., 2002) (Fig. S3, see Supporting Information). These two domains are associated with glycosyltransferase and assembly of the rhamnose‐glucose polysaccharide (RGP), respectively, implying a potent role of PXO_03177 in polysaccharide biosynthesis. A further sequence similarity search revealed that PXO_03177 is highly conserved in other sequenced Xanthomonas species, including X. oryzae, X. campestris, X. fuscans, X. perforans, X. vesicatoria, X. gardneri and X. albilineans, with over 63% amino acid identity (Table 2), revealing a potentially important role of the PXO_03177 homologue in each Xanthomonas species. Furthermore, our computational search also revealed a minor difference between PXO_03177 and its homologues, where a few of the PXO_03177 homologues were annotated as LPS biosynthesis proteins. Such a difference may be caused by the use of domain prediction to give a defined function for these homologues. Nevertheless, in most cases (Table 2), PXO_03177 and its homologues were defined as hypothetical proteins without described functions in Xanthomonas species.
Table 2.
PXO_03177 homologues in Xanthomonas spp.
| Homologue | |||||
|---|---|---|---|---|---|
| Strain* | Gene | Product | Size (amino acid) | Predicted domain† | Identity (%)‡ |
| Xoo PXO99A | PXO_03177 | Hypothetical protein | 695 | Rgpf (1); Glycos_transf_WbsX (1) | 100 |
| Xoo PXO86 | AZ54_20800 | Hypothetical protein | 695 | Rgpf (1); Glycos_transf_WbsX (1) | 100 |
| Xoo KACC10331 | XOO0800 | Lipopolysaccharide biosynthesis protein | 727 | Rgpf (1); Glycos_transf_WbsX (1) | 99 |
| Xoc BLS256 | Xoc_3838 | Hypothetical protein | 695 | Rgpf (1); Glycos_transf_WbsX (1) | 99 |
| Xpe 91–118 | XPE_16870 | Hypothetical protein | 695 | Rgpf (1); Glycos_transf_WbsX (1) | 91 |
| Xac A306 | J151_03761 | Hypothetical protein | 695 | Rgpf (1); Glycos_transf_WbsX (1) | 91 |
| Xcc B100 | XCC_b100_3726 | Uncharacterized protein | 695 | Rgpf (1); Glycos_transf_WbsX (1) | 80 |
| Xcc ATCC33913 | XCC0629 | Hypothetical protein | 695 | Rgpf (1); Glycos_transf_WbsX (1) | 80 |
| Xga ATCC19865 | XGA_2065 | Lipopolysaccharide biosynthesis protein | 706 | Rgpf (1); Glycos_transf_WbsX (1) | 79 |
| Xve ATCC35937 | XVE_2013 | Putative glycosyltransferase | 695 | Rgpf (1); Glycos_transf_WbsX (1) | 78 |
| Xan GPE PC73 | XALC_2689 | Hypothetical protein | 686 | Rgpf (1); Glycos_transf_WbsX (1) | 63 |
*Xoo PXO99A, Xanthomonas oryzae pv. oryzae PXO99A (GenBank accession number: CP000967); Xoo PXO86, X. oryzae pv. oryzae (CP007166); Xoo KACC10331, X. oryzae pv. oryzae KACC10331 (AE0135983); Xoc BLS256, X. oryzae pv. oryzicola BLS256 (AAQN00000000); Xpe 91–118, X. perforans 91–118 (AEQW00000000); Xac A306, X. axonopodis pv. citri strain A306 (CP006857); Xcc B100, X. campestris pv. campestris (AM920689); Xcc ATCC33913, X. campestris pv. campestris strain ATCC33913 (AE008922); Xga ATCC19865, X. gardneri ATCC19865 (AEQX00000000); Xve ATCC35937, X. vesicatoria ATCC35937 (AEQV00000000); Xan GPE PC73, X. albilineans GPE PC73(FP565176).
†Domains were predicted using the Pfam program http://pfam.xfam.org/. The total number of domains is indicated in parentheses.
‡According to a blastp search.
PXO_03177 regulates biofilm formation in Xoo
Bacterial biofilms are involved in adaptation to complex environments and are used by pathogenic bacteria to colonize host cells (Li et al., 2015; Pradhan et al., 2012). To test whether PXO_03177 has a role in biofilm formation, two independent assays were carried out. In the first assay, a well‐used crystal violet (CV) staining approach was applied. The results showed that inactivation of PXO_03177 caused a two‐fold reduction in biofilm formation on the polystyrene surface after staining by CV, compared with that of the wild‐type, and the complemented strain partially restored biofilm formation to wild‐type levels (Fig. 2). These data reveal that PXO_03177 is required for full biofilm formation. To validate this finding, we performed an additional assay using confocal laser scanning microscopy (CLSM). In this assay, the plasmid pUFZ75 expressing the green fluorescent protein (GFP) was transformed into the PXO_03177 mutant and wild‐type, enabling both strains to produce GFP. After 3 days of static incubation in chamber‐covered glass slides, the wild‐type PXO99A (pUFZ75) developed a structured biofilm in which bacteria were densely packed and organized, forming large aggregates that extended over the entire surface. In contrast, ΔPXO_03177 (pUFZ75) cells generated smaller dispersed micro‐colonies that were considerably less well organized than those produced by wild‐type PXO99A (pUFZ75) (Fig. 3). Taken together, the results from both assays strongly suggest that PXO_03177 controls Xoo biofilm formation.
Figure 2.
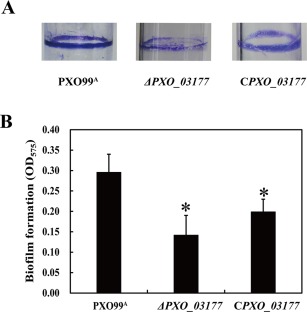
Biofilm formation of Xanthomonas oryzae pv. oryzae strains determined by crystal violet (CV) staining. (A) Representative images of CV‐stained biofilms produced by test strains. (B) Quantification of biofilm production corresponding to (A). PXO99A is wild‐type X. oryzae pv. oryzae; ΔPXO_03177 is the PXO_03177 deletion mutant; CPXO_03177 is the complemented strain of ΔPXO_03177 containing a plasmid‐borne PXO_03177. OD575, optical density at 575 nm. Data from triplicate experiments are shown. *P < 0.01.
Figure 3.
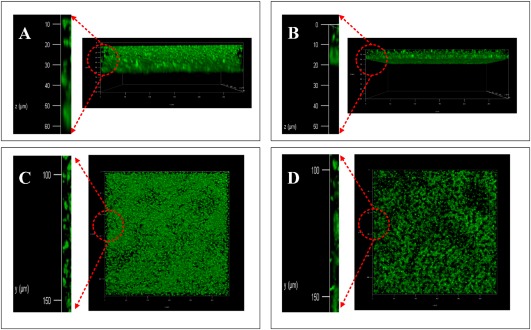
Confocal laser scanning microscopy (CLSM) of biofilms formed by Xanthomonas oryzae pv. oryzae strains. CLSM was performed to observe the three‐dimensional structure of biofilms. Images were obtained using a 20× objective after 72 h for green fluorescent protein (GFP)‐labelled cells grown on chamber‐covered glass slides. (A, C) Side (z and x axis) and top (y and x axis) faces of the three‐dimensional biofilm structure formed by PXO99A (pUFZ75), which is the wild‐type expressing GFP activity. (B, D) Side and top faces of the three‐dimensional biofilm structure formed by ΔPXO_03177 (pUFZ75), which is the PXO_03177 mutant expressing GFP activity. In each part, amplification of the red circle is presented on the left. At least four independent experiments were carried out for each strain with three replicates. The outcome of all experiments for each strain was similar.
PXO_03177 is required for full tolerance to sodium dodecyl sulfate (SDS) in Xoo
As documented previously, a biofilm is a type of sessile bacterial community that helps bacteria to survive under adverse conditions (Penesyan et al., 2015). This point, together with the key contribution of PXO_03177 in Xoo biofilm formation, encouraged us to explore whether PXO_03177 plays a crucial role in Xoo in adaptation to environmental stresses. For this purpose, the survival of the PXO_03177 mutant was investigated under various stresses, including saline, SDS and H2O2 oxidative stress. These stresses would probably be experienced at the stage of infection when Xoo attaches to the leaf surface and later survives inside the host plant (Li and Wang, 2012). Our results partly supported such an idea because the PXO_03177 mutant was more sensitive than the wild‐type to SDS exposure. In detail, the ΔPXO_03177 strain exhibited visually impaired tolerance to SDS compared with the wild‐type on NB agar (NA) plates containing 0.0125% SDS (w/v) (Fig. 4). Such a deficiency for the PXO_03177 mutant could be fully complemented in trans. However, the PXO_03177 mutant exhibited a wild‐type level of tolerance to saline and H2O2 oxidative stress under the test conditions used. These results demonstrate that PXO_03177 contributes importantly to SDS stress adaptation associated with Xoo infection. Taken together, the findings suggest that PXO_03177 mediates Xoo virulence probably via activities related to biofilm formation and stress tolerance, thus conferring the virulence‐associated functions of the conserved hypothetical protein PXO_03177 in Xanthomonas.
Figure 4.
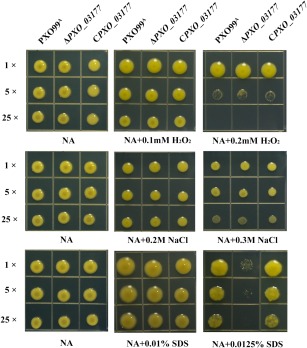
Determination of the roles of PXO99A, ΔPXO_03177 and CPXO_03177 strains in multiple stress tolerance using plate‐based assays. The Xanthomonas oryzae pv. oryzae strains were precultured to the exponential phase [optical density at 600 nm (OD600) = 1.0, initial culture) in nutrient broth (NB) medium, and the initial culture (1×) was diluted five‐fold (5×) and 25‐fold (25×). Three microlitres of the initial culture and diluted cultures for each strain were spotted onto NB agar (NA) plates and NA plates containing multiple stressors at the indicated concentrations. All plates were incubated at 28 °C for 3–5 days to observe growth ability. The experiment was performed at least three times, with similar results. SDS, sodium dodecyl sulfate.
The global regulator Clp controls the expression of PXO_03177
In order to understand how PXO_03177 expression (i.e. transcription) is controlled in Xoo, we made a detailed study of its promoter region via bioinformatics analyses. According to the recognized sequences of the Clp protein of X. campestris and the cyclic adenosine monophosphate receptor protein (CRP) of Escherichia coli (Berg and von Hippel, 1988; Chen et al., 2010; Hsiao et al., 2008), we found that the PXO_03177 promoter region contains a putative binding site potentially recognized by the Xoo Clp, a transcriptional regulator in this bacterium. As shown in Table 3, we found that the promoter of PXO_03177 possessed two Clp/CRP‐recognized semi‐conserved binding sites, as documented previously (Dong and Ebright, 1992; He et al., 2007). To better understand the promoter characteristics of PXO_03177, we performed a 5′ rapid amplification of cDNA ends (RACE) assay to map the transcription initiation site (TIS) of PXO_03177. This investigation led to the identification of TIS in the promoter region of PXO_03177 (Fig. 5A). Combing this and bioinformatics (http://www.prodoric.de), we further predicted the −10 and −35 sequences, and the ribosome binding site (RBS), upstream of the identified TIS (Fig. 5B). Interestingly, the predicted Clp binding site was mapped to downstream of the identified TIS (Fig. 5B). These findings collectively suggest that binding of Clp to the promoter of PXO_03177 may affect the mRNA elongation of this gene, further raising the possibility that Clp is likely to control PXO_03177 transcription in Xoo.
Table 3.
The reported cyclic adenosine monophosphate receptor protein (CRP)‐ and Clp‐recognized binding sequences
| Gene name | Species* | Binding sequence† | Source |
|---|---|---|---|
| ansB | Escherichia coli | 5′‐AAATGTGA‐TCTAGA‐TCACATTT‐3′ | Scott et al. (1995) |
| PehA | Xcc Xc17 | 5′‐CCACGTGG‐ATCTGC‐GCACTTGG‐3′ | Hsiao et al. (2005) |
| engA | Xcc Xc17 | 5′‐TTCTGTGG‐GGACGA‐TCACACCA‐3′ | Hsiao et al. (2009) |
| pelA1 | Xcc Xc17 | 5′‐GCGCGTGC‐ACATCG‐ACACATCG‐3′ | Hsiao et al. (2008) |
| xpsE | Xcc 8004 | 5′‐AACGGTGC‐GCTTCA‐CCGCACCC‐3′ | Ge and He (2008) |
| PXO_03177 | Xoo PXO99A | 5′‐TGGTCACA‐CCGGAG‐GGTGGTTC‐3′ | This study |
Figure 5.
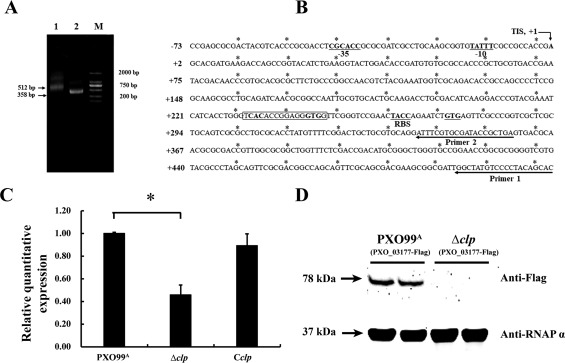
Promoter sequence characterization of PXO_03177 and its transcriptional regulation by Clp in Xanthomonas oryzae pv. oryzae. (A) Mapping of the transcription initiation site (TIS) of PXO_03177 using 5′ rapid amplification of cDNA ends (RACE). Fragments were amplified by polymerase chain reaction (PCR) using the abridged universal amplification primers described in Experimental procedures in combination with the gene‐specific primer 1 (lane 1) or primer 2 (lane 2). Lane M, standard markers. (B) Promoter sequence and characterization of PXO_03177. The identified transcription initiation site (TIS, +1) is indicated by a vertical arrow. The predicted −10 and −35 sequences are underlined. The putative ribosomal binding site (RBS) and the translation start codon (GTG) are in bold type and underlined. The putative Clp binding sequence is boxed. (C) Clp controls PXO_03177 transcription as determined by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). Data from triplicate experiments are shown. *P < 0.01. (D) Clp controls PXO_03177 protein levels. The 78‐kDa band corresponding to the predicted size of the PXO_03177‐Flag fusion was detected by the monoclonal antibody Anti‐Flag. The RNA polymerase α‐subunit (37 kDa) was used as the internal control for loading, which was detected by the specific antibody (Anti‐RNAP α).
To test the above hypothesis, we first determined the expression of PXO_03177 in the clp mutant, and the result showed that this mutation of clp led to a significant decrease in PXO_03177 transcription, whereas complementation of clp rescued PXO_03177 transcription to a wild‐type level (Fig. 5C). These results provide a piece of genetic evidence to support our hypothesis. In agreement, Western blot analyses showed that deletion of clp resulted in almost no detectable levels of the PXO_03177 protein, compared with the wild‐type, in which a clear band corresponding to the predicted size of the PXO_03177‐Flag fusion (78 kDa) was detected (Fig. 5D). Under similar test conditions, the internal control (the α subunit of the RNA polymerase; 37 kDa) was detected at similar levels in both test samples. As a result of the operational difficulties in introducing a plasmid‐expressed PXO_03177‐Flag fusion into the complemented strain, which already contains the same plasmid expressing clp for complementation, we could not perform the corresponding Western blot assay to test the protein level of the PXO_03177‐Flag fusion in the clp complemented strain. Nevertheless, our results reveal that Clp controls the transcription of PXO_03177 and, as a result, modulates PXO_03177 protein levels.
Clp could directly bind to the promoter region of PXO_03177
To test whether Clp controls PXO_03177 transcription via direct binding to its promoter, we employed a newly developed bacterial one‐hybrid system to test the potential direct interaction between Clp and the PXO_03177 promoter (P‐PXO_03177), according to our earlier reports (Xu et al., 2016). As shown in Fig. 6A, on selective medium, we clearly observed that the test E. coli strain containing both Clp and P‐PXO_03177 grew well, in a similar manner to the positive control, whereas the negative control failed to grow. This result indicates that direct binding of Clp to P‐PXO_03177 occurred under the test conditions.
Figure 6.
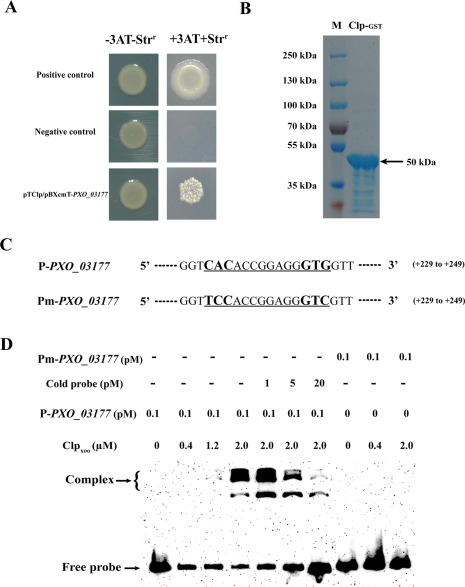
Clp can bind to the promoter region of PXO_03177. (A) The direct physical interaction between Clp and the PXO_03177 promoter region was detected in Escherichia coli. Positive control, co‐transformant containing pBX‐R2031 and pTRG‐R3133; negative control, co‐transformant containing pBXcmT‐PXO_03177 and empty pTRG; pTClp/p‐PXO_03177, co‐transformant possessing both pTRG‐Clp and pBXcmT‐PXO_03177; −3AT‐Strr, no selective Luria–Bertani (LB) medium plate; +3AT+Strr, M9‐based selective medium plate. (B) Sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) analysis of the purified glutathione transferase (GST)‐tagged Clp. (C) The probe and mutational probe sequence of PXO_03177. The putative Clp binding site and its mutations are underlined. (D) Clp binds to the PXO_03177 promoter (P‐PXO_03177) in vitro. Unlabelled PXO_03177 probe (code probe) was found to competitively inhibit the formation of the labelled Clp–DNA complex. Arrows indicate free DNA and Clp–DNA complexes. No binding of Clp with the mutational probe (Pm‐PXO_03177) was observed.
To verify the above results, an additional electrophoretic mobility shift assay (EMSA) was performed. We constructed a glutathione transferase (GST)‐fused clp clone, and overexpressed and purified the GST‐fused Clp protein (Fig. 6B). The use of the protein (Clp‐GST fusion) concentration (2.0 µm) in this work is based on, and in agreement with, a recent report (Chin et al., 2010), in which the authors also employed micromolar concentrations of Clp protein of X. campestris pv. campestris (Xcc) to test its DNA binding capacity via EMSA. Our EMSA revealed Clp binding with the P‐PXO_03177 probe (Fig. 6C). In detail, the protein–DNA complex was clearly detected when 2.0 µm protein (Clp‐GST fusion) was added, and this complex could be competitively inhibited by the addition of unlabelled probe (cold probe) at concentrations of 5 and 20 pm, where the corresponding free probe also increased, in particular, when 20 pm of cold probe was applied. These results clearly show that Clp could specifically bind to the promoter region of PXO_03177. Moreover, the protein–DNA complex did not appear clearly when 0.4 or 1.2 µm protein (Clp‐GST fusion) was applied, suggesting that a certain concentration of Clp protein is probably required to form a detectable protein–DNA complex in the applied EMSA system. To further validate the above findings, we mutated the predicted Clp recognition sequence in the promoter region of PXO_03177 by changing the conserved motif ‘CAC‘ to ‘TCC’, creating a mutated probe, Pm‐PXO_03177 (Fig. 6C). This mutated probe failed to bind Clp under similar test EMSA conditions (Fig. 6D). Finally, we performed an isothermal titration calorimetry (ITC) assay to quantify Clp binding affinity to P‐PXO_03177. The results showed that Clp bound to P‐PXO_03177 with a dissociation constant (K d) of approximately 2.0 µm (Fig. S5, see Supporting Information), in agreement with the concentration used in this work (Fig. 6D). Our data are also comparable with a previous report (Chin et al., 2010), in which the authors also found that Xcc Clp bound to DNA with a K d value of approximately 0.36 µm in high salt buffer, determined by surface plasmon resonance (SPR). Overall, our findings thus show the first molecular mechanism underlying the cellular regulation of the conserved hypothetical protein PXO_03177 in Xanthomonas.
Discussion
Hypothetical proteins are a large group of proteins without defined functions in the published genomes of a wide range of bacterial species (Alegria et al., 2005; Guo et al., 2012a; Laia et al., 2009). Understanding their roles and molecular details individually or collectively will provide fundamental insights into gene evolution, which is a basic question in genome biology. In this study, we provide evidence to define a conserved hypothetical protein as a new virulence determinant in Xoo, a model pathogenic bacterium for the study of molecular plant pathology. Notably, we found that this hypothetical protein has functions involved in the regulation of biofilm formation and stress tolerance. In addition, we found that the global regulator Clp binds to the promoter region of PXO_03177 and controls its transcription, thus dissecting the cellular transcription of this hypothetical protein gene in Xoo. Our findings collectively show that a Xoo hypothetical protein possesses defined virulence‐associated functions and cellular regulation, and may serve as a reference protein for annotation of the functionality of its homologues, not only in Xanthomonas, but also in other bacterial genomes.
The elucidation of the functionality of bacterial hypothetical proteins is a challenge, because no reference information is available. An alternative way to achieve this goal is to employ domain‐associated analyses, which could help to predict the potential functionality of hypothetical proteins. However, such a strategy does not seem to support the findings of this work. Based on the domain definition shown in Fig. S3, PXO_03177 should play a role in polysaccharide biosynthesis. However, this domain prediction was not consistent with the experimental data from this work, where we found that the PXO_03177 mutant showed a wild‐type level of production of EPS and LPS (Procedures S1, Fig. S4, see Supporting Information), two major polysaccharides formed by Xoo and related to its infection (Girija et al., 2017; Wang et al., 2013). We also provided a piece of experimental evidence to show that PXO_03177 is unlikely to induce modification or structural changes in LPS (Fig. S4A,B). Taken together, our findings may have implications for the modification of the functional annotation of PXO_03177 homologues in several Xanthomonas species, in which PXO_03177 homologues have been defined as LPS synthesis proteins. Importantly, we found that PXO_03177 could serve as a new virulence determinant and that it is associated with, and may control, several key virulence‐associated functions, including biofilm formation and stress tolerance. These findings undoubtedly show that PXO_03177 is a protein factor which promotes Xoo virulence, implying a key role of this hypothetical protein in the Xoo–rice interaction. However, the mechanisms by which PXO_03177 controls these virulence‐associated functions still remain unclear. For example, as mutation of PXO_03177 causes a greater sensitivity to SDS relative to the wild‐type, PXO_03177 may have an effect on the integrity of the membrane or may somehow alter the inner and/or outer membrane in Xoo. A detailed investigation is required in the future. Furthermore, as PXO_03177 has no DNA binding domain, it may employ a protein–protein interaction strategy to affect downstream gene/protein expression, resulting in the control of virulence‐associated functions, such as biofilm formation. The testing of such a hypothesis by combining co‐immunoprecipitation (Co‐IP) and RNA‐sequencing (RNA‐Seq) approaches is also in progress in our laboratory and will be reported in a separate article in the future.
In the present study, to investigate how PXO_03177 is transcriptionally regulated in Xoo, we carried out detailed bioinformatics analyses on the promoter region. This led to the finding that the downstream non‐coding region of PXO_03177 is a possible binding region for the global regulator Clp, a ‘star’ protein in Xcc, a Xoo‐related bacterium (Chen et al., 2010; Chin et al., 2010). Our molecular and biochemical results strongly supported this idea, as we found that Clp controlled the transcription of PXO_03177 by direct binding to its promoter region. Interestingly, we identified Xcc0629 as a homologue of PXO_03177 in Xcc ATCC33913; however, this homologous gene is not found to be regulated transcriptionally by Clp in this bacterium, according to the reported microarray analysis data (He et al., 2007). Sequence analysis also showed that the promoter of Xcc0629 did not contain a putative Clp binding sequence (He et al., 2007), indicating that the transcriptional regulation of Clp in PXO_03177 and its homologues may vary in different species of Xanthomonas.
As described previously, Clp is a downstream transcription factor of the DSF signalling pathway of Xcc (He et al., 2007), and is responsible for multiple DSF‐controlled functions. However, Clp can also bypass DSF signalling and carry out DSF‐independent modulation in several virulence‐related functions in Xcc (He et al., 2007). In Xoo, Clp is also required for virulence and controls EPS production, cell motility and tolerance to adverse stresses (Cho et al., 2011). Moreover, some Clp‐controlled phenotypes are also affected by PXO_03177 in Xoo, according to our results and earlier reports (Cho et al., 2011), suggesting that PXO_03177 is a downstream component of Clp in Xoo. Collectively, it is possible that PXO_03177 is a previously uncharacterized factor that is located downstream in the regulation of Clp and plays a key role in the Clp‐controlled functions in Xoo. However, it is still not clear whether PXO_03177 is the target employed by Clp to control the functions/phenotypes independent of the DSF signalling pathway in Xoo.
In summary, in this work, we have identified a conserved hypothetical protein as a new virulence determinant in Xoo and have dissected its functionality and cellular transcription mechanism. As a result, this hypothetical protein has been brought into the Xanthomonas–host interaction arena and can provide experimental reference information for the annotation of the function of its homologues in bacterial genomes.
Experimental Procedures
Strains, plasmids and growth conditions
The bacterial strains and plasmids used in this study are listed in Tables 1 and S1. The Xoo wild‐type strain PXO99A and its derivatives were cultured in NB medium or on NA plates at 28 °C. Escherichia coli was cultured in Luria–Bertani (LB) medium or on LB agar (LA) plates at 37 °C. XOM3 (1.8 g/L d‐xylose, 670 mm d,l‐methionine, 10 mm sodium l‐glutamate, 240 mm NaFe2+‐ethylenediaminetetraacetate, 5 mm MgCl2, 14.7 mm KH2PO4, 40 mm MnSO4, pH 6.0) was used as the plant cell‐mimic medium. When required, kanamycin (Km, 50 μg/mL), ampicillin (Amp, 100 μg/mL) and gentamicin (Gm, 25 μg/mL) were added to the growth media, as appropriate, for selection.
Plant material and bacterial virulence assays
The plant material and bacterial virulence assays have been described previously (Yang and Bogdanove, 2013). In brief, susceptible rice variety IR24 plants were grown in a growth chamber under a cycle of 12 h of light at 28 °C and 12 h of dark at 25 °C with ∼70% relative humidity. Plants were inoculated at 4–5 weeks with bacterial suspensions in sterile distilled water at a concentration of approximately OD600 = 0.5 (OD600, optical density at 600 nm) by immersing scissors in freshly prepared bacterial suspensions and clipping approximately 2 cm from the tips of fully expanded leaves. Lesion lengths were measured at 7 and 14 dpi, and representative images of IR24 leaves were photographed at 14 dpi.
Generation of mutants and complemented strains
The generation of the in‐frame deletion mutant was carried out using wild‐type PXO99A as the parental strain via allelic homologous recombination (Qian et al., 2013). Briefly, two flanking regions were generated by polymerase chain reaction (PCR) using the corresponding primer pairs (Table S2, see Supporting Information). The two PCR fragments were digested with the corresponding restriction enzymes and ligated into pK18mobsacB (Table 2). The resulting recombinant vectors for each target gene were validated by sequencing and introduced into wild‐type PXO99A via electroporation. Transformants were selected on NA without sucrose (NANS) plates containing 50 μg/mL Km for the first cross‐over event. Positive colonies were then plated onto NA plates containing 10% (w/v) sucrose to screen for a second cross‐over event. After two rounds of recombination, the resulting in‐frame deletion mutants were confirmed by PCR analysis. For complementation, each target gene with its predicted promoter region was amplified by PCR with specific primer sets (Table S2) and cloned into the broad‐host‐range vector pUFR047 (Andrade et al., 2014). The resulting plasmid was then transferred into the corresponding mutant via electroporation to generate the complemented strains.
Bioinformatics analyses
Each hypothetical protein sequence tested in this work was obtained from the PXO99A genome database (GenBank accession NC_010717.2). The resulting protein sequences were then subjected to Pfam (http://pfam.janelia.org/) and Smart (v4.0, http://smart.embl-heidelberg.de/) database search to identify potential domains. Cluster of orthologous groups (COG) analyses were performed using the Protein Basic Local Alignment Search Tool (blast). The PXO_03177‐containing genomic organization was drawn by the BioCyc Database Collection (https://biocyc.org/). The prediction of the theoretical pI (protein isoelectric point) and molecular weight for the selected proteins was performed using the ExPASy tool (https://www.expasy.org/). The promoter sequence and its details were predicted by BDGP (http://www.fruitfly.org/).
Biofilm formation assay
A biofilm formation assay was performed as described previously (Li and Wang, 2012) with some modifications. Briefly, bacterial cells were cultured in NB medium to a final concentration of OD600 = 1.0. Then, 4 mL of cell suspension were added to sterilized polystyrene tubes. These tubes were kept in a humidified chamber at 28 °C for 7 days without shaking. The cultures were then moved and the tubes were washed three times in tap water. Biofilm formation on the tubes was visualized by staining with 0.1% CV, followed by washing three times in tap water. The CV‐stained biofilm in polystyrene tubes was dissolved in methanol–acetic acid–water (4 : 1 : 5, v/v/v) and quantified by measuring OD575 using an Agilent 8453 UV–visible spectrophotometer (Agilent Technologies, Inc., Santa Clara, CA, USA). The average of three replicates was used for quantitative measurement. The biofilm assays were repeated three times, and showed similar results with three replicates each time.
The confocal‐based biofilm formation assay was carried out as described previously (Du et al., 2016; Tondo et al., 2016). In brief, the plasmid pUFZ75 expressing GFP was transformed into the PXO99A and ΔPXO_03177 strains. Overnight cultures of the GFP‐labelled strains in NB medium were adjusted to the same OD600 = 1.0, diluted 1 : 100 in fresh medium and 200‐μL cultures were placed into chamber‐covered glass slides (Nu155411, Lab‐Tek, NUNC, Naperville, IL, USA). Chambers were statically incubated in a humidified polyvinylchloride (PVC) box at 28 °C for 3 days. Biofilm formation was visualized by CLSM (Leica Microsystems Inc., Buffalo Grove, IL, USA) with a 20× objective. Excitation and emission lines were used in the observation, with excitation at 488 nm and emission at 500–545 nm. The images were analysed with LAS_X_Small_2.0.0_14332 software, which was provided by the confocal described (Leica Microsystems Inc., Buffalo Grove, IL, USA) above.
Stress tolerance assay
The stress tolerance assay on agar plates containing different harmful materials was performed as described previously with minor modifications (Qian et al., 2013). Briefly, Xoo strains were cultured in NB medium at 28 °C to a final concentration of OD600 = 1.0, which was defined as the initial culture. Then, 3 μL of the initial culture for each strain were spotted onto NA plates or NA plates containing SDS at concentrations of 0.01% (w/v) and 0.0125% (w/v), H2O2 at concentrations of 0.1 and 0.2 mm, and NaCl at concentrations of 0.2 and 0.3 m. The growth phenotype of the wild‐type PXO99A on each plate was used as a control. All plates were incubated at 28 °C for 3–5 days until the colony formed by strain PXO99A at the lowest dilution was visible on each plate. The experiments were performed three times and each involved three replicates.
Identification of TIS of PXO_03177 mRNA by 5′ RACE
A 5′ RACE approach was used to determine TIS with a SMARTer™ RACE cDNA Amplification Kit (Cat. Nos. 634923 & 63492; Clontech, Shanghai, China). Total RNA was isolated from PXO99A (OD600 = 1.0) with the E.Z.N.A.® Bacterial RNA Kit (Omega Bio‐Tek, Inc., Lilburn, GA, USA). The SMARTer™ II A Oligonucleotide (5′‐AAGCAGTGGTATCAACGCAGAGTACXXXXX‐3′), the universal primer A mix long (5′‐CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT‐3′) and the universal primer A mix short (5′‐CTAATACGACTCACTATAGGGC‐3′) were used in combination with the gene‐specific primers. The gene‐specific primers for reverse transcription‐polymerase chain reaction (RT‐PCR) and nested PCR were primer 1 (5′‐GTGCTGTAGGGGACATAGCC‐3′) and primer 2 (5′‐TCAGCGGTATCGCACGAAAT‐3′), respectively. The PCR products were ligated into the pMD19‐T vector (No. D102A; Takara, Shanghai, China) for sequence verification.
Bacterial one‐hybrid assay
The bacterial one‐hybrid reporter system was used to examine the potential interaction between the transcriptional regulator (Clp) and the target DNA (the PXO_03177 promoter) in the present study. According to our earlier work (Xu et al., 2016), the bacterial one‐hybrid reporter system contains three components: plasmids pBXcmT and pTRG, which are used to clone the target DNA and to express a target protein, respectively, and E. coli XL1‐Blue MRF′ kan strain, which is the host strain for the propagation of pBXcmT and pTRG recombinants. In this study, the PXO_03177 promoter region (285 bp) was cloned into pBXcmT, generating the recombinant vector pBXcmT‐PXO_03177. Similarly, the coding region of Clp (669 bp) was cloned into pTRG, creating the final construct pTRG‐Clp. The two recombinant vectors were transformed into the XL1‐Blue MRF′ kan strain. If direct physical binding occurs between Clp and the PXO_03177 promoter, the positive‐transformed E. coli strain containing both pBXcmT‐PXO_03177 and pTRG‐Clp should grow well on selective medium, which is minimal medium containing 5 mm 3‐amino‐1,2,4‐triazole, streptomycin at 8 μg/mL, tetracycline at 12.5 μg/mL, chloramphenicol at 34 μg/mL and Km at 30 μg/mL, as described previously (Xu et al., 2016). Furthermore, the co‐transformant containing pBX‐R2031/pTRG‐R3133 served as a positive control (Guo et al., 2009; Xu et al., 2016), and the co‐transformant containing empty pTRG and pBXcmT‐PXO_03177 was used as a negative control. All co‐transformants were spotted onto selective medium and grown at 28 °C for 3–4 days, and then photographed.
Protein expression and purification
The assay of protein expression and purification was performed as described previously (Xu et al., 2016). In brief, the coding region of Clp was amplified by PCR. The PCR product was purified, digested and cloned into pGEX‐6p‐1, creating the final construct pGEX‐6p‐1‐clp. This vector was transformed into E. coli strain BL21 (DE3) for protein expression. Briefly, the transformed strain was cultivated in LB medium containing Amp at 100 μg/mL overnight at 37 °C. Then, a 5‐mL overnight culture was transferred into 500 mL of fresh LB medium in the presence of Amp at 100 μg/mL and grown at 37 °C with shaking at 220 rpm until OD600 = 0.4. Subsequently, isopropyl β‐d‐1‐thiogalactopyranoside (IPTG) at a final concentration of 0.4 mm was added to the culture, followed by a further incubation at 28 °C for 4 h. Then, the cells were collected by centrifugation (3381 g) at 4 °C and resuspended in 15 mL of phosphate‐buffered saline (PBS) lysis buffer supplemented with phenyl methyl sulfonyl fluoride (PMSF) at a final concentration of 1 mm. The cells were lysed by a brief sonication and the crude cell extracts were centrifuged at 7000 g and 4 °C. Soluble protein fractions were collected and mixed with glutathione sepharose 4B (GE) for 12 h at 4 °C, which was placed into a column and extensively washed with PBS. The proteins were subsequently eluted using elution buffer which contained reduced glutathione. Protein purity was assessed by sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and protein concentration was determined using a Bradford protein assay kit (Bio‐Rad, Richmind, CA, USA).
EMSAs
The PXO_03177 promoter region (194 bp) was amplified by PCR using the 5′‐biotin‐labelled primers prober‐PXO_03177‐F/R (Table S2). EMSA was carried out using the LightShift™ EMSA Optimization & Control Kit (Thermo, Shanghai, China), as recommended by the manufacturer with some modifications. In brief, 0.1 pmol of the biotin‐labelled probe and a series of Clp‐GST fusion concentrations (0–2.0 μm) were added to the reaction solution. The cold probe (unlabelled PXO_03177 promoter region) and mutational probe were included as appropriate in the assays. After incubation at 28 °C for 30 min, the products were loaded onto a native 8% (w/v) polyacrylamide gel and electrophoresed in 0.5 × Tris‐borate‐EDTA buffer for about 1.5 h at 100 V, and then transferred to nylon membrane (Millipore, MA, USA). Protein–DNA complexes were visualized by VersaDoc (Bio‐Rad).
Quantitative RT‐PCR (qRT‐PCR) assay
The transcription of target genes was detected by qRT‐PCR as described previously (Song et al., 2017). Briefly, for total RNA isolation, wild‐type and Δclp were pre‐incubated in NB overnight, resuspended at OD600 = 1.0 in XOM3 broth and washed twice. The bacterial suspension was then grown at 28 °C for 12 h. The samples were then collected to isolate total RNA using the E.Z.N.A.® Bacterial RNA Kit (Omega Bio‐Tek, Inc.) according to the manufacturer's instructions. A Nano Drop 2000 (Thermo Fisher Scientific, Waltham, MA, USA) was used to evaluate the RNA concentration and purity. cDNA was then synthesized from each RNA sample (2 μg) using a TransScript All‐in‐One First‐Strand cDNA Synthesis SuperMix (with One‐Step gDNA Removal) Kit (TransGen Biotech, Beijing, China) according to the manufacturer's instructions. qRT‐PCR was performed using TransStart Top Green qPCR SuperMix (TransGen Biotech) on a QuantStudio™ 6 Flex Real‐Time PCR System (Applied Biosystems, Foster City, CA, USA) with the following thermal cycling parameters: denaturation at 95 °C for 30 s; 40 cycles of 95 °C for 5 s and 60 °C for 34 s. The gene expression analyses were performed using the 2–ΔΔCT method using 16S rRNA as the endogenous control, and the expression level in the wild‐type was mathematically designated as unity. The experiments were performed three times and each involved three replicates.
Western blot analysis
Western blot analysis was performed according to the laboratory protocol with a minor modification (Xu et al., 2016). In brief, the coding region of PXO_03177 was translationally fused with a Flag tag and cloned into pUFR047, in which the transcription of PXO_03177 was driven under the control of its native promoter. This final construct was introduced into wild‐type and Δclp. Then, the test strains were cultured in NB medium at 28 °C to the mid‐exponential phase (OD600 = 2.0). Cells were harvested by centrifugation at 12 000 g and frozen at −80 °C. Soluble proteins were harvested from these bacterial cells, further separated by SDS‐PAGE and immobilized onto a polyvinylidene difluoride (PVDF) membrane using a semi‐dry blot machine (Bio‐Rad). Membranes were probed by the monoclonal antibody specific for the Flag tag (1 : 5000; Abmart), followed by detection with a horseradish peroxidase (HRP)‐conjugated anti‐rabbit secondary antibody (No. M21002, Abmart, Shanghai, China). The α subunit of RNA polymerase was used as a control for sample loading, and the antibody of the α subunit of RNA polymerase was provided by Dr Wei Qian (Chinese Academy of Sciences) (Deng et al., 2014).
Data analysis
In this study, all analyses were conducted using SPSS 14.0; the hypothesis test of percentages (Duncan's test) was used to determine significant differences in disease lesion length, bacterial population and relative gene expression.
Supporting information
Additional supporting information may be found in the online version of this article at the publisher's web‐site
Fig. S1 Virulence phenotypes of wild‐type PXO99A and five hypothetical protein gene mutants on the susceptible rice cultivar IR24. (A) Representative images of IR24 leaves inoculated with mutant strains at 14 days post‐inoculation (dpi). (B, C) Lesion length caused by test strains on IR24 rice leaves at 7 dpi (B) and 14 dpi (C). Bars are means ± standard deviation (SD) (n = 20). PXO99A is wild‐type Xanthomonas oryzae pv. oryzae; Δ# indicates the deletion mutant of a given gene. Data from triplicate experiments are shown.
Fig. S2 Growth curves of PXO99A, ΔPXO_03177 and CPXO_03177 strains in nutrient broth (NB) and liquid plant cell‐mimic (XOM3) media. All tested strains were inoculated into 25 mL of liquid medium to the initial concentration [optical density at 600 nm (OD600) = 0.01] and cultivated at 28 °C with shaking at 220 rpm. Bacterial growth was determined by measuring OD600 against the medium blank at every 12 h in the plant‐cell mimic medium XOM3 (A) or every 4 h in NB medium (B) after inoculation. Values are the means ± standard deviation (SD) from three independent experiments.
Fig. S3 Genomic location and predicted domains of PXO_03177. Open arrows indicate length, location and orientation of PXO_03177 and its neighbouring genes. The bottom figure shows the predicted domains of PXO_03177 using the Pfam database (http://pfam.xfam.org/).
Fig. S4 PXO_03177 is not involved in Xanthomonas oryzae pv. oryzae lipopolysaccharide (LPS) and extracellular polysaccharide (EPS) production or LPS modification. (A) Tris–Tricine gel analysis of LPS from digested whole‐cell lysates of Escherichia coli, PXO99A, ΔPXO_03177 and CPXO_03177. S, LPS standard from Salmonella typhimurium (L1‐100MG; Sigma, CA, USA). The gel was run in Tris–Tricine–SDS buffer, followed by silver staining using a commercial kit (Bio‐Rad Laboratories, Inc., USA) according to the manufacturer's instructions. (B) Sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) of LPS from digested whole‐cell lysates of PXO99A, ΔPXO_03177 and CPXO_03177. S, LPS standard from Salmonella typhimurium. The gel was run in SDS–glycine buffer and silver stained using a silver staining kit (Bio‐Rad Laboratories, Inc.) following the manufacturer's instructions. (C) EPS production in nutrient broth (NB) medium by PXO99A, ΔPXO_03177 and CPXO_03177. The data presented are the means ± standard deviation (SD) of triplicate measurements from a representative experiment. Similar results were obtained from three independent experiments.
Fig. S5 An in vitro measurement of Clp binding to the promoter region of PXO_03177 by isothermal titration calorimetry. The data indicate that Clp binds to P‐PXO_03177 (the promoter region of PXO_03177) with a dissociation constant (K d) of 2.1 ± 1.3 μm. Clp‐GST (25 μm) was titrated with P‐PXO_03177 (1.6 μm) in phosphate‐buffered saline (PBS) at 25 °C. The power necessary to maintain a constant temperature during the titration experiment (microcalories per second), corresponding to the heat released on binding, was measured over a series of 20 injections of Clp‐GST (A) and integrated over time to obtain a plot of enthalpy versus ligand–protein molar ratio (B).
Table S1 Additional bacterial strains and plasmids used in this study.
Table S2 Primers used in this study.
Procedures S1 Growth assay, isolation and analysis of lipopolysaccharides, quantification of extracellular polysaccharides and isothermal titration calorimetry (ITC) measurements.
Acknowledgements
We thank Dr Ian Toth (The James Hutton Institute) for critical revision of the manuscript. We also thank Dr Wei Qian (Chinese Academy of Sciences) for providing the antibody of the α subunit of RNA polymerase as a gift, as well as for his technical support in EMSA. This work was supported by the National Natural Science Foundation of China (31571974; 31371906) and the Special Fund for Agro‐scientific Research in the Public Interest (No. 201303015). Innovation Team Program for Jiangsu Universities (2017).
Contributor Information
Guoliang Qian, Email: glqian@njau.edu.cn.
Fengquan Liu, Email: fqliu20011@sina.com.
References
- Alegria, M.C. , Souza, D.P. , Andrade, M.O. , Docena, C. , Khater, L. , Ramos, C.H.I. , da Silva, A.C.R. and Farah, C.S. (2005) Identification of new protein–protein interactions involving the products of the chromosome‐ and plasmid‐encoded type IV secretion loci of the phytopathogen Xanthomonas axonopodis pv. citri . J. Bacteriol. 187, 2315–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade, M. , Farah, S.C. and Wang, N. (2014) The post‐transcriptional regulator rsmA/csrA activates the T3SS in Xanthomonas citri by stabilizing the master regulator 5' UTR of hrpG. Phytopathology, 104, 8–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg, O.G. and von Hippel, P.H. (1988) Selection of DNA‐binding sites by regulatory proteins. 2. The binding‐specificity of cyclic‐amp receptor protein to recognition sites. J. Mol. Biol. 200, 709–723. [DOI] [PubMed] [Google Scholar]
- Chan, J.W.Y.F. and Goodwin, P.H. (1999) The molecular genetics of virulence of Xanthomonas campestris . Biotechnol. Adv. 17, 489–508. [DOI] [PubMed] [Google Scholar]
- Chen, C.H. , Lin, N.T. , Hsiao, Y.M. , Yang, C.Y. and Tseng, Y.H. (2010) Two non‐consensus Clp binding sites are involved in upregulation of the gum operon involved in xanthan polysaccharide synthesis in Xanthomonas campestris pv. campestris . Res. Microbiol. 161, 583–589. [DOI] [PubMed] [Google Scholar]
- Chin, K.H. , Lee, Y.C. , Tu, Z.L. , Chen, C.H. , Tseng, Y.H. , Yang, J.M. , Ryan, R.P. , McCarthy, Y. , Dow, J.M. , Wang, A.H.J. and Chou, S.H. (2010) The cAMP receptor‐like protein CLP is a novel c‐di‐GMP receptor linking cell–cell signaling to virulence gene expression in Xanthomonas campestris . J. Mol. Biol. 396, 646–662. [DOI] [PubMed] [Google Scholar]
- Cho, J.H. , Jeong, K.S. , Han, J.W. , Kim, W.J. and Cha, J.S. (2011) Mutation in clp(xoo4158) reduces virulence and resistance to oxidative stress in Xanthomonas oryzae pv. oryzae KACC10859. Plant Pathol. J. 27, 89–92. [Google Scholar]
- Deng, C.Y. , Deng, A.H. , Sun, S.T. , Wang, L. , Wu, J. , Wu, Y. , Chen, X.Y. , Fang, R.X. , Wen, T.Y. and Qian, W. (2014) The periplasmic PDZ domain‐containing protein Prc modulates full virulence, envelops stress responses, and directly interacts with dipeptidyl peptidase of Xanthomonas oryzae pv. oryzae . Mol. Plant–Microbe Interact. 27, 101–112. [DOI] [PubMed] [Google Scholar]
- Deng, Y.Y. , Schmid, N. , Wang, C. , Wang, J.H. , Pessi, G. , Wu, D.H. , Lee, J. , Aguilar, C. , Ahrens, C.H. , Chang, C.Q. , Song, H.W. , Eberl, L. and Zhang, L.H. (2012) Cis‐2‐dodecenoic acid receptor RpfR links quorum‐sensing signal perception with regulation of virulence through cyclic dimeric guanosine monophosphate turnover. Proc. Natl. Acad. Sci. USA, 109, 15 479–15 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, Q.P. and Ebright, R.H. (1992) DNA‐binding specificity and sequence of Xanthomonas campestris catabolite gene activator protein‐like protein. J. Bacteriol. 174, 5457–5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, H.C. , Pang, M.D. , Dong, Y.H. , Wu, Y.F. , Wang, N.N. , Liu, J. , Awan, F.Q. , Lu, C.P. and Liu, Y.J. (2016) Identification and characterization of an Aeromonas hydrophila oligopeptidase gene pepF negatively related to biofilm formation. Front. Microbiol. 7, 1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferluga, S. , Bigirimana, J. , Hofte, M. and Venturi, V. (2007) A LuxR homologue of Xanthomonas oryzae pv. oryzae is required for optimal rice virulence. Mol. Plant Pathol. 8, 529–538. [DOI] [PubMed] [Google Scholar]
- Ge, C. and He, C.Z. (2008) Regulation of the type II secretion structural gene xpsE in Xanthomonas campestris pathovar campestris by the global transcription regulator Clp. Curr. Microbiol. 56, 122–127. [DOI] [PubMed] [Google Scholar]
- Girija, A.M. , Kinathi, B.K. , Madhavi, M.B. , Ramesh, P. , Vungarala, S. , Patel, H.K. , Patel, H.K. and Sonti, R.V. (2017) Rice leaf transcriptional profiling suggests a functional interplay between Xanthomonas oryzae pv. oryzae lipopolysaccharide and extracellular polysaccharide in modulation of defense responses during infection. Mol. Plant–Microbe Interact. 30, 16–27. [DOI] [PubMed] [Google Scholar]
- Guo, M.M. , Feng, H. , Zhang, J. , Wang, W.Q. , Wang, Y. , Li, Y.Q. , Gao, C.H. , Chen, H.C. , Feng, Y. and He, Z.G. (2009) Dissecting transcription regulatory pathways through a new bacterial one‐hybrid reporter system. Genome Res. 19, 1301–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, W. , Cui, Y.P. , Li, Y.R. , Che, Y.Z. , Yuan, L. , Zou, L.F. , Zou, H.S. and Chen, G.Y. (2012a) Identification of seven Xanthomonas oryzae pv. oryzicola genes potentially involved in pathogenesis in rice. Microbiology, 158, 505–518. [DOI] [PubMed] [Google Scholar]
- Guo, Y.P. , Zhang, Y.P. , Li, J.L. and Wang, N.A. (2012b) Diffusible signal factor‐mediated quorum sensing plays a central role in coordinating gene expression of Xanthomonas citri subsp citri . Mol. Plant–Microbe Interact. 25, 165–179. [DOI] [PubMed] [Google Scholar]
- Ham, J.H. (2013) Intercellular and intracellular signalling systems that globally control the expression of virulence genes in plant pathogenic bacteria. Mol. Plant Pathol. 14, 308–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y.W. , Ng, A.Y.J. , Xu, M. , Lin, K. , Wang, L.H. , Dong, Y.H. and Zhang, L.H. (2007) Xanthomonas campestris cell–cell communication involves a putative nucleotide receptor protein Clp and a hierarchical signalling network. Mol. Microbiol. 64, 281–292. [DOI] [PubMed] [Google Scholar]
- Hood, R.D. , Singh, P. , Hsu, F.S. , Guvener, T. , Carl, M.A. , Trinidad, R.R.S. , Silverman, J.M. , Ohlson, B.B. , Hicks, K.G. , Plemel, R.L. , Li, M. , Schwarz, S. , Wang, W.Y. , Merz, A.J. , Goodlett, D.R. and Mougous, J.D. (2010) A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe, 7, 25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao, Y.M. , Liao, H.Y. , Lee, M.C. , Yang, T.C. and Tseng, Y.H. (2005) Clp upregulates transcription of engA gene encoding a virulence factor in Xanthomonas campestris by direct binding to the upstream tandem Clp sites. FEBS Lett. 579, 3525–3533. [DOI] [PubMed] [Google Scholar]
- Hsiao, Y.M. , Zheng, M.H. , Hu, R.M. , Yang, T.C. and Tseng, Y.H. (2008) Regulation of the pehA gene encoding the major polygalacturonase of Xanthomonas campestris by CIp and RpfF. Microbiology, 154, 705–713. [DOI] [PubMed] [Google Scholar]
- Hsiao, Y.M. , Fang, M.C. , Sun, P.F. and Tseng, Y.H. (2009) Clp and RpfF upregulate transcription of pelA1 gene encoding the major pectate lyase in Xanthomonas campestris pv. campestris . J. Agric. Food Chem. 57, 6207–6215. [DOI] [PubMed] [Google Scholar]
- Laia, M.L. , Moreira, L.M. , Dezajacomo, J. , Brigati, J.B. , Ferreira, C.B. , Ferro, M.I.T. , Silva, A.C.R. , Ferro, J.A. and Oliveira, J.C.F. (2009) New genes of Xanthomonas citri subsp citri involved in pathogenesis and adaptation revealed by a transposon‐based mutant library. BMC Microbiol. 9, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, Y. , Kang, S.K. , Gao, J.X. , Jia, X.S. and Chen, L.L. (2013) Improved annotation of a plant pathogen genome Xanthomonas oryzae pv. oryzae PXO99A . J. Biomol. Struct. Dyn. 31, 342–350. [DOI] [PubMed] [Google Scholar]
- Li, H. , Yu, C. , Chen, H. , Tian, F. and He, C. (2015) PXO_00987, a putative acetyltransferase, is required for flagellin glycosylation, and regulates flagellar motility, exopolysaccharide production, and biofilm formation in Xanthomonas oryzae pv. oryzae . Microb. Pathog. 85, 50–57. [DOI] [PubMed] [Google Scholar]
- Li, J.Y. and Wang, N. (2012) The gpsX gene encoding a glycosyltransferase is important for polysaccharide production and required for full virulence in Xanthomonas citri subsp citri . BMC Microbiol. 12, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield, J. , Genin, S. , Magori, S. , Citovsky, V. , Sriariyanum, M. , Ronald, P. , Dow, M. , Verdier, V. , Beer, S.V. , Machado, M.A. , Toth, I. , Salmond, G. and Foster, G.D. (2012) Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, M.P. , Park, J. , Cho, M.H. and Lee, S.W. (2016) Role of DetR in defence is critical for virulence of Xanthomonas oryzae pv. oryzae . Mol. Plant Pathol. 17, 601–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penesyan, A. , Gillings, M. and Paulsen, I.T. (2015) Antibiotic discovery: combatting bacterial resistance in cells and in biofilm communities. Molecules, 20, 5286–5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan, B.B. , Ranjan, M. and Chatterjee, S. (2012) XadM, a novel adhesin of Xanthomonas oryzae pv. oryzae, exhibits similarity to Rhs family proteins and is required for optimum attachment, biofilm formation, and virulence. Mol. Plant–Microbe Interact. 25, 1157–1170. [DOI] [PubMed] [Google Scholar]
- Qian, G.L. , Zhang, Y.B. , Zhou, Y.J. , Liu, C.H. , Zhao, Y.C. , Song, Z.W. , Fan, J.Q. , Hu, B.S. and Liu, F.Q. (2012) epv, encoding a hypothetical protein, is regulated by DSF‐mediating quorum sensing as well as global regulator Clp and is required for optimal virulence in Xanthomonas oryzae pv. oryzicola . Phytopathology, 102, 841–847. [DOI] [PubMed] [Google Scholar]
- Qian, G.L. , Liu, C.H. , Wu, G.C. , Yin, F.Q. , Zhao, Y.C. , Zhou, Y.J. , Zhang, Y.B. , Song, Z.W. , Fan, J.Q. , Hu, B.S. and Liu, F.Q. (2013) AsnB, regulated by diffusible signal factor and global regulator Clp, is involved in aspartate metabolism, resistance to oxidative stress and virulence in Xanthomonas oryzae pv. oryzicola . Mol. Plant Pathol. 14, 145–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid, M.M. , Ikawa, Y. and Tsuge, S. (2016) GamR, the LysR‐type galactose metabolism regulator, regulates hrp gene expression via transcriptional activation of two key hrp regulators, HrpG and HrpX, in Xanthomonas oryzae pv. oryzae . Appl. Environ. Microbiol. 82, 3947–3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, S.K. , Rajeshwari, R. and Sonti, R.V. (2000) Mutants of Xanthomonas oryzae pv. oryzae deficient in general secretory pathway are virulence deficient and unable to secrete xylanase. Mol. Plant–Microbe Interact. 13, 394–401. [DOI] [PubMed] [Google Scholar]
- Rott, P. , Fleites, L. , Marlow, G. , Royer, M. and Gabriel, D.W. (2011) Identification of new candidate pathogenicity factors in the xylem‐invading pathogen Xanthomonas albilineans by transposon mutagenesis. Mol. Plant–Microbe Interact. 24, 594–605. [DOI] [PubMed] [Google Scholar]
- Salzberg, S.L. , Sommer, D.D. , Schatz, M.C. , Phillippy, A.M. , Rabinowicz, P.D. , Tsuge, S. , Furutani, A. , Ochiai, H. , Delcher, A.L. , Kelley, D. , Madupu, R. , Puiu, D. , Radune, D. , Shumway, M. , Trapnell, C. , Aparna, G. , Jha, G. , Pandey, A. , Patil, P.B. , Ishihara, H. , Meyer, D.F. , Szurek, B. , Verdier, V. , Koebnik, R. , Dow, J.M. , Ryan, R.P. , Hirata, H. , Tsuyumu, S. , Won Lee, S. , Seo, Y.S. , Sriariyanum, M. , Ronald, P.C. , Sonti, R.V. , Van Sluys, M.A. , Leach, J.E. , White, F.F. and Bogdanove, A.J. (2008) Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A . BMC Genomics, 9, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, S. , Busby, S. and Beacham, I. (1995) Transcriptional coactivation at the ansB promoters – involvement of the activating regions of Crp and Fnr when bound in tandem. Mol. Microbiol. 18, 521–531. [DOI] [PubMed] [Google Scholar]
- Shibata, Y. , Yamashita, Y. , Ozaki, K. , Nakano, Y. and Koga, T. (2002) Expression and characterization of streptococcal rgp genes required for rhamnan synthesis in Escherichia coli . Infect. Immun. 70, 2891–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Z.W. , Zhao, Y.C. , Qian, G.L. , Odhiambo, B.O. and Liu, F.Q. (2017) Novel insights into the regulatory roles of gene hshB in Xanthomonas oryzae pv. oryzicola . Res. Microbiol. 168, 165–173. [DOI] [PubMed] [Google Scholar]
- Su, J.M. , Zou, X. , Huang, L.B. , Bai, T.L. , Liu, S. , Yuan, M. , Chou, S.H. , He, Y.W. , Wang, H.H. and He, J. (2016) DgcA, a diguanylate cyclase from Xanthomonas oryzae pv. oryzae regulates bacterial pathogenicity on rice. Sci. Rep. 6, 25 978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swings, J. , Van Den Mooter, M. , Vauterin, L. , Hoste, B. , Gillis, M. , Mew, T.W. and Kersters, K. (1990) Reclassification of the causal agents of bacterial‐blight (Xanthomonas‐campestris pv oryzae) and bacterial leaf streak (Xanthomonas‐campestris pv oryzicola) of rice as pathovars of Xanthomonas‐oryzae (Ex Ishiyama 1922) Sp‐Nov, Nom‐Rev. Int. J. Syst. Bacteriol. 40, 309–311. [Google Scholar]
- Tao, J. , Feng, L. , Guo, H. , Li, Y. and Wang, L. (2004) The O‐antigen gene cluster of Shigella boydii O11 and functional identification of its wzy gene. FEMS Microbiol Lett. 234, 125–132. [DOI] [PubMed] [Google Scholar]
- Tondo, M.L. , Delprato, M.L. , Kraiselburd, I. , Zenoff, M.V.F. , Farias, M.E. and Orellano, E.G. (2016) KatG, the bifunctional catalase of Xanthomonas citri subsp citri, responds to hydrogen peroxide and contributes to epiphytic survival on citrus leaves. PLoS One, 11, e0151657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Vinogradov, E.V. and Bogdanove, A.J. (2013) Requirement of the lipopolysaccharide O‐chain biosynthesis gene wxocB for type III secretion and virulence of Xanthomonas oryzae pv. Oryzicola . J. Bacteriol. 195, 1959–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H.Y. , Chen, H.F. , Shen, Y.M. , Du, L.C. , Chou, S.H. , Liu, H.X. , Qian, G.L. and Liu, F.Q. (2016) Direct regulation of extracellular chitinase production by the transcription factor LeClp in Lysobacter enzymogenes OH11. Phytopathology, 106, 971–977. [DOI] [PubMed] [Google Scholar]
- Yang, B. and Bogdanove, A. (2013) Inoculation and virulence assay for bacterial blight and bacterial leaf streak of rice. Methods Mol. Biol. 956, 249–255. [DOI] [PubMed] [Google Scholar]
- Yang, F. , Qian, S. , Tian, F. , Chen, H. , Hutchins, W. , Yang, C.H. and He, C. (2016) The GGDEF‐domain protein GdpX1 attenuates motility, exopolysaccharide production and virulence in Xanthomonas oryzae pv. oryzae . J. Appl. Microbiol. 120, 1646–1657. [DOI] [PubMed] [Google Scholar]
- Yang, F.H. , Tian, F. , Sun, L. , Chen, H.M. , Wu, M.S. , Yang, C.H. and He, C. (2012) A novel two‐component system PdeK/PdeR regulates c‐di‐GMP turnover and virulence of Xanthomonas oryzae pv. oryzae . Mol. Plant–Microbe Interact. 25, 1361–1369. [DOI] [PubMed] [Google Scholar]
- Yu, C. , Chen, H. , Tian, F. , Yang, F. , Yuan, X. , Yang, C.H. and He, C. (2017) A ten gene‐containing genomic island determines flagellin glycosylation: implication for its regulatory role in motility and virulence of Xanthomonas oryzae pv. oryzae . Mol. Plant Pathol. 19, 579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y.C. , Qian, G.L. , Fan, J.Q. , Yin, F.Q. , Zhou, Y.J. , Liu, C.H. , Shen, Q. , Hu, B.S. and Liu, F.Q. (2012) Identification and characterization of a novel gene, hshB, in Xanthomonas oryzae pv. oryzicola co‐regulated by quorum sensing and clp . Phytopathology, 102, 252–259. [DOI] [PubMed] [Google Scholar]
- Zheng, D.H. , Yao, X.Y. , Duan, M. , Luo, Y.F. , Liu, B. , Qi, P.Y. , Sun, M. and Ruan, L. (2016) Two overlapping two‐component systems in Xanthomonas oryzae pv. oryzae contribute to full fitness in rice by regulating virulence factors expression. Sci. Rep. 6, 22 768. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional supporting information may be found in the online version of this article at the publisher's web‐site
Fig. S1 Virulence phenotypes of wild‐type PXO99A and five hypothetical protein gene mutants on the susceptible rice cultivar IR24. (A) Representative images of IR24 leaves inoculated with mutant strains at 14 days post‐inoculation (dpi). (B, C) Lesion length caused by test strains on IR24 rice leaves at 7 dpi (B) and 14 dpi (C). Bars are means ± standard deviation (SD) (n = 20). PXO99A is wild‐type Xanthomonas oryzae pv. oryzae; Δ# indicates the deletion mutant of a given gene. Data from triplicate experiments are shown.
Fig. S2 Growth curves of PXO99A, ΔPXO_03177 and CPXO_03177 strains in nutrient broth (NB) and liquid plant cell‐mimic (XOM3) media. All tested strains were inoculated into 25 mL of liquid medium to the initial concentration [optical density at 600 nm (OD600) = 0.01] and cultivated at 28 °C with shaking at 220 rpm. Bacterial growth was determined by measuring OD600 against the medium blank at every 12 h in the plant‐cell mimic medium XOM3 (A) or every 4 h in NB medium (B) after inoculation. Values are the means ± standard deviation (SD) from three independent experiments.
Fig. S3 Genomic location and predicted domains of PXO_03177. Open arrows indicate length, location and orientation of PXO_03177 and its neighbouring genes. The bottom figure shows the predicted domains of PXO_03177 using the Pfam database (http://pfam.xfam.org/).
Fig. S4 PXO_03177 is not involved in Xanthomonas oryzae pv. oryzae lipopolysaccharide (LPS) and extracellular polysaccharide (EPS) production or LPS modification. (A) Tris–Tricine gel analysis of LPS from digested whole‐cell lysates of Escherichia coli, PXO99A, ΔPXO_03177 and CPXO_03177. S, LPS standard from Salmonella typhimurium (L1‐100MG; Sigma, CA, USA). The gel was run in Tris–Tricine–SDS buffer, followed by silver staining using a commercial kit (Bio‐Rad Laboratories, Inc., USA) according to the manufacturer's instructions. (B) Sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) of LPS from digested whole‐cell lysates of PXO99A, ΔPXO_03177 and CPXO_03177. S, LPS standard from Salmonella typhimurium. The gel was run in SDS–glycine buffer and silver stained using a silver staining kit (Bio‐Rad Laboratories, Inc.) following the manufacturer's instructions. (C) EPS production in nutrient broth (NB) medium by PXO99A, ΔPXO_03177 and CPXO_03177. The data presented are the means ± standard deviation (SD) of triplicate measurements from a representative experiment. Similar results were obtained from three independent experiments.
Fig. S5 An in vitro measurement of Clp binding to the promoter region of PXO_03177 by isothermal titration calorimetry. The data indicate that Clp binds to P‐PXO_03177 (the promoter region of PXO_03177) with a dissociation constant (K d) of 2.1 ± 1.3 μm. Clp‐GST (25 μm) was titrated with P‐PXO_03177 (1.6 μm) in phosphate‐buffered saline (PBS) at 25 °C. The power necessary to maintain a constant temperature during the titration experiment (microcalories per second), corresponding to the heat released on binding, was measured over a series of 20 injections of Clp‐GST (A) and integrated over time to obtain a plot of enthalpy versus ligand–protein molar ratio (B).
Table S1 Additional bacterial strains and plasmids used in this study.
Table S2 Primers used in this study.
Procedures S1 Growth assay, isolation and analysis of lipopolysaccharides, quantification of extracellular polysaccharides and isothermal titration calorimetry (ITC) measurements.


