Summary
The fungal pathogen Fusarium pseudograminearum causes important diseases of wheat and barley. During a survey of secondary metabolites produced by this fungus, a novel class of cytokinins, herein termed Fusarium cytokinins, was discovered. Cytokinins are known for their growth‐promoting and anti‐senescence activities, and the production of a cytokinin mimic by what was once considered as a necrotrophic pathogen that promotes cell death and senescence challenges the simple view that this pathogen invades its hosts by employing a barrage of lytic enzymes and toxins. Through genome mining, a gene cluster in the F. pseudograminearum genome for the production of Fusarium cytokinins was identified and the biosynthetic pathway was established using gene knockouts. The Fusarium cytokinins could activate plant cytokinin signalling, demonstrating their genuine hormone mimicry. In planta analysis of the transcriptional response to one Fusarium cytokinin suggests extensive reprogramming of the host environment by these molecules, possibly through crosstalk with defence hormone signalling pathways.
Keywords: cytokinin, Fusarium, Fusarium crown rot, Fusarium graminearum, Fusarium pseudograminearum, phytohormones, secondary metabolites
Introduction
Plant growth and development are coordinated by phytohormones, which are small signal molecules active at low (nanomolar) concentrations. Eight classes of phytohormones are recognized: abscisic acid, auxins, cytokinins, ethylene, gibberellins, brassinosteroids, jasmonates and salicylic acid (SA) (Creelman and Mullet, 1997; Kende and Zeevaart, 1997). Phytohormones not only regulate physiological functions and reproduction, but are also involved in complex interconnected immune responses on pathogen and insect attack (Pieterse et al., 2009). SA, jasmonic acid (JA) and ethylene are well‐established central hormones regulating plant defences against pathogens (De Vos et al., 2005). However, a number of recent reports have implicated other hormones in plant defence or susceptibility, including the cytokinins (Hann et al., 2014; Jameson, 2000; Jiang et al., 2013; Novak et al., 2013; Siemens et al., 2006). Cytokinins are involved in diverse processes, including cellular proliferation, differentiation, the control of the morphological balance between shoot and root tissue, modification of source–sink relationships and delay of leaf senescence (Choi et al., 2011; Sakakibara, 2006). They also appear to have anti‐apoptotic properties (Othman et al., 2016). Cytokinin signalling has also been well characterized in the model plant Arabidopsis thaliana, as reviewed by To and Kieber (2008), where three histidine kinase receptors potentiate cytokinin signalling through a phospho‐relay system to response regulators which bind DNA and regulate transcription (positively or negatively). The receptors and response regulators are conserved in the model monocot Brachypodium distachyon (hereafter Brachypodium) (Kakei et al., 2015).
Cytokinins are adenine derivatives, which differ at the side chain at the N 6 position, which can be either an isoprenoid or aromatic group (Strnad, 1997; Tarkowska et al., 2003). The biosynthesis of cytokinins occurs through the prenylation of adenosine monophosphate (AMP), adenosine diphosphate (ADP) or adenosine triphosphate (ATP), which are catalysed by adenylate isopentenyltransferase (IPT) using dimethylallyl diphosphate (DMAPP) as a substrate (Kakimoto, 2001; Takei et al., 2001). This enzymatic step, leading to the formation of isopentenyladenine riboside monophosphate (iPRMP), was first identified in the amoeba Dictyostelium discoideum (Taya et al., 1978) and in Agrobacterium tumefaciens (Akiyoshi et al., 1984; Barry et al., 1984). iPRMP is subsequently hydroxylated by a cytochrome P450 monooxygenase before the cytokinins are transformed in a two‐step reaction by a nucleotidase and a nucleosidase (Chen and Kristopeit, 1981) to the active forms trans‐zeatin (tZ) and isopentenyladenine (iP). The final activation can also be performed in a single step by an enzyme named Lonely Guy (LOG) after the mutant phenotype in which rice flowers often contain only one stamen but no pistil (Kurakawa et al., 2007).
An alternative route for cytokinin biosynthesis is through a transfer RNA (tRNA) pathway. In this pathway, an adenine residue in tRNA is N 6‐prenylated by a tRNA isopentenyltransferase (tRNA‐IPT) and subsequently released from the tRNA by hydrolysis (Gray et al., 1996; Koenig et al., 2002; Miyawaki et al., 2006). tRNA‐IPTs are found in all organisms except Archaea and the addition of DMAPP to a tRNA‐bound adenine nucleotide, followed by degradation in Arabidopsis, results in the synthesis of cis‐zeatin (cZ) (Miyawaki et al., 2006).
Plant‐pathogenic fungi have developed numerous strategies to prevail over their hosts, including the production of phytohormones which interfere with plant immune responses. The maize pathogen Colletotrichum graminicola is thought to alter cytokinin responses at infection sites, either through fungal production of a cytokinin mimic or via an indirect route (Behr et al., 2012). Cytokinin activity has also been reported in extracts from members of the Fusarium genus, including the cereal pathogen F. culmorum (Michniewicz et al., 1986; Vanstaden and Nicholson, 1989). The tRNA pathway for cytokinin biosynthesis is present in all fungi, including the plant pathogens Magnaporthe oryzae (Chanclud et al., 2016), Leptosphaeria maculans (Trdá et al., 2017) and Claviceps purpurea (Hinsch et al., 2016), resulting in the production of iP and cZ. Claviceps purpurea also possesses a second pathway similar to the AMP prenylation route that produces iP via an IPT‐LOG fusion enzyme (Hinsch et al., 2015). A cytochrome P450 monooxygenase, which catalyses the iP hydroxylation to tZ, is located next to the IPT‐LOG gene, forming a small cytokinin production gene cluster in this species. Two active copies of this gene cluster responsible for tZ production are also present in members of the Fusarium fujikuroi species complex (Niehaus et al., 2016).
A role for plant‐produced cytokinins in defence processes is emerging in model systems. In Arabidopsis, both exogenous and endogenous cytokinins can lead to decreased susceptibility to bacterial and oomycete pathogens. Mechanistically, cytokinins are thought to act via the potentiation of SA‐mediated responses [reviewed in Albrecht and Argueso (2016)]. In rice, cytokinins also potentiate SA responses (Jiang et al., 2013), where the expression of classical marker genes, such as PR1b, is induced only by SA–cytokinin co‐treatment in an SA signalling‐dependent manner. In contrast with the potentiation of SA responses, cytokinins can also induce susceptibility to pathogens. Cytokinin production by M. oryzae, via the tRNA pathway, attenuates host defence responses and alters the sugar and amino acid distribution to favour the pathogen (Chanclud et al., 2016). Similarly, a Pseudomonas effector protein activates pools of inactive plant cytokinin precursors which, in turn, reduce pattern‐triggered immunity responses (Hann et al., 2014). In C. purpurea, cytokinin production contributes to pathogen virulence (Hinsch et al., 2016), presumably by the induction of susceptibility, but the precise mechanism of action of cytokinins in this pathosystem is not yet clear.
Fusarium pseudograminearum causes both Fusarium crown rot and Fusarium head blight diseases of wheat and barley (Obanor et al., 2013). In this study, we report the identification of a new class of cytokinins produced by F. pseudograminearum, describe the underlying fungal genes for their biosynthesis, propose their biosynthetic route and go on to verify their genuine cytokinin activity through receptor activity assays and the analysis of host gene expression in response to the most highly produced compound.
Results
Fusarium pseudograminearum produces novel cytokinin‐like molecules in axenic culture and during host infection
During the profiling of secondary metabolite production by F. pseudograminearum (CS3096), three pyrrole‐substituted purine derivatives, hereafter collectively termed Fusarium cytokinins, were identified. The three compounds were structurally elucidated by nuclear magnetic resonance (NMR; for detailed spectroscopic data, see Data S1 in Supporting Information) and named fusatin (1; C10H9N5), 8‐oxo‐fusatin (2; C10H9N5O1) and fusatinic acid (3; C10H7N5O2) (Fig. 1A). Fusatinic acid and 8‐oxo‐fusatin are novel compounds, whereas fusatin has been obtained synthetically previously through the oxidation of zeatin (Haidoune et al., 1990). A fourth compound, 8‐oxo‐isopentenyladenine (4; C10H7N5O2), was also confirmed as being produced by F. pseudograminearum by high‐resolution mass spectroscopy and NMR (data not shown). Trace amounts of a mass (C10H7N5O3, [M + H]+: m/z 246.062) corresponding to a carboxylic acid form of 8‐oxo‐fusatin (= 8‐oxo‐fusatinic acid) were also observed (data not shown). Fusatin and fusatinic acid were detected in wheat samples 14 days after single floret infection (Fig. 1B). 8‐oxo‐fusatin could not be detected in planta, although its lower limit of detection was considerably higher than that of the other compounds. Fusatinic acid was the more abundant of the two detected Fusarium cytokinins with levels between 0.1 and 1 μg/g (average of 0.2 μg/g). The mycotoxin deoxynivalenol, which is up‐regulated by Fusarium spp. during infection (Mudge et al., 2006), was present at concentrations approaching 5 μg/g in these samples. A concentration of 0.2 μg/g of fusatinic acid is equivalent to approximately 0.1 μm, a concentration at which native plant cytokinins can activate cytokinin receptors and are physiologically active (Miwa et al., 2007). As these extracts were from whole heads, localized concentrations of fusatinic acid are likely to be well in excess of the measured levels.
Figure 1.
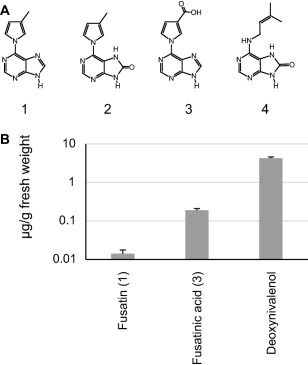
Fusarium pseudograminearum produces cytokinins in culture and in planta. (A) Structures of four Fusarium cytokinins isolated and elucidated from F. pseudograminearum wild‐type (CS3096) grown in yeast extract–sucrose medium: 1, fusatin; 2, 8‐oxo‐fusatin; 3, fusatinic acid; 4, 8‐oxo‐isopentenyladenine. (B) Quantification of Fusarium cytokinins and deoxynivalenol in infected wheat. 8‐Oxo‐fusatin (2) was not detected in planta and 8‐oxo‐isopentenyladenine (4) was not searched for. Values represent the average of 12 individual infected heads with error bars representing the standard error of the mean.
Biosynthesis of cytokinin molecules is encoded in a gene cluster expressed during infection
To identify the genes responsible for Fusarium cytokinin biosynthesis, the F. pseudograminearum genome was searched for homologues of the Arabidopsis LOG1 (AT2G28305.1). Two matches, both with ∼40% amino acid identity to LOG1, were identified (FPSE_07269 and FPSE_06372). FPSE_06372 is hereafter named FCK1. In addition to the LOG1 homology, FCK1 contained an IPT at its N‐terminus. FCK1 was deleted from F. pseudograminearum by homologous gene replacement (Fig. S1, see Supporting Information) and, in all four independent mutants generated, the production of the Fusarium cytokinins was abolished when analysed qualitatively by inspection of high‐performance liquid chromatography‐mass spectrometry (HPLC‐MS) traces (data not shown). Relative quantification analysis of Fusarium cytokinins of replicated cultures of a single FCK1 mutant confirmed the qualitative observations (Fig. S2, see Supporting Information). Furthermore, during the course of this study, a similar protein identified in C. purpurea (CPUR_04177) and two in F. fujikuroi (FFUJ_03536 and FFUJ_14354) were shown to contribute to a large proportion of the tZ pool produced by these species (Hinsch et al., 2015; Niehaus et al., 2016). This prompted an analysis of a broader suite of fungal genomes for sequence homologues of these genes. Twenty‐six additional IPT‐LOGs were identified in Fusarium spp. Eight F. oxysporum strains, as well as F. fujikuroi and F. verticillioides, contained two IPT‐LOG orthologues. Outside of Fusarium and Claviceps, there was a match in Zymoseptoria tritici, although this did not cover the full length of the protein. Phylogenetic analysis of IPT‐LOG suggests that three separate isoforms exist in fungal genomes (Fig. 2A).
Figure 2.
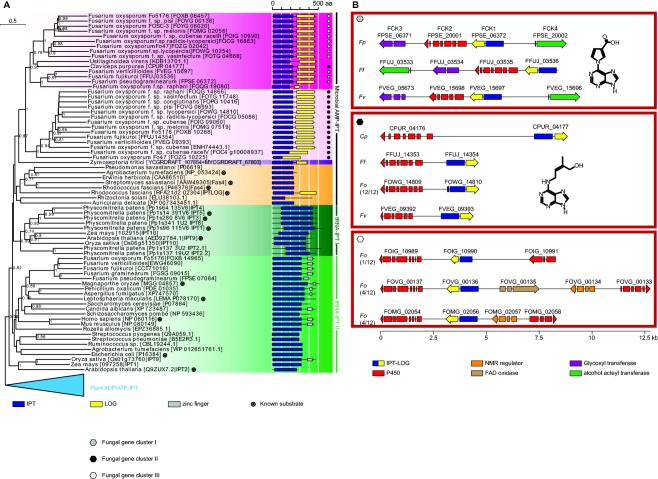
Fungal genomes encode three distinct cytokinin production clusters. (A) Phylogenetic analysis of isopentenyltransferase (IPT) proteins. (B) Three distinct cytokinin production gene clusters are found in various Fusarium spp. and Claviceps purpurea. aa, amino acid; FAD, flavin adenine dinucleotide; LOG, Lonely Guy; NMR, nuclear magnetic resonance.
Comparative genome analysis suggested that the F. pseudograminearum cluster consists of three additional genes [FCK2 (FPSE_20001), FCK3 (FPSE_06371) and FCK4 (FPSE_20002)] (Fig. 2B). FCK2 is a cytochrome P450 monooxygenase; FCK3 has a weak (e‐value, 6 × 10−8) hit to the capsule polysaccharide biosynthesis protein domain (pfam05704), which suggests that this protein might have a role as a glycosyl transferase; FCK4 has a domain match (e‐value, 2 × 10−15) to an alcohol acetyltransferase (pfam07247). The original annotation of the F. pseudograminearum genome (Gardiner et al., 2012) failed to predict FCK2 and FCK4, but RNA sequencing (RNAseq) data clearly demonstrated their presence (Fig. S3, see Supporting Information). The Fusarium cytokinin cluster is located 0.5 Mbp from the end of chromosome 3 in a region of high inter‐strain diversity (Gardiner et al., 2017).
As observed in the C. purpurea and F. fujikuroi clusters (Hinsch et al., 2015; Niehaus et al., 2016), cytochrome P450 monooxygenase‐encoding genes adjacent to IPT‐LOGs were observed for all analysed Fusarium strains. However, phylogenetic analysis of cytochrome P450 monooxygenases suggested that, like IPT‐LOG, there were three sequence types (Fig. S4, see Supporting Information). Taken together, the analyses of IPT‐LOG and the cytochrome P450 monooxygenases are suggestive of three separate cluster types which, when considered with the very different cytokinin‐like molecules produced by F. pseudograminearum and C. purpurea, are unlikely to be orthologous.
Biosynthesis of Fusarium cytokinins occurs through two parallel pathways
To determine the biosynthetic route (Fig. 3A) for Fusarium cytokinins, a combination of biochemical characterization of heterologously produced enzymes and analysis of culture extracts of mutant strains was employed. Strains of F. pseudograminearum carrying deletions of FCK1 were unable to produce any Fusarium cytokinins or iP, tZ or cZ (Fig. 3B), which were all detectable in the progenitor strain (CS3096). A role for FCK1 in the production of cZ was unexpected as this compound is commonly produced via the degradation of prenylated adenine moieties from tRNA, but, as tZ was also absent in the FCK1 mutant, it is possible that cZ is isomerized from tZ (Bassil et al., 1993). Heterologously expressed FCK1 was able to use AMP and DMAPP in vitro (Fig. 4), as the accumulation of both iPRMP and iP was observed. After extended incubation (18 h), almost no iPRMP was detectable and only iP was observed. The mutant analysis places FCK1 as the first committed step (from primary metabolites) in the biosynthesis of Fusarium cytokinins, and the biochemistry suggests that the reaction catalysed by the bifunctional FCK1 proceeds via prenylation, followed by the removal of the phosphoribose. The detection of iPRMP in these reactions is consistent with the release of this intermediate to the bulk solvent prior to utilization by the second active site.
Figure 3.
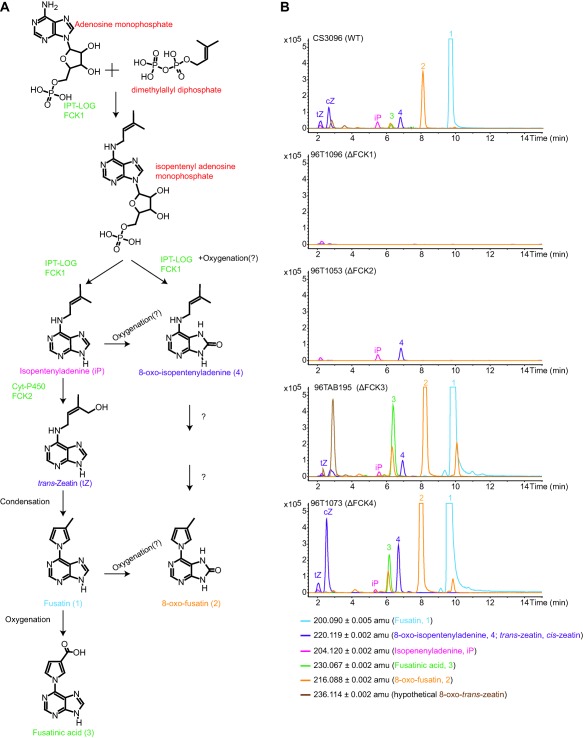
Biosynthesis of Fusarium cytokinins in Fusarium pseudograminearum. (A) Proposed biosynthetic pathway based on the accumulation of cytokinins in deletion mutants and biochemical characterization of FCK1. (B) Extracted ion chromatograms of Fusarium cytokinins and related molecules in F. pseudograminearum wild‐type (WT) and deletion mutants. Data are shown for one deletion mutant only in each gene, but are representative of traces for independent mutant generated for all genes.
Figure 4.
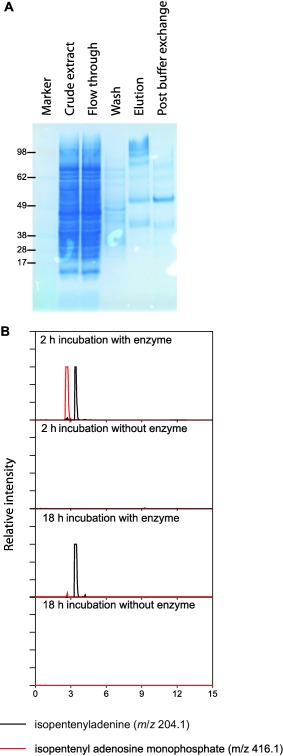
FCK1 catalyses the formation of isopentenyladenine in vitro from adenosine monophosphate and dimethylallyl pyrophosphate. (A) The FCK1 protein was expressed in Escherichia coli as an N‐terminally histidine (His)‐tagged fusion protein. The expected size of the protein is ∼55 kDa. (B) Extracted ion chromatograms of the final expected protonated product (isopentenyladenine, iP) and intermediate (isopentenyladenine riboside monophosphate, iPRMP) of the two reactions catalysed by the bifunctional FCK1.
Deletion of the cytochrome P450 monooxygenase‐encoding gene (FCK2) resulted in the production of only two cytokinins: iP and 8‐oxo‐isopentenyladenine (4). The accumulation of iP and 4 and the absence of all other cytokinins in the ΔFCK2 strains suggest that FCK2 is responsible for the step immediately downstream of these compounds, which we propose is tZ based on the knowledge that cytochrome P450 monooxygenases are responsible for this reaction in plants and fungi. Protein structural modelling and docking also support tZ as a product of FCK2 (Fig. S5, see Supporting Information). However, it is also possible that FCK2 (directly or indirectly) is responsible for the formation of the C–N bond in the production of the pyrrole ring, which might explain why the phylogenetic analysis suggests that the cytochrome P450 monooxygenase in the C. purpurea cluster and FCK2 are not orthologous. C–N bond formation by cytochrome P450 monooxygenases is unusual, but not unprecedented: a Streptomyces sp. cytochrome P450 monooxygenase is involved in the formation of a C–N bond in the biosynthesis of staurosporine (Onaka et al., 2005). The ΔFCK2 strain also provided the primary evidence for the parallel nature of the proposed biosynthesis pathway. Based on the accumulation of 4 in this mutant, it must arise from either the prenylation of 8‐oxo‐AMP (by FCK1) and/or oxygenation of iP. C‐8 carbonyl derivatives of adenine or cytokinins are uncommon in nature, although formation has been observed in DNA damaged by γ‐irradiation (Bonicel et al., 1980), and is also catalysed by a xanthine dehydrogenase in the rhizobacterium Serratia proteomaculans (Taylor et al., 2006). Several putative xanthine dehydrogenases are present in Fusarium spp.
Deletion of FCK3 resulted in enhanced production of cytokinins, including a compound with an m/z value of 236.1141 [M + H]+ (Fig. 3A), which could be 8‐oxo‐tZ (C10H13N5O2, theoretical [M + H]+: 236.1142), an obvious intermediate for the parallel biosynthesis of 8‐oxo‐fusatin, but repeated attempts to elucidate the structure failed, most probably as a result of insufficient amounts of compound. Deletion of FCK4 also resulted in the enhanced production of cytokinins compared with the wild‐type, which was most noticeable for cZ (Figs 3A and S2). The enhanced production in the ΔFCK3 and ΔFCK4 strains could suggest that both enzymes act downstream of the identified Fusarium cytokinins to produce as yet unidentified compounds. The mechanism for the oxidation of fusatin to fusatinic acid remains unknown. Furthermore, heterologous expression of the cytokinin cluster in F. graminearum resulted in the production of 1, 2, 3 and possibly 8‐oxo‐tZ, which were not observed in the wild‐type (Fig. S6, see Supporting Information).
Fusatinic acid shows weak senescence‐delaying activity in planta and can activate cytokinin signalling
Analysis of RNAseq data demonstrated that all four genes of the F. pseudograminearum cytokinin cluster are expressed during the infection of barley and Brachypodium. Expression levels for FCK1, FCK2 and FCK3 approached that of the first step (TRI5) of deoxynivalenol biosynthesis (Fig. S3), which is important for stem base infection of cereals by Fusarium spp. (Desmond et al., 2008; Scherm et al., 2011). Expression of FCK4 was at least an order of magnitude lower than that of the other genes in the cluster in barley and below the quantification limits in Brachypodium. In contrast, the only available dataset of F. verticillioides infecting plants (Lanubile et al., 2014) showed that only FCK4 was expressed during ear rot (data not shown). Fusarium cytokinins were also not detected in metabolite extracts of F. verticillioides or F. fujikuroi, suggesting that the cluster, with the exception of FCK4, may be inactive in culture, consistent with the analysis of transcriptional data for five different strains from this lineage by Niehaus et al. (2016).
Despite the structural similarities of Fusarium cytokinin molecules to compounds with cytokinin activity, small structural differences can convert cytokinin agonists into antagonists (Spichal et al., 2014). To test whether fusatinic acid had cytokinin activity, its ability to prevent the senescence of detached leaves was assessed and compared with that of the synthetic cytokinin 6‐benzyl aminopurine (BAP), which elicits plant growth and developmental responses (Chen and Yang, 2013; D'Aloia et al., 2011). The results showed that BAP prevented senescence in Brachypodium (Fig. 5A) with leaves appearing greener than dimethylsulfoxide (DMSO) controls. In contrast, fusatinic acid did not obviously reduce senescence based on visual inspection of the leaves (Fig. 5A). However, using a quantitative measure of leaf colour, the samples treated with fusatinic acid were slightly (∼7%) darker than the solvent control (P = 0.007) (Fig. 5B). BAP treatment caused leaves to be considerably darker (32%, P = 2 × 10−7). Thus, fusatinic acid may have very weak senescence‐inhibiting activities in Brachypodium.
Figure 5.
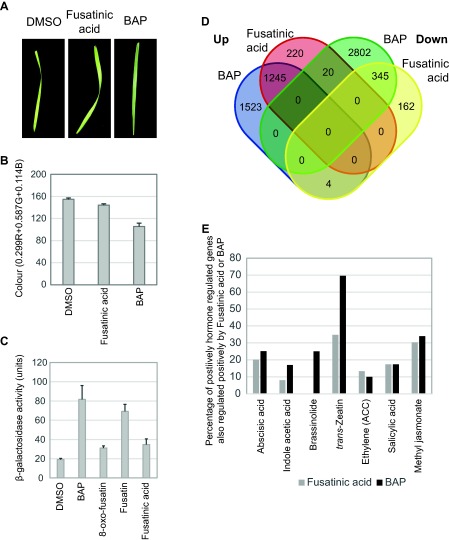
Fusarium cytokinin molecules have cytokinin activity. (A) Examples of gross effects of fusatinic acid and BAP (6‐benzyl aminopurine) treatment on detached Brachypodium leaves. Leaves were treated in 15 mL of 0.1% agarose with 10 μm of the hormone for 10 days on a laboratory bench. Dimethylsulfoxide (DMSO) (0.2% final concentration) was used as a control. (B) Relative leaf colour following treatment with fusatinic acid or BAP in Brachypodium, measured as the average red, green and blue pixel contents per unit leaf area. Lower numbers indicate darker leaves. The data consist of a minimum of 10 individual leaves per treatment with DMSO, fusatinic acid or BAP. (C) Fusarium cytokinins can activate the Arabidopsis histidine kinase 3 cytokinin receptor in a bacterial assay system. (D) RNA sequencing (RNAseq) analysis of gene expression changes in Brachypodium in response to BAP and fusatinic acid. Overlap between global gene expression changes in each treatment. All included genes were statistically significantly differentially regulated at P = 0.05 after Benjamini–Hochberg false discovery correction. (E) Overlap between the fusatinic acid or BAP positively regulated genes and the high‐stringency hormone regulated gene sets defined by Kakei et al. (2015). The positively hormone regulated high‐stringency gene sets contained the following numbers of genes: abscisic acid, 423; indole acetic acid, 112; brassinolide, 4; trans‐zeatin, 23; salicylic acid, 52; methyl jasmonate, 362. The overlap presented for ethylene was based on the low‐stringency set from Kakei et al. (2015) as these authors found no gene differentially regulated by ethylene when high‐stringency filtering was applied. ACC, 1‐aminocyclopropane‐1‐carboxylic acid.
To further investigate the potential for cytokinin agonistic activities of Fusarium cytokinins, their ability to activate a cytokinin receptor was assayed in a bacterial system. In this system, the Arabidopsis histidine kinase 3 (AHK3) cytokinin receptor controls the expression of β‐galactosidase (Mizuno and Yamashino, 2010), which has been used previously to demonstrate the signalling activity of a range of cytokinins and to functionally characterize the suite of cytokinin receptors in Arabidopsis (Inoue et al., 2001; Miwa et al., 2007; Spíchal et al., 2004; Suzuki et al., 2001). All Fusarium cytokinins activated AHK3 signalling (Fig. 5C). Taken together, the in vitro experiments suggest that Fusarium cytokinins are genuine cytokinin agonists, as fusatinic acid has the ability to weakly delay the senescence of detached leaves and to activate the AHK3 receptor.
Fusarium pseudograminearum cytokinin molecules are cytokinin signalling agonists in the model monocot Brachypodium
To further elucidate the effect of fusatinic acid on plants, a global analysis of Brachypodium gene expression in response to treatment with fusatinic acid or BAP was conducted. Fusatinic acid was chosen based on its terminal position in the proposed biosynthetic pathway and its measurement as the most abundant Fusarium cytokinin in planta (Figs 1 and 3). Treatment with fusatinic acid, when compared with the solvent control, resulted in statistically significant differential regulation of 1996 loci in Brachypodium (1485 up‐ and 511 down‐regulated). Of these fusatinic acid‐regulated genes, 81% were also differentially regulated under BAP treatment and, in 98.5% of these genes, the direction of regulation was the same under both fusatinic acid and BAP treatment (Fig. 5D). In the cytokinin signalling pathway, fusatinic acid up‐regulated the expression of four type A response regulators (Bradi2g6100, Bradi3g45930, Bradi4g43090 and Bradi5g25960; P = 5 × 10−5) between two‐ and four‐fold. Type A response regulators in Arabidopsis act as feedback regulators of not only cytokinin signalling, but also SA signalling (Argueso et al., 2012; To and Kieber, 2008). Two of the three cytokinin receptors were also significantly (P < 0.003) up‐regulated by fusatinic acid (Bradi1g10660 and Bradi2g59137), albeit by small fold changes of ∼1.3‐fold. The substantial overlap in the fusatinic acid and BAP regulon further supports the view that fusatinic acid has cytokinin agonistic activity in Brachypodium.
The role of cytokinins in plant resistance or susceptibility is poorly understood (Robert‐Seilaniantz et al., 2011). It has been proposed recently that pathogen‐produced cytokinins in the Magnaporthe–rice interaction act to suppress defence pathways, together with the alteration or maintenance of nutrient availability for the pathogen (Chanclud et al., 2016). However, in Arabidopsis, current evidence suggests that the opposite is true, where cytokinins contribute to resistance against biotrophic pathogens via an increase in SA responses, including the expression of the classical SA‐responsive gene PR1 (Choi et al., 2010). Following fusatinic acid treatment, PR1 (Bradi1g57590) was down‐regulated 2.1‐fold (P = 0.006). However, this was not observed under BAP treatment and under pathogen inoculation at a later time point, where this gene was, in fact, up‐regulated ∼20‐fold (? Powell et al., unpublished results). This observation led to the querying of whether other defence pathways were partially regulated by fusatinic acid. To this end, genes responding to fusatinic acid or BAP were compared with those responsive to other hormones presented by Kakei et al. (2015). Apart from the substantial overlap with genes in the response to the cytokinin tZ, more than 30% of all genes up‐regulated by methyl jasmonate were also up‐regulated by fusatinic acid and BAP (Fig. 5E). This included the up‐regulation of JA biosynthesis [lipoxygenase 3 (Bradi5g11590, 1.8‐fold, P = 5 × 10−5); three allene oxide synthases (Bradi1g69330, 1.5‐fold, P = 5 × 10−5; Bradi3g08160, 1.9‐fold, P = 5 × 10−5; Bradi3g23190, five‐fold, P = 5 × 10−5)] and response [nine different JAZ genes and Myc2 (Bradi3g34200, three‐fold, P = 1.3 × 10−3)] genes. The overlaps between responses to SA and fusatinic acid or BAP were lower at 18% of genes.
The observation of overlaps between the response to fusatinic acid and other plant hormones was consistent with a global analysis of gene ontologies conducted on the positively fusatinic acid‐regulated gene set (Data S2, see Supporting Information). In this analysis, terms corresponding to the response to JA, salicylate, auxin, gibberellin, abscisic acid and ethylene, but, surprisingly, not cytokinin, were all enriched [false discovery rate (FDR) ≤ 4 × 10−3]. When the log2 fold expression data comparing fusatinic acid with DMSO were mapped on to the Brachypodium metabolic pathways, 472 genes that were statistically significant (up or down) at the 0.05 level were assigned to known reactions. However, the majority of enzymes that could be assigned to reactions in the BrachyCyc database (Tello‐Ruiz et al., 2016) are yet to be assembled into larger pathways. Nonetheless, some trends were evident in the dataset. In this analysis, proteins related to cytokinin homeostasis were not observed. However, there appeared to be specific changes which suggested that SA and ethylene pathways might be modified by fusatinic acid. Ethylene is synthesized from 1‐aminocyclopropane‐1‐carboxylic acid (ACC), which is a methionine derivative. Methionine adenosyl transferase (Bradi2g12150, two‐fold, P = 5 × 10−5), three ACC synthases (Bradi1g10030, six‐fold, p = 5 × 10−5; Bradi2g05790, 1.9‐fold, P = 3 × 10−3; Bradi5g19100, 2.1‐fold, P = 5 × 10−5) and one ACC oxidase (Bradi2g35860, 1.5‐fold, P = 5 × 10−5) are up‐regulated by fusatinic acid. This suggests that methionine is preferentially converted to ACC and, in turn, may be converted to ethylene. A methyl‐salicylate esterase which releases SA (Bradi2g52110, 2.4‐fold, P = 2 × 10−4), important for converting biologically inactive methyl salicylate (MeSA) into active SA during systemic acquired resistance (Park et al., 2007), is also up‐regulated, suggesting a possible activation of SA responses, in contrast with the observation of the down‐regulation of PR1. Taken together, the host gene expression response, at the level of hormone regulatory pathways, to fusatinic acid is complex, but suggestive of substantive pathogen‐induced reprogramming of the plant environment. Presumably, this is to the benefit of the pathogen.
Discussion
The identification of novel cytokinins produced by F. pseudograminearum adds to the growing collection of plant pathogens that produce or directly manipulate plant hormones during infection. Indeed, based on its close relationship to F. graminearum, F. pseudograminearum is also likely to be able to produce auxin and interfere with ethylene synthesis (Adam et al., 2015). Cytokinin production by pathogens during infection has, in many cases, been shown to contribute to virulence or symptom development (Chanclud et al., 2016; Hinsch et al., 2016; Siddique et al., 2015). However, in initial experiments with an FCK1 mutant on wheat, a clear role for Fusarium cytokinins in pathogen virulence could not be ascertained (data not shown), although a comprehensive study has yet to be undertaken. Furthermore, the tRNA pathway for cytokinin (cZ) production is also active in F. pseudograminearum and it is possible that the abolishment of both pathways is required before a virulence‐related effect can be observed. Indeed, in C. purpurea, which also has two cytokinin production pathways, no role in virulence was observed until both pathways were mutated simultaneously (Hinsch et al., 2016).
The production of an active cytokinin during infection simultaneously with the cell death‐inducing toxin deoxynivalenol appears to be counterproductive at first impression. However, the production of Fusarium cytokinins in such high quantity during infection presumably imparts some selective advantage to the pathogen. The analysis of host gene expression following fusatinic acid treatment suggested the possibility that this may be via phytohormone crosstalk. However, the direct effect of fusatinic acid on the cytokinin responses of the host is an equally plausible evolutionary reason for the maintenance and activity of this cluster, and would suggest that F. pseudograminearum behaves like a hemi‐biotroph during infection. This has been proposed previously based on histopathological studies and biomass monitoring time course experiments, in which crown infection by F. pseudograminearum and F. graminearum of wheat and barley has distinct phases of fungal growth and symptom development (Knight and Sutherland, 2016; Stephens et al., 2008). Experiments to simultaneously monitor the spatial and temporal production of both deoxynivalenol and the Fusarium cytokinins will be necessary to further dissect the apparent co‐production of deoxynivalenol and Fusarium cytokinins during plant infection. Fusarium pseudograminearum infection is exacerbated in plants undergoing drought stress and, given that cytokinins can ameliorate drought symptoms in grasses, possibly via increasing sink strength at the site of production (Chang et al., 2016; Reguera et al., 2013), the Fusarium cytokinins may promote plant survival by direct activation of cytokinin responses. Promotion of host survival may be beneficial to the pathogen to ensure that sufficient plant biomass is available to the pathogen after host senescence or crop harvest. This strategy would be akin to ‘anti‐virulence’ loci in bacteria in that, although they reduce the measured virulence of a pathogen in laboratory assays, they nonetheless contribute to overall pathogen fitness by, for example, contributing positively to pathogen transmissibility or ecological survival between hosts (Foreman‐Wykert and Miller, 2003). This is a component of pathogen virulence underexplored in molecular studies of plant‐pathogenic fungi largely because of the difficulty in establishing suitable experimental systems (Preston, 2017). Such systems, which also consider the host–pathogen interaction over evolutionary timescales, are likely to be important for us to fully understand the intricacies of interactions between plant pathogens and their hosts. Cytokinin production by F. pseudograminearum, and secondary metabolism by fungi in general, represents one of these areas of exquisite intricacy that may be difficult to understand without probing the entire life cycle of the producing organisms in the context of their own pathogen population and environment.
Experimental Procedures
Fungal strains
Fusarium pseudograminearum isolate CS3096 was used for metabolite analyses and isolate RBG5266 (Bentley et al., 2008) was used for wheat head blight infections. Fusarium verticillioides strains BRIP14953a, BRIP53263a, BRIP53273b, BRIP53590a and BRIP54043a were provided by the Queensland Department of Primary Industries and originally isolated from maize or sorghum in New South Wales or Queensland, Australia. The genome sequenced F. verticillioides (FGSC7600), F. fujikuroi (IMI58289) and F. oxysporum (FGSC9935) have been described previously (Ma et al., 2010; Wiemann et al., 2013).
Identification of cytokinin biosynthetic genes
FCK1 was identified in the F. pseudograminearum genome based on its homology to the Arabidopsis LOG1 (AT2G28305.1). To conduct phylogenetic analyses, a set of IPT sequences from bacterial and eukaryotic sources was assembled from diverse sources using the Arabidopsis LOG1 as a query in a blastP analysis in GenBank. The set also included tRNA IPTs and is available in Data S3 (see Supporting Information). The amino acid sequences were aligned by multiple alignment using fast Fourier transform (MAFFT) employing the T‐REX web server (Boc et al., 2012). The alignments were analysed with MetaPIGA v2.0 (Helaers and Milinkovitch, 2010) using maximum likelihood with 100 bootstraps and visualized with EvolView (http://evolgenius.info/evolview) (Zhang et al., 2012). Protein sequences of fungal cytochrome P450 monooxygenases located adjacent to IPT‐LOGs were included in a phylogenetic analysis using the same procedure as described above.
Biochemical characterization of FCK1
FCK1 was expressed as an N‐terminal histidine (His) tag fusion protein in Escherichia coli. The coding sequence was synthesized and cloned into pET‐28a by GenScript (Piscataway, NJ, USA) using NheI and HindIII cloning sites. The expression and purification of the protein were performed as described previously (Kettle et al., 2015), except that the His tag was not removed from the protein prior to biochemical characterization. The final protein storage conditions were 50 mm sodium phosphate buffer, 300 mm NaCl and 20% glycerol, pH 7.5. Purification of the protein was assessed using sodium dodecylsulfate‐polyacrylamide gel electrophoresis employing a BOLT 4%–12% Bis‐Tris gel stained with SimplyBlue™ SafeStain (Thermo Fisher Scientific, Melbourne, Victoria, Australia). A SeeBlue® Plus2 prestained marker was used (Thermo Fisher Scientific). The enzyme assay was modified from those previously used to characterize an Arabidopsis IPT (Takei et al., 2001). The reaction mixture contained a final concentration of 880 mm betaine, 17.5 mm triethanolamine, 44 mm KCl, 8.8 mm MgCl2, 880 μm dithiothreitol (DTT), 880 μg/mL bovine serum albumin, 1 mm adenosine monophosphate, 286 μm dimethylallyl pyrophosphate, pH 8.0. The reaction was initiated by the addition of purified protein or buffer control. The reaction was incubated at room temperature, sampled at 2 and 18 h, stopped with an equal volume of 100% ethanol and heated to 90 °C for 10 min. Ten microlitres of the reactions were analysed via liquid chromatography‐mass spectrometry (LC‐MS) using system 1, as described in Methods S1 (see Supporting Information).
In silico FCK2 model building and docking analysis
A similar approach to model building and docking analysis was carried out as performed previously for another Fusarium cytochrome P450 monooxygenase (Droce et al., 2016). An initial homology model of FCK2 was generated with SWISS MODEL (Bordoli et al., 2008) (Basel, Switzerland) using PDB ID 4D6Z as the template structure. Following coordination of the deoxy‐haem group, subsequent energy minimization and a 500‐ns molecular dynamics model refinement with the md_refine macro were carried out in YASARA/WHAT IF Twinset (Vienna, Austria; version 16.7.22) using the Yasara2 force‐field in explicit water (TIP3P water model) (Krieger and Vriend, 2014; Krieger et al., 2004). Following energy minimizations of tZ, cZ, isopentenyladenine and fusatinic acid in YASARA (Yasara2 forcefield, TIP3P water model), each compound was globally docked in AutoDock VINA (Trott and Olson, 2010) to the best Z‐scoring, low‐energy, conformer of FCK2 (dock_run macro with 100 docking runs). Results were based on a comparison of the calculated dissociation constant of the high‐scoring clusters.
Heterologous expression in F. graminearum
The fungal cytokinin gene cluster was polymerase chain reaction (PCR) amplified from F. pseudograminearum CS3096 genomic DNA in three overlapping fragments with the primers provided in Table S1 (see Supporting Information). The primers used for the two outermost PCRs were each had a 30‐bp tail with homology to the plasmid multiple cloning site for integration through homologous recombination. A multi‐purpose shuttle vector was prepared for capture of the gene cluster and introduction to F. graminearum PH‐1 (Fig. S7, see Supporting Information). Vector construction was performed by transforming Saccharomyces cerevisiae strain BY4743 cells with BamHI/XhoI linearized plasmid DNA and three PCR products (Gietz and Schiestl, 2007). Transformants were plated on synthetic yeast drop‐out medium without uracil (Sigma, Sydney, Australia Cat. No. Y1501) and incubated at 28 ºC for 3 days. Plasmid DNA was purified from yeast colonies using Zymoprep Yeast Plasmid II (Zymoresearch, Irvine, CA, USA Cat No. D2004). The cytokinin cluster was transformed into F. graminearum through Agrobacterium tumefaciens‐mediated transformation (Malz et al., 2005). A transformant carrying the cytokinin gene cluster was identified by PCR with primers hybridizing outside the tubulin locus in combination with primers hybridizing inside the inserted gene cluster.
Targeted gene deletion
Vectors were synthesized by GenScript and consisted of: 1000 bp of sequence immediately upstream of the target gene's start codon, a 20‐bp sequence (GATGTCCACGAGGTCTCT), a unique 20‐bp sequence (see later), another 20‐bp sequence (CGTACGCTGCAGGTCGAC), the Aspergillus nidulans TrpC promoter and nourseothricin resistance cassette corresponding to nucleotide positions 437–1387 of GenBank accession AY631958.2 (Gardiner et al., 2005), and 993 bp of sequence immediately following the stop codon of each gene. The 20‐bp unique sequences were AGCTCAATATTGCGTGCGCA (FCK2), TCGCTGATCCACAGGTAGAT (FCK1), AGTGGACGTATCACATCTCG (FCK4) and ACTAGACGCTACGATCTTGG (FCK3). The synthesized fragments were amplified using Phusion DNA polymerase with M13 forward and reverse primers. Eight 50‐μL PCRs were pooled, polyethylene glycol precipitated and transformed into CS3096 protoplasts, as described previously (Gardiner et al., 2012). Transformants were screened using a triplex PCR assay with primers as listed in Table S1. Absence of the wild‐type band was used as confirmation of successful deletion of the target gene (Fig. S1).
Production and analyses of cytokinins and deoxynivalenol
Fusarium pseudograminearum (CS3096) was cultivated for 2 weeks at 25°C in the dark on yeast extract–sucrose (YES) agar medium (yeast extract from Scharlau, Barcelona, Spain) (Sørensen and Sondergaard, 2014). Secondary metabolites were extracted using the micro‐scale extraction procedure (Smedsgaard, 1997) modified as described previously (Sørensen et al., 2014). The extracts were initially examined on LC system 2 and then LC system 3 (Methods S1).
In planta quantification of Fusarium cytokinins and deoxynivalenol, and analysis of Australian isolates of F. verticillioides, were performed on LC system 4 (Methods S1). For plant samples, extracts were made from individual freeze‐dried heads that had been point inoculated with F. pseudograminearum isolate RBG5266 (Bentley et al., 2008) at 14 days prior to harvest. These were ground with a mortar and pestle and resuspended in 10 mL of extraction solution according to the modified micro‐scale extraction procedure, placed in a sonicator bath for 45 min, dried under nitrogen and resuspended in 400 μL of methanol. Standard curves for all compounds were constructed from pure compounds (verified by NMR) for the Fusarium cytokinins or deoxynivalenol purchased from Sigma. In planta molar concentration equivalents were calculated based on 1 g of fresh material being equivalent to 1 mL of solution and the water content of samples being about 90%.
Isolation and structural elucidation of cytokinins
Fusarium pseudograminearum CS3096 was grown on 50 Petri dishes (90 mm) with YES medium for 2 weeks at 25 °C in the dark, and cytokinins were isolated from the F. pseudograminearum extract on a semi‐preparative HPLC system (LC system 5 in Methods S1). The structures of the isolated cytokinins were elucidated by NMR spectroscopy. Fusatinic acid (3), fusatin (1), 8‐oxo‐fusatin (2) and 8‐oxo‐isopentenyladenine (4) were dissolved in methanol‐d4. 1H‐NMR, [1H,13C]‐heteronuclear single quantum coherence (HSQC), [1H,13C]‐heteronuclear multiple‐bond correlation (HMBC) (not molecule 4) and double‐quantum filtered correlation spectroscopy (DQF‐COSY) spectra were recorded on a Bruker (Billerica, MA, USA) AVIII‐600 MHz NMR spectrometer for all compounds. For fusatinic acid, additional [1H,15N]‐HMBC spectra optimized for J HN = 4, 8 and 16 Hz were recorded on a Bruker AVIII‐800 MHz spectrometer. Spectra were recorded and processed in TopSpin 3.2. The spectroscopic data and argument for structural elucidation are provided in Supporting Information (Data S1). Structures were checked against the CSEARCH Robot Referee (Haider and Robien, 2016) for consistency.
Detached leaf cytokinin activity assay
Stocks of fusatinic acid or BAP were dissolved in DMSO at 5 mm; 10–13 individual whole first leaves were treated in 15 mL of 0.1% agarose in 50‐mL tubes to which 30 μL of test compounds had been added. Leaves were maintained in the treatment solution on a slowly rotating tube roller in a laboratory maintained at 22 °C for 10 days prior to photographing using a 10‐megapixel digital camera on a black velvet background. The black background in the image was selected and deleted using the colour range selection feature set to shadows in Adobe Photoshop CS2. This area was replaced with a true black (RBG, 000) background and the image was saved and opened in ImageJ version 1.50i (Schneider et al., 2012). Each leaf was individually selected using the wand tool with default settings and the average red, green and blue pixel contents across each leaf were measured using the RBG Measure plugin with data exported to Microsoft Excel for statistical analysis.
An identical experimental system was used for gene expression analysis. Treatment occurred for 2 h prior to sampling by removing the leaves from the treatment solution and snap freezing. Each replicate consisted of a pool of 12 detached first leaves (treated in a single tube), and four biological replicates were used per treatment for RNA extraction and RNAseq analysis.
RNAseq analysis
RNA was extracted from freeze‐dried leaves using a QIAgen (Melbourne, Victoria, Australia) RNeasy spin plant RNA extraction kit according to the manufacturer's instructions. RNA was sent to the Australian Genome Research Facility (Melbourne, Australia) for Illumina (San Diego, CA, USA) TruSeq stranded mRNA library construction and sequencing using two lanes of a HiSeq2000 with 125‐bp paired‐end sequencing. Four biological replicates were used per treatment. All 12 libraries were sequenced in both lanes as technical replicates to eliminate lane effects, and output files for these technical replicates were combined during the analysis.
SolexaQA++ was used to trim reads to a minimum length of 50 nucleotides and trimmed to the longest contiguous segments with quality scores above Phred 30 (Cox et al., 2010). The Cufflinks package version 2.2.1 was used for RNAseq analysis (Trapnell et al., 2012). Trimmed reads were aligned using TopHat 2.1.0 (Kim et al., 2013) to the B. distachyon genome v3.0 downloaded from phytozome. Alignment was guided by a genome feature file (v3.1) and default parameters were used. Transcripts were assembled using Cufflinks for individual samples and then merged into a final transcriptome assembly using cuffmerge, both run with default parameters. Differential expression was identified using cuffdiff run with default parameters.
Gene ontology (GO) analysis was performed at AgriGO (Du et al., 2010). Although an annotation of Brachypodium is available, the Arabidopsis GO term annotation is considerably richer in the AgriGO database. Therefore, orthologues of the Brachypodium primary protein set of annotation version 3.1 were identified in the Arabidopsis (TAIR10) primary protein set. blastP version 2.2.25 was run on cluster computing resources available at the Commonwealth Scientific and Industrial Research Organization (CSIRO) with default blastP parameters and output limited to the best hit in tabular format. Reciprocal best hits were extracted using custom perl scripts.
Pathway analysis was conducted at Gramene (http://pathway.gramene.org/) in the BrachyCyc version 2 database (Tello‐Ruiz et al., 2016). The enzyme database used at BrachyCyc was downloaded and the corresponding entries in this database to that in Brachypodium annotation version 3.1 were identified using reciprocal best blast hits. Locus ids and their corresponding expression values for genes that were significantly up‐ or down‐regulated by fusatinic acid treatment were uploaded to the BrachyCyc query form and the resulting metabolic pathways were inspected manually.
Bacterial assay for cytokinin receptor activation
Arabidopsis histidine kinase 3 (AHK3) activation was assayed using a bacterial‐based assay employing β‐galactosidase activity as a read out of activity, as described previously (Mizuno and Yamashino, 2010), with the minor modifications as follows. The Arabidopsis histidine kinase 3 receptor was used in pSTV28 in E. coli KMI001; 400 μL of an overnight culture, which had been grown at room temperature with 10 μm of the test compounds (dissolved in DMSO with the final DMSO concentration in the culture of 0.1%), were pelleted by centrifugation and resuspended in 700 μL of Z‐buffer; 200 µL of this suspension were transferred to a 96‐well culture plate for measurement of the optical density at 600 nm (OD600) using a Perkin‐Elmer (Melbourne, Victoria, Australia) Envision multimode plate reader fitted with monochromators. To the remaining 500 μL, 10 μL of toluene were added and the mixture was incubated at 37 °C for 60 min; 100 μL of 4 mg/mL ortho‐Nitrophenyl‐β‐galactoside (ONPG) in Z‐buffer were added to the tubes and allowed to incubate at 28 °C for 50 min, after which 900 μL of 1 m NaCO3 were added to stop the reaction. This was centrifuged for 2 min at 20 000 g in a microcentrifuge to pellet the cellular debris; 200 μL of the supernatant were transferred to a 96‐well plate and the optical density at 420 nm (OD420) of this cleared reaction was measured. A simplified equation for the calculation of β‐galactosidase activity was also used, removing the approximation of light scatter caused by cellular debris as a cleared reaction was employed. The equation was: 1000 × OD420/(time × volume × OD600).
Accessions
The raw reads for the RNAseq experiment assaying the Brachypodium response to cytokinins can be found under BioProject PRJNA325847, SRA accession SRP076768. Fusarium pseudograminearum gene expression was quantified from RNAseq data that can be found under BioProject PRJNA326033, SRA accession SRP076777.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Generation and verification of mutant strains of Fusarium pseudograminearum. The design of the targeting vectors and the polymerase chain reaction (PCR) screening assays are shown for all genes. Central to the image is the entire Fusarium cytokinin biosynthetic gene cluster and two flanking genes labelled as ‘wild‐type locus’. Below or above this, for each of the four genes in the cluster, schematics of the synthesized targeting vectors are shown. These consist of upstream and downstream homologous flanking sequences of each gene, with the regions of homology to the wild‐type locus indicated by grey shading. The black box and arrow in each targeting vector represent the TrpC promoter‐driven nourseothricin acetyl transferase used to select transformants. Transformants were screened with a triplex PCR assay in which a vector‐specific and/or parental gene‐specific product can be generated depending on the genotype of the transformant. In each case, the vector‐specific products (depicted as blue lines) were designed to be larger than the parental gene‐specific products (red lines). Agarose gels are shown of the products of the triplex PCR assays for each gene. Sample nomenclatures are unique identifiers given to each transformant. In the case of the FCK1, FCK2 and FCK4 transformation screens, mutants at one of the other genes in the cluster were used as controls. The amplification of the wild‐type band in these strains demonstrates that, for these strains, deletion of the target gene does not impact other genes in the cluster. FCK1 mutants were generated at both Aalborg University and the Commonwealth Scientific and Industrial Research Organization (CSIRO). A single mutant (AAU2.2) was identified in transformations conducted at Aalborg University with other transformants with ectopic integrations of the targeting cassette (lower gel), and three mutants were identified in transformations conducted at CSIRO with only these selected mutants included in the screen depicted in the upper gel for FCK1. For AAU2.2, the flanking genes FCK2 and FCK4 showed amplification identical to the CS3096 wild‐type strain (data not shown).
Fig. S2 Quantification of aurofusarin, fusatin, 8‐oxo‐fusarin, fusatinic acid and 8‐oxo‐fusatinic acid using the protonated ion traces [M + H]+ for each compound. The columns represent the mean values of three replicates with the standard error of the mean error bars. A single mutant in each gene was selected for this analysis, but the loss of production of Fusarium cytokinins in independent mutants of FCK1 and FCK2, and enhanced production in FPSE_20002 and FPSE_06371 mutants, were consistently observed.
Fig. S3 Expression of the four genes of the Fusarium cytokinin cluster in Fusarium pseudograminearum during infection of barley and Brachypodium in comparison with the trichodiene synthase encoding gene (TRI5). FPKM, fragments per kilobase of transcript per million mapped reads. FCK4‐derived RNA sequencing (RNAseq) reads were observed in Brachypodium‐derived samples, but their abundance was lower than that required to calculate an FPKM value. Values represent the average of four biological replicates with error bars representing the standard error of the mean; 0.2% of all reads were of fungal origin from barley samples. In the Brachypodium set, this was 2.2%–3.6%.
Fig. S4 Phylogenetic analysis of cytochrome P450 monooxygenases encoded in fungal cytokinin production clusters. White, grey and black hexagons illustrate to which clade the neighbouring isopentenyltransferase‐Lonely Guy (IPT‐LOG) fusion enzyme belongs (Fig. 2A).
Fig. S5 Active site modelling of the cytochrome P450 monooxygenase FCK2. (A) Of the tested compounds [trans‐zeatin (tZ), cis‐zeatin, fusatin, isopentenyladenine and fusatinic acid], docking analyses suggests that tZ is the most likely product of this enzyme. Outputs of the docking are represented as relative binding strengths (dissociation constant). The model cannot exclude the possibility that other untested compounds are better fits for the active site, nor does it take into account the possibility of other conformational states. (B) Ribbon structural representation of the entire FCK2 protein. (C) Active site of FCK2 with tZ bound.
Fig. S6 Chromatograms of Fusarium graminearum wild‐type (PH‐1) and transformed with the cytokinin cluster (PH‐1::FCK). (A) UV (280 nm) chromatogram. (B) Extracted chromatograms ([M–H]–) for 1, 2, 3 and 8‐oxo‐trans‐zeatin.
Fig. S7 Illustration of the procedure for heterologous expression of the cytokinin cluster in Fusarium graminearum. Three overlapping polymerase chain reaction (PCR) fragments were cloned into the Y‐GOTL vector and transferred into the tubulin locus of F. graminearum (Josefsen et al., 2012).
Data S1 Structural elucidation of fusatin, 8‐oxo‐fusatin and fusatinic acid.
Data S2 Gene ontology analysis of genes differentially regulated by fusatinic acid in Brachypodium.
Data S3 Amino acid sequences used in the construction of the isopentenyltransferase (IPT) phylogeny.
Table S1 Primers for the screening of transformants for successful gene deletion and for heterologous expression of the cytokinin cluster in Fusarium graminearum.
Methods S1 Supplementary methods.
Acknowledgements
This work was partially supported by a CSIRO Office of the Chief Executive Julius Career Award to DMG. The authors wish to thank Kemal Kazan (CSIRO) for useful discussions and Chunji Liu and Ahsan Habib (CSIRO) for providing the F. pseudograminearum‐infected barley RNAseq dataset. We thank Wolfgang Bermel and Bruker Biospin for providing 15N‐HMBC spectra. The NMR laboratory at Aalborg University is supported by the Obel, SparNord and Carlsberg Foundations. DTU (Technical University of Denmark) is grateful to Agilent Technologies for the Thought Leader Donation of the 1290 UHPLC‐6550 quadrupole time‐of‐flight (QTOF) instrument.
References
- Adam, G. , Spörhase, P. , Bartholomäus, A. , Svoboda, T. , Wiesenberger, G. , Güldener, U. Schmeitzl, C. , Michlmayr, H. , Fruhmann, P. , Parich, A. , Kluger, B. , Mewes, H. , Krska, R. , Schuhmacher, R. , Beltran Iturat, E. and Berthiller, F. (2015) Fusarium graminearum is able to synthesize auxin and to inactivate the ethylene precursor 1‐aminocyclopropane‐1‐carboxylic acid (ACC). In: Fungal Genetics Conference, Monterey, CA, USA Available at http://www.genetics-gsa.org/fungal/2015/pdf/Fungal%20Abstract%20Book%202015.pdf
- Akiyoshi, D.E. , Klee, H. , Amasino, R.M. , Nester, E.W. and Gordon, M.P. (1984) T‐DNA of Agrobacterium tumefaciens encodes an enzyme of cytokinin biosynthesis. Proc. Natl. Acad. Sci. USA, 81, 5994–5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht, T. and Argueso, C.T. (2016) Should I fight or should I grow now? The role of cytokinins in plant growth and immunity and in the growth–defence trade‐off. Ann. Bot. 119, 725–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso, C.T. , Ferreira, F.J. , Epple, P. , To, J.P.C. , Hutchison, C.E. , Schaller, G.E. , Dangl, J.L. and Kieber, J.J. (2012) Two‐component elements mediate interactions between cytokinin and salicylic acid in plant immunity. PLOS Genet. 8, e1002448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry, G.F. , Rogers, S.G. , Fraley, R.T. and Brand, L. (1984) Identification of a cloned cytokinin biosynthetic gene. Proc. Natl. Acad. Sci. USA, 81, 4776–4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassil, N.V. , Mok, D.W.S. and Mok, M.C. (1993) Partial purification of a cis–trans‐isomerase of zeatin from immature seed of Phaseolus vulgaris L. Plant Physiol. 102, 867–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behr, M. , Motyka, V. , Weihmann, F. , Malbeck, J. , Deising, H.B. and Wirsel, S.G.R. (2012) Remodeling of cytokinin metabolism at infection sites of Colletotrichum graminicola on maize leaves. Mol. Plant–Microbe Interact. 25, 1073–1082. [DOI] [PubMed] [Google Scholar]
- Bentley, A.R. , Summerell, B.A. and Burgess, L.W. (2008) Sexual compatibility in Fusarium pseudograminearum (Gibberella coronicola). Mycol. Res. 112, 1101–1106. [DOI] [PubMed] [Google Scholar]
- Boc, A. , Diallo, A.B. and Makarenkov, V. (2012) T‐REX: a web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 40, W573–W579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonicel, A. , Mariaggi, N. , Hughes, E. and Teoule, R. (1980) In vitro γ irradiation of DNA: identification of radioinduced chemical modifications of the adenine moiety. Radiat. Res. 83, 19–26. [PubMed] [Google Scholar]
- Bordoli, L. , Kiefer, F. , Arnold, K. , Benkert, P. , Battey, J. and Schwede, T. (2008) Protein structure homology modeling using SWISS‐MODEL workspace. Nat. Protoc. 4, 1–13. [DOI] [PubMed] [Google Scholar]
- Chanclud, E. , Kisiala, A. , Emery, N.R.J. , Chalvon, V. , Ducasse, A. , Romiti‐Michel, C. , Gravot, A. , Kroj, T. and Morel, J.B. (2016) Cytokinin production by the rice blast fungus is a pivotal requirement for full virulence. PLoS Pathog. 12, e1005457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Z. , Liu, Y. , Dong, H. , Teng, K. , Han, L. and Zhang, X. (2016) Effects of cytokinin and nitrogen on drought tolerance of creeping bentgrass. PLoS One, 11, e0154005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, B. and Yang, H. (2013) 6‐Benzylaminopurine alleviates chilling injury of postharvest cucumber fruit through modulating antioxidant system and energy status. J. Sci. Food Agric. 93, 1915–1921. [DOI] [PubMed] [Google Scholar]
- Chen, C.‐M. and Kristopeit, S.M. (1981) Metabolism of cytokinin: deribosylation of cytokinin ribonucleoside by adenosine nucleosidase from wheat germ cells. Plant Physiol. 68, 1020–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, J. , Huh, S.U. , Kojima, M. , Sakakibara, H. , Paek, K.‐H. and Hwang, I. (2010) The cytokinin‐activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1‐dependent salicylic acid signaling in Arabidopsis . Dev. Cell, 19, 284–295. [DOI] [PubMed] [Google Scholar]
- Choi, J. , Choi, D. , Lee, S. , Ryu, C.M. and Hwang, I. (2011) Cytokinins and plant immunity: old foes or new friends? Trends Plant Sci. 16, 388–394. [DOI] [PubMed] [Google Scholar]
- Cox, M. , Peterson, D. and Biggs, P. (2010) SolexaQA: at‐a‐glance quality assessment of Illumina second‐generation sequencing data. BMC Bioinformatics, 11, 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creelman, R.A. and Mullet, J.E. (1997) Oligosaccharins, brassinolides, and jasmonates: nontraditional regulators of plant growth, development, and gene expression. Plant Cell, 9, 1211–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aloia, M. , Bonhomme, D. , Bouché, F. , Tamseddak, K. , Ormenese, S. , Torti, S. , Coupland, G. and Périlleux, C. (2011) Cytokinin promotes flowering of Arabidopsis via transcriptional activation of the FT paralogue TSF. Plant J. 65, 972–979. [DOI] [PubMed] [Google Scholar]
- De Vos, M. , Van Oosten, V.R. , Van Poecke, R.M.P. , Van Pelt, J.A. , Pozo, M.J. , Mueller, M.J. , Buchala, A.J. , Métraux, J.P. , Van Loon, L.C. , Dicke, M. and Pieterse, C.M. (2005) Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol. Plant–Microbe Interact. 18, 923–937. [DOI] [PubMed] [Google Scholar]
- Desmond, O.J. , Manners, J.M. , Schenk, P.M. , Maclean, D.J. and Kazan, K. (2008) Gene expression analysis of the wheat response to infection by Fusarium pseudograminearum . Physiol. Mol. Plant Pathol. 73, 40–47. [Google Scholar]
- Droce, A. , Saei, W. , Jørgensen, S. , Wimmer, R. , Giese, H. , Wollenberg, R. , Sondergaard, T.E. and Sørensen, J.L. (2016) Functional analysis of the fusarielin biosynthetic gene cluster. Molecules, 21, 1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, Z. , Zhou, X. , Ling, Y. , Zhang, Z. and Su, Z. (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38, W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman‐Wykert, A.K. and Miller, J.F. (2003) Hypervirulence and pathogen fitness. Trends Microbiol. 11, 105–108. [DOI] [PubMed] [Google Scholar]
- Gardiner, D.M. , Jarvis, R.S. and Howlett, B.J. (2005) The ABC transporter gene in the sirodesmin biosynthetic gene cluster of Leptosphaeria maculans is not essential for sirodesmin production but facilitates self‐protection. Fungal Genet. Biol. 42, 257–263. [DOI] [PubMed] [Google Scholar]
- Gardiner, D.M. , McDonald, M.C. , Covarelli, L. , Solomon, P.S. , Rusu, A.G. , Marshall, M. , Kazan, K. , Chakraborty, S. , McDonald, B.A. and Manners, J.M. (2012) Comparative pathogenomics reveals horizontally acquired novel virulence genes in fungi infecting cereal hosts. PLoS Pathog. 8, e1002952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner, D.M. , Benfield, A.H. , Stiller, J. , Stephen, S. , Aitken, K. , Liu, C. and Kazan, K. (2017) A high resolution genetic map of the cereal crown rot pathogen Fusarium pseudograminearum provides a near complete genome assembly. Mol. Plant Pathol. in press. doi:10.1111/mpp.12519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R.D. and Schiestl, R.H. (2007) High‐efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34. [DOI] [PubMed] [Google Scholar]
- Gray, J. , Gelvin, S.B. , Meilan, R. and Morris, R.O. (1996) Transfer RNA is the source of extracellular isopentenyladenine in a Ti‐plasmidless strain of Agrobacterium tumefaciens . Plant Physiol. 110, 431–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider, N. and Robien, W. (2016) CSEARCH Robot Referee. http://nmrpredict.orc.univie.ac.at/c13robot/robot.php
- Haidoune, M. , Mornet, R. and Laloue, M. (1990) Synthesis of 6‐(3‐methylpyrrol‐1‐yl)‐9‐β‐D‐ribofuranosyl purine, a novel metabolite of zeatin riboside. Tetrahedron Lett. 31, 1419–1422. [Google Scholar]
- Hann, D.R. , Domínguez‐Ferreras, A. , Motyka, V. , Dobrev, P.I. , Schornack, S. , Jehle, A. , Felix, G. , Chinchilla, D. , Rathjen, J.P. and Boller, T. (2014) The Pseudomonas type III effector HopQ1 activates cytokinin signaling and interferes with plant innate immunity. New Phytol. 201, 585–598. [DOI] [PubMed] [Google Scholar]
- Helaers, R. and Milinkovitch, M. (2010) MetaPIGA v2.0: maximum likelihood large phylogeny estimation using the metapopulation genetic algorithm and other stochastic heuristics. BMC Bioinformatics, 11, 379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinsch, J. , Vrabka, J. , Oeser, B. , Novák, O. , Galuszka, P. and Tudzynski, P. (2015) De novo biosynthesis of cytokinins in the biotrophic fungus Claviceps purpurea . Environ. Microbiol. 17, 2935–2951. [DOI] [PubMed] [Google Scholar]
- Hinsch, J. , Galuszka, P. and Tudzynski, P. (2016) Functional characterization of the first filamentous fungal tRNA‐isopentenyltransferase and its role in the virulence of Claviceps purpurea . New Phytol. 211, 980–992. [DOI] [PubMed] [Google Scholar]
- Inoue, T. , Higuchi, M. , Hashimoto, Y. , Seki, M. , Kobayashi, M. , Kato, T. , Tabata, S. , Shinozaki, K. and Kakimoto, T. (2001) Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature, 409, 1060–1063. [DOI] [PubMed] [Google Scholar]
- Jameson, P.E. (2000) Cytokinins and auxins in plant–pathogen interactions – an overview. Plant Growth Regul. 32, 369–380. [Google Scholar]
- Jiang, C.‐J. , Shimono, M. , Sugano, S. , Kojima, M. , Liu, X. , Inoue, H. , Sakakibara, H. and Takatsuji, H. (2013) Cytokinins act synergistically with salicylic acid to activate defense gene expression in rice. Mol. Plant–Microbe Interact. 26, 287–296. [DOI] [PubMed] [Google Scholar]
- Josefsen, L. , Droce, A. , Sondergaard, T.E. , Sorensen, J.L. , Bormann, J. , Schafer, W. , Giese, H. and Olsson, S. (2012) Autophagy provides nutrients for nonassimilating fungal structures and is necessary for plant colonization but not for infection in the necrotrophic plant pathogen Fusarium graminearum . Autophagy, 8, 326–337. [DOI] [PubMed] [Google Scholar]
- Kakei, Y. , Mochida, K. , Sakurai, T. , Yoshida, T. , Shinozaki, K. and Shimada, Y. (2015) Transcriptome analysis of hormone‐induced gene expression in Brachypodium distachyon . Sci. Rep. 5, 14 476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakimoto, T. (2001) Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate:ATP/ADP isopentenyltransferases. Plant Cell Physiol. 42, 677–685. [DOI] [PubMed] [Google Scholar]
- Kende, H. and Zeevaart, J.A.D. (1997) The five ''classical'' plant hormones. Plant Cell, 9, 1197–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettle, A.J. , Carere, J. , Batley, J. , Benfield, A.H. , Manners, J.M. , Kazan, K. and Gardiner, D.M. (2015) A γ‐lactamase from cereal infecting Fusarium spp. catalyses the first step in the degradation of the benzoxazolinone class of phytoalexins. Fungal Genet. Biol. 83, 1–9. [DOI] [PubMed] [Google Scholar]
- Kim, D. , Pertea, G. , Trapnell, C. , Pimentel, H. , Kelley, R. and Salzberg, S.L. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, N.L. and Sutherland, M.W. (2016) Histopathological assessment of Fusarium pseudograminearum colonization of cereal culms during crown rot infections. Plant Dis. 100, 252–259. [DOI] [PubMed] [Google Scholar]
- Koenig, R.L. , Morris, R.O. and Polacco, J.C. (2002) tRNA is the source of low‐level trans‐zeatin production in Methylobacterium spp. J. Bacteriol. 184, 1832–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger, E. and Vriend, G. (2014) YASARA View—molecular graphics for all devices—from smartphones to workstations. Bioinformatics, 30, 2981–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger, E. , Darden, T. , Nabuurs, S.B. , Finkelstein, A. and Vriend, G. (2004) Making optimal use of empirical energy functions: force‐field parameterization in crystal space. Proteins, 57, 678–683. [DOI] [PubMed] [Google Scholar]
- Kurakawa, T. , Ueda, N. , Maekawa, M. , Kobayashi, K. , Kojima, M. , Nagato, Y. , Sakakibara, H. and Kyozuka, J. (2007) Direct control of shoot meristem activity by a cytokinin‐activating enzyme. Nature, 445, 652–655. [DOI] [PubMed] [Google Scholar]
- Lanubile, A. , Ferrarini, A. , Maschietto, V. , Delledonne, M. , Marocco, A. and Bellin, D. (2014) Functional genomic analysis of constitutive and inducible defense responses to Fusarium verticillioides infection in maize genotypes with contrasting ear rot resistance. BMC Genomics, 15, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L.J. , van der Does, H.C. , Borkovich, K.A. , Coleman, J.J. , Daboussi, M.J. , Di Pietro, A. , Dufresne, M. , Freitag, M. , Grabherr, M. , Henrissat, B. and Houterman, P.M. (2010) Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium . Nature, 464, 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malz, S. , Grell, M.N. , Thrane, C. , Maier, F.J. , Rosager, P. , Felk, A. , Albertsen, K.S. , Salomon, S. , Bohn, L. , Schäfer, W. and Giese, H (2005) Identification of a gene cluster responsible for the biosynthesis of aurofusarin in the Fusarium graminearum species complex. Fungal Genet. Biol. 42, 420–433. [DOI] [PubMed] [Google Scholar]
- Michniewicz, M. , Rozej, B. and Bobkiewicz, W. (1986) The production of growth regulators by Fusarium culmorum (W. G. Sm.) Sacc. as related to the age of mycelium. Acta Physiol. Plant. 8, 85–91. [Google Scholar]
- Miwa, K. , Ishikawa, K. , Terada, K. , Yamada, H. , Suzuki, T. , Yamashino, T. and Mizuno, T. (2007) Identification of amino acid substitutions that render the Arabidopsis cytokinin receptor histidine kinase AHK4 constitutively active. Plant Cell Physiol. 48, 1809–1814. [DOI] [PubMed] [Google Scholar]
- Miyawaki, K. , Tarkowski, P. , Matsumoto‐Kitano, M. , Kato, T. , Sato, S. , Tarkowska, D. , Tabata, S. , Sandberg, G. and Kakimoto, T. (2006) Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl. Acad. Sci. USA, 103, 16 598–16 603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, T. and Yamashino, T. (2010) Biochemical characterization of plant hormone cytokinin‐receptor histidine kinases using microorganisms In: Methods in Enzymology (Melvin I. S., Brian R. C. and Alexandrine C., eds.), pp. 335–356. Academic Press. San Diego, CA, USA. [DOI] [PubMed] [Google Scholar]
- Mudge, A.M. , Dill‐Macky, R. , Dong, Y. , Gardiner, D.M. , White, R.G. and Manners, J.M. (2006) A role for the mycotoxin deoxynivalenol in stem colonisation during crown rot disease of wheat caused by Fusarium graminearum and Fusarium pseudograminearum . Physiol. Mol. Plant Pathol. 69, 73–85. [Google Scholar]
- Niehaus, E.‐M. , Münsterkötter, M. , Proctor, R.H. , Brown, D.W. , Sharon, A. , Idan, Y. , Oren‐Young, L. , Sieber, C.M. , Novák, O. , Pěnčík, A. and Tarkowská, D. (2016) Comparative “Omics” of the Fusarium fujikuroi species complex highlights differences in genetic potential and metabolite synthesis. Genome Biol. Evol. 8, 3574–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak, J. , Pavlu, J. , Novak, O. , Nozkova‐Hlavackova, V. , Spundova, M. , Hlavinka, J. , Koukalová, Š. , Skalák, J. , Černý, M. and Brzobohatý, B. (2013) High cytokinin levels induce a hypersensitive‐like response in tobacco. Ann. Bot. 112, 41–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obanor, F. , Neate, S. , Simpfendorfer, S. , Sabburg, R. , Wilson, P. and Chakraborty, S. (2013) Fusarium graminearum and Fusarium pseudograminearum caused the 2010 head blight epidemics in Australia. Plant Pathol. 62, 79–91. [Google Scholar]
- Onaka, H. , Asamizu, S. , Igarashi, Y. , Yoshida, R. and Furumai, T. (2005) Cytochrome P450 homolog is responsible for C–N bond formation between aglycone and deoxysugar in the staurosporine biosynthesis of Streptomyces sp. TP‐A0274. Biosci. Biotechnol. Biochem. 69, 1753–1759. [DOI] [PubMed] [Google Scholar]
- Othman, E.M. , Naseem, M. , Awad, E. , Dandekar, T. and Stopper, H. (2016) The plant hormone cytokinin confers protection against oxidative stress in mammalian cells. PLoS One, 11, e0168386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S.‐W. , Kaimoyo, E. , Kumar, D. , Mosher, S. and Klessig, D.F. (2007) Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science, 318, 113–116. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M.J. , Leon‐Reyes, A. , Van der Ent, S. and Van Wees, S.C.M. (2009) Networking by small‐molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316. [DOI] [PubMed] [Google Scholar]
- Preston, G.M. (2017) Profiling the extended phenotype of plant pathogens. Mol. Plant Pathol. 18, 443–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera, M. , Peleg, Z. , Abdel‐Tawab, Y.M. , Tumimbang, E.B. , Delatorre, C.A. and Blumwald, E. (2013) Stress‐induced cytokinin synthesis increases drought tolerance through the coordinated regulation of carbon and nitrogen assimilation in rice. Plant Physiol. 163, 1609–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert‐Seilaniantz, A. , Grant, M. and Jones, J.D.G. (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate–salicylate antagonism. Annu. Rev. Phytopathol. 49, 317–343. [DOI] [PubMed] [Google Scholar]
- Sakakibara, H. (2006) Cytokinins: activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 57, 431–449. [DOI] [PubMed] [Google Scholar]
- Scherm, B. , Orrù, M. , Balmas, V. , Spanu, F. , Azara, E. , Delogu, G. , Hammond, T.M. , Keller, N.P. and Migheli, Q. (2011) Altered trichothecene biosynthesis in TRI6‐silenced transformants of Fusarium culmorum influences the severity of crown and foot rot on durum wheat seedlings. Mol. Plant Pathol. 12, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, C.A. , Rasband, W.S. and Eliceiri, K.W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique, S. , Radakovic, Z.S. , De La Torre, C.M. , Chronis, D. , Novák, O. , Ramireddy, E. , Holbein, J. , Matera, C. , Hütten, M. , Gutbrod, P. and Anjam, M.S. (2015) A parasitic nematode releases cytokinin that controls cell division and orchestrates feeding site formation in host plants. Proc. Natl. Acad. Sci. USA, 112, 12 669–12 674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemens, J. , Keller, I. , Sarx, J. , Kunz, S. , Schuller, A. , Nagel, W. , Schmülling, T. , Parniske, M. and Ludwig‐Müller, J. (2006) Transcriptome analysis of Arabidopsis clubroots indicates a key role for cytokinins in disease development. Mol. Plant–Microbe Interact. 19, 480–494. [DOI] [PubMed] [Google Scholar]
- Smedsgaard, J. (1997) Micro‐scale extraction procedure for standardized screening of fungal metabolite production in cultures. J. Chromatogr. A, 760, 264–270. [DOI] [PubMed] [Google Scholar]
- Sørensen, J.L. and Sondergaard, T.E. (2014) The effects of different yeast extracts on secondary metabolite production in Fusarium . Int. J. Food Microbiol. 170, 55–60. [DOI] [PubMed] [Google Scholar]
- Sørensen, J.L. , Sondergaard, T.E. , Covarelli, L. , Fuertes, P.R. , Hansen, F.T. , Frandsen, R.J.N. , Saei, W. , Lukassen, M.B. , Wimmer, R. , Nielsen, K.F. and Gardiner, D.M. (2014) Identification of the biosynthetic gene clusters for the lipopeptides Fusaristatin A and W493 B in Fusarium graminearum and F. pseudograminearum . J. Nat. Prod. 77, 2619–2625. [DOI] [PubMed] [Google Scholar]
- Spichal, L. , Popa, I. , Voller, J. , Dolezal, K. , Strnad, M. , Werner, T. and Schmulling, T. (2014) Substituted 6‐(alkylbenzylamino)purine derivatives for use as cytokinin receptor antagonists and preparations containing these derivatives. Google Patents. EP2203451 Prague, Czech Republic.
- Spíchal, L. , Rakova, N.Y. , Riefler, M. , Mizuno, T. , Romanov, G.A. , Strnad, M. and Schmülling, T. (2004) Two cytokinin receptors of Arabidopsis thaliana, CRE1/AHK4 and AHK3, differ in their ligand specificity in a bacterial assay. Plant Cell Physiol. 45, 1299–1305. [DOI] [PubMed] [Google Scholar]
- Stephens, A.E. , Gardiner, D.M. , White, R.G. , Munn, A.L. and Manners, J.M. (2008) Phases of infection and gene expression of Fusarium graminearum during crown rot disease of wheat. Mol. Plant–Microbe Interact. 21, 1571–1581. [DOI] [PubMed] [Google Scholar]
- Strnad, M. (1997) The aromatic cytokinins. Physiol. Plant. 101, 674–688. [Google Scholar]
- Suzuki, T. , Miwa, K. , Ishikawa, K. , Yamada, H. , Aiba, H. and Mizuno, T. (2001) The Arabidopsis sensor His‐kinase, AHK4, can respond to cytokinins. Plant Cell Physiol. 42, 107–113. [DOI] [PubMed] [Google Scholar]
- Takei, K. , Sakakibara, H. and Sugiyama, T. (2001) Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana . J. Biol. Chem. 276, 26 405–26 410. [DOI] [PubMed] [Google Scholar]
- Tarkowska, D. , Dolezal, K. , Tarkowski, P. , Astot, C. , Holub, J. , Fuksova, K. , Schmülling, T. , Sandberg, G. and Strnad, M. (2003) Identification of new aromatic cytokinins in Arabidopsis thaliana and Populus x canadensis leaves by LC‐(+)ESI‐MS and capillary liquid chromatography frit‐fast atom bombardment mass spectrometry. Physiol. Plant. 117, 579–590. [DOI] [PubMed] [Google Scholar]
- Taya, Y. , Tanaka, Y. and Nishimura, S. (1978) 5′‐AMP is a direct precursor of cytokinin in Dictyostelium discoideum . Nature, 271, 545–547. [DOI] [PubMed] [Google Scholar]
- Taylor, J.L. , Zaharia, L.I. , Chen, H. , Anderson, E. and Abrams, S.R. (2006) Biotransformation of adenine and cytokinins by the rhizobacterium Serratia proteamaculans . Phytochemistry, 67, 1887–1894. [DOI] [PubMed] [Google Scholar]
- Tello‐Ruiz, M.K. , Stein, J. , Wei, S. , Preece, J. , Olson, A. , Naithani, S. , Amarasinghe, V. , Dharmawardhana, P. , Jiao, Y. , Mulvaney, J. and Kumari, S. (2016) Gramene 2016: comparative plant genomics and pathway resources. Nucleic Acids Res. 44, D1133–D1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To, J.P.C. and Kieber, J.J. (2008) Cytokinin signaling: two‐components and more. Trends Plant Sci. 13, 85–92. [DOI] [PubMed] [Google Scholar]
- Trapnell, C. , Roberts, A. , Goff, L. , Pertea, G. , Kim, D. , Kelley, D.R. , Pimentel, H. , Salzberg, S.L. , Rinn, J.L. and Pachter, L. (2012) Differential gene and transcript expression analysis of RNA‐seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trdá, L. , Barešová, M. , Šašek, V. , Nováková, M. , Zahajská, L. , Dobrev, P.I. , Motyka, V. and Burketová, L. (2017) Cytokinin metabolism of pathogenic fungus Leptosphaeria maculans involves isopentenyltransferase, adenosine kinase and cytokinin oxidase/dehydrogenase. Front Microbiol. 8, 1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott, O. and Olson, A.J. (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanstaden, J. and Nicholson, R.I.D. (1989) Cytokinins and mango flower malformation. II: the cytokinin complement produced by Fusarium moniliforme and the ability of the fungus to incorporate [8‐14C]adenine into cytokinins. Physiol. Mol. Plant Pathol. 35, 423–431. [Google Scholar]
- Wiemann, P. , Sieber, C.M.K. , von Bargen, K.W. , Studt, L. , Niehaus, E.‐M. , Espino, J.J. , Huß, K. , Michielse, C.B. , Albermann, S. , Wagner, D. and Bergner, S.V. (2013) Deciphering the cryptic genome: genome‐wide analyses of the rice pathogen Fusarium fujikuroi reveal complex regulation of secondary metabolism and novel metabolites. PLoS Pathog. 9, e1003475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Gao, S. , Lercher, M.J. , Hu, S. and Chen, W.‐H. (2012) EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 40, W569–W572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Generation and verification of mutant strains of Fusarium pseudograminearum. The design of the targeting vectors and the polymerase chain reaction (PCR) screening assays are shown for all genes. Central to the image is the entire Fusarium cytokinin biosynthetic gene cluster and two flanking genes labelled as ‘wild‐type locus’. Below or above this, for each of the four genes in the cluster, schematics of the synthesized targeting vectors are shown. These consist of upstream and downstream homologous flanking sequences of each gene, with the regions of homology to the wild‐type locus indicated by grey shading. The black box and arrow in each targeting vector represent the TrpC promoter‐driven nourseothricin acetyl transferase used to select transformants. Transformants were screened with a triplex PCR assay in which a vector‐specific and/or parental gene‐specific product can be generated depending on the genotype of the transformant. In each case, the vector‐specific products (depicted as blue lines) were designed to be larger than the parental gene‐specific products (red lines). Agarose gels are shown of the products of the triplex PCR assays for each gene. Sample nomenclatures are unique identifiers given to each transformant. In the case of the FCK1, FCK2 and FCK4 transformation screens, mutants at one of the other genes in the cluster were used as controls. The amplification of the wild‐type band in these strains demonstrates that, for these strains, deletion of the target gene does not impact other genes in the cluster. FCK1 mutants were generated at both Aalborg University and the Commonwealth Scientific and Industrial Research Organization (CSIRO). A single mutant (AAU2.2) was identified in transformations conducted at Aalborg University with other transformants with ectopic integrations of the targeting cassette (lower gel), and three mutants were identified in transformations conducted at CSIRO with only these selected mutants included in the screen depicted in the upper gel for FCK1. For AAU2.2, the flanking genes FCK2 and FCK4 showed amplification identical to the CS3096 wild‐type strain (data not shown).
Fig. S2 Quantification of aurofusarin, fusatin, 8‐oxo‐fusarin, fusatinic acid and 8‐oxo‐fusatinic acid using the protonated ion traces [M + H]+ for each compound. The columns represent the mean values of three replicates with the standard error of the mean error bars. A single mutant in each gene was selected for this analysis, but the loss of production of Fusarium cytokinins in independent mutants of FCK1 and FCK2, and enhanced production in FPSE_20002 and FPSE_06371 mutants, were consistently observed.
Fig. S3 Expression of the four genes of the Fusarium cytokinin cluster in Fusarium pseudograminearum during infection of barley and Brachypodium in comparison with the trichodiene synthase encoding gene (TRI5). FPKM, fragments per kilobase of transcript per million mapped reads. FCK4‐derived RNA sequencing (RNAseq) reads were observed in Brachypodium‐derived samples, but their abundance was lower than that required to calculate an FPKM value. Values represent the average of four biological replicates with error bars representing the standard error of the mean; 0.2% of all reads were of fungal origin from barley samples. In the Brachypodium set, this was 2.2%–3.6%.
Fig. S4 Phylogenetic analysis of cytochrome P450 monooxygenases encoded in fungal cytokinin production clusters. White, grey and black hexagons illustrate to which clade the neighbouring isopentenyltransferase‐Lonely Guy (IPT‐LOG) fusion enzyme belongs (Fig. 2A).
Fig. S5 Active site modelling of the cytochrome P450 monooxygenase FCK2. (A) Of the tested compounds [trans‐zeatin (tZ), cis‐zeatin, fusatin, isopentenyladenine and fusatinic acid], docking analyses suggests that tZ is the most likely product of this enzyme. Outputs of the docking are represented as relative binding strengths (dissociation constant). The model cannot exclude the possibility that other untested compounds are better fits for the active site, nor does it take into account the possibility of other conformational states. (B) Ribbon structural representation of the entire FCK2 protein. (C) Active site of FCK2 with tZ bound.
Fig. S6 Chromatograms of Fusarium graminearum wild‐type (PH‐1) and transformed with the cytokinin cluster (PH‐1::FCK). (A) UV (280 nm) chromatogram. (B) Extracted chromatograms ([M–H]–) for 1, 2, 3 and 8‐oxo‐trans‐zeatin.
Fig. S7 Illustration of the procedure for heterologous expression of the cytokinin cluster in Fusarium graminearum. Three overlapping polymerase chain reaction (PCR) fragments were cloned into the Y‐GOTL vector and transferred into the tubulin locus of F. graminearum (Josefsen et al., 2012).
Data S1 Structural elucidation of fusatin, 8‐oxo‐fusatin and fusatinic acid.
Data S2 Gene ontology analysis of genes differentially regulated by fusatinic acid in Brachypodium.
Data S3 Amino acid sequences used in the construction of the isopentenyltransferase (IPT) phylogeny.
Table S1 Primers for the screening of transformants for successful gene deletion and for heterologous expression of the cytokinin cluster in Fusarium graminearum.
Methods S1 Supplementary methods.


