Summary
To investigate its susceptibility to ergot infection, we inoculated Brachypodium distachyon with Claviceps purpurea and compared the infection symptoms with those on rye (Secale cereale). We showed that, after inoculation of Brachypodium with Claviceps, the same disease symptoms occurred in comparable temporal and spatial patterns to those on rye. The infection rate of Claviceps on this host was reduced compared with rye, but the disease could be surveyed by fungal genomic DNA quantification. Mutants of Claviceps which were virulence attenuated on rye were also affected on Brachypodium. We were able to show that pathogenesis‐related gene expression changed in a typical manner for biotrophic pathogen attack. Our results indicated that the Claviceps–Brachypodium interaction was dependent on salicylic acid, cytokinin and auxin. We consider Brachypodium to be a suitable and useful alternative host; the increased sensitivity compared with rye will be valuable for the identification of infection mechanisms. Future progess in understanding the Claviceps–plant interaction will be facilitated by the use of a well‐characterized model host system.
Keywords: biotrophic pathogen, host–pathogen interaction, host response, plant hormones
Introduction
Claviceps purpurea can infect more than 400 monocotyledonous species, including rye (Secale cereale). Fungal infection leads to poisoning of harvested cereal by Claviceps‐typical ergot alkaloids, making it unsuitable as food and feed (Haarmann et al., 2009). Claviceps has a biotrophic lifestyle and is restricted to unfertilized ovaries (Hinsch and Tudzynski, 2015). Spores germinate on stigmatic hairs, penetrate the plant tissue and form a hyphal bundle, which grows mainly intercellularly and strictly polar to the base of the ovary. After tapping into the vascular system, the fungal mycelium replaces the ovarian tissue to form the so‐called sphacelium, which induces the secretion of viscous honeydew enriched with conidia at about 7 days post‐infection (dpi). Finally, the sphacelium differentiates into a dark pigmented, ergot alkaloid‐containing sclerotium, the resistant overwintering structure. Although Claviceps infection causes severe economic losses (Klotz and Smith, 2015), studies of the infection process have so far been focused on the fungus, mainly as a result of the limited accessibility of the standard host plant rye to molecular analyses (Haarmann et al., 2009; Hinsch and Tudzynski, 2015). Recently, publication of the rye genome has allowed important progress in rye research and has enabled an in planta RNA‐sequencing (RNA‐Seq) analysis of Claviceps‐infected rye (Bauer et al., 2017; B. Oeser et al., 2017). Nevertheless, partly because of its complex genome, rye is poorly characterized at the molecular level, especially with respect to the transcriptome responses concerning, for example, biotic stress, hormone signalling and metabolism. This has impeded the characterization of the Claviceps–rye interaction, as the characterization of both pathogen and host participation is important in order to elucidate the infection mechanisms of successful biotrophic pathogens, such as Claviceps.
The challenge of deciphering the highly specific biotrophic infection mechanisms of Claviceps and the respective vulnerabilities in plant defence would be simplified by having a genetically characterized host with an increasing community‐derived pool of information, especially expression data, and the availability of mutants. A unique combination of several features (biotrophic, organ‐restricted, broad host range) characterizes the infection of Claviceps, thus indicating a strategy that differs from those of other pathogens. Hence, insight into the specific infection mechanisms of Claviceps will contribute to the understanding of the plant defence system.
Brachypodium distachyon is gaining relevance as a monocotyledonous model plant, because it has one of the smallest known plant genomes, undemanding growth requirements and is closely related to grass crops, such as wheat, barley and rye (Draper et al., 2001; The International Brachypodium Initiative, 2010). In addition, it is easily accessible for genetic manipulation, and a large collection of defined mutants and an increasing number of transcriptomic datasets are available (Bragg et al., 2012; Brkljacic et al., 2011; Dalmais et al., 2013; Kakei et al., 2015). As it has been described as a model host for other grass pathogens (e.g. Magnaporthe oryzae, Routledge et al., 2004; Fusarium graminearum, Peraldi et al., 2011; reviewed in Fitzgerald et al., 2015), it could represent an excellent alternative host for Claviceps.
Here, we demonstrate the usefulness of Brachypodium as an alternative host for Claviceps. We show that Brachypodium is generally susceptible to Claviceps infection and how the degree of infection can be studied, that virulence factors identified in rye are also relevant for the infection of Brachypodium, and that this host mounts a transcriptomic response typical of a biotrophic attack.
Results and Discussion
To investigate its susceptibility to ergot infection, we developed a protocol for B. distachyon with C. purpurea. As preliminary experiments indicated that Claviceps could induce disease symptoms on Brachypodium similar to those observed on rye, we optimized the infection protocol. The rye cultivar used by default for Claviceps virulence assays is male sterile, a factor which is known to increase susceptibility because infection only occurs on non‐fertilized florets. To our knowledge, a male‐sterile Brachypodium genotype is not yet available. To mimic male sterility, Brachypodium florets were emasculated manually prior to individual inoculation, as is performed for genetic crossing in Brachypodium. However, in some cases, necrotized ovaries were found instead of kernels, even in the water controls, probably caused by mechanical wounding by the emasculation. In contrast, the inoculation of young florets by the injection of spore suspensions with a syringe affected seed development only rarely and provided a rapid inoculation procedure. Hence, all experiments in this study are based on syringe inoculation of Brachypodium without emasculation.
To characterize the disease development on Brachypodium, we focused on symptoms which are typically used to evaluate virulence on rye, such as hyphal growth in isolated ovaries, honeydew secretion and sclerotia formation. Our results showed that all of these symptoms can be observed on inoculation of Brachypodium (Fig. 1C,H,E,J). The intercellular hyphae can be observed in the Brachypodium ovary at 5 dpi as prominently as seen in the rye ovary. Clearly visible copious amounts of honeydew containing massive numbers of conidia appeared at 10–18 dpi, several days later than on rye (7–9 dpi). This result is probably caused by the reduced amount of honeydew. Sclerotia were formed at 35 dpi and these were smaller than those typically formed on rye (compare Fig. 1E,J). Independent of the host plant, remains of the ovaries could be observed on top of the sphacelium and sclerotia (Fig. 1B,G,E,J). These data show that Brachypodium is susceptible to the ergot fungus and can be used as a host plant. However, a major difference between the inoculation of rye and Brachypodium is the frequency of the macroscopic symptoms of infection. Honeydew secretion was observed on 90%–100% of inoculated rye ears, whereas this only applied to 9%–33% of inoculated Brachypodium spikelets. In contrast with Brachypodium, rye is self‐sterile and anemophilous with exposed stigmatic hairs at anthesis. This probably explains why rye is naturally highly susceptible to Claviceps, whereas natural infection has rarely been described for Brachypodium (only for B. sylvaticum; Halbritter et al., 2012; Peach and Loveless, 1975). Apparently, this biological and physical barrier to infection can be overcome by injection of the spore suspension prior to pollen release. Nevertheless, the constantly high infection rates obtained in the rye system based on macroscopic criteria could not be achieved here. To allow reproducible quantitative virulence assays, we devised a more reliable method to follow the infection process that is independent of macroscopic symptoms of infection. A quantitative polymerase chain reaction (qPCR)‐based detection of the fungal genomic DNA (gDNA) portion has proved to be a valuable tool to assess the progress and degree of infection in several plant–fungus interactions (Brouwer et al., 2003; Weßling and Panstruga, 2012). This method is independent of subjective categorization and requires less inoculated material (i.e. 15 spikelets).
Figure 1.
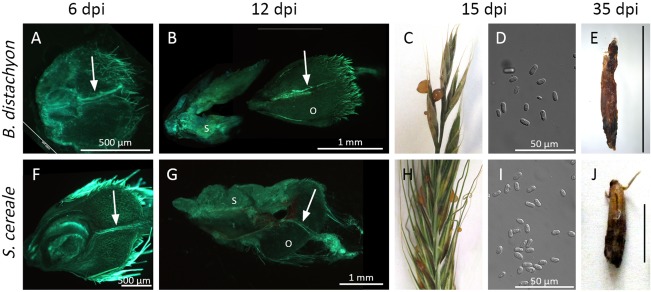
Comparison of disease symptoms of Claviceps purpurea on Brachypodium distachyon (A–E) and the established host Secale cereale (F–J). For microscopic observations (A, B, F, G), ovaries were cut longitudinally and stained with aniline blue to visualize fungal tissue when exposed to UV light. (A, F) Fungal hyphae in the transmitting tissue (indicated by arrows) at 6 days post‐infection (dpi). (B, G) Sphacelium (S) formed at the base of the ovary at 12 dpi with the ovary (O) remaining on top. (C, H) At 15 dpi, enough honeydew is secreted to form visible droplets that are enriched with conidia (D, I). (E, J) After 5 weeks, sclerotia developed (black scale bars, 1 cm).
To observe the gain of fungal biomass in inoculated spikelets, gDNA was extracted at different time points and the ratio of fungal to plant DNA was determined by quantitative real‐time PCR (Fig. 2A). Fungal DNA could not be detected in the H2O controls (mock). The ratio in the wild‐type (WT) sample did not alter significantly between 0 and 5 dpi, but increased multiple times from 5 to 10 dpi (2.6E‐4 to 7.8E‐3; +3000%). This fits with the microscopic observations of restricted growth in the early infection phase (0–5 dpi, few hyphae grow towards the base of the ovary, compare Fig. 1A,F). In the later infection stage (5–10 dpi), the fungus begins to replace the ovarian tissue and produces more biomass (Fig. 1B,G). This result confirms that the fungal gDNA portion reflects the portion of fungal biomass. The final increase in biomass in the WT sample requires extensive growth, which implies that Claviceps has obtained access to nutrients and thus has established a compatible interaction with this novel host. This was also confirmed by honeydew secretion on control plants inoculated with the same spore suspension (as shown in Fig. 1C).
Figure 2.
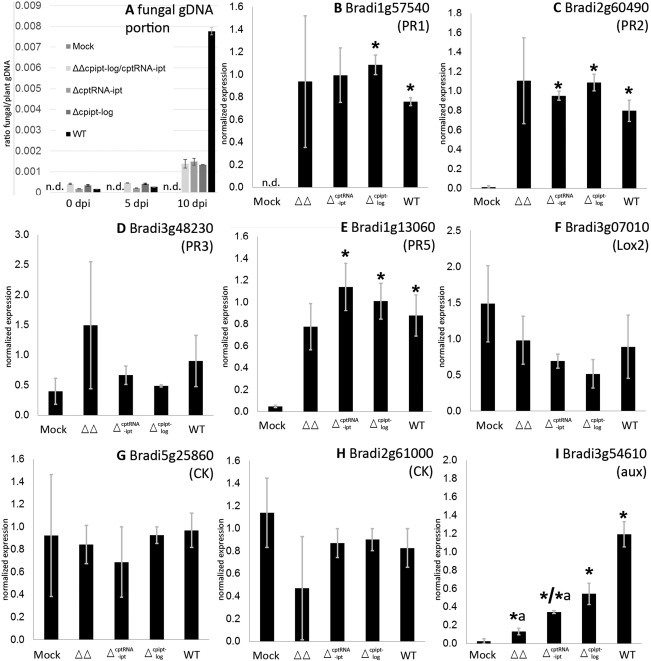
Fungal genomic DNA (gDNA) portion and plant gene expression during infection. (A) Fungal gDNA portion of inoculated florets was determined by quantitative polymerase chain reaction (qPCR) at different time points during the course of infection. The panel shows the numbers of one experiment; the standard deviation (SD) refers to technical replicates. A biological replicate gave similar results with an even greater portion in the wild‐type (WT) sample at 10 days post‐infection (dpi). (B–I) Expression of several plant genes was determined at 5 dpi by quantitative reverse transcription‐PCR. Bars represent the mean and SD of two biological replicates. PR, pathogenesis related like; CK/aux, expected responsiveness to cytokinin/auxin according to Kakei et al. (2015); n.d., not detectable; *, significant difference between mock and infected tissue according to Student's unpaired t‐test at P ≤ 0.05, *a, significant difference between WT and mutant‐infected tissue.
As a next step, we included mutants in this assay which show attenuated virulence on rye. A recently identified virulence factor of Claviceps is the biosynthesis and secretion of cytokinins (CKs, Hinsch et al., 2016). The fungus synthesizes CKs via two pathways: de novo and by the degradation of prenylated transfer RNA (tRNA). Deletion of the key genes of both pathways (cpipt‐log encoding a bifunctional enzyme of the de novo pathway and the tRNA prenyltransferase encoding cptRNA‐ipt) is required to completely abandon CK synthesis. Only double deletion strains (ΔΔcpipt‐log/tRNA‐ipt) are completely CK free and cannot establish a compatible interaction with rye. In order to test whether this is also true for Brachypodium, we included the double mutant (ΔΔcpipt‐log/tRNA‐ipt) and the corresponding single mutants (Δipt‐log, ΔtRNA‐ipt) in the assay. The fungal gDNA portion also increased when the mutants were used for inoculation, but to a much lesser degree than in the WT infection (Fig. 2A). At 10 dpi, the maximum colonization rate of the mutant strains was less than 20% of the WT (0.0014 compared with 0.0077). Apparently, the mutant strains did not grow extensively within the ovaries. The limited gain of biomass probably reflects the germination of the spores and the basipetal hyphal growth in the ovaries at 5 dpi (Fig. 3), but replacement of the ovarian tissue by fungal sphacelium was not initiated. Obviously, the mutants were unable to establish a compatible interaction. This finding was further supported by the absence of honeydew formation in all infections with the CK mutant strains (20 spikelets tested per strain). Taken together, these results confirm that virulence defects of the Claviceps mutants observed on rye are also detectable on Brachypodium, and that the fungal gDNA quantification allows reproducible virulence assays of Claviceps strains.
Figure 3.
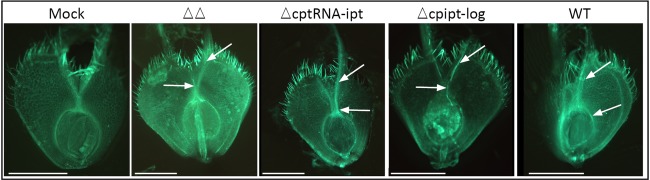
Hyphal growth of different Claviceps purpurea strains in isolated Brachypodium distachyon ovaries at 5 days post‐infection (dpi). ΔΔ, double deletion strain of cpipt‐log and cptRNA‐ipt. For microscopic observations, ovaries were cut longitudinally and stained with aniline blue to visualize fungal tissue when exposed to UV light. Hyphal bundles are indicated with arrows. Scale bar, 500 µm.
To further understand the interaction between Claviceps and Brachypodium and to identify the pathways that are altered by the infection, the expression of several Brachypodium genes and their alteration on inoculation were determined. The genes were chosen based on previous findings about their regulation. Brachypodium orthologues of the Arabidopsis pathogenesis‐related (PR) genes 1, 2, 3 and 5, and of lox2 [involved in jasmonic acid (JA) biosynthesis], have been described previously as differentially regulated in compatible interactions (Blümke et al., 2015; Mandadi and Scholthof, 2012). During infection, rye is assumed to receive a CK signal, most probably from fungal‐derived CKs (Hinsch et al., 2015), and Claviceps also synthesizes auxins (P. Galuszka et al., unpublished data). Accordingly, we were interested to determine whether Brachypodium transcription indicates CK or auxin signal perception, because the genes Bradi5g25860, Bradi2g61000 and Bradi3g54610 were found to be induced by CK or auxin treatment (Kakei et al., 2015).
As shown in Fig. 2B,C,E, expression of the PR1‐, PR2‐ and PR5‐like genes was increased significantly in the inoculated samples compared with the mock control (i.e. not caused by mechanical wounding), demonstrating that Brachypodium induces a specific defence response on Claviceps inoculation. Although the fungal attack was recognized and the defence response was initiated, Brachypodium was unable to repel the infection by WT. A similar induction of PR genes has been described during infection by F. graminearum (PR1 and PR2; Blümke et al., 2015) and the Panicum mosaic virus (PMV, PR1–3, PR5; Mandadi and Scholthof, 2012). In contrast, lox2 was down‐regulated on PMV infection, a result which could not be confirmed for Claviceps infection (Fig. 2F). In addition, we found that there was no significant induction of PR3 (Fig. 2D). Opposing regulation patterns for PR1, PR2 and PR5, on the one hand, and PR3, on the other, have also been described in Arabidopsis (Thomma et al., 1998). The regulation of PR1, PR2 and PR5 has been shown to be salicylic acid (SA) dependent, whereas PR3 is JA dependent. Lox2 is also associated with JA as it is part of its biosynthesis pathway. According to these findings, SA appears to be more relevant than JA for the defence response of Brachypodium to Claviceps, although statements about PR protein families regarding regulation usually do not apply to all members of a particular family. However, the observation that SA is more relevant than JA would conform to the biotrophic infection strategy of Claviceps. There is general acceptance that SA is the fundamental mediator for defence responses to biotrophic (and hemibiotrophic) pathogens, whereas JA (and ethylene) are more important for reactions to necrotrophic pathogens (Glazebrook, 2005). To summarize, the alteration of PR gene expression levels shows that Brachypodium detects Claviceps and responds to its inoculation in a typical manner for biotrophic pathogens.
There were no significant differences in PR gene expression between WT and the tested mutants (Fig. 2B–F). The expression of PR1, PR2 and PR5 in plants inoculated with the single mutants was comparable with that of the WT sample and different from the mock control. The expression levels, however, did not allow firm conclusions to be drawn about the virulence of the mutants as they induced similar responses to the WT. According to the fungal gDNA portion, they were not able to establish a compatible interaction.
The expression of CK‐responsive genes (Bradi5g25860 and Bradi2g61000) was not significantly different for any sample and did not indicate an alteration in CK signalling in Brachypodium during the infection, a result in marked contrast with the work on rye (Hinsch et al., 2015). Fungal interference with Brachypodium CK signalling cannot be excluded, as there is a much broader spectrum of CK‐responsive genes and those tested so far have only been shown to respond to one specific CK (trans‐zeatin) treatment as seedlings (Kakei et al., 2015). The incapacity of the CK biosynthesis‐attenuated mutants Δcpipt‐log, ΔtRNA‐ipt and ΔΔcpipt‐log/tRNA‐ipt to colonize Brachypodium also suggests that fungal‐derived CKs are essential for the infection process. As even the single deletion strains were affected, it may be even more fundamental than for the infection of rye. As a result of the reduced susceptibility of Brachypodium compared with rye to Claviceps, it could also be considered more sensitive than the rye system. Even slight alterations in the Claviceps secretome, such as different ratios of CK types in the single CK deletion strains, could impair the infection of Brachypodium, whereas infection of rye was only affected when CK biosynthesis was completely blocked (Hinsch et al., 2016).
Surprisingly, the expression of the auxin‐responsive gene (Bradi3g54610) differed not only between the mock and WT, but also between WT and the ΔcptRNA‐ipt and ΔΔcpipt‐log/cptRNA‐ipt mutants (Fig. 2I). Assuming that the differential expression of Bradi3g54610 is indeed auxin signal dependent, its expression level could influence the virulence of the invading strain, indicating that auxin signalling is involved in the interaction, as in rye (B. Oeser et al., 2017). As Claviceps synthesizes auxins (P. Galuszka et al., unpublished data), the expression level of Bradi3g54610 could also quantitatively reflect the amount of fungal‐derived auxin.
We have shown here that Brachypodium can serve as a model host for the study of Claviceps virulence and resistance mechanisms. Mutual phytohormone signalling appears to be involved in the Claviceps–host interaction, and the unique technical options provided by the new model host Brachypodium will facilitate a detailed understanding of this biotrophic interaction.
Experimental Procedures
Strains, media and growth conditions
The Claviceps purpurea (Fr.) Tul. strain 20.1 (Hüsgen et al., 1999) was used for this work. The mutants originate from this strain and it served as a WT control in all experiments. Δcpipt‐log was derived from Hinsch et al. (2015) and ΔΔcpipt‐log/tRNA‐ipt from Hinsch et al. (2016). Mycelia were grown on complete medium BII (Esser and Tudzynski, 1978) for cultivation and on Mantle medium (Mantle and Nisbet, 1976) to obtain conidia. All strains were cultivated in the dark at 26 °C.
Nucleic acid extraction and analysis
Prior to DNA preparation, mycelium was lyophilized, plant material was shock frozen with liquid nitrogen, and 50 mg of fresh florets were ground with metal beads at 30 Hz for 2 × 90 s in a Retsch MM400 (Haan, Germany) swing mill. gDNA from C. purpurea and B. distachyon florets was prepared according to Cenis (1992). For total RNA preparation, material was prepared in the same way. RNA was extracted using the Qiagen (Hilden, Germany) RNeasy plant mini kit following the manufacturer's protocol. PCR was performed as described in Sambrook et al. (1989) using BioTherm Polymerase (GeneCraft, Lüdinghausen, Germany). All primers used are listed in Table S1 (see Supporting Information) and were synthesized by Biolegio (Nijmegen, the Netherlands). Reverse transcription‐PCR was performed using Superscript II (Invitrogen, Carlsbad, CA, USA), oligo d(T) primer and 1 µg of total DNase‐treated RNA as template, according to the manufacturer's instructions.
qPCR
qPCRs were performed with BioRad iTaq Universal SYBR Green Supermix and the CFX96 Touch Cycler (BioRad, Hercules, CA, USA). Programming, data collection and analyses were performed with CFX Manager Software Version 3.1 (BioRad). The portion of fungal gDNA was calculated as a ratio of the fungal β‐tubulin gene [National Center for Biotechnology Information (NCBI) accession KP689578.1] to the actin gene from Brachypodium (Bradi4g41850) using the ΔCt method (Livak and Schmittgen, 2001). The expression of the tested genes was normalized to the expression of UBIQUITIN18 (Bd4g00660) according to Hong et al. (2008). The expression of all genes was calculated according to the ΔΔCt (cycle threshold) method. Expression was verified in two independent biological replicates.
Plant growth conditions, infection and sampling
Pathogenicity assays were performed using the cytoplasmic male‐sterile Secale cereale Lo37‐PxLo55‐N (KWS Lochow GmbH, Bergen, Germany) cultivar, which was cultivated and inoculated as described previously (Giesbert et al., 2008; Smit and Tudzynski, 1992).
Brachypodium distachyon (L.) Beauv. inbred line Bd21 (Vogel et al., 2006) was cultivated in sowing substrate under conditions of 20 h of light (24 °C)/4 h of darkness (18 °C). Approximately 6 weeks after sowing, the spikelets were inoculated before anthesis. Inoculation was performed by needle infiltration with a spore suspension [(6–8) × 106 conidia/mL] or sterile tap water (mock control). To avoid cross‐contamination, the ears were covered with paper bags directly after inoculation. Twenty‐four inoculated florets were collected at 0 and 10 dpi and 48 at 5 dpi, shock frozen with liquid nitrogen and stored at −80 °C before further processing.
The in vitro pathogenicity assay of isolated ovaries was performed as described previously (Scheffer and Tudzynski, 2006).
Microscopic analyses
For microscopic studies in planta, ovaries were embedded in 8% agarose to enable longitudinal bisection, stained with KOH–aniline blue, as described previously (Scheffer and Tudzynski, 2006), and examined with a Zeiss (Jena, Germany) Discovery V20 stereomicroscope fitted with an AxioCam MRc camera. Image analysis was performed with Axiovision Rel 4.8 software.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Primers used in this study.
Acknowledgements
We thank Brian Williamson for critical reading of the manuscript, Daniela Odinius for excellent technical assistance, Wilhem Schäfer for providing Brachypodium seeds, and the Chinese–German Science Center for funding (GZ928).
References
- Bauer, E. , Schmutzer, T. , Barilar, I. , Mascher, M. , Gundlach, H. , Martis, M.M. , Twardziok, S.O. , Hackauf, B. , Gordillo, A. , Wilde, P. , Schmidt, M. , Korzun, V. , Mayer, K.F.X. , Schmid, K. , Schön, C. and Scholz, U. (2017) Towards a whole‐genome sequence for rye (Secale cereale L.). Plant J. 89, 853–869. [DOI] [PubMed] [Google Scholar]
- Blümke, A. , Sode, B. , Ellinger, D. and Voigt, C.A. (2015) Reduced susceptibility to Fusarium head blight in Brachypodium distachyon through priming with the Fusarium mycotoxin deoxynivalenol. Mol. Plant Pathol. 16, 472–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragg, J.N. , Wu, J. , Gordon, S.P. , Guttman, M.E. , Thilmony, R. , Lazo, G.R. , Gu, Y.Q. and Vogel, J.P. (2012) Generation and characterization of the Western Regional Research Center Brachypodium T‐DNA insertional mutant collection. PLoS One, 7, e41916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brkljacic, J. , Grotewold, E. , Scholl, R. , Mockler, T. , Garvin, D.F. , Vain, P. , Brutnell, T. , Sibout, R. , Bevan, M. , Budak, H. , Caicedo, A.L. , Gao, C. , Gu, Y. , Hazen, S.P. , Holt, B.F., 3rd , Hong, S.Y. , Jordan, M. , Manzaneda, A.J. , Mitchell‐Olds, T. , Mochida, K. , Mur, L.A. , Park, C.M. , Sedbrook, J. , Watt, M. , Zheng, S.J. and Vogel, J.P. (2011) Brachypodium as a model for the grasses: today and the future. Plant Physiol. 157, 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer, M. , Lievens, B. , Van Hemelrijck, W. , Van den Ackerveken, G. , Cammue, B.P. and Thomma, B.P. (2003) Quantification of disease progression of several microbial pathogens on Arabidopsis thaliana using real‐time fluorescence PCR. FEMS Microbiol. Lett. 228, 241–248. [DOI] [PubMed] [Google Scholar]
- Cenis, J.L. (1992) Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids Res. 20, 2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmais, M. , Antelme, S. , Ho‐Yue‐Kuang, S. , Wang, Y. , Darracq, O. , D'Yvoire, M.B. , Cézard, L. , Légée, F. , Blondet, E. and Oria, N. (2013) A TILLING platform for functional genomics in Brachypodium distachyon . PLoS One, 8, e65503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper, J. , Mur, L.A.J. , Jenkins, G. , Ghosh‐Biswas, G.C. , Bablak, P. , Hasterok, R. and Routledge, A.P.M. (2001) Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiol. 127, 1539–1555. [PMC free article] [PubMed] [Google Scholar]
- Esser, K. and Tudzynski, P. (1978) Genetics of the ergot fungus Claviceps purpurea . Theor. Appl. Genet. 53, 145–149. [DOI] [PubMed] [Google Scholar]
- Fitzgerald, T.L. , Powell, J.J. , Schneebeli, K. , Hsia, M.M. , Gardiner, D.M. , Bragg, J.N. , McIntyre, C.L. , Manners, J.M. , Ayliffe, M. , Watt, M. , Vogel, J.P. , Henry, R.J. and Kazan, K. (2015) Brachypodium as an emerging model for cereal–pathogen interactions. Ann. Bot. 115, 717–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesbert, S. , Schürg, T. , Scheele, S. and Tudzynski, P. (2008) The NADPH oxidase Cpnox1 is required for full pathogenicity of the ergot fungus Claviceps purpurea . Mol. Plant Pathol. 9, 317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Haarmann, T. , Rolke, Y. , Giesbert, S. and Tudzynski, P. (2009) Ergot: from witchcraft to biotechnology. Mol. Plant Pathol. 10, 563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbritter, A.H. , Carroll, G.C. , Gusewell, S. and Roy, B.A. (2012) Testing assumptions of the enemy release hypothesis: generalist versus specialist enemies of the grass Brachypodium sylvaticum . Mycologia, 104, 34–44. [DOI] [PubMed] [Google Scholar]
- Hinsch, J. and Tudzynski, P. (2015) Claviceps: the ergot fungus In: Molecular Biology of Food and Water Borne Mycotoxigenic and Mycotic Fungi (Russell R. and Paterson M., eds), pp. 229–250. Boca Raton, FL: CRC Press. [Google Scholar]
- Hinsch, J. , Vrabka, J. , Oeser, B. , Novák, O. , Galuszka, P. and Tudzynski, P. (2015) De novo biosynthesis of cytokinins in the biotrophic fungus Claviceps purpurea . Environ. Microbiol. 17, 2935–2951. [DOI] [PubMed] [Google Scholar]
- Hinsch, J. , Galuszka, P. and Tudzynski, P. (2016) Functional characterization of the first filamentous fungal tRNA‐isopentenyltransferase and its role in the virulence of Claviceps purpurea . New Phytol. 11, 9801–9992. [DOI] [PubMed] [Google Scholar]
- Hong, S. , Seo, P.J. , Yang, M. , Xiang, F. and Park, C. (2008) Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real‐time PCR. BMC Plant Biol. 8, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüsgen, U. , Büttner, P. , Müller, U. and Tudzynski, P. (1999) Variation in karyotype and ploidy level among field isolates of Claviceps purpurea . J. Phytopathol. 147, 591–597. [Google Scholar]
- Kakei, Y. , Mochida, K. , Sakurai, T. , Yoshida, T. , Shinozaki, K. and Shimada, Y. (2015) Transcriptome analysis of hormone‐induced gene expression in Brachypodium distachyon . Sci. Rep. 5, 14 476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz, J.L. and Smith, D.L. (2015) Recent investigations of ergot alkaloids incorporated into plant and/or animal systems. Front. Chem. 3, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔCT method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Mandadi, K.K. and Scholthof, K.B. (2012) Characterization of a viral synergism in the monocot Brachypodium distachyon reveals distinctly altered host molecular processes associated with disease. Plant Physiol. 160, 1432–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantle, P.G. and Nisbet, L.J. (1976) Differentiation of Claviceps purpurea in axenic culture. J. Gen. Microbiol. 93, 321–334. [DOI] [PubMed] [Google Scholar]
- Oeser, B. , Kind, S. , Schurack, S. , Schmutzer, T. , Tudzynski, P. and Hinsch, J. (2017) Cross‐talk of the biotrophic pathogen Claviceps purpurea and its host Secale cereale . BMC Genomics, 18, 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peach, J.M. and Loveless, A.R. (1975) A comparison of two methods of inoculating Triticum aestivum with spore suspensions of Claviceps purpurea . Br. Mycol. Soc. 64, 328–331. [Google Scholar]
- Peraldi, A. , Beccari, G. , Steed, A. and Nicholson, P. (2011) Brachypodium distachyon: a new pathosystem to study Fusarium head blight and other Fusarium diseases of wheat. BMC Plant Biol. 11, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routledge, A.P.M. , Shelley, G. , Smith, J.V. , Talbot, N.J. , Draper, J. and Mur, L.A.J. (2004) Magnaporthe grisea interactions with the model grass Brachypodium distachyon closely resemble those with rice (Oryza sativa). Mol. Plant Pathol. 5, 253–265. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. , Fritsch, E.F. and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Scheffer, J. and Tudzynski, P. (2006) In vitro pathogenicity assay for the ergot fungus Claviceps purpurea . Mycol. Res. 110, 465–470. [DOI] [PubMed] [Google Scholar]
- Smit, R. and Tudzynski, P. (1992) Efficient transformation of Claviceps purpurea using pyrimidine auxotrophic mutants: cloning of the OMP decarboxylase gene. Mol. Gen. Genet. 234, 297–305. [DOI] [PubMed] [Google Scholar]
- The International Brachypodium Initiative (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon . Nature, 463, 763–768. [DOI] [PubMed] [Google Scholar]
- Thomma, B.P. , Eggermont, K. , Penninckx, I.A. , Mauch‐Mani, B. , Vogelsang, R. , Cammue, B.P. and Broekaert, W.F. (1998) Separate jasmonate‐dependent and salicylate‐dependent defense‐response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA, 95, 15 107–15 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, J.P. , Garvin, D.F. , Leong, O.M. and Hayden, D.M. (2006) Agrobacterium‐mediated transformation and inbred line development in the model grass Brachypodium distachyon . Plant Cell Tissue Organ Cult. 84, 199–211. [Google Scholar]
- Weßling, R. and Panstruga, R. (2012) Rapid quantification of plant–powdery mildew interactions by qPCR and conidiospore counts. Plant Methods, 8, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Primers used in this study.


