Summary
Nucleorhabdoviruses such as Sonchus yellow net virus (SYNV) replicate in the nuclei and undergo morphogenesis at the inner nuclear membrane (IM) in plant cells. Mature particles are presumed to form by budding of the Matrix (M) protein‐nucleocapsid complexes through host IMs to acquire host phospholipids and the surface glycoproteins (G). To address mechanisms underlying nucleorhabdovirus budding, we generated recombinant SYNV G mutants containing a truncated amino‐terminal (NT) or carboxyl‐terminal (CT) domain. Electron microscopy and sucrose gradient centrifugation analyses showed that the CT domain is essential for virion morphogenesis whereas the NT domain is also required for efficient budding. SYNV infection induces IM invaginations that are thought to provide membrane sites for virus budding. We found that in the context of viral infections, interactions of the M protein with the CT domain of the membrane‐anchored G protein mediate M protein translocation and IM invagination. Interestingly, tethering the M protein to endomembranes, either by co‐expression with a transmembrane G protein CT domain or by artificial fusion with the G protein membrane targeting sequence, induces IM invagination in uninfected cells. Further evidence to support functions of G‐M interactions in virus budding came from dominant negative effects on SYNV‐induced IM invagination and viral infections that were elicited by expression of a soluble version of the G protein CT domain. Based on these data, we propose that cooperative G‐M interactions promote efficient SYNV budding.
Keywords: budding, glycoprotein, matrix protein, nuclear membrane invagination, nucleorhabdovirus, plant rhabdovirus, sonchus yellow net virus
Introduction
Budding processes are final events that are critical for the life cycles of enveloped viruses. During this phase, viruses bud into host membranes containing viral glycoproteins (G proteins) and host lipids to provide envelopes that function during entry into new cells. Without competent budding, enveloped viruses often undergo abortive infections and fail to release mature infectious virions.
The budding mechanisms of animal rhabdoviruses have been studied extensively with the prototype members Vesicular stomatitis virus (VSV) and Rabies virus (RV). Rhabdoviruses represent one of the most diverse families of enveloped viruses with negative stranded RNA genomes that are mostly monopartite, although some viruses with bipartite genomes are included in the family (Dietzgen et al., 2017). Members in the family Rhabdoviridae infect a broad range of hosts including vertebrates, invertebrates, and plants (Dietzgen et al., 2017; Jackson et al., 2005). All rhabdoviruses encode at least five conserved proteins, the nucleoprotein (N), the phosphoprotein (P), the matrix protein (M), the transmembrane spike glycoprotein (G), and the ‘large’ RNA dependent RNA polymerase protein (L). A general model outlining several steps in animal rhabdovirus morphogenesis has been proposed (Dancho et al., 2009; Finke and Conzelmann, 1997; Luan et al., 1995; Lyles, 2013; Mebatsion et al., 1996, 1999; Okumura and Harty, 2011; Robison and Whitt, 2000; Simons and Garoff, 1980): (i) Nucleocapsids (NCs) are formed in the cytoplasm by binding of N, P, and L proteins to nascent viral antigenomic (ag) and genomic (g) RNAs. (ii) During the intermediate to late stages of replication, accumulating M proteins condense the gNCs into tightly coiled compact structures. (iii) Simultaneously plasma membrane budding sites containing inserted G proteins begin to accumulate. (iv) The condensed gNCs are recruited to budding sites via interactions with viral and host proteins. (v) The gNCs undergo envelopment at the budding sites to form bullet‐shaped virions that are released from plasma membrane into the surrounding media (Jayakar et al., 2004).
In this general model, the M protein is considered to be a primary factor regulating animal rhabdoviruses budding. It has been reported that temperature‐sensitive M mutants of VSV produce low levels of virions at the restrictive temperature (Lyles et al., 1996). Moreover, direct evidence of the importance of M in virus budding was provided in RV infections with a recombinant M protein deletion mutant in which the mutant virus budding was reduced by 500,000‐fold compared with wild‐type RV (Mebatsion et al., 1999). In addition to the M protein, the G protein also has an important role in RV and VSV budding. When recombinant RV or VSV G deletion mutants were rescued in cells, the budding efficiency was reduced by approximately 30‐fold compared to the respective wild‐type viruses, but typical bullet‐shaped virions were still observed in infected cells (Mebatsion et al., 1996; Schnell et al., 1997). These results indicate that the G protein is important, but is not the primary determinant specifying animal rhabdovirus morphogenesis.
Budding of plant rhabdovirus particles has also been observed in infected plant and insect vector cells (Jackson et al., 1987; McDaniel et al., 1985; Van Beek et al., 1985) and the morphogenesis sites have been used to separate the monopartite members into the Nucleorhabdovirus and Cytorhabdovirus genera (Dietzgen et al., 2017; Jackson et al., 2005). Transmission electron microscopy (TEM) analyses support the hypothesis that during nucleorhabdovirus morphogenesis, condensed NCs bud through the inner nuclear membrane (IM) followed by release of enveloped bacilliform virus particles into perinuclear spaces (Ismail et al., 1987; Martins et al., 1998; Van Beek et al., 1985). In the case of two most extensively studied nucleorhabdovirus, Sonchus yellow net virus (SYNV) and Potato yellow dwarf virus (PYDV), infections result in invagination of the IM into the nucleus, which is thought to be a precondition for virus budding (Goodin et al., 2007, 2005; Martins et al., 1998). Although budding and envelopment processes of plant rhabdoviruses have been observed for more than 30 years, the mechanisms underlying these processes are poorly understood, compared with those of RV and VSV (Lyles, 2013; Okumura and Harty, 2011).
Recently, we established a reverse genetic system for SYNV that permits studies of nucleorhabdovirus replication and morphogenesis, and preliminary studies of a G protein deletion mutant suggested that NCs participate in cell‐to‐cell and vascular movement, but that the mutant virus undergoes aberrant morphogenesis (Wang et al., 2015). To further investigate SYNV morphogenesis, we have conducted an analysis of recombinant SYNV (rSYNV) G protein mutants and interactions with the M protein. Based on our results, we propose a revised model for plant nucleorhabdovirus budding requiring interactions between the M protein and the G protein carboxyl (C)‐terminal tail (CT) domain. These interactions are envisioned to mediate translocation of the M protein from the nucleoplasm to the IM, elicitation of nuclear membrane invagination, and promotion of SYNV budding.
Results
Rescue and phenotypic properties of rSYNV G protein mutants
Sequence predictions by Protter software (https://wlab.ethz.ch/protter ) (Omasits et al., 2014) have shown that the SYNV G protein is a 632‐amino acid (aa) transmembrane protein consisting of a 23‐aa signal peptide (SP) preceding a 536‐aa amino (N)‐terminal (NT) luminal domain, a 25‐aa transmembrane (TM) domain and a 48‐aa CT domain (Fig. 1A, top panel). The SP is thought to be cleaved by signal peptidases during translation and translocation to the endoplasmic reticulum (ER) to yield a “mature” G protein consisting of the NT, TM and CT domains that is inserted into ER membranes and the contiguous nuclear envelope (Goodin et al., 2007, 2005; Martins et al., 1998).
Figure 1.
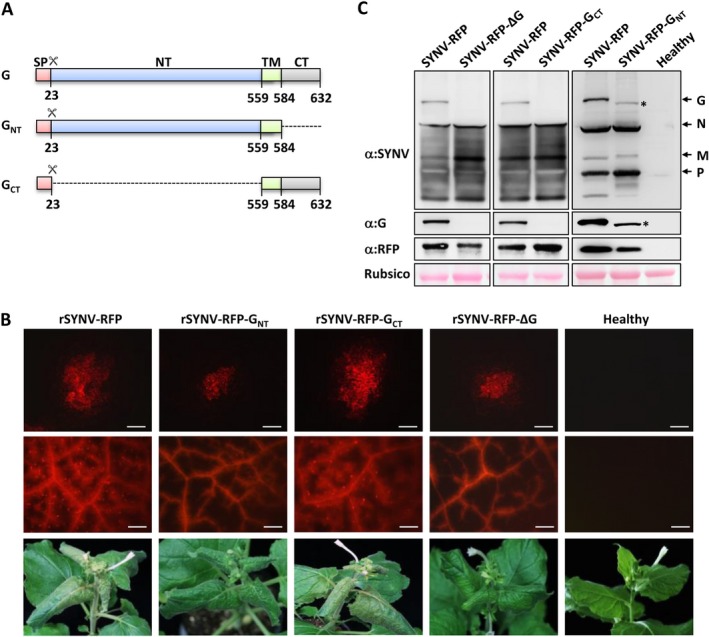
In planta rescue and characterization of recombinant SYNV (rSYNV) glycoprotein (G) mutants from cloned cDNAs. (A) Schematic representation of the G protein domains and mutants. The G protein domains and amino acid positions are indicated. The scissors symbol represents the predicted signal peptide cleavage site. SP: signal peptide sequence; NT: N‐terminal domain; TM: transmembrane domain; CT: C‐terminal domain. (B) Infection of Nicotiana benthamiana plants with rSYNV‐RFP derivatives. Top panels: RFP patterns in agroinfiltrated leaves at 14 days post infiltration (dpi). (Scale bar = 200 μm). Middle panels: RFP expression patterns in systemically infected leaves at 35 dpi. (Scale bar = 1 mm). Bottom panels: Typical systemic symptoms including stunting and vein clearing in infected N. benthamiana plants at 35 dpi. (C) Western blot analyses of upper uninoculated leaf extracts with antibodies against disrupted SYNV virions, G, and RFP proteins. The bottom panel shows the Rubisco large subunit used as a loading control. The positions of SYNV structural proteins are indicated along the right side of the gel and the position of GNT is indicated with an asterisk.
To map the G protein domains involved in plant infection and morphogenesis processes, we first engineered an SYNV reporter virus (rSYNV‐RFP) in which a red fluorescence protein (RFP) gene flanked by a duplicated N/P gene junction transcriptional cis‐elements was inserted between the N and P genes to permit viral replication and movement to be tracked by RFP expression. The G protein gene from rSYNV‐RFP was next deleted to produce the rSYNV‐RFP‐ΔG plasmid, and we also constructed rSYNV‐RFP‐GNT and rSYNV‐RFP‐GCT mutants in which the G protein CT and NT domains were deleted, respectively (Fig. 1A). The rSYNV derivatives were recovered by infiltration of Nicotiana benthamiana leaves with Agrobacterium tumefaciens mixtures harboring each of the antigenome transcription plasmids along with plasmids for expression of the SYNV N, P and L core proteins, and viral suppressors of RNA silencing (Wang et al., 2015).
Agroinfiltrated regions of the N. benthamiana leaves were evaluated for rescue of rSYNV derivatives by fluorescence microscopy observations of RFP accumulation. RFP foci were observed in single cells by 8 dpi, and the fluorescence subsequently spread into adjacent mesophyll and vascular cells of the leaves at 14 dpi (Fig. 1B, top panel). Areas of 20 fluorescent foci were measured for each rSYNV derivative and the mean sizes were calculated to determine the cell‐to‐cell movement rates. The data revealed that rSYNV‐RFP (0.393 ± 0.093 mm2) and rSYNV‐RFP‐GCT (0.408 ± 0.091 mm2) had similar localized rates of movement, which are ~ 2‐ to 2.4‐fold higher than those of the rSYNV‐RFP‐ΔG (0.170 ± 0.025 mm2) and rSYNV‐RFP‐GNT (0.186 ± 0.038 mm2) mutants. The differences in localized movement correlated with the proportion of plants developing visual systemic symptoms. By 36 dpi, systemic symptoms appeared in 33 of 150 rSYNV‐RFP agroinfiltrated plants (22%), and on 35 of 150 rSYNV‐RFP‐GCT infiltrated plants (23.3%), but only approximately 3% of rSYNV‐RFP‐ΔG (5/180) and rSYNV‐RFP‐GCT (5/150) infiltrated plants developed systemic symptoms. Regardless of the different systemic infection levels, all plants infected by the SYNV G mutants exhibited symptoms similar to those of the SYNV‐RFP infections (Fig. 1B, bottom row).
The different localized movement rates of the rSYNV derivatives also correlated with their systemic invasiveness in the upper leaf tissues as assessed by RFP fluorescence patterns. The rSYNV‐RFP‐ and rSYNV‐RFP‐GCT‐infected upper leaves exhibited extensive fluorescence in the veins and in mesophyll tissues surrounding the veins. However, fluorescence from the rSYNV‐RFP‐ΔG and rSYNV‐RFP‐GNT mutants was mostly confined to the veins of the systemic infected leaves (Fig. 1B, middle row). The mutant infections were confirmed by immunoblotting of systemically infected leaf extracts with antibodies raised against disrupted SYNV particles, G protein, or RFP (Fig. 1C). As expected, G protein bands were detected in leaf tissue infected by rSYNV‐RFP, but not rSYNV‐RFP‐ΔG (Fig. 1C, left panels), and the rSYNV‐RFP‐GNT infected tissue contained a rapidly migrating G protein band that correlates with the 48‐aa CT domain deletion (Fig. 1C, right panel; Note the asterisk identifying the GNT protein). However, G protein‐specific bands were not observed in the rSYNV‐RFP‐GCT extracts (Fig. 1C, middle panel), probably due to the lack of polyclonal antibodies targeting the G protein CT domain during immunization.
These data are consistent with our previously report indicating that the glycoprotein is dispensable for systemic SYNV infection, but contributes in undefined ways to cell‐to‐cell movement (Wang et al., 2015). Because rSYNV‐RFP and rSYNV‐RFP‐GCT have greater cell‐to‐cell efficiencies and systemic infection rates than the rSYNV‐RFP‐ΔG and rSYNV‐RFP‐GNT mutants, the results suggest that the CT domain has an important role in facilitating SYNV cell‐to‐cell transport and pathogenesis.
Contributions of the G protein NT and CT domains in SYNV morphogenesis
Our previous studies have shown that the G protein is required for morphogenesis of enveloped SYNV particles (Wang et al., 2015). To assess the G protein domains involved in this process, we investigated morphogenesis of the rSYNV‐RFP, rSYNV‐RFP‐GNT, rSYNV‐RFP‐GCT and rSYNV‐RFP‐ΔG virions in thin sections of infected leaf cells by TEM (Fig. 2A). Tissue sections from rSYNV‐RFP infected plants contained enlarged nuclei and bacilliform particles of ~72 nm in diameter. These particles appeared to accumulate in membrane spherules within the nuclei that are believed to be derived from the invaginated IM (Fig. 2A; upper left panel, red arrows). However, although the rSYNV‐RFP‐ΔG infected cells also had abnormally enlarged nuclei, only smaller virus‐like particles (~52 nm in diameter) that are typical of nucleocapsids were scattered throughout the nuclei (Fig. 2A; upper middle panel). The nuclear pathology and nucleocapsid sized particles (~53 nm diameter) were observed in the rSYNV‐RFP‐GNT infected cells (Fig. 2A, upper right panel). In the case of rSYNV‐RFP‐GCT infections, most of the particles had diameters similar to nucleocapsids (Fig. 2A, bottom left panel), but particles with size similar to mature virions were also occasionally observed within membrane spherules in some tissue sections (Fig. 2A, bottom middle panel). Of those that were subjected to careful measurement, 20 of the smaller particles had diameters of 54.4 ± 1.9 nm, and 10 of the 11 larger particles detected were similar in size (73.3 ± 2.4 nm diameter; n = 10) to those of rSYNV‐RFP particles. Our interpretation of these results is that a small proportion of the rSYNV‐RFP‐GCT nucleocapsids are able to complete morphogenesis.
Figure 2.

The G protein CT domain is required for maturation of SYNV virions. (A) Budding of rSYNV‐RFP and derived mutants. Thin sections of systemically infected N. benthamiana leaves at 35 dpi were observed by transmission electron microscopy. The enlarged rectangles show mature virions (~72 nm diameter) or nucleocapsid core particles (~52 nm diameter). Red arrows in the rSYNV‐RFP and rSYNV‐RFP‐GCT panels show the invaginated inner nuclear envelope surrounding the mature virions. C: core; V: virion; Vp: viroplasm; IM: inner nuclear membranes; M: mitochondria; Ch: chloroplast. Scale bar = 1 μm. (B) Sedimentation of SYNV structural proteins extracted from N. benthamiana plants infected with rSYNV‐RFP and mutant derivatives. Systemically infected leaf extracts recovered at 35 dpi were centrifuged in continuous 5%–30% sucrose gradients to separate low molecular weight proteins and nucleocapsid cores from virions. Twelve fractions of 1.0 mL each were recovered and analyzed by Western blotting with SYNV polyclonal antibodies. (C) Bacilliform particles in negative stained gradient fraction 9 from the SYNV‐RFP preparations. Scale bar = 200 nm.
To determine the sedimentation rates of viral particles in cells infected with the rSYNV‐RFP derivatives, we subjected systemically infected tissue extracts to centrifugation through 5% to 30% continuous sucrose gradients. Twelve fractions recovered from the gradients were analyzed by western blotting with SYNV antibodies (Fig. 2B). After centrifugation, we expected that soluble proteins and nucleocapsids, which have sedimentation coefficients between 200 to 250 S20,W (Jackson, 1978), would be located within fractions near the top of the gradient, whereas virions estimated to be about 1050 S20,W (Jackson and Christie, 1977) would sediment in fractions near the bottom of the gradient. In tissues infected with rSYNV‐RFP, the highest concentrations of the N and P core proteins were located in the top fractions (1 to 4), but the G and M proteins were enriched in fractions 9 and 10 that also contained substantial amounts of the N proteins and smaller amounts of the P proteins (Fig. 2B, top left panel). The L proteins were barely detectable due to low abundance. As anticipated, the presence of virions in fraction 9 was confirmed by TEM (Fig. 2C). In contrast, rSYNV‐RFP‐ΔG structural proteins were mostly present in fractions 1 to 4 and the sedimenting G and M proteins characteristic of virion components were absent in the bottom gradient fractions (Fig. 2B, bottom left panel). The lack of virions in fractions 9 and 10 was also confirmed by TEM (data not shown). Similar results were obtained in both the rSYNV‐RFP‐GNT and rSYNV‐RFP‐GCT infections, suggesting that morphogenesis was either abrogated or greatly reduced. Alternatively, enveloped particles may have been produced in plants but were not sufficiently stable to withstand the sample preparation process. These results verify that the G protein is required for morphogenesis of stable SYNV particles, and suggests that the CT domain is required for virion formation, and that the NT domain contributes to efficient budding.
Nuclear envelope invagination in SYNV infections requires both the M and G proteins
Fluorescence in N. benthamiana ‘16c’ plants constitutively expressing a GFP gene is targeted to the ER (ER:GFP) by fusion with an SP sequence and the HDEL ER‐retention sequence (mGFP5; Haseloff et al., 1997), and is not present in the nuclei of uninfected cells (Goodin et al., 2005). In contrast, in 16c cells infected with the nucleorhabdoviruses SYNV and PYDV, GFP foci are frequently observed in the nuclei in apparent association with viroplasm sites that support replication of the two viruses (Goodin et al., 2007, 2005 ). To determine the viral protein(s) responsible for induction of nuclear membrane invagination, we infected 16c plants by agroinfiltration with rSYNV‐RFP or rSYNV‐RFP‐ΔG, or the sc4 movement protein (rSYNV‐RFP‐Δsc4) or the M (rSYNV‐RFP‐ΔM) deletion mutants (Wang et al., 2015). Confocal microscopy at 14 dpi revealed that the rSYNV‐RFP derivatives replicated in the 16c agroinfiltrated leaves as indicated by RFP fluorescence (Fig. 3, upper panels). As anticipated from previous studies (Goodin et al., 2005), rSYNV‐RFP infection resulted in intense intranuclear accumulation of GFP foci, as did the rSYNV‐RFP‐Δsc4 mutant (Fig. 3B, middle panels). In marked contrast, leaves agroinfiltrated with the rSYNV‐RFP‐ΔM and the rSYNV‐RFP‐ΔG mutants were similar to those of mock infiltrated cells by exhibiting green fluorescent around the nuclear periphery and cytosolic membranes, but not within the nuclei. To obtain additional information relevant to the requirements for the G protein CT and NT domains for IM invagination, we agroinfiltrated 16c leaves with the rSYNV‐RFP‐GNT and rSYNV‐RFP‐GCT derivatives. The results show that membrane invagination associated with the rSYNV‐RFP‐GNT mutant is minimal, but that fluorescence patterns elicited by the rSYNV‐RFP‐GCT mutant more closely resembles those associated with rSYNV‐RFP infections (Fig. 3). To summarize, these results verify and extend previous results (Goodin et al., 2007, 2005 ) by showing that the sc4 protein has minimal, if any direct effects, on IM invagination. In contrast, both the M protein and the G protein CT domain are required for extensive invagination of the nuclear envelope during SYNV infection. Moreover, the consistent levels of expression of RFP from the viral genome suggests that invagination per se has an insignificant role in SYNV replication and gene expression.
Figure 3.
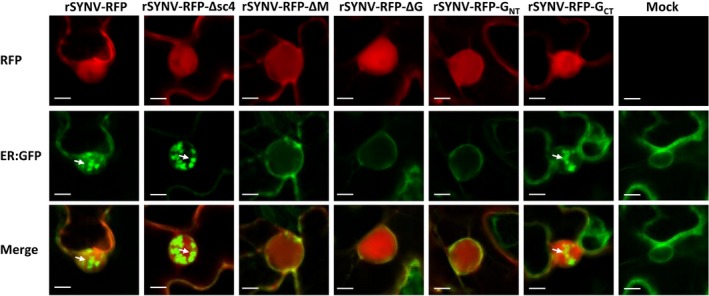
The M protein and the G protein CT domain are required for SYNV induced inner nuclear membrane invagination. Leaves of 16c transgenic N. benthamiana plants expressing the ER:GFP maker were agroinfiltrated with mixtures required for recovery of rSYNV‐RFP and each of the indicated deletion derivatives. The infected cells and cells of a mock treated plant at 14 dpi were examined under confocal microscope. White arrows indicate the invaginated ER:GFP foci. Scale bar = 10 μm.
Co‐expression of the SYNV M protein and the G protein CT domain elicits nuclear membrane invagination in uninfected cells
The results in Fig. 3 do not exclude the possibility that viral factors other than the M protein and the G protein may also be involved in elicitation of IM invagination during SYNV infection. To address this issue, we fused a cyan fluorescent protein (CFP) tag to the N‐terminal of the M protein (CFP:M) (Fig. 4A). As shown previously (Goodin et al., 2007; Martin et al., 2009), the CFP:M fusion is targeted to the nuclei and co‐localizes with the nucleoplasm marker, RFP‐histone 2B (RFP:H2B) (Supporting Information Fig. S1A, top panels). In addition, the G, GNT, and GCT proteins, as well as the GTM protein mutant in which both the NT and CT domains were deleted, were tagged with mCherry to determine subcellular localization of the proteins and their effects on membrane invagination. Note that the mCherry tags were fused immediately downstream of the predicted SP in order to preserve the membrane‐targeting signal of the fusion proteins (Fig. 4A). Confocal microscopy showed that the mCherry:G fusion protein and the deletion derivatives all co‐localized with the ER:GFP signal in cytoplasmic endomembranes and around the nuclear envelope (Supporting Information Fig. S1C). Membrane flotation assays also revealed that the mCherry:G and the deletion derivatives floated to the 65% sucrose interface fractions in discontinuous gradients that contained the membrane marker PIP2A‐DsRed, whereas the soluble GFP protein equilibrated in the 72% interface fractions (Supporting Information Fig. S1D). As anticipated from a previous study (Goodin et al., 2007), expression of either CFP‐M or each of the mCherry:G derivatives alone failed to induce pronounced intranuclear accumulation of ER:GFP (Supporting Information Fig. S1B,C).
Figure 4.
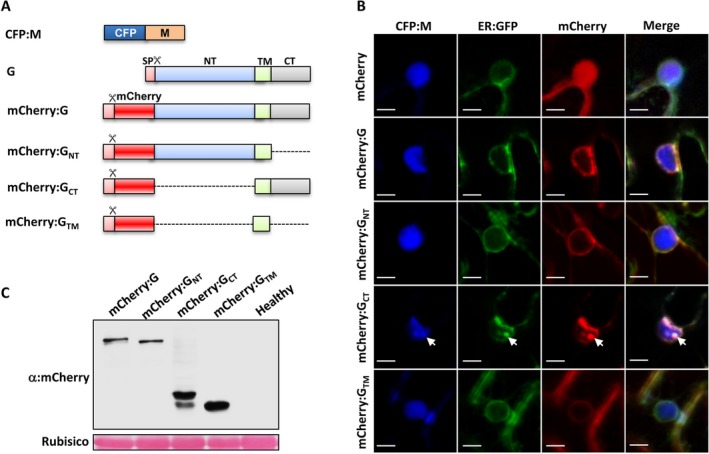
Elicitation of nuclear membrane invagination in uninfected N. benthamiana by ectopic expression of the M protein and G protein CT domain. (A) Schematic representation of the fluorescent protein fusions of the M and G protein derivatives. Dotted lines indicate regions deleted from the G protein. The G protein symbols were described in the Fig.1 legends. (B) Subcellular localization of the ER:GFP maker after transient expression of the CFP:M and mCherry:G fusion derivatives. Confocal images of the nuclear membranes of N. benthamiana 16c plant cells showed that IM invaginations (indicated by white arrows) are present only in cells co‐expressing the CFP:M and mCherry:GCT proteins. Scale bars = 10 μm. (C) Western blot analyses of the expression levels of the mCherry:G fusion derivatives. The stained Rubisco large subunit was used as a loading control.
We next co‐expressed CFP:M with each of the mCherry:G derivatives in N. benthamiana 16c leaves. As showed in Fig. 4B, co‐expression of the CFP:M and the mCherry:G protein failed to induce membrane invaginations into the nuclei, and similar results were also observed in cells expressing the CFP:M and mCherry:GNT or mCherry:GTM proteins. Unexpectedly, however, co‐expression of the CFP:M protein with mCherry:GCT formed intranuclear punctate GFP foci that appear to be similar to those elicited during SYNV infection. The CFP:M protein, which has a nuclear localization when expressed alone (Fig. 4B, top row), shifted to co‐localize with invaginated ER‐GFP and mCherry:GCT (Fig. 4B, the fourth row; indicated by white arrows). Cell counts showed that 64% of the infiltrated cells contained nuclear membrane invaginations. However, western blot analyses revealed that the mCherry:GCT was expressed at considerable higher levels than mCherry:G (Fig. 4C), which could explain the failure of co‐expressed mCherry:G and CFP:M proteins to induce membrane invagination. Alternatively, it is possible that the NT domain may have an as yet unidentified role in down‐regulating nuclear membrane invagination.
Interactions with the G protein CT domain redirect the M protein from the nucleoplasm to invaginated nuclear membranes
In SYNV infected plant cells, the G and M proteins are co‐localized at invaginated IM (Goodin et al., 2007, 2005; Martin et al., 2009). To explore the relationships of M protein relocalization and IM invagination, CFP:M was expressed by agroinfiltration into leaves of N. benthamiana 16c plants that had been infected by rSYNV derivatives and subcellular locations of the ER:GFP marker and the CFP:M fusion proteins were evaluated by confocal microscopy. The results shown in Fig. 5A reveal that in rSYNV and rSYNV‐GCT infected cells, the CFP:M protein forms punctate foci in the nuclei that co‐localize with ER:GFP‐labeled invaginated IM. As a negative control, the unfused CFP was found to distribute uniformly throughout the nucleoplasm in rSYNV infected cells even though the intranuclear ER:GFP foci from the 16c plants indicated IM invagination. Careful examination of the fluorescent signals indicated that the CFP:M protein is recruited to the periphery of the invaginated ER:GFP inclusions. In contrast, fluorescence from the CFP:M protein was completely localized within the nucleoplasm in rSYNV‐GNT and rSYNV‐ΔG infected nuclei, which lack pronounced intranuclear ER:GFP accumulation. These results suggest that the G protein CT domain is critical for redistribution of the M protein from the nucleoplasm to invaginated IM regions. In addition, these data also reveal a correlation between M protein relocalization and induction of membrane invagination.
Figure 5.

Requirement of the G protein CT domain for interactions with the M protein and translocation to invaginated membranes. (A) Localization of CFP:M in rSYNV, rSYNV‐GNT, rSYNV‐GCT, and rSYNV‐ΔG infected nuclei. Upper symptomatic leaves of 16c N. benthamiana plants infected by rSYNV derivatives were agroinfiltrated for expression of the CFP or CFP:M proteins and examined by confocal microscopy at 48h post infiltration (hpi). The white arrows indicate the invaginated ER:GFP foci. Scale bar = 10 μm. (B) Bimolecular fluorescence complementation (BiFC) analysis of the G‐M protein interactions. YFP C‐terminal‐fusions to the G, GNT, mCherry:GCT and mCherry:GTM were co‐expressed with YFP N‐terminal fusions to the M protein in transgenic CFP‐H2B N. benthamiana cells. Confocal microscopy images of representative cells were taken under CFP and YFP channels at 36 hpi. The right panels show higher magnifications of the boxed areas in the left panels to show the nuclear regions of selected cells. Scale bars = 25 μm.
We next tested interactions of the M protein with the G protein derivatives by bimolecular fluorescence complementation (BiFC) assays in leaves of transgenic N. benthamiana plant expressing the H2B:CFP protein. In these experiments, co‐expression of the M protein fusions to the N‐terminal fragment of yellow fluorescence protein (YFP) (M:YFPN) with the G protein fusions to the C‐terminal fragment of YFP (G:YFPC) in H2B‐CFP leaf cells resulted in strong YFP foci in the cytoplasm and the nuclear periphery (Fig. 5B, top row). Similar YFP patterns were observed when M:YFPN and mCherry:GCT:YFPC were co‐expressed in leaf cells (Fig. 5B, the third row). However, as expected, CFP:M failed to interact with mCherry:GNT or mCherry:GTM (Fig. 5B, the second and forth rows). These data indicate that the G protein CT domain interacts with the M protein in vivo and redirects it to IM surfaces where invagination occurs.
Tethering the M protein to endomembranes by fusion with the G protein membrane targeting sequence induces membrane rearrangement
To provide additional information about G‐M protein interactions and M protein relocalization leading to nuclear membrane invaginations, we generated a chimeric G:M protein (mCherry:GCT:M) by fusing the mCherry:GCT protein to the N‐terminus of the M protein (Fig. 6A). As a control, we generated a mCherry:GCT:CFP fusion in which the mCherry:GCT was fused to the CFP protein. These derivatives were transiently expressed in N. benthamiana 16c leaves and the GFP and mCherry fluorescent signals were examined by confocal microscopy. The mCherry fluorescence from both the mCherry:GCT:M and mCherry:GCT:CFP co‐localized with the ER:GFP, suggesting proper targeting of these fusion proteins to endomembranes (Fig. 6B). Interestingly, expression of the mCherry:GCT:M resulted in intranuclear accumulation of overlapping ER:GFP and mCherry foci (Fig. 6B, upper row; see white arrows), as well as large fluorescent inclusions in the cytoplasm (see red arrows). In contrast, expression of the mCherry:GCT:CFP fusion failed to induce membrane invaginations and the ER:GFP patterns appeared to be similar to cells in mock infiltrated tissue (Fig. 6B, compare middle and bottom rows). The fluorescent nuclear inclusions triggered by mCherry:GCT:M expression are similar to the intranuclear ER:GFP foci induced in SYNV infected cells, and likely represent invaginated IM. However, large cytoplasmic fluorescent inclusions were not observed in SYNV infected cells, so the nature of these structures is unclear. Because the G protein targeting signal should direct the mCherry:GCT:M fusion to nuclear envelopes as well as ER‐derived endomembranes, we surmise that these cytoplasmic fluorescent aggregates may represent rearranged ER‐derived membrane subdomains. Taken together, these findings show that membrane association of the M protein induces nuclear membrane invagination and remodeling of endomembrane systems.
Figure 6.
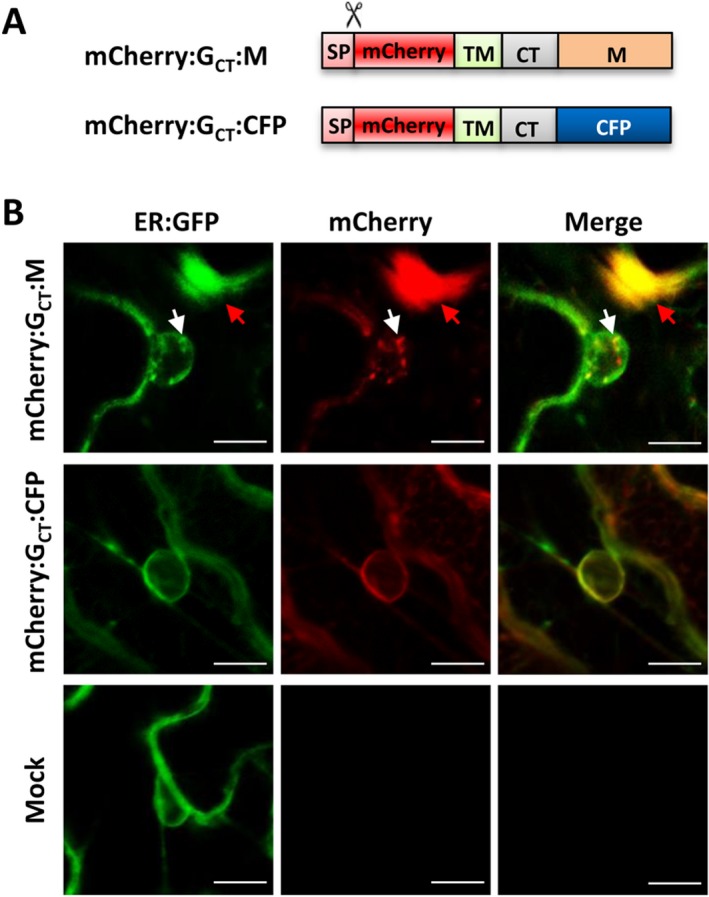
Membrane‐anchored M protein induces nuclear membrane invagination. (A) Schematic representation of the membrane anchored mCherry:GCT:M and mCherry:GCT:CFP fusions. Symbols and abbreviations are described in the Fig. 1 legends. (B) Subcellular localization of the ER:GFP and mCherry fusion proteins at 36 hpi. N. benthamiana 16c leaves were agroinfiltrated to express the mCherry:GCT:M or mCherry:GCT:CFP proteins. White arrows highlight nuclear membrane invagination induced by the mCherry:GCT:M protein, and the red arrows identify cytoplasmic inclusion bodies. Scale bar = 10 μm.
Ectopic expression of the G protein CT domain inhibits nuclear membrane invagination during SYNV infections
The results above suggested that the membrane‐anchored G protein interacts with the M protein through the CT domain to relocate the M protein from the nucleoplasm to the inner‐nuclear membrane to provide sites for SYNV morphogenesis. Hence, we tested whether expression of an unanchored G protein CT domain targeted artificially to the nucleoplasm could have a dominant negative effect on SYNV infection by interfering with the G‐M interactions. To permit expression of the CT domain in the nuclei of infected cells, we generated a recombinant SYNV mutant (rSYNV‐NLS:mCherry:GCT). This mutant was engineered by fusing the mCherry:GCT protein sequence to the C‐terminus of the Simian virus 40 (SV40) nuclear localization signal (NLS) (Fig. 7A), and substituting the derivative for the RFP gene in the SYNV‐RFP genome. As a control, an rSYNV‐NLS:mCherry containing the nuclear‐targeted mCherry fusion (NLS:mCherry) was also substituted for RFP (Fig. 7A).
Figure 7.

Dominant negative effects of a nuclear‐targeted G protein CT domain on induction of nuclear membrane invagination and SYNV infections. (A) Schematic representation of the rSYNV‐NLS:mCherry and rSYNV‐NLS:mCherry:CT constructs. NLS: Simian virus 40 nuclear localization sequence; CT: G protein CT domain. (B) Confocal micrographs of ER:GFP localization in N. benthamiana 16c plants infected with rSYNV‐NLS:mCherry and rSYNV‐NLS:mCherry:CT. White arrows indicate the invaginated ER:GFP foci. Scale bar = 10 μm. (C) Symptoms induced by rSYNV‐NLS:mCherry and rSYNV‐NLS:mCherry:CT at 35 dpi. Two different sized systemically infected leaves are shown in the bottom panels. (D) Western blot analyses of upper leaf extracts with antibodies against SYNV, G protein, and mCherry. The stained Rubisco large subunit was used as a loading control. The positions of the SYNV structural proteins are indicated along the right side of the gel.
After agroinoculation of N. benthamiana 16c leaves, infected leaves were examined by confocal microscopy. Typical nuclear invaginations were induced in 73 of 80 (91.2%) of rSYNV‐NLS:mCherry infected cells (Fig. 7B, upper panels). In contrast, only 17 of 80 cells (21.2%) infected with the rSYNV‐NLS:mCherry:GCT exhibited nuclear membrane invagination, and even in these cases, the intranuclear ER‐GFP foci appeared to be smaller than those in SYNV‐NLS:mCherry infected cells (Fig. 7B, compare the top and middle panels). In addition, the percentages of plants systemically infected with rSYNV‐NLS:mCherry:GCT (8/90, 8.9%) were substantially lower than those of the rSYNV‐NLS:mCherry infections (17/95, 17.8%). Moreover, rSYNV‐NLS:mCherry:GCT infected plants exhibited milder leaf symptoms (Fig. 7C), and produced slight lower levels of structure proteins (Fig. 7D) than the rSYNV‐NLS:mCherry infections. These results suggest that expression of the nuclei‐targeted soluble G protein CT domain inhibits SYNV induced nuclear membrane invagination in infected leaf cells and attenuates SYNV virulence.
Discussion
The infection cycle of nucleorhabdoviruses differ from most other Mononegavirales members by replicating in the nuclei and undergoing budding at the IM in plant cells (Jackson et al., 2005; Martins et al., 1998; Van Beek et al., 1985). Although budding of animal rhabdoviruses has been studied extensively (Lyles, 2013; Okumura and Harty, 2011), little is known about the molecular events leading to morphogenesis of plant rhabdovirus counterparts, or whether budding contributes to infections of plant hosts. This is mainly due to the fact that plant rhabdovirus genomes have not previously been amenable to genetic manipulation.
Using our recently devised SYNV reverse genetics system, we have shown a rSYNV mutant completely lacking the G protein is blocked in morphogenesis, and that infected plants accumulate nuclear localized nucleocapsids and fail to produce mature virions (Wang et al., 2015). To further evaluate the role of SYNV G protein domains in morphogenesis, we generated rSYNV mutants containing a truncated G protein lacking either the NT or the CT domain. TEM and sucrose gradient centrifugation analyses revealed that the G protein CT domain is essential for morphogenesis, and that the NT domain also contributes to efficient budding (Fig. 2). The important roles of the CT domains of animal rhabdovirus G proteins in efficient budding have also been documented previously. Recombinant VSV and RV mutants encoding a G protein with truncated CT domains are compromised in budding efficiencies, but these truncations only reduced virion production by about 5‐ to 10‐fold compared with their respective wild‐type virus (Mebatsion et al., 1996; Schnell et al., 1998). In contrast, complete deletion of the G genes of RV and VSV reduced particle yields by ~30‐ to 50‐ fold (Mebatsion et al., 1996; Schnell et al., 1997), suggesting that the NT domain, or other regions of the G protein, are also required for optimal budding. The luminal NT domain of the SYNV G protein is glycosylated (Jackson, 1978; Jones and Jackson, 1990), and contains six potential glycosylation sites within the NT domain (Goldberg et al., 1991). It was previously shown that SYNV budding could be interrupted by treatment of infected protoplasts with tunicamycin, a glycosylation inhibitor (Van Beek et al., 1985). In tunicamycin treated protoplasts, mature virions were not observed, but instead numerous naked core particles accumulated throughout the nuclei. These results suggested that G protein glycosylation may be required for morphogenesis, which is consistent with our finding that the NT domain deletion mutant is deficient in budding.
The SYNV and PYDV nucleorhabdoviruses are unique in that replication of these viruses elicit IM invaginations into infected nuclei (Anderson et al., 2018; Goodin et al., 2007, 2005; Martins et al., 1998). During morphogenesis, condensed SYNV NCs bud from the IM and enveloped bacilliform viral particles accumulate in spherule‐like structures derived from invaginated IM (Ismail et al., 1987; Martins et al., 1998; Van Beek et al., 1985). Although the precise function of IM invagination in SYNV and PYDV infections is unknown, it has been postulated that the invaginated membranes may provide accessible sites for viral morphogenesis (Goodin et al., 2005). Because rSYNV lacking the M protein was not recovered from plasmids, the role of the M protein in virus budding can not be readily dissected by analysis of mutant progenies. We therefore used SYNV‐induced ER:GFP nuclear invagination in N. benthamiana 16c plants as a facile reporter system to provide insights into SYNV budding requirements. Our recombinant studies reveal that in the context of SYNV infections, the M and G proteins, and more specifically the G protein CT domain, are required for induction of IM invagination (Fig. 3). Moreover, specific interaction between the M and G protein CT domain mediate relocalization of the M protein from nucleoplasm to membrane sites where IM invagination occurs (Fig. 5). These results are compatible with a model in which the SYNV G protein CT domain that protrudes through the IM interacts with M proteins to induce associated nucleocapsids to the invaginated IM for virus budding. Further experimental support for this notion comes from our observations that the rSYNV‐NLS:mCherry:GCT expressing an unanchored CT domain fusion have a dominant negative effect on induction of IM invagination and virus infections (Fig. 7). Our SYNV budding model differs somewhat from that of VSV, in which specific G‐M interactions are not required for efficient budding (Jayakar et al., 2004). For example, recombinant VSVs encoding a chimeric G protein with the CT domain substituted for unrelated foreign sequences were still able to produce near wild‐type amounts of extracellular viral particles, even though complete truncation of the CT domain reduced virion production (Schnell et al., 1998).
Interestingly, we found that simultaneous, but not individual, expression of the mCherry:GCT and CFP‐M protein in uninfected cells induces nuclear membrane invaginations where the G and M fusion protein are co‐localized (Fig. 4). In addition, anchoring the M protein to endomembranes by expression of an artificial fusion protein (mCherry:GCT:M) also induces IM invaginations and endomembrane rearrangements (Fig. 6). In contrast to the SYNV M protein, expression of the PYDV M protein alone is sufficient to trigger invagination of nuclear envelope and the PYDV M protein appears to be associated with the invaginated membranes (Anderson et al., 2018; Bandyopadhyay et al., 2010; Jang et al., 2017). These observations suggest that the PYDV M protein may have an intrinsic ability to elicit endomembrane invagination, whereas the SYNV M protein requires interactions with the G protein to associate with membranes and induce endomembrane remodeling. In the case of the VSV M protein, a small fraction (approximately 10%) was found to associate with the plasma membrane in vivo (Chong and Rose, 1993; Ye et al., 1994), and this fraction of the M protein is believed to be responsible for initiation of virion assembly and budding (Jayakar et al., 2004). In addition, the purified VSV M protein has a strong propensity to bind to unilamellar vesicles containing negatively charged phospholipids in vitro, and the binding imposes membrane curvature (Solon et al., 2005). Notably, both the VSV M and PYDV M are basic and positively charged proteins with isoelectric points (PI) of 9.08 and 9.07, respectively, whereas the SYNV M protein approach neutrality (PI of 7.76) under physiological conditions. These differences in the M protein charges may account for discrepancies in their abilities to interact with the negatively charged lipids of the membrane.
Another interesting observation is that the abilities of the rSYNV G deletion mutants to induce IM invagination correlates with their movement rates and tissue invasiveness (Figs 1B and 3). This observation suggests that mature virions may be involved in infection processes in an undefined way, even though nucleocapsids are able to participate in limited local movement in the absence of the M and G proteins (Wang et al., 2015). The fact that rSYNV‐RFP‐GCT is capable of only limited budding but still exhibits wild‐type of infection properties indicates that a small amount of mature or partially budded particles may be sufficient to support efficient infections. The enveloped particles of SYNV accumulate in perinuclear space surrounded by invaginated IM, which is continuous with the ER lumen as shown by fluorescence recovery after photobleaching analysis (Goodin et al., 2005). It was therefore proposed that the mature virions may participate in movement by either passing through the ER lumen and the contiguous desmotubules, or by budding out from the ER lumen to release the NC cores into cytoplasm (Goodin et al., 2005). Testing these models requires sophisticated electron and confocal microscopy studies coupled with recombinant labelling of SYNV structural proteins.
Experimental Procedures
Preparation of plasmids
SYNV antigenomic cDNA clones designed to recover rSYNV‐RFP and all the deletion derivatives were generated by engineering of previously described pSYNV vector (Wang et al., 2015). Detailed construction procedures for these clones, as well as for the transient expression vectors and BiFC vectors are described in the Supporting Information Protocols, and the primers used for cloning are listed in Supporting Information Table S1.
Recovery of recombinant SYNV derivatives and agroinfiltration
Rescue of rSYNV derivatives was carried out as previously described (Wang et al., 2015). Leaves of six to eight week‐old N. benthamiana plants were infiltrated with mixture of A. tumefaciens strain EHA 105 harboring each of the pSYNV plasmid derivatives, the pGD‐NPL plasmid and, plasmids for expression of viral suppressor of RNA silencing. Infiltrated plants were grown in a growth room at 25°C constant temperature.
Fluorescence microscopy and confocal laser scanning microscopy
A Zeiss SteREO Lumar V12 epifluorescence microscope was used to determine fluorescence in infiltrated leaves and in leaves systemically infected with the rescued rSYNV derivatives. Images were captured with the Lumar 31 filter set for RFP detection (excitation 565/30; emission 620/60). Subcellular localization and BiFC assays were performed with a Zeiss 780 confocal microscope. The GFP, RFP or mCherry, CFP, and YFP were excited by using 470, 545, 405, and 488 nm laser lines, respectively. All images were processed using LSM software Zen 2009 (Carl Zeiss, Germany).
Electron microscopy
Tissue sections (1 mm × 4 mm) were excised from leaves systemically infected with SYNV‐RFP, rSYNV‐RFP‐GNT, rSYNV‐RFP‐GCT or rSYNV‐RFP‐ΔG. Tissue fixation, embedding, sectioning and TEM were carried out essentially as described previously (Wang et al., 2015). For negative staining of purified SYNV virions, samples from sucrose gradients were placed on grids and stained with 1% potassium phosphotungstate (PTA), pH 7.0 for 1 min as described previously (Jackson and Christie, 1977).
Western blot analysis
Proteins were separated by 12% SDS‐PAGE and transferred to nitrocellulose membranes for immunoblotting analyses. Antigens bound to membranes were detected with polyclonal antibodies specific to disrupted SYNV virions (Jackson and Christie, 1977) or SYNV G protein (Jones and Jackson, 1990), or monoclonal antibodies against GFP or RFP (Abcam, Cambridge, UK).
BiFC assay
The coding sequences of the M protein and G protein derivatives were fused to the N‐terminal and C‐terminal fragment of enhanced YFP in the p2YN and p2YC vectors, respectively (Yang et al., 2007). BiFC plasmids were individually introduced into A. tumefaciens strain EHA105 by electroporation, and equal volumes of bacterial cultures were mixed and infiltrated into leaves of transgenic H2B‐CFP N. benthamiana plants that express CFP fused to histone 2B (Martin et al., 2009). YFP and CFP fluorescence was visualized by Zeiss 780 confocal microscopy (Carl Zeiss, Germany) at 48 to 72 hpi.
Sucrose gradient centrifugation
Sedimentation analyses was carried out essentially as described previously (Jackson and Christie, 1977) with minor modifications. Systemically infected leaves (0.5 g) were ground in 1 mL of fresh extraction buffer (100 mM Tris base, PH 8.4; 10 mM Mg Acetate; 1 mM MnCl2; 40 mM Na2SO3), and then centrifuged at 12,000 rpm for 2 min in a microfuge. 500 μL of the supernatant fractions was loaded onto 5 to 30% sucrose gradients in maintenance buffer (0.1 M Tris base, PH 7.5; 10 mM Mg Acetate; 1 mM MnCl2; 40 mM Na2SO3). Gradients were centrifuged in a Beckman SW41 rotor at 20,000 rpm for 50 min at 4°C. Twelve 1‐mL fractions were analyzed by western blotting with polyclonal antibodies specific to disrupted SYNV virions (Jackson and Christie, 1977).
Membrane‐flotation assay
Membrane‐flotation assays were carried out essentially as previously described (Sanfaçon and Zhang, 2008). Leaf samples (0.5 g) from Agrobacterium‐infiltrations were collected at 2 dpi, gently ground into a fine powder with a mortar and pestle in liquid nitrogen, followed by extraction in three volumes of homogenization buffer [50 mM Tris–HCl, pH 8.0, 10 mM KCl, 3 mM MgCl2, 1 mM EDTA, 1 mM DTT, 0.1% BSA, 0.3% dextran, 13% (w/v) sucrose, and Complete Mini Protease Inhibitor Cocktail (Roche, Germany)]. The samples were then centrifuged at 3,700 g for 10 min at 4°C. 300 μL supernatants were collected and transferred to clean centrifuge tubes and then mixed with 1.6 mL of 85% sucrose to a final concentration of 71.5% sucrose. These mixtures were added to 13.2 mL swinging‐bucket centrifuge tubes (Ultra‐Clear tube, Beckman Coulter, California, USA) and overlaid with 7 mL of 65% sucrose and 3.1 mL of 10% sucrose. After centrifugation at 100,000 g for 18 h at 4°C, eight 1.5‐mL fractions were collected from the top of the tubes and subjected to SDS‐PAGE/western blot analysis.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's web‐site:
Protocols Descriptions of plasmid construction details.
Table S1 Oligonucleotide primers used for cloning.
Fig. S1 Subcellular localization and discontinuous sucrose gradient fractionation of CFP:M and mCherry:G protein fusion and deletion derivatives. (A) Expression of CFP:M in RFP:H2B transgenic N. benthamiana plants. (B) Expression of CFP:M and (C) Cherry:G protein derivatives in N. benthamiana 16c plants. (Scale bar = 50 μm). (D) Membrane‐flotation analyses of mCherry:G fusion and deletion derivatives. Total protein extracts were centrifuged through the discontinuous sucrose gradients, and each fraction were subjected to western blot analysis using RFP or GFP specific monoclonal antibody.
Acknowledgements
We thank Professor Michael Goodin (University of Kentucky, USA) for providing RFP:H2B and CFP:H2B transgenic N. benthamiana seeds, and Professor Sir David Baulcombe (University of Cambridge, UK) for providing 16c transgenic N. benthamiana seeds. This work was supported by National Science Foundation of China (31671996 and 31470255) and the National Basic Research Program of China (2014CB138400). The authors declare no conflict of interest.
References
- Anderson, G. , Jang, C. , Wang, R. and Goodin, M. (2018) Mapping the nuclear localization signal in the matrix protein of potato yellow dwarf virus. J. Gen. Virol. 99, 743–752. [CrossRef][10.1099/jgv.0.001051]. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay, A. , Kopperud, K. , Anderson, G. , Martin, K. and Goodin, M. (2010) An integrated protein localization and interaction map for Potato yellow dwarf virus, type species of the genus Nucleorhabdovirus . Virology 402, 61–71. [CrossRef][10.1016/j.virol.2010.03.013]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong, L.D. and Rose, J.K. (1993) Membrane association of functional vesicular stomatitis virus matrix protein in vivo. J. Virol. 67, 407–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancho, B. , McKenzie, M.O. , Connor, J.H. and Lyles, D.S. (2009) Vesicular stomatitis virus matrix protein mutations that affect association with host membranes and viral nucleocapsids. J. Biol. Chem. 284, 4500–4509. [CrossRef][10.1074/jbc.M808136200]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzgen, R.G. , Kondo, H. , Goodin, M.M. , Kurath, G. and Vasilakis, N. (2017) The family Rhabdoviridae: mono‐ and bipartite negative‐sense RNA viruses with diverse genome organization and common evolutionary origins. Virus Res. 227, 158–170. [CrossRef][10.1016/j.virusres.2016.10.010]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finke, S. and Conzelmann, K.K. (1997) Ambisense gene expression from recombinant rabies virus: random packaging of positive‐ and negative‐strand ribonucleoprotein complexes into rabies virions. J. Virol. 71, 7281–7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, K.B. , Modrell, B. , Hillman, B.I. , Heaton, L.A. , Choi, T.J. and Jackson, A.O. (1991) Structure of the glycoprotein gene of sonchus yellow net virus, a plant rhabdovirus. Virology 185, 32–38. [PMC][1926779]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodin, M.M. , Chakrabarty, R. , Yelton, S. , Martin, K. , Clark, A. and Brooks, R. (2007) Membrane and protein dynamics in live plant nuclei infected with Sonchus yellow net virus, a plant‐adapted rhabdovirus. J. Gen. Virol. 88, 1810–1820. [CrossRef][10.1099/vir.0.82698‐0]. [DOI] [PubMed] [Google Scholar]
- Goodin, M. , Yelton, S. , Ghosh, D. , Mathews, S. and Lesnaw, J. (2005) Live‐cell imaging of rhabdovirus‐induced morphological changes in plant nuclear membranes. Mol. Plant Microbe Interact. 18, 703–709. [CrossRef][10.1094/MPMI‐18‐0703]. [DOI] [PubMed] [Google Scholar]
- Haseloff, J. , Siemering, K.R. , Prasher, D.C. and Hodge, S. (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94, 2122–2127. [CrossRef][10.1073/pnas.94.6.2122]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail, I.D. , Hamilton, I.D. , Robertson, E. and Milner, J.J. (1987) Movement and intracellular location of sonchus yellow net virus within infected Nicotiana edwardsonii . J. Gen. Virol. 68, 2429–2438. [CrossRef][10.1099/0022‐1317‐68‐9‐2429]. [Google Scholar]
- Jackson, A.O. (1978) Partial characterization of the structural proteins of sonchus yellow net virus. Virology 87, 172–181, [PMC][664251]. [DOI] [PubMed] [Google Scholar]
- Jackson, A.O. and Christie, S.R. (1977) Purification and some physicochemical properties of sonchus yellow net virus. Virology 77, 344–355. [PMC][841865]. [DOI] [PubMed] [Google Scholar]
- Jackson, A.O. , Francki, R. and Zuidema, D. (1987) Biology, structure and replication of plant rhabdoviruses In: The rhabdoviruses (Wagner R.R., ed.), pp 427–508. New York: Plenum Press. [Google Scholar]
- Jackson, A.O. , Dietzgen, R.G. , Goodin, M.M. , Bragg, J.N. and Deng, M. (2005) Biology of plant rhabdoviruses. Annu. Rev. Phytopathol. 43, 623–660, [PMC][10.1146/annurev.phyto.43.011205.141136] [16078897]. [DOI] [PubMed] [Google Scholar]
- Jang, C. , Wang, R. , Wells, J. , Leon, F. , Farman, M. , Hammond, J. and Goodin, M.M. (2017) Genome sequence variation in the constricta strain dramatically alters the protein interaction and localization map of Potato yellow dwarf virus. J. Gen. Virol. 98, 1526–1536. [CrossRef][10.1099/jgv.0.000771]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayakar, H.R. , Jeetendra, E. and Whitt, M.A. (2004) Rhabdovirus assembly and budding. Virus Res. 106, 117–132. [PMC][10.1016/j.virusres.2004.08.009] [15567492]. [DOI] [PubMed] [Google Scholar]
- Jones, R.W. and Jackson, A.O. (1990) Replication of sonchus yellow net virus in infected protoplasts. Virology 179, 815–820. [PMC][2238470]. [DOI] [PubMed] [Google Scholar]
- Luan, P. , Yang, L. and Glaser, M. (1995) Formation of membrane domains created during the budding of vesicular stomatitis virus. A model for selective lipid and protein sorting in biological membranes. Biochemistry 34, 9874–9883. [CrossRef][10.1021/bi00031a008]. [DOI] [PubMed] [Google Scholar]
- Lyles, D.S. (2013) Assembly and budding of negative‐strand RNA viruses. Adv. Virus Res. 85, 57–90. [PMC][10.1016/B978‐0‐12‐408116‐1.00003‐3] [23439024]. [DOI] [PubMed] [Google Scholar]
- Lyles, D.S. , McKenzie, M.O. , Kaptur, P.E. , Grant, K.W. and Jerome, W.G. (1996) Complementation of M gene mutants of vesicular stomatitis virus by plasmid‐derived M protein converts spherical extracellular particles into native bullet shapes. Virology 217, 76–87. [CrossRef][10.1006/viro.1996.0095]. [DOI] [PubMed] [Google Scholar]
- Martin, K. , Kopperud, K. , Chakrabarty, R. , Banerjee, R. , Brooks, R. and Goodin, M.M. (2009) Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J. 59, 150–162. [CrossRef][10.1111/j.1365‐313X.2009.03850.x]. [DOI] [PubMed] [Google Scholar]
- Martins, C.R. , Johnson, J.A. , Lawrence, D.M. , Choi, T.J. , Pisi, A.M. , Tobin, S.L. , Lapidus, D. , Wagner, J.D. , Ruzin, S. , McDonald, K. and Jackson, A.O. (1998) Sonchus yellow net rhabdovirus nuclear viroplasms contain polymerase‐associated proteins. J. Virol. 72, 5669–5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcdaniel, L.L. , Ammar, E.D. and Gordon, D.T. (1985) Assembly, morphology, and accumulation of a Hawaiian isolate of maize mosaic virus in maize. Phytopathology 75, 1167–1172. [CrossRef][10.1094/Phyto‐75‐1167]. [Google Scholar]
- Mebatsion, T. , Konig, M. and Conzelmann, K.K. (1996) Budding of Rabies Virus Particles in the Absence of the Spike Glycoprotein. Cell 84, 941–951. 10.1016/S0092-8674(00)81072-7. [DOI] [PubMed] [Google Scholar]
- Mebatsion, T. , Weiland, F. and Conzelmann, K.K. (1999) Matrix protein of rabies virus is responsible for the assembly and budding of bullet‐shaped particles and interacts with the transmembrane spike glycoprotein G. J. Virol. 73, 242–250. [PMC][9847327]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura, A. and Harty, R.N. (2011) Rabies virus assembly and budding. Adv. Virus Res. 79, 23–32. [PMC][10.1016/B978‐0‐12‐387040‐7.00002‐0] [21601040]. [DOI] [PubMed] [Google Scholar]
- Omasits, U. , Ahrens, C.H. , Muller, S. and Wollscheid, B. (2014) Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30, 884–886. [CrossRef][10.1093/bioinformatics/btt607]. [DOI] [PubMed] [Google Scholar]
- Robison, C.S. and Whitt, M.A. (2000) The membrane‐proximal stem region of vesicular stomatitis virus G protein confers efficient virus assembly. J. Virol. 74, 2239–2246. [CrossRef][10.1128/JVI.74.5.2239‐2246.2000]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanfaçon, H. and Zhang, G. (2008) Analysis of interactions between viral replicase proteins and plant intracellular membranes. Methods Mol. Biol. 451, 361–375. [DOI] [PubMed] [Google Scholar]
- Schnell, M.J. , Johnson, J.E. , Buonocore, L. and Rose, J.K. (1997) Construction of a novel virus that targets HIV‐1‐infected cells and controls HIV‐1 infection. Cell 90, 849–857. [PMC][9298897]. [DOI] [PubMed] [Google Scholar]
- Schnell, M.J. , Buonocore, L. , Boritz, E. , Ghosh, H.P. , Chernish, R. and Rose, J.K. (1998) Requirement for a non‐specific glycoprotein cytoplasmic domain sequence to drive efficient budding of vesicular stomatitis virus. EMBO J. 17, 1289–1296. [CrossRef][10.1093/emboj/17.5.1289]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, K. and Garoff, H. (1980) The budding mechanisms of enveloped animal viruses. J. Gen. Virol. 50, 1–21, [PMC][10.1099/0022‐1317‐50‐1‐1] [6255080]. [DOI] [PubMed] [Google Scholar]
- Solon, J. , Gareil, O. , Bassereau, P. and Gaudin, Y. (2005) Membrane deformations induced by the matrix protein of vesicular stomatitis virus in a minimal system. J. Gen. Virol. 86, 3357–3363. [CrossRef][10.1099/vir.0.81129‐0]. [DOI] [PubMed] [Google Scholar]
- VanBeek, N.A.M.V. , Lohuis, D. , Dijkstra, J. and Peters, D. (1985) Morphogenesis of Sonchus yellow net virus in cowpea protoplasts. J. Ultrastruct. Res. 90, 294–303. 10.1016/S0022-5320(85)80007-1. [DOI] [Google Scholar]
- Wang, Q. , Ma, X. , Qian, S. , Zhou, X. , Sun, K. , Chen, X. , Zhou, X. , Jackson, A.O. and Li, Z. (2015) Rescue of a plant negative‐strand RNA virus from cloned cDNA: insights into enveloped plant virus movement and morphogenesis. PLoS Pathog. 11, e1005223. [CrossRef][10.1371/journal.ppat.1005223]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. , Baliji, S. , Buchmann, R.C. , Wang, H. , Lindbo, J.A. , Sunter, G. and Bisaro, D.M. (2007) Functional modulation of the geminivirus AL2 transcription factor and silencing suppressor by self‐interaction. J. Virol. 81, 11972–11981. [CrossRef][10.1128/JVI.00617‐07]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, Z. , Sun, W. , Suryanarayana, K. , Justice, P. , Robinson, D. and Wagner, R.R. (1994) Membrane‐binding domains and cytopathogenesis of the matrix protein of vesicular stomatitis virus. J. Virol. 68, 7386–7396. [CrossRef][10.1099/0022‐1317‐73‐3‐743]. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's web‐site:
Protocols Descriptions of plasmid construction details.
Table S1 Oligonucleotide primers used for cloning.
Fig. S1 Subcellular localization and discontinuous sucrose gradient fractionation of CFP:M and mCherry:G protein fusion and deletion derivatives. (A) Expression of CFP:M in RFP:H2B transgenic N. benthamiana plants. (B) Expression of CFP:M and (C) Cherry:G protein derivatives in N. benthamiana 16c plants. (Scale bar = 50 μm). (D) Membrane‐flotation analyses of mCherry:G fusion and deletion derivatives. Total protein extracts were centrifuged through the discontinuous sucrose gradients, and each fraction were subjected to western blot analysis using RFP or GFP specific monoclonal antibody.


