Summary
Amongst the various post‐translational modifications (PTMs), SUMOylation is a conserved process of attachment of a small ubiquitin‐related modifier (SUMO) to a protein substrate in eukaryotes. This process regulates many important biological mechanisms, including transcriptional regulation, protein stabilization, cell cycle, DNA repair and pathogenesis. However, the functional role of SUMOylation is not well understood in plant‐pathogenic fungi, including the model fungal pathogen Magnaporthe oryzae. In this study, we elucidated the roles of four SUMOylation‐associated genes that encode one SUMO protein (MoSMT3), two E1 enzymes (MoAOS1 and MoUBA2) and one E2 enzyme (MoUBC9) in fungal development and pathogenicity. Western blot assays showed that SUMO modification was abolished in all deletion mutants. MoAOS1 and MoUBA2 were mainly localized in the nucleus, whereas MoSMT3 and MoUBC9 were localized in both the nucleus and cytoplasm. However, the four SUMOylation‐associated proteins were predominantly localized in the nucleus under oxidative stress conditions. Deletion mutants for each of the four genes were viable, but showed significant defects in mycelial growth, conidiation, septum formation, conidial germination, appressorium formation and pathogenicity. Several proteins responsible for conidiation were predicted to be SUMOylated, suggesting that conidiation is controlled at the post‐translational level by SUMOylation. In addition to infection‐related development, SUMOylation also played important roles in resistance to nutrient starvation, DNA damage and oxidative stresses. Therefore, SUMOylation is required for infection‐related fungal development, stress responses and pathogenicity in M. oryzae. This study provides new insights into the role of SUMOylation in the molecular mechanisms of pathogenesis of the rice blast fungus and other plant pathogens.
Keywords: Magnaporthe oryzae, pathogenicity, post‐translational modification, rice blast fungus, SUMOylation, ubiquitination
Introduction
Post‐translational modifications (PTMs) regulate diverse biological processes by the attachment of specific proteins or functional groups to various substrates (Gill, 2004). PTMs include diverse protein‐modifying processes that are widely conserved in eukaryotic organisms (Watts, 2013). In fungal plant pathogens, the most comprehensively studied PTM is phosphorylation, which is essential for appressorium formation (Dean, 1997; Flaishman et al., 1995; Kang et al., 1999, Zhao et al., 2007). Methylation in fungi has also been studied as a post‐translational epigenetic modification (Jeon et al., 2015; Martienssen and Colot, 2001). Ubiquitination is a major mechanism of protein degradation in eukaryotic organisms (Liu and Xue, 2011) and has been actively studied in various fungi, including plant pathogens (Guo et al., 2015; Oh et al., 2012; Prakash et al., 2016). In contrast, in‐depth studies of SUMOylation, which has components different from those of ubiquitination, are limited to the model fungus Saccharomyces cerevisiae and human fungal pathogens (Denison et al., 2005; Leach et al., 2011; Park et al., 2011).
SUMOylation involves the attachment of small ubiquitin‐related modifiers (SUMOs) to substrate proteins. This process has several important biological roles, including transcriptional regulation, protein localization and stability, cell cycle, DNA repair and stress resistance (Alonso et al., 2015; Johnson and Blobel, 1999; Lomeli and Vazquez, 2011). The main components of SUMOylation, other than SUMO, are SUMO‐activating enzymes (E1), SUMO‐conjugating enzymes (E2), SUMO ligases (E3) and SUMO proteases. In the SUMOylation process, the SUMO precursor is cleaved by SUMO protease (hydrolase) to reveal the diglycine motif at its C‐terminal end and to become a mature form (Hickey et al., 2012). SUMO is then activated by the formation of a thioester bond between a cysteine residue of E1 and a glycine residue of SUMO. SUMO is transferred from E1 to a cysteine residue of E2 by a thioester bond (Desterro et al., 1999; Johnson and Blobel, 1997). E3 facilitates the formation of an isopeptide bond between a glycine residue of SUMO and lysine residues of the consensus motif ΨKXE/D (Ψ and X represent a hydrophobic amino acid and any amino acid, respectively) or a non‐consensus motif on the substrate (Tatham et al., 2001). This isopeptide bond is cleaved by a SUMO protease (isopeptidase) (Hickey et al., 2012). SUMO is then recycled for use in SUMOylation (Dohmen, 2004; Lomeli and Vazquez, 2011; Sriramachandran and Dohmen, 2014). This process is similar to ubiquitination, but E1 acts as a heterodimer and E3 is not essential for the attachment of SUMO to substrates (Gill, 2004; Müller et al., 2001; Saracco et al., 2007).
SUMOylation regulates the inflammatory response in mammals, flowering time and resistance to abiotic stress in plants, and sporulation and virulence in fungi (Dohmen, 2004; Gujjula et al., 2016; Harting et al., 2013; Leach et al., 2011; Raorane et al., 2013; Wong et al., 2008). In S. cerevisiae, SMT3 plays a role as SUMO, AOS1 and UBA2 as E1 SUMO‐activating enzymes, UBC9 as an E2 SUMO‐conjugating enzyme, SIZ1, SIZ2, CST9 and MMS21 as E3 SUMO ligases, and ULP1, ULP2 and WSS1 as SUMO proteases (Mullen et al., 2010; Pasupala et al., 2012). Amongst the SUMOylation components, SMT3, AOS1, UBA2, UBC9, MMS21 and ULP1 are essential for viability (Johnson and Blobel, 1999). However, in other fungal species, such as Schizosaccharomyces pombe, Aspergillus nidulans, A flavus and Candida albicans, SUMO is not essential for viability (Leach et al., 2011; Nie et al., 2016; Tanaka et al., 1999; Wong et al., 2008). Despite these functional studies, the functional roles of the SUMOylation pathway in plant‐pathogenic fungi are unclear.
Magnaporthe oryzae is an ascomycete plant pathogen which causes rice blast disease and is associated with a considerable socioeconomic burden (Kim et al., 2009). Magnaporthe oryzae is an important model organism for the study of plant–fungal pathogen interactions because the host and fungal genomes have been sequenced and the genomes are now applicable to molecular biological studies (Dean et al., 2005; Goff et al., 2002; Valent, 1990). The disease cycle starts when a conidium (asexual spore) attaches to a hydrophobic surface of the host plant, and germinates and forms an appressorium, a specialized infection structure, from the tip of the germ tube. The appressorium penetrates the host surface by exerting a pressure of >8 MPa (Ebbole, 2007). After colonizing the host plant, secondary inoculum is produced by conidiogenesis (Goh et al., 2011b). The secondary inoculum is a key factor for epidemics as M. oryzae is a polycyclic pathogen (Kim et al., 2009). A better understanding of SUMOylation in the rice blast fungus will provide new insights into infection‐related fungal development.
In this study, we identified the components of SUMOylation amongst ubiquitin‐like modifiers (UBLs) by phylogenetic analysis, and determined their functions in M. oryzae. The SUMOylation machinery is conserved in diverse fungal phyla. Deletion of the four genes encoding SUMO, two E1 enzymes and one E2 enzyme led to pleiotropic phenotypes, including conidiation, septum formation, sensitivity to stress and pathogenicity. Further analysis indicated that infection‐related fungal development and pathogenicity were regulated by SUMOylation. This study improves our understanding of SUMOylation in the rice blast pathogen and other plant‐pathogenic fungi.
Results
Phylogenetic analysis of UBLs in fungi
We identified S. cerevisiae SUMOylation homologues in fungi by protein blast and Pfam domain analysis. The domain analysis of S. cerevisiae showed that SMT3 possesses a SUMO domain (PF11976), AOS1 and UBC2 possess a ThiF domain (PF00899), UBC9 contains a ubiquitin‐conjugating domain (PF00179) and the E3 enzymes possess various zinc finger domains (PF02891 and PF14634) (Table S1, see Supporting Information). Proteins with these domains were identified from the selected model organisms and fungal species. This retrieved not only SUMOylation‐associated proteins, but all UBLs, including those for neddylation and urmylation (Fig. 1A and Table S2, see Supporting Information). Fungi showed relatively fewer UBLs than animals and plants. For SUMO, only one gene was widely conserved in most fungal species, but some had more than one polyphyletic gene (Fig. S1A, see Supporting Information). Unlike SUMO, E1 enzymes were divided into six clades with specific functions, annotated on the basis of S. cerevisiae proteins, including UBA1 for ubiquitination, UBA3 for neddylation, UBA4 for urmylation, the two SUMOylation clades (AOS1 and UBA2) and one clade with unknown function (Figs 1B and S1B). UBC9 showed the highest diversity, but its homologues were well conserved in Ascomycetes (Fig. S1C). SIZ2 homologues were identified in all classes, but SIZ1 was identified only in fungi and animals, and MMS21 only in fungi and oomycota (Fig. S1D,E). CST9 was found only in S. cerevisiae. CST9 homologues were also found in Allomyces macrogynus and Candida albicans by blast, but these lacked the E3 ligase zinc‐RING finger domain (PF14634). ULP1 and ULP2 were also detected in all classes, but WSS1 was absent from animals (Fig. S1F,G). Unlike the SIZ1 and SIZ2 homologues, which were polyphyletic, the fungal ULP1 and ULP2 homologues were monophyletic (Fig. S1D,F). In M. oryzae, the domains of all SUMOylation components showed ≥42% sequence similarity with corresponding S. cerevisiae components (Table S3, see Supporting Information). The homologues of M. oryzae SUMOylation components with the highest levels of similarity were named after the S. cerevisiae genes.
Figure 1.
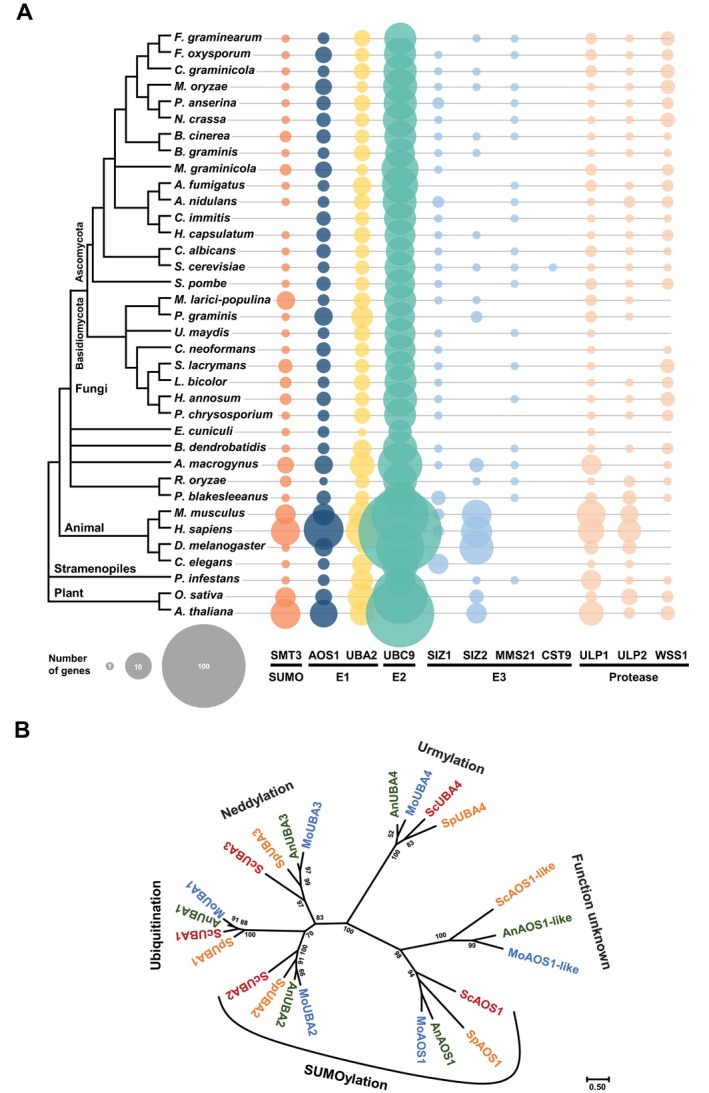
Yeast homologues of ubiquitin‐like modifiers (UBLs) in fungi. (A) A schematic species tree of the Fungal Genome Gold Standard from the Comparative Fungal Genomics Platform (CFGP; http://cfgp.riceblast.snu.ac.kr) and model organisms was constructed on the basis of the National Center for Biotechnology Information (NCBI) taxonomy. The number of yeast homologues in each species is represented by the areas of the circles. (B) Unrooted E1 domain tree of Magnaporthe oryzae (Mo), Aspergillus nidulans (An), Schizosaccharomyces pombe (Sp) and Saccharomyces cerevisiae (Sc). The tree was constructed by the maximum‐likelihood method with 1000 bootstraps.
SUMO activation and conjugation enzymes are essential for SUMOylation in M. oryzae
To confirm that the identified SUMOylation‐associated genes are important for SUMOylation in M. oryzae, we generated targeted deletion mutants in the genes encoding SUMO (MoSMT3), E1 (MoAOS1 and MoUBA2) and E2 (MoUBC9) by double‐joint polymerase chain reaction (PCR) and homologous recombination. The single‐gene deletion mutants were generated by inserting a hygromycin resistance cassette, and a targeted double‐gene deletion mutant was generated by inserting a geneticin resistance cassette into the single‐gene deletion mutants. Deletion of the target genes was confirmed by Southern blotting and quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) (Fig. S2, see Supporting Information). In previous studies, the levels of SUMO‐modified substrates increased on exposure to high temperature, nutrient, DNA damage and oxidative stresses (Leach et al., 2011; Lewicki et al., 2015). Therefore, thermal stress was applied to the wild‐type and to ΔMoaos1, ΔMouba2 and ΔMoubc9. A plasmid (pHA‐SMT3) was constructed by fusing MoSMT3 with a haemagglutinin (HA) tag and the native promoter of MoSMT3, and was transformed into the wild‐type and single‐gene deletion mutants. The phenotypes of the deletion mutants and the wild‐type were unaffected by transformation. This suggests that HA‐tagged MoSMT3 does not affect any other biological processes. Western blotting indicated that SUMOylation in the wild‐type was dramatically greater during mycelial growth and conidiation at high temperatures than at 25 °C (Figs 2A and S3, see Supporting Information). The level of SUMOylation was also increased under oxidative stress (100 mm H2O2) (Fig. S4, see Supporting Information). However, the levels of SUMOylated proteins in the deletion mutants were significantly decreased at high temperatures (Fig. 2A). These results indicate that MoSMT3 acts as SUMO, and that MoAOS1, MoUBA2 and MoUBC9 are indispensable for SUMOylation in M. oryzae. Ubiquitination‐associated components were included in the predicted SUMOylation machineries (Table S2). In contrast, deletion of MoSMT3, MoAOS1, MoUBA2 and MoUBC9 did not affect the level of ubiquitination, suggesting that they are specific to SUMOylation (Fig. S5, see Supporting Information).
Figure 2.
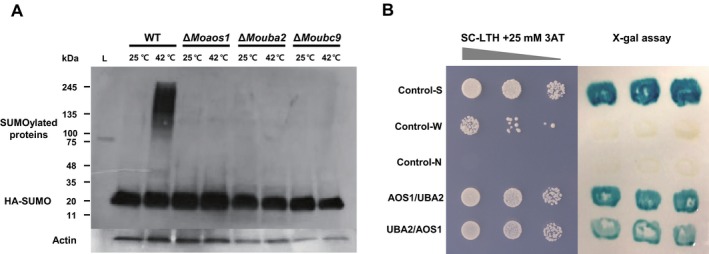
SUMOylated proteins in wild‐type (WT), ΔMoaos1, ΔMouba2 and ΔMoubc9, and yeast two‐hybrid (Y2H) assay of MoAOS1 and MoUBA2. (A) Protein extracts from the WT and deletion mutants were separated by sodum dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and subjected to Western blot analysis using an anti‐haemagglutinin (HA) antibody. (B) Y2H assay of the interaction of MoAOS1 and MoUBA2 (Control‐S, strong interaction; Control‐W, weak interaction; Control‐N, no interaction). X‐gal, 5‐Bromo‐4‐Chlroro‐3‐Indolyl‐D‐Galactopyranoside.
Interaction of the AOS1 and UBA2 subunits of E1 heterodimers is required for SUMO activation in eukaryotic organisms, including humans, Arabidopsis thaliana and S. cerevisiae (Gill, 2004). To determine whether MoAOS1 and MoUBA2 interact as heterodimers, we performed a yeast two‐hybrid (Y2H) assay. AOS1 and UBA2 were cloned into both AD and BD vectors. After co‐transformation, MoAOS1/MoUBA2 and MoUBA2/MoAOS1 were selected on synthetic complete medium lacking leucine and tryptophan (SC‐Leu/Trp) medium and confirmed by PCR amplification. AOS1 and UBA2 strongly interacted on synthetic complete medium lacking leucine, tryptophan and histidine (SC‐Leu/Trp/His) + 25 mm 3‐amino‐1,2,4‐triazole (3AT) medium. The interactions of MoAOS1 and MoUBA2 were confirmed by 5‐Bromo‐4‐Chlroro‐3‐Indolyl‐D‐Galactopyranoside (X‐gal) assay (Fig. 2B). These results indicate that MoAOS1 and MoUBA2 are subunits of the E1 activating enzyme complex, similar to ScAOS1 and ScUBA2.
The SUMOylation machinery is essential for fungal development
To assess the biological function of SUMOylation in M. oryzae, we generated five ΔMosmt3, two ΔMoaos1, two ΔMouba2, eight ΔMoaos1ΔMouba2 and three ΔMoubc9, as explained above. The phenotype of each deletion mutant was the same. We also generated complemented strains for all deletion mutants. In comparison with the wild‐type, all deletion mutants showed significant defects in mycelial growth, conidial size, conidiation, conidial germination and appressorium formation (Table S4, see Supporting Information). Mycelial growth of the deletion mutants was reduced by 10%–25% compared with the wild‐type (Table S4). The number of conidia was counted to quantitatively confirm the production of conidia. All of the deletion mutants produced (4–8) × 104 conidia/mL, which was markedly lower than that of the wild‐type (4.8 × 105 conidia /mL) (Fig. 3A). To identify the reason for the reduced conidial formation, we observed conidiophores under a microscope. In the wild‐type, conidia were produced in a sympodial pattern on the conidiophores, but the deletion mutants produced none, one or two conidia on the conidiophores (Fig. 3B). To further assess the defects in conidiation, the expression levels of conidiation‐related genes (Cos1, Com1, Con7 and Hox2) were examined in the wild‐type and deletion mutants (Kim et al., 2009; Odenbach et al., 2007; Yang et al., 2010; Zhou et al., 2009). However, the expression levels of Cos1, Com1, Con7 and Hox2 did not differ between the wild‐type and deletion mutants (Fig. 3C).
Figure 3.
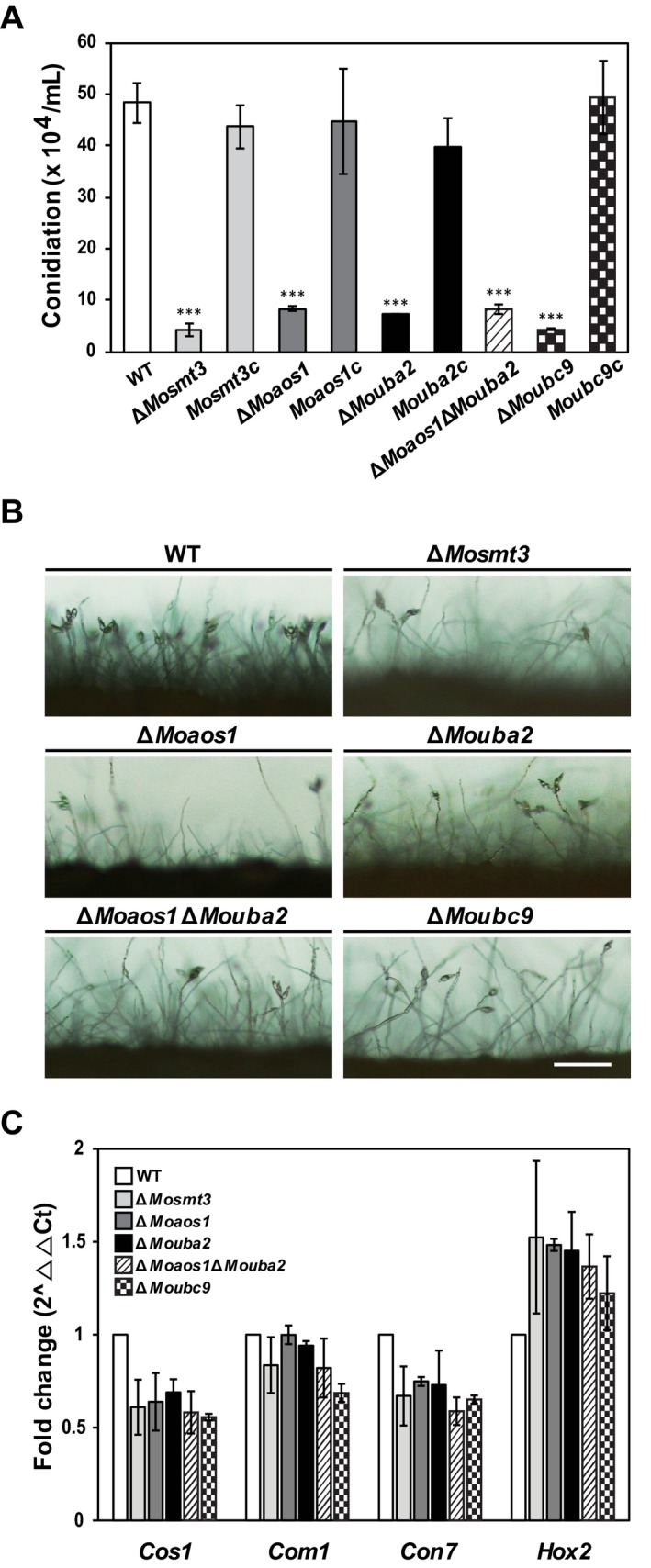
Conidiation of the wild‐type (WT), ΔMosmt3, ΔMoaos1, ΔMouba2 and ΔMoubc9 in Magnaporthe oryzae. (A) Conidia of the WT, deletion mutants and complemented strains were collected from V8 agar after incubation for 7 days. Significance was determined by t‐test (***P < 0.001). (B) Conidiogenesis on conidiophores was observed under a microscope. Scale bar, 100 µm. (C) Expression of conidiation‐related genes during conidiation was quantified by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR).
In the wild‐type, 95% conidial germination and 92% appressorium formation were observed on a hydrophobic surface after 2 and 8 h of incubation, respectively. In contrast, the frequencies of conidial germination and appressorium formation were 69%–79% and 65%–71% in the deletion mutants at 2 and 8 h, respectively (Fig. 4). However, following incubation for a further 24 h, 84%–89% of the deletion mutant conidia germinated and 70%–77% of the germ tubes formed appressoria (Table S5, see Supporting Information). Thus, we performed a conidial adhesion test to assess the cause of the delayed conidial germination and appressorium formation. Compared with the wild‐type (91%), conidia of the deletion mutants (54%–76%) were ineffectively attached to the hydrophobic surface of gelbond film. These results suggest that ineffective conidial adhesion is responsible for delayed conidial germination and appressorium formation in the deletion mutants. These pleiotropic phenotypes were recovered in the complemented strains (Fig. S6, Tables S4 and S5, see Supporting Information).
Figure 4.
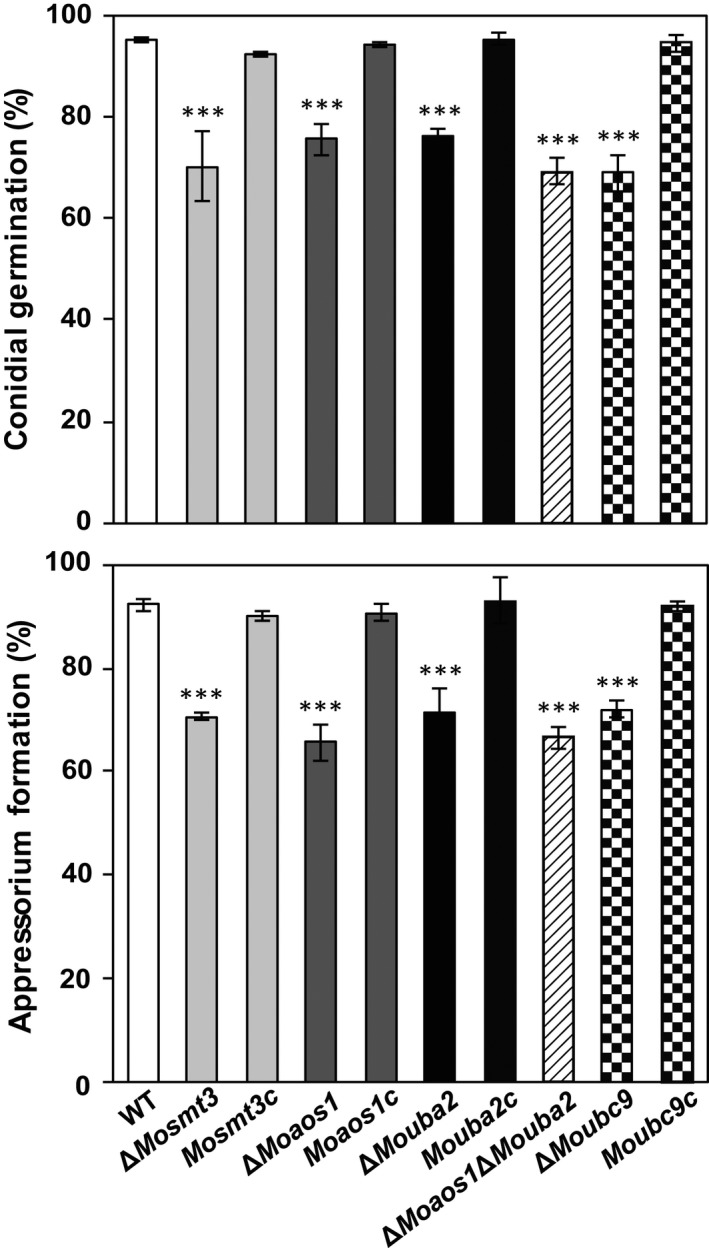
Conidial germination and appressorium formation of the wild‐type (WT), ΔMosmt3, ΔMoaos1, ΔMouba2 and ΔMoubc9 in Magnaporthe oryzae. Percentage of conidial germination and appressorium formation on a hydrophobic surface after incubation for 2 and 8 h, respectively. Significance was determined by t‐test (***P < 0.001).
SUMOylation‐associated genes are involved in septum formation
To examine the roles of SUMOylation‐associated genes in septum formation in M. oryzae, conidia septa were observed under a microscope. In the wild‐type, most conidia (93%) had three cells, compared with 58%–64% in the deletion mutants, in which 23%–28% and 11%–18% had two and one cell, respectively (Fig. 5A). Moreover, conidia of the deletion mutants were significantly shorter than those of the wild‐type (Table S4). The abnormal conidial morphology was recovered in the complemented strains (Fig. 5A and Table S4). Hyphal growth of the deletion mutants was observed under a microscope after staining with Calcofluor white (CFW). In contrast with the wild‐type (60 μm), the cell length of hyphae between two septa was markedly shorter in the deletion mutants (45–50 μm) (Fig. 5B,C). These results suggest that SUMOylation‐associated genes are involved in septum formation of conidia and hyphae in M. oryzae.
Figure 5.
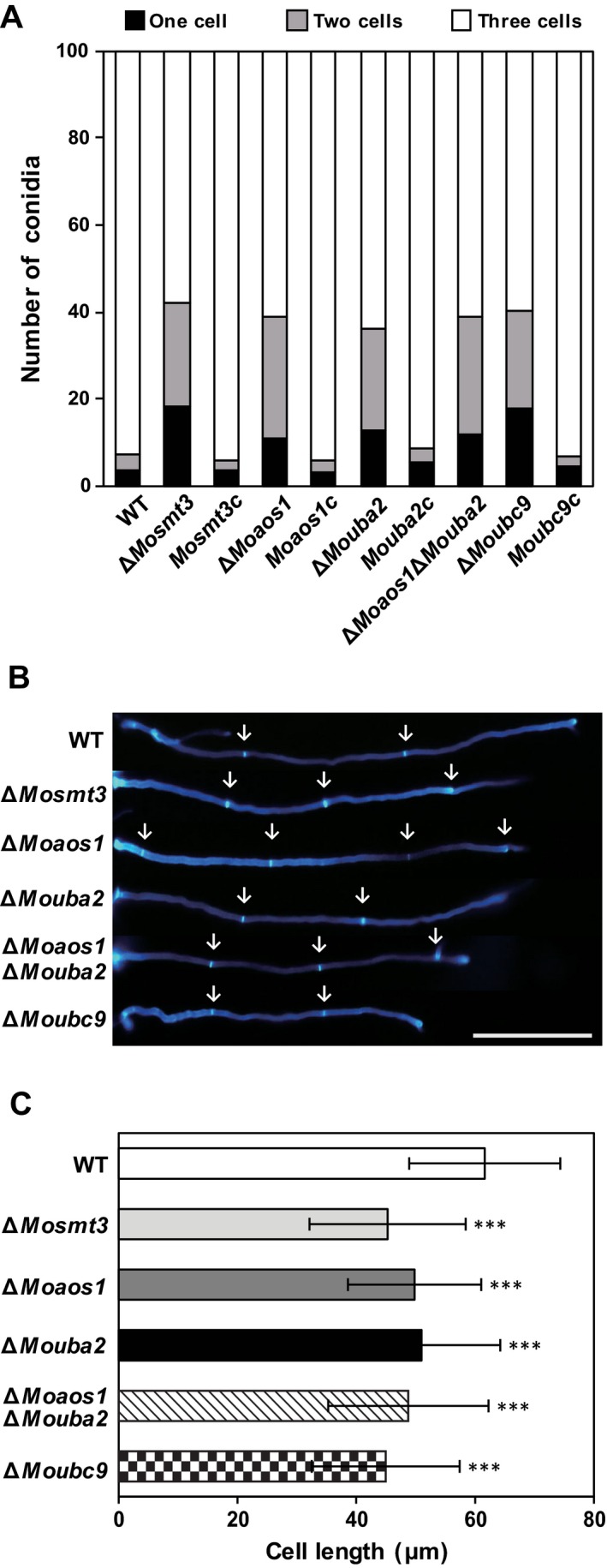
Number of cells in conidia and cell length in hyphae. (A) Percentage of conidia with one, two or three cells in the wild‐type (WT), deletion mutants and complemented strains. (B) Hyphae were stained with Calcofluor white (CFW) and observed under a fluorescence microscope. White arrows indicate septa of hyphae; scale bar, 200 μm. (C) Cell length of hyphae was measured using ImageJ. Significance was determined by t‐test (***P < 0.001).
The SUMOylation components are important for stress tolerance
SUMO deletion mutants of S. pombe and A. nidulans are sensitive to heat and DNA damage stresses (Tanaka et al., 1999; Wong et al., 2008). To investigate the role of SUMOylation‐associated genes in stress resistance in M. oryzae, we measured mycelial growth after culture of the wild‐type and deletion mutants with the following stresses: minimal agar medium (MMA) for nutrient starvation stress, 10 mm hydroxyurea (HU) and 0.05% methyl methanesulfonate (MMS) for DNA damage stress, and 5 mm H2O2 and 3 mm methyl viologen (MV) for oxidative stress (Fig. S7, see Supporting Information). The sensitivity of ΔMosmt3, ΔMoaos1, ΔMouba2 and ΔMoaos1ΔMouba2, but not ΔMoubc9, to the nutrient starvation and oxidative stresses was greater than that of the wild‐type (Figs 6A, B and S7). The sensitivity of all of the deletion mutants to DNA damage stress was also higher than that of the wild‐type (Figs 6A, B and S7). These sensitivities to stress conditions were recovered in the complemented strains (Fig. S7). These results indicate that SUMOylation components are involved in stress resistance.
Figure 6.
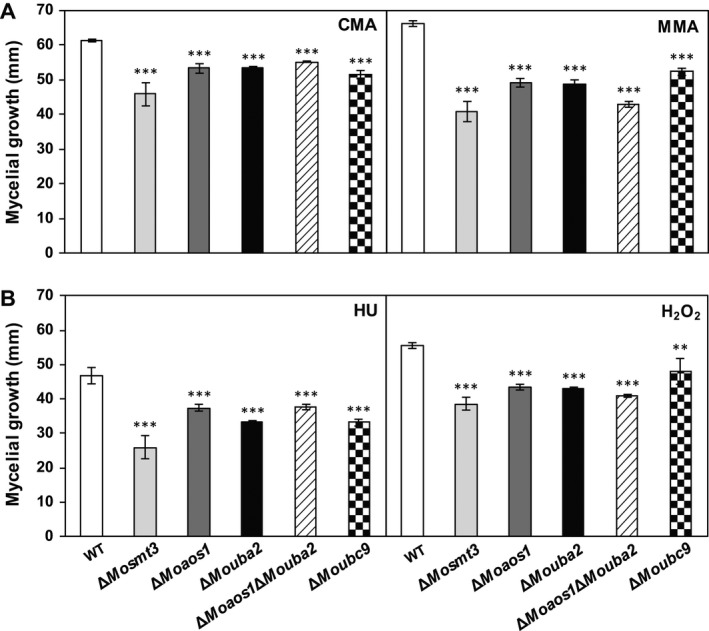
Mycelial growth under stress conditions. (A) Nutrient starvation stress on minimal agar medium (MMA) and complete agar medium (CMA). (B) DNA damage and oxidative stress on CMA containing 10 mm hydroxyurea (HU) and 5 mm H2O2. Significance was determined by t‐test (**P < 0.05 and ***P < 0.001). WT, wild‐type.
SUMO, E1 and E2 are required for functional appressoria and fungal pathogenicity
SUMOylation is necessary for virulence in Candida glabrata, an opportunistic pathogen of bloodstream infections (BSIs) (Gujjula et al., 2016). To investigate whether SUMOylation is required for the pathogenicity of M. oryzae, we performed spray and sheath inoculation assays. Conidial suspensions (5 × 104/mL) of the wild‐type and deletion mutants were sprayed onto 4‐week‐old rice plants of cultivar Nakdongbyeo. The wild‐type induced typical lesions, but the deletion mutants showed smaller and fewer lesions (Fig. 7A). The disease lesion type was evaluated on the basis of lesion size (Valent et al., 1991). In the wild‐type and complemented strains, the disease lesion type was 4, but the disease lesion type of deletion mutants was 1.5–2. The reduced pathogenicity of the deletion mutants was recovered in the complemented strains (Fig. 7A). Penetration sites of appressoria were observed in rice sheath cells at 24 h post‐inoculation (hpi) under a microscope. In comparison with the wild‐type, collapsed appressoria were frequently observed in the rice sheath cells inoculated by the deletion mutants. In the wild‐type, 62% of appressoria penetrated host cells, compared with 1%–10% in the deletion mutants. In particular, the penetration function of ΔMoaos1ΔMouba2 was reduced to the greatest extent (Fig. 8A,B). These results suggest that the reduced pathogenicity was a result of defective penetration of appressoria. Therefore, we performed a cytorrhysis test to estimate the turgor pressure in appressoria. Matured appressoria of the wild‐type and deletion mutants were treated with glycerol solution (1, 3 and 5 m). In the wild‐type, the number of collapsed appressoria increased from 49% to 74% with increasing glycerol concentration. However, the ΔMosmt3, ΔMouba2, ΔMoaos1ΔMouba2 and ΔMoubc9 mutants showed fewer collapsed appressoria than the wild‐type. (Fig. 8C). These data suggest that the turgor pressure in the appressoria of the deletion mutants was not less than that in the wild‐type. Therefore, to determine the cell wall integrity of appressoria, we performed plasmolysis assay using polyethylene glycol (PEG) of different molecular sizes. This showed that the plasmolysis to cytorrhysis ratios of the deletion mutants did not differ significantly from that of the wild‐type (data not shown).
Figure 7.
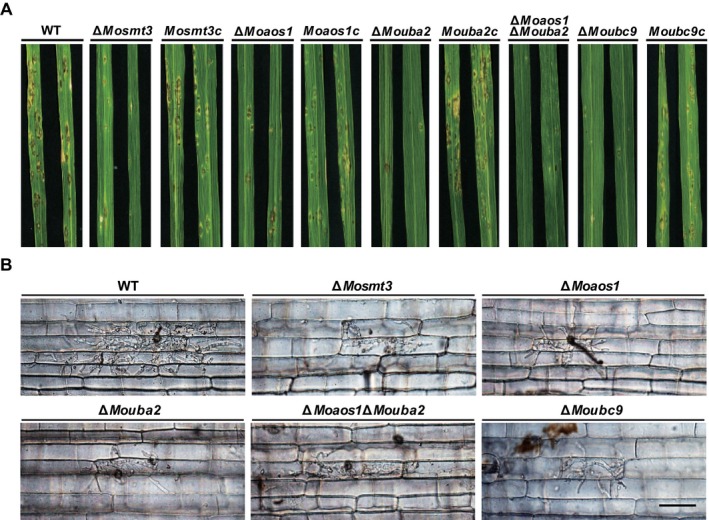
Pathogenicity assays by spray and sheath inoculation. (A) Conidial suspensions (5 × 104/mL) were sprayed onto 4‐week‐old rice seedlings, and lesions were observed at 6 days post‐inoculation (dpi). (B) Conidial suspensions (2 × 104/mL) were inoculated onto rice sheath cells. The growth of invasive hyphae (IH) was observed under a microscope at 48 h post‐inoculation (hpi). Scale bar, 50 μm. WT, wild‐type.
Figure 8.
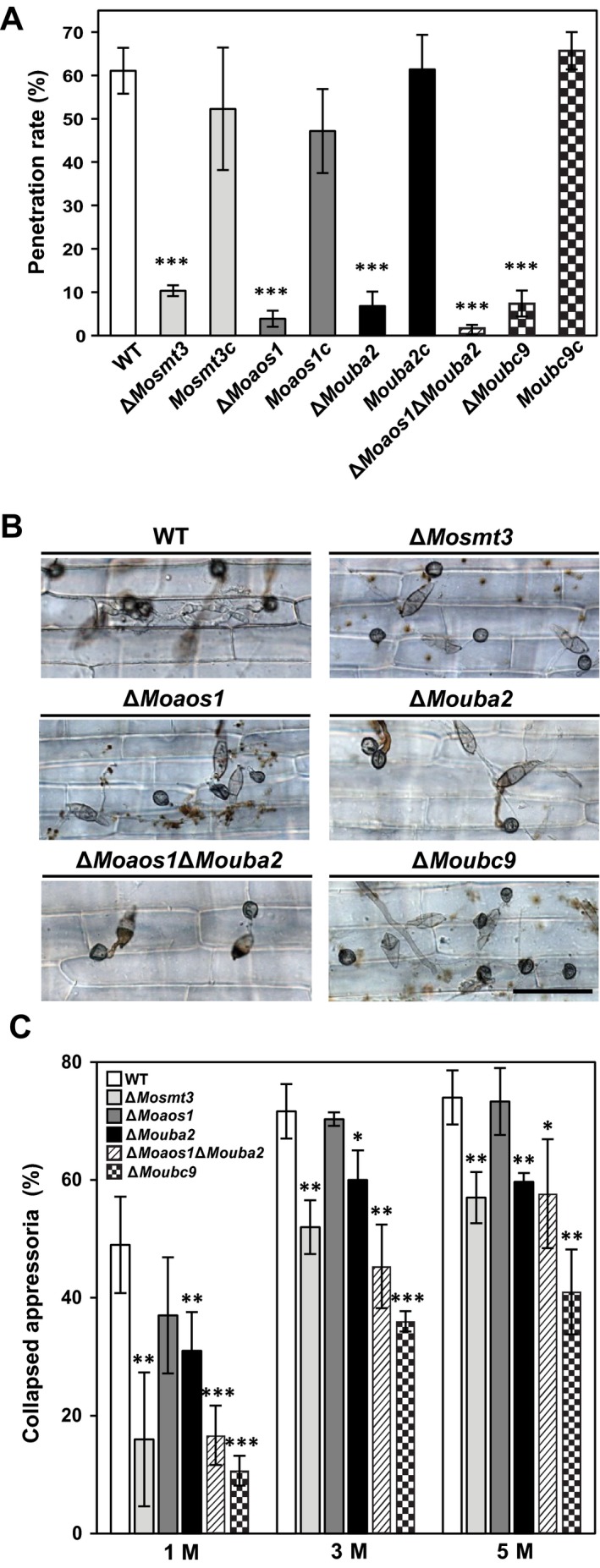
Penetration function of appressoria. (A, B) Rate of penetration of rice sheath cells by appressoria at 24 h post‐inoculation (hpi). Scale bar, 50 μm. (C) The number of collapsed appressoria after treatment with 1, 3 and 5 m glycerol. Significance was determined by t‐test (*P < 0.01, **P < 0.05 and ***P < 0.001). WT, wild‐type.
Next, we performed a sheath inoculation assay to evaluate the cause of restricted lesion development on rice cells. Growth of invasive hyphae (IH) at 48 hpi was classified into three types: Type I for IH restricted to the primary infection cell, Type II for IH growth to adjacent cells and Type III for extensive growth of IH over adjacent cells. Although 82% of IH from the appressoria of the wild‐type were Type II and III, the corresponding values in the deletion mutants were only 31%–43% (Fig. S8B, see Supporting Information). Remarkably, IH growth was significantly reduced in ΔMoaos1ΔMouba2 compared with the single‐gene deletion mutants (Figs 7B and S8B). The reduced penetration rate and invasive growth were recovered in the complemented strains (Figs S8A and S9, see Supporting Information). These results indicate that SUMOylation is important for functional appressoria and the pathogenicity of M. oryzae.
SUMOylation‐associated proteins are localized to the nucleus and cytoplasm
Most SUMOylation components are localized to the nucleus and, to a lesser extent, to the cytoplasm (Gill, 2004; Gillies et al., 2016; Truong et al., 2012). To observe the localization of SUMOylation components, we transformed the deletion mutants with plasmids containing monomeric red fluorescent protein (mRFP) fused with MoSMT3, MoAOS1, MoUBA2 or MoUBC9. Intracellular localization of MoSMT3, MoAOS1, MoUBA2 and MoUBC9 was observed under a fluorescence microscope. MoAOS1 and MoUBA2 were largely localized to the nucleus, but MoSMT3 and MoUBC9 were present in the nucleus and cytoplasm (Fig. S10, see Supporting Information). However, all four SUMOylation components were predominantly localized in the nucleus under oxidative stress conditions (100 mm H2O2) (Fig. 9B).
Figure 9.
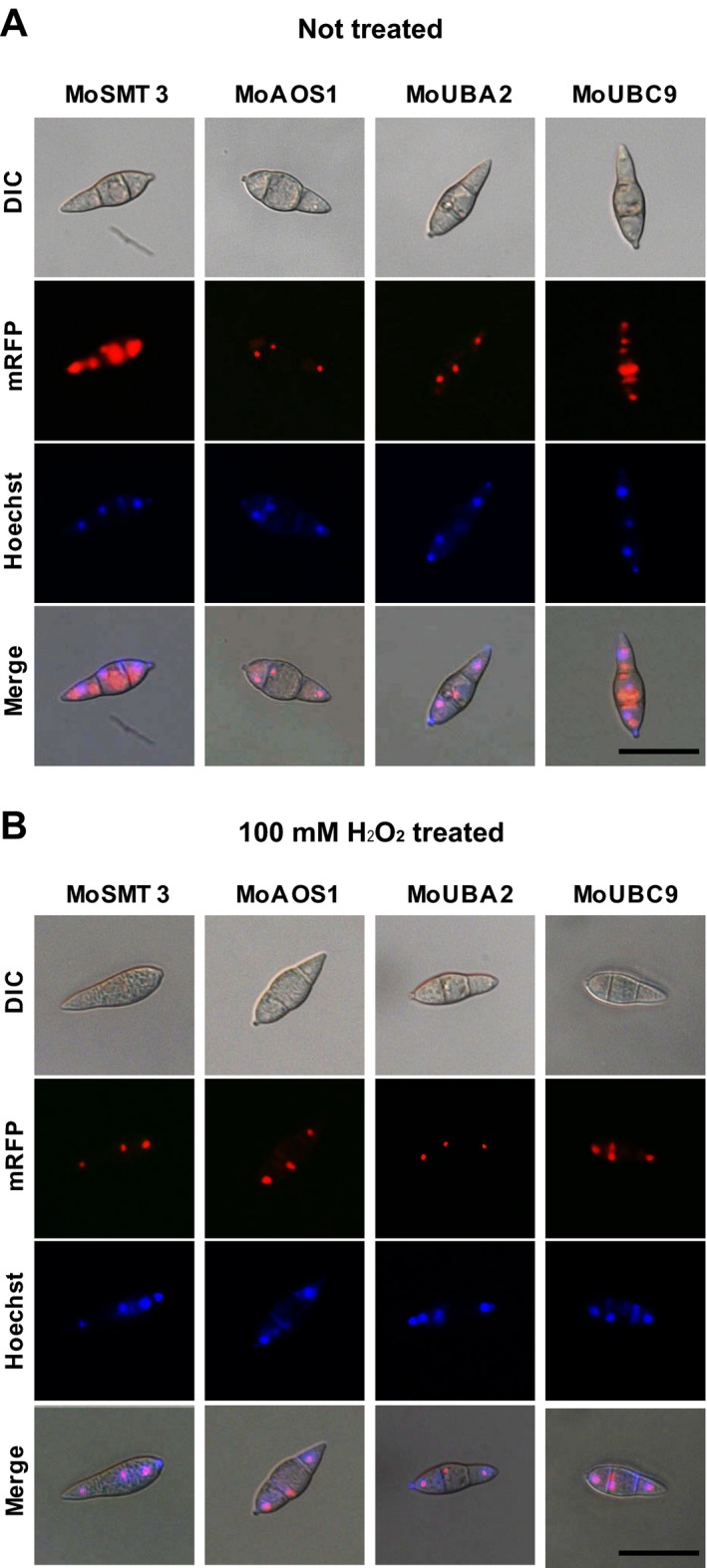
Intracellular localization of SUMOylation components in Magnaporthe oryzae. (A) MoAOS1 and MoUBA2 were localized predominantly in the nucleus, but MoSMT3 and MoUBC9 were localized to both the nucleus and the cytoplasm. Nuclei were stained with Hoechst 33342. Scale bar, 25 μm. (B) The four SUMOylation components were predominantly localized to the nucleus under oxidative stress conditions (100 mm H2O2). Scale bar, 25 μm. Differential interference contrast (DIC), mRFP, monomeric red fluorescent protein.
Discussion
Since the discovery of SUMO in S. cerevisiae in 1995, SUMOylation has been investigated for over 20 years in other model organisms (Broday et al., 2004; Ihara et al., 2008; Meluh and Koshland, 1995; Nacerddine et al., 2005; Nie et al., 2009; Nowak and Hammerschmidt, 2006; Park et al., 2011). However, the machinery and functional roles of SUMOylation in plant‐pathogenic fungi are unclear. Recent advances in genome sequencing and molecular tools have enabled the identification and functional characterization of SUMOylation components. We comprehensively analysed SUMOylation in the model fungal plant pathogen M. oryzae.
The SUMOylation machinery in fungi differs from other UBLs
An understanding of the function of the SUMOylation machinery is limited to model species. Unlike ubiquitination, SUMOylation does not involve protein degradation, but instead enhances protein stability (Müller et al., 2001). A previous study on ubiquitination in M. oryzae identified SUMO, E1 and E2 enzymes as ubiquitination components using a domain search method (Oh et al., 2012). However, SUMOylation and other PTMs in yeast utilize different machineries (Johnson et al., 1997; Müller et al., 2001). Our phylogenetic analysis showed that MoAOS1 and MoUBA2 are clearly separated from the other E1 UBLs. The ubiquitin E2 enzyme (MGG_01756) in M. oryzae (Shi et al., 2016) was not grouped with either MoUBC9 or ScUBC9, therefore, ubiquitination‐related proteins formed their own clade. In addition, the MoSTM3, MoAOS1, MoUBA2 and MoUBC9 deletion mutants exhibited normal ubiquitination. Although all UBLs have the same ubiquitin‐like domains, their diverse functions may be a result of differences in their sequences. For example, the three‐dimensional structure of human SUMO protein (12 kDa) is similar to that of ubiquitin (8.5 kDa) despite their 18% sequence similarity, and SUMO has an additional 15 amino acids at its N‐terminal end (Melchior, 2000; Müller et al., 2001). Overall, our data indicated that SUMOylation components are not involved in ubiquitination, instead being involved in a unique PTM.
SUMOylation does not affect viability, but is essential for the development of M. oryzae
Saccharomyces cerevisiae deletion mutants Δsmt3, Δaos1, Δuba2 and Δubc9 were non‐viable (Schwarz et al., 1998). However, deletion of MoSmt3, MoAos1, MoUba2 and MoUbc9 did not affect the viability of M. oryzae. SUMO deletion mutants of A. nidulans, A. flavus, C. albicans and S. pombe were also viable (Leach et al., 2011; Nie et al., 2016; Shayeghi et al., 1997; Tanaka et al., 1999; Wong et al., 2008). Although fungi with deletions of SUMOylation‐associated genes are viable, they are defective in development. This phenomenon may be a result in part of the presence of structural paralogues of the SUMOylation‐associated proteins. Our phylogenetic analysis showed that the AOS1 clade contains only one protein in S. cerevisiae, but two proteins in other fungal species, such as M. oryzae, A. nidulans and S. pombe. This additional gene may have the same function as AOS1 for SUMOylation. However, this speculation does not fully support the viability of other SUMOylation‐defective mutants of M. oryzae, such as ΔMosmt3. Alternatively, this may be explained by the ‘centrality–lethality’ rule (Jeong et al., 2001). Further study is required to understand the non‐lethality of SUMOylation‐defective mutants, including in fungi such as M. oryzae.
Like the other mutants of SUMOylation components in other organisms (Gujjula et al., 2016; Leach et al., 2011; Nie et al., 2016; Wong et al., 2008), ΔMosmt3, ΔMoaos1, ΔMouba2, ΔMoaos1ΔMouba2 and ΔMoubc9 showed significantly reduced mycelial growth, conidiation, conidial germination, appressorium formation, stress resistance and pathogenicity. Deletion of the SUMOylation‐associated genes was not lethal, unlike in S. cerevisiae and C. glabrata (Gujjula et al., 2016; Schwarz et al., 1998), but deletion of even one of these genes almost abolished SUMOylation in M. oryzae. These data indicate that all SUMOylation components are indispensable for SUMOylation. The pleiotrophic phenotypes of the deletion mutants may be a result of the dysfunction of SUMOylation, because SUMOylated proteins play roles in chromatin organization, transcriptional regulation, RNA biosynthesis and ribosome biogenesis (Nie et al., 2015).
SUMOylation is involved in conidiation
Conidiation is a key factor in the outbreak of rice blast epidemics (Kim et al., 2009). In this study, conidiation was defective in the SUMOylation‐associated gene deletion mutants. However, the expression of conidiation‐related genes (Cos1, Com1, Con7 and Hox2), reported by others (Kim et al., 2009; Odenbach et al., 2007; Yang et al., 2010; Zhou et al., 2009), was not significantly increased or decreased in the deletion mutants. These data suggest that conidiation is regulated not only at the transcriptional level, but also at the post‐translational level. This is supported by the fact that SUMO acts as a stabilizer of transcription factors (Rosonina et al., 2017). Subsequent identification of SUMOylated motifs in conidiation‐related genes further supports the hypothesis that the modification of conidiation‐related genes by SUMO controls conidiation in M. oryzae. Similar conidiation phenotypes were reported in SUMOylation mutants of A. nidulans and A. flavus (Nie et al., 2016; Wong et al., 2008). These results suggest that SUMOylation is crucial for conidiation in filamentous fungi.
SUMOylation provides new insights into the function of appressoria and pathogenicity
In this study, spray inoculation of the deletion mutants resulted in the generation of fewer and smaller lesions. The rates of penetration by appressoria of the deletion mutants were significantly reduced, suggesting the dysfunctionality of appressoria. A cytorrhysis assay indicated that the turgor pressure generation in the appressoria of deletion mutants was abnormal, but not less than that of the wild‐type. Furthermore, ratios of plasmolysis to cytorrhysis, indicating the cell wall integrity, were not significantly different between the deletion mutants and the wild‐type. Remodelling of the actin cytoskeleton, together with turgor pressure and cell wall integrity, has been reported as another important factor for appressoria function (Dagdas et al., 2012). Deletion mutants of septin GTPase showed defects in penetration and pathogenicity, because of the abnormal assembly of the actin cytoskeleton, in M. oryzae (Gupta et al., 2015). These septin proteins, MoSEP3, MoSEP4, MoSEP5 and MoSEP6, are predicted to be SUMOylated according to GPS‐SUMO 2.0 (Zhao et al., 2014). This suggests that SUMOylation may affect other factors, such as remodelling of the actin cytoskeleton, rather than turgor generation and the cell wall integrity of appressoria.
IH growth of the deletion mutants was significantly lower than that of the wild‐type. This may be a result of the role of SUMOylation conferring tolerance to oxidative stress induced by the plant (Feligioni and Nisticò, 2013). The degree of inhibition of mycelial growth of the deletion mutants by oxidative stress was greater than that of the wild‐type. Further study should evaluate the mechanism by which SUMOylation affects oxidative stress resistance and IH growth.
In this study, we found that the SUMOylation machinery differs from that involved in ubiquitination, and evaluated the roles of SUMOylation‐associated genes in infection‐related development, the stress response and the pathogenicity of M. oryzae. These findings provide not only new insights into SUMOylation as a unique PTM in fungi, but also facilitate the identification of the mechanisms of pathogenesis of M. oryzae and other fungal plant pathogens.
Experimental Procedures
Identification of SUMOylation machinery in fungi
SUMOylation components in fungi and model organisms were identified by whole‐protein blast search of the S. cerevisiae SUMO (SMT3), E1 (AOS1 and UBA2), E2 (UBC9), E3 (SIZ1, SIZ2, MMS21 and CST9) and SUMO protease (ULP1, ULP2 and WSS1) sequences using an e‐value of 1e−5. The fungal proteome datasets used were the Fungal Genome Gold Standard from CFGP 2.0 (Choi et al., 2013), the model organisms were from Ensembl (Aken et al., 2016) and the S. cerevisiae query proteins were from Saccharomyces Genome Database (SGD, https://www.yeastgenome.org/) (Cherry, 2015). A domain analysis was performed using InterProScan‐5.25–64.0 software to eliminate sequences without characterized domains. The domain sequences were retrieved and aligned by MAFFT software (Katoh and Standley, 2013) and sparsely aligned regions were trimmed using trimAl software (Capella‐Gutierrez et al., 2009). For phylogenetic analysis, the best protein evolution model for each component alignment was determined, and phylogenetic trees were constructed using RAxML v8.2 software with 1000 bootstraps (Stamatakis, 2014).
Targeted gene deletion and complementation
Magnaporthe oryzae KJ201 (wild‐type) was obtained from the Center for Fungal Genetic Resources (CFGR) at Seoul National University, Seoul, South Korea (Table S6, see Supporting Information). Protoplasts were generated from mycelia grown in liquid complete medium (CM) at 25 °C for 3 days. The upstream (1.0–1.5 kb) and downstream (1.0–1.5 kb) flanking regions of each SUMOylation‐associated gene (MoSMT3, MoAos1, MoUba2 and MoUbc9) were amplified from the genomic DNA (gDNA) of the wild‐type (Table S7, see Supporting Information). The 1.4‐kb hygromycin B phosphotransferase gene (HPH) cassette was amplified from pBCATPH (Yun, 1998). Constructs were produced by double‐joint PCR using three amplicons (upstream flanking, downstream flanking and HPH cassette) for targeted gene deletion. Targeted gene deletion mutants were generated by transforming the constructs into protoplasts using PEG. The targeted gene deletion mutants were selected on TB3 agar supplemented with 200 ppm hygromycin B, and screened by PCR analysis of mycelia using the open reading frame (ORF) primers. To generate the double‐gene deletion mutant, a 1.2‐kb geneticin resistance cassette was amplified from pII99 (Lee et al., 2003), and transformed into ΔMoaos1 protoplasts. To produce complementation strains, constructs containing each ORF and promoter were amplified using upstream forward and downstream reverse primers for the gDNA of the wild‐type. The complemented strains were generated by co‐transformation of these constructs and a geneticin cassette into protoplasts of the single‐gene deletion mutants. The complemented strains were selected on TB3 agar supplemented with 800 ppm geneticin, and screened by PCR of mycelia using the ORF primers. All strains used in this study have been deposited in the Center for Fungal Genetic Resources (http://genebank.snu.ac.kr) at Seoul National University, Seoul, South Korea.
Southern blotting and RT‐PCR
The wild‐type and deletion mutants were cultured in liquid CM at 25 °C for 3 days to extract gDNA and total RNA, as described previously (Kong et al., 2015). The gDNA was digested at 37 °C with restriction enzymes, separated on a 1% agarose gel and transferred to a nitrocellulose membrane. The membrane was hybridized with p32‐labelled probe generated using a Random Primers DNA Labeling System kit (Invitrogen, California, USA), and exposed to X‐ray film. Total RNA (5 μg) was reverse transcribed to complementary DNA (cDNA) using an ImProm‐II Reverse Transcription System kit (Promega, Wisconsin, USA), and subjected to qRT‐PCR using 50 ng of cDNA, 3 μL of primers and 5 μL of SYBR Green PCR Master Mix in a Rotor‐Gene Q 2plex (Qiagen, Hilden, Germany). Transcript levels were quantified by RT‐PCR using 100 ng of cDNA, 3 μL of primers and 10 μL of 2X master mix in a C1000 thermal cycler (Bio‐Rad, California, USA).
Mycelial growth, conidiation and appressorium formation
All strains were cultured in modified complete agar medium (CMA) at 25 °C for 9 days for assessment of mycelial growth and colony morphology (Iyer and Chattoo, 2003). Conidia were collected from cultures on V8 agar after incubation for 7 days, and conidiation was measured under a microscope using a haemacytometer. To assess conidial germination and appressorium formation, 70 μL of conidial suspension (3 × 104/mL) was dropped onto a hydrophobic cover glass at 25 °C. Conidial germination and appressorium formation were evaluated under a microscope after incubation for 2, 8 and 24 h. Conidiogenesis and conidial adhesion assays were performed as described previously (Goh et al., 2011a; Lau and Hamer, 1998). These experiments were performed in triplicate three times.
The morphology of conidia was observed using at least 100 conidia. Hyphae from conidia were incubated with CFW (10 μg/mL; Sigma Aldrich, Missouri, USA) at 25 °C for 5 min to stain the septa, and visualized using a fluorescence microscope (Carl Zeiss, Oberkochen, Germany). Cell length was measured using ImageJ software. These experiments were performed in triplicate three times.
Cytorrhysis and plasmolysis assays
Conidial suspensions (3 × 104/mL) was incubated on a hydrophobic cover glass at 25 °C for 48 h. The sterilized distilled water (SDW) was removed, and the appressoria were treated with glycerol (1, 3 and 5 m) or PEG (400, 1000, 3350 and 8000 Da). At least 100 collapsed and plasmolysed appressoria were counted under a microscope. These experiments were performed in triplicate three times.
Sensitivity assessment to stress conditions
MMA was used to test sensitivity to nutrient starvation stress (Choi et al., 2015). To assess sensitivity to DNA damage stress, CMA was supplemented with 10 mm HU (DNA synthesis inhibitor) and 0.05% MMS (DNA alkylating agent). For oxidative stress, CMA was supplemented with 5 mm H2O2 and 3 mm MV. The wild‐type, deletion mutants and complemented strains were cultured in triplicate under the above five stress conditions and CMA (control) at 25 °C for 9 days.
Pathogenicity tests
A pathogenicity test was performed using the susceptible rice cultivar Nakdongbyeo. For spray inoculation assays, 10 mL of conidial suspension (5 × 104/mL, containing 250 ppm Tween‐20) was inoculated onto 4‐week‐old rice seedlings. Inoculated rice plants were incubated at 25 °C for 1 day at a relative humidity (RH) of 100% in the dark, and then at 28 °C for 5–6 days in a growth chamber. For sheath inoculation assays, conidial suspension (2 × 104/mL) was inoculated onto sheaths of 6‐week‐old rice seedlings, which were then incubated at 25 °C for 24 or 48 h. At least 50 infection sites were observed under a microscope. These experiments were performed in triplicate three times.
Western blot
The promoters of MoSmt3 (EcoRI‐SalI) and HA‐fused MoSmt3 (SalI‐ApaI) were thymine and adenine (TA) cloned into pGEMT‐easy (Promega) (Table S7). The cloned vectors were digested with the corresponding restriction enzymes and ligated into pCB1004 (Wang et al., 1999). The pCB1004:native promoter:HA:SMT3 (pHA‐SMT3) and geneticin resistance cassette were co‐transformed into the wild‐type and single‐gene deletion mutants. Transformants were cultured in liquid CM at 25 °C for 3 days to harvest the mycelia. Proteins were extracted using 600 μL of PRO‐PREP protein extraction solution (Intron Biotechnology, Seongnam, South Korea). Proteins (75 μg) were separated on a 4%–20% gradient SDS‐PAGE gel (Bio‐Rad) and transferred to an Immun‐Blot LF Polyvinylidene difluoride (PVDF) membrane using transfer buffer [20% methanol (v/v), 0.25 m Tris and 2 m glycine). The membrane was probed with an anti‐HA antibody (1 : 1000, Bethyl, Alabama, USA) and anti‐ubiquitin antibody (P4D1) (1 : 1000, Cell Signaling Technology, Massuchusetts, USA) using a Pierce Fast Western Blot Kit and ECL Substrate (Pierce Biotechnology, Rockford, IL, USA). An anti‐actin antibody (1 : 1000, Cell Signaling Technology) was used as the loading control. The probed membrane was exposed to X‐ray film.
Y2H assay
The coding sequences of MoAOS1 and MoUBA2 were amplified from cDNA of the wild‐type (Table S7). A Y2H assay was performed using the ProQuest Two‐Hybrid System kit (Invitrogen). The coding sequences of MoAOS1 and MoUBA2 were inserted into both pDEST22 and pDEST32 using a Gateway system. The MaV203 yeast strain was cultured in Yeast extract‐Peptone‐Adenine‐Dextrose (YPAD) liquid medium at 30 °C until an optical density at 600 nm of 0.4 was reached (Table S6). Yeast competent cells were generated using lithium acetate (LiAc). Next, 1 μg of MoAOS1 and MoUBA2 inserted vectors, 100 μg of denatured salmon sperm DNA and 100 μL of competent cells were co‐transformed using LiAc and PEG. The control plasmids (pEXP22RalGDS‐wt, pEXP22RalGDS‐m1 and pEXP22RalGDS‐m2) were co‐transformed with pEXP32Krev1 using the same method and selected on SC‐Leu/Trp (SC‐LT) agar. Selected co‐transformants were confirmed by amplifying the inserted coding sequence. Interaction of MoAOS1 and MoUBA2 was confirmed on SC‐Leu/Trp/His + 25 mm 3AT (SC‐LTH + 3AT) agar and validated by X‐gal assay.
Intracellular localization
The EF1α promoter (originating from Fusarium verticillioides) was amplified from YL1320. The coding sequences of MoSMT3, MoAOS1, MoUBA2 and MoUBC9 were amplified from the cDNA of the wild‐type. Fusion constructs of the EF1α promoter and each coding sequence were generated using double‐joint PCR and fused in pFPL‐rh containing the coding sequence of mRFP. Next, the coding sequence of mRFP was conjugated to the C‐terminal coding sequence. The C‐terminus of SMT3 is cleaved on reaching maturity. Therefore, another plasmid was generated to observe the localization of SMT3. mRFP and the TrpC terminator were amplified from pFPL‐rh and pBCATPH. The EF1α promoter:mRFP (XbaI‐HindIII) and SMT3:TrpC terminator (HindIII‐XhoI) fusion constructs were generated using double‐joint PCR and TA cloned into pGEMT‐easy. The cloned vectors were digested with the corresponding restriction enzymes and fused in pCB1004. The pFPL‐rh:EF1α promoter:AOS1 (pLOCAL01669), pFPL‐rh:EF1α promoter:UBA2 (pLOCAL06733), pFPL‐rh:EF1α promoter:UBC9 (pLOCAL00970) and pCB1004:EF1α promoter:mRFP:SMT3:TrpC terminator (pLOCAL05737) constructs were co‐transformed with a geneticin resistance cassette into all of the deletion mutants. The localization of MoSMT3, MoAOS1, MoUBA2 and MoUBC9 in conidia and developmental stages, including conidial germination and appressorium formation, was observed using a fluorescence microscope (Carl Zeiss). Conidia were incubated in Hoechst33342 solution (10 mg/mL, Invitrogen) at 25 °C for 30 min to stain the nuclei.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Phylogenetic trees of SUMOylation components in fungi and model organisms. Phylogenetic trees of SUMOylation components in fungi and model organisms that were homologous to Saccharomyces cerevisiae SUMOylation components were constructed by the maximum‐likelihood method with 1000 bootstraps and the LG or BLOSUM62 protein substitution method. The sequences used for alignments were: (A) PF11976 for SUMO, SMT3; (B) PF00899 for E1, AOS1 and UBA2; (C) PF00179 for E2, UBC9; (D) PF02891 for E3, SIZ1 and SIZ2; (E) PF11789 for E3, MMS21; (F) PF02902 for protease, ULP1 and ULP2; (G) PF08325 for protease and WSS1. Clades that contained more than three species of the same fungal phylum, animal and plant are shown without genus/species names. The yeast components are marked with a star and Magnaporthe oryzae homologues are shown in bold. Only bootstrap values >50% are shown.
Fig. S2 Southern blot analysis and reverse transcription‐polymerase chain reaction (RT‐PCR) of the deletion mutants. Genomic DNA of wild‐type (WT) and deletion mutants was extracted and digested with PstI, SalI, HindIII or XhoI. The upstream or downstream construct of each gene was used as a probe for Southern blot analysis. Complementary DNA was synthesized from total RNA. β‐tubulin was used for normalization.
Fig. S3 SUMOylation in wild‐type (WT) and ΔMoaos1 during conidiation. Protein extract from the WT during conidiation was separated by sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and subjected to Western blot analysis using an anti‐haemagglutinin (HA) antibody.
Fig. S4 SUMOylated proteins in wild‐type (WT) under oxidative stress. Protein extract from the WT treated with oxidative stress (100 mm H2O2) was separated by sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and subjected to Western blot analysis using an anti‐haemagglutinin (HA) antibody.
Fig. S5 Ubiquitination in the wild‐type (WT), ΔMosmt3, ΔMoaos1, ΔMouba2 and ΔMoubc9. Protein extracts of WT, ΔMosmt3, ΔMoaos1, ΔMouba2 and ΔMoubc9 were separated by sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and subjected to Western blot analysis using an anti‐ubiquitin (P4D1) antibody. Ubiquitination was not affected in all of the deletion mutants.
Fig. S6 Defective conidiogenesis was recovered in complemented strains. Conidiogenesis on conidiophores was observed under a microscope. Scale bar, 100 µm.
Fig. S7 Mycelial growth under stress conditions. Strains were inoculated on nutrient starvation stress [minimal agar medium (MMA)], DNA damage stress [10 mm hydroxyurea (HU) and 0.05% methyl methanesulfonate (MMS)] and oxidative stress [5 mm H2O2 and 3 mm methyl viologen (MV)]. Mycelial growth was measured 9 days after inoculation. CMA, complete agar medium.
Fig. S8 Growth of invasive hyphae (IH) in the complemented strains. (A) Conidial suspensions (2 × 104/mL) of the complemented strains were inoculated onto 6‐week‐old rice sheath cells. IH growth was observed under a microscope at 48 h post‐inoculation (hpi). Scale bar, 50 μm. (B) Growth of IH into neighbouring cells was classified into three types: Type I for IH restricted to the primary infection cell; Type II for IH growth to adjacent cells; Type III for extensive growth of IH over adjacent cells.
Fig. S9 Penetration assay of the complemented strains. Conidial suspensions (2 × 104/mL) of the complemented strains were inoculated onto 6‐week‐old rice sheath cells. Penetration by appressoria at 24 h post‐inoculation (hpi) was observed under a microscope. Scale bar, 50 μm.
Fig. S10 Intracellular localization of SUMOylation components in Magnaporthe oryzae. MoAOS1, MoUBA2, MoSMT3 and MoUBC9 were predominantly localized in the nuclei during conidial germination and appressorium formation. Scale bar, 50 μm.
Table S1 Domains of Saccharomyces cerevisiae SUMOylation components.
Table S2 The number of Saccharomyces cerevisiae homologues of SUMOylation components in the selected species.
Table S3 The closest Saccharomyces cerevisiae homologues of SUMOylation components in Magnaporthe oryzae.
Table S4 Developmental phenotypes of the wild‐type (WT), deletion mutants and complemented strains.
Table S5 Conidial germination and appressorium formation of the wild‐type (WT), deletion mutants and complemented strains after incubation for 2, 8 and 24 h.
Table S6 List of the strains used in this study.
Table S7 Primer sequences used in this study.
Acknowledgements
This work was supported by grants from National Research Foundation of Korea (NRF‐2017R1A2A1A17069504, NRF‐2015M3A9B8028679). Y‐J L is grateful for a graduate fellowship through the Brain Korea 21 Plus Program.
References
- Aken, B.L. , Ayling, S. , Barrell, D. , Clarke, L. , Curwen, V. , Fairley, S. , Fernandez Banet, J. , Billis, K. , García Girón, C. , Hourlier, T. , Howe, K. , Kähäri, A. , Kokocinski, F. , Martin, F.J. , Murphy, D.N. , Nag, R. , Ruffier, M. , Schuster, M. , Tang, Y.A. , Vogel, J.‐H. , White, S. , Zadissa, A. , Flicek, P. and Searle, S.M.J. (2016) The Ensembl gene annotation system. Database (Oxford), 1, baw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, A. , Greenlee, M. , Matts, J. , Kline, J. , Davis, K.J. and Miller, R.K. (2015) Emerging roles of sumoylation in the regulation of actin, microtubules, intermediate filaments, and septins. Cytoskeleton, 72, 305–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broday, L. , Kolotuev, I. , Didier, C. , Bhoumik, A. , Gupta, B.P. , Sternberg, P.W. , Podbilewicz, B. and Ronai, Z. (2004) The small ubiquitin‐like modifier (SUMO) is required for gonadal and uterine‐vulval morphogenesis in Caenorhabditis elegans . Genes Dev. 18, 2380–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella‐Gutierrez, S. , Silla‐Martinez, J.M. and Gabaldon, T. (2009) trimAl: a tool for automated alignment trimming in large‐scale phylogenetic analyses. Bioinformatics, 25, 1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry, J.M. (2015) The Saccharomyces genome database: a tool for discovery. Cold Spring Harb. Protoc. 12, pdb top083840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, J. , Cheong, K. , Jung, K. , Jeon, J. , Lee, G.‐W. , Kang, S. , Kim, S. , Lee, Y.‐W. and Lee, Y.‐H. (2013) CFGP 2.0: a versatile web‐based platform for supporting comparative and evolutionary genomics of fungi and oomycetes. Nucleic Acids Res. 41, D714–D719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, J. , Chung, H. , Lee, G.‐W. , Koh, S.‐K. , Chae, S.‐K. and Lee, Y.‐H. (2015) Genome‐wide analysis of hypoxia‐responsive genes in the rice blast fungus, Magnaporthe oryzae . PLoS One, 10, e0134939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagdas, Y.F. , Yoshino, K. , Dagdas, G. , Ryder, L.S. , Bielska, E. , Steinberg, G. and Talbot, N.J. (2012) Septin‐mediated plant cell invasion by the rice blast fungus, Magnaporthe oryzae . Science, 336, 1590–1595. [DOI] [PubMed] [Google Scholar]
- Dean, R.A. (1997) Signal pathways and appressorium morphogenesis. Annu. Rev. Phytopathol. 35, 211–234. [DOI] [PubMed] [Google Scholar]
- Dean, R.A. , Talbot, N.J. , Ebbole, D.J. , Farman, M.L. , Mitchell, T.K. , Orbach, M.J. , Thon, M. , Kulkarni, R. , Xu, J.‐R. , Pan, H. , Read, N.D. , Lee, Y.‐H. , Carbone, I. , Brown, D. , Oh, Y.Y. , Donofrio, N. , Jeong, J.S. , Soanes, D.M. , Djonovic, S. , Kolomiets, E. , Rehmeyer, C. , Li, W. , Harding, M. , Kim, S. , Lebrun, M.‐H. , Bohnert, H. , Coughlan, S. , Butler, J. , Calvo, S. , Ma, L.‐J. , Nicol, R. , Purcell, S. , Nusbaum, C. , Galagan, J.E. and Birren, B.W. (2005) The genome sequence of the rice blast fungus Magnaporthe grisea . Nature, 434, 980. [DOI] [PubMed] [Google Scholar]
- Denison, C. , Rudner, A.D. , Gerber, S.A. , Bakalarski, C.E. , Moazed, D. and Gygi, S.P. (2005) A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol. Cell. Proteomics, 4, 246–254. [DOI] [PubMed] [Google Scholar]
- Desterro, J.M. , Rodriguez, M.S. , Kemp, G.D. and Hay, R.T. (1999) Identification of the enzyme required for activation of the small ubiquitin‐like protein SUMO‐1. J. Biol. Chem. 274, 10 618–10 624. [DOI] [PubMed] [Google Scholar]
- Dohmen, R.J. (2004) SUMO protein modification. Biochim. Biophys. Acta, 1695, 113–131. [DOI] [PubMed] [Google Scholar]
- Ebbole, D.J. (2007) Magnaporthe as a model for understanding host–pathogen interactions. Annu. Rev. Phytopathol. 4, 437–456. [DOI] [PubMed] [Google Scholar]
- Feligioni, M. and Nisticò, R. (2013) SUMO: a (oxidative) stressed protein. Neuromolecular Med. 15, 707–719. [DOI] [PubMed] [Google Scholar]
- Flaishman, M. , Hwang, C.‐S. and Kolattukudy, P. (1995) Involvement of protein phosphorylation in the induction of appressorium formation in Colletotrichum gloeosporioides by its host surface wax and ethylene. Physiol. Mol. Plant Pathol. 47, 103–117. [Google Scholar]
- Gill, G. (2004) SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 18, 2046–2059. [DOI] [PubMed] [Google Scholar]
- Gillies, J. , Hickey, C.M. , Su, D. , Wu, Z. , Peng, J. and Hochstrasser, M. (2016) SUMO pathway modulation of regulatory protein binding at the ribosomal DNA locus in Saccharomyces cerevisiae . Genetics, 202, 1377–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff, S.A. , Ricke, D. , Lan, T.H. , Presting, G. , Wang, R. , Dunn, M. , Glazebrook, J. , Sessions, A. , Oeller, P. , Varma, H. , Hadley, D. , Hutchison, D. , Martin, C. , Katagiri, F. , Lange, B.M. , Moughamer, T. , Xia, Y. , Budworth, P. , Zhong, J. , Miquel, T. , Paszkowski, U. , Zhang, S. , Colbert, M. , Sun, W.L. , Chen, L. , Cooper, B. , Park, S. , Wood, T.C. , Mao, L. , Quail, P. , Wing, R. , Dean, R. , Yu, Y. , Zharkikh, A. , Shen, R. , Sahasrabudhe, S. , Thomas, A. , Cannings, R. , Gutin, A. , Pruss, D. , Reid, J. , Tavtigian, S. , Mitchell, J. , Eldredge, G. , Scholl, T. , Miller, R.M. , Bhatnagar, S. , Adey, N. , Rubano, T. , Tusneem, N. , Robinson, R. , Feldhaus, J. , Macalma, T. , Oliphant, A. and Briggs, S. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science, 296, 92–100. [DOI] [PubMed] [Google Scholar]
- Goh, J. , Jeon, J. , Kim, K.S. , Park, J. , Park, S.‐Y. and Lee, Y.‐H. (2011a) The PEX7‐mediated peroxisomal import system is required for fungal development and pathogenicity in Magnaporthe oryzae . PLoS One, 6, e28220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh, J. , Kim, K.S. , Park, J. , Jeon, J. , Park, S.‐Y. and Lee, Y.‐H. (2011b) The cell cycle gene MoCDC15 regulates hyphal growth, asexual development and plant infection in the rice blast pathogen Magnaporthe oryzae . Fungal Genet. Biol. 48, 784–792. [DOI] [PubMed] [Google Scholar]
- Gujjula, R. , Veeraiah, S. , Kumar, K. , Thakur, S.S. , Mishra, K. and Kaur, R. (2016) Identification of components of the SUMOylation machinery in Candida glabrata: role of the deSUMOylation peptidase CgUlp2 in virulence. J. Biol. Chem. 291, 19 573–19 589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, M. , Gao, F. , Zhu, X. , Nie, X. , Pan, Y. and Gao, Z. (2015) MoGrr1, a novel F‐box protein, is involved in conidiogenesis and cell wall integrity and is critical for the full virulence of Magnaporthe oryzae . Appl. Microbiol. Biotechnol. 99, 8075–8088. [DOI] [PubMed] [Google Scholar]
- Gupta, Y.K. , Dagdas, Y.F. , Martinez‐Rocha, A.L. , Kershaw, M.J. , Littlejohn, G.R. , Ryder, L.S. , Sklenar, J. , Menke, F. and Talbot, N.J. (2015) Septin‐dependent assembly of the exocyst is essential for plant infection by Magnaporthe oryzae . Plant Cell, 27, 3277–3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harting, R. , Bayram, Ö. , Laubinger, K. , Valerius, O. and Braus, G.H. (2013) Interplay of the fungal sumoylation network for control of multicellular development. Mol. Microbiol. 90, 1125–1145. [DOI] [PubMed] [Google Scholar]
- Hickey, C.M. , Wilson, N.R. and Hochstrasser, M. (2012) Function and regulation of SUMO proteases. Nat. Rev. Mol. Cell Biol. 13, 755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara, M. , Stein, P. and Schultz, R.M. (2008) UBE2I (UBC9), a SUMO‐conjugating enzyme, localizes to nuclear speckles and stimulates transcription in mouse oocytes. Biol. Reprod. 79, 906–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer, G. and Chattoo, B.B. (2003) Purification and characterization of laccase from the rice blast fungus, Magnaporthe grisea . FEMS Microbiol. Lett. 227, 121–126. [DOI] [PubMed] [Google Scholar]
- Jeon, J. , Choi, J. , Lee, G.‐W. , Park, S.‐Y. , Huh, A. , Dean, R.A. and Lee, Y.‐H. (2015) Genome‐wide profiling of DNA methylation provides insights into epigenetic regulation of fungal development in a plant pathogenic fungus, Magnaporthe oryzae . Sci. Rep. 5, 8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, H. , Mason, S.P. , Barabási, A.L. and Oltvai, Z.N. (2001) Lethality and centrality in protein networks. Nature, 411, 41. [DOI] [PubMed] [Google Scholar]
- Johnson, E.S. and Blobel, G. (1997) Ubc9p is the conjugating enzyme for the ubiquitin‐like protein Smt3p. J. Biol. Chem. 272, 26 799–26 802. [DOI] [PubMed] [Google Scholar]
- Johnson, E.S. and Blobel, G. (1999) Cell cycle‐regulated attachment of the ubiquitin‐related protein SUMO to the yeast septins. J. Cell Biol. 147, 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, E.S. , Schwienhorst, I. , Dohmen, R.J. and Blobel, G. (1997) The ubiquitin‐like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 16, 5509–5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, S.H. , Khang, C.H. and Lee, Y.‐H. (1999) Regulation of cAMP‐dependent protein kinase during appressorium formation in Magnaporthe grisea . FEMS Microbiol. Lett. 170, 419–423. [Google Scholar]
- Katoh, K. and Standley, D.M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. , Park, S.‐Y. , Kim, K.S. , Rho, H.‐S. , Chi, M.‐H. , Choi, J. , Park, J. , Kong, S. , Park, J. , Goh, J. and Lee, Y.‐H. (2009) Homeobox transcription factors are required for conidiation and appressorium development in the rice blast fungus Magnaporthe oryzae . PLoS Genet. 5, e1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, S. , Park, S.‐Y. and Lee, Y.‐H. (2015) Systematic characterization of the bZIP transcription factor gene family in the rice blast fungus, Magnaporthe oryzae . Environ. Microbiol. 17, 1425–1443. [DOI] [PubMed] [Google Scholar]
- Lau, G.W. and Hamer, J.E. (1998) Acropetal: a genetic locus required for conidiophore architecture and pathogenicity in the rice blast fungus. Fungal Genet. Biol. 24, 228–239. [DOI] [PubMed] [Google Scholar]
- Leach, M.D. , Stead, D.A. , Argo, E. and Brown, A.J. (2011) Identification of sumoylation targets, combined with inactivation of SMT3, reveals the impact of sumoylation upon growth, morphology, and stress resistance in the pathogen Candida albicans . Mol. Biol. Cell, 22, 687–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. , Lee, T. , Lee, Y.‐W. , Yun, S.‐H. and Turgeon, B.G. (2003) Shifting fungal reproductive mode by manipulation of mating type genes: obligatory heterothallism of Gibberella zeae . Mol. Microbiol. 50, 145–152. [DOI] [PubMed] [Google Scholar]
- Lewicki, M.C. , Srikumar, T. , Johnson, E. and Raught, B. (2015) The S. cerevisiae SUMO stress response is a conjugation–deconjugation cycle that targets the transcription machinery. J. Proteomics, 118, 39–48. [DOI] [PubMed] [Google Scholar]
- Liu, T.‐B. and Xue, C. (2011) The ubiquitin‐proteasome system and F‐box proteins in pathogenic fungi. Mycobiology, 39, 243–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomeli, H. and Vazquez, M. (2011) Emerging roles of the SUMO pathway in development. Cell. Mol. Life Sci. 68, 4045–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen, R.A. and Colot, V. (2001) DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science, 293, 1070–1074. [DOI] [PubMed] [Google Scholar]
- Melchior, F. (2000) SUMO‐nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 16, 591–626. [DOI] [PubMed] [Google Scholar]
- Meluh, P.B. and Koshland, D. (1995) Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP‐C. Mol. Biol. Cell. 6, 793–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen, J.R. , Chen, C.F. and Brill, S.J. (2010) Wss1 is a SUMO‐dependent isopeptidase that interacts genetically with the Slx5‐Slx8 SUMO‐targeted ubiquitin ligase. Mol. Cell Biol. 30, 3737–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, S. , Hoege, C. , Pyrowolakis, G. and Jentsch, S. (2001) SUMO, ubiquitin's mysterious cousin. Nat. Rev. Mol. Cell Biol. 2, 202. [DOI] [PubMed] [Google Scholar]
- Nacerddine, K. , Lehembre, F. , Bhaumik, M. , Artus, J. , Cohen‐Tannoudji, M. , Babinet, C. , Pandolfi, P.P. and Dejean, A. (2005) The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev. Cell, 9, 769–779. [DOI] [PubMed] [Google Scholar]
- Nie, M. , Xie, Y. , Loo, J.A. and Courey, A.J. (2009) Genetic and proteomic evidence for roles of Drosophila SUMO in cell cycle control, Ras signaling, and early pattern formation. PLoS One, 4, e5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie, M. , Vashisht, A.A. , Wohlschlegel, J.A. and Boddy, M.N. (2015) High confidence fission yeast SUMO conjugates identified by tandem denaturing affinity purification. Sci. Rep. 5, 14 389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie, X. , Yu, S. , Qiu, M. , Wang, X. , Wang, Y. , Bai, Y. , Zhang, F. and Wang, S. (2016) Aspergillus flavus SUMO contributes to fungal virulence and toxin attributes. J. Agric. Food Chem. 64, 6772–6782. [DOI] [PubMed] [Google Scholar]
- Nowak, M. and Hammerschmidt, M. (2006) Ubc9 regulates mitosis and cell survival during zebrafish development. Mol. Biol. Cell, 17, 5324–5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odenbach, D. , Breth, B. , Thines, E. , Weber, R.W. , Anke, H. and Foster, A.J. (2007) The transcription factor Con7p is a central regulator of infection‐related morphogenesis in the rice blast fungus Magnaporthe grisea . Mol. Microbiol. 64, 293–307. [DOI] [PubMed] [Google Scholar]
- Oh, Y. , Franck, W.L. , Han, S.‐O. , Shows, A. , Gokce, E. , Muddiman, D.C. and Dean, R.A. (2012) Polyubiquitin is required for growth, development and pathogenicity in the rice blast fungus Magnaporthe oryzae . PLoS One, 7, e42868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, H.J. , Kim, W.Y. , Park, H.C. , Lee, S.Y. , Bohnert, H.J. and Yun, D.J. (2011) SUMO and SUMOylation in plants. Mol. Cells, 32, 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasupala, N. , Easwaran, S. , Hannan, A. , Shore, D. and Mishra, K. (2012) The SUMO E3 ligase Siz2 exerts a locus‐dependent effect on gene silencing in Saccharomyces cerevisiae . Eukaryot. Cell, 11, 452–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash, C. , Manjrekar, J. and Chattoo, B.B. (2016) Skp1, a component of E3 ubiquitin ligase, is necessary for growth, sporulation, development and pathogenicity in rice blast fungus (Magnaporthe oryzae). Mol. Plant Pathol. 17, 903–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raorane, M.L. , Mutte, S.K. , Varadarajan, A.R. , Pabuayon, I.M. and Kohli, A. (2013) Protein SUMOylation and plant abiotic stress signaling: in silico case study of rice RLKs, heat‐shock and Ca2+‐binding proteins. Plant Cell Rep. 32, 1053–1065. [DOI] [PubMed] [Google Scholar]
- Rosonina, E. , Akhter, A. , Dou, Y. , Babu, J. and Sri Theivakadadcham, V.S. (2017) Regulation of transcription factors by sumoylation. Transcription, 8, 220–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saracco, S.A. , Miller, M.J. , Kurepa, J. and Vierstra, R.D. (2007) Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol. 145, 119–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, S.E. , Matuschewski, K. , Liakopoulos, D. , Scheffner, M. and Jentsch, S. (1998) The ubiquitin‐like proteins SMT3 and SUMO‐1 are conjugated by the UBC9 E2 enzyme. Proc. Natl. Acad. Sci. USA, 95, 560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shayeghi, M. , Doe, C.L. , Tavassoli, M. and Watts, F.Z. (1997) Characterisation of Schizosaccharomyces pombe rad31, a UBA‐related gene required for DNA damage tolerance. Nucleic Acids Res. 25, 1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H.‐B. , Chen, G.‐Q. , Chen, Y.‐P. , Dong, B. , Lu, J.‐P. , Liu, X.‐H. and Lin, F.‐C. (2016) MoRad6‐mediated ubiquitination pathways are essential for development and pathogenicity in Magnaporthe oryzae . Environ. Microbiol. 18, 4170–4187. [DOI] [PubMed] [Google Scholar]
- Sriramachandran, A. M. and Dohmen, R. J. (2014) SUMO‐targeted ubiquitin ligases. Biochim. Biophys. Acta, 1843, 75–85. [DOI] [PubMed] [Google Scholar]
- Stamatakis, A. (2014) RAxML version 8: a tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics, 30, 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, K. , Nishide, J. , Okazaki, K. , Kato, H. , Niwa, O. , Nakagawa, T. , Matsuda, H. , Kawamukai, M. and Murakami, Y. (1999) Characterization of a fission yeast SUMO‐1 homologue, pmt3p, required for multiple nuclear events, including the control of telomere length and chromosome segregation. Mol. Cell. Biol. 19, 8660–8672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham, M.H. , Jaffray, E. , Vaughan, O.A. , Desterro, J.M. , Botting, C.H. , Naismith, J.H. and Hay, R.T. (2001) Polymeric chains of SUMO‐2 and SUMO‐3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276, 35 368–35 374. [DOI] [PubMed] [Google Scholar]
- Truong, K. , Lee, T.D. , Li, B. and Chen, Y. (2012) Sumoylation of SAE2 C terminus regulates SAE nuclear localization. J. Biol. Chem. 287, 42 611–42 619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent, B. (1990) Rice blast as a model system for plant pathology. Phytopathology, 80, 33–36. [Google Scholar]
- Valent, B. , Farrall, L. and Chumley, F.G. (1991) Magnaporthe grisea genes for pathogenicity and virulence identified through a series of backcrosses. Genetics, 127, 87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H.L. , Kim, S.H. , Siu, H. and Breuil, C. (1999) Transformation of sap staining fungi with hygromycin B resistance plasmids pAN 7‐1 and pCB1004. Mycol. Res. 103, 77–80. [Google Scholar]
- Watts, F.Z. (2013) Starting and stopping SUMOylation. What regulates the regulator? Chromosoma, 122, 451–463. [DOI] [PubMed] [Google Scholar]
- Wong, K.H. , Todd, R.B. , Oakley, B.R. , Oakley, C.E. , Hynes, M.J. and Davis, M.A. (2008) Sumoylation in Aspergillus nidulans: sumO inactivation, overexpression and live‐cell imaging. Fungal Genet. Biol. 45, 728–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. , Zhao, X. , Sun, J. , Kang, Z. , Ding, S. , Xu, J.‐R. and Peng, Y.L. (2010) A novel protein Com1 is required for normal conidium morphology and full virulence in Magnaporthe oryzae . Mol. Plant–Microbe Interact. 23, 112–123. [DOI] [PubMed] [Google Scholar]
- Yun, S.‐H. (1998). Molecular genetics and manipulation of pathogenicity and mating determinants in Mycosphaerella zeae‐maydis and Cochliobolus heterostrophus. Ithaca, NY: Cornell University.
- Zhao, Q. , Xie, Y. , Zheng, Y. , Jiang, S. , Liu, W. , Mu, W. , Liu, Z. , Zhao, Y. , Xue, Y. and Ren, J. (2014) GPS‐SUMO: a tool for the prediction of sumoylation sites and SUMO‐interaction motifs. Nucleic Acids Res. 42, W325–W330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X. , Mehrabi, R. and Xu, J.‐R. (2007) Mitogen‐activated protein kinase pathways and fungal pathogenesis. Eukaryot. Cell, 6, 1701–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z. , Li, G. , Lin, C. and He, C. (2009) Conidiophore stalk‐less1 encodes a putative zinc‐finger protein involved in the early stage of conidiation and mycelial infection in Magnaporthe oryzae . Mol. Plant–Microbe Interact. 22, 402–410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Phylogenetic trees of SUMOylation components in fungi and model organisms. Phylogenetic trees of SUMOylation components in fungi and model organisms that were homologous to Saccharomyces cerevisiae SUMOylation components were constructed by the maximum‐likelihood method with 1000 bootstraps and the LG or BLOSUM62 protein substitution method. The sequences used for alignments were: (A) PF11976 for SUMO, SMT3; (B) PF00899 for E1, AOS1 and UBA2; (C) PF00179 for E2, UBC9; (D) PF02891 for E3, SIZ1 and SIZ2; (E) PF11789 for E3, MMS21; (F) PF02902 for protease, ULP1 and ULP2; (G) PF08325 for protease and WSS1. Clades that contained more than three species of the same fungal phylum, animal and plant are shown without genus/species names. The yeast components are marked with a star and Magnaporthe oryzae homologues are shown in bold. Only bootstrap values >50% are shown.
Fig. S2 Southern blot analysis and reverse transcription‐polymerase chain reaction (RT‐PCR) of the deletion mutants. Genomic DNA of wild‐type (WT) and deletion mutants was extracted and digested with PstI, SalI, HindIII or XhoI. The upstream or downstream construct of each gene was used as a probe for Southern blot analysis. Complementary DNA was synthesized from total RNA. β‐tubulin was used for normalization.
Fig. S3 SUMOylation in wild‐type (WT) and ΔMoaos1 during conidiation. Protein extract from the WT during conidiation was separated by sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and subjected to Western blot analysis using an anti‐haemagglutinin (HA) antibody.
Fig. S4 SUMOylated proteins in wild‐type (WT) under oxidative stress. Protein extract from the WT treated with oxidative stress (100 mm H2O2) was separated by sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and subjected to Western blot analysis using an anti‐haemagglutinin (HA) antibody.
Fig. S5 Ubiquitination in the wild‐type (WT), ΔMosmt3, ΔMoaos1, ΔMouba2 and ΔMoubc9. Protein extracts of WT, ΔMosmt3, ΔMoaos1, ΔMouba2 and ΔMoubc9 were separated by sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and subjected to Western blot analysis using an anti‐ubiquitin (P4D1) antibody. Ubiquitination was not affected in all of the deletion mutants.
Fig. S6 Defective conidiogenesis was recovered in complemented strains. Conidiogenesis on conidiophores was observed under a microscope. Scale bar, 100 µm.
Fig. S7 Mycelial growth under stress conditions. Strains were inoculated on nutrient starvation stress [minimal agar medium (MMA)], DNA damage stress [10 mm hydroxyurea (HU) and 0.05% methyl methanesulfonate (MMS)] and oxidative stress [5 mm H2O2 and 3 mm methyl viologen (MV)]. Mycelial growth was measured 9 days after inoculation. CMA, complete agar medium.
Fig. S8 Growth of invasive hyphae (IH) in the complemented strains. (A) Conidial suspensions (2 × 104/mL) of the complemented strains were inoculated onto 6‐week‐old rice sheath cells. IH growth was observed under a microscope at 48 h post‐inoculation (hpi). Scale bar, 50 μm. (B) Growth of IH into neighbouring cells was classified into three types: Type I for IH restricted to the primary infection cell; Type II for IH growth to adjacent cells; Type III for extensive growth of IH over adjacent cells.
Fig. S9 Penetration assay of the complemented strains. Conidial suspensions (2 × 104/mL) of the complemented strains were inoculated onto 6‐week‐old rice sheath cells. Penetration by appressoria at 24 h post‐inoculation (hpi) was observed under a microscope. Scale bar, 50 μm.
Fig. S10 Intracellular localization of SUMOylation components in Magnaporthe oryzae. MoAOS1, MoUBA2, MoSMT3 and MoUBC9 were predominantly localized in the nuclei during conidial germination and appressorium formation. Scale bar, 50 μm.
Table S1 Domains of Saccharomyces cerevisiae SUMOylation components.
Table S2 The number of Saccharomyces cerevisiae homologues of SUMOylation components in the selected species.
Table S3 The closest Saccharomyces cerevisiae homologues of SUMOylation components in Magnaporthe oryzae.
Table S4 Developmental phenotypes of the wild‐type (WT), deletion mutants and complemented strains.
Table S5 Conidial germination and appressorium formation of the wild‐type (WT), deletion mutants and complemented strains after incubation for 2, 8 and 24 h.
Table S6 List of the strains used in this study.
Table S7 Primer sequences used in this study.


