Summary
Tandem CCCH zinc finger (TZnF) proteins have been implicated in plant defence, but their role in pepper (Capsicum annuum) is unclear. In the present study, the role of CaC3H14, a pepper TZnF protein, in the immune response of pepper plants to Ralstonia solanacearum infection was characterized. When fused to the green fluorescent protein, CaC3H14 was localized exclusively to the nuclei in leaf cells of Nicotiana benthamiana plants transiently overexpressing CaC3H14. Transcript abundance of CaC3H14 was up‐regulated by inoculation with R. solanacearum. Virus‐induced silencing of CaC3H14 increased the susceptibility of the plants to R. solanacearum and down‐regulated the genes associated with the hypersensitive response (HR), specifically HIR1 and salicylic acid (SA)‐dependent PR1a. By contrast, silencing resulted in the up‐regulation of jasmonic acid (JA)‐dependent DEF1 and ethylene (ET) biosynthesis‐associated ACO1. Transient overexpression of CaC3H14 in pepper triggered an intensive HR, indicated by cell death and hydrogen peroxide (H2O2) accumulation, up‐regulated PR1a and down‐regulated DEF1 and ACO1. Ectopic overexpression of CaC3H14 in tobacco plants significantly decreased the susceptibility of tobacco plants to R. solanacearum. It also up‐regulated HR‐associated HSR515, immunity‐associated GST1 and the SA‐dependent marker genes NPR1 and PR2, but down‐regulated JA‐dependent PR1b and ET‐dependent EFE26. The CaC3H14 promoter and was bound and its transcription was up‐regulated by CaWRKY40. Collectively, these results indicate that CaC3H14 is transcriptionally targeted by CaWRKY40, is a modulator of the antagonistic interaction between SA and JA/ET signalling, and enhances the defence response of pepper plants to infection by R. solanacearum.
Keywords: Capsicum annuum, CCCH‐type zinc finger, immunity, Ralstonia solanacearum
Introduction
Through the co‐evolution of plants and their potential pathogens, plants have developed efficient defences against micro‐organisms, enabling them to effectively limit pathogen infection. Many of the genes and proteins associated with plant defence modify the expression patterns of the plant's transcriptomes and proteomes, thereby conferring resistance to specific pathogens (Attard et al., 2014; Dörmann et al., 2014; Grewal et al., 2012; Laura et al., 2015; Rudd et al., 2015). A zag–zig–zag model has been established that consists of two layers of immunity: effector‐triggered immunity (ETI) and pathogen‐associated molecular pattern (PAMP)‐triggered immunity (PTI) (Hein et al., 2009; Jones and Dangl, 2006). The difference between PTI and ETI is quantitative, and they share overlapping signalling mechanisms, including Ca2+, reactive oxygen species, hormones [e.g. salicylic acid (SA), jasmonic acid (JA) and ethylene (ET)], mitogen‐activated protein kinase (MAPK) cascades and various transcription factors (TFs). However, the mechanism by which plants perceive stress and translate the signals into an appropriate defence response is unknown. This is particularly true in plants that do not respond in a manner similar to well‐studied model systems.
Plant defences appear to be primarily regulated transcriptionally through the action of various TFs (Riechmann and Ratcliffe, 2000; Wray, 2003). The action of these TFs may interconnect, creating transcriptional networks (Garner et al., 2016; Moore et al., 2011; Tsuda and Somssich, 2015; Wang et al., 2006). Zinc finger (Znf) proteins are a large and diverse family of TFs, characterized by the Znf motif. The protein motif consists of cysteine (C) or histidine (H) residues coordinated with several zinc ions (Hall, 2005). Znf proteins have been classified into several different types based on the number and order of the C and H residues binding the zinc ions. These types include C2H2, C2C2, C2HC, C2C2C2C2, C2HCC2C2 and C3H1 (also called CCCH) (Berg and Shi, 1996; Mackay and Crossley, 1998; Moore and Ullman, 2003; Schumann et al., 2007). The CCCH Znf family is unique because it regulates gene expression by direct binding of mRNA or DNA (Li et al., 2001; Pomeranz et al., 2010; Wang D et al., 2008) and possesses nuclease activity (Addepalli and Hunt, 2008). Other Znf families regulate gene expression with the aid of DNA‐binding or protein‐binding proteins. Genome‐wide annotation analyses identified 67 genes coding for CCCH Znf proteins in rice (Oryza sativa), 68 in Arabidopsis (Wang D et al., 2008), 34 in Medicago truncatula (Dang et al., 2013), 68 in maize (Peng et al., 2012) and 91 in Populus (Chai et al., 2012). The CCCH family can be divided into seven groups based on phylogenetic analysis (Peng et al., 2012). Members of the CCCH family appear to be involved in many aspects of plant regulation, including growth (von Saint Paul et al., 2011), development (Grabowska et al., 2009), plant architecture (Wang L et al., 2008) and plant immunity (Deng et al., 2012; Guo et al., 2009; Lee et al., 2013). The proteins involved in these aspects of plant physiology include PEI1, AtSZF1/AtSZF2 (Sun et al., 2007), SOMNUS (Wang L et al., 2008), AtTZF1, AtOZF1 (Huang et al., 2011) and AtOZF2 in Arabidopsis; OsDOS (Kong et al., 2006), OsTZF1, Ehd4, OsGZF1 and OsLIC (Wang L et al., 2008) in rice; GhZFP1 in cotton (Guo et al., 2009); and CsSEF1 in cucumber (Grabowska et al., 2009). However, the majority of the members in this family remain functionally unidentified, especially in non‐model plants.
Another important group of plant TFs is the WRKY family. Its members have one or two highly conserved WRKY domain(s) and a Znf‐like motif (Ulker and Somssich, 2004). These conserved WRKY domains recognize and bind the highly conserved W‐box (TTGACC/T) (Eulgem et al., 2000); however, this binding can be affected by the flanking sequences of the W‐box (Agarwal et al., 2011; Ciolkowski et al., 2008). DNA binding allows these TFs to act as regulators of plant immunity (Buscaill and Rivas, 2014; Garner et al., 2016; Pandey and Somssich, 2009). The W‐box is highly enriched in promoters of immunity‐associated genes, including genes that encode various TFs, indicating that WRKY TFs may fulfil their function by complexing with various TFs to form different transcriptional pathways or networks (Birkenbihl et al., 2017; Liu S et al., 2015). However, the precise position of most of the WRKY TFs in the transcriptional networks is unknown.
Pepper (Capsicum annuum) is an important vegetable worldwide and a prototypical member of the family Solanaceae. It is susceptible to several soil‐borne pathogens, specifically Ralstonia solanacearum and Phytophthora capsici, the causal agents of bacterial wilt and Phytophthora blight, respectively. In pepper production, these diseases frequently cause heavy yield losses. Plant breeding is a powerful tool for the development of crop varieties with enhanced performance and superior traits. A better understanding of the molecular mechanisms underlying plant immunity could lead to genetic strategies to improve plant disease resistance. However, few genes have been functionally characterized in pepper, and the molecular mechanisms of pepper immunity have not been characterized completely. In a previous study, we concluded that, in pepper, CaWRKY40 promotes an immune response to R. solanacearum infection, but we did not identify its mechanism of action. Here, we have obtained a full‐length cDNA of a CCCH‐type gene, designated CaC3H14, through random sequencing of a normalized cDNA library from pepper. Our data are consistent with the hypothesis that CaC3H14 is directly targeted and transcriptionally regulated by CaWRKY40, and that CaC3H14 enhances pepper resistance to R. solanacearum infection.
Results
Cloning and sequence analysis of CaC3H14 cDNA
Tandem CCCH zinc finger (TZnF) proteins have been implicated in plant growth and development, as well as plant defence responses. To date, no TZnF protein has been functionally characterized in pepper. Using cDNA‐amplified fragment length polymorphism, a transcript‐derived fragment (TDF), which was transcriptionally up‐regulated by inoculation with R. solanacearum and contained two conserved CCCH domains (data not shown), was identified, and its corresponding full‐length cDNA was cloned from a cDNA library. The cDNA was 1860 bp in length and harboured an open reading frame (ORF) that encoded a protein with three Znf CCCH domains. We designated it CaC3H14, because its amino acid sequence had the highest similarity (68%–83%) to the protein encoded by the C3H14 gene when compared with other members in C. annuum and several known CCCH subfamily members from Solanum lycopersicum and Nicotiana attenuata (Fig. S1, see Supporting Information).
Expression pattern of CaC3H14 in pepper plants
To confirm CaC3H14 was induced following R. solanacearum infection, plants were inoculated with the highly pathogenic R. solanacearum strain FJC100301. Infected leaves were harvested to isolate total RNA. CaC3H14 transcript levels were assessed using real‐time reverse transcription‐polymerase chain reaction (RT‐PCR). Figure 1 shows that CaC3H14 transcription is significantly enhanced following inoculation with the pathogen compared with control plants. Transcription of CaPR1a, an SA‐dependent pathogenesis‐related (PR) gene in plants, is also induced. Expression of a homologous gene is up‐regulated during infection of Nicotiana benthamiana by R. solanacearum (Zhang et al., 2012).
Figure 1.
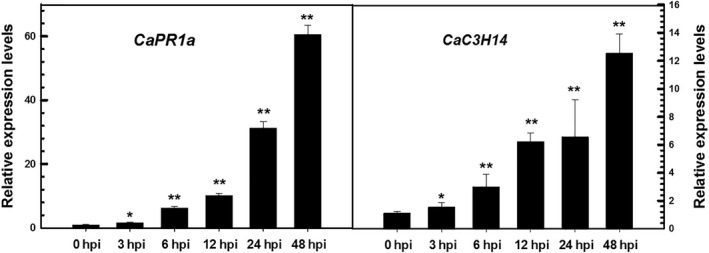
Relative expression levels of CaC3H14 and CaPR1a in pepper plants infected with Ralstonia solanacearum at various hours post‐inoculation (hpi) from quantitative real‐time polymerase chain reaction (PCR) analysis. The relative transcript levels of CaC3H14 were compared with those in control plants, which were assigned as unity. CaPR1a was used as a positive control. Data represent the mean ± standard deviation (SD) of two independent experiments, each with three replicates (n = 6). *P < 0.05; **P < 0.01 (Student–Newman–Keuls test).
Subcellular localization of CaC3H14
The subcellular localization of a protein usually provides a clue to its function, and so we identified the subcellular localization of CaC3H14. We transiently overexpressed (OE) a CaC3H14‐green fluorescence protein (GFP) fusion protein in epidermal cells of N. benthamiana leaves. GFP images were visualized using a confocal microscope. CaC3H14‐GFP was exclusively observed in the nuclei based on co‐localization with 4,6‐diamidino‐2‐phenylindole (DAPI) staining (Fig. 2). Thus, we concluded that CaC3H14 was localized to the nucleus.
Figure 2.
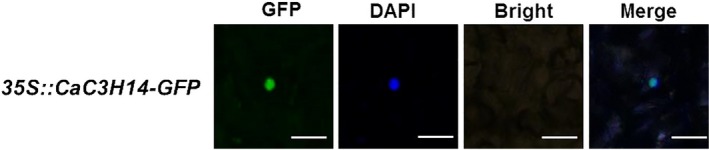
Subcellular localization of CaC3H14. Nicotiana benthamiana leaves were infiltrated with cells of Agrobacterium tumefaciens strain GV3101 containing the 35S::CaC3H14‐GFP construct. Fluorescence of the green fluorescent protein (GFP) was detected 48 h after inoculation. Bar, 50 μm. DAPI, 4,6‐diamidino‐2‐phenylindole.
Silencing of CaC3H14 enhances susceptibility of pepper to R. solanacearum
To further study the role of CaC3H14 in the response to infection by R. solanacearum strain FJC100301, we performed a loss‐of‐function experiment in pepper seedlings using virus‐induced gene silencing (VIGS) of CaC3H14. Quantitative PCR analysis showed that CaC3H14 was effectively silenced in TRV::CaC3H14 pepper plants. The number of CaC3H14 transcripts in TRV::CaC3H14 (CaC3H14‐silenced) plants was reduced to approximately 30% of that in TRV::00 (control) plants, and CaC3H14 transcript levels in R. solanacearum‐infected CaC3H14‐silenced plants were approximately 5%–15% of those in R. solanacearum‐infected control plants at 24 and 48 h post‐inoculation (hpi; Fig. 3a). Ralstonia solanacearum replication was significantly enhanced in CaC3H14‐silenced pepper plants compared with that in control plants (Fig. 3b). When roots were inoculated with the pathogen, CaC3H14‐silenced plants consistently showed more severe disease symptoms than control plants (Fig. 3c). A dynamic disease index of inoculated control and CaC3H14‐silenced plants, performed from 6 to 12 days post‐inoculation (dpi), confirmed the increased bacterial growth in CaC3H14‐silenced plants relative to that in control plants (Fig. 3d). Quantitative RT‐PCR was performed to evaluate the expression levels of defence‐related genes in inoculated, CaC3H14‐silenced plants. CaC3H14 silencing significantly down‐regulated the expression of hypersensitive cell death marker genes, such as CaHIR1, and the immunity‐associated gene CaPR1a, whereas it up‐regulated the expression of the defence‐related genes CaDEF1 and CaACO1 (Fig. 3e). We concluded that CaC3H14 silencing enhanced the susceptibility of pepper to R. solanacearum.
Figure 3.
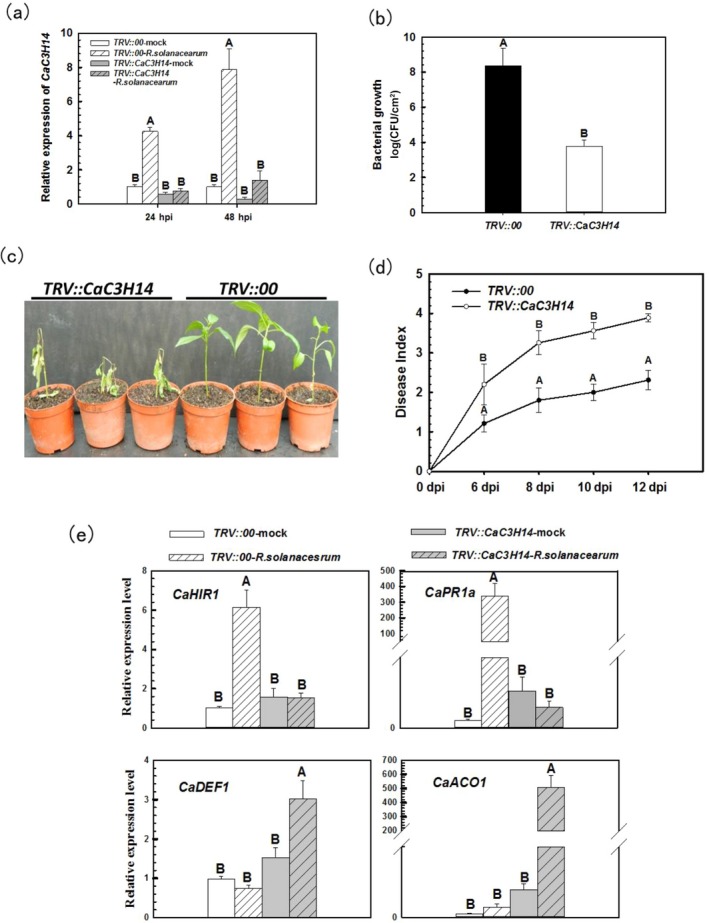
Virus‐induced gene silencing (VIGS) of CaC3H14 enhanced the susceptibility of pepper plants to Ralstonia solanacearum. (a) The silencing efficiency of CaC3H14 in TRV::CaC3H14 plants, with or without inoculation, detected by real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) at 24 and 48 h post‐inoculation (hpi). The relative transcription level of CaC3H14 in TRV::CaC3H14 plants was compared with that in mock‐treated TRV::00 plants, which was set as unity. (b) Ralstonia solanacearum growth in inoculated CaC3H14‐silenced or control plants at 48 hpi. CFU, colony‐forming unit. (c) Disease symptoms of TRV::00 and TRV::CaC3H14 plants at 6 days post‐inoculation (dpi) by root irrigation. (d) Dynamic disease index of TRV::00 and TRV::CaC3H14 pepper plants from 6 to 12 dpi; data represent the mean ± standard deviation (SD) of three biological replicates each comprising five plants. (e) The effect of CaC3H14 silencing on the relative expression of defence‐related genes in inoculated plants at 24 hpi. The relative transcription of immunity‐associated marker genes in R. solanacearum‐inoculated TRV::00 and TRV::CaC3H14 and mock‐treated TRV::CaC3H14 plants was compared with that in mock‐treated TRV::00 plants, which was set as unity. In (a), (b), (d) and (e), data represent the mean ± SD of two independent experiments, each with three replicates (n = 6). Different letters indicate significant difference as determined by Student–Newman–Keuls test (P < 0.01).
Transient overexpression of CaC3H14 induces cell death and defence‐related gene expression in pepper plants
As transient overexpression of Myc‐ or HA‐tagged immunity‐associated proteins has been frequently employed to define the roles of these proteins in plant immunity(Choi and Hwang, 2011; Kim et al., 2014), we used an HA‐tagged transient overexpression approach to define the role of CaC3H14 in pepper immunity. GV3101 cells containing 35S::CaC3H14‐HA or 35S::00 were infiltrated into pepper leaves. Transient CaC3H14 overexpression was detected by immunoblotting against human influenza haemagglutinin antibody (HA; data not shown) (Fig. S3, see Supporting Information). Overexpression of CaC3H14‐HA visibly triggered cell death after 4 dpi, whereas controls did not (Fig. 4a, left panel). Hypersensitive response (HR)‐associated cell death was assessed by staining with trypan blue to identify necrotic cells. Control leaves produced a weak to non‐existent HR‐mediated necrotic response, whereas leaves transiently overexpressing CaC3H14‐HA clearly induced a necrotic response (Fig. 4a, right panel). Hydrogen peroxide (H2O2) production was detected using 3,3′‐diaminobenzidine (DAB) staining. There was visible DAB staining in the leaves transiently expressing CaC3H14‐HA, but not in control leaves (Fig. 4a, middle panel). We also performed an ion leakage test to analyse cell necrosis severity caused by plasma membrane damage in leaves transiently overexpressing CaC3H14‐HA (Fig. 4b). There was more ion leakage in leaves transiently overexpressing CaC3H14‐HA compared with that in control leaves at 48 and 72 h after agro‐infiltration with GV3101 cells.
Figure 4.
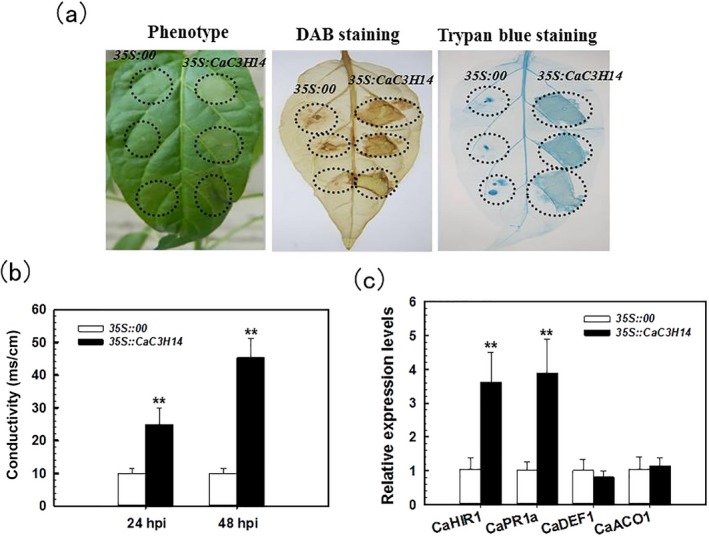
Transient overexpression of CaC3H14 induced cell death and defence‐related gene expression in pepper plants. (a) Photographs of pepper leaves which had been infiltrated with Agrobacterium tumefaciens strain GV3101 carrying the 35S::00 (empty vector) or 35S::CaC3H14 construct. Left panel: infiltrated pepper leaves at 4 days post‐inoculation (dpi); middle panel: after staining with 3,3′‐diaminobenzidine (DAB) to detect hydrogen peroxide; right panel: after staining with trypan blue to assess cell death. (b) Electrolyte leakage of pepper leaves measured by conductivity after agro‐infiltration with A. tumefaciens strain GV3101 cells containing the 35S::00 or 35S::CaC3H14 construct. (c) Relative expression levels of defence‐related genes, assessed by quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) analysis, in leaves transiently overexpressing 35S::CaC3H14 or 35S::00 at 24 h post‐inoculation (hpi). The transcript levels of defence‐related genes in CaC3H14 transient overexpressing leaves were compared with those in control plants, which were assigned as unity. In (b) and (c), data represent the mean ± standard deviation (SD) of two independent experiments, each with three replicates (n = 6). **P < 0.01 (Student–Newman–Keuls test).
To test whether transient overexpression of CaC3H14 could alter the expression of defence‐related genes in pepper plants, we examined the transcript levels of CaHIR1, CaPR1a, CaDEF1 and CaACO1. The relative transcript levels of CaHIR1 and CaPR1a were significantly higher in CaC3H14 transiently overexpressing leaves than in control leaves, whereas the relative transcript levels of CaDEF1 and CaACO1 were not significantly changed (Fig. 4c).
Ectopic overexpression of CaC3H14 enhances resistance of tobacco to R. solanacearum attack
To confirm the role of CaC3H14 in plant immunity, transgenic tobacco plants overexpressing CaC3H14 were generated, and the effect of ectopic CaC3H14 overexpression on tobacco immunity was assayed. Nineteen independent T0 transgenic tobacco lines constitutively expressing CaC3H14 driven by the CaMV35S promoter were acquired, and their corresponding T1 and T2 lines were generated. No significant altered growth and development were found between the plants of the acquired T2 lines and wild‐type (WT) plants. Two T2 transgenic lines, CaC3H14‐OE‐2 and CaC3H14‐OE‐8, with average numbers of CaC3H14 transcripts and normal growth and development were selected for further analysis. The two transgenic plant lines and WT plants were inoculated with R. solanacearum strain FJC100301 using root irrigation. At 7 dpi, we observed wilting symptoms in all three types of plant, but these symptoms were more severe in WT plants than in the two transgenic lines (Fig. 5a). Bacterial growth in the inoculated leaves of the plants was assessed by measuring the number of colony‐forming units (CFUs) at 36 hpi. At this time point, there were significantly fewer CFUs in transgenic plants than in WT plants (Fig. 5b). The disease index was assessed from 6 to 12 dpi and was significantly higher in WT plants than in either transgenic line (Fig. 5c).
Figure 5.
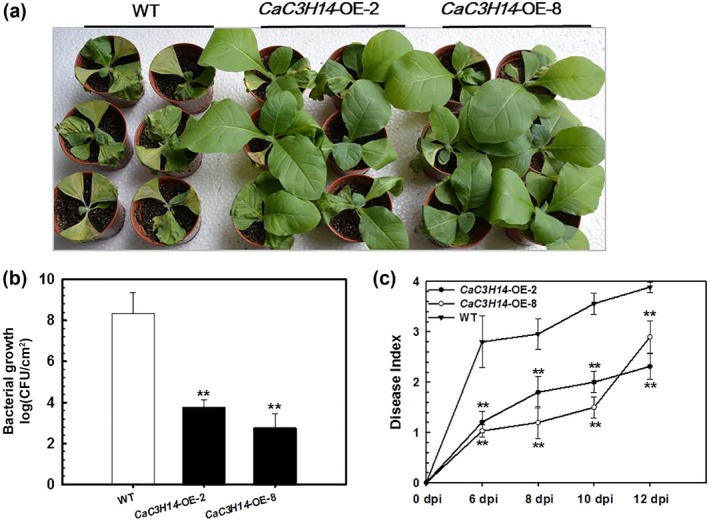
Ectopic overexpression of CaC3H14 decreased susceptibility of the transgenic tobacco plants to Ralstonia solanacearum strain FJC100301 compared with wild‐type (WT) plants. (a) Disease symptoms of R. solanacearum‐inoculated tobacco plants at 7 days post‐inoculation (dpi) by root irrigation. (b) Growth of R. solanacearum in the third leaves of R. solanacearum‐inoculated plants of transgenic lines (CaC3H14‐OE‐2 and CaC3H14‐OE‐8) and WT control at 36 h post‐inoculation (hpi). (c) Dynamic disease index of plants inoculated with R. solanacearum using root irrigation, from 6 to 12 dpi. Data represent the mean ± standard deviation (SD) of three biological replicates, each comprising five plants. **P < 0.01 (Student–Newman–Keuls test).
To confirm the effects of CaC3H14 overexpression observed phenotypically in infected plants and to investigate the possible modes of action, we measured the transcription of defence‐related genes in CaC3H14‐OE transgenic and WT tobacco plants using quantitative RT‐PCR. We examined the transcript levels of HR‐associated genes NtHSR515 (Czernic et al., 1996) and NtGST1 (Peng et al., 2004), the SA‐responsive genes NtNPR1 (Ghanta et al., 2011) and NtPR2, the JA‐responsive gene PR1b (Sohn et al., 2007) and the ET‐associated gene NtEFE26 (Sohn et al., 2007). There were significantly more transcripts of the HR‐associated genes NtHSR515 and NtGST1 in CaC3H14‐OE transgenic plants than in WT plants, whereas the numbers of NtNPR1, NtPR2, NtPR1b and NtEFE26 transcripts were not significantly different between transgenic and WT plants. By contrast, there were fewer NtPR1b transcripts in transgenic plants than WT plants (Fig. 6).
Figure 6.
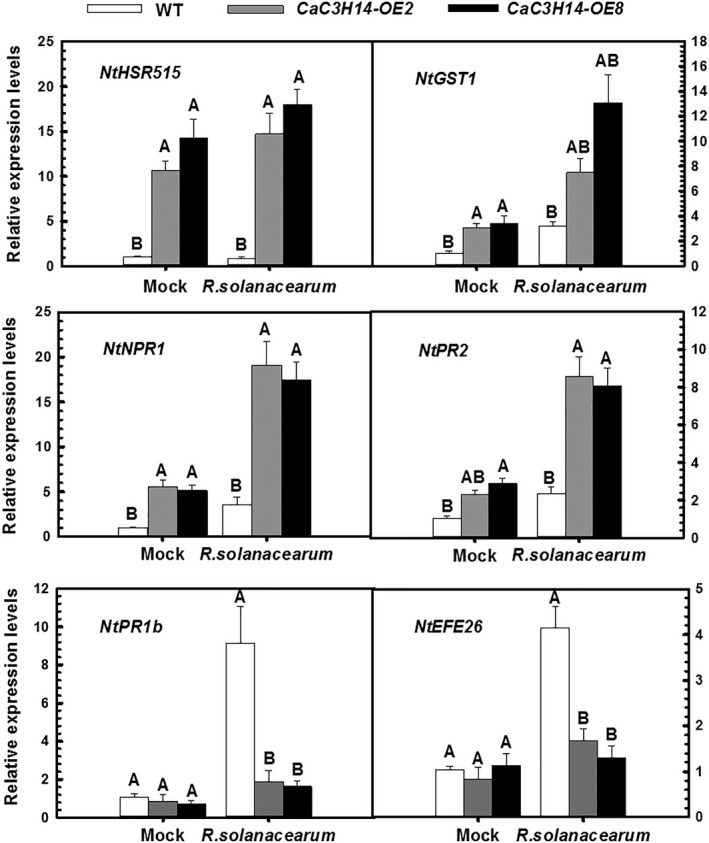
Relative expression of defence‐related genes in transgenic tobacco lines overexpressing CaC3H14 or wild‐type (WT) tobacco plants, assessed using quantitative reverse transcription‐polymerase chain reaction (RT‐PCR). Plants were inoculated with Ralstonia solanacearum using root irrigation and assessed 48 h later. Controls were mock‐treated WT plants and transgenic plants. Expression in the former was set to unity. Data represent the mean ± standard deviation (SD) of two independent experiments, each with three replicates (n = 6). Different letters indicate significant difference as determined by Student–Newman–Keuls test (P < 0.01).
We also evaluated the transcript levels of these defence‐related genes in tobacco plants 48 h after inoculation with R. solanacearum. Transcription of NtHSR515, NtGST1, NtNPR1 and NtPR2 in WT plants was up‐regulated to different degrees, and up‐regulation was potentiated by CaC3H14 overexpression in transgenic plants. Transcription of NtPR1b and NtEFE26 was significantly up‐regulated in inoculated WT plants, but CaC3H14 overexpression significantly reduced the up‐regulation in inoculated transgenic plants (Fig. 6).
Chromatin immunoprecipitation (ChIP) analysis of CaWRKY40 binding to the CaC3H14 promoter
As both CaC3H14 and CaWRKY40 are transcriptionally up‐regulated following R. solanacearum infection, and three typical W‐boxes were identified in the promoter region of CaC3H14 (Fig. 7a), we hypothesized that CaWRKY40 acts as a regulator of CaC3H14. We performed ChIP to determine if CaWRKY40 binds the CaC3H14 promoter. Cells of Agrobacterium tumefaciens strain GV3101 containing the 35S::CaWRKY40‐HA or 35S::00 construct were infiltrated into pepper leaves, and the leaves were harvested 48 h later for chromatin isolation. The isolated chromatin was randomly sheared into fragments of 300 − 500 bp, and the fragments bound to the target proteins were immunoprecipitated with HA. The resulting DNA fragments were used as templates for PCR analysis with specific primer pairs. The results showed that only PW3 produced amplified product from immunoprecipitated DNA template derived from CaWRKY40‐HA transiently overexpressing pepper leaves; however, PW1–2 and the control primer pair (CP), which was designed as a negative control based on a promoter region without a W‐box, did not amplify any product from the immunoprecipitated DNA template derived from CaWRKY40‐HA (Fig. 7a,b), indicating that the W3‐box might be responsible for CaWRKY40 targeting. This result was further confirmed by microscale thermophoresis (MST) in solution; the K d value of binding of the DNA fragment to CaWRKY40 was 8.8142E‐07, whereas that of the GFP control was 2.8E‐08 (Fig. 7c); all of these data suggest the direct targeting of CaWRKY40 to CaC3H14, probably via the W3‐box. To test the effect of R. solanacearum inoculation on the binding of CaWRKY40 to the CaC3H14 promoter, ChIP analysis and real‐time PCR were performed following infection. The results showed that the CaC3H14 promoter of infected pepper leaves was more enriched with CaWKRY40 than that of mock‐treated control plants (Fig. 7d). To test the specificity or selectivity of the targeting of CaWRKY40 to CaC3H14, the binding of CaWRKY27 and CaWRKY58, the two other WRKY TFs implicated in pepper immunity by our previous studies (Dang et al., 2014; Wang et al., 2013), to the promoter of CaC3H14 was tested by ChIP analysis. The results showed that the primer pair PW3 amplified clear product from immunoprecipitated DNA templates derived from CaWRKY40‐HA, but not from CaWRKY27‐HA or CaWRKY58‐HA transiently overexpressing pepper leaves, indicating the selectivity of targeting of CaC3H14 by CaWRKY40 (Fig. 7e).
Figure 7.
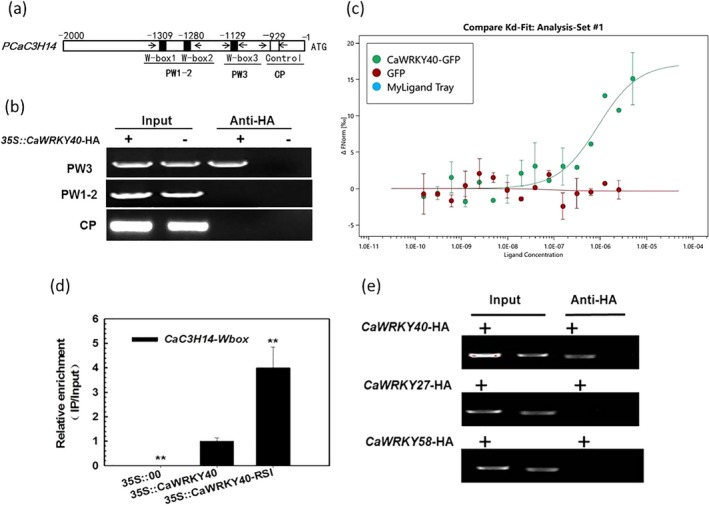
CaC3H14 is directly targeted and transcriptionally regulated by CaWRKY40. (a) The distribution of W‐boxes in the CaC3H14 promoter and the primer pairs used in chromatin immunoprecipitation (ChIP) analyses. PW1–2, PW3, primer pairs of the fragment containing W‐box 1 and 2, which are close to each other, and W‐box 3, respectively. CP, control primer pair (negative control), which was designed based on a region without a W‐box. (b) Assay of binding of CaWRKY40 to the CaC3H14 promoter determined by ChIP‐polymerase chain reaction (PCR) with different specific primer pairs. Pepper leaves were inoculated with Agrobacterium tumefaciens strain GV3101 cells containing the construct of 35S::CaWRKY40‐HA and harvested 48 h after inoculation. The immunoprecipitated DNA was used as a template for PCR. (c) Interaction of CaWRKY40 with the promoter fragment of CaC3H14 by microscale thermophoresis in solution. Data represent the mean ± standard deviation (SD) of three replicates. (d) The binding of CaWRKY40 to the CaC3H14 promoter was enhanced by Ralstonia solanacearum inoculation. Pepper leaves were inoculated with Agrobacterium tumefaciens strain GV3101 cells containing a construct of 35S::CaWRKY40‐HA; at 24 h post‐inoculation, they were inoculated with R. solanacearum, and 24 h later they were harvested. The PW3 primer pair was used for real‐time PCR; the data represent the mean ± SD of two independent experiments, each with three replicates (n = 6). **P < 0.01 (Student–Newman–Keuls test). (e) Assay of binding of CaWRKY27 and CaWRKY58 to the CaC3H14 promoter determined by ChIP‐PCR with different specific primer pairs using CaWRKY40 as positive control; the Anti‐HA+ derived from CaWRKY27 or CaWRKY58 transient overexpressing leaves was used as template with PW3 as the primer pair for PCR. Input, total DNA–protein complex; HA, human influenza haemagglutinin; Anti‐HA+, DNA–protein complex immunoprecipitated with anti‐HA antibody (α‐HA).
Effect of transient CaWRKY40 expression on CaC3H14 transcriptional expression
The data from the ChIP analysis were consistent with the hypothesis that CaC3H14 is a direct target gene of CaWKRY40. Thus, we explored the effect of CaWRKY40 on CaC3H14 transcription. We first assessed the transcript levels of CaC3H14 in CaWRKY40‐overexpressing, CaWRKY40‐silenced and control pepper leaves using real‐time RT‐PCR analysis. Following inoculation, transient overexpression of CaWRKY40 significantly activated CaC3H14 expression in leaves at both 24 and 48 hpi (Fig. 8a). The silencing of CaWRKY40 was performed by VIGS; the success of CaWRKY40 silencing was measured by detection of the transcript level of CaWRKY40 in R. solanacearum‐inoculated TRV::CaWRKY40 plants. The specificity of CaWRKKY40 silencing was tested by detecting the transcription of CaWRKY40b in mock‐treated TRV::CaWRKY40 pepper plants. CaWRKY40b is a negative regulator down‐regulated by R. solanacearum inoculation and shares the highest deduced amino acid sequence identity (64.27%) to that of CaWRKY40 amongst all of the pepper WRKYs; CaWRKY40b is not transcriptionally regulated by CaWRKY40 under room temperature (our unpublished data). The results showed that the transcript levels of CaWRKY40 in R. solanacearum‐inoculated TRV::CaWRKY40 plants were significantly decreased compared with those in control plants (Fig. S2a, see Supporting Information), whereas the transcript levels of CaWRKY40b did not decrease in TRV::CaWRKY40; instead, the transcript levels of CaWRKY40b were enhanced in TRV::CaWRKY40 plants compared with TRV::00 plants, indicating the specific silencing of CaWRKY40 by VIGS (Fig. S2b). From our unpublished data, CaWRKY40b is a negative regulator and is down‐regulated by Ralstonia infection, and this gene is also negatively regulated by CaWRKY40; therefore, the specific silencing of CaWRKY40 de‐repressed the transcription of CaWRKY40b. The transcription of CaC3H14 was detected in TRV::CaWRKY40 plants. The results showed that CaC3H14 transcript levels in CaWRKY40‐silenced pepper leaves were significantly lower than those in control plant leaves (Fig. 8b). These data indicate that CaWRKY40 directly targets CaC3H14 and positively regulates its transcription in R. solanacearum‐infected pepper plants.
Figure 8.
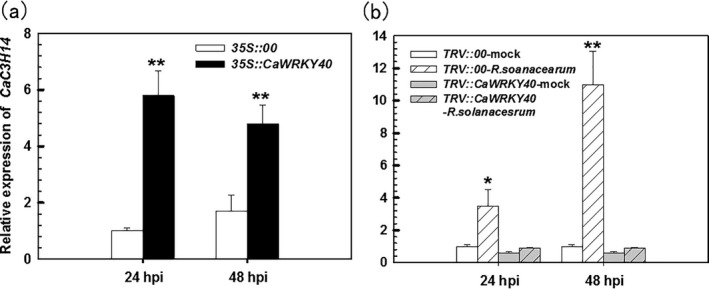
The transcription of CaC3H14 in pepper was up‐regulated by transient overexpression of CaWRKY40 and down‐regulated by CaWRKY40‐silencing using virus‐induced gene silencing (VIGS). (a) Relative expression levels of CaC3H14 in plants in which CaWRKY40 was transiently overexpressed. Expression in mock‐treated controls (35S::00) was set to 1. (b) Relative expression levels of CaC3H14 in CaWRKY40‐silenced plants. Expression in mock‐treated control plants (35S::00 or TRV::00) was set to 1. dpi; days post inoculation with R. solanacearum; data represent mean ± SD of two independent experiments, each with three replicates (n = 6); *, p < 0.05; **, p < 0.01 (Student–Newman–Keuls test).
Discussion
CCCH genes are important for plant growth, development and environmental responses (Bogamuwa and Jang, 2013; Higuera et al., 2014; Huang et al., 2012; Jan et al., 2013; Kim et al., 2008; Kong et al., 2006; Lin et al., 2011; Lu et al., 2014; Yang et al., 2013; Zhou et al., 2014). Studies of these genes have mainly focused on model plants, such as rice and Arabidopsis, and the determination of their role in members of the Solanaceae family, such as pepper, is limited. Here, we characterized the role of one of these genes, CaC3H14, in the immune response of pepper to infection by R. solanacearum. We concluded that CaC3H14 positively regulates the immune response in pepper to R. solanacearum and is targeted by CaWRKY40.
We isolated CaC3H14 from a pepper cDNA library. Its deduced amino acid sequence contains three typical CCCH Znf motifs: Cx8Cx5Cx3H, Cx8Cx5Cx3H and Cx7Cx5Cx3H. When this protein was fused with GFP, the fusion products were present exclusively in the nucleus, similar to the CCCH Znf proteins OsGZF1 and GhZFP1 in their respective plants (Guo et al., 2009; Yang et al., 2013). However, other CCCH proteins, including OsTZF1 and AtTZF1, have dynamic subcellular localization patterns in the cytoplasm and the nucleus (Jan et al., 2013; Pomeranz et al., 2011, 2010), indicating that CCCH Znf proteins have diverse subcellular localization, probably related to their functions.
When plants are infected by pathogens, they must mount a large‐scale transcriptional reprogramming of defence‐associated genes (Bartsch et al., 2006; Ramonell et al., 2005; Rowland et al., 2005). In this study, CaC3H14 was significantly up‐regulated in response to R. solanacearum inoculation, and we hypothesized that this protein is important for the pepper immune response. Several lines of evidence supported this hypothesis. First, we performed several loss‐ and gain‐of‐function analyses. Silencing of CaC3H14 using VIGS significantly enhanced the susceptibility of pepper plants to R. solanacearum, whereas its transient overexpression triggered an intensive HR. Evidence for the HR was twofold: transient overexpression of CaC3H14 was accompanied by enhanced trypan blue staining, indicative of cell death, a common event in ETI which may also occur in PTI (Tsuda and Katagiri, 2010). Second, there was an accumulation of H2O2 in overexpressing plants. Potentiated H2O2 production is believed to confer cell death (Dang et al., 2013). We also observed that ectopic overexpression of CaC3H14 consistently enhanced the resistance of tobacco plants to R. solanacearum inoculation. Our hypothesis was also supported by the results of an expression assay of some immunity‐associated marker genes. Specifically, in pepper plants, the HR‐associated gene HIR1 and the SA‐dependent PR gene PR1a were significantly down‐regulated by silencing of CaC3H14, but up‐regulated by its transient overexpression. We also tested the HR‐associated genes NtHSR515 (Czernic et al., 1996; Takahashi et al., 2004; Tronchet et al., 2001) and NtGST1 (Peng et al., 2004), as well as the SA‐dependent genes NtPR2 (Li et al., 2015; Naoumkina et al., 2008) and NtNPR1 (Ghanta et al., 2011). These genes were consistently up‐regulated by ectopic overexpression of CaC3H14 in transgenic tobacco plants. All of these data suggest that CaC3H14 acts as a positive regulator of pepper immunity against R. solanacearum attack.
We next reasoned that R. solanacearum‐induced CaC3H14 transcription would require regulation. We have observed previously that CaWRKY40 is induced by inoculation with R. solanacearum and enhances the immune response in pepper (Dang et al., 2013). Because there are three W‐boxes in the promoter of CaC3H14, we hypothesized that CaWRKY40 regulates the transcription of CaC3H14 by binding one of these W‐boxes. By ChIP assay, it was found that, amongst the three typical W‐boxes, only the W3‐box appeared to be bound by CaWRKY40, which was further supported by the data from MST analysis. However, the W3‐box was not found to be bound by CaWRKY27 and CaWRKY58, the two other pepper WRKY TFs implicated in pepper immunity against R. solanacearum by our previous studies (Dang et al., 2014; Wang et al., 2013); this suggests that the binding of CaWRKY40 to the W3‐box containing the CaC3H14 promoter is selective; however, its possible binding by other WRKY TFs cannot be ruled out at present. We also observed that, on R. solanacearum infection, pepper plants transiently overexpressing CaWRKY40 exhibited significantly enhanced CaC3H14 transcription, whereas TRV::CaWRKY40 plants exhibited significantly down‐regulated CaC3H14 transcript abundance, compared with control plants. These data provide solid evidence that CaC3H14 is a direct target of CaWRKY40 during the response of pepper to R. solanacearum infection.
Notably, the binding of CaWRKY40 to the CaC3H14 promoter was enhanced by R. solanacearum infection, and the transcription of immunity‐associated genes, which were modulated by CaC3H14, was similarly affected by R. solanacearum infection. It is possible that both CaWRKY40 and CaC3H14 are modulated by some unidentified components activated by R. solanacearum infection. WRKY proteins have been found to interact with many other proteins, such as MAPK, VQ proteins, calmodulin, 14‐3‐3, chromatin remodelling proteins and other TFs (e.g. WRKY, NAC), and modify the target specificity or transcriptional activity of these proteins (Alves et al., 2014; Chi et al., 2013; Giri et al., 2013; Shan et al., 2016). Other CCCH proteins include AtTZF5, which interacts with MARD1 and RD21A (Bogamuwa and Jang, 2016), and GhZFP1, which enhances salt stress tolerance and fungal disease resistance in transgenic tobacco by interacting with GZIRD21A and GZIPR5 (Guo et al., 2009). Further characterization of the proteins that interact with CaWRKY40 and CaC3H14 during the response of pepper to R. solanacearum infection should clarify the underlying mechanism of the pepper immune response to R. solanacearum.
The plant hormones SA, JA and ET are important for plant defence (Kunkel and Brooks, 2002; Robert‐Seilaniantz et al., 2011) and are produced in response to infection by a variety of pathogens. The responses to pathogen infection differ depending on the nature of the pathogen and its infection strategy (De Vos et al., 2005). SA was originally shown to be involved in defence against biotrophic pathogens, and JA was shown to be involved in defence responses to necrotrophic pathogens (Glazebrook, 2005). However, there is intensive crosstalk between SA and JA/ET signalling pathways, and this crosstalk is an important regulatory mechanism in itself. The crosstalk is believed to provide regulation so that the immune response is tailored to the type of pathogen encountered (Pieterse et al., 2012). Complex antagonistic and synergistic regulatory relationships between the SA and JA signalling sectors of the plant immune signalling network have been found in Arabidopsis in response to different pathogens (De Vos et al., 2005; Glazebrook et al., 2003; Sato et al., 2010). An additive or synergistic interaction between JA and SA signalling sectors was revealed during PTI, and they are partially antagonistic to each other during ETI (Tsuda et al., 2009). The latter phenomenon seems to allow for compensatory effects if a defined sector is disabled as a result of interference with pathogen effectors. In our study, overexpression of CaC3H14 in either pepper or tobacco triggered the expression of SA‐dependent immunity‐associated genes, but not JA‐ or ET‐dependent immunity‐associated marker genes, whereas silencing of CaC3H14 down‐regulated SA‐dependent PR genes, but enhanced JA‐ and ET‐dependent PR genes in pepper plants. We interpret this as evidence that CaC3H14 regulates the antagonistic interaction between SA and JA/ET signalling, similar to the role of AtWRKY70. The overexpression of AtWRKY70 increases resistance to virulent pathogens and results in the constitutive expression of SA‐induced PR genes, whereas its suppression activates JA‐responsive/coronatine insensitive 1 (COI1)‐dependent genes (Li et al., 2004, 2006). We speculate that the up‐regulation of CaC3H14 with a biotrophic lifestyle in the early stages of infection by R. solanacearum might initiate the HR and activate SA‐dependent PR genes, whilst blocking unnecessary JA/ET‐dependent defence genes. Notably, although CaC3H14 was directly and transcriptionally regulated by CaWRKY40 during the response of pepper to R. solanacearum infection, the apparent relationship of CaC3H14 to SA, JA and ET signalling is different from that of CaWRKY40. The latter appears to regulate the synergistic interaction between SA and JA/ET signalling (Dang et al., 2013). Although the underlying mechanism is not clear, we hypothesize that unknown proteins might contribute to this difference, either at the transcriptional or post‐transcriptional level. Further investigation may reveal the mechanism.
It is worth noting that CaC3H14‐overexpressing transgenic tobacco plants exhibited significantly enhanced resistance to R. solanacearum inoculation with little apparent effect on growth or development, consistent with other reports (Dang et al., 2011; Hong et al., 2017; Shen et al., 2015; Vaid et al., 2015). For example, overexpression of MoSM1 by the rice blast fungus Magnaporthe oryzae induces broad‐spectrum resistance against fungal and bacterial diseases in rice plants without reducing abiotic stress tolerance or grain yield (Hong et al., 2017). Also, overexpression of the receptor‐like kinase ERECTA improves thermotolerance in rice and tomato without affecting growth (Shen et al., 2015). A possible explanation is that the random insertion of the expression cassette of foreign genes results in many transgenic clones that carry genes with adverse effects on growth or development, but only the clones with low or moderate expression levels that exhibit no significant growth penalty can generate transgenic plants. For example, transgenic plants expressing high levels of AtWRKY18, a pathogen‐ and SA‐induced Arabidopsis TF, were stunted in growth, but AtWRKY18 expressed at moderate levels did not cause substantial negative effects on plant growth (Chen and Chen, 2002). Another possibility is that the adverse effects of the activation of SA‐dependent genes following CaC3H14 overexpression could be counteracted to some degree by depression of JA‐ and ET‐dependent defence pathways. In addition, there was no difference in the transcript levels of marker genes of plant immunity, such as NtPR1a and HSR515, in WT tobacco plants and CaC3H14‐OE lines; however, when the plants were inoculated with R. solanacearum, these marker genes were significantly up‐regulated. It seems likely that CaC3H14 mediates signalling components that are activated by infection with R. solanacearum.
Based on the collective results of this study, we propose that CaC3H14 is transcriptionally targeted by CaWRKY40. It appears to modulate the antagonistic interaction between SA and JA/ET signalling and to enhance the defence response of pepper plants to infection by R. solanacearum.
Experimental Procedures
Plant material and growth conditions
Seeds of pepper (C. annuum cultivar GZ03) and tobacco (Nicotiana tabacum cultivar Honghuadajinyuan and N. benthamiana) were sown in a soil mix [peat moss : perlite, 2 : 1 (v/v)] in plastic pots and placed in a growth room at 25 °C, 70% relative humidity, with 60–70 mmol photons/m2/s for 16 h, followed by 8 h in the dark.
Inoculation with bacteria
Bacteria of R. solanacearum strain FJC100301, which is highly virulent to pepper, were isolated from wilted samples of pepper grown in Fujian Province (China). Bacteria were cultured as described previously by Dang et al. (2013). The bacteria were incubated at 28 °C in potato sucrose agar (PSA) medium (200 g/L of potato extract, 20 g/L of sucrose, 3 g/L of beef extract, 5 g/L of tryptone) for 36 h with shaking at 200 rpm, and were pelleted by spinning for 10 min at 10 000 g and resuspended in 10 mm MgCl2 solution. The bacterial suspension was diluted to 108 CFU/mL [equivalent to an optical density at 600 nm (OD600) of 0.8] and used for inoculation.
Bacterial inoculations were performed when the pepper or tobacco plants had eight leaves, unless otherwise indicated. Roots were inoculated by irrigating the soil with 1 mL of suspension. For leaf inoculation, the third leaf from the plant apex was inoculated with 10 μL of suspension by infiltrating it on the leaf using a needle‐less syringe once the leaf was fully expanded. Control plants were inoculated with 10 mm MgCl2. Samples were collected at various times after inoculation for further analysis.
Disease index measurement
Ralstonia solanacearum‐inoculated pepper or tobacco plants were scored every 3 days following the method of Dang et al. (2013).
Isolation and sequence analysis of cDNA of CaC3H14
A full‐length cDNA with high sequence similarity to C3H14 of other plant species was acquired by random sequencing of a normalized cDNA library of C. annuum constructed in our laboratory. The deduced amino acid sequence served as a query to search for homologues from other plant species at the website: http://www.ncbi.nlm.nih.gov/. The alignment analysis of homologous sequences was performed using DNAMAN5 software.
Construction of the vectors
All of the vectors in the present study were constructed by Gateway cloning technology (Invitrogen, Carlsbad, CA, USA).To construct vectors for overexpression and subcellular localization analysis, the full‐length ORF of CaC3H14 was cloned into the entry vector pDONR207 using a BP reaction. It was then cloned into the destination vectors pK7WG2 for overexpression and pMDC103 for subcellular localization by LR reactions. To construct the vector of 35S::CaWRKY27‐HA, 35S::CaWRKY40‐HA or 35S::CaWRKY58‐HA, the full‐length ORFs of CaWRKY27, CaWRKY40 and CaWRKY58 without the stop codon were cloned into entry vector pDONR207 by BP reaction, also by Gateway cloning technology, and then into destination vector pEarleyGate 201 by LR reaction. To construct a vector for VIGS, a specific 351‐bp fragment of the ORF of CaC3H14 and a 300‐bp fragment of the 3′‐untranslated region (3′‐UTR) of CaWKRY40 were employed for vector construction. The specificities of these two fragments were confirmed by homology sequence searching by blast against the database genome sequences of CM334 (http://peppergenome.snu.ac.kr/) and Zunla‐1 (http://peppersequence.genomics.cn/page/species/blast.jsp). The specific fragments were cloned into the entry vector pDONR207, and then cloned into the pYL279 vector. All of the primers used in vector construction are listed in Table S1 (see Supporting Information).
Subcellular localization
The A. tumefaciens strain GV3101 containing 35S::CaC3H14‐GFP was grown overnight and resuspended in an induction medium (10 mm MES, 10 mm MgCl2, 200 µm acetosyringone, pH 5.6). Bacterial suspensions of OD600 = 0.8 were used to inoculate N. benthamiana leaves. At 48 hpi, the fluorescence of GFP was assessed by imaging with a laser scanning confocal microscope (TCS SP8, Leica, Solms, Germany), using an excitation wavelength of 488 nm and a 505–530‐nm bandpass emission filter.
VIGS of CaC3H14 and CaWRKY40 in pepper plants
For VIGS of CaC3H14 or CaWRKY40 in pepper plants, A. tumefaciens strain GV3101 harbouring pYL192 with pYL279‐CaC3H14, pYL279‐CaWRKY40 or pYL279 (negative control) was resuspended in induction medium at a 1 : 1 ratio (OD600 = 0.6) and co‐infiltrated into cotyledons of 2‐week‐old pepper plants. Details of the procedure have been published previously (Cai et al., 2015; Cheng et al., 2016; Dang et al., 2013; Shen et al., 2016a, 2016b). The specificity of CaWRKY40 silencing was assayed by the detection of transcript levels of CaWRKY40b in CaWRKY40‐silenced pepper plants.
Transient expression of CaC3H14 in pepper leaves
For transient expression analysis, A. tumefaciens strain GV3101 harbouring either 35S::CaC3H14 or the empty vector 35S::00 was grown overnight and resuspended in induction medium. Bacterial suspension (OD600 = 0.8) was infiltrated into leaves of pepper plants.
Generation of transgenic CaC3H14‐overexpressing tobacco plants
Tobacco cultivar Honghuadajinyuan was used to generate CaC3H14‐overexpressing tobacco plants by transforming leaf discs with A. tumefaciens strain GV3101 harbouring the 35S:CaC3H14 vector. Twenty independent T0 transgenic tobacco lines were selected by kanamycin, and further confirmed by PCR and quantitative PCR. Two T2 transgenic lines that exhibited mid‐range numbers of CaC3H14 transcripts were selected for analyses in this study.
Histochemical staining
Leaves were stained for histological analysis by trypan blue or DAB as described by Choi et al. (2012) and our previous studies (Cai et al., 2015; Dang et al., 2014; Liu Z‐Q et al., 2015; Zhang et al., 2015).
Measurement of electrolyte leakage
Electrolyte leakage of the inoculated sites was measured to evaluate cell death. Leaf discs of 6 mm in diameter were taken at 24 and 48 hpi, placed into sterilized distilled water and slowly shaken at room temperature. Electrolyte leakage was measured using a Mettler Toledo 326 (Mettler, Zurich, Switzerland).
Overproduction and purification of the recombinant CaWRKY40‐GFP
For the overproduction of CaWRKY40‐GFP, the pET‐11a‐CaWRKY40‐GFP plasmid was used to transform the Escherichia coli expression strain BL21(DE3). The protein expression of the two proteins and their purification were carried out following the method of Papageorgiou et al. (2016).
Interaction of CaWRKY40 with promoter fragment of CaC3H14 by MST in solution
The interaction of CaWRKY40 with the promoter fragment of CaC3H14 was performed by MST (Zillner et al., 2012). In this experiment, GFP in the fused protein CaWRKY40‐GFP was used as fluorescent label, and a fragment containing a W‐box within the promoter of CaC3H14, which was amplified by PCR with a specific primer pair (PW3) and further purified, was used as the non‐fluorescent molecule. The protein–DNA interactions were measured at various DNA concentrations keeping the protein concentration constant at 20 μm. The DNA used in 16 different concentrations was prepared using serial dilutions. The initial concentration of the DNA was 0.08 nm. The interaction buffer was 20 mm sodium phosphate, pH 8.0, 1 mm ethylenediaminetetraacetic acid (EDTA) and 100 mm NaCl. The protein and DNA were mixed and balanced for 10 min, after which the samples were loaded into a hydrophilic capillary for DNA–protein interaction measurement in a monolith NT.115 5 instrument (NanoTemper Technologies GmbH, Munich, Germany) using 50% IR laser power and an LED excitation source with λ = 470 nm at ambient temperature. The NanoTemperAnalysis 1.2.20 software was used to fit the data and determine the apparent K d values (Papageorgiou et al., 2016; Zillner et al., 2012).
ChIP analysis
ChIP assays were performed as described previously (Sun et al., 2015) with slight modification. Leaves were inoculated with GV3101 cells containing 35S::CaWRKY27‐HA, 35S::CaWRKY40‐HA or 35S::CaWRKY58‐HA and 35S::HA, and collected at 24 hpi. For each sample, about 4 g of leaves were treated with 1.0% formaldehyde for 8 min. Glycine (3 m) was added to a final concentration of 0.125 m glycine. The sample was then vacuum infiltrated for an additional 5 min to stop crosslinking. Nuclear extracts were isolated and resuspended sequentially with extraction buffer I (0.4 m sucrose, 10 mm Tris‐Cl, pH 8.0, 10 mm MgCl2, 5 mm β‐mercaptoethanol, 1 U protease inhibitors), buffer II (0.25 m sucrose, 10 mm Tris‐Cl, pH 8.0, 10 mm MgCl2, 1% Triton X‐100, 5 mm β‐mercaptoethanol, 1 U protease inhibitors) and buffer III (1.7 m sucrose, 10 mm Tris‐Cl, pH 8.0, 2 mm MgCl2, 0.15% Triton X‐100, 5 mm β‐mercaptoethanol, 1 U protease inhibitors). After this series, they were digested with micrococcal nuclease (Takara, Dalian, China) according to the manufacturer's instructions. Magnetic beads (Invitrogen) linked with antibody against HA (anti‐HA tag, rabbit polyclonal antibody, Sigma, St Louis, Missouri, The United States) or anti‐FLAG (negative control) were added to digested samples and eluted following the manufacturer's instructions. Two millilitres of 10 mg/mL proteinase K were added to each sample, incubated at 45 °C for 1 h, extracted twice with the same volume of Tris‐saturated phenol : chloroform : isoamyl alcohol (25 : 24 : 1, v/v), centrifuged at 4 °C and 20 000 g for 15 min and the aqueous phase was moved to another tube. DNA was then precipitated by adding twice the volume of 100% ethanol, 1/10 volume of 3 m sodium acetate and 1 mL of 2 m glycogen, and incubated at −20 °C overnight. DNA was pelleted by spinning for 20 min at 16 700 g, and the pellets were washed with 70% ethanol, dried at room temperature, resuspended in 50 μL of TE buffer and stored at −20 °C for further use. The immunoprecipitated DNA was analysed for the enrichment of CaWRKY40 at the promoter region of the target genes using semi‐quantitative PCR or quantitative real‐time PCR. For quantitative real‐time PCR, fold increases in immunoprecipitated DNA were calculated relative to the input DNA (relative enrichment) and the internal control, CaActin. The primers used for real‐time PCR analysis in ChIP assays are listed in Table S1.
Immunoblot analysis
Protein extraction buffer was used to extract the total protein of pepper samples. At 4 °C, total extracted protein was incubated together with anti‐HA agarose (Thermo Fisher Scientific, Waltham, MA, USA) overnight. Beads were collected using a magnetic rack and washed three times with Tris‐buffered saline and Tween‐20 (0.05%). Eluted protein was examined by immunoblotting using anti‐HA–peroxidase antibodies (Abcam, Cambridge, UK).
Quantitative real‐time RT‐PCR
To determine the relative transcription levels of selected genes, real‐time PCR was performed with specific primers (Table S1) according to the manufacturer's instructions for the BIO‐RAD Real‐time PCR system (Foster City, CA, USA) and the SYBR Premix Ex Taq II system (TaKaRa, Dalian China). Total RNA preparation and real‐time RT‐PCR were carried out as described previously (Cai et al., 2015; Dang et al., 2014; Zhang et al., 2015). Two independent biological experiments were performed in triplicate. Data were analysed using the Livak method (Livak and Schmittgen, 2001) and expressed as a normalized relative expression level (2‐ΔΔCT) of the respective genes (mock‐treated or control plants). The relative transcript level of each sample was normalized to CaActin or NtEF1a.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 The primers used in vector construction and gene expression analyses by polymerase chain reaction (PCR) in the present study
Fig. S1 The structural domains of CaC3H14 and comparison of its amino acid sequence with that of homologues in other plant species. The accession number of CaC3H14 is XP_016543877, highlighted by a red ellipse. The three ZnF_C3H1 domains, CX8CX5CX3H, CX8CX5CX3H and CX7CX5CX3H, are indicated with black lines. The homologues of CaC3H14 in other plant species include: XP_016543677 (Capsicum annuum), XP_019245694 (Nicotiana attenuata), XP_019266162 (N. attenuata), XP_010312058 (Solanum lycopersicum) and XP_004249283 (S. lycopersicum). Blue shading, 50%–75% identity; red shading, 75% identity; black shading, 100% identity. The blue ellipse indicates the specific sequence of CaC3H14. The alignment analysis was made using DNAMAN5 software.
Fig. S2 The success and specificity of CaWRKY40 silencing by virus‐induced gene silencing (VIGS). (a) The transcript levels of CaWRKY40 in TRV::CaWRKY40 and control pepper plants with or without Ralstonia solanacearum inoculation; the transcript level in mock‐treated control plants (TRV::00) was set to unity. (b) The relative transcript levels of CaWRKY40b in TRV::CaWRKY40 and control pepper plants; the transcript level of TRV::00 was set to unity. dpi, days post‐inoculation with R. solanacearum; data represent mean ± standard deviation (SD) of six replicates (n = 6). Different letters indicate significant difference as determined by Student–Newman–Keuls test (P < 0.01).
Fig. S3 The detection of the success of CaC3H14‐HA transient overexpression in pepper leaves by western blot with haemagglutinin antibody.
Acknowledgements
We thank Mark D. Curtis for kindly providing the Gateway destination vectors and Dr S. P. Dinesh‐Kumar of Yale University for the pTRV1 and pTRV2 vectors. This work was supported by grants from the National Natural Science Foundation of China (31401890, 31401312, 30971718, 31372061). The authors declare that there are no competing interests.
References
- Addepalli, B. and Hunt, A.G. (2008) Ribonuclease activity is a common property of Arabidopsis CCCH‐containing zinc‐finger proteins. FEBS Lett. 582, 2577–2582. [DOI] [PubMed] [Google Scholar]
- Agarwal, P. , Reddy, M.P. and Chikara, J. (2011) WRKY: its structure, evolutionary relationship, DNA‐binding selectivity, role in stress tolerance and development of plants. Mol. Biol. Rep. 38, 3883–3896. [DOI] [PubMed] [Google Scholar]
- Alves, M.S. , Dadalto, S.P. , Goncalves, A.B. , de Souza, G.B. , Barros, V.A. and Fietto, L.G. (2014) Transcription factor functional protein–protein interactions in plant defense responses. Proteomes, 2, 85–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attard, A. , Evangelisti, E. , Kebdani‐Minet, N. , Panabières, F. , Deleury, E. , Maggio, C. , Ponchet, M. and Gourgues, M. (2014) Transcriptome dynamics of Arabidopsis thaliana root penetration by the oomycete pathogen Phytophthora parasitica . BMC Genomics, 15, 538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch, M. , Gobbato, E. , Bednarek, P. , Debey, S. , Schultze, J.L. , Bautor, J. and Parker, J.E. (2006) Salicylic acid‐independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell, 18, 1038–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg, J.M. and Shi, Y. (1996) The galvanization of biology: a growing appreciation for the roles of zinc. Science, 271, 1081–1085. [DOI] [PubMed] [Google Scholar]
- Birkenbihl, R.P. , Kracher, B. , Roccaro, M. and Somssich, I.E. (2017) Induced genome‐wide binding of three Arabidopsis WRKY transcription factors during early MAMP‐triggered immunity. Plant Cell, 29, 20–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogamuwa, S. and Jang, J.C. (2013) The Arabidopsis tandem CCCH zinc finger proteins AtTZF4, 5 and 6 are involved in light‐, abscisic acid‐ and gibberellic acid‐mediated regulation of seed germination. Plant Cell Environ. 36, 1507–1519. [DOI] [PubMed] [Google Scholar]
- Bogamuwa, S. and Jang, J.C. (2016) Plant tandem CCCH zinc finger proteins interact with ABA, drought, and stress response regulators in processing‐bodies and stress granules. PLoS One, 11, e0151574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaill, P. and Rivas, S. (2014) Transcriptional control of plant defence responses. Curr. Opin. Plant Biol. 20, 35–46. [DOI] [PubMed] [Google Scholar]
- Cai, H. , Yang, S. , Yan, Y. , Xiao, Z. , Cheng, J. , Wu, J. , Qiu, A. , Lai, Y. , Mou, S. , Guan, D. , Huang, R. and He, S. (2015) CaWRKY6 transcriptionally activates CaWRKY40, regulates Ralstonia solanacearum resistance, and confers high‐temperature and high‐humidity tolerance in pepper. J. Exp. Bot. 66, 3163–3174. [DOI] [PubMed] [Google Scholar]
- Chai, G. , Hu, R. , Zhang, D. , Qi, G. , Zuo, R. , Cao, Y. , Chen, P. , Kong, Y. and Zhou, G. (2012) Comprehensive analysis of CCCH zinc finger family in poplar (Populus trichocarpa). BMC Genomics, 13, 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. and Chen, Z. (2002) Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen‐induced Arabidopsis transcription factor. Plant Physiol. 129, 706–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, W. , Xiao, Z. , Cai, H. , Wang, C. , Hu, Y. and Xiao, Y. (2016) A novel leucine‐rich repeat protein, CaLRR51, acts as a positive regulator in the response of pepper to Ralstonia solanacearum infection. Mol. Plant Pathol. 18, 1089–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi, Y. , Yang, Y. , Zhou, Y. , Zhou, J. , Fan, B. , Yu, J.‐Q. and Chen, Z. (2013) Protein–protein interactions in the regulation of WRKY transcription factors. Mol. Plant, 6, 287–300. [DOI] [PubMed] [Google Scholar]
- Choi, D.S. and Hwang, B.K. (2011) Proteomics and functional analyses of pepper abscisic acid‐responsive 1 (ABR1), which is involved in cell death and defense signaling. Plant Cell, 23, 823–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, D.S. , Hwang, I.S. and Hwang, B.K. (2012) Requirement of the cytosolic interaction between PATHOGENESIS‐RELATED PROTEIN10 and LEUCINE‐RICH REPEAT PROTEIN1 for cell death and defense signaling in pepper. Plant Cell, 24, 1675–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciolkowski, I. , Wanke, D. , Birkenbihl, R.P. and Somssich, I.E. (2008) Studies on DNA‐binding selectivity of WRKY transcription factors lend structural clues into WRKY‐domain function. Plant Mol. Biol. 68, 81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czernic, P. , Huang, H.C. and Marco, Y. (1996) Characterization of hsr201 and hsr515, two tobacco genes preferentially expressed during the hypersensitive reaction provoked by phytopathogenic bacteria. Plant Mol. Biol. 31, 255–265. [DOI] [PubMed] [Google Scholar]
- Dang, F. , Wang, Y. , She, J. , Lei, Y. , Liu, Z. , Eulgem, T. , Lai, Y. , Lin, J. , Yu, L. , Lei, D. , Guan, D. , Li, X. , Yuan, Q. and He, S. (2014) Overexpression of CaWRKY27, a subgroup IIe WRKY transcription factor of Capsicum annuum, positively regulates tobacco resistance to Ralstonia solanacearum infection. Physiol. Plant, 150, 397–411. [DOI] [PubMed] [Google Scholar]
- Dang, F.‐F. , Wang, Y.‐N. , Yu, L. , Eulgem, T. , Lai, Y. , Liu, Z.‐Q. , Wang, X. , Qiu, A.‐L. , Zhang, T.‐X. , Lin, J. , Chen, Y.‐S. , Guan, D.‐Y. , Cai, H.‐Y. , Mou, S.‐L. and He, S.‐L. (2013) CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 36, 757–774. [DOI] [PubMed] [Google Scholar]
- Dang, H.Q. , Tran, N.Q. , Gill, S.S. , Tuteja, R. and Tuteja, N. (2011) A single subunit MCM6 from pea promotes salinity stress tolerance without affecting yield. Plant Mol. Biol. 76, 19–34. [DOI] [PubMed] [Google Scholar]
- De Vos, M. , Van Oosten, V.R. , Van Poecke, R.M.P. , Van Pelt, J.A. , Pozo, M.J. , Mueller, M.J. , Buchala, A.J. , Métraux, J.‐P. , Van Loon, L.C. , Dicke, M. and Pieterse, C.M.J. (2005) Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol. Plant–Microbe Interact. 18, 923–937. [DOI] [PubMed] [Google Scholar]
- Deng, H. , Liu, H. , Li, X. , Xiao, J. and Wang, S. (2012) A CCCH‐type zinc finger nucleic acid‐binding protein quantitatively confers resistance against rice bacterial blight disease. Plant Physiol. 158, 876–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörmann, P. , Kim, H. , Ott, T. , Schulze‐Lefert, P. , Trujillo, M. , Wewer, V. and Hückelhoven, R. (2014) Cell‐autonomous defense, re‐organization and trafficking of membranes in plant–microbe interactions. New Phytol. 204, 815–822. [DOI] [PubMed] [Google Scholar]
- Eulgem, T. , Rushton, P.J. , Robatzek, S. and Somssich, I.E. (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. [DOI] [PubMed] [Google Scholar]
- Garner, C.M. , Kim, S.H. , Spears, B.J. and Gassmann, W. (2016) Express yourself: transcriptional regulation of plant innate immunity. Semin. Cell Dev. Biol. 56, 150–162. [DOI] [PubMed] [Google Scholar]
- Ghanta, S. , Bhattacharyya, D. , Sinha, R. , Banerjee, A. and Chattopadhyay, S. (2011) Nicotiana tabacum overexpressing gamma‐ECS exhibits biotic stress tolerance likely through NPR1‐dependent salicylic acid‐mediated pathway. Planta, 233, 895–910. [DOI] [PubMed] [Google Scholar]
- Giri, P. , Taj, G. , Tasleem, M. and Kumar, A. (2013) In silico‐prediction of downstream WRKY interacting partners of MAPK3 in Brassica. Bioinformation, 9, 1036–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. , Chen, W. , Estes, B. , Chang, H.‐S. , Nawrath, C. , Metraux, J.‐P. , Zhu, T. and Katagiri, F. (2003) Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J. 34, 217–228. [DOI] [PubMed] [Google Scholar]
- Grabowska, A. , Wisniewska, A. , Tagashira, N. , Malepszy, S. and Filipecki, M. (2009) Characterization of CsSEF1 gene encoding putative CCCH‐type zinc finger protein expressed during cucumber somatic embryogenesis. J. Plant Physiol. 166, 310–323. [DOI] [PubMed] [Google Scholar]
- Grewal, R.K. , Gupta, S. and Das, S. (2012) Xanthomonas oryzae pv oryzae triggers immediate transcriptomic modulations in rice. BMC Genomics, 13, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Y.‐H. , Yu, Y.‐P. , Wang, D. , Wu, C.‐A. , Yang, G.‐D. , Huang, J.‐G. and Zheng, C.‐C. (2009) GhZFP1, a novel CCCH‐type zinc finger protein from cotton, enhances salt stress tolerance and fungal disease resistance in transgenic tobacco by interacting with GZIRD21A and GZIPR5. New Phytol. 183, 62–75. [DOI] [PubMed] [Google Scholar]
- Hall, T.M. (2005) Multiple modes of RNA recognition by zinc finger proteins. Curr. Opin. Struct. Biol. 15, 367–373. [DOI] [PubMed] [Google Scholar]
- Hein, I. , Gilroy, E.M. , Armstrong, M.R. and Birch, P.R. (2009) The zig–zag–zig in oomycete–plant interactions. Mol. Plant Pathol. 10, 547–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuera, J.J. , Fernandez, E. and Galvan, A. (2014) Chlamydomonas NZF1 a tandem repeated zinc finger factor involved in nitrate signaling by controlling the regulatory gene NIT2. Plant Cell Environ. 37, 2139–2150. [DOI] [PubMed] [Google Scholar]
- Hong, Y. , Yang, Y. , Zhang, H. , Huang, L. , Li, D. and Song, F. (2017) Overexpression of MoSM1, encoding for an immunity‐inducing protein from Magnaporthe oryzae, in rice confers broad‐spectrum resistance against fungal and bacterial diseases. Sci. Rep. 7, 41 037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, P. , Chung, M.‐S. , Ju, H.‐W. , Na, H.‐S. , Lee, D.J. , Cheong, H.‐S. and Kim, C.S. (2011) Physiological characterization of the Arabidopsis thaliana oxidation‐related zinc finger 1, a plasma membrane protein involved in oxidative stress. J. Plant Res. 124, 699–705. [DOI] [PubMed] [Google Scholar]
- Huang, P. , Ju, H.‐W. , Min, J.‐H. , Zhang, X. , Chung, J.‐S. , Cheong, H.‐S. and Kim, C.S. (2012) Molecular and physiological characterization of the Arabidopsis thaliana Oxidation‐related Zinc Finger 2, a plasma membrane protein involved in ABA and salt stress response through the ABI2‐mediated signaling pathway. Plant Cell Physiol. 53, 193–203. [DOI] [PubMed] [Google Scholar]
- Jan, A. , Maruyama, K. , Todaka, D. , Kidokoro, S. , Abo, M. , Yoshimura, E. , Shinozaki, K. , Nakashima, K. and Yamaguchi‐Shinozaki, K. (2013) OsTZF1, a CCCH‐tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress‐related genes. Plant Physiol. 161, 1202–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kim, D.H. , Yamaguchi, S. , Lim, S. , Oh, E. , Park, J. , Hanada, A. , Kamiya, Y. and Choi, G. (2008) SOMNUS, a CCCH‐type zinc finger protein in Arabidopsis, negatively regulates light‐dependent seed germination downstream of PIL5. Plant Cell, 20, 1260–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, N.H. , Kim, D.S. , Chung, E.H. and Hwang, B.K. (2014) Pepper suppressor of the G2 allele of skp1 interacts with the receptor‐like cytoplasmic kinase1 and type III effector AvrBsT and promotes the hypersensitive cell death response in a phosphorylation‐dependent manner. Plant Physiol. 165, 76–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, Z. , Li, M. , Yang, W. , Xu, W. and Xue, Y. (2006) A novel nuclear‐localized CCCH‐type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol. 141, 1376–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel, B.N. and Brooks, D.M. (2002) Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5, 325–331. [DOI] [PubMed] [Google Scholar]
- Laura, M. , Borghi, C. , Bobbio, V. and Allavena, A. (2015) The effect on the transcriptome of Anemone coronaria following infection with rust (Tranzschelia discolor). PLoS One, 10, e0118565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.H. , Kim, Y.C. , Choi, D. and Park, J.M. (2013) Identification of novel pepper genes involved in Bax‐ or INF1‐mediated cell death responses by high‐throughput virus‐induced gene silencing. Int. J. Mol. Sci. 14, 22 782–22 795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Jia, D. and Chen, X. (2001) HUA1, a regulator of stamen and carpel identities in Arabidopsis, codes for a nuclear RNA binding protein. Plant Cell, 13, 2269–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Brader, G. and Palva, E.T. (2004) The WRKY70 transcription factor: a node of convergence for jasmonate‐mediated and salicylate‐mediated signals in plant defense. Plant Cell, 16, 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Brader, G. , Kariola, T. and Palva, E.T. (2006) WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 46, 477–491. [DOI] [PubMed] [Google Scholar]
- Li, J.B. , Luan, Y.S. and Liu, Z. (2015) Overexpression of SpWRKY1 promotes resistance to Phytophthora nicotiana and tolerance to salt and drought stress in transgenic tobacco. Physiol. Plant. 155, 248–266. [DOI] [PubMed] [Google Scholar]
- Lin, P.‐C. , Pomeranz, M.C. , Jikumaru, Y. , Kang, S.G. , Hah, C. , Fujioka, S. , Kamiya, Y. and Jang, J.‐C. (2011) The Arabidopsis tandem zinc finger protein AtTZF1 affects ABA‐ and GA‐mediated growth, stress and gene expression responses. Plant J. 65, 253–268. [DOI] [PubMed] [Google Scholar]
- Liu, S. , Kracher, B. , Ziegler, J. , Birkenbihl, R.P. and Somssich, I.E. (2015) Negative regulation of ABA signaling by WRKY33 is critical for Arabidopsis immunity towards Botrytis cinerea 2100. Elife, 4, e07295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z.‐Q. , Qiu, A.‐L. , Shi, L.‐P. , Cai, J.‐S. , Huang, X.‐Y. , Yang, S. , Wang, B. , Shen, L. , Huang, M.‐K. , Mou, S.‐L. , Ma, X.‐L. , Liu, Y.‐Y. , Lin, L. , Wen, J.‐Y. , Tang, Q. , Shi, W. , Guan, D.‐Y. , Lai, Y. and He, S.‐L. (2015) SRC2‐1 is required in PcINF1‐induced pepper immunity by acting as an interacting partner of PcINF1. J. Exp. Bot. 66, 3683–3698. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(–Delta Delta C(T)) method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lu, P. , Chai, M. , Yang, J. , Ning, G. , Wang, G. and Ma, H. (2014) The Arabidopsis CALLOSE DEFECTIVE MICROSPORE1 gene is required for male fertility through regulating callose metabolism during microsporogenesis. Plant Physiol. 164, 1893–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay, J.P. and Crossley, M. (1998) Zinc fingers are sticking together. Trends Biochem. Sci. 23, 1–4. [DOI] [PubMed] [Google Scholar]
- Moore, J.W. , Loake, G.J. and Spoel, S.H. (2011) Transcription dynamics in plant immunity. Plant Cell, 23, 2809–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, M. and Ullman, C. (2003) Recent developments in the engineering of zinc finger proteins. Brief. Funct. Genomic. Proteomic. 1, 342–355. [DOI] [PubMed] [Google Scholar]
- Naoumkina, M.A. , He, X. and Dixon, R.A. (2008) Elicitor‐induced transcription factors for metabolic reprogramming of secondary metabolism in Medicago truncatula . BMC Plant Biol. 8, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, S.P. and Somssich, I.E. (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol. 150, 1648–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papageorgiou, A.C. , Adam, P.S. , Stavros, P. , Nounesis, G. , Meijers, R. , Petratos, K. and Vorgias, C.E. (2016) HU histone‐like DNA‐binding protein from Thermus thermophilus: structural and evolutionary analyses. Extremophiles, 20, 695–709. [DOI] [PubMed] [Google Scholar]
- Peng, J.L. , Bao, Z.L. , Ren, H.Y. , Wang, J.S. and Dong, H.S. (2004) Expression of harpin(xoo) in transgenic tobacco induces pathogen defense in the absence of hypersensitive cell death. Phytopathology, 94, 1048–1055. [DOI] [PubMed] [Google Scholar]
- Peng, X. , Zhao, Y. , Cao, J. , Zhang, W. , Jiang, H. , Li, X. , Ma, Q. , Zhu, S. and Cheng, B. (2012) CCCH‐type zinc finger family in maize: genome‐wide identification, classification and expression profiling under abscisic acid and drought treatments. PLoS One, 7, e40120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M. , Van der Does, D. , Zamioudis, C. , Leon‐Reyes, A. and Van Wees, S.C. (2012) Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. [DOI] [PubMed] [Google Scholar]
- Pomeranz, M. , Zhang, L. , Finer, J. and Jang, J.C. (2011) Can AtTZF1 act as a transcriptional activator or repressor in plants? Plant Signal. Behav. 6, 719–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz, M.C. , Hah, C. , Lin, P.‐C. , Kang, S.G. , Finer, J.J. , Blackshear, P.J. and Jang, J.‐C. (2010) The Arabidopsis tandem zinc finger protein AtTZF1 traffics between the nucleus and cytoplasmic foci and binds both DNA and RNA. Plant Physiol. 152, 151–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramonell, K. , Berrocal‐Lobo, M. , Koh, S. , Wan, J. , Edwards, H. and Stacey, G. (2005) Loss‐of‐function mutations in chitin responsive genes show increased susceptibility to the powdery mildew pathogen Erysiphe cichoracearum . Plant Physiol. 138, 1027–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann, J.L. and Ratcliffe, O.J. (2000) A genomic perspective on plant transcription factors. Curr. Opin. Plant Biol. 3, 423–434. [DOI] [PubMed] [Google Scholar]
- Robert‐Seilaniantz, A. , Grant, M. and Jones, J.D. (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate–salicylate antagonism. Annu. Rev. Phytopathol. 49, 317–343. [DOI] [PubMed] [Google Scholar]
- Rowland, O. , Ludwig, A.A. , Merrick, C.J. , Baillieul, F. , Tracy, F.E. , Durrant, W.E. , Fritz‐Laylin, L. , Nekrasov, V. , Sjölander, K. , Yoshioka, H. and Jones, J.D. (2005) Functional analysis of Avr9/Cf‐9 rapidly elicited genes identifies a protein kinase, ACIK1, that is essential for full Cf‐9‐dependent disease resistance in tomato. Plant Cell, 17, 295–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd, J.J. , Kanyuka, K. , Hassani‐Pak, K. , Derbyshire, M. , Andongabo, A. , Devonshire, J. , Lysenko, A. , Saqi, M. , Desai, N.M. , Powers, S.J. , Hooper, J. , Ambroso, L. , Bharti, A. , Farmer, A. , Hammond‐Kosack, K.E. , Dietrich, R.A. and Courbot, M. (2015) Transcriptome and metabolite profiling of the infection cycle of Zymoseptoria tritici on wheat reveals a biphasic interaction with plant immunity involving differential pathogen chromosomal contributions and a variation on the hemibiotrophic lifestyle definition. Plant Physiol. 167, 1158–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Saint Paul, V. , Zhang, W. , Kanawati, B. , Geist, B. , Faus‐Keßler, T. , Schmitt‐Kopplin, P. and Schäffner, A.R. (2011) The Arabidopsis glucosyltransferase UGT76B1 conjugates isoleucic acid and modulates plant defense and senescence. Plant Cell, 23, 4124–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, M. , Tsuda, K. , Wang, L. , Coller, J. , Watanabe, Y. , Glazebrook, J. and Katagiri, F. (2010) Network modeling reveals prevalent negative regulatory relationships between signaling sectors in Arabidopsis immune signaling. PLoS Pathog. 6, e1001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann, U. , Prestele, J. , O'Geen, H. , Brueggeman, R. , Wanner, G. and Gietl, C. (2007) Requirement of the C3HC4 zinc RING finger of the Arabidopsis PEX10 for photorespiration and leaf peroxisome contact with chloroplasts. Proc. Natl. Acad. Sci. USA, 104, 1069–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, W. , Chen, J.Y. , Kuang, J.F. and Lu, W.J. (2016) Banana fruit NAC transcription factor MaNAC5 cooperates with MaWRKYs to enhance the expression of pathogenesis‐related genes against Colletotrichum musae . Mol. Plant Pathol. 17, 330–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, H. , Zhong, X. , Zhao, F. , Wang, Y. , Yan, B. , Li, Q. , Chen, G. , Mao, B. , Wang, J. , Li, Y. , Xiao, G. , He, Y. , Xiao, H. , Li, J. and He, Z. (2015) Overexpression of receptor‐like kinase ERECTA improves thermotolerance in rice and tomato. Nat. Biotechnol. 33, 996–1003. [DOI] [PubMed] [Google Scholar]
- Shen, L. , Liu, Z. , Yang, S. , Yang, T. , Liang, J. , Wen, J. , Liu, Y. , Li, J. , Shi, L. , Tang, Q. , Shi, W. , Hu, J. , Liu, C. , Zhang, Y. , Lin, W. , Wang, R. , Yu, H. , Mou, S. , Hussain, A. , Cheng, W. , Cai, H. , He, L. , Guan, D. , Wu, Y. and He, S. (2016a) Pepper CabZIP63 acts as a positive regulator during Ralstonia solanacearum or high temperature–high humidity challenge in a positive feedback loop with CaWRKY40. J. Exp. Bot. 67, 2439–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, L. , Yang, S. , Yang, T. , Liang, J. , Cheng, W. , Wen, J. , Liu, Y. , Li, J. , Shi, L. , Tang, Q. , Shi, W. , Hu, J. , Liu, C. , Zhang, Y. , Mou, S. , Liu, Z. , Cai, H. , He, L. , Guan, D. , Wu, Y. and He, S. (2016b) CaCDPK15 positively regulates pepper responses to Ralstonia solanacearum inoculation and forms a positive‐feedback loop with CaWRKY40 to amplify defense signaling. Sci. Rep. 6, 22439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn, S.I. , Kim, Y.H. , Kim, B.R. , Lee, S.Y. , Lim, C.K. , Hur, J.H. and Lee, J.Y. (2007) Transgenic tobacco expressing the hrpN(EP) gene from Erwinia pyrifoliae triggers defense responses against Botrytis cinerea . Mol. Cells, 24, 232–239. [PubMed] [Google Scholar]
- Sun, J. , Jiang, H. , Xu, Y. , Li, H. , Wu, X. , Xie, Q. and Li, C. (2007) The CCCH‐type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis. Plant Cell Physiol. 48, 1148–1158. [DOI] [PubMed] [Google Scholar]
- Sun, T. , Zhang, Y. , Li, Y. , Zhang, Q. , Ding, Y. and Zhang, Y. (2015) ChIP‐seq reveals broad roles of SARD1 and CBP60g in regulating plant immunity. Nat. Commun. 6, 10 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, Y. , Uehara, Y. , Berberich, T. , Ito, A. , Saitoh, H. , Miyazaki, A. , Terauchi, R. and Kusano, T. (2004) A subset of hypersensitive response marker genes, including HSR203J, is the downstream target of a spermine signal transduction pathway in tobacco. Plant J. 40, 586–595. [DOI] [PubMed] [Google Scholar]
- Tronchet, M. , Ranty, B. , Marco, Y. and Roby, D. (2001) HSR203 antisense suppression in tobacco accelerates development of hypersensitive cell death. Plant J. 27, 115–127. [DOI] [PubMed] [Google Scholar]
- Tsuda, K. and Katagiri, F. (2010) Comparing signaling mechanisms engaged in pattern‐triggered and effector‐triggered immunity. Curr. Opin. Plant Biol. 13, 459–465. [DOI] [PubMed] [Google Scholar]
- Tsuda, K. and Somssich, I.E. (2015) Transcriptional networks in plant immunity. New Phytol. 206, 932–947. [DOI] [PubMed] [Google Scholar]
- Tsuda, K. , Sato, M. , Stoddard, T. , Glazebrook, J. and Katagiri, F. (2009) Network properties of robust immunity in plants. PLoS Genet. 5, e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulker, B. and Somssich, I.E. (2004) WRKY transcription factors: from DNA binding towards biological function. Curr. Opin. Plant Biol. 7, 491–498. [DOI] [PubMed] [Google Scholar]
- Vaid, N. , Pandey, P. , Srivastava, V.K. and Tuteja, N. (2015) Pea lectin receptor‐like kinase functions in salinity adaptation without yield penalty, by alleviating osmotic and ionic stresses and upregulating stress‐responsive genes. Plant Mol. Biol. 88, 193–206. [DOI] [PubMed] [Google Scholar]
- Wang, D. , Amornsiripanitch, N. and Dong, X. (2006) A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2, e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D. , Guo, Y. , Wu, C. , Yang, G. , Li, Y. and Zheng, C. (2008) Genome‐wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genomics, 9, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Xu, Y. , Zhang, C. , Ma, Q. , Joo, S.‐H. , Kim, S.‐K. , Xu, Z. and Chong, K. (2008) OsLIC, a novel CCCH‐type zinc finger protein with transcription activation, mediates rice architecture via brassinosteroids signaling. PLoS One, 3, e3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Dang, F. , Liu, Z. , Wang, X. , Eulgem, T. , Lai, Y. , Yu, L. , She, J. , Shi, Y. , Lin, J. , Chen, C. , Guan, D. , Qiu, A. and He, S. (2013) CaWRKY58, encoding a group I WRKY transcription factor of Capsicum annuum, negatively regulates resistance to Ralstonia solanacearum infection. Mol. Plant Pathol. 14, 131–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray, G.A. (2003) Transcriptional regulation and the evolution of development. Int. J. Dev. Biol. 47, 675–684. [PubMed] [Google Scholar]
- Yang, H. , Yuan, S. , Luo, Y. , Huang, J. , Chen, Y.‐E. , Sun, X. , Huang, Y. , Xid, D.‐H. and Lin, H.‐H. (2013) Diverse responses are involved in the defence of Arabidopsis thaliana against Turnip crinkle virus. Z. Naturforsch C, 68, 148–154. [PubMed] [Google Scholar]
- Zhang, L. , Li, Y. , Lu, W. , Meng, F. , Wu, C.A. and Guo, X. (2012) Cotton GhMKK5 affects disease resistance, induces HR‐like cell death, and reduces the tolerance to salt and drought stress in transgenic Nicotiana benthamiana . J. Exp. Bot. 63, 3935–3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q. , Kuang, H. , Chen, C. , Yan, J. , Do‐Umehara, H.C. , Liu, X.Y. , Dada, L. , Ridge, K.M. , Chandel, N.S. and Liu, J. ( 2015) The kinase Jnk2 promotes stress‐induced mitophagy by targeting the small mitochondrial form of the tumor suppressor ARF for degradation. Nat. Immunol. 16, 458–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, T. , Yang, X. , Wang, L. , Xu, J. and Zhang, X. (2014) GhTZF1 regulates drought stress responses and delays leaf senescence by inhibiting reactive oxygen species accumulation in transgenic Arabidopsis. Plant Mol. Biol. 85, 163–177. [DOI] [PubMed] [Google Scholar]
- Zillner, K. , Jerabek‐Willemsen, M. , Duhr, S. , Braun, D. , Langst, G. and Baaske, P. (2012) Microscale thermophoresis as a sensitive method to quantify protein:nucleic acid interactions in solution. Methods Mol. Biol. 815, 241–252. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 The primers used in vector construction and gene expression analyses by polymerase chain reaction (PCR) in the present study
Fig. S1 The structural domains of CaC3H14 and comparison of its amino acid sequence with that of homologues in other plant species. The accession number of CaC3H14 is XP_016543877, highlighted by a red ellipse. The three ZnF_C3H1 domains, CX8CX5CX3H, CX8CX5CX3H and CX7CX5CX3H, are indicated with black lines. The homologues of CaC3H14 in other plant species include: XP_016543677 (Capsicum annuum), XP_019245694 (Nicotiana attenuata), XP_019266162 (N. attenuata), XP_010312058 (Solanum lycopersicum) and XP_004249283 (S. lycopersicum). Blue shading, 50%–75% identity; red shading, 75% identity; black shading, 100% identity. The blue ellipse indicates the specific sequence of CaC3H14. The alignment analysis was made using DNAMAN5 software.
Fig. S2 The success and specificity of CaWRKY40 silencing by virus‐induced gene silencing (VIGS). (a) The transcript levels of CaWRKY40 in TRV::CaWRKY40 and control pepper plants with or without Ralstonia solanacearum inoculation; the transcript level in mock‐treated control plants (TRV::00) was set to unity. (b) The relative transcript levels of CaWRKY40b in TRV::CaWRKY40 and control pepper plants; the transcript level of TRV::00 was set to unity. dpi, days post‐inoculation with R. solanacearum; data represent mean ± standard deviation (SD) of six replicates (n = 6). Different letters indicate significant difference as determined by Student–Newman–Keuls test (P < 0.01).
Fig. S3 The detection of the success of CaC3H14‐HA transient overexpression in pepper leaves by western blot with haemagglutinin antibody.


