Summary
LysR‐type transcriptional regulators (LTTRs) are ubiquitous and abundant amongst bacteria and control a variety of cellular processes. Here, we investigated the effect of Rsc1880 (a putative LTTR, hereafter designated as PrhO) on the pathogenicity of Ralstonia solanacearum. Deletion of prhO substantially reduced the expression of the type III secretion system (T3SS) both in vitro and in planta, and resulted in significantly impaired virulence in tomato and tobacco plants. Complementary prhO completely restored the reduced virulence and T3SS expression to that of the wild‐type. Moreover, PrhO‐dependent T3SS and virulence were conserved amongst R. solanacearum species. However, deletion of prhO did not alter biofilm formation, swimming mobility and in planta growth. The expression of some type III effectors was significantly reduced in prhO mutants, but the hypersensitive response was not affected in tobacco leaves. Consistent with the key regulatory role of HrpB on T3SS, PrhO positively regulated the T3SS through HrpB. Furthermore, PrhO regulated hrpB expression via two close paralogues, HrpG and PrhG, which are two‐component response regulators and positively regulate hrpB expression in a parallel manner. However, deletion of prhO did not alter the expression of phcA, prhJ and prhN, which are also involved in hrpB regulation. In addition, PrhO was expressed in a cell density‐dependent manner, but negatively repressed by itself. No regulation was observed for HrpB, PhcA and PrhN on prhO expression. Taken together, we genetically demonstrated that PrhO is a novel virulence regulator of R. solanacearum, which positively regulates T3SS expression through HrpG, PrhG and HrpB and contributes to virulence.
Keywords: hrp regulation, LTTRs, pathogenicity, Ralstonia solanacearum, type III secretion system
Introduction
The LysR‐type transcriptional regulators (LTTRs), which contain an N‐terminal helix‐turn‐helix (HTH) DNA‐binding domain and a C‐terminal co‐inducer‐binding domain, are one of the largest families of regulators (more than 4000 orthologues) in diverse bacteria, archaea and algae (Maddocks and Oyston, 2008; Reen et al., 2013; Schell, 1993). Extensive research has generally characterized LTTRs as extremely abundant, similar sized (300–350 amino acids), positively or negatively auto‐regulated, and global regulators of diverse cellular processes (Heroven et al., 2004; Maddocks and Oyston, 2008). LTTRs activate or repress transcription by affecting the efficiency of transcription initiation, and hence control a great variety of cellular processes, including metabolism, the oxidative stress response, antibiotic resistance, quorum sensing, cell motility and virulence (Hernández‐Lucas et al., 2008; Maddocks and Oyston, 2008; Sperandio et al., 2002). Most LTTRs regulate the transcription of closely linked or genes in operon, whereas some regulate multiple unlinked genes (Maddocks and Oyston, 2008; Schell, 1993). To date, numerous LTTRs have been identified to be important for the virulence of many pathogenic bacteria of humans, animals and plants (Habdas et al., 2010; Huang et al., 1998; Jin et al., 2011; Rashid et al., 2016). For instance, BexR, MexT and MvfR regulate genes for virulence and antibiotic resistance in Pseudomonas aeruginosa (Deziel et al., 2005; Jin et al., 2011; Keith et al., 2009; Reen et al., 2013). In Escherichia coli, O157, QseA and QseD integrate several signalling networks and contribute to virulence (Habdas et al., 2010; Kendall et al., 2010). In the phytopathogenic bacterium, Ralstonia solanacearum, PhcA, a quorum‐sensing‐dependent LTTR, has been well characterized as a global regulator that controls the expression of diverse virulence‐related genes, including those involved in plant cell wall degradation, motility, synthesis of extracellular polysaccharide (EPS) and the type III secretion system (T3SS) (Bhatt and Denny, 2004; Brumbley et al., 1993; Genin et al., 2005; Huang et al., 1995).
Ralstonia solanacearum, the causal agent of bacterial wilt of plants, is a Gram‐negative, soil‐borne, vascular bacterium which causes severe losses in many economically important plants worldwide (Genin, 2010). As a vascular bacterium, it generally invades the xylem vessels through root wounds or natural openings (Vasse et al., 1995). Once inside the xylem vessels, R. solanacearum proliferates extensively and produces a huge amount of EPS slime, which severely blocks sap flow in xylem vessels and causes rapid stunting and wilting of plants (Denny, 1995; Roberts et al., 1988). In addition to EPS, the syringe‐like T3SS is essential for the infection process of R. solanacearum in host plants (Boucher et al., 1987; Galán and Wolf‐Watz, 2006; Lindgren, 1997). Bacteria use it to inject virulence factors, called type III effectors (T3Es), into host cytosol to subvert host defence and cause diseases (Cunnac et al., 2004; Fujiwara et al., 2016; Popa et al., 2016; Tasset et al., 2010). Ralstonia solanacearum is highly heterogeneous, which might be responsible for its extremely broad range of host species (Genin and Denny, 2012). The T3SS in R. solanacearum is encoded by approximately 20 genes located in the hypersensitive response and pathogenicity (hrp) gene cluster (Arlat et al., 1992), and is highly conserved amongst R. solanacearum species (Coll and Valls, 2013; Genin and Denny, 2012). Moreover, R. solanacearum harbours an unusually large repertoire of T3Es (average of more than 70 T3Es per strain and total of more than 110 T3Es amongst R. solanacearum species), which is extremely more abundant than those of any closely related phytopathogenic bacterium, i.e. 20–30 T3Es in Pseudomonas syringae, 10–15 T3Es in Erwinia spp. and approximately 40 T3Es in Xanthomonas spp. To date, about 30 T3Es have been functionally characterized in the interaction between R. solanacearum and host cells (Coll and Valls, 2013; Peeters et al., 2013; Tasset et al., 2010).
The T3SS and entire T3Es in R. solanacearum are directly controlled by the master regulator HrpB, an AraC family transcriptional regulator (Angot et al., 2006; Mukaihara et al., 2004, 2010). The expression of hrpB, T3SS and T3Es is repressed in nutrient‐rich medium, but activated in nutrient‐limited medium, which might mimic plant apoplastic fluids, and can be increased to much higher levels when in contact with host plants or when invading host plants (Arlat et al., 1992; Clough et al., 1997; Marenda et al., 1998; Yoshimochi et al., 2009b). Two close paralogues, HrpG and PrhG, which belong to the OmpR/PhoB family of two‐component response regulators, positively regulate hrpB expression in a parallel manner (Plener et al., 2010; Zhang et al., 2013). Plant signals or certain mimic signals are presumed to be recognized by an outer membrane protein PrhA, and transferred to HrpG through the PrhA–PrhR/I–PrhJ–HrpG signalling cascade (Marenda et al., 1998; Valls et al., 2006). HrpG and PrhG respond to host signals by phosphorylation at certain residues and greatly enhance hrpB expression, but the regulation mechanism remains to be further elucidated (Yoshimochi et al., 2009b). Moreover, a well‐characterized global LTTR regulator, PhcA, negatively regulates hrpB expression, which is activated at high cell density and binds to the promoter of prhI/R genes to repress their expression; this, in turn, shuts down the expression of prhJ, hrpG, hrpB and T3SS (Genin et al., 2005; Yoshimochi et al., 2009a). Different from HrpG, PrhG is independent of the PrhA cascade, and is positively regulated by PhcA and PrhN (Zhang et al., 2011, 2017).
In order to further elucidate the regulation of T3SS, we generated a popA‐lacZYA fusion, which belongs to T3Es, to monitor the expression profiles of T3SS in R. solanacearum. The generated reporter strains exhibited identical virulence and T3SS expression to those of wild‐type strains under different conditions (Yoshimochi et al., 2009b; Zhang et al., 2013). We screened 43 T3SS‐related candidates in the OE1‐1 strain with transposon mutagenesis (Zhang et al., 2013). Amongst them is Rsc1880, a putative LTTR (hereafter designated as PrhO). The expression level of the T3SS was substantially reduced in prhO transposon mutants. In total, 63 transcriptional regulators are predicted to be LTTRs in the genome of the OE1‐1 strain, but only PhcA has been functionally characterized to date. Here, we investigated the role of PrhO in the regulation of T3SS expression and virulence in R. solanacearum.
Results
PrhO positively regulates the T3SS expression of R. solanacearum species both in vitro and in planta
Previously, we used a popA‐lacZYA fusion in OE1‐1 (RK5050) to monitor T3SS expression and screened several T3SS‐related candidates (Zhang et al., 2013). Here, we generated a prhO in‐frame‐deleted mutant RK5738 (RK5050, ΔprhO) to confirm its effect on T3SS expression. Consistent with observations in transposon mutants, popA expression was substantially reduced in RK5738 (97 versus 293 Miller units of RK5050) in hrp‐inducing (sucrose) medium, and complementary prhO completely restored the reduced popA expression to that of RK5050 (Fig. 1a), confirming that PrhO positively regulates T3SS expression of OE1‐1 in hrp‐inducing medium.
Figure 1.
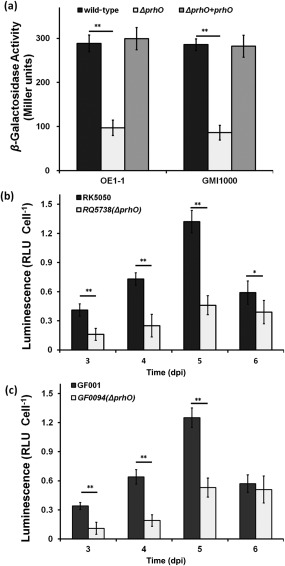
Expression of popA in Ralstonia solanacearum strains. (a) Expression of popA in OE1‐1 (left) and GMI1000 (right) derivatives grown in hrp‐inducing medium. (b) Expression of popA in OE1‐1 derivatives in planta. (c) Expression of popA in GMI1000 derivatives in planta. Black column, RK5050 (OE1‐1, popA‐lacZYA) or GF001 (GMI1000, popA‐lacZYA); white column, prhO mutants RQ5738 (RK5050, ΔprhO) or GF0094 (GF001, ΔprhO); gray column, prhO complemented strains RQC0090 (RQ5738 + prhO) or RQC0095 (GF0094 + prhO). (a) Cells were grown in hrp‐inducing medium to an optical density at 600 nm (OD600) of approximately 0.1 and subjected to β‐galactosidase assay. Enzymatic activities are presented in Miller units. (b, c) Tomato plants were inoculated with R. solanacearum strains using petiole inoculation and stem species were removed at 3–6 days post‐inoculation (dpi) for enzyme assay in planta with a Galacto‐Light Plus kit (Life, USA). Cell numbers were quantified by dilution plating and luminescence was evaluated using a GloMax20 luminometer (Promega, USA). Enzymatic activity was presented as luminescence normalized by the cell number. RLU, relative luminescence unit. The mean values of four independent experiments with four replications per trial were averaged and presented with standard deviation (SD) (error bars). Significance level: *P < 0.05; **P < 0.01 (t‐test).
Ralstonia solanacearum species are highly heterogeneous, and strains usually show different properties in pathogenicity. For example, strain OE1‐1 is virulent in both tomato and tobacco plants, whereas strain GMI1000 is virulent in tomato plants, but elicits a hypersensitive response (HR) in tobacco leaves. PrhOs are highly conserved in R. solanacearum species that exhibit more than 90% identity at the amino acid level. We therefore deleted prhO from GF001 (GMI1000, popA‐lacZYA) to evaluate its role in GMI1000. Consistent with the observations in OE1‐1, popA expression was substantially reduced in GF0094 (GF001, ΔprhO) (Fig. 1a). As expected, OE1‐1 PrhO completely restored the reduced popA expression in GF0094 to that of GF001 (Fig. 1a), suggesting that the PrhO‐dependent expression of T3SS is conserved amongst R. solanacearum species.
T3SS expression can be increased to a much higher level in planta than that in hrp‐inducing medium. We therefore evaluated whether T3SS expression in planta was altered with prhO deletion. When directly inoculated into xylem by petiole inoculation, R. solanacearum causes wilting and death in tomato plants at 3 and 7 days post‐inoculation (dpi), respectively. Hence, bacterial cells were collected daily from petiole‐inoculated tomato stems at 3–6 dpi and subjected to enzyme assay. The two prhO mutants, including derivatives of OE1‐1 (RK5738) (Fig. 1b) and GMI1000 (GF0094) (Fig. 1c), exhibited significantly reduced T3SS expression at 3–5 dpi (P < 0.01) compared with that of wild‐type strains in tomato stems (Fig. 1b,c). At 6 dpi, T3SS expression in GF0094 was not changed (P = 0.31) (Fig. 1c), but slightly reduced in RK5738 (P < 0.05) (Fig. 1b). Our data indicate that the in planta expression of R. solanacearum T3SS is also positively regulated by PrhO.
PrhO positively regulates the expression of some T3Es in R. solanacearum
The R. solanacearum species complex harbours more than 110 T3Es, with some playing important roles during the infection process in host plants. PopA is a T3E, and we therefore evaluated whether the expression of T3Es was altered with prhO deletion. Total RNA was isolated from GF001 (GMI1000, popA‐lacZYA) and GF0094 (GF001, ΔprhO), and mRNA levels of ripAA, ripB, ripD, ripE, ripO, ripPI, ripR, ripTAL, ripW and ripX (popA) were quantified by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). Consistent with results from the enzyme assay, the mRNA level of ripX was significantly reduced in GF0094 relative to that of the wild‐type strain GF001 (P < 0.01) (Fig. 2). The mRNA levels of ripAA, ripB, ripD, ripE, ripO, ripPI and ripW were also significantly reduced (P < 0.01), and those of ripR and ripTAL were slightly reduced (P < 0.05), compared with that of the wild‐type strain GF001 (Fig. 2), suggesting that the expression of some T3Es is also positively regulated by PrhO in R. solanacearum.
Figure 2.
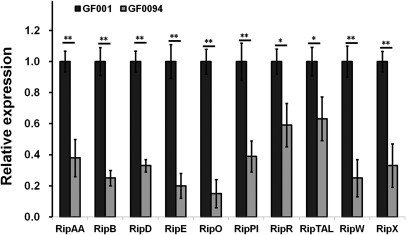
Relative expression of selected type III effector (T3E) genes in Ralstonia solanacearum. Total RNA was isolated from GF001 and prhO mutant (GF0094), and mRNA levels of representative T3E genes in hrp‐inducing medium were determined by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). serC was selected as reference for normalization. Normalized values of GF0094 were divided by those of GF001 and relative values (relative expression) are presented. Mean values of at least three independent trials were averaged and are presented with standard deviation (SD) (error bars). Statistical significance was assessed between normalized values of GF001 and GF0094. Significance level: *P < 0.05; **P < 0.01 (t‐test).
PrhO contributes to the virulence of R. solanacearum in host plants
T3SS and some T3Es play important roles in the pathogenicity of R. solanacearum, and we therefore evaluated the contribution of PrhO to pathogenicity. With soil‐soaking inoculation, which mimics natural invasion through roots, wild‐type strains caused wilting and death of most tomato plants within 10 dpi (Fig. 3a,b). In contrast, the two prhO mutants, including derivatives of OE1‐1 (Fig. 3a) and GMI1000 (Fig. 3b), caused the eventual death of only about half of the tested tomato plants, and about half of the tomato plants remained healthy or only slightly wilted up to 15 dpi (Fig. 3a,b). With petiole inoculation, by which bacteria could invade xylem vessels directly, significantly reduced virulence was also observed in RQ5738 (Fig. 3c). When complemented with PrhO, the reduced virulence in prhO mutants was completely restored to that of the wild‐type (Fig. 3a–c). We also evaluated the effect of PrhO on the virulence of OE1‐1 in tobacco plants. With leaf infiltration, significantly reduced virulence was also observed in RQ5738, as about half of the RK5Q38‐inoculated tobacco plants remained healthy or slightly wilted even up to 24 dpi (Fig. 3d). These results confirm that PrhO is important for the virulence of R. solanacearum species in different host plants.
Figure 3.
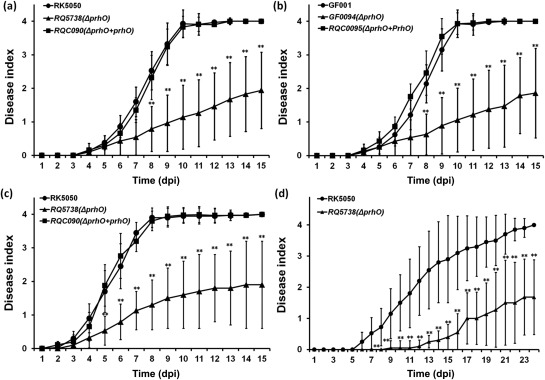
Pathogenicity test. Ralstonia solanacearum strains used: (a, c, d) OE1‐1 derivatives; (b) GMI1000 derivatives. Test plants: (a, b, c) tomato plants; (d) tobacco plants. Inoculation methods: (a, b) soil‐soaking inoculation; a bacterial suspension was poured into the soil of plants at a final concentration of 107 colony‐forming units (cfu)/g of soil; (c) petiole inoculation; about 3 μL of bacterial suspension at 108 cfu/mL was dropped onto the freshly cut petiole surface; (d) leaf infiltration in tobacco leaves; about 50 μL of bacterial suspension at 108 cfu/mL was infiltrated into tobacco leaves with a blunt‐end syringe. Filled circles, wild‐type strains; filled triangles, prhO mutants; filled squares, prhO mutants complemented with OE1‐1 prhO. Plants were inspected daily for wilting symptoms and scored on a disease index scale of 0–4 (0, no wilting; 1, 1%–25% wilting; 2, 26%–50% wilting; 3, 51%–75% wilting; 4, 76%–100% wilting or death). dpi, days post‐inoculation. Each assay was repeated in four independent trials and each trial contained at least 12 plants. Mean values of all results were averaged and are presented with standard deviation (SD) (error bars). Significance level: *P < 0.05; **P < 0.01 (t‐test).
PrhO is not required for bacterial growth in planta or HR elicitation in tobacco leaves
As an avascular pathogen, extensive proliferation in xylem vessels is one of the most important pathogenicity determinants in R. solanacearum. We therefore evaluated whether the impaired virulence of prhO mutants was a result of defective bacterial growth in host plants. With petiole inoculation of wild‐type strains, tomato plants normally started to wilt at 3 dpi and died at 7 dpi; hence, we collected and quantified RK5738 cells from tomato stems at 4 and 6 dpi, respectively. RQ5738 exhibited similar bacterial growth to that of RK5050 in tomato stems (Fig. 4a). With soil‐soaking inoculation, no difference was observed in bacterial growth between RQ5738 and RK5050 in tomato stems (Fig. 4b), suggesting that PrhO is not required for the bacterial growth of R. solanacearum in host plants.
Figure 4.
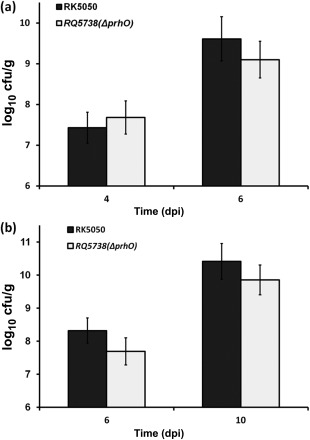
Bacterial growth in planta. Tomato plants were inoculated with OE1‐1 derivatives using petiole inoculation (a) and soil‐soaking inoculation (b), and stem species were removed for quantification by dilution plating. Black column, RK5050; gray column, prhO mutant (RQ5738). cfu, colony‐forming unit; dpi, days post‐inoculation. Each assay was repeated in four independent trials and each trial contained at least 12 plants. Mean values of all results were averaged and are presented with standard deviation (SD) (error bars).
GMI1000 elicits HR in tobacco leaves, and some T3Es are responsible for HR elicitation. We therefore evaluated whether prho deletion could alter the HR elicitation of GMI1000 in tobacco leaves. The generated popA‐lacZYA strain (GF001) exhibited identical HR development to GMI1000 in tobacco leaves (Zhang et al., 2017). Moreover, tobacco (Nicotiana tabacum BY) leaves were infiltrated with GF0094 and GF001, and the development of necrotic lesions was investigated periodically. It was intriguing that GF0094 exhibited almost identical development of necrotic lesions to that of GF001 in tobacco leaves (Fig. 5), indicating that PrhO is not involved in the HR elicitation of GMI1000 in tobacco leaves.
Figure 5.

Hypersensitive response (HR) test. Approximately 50 μL of bacterial suspension at 108 colony‐forming units (cfu)/mL was infiltrated into tobacco leaves with a blunt‐end syringe. (A) GF001 (GMI1000, popA‐lacZYA); (B) GF0094 (GF001, ΔprhO); (C) RQC0095 (GF0094 complemented with prhO); (D) distilled water. The development of necrotic lesions was observed periodically and photographs were taken. Each experiment was repeated four times and each treatment contained four plants. The results presented are from a representative experiment, and similar results were obtained in all experiments.
PrhO positively regulates T3SS expression through HrpG and PrhG to HrpB
The expression of T3SS and T3Es is directly controlled by HrpB. We thus deleted prhO from RK5046 (OE1‐1, hrpB‐lacZYA) to evaluate its impact on hrpB expression. Our results showed that hrpB expression was significantly reduced in RQ5940 (hrpB‐lacZYA, ΔprhO) in hrp‐inducing medium, and complementary PrhO completely restored hrpB expression to that of RK5046 (Fig. 6a), suggesting that the regulation of PrhO on T3SS is mediated through HrpB.
Figure 6.
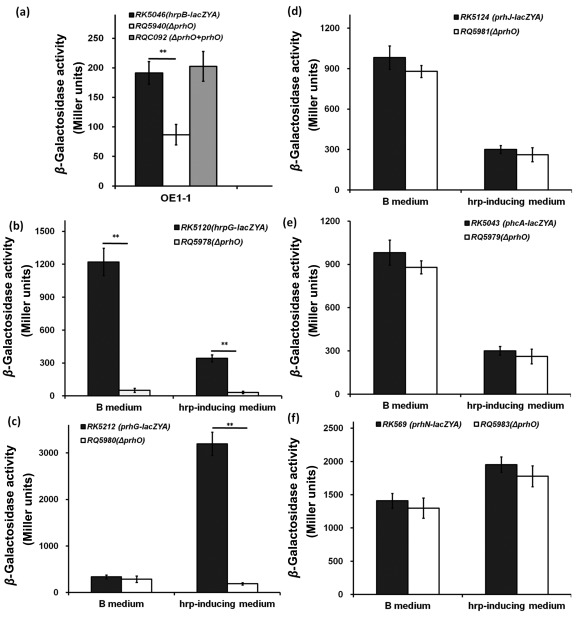
Expression of certain known regulators. (a) hrpB; (b) hrpG; (c) prhG; (d) prhJ; (e) phcA; (f) prhN. Black column, reporter strains RK5046 (OE1‐1, hrpB‐lacZYA), RK5120 (OE1‐1, hrpG‐lacZYA), RK5212 (OE1‐1, prhG‐lacZYA), RK5124 (OE1‐1, prhJ‐lacZYA), RK5043 (OE1‐1, phcA‐lacZYA) and RK5619 (OE1‐1, prhN‐lacZYA); white column, prhO mutants RQ5940 (RK5046, ΔprhO), RQ5978 (RK5120, ΔprhO), RQ5980 (RK5212, ΔprhO), RQ5971 (RK5124, ΔprhO), RQ5979 (RK5043, ΔprhO) and RQ5983 (RK5619, ΔprhO); gray column, complemented strain RQC092 (RQ5940 + prhO). Cells were grown in rich medium or hrp‐inducing medium to an optical density at 600 nm (OD600) of approximately 0.1, and subjected to β‐galactosidase assay. The mean values of all results were averaged and are presented in Miller units with standard deviation (SD) (error bars). Significance level: **P < 0.01 (t‐test).
The expression of hrpB is positively regulated by both HrpG and PrhG in a parallel manner. Furthermore, we deleted prhO from RK5120 (OE1‐1, hrpG‐lacZYA) and RK5212 (OE1‐1, prhG‐lacZYA) to evaluate its impact on the expression of hrpG and prhG. hrpG expression was almost abolished in RQ5978 (RK5120, ΔprhO) in both rich medium (51 versus 1222 Miller units of RK5120) and hrp‐inducing medium (32 versus 343 Miller units of RK5120) (P < 0.01) (Fig. 6b). Similarly, the expression of prhG was also significantly reduced in RQ5980 (RK5212, ΔprhO), but only in hrp‐inducing medium (190 versus 3197 Miller units of RK5212) (P < 0.01) (Fig. 6c).
The expression of hrpG is positively regulated by the PrhA–PrhI/R–PrhJ signalling cascade, but negatively regulated by PhcA‐mediated PrhI/R. However, the expression of prhG is positively regulated by PhcA and PrhN, but independent of the PrhA cascade. We thus deleted prhO from RK5043 (OE1‐1, phcA‐lacZYA), RK5124 (OE1‐1, prhJ‐lacZYA) and RK5619 (OE1‐1, prhN‐lacZYA) to evaluate its impact on the expression of phcA, prhJ and PrhN. However, deletion of prhO did not alter the expression levels of prhJ, phcA or prhN in either rich or hrp‐inducing medium (Fig. 6d–f). Taken together, these data indicate that PrhO positively regulates T3SS expression through HrpG and PrhG to HrpB, but not through the PrhA–PrhI/R–PrhJ signalling cascade, PhcA or PrhN.
PrhO is not required for biofilm formation and swimming motility in R. solanacearum
EPS production, biofilm formation and swimming motility are also important for the virulence of R. solanacearum in host plants and are globally controlled by PhcA (a representative LTTR in R. solanacearum). The prhO mutants remained mucoid on agar plates, indicating that EPS production was not abolished with prhO deletion. In polystyrene microtitre plates, RQ5738 exhibited biofilm formation similar to that of the wild‐type in rich medium (Fig. 7a). On semi‐solid motility agar plates, RQ5738 produced swimming halos similar to those of the wild‐type strain. All of these results suggest that PrhO is not required for EPS production, biofilm formation or swimming motility in R. solanacearum.
Figure 7.
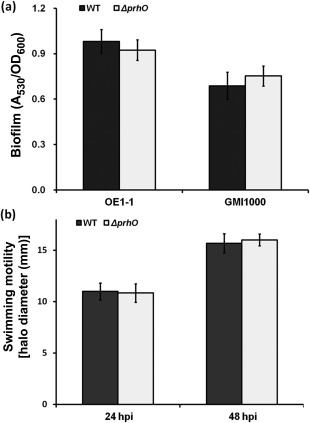
Effect of PrhO on biofilm formation and swimming motility. (a) Biofilm formation. Left, OE1‐1 derivatives; right, GMI1000 derivatives. Twenty microlitres of bacterial suspension at an optical density at 600 nm (OD600) of 0.1 were inoculated into 180 μL of fresh rich medium and kept at 28 °C for 24 h without shaking. After staining with 0.1% crystal violet, biofilm formation was quantified with the absorbance at 530 nm (A 530) and normalized with OD600. (b) Swimming motility. Three microlitres of bacterial suspension at OD600 = 1.0 were dropped onto 0.3% agar plates and kept at 28 °C for 48 h. Swimming motility was quantified as the halo diameter in millimetres. hpi, hours post‐inoculation. Mean values of all results were averaged and are presented with standard deviation (SD) (error bars).
PrhO is expressed in a cell density‐dependent manner and negatively represses it expression, but there is no feedback regulation from PhcA, PrhN or HrpB
PhcA is a well‐characterized LTTR in R. solanacearum which is quorum‐sensing dependent. We constructed a prhO‐lacZYA reporter strain (RQ5985) to evaluate its expression profile at different cell densities. The expression of prhO increased constantly with cell density in both rich medium (Fig. 8a) and hrp‐inducing medium (data not shown), indicating that PrhO is expressed in a cell density‐dependent manner. As PhcA globally regulates the expression of several virulence‐related genes, we thus generated a pchA deletion mutant (RQ5987) from RQ5985 (OE1‐1, prhO‐lacZYA) to evaluate its impact on prhO expression. RQ5987 and RQ5985 exhibited similar expression levels of prhO in both rich and hrp‐inducing medium (Fig. 8b). Moreover, prhO expression was not changed with prhN deletion in both rich and hrp‐inducing medium (Fig. 8b), suggesting that prhO expression is independent of PhcA and PrhN.
Figure 8.
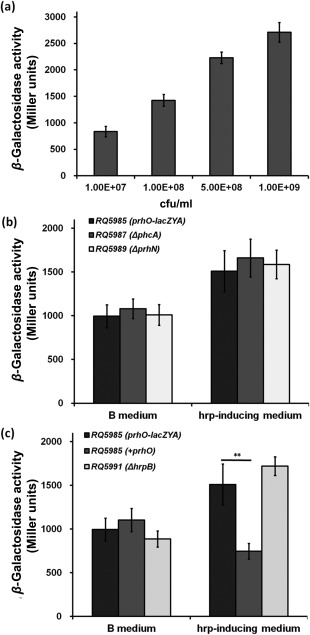
prhO expression at different cell densities (a), in phcA and prhN mutants (b) and in the prhO complemented strain or hrpB mutant (c) in rich medium (left) and hrp‐inducing medium (right). Mean values of all results were averaged and are presented with standard deviation (SD) (error bars). **P < 0.01 (t‐test).
Many LTTRs are auto‐regulated, and we thus introduced the complementary prhO into RQ5985 (OE1‐1, prhO‐lacZYA) to evaluate its auto‐regulation. Interestingly, the complementary prhO significantly decreased prhO expression, but only in hrp‐inducing medium (Fig. 8c), suggesting that prhO expression is negatively regulated by itself under hrp‐inducing conditions. Plener et al. (2010) reported that HrpB could regulate its expression through certain unknown mediators. We thus generated a hrpB deletion mutant (RQ5991) from RQ5985 (OE1‐1, prhO‐lacZYA) to evaluate whether PrhO is involved in the feedback regulation of HrpB. However, Q5991 and RQ5985 exhibited similar expression levels of prhO in both rich and hrp‐inducing medium (Fig. 8c), suggesting that prhO expression is negatively regulated by itself, but there is no feedback regulation from PhcA, PrhN or HrpB.
Discussion
LTTRs are abundant regulators in diverse bacteria, archaea and algae, which globally control a great variety of cellular processes, including pathogenicity (Maddocks and Oyston, 2008; Reen et al., 2013; Schell, 1993). With genome searching, 63 transcriptional regulators were predicted to be LTTRs in OE1‐1, but only PhcA has been functionally well characterized in R. solanacearum (Brumbley et al., 1993; Genin et al., 2005; Yoshimochi et al., 2009a). In the present study, we provide genetic evidence to demonstrate that PrhO, a putative LTTR, is a novel virulence regulator, which positively regulates T3SS expression and contributes to the virulence of R. solanacearum in host plants.
T3SS is essential for pathogenicity in many pathogenic bacteria of humans, animals and plants (Galan and Wolf‐Watz, 2006; Hueck, 1998). To date, numerous LTTRs have been identified to control the T3SS in diverse pathogenic bacteria, such as BexR, MexT and MvfR in Pseudomonas aeruginosa (Deziel et al., 2005; Jin et al., 2011; Keith et al., 2009; Reen et al., 2013), QseA and QseD in enterohaemorrhagic E. coli K12 (Habdas et al., 2010; Kendall et al., 2010), GamR in Xanthomonas oryzae pv. oryzae (Rashid et al., 2016) and PhcA in R. solanacearum (Huang et al., 1998; Yoshimochi et al., 2009a). The LTTR PrhO positively controls the T3SS in R. solanacearum, which is consistent with the impact of LTTRs on T3SS regulation. Deletion of prhO significantly reduced the T3SS expression of R. solanacearum both in vitro and in planta, but not the bacterial growth in host plants. However, the deletion of prhO did not alter EPS production, biofilm formation and swimming mobility, which are also important for the pathogenicity of R. solanacearum. Moreover, the extensive proliferation of R. solanacearum in the xylem vessels of host plants was not altered in prhO mutants, which is one of the most important pathogenicity determinants in R. solanacearum. All of these data suggest that significantly reduced T3SS expression in planta is the probable cause of the impaired virulence of prhO mutants in host plants, not the defective growth in host plants. RNA sequencing (RNA‐seq) is planned to further ascertain whether novel pathways are involved in the regulation of PrhO on the pathogenicity of R. solanacearum in host plants.
The expression levels of some T3Es were also significantly reduced in prhO mutants and some were responsible for the HR elicitation of R. solanacearum in resistant plants (Peeters et al., 2013). It is intriguing that prhO mutants caused almost identical HR elicitation as GMI1000 in tobacco leaves. It should be noted that the expression of popA and some T3Es was significantly reduced, but not completely eliminated, in prhO mutants. This might be the result of a certain amount of residual hrpB expression in prhO mutants, and these T3Es could continue to elicit HR in resistant plants; this might be the reason why significantly impaired T3E expression did not delay HR elicitation of GMI100 in tobacco leaves.
The expression of the T3SS and entire T3Es in R. solanacearum is directly controlled by the key regulator HrpB which binds directly to the plant‐inducible promoter (PIP) motif present in the promoters of the target genes (Cunnac et al., 2004; Mukaihara et al., 2010). PrhO positively regulates T3SS expression via HrpB, which is consistent with the key regulatory role of HrpB on T3SS. Expression of hrpB is activated by two close paralogues, HrpG and PrhG, in a parallel manner. Our results showed that the expression of hrpG and prhG was almost eliminated with prhO deletion, suggesting that PrhO positively regulates the expression of both hrpG and prhG, and consequently regulates the expression of hrpB. However, it remains to be further ascertained whether PrhO regulates directly the expression of both hrpG and prhG. Further experiments are planned to identify the direct regulating targets of PrhO and to reveal its linear regulation on T3SS. Moreover, it is intriguing how PrhO simultaneously regulates the expression of hrpG and prhG, as they exhibit distinctly different expression properties. The most remarkable is that the expression level of hrpG is much higher in rich medium than in hrp‐inducing medium or when co‐cultivated with Arabidopsis thaliana seedlings (Yoshimochi et al., 2009b), whereas the expression level of prhG is much lower in rich medium than in hrp‐inducing medium (Zhang et al., 2013). Moreover, the global regulator, PhcA, negatively regulates hrpG expression, but positively regulates prhG expression (Yoshimochi et al., 2009b; Zhang et al., 2013). prhO deletion did not alter the expression of prhJ, phcA and prhN, which are known to be important for the expression of hrpG and prhG (Hikichi et al., 2007; Zhang et al., 2015), indicating that the regulation of PrhO on the expression of hrpG and prhG is independent of these known regulating pathways. Recently, we have identified a plant‐derived compound, umbelliferone, which impairs T3SS expression via HrpG and PrhG, but independent of these known regulators (Yang et al., 2017), supporting our speculation that certain novel pathways should be integrated for the expression of hrpG and prhG in R. solanacearum. It should be noted that HrpB is not expressed until contact with hrp‐inducing medium or with host plants, whereas HrpG is best expressed in rich medium. As members of the OmpR/PhoB family of two‐component response regulators, HrpG and PrhG respond to host signals by phosphorylation at certain residues and sequentially activate hrpB expression (Yoshimochi et al., 2009b). It remains to be further ascertained whether PrhO is involved in the host signal response with HrpG and PrhG.
The LTTR PhcA globally controls many virulence‐related factors other than the T3SS, including EPS synthesis, biofilm formation and cell motility, which are important for the pathogenicity of R. solanacearum in host plants (Brumbley et al., 1993; Genin et al., 2005). However, prhO deletion did not alter these phenomena, except the T3SS, reflecting the complexity of LTTRs in diverse cellular processes. LTTR PhcA expression is quorum‐sensing dependent, and its activity is triggered at high cell density by a unique autoinducer, 3‐hydroxy palmitic acid methyl ester (3‐OH PAME) or methyl 3‐hydroxymyristate (3‐OH MAME) (Flavier et al., 1997; Mori et al., 2017). prhO expression increases constantly with increasing cell density, suggesting that PrhO expression is also quorum‐sensing dependent. It remains to be further elucidated how PrhO responds to cell density. Most LTTRs have been reported to be negatively auto‐regulated and to repress their own expression, frequently with divergent promoters mediating their regulating targets (Maddocks and Oyston, 2008; Schell, 1993). prhO expression was significantly decreased by itself, but unchanged with the deletion of hrpB and phcA, indicating that PrhO was negatively regulated by itself, but the regulation pathway remains to be further elucidated.
The R. solanacearum species complex is extremely heterogeneous (Genin and Denny, 2012), but PrhO is highly conserved (more than 90% amino acid identity) in all R. solanacearum species. Indeed, PrhO of OE1‐1 could functionally substitute for that of GMI1000 in T3SS expression and virulence in different host plants, indicating that the function of PrhO in T3SS expression and virulence is conserved amongst R. solanacearum species. With National Center for Biotechnology Information (NCBI) blast, PrhO orthologues were also found in certain bacteria, such as Pandoraea species (about 70% similarity), Bordetella holmesii (about 60%) and Pseudomonas fuscovaginae (about 50%), whereas no orthologues (more than 30% similarity) were found in Pseudomonas syringae, Xanthomonas campestris or Xanthomonas oryzae species, which are known to be much more closely related to phytopathogenic R. solanacearum.
In summary, our genetic results demonstrate that PrhO, a putative LTTR, is a novel virulence regulator of R. solanacearum. PrhO positively regulates the expression of T3SS through HrpG and PrhG to HrpB, and contributes to virulence. Future investigations will aim to ascertain the biochemical role of LTTR PrhO and its regulation mechanism on the expression of hrpG and PrhG.
Experimental Procedures
Bacterial strains and culture conditions
The R. solanacearum strains used in this study are listed in Table 1. It should be noted that strain OE1‐1 (Kanda et al., 2003) is virulent in both tomato and tobacco plants, whereas strain GMI1000 (Salanoubat et al., 2002) is virulent in tomato plants, but causes HR in tobacco leaves. Ralstonia solanacearum strains were grown at 28 °C in nutrient‐rich medium (B medium) or in hrp‐inducing medium (sucrose medium) (Yoshimochi et al., 2009b). Escherichia coli DH12S and S17‐1 were grown in Luria–Bertani (LB) medium at 37 °C for plasmid construction and conjugational transfer, respectively.
Table 1.
Strains used in this study.
| Strain | Relative characteristics | Reference source |
|---|---|---|
| OE1‐1 | Wild‐type, race 1, biovar 3 | Kanda et al. (2003) |
| RK5050 | OE1‐1 popA‐lacZYA | Yoshimochi et al. (2009b) |
| RQ5738 | OE1‐1 popA‐lacZYA, ΔprhO | This study |
| RQ5739 | OE1‐1, ΔprhO | This study |
| RQC090 | RQ5738, prhO complementation | This study |
| RK5046 | OE1‐1 hrpB‐lacZYA | Yoshimochi et al. (2009b) |
| RQ5940 | OE1‐1 hrpB‐lacZYA, ΔprhO | This study |
| RQC092 | RQ5940, prhO complementation | This study |
| GMI1000 | Wild‐type, race 1, biovar 4 | Salanoubat et al. (20022) |
| GF001 | GMI1000, popA‐lacZYA | Zhang et al. (2017) |
| GF0094 | GMI1000, popA‐lacZYA, ΔprhO | This study |
| RQC0095 | GF0094, prhO complementation | This study |
| RK5120 | OE1‐1 hrpG‐lacZYA | Yoshimochi et al. (2009b) |
| RQ5978 | OE1‐1 hrpG‐lacZYA, ΔprhO | This study |
| RK5212 | OE1‐1 prhG‐lacZYA | Zhang et al. (2013) |
| RQ5980 | OE1‐1 prhG‐lacZYA, ΔprhO | This study |
| RK5043 | OE1‐1 phcA‐lacZYA | Yoshimochi et al. (2009b) |
| RQ5979 | OE1‐1 phcA‐lacZYA, ΔprhO | This study |
| RK5124 | OE1‐1 prhJ‐lacZYA | Yoshimochi et al. (2009b) |
| RQ5981 | OE1‐1 prhJ‐lacZYA, ΔprhO | This study |
| RK5619 | OE1‐1 prhN‐lacZYA | Zhang et al. (2015) |
| RQ5983 | OE1‐1 prhN‐lacZYA, ΔprhO | This study |
| RQ5985 | OE1‐1 prhO‐lacZYA | This study |
| RQ5987 | OE1‐1 prhO‐lacZYA, ΔphcA | This study |
| RQ5989 | OE1‐1 prhO‐lacZYA, ΔprhN | This study |
| RQ5991 | OE1‐1 prhO‐lacZYA, ΔhrpB | This study |
| RQC0096 | RQ5983, prhO complementation | This study |
Construction of prhO in‐frame‐deleted mutants
Mutants with in‐frame deletion of genes were generated via pK18mobsacB‐based homologous recombination, as described previously (Zhang et al., 2015). For plasmid construction, two DNA fragments (about 600 bp) flanking the prhO gene were PCR amplified from OE1‐1 genomic DNA with the primer pairs 1880A1B‐1880B1C and 1880A2C‐1880B2H, respectively. It should be noted that 1880B1C and 1880A2C are fully complemented and these two DNA fragments were mixed for the second round of PCR to generate the DNA fragment (about 1.2 kb) with primers 1880A1B and 1880B2H, in which the open‐reading frame of prhO was absent. The generated DNA fragment was first cloned into pBluescript KS (+) to obtain pKSd1880, and subcloned into pK18mobsacB to obtain pK18d1880. After validating the sequence, pK18d1880 was transferred from E. coli S17‐1 into R. solanacearum strains, and prhO mutants (listed in Table 1) were generated and confirmed by colony PCR with the primer pairs 1880A1B and 1880B2H. The primers used in this study are listed in Table S1 (see Supporting Information).
Complementation analyses
Complementation analyses were performed with the pUC18‐mini‐Tn7T‐Gm‐based site‐specific chromosome integration system (Choi et al., 2005) with some minor modifications (Zhang et al., 2015). The prhO gene containing a 500‐bp upstream region, empirically harbouring its native promoter, was PCR amplified from OE1‐1 genomic DNA with the primer pairs 1880A1B and 1880B3H. The DNA fragment was first cloned into pBluescript KS (+) to obtain pKS1880C, and subcloned into pUC18‐mini‐Tn7T‐Gm to obtain pUCprhO. After validating the sequence, complementary prhO was specifically integrated into the R. solanacearum chromosome at a single attTn7 site (25 bp downstream of the glmS gene) and confirmed by colony PCR with the primer pairs glmsdown and Tn7R (Zhang et al., 2011).
β‐Galactosidase assay
The expression levels of genes fused with promoterless lacZYA were evaluated by the β‐galactosidase assay (both in vitro and in planta), as described previously (Zhang et al., 2013). Enzyme activity in vitro was expressed in Miller units (Miller, 1992), and in planta was normalized by the luminescence divided by the cell number. Each assay was independently repeated in at least four experiments, and each treatment included four replications. The mean values of all experiments were averaged with the standard deviation (SD) and the statistical significance was assessed using a post‐hoc Dunnett test following analysis of variance (ANOVA).
qRT‐PCR analysis
The expression level of genes without lacZYA fusion was quantified by qRT‐PCR analysis, as described previously (Zhang et al., 2017). Representative T3E genes, ripAA (avrA), ripB, ripD, ripE, ripO, ripPI (popPI), RipR, ripTAL, ripW and popA (ripX), were selected for qRT‐PCR analysis in this study. It should be noted that RipAA and RipPI have been well characterized to be responsible for HR development of GMI1000 in tobacco plants (Lavie et al., 2002; Poueymiro et al., 2009). serC was selected as reference for normalization and ripX was selected as positive control (Monteiro et al., 2012). Total RNA was isolated from GF001 and prhO mutant (GF0094) and subjected to qRT‐PCR analysis. Each assay was repeated independently from RNA isolation at least three times, and each trial included four replications. The mean values of all experiments were averaged with SD and the statistical significance was assessed using a post‐hoc Dunnett test following ANOVA.
Virulence assay and HR test
Virulence assay and HR test were performed as described previously (Yao and Allen, 2007). Wilt‐susceptible tomato plants (Solanum lycopersicum cv. Moneymaker) and tobacco plants (Nicotiana tabacum cv. Bright Yellow) were subjected to virulence assay with soil soaking, which mimics the natural invasion through the roots, and petiole inoculation, by which bacteria can invade directly the xylem vessels. Each assay was repeated independently four times with 12 plants per trial. Wilt symptoms of plants were inspected as the disease index (scale of 1–4) and the mean values of all experiments were averaged with SD. The statistical significance was assessed using a post‐hoc Dunnett test following ANOVA.
The HR test was carried out in leaves of N. tabacum with leaf infiltration, and the symptom development of HR was recorded periodically. Each test was repeated independently four times with four plants per treatment. A representative result was presented.
Bacterial growth in planta
Bacterial growth in planta was assessed as described previously (Zhang et al., 2013). Bacterial cells were collected daily from tomato stems and quantified by dilution plating. Each assay was repeated independently four times with 12 plants per treatment. The mean values of all experiments were averaged with SD and the statistical significance was assessed using a post‐hoc Dunnett test following ANOVA.
Biofilm and swimming motility assay
Biofilm formation was performed in 96‐well polystyrene microtitre plates as reported by Yao and Allen (2007) with some minor modifications (Mori et al., 2016). Briefly, 20 μL of bacterial suspension at an optical density at 600 nm (OD600) of 1.0 was inoculated into 180 μL of fresh B medium and kept at 28 °C for 24 h without shaking. After staining with crystal violet, biofilm formation was quantified by measuring the absorbance at 530 nm (A 530), and normalized by the cell number (OD600). The swimming motility assay was carried out on semi‐solid medium (0.3% agar plates), as reported previously (Kelman and Hruschka, 1973). The diameters of the swimming halos on semi‐solid medium (28 °C for 48 h) were measured. Each assay was repeated independently at least four times and each trial included three replications. The mean values of all experiments were averaged with SD and the statistical significance was assessed using a post‐hoc Dunnett test following ANOVA.
Construction of prhO‐lacZYA reporter strain and auto‐regulation analyses
A promoterless lacZYA fragment was inserted 22 bp downstream of the start codon of prhO to construct the prhO‐lacZYA fusion, in which six nucleotides (GCCTTT) were replaced by GGTACC (KpnI) by PCR for lacZYA insertion. Two DNA fragments flanking the insertion site were amplified from OE1‐1 genomic DNA with the primer pairs 1880A1B and 1880B1K, and 1880A2K and 1880B3H, respectively. It should be noted that the primers 1880b1K and 1880A2K were fully complemented, and these two DNA fragments were mixed for the second round of PCR with the primer pairs 1880A1B and 1880B3H to generate the DNA fragment which contained the KpnI site for lacZYA insertion. The generated DNA fragment was finally subcloned into pk18mobSacB to obtain pK18prhO. After validating the sequence, the KpnI‐digested promoterless lacZYA from pUClacZYA was inserted into pK18prhO to obtain pK18prhO‐lacZYA. It should be noted that lacZYA maintains the same transcription direction as prhO and that they share the same promoter. After validating the sequence, pK18prhO‐lacZYA was transferred from E. coli S17‐1 into R. solanacearum, and the reporter strain RQ5985 (OE1‐1 prhO‐lacZYA) was generated and confirmed by colony PCR with the primer pairs 1880A1B and lacZR1. Based on RQ5985, mutants with deletion of phcA, prhN or hrpB were generated to evaluate their impact on prhO expression. Complementary PrhO was introduced into RQ5985 for the auto‐regulation analyses.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Primers used in this study.
Acknowledgements
We express our thanks for funding support from the National Natural Science Foundation of China (31670082) and the Fundamental Research Foundation for the Central Universities (SWU113016 and XDJK2016B41) to Y.Z.
Contributor Information
Yong Zhang, Email: bioyongzhang@swu.edu.cn.
Kouhei Ohnishi, Email: kouheio@kochi-u.ac.jp.
References
- Angot, A. , Peeters, N. , Lechner, E. , Vailleau, F. , Baud, C. , Gentzbittel, L. , Sartorel, E. , Genschik, P. , Boucher, C. and Genin, S. (2006) Ralstonia solanacearum requires F‐box‐like domain‐containing type III effectors to promote disease on several host plants. Proc. Natl. Acad. Sci. USA, 103, 14 620–14 625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlat, M. , Gough, C.L. , Zischek, C. , Barberis, P.A. , Trigalet, A. and Boucher, C. (1992) Transcriptional organization and expression of the large hrp gene cluster of Pseudomonas solanacearum . Mol. Plant–Microbe Interact. 5, 187–193. [DOI] [PubMed] [Google Scholar]
- Bhatt, G. and Denny, T.P. (2004) Ralstonia solanacearum iron scavenging by the siderophore staphyloferrin B is controlled by PhcA, the global virulence regulator. J. Bacteriol. 23, 7896–7904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher, C.A. , Van Gijsegem, F. , Barberis, P.A. , Arlat, M. and Zischek, C. (1987) Pseudomonas solanacearum genes controlling both pathogenicity on tomato and hypersensitivity on tobacco are clustered. J Bacteriol. 169, 5626–5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbley, S.M. , Carney, B.F. and Denny, T.P. (1993) Phenotype conversion in Pseudomonas solanacearum due to spontaneous inactivation of PhcA, a putative LysR transcriptional activator. J. Bacteriol. 175, 5477–5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, K.H. , Gaynor, J.B. , White, K.G. , Lopez, C. , Bosio, C.M. , Karkhoff‐ Chweizer, R.R. and Schweizer, H.P. (2005) A Tn7‐based broad range bacterial cloning and expression system. Nat. Methods, 2, 443–448. [DOI] [PubMed] [Google Scholar]
- Clough, S.J. , Flavier, A.B. , Schell, M.A. and Denny, T.P. (1997) Differential expression of virulence genes and motility in Ralstonia (Pseudomonas) solanacearum during exponential growth. Appl. Environ. Microbiol. 63, 844–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll, N.S. and Valls, M. (2013) Current knowledge on the Ralstonia solanacearum type III secretion system. Microb. Biotechnol. 6, 614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunnac, S. , Occhialini, A. , Barberis, P. , Boucher, C. and Genin, S. (2004) Inventory and functional analysis of the large Hrp regulon in Ralstonia solanacearum: identification of novel effector proteins translocated to plant host cells through the type III secretion system. Mol. Microbiol. 53, 115–128. [DOI] [PubMed] [Google Scholar]
- Denny, T.P. (1995) Involvement of bacterial polysaccharides in plant pathogenesis. Annu. Rev. Phytopathol. 33, 173–197. [DOI] [PubMed] [Google Scholar]
- Deziel, E. , Gopalan, S. , Tampakaki, A.P. , Lepine, F. , Padfield, K.E. , Saucier, M. , Xiao, G. and Rahme, L.G. (2005) The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing‐regulated genes are modulated without affecting lasRI, rhlRI or the production of N‐acyl‐L‐homoserine lactones. Mol. Microbiol. 55, 998–1014. [DOI] [PubMed] [Google Scholar]
- Flavier, A.B. , Clough, S.J. , Schell, M.A. and Denny, T.P. (1997) Identification of 3‐hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia solanacearum . Mol. Microbiol. 26, 251–259. [DOI] [PubMed] [Google Scholar]
- Fujiwara, S. , Kawazoe, T. , Ohnishi, K. , Kitagawa, T. , Popa, C. , Valls, M. , Genin, S. , Nakamura, K. , Kuramitsu, Y. , Tanaka, N. and Tabuchi, M. (2016) RipAY, a plant pathogen effector protein, exhibits robust γ‐glutamyl cyclotransferase activity when stimulated by eukaryotic thioredoxins. J. Biol. Chem. 291, 6813–6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán, J.E. and Wolf‐Watz, H. (2006) Protein delivery into eukaryotic cells by type III secretion machines. Nature, 444, 567–573. [DOI] [PubMed] [Google Scholar]
- Genin, S. (2010) Molecular traits controlling host range and adaptation to plants in Ralstonia solanacearum . New. Phytol. 187, 920–928. [DOI] [PubMed] [Google Scholar]
- Genin, S. and Denny, T.P. (2012) Pathogenomics of the Ralstonia solanacearum species complex. Annu. Rev. Phytopathol. 50, 67–89. [DOI] [PubMed] [Google Scholar]
- Genin, S. , Brito, B. , Denny, T.P. and Boucher, C. (2005) Control of the Ralstonia solanacearum type III secretion system (Hrp) genes by the global virulence regulator PhcA. FEBS Lett. 579, 2077–2081. [DOI] [PubMed] [Google Scholar]
- Habdas, B.J. , Smart, J. , Kaper, J.B. and Sperandio, V. (2010) The LysR‐type transcriptional regulator QseD alters type three secretion in enterohemorrhagic Escherichia coli and motility in K‐12 Escherichia coli . J. Bacteriol. 192, 3699–3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández‐Lucas, I. , Gallego‐Hernandez, A.L. , Encarnacion, S. , Fernandez‐Mora, M. , Martinez‐Batallar, A.G. , Salgado, H. , Oropeza, R. and Calva, E. (2008) The LysR‐type transcriptional regulator LeuO controls expression of several genes in Salmonella enterica serovar typhi . J. Bacteriol. 190, 1658–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heroven, A.K. , Nagel, G. , Tran, H.J. , Parr, S. and Dersch, P. (2004) RovA is autoregulated and antagonizes H‐NS‐mediated silencing of invasion and rovA expression in Yersinia pseudotuberculosis . Mol. Microbiol. 53, 871–888. [DOI] [PubMed] [Google Scholar]
- Hikichi, Y. , Yoshimochi, T. , Tsujimoto, S. , Shinohara, R. , Nakaho, K. , Kanda, A. , Kiba, A. and Ohnishi, K. (2007) Global regulation of pathogenicity mechanism of Ralstonia solanacearum . Plant Biotechnol. 24, 149–154. [Google Scholar]
- Huang, J. , Carney, B.F. , Denny, T.P. , Weissinger, A.K. and Schell, M.A. (1995) A complex network regulates expression of eps and other virulence genes of Pseudomonas solanacearum . J. Bacteriol. 177, 1259–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J. , Yindeeyoungyeon, W. , Garg, R.P. , Denny, T.P. and Schell, M.A. (1998) Joint transcriptional control of xpsR, the unusual signal integrator of the Ralstonia solanacearum virulence gene regulatory network, by a response regulator and a LysR‐type transcriptional activator. J. Bacteriol. 180, 2736–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueck, C.J. (1998) Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62, 379–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Y. , Yang, H. , Qiao, M. and Jin, S. (2011) MexT regulates the type III secretion system through MexS and PtrC in Pseudomonas aeruginosa . J. Bacteriol. 193, 399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda, A. , Ohnishi, S. , Tomiyama, H. , Hasegawa, H. , Yasukohchi, M. , Kiba, A. , Ohnishi, K. , Okuno, T. and Hikichi, Y. (2003) Type III secretion machinery‐deficient mutants of Ralstonia solanacearum lose their ability to colonize resulting in loss of pathogenicity. J. Gen. Plant Pathol. 69, 250–257. [Google Scholar]
- Keith, H.T. , Vallet‐Gely, I. and Simon, L.D. (2009) Epigenetic control of virulence gene expression in Pseudomonas aeruginosa by a LysR‐type transcription regulator. PLoS Genet. 5, e1000779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelman, A. and Hruschka, J. (1973) The role of motility and aerotaxis in the selective increase of avirulent bacteria in still broth cultures of Pseudomonas solanacearum . Microbiology, 76, 177–188. doi: 10.1099/00221287-76-1-177. [DOI] [PubMed] [Google Scholar]
- Kendall, M.M. , Rasko, D.A. and Sperandio, V. (2010) The LysR‐type regulator QseA regulates both characterized and putative virulence genes in enterohemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 76, 1306–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavie, M. , Shillington, E. , Eguiluz, C. , Grimsley, N. and Boucher, C. (2002) PopP1, a new member of the YopJ/AvrRxv family of type III effector proteins, acts as a host‐specificity factor and modulates aggressiveness of Ralstonia solanacearum . Mol. Plant–Microbe Interact. 15, 1058–1068. [DOI] [PubMed] [Google Scholar]
- Lindgren, P.B. (1997) The role of hrp genes during plant–bacterial interactions. Annu. Rev. Phytopathol. 35, 129–152. [DOI] [PubMed] [Google Scholar]
- Maddocks, S.E. and Oyston, P.C. (2008) Structure and function of the LysR‐type transcriptional regulator (LTTR) family proteins. Microbiology, 154, 3609–3623. [DOI] [PubMed] [Google Scholar]
- Marenda, M. , Brito, B. , Callard, D. , Genin, S. , Barberis, P. , Boucher, C. and Arlat, M. (1998) PrhA controls a novel regulatory pathway required for the specific induction of Ralstonia solanacearum hrp genes in the presence of plant cells. Mol. Microbiol. 27, 437–453. [DOI] [PubMed] [Google Scholar]
- Miller, J.H. (1992) The lac system In: A Short Course in Bacterial Genetics. A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria (Miller J.H., ed), pp. 43–80. Plainview, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Monteiro, F. , Genin, S. , Van Dijk, I. and Valls, M. (2012) A luminescent reporter evidences active expression of Ralstonia solanacearum type III secretion system genes throughout plant infection. Microbiology, 158, 2107–2116. [DOI] [PubMed] [Google Scholar]
- Mori, Y. , Inoue, K. , Ikeda, K. , Nakayashiki, H. , Higashimoto, C. , Ohnishi, K. , Kiba, A. and Hikichi, Y. (2016) The vascular plant‐pathogenic bacterium Ralstonia solanacearum produces biofilms required for its virulence on the surfaces of tomato cells adjacent to intercellular spaces. Mol. Plant Pathol. 17, 890–902. doi: 10.1111/mpp.12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, Y. , Ishikawa, S. , Ohnishi, H. , Shimatani, M. , Morikawa, Y. , Hayashi, K. , Ohnishi, K. , Kiba, A. , Kai, K. and Hikichi, Y. (2017) Involvement of ralfuranones in the quorum sensing signalling pathway and virulence of Ralstonia solanacearum strain OE1‐1. Mol. Plant Pathol. 19, 454–463. doi: 10.1111/mpp.12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukaihara, T. , Tamura, N. , Murata, Y. and Iwabuchi, M. (2004) Genetic screening of Hrp type III‐related pathogenicity genes controlled by the HrpB transcriptional activator in Ralstonia solanacearum . Mol. Microbiol. 54, 863–875. [DOI] [PubMed] [Google Scholar]
- Mukaihara, T. , Tamura, N. and Iwabuchi, M. (2010) Genome‐wide identification of a large repertoire of Ralstonia solanacearum type III effector proteins by a new functional screen. Mol. Plant–Microbe Interact. 23, 251–262. [DOI] [PubMed] [Google Scholar]
- Peeters, N. , Carrère, S. , Anisimova, M. , Plener, L. , Cazalé, A.‐C. and Genin, S. (2013) Repertoire, unified nomenclature and evolution of the Type III effector gene set in the Ralstonia solanacearum species complex. BMC Genomics, 14, 859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plener, L. , Manfredi, P. , Valls, M. and Genin, S. (2010) PrhG, a transcriptional regulator responding to growth conditions, is involved in the control of the type III secretion system regulon in Ralstonia solanacearum . J. Bacteriol. 192, 1011–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popa, C. , Li, L. , Gil, S. , Tatjer, L. , Hashii, K. , Tabuchi, M. , Coll, N.S. , Ariño, J. and Valls, M. (2016) The effector AWR5 from the plant pathogen Ralstonia solanacearum is an inhibitor of the TOR signalling pathway. Sci. Rep. 6, 27 058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poueymiro, M. , Cunnac, S. , Barberis, P. , Deslandes, L. , Peeters, N. , Cazale‐Noel, A.‐C. , Boucher, C. and Genin, S. (2009) Two type III secretion system effectors from Ralstonia solanacearum GMI1000 determine host‐range specificity on tobacco. Mol. Plant–Microbe Interact. 14, 538–550. doi: 10.1094/MPMI-22-5-0538. [DOI] [PubMed] [Google Scholar]
- Rashid, M.M. , Ikawa, Y. and Tsuge, S. (2016) GamR, the LysR‐type galactose metabolism regulator, regulates hrp gene expression via transcriptional activation of two key hrp regulators, HrpG and HrpX, in Xanthomonas oryzae pv. oryzae . Appl. Environ. Microbiol. 82, 3947–3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reen, F.J. , Barret, M. , Fargier, E. , O'Muinneacháin, M. and O'Gara, F. (2013) Molecular evolution of LysR‐type transcriptional regulation in Pseudomonas aeruginosa . Mol. Phylogenet. Evol. 66, 1041–1049. [DOI] [PubMed] [Google Scholar]
- Roberts, D.P. , Denny, T.P. and Schell, M.A. (1988) Cloning of the egl gene of Pseudomonas solanacearum and analysis of its role in phytopathogenicity. J. Bacteriol. 170, 1445–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanoubat, M. , Genin, S. , Artiguenave, F. , Gouzy, J. , Mangenot, S. , Arlat, M. , Billault, A. , Brottier, P. , Camus, J.C. , Cattolico, L. , Chandler, M. , Choisne, N. , Claudel‐Renard, C. , Cunnac, S. , Demange, N. , Gaspin, C. , Lavie, M. , Moisan, A. , Robert, C. , Saurin, W. , Schiex, T. , Siguier, P. , Thébault, P. , Whalen, M. , Wincker, P. , Levy, M. , Weissenbach, J. and Boucher, C.A. (2002) Genome sequence of the plant pathogen Ralstonia solanacearum . Nature, 415, 497–502. [DOI] [PubMed] [Google Scholar]
- Schell, M.A. (1993) Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47, 597–626. [DOI] [PubMed] [Google Scholar]
- Sperandio, V. , Li, C.C. and Kaper, J.B. (2002) Quorum‐sensing Escherichia coli regulator A: a regulator of the LysR family involved in the regulation of the locus of enterocyte effacement pathogenicity island in enterohemorrhagic E. coli . Infect. Immun. 70, 3085–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasset, C. , Bernoux, M. , Jauneau, A. , Pouzet, C. , Brière, C. , Kieffer‐Jacquinod, S. , Rivas, S. , Marco, Y. , Deslandes, L. and Dangl, J.L. (2010) Autoacetylation of the Ralstonia solanacearum effector PopP2 targets a lysine residue essential for RRS1‐R‐mediated immunity in Arabidopsis . PLoS Pathog. 6, e1001202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valls, M. , Genin, S. and Boucher, C. (2006) Integrated regulation of the type III secretion system and other virulence determinants in Ralstonia solanacearum . PLoS Pathog. 2, e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasse, J. , Frey, P. and Trigalet, A. (1995) Microscopic studies of intercellular infection and protoxylem invasion of tomato roots by Pseudomonas solanacearum . Mol. Plant–Microbe Interact. 8, 241–251. [Google Scholar]
- Yang, L. , Li, S. , Qin, X. , Jiang, G. , Chen, J. , Li, B. , Yao, X. , Liang, P. , Zhang, Y. and Ding, W. (2017) Exposure to umbelliferone reduces Ralstonia solanacearum biofilm formation, transcription of type III secretion system regulators and effectors and virulence on tobacco. Front. Microbiol. 8, 1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, J. and Allen, C. (2007) The plant pathogen Ralstonia solanacearum needs aerotaxis for normal biofilm formation and interactions with its tomato host. J. Bacteriol. 189, 6415–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimochi, T. , Hikichi, Y. , Kiba, A. and Ohnishi, K. (2009a) The global virulence regulator PhcA negatively controls the Ralstonia solanacearum hrp regulatory cascade by repressing expression of the PrhIR signaling proteins. J. Bacteriol. 191, 3424–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimochi, T. , Zhang, Y. , Kiba, A. , Hikichi, Y. and Ohnishi, K. (2009b) Expression of hrpG and activation of response regulator HrpG are controlled by distinct signal cascades in Ralstonia solanacearum . J. Gen. Plant. Pathol. 75, 196–204. [Google Scholar]
- Zhang, Y. , Kiba, A. , Hikichi, Y. and Ohnishi, K. (2011) prhKLM genes of Ralstonia solanacearum encode novel activators of hrp regulon and are required for pathogenesis in tomato. FEMS Microbiol. Lett. 317, 75–82. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Chen, L. , Yoshimochi, T. , Kiba, A. , Hikichi, Y. and Ohnishi, K. (2013) Functional analysis of Ralstonia solanacearum PrhG regulating the hrp regulon in host plants. Microbiology, 159, 1695–1704. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Luo, F. , Wu, D. , Hikichi, Y. , Kiba, A. , Igarashi, Y. , Ding, W. and Ohnishi, K. (2015) PrhN, a putative marR family transcriptional regulator, is involved in positive regulation of type III secretion system and full virulence of Ralstonia solanacearum . Front. Microbiol. 6, 357. doi: 10.3389/fmicb.2015.00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Li, J. , Zhang, W. , Wang, R. , Qiu, Q. , Luo, F. , Hikichi, Y. , Ohnishi, K. and Ding, W. (2017) Ferulic acid, but not all hydroxycinnamic acids, is a novel T3SS inducer of Ralstonia solanacearum and promotes its infection process in host plants under hydroponic condition. Front. Plant Sci. 8, 1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Primers used in this study.


