SUMMARY
Salicylic acid (SA) plays an important role in signal transduction and disease resistance. In Arabidopsis, SA can be made by either of two biosynthetic branches, one involving isochorismate synthase (ICS) and the other involving phenylalanine ammonia‐lyase (PAL). However, the biosynthetic pathway and the importance of SA remain largely unknown in Triticeae. Here, we cloned one ICS and seven PAL genes from barley, and studied their functions by their overexpression and suppression in that plant. Suppression of the ICS gene significantly delayed plant growth, whereas PAL genes, both overexpressed and suppressed, had no significant effect on plant growth. Similarly, suppression of ICS compromised plant resistance to Fusarium graminearum, whereas similar suppression of PAL genes had no significant effect. We then focused on transgenic plants with ICS. In a leaf‐based test with F. graminearum, transgenic plants with an up‐regulated ICS were comparable with wild‐type control plants. By contrast, transgenic plants with a suppressed ICS lost the ability to accumulate SA during pathogen infection and were also more susceptible to Fusarium than the wild‐type controls. This suggests that ICS plays a unique role in SA biosynthesis in barley, which, in turn, confers a basal resistance to F. graminearum by modulating the accumulation of H2O2, and reactive oxygen‐associated enzymatic activities. Although SA mediates systemic acquired resistance (SAR) in dicots, there was no comparable SAR response to F. graminearum in barley. This study expands our knowledge about SA biosynthesis in barley and proves that SA confers basal resistance to fungal pathogens.
Keywords: Hordeum vulgare, isochorismate synthase, phenylalanine ammonia‐lyase, systemic acquired resistance
INTRODUCTION
Salicylic acid (SA) is a small phenolic compound that plays an important role in plant immunity (An and Mou, 2011), plant growth and development (Gallego‐Giraldo et al., 2011; Rivas‐San Vicente and Plasencia, 2011). To date, the majority of studies, particularly those on eudicot species, have classified SA as a defence‐related plant hormone.
Plants possess complex mechanisms to cope with both biotic and abiotic factors in nature. To respond to invading pathogens, plants have evolved two cross‐talking defence cascades: pathogen‐associated molecular pattern (PAMP)‐triggered immunity (PTI), which confers basal resistance, and effector‐triggered immunity (ETI), which induces the expression of resistance (R) genes (Dodds and Rathjen, 2010). Both PTI and ETI promote the accumulation of reactive oxygen species (ROS) (Yoshioka et al., 2015). For biotrophic pathogens, ETI effectively limits pathogen spread by inducing a hypersensitive response (HR) at the site of pathogen entry (Jones and Dangl, 2006).
Systemic acquired resistance (SAR) is an induced systemic resistance after a pathogen has infected a plant locally (Fu and Dong, 2013). SAR is activated by several phloem‐mobile signals, e.g. methyl salicylate (MeSA), dicarboxylic acid, azelaic acid (AzA) and jasmonic acid (JA) (Dempsey and Klessig, 2012; Fu and Dong, 2013), which form two SAR‐inducing branches: one mediated by SA and its signalling component NON‐EXPRESSER OF PATHOGENESIS‐RELATED GENE 1 (NPR1), and the other mediated by several free radicals, specifically nitric oxide (NO), ROS and AzA (Gao et al., 2015). Once operational, SAR promotes the accumulation of pathogenesis‐related (PR) proteins to confer disease resistance.
SA is synthesized via the isochorismate synthase (ICS)‐ and/or phenylalanine ammonia‐lyase (PAL)‐based branches of the shikimic acid pathway (Gao et al., 2015; Sadeghi et al., 2013; Vlot et al., 2009; Wildermuth et al., 2001). For example, tobacco can synthesize SA via the PAL branch (Lee et al., 1995), and Arabidopsis can synthesize SA via the ICS branch (Abreu and Munné‐Bosch, 2009; Wildermuth et al., 2001). Both PAL and ICS branches are important for pathogen‐induced SA synthesis in soybean (Shine et al., 2016). In Arabidopsis, there are two ICS genes, of which ICS1 accounts for 90% of the SA products induced by pathogens or UV light (Garcion et al., 2008). Both SA production and pathogen resistance are severely compromised in the ics1 mutant (Garcion et al., 2008). However, there are still some SA products in the ics1/ics2 double mutant in Arabidopsis (Garcion et al., 2008). SA is normally stored as inactive derivatives that can be converted back to free SA at the site of action. MeSA is a volatile SA derivative that is converted back to SA to induce SAR in the distal tissues between 48 and 72 h after primary infection (Gao et al., 2015).
Fusarium graminearum causes the destructive Fusarium head blight (FHB) in wheat and barley. The fungus infects florets, spreads to adjacent spikelets, and causes necrosis and bleaching of the infected spikelets (Goswami and Kistler, 2004). SA provides basal resistance to multiple pathogens (Murray et al., 2007; Thomma et al., 1998). For example, SA confers both local and systemic defence responses to virus infection in tobacco (Yalpani et al., 1991, 1993). The disruption of SA biosynthesis in Arabidopsis causes an increased susceptibility to F. graminearum (Makandar et al., 2010). Similarly, in wheat, SA treatments have been shown to induce resistance to F. graminearum through a process that involves the increased expression of a number of anti‐pathogen genes, including PR1 (Makandar et al., 2012). SA treatments can also cause disease‐mimicking lesion formation in wheat in the absence of any pathogen (Anand et al., 2003). In Arabidopsis, NPR1 causes SAR, and the expression of Arabidopsis NPR1 in wheat confers improved resistance to F. graminearum (Makandar et al., 2006). However, there are very few reports of a cisgenic study of SA biosynthesis and disease resistance in Triticeae.
Here, we use barley, which is both an economically important cereal and a model member of the Triticeae family, to answer three questions about SA biosynthesis and its function in the Triticeae. (i) Can SA be synthesized by the ICS branch? (ii) Does SA contribute to basal resistance to F. graminearum? (iii) Does SA mediate SAR to the hemibiotrophic pathogen?
RESULTS
Low levels of SA promote plant growth
A gradient of SA, from 10 to 500 μm, was supplied during barley germination. Interestingly, although low levels of SA (10–100 μm, 50 μm in particular) promoted germination, and shoot and root growth, high levels of SA (200–500 μm) suppressed germination and plant growth (Fig. 1). Based on this complex response, SA appears to function as a growth factor or phytohormone in barley.
Figure 1.

Low concentrations of salicylic acid (SA) promote seedling growth in barley. Golden Promise seeds were treated with a gradient of SA (0–500 μm) for 1 week, when the photographs were taken. The micromolar (μm) concentration is indicated for each treatment. Scale bar, 1 cm.
ICS and PAL are differentially organized in barley
As in rice (Yuan et al., 2009), barley has a single ICS gene (GenBank AK358208; Table S1, see Supporting Information). The barley ICS gene is located on chromosome 5H and is orthologous to the rice ICS gene on chromosome 9. The cDNA of barley ICS is approximately 1.7 kb and is derived from 15 exons. The ICS proteins of barley, rice and Brachypodium share 77%–85% identity, but they show less than 51% identity when compared with the ICS proteins of Arabidopsis (Fig. 2a). Despite this, the ICS proteins of Arabidopsis, barley and rice fold into a similar predicted three‐dimensional structure (Fig. S1a, see Supporting Information), suggesting a similar function.
Figure 2.
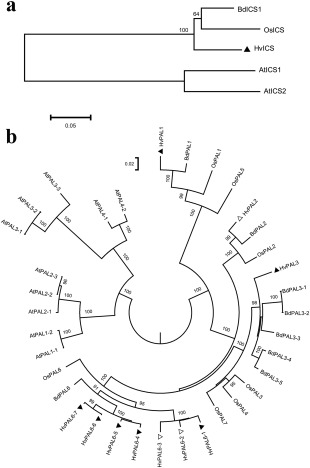
Phylogenetic analysis of isochorismate synthase (ICS) and phenylalanine ammonia‐lyase (PAL) proteins in barley. Phylogenetic trees of ICS (a) and PAL (b). We aligned the protein entries using ClustalW in MEGA6.06 (Tamura et al., 2013), removed poor alignments on terminal regions and constructed a neighbour‐joining tree with 1000 bootstrap iterations. The distance scale bar indicates amino acid differences per unit length. Bootstrap numbers above 50 and 80 are shown for ICS and PAL, respectively. The nomenclature of PAL is based on the rice homologues (Tonnessen et al., 2015). Barley PALs are highlighted by triangles (▲ or Δ), and a solid triangle (▲) indicates a PAL gene under investigation. Protein alignment refers to Figs S1, S10 and S11 (see Supporting Information). Abbreviations: At, Arabidopsis thaliana; Bd, Brachypodium distachyon; Hv, Hordeum vulgare; Os, Oryza sativa. GenBank Accession Numbers: AtICS1 (AAL17715), AtICS2 (ACC60228), BdICS (XM_010239787), HvICS (AK358208), OsICS (AK120689), AtPAL1‐1 (AY045919), AtPAL1–2 (XM002881442), AtPAL2‐1 (XM002877863), AtPAL2‐2 (NM115186), AtPAL2–3 (AY092957), AtPAL3‐1 (NM120505), AtPAL3‐2 (NM001203294), AtPAL3‐3 (XM002871035), AtPAL4‐1 (NP187645), AtPAL4‐2 (XM002882616), BdPAL1 (XM003575348), BdPAL2 (XM003575352), BdPAL3‐1 (XM003575190), BdPAL3‐2 (XM003575192), BdPAL3‐3 (XM003575355), BdPAL3–4 (XM003575356), BdPAL3–5 (XM003575317), BdPAL6 (XM003580096), HvPAL1 (AK253101), HvPAL2 (AK356545), HvPAL3 (AK251356), HvPAL6‐1 (AK248831), HvPAL6‐2 (AK361880), HvPAL6‐3 (AK371460), HvPAL6‐4 (AK249266), HvPAL6‐5 (AK248841), HvPAL6‐6 (AK250690), HvPAL6–7 (AK250100), OsPAL1 (Os02g41630), OsPAL2 (Os02g41650), OsPAL3 (Os02g41670), OsPAL4 (Os02g41680), OsPAL5 (Os04g43760), OsPAL6 (Os04g43800) and OsPAL7 (Os05g35290).
There are multiple PAL genes in Arabidopsis, barley, Brachypodium and rice (Fig. 2b). The Arabidopsis PAL genes are more divergent from those in grasses. In barley, at least 10 PAL genes are found on chromosomes 1H, 2H and 6H (Table S1). The barley PAL genes are highly conserved with two exons and an approximate length of 2.1 kb, and their protein products share approximately 90% identity. Seven barley PAL genes form an independent clade similar to OsPAL6, but HvPAL1 (AK253101), HvPAL2 (AK356545) and HvPAL3 (AK251356) were grouped in other clades with Brachypodium and rice homologues (Fig. 2b). The barley proteins have an alanine‐serine‐glycine (Ala‐Ser‐Gly) peptide (Fig. S1b) similar to the sequence found in the active site of other PAL proteins (Calabrese et al., 2004).
ICS probably accounts for SA biosynthesis in barley
The barley ICS displayed ubiquitous expression in leaves, stem and sheath, but relatively low expression in spikes (Fig. 3a), suggesting that the plant's demands for SA might differ in different tissues. In the barley database, the ICS transcripts are also enriched in leaf tissue, but poorly represented in spikes, inflorescences or developing grains (Fig. S2, see Supporting Information). The seven PAL genes selected showed an equally biased organ‐specific pattern of expression. Although the expression pattern of each PAL gene was unique, all showed lower expression in flag leaves (Figs 3b–f and S3, see Supporting Information). For example, PAL1 is enriched in spikes, whereas PAL6‐4 and PAL6‐6 are enriched in stems. PAL6‐1 displays poor expression in the current study and in the barley database (Figs 3 and S2). Collectively, the expression of PAL genes was organ specific, most probably because of organ‐specific differences in the flux of trans‐cinnamic acid through secondary metabolism.
Figure 3.
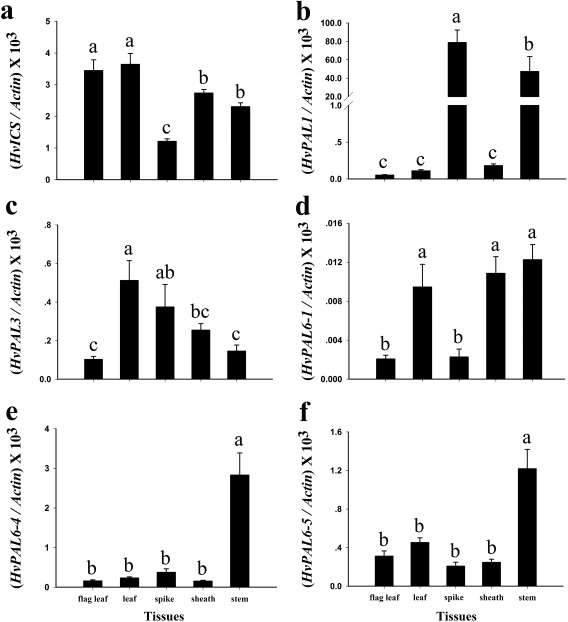
Expression of ICS and PAL genes in barley. The mRNA expression of HvICS (a), HvPAL1 (b), HvPAL3 (c), HvPAL6‐1 (d), HvPAL6‐4 (e) and HvPAL6‐5 (f). The expression of these genes was analysed in the wild‐type ‘Golden Promise’ at the heading stage. The tested organs were the flag leaf, penultimate leaf (leaf), peduncle (stem), sheath of the flag leaf (sheath) and inflorescence (spike). All measurements were normalized to the expression of Actin. Using SAS version 9.0 (SAS Institute Inc., Cary, NC, USA), comparisons were made using Duncan multiple range tests at a significance level of 0.05. Error bars represent the standard error of the mean (SEM).
To study the function of ICS and PAL genes, we prepared both overexpression (OE) and RNA interference (RNAi) constructs (Fig. 4a), and transformed them into barley ‘Golden Promise’ (Table S2, see Supporting Information). Because the barley ICS is a single‐copy gene, complete or near‐complete silencing of this gene could be deleterious. As a precaution, we employed two different promoters to express ICS RNAi. The maize Ubiquitin (Ubi) promoter (Christensen et al., 1992) is a constitutive promoter widely used in research. The wheat Wks1 promoter (Fu et al., 2009), on the other hand, is an alternative promoter that has been successfully used to express transgenes (Yuan et al., 2018). All of the remaining constructs derived from the ICS and PAL genes were based on the Ubi promoter. All OE transgenic plants were generated using standard protocols. In the T1 generation, plants segregated for the target transgene (Table S3, see Supporting Information). Except for Wks1::ICS RNAi2, most transgenic plants were comparable with non‐transgenic controls in terms of flowering time, tiller number and other tangible agronomic traits (Table S4, see Supporting Information). They behaved similarly in the T2 generation (Fig. 4b). For RNAi studies, we generated transgenic plants for six PAL genes using standard procedures. The PAL6‐6 construct actually targeted both PAL6‐6 and PAL6–7. All of the PAL RNAi plants were morphologically comparable with the non‐transgenic control. However, we were unable to produce RNAi plants for the ICS gene using the established transformation procedure (Table S2). Both callus proliferation and plantlet regeneration were arrested, probably because the silencing of ICS led to inadequate SA to sustain normal cellular activity on the medium. We then modified the medium composition by the addition of 100 μm SA. As expected, the SA supplement partially rescued transgenic ICS RNAi cells, allowing them to proliferate and regenerate (Table S2). T0 ICS RNAi plantlets, once established in soil, managed to flower and to set seed when they were supplied with water containing 100 μm SA. In the T1 generation, additional SA was not essential to allow ICS RNAi plants to flower and set seed. The overexpression and silencing of three target genes were validated in the tested T1 plants using quantitative real‐time polymerase chain reaction (qRT‐PCR) (Fig. S4, see Supporting Information). Based on these observations, the ICS gene probably accounts for the majority of SA biosynthesis in barley.
Figure 4.
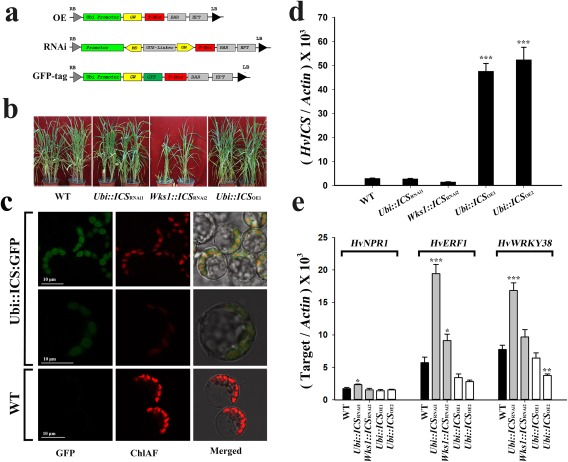
Characterization of transgenic barley plants for the ICS gene. (a) Maps of the T‐DNAs used to make constructs to overproduce (OE), silence (RNAi) or GFP‐tag the target genes. (b) Typical phenotypes of wild‐type (WT) barley, and transgenic T2 barley with the ICS gene. (c) Subcellular localization of ICS‐GFP and autofluorescing chlorophyll (ChlAF) in transgenic T1 Brachypodium (top two rows). (d) The expression of ICS in transgenic T2 barley with the ICS gene. (e) The expression of HvNPR1, HvERF1 and HvWRKY38 in transgenic T2 barley with altered expression of ICS. The overexpression (OE) and RNA interference (RNAi) vectors were used for the ICS and PAL genes, but the green fluorescent protein (GFP) tag vector was only used for the ICS gene. The maize Ubiquitin (Ubi) promoter (Christensen et al., 1992) and the wheat Wks1 promoter (Fu et al., 2009) were used to express the RNAi matrix. The Ubi::ICS RNAi1 and Wks1::ICS RNAi2 plants were derived from the Ubi‐ and Wks1‐based constructs, respectively. All measurements were normalized to the expression of Actin. Comparisons were made using Dunnett's tests (transgenic lines vs. WT; *P < 0.05, **P < 0.01, ***P < 0.001). Error bars are the standard error of the mean (SEM). GenBank Accession Numbers: NPR1 (AM050559), ERF1 (AK354662) and WRKY38 (AK360269).
For the ICS gene, we obtained at least 11 independent OE lines, all employing the Ubi promoter. We only obtained three independent RNAi lines: one with Ubi and two with Wks1 promoters (Table S2). In order to limit the breadth of the follow‐up studies, we focused on two representative OE lines (Ubi::ICS OE1 and Ubi::ICS OE2) and two representative RNAi lines (Ubi::ICS RNAi1 and Wks1::ICS RNAi2) (Table S3). Except for the Wks1::ICS RNAi2 line, all displayed a simple Mendelian segregation for one transgene in the T1 generation (Table S3). Further phenotyping showed that both Ubi::ICS OE1 and Ubi::ICS OE2 lines were comparable in many key measures of this study. Similarly, the Ubi::ICS RNAi1 line was comparable with plants of the Wks1::ICS RNAi2 line. Of the three spike lines of Wks1::ICS RNAi2 (Table S3), both the W4‐ and W6‐derived plants were comparable in plant height (P = 0.6) and tiller number (P = 0.7) in the T1 generation. We then selected the spike line W4–8 as the representative source material for Wks1::ICS RNAi. In the T2 generation, we analysed plants of the Ubi::ICS OE1 and Wks1::ICS RNAi2 lineages, but also documented data from the T2 plants of Ubi::ICS OE2 and Ubi::ICS RNAi1 in either the main text or Supporting Information.
We also prepared a tagged construct for ICS:GFP (Fig. 4a), and transformed it into Brachypodium ‘Bd21‐3’. In transgenic Brachypodium, ICS:GFP was localized to the chloroplast (Fig. 4c), which indicates that SA is most probably synthesized in chloroplasts in grass species as it is in Arabidopsis (Garcion et al., 2008).
ICS regulates SA content and responsive genes in barley
In barley, overexpression increased the ICS mRNA by about 16‐ to 18‐fold in T2 plants (e.g. Ubi::ICS OE1 and Ubi::ICS OE2 derived from independent calli; Table S3) compared with endogenous ICS in wild‐type controls (Fig. 4d). Interestingly, this correlated with a comparatively small increase in SA in T2 plants (e.g. 24 ppb in Ubi::ICS OE1; 1 ppb = 1 ng/g) compared with the WT control (15 ppb). As described above, we used both the Ubi and Wks1 promoters to express ICS RNAi. It appeared that both Ubi‐ and Wks1‐based RNAi reduced the ICS mRNA in T2 plants (e.g. Ubi::ICS RNAi1 and Wks1::ICS RNAi2 derived from independent calli; Table S3) to about 49% (in Wks1::ICS RNAi2) to 73% (in Ubi::ICS RNAi1) of that in the WT control (Fig. 4d). However, the reduction was lower than that in the T1 generation, which was about 13%–20% of that in the WT control (Fig. S4). Presumably, the recipient plants have certain mechanisms to counteract the engineered RNAi. The SA content in the RNAi T2 plants (e.g. Wks1::ICS RNAi2) was 13 ppb, which was lower but comparable with that of the WT control (e.g. 15 ppb). Collectively, the expression of ICS correlated with the SA content in barley, but evidently was not the only gene product responsible for the determination of SA accumulation and turnover.
Even these somewhat moderate perturbations in SA metabolism had a significant impact on the expression of SA‐regulated genes. Using ICS‐based transgenic barley, we examined NPR1, which encodes an SA signal transducer in Arabidopsis (Pieterse and Van Loon, 2004), ERF1, which encodes a positive regulator for disease resistance in JA/ethylene (ET) pathways (Lorenzo et al., 2003), and WRKY38, which encodes a repressor targeting both SA and JA signalling (Kim et al., 2008; Vlot et al., 2009). The barley NPR1 was not responsive to the level of expression of the ICS gene, but ERF1 and WRKY38 displayed a negative correlation with the mRNA content of ICS (Fig. 4e).
Suppression of barley ICS compromises foliar resistance to F. graminearum
Fusarium graminearum causes FHB in wheat and barley; it is a necrotic pathogen, but with a biotrophic phase (Kazan et al., 2012). Using a leaf‐based test, we compared the F. graminearum lesions on infected leaves at 72 h post‐inoculation (hpi). The RNAi T2 plants (e.g. Wks1::ICS RNAi2) showed the largest lesions, but those on the OE line (Ubi::ICS OE1) were comparable with those on the WT control (Fig. 5a,b). The silencing of ICS compromised the basal resistance levels to F. graminearum in barley. We also tested the RNAi T2 plants for their responses to Blumeria graminis f. sp. hordei, which is a biotrophic pathogen. At 5 days post‐inoculation (dpi), the RNAi T2 plants (e.g. Wks1::ICS RNAi2) showed longer mycelia relative to those of the other lines (Fig. S5, see Supporting Information).
Figure 5.
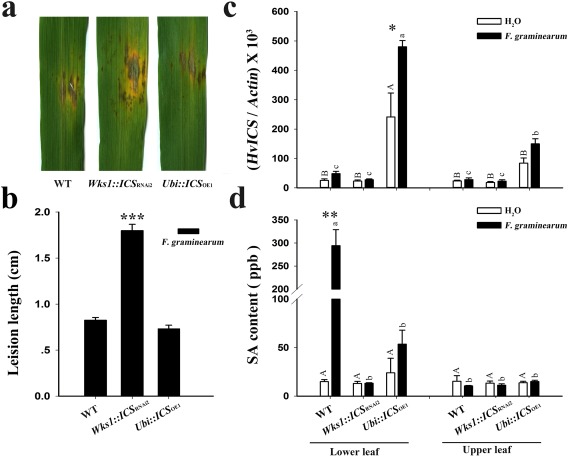
ICS impacts barley foliar resistance to Fusarium graminearum. This figure illustrates lesion formation by F. graminearum on barley leaves (a), the length of developing lesions (b, n = 10), the mRNA level of ICS (c, n = 6) and the salicylic acid (SA) content (d, n = 3) in the wild‐type (WT) and T2 transgenic plants at 72 h post‐inoculation (hpi). In (c) and (d), the lower leaves were treated using water or AmCyanPH‐1; the upper leaves were untreated; the samples in (c) are arranged in the same order as those in (d). Dunnett's tests were used to compare transgenic vs. WT plants in (b), and the H2O‐ vs. AmCyanPH‐1‐treated samples in (c) and (d) (*P < 0.05, **P < 0.01, ***P < 0.001). Duncan's multiple range tests were used to compare the H2O‐treated samples (capital letters at P < 0.05) and the AmCyanPH‐1‐treated samples (small letters at P < 0.05) in (c) and (d). Actin was used as the internal control. Error bars are the standard error of the mean (SEM). Relative data of independent transgenic events refer to Fig. S12 (see Supporting Information).
In Arabidopsis, pathogen infection induces expression levels of the ICS1 gene (Wildermuth et al., 2001). Similarly, F. graminearum infection in barley at 72 hpi up‐regulates the expression of ICS (Fig. 5c), NPR1 and WRKY38 (Fig. S6, see Supporting Information). For all three tested genes, the OE line (Ubi::ICS OE1) displayed the highest up‐regulation ratio (2 : 1) when compared with the RNAi line (Wks1::ICS RNAi2) and the WT control. In the infected tissue at 72 hpi, we also found that F. graminearum infection increased the localized SA content in the OE line (Ubi::ICS OE1) and the WT control (Fig. 5d). The WT control was unexpectedly high with regard to localized SA. It remains unknown what caused an acute increase in SA in the WT control. However, the RNAi T2 plant (e.g. Wks1::ICS RNAi2) retained a low SA content despite F. graminearum infection (Fig. 5d). Apparently, barley ICS regulates the SA content under pathogen attack and, as a result, leads to higher defensive responses.
Overexpression of barley ICS enhances floral resistance to F. graminearum
Using a green fluorescent protein (GFP)‐labelled strain, we then tested the floral responses to F. graminearum on transgenic plants with the ICS gene. The OE line (Ubi::ICS OE1) was associated with the least fluorescence relative to the RNAi T2 plants and WT control at an early infection stage (e.g. 24 and 48 hpi, Fig. 6a). However, there was no significant difference between the OE plants and other lines at the late infection stage (e.g. 72 hpi, Fig. 6a). We then estimated F. graminearum growth by quantification of the integrated fluorescence intensity (IFI) of the infected florets. Again, the OE plant was associated with the lowest IFI reading at 24 and 48 hpi (P < 0.001), but not at 72 hpi (Fig. 6b). Therefore, barley ICS, when overexpressed (Fig. 6c), enhances floral resistance to F. graminearum, but only during the initial stages of infection.
Figure 6.
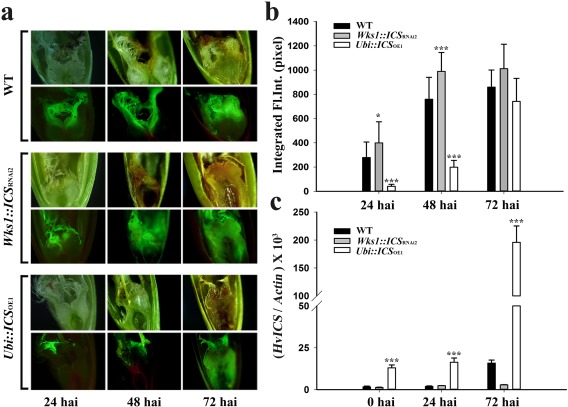
ICS regulates floral resistance to Fusarium head blight (FHB) in T2 transgenic barley. (a) AmCyanPH‐1‐based green fluorescence in infected florets of three barley lines labelled on the left. Each line was photographed in both bright field (top images of each set) and fluorescence (bottom images of each set). (b) Integrated fluorescence intensity (Integrated Fl. Int.) of the infected florets: at least 10 florets were analysed for each genotype using ImageJ. (c) The expression of ICS in infected florets. Tukey–Kramer's tests plus a pdiff option were used to compare transgenic vs. wild‐type (WT) plants in (b), and Dunnett's tests were used to compare transgenic vs. WT plants in (c) (*P < 0.05, ***P < 0.001). Actin was used as the internal control. Error bars are the standard error of the mean (SEM). Relative data of independent transgenic events refer to Fig. S13 (see Supporting Information). hai, hours after inoculation.
ICS modulates antioxidant enzymes and ROS in Fusarium‐infected barley
ROS are responsive to pathogen infection (Torres et al., 2006). We estimated the levels of superoxide radical ( ) and hydrogen peroxide (H2O2) in F. graminearum‐infected spikes from transgenic barley with altered ICS expression. After inoculation, the OE plant (e.g. Ubi::ICS OE1) was lighter in colour when stained with diaminobenzidine (DAB) (Fig. 7a) and nitroblue tetrazolium (NBT) (Fig. 7b) than the WT control. This indicated that there were lower concentrations of and H2O2 in the OE plant when compared with the unaltered plant. We further confirmed this pattern by quantifying the content of H2O2 and the production rate in different genotypes (Fig. 7c,d). We then estimated the activity of antioxidant enzymes. The OE plant (e.g. Ubi::ICS OE1) was associated with higher activity of ascorbate peroxidase (APX), catalase (CAT), guaiacol peroxidase (POD) and superoxide dismutase (SOD) at both the infection site (central spikelet) and in an uninfected site (upper spikelet) (Fig. 8). APX and SOD showed maximum expression at 24 hpi, but CAT and POD (of the upper spikelet) peaked at 72 hpi. It is likely that higher activities of antioxidant enzymes were responsible for reducing and H2O2 accumulation in the OE plants (e.g. Ubi::ICS OE1). However, the RNAi plants either accumulated WT, or less than WT, levels of APX, CAT, POD and SOD (except for the upper spikelet at 72 hpi) (Figs 7 and 8).
Figure 7.
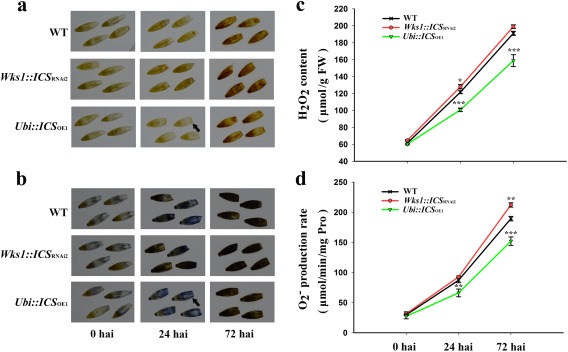
Accumulation of reactive oxygen species in T2 transgenic barley after Fusarium graminearum infection. (a) Diaminobenzidine (DAB) staining for H2O2. (b) Nitroblue tetrazolium (NBT) staining for . (c) Increase in H2O2 levels over 72 h after infection (hai). (d) Increased rate of production of over 72 hai. Dunnett's tests were used to compare transgenic vs. wild‐type (WT) plants (*P < 0.05, **P < 0.01, ***P < 0.001). Arrows indicate a low stain in the Ubi::ICS OE1 plant at 24 hai. Error bars are standard error of the mean (SEM). FW, fresh weight; hai, hours after inoculation; Pro, proline.
Figure 8.
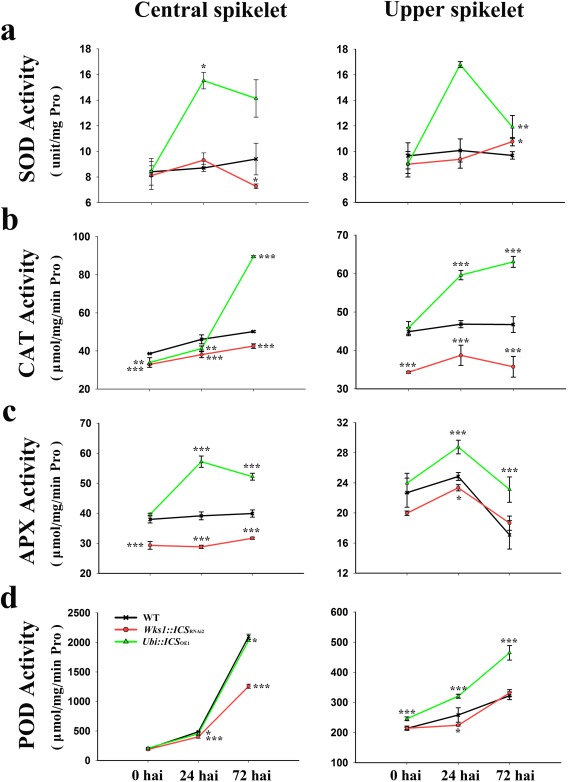
Characterization of antioxidant enzymes in T2 transgenic barley after Fusarium infection. Enzymatic activities of superoxide dismutase (SOD) (a), catalase (CAT) (b), ascorbate peroxidase (APX) (c) and guaiacol peroxidase (POD) (d). Central spikelets were inoculated with F. graminearum; upper spikelet represents a secondary uninfected site. Dunnett's tests were used to compare transgenic vs. wild‐type (WT) plants (*P < 0.05, **P < 0.01, ***P < 0.001). Error bars are standard error of the mean (SEM). hai, hours after inoculation; Pro, proline.
SAR is not significant in barley
SA plays an important role in SAR in numerous plants (Vlot et al., 2009). In barley, exogenous SA reduces Fusarium seedling blight (Wiśniewska and Chełkowski, 1999). However, whether SA induces a practical SAR to one or multiple pathogens in barley remains to be answered. In order to investigate this, we examined the expression of some relevant genes and measured the SA content at a primary F. graminearum infection site and at an uninfected site (upper leaves). In the primary infection site, ICS expression in the OE plant (e.g. Ubi::ICS OE1) was up‐regulated when compared with the H2O control. No such up‐regulation occurred in Wks1::ICS RNAi2 (Fig. 5c). NPR1 and WRKY38 similarly showed increased expression at the primary infection sites in OE plants (Fig. S6a,b). However, in uninfected sites, only WRKY38, and not ICS and NPR1, showed a significant increase in expression in OE plants (Figs 5c and S6a,b). Silencing of the ICS gene abolished the systemic up‐regulation of ICS, NPR1 and WRKY38.
We then compared the hormone contents at both primary infected and secondary uninfected sites. At the primary infection site, F. graminearum triggered an increase in SA content in the WT, but not in the RNAi and OE genotypes (Fig. 5d). These same infections appeared to increase JA in all genotypes, but not abscisic acid (ABA) (Fig. S6c,d). However, comparable increases in the hormones did not occur at the secondary uninfected site (Figs 5d and S6c,d). Phenotypically, SAR was also not detected on barley leaves tested with F. graminearum (Fig. S7, see Supporting Information).
DISCUSSION
SA is an essential hormone in barley development
In plants, SA is normally classified as a signal molecule that promotes anti‐pathogen defences in plants (An and Mou, 2011). However, SA also plays an important role in abiotic adaptation, growth and development (Rivas‐San Vicente and Plasencia, 2011). In this study, SA treatment significantly impacted seed growth in barley (Fig. 1). In Arabidopsis, low levels of endogenous SA led to increased leaf biomass production at the early stages of reproduction and increased seed yield in NahG‐expressing transgenic lines and sid2 mutants (Abreu and Munné‐Bosch, 2009). In the current study, low levels of endogenous SA strongly delayed growth and flowering. For instance, RNAi‐based repression of the ICS gene diminished the ability of transformed cells to regenerate. These transgenic cells could be rescued by supplementation of their medium with 100 μm SA. However, how the ICS RNAi progenies (T1 and T2) bypassed the need for SA supplementation in order to grow, flower and set seed remains unclear. It appears that tissue culture and regeneration are more sensitive than the whole plant to the effects of SA reduction. It was noted that, although the RNAi plants developed without SA‐supplied watering, Wks1::ICS RNAi2 plants appeared to be slow in germination (Fig. S8, see Supporting Information) and in the other developmental processes. Similarly, a knock‐out of the ICS‐A locus led to dwarfism and late flowering in the durum wheat ‘Kronos’ (Fig. S9, see Supporting Information). Too much SA can also be inhibitory: the overaccumulation of SA caused a stunted rosette growth in Arabidopsis (Janda et al., 2014). The present article shows that high levels of exogenous SA delay seed germination and shoot growth in barley (Fig. 1).
In Arabidopsis, chorismate is a substrate for producing isochorismate and, subsequently, SA through the actions of ICS and pyruvate lyase (Wildermuth et al., 2001). Most pathogen‐induced SA is synthesized by this pathway in Nicotiana benthamiana (Catinot et al., 2008). Arabidopsis ICS1 accounts for approximately 90% of SA and its derivatives that are induced by pathogens or UV light (Garcion et al., 2008). The barley ICS, encoded by a single gene, shares high similarity with the isoforms found in Arabidopsis and rice (Orvza sativa). The current study proposes that ICS is required to synthesize SA in barley, and probably in other grass species.
SA confers basal resistance to F. graminearum in barley
In Arabidopsis, Erysiphe orontii infection increased the SA content about five‐fold (Wildermuth et al., 2001). Similarly, F. graminearum infection resulted in extensive SA induction in wild‐type barley (Fig. 5d). Unexpectedly, this induction was not seen in OE lines. Why the OE lines did not respond in a similar manner to WT barley remains unclear. Apparently, the SA pathway and its regulatory network are more complicated than expected, and it is already known that SA interacts with the biosynthetic pathways of other phytohormones (Robert‐Seilaniantz et al., 2011). Therefore, the conclusions of this study are viewed as being based on the ‘perturbation’ of SA accumulation rather than on simple decreases/increases in SA levels.
In Arabidopsis, the expression of ICS positively responds to pathogen infection (Garcion et al., 2008), consistent with the suggested role of SA biosynthesis in biotic stress responses. Here, we demonstrated that F. graminearum infection increased the SA content in the wild‐type, but not in the ICS‐silenced line (Fig. 5d). This suggests that barley synthesizes SA via an ICS‐dependent pathway when responding to pathogen attack.
Traditionally, SA has been assumed to protect plants from biotrophic pathogens, whereas JA and ET protect plants from necrotrophic pathogens (Glazebrook, 2005). Fusarium graminearum is hemibiotrophic, that is, biotrophic initially, but necrotrophic during pathogenesis when cell death is induced (Kazan et al., 2012). In wheat, F. graminearum ‘true’ resistance is probably mediated by JA and ET, and not SA (Li and Yen, 2008). However, SA contributes to this resistance by inhibiting mycelial growth and conidial germination of F. graminearum (Qi et al., 2012). In the current study, overexpression of ICS increased SA concentration, which repressed the initial infection at 24 and 48 hpi (Fig. 6a), but had no significant effect on the repression of pathogenesis at a later stage of infection (Fig. 6a,b). Apparently, SA enhanced the basal resistance during the biotrophic stage of F. graminearum attack. Here, we have further confirmed that the down‐regulation of SA biosynthesis promotes the growth of Blumeria graminis f. sp. hordei – a biotrophic pathogen. Therefore, SA regulates plant basal resistance to some biotrophic pathogens or to some pathogens at the biotrophic stages of their life cycle.
The accumulation of ROS is one of the earliest events in plants under pathogen attack. A ROS burst could be caused by or, through a feed‐back loop, cause SA signalling (Chen et al., 1993; Leon et al., 1995). In this study, ICS‐overexpressing lines were associated with higher antioxidant enzyme activity and lower levels of H2O2 and than those in the wild‐type controls and the ICS‐silenced lines (Fig. 7c,d). However, how an improved antioxidant activity contributes to F. graminearum resistance remains unclear.
SAR is ineffective against F. graminearum in barley
SAR, as a form of broad‐spectrum resistance, is induced in response to local infections and protects uninfected parts against subsequent secondary infections by related or unrelated pathogens (Singh et al., 2017). SAR has been well documented in dicotyledonous plants (Fu and Dong, 2013; Vlot et al., 2009), but is poorly understood in monocotyledonous plants.
In wheat, a number of reports have addressed SAR (Moya‐Elizondo and Jacobsen, 2016; Plotnikova, 2009; Yang et al., 2013). For example, foliar applications of Bacillus mycoides reduced the severity of Fusarium crown rot, which was assumed to be caused by B. mycoides‐induced SAR (Moya‐Elizondo and Jacobsen, 2016). In barley, foliar application of Pseudomonas syringae induced broad‐spectrum acquired resistance (AR) at a distance from the local lesion (Colebrook et al., 2012). Other studies using the biotrophic pathogen Blumeria graminis have revealed that the barley AR is strictly localized to the inoculated cell or to the cells immediately adjacent to the site of attempted penetration (Lyngkjær and Carver, 1999; Ouchi et al., 1976). Because these results are so contradictory, it remains unclear whether a classic SAR protective against varied pathogens exists in this group of grasses.
To test the F. graminearum infection in barley, we performed a double inoculation procedure (‘inducer’ followed by ‘challenger’) (Lyngkjær and Carver, 1999). We found that the infection of the inducer on one leaf did not induce SAR against the challenger on a different leaf (Fig. S7). There was no significant change in ICS expression and SA level at uninfected sites (Fig. 5c,d), even though peroxidase activity was up‐regulated at uninfected sites in the ICS OE line (Fig. 8). Thus, although barley appears to have a basal systemic defence response, as indicated by the detectable cellular responses of SAR at a secondary uninfected site, this is inadequate to confer a practical SAR for secondary infections, at least for F. graminearum tested here.
EXPERIMENTAL PROCEDURES
Plant material and Fusarium strain
This study was performed on spring barley (Hordeum vulgare L., cultivar ‘Golden Promise’) and purple false brome (Brachypodium distachyon L., inbred line ‘Bd21‐3’). Wild‐type and transgenic plants were grown in glasshouses with a 16‐h photoperiod (light intensity, 105 μmol/m2/s) and a daytime temperature of 25 ºC.
Fusarium tests were performed using the F. graminearum strain ‘AmCyanPH‐1’, which is an engineered strain expressing the AmCyan fluorescent protein (Zhang et al., 2012). AmCyanPH‐1 was provided by Dr W. Tang (Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China). Powdery mildew tests were performed using the Blumeria graminis f. sp. hordei isolate ‘K1’ (Liu et al., 2014), which is virulent to Golden Promise. Blumeria graminis f. sp. hordei isolate K1 was provided by Dr Q. Shen (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China).
Plasmid construction
In barley, there is a single ICS gene (GenBank AK358208) and at least seven PAL homologues: PAL1 (AK253101), PAL3 (AK251356), PAL6‐1 (AK248831), PAL6‐4 (AK249266), PAL6‐5 (AK248841), PAL6‐6 (AK250690) and PAL6‐7 (AK250100). Their full‐length cDNAs were isolated from the barley cultivar ‘Tamalpais’, and cloned into the Gateway (GW)‐compatible OE vector PC186 (pUbi::GWOE::Nos). To trigger specific RNAi of ICS, the 80–385‐bp region (counted from the start codon) was cloned into the RNAi vectors PC336 (Ubi::GWRNAi::Nos) and PC691 (Wks1::GWRNAi::Nos), where Ubi, Wks1 and Nos are the maize Ubi promoter (Christensen et al., 1992), wheat Wks1 gene promoter (Fu et al., 2009) and Agrobacterium Nos terminator (Wood et al., 2001), respectively. For PAL genes, we cloned a 1792–1990‐bp region (counted from the start codon) into the RNAi vector PC336 (Ubi::GWRNAi::Nos). The barley ICS gene was also cloned into the vector PC611 (Ubi::GW:GFP::Nos), where it gained a C‐terminal GFP. PCR primers are described in Table S5 (see Supporting Information).
Generation of transgenic plants
Agrobacterium‐mediated barley transformation
Agrobacterium‐mediated gene transfer was performed using the published protocol (Harwood et al., 2009) with slight modifications. Plant expression constructs were first introduced into the A. tumefaciens strain ‘AGL1’ using electroporation. A single AGL1 colony was amplified in liquid MG/L medium containing 50 mg/L kanamycin, 40 mg/L rifampicin and 100 mg/L carbenicillin, and the culture was shaken at 200 rpm and 28 °C for 48 h. The AGL1 suspension, about 200 µL, was spread on solid MG/L medium with the same supplements, and set in a 28 °C incubator for 48 h. AGL1 colonies were suspended in liquid callus induction medium (CIM), adjusted to an optical density at 600 nm (OD600) of 0.6, supplied with 200 µm acetosyringone and 0.1% (v/v) PE/F68 (Sigma, St Louis, MO, USA), and subjected to co‐cultivation with barley explants.
To prepare immature embryos, caryopses were collected from developing spikes that were approximately 14 days post‐pollination, sterilized in a solution of ethanol (75%, v/v) and Tween‐20 (5%, v/v) for 5 min, plus an ultrasonic treatment at room temperature (stimulating time, 6 s; interval, 30 s; repeated six times), sterilized again in 20% (v/v) sodium hypochlorite for 15 min with shaking (200 rpm), and rinsed five times using sterile deionized water. Immature embryos (1.5–2.5 mm in length) were collected from sterilized caryopses using fine forceps; after removing the embryonic axis, immature embryos were placed with the scutellum side up onto CIM supplied with 2.5 mg/L Dicamba and incubated in the dark at 24 °C for 2 days.
A drop of 10 µL of fresh AGL1 inoculum (OD600 = 0.6) was added to the embryo scutellum. After the surface had been air dried (about 60 min at room temperature), the infected embryos (scutellum side up) were co‐cultured on fresh CIM medium supplied with 2.5 mg/L Dicamba and 200 µm acetosyringone at 24 °C in the dark for 3 days, and then transferred to CIM medium supplemented with 200 mg/L Timentin and 25 mg/L hygromycin at 24 °C. The infected material was maintained in the dark, and subcultured every 14 days for a total of 12 weeks. During/after subculture, the most vigorously growing embryogenic callus was transferred to shoot induction medium (SIM) supplied with 2.5 mg/L 2,4‐Dichlorophenoxyacetic acid (2,4‐D), 0.1 mg/L 6‐Benzylaminopurine (6‐BA), 200 mg/L Timentin and 25 mg/L hygromycin, and incubated at 24 °C under 16 h of fluorescent light (light intensity, 2 μmol/m2/s) for 2 weeks. Vigorous embryogenic callus with green tissue was moved to SIM medium supplemented with 200 mg/L Timentin and 25 mg/L hygromycin with no additional hormones, and incubated at 24 °C under 16 h of fluorescent light (light intensity, 25 μmol/m2/s) for 3–4 weeks. Regenerated plantlets were transplanted to root induction medium (RIM). Plantlets with healthy roots were maintained in an open tissue culture bottle for 3 days, and then transferred to soil in a glasshouse.
Transgenic plants were confirmed by PCR amplification of the selection markers (BAR or HPT) and/or vector‐specific fragments. Transgenic plants were also wipe tested for their resistance to 0.3% (v/v) Finale herbicide (undiluted herbicide containing 11.33% glufosinate ammonium). The T2 transgenic plants were prioritized for the current study.
Agrobacterium‐mediated transformation in Brachypodium
Tissue culture and Agrobacterium‐mediated transformation of Brachypodium were conducted as reported by Bragg et al. (2012).
Quantitative transcription analysis
Total RNA was extracted using Trizol reagent (Life Technologies, Grand Island, NY, USA). cDNA was synthesized using a Fermentas First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA). For qRT‐PCR, primer efficiency (93–100%, Table S5) was tested on 10‐fold serial dilutions of cDNA templates using StepOnePlus Systems (Life Technologies) and FastStart SYBR Green Master (Roche Applied Science, Indianapolis, IN, USA). qRT‐PCR cycles were 10 min at 95 °C, 40 cycles of two consecutive steps of 15 s at 95 °C and 1 min at 60 °C, and a standard dissociation procedure. Quantitative transcription was averaged from six biological replicates. Actin and Cyclophilin were used as endogenous controls (Table S5).
Measurement of SA and JA contents
An established procedure (Xu et al., 2016) was used to quantify SA and JA in barley leaves and spikes. About 150 mg of fresh leaves or spikes were harvested and ground in liquid nitrogen for analysis. Hormones were extracted in 1.9 mL of 70% acetone: 30% 50 mm citric acid solution supplemented with 10 ng each of the internal standards dihydrojasmonic acid (Sigma) and [2H4]‐SA (Olchemim, Olomouc, Czech Republic). After removing acetone by overnight evaporation, samples were re‐extracted with diethyl ether. Dry samples were resuspended in methanol, filtered through a 0.22‐mm Polyvinylidene fluoride (PVDF) membrane (Millipore, Bedford, MA, USA) and analysed using a liquid chromatography–tandem mass spectrometry system (TSQ Quantiva, Thermo Fisher Scientific, Waltham, MA, USA) with an optimized ion source. Each compound and its internal standard were quantified by tandem mass spectrometry using the scan mode at m/z 137.12 (SA) and 141.27 ([2H4]‐SA), 209.3 (JA) and 211.09 (dihydrojasmonic acid). Electrospray ionization (ESI) analysis was performed in negative ionization mode. Each extraction and analysis were replicated four times.
Subcellular localization
Transgenic Brachypodium plants with a GFP tag were germinated in a dark chamber at 28 °C. Protoplasts were isolated from 2‐week‐old seedlings using a published protocol (Hong et al., 2012). In brief, the first two leaves were excised and cut at intervals of 0.1 mm using a sharp razor blade. Leaf cuts were immediately soaked in 20 mL of filter‐sterilized solution containing 0.6 m mannitol, 10 mm 2‐(N‐morpholino)ethanesulfonic acid (MES, pH 5.7), 1.5% cellulase ‘Onozuka R‐10’ (Yakult Pharmaceutical, Tokyo, Japan), 0.75% macerozyme ‘R‐10’ (Yakult Pharmaceutical), 0.1% bovine serum albumin, 1 mm CaCl2 and 5 mm β‐mercaptoethanol. A vacuum (–68.95 kPa) was applied for 30 min to enhance enzyme filtration into the tissues. The leaf–enzyme mixture was first shaken at 40 rpm in the dark at room temperature for 3 h, and was then shaken at 60 rpm for 30 min to release the protoplasts. Protoplasts were inspected under a Leica TCS SP5 II laser scanning confocal microscope (Leica Microsystems, Wetzlar, Hesse, Germany).
Accumulation of superoxide radicals ( ) and hydrogen peroxide (H2O2)
The accumulation of and H2O2 in leaves was visually evaluated by staining with NBT (0.5 mg/mL, pH 7.6) for and DAB (1 mg/mL, pH 3.8) for H2O2. Chlorophyll was removed by infiltrating leaves with lactol–glycerol–ethanol (1 : 1 : 4, v/v/v).
The concentration of was measured as reported previously (Sui et al., 2007). Leaf tissues were thoroughly ground in potassium phosphate buffer (50 mm, pH 7.8) on ice. A 0.5‐mL aliquot of supernatant was removed and mixed with 0.5 mL of phosphate buffer (50 mm, pH 7.8) and 1 mL of 10 mm hydroxylammonium chloride. The mixture was incubated at 25 °C for 20 min. Finally, 4 mL of ethyl ether was added to the mixture. The absorbance of the water phase was measured at 530 nm. The content of H2O2 was determined by estimating the titanium‐hydroperoxide complex (Mukherjee and Choudhuri, 1983).
Antioxidant enzyme activity
The activities of selected enzymes were determined as described previously, including SOD (He et al., 2005), APX (Cakmak and Marschner, 1992), CAT (Durner and Klessig, 1996) and POD (Scebba et al., 2001). Enzymatic activity was quantified using a UV–visible spectrophotometer (UV‐2550, Shimadzu, Japan) at 25 °C.
Fusarium graminearum test
Both leaf‐ and spike‐based tests were performed. The harvested conidia were resuspended in sterile water and adjusted to 107/mL. For the leaf‐based bioassay, the conidial suspension was pressure infiltrated with a needle‐less syringe on the abaxial side to produce a 15‐mm‐long water‐soaked area. The infiltrated leaf was enclosed in a size‐compatible plastic bag with wet cotton to maintain 100% humidity. The lesion length was measured at 4 dpi using the ImageJ program (ImageJ 1.44p, Wayne Rasband, National Institutes of Health, Bethesda, MD, USA; http://imagej.nih.gov/ij).
For the spike‐based bioassay, 10 μL of fresh conidial suspension (106/mL) was added between the lemma and palea in the middle florets of a spike (Kang and Buchenauer, 1999), whereas the control plants were mock inoculated with distilled water. After inoculation, wheat heads were misted with water and covered with plastic bags to maintain high humidity for 72 h. For each fungal strain, two independent experiments were performed. At least six barley heads were examined in each replicate. Green fluorescence was observed at 24, 48, 72 and 96 hpi.
To test SAR in barley, we performed a double inoculation procedure (‘inducer’ followed by ‘challenger’) (Lyngkjær and Carver, 1999). Three experiments were designed: (I) inducer of F. graminearum on lower leaves and challenger of F. graminearum on upper leaves; (II) inducer of F. graminearum on upper leaves and challenger of F. graminearum on lower leaves; and (III) inducer of F. graminearum on middle leaves of tiller one and challenger of F. graminearum on middle leaves of tiller two. Water ‘inducer’ controls and F. graminearum ‘challenger’ were performed separately. The challenger infection was conducted 4 days after the inducer treatment on the same plant. Experiments were repeated 10 times, with one replication per plant.
Powdery mildew bioassay
Barley seedlings at the tillering stage were infiltrated with a solution of spores of B. graminis f. sp. hordei isolate K1 (density, 5 conidia/mm2) (Lumbroso et al., 1982), and maintained in a humid glasshouse for 3 days. Leaf tissues were then boiled for 10 min in a fresh solution containing phenol, lactic acid, glycerol and distilled water (1 : 1 : 1 : 1, v/v/v/v) with 0.5 mg/mL trypan blue, and maintained at room temperature for 6–8 h. Leaf tissues were then clarified overnight in 2.5 mg/mL chloral hydrate. Leaf samples were examined using an Olympus BX50 light microscope (Olympus, Tokyo, Japan).
Phylogenetic analysis
Phylogenetic analysis was performed on the protein sequences. The program MEGA 6.06 (Tamura et al., 2013) was used to align entries, to remove InDels and to construct a neighbour‐joining tree with 1000 bootstrap iterations. The distance scale bar indicates the amino acid differences per unit length. Bootstrap numbers above 80 and 50 are shown for PAL and ICS, respectively.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Protein structure of isochorismate synthase (ICS) and phenylalanine ammonia‐lyase (PAL). (a) Predicted three‐dimensional structure of ICS proteins. (b) The alanine‐serine‐glycine (Ala‐Ser‐Gly, ASG) signature of the identified PAL proteins. Proteins are from Arabidopsis thaliana (At), Brachypodium distachyon (Bd), Hordeum vulgare (Hv) and Oryza sativa (Os).
Fig. S2 Barley ICS and PAL transcripts in the IPK database. In the IPK server (http://webblast.ipk-gatersleben.de/barley_ibsc/viroblast.php), we searched for the ICS gene in the HC_genes_CDS_Seq_2012 database, and for the PAL genes in the Barley CDS HC May2016 database. The x axes (in b–g) share the same labels, and tissues were numbered from 1 to 16 as shown in (g). CAR5/CAR15, developing grain (5 or 15 DAP); EMB, 4‐day embryo; EPI, epidermal strips (28 DAP); ETI, etiolated seedling, dark condition (10 DAP); FPKM, fragments per kilobase million; INF1/INF2, developing inflorescences (5 mm or 1–1.5 cm); LEA, shoots from seedlings (10‐cm shoot stage); LEM/LOD, inflorescences, lemma (42 DAP) or lodicule (42 DAP); NOD, developing tillers, third internode (42 DAP); PAL/RAC, dissected inflorescences, palea (42 DAP) or rachis (35 DAP); ROO1/ROO2, roots (from 10‐cm shoot stage or 28 DAP); SEN, senescing leaves (56 DAP). DAP, days after pollination.
Fig. S3 Expression of PAL6‐6 and PAL6‐7 in barley. HvPAL6‐6 and HvPAL6‐7 were collectively determined in ‘Golden Promise’ at the heading stage. The tested organs were the flag leaf, penultimate leaf (leaf), peduncle (stem), sheath of the flag leaf (sheath) and inflorescence (spike). All measurements were normalized to the expression of Actin. Differences between different parts of the plant were compared using Duncan's multiple range tests at a significance level of 0.05. Error bars represent the standard error of the mean (SEM).
Fig. S4 Expression of ICS and PAL in transgenic T1 plants. In the T1 generation, we measured the expression of ICS, HvPAL1 and HvPAL6‐4 in the respective transgenic plants. The genetic background of the PAL transgenic lines is provided in Table S3. All measurements were normalized to the expression of Cyclophilin. Comparisons were made using Dunnett's tests [transgenic lines vs. wild‐type (WT); *P < 0.05, ***P < 0.001]. Error bars are the standard error of the mean (SEM).
Fig. S5 ICS affects powdery mildew resistance in barley. (a) Trypan blue staining of developing hyphae of Blumeria graminis f. sp. hordei. (b) Length of fungal hyphae in infected plants. Error bars represent the standard error of the mean (SEM). Tukey–Kramer's tests plus a pdiff option were used to compare hyphal length in the T2 transgenic vs. wild‐type (WT) plants (**P < 0.01). Scale bars, 100 μm (a).
Fig. S6 Gene expression and hormone accumulation in the ICS‐based T2 transgenic plants. This figure illustrates the expression of HvNPR1 (a) and HvWRKY (b) and the content of jasmonic acid (JA) (c) and abscisic acid (ABA) (d) in wild‐type and transgenic plants at 72 h post‐inoculation (hpi). Samples in (a) were arranged in the same order as those in (b), and likewise for (c) and (d). Duncan's multiple range tests were used to compare the H2O‐treated samples (capital letters at P < 0.05) and AmCyanPH‐1‐treated samples (small letters at P < 0.05). Dunnett's tests were used to compare the H2O‐ vs. AmCyanPH‐1‐treated samples (*P < 0.05, **P < 0.01, ***P < 0.001). Actin was used as the internal control. Error bars are the standard error of the mean (SEM). Open bars, water control; black bars, F. graminearum infected.
Fig. S7 Barley leaves showing insignificant systemic acquired resistance (SAR) response to Fusarium graminearum. The figure illustrates necrotic spots (a) and lesion sizes (b, c). The three experiments were as follows: (I) inducer of F. graminearum (fg) or water (CK) on lower leaves (LL) and challenger of F. graminearum on upper leaves (UL); (II) inducer of F. graminearum or water on upper leaves and challenger of F. graminearum on lower leaves; and (III) inducer of F. graminearum or water on middle leaves of tiller one (TL1) and challenger of F. graminearum on middle leaves of tiller two (TL2). Inducer and challenger responses from the same plant are highlighted by labels with or without an underline. The challenger responses in each experiment were compared using Dunnett's tests. Error bars are the standard error of the mean (SEM).
Fig. S8 Germination of the transgenic T2 seeds. Barley grains were hand‐threshed and treated in tap water for 2 days at room temperature with 12 h light. WT, wild‐type.
Fig. S9 ICS mutants of tetraploid wheat ‘Kronos’. (a) Growth of the wild‐type (WT) Kronos and two ICS‐A mutants (3735, G593A, missense mutation; 2561, G1905A, splicing mutation). (b) TILLING (Targeting Induced Local Lesions IN Genomes) analysis of sister plants of the selected mutant lines. Among the sister plants, white and red IDs correspond to segregants with and without the target mutations, respectively. (c) Chlorophyll (Chl) content. (d) Plant height. (e) Heading date. The Kronos mutants were identified by blast search (Krasileva et al., 2017) or by traditional TILLING (Ni et al., 2017). Dunnett's tests were used to compare transgenic vs. WT plants in (c) (***P < 0.001). Tukey–Kramer's tests plus a pdiff option were used to compare transgenic vs. WT plants in (d) and (e) (*P < 0.05, ***P < 0.001). Error bars are the standard error of the mean (SEM). Scale bars, 10 cm (in a); M, DNA ladder (in b). Note that ICS‐A mutants grew more slowly, matured to heading more slowly and, in the more severe mutant (2561), had less chlorophyll in their leaves. FW, fresh weight.
Fig. S10 Sequence alignment of the isochorismate synthase (ICS) proteins. Protein sequences were aligned using CustalW in MEGA6.06 (Tamura et al., 2013) and illustrated by GeneDoc (Nicholas et al., 1997). GenBank Accession Numbers refer to Fig. 2. At, Arabidopsis thaliana; Bd, Brachypodium distachyon; Hv, Hordeum vulgare; Os, Oryza sativa.
Fig. S11 Sequence alignment of the phenylalanine ammonia‐lyase (PAL) proteins. Protein sequences were aligned using ClustalW in MEGA6.06 (Tamura et al., 2013) and illustrated by GeneDoc. GenBank Accession Numbers refer to Fig. 2. At, Arabidopsis thaliana; Bd, Brachypodium distachyon; Hv, Hordeum vulgare; Os, Oryza sativa.
Fig. S12 ICS impacts barley foliar resistance to Fusarium graminearum. This figure illustrates lesion formation by F. graminearum on barley leaves (a), the length of developing lesions (b, n = 10) and the mRNA level of ICS (c, n = 6) in the wild‐type (WT) and T2 transgenic plants at 72 h post‐inoculation (hpi). In (c), the lower leaves were treated using water or AmCyanPH‐1; the upper leaves were untreated. Dunnett's tests were used to compare transgenic vs. WT plants in (b), and the H2O‐ vs. AmCyanPH‐1‐treated samples in (c) (*P < 0.05, ***P < 0.001). Duncan's multiple range tests were used to compare the H2O‐treated samples (capital letters at P < 0.05) and the AmCyanPH‐1‐treated samples (small letters at P < 0.05) in (c). Actin was used as the internal control. Error bars are the standard error of the mean (SEM). Relative data of independent transgenic events refer to the main text (Fig. 5).
Fig. S13 ICS regulates floral resistance to Fusarium head blight (FHB) in T2 transgenic barley. (a) AmCyanPH‐1‐based green fluorescence in infected florets of three barley lines labelled on the left. Each line was photographed in both bright field (top images of each set) and fluorescence (bottom images of each set). (b) Integrated fluorescence intensity (Integrated Fl. Int.) of the infected florets: at least 10 florets were analysed for each genotype using ImageJ. (c) The expression of ICS in infected florets. Tukey–Kramer's tests plus a pdiff option were used to compare transgenic vs. wild‐type (WT) plants in (b), and Dunnett's tests were used to compare transgenic vs. WT plants in (c) (*P < 0.05, ***P < 0.001). Actin was used as the internal control. Error bars are the standard error of the mean (SEM). Relative data of independent transgenic events refer to the main text (Fig. 6). hai, hours after inoculation.
Table S1 Barley genes used in the current study.
Table S2 Genetic transformations of different target genes in barley.
Table S3 Transgene segregation in the T1 generation.
Table S4 Plant height and tiller number of assayed transgenic events.
Table S5 Polymerase chain reaction (PCR) primers used in the current study.
Acknowledgements
We thank Dr Weihua Tang for kindly providing the F. graminearum strain AmCyanPH‐1 and Dr Qianhua Shen for providing Blumeria graminis f. sp. hordei isolate K1. We also thank Dr Lynn Epstein for reviewing the manuscript. This work was supported by the National Key Research and Development Program of China (2016YFD0100602 and 2016YFD0101004), the National Key Basic Research Program in China (2013CB1277002), the Hatch project IDA01587 and the Agriculture and Food Research Initiative Competitive Grant 2017‐67007‐25939 from the USDA National Institute of Food and Agriculture. There were no conflicts of interest.
Contributor Information
Qian Xu, Email: xuqian@sdau.edu.cn.
Daolin Fu, Email: dlfu@uidaho.edu.
References
- Abreu, M.E. and Munné‐Bosch, S. (2009) Salicylic acid deficiency in NahG transgenic lines and sid2 mutants increases seed yield in the annual plant Arabidopsis thaliana . J. Exp. Bot. 60, 1261–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, C. and Mou, Z. (2011) Salicylic acid and its function in plant immunity. J. Integr. Plant Biol. 53, 412–428. [DOI] [PubMed] [Google Scholar]
- Anand, A. , Schmelz, E.A. and Muthukrishnan, S. (2003) Development of a lesion‐mimic phenotype in a transgenic wheat line overexpressing genes for pathogenesis‐related (PR) proteins is dependent on salicylic acid concentration. Mol. Plant–Microbe Interact. 16, 916–925. [DOI] [PubMed] [Google Scholar]
- Bragg, J.N. , Wu, J. , Gordon, S.P. , Guttman, M.E. , Thilmony, R. , Lazo, G.R. , Gu, Y.Q. and Vogel, J.P. (2012) Generation and characterization of the Western Regional Research Center Brachypodium T‐DNA insertional mutant collection. PLoS One, 7, e41916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakmak, I. and Marschner, H. (1992) Magnesium deficiency and high light intensity enhance activities of superoxide dismutase, ascorbate peroxidase, and glutathione reductase in bean leaves. Plant Physiol. 98, 1222–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese, J.C. , Jordan, D.B. , Boodhoo, A. , Sariaslani, S. and Vannelli, T. (2004) Crystal structure of phenylalanine ammonia lyase: multiple helix dipoles implicated in catalysis. Biochemistry, 43, 11 403–11 416. [DOI] [PubMed] [Google Scholar]
- Catinot, J. , Buchala, A. , Abou‐Mansour, E. and Métraux, J.‐P. (2008) Salicylic acid production in response to biotic and abiotic stress depends on isochorismate in Nicotiana benthamiana . FEBS Lett. 582, 473–478. [DOI] [PubMed] [Google Scholar]
- Chen, Z. , Silva, H. and Klessig, D. (1993) Active oxygen species in the induction of plant systemic acquired resistance by salicylic acid. Science, 262, 1883–1886. [DOI] [PubMed] [Google Scholar]
- Christensen, A.H. , Sharrock, R.A. and Quail, P.H. (1992) Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol. Biol. 18, 675–689. [DOI] [PubMed] [Google Scholar]
- Colebrook, E.H. , Creissen, G. , McGrann, G.R.D. , Dreos, R. , Lamb, C. and Boyd, L.A. (2012) Broad‐spectrum acquired resistance in barley induced by the Pseudomonas pathosystem shares transcriptional components with Arabidopsis systemic acquired resistance. Mol. Plant–Microbe Interact. 25, 658–667. [DOI] [PubMed] [Google Scholar]
- Dempsey, D.M.A. and Klessig, D.F. (2012) SOS – too many signals for systemic acquired resistance? Trends Plant Sci. 17, 538–545. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N. and Rathjen, J.P. (2010) Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Durner, J. and Klessig, D.F. (1996) Salicylic acid is a modulator of tobacco and mammalian catalases. J. Biol. Chem. 271, 28 492–28 501. [DOI] [PubMed] [Google Scholar]
- Fu, D. , Uauy, C. , Distelfeld, A. , Blechl, A. , Epstein, L. , Chen, X. , Sela, H. , Fahima, T. and Dubcovsky, J. (2009) A kinase‐START gene confers temperature‐dependent resistance to wheat stripe rust. Science, 323, 1357–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Z. and Dong, X. (2013) Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. [DOI] [PubMed] [Google Scholar]
- Gallego‐Giraldo, L. , Escamilla‐Trevino, L. , Jackson, L.A. and Dixon, R.A. (2011) Salicylic acid mediates the reduced growth of lignin down‐regulated plants. Proc. Natl. Acad. Sci. USA, 108, 20 814–20 819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Q.‐M. , Zhu, S. , Kachroo, P. and Kachroo, A. (2015) Signal regulators of systemic acquired resistance. Front. Plant Sci. 6, 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcion, C. , Lohmann, A. , Lamodiere, E. , Catinot, J. , Buchala, A. , Doermann, P. and Metraux, J.‐P. (2008) Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis . Plant Physiol. 147, 1279–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Goswami, R.S. and Kistler, H.C. (2004) Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 5, 515–525. [DOI] [PubMed] [Google Scholar]
- Harwood, W.A. , Bartlett, J.G. , Alves, S.C. , Perry, M. , Smedley, M.A. , Leyland, N. and Snape, J.W. (2009) Barley transformation using Agrobacterium‐mediated techniques In: Transgenic Wheat, Barley and Oats: Production and Characterization Protocols (Jones D. H. and Shewry R. P., eds), pp. 137–147. Totowa, NJ: Humana Press. [DOI] [PubMed] [Google Scholar]
- He, P. , Osaki, M. , Takebe, M. , Shinano, T. and Wasaki, J. (2005) Endogenous hormones and expression of senescence‐related genes in different senescent types of maize. J. Exp. Bot. 56, 1117–1128. [DOI] [PubMed] [Google Scholar]
- Hong, S.‐Y. , Seo, P.J. , Cho, S.‐H. and Park, C.‐M. (2012) Preparation of leaf mesophyll protoplasts for transient gene expression in Brachypodium distachyon . J. Plant Biol. 55, 390–397. [Google Scholar]
- Janda, M. , Šašek, V. and Ruelland, E. (2014) The Arabidopsis pi4kIIIβ1β2 double mutant is salicylic acid‐overaccumulating: a new example of salicylic acid influence on plant stature. Plant Signal. Behav. 9, e977210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kang, Z. and Buchenauer, H. (1999) Immunocytochemical localization of fusarium toxins in infected wheat spikes by Fusarium culmorum . Physiol. Mol. Plant Pathol. 55, 275–288. [Google Scholar]
- Kazan, K. , Gardiner, D.M. and Manners, J.M. (2012) On the trail of a cereal killer: recent advances in Fusarium graminearum pathogenomics and host resistance. Mol. Plant Pathol. 13, 399–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K.‐C. , Lai, Z. , Fan, B. and Chen, Z. (2008) Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell, 20, 2357–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasileva, K.V. , Vasquez‐Gross, H.A. , Howell, T. , Bailey, P. , Paraiso, F. , Clissold, L. , Simmonds, J. , Ramirez‐Gonzalez, R.H. , Wang, X. , Borrill, P. , Fosker, C. , Ayling, S. , Phillips, A.L. , Uauy, C. and Dubcovsky, J. (2017) Uncovering hidden variation in polyploid wheat. Proc. Natl. Acad. Sci. USA, 114, E913–E921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H.I. , León, J. and Raskin, I. (1995) Biosynthesis and metabolism of salicylic acid. Proc. Natl. Acad. Sci. USA, 92, 4076–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon, J. , Lawton, M.A. and Raskin, I. (1995) Hydrogen peroxide stimulates salicylic acid biosynthesis in tobacco. Plant Physiol. 108, 1673–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G. and Yen, Y. (2008) Jasmonate and ethylene signaling pathway may mediate Fusarium head blight resistance in wheat. Crop Sci. 48, 1888–1896. [Google Scholar]
- Liu, J. , Cheng, X. , Liu, D. , Xu, W. , Wise, R. and Shen, Q.‐H. (2014) The miR9863 family regulates distinct Mla alleles in barley to attenuate NLR receptor‐triggered disease resistance and cell‐death signaling. PLOS Genet. 10, e1004755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo, O. , Piqueras, R. , Sánchez‐Serrano, J.J. and Solano, R. (2003) ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell, 15, 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumbroso, E. , Fischbeck, G. and Wahl, I. (1982) Infection of barley with conidia suspensions of Erysiphe graminis f. sp. hordei . J. Phytopathol. 104, 222–233. [Google Scholar]
- Lyngkjær, M.F. and Carver, T.L.W. (1999) Induced accessibility and inaccessibility to Blumeria graminis f.sp. hordei in barley epidermal cells attacked by a compatible isolate. Physiol. Mol. Plant Pathol. 55, 151–162. [Google Scholar]
- Makandar, R. , Essig, J.S. , Schapaugh, M.A. , Trick, H.N. and Shah, J. (2006) Genetically engineered resistance to Fusarium head blight in wheat by expression of Arabidopsis NPR1. Mol. Plant–Microbe Interact. 19, 123–129. [DOI] [PubMed] [Google Scholar]
- Makandar, R. , Nalam, V. , Chaturvedi, R. , Jeannotte, R. , Sparks, A.A. and Shah, J. (2010) Involvement of salicylate and jasmonate signaling pathways in Arabidopsis interaction with Fusarium graminearum . Mol. Plant–Microbe Interact. 23, 861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makandar, R. , Nalam, V.J. , Lee, H. , Trick, H.N. , Dong, Y. and Shah, J. (2012) Salicylic acid regulates basal resistance to Fusarium head blight in wheat. Mol. Plant–Microbe Interact. 25, 431–439. [DOI] [PubMed] [Google Scholar]
- Moya‐Elizondo, E.A. and Jacobsen, B.J. (2016) Integrated management of Fusarium crown rot of wheat using fungicide seed treatment, cultivar resistance, and induction of systemic acquired resistance (SAR). Biol. Control. 92, 153–163. [Google Scholar]
- Mukherjee, S.P. and Choudhuri, M.A. (1983) Implications of water stress‐induced changes in the levels of endogenous ascorbic acid and hydrogen peroxide in Vigna seedlings. Physiol. Plant. 58, 166–170. [Google Scholar]
- Murray, S.L. , Ingle, R.A. , Petersen, L.N. and Denby, K.J. (2007) Basal resistance against Pseudomonas syringae in Arabidopsis involves WRKY53 and a protein with homology to a nematode resistance protein. Mol. Plant–Microbe Interact. 20, 1431–1438. [DOI] [PubMed] [Google Scholar]
- Ni, F. , Qi, J. , Hao, Q. , Lyu, B. , Luo, M.‐C. , Wang, Y. , Chen, F. , Wang, S. , Zhang, C. , Epstein, L. , Zhao, X. , Wang, H. , Zhang, X. , Chen, C. , Sun, L. and Fu, D. (2017) Wheat Ms2 encodes for an orphan protein that confers male sterility in grass species. Nat. Commun. 8, 15 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas, K.B. , Nicholas, H.B. Jr. and Deerfield, D.W. II. (1997) GeneDoc: Analysis and visualization of genetic variation. Embnet News 4, 1–4. [Google Scholar]
- Ouchi, S. , Oku, H. and Hibino, C. (1976) Localization of induced resistance and susceptibility in barley leaves inoculated with the powdery mildew fungus. Phytopathology, 66, 901–905. [Google Scholar]
- Pieterse, C.M.J. and Van Loon, L.C. (2004) NPR1: the spider in the web of induced resistance signaling pathways. Curr. Opin. Plant Biol. 7, 456–464. [DOI] [PubMed] [Google Scholar]
- Plotnikova, L.Y. (2009) Effect of benzothiadiazole, an inducer of systemic acquired resistance, on the pathogenesis of wheat brown rust. Russ. J. Plant Physiol. 56, 517–526. [Google Scholar]
- Qi, P.‐F. , Johnston, A. , Balcerzak, M. , Rocheleau, H. , Harris, L.J. , Long, X.‐Y. , Wei, Y.‐M. , Zheng, Y.‐L. and Ouellet, T. (2012) Effect of salicylic acid on Fusarium graminearum, the major causal agent of fusarium head blight in wheat. Fungal Biol. 116, 413–426. [DOI] [PubMed] [Google Scholar]
- Rivas‐San Vicente, M. and Plasencia, J. (2011) Salicylic acid beyond defence: its role in plant growth and development. J. Exp. Bot. 62, 3321–3338. [DOI] [PubMed] [Google Scholar]
- Robert‐Seilaniantz, A. , Grant, M. and Jones, J.D.G. (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate–salicylate antagonism. Annu. Rev. Phytopathol. 49, 317–343. [DOI] [PubMed] [Google Scholar]
- Sadeghi, M. , Dehghan, S. , Fischer, R. , Wenzel, U. , Vilcinskas, A. , Kavousi, H.R. and Rahnamaeian, M. (2013) Isolation and characterization of isochorismate synthase and cinnamate 4‐hydroxylase during salinity stress, wounding, and salicylic acid treatment in Carthamus tinctorius . Plant Signal. Behav. 8, e27335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scebba, F. , Sebastiani, L. and Vitagliano, C. (2001) Activities of antioxidant enzymes during senescence of Prunus armeniaca leaves. Biol. Plant. 44, 41–46. [Google Scholar]
- Shine, M.B. , Yang, J.‐W. , El‐Habbak, M. , Nagyabhyru, P. , Fu, D.‐Q. , Navarre, D. , Ghabrial, S. , Kachroo, P. and Kachroo, A. (2016) Cooperative functioning between phenylalanine ammonia lyase and isochorismate synthase activities contributes to salicylic acid biosynthesis in soybean. New Phytol. 212, 627–636. [DOI] [PubMed] [Google Scholar]
- Singh, A. , Lim, G.‐H. and Kachroo, P. (2017) Transport of chemical signals in systemic acquired resistance. J. Integr. Plant Biol. 59, 336–344. [DOI] [PubMed] [Google Scholar]
- Sui, N. , Li, M. , Liu, X.‐Y. , Wang, N. , Fang, W. and Meng, Q.‐W. (2007) Response of xanthophyll cycle and chloroplastic antioxidant enzymes to chilling stress in tomato over‐expressing glycerol‐3‐phosphate acyltransferase gene. Photosynthetica, 45, 447–454. [Google Scholar]
- Tamura, K. , Stecher, G. , Peterson, D. , Filipski, A. and Kumar, S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P.H.J. , Eggermont, K. , Penninckx, I.A.M.A. , Mauch‐Mani, B. , Vogelsang, R. , Cammue, B.P.A. and Broekaert, W.F. (1998) Separate jasmonate‐dependent and salicylate‐dependent defense‐response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA, 95, 15 107–15 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonnessen, B.W. , Manosalva, P. , Lang, J.M. , Baraoidan, M. , Bordeos, A. , Mauleon, R. , Oard, J. , Hulbert, S. , Leung, H. and Leach, J.E. (2015) Rice phenylalanine ammonia‐lyase gene OsPAL4 is associated with broad spectrum disease resistance. Plant Mol. Biol. 87, 273–286. [DOI] [PubMed] [Google Scholar]
- Torres, M.A. , Jones, J.D.G. and Dangl, J.L. (2006) Reactive oxygen species signaling in response to pathogens. Plant Physiol. 141, 373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot, A.C. , Dempsey, D.M.A. and Klessig, D.F. (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. [DOI] [PubMed] [Google Scholar]
- Wildermuth, M.C. , Dewdney, J. , Wu, G. and Ausubel, F.M. (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature, 414, 562–565. [DOI] [PubMed] [Google Scholar]
- Wiśniewska, H. and Chełkowski, J. (1999) Influence of exogenic salicylic acid on Fusarium seedling blight reduction in barley. Acta Physiol. Plant. 21, 63–66. [Google Scholar]
- Wood, D.W. , Setubal, J.C. , Kaul, R. , Monks, D.E. , Kitajima, J.P. , Okura, V.K. , Zhou, Y. , Chen, L. , Wood, G.E. , Almeida, NF. Jr. , Woo, L., Chen, Y. , Paulsen, I.T. , Eisen, J.A. , Karp, P.D. , Bovee, D. Sr. , Chapman, P. , Clendenning, J. , Deatherage, G. , Gillet, W. , Grant, C. , Kutyavin, T. , Levy, R. , Li, M.J. , McClelland, E. , Palmieri, A. , Raymond, C. , Rouse, G. , Saenphimmachak, C. , Wu, Z. , Romero, P. , Gordon, D. , Zhang, S. , Yoo, H. , Tao, Y. , Biddle, P. , Jung, M. , Krespan, W. , Perry, M. , Gordon‐Kamm, B. , Liao, L. , Kim, S. , Hendrick, C. , Zhao, Z.Y. , Dolan, M. , Chumley, F. , Tingey, S.V. , Tomb, J.F. , Gordon, M.P. , Olson, M.V. and Nester, E.W. (2001) The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science, 294, 2317–2323. [DOI] [PubMed] [Google Scholar]
- Xu, Q. , Truong, T.T. , Barrero, J.M. , Jacobsen, J.V. , Hocart, C.H. and Gubler, F. (2016) A role for jasmonates in the release of dormancy by cold stratification in wheat. J. Exp. Bot. 67, 3497–3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalpani, N. , Silverman, P. , Wilson, T.M. , Kleier, D.A. and Raskin, I. (1991) Salicylic acid is a systemic signal and an inducer of pathogenesis‐related proteins in virus‐infected tobacco. Plant Cell, 3, 809–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalpani, N. , Shulaev, V. and Raskin, I. (1993) Endogenous salicylic acid levels correlate with accumulation of pathogenesis‐related proteins and virus resistance in tobacco. Phytopathology, 83, 702–708. [Google Scholar]
- Yang, Y. , Zhao, J. , Liu, P. , Xing, H. , Li, C. , Wei, G. and Kang, Z. (2013) Glycerol‐3‐phosphate metabolism in wheat contributes to systemic acquired resistance against Puccinia striiformis f. sp. tritici . PLoS One, 8, e81756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka, H. , Adachi, H. , Ishihama, N. , Nakano, T. , Shiraishi, Y. , Miyagawa, N. , Nomura, H. , Yoshioka, M. and Asai, S. (2015) Molecular mechanisms of ROS burst conferred by protein phosphorylation. Jpn. J. Phytopathol. 81, 1–8. [Google Scholar]
- Yuan, C. , Wu, J. , Yan, B. , Hao, Q. , Zhang, C. , Lyu, B. , Ni, F. , Caplan, A. , Wu, J. and Fu, D. (2018) Remapping of the stripe rust resistance gene Yr10 in common wheat. Theor. Appl. Genet. DOI: 10.1007/s00122-00018-03075-00129; [DOI] [PubMed] [Google Scholar]
- Yuan, Y. , Chung, J.‐D. , Fu, X. , Johnson, V.E. , Ranjan, P. , Booth, S.L. , Harding, S.A. and Tsai, C.‐J. (2009) Alternative splicing and gene duplication differentially shaped the regulation of isochorismate synthase in Populus and Arabidopsis . Proc. Natl. Acad. Sci. USA, 106, 22 020–22 025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X.‐W. , Jia, L.‐J. , Zhang, Y. , Jiang, G. , Li, X. , Zhang, D. and Tang, W.‐H. (2012) In planta stage‐specific fungal gene profiling elucidates the molecular strategies of Fusarium graminearum growing inside wheat coleoptiles. Plant Cell, 24, 5159–5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Protein structure of isochorismate synthase (ICS) and phenylalanine ammonia‐lyase (PAL). (a) Predicted three‐dimensional structure of ICS proteins. (b) The alanine‐serine‐glycine (Ala‐Ser‐Gly, ASG) signature of the identified PAL proteins. Proteins are from Arabidopsis thaliana (At), Brachypodium distachyon (Bd), Hordeum vulgare (Hv) and Oryza sativa (Os).
Fig. S2 Barley ICS and PAL transcripts in the IPK database. In the IPK server (http://webblast.ipk-gatersleben.de/barley_ibsc/viroblast.php), we searched for the ICS gene in the HC_genes_CDS_Seq_2012 database, and for the PAL genes in the Barley CDS HC May2016 database. The x axes (in b–g) share the same labels, and tissues were numbered from 1 to 16 as shown in (g). CAR5/CAR15, developing grain (5 or 15 DAP); EMB, 4‐day embryo; EPI, epidermal strips (28 DAP); ETI, etiolated seedling, dark condition (10 DAP); FPKM, fragments per kilobase million; INF1/INF2, developing inflorescences (5 mm or 1–1.5 cm); LEA, shoots from seedlings (10‐cm shoot stage); LEM/LOD, inflorescences, lemma (42 DAP) or lodicule (42 DAP); NOD, developing tillers, third internode (42 DAP); PAL/RAC, dissected inflorescences, palea (42 DAP) or rachis (35 DAP); ROO1/ROO2, roots (from 10‐cm shoot stage or 28 DAP); SEN, senescing leaves (56 DAP). DAP, days after pollination.
Fig. S3 Expression of PAL6‐6 and PAL6‐7 in barley. HvPAL6‐6 and HvPAL6‐7 were collectively determined in ‘Golden Promise’ at the heading stage. The tested organs were the flag leaf, penultimate leaf (leaf), peduncle (stem), sheath of the flag leaf (sheath) and inflorescence (spike). All measurements were normalized to the expression of Actin. Differences between different parts of the plant were compared using Duncan's multiple range tests at a significance level of 0.05. Error bars represent the standard error of the mean (SEM).
Fig. S4 Expression of ICS and PAL in transgenic T1 plants. In the T1 generation, we measured the expression of ICS, HvPAL1 and HvPAL6‐4 in the respective transgenic plants. The genetic background of the PAL transgenic lines is provided in Table S3. All measurements were normalized to the expression of Cyclophilin. Comparisons were made using Dunnett's tests [transgenic lines vs. wild‐type (WT); *P < 0.05, ***P < 0.001]. Error bars are the standard error of the mean (SEM).
Fig. S5 ICS affects powdery mildew resistance in barley. (a) Trypan blue staining of developing hyphae of Blumeria graminis f. sp. hordei. (b) Length of fungal hyphae in infected plants. Error bars represent the standard error of the mean (SEM). Tukey–Kramer's tests plus a pdiff option were used to compare hyphal length in the T2 transgenic vs. wild‐type (WT) plants (**P < 0.01). Scale bars, 100 μm (a).
Fig. S6 Gene expression and hormone accumulation in the ICS‐based T2 transgenic plants. This figure illustrates the expression of HvNPR1 (a) and HvWRKY (b) and the content of jasmonic acid (JA) (c) and abscisic acid (ABA) (d) in wild‐type and transgenic plants at 72 h post‐inoculation (hpi). Samples in (a) were arranged in the same order as those in (b), and likewise for (c) and (d). Duncan's multiple range tests were used to compare the H2O‐treated samples (capital letters at P < 0.05) and AmCyanPH‐1‐treated samples (small letters at P < 0.05). Dunnett's tests were used to compare the H2O‐ vs. AmCyanPH‐1‐treated samples (*P < 0.05, **P < 0.01, ***P < 0.001). Actin was used as the internal control. Error bars are the standard error of the mean (SEM). Open bars, water control; black bars, F. graminearum infected.
Fig. S7 Barley leaves showing insignificant systemic acquired resistance (SAR) response to Fusarium graminearum. The figure illustrates necrotic spots (a) and lesion sizes (b, c). The three experiments were as follows: (I) inducer of F. graminearum (fg) or water (CK) on lower leaves (LL) and challenger of F. graminearum on upper leaves (UL); (II) inducer of F. graminearum or water on upper leaves and challenger of F. graminearum on lower leaves; and (III) inducer of F. graminearum or water on middle leaves of tiller one (TL1) and challenger of F. graminearum on middle leaves of tiller two (TL2). Inducer and challenger responses from the same plant are highlighted by labels with or without an underline. The challenger responses in each experiment were compared using Dunnett's tests. Error bars are the standard error of the mean (SEM).
Fig. S8 Germination of the transgenic T2 seeds. Barley grains were hand‐threshed and treated in tap water for 2 days at room temperature with 12 h light. WT, wild‐type.
Fig. S9 ICS mutants of tetraploid wheat ‘Kronos’. (a) Growth of the wild‐type (WT) Kronos and two ICS‐A mutants (3735, G593A, missense mutation; 2561, G1905A, splicing mutation). (b) TILLING (Targeting Induced Local Lesions IN Genomes) analysis of sister plants of the selected mutant lines. Among the sister plants, white and red IDs correspond to segregants with and without the target mutations, respectively. (c) Chlorophyll (Chl) content. (d) Plant height. (e) Heading date. The Kronos mutants were identified by blast search (Krasileva et al., 2017) or by traditional TILLING (Ni et al., 2017). Dunnett's tests were used to compare transgenic vs. WT plants in (c) (***P < 0.001). Tukey–Kramer's tests plus a pdiff option were used to compare transgenic vs. WT plants in (d) and (e) (*P < 0.05, ***P < 0.001). Error bars are the standard error of the mean (SEM). Scale bars, 10 cm (in a); M, DNA ladder (in b). Note that ICS‐A mutants grew more slowly, matured to heading more slowly and, in the more severe mutant (2561), had less chlorophyll in their leaves. FW, fresh weight.
Fig. S10 Sequence alignment of the isochorismate synthase (ICS) proteins. Protein sequences were aligned using CustalW in MEGA6.06 (Tamura et al., 2013) and illustrated by GeneDoc (Nicholas et al., 1997). GenBank Accession Numbers refer to Fig. 2. At, Arabidopsis thaliana; Bd, Brachypodium distachyon; Hv, Hordeum vulgare; Os, Oryza sativa.
Fig. S11 Sequence alignment of the phenylalanine ammonia‐lyase (PAL) proteins. Protein sequences were aligned using ClustalW in MEGA6.06 (Tamura et al., 2013) and illustrated by GeneDoc. GenBank Accession Numbers refer to Fig. 2. At, Arabidopsis thaliana; Bd, Brachypodium distachyon; Hv, Hordeum vulgare; Os, Oryza sativa.
Fig. S12 ICS impacts barley foliar resistance to Fusarium graminearum. This figure illustrates lesion formation by F. graminearum on barley leaves (a), the length of developing lesions (b, n = 10) and the mRNA level of ICS (c, n = 6) in the wild‐type (WT) and T2 transgenic plants at 72 h post‐inoculation (hpi). In (c), the lower leaves were treated using water or AmCyanPH‐1; the upper leaves were untreated. Dunnett's tests were used to compare transgenic vs. WT plants in (b), and the H2O‐ vs. AmCyanPH‐1‐treated samples in (c) (*P < 0.05, ***P < 0.001). Duncan's multiple range tests were used to compare the H2O‐treated samples (capital letters at P < 0.05) and the AmCyanPH‐1‐treated samples (small letters at P < 0.05) in (c). Actin was used as the internal control. Error bars are the standard error of the mean (SEM). Relative data of independent transgenic events refer to the main text (Fig. 5).
Fig. S13 ICS regulates floral resistance to Fusarium head blight (FHB) in T2 transgenic barley. (a) AmCyanPH‐1‐based green fluorescence in infected florets of three barley lines labelled on the left. Each line was photographed in both bright field (top images of each set) and fluorescence (bottom images of each set). (b) Integrated fluorescence intensity (Integrated Fl. Int.) of the infected florets: at least 10 florets were analysed for each genotype using ImageJ. (c) The expression of ICS in infected florets. Tukey–Kramer's tests plus a pdiff option were used to compare transgenic vs. wild‐type (WT) plants in (b), and Dunnett's tests were used to compare transgenic vs. WT plants in (c) (*P < 0.05, ***P < 0.001). Actin was used as the internal control. Error bars are the standard error of the mean (SEM). Relative data of independent transgenic events refer to the main text (Fig. 6). hai, hours after inoculation.
Table S1 Barley genes used in the current study.
Table S2 Genetic transformations of different target genes in barley.
Table S3 Transgene segregation in the T1 generation.
Table S4 Plant height and tiller number of assayed transgenic events.
Table S5 Polymerase chain reaction (PCR) primers used in the current study.


