Summary
Plant elicitor peptides (Peps) are widely distributed among angiosperms, and have been shown to amplify immune responses in multiple plant families. Here, we characterize three Peps from soybean (Glycine max) and describe their effects on plant defences against two damaging agricultural pests, the root‐knot nematode (Meloidogyne incognita) and the soybean cyst nematode (Heterodera glycines). Seed treatments with exogenous GmPep1, GmPep2 or GmPep3 significantly reduced the reproduction of both nematodes. Pep treatment also protected plants from the inhibitory effects of root‐knot nematodes on above‐ground growth, and up‐regulated basal expression levels of nematode‐responsive defence genes. GmPep1 induced the expression of its propeptide precursor (GmPROPEP1), a nucleotide‐binding site leucine‐rich repeat protein (NBS‐LRR), a pectin methylesterase inhibitor (PMEI), Respiratory Burst Oxidase Protein D (RBOHD) and the accumulation of reactive oxygen species (ROS) in leaves. In addition, GmPep2 and GmPep3 seed treatments up‐regulated RBOHD expression and ROS accumulation in roots and leaves. These results suggest that GmPeps activate plant defences through systemic transcriptional reprogramming and ROS signalling, and that Pep seed treatments represent a potential strategy for nematode management.
Keywords: Heterodera glycines, Meloidogyne incognita, plant elicitor peptides (Peps), propeptide, RBOHD, ROS, seed treatment
Introduction
Plant elicitor peptides (Peps) are widely distributed plant signalling molecules that contribute to broad‐spectrum defences against insects and pathogens. Bioactive Peps are 23–36‐amino‐acid‐long sequences that are generated from the carboxyl termini of longer propeptide precursors (PROPEPs), and typically contain a glycine‐enriched motif: (S/G)(S)Gxx(G/P)xx(N) (Tavormina et al., 2015). A family of eight Peps was first discovered in Arabidopsis thaliana, and orthologues have since been identified through amino acid sequence homology in most sequenced angiosperm genomes, including those of staple crops such as maize, wheat, rice, potato and soybean (Huffaker et al., 2006; Lori et al., 2015). Several lines of evidence have indicated that Peps participate in defence signalling. Certain PROPEP genes are up‐regulated in response to pathogen infection and insect oral secretions (Huffaker et al., 2006, 2013), and mature Peps interact with plant elicitor peptide receptors (PEPRs) to trigger calcium signalling, the accumulation of reactive oxygen species (ROS) and nitric oxide, phytohormone production, transcriptional reprogramming and the synthesis of defensive proteins and metabolites (Huffaker, 2015; Huffaker et al., 2011; Klauser et al., 2013; Ma et al., 2012, 2013; Qi et al., 2010; Yamaguchi and Huffaker, 2011). Transgenic Arabidopsis plants that constitutively overexpress AtPROPEP1 display enhanced disease resistance against a necrotrophic root pathogen, Pythium irregulare (Huffaker et al., 2006), and exogenous application of AtPep1 to Arabidopsis foliage reduces the proliferation of the hemibiotrophic foliar pathogen, Pseudomonas syringae pv. tomato (Yamaguchi et al., 2010). Application of ZmPeps to maize leaves and stems also protects against southern leaf blight and anthracnose stalk rot, limits larval growth of the beet armyworm Spodoptera exigua and attracts beneficial parasitic wasps that attack this herbivore (Huffaker et al., 2011, 2013). Moreover, the Arabidopsis pepr1pepr2 mutant, which is unable to respond to Peps, displays enhanced susceptibility to the cotton leafworm, Spodoptera littoralis (Klauser et al., 2015). These effects appear to be mediated in part by the release of green leaf volatiles (GLVs) and the production of defensive allelochemicals, and treatment of maize, soybean or eggplant with their own Peps induces the production of GLVs and other metabolites associated with plant defences against insects (Huffaker et al., 2013). Together, these results indicate that Peps and PEPRs modulate plant defence responses in multiple plant species, and can be manipulated to confer resistance to insects and diverse pathogens of roots and leaves.
The goal of the current study was to investigate whether Peps from soybean could confer resistance to two agricultural pests that attack roots: the root‐knot nematode (RKN) Meloidogyne incognita and the soybean cyst nematode (SCN) Heterodera glycines. SCN has been estimated to cause nearly $1 billion in annual yield losses on soybean (Davis and Tylka, 2000). RKNs (Meloidogyne spp.) are not only a major pest on soybean, but also attack hundreds of other important crops, placing them amongst the most common and damaging crop pests world‐wide (Mitkowski and Abawi, 2003). Although sources of resistance to cyst nematodes and RKNs have been identified in soybean, they are unlinked, and elite cultivars with resistance to both nematodes are currently lacking (Vuong et al., 2013). Thus, in soybean production areas in which cyst nematodes and RKNs coexist, these pests pose a particular challenge for growers because they are typically managed with different cultivars as well as different rotation strategies.
Peps represent a potential source of broad‐spectrum resistance against nematodes because they activate multiple defensive pathways, including defences previously implicated in nematode resistance. Transcript profiling in Arabidopsis has suggested that Peps coactivate salicylate (SA), jasmonate (JA) and ethylene signalling (Huffaker and Ryan, 2007; Ross et al., 2014). All three pathways are also activated by nematode infection, and each has been implicated in plant defences against nematodes in at least certain host plant–nematode combinations and infection stages (Branch et al., 2004; Manosalva et al., 2015; Mantelin et al., 2013; Nahar et al., 2011; Xie et al., 2016; Zhao et al., 2015). Peps also activate signal transduction events and oxidative responses that overlap with plant defence responses against nematodes. Pep perception by the PEPR1 receptor stabilizes a physical interaction between PEPR1 and a Brassinosteroid Receptor‐Associated Kinase1 (BAK1) (Yamada et al., 2016), stimulates the phosphorylation of Botrytis‐Induced Kinase1 (BIK1) (Liu et al., 2013) and promotes the accumulation of ROS in a BAK1‐ and BIK1‐dependent manner (Kadota et al., 2014; Roux et al., 2011). BIK1 is known to phosphorylate Respiratory Burst Oxidase Protein D (RBOHD), an NADPH oxidase that is responsible for the majority of the ROS produced during the oxidative burst associated with race‐specific effector‐triggered immunity (ETI) (Torres et al., 2002) and with broader spectrum pattern‐triggered immunity (PTI) (Macho and Zipfel, 2014). Moreover, mutations in RBOHD that prevent phosphorylation by BIK1 inhibit ROS induction by Pep1 (Kadota et al., 2014). Together, these data suggest that receptor‐mediated perception of Peps triggers RBOHD‐dependent ROS production via a BAK1/BIK1 phosphorylation cascade. Previous studies have shown that PTI against RKNs in Arabidopsis is compromised in mutants with impairments in BAK1, BIK1 or NADPH oxidase genes (RBOHD/F) (Teixeira et al., 2016). Comparison of transcript profiles induced by SCN in resistant versus susceptible soybean cultivars also indicates that BIK1 and RBOHD induction correlate with cyst nematode resistance (Matsye et al., 2011; Wan et al., 2015). Given that Peps induce defence responses that are also associated with PTI or ETI against nematodes, Peps could potentially be utilized to induce nematode resistance in susceptible genotypes.
To test this hypothesis, the present study examined the effects of Peps on nematode infestation and plant defence responses in a soybean cultivar susceptible to both SCN and RKN. The study was conducted with Peps derived from soybean, because sequence divergence amongst Peps and PEPRs limits the responsiveness of plants to Peps from other plant families (Huffaker et al., 2013; Lori et al., 2015). Our results indicate that seed treatments with exogenous Peps activate defence gene expression and ROS accumulation, and diminish nematode reproduction and damage.
Results
The soybean genome encodes eight putative Peps
The analysis of predicted amino acid sequences from the soybean genome (Schmutz et al., 2010) identified six putative plant elicitor propeptide (PROPEP) genes: GmPROPEP1 (on chromosome 10), GmPROPEP2 (chromosome 20), GmPROPEP3, GmPROPEP4 and GmPROPEP5 (grouped within a 7‐kb region of chromosome 13), and GmPROPEP6 (chromosome 4). These genes encode ∼70 to ∼120 residue precursor proteins that are predicted to be post‐translationally processed, releasing 23‐residue elicitor peptides (Fig. 1A). GmPROPEP4 and GmPROPEP5 are each predicted to encode two alternative mature peptides. GmPROPEP6 is somewhat degenerate, and appears to have a potential amino acid deletion at position 20, shifting the motif sequence. We have named this as a potential PROPEP orthologue, but future studies will determine whether it has bioactivity similar to other PROPEPs. The comparison of the amino acids of 23‐amino‐acid bioactive GmPeps shown in Fig. 1A indicates that each GmPep contains several conserved motifs in the carboxyl region, especially in position Gln21 and Asn23 (Fig. 1B). The analysis of the amino acid composition shows that seven of eight GmPeps are highly enriched in glycine. Phylogenetic analysis also demonstrated homology between GmPeps and Arabidopsis elicitor peptide AtPep1. The deduced amino acid sequence of AtPep1 shows more similarity to GmPep6 than to any other GmPep. Interestingly, GmPep1, GmPep2 and GmPep3 show distinctive similarity to each other and, in addition, GmPep4 has more similarity with GmPep5 than with any other GmPep, indicating that GmPeps can be classified into three groups which might be involved in different functions (Fig. 1C).
Figure 1.
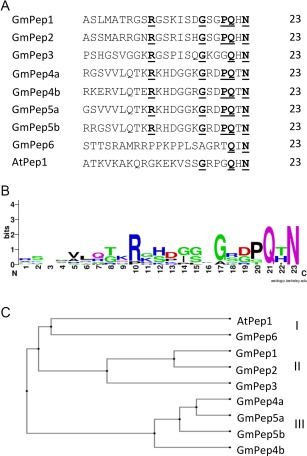
The soybean genome encodes a family of plant elicitor peptides (GmPeps). (A) Conserved motifs of GmPeps are underlined and in bold. GmPROPEP4 and GmPROPEP5 precursors are each predicted to generate two encoded peptides, designated ‘a’ and ‘b’. (B) Carboxyl terminus of the peptide confirmed to be the following motif beginning at residue 15 (S/GxGxxQxN). (C) A phylogenetic tree was generated by ClustalW2. Included in the alignment are GmPep1 (Glyma10g36290), GmPep2 (Glyma20g31306), GmPep3 (Glyma13g34221), GmPep4a,b (Glyma13g34210), GmPep5a,b (Glyma13g34235), GmPep6 (Glyma04g39760) and AtPep1 (At5g64900).
To explore the expression patterns of these GmPROPEP genes in different plant tissues and growth stages, we queried an RNA‐sequencing (RNA‐seq) atlas available through SoyBase (www.soybase.org) which reports the transcript profiles of leaves, roots, nodules, flowers, pods and seven stages of seed development from healthy, uninfected soybean plants (Severin et al., 2010). Transcripts for three of the six GmPROPEP genes were detected in the atlas, indicating some constitutive expression in the absence of pests, pathogens or wounding. GmPROPEP1 (Glyma10g36290) was expressed in all of the tested tissues, including roots, leaves, flowers, seeds, nodules and pods. GmPROPEP4 (Glyma13g34120) was expressed in seeds, and GmPROPEP6 (Glyma04g39760) was expressed in seeds and roots at a very low level (Fig. S1, see Supporting Information). We chose to focus subsequent experiments on GmPROPEP1, GmPROPEP2 and GmPROPEP3 because they constitute a distinct subgroup based on sequence homology (Fig. 1C), but differ in whether they are constitutively expressed (GmPROPEP1) or not (GmPROPEP2 and GmPROPEP3). Moreover, a previous study has shown that exogenous application of synthetic GmPep3 to soybean foliage could induce the production of volatile organic compounds (VOCs) associated with the recruitment of the natural enemies of herbivores (Huffaker et al., 2013).
Seed treatments with soybean Peps reduce nematode reproduction
To assess whether GmPeps could limit nematode infection, we analysed RKN reproduction on plants that had received seed treatments with GmPep1, GmPep2 or GmPep3. Peptide treatment caused significant differences in RKN reproduction amongst treatment groups [Fig. 2A; one‐way analysis of variance (ANOVA), P = 0.036]. Egg mass numbers per gram of root mass were significantly lower in response to each of the peptide treatments compared with water‐treated controls, with 40%–50% lower reproduction on the peptide treatments than on the water‐treated control group (Fig. 2A; Student's t‐tests, P < 0.05). To assess whether the nematode resistance induced by GmPeps was limited to RKNs or extended to other nematode species, reproduction of SCN was also compared on GmPep‐treated and untreated plants. Cyst production varied significantly amongst treatment groups (one‐way ANOVA, P = 0.0057), and was significantly lower on GmPep1‐, GmPep2‐ or GmPep3‐treated plants than on water‐treated controls (Student's t‐tests, P < 0.05), displaying ∼40% to ∼70% reduction in nematode reproduction (Fig. 2B). These results indicate that GmPep treatments can induce broad‐spectrum nematode resistance.
Figure 2.
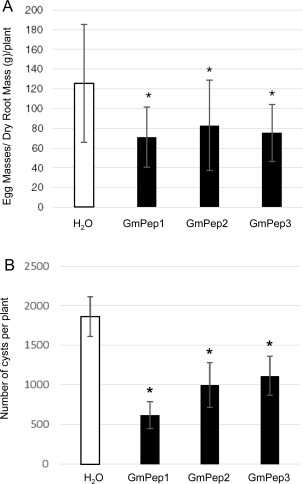
Plant elicitor peptide (Pep) seed treatments limit nematode reproduction on soybean. (A) Reproduction by the root‐knot nematode (Meloidogyne incognita) was compared by measuring the number of egg masses per gram of dry root mass at 7 weeks after inoculation (n = 10 for H2O, n = 11 for GmPep1, n = 11 for GmPep2, n = 6 for GmPep3). (B) Reproduction of the soybean cyst nematode (Heterodera glycines) was also compared on plants that had received seed treatments with GmPep1, GmPep2 or GmPep3 (1 µm) or water by counting the number of cysts per plant at 2 months after inoculation (n = 15 for all treatments). For both nematode species, there were significant differences in reproduction amongst the treatment groups [one‐way analyses of variance (ANOVAs), P < 0.05]. Treatments labelled with asterisks are significantly different from the water‐treated control group according to Student's t‐tests (α = 0.05). Error bars represent the standard deviations (SDs).
Seed treatments with soybean Peps have neutral or positive effects on plant growth
Certain forms of induced defence in plants are associated with delays in growth and, if they are induced in the absence of strong pest pressure, they may have fitness costs (Huot et al., 2014). To assess whether GmPep seed treatments might cause such tradeoffs, above‐ and below‐ground biomasses of plants treated with H2O, GmPep1, GmPep2 and GmPep3 were compared in the absence of nematode infection. None of the three GmPep treatments had any significant effect on shoot or root biomass (Fig. 3A,B; one‐way ANOVA, P > 0.50), indicating that no negative side effects on plant growth were observed in the absence of nematodes. To follow up on this, we also compared the influence of GmPep1 on plant growth in RKN‐inoculated versus uninoculated plants. Nematode reproduction was relatively low on inoculated plants because of cool glasshouse conditions (24.37 ± 17.36 egg masses/g dry root weight on water‐treated plants and 17.36 ± 12.68 egg masses/g dry root weight on GmPep1‐treated plants), but reproduction was significantly lower on GmPep1‐treated plants than on water‐treated controls (one‐way ANOVA, P = 0.0090). For above‐ground biomass (Fig. 4A), the interaction between seed treatments and inoculation was statistically significant (two‐way ANOVA, P = 0.0218), suggesting that the effect of nematode infection on above‐ground biomass varied depending on whether or not the plants had received a seed treatment. Contrast statements revealed that nematode infestation reduced the shoot growth of water‐treated plants; the above‐ground dry weight of nematode‐inoculated, water‐treated plants was significantly lower than that of water‐treated, mock‐inoculated plants (P = 0.0008). In comparison, GmPep1 seed treatment appeared to protect plants from the negative effects of infection on shoot weight; the above‐ground biomass of nematode‐infested, elicitor‐treated plants was significantly higher than that of nematode‐infested, water‐treated plants (P = 0.0256), and was similar to that of uninfested treatment groups (P > 0.4). These data indicate that GmPep seed treatment can limit the damaging effects of nematode infection on foliar growth. Consistent with our previous assay with unchallenged plants (Fig. 3A), there was also no difference in above‐ground weight between elicitor‐treated uninfested plants and water‐treated, uninfested controls (P = 0.6413). Thus, the effects of Pep seed treatment on above‐ground biomass were beneficial when nematodes were present and neutral when nematodes were absent.
Figure 3.
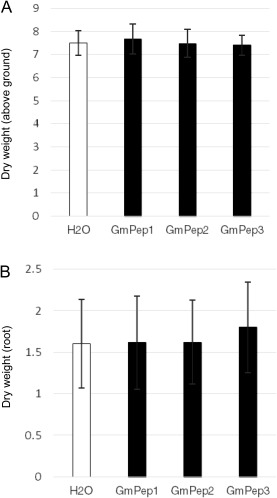
GmPep seed treatments do not affect above‐ or below‐ground biomass. Above‐ground (A) and below‐ground (B) biomasses of plants treated with H2O, GmPep1, GmPep2 or GmPep3 were compared in the absence of nematode infection. Data were collected 6 weeks after treatment. For both above‐ground and root biomass, there were no significant differences amongst the four treatment groups [one‐way analysis of variance (ANOVA), P > 0.50]. Error bars indicate standard deviations (SDs).
Figure 4.
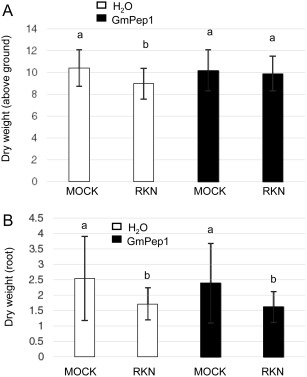
GmPep1 treatment limits the impact of root‐knot nematode (RKN) infection on above‐ground plant growth. Above‐ground dry weights (A) and below‐ground dry weights (B) were compared in plants that had received a GmPep1 seed treatment (1 µm) or water (H2O), and had been inoculated with RKN or mock inoculated (MOCK) with water. Data were collected 8 weeks after inoculation. For above‐ground biomass (A), a two‐way analysis of variance (ANOVA) indicated a significant interaction between the main effects of seed treatment and inoculation (P = 0.0218). Values labelled with different letters are significantly different at P < 0.05 according to contrast statements. For root biomass (B), there was no significant interaction between treatment and inoculation (P = 0.84), and no significant effect of elicitor treatment (P = 0.32), indicating that seed treatment has no effect on root weight. Error bars indicate standard deviations (SDs).
Although biotic stress often impacts chlorophyll content (Barry, 2009), at 6 weeks after inoculation this parameter was similar in the first fully expanded leaf of water‐treated uninoculated plants (26.49 ± 1.42 SPAD units), water‐treated infected plants (27.41 ± 1.13 SPAD units), elicitor‐treated uninoculated plants (29.24 ± 1.13 SPAD units) and elicitor‐treated inoculated plants (27.39 ± 0.89 SPAD units). We did not detect any significant effect of seed treatment (two‐way ANOVA, P = 0.56), nematode inoculation (two‐way ANOVA, P = 0.87) or the interaction between these two variables (two‐way ANOVA, P = 0.83) on chlorophyll content. Similar patterns were observed in the chlorophyll content of mature foliage (the middle third and top fifth expanded leaves from the apical meristem) and, for all leaf positions tested, we also did not detect any significant differences in instantaneous chlorophyll fluorescence (data not shown), a common marker of plant stress (Brestic and Zivcak, 2013).
In addition to the measurement of above‐ground growth and chlorophyll content, we also examined root biomass at the end of the assay, when nematode infestation was scored (Fig. 4B). Root weights were significantly lower on inoculated samples than on uninoculated controls in water‐treated plants (two‐way ANOVA, main effect of inoculation: P < 0.0001), but there was no significant effect of elicitor treatment (Fig. 4B; P = 0.32), or any significant interaction between seed treatment and inoculation (P = 0.84). In other words, nematode inoculation reduced root weights and seed treatments had no effect on root weight.
Together, these data indicate that GmPep1 treatment does not deter plant growth; moreover, it may protect plants against the negative impacts of nematode infection on above‐ground biomass. The benefits of GmPep treatment appeared to be greater for above‐ground rather than below‐ground growth.
GmPeps induce defence genes regulating soybean resistance in roots
To investigate whether Pep seed treatments limit nematode infestation through transcriptional reprogramming of the roots, we examined the expression of several defence genes previously implicated in resistance to SCN or RKN. Genes encoding a putative respiratory burst oxidase protein (RBOHD) and a nucleotide‐binding site leucine‐rich repeat‐type disease resistance protein (NBS‐LRR) have been reported previously to be up‐regulated by SCN infestation on resistant, but not susceptible, plants (Wan et al., 2015). In addition, a gene encoding a putative pectin methylesterase inhibitor (PMEI, Glyma10g02150) was identified as a candidate gene for RKN resistance through quantitative trait locus (QTL) mapping (Xu et al., 2013). The expression of these genes was analysed in root tissues grown from seeds treated with GmPep or with a water control (Fig. 5). To correct for unequal variances, relative expression values were log‐transformed before statistical analysis. Seed treatment with any of the three Peps was sufficient to induce strong expression of RBOHD (Fig. 5A–C; one‐way ANOVAs, P < 0.05). NBS‐LRR was up‐regulated in response to GmPep1 and GmPep3 (one‐way ANOVAs, P < 0.05), but not GmPep2 (P = 0.59), and PMEI was induced by GmPep1 (P = 0.0012), but not GmPep2 or GmPep3 (P > 0.10) (Fig. 5A–C). In addition to up‐regulating all three defence genes, GmPep1 seed treatment also up‐regulated its own precursor, GmPROPEP1 (Fig. 5A; P = 0.0438), although GmPep2 and GmPep3 did not induce their respective precursors GmPROPEP2 and GmPROPEP3 (P > 0.10).
Figure 5.
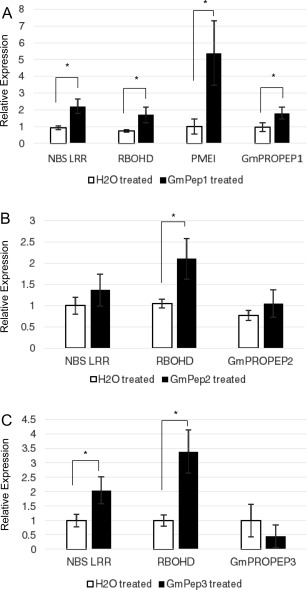
Seed treatments with GmPeps induce the expression of defence‐associated genes in roots. Transcript levels of NBS‐LRR (nucleotide‐binding site leucine‐rich repeat gene, Glyma06g26800), RBOHD (Respiratory Burst Oxidase Protein D, Glyma15g26790), PMEI (pectin methylesterase inhibitor, Glyma10g02150), GmPROPEP1, GmPROPEP2 and GmPROPEP3 were analysed by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) after treatment of soybean seeds with water (control) or 1 µm synthetic GmPep1 (A), GmPep2 (B) or GmPep3 (C). Relative expression values for each gene were normalized using the ELF1b housekeeping gene and analysed by one‐way analyses of variance (ANOVAs). Treatments labelled with an asterisk are significantly different at α = 0.05; for all other pairs, P > 0.1. Error bars indicate the standard errors of the mean (SEMs).
Because RBOHD was induced by all three GmPeps, we also examined the effect of nematode infection on the expression of this gene singly and in combination with GmPep1, GmPep2 or GmPep3 seed treatments (Fig. 6). Relative expression data were log‐transformed prior to statistical analysis to correct for unequal variances. As shown in Fig. 6A, GmPep1 seed treatment and RKN infection each significantly up‐regulated RBOHD expression (two‐way ANOVA; main effect of GmPep1 treatment: P = 0.012; main effect of nematode infection: P < 0.0001), and nematodes and GmPep1 appeared to have an additive effect on gene expression, with no significant interaction between the two factors (P = 0.94). In experiments with GmPep2 and GmPep3, peptide treatment and nematode infection also strongly induced RBOHD, but there was a significant interaction between the two factors (two‐way ANOVAs, P < 0.05 for interaction factors and main effects). Mean separations revealed that, although RBOHD expression was higher in plants treated with GmPep2, GmPep3 or nematodes compared with water‐treated plants (P < 0.02), nematode inoculation did not lead to additional increases in RBOHD expression in plants that had also received GmPep2 or GmPep3 seed treatments (P > 0.1).
Figure 6.
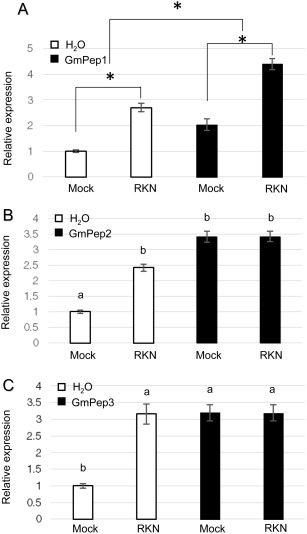
GmPep treatment and root‐knot nematode (RKN) inoculation both induce Respiratory Burst Oxidase Protein D (RBOHD) expression in soybean roots. Three days after seeds had been treated with water (H2O) or 1 µm GmPep1 (A), GmPep2 (B) or GmPep3 (C), seedlings were inoculated with RKN or mock inoculated with water (Mock). Twenty‐four hours after inoculation, root tissue was collected and used to measure RBOHD expression by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR). For GmPep1 (A), both seed treatment and nematode inoculation significantly up‐regulated RBOHD [two‐way analysis of variance (ANOVA), P < 0.05; significant main effects indicated with asterisks], but there was no significant interaction between Pep treatment and RKN inoculation (two‐way ANOVA, P = 0.94). For GmPep2 and GmPep3, there was a significant interaction between seed treatment and inoculation (two‐way ANOVA, P < 0.05), and so contrast statements were used to separate the means. In (B) and (C), values labelled with different letters are significantly different at P < 0.05 according to contrast statements. Error bars indicate the standard errors of the mean (SEMs).
GmPeps trigger defence gene expression in soybean leaves
The expression patterns of defence‐associated genes and PROPEP genes were also analysed in leaves of soybean plants that had received GmPep seed treatments or a water mock treatment (Fig. 7). Analysis of log‐transformed data showed that RBOHD expression in foliage was induced by all three peptides (one‐way ANOVAs, P < 0.05). Foliar expression of NBS‐LRR was up‐regulated by GmPep2 (P = 0.004), but not by GmPep1 or GmPep3 (P > 0.1). GmPep1 up‐regulated the expression of its own precursor, GmPROPEP1 (P = 0.0094), but GmPep2 and GmPep3 did not significantly up‐regulate their own expression (one‐way ANOVAs, P > 0.1). Expression of PMEI was not detected in the foliage. These results highlight that GmPep seed treatments promote defence gene induction not only in roots, but also systemically in leaves, and that, although the three peptides trigger distinct transcript profiles, they all promote RBOHD expression.
Figure 7.
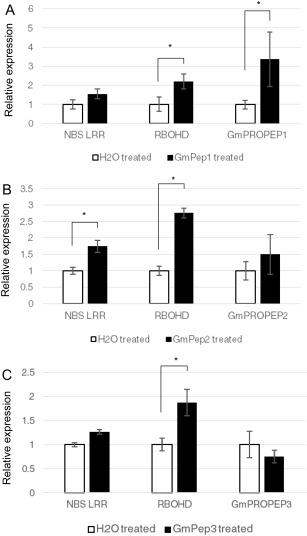
Seed treatments with GmPeps induce the expression of defence‐associated genes in foliage. Transcript levels of NBS‐LRR (nucleotide‐binding site leucine‐rich repeat gene) and RBOHD (Respiratory Burst Oxidase Protein D) were analysed by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) in leaf samples collected 3 weeks after treatment of soybean seeds with water (control) or 1 µm synthetic GmPep1 (A), GmPep2 (B) or GmPep3 (C). Relative expression values for each gene were analysed by one‐way analyses of variance (ANOVAs). Treatments labelled with an asterisk are significantly different at α = 0.05. For all other pairs, P > 0.1. Error bars indicate the standard errors of the mean (SEMs).
ROS production in soybean roots or leaves in response to GmPeps
The expression of RBOHD in response to all three GmPeps tested suggests that GmPep seed treatments might stimulate the production of ROS, much like foliar treatments with Arabidopsis elicitor peptide AtPep1 induce H2O2 in Arabidopsis leaves (Huffaker et al., 2006). We measured ROS production in soybean roots or leaves in seedlings that had received a seed treatment with GmPep1, GmPep2, GmPep3 or water (control) through a luminescence‐based assay. Luminescence values were log‐transformed because of unequal variances. ROS levels in both roots and leaves differed significantly amongst our treatment groups (Fig. 8A; one‐way ANOVAs, P < 0.05). ROS accumulation in roots was significantly higher in plants that had been treated with GmPep2 or GmPep3 than in water‐treated controls, and all three GmPep treatments led to significant increases in foliar ROS levels (Student's t‐tests, P < 0.05). These data indicate that the enhanced RBOHD levels observed were predictive of enhanced ROS accumulation, and that GmPep seed treatments can induce defence signalling in both roots and leaves.
Figure 8.
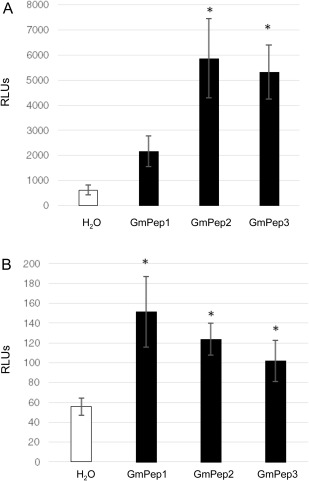
Seed treatments with GmPeps stimulate reactive oxygen species (ROS) production in soybean roots and leaves. A luminescence‐based assay was used to measure ROS content in soybean roots 4 days after treatment of seeds with H2O or synthetic 1 µm GmPeps (A), or in foliage at the first true leaf stage, approximately 10 days after seed treatment (B). RLUs, relative light units. According to one‐way analyses of variance (ANOVAs), luminescence varied significantly amongst treatments in both roots and leaves (P < 0.05). Asterisks represent significant differences between H2O and Pep treatments at α = 0.05 according to Student's t‐tests. Error bars indicate standard deviations (SDs).
Discussion
Peps have been identified through in silico analysis in a taxonomically diverse array of plants, including soybean and other crops, but measures of their impact on colonization by pests or pathogens are primarily limited to studies in Arabidopsis (Huffaker, 2015; Huffaker et al., 2006; Lori et al., 2015). GmPep3, a Pep from soybean, has been shown previously to induce the production of VOCs associated with defence when it is exogenously applied to soybean foliage (Huffaker et al., 2013), but its influence on soybean pests was not tested. Here, we demonstrate that GmPep3 and two closely related homologues, GmPep1 and GmPep2, can activate plant defences to limit infection by two agronomically important pests of soybean, RKN and SCN (Fig. 2). These results indicate that the defensive pathways activated by Peps are active against biotrophic endoparasites, and add to previous reports of Pep‐mediated resistance against necrotrophic (Pythium irregulare), hemibiotrophic (Pseudomonas syringae) and herbivorous (Spodoptera littoralis) pests in Arabidopsis (Huffaker et al., 2006; Klauser et al., 2015; Yamaguchi et al., 2010). Our findings also suggest that seed treatments with Peps can immunize plants against subsequent infections during the seedling stage.
Seed treatments with GmPep1, GmPep2 or GmPep3 all reduced SCN and RKN infestation on soybean and up‐regulated defence gene expression in roots and shoots. The three different GmPeps varied in their effects on the expression of certain defence genes, such as a pectin methylesterase inhibitor (PMEI) involved in cell wall modification and a putative NBS‐LRR gene. However, a commonality was that, in both roots and leaves, all three peptides up‐regulated RBOHD, which encodes an NADPH oxidase that plays a critical role in ROS generation in the apoplast. Furthermore, all three GmPep treatments enhanced ROS accumulation in the foliage (Fig. 8), which is consistent with patterns of RBOHD expression and with previous evidence that the recognition of AtPep1 in Arabidopsis triggers an oxidative burst (Krol et al., 2010). Interestingly, although GmPep2 and GmPep3 promoted ROS accumulation in the roots as well as in the leaves, the effects of GmPep1 were statistically significant only in the leaves. It is possible that, in roots, the timing of ROS induction by GmPep1 differs from that of GmPep2 or GmPep3.
Rapid production of ROS in the apoplast by RBOHD is an early defence response that occurs after the successful recognition of pathogens by the plant immune system, and that can kill pathogens, trigger hypersensitive cell death, activate the expression of defence‐related genes and interact with plant signalling networks to modulate local and systemic immunity (Apel and Hirt, 2004). Studies in several different plant species have also indicated that ROS play important roles in plant–nematode interactions (Li et al., 2015). In certain compatible interactions, nematodes appear to utilize effector molecules to manipulate ROS levels and to suppress effective defence responses in the host plant (Dubreuil et al., 2011; Lin et al., 2016; Siddique et al., 2014), whereas, in other interactions, ROS activation contributes to basal defences or is associated with incompatibility (Kandoth et al., 2011; Kong et al., 2015; Melillo et al., 2006; Teixeira et al., 2016). For example, in soybean, transcripts associated with ROS generation are more highly expressed at SCN infection sites in a resistant cultivar than in a near‐isogenic susceptible line (Kandoth et al., 2011). Therefore, the induction of RBOHD expression by exogenous GmPeps may potentially contribute to the suppressive effects of GmPep treatments on nematode infestations through local (GmPep2 and GmPep3) and/or systemic (GmPep1, GmPep2, and GmPep3) ROS accumulation.
In addition to demonstrating bioactivity for GmPeps, our results also raise the possibility that Pep seed treatments could be utilized as plant activators for the management of nematodes and other pests. Plant activators are naturally derived or synthetic small molecules that are applied to plants to stimulate immunity, and several of these elicitors have been commercialized for pest management (Bektas and Eulgem, 2015). They offer the advantage that they can be combined with any agronomically desirable germplasm to stimulate plant defences in susceptible cultivars or, potentially, to broaden or enhance defences in resistant cultivars. Nematode infections on some crops, such as tomato, are inhibited by root drenches or foliar applications of certain plant activators, including dl‐β‐amino‐n‐butyric acid (BABA), benzo(1,2,3)thiadiazole‐7‐carbothionic acid‐S‐methyl ester (BTH or Actigard), 2,6‐dichloroisonicotinic acid (INA) and JAs (Cooper et al., 2005; Molinari, 2016; Oka et al., 1999; Vieira dos Santos et al., 2013). The fact that Peps can induce plant defences when applied as a seed treatment offers several advantages over drenches or sprays, because seed treatments cut labour costs for application, reduce the amount of active ingredient required and allow for earlier protection during germination and establishment. Pep seed treatments could also potentially provide protection against above‐ground pests in addition to below‐ground nematodes, given that GmPeps induce systemic defence gene expression and ROS accumulation in foliage (Fig. 8), and that Peps in Arabidopsis impact diverse attackers (Huffaker et al., 2006; Klauser et al., 2015; Yamaguchi et al., 2010). Another positive aspect of GmPep seed treatments is that they do not impair soybean growth in the absence of pests, and could potentially help protect against growth inhibition by nematodes (Fig. 4). This is encouraging because certain other forms of induced defence result in tradeoffs with plant growth (Huot et al., 2014). In conclusion, soybean Peps can activate broad‐spectrum nematode resistance, and Pep seed treatments merit further study as a potential tool to decrease management costs and simplify nematode management decisions.
Experimental Procedures
Identification of soybean propeptide genes
Candidate PROPEPs from soybean were identified by the application of tblastn algorithms to expressed sequence tag (EST) data and genomic sequences available through the National Center for Biotechnology Information (NCBI) and SoyBase, with expected values set to 10,000 or more. Candidate PROPEPs were selected if they contained Pep‐like 23mers that conformed to the following criteria: (i) the amino‐terminal end contained a number of basic residues; and (ii) the carboxyl terminus of the peptide conformed to the following motif beginning at residue 15: S/GxGxxQxN (Fig. 1B). The relatedness of the putative PROPEPs was analysed by generating a phylogenetic tree with ClustalW2.
Nematode culture
SCNs
An SCN H. glycines isolate originally collected from soybean in Arkansas was reared under glasshouse conditions on soybean (cv. Lee), and cysts were extracted from infested soil by flotation in water and collection on a No. 60 (250 µm) sieve. Eggs of SCN were obtained by gently crushing cysts that had been extracted from glasshouse cultures (Davis et al., 1996)
RKNs
An RKN M. incognita isolate collected from soybean fields in Arkansas was kindly provided by Terry Kirkpatrick (University of Arkansas Extension Center, Fayetteville, AR, USA), and was cultured on susceptible tomato (Solanum lycopersicum cv. MoneyMaker) in a glasshouse at 23–27 °C with a 16‐h light/8‐h dark photoperiod. RKN eggs were extracted from infected roots by homogenization of roots in 10% bleach solution (0.6% sodium hypochlorite), rinsing with water and collection of eggs with a No. 500 (25 µm) sieve (Hussey and Barker, 1973). To prepare second‐stage juveniles (J2s), extracted eggs were surface sterilized with 10% bleach solution three times and placed in mesh baskets lined with two layers of Kim wipes suspended above a solution of 0.01% sodium dodecyl sulfate (SDS) and 0.1% plant preservative mixture (PPM, Plant Cell Technology, Washington DC, USA) at room temperature. After 48 h, J2s that crawled through the mesh were collected by filtering the solution with an autoclaved vacuum filtration system with a 0.22‐µm membrane filter (Millipore Corp., Burlington, MA, USA) (Atamian et al., 2012).
Peptide synthesis and seed treatments
In vitro synthesis of the 23‐amino‐acid peptides GmPep1 (amino acid sequence: ASLMATRGSRGSKISDGSGPQHN), GmPep2 (ASSMARRGNRGSRISHGSGPQHN) and GmPep3 (PSHGSVGGKRGSPISQGKGGQHN) was performed by Biomatik Corporation (Cambridge, ON, Canada), and their purity was verified by C18 high‐performance liquid chromatography (HPLC) and mass spectrometry. Soybean seeds (Glycine max cv. Williams82) were imbibed in Petri dishes at room temperature (24 °C) overnight in a solution of 0.05% Tween 20 and 1 µm of GmPep1, GmPep2 or GmPep3. Control seeds were treated with water and Tween 20 only. The next day, seeds were wrapped in wet paper towel to improve germination, and kept at 23 °C for 2–3 days prior to transfer of the seeds to sandy soil for subsequent bioassays or tissue collection experiments.
Bioassays for nematode resistance
Pep‐ or water‐treated seeds germinated on paper were planted in an autoclaved 2 : 1 mixture of coarse sand (Quikrete, Atlanta, GA, USA) and field‐collected sandy loam in 1‐L Styrofoam pots. Plants were grown under glasshouse conditions (16‐h light/8‐h dark photoperiod, 21–27 °C), and fertilized three times per day, 6 days a week, with a dilute fertilizer solution [Mixture of Grow More (Hydrobuilder), MgSO4 and CaNO3]. When the plants had two true leaves, they were inoculated with either SCN (5000 eggs per plant) or RKN (∼8000 M. incognita eggs). Plants were inoculated by pipetting eggs suspended in water into the soil via three holes about 3 cm deep. Infection levels were measured 4 weeks after inoculation for SCN assays, and 8 weeks after inoculation for RKN assays. For SCN assays, roots were washed in water and cysts were separated from the root systems using 250 µm sieve (Ithal et al., 2007). The number of cysts collected from each root system was counted under a dissecting microscope. Two independent experiments were conducted and 15 replicates of each treatment were included. To assess RKN reproduction, roots were washed gently in water to remove soil and debris, stained with Phloxine B (100 µg/mL) and observed under a dissecting microscope to count stained egg masses on the surface of the root systems (Daykin and Hussey, 1985). Root systems were then dried at 50 °C and weighed to calculate the average number of egg masses per unit of dry root mass. Each assay was repeated at least once with similar results.
Assessment of the effects of Pep seed treatments and nematode inoculation on plant growth and chlorophyll content
To test the effects of GmPeps on plant growth in the absence of pests, seeds were treated with water or 1 µm of GmPep1, GmPep2 or GmPep3, and then grown in sandy loam under glasshouse conditions, as described for nematode bioassays for nematode resistance. Six weeks after treatment, above‐ground portions of the plants were harvested, root systems were collected and washed of soil, samples were oven dried at 50 °C for 2 days, and weighed.
To compare the effects of GmPep1 on plant growth in the presence or absence of nematodes, GmPep1‐ or water‐treated seeds were inoculated with RKNs (∼8000 M. incognita eggs). At 5 weeks after inoculation, the chlorophyll content of intact soybean leaves was measured spectrophotometrically using a SPAD 502 chlorophyll meter (Konica Minolta, Osaka, Japan). Briefly, the chlorophyll content was assessed at three different leaf positions (the first, third and fifth fully expanded leaves below the meristem) by taking the mean of three readings at each position. SPAD values were measured at the midpoint of the leaf next to the main leaf vein. Instantaneous chlorophyll fluorescence was also measured in the same leaf positions using a FluorPen FP100 (Photon Systems Instruments, Drasov, Czech Republic). Eight weeks after inoculation, above‐ground portions of the plants were harvested, oven dried at 50 °C for 2 days and weighed. Root systems were examined to count RKN egg masses as described above, and were also dried and weighed. As a result of space constraints, the assay to measure biomass and nematode reproduction was replicated in time, and the different runs of the experiment were treated as blocks in the statistical analysis.
Tissue collection for gene expression analysis
To test the influence of Pep treatments on gene expression in roots and leaves, Pep‐treated or water‐treated seeds were germinated on wet paper. Three days after seed treatment, seedlings were either used for the collection of root tissues (roots from eight replicate seedlings/treatment were flash frozen in liquid nitrogen and stored at −80 °C) or transplanted in sandy soil and grown in a 16‐h light/8‐h dark cycle. When the transplanted seedlings had two fully expanded leaves (∼3 weeks after seed treatment), leaves from eight replicate seedlings per treatment were collected and flash frozen for RNA extraction. To test how Pep treatment and nematode infection interact to influence gene expression in roots, Pep‐treated or water‐treated seeds were germinated on wet paper and, at 3 days after Pep treatment, were transferred to wet filter paper in a Petri dish and either inoculated with RKN‐infective J2s (eight seedlings per Petri dish, 4000 J2s per Petri dish) or mock treated with water in a full factorial design. Root tissue (eight biological replicates per treatment) was collected and flash frozen 24 h after inoculation. Each gene expression analysis experiment was repeated at least once with similar results starting from independent tissue collection assays.
Quantification of gene expression
RNA extraction from root tissue was performed as described previously (Das et al., 2013). RNA from foliar tissues was isolated using Trizol reagent according to the manufacturer's instructions (Invitrogen, Carlsbad, CA, USA). cDNA was generated with Superscript III reverse transcriptase and oligo‐dT primers, and qPCR was performed with an Applied Biosystems StepOnePlus thermal cycler (Foster city, CA, USA) using a QuantiTect SYBR Green PCR kit (Valencia, CA, USA), as described previously (Avila et al., 2013), with a final primer concentration of 0.3 µm. The PCR conditions were 95 °C for 15 min, followed by 40 cycles of 95 °C for 15 s, 55 °C for 30 s and 72 °C for 30 s, with data acquisition at the end of each cycle. The specificity of the PCR amplification was monitored by melting curve analysis following the final step of the PCR from 65 °C to 95 °C every 0.3 °C. Data from our genes of interest were normalized to the expression levels of the endogenous control Translational elongation factor 1 subunit β (ELF1b) (Glyma02g44460). The primer pairs used for RT‐qPCR were as follows: NBS LRR (Glyma06g268600), forward (5′‐TGGGAATATGCTGTGGAACA‐3′); reverse (5′‐CAGCCTCACAAAATCTGCAA‐3′); RBOHD (Glyma06g17030), forward (5′‐ACATGAAGGCCATGGAAGAG‐3′); reverse (5′‐CCGAAGCTAACTGACCAAGC‐3′); PMEI (Glyma10g02150), forward (5′‐CTGCTGGTTTGATAAGAATG‐3′); reverse (5′‐AATGTGAGACACCAATAGAA‐3′); GmPROPEP1 (NM_001248225), forward (5′‐AGGGTCTTCACCATCCATTG‐3′); reverse (5′‐AATTATGCTGAGGGCCTGAC‐3′); GmPROPEP2 (XP_420382), forward (5′‐AAGGGTCTTCAGCATCATCG‐3′); reverse (5′‐CCTGACCCATGGCTAATTCT‐3′); GmPROPEP3 (NM_001248158), forward (5′‐CTCGCCTATTGGGAAACCTT‐3′); reverse (5′‐TCAACCCTAGCCTCGTCATT‐3′); ELF1b (Glyma02g44460), forward (5′‐GTTGAAAAGCCAGGGGACA‐3′); reverse (5′‐TCTTACCCCTTGAGCGTGG‐3′). PCR efficiency was calculated using the E = 10[–1/ Ct slope] formula (Rasmussen, 2000) and relative gene expression levels were calculated following the method of Pfaffl (2001).
Measurement of ROS level in roots and leaves
To test the effects of Pep treatments on ROS accumulation in roots, Pep‐ or water‐treated seeds were germinated on wet paper towel, and an approximately 10‐mg root section per sample was harvested 4 days after H2O or GmPep treatment. ROS accumulation in root segments was measured by a chemiluminescence assay as described previously (Hilbert et al., 2013). In brief, roots were incubated in water in 96‐well plates (Costar, Kennebunk, ME, USA) to recover from wounding during tissue collection. After a 4‐h recovery period, water was replaced with 100 µL of an elicitation solution containing 34 µg/mL luminol (Sigma, St. Louis, MO, USA) and 20 µg/mL horseradish peroxidase (HRP; Alfa Aesar, Haverhill, MA, USA) dissolved in water. The luminescence of each root sample was immediately measured using a Cytation3 plate reader (BioTeK, Winooski, VT, USA). To test the effects of Pep treatments on ROS accumulation in leaves, after a 3‐day germination period on wet paper towel, soybean seedlings were transplanted into sandy soil and grown at 23 °C, using the same soil and growth conditions as employed for nematode bioassays. Three weeks after transplanting, fully expanded leaves of uniform age were used for a chemiluminescent assay for ROS measurement (Smith and Heese, 2014), similar to the assay employed in roots. In brief, leaf discs from three plants per treatment were collected using a 5‐mm‐diameter plant tissue cork borer and cut into equal halves using razor blades, and tissue sections were incubated in H2O in 96‐well plates at 23 °C for a 20‐h recovery period. After the recovery period, the water was removed, elicitation solution containing 34 µg/mL luminol and 20 µg/mL HRP was added, and luminescence was measured with a Cytation3 plate reader (BioTeK) 10 min after the addition of elicitation solution.
Statistical analysis
All experiments were analysed using JMP Genomics 7.0 (SAS Institute, Cary, NC, USA). For datasets in which variances were unequal, the data were log‐transformed before analysis. ANOVAs were performed to detect differences among treatments and, for parameters that differed significantly (P < 0.05) according to ANOVAs, mean separations of multiple treatment groups were performed using contrast statements. Student's t‐tests were used for experimental designs in which we wished to make pairwise comparisons between treatments and their respective controls. All nematode bioassays, gene expression analyses and ROS measurements were performed at least twice with similar results.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 In silico analysis of GmPROPEP gene expression in various soybean plant tissues including leaf, flower, pod, seed, root and nodules. To examine the expression levels for each GmPROPEP, we took advantage of the soybean RNA‐sequencing (RNA‐seq) atlases described by Severin et al. (2010, http://soybase.org/soyseq). The Glyma identifiers (Soybase: http://soybase.org/soyseq/) of all six soybean GmPROPEPs were used as queries; however, only the expression of three GmPROPEPs were detected. GmPROPEP1 was the most highly and widely expressed GmPROPEP, with greatest expression in leaf, flower and seed. DAF, days after flowering.
Acknowledgements
This research was supported by the Arkansas Soybean Board, National Science Foundation (NSF) grant #11A‐1430427 and the Arkansas Experiment Station. We thank Dr Junhuan Xu for technical assistance, Dr Jung Ae Lee for statistical advice, Dr. Antje Heese for advice on ROS measurement and Dhaval Shah and Jessica Kivett for support with laboratory and glasshouse facilities.
References
- Apel, K. and Hirt, H. (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant. Biol. 55, 373–399. [DOI] [PubMed] [Google Scholar]
- Atamian, H.S. , Roberts, P.A. and Kaloshian, I. (2012) High and low throughput screens with root‐knot nematodes Meloidogyne spp. J. Vis. Exp. 61, e3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila, C.A. , Arevalo‐Soliz, L.M. , Lorence, A. and Goggin, F.L. (2013) Expression of alpha‐DIOXYGENASE 1 in tomato and Arabidopsis contributes to plant defenses against aphids. Mol. Plant–Microbe Interact. 26, 977–986. [DOI] [PubMed] [Google Scholar]
- Barry, C.S. (2009) The stay‐green revolution: recent progress in deciphering the mechanisms of chlorophyll degradation in higher plants. Plant Sci. 176, 325–333. [Google Scholar]
- Bektas, Y. and Eulgem, T. (2015) Synthetic plant defense elicitors. Front. Plant. Sci. 5, 804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branch, C. , Hwang, C.F. , Navarre, D.A. and Williamson, V.M. (2004) Salicylic acid is part of the mi‐1‐mediated defense response to root‐knot nematode in tomato. Mol. Plant–Microbe Interact. 17, 351–356. [DOI] [PubMed] [Google Scholar]
- Brestic, M. and Zivcak, M. (2013) PSII fluorescence techniques for measurements of drought and high temperature stress signal of crop plants: protocols and applications In: Molecular Stress Physiology of Plants (Rout G.R. and Das A.B, eds), pp. 87–131. Dordrecht: Springer. [Google Scholar]
- Cooper, W.R. , Jia, L. and Goggin, L. (2005) Effects of jasmonate‐induced defenses on root‐knot nematode infection of resistant and susceptible tomato cultivars. J. Chem. Ecol. 31, 1953–1967. [DOI] [PubMed] [Google Scholar]
- Das, A. , Saha, D. and Mondal, T. (2013) An optimized method for extraction of RNA from tea roots for functional genomics analysis. Indian J. Biotechnol. 12, 129–132. [Google Scholar]
- Davis, E.L. and Tylka, G.L. (2000) Soybean cyst nematode disease. Plant Health Instr. doi: 10.1094/PHI-I-2000-0725-01. [DOI] [Google Scholar]
- Davis, E.L. , Koenning, S.R. , Burton, J.W. and Barker, K.R. (1996) Greenhouse evaluation of selected soybean germplasm for resistance to North Carolina populations of Heterodera glycines, Rotylenchulus reniformis, and Meloidogyne species. J. Nematol. 28, 590–598. [PMC free article] [PubMed] [Google Scholar]
- Daykin, M.E. and Hussey, R.S. (1985) Staining and histopathological techniques in nematology. An advanced treatise on Meloidogyne . Methodology, II, 39–48. [Google Scholar]
- Dubreuil, G. , Deleury, E. , Magliano, M. , Jaouannet, M. , Abad, P. and Rosso, M.N. (2011) Peroxiredoxins from the plant parasitic root‐knot nematode, Meloidogyne incognita, are required for successful development within the host. Int. J. Parasitol. 41, 385–396. [DOI] [PubMed] [Google Scholar]
- Hilbert, M. , Nostadt, R. and Zuccaro, A. (2013) Exogenous auxin affects the oxidative burst in barley roots colonized by Piriformospora indica . Plant Signal. Behav. 8, e23572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker, A. (2015) Plant elicitor peptides in induced defense against insects. Curr. Opin. Insect Sci. 9, 44–50. [DOI] [PubMed] [Google Scholar]
- Huffaker, A. and Ryan, C.A. (2007) Endogenous peptide defense signals in Arabidopsis differentially amplify signaling for the innate immune response. Proc. Natl. Acad. Sci. USA, 104, 10 732–10 736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker, A. , Pearce, G. and Ryan, C.A. (2006) An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl. Acad. Sci. USA, 103, 10 098–10 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker, A. , Dafoe, N.J. and Schmelz, E.A. (2011) ZmPep1, an ortholog of Arabidopsis elicitor peptide 1, regulates maize innate immunity and enhances disease resistance. Plant Physiol. 155, 1325–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker, A. , Pearce, G. , Veyrat, N. , Erb, M. , Turlings, T.C. , Sartor, R. , Shen, Z. , Briggs, S.P. , Vaughan, M.M. , Alborn, H.T. , Teal, P.E. and Schmelz, E.A. (2013) Plant elicitor peptides are conserved signals regulating direct and indirect antiherbivore defense. Proc. Natl. Acad. Sci. USA, 110, 5707–5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huot, B. , Yao, J. , Montgomery, B.L. and He, S.Y. (2014) Growth‐defense tradeoffs in plants: a balancing act to optimize fitness. Mol. Plant, 7, 1267–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey, R.S. and Barker, B.K. (1973) A comparison of methods for collecting inocula for Meloidogyne sp., including a new technique. Plant Dis. Rep. 57, 1025–1028. [Google Scholar]
- Ithal, N. , Recknor, J. , Nettleton, D. , Hearne, L. , Maier, T. , Baum, T.J. and Mitchum, M.G. (2007) Parallel genome‐wide expression profiling of host and pathogen during soybean cyst nematode infection of soybean. Mol. Plant–Microbe Interact. 20, 293–305. [DOI] [PubMed] [Google Scholar]
- Kadota, Y. , Sklenar, J. , Derbyshire, P. , Stransfeld, L. , Asai, S. , Ntoukakis, V. , Jones, J.D. , Shirasu, K. , Menke, F. , Jones, A. and Zipfel, C. (2014) Direct regulation of the NADPH oxidase RBOHD by the PRR‐associated kinase BIK1 during plant immunity. Mol. Cell, 54, 43–55. [DOI] [PubMed] [Google Scholar]
- Kandoth, P.K. , Ithal, N. , Recknor, J. , Maier, T. , Nettleton, D. , Baum, T.J. and Mitchum, M.G. (2011) The soybean Rhg1 locus for resistance to the soybean cyst nematode Heterodera glycines regulates the expression of a large number of stress‐ and defense‐related genes in degenerating feeding cells. Plant Physiol. 155, 1960–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauser, D. , Flury, P. , Boller, T. and Bartels, S. (2013) Several MAMPs, including chitin fragments, enhance AtPep‐triggered oxidative burst independently of wounding. Plant. Signal. Behav. 8, e25346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauser, D. , Desurmont, G.A. , Glauser, G. , Vallat, A. , Flury, P. , Boller, T. , Turlings, T.C. and Bartels, S. (2015) The Arabidopsis pep‐PEPR system is induced by herbivore feeding and contributes to JA‐mediated plant defence against herbivory. J. Exp. Bot. 66, 5327–5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, L.A. , Wu, D.Q. , Huang, W.K. , Peng, H. , Wang, G.F. , Cui, J.K. , Liu, S.M. , Li, Z.G. , Yang, J. and Peng, D.L. (2015) Large‐scale identification of wheat genes resistant to cereal cyst nematode Heterodera avenae using comparative transcriptomic analysis. BMC Genomics, 16, 801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol, E. , Mentzel, T. , Chinchilla, D. , Boller, T. , Felix, G. , Kemmerling, B. , Postel, S. , Arents, M. , Jeworutzki, E. , Al‐Rasheid, K.A. , Becker, D. and Hedrich, R. (2010) Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J. Biol. Chem. 285, 13471–13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R. , Rashotte, A.M. , Singh, N.K. , Weaver, D.B. , Lawrence, K.S. and Locy, R.D. (2015) Integrated signaling networks in plant responses to sedentary endoparasitic nematodes: a perspective. Plant Cell Rep. 34, 5–22. [DOI] [PubMed] [Google Scholar]
- Lin, B. , Zhuo, K. , Chen, S. , Hu, L. , Sun, L. , Wang, X. , Zhang, L.H. and Liao, J. (2016) A novel nematode effector suppresses plant immunity by activating host reactive oxygen species‐scavenging system. New Phytol. 209, 1159–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z. , Wu, Y. , Yang, F. , Zhang, Y. , Chen, S. , Xie, Q. , Tian, X. and Zhou, J.M. (2013) BIK1 interacts with PEPRs to mediate ethylene‐induced immunity. Proc. Natl. Acad. Sci. USA, 110, 6205–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lori, M. , van Verk, M.C. , Hander, T. , Schatowitz, H. , Klauser, D. , Flury, P. , Gehring, C.A. , Boller, T. and Bartels, S. (2015) Evolutionary divergence of the plant elicitor peptides (peps) and their receptors: interfamily incompatibility of perception but compatibility of downstream signalling. J. Exp. Bot. 66, 5315–5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Y. , Walker, R.K. , Zhao, Y. and Berkowitz, G.A. (2012) Linking ligand perception by PEPR pattern recognition receptors to cytosolic Ca2+ elevation and downstream immune signaling in plants. Proc. Natl. Acad. Sci. USA, 109, 19 852–19 857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Y. , Zhao, Y. , Walker, R.K. and Berkowitz, G.A. (2013) Molecular steps in the immune signaling pathway evoked by plant elicitor peptides: Ca2+‐dependent protein kinases, nitric oxide, and reactive oxygen species are downstream from the early Ca2+ signal. Plant Physiol. 163, 1459–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho, A.P. and Zipfel, C. (2014) Plant PRRs and the activation of innate immune signaling. Mol. Cell, 54, 263–272. [DOI] [PubMed] [Google Scholar]
- Manosalva, P. , Manohar, M. , von Reuss, S.H. , Chen, S. , Koch, A. , Kaplan, F. , Choe, A. , Micikas, R.J. , Wang, X. , Kogel, K.H. , Sternberg, P.W. , Williamson, V.M. , Schroeder, F.C. and Klessig, D.F. (2015) Conserved nematode signalling molecules elicit plant defenses and pathogen resistance. Nat. Commun. 6, 7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantelin, S. , Bhattarai, K.K. , Jhaveri, T.Z. and Kaloshian, I. (2013) Mi‐1‐mediated resistance to Meloidogyne incognita in tomato may not rely on ethylene but hormone perception through ETR3 participates in limiting nematode infection in a susceptible host. PLoS One, 8, e63281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsye, P.D. , Kumar, R. , Hosseini, P. , Jones, C.M. , Tremblay, A. , Alkharouf, N.W. , Matthews, B.F. and Klink, V.P. (2011) Mapping cell fate decisions that occur during soybean defense responses. Plant Mol. Biol. 77, 513–528. [DOI] [PubMed] [Google Scholar]
- Melillo, M.T. , Leonetti, P. , Bongiovanni, M. , Castagnone‐Sereno, P. and Bleve‐Zacheo, T. (2006) Modulation of reactive oxygen species activities and H2O2 accumulation during compatible and incompatible tomato–root‐knot nematode interactions. New Phytol. 170, 501–512. [DOI] [PubMed] [Google Scholar]
- Mitkowski, N.A. and Abawi, G.S. (2003) Root‐knot nematodes. Plant Health Instr. doi: 10.1094/PHI-1-2003-0917-01. [DOI]
- Molinari, S. (2016) Systemic acquired resistance activation in Solanaceous crops as a management strategy against root‐knot nematodes. Pest Manag. Sci. 72, 888–896. [DOI] [PubMed] [Google Scholar]
- Nahar, K. , Kyndt, T. , De Vleesschauwer, D. , Hofte, M. and Gheysen, G. (2011) The jasmonate pathway is a key player in systemically induced defense against root knot nematodes in rice. Plant Physiol. 157, 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka, Y. , Cohen, Y. and Spiegel, Y. (1999) Local and systemic induced resistance to the root‐knot nematode in tomato by DL‐beta‐amino‐n‐butyric acid. Phytopathology, 89, 1138–1143. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M.W. (2001) A new mathematical model for relative quantification in real‐time RT‐PCR. Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, Z. , Verma, R. , Gehring, C. , Yamaguchi, Y. , Zhao, Y. , Ryan, C.A. and Berkowitz, G.A. (2010) Ca2+ signaling by plant Arabidopsis thaliana pep peptides depends on AtPepR1, a receptor with guanylyl cyclase activity, and cGMP‐activated Ca2+ channels. Proc. Natl. Acad. Sci. USA, 107, 21 193–21 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen, R. (2000) Quantification on the LightCycler In: Rapid Cycle Real‐Time PCR. Methods and Applications (Meuer S., Wittwer C. and Nakagawara K., eds), pp. 21–34. Heidelberg: Springer. [Google Scholar]
- Ross, A. , Yamada, K. , Hiruma, K. , Yamashita‐Yamada, M. , Lu, X. , Takano, Y. , Tsuda, K. and Saijo, Y. (2014) The Arabidopsis PEPR pathway couples local and systemic plant immunity. EMBO J. 33, 62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux, M. , Schwessinger, B. , Albrecht, C. , Chinchilla, D. , Jones, A. , Holton, N. , Malinovsky, F.G. , Tör, M. , De Vries, S. and Zipfel, C. (2011) The Arabidopsis leucine‐rich repeat receptor‐like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell, 23, 2440–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz, J. , Cannon, S.B. , Schlueter, J. , Ma, J. , Mitros, T. , Nelson, W. , Hyten, D.L. , Song, Q. , Thelen, J.J. , Cheng, J. , Xu, D. , Hellsten, U. , May, G.D. , Yu, Y. , Sakurai, T. , Umezawa, T. , Bhattacharyya, M.K. , Sandhu, D. , Valliyodan, B. , Lindquist, E. , Peto, M. , Grant, D. , Shu, S. , Goodstein, D. , Barry, K. , Futrell‐Griggs, M. , Abernathy, B. , Du, J. , Tian, Z. , Zhu, L. , Gill, N. , Joshi, T. , Libault, M. , Sethuraman, A. , Zhang, X.C. , Shinozaki, K. , Nguyen, H.T. , Wing, R.A. , Cregan, P. , Specht, J. , Grimwood, J. , Rokhsar, D. , Stacey, G. , Shoemaker, R.C. and Jackson, S.A. (2010) Genome sequence of the palaeopolyploid soybean. Nature, 463, 178–183. [DOI] [PubMed] [Google Scholar]
- Severin, A.J. , Woody, J.L. , Bolon, Y.T. , Joseph, B. , Diers, B.W. , Farmer, A.D. , Muehlbauer, G.J. , Nelson, R.T. , Grant, D. , Specht, J.E. , Graham, M.A. , Cannon, S.B. , May, G.D. , Vance, C.P. and Shoemaker, R.C. (2010) RNA‐seq atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol. 10, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique, S. , Matera, C. , Radakovic, Z.S. , Hasan, M.S. , Gutbrod, P. , Rozanska, E. , Sobczak, M. , Torres, M.A. and Grundler, F.M. (2014) Parasitic worms stimulate host NADPH oxidases to produce reactive oxygen species that limit plant cell death and promote infection. Sci. Signal. 7, ra33. [DOI] [PubMed] [Google Scholar]
- Smith, J.M. and Heese, A. (2014) Rapid bioassay to measure early reactive oxygen species production in Arabidopsis leaf tissue in response to living Pseudomonas syringae . Plant Methods, 10, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavormina, P. , De Coninck, B. , Nikonorova, N. , De Smet, I. and Cammue, B.P. (2015) The plant peptidome: an expanding repertoire of structural features and biological functions. Plant Cell, 27, 2095–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira, M.A. , Wei, L. and Kaloshian, I. (2016) Root‐knot nematodes induce pattern‐triggered immunity in Arabidopsis thaliana roots. New Phytol. 211, 276–287. [DOI] [PubMed] [Google Scholar]
- Torres, M.A. , Dangl, J.L. and Jones, J.D. (2002) Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA, 99, 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira dos Santos, M.C. , Curtis, R.H.C. and Abrantes, I. (2013) Effect of plant elicitors on the reproduction of the root‐knot nematode Meloidogyne chitwoodi on susceptible hosts. Eur. J. Plant Pathol. 136, 193–202. [Google Scholar]
- Vuong, T.D. , Jiao, Y. , Shannon, J.G. and Nguyen, H. (2013) Nematode resistance in soybean. In: Translational Genomics for Crop Breeding, Vol. 1 (Varshney R. and Roberto T., eds), pp. 95–124. John wiley & sons, Inc; Hoboken, NJ. [Google Scholar]
- Wan, J. , Vuong, T. , Jiao, Y. , Joshi, T. , Zhang, H. , Xu, D. and Nguyen, H.T. (2015) Whole‐genome gene expression profiling revealed genes and pathways potentially involved in regulating interactions of soybean with cyst nematode (Heterodera glycines ichinohe). BMC Genomics, 16, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, J. , Li, S. , Mo, C. , Wang, G. , Xiao, X. and Xiao, Y. (2016) A novel Meloidogyne incognita effector Misp12 suppresses plant defense response at latter stages of nematode parasitism. Front. Plant. Sci. 7, 964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X. , Zeng, L. , Tao, Y. , Vuong, T. , Wan, J. , Boerma, R. , Noe, J. , Li, Z. , Finnerty, S. , Pathan, S.M. , Shannon, J.G. and Nguyen, H.T. (2013) Pinpointing genes underlying the quantitative trait loci for root‐knot nematode resistance in palaeopolyploid soybean by whole genome resequencing. Proc. Natl. Acad. Sci. USA, 110, 13 469–13 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, K. , Yamashita‐Yamada, M. , Hirase, T. , Fujiwara, T. , Tsuda, K. , Hiruma, K. and Saijo, Y. (2016) Danger peptide receptor signaling in plants ensures basal immunity upon pathogen‐induced depletion of BAK1. EMBO J. 35, 46–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, Y. and Huffaker, A. (2011) Endogenous peptide elicitors in higher plants. Curr. Opin. Plant Biol. 14, 351–357. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, Y. , Huffaker, A. , Bryan, A.C. , Tax, F.E. and Ryan, C.A. (2010) PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell, 22, 508–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, W. , Li, Z. , Fan, J. , Hu, C. , Yang, R. , Qi, X. , Chen, H. , Zhao, F. and Wang, S. (2015) Identification of jasmonic acid‐associated microRNAs and characterization of the regulatory roles of the miR319/TCP4 module under root‐knot nematode stress in tomato. J. Exp. Bot. 66, 4653–4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 In silico analysis of GmPROPEP gene expression in various soybean plant tissues including leaf, flower, pod, seed, root and nodules. To examine the expression levels for each GmPROPEP, we took advantage of the soybean RNA‐sequencing (RNA‐seq) atlases described by Severin et al. (2010, http://soybase.org/soyseq). The Glyma identifiers (Soybase: http://soybase.org/soyseq/) of all six soybean GmPROPEPs were used as queries; however, only the expression of three GmPROPEPs were detected. GmPROPEP1 was the most highly and widely expressed GmPROPEP, with greatest expression in leaf, flower and seed. DAF, days after flowering.


